- 1Department of Microbiology, Faculty of Science, Mahidol University, Bangkok, Thailand
- 2Center of Excellence for Shrimp Molecular Biology and Biotechnology, Faculty of Science, Mahidol University, Bangkok, Thailand
- 3Department of Morphophysiology and Pharmacology, College of Veterinary Science and Medicine, Central Luzon State University, Muñoz, Philippines
- 4Department of Biochemistry, Faculty of Science, Mahidol University, Bangkok, Thailand
- 5Integrative Computational BioScience (ICBS) Center, Mahidol University, Nakhon Pathom, Thailand
- 6Systems Biology of Diseases Research Unit, Faculty of Science, Mahidol University, Bangkok, Thailand
- 7National Center of Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailand
Background
Early mortality syndrome (EMS) has emerged as a devastating disease in shrimp production aquaculture since it was firstly discovered in China (2009), and onward in Malaysia (2011), Thailand (2012), and Mexico (2013) (Thitamadee et al., 2016). This disease leads to 100% shrimp mortality within 30 days after the infection and thus, economic loss of over USD 1 billion per annum worldwide (de Schryver et al., 2014; Zorriehzahra and Banaederakhshan, 2015). Penaeus monodon and P. vannamei shrimps in the post larva stage are the hosts of the EMS causative Gram-negative bacterium, V. parahaemolyticus (Flegel, 2012).
V. parahaemolyticus is a Gram-negative bacterium inhabiting in marine and estuarine environments (Tran et al., 2013). The same bacterium is also one of human foodborne pathogens that cause gastroenteritis after consumption of contaminated seafood (Letchumanan et al., 2014). Toxins of V. parahaemolyticus acting as porins in enterocyte plasma membrane and interrupting ion flux in intestine are thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH). TDH and TRH associate with V. parahaemolyticus hemolytic and cytotoxicity activities in the host. Another hemolysin is thermolabile hemolysin (TLH), which also causes red blood cell lysis (Wang et al., 2015). Moreover, asymptomatic infection or presence of pathogenic strains in shrimp farms or marine environments can be a reservoir for human infection.
Nowadays, EMS-causing V. parahaemolyticus isolates have been classified into two groups based on their histological signs: acute hepatopancreatic necrosis syndrome (AHPND) and non-AHPND (Joshi et al., 2014). AHPND V. parahaemolyticus, e.g., the 5HP strain, carries a 69-kb plasmid named pVA1 that contains V. parahaemolyticus Photorhabdus, insect-related (Pir) toxin-like genes, pirAvp and pirBvp (Lee et al., 2015; Lin et al., 2017). These PirABvp toxins damage the hepatopancreatic cells by necrosis, called sloughing cells. The shrimps infected by the AHPND V. parahaemolyticus normally completely die within 30 days. Gross signs of infected shrimps by AHPND strains show empty stomach and midgut, and pale hepatopancreas (Zorriehzahra and Banaederakhshan, 2015). On the other hand, the non-AHPND strains, e.g., the 2HP strain, cannot produce PirABvp toxins (Joshi et al., 2014). Infected shrimps by the non-AHPND strains still show empty stomach and pale hepatopancreas. Hepatopancreas tubule epithelia show thinness and numerous vacuoles in E-cell or collapsed cells. In immersion challenge testing, 2HP causes 75% of shrimps to die within 96 h-post infection; in contrast, the AHPND isolates (1D, 3HP, 5HP, and “China”) cause 100% mortality in shrimp in the same period of time (Joshi et al., 2014). 2HP may maintain other virulence factors that causes collapsed lesion in shrimp. Recently, we learned that the non-AHPND V. parahaemolyticus 2HP carries three plasmids: 183-kb pVP2HP, 65-kb pVP2HPa, and 50-kb pVP2HPb (Theethakaew et al., 2017). The 183-kb pVP2HP shares genetic element of combined 69-kb pVPE61a (containing pirAvp and pirBvp genes) and 118-kb pVPE61b, but pirAvp and pirBvp genes are absent (Theethakaew et al., 2017). However, virulence factors in non-AHPND V. parahaemolyticus that cause collapsed lesion and lead to EMS pathogenesis are still unclear. The virulence factor candidates in the non-AHPND V. parahaemolyticus 2HP might be maintained in its chromosomes (I and II). In order to provide better insights into the virulence factors in the non-AHPND V. parahaemolyticus, for the first time to the best of our knowledge, we performed whole genome sequencing of V. parahaemolyticus 2HP, and comprehensively compare with the existing genomes of the AHPND V. parahaemolyticus isolates (3HP, 5HP, and China), as well as non-pathogenic V. parahaemolyticus S02 (Yang et al., 2014). The virulence genes and pathogenicity islands were predicted in order to explore putative factors that might be involved in non-AHPND EMS pathogenesis. In addition to the pathogenic protein-coding genes, non-coding RNAs (ncRNAs) are one of the factors regulating many processes of cell and affect protein expression or activity (Gottesman and Storz, 2011). Some ncRNAs involve in regulation of virulence factors and pathogenesis (Huang et al., 2015; Wang et al., 2016; Pappesch et al., 2017). Thus, we also predicted ncRNAs from the newly sequenced non-AHPND V. parahaemolyticus 2HP to investigate its involvement in EMS pathogenesis. In addition, antibiotic resistant genes were predicted which suggested the incident of drug resistance in the shrimp pathogen. The genome sequences of non-AHPND and AHPND isolates from Thailand and other Asian countries were compared to elucidate common and unique mechanisms of pathogenesis in EMS-causing V. parahaemolyticus.
Data Description
Bacterial Strain, Culture Conditions, and Genomic DNA Extraction
V. parahaemolyticus 2HP was cultured in tryptic soy broth (TSB) supplemented with 1.5% NaCl. The bacterial culture was incubated at 30°C and shaken at 200 rpm overnight. Then, bacterial cells were collected by centrifugation. Genomic DNA of V. parahaemolyticus 2HP was extracted by the QIAamp DNA Micro kit (Qiagen, Hilden, Germany). Quantity and quality of extracted genomic DNA was measured by the Denovix spectrophotometer (Denovix, DE, USA) and 1.5% agarose gel electrophoresis.
Genomic Sequencing and Annotation
Whole genome sequencing library of genomic DNA was prepared by the QIAseq FX DNA library kit (Qiagen, Hilden, Germany) and the library was sequenced on the Illumina HiSeq 2500 platform (Illumina, CA, UAS). Sequences were trimmed and assembled de novo by the CLC genomic workbench version 11.0.1 (CLC bio, Denmark). De novo assembled contigs were evaluated for quality, and genome size was estimated against V. parahaemolyticus RIMD2210633 (Makino et al., 2003) used as the reference genome using QUAST (Gurevich et al., 2013). Non-coding RNA, rRNA, and tRNA were predicted using Infernal (Kolbe and Eddy, 2011), RNAmmer (Lagesen et al., 2007) and Aragorn (Laslett and Canback, 2004) available from the PROKKA pipeline (Seemann, 2014) via the Galaxy server (Afgan et al., 2018). Genes and virulence factors were predicted using Rapid Annotation using Subsystem Technology tool kit (RASTtk) (Brettin et al., 2015). Pathogenicity islands (PAIs) and mobile elements were predicted using VRprofile 2.0 (Li et al., 2018a). HMMer E-value at 0.0001 is set as a cut-off. Minimum number of the significant hits for each prophage-like region was 5. Other strains of AHPND and non-pathogenic V. parahaemolyticus were selected to analyze and against 2HP as well. The V. parahaemolyticus 3HP, 5HP, and China isolates were all AHPND strains, obtained from GenBank using the accession numbers JPKS00000000, JPKT00000000, and JPKU00000000, respectively (Yang et al., 2014). The V. parahaemolyticus S02 (accession number JPKV00000000) was a non-pathogenic strain used in this comparison (Yang et al., 2014). Antibiotic resistant genes were predicted by ResFinder 3.1 (Zankari et al., 2012) and PATRIC 3.5.39 (Wattam et al., 2017). The comparative genomics were performed by protein family sorter program with PATRIC genus-specific families (PLfams) as a filter in the PATRIC 3.5.39 server (Wattam et al., 2017). This program uses k-mer-based functional assessment to categorize the protein family formation and differentiates the cluster of protein families across different genomes using Markov Cluster algorithm (MCL), MCL inflation value = 3 as a cut-off (Davis et al., 2016). The comparison of zot and ace nucleotide sequences were performed by Multiple Sequence Alignment, EMBL-EBI (Madeira et al., 2019) and BLASTN, NCBI (Johnson et al., 2008).
Genome Characteristics of V. parahaemolyticus Strain 2HP
The genome assembly of V. parahaemolyticus 2HP is consisted of 5,501,309 bp in 194 contigs. There are 97 RNAs i.e., 4 rRNA, and 93 tRNA. The G+C content of this assembled genome is 45.2%. The quality assessment of draft genome from QUAST showed that N50 (representing the contig length that longer or equal length contigs presents half of the bases of the assembly) and L50 (representing the minimum number of contigs that present half of the bases of the assembly) were 183,055 bp and 10, respectively. Two chromosomes of 2HP were estimated to be 2,996,005 and 1,724,740 bp for chromosome I and II, respectively. RASTtk analysis showed that, from the whole genome, the number of coding sequences was 5,424 and the number of functional subsystems was 406 (Figure 1).
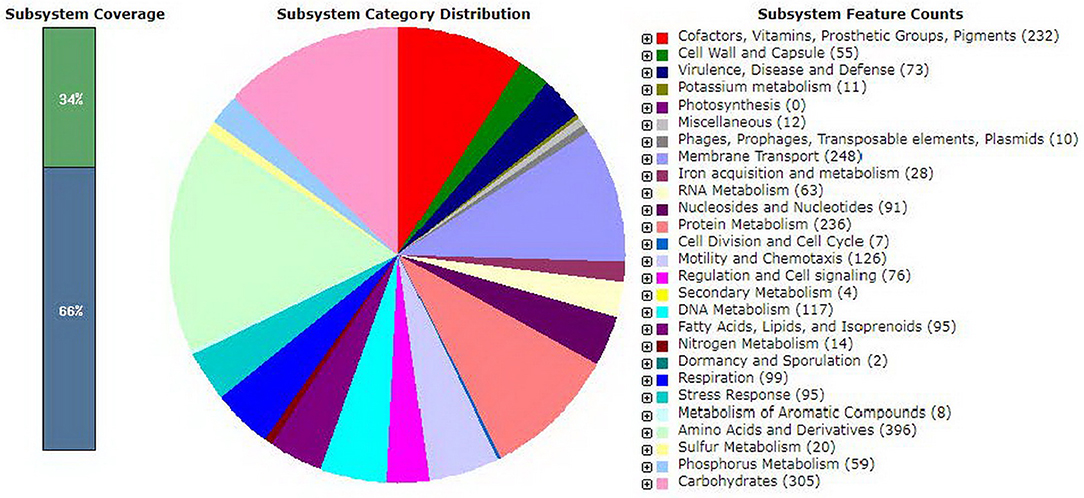
Figure 1. Subsystem category distribution of V. parahaemolyticus 2HP (results of RASTtk annotation). Subsystem coverage shows sets of functional roles in specific biological processes (green box) and non-specific processes (blue box). Overall genome annotation is represented in a pie chart of subsystem category distribution. Different colors in pie chart explain details and numbers of genes in subsystem feature counts.
Virulence and Antibiotic Resistance Genes in V. parahaemolyticus Strain 2HP
From the RASTtk analysis, we found four encoding homologs of V. cholerae toxins, which are also similar to the V. parahaemolyticus AHPND strains (Yang et al., 2014). Six toxin genes are two zona occluden toxins (zot), two accessory cholera toxins (ace), the gene encoding transcriptional activator ToxR (toxR), and the gene encoding transmembrane regulatory protein ToxS (toxS). 2HP carries two different sizes of putative ace, 345 and 333 bp, renamed putative ace1 and ace2, respectively. 3HP and 5HP also have two types of putative ace, but the China strain has only one putative ace1 (345 bp). The 2HP zot is also consisted of two types with different sizes, 1,401 and 1,425 bp, called putative zot1 and zot2, respectively. Similar with ace, 3HP and 5HP carry two types of zot. A unique gene found only in the China strain is a putative zot (1,386 bp), which has a different size when compared to the Thai isolates. The zot1 and ace1 genes are maintained on chromosome I while zot2 and ace2 are carried on chromosome II. We also found only one type of hemolysin in the current draft genome, the thermolabile hemolysin or tlh gene. These toxin genes might play a role in the EMS pathogenesis in shrimps. In addition, the presence of zot and ace genes, which are virulence factors in clinical field (Perez-Reytor et al., 2018), indicates that this 2HP strain could cause disease in human, despite lacking thermostable directed hemolysin (tdh) and TDH-related hemolysin (trh) genes (Castillo et al., 2018). Presence of zot1, zot2, ace1, and ace2 genes was confirmed by PCR (Figures S1–S3).
VRprofile predicted 19 mobile elements and secretion systems. We discovered one integrative and conjugation element (ICE), 12 insertion sequence elements, one type-3 secretion system, one type-4 secretion system, two type-6 secretion systems, and two prophages. Interestingly, one of two prophages in 2HP contains zot1 and ace1 (Figure 2A). This prophage contains 12 encoding genes for RstB phage-related integrase, seven hypothetical proteins, accessory cholera toxin 1, zona occluden toxin 1, putative plasmid partitioning protein Soj or ParA-like protein, and bacteriophage f237 repressor 1 (Table S1).
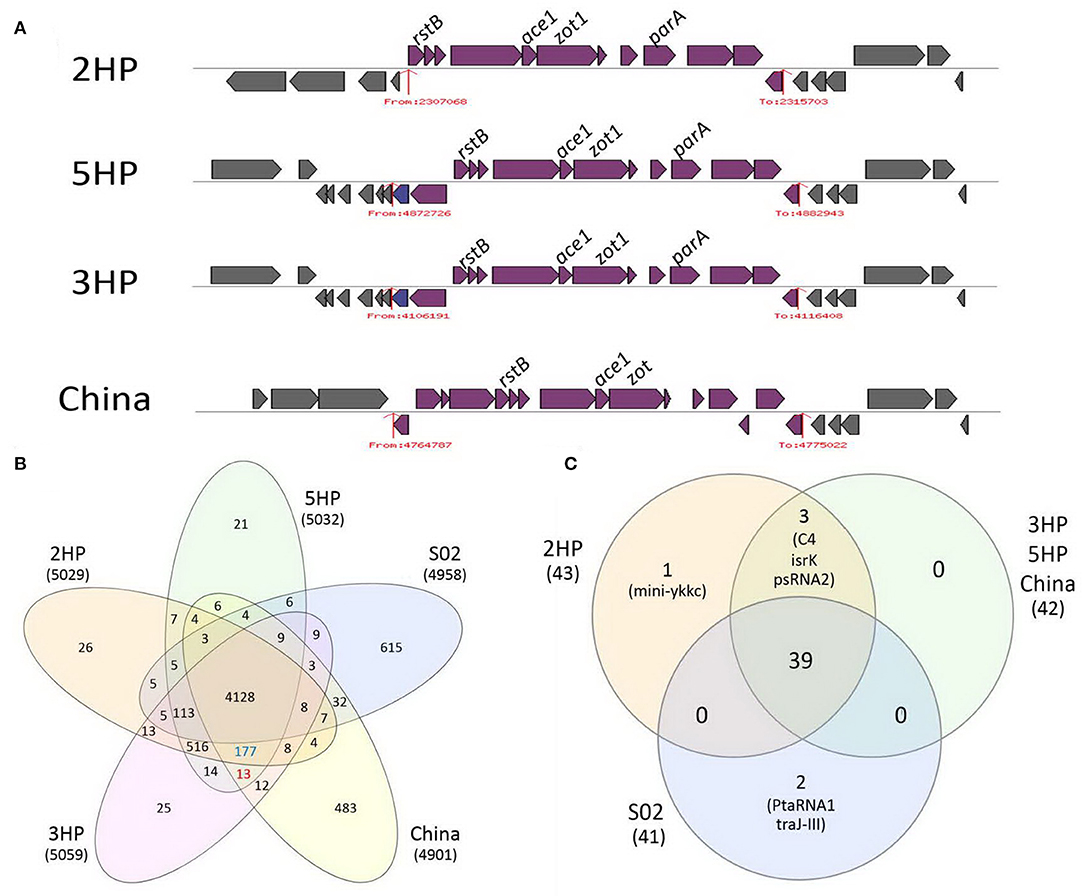
Figure 2. Comparative analyses of (A) prophage structures based on VRprofile prediction, (B) protein families based on PATRIC genus-specific families, PLfams, and (C) ncRNAs (C) (based on Infernal prediction) among non-AHPND V. parahaemolyticus (2HP), V. parahaemolyticus AHPND (3HP, 5HP, and China), and non-pathogenic V. parahaemolyticus (S02). (A) The gray, purple, and dark blue blocks represent genes of outer structure of prophage, prophage, and PAI, respectively. Red arrows and numbers show boundary and start/stop positions of prophage structures. Labeled regions show regions of rstB, zot, ace, and parA-like genes. (B) shows five-circle Venn diagram of distribution of protein families across different V. parahaemolyticus genomes. The orange, purple, green, yellow, and blue circles represent 2HP, 3HP, 5HP, China, and, S02, respectively, with number of total genes in each strain. The overlapped areas show shared protein families among strains, and numbers of proteins found in all strains are shown. Numbers of specific features in AHPND (3HP, 5HP, and China) and EMS (AHPND strains and 2HP) areas show in red and blue, respectively. (C) shows ncRNA prediction in V. parahaemolyticus isolates. Orange, green, and blue circles represent 2HP, AHPND strains, and S02, respectively, with the number of predicted ncRNAs of each strain. The overlapped areas represent the shared predicted ncRNAs among strains, and numbers of ncRNAs found in all strains are shown.
In addition to the virulence genes and putative PAIs described, RASTtk also annotated 34 genes that are putatively responsible for resistance to antibiotics and toxic compounds: four genes for resistance to fluoroquinolones, four genes for multidrug resistance efflux pumps, one gene for resistance to chromium compounds, 11 genes for cobalt-zinc-cadmium resistance, 14 genes for copper homeostasis (Table S5). The genome analysis on ResFinder 3.1 (% identity threshold and minimized length were 80 and 60% as cut-offs) and PATRIC (based on database of National Database of Antibiotic Resistant Organisms) found presence of one beta-lactam resistant gene, beta-lactamase CARB-type (blaCARB-41), and two tetracycline resistant genes, tet-34 and tet-35. In aquaculture, tetracycline is one of common drugs for the treatment of bacterial diseases, e.g., vibriosis (Neela et al., 2007), and beta-lactam drugs (such as ampicillin and penicillin) have been applied in human and animal treatment, (Fernandes et al., 2013). Our finding supports the significance of antimicrobial resistance gene surveillance in aquaculture.
Comparisons of Pathogenicity Islands Prediction With V. parahaemolyticus AHPND Isolates
We next investigated the common and unique PAIs in our newly sequence non-AHPND 2HP and other existing EMS V. parahaemolyticus AHPND 3HP, 5HP, China, and the reference S02 strains. We found the consistency of predicted prophage in the EMS-causing strains on chromosome I, but not in the non-pathogenic strain (Figure 2A and Table S1). Prophage carrying zot1 and ace1 genes was found in 3HP and 5HP strains, while China strain was presented with zot and ace1 in prophage. Those prophage structures are closely related to pre-CTX prophage (lacking ctxAB in CTX prophage) (Pant et al., 2019). In V. mimicus, putative pre-CTX prophage was found in a clinical strain MB451, not in an environmental strain VM223 (Hasan et al., 2010). The putative pre-CTX prophage is consistently found in EMS strains and might be involved with EMS pathogenesis, in addition to pirABvp.
Non-coding RNA Prediction and Comparison
The prediction of ncRNAs by the Infernal program (Kolbe and Eddy, 2011) showed 43 putative ncRNAs in 2HP, 42 in AHPND strains, and 41 in S02 (Figure 2C). We found that the C4, IsrK, and psRNA2 ncRNAs are present only in the EMS-causing strains (2HP, 3HP, 5HP, and China), but not in S02. C4 targets incoming bacteriophage and/or interact with bacteriophage transcription (Weiss et al., 2016) while IsrK interferes with production of AntQ, an antitermination protein for transcription (Hershko-Shalev et al., 2016). The function of psRNA2; however, is still unclear (Gvakharia et al., 2010). For unique ncRNAs, mini-ykkc ncRNA, a class II guanidine riboswitch (Sherlock et al., 2017), is only found in 2HP, while PtaRNA1 and TraJ-II ncRNAs are found only in S02. PtaRNA1 is an antisense antitoxin ncRNA in XCV2162-ptaRNA1 system belonging to class I toxin-antitoxin system (Findeiss et al., 2010). TraJ-II is found in 5′-UTR of traJ gene and functions in bacterial conjugation process (Li et al., 2018b). We validated the presence of predicted ncRNA in 2HP by RT-qPCR. Spot42, RyhB, and mini-ykkc were chosen as representatives and their transcript levels shown in (Figure S4) confirmed expression of the selected ncRNAs.
Comparative Pangenomic Analyses of EMS-Causing V. parahaemolyticus Isolates
Not only virulence factors lead to pathogenesis, but some genes known to be involved in the bacterial fitness, which are specific to each strain, may also play crucial roles in disease establishment (Diard and Hardt, 2017). Hence, 2HP may carry some features, in addition to zot and ace, which overlap with AHPND strains or features unique to 2HP which lead to EMS in shrimp. To investigate the strain-specific features and unique features in each group of AHPND and EMS, we selected 2HP, 3HP, 5HP, China, and S02 for pan-genomic analysis by the protein family sorter program in PATRIC (Figure 2B). The number of unique genes in 2HP, 3HP, 5HP, China, and S02 are 26, 25, 21, 483, and 615 genes, respectively (Table S4). The 2HP-specifc genes include one encoding a transposase and 25 encode hypothetical proteins.
Pangenome (all genes from all genomes) and core genomes (present in all the genomes) contained 6,287 and 4,128 genes, respectively. Accessory genome (genes present in specific strains) in AHPND (3HP, 5HP, and China) and EMS strains (AHPND strains and 2HP) contained 13 and 177 genes, respectively (Tables S2, S3). AHPND-specific genes are pirAvp, pirBvp, one gene encoding peptidase M20, and 10 hypothetical genes. EMS strain-specific genes compose of many genes encoding functional proteins in methylation and acetylation e.g., S-adenosylmethionine (SAM)-dependent methyltransferase, acetyltransferase, histone acetyltransferase HPA2. The rsbW gene, which encodes a serine kinase RsbW, is specific to the EMS groups. Free RsbW phosphorylates SypA leading to regulation of biofilm formation (Morris and Visick, 2010). The ORFs for capsular polysaccharide biosynthesis protein and YjbG polysaccharide synthesis-related protein, important in capsule formation and adherence to surface (Ferrieres et al., 2007; Broberg et al., 2011), are also EMS-specific.
In terms of key virulence factors, nucleotide sequences of accessory cholera toxin and zona occluden toxin in China strain are different from those of EMS strains from Thailand (2HP, 3HP, and 5HP). The zot and ace1 from China strain are closely related to zot and ace in V. parahaemolyticus RIMD2210633 (clinical strain). The zot gene from China strain shows 97.62% similarity to RIMD2210633 strain while the zot1 and zot2 genes from Thai isolates share 50.73 and 51.06% nucleotide similarity, respectively, to RIMD2210633 strain. The ace1 gene from China strain shows 99.13% nucleotide similarity to RIMD2210633 strain while ace1 and ace2 genes of Thai isolates show 49.86 and 47.17% nucleotide similarity, respectively, to RIMD2210633. Therefore, zot1, zot2, ace1 (Thai), and ace2 genes are considered members of 2HP-3HP-5HP accessory genome which are different from those of RIMD2210633 and China strains.
In summary, we have for the first time sequenced the non-AHPND 2HP V. parahaemolyticus and comprehensively annotated the newly sequence genome. Our comparative genomic analysis revealed the “conserved” virulence genes and prophage commonly found in both pathogenic AHPND and non-AHPND strains. In V. cholerae, Zot increases permeability of epithelial barrier by disassembling the intercellular tight junctions, and Ace increases ion transport and water secretion (Perez-Reytor et al., 2018). Sequences of zot and ace in 2HP, 3HP, and 5HP are different from those of clinical strain RIMD2210633. Therefore, the activities of Zot and Ace might be more specific to shrimp than human. However, zot and ace in the China strain are closely similar to those of RIMD2210633. Zot and Ace in this China strain might affect both human and shrimp. Both toxins in non-AHPND and AHPND strains might have similar functions in shrimp, similar to other hepatopancreatic toxins such as to PirABvp toxins (Prachumwat et al., 2019). Prophage carrying zot and ace might be necessary for EMS pathogenesis in both AHPND and non-AHPND. Moreover, this prophage may be used as biomarker for diagnosis of EMS-causing V. parahaemolyticus. Not only virulence genes and prophage, but ncRNA also showed specific patterns among the EMS strains. We also identified conserved ncRNAs in EMS strains involved in transcription process. These might be transcriptional factors for enhancement of EMS pathogenesis. From comparative genomic analysis, we found that most of the strains-specific genes were hypothetical genes. Interestingly, we found ORFs which encode regulatory protein and capsule formation proteins in EMS group. This finding suggests that EMS strains, AHPND and non-AHPND, carry the special features that S02 lacks. These genes might enhance EMS strains to induce the virulence and pathogenesis. The 2HP-specific genes contain a lot of hypothetical genes. 2HP may maintain some unique features that involve in EMS mechanism of the non-AHPND strain. The functions of these gene are still unknown. Further investigation is required to identify mechanisms of pathogenesis in AHPND and non-AHPND strain for EMS.
Data Availability Statement
The genome data of V. parahaemolyticus 2HP from this study had been submitted to DDBJ/EMBL/GenBank under the accession number SRKW00000000. The version of genome sequence described in this work is the first version, SRKW01000000.
Author Contributions
NT performed the experiments, analyzed the data analysis, and prepared the manuscript. MS assisted with antibiotic resistant gene prediction. VC, VS, and SC designed the study and prepared the manuscript. All authors proofread the manuscript.
Funding
NT was supported by funding from Thailand Graduate Institute of Science and Technology (TGIST), National Science and Technology Development Agency (NSTDA), Thailand (SCA-CO-2559-2502-TH). This study was supported by Mahidol University. VC was supported by the Thailand Research Fund (TRF) grants MRG6080235 and DBG6080003.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors thank Dr. Kallaya Sritunyalucksana Dangtip for kindly providing V. parahaemolyticus 2HP.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00290/full#supplementary-material
References
Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Cech, M., et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Castillo, D., Perez-Reytor, D., Plaza, N., Ramirez-Araya, S., Blondel, C. J., Corsini, G., et al. (2018). Exploring the genomic traits of non-toxigenic Vibrio parahaemolyticus strains isolated in Southern chile. Front. Microbiol. 9:161. doi: 10.3389/fmicb.2018.00161
Davis, J. J., Gerdes, S., Olsen, G. J., Olson, R., Pusch, G. D., Shukla, M., et al. (2016). PATtyFams: protein families for the microbial genomes in the PATRIC database. Front. Microbiol. 7:118. doi: 10.3389/fmicb.2016.00118
de Schryver, P., Defoirdt, T., and Sorgeloos, P. (2014). Early mortality syndrome outbreaks: a microbial management issue in shrimp farming? PLoS Pathog. 10:e1003919. doi: 10.1371/journal.ppat.1003919
Diard, M., and Hardt, W. D. (2017). Evolution of bacterial virulence. FEMS Microbiol. Rev. 41, 679–697. doi: 10.1093/femsre/fux023
Fernandes, R., Amador, P., and Prudêncio, C. (2013). β-Lactams: chemical structure, mode of action and mechanisms of resistance. Rev. Med Microbiol. 24, 7–17. doi: 10.1097/MRM.0b013e3283587727
Ferrieres, L., Aslam, S. N., Cooper, R. M., and Clarke, D. J. (2007). The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153, 1070–1080. doi: 10.1099/mic.0.2006/002907-0
Findeiss, S., Schmidtke, C., Stadler, P. F., and Bonas, U. (2010). A novel family of plasmid-transferred anti-sense ncRNAs. RNA Biol. 7, 120–124. doi: 10.4161/rna.7.2.11184
Flegel, T. W. (2012). Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 110, 166–173. doi: 10.1016/j.jip.2012.03.004
Gottesman, S., and Storz, G. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. doi: 10.1101/cshperspect.a003798
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Gvakharia, B. O., Tjaden, B., Vajrala, N., Sayavedra-Soto, L. A., and Arp, D. J. (2010). Computational prediction and transcriptional analysis of sRNAs in Nitrosomonas europaea. FEMS Microbiol. Lett. 312, 46–54. doi: 10.1111/j.1574-6968.2010.02095.x
Hasan, N. A., Grim, C. J., Haley, B. J., Chun, J., Alam, M., Taviani, E., et al. (2010). Comparative genomics of clinical and environmental Vibrio mimicus. Proc. Natl. Acad. Sci. U. S. A. 107, 21134–21139. doi: 10.1073/pnas.1013825107
Hershko-Shalev, T., Odenheimer-Bergman, A., Elgrably-Weiss, M., Ben-Zvi, T., Govindarajan, S., Seri, H., et al. (2016). Gifsy-1 prophage IsrK with dual function as small and messenger RNA modulates vital bacterial machineries. PLoS Genet. 12:e1005975. doi: 10.1371/journal.pgen.1005975
Huang, L., Hu, J., Su, Y., Qin, Y., Kong, W., Ma, Y., et al. (2015). Identification and characterization of three Vibrio alginolyticus non-coding RNAs involved in adhesion, chemotaxis, and motility processes. Front. Cell Infect. Microbiol. 5:56. doi: 10.3389/fcimb.2015.00056
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Joshi, J., Srisala, J., Truong, V. H., Chen, I. T., Nuangsaeng, B., Suthienkul, O., et al. (2014). Variation in Vibrio parahaemolyticus isolates from a single thai shrimp farm experiencing an outbreak of acute hepatopancreatic necrosis disease (AHPND). Aquaculture 428–429, 297–302. doi: 10.1016/j.aquaculture.2014.03.030
Kolbe, D. L., and Eddy, S. R. (2011). Fast filtering for RNA homology search. Bioinformatics 27, 3102–3109. doi: 10.1093/bioinformatics/btr545
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Laslett, D., and Canback, B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. doi: 10.1093/nar/gkh152
Lee, C. T., Chen, I. T., Yang, Y. T., Ko, T. P., Huang, Y. T., Huang, J. Y., et al. (2015). The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. U. S. A. 112, 10798–10803. doi: 10.1073/pnas.1503129112
Letchumanan, V., Chan, K. G., and Lee, L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Li, J., Tai, C., Deng, Z., Zhong, W., He, Y., and Ou, H. Y. (2018a). VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief Bioinform. 19, 566–574. doi: 10.1093/bib/bbw141
Li, Y., da Silva, G. C., Li, Y., Rossi, C. C., Fernandez Crespo, R., Williamson, S. M., et al. (2018b). Evidence of illegitimate recombination between two pasteurellaceae plasmids resulting in a novel multi-resistance replicon, pM3362MDR, in Actinobacillus pleuropneumoniae. Front. Microbiol. 9:2489. doi: 10.3389/fmicb.2018.02489
Lin, S. J., Hsu, K. C., and Wang, H. C. (2017). Structural insights into the cytotoxic mechanism of Vibrio parahaemolyticus PirA(vp) and PirB(vp) toxins. Mar. Drugs 15:373. doi: 10.3390/md15120373
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749. doi: 10.1016/S0140-6736(03)12659-1
Morris, A. R., and Visick, K. L. (2010). Control of biofilm formation and colonization in Vibrio fischeri: a role for partner switching? Environ. Microbiol. 12, 2051–2059. doi: 10.1111/j.1462-2920.2010.02269.x
Neela, F. A., Nonaka, L., and Suzuki, S. (2007). The diversity of multi-drug resistance profiles in tetracycline-resistant Vibrio species isolated from coastal sediments and seawater. J. Microbiol. 45, 64–68.
Pant, A., Das, B., and Bhadra, R. K. (2019). CTX phage of Vibrio cholerae: genomics and applications. Vaccine 38(Suppl.1), A7–A12. doi: 10.1016/j.vaccine.2019.06.034
Pappesch, R., Warnke, P., Mikkat, S., Normann, J., Wisniewska-Kucper, A., Huschka, F., et al. (2017). The regulatory small RNA MarS supports virulence of Streptococcus pyogenes. Sci. Rep. 7:12241. doi: 10.1038/s41598-017-12507-z
Perez-Reytor, D., Jana, V., Pavez, L., Navarrete, P., and Garcia, K. (2018). Accessory toxins of Vibrio pathogens and their role in epithelial disruption during infection. Front. Microbiol. 9:2248. doi: 10.3389/fmicb.2018.02248
Prachumwat, A., Taengchaiyaphum, S., Mungkongwongsiri, N., Aldama-Cano, D. J., Flegel, T. W., and Sritunyalucksana, K. (2019). Update on early mortality syndrome/acute hepatopancreatic necrosis disease by April 2018. J. World Aquaculture Soc. 50, 5–17. doi: 10.1111/jwas.12559
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sherlock, M. E., Malkowski, S. N., and Breaker, R. R. (2017). Biochemical validation of a second guanidine Riboswitch class in bacteria. Biochemistry 56, 352–358. doi: 10.1021/acs.biochem.6b01270
Theethakaew, C., Nakamura, S., Motooka, D., Matsuda, S., Kodama, T., Chonsin, K., et al. (2017). Plasmid dynamics in Vibrio parahaemolyticus strains related to shrimp Acute Hepatopancreatic Necrosis Syndrome (AHPNS). Infect. Genet. Evol. 51, 211–218. doi: 10.1016/j.meegid.2017.04.007
Thitamadee, S., Prachumwat, A., Srisala, J., Jaroenlak, P., Salachan, P. V., Sritunyalucksana, K., et al. (2016). Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 452, 69–87. doi: 10.1016/j.aquaculture.2015.10.028
Tran, L., Nunan, L., Redman, R. M., Mohney, L. L., Pantoja, C. R., Fitzsimmons, K., et al. (2013). Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis Aquat. Organ. 105, 45–55. doi: 10.3354/dao02621
Wang, L., Yang, G., Qi, L., Li, X., Jia, L., Xie, J., et al. (2016). A novel small RNA regulates tolerance and virulence in Shigella flexneri by responding to acidic environmental changes. Front. Cell Infect. Microbiol. 6:24. doi: 10.3389/fcimb.2016.00024
Wang, R., Zhong, Y., Gu, X., Yuan, J., Saeed, A. F., and Wang, S. (2015). The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front. Microbiol. 6:144. doi: 10.3389/fmicb.2015.00144
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. doi: 10.1093/nar/gkw1017
Weiss, A., Broach, W. H., Lee, M. C., and Shaw, L. N. (2016). Towards the complete small RNome of Acinetobacter baumannii. Microb. Genom 2:e000045. doi: 10.1099/mgen.0.000045
Yang, Y. T., Chen, I. T., Lee, C. T., Chen, C. Y., Lin, S. S., Hor, L. I., et al. (2014). Draft genome sequences of four strains of Vibrio parahaemolyticus, three of which cause early mortality syndrome/acute hepatopancreatic necrosis disease in shrimp in China and Thailand. Genome Announc. 2:00816–14. doi: 10.1128/genomeA.00816-14
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Vibrio parahaemolyticus, non-acute hepatopancreatic necrosis disease, early mortality syndrome, shrimp disease, comparative genomics
Citation: Thadtapong N, Salinas MBS, Charoensawan V, Saksmerprome V and Chaturongakul S (2020) Genome Characterization and Comparison of Early Mortality Syndrome Causing Vibrio parahaemolyticus pirABvp− Mutant From Thailand With V. parahaemolyticus pirABvp+ AHPND Isolates. Front. Mar. Sci. 7:290. doi: 10.3389/fmars.2020.00290
Received: 10 November 2019; Accepted: 09 April 2020;
Published: 28 April 2020.
Edited by:
Weiqi Fu, New York University Abu Dhabi, United Arab EmiratesReviewed by:
Kumari Kavita, National Institutes of Health (NIH), United StatesFudan Gao, Shanghai Normal University, China
Copyright © 2020 Thadtapong, Salinas, Charoensawan, Saksmerprome and Chaturongakul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soraya Chaturongakul, c29yYXlhLmNoYUBtYWhpZG9sLmFjLnRo
 Nalumon Thadtapong
Nalumon Thadtapong Marvin Bryan Segundo Salinas
Marvin Bryan Segundo Salinas Varodom Charoensawan
Varodom Charoensawan Vanvimon Saksmerprome
Vanvimon Saksmerprome Soraya Chaturongakul
Soraya Chaturongakul