
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 02 August 2019
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 6 - 2019 | https://doi.org/10.3389/fmars.2019.00494
This article is part of the Research Topic Exploring the Genetic Diversity of Marine Organisms Based on the Analysis of Chromosome and Genomic DNA Markers View all 10 articles
The commercially important brown alga Saccharina japonica in China has been believed to be accidentally introduced from Japan in 1920s. Meanwhile, spontaneous populations in the wild are assumed to be derived from the locally farmed populations. Spontaneous populations are often observed in the subtidal zones and on the cultivation infrastructure near farmed populations in the north of China. However, the genetic connectivity between these sympatric spontaneous and farmed populations remains unclear. Here, three commonly farmed cultivars (farmed populations) and three spontaneous populations (two from subtidal zones and one from cultivation rafts) were sampled from a typical kelp farm in Dalian, China, and analyzed with ten polymorphic microsatellite markers. Genetic diversity of farmed populations was found to be higher than that of the subtidal spontaneous populations. Neighbor joining cluster analysis based on genetic distance, Bayesian model-based structure analysis, and discriminant analysis of principal components revealed significant genetic divergence between the farmed populations and the subtidal spontaneous ones. Gene flow out of farmed populations to the subtidal spontaneous populations was revealed to be very limited, but gene flow in the contrary direction was more prominent. The spontaneous sporophytes on the structural rafts contained pedigree from both farmed and subtidal spontaneous populations. Results of this study may help us to understand reciprocal impacts between sympatric spontaneous and farmed populations of S. japonica.
The important economic kelp species Saccharina japonica (J. E. Areschoug) C. E. Lane, C. Mayes, Druehl, and G.W. Saunders has been assumed to be non-native to China, but unintentionally introduced from the north of Japan in 1920s (Tseng and Zhang, 1952; Tseng, 2001). This assumption is supported by the recent genetic diversity analysis of S. japonica populations from Japan and China as founder effect is revealed to be evident in Chinese populations (Shan et al., 2017). Originally, small scale cultivation experiments of this kelp was conducted using traditional Japanese method by throwing stones with attached spores to the sea (Hasegawa, 1976; Tseng, 2001). The experiments were first performed in Dalian, and later expanded to Yantai, Weihai and Qingdao in Shandong Province. Original spontaneous (wild) populations of S. japonica in rocky shores of these regions were thought to be derived from the zoospores escaping from cultivated individuals (Tseng and Zhang, 1952). Now the southernmost distribution of wild S. japonica populations in China is around 36°N latitude (Pang et al., 2007).
Conspicuous spontaneous populations of S. japonica are often observed in rocky subtidal zones and on the cultivation infrastructures adjacent to farmed populations in northern China (Figure 1). The sympatric distribution of farmed and spontaneous populations provides possibility of gene flow between them. On one hand, farmers and breeders are most concerned with the potential for contamination of wild pedigree into the farmed populations, which may alter the agronomical traits of the latter and thus degrade their quality or/and yield. On the other hand, spontaneous populations play important ecological roles in near shore rocky ecosystem, and may represent specific gene repertoire which can be very valuable materials in breeding programs (Fei, 2004; Zhang et al., 2017). From these perspectives, measures should be taken to prevent gene flow between them. It is therefore very important to evaluate the level of genetic connectivity between these sympatric farmed and spontaneous populations. However, no such studies have been conducted for S. japonica in China.
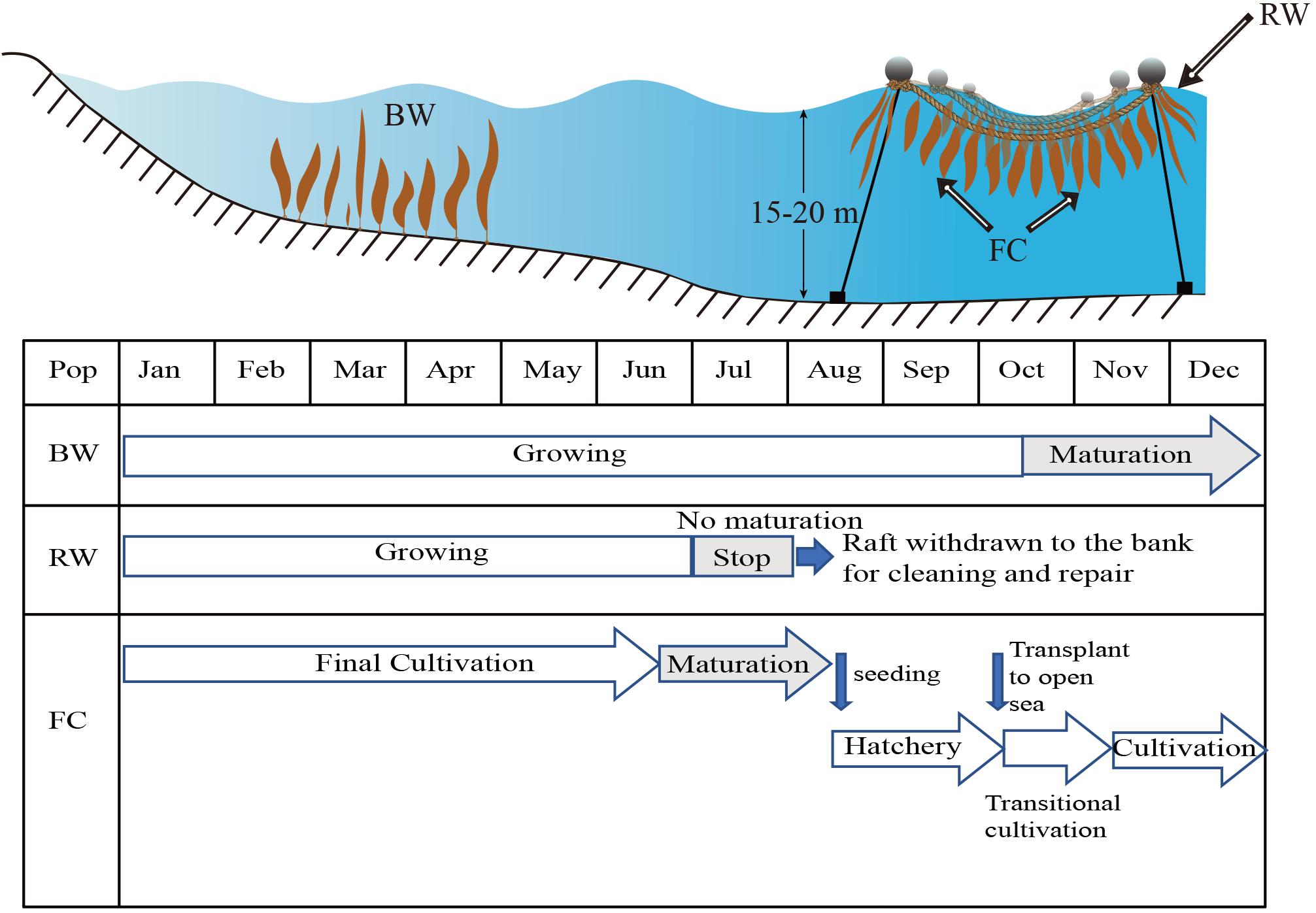
Figure 1. Schematic drawing of the sympatric spontaneous populations in subtidal benthic zone (BW) and on the cultivation rafts (RW), and farmed cultivars (FC) of Saccharina japonica on a typical kelp farm in Dalian China. Their growing and maturation periods are also shown.
Supposedly at least two alternative prerequisites should be satisfied for the possibility of reciprocal gene flow between farmed and subtidal spontaneous populations. One is overlap of the reproduction season which makes their crossing possible. Otherwise, the alternative one is that zoospores released from the spontaneous or farmed populations are able to colonize the cultivation longlines or subtidal zones, respectively, and grow into mature sporophytes. Subtidal spontaneous sporophytes of S. japonica are biennial. They usually reproduce twice in their life history, in autumns when they are 1 year and 2 years old, respectively, and then the sporophytes die away (Tseng and Wu, 1962). In earlier times the “autumn sporeling” method was used, in which seedlings are produced by employing mature subtidal spontaneous individuals as parents (Shan et al., 2011). After it was devised in the 1950s, the “summer sporeling” method was gradually accepted by the cultivation industry and has become the dominant method for large-scale production of seedlings in China (Tseng et al., 1955; Tseng, 2001). The farmed populations derived from this method produce sori in summer (mainly from July to August), different from the reproductive time of subtidal spontaneous populations (Shan et al., 2011). Most of the farmed populations are harvested before July and only hundreds of selected parental sporophytes are hung in deep waters until August for the purpose of seedling production. Even though the zoospores released from these parental sporophytes are able to colonize in the subtidal regions, their derived gametophytes and sporophytes are unlikely to survive the high temperature (up to 27–28°C) in summer (Pang et al., 2007). Strings, on which grow young seedlings about 3–4 cm long and the remnant gametophytes, are transported from hatcheries to the open sea for transitional cultivation when seawater temperature drops below 20°C in late October. The transitional cultivation lasts for 1 month until the young sporophytes reach an average length of 20 cm. The subtidal spontaneous populations reproduce at this period (Figure 1). Zoospores released from them are likely to attach to the seedling strings and their derived gametophytes might self-cross or cross with the remnant gametophytes of the farmed populations. However, selection of larger sporophytes in the insertion process for final cultivation is expected to minimize the pedigree contamination from spontaneous populations. According to the above analysis, the genetic connectivity between subtidal spontaneous and farmed populations is expected to be very limited. Spontaneous sporophytes are also often observed growing on the structural rafts of longline cultivation system (Figure 1). They usually become conspicuous from January and February. They are hypothesized to be derived from the zoospores released by the subtidal spontaneous populations in autumn.
Microsatellites have been the markers of choice in genetic structure analysis due to its codominant nature and high polymorphism information content per locus owing to its multiallelic nature (Liu and Cordes, 2004). They were successfully used to assess the genetic connectivity between farmed and the adjacent spontaneous populations of Undaria pinnatifida on the cultivation infrastructure in our previous study (Shan et al., 2018). By employing ten highly polymorphic microsatellites, our objective in the present study is to assess the genetic connectivity between farmed populations on a typical kelp farm and the adjacent spontaneous populations of S. japonica in China.
Sampling of S. japonica were conducted from a typical kelp farm and the adjacent subtidal regions on May 10, 2018 in a bay of Lüshun district, Dalian city, China (Figure 2). The sampling range was from 38°47′N, 121°15′E to 38°47′N, 121°16′E. Three commonly farmed cultivars, which are designated as FB, FJ, and XS, were sampled from longlines. All of them were produced through “summer sporeling” method and grown on 8-m long cultivation ropes, with both ends tied to the structural rafts (Figure 1 in Shan et al., 2018). Subtidal spontaneous populations were collected from two rocky locations adjacent to the farmed populations, with the individuals sampled from the west and east regarded as two separate populations, designated as SW and SE, respectively (Figure 2). The distance between them was ca. 400 m. Spontaneous sporophytes were also collected from structural rafts adjacent to the farmed populations and designated as population RW. The distance between any two sampled individuals was at least 3 m to reduce the chance of collecting siblings or close relatives. Thirty individuals were sampled from each population except SE population, which included 24 individuals. A small part of the blade was cut from each individual, cleaned by sterilized seawater, blotted with paper towels, and then dried in silica gels in plastic bags.
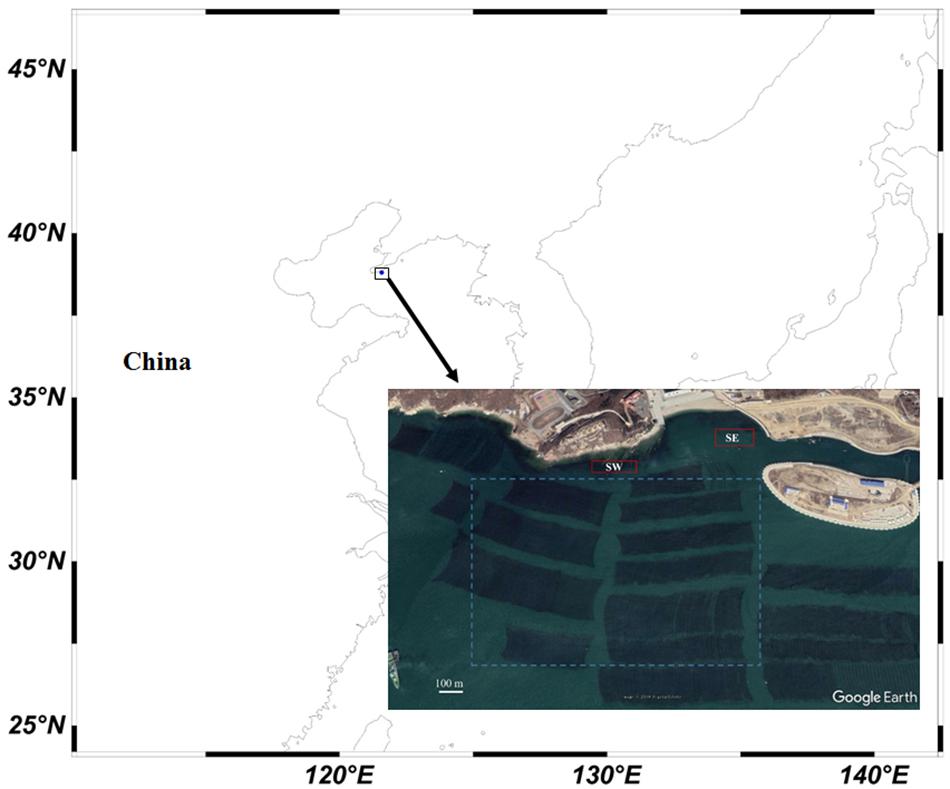
Figure 2. Sampling site of the farmed populations and the adjacent spontaneous populations of Saccharina japonica. Red rectangles indicate the sampling range of the subtidal spontaneous populations SW and SE. Blue dashed rectangle indicates the sampling range of farmed populations and the spontaneous population on the cultivation structural rafts. The dark rectangular areas in the sea are the cultivation infrastructure.
Genomic DNA was isolated using Plant Genomic DNA kit Ca.#DP305 (TIANGEN, Beijing, China).Ten polymorphic microsatellite markers H123 (Shi et al., 2007), Zspj6, Zspj9, Zspj14, Zspj17, Zspj20, Zspj26, Zspj28, Zspj39, and Zspj40 (Zhang et al., 2014) were employed for genetic analysis. The polymerase chain reaction (PCR) and microsatellite genotyping were conducted as described in Shan et al. (2019).
The number of alleles (Na), observed and expected heterozygosities (Ho and He), inbreeding coefficient Fis and Nei’s standard genetic distance (Nei, 1972) were computed with GenAlEx 6.5 (Peakall and Smouse, 2006, 2012). The deviation from Hardy–Weinberg equilibrium (HWE) was evaluated by GENEPOP version 4.7.0 (Rousset, 2008). The adjusted P-value < 0.05 after Bonferroni correction (Rice, 1989) was regarded to be significant. The population selfing rate was estimated by the index g2 using the inbreedR package (Stoffel et al., 2016). A neighbor-joining (NJ) unrooted phylogenetic tree was constructed with calculated genetic distance between populations by the POPTREE software (Takezaki et al., 2010). During bootstrapping, 1000 permutations were performed to assess the robustness of the clusters. The Fst values were computed by ARLEQUIN version 3.11 with 1000 permutations to assess pairwise population genetic differentiation (Laurent et al., 2005). To adjust for multiple comparisons, the false discovery rate (FDR) was controlled by using “BH” method (Benjamini and Hochberg, 1995) in the p. adjust function in R. The adjusted P-value < 0.05 was considered to be significant.
A Bayesian model-based clustering analysis was conducted with STRUCTURE 2.3.4 to evaluate the most possible number of genetic clusters (Pritchard et al., 2000). This clustering method was applied in identification of genetically distinct subpopulations based on the allele frequencies. The number of clusters (K value) was set from 1 to 6, and 20 independent runs were carried out for each fixed K value using the admixture model and allele frequencies correlated model. Each run included a burn-in length of 100,000 followed by 1,000,000 Mote Carlo Markov Chain (MCMC) repetitions. The optimal K value was determined by submitting all results files of K = 1–6 to STRUCTURE HARVESTER (Earl and Vonholdt, 2012) according to the method of Evanno et al. (2005). The run with the highest Ln Pr (X|K) value (log probability) among the 20 independent runs was chosen and the graphical result was displayed with DISTRUCT 1.1 (Rosenberg, 2004). A multivariate discriminant analysis of principal components (DAPC) was also performed using the adegenet package in R in order to infer genetic relationships among populations (Jombart, 2008; Jombart et al., 2010).
The data of microsatellite genotyping and allele frequencies was provided in Supplementary Tables S1, S2, respectively. Both the average Na and He across the 10 microsatellite loci were found to be highest in FJ (Na = 3.8, He = 0.496) and lowest in SW (Na = 2.6, He = 0.328) (Table 1). The genetic diversity of farmed populations was higher than that of the subtidal spontaneous populations in terms of average Na and He. No significant deviation from HWE was detected for all populations either at each locus or across all the 10 loci (Table 1 and Supplementary Table S3). The average Fis values across loci were close to zero for all populations (Table 1). The selfing rate g2 was only detected to be significant in RW (P = 0.036).
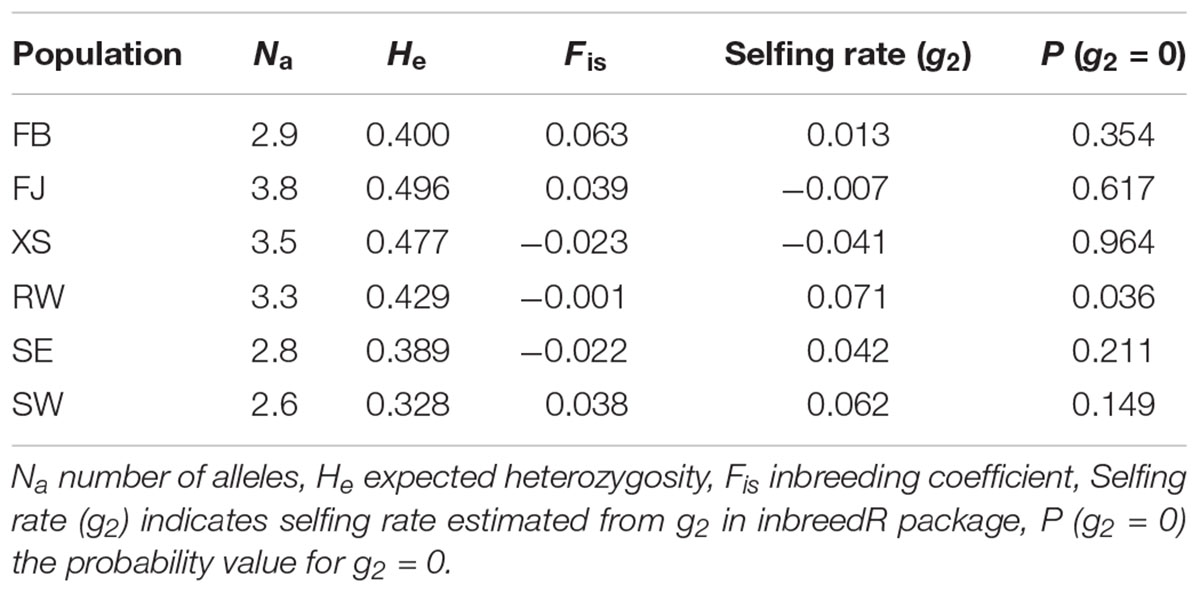
Table 1. Genetic diversity and selfing rates of the populations of Saccharina japonica from a typical farm and the adjacent subtidal zones in Dalian, China.
Fst values were detected to be significant among all populations except that between FJ and XS (Table 2). Values of both Fst and Nei’s genetic distance showed that the genetic differentiation between the farmed and the subtidal spontaneous populations was much higher than those within the farmed or subtidal spontaneous populations. The NJ phylogenetic tree, which was constructed based on pair-wise genetic distance, exhibited that SW and SE were grouped into a cluster, and FJ, XS, and FB were grouped into another cluster (Figure 3). These two clusters were clearly separated with large distance. RW was shown to be intermediate between them.
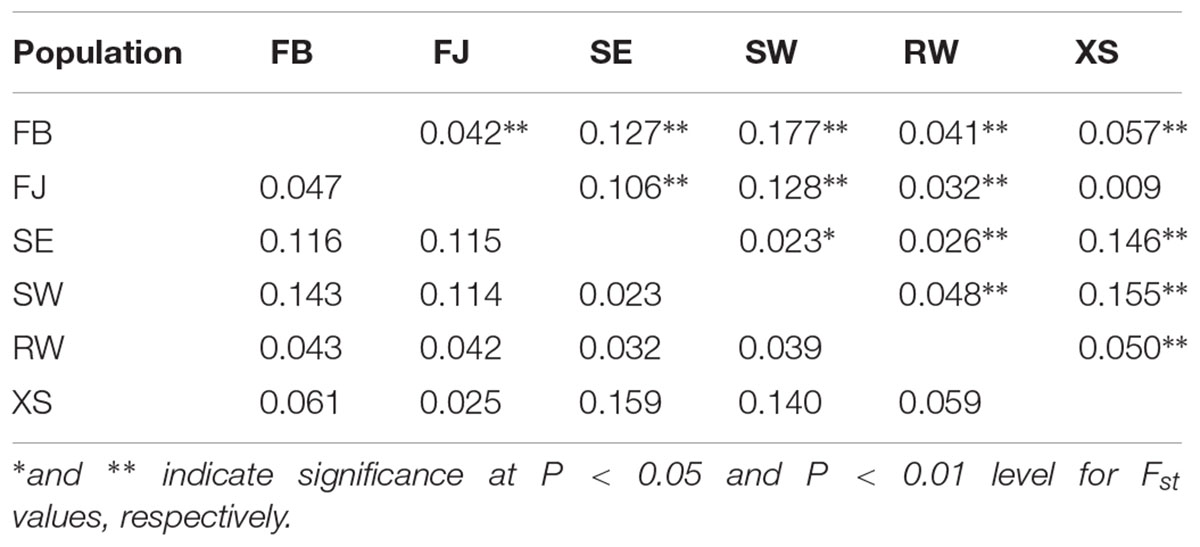
Table 2. Pairwise genetic distance (below diagonal) and Fst values (above diagonal) in six populations of Saccharina japonica from a kelp farm and the adjacent subtidal zones in Dalian, China.
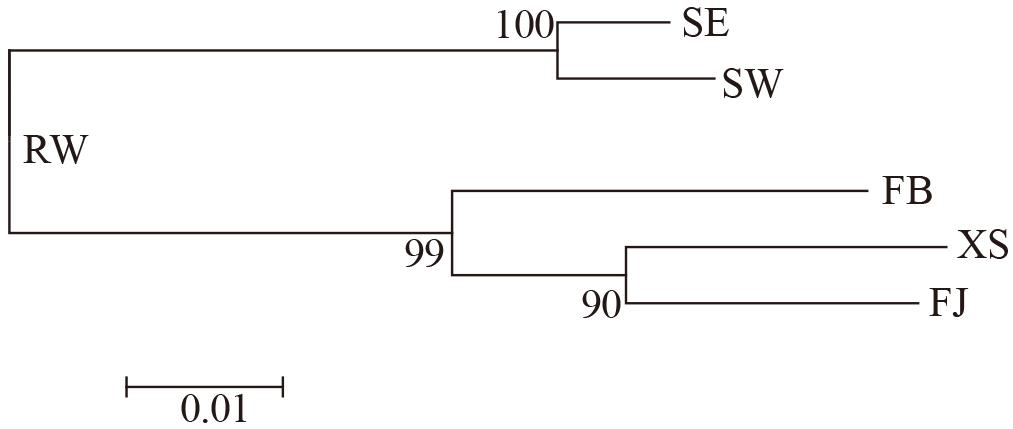
Figure 3. Neighbor-joining phylogenetic tree based on genetic distance among the farmed and spontaneous populations of Saccharina japonica from a kelp farm in China. The bar indicates the genetic distance.
The most likely number of K was identified to be 2 by the use of STRUCTURE HARVESTER according to the ΔK values (Figure 4 and Supplementary Figure S1). Subtidal spontaneous populations were assigned to the first cluster with high proportion of membership (>0.90), with only three individuals containing a proportion of membership >20% from farmed populations. Most individuals of the farmed populations were assigned to the second cluster, but some individuals of FB and FJ possessed high proportion of membership originated from the subtidal spontaneous populations. The total proportion of membership originated from subtidal spontaneous populations was 0.28 and 0.19 in FB and FJ, respectively. By comparison, XS contained less membership derived from subtidal spontaneous populations (<0.10). Admixture of membership was most marked in RW, with the membership proportion from the first and second clusters being 0.62 and 0.39, respectively. The genetic relationships among populations revealed by the DAPC analysis were very consistent with those revealed by STURCTURE and the NJ phylogeny (Figure 5).
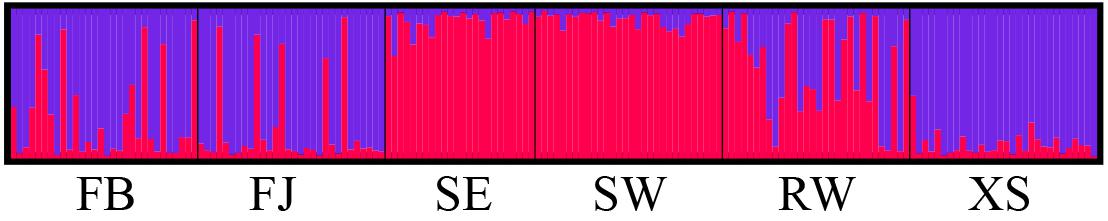
Figure 4. Genetic structure revealed by the Bayesian model-based analysis using STRUCTURE 2.3.4 for farmed and spontaneous populations of Saccharina japonica from a kelp farm in China. Each individual is indicated by a vertical colored bar, and the proportion of the color in each bar represents the probability of membership in the corresponding cluster.
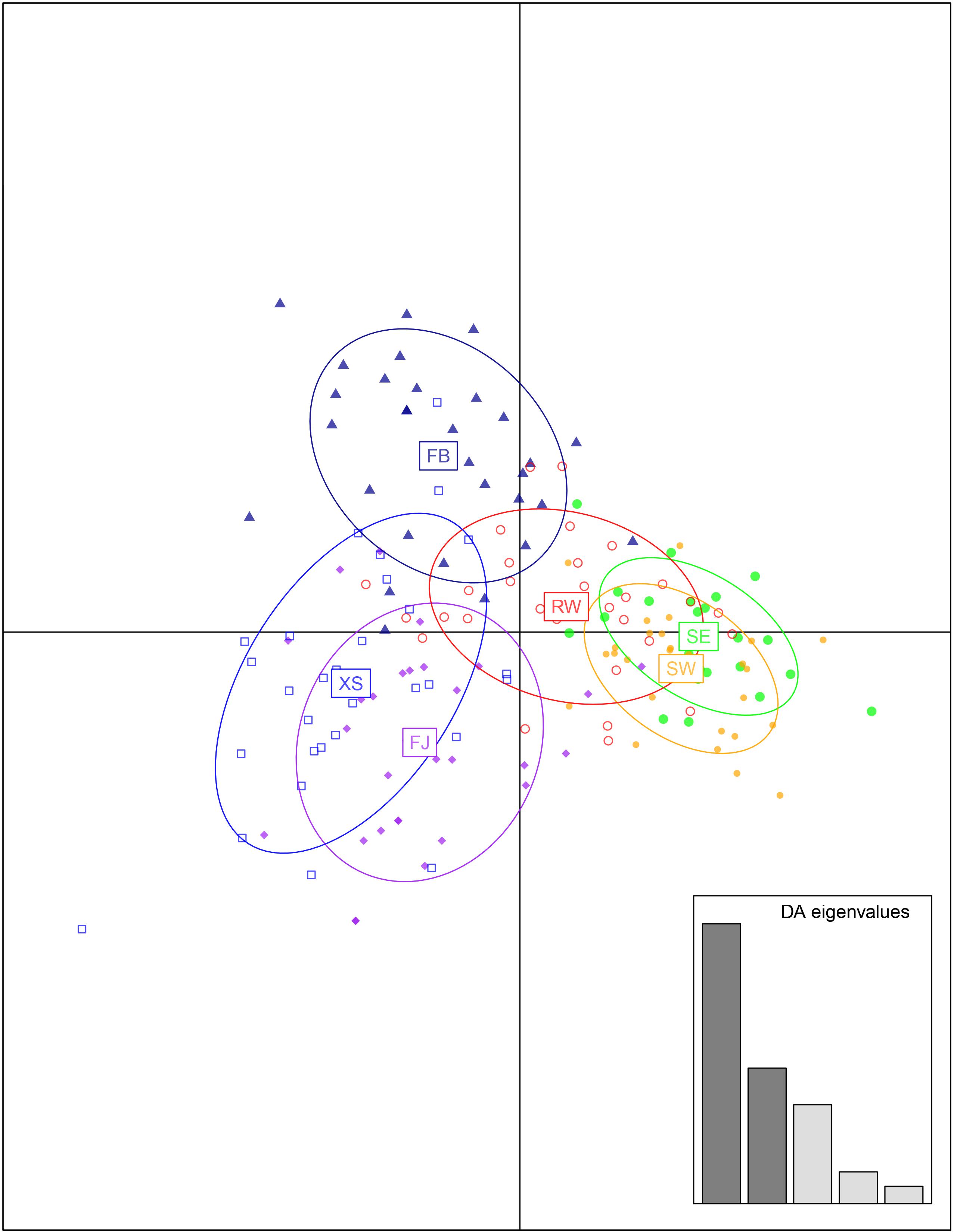
Figure 5. Discriminant analysis of principal components for farmed and spontaneous populations of Saccharina japonica from a kelp farm in China.
As expected, farmed populations were found to have exerted very limited impact on the genetic composition of the sympatric subtidal spontaneous populations. This was consistent with the results found by Zhang et al. (2017), although the spontaneous population appeared to be far from the farmed populations investigated in that study. The subtidal spontaneous populations are therefore speculated to evolve almost without the interference of the farmed population under the current farming system. They are potentially valuable breeding materials for they can integrate such characteristics as tolerance to high temperature that has evolved to adapt to the local environment (Pang et al., 2007). However, the gene flow from subtidal spontaneous populations to the farmed ones was revealed to be larger than we had expected. As is expected in the section “Introduction,” the zoospores discharged by the subtidal spontaneous populations may attach to the seedling ropes during the 1-month transitional cultivation of the young sporelings in the open sea (Figure 1). The derived gametophytes can produce sporophytes through selfing or crossing with the remnant gametophytes of the farmed cultivars on the ropes. On one hand, growth of some of these sporophytes are likely to catch up with that of the earlier born larger sporelings during the 1-month period, and thus are exploited in the sporeling-insertion process for final farming. On the other hand, because some sporelings usually detach from the cultivation rope after insertion due to the loose attachment of the holdfast, farmers need to supplement them by inserting new sporelings from the back-up, and thus the smaller sporophytes will obtain the opportunity of being used. These scenarios may account for the relatively high level of gene flow from subtidal spontaneous populations to the farmed ones. The existence of more gene flow from subtidal spontaneous populations to the farmed ones is likely to be one reason that results in higher genetic diversity in farmed populations.
What is also unexpected is that the spontaneous population on the structural rafts (RW) was revealed to contain a large proportion of pedigree from farmed populations (39%), in addition to that from subtidal spontaneous populations. From June to July, zoospores released from the mature farmed sporophytes may attach to the cultivation infrastructure. After the harvest of S. japonica is finished, however, the cultivation infrastructure including the cultivation ropes and structural rafts are withdrawn to the bank for cleaning and repair. The attached spores and gametophytes are impossible to survive this long period of drought. Furthermore, the sporophytes of RW were observed to be conspicuous from January and February. So they were supposedly established in last November and December. But during this period there were no mature farmed sporophytes on longlines. One potential explanation is that they were derived from the farmed sporophytes that detached from the cultivation rope and sank in the sea bottom. From the very beginning of transitional cultivation of young sporelings to the time when they grow into adults, many of sporophytes detach from the ropes due to currents, storm and unsteady fastening of their hapteras. The detachment rate of the adult sporophytes was especially higher, up to 50% at the most severe situation in recent years (M. F. Zhang, 2015–2019). Some of them might be tangled with the substratum or the flora at the bottom of the sea, and became sessile. Some might go back and forth near the farm under driving force of the tides and currents. It has been revealed that low temperature and low light intensity can delay the mature time of sporophytes (Tseng and Wu, 1962; Mizuta et al., 1999). This principle has been exploited by farmers to adjust the reproductive time in farming and seedling production practice. In the north of China, farmers usually select desirable sporophytes as parental individuals during the harvest season and hang them deeply in remote areas where the currents are fast, and both the temperature and light intensity are lower. In the south of China (mainly in Fujian) where the high summer temperature makes it unfeasible to preserve parental sporophytes in the sea, the selected parental sporophytes are transported to hatchery rooms, and low temperature and light are given (Pang et al., 2007). Likewise, the sporophytes that have sunken in the sea bottom experience poor light and low temperature, and thus their mature time is likely to be delayed to autumn, similar to that of the subtidal spontaneous populations. These detached sporophytes could also be potential media for the gene flow from the farmed populations to the subtidal spontaneous ones, which may account for the small amount of pedigree of the former detected in the latter in this study.
The genetic diversity of the farmed and spontaneous populations was consistent with those detected in the previous studies (Li et al., 2017; Shan et al., 2017; Zhang et al., 2017). Interestingly, the subtidal spontaneous populations exhibited lower genetic diversity than the farmed populations. As suggested by Zhang et al. (2017), introduction of alien germplasm, intra- and inter-specific crossing in the breeding process might increase the genetic diversity of farmed populations. By comparison, the subtidal spontaneous populations have supposedly been isolated from immigrants since their original foundation. Reduction in genetic diversity is potentially caused by founder effect and the subsequent genetic drift. Meanwhile, the three farmed cultivars were demonstrated to be genetically close to each other. As was suggested and revealed in our previous studies, mixing among different cultivars were most likely caused by sharing of refrigerated seawaters during seedling production in the hatchery (Shan et al., 2011; Li et al., 2017). The result was the homogenization of genetic composition and agronomical features of the cultivars.
In conclusion, very significant genetic divergence was demonstrated to exist between the farmed populations on a typical kelp farm and the adjacent subtidal spontaneous populations of S. japonica in China. Gene flow out of farmed populations to the subtidal spontaneous populations was revealed to be very limited, but gene flow in the contrary direction was more prominent. The pedigree of spontaneous sporophytes occurring on the structural rafts was exhibited to be admixture of both farmed and subtidal spontaneous populations. This study provides useful information for management of breeding programs and conservation of stock resources. In the long run, genetic diversity of farmed and spontaneous populations, and the genetic connectivity between them ought to be monitored continuously for the benefit of sustainable development of cultivation industry of S. japonica.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
SP and TS conceived the study. TS, QL, XW, and LS did the sampling work in the field and the analysis in the lab. All authors contributed to the writing of the manuscript.
This research was supported by grants from China Agriculture Research System (CARS-50), the State’s Key Project of Research and Development Plan (2016YFC1402507), the National Key Technology Support Program (2015BAD13B05), a special supporting program for maintaining seaweed stock culture from the Chinese Academy of Sciences (CZBZX-1), the National Infrastructure of Fishery Germplasm Resource (2019DKA30470), the Taishan Scholar Program of Shandong Province, and Huiquan Scholar Program of Institute of Oceanology, Chinese Academy of Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Mingfu Zhang for his kind help in samples collection.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00494/full#supplementary-material
FIGURE S1 | Delta-K graph obtained by submitting all results files of K = 1–6 to STRUCTURE HARVESTER.
TABLE S1 | Microsatellite genotyping data at 10 loci in six populations of Saccharina japonica.
TABLE S2 | Allele frequencies at 10 microsatellite loci in six populations of Saccharina japonica.
TABLE S3 | Genetic diversity of the farmed and spontaneous populations of Saccharina japonica from a kelp farm in China at each microsatellite locus.
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Earl, D. A., and Vonholdt, B. M. (2012). Structure harvester: a website and program for visualizing structure output and implementing the evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294x.2005.02553.x
Fei, X. G. (2004). Solving the coastal eutrophication problem by large scale seaweed cultivation. Hydrobiologia 512, 145–151. doi: 10.1007/978-94-007-0944-7_19
Hasegawa, Y. (1976). Progress of Laminaria cultivation in Japan. J. Fish. Board Can. 33, 1002–1006. doi: 10.1139/f76-127
Jombart, T. (2008). Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Jombart, T., Devillard, S., and Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. doi: 10.1186/1471-2156-11-94
Laurent, E., Guillaume, L., and Stefan, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50.
Li, X., Pang, S. J., and Shan, T. F. (2017). Genetic diversity and population structure among cultivars of Saccharina japonica currently farmed in northern China. Phycol. Res. 65, 111–117. doi: 10.1111/pre.12167
Liu, Z. J., and Cordes, J. (2004). DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238, 1–37. doi: 10.1016/j.aquaculture.2004.05.027
Mizuta, H., Nimura, K., and Yamamoto, H. (1999). Inducible conditions for sorus formation of the sporophyte discs of Laminaria japonica areschoug (Phaeophyceae). Fish. Sci. 65, 104–108. doi: 10.2331/fishsci.65.104
Pang, S. J., Jin, Z. H., Sun, J. Z., and Gao, S. Q. (2007). Temperature tolerance of young sporophytes from two populations of Laminaria japonica revealed by chlorophyll fluorescence measurements and short-term growth and survival performances in tank culture. Aquaculture 262, 493–503. doi: 10.1016/j.aquaculture.2006.11.018
Peakall, R., and Smouse, P. E. (2006). genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Resour. 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x
Rosenberg, N. A. (2004). Distruct: a program for the graphical display of population structure. Mol. Ecol. Resour. 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
Rousset, F. (2008). Genepop’007: a complete re-implementation of the genepop software for windows and linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Shan, T., Liu, F., Liu, Q., and Pang, S. (2011). Review and prospect of “summer sporeling” technique of Saccharina japonica in China. J. Agric. Sci. Technol. 13, 129–134. (in Chinese with English abstract)
Shan, T., Pang, S., Wang, X., Li, J., and Su, L. (2018). Assessment of the genetic connectivity between farmed and wild populations of Undaria pinnatifida (Phaeophyceae) in a representative traditional farming region of China by using newly developed microsatellite markers. J. Appl. Phycol. 30, 2707–2714. doi: 10.1007/s10811-018-1449-1447
Shan, T., Yotsukura, N., and Pang, S. (2017). Novel implications on the genetic structure of representative populations of Saccharina japonica (Phaeophyceae) in the northwest pacific as revealed by highly polymorphic microsatellite markers. J. Appl. Phycol. 29, 631–638. doi: 10.1007/s10811-016-0888-882
Shan, T. F., Pang, S. J., Wang, X. M., Li, J., Su, L., Schiller, J., et al. (2019). Genetic analysis of a recently established Undaria pinnatifida (Laminariales: Alariaceae) population in the northern wadden sea reveals close proximity between drifting thalli and the attached population. Eur. J. Phycol. 54, 154–161. doi: 10.1080/09670262.2018.1532116
Shi, Y., Yang, G., Liu, Y., Liao, M., Li, X., and Cong, Y. (2007). Development of 18 polymorphic microsatellite DNA markers of Laminaria japonica (Phaeophyceae). Mol. Ecol. Notes 7, 620–622. doi: 10.1111/j.1471-8286.2006.01652.x
Stoffel, M. A., Esser, M., Kardos, M., Humble, E., Nichols, H., David, P., et al. (2016). inbreedR: an R package for the analysis of inbreeding based on genetic markers. Methods Ecol. Evol. 7, 1331–1339. doi: 10.1111/2041-210x.12588
Takezaki, N., Nei, M., and Tamura, K. (2010). Poptree2: software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol. Biol. Evol. 27, 747–752. doi: 10.1093/molbev/msp312
Tseng, C., Sun, K., and Wu, C. (1955). On the cultivation of Haitai (Laminaria japonica Aresch.) by summering young sporophytes at low temperature. Acta Bot. Sin. 4, 255–264. (in Chinese with English abstract)
Tseng, C. K. (2001). Algal biotechnology industries and research activities in China. J. Appl. Phycol. 13, 375–380.
Tseng, C. K., and Wu, C. Y. (1962). Mariculture of Saccharina Japonica. Beijing: Beijing Science Press, (in Chinese).
Tseng, C. K., and Zhang, J. F. (1952). Economic seaweeds in the north of China. J. Shandong Univ. 2, 57–82. doi: 10.1111/j.1529-8817.2012.01229.x (in Chinese)
Zhang, J., Wang, X., Yao, J., Li, Q., Liu, F., Yotsukura, N., et al. (2017). Effect of domestication on the genetic diversity and structure of Saccharina japonica populations in China. Sci. Rep. 7:42158. doi: 10.1038/srep42158
Keywords: kelp, seaweed farming, simple sequence repeat, genetic structure, gene flow
Citation: Shan T, Li Q, Wang X, Su L and Pang S (2019) Assessment of the Genetic Connectivity Between Farmed Populations on a Typical Kelp Farm and Adjacent Spontaneous Populations of Saccharina japonica (Phaeophyceae, Laminariales) in China. Front. Mar. Sci. 6:494. doi: 10.3389/fmars.2019.00494
Received: 10 June 2019; Accepted: 22 July 2019;
Published: 02 August 2019.
Edited by:
Zhijun Dong, Yantai Institute of Coastal Zone Research (CAS), ChinaReviewed by:
Naihao Ye, Yellow Sea Fisheries Research Institute (CAFS), ChinaCopyright © 2019 Shan, Li, Wang, Su and Pang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tifeng Shan, c2hhbnRpZmVuZ0BxZGlvLmFjLmNu; Shaojun Pang, c2pwYW5nQHFkaW8uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.