- Coastal Ecology Section, Alfred Wegener Institute, Helmholtz Centre for Polar and Marine Research, Wadden Sea Station Sylt, Bremerhaven, Germany
Ocean environments are changing rapidly and marine organisms need to cope with these changes in order to survive, develop, and reproduce. To do so, organisms can either migrate, adapt in situ or acclimate via phenotypic plasticity. In this context, the emerging field of environmental epigenetics investigates the contribution of genetic and epigenetic information to adaptive potential of wild populations. Epigenetic modifications are based on the highly dynamic combination of DNA methylation, histone modifications, and non-coding RNAs, which may facilitate phenotypic plasticity through genotype-epigenotype-environment interactions, and can drive rapid evolution in wild populations. However, while knowledge of epigenetic contributions to phenotypes across different developmental and generational timescales is increasing for medical research model species, the mechanistic and synergistic action of these modifications remain comparatively understudied in ecological models such as teleost fishes. Here, we characterized the evolution of the gene toolkit involved in key molecular epigenetic pathways including DNA methylation, histone modifications, macroH2A histone, and miRNA biogenesis/turnover in threespine stickleback, a model species in evolution and ecology. We then investigated these genes within a phylogenetic context by comparing them in stickleback to human, mouse, chicken, tropical clawed frog, zebrafish, medaka, green spotted puffer, channel catfish, and mangrove rivulus. We found that, in general, conserved domains, in conjunction with their phylogenetic positions, suggest evolutionary conservation of putative enzyme activity in stickleback. However, molecular epigenetic pathways also revealed that teleost gene evolution is diversified and complex. Specifically, the number of genes, gene loss/duplication events, identified conserved domains, and putative protein lengths vary greatly from one species to another, particularly within fishes, which exhibit a potentially new class of histone deacetylases. This suggests different biological functions specific to fish species, and that the action of genes regulating epigenetic modifications in model species are not necessarily applicable to other related species. We integrate our results into recent advances concerning epigenetic mechanisms in teleosts, and conclude by discussing the necessity to delve deeper into the fundamental mechanics of epigenetic modifications in a wide array of taxa, particularly those relevant for assisted evolution, conservation, aquaculture, fisheries, and climate change-adaptation studies.
Introduction
Epigenetic modifications are based on the highly dynamic combination of DNA methylation, histone modifications, and non-coding RNAs that control transcription (Best et al., 2018; Eirin-lopez and Putnam, 2019). They can be broadly summarized as potentially heritable molecular mechanisms that change gene expression without necessarily being associated with modifications of the DNA sequence (Fincham, 1997). Importantly, they are known to facilitate phenotypic plasticity through genotype-epigenotype-environment interactions (Cosseau et al., 2017). In the context of anthropogenic climate change, understanding the specific contributions of genetic and epigenetic information to adaptive potential of populations and the underlying mechanisms are crucial to assess species’ evolution in fast-changing and often unstable environments, in particular when applied to ecological and aquaculture challenges (Gavery and Roberts, 2017; Eirin-lopez and Putnam, 2019). For instance, recent studies in environmental epigenetics applied to marine species (e.g., bivalves, fish) have brought the concept of assisted evolution via epigenetically mediated adaptation to light for aquaculture and conservation applications. The possibility to generate environment-specific phenotypes mediated through heritable epigenetic-based events has been suggested in several species and for a number of traits (e.g., sex ratio, growth, immunity) relevant for the aquaculture industry, even if the precise mechanisms underlying these are poorly described (Gavery and Roberts, 2017; Le Luyer et al., 2017; Panserat et al., 2017; Balasubramanian et al., 2019; Gavery et al., 2019).
Although fish are widely used as model organisms in aquaculture, medicine, ecology, evolution, and ecotoxicology (Cossins and Crawford, 2005), most of our knowledge of their epigenetic mechanisms is mainly focused on laboratory-cultured zebrafish (Horsfield, 2019). However, studies of enzyme evolution (Best et al., 2018) and epigenetic regulation of fish development (Potok et al., 2013; Fellous et al., 2018; Wang and Bhandari, 2019) support the idea of species-specific mechanisms. For instance, an increasing number of studies show an influence of epigenetic modifications in developmental plasticity of trout and salmon, which have implications for species-specific conservation through hatchery rearing programs (Le Luyer et al., 2017; Gavery et al., 2019). Despite the increasing availability of teleost genomic resources, the comparative biology of teleost epigenetic mechanisms remains limited, particularly for non-model species and wild populations (Metzger and Schulte, 2016; Firmino et al., 2017; Labbé et al., 2017; Best et al., 2018; Fellous et al., 2018, 2019a,b; Todd et al., 2019). Hence, our ability to assess the role of epigenetic modifications in adaptive potential and population persistence of ecologically relevant species is still in its infancy.
An ecologically important fish species of interest for epigenetic studies in wild populations is threespine stickleback (Gasterosteus aculeatus), as it has been widely used as a model system to investigate the genetic and non-genetic basis of adaptation to novels environments (Shama and Wegner, 2014; Shama et al., 2016; Shama, 2017; Heckwolf et al., 2018; Metzger and Schulte, 2018; Kitano et al., 2019). After the last glaciation, marine stickleback colonized freshwater habitats of the north temperate zone, leading to local adaptation of populations that exhibit different phenotypes based on, for example, behavior, armor plate number, body shape, and gene expression plasticity (Kitano et al., 2019). Furthermore, whole genome sequence comparisons between freshwater and marine individuals revealed that differential gene expression contributed more to adaptive evolution than protein sequence evolution (Kitano et al., 2019). This, in combination with the ability to study the evolution of sex chromosome systems make stickleback an ideal candidate to elucidate the role of epigenetic mechanisms in reproduction, development, and adaptation in fishes (Figure 1). Nevertheless, our knowledge about the importance of epigenetic modifications, their precise mechanistic basis, and the number, conservation, and evolution of enzymes implicated in the regulation of these modifications remains scarce. Several recent studies have indeed suggested that phenotypic divergence observed between marine and freshwater populations (Teigen et al., 2015; Artemov et al., 2017; Metzger and Schulte, 2018; Smith et al., 2018), and transgenerational plasticity in response to environmental change (Shama and Wegner, 2014; Shama et al., 2016; Heckwolf et al., 2018) together with local adaptation of populations (Heckwolf et al., 2019) might be due to underlying differences in DNA methylation patterns, but have not yet explicitly demonstrated how epigenetic modifications are regulated in the species.
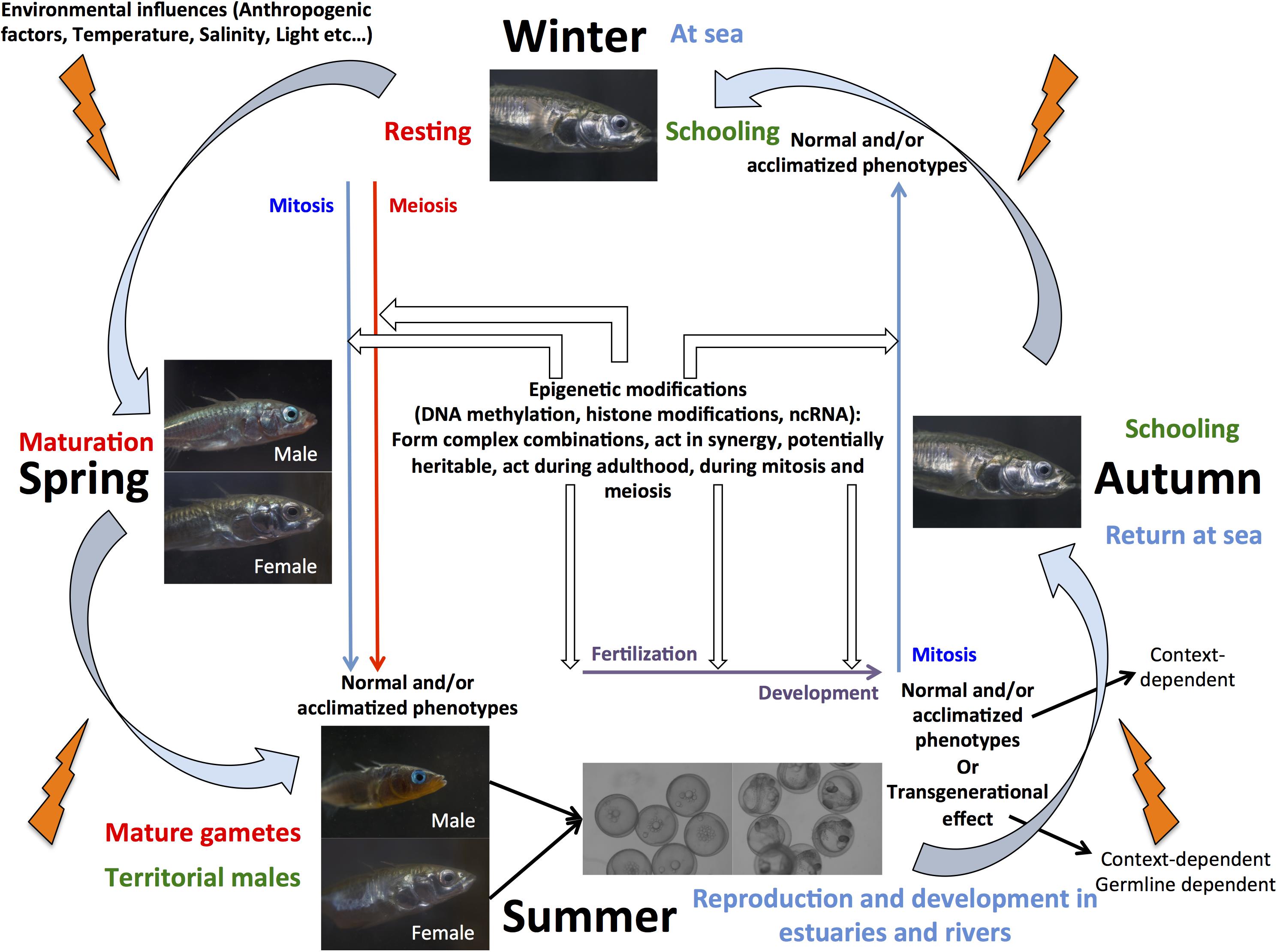
Figure 1. Schmatic view of marine stickleback life cycle illustrating the potential for this species as a model to elucidate the synergistic role of epigenetic mechanisms in aquaculture and eco-evolutionary studies.
One of the key challenges identified for our understanding of marine environmental epigenetics is to generate detailed knowledge of epigenetic mechanisms and understanding of synergistic action among epigenetic actors (Eirin-lopez and Putnam, 2019). This information will help to determine the relative weight of these modifications in adaptation, to understand how epigenetic landscapes are generated according to different environmental conditions, as well as if and how they can be transmitted across multiple generations. Indeed, the role of epigenetic modifications in the organism-environment interface or in response to anthropogenic or natural stressors, their mechanistic basis, and the evolution of genes underlying key pathways have been extensively explored in plant and medical model organisms (Baulcombe and Dean, 2014; Yang, 2015; Wang and Köhler, 2017; Shanmugam et al., 2018). These studies were crucial to advances in cancer diagnostics and prognostics (Costa-pinheiro and Montezuma, 2015), to understanding of plant production and plant responses to the environment (Baulcombe and Dean, 2014; Wang and Köhler, 2017), and to understanding of the mechanistic basis of transgenerational epigenetic inheritance in mammals (Van Otterdijk and Michels, 2016). Importantly, recent studies of the DNA methyltransferase family have also revealed that changes in gene copy numbers and molecular interactions can modulate enzyme activity and their role in transcriptional regulation (Lyko, 2018), and that a vast diversity of epigenetic and transcriptional patterns during development are observed among species (Riviere et al., 2013; Eckersley-maslin et al., 2018; Fellous et al., 2018, 2019c; Horsfield, 2019; Vastenhouw et al., 2019). This diversity, coupled with a variable number of genes and duplication events among species reflects the importance of precisely elucidating species-specific epigenetic toolkits, how the different epigenetic pathways behind are regulated, and how they vary in their biological functions. However, this key knowledge is mostly lacking in fishes (including threespine stickleback; Best et al., 2018; Metzger and Schulte, 2018; Fellous et al., 2019a, b), and marine organisms in general (Gavery and Roberts, 2017).
Here, we performed a genomic survey of genes coding for putative enzymes implicated in key epigenetic pathways: DNA methylation, DNA hydroxymethylation, histone methylation, acetylation, phosphorylation, ubiquitination, sumoylation, poly-ADP ribosylation, and glycosylation, as well as genes coding for putative macroH2A, and genes implicated in the biogenesis/turnover of miRNAs in threespine stickleback. We used in silico analyses to identify the epigenetic toolkit in stickleback and characterize the putative proteins molecularly. We then performed phylogenetic analyses of genes potentially implicated in the different known epigenetic pathways by comparing them in stickleback to human, mouse, chicken, tropical clawed frog, zebrafish, medaka, green spotted puffer, channel catfish, and mangrove rivulus. In doing so, we were able to precisely compare the different species’ epigenetic toolkits and any duplication/loss events that may have occurred. Finally, based on our results, we discuss the biological role of these putative epigenetic genes in animals, and particularly in other fish species, to provide a fundamental basis for further studies of epigenetic mechanisms potentially underlying adaptive responses of non-model and wild fish populations.
Materials and Methods
Identification of Putative Epigenetic Genes Orthologs
Genomic resources of threespine stickleback were screened on the Ensembl genome server1. Homology-producing sequences were identified based on the HUGO Gene Nomenclature Committee. Protein conserved domains were identified using Blast2 and SMART software3. All sequences and Ensembl accession numbers are provided in Supplementary Data Folder 1.
Phylogenetic Analyses
Sequences encoding the different putative proteins from human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), western clawed frog (Xenopus tropicalis), zebrafish (Danio rerio), medaka (Oryzias latipes), mangrove rivulus (Kryptolebias marmoratus), green spotted puffer (Tetraodon nigroviridis), and channel catfish (Ictalurus punctatus) were obtained from the Ensembl genome server4, and were aligned with the Muscle algorithm (Edgar, 2004). Phylogenetic analyses were performed using a Neighbor-joining method (Bootstrap method: 500 bootstraps, complete or partial deletion of gaps). Results were compared using Minimum Evolution and Maximum Likelihood methods (Bootstrap method: 500 bootstraps, complete or partial deletion of gaps). All analyses were conducted with MEGA software version 7 (Kumar et al., 2016). Sequences used in the analyses and resulting phylogenetic trees are provided in the Supplementary Data Folder 2.
Results
Putative Stickleback Enzymes Implicated in DNA Methylation/Hydroxymethylation
Our approach led to the characterization of 14 cDNAs coding for 6 DNA-methyltransferase (DNMT), 5 Methyl DNA-Binding (MDB), and 3 Ten-Eleven Translocated enzyme (TET) putative proteins in stickleback (Table 1). While the number of DNMT and TET proteins was fairly constant, the number of MBDs was more variable among the species examined (Table 2). DNMT sequences contained a DCM domain that potentially confers methyltransferase activity (Supplementary Figure 1A). In addition, they exhibited additional conserved domains such as DNMT1-RFD or PWWP, which recognize and bind the correct residue (Supplementary Figure 1A). Phylogenetic analyses (Supplementary Figure 1B) revealed that DNMTs cluster into four groups: DNMT1 (maintenance), DNMT3A-DNMT3Bb (de novo), DNMT3Ba (de novo), and DNMT3l (imprinting). Interestingly, DNMT3A was duplicated once in stickleback, while in zebrafish and medaka, DNMT3A experienced more than one duplication. DNMT3Ab and DNMT3Bb appear to be more closely related to other vertebrate orthologs, whereas DNMT3Aa and DNMT3Ba formed two teleost-specific groups (Supplementary Figure 1B). Finally, DNMT3Bb seems to be more closely related to DNMT3A than DNMT3Ba, and no DNMT3l putative proteins were found outside of mammals.
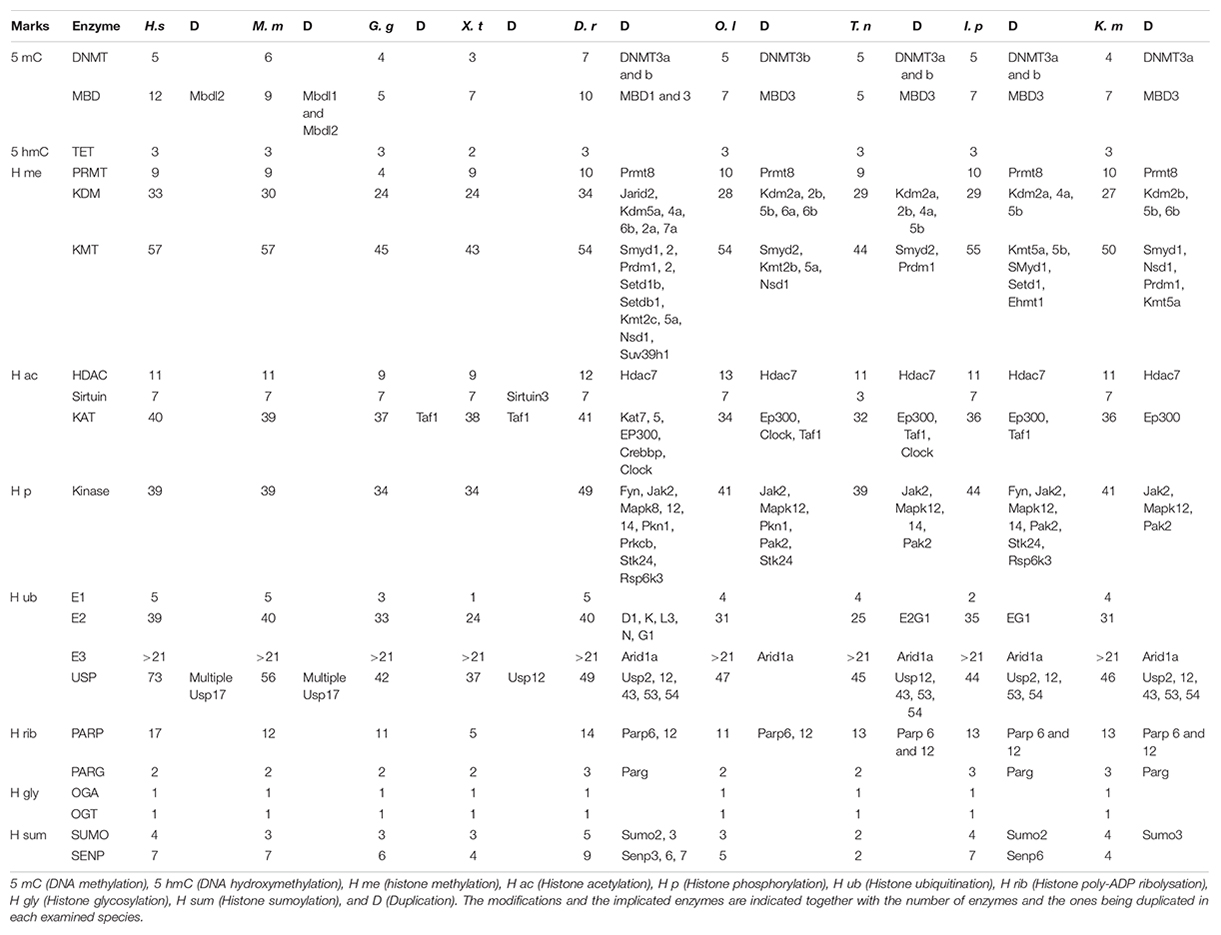
Table 2. Epigenetic toolkit of human (Homo sapiens, H. s), mouse (Mus musculus, M. m), chicken (Gallus gallus, G. g), tropical clawed frog (Xenopus tropicalis, X. p), zebrafish (Danio rerio, D. r), medaka (Oryzias latipes, O. l), green spotted puffer (Tetraodon nigroviridis, T, n), channel catfish (Ictalurus punctatus, I. p), and mangrove rivulus (Kryptolebias marmoratus, K. m).
The identified Methyl CpG-binding (MBD) proteins (except Mec2P without conserved domain) contained the MBD domain that binds methylated DNA, while MBD2 and MBD3 had an MBD-C domain known to interact with the NuRD/Mi2 deacetylase complex (Supplementary Figure 1A). Stickleback MBD seems to be conserved in vertebrate evolution, whereas the gene coding for the MBD3 protein was duplicated, as also found for the other fish species examined (Supplementary Figure 1C). Interestingly, stickleback MDB3a and b are closely related, whereas they form two separate groups in the other fish species examined. The three TET enzymes contained the TET_JBP domain responsible for conversion of 5 methyl-cytosine (5-mC) to 5 hydroxymethyl-cytosine (5 hmC), or further oxidation to 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Supplementary Figure 1A). Phylogenetic analyses showed that TETs seem to be highly conserved in vertebrate evolution, and no duplication events occurred in stickleback (or the other fish species examined) (Supplementary Figure 1D).
Putative Stickleback Enzymes Implicated in Histone Methylation
In silico analyses revealed 10 N-arginine methyltransferase (PRMT) family members in stickleback (Table 1), similar to zebrafish, medaka, channel catfish, and mangrove rivulus (Table 2). The PRMTs (1-2-3-4-6-8-8b-9) exhibited an AdoMet-MTase conserved domain putatively responsible for methyltransferase activity, while PRMT7 did not contain a conserved domain (Supplementary Figure 2A). In contrast to the other PRMTs, PRMT5 had gene-specific domains (PRMT5-TIM, PRMT5, PRMT5-C), which are also responsible for methyltransferase activity (Supplementary Figures 2A,B). PRMT4 displayed an additional methyltransferase domain, Carm1, and PRMT2 beared a binding domain, SH3 (Supplementary Figure 2A). Stickleback PRMT phylogeny seems to demonstrate that the different classes of enzymes are conserved among vertebrates, with PRMT8 being duplicated in fish (except in the green spotted puffer; Supplementary Figure 2C).
Furthermore, 54 cDNAs coding for putative Histone Lysine Methyltransferase (KMTs) were found in stickleback (Table 1). While the same number of KMTs was found between human and mouse, it was more variable in the other species examined (Table 2). Except for Eef1akmt (Eef1a Lysine methyltransferase) and Dot1 [Histone (H) 3 Lysine (K) 79 N-methyltransferase specific], they all contained the conserved domain SET (Su(var)3–9, Enhancer-of-zeste, Trithorax) responsible for methyltransferase activity, but also additional domains such as SANT (Sswi3, Ada2, N-Cor, TFIIIB), or Chromo (CHRomatin Organization Modifier), potentially responsible for chromatin remodeling protein interactions with histones, and chromatin targeting, respectively (Supplementary Figures 3A,D). Dot1l contained a Dot1 domain, which is also responsible for methyltransferase activity (Supplementary Figure 3A). As seen in Tables 1, 2 and Supplementary Figures 3B,C KMT subfamilies seem to be conserved in vertebrates, but duplication events differ among the fish species examined. For example, Smyd1 (SET and MYND Domain Containing 1) was duplicated in stickleback, zebrafish, mangrove rivulus and channel catfish, but not in medaka and green spotted puffer (Supplementary Figure 3B).
In addition, 32 Histone Lysine Demethylases (KDM) appear to be present in the stickleback genome (Table 1). Except for the Kdm1 and Kdm5 enzymes, they all contained the Jumonji-C conserved domain (Jmj-C) (Supplementary Figure 4A), putatively responsible for demethylase activity (Supplementary Figure 4B). KDMs also had other conserved domains such as JmjN (JumonjiN) or ARID (AT-Rich Interaction Domain), implicated as a co-unit with JmjC and in DNA binding, respectively (Supplementary Figure 4A). The different subfamilies of KDMs seem to be conserved in stickleback (Supplementary Figure 4C). Interestingly, six KDMs (Kdm1, Kdm2a, Kdm2b, Kdm4a, Kdm5b, Kdm6a) were duplicated in stickleback (Supplementary Figure 4C), whereas more variability in terms of duplication events was observed for the other fish species examined (Table 1 and Supplementary Figure 4C). Finally, Kdm3a appears to be absent in stickleback and in fish in general (Supplementary Figure 4C).
Putative Stickleback Enzymes Implicated in Histone Acetylation
Our genome survey of stickleback revealed 10 genes coding for Histone Deacetylase enzymes (HDAC) and 6 coding for SIlent mating-information Regulation (SIR2)/Sirtuin (Table 1). The number of HDAC appears to be lower in stickleback than in the other fish species examined (Tables 1, 2), whereas the number of sirtuins was constant among the examined species (except green spotted puffer) (Table 2). Each HDAC member had an Arginase_Hdac superfamily conserved domain, and each SIR2/Sirtuin member exhibited the SIR2 superfamily conserved domain (Supplementary Figure 5A), which are both putatively responsible for deacetylase activity (Supplementary Figure 5B). Furthermore, Hdac4, Hdac5, and Hdac9b contained a Hdac_Gln (Glutamine Rich N Terminal of Histone Deacetylase) domain that is thought to deacetylate non-histone proteins (Supplementary Figure 5A). Phylogenetic analyses of stickleback HDAC/Sirtuin demonstrated that the different classes and subfamilies are conserved in vertebrates and among fish species, with Hdac12 potentially specific to teleosts (Supplementary Figures 5C,D). These analyses also showed that Hdac2, Hdac6 (Supplementary Figure 5C), and Sirt2 (Supplementary Figure 5D) are not present in stickleback, with Hdac2 also absent in zebrafish and mangrove rivulus (Supplementary Figure 5C). Interestingly, while Hdac7 was duplicated in medaka and zebrafish, stickleback exhibited only Hdac7b (Supplementary Figure 5C).
Forty putative Histone Acetyltransferases (KATs/HATs) were found in the stickleback genome (Table 1), as also found in the human genome (Table 2). However, specific loss/duplication events occurred in stickleback (Table 1) and others fishes (Table 2), resulting in a difference between fish and mammals, but also high variability among the teleost species examined. Based on HUGO nomenclature, all KATs/HATs exhibited conserved domains (Supplementary Figure 6A) that conferred putative activity and specificity to each member (Supplementary Figure 6B). For example, Kat1 (Hat1) contained an acetyltransferase domain, and the Kat2 enzymes displayed the N-acetyltransferase domain (Nat) (Supplementary Figure 6A). Phylogenetic reconstruction of KATs showed that the different subfamilies are conserved in stickleback, with EP300 and CREBBP being duplicated in teleost species (Tables 1, 2 and Supplementary Figure 6C). However, Clockb was not duplicated in stickleback, whereas this was the case in zebrafish, medaka, and green spotted puffer (Tables 1, 2 and Supplementary Figure 6C).
Putative Stickleback Enzymes Implicated in Histone Phosphorylation
Forty sequences coding for putative kinases implicated in histone phosphorylation were characterized in stickleback (Table 1), and a variable number of genes and duplication events were observed among species (Table 2). Except for Baz1b (Bromodomain adjacent to zinc finger domain protein 1B) and Bub1b (“budding uninhibited by benzimidazoles 1” mitotic checkpoint serine/threonine kinase B), they all exhibited the pKC (Protein Kinase C) domain (Supplementary Figure 7A) responsible for serine/threonine phosphorylation (Supplementary Figure 7B). Some sequences also contained other conserved domains such as B41 (Band 4.1 homologs) on Jak2 (Janus tyrosine kinases 2), known as a plasma membrane-binding domain, or BROMO (Bromodomain) on Baz1B that recognizes acetylated residues (Supplementary Figure 7A). While the main subfamilies of kinases seem to be conserved in vertebrates, some enzymes such as Jak2 appear to be duplicated in the fish species examined (Table 2 and Supplementary Figure 7C). However, there were exceptions, such as Mapk8b (Mitogen-Activated Protein Kinase 8 B), which was only duplicated in zebrafish (Table 2 and Supplementary Figure 7C).
Putative Stickleback Enzymes Implicated in Histone Ubiquitination
In silico analyses revealed genes coding for (at least) 21 ligases E3, 27 ligases E2, and 3 SWI/SNF (Switch/Sucrose Non-Fermentable) E3 proteins (Table 1). Four Ubiquitin activating enzymes whose Ube1 containing the conserved catalytic domain Ube1 (Supplementary Figure 8A) were detected in stickleback (Table 1). All examined stickleback E2s possessed the UbC (Ubiquitin C) domain, putatively active in ubiquitination under stressful conditions (Supplementary Figure 8A). While the three ARID (AT-Rich Interactive domain containing) proteins contained the ARID domain implicated in DNA-Protein and/or Protein–Protein interactions, only ARID1aa and ARID1ab had the BAF250 domain responsible of H2B ubiquitination (Supplementary Figures 8A,C). The ligases E3 contained a large variety of conserved domains such as the HECTc (Homologous to the E6-AP Carboxyl Terminus c) on E3A (Enzyme Ligase 3A) or Huwe1 (HECT, UBA (Ubiquitin-Associated domain) and WWE domain-containing protein 1) (Supplementary Figure 8A), reflecting a diversity of substrates and functions (Supplementary Figure 8B). The different ligase subfamilies appear to be conserved in vertebrates (Supplementary Figures 8C–E), while, particularly for the E2 ligases, the number of genes and duplication events varied among the vertebrates examined (Table 2).
Among the 51 enzymes potentially capable of histone deubiquitination (Table 1), 48 were identified as Usp (Ubiquitin carboxyl-terminal hydrolase) (Supplementary Figure 9). Thus, all Usp proteins had the peptidase_C19 domain (Supplementary Figure 9A), conferring deubiquitination activity against histone and non-histone proteins (Supplementary Figure 9B), and many (e.g., Usp25), had additional conserved domains like UBA that bind ubiquitin (Supplementary Figure 9A). Finally, the peptidase_C12 domain on BAP1 (BRCA1: Breast cancer type 1 susceptibility protein associated protein 1), MPN (Mpr1, Pad1 N-terminal) on BRCC36 [Lysine-63-specific deubiquitinase BRCC36 (BRCA1/BRCA2-containing complex subunit 36)], and MYSM1 [Histone H2A deubiquitinase MYSM1 (Myb-like, SWIRM, and MPN domain-containing protein 1)] were also found, and are implicated in deubiquitination (Supplementary Figure 9A). The main subfamily classification seems to be conserved in vertebrates (Supplementary Figures 9C–E), despite high variability among species in the number of genes and duplication/loss events (Table 2).
Putative Stickleback Enzymes Implicated in Histone Poly-ADP Ribosylation
Eleven Poly (ADP-ribose) polymerases (Parp) were found in stickleback (Table 1), whereas 17 have been characterized in humans (Table 2). Specific loss/duplication events were also observed for the other fish species examined (Table 2). Except for Parp9 and Parp14, they all contained the conserved domain Parp responsible for Poly (ADP-ribose) polymerization often implicated in DNA repair (Supplementary Figure 10A). Parp9 and Parp14 exhibited a Macro domain that putatively binds ADP-ribose (Supplementary Figure 10A). Other conserved domains were also detected, such as the WGR (named after its most conserved motif) domain on Parp1-2-3 (Supplementary Figure 10A), which may be a nucleic acid domain. Well-conserved among vertebrates, the different Parp subfamilies were also found in our phylogenetic analyses (Supplementary Figure 10B). Interestingly, while most of the discovered Parps were found only once in each species, Parp6 and Parp12 seem to be duplicated in fishes but not in stickleback (Tables 1, 2 and Supplementary Figure 10B). In addition to the Parps, two Poly (ADP-ribose) glycohydrolases [Parga and Adprhl2 (ADP-ribose glycohydrolase ARH3 (ADP-Ribohydrolase 3)] were found in stickleback (Table 1). They both exhibited a conserved domain responsible for poly (ADP-ribose) glycohydrolase (Supplementary Figure 10A). Also conserved in vertebrates, Adprhl2 does not seem to be duplicated in fish, whereas Parga may have been duplicated in zebrafish and mangrove rivulus (Table 2 and Supplementary Figure 10C).
Putative Stickleback Enzymes Implicated in Histone Glycosylation
One glycosidase (Oga; nuclear cytoplasmic O-GlcNAcase and acetyltransferase) and one glycosyltransferase (Ogt; UDP -N-acetylglucosaminepeptide N-acetylglucosaminyltransferase 110 kDa subunit) were found in stickleback (Table 1). A catalytic domain was identified for each of the putative proteins (Supplementary Figure 11). While the stickleback enzymes seem to be conserved among vertebrates, there were no apparent duplications in the other fish species examined (Table 2 and Supplementary Figure 11).
Putative Stickleback Enzymes Implicated in Histone Sumoylation
The SUMO activating enzyme E1 [SAE1 (SUMO1 activating enzyme subunit 1)], conjugating enzyme E2 (UBC9), and two SUMO ligases E3 (NSE2 and EGR2) were identified (Supplementary Data Folder 1). Three Small Ubiquitin-like MOdifers (SUMO) were also found in the stickleback genome (Table 1). This number appears to be variable in teleosts (Table 2), whereas four SUMOs are known in humans (Table 2). The SUMO proteins all exhibited a conserved domain (Supplementary Figure 12A), and the different subfamilies appear to be conserved in vertebrates (Supplementary Figure 12B). While Sumo2 and/or Sumo3 were duplicated in zebrafish, channel catfish, and mangrove rivulus (Table 2), there were no apparent duplications in stickleback (Supplementary Figure 12B). Furthermore, five SUMO/Sentrin specific Peptidases (Protease) (SENP), corresponding to each vertebrate subfamilies, were detected in stickleback (Table 1), in comparison to seven in mammals and a variable number in the other species examined (Table 2). They all exhibited a catalytic domain responsible for peptidase activity (Supplementary Figure 12C). Interestingly, outside of human and mouse, this number was also variable among the species examined (Table 2). SENP6 and SENP7 were duplicated in zebrafish, whereas they were not duplicated in stickleback (Supplementary Figure 12D). Finally, while all of the SENPs clustered with their vertebrate homologs, stickleback SENP6 was found to be more closely related to zebrafish SENP7a (Supplementary Figure 12D).
Stickleback H2A
Our in silico analyses identified two genes coding for putative H2A proteins [Supplementary Figure 13(1)]. Interestingly, while these genes seem to be duplicated in vertebrates for both H2As, this was apparently not the case for stickleback, channel catfish, or green spotted puffer [Supplementary Figure 13(2)].
Detection of Stickleback Genes Coding for Proteins Implicated in miRNA Biogenesis/Turnover
Finally, we looked for different putative proteins implicated in miRNA biogenesis and turnover. While all of the genes coding for proteins involved in miRNA biogenesis (Supplementary Figure 14A) and turnover (Supplementary Figure 14B) pathways were identified in stickleback, no corresponding sequence for Tut4 (Terminal uridylyltransferase 4) could be retrieved. For each of these genes, no duplication events were detected.
Discussion
We conducted a comprehensive characterization of putative proteins implicated in the large variety of epigenetic pathways in threespine stickleback in order to provide a knowledge base for future studies. Despite stickleback being an excellent candidate fish species for evolutionary and environmental epigenetic studies (Shama et al., 2016; Metzger and Schulte, 2017, 2018; Heckwolf et al., 2018, 2019; Smith et al., 2018), mechanisms underlying different DNA methylation patterns and histone modification profiles, how they are modulated, and how they interact with non-coding RNAs have not yet been described. Here, we showed that stickleback possess a diversified epigenetic toolkit which shares similarities with other fishes and vertebrates, but also differs in the number of genes, loss/duplication events, and putative protein structures, indicating that even if epigenetic marks seem to be conserved across taxa, they might be differentially regulated among species and/or have potentially unknown functions. Our study underlines the importance of considering species-specific gene regulation pathways, and that patterns observed in model species may not always be applicable to other related species. Below, we discuss the biological role of these epigenetic actors and highlight the potential utility of some in assisted evolution, conservation, aquaculture, and hypothesis testing of species adaptive responses to rapid climate change.
DNA Methylation/Hydroxymethylation
DNA methylation is currently the most studied mechanism in environmental epigenetics. In vertebrates, DNA methylation mainly refers to the transfer of a methyl group to position 5 of cytosine residues to form 5-methylcytosine (5 mC) in a CpG dinucleotide context (Feng et al., 2010). A methyltransferase family, the DNMTs, catalyzes this reaction and is composed of three conserved proteins (DNMT1, DNMT3A, DNMT3B) (Okano et al., 1999; Campos et al., 2012). DNMT1 is called a maintenance enzyme, while DNMT3A and DNMT3B are de novo methyltransferases (Li et al., 1992; Okano et al., 1999). DNA methylation also influences chromatin compaction and the associated gene expression via interactions through MBD proteins (six members in vertebrates) (Ballestar and Wolffe, 2001; Lindeman et al., 2010). In addition, DNA 5-hydroxymethylation (5 hmC), a possible stage of DNA demethylation, was recently characterized in vertebrates (Zhao and Chen, 2013), and is catalyzed by the TET enzymes (TET1, TET2, TET3) (Santiago et al., 2014). Nevertheless, the precise relationship between 5 mC and 5 hmC has not yet been established (Kamstra et al., 2015).
Our phylogenetic analyses and the presence of a catalytic domain and additional motifs suggest species-specific differences in genes underlying DNA methylation/hydroxymethylation. As also shown in zebrafish (Campos et al., 2012), bluehead wrasses (Todd et al., 2019), and mangrove rivulus (Fellous et al., 2018), DNMT3A and DNMT3B show duplication events, and DNMT3l is also absent in stickleback. This calls into question the generality of evolutionary conservation of de novo methylation and genomic imprinting genes in fishes. Furthermore, the number of MBDs in stickleback suggests a loss of some genes and a duplication of MBD3 due to 3R genome duplication (Best et al., 2018; Balasch and Tort, 2019). In addition, the absence of identified conserved domains on MecP2 questions its functionality (Fatemi and Wade, 2006) in stickleback, since it does exhibit a conserved domain in mangrove rivulus (Fellous et al., 2018). MDBs are still not well known in fishes, and so far, only MeCP2 has been studied in zebrafish (Gao et al., 2015; Nozawa et al., 2017) and mangrove rivulus (Fellous et al., 2018). Furthermore, while the TET family appears to be well conserved in vertebrates, 5 mC/5 hmC patterns during embryonic development and their inheritance are still controversial among fishes (Potok et al., 2013; Fellous et al., 2018; Wang and Bhandari, 2019), illustrating the need to delve deeper into the mechanistic basis of 5 mC/5 hmC regulation.
The biological function of DNMT enzymes appears much more complex than “traditional” maintenance and de novo DNA methylation (Lyko, 2018). For example, a recent study supports the role of DNA methylation during reprogramming in the regulation of transposable elements in whitefish that contributes to the survival of nascent species (Laporte et al., 2019). Also, together with an implication of 5 mC in sex ratio determination (Ellison et al., 2015) and environmental sex changes (Todd et al., 2019), a role for DNA methylation in intra- and inter-generational acclimation through de novo methylation has been suggested in fish (Fellous et al., 2018) and corals (Putnam and Gates, 2015; Putnam et al., 2016). In stickleback, one study supports this function for de novo methylation, but also underlies the possibility of “deleterious” evolution (Mcghee and Bell, 2014), even if the precise regulation of 5 mC remains unknown (Teigen et al., 2015; Artemov et al., 2017; Metzger and Schulte, 2018; Smith et al., 2018; Heckwolf et al., 2019). Thus, future studies would benefit from a more precise understanding of potential interactions between DNMT, MBD, and TET proteins. This, together with investigations of the importance of 5 mC/5 hmC will allow for better mechanistic understanding of the functional significance of the observed DNA methylation patterns in stickleback adaptation and evolution (Metzger and Schulte, 2018; Smith et al., 2018; Heckwolf et al., 2019).
Histone Methylation
Histone methylation is a key element of the “epigenetic code”, and regulation of this modification plays important roles in transcription, cell cycles, and genome integrity (Black et al., 2013). Histone proteins can be methylated on arginine (R) (Zhang et al., 2019) or on lysine (K) residues, with lysine potentially mono- (me1), di- (me2), or tri- (me3) methylated (Martin and Zhang, 2005). Arginines are methylated by PRMT enzymes that comprise a group of nine members classified into three types [I (PRMT1-2-3-4-6-8), II (PRMT5-9), III (PRMT7)] with different specificities in vertebrates (Zhang et al., 2019). Lysine methyltransferases (KMTs), specific to different residues, are categorized into two families. The first family contains the SET domain-containing enzymes and is the largest with at least forty members, while the second family has only one member (Dot1l) and is characterized by the lack of a SET domain (Van Leeuwen et al., 2002; Dupret et al., 2017). Both arginine and lysine seem to be demethylated by a major class of enzymes (>100 members), the Jumonji C domain, containing histone demethylases (JHDMs) or Lysine demethylases (KDMs) (Shi et al., 2011) which exhibit a large diversity of substrate and biochemical activities related to the number of methyl groups carried on target residues (Zhang et al., 2019).
Specific knowledge related to PRMT enzymes in fish biology is focused mainly on zebrafish (Lei et al., 2012), and to a lesser extent on channel catfish (Yeh and Klesius, 2012), and our results support the idea that some PRMTs are restricted among metazoans (Wang and Li, 2012). Interestingly, PRMT8, which is duplicated in stickleback, plays a critical role during zebrafish embryonic and neural development (Wang and Li, 2012), and is the only enzyme with tissue-restricted expression. Whereas they are less known outside of other non-model species (Best et al., 2018), the KMTs seem to be conserved in stickleback, as well as in mangrove rivulus (Fellous et al., 2019b), insects (Jiang et al., 2017), and humans (Petrossian and Clarke, 2011), although species-specific loss/duplication events have occurred. The number of KDMs in stickleback differs slightly from other fish species (Qian et al., 2015; Fellous et al., 2019b). However, despite being well documented in taxa ranging from fruit flies to mammals and crucial in development and adaptation of a number of organisms, as well as being temperature sensitive (Takeuchi et al., 1999; Sasai et al., 2007; Fellous et al., 2014, 2015; Zhang et al., 2019), these fundamental proteins remain understudied in fishes, with the exception of zebrafish (Stewart et al., 2009; Akerberg et al., 2017). Furthermore, the dynamics of histone modifications are particularly important to study in the context of epigenetic inheritance and chromatin configurations in gametes of fishes, but remain largely under studied (Labbé et al., 2017; Best et al., 2018).
Histone Acetylation
Histone acetylation is one of the best studied epigenetic modifications, and plays important roles in the control of chromatin assembly, cell reprogramming, DNA repair, and gene expression (Shin et al., 2018). Histone acetyltransferases (HATs or recently KATs) add acetyl groups to lysine residues and have been classified into two types, A (main chromatin regulator) and B. Type A uses nucleosomal histones only, while type B uses non-nucleosomal histone associated with a partial cytoplasmic localization, and includes five distinct multi-gene families: Myst (Myst1-2-3-4), GNAT (GCN5-related N-acetyltransferase), Basal transcription/nuclear receptor cofactors, P300-CBP (CREB-binding protein) and Camello (Karmodiya et al., 2014; Sheikh and Akhtar, 2018). While many of these enzymes are well characterized in mammals, some new members are not yet established as real KATs (Sheikh and Akhtar, 2018). On the other hand, histone deacetylases are classified into two families: the “classic” HDAC which is divided into three groups [Class I (HDAC1-2-3-8), Class II (HDAC4-5-6-7-9-10), Class IV (HDAC11)], and the Sirtuins (Class III) (Greiss and Gartner, 2013; Seto and Yoshida, 2014).
The different KAT subfamilies and HDAC/Sirtuins classes are present in stickleback, but the number of genes and loss/duplication events differ among fish species. Interestingly, while sirtuin 1 is mentioned in stickleback by Teigen et al. (2015), no corresponding sequence was detected during our in silico analyses. Furthermore, the putative protein structure of HDAC9 questions its specificity toward histones (Petrie et al., 2003), since the arginase-HDAC domain was not found in stickleback, and different biochemical activities were observed in human and western-clawed frog (Petrie et al., 2003). Our analyses also revealed a possible new class of HDAC, HDAC12, which may be specific to teleosts. However, a complete molecular characterization is needed to confirm this. Nevertheless, the presence of a catalytic domain and their protein architecture suggest that these proteins are active, potentially filling a gap of knowledge regarding histone acetylation in fishes (Best et al., 2018). Whereas most studies are primarily focused on zebrafish (He et al., 2014; Román et al., 2018), KAT and HDAC/Sirtuin mRNA expression was recently described in mangrove rivulus during gonad-embryogenesis (Fellous et al., 2019a), and in trout, where histones undergo acetylation during spermatogenesis (Best et al., 2018). In the context of climate-driven range shifts, for species considered to be climate migrants (including stickleback), the precise mechanistic basis of cold acclimation through sirtuin(s) expression (Teigen et al., 2015) should be elucidated, since histone deacetylase activity is known to mediate thermal plasticity in zebrafish (Seebacher and Simmonds, 2019).
Histone Phosphorylation
Response to DNA damage is the best known function of histone phosphorylation, where phosphorylated H2A(X) delimits chromatin domains around the damaged DNA (Rossetto et al., 2012). However, this is not its only function, as it has been shown that a single histone phosphorylation event might be involved in different cellular processes (Rossetto et al., 2012). Histone serine (S) and tyrosine (Y) are part of the target of kinases, but little is known about their specificities (Rossetto et al., 2012; Best et al., 2018). In our study, most of the histone kinases known from mammals were also found in stickleback. However, as for other epigenetic pathways, most of our knowledge comes from zebrafish (Best et al., 2018), and the complexity of this mark is understudied. Thus, the specific loss/duplication of some members of this gene family raises the possibility of additional functions, for example phosphorylation of HDAC1 by aurora in zebrafish embryos (Loponte et al., 2016), or in diapause of the annual killifish (Toni and Padilla, 2016). Finally, phosphorylation of histone H2A.XF, a distinct isoform of H2A only present in very early development of Xenopus and zebrafish (Shechter et al., 2009), suggests a role for this modification in establishing particular transient chromatin conformations necessary for oocyte maturation of rapidly developing aquatic organisms facing environmental changes and/or perturbations.
Histone Ubiquitination
Ubiquitination is one of the main histone post-translational modifications. As in methylation, this mark is associated with both transcriptional activation and repression, whereas, in general, acetylation and phosphorylation are associated with gene expression, and sumoylation with gene repression (Sheng et al., 2014). Histone ubiquitination has been implicated in various cellular processes and cancers (Cao and Yan, 2012), was shown to be crucial to spermatogenesis (Sheng et al., 2014), and is also an aging biomarker (Wang et al., 2019) in mammals. In fish, this mark is largely undescribed (Best et al., 2018), but recent zebrafish studies have demonstrated the importance of ubiquitination in brain patterning (Kumar et al., 2019), and of deubiquination in craniofacial development (Ka and Tse, 2017). The general model of the ubiquitination process requires three enzymes: the ubiquitin activation enzyme E1, the ubiquitin conjugating enzyme (E2), and the ubiquitin ligase enzymes (E3), these last enzymes being very diverse (Sheng et al., 2014; Zheng and Shabek, 2017). Similar to many other epigenetic marks, ubiquitination is reversible, and deubiquitination enzymes (DUBs) deconjugate ubiquitin (Belle and Nijnik, 2014). In stickleback, conserved E1, E2, and E3 enzymes were identified but their enzymatic activity and biological functions are unknown. The number of characterized E3 may not be fixed, as an increasing number of studies (in mammals) have shown not only new substrates but also new E3 enzymes (Zheng and Shabek, 2017). Our results suggest that many loss/duplication events in DUBs may have occurred in stickleback and in fishes in general (Tse et al., 2009) compared to humans (Nijman et al., 2005).
Histone Poly-ADP Ribosylation
Histone poly-ADP ribosylation is a modification catalyzed by only one known family, the PARP (Poly(ADP-ribose)polymerase) enzymes (Perina et al., 2014). Also called PARylation, this mark is important in many biological processes such as DNA repair, telomere length regulation, apoptosis, aging, and protein degradation (Perina et al., 2014), and can also affect other epigenetic marks such as acetylation (Verdone et al., 2015). Only a small number of eukaryotic species do not possess PARP genes, and the human genome encodes 17 PARP proteins distributed in five classes (Perina et al., 2014). Interestingly, only 11 putative PARPs distributed in four classes were found in stickleback, and only PARP9/PARP14 contain the macro domain, likely reflecting the different evolutionary histories of stickleback and humans (Best et al., 2018). Histone poly-ADP ribosylation is reversible and is degraded by Poly(ADP-ribose)glycohydrolase, which necessitates the complementary action of (ADP-ribosyl)hydrolases (ARHs: two members ARH1/ARH3) (Rack et al., 2018). Thus, while PARGa has a conserved protein architecture, only ARH3 seems to be present in stickleback, suggesting a potential loss of ARH1 in stickleback (Best et al., 2018; Rack et al., 2018). Together, these results suggest that histone poly-ADP ribosylation, implicated in, for example, tumor suppression and defense against bacterial toxins that are specifically associated to ARH1, might be differentially regulated in stickleback compared to humans. Moreover, the biochemical activity of ARH3 should be studied in order to determine if it can compensate for the loss of ARH1 or if different mechanisms occur in stickleback, and if specific interactions with other epigenetic marks occur.
Histone Glycosylation
O-GlcNAcylation seems to contribute greatly to epigenetic reprogramming, and acts as a nutrient sensor implicated in some cancers (Dehennaut et al., 2014). Interests for nutritional programming through, for example, glucose metabolism, are increasing for cultivated fish species, and it has been demonstrated that rainbow trout juveniles may be reprogrammed through epigenetic modifications (DNA methylation and histone acetylation) (Panserat et al., 2017). Nevertheless, O-GlcNAcylation is poorly understood in teleosts, and has so far only been investigated in zebrafish (Sohn and Do, 2005) as a potential mechanism to confer heat resistance (Radermacher et al., 2014). Recent studies have described O-GlcNAcylation as a very important part of the “histone code”, and identified OGT as an interacting partner of the TET enzymes (Dehennaut et al., 2014). In stickleback, we identified OGT and OGA regulating O-GlcNAcylation (Dehennaut et al., 2014), and they seem to be conserved among vertebrates. Together with an absence of duplication/loss events, their putative protein architectures suggest strong conservation of their biochemical activity, making them particularly interesting candidates for epigenetically mediated phenotypes used in assisted evolution, aquaculture, and eco-evolutionary studies (Gavery and Roberts, 2017; Eirin-lopez and Putnam, 2019).
Histone Sumoylation
Histone sumoylation has been recently characterized in mammalian cells and is associated with transcriptional repression (Shiio and Eisenman, 2003). This modification is directed by an enzymatic cascade (E1-E2-E3, similar to ubiquitination) that attaches the three known SUMO proteins (Deyrieux and Wilson, 2017) and is crucial for sperm differentiation, oocyte maturation, and embryogenesis (Deyrieux and Wilson, 2017). On the other hand, deSUMOylation is performed by a family of enzymes called SUMO-specific proteases (SENP). While eight members comprise the human SENP, it seems that only six of them are considered true SENPs (Kim and Baek, 2009). SENPs can be separated into two groups: I containing SENP1-2-3-5, and II containing SENP6-7. In humans, SENP3 and SENP4 are identical, and SENP8 is not specific against SUMO (Kim and Baek, 2009). In stickleback, we found that E1 and E2 seem to be conserved, but only two E3 members were identified. Interestingly, the three stickleback SUMO proteins are conserved among the examined vertebrates, even if SUMO3 underwent different duplication events among the fish species examined. In stickleback, the two groups of SENPs were found, but SENP1 (I) or SENP7 (II) may have been lost. In zebrafish development, histone sumoylation is essential but enzymatic activity of each SUMO protein is redundant (Yuan et al., 2010). Consequently, the loss of SENP1/SENP7 in stickleback might be compensated for and/or the underlying molecular mechanisms as well as interactions with other epigenetic pathways may differ.
Stickleback H2A
Histones (H) are small, basic, and conserved proteins, which constitute a key element to pack DNA and to regulate genome accessibility to transcriptional machinery. In eukaryotes, DNA is wrapped around an octamer of histones constituted by H2A, H2B, H3, and H4 to form the nucleosome, the fundamental subunit of chromatin (Biterge and Schneider, 2014). While histones undergo multiple post-translational modifications on their residues, they can also be replaced by variants (Biterge and Schneider, 2014). Some of these variants such as macroH2A have been studied in marine organisms outside of model species (from diatoms to mollusks and fish) and play a role in environmental responses (Eirin-lopez and Putnam, 2019). For instance, a novel H2A variant (H2Af1o) was discovered in fish oocytes (gibel carp, crucian carp, and zebrafish), and its photophosphorylation seems to be implicated in oocyte maturation (Wu et al., 2009). Despite high expression in early embryogenesis and a higher mobility in the nucleosome than other H2A variants, more research is needed to better understand the underlying mechanisms and their physiological function during gametogenesis and development (Wu et al., 2009). In stickleback, we found multiple genes coding for H2A variants such as H2Afv, while only one gene codes for macroH2A that can give rise to three isoforms. This differs from mammals, where macroH2A was duplicated and gives rise to three isoforms (2 proteins gene 1, 1 protein gene 2) (Kozlowski et al., 2018). This, in conjunction with its duplication in a number of fish species, suggests potential teleost-specific biological functions. Furthermore, in our opinion, macroH2A might be of particular interest for eco-evolutionary studies as it has been linked to embryonic defects in mouse and zebrafish (Kozlowski et al., 2018), and is essential to cold acclimation in fish (Pinto et al., 2005).
Stickleback Genes Coding for Proteins Implicated in miRNA Biogenesis/Turnover
MicroRNAs, or miRNAs, are a class of non-coding RNA found from plants to animals that play important roles in mRNA destabilization and/or translation. With a size of approximately 22 nucleotides and being highly conserved among eukaryotes, they are typically targets for hundreds of genes making them very important for transcription regulation in time and space (Lim et al., 2005). As described in Best et al. (2018), genes implicated in mRNA biogenesis/turnover are well conserved in fish and vertebrates, even though they were unstudied in stickleback at that time. Our genomic survey supports their conclusion, and further shows that all genes implicated in the different miRNA pathways also occur in stickleback. Also, we did not detect any duplication events among the genes observed in the examined fish species, except for Drosha which was duplicated in salmonids (Best et al., 2018). Understanding their modulation and role in particular pathways appears to be important, since miRNAs are conserved in zebrafish (Desvignes et al., 2019), and they have a function in species acclimation to freshwater conditions (Rastorguev et al., 2016, 2017). Moreover, an increasing number of studies in different fish species suggest that miRNA are relevant for adaptation, gametogenesis and development (Tse et al., 2016; Baumgart et al., 2017; Gay et al., 2018), making investigations of miRNA biogenesis/turnover important for potential aquaculture applications, eco-evolutionary studies, and in particular, studies of transgenerational plasticity in the context of rapid climate change (Shama et al., 2016; Gavery and Roberts, 2017; Eirin-lopez and Putnam, 2019).
Conclusion
Interest in the interplay between genetic and epigenetic components of adaptive potential in marine aquaculture and wild populations is growing fast, and sticklebacks are an excellent model to address these questions. However, as in many other fish species, we suffer from a lack of knowledge concerning the mechanistic basis of epigenetic responses in comparison to well established model organisms. Recent advances using model species (e.g., zebrafish) demonstrate that epigenetic actors, their enzymatic activities, molecular interactions and biological functions are much more diversified and complex than previously thought. Here, we show that the variable number of genes and loss/duplication events among species could have considerable impacts on the different epigenetic pathways, potentially modifying their “traditional” function. Although most of the epigenetic toolbox seems to be evolutionarily conserved, specific loss/duplication events may have occurred, and underlie the need to precisely characterize the mechanistic basis of observed epigenetic variation in order to understand the role of these modifications and their importance in species adaptation. For this, molecular characterization of each enzyme using classic molecular biology tools (sequencing, expression, localization, enzymatic assays, and inhibition) but also new technologies in a collaborative setting are needed to explore the complex synergy of epigenetic mechanisms in teleosts and other taxonomic groups. This will allow us to tease apart the relative contributions of the “general” epigenetic toolkit (which appears to be more complex than previously thought) and species-specific mechanisms, leading to more precise and efficient use of epigenetic modifications in assisted evolution, aquaculture or conservation, and eco-evolutionary studies.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://www.ensembl.org/Gasterosteus_aculeatus.
Ethics Statement
This study was conducted in accordance with German animal welfare standards [Schleswig-Holstein Ministerium für Energiewende, Landwirtschaft, Umwelt, Natur und Digitalisierung (Tierschutz), permit no. V244-17922/2018(38-4/18)].
Author Contributions
AF conceived, designed, and performed the experiments, and analyzed the data. AF and LS wrote the manuscript.
Funding
This study was funded by a Strategy Funds Grant of the Alfred-Wegener-Institut, Helmholtz-Zentrum für Polar- und Meeresforschung awarded to LS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00721/full#supplementary-material
Footnotes
- ^ http://www.ensembl.org/Gasterosteus_aculeatus
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.ensembl.org
References
Akerberg, A. A., Henner, A., Stewart, S., and Stankunas, K. (2017). Histone demethylases Kdm6ba and Kdm6bb redundantly promote cardiomyocyte proliferation during zebrafish heart ventricle maturation. Dev. Biol. 426, 84–96. doi: 10.1016/j.ydbio.2017.03.030
Artemov, A. V., Mugue, N. S., Rastorguev, S. M., Zhenilo, S., Mazur, A. M., Tsygankova, S. V., et al. (2017). Genome-Wide DNA methylation profiling reveals epigenetic adaptation of stickleback to marine and freshwater conditions. Mol. Biol. Evol. 34, 2203–2213. doi: 10.1093/molbev/msx156
Balasch, J. C., and Tort, L. (2019). Netting the stress responses in fish. Front. Endrocrinol. 10:62. doi: 10.3389/fendo.2019.00062
Balasubramanian, S., Raghunath, A., and Perumal, E. (2019). Role of epigenetics in zebrafish development. Gene 18:144049. doi: 10.1016/j.gene.2019.144049
Ballestar, E., and Wolffe, A. P. (2001). Methyl-CpG-binding proteins Targeting specific gene repression. Eur. J. Biochem. FEBS 6, 1–6. doi: 10.1046/j.1432-1327.2001.01869.x
Baulcombe, D. C., and Dean, C. (2014). Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 6, a019471. doi: 10.1101/cshperspect.a019471
Baumgart, M., Barth, E., Savino, A., Groth, M., Koch, P., Petzold, A., et al. (2017). A miRNA catalogue and ncRNA annotation of the short-living fish Nothobranchius furzeri. BMC Genomics 18:693. doi: 10.1186/s12864-017-3951-3958
Belle, J. I., and Nijnik, A. (2014). The international journal of biochemistry H2A-DUBbing the mammalian epigenome: expanding frontiers for histone H2A deubiquitinating enzymes in cell biology and physiology. Int. J. Biochem. Cell Biol. 50, 161–174. doi: 10.1016/j.biocel.2014.03.004
Best, C., Ikert, H., Kostyniuk, D. J., Craig, P. M., Navarro-martin, L., Marandel, L., et al. (2018). Epigenetics in teleost fish: from molecular mechanisms to physiological phenotypes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 224, 210–244. doi: 10.1016/j.cbpb.2018.01.006
Biterge, B., and Schneider, R. (2014). Histone variants: key players of chromatin. Cell Tissue Resp. 356, 457–466. doi: 10.1007/s00441-014-1862-1864
Black, J. C., Rechem, C. V., and Whetstine, J. R. (2013). Histone lysine methylation dynamics: establishment. Regulation Biol. Impact. Mol. Cell 48, 1–32. doi: 10.1016/j.molcel.2012.11.006.Histone
Campos, C., Valente, L. M. P., and Fernandes, J. M. O. (2012). Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene 500, 93–100. doi: 10.1016/j.gene.2012.03.041
Cao, J., and Yan, Q. (2012). Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2:26. doi: 10.3389/fonc.2012.00026
Cosseau, C., Wolkenhauer, O., Padalino, G., Geyer, K. K., Hoffmann, K. F., and Grunau, C. (2017). (Epi)genetic Inheritance in Schistosoma mansoni: a systems approach. Trends Parasitol. 33, 285–294. doi: 10.1016/j.pt.2016.12.002
Cossins, A. R., and Crawford, D. L. (2005). Fish as models for environmental genomics. Nat. Rev. Genet. 6, 324–340.
Costa-pinheiro, P., and Montezuma, D. (2015). Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics 7, 1003–1015. doi: 10.2217/epi.15.56
Dehennaut, V., Leprince, D., and Lefebvre, T. (2014). O -GlcNAcylation, an epigenetic mark. Focus on the histone code, TET family proteins, and polycomb group proteins. Front. Endrocrinol. 5:155. doi: 10.3389/fendo.2014.00155
Desvignes, T., Batzel, P., Sydes, J., Eames, B. F., and Postlethwait, J. H. (2019). miRNA analysis with Prost! reveals evolutionary conservation of organ-enriched expression and post-transcriptional modifications in three-spined stickleback and zebrafish. Sci. Rep. 9:3913. doi: 10.1038/s41598-019-40361-40368
Deyrieux, A. F., and Wilson, V. G. (2017). Sumoylation in development and differentiation. Adv. Exp. Med. Biol. 963, 197–214. doi: 10.1007/978-3-319-50044-7
Dupret, B., Völkel, P., Vennin, C., Toillon, R., Le, X., and Angrand, P. (2017). The histone lysine methyltransferase Ezh2 is required for maintenance of the intestine integrity and for caudal fin regeneration in zebrafish. BBA Gene. Regul. Mech. 1860, 1079–1093. doi: 10.1016/j.bbagrm.2017.08.011
Eckersley-maslin, M. A., Alda-catalinas, C., and Reik, W. (2018). Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450. doi: 10.1038/s41580-018-0008-z
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eirin-lopez, J. M., and Putnam, H. M. (2019). Marine environmental epigenetics. Ann. Rev. Mar. Sci. 11, 335–368. doi: 10.1146/annurev-marine-010318-095114
Ellison, A., Rodrı, C. M., Moran, P., Breen, J., Swain, M., Megias, M., et al. (2015). Epigenetic regulation of sex ratios may explain natural variation in self-fertilization rates. Proc. R. Soc. B 282:20151900. doi: 10.1098/rspb.2015.1900
Fatemi, M., and Wade, P. A. (2006). MBD family proteins: reading the epigenetic code. J. Cell Sci. 119(Pt 15), 3033–3037. doi: 10.1242/jcs.03099
Fellous, A., Earley, R. L., and Silvestre, F. (2019a). Identification and expression of mangrove rivulus (Kryptolebias marmoratus) histone deacetylase (HDAC) and lysine acetyltransferase (KAT) genes. Gene 691, 56–69. doi: 10.1016/j.gene.2018.12.057
Fellous, A., Earley, R. L., and Silvestre, F. (2019b). The Kdm/Kmt gene families in the self-fertilizing mangrove rivulus fish, Kryptolebias marmoratus, suggest involvement of histone methylation machinery in development and reproduction. Gene 687, 173–187. doi: 10.1016/j.gene.2018.11.046
Fellous, A., Franc, L. L. E., Jouaux, A., Goux, D., and Favrel, P. (2019c). Histone methylation participates in gene expression control during the early development of the pacific oyster Crassostrea gigas. Genes 10:695. doi: 10.3390/genes10090695
Fellous, A., Favrel, P., Guo, X., and Riviere, G. (2014). The Jumonji gene family in Crassostrea gigas suggests evolutionary conservation of Jmj-C histone demethylases orthologues in the oyster gametogenesis and development. Gene 538, 164–175. doi: 10.1016/j.gene.2013.12.016
Fellous, A., Favrel, P., and Riviere, G. (2015). Temperature influences histone methylation and mRNA expression of the Jmj-C histone-demethylase orthologues during the early development of the oyster Crassostrea gigas. Mar. Genomics 19, 23–30. doi: 10.1016/j.margen.2014.09.002
Fellous, A., Labed-Veydert, T., Locrel, M., Voisin, A.-S., Earley, R. L., and Silvestre, F. (2018). DNA methylation in adults and during development of the self-fertilizing mangrove rivulus, Kryptolebias marmoratus. Ecol. Evol. 8, 6016–6033. doi: 10.1002/ece3.4141
Feng, S., Cokus, S. J., Zhang, X., Chen, P.-Y., Bostick, M., Goll, M. G., et al. (2010). Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U.S.A. 107, 8689–8694. doi: 10.1073/pnas.1002720107
Fincham, J. (1997). Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Firmino, J., Carballo, C., Armesto, P., Campinho, M. A., Power, D. M., and Manchado, M. (2017). Phylogeny, expression patterns and regulation of DNA Methyltransferases in early development of the flatfish, Solea senegalensis. BMC Dev. Biol. 17:11. doi: 10.1186/s12861-017-0154-0
Gao, H., Bu, Y., Wu, Q., Wang, X., Chang, N., Lei, L., et al. (2015). Mecp2 regulates neural cell differentiation by suppressing the Id1 to Her2 axis in zebrafish. J. Cell Sci. 128, 2340–2350. doi: 10.1242/jcs.167874
Gavery, M. R., Nichols, K. M., Berejikian, B. A., Tatara, C. P., Goetz, G. W., Dickey, J. T., et al. (2019). Temporal dynamics of DNA methylation patterns in response to rearing juvenile steelhead (Oncorhynchus mykiss) in a hatchery versus simulated stream environment. Genes 10:E356. doi: 10.3390/genes10050356
Gavery, M. R., and Roberts, S. B. (2017). Epigenetic considerations in aquaculture. PeerJ 5:e4147. doi: 10.7717/peerj.4147
Gay, S., Bugeon, J., Bouchareb, A., Henry, L., Delahaye, C., Legeai, F., et al. (2018). MiR-202 controls female fecundity by regulating medaka oogenesis. LoS Genet. 14:e1007593. doi: 10.1371/journal.pgen.1007593
Greiss, S., and Gartner, A. (2013). Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cells 28, 407–415. doi: 10.1007/s10059-009-0169-x.Sirtuin/Sir2
He, Y., Wu, J., Mei, H., Yu, H., Sun, S., Shou, J., et al. (2014). Histone deacetylase activity is required for embryonic posterior lateral line development. Cell Prolif. 47, 91–104. doi: 10.1111/cpr.12081
Heckwolf, M. J., Eizaguirre, C., Meyer, B. S., Döring, T., and Reusch, T. B. H. (2018). Transgenerational plasticity and selection shape the adaptive potential of sticklebacks to salinity change. Evol. Appl. 11, 1873–1885. doi: 10.1111/eva.12688
Heckwolf, M. J., Meyer, B. S., Häsler, R., and Höppner, M. P. (2019). DNA methylation facilitates local adaptation and adaptive transgenerational plasticity. Biorxiv. [Preprint]. doi: 10.1101/649574
Horsfield, J. A. (2019). Packaging development: how chromatin controls transcription in zebrafish embryogenesis. Biochem. Soc. Trans. 47, 713–724. doi: 10.1042/BST20180617
Jiang, F., Liu, Q., Wang, Y., Zhang, J., Wang, H., Song, T., et al. (2017). Comparative genomic analysis of SET domain family reveals the origin, expansion, and putative function of the arthropod-specific SmydA genes as histone modifiers in insects. Giga Sci. 6, 1–16. doi: 10.1093/gigascience/gix031
Ka, W., and Tse, F. (2017). Importance of deubiquitinases in zebrafish craniofacial development. Biochem. Biophys. Res. Commun. 487, 813–819. doi: 10.1016/j.bbrc.2017.04.132
Kamstra, J. H., Aleström, P., Kooter, J. M., and Legler, J. (2015). Zebrafish as a model to study the role of DNA methylation in environmental toxicology. Environ. Sci. Pollut. Res. 22, 16262–16276. doi: 10.1007/s11356-014-3466-7
Karmodiya, K., Anamika, K., Muley, V., Pradhan, S. J., Bhide, Y., and Galande, S. (2014). Camello, a novel family of histone acetyltransferases that acetylate histone H4 and is essential for zebrafish. Sci. Rep. 4, 1–9. doi: 10.1038/srep06076
Kim, J. H., and Baek, S. H. (2009). Emerging roles of desumoylating enzymes. BBA Mol. Basis Dis. 1792, 155–162. doi: 10.1016/j.bbadis.2008.12.008
Kitano, J., Ishikawa, A., and Kusakabe, M. (2019). Parallel transcriptome evolution in stream threespine sticklebacks. Dev. Growt. Differ. 61, 104–113. doi: 10.1111/dgd.12576
Kozlowski, M., Corujo, D., Hothorn, M., Guberovic, I., Mandemaker, I. K., Blessing, C., et al. (2018). MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep. 19:e44445. doi: 10.15252/embr.201744445
Kumar, A., Anuppalle, M., Maddirevula, S., Huh, L., Choe, J., Rhee, M., et al. (2019). Peli1b governs the brain patterning via ERK signaling pathways in zebrafish embryos. Gene. 694, 1–6. doi: 10.1016/j.gene.2018.12.078
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Labbé, C., Robles, V., and Paz, M. (2017). Epigenetics in fish gametes and early embryo. Aquaculture 472, 93–106. doi: 10.1016/j.aquaculture.2016.07.026
Laporte, M., Luyer, J., Le Rougeux, C., Krick, M., and Bernatchez, L. (2019). DNA methylation reprogramming, TE derepression, and postzygotic isolation of nascent animal species. Sci. Adv. 5, 1–9. doi: 10.1126/sciadv.aaw1644
Le Luyer, J., Laporte, M., Beacham, T. D., Kaukinen, K. H., Withler, R. E., and Leong, J. S. (2017). Parallel epigenetic modifications induced by hatchery rearing in a Pacific salmon. Proc. Natl. Acad. Sci. U.S.A. 114, 12964–12969. doi: 10.1073/pnas.1711229114
Lei, L., Zhou, S., Ma, H., and Zhang, L. (2012). Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 12:51. doi: 10.1186/1471-2148-12-51
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. doi: 10.1016/0092-8674(92)90611-f
Lim, L. P., Lau, N. C., Garrett-engele, P., and Grimson, A. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 292, 288–292.
Lindeman, L. C., Winata, C. L., Aanes, H., Mathavan, S., Aleström, P., and Collas, P. (2010). Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int. J. Dev. Biol. 813, 803–813. doi: 10.1387/ijdb.103081ll
Loponte, S., Segré, C. V., Senese, S., Miccolo, C., Santaguida, S., De, G., et al. (2016). Dynamic phosphorylation of histone deacetylase 1 by aurora kinases during mitosis regulates zebrafish embryos development. Sci. Rep. 6:30213. doi: 10.1038/srep30213
Lyko, F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. doi: 10.1038/nrg.2017.80
Martin, C., and Zhang, Y. (2005). The diverse functions of histone lysine methylation. Mol. Cell Biol. 6, 838–849. doi: 10.1038/nrm1761
Mcghee, K. E., and Bell, A. M. (2014). Paternal care in a fish: epigenetics and fitness enhancing effects on offspring anxiety. Proc. R. Soc. B 281, 2–7.
Metzger, D. C. H., and Schulte, P. M. (2016). Epigenomics in marine fishes. Mar. Genomics 30, 43–54. doi: 10.1016/j.margen.2016.01.004
Metzger, D. C. H., and Schulte, P. M. (2017). Persistent and plastic effects of temperature on DNA methylation across the genome of threespine stickleback (Gasterosteus aculeatus). Proc. R. Soc. B 284:20171667. doi: 10.1098/rspb.2017.1667
Metzger, D. C. H., and Schulte, P. M. (2018). The DNA methylation landscape of stickleback reveals patterns of sex chromosome evolution and effects of environmental salinity. Genome Biol. Evol. 10, 1–30. doi: 10.1093/gbe/evy034/4840697
Nijman, S. M. B., Luna-vargas, M. P. A., Velds, A., Brummelkamp, T. R., Dirac, A. M. G., Sixma, T. K., et al. (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786. doi: 10.1016/j.cell.2005.11.007
Nozawa, K., Lin, Y., Kubodera, R., Shimizu, Y., and Tanaka, H. (2017). Zebrafish Mecp2 is required for proper axonal elongation of motor neurons and synapse formation. Dev. Neurobiol. 77, 1101–1113. doi: 10.1002/dneu.22498
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. doi: 10.1016/s0092-8674(00)81656-6
Panserat, S., Marandel, L., Geurden, I., Veron, V., Dias, K., Plagnes-juan, E., et al. (2017). Muscle catabolic capacities and global hepatic epigenome are modi fi ed in juvenile rainbow trout fed different vitamin levels at fi rst feeding. Aquaculture 468, 515–523. doi: 10.1016/j.aquaculture.2016.11.021
Perina, D., Mikoè, A., Ahel, J., Cetkovi, H., and Ahel, I. (2014). Distribution of protein poly (ADP-ribosyl) ation systems across all domains of life. DNA Repair. 23, 4–16. doi: 10.1016/j.dnarep.2014.05.003
Petrie, K., Guidez, F., Howell, L., Healy, L., Waxman, S., Greaves, M., et al. (2003). The histone deacetylase 9 gene encodes multiple protein isoforms. J. Biol. Chem. 278, 16059–16072. doi: 10.1074/jbc.M212935200
Petrossian, T. C., and Clarke, S. G. (2011). Uncovering the human methyltransferasome. Mol. Cell. Proteomics 10:M110.000976. doi: 10.1074/mcp.M110.000976
Pinto, R., Ivaldi, C., Reyes, M., Mietton, F., Mongelard, F., Alvarez, M., et al. (2005). Seasonal environmental changes regulate the expression of the histone variant macroH2A in an eurythermal fish. FEBS Lett. 579, 5553–5558. doi: 10.1016/j.febslet.2005.09.019
Potok, M., Nix, D., Parnell, T., and Cairns, B. (2013). Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772. doi: 10.1016/j.cell.2013.04.030
Putnam, H. M., Davidson, J. M., and Gates, R. D. (2016). Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evol. Appl. 9, 1165–1178. doi: 10.1111/eva.12408
Putnam, H. M., and Gates, R. D. (2015). Preconditioning in the reef-building coral Pocillopora damicornis and the potential for trans-generational acclimatization in coral larvae under future climate change conditions. J. Exp. Biol. 218, 2365–2372. doi: 10.1242/jeb.123018
Qian, S., Wang, Y., Ma, H., and Zhang, L. (2015). Expansion and functional divergence of jumonji C-Containing histone demethylases: signi fi cance of duplications in ancestral angiosperms and vertebrates. Plant Physiol. 168, 1321–1337. doi: 10.1104/pp.15.00520
Rack, J. G. M., Ariza, A., Drown, B. S., Shirai, T., Hergenrother, P. J., Shirai, T., et al. (2018). (ADP-ribosyl) hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition Basis for Differential Substrate Recognition and Inhibition. Cell Chem. Biol. 25, 1533.e12–1546.e12. doi: 10.1016/j.chembiol.2018.11.001
Radermacher, P. T., Myachina, F., Bosshardt, F., Pandey, R., Mariappa, D., and Müller, H. J. (2014). O- GlcNAc reports ambient temperature and confers heat resistance on ectotherm development. Proc. Natl. Acad. Sci. U.S.A. 111, 5592–5597. doi: 10.1073/pnas.1322396111
Rastorguev, S. M., Nedoluzhko, A. V., Gruzdeva, N. M., Boulygina, E. S., and Sharko, F. S. (2017). Differential miRNA expression in the three-spined stickleback, response to environmental changes. Sci. Rep. 7:18089. doi: 10.1038/s41598-017-18128-w
Rastorguev, S. M., Nedoluzhko, A. V., Sokolov, A. S., Gruzdeva, N. M., Skryabin, K. G., and Prokhortchouk, E. B. (2016). Identification of novel microRNA genes in freshwater and marine ecotypes of the three- spined stickleback (Gasterosteus aculeatus). Mol. Ecol. Resour. 16, 1491–1498. doi: 10.1111/1755-0998.12545
Riviere, G., Wu, G. C., Fellous, A., Goux, D., Sourdaine, P., and Favrel, P. (2013). DNA methylation is crucial for the early development in the Oyster C. gigas. Mar. Biotechnol. 15, 739–753. doi: 10.1007/s10126-013-9523-9522
Román, A.-C., Vicente-Page, J., Pérez-Escudero, A., Carvajal-González, J. M., Fernández-Salguero, P. M., and de Polavieja, G. G. (2018). Histone H4 acetylation regulates behavioral inter-individual variability in zebrafish. Genome Biol. 19:55. doi: 10.1186/s13059-018-1428-y
Rossetto, D., Avvakumov, N., and Côté, J. (2012). Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigen 7, 1098–1108. doi: 10.4161/epi.21975
Santiago, M., Antunes, C., Guedes, M., Sousa, N., and Marques, C. J. (2014). Genomics TET enzymes and DNA hydroxymethylation in neural development and function – How critical are they? Genomics 104, 334–340. doi: 10.1016/j.ygeno.2014.08.018
Sasai, N., Kato, Y., Kimura, G., and Takeuchi, T. (2007). The Drosophila jumonji gene encodes a JmjC-containing nuclear protein that is required for metamorphosis. FEBS J. 274, 6139–6151. doi: 10.1111/j.1742-4658.2007.06135.x
Seebacher, F., and Simmonds, A. I. M. (2019). Histone deacetylase activity mediates thermal plasticity in zebrafish (Danio rerio). Sci. Rep. 9:8216. doi: 10.1038/s41598-019-44726-x
Seto, E., and Yoshida, M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6, 1–26. doi: 10.1101/cshperspect.a018713
Shama, L. N. S. (2017). The mean and variance of climate change in the oceans: hidden evolutionary potential under stochastic environmental variability in marine sticklebacks. Sci. Rep. 7:8889. doi: 10.1038/s41598-017-07140-7149
Shama, L. N. S., Mark, F. C., Strobel, A., Lokmer, A., John, U., and Wegner, K. M. (2016). Transgenerational effects persist down the maternal line in marine sticklebacks: gene expression matches physiology in a warming ocean. Evol. Appl. 9, 1096–1111. doi: 10.1111/eva.12370
Shama, L. N. S., and Wegner, K. M. (2014). Grandparental effects in marine sticklebacks: transgenerational plasticity across multiple generations. J. Evol. Biol. 27, 2297–2307. doi: 10.1111/jeb.12490
Shanmugam, M. K., Arfuso, F., and Arumugam, S. (2018). Role of novel histone modifications in cancer. Oncotarget 9, 11414–11426.
Shechter, D., Chitta, R. K., Xiao, A., Shabanowitz, J., Hunt, D. F., and Allis, C. D. (2009). A distinct H2A.X isoform is enriched in Xenopus laevis eggs and early embryos and is phosphorylated in the absence of a checkpoint. PNAS 106, 749–754. doi: 10.1073/pnas.0812207106
Sheikh, B. N., and Akhtar, A. (2018). The many lives of KATs – detectors, integrators and modulators of the cellular environment. Nat. Rev. Genet. 20, 7–23. doi: 10.1038/s41576-018-0072-74
Sheng, K., Liang, X., Huang, S., and Xu, W. (2014). The role of histone ubiquitination during spermatogenesis. Biomed Res. Int. 2014:870695. doi: 10.1155/2014/870695
Shi, L., Sun, L., Li, Q., Liang, J., Yu, W., Yi, X., et al. (2011). Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 7541–7546. doi: 10.1073/pnas.1017374108
Shiio, Y., and Eisenman, R. N. (2003). Histone sumoylation is associated with transcriptional repression. PNSA 100, 13225–13230. doi: 10.1073/pnas.1735528100
Shin, J., Kim, J., Park, H., and Kim, J. (2018). Investigating the role of Sirtuins in cell reprogramming. BMB Rep. 51, 500–507. doi: 10.5483/bmbrep.2018.51.10.172
Smith, G., Smith, C., Kenny, J. G., Chaudhuri, R. R., and Ritchie, M. G. (2018). Genome-Wide DNA methylation patterns in wild samples of two morphotypes of threespine stickleback (Gasterosteus aculeatus). Mol. Biol. Evol. 32, 888–895. doi: 10.1093/molbev/msu344
Sohn, K., and Do, S. (2005). Transcriptional regulation and O-GlcNAcylation activity of zebrafish OGT during embryogenesis. Biochem. Biophys. Res. Commun. 337, 256–263. doi: 10.1016/j.bbrc.2005.09.049
Stewart, S., Tsun, Z., Carlos, J., and Belmonte, I. (2009). A histone demethylase is necessary for regeneration in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 106, 19889–19894. doi: 10.1073/pnas.0904132106
Takeuchi, T., Kojima, M., Nakajima, K., and Kondo, S. (1999). jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech. Dev. 86, 29–38. doi: 10.1016/s0925-4773(99)00100-8
Teigen, L. E., Orczewska, J. I., McLaughlin, J., and O’Brien, K. M. (2015). Cold acclimation increases levels of some heat shock protein and sirtuin isoforms in threespine stickleback. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 188, 139–147. doi: 10.1016/j.cbpa.2015.06.028
Todd, E. V., Ortega-recalde, O., Liu, H., Lamm, M. S., Rutherford, K. M., Cross, H., et al. (2019). Stress, novel sex genes, and epigenetic reprogramming orchestrate socially controlled sex change. Sci. Adv. 5, 1–14.
Toni, L. S., and Padilla, P. A. (2016). Developmentally arrested austrofundulus limnaeus embryos have changes in post-translational modifications of histone H3. Co. Biol. 219, 544–552. doi: 10.1242/jeb.131862
Tse, A. C., Li, J., Yuan, S., Chan, T., Po, K., and Wu, R. S. (2016). Hypoxia alters testicular functions of marine medaka through microRNAs regulation. Aquat. Toxicol. 180, 266–273. doi: 10.1016/j.aquatox.2016.10.007
Tse, W. K. F., Eisenhaber, B., Ho, S. H. K., Ng, Q., Eisenhaber, F., and Jiang, Y. (2009). Genome-wide loss-of-function analysis of deubiquitylating enzymes for zebrafish development. BMC Genomics 15:637. doi: 10.1186/1471-2164-10-637
Van Leeuwen, F., Gafken, P. R., and Gottschling, D. E. (2002). Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756. doi: 10.1016/S0092-8674(02)00759-756
Van Otterdijk, S. D., and Michels, K. B. (2016). Transgenerational epigenetic inheritance in mammals: how good is the evidence? FASEB J. 30, 2457–2465. doi: 10.1096/fj.201500083
Vastenhouw, N. L., Cao, W. X., and Lipshitz, H. D. (2019). The maternal-to-zygotic transition revisited. Development 146, dev161471. doi: 10.1242/dev.161471
Verdone, L., Fortezza, M., La Ciccarone, F., and Caiafa, P. (2015). Poly (ADP-Ribosyl) ation affects histone acetylation and transcription. PLoS One 3:e0144287. doi: 10.1371/journal.pone.0144287
Wang, G., and Köhler, C. (2017). Epigenetic processes in flowering plant reproduction. J. Exp. Bot. 68, 797–807. doi: 10.1093/jxb/erw486
Wang, X., and Bhandari, R. K. (2019). DNA methylation dynamics during epigenetic reprogramming of medaka embryo embryo. Epigenetics 0, 1–12. doi: 10.1080/15592294.2019.1605816
Wang, Y., and Li, C. (2012). Evolutionarily conserved protein arginine methyltransferases in non-mammalian animal systems. FEBS J. 279, 932–945. doi: 10.1111/j.1742-4658.2012.08490.x
Wang, Y., Liu, Q., Tang, F., Yan, L., and Qiao, J. (2019). Epigenetic regulation and risk factors during the development of human gametes and early embryos. Annu. Rev. Genomics Hum. Genet. 20, 1–20. doi: 10.1146/annurev-genom-083118-015143
Wu, N., Yue, H., Chen, B., and Gui, J. (2009). Histone H2A has a novel variant in fish oocytes 1. Biol. Reprod. 283, 275–283. doi: 10.1095/biolreprod.108.074955
Yang, X. (2015). MOZ and MORF acetyltransferases: molecular interaction, animal development and human disease. BBA Mol. Cell Res. 1853, 1818–1826. doi: 10.1016/j.bbamcr.2015.04.014
Yeh, H., and Klesius, P. H. (2012). Molecular characterization, phylogenetic analysis and expression patterns of five protein arginine methyltransferase genes of channel catfish, Ictalurus punctatus (Rafinesque). Fish Physio. Bioch. 38, 1083–1098. doi: 10.1007/s10695-011-9593-x
Yuan, H., Zhou, J., Deng, M., Liu, X., Le Bras, M., De The, H., et al. (2010). Small ubiquitin-related modifier paralogs are indispensable but functionally redundant during early development of zebrafish. Cell Res. 20, 185–196. doi: 10.1038/cr.2009.101
Zhang, J., Jing, L. I., Li, M., He, L., and Guo, Z. (2019). Regulation of histone arginine methylation/demethylation by methylase and demethylase (Review). Mol. Med. Rep. 19, 3963–3971. doi: 10.3892/mmr.2019.10111
Zhao, H., and Chen, T. (2013). Tet family of 5-methylcytosine dioxygenases in mammalian development. J. Hum. Genet. 58, 421–427. doi: 10.1038/jhg.2013.63
Keywords: evolution, fish, stickleback, DNA methylation/hydroxymethylation, histone modifications, miRNA, global climate change, aquaculture
Citation: Fellous A and Shama LNS (2019) Genome Survey of Chromatin-Modifying Enzymes in Threespine Stickleback: A Crucial Epigenetic Toolkit for Adaptation? Front. Mar. Sci. 6:721. doi: 10.3389/fmars.2019.00721
Received: 02 September 2019; Accepted: 07 November 2019;
Published: 20 November 2019.
Edited by:
Jose M. Eirin-Lopez, Florida International University, United StatesReviewed by:
James Dimond, University of Washington, United StatesHans J. Lipps, Universität Witten/Herdecke, Germany
Copyright © 2019 Fellous and Shama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandre Fellous, YWxleGFuZHJlLmZlbGxvdXNAYXdpLmRl
 Alexandre Fellous
Alexandre Fellous Lisa N. S. Shama
Lisa N. S. Shama