- 1Protistology & Aquatic Ecology, Ghent University, Ghent, Belgium
- 2Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
- 3VIB Center for Plant Systems Biology, VIB, Ghent, Belgium
- 4UMI3376 Takuvik Joint International Laboratory, Département de Biologie, Centre National de la Recherche Scientifique/Université Laval, Québec, QC, Canada
- 5Pflanzliche Ökophysiologie, Zukunftskolleg, Universität Konstanz, Konstanz, Germany
- 6Cell, Molecular Biology and Genomics Group, Department of Biology, Norwegian University of Science and Technology, Trondheim, Norway
Intertidal benthic diatoms experience a highly variable light regime, which especially challenges these organisms to cope with excess light energy during low tide. Non-photochemical quenching of chlorophyll fluorescence (NPQ) is one of the most rapid mechanisms diatoms possess to dissipate excess energy. Its capacity is mainly defined by the xanthophyll cycle (XC) and Light-Harvesting Complex X (LHCX) proteins. Whereas the XC and its relation to NPQ have been relatively well-studied in both planktonic and benthic diatoms, our current knowledge about LHCX proteins and their potential involvement in NPQ regulation is largely restricted to planktonic diatoms. While recent studies using immuno-blotting have revealed the presence of light regulated LHCX proteins in benthic diatom communities and isolates, nothing is as yet known about the diversity, identity and transcriptional regulation or function of these proteins. We identified LHCX genes in the draft genome of the model benthic diatom Seminavis robusta and followed their transcriptional regulation during a day/night cycle and during exposure to high light conditions. The S. robusta genome contains 17 LHCX sequences, which is much more than in the sequenced planktonic model diatoms (4–5), but similar to the number of LHCX genes in the sea ice associated diatom Fragilariopsis cylindrus. LHCX diversification in both species, however, appears to have occurred independently. Interestingly, the S. robusta genome contains LHCX genes that are related to the LHCX6 of the model centric diatom Thalassiosira pseudonana, which are lacking in the well-studied pennate model diatom Phaeodactylum tricornutum. All investigated LHCX genes, with exception of SrLHCX6, were upregulated during the daily dark-light transition. Exposure to 2,000 μmol photons m–2 s–1, furthermore, increased transcription of all investigated LHCX genes. Our data suggest that the diversification and involvement of several light regulated LHCX genes in the photophysiology of S. robusta may represent an adaptation to the complex and highly variable light environment this benthic diatom species can be exposed to.
Introduction
Due to the complex interplay of diurnal and tidal cycles and weather conditions, the surface sediments of tidal flats experience highly variable light conditions. Nevertheless, they are very productive ecosystems thanks to the presence of biofilms dominated by benthic microalgae (microphytobenthos) and especially diatoms (Underwood and Kromkamp, 1999). The fluctuating light conditions challenge these diatoms to maximize light harvesting under low light (LL) conditions while avoiding oxidative damage to their photosynthetic apparatus under high light (HL), either by minimizing light absorbance or by the dissipation of excess light energy. Benthic diatoms possess two main strategies which are fast enough to track rapid fluctuations in light intensity, namely, vertical migration and dissipation of excess energy as heat (Lavaud and Goss, 2014; Laviale et al., 2016). Raphid pennate diatoms possess a special cell wall structure called the raphe through which mucilage is secreted, allowing the diatoms to move. Such motile diatoms, often referred to as epipelic diatoms, can form dense biofilms on fine-grained sediments (Sabbe, 1993; Ribeiro et al., 2013) and are able to position themselves within the sediment light gradient via vertical migration (Admiraal, 1984; Consalvey et al., 2004; Serôdio et al., 2006; Cartaxana et al., 2016). Dissipation of excess light energy as heat can be measured as Non-Photochemical Quenching of chlorophyll a fluorescence (NPQ). In diatoms, NPQ comprises a quickly and a slowly relaxing component (Lavaud and Goss, 2014). We will refer to the quickly relaxing component as “flexible NPQ” (Niyogi and Truong, 2013) and to the slowly relaxing component as “sustained NPQ” or NPQs (Lavaud and Goss, 2014). The capacity for flexible NPQ is mainly defined by the xanthophyll cycle (XC) pigment diatoxanthin (Dtx) produced via de-epoxidation of diadinoxanthin (Ddx) (Lavaud and Goss, 2014; Barnett et al., 2015; Goss and Lepetit, 2015; Blommaert et al., 2017), and the presence of Light-Harvesting Complex X (LHCX) proteins (Bailleul et al., 2010; Ghazaryan et al., 2016; Taddei et al., 2016, 2018; Lepetit et al., 2017). While the XC in benthic diatoms has been well-studied in natural communities (van Leeuwe et al., 2008; Jesus et al., 2009; Serôdio et al., 2012; Laviale et al., 2015) and more recently also using unialgal isolates (Barnett et al., 2015; Blommaert et al., 2017), our current knowledge about LHCX proteins as an NPQ regulator is mostly based on studies of planktonic diatoms (Nymark et al., 2009, 2013; Zhu and Green, 2010; Zhu et al., 2010; Büchel, 2014; Lavaud and Goss, 2014; Valle et al., 2014; Dong et al., 2015; Goss and Lepetit, 2015; Ghazaryan et al., 2016; Grouneva et al., 2016; Lepetit et al., 2017; Taddei et al., 2018, 2016). The latter includes studies on the pennate model diatom Phaeodactylum tricornutum which to date has only been isolated from water samples from various coastal environments, but may have a benthic growth phase as well (De Martino et al., 2007).
Light-Harvesting Complex X proteins are closely related to the Light-Harvesting Complex Stress-Related (LHCSR) proteins that are present in most eukaryotic algae and mosses, but absent in flowering plants (Niyogi and Truong, 2013; Goss and Lepetit, 2015). Even though LHCX/LHCSR proteins are Light Harvesting Proteins, they have an energy dissipating rather than a light harvesting function (Niyogi and Truong, 2013). LHCSR proteins appear to function both as excess light sensors and quenching sites (Bonente et al., 2011; Ballottari et al., 2016). Therefore, they possess protonatable amino-acid residues that sense a low luminal pH (ΔpH) (Ballottari et al., 2016). A similar function as NPQ regulators has been proposed for LHCX proteins in planktonic diatoms as HL induces LHCX transcription and enhances LHCX protein content (Oeltjen et al., 2002; Nymark et al., 2009, 2013; Bailleul et al., 2010; Park et al., 2010; Zhu and Green, 2010; Lepetit et al., 2013, 2017; Taddei et al., 2016; Hippmann et al., 2017). LHCX proteins are hypothesized to bind the XC pigments Ddx and Dtx (Beer et al., 2006; Lepetit et al., 2013) and change the supramolecular organization of antenna complexes (Ghazaryan et al., 2016). Furthermore, excess energy dissipation, mediated by LHCX proteins, was linked to a decrease in the functional absorption cross-section of photosystem II (Buck et al., 2019).
Light-Harvesting Complex X function and transcriptional regulation has been intensively studied in the diatom P. tricornutum (Nymark et al., 2009, 2013; Bailleul et al., 2010; Lepetit et al., 2013, 2017; Valle et al., 2014; Taddei et al., 2016; Buck et al., 2019). The P. tricornutum genome contains four LHCX genes (LHCX1-4). Of these four genes, LHCX1 is highly expressed in non-stressful light conditions, whilst additional expression upon HL exposure is low (Nymark et al., 2009; Lepetit et al., 2013; Taddei et al., 2016, 2018). Its corresponding protein is consequently present in LL conditions, where it provides the diatom with a basal capacity for NPQ, localized mainly near the PSII core in P. tricornutum, to cope with sudden changes in light climate (Taddei et al., 2018). In addition, the different content in LHCX1 between different P. tricornutum ecotypes has been related to their natural variability in NPQ capacity (Bailleul et al., 2010). In (prolonged) HL conditions, both transcription of LHCX2 and LHCX3 is strongly induced (Nymark et al., 2009; Lepetit et al., 2013, 2017; Taddei et al., 2016). As both proteins accumulate in concert with the de novo synthesis of Ddx + Dtx, they may provide additional Ddx/Dtx binding sites to activate NPQ in the antenna to enhance the basal NPQ provided by LHCX1 (Lepetit et al., 2013, 2017; Taddei et al., 2018; Buck et al., 2019). Indeed, overexpression of both LHCX2&3 has been shown to rescue NPQ in a low-NPQ ecotype of P. tricornutum (Pt4) (Taddei et al., 2016).
LHCX4 gene expression is inhibited by light, whilst its transcript accumulates in prolonged darkness, questioning its role in photoprotection (Lepetit et al., 2013; Nymark et al., 2013; Taddei et al., 2016; Buck et al., 2019).
The findings for P. tricornutum may not be directly transferable to other pennate diatoms, as for instance 11 LHCX genes were discovered in the genome of the sea ice diatom Fragilariopsis cylindrus, none of which could readily be related to the four LHCX genes in P. tricornutum (Mock et al., 2017). In addition, the F. cylindrus genome contains an LHCX gene that is related to the LHCX6 in Thalassiosira pseudonana, whereas a similar sequence is absent in the P. tricornutum genome. The T. pseudonana LHCX6 protein has been hypothesized to be associated with Dtx binding and as such play a direct role in excess energy dissipation via NPQs during acclimation to prolonged HL stress (Zhu and Green, 2010). This protein, furthermore, is downregulated in fluctuating light conditions (Grouneva et al., 2016), underscoring its potential role in more prolonged oversaturating light conditions.
Recent studies using immuno-blotting revealed the presence of several light-regulated LHCX-proteins in natural communities and isolates of the microphytobenthic diatom S. robusta, of which some differ in size from P. tricornutum homologs (Laviale et al., 2015; Blommaert et al., 2017). To date, however, nothing is known about the diversity, organization and transcriptional regulation of these LHCX proteins in truly benthic diatoms. In the present study, we therefore identified LHCX genes in the S. robusta draft genome and followed their transcriptional regulation during a day/night cycle and during exposure to HL conditions. In addition, we investigated the conservation of potential ΔpH sensing amino-acid residues.
Materials and Methods
Culture Conditions
Seminavis robusta strain 85A was obtained from the diatom culture collection (BCCM/DCG) of the Belgian Coordinated Collection of Micro-organisms1, accession number (DCG 0105). Diatom cultures were grown at 20°C in semi-continuous batch culture in 1.8 L glass Fernbach flasks (Schott) under a day/night rhythm of 16/8 h with a light intensity of 20 μmol photons m–2 s–1. Cells were cultured in Provasoli’s enriched f/2 seawater medium (Guillard, 1975) using Tropic Marin artificial sea salt (34.5 g L–1) enriched with NaHCO3 (80 mg L–1 final concentration). Cultures were acclimated to these culturing conditions for at least 2 weeks.
Day/Night Cycle and Prolonged Darkness
After acclimation, S. robusta was grown in 650 mL culture flasks (Greiner bio-one) to monitor LHCX expression during a 16/8-h day/night cycle and in parallel during an extended dark cycle (during which cultures were kept in the dark for 24 h). Three biological replicates were sampled independently at 0:00, the start of the daily dark period, at 6:00, 2 h before the start of the light period, at 15 min, 1 h and 4 h after the start of the day period, and at the end of the light period (24:00). Gene expression was compared to gene expression in the samples at 6:00 h.
High Light Exposure
High light exposure was identical to the conditions described in Blommaert et al. (2017) and imposed about 6–8 h after the onset of the daily light period. Cultures in exponential growth were concentrated to 10 mg L–1 Chl a (determined spectrophotometrically, Jeffrey and Humphrey, 1975) by centrifugation at 4,000 RCF for 5 min and were allowed to recover in growth conditions (20°C, 20 μmol photons m–2 s–1) for 2 h. Immediately before the start of the experiment, NaHCO3 (4 mM) was added from a 2 M stock to prevent carbon limitation during the experiment. Four 65 W white light energy-saving lamps (Lexman) were used to provide HL conditions (2,000 μmol photons m–2 s–1) as used by Lepetit et al. (2013). Cells were continuously stirred in a glass test tube to obtain a homogenous cell suspension. The glass test tube was continuously cooled in a custom-made glass cooler by a water bath at 20°C. Three biological replicates were sampled immediately before the onset of 2,000 μmol photons m–2 s–1 and after 15, 30, and 60 min of HL. Gene expression in the treated samples was compared to the samples before HL (0 min).
RNA Extraction and cDNA Synthesis
Four milliliters of cell culture was sampled each time on 3 μm Versapor filters (PALL Corporation). The filter was washed with ice-cooled phosphate buffered saline (PBS) and immediately frozen in liquid nitrogen. Samples were stored at -80°C before RNA extraction. RNA extraction was based on Le Bail et al. (2008). Frozen samples were immediately incubated in 500 μL extraction buffer (100 mM Tris–HCl pH 7.5, 2% CTAB, 1.5 M NaCl, 50 mM EDTA, and 10% β-mercapto-ethanol) and subsequently beaten with carbid beads for 30 min in a bead-beater at 30 Hz. One hundred microliters of 10% Chelex-100 was added before the samples were incubated for 15 min at 56°C with occasional vortexing. One volume of chloroform:isoamyl alcohol (24:1, Vol/Vol) was subsequently added before shaking the samples for 25 min at 5 Hz. After centrifugation, the upper phase was transferred to a new tube and mixed with 0.3 volume of absolute ethanol to precipitate polysaccharides. One volume of chloroform was added and after centrifugation the upper phase was transferred to a fresh tube. RNA was precipitated overnight at −20°C, by adding 0.25 volumes of 12M LiCl and 1% (of final volume) β-mercapto-ethanol. The next day, the RNA was pelleted, dried and washed with 70% ethanol. Residual DNA was eliminated with DNAse I (Turbo DNAse, Ambion) according to the manufacturer’s instructions. Extraction was performed with 1 volume Phenol-Chloroform (1:1, Vol/Vol). After centrifugation the upper phase was transferred to a fresh tube, extracted with one volume of chloroform:isoamylic alcohol (24:1, Vol/Vol) and centrifuged again. The upper phase was precipitated with 0.3 M NaOAc (pH 5.5) and 100% ice-cold ethanol by incubating for 1 h at −80°C. After the samples were centrifuged for 20 min at 4°C, the supernatant was discarded, and the pellet washed with 70% ethanol. The pellet was finally resuspended in RNAse-free water. The samples were reverse transcribed using Bio-Rad iScript cDNA kit.
Identification of LHCX Genes in the S. robusta Draft Genome
LHCX sequences from T. pseudonana, P. tricornutum, F. cylindrus and Pseudo-nitzschia multiseries (with kind permission of E. V. Armbrust) were obtained from the JGI database2. These LHCX sequences were used to build an amino-acid HMM (Hidden Markov Model) profile using HMMer (version 3.1b1; Mistry et al., 2013), which was used to search all genes (annotation version 1.0) predicted for the in-house draft genome of the D6 strain of S. robusta (DCG 0489). Only hits having an E-value ≤ e–50 were retained. This way, 17 S. robusta LHCX (SrLHCX) genes were found. GenBank accession numbers are provided (Table 1). Similarly, LHCX/LHCSR were retrieved from Thalassiosira oceanica, Cyclotella cryptica, Fistulifera solaris, Synedra acus, Pseudo-nitzschia multistriata, the moss Physcomitrella patens and the brown alga Ectocarpus siliculosus.
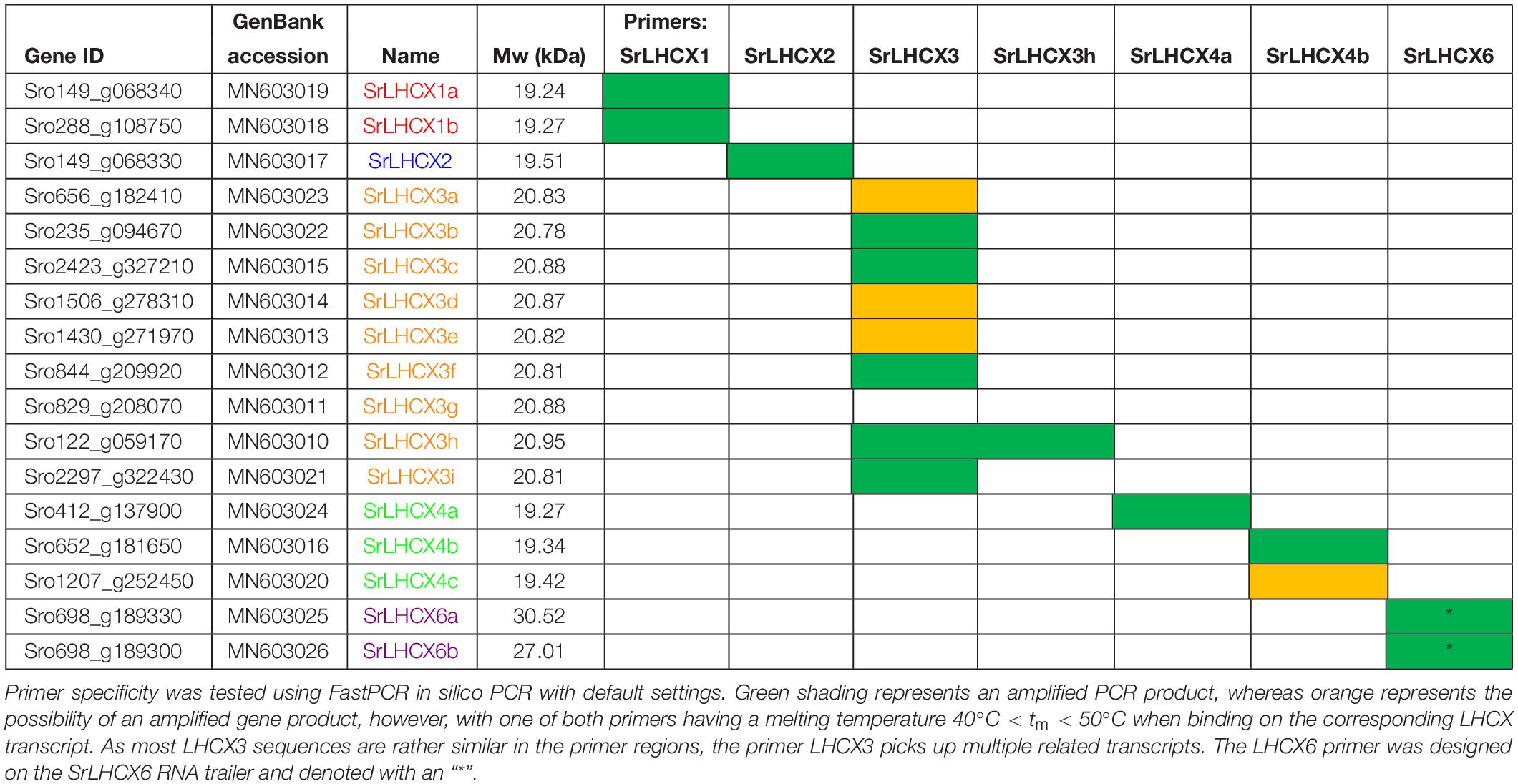
Table 1. Gene ID, GenBank accession number, Name, calculated molecular weight (Mw) and primer specificity. Name colours match the clade colours used in Figure 1.
A multiple alignment was made using MAFFT L-INS-i (version 7.187; Katoh and Standley, 2013) and visualized using ESPript (Robert and Gouet, 2014; Supplementary Figure S1; coding sequences are provided in Supplementary Figure S2). The signal peptide of each LHCX (between 17 AA and 22 AA) was found using ASAFind (Gruber et al., 2015) after running SignalP 3.0 (Bendtsen et al., 2004). It was removed to calculate the approximate molecular weight (Mw) using the Compute pI/Mw tool (ExPASy, Swiss Bioinformatics Institute).
This unedited alignment (383 amino-acid positions) was used for phylogenetic tree construction using IQ-tree (version 1.5.5; Nguyen et al., 2015) under the best amino-acid substitution model selected by the build-in model-selection function (ModelFinder) (Kalyaanamoorthy et al., 2017) allowing for the following set of potential models: JTT, LG, WAG, Blosum62, VT and Dayhoff. The FreeRate model (Yang, 1995) was chosen to account for rate-heterogeneity across sites and empirical amino-acid frequencies were calculated from the data. 10,000 ultra-fast bootstrapping cycles (Minh et al., 2013) were performed to validate clustering. LHCF sequences were used as an outgroup to root the tree, based on Nymark et al. (2013).
RT-qPCR
RT-qPCR was performed with a Light Cycler® 480II (ROCHE). Primer sets were designed using Primer3 (Supplementary Table S1). Primer specificity was tested in silico with FastPCR (PrimerDigital). Single nucleotide polymorphisms (SNPs) between the whole genome sequenced strain (D6) and the strain used in the experiments (85A) were identified using in-house RNAseq data (Bilcke et al., 2020) using Integrative Genomics Viewer (IGV, Broad Institute) and did not affect primer specificity. CDKA1, V4 and V1 (Moeys et al., 2016) were used for normalization as these were most stably expressed (Qbase + software). Qbase + normalized data is shown in Supplementary Figure S3. Log2 expression ratios were compared with REST2013 software. The RT-qPCR program contained the following steps: pre-incubation: 95°C – 5 min, amplification: 95°C – 10 s, 58°C – 10 s, 72°C – 20 s (40 cycles), melting curve: 95°C – 5 min, 65°C – 1 min, 97°C.
Results
LHCX Presence in the Genome of S. robusta
A HMMER search, with a profile based on annotated LHCX genes from T. pseudonana, P. tricornutum, F. cylindrus and P. multiseries, yielded 17 putative LHCX sequences in the draft S. robusta genome (SrLHCX), all with a calculated Mw of about 20 kDa (Table 1), with exception of SrLHCX6a and SrLHCX6b, the latter being truncated on the C-terminus and possibly being a pseudogene, see Supplementary Figure S1. The resulting maximum-likelihood phylogenetic tree (Figure 1) shows two main well-supported clades, the uppermost diatom-specific clade comprising centric as well as pennate diatom sequences, but lacking sequences from the pennate model P. tricornutum and the araphid pennate Synedra acus. SrLHCX6a and b are found in a subcluster containing also TpLHCX6 and FcLHCX6.
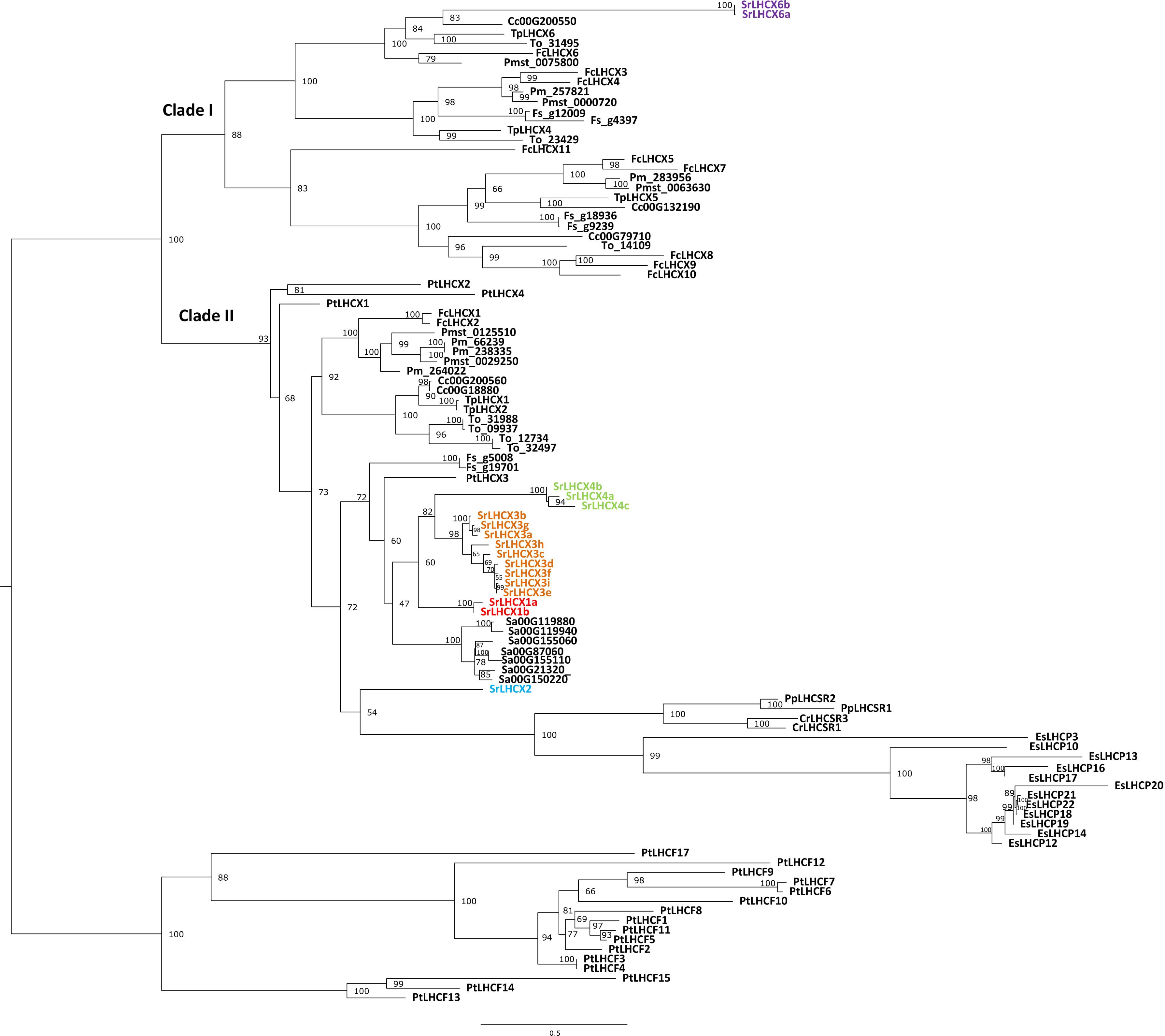
Figure 1. Phylogenetic tree of LHCX genes in Seminavis robusta (Sr, colored), based on a multiple alignment made using MAFFT L-INS-i and constructed using IQ-tree. Total of 10,000 ultra-fast bootstrap approximation iterations were run (values are shown at the nodes). LHCX/LHCSR sequences are shown for the diatoms Thalassiosira pseudonana (Tp), Thalassiosira oceanica (To), Cyclotella cryptica (Cc), Phaeodactylum tricornutum (Pt), Fragilariopsis cylindrus (Fc), Fistulifera solaris (Fs), Synedra acus (Sa), Pseudo-nitzschia multistriata (Pmst) and Pseudo-nitzschia multiseries (Pm, with kind permission of E. V. Armbrust), the green alga Chlamydomonas reinhardtii (Cr), the moss Physcomitrella patens (Pp) and the brown alga Ectocarpus siliculosus (Es).
The lower cluster contains all other SrLHCX proteins, all P. tricornutum proteins and proteins from the green alga Chlamydomonas reinhardtii, the moss P. patens and the brown algae E. siliculosus. Most SrLHCX proteins (apart from SrLHCX6a and b and LHCX2) cluster together with PtLHCX3 in one relatively well supported cluster. Within this cluster, three well-supported subclusters can be distinguished (grouping the SrLHCX1, three and four sequences). The seven Synedra acus LHCX proteins also cluster closely together and are related to SrLHCX1,3&4.
On a lower hierarchical level, the relatedness between both Pseudo-nitzchia species and F. cylindrus is evident, as is the case for both Thalassiosira species and C. cryptica.
LHCX Gene Expression
We studied LHCX expression in S. robusta during 24 h of a 16 h light (20 μmol photons m2 s–1)–8 h dark cycle (Figure 2A). Cultures kept in prolonged darkness were sampled in parallel (Figure 2B). Note that for several primer sets multiple related transcripts can be amplified; the specificity of primer sets is given in Table 1. We will refer to the primer pair that amplifies multiple SrLHCX3 transcripts as “LHCX3.” Gene expression levels were compared to expression levels in samples 2 h before the light period (time point 6:00 in Figures 2A,B) and samples taken during the light period, compared to samples taken in parallel during the extended darkness treatment (Figure 2C). As in some replicates SrLHCX2 and SrLHCX4a transcripts were not detected at 0:00, data for this timepoint is not shown. Due to considerable variance between technical replicates of SrLHCX6 throughout the 24-h cycle, these data are not shown.
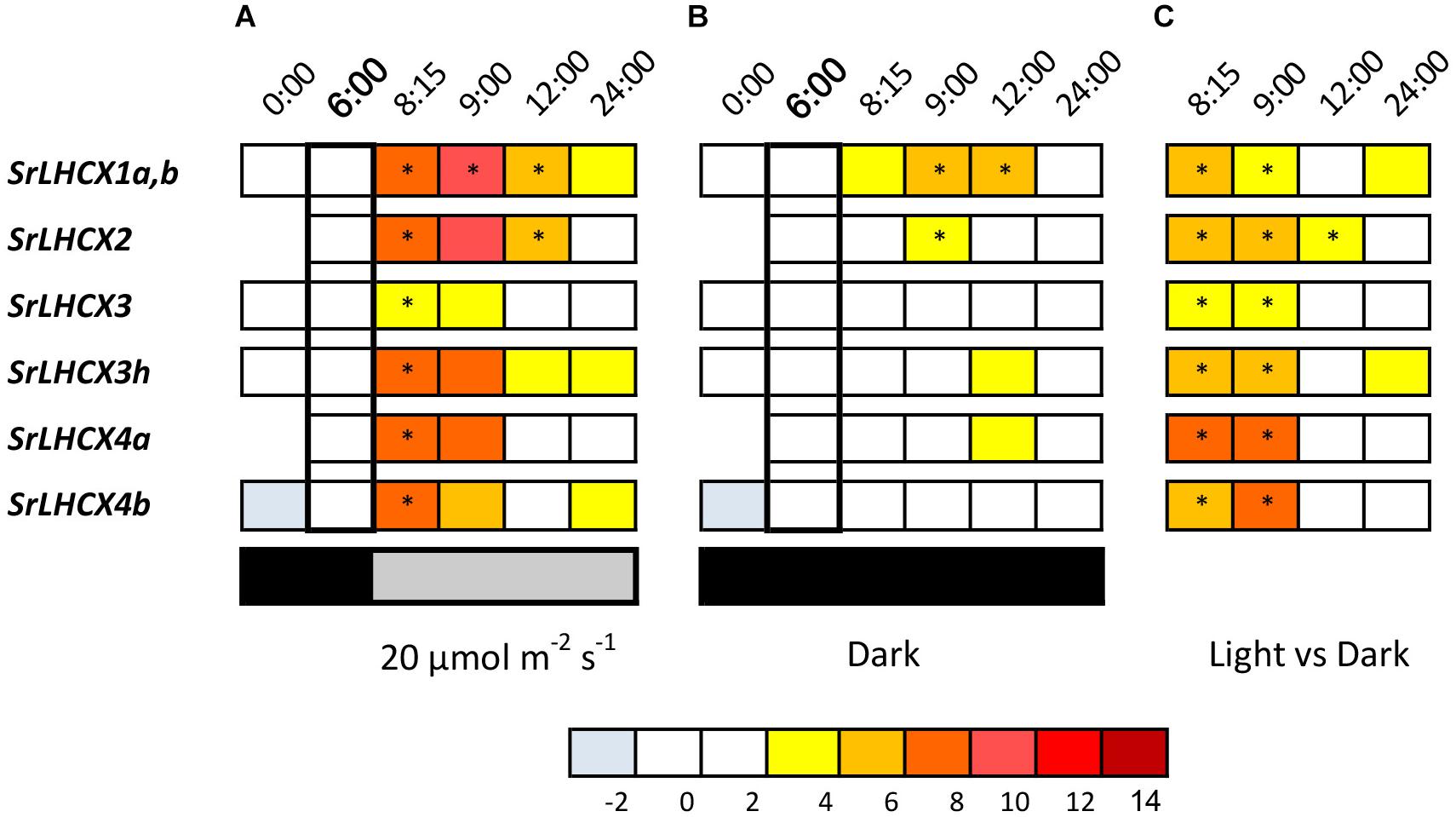
Figure 2. LHCX transcription levels, compared to the 6:00 time point (in bold) during a 24-h 16/8 h day/night regime with light onset at 8:00 (A), during extended darkness (B) and the transcript levels during the dark/light cycle compared to the extended dark period (C). Expression ratios are log2 transformed and values are indicated by the color chart. Values are averages of three independent biological replicates. As for SrLHCX2 and SrLHCX4a no transcripts were detected at (0:00) in some replicates, no values are shown. Significant changes (p < 0.05, Pairwise Fixed Reallocation Randomization Test performed by REST2006) are indicated with an asterisk.
All investigated SrLHCX genes showed a significant upregulation 15 min after the dark/light transition compared to 2 h before light onset (Figure 2B), which was not the case for samples kept in prolonged darkness (Figure 2B). For samples kept in prolonged darkness only SrLHCX1a,b and SrLHCX2 were significantly upregulated 1 h after the light period would have started, compared to 6:00. Transcript levels for all investigated SrLHCX genes were significantly higher in the cells sampled 15 and 60 min after the dark/light transition, compared to cells which were kept in prolonged darkness (Figure 2C).
In addition, we studied the expression of SrLHCX genes in S. robusta in response to HL (Figure 3). All investigated SrLHCX genes were highly upregulated after 15, 30, and 60 min of HL, compared to the LL before HL exposure, except for LHCX3h at 60 min. SrLHCX4b and SrLHCX6 showed significantly higher expression at 30 min, compared to 15 min of HL, whereas SrLHCX2 declined significantly in expression between 15 and 30 min of HL. Between 30 and 60 min of HL, LHCX2, LHCX3, and LHCX3h declined significantly in expression.
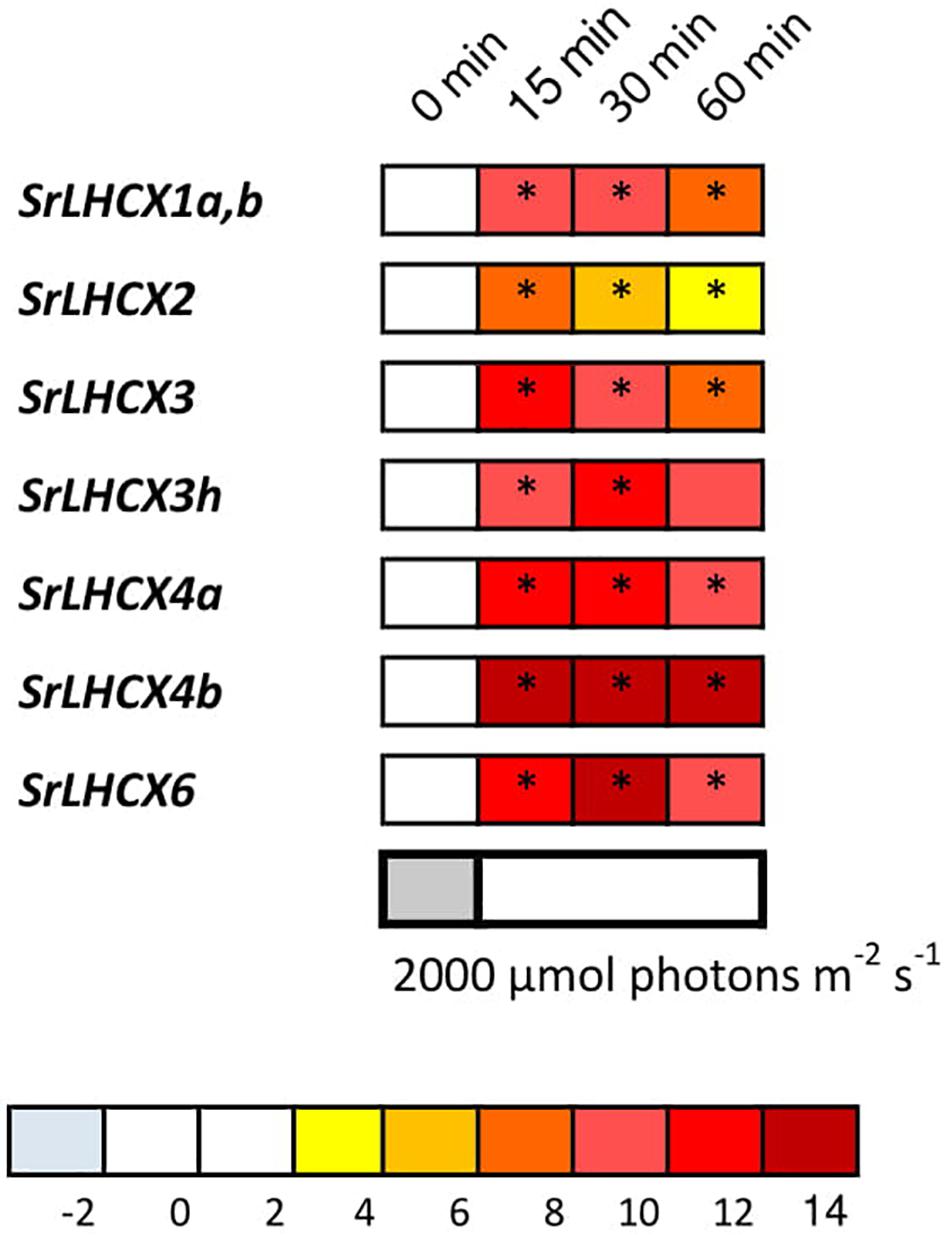
Figure 3. LHCX transcription levels during exposure to high light (2,000 μmol photons m–2 s–1). Expression ratios are log2 transformed and indicated by the color chart. Values are averages of three independent biological replicates and relative to the respective initial values (LL). Significant changes at p < 0.05 (Pairwise Fixed Reallocation Randomization Test performed by REST2006) are indicated with an asterisk.
We investigated the presence of three amino-acid residues which are known to function as sensors of the thylakoid lumen pH in the LHCSR3 in Chlamydomonas reinhardtii and which are indispensable for NPQ functioning (Ballottari et al., 2016). Two of these are also present in P. tricornutum LHCX1-3 sequences, but only one in LHCX4 (Figure 4). SrLHCX6 contains none of the protonatable residues in C. reinhardtii as is the case for LHCX6 in T. pseudonana. The same residues are conserved in all SrLHCX sequences as in PtLHCX1-3, with the exception of SrLHCX4a,b,c which lack the same residue as PtLHCX4. However, unlike the PtLHCX4 sequence, the SrLHCX4a-c sequences contain a glutamate residue (E, highlighted in yellow), which may have a protonatable function.
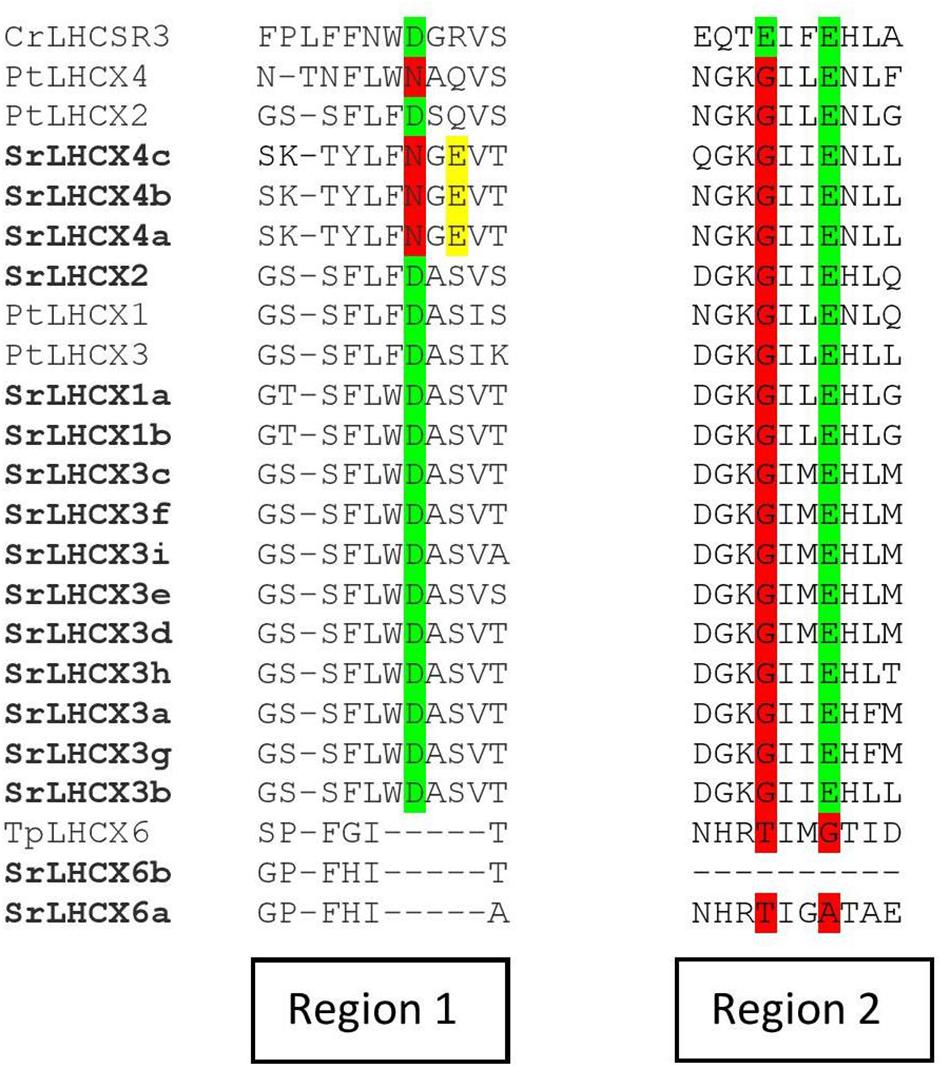
Figure 4. Amino-acid alignment of regions 1 and 2 of Chlamydomonas reinhardtii CrLHCSR3, P. tricornutum LHCX1-4, T. pseudonana LHCX6, and all LHCX sequences of S. robusta. The highlights in green represent conserved pH-sensing residues, whereas red highlights represent the absence of a conserved pH-sensing residue. Glutamate residues (E) in SrLHCX4a-c are highlighted in yellow.
Discussion
As LHCX proteins play a central role in the NPQ mechanism of planktonic diatoms (Bailleul et al., 2010; Zhu and Green, 2010; Lepetit et al., 2013, 2017; Taddei et al., 2016) and light responsive LHCX-proteins have been observed in benthic diatom isolates and communities (Laviale et al., 2015; Blommaert et al., 2017), we investigated the presence of LHCX genes in the benthic diatom S. robusta and studied their transcriptional regulation during HL conditions and a darkness/LL transition.
We detected 17 LHCX genes in S. robusta, which is a high number compared to the model diatoms P. tricornutum (4) and T. pseudonana (5) but in the same range as in the psychrophilic sea ice diatom F. cylindrus (11) (Armbrust et al., 2004; Bowler et al., 2008; Mock et al., 2017), the brown alga E. siliculosus (13) and the haptophyte Emiliania huxleyi (15) (see Dittami et al., 2010 for an overview and a comprehensive phylogenetic tree). Even though a comparable number of LHCX genes was discovered in both S. robusta and F. cylindrus, LHCX diversification in both raphid diatoms seems to have occurred independently as LHCX genes of both species were found to belong to different clades, with most diversity in S. robusta being related to PtLHCX3. Interestingly, a diversification of LHCX genes related to PtLHCX3 seems to have occurred as well in the araphid diatom Synedra acus. There seems, however, no general trend in the amount of LHCX genes in the genomes of centric and pennate diatoms.
Even though a certain degree of functional redundancy can be expected due to the high number of LHCX genes in S. robusta, transcription appears to be light regulated for all the studied genes and all investigated LHCX transcripts were strongly upregulated in HL conditions. However, the limited specificity of the LHCX3 primer combination makes it difficult to generalize about LHCX genes in this clade. Our results, as such, do not allow to conclude why S. robusta has such a high number of LHCX genes. The expansion of gene families seems to be a general feature of S. robusta and is not exclusive to the LHCX genes. The large genome size of S. robusta allows for duplication events which could in turn lead to adaptive evolution (Osuna-Cruz et al., 2020). A large set of LHCX genes could be required to cope with highly variable light conditions. However, P. tricornutum only possesses four LHCX genes, and these still enable the species to rapidly adjust to a highly fluctuating light climate (Lepetit et al., 2017). In addition, the ability of motile epipelic diatoms to rapidly migrate away from strong light conditions could minimize the need for strong physiological photoprotection (Serôdio et al., 2001; Barnett et al., 2015; Laviale et al., 2016; Blommaert et al., 2018) and hence also the need for so many functional genes in S. robusta. Lastly, the presence of many LHCX genes may allow to integrate various environmental signals and stresses to finely regulate LHCX content (Taddei et al., 2016; Buck et al., 2019).
LHCX transcripts are present during a LL day/night cycle in S. robusta. This is consistent with the presence of an LHCX protein in LL conditions, as reported by Blommaert et al. (2017). This protein, as PtLHCX1, could provide a basal NPQ capacity localized near the PSII core (Nymark et al., 2009; Bailleul et al., 2010; Lepetit et al., 2013, 2017; Taddei et al., 2018). As all SrLHCX transcripts (except SrLHCX6 for which data are lacking) were induced upon a dark/light transition, we cannot rule out that multiple SrLHCX proteins fulfill a similar role as PtLHCX1. Linking transcriptional data and immuno-blotting, in this case, is not straightforward as the used antibody (anti-LHCSR3, Bonente et al., 2011) was not specifically designed to recognize diatom LHCX isoforms. In addition, the large number of LHCX genes of similar sizes (Table 1) and differences in actual and predicted LHCX/LHCSR protein size, because of posttranslational modifications (Bonente et al., 2011), complicate the comparison of both datasets as was also observed for the discrepancies in transcriptional and translational regulation of LHCXs in P. tricornutum and T. pseudonana (Zhu and Green, 2010; Lepetit et al., 2017).
High light-induced transcription of LHCX genes as observed in S. robusta was reported for planktonic diatoms (Nymark et al., 2009; Zhu and Green, 2010; Lepetit et al., 2013; Taddei et al., 2018) and may enhance the basal NPQ level provided by the LHCX protein(s) already present in LL (Taddei et al., 2018). HL induced transcription of LHCX genes in S. robusta is indeed reflected on the protein level after 1 h (Blommaert et al., 2017). Even though we did not study the effect of prolonged HL on LHCX accumulation, S. robusta is able to increase its NPQ capacity together with its XC features during long-term acclimation upon a shift from 20 to 75 μmol photons m–2 s–1 (Barnett et al., 2015). The accumulation of LHCX proteins during HL was also reported for natural benthic communities and in the benthic diatom Navicula phyllepta (Laviale et al., 2015) and may together with XC pigments allow epipelic species to acclimate to prolonged higher light conditions (Barnett et al., 2015; Ezequiel et al., 2015).
SrLHCX6, interestingly, is not closely related to the majority of LHCX genes in S. robusta: SrLHCX6 clusters in a clade containing LHCX6 of the centric diatom T. pseudonana and of the pennate diatom F. cylindrus. As SrLHCX6 was strongly upregulated in HL, it may play a role in sustained quenching (NPQs), as proposed for the LHCX6 protein in T. pseudonana (Zhu and Green, 2010; Grouneva et al., 2016). This matches the observation of sustained quenching and de novo Dtx synthesis in S. robusta under identical HL conditions (Blommaert et al., 2017). In N. phyllepta, which is phylogenetically closely related to S. robusta (Chepurnov et al., 2008), the anti-LHCX6 antibody raised against LHCX6 in T. pseudonana recognized a HL inducible LHCX protein, whose size (∼33 kDa) is similar to the calculated size of SrLHCX6 (Table 1; Laviale et al., 2015). The same antibody, nonetheless, failed to recognize a protein of any size in S. robusta (Blommaert et al., 2017).
SrLHCX4b was highly expressed upon HL exposure and remained highly expressed even after 60 min of HL exposure. This gene is closely related to SrLHCX4a and SrLHCX4c and seems to be differentiated from the majority of SrLHCX3 sequences. A major difference between the SrLHCX4 proteins and the other SrLHCX proteins (except for SrLHCX6) is that only one instead of two protonatable amino-acid residues (compared to the three amino-acids responsible for the switch to energy-dissipating mode in C. reinhardtii) is conserved. A similar difference in amino-acid sequence has been reported for PtLHCX4, compared to the other P. tricornutum proteins, the former, however, being induced only in prolonged darkness (Nymark et al., 2013; Taddei et al., 2016). Even though NPQ regulation by a light-induced luminal pH change in diatoms is less clear than in green algae and plants, which relax NPQ immediately after ΔpH breakdown (Goss and Lepetit, 2015), the above findings may suggest that SrLHCX4a–c as PtLHCX4 are not, or only to a lesser degree, controlled by a trans-thylakoidal proton gradient. They could contribute to a more sustained NPQ component as was suggested for TpLHCX6 (Zhu and Green, 2010), which completely lacks these residues and may provide binding sites for Dtx.
In this study, we demonstrated the presence of multiple light-regulated LHCX genes, which may allow epipelic species to respond and/or acclimate to prolonged higher light conditions (Barnett et al., 2015; Ezequiel et al., 2015), either through an increase of the flexible NPQ component or through more sustained quenching (Lavaud and Goss, 2014). Our study paves the way for future detailed investigations with defined experimental approaches in order to dissect the specific regulation and function of individual SrLHCX genes and proteins, involving e.g. experiments including natural light simulation conditions such as gradual increasing fluctuating light (Grouneva et al., 2016; Lepetit et al., 2017) or nutrient starvation (Taddei et al., 2016; Hippmann et al., 2017), the localization of NPQ (Taddei et al., 2018) as well as reverse genetics approaches (Bailleul et al., 2010) as soon as S. robusta becomes transformable.
Data Availability Statement
The datasets for this study can be found in GenBank: https://www.ncbi.nlm.nih.gov/genbank/. Accession numbers are provided in Table 1.
Author Contributions
LB: experimental design, practical work, and manuscript writing. EV and MH: bioinformatics and manuscript writing. CO-C: bioinformatics. SD: molecular lab work. JL, BL, and KS: experimental design and manuscript writing. PW and AB: S. robusta sequencing, gene annotation, and bioinformatics. KV: supervision bioinformatics. WV: manuscript writing.
Funding
The authors would like to thank the Research Foundation Flanders (FWO project G.0222.09N), Ghent University (BOF-GOA 01G01911 and BOF-GOA 01G01715) and the Egide/Campus France-PHC Tournesol (n128992UA) exchange program for their financial support. JL thanks the CNRS for their financial support. MH acknowledges a Postdoctoral Fellowship of the Research Foundation Flanders. AB and PW were supported by grant 267474 from the Norwegian Research Council.
Conflict of Interest
MH is currently an employee of the European Research Council Executive Agency. The views expressed are purely those of the writer and may not in any circumstances be regarded as stating an official position of the European Commission.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank E. V. Armbrust (University of Washington, United States) for granting us access to the Pseudo-nitzschia multiseries genome. A preliminary version of this manuscript was part of the doctoral thesis “Photoprotection in intertidal benthic diatoms” of LB (Blommaert, 2017), with promotors KS and WV and published by Ghent University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00192/full#supplementary-material
Footnotes
References
Admiraal, W. (1984). The ecology of estuarine sediment-inhabiting diatoms. Prog. Phycol. Res. 3, 269–322.
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Bailleul, B., Rogato, A., De Martino, A., Coesel, S., Cardol, P., Bowler, C., et al. (2010). An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc. Natl. Acad. Sci. U.S.A. 107, 18214–18219. doi: 10.1073/pnas.1007703107
Ballottari, M., Truong, T. B., De Re, E., Erickson, E., Stella, G. R., Fleming, G. R., et al. (2016). Identification of pH-sensing sites in the light harvesting complex stress-related 3 protein essential for triggering non-photochemical quenching in Chlamydomonas reinhardtii. J. Biol. Chem. 291, 7334–7346. doi: 10.1074/jbc.M115.704601
Barnett, A., Méléder, V., Blommaert, L., Lepetit, B., Gaudin, P., Vyverman, W., et al. (2015). Growth form defines physiological photoprotective capacity in intertidal benthic diatoms. ISME J. 9, 32–45. doi: 10.1038/ismej.2014.105
Beer, A., Gundermann, K., Beckmann, J., and Büchel, C. (2006). Subunit composition and pigmentation of fucoxanthin-chlorophyll Proteins in diatoms: evidence for a subunit involved in diadinoxanthin and diatoxanthin binding. Biochemistry 45, 13046–13053. doi: 10.1021/bi061249h
Bendtsen, J. D., Nielsen, H., Von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Bilcke, G., Van den Berge, K., De Decker, S., Bonneure, E., Poulsen, N., Bulankova, P., et al. (2020). Mating type specific transcriptomic response to sex inducing pheromone in the pennate diatom Seminavis robusta. BioRxiv [Preprint]. Available online at: https://www.biorxiv.org/content/10.1101/2020.03.16.987719v1 (accessed March 19, 2020).
Blommaert, L., Huysman, M. J. J., Vyverman, W., Lavaud, J., and Sabbe, K. (2017). Contrasting NPQ dynamics and xanthophyll cycling in a motile and a non-motile intertidal benthic diatom. Limnol. Oceanogr. 62, 1466–1479. doi: 10.1002/lno.10511
Blommaert, L., Lavaud, J., Vyverman, W., and Sabbe, K. (2018). Behavioural versus physiological photoprotection in epipelic and epipsammic benthic diatoms. Eur. J. Phycol. 53, 146–155. doi: 10.1080/09670262.2017.1397197
Bonente, G., Ballottari, M., Truong, T. B., Morosinotto, T., Ahn, T. K., Fleming, G. R., et al. (2011). Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 9:e1000577. doi: 10.1371/journal.pbio.1000577
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Büchel, C. (2014). Non-Photochemical Quenching and Energy Dissipation in Plants, Algae and Cyanobacteria. Berlin: Springer.
Buck, J. M., Bártulos, C. R., Serif, M., Halder, M., Henkel, J., Kroth, P. G., et al. (2019). Lhcx proteins provide photoprotection via thermal dissipation of absorbed light in the diatom Phaeodactylum tricornutum. Nat. Commun. 10:4167. doi: 10.1038/s41467-019-12043-12046
Cartaxana, P., Cruz, S., Gameiro, C., and Kuhl, M. (2016). Regulation of intertidal microphytobenthos photosynthesis over a diel emersion period is strongly affected by diatom migration patterns. Front. Microbiol. 7:872. doi: 10.3389/fmicb.2016.00872
Chepurnov, V. A., Mann, D. G., Von Dassow, P., Vanormelingen, P., Gillard, J., Inzé, D., et al. (2008). In search of new tractable diatoms for experimental biology. Bioessays 30, 692–702. doi: 10.1002/bies.20773
Consalvey, M., Paterson, D. M., and Underwood, G. J. C. (2004). The ups and downs of life in a benthic biofilm: migration of benthic diatoms. Diatom Res. 19, 181–202. doi: 10.1080/0269249x.2004.9705870
De Martino, A., Meichenin, A., Shi, J., Pan, K., and Bowler, C. (2007). Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. J. Phycol. 43, 992–1009. doi: 10.1111/j.1529-8817.2007.00384.x
Dittami, S. M., Michel, G., Collén, J., Boyen, C., and Tonon, T. (2010). Chlorophyll-binding proteins revisited - a multigenic family of light-harvesting and stress proteins from a brown algal perspective. BMC Evol. Biol. 10:365. doi: 10.1186/1471-2148-10-365
Dong, Y.-L., Jiang, T., Xia, W., Dong, H.-P., Lu, S.-H., and Cui, L. (2015). Light harvesting proteins regulate non-photochemical fluorescence quenching in the marine diatom Thalassiosira pseudonana. ALGAL 12, 300–307. doi: 10.1016/j.algal.2015.09.016
Ezequiel, J., Laviale, M., Frankenbach, S., Cartaxana, P., and Serôdio, J. (2015). Photoacclimation state determines the photobehaviour of motile microalgae: the case of a benthic diatom. J. Exp. Mar. Bio. Ecol. 468, 11–20. doi: 10.1016/j.jembe.2015.03.004
Ghazaryan, A., Akhtar, P., Garab, G., Lambrev, P. H., and Büchel, C. (2016). Involvement of the Lhcx protein Fcp6 of the diatom Cyclotella meneghiniana in the macro-organisation and structural flexibility of thylakoid membranes. Biochim. Biophys. Acta Bioenerg. 1857, 1373–1379. doi: 10.1016/j.bbabio.2016.04.288
Goss, R., and Lepetit, B. (2015). Biodiversity of NPQ. J. Plant Physiol. 172, 13–32. doi: 10.1016/j.jplph.2014.03.004
Grouneva, I., Muth-Pawlak, D., Battchikova, N., and Aro, E.-M. (2016). Changes in relative thylakoid protein abundance induced by fluctuating light in the diatom Thalassiosira pseudonana. J. Proteome Res. 15, 1649–1658. doi: 10.1021/acs.jproteome.6b00124
Gruber, A., Rocap, G., Kroth, P. G., Armbrust, E. V., and Mock, T. (2015). Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 81, 519–528. doi: 10.1111/tpj.12734
Guillard, R. L. (1975). Culture of Marine Invertebrate Animals, eds W. L. Smith and M. H. Chanley (Boston, MA: Springer), doi: 10.1007/978-1-4615-8714-9
Hippmann, A. A., Schuback, N., Moon, K.-M., McCrow, J. P., Allen, A. E., Foster, L. J., et al. (2017). Contrasting effects of copper limitation on the photosynthetic apparatus in two strains of the open ocean diatom Thalassiosira oceanica. PLoS One 12:e0181753. doi: 10.1371/journal.pone.0181753
Jeffrey, S. W., and Humphrey, G. S. (1975). New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. Bd 167, 191–194. doi: 10.1016/s0015-3796(17)30778-3
Jesus, B., Brotas, V., Ribeiro, L., Mendes, C. R., Cartaxana, P., and Paterson, D. M. (2009). Adaptations of microphytobenthos assemblages to sediment type and tidal position. Cont. Shelf Res. 29, 1624–1634. doi: 10.1016/j.csr.2009.05.006
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lavaud, J., and Goss, R. (2014). “The peculiar features of the non-photochemical fluorescence quenching in diatoms and brown algae,” in Non-Photochemical Quenching and Energy Dissipation in Plants, Algae and Cyanobacteria, eds B. Demmig-Adams, G. Garab, W. Adams, and Govindjee (Berlin: Springer), 421–443. doi: 10.1007/978-94-017-9032-1_20
Laviale, M., Barnett, A., Ezequiel, J., Lepetit, B., Frankenbach, S., Méléder, V., et al. (2015). Response of intertidal benthic microalgal biofilms to a coupled light-temperature stress: evidence for latitudinal adaptation along the Atlantic coast of Southern Europe. Environ. Microbiol. 17, 3662–3677. doi: 10.1111/1462-2920.12728
Laviale, M., Frankenbach, S., and Serôdio, J. (2016). The importance of being fast: comparative kinetics of vertical migration and non-photochemical quenching of benthic diatoms under light stress. Mar. Biol. 163:10. doi: 10.1007/s00227-015-2793-2797
Le Bail, A., Dittami, S. M., de Franco, P.-O., Rousvoal, S., Cock, M. J., Tonon, T., et al. (2008). Normalisation genes for expression analyses in the brown alga model Ectocarpus siliculosus. BMC Mol. Biol. 9:75. doi: 10.1186/1471-2199-9-75
Lepetit, B., Gélin, G., Lepetit, M., Sturm, S., Vugrinec, S., Rogato, A., et al. (2017). The diatom Phaeodactylum tricornutum adjusts non-photochemical fluorescence quenching capacity in response to dynamic light via fine-tuned Lhcx and xanthophyll cycle pigment synthesis. New Phytol. 214, 205–218. doi: 10.1111/nph.14337
Lepetit, B., Sturm, S., Rogato, A., Gruber, A., Sachse, M., Falciatore, A., et al. (2013). High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol. 161, 853–865. doi: 10.1104/pp.112.207811
Minh, B. Q., Nguyen, M. A. T., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121. doi: 10.1093/nar/gkt263
Mock, T., Otillar, R. P., Strauss, J., McMullan, M., Paajanen, P., Schmutz, J., et al. (2017). Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541, 536–540. doi: 10.1038/nature20803
Moeys, S., Frenkel, J., Lembke, C., Gillard, J. T. F., Devos, V., Van den Berge, K., et al. (2016). A sex-inducing pheromone triggers cell cycle arrest and mate attraction in the diatom Seminavis robusta. Sci. Rep. 6:19252. doi: 10.1038/srep19252
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Niyogi, K. K., and Truong, T. B. (2013). Evolution of flexible non-photochemical quenching. Curr. Opin. Plant Biol. l16, 307–314. doi: 10.1016/j.pbi.2013.03.011
Nymark, M., Valle, K. C., Brembu, T., Hancke, K., Winge, P., Andresen, K., et al. (2009). An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS One 4:e7743. doi: 10.1371/journal.pone.0007743
Nymark, M., Valle, K. C., Hancke, K., Winge, P., Andresen, K., Johnsen, G., et al. (2013). Molecular and photosynthetic responses to prolonged darkness and subsequent acclimation to re-illumination in the diatom Phaeodactylum tricornutum. PLoS One 8:e58722. doi: 10.1371/journal.pone.0058722
Oeltjen, A., Krumbein, W. E., and Rhiel, E. (2002). Investigations on transcript sizes, steady state mRNA concentrations and diurnal expression of genes encoding fucoxanthin chlorophyll a/c light harvesting polypeptides in the centric diatom Cyclotella cryptica. Plant Biol. 4, 250–257. doi: 10.1055/s-2002-25737
Osuna-Cruz, C. M., Bilcke, G., Vancaester, E., De Decker, S., Poulsen, N., Bulankova, P., et al. (2020). The Seminavis robusta genome provides insights into the evolutionary adaptations of benthic diatoms. BioRxiv [Preprint]. Available online at: https://www.biorxiv.org/content/10.1101/2020.02.11.942037v1 (accessed February 12, 2020).
Park, S., Jung, G., Hwang, Y., and Jin, E. (2010). Dynamic response of the transcriptome of a psychrophilic diatom. Chaetoceros neogracile, to high irradiance. Planta 231, 349–360. doi: 10.1007/s00425-009-1044-x
Ribeiro, L., Brotas, V., Rincé, Y., and Jesus, B. (2013). Structure and diversity of intertidal benthic diatom assemblages in contrasting shores: a case study from the Tagus estuary. J. Phycol. 49, 258–270. doi: 10.1111/jpy.12031
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Sabbe, K. (1993). Short-term fluctuations in benthic diatom numbers on an intertidal sandflat in the Westerschelde estuary (Zeeland. The Netherlands). Hydrobiologia 26, 275–284. doi: 10.1007/BF00028026
Serôdio, J., Ezequiel, J., Barnett, A., Mouget, J.-L., Méléder, V., Laviale, M., et al. (2012). Efficiency of photoprotection in microphytobenthos: role of vertical migration and the xanthophyll cycle against photoinhibition. Aquat. Microb. Ecol. 67, 161–175. doi: 10.3354/ame01591
Serôdio, J., Marques da Silva, J., and Catarino, F. (2001). Use of in vivo chlorophyll a fluorescence to quantify short-term variations in the productive biomass of intertidal microphytobenthos. Mar. Ecol. Prog. Ser. 218, 45–61. doi: 10.3354/meps218045
Serôdio, J., Vieira, S., Cruz, S., and Coelho, H. (2006). Rapid light-response curves of chlorophyll fluorescence in microalgae: relationship to steady-state light curves and non-photochemical quenching in benthic diatom-dominated assemblages. Photosynth. Res. 90, 29–43. doi: 10.1007/s11120-006-9105-9105
Taddei, L., Chukhutsina, V. U., Lepetit, B., Stella, G. R., Bassi, R., van Amerongen, H., et al. (2018). Dynamic changes between two LHCX-related energy quenching sites control diatom photoacclimation. Plant Physiol. 177, 953–965. doi: 10.1104/pp.18.00448
Taddei, L., Stella, G. R., Rogato, A., Bailleul, B., Fortunato, A. E., Annunziata, R., et al. (2016). Multisignal control of expression of the LHCX protein family in the marine diatom Phaeodactylum tricornutum. J. Exp. Bot. 67, 3939–3951. doi: 10.1093/jxb/erw198
Underwood, G. J. C., and Kromkamp, J. (1999). Primary production by phytoplankton and microphytobenthos in estuaries estuaries. Adv. Ecol. Res. 29, 93–153. doi: 10.1016/S0065-2504(08)60192-60190
Valle, K. C., Nymark, M., Aamot, I., Hancke, K., Winge, P., Andresen, K., et al. (2014). System responses to equal doses of photosynthetically usable radiation of blue, green, and red light in the marine diatom Phaeodactylum tricornutum. PLoS One 9:e114211. doi: 10.1371/journal.pone.0114211
van Leeuwe, M., Brotas, V., Consalvey, M., Forster, R., Gillespie, D., Jesus, B., et al. (2008). Photoacclimation in microphytobenthos and the role of xanthophyll pigments. Eur. J. Phycol. 43, 123–132. doi: 10.1080/09670260701726119
Yang, Z. (1995). A space-time process model for the evolution of DNA sequences. Genetics 139, 993–1005.
Zhu, S.-H., and Green, B. R. (2010). Photoprotection in the diatom Thalassiosira pseudonana: role of LI818-like proteins in response to high light stress. Biochim. Biophys. Acta 1797, 1449–1457. doi: 10.1016/j.bbabio.2010.04.003
Keywords: diatom, microphytobenthos, light stress, LHCX, physiology
Citation: Blommaert L, Vancaester E, Huysman MJJ, Osuna-Cruz CM, D’hondt S, Lavaud J, Lepetit B, Winge P, Bones AM, Vandepoele K, Vyverman W and Sabbe K (2020) Light Regulation of LHCX Genes in the Benthic Diatom Seminavis robusta. Front. Mar. Sci. 7:192. doi: 10.3389/fmars.2020.00192
Received: 30 October 2019; Accepted: 12 March 2020;
Published: 15 April 2020.
Edited by:
Ciro Rico, The University of the South Pacific, FijiReviewed by:
Yusuke Matsuda, Kwansei Gakuin University, JapanXin Lin, Xiamen University, China
Radek Litvin, University of South Bohemia, Czechia
Copyright © 2020 Blommaert, Vancaester, Huysman, Osuna-Cruz, D’hondt, Lavaud, Lepetit, Winge, Bones, Vandepoele, Vyverman and Sabbe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lander Blommaert, bGFuZGVyLmJsb21tYWVydEBnbWFpbC5jb20=
†Present address: Lander Blommaert, Department of Estuarine and Delta Systems, NIOZ Royal Netherlands Institute for Sea Research, Utrecht University, Yerseke, Netherlands
 Lander Blommaert
Lander Blommaert Emmelien Vancaester
Emmelien Vancaester Marie J. J. Huysman
Marie J. J. Huysman Cristina M. Osuna-Cruz
Cristina M. Osuna-Cruz Sofie D’hondt
Sofie D’hondt Johann Lavaud
Johann Lavaud Bernard Lepetit
Bernard Lepetit Per Winge
Per Winge Atle M. Bones
Atle M. Bones Klaas Vandepoele
Klaas Vandepoele Wim Vyverman
Wim Vyverman Koen Sabbe
Koen Sabbe