- 1Life Sciences Department, Natural History Museum, London, United Kingdom
- 2Australian National Centre for Ocean Resources and Security, University of Wollongong, North Wollongong, NSW, Australia
- 3ABS-int, Bruges, Belgium
- 4Faculty of Pharmaceutical Sciences, Clinical Pharmacology and Pharmacotherapy, KU Leuven, Leuven, Belgium
- 5Institute of Marine and Environmental Law, University of Cape Town, Cape Town, South Africa
- 6Intergovernmental Oceanographic Commission of UNESCO, IOC Project Office for IODE, Oostende, Belgium
- 7Botanic Garden and Botanical Museum Berlin, Freie Universität Berlin, Berlin, Germany
- 8Senckenberg Research Institute and Natural History Museum, Frankfurt, Germany
- 9Institute for Ecology, Evolution and Diversity, Goethe University Frankfurt, Frankfurt, Germany
- 10Instituto de Biotecnología, Universidad Nacional Autónoma de México, Cuernavaca, Mexico
- 11Department of Marine Sciences, University of Gothenburg, Gothenburg, Sweden
- 12NORCE Norwegian Research Centre, Bergen, Norway
- 13National Oceanography Centre, Southampton, United Kingdom
Better scientific knowledge of the poorly-known deep sea and areas beyond national jurisdiction (ABNJ) is key to its conservation, an urgent need in light of increasing environmental pressures. Access to marine genetic resources (MGR) for the biodiversity research community is essential to allow these environments to be better characterised. Negotiations have commenced under the auspices of the United Nations Convention on the Law of the Sea (UNCLOS) to develop a new treaty to further the conservation and sustainable use of marine biological diversity in ABNJ. It is timely to consider the relevant issues with the development of the treaty underway. Currently uncertainties surround the legal definition of MGR and scope of related benefit-sharing, against a background of regional and global governance gaps in ABNJ. These complications are mirrored in science, with recent major advances in the field of genomics, but variability in handling of the resulting increasing volumes of data. Here, we attempt to define the concept of MGR from a scientific perspective, review current practices for the generation of and access to MGR from ABNJ in the context of relevant regulations, and illustrate the utility of best-practice with a case study. We contribute recommendations with a view to strengthen best-practice in accessibility of MGR, including: funder recognition of the central importance of taxonomy/biodiversity research; support of museums/collections for long-term sample curation; open access to data; usage and further development of globally recognised data standards and platforms; publishing of datasets via open-access, quality controlled and standardised data systems and open access journals; commitment to best-practice workflows; a global registry of cruises; and lastly development of a clearing house to further centralised access to the above. We argue that commitment to best-practice would allow greater sharing of MGR for research and extensive secondary use including conservation and environmental monitoring, and provide an exemplar for access and benefit-sharing (ABS) to inform the biodiversity beyond national jurisdiction (BBNJ) process.
Introduction
Areas beyond national jurisdiction (ABNJ), the vast majority of which are deep sea, represent the largest environments on the planet, yet are the least understood (Ramirez-Llodra et al., 2010). The vast majority of the seafloor is unmapped at high-resolution, and the deep sea very poorly characterised compared to other marine ecosystems (Higgs and Attrill, 2015; Glover et al., 2018). Sampling these environments presents substantial technical challenges, and, species discovery rates are significantly lower as a result (Webb et al., 2010; Higgs and Attrill, 2015). High proportions of species collected from these environments are new to science, with estimates varying from 35 to 95% (Poore et al., 2015). Taxonomy, the science of describing, naming and classifying biodiversity underpins all biological research; and is therefore necessary before other research can take place (Glover et al., 2018). Streamlined access to deep-sea collections and data is critical to allow taxonomic and other biological research in these poorly known environments and enable their future management in the light of increasing environmental pressures.
The deep sea faces a multitude of environmental challenges, such as cumulative impacts of climate change (Levin, 2018; IPBES, 2019); and potential new ones from seabed mining, with recent reviews suggesting mining operations could result in net biodiversity losses (Niner et al., 2018). While some mechanisms exist for monitoring and environmental protection in ABNJ, including environmental impact assessments and area-based management tools including marine protected areas, governance gaps are evident (Wright et al., 2018b), and no overarching framework exists for the allocation of marine protected areas in ABNJ (De Santo, 2018). In addition, there is a disconnect between regional and global governance in ABNJ (Gjerde et al., 2018). For example, the South Pacific Regional Fisheries Management Organisation (SPRFMO) recently revised bottom fishing rules to allow fishing to potentially continue even when encountering vulnerable marine ecosystems as assessed by observers, rather than implementing United Nations General Assembly recommendations. Overall, the current international ocean governance framework is poorly equipped to conserve and protect biodiversity beyond national jurisdiction (BBNJ) (Gjerde et al., 2018). Recognising these legal gaps, the United Nations General Assembly established a Preparatory Committee by resolution 69/292 for the development of an international legally binding treaty under United Nations Convention on the Law of the Sea (UNCLOS) for ‘the conservation and sustainable use of marine biological diversity of areas beyond national jurisdiction’. Further to these aims and in accordance with UN resolution 72/249, negotiations have commenced, and the third Intergovernmental Conference on BBNJ, will be held at the UN in August 2019. Here, deliberations on the recently released draft treaty text will take place.
These negotiations, also known as the BBNJ process, are focussed on four main components: marine genetic resources (MGR), area-based management tools, capacity building and technology transfer, and environmental impact assessments (Tiller et al., 2019). Taxonomy provides a unifying element to all of these components: as MGRs are in essence marine biodiversity, environmental impact assessments require knowledge of the species that live in a habitat being impacted/under assessment, and area based management tools depend on knowledge of species connectivity to inform spatial planning as require knowledge of the biodiversity present in a given region. Taxonomy is also key to capacity development as it is fundamental to all scientific research and therefore to building research capacity. While MGR are only one item of four in the negotiations, they are currently receiving the most attention. MGR and the sharing of benefits arising from their utilisation have long presented a challenge in the BBNJ process (Jorem and Tvedt, 2014; Tladi, 2015; Leary, 2019), key questions being the inclusion or exclusion of genetic or sequence data in the definition of MGR, and the scope of benefit-sharing, whether monetary or non-monetary.
Meanwhile, there are complexities in terms of current processes for the handling of marine samples and associated data in the scientific community. Biological research is becoming increasingly computationally intensive, and both capacity of genomics and the resulting generation of data are ever expanding. The accessibility of samples and data has been improving in response to an increased need for access. However, data are not always fully open, if published in restricted access papers, or in formats which limit their use, and in some cases not available at all (Bax et al., 2016). In the science community, data standards and sampling protocols are available (Wieczorek et al., 2012; Glover et al., 2015; Clark et al., 2016), but awareness and adoption is patchy. Poorly managed samples and data can hamper research progress through loss of knowledge and potentially lead to unsupported policy decisions. Further, commercial entities like the deep-sea mining and fisheries sectors often have a culture of withholding data (Wright et al., 2018a). While access and benefit-sharing (ABS) is happening now through current data and sample availability, there is room to improve. Best practice, or usage of standardised workflows and quality assurance/quality control measures which assure quality of output, can assist here.
There is a recognised need to build a repository of best-practices to support both biodiversity research and governance for BBNJ (Pearlman et al., 2017; Muller-Karger et al., 2018). Synergies across key organisations could be achieved here through best-practice marine sample and data collection.
To support marine biodiversity research and inform the BBNJ process, we discuss current practices in data and sample management from ABNJ, and outline recommendations to strengthen and raise awareness of best-practices. In section ‘Conceptualising MGR’, we conceptualise MGR samples and data from a scientific perspective, and discuss the BBNJ process in relation to these concepts. In section ‘From Sample Collection to Data Repository: Current Practices and Regulations’, we examine the current scientific research process from sample collection to data repository, highlighting current protocols and discussing challenges. In section ‘Current Best-Practices for MGR Sample and Data Collection and Archiving’, we describe current best-practices for MGR sample and data collection and archiving, and in section ‘How Could Best-Practice Regarding MGR Samples and Data Be Further Strengthened in Terms of Access, Sharing, and Transparency’ how best-practice could be further strengthened. We argue that commitment to a best-practice approach would increase sharing of samples and data from ABNJ, supporting marine scientific research and extensive secondary use including conservation, and therefore also provide a pathway for ABS in the BBNJ process.
Conceptualising MGR
What Are MGR Samples?
There is currently no internationally agreed legal definition of MGR, but a meaning for this term can be inferred from related definitions provided in the 1992 Convention on Biological Diversity (CBD), and the 2010 Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of the Benefits Arising from their Utilisation (Vierros et al., 2016; Harden-Davies, 2017). MGR therefore can be described as ‘material from marine plants, algae, animals, and microbial or other organisms, and parts thereof containing functional units of heredity of actual or potential value (CBD, Article 2)’. The Nagoya Protocol also provides the definition of derivatives (not linked to the definition of genetic resources but to their utilisation), and includes any ‘naturally occurring biochemical compound resulting from the genetic expression or metabolism of biological or genetic resources, even if it does not contain functional units of heredity’, therefore also encompassing secondary metabolites, enzymes, and natural products. These processes are depicted in Figure 1.
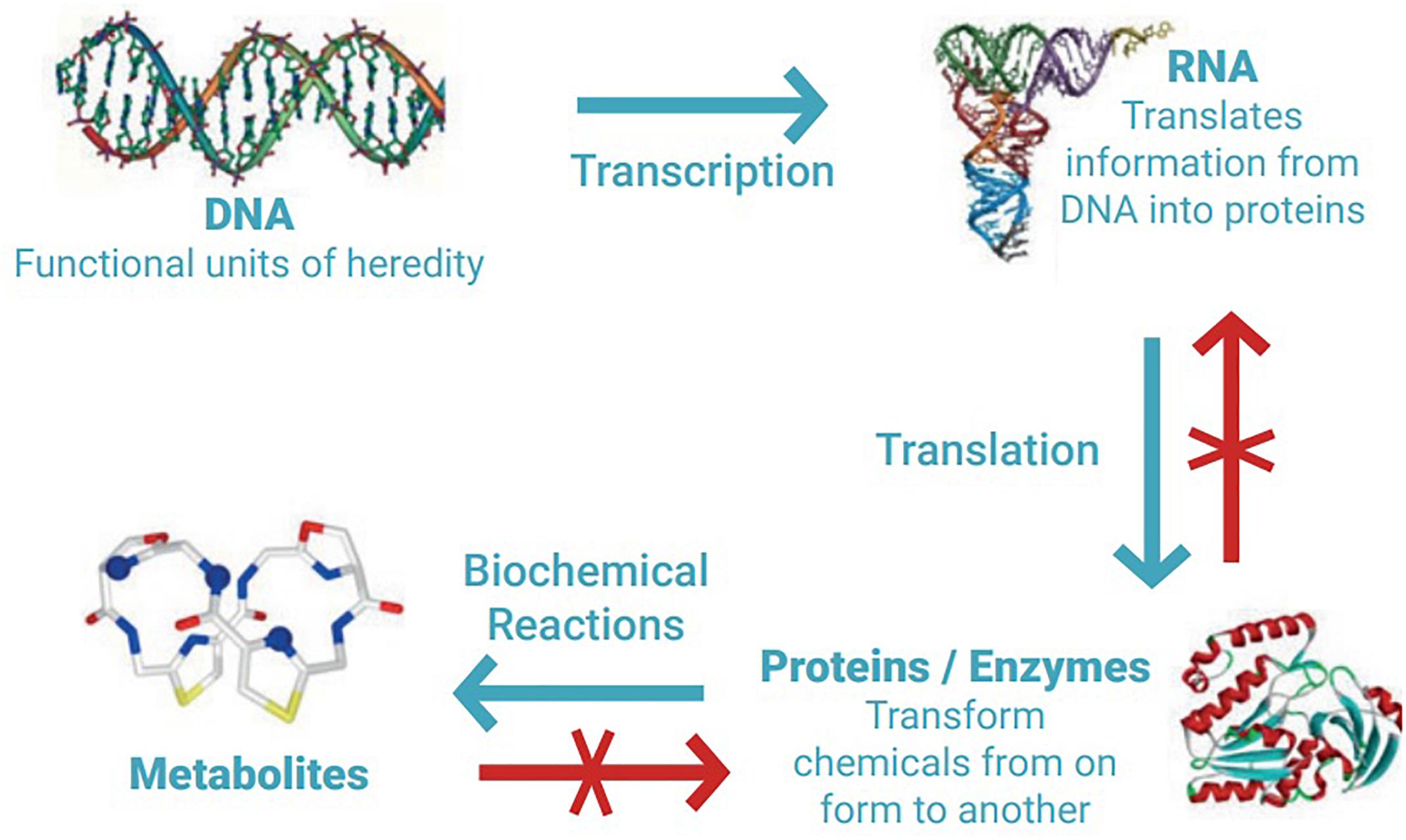
Figure 1. DNA is the cell’s master ‘instruction manual’; segments of the DNA (genes) are read off (transcribed) to build a corresponding piece of RNA, which ultimately helps to create a protein. Proteins are the functional unit of the cell, used for cell repair or to make hormones, for example. By turning one chemical into another, proteins make secondary metabolites, or biochemical derivatives. Biomolecule data cannot be ‘reverse engineered’; knowing a metabolite’s formula or a protein’s sequence is insufficient to predict the sequence of the underlying gene. Figure and caption reproduced with permission of Jeff Marlow (Marlow et al., 2019).
In terms of utilisation and intent, biological samples used in marine scientific research are collected for a wide variety of scientific fields, including taxonomy, ecology, biogeography, conservation biology, and climate change research. Such research may generate samples containing MGR that may be of interest for bioprospecting, i.e., the development of commercially valuable products for pharmaceutical, cosmetic and/or other applications (Jaspars et al., 2016). To date, seven commercial products on the market have been derived from MGR, including one from a species found both in coastal regions and open water, i.e., ABNJ (Broggiato et al., 2018). Increasingly, environmental samples are the focus of research activities in marine environments, such as those collected for microbial metagenomic studies and often represent mixed communities of Operational Taxonomic Units (OTU’s) rather than species assemblages (Walls et al., 2014; Godoy-Lozano et al., 2018). Marine microbes are often the focus for bioprospecting activities, and represent the majority of patent applications from ABNJ (Blasiak et al., 2018).
Samples not collected for molecular work, but retained in a preserved state for later research still contain MGR. These could include sea water, sea ice or sediment samples from ABNJ, collected for environmental chemistry or physical oceanography research. While most utilisation of MGR will come from live cultured, frozen or ethanol-preserved materials, sequencing technologies are advancing very rapidly, and extraction of DNA from formalin-fixed and ancient materials is now an established practice (Palero et al., 2010; Cook et al., 2015; Ruane and Austin, 2017). Therefore, MGR could also be obtained from samples neither originally collected nor preserved for molecular work. Derived samples are often generated from an MGR, such as a tissue subsample from a specimen, or extracted DNA. Any ‘child’ preparation or sample derived from a ‘parent’ specimen must retain the link to the original specimen, and can be considered an MGR sample. Specimen parts which may not contain genetic information e.g., molluscan shells, but are crucial in providing key taxonomic information for a specimen also need to stay associated with the MGR in question.
An MGR sample can therefore encompass a very wide range of sample types, from environmental samples of water, ice or sediment that (may) contain whole or partial organisms; through to whole organisms, e.g., single identified specimens, or mixed samples of specimens; to samples derived from any of these, such as extracted DNA or tissue preparations; preserved in such a way as to enable utilisation (defined in Article 2 of the Nagoya Protocol as ‘conducting research and development on the genetic/and or biochemical composition of genetic resources’). Any of these sample types should, in scientific terms, be considered an MGR sample.
What Are MGR Data?
Legal definitions related to MGR data also remain unclear and lack a standard definition. The term ‘digital sequence information’ (DSI) on genetic resources was introduced in decisions CBD XIII/16 and the Nagoya Protocol NP-2/14. But this term is not used by the scientific community, and its usage remains heatedly debated under the auspices of the CBD (Laird and Wynberg, 2018). In a BBNJ context of the discussions of the Preparatory Committee, the terms ‘in silico’ and ‘digital sequence information’ have been used by different delegations. MGR data, in simple scientific terms however is genetic or genomic sequence information obtained originally from a marine sample or MGR, as outlined above. It could encompass data from: mitochondrial or nuclear genomes for eukaryotic DNA, chromosomes and plasmids for prokaryotic DNA; protein structure, and/or secondary metabolites (Figure 1; Marlow et al., 2019). Raw genetic or sequence data are reads of a sequence: an arrangement of nucleotides on a strand of DNA or RNA (Consortium of European Taxonomic Facilities [CETAF], 2019). Raw sequence data files – direct output from a sequencing machine may be specific to the sequencing technology or platform used, or in generic formats such as FASTA or FASTQ: text files with sequence data represented by single-letter codes, and also containing data quality information.
Processed sequence data in contrast have undergone various degrees of analysis (Laird and Wynberg, 2018). Sequence data are generally analysed using the Basic Local Alignment Search Tool (BLAST) algorithm against reference sequences in sequence databases (listed in Table 1). Analysis methods may also include scripts for bioinformatics analysis pipelines, and/or parameters used in software such as Geneious or Bowtie (Langmead, 2010; Kearse et al., 2012). These methods, together with interpretations, e.g., phylogenetic trees, or population genetic inferences are likely to be detailed in a resulting publication. Methodologies are sometimes incomplete however, and lacking references to pipelines used for bioinformatic analysis. If version information for a reference dataset used in a metagenomics study is missing for example, reproducibility is undermined (ten Hoopen et al., 2017). There are multiple points in the process of generation of MGR data where variability is introduced, so comprehensive methodology is critical to reproducibility (Escobar-Zepeda et al., 2018). Overall, MGR data could therefore represent a wide range of states: a raw mitochondrial DNA sequence, or multiple processed sequence alignments; reconstructed genomic fragments, to fully assembled reference genomes, and downstream analysis such as functional annotations and full identifications of putative genes.
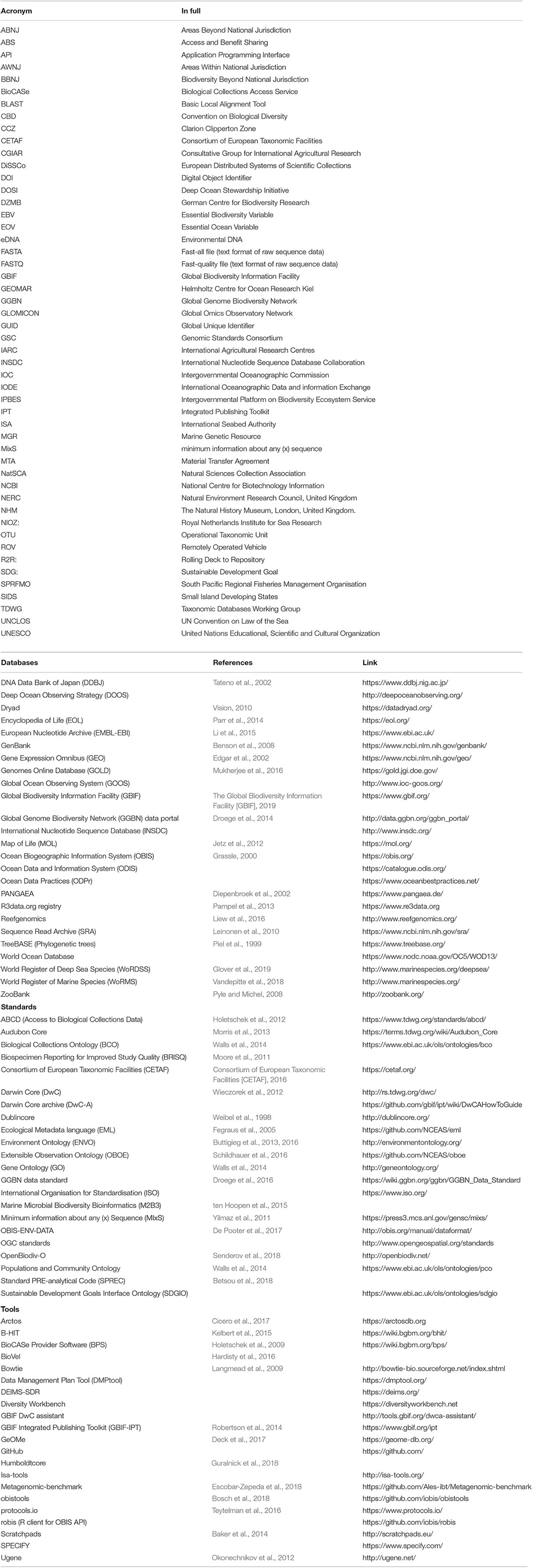
Table 1. Databases, data standards, and open-source data handling tools/software relevant to MGR data (if acronyms are included in the manuscript only, they are in the first table section, otherwise they are included with the relevant item in the table).
MGR data, whether in raw or processed form, cannot be viewed in isolation. It is critical that the data stay associated with its contextual information, i.e., the data describing all aspects of the MGR sample it was derived from. Without the integration of sequence and associated data for an MGR the sequence data is of minimal scientific use, as cannot be placed in its context – i.e., how it originated. Depending on the sample type, these data can include the current taxonomy/identification of the sample, physical location and preservation method of a sample (including sample preservation history); occurrence and sampling data (where, when and how the sample was collected), associated environmental data (e.g., oceanographic data); and derived sample information (e.g., extractions isolated from a parent sample). Associated laboratory data also need to be recorded with each sample, and could include DNA quality information, extraction technique, library preparation protocol, marker information and sequencing technology used (ten Hoopen et al., 2017). Associated data can also span a variety of types and formats, and include image files, video e.g., remotely operated vehicle (ROV) footage, acoustic data e.g., telemetry, and spatial data, such as bathymetry. It is important to ensure there are clear links between all these types of data. MGR data and the sample it is derived from are intrinsically linked, and must remain so to safeguard the scientific value of the MGR in question. If linkages are lost, a sample is at real risk of disposal. In summary, we consider MGR data to encompass all information associated with or extracted from a physical MGR sample, specifically including any genetic sequence information, in both raw and processed form.
BBNJ Background
The accessibility of MGR from sources in situ (on site), ex situ (samples in collections) and in silico (information in databases) is key to the functioning of the deep-sea research community, as outlined above, and pertinent to the ABS debate for BBNJ. Much of the debate on the legal status of MGR from ABNJ has been concerned with monetary benefits that could arise from any commercial utilisation. A few commentators have pointed out that these monetary benefits have gained a prominence out of scale to their likelihood and without any evidence of their importance to date, similar to the anticipated ‘green-gold’ of the Nagoya Protocol discussions that failed to eventuate (Leary, 2019). Recently, more attention has been paid to the merits of non-monetary benefits (Leary and Juniper, 2013; Broggiato et al., 2018; Morgera, 2018). The so called ‘non-monetary’ benefits of participation in marine scientific research, access to research results, data and collections, and other elements of technology transfer and capacity building present elements of a solution to the MGR ‘problem’ (IISD, 2018). However, the majority of the focus remains on monetary benefits. This urgently needs redressing given the key importance of biodiversity research to conservation and benefit-sharing/building capacity.
Given significant research infrastructure requirements, and the substantial expense of undertaking deep-sea investigations, difficulties in conducting such research are amplified for developing countries (DOSI, 2016; Harden-Davies, 2017). Discussions on benefit-sharing measures for ABNJ have recognised the specific circumstances of both developing countries and Small Island Developing States (SIDS) accordingly (President’s Aid to Negotiations, 2019). There is also a disconnect between availability of MGR (data or samples) in developed countries and its actual accessibility by developing ones: while MGR may be made available by scientific efforts, in reality actual utilisation is often not feasible by developing countries, where there is often reduced technical infrastructure, or even research capacity. For example, bioinformatics can be computationally intensive, representing a potential barrier in countries with poor IT infrastructure. As a result, capacity building and technology transfer discussions in the BBNJ negotiations are addressing how to improve access to MGR for developing countries.
Another key question in the BBNJ process is the inclusion of genetic sequence data in a definition of MGR. For the Nagoya Protocol, the potential inclusion of DSI within the definition of genetic resource is currently being debated, with concern that it could lead to restrictions on access to data that are currently open, hampering research (GGBN, 2017; International Chamber of Commerce [ICC], 2017). The scientific community has pointed to current barriers to taxonomic related biodiversity research arising from ABS regulations within national jurisdictions (Pethiyagoda, 2004; Kumar, 2018; Neumann et al., 2018; Prathapan et al., 2018). Similar concerns have been raised by several States that any future regulation of access to MGR in ABNJ could hinder marine scientific research (IISD, 2018). Yet, there is also concern that its exclusion could lead to biotechnology companies profiting from use of the ‘global commons’ without redistribution to those states with a reduced capacity to undertake such work themselves (Laird and Wynberg, 2018). Most commercial products derived from genetic resources, marine or otherwise, originate from publically funded scientific research. There is a perceived trend of developing countries in favour of including the term and developed countries opposed, but Kumar (2018) points out that all researchers are united in concern about any potential restriction to access for these openly available data.
From a scientific perspective, MGR data and samples are intrinsically linked, but wide definitions have legal and policy implications. For the most part, biodiversity research/taxonomy does not utilise derivatives, such as metabolites (Figure 1), often a target for biotechnology applications. Similarly, functional annotations are generally not directly relevant for taxonomic research. For this reason, the Consortium of European Taxonomic Facilities (CETAF) has recently proposed a definition to replace DSI with NSD: Nucleotide Sequence Data, a more precise term specifically referring to raw sequences (Consortium of European Taxonomic Facilities [CETAF], 2019). More accurate legal terminology reflecting scientific realities would be of benefit but the necessity of maintaining open access to MGR data must be emphasised.
The BBNJ negotiations are further complicated by additional uncertainties around definitions. For example, the water column beyond national jurisdiction is known as the ‘deep sea’ or ‘open-ocean’ to those who conduct research there, but in legal terms it is called the ‘High Seas’ (UNCLOS, Article 87). Scientists generally use the term ‘deep sea’ to refer to any part of the ocean, pelagic or benthic, deeper than 200 m, without reference to national boundaries. In reality, marine scientific research often conducts sampling across such boundaries, even during a single expedition (Figure 2). The ‘Area’ is the term used to describe the seabed in ABNJ (defined in UNCLOS as ‘the seabed and ocean floor and subsoil thereof, beyond the limits of national jurisdiction’). A legal dichotomy further confusing matters is that while the ‘High Seas’ are open, and largely unregulated, the mineral resources of the ‘Area’, or seafloor in ABNJ are administered by the International Seabed Authority (ISA), established by UNCLOS in 1994.
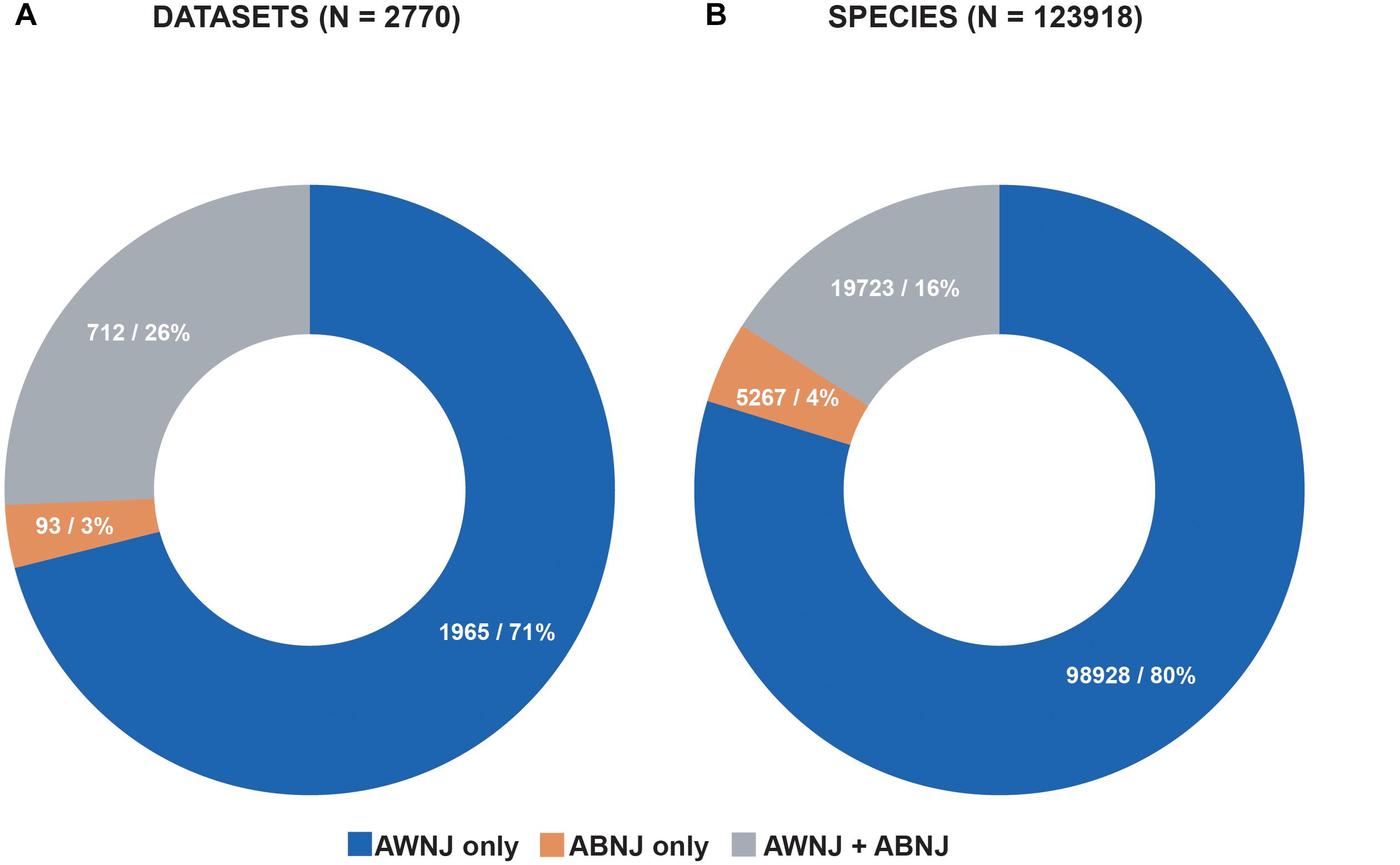
Figure 2. (A) 3% of the datasets in OBIS only hold data from ABNJ and 26% of the datasets hold data from both AWNJ and ABNJ, 71% of the datasets in OBIS hold data from AWNJ. (B) 4% or more than 5000 species occur exclusively in ABNJ, 16% occur in both AWNJ and ABNJ, and 80% occur only in AWNJ. Source: OBIS, 1 February 2019.
From Sample Collection to Data Repository: Current Practices and Regulations
Sample and Data Collection
To illustrate how current practices and regulations can be streamlined, we will discuss how the process is already managed and regulated from the point of MGR collection, through to the subsequent research, to sample and data archiving and third party access.
The initiation point of this process is a cruise or expedition to collect an MGR sample. Cruises are generally undertaken by international partnerships of various research institutes, and the ships that conduct them are managed and owned largely by institutes and governments, but increasingly also by private industry and philanthropic individuals (León-Zayas et al., 2017). Both pre-cruise planning and post-cruise information logging mechanisms are available in many countries. In the United Kingdom, the Natural Environment Research Council (NERC) managed research vessel programme can be viewed in a jointly managed database1 with those of Royal Netherlands Institute for Sea Research (NIOZ) and the Helmholtz Centre for Ocean Research Kiel (GEOMAR), which allows for comprehensive cruise planning. In some countries, research cruises are registered, for example, the Rolling Deck to Repository (R2R2) database records data from all US-based academic vessels. Cruise reports usually contain relevant navigational, oceanographic and environmental sample data, and these are openly available on institutional websites3 in many countries. However, the information regarding all research cruises taking place in ABNJ globally, where the eventual deposit of the collected samples and data will be, and who is responsible for them is not currently housed anywhere centrally.
Regarding on-board sample collection, deep-sea sampling methodologies are complicated by unique technical challenges, and can be very challenging to standardise (Danovaro, 2009; Glover et al., 2015). As deep-sea sampling has been extensively reviewed in recent publications (Glover et al., 2015; Kopf et al., 2015; ten Hoopen et al., 2015; Clark et al., 2016), it will not be covered here. In most cases, at the demobilisation of a research expedition, the samples will be transferred from ship to numerous laboratories for the next phase of research to take place. It is standard practice for Principal Investigators of a research cruise to keep a record of which samples have been transferred where and for what purpose. As such, there is no current national or international standardisation of the process, and significant improvements could be made in the management of collections from cruise to laboratory.
Data collection on ship is also undertaken in a variety of ways. The interdisciplinary nature of marine scientific research often requires integration of a range of data types. This may be done on board, via databases linking imagery and bathymetry with sampling sites for example (Clark et al., 2016), but this is likely a best-case scenario. Data are collected in a range of formats, and while data standards are available they are not consistently employed. There also often a lack of integration between species-occurrence data (where species occur in time and space), and associated oceanographic or environmental datasets. This is partly an artefact of data-recording practices, since occurrences can be recorded in a varied fashion and subject to change over time with taxonomic revisions, but oceanographic data is typically generated in a final form on board ship via instrument readouts. Provision of data in ‘real-time’ is often proposed as a solution for benefit sharing, but this is not currently possible with biodiversity research, given the time between collection and identifying a specimen can be considerable. Sound collation of high quality data requires considerable time and effort – from this initial stage to throughout the data and sample lifecycle.
Sample Archiving and Related Challenges
When primary research work is completed, samples and associated data should be archived accordingly. Voucher specimens are central to taxonomy and reproducible science (Huber, 1998; Beaman and Cellinese, 2012; Swiss Academies of Arts and Sciences, 2019). Despite the importance of vouchering, there are very few specific regulations for long-term sample archiving and management. According to the ‘best scientific practice’ of the German Research Foundation, samples have to be stored and kept for only 10 years, and the oil and gas industry in the EU is only required to archive samples for 5 years post collection (Bennear, 2015), compromising the long-term availability of collections and endangering the repeatability of research. Such archival practices however may be a best-case scenario. While museums generally have robust practices for long-term sample curation for collections, universities often have no equivalent procedures. Increasingly, shorter-term projects not involving museums are funded, and MGR samples are retained in disparate university or government-funded laboratories. Samples collected as part of large programmes at great expense, and which likely include numerous undescribed taxa are at real risk of disposal when a project is considered completed, or a researcher leaves the institute. There have been recent improvements to recognise the importance of archiving with journals and databases requiring information on where vouchers are stored, although this is by no means established practice.
There are significant implications of storage of physical specimens, including substantial curation costs but also those for maintenance of collections buildings, their contents, and associated databases, alongside the key cost of staff time. It is important to recognise that non-monetary benefits are subsidised in this way by museums and similar collections-based institutions. With the adoption by many countries of the Nagoya Protocol in 2010 (which came in to force in 2014), several reports and papers address the need for standardisation in curation of collections that fall under its scope (although there are no specific rules on this in the Protocol itself). For example the (Consortium of European Taxonomic Facilities [CETAF], 2018)4 ABS Code of Conduct provides a set of basic collection management principles to abide by.
In museums, recording of acquisitions which specify both what materials have been acquired and the terms of acquisition, is an established practice. Larger natural history collections in general will require transfer of ownership to the museum at the point of acquisition, and Material Transfer Agreements (MTA’s) to reflect this, in exchange for the absorption of long-term curation and data management costs. For smaller non-museum institutes, recording of the acquisition may be incomplete or even lacking. In the context of ABS, transfer of ownership is a potential challenge in the negotiation of an MTA. As such, the practice of title transfer is currently under debate (Sarah Long, personal communication). Distributed collections, or where some collections are held by another institute and linked by a common database could provide a potential solution here, for example as proposed by the recently established European Distributed Systems of Scientific Collections (DiSSCo) programme (Hobern et al., 2019). This would still require some institutions to take on long term curatorial responsibility. The Nagoya Protocol has placed increased pressure on both museum and non-museum institutes to comply with these new regulations when working in areas within national jurisdiction (AWNJ). Institutes dealing largely with open-ocean and deep-sea ecosystems may be unaware of these regulations. There is also potential for confusion and unintentional non-compliance where cruises collect samples from both ABNJ and AWNJ (Figure 2).
Accessing Samples
Samples held in museums have long been available for research by external parties and the associated datasets are becoming increasingly accessible as the need and the technology to allow it have evolved. Access to deep-sea collections is currently mainly through contact with museums or biorepositories and larger institutes who conduct regular research cruises, and databases or publications. The Global Registry of Biodiversity Repositories, GRBio (Schindel et al., 2016), a collation of national and global registers of sample collections, has recently been incorporated into GBIF, the Global Biodiversity Information Facility (The Global Biodiversity Information Facility [GBIF], 2019). National registers may also exist e.g., NatSCA in the United Kingdom,5 and specific sample collections are also displayed on institutional databases and in data aggregators including GBIF and OBIS (Ocean Biogeographic Information System) with holding institutions displayed alongside records. The Global Genome Biodiversity Network (GGBN) aggregates records of molecular/genetic collections housed in various institutes that are available for research6.
Data Archiving and Access, and Related Challenges
Marine genetic resources data are generally accessed through online data repositories and research publications. There are many databases relevant for access and publishing of MGR data (Table 1), with differing applications, focussing separately on sequence data or species occurrences for example. Key databases include the International Nucleotide Sequence Database Collaboration (INSDC) group, the World Register of Marine Species (WoRMS), and OBIS. Most databases listed provide additional functionality to their core remit as data repositories, and are therefore useful for both curating as well as sourcing data. WoRMS is a comprehensive checklist of marine species names, curated by around 300 taxonomists in accordance with best-practice (Horton et al., 2017; WoRMS Editorial Board, 2019). It also provides data on species traits as well as distributions (Vandepitte et al., 2018). The WoRMS taxon match tool for resolving names, provides crucial quality control support for taxonomic data for the research community and for biodiversity platforms (Vandepitte et al., 2018). The OBIS platform has mapping functionality including the ability to search within ABNJ, and a deep-sea node (O’Hara et al., 2015). Research publications – while a useful source of information regarding locations of MGR samples and data, are not always openly available, with many being retained behind a paywall, and access is therefore limited. Many governments and funders now require research to be either published in open-access journals, or included in an open-access repository to overcome this issue.
In terms of research outputs, practices of depositing genetic data in open access databases prior to publication are well-established in the scientific community, and required for peer-reviewed scientific journals. Funding bodies usually also require a data management plan7 with commitments to making data available through open access platforms, and open data policies following an embargo period 2 or 3 years from collection are a standard practice. There are legal requirements underpinning this, for example, the United Kingdom government requires public institutions to provide access to their data. This is a substantial contribution to benefit-sharing in the context of the BBNJ negotiations (Laird and Wynberg, 2018).
However, there are a number of issues with data available in genetic databases, a key one being the increasing number of sequences deposited without reference to formal scientific names, which has resulted in an explosion of ‘dark taxa’ (Page, 2016). This is a substantial problem resulting in the generation of additional ‘taxonomic entities’, with limited scientific meaning. Another issue is a lack of site and other core associated data connected to genetic data (Pope et al., 2015). While guidelines often recommend this contextual data is uploaded with sequences8, there is no obligation to provide any more data about the specimen than a mandatory specimen ID number. This has resulted in a proliferation of sequences deposited at genetic data repositories (e.g., GenBank) without sample collection information. This disconnect between sequence and contextual data is a significant problem (Pope et al., 2015; Gratton et al., 2016; Deck et al., 2017). This can lead to confusion about which sequence is linked to a type locality for example. Current practice is improving following efforts of organisations like the Genomic Standards Consortium (GSC), GGBN (Droege et al., in preparation), and the marine microbial research community (Kopf et al., 2015). Data storage for genetic data from MGR research is another complicating factor. In light of research reproducibility, archiving raw and processed genetic data is important, but in practice, this could mean storing terabytes of data for just a single study.
Considering occurrence data, there is a well-recognised data gap in OBIS for deep-sea species, resulting from the aforementioned substantial technical challenges and costs in accessing these environments (Appeltans and Webb, 2014; Glover et al., 2018). There are also issues arising from data quality where mis-identified species are uploaded to online repositories as valid species occurrences. This is partly due to limited taxonomic expertise arising from a shortage of adequately trained taxonomists. Another issue, particularly relevant for certain taxa such as marine microbes, is that the highest level of species description may be an OTU, but currently aggregators such as GBIF and OBIS are limited to species occurrences only. According to Bingham et al. (2017), four of the ten most connected databases in the biodiversity informatics space are primarily marine in focus (namely OBIS, WoRMS, FishBase and AquaMaps). A decentralised system of many connected nodes currently exists in the biodiversity informatics space, which provides resilience in the system (Bingham et al., 2017). While this also results in some overlap and duplication, improved links between initiatives will lead to greater interoperability (Costello et al., 2018; Kroh et al., 2019). In summary, access, and therefore benefit-sharing of MGR is happening now, but there is certainly scope to improve.
To demonstrate other deep-sea records currently available from ABNJ, we have extracted data from ABNJ at depths of 500 m and greater from OBIS, which gave 371,890 records of 10,437 species, observed between 1866 and 2018 (Figure 3).9 The data clearly presents geographic biases, extensive sampling in the North East Atlantic for example. Taxonomic biases and gaps are also evident. The data can be downloaded and viewed at https://mapper.obis.org/?areaid(=1&startdepth(=500).
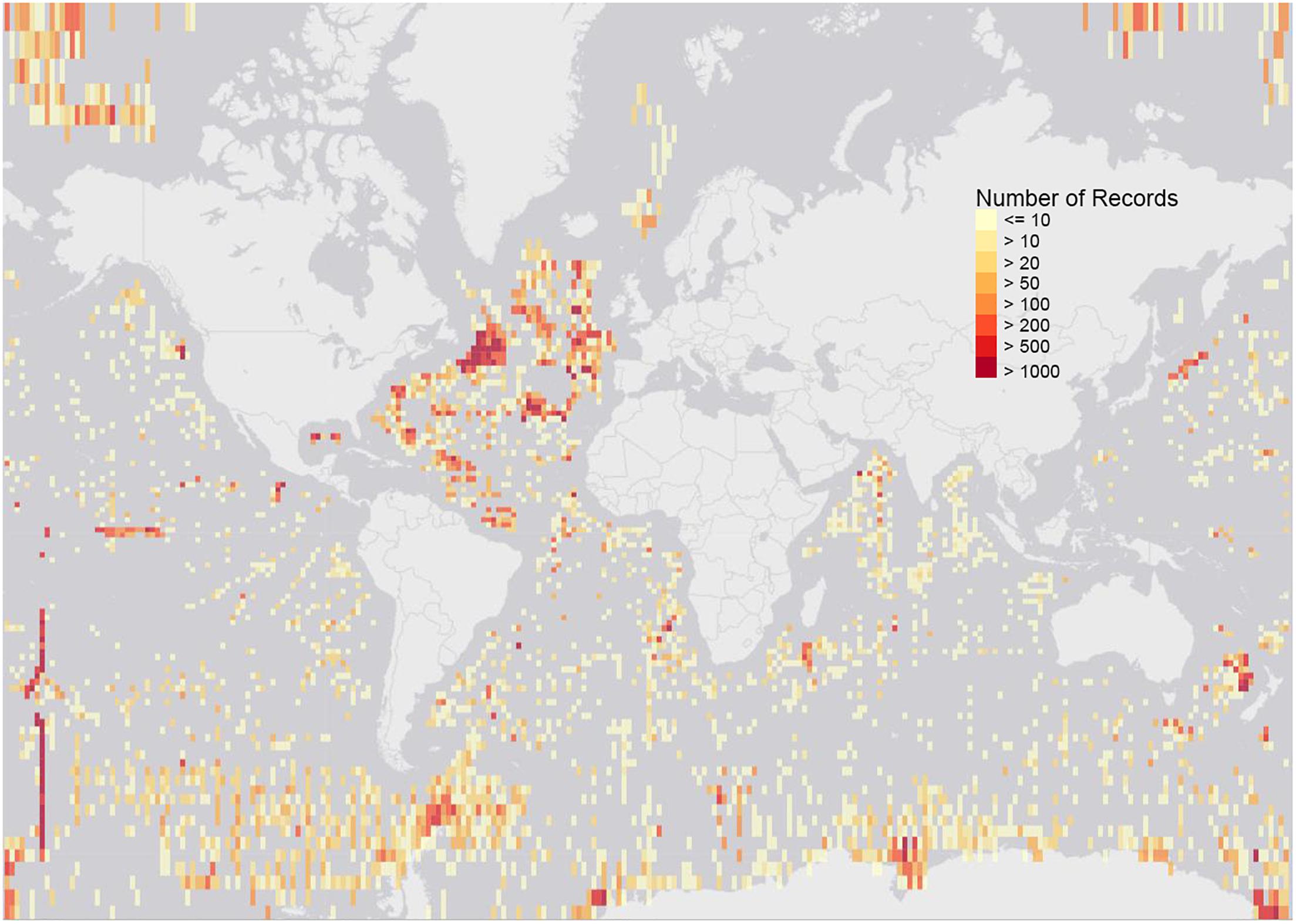
Figure 3. Map of all OBIS records from ABNJ from depths of 500m and greater, 371,890 records of 10,437 species in total, observed between 1866 and 2018 (data at 12 May 2019). The dataset can be downloaded and viewed at https://mapper.obis.org/?areaid=1&startdepth=500.
Current Best-Practices for MGR Sample and Data Collection and Archiving
Data Standards
Use of data standards is critical to data sharing, by enabling database interoperability, simplifying downstream applications and allowing comparison of data across studies. Standards are therefore key to making data FAIR: Findable, Accessible, Interoperable and Reusable (Wilkinson et al., 2016). Key data standards and ontologies for biodiversity data are listed in Table 1. These standards have different aims, for example OBIS-ENV-DATA allows integration of environmental/oceanographic data and species occurrences (De Pooter et al., 2017), averting potential loss of crucial contextual information. Alongside data standards, persistent identifiers are critical to accurately resolving data records (Hobern et al., 2019). Identifiers come in a range of formats, such as DOIs (Digital Object Identifiers), and have varying attributes and levels of adoption, but the key factors are global uniqueness, resolvability, persistence, discoverability, and authority (Guralnick et al., 2018). Without these characteristics, communication and interoperability between databases is compromised (Güntsch et al., 2017), and the records are at risk of duplication or even loss. Therefore, awareness and usage of persistent identifiers by the research community is critical.
Networks, such as GGBN, CETAF and TDWG (the Taxonomic Databases Working Group), are key to adoption and development of ontologies and data standards. Development is generally harmonised so standards complement each other rather than overlap, for example the Biological Collections Ontology (BCO) reuses Darwin Core (DwC) terms (Walls et al., 2014), and the GGBN Data Standard was developed to complement existing standards such as DwC, and covers the requirements of tissue, DNA and environmental sample collections (Droege et al., 2014). GGBN has also developed a High Throughput Sequencing library data standard to be reviewed by the community. These activities are crucial not only to data interoperability but also streamlining sample sharing and curation processes (Benson et al., 2016; Nussbeck et al., 2016). These networks are also important to both raising awareness and further development of existing tools for use with data standards and databases (Table 1).
Standardisation Frameworks: EOVs and EBVs
Essential ocean variables and essential biodiversity variables (EOVs and EBVs) represent frameworks which aim to standardise monitoring of biodiversity at global scales (Wetzel et al., 2015; Constable et al., 2016; Proença et al., 2017; Miloslavich et al., 2018). EOVs/EBVs therefore provide a mechanism to apply scientific findings to conservation and environmental monitoring and policy objectives. For example EOVs/EBVs aim to progress the UN Sustainable Development Goals (SDGs), including SDG1410 (Anderson et al., 2017). As such these frameworks directly link science and policy, and further development of these links is being discussed (Geijzendorffer et al., 2016; Anderson et al., 2017; Weatherdon et al., 2017; Benson et al., 2018; Wetzel et al., 2018). Quality and timely data is critical to environmental monitoring (Benson et al., 2018). Therefore, accurate taxonomic data and usage of data standards are necessary for these frameworks (Kissling et al., 2018). However, there are often delays between incorporation of scientific findings into policy. Potential issues of incomplete information in employment of EBVs have been addressed in recent reviews (Weatherdon et al., 2017; Muller-Karger et al., 2018). EOVs and EBVs address species distribution and abundance, but in a deep-sea context baseline biodiversity data are often lacking, which underscores the need for fundamental taxonomic research to be undertaken in these environments, particularly if these variables are shaping policy.
Case Study: Clarion Clipperton Zone ABYSSLINE Programme
While adoption and usage of data standards and platforms is key to data sharing, there is room for improvement both in the awareness of existing data standards and platforms within the biodiversity research community. Compounding this are potential difficulties in navigating the array of tools and software available to assist with data handling. With these challenges in mind, here, we illustrate the utility of data standards with a case study from recently published datasets of MGR from ABNJ. The ABYSSLINE (ABYSSal BaseLINE) programme coordinated by the University of Hawaii for the contractor UK Seabed Resources, has a DNA taxonomy component that provides a case where best-practice has been attempted in taxonomic methods and data and sample management (Glover et al., 2015). One of the key aims of this research programme is to characterise abundance and diversity of abyssal fauna from the UK-1 exploration contract area in the eastern Clarion Clipperton Zone (CCZ) in the Central Pacific, a region undergoing intensive mineral exploration for polymetallic nodules. The programme aims to address the chronic lack of data from the region, despite several decades of cruises associated with mineral exploration activities (Amon et al., 2016; Glover et al., 2018).
Use of Darwin Core is central to the data workflow, where data are mapped to the terms and current taxonomic identifications and occurrence data are exported to a Darwin Core archive. Data and sample handling workflows are detailed in Glover et al. (2015). Following specimen identification (via morphology and molecular methods), species names are recorded and validated against WoRMS. Records are published on the Natural History Museum London data portal (Scott et al., 2019), with global unique identifiers (GUIDS) linking the museum database to the specimen permanent record. Taxonomic data papers are published in the open access journals, Biodiversity Data Journal and ZooKeys (Dahlgren et al., 2016; Glover et al., 2016; Wiklund et al., 2017; Wiklund et al., accepted). These journals are formatted to maximise interoperability: semantically enhanced with embedded links to databases such as GenBank. The datasets are also registered with GBIF, allocated a DOI and published on the OBIS deep-sea node. This allows tracking of use, citation and versioning of the data underlying the publications.
The ABYSSLINE collections include preparations from specimens that do not represent MGR but are derived from it, i.e., slides and scanning electron microscopy stubs, which need to stay in association with the specimen record as they provide additional taxonomic data. Here, relationships between parent specimens and derived samples or preparations (i.e., tissue and DNA vouchers) are captured using the GGBN data standard (Droege et al., 2016). The OBIS deep-sea node then converts the dataset in GGBN format to the OBIS-ENV-DATA data standard (De Pooter et al., 2017). Usage of these standards has allowed increased integration of genetic data with other specimen information. The macrofaunal tissue samples and DNA vouchers are housed in the Natural History Museum’s Molecular Collection facility, and are available for research use, as displayed on the GGBN portal11.
The project findings, including the description of several species new to science, have been a major contribution to faunal records in the CCZ (Wiklund et al., in press; Glover et al., 2018). This new knowledge also includes discovery of a new genus and species of sponge Plenaster craigii (Lim et al., 2017), a potential indicator species in environmental impact assessments (Glover et al., 2018). Further, connectivity studies on this sponge have provided the first evidence-based recommendations for area based management tools in the region (Glover et al., 2018). The CCZ has recently been proposed as a demonstration project for the Deep Ocean Observing Strategy (DOOS) (Levin et al., 2019). The findings described here would directly contribute to such a project, and to the emerging EOV of marine benthic invertebrate diversity. The application of best-practice using available data standards, facilitating interoperability of existing databases provides a clear example for future work and a potential model for use in the BBNJ process.
How Could Best-Practice Regarding MGR Samples and Data Be Further Strengthened in Terms of Access, Sharing, and Transparency
Sample Archiving
Greater open access to samples is critical for the research community, and would facilitate collaboration between deep-sea investigators and expert taxonomists from understudied groups, and would be of great benefit to deep-sea research. For example, The German Centre for Biodiversity Research (DZMB) tracks specimens and data obtained during German expeditions (including third party usage), and following species identification, sends vouchers to the natural history collections of Senckenberg (Brandt et al., 2018). In some cases, archive of a physical sample is not possible, for example, where a specimen is completely consumed during analysis and all that is left is resulting data, not even a DNA voucher. This is sometimes the case for environmental DNA (eDNA) collections and molecular collections more widely. Where possible, Whole Genome Amplification could be undertaken on a subset of critical specimens to allow more sustainable usage. While there are issues with the technique, such as potential amplification bias of non-target DNA, recent methodologies address this (Dagnall et al., 2018). Destructive sampling illustrates the importance of considered decision-making for usage of highly valuable and difficult to collect material, particularly relevant in the context of deep-sea collections from ABNJ. Protocols for sustainable, responsible usage of material, and commitments to good data practice and comprehensive data archiving for those collections where some destructive sampling is unavoidable, are crucial. In other cases, video footage, such as seafloor imagery of putative morphotypes may represent the only ‘sample’ – in these cases robust data annotation and archiving processes is critical as these data represent ‘virtual’ vouchers (Levin et al., 2019).
Data Publishing
Good-data management practice is of benefit to the data creators, as well as the scientific community as a whole. Publishing of datasets independent of a publication is a key mechanism to not only increase data accessibility, but also encourage best-practice, as transparency will encourage good data management. Further, reuse of data multiplies its value. Datasets can be allocated a DOI, enabling citation and dynamic updates of data, as in the ABYSSLINE case study. This allows datasets to be cited and versioned, increases discoverability, and provides recognition for data creators outside of traditional publications (Michener, 2015; Wetzel et al., 2018). Where possible, an open license such as creative commons should be used for datasets to safeguard open access (Michener, 2015). Semantically linked publications in journals such as ZooKeys provide an existing framework to maximise accessibility, interoperability and traceability of data (Penev et al., 2010). High-quality metadata is essential to both data longevity and tracking provenance, and as such requires special attention. Metadata are often mischaracterised as the associated or contextual data of a sample, such as site information, but in fact are data that document and describe a dataset: data ‘about’ data (Stow et al., 2018). Good metadata allows interoperability, and enhances data and sample sharing e.g., by recording of the institute archiving the data. Comprehensive guidance on publishing data and metadata are available12 (Penev et al., 2017; Stow et al., 2018).
Further Developing Existing Data Systems
In terms of tools available for data management, gaps in functionality and potential for further development are evident. Some existing tools and software could benefit from improvement. GBIF’s Integrated Publishing Toolkit (IPT13), an open source software tool used to publish and share biodiversity datasets, requires significant input from the researcher that would be better handled by systems. In contrast, OBIS works on a system of nodes to support researchers in publishing their data and metadata. The less onerous the system, the more likely data creators are to publish their datasets. Databases require curated, cleaned data and ideally, these processes should be automated as much as possible, via an application or software. While mechanisms to ensure overall data quality, as well as for feedback of data quality between data provider and platform are currently in place (Vandepitte et al., 2014), they could be further expanded. GBIF and OBIS now provide data quality metrics. For example, the new OBIS portal provides a feedback function at the dataset level, and an annotation tool at the record level is in development. This would allow users to flag errors and OBIS could record the reason (e.g., species X does not occur in area Y), which would further contribute to data quality control.
Open Source
While various software and tools are available for annotating data, processing a full data lifecycle of a specimen generally requires moving between tools and systems rather than working within an integrated one. Technical innovations require an active community and efforts on open source code libraries. Open source accompanies open data, and future software developments should archive code (e.g., on code sharing sites like GitHub14), which is becoming commonplace in the biodiversity informatics community (Wilson et al., 2017). A good example is the work done by TDWG’s data quality interest group15. Based on code libraries, data dashboards could be developed, such as those proposed in the Humboldt core initiative (Guralnick et al., 2018). This allows users to annotate and visualise their data, similar to web-based data management platforms like BioVel, which integrates key aspects of a sample lifecycle (Hardisty et al., 2016). OBIS is a key platform in the context of MGR in ABNJ and the deep-sea OBIS node provides a platform for researchers to both access and publish deep-sea data. However, success depends on community adoption and involvement. OBIS provides data access to their Application Programming Interface (API), allowing machine to machine communication, and development of data products or applications without the need to physically store the data. These processes align with ABS as open data enhances collaboration, transparency and also potentially technology transfer via sharing of knowledge.
Increased Integration Between Data Systems and Observation Frameworks
There are existing links between sequence-data platforms like GenBank and those housing occurrence data, like OBIS or taxonomic and species-trait data such as WoRMS (e.g., WoRMS provides links between individual species pages and records available in GenBank). However, visibility and reciprocal links could be improved and greater integration is needed (Muller-Karger et al., 2018; Kroh et al., 2019). This could be facilitated by greater visibility of associated sequence data in OBIS, and links to GenBank via accession numbers. Future adoption and usage of data standards, including making them genomic standards such as MIxS interoperable with Darwin Core will contribute here. There is scope to address this via integration of genetic and occurrence databases (such as GenBank and OBIS) and via new initiatives such as the Global Omics Observatory Network (GLOMICON) (see footnote 9). Together with the Genomic Observatories Network (GON), GLOMICON and OBIS are prototyping a molecular biodiversity data pipeline directly integrated with occurrence databases16. These efforts would be a significant contribution to streamlining access for MGR.
Essential biodiversity variables specifically address the monitoring of genetic diversity via the EBV class genetic composition, which has been identified as a data gap (Geijzendorffer et al., 2016). DNA sequencing and metabarcoding has recently been piloted as a monitoring tool (Keeling et al., 2014; Goodwin et al., 2017), and eDNA metabarcoding pipelines for biomonitoring are being developed (Andruszkiewicz et al., 2017). Levin et al. (2019) suggest ‘a large-scale metabarcoding, metagenomic, and transcriptomic census, as has been conducted for marine plankton’. This must be undertaken with commitment to baseline reference data, and generation of morphological and molecular reference data libraries. Also it is important to note that metabarcoding for monitoring cannot be used as a replacement for species description with so many undescribed species in the deep sea. It is also important to recognise that since many of deep-sea species are new to science they may only be identified to genus or higher taxonomic ranks, and will likely remain as OTUs. The characterisation of OTUs is critically important to provide workable ecological information, and must be very carefully managed to ensure comparability between surveys in the future.
Future Scenarios
Future scenarios may include an increased need for data and sample processing from in situ deep-sea observatories and increased use of methods such as machine learning for video and image data. Such datasets are particularly important in cases where collection of a physical voucher specimen is not feasible. MGR may be increasingly studied and monitored remotely by mechanisms such as robotic samplers with inbuilt sequencers (Kissling et al., 2018). Overall as in lab technique development there is scope for much innovation here. Future development should be integrated as far as possible with existing systems and platforms that have a global user community. A current key initiative is the General Bathymetric Chart of the Oceans (GEBCO) Seabed 2030 Project (Mayer et al., 2018), aiming to map the seafloor by 2030. Such efforts are critical to understanding biodiversity and characterising biogeography in the deep sea and present an opportunity to set standards in these previously un-mapped regions that are not available for the already mapped terrestrial areas. There are potential issues with making this data available as with open access as vulnerable ecosystems such as seamounts, or rare and threatened species, may be further targeted. Potential solutions here could draw on experience in terrestrial systems, such adding randomisation to coordinates of species occurrences. In terms of data access in developing countries, public-private partnerships may also be required to further develop IT infrastructures to address barriers to data accessibility. Alternatively, bioinformatics capacity could be leveraged by access to a system of distributed nodes to run analyses on, such as the PlanetLab model.17
The Role of Networks
Community involvement and networks are important to progressing data and sample accessibility in the deep-sea research community, and to building consensus on standardisation. Strong networks also have also been identified as a key factor in sharing data across science and policy (Weatherdon et al., 2017). The deep-sea research community is already international and collaborative, with global networks such as INDEEP and the Deep-Ocean Stewardship Initiative (DOSI)18 (Mengerink et al., 2014), Other non-marine focussed networks are also relevant here. CETAF, while a European network, provides documentation on best-practice for taxonomic sample collections19. The GGBN network provides guidance on best-practice for managing genomic/molecular biodiversity collections and, as discussed earlier, provides established data standards and pipelines. GGBN supports best-practice through initiatives like the GGBN Document Library representing a knowledge platform for the biodiversity research community20. Samples of all kinds, including environmental samples, are a core focus of GGBN, and its members’ collections provide long-term storage for high quality DNA, tissue and environmental samples, key to the current context of how to facilitate access of MGR from ABNJ. Protocols for collection, preservation and analysis of environmental samples for genetic/genomic purposes is still very much in development, however, GGBN can play an important role in organising workshops on this topic. These activities have the potential to support the BBNJ process in finding solutions for sampling in ABNJ and accessibility of MGR.
Capacity Building
Like ABS, capacity development is already happening, but there is room to improve in terms of strengthening cooperation and awareness of existing efforts and initiatives (Stephenson et al., 2017; Harden-Davies and Gjerde, 2019). The treaty can help to coordinate and raise awareness, highlighting the value of non-monetary benefits in building research capacity and in turn, the importance of capacity building to conservation. Networks can significantly progress capacity building, for example, through facilitating voluntary mentorship/pairing schemes, where individuals from developing and developed states in the scientific community are paired up. A number of such schemes are already in existence. The OceanTeacher Global Academy runs marine science and data literacy training workshops in developing countries, which both develop capacity and provide a framework whereby host countries can build their own training centres, making efforts sustainable. Another example is the POGO/SCOR21 fellowship programme, which enables scientists from developing countries to visit oceanographic centres for training. MGR collected during a project could also be used for capacity building. Agreements between countries and institutions involved could facilitate the exchange of students and early career researchers between countries, the sharing of samples and data and lead to joint expeditions with shared distribution of the scientific material obtained. Enabling early career researchers to participate in research cruises has been identified as a key mechanism to support capacity development for deep-sea scientists.
Role of Museums and Repositories
One of the key questions is who will facilitate improved access to data and samples. As described here, those involved will include individual scientists, working in research institutions and sometimes linked either formally or informally by international networks. Collections and biorepositories such as museums are the main suppliers of MGR, and also lead in standardising procedures for curation and tracking collections (Harden-Davies, 2017; UN Ocean Decade Report, 2018). Under the Nagoya Protocol, collections such as museums are recognised as a key part of benefit-sharing. While a formalised role and recognition of collections could streamline management of MGR from ABNJ, the scientific community has warned that without additional financial resources to meet heightened administrative and other requirements, such regulations could actually hinder biodiversity research (Kumar, 2018; Neumann et al., 2018). This is a real possibility that must be avoided by the treaty at all costs. Currently, while museums and non-museum institutes support non-monetary benefits, they absorb substantial curation and data management costs in the process. It is key that governments are aware of the need for additional infrastructure and funds to support any requirements for sample curation and data management in the future treaty. The treaty itself could assist here in raising awareness of this need.
Learning Lessons From Nagoya
The negotiations leading to the Nagoya Protocol and its subsequent implementation at national levels provide substantive lessons for the BBNJ process. In particular, it is clear that problems remain with regards to the definitions of what constitutes MGR, MGR data and utilisation and that these issues must be overcome for incorporation into the new treaty for ABNJ. One of the issues of the Nagoya Protocol negotiations was a lack of engagement of the scientific community in providing the understanding of these terms and explaining the related processes. As has been illustrated here, the management of MGR data is currently conducted in an open access arena, and any attempt to restrict the sharing of data from ABNJ will be highly problematic to the scientific community, and the funding bodies who support them. Regulation must ensure to not stifle innovation or impede research progress in any way. Input from the scientific community is therefore crucial to the BBNJ discussions (Vierros et al., 2016; Harden-Davies, 2018). This consultation process, drawing on lessons learnt from the Nagoya Protocol is also central for capacity building discussions, from how to implement at the practical level (Vierros et al., 2016), to a broad scope on what is needed, with input from both North and South scientific communities (Gjerde et al., 2018). It will be key to road test any potential benefit-sharing measures, which could be achieved by building iterative stages into the BBNJ process. Science advances much faster than resulting legislation, one of the pitfalls currently facing the Nagoya Protocol (Kumar, 2018), so this approach would have the additional benefit of accounting for the rapid pace of development in science.
Recommendations (and How to Streamline Processes Through the BBNJ Treaty?)
The BBNJ treaty has at least significant capacity to be of benefit to the science community with its global scope, and considering that the negotiations so far have proved to be constructive in the main part (Tiller et al., 2019), and acknowledging the important role of science in any governance mechanism for ABNJ. Here, we provide recommendations which could contribute to treaty discussions, addressing how to strengthen best-practice, to support access, sharing and transparency in relation to MGR.
Sample Archiving
Archiving samples and maintaining collections for as wide as possible future use is crucial for the research community, supporting taxonomy, reproducibility of research and reducing the need for repeat collection. Long-term specimen storage must be planned prior to sample collection and included in funding proposals, with requirements from funding bodies for archiving of material in national collections. Because cruises are so expensive, the post-cruise proportion of the budget is often contracted as a result (Glover et al., 2018). Data management and sample archiving must therefore be fully costed into a grant application, and samples and data made fully available from national collections and data repositories. Journals and genetic databases could require archiving of specimens in national collections before publishing of data. Much of the onus is on funders, whether public or private and regulatory bodies recognise the key importance of taxonomy and related archiving of voucher samples. Museums play a key role in ABS of MGR as well as supporting biodiversity research: the BBNJ agreement should support and recognise the importance of biorepositories and sample archiving.
Data Standards and Open-Access
In terms of data practices, the treaty with its global scope could support best-practice and open access by building consensus on standardisation. usage of currently available global data standards It could also make recommendations to that data standards are imbedded into protocols in sample and data collection, that all datasets are published in open-access journals and via the OBIS deep-sea node, that sequence data are deposited with reference to voucher material and associated site and collection information. A potential workflow illustrating these principles from cruise to data publication is suggested in Figure 4. While commercial entities are generally less willing to openly share their data, to protect their intellectual property, there are examples of companies showing commitment to open source, for example Geneious bioinformatics software provides access to their API (Kearse et al., 2012). In terms of biotechnology applications on MGR, Blasiak et al. (2019) state that scientists should disclose the origin of an MGR when a patent is filed. This could be adopted as common practice, and companies committing to transparency here would build goodwill and trust. Also site information is paramount for ABS to distinguish between AWNJ and ABNJ, and therefore which legal regime would be applicable to an MGR sample. In a data collection context, privately funded research expeditions should also take heed of best practices in data and sample collection, and provide access to their data. Similarly, sectors like deep-sea mining, oil and gas and fisheries should make their data available for research and monitoring purposes as far as feasible, particularly samples and data from environmental impact assessments. Journals could increase requirements of data and code publishing (bioinformatics pipelines and workflows for analysis of genomic data).
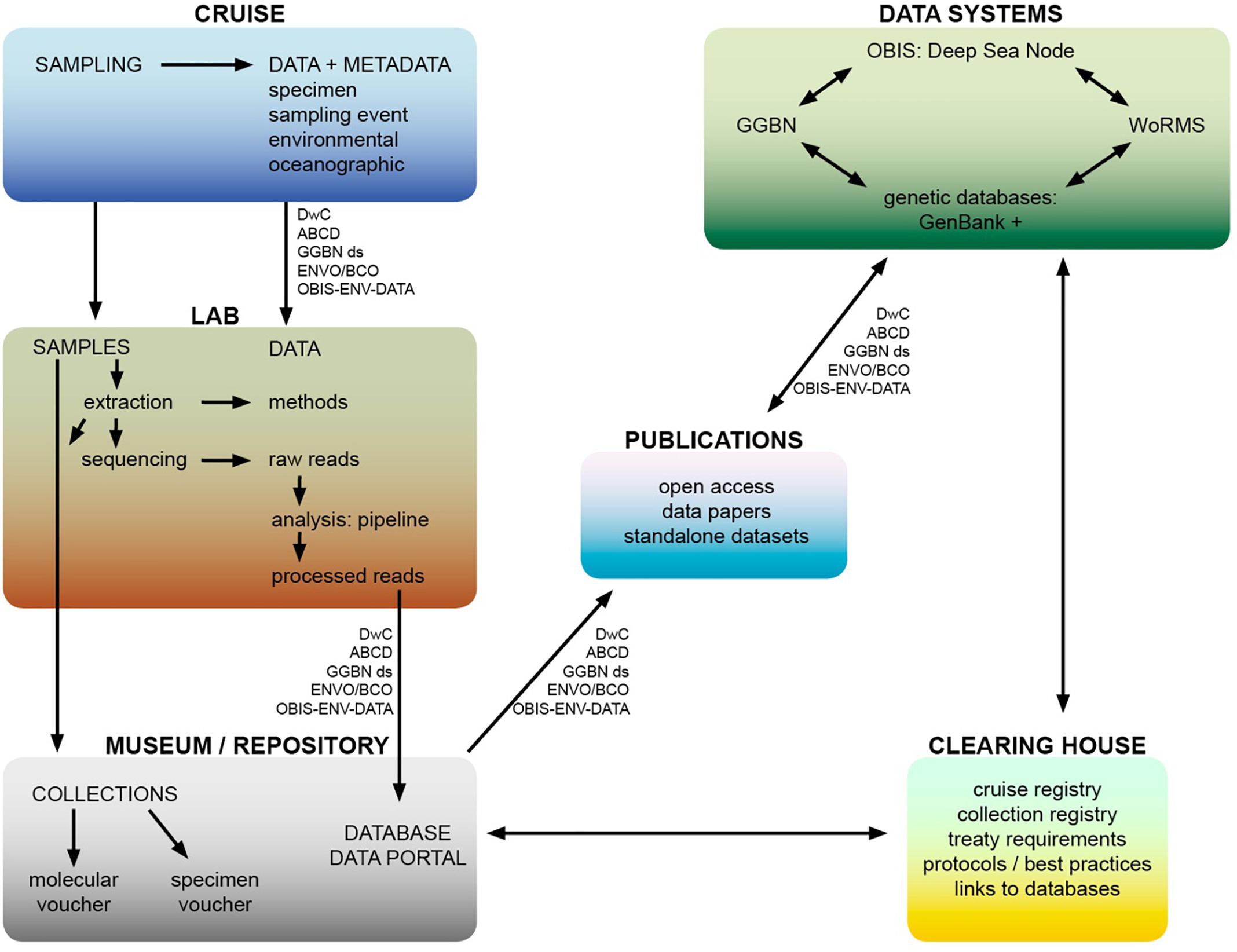
Figure 4. Potential workflow from cruise to data publication of access to MGR from ABNJ, including the scope of a potential clearing-house mechanism. Data standards are also shown, acronyms are as follows – Dwc: Darwin Core, ABCD: Access to Biological Collection Data GGBN ds: Global Genome Biodiversity Network data standard, ENVO: Environment Ontology, BCO: Biological Collections Ontology, OBIS-ENV-DATA: OBIS extendedMeasurementorFact DarwinCore Extension. These data standards are also listed in Table 1.
Registration of Cruises
Open access to genetic data and samples is critical to the functioning of the deep-sea research community and a key improvement to our current management is a global system to hold information on research cruises that are taking place and collecting samples in ABNJ (and AWNJ). This would be of great benefit to the deep-sea research community (Rabone et al., 2019). It is recommended therefore that the registration of cruises and a mechanism to manage this should be encouraged by the treaty. This mechanism should also aim to take into account privately-funded cruises together with their respective collections and datasets. A global registry of cruises also could facilitate greater shared research planning to identify gaps and/or overlaps as suggested by Oldham et al. (2014).
Clearing-House
A clearing-house mechanism has been proposed to facilitate the sharing of benefits from MGRs from ABNJ (President’s Aid to Negotiations, 2019). How the clearing-house will be administered remains up for debate, but one possibility is through the International Oceanographic Data and Information Exchange (IODE) Programme of IOC-UNESCO (UN Ocean Decade Report, 2018). The clearing-house could also leverage emerging data systems such as Ocean Data Information System (ODIS22), and Ocean Best Practices23. However, questions remain about the functions such a mechanism would need to include (President’s Aid to Negotiations, 2019). A centralised repository/ABS clearing-house mechanism could streamline processes by providing documentation, guidance, and links to existing platforms and databases relevant to MGR (Figure 4). The recommendations outlined in our paper could be streamlined by such a mechanism, including provisions for a global registry of cruises. The clearing-house could also include information about which institutes hold collections, their availability, and how they could be accessed. The Obligatory Prior Electronic Notification (OPEN) system proposes archiving of samples and data through track and trace compliance (Broggiato et al., 2018). While the OPEN system could ensure comprehensive tracking of samples and data, it may be a considerable administrative burden to undertake for all MGR samples collected from ABNJ. A more flexible approach would be a commitment to a common set of principles, such as FAIR data and open access as described here. Alternatively, exemptions for data and samples collected for research purposes could be considered, so that systems track and trace commercial applications only.
A clearing-house could also help with unworked samples. Numerous unsorted faunal samples from ABNJ currently exist (in museums and research institutes globally) and these are invisible to other researchers, since they are neither mentioned in peer-reviewed publications nor included in datasets uploaded to open-access database repositories. These include unsorted bulk specimens, archived but not worked on further, generally owing to funding and time constraints. A mechanism to make such unworked samples visible and their availability known to other researchers globally (Harden-Davies, 2017) would be efficient and potentially reduce the need for new costly sample collections in ABNJ, thereby supporting ABS. These collections could be published on existing platforms i.e., OBIS, using existing functionality. There are potential implications here for the home institution, so availability of these samples and associated data would require additional resourcing. The clearing-house would in essence be a mechanism not to create a ‘new’ data system – but a means to strengthen and improve access to and awareness of existing systems and encourage best-practices. Having a central collation of available collections, relevant documentation and a repository of best-practice protocols would be of great benefit to the scientific community.
Conclusion
The inherent difficulties in undertaking work in deep-sea environments and the resulting legacy data-gaps for the deep-sea mean that access to MGR from ABNJ is of crucial importance to the scientific community. Fundamental taxonomic research provides a unifying element to the BBNJ negotiations, as the four main components of the future treaty all depend on knowledge of the species that live in ABNJ. Support for taxonomy/biodiversity research can therefore help to achieve goals of the treaty, allowing sound implementation of conservation and environmental management. ABS through access to samples and data is happening now, but there is room for improvements in current processes. Commitment to open access and best practice provides a pathway to achieve these improvements, for example though usage of robust and FAIR data and sample management pipelines, as discussed in our case study in the CCZ. We have outlined recommendations to streamline access to MGR from ABNJ. In particular we recommend the following: funders (both public and private) and regulators recognise the central importance of taxonomy, through support of museums/collections for long-term sample curation; open access to data (including that generated by private industry/companies); usage of globally recognised data standards; usage (and further development) of existing platforms, commitment to best-practice workflows; publishing of datasets via global online repositories and open access journals; increased integration of observation initiatives and biodiversity research, a global registry of cruises; and lastly, development of a clearing house to allow centralised access to the above. It is important that the treaty is flexible enough to adapt as science and technology evolves, and recognises both the realities of undertaking biological research as well as fundamental biological realities themselves, Ongoing consultation and communication between the science community and policy community will be key here. The future treaty has the potential to support science through improved awareness of best-practice, access to data and samples, and benefit-sharing, drawing on lessons from the Nagoya Protocol. Together with recent advancements in best-practice in the research community, existing platforms and data systems with robust data quality processes, current networks, and discussions at the recent Intergovernmental Conference at UN, the elements are in place to build an agreement that can support science and society.
Author Contributions
MR and HH-D conceived the idea for the manuscript. TH, JC, HH-D, and MR defined the scope of the manuscript. JC and HH-D wrote the initial outline. WA and MR undertook the data analysis and prepared the figures. MR and TH led the writing and editing of the manuscript. All authors contributed to this process and approved the final version of the manuscript.
Funding
This work was supported by the Swedish Research Council FORMAS, the German Research Foundation and the OpenAccess Publication Fund of Freie Universität Berlin. The ABYSSLINE UK-1 benthic taxonomy research undertaken at the Natural History Museum has been funded by the UK Seabed Resources. While no funding was received for the writing of this work, MR was supported by the Wellcome Trust Grant (104958/Z/14/Z), at the NHM and TH was supported by the Natural Environment Research Council (NERC) National Capability funding to the National Oceanography Centre, as part of the Climate Linked Atlantic Section Science (CLASS) programme (Grant Number NE/R015953/1). JC was supported by the Marie Curie H2020 fund (H2020-MSCA-ITN-ETN: MarPipe GA 721421). HH-D was supported by the Nereus Fellow Program. This work was also supported by the National Council of Science and Technology of Mexico (201441) and the Gulf of Mexico Research Consortium (CIGoM).
Conflict of Interest Statement
JC is employed by ABS-int.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Deep Ocean Stewardship Initiative (DOSI) particularly Maria Baker (University of Southampton), the DOSI Marine Genetic Resources (MGR) working group particularly Marcel Jaspars, and also Jeff Marlow for provision of Figure 1; Sarah Long, Chris Lyal, and Gordon Paterson at the Natural History Museum for discussions about topics relevant to the manuscript, Jackie Mackenzie-Dodds at the NHM Molecular Collections Facility, and Chiho Ikebe (NHM) for support in curation of the UK-1 benthic macrofauna collections at the NHM, the attendees of the Senkenberg ‘Deep-Sea taxonomy under pressure’ workshop organised by Torben Riehl for relevant discussions, and Daniel L. Geiger (Santa Barbara Museum of Natural History), and the three reviewers for their helpful comments on the manuscript.
Footnotes
- ^ https://nerc.marinefacilitiesplanning.com/programme
- ^ http://www.rvdata.us/overview
- ^ https://www.bodc.ac.uk/resources/inventories/cruise_inventory
- ^ https://cetaf.org/services/natural-science-collections-and-access-and-benefit-sharing
- ^ http://natsca.org/NHNearYou
- ^ http://www.ggbn.org/
- ^ https://nerc.ukri.org/research/sites/data/policy/
- ^ https://www.ncbi.nlm.nih.gov/WebSub/?tool=genbank
- ^ Data extracted from OBIS on 12 May 2019.
- ^ SDG14: to ‘conserve and sustainably use the oceans, seas and marine resources for sustainable development’.
- ^ http://www.ggbn.org/ggbn_portal/search/record?unitID=8056784&collectioncode=ZOO&institutioncode=NHMUK
- ^ http://obis.org/manual/eml/
- ^ https://www.gbif.org/ipt
- ^ https://github.com/
- ^ https://www.tdwg.org/community/bdq/
- ^ https://github.com/GLOMICON/asvBiomXchange
- ^ ‘A global research network that supports the development of new network services’ https://www.planet-lab.org/.
- ^ http://dosi-project.org/
- ^ https://cetaf.org/taxonomy/publications
- ^ https://library.ggbn.org/
- ^ http://ocean-partners.org/pogo-scor-fellowship
- ^ https://catalogue.odis.org/
- ^ http://www.oceandatapractices.net/
References
Amon, D. J., Ziegler, A. F., Dahlgren, T. G., Glover, A. G., Goineau, A., Gooday, A. J., et al. (2016). Insights into the abundance and diversity of abyssal megafauna in a polymetallic-nodule region in the eastern Clarion-Clipperton Zone. Sci. Rep. 6:30492. doi: 10.1038/srep30492
Anderson, K., Ryan, B., Sonntag, W., Kavvada, A., and Friedl, L. (2017). Earth observation in service of the 2030 agenda for sustainable development. Geo Spat. Inform. Sci. 20, 77–96. doi: 10.1080/10095020.2017.1333230
Andruszkiewicz, E. A., Starks, H. A., Chavez, F. P., Sassoubre, L. M., Block, B. A., and Boehm, A. B. (2017). Biomonitoring of marine vertebrates in Monterey Bay using eDNA metabarcoding. PLoS One 12:e0176343. doi: 10.1371/journal.pone.0176343
Appeltans, W., and Webb, T. J. (2014). Biodiversity baselines in the deep sea. Deep Sea Life 4, 45–46.
Baker, E., Rycroft, S., and Smith, V. (2014). Linking multiple biodiversity informatics platforms with Darwin Core Archives. Biodivers. Data J. 2:e1039. doi: 10.3897/BDJ.2.e1039
Bax, N., Harden-Davies, H., Thiele, T., and Halpin, P. (2016). Open data: enabling conservation and sustainable use of biodiversity in areas beyond national jurisdiction. Nereus policy Brief. 1–3.
Beaman, R. S., and Cellinese, N. (2012). Mass digitization of scientific collections: new opportunities to transform the use of biological specimens and underwrite biodiversity science. ZooKeys 209, 7–17. doi: 10.3897/zookeys.209.3313
Bennear, L. S. (2015). Offshore oil and gas drilling: a review of regulatory regimes in the United States, United Kingdom, and Norway. Rev. Environ. Econ. Policy 9, 2–22. doi: 10.1093/reep/reu013
Benson, A., Brooks, C. M., Canonico, G., Duffy, E., Muller-Karger, F., Sosik, H. M., et al. (2018). Integrated observations and informatics improve understanding of changing marine ecosystems. Front. Mar. Sci. 5:428. doi: 10.3389/fmars.2018.00428
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Wheeler, D. L. (2008). GenBank. Nucleic Acids Res. 36, D25–D30. doi: 10.1093/nar/gkm929
Benson, E. E., Harding, K., and Mackenzie-Dodds, J. (2016). A new quality management perspective for biodiversity conservation and research: investigating biospecimen reporting for improved study quality (BRISQ) and the standard PRE -analytical Code (SPREC) using natural history museum and culture collections as case studies. Syst. Biodivers. 14, 1–23. doi: 10.1080/14772000.2016.1201167
Betsou, F., Bilbao, R., Case, J., Chuqui, R., Clements, J. A., and De Souza, Y. (2018). Standard PREanalytical code version 3.0. Biopreserv. Biobank. 16, 9–12.
Bingham, H., Doudin, M., Weatherdon, L., Despot-Belmonte, K., Wetzel, F., Groom, Q., et al. (2017). The biodiversity informatics landscape: elements, connections and opportunities. Res. Ideas Outcomes 3:e14059. doi: 10.3897/rio.3.e14059
Blasiak, R., Jouffray, J.-B., Wabnitz, C. C. C., and Österblom, H. (2019). Scientists should disclose origin in marine gene patents. Trends Ecol. Evol. 34, 392–395. doi: 10.1016/j.tree.2019.02.004
Blasiak, R., Jouffray, J.-B., Wabnitz, C. C. C., Sundström, E., and Österblom, H. (2018). Corporate control and global governance of marine genetic resources. Sci. Adv. 4:eaar5237. doi: 10.1126/sciadv.aar5237
Bosch, S., Provoost, P., and Appeltans, W. (2018). Fit for purpose? Identifying and resolving quality issues with marine biodiversity datasets in R. PeerJ Preprints 6:e26776v1. doi: 10.7287/peerj.preprints.26776
Brandt, A., Scholz, J., Allspach, A., Brenke, N., Brix, S., George, K. H., et al. (2018). 200 Years of marine research at Senckenberg – selected highlights. Mar. Biodivers. 48, 159–178. doi: 10.1007/s12526-017-0839-4
Broggiato, A., Vanagt, T., Lallier, L. E., Jaspars, M., Burton, G., and Muyldermans, D. (2018). Mare geneticum: balancing governance of marine genetic resources in international waters. Int. J. Mar. Coastal Law 33, 3–33. doi: 10.1163/15718085-13310030
Buttigieg, P. L., Morrison, N., Smith, B., Mungall, C. J., and Lewis, S. E. (2013). The environment ontology - contextualising biological and biomedical entities. J. Biomed. Semant. 4:43. doi: 10.1186/2041-1480-4-43
Buttigieg, P. L., Pafilis, E., Lewis, S. E., Schildhauer, M. P., Walls, R. L., and Mungall, C. J. (2016). The environment ontology in 2016 - bridging domains with increased scope, semantic density, and interoperation. J. Biomed. Semant. 7:57. doi: 10.1186/s13326-016-0097-6
Cicero, C., Cook, J., Campbell, M., Hildebrandt, K., Mayfield, T., and Wieczorek, J. (2017). The arctos community model for sustaining and enriching access to biodiversity data. Biodivers. Inform. Sci. Stand. 1:e20466. doi: 10.3897/tdwgproceedings.1.20466
Clark, M. R., Consalvey, M., and Rowden, A. A. (2016). Biological Sampling in the Deep Sea. Chichester: John Wiley & Sons.
Consortium of European Taxonomic Facilities [CETAF] (2016). Strategy and Strategic Development Plan 2015-2025. Belgique: Consortium of European Taxonomic Facilities.
Consortium of European Taxonomic Facilities [CETAF] (2018). Access and Benefit Sharing Code of Conduct. Belgique: Consortium of European Taxonomic Facilities.
Consortium of European Taxonomic Facilities [CETAF] (2019). Digital Sequence Information on Genetic Resources – Concept and Benefit-Sharing Submission of Views. Belgique: Consortium of European Taxonomic Facilities.
Constable, A. J., Costa, D. P., Schofield, O., Newman, L., Urban, E. R. Jr., Fulton, E. A., et al. (2016). Developing priority variables (“ecosystem essential ocean variables” - eEOVs) for observing dynamics and change in Southern Ocean ecosystems. J. Mar. Syst. 161, 26–41. doi: 10.1016/j.jmarsys.2016.05.003
Cook, S., Dodge, C., Morgan, R., and Sandusky, G. E. (2015). DNA/RNA degradation rate in long term fixed museum specimens. Forensic Med. Anat. Res. 3, 1–6. doi: 10.4236/fmar.2015.31001
Costello, M. J., Horton, T., and Kroh, A. (2018). Sustainable biodiversity databasing: international, collaborative, dynamic, centralised. Trends Ecol. Evol. 33, 803–805. doi: 10.1016/j.tree.2018.08.006
Dagnall, C. L., Morton, L. M., Hicks, B. D., Li, S., Zhou, W., Karlins, E., et al. (2018). Successful use of whole genome amplified DNA from multiple source types for high-density Illumina SNP microarrays. BMC Genomics 19:182. doi: 10.1186/s12864-018-4572-6
Dahlgren, T. G., Wiklund, H., Rabone, M., Amon, D., Ikebe, C., Watling, L., et al. (2016). Abyssal fauna of the UK-1 polymetallic nodule exploration area, Clarion-Clipperton Zone, central Pacific Ocean: Cnidaria. Biodivers. Data J. 4:e9277. doi: 10.3897/BDJ.4.e9277
Danovaro, R. (2009). Methods for the Study of Deep-Sea Sediments, Their Functioning and Biodiversity. Boca Raton, FL: CRC Press, 458.
De Pooter, D., Appeltans, W., Bailly, N., Bristol, S., Deneudt, K., Eliezer, M., et al. (2017). Toward a new data standard for combined marine biological and environmental datasets - expanding OBIS beyond species occurrences. Biodivers. Data J. 5:e10989. doi: 10.3897/BDJ.5.e10989
De Santo, E. M. (2018). Implementation challenges of area-based management tools (ABMTs) for biodiversity beyond national jurisdiction (BBNJ). Mar. Policy 97, 34–43. doi: 10.1016/j.marpol.2018.08.034
Deck, J., Gaither, M. R., Ewing, R., Bird, C. E., Davies, N., Meyer, C., et al. (2017). The genomic observatories metadatabase (GeOMe): a new repository for field and sampling event metadata associated with genetic samples. PLoS Biol. 15:e2002925. doi: 10.1371/journal.pbio.2002925
Diepenbroek, M., Grobe, H., Reinke, M., Schindler, U., Schlitzer, R., and Sieger, R. (2002). PANGAEA—an information system for environmental sciences. Comput. Geosci. 28, 1201–1210. doi: 10.1016/S0098-3004(02)00039-0
DOSI (2016). Deep-Sea Marine Scientific Research and Genetic Resources in Areas Beyond National Jurisdiction: Submission. Available at: http://www.un.org/depts/los/biodiversity/prepcom_files/DOSI.pdf (accessed March 22, 2016).
Droege, G., Barker, K., Astrin, J. J., Bartels, P., Butler, C., Cantrill, D., et al. (2014). The global genome biodiversity network (GGBN) data portal. Nucleic Acids Res. 42, D607–D612. doi: 10.1093/nar/gkt928
Droege, G., Barker, K., Seberg, O., Coddington, J. A., Benson, E., Berendsohn, W. G., et al. (2016). The global genome biodiversity network (GGBN) data standard specification. Database 2016:baw125. doi: 10.1093/database/baw125
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. doi: 10.1093/nar/30.1.207
Escobar-Zepeda, A., Godoy-Lozano, E. E., Raggi, L., Segovia, L., Merino, E., Gutierrez-Rios, R. M., et al. (2018). Analysis of sequencing strategies and tools for taxonomic annotation: defining standards for progressive metagenomics. Sci. Rep. 8:12034. doi: 10.1038/s41598-018-30515-5
Fegraus, E. H., Andelman, S., Jones, M. B., and Schildhauer, M. (2005). Maximizing the value of ecological data with structured metadata: An introduction to Ecological Metadata Language (EML) and principles for metadata creation. Bulletin Ecol. Soc. Am. 86, 158–168. doi: 10.1890/0012-9623(2005)86[158:MTVOED]2.0.CO;2
Geijzendorffer, I. R., Regan, E. C., Pereira, H. M., Brotons, L., Brummitt, N., Gavish, Y., et al. (2016). Bridging the gap between biodiversity data and policy reporting needs: an essential biodiversity variables perspective. J. Appl. Ecol. 53, 1341–1350. doi: 10.1111/1365-2664.12417
GGBN (2017). GGBN Letter to CBD on Digital Sequence Information. Available at: https://library.ggbn.org/share/s/K_4MeUeQQXC92Q3BTSoMEg (accessed May 11, 2019).
Gjerde, K., Boteler, B., Durussel, C., Rochette, J., Unger, S., and Wright, G. (2018). Conservation and sustainable use of marine biodiversity in areas beyond national jurisdiction: options for underpinning a strong global BBNJ agreement through regional and sectoral governance. Strong High Seas Project
Glover, A. G., Dahlgren, T. G., Wiklund, H., Mohrbeck, I., and Smith, C. R. (2015). An end-to-end DNA taxonomy methodology for benthic biodiversity survey in the Clarion-Clipperton Zone, central Pacific abyss. J. Mar. Sci. Eng. 4:2. doi: 10.3390/jmse4010002
Glover, A. G., Higgs, N., and Horton, T. (2019). World Register of Deep-Sea species (WoRDSS). Available at: http://www.marinespecies.org/deepsea (accessed Feberuary 15, 2019).
Glover, A. G., Wiklund, H., Chen, S., and Dahlgren, T. G. (2018). Managing a sustainable deep-sea “blue economy” requires knowledge of what actually lives there. eLife 7:e41319. doi: 10.7554/eLife.41319
Glover, A. G., Wiklund, H., Rabone, M., Amon, D., Smith, C., O’Hara, T., et al. (2016). Abyssal fauna of the UK-1 polymetallic nodule exploration claim, Clarion-Clipperton Zone, central Pacific Ocean: Echinodermata. Biodivers. Data J. 4:e7251. doi: 10.3897/BDJ.4.e7251
Godoy-Lozano, E. E., Escobar-Zepeda, A., Raggi, L., Merino, E., Gutierrez-Rios, R. M., Juarez, K., et al. (2018). Bacterial diversity and the geochemical landscape in the southwestern Gulf of Mexico. Front. Microbiol. 9:2528. doi: 10.3389/fmicb.2018.02528
Goodwin, K. D., Thompson, L. R., Duarte, B., Kahlke, T., Thompson, A. R., Marques, J. C., et al. (2017). DNA sequencing as a tool to monitor marine ecological status. Front. Mar. Sci. 4:107. doi: 10.3389/fmars.2017.00107
Grassle, F. (2000). The ocean biogeographic information system (OBIS): an on-line, worldwide atlas for accessing, modeling and mapping marine biological data in a multidimensional geographic context. Oceanography 13, 5–7. doi: 10.5670/oceanog.2000.01
Gratton, P., Marta, S., Bocksberger, G., and Winte, M. (2016). A world of sequences: can we use georeferenced nucleotide databases for a robust automated phylogeography? J. Biogeogr. 44, 475–486. doi: 10.1111/jbi.12786
Güntsch, A., Hyam, R., Hagedorn, G., Chagnoux, S., Röpert, D., Casino, A., et al. (2017). Actionable, long-term stable and semantic web compatible identifiers for access to biological collection objects. Database 2017:bax003. doi: 10.1093/database/bax003
Guralnick, R., Walls, R., and Jetz, W., (2018). Humboldt Core – toward a standardized capture of biological inventories for biodiversity monitoring, modeling and assessment. Ecography 41, 713–725. doi: 10.1111/ecog.02942
Harden-Davies, H. (2017). Deep-sea genetic resources: new frontiers for science and stewardship in areas beyond national jurisdiction. Deep Sea Res. II 137, 504–513. doi: 10.1016/j.dsr2.2016.05.005
Harden-Davies, H. (2018). The next wave of science diplomacy: marine biodiversity beyond national jurisdiction. ICES J. Mar. Sci. 75, 426–434. doi: 10.1093/icesjms/fsx165
Harden-Davies, H., and Gjerde, K. (2019). Building scientific and technological capacity: a role for benefit-sharing in the conservation and sustainable use of marine biodiversity beyond National Jurisdiction. Ocean Yearb. 33, 377–400. doi: 10.1163/9789004395633_015
Hardisty, A. R., Bacall, F., Beard, N., Balcázar-Vargas, M.-P., Balech, B., Barcza, Z., et al. (2016). BioVeL: a virtual laboratory for data analysis and modelling in biodiversity science and ecology. BMC Ecol. 16:49. doi: 10.1186/s12898-016-0103-y
Higgs, N. D., and Attrill, M. J. (2015). Biases in biodiversity: wide-ranging species are discovered first in the deep sea. Front. Mar. Sci. 2:61. doi: 10.3389/fmars.2015.00061
Hobern, D., Baptiste, B., Copas, K., Guralnick, R., Hahn, A., van Huis, E., et al. (2019). Connecting data and expertise: a new alliance for biodiversity knowledge. Biodivers. Data J. 7:e33679. doi: 10.3897/BDJ.7.e33679
Holetschek, J., Droege, G., Güntsch, A., and Berendsohn, W. G. (2012). The ABCD of primary biodiversity data access. Plant Biosyst. Int. J. Deal. Aspects Plant Biol. 146, 771–779. doi: 10.1080/11263504.2012.740085
Holetschek, J., Kelbert, P., Muller, A., Ciardelli, P., Güntsch, A., and Berendsohn, W. G. (2009). “International networking of large amounts of primary biodiversity data,” in Proceedings Informatik 2009 - Im Focus das Leben, Vol. 26, (Lübeck), 552–564.
Horton, T., Gofas, S., Kroh, A., Poore, G. C. B., Read, G., Rosenberg, G., et al. (2017). Improving nomenclatural consistency: a decade of experience in the World Register of Marine Species. Eur. J. Taxon. 389, 1–24. doi: 10.5852/ejt.2017.389
Huber, J. T. (1998). The importance of voucher specimens, with practical guidelines for preserving specimens of the major invertebrate phyla for identification. J. Nat. History 32, 367–385. doi: 10.1080/00222939800770191
International Chamber of Commerce [ICC] (2017). Commission on Intellectual Property Digital Sequence Information Report. Available at: https://cdn.iccwbo.org/content/uploads/sites/3/2017/05/ICC-IP-position-paper-on-digital-sequence-information.pdf (accessed May 11, 2019).
Jaspars, M., De Pascale, D., Andersen, J. H., Reyes, F., Crawford, A. D., and Ianora, A. (2016). The marine biodiscovery pipeline and ocean medicines of tomorrow. J. Mar. Biol. Assoc. United Kingdom 96, 151–158. doi: 10.1017/S0025315415002106
Jetz, W., McPherson, J. M., and Guralnick, R. P. (2012). Integrating biodiversity distribution knowledge: toward a global map of life. Trends Ecol. Evol. 27, 151–159. doi: 10.1016/j.tree.2011.09.007
Jorem, A., and Tvedt, M. W. (2014). Bioprospecting in the high seas: existing rights and obligations in view of a new legal regime for marine areas beyond National Jurisdiction. Int. J. Mar. Coastal Law 29, 321–324. doi: 10.1163/15718085-12341319
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Keeling, P. J., Burki, F., Wilcox, H. M., Allam, B., Allen, E. E., Amaral-Zettler, L. A., et al. (2014). The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. doi: 10.1371/journal.pbio.1001889
Kelbert, P., Droege, G., Barker, K., Braak, K., Cawsey, E. M., Coddington, J. A., et al. (2015). B-HIT - a tool for harvesting and indexing biodiversity data. PLoS One 10:e0142240. doi: 10.1371/journal.pone.0142240
Kissling, W. D., Ahumada, J. A., Bowser, A., Fernandez, M., Fernández, N., García, E. A., et al. (2018). Building essential biodiversity variables (EBVs) of species distribution and abundance at a global scale. Biol. Rev. Camb. Philos. Soc. 93, 600–625. doi: 10.1111/brv.12359
Kopf, A., Bicak, M., Kottmann, R., Schnetzer, J., Kostadinov, I., Lehmann, K., et al. (2015). The ocean sampling day consortium. GigaScience 4:27. doi: 10.1186/s13742-015-0066-5
Kroh, A., Costello, M. J., and Horton, T. (2019). Biodiversity databases in the future: reply to Cene Fišer. Trends Ecol. Evol. 34, 185–186. doi: 10.1016/j.tree.2018.12.011
Kumar, D. (2018). The Nagoya protocol: legal protections for genetic resources and ramifications for aquatic science. Assoc. Sci. Limnol. Oceanogr. 27, 31–35. doi: 10.1002/lob.10235
Laird, S., and Wynberg, R. (2018). A Fact-Finding and Scoping Study on Digital Sequence Information on Genetic Resources in the Context of the Convention on Biological Diversity and the Nagoya Protocol. Rio de Janeiro: Convention on Biological Diversity.
Langmead, B. (2010). Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics 32, 11.7.1–11.7.14. doi: 10.1002/0471250953.bi1107s32
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Leary, D. (2019). Agreeing to disagree on what we have or have not agreed on: the current state of play of the BBNJ negotiations on the status of marine genetic resources in areas beyond national jurisdiction. Mar. Policy 99, 21–29. doi: 10.1016/j.marpol.2018.10.031
Leary, D., and Juniper, S. K. (2013). “Addressing the marine genetic resources issue: is the bdebate heading in the wrong direction?,” in The Limits of Maritime Jurisdiction, eds C. H. Schofield, S. Lee, and M. Kwon (Leiden: BRILL), 769–785. doi: 10.1163/9789004262591_037
Leinonen, R., Sugawara, H., and Shumway, M. (2010). The sequence read archive. Nucleic Acids Res. 39, 19–21. doi: 10.1093/nar/gkq1019
León-Zayas, R., Peoples, L., Biddle, J. F., Podell, S., Novotny, M., Cameron, J., et al. (2017). The metabolic potential of the single cell genomes obtained from the Challenger Deep, Mariana Trench within the candidate superphylum Parcubacteria (OD1). Environ. Microbiol. 19, 2769–2784. doi: 10.1111/1462-2920.13789
Levin, L. (2018). Manifestation, drivers, and emergence of open ocean deoxygenation. Annu. Rev. Mar. Sci. 10, 229–260. doi: 10.1146/annurev-marine-121916-163359
Levin, L. A., Bett, B. J., Gates, A. R., Heimbach, P., Howe, B. M., Janssen, F., et al. (2019). Global observing needs in the Deep Ocean. Front. Mar. Sci. 6:241. doi: 10.3389/fmars.2019.00241
Li, W., Cowley, A., Uludag, M., Gur, T., McWilliam, H., Squizzato, S., et al. (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, 580–584. doi: 10.1093/nar/gkv279
Liew, Y. J., Aranda, M., and Voolstra, C. R. (2016). Reefgenomics.Org - a repository for marine genomics data. Database 2016:baw152. doi: 10.1093/database/baw152
Lim, S.-C., Wiklund, H., Glover, A. G., Dahlgren, T. G., and Tan, K.-S. (2017). A new genus and species of abyssal sponge commonly encrusting polymetallic nodules in the Clarion-Clipperton Zone, East Pacific Ocean. Syst. Biodivers. 15, 507–519. doi: 10.1080/14772000.2017.1358218
Marlow, J., Harden-Davies, H., Snelgrove, P., Jaspars, M., Blasiak, R., and Wabnitz, C. (2019). The Full Value of Marine Genetic Resources (MGR). Deep Ocean Stewardship Initiative Policy Brief. Available at: http://dosi-project.org/wp-content/uploads/2018/05/Full-value-mgr-March2019.pdf (accessed May 11, 2019).
Mayer, L., Jakobsson, M., Allen, G., Dorschel, B., Falconer, R., Ferrini, V., et al. (2018). The Nippon Foundation—GEBCO Seabed 2030 Project: the quest to see the world’s oceans completely mapped by 2030. Geosciences 8:63. doi: 10.3390/geosciences8020063
Mengerink, K. J., Van Dover, C. L., Ardron, J., Baker, M., Escobar-Briones, E., Gjerde, K., et al. (2014). A call for deep-ocean stewardship. Science 344, 696–698. doi: 10.1126/science.1251458
Michener, W. (2015). Ecological data sharing. Ecol. Inform. 29, 33–44. doi: 10.1016/j.ecoinf.2015.06.010
Miloslavich, P., Bax, N. J., Simmons, S. E., Klein, E., Appeltans, W., and Aburto-Oropeza, O. (2018). Essential ocean variables for global sustained observations of biodiversity and ecosystem changes. Glob. Change Biol. 24, 2416–2433. doi: 10.1111/gcb.14108
Moore, H. M., Kelly, A. B., Jewell, S. D., McShane, L. M., Clark, D. P., Greenspan, R., et al. (2011). Biospecimen reporting for improved study quality (BRISQ). J. Proteome Res. 10, 3429–3438. doi: 10.1021/pr200021n
Morgera, E. (2018). Fair and equitable benefit-sharing in a new treaty on marine biodiversity: a principled approach towards partnership building? Maritime Saf. Secur. Law J. 5, 48–77.
Morris, R. A., Barve, V., Carausu, M., Chavan, V., Cuadra, J., and Freeland, C. (2013). Discovery and publishing of primary biodiversity data associated with multimedia resources: the Audubon Core strategies and approaches. Biodivers. Inform. 8, 185–197. doi: 10.17161/bi.v8i2.4117
Mukherjee, S., Stamatis, D., Bertsch, J., Ovchinnikova, G., Verezemska, O., Isbandi, M., et al. (2016). Genomes OnLine Database (GOLD) v.6: data updates.and feature enhancements. Nucleic Acids Res. 45, D446–D456. doi: 10.1093/nar/gkw992
Muller-Karger, F. E., Miloslavich, P., Bax, N. J., Simmons, S., Costello, M. J., Sousa Pinto, I., et al. (2018). Advancing marine biological observations and data requirements of the complementary essential ocean variables (EOVs) and essential biodiversity variables (EBVs) frameworks. Front. Mar. Sci. 5:211. doi: 10.3389/fmars.2018.00211
Neumann, D., Borisenko, A. V., Coddington, J. A., Hauser, C. I, Butler, C. R., Casino, A., et al. (2018). Global biodiversity research tied up by juridical interpretations of access and benefit-sharing. Organ. Divers. Evol. 18, 1–12. doi: 10.1007/s13127-017-0347-1
Niner, H. J., Ardron, J. A., Escobar, E. G., Gianni, M., Jaeckel, A., Jones, D. O. B., et al. (2018). Deep-sea mining with no net loss of biodiversity—an impossible aim. Front. Mar. Sci. 5:53. doi: 10.3389/fmars.2018.00053
Nussbeck, S. Y., Rabone, M., Benson, E. E., Droege, G., Mackenzie-Dodds, J., and Lawlor, R. T. (2016). Life in data’- outcome of a multi-disciplinary, interactive biobanking conference session on sample data. Biopreserv. Biobank. 14, 1–8. doi: 10.1089/bio.2015.0061
O’Hara, T., Tittensor, D., Baker, M., Stocks, K., and Appeltans, W. (2015). OBIS: a global deep sea biodiversity data sharing platform. Deep Sea Life 6:12.
Okonechnikov, K., Golosova, O., and Fursov, M., and The Ugene team (2012). Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. doi: 10.1093/bioinformatics/bts091
Oldham, P., Hall, S., Barnes, C., Oldham, C., Cutter, M., Burns, N., et al. (2014). Valuing the Deep: Marine Genetic Resource in Areas Beyond National Jurisdiction Defra Contract MB0128– A Review of Current Knowledge Regarding Marine Genetic Resources and their Current and Projected Economic Value to the UK economy. Final Report Version One. London: DEFRA.
Page, R. D. M. (2016). DNA barcoding and taxonomy: dark taxa and dark texts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150334. doi: 10.1098/rstb.2015.0334
Palero, F., Hall, S., Clark, P. F., Johnston, D., Mackenzie-Dodds, J., and Thatje, S. (2010). DNA extraction from formalin-fixed tissue: new light from the deep sea. Sci. Mar. 74, 465–470. doi: 10.3989/scimar.2010.74n3465
Pampel, H., Vierkant, P., Scholze, F., Bertelmann, R., Kindling, M., Klump, J., et al. (2013). Making research data repositories visible: the re3data.org registry. PLoS One 8:e78080. doi: 10.1371/journal.pone.0078080
Parr, C., Wilson, N., Leary, P., Schulz, K., Lans, K., Walley, L., et al. (2014). The encyclopedia of life v2: providing global access to knowledge about life on earth. Biodivers. Data J. 2:e1079. doi: 10.3897/BDJ.2.e1079
Pearlman, J. S., Buttigieg, P. L., Simpson, P., Mas, C. M., Heslop, E., and Hermes, J. (2017). “Accessing existing and emerging best practices for ocean observation a new approach for end-to-end management of best practices,” in Proceedings of the OCEANS 2017 - Anchorage, (Anchorage, AK), 1–7.
Penev, L., Agosti, D., Georgiev, T., Catapano, T., Miller, J., Blagoderov, V., et al. (2010). Semantic tagging of and semantic enhancements to systematics papers: ZooKeys working examples. ZooKeys 50, 1–16. doi: 10.3897/zookeys.50.538
Penev, L., Mietchen, D., Chavan, V., Hagedorn, G., Smith, V., Shotton, D., et al. (2017). Strategies and guidelines for scholarly publishing of biodiversity data. Res. Ideas Outcomes 3:e12431. doi: 10.3897/rio.3.e12431
Pethiyagoda, R. (2004). Biodiversity law has had some unintended effects. Nature 429:129. doi: 10.1038/429129a
Piel, W. H., Donoghue, M. J., and Sanderson, M. J. (1999). “TreeBASE: a database of phylogenetic information,” in Proceedings of the Abstracts of the 16th International Botanical Congress, (St. Louis).
Poore, G. C. B., Avery, L., Błażewicz-Paszkowycz, M., Browne, J., Bruce, N. L., Gerken, S., et al. (2015). Invertebrate diversity of the unexplored marine western margin of Australia: taxonomy and implications for global biodiversity. Mar. Biodivers. 45:271. doi: 10.1007/s12526-014-0255-y
Pope, L. C., Liggins, L., Keyse, J., Carvalhos, S., and Riginos, C. (2015). Not the time or the place: the missing spatio-temporal link in publicly available genetic data. Mol. Ecol. 24, 3802–3809. doi: 10.1111/mec.13254
Prathapan, K. D., Pethiyagoda, R., Bawa, K. S., Raven, P. H., and Rajan, D. P., and 172 co-signatories from 35 countries (2018). When the cure kills–CBD limits biodiversity research. Science 360, 1405–1406. doi: 10.1126/science.aat9844
President’s aid to negotiations (2019). “Intergovernmental conference on an international legally binding instrument under the United Nations Convention on the Law of the Sea on the conservation and sustainable use of marine biological diversity of areas beyond national jurisdiction. Second session,” in Proceedings of the Intergovernmental Conference On Marine Biodiversity Of Areas Beyond National Jurisdiction, (New York, NY).
Proença, V., Martin, L. J., Pereira, H. M., Fernandez, M., McRae, L., Belnap, J., et al. (2017). Global biodiversity monitoring: from data sources to essential biodiversity variables. Biol. Conserv. 213, 256–263. doi: 10.1016/j.biocon.2016.07.014
Pyle, R. L., and Michel, E. (2008). ZooBank: developing a nomenclatural tool for unifying 250 years of biological information. Zootaxa 1950, 39–50. doi: 10.11646/zootaxa.1950.1.6
Rabone, M., Harden-Davies, H., Horton, T., Jaspars, M., Gjerde, K., Baker, M., et al. (2019). Accessing and Sharing Benefits from Marine Genetic Resources from Areas Beyond National Jurisdiction: Building on Best Practices in the Scientific Community. Available at: http://dosi-project.org/wp-content/uploads/2018/05/027-DOSI-MGR-2-print-2-2.pdf (accessed May 11, 2019).
Robertson, T., Döring, M., Guralnick, R., Bloom, D., Wieczorek, J., Braak, K., et al. (2014). The GBIF integrated publishing toolkit: facilitating the efficient publishing of biodiversity data on the Internet. PLoS One 9:e102623. doi: 10.1371/journal.pone.0102623
Ramirez-Llodra, E., Brandt, A., Danovaro, R., De Mol, B., Escobar, E., German, C. R., et al. (2010). Deep, diverse and definitely different: unique attributes of the world’s largest ecosystem. Biogeosciences 7, 2851–2899.
Ruane, S., and Austin, C. C. (2017). Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Mol. Ecol. Resourc. 17, 1003–1008. doi: 10.1111/1755-0998.12655
Schildhauer, M., Jones, M. B., Bowers, S., Madin, J., Krivov, S., Pennington, D., et al. (2016). OBOE: the Extensible Observation Ontology, version 1.1. KNB Data Repository.
Schindel, D., Miller, S., Trizna, M., Graham, E., and Crane, A. (2016). The global registry of biodiversity repositories: a call for community curation. Biodivers. Data J. 4:e10293. doi: 10.3897/BDJ.4.e10293
Scott, B., Baker, E., Woodburn, M., Vincent, S., Hardy, H., and Smith, V. S. (2019). The natural history museum data portal. Database 2019:baz038. doi: 10.1093/database/baz038
Senderov, V., Simov, K., Franz, N., Stoev, P., Catapano, T., Agosti, D., et al. (2018). OpenBiodiv-O: ontology of the OpenBiodiv knowledge management system. J. Biomed. Semant. 9:5. doi: 10.1186/s13326-017-0174-5
Stephenson, P. J., Bowles-Newark, N., Regan, E., Stanwell-Smith, D., Diagana, M., and Höft, R. (2017). Unblocking the flow of biodiversity data for decision-making in Africa. Biol. Conserv. 213, 335–340.
Stow, C. A., Webster, K. E., Wagner, T., Lottig, N., Soranno, P. A., and Cha, Y. (2018). Small values in big data: the continuing need for appropriate metadata. Ecol. Inform. 45, 26–30. doi: 10.1016/j.ecoinf.2018.03.002
Swiss Academies of Arts and Sciences (2019). National significance of natural history collections in Switzerland. Swiss Acad. Rep. 14, 1–36.
Tateno, Y., Imanishi, T., Miyazaki, S., Fukami-Kobayashi, K., Saitou, N., Sugawara, H., et al. (2002). DNA Data Bank of Japan (DDBJ) for genome scale research in science. Nucleic Acids Res. 30, 27–30. doi: 10.1093/nar/30.1.27
ten Hoopen, P., Finn, R. D., Bongo, L. A., Corre, E., Fosso, B., Meyer, F., et al. (2017). The metagenomic data life-cycle: standards and best-practices. GigaScience 6, 1–11. doi: 10.1093/gigascience/gix047
ten Hoopen, P., Pesant, S., Kottmann, R., Kopf, A., Bicak, M., Claus, S., et al. (2015). Marine microbial biodiversity, bioinformatics and biotechnology (M2B3) data reporting and service standards. Stand. Genomic Sci. 10:20. doi: 10.1186/s40793-015-0001-5
Teytelman, L., Stoliartchouk, A., Kindler, L., and Hurwitz, B. L. (2016). Protocols.io: virtual communities for protocol development and discussion. PLoS Biol. 14:e1002538. doi: 10.1371/journal.pbio.10025
The Global Biodiversity Information Facility [GBIF] (2019). What is GBIF?. Copenhagen: Global Biodiversity Information Facility.
Tiller, R., De Santo, E., Mendenhall, E., and Nyman, E. (2019). The once and future treaty: towards a new regime for biodiversity in areas beyond national jurisdiction. Mar. Policy 99, 239–242. doi: 10.1016/j.marpol.2018.10.046
Tladi, D. (2015). The proposed implementing agreement: options for coherence and consistency in the establishment of protected areas beyond National Jurisdiction. Int. J. Mar. Coastal Law 30:4. doi: 10.1163/15718085-12341375
UN Ocean Decade Report (2018). Revised Roadmap for the UN Decade of Ocean Science for Sustainable Development. Updated Version 2.0. Paris: UNESCO.
Vandepitte, L., Bosch, S., Tyberghein, L., Waumans, F., Vanhoorne, B., Hernandez, F., et al. (2014). Fishing for data and sorting the catch: assessing the data quality, completeness and fitness for use of data in marine biogeographic databases. Database 2015:bau125. doi: 10.1093/database/bau125
Vandepitte, L., Vanhoorne, B., Decock, W., Vranken, S., Lanssens, T., Dekeyzer, S., et al. (2018). A decade of the world register of marine species - general insights and experiences from the data management team: where are we, what have we learned and how can we continue? PLoS One 13:e0194599. doi: 10.1371/journal.pone.0194599
Vierros, M., Suttle, C. A., Harden-Davies, H., and Burton, G. (2016). Who owns the ocean? policy issues surrounding marine genetic resources. Assoc. Sci. Limnol. Oceanogr. 25, 29–35. doi: 10.1002/lob.10108
Vision, T. (2010). The Dryad digital repository: published evolutionary data as part of the greater data ecosystem. Nat. Proc. doi: 10.1038/npre.2010.4595.1
Walls, R. L., Deck, J., Guralnick, R., Baskauf, S., Beaman, R., Blum, S., et al. (2014). Semantics in support of biodiversity knowledge discovery: an introduction to the biological collections ontology and related ontologies. PLoS One 9:e89606. doi: 10.1371/journal.pone.0089606
Weatherdon, L. V., Appeltans, W., Bowles-Newark, N., Brooks, T. M., Davis, F. E., Despot-Belmonte, K., et al. (2017). Blueprints of effective biodiversity and conservation knowledge products that support marine policy. Front. Mar. Sci. 4:96. doi: 10.3389/fmars.2017.00096
Webb, T. J., Vanden Berghe, E., and O’Dor, R. (2010). Biodiversity’s big wet secret: the global distribution of marine biological records reveals chronic under-exploration of the deep pelagic ocean. PLoS One 5:e10223. doi: 10.1371/journal.pone.0010223
Weibel, S., Kunze, J., Lagoze, C., and Wolf, M. (1998). Dublin Core Metadata for Resource Discovery. RFC 2413. Tempe, AZ: RFC.
Wetzel, F. T., Bingham, H. C., Groom, Q., Haase, P., Kõljalg, U., Kuhlmann, M., et al. (2018). Unlocking biodiversity data: prioritization and filling the gaps in biodiversity observation data in Europe. Biol. Conserv. 221, 78–85. doi: 10.1016/j.biocon.2017.12.024
Wetzel, F. T., Saarenmaa, H., Regan, E., Martin, C. S., Mergen, P., Smirnova, L., et al. (2015). The roles and contributions of biodiversity observation networks (BONs) in better tracking progress to 2020 biodiversity targets: a European case study. Biodiversity 16, 137–149. doi: 10.1080/14888386.2015.1075902
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., et al. (2012). Darwin core: an evolving community-developed biodiversity data standard. PLoS One 7:e29715. doi: 10.1371/journal.pone.0029715
Wiklund, H., Neal, L., Glover, A. G. G., Drennan, R., Rabone, M., and Dahlgren, T. G. (in press). Abyssal fauna. (of)polymetallic nodule exploration areas, eastern Clarion-Clipperton Zone, central Pacific Ocean: Annelida: Capitellidae, Opheliidae, Scalibregmatidae and Travisiidae. Zookeys.
Wiklund, H., Taylor, J. D., Dahlgren, T. G., Todt, C., Ikebe, C., Rabone, M., et al. (2017). Abyssal fauna of the UK-1 polymetallic nodule exploration area, Clarion-Clipperton Zone, central Pacific Ocean: Mollusca. ZooKeys 707, 1–46. doi: 10.3897/zookeys.707.13042
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 3:160018. doi: 10.1038/sdata.2016.18
Wilson, G., Bryan, J., Cranston, K., Kitzes, J., Nederbragt, L., and Teal, T. K. (2017). Good enough practices in scientific computing. PLoS Comput. Biol. 13:e1005510. doi: 10.1371/journal.pcbi.1005510
WoRMS Editorial Board (2019). World Register of Marine Species. Available at: http://www.marinespecies.org at VLIZ (accessed Feberuary 15, 2019).
Wright, G., Rochette, J., Gjerde, K. M., and Seeger, I. (2018b). The Long and Winding Road: Negotiating a Treaty for the Conservation and Sustainable Use of Marine Biodiversity in Areas Beyond National Jurisdiction. Paris: Institut du développement durable et des relations internationales (IDDRI) Studies, 82.
Wright, G., Gjerde, K. M., Johnson, D. E., Finkelstein, A., Adelaide Ferreira, M., Dunn, D. C., et al. (2018a). Marine spatial planning in areas beyond national jurisdiction. doi: 10.1016/j.marpol.2018.12.003 [in press]. Mar. Policy
Keywords: marine genetic resources (MGR), data standards, open access, taxonomy, biodiversity, access and benefit-sharing (ABS), conservation, areas beyond national jurisdiction (ABNJ)
Citation: Rabone M, Harden-Davies H, Collins JE, Zajderman S, Appeltans W, Droege G, Brandt A, Pardo-Lopez L, Dahlgren TG, Glover AG and Horton T (2019) Access to Marine Genetic Resources (MGR): Raising Awareness of Best-Practice Through a New Agreement for Biodiversity Beyond National Jurisdiction (BBNJ). Front. Mar. Sci. 6:520. doi: 10.3389/fmars.2019.00520
Received: 22 February 2019; Accepted: 09 August 2019;
Published: 03 September 2019.
Edited by:
Rob Harcourt, Macquarie University, AustraliaReviewed by:
Porter Hoagland, Woods Hole Oceanographic Institution, United StatesRachel Gjelsvik Tiller, SINTEF, Norway
Di Jin, Woods Hole Oceanographic Institution, United States
Copyright © 2019 Rabone, Harden-Davies, Collins, Zajderman, Appeltans, Droege, Brandt, Pardo-Lopez, Dahlgren, Glover and Horton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muriel Rabone, bS5yYWJvbmVAbmhtLmFjLnVr; Gabi Droege, Ry5Ecm9lZ2VAYmdibS5vcmc=
†ORCID: Muriel Rabone, orcid.org/0000-0002-8351-2313
 Muriel Rabone
Muriel Rabone Harriet Harden-Davies2
Harriet Harden-Davies2 Sabine Zajderman
Sabine Zajderman Ward Appeltans
Ward Appeltans Gabi Droege
Gabi Droege Angelika Brandt
Angelika Brandt Liliana Pardo-Lopez
Liliana Pardo-Lopez Adrian G. Glover
Adrian G. Glover Tammy Horton
Tammy Horton