
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 11 September 2019
Sec. Marine Molecular Biology and Ecology
Volume 6 - 2019 | https://doi.org/10.3389/fmars.2019.00555
Stable isotope probing (SIP) was used to track prokaryotic and eukaryotic carbon uptake along a meridional transect (Long. 52°W) in the North Atlantic to assess if 13C-resource partitioning between bacteria and archaea and 13C-labeled eukaryotic predators could be detected. One-liter SIP microcosms were amended with 13C-acetate or 13C-urea and incubated for 48 h. Our data indicated archaea often outcompeted bacteria for 13C-urea while both archaea and bacteria could incorporate 13C-acetate. This 13C label could also be tracked into eukaryotic microbes. The largest number of 13C-labeled eukaryotic OTUs, and the greatest percentage of eukaryotic 13C signal, were observed in conjunction with both archaeal and bacterial 13C incorporation, suggesting that most eukaryotic predators do not distinguish between archaeal and bacterial prey. However, other 13C-eukaryotic OTUs were exclusively associated with either 13C-archaeal or 13C-bacterial OTUs. These archaeal-specific and bacterial-specific 13C-eukaryotic OTUs were related to known bactivorous predators including Ancyromonas, Amastigomonas, Cafeteria, and Caecitellus. Our SIP findings suggest both resource partitioning between bacteria and TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota) archaea and selective predation by eukaryotic predators. Determining the equalizing mechanisms for co-existence in the marine environment can help map predator/prey interactions to better estimate carbon flow in the deep ocean.
G. E. Hutchinson confronted a major contradiction between ecological theory and the natural world in “The Paradox of the Plankton” (1961). According to the competitive exclusion principle, two species competing for the same resources cannot stably coexist. However, hundreds/thousands of phytoplankton species can be found within small volumes of water in lakes, despite limited room for niche specialization and intense competition for the same resources (Scheffer and Carpenter, 2003). Hutchinson surmised that because environmental conditions can change very rapidly in aquatic systems, no one species has the advantage long enough to exclude others (Hutchinson, 1961; Richardson et al., 1970). Additionally, stabilizing forces may also exist that reduce intra- and inter-specific competition (Chesson and Huntley, 1997; Chesson, 2000). Multiple mechanisms have been proposed to account for this co-occurrence of highly similar species, including temporal separation of activity, resource partitioning, immigration/emigration, selective predation, and dormancy.
These ecological theories of coexistence can also be tested in deep ocean environments containing both archaea and bacteria. For many years archaea were believed to be solely extremophiles. Specifically, these microorganisms were thought to grow under conditions of very high temperature, high salinity, or extreme anaerobiosis where CO2 is used as a terminal electron acceptor, yielding methane. Because archaea occurred in habitats that would not support the growth of eukaryotes or prokaryotes, no apparent conflict was perceived between these various life forms. However, the concept of archaea being obligate extremophiles changed when these microbes were found to inhabit coastal oceans (DeLong, 1992; Fuhrman et al., 1992). Since then, mesothermal archaeal diversity has been revealed in open ocean areas around the globe (Massana et al., 2000) and in deep-sea sediments (Vetriani et al., 1999; Teske et al., 2002; Søresnson and Teske, 2006; Biddle et al., 2008). Archaea have also been found in waters between 4 and 8°C (Massana et al., 1997), polar seas (Murray et al., 1998), soils (Birtrim et al., 1997), caves (Gonzalez et al., 2006), salt marshes (Nelson et al., 2009), and in estuarine settings (Abreu et al., 2001). The discovery of marine mesophilic archaea represents an entire clade of organisms that, up until 30 years ago, had gone completely unnoticed because of their resistance to laboratory culturing, yet are apparently ubiquitous and frequently abundant in the ocean (DeLong et al., 1994; Stein and Simon, 1996; Karner et al., 2001).
Exactly what strategies marine archaea are using to remain relatively abundant and ubiquitous remains largely unknown. Surface ocean prokaryotic communities tend to be dominated by bacteria and a small percentage of euryarchaea, but below the euphotic zone and in sediments, thaumarchaea and bathyarchaea of the TACK superphylum (Guy and Ettema, 2011) equal or outnumber bacteria in abundance (Karner et al., 2001; Herndl et al., 2005; Amano-Santo et al., 2013). Both thaumarchaea (Ouverney and Fuhrman, 2000; Seyler et al., 2014, 2018) and bathyarchaea (Seyler et al., 2014; Lazar et al., 2016) appear to be capable of organic carbon uptake, competing with bacteria for the same organic substrates. Metagenomic evidence suggests that thaumarchaea may also be capable of both autotrophy and mixotrophy (Hallam et al., 2006), and radiocarbon data has suggested that autotrophy is the dominant lifestyle of thaumarchaea below the photic zone (Ingalls et al., 2006). Thaumarchaea, however, are most frequently studied with respect to their role in ammonia oxidation in the ocean. In 2004, a unique ammonia monooxygenase (amoA) gene was discovered on an archaeal-associated genomic scaffold (Venter et al., 2004); a year later amoA genes were found in thaumarchaea (Treusch et al., 2005) and examples of archaeal amoA genes were observed in a variety of other marine environments (Francis et al., 2005). Currently, few thaumarchaea have been cultured, and all are ammonia oxidizers (Könneke et al., 2005; Park et al., 2012; Qin et al., 2014). However, a nitrifying lifestyle for archaea would also directly compete with nitrifying bacteria in the marine environment. It has been suggested that ammonia-oxidizing archaea are of particular importance in low-oxygen environments such as sediments and oxygen minimum zones (Francis et al., 2005; Mosier and Francis, 2008; Moin et al., 2009; Park et al., 2010). These nitrifying archaea may also be well-adapted to oligotrophic environments where the amount of available nitrogen is comparatively low (Martens-Habbena et al., 2009) which could be considered as a stabilizing force regarding preference of low rather than high ammonium concentrations.
All this conflicting potential metabolic data indicates that if the TACK-archaeota and bacteria are living in the same environment, using similar carbon and energy sources, there should be competitive exclusion and one group would eventually out-compete the other. Yet, we find both living together in the same oceanic samples. In this report, resource partitioning and selective predation were investigated as two possible mechanisms that may allow archaea and bacteria to stably coexist in the deep ocean. We chose to utilize DNA stable-isotope probing techniques (SIP; Radajewski et al., 1999) as a means to create different pools of 13C-microbial prey items in marine samples (by labeling either bacteria or archaea with different 13C-carbon sources). Prior research had demonstrated that archaea could successfully outcompete bacteria for 13C-urea in a coastal salt marsh at low concentrations (<30 μM) during short term incubations (5 days) (Seyler et al., 2014). However, it is unclear if deep ocean archaea would also outcompete bacteria under comparable conditions to our salt marsh study. We initially set out to determine if similar resource partitioning results could also be observed in oceanic water column samples. If resource partitioning could be demonstrated, we could then test a second mechanism of co-existence (selective predation) by determining if eukaryotic predators consumed 13C-archaeal or 13C-bacterial prey in a specific fashion. Our findings suggested there was specific uptake of 13C-urea by TACK archaea in many of our SIP experiments. Furthermore, particular 13C-eukaryotes were associated exclusively with 13C-archaea (archaevores), with 13C-bacteria (bactivores) or detected in samples without any archaea or bacterial 13C uptake (osmotrophs). This information on 13C-resource partitioning and specific predation by eukaryotes provides insight into the competitive relationships between archaea and bacteria. Finally, this SIP approach also demonstrates how the deep sea can be a model system for studying competition between other microbial groups in natural settings to further our understanding of microbial ecology.
In April 2012, water samples were collected onboard the R/V Atlantis as part of the US CLIVAR/CO2 Repeat Hydrography program, along World Ocean Circulation Experiment (WOCE) line A20. This was a cruise of opportunity, done in conjunction with a larger effort to characterize water chemistry and marine microbial productivity. Therefore, the volumes of water available for sampling were often limited (generally <5 L at each station/depth per cast). As such, a total of 36 SIP microcosms were established at three stations (Figure 1A) from three water column zones: the mixed layer (euphotic zone, 65–115 m), the local oxygen minimum (mesopelagic zone, 365–835 m), and the bathypelagic zone (2335–2585 m; Figure 1B), as part of a larger effort to determine autotrophy, heterotrophy and mixotrophy in N. Atlantic waters (Seyler et al., 2018). Duplicate 1-L samples of water from each depth were amended with 13C-acetate/12C-urea or 13C-urea/12C-acetate at equimolar concentrations (20 μM), to stimulate the same populations of microbes in all microcosms from the same station/depth and maximize the number of 13C-carbon sources that could be tested. Microcosms were incubated in covered containers on deck for 48 h, with a constant inflow of surface seawater to maintain stable temperature (∼ 22°C). Early in the cruise, duplicate 12C-acetate and 13C-acetate microcosms were established and incubated for 12–72 h to verify the minimum time necessary to obtain a positive 13C-signal (Seyler et al., 2018). Negative SIP controls using 12C sodium acetate were incubated for 72 h. Biomass was collected on a 0.2-μm filter using vacuum filtration.
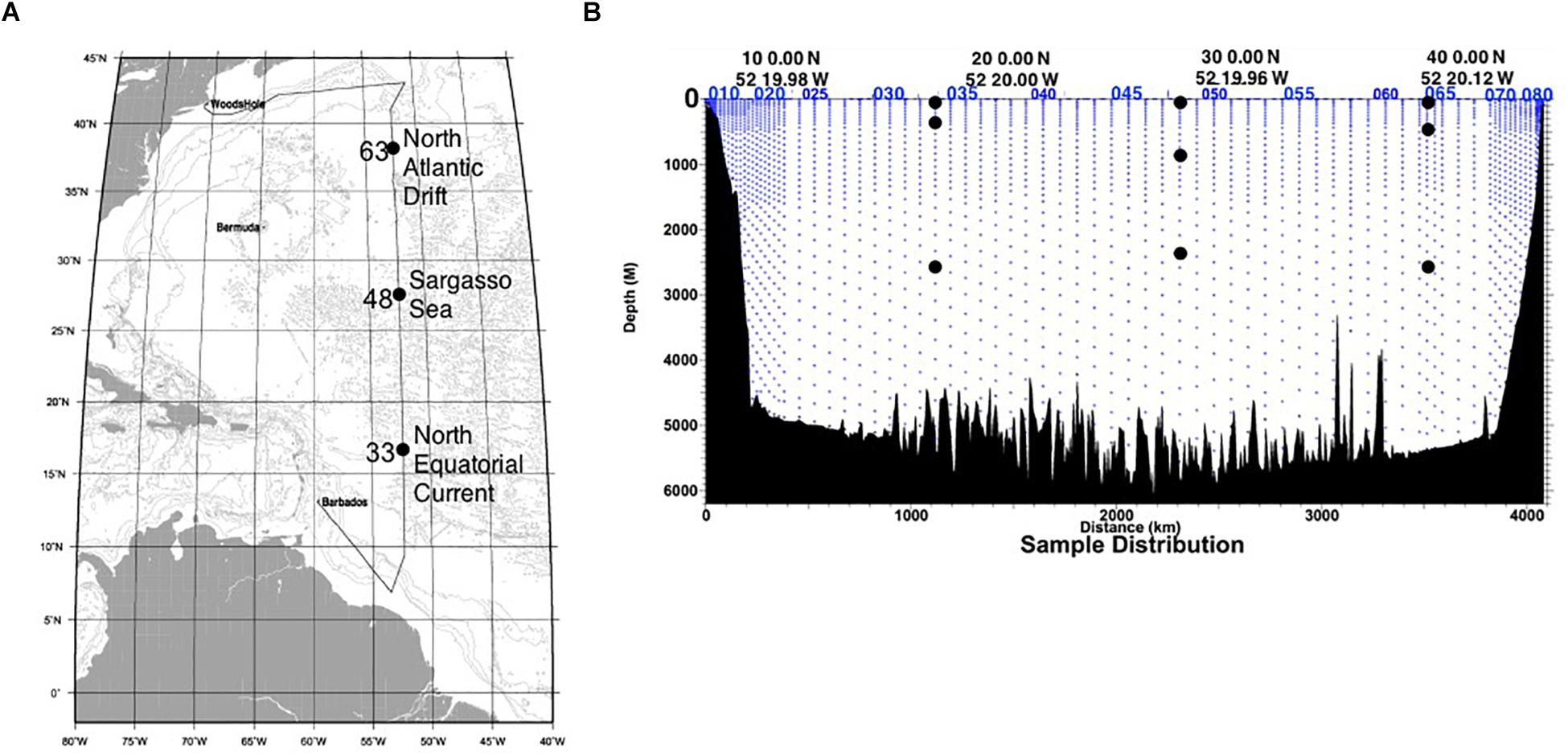
Figure 1. (A) Map of the CLIVAR A20 cruise track, with each SIP sampling station indicated. (B) Cross section of the sampling depths of the CLIVAR 20 cruise (blue dots). The sites of the water collected for SIP microcosms are indicated (black dots). Note: Water samples were not available at all depths and stations during this cruise of opportunity. These figures are modified from the CLIVAR A20 2012 cruise report (http://cchdo.ucsd.edu, Expocode: 33AT20120419do).
DNA was purified using phenol-chloroform methods and 12C/13C-DNA was separated by isopycnic cesium chloride gradient ultracentrifugation at 200,000 × g for 36 h, using 13C-labeled H. salinarum or E. coli DNA as a carrier (Gallagher et al., 2005). Carrier DNA produces a visible 13C-lower band in the gradient, removing the need to fractionate the gradient and increasing sensitivity (Gallagher et al., 2010; Kerkhof et al., 2011; Seyler et al., 2014, 2018; Tuorto et al., 2014). Fluorescently-labeled (using ethidium bromide) 12C-(upper) and 13C-(lower) bands were collected by pipette. Upper and lower bands (1 μl each) were amplified using nested PCR with 5′-fluorescently- labeled, 16S rRNA archaea-specific 21F (5′-TCCGGTTGATCCYGCCGG)/958R (5′-YCCGGCGTTGAMTCCAATT) primers using the following protocol: 94°C for 5 min, 23 cycles of (94°C for 1 min, 55°C for 2 min, 72°C for 3 min). A 1 μl aliquot was then taken from this reaction and amplified using thaum/cren/bathyarchaeota-specific 7F (5′-TTCCGGTTGATCCYGCCGGACC)/518R (5′-GCTGGTWTTACCGCGGCGGCTGA) primers using the following protocol: 94°C for 5 min, 30 cycles of (94°C for 1 min, 73°C for 2 min, 72°C for 3 min), 72°C for 10 min (Perevalova et al., 2003; Ionescu et al., 2008; Seyler et al., 2018). Upper and lower bands were also amplified using bacteria-specific 27F (5′AGAGTTTGATCMTGGCTCAG)/1100R (5′-GGGTTG CGCTCGTTG) primers (Ludwig, 2007) in a 2-step process: step 1 at 94°C for 1 min 30 s, 20 cycles of (94°C for 30 s, 57°C for 30 s, 72°C for 1 min 10 s); 1 μl aliquot amplified in step 2 at 94°C for 1 min 30 s, 25 cycles of (94°C for 30 s, 57°C for 30 s, 72°C for 1 min 10 s), 72°C for 10 min. Finally, upper and lower bands were amplified for eukaryotes using 18S rRNA universal eukaryote 328F (5′-ACCTGGTTGATCCTGCCAG) (Medlin et al., 1988)/516R (5′-ACCAGACTTGCCCTCC) (Amann et al., 1990) primers in a 2-step PCR: 94°C for 1 min 30 s, 15 cycles of [94°C for 45 s, 56°C for 45 s, 72°C for 2 min], followed by 94°C for 1 min 30 s, 30 cycles of [94°C for 45 s, 56°C for 45 s, 72°C for 2 min], 72°C for 10 min. Amplicons were digested with MnlI (prokaryotic amplicons) or HaeIII (eukaryotic amplicons) in 20 μl volumes for 6 h at 37°C, then precipitated using sodium acetate and 95% ethanol (McGuinness et al., 2006). Precipitated DNA was dried and re-suspended in 19.7 μl de-ionized formamide with 0.3 μl ROX 500 size standard (Applied Biosystems). TRFLP fingerprinting (Avaniss-Aghajani et al., 1994) was carried out on an ABI 310 genetic analyzer (Applied Biosystems, Foster City, CA, United States) using GENESCAN software. Peak detection was set at 25 arbitrary fluorescent units. Each resulting peak was considered an operational taxonomic unit (OTU).
An 18S amplicon clonal library was constructed using the Topo TA cloning kit according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, United States). 192 recombinant clones from the 13C-labeled bands were screened in a multiplex format (Babcock et al., 2007), to determine portions of the 18S rRNA genes which corresponded to the TRF of the 13C-eukaryotic profile. Thirteen 18S rRNA genes that matched TRFs of interest were sequenced via Sanger methods using M13 primers (GENEWIZ, Inc., NJ, United States), producing 9 unique sequences (<99% similarity to each other) between 520 and 603 bp. These sequences were compared to the Silva-ARB Eukaryotic database by discontinuous MEGABLAST. Best hits were used to construct a phylogenetic tree using 460 unambiguously aligned base pairs and maximum likelihood methods (Guindon et al., 2010) in Geneious 11.1.4. Clone sequences were submitted to GenBank (accession numbers KT749976–KT749984).
A minimum incubation time of 48 h was required to unambiguously detect 13C-archaeal DNA synthesis in the 13C-carrier band (Seyler et al., 2018). In 7 microcosms (6 epipelagic, 1 bathypelagic), no signal could be detected in the 13C-band (no PCR amplicon could be detected by electrophoresis gel, and the resulting digests produced no TRFLP peaks). In 7 microcosms from mesopelagic or bathypelagic samples, signal from 13C-urea was only detected in archaea. In one bathypelagic microcosm (Station 63), signal from 13C-urea was only detected in bacteria. Three mesopelagic microcosms had both archaea and bacteria 13C signals resulting from 13C-urea amendments. In all twelve 13C-acetate mesopelagic and bathypelagic microcosms, both archaea and bacteria incorporated 13C, except bathypelagic microcosms at Station 63, where only bacterial uptake of 13C-acetate was detected (Figure 2). No 13C uptake was detected in 12C controls.
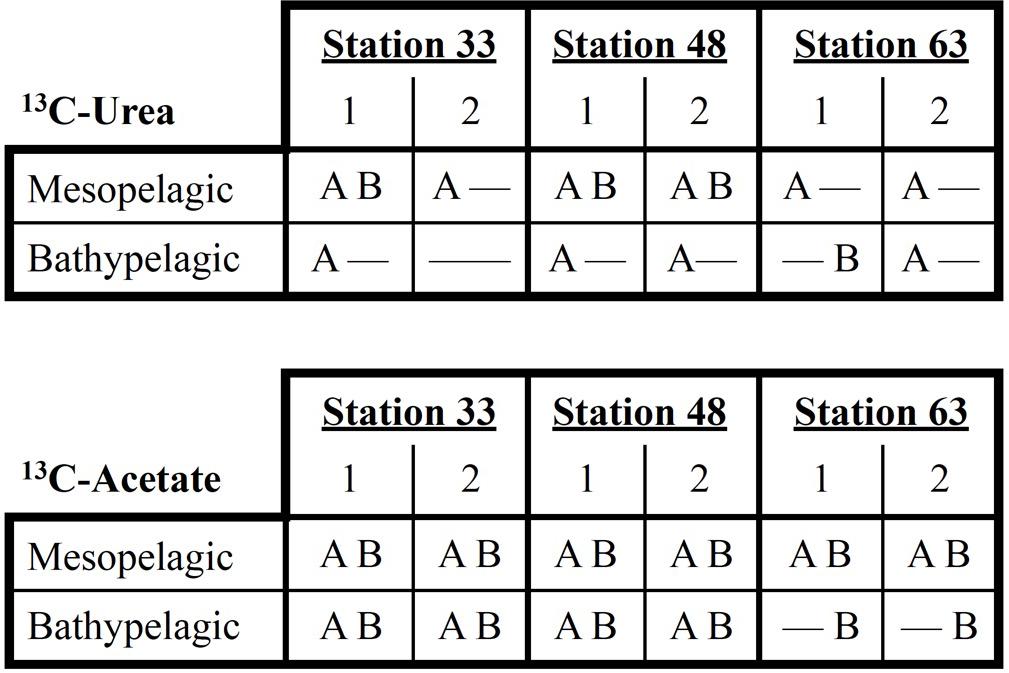
Figure 2. Patterns of 13C-incorporation by archaea (A) and bacteria (B) based on 13C-urea or 13C-acetate additions in meso-bathypelagic samples. No incorporation of 13C was detected in any microcosms established with water from the epipelagic zone.
TRFLP analysis detected 37 eukaryotic OTUs, grouped based on whether the 13C signal was also detected in the 13C-archaea and 13C-bacteria, in the 13C-archaea alone, in the 13C-bacteria alone, or without a 13C signal from either prokaryotic group (Figure 3). Seventeen eukaryotic OTUs were detected in SIP microcosms with both bacterial and archaeal 13C uptake. Ten eukaryotic OTUs were associated exclusively with 13C-archaea (archaevores), and 7 eukaryotic OTUs were only observed with 13C-bacteria (bacterivores). Three eukaryotic OTUs were detected in samples without any archaea or bacterial 13C uptake (osmotrophs). The total number of observed archaeal-specific predator OTUs was higher than that of bacterial-specific predators, however, if the total adjusted TRFLP peak area was considered a proxy for relative abundance, the non-specific predators tended to constitute a greater percentage of the overall 13C-eukaryotic community profile (Figure 3).
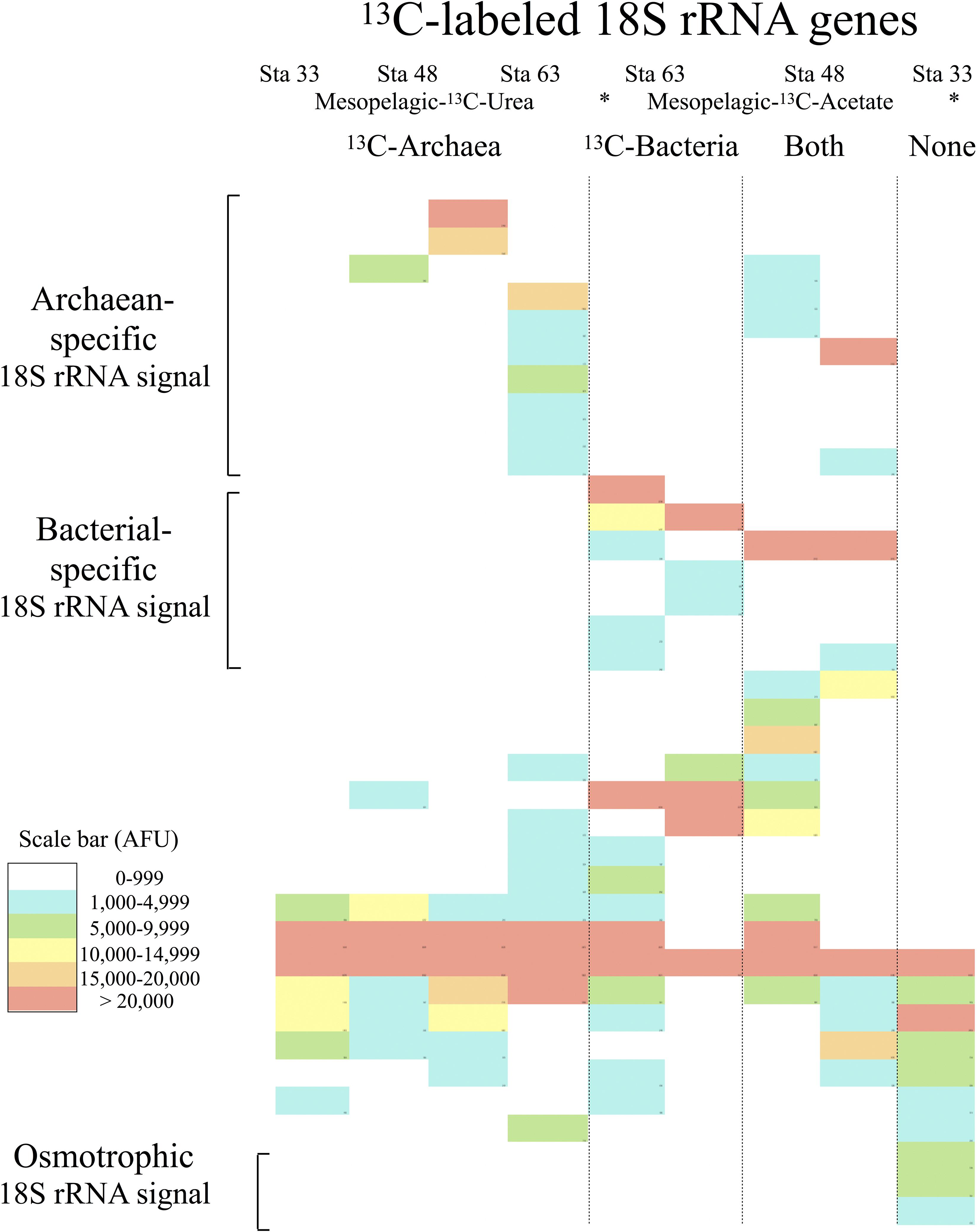
Figure 3. Heat map of TRFLP peaks from 13C-labeled 18S rRNA genes detected in SIP microcosms after 48 h. The station numbers and the 13C substrates used for labeling the archaea or bacteria in the microcosms are indicated on top. TRFLP peaks are grouped by predation pattern (i.e., only 13C-archaea, only 13C-bacteria, both 13C-archaea and 13C-bacteria, and neither the 13C-archaea nor 13C-bacteria). Intensity of cell shading corresponds to the normalized TRFLP peak area for the overall community profiles in arbitrary fluorescence units (scale bar-AFUs).
Nine rRNA genes with 13C-eukaryotic TRF peaks were identified in the clonal library. Of these, three were found exclusively in microcosms where bacteria incorporated 13C signal (TRFs 182, 201, 269), three were found only in microcosms where archaea incorporated 13C signal (TRFs 329, 333, 339), two were found in both (TRFs 273, 305), and one was found in the microcosm where neither bacteria nor archaea incorporated 13C signal (TRF 293). An 18S phylogenetic tree (Figure 4) indicated bacterial uptake-associated eukaryotes were associated with relatives of the parasitic dinoflagellate Syndiniales group, a flagellate (Ancyromonas sp.), and an apuzoan (Amastigomonas sp.). The archaea uptake-associated eukaryotes were associated with radiolarians or a dinoflagellate group (Heterocapsa sp.). Non-specific prokaryotic predators were related to a bicosoecida group (Cafeteria sp.) and a marine fungus (Paraphysoderma sedebokerense). The 18S rRNA gene from the eukaryote believed to be osmotrophic aligned to another flagellate group (Caecitellus paraparvulus).
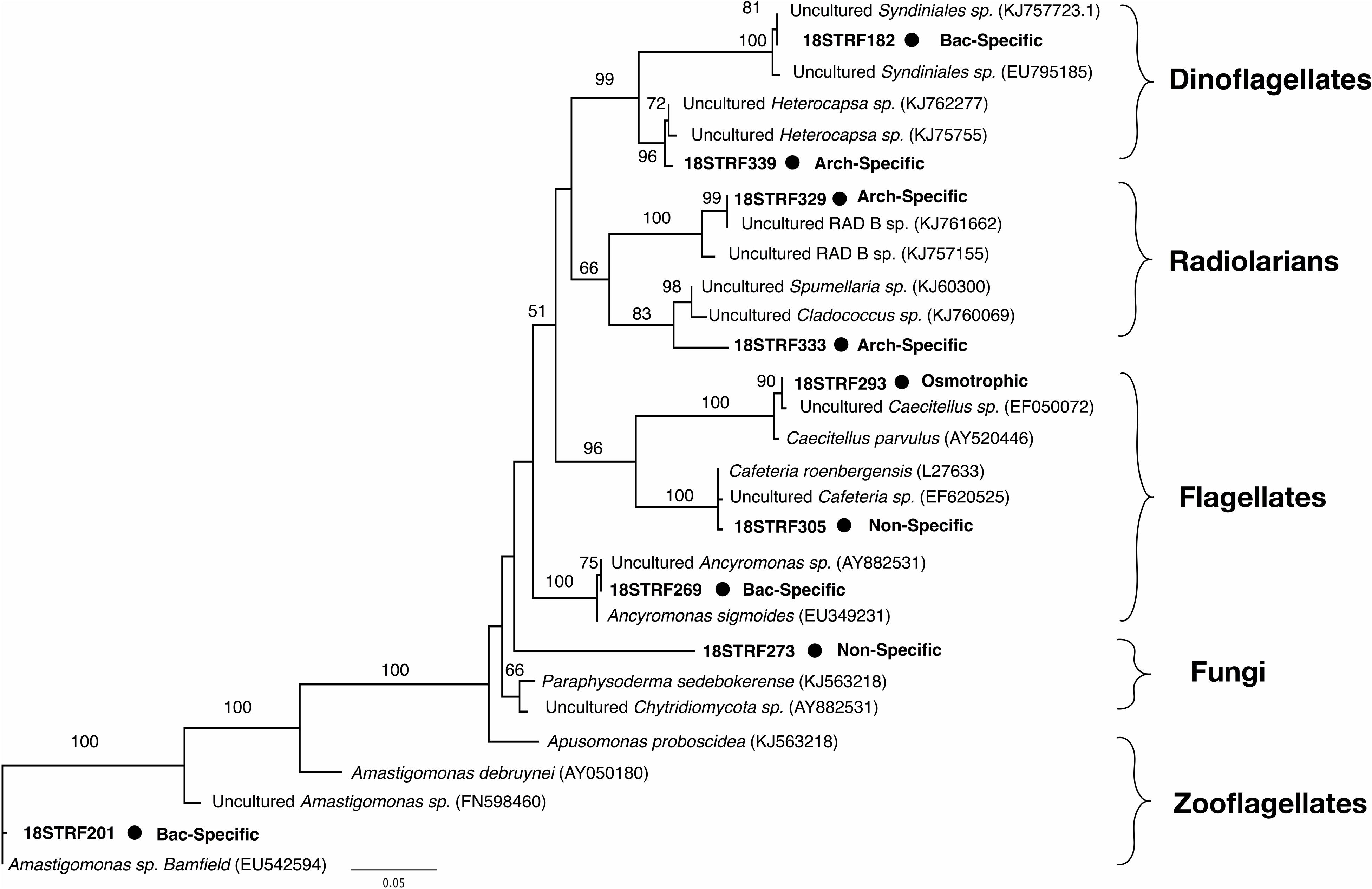
Figure 4. Maximum likelihood phylogenetic tree of 13C-labeled 18S rRNA genes corresponding with nine of the TRFLP peaks shown in Figure 3. The tree is based on 460 unambiguously aligned base pairs. The size of the TRFLP peak, the 13C-predation pattern, and the bootstrap values for 100 iterations are all indicated.
Interactions between prokaryotes, protist predators, and dissolved organic carbon pools in the world’s oceans may have far-reaching effects on global biogeochemical cycles (Kujawinski, 2011). Deep ocean waters represent the largest pool of dissolved organic carbon (Hansel and Carlson, 1998), the respiration of which accounts for up to one third of oceanic biological CO2 production (Del Giorgio and Duarte, 2002; Arístegui et al., 2005). The factors which control microbial community dynamics, carbon respiration, and mortality in the deep ocean are therefore of great importance to the global carbon cycle. Archaea, particularly thaumarchaea belonging to the TACK superphylum, have been found to dominate prokaryotic communities in the deep ocean (Herndl et al., 2005; Varela et al., 2008). Yet, archaeal utilization of dissolved and particulate organic matter in the deep sea remains poorly constrained, and even less is known about the relationship of predator grazing to archaeal function in the deep sea.
“Bottom-up” controls such as the availability of nutrients (Wilkins et al., 2012) and the character of dissolved organic matter (Smriga et al., 2016) may have significant effects on the community structure of prokaryotes. Past studies demonstrated that archaea take up certain carbon sources at an equal or higher rate than bacteria in deep ocean waters (e.g., Ouverney and Fuhrman, 2000; Teira et al., 2006; Kirchman et al., 2007). Thaumarchaea are known to incorporate urea as a carbon source (Seyler et al., 2014, 2018) and they may use it as an alternative nitrogen source to ammonium to fuel nitrification (Alonso-Sáez et al., 2012). In many of our mesopelagic and bathypelagic microcosms, 13C-urea uptake was detected in archaea but not bacteria, suggesting that archaea can outcompete bacteria for urea at those particular samples, at the concentration tested here. Competition for substrates in oligotrophic environments where nutrients and energy are limiting seems to favor archaea over bacteria (Jardillier et al., 2005; Vuillemin et al., 2019). Additionally, PCR amplification of key genes in the Thaumarchaeota suggest that ecological functioning of this group may shift with depth (Hu et al., 2011). One possibility of our observed resource partitioning for 13C-urea by archaea may be our SIP incubation conditions. That is, by conducting the SIP incubations at sea surface temperature, rather than at in situ mesopelagic and bathypelagic temperatures, we may have stimulated or accelerated activity in some clades. However, an increase in temperature cannot bestow metabolic capability where none had previously existed; we therefore feel confident in stating that archaea are capable of outcompeting bacteria for urea under the conditions provided in our microcosms [For further discussion, see Seyler et al. (2018)].
In addition to resource partitioning, our SIP experiment indicated that selective predation may partly account for archaeal/bacterial co-existence. The main causes of mortality in marine microorganisms are viruses and grazing by phagotrophic protists (Jardillier et al., 2005; Arístegui et al., 2009). Phagotrophic grazing- “top-down” population control- can structure the microbial community, activity, and physiology of prokaryotic prey (Šimek et al., 1997; Lin et al., 2007; Saleem et al., 2012), including shifts in community assemblages as a result of the prey preferences of protists (Jardillier et al., 2005; Jezbera et al., 2006). Protistan species can act as bacterivore, herbivore, carnivore of other protozoa and/or metazoans (e.g., nauplii, copepodite, and metazoan larvae), and can even consume dissolved organic matter through saprotrophy or osmotrophy (Swanberg and Caron, 1991; Caron et al., 2009; Worden et al., 2015). Specific predation of archaea has been addressed in very few studies, e.g., at marine oxic-anoxic interfaces (Anderson et al., 2012) and in freshwater (Ballen-Segura et al., 2017). Here, we demonstrate both archaeal and bacterial-specific predation by a subset of eukaryotic predators in the mesopelagic and bathypelagic water column. However, our data suggest the archaea-specific predators represent a small percentage of the overall the eukaryotic predatory community compared to non-specific microbial predators.
This study demonstrates that certain 13C-eukaryotic OTUs are associated with either 13C-archaeal or 13C-bacterial signals. This transfer of 13C-carbon from prokaryote to eukaryote could result from predation or by exudation/episymbiosis. Often ciliates or flagellates have surface associations with bacteria for detoxification or defense (Petroni et al., 2000; Edgcomb et al., 2011). Although we did not test for the presence of prokaryotes in food vacuoles or on the external surfaces of deep-sea eukaryotes by microscopy, an assessment of whether predation or symbiosis is the predominant mechanism for carbon transfer can be made using the SIP results directly. To a first order, if predation is the dominant mechanism, we can presume that in many microcosms, the 13C-carbon will pass directly from substrate to prokaryote to eukaryote predator. Therefore, a much higher 13C-eukaryotic signal compared to a 13C-prokaryotic signal will be observed in the PCR product from the 13C-labeled DNA. On the other hand, if growth of the eukaryote is predicated on growth of an epibiont, there should always be a comparable PCR signal from both prokaryote and eukaryote within the 13C-band. All of our SIP microcosms had significantly higher eukaryotic signal than prokaryotic signal in the 13C-band (Supplementary Figure S1), suggesting a tight coupling between these groups that is likely a predation signal.
If differential predation influences the coexistence of archaea and bacteria, the mechanisms used by predators to discern between the archaea and bacteria are largely unexplored. It is known that prey item size, surface properties, concentration, and particle attachment can be major controlling elements selecting for prey items by eukaryotic grazers (Lampert, 1987; Gonzalez et al., 1990, 1993; Jürgens and Matz, 2002; Thurman et al., 2010; Tuorto and Taghon, 2014). Recent studies suggest that marine archaea are significantly smaller than bacteria on average (Stepanauskas et al., 2017; Santoro et al., 2018). Aggregation on detrital particles, as observed in euryarchaea (Orsi et al., 2015), may provide refuge from predation (Langenheder and Jürgens, 2001; Šimek et al., 2001), as well as a host of biochemical interactions, exopolymers, and toxicity (Jürgens and Matz, 2002). Further studies will be necessary to explore the mechanisms of selective predation in deep ocean settings.
Predation by protists is also a key mechanism for the transfer of dissolved organic carbon to higher trophic levels in the microbial loop (Azam et al., 1983; Jürgens and Massana, 2008; Worden et al., 2015). The structure of the marine microbial food web and efficiency of carbon transfer are major determinants in the rate of oceanic carbon sequestration and the type/amount of higher consumers in the ecosystem (Limardo and Worden, 2015). Although prokaryotes mobilize dissolved carbon into biomass, it is the heterotrophic strategies of small eukaryotic consumers that determine whether the net flow of organic matter will ultimately end up as a trophic “link” (i.e., trophic transfer to top predators and export) versus a trophic “sink” (i.e., be remineralized as dissolved organic matter or CO2). The archaeal contribution to the microbial loop is still poorly understood and, consequently, often easy to ignore. SIP studies such as described here can map trophic interactions between microbial predators and prey to predict the fate of organic carbon in marine ecosystems.
The datasets generated for this study can be found in GenBank, KT749976–KT749984.
LS and LK wrote the original manuscript. LS, LM, and LK designed the SIP experiments. LS set up microcosms and performed all the laboratory work. ST and LM performed the data analysis and interpretation of data. DG was co-chief scientist on R/V Atlantis during CLIVAR leg A20 and supervised all onboard research. LK conceived the original idea, oversaw all laboratory work, and provided data analysis.
This research was made possible through the support of the U.S Global Ocean Carbon and Repeat Hydrography Program and NSF Ocean Technology and Interdisciplinary Program grant (#1131022) to LK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank Dr. Michael McCartney for allowing LS to participate as a graduate student researcher onboard the R/V Atlantis during CLIVAR leg A20.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00555/full#supplementary-material
FIGURE S1 | Top half of gel showing amplification of 18S rRNA genes from either the 12C-band or the 13C-band within the SIP microcosms indicated. Lanes are (A) empty; (B) lambda HinDIII ladder; (C) negative PCR control; (D) 13C-Halobacterium carrier; (E) station 63–2585 m, 13C-acetate amended, 12C-band; (F) station 63–2585 m, 13C-acetate amended, 13C-band; (G) station 48–2335 m, 13C-acetate amended, 12C-band; (H) station 48–2335 m, 13C-acetate amended, 13C-band; (I) station 48–2335 m, 13C acetate-amended, 13C-band (duplicate Niskin cast); (J) empty; (K) positive control; (L) empty. Bottom half gel of indicates amplification of 16S rRNA genes from the identical samples as the 18S rRNA amplification above with the exception that the 12C-bands (Lanes E, G) were diluted 1:10 prior to amplification of the 16S rRNA genes. All 13C-bands were all amplified for 16S rRNA genes without dilution as above. The concentration of PCR product from each amplification is indicated above the band.
Abreu, C., Jurgens, G., De Marco, P., Saana, A., and Bordalo, A. A. (2001). Crenarchaeota and euryarchaeota in temperate estuarine sediments. J. Appl. Microbio. 90, 713–718. doi: 10.1046/j.1365-2672.2001.01297.x
Alonso-Sáez, L., Waller, A. S., Mende, D. R., Bakker, K., Farnelid, H., Yager, P. L., et al. (2012). Role for urea in nitrification by polar marine Archaea. PNAS 109, 17989–17994. doi: 10.1073/pnas.1201914109
Amann, R. I., Binder, B. J., Olson, R. J., Chisholm, S. W., Devereux, R., and Stahl, D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925.
Amano-Santo, C., Akiyama, S., Uchida, M., Shimada, K., and Utsumi, M. (2013). Archaeal distribution and abundance in water masses of the Arctic Ocean, Pacific sector. Aquat. Microb. Ecol. 69, 101–112. doi: 10.3354/ame01624
Anderson, R., Winter, C., and Jürgens, K. (2012). Protist grazing and viral lysis as prokaryotic mortality factors at Baltic Sea oxic-anoxic interfaces. MEPS 467, 1–14. doi: 10.3354/meps10001
Arístegui, J., Duarte, C. M., Gasol, J. M., and Alonso-Sáez, L. (2005). Active mesopelagic prokaryotes support high respiration in the subtropical northeast Atlantic Ocean. Geophys. Res. Lett. 32, 1–4.
Arístegui, J., Gasol, J. M., Duarte, C. M., and Herndl, G. J. (2009). Microbial oceanography of the dark ocean’s pelagic realm. Limnol. Oceanogr. 54, 1501–1529. doi: 10.4319/lo.2009.54.5.1501
Avaniss-Aghajani, E., Jones, K., Chapman, D., and Brunk, C. (1994). A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. Biotechniques 17, 144–146.
Azam, F., Fenchel, T., Field, J., Gray, J., Meyer-Reil, L. A., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Babcock, D. A., Wawrik, B., Paul, J. H., McGuinness, L., and Kerkhof, L. J. (2007). Rapid screening of a large insert BAC library for specific 16S rRNA genes using TRFLP. J. Microbiol. Methods 71, 156–161. doi: 10.1016/j.mimet.2007.07.015
Ballen-Segura, M., Felip, M., and Catalan, J. (2017). Some mixotrophic flagellate species selectively graze on archaea. Appl. Env. Microbiol. 83:e2317-16. doi: 10.1128/AEM.02317-16
Biddle, J. F., Fitz-Gibbon, S., Schuster, S. C., Brenchley, J. E., and House, C. H. (2008). Metagenomic signatures of the peru margin susbeafloor biosphere show a genetically distinct environment. PNAS 105, 10583–10588. doi: 10.1073/pnas.0709942105
Birtrim, S. B., Donohue, T. J., Handelsman, J., Roberts, G. P., and Goodman, R. M. (1997). Molecular phylogeny of Archaea from soil. PNAS 94, 277–282. doi: 10.1073/pnas.94.1.277
Caron, D. A., Worden, A. Z., Countway, P. D., Demir, E., and Heidelberg, K. B. (2009). Protists are microbes too: a perspective. ISME J. 3, 4–12. doi: 10.1038/ismej.2008.101
Chesson, P. (2000). Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. doi: 10.1146/annurev.ecolsys.31.1.343
Chesson, P., and Huntley, N. (1997). The roles of harsh conditions in the dynamics of ecological communities. Am. Nat. 150, 519–553. doi: 10.1086/286080
Del Giorgio, P. A., and Duarte, C. M. (2002). Respiration in the open ocean. Nature 420, 379–384. doi: 10.1038/nature01165
DeLong, E. F. (1992). Archaea in coastal marine environments. PNAS 89, 5685–5689. doi: 10.1073/pnas.89.12.5685
DeLong, E. F., Wu, K. Y., Prézelin, B. B., and Jovine, R. V. M. (1994). High abundance of Archaea in Antarctic marine picoplankton. Nature 37, 695–697. doi: 10.1038/371695a0
Edgcomb, V. P., Breglia, S. A., Yubuki, N., Beaudoin, D., Patterson, D. J., Leander, B. S., et al. (2011). Identity of epibiotic bacteria on symbiontid euglenozoans in O2-depleted marine sediments: evidence for symbiont and host co-evolution. ISME J. 5, 231–243. doi: 10.1038/ismej.2010.121
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. PNAS 102, 14683–14688. doi: 10.1073/pnas.0506625102
Fuhrman, J. A., McCallum, K., and Davis, A. A. (1992). Novel major archaebacterial group from marine plankton. Nature 356, 148–149. doi: 10.1038/356148a0
Gallagher, E., McGuinness, L., Phelps, C., Young, L. Y., and Kerkhof, L. J. (2005). 13C-carrier DNA shortens the incubation time needed to detect benzoate-utilizing denitrifying bacteria by stable-isotope probing. Appl. Environ. Microbiol. 71, 5192–5196. doi: 10.1128/aem.71.9.5192-5196.2005
Gallagher, E. M., Young, L. Y., McGuinness, L. M., and Kerkhof, L. J. (2010). Detection of 2,4,6-trinitrotoluene anaerobic bacteria by 15N and 13C incorporation. Appl. Environ. Microbiol. 76, 1695–1698. doi: 10.1128/AEM.02274-09
Gonzalez, J. M., Portilo, M. C., and Saiz-Jimenez, C. (2006). Metabolically active Crenarchaeota in Altamira cave. Naturwissenschaften 93, 42–45. doi: 10.1007/s00114-005-0060-3
Gonzalez, J. M., Sherr, E. B., and Sherr, B. F. (1990). Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl. Environ. Microbiol. 56, 583–589.
Gonzalez, J. M., Sherr, E. B., and Sherr, B. F. (1993). Differential feeding by marine flagellates on growing versus starving, and on motile versus nonmotile, bacterial prey. Mar. Ecol. Prog. Ser. 102, 257–267. doi: 10.3354/meps095257
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms, and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guy, L., and Ettema, T. J. G. (2011). The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 19, 580–587. doi: 10.1016/j.tim.2011.09.002
Hallam, S. J., Mincer, T. J., Schleper, C., Preston, C. M., Roberts, K., Richardson, P. M., et al. (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:e95. doi: 10.1371/journal.pbio.0040095
Hansel, D. A., and Carlson, C. A. (1998). Deep-ocean gradients in the concentration of dissolved organic carbon. Nature 395, 263–266. doi: 10.1038/26200
Herndl, G. J., Reinthaler, T., Teira, E., van Aken, H., Veth, C., Pernthaler, A., et al. (2005). Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71, 2303–2309. doi: 10.1128/aem.71.5.2303-2309.2005
Hu, A., Jiao, N., Zhang, R., and Yang, Z. (2011). Niche partitioning of marine group I Crenarchaeota in the euphotic and upper mesopelagic zones of the East China Sea. Appl. Environ. Microbiol. 77, 7469–7478. doi: 10.1128/AEM.00294-11
Ingalls, A. E., Shah, S. R., Hansman, R. L., Aluwihare, L. I., Santos, G. M., Druffel, E. R. M., et al. (2006). Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. PNAS 103, 6442–6447. doi: 10.1073/pnas.0510157103
Ionescu, D., Penno, S., Haimovich, M., Rihtman, B., Goodwin, A., Schwartz, D., et al. (2008). Archaea in the Gulf of Aqaba. FEMS Microbiol. 69, 425–438. doi: 10.1111/j.1574-6941.2009.00721.x
Jardillier, L., Bettarel, Y., Richardot, M., Bardot, C., Amblard, C., Sime-Ngando, T., et al. (2005). Effects of viruses and predators on prokaryotic community composition. Microbial. Ecol. 50, 557–569. doi: 10.1007/s00248-005-5030-y
Jezbera, J., Hornák, K., and Šimek, K. (2006). Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ. Microbiol. 8, 1330–1339. doi: 10.1111/j.1462-2920.2006.01026.x
Jürgens, K., and Massana, R. (2008). “Protist grazing on marine bacterioplankton,” in Microbial Ecology of the Oceans, 2 Edn, ed. D. L. Kirchman (Hoboken, NJ: Wiley), 383–441. doi: 10.1002/9780470281840.ch11
Jürgens, K., and Matz, C. (2002). Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Leeuwenhoek 81, 413–434.
Karner, M. B., DeLong, E. F., and Karl, D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
Kerkhof, L. J., Williams, K. H., Long, P. E., and McGuinness, L. R. (2011). Phase preference by active, acetate-utilizing bacteria at the rifle, CO integrated field research challenge site. Environ. Sci. Technol. 45, 1250–1256. doi: 10.1021/es102893r
Kirchman, D. L., Elifantz, H., Dittel, A. I., Malmstrom, R. R., and Cottrell, M. T. (2007). Standing stocks and activity of Archaea and Bacteria in the western Arctic Ocean. Limnol. Oceanogr. 52, 495–507. doi: 10.4319/lo.2007.52.2.0495
Könneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Kujawinski, E. B. (2011). The impact of microbial metabolism on marine dissolved organic matter. Annu. Rev. Mar. Sci. 3, 567–599. doi: 10.1146/annurev-marine-120308-081003
Lampert, W. (1987). “Predictability in lake ecosystems: the role of biotic interactions,” in Potential and Limitations of Ecosystem Analysis (Ecological Studies 61), eds E. D. Schultz and H. Zwölfer (Berlin: Springer-Verlag), 333–346. doi: 10.1007/978-3-642-71630-0_16
Langenheder, S., and Jürgens, K. (2001). Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol. Oceanogr. 46, 121–134. doi: 10.4319/lo.2001.46.1.0121
Lazar, C. S., Baker, B. J., Seitz, K., Hyde, A. S., Dick, G. J., Hinrichs, K. U., et al. (2016). Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ. Microbiol. 18, 1200–1211. doi: 10.1111/1462-2920.13142
Limardo, A. J., and Worden, A. Z. (2015). Microbiology: exclusive networks in the sea. Nature 522, 36–37. doi: 10.1038/nature14530
Lin, X., Scranton, M. I., Varela, R., Chistoserdov, A., and Taylor, G. T. (2007). Compositional responses of bacterial communities to redox gradients and grazing in the anoxic Cariaco Basin. Aquat. Microb. Ecol. 47, 57–72. doi: 10.3354/ame047057
Ludwig, W. (2007). Nucleic acid techniques in bacterial systematics and identification. Int. J. Food Microbiol. 120, 225–236. doi: 10.1016/j.ijfoodmicro.2007.06.023
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
Massana, R., DeLong, E. F., and Pedrós-Alió, C. (2000). A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Environ. Microbiol. 66, 1777–1787. doi: 10.1128/aem.66.5.1777-1787.2000
Massana, R., Murray, A. E., Preston, C. M., and DeLong, E. F. (1997). Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara channel. Appl. Environ. Microbiol. 63, 50–56.
McGuinness, L. M., Salganik, M., Vega, L., Pickering, K. D., and Kerkhof, L. J. (2006). Replicability of bacterial communities in denitrifying bioreactors as measured by PCR/T-RFLP analysis. Environ. Sci. Technol. 40, 509–515. doi: 10.1021/es050900l
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Moin, N. S., Nelson, K. A., Bush, A., and Bernhard, A. E. (2009). Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl. Environ. Microbiol. 75, 7461–7468. doi: 10.1128/AEM.01001-09
Mosier, A. C., and Francis, C. A. (2008). Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10, 3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x
Murray, A. E., Preston, C. M., Massana, R., Taylor, L. T., Blakis, A., Wu, K., et al. (1998). Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island. Antarctica. Appl. Environ. Microbiol. 64, 2585–2595.
Nelson, K. A., Moin, N. S., and Bernhard, A. E. (2009). Archaeal diversity and the prevalence of Crenarchaeota in salt marsh sediments. Appl. Environ. Mircobiol. 75, 4211–4215. doi: 10.1128/AEM.00201-09
Orsi, W. D., Smith, J. M., Wilcox, H. M., Swalwell, J. E., Carini, P., Worden, A. Z., et al. (2015). Ecophysiology of uncultivated marine euryarchaea is linked to particulate organic matter. ISME J. 9, 1747–1763. doi: 10.1038/ismej.2014.260
Ouverney, C. C., and Fuhrman, J. A. (2000). Marine planktonic archaea take up amino acids. Appl. Environ. Microbiol. 66, 4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000
Park, B. J., Park, S. J., Yoon, D. N., Schouten, S., Sinninghe Damsté, J. S., and Rhee, S. K. (2010). Cultivation of autotrophic ammonia-oxidzing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl. Environ. Microbiol. 76, 7575–7587. doi: 10.1128/AEM.01478-10
Park, S. J., Kim, J. G., Jung, M. Y., Kim, S. J., Cha, I. T., Kwon, K. K., et al. (2012). Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus koreensis” AR1, from marine sediment. J. Bacteriol. 194, 6940–6941. doi: 10.1128/jb.01857-12
Perevalova, A. A., Lebedinsky, A. V., Bonch-Osmolovskaya, E. A., and Chernyh, N. A. (2003). Detection of hyperthermophilic Archaea of the genus Desulfurococcus by hybridization with oligonucleotide probes. Microbiology 72, 340–346.
Petroni, G., Spring, S., Schleifer, K. H., Verni, F., and Roasati, G. (2000). Defensive extrusive ectosymbionts of Euplotidium (Ciliophora) that contain microtubule-like structures are bacteria related to Verrucomicrobia. PNAS 97, 1813–1817. doi: 10.1073/pnas.030438197
Qin, W., Amin, S. A., Martins-Habbena, W., Walker, C. B., Urakawa, H., Devol, A. H., et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. PNAS 111, 12504–12509. doi: 10.1073/pnas.1324115111
Radajewski, S., Ineson, P., Parekh, N. R., and Murrell, J. C. (1999). Stable-isotope probing as a tool in microbial ecology. Nature 403, 646–649. doi: 10.1038/35001054
Richardson, P., Armstrong, R., and Goldman, C. R. (1970). Contemporaneous disequilibrium, a new hypothesis to explain the “Paradox of the Plankton.” PNAS 67, 1710–1714. doi: 10.1073/pnas.67.4.1710
Saleem, M., Fetzer, I., Dormann, C. F., Harms, H., and Chatzinotas, A. (2012). Predator richness increases the effect of prey diversity on prey yield. Nat. Commun. 3:1305. doi: 10.1038/ncomms2287
Santoro, A. E., Richter, R. A., and Dupont, C. L. (2018). Planktonic marine archaea. Ann. Rev. Mar. Sci. 11, 131–158. doi: 10.1146/annurev-marine-121916-063141
Scheffer, M., and Carpenter, S. R. (2003). Catastrophic regime shifts in ecosystems: linking theory to observation. Trends Ecol. Evol. 18, 648–656. doi: 10.1016/j.tree.2003.09.002
Seyler, L. M., McGuinness, L. M., and Kerkhof, L. J. (2014). Crenarchaeal heterotrophy in salt marsh sediments. ISME J. 8, 1534–1543. doi: 10.1038/ismej.2014.15
Seyler, L. M., McGuinness, L. R., Gilbert, J. A., Biddle, J. F., Gong, D., and Kerkhof, L. J. (2018). Discerning autotrophy, mixotrophy, and heterotrophy in marine TACK archaea from the North Atlantic. FEMS Microbiol. Ecol. 94:fiy014. doi: 10.1093/femsec/fiy014
Šimek, K., Pernthaler, J., Weinbauer, M. G., Hornák, K., Dolan, J. R., Nedoma, J., et al. (2001). Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67, 2723–2733. doi: 10.1128/aem.67.6.2723-2733.2001
Šimek, K., Vrba, J., Pernthaler, J., Posch, T., Hartman, P., Nedoma, J., et al. (1997). Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Environ. Microbiol. 63, 587–595.
Smriga, S., Fernandez, V. I., Mitchell, J. G., and Stocker, R. (2016). Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. PNAS 113, 1576–1581. doi: 10.1073/pnas.1512307113
Søresnson, K. B., and Teske, A. (2006). Stratified communities of active archaea in deep marine subsurface sediments. Appl. Environ. Microbiol. 72, 4596–4603. doi: 10.1128/aem.00562-06
Stein, J. L., and Simon, M. I. (1996). Archaeal ubiquity. PNAS 93, 6228–6230. doi: 10.1073/pnas.93.13.6228
Stepanauskas, R., Fergusson, E. A., Brown, J., Poulton, N., Tupper, B., Labonté, J. M., et al. (2017). Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and vial particles. Nat. Commun. 8:84.
Swanberg, N. R., and Caron, D. A. (1991). Patterns of sardine feeding in epipelagic oceanic plankton. J. Plankton Res. 13, 287–312. doi: 10.1093/plankt/13.2.287
Teira, E., van Aken, H., Veth, C., and Herndl, G. J. (2006). Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol. Oceanogr. 51, 60–69. doi: 10.4319/lo.2006.51.1.0060
Teske, A., Hinrichs, K.-U., Edgcomb, V., Gomez, A. D. V., Kysela, D., Sylva, S. P., et al. (2002). Microbial diversity of hydrothermal sediments in the guaymas basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68, 1994–2007. doi: 10.1128/aem.68.4.1994-2007.2002
Thurman, J., Parry, J. D., Hill, P. J., and Laybourn-Parry, J. (2010). The filter-feeding ciliates Colpidium striatum and Tetrahymena pyriformis display selective feeding behaviors in the presence of mixed, equally-sized, bacterial prey. Protist 161, 577–588. doi: 10.1016/j.protis.2010.04.001
Treusch, A. H., Leininger, S., Kletzin, A., Schuster, S. C., Klenk, H. P., and Schleper, C. (2005). Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Envrion. Microbiol. 7, 1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x
Tuorto, S. J., Darias, P., McGuinness, L. R., Panikov, N., Zhang, T., Häggblom, M. M., et al. (2014). Bacterial genome replication at subzero temperatures in permafrost. ISME J. 8, 139–149. doi: 10.1038/ismej.2013.140
Tuorto, S. J., and Taghon, G. L. (2014). Rates of benthic bactivory of marine ciliates as a function of prey concentration. J. Exp. Mar. Biol. Ecol. 460, 129–134. doi: 10.1016/j.jembe.2014.06.014
Varela, M. M., van Aken, H. M., Sintes, E., and Herndl, G. J. (2008). Latitudinal trends of Crenarchaeota and bacteria in the meso- and bathypelagic water masses of the Eastern North Atlantic. Environ. Microbiol. 10, 110–124. doi: 10.1111/j.1462-2920.2007.01437.x
Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74. doi: 10.1126/science.1093857
Vetriani, C., Jannasch, H. W., MacGregor, B. J., Stahl, D. A., and Reysenbach, A. L. (1999). Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65, 4375–4384.
Vuillemin, A., Wankel, S. D., Coskun, O. K., Magritsch, T., Vargas, S., Estes, E. R., et al. (2019). Archaea dominate oxic subseafloor communities over multimillion-year time scales. Sci. Adv. 5:eaaw4108. doi: 10.1126/sciadv.aaw4108
Wilkins, D., Lauro, F. M., Williams, T. J., Demaere, M. Z., Brown, M. V., Hoffman, J. M., et al. (2012). Biogeographic partitioning of southern ocean microorganisms revealed by metagenomics. Environ. Microbiol. 15, 1318–1333. doi: 10.1111/1462-2920.12035
Keywords: archaea, bacteria, predation, competition, stable isotope probing, deep ocean
Citation: Seyler LM, Tuorto S, McGuinness LR, Gong D and Kerkhof LJ (2019) Bacterial and Archaeal Specific-Predation in the North Atlantic Basin. Front. Mar. Sci. 6:555. doi: 10.3389/fmars.2019.00555
Received: 02 May 2019; Accepted: 23 August 2019;
Published: 11 September 2019.
Edited by:
Fengping Wang, Shanghai Jiao Tong University, ChinaCopyright © 2019 Seyler, Tuorto, McGuinness, Gong and Kerkhof. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lauren M. Seyler, bG1zZXlsZXJAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.