- 1Institute of Zoology, Zoological Society of London, Regent's Park, London, United Kingdom
- 2Imperial College London, London, United Kingdom
- 3Natural History Museum, London, United Kingdom
- 4School of Natural Sciences and Ryan Institute, National University of Ireland, Galway, Ireland
- 5P.P. Shirshov Institute of Oceanology, Russian Academy of Sciences, Moscow, Russia
- 6School of Biological, Earth and Environmental Sciences/iCRAG/MaREI/ERI, University College Cork, Cork, Ireland
Black corals (Anthozoa: Antipatharia) are an ecologically and culturally important group of deep-sea cnidarians. However, as the majority of species inhabit depths >50 m, they are relatively understudied. The inaccessibility of well-preserved tissue for species of interest has limited the scope of molecular analysis, and as a result only a small number of antipatharian mitochondrial genomes have been published. Using next generation sequencing, 18 complete and five partial antipatharian mitochondrial genomes were assembled, increasing the number of complete mitochondrial genomes to 22. This includes species from six antipatharian families, four of which were previously unrepresented, enabling the first family-level, full mitochondrial gene analysis over the whole order. The circular mitogenomes ranged in size from 17,681 to 21,669 bp with the large range in size due to the addition of an intron in COX1 in some species and size variation of intergenic regions. All mitogenomes contained the genes standard to all hexacoral mitogenomes (13 protein coding genes, two rRNAs and two tRNAs). The only difference in gene content is the presence of the COX1 intron in five families. The most variable mitochondrial gene is ND4 which may have implications for future barcoding studies. Phylogenetic analysis confirms that Leiopathidae is sister to all other families. Families Antipathidae, Cladopathidae and Schizopathidae are polyphyletic, supporting previous studies that call for a taxonomic revision.
Introduction
The order Antipatharia Milne Edwards and Haime, 1857, also known as black or thorny corals, is an ecologically and culturally important group of deep-sea anthozoans in the hexacoral subclass (Cnidaria: Anthozoa: Hexacorallia). The order includes 273 species in 45 genera and seven families (Molodtsova and Opresko, 2020), with many new species expected. Black corals are found in every ocean occupying depths from 2–8,600 m (Wagner et al., 2012), but are primarily concentrated at depths >50 m (Cairns, 2007), with ~50% of known genera inhabiting depths >500 m (Molodtsova and Opresko, 2017). Anipatharians are exclusively colonial and exhibit a diverse range of polyp, skeletal and spine morphologies. They range in height from 5 to 10 cm to >3 m (Wagner et al., 2012). Unlike scleractinian corals, the skeletons of antipatharians are not calcified but are instead rich in protein and chitin [(Bo et al., 2012b); see Ehrlich (2010) for a review] and often appear black when visible. Although not reef forming, antipatharians can play a crucial ecological role in forming coral garden habitats (Bo et al., 2009b) and individuals can support significant epifaunal populations (Love et al., 2007; Roberts et al., 2009). From China to Hawaii, black coral has been utilized in traditional medicine for a wide variety of therapeutic and spiritual uses, while its common use in jewelery has seen commercial harvesting established in the Caribbean, Hawaii and parts of Asia (Wagner et al., 2012). This has resulted in all species of black coral being listed on CITES appendix II. Their use in traditional medicine has led to interest from pharmacologists, with studies showing potential roles in cancer treatments (Qi et al., 2009; Ghandourah and Alorfi, 2019) and the presence of antibacterial properties (Al-Lihaibi et al., 2010).
The use of complete mitochondrial genomes has become an increasingly popular tool for taxonomic and phylogenetic research due to the advances and availability of next generation sequencing technology (Kayal et al., 2013; Crampton-Platt et al., 2016). In addition, metazoan mitogenomes provide a number of genome-level characters that can support phylogenetic hypotheses. These include gene order arrangements, position of introns, insertions and inferred secondary structures of transfer RNAs (tRNA) and ribosomal RNAs (rRNA) (Boore, 2006). In particular, gene order arrangements have proved useful in anthozoan studies by furthering understanding of evolutionary relationships (Brugler and France, 2007; Brockman and McFadden, 2012; Figueroa and Baco, 2014; Hogan et al., 2019) and provided additional resolution when nucleotide and protein sequence phylogenies have proved contradictory (Lin et al., 2014). This is in part due to a number of advantageous features of the mitogenome, including the presence of a near universal homology of mitochondrial genes found across the animal kingdom (Wolstenholme, 1992; Boore and Fuerstenberg, 2008). In addition, the high number of possible gene arrangements suggests that the observed gene order is most likely to be a derived synapomorphy shared among related taxa, rather than the result of a convergent rearrangement (Clayton, 1992; Falkenberg et al., 2007) [however, ancestral state reversions have been observed, see Figueroa and Baco (2014)].
Hexacoral mitogenomes share some unique properties amongst metazoans. Complete mitogenomes have been sequenced for species from each of the five hexacoral orders: Actiniaria (Foox et al., 2015; Zhang et al., 2017), Antipatharia (Brugler and France, 2007; Kayal et al., 2013; Figueroa et al., 2019), Corallimorpharia (Lin et al., 2014), Scleractinia (Medina et al., 2006), and Zoantharia (Sinniger et al., 2007; Chi and Johansen, 2017). Consistent is a reduced set of the 22 tRNAs necessary for mitochondrial translation, with only tRNA−met (Methionine) and tRNA−trp (Tryptophan) being present. In addition, group 1 introns are found within the NADH dehydrogenase subunit 5 gene (ND5) with varying numbers of embedded energy pathway genes specific to each order. A second group 1 intron within the cytochrome c oxidase subunit I gene (COX1), with an embedded homing endonuclease gene (HEG), has also been identified for some species in each of the hexacoral orders (Celis et al., 2017).
The first sequenced antipatharian mitochondrial genome, Chrysopathes formosa was circular and contained 13 energy pathway genes, two ribosomal RNA genes and a reduced number of tRNA genes consistent with all hexacorals (Medina et al., 2006; Brugler and France, 2007; Lin et al., 2014; Foox et al., 2015; Chi and Johansen, 2017). The gene order was described as most similar to the sea anemone Metridium senile, with the exception of a transposition of three continuous genes COX2-ND4-ND6 and lack of a COX1 intron. However, the subsequent fully sequenced antipatharian mitogenomes, Stichopathes luetkeni (Kayal et al., 2013), Myriopathes japonica (GenBank NC_027667.1), Tanacetipathes thamnea (Figueroa et al., 2019) and the partially sequenced Leiopathes glaberrima (Sinniger and Pawloski, 2009) had a COX1 intron containing a HEG gene. With the exception of the COX1 intron, gene content was the same as in C. formosa. The ND5 intron contains two embedded genes; the only copies of ND1 and ND3, as in actiniarians (Foox et al., 2015; Zhang et al., 2017) and zoantharians (Sinniger et al., 2007; Chi and Johansen, 2017).
Molecular approaches to examining phylogenetic relationships within Antipatharia are a relatively recent addition to systematic studies (Lapian et al., 2007; Wagner et al., 2010; Bo et al., 2012a). Nuclear ribosomal genes (rRNA) and their associated internal transcribed spacers (ITS1 and ITS2) have been effective in resolving phylogenetic relationships within Antipatharia (Lapian et al., 2007; Lapian, 2009; Bo et al., 2012a, 2018). Mitochondrial DNA has been used in a number of antipatharian phylogenetic studies in addition to nuclear DNA, focusing on intergenic regions (IGRs) spanning trnW-ND2 and COX3-COX1 (Wagner et al., 2010; Brugler et al., 2013; MacIsaac et al., 2013) and also ND5-ND1 (Brugler et al., 2013; MacIsaac et al., 2013). Most studies have focused on clarifying the relationships within or between the families Antipathidae, Aphanipathidae, and Myriopathidae (Lapian et al., 2007; Bo et al., 2009a, 2012a; Lapian, 2009; Wagner et al., 2010), while MacIsaac et al. (2013) was the first to examine relationships within Schizopathidae. To date, only one molecular study has included species from all seven antipatharian families, providing the first full family phylogeny (Brugler et al., 2013). However, the regions examined in that study were unable to fully resolve the phylogeny, particularly at basal nodes.
This study aims to expand our knowledge of antipatharians through sequencing and examining complete mitochondrial genomes from species representing six families. Previously sequenced antipatharian mitogenomes downloaded from GenBank are re-analyzed alongside those from this study, expanding the analysis to cover all seven families.
Materials and Methods
Sampling and DNA Extraction
Antipatharian coral samples were collected from surveys around Whittard Canyon, north of the Porcupine Bank and adjacent to the Belgica Mound Province SAC in the NE Atlantic (Irish Margin) at depths of 669–2,817 m, between 2013 and 2017 using a remotely operated vehicle (ROV) Holland I onboard the R/V Celtic Explorer. Species identifications were made by examining morphological traits based on in situ and ex situ photographs with reference to classic and current taxonomical literature. Whole genomic DNA was extracted from tissue using the NucleoSpinVR™ tissue Kit (Macherey-Nagel, Düren, Germany) following manufacturer's instructions.
Sequencing Preparation
Samples were prepared for sequencing following the TruSeq DNA PCR-Free Library Preparation Kit protocol (Illumina Inc., San Diego, CA), with the exception of Tylopathes n. sp. which was prepared following the QIAseq FX DNA Library protocol (QIAGEN). For the TrueSeq samples DNA was fragmented to a target insert size of 550 bp using a Covaris Focused-ultrasonicator M220™. Twenty-three samples were sequenced on a single run of an Illumina MiSeq (MiSeq Reagent Kit v3 600-cycle).
Clean up and Assembly
Read quality was assessed using FastQC (Andrews, 2010). Illumina TruSeq index adapters were trimmed and low quality reads (Q <28) removed using cutadapt 2.3 (Martin, 2011). Mitogenome assembly was carried out de novo in MITObim 1.9.1 (Hahn et al., 2013) utilizing the COX1 seed sequence from Chrysopathes formosa (GenBank accession number: NC_008411.1). Complete circular read coverage was confirmed by mapping the clean reads onto the assembled circular sequence in Geneious Prime 2019.2.1 (Kearse et al., 2012) (see Supplementary Material for additional detail).
Genome Annotation
Protein coding regions, tRNAs and rRNAs were identified in Geneious using the live annotate function based on three published antipatharian mitogenomes (GenBank accession numbers: Chrysopathes formosa NC_008411.1, Stichopathes luetkeni JX023266.1, Myriopathes japonica NC_027667.1). Additionally, confirmation was sought using the online tool Mitos2 (Bernt et al., 2013) with NCBI RefSeq 81 Metazoa and genetic code 4 (mold, protozoan and coelenterate mitochondrial code). Annotations were manually edited using the widest possible open reading frame for protein coding regions based on genetic code 4 and cnidarian start (ATG) and stop (TAG, TAA) codons. Intron boundaries were confirmed manually using homologous regions in non-intron containing taxa. The position of tRNAs were confirmed using tRNAscan-SE (Lowe and Chan, 2016).
Genome Composition
Nucleotide composition and GC content were calculated in Geneious. Relative GC and AT skew was calculated using the skew formula of Perna and Kocher (1995). Origins of replication were predicted using the DNA walker method (Lobry, 1996) within GraphDNA (Thomas et al., 2007) which assesses for abrupt switches in polarity in the asymmetry of strand specific frequencies of nucleotide composition. All genomes were assessed for tandem repeats using Tandem Repeats Finder (Benson, 1999) and inverted repeats using EMBOSS einverted (Rice et al., 2000).
Nuclear DNA
Nuclear ribosomal DNA spanning the region partial 18S rRNA, ITS1, 5.8S rRNA, ITS2 and partial 28S rRNA were assembled from the cleaned reads using a combination of MITObim (see above) and read mapping onto a reference sequence (Chrysopathes formosa, GenBank accession number MG023168.1).
Alignment for Phylogenetic Analysis
The 13 mitochondrial protein coding genes and five nuclear regions were aligned separately in MAFFT v7.407 (Katoh and Standley, 2013) using the L-INS-i method; maxiterate: 1000, as this showed the highest pairwise identity (PI) and percentage of identical sites (IS). Each alignment was processed through Gblocks 0.91b (Castresana, 2000) which improves alignments for phylogenetic analysis by removing poorly aligned and divergent regions (see Table S1 in Supplementary Material for Gblock parameters). Concatenated sequences were created for (a) the 13 mitochondrial genes; (b) five nuclear regions; and (c) combined mitochondrial and nuclear regions, using a seqCAT.pl (Bininda-Emonds, 2005) script. Published genomes used in this study were re-analyzed alongside the newly sequenced genomes.
Phylogenetic Analysis
Three phylogenies were constructed from the concatenated sequences; (a) the 13 mitochondrial genes (30 taxa; 12,227 characters); (b) five nuclear regions (29 taxa; 10,28 characters); (c) combined mitochondrial and nuclear regions (29 taxa; 13,255 characters), using maximum likelihood (ML) implemented in IQ tree (Nguyen et al., 2014) and Bayesian inference (BI) statistics implemented in MrBayes 3.2.6 (Ronquist et al., 2012). Due to the lack of a suitable nuclear sequence, Stichopathes luetkeni and Tanacetipathes thamnea were left out of the nuclear dataset phylogeny. As nuclear regions for Myriopathes japonica were unavailable, the congeneric Myriopathes myriophylla (GenBank accession number AM404328.1) was used to create a chimeric sequence for Myriopathes in the combined dataset phylogeny, and as the single representative of Myriopathes in the nuclear phylogenies. Edwardsia timida replaced Nematostella sp. within the outgroup of the nuclear phylogeny, as suitable nuclear sequences were unavailable for Nematostella sp. A chimeric sequence consisting of nuclear regions for E. timida and 13 mitochondrial genes of Nematostella sp. represented the family Edwardsiidae as an actiniarian outgroup in the combined phylogeny. Models of sequence evolution were determined for each Gblock-edited region using jModelTest 2.1.10 (Guindon and Gascuel, 2003; Darriba et al., 2012) based on AIC scores, and implemented in Bayesian analysis (see Table S2 for models). Partitions for each region were also input to IQtree which runs an automatic model testing and selection using ModelFinder (Kalyaanamoorthy et al., 2017) (Table S2). Both IQtree and MrBayes analyses were carried out on the CIPRES Science Gateway V. 3.3 (Miller et al., 2010). For ML analysis, 1,000 bootstrap replicates were performed within IQtree using the ultrafast bootstrap (Hoang et al., 2017). Bayesian analysis ran for 10 million generations, sampling every 1000 generations on two separate runs and four chains. All runs were visualized in Tracer v1.7.1 (Rambaut et al., 2018). All runs showed an effective sample size >6,000. A burn-in of 1 million was applied and a single target tree obtained using TreeAnnotator v1.10.4 (Bouckaert et al., 2014). Tests of monophyly were performed by running constrained analyses fixing the monophyly of selected groups and comparing likelihoods with the unconstrained tree using an approximately unbiased (AU) test (Shimodaira, 2002).
Outgroup Selection
We have included a representative from each of the four additional hexacoral orders, including three from Actiniaria as outgroups. Phylogenies are rooted at a midpoint between Antipatharia and the hexacoral outgoups to examine Antipatharia monophyly rather than examine hexacoral relationships. (see Table S3 for GenBank accession numbers). Phylogenies with different outgroup combinations were explored to assess ingroup topology consistency (see Figures S1–S3).
Results
Genome Organisation
Eighteen complete and five partial mitochondrial genomes of 23 deep-sea antipatharian morphospecies from the Northeast Atlantic, were assembled and annotated (Table 1). These represent 14 genera and six families. The 18 complete genomes are circular and range in size from 17,679 (Tylopathes n. sp.) to 21,669 bp (Leiopathes cf. glaberrima) with the size range primarily due to the addition of the COX1 intron and size variation of IGRs. Partial mitogenomes were recovered for five morphospecies. Two of these were almost complete, with Leiopathes montana missing a 156 bp gap (predicted based on genome similarity to L. cf. glaberrima) and Antipathes cf. dichotoma missing a 9 bp gap (predicted based on genome similarities to the three additional Antipathidae samples) and possessing 16 ambiguous sites. Three samples (Chrysopathes cf. micracantha; Cladopathes cf. plumosa; Trissopathes grasshoffi) yielded very limited coverage, resulting in the recovery of 3.2, 48.2, 1.9%, respectively, of a full-size genome (based on the full genome size of Trissopathes cf. tetracrada and Chrysopathes formosa).
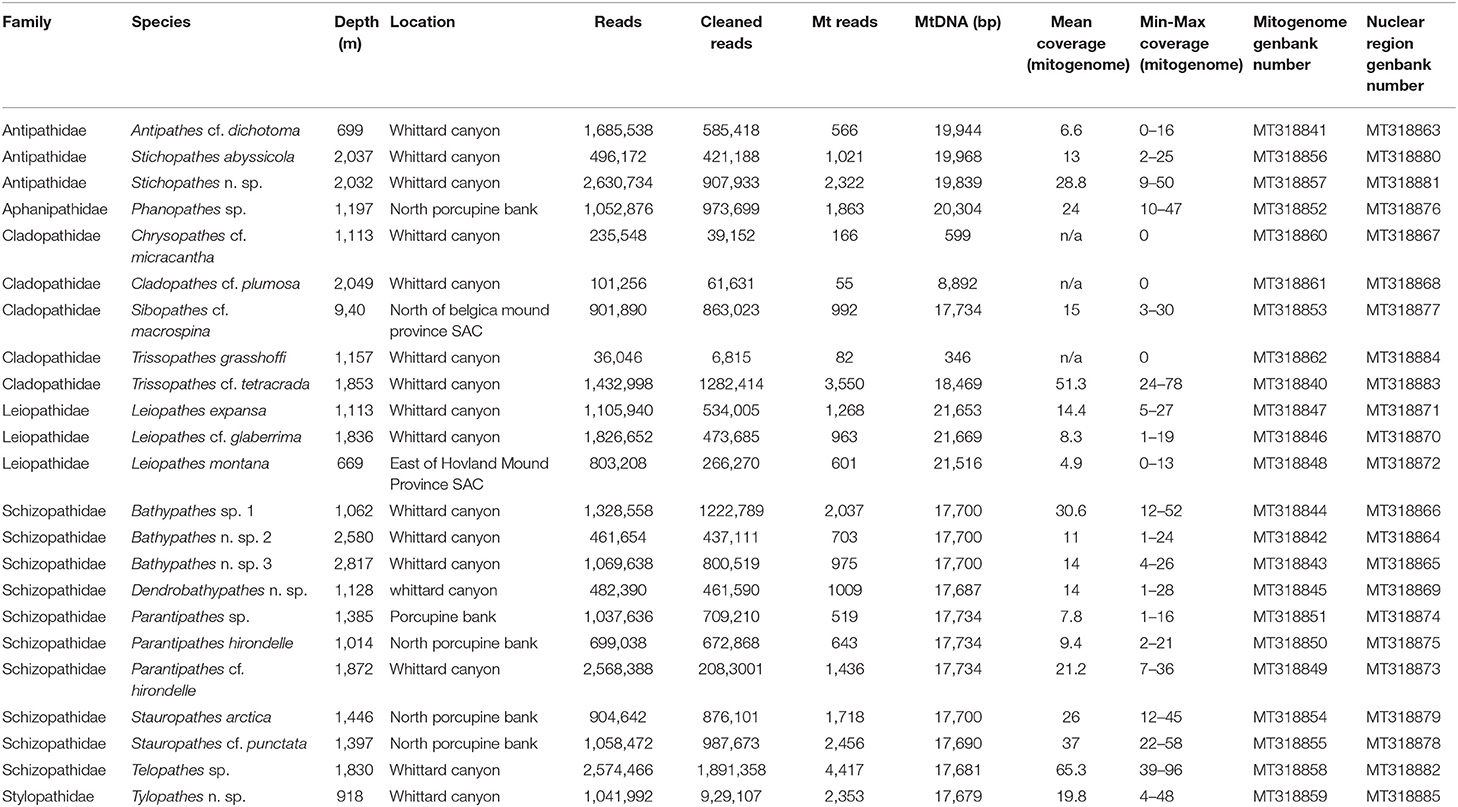
Table 1. Antipatharian samples, GenBank accession numbers, collection information and sequencing data used in this study.
All recovered genomes contain 13 energy pathway genes (COX1-3, ND1-6, ND4L, CYTB, ATP6, and ATP8), two tRNAs (tRNA−met and tRNA−trp) and two subunit rRNAs (12S and 16S) (Table 2). Genomes from eight samples are interrupted by a group 1 intron in COX1 at the same insertion site, the ‘3 end of position 884 [see Emblem et al. (2011) for intron nomenclature]. Additionally, all taxa with COX1 introns contain a HEG gene which varies in length from 657 (Myriopathes japonica, Tanacetipathes thamnea and Tylopathes n. sp.) to 1,062 bp (Stichopathes luetkeni). All genomes contain a group 1 intron in ND5 with the same insertion site at the ‘3 end of position 717, containing the only genome-wide copies of ND1 and ND3. All genes are separated by IGRs which show large size variation (Table 2; see Table S4 for full species table).
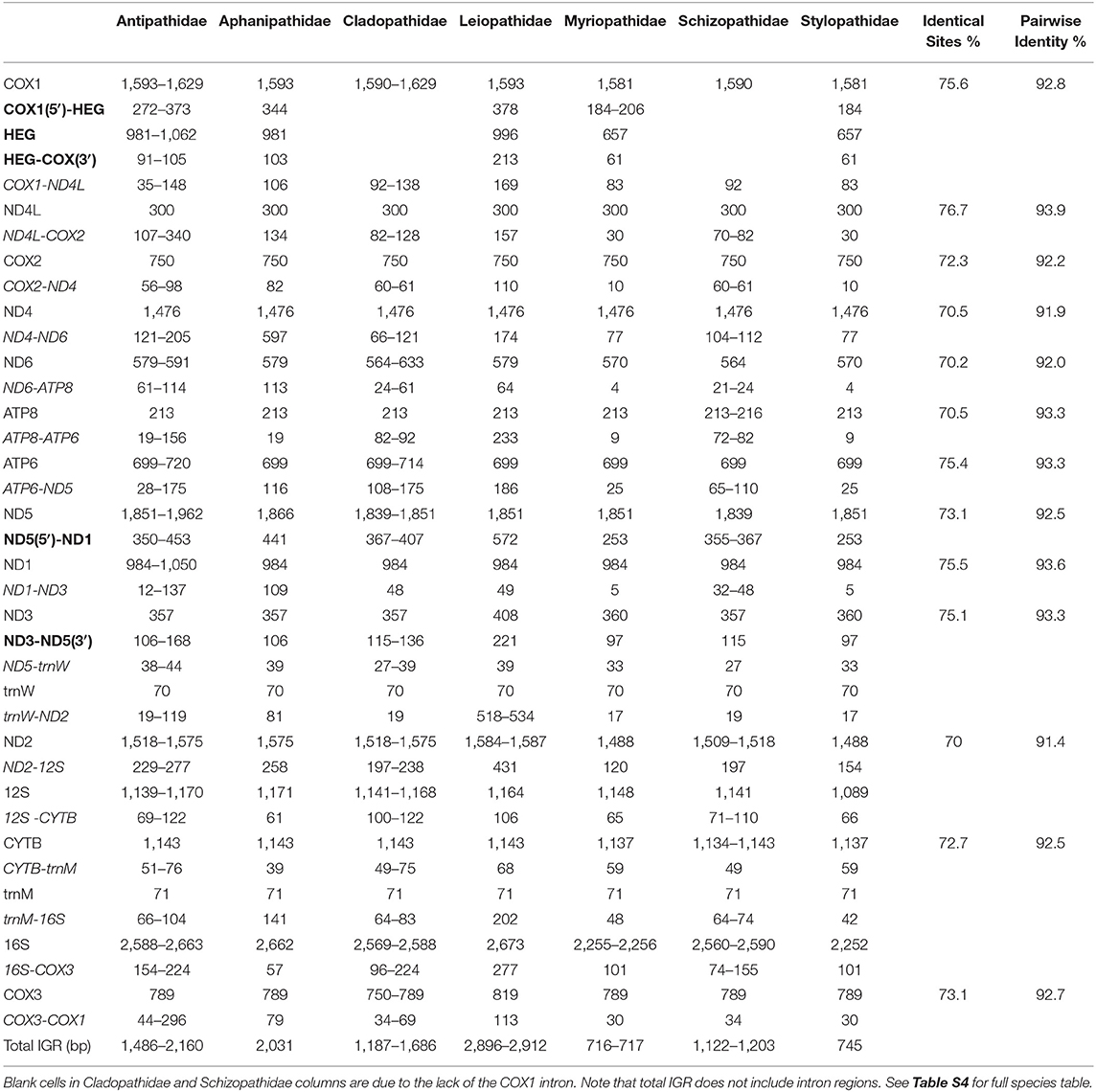
Table 2. Antipatharian family gene lengths (bp), intergenic regions (italicized) (bp), intron regions (bold) (bp) and gene variation from Gblock alignments (percentage identical sites and percentage pairwise identity).
Across all samples the largest IGR is found between ND4 and ND6 in Phanopathes sp. and extends for 597 bp. However, the range for this region between samples is 531 bp, extending for only 66 bp in C. formosa. The smallest IGR is found between ND6 and ATP8 in M. japonica, T. thamnea and Tylopathes n. sp. and extends for 4 bp. The genomes with the largest total intergenic space are L. cf. glaberrima and L. montana both with a length of 2912 bp, while M. japonica has the smallest at 716 bp.
A number of novel ORFs were identified within the IGRs (see Table S5) The largest ORF is found exclusively in Phanopathes sp. in the IGR ND4-ND6 spanning 477 bp. No single ORF was found consistently across all samples, however within families there are conserved ORFs of the same length and at the same location.
Gene content within the sample genomes shows two variations (Figure 1), with the only variation due to the absence of the HEG gene in the families Schizopathidae and Cladopathidae. All genes are transcribed in the same direction and on the same strand.
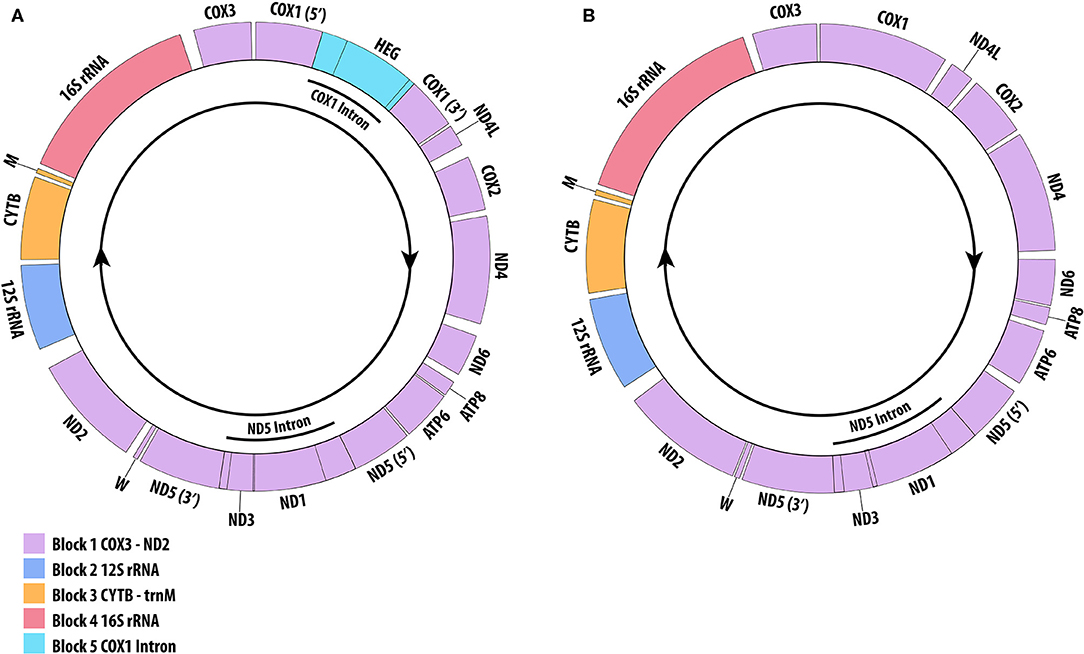
Figure 1. Antipatharian mitogenome graphical map showing gene blocks and gene orders. tRNAs are displayed according to their one letter code. (A) Representative of families Antipathidae, Aphanipathidae, Leiopathidae, Myriopathidae and Stylopathidae with the COX1 intron (relative size based on Stichopathes abyssicola). (B) Representative of families Cladopathidae and Schizopathidae with the absence of the COX1 intron (relative size based on Stauropathes cf. punctata). Arrow indicates direction of transcription. Gene map created using OGDraw (Greiner et al., 2019).
Analysis of Base Composition
Whole genome base composition is A+T rich ranging from 64.8 (Stichopathes luetkeni) to 58.3% (Antipathes cf. dichotoma and Stichopathes abyssicola) (Table 3). GC skew was positive ranging from 0.084 (Tylopathes n. sp.) to 0.129 (Stichopathes luetkeni), while AT skew was negative ranging from −0.116 (Myriopathes japonica) to −0.097 (Leiopathes montana).
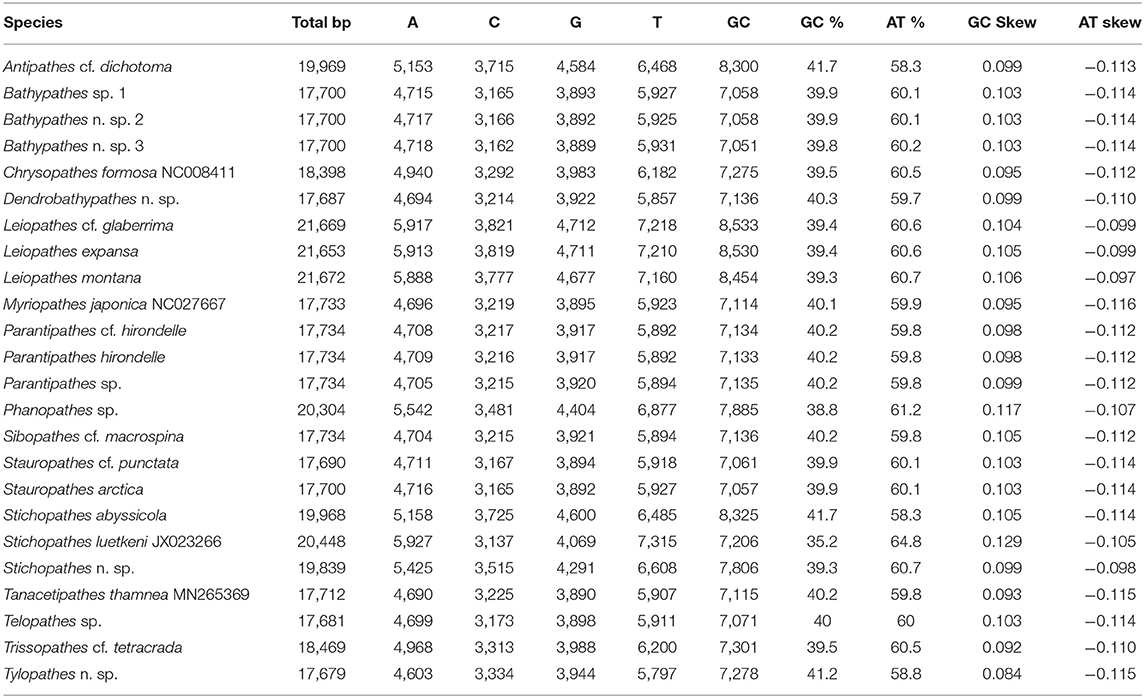
Table 3. Antipatharian mitochondrial genome nucleotide compositions (from this study and GenBank species).
The DNA walk method found four abrupt changes in base composition across all genomes (Figure 2). These are found in the intergenic regions ND2-12S, 12S-CYTB, trnM-16S and 16S-COX3. An additional reversal in polarity is present only in samples with the COX1 intron, marking the boundaries of the intron. These junctions suggest the genome is split into four blocks (Figure 1); block 1: COX3-ND2, block 2: 12S, block 3: CYTB-trnM, block 4: 16S, and an additional block for samples with the COX1 intron; block 5: COX1 intron. Three of the IGR which show abrupt changes in polarity are rich in A+T content across all genomes; ND2-12S A+T range of 60.5 to 70.5%, trnM-16S A+T range of 58.4 to 71.4% and 16S-COX3 A+T range of 56.4 to 69.4% (data not shown). The only tandem repeats (>15 bp) were a 37 bp sequence found in the IGR COX2-ND4 in two samples S. abyssicola and A. cf. dichotoma, and a 22 bp repeat found in M. japonica within the COX1 intron. Inverted repeats (>15 bp) were found in six samples (Table S6), with the only consistent repeats being found in the three Leiopathes samples.
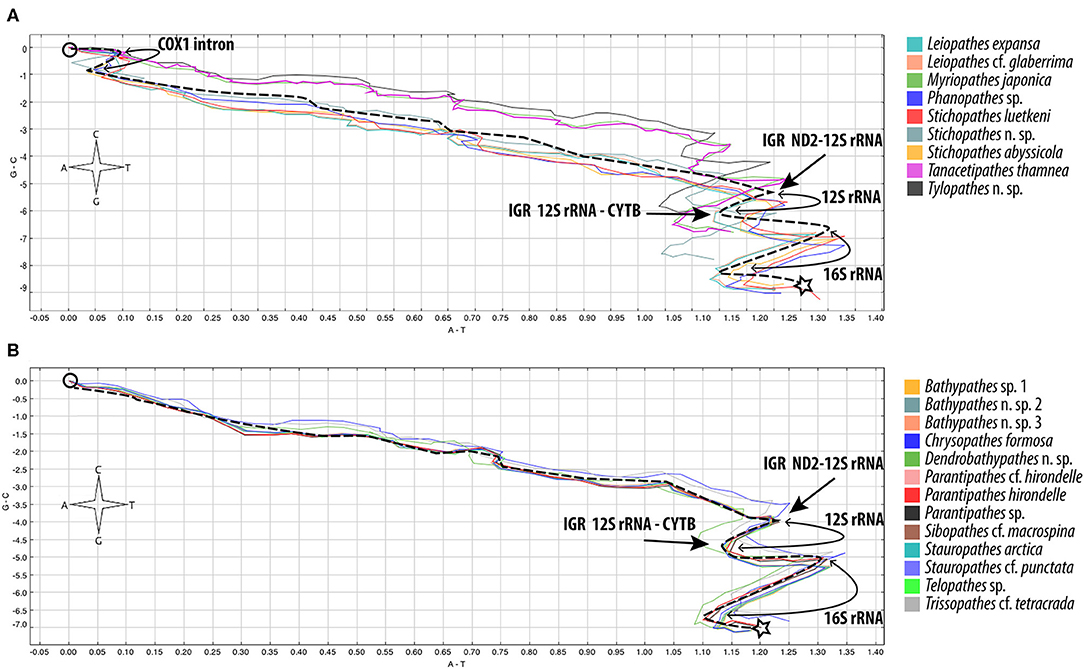
Figure 2. DNA Walk [GraphDNA (Thomas et al., 2007)] results from vectorial representation of a linearized genome beginning at COX1 (circular symbol) and ending at IGR COX3-COX1 (star symbol). Each of the four nucleotides in the genome sequence is designated a direction as displayed on the compass. Units on axis are cumulative skew measured as number of nucleotides X 103 (Window size is displayed at 400 for ease of viewing). (A) Genomes with the COX1 intron. (B) Genomes with the COX1 intron absent. The dotted line represents the general shape of the walk. Abrupt changes in direction are characteristic of a separate evolutionary origin for that particular region.
Protein Coding Genes and rRNAs
Protein coding gene length was similar across all genomes (Table 2), with ND5 varying most in length, while the lengths of COX2 and ND4L were conserved across all antipatharians. The ND4L gene varied least (93.9% PI; 76.7% IS) while ND2 varied most with 91.4% PI and 70% IS, however the variation is due to multiple indels. The most variable gene free of indels was ND4 (91.6% PI; 70.4% IS). The large ribosomal subunit 16S varied widely in length across species and was the largest assigned region of the genome.
Phylogenies
Phylogenetic analyses (mitochondrial, nuclear and combined) produced largely congruent trees (Figure 3; see Figures S4–S7 for nuclear regions and combined trees). Various combinations of hexacoral outgroups (see Figures S1–S3) confirm the monophyly of Antipatharia and maintain the same ingroup topology. The family Leiopathidae formed a monophyletic group sister to all other antipatharians. The families Antipathidae (in the mitochondrial and combined phylogenies only), Cladopathidae and Schizopathidae (due to the placement of Sibopathes cf. macrospina nesting within a clade containing three Parantipathes spp) did not form monophyletic groups. The genera Bathypathes, Parantipathes, Stauropathes, Stichopathes and Trissopathes did not form monophyletic groups. Constraining the monophyly of each polyphyletic group produced a significantly worse likelihood value tree compared to the unconstrained tree (P < 0.005; see Figure S8).
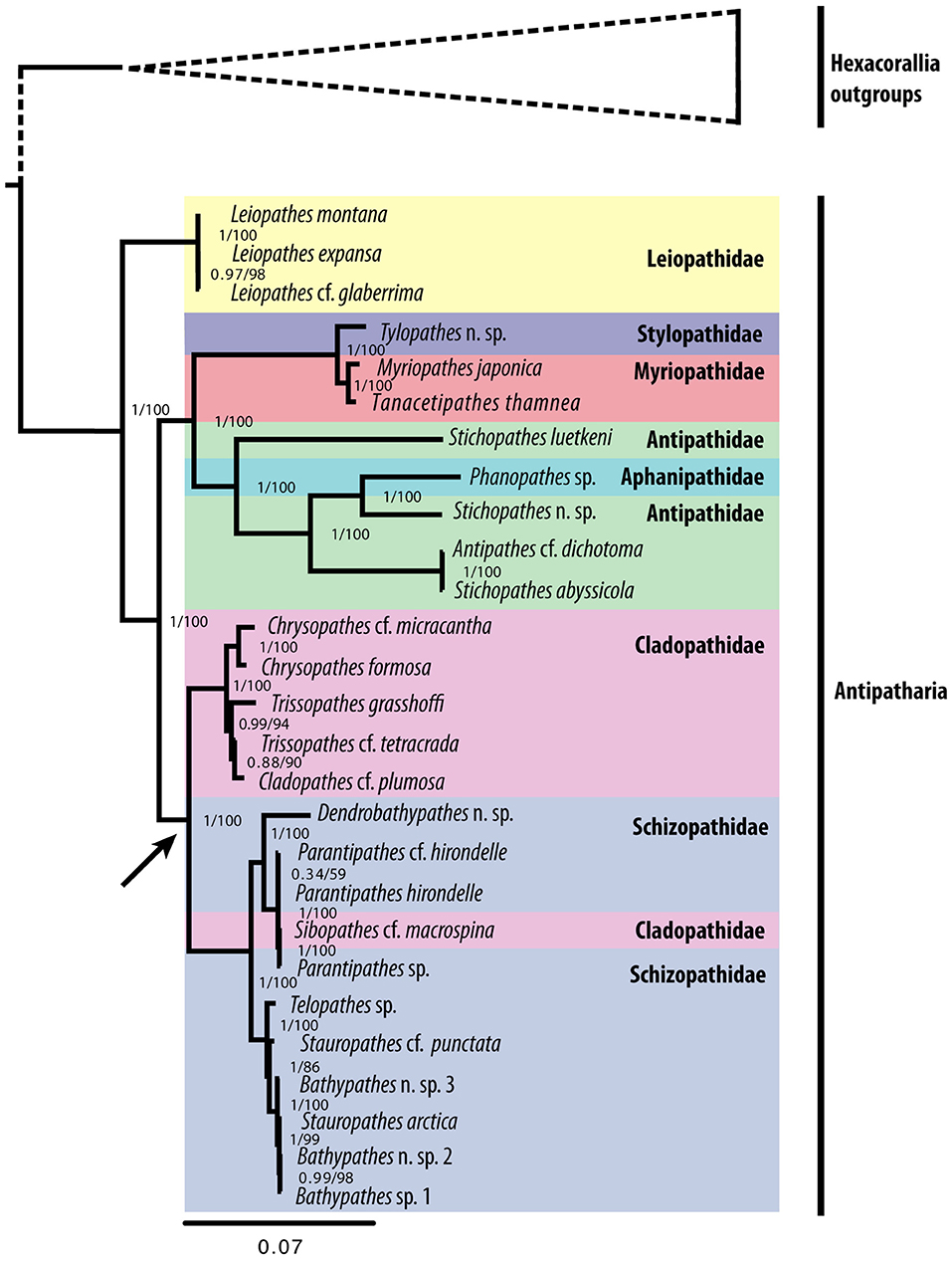
Figure 3. Phylogenetic tree reconstruction based on all available (newly sequenced from this study and GenBank samples) antipatharian mitochondrial genomes using the 13 energy pathway genes based on a Bayesian inference (BI) and maximum likelihood (ML) analysis. The trees are rooted at a midpoint between Antipatharia and the hexacoral outgroups (See Figure S1 for outgroup relationships). BI posterior probabilities and ML bootstrap values shown at nodes (BI/ML). The arrow indicates the apparent loss of the COX1 intron at an internal node.
Discussion
This study increases the total number of complete antipatharian mitochondrial genomes represented in the literature from four to 22, with five additional partial sequences. This includes species from four families previously unrepresented, enabling for the first time, a family-level full mitochondrial gene analysis over the whole order.
Genome Organization, Gene Order, and Variable Regions
Based on our analysis, antipatharian mitogenomes have the same features typical of all published hexacoral order mitogenomes (Lin et al., 2014; Chi and Johansen, 2017; Zhang et al., 2017). Gene order and content are highly conserved across the antipatharian order, consistent with previous findings (Brugler and France, 2007; Kayal et al., 2013) with the absence of the COX1 intron as the main notable variation. Internal gene rearrangements have been reported within all hexacoral orders (Emblem et al., 2011; Lin et al., 2014; Foox et al., 2015), but it appears that antipatharians are an exception, along with zoantharians (Poliseno et al., 2020). The stable structural arrangement in these two orders may be linked to the slow rate of nucleotide substitution in anthozoans (Hellberg, 2006).
Metazoan mitochondrial DNA, and specifically COX1, was originally viewed as an attractive target for barcoding as it was thought to be almost entirely free of introns and showed a greater range of diversity than other mitochondrial genes (Hebert et al., 2003). In the present analysis, COX1 has more identical sites and a higher PI (92.8% PI; 75.6% IS), than seven other mitochondrial genes (see Table 2). ND2 varies most (91.4% PI; 70% IS), however, the presence of indels, ranging from 6 to 20 bp at multiple locations, drives this variability. The highest variability seen in an indel-free alignment is ND4 spanning 1476 conserved bp (91.9% PI; 70.5% IS). Additional ML phylogenetic analysis of the ND4 Gblock alignment recovered a tree with identical topology to the full mitochondrial tree, with high bootstrap nodal support for all major clades (>82% BS). A comparative COX1 tree revealed weaker BS support values for all major clades (>57% BS). While COX1 continues to be an effective marker for barcoding studies in Antipatharia (Brugler et al., 2013), future researchers may wish to consider the addition of ND4 to further increase the resolving power of taxonomic identification.
Origins of Replication and Intergenic Regions
The origins of replication in mitogenomes are often associated with large intergenic spaces, inverted repeats, A+T rich regions and at the junction of conserved gene blocks (Pearson et al., 1996; Brugler and France, 2008; Hogan et al., 2019). Additionally, mitogenomes have two separate origins of replication (Clayton, 1992), one found on each strand; OriH found on the heavy strand and OriL found on the light strand. Brugler and France (2007) investigated the ND2-12S IGR as a possible location of OriH due to its large size. With only one complete antipatharian genome for their analysis, comparisons were made with other hexacoral species, and they ultimately rejected this location as the OriH. However, with the larger data set provided by the current study, we note abrupt compositional changes around this region, the presence of inverted repeats and relatively high conservation (aligning all ND2-12S IGRs produces a 278 bp region with 67.3% PI), all of which suggest the IGR ND2-12S is the most likely candidate for the OriH in Antipatharia.
Defining the exact location of OriL has proved difficult in invertebrates (Wolstenholme, 1992). Brugler and France (2008) proposed candidate regions between 12S-CYTB and trnM-16S for Chrysopathes formosa based on the abrupt changes in base composition at these locations. In the present study 12S-CYTB has a relatively balanced A+T/G+C base content (average 48.6% A+T) and a relatively small IGR (mean 86 bp). In contrast the IGRs trnM-16S and 16S-COX3 both show an abrupt change in base composition and have a high A+T content (mean 65.4 and 64.5%, respectively), while IGR 16S-COX3 has a relatively large IGR (mean 136 bp), making it the preferred candidate for OriL. Further investigation into the secondary structures of the IGRs, including presence of A+T stable stem loops, would be necessary to provide additional evidence for the location of OriH and OriL.
Open read frames of unknown function have not previously been reported for antipatharian mitogenomes. However, they are common in other hexacoral orders (Flot and Tillier, 2007; Emblem et al., 2014; Foox et al., 2015; Chi and Johansen, 2017). Their functionality in actinarians has been speculated to be that of transposon-like elements capable of influencing nearby genes, as possible enhancers or promotors (Zhang et al., 2017). Emblem et al. (2014) demonstrated that ORFs within actinarian mitogenomes are likely functional units. The lack of conserved ORFs across antipatharians suggests that many are likely to be small pseudogenes. However, small proteins, often defined as <100 amino acid residues (Su et al., 2013), have been shown to be responsible for important biological processes in other metazoans (Galindo et al., 2007). The largest ORF found in this study, 477 bp in Phanopathes sp. is the most likely candidate for a functional protein due to its size. Transposon-like elements have been demonstrated to play important roles in adaptation to environmental stress (Casacuberta and González, 2013). Considering the environmental extremes of antipatharian deep-sea habitats, the function of these ORFs may well be similar to transposon-like elements offering beneficial metabolic adaptations to the deep-sea.
The potential for non-homologous cross mapping of nuclear DNA of mitochondrial origin (nuMT) to mitochondrial DNA (mtDNA) assemblies [see Maude et al. (2019) for a discussion] was considered in light of the large number of varied ORFs found throughout the mitogenomes. BLAST analysis of the larger ORF (>70 bp) revealed no matches to hexacoral nuclear regions. Read coverage for ORF loci was consistent with coverage for the overall mitogenome (i.e., if similar copies were present in the nuclear genome, we might expect higher coverage). Additionally, we might expect higher sequence variability in these ORFs compared with other coding regions (e.g., greater disagreement of mapped reads in these ORFs), however this was not the case.
Introns
A cyclic evolutionary model of intron gain and loss has previously been proposed for the presence of the COX1 intron in cnidarians (Goddard et al., 2006). Celis et al. (2017) suggest the COX1 intron was inserted early in cnidarian history and rapidly invaded intron-less alleles via horizontal transfer due to the HEG gene. After becoming fixed in the population, it was subject to mutational degradation, losing HEG functionality, but continued to be inherited vertically as a HEG remnant. The effects of purifying selection resulted in the complete deletion of the intron and HEG from COX1. Support for this hypothesis in Antipatharia is evident in the variation of HEG sizes observed across families, with the presence of shorter HEG genes in Myriopathidae and Stylopathidae (657 bp) indicating they are in a later stage of the degradation period as compared to Antipathidae (1,062 bp), Aphanipathidae (981 bp) and Leiopathidae (996 bp). The absence of the COX1 intron within the Cladopathidae and Schizopathidae suggest a complete evolutionary cyclic history of intron invasion, degradation and deletion has occurred within antipatharians, as has previously been shown in Actinaria (Emblem et al., 2014), and in Scleractinia and Corallimorpharia (Celis et al., 2017). This is further implied by the phylogenetic analysis which suggests the COX1 intron was present in the ancestral state due to its presence in Leiopathidae, which is sister to all other antipatharian families. It was then subject to a single loss in the branch leading to the Cladopathidae + Schizopathidae clade (see Figure 3), suggesting a close evolutionary relationship between these two families as shown by Brugler et al. (2013). Interestingly, species of Cladopathidae and Schizopathidae are the only antipatharians found in the abyssal zone >3,000 m (Molodtsova and Opresko, 2017). Whether the loss of the COX1 intron is an adaptation to the deep-sea environment warrants further investigation.
In contrast to the COX1 intron, the ND5 intron is present within all sequenced genomes. The insertion position is at the same location as found in all hexacoral orders (Chi and Johansen, 2017), referred to as position 717 (Emblem et al., 2011). Emblem et al. (2011) propose that the ND5 intron originally contained a HEG gene and was subject to horizonal transfer and a similar cyclic scenario as the COX1 intron. However, at one stage, the insertion of ND1 and ND3 resulted in the intron becoming a compulsory element, resulting in a vertical pattern of inheritance, supported by the conserved intron position. In contrast, three insertion sites have been identified for the COX1 intron across the hexacorals (Celis et al., 2017), suggesting a lack of positive selection.
Phylogenies and Interfamilial Relationships
Previous molecular studies show consistent congruent sister groupings between the following sets of antipatharian families: Cladopathidae + Schizopathidae (Brugler et al., 2013; Bo et al., 2018), Myriopathidae + Stylopathidae (Brugler et al., 2013), Antipathidae + Aphanipathidae (Lapian et al., 2007; Bo et al., 2009a, 2018; Lapian, 2009; Wagner et al., 2010; Brugler et al., 2013). The phylogenies presented in this study are in agreement with these family groupings. Based on morphology, Opresko (1998) considered Leiopathidae to be a distinctive family and suggested it may warrant a higher level classification, while Brugler et al. (2013) suggested it may be the most primitive family. However, molecular studies have shown conflicting results with both Antipathidae and Leiopathidae being recovered as sister to the remaining antipatharian families (Brugler et al., 2013; Bo et al., 2018). As the ingroup topology is sensitive to the choice of outgroup and number of outgroup samples (Smith, 1994), we have chosen to include three samples from a close sister order, Actiniaria, and a single representative from each of the three remaining hexacoral orders, Corallimorpharia, Scleractinia and Zoantharia. This study presents the largest molecular character set to date (13,255 characters) and supports Leiopathidae as sister to all other families, reconciling the morphological/molecular positions. This study should be considered a preliminary assessment of the interfamilial relationships within Antipatharia until broader geographic and taxonomic representation can be achieved using full mitogenomes.
The species reported here as Sibopathes cf. macrospina (family Cladopathidae) clustered within a clade with three Parantipathes species (family Schizopathidae) with marginal genetic differences. Similar results were reported by Brugler et al. (2013) for material from the Gulf of Mexico and Western Atlantic. This suggests S. cf. macrospina may indeed belong to the genus Parantipathes. There are a number of morphological differences reported between S. cf. macrospina and the type species of the genus Sibopathes [S. gephura van Pesch, 1914], that include pinnulation pattern, form of spines and morphology of polyps (van Pesch, 1914; Opresko, 1993; unpubl. observations T.M.). Molecular studies including S. gephura are necessary to validate the phylogenetic position of the genus Sibopathes and resolve the apparent polyphyly in Cladopathidae and Schizopathidae. In addition, species in the genera Trissopathes and Cladopathes formed a well-supported clade with C. cf. plumosa and T. cf. tetracrada in particular showing almost no evolutionary distance between them in the nuclear phylogenies (Figures S4, S5). Further studies may conclude that they represent the same genus, however it is significant to note that the mitogenomes of C. cf. plumosa and T. grasshoffi are currently incomplete.
Polyphyly in Antipathidae has been noted in previous studies (Brugler et al., 2013; Bo et al., 2018) with the suggestion that a taxonomic revision is necessary. In particular the genera Antipathes and Stichopathes have been posited for revision and splitting (Bo et al., 2012a, 2018). The present study found the single Antipathes sample A. cf. dichotoma, formed a 100% BS (BPP=1) supported clade with S. abyssicola, with little evolutionary distance between them. A similar lack of phylogenetic signal between the genera Antipathes and Stichopathes has been reported by previous studies (Bo et al., 2012a, 2018; Brugler et al., 2013). Bo et al. (2012a) highlights that the single-stem morphology (attributed to Stichopathes-like specimens), is an unreliable characteristic as it may have evolved independently in different taxa. This would account for the lack of grouping between the three Stichopathes species in this study.
With the current study, only one sample is representative of Aphanipathidae, Phanopathes sp., which forms a well-supported clade with the antipathid, Stichopathes n. sp. This is consistent with the findings of previous studies (Brugler et al., 2013; Bo et al., 2018), which have found Phanopathes species clustering with A. dichotoma and various Stichopathes species. With only one full genome representing Aphanipathidae, the current analysis is limited in scope, however the high support values for the Phanopathes sp. + Stichopathes n. sp. clade is noteworthy, although the large branch lengths indicate high genetic differentiation between these species. Bo et al. (2018) suggests that the name bearing type for the genus Antipathes and ultimately the family Antipathidae, A. dichotoma Pallas, 1766, is likely to be unrepresentative of the genus and therefore the family. Our study supports the need for taxonomic revision within Antipathidae and Aphanipathidae (Brugler et al., 2013; Bo et al., 2018), which would benefit from further sequencing of full mitogenomes representative of a greater range of diversity within these families.
The polyphyly and particularly short genetic distances found within two genera of the Schizopathidae samples; Bathypathes and Stauropathes, support the morphological similarities observed between Telopathes, Stauropathes and Bathypathes (MacIsaac et al., 2013). They form a tight clade in the current and previous studies (MacIsaac et al., 2013; Bo et al., 2018), warranting further morphological analysis to validate these genera.
Conclusions and Future Investigations
Antipatharia appears to have a stable gene order and content, with the exception of the absence of the COX1 intron in two families. The most variable gene region is found to be ND4, in contrast to the commonly recommended COX1 ‘barcode of life'. The variability in size range of IGRs, presence of multiple ORFs and HEG gene length variations across families, offer an avenue for future studies to investigate additional evolutionary scenarios and possible adaptations to metabolic functions. Phylogenetically we find that Leiopathidae is sister to all other antipatharian families. Families Antipathidae, Cladopathidae and Schizopathidae are polyphyletic, and several genera may be in need of taxonomic revision.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
The project was conceived by AA, RH, and CY and designed by CY with input from NB. Sample collection, initial taxonomic identification, and DNA extraction was carried out by AA and RH, with the exception of Sibopathes cf. macrospina, which was collected by AW. Final taxonomic identification was carried out by TM. DNA library preparation for sequencing was carried out by KH and NB. The pipeline assembly including data cleaning, genome assembly, annotation, and all downstream analysis was carried out by NB with advice from CY. The manuscript was written by NB with contributions from all authors. All authors edited the manuscript before submission.
Funding
Research surveys from 2013 to 2016 were supported by the Marine Institute and funded under the Marine Research Programme by the Irish Government. Research Survey CE15009 contributes to outputs of the Irish Center for Research in Applied Geosciences funded through Science Foundation Ireland Grant Number 13/RC/2092 and co-funded under the European Regional Development Fund 2014–2020. Research Survey CE17008 was conducted as part of the project Exploiting and conserving deep-sea genetic resources funded by Science Foundation Ireland and the Marine Institute under the Investigators Programme Grant No. SFI/15/1A/3100 and co-funded under the European Regional Development Fund 2014-2020. RH was supported by Brazilian National Council for Scientific and Technological Development (CNPq) Grant No. 248444/2013-1. TM was funded by RF State Assignment (0149-2019-0009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Samples were collected using ROV Holland I deployed from RV Celtic Explorer with support from the officers, crew and ROV technical team. NB and CY would like to thank Nadia Santodomingo for her support and advice.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00440/full#supplementary-material
References
Al-Lihaibi, S. S., Ayyad, S.-E. N., Shaher, F., and Alarif, W. M. (2010). Antibacterial sphingolipid and steroids from the black coral Antipathes dichotoma. Chem. Pharm. Bull. 58, 1635–1638. doi: 10.1248/cpb.58.1635
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed August, 2019).
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., et al. (2013). MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. doi: 10.1016/j.ympev.2012.08.023
Bininda-Emonds, O. R. P. (2005). seqCat.pl. Available online at: http://users-cs.au.dk/kmt/scriptlist/seqCat.txt (accessed March, 2020).
Bo, M., Barucca, M., Biscotti, M. A., Brugler, M. R., Canapa, A., Canese, S., et al. (2018). Phylogenetic relationships of mediterranean black corals (Cnidaria: Anthozoa: Hexacorallia) and implications for classification within the order Antipatharia. Invertebr. Syst. 32, 1102–1110. doi: 10.1071/IS17043
Bo, M., Barucca, M., Biscotti, M. A., Canapa, A., Lapian, H. F. N., Olmo, E., et al. (2009a). Description of Pseudocirrhipathes (Cnidaria: Anthozoa: Hexacorallia: Antipathidae), a new genus of whip black corals from the Indo-Pacific. Ital. J. Zool. 76, 392–402. doi: 10.1080/11250000802684104
Bo, M., Bavestrello, G., Barucca, M., Makapedua, D. M., Poliseno, A., Forconi, M., et al. (2012a). Morphological and molecular characterization of the problematic whip black coral genus stichopathes (Hexacorallia: Antipatharia) from indonesia (north sulawesi, celebes sea). Zool. J. Linn. Soc. 166, 1–13. doi: 10.1111/j.1096-3642.2012.00834.x
Bo, M., Bavestrello, G., Canese, S., Giusti, M., Salvati, E., Angiolillo, M., et al. (2009b). Characteristics of a black coral meadow in the twilight zone of the central mediterranean sea. Mar. Ecol. Progr. Ser. 397, 53–61. doi: 10.3354/meps08185
Bo, M., Bavestrello, G., Kurek, D., Paasch, S., Brunner, E., Born, R., et al. (2012b). Isolation and identification of chitin in the black coral parantipathes larix (Anthozoa: Cnidaria). Int. J. Biol. Macromol. 51, 129–137. doi: 10.1016/j.ijbiomac.2012.04.016
Boore, J. L. (2006). The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 21, 439–446. doi: 10.1016/j.tree.2006.05.009
Boore, J. L., and Fuerstenberg, S. I. (2008). Beyond linear sequence comparisons: the use of genome-level characters for phylogenetic reconstruction. Philos. Trans. R. Soc. B Biol. Sci. 363, 1445–1451. doi: 10.1098/rstb.2007.2234
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Brockman, S. A., and McFadden, C. S. (2012). The mitochondrial genome of Paraminabea aldersladei (Cnidaria: Anthozoa: Octocorallia) supports intramolecular recombination as the primary mechanism of gene rearrangement in octocoral mitochondrial genomes. Genome Biol. Evol. 4, 994–1006. doi: 10.1093/gbe/evs074
Brugler, M., Opresko, D. M., and France, S. C. (2013). The evolutionary history of the order Antipatharia (Cnidaria : Anthozoa : Hexacorallia) as inferred from mitochondrial and nuclear DNA : implications for black coral taxonomy and systematics. Zool. J. Linn. Soc. 169, 312–361. doi: 10.1111/zoj.12060
Brugler, M. R., and France, S. C. (2007). The complete mitochondrial genome of the black coral Chrysopathes formosa (Cnidaria : Anthozoa : Antipatharia) supports classification of antipatharians within the subclass Hexacorallia. Mol. Phylogenetics Evol. 42, 776–788. doi: 10.1016/j.ympev.2006.08.016
Brugler, M. R., and France, S. C. (2008). The mitochondrial genome of a deep-sea bamboo coral (Cnidaria, Anthozoa, Octocorallia, Isididae): genome structure and putative origins of replication are not conserved among octocorals. J. Mol. Evol. 67, 125–136. doi: 10.1007/s00239-008-9116-2
Cairns, S. D. (2007). Deep-water corals: an overview with special reference to diversity and distribution of deep-water scleractinian corals. Bull. Mar. Sci. 81, 311–322. Available online at: https://repository.si.edu/handle/10088/7536
Casacuberta, E., and González, J. (2013). The impact of transposable elements in environmental adaptation. Mol. Ecol. 22, 1503–1517. doi: 10.1111/mec.12170
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Celis, J. S., Edgell, D. R., Stelbrink, B., Wibberg, D., Hauffe, T., Blom, J., et al. (2017). Evolutionary and biogeographical implications of degraded LAGLIDADG endonuclease functionality and group i intron occurrence in stony corals (Scleractinia) and mushroom corals (Corallimorpharia). PLoS ONE 12:e0173734. doi: 10.1371/journal.pone.0173734
Chi, S. I., and Johansen, S. D. (2017). Zoantharian mitochondrial genomes contain unique complex group I introns and highly conserved intergenic regions. Gene 628, 24–31. doi: 10.1016/j.gene.2017.07.023
Clayton, D. A. (1992). Transcription and replication of mitochondrial DNA. Hum. Reprod. 15, 11–17. doi: 10.1093/humrep/15.suppl_2.11
Crampton-Platt, A., Yu, D. W., Zhou, X., and Vogler, A. P. (2016). Mitochondrial metagenomics : letting the genes out of the bottle. Gigascience 5:15. doi: 10.1186/s13742-016-0120-y
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Ehrlich, H. (2010). Biological Materials of Marine Origin. Invertebrates. Dordrecht: Springer-Verlag, 569. doi: 10.1007/978-94-007-5730-1
Emblem, Å., Karlsen, B. O., Evertsen, J., and Johansen, S. D. (2011). Mitogenome rearrangement in the cold-water scleractinian coral Lophelia pertusa (Cnidaria, Anthozoa) involves a long-term evolving group I intron. Mol. Phylogenet. Evol. 61, 495–503. doi: 10.1016/j.ympev.2011.07.012
Emblem, Å., Okkenhaug, S., Weiss, E. S., Denver, D. R., Karlsen, B. O., Moum, T., et al. (2014). Sea anemones possess dynamic mitogenome structures. Mol. Phylogenet. Evol. 75, 184–193. doi: 10.1016/j.ympev,.2014.02.016
Falkenberg, M., Larsson, N.-G., and Gustafsson, C. M. (2007). DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76, 679–699. doi: 10.1146/annurev.biochem.76.060305.152028
Figueroa, D. F., and Baco, A. R. (2014). Octocoral mitochondrial genomes provide insights into the phylogenetic history of gene order rearrangements, order reversals, and cnidarian phylogenetics. Genome Biol. Evol. 7, 391–409. doi: 10.1093/gbe/evu286
Figueroa, D. F., Hicks, D., and Figueroa, N. J. (2019). The complete mitochondrial genome of Tanacetipathes thamnea Warner, 1981 (Antipatharia: Myriopathidae). Mitochondrial DNA Part B Resour. 4, 4109–4110. doi: 10.1080/23802359.2019.1692701
Flot, J. F., and Tillier, S. (2007). The mitochondrial genome of Pocillopora (Cnidaria: Scleractinia) contains two variable regions: the putative D-loop and a novel ORF of unknown function. Gene 401, 80–87. doi: 10.1016/j.gene.2007.07.006
Foox, J., Brugler, M., Siddall, M. E., and Rodríguez, E. (2015). Multiplexed pyrosequencing of nine sea anemone (Cnidaria: Anthozoa: Hexacorallia: Actiniaria) mitochondrial genomes. Mitochondrial DNA 27, 2826–2832. doi: 10.3109/19401736.2015.1053114
Galindo, M. I., Pueyo, J. I., Fouix, S., Bishop, S. A., and Couso, J. P. (2007). Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLOS Biol. 5:e106. doi: 10.1371/journal.pbio.0050106
Ghandourah, M. A., and Alorfi, H. S. (2019). New cytotoxic fatty acid esters from the black coral, Antipathes dichotoma. Trop. J. Pharm. Res. 18, 69–74. doi: 10.4314/tjpr.v18i1.11
Goddard, M. R., Leigh, J., Roger, A. J., and Pemberton, A. J. (2006). Invasion and persistence of a selfish gene in the Cnidaria. PLoS ONE 1:e3. doi: 10.1371/journal.pone.0000003
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic. Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Hahn, C., Bachmann, L., and Chevreux, B. (2013). Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129. doi: 10.1093/nar/gkt371
Hebert, P. D. N., Cywinska, A., Ball, S. L., and DeWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hellberg, M. E. (2006). No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evol. Biol. 6:24. doi: 10.1186/1471-2148-6-24
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2017). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hogan, R. I., Hopkins, K., Wheeler, A. J., Allcock, A. L., and Yesson, C. (2019). Novel diversity in mitochondrial genomes of deep-sea Pennatulacea (Cnidaria: Anthozoa: Octocorallia). Mitochondrial DNA A 30, 764–777. doi: 10.1080/24701394.2019.1634699
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14:587. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kayal, E., Roure, B., Philippe, H., Collins, A. G., and Lavrov, D. V. (2013). Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evol. Biol. 13:5. doi: 10.1186/1471-2148-13-5
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Lapian, H. F. N. (2009). Biodiversity study of black coral (Order: Antipatharia) collected from Manado, Indonesia based on rDNA internal transcribed spacer (ITS) sequences analysis. Biodiversitas J. Biol. Divers. 10, 1–5. doi: 10.13057/biodiv/d100101
Lapian, H. F. N., Barucca, M., Maria, B., Marzia, A. B., Canapa, A., Tazioli, S., et al. (2007). A systematic study of some black corals species (Antipatharia, Hexacorallia) based on rDNA internal transcribed spacers sequences. Mar. Biol. 151, 785–792. doi: 10.1007/s00227-006-0525-8
Lin, M. F., Kitahara, M. V., Luo, H., Tracey, D., Geller, J., Fukami, H., et al. (2014). Mitochondrial genome rearrangements in the scleractinia/corallimorpharia complex: implications for coral phylogeny. Genome Biol. Evol. 6, 1086–1095. doi: 10.1093/gbe/evu084
Lobry, J. R. (1996). A simple vectorial representation of DNA sequences for the detection of replication origins in bacteria. Biochimie 78, 323–326. doi: 10.1016/0300-9084(96)84764-X
Love, M. S., Yoklavich, M. M., Black, B. A., and Andrews, A. H. (2007). Age of black coral (Antipathes dendrochristos) colonies, with notes on associated invertebrate species. Bull. Mar. Sci. 80, 391–399. Available online at: https://www.ingentaconnect.com/contentone/umrsmas/bullmar/2007/00000080/00000002/art00008
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
MacIsaac, K. G., Anstey, L. J., Best, M., Kenchington, E. L., Brugler, M. R., and Jordan, T. (2013). Telopathes magna gen. nov., spec. nov. (Cnidaria: Anthozoa: Antipatharia: Schizopathidae) from deep waters off Atlantic Canada and the first molecular phylogeny of the deep-sea family schizopathidae. Zootaxa. 3700, 237–258. doi: 10.11646/zootaxa.3700.2.3
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17, 10–12. doi: 10.14806/ej.17.1.200
Maude, H., Davidson, M., Charitakis, N., Diaz, L., Bowers, W. H. T., Gradovich, E., et al. (2019). NUMT confounding biases mitochondrial heteroplasmy calls in favor of the reference allele. Front. Cell Dev. Biol. 7:201. doi: 10.3389/fcell.2019.00201
Medina, M., Collins, A. G., Takaoka, T. L., Kuehl, J. V., and Boore, J. L. (2006). Naked corals: skeleton loss in Scleractinia. Proc. Natl. Acad. Sci. U S A. 103, 9096–9100. doi: 10.1073/pnas.0602444103
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees. Proc. Gatew. Comput. Environ. Work. 89, 1–8. doi: 10.1109/GCE.2010.5676129
Molodtsova, T., and Opresko, D. (2020). World List of Antipatharia. Antipatharia. Accessed through: world register of marine species Available Online at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=22549 (accessed 05 January 2020).
Molodtsova, T. N., and Opresko, D. M. (2017). Black corals (Anthozoa: Antipatharia) of the clarion-clipperton fracture zone. Mar. Biodivers. 47, 349–365. doi: 10.1007/s12526-017-0659-6
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Opresko, D. M. (1993). A new species of sibopathes (Cnidaria: Anthozoa: Antipatharia: Antipathidae) from the gulf of Mexico. Proc. Biol. Soc. Washington. 106, 195–203.
Opresko, D. M. (1998). Three new species of Leiopathes (Cnidaria: Anthozoa: Antipatharia) from southern Australia. Rec. South Aust. Museum 31, 99–111. Available Online at: https://www.biodiversitylibrary.org/part/78875 (accessed March, 2020).
Pearson, C. E., Zorbas, H., Price, G. B., and Zannis-Hadjopoulos, M. (1996). Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication. J. Cell. Biochem. 63, 1–22. doi: 10.1002/(SICI)1097-4644(199610)63:1<1::AID-JCB1>3.0.CO;2-3
Perna, N. T., and Kocher, T. D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41, 353–358. doi: 10.1007/BF00186547
Poliseno, A., Santos, M. E. A., Kise, H., Macdonald, B., Quattrini, A. M., McFadden, C. S., et al. (2020). Evolutionary implications of analyses of complete mitochondrial genomes across order Zoantharia (Cnidaria: Hexacorallia). J. Zool. Syst. Evol. Res. 1–11. doi: 10.1111/jzs.12380
Qi, S.-H., Su, G.-C., Wang, Y.-F., Liu, Q.-Y., and Gao, C.-H. (2009). Alkaloids from the South China Sea black coral Antipathes dichotoma. Chem. Pharm. Bull. 57, 87–88. doi: 10.1248/cpb.57.87
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Rice, P., Longden, L., and Bleasby, A. (2000). EMBOSS: The European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/S0168-9525(00)02024-2
Roberts, J. M., Wheeler, A. J., Freiwald, A., and Cairns, S. (2009). Cold-Water Corals: The Biology and Geology of Deep-Sea Coral Habitats. Cambridge: Cambridge University Press, 334. doi: 10.1017/CBO9780511581588
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shimodaira, H. (2002). An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508. doi: 10.1080/10635150290069913
Sinniger, F., Chevaldonn,é, P., and Pawlowski, J. (2007). Mitochondrial genome of Savalia savaglia (Cnidaria, Hexacorallia) and early metazoan phylogeny. J. Mol. Evol. 64, 196–203. doi: 10.1007/s00239-006-0015-0
Sinniger, F., and Pawloski, J. (2009). The partial mitochondrial genome of Leiopathes glaberrima (Hexacorallia: Antipatharia) and the first report of the presence of an intron in COI in black corals. Galaxea, J. Coral Reef Stud. 11, 21–26. doi: 10.3755/galaxea.11.21
Smith, A. B. (1994). Rooting molecular trees: problems and strategies. Biol. J. Linn. Soc. 51, 279–292. doi: 10.1111/j.1095-8312.1994.tb00962.x
Su, M., Ling, Y., Yu, J., Wu, J., and Xiao, J. (2013). Small proteins: untapped area of potential biological importance. Front. Genet. 4:286. doi: 10.3389/fgene.2013.00286
Thomas, J. M., Horspool, D., Brown, G., Tcherepanov, V., and Upton, C. (2007). GraphDNA: a java program for graphical display of DNA composition analyses. BMC Bioinformatics 8:21. doi: 10.1186/1471-2105-8-21
van Pesch, A. J. (1914). The antipatharia of the siboga expedition. Siboga-Expeditie Monographe 17, 1–258.
Wagner, D., Brugler, M. R., Opresko, D. M., France, S. C., Montgomery, A. D., and Toonen, R. J. (2010). Using morphometrics, in situ observations and genetic characters to distinguish among commercially valuable Hawaiian black coral species. Invertebr. Syst. 24, 271–290. doi: 10.1071/IS10004
Wagner, D., Luck, D. G., and Toonen, R. J. (2012). The biology and ecology of black corals (Cnidaria: Anthozoa: Hexacorallia: Antipatharia). Adv. Mar. Biol. 63, 67–132. doi: 10.1016/B978-0-12-394282-1.00002-8
Wolstenholme, D. R. (1992). Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 141, 173–216. doi: 10.1016/S0074-7696(08)62066-5
Keywords: cnidaria, gene order, next generation sequencing, NGS, HEG, barcoding, deep-sea, COX1 intron
Citation: Barrett NJ, Hogan RI, Allcock AL, Molodtsova T, Hopkins K, Wheeler AJ and Yesson C (2020) Phylogenetics and Mitogenome Organisation in Black Corals (Anthozoa: Hexacorallia: Antipatharia): An Order-Wide Survey Inferred From Complete Mitochondrial Genomes. Front. Mar. Sci. 7:440. doi: 10.3389/fmars.2020.00440
Received: 23 January 2020; Accepted: 19 May 2020;
Published: 23 June 2020.
Edited by:
Luisa Fernanda Dueñas, National University of Colombia, ColombiaReviewed by:
Mercer Robert Brugler, New York City College of Technology (CUNY), United StatesJaret Bilewitch, The University of Queensland, Australia
Copyright © 2020 Barrett, Hogan, Allcock, Molodtsova, Hopkins, Wheeler and Yesson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nick J. Barrett, bi5qLmJhcnJldHRAb3V0bG9vay5jb20=
 Nick J. Barrett
Nick J. Barrett Raissa I. Hogan
Raissa I. Hogan A. Louise Allcock
A. Louise Allcock Tina Molodtsova
Tina Molodtsova Kevin Hopkins
Kevin Hopkins Andrew J. Wheeler
Andrew J. Wheeler Chris Yesson
Chris Yesson