- Department of Earth, Ocean and Atmospheric Sciences, The University of British Columbia, Vancouver, BC, Canada
Marine heterotrophic bacteria play a crucial role in the cycling of energy and nutrients in the ocean. Copper (Cu) belongs to the repertoire of essential trace nutrients for bacterial growth, yet physiological responses of marine heterotrophic bacteria to Cu deficiency remain unexplored. Here, we examined these responses in oceanic and coastal isolates of heterotrophic bacteria from ecologically significant microbial clades (Flavobacteriia class from Bacteroidetes phylum, and marine Roseobacter clade within Alphaproteobacteria class and Alteromonadales within Gammaproteobacteria class, both from Proteobacteria phylum). Bacterial growth, Cu quotas (Cu:P), macronutrient content and stoichiometry (cellular C, N, P, S, and C:N, S:P), as well as carbon metabolism (respiration, productivity, carbon demand, growth efficiency) were monitored across a gradient of Cu conditions, characteristic of coastal and open-ocean surface waters. Cu deficiency had most severe effects on a Flavobacteriia member Dokdonia sp. strain Dokd-P16 for which we observed significant reductions in growth, C metabolism and Cu quotas. Other strains did not significantly reduce their growth rate, but adjusted their Cu content and some C metabolic rates (Ruegeria pomeroyi DSS-3, Roseobacter clade) or were unaffected (Pseudoalteromonas sp. strains PAlt-P26 and PAlt-P2, Alteromonadales clade). These diverse bacterial responses were accompanied by constant cellular composition of major elements and stoichiometric ratios. Changes in bacterial Cu quotas occurred within a modest range (∼5-fold range) relative to the 50-fold variation in total Cu in the media. We hypothesize that this may reflect a well-controlled Cu homeostasis in marine heterotrophic bacteria. In a preliminary assessment, we found that Cu quotas of bacteria and those of eukaryotic phytoplankton are not statistically different. However, compared to eukaryotic phytoplankton, the variability of Cu quotas in marine heterotrophic bacteria is smaller, which could reflect differences in their Cu homeostasis. Using Cu quotas obtained in our study, we assessed the contribution of bacterial Cu to the biogenic Cu pool in an oceanic euphotic zone in the NE Pacific. These preliminary estimates suggest that up to 50% of the biogenic Cu could be contained in the biomass of marine heterotrophic bacteria. Our study sheds light on the interactions between Cu and marine heterotrophic bacteria, demonstrating the potential for Cu to influence microbial ecology and for microbes to play role in Cu biolgeochemical cycle.
Introduction
Planktonic bacteria represent most of the living biomass in the world’s oceans (Whitman et al., 1998). In surface waters, prokaryotic assemblages are dominated by autotrophic and heterotrophic bacteria, while archaea are more abundant in the ocean’s interior (Karner et al., 2001). Heterotrophic members of the bacterioplankton community play a critical role as the recyclers of organic material, thus influence fluxes of energy and essential elements within oceanic ecosystems. Their activity is governed by the availability of organic substrates as well as inorganic nutrients. The latter group includes bioactive metals, such as iron (Fe), copper (Cu), manganese (Mn), zinc (Zn), and cobalt (Co), that facilitate a variety of biological pathways in a prokaryotic cell. In this group, iron has been studied most intensively and has a well-established role as a factor limiting growth and metabolism of marine heterotrophic bacteria (Pakulski et al., 1996; Tortell et al., 1996; Kirchman et al., 2003a; Bertrand et al., 2011; Fourquez et al., 2014). Furthermore, previous studies have linked heterotrophic bacterioplankton to the marine biogeochemical cycle of Fe (Tortell et al., 1996; Maldonado and Price, 1999; Maldonado et al., 2016). By contrast, there is a paucity of information on how the availability of other essential metals regulate fundamental processes of heterotrophic bacteria, nor is it clear how these metals shape bacterial ecophysiology in the sea.
Copper plays a unique biological role because even though it is required for growth, it can be extremely toxic at very low concentrations. Its toxicity in marine bacteria and phytoplankton was found to be dependent on the abundance of the free Cu (II) ion (Sunda and Lewis, 1978; Schreiber et al., 1985), which is present in pM levels in seawater due to complexation with natural organic chelators (Santos-Echeandía et al., 2008; Heller and Croot, 2015). Copper has the potential to induce oxidative stress as it easily enters Fenton-like reactions that generate harmful hydroxyl radicals in the cell (Rowley and Halliwell, 1983). Another mode of Cu-induced toxicity is the destruction of Fe–S clusters of enzymes by direct complexation of Cu(I) ion to the coordinating sulfur atom (Macomber and Imlay, 2009). It is therefore essential to strictly control cellular Cu levels so that bacterial nutritional requirements are met while Cu toxicity is minimized. Prokaryotic Cu homeostasis has been the subject of extensive research in model bacteria (see reviews of: Rensing and Grass, 2003; Solioz and Stoyanov, 2003; Solioz et al., 2010; Argüello et al., 2013; Bondarczuk and Piotrowska-Seget, 2013), with a focus on elucidating mechanisms for Cu detoxification. In this context, the interactions between Cu and marine bacteria have also been explored (Gordon et al., 1993, 1994; Moffett and Brand, 1996; Mann et al., 2002). However, far less is known of the impacts of low Cu availability on prokaryotic cells (bacteria and archaea) and what adaptations these microorganisms have for dealing with Cu shortage in their environment.
To our best knowledge, the effects of Cu deficiency have only been investigated in model strains from two groups of marine microbes: heterotrophic denitrifying bacteria and autotrophic ammonia-oxidizing archaea (Granger and Ward, 2003; Moffett et al., 2012; Amin et al., 2013); whereas aerobic heterotrophic bacteria are yet to be explored. Copper serves as a catalyst in thirteen known prokaryotic cuproenzymes (as reviewed by Argüello et al., 2013), with the respiratory enzyme cytochrome c oxidase (COX) having the most prominent use in aerobic bacteria (Ridge et al., 2008). Hence, Cu starvation is likely to affect C metabolism in these microorganisms, impacting a cell’s energetic status and growth.
The aim of our study was to gain insight into the role of Cu as a nutrient in marine heterotrophic bacteria. We focused on examining key aspects of bacterial physiology: growth, carbon metabolism, cellular Cu quotas, as well as macronutrient content and stoichiometry across a gradient of Cu availability typical of coastal and open-ocean surface waters. We investigated four marine heterotrophic bacteria, which included three isolates (Dokdonia sp. strain Dokd-P16; Pseudoalteromonas sp. strains PAlt-P2 and PAlt-P26) from surface waters of the Northeast Pacific Ocean (including Fe-limited waters), and the model bacterium Ruegeria pomeroyi DSS-3. These strains belong to ecologically important microbial groups within the world’s ocean, namely Flavobacteriia class within the phylum of Bacteroidetes, Roseobacter clade within the Alphaproteobacteria class (Proteobacteria phylum), and the order Alteromonadales within the Gammaproteobacteria class (Proteobacteria phylum). These bacterial groups are known for their importance in the transformation of phytoplankton-derived organic matter during bloom events (as reviewed in Buchan et al., 2014).
Materials and Methods
Study Organisms
The heterotrophic bacteria used in this study were isolated from different locations along Line P, a transect extending from the coast of British Columbia, Canada, into the open-ocean waters of the Gulf of Alaska, 1,500 km offshore. Near-surface seawater (∼25 m) was collected at four different stations along the transect (P2, P4, P16, and P26) during the June cruise in 2012 (Line P program, cruise 2012-12)1. The seawater was then plated onto marine agar supplemented with either 0.5 g L-1 (for seawater from the coastal stations P2 and P4) or 0.05 g L-1 (for seawater from oceanic stations P16 and P26) of organic substrate (bactopeptone and casein hydrolysate in a 1:1 ratio). The agar plates were prepared using filtered seawater (0.22 μm) collected from ∼40 m at station P26. The colonies which grew on the plates were purified by sequential plating on marine agar. Pure cultures were preserved and stored in 15% glycerol stock at -80°C and plated as needed for the experiments.
Line P bacteria isolates were identified using 16S ribosomal DNA PCR amplification. PCR was performed using liquid cultures from colonies originally grown on agar plates. Liquid cultures were grown by adding a single colony to 2 mL of sterile liquid media made with station P26 seawater (from 10 m depth) supplemented with 0.5 g DOC (0.25 g L-1 bactopeptone and 0.25 g L-1 casein hydrolysate). Test tubes were placed in a shaker for 24 h at room temperature. Following culture growth, the full-length 16S rRNA gene was PCR amplified with primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). PCR amplification was performed in a 25 μL reaction volume with 16.7 μL of nuclease free water, 2.5 μL of buffer, 1.5 μL MgCl2 (Sigma Aldrich), 2 μL dNTPs, 0.5 μL forward and reverse primers, 0.3 μL of Taq polymerase and 1 μL of liquid culture. The reaction mixture was placed in the Thermocycler and run under the following conditions 95°C for 10 min, followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 90 s, followed by a final extension for 10 min at 72°C. The amplicon size was verified with an agarose gel, and the amplicons were purified with the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel). Samples were diluted to ∼4 ng μl-1 and sent to the GeneWiz sequencing center (GeneWiz, Inc.) for sequencing using three primers, 27F, 1492R, and a center primer, 519R (5′-GNTTTACCGCGGCKGCTG-3′). The three sequences for each clone were aligned using Sequencher 5.1 and a consensus sequence for each clone was obtained. The sequences were compared to the SILVA database version 1152 and isolates were taxonomically identified to the genus level (Table 1). The sequence data can be found in publicly available repository on Github3.
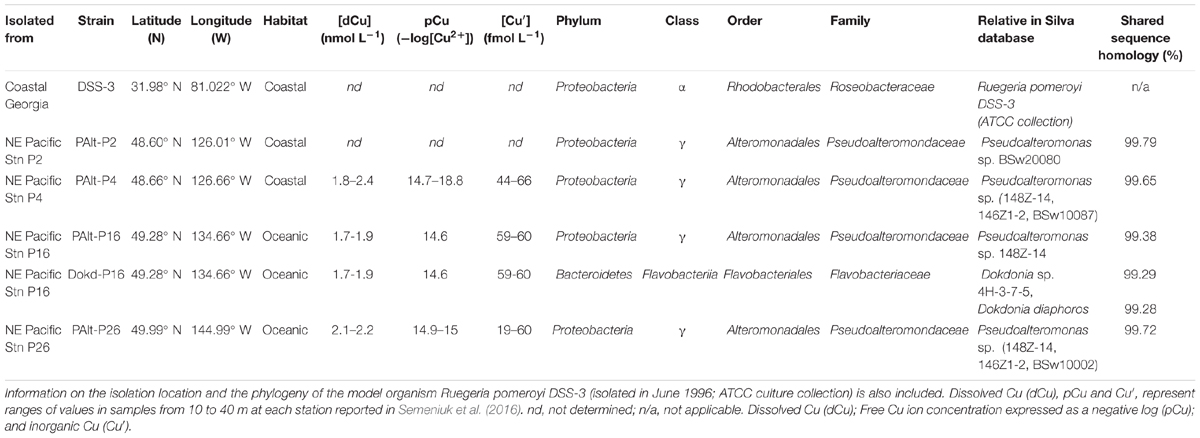
Table 1. Phylogenetic affiliation of gram-negative bacterial strains isolated from 25 m at various stations along Line P (June 2012 cruise).
With the exception of one strain, bacteria isolated from Line P surface waters belonged to the genus Pseudoalteromonas within the Gammaproteobacteria class (Table 1). Members of this genus may play a role in processing of organic carbon during phytoplankton blooms (e.g., transient peaks in Pseudolateromonas sp. in North Sea bloom reported by Lucas et al., 2015) and influencing Fe speciation and bioavailability in the ocean given their ability to produce Fe-binding siderophores (Granger and Price, 1999; Armstrong et al., 2004; Sijerčić and Price, 2015). One isolate from Line P belonged to genus Dokdonia sp. within the Flavobacteriia class. Many of Dokdonia sp. isolates from previous studies carry out rhodopsin-based bacterial phototrophy (Gómez-Consarnau et al., 2007; Kimura et al., 2011; Riedel et al., 2013; Bogachev et al., 2016), suggesting that this taxon may play a role in the cycling of carbon and energy in the ocean. In addition to the Line P isolates, we studied the model marine bacterium Ruegeria pomeroyi DSS-3, DMS 15171 (isolated from coastal Georgia), which was obtained from the American Type Culture Collection (ATCC, Manassas, VA, United States). The Roseobacter member Ruegeria pomeroyi DSS-3 has a range of adaptations that have been implicated in various biogeochemical cycles (e.g., Green et al., 2012; Cunliffe, 2013; Durham et al., 2015), such as an ability to degrade the algal osmolite dimethylsulphoniopropionate (DMSP, Reisch et al., 2011; Todd et al., 2012). The bacterial strains examined here are diverse in terms of their provenance and phylogeny (Table 1), and thus are likely to have distinct Cu requirements and adaptations to changing Cu availability. In our preliminary studies, we found a similar growth response to low and high Cu availability in different Pseudoalteromonas sp. isolates (Supplementary Figure 1). Ultimately, we selected one coastal (station P2, strain PAlt-P2) and one oceanic isolate (station P26, strain PAlt-P26) of Pseudoalteromonas sp. for more detailed investigations.
Limitations of Using Isolates as Models for Bacteria in the Sea
Using readily cultivable bacteria (as those in our study) to describe the ecophysiology of marine microbes is challenging as it remains unclear if such bacteria are good models for natural microbial populations. This can be inferred from low abundance of the types of bacteria commonly isolated using traditional plate methods in natural communities (e.g., 16S RNA surveys by Eilers et al., 2000). Furthermore, these bacteria are commonly characterized by having a copiotrophic lifestyle, thus a preference for high nutrient availability, a condition that is typically episodic in surface open-ocean. These considerations have put the ecological relevance of bacterial isolates and the rare bacterial biosphere into question.
However, there is increasing evidence that rare bacteria can have a significant impact on the ecosystem dynamics, which may be in part related to their high activity when conditions favor growth. For example, a large, fast-growing single strain of Alteromonas sp. AltSIO (Alteromonadaceae) was shown to consume the entire pool of dissolved organic carbon in a coastal ecosystem (Pedler et al., 2014). Furthermore, peaks in abundance of bacteria classically considered rare, such as Ulvibacter and Formosa sp. of Flavobacteriia, were observed during phytoplankton blooms (Teeling et al., 2012). There are also some reports of culturable bacteria accounting for 12–14% of the indigenous bacterial populations (Lebaron et al., 2001). Collectively, these observations are beginning to challenge perceptions of the ecological irrelevance of rare and cultivable bacteria. Our views on the ecology of culturable bacteria may change as we begin to accumulate information on their identity, physiology and genetic characteristics and integrate this information with culture-independent studies (Hagström et al., 2017).
Experimental Design and Culture Conditions
Bacterial physiology and metabolism were examined at five dissolved Cu levels (dCu) from limiting to replete, with a 50-fold variation in the total dCu (Table 2). At each Cu concentration, the following measurements were obtained: (1) growth rates; (2) cellular Cu quota; (3) major cellular elemental composition [carbon (C), nitrogen (N), phosphorus (P) and sulfur (S)]; and (4) bacterial O2 consumption rates. Bacteria were grown at 19 ± 1°C as semi-continuous batch cultures in the chemically well-defined artificial seawater medium Aquil (Price et al., 1989) modified for culturing marine heterotrophic bacteria (Granger and Price, 1999). Organic substrates were added to a final concentration of 0.5 g L-1 and consisted of casein hydrolysate and bactopeptone (1:1 ratio, Sigma-Aldrich). Organic substrate stocks were purified separately with Chelex 100 resin (25 g per 100 mL of organic substrate, Bio-Rad) prepared following the protocol of Price et al. (1989). These purified stocks were then used to amend 250 mL of artificial seawater (SOW) in trace metal-clean polycarbonate bottles and the media were microwave-sterilized for approximately 7 min. Once cooled, the SOW with organic substrates was amended with filter-sterilized additions of vitamins and trace metal stocks. Trace metals were buffered with 100 μM ethylenediaminetetraacetic acid (EDTA), and except for Cu, their total concentrations in the media were identical to those in Maldonado et al. (2006) and as reported in Supplementary Table 1. Copper was either omitted or added separately as Cu-EDTA complex (1:1.1) to create 5 different Cu treatments (Table 2). All plastics used in the growth experiments were sterilized and rigorously cleaned prior to use by storage in Extran, followed by 10% HCl for at least 24 h. All bacterial culture manipulations were done under sterile and trace metal clean conditions using a laminar flow hood.
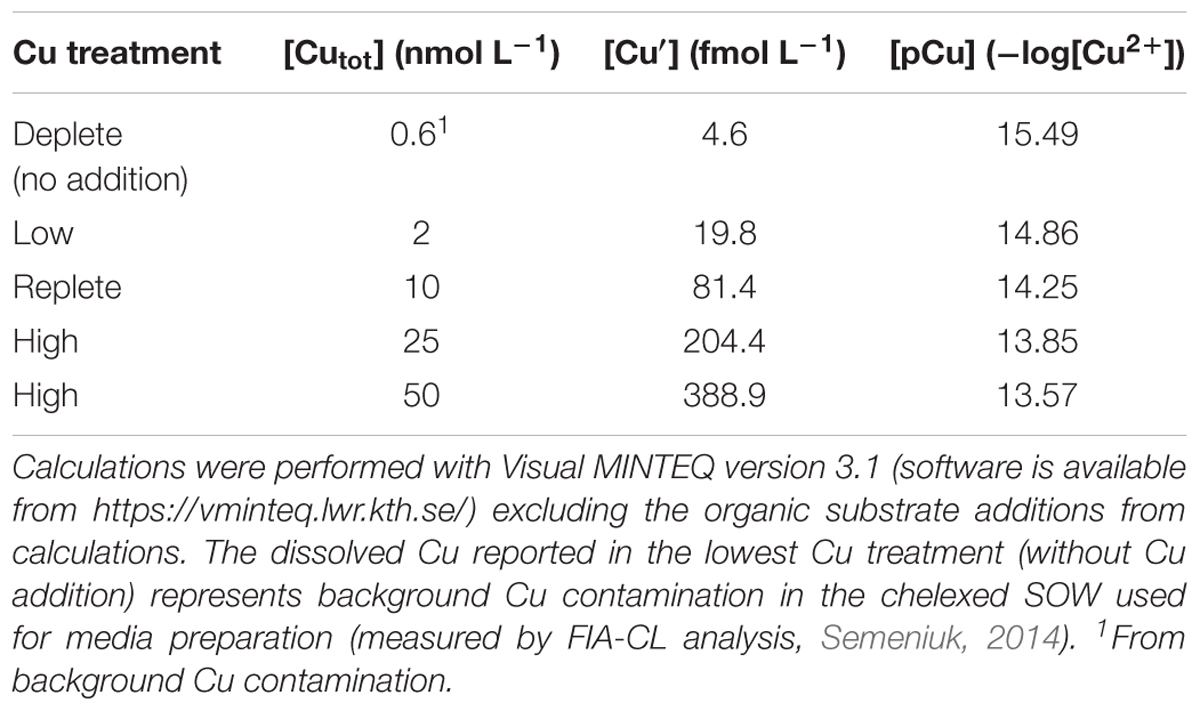
Table 2. Total dissolved copper (Cutot), inorganic Cu (Cu′) and free Cu ion concentrations (pCu, -log[Cu2+]) in bacterial growth media.
In Aquil medium, EDTA and trace metals are manipulated to buffer the concentrations of the free metal ions—typically the most bioavailable metal species—at environmentally relevant concentrations. Because of the unknown affinity of Cu for the organic substrate added to the bacterial growth media, Cu speciation in these media cannot be precisely estimated. For this reason, Cu treatments are primarily reported here in terms of the total dissolved Cu (Cutot). Nevertheless, we also provide the estimates for the concentrations of free Cu (pCu = -log [Cu2+]) and of inorganic Cu (Cu′) in the growth media (ignoring the organic substrate additions) using the chemical equilibrium model Visual MINTEQ (version 3.1, Gustafsson, 2016, Table 2). These values are only approximate but are included here as a reference, as previously done for other studies of marine heterotrophic bacteria involving organically amended Aquil (Granger and Price, 1999; Fourquez et al., 2014).
Granger and Price (1999) found that the concentration of kinetically labile Fe was higher in Aquil modified for heterotrophic bacteria compared to the traditional Aquil (with the same amount of Fe and EDTA) and suggested that the additions of organic substrates for bacterial growth may have enhanced Fe lability. This may be related to the competition between the stronger (EDTA) and the weaker ligands for trace metals (i.e., organics such as bactopeptone and casein hydrolysate) in bacterial culture media. Indeed, there is some evidence that the presence of weaker ligands (e.g., cysteine and histidine) in seawater media containing a stronger ligand (e.g., EDTA) can increase Cu and Zn bioavailability (Aristilde et al., 2012; Kim et al., 2015; Semeniuk et al., 2015; Walsh et al., 2015). Unfortunately, the kinetically labile (bioavailable) concentrations of Cu, Co, Mn, and Zn in organics-amended Aquil have not been evaluated, but it is possible that, as with Fe, their levels are higher than in the traditional Aquil media. Therefore, the levels of Cu′ and pCu we report for our organically enriched Aquil media are only approximate values that probably underestimate the true values of Cu bioavailability. Nevertheless, the approximate levels of inorganic Cu [Cu′] in our experiments encompass a range of concentrations that may be experienced by bacteria in both coastal and open-ocean environments (Moffett, 1995; Jacquot et al., 2013; Semeniuk et al., 2016).
Growth Rate Measurements
Bacterial growth rates were determined as previously described (Granger and Price, 1999). Briefly, cells acclimated to different Cu treatments (for eight generations) were inoculated into trace metal clean, sterile polystyrene cuvettes with caps and a stir bar (all sterilized by microwaving). Cultures were incubated in the spectrophotometer (Cary 1E UV-Vis, Varian) at 19°C and their optical density (A = 600 nm) was measured at 5 min interval. Absolute growth rates were determined during the exponential phase from log-linear regressions of absorbance versus time.
Enumeration of Bacteria
Bacterial cell numbers were estimated using flow cytometry analysis as described in Brussaard (2004). Cultures were diluted (x5) with pre-filtered SOW (0.22 μm polycarbonate filters) before the addition of glutaraldehyde (Electron Microscopy Grade, Sigma-Aldrich) to a final concentration of 0.5 % (i.e., 980 μL of culture sample containing 20 μL of 25% glutaraldehyde). Samples were taken in duplicate, flash frozen with liquid nitrogen and stored at -80°C until analysis. For flow cytometry analysis with FACS Calibur (Becton-Dickinson, Franklin Lakes, NJ, United States) samples were diluted up to 1:1000 in sterile 0.1 μm filtered 10:1 TE buffer (10 mM Tris-HCL and 1 mM EDTA, pH 8), stained with SYBR Green I (x10000, Invitrogen, Waltham, MA, United States) at a final concentration of 0.5 × 10-4 of commercial stock. Samples were incubated for 15 min in the dark before the processing run for 1 min at a high rate. Event rates were kept between 100 and 1000 per second and green fluorescence and side scatter detectors were used. Data were processed and gated using Cell-Quest software (Becton-Dickson).
Intracellular Carbon and Nitrogen Analysis
Samples in mid- to late-exponential growth phase (10–20 mL) were filtered by gravity onto 25 mm pre-combusted Whatman GF/F filters (>4 h, 450°C). Cells were rinsed with 2 mL of 0.6 M NaCl to remove extracellular organics and the filters were preserved at -20°C until analysis. To normalize the C and N data to per cell basis, samples for flow cytometry analysis were collected from the bacterial cultures before filtration onto GF/F filters, as well as from the filtrate. This allowed us to correct the cell numbers for any cell losses during filtration. However, on average, more than 99% of the bacteria were retained on the filters, similarly to a previous study using gravity filtration to collect cultured heterotrophic bacteria for C analysis (Pedler et al., 2014). Procedural blanks consisted of filters that were rinsed with media containing no bacterial cells. These blanks were typically < 5 % of the signal of the sample and the precision of the analysis ranged from 1 to 6% (range in RSD of 20 duplicate samples). Filters were dried overnight at 50°C, fumed with HCl, and dried again at 50°C before analysis on Perkin Elmer analyzer.
Intracellular P, S, and Trace Metal Analysis
Samples for trace metal analysis were collected in mid-exponential growth phase. From each biological replicate, 50 mL of culture was collected and centrifuged at 9,000 × g for 10 min. The supernatant was decanted, and cells were rinsed with cold (4°C) 20 mL of oxalate wash (pH 7.0, Tang and Morel, 2006) for 15 min at 4°C. Preliminary experiments suggested that washing bacterial cells with the oxalate solution does not lead to significant leakage of intracellular material, as described in Methods Section “Assessment of Cell Leakage Due to Washes.” Cells were then centrifuged for 10 min (9,000 × g, 4°C) and rinsed with 20 mL of chelexed 0.6 M NaCl for 5 min at 4°C and centrifuged again. The pellet was then re-suspended in 1 mL of chelexed 0.6 M NaCl and transferred to a clean microcentrifuge tube. An aliquot (5 μL) was taken for cell enumeration by flow cytometry. Subsequently, the sample was centrifuged for 10 min (10,000 × g, 4°C), the supernatant was removed, and the pellets were immediately stored at -20°C until analysis. The metal content of bacterial pellets was determined via inductively-coupled plasma-mass spectrometry (ICP-MS) using an instrument equipped with a hyperbolic quadrupole mass analyzer and octopole collision cell (Agilent, 7700). Bacterial pellets were digested in 5 mL Teflon digestion vials, which were cleaned following these steps: (1) detergent Extran for 24 h (50°C); (2) 6N HCl for 1-week (50°C); and (3) 1N HNO3 for 1-week (50°C). Vials were rinsed in between the cleaning steps with MilliQ water (x 6 rinses) and were leached in MilliQ for a week at 50°C before use. Pellets were digested in 3 mL of ultrapure HNO3 at 120°C for 7 h, cooled down, and evaporated to dryness overnight at 120°C. Subsequently, 1 mL of ultrapure H2O2 was added to each vial and evaporated to dryness at 120°C for ∼ 4 h. Procedural blanks consisted of vials with no pellets treated in the same way as the samples. For ICP-MS analysis, vials were filled with a matrix solution, consisting of 2% HNO3 spiked with 10 ppb In as an internal standard, and allowed to stand for 10 min before being transferred to 15 mL trace metal clean polyethylene tube. The final volume of samples was subsequently adjusted to ∼10 mL.
The instrument was calibrated using multi-element standard (High-Purity Standards, Charleston, SC, United States) prepared in the same matrix solution as the samples. Two isotopes of copper (Cu 63/65) were measured to ensure that background and interference were acceptably low. Average procedural blank was 0.05 nmol Cu L-1 and the limit of detection calculated as 3 × SD of the blank was 0.07 nmol L-1, with samples values on average > 40–100 times higher than the detection limit. Determination of phosphorus (31P) by quadrupole ICP-MS is challenging due to high background and interferences from polyatomic ions such as 15N16O+; 14N17O+; 14N16O1H+ and 30Si H+. We assessed P data obtained using ICP-MS by comparison with measurements of P in the same samples by ICP-Optical Emission Spectroscopy (OES). There was an excellent agreement between the P measurements made by these two different instruments (Supplementary Figure 2), thus confirming the accuracy of the P concentrations obtained in our samples with ICP-MS. Sulfur data were obtained using ICP-OES. Trace metal data were normalized using P, cell numbers or cellular carbon data. We focus on P normalized quotas because this element was measured simultaneously with Cu during ICP-MS analysis, thus eliminating errors due to different analytical techniques. Phosphorous normalization allows for comparison of metal contents independently of bacterial cell volume, which was not measured during this study. While bacterial P content, can be plastic depending on environmental conditions (e.g., organic substrate quality, Godwin and Cotner, 2015), we found no statistically significant changes in cell normalized P content in response to Cu (see Results, Section “Macronutrient Cellular Composition in Response to Cu”). Indeed, P-normalized Cu quotas followed the same trends as those of the cell- and C-normalized Cu quotas (Supplementary Figure 3), suggesting that organic P was an appropriate biomass proxy.
Assessment of Cell Leakage Due to Washes
Determination of intracellular elements requires a washing step to remove any elements adsorbed to the outside of the cells. There are various washing protocols, depending on the element of interest (e.g., Fe vs. Cu) and the method used for metal quota determination (e.g., radioisotope techniques vs. ICP-MS). However, washing solutions could affect cell membrane integrity and lead to a loss of intracellular elements, resulting in underestimated elemental quotas.
In this study, we examined whether two commonly used washes, oxalate-EDTA (Tang and Morel, 2006) and DTPA (Croot et al., 2003), induce cell leakage in bacteria using 14C radiolabeled glucose as a tracer of intracellular material. For these experiments, the oceanic Pseudoalteromonas sp. PAlt-P26 strain was selected as it grew successfully in modified media containing glucose as a C source (μ ∼ 3 d-1), unlike Dokdonia sp. strain Dokd-P16, which we were unable to grow in this medium. The modified medium contained 35 mM glucose, 6 mM ammonium and 0.1 mM phosphate in filtered seawater (Weaver et al., 2003). We used water from Ocean Station Papa (OSP, station P26, ∼10 m) amended with vitamins and trace metals, as for the Aquil media recipe. A small inoculum of exponentially growing PAlt-P26 acclimated to the modified media was transferred into 250 mL medium containing 20 μCi 14C-glucose and cultured overnight at room temperature. Wash test experiments involved filtering two aliquots (2.5 mL) of exponentially growing bacterial culture (0.22 μm polycarbonate filters) and incubating one aliquot with a wash (2.5 mL, oxalate or DTPA solutions) and the other aliquot with sterile SOW (2.5 mL) for 15 min in the dark at room temperature. This procedure was performed in triplicates. The oxalate and the DTPA incubated aliquots of cell culture were rinsed with sterile SOW before final filtration. Filter blanks (n = 3) were collected by subjecting filters to the same washing procedure but without bacterial cells and were used to correct the 14C activity of sample filters. Cell leakage was determined by comparing the 14C activity of wash-treated (oxalate or DTPA) and SOW only treated cells.
A paired Student’s t-test was used to determine whether the differences between the washes (oxalate or DTPA) and SOW were statistically significant. There was no statistically significant difference between wash-treated and SOW-treated cells (oxalate, T-value = 3.85; p = 0.061; DTPA, T-value = 1.6; p = 0.24), supporting our conclusion that the washing solutions do not induce a significant loss of cellular contents (Supplementary Figure 4). We thus assumed that just like with 14C-glucose, the loss of cellular Cu would be minimal. We do acknowledge, however, that the lability of the intracellular C and Cu pools may not be the same. In gram-negative bacteria, Cu-containing proteins are believed to be mostly located in the inner membrane and/or the periplasmic space (Tottey et al., 2005). We suspect that damage to the cell’s membranes would lead to a loss of cytoplasmic C constituents, as well as membrane-bound Cu and Cu ions in the periplasm. Ideally, to assess Cu leakage, cellular Cu pools in the cells would have been labeled with a radioactive Cu tracer (e.g., 64Cu or 67Cu). Unfortunately, these tracers are not commercially available. Ultimately, oxalate solution was selected for cell washing during the collection of samples for ICP-MS analysis (as described in Section “Intracellular P, S, and Trace Metal Analysis”) due to its previous use in the removal of multiple biogenic elements (e.g., Wilhelm et al., 2013), as opposed to the DTPA wash, which was developed specifically for extracellular Cu removal.
Oxygen Consumption Rates
Bacterial respiration was measured using a S1 Clark type oxygen electrode (Hansatech Instruments, Norfolk, England). Cultures were sampled in mid-exponential phase as 1 mL aliquots that were transferred to a clean O2 electrode chamber connected to a 19°C water bath. Oxygen consumption rates were derived as the slope of O2 consumption over time (10 min) in the chamber containing the bacterial culture. Prior to these measurements, the electrode was calibrated by filling the electrode chamber with O2-saturated synthetic seawater (SOW, bubbled with O2 for 1 h prior to experiment in a water bath at 19°C). Subsequently, the SOW in the electrode chamber was gently bubbled with N2 to record the O2-minimum signal. The electrode was controlled by a CB1D O2 electrode control box (Hansatech) and the data were collected and processed with the Labjackoxy software (Hansatech). For each biological replicate, two or more technical replicates were collected during the exponential phase and averaged. The O2 consumption rates were normalized to bacterial cell abundance.
Bacterial Carbon Metabolism Calculations
Growth and respiration rates, as well as C content data, were used to derive different estimates of heterotrophic C metabolism as previously described (del Giorgio and Cole, 1998):
where BP stands for bacterial productivity (fmol C cell-1 d-1), BCD for bacterial carbon demand (fmol C cell-1 d-1), BR for bacterial respiration (fmol C cell-1 d-1), and BGE for bacterial growth efficiency (ratio, unitless).
These metabolic rates were derived for Dokdonia sp. strain Dokd-P16, oceanic Pseudoalteromonas sp. strain PAlt-P26 and R. pomeroyi DSS-3 using triplicates measurements. For these estimates, bacterial respiration rates were converted from the amount of oxygen consumed (fmol O2 cell-1 d-1) to the amount of carbon respired (fmol C cell-1 d-1) using a respiratory quotient (RQ – ratio of O2 consumed to CO2 produced) of 1, as has been assumed in other studies (del Giorgio and Cole, 1998; Reinthaler and Herndl, 2005; Hörtnagl et al., 2011; Pradeep Ram et al., 2016). While RQ can vary depending on the organic substrate quality (0.7–1.2, Berggren et al., 2012), it is less clear if changes in inorganic nutrients affect RQ. Fourquez et al. (2014) found that Fe-limited marine heterotrophic bacterium Alteromonas macleodii alters certain C metabolic pathways, which may have an effect on RQ. However, no studies have yet quantified the magnitude of this effect on RQ due to Fe limitation or any other metal.
Statistical Analysis and Figures
The statistical analyses were conducted using an open-source programming language R (R Core Team, 2016). Data processing was done using package “dplyr” (Wickham, 2016), while all figures were generated using “ggplot” (Wickham, 2009) and “cowplot” (Wilke and Wickham, 2016) packages in R. All the data presented were calculated as means ± standard deviation. One-way Analysis of Variance (ANOVA) was used to determine the significance among different Cu treatments for individual bacterial strains with a significance level α of 0.05. Post hoc analyses were performed using a pairwise t-test with Bonferroni correction of α (at 0.05) when the variance was homogeneous using the R base package “stats”. For data that displayed heterogeneity of variance, we applied Tukey’s honest significance difference test, using “multcomp” package in R (version 1.4–6, Herberich et al., 2010). Data and code used to produce figures, summary tables and statistical tests in this manuscript can be found in publicly available repository on Github4.
Results
Growth Rates and Cu Quotas in Response to Changing Cu Availability
We found diverse responses of bacterial growth rates and Cu quotas to changing Cu availability (Figure 1). Both coastal and oceanic strains of Pseudoalteromonas sp. (PAlt-P2 and PAlt-P26, respectively) had the fastest growth rates, and minor reductions in growth rates in the lowest Cu treatments (Cutot = 0.6 nmol L-1, μ/μmax of 0.72 and 0.84 for Pseudoalteromonas sp. PAlt-P2, and PAlt-P26, respectively, Table 3). These strains also had similar Cu quotas (Cu:P, mmol:mol) across all Cu treatments (Figure 1). Statistical analysis confirmed that neither their growth rates nor the Cu:P ratio were significantly affected by Cu concentrations (Table 4). Interestingly, lack of response in stoichiometry when growth rates are above 0.6 μ/μmax has been previously reported for freshwater bacterial isolates, although for macronutrients (C, N, P, Godwin and Cotner, 2018). In contrast, while the growth rate of the Roseobacter member, R. pomeroyi DSS-3, was not affected by changes in Cu availability (F = 2.1, p = 0.158, Table 4 and Figure 1), its Cu quota (Cu:P, mmol:mol) varied significantly (F = 20, p < 0.001, Table 4 and Figure 1) showing a 2-fold reduction from the highest to the lowest Cu treatment (Cu:P = 0.15 ± 0.01 versus 0.05 ± 0.01 mmol:mol, respectively; Table 3). The growth rate of the Flavobacteriia member, Dokdonia sp. strain Dokd-P16, was significantly affected by Cu availability (F = 79, p < 0.001, Table 4), and we found up to ∼ 80% reduction in growth when the cultures were most deprived of Cu (at 0.6 nmol Cu L-1, Table 3) compared to the maximal growth rate under Cu replete conditions. For this strain, reductions in growth rates were accompanied by ∼50% decrease in Cu quotas at the two lowest Cu levels (average 0.7 mmol Cu:mol P for the 0.6 and 2 nM Cu cultures) compared to the two highest Cu replete treatments (average 0.13 mmol Cu:mol P for 25 and 50 nM Cu treatments, Figure 1 and Table 3). Interestingly, similar decreases in Cu quota were observed at moderate Cu levels in the media (10 nmol Cu L-1) even though the growth rate was unaffected (Figure 1 and Table 3).
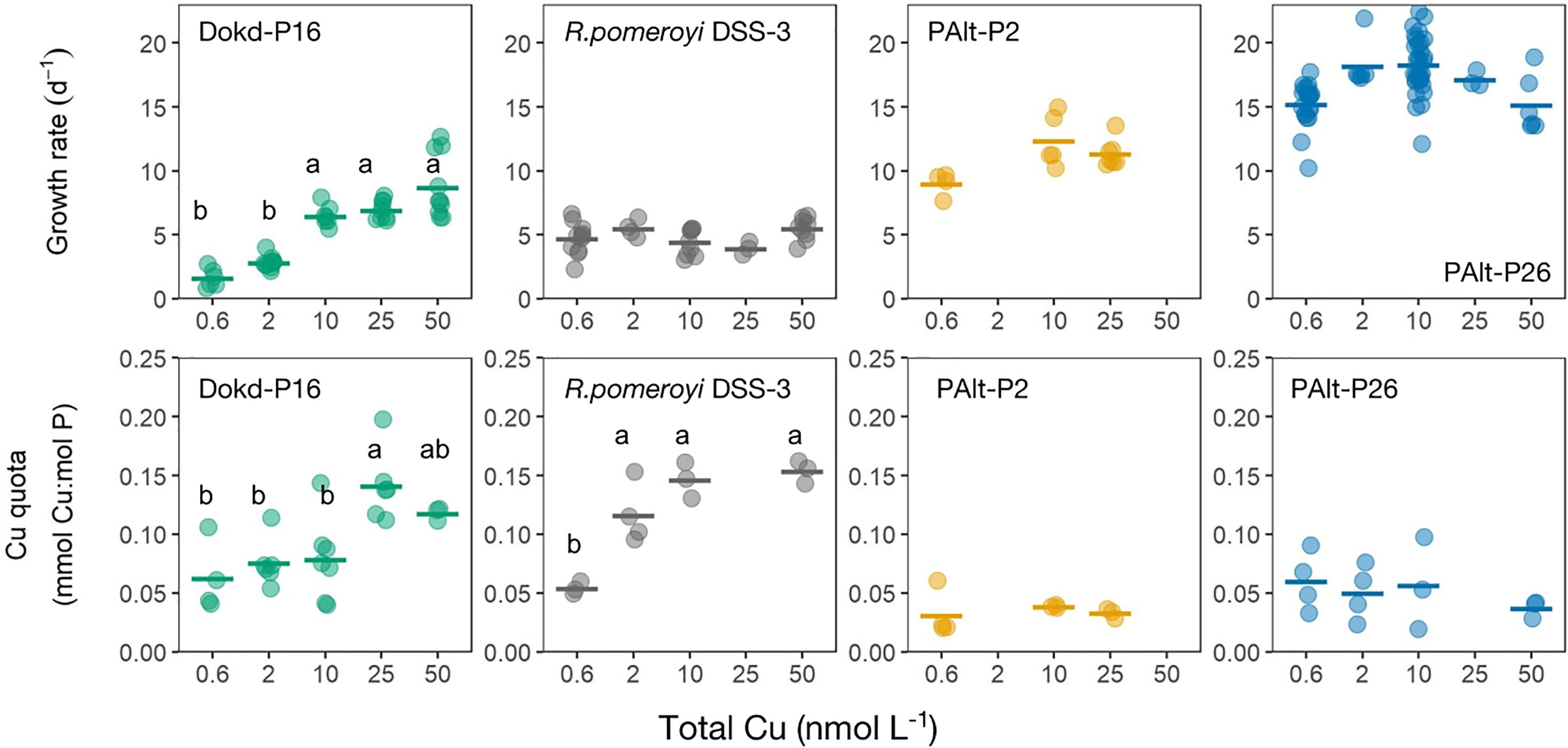
Figure 1. Bacterial growth rates (d-1) and Cu quota (Cu:P ratio, mmol: mol) at different levels of total dissolved Cu (nmol L-1). The horizontal lines represent the mean and the superimposed colored circles are data for each biological replicate that was used in the calculation of mean. Within a single graph, values with the same letter were not significantly different from each other (pairwise t-test with Bonferroni correction, p < 0.05). All data are presented in Table 3 and the ANOVA results are presented in Table 4. Note x-axis is not linear.
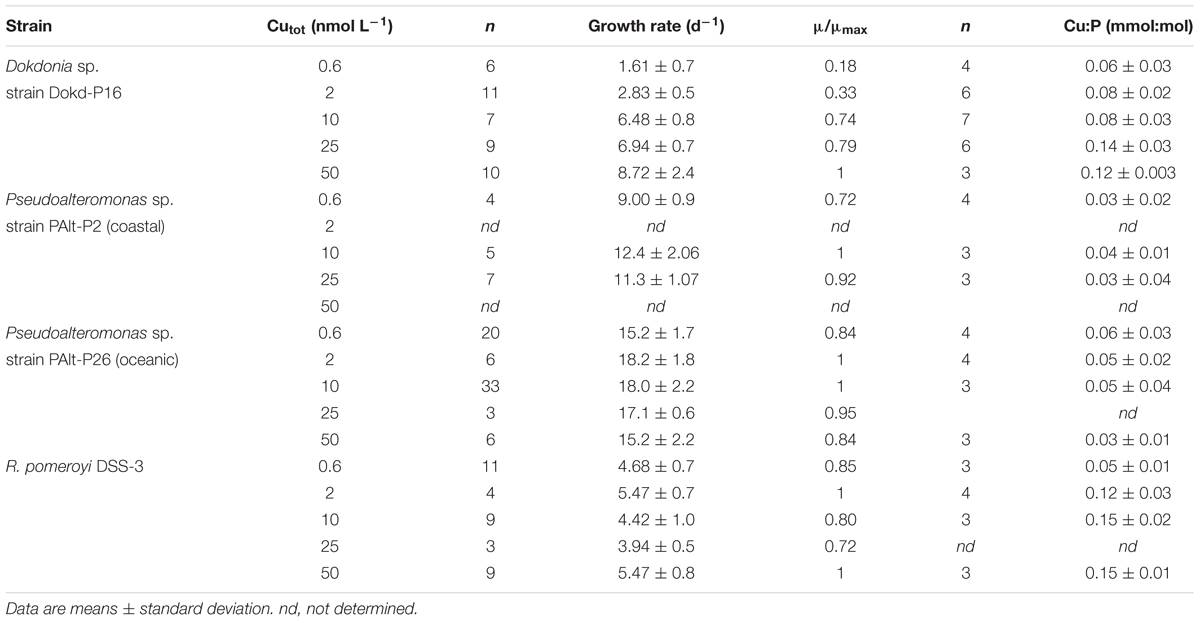
Table 3. Specific growth rates (d-1), relative growth rates (μ/μmax), and phosphorus normalized Cu quotas (Cu:P, mmol:mol) of bacterial strains at various levels of Cu in the growth media (nmol Cu L-1).
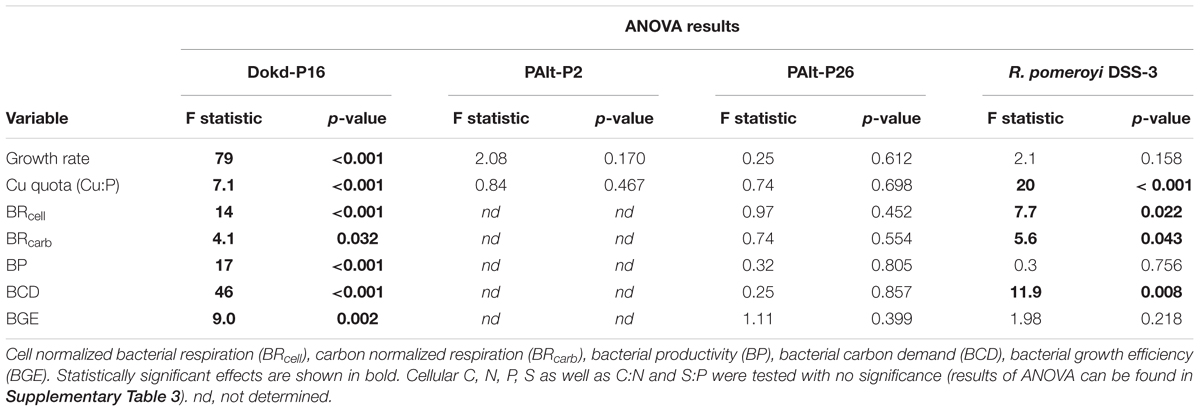
Table 4. One-way ANOVA values for the effect of Cu concentration in the media on various metabolic variables of the bacterial strains.
Macronutrient Cellular Composition in Response to Cu
The major element composition of bacterial strains at varying Cu levels were assessed as cell-normalized carbon (C), nitrogen (N), phosphorus (P) and sulfur (S) (Figure 2 and Supplementary Table 2). In general, cellular macronutrients did not vary significantly in response to the Cu treatments (Figure 2; ANOVA results, Supplementary Table 3). When averaged across all treatments, the oceanic Pseudoalteromonas sp. strain PAlt-P26 had the highest macronutrient quotas [cellular fmolar quota (fmol cell-1) of 27 C, 6 N, 0.5 P, and 0.2 S], followed by the coastal Pseudoalteromonas sp. strain PAlt-P2 (fmol cell-1 of 22 C, 6 N, 0.5 P, and 0.2 S), R. pomeroyi DSS-3 (fmol cell-1 of 16 C, 4 N, 0.2 P, and 0.15 S) and Dokdonia sp. strain Dokd-P16 (fmol cell-1 of 10 C, 2 N, 0.2 P, and 0.12 S). The larger cellular macronutrient content of the Pseudoalteromonas sp. strains may reflect their larger cell size compared to R. pomeroyi DSS-3 and Dokd-P16. To further assess the effects of Cu on the elemental composition of bacteria, we determined the stoichiometric ratios of major elements (C:N and S:P, Figure 2). We consider ratios only for those elements that were analyzed simultaneously (C and N, P, and S) to minimize the error due to differences in analytical techniques. In all bacterial strains, the molar ratios of both C:N and S:P were not significantly affected by changing Cu availability (ANOVA results, Supplementary Table 3). The elemental ratios determined in this study are on par with those of native and cultured marine and freshwater bacteria reported in Fagerbakke et al. (1996) (Figure 2).
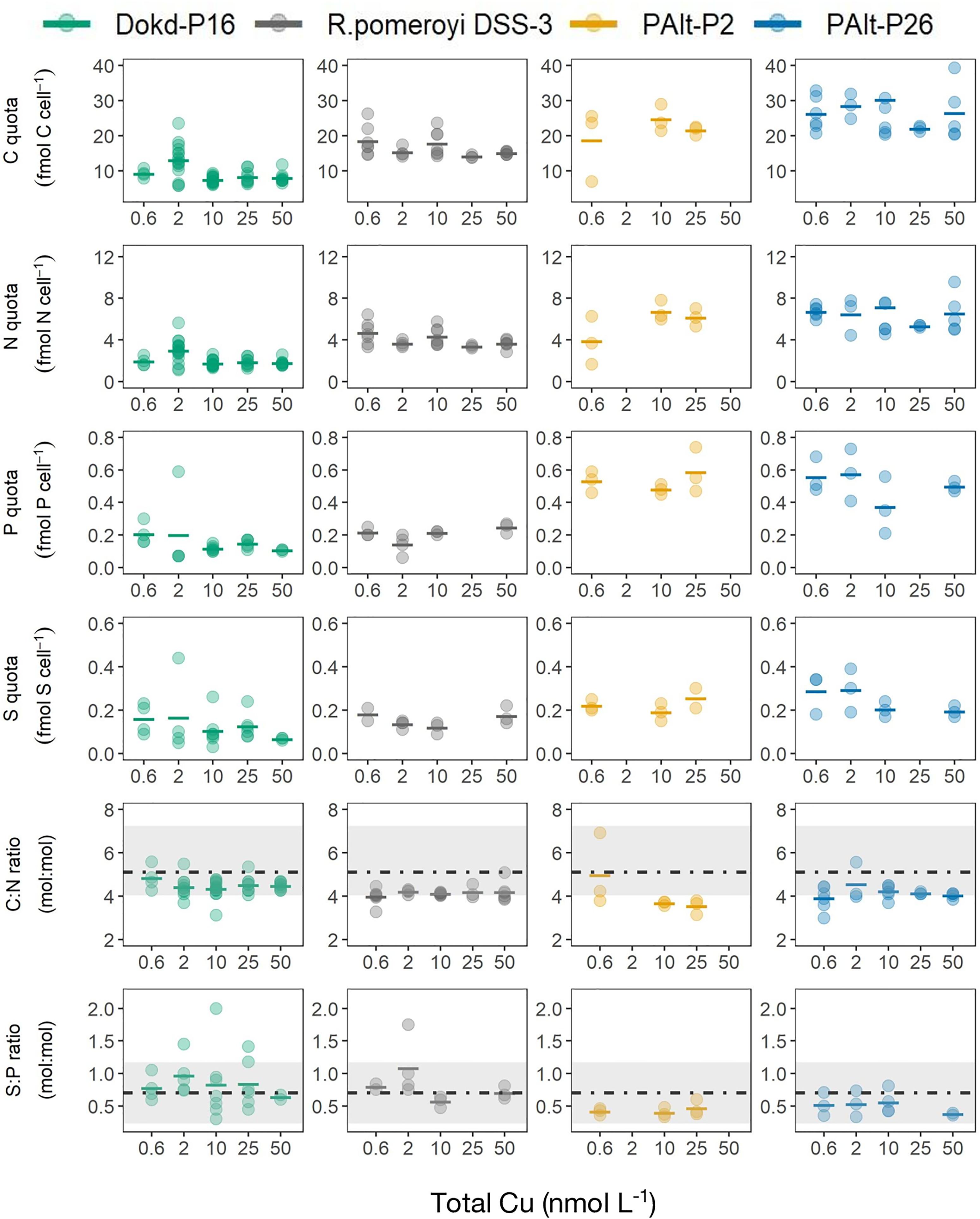
Figure 2. Cellular carbon (fmol C cell-1), nitrogen (fmol N cell-1), phosphorus (fmol P cell-1), sulfur (fmol S cell-1), and molar macronutrient ratios (C:N and S:P) in Dokdonia sp. strain Dokd-P16, R. pomeroyi DSS-3, coastal Pseudoalteromonas sp. strain PAlt-P2, oceanic Pseudoalteromonas sp. strain PAlt-P26 at different levels of Cu (nmol L-1). The horizontal line represent the mean and the superimposed colored circles are data for each biological replicate that was used in the calculation of mean. The horizontal dashed lines indicate the average C:N and S:P ratios for native and cultured bacteria reported in a study by Fagerbakke et al. (1996), with the shaded regions spanning the range of each of the macro-element ratios reported in Fagerbakke et al. (1996). The data used to produce the figures can be found in Supplementary Table 2, and the ANOVA results for the effect of Cu on major elemental composition in the Supplementary Table 3. Note x-axis is not linear.
Regulation of Bacterial Carbon Metabolism by Cu Availability
Using growth rate, cellular C, and bacterial respiration data, we estimated various aspects of heterotrophic C metabolism and examined its sensitivity to changes in Cu availability for three strains: Dokd-P16, oceanic PAlt-P26, and R. pomeroyi DSS-3 (Figure 3). Carbon metabolism of bacteria can be distinguished into anabolic (bacterial productivity, BP) and catabolic reactions (bacterial respiration, BR), which can be used to derive bacterial carbon demand (BCD; the carbon biomass that is required to sustain net bacterial metabolic needs) and bacterial growth efficiency (BGE; carbon assimilation efficiency into biomass) (del Giorgio and Cole, 1998). Of all strains, only Dokd-P16 was found to significantly reduce all metabolic rates (BR, BP, and BCD) and BGE in response to low Cu availability (One-way ANOVA, Table 4). Copper limitation (0.6 and 2 nmol Cu L-1 treatments) was associated with ∼20% reduction in cellular respiration (BRcell), ∼ 40% reduction in productivity, and ∼40–50% reduction in both BCD and BGE, compared to Cu-replete conditions (25 and 50 nmol Cu L-1, Figure 3 and Supplementary Table 4). Furthermore, BR and BCD were significantly reduced at moderate Cu levels (10 nmol Cu L-1) compared to the two highest Cu treatments (Figure 3), yet, BGE estimates were similar across these treatments. In R. pomeroyi DSS-3, BR and BCD were also significantly affected by Cu availability (Table 4), but in contrast to Dokd-P16, these rates were substantially elevated under Cu limitation (higher at 0.6 than at 50 nmol Cu L-1 treatment; p < 0.05, Figure 3 and Supplementary Table 4). Finally, none of the carbon metabolism parameters varied significantly for PAlt-P26 in response to changing Cu availability (Table 4 and Figure 3).
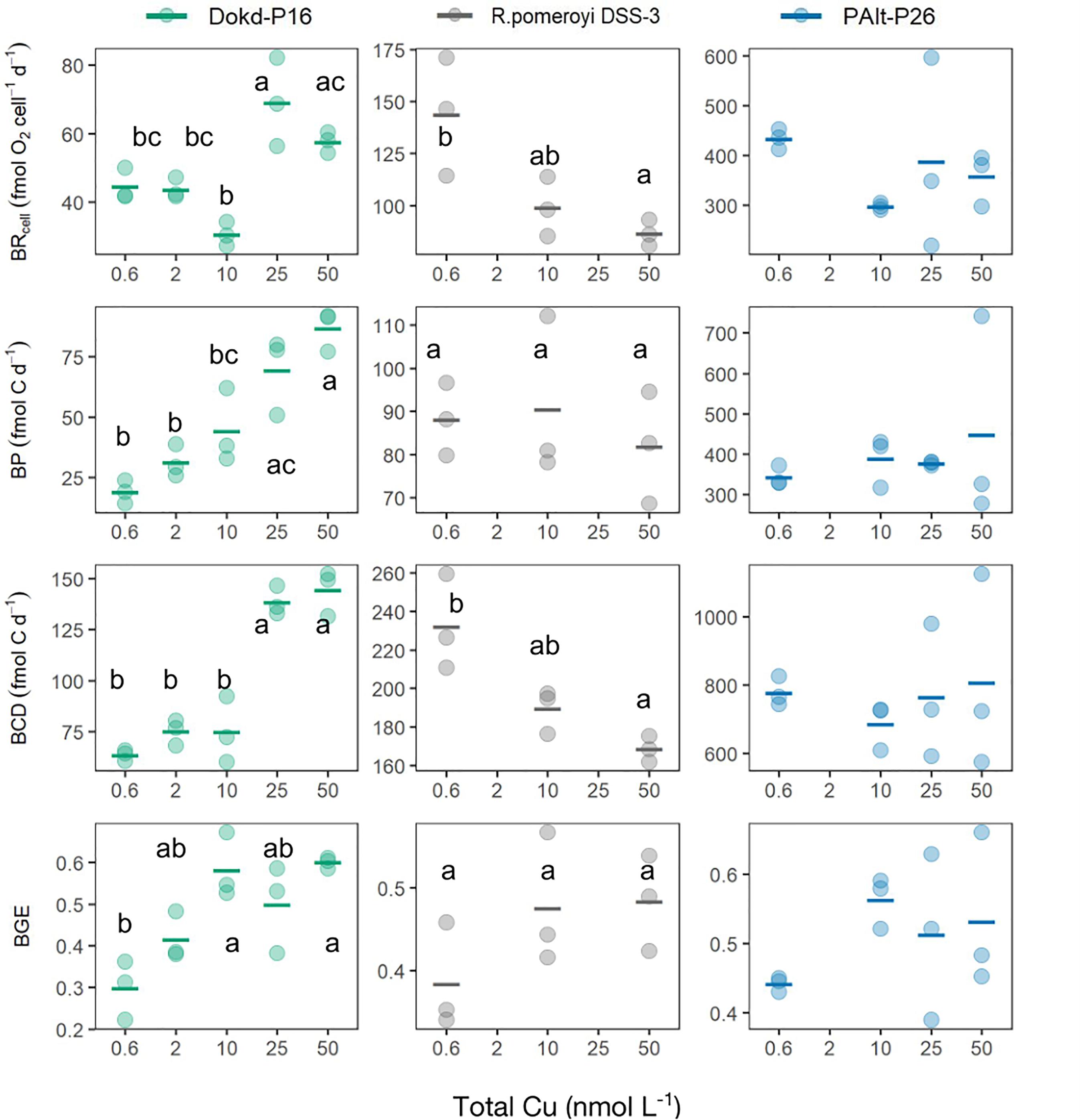
Figure 3. Carbon metabolism of marine heterotrophic bacteria at different levels of Cu (nmol L-1). Cell-normalized respiration (BRcell, fmol O2 cell-1 d-1), bacterial productivity (BP, fmol C d-1), bacterial carbon demand (BCD, fmol C d-1) and bacterial growth efficiency (BGE, unitless) of Dokdonia sp. strain Dokd-P16, Ruegeria pomeroyi DSS-3, Pseudoalteromonas sp. strain PAlt-P26. The horizontal line represents the mean and the superimposed colored circles are data for each biological replicate that was used in the calculation of mean. Significant effects (p < 0.05) are shown with different letters for Dokd-P16 and R. pomeroyi DSS-3 (pairwise t-test with Bonferroni correction of α at 0.05). The data used to produce the figure can be found in the Supplementary Table 4. Note x-axis is not linear.
The BGE estimates in this study varied from 0.3 to 0.6 and are in the range of those reported for Vibrio harveyi (<0.1-0.5, Kirchman et al., 2003a), and are typical of bacterial communities in eutrophic systems (as reviewed in del Giorgio and Cole, 1998). Respiration rates obtained for PAlt-P26 (ranging from 344 to 449 fmol O2 cell-1 d-1, Supplementary Table 4) are higher than those of coastal and oceanic Alteromonas macleodii strains (order Alteromonadales), cultured in Aquil modified media with replete Fe concentrations (226 fmol O2 cell-1 d-1, Fourquez et al., 2014).
Discussion
Responses of Bacterial Isolates to Changing Cu Availability
Bacterial Growth and Cellular Cu Quotas
We investigated four strains of marine heterotrophic bacteria from three distinct taxa, and found that these bacteria showed three contrasting responses to changes in Cu availability. First, with the exception of macronutrient content and stoichiometry, all physiological assessments of the Flavobacteriia member Dokdonia sp. Dokd-P16 (oceanic) were affected by low Cu, suggesting that this bacterium has a metabolism highly dependent on Cu availability. The growth rate of Cu-limited Dokd-P16 was severely reduced, as well as its cellular Cu. Second, the Roseobacter member R. pomeroyi DSS-3 (coastal) also reduced its Cu content under low Cu concentrations but its growth rate remained unchanged. Interestingly, both Dokd-P16 and R. pomeroyi DSS-3 had similar cellular Cu quotas at various Cu levels (mean 0.05–0.15 mmol Cu:mol P, Figure 1 and Table 3). Thus, their contrasting growth responses to Cu deficiency cannot be readily explained by their cellular Cu. The third response characterizes both strains of Pseudoalteromonas sp. These bacteria had unaffected growth rates, as well as low and invariant average cellular Cu quotas despite a 50-fold variation in Cu availability (mean 0.03–0.06 mmol Cu:mol P, Table 3, One-way ANOVA, Table 4), indicating tightly controlled intracellular Cu levels. Our results suggest that both strains of Pseudoalteromonas sp., as well as R. pomeroyi DSS-3 would be more successful under low Cu availability than Dokd-P16. While future genomic and proteomic studies are needed to identify the metabolic pathways affected by low Cu in Dokd-P16, our findings indicate that this strain may have unique Cu requirements.
One important observation in our study is that, although there was some strain-specific variability in bacterial Cu quotas, the range of Cu:P ratios was narrow (∼5-fold, 0.03–0.15 mmol Cu:mol P) relative to the 50-fold variation of Cu concentrations in the growth media. Much of our understanding of bacterial Cu homeostasis comes from studies with model bacteria such as Escherichia coli. These studies suggest that bacteria are highly sensitive to Cu toxicity and that their Cu trafficking and cellular inventories are under strict management via a variety of mechanisms (as reviewed by: Rensing and Grass, 2003; Solioz and Stoyanov, 2003; Solioz et al., 2010; Argüello et al., 2013; Bondarczuk and Piotrowska-Seget, 2013). In E.coli, Cu efflux (via a regulatory protein CueR) is activated at zeptomolar (10-21) Cu2+ levels (<600 atoms per cell), suggesting that this bacterium is extremely sensitive to free Cu in the cytosol, and operates Cu-detoxification mechanisms even under Cu deprivation (Changela et al., 2003). Furthermore, with the exception of photosynthetic bacteria, no cytosolic Cu-containing proteins have been reported in bacteria, and in E. coli these proteins are confined to the periplasm or the cytoplasmic membrane (Tottey et al., 2005). Taken together, bacteria are required to maintain low cellular Cu in order to avoid Cu toxicity, which can cause oxidative stress and destruction of Fe–S clusters in proteins (reviewed in Dupont et al., 2011). Therefore, the narrow range of Cu quotas displayed by marine heterotrophic bacteria from this study relative to the broad Cu availability in the growth media appears to fit with the model of strict Cu regulation in gram-negative bacteria.
Copper Availability and Dokdonia sp. Dokd-P16
As shown in this study, Cu is a critical micronutrient for the Flavobacteriia member Dokdonia sp. Dokd-P16. Indeed, Dokd-P16 significantly reduces its growth rates (33% of μmax at 2 nmol Cu L-1) at free divalent Cu (Cu2+) concentrations (1.38 × 10-15 mol Cu2+ L-1) that are similar to those (1 × 10-15mol Cu2+ L-1) limiting several coastal phytoplankton (e.g., Thalassiosira weissflogii and Chaetoceros decipiens, the coccolithophore Emiliania huxleyi, and the prymnesiophyte Phaeocystis cordata; Annett et al., 2008; Guo et al., 2012). In contrast to eukaryotic phytoplankton, little is known about Cu conditions that limit the growth of marine prokaryotes; so far responses to Cu deprivation have only been explored in some species of marine denitrifying bacteria and one ammonia oxidizing archaeon (Matsubara et al., 1982; Granger and Ward, 2003; Amin et al., 2013; Moffett et al., 2012). The levels of Cu2+ that limit the growth of Dokd-P16 are one order of magnitude higher ([Cu2+] = 10-15 mol L-1) than those found to reduce the growth of some marine denitrifiers in very similar media ([Cu2+] < 10-16 mol L-1, Moffett et al., 2012). In contrast, free divalent Cu2+ levels (10-15 mol L-1) are two orders of magnitute lower than those limiting ammonia oxidizing archaeon (10-13 mol L-1, Amin et al., 2013). These results suggest that the Flavobacteriia member Dokd-P16 may have Cu requirements higher than those of denitrifying bacteria but lower than those of AOA.
Furthermore, the levels of Cu2+ reported for surface waters at station P16 in the NE Pacific [Cu2+ = 2.5 × 10-15 mol L-1 (10 and 35 m), Semeniuk et al., 2016], where Dokd-P16 was isolated from, fall within the range of Cu levels in our study that resulted in relative growth rates of 74 and 33% μmax in this strain (Cutot treatments of 10 nmol Cu L-1, Cu2+ = 5.6 × 10-15 nmol L-1 and 2 nmol Cu L-1, Cu2+ = 1.38 × 10-15 nmol L-1, respectively). Hence, this bacterium could be experiencing Cu limitation in situ.
At present, little information is available on the ecology of Dokd-P16 isolate and whether it serves as a good model for natural Flavobacteriia populations. However, it is promising that the members of Flavobacteriia responded preferentially to an increase in Cu availability in a recent microcosm study in the Southern Ocean (Flavobacteria–Cytophagia cluster; Ramaiah et al., 2015), supporting our hypothesis of an important role for Cu in Flavobacteriia. However, the causes for the positive response of Flavobacteriia to Cu were not explored in study of Ramaiah et al. (2015). It remains to be determined if Flavobacteriia in wild populations favor high Cu levels for their growth and metabolism as observed for Dokdonia sp. in our study. Given the significant representation of Flavobacteriia in marine surface waters (10–70%, Glöckner et al., 1999; Eilers et al., 2000; Kirchman et al., 2003b; Abell and Bowman, 2005; Williams et al., 2013), as well as their significance in organic matter transformation (particularly the complex, high molecular weight compounds), evaluating the role of Cu in this group is of interest.
Composition of Major Elements in Response to Cu
Microbial biosynthesis requires a balanced stoichiometry of carbon (C), nitrogen (N), and phosphorus (P) (Herbert, 1976). Since bacterial biomass represents a major sink for these elements in the aquatic environment, understanding factors that influence bacterial stoichiometry is critical to predicting the fate of C, N, and P in the ocean. Sulfur is not naturally limiting in the ocean (seawater sulfate concentrations are approximately 28 mmol SO4 L-1; Emerson and Hedges, 2008), and rarely considered in stoichiometric studies. However, bacterial S content is of interest given the involvement of S-containing compounds (e.g., glutathione) in intracellular Cu binding and trafficking (Helbig et al., 2008; Solioz et al., 2010).
In the present study, bacterial C:N and S:P molar ratios did not vary significantly across various Cu growth conditions. In agreement with our findings, C:N ratios were also constant for marine heterotrophic bacteria grown under different Fe regimes (Tortell et al., 1996; Fourquez et al., 2014). These invariant bacterial C:N ratios indicate that assimilatory pathways of C and N in bacteria remain highly interconnected under changing availability of bioactive metals, such as Fe and Cu. The molar ratios of all elements normalized to P (C:N:S:P) and averaged across all Cu levels for different bacterial strains are summarized in Table 5 and compared to the data from literature. Surprisingly, there are only few reports on the major elemental stoichiometry of native marine heterotrophic bacteria (i.e., Fagerbakke et al., 1996; Gundersen et al., 2002). In general, our data are comparable with those studies. The elemental stoichiometry of bacteria investigated here, for both major and minor elements, will be discussed in an upcoming publication.
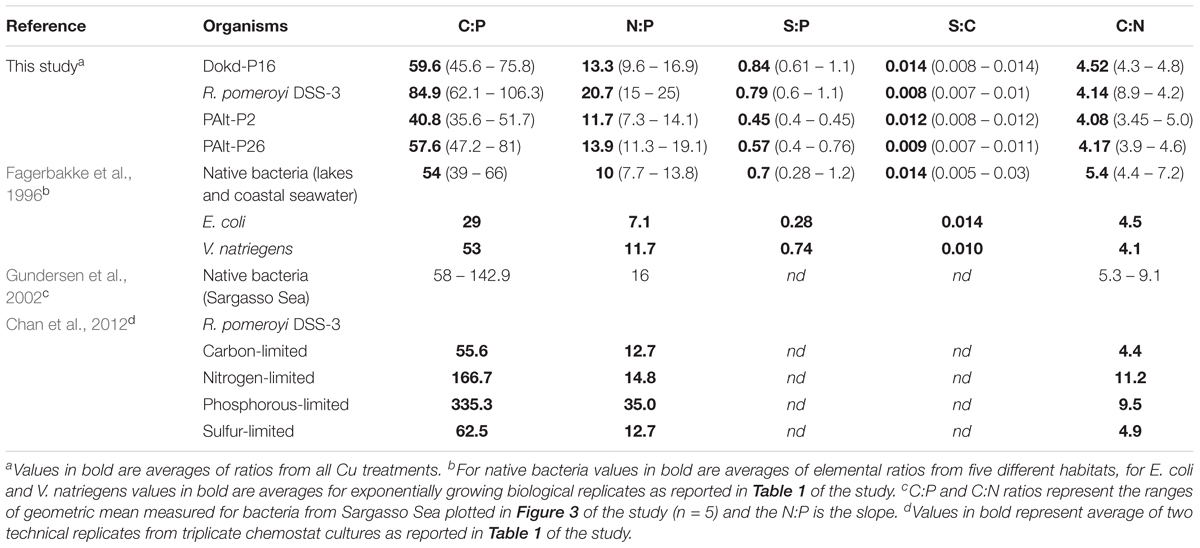
Table 5. Comparison of the molar macronutrient ratios of bacteria used in this study and values reported in literature.
Cu Regulation of Microbial Carbon Metabolism
Microbial growth and metabolism is dependent on the consumption of organic carbon, which at a cellular level is partitioned between biosynthetic (anabolic – bacterial production) and energy-yielding (catabolic – bacterial respiration) processes (del Giorgio and Cole, 1998; Carlson et al., 2007). This partitioning is flexible (Teixeira de Mattos and Neijssel, 1997), a strategy that allows bacteria to maximize growth depending on environmental conditions (Tempest and Neijssel, 1992; Russell and Cook, 1995). One way to parameterize the relationship between cellular catabolism and anabolism is to estimate bacterial growth efficiency (BGE - sometimes termed carbon use efficiency, CUE), which represents the ratio of C allocated toward bacterial biomass production relative to the total amount of C consumed to support bacterial respiration and growth [BGE = BP/(BP + BR)] (del Giorgio and Cole, 1998; Rivkin and Legendre, 2001). The higher the BGE values the more efficient Cu use (i.e., allocation toward bacterial growth) and lower portion of the organic C consumed being respired (as CO2). In contrast, low BGE’s point to a greater loss of C (as CO2) and inefficient energy metabolism, which has been linked to imbalanced substrate stoichiometry (Keiblinger et al., 2010), viral infection (Pradeep Ram et al., 2016), UV exposure (Hörtnagl et al., 2011), and iron deficiency (Tortell et al., 1996; Kirchman et al., 2003a). However, the role of bioactive trace metals, such as Cu, in controlling BGE remains unexplored.
Here, we found a strong connection between C metabolism and Cu availability in a Flavobacteriia member Dokdonia sp. strain Dokd-P16. In this bacterium, Cu starvation led to drastically reduced growth rates (Figure 1), as well as BCD, and slower BR and BP rates (Figure 3). We suspect that slower BR rates could be partly related to the impairment of the respiratory system, since in aerobic bacteria this is the major repository of cellular Cu (in the form of COX; Ridge et al., 2008). On average, the reduction in BP (∼40%) was greater than that in BR (∼20%), indicating a higher flux of C for energy production than for biomass in Cu-limited Dokd-P16, which was confirmed by a reduction in BGE (by 50%). We also found a poor correlation between its growth and respiration rates (R2 = 0.12, Supplementary Figure 5), which has been similarly noted for bacteria growing under Fe limitation, although with a higher statistical strength (R2 = 0.45, Fourquez et al., 2014). This may be explained by the fact that bacteria allocate energy to multiple processes that are independent of cell biomass formation, notably, overflow metabolism, futile cycles and maintenance metabolism (reviewed by Russell and Cook, 1995). The energetic requirements for processes such as cell maintenance (e.g., activation of nutrient uptake system) are likely to increase under constrained growth (Russell and Cook, 1995), which may explain the trends we observed in BR and BP, and ultimately reduced efficiency of C use in Cu-limited Dokd-P16. Iron limitation led to a similar response, lowering BGE in 3 out of 5 marine heterotrophic bacteria (up to 70%, Tortell et al., 1996). Our results suggest a link between Cu availability and the fate of organic C transformed by Dokdonia sp. strain Dokd-P16.
In contrast, the growth rate of R. pomeroyi DSS-3 was not affected by Cu limitation, but its BR and BCD increased substantially (Figure 3 and Supplementary Table 4). Given that BCD provides an estimate of the C needed to support bacterial production (Carlson et al., 2007), its increase in R. pomeroyi DSS-3 under low Cu suggest that the energetic demand to support BP under these conditions is higher than when Cu is replete. Perhaps this reflects the energetic investment in mechanisms allowing R. pomeroyi DSS-3 to maintain constant BP even when Cu is scarce (Figure 3). In this regard, our data suggested that this strain is lowering its Cu use under Cu-limitation (Figure 1). On the other hand, strain PAlt-P26 did not change either its growth nor its Cu quota under varying Cu availability, which may explain why we did not observe any significant modifications to its C metabolism.
Comparison of Bacterial Cu Quotas From This Study With Other Bacteria and Eukaryotic Phytoplankton
The trace metal content of microorganisms reflects the trace metal availability in their growth environment (varying in time and space) and their physiological requirements (as reviewed by Twining and Baines, 2013). There is also some evidence that trace metal availability during microbial evolution played a role in shaping the metal composition of phytoplankton superfamilies (Quigg et al., 2011). Phytoplankton and bacteria originally evolved under drastically different Cu availabilities, but presently little is known about whether their evolutionary histories may be reflected in their present-day metal quotas. To explore this, we compared the Cu quotas of marine heterotrophic bacteria with published values for other prokaryotes (as discussed below), as well as eukaryotic phytoplankton (compared in Figure 4).
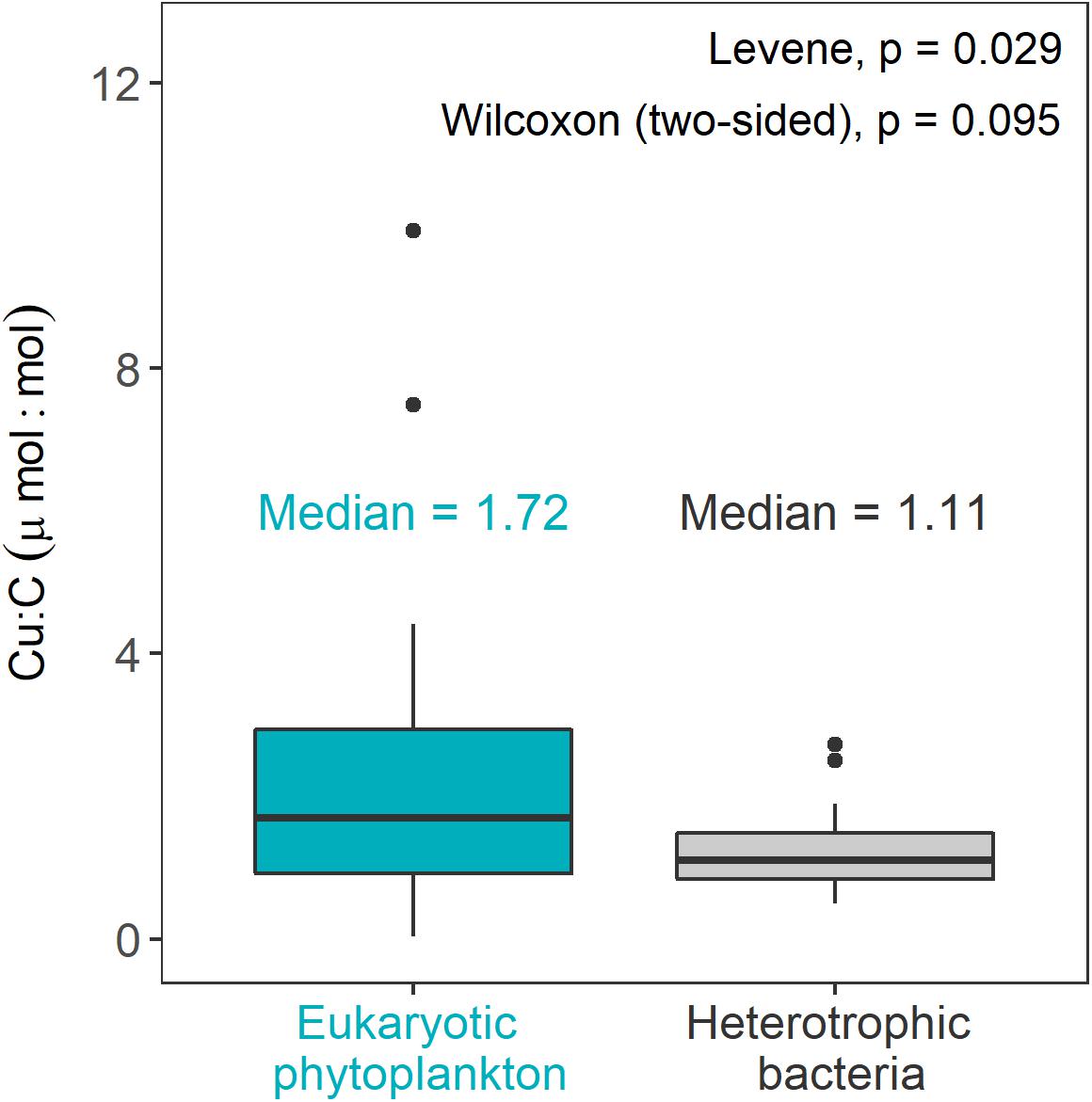
Figure 4. Comparison of Cu:C ratios (μmol:mol) of marine heterotrophic bacteria from this study (data from all Cu treatments, averages of each bacterial strain at each level of Cu) with published Cu:C ratios for eukaryotic phytoplankton (cultured in EDTA-buffered media also at various Cu levels). On each boxplot, the central mark indicates the median, and the box extends to the 25th and 75th percentiles, respectively. Values for eukaryotic phytoplankton are those reported in Sunda and Huntsman (1995), Ho et al. (2003), Annett et al. (2008), Quigg et al. (2011), Guo et al., 2012 (Fe-replete treatments). For studies where eukaryotic phytoplankton Cu quotas were reported as Cu:P (Ho et al., 2003; Quigg et al., 2011) we used C:P ratios accompanying these data to estimate Cu:C for each species. For clarity, errors form different studies were not included. An outlier value of 28 μmol Cu:mol C in the phytoplankton dataset (not shown in the plot) was excluded from statistical analysis. The data used to produce this figure along with the metadata can be found at https://github.com/aposacka/Marine_het_bacteria-copper.
Copper quotas of marine heterotrophic bacteria agree well with those measured for the model heterotroph E. coli, despite differences in culturing conditions (1.8–5.3 μmol Cu:mol C, Outten and O’halloran, 2001; 0.5–3.3 μmol Cu:mol C, Cameron et al., 2012). The range of Cu quotas of our strains and E. coli (0.6–5 μmol Cu:mol C) may, therefore, provide a good approximation of Cu requirements in aerobic heterotrophic prokaryotes.
Copper quotas of heterotrophic bacteria (data pooled from all Cu treatments) are also similar to those of eukaryotic phytoplankton cultured in EDTA-buffered growth Aquil media under various Cu conditions (not statistically significant, Wilcoxon test, p = 0.095; Figure 4). However, relative to marine heterotrophic bacteria Cu quotas of eukaryotic phytoplankton (both cultured in EDTA-buffered growth medium Aquil) are significantly more variable (Levene test, p = 0.026, Figure 4). This comparison should be considered preliminary as the heterotrophic bacteria dataset consists of considerably fewer measurements than the well-studied phytoplankton.
The larger range of Cu quotas in eukaryotic phytoplankton may reflect greater tolerance to a wider range of intracellular Cu or differences in Cu availability in culture media. To explore these factors, we plotted Cu quota data in Figure 4 versus bioavailable Cu concentrations in culture media (inorganic Cu [Cu′]) of both marine heterotrophic bacteria and eukaryotic phytoplankton (Figure 5). In general, heterotrophic bacteria maintain a well-controlled inventory of Cu across varying levels of Cu′, and their average Cu content agrees particularly well with that of eukaryotic phytoplankton and cyanobacteria under low Cu availability (-log [Cu′] < 13, Figure 5). However, as Cu availability increases there is a larger variation in Cu quotas in eukaryotic phytoplankton compared to quotas of heterotrophs. The larger variation in phytoplankton may reflect greater taxonomic variability in their Cu requirements and/or cell size, greater ability to store and/or detoxify Cu intracellularly. In eukaryotic phytoplankton, excess intracellular metals are sequestered by phytochelatins and metallothioneins (cysteine-rich peptides binding metals) and delivered to vacuoles, where these metals can be safely stored (Cobbett and Goldsbrough, 2002). In contrast, bacteria detoxify metals mainly by efflux (see reviews of Nies, 1999; Rensing and McDevitt, 2013), suggesting a tight intracellular trace metal control over a range of concentrations outside the cell. Furthermore, as opposed to eukaryotic phytoplankton and cyanobacteria that have Cu-dependent proteins in the cytosol (e.g., cytochrome c oxidase in the thylakoid plasma and membrane of cyanobacteria, Paumann et al., 2003), there are no known cytosolic Cu proteins in heterotrophic bacteria (Tottey et al., 2005).
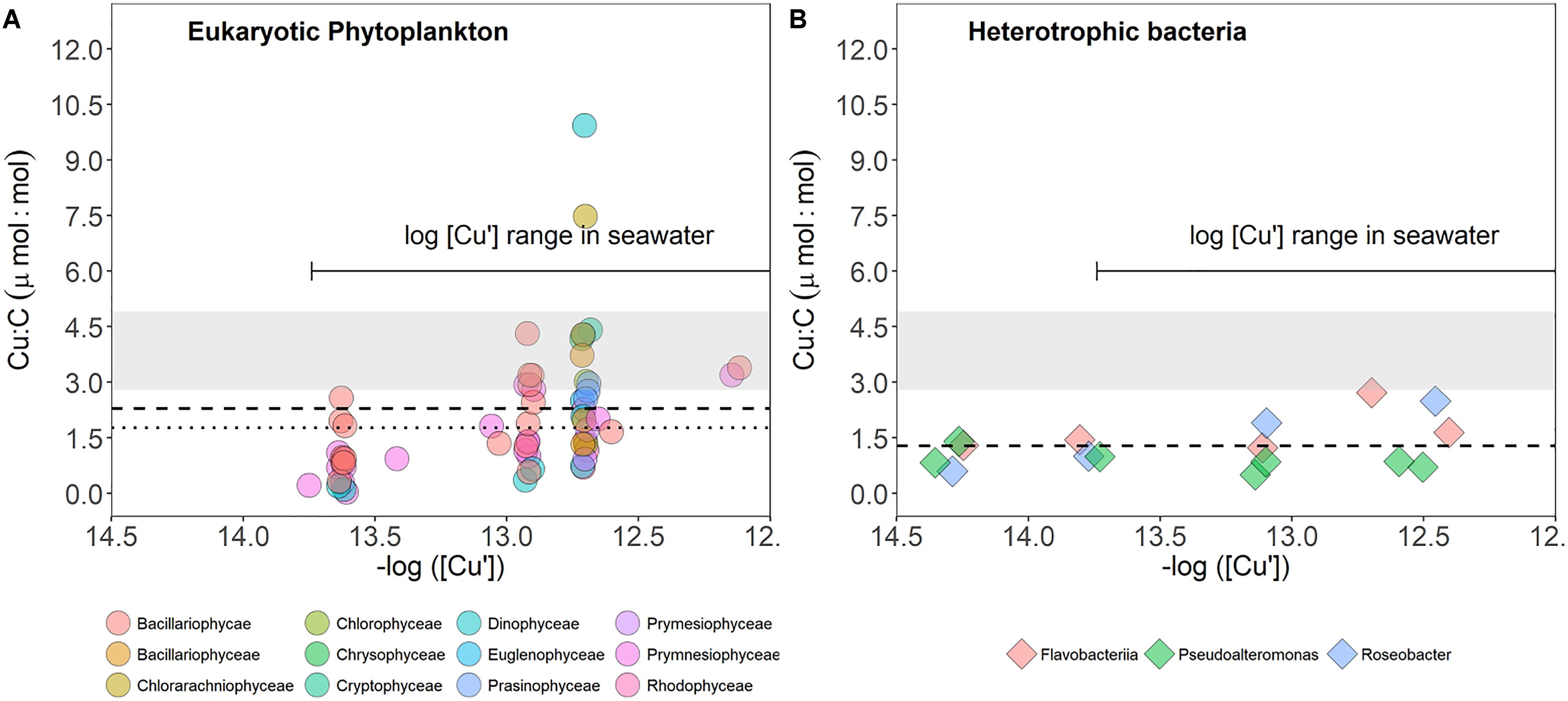
Figure 5. Comparison of Cu quotas (Cu:C, μmol:mol) of cultured marine eukaryotic phytoplankton (A) and marine heterotrophic bacteria from this study (B). Eukaryotic phytoplankton quotas are for cultures grown in Aquil media (Sunda and Huntsman, 1995; Ho et al., 2003; Annett et al., 2008; Quigg et al., 2011; Guo et al., 2012). Data from Ho et al. (2003) and Quigg et al. (2011) were converted from Cu:P to Cu:C using the C:P ratios reported for each species in both studies. Colors in all figures represent different taxa. Dashed lines in both plots show the mean quota averaged across all Cu levels, and dotted line in phytoplankton plot shows the median. The shaded region represents the range of Cu:C remineralization ratios obtained by converting Cu:P ratios (regressions for upper waters, <800 m) reported for different oceans assuming the 106C:1P stoichiometry [from Annett et al., 2008 (Fe-replete waters) and Twining and Baines, 2013]. The log [Cu′] is based on data of Moffett (1995) and Semeniuk et al. (2016). For clarity, errors are not shown and the figure with phytoplankton data does not display an outlier quota of 28 μmol Cu:mol C (Glaucocytophyceae member; Quigg et al., 2011). The data values and the metadata for this figure can be found at https://github.com/aposacka/Marine_het_bacteria-copper. Cu′ represents the concentration of inorganic copper in growth media.
It has been hypothesized that trace metal availability during the evolution of various organisms influenced metal selection for biological usage (Williams and Fraústo da Silva, 2003). Early prokaryotic cells (i.e., cyanobacteria) evolved in an anoxic ocean (Archean, ∼2750 Myr) where Cu chemistry, as indicated by speciation models, was dominated by complexation with sulfide, rendering Cu less bioavailable (Saito et al., 2003). This contrasts with the evolutionary environment of eukaryotic phytoplankton (Paleozoic ∼1000 Myr to modern ocean ∼540 to 120 Myr), which was characterized by higher Cu availability as a result of Earth’s oxygenation (Saito et al., 2003). These considerations have been used to explain the greater sensitivity of modern cyanobacteria to Cu toxicity (Brand et al., 1986; Saito et al., 2003). The narrow range of Cu quotas of heterotrophic bacteria relative to eukaryotic phytoplankton in our preliminary comparison can be linked to greater sensitivity of bacteria to Cu and their strict Cu homeostasis. This may be a result of the contrasting early evolutionary metal conditions of those groups.
Preliminary Assessment of the Role of Marine Heterotrophic Bacteria in the Cu Cycle
Bacteria dominate the living carbon biomass in the ocean (Whitman et al., 1998) and may equate or even exceed phytoplankton biomass in euphotic zones, particularly in oligotrophic regions (Cho and Azam, 1990; Ducklow, 1999). Hence, Cu in these microbes might represent a significant portion of Cu associated with the living biomass (biogenic Cu) in the upper ocean. To explore this, we calculated biogenic Cu associated with heterotrophic bacteria and phytoplankton (algae and cyanobacteria) biomass, using mean Cu:C ratios from culture studies of both groups (Table 6, using 1.36 and 2.4 μmol Cu: for heterotrophic bacteria and phytoplankton, respectively, values represented by dashed lines in Figure 5). For these estimates, we employed euphotic zone integrated heterotrophic bacterial and phytoplankton biomass measurements at station P26 in the NE Pacific in different seasons (Sherry et al., 1999, Table 6). As exemplified using this model system, heterotrophic bacteria may account for ∼50% of the total biogenic Cu (associated with planktonic organisms), and the total biogenic Cu (5.6–34 pM Cu, Table 6) might be a significant fraction (5.9–36%) of the particulate Cu pool in surface waters of the North Pacific (∼94 pM, Nakatsuka et al., 2009). Similar findings have been reported for the North Atlantic by Twining et al. (2015)—the biogenic Cu pool contributing up to ∼10% to the total particulate Cu pool. Furthermore, the impact of heterotrophic bacteria on the dissolved Cu pool can be estimated by calculating their biological Cu demand relative to the dissolved Cu supply. Using conservative estimates of heterotrophic bacterial growth rates (1–5 d-1, Table 3) and an average biogenic heterotrophic bacteria Cu pool of 1 pM (Table 6), the annual heterotrophic bacterial Cu demand would range between 0.36 and 1.8 nM per year. This biogenic Cu demand could be significant relative to the supply rate of Cu mediated by upwelling at P26 (0.58–1.95 nM, Posacka et al., 2017). However, bacterially assimilated Cu is unlikely to be exported into the deep ocean as bacteria are not dense enough to sink on their own. Therefore, accumulation of Cu by heterotrophic bacteria is probably more important in retaining Cu in surface waters, where it could be subject to internal cycles (e.g., recycling through viral lysis) or enter the food web via protozoans that graze upon them.
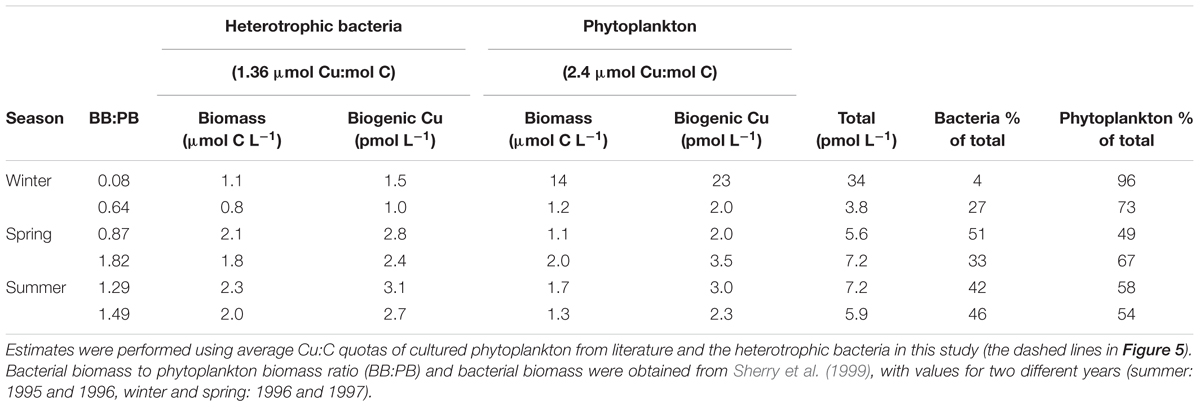
Table 6. Calculations of biogenic Cu concentration (pmol L-1) for marine bacteria and phytoplankton (algae and cyanobacteria) in the seasonal euphotic zone (winter, spring, summer, Sherry et al., 1999) at station P26 in the subarctic NE Pacific.
Data Availability Statement
Data and code for reproducing analyses can be found in publicly available repository on Github: https://github.com/aposacka/Marine_het_bacteria-copper.
Author Contributions
AP, MM, and DS carried out the experimental design. AP conducted the sample collection and analysis and mainly wrote the manuscript. All authors contributed to data interpretation.
Funding
AP and DS were funded by a Natural Sciences and Engineering Research Council of Canada (261521-13) Discovery grant to MM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Nina Schuback for isolation of bacterial strains from the Line P transect, subarctic NE Pacific. We are grateful to Jan Finke and the Suttle Lab for help with flow cytometry analysis of bacterial cells. Thanks to Maureen Soon and Vivian Lai for assistance with elemental analyses with ICP-OES and ICP-MS. Also, thank you to Marion Fourquez, Steven Hallam, Robert Sherell, Yeala Shaked for helpful discussions, as well as JC and two reviewers for their comments on earlier versions of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00523/full#supplementary-material
Footnotes
- ^http://linep.waterproperties.ca/2012-12/index.php
- ^http://www.arb-silva.de/
- ^https://github.com/aposacka/Marine_het_bacteria-copper
- ^https://github.com/aposacka/Marine_het_bacteria-copper
References
Abell, G. C. J., and Bowman, J. P. (2005). Colonization and community dynamics of class Flavobacteria on diatom detritus in experimental mesocosms based on Southern Ocean seawater. FEMS Microbiol. Ecol. 53, 379–391. doi: 10.1016/j.femsec.2005.01.008
Amin, S. A., Moffett, J. W., Martens-Habbena, W., Jacquot, J. E., Han, Y., Devol, A., et al. (2013). Copper requirements of the ammonia-oxidizing archaeon Nitrosopumilus maritimus SCM1 and implications for nitrification in the marine environment. Limnol. Oceanogr. 58, 2037–2045. doi: 10.4319/lo.2013.58.6.2037
Annett, A. L., Lapi, S., Ruth, T. J., and Maldonado, M. T. (2008). The effects of Cu and Fe availability on the growth and Cu:C ratios of marine diatoms. Limnol. Oceanogr. 53, 2451–2461. doi: 10.4319/lo.2008.53.6.2451
Argüello, J. M., Raimunda, D., and Padilla-benavides, T. (2013). Mechanisms of copper homeostasis in bacteria. Front. Cell. Infect. Microbiol. 3:73. doi: 10.3389/fcimb.2013.00073
Aristilde, L., Xu, Y., and Morel, F. M. M. (2012). Weak organic ligands enhance zinc uptake in marine phytoplankton. Environ. Sci. Technol. 46, 5438–5445. doi: 10.1021/es300335u
Armstrong, E., Granger, J., Mann, E. L., and Price, N. M. (2004). Outer-membrane siderophore receptors of heterotrophic oceanic bacteria. Limnol. Oceanogr. 49, 579–587. doi: 10.4319/lo.2004.49.2.0579
Berggren, M., Lapierre, J. F., and del Giorgio, P. A. (2012). Magnitude and regulation of bacterioplankton respiratory quotient across freshwater environmental gradients. ISME J. 6, 984–993. doi: 10.1038/ismej.2011.157
Bertrand, E. M., Saito, M. A., Lee, P. A., Dunbar, R. B., Sedwick, P. N., and Ditullio, G. R. (2011). Iron limitation of a springtime bacterial and phytoplankton community in the Ross Sea: implications for vitamin B12 nutrition. Front. Microbiol. 2:160. doi: 10.3389/fmicb.2011.00160
Bogachev, A. V., Bertsova, Y. V., Verkhovskaya, M. L., Mamedov, M. D., and Skulachev, V. P. (2016). Real-time kinetics of electrogenic Na+ transport by rhodopsin from the marine flavobacterium Dokdonia sp. PRO95. Sci. Rep. 6:21397. doi: 10.1038/srep21397
Bondarczuk, K., and Piotrowska-Seget, Z. (2013). Molecular basis of active copper resistance mechanisms in Gram-negative bacteria. Cell Biol. Toxicol. 29, 397–405. doi: 10.1007/s10565-013-9262-1
Brand, L. E., Sunda, W. G., and Guillard, R. R. L. (1986). Reduction of marine phytoplankton reproduction rates by copper and cadmium. J. Exp. Mar. Bio. Ecol. 96, 225–250. doi: 10.1016/0022-0981(86)90205-4
Brussaard, C. P. D. (2004). Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70, 1506–1513. doi: 10.1128/AEM.70.3.1506
Buchan, A., LeCleir, G. R., Gulvik, C. A., and González, J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Cameron, V., House, C. H., and Brantley, S. L. (2012). A first analysis of metallome biosignatures of hyperthermophilic archaea. Archaea 2012:789278. doi: 10.1155/2012/789278
Carlson, C., del Giorgio, P., and Herndl, G. (2007). Microbes and the dissipation of energy and respiration: from cells to ecosystems. Oceanography 20, 89–100. doi: 10.5670/oceanog.2007.52
Chan, L.-K., Newton, R. J., Sharma, S., Smith, C. B., Raypati, P., Limardo, A. J., et al. (2012). Transcriptional changes underlying elemental stoichiometry shifts in a marine heterotrophic bacterium. Front. Microbiol. 3:159. doi: 10.3389/fmicb.2012.00159
Changela, A., Chen, K., Xue, Y., Holschen, J., Outten, C. E., and Halloran, T. V. O. (2003). Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1386. doi: 10.1126/science.1085950
Cho, B., and Azam, F. (1990). Biogeochemical significance of bacterial biomass in the ocean’s euphotic zone. Mar. Ecol. Prog. Ser. 63, 253–259. doi: 10.3354/meps063253
Cobbett, C., and Goldsbrough, P. (2002). Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53, 159–182. doi: 10.1146/annurev.arplant.53.100301.135154
Croot, P. L., Karlson, B., ven Elteren, J. T., and Kroon, J. J. (2003). Uptake and efflux of 64Cu by the marine cyanobacterium Synechococcus (WH7803). Limnol. Oceanogr. 48, 179–188. doi: 10.4319/lo.2003.48.1.0179
Cunliffe, M. (2013). Physiological and metabolic effects of carbon monoxide oxidation in the model marine bacterioplankton Ruegeria pomeroyi DSS-3. Appl. Environ. Microbiol. 79, 738–740. doi: 10.1128/AEM.02466-12
del Giorgio, P. A., and Cole, J. J. (1998). Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst. 29, 503–541. doi: 10.1146/annurev.ecolsys.29.1.503
Ducklow, H. W. (1999). Minireview: the bacterial content of the oceanic euphotic zone. FEMS Microbiol. 30, 1–10. doi: 10.1016/S0168-6496(99)00031-8
Dupont, C. L., Grass, G., and Rensing, C. (2011). Copper toxicity and the origin of bacterial resistance — new insights and applications. Metallomics 3, 1109–1118. doi: 10.1039/c1mt00107h
Durham, B. P., Sharma, S., Luo, H., Smith, C. B., Amin, S. A., Bender, S. J., et al. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 112, 453–457. doi: 10.1073/pnas.1413137112
Eilers, H., Pernthaler, J., Glöckner, F., and Amann, R. (2000). Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66, 3044–3051. doi: 10.1128/AEM.66.7.3044-3051.2000
Emerson, S., and Hedges, J. (2008). Chemical Oceanography and the Marine Carbon Cycle, 1st Edn. Cambridge: Cambridge University Press, 13. doi: 10.1017/CBO9780511793202
Fagerbakke, K. M., Heldal, M., and Norland, S. (1996). Content of carbon, nitrogen, oxygen, sulfur and phosphorus in native aquatic and cultured bacteria. Aquat. Microb. Ecol. 10, 15–27. doi: 10.3354/ame010015
Fourquez, M., Devez, A., Schaumann, A., Guéneuguès, A., Jouenne, T., Obernosterer, I., et al. (2014). Effects of iron limitation on growth and carbon metabolism in oceanic and coastal heterotrophic bacteria. Limnol. Oceanogr. 59, 349–360. doi: 10.4319/lo.2014.59.2.0349
Glöckner., F. O., Fuchs, B. M., and Amann, R. (1999). Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65, 3721–3726.
Godwin, C. M., and Cotner, J. B. (2015). Aquatic heterotrophic bacteria have highly flexible phosphorus content and biomass stoichiometry. ISME J. 9, 1–4. doi: 10.1038/ismej.2015.34
Godwin, C. M., and Cotner, J. B. (2018). What intrinsic and extrinsic factors explain the stoichiometric diversity of aquatic heterotrophic bacteria? ISME J. 12, 598–609. doi: 10.1038/ismej.2017.195
Gómez-Consarnau, L., González, J. M., Coll-Lladó, M., Gourdon, P., Pascher, T., Neutze, R., et al. (2007). Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445, 210–213. doi: 10.1038/nature05381
Gordon, A. S., Harwood, V. J., and Sayyar, S. (1993). Growth, copper-tolerant cells, and extracellular protein production in copper-stressed chemostat cultures of Vibrio alginolyticus. Appl. Environ. Microbiol. 59, 60–66.
Gordon, A. S., Howell, L. D., and Harwood, V. (1994). Responses of diverse heterotrophic bacteria to elevated copper concentrations. Can. J. Microbiol. 40, 408–411. doi: 10.1139/m94-067
Granger, J., and Price, N. M. (1999). The importance in iron nutrition of siderophores of heterotrophic marine bacteria. Limnol. Oceanogr. 44, 541–555. doi: 10.4319/lo.1999.44.3.0541
Granger, J., and Ward, B. B. (2003). Accumulation of nitrogen oxides in copper-limited cultures of denitrifying bacteria. Limnol. Oceanogr. 48, 313–318. doi: 10.4319/lo.2003.48.1.0313
Green, R. T., Todd, J. D., and Johnston, A. W. B. (2012). Manganese uptake in marine bacteria; the novel MntX transporter is widespread in Roseobacters, Vibrios, Alteromonadales and the SAR11 and SAR116 clades. ISME J. 7, 581–591. doi: 10.1038/ismej.2012.140
Gundersen, K., Heldal, M., Norland, S., Purdie, D. A., and Knap, A. H. (2002). Elemental C, N, and P cell content of individual bacteria collected at the Bermuda Atlantic Time-Series Study (BATS) site. Limnol. Oceanogr. 47, 1525–1530. doi: 10.4319/lo.2002.47.5.1525
Guo, J., Lapi, S., Ruth, T. J., and Maldonado, M. T. (2012). The effects of iron and copper availability on the copper stoichiometry of marine phytoplankton. J. Phycol. 48, 312–325. doi: 10.1111/j.1529-8817.2012.01133.x
Gustafsson, J. P. (2016). Visual MINTEQ (v3.0). A Windows version of MINTEQA2. Available at: http://vminteq.lwr.kth.se/
Hagström,Å., Azam, F., Berg, C., and Zweifel, U. (2017). Isolates as models to study bacterial ecophysiology and biogeochemistry. Aquat. Microb. Ecol. 80, 15–27. doi: 10.3354/ame01838
Helbig, K., Bleuel, C., Krauss, G. J., and Nies, D. H. (2008). Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 190, 5431–5438. doi: 10.1128/JB.00271-08
Heller, M. I., and Croot, P. L. (2015). Copper speciation and distribution in the Atlantic sector of the Southern Ocean. Mar. Chem. 173, 253–268. doi: 10.1016/j.marchem.2014.09.017
Herberich, E., Sikorski, J., and Hothorn, T. (2010). A robust procedure for comparing multiple means under heteroscedasticity in unbalanced designs. PLoS One 5:e9788. doi: 10.1371/journal.pone.0009788
Herbert, D. (1976). “Stoichiometric aspects of microbial growth,” in Continuous Culture: 6 Applications and New Fields, eds A. C. R. Dean, D. C. Ellwood, C. G. T. Evans, and J. Melling (Chichester: Ellis Horwood), 1–30.
Ho, T.-Y., Quigg, A., Finkel, Z. V., Milligan, A. J., Wyman, K., Falkowski, P. G., et al. (2003). The elemental composition of some marine phytoplankton. J. Phycol. 39, 1145–1159. doi: 10.1111/j.0022-3646.2003.03-090.x
Hörtnagl, P., Pérez, M. T., and Sommaruga, R. (2011). Contrasting effects of ultraviolet radiation on the growth efficiency of freshwater bacteria. Aquat. Ecol. 45, 125–136. doi: 10.1007/s10452-010-9341-9
Jacquot, J. E., Kondo, Y., Knapp, A. N., and Moffett, J. W. (2013). The speciation of copper across active gradients in nitrogen-cycle processes in the eastern tropical South Pacific. Limnol. Oceanogr. 58, 1387–1394. doi: 10.4319/lo.2013.58.4.1387
Karner, M. B., DeLong, E. F., and Karl, D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
Keiblinger, K. M., Hall, E. K., Wanek, W., Szukics, U., Hämmerle, I., Ellersdorfer, G. G., et al. (2010). The effect of resource quantity and resource stoichiometry on microbial carbon-use-efficiency. FEMS Microbiol. Ecol. 73, 430–440. doi: 10.1111/j.1574-6941.2010.00912.x
Kim, J.-M., Baars, O., and Morel, M. M. (2015). Bioavailability and electroreactivity of zinc complexed to strong and weak organic ligands. Environ. Sci. Technol. 49, 10894–10902. doi: 10.1021/acs.est.5b02098
Kimura, H., Young, C. R., Martinez, A., and Delong, E. F. (2011). Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium. ISME J. 5, 1641–1651. doi: 10.1038/ismej.2011.36
Kirchman, D. L., Hoffman, K. A., Weaver, R., and Hutchins, D. A. (2003a). Regulation of growth and energetics of a marine bacterium by nitrogen source and iron availability. Mar. Ecol. Prog. Ser. 250, 291–296. doi: 10.3354/meps250291
Kirchman, D. L., Yu, L., and Cottrell, M. T. (2003b). Diversity and abundance of uncultured Cytophaga -like bacteria in the Delaware Estuary. Appl. Environ. Microbiol. 69, 6587–6596. doi: 10.1128/AEM.69.11.6587
Lebaron, P., Servais, P., Troussellier, M., Courties, C., Muyzer, G., Bernard, L., et al. (2001). Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34, 243–253. doi: 10.1111/j.1574-6941.2001.tb00776.x
Lucas, J., Wichels, A., Teeling, H., Chafee, M., Scharfe, M., and Gerdts, G. (2015). Annual dynamics of North Sea bacterioplankton: seasonal variability superimposes short-term variation. FEMS Microbiol. Ecol. 91, 1–11. doi: 10.1093/femsec/fiv099
Macomber, L., and Imlay, J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349. doi: 10.1073/pnas.0812808106
Maldonado, M. T., Allen, A. E., Chong, J. S., Lin, K., Leus, D., Karpenko, N., et al. (2006). Copper-dependent iron transport in coastal and oceanic diatoms. Limnol. Oceanogr. 51, 1729–1743. doi: 10.4319/lo.2006.51.4.1729
Maldonado, M. T., and Price, N. M. (1999). Utilization of Fe bound to strong organic ligands by phytoplankton communities in the subarctic Pacific Ocean. Deep Res. II 46, 2447–2473. doi: 10.1016/S0967-0645(99)00071-5
Maldonado, M. T., Surma, S., and Pakhomov, E. A. (2016). Southern Ocean biological iron cycling in the pre-whaling and present ecosystems. Philos. Trans. R. Soc. A 374:20150292. doi: 10.1098/not
Mann, E. L., Ahlgren, N., Moffett, J. W., and Chisholm, S. W. (2002). Copper toxicity and cyanobacteria ecology in the Sargasso Sea. Limnol. Oceanogr. 47, 976–988. doi: 10.4319/lo.2002.47.4.0976
Matsubara, T., Frunzke, K., and Zumft, W. G. (1982). Modulation by copper of the products of nitrite respiration in Pseudomonas perfectomarinus. J. Bacteriol. 149, 816–823.
Moffett, J. W. (1995). Temporal and spatial variability of copper complexation by strong chelators in the Sargasso Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 42, 1273–1295. doi: 10.1016/0967-0637(95)00060-J
Moffett, J. W., and Brand, L. E. (1996). Production of strong, extracellular Cu chelators by marine cyanobacteria in response to Cu stress. Limnol. Oceanogr. 41, 388–395. doi: 10.4319/lo.1996.41.3.0388
Moffett, J. W., Tuit, C. B., and Ward, B. B. (2012). Chelator-induced inhibition of copper metalloenzymes in denitrifying bacteria. Limnol. Oceanogr. 57, 272–280. doi: 10.4319/lo.2012.57.1.0272
Nakatsuka, S., Okamura, K., Takeda, S., Nishioka, J., Lutfi Firdaus, M., Norisuye, K., et al. (2009). Behaviors of dissolved and particulate Co, Ni, Cu, Zn, Cd and Pb during a mesoscale Fe-enrichment experiment (SEEDS II) in the western North Pacific. Deep Res. Part II Top. Stud. Oceanogr. 56, 2822–2838. doi: 10.1016/j.dsr2.2009.06.008
Nies, D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–751. doi: 10.1007/s002530051457
Outten, C. E., and O’halloran, T. V. (2001). Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492. doi: 10.1126/science.1060331
Pakulski, J. D., Coffin, R. B., Kelley, C. A., Holder, S. L., Downer, R., Aas, P., et al. (1996). Iron stimulation of Antarctic bacteria. Nature 383, 133–134. doi: 10.1038/383133b0
Paumann, M., Lubura, B., Regelsberger, G., Feichtinger, M., Köllensberger, G., Jakopitsch, et al. (2003). Soluble CuA domain of cyanobacterial cytochrome c oxidase. J. Biol. Chem. 279, 10293–10303. doi: 10.1074/jbc.M308903200
Pedler, B. E., Aluwihare, L. I., and Azam, F. (2014). Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 7202–7207. doi: 10.1073/pnas.1401887111
Posacka, A. M., Semeniuk, D. M., Whitby, H., van den Berg, C. M. G., Cullen, J. T., Orians, K., et al. (2017). Dissolved copper (dCu) biogeochemical cycling in the subarctic Northeast Pacific and a call for improving methodologies. Mar. Chem. 196, 47–61. doi: 10.1016/j.marchem.2017.05.007
Pradeep Ram, A. S. P., Colombet, J., Perriere, F., Thouvenot, T., and Sime-Ngando, T. (2016). Viral regulation of prokaryotic carbon metabolism in a hypereutrophic freshwater reservoir ecosystem (Villerest. France). Front. Microbiol. 7:81. doi: 10.3389/fmicb.2016.00081
Price, N. M., Harrison, G. I., Hering, J., Hudson, R. J., Pascale, M. V. N., Palenik, B., et al. (1989). Preparation and chemistry of the artificial algal culture medium aquil. Biol. Oceanogr. 6, 443–461. doi: 10.1080/01965581.1988.10749544
Quigg, A., Irwin, A. J., and Finkel, Z. V. (2011). Evolutionary inheritance of elemental stoichiometry in phytoplankton. Proc. Biol. Sci. 278, 526–534. doi: 10.1098/rspb.2010.1356
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ramaiah, N., Jain, A., Meena, R. M., Naik, R. K., Verma, R., Bhat, M., et al. (2015). Response of bacteria and phytoplankton from a subtropical front location Southern Ocean to micronutrient amendments ex-situ. Deep Sea Res. Part II Top. Stud. Oceanogr. 118, 209–220. doi: 10.1016/j.dsr2.2015.03.013
Reinthaler, T., and Herndl, G. J. (2005). Seasonal dynamics of bacterial growth efficiencies in relation to phytoplankton in the southern North Sea. Aquat. Microb. Ecol. 39, 7–16. doi: 10.3354/ame039007
Reisch, C. R., Moran, M. A., and Whitman, W. B. (2011). Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2:172. doi: 10.3389/fmicb.2011.00172
Rensing, C., and Grass, G. (2003). Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213. doi: 10.1016/S0168-6445(03)00049-4
Rensing, C., and McDevitt, S. F. (2013). “The copper metallome in prokaryotic cells,” in Metallomics and the Cell, ed. L. Banci (Dordrecht: Springer), 417–450.
Ridge, P. G., Zhang, Y., and Gladyshev, V. N. (2008). Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One 3:e1378. doi: 10.1371/journal.pone.0001378
Riedel, T., Gómez-Consarnau, L., Tomasch, J., Martin, M., Jarek, M., González, J. M., et al. (2013). Genomics and physiology of a marine flavobacterium encoding a proteorhodopsin and a xanthorhodopsin-like protein. PLoS One 8:e57487. doi: 10.1371/journal.pone.0057487
Rivkin, R. B., and Legendre, L. (2001). Biogenic carbon cycling in the upper ocean: effects of microbial respiration. Science 291, 2398–2400. doi: 10.1126/science.291.5512.2398
Rowley, D. A., and Halliwell, B. (1983). Superoxide-dependent and ascorbate-dependent formation of hydroxyl radicals in the presence of copper salts: a physiologically significant reaction? Arch. Biochem. Biophys. 225, 279–284. doi: 10.1016/0003-9861(83)90031-0
Russell, J. B., and Cook, G. M. (1995). Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Mol. Biol. Rev. 59, 48–62.
Saito, M. A., Sigman, D. M., and Morel, F. M. M. (2003). The bioinorganic chemistry of the ancient ocean: the co-evolution of cyanobacterial metal requirements and biogeochemical cycles at the Archean-Proterozoic boundary? Inorganica Chim. Acta 356, 308–318. doi: 10.1016/S0020-1693(03)00442-0
Santos-Echeandía, J., Laglera, L. M., Prego, R., and van den Berg, C. M. G. (2008). Copper speciation in estuarine waters by forward and reverse titrations. Mar. Chem. 108, 148–158. doi: 10.1016/j.marchem.2007.11.004
Schreiber, D. R., Gordon, A. S., and Millero, F. J. (1985). The toxicity of copper to the marine bacterium Vibrio alginolyticus. Can. J. Microbiol. 31, 83–87. doi: 10.1016/j.marpolbul.2016.07.031
Semeniuk, D., Bundy, R., Payne, C., and Barbeau, K. (2015). Acquisition of organically complexed copper by marine phytoplankton and bacteria in the northeast subarctic Pacific Ocean. Mar. Chem. 173, 222–233. doi: 10.1016/j.marchem.2015.01.005
Semeniuk, D. M. (2014). Copper Nutrition and Transport Mechanisms in Plankton Communities in the Northeast Pacific Ocean. Available at: https://open.library.ubc.ca/cIRcle/collections/ubctheses/24/items/1.0165543
Semeniuk, D. M., Bundy, R. M., Posacka, A. M., Robert, M., Barbeau, K. A., and Maldonado, M. T. (2016). Using 67Cu to study the biogeochemical cycling of copper in the Northeast subarctic Pacific Ocean. Front. Mar. Sci. 3:78. doi: 10.3389/fmars.2016.00078
Sherry, N. D., Boyd, P. W., Sugimoto, K., and Harrison, P. J. (1999). Seasonal and spatial patterns of heterotrophic bacterial production, respiration, and biomass in the subarctic NE Pacific. Deep Sea Res. Part II 46, 2557–2578. doi: 10.1016/S0967-0645(99)00076-4
Sijerčić, A., and Price, N. M. (2015). Hydroxamate siderophore secretion by Pseudoalteromonas haloplanktis during steady-state and transient growth in Fe-limited culture. Mar. Ecol. Prog. Ser. 531, 105–120. doi: 10.3354/meps11338
Solioz, M., Abicht, H. K., Mermod, M., and Mancini, S. (2010). Response of gram-positive bacteria to copper stress. J. Biol. Inorg. Chem. 15, 3–14. doi: 10.1007/s00775-009-0588-3
Solioz, M., and Stoyanov, J. V. (2003). Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27, 183–195. doi: 10.1016/S0168-6445(03)00053-6
Sunda, W. G., and Huntsman, S. A. (1995). Regulation of copper concentration in the oceanic nutricline by phytoplankton uptake and regeneration cycles. Limnol. Oceanogr. 40, 132–137. doi: 10.4319/lo.1995.40.1.0132
Sunda, W. G., and Lewis, J. A. M. (1978). Effect of complexation by natural organic ligands on the toxicity of copper to a unicellular alga, Monochrysis lutheri. Limnol. Oceanogr. 23, 870–876. doi: 10.4319/lo.1978.23.5.0870
Tang, D., and Morel, M. M. (2006). Distinguishing between cellular and Fe-oxide-associated trace elements in phytoplankton. Mar. Chem. 98, 18–30. doi: 10.1016/j.marchem.2005.06.003
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Teixeira de Mattos, M. J., and Neijssel, O. M. (1997). Bioenergetic consequences of microbial adaptation to low-nutrient environments. J. Biotechnol. 59, 117–126. doi: 10.1016/S0168-1656(97)00174-0
Tempest, D. W., and Neijssel, O. M. (1992). Physiological and energetic aspects of bacterial metabolite overproduction. FEMS Microbiol. Lett. 100, 169–170. doi: 10.1016/0378-1097(92)90205-3
Todd, J. D., Kirkwood, M., Newton-Payne, S., and Johnston, A. W. B. (2012). DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 6, 223–226. doi: 10.1038/ismej.2011.79
Tortell, P. D., Maldonado, M. T., and Price, N. M. (1996). The role of heterotrophic bacteria in iron-limited ocean ecosystems. Nature 383, 330–331. doi: 10.1038/383330a0
Tottey, S., Harvie, D. R., and Robinson, N. J. (2005). Understanding how cells allocate metals using metal sensors and metallochaperones. Acc. Chem. Res. 38, 775–783. doi: 10.1021/ar0300118
Twining, B. S., and Baines, S. B. (2013). The trace metal composition of marine phytoplankton. Ann. Rev. Mar. Sci. 5, 191–215. doi: 10.1146/annurev-marine-121211-172322
Twining, B. S., Rauschenberg, S., Morton, P. L., and Vogt, S. (2015). Metal contents of phytoplankton and labile particulate material in the North Atlantic Ocean. Prog. Oceanogr. 137, 261–283. doi: 10.1016/j.pocean.2015.07.001
Walsh, M. J., Goodnow, S. D., Vezeau, G. E., Richter, L. V., and Ahner, B. A. (2015). Cysteine enhances bioavailability of copper to marine phytoplankton. Environ. Sci. Technol. 49, 12145–12152. doi: 10.1021/acs.est.5b02112
Weaver, R. S., Kirchman, D. L., and Hutchins, D. A. (2003). Utilization of iron/organic ligand complexes by marine bacterioplankton. Aquat. Microb. Ecol. 31, 227–239. doi: 10.3354/ame031227
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Perspective Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. doi: 10.1016/j.cbpa.2016.05.002
Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer. doi: 10.1007/978-0-387-98141-3
Wickham, H. (2016). Dplyr: a Grammar of Data Manipulation, version 0.5.0 (R package). Available at: https://github.com/tidyverse/dplyr
Wilhelm, S. W., King, A. L., Twining, B. S., LeCleir, G. R., DeBruyn, J. M., Strzepek, R. F., et al. (2013). Elemental quotas and physiology of a southwestern Pacific Ocean plankton community as a function of iron availability. Aquat. Microb. Ecol. 68, 185–194. doi: 10.3354/ame01611
Wilke, C. O., and Wickham, H. (2016). cowplot: Streamlined Plot Theme and Plot Annotations for “ggplot2” (R package). Available at: https://cran.r-project.org/web/packages/cowplot/index.html
Williams, R. J. P., and Fraústo da Silva, J. J. R. (2003). Evolution was chemically constrained. J. Theor. Biol. 220, 323–343. doi: 10.1006/jtbi.2003.3152
Williams, T. J., Wilkins, D., Long, E., Evans, F., Demaere, M. Z., Raftery, M. J., et al. (2013). The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 15, 1302–1317. doi: 10.1111/1462-2920.12017
Keywords: copper, Cu quota, limitation, marine heterotrophic bacteria, Flavobacteriia, Roseobacteria, Alteromonadales, Dokdonia sp.
Citation: Posacka AM, Semeniuk DM and Maldonado MT (2019) Effects of Copper Availability on the Physiology of Marine Heterotrophic Bacteria. Front. Mar. Sci. 5:523. doi: 10.3389/fmars.2018.00523
Received: 18 May 2018; Accepted: 21 December 2018;
Published: 05 February 2019.
Edited by:
James Cotner, University of Minnesota Twin Cities, United StatesReviewed by:
Anne W. Thompson, Portland State University, United StatesChristian Schlosser, GEOMAR Helmholtz Centre for Ocean Research Kiel, Germany
Copyright © 2019 Posacka, Semeniuk and Maldonado. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna M. Posacka, cG9zYWNrYWFubmFAZ21haWwuY29t
 Anna M. Posacka
Anna M. Posacka David M. Semeniuk
David M. Semeniuk Maria T. Maldonado
Maria T. Maldonado