- State Key Laboratory of Marine Environmental Science, Fujian Key Laboratory of Marine Carbon Sequestration, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
Organic sulfur compounds are not only essential for organismal survival but also indispensable for the sulfur cycle. Over the past few decades, dimethylsulfoniopropionate (DMSP) and dimethyl sulfide (DMS) cycling in the upper ocean have been well characterized from the genetic to the ecosystem level. Recent advances in the study of marine sulfonate transformation have indicated that phytoplankton and microbes play key roles in oceanic sulfur and carbon fluxes. This review provides biochemical details of the major sulfur metabolites, and presents an interlinked reaction network with genetic information on the microbial transformation and mineralization of sulfur compounds. This review also discusses future prospects for the discovery and characterization of novel substrates and enzymes involved in organosulfur cycling, as well as for investigations of deep sea and sedimentary organic sulfur.
Introduction
Sulfur is an essential component of all life forms. Over the past three decades, our understanding of the transformation of inorganic sulfur species with oxidation states ranging from −2 to +6 in the marine environment has dramatically improved, including the microbial oxidation, reduction, or disproportionation of sulfide (S2–), elemental sulfur (S0), polysulfides (Sn2–), thiosulfate (S2O32–), sulfite (SO32–), and sulfate (SO42–) (Friedrich et al., 2001; Pro et al., 2007; Klotz et al., 2011). In the ocean, most sulfur is present as inorganic sulfate (approximately 28–29 mM) (Canfield and Farquhar, 2009). Because phytoplankton and microbes assimilate inorganic sulfate into the amino acid cysteine (Cys) and methionine (Met) (Kiene et al., 1999), organic sulfur metabolism is ubiquitous in living organisms. Organic sulfur compounds constitute the major sinks for sulfate in the ocean; these include dissolved and particulate sulfur compounds in seawater, and biomass sulfur generated by phytoplankton and microbes (Ksionzek et al., 2016). The biomineralization and recycling of organic sulfur are critically important for the sustainment of the sulfur cycle. The fundamental biogeochemical processes of organic sulfur have been described in detail elsewhere (Moran and Durham, 2019). This review summarizes current knowledge of the chemistry and biochemistry of sulfur metabolites in the ocean, and constructs a sulfur metabolic network using reaction data cataloged in the MetaCyc database (Caspi et al., 2018). Next, the genetic capabilities of marine microbes with respect to the transformation and mineralization of organic sulfur are examined using a publicly available genomic database, as well as the Tara Oceans metagenomic datasets (Sunagawa et al., 2015). The review also discusses the prospective future chemical, biochemical and biogeochemical studies of organic sulfur compounds.
Chemical Diversity and Redox Activity of Biogenic Organic Sulfur
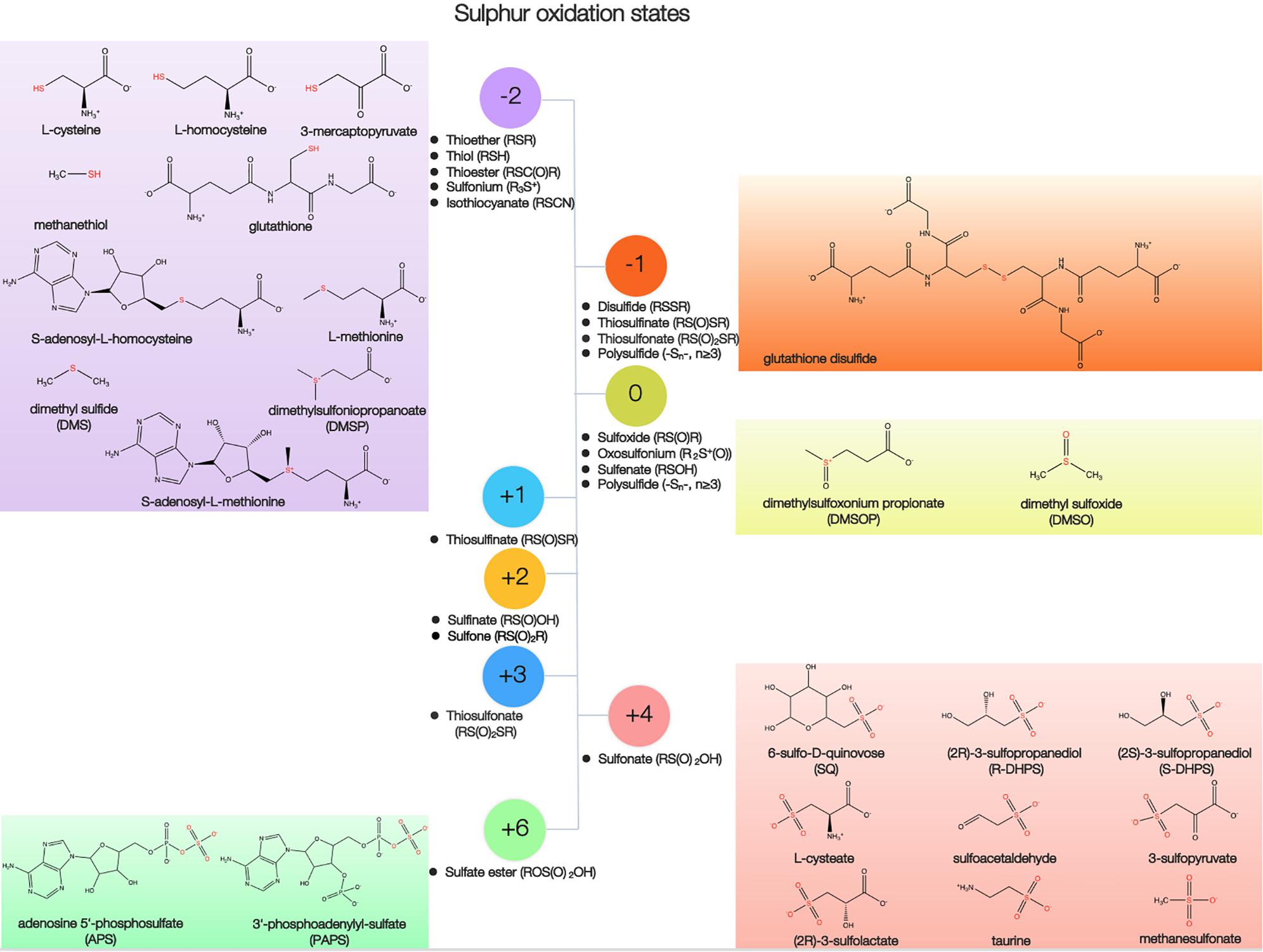
Sulfur is found in various organic molecules in the cell, including amino acids, lipids, carbohydrates, coenzymes, cofactors, vitamins, steroids, and secondary metabolites. More than 20 sulfur-containing functional groups are present in biogenic organic compounds (Figure 1 and Supplementary Table S1). Sulfur in organosulfur compounds can be directly linked to the carbon chain (e.g., the sulfonate moiety, -C-SO3), or can be attached to either an oxygen atom (e.g., sulfate ester, -O-SO3) or a nitrogen atom (e.g., sulfonamide, -NH-S-). As Supplementary Table S1 shown, the most abundant class of sulfur compounds is thioether (-S-), followed by sulfate ester, thioester [-(O)C-S-], thiol (-SH), sulfonates, sulfoxide (-SO-), disulfide (-S-S-), and sulfonium ([R3S]+) classes.
Sulfur in organosulfur compounds exists in several oxidation states, ranging from −2 to +6, and undergoes formal oxidation state transformations via electron transfer, hydrogen atom transfer, radical processes, and exchange reactions (Figure 1 and Supplementary Table S1). The distinct redox properties of sulfur species ensure that they play vital roles within a cell, including antioxidation and maintenance of the redox state (Jacob and Anwar, 2008). The compounds containing sulfur in its most reduced state (−2) represent the largest part of the overall antioxidant pool in cells, which include Cys, Met, glutathione (GSH, L-γ-glutamyl-L-cysteinl-glycine), dimethylsulfoniopropionate (DMSP), and dimethyl sulfide (DMS) (Sunda et al., 2002; Fahey, 2013; Mukwevho et al., 2014). An antioxidant role for DMSP is further supported based on evidence that this compound can be oxidized to dimethylsulfoxonium propionate (DMSOP, a sulfur atom with an oxidation state of 0) (Figure 1) (Thume et al., 2018). Compounds containing more oxidized forms of sulfur also have antioxidative properties, including dimethylsulfoxide (DMSO, sulfoxide, 0) and taurine (sulfonate, +4) (Figure 1).
Major Biogenic Organic Sulfur Compounds in the Ocean
Dissolved organic sulfur (DOS) concentrations are generally around 200–300 nM in oceanic surface waters, and decrease with depth to approximately 160–200 nM in bathypelagic waters (Ksionzek et al., 2016). Phytoplankton was estimated to assimilated ∼1,360 Tg S/per year (1 Tg = 106 tons) into organic matter and support a major fraction of sulfur flux in the surface ocean (Ksionzek et al., 2016). Sulfate is converted into organic matter by phytoplankton, including proteins (∼35%), sulfate esters (∼21%), sulfolipids (∼4%) and other low-weight compounds (∼40%) (Bates et al., 1994; Giordano et al., 2005). Sulfur metabolites are part of the molecules exchanged between phytoplankton and marine microbes (Seymour et al., 2017). For example, heterotrophic bacteria can utilize phytoplankton-derived DOS such as DMSP, 3-sulfopropanediol (DHPS), sulfur-containing cofactors vitamin B1 and B7 for growth requirement and can produce secondary metabolite such as tropodithietic acid, which is thought to protect the algal growth (Wang et al., 2016). Clearly, many different organic sulfur biosynthesis contributes to the pool of sulfur in the marine environment. Of those compounds, sulfur-containing amino acids and derivatives, methyl-sulfur compounds, sulfonates and sulfate esters as the primary sulfur sources are rapidly cycled by the upper-ocean planktonic microbes (Figure 1), and have low concentrations in water columns (commonly in the nanomole range per liter) (Moran and Durham, 2019). Those naturally occurring organic sulfur available to microbes as sources of sulfur or carbon are described below.
Sulfur-Containing Amino Acids and Derivatives
The sulfur-containing amino acid Cys and Met contain thiol and thioether functional groups, respectively. Apart from their function as a structural component of proteins, Cys and Met also plays fundamental roles in cellular metabolism. Cys is a precursor for biomolecules such as L-homocysteine, coenzyme A and GSH (Vermeij and Kertesz, 1999). Met is an initiator of protein synthesis and a precursor in many metabolic processes, such as methylation and formylation via its derivatives S-adenosyl-L-homomethione and N-formyl methionine-tRNA, respectively (Ferla and Patrick, 2014). The intracellular free Cys, Met, L-homocysteine, and S-adenosyl-L-homomethione in bacteria are maintained at low (sub-millimolar) levels but are sufficient for protein synthesis and production of essential metabolites (Bennett et al., 2009). GSH, one of the most abundant naturally occurring thiols, is the major antioxidant found in many organisms in millimolar concentration (Bennett et al., 2009; Fahey, 2013). Thiol compounds, such as Cys and GSH, may bind metal ions and thus increase the uptake of metal by marine phytoplankton (Ahner et al., 2002; Walsh et al., 2015). Glutathione disulfide (GSSG) is also present in cells in millimolar concentration (Bennett et al., 2009), which is formed as a result of GSH oxidation. In conjunction with GSH, GSSG helps to prevent oxidative damage and detoxify harmful substances (Diaz et al., 2019).
Concentrations of dissolved thiol are commonly ∼10 nM in the open ocean, but can be larger in regions of high productivity (Al-Farawati and van den Berg, 2001; Dupont et al., 2006). Cys, GSH, and γ-glutamylcysteine (γ-Glu–Cys) are the low-molecular-weight thiols that are frequently detected in the marine environments (Dupont et al., 2006). GSH is the most abundant particulate thiol in the ocean, while Cys and γ-Glu–Cys are primarily found dissolved (Dupont et al., 2006). Dissolved γ-Glu–Cys concentrations up to 15 nM have been measured in the northeast Pacific Ocean (an area of high productivity); in this region, Cys and GSH concentrations ranged from below detection limits (∼0.01 nM) up to 1 nM, consistent with levels reported in other ocean regions (Swarr et al., 2016). Previous estimates of dissolved Met concentrations ranged from 0.01 to 1 nM in surface waters (Valle et al., 2015). However, Cys and Met in dissolved combined amino acids is 10–100 more abundant than dissolved free ones.
Methyl-Sulfur Compounds
The sulfonium compound DMSP is a secondary metabolite; abundant DMSP concentrations are produced and stored by micro- and macroalgae (Curson et al., 2018). DMSP may function as a compatible solute, a cryoprotectant, an antioxidant, or a chemoattractant, and may also act as transient pool for excess intracellular sulfur (Kiene et al., 2000; Sunda et al., 2002; Yoch, 2002; Otte et al., 2004; Seymour et al., 2010). Approximately one billion tons of DMSP is generated annually by marine phytoplankton (Galí et al., 2015). DMSP concentrations in diatom cells are typically 1–50 mM, while DMSP concentrations in dinoflagellate and coccolithophore cells are typically 100–300 mM (Matrai and Keller, 1994; Ho et al., 2003). Concentrations of dissolved DMSP are typically 0.5–3 nM in the open ocean, but dissolved DMSP is occasionally measured at high concentrations (>100 nM) (Kiene and Slezak, 2006; Jarníková et al., 2018). In addition, particulate DMSP concentrations are consistently higher than dissolved DMSP concentrations in field samples (Archer et al., 2001; Kiene and Slezak, 2006). The concentrations of particulate DMSP were up to 4,240 nM during a massive bloom of the dinoflagellate Akashiwo sanguinea in estuarine waters, however, the extracellular dissolved DMSP remained below 7.2 nM due to microbial consumption (Kiene et al., 2019).
DMSP is a globally important substrate in the sulfur cycle as it is the precursor of the DMS (Simó, 2001; Yoch, 2002), methanethiol (MeSH) (Curson et al., 2011b), DMSO and DMSOP (Thume et al., 2018). DMS is the main source of biogenic sulfur in the atmosphere, and may influence the Earth’s climate due to its atmospheric oxidation products, which form cloud condensation nuclei (Andreae and Crutzen, 1997; Barnes et al., 2006). Approximately 107 tons of DMS are produced annually, with a typical sea surface concentration of 1–7 nM (Kloster et al., 2006; Lana et al., 2011; Alcolombri et al., 2014). It was estimated that approximately 25–70% of DMS flux is oxidized to methanesulfonate (MSA, CH3SO3H), which is the most abundant biogenic sulfur compound in the atmosphere (Barnes et al., 2006). DMSP degradation may yield MeSH, a reduced sulfur source that may potentially provide 30–100% of the total sulfur demand of marine bacteria (Curson et al., 2011b). Intracellular concentrations of MeSH in diatoms and dinoflagellates vary from ∼0.1 mM –1 mM (Giordano et al., 2005). MeSH concentrations are uniformly low in the open ocean (∼1 nM) and decline with depth (Henriques and Marco, 2015). DMSO may protect algal cells from photo-generated oxidants and cryogenic damage (Lee and De Mora, 1999). DMSO represents another pool of dissolved dimethylated sulfur in seawater, with concentrations of ∼1–10 nM (Vila-Costa et al., 2008). DMSOP concentrations are much lower than DMSO concentration; average DMSOP concentration in coastal marine waters is 0.14 nM (Thume et al., 2018).
Sulfonates
Sulfonates have a sulfonic acid group covalently attached to carbon, and are generated by plants, animals, and the microbial conversion of inorganic sulfate (Kertesz, 1999). Chirality is common among sulfonates (Figure 1). Sulfoquinovose (SQ) is a sulfonated hexose with a stable carbon-sulfur bond (Figure 1) (Goddard-borger and Williams, 2017). SQ is considered one of the most abundant sulfur compounds in the biosphere following Cys and Met, with an estimated annual global production of approximately 1010 tons (Goddard-borger and Williams, 2017). SQ is the polar head group of the plant sulfolipid sulfoquinovosyldiacylglycerol (SQDG) in variety of photosynthetic membranes from marine cyanobacteria (Synechococcus and Prochlorococcus) bacteria and algae (Goddard-borger and Williams, 2017), as well as of the cell wall in the Archaea (Palmieri et al., 2013). SQDG may maintain anionic charge homeostasis in the cell membrane by acting as a phosphatidylglycerol lipid substitute under very low phosphate availability conditions (Van Mooy et al., 2006). Concentrations of SQDG in the phosphate-depleted Sargasso Sea and in the South Pacific were 0.8 μg liter–1 and 0.3 μg liter–1, respectively (Van Mooy et al., 2009).
Besides SQDG, SQ is also the precursor for the synthesis of DHPS (Roy et al., 2003; Denger et al., 2014). The three-carbon (C3) component of sulfonates, including DHPS, sulfolactate, sulfopyruvate and cysteate (Figure 1), as well as the C2-sulfonates taurine and sulfoacetaldehyde (Figure 1), is found in marine organisms (Goddard-borger and Williams, 2017). DHPS has been hypothesized to perform osmotic pressure or redox balancing in cells (Götz et al., 2018; Williams and Todd, 2019). High concentrations of DHPS can accumulate within diatom and coccolithophore cells in the millimolar range, on a scale comparable with that of DMSP (Durham et al., 2019). Furthermore, the concentration of particulate DHPS at multiple coastal sites was shown to actually exceed that of particulate DMSP (Durham et al., 2015). Most phytoplankton and cyanobacteria contained micromolar concentrations of cysteate, which can be formed in the oxidation of cysteine (Durham et al., 2019). Taurine, an amino sulfonic acid, has many biological roles in the cell, including bile acid chelation, osmoregulation, and membrane stabilization (Lambert et al., 2015). Taurine is widely produced by marine metazoans and some phytoplankton at intracellular concentrations that may exceed 200 mM (Visscher et al., 1999; Tevatia et al., 2015; Clifford et al., 2017). In the North Atlantic Ocean, taurine concentrations were ∼0.1–10 nM in epipelagic waters, and ∼0.01–1 nM in bathypelagic waters (Clifford et al., 2019).
Sulfate Esters
Besides sulfonates, sulfate esters are common forms of sulfur present in the marine sediments (Kertesz, 1999). Furthermore, a number of sulfate esters in marine organisms are known, ranging from aliphatic sulfate to sulfated polysaccharides (Kertesz, 1999; Kellner Filho et al., 2019). Sulfated polysaccharides are primarily found in marine eukaryotes: as fucoidan, ulvans, carragenans, agarans, or mucin in marine algae, and as chondroitin sulfate or heparan sulfate in marine invertebrates (Jiao et al., 2011; Wang et al., 2014). Fucoidan and ulvans usually constitutes about 5–10% of the dry biomass of brown algae and 15-65% of the dry biomass of green algae, receptively (Holdt and Kraan, 2011; Cunha and Grenha, 2016). Sulfated polysaccharides are located at the cell wall, on the cell surface, in the extracellular matrix, or within the intracellular compartment, and are primarily involved in cell growth, differentiation, adhesion, signaling, and extracellular matrix interactions (Jiao et al., 2011).
Biochemical Reaction Network for Organic Sulfur
To understand the fate and transformations of organic sulfur compounds listed above in organisms, a sulfur metabolic network was constructed using reaction data cataloged in the MetaCyc database (Caspi et al., 2018). The sulfur metabolic map consisted of nodes, representing 432 sulfur metabolites (Supplementary Table S1), and edges, representing 1,598 enzymatic reactions (Supplementary Table S1). This network described the transfer of sulfur among organic compounds (Figure 2). Nodes with many connectors can be considered putative metabolic hubs. At these locations, many metabolic processes occur, enabling substances such as reaction intermediates to transfer between reaction processes on an as-needed basis. The sulfur metabolite hubs were generated by the most widespread, essential biochemical processes linked to sulfur-containing amino acid biosynthesis (Cys, Met, L-homocysteine, S-adenosyl-L-homocysteine, and S-adenosyl-L-homomethione), sulfate reduction (3′-phosphoadenosyl-5′-phosphosulfate, PAPS), and antioxidant agent biosynthesis (GSH and GSSG) (Figure 2).
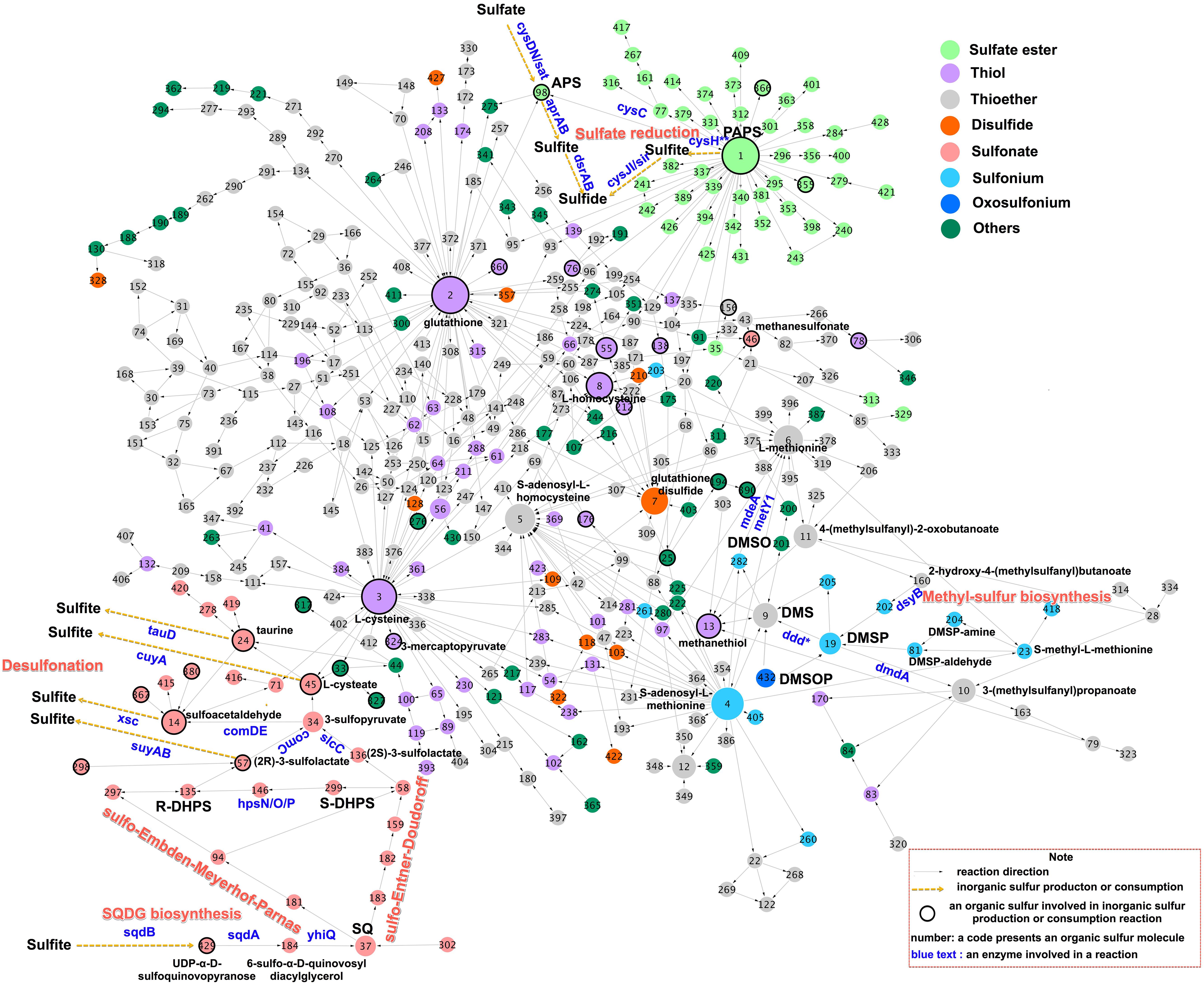
Figure 2. A biochemical network of sulfur-containing metabolites. The network was constructed using Cytoscape 3.6 (Otasek et al., 2019) based on data from MetaCyc (https://metacyc.org) (Caspi et al., 2018), which encompasses all known sulfur metabolic reactions. Nodes representing the sulfur-containing compounds are linked if they are interconverted through reactions. Node color indicates the functional group of the sulfur metabolite, while node size indicates the number of reactions added in the subsequent iteration. Black circles indicate nodes representing substrates, which directly produce or consume inorganic sulfur species via enzymatic reactions. Arrows indicate the directions of the enzymatic reaction. Essential substrates, genes, and metabolic pathways are highlighted using the colored text. Sulfur compounds containing coenzyme A were excluded from the network. Node structures and enzymatic reactions are given in Supplementary Table S1; this table may be searched using the node numbers given in the figure.
The most widespread mechanism of sulfur assimilation into biomass is the sulfate reduction pathway. However, this pathway is energetically costly compared to the nitrate assimilation pathway, consuming 732 kJ mol–1 instead of 347 kJ mol–1 (Leustek and Saito, 1999). Methyl-sulfur compounds and sulfonates are generated by other critical sulfur-reducing transformation pathways (Figure 2). The complete DHPS desulfonation pathway converts sulfonates to sulfite (Mayer et al., 2010), an intermediate in the sulfate reduction pathway. These pathways provide a mechanism by which microorganisms can generate reduced sulfur from organic matter in order to satisfy cellular growth requirements, instead of depending on sulfate- or sulfur-containing amino acids as sulfur sources.
Sulfur-Containing Amino Acids and Derivatives Metabolisms
Microorganismal sulfate assimilation classically revolve around the activation and reduction of sulfate to sulfide, the incorporation of reduced sulfur into Cys, and the transfer of reduced sulfur from Cys into the downstream metabolites such as L-homocysteine, Met and GSH (Figure 2). Adenosine 5-phosphosulfate (APS) and PAPS contain sulfate ester functional groups (Figure 2), which are key intermediates in the primary assimilation of sulfate into Cys (Klaassen and Boles, 1997). APS is synthesized via the adenylation of sulfate by sulfate adenylyltransferase (sat/cysD), a metabolic branchpoint in the sulfate reduction pathway (Figure 2). In the canonical microbial assimilatory sulfate reduction pathway, APS is phosphorylated by APS kinase (cysC) to generate PAPS, PAPS is reduced to sulfide, and organic sulfur compounds are biosynthesized by PAPS reductase and assimilatory sulfite reductases (Figure 2). However, APS can also be reduced to sulfite by adenylyl-sulfate reductase (aprAB) and further reduced to sulfide for energy purposes by reversible dissimilatory sulfite reductase (dsrAB) in the dissimilatory sulfate reduction pathway (Figure 2).
Met can be synthesized from homoserine and Cys via a transsulfuration pathway, which produces an intermediate metabolite L-homocysteine (Vermeij and Kertesz, 1999; Ferla and Patrick, 2014). Homocysteine can also be produced directly from sulfide and acylhomoserine by an O-acylhomoserine sulfhydrylase (metY) (Hacham et al., 2003). The conversion Met to Cys involves the synthesis of L-homocysteine via S-adenosyl-L-methionine and S-adenosyl-L-homocysteine (Wheeler et al., 2005). Cys is generally the rate-limiting substrate for GSH synthesis (Lu, 2013), in turn, GSH can be broken down by peptidases to give Cys (Suzuki et al., 2001). The major catabolic pathways of Cys are the transamination pathway and the direct oxidation pathway, finally yielding 3-mercaptopyruvate and cysteate, respectively (Christophersen and Anthoni, 1986). Met can be directly catabolized to MeSH by a methionine γ-lyase (mdeA) (Amarita et al., 2004), showing that sulfur-containing amino acids themselves serve as precursors of methyl-sulfur and sulfonate compounds.
Methyl-Sulfur Compounds Metabolisms
The biochemical and genetic mechanisms of DMSP biosynthesis and degradation have been reviewed in detail elsewhere (Moran et al., 2012; Bullock et al., 2017). Briefly, the biosynthesis of DMSP from Met by phytoplankton may proceed along a transamination pathway, a methylation pathway, or a decarboxylation pathway (Stefels, 2000). In the transamination pathway, Met first undergoes transamination to 4-methylthio-2-oxobutyrate, is then reduced to 4-methylthio-2-hydroxybutyrate (MTHB), followed by S-methylation to 4-dimethylsulphonio-2-hydroxybutyrate catalyzed by the MTHB methyltransferase enzyme (DYSB) (Curson et al., 2018), and finally undergoes oxidative decarboxylation to DMSP (Figure 2). DMSP can be degraded via two enzymatically-mediated pathways: a cleavage pathway that forms DMS, catalyzed by DMSP lyases (ddd), and a demethylation pathway that produces MeSH via series of enzymes including DMSP demethylase (dmdA) (Howard et al., 2006). MeSH can be converted to Met by O-acylhomoserine (Thiol)-lyases (metY1) (Ferla and Patrick, 2014). DMS is oxidized to DMSO by photochemical oxidation in the atmosphere and by microbial oxidation with monooxygenases in surface seawater; DMSO can be recycled back to DMS via a reduction pathway (Del Valle et al., 2009). DMSOP, derived from DMSP, is taken up and metabolized to DMSO by bacteria (Thume et al., 2018). MSA is one of the main products of DMS oxidation, generated via the reaction between ⋅OH and NO3⋅ radicals. Methanesulfonate monooxygenase (msmA) mediates the primary degradation of MSA to produce inorganic sulfite (Henriques and Marco, 2015).
Sulfonate Metabolisms
Sulfonation and desulfonation routes regulate the incorporation and release of sulfite via sub-pathways that generate a set of sulfonated compounds (Figure 2) (Cook et al., 1998). The biogenesis of SQDG involves the assembly of uridine 5′-diphospho (UDP)-SQ from UDP-glucose and sulfite by UDP-SQ synthase (sqdB) (Benning and Somerville, 1992b), and the glycosyltransferase-catalyzed conjugation of UDP-SQ to diacylglycerol by an acyltransferase (sqdA) (Figure 2) (Benning and Somerville, 1992a). A sulfoquinovosidase (yihQ) identified in Escherichia coli catalyzes the hydrolysis of SQDG to SQ, which is the gateway enzyme for SQ degradation (Speciale et al., 2016). SQ has been shown to be degraded by certain aerobic as well as anaerobic bacteria (Roy et al., 2003; Denger et al., 2012, 2014; Felux et al., 2015; Burrichter et al., 2018) and two major sulfoglycolytic pathways, the sulfo-Embden-Meyerhof-Parnas pathway (originally found in E. coli) and the sulfo-Entner-Doudoroff pathway (originally found in Pseudomonas putida SQ1) have been described (Denger et al., 2014; Felux et al., 2015). These pathways are analogous to the classic glycolysis pathways (Figure 2) (Felux et al., 2015). Bacterial SQ degradation yields the C3-organosulfonate products DHPS and sulfolactate (Figure 2) (Roy et al., 2003; Denger et al., 2012, 2014; Felux et al., 2015; Burrichter et al., 2018). These products can be excreted for further bacterial utilization.
The DHPS degradation pathway in aerobic bacteria involves a series of hydrogenase enzymes (hpsO, hpsP, and hpsN) that produce sulfolactate (Figure 2) (Mayer et al., 2010). Sulfolactate is metabolized by bacteria via three enzymatic pathways (Figure 2) (Graupner et al., 2000; Denger et al., 2009; Mayer et al., 2010). The first pathway converts sulfolactate into pyruvate and sulfite using sulfolactate sulfo-lyase (suyAB) (Figure 2). The second pathway oxidizes sulfolactate to sulfopyruvate using sulfolactate dehydrogenase (comC or slcC) (Mayer et al., 2010), transaminates sulfopyruvate to L-cysteate, and desulfonates L-cysteate using L-cysteate sulfo-lyase (cuyA) to produce pyruvate and sulfite (Figure 2) (Denger et al., 2009). The third pathway decarboxylates sulfopyruvate to sulfoacetaldehyde using sulfopyruvate decarboxylase (comD and comE), and then desulfonates sulfoacetaldehyde using sulfoacetaldehyde acetyltransferase (xsc) to yield acetyl-CoA and sulfite (Figure 2) (Graupner et al., 2000). The produced sulfite may either be exported by a specific exporter, or act as a source for other sulfur metabolites (Durham et al., 2015). DHPS also can be degraded completely by Desulfovibrio sp. DF1 under anaerobic conditions, yielding acetate and sulfide (Burrichter et al., 2018).
Taurine can be derived from Cys via the activities of Cys dioxygenase and Cys sulfinic acid decarboxylase in phytoplankton cells (Tevatia et al., 2015). Alternatively, taurine may be produced from cysteate via a decarboxylation step (Agnello et al., 2013). Taurine can then be degraded by taurine dioxygenase (tauD) into either aminoacetaldehyde plus sulfite or sulfoacetaldehyde (Figure 2) (Van der Ploeg et al., 1996).
Sulfate Esters Metabolisms
PAPS is an important sulfuryl donor for various metabolites via specific sulfotransferases (Figure 2); for example, PAPS may donate sulfur for the grafting of a sulfate ester onto aliphatic compounds (e.g., choline sulfate) or carbohydrates (e.g., heparan sulfate and chondroitin sulfate) (Chapman et al., 2004). Sulfate esters provide bacteria with either sulfur or carbon source, and microorganisms have genes encoding sulfatase enzymes that catalyze the hydrolysis of sulfates from various sulfate esters. The choline-sulfatase (betC) catalyzes the hydrolysis of choline sulfate to choline (Østerås et al., 1998), while N-acetylglucosamine-6-sulfatase (GNS) and N-acetylgalactosamine-6-sulfatase (GALNS) catalyze the hydrolysis of ester-bonded sulfur from heparan sulfate and chondroitin sulfate, respectively (Helbert, 2017).
Genes for the Interconversion Between Organic and Inorganic Sulfur in the Ocean
In the sulfur metabolic network (Figure 2), thiols (e.g., Cys, GSH, L-homocysteine, and MeSH), sulfate esters (e.g., APS), and sulfonated compounds (e.g., taurine, cysteate, SL, sulfoacetadehyde, and MSA) are directly desulfurized to produce inorganic sulfur (Figure 2). These compounds are thus key intermediates in the recycling and mineralization of organosulfur to form inorganic sulfur species. Of these compounds, Cys, L-homocysteine, MeSH and 3-mercaptopyruvate may be sources of sulfide, and the desulfonation of sulfoacetadehyde, sulfolactate, taurine and MSA releases sulfite (Figure 3), catalyzed predominantly by microbial enzymes. The corresponding genes were distributed among a variety of divergent bacterial taxa (Figure 3 and Supplementary Datasheet S1). The frequency of these sulfur metabolism genes in the Tara Oceans database provided a rough estimate of the prevalence of genes in ocean waters, ranging from a low of approximately 2 per million sequences for mstA (encoding methylthiol: coenzyme M methyltransferase) to high of approximately 31 per million sequences for cysK/cysM (encoding cysteine synthase) (Figure 3).
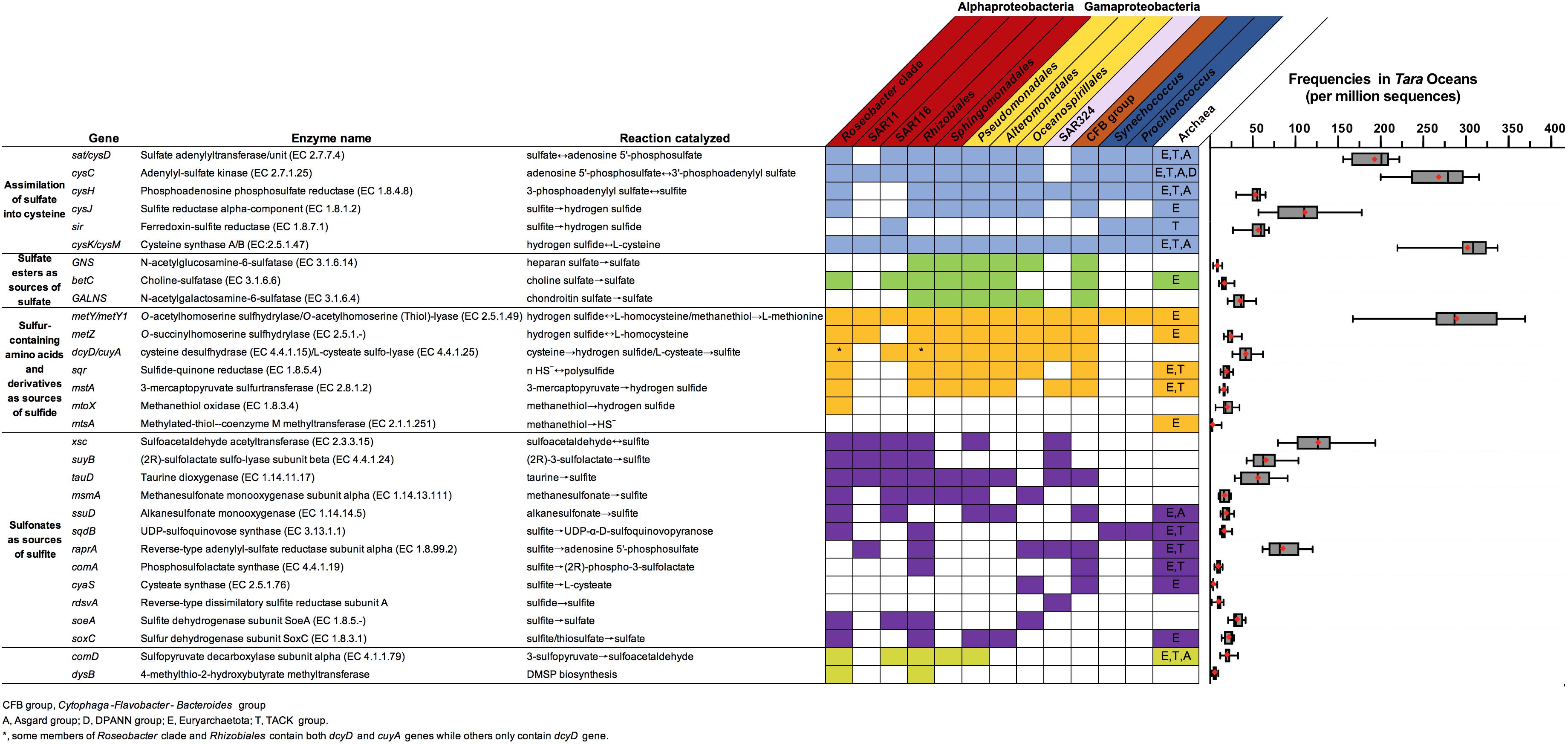
Figure 3. Genes involved in the conversion between inorganic and organic sulfur compounds. Colored boxes highlight genes found in at least five representatives of marine group. The corresponding enzymatic reactions, genetic distributions in marine microbes, and frequencies in the Tara Oceans datasets are presented. Colored boxes highlight genes found in at least five representatives of the group. Thick black lines indicate the median value; red diamonds represent the mean gene frequency across the global ocean. The sulfur genes homologous search with reference sequences (Supplementary Table S2) were performed against IMG genomes (https://img.jgi.doe.gov). To identify sulfur genes distributions in the marine environment, a BLASTP against a refined Tara Oceans database (Delmont et al., 2018) with reference sequences was performed with pre-determined E-value and identity cutoff values as Supplementary Table S2.
Cys as a Sink of Sulfate
A genetic basis for sulfate assimilation has been identified in diverse bacterial and archeal taxa, including Proteobacteria, Cyanobacteria, Bacteroides, and Archea (Figure 3). Taxa in the SAR11 clade, which are important for organic sulfur turnover, make up 33% of all euphotic-zone bacteria and 25% of all mesopelagic bacteria (Giovannoni, 2017). Genes encoding the cysC and cysM proteins in the sulfate assimilation pathway have been identified in all available SAR11 genomes (Tripp et al., 2008). However, other essential genes for assimilation of sulfur into Cys are lacking in SAR11, including sat/cysD, cysH and cysJ/sir (encoding sulfite reductase) (Figure 3), and hence they require exogenous sources of reduced sulfur to support their growth (Tripp et al., 2008). In contrast to the Euryarchaetota and the TACK, the Asgard and DPANN superphyla of the Archaea lack sulfate assimilation abilities (Figure 3).
Sulfate Esters as Sources of Sulfate
The average frequencies of three sulfatases genes (betC, GNS and GALNS) are low in the global ocean (Figure 3). Although SAR11, SAR324, and cyanobacteria lack sulfatase genes (Figure 3), betC genes have been identified in the Rosebacter, SAR116, and Euryarchaetota (Figure 3). Members of the Cytophaga-Flavobacteria-Bacteroides (CFB) group have an unusually high number of sulfatase-encoding genes, which are associated with the degradation of sulfated marine algal polysaccharides; these genes may be upregulated in CFB taxa when concentrations of sulfated organics increase significantly (e.g., during the phytoplankton blooms) (Teeling et al., 2016).
Sulfur-Containing Amino Acids and Derivatives as Sources of Sulfide
The absence of aprAB and dsrAB genes in the dissimilatory sulfate reduction pathway in Tara Oceans datasets shows that the reduction of sulfate to sulfide occurs rarely in the seawaters; in contrast, Cys and its derivatives are regarded as potential sources of sulfide (Figure 3). Genes metY and metZ (encoding O-succinylhomoserine sulfhydrylase) have been identified in most bacteria and Euryarchaetota (Figure 3), suggesting that these taxa have the capacity to use homocysteine as sulfide sources (Figure 3). In bacteria, Cys desulfhydrase (dycD) and 3-mercaptopyruvate sulfurtransferase (mstA) are believed to be chiefly responsible for sulfide biogenesis from Cys and 3-mercaptopyruvate (Carbonero et al., 2012), respectively. However, the corresponding genes have not been identified in SAR11 or cyanobacterial genomes. The microbial catabolism of MeSH releases sulfide through the enzymatic activity of methanethiol oxidase (mtoX) (Eyice et al., 2018) or mtsA (Carrión et al., 2017). The mtoX genes are widely distributed in marine environments and are more abundant than mstA genes (Figure 3). The mtoX gene is found not only in methylotrophic bacteria (Carrión et al., 2019) but also taxa in the Roseobacter clade, including Ruegeria pomeroyi DSS-3, a DMSP-degrading bacterium (Eyice et al., 2018). This suggests a possible pathway for sulfide production from DMSP in the marine environment. Some Oceanospirillales and Euryarchaetota genomes harbor the mstA gene (Figure 3). Sulfide:quinone oxidoreductase (sqr), which is an important enzyme for microbial sulfide oxidation (Friedrich et al., 2001), is absent in major bacterial taxa in marine environments (e.g., SAR11 and cyanobacteria) (Figure 3).
Sulfonates as Sources and Sinks of Sulfite
Besides sulfite from assimilatory sulfate reduction pathway, sulfonates were the main sources of endogenous production of sulfite in microbes. Several genes associated with the desulfonation of sulfoacetadehyde (xsc), sulfolactate (suyB), taurine (tauD) are highly abundant in the Tara Oceans datasets (Figure 3). This suggests that many marine microbes have the potential to degrade sulfonate. For example, such genes are present in Alphaproteobacteria (mainly in the order Rhizobiales, the genus Roseobacter, and the clades SAR11 and SAR116), but are absent in the cyanobacteria and archaea (Figure 3). The gene tauD was also identified in the bacterial orders Pseudomonadales, Alteromonadales, Oceanospirillales (in the Gammaproteobacteria; Figure 3). Members of the ubiquitous alphaproteobacterial SAR11 clade account for a large fraction of the cells that take up taurine, especially in surface waters (Clifford et al., 2019). Genes important for the desulfonation of cysteate (cuyA), methanesulfonate (msmA) and alkanesulfonate (ssuD) have been identified in Roseobacter species, and may allow these bacteria to use a broad range of sulfonates for growth (Figure 3). Members of the SAR116 and Pseudomonadales clades harbored the genes msmA and ssuD (Figure 3). The rdsvA gene, which was identified in SAR324 (Figure 3), codes for a reverse operating siroheme dissimilatory sulfite reductase subunit that catalyzes the direct oxidation of sulfide into sulfite (Swan et al., 2011).
The involvement of sqdB in SQDG biosynthesis is not ubiquitous across the heterotrophic bacteria, with the exception of the Roseobacter clade and the order Rhizobiales (Figure 3). Members of the cyanobacteria and archaea harbor sqdB genes (Figure 3). Most of the sqdB genes known to be involved in SQDG synthesis identified in the surface waters of the Sargasso Sea and the North Sea were affiliated with cyanobacteria, not heterotrophic bacteria (Van Mooy et al., 2006; Villanueva et al., 2014). The gene soeA, encoding a subunit of the sulfite-oxidizing enzyme, is found in the Roseobacter clade; this gene is involved in sulfite oxidation (Boughanemi et al., 2016). The gene soxC. from the sox system, is implicated in sulfite oxidation and is frequently identified in sulfur-oxidizing bacteria (Kappler and Dahl, 2001). Another sulfite oxidation reaction is mediated by cytoplasmic reverse APS reductase (encoded by raprA and raprB); in SAR11, cytoplasmic reverse APS reductase may utilize AMP and sulfite to generate APS (Kappler and Dahl, 2001), as a mechanism for the intracellular detoxification of sulfite (Meyer et al., 2007). The raprA gene was common in the Tara Oceans datasets, indicating that the potential for sulfite oxidation is ubiquitous in seawater (Figure 3). It has been proposed that dissimilatory sulfur oxidation may be used for the energetic support of carbon fixation in autotrophic SAR324 populations based on the presence of both raprA and rdsvA genes (Swan et al., 2011). Phosphosulfolactate synthase (comA) and cysteate synthase (cyaS) in some archaeal methanogens incorporate sulfite into phosphoenolpyruvate and phosphoserine to form phosphosulfolactate and cysteate, respectively (Graham et al., 2002, 2009).
Genomic Insight Into Dmsp and Dhps Transformation in the Ocean
Genes involved in DMSP and DHPS transformation play crucial roles in the sulfur cycling of the oceans (Howard et al., 2006; Durham et al., 2019). The key intermediates MTHB and the dysB enzyme were identified in the marine bacteria Labrenzia aggregata LZB033, indicating that DMSP biosynthesis via Met transamination may operate in heterotrophic bacteria (Curson et al., 2017). Only 140 sequences homologous to dysB were identified in the Alphaproteobacteria genomes, and the frequency of the dysB gene was low in the Tara Oceans datasets, indicating that heterotrophic bacteria were not major DMSP producers in the surface ocean, as compared to the large phytoplankton. Seven distinct functional DMSP lyase genes (dddD, dddK, dddL, dddP, dddQ, dddY, and dddW) are found in the proteobacteria (Todd et al., 2009, 2010, 2011; Curson et al., 2011a, b; Sun et al., 2016). An additional functional DMSP lyase gene, Alma1, is firstly annotated and the most highly expressed in the alga Emiliania huxleyi (Alcolombri et al., 2015). The dddK genes are the most abundant bacterial DMSP lyase genes found in the marine metagenome (Landa et al., 2019). The ddd genes have been identified in the marine Alphaproteobacteria (e.g., the Roseobacter clade, SAR11, and SAR116), as well as in the Gammaproteobacteria (Moran et al., 2012; Sun et al., 2016; Nowinski et al., 2019). Homologs of the DMSP demethylase gene dmdA appeared to be more abundant in marine environments than the DMSP lyase genes or the DHPS utilization gene hpsN (Figure 4). The genes dmdA and hpsN are known to be harbored by the members of the Roseobacter, SAR11, SAR116, SAR324, and Gammaproteobacteria. SAR11 and Roseobacter carry most of the dmdA and hspN genes identified in the euphotic zones, whereas SAR324 and Gammaproteobacteria carry the majority of the hspN genes identified in the mesopelagic zones (Landa et al., 2019). The relative global abundance of dmdA, ddd, and hpsN in the Tara Oceans datasets was 0.09–0.65 genes cell–1 (30–300 per million reads), 0.05–0.51 genes cell–1 (18–160 per million reads), and 0.01–0.50 genes cell–1 (25–210 per million reads), respectively (Figure 4). These genes are ubiquitous in the global euphotic and mesopelagic zones (Figure 4). Surface water and water at the depth of the deep chlorophyll maximum had significantly higher relative abundances of these three genes than water in the mesopelagic zones (p < 0.05, Figure 4). The highest average relative abundances of dmdA and hpsN (genes per cell) in the eutrophic zones were observed in the Mediterranean Sea, and the lowest abundances were observed in the Southern Ocean (Figure 4). However, abundances were highly variable in each oceanic region.
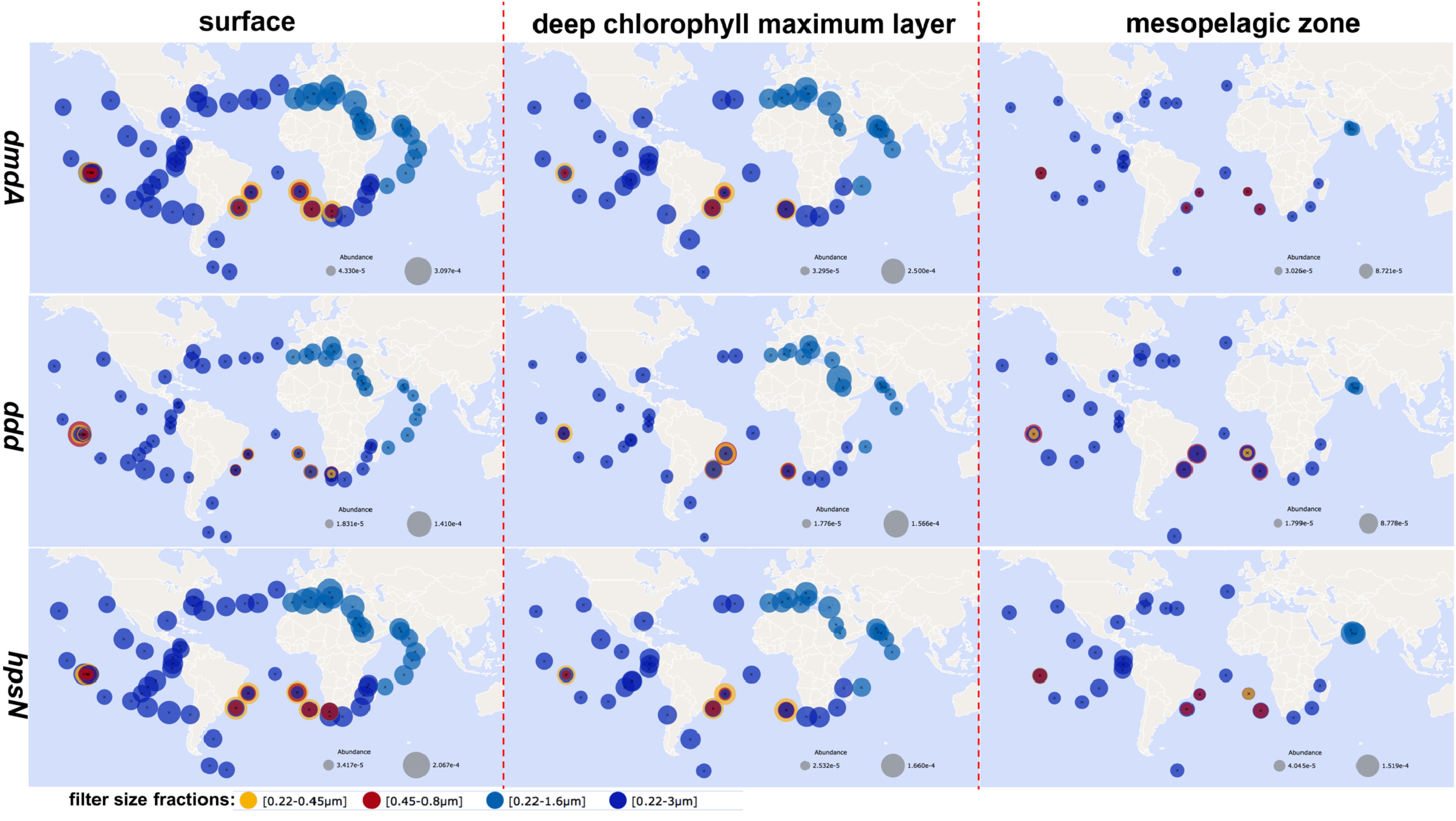
Figure 4. Biogeography of dmdA, ddd, and hpsN. The relative abundances of these genes at each station (gene sequences/total sequences) were derived from the Ocean Gene Atlas across the different size fractions and sampling depths (http://tara-oceans.mio.osupytheas.fr/ocean-gene-atlas/) (Villar et al., 2018) using reference sequences via BLASTP (E-value: dmdA (Q4FP21) 1 × 10–20; ddd (Q5LSR3) 1 × 10–20; hpsN (Q5LVV1) 1 × 10–40).
Future Perspectives
The annual production of DMSP, which is well studied, accounts for approximately 18% of yearly sulfur assimilation by marine primary producers (Curson et al., 2017). However, other relevant compounds in the sulfur cycle are far less studied, leaving many questions regarding the biochemistry of sulfur metabolites unanswered. Furthermore, the relative importance of the processes affecting organic sulfur dynamics, especially in the deep ocean and sediments, are relatively poorly constrained. Recent studies of sulfonates, which have combined ‘omic’ and chemical detection methods, deserve careful attention as there is a crucial link between marine heterotrophic bacteria and primary producers (Durham et al., 2019; Landa et al., 2019). Here, current research is synthesized, gaps in our understanding of organic sulfur are highlighted, and future research directions are recommended.
Characterizations of Substrates and Specific Enzymes
Details of the chemistry and biochemical machinery of organic sulfur transformation can help us to untangle intriguing and hitherto unresolved sulfur biogeochemical processes. Despite the successful detection of cellular sulfur metabolites and DOS in seawater using mass spectrometry-based techniques, such as Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS), these compounds remain largely undefined at barely detectable levels. However, DOS extraction methods, such as solid phase extraction, are likely to remove many small polar compounds prior to mass spectrometry analyses; for example, solid phase extraction may remove dissolved DHPS with an extraction efficiency of ∼1% (Johnson et al., 2017). The quantification of sulfonates in seawater, including SQ, SQDG, and DHPS, is an important challenge for sulfur chemical ecology. Genomic data for reactions enzymatically catalyzed by organic sulfur in eukaryotic phytoplankton are severely limited. Recently proposed sulfonate metabolic pathways showed that the sulfur in DHPS might originally arise from Cys and PAPS, or might be directly derived from the sulfonation of glycerol phosphate (Durham et al., 2019). This helps to explain the high levels of DHPS inside phytoplankton cells. However, the enzymes that catalyze DHPS synthesis are still unknown. All cyanobacteria were believed to lack dmdA gene (González et al., 2019), however, they can take up and assimilate DMSP via an as-yet-unidentified pathway (Malmstrom et al., 2005). In addition, the enzymatic or chemical mechanisms of DMSOP synthesis in DMSP and DMS cycling remained unclear (Thume et al., 2018). The microbial uptake of exogenous sulfur-containing compounds, such as Cys, Met, GSH, DHPS, DMSP and taurine, requires transporter proteins (Supplementary Table S3), but specific transporters for many known sulfur metabolites still await discovery. Enantioselective and diastereoselective interactions may occur between chiral sulfonates and enzymes. For example, the enzymes hpsN and hspO may act on R-DHPS and S-DHPS, respectively, but experimental evidence for these interactions are lacking. Although the molecular configuration of DHPS in natural environments is unknown, SAR11 seems to only utilize one enantiomeric form of DHPS as SAR11 taxa possesses only the hpsN gene. In contrast, Roseobacter species metabolize two enantiomeric forms of DHPS and possess both hpsN and hpsO. Thus, the future discovery and characterization of novel substrates and enzymes is essential for a better understanding of a key step in the sulfur cycle.
Revealing Organosulfur Cycling in the Dark Ocean and Sediment
Although the pelagic realm of the dark ocean is the largest reservoir of heterogeneous DOS in the ocean (>6,700 Tg S), few details are known about the source and sink of biogenic sulfur in this region (Ksionzek et al., 2016). Deep-ocean DOS is thought mainly to originate from biological processes in the upper ocean and from organic matter transported from the upper ocean to the deep ocean and sediments; DOS in the deep ocean may persist for thousands of years (Koch et al., 2017). The most important biologically mediated processes regulating the long term storage of organic matter in the ocean are the biological carbon pump (Ducklow et al., 2001) and the microbial carbon pump (Jiao et al., 2010, 2014, 2018). After the long-term incubation (110 days) of a microbial community in natural seawater, more sulfur metabolites may be produced, suggesting that biotic process lead to the formation of refractory compounds (Figure 5A) (Cai et al., 2019). SQDG is efficiently removed as it sinks to the deep sea; in contrast, around 40 sulfur-containing lipids, likely produced by photoautotrophs in the upper ocean, are potentially important as deep-ocean carbon storage compounds (Schubotz et al., 2018). However, the hundreds of DOS identified in the deep sea far exceeds the number of DOS substrates taken up by microbial transporters (Supplementary Table S3). In addition, although the affinity of transporter constants (Ks) varies greatly from millimolar to nanomolar, this affinity is generally higher than the apparent picomolar concentrations of DOS. Thus, DOS in the deep ocean is either intrinsically refractory or present in extremely low concentrations, resulting in its resistance to microbial utilization. Besides describing the molecular composition of deep-sea DOS, future work should investigate whether the microbial carbon pump combined with biological pump carbon transforms organic sulfur into refractory one. If so, this might help explain the observed huge DOS pool in the dark ocean.
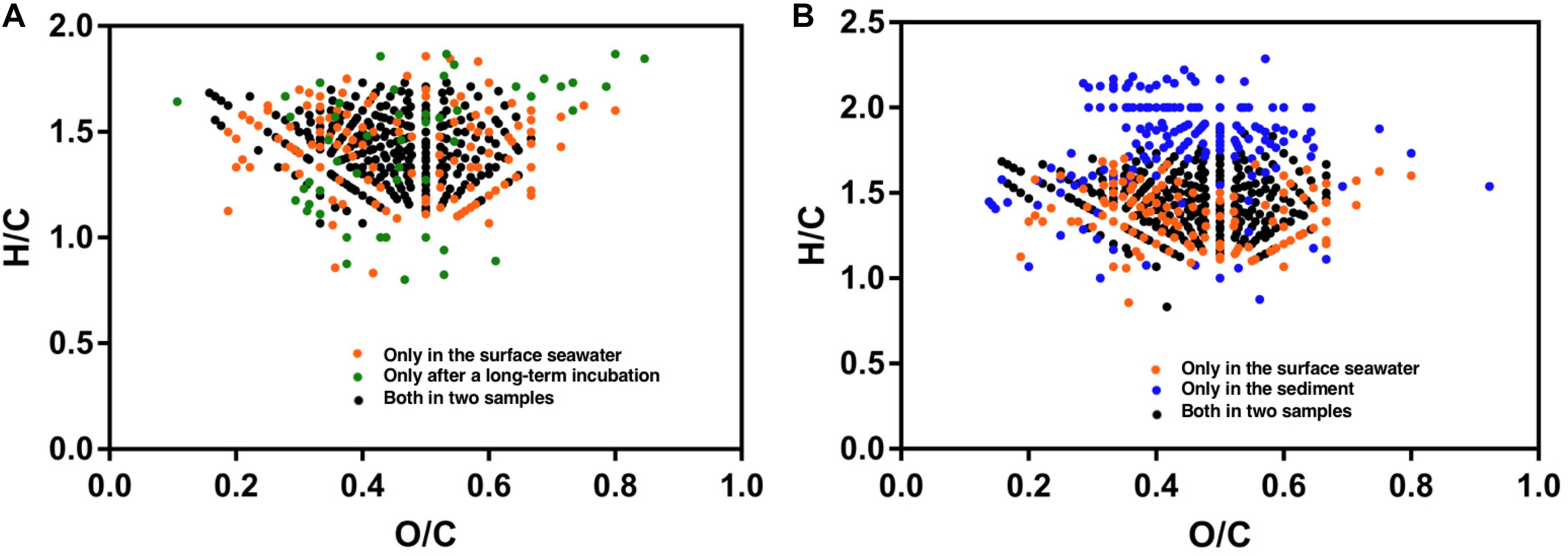
Figure 5. Van Krevelen diagrams [hydrogen-to-carbon (H/C) and oxygen-to-carbon (O/C) ratios] showing the S-containing formulae derived from FT-ICR-MS (data derived from Cai et al., 2019). (A) Samples from the surface water and incubation experiments using surface water (110 days at 20°C). (B) Samples from the surface water and surficial sediment.
Organic sulfur is the second most important reduced sulfur pool in sedimentary environments. FT-ICR-MS has shown that organic sulfur in surficial sediments is composed of a heterogeneous mixture of sulfur compounds (Cai et al., 2019). These compounds are considerably different from sulfur compounds identified in seawater. For example, unique saturated compounds with relatively high hydrogen to carbon ratios have been identified in sediment (Figure 5B). Bacteria have recently been shown to be an important source of DMSP; high concentrations of DMSP are produced by certain bacterial species (Williams et al., 2019). Moreover, MeSH and DMS consumption processes, mediated by bacteria, were observed in the surficial sediments of a coastal saltmarsh (Williams et al., 2019). Although sulfonated organics generally account for 20–40% of all organic sulfur in marine sediments (Vairavamurthy et al., 1994), their sources and fates remain unclear. Both biotic and abiotic processes of organic matter diagenetic sulfurization may occur in the sulfidic sediments (Pohlabeln et al., 2017), whereby sulfate-reducing microorganisms might substantially contribute to sulfide incorporation into organic matter through the dissimilatory sulfur reduction pathway. Such diagenetically formed organic sulfur compounds might be buried during carbon sequestration as they may not be recognized by microbial enzymes (Wasmund et al., 2017). Future studies of the biogeochemical processes of sedimentary organic sulfur might substantially expand our understanding of the marine sulfur cycle.
Summary
Biogenic organic sulfur compounds have diverse chemical forms and play essential biological roles in marine organisms. Methyl-sulfur compounds, sulfonates, thiols, sulfolipids, and sulfate esters are widespread and represent a significant fraction of the DOS pool in the upper ocean. Phytoplankton, which produce much labile organic sulfur, together with heterotrophic bacteria, plays a major role in the carbon and sulfur fluxes in the pelagic ocean via a large set of sulfur metabolites that are intertwined in the biochemical reaction network. Methylated sulfur compounds and sulfonate catabolisms allow the microbial acquisition of reduced sulfur and carbon from organic sulfur compounds. Taxa in the Roseobacter, SAR11, SAR116, SAR324, and Gammaproteobacteria constitute an important gene pool for the transformation and mineralization of organic sulfur in the upper ocean. Future research on the characterization of novel substrates and enzymes involved in sulfur metabolism, and the investigation of organic sulfur transformation in the deep sea and in sediments, are essential steps toward a comprehensive understanding of the microbial contribution to the marine sulfur cycle.
Author Contributions
KT conceived of and wrote the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China project (41776167, 91751207, U1805242, and 41861144018) and the National Key Research and Development Program of China (2016YFA0601100 and 2016YFA0601400).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00068/full#supplementary-material
Abbreviations
γ-Glu –Cys, γ-glutamylcysteine; Alma1, DMSP lyase in Emiliania huxleyi;; aprAB, adenylyl-sulfate reductase; APS, adenosine 5-phosphosulfate; betC, choline-sulfatase; CFB, Cytophaga-Flavobacteria-Bacteroides; comA, phosphosulfolactate synthase; comC or slcC, sulfolactate dehydrogenase; comD, sulfopyruvate decarboxylase subunit alpha; comE, sulfopyruvate decarboxylase subunit; cuyA, L-cysteate sulfo-lyase; cyaS, cysteate synthase; Cys, cysteine; cysC, adenylyl-sulfate kinase; cysH, phosphoadenosine phosphosulfate reductase; cysJ, sulfite reductase alpha-component; cysK/cysM, cysteine synthase A/B; dcyD, cysteine desulfhydrase; ddd, DMSP lyases (including dddD, dddK, dddL, dddP, dddQ, dddY and dddW); DHPS, 3-sulfopropanediol; dmdA, DMSP demethylase; DMS, dimethyl sulfide; DMSO, dimethylsulfoxide; DMSOP, dimethylsulfoxonium propionate; DMSP, dimethylsulfoniopropionate; DOS, dissolved organic sulfur; dsrAB, reversible dissimilatory sulfite reductase; DYSB/dysB (eukaryotic/prokaryotic gene name), 4-methylthio-2-hydroxybutyrate methyltransferase; FT-ICR-MS, Fourier transform ion cyclotron resonance mass spectrometry; GALNS, N-acetylgalactosamine -6-sulfatase; GNS, N-acetylglucosamine -6-sulfatase; GSH, glutathione; GSSG, glutathione disulfide; hpsO, hpsP and hpsN, DHPS hydrogenase enzymes; mdeA, methionine γ-lyase; MeSH, methanethiol; Met, methionine; metY, O-acetylhomoserine sulfhydrylase; metY1, O-acetylhomoserine; metZ, O-succinylhomoserine sulfhydrylase; MSA, methanesulfonate; MSA, methanesulfonate; msmA, methanesulfonate monooxygenase subunit alpha; mstA, 3-mercaptopyruvate sulfurtransferase; MTHB, 4-methylthio-2-hydroxybutyrate; mtoX, methanethiol oxidase; mtsA, methylated-thiol–coenzyme M methyltransferase; PAPS, 3′ -phosphoadenosyl-5′ -phosphosulfate; raprA, reverse-type adenylyl-sulfate reductase subunit alpha; raprB, reverse-type adenylyl-sulfate reductase subunit beta; rdsvA, reverse-type dissimilatory sulfite reductase subunit A; sat/cysD, sulfate adenylyltransferase; sir, ferredoxin-sulfite reductase; soeA, sulfite dehydrogenase subunit SoeA; soxC, Sulfur dehydrogenase subunit SoxC; SQ, sulfoquinovose; sqdA, UDP-sulfoquinovose acyltransferase; sqdB, UDP-sulfoquinovose synthase; SQDG, sulfoquinovosyldiacylglycerol; sqr, sulfide-quinone reductase; ssuD, alkanesulfonate monooxygenase; suyAB, (2R)-sulfolactate sulfo-lyase; suyB, (2R)-sulfolactate sulfo-lyase subunit beta; tauD, taurine dioxygenase; UDP, uridine diphosphate; xsc, sulfoacetaldehyde acetyltransferase; yihQ, sulfoquinovosidase.
References
Agnello, G., Chang, L. L., Lamb, C. M., Georgiou, G., and Stone, E. M. (2013). Discovery of a substrate selectivity motif in amino acid decarboxylases unveils a taurine biosynthesis pathway in Prokaryotes. ACS. Chem. Biol. 8, 2264–2271. doi: 10.1021/cb400335k
Ahner, B. A., Wei, L., Oleson, J. R., and Ogura, N. (2002). Glutathione and other low molecular weight thiols in marine phytoplankton under metal stress. Mar. Ecol. Prog. Ser. 232, 93–103. doi: 10.3354/meps232093
Alcolombri, U., Ben-Dor, S., Feldmesser, E., Levin, Y., Tawfik, D. S., and Vardi, A. (2015). Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348, 1466–1469. doi: 10.1126/science.aab1586
Alcolombri, U., Laurino, P., Lara-Astiaso, P., Vardi, A., and Tawfik, D. S. (2014). DddD is a CoA-transferase/lyase producing dimethyl sulfide in the marine environment. Biochemistry 53, 5473–5475. doi: 10.1021/bi500853s
Al-Farawati, R., and van den Berg, C. M. (2001). Thiols in coastal waters of the western North Sea and english channel. Environ. Sci. Technol. 35, 1902–1911. doi: 10.1021/es000073i
Amarita, F., Yvon, M., Nardi, M., Chambellon, E., Delettre, J., and Bonnarme, P. (2004). Identification and functional analysis of the gene encoding methionine-γ -lyase in Brevibacterium linens. Appl. Environ. Microbiol. 70, 7348–7354. doi: 10.1128/AEM.70.12.7348-7354.2004
Andreae, M. O., and Crutzen, P. J. (1997). Atmospheric aerosols: biogeochemical sources and role in atmospheric chemistry. Science. 276, 1052–1058. doi: 10.1126/science.276.5315.1052
Archer, S. D., Widdicombe, C. E., Tarran, G. A., Rees, A. P., and Burkill, P. H. (2001). Production and turnover of particulate dimethylsulphoniopropionate during a coccolithophore bloom in the northern North Sea. Aquat. Microb. Ecol. 24, 225–241. doi: 10.3354/ame024225
Barnes, I., Hjorth, J., and Mihalapoulos, N. (2006). Dimethyl sulfide and dimethyl sulfoxide and their oxidation in the atmosphere. Chem. Rev. 106, 940–975. doi: 10.1021/cr020529
Bates, S., Kiene, P., Wolfe, V., Matrai, A., Chavez, P., Buck, R., et al. (1994). The cycling of sulfur in surface seawater of the northeast pacific. J. Geophys. Res. 99, 7835–7843. doi: 10.1029/93jc02782
Bennett, B. D., Kimball, E. H., Gao, M., Osterhout, R., Van Dien, S. J., and Rabinowitz, J. D. (2009). Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599. doi: 10.1038/nchembio.186
Benning, C., and Somerville, C. R. (1992a). Identification of an operon involved in sulfolipid biosynthesis in Rhodobacter sphaeroides. J. Bacteriol. 174, 6479–6487. doi: 10.1128/jb.174.20.6479-6487.1992
Benning, C., and Somerville, C. R. (1992b). Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J. Bacteriol. 174, 2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992
Boughanemi, S., Lyonnet, J., Infossi, P., Bauzan, M., Lignon, S., and Guiral, M. (2016). Microbial oxidative sulfur metabolism: biochemical evidence of the membrane-bound heterodisulfide reductase-like complex of the bacterium Aquifex aeolicus. FEMS. Microbio. Lett. 363:fnw156. doi: 10.1093/femsle/fnw156
Bullock, H. A., Luo, H., and Whitman, W. B. (2017). Evolution of dimethylsulfoniopropionate metabolism in marine phytoplankton and bacteria. Front. Microbiol. 8:637. doi: 10.3389/fmicb.2017.00637
Burrichter, A., Denger, K., Franchini, P., Huhn, T., Müller, N., Spiteller, D., et al. (2018). Anaerobic degradation of the plant sugar sulfoquinovose concomitant with H2S production: Escherichia coli K-12 and Desulfovibrio sp. strain DF1 as co-culture model. Front. Microbiol. 9:2792. doi: 10.3389/fmicb.2018.02792
Cai, R., Zhou, W., He, C., Tang, K., Guo, W., Shi, Q., et al. (2019). Microbial processing of sediment-derived dissolved organic matter: implications for its subsequent biogeochemical cycling in overlying seawater. J. Geophys. Res. Biogeosci. 124, 3479–3490. doi: 10.1029/2019JG005212
Canfield, D. E., and Farquhar, J. (2009). Animal evolution, bioturbation, and the sulfate concentration of the oceans. Proc. Natl. Acad. Sci. U.S.A. 106, 8123–8127. doi: 10.1073/pnas.0902037106
Carbonero, F., Benefiel, A. C., Alizadeh-ghamsari, A. H., and Gaskins, H. R. (2012). Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 3:448. doi: 10.3389/fphys.2012.00448
Carrión, O., Pratscher, J., Curson, A. R. J., Williams, B. T., Rostant, W. G., Murrell, J. C., et al. (2017). Methanethiol-dependent dimethylsulfide production in soil environments. ISME J. 11, 2379–2390. doi: 10.1038/ismej.2017.105
Carrión, O., Pratscher, J., Richa, K., Rostant, W. G., Farhan, M., Haque, U., et al. (2019). Methanethiol and dimethylsulfide cycling in stiffkey saltmarsh. Front. Microbiol. 10:1040. doi: 10.3389/fmicb.2019.01040
Caspi, R., Billington, R., Fulcher, C. A., Keseler, I. M., Kothari, A., Krummenacker, M., et al. (2018). The metacyc database of metabolic pathways and enzymes. Nucleic Acids Res. 46, D633–D639. doi: 10.1093/nar/gkx935
Chapman, E., Best, M. D., Hanson, S. R., Wong, C. -H., and Wong, C. H. (2004). Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. Int. Ed Eng. 42, 3526–3548. doi: 10.1002/anie.200300631
Christophersen, C., and Anthoni, U. (1986). Organic sulfur compounds from marine organisms. Sulfur Rep. 4, 365–442. doi: 10.1080/01961778608082487
Clifford, E. L., Hansell, D. A., Varela, M. M., Nieto-Cid, M., Herndl, G. J., and Sintes, E. (2017). Crustacean zooplankton release copious amounts of dissolved organic matter as taurine in the ocean. Limnol. Oceanogr. 62, 2745–2758. doi: 10.1002/lno.10603
Clifford, E. L., Varela, M. M., De Bode, A., Ortiz, V., Herndl, G. J., et al. (2019). Taurine is a major carbon and energy source for marine prokaryotes in the North Atlantic Ocean off the Iberian Peninsula. Microb. Ecol. 78, 299–312. doi: 10.1007/s00248-019-01320-y
Cook, A. M., Laue, H., and Junker, F. (1998). Microbial desulfonation. FEMS Microbiol. Rev. 22, 399–419. doi: 10.1016/S0168-6445(98)00028-X
Cunha, L., and Grenha, A. (2016). Sulfated seaweed polysaccharides as multifunctional materials in drug delivery applications. Mar. Drugs 14:42. doi: 10.3390/md14030042
Curson, A. R. J., Liu, J., Martínez, A. B., Green, R. T., Chan, Y., Carrión, O., et al. (2017). Dimethylsulfoniopropionate biosynthesis in marine bacteria and identifi cation of the key gene in this process. Nat. Microbiol. 2:17009. doi: 10.1038/nmicrobiol.2017.9
Curson, A. R. J., Sullivan, M. J., Todd, J. D., and Johnston, A. W. B. (2011a). DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME. J. 5, 1191–1200. doi: 10.1038/ismej.2010.203
Curson, A. R. J., Todd, J. D., Sullivan, M. J., and Johnston, A. W. B. (2011b). Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9, 849–859. doi: 10.1038/nrmicro2653
Curson, A. R. J., Williams, B. T., Pinchbeck, B. J., Sims, L. P., Martínez, A. B., Rivera, P. P. L., et al. (2018). DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat. Microbiol. 3, 430–439. doi: 10.1038/s41564-018-0119-5
Del Valle, D. A., Kieber, D. J., Toole, D. A., Bisgrove, J., and Kiene, R. P. (2009). Dissolved DMSO production via biological and photochemical oxidation of dissolved DMS in the Ross Sea, Antarctica. Deep Sea Res. Part I 56, 166–177. doi: 10.1016/j.dsr.2008.09.005
Delmont, T. O., Quince, C., Shaiber, A., Esen, Ö. C., Lee, S. T., Rappé, M. S., et al. (2018). Nitrogen-fixing populations of planctomycetes and proteobacteria are abundant in the surface ocean. Nat. Microbiol. 3, 804–813. doi: 10.1038/s41564-018-0176-9
Denger, K., Huhn, T., Hollemeyer, K., Schleheck, D., and Cook, A. M. (2012). Sulfoquinovose degraded by pure cultures of bacteria with release of C3-organosulfonates: complete degradation in two-member communities. FEMS. Microbiol. Lett. 328, 39–45. doi: 10.1111/j.1574-6968.2011.02477.x
Denger, K., Mayer, J., Buhmann, M., Weinitschke, S., Smits, T. H. M., and Cook, A. M. (2009). Bifurcated degradative pathway of 3-sulfolactate in Roseovarius nubinhibens ISM via sulfoacetaldehyde acetyltransferase and (S)-cysteate sulfolyase. J. Bacteriol. 191, 5648–5656. doi: 10.1128/JB.00569-09
Denger, K., Weiss, M., Felux, A., Schneider, A., Mayer, C., Spiteller, D., et al. (2014). Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature 507, 114–117. doi: 10.1038/nature12947
Diaz, J. M., Plummer, S., Hansel, C. M., Andeer, P. F., Saito, M. A., and McIlvin, M. R. (2019). NADPH-dependent extracellular superoxide production is vital to photophysiology in the marine diatom Thalassiosira oceanica. Proc. Natl. Acad. Sci. U.S.A. 116, 16448–16453. doi: 10.1073/pnas.1821233116
Ducklow, H. W., Steinberg, D. K., and Buesseler, K. O. (2001). Upper ocean carbon export and the biological pump. Oceanography 14, 50–58. doi: 10.5670/oceanog.2001.06
Dupont, C. L., Moffett, J. W., Bidigare, R. R., and Ahner, B. A. (2006). Distributions of dissolved and particulate biogenic thiols in the subartic Pacific Ocean. Deep Sea. Res. Part I 53, 1961–1974. doi: 10.1016/j.dsr.2006.09.003
Durham, B. P., Boysen, A. K., Carlson, L. T., Groussman, R. D., Heal, K. R., Cain, K. R., et al. (2019). Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean. Nat. Microbiol. 4, 1706–1715. doi: 10.1038/s41564-019-0507-5
Durham, B. P., Sharma, S., Luo, H., Smith, C. B., Amin, S. A., and Bender, S. J. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 112, 453–457. doi: 10.1073/pnas.1413137112
Eyice, Ö., Myronova, N., Pol, A., Carrión, O., Todd, J. D., Smith, T. J., et al. (2018). Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME. J. 12, 145–160. doi: 10.1038/ismej.2017.148
Fahey, R. C. (2013). Glutathione analogs in prokaryotes. Biochim. Biophys. Acta Gen. Subj. 1830, 3182–3198. doi: 10.1016/j.bbagen.2012.10.006
Felux, A., Spiteller, D., Klebensberger, J., and Schleheck, D. (2015). Entner– doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1. Proc. Natl. Acad. Sci. U. S. A. 4298–4305. doi: 10.1073/pnas.1507049112
Ferla, M. P., and Patrick, W. M. (2014). Bacterial methionine biosynthesis. Microbiology 160, 1571–1584. doi: 10.1099/mic.0.077826-0
Friedrich, C. G., Rother, D., Bardischewsky, F., and Quentmeier, A., and Jörg, F. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microb. 67, 2873–2882. doi: 10.1128/AEM.67.7.2873
Galí, M., Devred, E., Levasseur, M., Royer, S. J., and Babin, M. (2015). A remote sensing algorithm for planktonic dimethylsulfoniopropionate (DMSP) and an analysis of global patterns. Remote Sens. Environ. 171, 171–184. doi: 10.1016/j.rse.2015.10.012
Giordano, M., Norici, A., and Hell, R. (2005). Sulfur and phytoplankton: acquisition, metabolism and impact on the environment. New Phytol. 166, 371–382. doi: 10.1111/j.1469-8137.2005.01335.x
Giovannoni, S. J. (2017). SAR11 bacteria: the most abundant plankton in the oceans. Ann. Rev. Mar. Sci. 9, 231–258. doi: 10.1146/annurev-marine-010814-015934
Goddard-borger, E. D., and Williams, S. J. (2017). Sulfoquinovose in the biosphere: occurrence, metabolism and functions. Biochem. J. 474, 827–849. doi: 10.1042/BCJ20160508
González, J. M., Hernández, L., Manzano, I., and Pedrós-Alió, C. (2019). Functional annotation of orthologs in metagenomes: a case study of genes for the transformation of oceanic dimethylsulfoniopropionate. ISME J. 13, 1183–1197. doi: 10.1038/s41396-019-0347-6
Götz, F., Longnecker, K., Kido, M. C., Kevin, S., Mcnichol, J., Kujawinski, E. B., et al. (2018). Targeted metabolomics reveals proline as a major osmolyte in the chemolithoautotroph Sulfurimonas denitrificans. Microbiologyopen 7:e586. doi: 10.1002/mbo3.586
Graham, D. E., Taylor, S. M., Wolf, R. Z., and Namboori, S. C. (2009). Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase. Biochem. J. 424, 467– 478. doi: 10.1042/BJ20090999
Graham, D. E., Xu, H., and White, R. H. (2002). Identification of coenzyme M biosynthetic phosphosulfolactate synthase. J. Biol. Chem. 277, 13421–13429. doi: 10.1074/jbc.M201011200
Graupner, M., Xu, H., and White, R. H. (2000). Identification of the gene encoding sulfopyruvate decarboxylase, an enzyme involved in biosynthesis of coenzyme M. J. Bacteriol. 182, 4862–4867. doi: 10.1128/JB.182.17.4862-4867.2000
Hacham, Y., Gophna, U., and Amir, R. (2003). In vivo analysis of various substrates utilized by cystathionine γ-synthase and O-acetylhomoserine sulfhydrylase in methionine biosynthesis. Mol. Biol. Evol. 20, 1513–1520. doi: 10.1093/molbev/msg169
Helbert, W. (2017). Marine polysaccharide sulfatases. Front. Mar. Sci. 4:6. doi: 10.3389/fmars.2017.00006
Henriques, A. C., and Marco, P. D. (2015). Methanesulfonate (MSA) catabolic genes from marine and estuarine bacteria. PLoS One 10:e0125735. doi: 10.1371/journal.pone.0125735
Ho, T., Quigg, A., Finkel, Z. V., Milligan, A. J., Wyman, K., Falkowski, P. G., et al. (2003). The elemental composition of some marine phytoplankton. J. Phycol. 39, 1145–1159. doi: 10.1111/j.0022-3646.2003.03-090.x
Holdt, S. L., and Kraan, S. (2011). Bioactive compounds in seaweed: functional food applications and legislation. J. Appl. Phycol. 23, 543–597. doi: 10.1007/s10811-010-9632-5
Howard, E. C., Henriksen, J. R., Buchan, A., Reisch, C. R., Bürgmann, H., Welsh, R., et al. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–652. doi: 10.1126/science.1130657
Jacob, C., and Anwar, A. (2008). The chemistry behind redox regulation with a focus on sulphur redox systems. Physiol. Plant 133, 469–480. doi: 10.1111/j.1399-3054.2008.01080.x
Jarníková, T., Dacey, J., Lizotte, M., Levasseur, M., and Tortell, P. (2018). The distribution of methylated sulfur compounds, DMS and DMSP, in Canadian subarctic and Arctic marine waters during summer, 2015. Biogeosciences 15, 2449–2465. doi: 10.5194/bg-15-2449-2018
Jiao, G., Yu, G., Zhang, J., and Ewart, H. S. (2011). Chemical structures and bioactivities of sulfated polysaccharides from marine algae. Mar. Drugs 9, 196–233. doi: 10.3390/md9020196
Jiao, N., Cai, R., Zheng, Q., Tang, K., Liu, J., Jiao, F., et al. (2018). Unveiling the enigma of refractory carbon in the ocean. Natl. Sci. Rev. 5, 459–463. doi: 10.1093/nsr/nwy020
Jiao, N., Herndl, G. J., Hansell, D. A., Benner, R., Kattner, G., Wilhelm, S. W., et al. (2010). Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599. doi: 10.1038/nrmicro2386
Jiao, N., Robinson, C., Azam, F., Thomas, H., Baltar, F., Dang, H., et al. (2014). Mechanisms of microbial carbon sequestration in the ocean future research directions. Biogeosciences 11, 5285–5306. doi: 10.5194/bg-11-5285-2014
Johnson, W. M., Kido Soule, M. C., and Kujawinski, E. B. (2017). Extraction efficiency and quantification of dissolved metabolites in targeted marine metabolomics. Limnol. Oceanogr. Methods 15, 417–428. doi: 10.1002/lom3.10181
Kappler, U., and Dahl, C. (2001). Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS. Microbiol. Lett. 203, 1–9. doi: 10.1016/S0378-1097(01)00304-4
Kellner Filho, L. C., Picão, B. W., Silva, M. L. A., Cunha, W. R., Pauletti, P. M., Dias, G. M., et al. (2019). Bioactive aliphatic sulfates from marine invertebrates. Mar. Drugs 17:527. doi: 10.3390/md17090527
Kertesz, M. A. (1999). Riding the sulfur cycle- metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS. Microbiol. Rev. 24, 135–175. doi: 10.1016/S0168-6445(99)00033-9
Kiene, R. P., Linn, L. J., and Bruton, J. A. (2000). New and important roles for DMSP in marine microbial communities. J. Sea. Res. 43, 209–224. doi: 10.1016/s1385-1101(00)00023-x
Kiene, R. P., Linn, L. J., González, J., Moran, M. A., and Bruton, J. A. (1999). Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65, 4549–4558. doi: 10.1128/aem.65.10.4549-4558.1999
Kiene, R. P., Nowinski, B., Esson, K., Preston, C., Marin, R., Birch, J., et al. (2019). Unprecedented DMSP concentrations in a massive dinoflagellate bloom in Monterey Bay, CA. Geophys. Res. Lett. 12, 279–288. doi: 10.1029/2019gl085496
Kiene, R. P., and Slezak, D. (2006). Low dissolved DMSP concentrations in seawater revealed by small-volume gravity filtration and dialysis sampling. Limnol. Oceanogr. Methods 4, 80–95. doi: 10.4319/lom.2006.4.80
Klaassen, C. D., and Boles, W. (1997). Sulfation and sulfotransferases 5: the importance phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 11, 404–418. doi: 10.1096/fasebj.11.6.9194521
Kloster, S., Feichter, J., Maier-Reimer, E., Six, K. D., Stier, P., and Wetzel, P. (2006). DMS cycle in the marine ocean-atmosphere system – a global model study. Biogeosciences 3, 29–51. doi: 10.5194/bg-3-29-2006
Klotz, M. G., Bryant, D. A., and Hanson, T. E. (2011). The microbial sulfur cycle. Front. Microbiol. 2:241. doi: 10.3389/fmicb.2011.00241
Koch, B. P., Ksionzek, K. B., Lechtenfeld, O. J., Mccallister, S. L., Schmitt-kopplin, P., Geuer, J. K., et al. (2017). Response to comment on “Dissolved organic sulfur in the ocean: biogeochemistry of a petagram inventory.” Science 356:813. doi: 10.1126/science.aam6328
Ksionzek, K. B., Ksionzek, K. B., Lechtenfeld, O. J., Mccallister, S. L., Schmitt-kopplin, P., Geuer, J. K., et al. (2016). Dissolved organic sulfur in the ocean: biogeochemistry of a petagram inventory. Science 354, 456–459. doi: 10.1126/science.aaf7796
Lambert, I. H., Kristensen, D. M., Holm, J. B., and Mortensen, O. H. (2015). Physiological role of taurine–from organism to organelle. Acta. Physiol. (Oxf.) 213, 191–212. doi: 10.1111/apha.12365
Lana, A., Bell, T. G., Simó, R., Vallina, S. M., Ballabrera-Poy, J., Kettle, A. J., et al. (2011). An updated climatology of surface dimethlysulfide concentrations and emission fluxes in the global ocean. Glob. Biogeochem. Cycles 25, 1–17. doi: 10.1029/2010GB003850
Landa, M., Burns, A. S., Durham, B. P., Esson, K., Nowinski, B., Sharma, S., et al. (2019). Sulfur metabolites that facilitate oceanic phytoplankton – bacteria carbon flux. ISME. J. 13, 2536–2550 doi: 10.1038/s41396-019-0455-3
Lee, P. A., and De Mora, S. J. (1999). Intracellular dimethylsulfoxide (DMSO) in unicellular marine algae: speculations on its origin and possible biological role. J. Phycol. 35, 8–18. doi: 10.1046/j.1529-8817.1999.3510008.x
Leustek, T., and Saito, K. (1999). Sulfate transport and assimilation in plants. Plant Physiol. 120, 637–643. doi: 10.2307/4278843
Lu, S. C. (2013). Glutathione synthesis. Biochim. Biophys. Acta 1830, 3143–3153. doi: 10.1016/j.bbagen.2012.09.008
Malmstrom, R. R., Kiene, R. P., Csic, C., and Kirchman, D. L. (2005). Dimethylsulfoniopropionate (DMSP) assimilation by Synechococcus in the Gulf of Mexico and northwest Atlantic Ocean. Limnol. Oceanogr. 50, 1924–1931. doi: 10.4319/lo.2005.50.6.1924
Matrai, P. A., and Keller, M. D. (1994). Total organic sulfur and dimethylsulfoniopropionate in marine phytoplankton: intracellular variations. Mar. Bio. 119, 61–68. doi: 10.1007/BF00350107
Mayer, J., Huhn, T., Habeck, M., Denger, K., Hollemeyer, K., and Cook, A. M. (2010). Cupriavidus pinatubonensis JMP134: purification of dihydroxypropanesulfonate 3-dehydrogenase. Microbiology 156, 1556–1564. doi: 10.1099/mic.0.037580-0
Meyer, B., Imhoff, J. F., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria – evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9, 2957–2977. doi: 10.1111/j.1462-2920.2007.01407.x
Moran, M. A., and Durham, B. P. (2019). Sulfur metabolites in the pelagic ocean. Nat. Rev. Microbiol. 17, 665–678. doi: 10.1038/s41579-019-0250-1
Moran, M. A., Reisch, C. R., Kiene, R. P., and Whitman, W. B. (2012). Genomic insights into bacterial DMSP transformations. Ann. Rev. Mar. Sci. 4, 523–542. doi: 10.1146/annurev-marine-120710-100827
Mukwevho, E., Ferreira, Z., and Ayeleso, A. (2014). Potential role of sulfur-containing antioxidant systems in highly oxidative environments. Molecules 19376–19389. doi: 10.3390/molecules191219376
Nowinski, B., Motard-Côté, J., Landa, M., Preston, C. M., Scholin, C. A., Birch, J. M., et al. (2019). Microdiversity and temporal dynamics of marine bacterial dimethylsulfoniopropionate genes. Environ. Microbiol. 21, 1687–1701. doi: 10.1111/1462-2920.14560
Østerås, M., Boncompagni, E., Vincent, N., Poggi, M. C., and Le Rudulier, D. (1998). Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: choline-O-sulfate is metabolized into glycine betaine. Proc. Natl. Acad. Sci. U.S.A. 95, 11394–11399. doi: 10.1073/pnas.95.19.11394
Otasek, D., Morris, J. H., Bouças, J., Pico, A. R., and Demchak, B. (2019). Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 20, 1–15. doi: 10.1186/s13059-019-1758-4
Otte, M. L., Wilson, G., Morris, J. T., and Moran, B. M. (2004). Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. J. Exp. Bot. 55, 1919–1925. doi: 10.1093/jxb/erh178
Palmieri, G., Balestrieri, M., Peter-Katalinić, J., Pohlentz, G., Rossi, M., Fiume, I., et al. (2013). Surface-exposed glycoproteins of hyperthermophilic sulfolobus solfataricus P2 show a common N- glycosylation profile. J. Proteome Res. 12, 2779–2790. doi: 10.1021/pr400123z
Pohlabeln, A. M., Gomez-saez, G. V., and Noriega-ortega, B. E. (2017). Experimental evidence for abiotic sulfurization of marine dissolved organic matter. Front. Mar. Sci. 4:364. doi: 10.3389/fmars.2017.00364
Pro, V., St, B., Sea, B., Sievert, S. M., Kiene, R. P., and Schulz-vogt, H. N. (2007). The sulfur cycle. Oceanography 20, 117–123. doi: 10.5670/oceanog.2007.55
Roy, A. B., Hewlins, M. J. E., Ellis, A. J., Harwood, J. L., and White, G. F. (2003). Glycolytic breakdown of sulfoquinovose in bacteria: a missing link in the sulfur cycle. Appl. Environ. Microbiol. 69, 6434–6441. doi: 10.1128/AEM.69.11.6434
Schubotz, F., Xie, S., Lipp, J. S., Hinrichs, K., and Wakeham, S. G. (2018). Intact polar lipids in the water column of the eastern tropical North Pacific: abundance and structural variety of non-phosphorus lipids. Biogeosciences 15, 6481–6501. doi: 10.5194/bg-15-6481-2018
Seymour, J. R., Amin, S. A., Raina, J. B., and Stocker, R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2:17065. doi: 10.1038/nmicrobiol.2017.65
Seymour, J. R., Simo, R., Ahmed, T., and Stocker, R. (2010). Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–346. doi: 10.1126/science.1188418
Simó, R. (2001). Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16, 287–294. doi: 10.1016/S0169-5347(01)02152-8
Speciale, G., Jin, Y., Davies, G. J., Williams, S. J., and Goddard-borger, E. D. (2016). YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids. Nat. Chem. Biol. 12, 1–4. doi: 10.1038/nchembio.2023
Stefels, J. (2000). Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43, 183–197. doi: 10.1016/s1385-1101(00)00030-7
Sun, J., Todd, J. D., Thrash, J. C., Qian, Y., Qian, M. C., Temperton, B., et al. (2016). The abundant marine bacterium Pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol. Nat. Microbiol. 1:16065. doi: 10.1038/nmicrobiol.2016.65
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348, 1–10. doi: 10.1126/science.1261359
Sunda, W. G., Kieber, D., Kiene, R. P., and Huntsman, S. (2002). An antioxidant function of DMSP and DMS in marine algae oceanic dimethylsulfide (DMS) photolysis View project. Nature 418, 317–320. doi: 10.1038/nature00851
Suzuki, H., Kamatani, S., Kim, E. S., and Kumagai, H. (2001). Aminopeptidases A, B, and N and dipeptidase D are the four cysteinylglycinases of Escherichia coli K-12. J. Bacteriol. 183, 1489–1490. doi: 10.1128/JB.183.4.1489-1490.2001
Swan, B. K., Martinez-Garcia, M., Preston, C. M., Sczyrba, A., Woyke, T., Lamy, D., et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333, 1296–1300. doi: 10.1126/science.1203690
Swarr, G. J., Kading, T., Lamborg, C. H., Hammerschmidt, C. R., and Bowman, K. L. (2016). Dissolved low-molecular weight thiol concentrations from the U. S. GEOTRACES North Atlantic Ocean zonal transect. Deep Res. Part I 116, 77–87. doi: 10.1016/j.dsr.2016.06.003
Teeling, H., Fuchs, B. M., Bennke, C. M., Kru, K., Chafee, M., Kappelmann, L., et al. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 5:e11888. doi: 10.7554/eLife.11888
Tevatia, R., Allen, J., Rudrappa, D., White, D., Clemente, T. E., Cerutti, H., et al. (2015). The taurine biosynthetic pathway of microalgae. Algal Res. 9, 21–26. doi: 10.1016/j.algal.2015.02.012
Thume, K., Chen, L., Meyer, N., Kieber, D. J., and Pohnert, G. (2018). The metabolite dimethylsulfoxonium propionate extends the marine organosulfur cycle. Nature 563, 412–415. doi: 10.1038/s41586-018-0675-0
Todd, J. D., Curson, A. R. J., Dupont, C. L., Nicholson, P., and Johnston, A. W. B. (2009). The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some ascomycete fungi. Environ. Microbiol. 11, 1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x
Todd, J. D., Curson, A. R. J., Kirkwood, M., Sullivan, M. J., Green, R. T., and Johnston, A. W. B. (2011). DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine Roseobacters and in uncultured marine bacteria. Environ. Microbiol. 13, 427–438. doi: 10.1111/j.1462-2920.2010.02348.x
Todd, J. D., Curson, A. R. J., Nikolaidou-Katsaraidou, N., Brearley, C. A., Watmough, N. J., Chan, Y., et al. (2010). Molecular dissection of bacterial acrylate catabolism – unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ. Microbiol. 12, 327–343. doi: 10.1111/j.1462-2920.2009.02071.x
Tripp, H. J., Kitner, J. B., Schwalbach, M. S., Dacey, J. W. H., Wilhelm, L. J., and Giovannoni, S. J. (2008). SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–744. doi: 10.1038/nature06776
Vairavamurthy, A., Zhou, W., Eglinton, T., and Manowitz, B. (1994). Sulfonates: a novel class of organic sulfur compounds in marine sediments. Geochim. Cosmochim. Acta 58, 4681–4687. doi: 10.1016/0016-7037(94)90200-3
Valle, D. A., Martínez-garcía, S., Sañudo-wilhelmy, S. A., Kiene, R. P., and Karl, D. M. (2015). Methionine and dimethylsulfoniopropionate as sources of sulfur to the microbial community of the North Pacific subtropical gyre. Aquat. Microb. Ecol. 75, 103–116. doi: 10.3354/ame01750
Van der Ploeg, J. R., Weiss, M. A., Saller, E., Nashimoto, H., Saito, N., Kertesz, M. A., et al. (1996). Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 178, 5438–5446. doi: 10.1128/jb.178.18.5438-5446.1996
Van Mooy, B. A. S., Fredricks, H. F., Pedler, B. E., Dyhrman, S. T., Karl, D. M., Lomas, M. W., et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458, 69–72. doi: 10.1038/nature07659
Van Mooy, B. A. S., Rocap, G., Fredricks, H. F., Evans, C. T., and Devol, A. H. (2006). Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc. Natl. Acad. Sci. U.S.A. 103, 8607–8612. doi: 10.1073/pnas.0600540103
Vermeij, P., and Kertesz, M. A. (1999). Pathways of assimilative sulfur metabolism in Pseudomonas putida. J. Bacteriol. 181, 5833–5837. doi: 10.1128/jb.181.18.5833-5837.1999
Vila-Costa, M., Kiene, R. P., and Simó, R. (2008). Seasonal variability of the dynamics of dimethylated sulfur compounds in a coastal northwest Mediterranean site. Limnol. Oceanogr. 53, 198–211. doi: 10.4319/lo.2008.53.1.0198
Villanueva, L., Bale, N., Hopmans, E. C., Schouten, S., Damsté, J. S. S., and Box, P. O. (2014). Diversity and distribution of a key sulpholipid biosynthetic gene in marine microbial assemblages. Environ. Microbiol. 16, 774–787. doi: 10.1111/1462-2920.12202
Villar, E., Vannier, T., Vernette, C., Lescot, M., Cuenca, M., Bachelerie, P., et al. (2018). The Ocean Gene Atlas: exploring the biogeography of plankton genes online. Environ. Microbiol. 46, 289–295. doi: 10.1093/nar/gky376
Visscher, P. T., Gritzer, R. F., and Leadbetter, E. R. (1999). Low-molecular-weight sulfonates, a major substrate for sulfate reducers in marine microbial mats. Appl. Environ. Microbiol. 65, 3272–3278. doi: 10.1128/aem.65.8.3272-3278.1999
Walsh, M. J., Goodnow, S. D., Vezeau, G. E., Richter, L. V., and Ahner, B. A. (2015). Cysteine enhances bioavailability of copper to Marine Phytoplankton. Environ. Sci. Technol. 49, 12145–12152. doi: 10.1021/acs.est.5b02112
Wang, L., Wang, X., Wu, H., and Liu, R. (2014). Overview on biological activities and molecular characteristics of sulfated polysaccharides from marine green algaein recent years. Mar. Drugs 2, 4984–5020 doi: 10.3390/md12094984
Wang, R., Gallant, É., and Seyedsayamdost, M. R. (2016). Investigation of the genetics and biochemistry of roseobacticide production in the Roseobacter clade bacterium Phaeobacter inhibens. mBio 7:e02118. doi: 10.1128/mBio.02118-15
Wasmund, K., Mußmann, M., and Loy, A. (2017). The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 9, 323–344. doi: 10.1111/1758-2229.12538
Wheeler, P. R., Coldham, N. G., Keating, L., Gordon, S. V., Wooff, E. E., Parish, T., et al. (2005). Functional demonstration of reverse transsulfuration in the Mycobacterium tuberculosis complex reveals that methionine is the preferred sulfur source for pathogenic mycobacteria. J. Biol. Chem. 280, 8069–8078. doi: 10.1074/jbc.M412540200
Williams, B. T., Cowles, K., Bermejo Martínez, A., Curson, A. R. J., Zheng, Y., Liu, J., et al. (2019). Bacteria are important dimethylsulfoniopropionate producers in coastal sediments. Nat. Microbiol. 4, 1815–1825. doi: 10.1038/s41564-019-0527-1
Williams, B. T., and Todd, J. D. (2019). A day in the life of of marine sulfonates. Nat. Microbiol. 4, 1610–1611. doi: 10.1038/s41564-019-0576-5
Keywords: microbe, organic sulfur, gene, sulfur cycle, transformation
Citation: Tang K (2020) Chemical Diversity and Biochemical Transformation of Biogenic Organic Sulfur in the Ocean. Front. Mar. Sci. 7:68. doi: 10.3389/fmars.2020.00068
Received: 22 October 2019; Accepted: 29 January 2020;
Published: 20 February 2020.
Edited by:
Jose M. Gonzalez, University of La Laguna, SpainReviewed by:
Jean-Baptiste Raina, University of Technology Sydney, AustraliaMary Ann Moran, University of Georgia, United States
Copyright © 2020 Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Tang, dGFuZ2thaUB4bXUuZWR1LmNu
 Kai Tang
Kai Tang