Corrigendum: Diversity, Distribution and Phylogeny of Hesionidae (Annelida) Colonizing Whale Falls: New Species of Sirsoe and Connections Between Ocean Basins
- 1Departamento de Oceanografia Biológica, Instituto Oceanográfico, Universidade de São Paulo, São Paulo, Brazil
- 2Laboratório de Zoologia de Invertebrados, Departamento de Zoologia, Instituto de Biologia, Universidade do Estado do Rio de Janeiro, Rio de Janeiro, Brazil
- 3Molette Biology Laboratory for Environmental and Climate Change Studies, Department of Biological Sciences, Auburn University, Auburn, AL, United States
Whale falls are important environments contributing to biodiversity, connectivity and evolutionary novelty in deep-sea ecosystem. Notwithstanding, most of this knowledge is based in studies from NE Pacific basin. Interestingly, the only known natural whale fall on the SW Atlantic has faunal composition affinities with carcasses from other deep-ocean basins. In this carcass, annelid worms belonging to Hesionidae are abundant and species-rich, and include some shared species with NE Pacific Ocean. Here we evaluate the diversity of Hesionidae on the SW Atlantic using new information of implanted whale bones and explore whether some species have interbasin distribution or if they represent cryptic species in different basins. We described, using morphological and molecular data, a total of 10 new hesionid species and report of a new lineage Sirsoe ‘BioSuOr,’ not formally described herein. Two hesionids found exclusively in deep-sea chemosynthetic environments, Sirsoe Pleijel (1998) and Vrijenhoekia Pleijel et al. (2008), are primarily distinguished from each other by the presence of a median antenna on the former and its absence on the latter. However, our analyses showed that Vrijenhoekia should be synonymized with Sirsoe and for this reason we emended the diagnosis of Sirsoe. We also emphasized the presence of Sirsoe balaenophila comb. nov. and S. sirikos in SW Atlantic whale falls confirming their interbasin distribution. Moreover, COI and 16S rDNA data reveal that S. balaenophila comb. nov. also comprises cryptic species on the SW Atlantic (S. pirapuan sp. nov. and S. ypupiara sp. nov) and perhaps also in the Pacific Ocean (herein named as S. balaenophila lineage-2). The new species, S. maximiano, is shared between whale falls from SW Atlantic and vent sites from Mid-Cayman Spreading Center. Our data adds to the growing literature showing species are shared between deep ocean basins and among cognate deep-sea environments. Zoobank registration publication LSID - urn:lsid:zoobank.org:pub:7E891B1D-DCE4-45C8-83F4-8586D286B327.
Introduction
Whale carcasses impact the food-limited deep sea (>200 m) creating remarkable island-like habitats considered hotspots of biodiversity and evolutionary novelty (Smith and Baco, 2003; Smith et al., 2015). Assemblages of opportunist and specialist fauna colonize carcasses during overlapping successional stages and also on the mosaic of microhabitats formed on the skeleton and surrounding sediments (Baco and Smith, 2003; Smith and Baco, 2003; Fujiwara et al., 2007; Lundsten et al., 2010b; Smith et al., 2014, 2015; Alfaro-Lucas et al., 2017). Whale falls have been hypothesized to act as ecological and evolutionary stepping stones for some chemosymbiotic hydrothermal vent and cold seep fauna, such as vesicomyid and bathymodiolin bivalves (Smith et al., 1989, 2015, 2017; Distel et al., 2000; Smith and Baco, 2003; Lorion et al., 2009, 2013; Miyazaki et al., 2010; Kiel, 2017). During degradation, whale falls become chemosynthetic-based habitats, similar to hydrothermal vents and cold seeps in which part of the faunal composition is reliant on chemosynthetic production (Smith et al., 1989, 2015; Smith and Baco, 2003; Alfaro-Lucas et al., 2018). In this scenario, organisms dependent on chemosynthesis could use whale falls as intermediate habitats facilitating dispersion among spatially isolated cognate habitats (Smith et al., 1989).
Whale falls are found worldwide mainly along whale migratory corridors at continental margins and around equatorial latitudes (Smith and Baco, 2003; Sumida et al., 2016). The biodiversity and species distribution of whale-fall fauna remain poorly known, especially in areas other than the NE Pacific, limiting fundamental knowledge such as biogeographic patterns of species associated with these environments (Smith et al., 2015, 2017; Sumida et al., 2016). Specialist whale-fall fauna has already likely experienced extinction in regions that were subject to heavy commercial whaling, such as in the Atlantic (Roman et al., 2014). Therefore, addressing patterns of whale-fall biodiversity is crucial for the effective protection and understanding of these hotspots of deep-sea biodiversity.
Annelids are the most diverse and abundant taxa in whale-fall communities, including some of the most iconic specialists (Rouse et al., 2004, 2008, 2009), and comprises the most shared taxa among other chemosynthetic habitats (Smith et al., 1989, 2014, 2015, 2017; Smith and Baco, 2003; Braby et al., 2007; Fujiwara et al., 2007; Lundsten et al., 2010a,b; Amon et al., 2013; Sumida et al., 2016; Alfaro-Lucas et al., 2017). In particular, some annelid families are species-rich on whale falls, such as Hesionidae Grube 1850, which has five genera reported to date from these environments (Summers et al., 2015; Sumida et al., 2016; Alfaro-Lucas et al., 2017). Among them, Sirsoe Pleijel (1998) and Vrijenhoekia Pleijel et al. (2008) are exclusively known from deep-sea chemosynthetic-based ecosystems, including whale falls, where they represent important constituent of the fauna (Summers et al., 2015). Some hesionid species are potentially shared between the SW Atlantic and NE Pacific basins, such as V. balaenophila Pleijel et al. (2008), suggesting an interbasin distribution (Sumida et al., 2016).
Despite their ecological importance, the taxonomy of Sirsoe and Vrijenhoekia is complex. Members of Sirsoe were originally described within Orseis Ehlers (1864), which was subsequently considered as a junior synonym of Oxydromus (Grube, 1855; Pleijel, 1998). Aiming to accommodate Orseis grasslei (Blake, 1985), Pleijel (1998) erected a new genus named Sirsoe. Re-examination of S. grasslei revealed similarities with the “iceworm,” S. methanicola (Desbruyères and Toulmond, 1998), such as the enlarged ventral cirri on the first three segments and presence of a median antenna (Pleijel, 1998). Interestingly, the iceworm was originally described as member of Hesiocaeca (Hartman, 1965; Desbruyères and Toulmond, 1998). Pleijel (1998) assigned S. methanicola within Sirsoe since he considered Hesiocaeca as incertae sedis due to the poor condition of H. bermudensis Hartman (1965). This taxonomic move created confusion as Pleijel placed the iceworm into Sirsoe prior to the formal description by Desbruyères and Toulmond (1998), and thus creating questions about the applicability of taxonomic rules. Pleijel (1998) and Ruta et al. (2007) considered the diagnostic feature of Sirsoe to be the presence of median dorsally inserted antenna.
Since the discovery of the first species of Vrijenhoekia, V. balaenophila, analyses of molecular and morphological data revealed a close affinity to Sirsoe (Pleijel et al., 2008). Pleijel et al. (2008) acknowledged the similarity between Sirsoe and Vrijenhoekia but cited some morphological differences, such as the absence of a median antenna, the presence of glandular lip pads (GLP) and a well-developed neuropodial lobe starting on the fourth segment, in Vrijenhoekia but not Sirsoe (Pleijel et al., 2008). Importantly, Pleijel et al. (2008) considered the erection of Vrijenhoekia as “largely arbitrary” but did so to avoid the use of polymorphisms in the generic diagnosis of Sirsoe. The discovery of Vrijenhoekia ketea Summers et al. (2015) species complex, however, made the acceptance of Vrijenhoekia more doubtful, given that this complex group bears median antenna. Currently, both genera, Sirsoe and Vrijenhoekia, are recognized being the sister clades to each other (Pleijel et al., 2008; Summers et al., 2015) even though the diagnosis of the genera is overlapping. To the best of our knowledge there are four named Sirsoe species and four Vrijenhoekia species (Read and Fauchald, 2019).
Here we aim to evaluate (a) the diversity of Hesionidae in whale falls from SW Atlantic Ocean using traditional and molecular taxonomy; (b) the phylogenetic relationships between Sirsoe and Vrijenhoekia species relative to the diagnosis of Vrijenhoekia and Sirsoe; and (c) whether V. balaenophila presents an interbasin distribution ranging from the NE Pacific to the SW Atlantic Ocean or, alternatively, if it is a cryptic species-complex.
Materials and Methods
Sample Collection
Samples were collected for molecular and morphological analyses from seven sampling sites (Table 1) during two projects: the BioSuOr project (Biodiversity and Connectivity of Benthic Communities on Organic Substrates in the Deep SW Atlantic Ocean) and the Iatá-Piuna expedition, a collaborative scientific partnership between Brazil and Japan, as part of the around-the-world project QUELLE 2013 (Quest for the Limit of Life) from JAMSTEC (Japan Agency for Marine-Earth Science and Technology). BioSuOr project samples were collected using six deep-sea landers (Table 1) equipped with acoustic releases. One lander was deployed on the upper slope (∼500 m depth) in the Santos Basin, a region with pockmarks and salt diapirs (Mahiques et al., 2017). Two other landers were deployed at ∼1,500 m and three at ∼3,300 m, along an area between 21–28° S and 38–45° W (Table 1). Three sets of humpback whale vertebrae (Megaptera novaeangliae) and cross-sectional wood-log pieces of slash pine tree (Pinus elliottii) were used as organic substrate in each lander. Substrates were attached inside a meshed cage (500 μM) and on the inner part of each lid. The meshed cages allowed water circulation around the substrate and, upon recovery, the lid closed over the mesh cage to prevent the loss of organisms during ascent. For more details of the experimental deployment and sampling design of landers see Saeedi et al. (2019). Samples from the Iatá-Piuna expedition were obtained from a natural minke whale carcass (Balaenoptera bonaerensis) found at the base of São Paulo Ridge at 4,204 m depth and were sampled using the HOV Shinkai 6500 (Sumida et al., 2016) (Table 1).
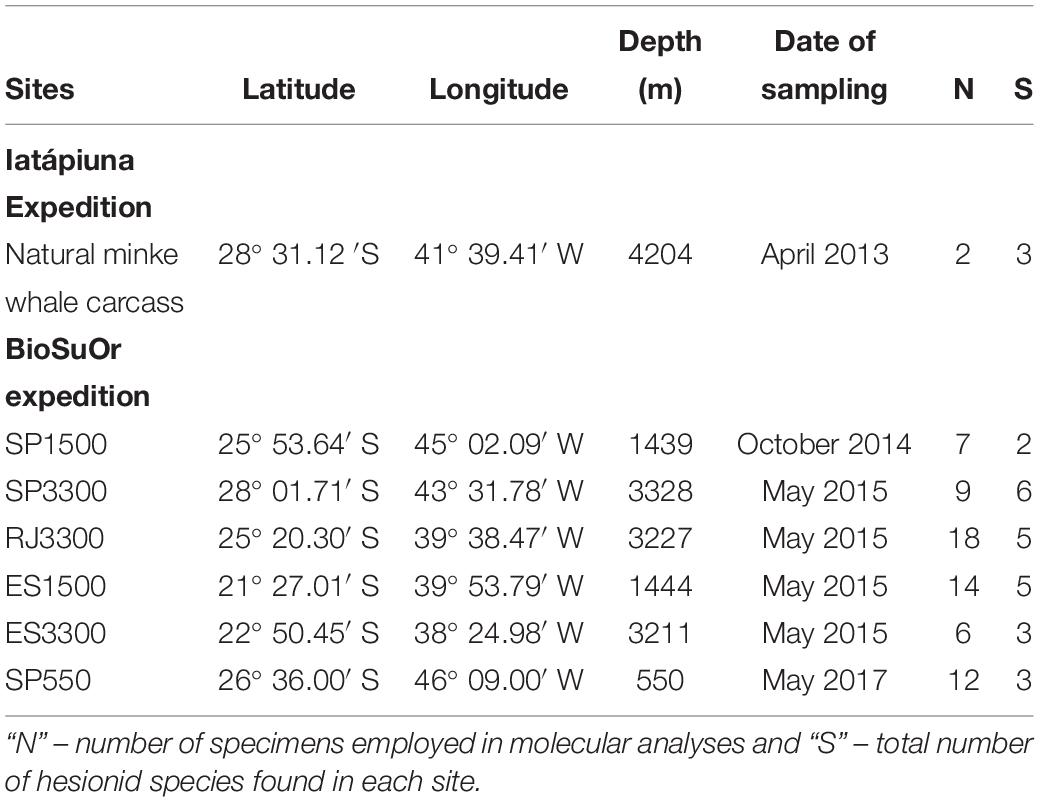
Table 1. Sampling station of natural whale-fall (Iatá-Piuna Expedition) and lander deployment (BioSuOr project).
Samples from the BioSuOr project were initially identified on board and separated for morphological and molecular analyses. Organisms for morphological analysis were fixed in 4% formalin, later replaced by 70% ethanol, and were used for photography under stereomicroscope, light microscopy and SEM. The posterior part of some specimens were preserved in non-denatured 99% ethanol (Sigma-Aldrich®, molecular grade) for molecular analysis. After the cruise, whalebones preserved in 96% ethanol were sorted at the laboratory, to extract more individuals and include specimens from the bone endofauna. Organisms collected during the Iatá-Piuna expedition were preserved in 96% ethanol on board and identified subsequently in the laboratory.
Morphological Examination
We used the presence of median antenna to distinguish Sirsoe from Vrijenhoekia morphotypes, a priori. According Pleijel (1998) and Pleijel et al. (2008), Sirsoe possesses a dorsal median antenna on prostomium that Vrijenhoekia lacks. Conversely, Vrijenhoekia has GLPs, which are lacking in Sirsoe. We used the papilla-shaped neuropodial on segment 3 and the presence of three pairs of large GLPs around the mouth to distinguish V. balaenophila in our material from the other Vrijenhoekia morphotypes (Pleijel et al., 2008; Summers et al., 2015).
The morphological analyses were made under stereo- and light microscopes and scanning electron microscopy (SEM). Some specimens were stained with methyl green or Shirlastain® A to improve the observation of some morphological features. Photographs were taken using Leica M205 C stereomicroscope and Leica MC170 HD camera at LAMP (IOUSP). Specimens for SEM were dehydrated in a progressive series of ethanol (70, 80, 90, 95, and 3x 100%) and critical-point dried (Critical Point Dryer model Balzers CPD-030). Some individuals were dehydrated in a grade series of hexamethyldizilazane (HMDS, Sigma-Aldrich) and then air dried; the ascending series was: 1:1 (HMDS:Ethanol) followed by 100% HMDS (two times), ∼5 min in each step. The prepared samples were mounted on stubs and sputter-coated with gold (∼25 nm thickness) (Sputter Coater model Balzers SCD-050) for examination under SEM. SEM images were taken using Sigma VP Carl Zeiss SEM at the Instituto de Biociências, Universidade de São Paulo and a Jeol JSM 5800LV SEM at the Universidade de Campinas. Specimens were deposited at the Museu de Zoologia, Universidade de São Paulo and at the ColBio (Coleção Biológica Prof. Edmundo F. Nonato), Instituto Oceanográfico, Universidade de São Paulo.
Molecular Data Collection
For most specimens we extracted DNA from the posterior region using the QIAGEN DNeasy Blood & Tissue kit and following the manufacture’s protocol, except that DNA was eluted in 50–100 μl of ddH2O. In cases, where the animal was small (the putative new species, Sirsoe sp. ‘BioSuOr,’ and some individuals of S. alphacrucis sp. nov. and S. yokosuka sp. nov.) we used the entire specimen for DNA extraction. For all new species, we retained voucher specimens (see Museum numbers in Systematic section). The fragment of the cytochrome oxidase subunit-I gene (COI) was amplified using the primers: LCO1490 (5′ – GGT CAA CAA ATC ATA AAG ATA TTG G – 3′) and HCO2198 (5′ – TAA ACT TCA GGG TGA CCA AAA AAT CA – 3′) (Folmer et al., 1994). PCRs for COI contained 12.5 μL of GoTaq® Green Master Mix (Promega), 0.125 μL of each primer (20 μM), 2 μL DNA template (20–100 ng) and 10.25 μL of nuclease-free water (Promega). The thermal cycling profile for COI was: 95°C for 2 min, 30 cycles of 94°C for 45 s, 51°C for 1 min, 72°C, 1 min and a final step of 72°C for 7 min. Additionally, a fragment of the 16S gene was amplified using the primers: 16SarL (5′ – CGC CTG TTT ATC AAA AAC AT – 3′) and 16SbrH (5′ – CCG GTC TGA ACT CAG ATC ACG T – 3′) (Palumbi et al., 1991). PCRs for 16S contained 6.3 μL of GoTaq® Green Master Mix (Promega), 0.1 μL of each primer (20 μM), 2 μL DNA template (20–100 ng) and 4 μL of nuclease-free water (Promega). The thermal cycling profile for 16S was: 95°C for 2 min, 35 cycles of 94°C for 45 s, 50°C for 1 min, 72°C for 1 min and a final step of 72°C for 7 min. Amplified DNA was sequenced at the CEGH facility (Centro de Pesquisa sobre o Genoma Humano e Células-Tronco) at the University of São Paulo using an ABI 3730 DNA Analyzer. Sequencing reactions were made with BigDye Terminator v3.1 cycle sequencing Kit. Fragments of each gene were bi-directly sequenced and assembled in Geneious V.10.2.2 (Kearse et al., 2012). These sequences were deposited at GenBank (Supplementary Table S1).
Phylogenetic Analysis
Phylogenetic analyses were undertaken using three datasets: COI data, 16S data, and a combined matrix with both genes (COI+16S). Additionally, Sirsoe and Vrijenhoekia sequences were retrieved from GenBank (Supplementary Table S1). Nereimyra was used as outgroup based on the current phylogenetic understanding (Pleijel et al., 2008; Summers et al., 2015; Rouse et al., 2018a). Sequences were aligned with MAFFT (Katoh et al., 2002), using G-INS-i option for COI and E-INS-i for 16S. COI sequences were translated into amino acid sequences to check the presence of stop codons. We examined the 16S alignment manually and no gap was observed. We also used Gblocks 0.91b (Castresana, 2000) in order to delete possible poorly aligned positions and too divergent regions. Gblocks only trimmed the edges of the sequence data due to differences in sequence read length; internal regions were not removed. The COI and 16S combined dataset was obtained by concatenating the trimmed alignment of each gene.
Phylogenetic analyses were performed using Maximum Likelihood (ML) and Bayesian Inference (BI). The ML analysis was performed in RAxML v.8.2.7 (Stamatakis, 2014). PartitionFinder v.2 (Lanfear et al., 2016) was used to determine the model choice, for COI data returning the GTR+G model for the 1st and 2nd position of COI sequences and the GTR+G+I model for 3rd position. For 16S, the best model selected was the GTR+G+I. In ML we used GTR+G for COI and GTR+G+I for 16S. Statistical support of ML nodes was obtained by bootstraps of 1000 replicates. BI analysis was conducted in MrBayes v. 3.2 (Ronquist et al., 2012), using GTR+G for 1st and 2nd positions of COI and GTR+G+I for 3rd position of COI and for 16S. BI analyses used two independent simultaneous runs of four chains run for 107 Markov chain Monte Carlo generations sampled every 1000th generation. The first 25% of the generations were discarded as burn-in. ML and BI for the COI and 16S combined dataset were performed using the GTR+G+I model for each of the two gene partitions. Genetic distances were estimated without correction (p-distance) and corrected by K2P model to allow comparison with literature. Distances were calculated in MEGA v.7 (Kumar et al., 2016).
Intraspecific Analysis of Vrijenhoekia balaenophila
The relationships among V. balaenophila lineages were investigated using a 564 bp COI alignment. Given that this was a potentially intraspecific dataset, we calculated the total number of parsimony informative sites, number of haplotypes (h), haplotype diversity, and nucleotide diversity (π) using DNAsp 5.10.1 (Librado and Rozas, 2009) for each recovered clade and for the V. balaenophila complex. In addition, a haplotype network was created including all V. balaenophila lineages using the TCS method (Clement et al., 2000) in PopART1.
Results
Species Delimitation Based on Morphological and DNA Barcoding Data and Distribution in the SW Atlantic
Based on initial morphological examination, we identified Sirsoe sirikos and a Vrijenhoekia balaenophila complex, as well as 7 putative new species in our samples from the SW Atlantic (Supplementary Figure S1), all of which do not a median antenna. All species had reproductive mature specimens since we observed eggs in some individuals. The COI dataset consisted in 86 sequences of Sirsoe/Vrijenhoekia, which were 602-bp in length with 215 variable and 190 parsimony informative sites. The 16S dataset consisted of 57 terminal taxa and 442-bp containing 84 variable and 69 parsimony informative sites. The COI and 16S datasets agree with our morphological identification confirming the presence of S. sirikos and V. balaenophila in SW Atlantic. Notwithstanding, both datasets also highlight that V. balaenophila and our morphotype Vrijenhoekia sp. 4 contain cryptic species (Supplementary Figure S1).
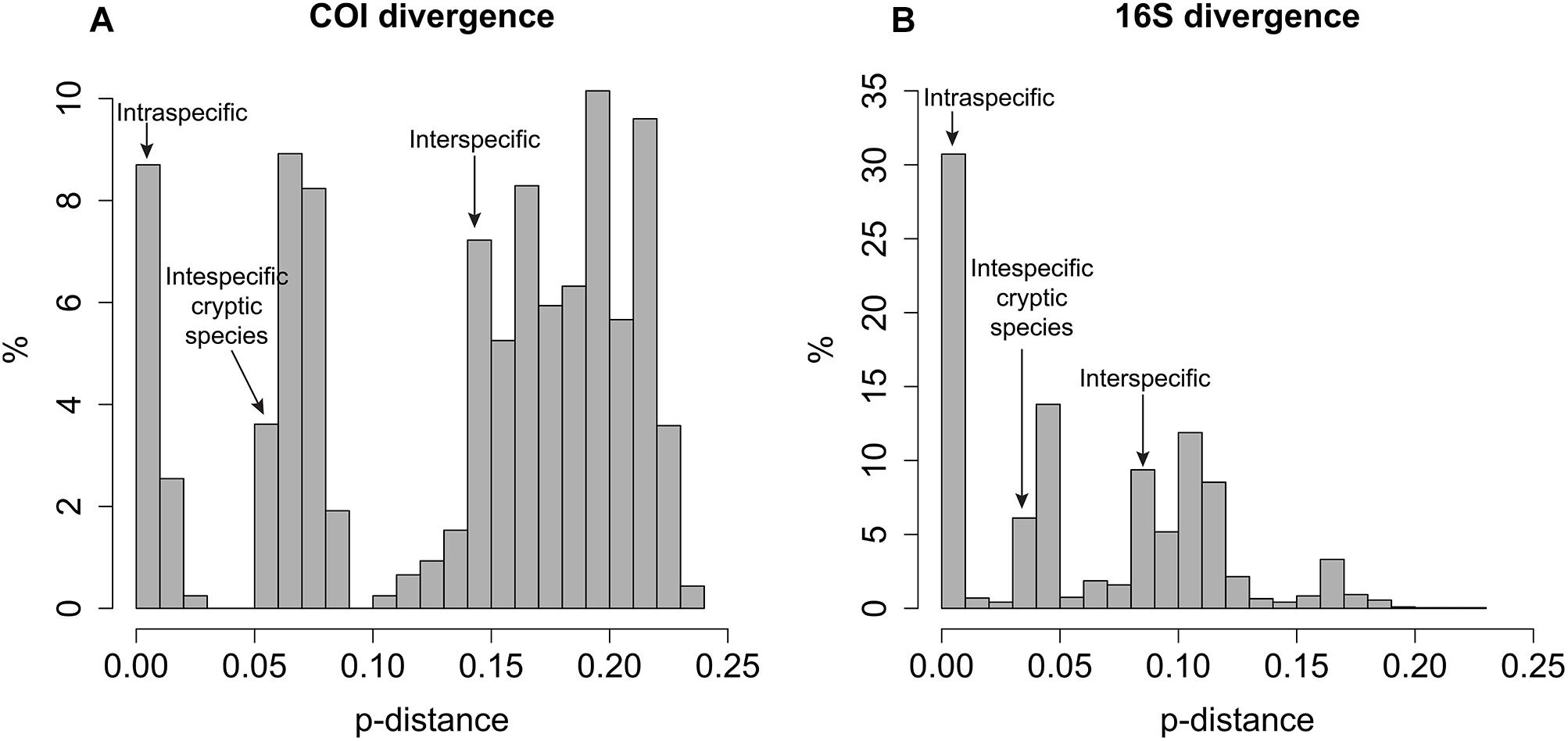
In general, intraspecific divergence of COI and 16S was close to zero (usually less than 1%, Figure 1). The interspecific divergence of COI averaged 18% (ranging from 6.1 to 23.8%) and 16S averaged 9.1% (ranging from 0.3 to 17.2%) and the interspecific divergence range among cryptic species is 5–10% for COI and <5% for 16S (Figure 1). COI divergence between Sirsoe maximiano sp. nov. and Sirsoe ‘SP-2014’ (GenBank Accession Number KJ566956) was 0% (Supplementary Table S2). Through the integrating of molecular and morphological information, we delimited 13 species in whale falls from the SW Atlantic (see descriptions in “Systematic” Section) (Supplementary Figure S1). The highest number of species occurred at SP3300 site (six species), while the lowest number of species occurred at SP1500 site (Table 1 and Figure 2). Three species present eurybathic distribution (1500-3300/4204 m depth) (Figure 2). The new species S. alphacrucis sp. nov. is restrict to the upper slope (550–1500 m depth), while its sister-clade S. yokosuka sp. nov. was only found in abyssal depths (3300–4204 m) (Figure 2).
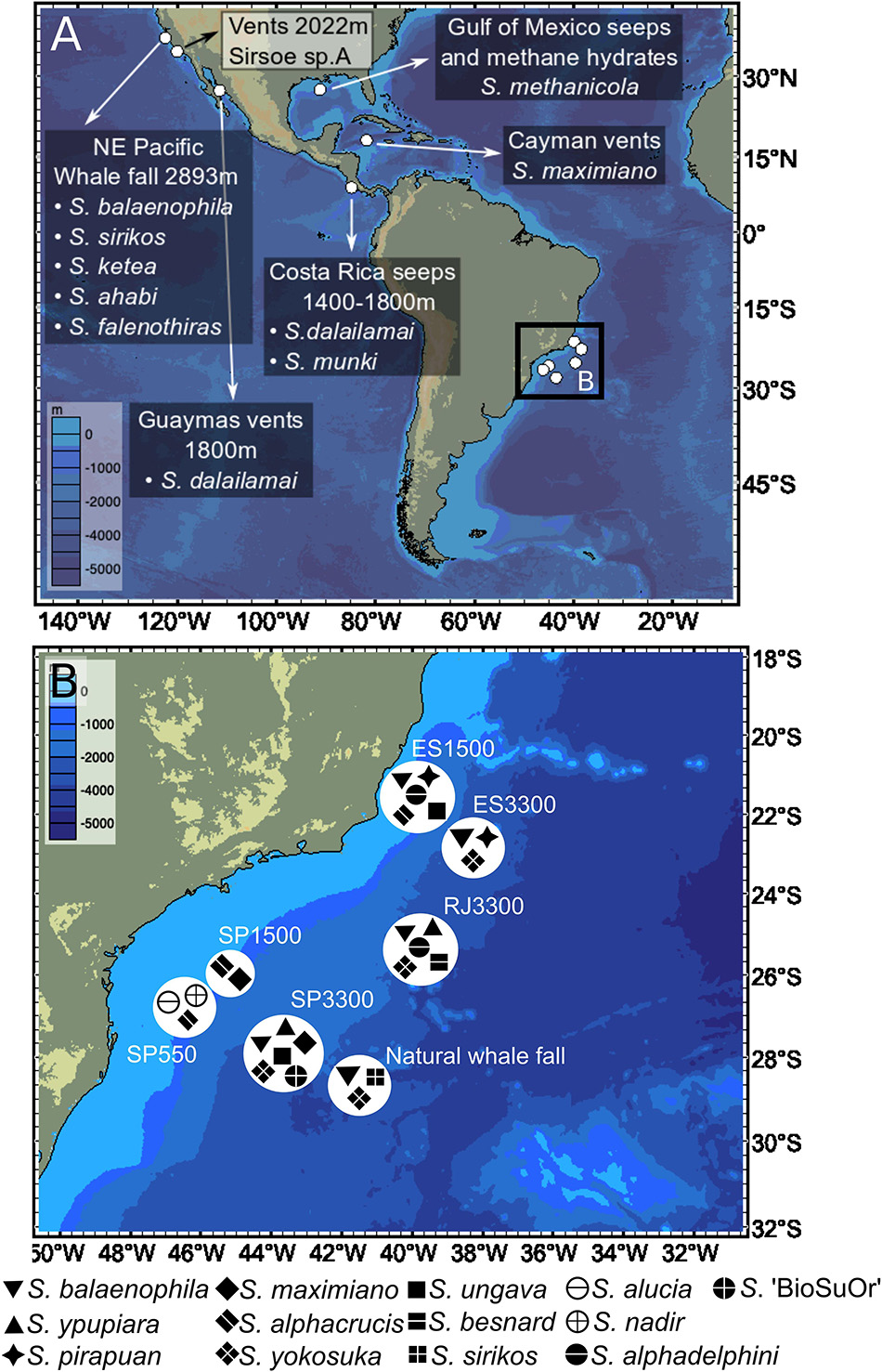
Figure 2. World-wide distribution of Sirsoe species in: (A) Previous records from NE Pacific, Gulf of Mexico, and Caribbean Sea; and (B) New data (this study) from the SW Atlantic Ocean. We omitted the occurrence of Sirsoe hessleri (Blake, 1991) in hydrothermal vents from Mariana back-arc basin (18° 12.6′N, 144° 42.4′E, 3640 m).
Phylogeny of Sirsoe and Vrijenhoekia
The concatenated COI+16S dataset consisted of 22 terminal taxa (three outgroups) with 1044-bp, 318 variable sites and 267 parsimony informative sites. The recovered topology (Figure 3) included three well-supported clades (BS: 83–100% PP: 1.0). Clade-I clustered (BS: 88%, PP: 1) the previous known Sirsoe species (S. methanicola, S. sirikos, S. dalailamai and S. munki), and two new species, S. alucia and S. maximiano. S sirikos was monophyletic (BI: 0.93), and S. maximiano close related to S. sirikos (BI: 1) (Figure 3B). Within clade-I, the previous known species bear a median antenna, while the new species do not. Clade-II was formed by V. balaenophila species complex and five new species: S. alphacrucis, S. yokosuka, S. alphadelphini, S. besnard, and S. nadir (BS: 76%). All species in clade-II lack median antennae on prostomium. Clade-III (BS:48%, BI: 1) joined the V. ketea species complex, with S. ungava sp. nov. and two unnamed morphotypes: Vrijenhoekia sp. A (from Summers et al., 2015) and Sirsoe sp. ‘BioSuOr’ (from this study). Sirsoe ungava sp. nov. lacks a median antenna even though this structure is present on prostomium of V. ketea species complex and Vrijenhoekia sp. A. We do not describe Sirsoe sp. ‘BioSuOr,’ since only one small specimen was found and the whole individual was used for DNA extraction; we did not observe a median antenna in our initial morphological examination of this individual, and the proboscis was not observed neither. The presence or absence of a median antenna does not seem to distinguish Sirsoe from Vrijenhoekia and for this reason, from hereafter we use Vrijenhoekia as a junior synonym of Sirsoe (see Supplementary Information, Systematic Account, remarks of Sirsoe).
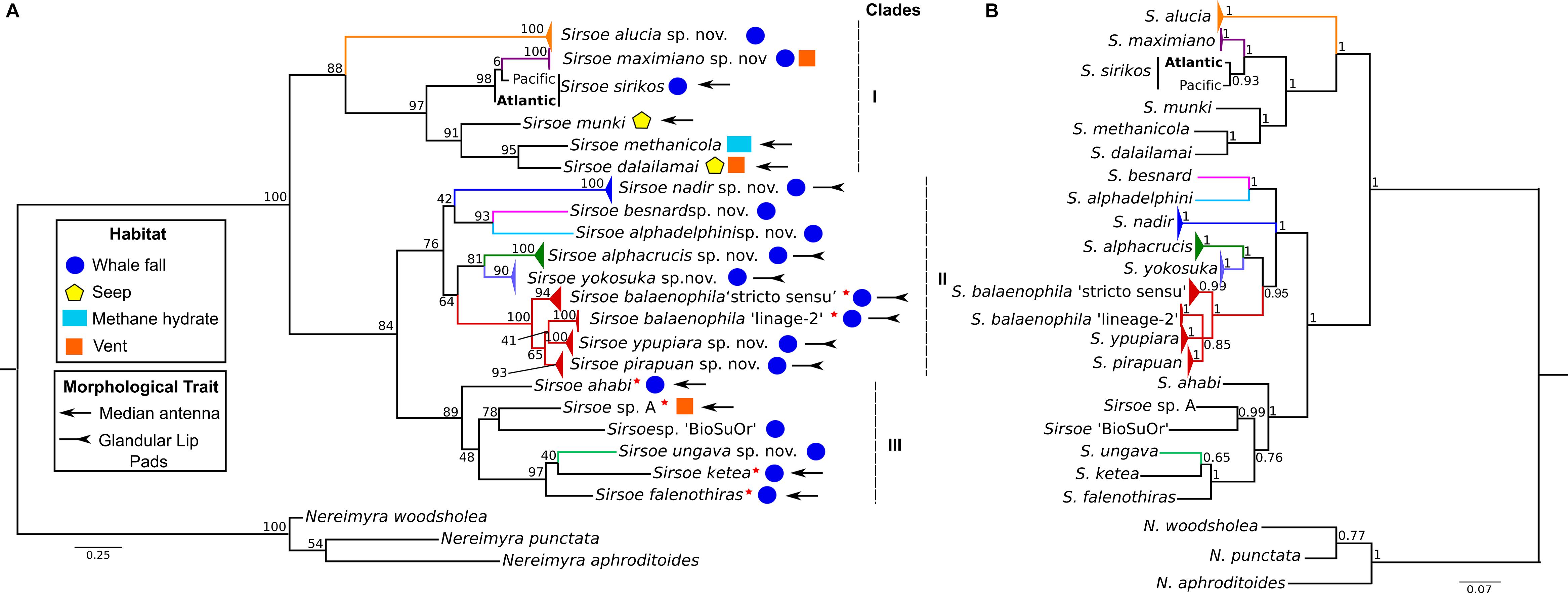
Figure 3. Phylogenetic relationships of Sirsoe of the concatenated dataset (COI+16S). (A) Phylogeny based on Maximum Likelihood (ML); (B) Phylogeny based on Bayesian inference (BI). Values close to nodes are bootstrap supports of ML and posterior probabilities supports of BI. Red star close to species name indicates former species within Vrijenhoekia genus. Habitat use: circle – whale fall; pentagon – seep; rectangle – methane hydrate; square – vent. Morphological trait: arrow – median antenna present; inverted arrow – glandular lip pads present; species without symbol for morphological trait indicate absence of median antenna and glandular lip pads.
Glandular lip pads (GLP) were only observed in some species of clade-II, although this clade also clustered species without any GLP, such as S. besnard sp. nov. and S. alphadelphini sp. nov. (Figure 3). Rudimentary neuropodial lobes with papilliform shape were found only in V. balaenophila species complex (see Systematic accounts). All clades have species in which the neuropodial lobes start on the third segment. Nevertheless, in clade-I, neuropodial lobes start on the fourth segment for S. dalailamai and for Pacific specimens of S. sirikos. For, specimens of S. sirikos from the Atlantic Ocean, neuropodia start on the third segment (see in Systematic accounts). Sirsoe balaenophila comb. nov., a clade-II species, has a papilliform lobe on the third segment, with or without acicula and chaetae. In clade-III, S. ketea comb. nov. species-complex bear neuropodial lobes since the fourth segment.
Clade-I clusters species that inhabit whale falls, seeps, methane hydrates and hydrothermal vents (Figure 3) with S. maximiano sp. nov. and S. dalailamai inhabiting vents/whale falls and seeps/whale falls, respectively (Figure 3). All species from clade-II and clade-III inhabit whale falls, except Sirsoe sp. A sensu Summers et al. (2015), which inhabits vents (Figure 3).
Sirsoe balaenophila comb. nov. - Interbasin Distribution and Related Species
The S. balaenophila comb. nov. species complex COI dataset consisted in 564-bp and 47 terminal specimens. This dataset exhibits a total of 81 segregating sites, 73 of them parsimony informative sites (Table 2). Sirsoe balaenophila species complex is composed by S. balaenophila ‘stricto sensu’ (Pleijel et al., 2008), two new cryptic species, S. pirapuan and S. ypupiara, and a putatively new cryptic species here named as S. balaenophila ‘lineage-2’ (Figures 3, 4). Nucleotide and haplotype diversities were higher in S. balaenophila ‘stricto sensu’ in comparison with both new cryptic species: S. pirapuan and S. ypupiara (Table 2). Sirsoe balaenophila ‘stricto sensu’ was present on the Pacific and the Atlantic basins, although S. pirapuan sp. nov. and S. ypupiara sp. nov. were sampled only in the Atlantic Ocean. In the Atlantic, S. balaenophila ‘stricto sensu’ was sampled in four sites at two different depths (1500 and 3300 m depth) (Figures 2, 4). Sirsoe pirapuan sp. nov. was found at 1500 and 3300 m depth, but only in the Espírito Santo landers, and S. ypupiara sp. nov. was found only at SP3300 and RJ3300 sites (Figures 2, 4). Sirsoe balaenophila ‘stricto sensu’ comprised 10 haplotypes from the Pacific and four from the Atlantic Ocean (Figure 3). No haplotypes were shared between both basins with three substitutions sites separating the closest haplotypes (Figure 4). Three S. balaenophila ‘stricto sensu’ haplotypes from the Atlantic were shared between different sites and one of them was found at both ∼1500 m to ∼3300 m depths (Figure 4). Sirsoe pirapuan sp. nov. exhibited 5 haplotypes, two of them shared between ∼1500 m and ∼3300 m-depth sites (Figure 4). Sirsoe ypupiara sp. nov. occurred only at 3300 m depth at two sites (Figure 4). One haplotype of this clade is shared between different locations. The S. balaenophila ‘lineage-2’ was formed by two single haplotypes from the Pacific.
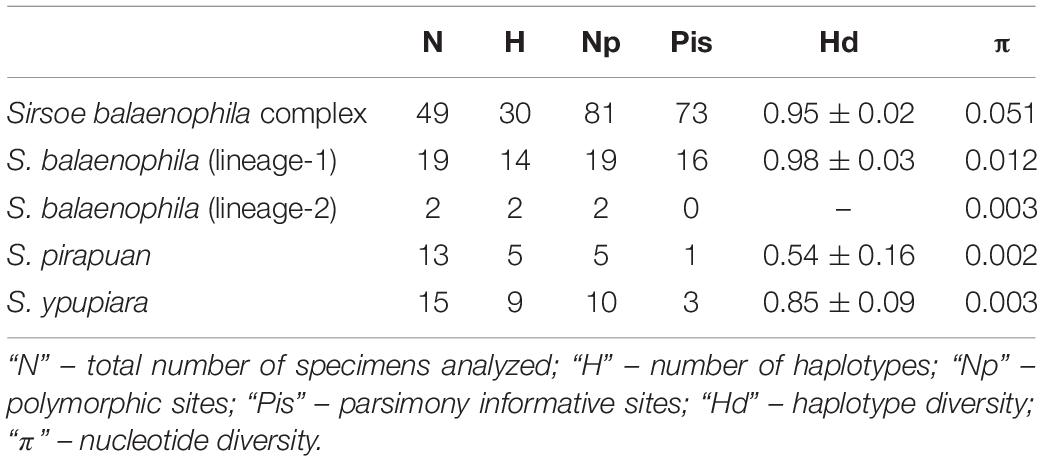
Table 2. Summary of genetic variability of COI for Sirsoe balaenophila species complex and for each lineage.
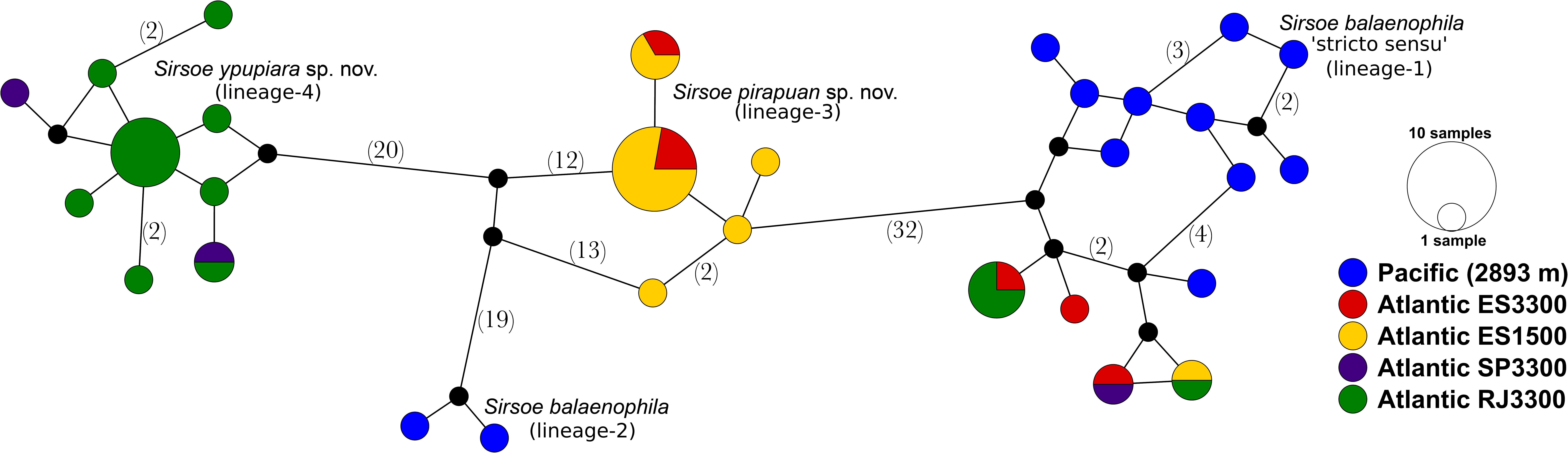
Figure 4. Haplotype network of Sirsoe balaenophila species complex based on TCS analyses (Clement et al., 2000). The network was produced in PopART (Leigh and Bryant, 2015). Numbers in parentheses are numbers of substitution sites between haplotypes. Only one substitution was omitted. Black circle represents a missing inferred haplotype.
Systematic Account
Hesionidae Grube, 1850
Psamathinae Pleijel, 1998
Remarks: Pleijel (1998) defined Psamathini sub-tribe as Hesioninae with facial tubercle absent and prolonged teeth of chaetal blades. The dorsal cirri pattern is distinct without elevated dorsal cirri in segment 19th (Ruta and Pleijel, 2006). The Psamathini sub-tribe was elevated to sub-family status by Summers et al. (2015), using the same definition of Psamathini as Pleijel (1998).
Sirsoe Pleijel, 1998 emended urn:lsid:zoobank.org:act:AED41380-D297-4F21-A119-3B55DEDC7A2B Type species: Sirsoe grasslei (Blake, 1985)
Type species: Sirsoe grasslei (Blake, 1985)
Vrijenhoekia Pleijel et al. (2008), new synonymy. Type species Vrijenhoekia balaenophila (new combination Sirsoe balaenophila).
Material examined: S. grasslei: holotype from Guaymas basin, 27.017°N, 111.417°W, 2022 m (USNM 81808); S. hessleri: holotype from N Pacific, 18.21°N, 144.707°E, 3640 m (USNM132660); S. methanicola: holotype from Gulf of Mexico, 27.747°N, 91.225°W, 538 m (USNM 182243); S. sirikos two specimens from SW Atlantic, 28.519°S, 41.657°W, 4204 m (ColBio DS00140); S. balaenophila seven specimens from SW Atlantic, 21.450°S, 39.897°W, 1444 m (ColBio DS0014) and 25.338°S, 39.642°W, 3227 m (ColBio DS00143); S. maximiano: holotype from SW Atlantic, 25.894°S, 45.035°W, 1439 m (MZUSP 3371); S. alphacrucis: holotype from SW Atlantic, 25.894°S, 45.035°W, 1439 m (MZUSP 3363); S. yokosuka: holotype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3366); S. alucia: holotype from SW Atlantic, 26.600°S, 46.150°W, 550 m (MZUSP 3359); S. nadir: holotype from SW Atlantic, 26.600°S, 46.150°W, 550 m (MZUSP 3361); S. alphadelphini: holotype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3373); S. ungava: holotype from SW Atlantic, 21.450°S, 39.897°W, 1444 m (MZUSP3368); S. besnard: holotype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3370).
Diagnosis: Psamathinae with reduced small-depression-like nuchal organs not projected on posterior margin of prostomium (Figure 4C), frontal tubercle present (Figures 5E, 6C, 7B), with (Figures 4A,C) or without (Figures 5F, 6C, 7B, 8A, 9C, 10B, 11D, 12A) dorsally median antenna, eyes absent, with or without glandular lip pads.
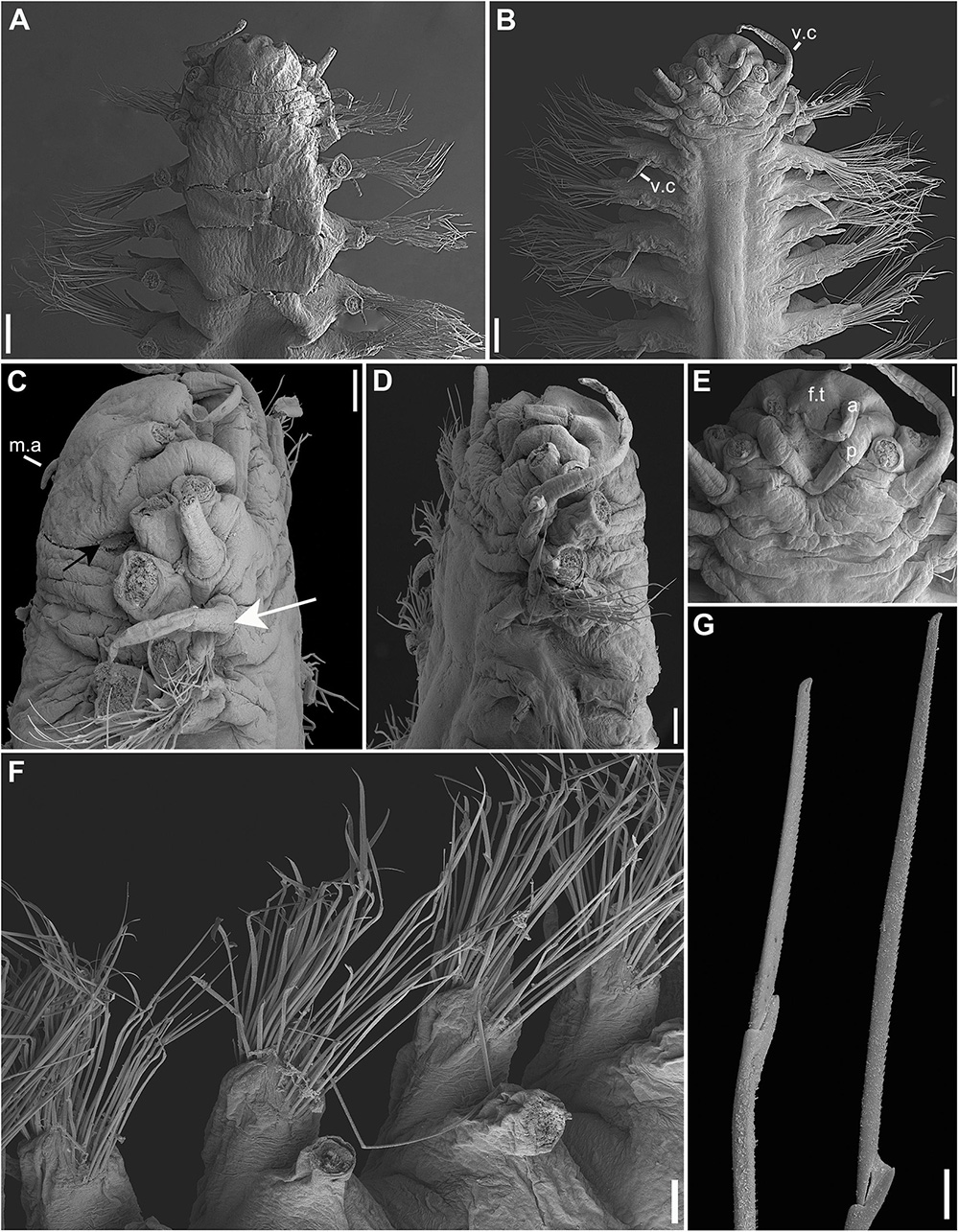
Figure 5. (A–G) Sirsoe sirikos. (A) anterior end, dorsal view; (B) anterior end, ventral view; (C,D) anterior end, lateral view; (E) detail of anterior end, ventral view; (F) median neuropodia; (G) supra- and sub-acicular neurochaetae. a, lateral antenna; f.t., frontal tubercle; m.a, median antenna; p, palps; v.c., ventral cirri; white arrow on C: indicate neuropodial lobe on 3rd segment; black arrow on C: indicate small-depression-like nuchal organs. Scales: (A,B) 200 μm; (C,G) 100 μm.
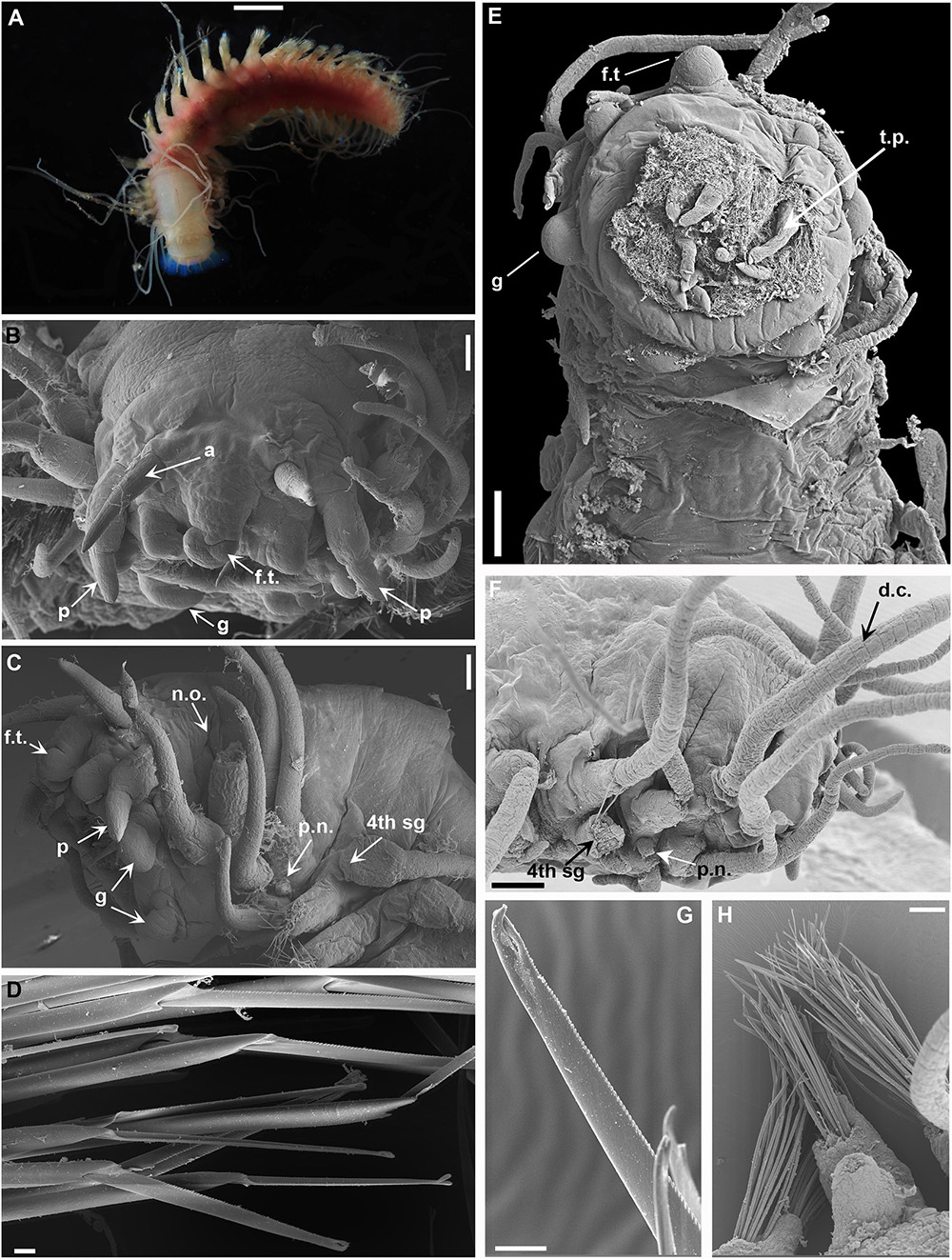
Figure 6. (A) Sirsoe balaenophila comb. nov., specimen from SW Atlantic. (B–D) Sirsoe ypupiara sp. nov.; (B) anterior end, dorsal view; (C) anterior end, lateral view; (D) neurochaetae. (E–H): S. pirapuan sp. nov.: (E) anterior end, ventral view; (F) anterior end, lateral view; (G) neurochaetae; (H) median neuropodia. 4th sg, fourth segment; a, lateral antenna; d.c., dorsal cirri; f.t, frontal tubercle; g, glandular lip pad; p, palps; p.n., papilliform neuropodial lobe; n.o., nuchal organs. Scales: (A) 3 mm; (B,E,F) 200 μm; (C,H) 100 μm; (D,G) 10 μm.
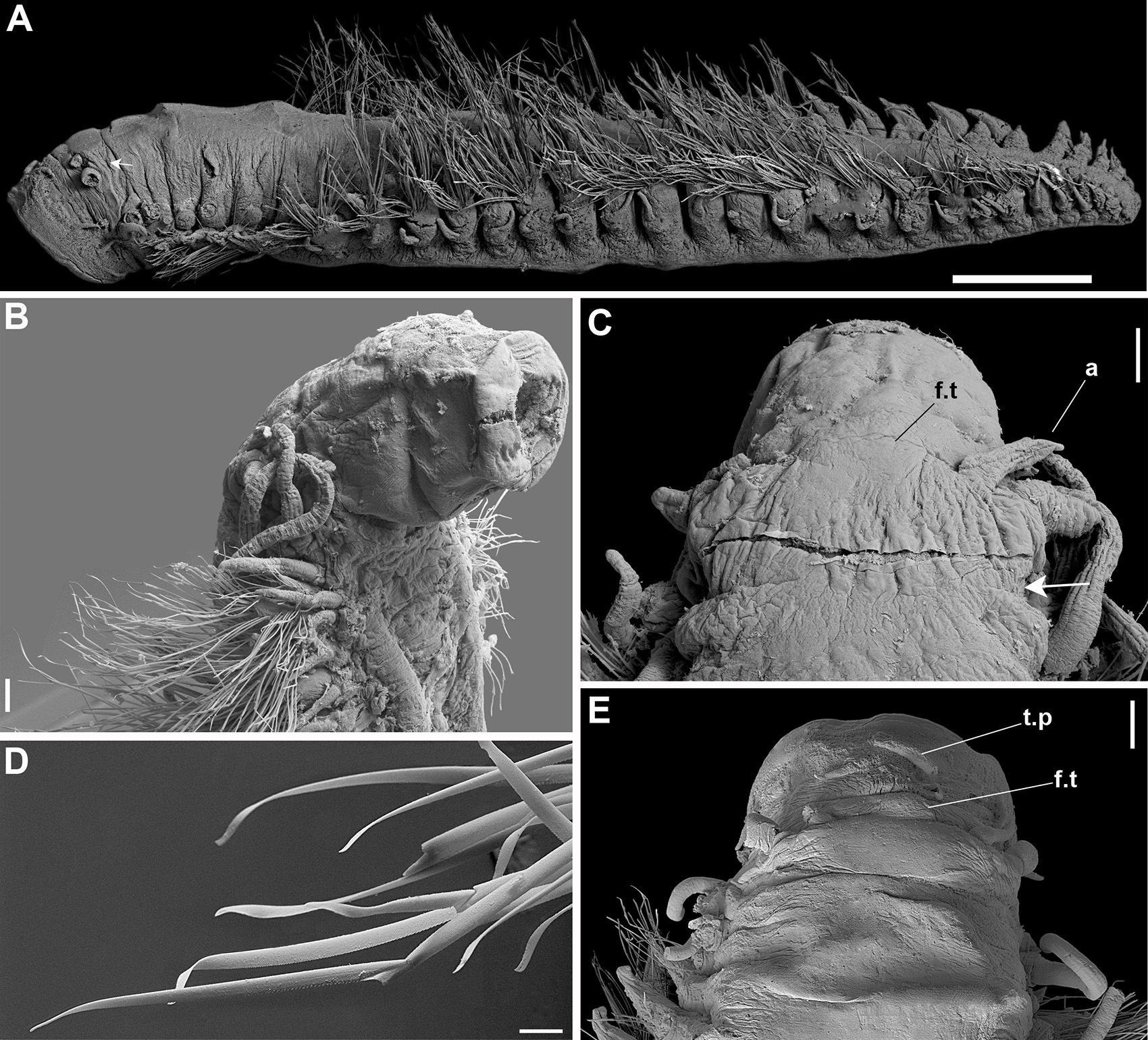
Figure 7. (A–E) Sirsoe maximiano sp. nov. (A) complete specimen, lateral view; (B) anterior end, lateral view; (C) anterior end, dorsal view; (D) neurochaetae; (E) anterior end, dorsal view. a, lateral antenna; f.t, frontal tubercle; t.p, terminal papilla; white arrows on (A,C) indicate nuchal organs. Scales: (A) 1 mm; (B,C) 100 μm; (D) 20 μm; (E) 200 μm.
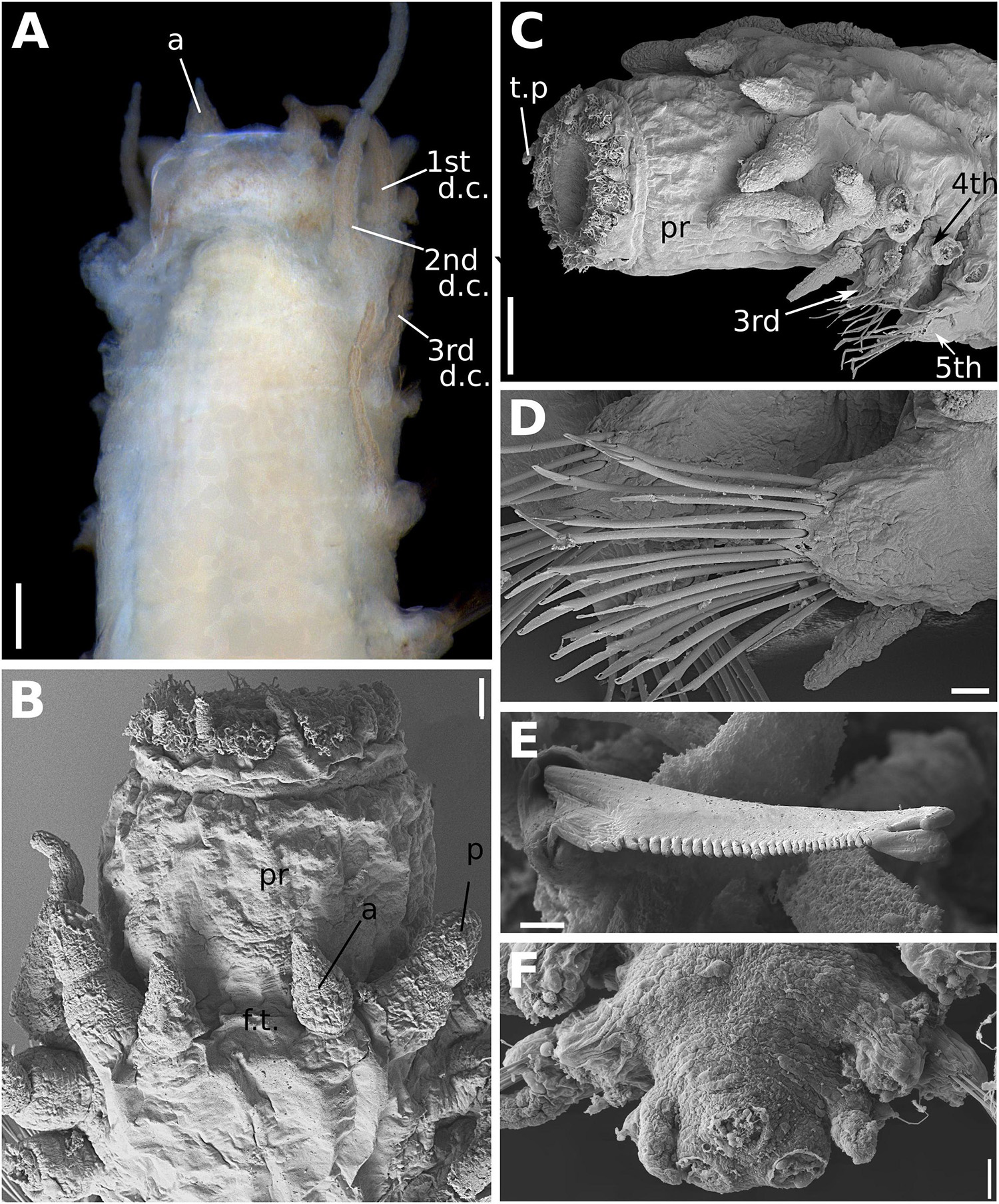
Figure 8. (A–F) Sirsoe alphacrucis sp. nov. (A) anterior end, dorsal view; (B) anterior end, dorsal view; (C) anterior end, lateral view; (D) median neuropodia; (E) detail of neurochaeta; (F) posterior end, dorsal view, showing the insertions of pygidial cirri and papilla. 1st d.c., first dorsal cirrus; 2nd d.c., second dorsal cirrus; 3rd d.c., third dorsal cirrus; a, lateral antenna; ci, ciliation; c.v, ventral cirri; p, palp: pr, proboscis; t.p, terminal papilla. Numbers in (C) referring to the segment. Scales: (A) 100 μm; (B) 30 μm; (C,D,F) 20 μm; (E) 2 μm.
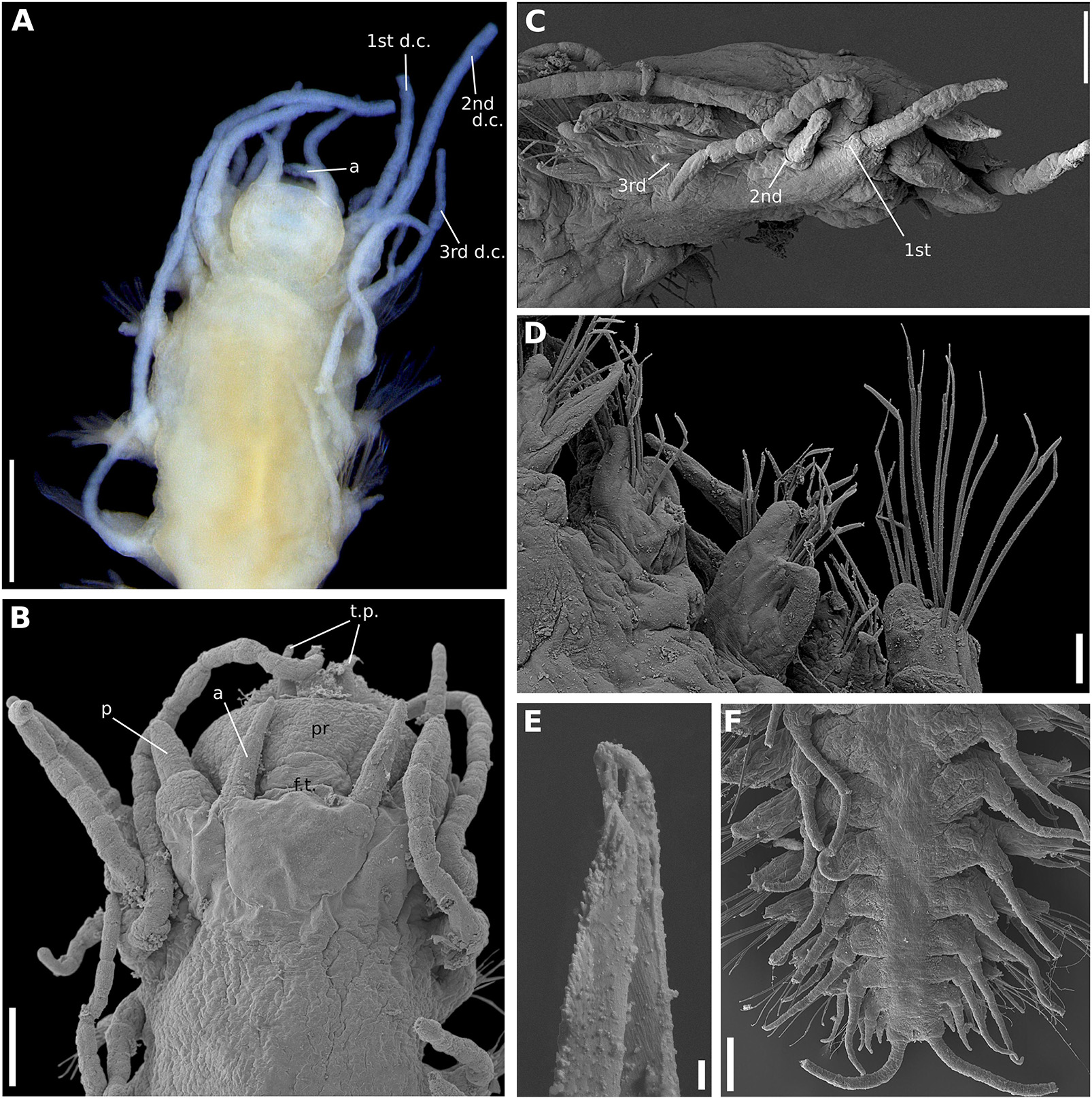
Figure 9. (A–F) Sirsoe yokosuka sp. nov. (A) anterior end, dorsal view; (B) anterior end, dorsal view; (C) anterior end, lateral view; (D) median neuropodia, ventral view; (E) detail of neurochaeta; (F) posterior end, dorsal view. 1st d.c., first dorsal cirrus; 2nd d.c., second dorsal cirrus; 3rd d.c., third dorsal cirrus; a, lateral antenna; ci, ciliation; d.c, dorsal cirri; f.t, frontal tubercle; p, palps; p.c, pygidial cirri; p.p, pygidial papilla; t.p, terminal papilla; v.c, ventral cirri. Numbers in (C) referring to the segment. Scales: (A) 200 μm; (B,F) 100 μm; (C) 20 μm; (D) 40 μm; (E) 2 μm.
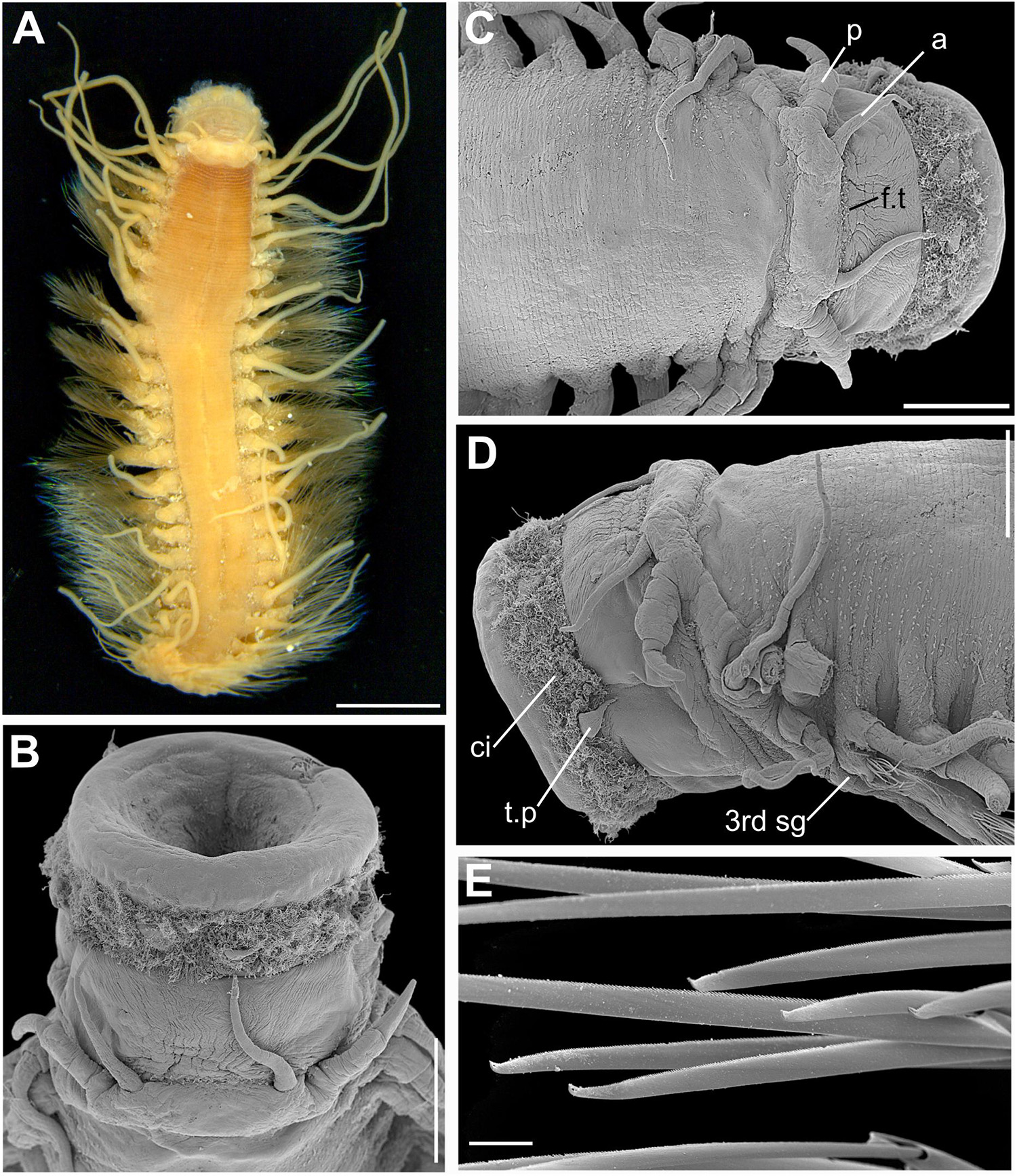
Figure 10. (A–E) Sirsoe alucia sp. nov. (A) complete specimen, dorsal view; (B) anterior end, dorsal view with proboscis extended; (C) anterior end, dorsal view; (D) anterior end, lateral view; (E) detail of neurochaetae. 3rd sg, third segment; a, lateral antenna; ci, ciliation; f.t, frontal tubercle; p, palps. Scales: (A) 1.5 mm; (B–D) 500 μm; (E) 20 μm.
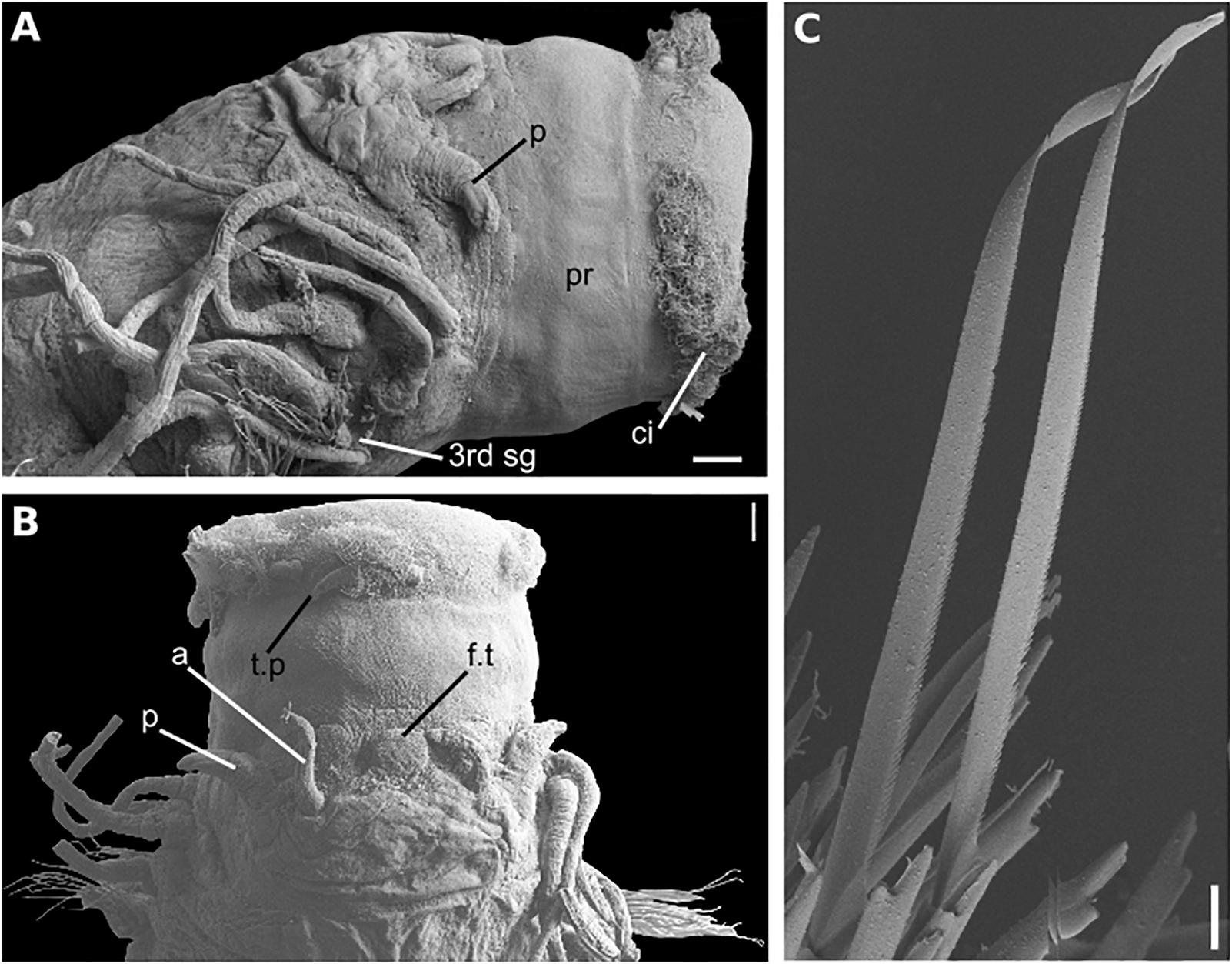
Figure 11. Sirsoe nadir sp. nov. (A) anterior end, lateral view; (B) anterior end, dorsal view; (C) neurochaetae from median neuropodia. Abbreviations: 3rd sg: third segment; a, lateral antenna; ci, ciliation; f.t, frontal tubercle; n.o, nuchal organs; p, palps; pr, proboscis. Scales: (A,B): 100 μm; (C): 20 μm.
Figure 12. (A–C) Sirsoe alphadelphini sp. nov. (A) anterior end, dorsal view; (B) detail of proboscis; (C) neurochaetae. (D–F) Sirsoe ungava sp. nov. (D) complete specimen, dorsal view; (E) anterior end, lateral view; (F) neurochaetae. Scales: (A,B) 500 μm; (C) 50 μm; (D) 2 mm; (E) 250 μm; (F) 30 μm.
Remarks: Sirsoe was originally defined to include Psamathinae with a dorsal median antenna and absence of eyes (Pleijel, 1998), whereas Vrijenhoekia includes those without median antenna (Pleijel et al., 2008). Moreover, the diagnostic features of Vrijenhoekia, as originally described by Pleijel et al. (2008) based on a single species, i.e., V. balaenophila, are not present in any other species subsequently described. This includes the work of Summers et al. (2015) describing the Vrijenhoekia ketea species complex with median antenna. Vrijenhoekia is the sister clade to Sirsoe and up to now considered both reciprocally monophyletic (Pleijel et al., 2008; Summers et al., 2015; Rouse et al., 2018a). Our molecular phylogeny shows that species in clade I, such as Sirsoe maximiano sp. nov. (without median antenna) is closely related to Sirsoe sirikos (with median antenna). Moreover, Sirsoe ungava sp. nov. (without median antenna) is close to V. ketea complex species in clade III, which bear a median antenna. The presence and placement of a median antenna varies during the ontogeny of many hesionids, mainly within Psamathinae (such as Nereimyra, Micropodarke and Hesiospina) and becomes reduced or absent in adults (Pleijel, 1998, 2004; Pleijel and Rouse, 2005; Pleijel et al., 2012). An argument could be made to maintain Vrijenhoekia as restricted to clade II (recovered in our phylogeny), since this is the only clade including species without median antenna, but based on our phylogeny and the morphological examination, as well as evidence from the literature, the presence/absence of median antenna not seem to be a diagnostic feature to distinguish hesionid genera within Psamathinae.
According Pleijel et al. (2008), Vrijenhoekia is the only genus in Psamathinae with three pairs of GLPs. Nereimyra also presents GLPs, but only a ventro-lateral pair (Pleijel et al., 2012). The clade II of our phylogeny is the only that includes species with GLPs however, it also includes species without any GLP, such as S. alphadelphini sp. nov and S. besnard sp. nov. The new species, S. alphacrucis sp. nov. and S. yokosuka sp. nov. (also clustered into clade II) bear GLPs, but only possess two pairs, one dorsal and one ventro-lateral, and are reduced compared to those of Vrijenhoekia balaenophila (see Remarks of S. alphacrucis sp. nov.). Thus, our results suggest that the presence or no of GLPs, as well as the number of these structures, do not help to distinguish both genera.
The presence of a rudimentary papilliform neuropodial lobe on the third segment being completely developed only on segment four is diagnostic of V. balaenophila (Pleijel et al., 2008). Up to now, the V. ketea species complex also present neuropodial lobes starting on segment four without any rudimentary neuropodial lobe in the previous segments (Summers et al., 2015). In Sirsoe, well developed neuropodial lobe usually begins in segment two or three (S. grasslei and S. methanicola, respectively) (Pleijel, 1998). Recently, Rouse et al. (2018a) described S. dalailamai, which also bear a rudimentary neuropodial lobe on segment 3. Although not formerly named, Vrijenhoekia sp. A bears a neuropodial lobe on segment 3 (Summers et al., 2015). The new species, S. alphacrucis and S. yokosuka, present neuropodial lobe on segment 3, and both form a sister clade with S. balaenophila species complex. Specimens of S. sirikos from the Pacific have neuropodial lobe starting on the fourth segment (Summers et al., 2015), although we found specimens from the Atlantic with neuropodia on the third segment (see more in remarks of S. sirikos). The presence/absence of rudimentary neuropodial lobe and/or the segment where the neuropodial lobe starts do not seem stable in Sirsoe or even in species formerly described in Vrijenhoekia.
Analyses performed here clearly indicate that Sirsoe constitutes a monophyletic group only with the inclusion of Vrijenhoekia species. In other words, recognition of Vrijenhoekia renders both genera non-monophyletic. Alternatively, we could propose a new genus to include the species of clade III, and restrict Vrijenhoekia only for clade II and Sirsoe in clade I. However, as discussed above, to maintain both genera valid and to erect a new genus, we not found any diagnostic character to split them in three distinct genera. The choice to erect Vrijenhoekia was made to avoid the use of polymorphism, however with descriptions of Summers et al. (2015) and our species, the diagnosis of Vrijenhoekia include these polymorphisms and the erection of a new genus will not resolve the problem. Additionally, Pleijel et al. (2008) recognized that the erection of Vrijenhoekia was “arbitrary” and thus we opt not to needlessly create additional taxonomic terms. We propose Sirsoe over Vrijenhoekia based on Principle of Priority (Article 23, International Commission on Zoological Nomenclature [ICZN], 1999). We conclude that the diagnostic characters, which define Sirsoe genus, are: (1) the presence of a frontal tubercle (reduced or developed); and (2) a reduced small-depression-like nuchal organs not projected in posterior margin of prostomium. As discussed above, the presence of a median antenna in the prostomium and of the GLPs, and the segment in which the neuropodial lobe begins are variable features within Sirsoe and even for those species formerly in Vrijenhoekia.
The position, form and size of nuchal organs, and if they are coalescent or not, are morphological features used to distinguish genera within Hesionidae (Pleijel, 1998). Nuchal organs are well developed in most Psamathinae taxa, with the exception of Sirsoe, Micropodarke, and Syllidia (Pleijel, 1998). Micropodarke and Syllidia bear nuchal organs as slightly curved bands, projected in the posterior and lateral margins of the prostomium even though it not coalescent (Pleijel and Rouse, 2005; Ruta and Pleijel, 2006; Rizzo and Salazar-Vallejo, 2014). In contrast, Sirsoe bears reduced small depression-like nuchal organs, usually in an oblique rows, located on the corner of the posterior margin of the prostomium, but not projected dorsally on the posterior margin. Micropodarke has a proboscis diaphragm and basal spurs on the neurochaetae blades (Pleijel and Rouse, 2005), both features are not present in Sirsoe. The presence of jaws only occurs in Syllidia, distinguishing this genus from the others (Ruta and Pleijel, 2006; Rizzo and Salazar-Vallejo, 2014). Based on taxonomic revisions herein, Sirsoe includes 20 valid species (Supplementary Table S3).
Sirsoe sirikos Summers et al., 2015 (Figures 5A–G)
Material examined: Two specimens from SW Atlantic, 28.519°S, 41.657°W, 4204 m (ColBio DS00140; GenBank: MH935152 and MH935153) specimen-1 with 8.0 mm length and 0.8 mm wide, 28 chaetigers; specimen-2 8.0 mm length and 1.0 mm, 22 chaetigers).
Description: Body cylindrical, slightly flattened but not tapered; preserved specimens whitish. Prostomium slightly wider than long, nearly rectangular (Figure 5A). Eyes absent. One pair of small-depression-like nuchal organs positioned posteriorly to the prostomium (Figure 5C). Frontal tubercle distinct, short and cushion-like (Figures 5B,E). Palps biarticulated; palpophore rhomboid, slightly wider than long; palpostyle conical and longer than palpophore (Figure 5E). Lateral antennae slender and slightly longer than palps; median antenna short, delicate, inserted near of posterior margin (Figures 5A–E). GLPs absent. Proboscis not totally everted. Dorsal cirri of the first two segments enlarged with distinct cirrophores. Ventral cirri slender and shorter than dorsal ones; cirri of segments 1–3 enlarged with distinct cirrophores (Figure 5C); following ventral cirri attached in the mid-ventral margin of neuropodium without distinct cirrophores (Figure 5B). Noto- and neuropodial lobes and chaetae absent on segments 1 and 2; following segments with sub-birramous parapodia; notopodia reduced to dorsal cirrophores with two notoaciculae (not easily observed); neuropodia stout with pointed acicular lobe bearing up to three robust neuroaciculae. Anterior neuropodia with ∼40 compound chaetae, 15 supra-acicular and 25 sub-acicular (Figure 5F); posterior neuropodia with fewer chaetae than anterior ones, ∼10 supra-acicular and 10 sub-acicular (chaetigers 22); blades of supra-acicular chaetae ca. 15 times longer than wide, sub-acicular ones progressively shorter, nearly half the length of supra-acicular. Cutting edge of blades finely serrated, distally unidentate, with a sub-distal spine-like prolongation (Figure 5G). Pygidium small; pygidial cirri and papilla lost, but with three scars of insertion.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935152 and MH935153, respectively.
Distribution, habitat and depth: NE Pacific, whale falls, 2893 m depth (Summers et al., 2015); SW Atlantic, whale falls, 4204 m depth (Figure 2).
Remarks: The absence of neuropodial lobe on the third segment distinguishes S. sirikos from other Sirsoe species (Summers et al., 2015). However, it is possible to observe a lobe on the third segment of S. sirikos in the Figure 4c from Summers et al. (2015). Specimens studied herein from Atlantic Ocean present a neuropodial lobe on the segment 3. The first neuropodia is about half of the size of the second neuropodial lobe (Figures 5B,C). S. sirikos from Atlantic Ocean presents two noto- and three neuroaciculae, although according Summers et al. (2015) S. sirikos presents only one noto- and one neuroacicula. All other morphological features of S. sirikos from Atlantic Ocean agree with Summers et al. (2015) description, including the presence of enlarged ventral cirrus on the first three segments, inserted in a basal cirrophore, being absent in following segments. Although we observed some morphological variances between specimens from Atlantic and Pacific, the intraspecific divergences of COI and 16S were very low supporting the interbasin distribution of S. sirikos.
Intraspecific divergences were 2.9 and 0% (p-distance) for COI and 16S, respectively (Supplementary Table S2). Interspecific divergence ranged from 7.7% with S. maximiano sp. nov. to 22.7% with S. alphadelphini sp. nov. for COI, whereas 16S divergence ranged from 0.7% with S. maximiano sp. nov. to 13.0% with S. alphacrusis sp. nov. (Supplementary Table S2).
Sirsoe balaenophila (Pleijel et al., 2008) comb. nov. urn:lsid:zoobank.org:act:42715732-148D-47EE-B7FC-AAAC5613AA90 (Figure 6A)
Material examined: One specimen from SW Atlantic, 21.450°S, 39.897°W, 1444 m (ColBio DS00141; GenBank: MH935085 and MH935162), incomplete, 19 chaetigers, 14 mm length, 2.5 mm width; One specimen from SW Atlantic, 25.338°S, 39.642°W, 3227 m (ColBio DS00142; GenBank: MH935092 and MH935165), incomplete, 9 chaetigers, 7 mm length, 1.8 mm width; Six specimens from SW Atlantic, 25.338°S, 39.642°W, 3227 m (ColBio DS00143), incomplete; One specimen from SW Atlantic, 28.029°S, 43.530°W, 3328 m (ColBio DS00144), incomplete.
Description: Body stout and tapering to posterior region (Figure 6A). Live specimens reddish (Figure 6A); preserved specimens whitish to pale yellowish. Prostomium wider than long, nearly rectangular; prostomium forming a swollen ridge basally to the prostomium, extending laterally to the first parapodia. Eyes absent. One pair of small-depression-like nuchal organs posteriorly on the prostomium. Frontal tubercle distinct, robust and directed toward anterior end. Palps biarticulated; palpophore cylindrical; palpostyle conical, short, up to half of the length of palpophore. Terminal proboscis with ten filiform papillae and dense ciliation among each papilla. Three pairs of distinct well-developed glandular lip pads. Lateral antennae slender and slightly longer than palps; median antenna absent. Dorsal cirri of first five segments enlarged with distinct cirrophores. Ventral cirri slender, shorter than dorsal ones; ventral cirri enlarged with distinct cirrophores on first three segments; following ventral cirri with inconspicuous cirrophores as basal swollen on ventral margin of neuropodia. Noto- and neuropodial lobes and chaetae absent on first two segments; neuropodial lobe on third segment papilliform, usually without chaetae; following segments with distinct neuropodial lobes. Anterior neuropodia with 5–8 supra-acicular and sub-acicular neurochaetae; mid- to posterior neuropodia with 25–30 supra-acicular and 20–25 sub-acicular neurochaetae, last 4–5 neuropodia as anterior ones; supra- and sub-acicular chaetae with same shape; chambered shafts and long blades; blades ca. 15 times longer than wide. Cutting edge of blades finely serrated, distally unidentate, distal teeth sharply curved with sub-distal enlarged prolongation, resembling an empty balloon. Pygidium small, with a pair of long pygidial cirri as long as last dorsal cirri.
Molecular identity: New COI and 16S sequences were deposited in GenBank under the accession numbers MH935085-MH935093 – MH935162-MH935168, respectively.
Distribution, Habitat, depth: NE Pacific, whale fall, 2893 m depth (Pleijel et al., 2008); SW Atlantic, whale falls, 1491–4204 m depth (Figure 2).
Remarks: The morphology of specimens from the Atlantic is consistent with the original description given by Pleijel et al. (2008) for the Pacific specimens. Therefore, we confirm that the main diagnostic characters of S. balaenophila comb. nov. are the presence of papilla-shaped on segment three, long dorsal cirri (mainly in the five first segments), sometimes exceeding the length of the body, three pairs of GLPs and presence of swelling-globular-shape cirrophores. These features are restricted for species comprising the S. balaenophila complex. Our results provide robust evidence that S. balaenophila species complex, as originally described, includes at least three cryptic species (see above), two of them described here. Two molecular distinct lineages (S. pirapuan sp. nov. and S. ypupiara sp. nov, see description below) were found living in the same localities (Figure 2). The cryptic species found in Pacific Ocean was not described here since we have not examined the specimens morphologically. However, Pleijel et al. (2008) found no morphological variation within Pacific specimens.
Intraspecific divergence was 1.1 and 0% for COI and 16S, respectively (Supplementary Table S2). The interspecific divergence among cryptic species of S. balaenophila comb. nov. ranged from 6.5–7.9% for COI and from 0.5–0.7% for 16S. Despite of that, the minimum interspecific COI divergence found was 13.1% compared to S. yokosuka sp. nov., and the maximum interspecific COI divergence found was 20.3% compared with S. methanicola (Supplementary Table S2). Interspecific 16S divergence ranged from 3.1 to 14.3% (Supplementary Table S2).
Sirsoe ypupiara sp. nov. urn:lsid:zoobank.org:act:4DAB33F7-CC20-4665-88B1-11B5AA9ABC43 (Figures 6B–D)
Material examined: Holotype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3357; GenBank: MH935119 and MH935179), incomplete 10 chaetigers, 10 mm length, 3.2 mm width; Paratype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3358; GenBank: MH935111 and MH935187), incomplete 12 chaetigers, 19 mm length, 3.2 mm width; Paratype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (ColBio DS00150; GenBank: MH935117 and MH935190), incomplete 11 chaetigers, 12 mm length, 2 mm width; One specimen on SEM stub from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3578).
Description: Morphologically identical to S. balaenophila. Frontal tubercle longer than wide with rounded anterior end (Figures 6B,C). The base of frontal tubercle wider than anterior end. Three pairs of glandular lip pads being the ventral pair wider than long while the dorsal and lateral ones as wide as long (Figures 6B,C). Papilliform neuropodial lobe on third segment present, without chaetae (Figure 6D). Mid-body neuropodia with approximately 35 supra-acicular and sub-acicular neurochaetae. Neurochaetae with chambered shafts and blades ca. 13 times longer than wide. Cutting edge of blades finely serrated, distally unidentate, distal teeth curved with sub-distal enlarged prolongation, resembling an empty balloon (Figure 6D).
Etymology: Ypupîara is a marine demon from the Tupi-Guarani mythology (Tupi-Guarani is the largest indigenous nation from Brazil before colonization), also called as seaman, whose muzzle was hairy and presented a caudal fin instead of legs.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935107-MH935121 and MH935179-MH935188/MH935190, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 3285-3358 m depth (Figure 2).
Remarks: See remarks of S. balaenophila comb. nov. Intraspecific divergence was 0.3 and 0.1% for COI and 16S, respectively (Supplementary Table S2). Divergence between S. ypupiara sp. nov. and S. balaenophila comb. nov. was 6.1 and 0.3% for COI and 16S, respectively. Among other species COI divergence range was 6.1–21.8%, while 16S range was 0.3–14.6% (Supplementary Table S2).
Sirsoe pirapuan sp. nov. urn:lsid:zoobank.org:act:A6AB0868-4211-40D8-B776-0A68F22EB4F9 (Figures 6E–H)
Material examined: Holotype from SW Atlantic, 21.450°S, 39.897°W, 3211 m (MZUSP 3355; GenBank: MH935102 and MH935189), incomplete, 12 chaetigers, 8 mm length, 1.8 mm width; Paratype from SW Atlantic, 21.450°S, 39.897°W, 3211 m (MZUSP 3356; GenBank: MH935096 and MH935178), incomplete, 7 chaetigers, 8 mm length, 2 mm width; Three specimens from SW Atlantic, 21.450°S, 39.897°W, 3211 m (ColBio DS00147); Two specimens from SW Atlantic, 21.450°S, 39.897°W, 1444 m (MZUSP 3573); One specimen on SEM stub from SW Atlantic, 21.450°S, 39.897°W, 1444 m (MZUSP 3577).
Description: Morphologically with diagnostic features of S. balaenophila. Three pairs of glandular lip pads being the ventral pair wider than long while the dorsal and lateral ones as wide as long (Figure 6E). Papilliform neuropodial lobe on third segment present, without chaetae (Figure 6F). Mid-body neuropodia withapproximately 30 supra-acicular and sub-acicular neurochaetae (Figure 6H). Neurochaetae with chambered shafts and blades ca. 17 times longer than wide. Cutting edge of blades finely serrated, distally unidentate, distal teeth curved with sub-distal enlarged prolongation, resembling an empty balloon (Figure 6G).
Etymology: The word “Pirapuã” means whale in Tupi language (language spoken by Tupi-Guarani people).
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935094-MH935106 and MH935169-MH935178/MH935189, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 1491–3322 m depth (Figure 2).
Remarks: See remarks of S. balaenophila comb. nov. Intraspecific divergence was 0.2 and 0% for COI and 16S, respectively (Supplementary Table S2). Divergence between S. pirapuan sp. nov. and S. balaenophila sp. nov. was 5.9 and 0.2% for COI and 16S, respectively. Interspecific COI divergence with other species ranged between 5.9 and 21.9%, while 16S divergence varied between 0.2 and 14.6% (Supplementary Table S2).
Sirsoe maximiano sp. nov. urn:lsid:zoobank.org:act:73406A47-82FB-4183-A274-402F4ED6622C (Figures 7A–E)
Material examined: Holotype from SW Atlantic, 25.894°S, 45.035°W, 1439 m (MZUSP 3371; GenBank: MH935151 and MH935157), incomplete 17 chaetigers, 6 mm length, 1 mm width;; Paratypes: two specimens from SW Atlantic, 25.894°S, 45.035°W, 1439 m (ColBio DS00152); Paratypes: three specimens from SW Atlantic, 28.029°S, 43.530°W, 3328 m (MZUSP 3372); Two specimens on SEM stub from SW Atlantic, 25.894°S, 45.035°W, 1439 m (MZUSP 3579 and MZUSP 3580).
Description: Body stout, highly tapering from mid- to posterior body (Figure 7A); preserved specimens whitish to pale yellowish. Prostomium slightly wider than long. Eyes absent. Paired small-depression-like nuchal organs posteriorly on prostomium (Figure 7C). Frontal tubercle restricted to small pad, with the margin slightly crenulated (Figures 7C,E). Palps biarticulated; palpophores cylindrical, short and robust; palpostyles conical, twice or more the length of palpophores. Lateral antennae cirriform, longer than palps; median antenna absent. Proboscis with ten filiform terminal papillae covered by dense ciliation among each papilla (Figure 7B). Glandular lip pads absent. First three segments totally fused, with enlarged dorsal cirri longer than the following ones; dorsal cirrophores cylindrical throughout the body, shorter in anterior and posterior regions. Ventral cirri slender, progressively more distally inserted on neuropodia in posterior region; cirri of segments 1–3 enlarged, more robust than in following segments, inserted in distinct cirrophores; following ventral cirri not exceed the length of neuropodia. Parapodia sub-birramous with notopodia reduced to the dorsal cirrophores and neuropodia well developed, with 2–3 noto- and neuroaciculae. Neuropodia and neurochaetae starting on segment 3, with long and tapering acicular lobule. Anterior and posterior neuropodia with 10–15 supra-acicular and 15–20 sub-acicular neurochaetae, mid-body with 25–28 supra-acicular and 15–20 sub-acicular neurochaetae; all with chambered shafts and long blades; supra-acicular chaetae from anterior and posterior region with blades ca. 8 times shorter than those blades from mid-body; blades of supra-acicular chaetae ca. 4 times the longer than subacicular ones, in anterior and posterior regions, from median region ca. 20 times longer. Neurochaetae shafts from midbody longer than those from anterior and posterior regions; cutting edges of all blades are unidentate, finely serrate with sub-distal prolongation like an arista (Figure 7D). Pygidium not observed.
Etymology: Named (noun in apposition case) in honor to R/V Almirante Maximiano, the Brazilian Navy vessel used to recover the BioSuOr project landers.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935148-MH935151 and MH935154-MH935157, respectively.
Distribution, Habitat, depth: Caribbean Sea, hydrothermal vents, Von Damm field, 2290 m depth (Plouviez et al., 2015); SW Atlantic, whale falls, 1508–3358 m depth (Figure 2).
Remarks: The new species, S. maximiano sp. nov., is characterized by the absence of a distinct frontal tubercle and by bear dorsal cirri longer on first two segments than following ones, features also found in S. methanicola. In addition, the phylogeny shows S. maximiano sp. nov. close related to S. sirikos. However, S. maximiano sp. nov. can be distinguished from S. methanicola and S. sirikos by the absence of a median antenna on the prostomium.
Intraspecific divergence was 0.9 and 0% for COI and 16S, respectively (Supplementary Table S2). Interspecific COI divergence ranged between 7.7 and 23.4%, while 16S divergence range was 0.7–13.4% (Supplementary Table S2). According the COI divergence between S. maximiano and Sirsoe ‘SP-2014’ (KJ566956), the distribution of the new species range from SW Atlantic, close to the Brazilian coast, up to the Caribbean Sea. Moreover, S. maximiano sp. nov. colonizes vents (in the Mid-Cayman Spreading Center, see Plouviez et al., 2015 refering as Sirsoe ‘SP-2014’) and also in whale falls (this study).
Sirsoe alphacrucis sp. nov. urn:lsid:zoobank.org:act:7992F3BC-08F6-4B9F-B608-C08E8807E268 (Figures 8A–F)
Material examined: Holotype from SW Atlantic, 25.894°S, 45.035°W, 1439 m (MZUSP 3363), complete 23 chaetigers, 4 mm length, 0.5 mm width; Paratype from SW Atlantic, 25.894°S, 45.035°W, 1439 m (MZUSP 3364; GenBank: MH935140 and MH935191), complete 19 chaetigers, 3.5 mm length, 0.3 mm width; Paratype from SW Atlantic, 25.894°S, 45.035°W, 1439 m (ColBio DS00156); Paratypes from SW Atlantic, 21.450°S, 39.897°W, 1444 m (eight specimens, MZUSP 3570), (two specimens, MZUSP 3571), (three specimens, MZUSP 3572); 19 specimens from SW Atlantic, 25.894°S, 45.035°W, 1439 m (MZUSP 3365); One specimen in SEM stub from SW Atlantic 25.894°S, 45.035°W, 1439 m (MZUSP 3581).
Description: Thin cylindrical body, usually not flattened. Preserved specimens with brownish pigmentation on anterior segments, concentrated laterally between prostomium and first segments, forming a mask or covering the entire prostomium. Prostomium as wide as long, nearly squared (Figures 8A,B). Eyes absent, even though sometimes the pigmentation on the prostomium resembles two big eyes. Nuchal organs as small-depression, posteriorly on prostomium. Frontal tubercle inconspicuous as mild swelling (Figure 8B). Palps biarticulated; palpophores robust and cylindrical, as wide as long, and slightly shorter than palpostyles; palpostyles conical (Figures 8A,B). Lateral antennae slender, cirriform to conical, thinner than palps; median antenna absent (Figures 8A,B). Glandular lip pads present; one dorsal and one ventro-lateral pairs, not easily distinguishable. Everted proboscis with ten cirriform terminal papillae, densely ciliated among each papilla (Figures 8A,B). First two segments totally fused, the third partially fused with the second. Dorsal cirri of the first two segments enlarged, longer and more robust than following ones. Ventral cirri enlarged on first two segments and inserted in small cirrophores, being the first ventral cirri longer than the second; following ventral cirri attached to the base of neuropodia, as long as or slightly longer than the acicular lobule. Parapodia subbirramous with notopodia reduced to dorsal cirrophores bearing 1–2 notoaciculae. First neuropodia on segment 3 (Figure 8A), 2–3 times smaller than second neuropodial, following segments with neuropodia about same size; neuropodia short with slender acicular lobule and 2–3 neuroaciculae. Anterior and posterior neuropodia with ∼7–9 supra-acicular and 5–7 sub-acicular neurochaetae (Figure 8C); all compound neurochaetae with shafts all chambered. Blades up to 10 times longer than wide, with about same length along the body (Figure 8D). Cutting edge of blades finely serrated, distally unidentate, curved, with a sub-distal spoon-like prolongation, distally inflated, projecting toward to the distal tooth (Figure 8E); under light microscopy the sub-distal prolongation seems a hood. A pair of pygidial cirri and dorsal pygidial papilla present (Figure 8F).
Etymology: In honor to the R/V Alpha-Crucis (noun in apposition case), the IO-USP vessel used to deploy the landers during the BioSuOr project. Alpha-Crucis is also the brightest star from Crux constellation and represent São Paulo State in the Brazilian flag.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935133-MH935140 and MH935191-MH935195, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 550–1508 m depth (Figure 2).
Remarks: The cylindrical shape of the body as well as the prostomium morphology, i.e., as wide as long and densely pigmented, are features that easily distinguished S. alphacrucis sp. nov. from all other Sirsoe species. The short blades of neurochaetae and the spoon-like sub-distal prolongation of chaetae are also only found in S. alphacrucis sp. nov. Moreover, S. alphacrucis sp. nov. has neuropodial lobe starting on segment 3 while in S. balaenophila complex, S. ketea, S. falenothiras, and S. ahabi starts on segment 4. Sirsoe alphacrucis sp. nov. is very close related to S. yokosuka sp. nov., being almost morphologically indistinguishable. Both species have neuropodial lobe starting on segment 3 (Figures 8A, 9C), bear ventral cirri as long as the acicular lobe along the body (Figures 8C, 9D), bear a pygidial papilla between the pygidial cirri (Figures 8F, 9E) and have blades of compound neurochaetae with a sub-distal spoon-like prolongation, distally inflated, projecting toward to the distal tooth (Figures 8E, 9F). Sirsoe yokosuka sp. nov. have darker pigmentation on prostomium in comparison with S. alphacrucis sp. nov. (see description below), even though some individuals can present the pigmentation as well as S. alphacrucis sp. nov. The shape of lateral antennae is also slightly different; in S. alphacrucis sp. nov. some specimens have a conical lateral antennae, while in S. yokosuka sp. nov. is always cirriform (Figures 8A,B, 9A). However, the interspecific divergence of COI and 16S can easily distinguish both species.
Intraspecific divergence was 0.2 and 1.0% for COI and 16S, respectively (Supplementary Table S2). Interspecific COI divergence ranged from 11.2 to 22.5%, while for 16S ranged from 2.4 to 16.7% (Supplementary Table S2).
Sirsoe yokosuka sp. nov. urn:lsid:zoobank.org:act:F58CBAF8-F227-43A7-AA62-87E590598A39 (Figures 9A–F)
Material examined: Holotype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3366), complete 19 chaetigers, 3.5 mm length, 0.4 mm width; Paratype from SW Atlantic, 22.841°S, 38.416°W, 3211 m (MZUSP 3367; GenBank: MH935141 and MH935196), incomplete 14 chaetigers, 4 mm length, 0.4 mm width; Paratypes: five specimens, SW Atlantic, 25.338°S, 39.642°W, 3227 m (ColBio DS00160); Three specimens from SW Atlantic, 28.519°S, 41.657°W, 4204 m (ColBio DS00161); Four specimens from SW Atlantic, 28.029°S, 43.530°W, 3328 m (MZUSP 3567); 13 specimens from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3568); Nine specimens from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3569). Three specimens on SEM stub from SW Atlantic, 28.519°S, 41.657°W, 4204 m (MZUSP 3575); One specimen in SEM stub from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3576).
Description: Thin cylindrical body, not flattened. Prostomium rounded slightly wider than long (Figure 9A), covered by a spread dark pigmentation. Palps biarticulated; palpophores robust and cylindrical, as wide as long, and slightly shorter than palpostyles; palpostyles conical (Figure 9A). Lateral antennae slender, conical, thinner than palps; median antenna absent (Figure 9A). Everted proboscis with ten cirriform terminal papillae, densely ciliated among each papilla; cilia long, exceeding the length of terminal papillae (Figure 9B). Other morphological features as S. alphacrucis sp. nov.
Etymology: In honor to R/V Yokosuka (noun in apposition case), the mothership vessel of the HOV Shinkai 6500 of JAMSTEC used in the Iata-Piúna expedition during the discovery of the natural whale-fall off Brazil.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935141–MH935144 and MH935196–MH935198, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 3322–4204 m depth (Figure 2).
Remarks: Although S. yokosuka sp. nov. is morphologically similar to S. alphacrucis sp. nov., the former has the pigmentation on prostomium always spread, sometimes the pigmentation is dark, while the latter has pigmentation usually concentrated in postero-lateral region, resembling a mask. In addition, the bathymetric distribution of both species is different; S. yokosuka sp. nov. was found at the lower slope and abyssal regions of SW Atlantic, whereas S. alphacrucis sp. nov. was only found at the upper slope.
Intraspecific divergences were 0.4 and 0% for COI and 16S, respectively (Supplementary Table S2). Interspecific COI divergence ranged between 11.2 and 23%, while for 16S ranged between 2.1 and 14.8% (Supplementary Table S2).
Sirsoe alucia sp. nov. urn:lsid:zoobank.org:act:91E91094-DF25-4F96-9AD8-F48212DA92BB (Figures 10A–E)
Material examined: Holotype from SW Atlantic, 26.600°S, 46.150°W, 550 m (MZUSP 3359; GenBank: MH935128 and MH935204), incomplete 14 chaetigers, 4 mm length, 1.3 mm width; Paratype from SW Atlantic, 26.600°S, 46.150°W, 550 m (MZUSP 3360; GenBank: MH935127 and MH935158), incomplete 12 chaetigers, 6 mm length, 2 mm width; Paratypes: Seven specimens from SW Atlantic, 26.600°S, 46.150°W, 550 m (ColBio DS00164); One specimen in SEM stub from SW Atlantic, 26.600°S, 46.150°W, 550 m (MZUSP 3582).
Description: Body stout, tapering from median to posterior region (Figure 10A); preserved specimens pale brownish to reddish on body and parapodia with darker coloration. Prostomium wider than long (Figure 10B), with region between prostomium and first segment densely pigmented, pigmentation brownish-red to purple (Figure 10A); prostomium basally forming a swollen ridge, extending laterally to the first parapodia (Figure 10C). Eyes absent. One pair of smalldepression- like nuchal organ posteriorly on the prostomium (Figure 10C). Frontal tubercle inconspicuous as small pad with crenulated margin (Figures 10B,C). Palps biarticulated; palpophores cylindrical; palpostyles conical, usually half of the length of palpophores (Figures 10B,C). Lateral antennae filiform and slightly longer than palps; median antenna absent (Figure 10C). Glandular lip pads absent. Proboscis with ten foliaceous terminal papillae (Figure 10D), covered by dense ciliation among each papilla; ciliation short, not exceeding the length of papillae (Figures 10B–D). First three segments fused. Dorsal cirri of first five segments longer than the following ones, usually exceeding half of the length of the body (Figure 10A). Enlarged ventral cirri on segments 1–3, with distinct cirrophore (Figure 10D); following segments with ventral cirri up to the length of acicular lobule; cirrophores of these ventral cirri as mild swellings, not easily distinguishable. Parapodia sub-biramous with notopodia reduced to the dorsal cirrophores and neuropodia well developed, with 3–4 noto- and neuroaciculae. Neuropodia and neurochaetae starting on segment 3, with long and tapered acicular lobule. Anterior and posterior neuropodia with 7–15 supra-acicular and 10–20 sub-acicular neurochaetae, mid-body neuropodia with 20–35 supra- and sub-acicular ones; neurochaetae dark, from grayish amber to black color; supra-acicular neurochaetae with long blades (up to 50 times longer than wide) (Figure 10E) and sub-acicular with short blades (usually less than 25 times longer than wide). The difference in the length of the supra-acicular and sub-acicular blades are more distinct in median region of the body. Cutting edges of all blades are finely serrated, distally unidentate, curved, with a thin sub-distal spine-like prolongation (Figure 10E). Pygidial cirri paired and terminal.
Etymology: In honor to R/V Alucia (noun in apposition case) a vessel used to recover the lander at the Santos Basin (Brazil) at 560 m depth.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935122–MH935128 and MH935158–MH935159/MH935204, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 550 m depth (Figure 2).
Remarks: The enlarged dorsal cirri in the first five segments and cylindrical palpophores forming a swollen ridge basally to the prostomium of S. alucia sp. nov. resemble those features of S. balaenophila complex species. However, the new species can be distinguished by the presence of foliaceous terminal papillae of proboscis, absence of GLPs and by the dark coloration of neurochaetae.
Intraspecific COI divergence was 0.05% (Supplementary Table S2). Interspecific COI divergence range between 20 and 23%, while 16S range between 5.5 and 12.6% (Supplementary Table S2).
Sirsoe nadir sp. nov. urn:lsid:zoobank.org:act:F264A645-D68C-442B-9D4DB0DBFA85735D (Figures 11A–C)
Material examined: Holotype from SW Atlantic, 26.600°S, 46.150°W, 550 m (MZUSP 3361), complete 25 chaetigers, 4.5 mm length, 1 mm width; Paratype from SW Atlantic, 26.600°S, 46.150°W, 550 m (MZUSP 3362; GenBank: MH935129 and MH935200), incomplete 19 chaetigers, 5 mm length, 1.3 mm width; Three specimen from SW Atlantic, 26.600°S, 46.150°W, 550 m (ColBio DS00167); One specimen in SEM stub from SW Atlantic, 26.600°S, 46.150°W, 550 m (MZUSP 3583).
Description: Body short and slender with no evidence of posterior tapering; preserved specimens pale brownish to yellowish. Prostomium at least twice wider than long (Figures 11A,B). Eyes absent. One pair of small depression-like nuchal organ posteriorly on the prostomium. Frontal tubercle present as distinct rounded prolongation. Palps biarticulated (Figures 11A,B); palpophores cylindrical, about same length of palpostyle; palpostyles conical. Lateral antennae cirriform about same length of palps; median antenna absent (Figures 11A,B). Two pairs of indistinct glandular lip pads; one dorsal and one lateral (Figure 11B). Proboscis with ten cirriform terminal papillae, covered by dense ciliation among each papilla (Figures 11B,C). First two segments fused. Dorsal cirri of first two segments longer than the following ones. Ventral cirri of first three segments inserted in distinct cirrophores and more robust than the following ones; from the fourth segment ventral cirri inserted on neuropodial lobe, not exceeding the length of neuropodia in anterior and mid-body; ventral cirri of posterior region exceed the length of neuropodia. Parapodia sub-birramous with notopodia reduced to dorsal cirrophores with 2–3 notoaciculae and neuropodia well developed starting on segment 3 (Figure 11A), bearing 3–4 neuroaciculae. Neuropodia with acicular lobule long and tapering, distally pointed, with a mid-neuroacicula. Neurochaetae with shafts chambered; 7–10 supra-acicular and 5–10 sub-acicular in anterior and posterior region; 12–15 supra-acicular and 15–20 sub-acicular in mid-body. Shafts of supra-acicular chaetae usually one third longer than shafts of sub-acicular ones; supra-acicular chaetae with blades 20–25 times longer than wide; sub-acicular ones 10–15 times longer than wide. Cutting edges of all blades finely serrate, distally unidentate, curved, with sub-distal spine-like prolongation; sub-distal ones projected like an arista to distal teeth (Figure 11D). Pygidium and pygidial cirri not observed.
Etymology: From the HOV Nadir (noun in apposition case) used to film the lander SP550 on the seafloor.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935129-MH935131 and MH935200-MH935202, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 550 m depth (Figure 2).
Remarks: Sirsoe nadir sp. nov. shares with S. sirikos, S. alphacrucis complex species and S. alphadelphini sp. nov., the presence of long dorsal cirri on segments 1–2. In contrast, the absence of median antenna in the new species distinguished it from S. sirikos. Members of S. alphacrucis complex have sub-distal inflated-like-a-spoon prolongation, distinguishing them from S. nadir sp. nov., which present a spine-like prolongation. S. nadir sp. nov. has supra-acicular neurochaetae 20–25 times longer than wide along the body, whereas in S. alphadelphini sp. nov. these chaetae are eight times longer than wide in anterior region.
Intraspecific divergences were 0.9 and 0% for COI and 16S, respectively (Supplementary Table S2). Interspecific COI divergence ranged between 17.5 and 22.5%, while 16S divergence ranged between 4.7 and 16.2% (Supplementary Table S2).
Sirsoe alphadelphini sp. nov. urn:lsid:zoobank.org:act:69616969-9477-40FA-949F-773EECA1E4E1 (Figures 12A,B)
Material examined: Holotype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3373), incomplete four chaetiger, 3 mm length, 1.8 mm width; Paratype from SW Atlantic, 25.338°S, 39.642°W, 3227 m (ColBio DS00168; GenBank: MH935132 and MH935203); Paratype from SW Atlantic, 21.450°S, 39.897°W, 1444 m (MZUSP 3574).
Description: Body slender and short, slightly flattened in anterior region, posterior region not observed. Preserved specimen whitish. Prostomium slightly wider than long (Figure 12A). Eyes absent. Nuchal organs as small-depression, positioned posteriorly on the prostomium. Frontal tubercle as a small pad, inconspicuous. Palps biarticulated; palpophores cylindrical, longer than palpostyles; palpostyles conical. Lateral antennae cirriform as long as palps; median antenna absent. Glandular lip pads absent. Proboscis with 11 cirriform terminal papillae, covered ciliation among each papilla (Figure 12B). First two segments fused. Dorsal cirri longer on first two segments. Ventral cirri enlarged on first three segments, inserted in cirrophores in segments 1–2; third ventral cirri not inserted on neuropodium but cirrophore not evident; following ventral cirri inserted on neuropodia. Parapodia sub-birramous with notopodia reduced to dorsal cirrophores bearing 1–2 notoaciculae and neuropodia well developed with 2–3 neuroaciculae. Neurochaetae with chambered shafts and long blades along the body, shafts from mid-body longer than those from anterior region; supra-acicular chaetae from anterior region with blades ca. 8 times shorter than those blades from middle body region; blades of supra-acicular chaetae from middle body region ca. 20 times longer than sub-acicular ones. Cutting edge of all blades finely serrated, distally unidentate, curved, with a sub-distal spine-like prolongation. Pygidium not observed.
Etymology: In honor to the R/V Alpha Delphini (noun in apposition case), the IO-USP vessel used to recover the lander at 1500 m depth off Brazil during the BioSuOr project.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935132 and MH935203, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 3285 m depth (Figure 2).
Remarks: S. alphadelphini sp. nov. shares the presence of dorsal cirri elongated on the first two segments with S. alphacrucis sp. nov., S. yokosuka sp. nov. and S. sirikos. However, S. alphadelphini sp. nov. can be distinguished from these species as it has blades of neurochaetae ca. 20 times longer than wide, while S. alphacrucis sp. nov. and S. yokosuka sp. nov. both have short blades (up to 10 times longer than wide). Moreover, the sub-distal prolongation of the blades is simple in S. alphadelphini sp. nov. while in S. alphacrucis sp. nov. and S. yokosuka sp. nov. is inflated resembling a spoon. The presence of median antenna in S. sirikos can distinguishes it from S. alphadelphini sp. nov.
Minimum average interspecific COI divergence was 16.1% for Sirsoe ‘Ze50,’ while the maximum divergence was 23.8% for S. methanicola (Supplementary Table S2). Interspecific 16S divergence range was 2.1–11.6%, with S. yokosuka and S. maximiano, respectively (Supplementary Table S2).
Sirsoe ungava sp. nov. urn:lsid:zoobank.org:act:0D1137E6-B44A-476E-BFAE-4A52EF0A9B99 (Figures 12C–E)
Material examined: Holotype from SW Atlantic, 21.450°S, 39.897°W, 1444 m (MZUSP 3368), complete 30 chaetigers, 9.3 mm length, 1.1 mm width, without chaetigers; Paratype from SW Atlantic, 28.029°S, 43.530°W, 3328 m (MZUSP 3369; GenBank: MH935145 and MH935160), incomplete 12 chaetigers, 2.7 mm length, 1.0 mm width, without chaetigers; Paratype from SW Atlantic, 21.450°S, 39.897°W, 1444 m (ColBio DS00170), incomplete 5 segments, 2.1 mm length and 1.0 mm width, without chaetigers; Paratype from SW Atlantic, 21.450°S, 39.897°W, 1444 m (ColBio DS00171).
Description: Body stout and cylindrical, tapering from mid-body to posterior region (Figures 12C,D); preserved specimens yellowish. Prostomium wider than long (Figure 12D). Eyes absent. One pair of small-depression-like nuchal organ, posteriorly on the prostomium. Frontal tubercle distinct as rounded triangular prolongation (Figures 12C,D). Palps biarticulated; palpophores cylindrical slightly shorter or as long as palpostyles; palpostyles conical. Lateral antennae filiform slightly longer than palps (Figure 11C); median antenna absent. Glandular lip pads absent. Proboscis with ten minute terminal papillae, not densely ciliated among each papilla (Figures 12C,E). First two segments fused. Dorsal cirri of first three segments more robust than the following ones; third dorsal cirri twice longer than the previous ones. Ventral cirri inserted in distinct cirrophores on first three segments. Following ventral cirri inserted from mid to distal end of neuropodial lobe and slightly longer than acicular lobule. Parapodia sub-biramous with notopodia reduced to dorsal cirrophores and neuropodia well developed with 1–2 noto- and neuroaciculae. Neuropodia and neurochaetae starting on third segment (Figure 12C); neuropodial lobe short and stout with pointed acicular lobule. Each neuropodia with 10–15 supra-acicular and 5–10 subacicular neurochaetae along the body. Pygidium simple, pygidial cirri not observed.
Etymology: In honor to R/V Ungava (noun in apposition case), the first Brazilian research vessel from IO-USP.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935145 and MH935160, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 1491–3358 m depth (Figure 2).
Remarks: The new species, S. ungava, is close to S. ketea and S. falaenothiras, according to the phylogenetic tree and the divergence of COI and 16S (Figure 3 and Supplementary Table S2). Sirsoe ungava sp. nov. can be distinguished from S. ketea and S. falaenothiras by the lack of a median antenna. Moreover, S. ungava sp. nov. has a ring of 10 min terminal papillae on proboscis, while S. ketea proboscis is smooth. The proboscis morphology of S. falaenothiras is unknown (Summers et al., 2015). Interspecific divergence range between 10.7–23% and 4–16.4% for COI and 16S, respectively (Supplementary Table S2).
Sirsoe besnard sp. nov. urn:lsid:zoobank.org:act:D1272F00-3CF6-4818-BE71-B5617FE75FF4 (Figures 13A–C)
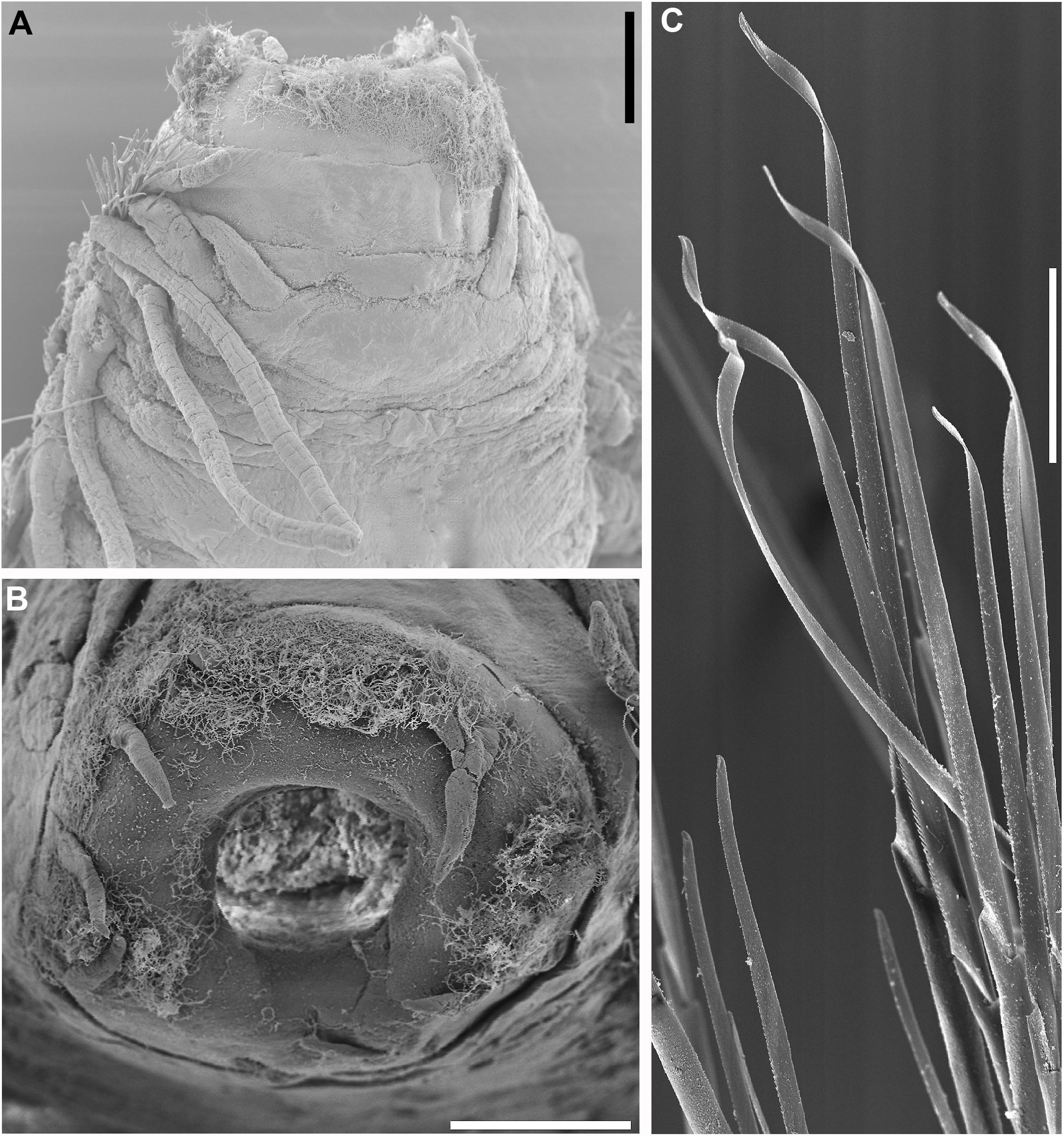
Figure 13. Sirsoe besnard sp. nov. (A) anterior end, dorsal view; (B) detail of proboscis; (C) neurochaetae from median neuropodia. (A,B) 200 μm; (C) 50 μm.
Material examined: Holotype in SEM stub from SW Atlantic, 25.338°S, 39.642°W, 3227 m (MZUSP 3370; GenBank: MH935147 and MH935199).
Description: Body stout, short and cylindrical, not tapered; preserved specimen pale yellowish. Prostomium wider than long (Figure 13A). Eyes absent. Nuchal organs as small-depression, positioned posteriorly on the prostomium. Frontal tubercle present, but inconspicuous (Figure 13A). Palps biarticulated; palpophores cylindrical longer than palpostyles; palpostyles conical. Lateral antennae cirriform slightly longer than palps; median antenna absent. Glandular lip pads absent. Proboscis with ten filiform terminal papillae, covered by ciliation among each papilla (Figure 13B). First two segments fused. Dorsal cirri of segments 1–2 longer than the following ones. Ventral cirri from segments 1–3 inserted in distinct cirrophores longer than the followings ones. Parapodia sub-biramous with notopodia reduced to dorsal cirrophores and neuropodia well developed starting on segment 3 bearing 1–2 noto- and neuroaciculae. Neuropodia with slender acicular lobule and 15–20 supra-acicula and sub-acicular neurochaetae; blades of neurochaetae usually 20–25 times longer than wide. Cutting edge of all blades basally finely serrated and distally smooth and unidentate, tooth curved, with a sub-distal spine-like prolongation (Figure 13C). Pygidium not observed.
Etymology: In honor to R/V Prof. W. Besnard (noun in apposition case) a vessel from IO-USP. The name of the vessel was after Dr. Wladimir Besnard, the first director of IO-USP and considered to be the father of the Brazilian oceanography.
Molecular identity: COI and 16S sequences were deposited in GenBank under the accession numbers MH935147 and MH935199, respectively.
Distribution, Habitat, depth: SW Atlantic, whale falls, 3285 m depth (Figure 2).
Remarks: Sirsoe besnard sp. nov. is very similar to S. alphadelphini and S. nadir, however can be distinguished by the blades of neurochaetae. S. besnard sp. nov. has neurochaetae blades almost smooth in the distal end, while in S. alphadelphini sp. nov. and S. nadir sp. nov. the blades are finely serrated in the distal end. The length of neurochaetae of S. besnard sp. nov. is uniform along the body and between supra- and sub-acicular bunches, while in S. alphadelphini sp. nov. the supra-acicular midbody neurochaetae are the longest ones and in S. nadir sp. nov. supra-acicular neurochaetae are longer than sub-acicular ones. Interspecific COI divergence range was 16.1–22.5%, while 16S divergence range was 4–16.1% (Supplementary Table S2).
Discussion
The species richness of deep-sea whale falls is much higher than as previously thought as evidenced here by the discovery and description of 10 new species of Sirsoe. Moreover, we reported for the very first time the interbasin distribution of Sirsoe balaenophila comb. nov. and Sirsoe sirikos (SW Atlantic and NE Pacific). We also found one new lineage, Sirsoe “BioSuOr,” not described here, summing up to now 13 Sirsoe species in the deep SW Atlantic Ocean. The majority of deep-sea hesionids have been reported at energy-rich habitats including hydrothermal vents and whale falls (Summers et al., 2015). Sirsoe has only been found at deep-sea chemosynthetic-based ecosystems. Summers et al. (2015) showed that the ancestors of Sirsoe inhabited the deep sea and subsequently diversified in high-energy habitats. Our results show that a high percentage (∼80%) of Sirsoe species are associated with whale falls (Figures 2, 3) suggesting that this environment could be a hotspot for the diversification of this genus.
Some annelid taxa are particularly abundant and diverse at whale falls, such as dorvilleids, ampharetids and the bone-eating worms Osedax (Vrijenhoek et al., 2009; Wiklund et al., 2012; Ravara et al., 2015; Eilertsen et al., 2017). The two former clades, also found in chemosynthetic-based ecosystems, demonstrate tolerance to toxic compounds and show trophic niche partitioning between species and genus, which it has been hypothesized to promote their high species diversity (Thornhill et al., 2012; Levin et al., 2013; Eilertsen et al., 2017). Alfaro-Lucas et al. (2018) showed trophic niche partitioning between some hesionid species in a SW Atlantic whale fall. These authors suggested that Sirsoe sirikos and ‘Vrijenhoekia’ sp. are probably omnivores/scavengers in whale falls, whereas another hesionid genus, Pleijelius, relies on chemosynthetic bacterial mats. The high diversity of Sirsoe in whale falls could also be related with trophic specialization avoiding comfamilial competition. Further studies are necessary on the trophic ecology of Sirsoe, including more species and also from different environments.
The distributions of Sirsoe balaenophila comb. nov. and Sirsoe sirikos were extended to the Atlantic Ocean. Both species were previously found only in whale falls from the deep NE Pacific Ocean (Pleijel et al., 2008; Summers et al., 2015). The interbasin distribution of these species supports the hypothesis that some whale-fall fauna may have a worldwide distribution (Sumida et al., 2016). We found limited COI divergence (∼1%) between Atlantic and Pacific individuals and COI haplotypes of different populations of S. balaenophila ‘stricto sensu’ were separated by a maximum of seven mutational steps. However, no shared haplotypes were found between both populations. A few whale-fall specialists have a widespread distribution, such as the bone-eating worm Osedax rubiplummus, reported in both sides of the Pacific and in the Southern Ocean (Smith et al., 2015). Moreover, five additional Osedax species show a trans-Pacific distribution (Rouse et al., 2018b). Studies showed that Osedax larval lifespan is about ∼10–20 days (Rouse et al., 2009; Miyamoto et al., 2013), suggesting that the dispersion from margin to margin of the Pacific Ocean in one generation is not feasible. Accordingly, Rouse et al. (2018b) hypothesized an abundant supply of adequate habitats facilitating a stepping-stone dispersal of these bone-eating worms. There is no information available regarding the larva of S. balaenophila. However, a close related species, S. methanicola, produces planktotrophic larvae with dispersal phases of ∼21 days (Fisher et al., 2000; Eckelbarger et al., 2001) and presents maximum dispersion distances of 300 km (Young et al., 2012). The distance between SW Atlantic and NE Pacific S. balaenophila populations is more than 15,000 km, supposing that the possible route to disperse is along the SE Pacific margin. In this way, intermediate whale falls along SE Pacific could connect SW Atlantic and NE Pacific populations of S. balaenophila.
Molecular data also suggest that S. balaenophila (as originally morphologically described) hosts two cryptic species in the Atlantic (S. pirapuan sp. nov. and S. ypupiara sp. nov.) and possibly more in the Pacific (e.g., S. balaenophila ‘lineage-2’). High interspecific genetic divergences within the S. balaenophila species complex observed here (6–8% and 0.3–0.7% for COI and 16S, respectively) agree with previous findings (Pleijel et al., 2008; Summers et al., 2015). The absence of morphological differences and the low ITS1 divergence were used to maintain S. balaenophila as a single species (Pleijel et al., 2008; Summers et al., 2015). Species complex can have low ITS1 divergence, as observed in Archinome species (Borda et al., 2013). Moreover, 16S gene data split S. balaenophila into four distinct lineages, although with low divergence among them. Notably, 16S tends to have slower rates of substitution than other mtDNA genes (Thornhill et al., 2012; Brasier et al., 2016).
The intraspecific COI divergence for S. balaenophila ‘stricto sensu’ (see Pleijel et al., 2008) was about 10 times less, even with the inclusion of specimens from the Atlantic, than the divergence among the cryptic complex (S. ypupiara sp. nov. and S. pirapuan sp. nov.). However, several studies have reported lower COI divergence within cryptic species (between 5 and 15%), as in the case of Syllis gracilis species complex in the N Atlantic and the Mediterranean Sea (Álvarez-Campos et al., 2017), Archinome cryptic species in chemosynthetic-based ecosystems (Borda et al., 2013), sympatric lineages of Eumida sanguinea species complex (Nygren and Pleijel, 2011), and Notophyllum foliosum cryptic species separated by depth (Nygren et al., 2010). Thus COI divergence found here may well highlight the occurrence of cryptic species within both Pacific and Atlantic S. balaenophila populations.
The sister relationship presented between S. alphacrucis sp. nov. and S. yokosuka sp. nov. may be an example of cryptic species isolated by depth, or allopatric speciation. The former species was found only in the upper slope, 550–1500 m, while the latter was found in the abyssal region (∼3300–4204 m depth) (Figure 2). More studies should be conducted to better understand the bathymetric isolation between these two species. However, depth has been advocated as one of the main drivers of speciation for deep-sea organisms, including many annelids (Zardus et al., 2006; Schüller, 2011; Cowart et al., 2014; Kobayashi et al., 2018). The different water-column masses found in the SW Atlantic could play an important role for larval dispersal leading bathymetric isolation. The SW Atlantic, between 20° and 40° S latitude, is characterized by the southward flowing of North Atlantic Deep Water (NADW) around 1500 m depth, while in the region close to 3300 m depth the Antarctic Deep Water (AABW) flows northward (De Madron and Weatherly, 1994; Stramma and England, 1999; Silveira et al., 2000). Although in areas further north within our sampled region (ES3300 and RJ3300, where S. yokosuka sp. nov. was found) exists a mixing zone of the AABW and the NADW, potentially connecting both bathymetric zones (Saeedi et al., 2019), each water mass and its flowing direction could confine the larval pool in each depth explaining the isolation of these Sirsoe species.
The presence of Sirsoe maximiano sp. nov. in the SW Atlantic whale falls, which was previously found in vents from the Mid-Cayman Spreading Center (as Sirsoe SP-2014 in Genbank, Accession number KJ566956), is consistent with the hypothesis of whale falls acting as stepping stones for dispersion of some vent and seep fauna (Smith et al., 1989; Smith and Baco, 2003). This hypothesis has been recently under debate (Kiel, 2016, 2017; Smith et al., 2017). According to Kiel (2016) whale falls were not likely to act as intermediate habitats for chemosynthetic mollusks inhabiting vents and seeps, although organic falls likely serve as stepping-stones for some annelids (Kiel, 2017; Smith et al., 2017). Vertebrate carcasses support stepping-stone dispersal mechanism for some organisms from other deep-sea reducing habitats (Sumida et al., 2016; Rouse et al., 2018b, this study). There is substantial evidence that whale falls provide intermediate habitats for some species to disperse between and among vents, seeps and other organic falls (Naganuma et al., 1996; Baco et al., 1999; Glover et al., 2005; Lundsten et al., 2010a; Amon et al., 2013; Hilário et al., 2015; Ravara et al., 2015; Smith et al., 2015, 2017; Sumida et al., 2016). For example, some siboglinid annelids, such as Escarpia spicata, Lamellibrachia satsuma and L. barhami and also the alvinocaridid shrimp Alvinocaris muricola, are species typical of vents and/or seeps but have been also found at whale falls (Fujiwara et al., 2007; Lundsten et al., 2010a; Teixeira et al., 2013).
In summary, this study showed that presence/absence of median antenna, GLPs and the first segment where the neuropodial lobe starts are not useful characteristics to distinguish Sirsoe and Vrijenhoekia. For this reason we conclude that Vrijenhoekia should be synonymized to Sirsoe. All Sirsoe species bear a reduced small depression-like nuchal organ (in comparison with other Psamathinae hesionids) and a frontal tubercle on the prostomium, which can be robust or reduced. The re-description of Hesiocaeca bermudensis is necessary to include the genus Hesiocaeca in the analysis and to clarify the relationship between Sirsoe and Hesiocaeca. From the 10 new described Sirsoe species we observed that, up to date, most of Sirsoe species inhabit deep-sea whale fall habitats. Moreover, we report the interbasin distribution of two species, S. sirikos and S. balaenophila comb. nov., occurring at the NE Pacific and SW Atlantic Ocean. There are probably many unknown populations to be discovered of both species connecting the NE Pacific and SW Atlantic populations, and future studies covering a wider geographical range, mainly along the SE Pacific margin, are needed to better understand the distribution of these species.
Sampling and Field Studies
All necessary permits for sampling were obtained by the authors from the competent authorities.
Data Availability
The datasets generated for this study can be found in the GenBank.
Author Contributions
MS and PS designed the study. MS, PS, and JA-L collected samples and did initial morphological examination with AR. MS conducted the extraction of DNA and edited sequences and alignment. MS and KH performed the molecular phylogeny. MS, PS, and OC took pictures under light microscopy and SEM. MS prepared the first draft. MS, PS, OC, and AR contributed to final description of new species. All authors contributed to species descriptions and the final version of the manuscript.
Funding
This study was funded by the São Paulo Research Foundation (FAPESP) into the Research Program on Biodiversity Characterization, Conservation, Restoration and Sustainable Use (BIOTA Program) – Grant number 2011/50185-1 to PS. MS was supported by CAPES/Proex Ph.D. scholarship and CAPES/PDSE doctoral internship at Auburn University. KH was supported by NSF/OCE-1155188.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Masters and crews of R/V Alpha-Crucis and R/V Alpha Delphini from IOUSP during the deployment and the recovering of the SP-1500 lander. We are also indebted to the Brazilian Navy and the Master and crew of PRV Almirante Maximiano and M. V. Alucia for the help during the recovering of the other landers. We are deeply grateful to Dr. H. Kitazato and the Master and crew of R/V Yokosuka and HOV Shinkai-6500 for the Iatá-Piuna expedition in which PS collected specimens of S. sirikos and S. yokosuka. We would like to thanks to Prof. M. M. de Mahiques by the invitation to PS and MS to participate in the Talude Expedition on board of R/V Alpha-Crucis, during this cruise was deployed the lander SP550. MS thanks Karen Osborn for providing the material of S. grasslei, S. hessleri, and S. methanicola for comparison, and also thanks Antônio J. M. Peixoto for taken the SEM pictures of these species. MS was supported by CAPES/Proex Ph.D. scholarship and by a CAPES/PDSE 88881.133024/2016-01 for a doctoral internship at Auburn University. KH was supported by NSF/OCE-1155188. This work was funded by BIOTA-FAPESP grant number 2011/50185-1 to PS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00478/full#supplementary-material
FIGURE S1 | Phylogenetic relationship of Sirsoe based on maximum likelihood (ML) analysis of: (A) COI and (B) 16S datasets. Values before slash are bootstrap supports of ML (only >50%) and after are posterior probabilities of Bayesian inference. Asterisks indicate 100% of support for ML and BI; ‘-‘ indicates node absent on BI.
TABLE S1 | Locality, depth and GenBank accession number of sequences employed in this study.
TABLE S2 | Average of uncorrected genetic distance (p-distance) among Sirsoe species. Values above the diagonal are interspecific 16S distance and below are COI distance. Diagonal values (in bold) are intraspecific distances; top is 16S and inferior is COI. Dark red and green represent high values and light colors low values.
TABLE S3 | List of valid Sirsoe species up to date, including type locality, other occurrences, depth range (in meters) and type of chemosynthetic-based ecosystem used by each species.
Footnotes
References
Alfaro-Lucas, J. M., Shimabukuro, M., Ferreira, G. D., Kitazato, H., Fujiwara, Y., and Sumida, P. Y. G. (2017). Bone-eating Osedax worms (Annelida: Siboglinidae) regulate biodiversity of deep-sea whale-fall communities. Deep Sea Res. II 146, 4–12. doi: 10.1016/j.dsr2.2017.04.011
Alfaro-Lucas, J. M., Shimabukuro, M., Ogata, I. V., Fujiwara, Y., and Sumida, P. Y. G. (2018). Trophic structure and chemosynthesis contributions to heterotrophic fauna inhabiting an abyssal whale carcass. Mar. Ecol. Prog. Ser. 596, 1–12. doi: 10.3354/meps12617
Álvarez-Campos, P., Giribet, G., and Riesgo, A. (2017). The Syllis gracilis species complex: a molecular approach to a difficult taxonomic problem (Annelida, Syllidae). Mol. Phylogenet. Evol. 109, 138–150. doi: 10.1016/j.ympev.2016.12.036
Amon, D. J., Glover, A. G., Wiklund, H., Marsh, L., Linse, K., Rogers, A. D., et al. (2013). The discovery of a natural whale fall in the Antarctic deep sea. Deep Sea Res. II 92, 87–96. doi: 10.1038/srep22139
Baco, A. R., and Smith, C. R. (2003). High species richness in deep-sea chemoautotrophic whale skeleton communities. Mar. Ecol. Prog. Ser. 260, 106–114.
Baco, A. R., Smith, C. R., Peek, A. S., Roderick, G. K., and Vrijenhoek, R. C. (1999). The phylogenetic relationships of the whale-fall vesicomyid clams based on mitochondrial COI DNA sequences. Mar. Ecol. Prog. Ser. 182, 205–223.
Blake, J. A. (1985). Polychaeta from the vicinity of the deep-sea geothermal vents in the eastern Pacific—I: Euphrosinidae, Phyllodocidae, Hesionidae, Nereididae, Glyceridae, Dorvilleidae, Orbiniidae, and Maldanidae. Biol. Soc. Wash. Bull. 6, 67–101.
Blake, J. A. (1991). A new species of Hesiocaeca (Polychaeta: Hesionidae) from hydrothermal vents at the mariana back-arc basin with notes on other polychaetes. Proc. Biol. Soc. Wash. 104, 175–180.
Borda, E., Kudenov, J. D., Chevaldonné, P., Blake, J. A., Desbruyères, D., Fabri, M. C., et al. (2013). Cryptic species of Archinome (Annelida: Amphinomida) from vents and seeps. Proc. Biol. Sci. 280:20131876. doi: 10.1098/rspb.2013.1876
Braby, C. E., Rouse, G. W., Johnson, S. B., Jones, W. J., and Vrijenhoek, R. C. (2007). Bathymetric and temporal variation among Osedax boneworms and associated megafauna on whale-falls in Monterey Bay, California. Deep Sea Res. I 54, 1773–1791. doi: 10.1016/j.dsr.2007.05.014
Brasier, M. J., Wiklund, H., Neal, L., Jeffreys, R., Linse, K., Ruhl, H., et al. (2016). DNA barcoding uncovers cryptic diversity in 50% of deep-sea Antarctic polychaetes. R. Soc. Open Sci. 3:160432. doi: 10.1098/rsos.160432
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Cowart, D. A., Halanych, K. M., Schaeffer, S. W., and Fisher, C. R. (2014). Depth-dependent gene flow in Gulf of Mexico cold seep Lamellibrachia tubeworms (Annelida, Siboglinidae). Hydrobiologia 736, 139–154. doi: 10.1007/s10750-014-1900-y
De Madron, X. D., and Weatherly, G. (1994). Circulation, transport and bottom boundary layers of the deep currents in Brazil Basin. J. Mar. Res. 52, 583–638. doi: 10.1357/0022240943076975
Desbruyères, D., and Toulmond, A. (1998). A new species of hesionid worm, Hesiocaeca methanicola sp. nov. (Polychaeta: Hesionidae), living in ice-like methane hydrates in the deep Gulf of Mexico. Cah. Biol. Mar. 39, 93–98.
Distel, D. L., Baco, A. R., Chuang, E., Morrill, W., Cavanaugh, C., and Smith, C. R. (2000). Marine ecology: do mussels take wooden steps to deep-sea vents? Nature 403, 725–726. doi: 10.1038/35001667
Eckelbarger, K. J., Young, C. M., Llodra, E. R., Brooke, S., and Tyler, P. (2001). Gametogenesis, spawning behavior, and early development in the “iceworm” Hesiocaeca methanicola (Polychaeta: Hesionidae) from methane hydrates in the Gulf of Mexico. Mar. Biol. 138, 761–775. doi: 10.1007/s002270000510
Ehlers, E. H. (1864). Die Borstenwürmer (Annelida Chaetopoda) Nach Systematischen Und Anatomischen Untersuchungen Dargestellt, Leipzig: Wilhelm Engelmann, 269–748.
Eilertsen, M. H., Kongsrud, J. A., Alvestad, T., Stiller, J., Rouse, G. W., and Rapp, H. T. (2017). Do ampharetids take sedimented steps between vents and seeps? Phylogeny and habitat-use of Ampharetidae (Annelida: Terebelliformia) in chemosynthesis-based ecosystems. BMC Evol. Biol. 17:222. doi: 10.1186/s12862-017-1065-1
Fisher, C. R., MacDonald, I. R., Sassen, R., Young, C. M., Macko, S. A., Hourdez, S., et al. (2000). Methane ice worms: Hesiocaeca methanicola colonizing fossil fuel reserves. Naturwissenschaften 87, 184–187. doi: 10.1007/s001140050700
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers from aplication of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotecnol. 3, 294–299.
Fujiwara, Y., Kawato, M., Yamamoto, T., Yamanaka, T., Sato-Okoshi, W., Noda, C., et al. (2007). Three-year investigations into sperm whale-fall ecosytems in Japan. Mar. Ecol. 28, 219–232. doi: 10.1111/j.1439-0485.2007.00150.x
Glover, A. G., Goetze, E., Dahlgren, T. G., and Smith, C. R. (2005). Morphology, reproductive biology and genetic structure of the whale-fall and hydrothermal vent specialist, Bathykurila guaymasensis (Annelida: Polynoidae). Mar. Ecol. 26, 223–234. doi: 10.1111/j.1439-0485.2005.00060.x
Grube, A. E. (1855). Beschreibung neuer oder wenig bekannter anneliden. Archiv. für Naturgeschichte 21, 81–136. doi: 10.5962/bhl.part.13989
Hartman, O. (1965). Deep-water benthic polychaetous annelids off New England to Bermuda and other North Atlantic areas. Occas. Paper Allan Hancock Fdn. Publ. 28, 1–378.
Hilário, A., Cunha, M. R., Génio, L., Marçal, A. R., Ravara, A., Rodrigues, C. F., et al. (2015). First clues on the ecology of whale falls in the deep Atlantic Ocean: results from an experimente using cow carcasses. Mar. Ecol. 36, 82–90. doi: 10.1111/maec.12246
International Commission on Zoological Nomenclature [ICZN] (1999). International Code of Zoological Nomenclature, 4th Edn. London: International Trust dor Zoological Nomemnclature, 306.
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method dor rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kiel, S. (2016). A biogeographic network reveals evolutionary links between deep-sea hydrothermal vent and methane seep faunas. Proc. Biol. Sci. 83:20162337. doi: 10.1098/rspb.2016.2337
Kiel, S. (2017). Reply to smith et al.: network analysis reveals connectivity patterns in the continuum of reducing ecosystems. Proc. Biol. Sci. 284:20171644. doi: 10.1098/rspb.2017.1644
Kobayashi, G., Goto, R., Takano, T., and Kojima, S. (2018). Molecular phylogeny of Maldanidae (Annelida): multiple losses of tube-capping plates and evolutionary shifts in habitat depth. Mol. Phylog. Evol. 127, 332–344. doi: 10.1016/j.ympev.2018.04.036
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2016). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Leigh, J. W., and Bryant, D. (2015). PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Levin, L. A., Ziebis, W., Mendonza, G. F., Bertics, V. J., Washington, T., Gonzalez, J., et al. (2013). Ecological release and niche partitioning under stress: lessons from dorvilleid polychaetes in sulfidic sediments at methane seeps. Deep Sea Res. II 92, 214–233. doi: 10.016/j/dsr2/2013/01/006
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lorion, J., Duperron, S., Gros, O., Cruaud, C., and Samadi, S. (2009). Several deep-sea mussels and their associated symbionts are able to live both on wood and on whale falls. Proc. Biol. Sci. 276, 177–185. doi: 10.1098/rspb.2008.1101
Lorion, J., Kiel, S., Faure, B., Kawato, M., Ho, S. Y. W., Marshall, B., et al. (2013). Adaptive radiation of chemosymbiotic deep-sea mussels. Proc. Biol. Sci. 280:20131243. doi: 10.1098/rspb.2013.1243
Lundsten, L., Paull, C. K., Schlining, K. L., McGann, M., and Ussler, W. III (2010a). Biological characterization of a whale-fall near Vancouver Island, British Columbia, Canada. Deep Sea Res. I 57, 918–922. doi: 10.1016/j.dsr.2010.04.006
Lundsten, L., Schlining, K. L., Frasier, K., Johnson, S. B., Kuhnz, L. A., Harvey, J. B. J., et al. (2010b). Times-series analysis of six whale-fall communities in Monterey Canyon, California, USA. Deep Sea Res. I 57, 1573–1584. doi: 10.1016/j.dsr.2010.09.003
Mahiques, M. M., Schattner, U., Lazar, M., Sumida, P. Y. G., and Souza, L. A. P. (2017). An extensive pockmark field on the upper Atlantic margin of Southeast Brazil: spatial analysis and its relationship with salt diapirism. Heliyon 3:e00257. doi: 10.1016/j.heliyon.2017.e00257
Miyamoto, N., Yamamoto, T., Yusa, Y., and Fujiwara, Y. (2013). Postembryonic development of the bone-eating worm Osedax japonicus. Naturwissenschaften 100, 285–289. doi: 10.1007/s00114-013-1024-7
Miyazaki, J.-I., de oliveira, M. L., Fujita, Y., Matsumoto, H., and Fujiwara, Y. (2010). Evolutionary process of deep-sea Bathymodiolus mussels. PLoS One 5:e10363. doi: 10.1371/journal.pone.0010363
Naganuma, T., Wada, H., and Fujioka, K. (1996). Biological community and sediment fatty acids associated with the deep-sea whale skeleton at the Torishima seamount. J. Oceanogr. 52, 1–15. doi: 10.1007/bf02236529
Nygren, A., Eklöf, J., and Pleijel, F. (2010). Cryptic species of Notophyllum (Polychaeta: Phyllodocidae) in Scandinavian waters. Org. Divers. Evol. 10, 193–204. doi: 10.1007/s13127-010-0014-2
Nygren, A., and Pleijel, F. (2011). From one to ten in a single stroke - resolving the European Eumida sanguinea (Phyllodocidae, Annelida) species complex. Mol. Phylog. Evol. 58, 132–141. doi: 10.1016/j.ympev.2010.10.010
Palumbi, S. R., Martin, A. P., Romano, S., McMillan, W. O., Stice, L., and Grabowski, G. (1991). The Simple Fool’s Guide to PCR. Honolulu, HI: Department of Zoology Special Publication, University of Hawaii.
Pleijel, F. (1998). Phylogeny and classification of Hesionidae (Polychaeta). Zool. Scr. 27, 89–163. doi: 10.1111/j.1463-6409.1998.tb00433.x
Pleijel, F. (2004). A revision of Hesiospina (Psamathini, Hesionidae, Polychaeta). J. Nat. Hist. 38, 2547–2566. doi: 10.1080/00222930310001647433
Pleijel, F., and Rouse, G. (2005). A revision of Micropodarke (Psamathini, Hesionidae, Polychaeta). J. Nat. Hist. 39, 1313–1326. doi: 10.1080/00222930400020129
Pleijel, F., Rouse, G. W., and Nygren, A. (2012). A revision of Nereimyra (Psamathini, Hesionidae, Aciculata, Annelida). Zool. J. Linn. Soc. 164, 36–51. doi: 10.1111/j.1096-3642.2011.00756.x
Pleijel, F., Rouse, G. W., Ruta, C., Wiklund, H., and Nygren, A. (2008). Vrijenhoekia balaenophila, a new hesionid polychaete from a whale fall off California. Zool. J. Linn. Soc. 152, 625–634. doi: 10.1111/j.1096-3642.2007.00360.x
Plouviez, S., Jacobson, A., Wu, M., and Van Dover, C. L. (2015). Characterization of vent fauna at the mid-cayman spreading center. Deep Sea Res. I 97, 124–133. doi: 10.1016/j.dsr.2014.11.011
Ravara, A., Marçal, A. R., Wiklund, H., and Hilário, A. (2015). First account on the diversity of Ophryotrocha (Annelida, Dorvilleidae) from a mammal-fall in the deep-Atlantic Ocean with the description of three new species. Syst. Biod. 13, 555–570. doi: 10.1080/14772000.2015.1047428
Read, G., and Fauchald, K. (eds) (2019). World Polychaeta Database. Sirsoe Pleijel, 1998. Available at: http://www.marinespecies.org/aphia.php?p = taxdetails&id = 324833 on 2019-04-22 doi: 10.1080/14772000.2015.1047428 (accessed March 13, 2019).
Rizzo, A. E., and Salazar-Vallejo, S. I. (2014). Hesionidae Grube, 1850 (Annelida: Polychaeta) from South-Southeastern Brazil, with descriptions of four new species. Zootaxa 3856, 267–291. doi: 10.11646/zootaxa/3856.2.7
Roman, J., Estes, J. A., Morissette, L., Smith, C., Costa, D., McCarthy, J., et al. (2014). Whales as marine ecosystem engineers. Front. Ecol. Environ. 12, 377–385. doi: 10.1890/130220
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes v. 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rouse, G. W., Carvajal, J. I., and Pleijel, F. (2018a). Phylogeny of Hesionidae (Aciculata, Annelida), with four new species from deep-sea eastern Pacific methane seeps, and resolution of the affinity of Hesiolyra. Invert. Syst. 32, 1050–1068. doi: 10.1071/IS17092
Rouse, G. W., Goffredi, S. K., Johnson, S. B., and Vrijenhoek, R. C. (2018b). An inordinate fondness for Osedax (Siboglinidae: Annelida): fourteen new species of bone worms from California. Zootaxa 4377, 451–489. doi: 10.11646/zootaxa.4377.4.1
Rouse, G. W., Goffredi, S. K., and Vrijenhoek, R. C. (2004). Osedax: bone-eating marine worms with dwarf males. Science 30, 668–671. doi: 10.1126/science.1098650
Rouse, G. W., Wilson, N. G., Goffredi, S. K., Johnson, S. B., Smart, T., Widmer, C., et al. (2009). Spawning and development in Osedax boneworms (Siboglinidae, Annelida). Mar. Biol. 156, 395–405. doi: 10.1007/s00227-008-1091-z
Rouse, G. W., Worsaae, K., Johnson, S. B., Jones, W. J., and Vrijenhoek, R. C. (2008). Acquisition of dwarf male "harems" by recently settled females of Osedax rouseus n. sp. (Siboglinida; Annelida). Biol. Bull. 214, 67–82. doi: 10.2307/25066661
Ruta, C., Nygren, A., Rousset, V., Sundberg, P., Tillier, A., Wiklund, H., et al. (2007). Phylogeny of Hesionidae (Aciculata, Polychaeta), assessed from morphology, 18S rDNA, 28S rDNA, 16S rDNA and COI. Zool. Scr. 36, 99–107. doi: 10.1111/j.1463-6409.2006.00255.x
Ruta, C., and Pleijel, F. (2006). A revision of Syllidia (Psamathini, Hesionidae, Polychaeta). J. Nat. Hist. 40, 503–521. doi: 10.1080/00222930600727291
Saeedi, H., Bernardino, A. F., Shimabukuro, M., Falchetto, G., and Sumida, P. Y. G. (2019). Macrofaunal community structure and biosiversity patterns based on a wood-fall experiment in the deep South-west Atlantic. Deep Sea Res. I 145, 73–82. doi: 10.1016/j.dsr.2019.01.008
Schüller, M. (2011). Evidence for a role of bathymetry and emergence in speciation in the genus Glycera (Glyceridae, Polychaeta) from the deep Eastern Weddell Sea. Polar Biol. 34, 549–564. doi: 10.1007/s00300-010-0913-x
Silveira, I. C. A., Schmidt, A. C. K., Campos, E. J. D., Godoi, S. S., and Ikeda, Y. (2000). A corrente do Brasil ao largo da costa leste brasileira. Rev. Bras. Oceanogr. 48, 171–183. doi: 10.1590/s1413-77392000000200008
Smith, C. R., Amon, D. J., Higgs, N. D., Glover, A. G., and Young, E. L. (2017). Data are inadequate to test whale falls as chemosynthetic stepping-stones using network analysis: faunal overlaps do support a stepping-stone role. Proc. Biol. Sci. 284:20171281. doi: 10.1098/rspb.2017.1281
Smith, C. R., and Baco, A. R. (2003). Ecology of whale falls at the deep-sea floor. Oceanogr. Mar. Biol. Ann. Rev. 41, 311–354.
Smith, C. R., Bernardino, A. F., Baco, A., Hannides, A., and Altamira, I. (2014). Seven-year enrichment: macrofaunal succession in deep-sea sediments around a 30 tonne whale fall in the Northeast Pacific. Mar. Ecol. Prog. Ser. 515, 133–149. doi: 10.3354/meps10955
Smith, C. R., Glover, A. G., Treude, T., Higgs, N. D., and Amon, D. J. (2015). Whale-fall ecosystems: recent insights into ecology, paleoecology, and evolution. Annu. Rev. Mar. Sci. 7, 571–596. doi: 10.1146/annurev-marine-010213-135144
Smith, C. R., Kukert, H., Wheatcroft, R. A., Jumars, P. A., and Deming, J. W. (1989). Vent fauna on whale remains. Nature 341, 27–28. doi: 10.1038/341027a0
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stramma, L., and England, M. (1999). On the water masses and mean circulation of the South Atlantic Ocean. J. Geophys. Res 104, 20863–20883. doi: 10.1029/1999jc900139
Sumida, P. Y. G., Alfaro-Lucas, J. M., Shimabukuro, M., Kitazato, H., Perez, J. A. A., Soares-Gomes, A., et al. (2016). Deep-sea whale fall fauna from the Atlantic resembles that of the Pacific Ocean. Sci. Rep. 6:22139. doi: 10.1038/srep22139
Summers, M., Pleijel, F., and Rouse, G. W. (2015). Whale falls, multiple colonization of the deep, and the phylogeny of Hesionidae (Annelida). Invert. Syst. 29, 105–123.
Teixeira, S., Olu, K., Decker, C., Cunha, R. L., Fuchs, S., Hourdez, S., et al. (2013). High connectivity across the fragmented chemosynthetic ecosystems of the deep Atlantic Equatorial Belt: efficient dispersal mechanisms or questionable endemism? Mol. Ecol. 22, 4663–4680. doi: 10.1111/mec.12419
Thornhill, D. J., Struck, T. H., Ebbe, B., Lee, R. W., Mendoza, G. F., Levin, L. A., et al. (2012). Adaptive radiation in extremophilic Dorvilleidae (Annelida): diversification of a single colonizer or multiple independent lineages? Ecol. Evol. 2, 1958–1970. doi: 10.1002/ece3.314
Vrijenhoek, R. C., Johnson, S. B., and Rouse, G. W. (2009). A remarkable diversity of bone-eating worms (Osedax; Siboglinidae; Annelida). BMC Biol 7:74. doi: 10.1186/1741-7007-7-74
Wiklund, H., Altamira, I. V., Glover, A. G., Smith, C. R., Baco, A. R., and Dahlgren, T. G. (2012). Systematics and biodiversity of Ophryotrocha (Annelida, Dorvilleidae) with descriptions of six new species from deep-sea whale-fall and wood-fall habitats in the north-east Pacific. Syst. Biodivers. 10, 243–259. doi: 10.1080/14772000.2012.693970
Young, C. M., He, R., Emlet, R. B., Li, Y., Qian, H., Arellano, S. M., et al. (2012). Dispersal of deep-sea larvae from the intra-American seas: simulations of trajectories using ocean models. Integr. Comp. Biol. 52, 483–496. doi: 10.1093/icb/ics090
Keywords: deep sea, biodiversity, cryptic species, biogeography, connectivity
Citation: Shimabukuro M, Carrerette O, Alfaro-Lucas JM, Rizzo AE, Halanych KM and Sumida PYG (2019) Diversity, Distribution and Phylogeny of Hesionidae (Annelida) Colonizing Whale Falls: New Species of Sirsoe and Connections Between Ocean Basins. Front. Mar. Sci. 6:478. doi: 10.3389/fmars.2019.00478
Received: 20 May 2019; Accepted: 16 July 2019;
Published: 08 August 2019.
Edited by:
Chong Chen, Japan Agency for Marine-Earth Science and Technology, JapanReviewed by:
Anja Schulze, Texas A&M University at Galveston, United StatesNaoto Jimi, National Institute of Polar Research, Japan
Copyright © 2019 Shimabukuro, Carrerette, Alfaro-Lucas, Rizzo, Halanych and Sumida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mauricio Shimabukuro, bWF1c2hpbWFAdXNwLmJy; bXNoaW1hODRAZ21haWwuY29t
†Present address: Mauricio Shimabukuro and Joan Manel Alfaro-Lucas, Institut Français de Recherche pour l’Exploitation de la Mer (IFREMER), Centre de Bretagne, REM/EEP, Institut Carnot EDROME, Plouzané, France
 Mauricio Shimabukuro
Mauricio Shimabukuro Orlemir Carrerette
Orlemir Carrerette Joan Manel Alfaro-Lucas
Joan Manel Alfaro-Lucas Alexandra Elaine Rizzo
Alexandra Elaine Rizzo Kenneth M. Halanych
Kenneth M. Halanych Paulo Yukio Gomes Sumida
Paulo Yukio Gomes Sumida