- 1Marine Science Institute, The University of Texas at Austin, Port Aransas, TX, United States
- 2Laboratoire d’Océanologie et des Geosciences, UMR LOG 8187, Université du Littoral Côte d’Opale, Wimereux, France
- 3Coastal Watershed Institute, Florida Gulf Coast University, Fort Myers, FL, United States
- 4Center for Marine and Environmental Studies, University of the Virgin Islands, St. Thomas, VI, United States
Population genetic studies provide insights into intraspecific diversity and dispersal patterns of microorganisms such as protists, which help understanding invasions, harmful algal bloom development and occurrence of seafood poisoning. Spatial genetic differentiation has been reported in many microbial species indicating significant dispersal barriers among different habitats. Temporal differentiation has been less studied and its frequency, drivers, and magnitude are thus relatively poorly understood. The toxic dinoflagellate species Gambierdiscus caribaeus was sampled during 2 years in the Florida Keys, and repeatedly from 2006 to 2016 at St. Thomas, US Virgin Islands (USVI), including a 3-year period with monthly sampling, enabling a comparison of spatial and temporal genetic differentiation. Samples from the USVI site showed high temporal variability in local population structure, which correlated with changes in salinity and benthic habitat cover. In some cases, temporal variability exceeded spatial differentiation, despite apparent lack of connectivity and dispersal across the Greater Caribbean Region based on the spatial genetic data. Thus, local processes such as selection might have a stronger influence on population structure in microorganisms than geographic distance. The observed high temporal genetic diversity challenges the prediction of harmful algal blooms and toxin concentrations, but illustrates also the evolutionary potential of microalgae to respond to environmental change.
Introduction
Population genetic studies of microorganisms provide valuable insights into intraspecific variability and evolutionary processes due to their short generation times and large population sizes (Elena and Lenski, 2003). In most species, successful genotypes can quickly increase in number due to asexual reproduction and impact the population structure on relatively short time scales, demonstrating microevolution in “fast motion” (e.g., Richlen et al., 2012). Microorganisms such as microalgae can thus be useful study organisms to investigate evolutionary response to different environmental conditions, e.g., after dispersal or due to climate change, and its impact on population structure (Lynch et al., 1991). Recent population genetic studies on microalgae have described patterns that are also found in multicellular organisms. The majority of the investigated, mainly planktonic microalgal species display genetically distinct populations and exhibit significant divergence over different spatial scales despite the absence of obvious dispersal barriers in most aquatic habitats (Foissner, 2006; Vanormelingen et al., 2008; Lowe et al., 2010; Rengefors et al., 2012; Godhe et al., 2016). This divergence can result from limited gene flow between populations due to isolation by geographic distance (Casteleyn et al., 2010; Orsini et al., 2013). Additionally, environmental variables have been shown to significantly impact genetic differentiation in microalgae (Boenigk et al., 2007; Souffreau et al., 2013; Rengefors et al., 2014) due to natural selection and local adaptation of populations (Hendry, 2004).
Compared to the multitude of studies examining geographic genetic structure, temporal genetic differentiation in microalgae has received less attention despite the rapid temporal fluctuation of environmental conditions in most habitats. This variation can cause frequent changes in intraspecific diversity and population structure in organisms with short generation times. For example, shifts in decadal population structure, assessed from resting stages in sediment cores, were observed in the dinoflagellate Pentapharsodinium dalei, and were likely linked to changes in hydrographic conditions (Lundholm et al., 2017). Investigations of temporal variability of the marine diatom Pseudo-nitzschia multistriata in the Gulf of Naples found that two genetically distinct populations were present over the course of 4 years. This succession over consecutive years might be due to differing reproductive success and dispersal of regionally distinct populations (Tesson et al., 2014). Similar patterns of genetic differentiation between years were observed in marine dinoflagellates (Dia et al., 2014) and freshwater raphidophytes (Lebret et al., 2012). Other studies showed that genetic diversity can rapidly change over the course of microalgal blooms, causing significant intra-annual genetic differentiation (Rynearson et al., 2006; Erdner et al., 2011; Richlen et al., 2012; Tammilehto et al., 2016; Ruggiero et al., 2017). Such changes in population structure can occur on temporal scales from a few days to a few months. High-resolution sampling over several years is needed to capture short-term and long-term trends in intraspecific genetic diversity and population structure of microalgae, and to directly compare genetic differentiation on these different temporal scales.
The benthic dinoflagellate genus Gambierdiscus occurs globally in tropical and sub-tropical ecosystems preferentially growing on macroalgae. Several species within this genus produce potent toxins (Bagnis et al., 1980; Litaker et al., 2017), which cause ciguatera fish poisoning worldwide (Van Dolah, 2000; Chateau-Degat et al., 2005; Friedman et al., 2017). The incidence and range of ciguatera appear to be increasing due to a decline in coral reefs and spreading of macroalgae, which provide suitable habitat for Gambierdiscus (Hallegraeff, 2010; Kohli et al., 2015). Best estimates indicate that more than 50,000 people are affected globally every year making it the most prominent and widespread human illness caused by harmful algae (Fleming et al., 1998; Van Dolah, 2000). The toxins produced by Gambierdiscus enter the food web when herbivorous fish consume these epiphytic microalgae during grazing on macroalgae, and are subsequently passed up the food chain. Ciguatera fish poisoning is very difficult to predict, as cell concentrations vary significantly over time and space due to changes in environmental conditions such as nutrient concentrations and sea surface temperature (Parsons et al., 2010). Furthermore, toxin contents differ significantly among Gambierdiscus species, and to a lesser extent among strains of the same species (Litaker et al., 2010, 2017; Pisapia et al., 2017). Community composition and population structure could thus influence the incidence, magnitude and duration of toxic outbreaks (Litaker et al., 2010). Intraspecific diversity and population structure, however, have not been investigated in any Gambierdiscus species. Genetic differentiation could be very pronounced in such epiphytic dinoflagellates, as they might experience less dispersal through ocean currents compared with planktonic microalgae. Additionally, locally anchored populations might exhibit strong adaptations to native environmental conditions, which could further enhance genetic differentiation from other geographically separated populations. Gambierdiscus occurs in varying concentrations year round in the tropics and subtropics (Chinain et al., 1999; Parsons et al., 2010) and is thus exposed to seasonal variability in environmental conditions. Natural selection may therefore regularly alter the population structure in this dinoflagellate species.
We hypothesized that natural selection of specific genotypes may contribute to genetic differentiation of Gambierdiscus populations over time. Meanwhile, dispersal barriers such as the large geographic distances across the Greater Caribbean Region (GCR), may however, overshadow this short-term variability and subsequently drive the population structure. To assess temporal and spatial genetic differentiation, we focused on Gambierdiscus caribaeus, a common species at our study sites in the GCR, and investigated its population structure using isolates from the Florida Keys and St. Thomas in the US Virgin Islands. Furthermore, the isolates from St. Thomas included a time series from 2006, 2007 and 2008, and monthly samples from August 2013 to April 2016. With this approach, we were able to directly compare genetic divergence over large spatial scales with continuous temporal differentiation.
Materials and Methods
Sampling and Establishment of Cultures
Macroalgal epiphyte communities were collected at St. Thomas, USVI, in 2006, 2007 and 2008, and at the Florida Keys (FlK) in 2013 to 2014, as described previously in Xu et al. (2014) and Lozano-Duque et al. (2018). SCUBA divers collected multiple samples of dominant macroalgal species, which varied by sampling date and site, along with some sea water in one plastic bag per site. Afterward, the bags were agitated to dislodge the epiphytic dinoflagellate cells from the macroalgae. The contents from the bags were sequentially filtered through 200 and 20 μm sieves to concentrate the microalgal cells. To increase temporal and spatial resolution, samples were also taken monthly from August 2013 to April 2016 at four St. Thomas, US Virgin Islands coral reef sampling sites (Black Point – nearshore, 9 m depth; Coculus Rock – nearshore, 7 m depth; Flat Cay – offshore, 12 m depth; Seahorse – offshore, 20 m depth; Figure 1). Parallel to the monthly sampling of cells for cultivation, dissolved nutrients, benthic coverage, and coral health (Smith et al., 2016) were assessed at the same sampling sites. Additionally, two buoys (Mooring VI104, CariCOOS Data Buoy C and Station 41052, National Buoy Data Center) provided hourly hydrological data from nearby locations (Supplementary Table S1).
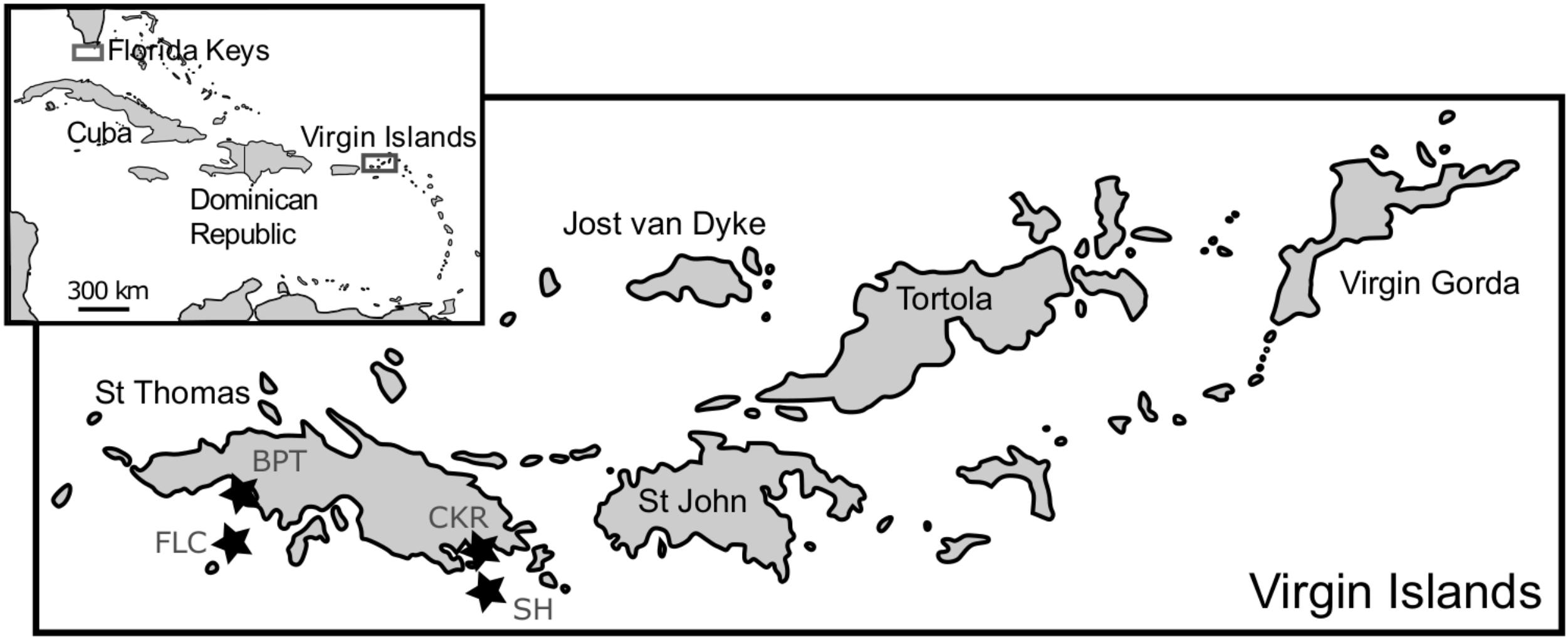
FIGURE 1. Map of sampling locations in the Florida Keys and St. Thomas, US Virgin Islands. The four sampling sites Black Point (BPT, Lat: 18.34450, Long: –64.98595), Coculus Rock (CKR, Lat: 18.31257, Long: –64.86058), Flat Cay (FLC, Lat: 18.31822, Long: –64.99104), and Seahorse (SH, Lat: 18.29467, Long: –64.86750) at St. Thomas are indicated with stars and are part of the Territorial Coral Reef Monitoring Program, with details and ancillary data found in Smith et al. (2016).
To obtain the individuals needed for the genetic analysis, single Gambierdiscus cells were isolated from the epiphyte collections. From each sampling site and date, at least 10 Gambierdiscus cells were isolated by micropipetting under an inverted microscope (Olympus CK40). Each cell was washed by serial transfers through sterile seawater, and transferred individually into a well of a 96-well plate containing 300 μl of K medium (Keller et al., 1987). The cells were cultivated in a 25°C climate chamber with a light:dark cycle of 14:10 h and a photon flux ca. 100 μmol photons m-2 s-1. Cells that divided into more than 8 cells per well were transferred into small glass tubes with 5 ml medium. After three more weeks, they were transferred into the final culture volume of 25 ml and maintained under the conditions described above. As six different Gambierdiscus species are reported from the Caribbean region, which cannot be microscopically distinguished, all strains were taxonomically identified using Restriction Fragment Length Polymorphisms of amplified fragments from 23S rDNA (Lozano-Duque et al., 2018). Isolates of the most common species G. caribaeus were used in all further analyses. Each clonal strain of G. caribeaus is one individual in our genetic analyses.
DNA Extraction
Vegetative, haploid algal strains were harvested by centrifuging 2 ml of aggregated cell culture at 1000 rpm for 15 min. The cell pellets were frozen at -20°C until further processing. DNA was extracted from all G. caribaeus strains using the DNeasy Blood and Tissue kit from Qiagen (Hilden, Germany). We slightly modified the original protocol by washing the cell pellets with PBS buffer at the beginning to remove extrapolymeric substances, disrupting the cells by vortexing with silica-zirconium beads (Biospec Products, Bartlesville, OK, United States) in ATL buffer for 2 min, and conducting the Proteinase K lysis step over night at 56°C on a shaker.
Amplification of Microsatellites
Seven microsatellites, four loci (G.carib1, G.carib2, G.carib3, and G.carib4) developed from shot-gun sequencing of the genome and three (G.caribT5, G.caribT6, and G.caribT7) discovered from a transcriptome (Sassenhagen and Erdner, 2017), were amplified in four separate PCR reactions. G.carib1, G.carib2, and G.caribT5 were amplified together in a 40 μl PCR reaction using an annealing temperature of 60°C. G.caribT6 and G.caribT7 were amplified in duplex in 30 μl PCR reactions with an annealing temperature of 54°C, and G.carib3 and G.carib4 were amplified individually in 15 μl PCR reactions (Sassenhagen and Erdner, 2017). The final PCR products were analyzed via capillary electrophoresis at the Genomics Core Lab, Texas A&M University, Corpus Christi.
Analysis of Microsatellite Data
Microsatellite peaks were scored in Geneious 8.1.7 (Kearse et al., 2012) based on a LIZ 600 size standard. Allele frequencies, genetic diversity, and fixation indices of genetic differences (FST) between populations were calculated using the software GenoDive 2.0b27 (Meirmans and Van Tienderen, 2004). FST values and their significance based on 9999 permutations were assessed for three different datasets: all seven loci, only the three loci from the transcriptome and only the five loci that did not show any sudden changes in allele frequencies. Allele diversity (based on all seven markers) was calculated for individual temporal groups including all microalgal strains as well as only unique ones, meaning that reoccurring genotypes were excluded. To assess genotype diversity, clones were assigned based on a step-wise mutation model in GenoDive. For this analysis, missing data were replaced by average allele length and a threshold of two bases was chosen to distinguish clones. Genotype diversity was also calculated after equalizing the sample size of each genetic cluster to N = 54 with the R function “sample.” Pairwise Jost’s D values between genetic clusters were calculated in the R package DEMEtics 0.8-7 (Gerlach et al., 2010) and significance of each value was assessed with 1000 bootstrap resamplings. Genotypic linkage disequilibrium between pairs of loci was calculated with the web version of GenePop (Rousset, 2008) using 1000 dememorizations, 100 batches, and 1000 iterations per batch.
Clustering of algal strains, roughly grouped by month, was determined via principal component analysis (PCA), which was calculated in Genodive and visualized in R. The significance of the axes was assessed using 1000 permutations. The number of distinct genetic clusters was determined by the program STRUCTURE, version 2.3.2 (Pritchard et al., 2000). Default parameters as well as prior information about sampling time points, admixture and non-admixture ancestry models with either correlated or independent allele frequencies were tested in STRUCTURE. Each run had a burn-in of 70000 iterations followed by 200000 iterations of data collection. Up to 12 populations, generously exceeding the number of identified clusters by PCA, were tested with 10 iterations at each level. The results were analyzed using the web based program STRUCTURE Harvester (Earl and vonHoldt, 2011) to identify the number of clusters yielding the best fit for this dataset. Genetic clustering in the high-resolution sampling period at USVI (1308-1604) based on the STRUCTURE results as well as other putative groupings was further investigated with an AMOVA using the program Genodive.
Analyses of Environmental Variables
All environmental data (Supplementary Table S1) expressed as percent benthic cover or prevalence, including habitat coverage and coral health, were arcsine transformed. The mean and standard deviation of all environmental variables were homogenized by subtracting the mean from each value and dividing this by the standard deviation [(value-mean)/SD]. The averaged environmental data from 1 month prior to sampling of Gambierdiscus displayed the strongest relation with the genetic data likely due to a delay in acclimatization of the microalgae and were thus used for all subsequent analyses. Percent zoanthid cover of the habitat correlated with coral cover and was therefore excluded in the following constrained ordination analysis. Percent macroalgal cover was also removed to avoid autocorrelation with benthic cover by the dominant macroalgal species Dictyota spp.
The relationship between genetic structure and environmental variables was assessed using a distance based redundancy analysis (dbRDA) (Legendre and Anderson, 1999). Distances between approximate monthly algal groups based on the microsatellite data were calculated with a principal coordinate analysis (PCoA) using the R package adegenet 1.0.1 (Jombart, 2008). The relationship between the coordinates of algal groups (row scores normed to the square root of the eigenvalue = li) from the PCoA and the environmental variables was tested with dbRDA using the R package vegan 2.4-1 (Oksanen et al., 2016). The best model was selected based on automatic stepwise model building using the function “ordistep” in the R package vegan starting with a model that included only the intercept, and adding all environmental variables afterward. Significant differences between characteristic environmental variables for each temporal genetic cluster were assessed with one-way ANOVA and Tukey HSD post hoc tests in R.
Results
Genetic Population Structure of G. caribaeus
Using seven microsatellite markers, 405 G. caribaeus strains were genotyped for this study. Individual strains lacked at most two loci out of the seven microsatellites. Due to very low sample sizes (<8 strains) in some months during the high-resolution sampling period at USVI, strains from 32 months (August 2013–April 2016) were pooled into 20 temporal groups each consisting in the end of at least eight individuals. Identifiers of these groups consist of the sampling region (USVI), year and predominant month (yymm: 1308, 1502, 1604, etc.). Samples collected at the Florida Keys and at USVI before the high-resolution sampling period represent yearly collections and are denoted with sampling region (USVI or FlK) and year (yy: 06, 07, 13, 14, etc.) There were 293 unique seven-locus genotypes in the entire data set resulting in an overall genotype diversity of 0.72 (G/N). No linkage disequilibrium was detected across the dataset among all pairs of loci.
To detect distinct genetic clusters in the data set, we used Bayesian analysis implemented in the program STRUCTURE. This program gave the most biologically sensible results using prior sampling information, as this reduces the bias due to uneven sample sizes (Falush et al., 2003), admixed ancestry and independent allele frequency settings. Using Ln(K) values and the Evanno method, K = 4 was identified as the most likely number of genetic clusters in the entire data set (Figure 2A). The STRUCTURE results indicated succession of four different genetic clusters at USVI, while FlK_13 and FlK_14 were very similar to USVI_06, USVI_07, and USVI_08. All of these clusters comprised mixed genotypes.
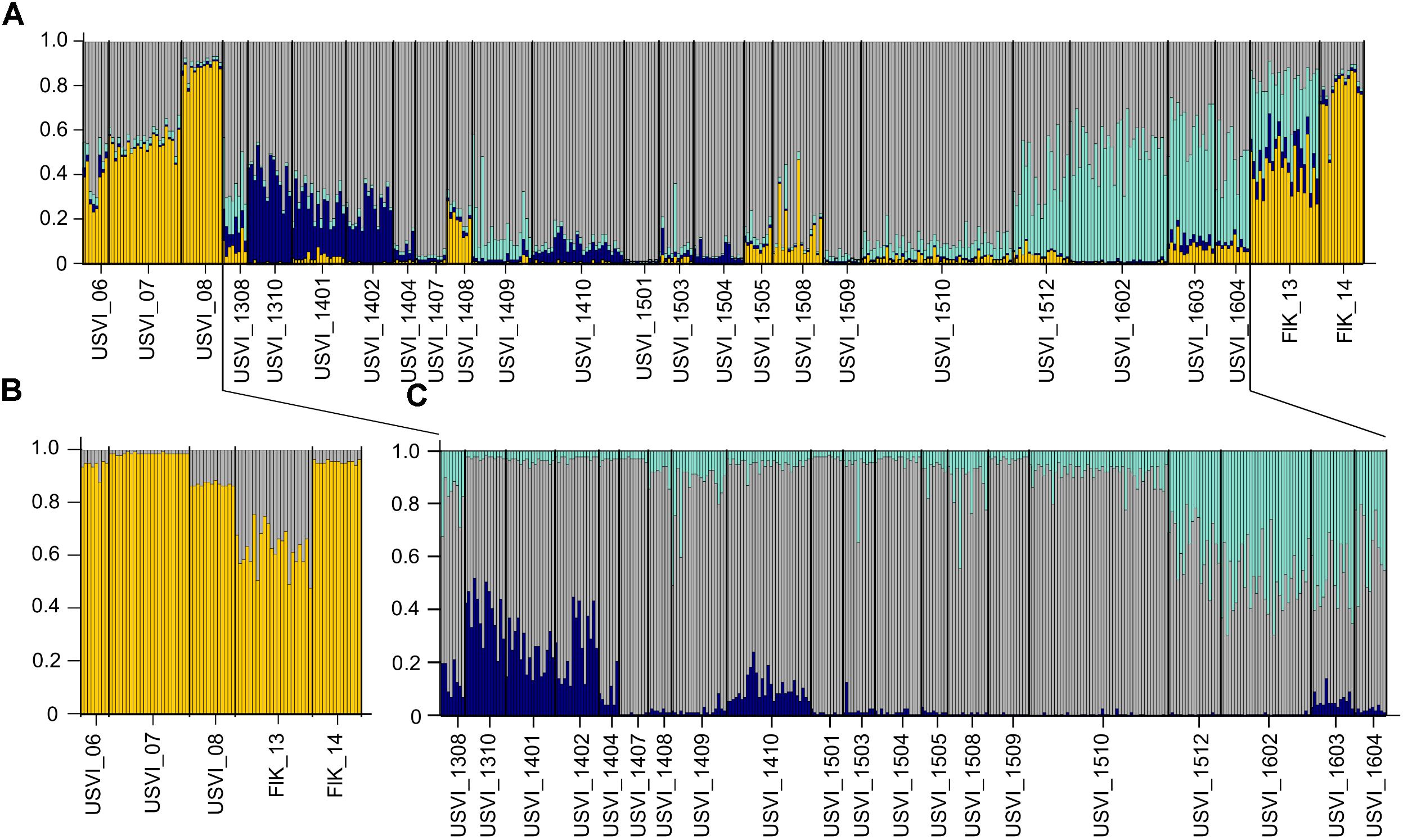
FIGURE 2. STRUCTURE plots illustrating the number of genetic clusters in the data set based on all seven microsatellite loci. Genotypes are represented as single vertical lines. (A) K = 4 in the entire data set include all samples from the Florida Keys and USVI. (B) K = 2 in a subset of the data: USVI 2006-2008 and Florida Keys 2013 and 2014. (C) K = 3 in the high-resolution sampling period (August 2013–April 2016) at USVI.
Using a hierarchal approach, the STRUCTURE analysis was repeated for the high-resolution sampling period at USVI, which comprised August 2013–April 2016 (1308-1604). No evidence for spatial genetic divergence among the four sampling locations at USVI was found. However, the results confirmed the differentiation of the high-resolution temporal data set into three distinct temporal groups, which we refer to as early (August 2013 through March 2014; 1308-1403), middle (April 2014 through October 2015; 1404-1510), and late (November 2015 through April 2016; 1511-1604) (Figure 2C). STRUCTURE analysis was then conducted for algal strains isolated in 2006, 2007, and 2008 at USVI, and in 2013 and 2014 at the Florida Keys to clarify the population structure of this group. The results indicated differentiation of the FlK_13 sample from all other strain groups (Figure 2B). However, the ability to detect additional divergence among USVI_06, USVI_07, USVI_08, and FlK_14 by STRUCTURE might be limited due to small sample sizes (Table 1).
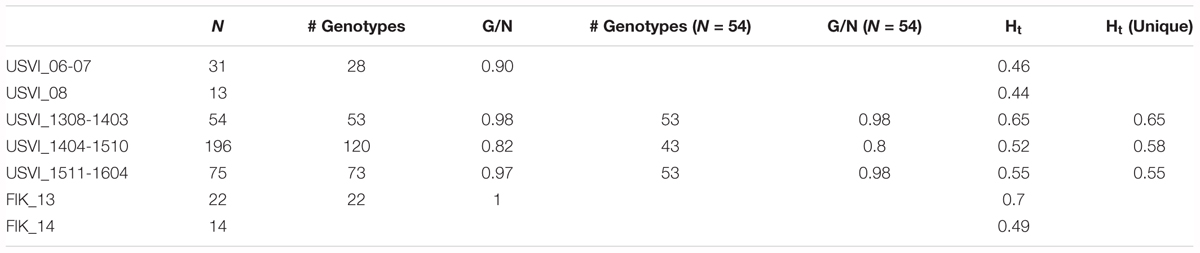
TABLE 1. Number of strains per genetic cluster (N), number of different genotypes (# genotypes), genotype diversity (G/N), allele diversity calculated with all strains (Ht), and excluding replicated genotypes (Ht unique).
Principal component analysis clustering was similar to the STRUCTURE plots, but highlighted the divergence of samples from the Florida Keys (2013 and 2014), and of USVI_08 (Figure 3). Pairwise FST values were calculated between seven different clusters representing USVI_06-07, USVI_08, USVI_1308-1403, USVI_1404-1510, USVI_1511-1604, FlK _13, and FlK _14. Significant pairwise FST values (calculated using all seven loci) between the genetic clusters ranged from 0.052 to 0.251 (Table 2). Most clusters displayed significant divergence from each other after Bonferroni correction except for FlK_2013 and FlK_2014. The degree of differentiation based on Jost’s D indices (Jost, 2008), which removes bias due to differences in allele diversity among populations, were as high as 0.443. Both FST and Jost’s D values indicated very high differentiation of USVI_08 from all other samples, especially from USVI_1308-1403. Furthermore, Jost’s D values suggested very high divergence between USVI_1308-1403 and FlK_14 (Jost’s D = 0.307), which was not as evident in the FST analysis (Table 2).
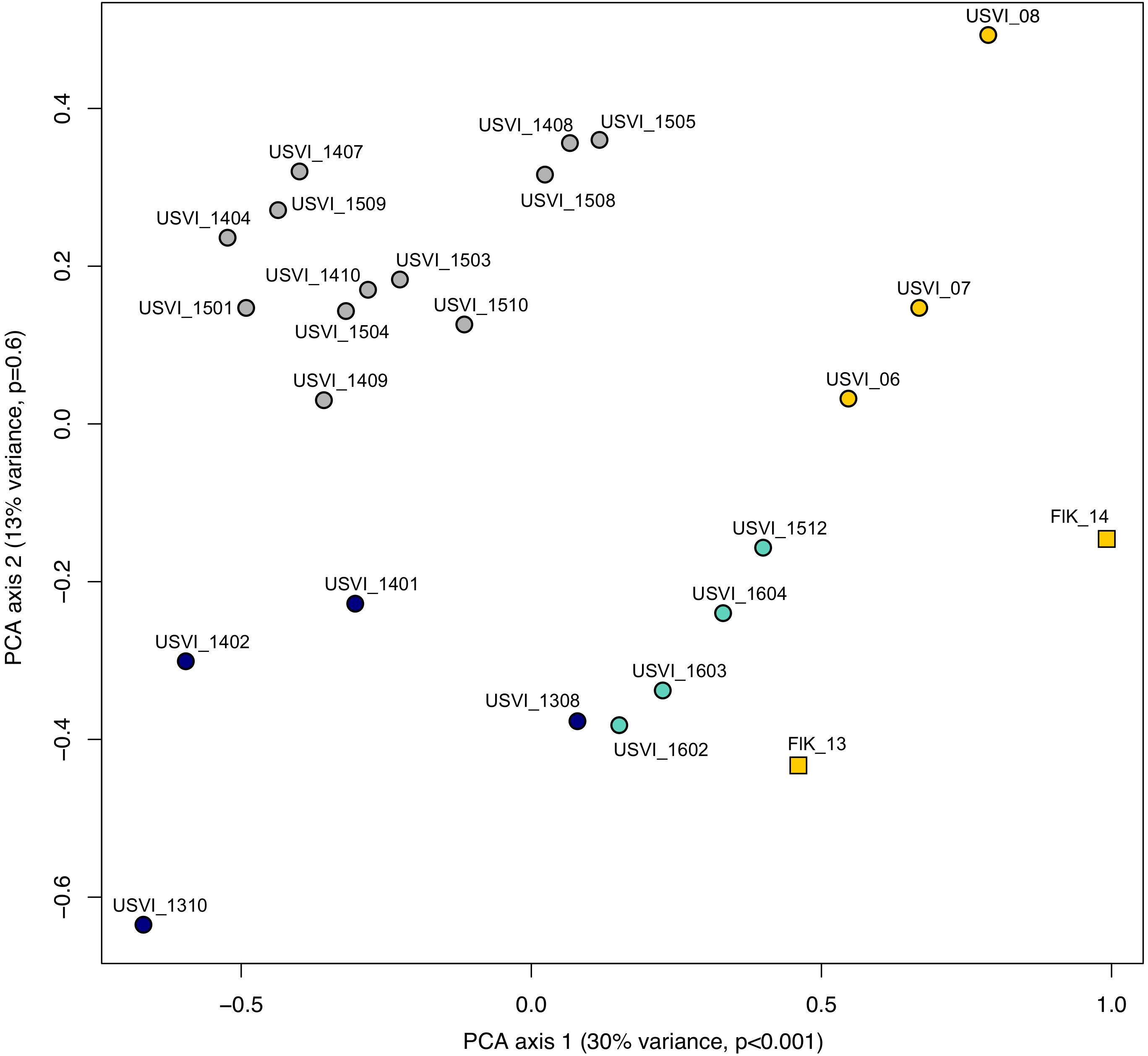
FIGURE 3. Principal component analysis (PCA) of genetic data from the Florida Keys (FlK) 2013 and 2014, USVI 2006, 2007 and 2008, and approximate monthly groups from the high-resolution sampling period at USVI (1308-1604). Colors correspond to the clusters suggested by the STRUCTURE analyses.
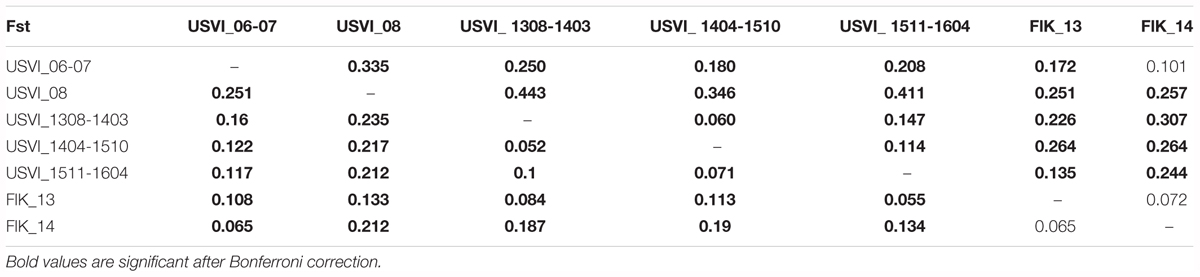
TABLE 2. FST values below the diagonal and Jost’s D values above the diagonal (based all seven microsatellites).
FST values were in general higher (2 to 10x) when calculated based on transcriptomic loci in comparison to neutral loci from the genome (Table 3). In particular, the differentiation between USVI_1308-1403 and USVI_1404-1510 was driven by transcriptomic loci, as this divergence was not significant based on genomic microsatellites alone. Additionally, differentiation between FlK_13 and FlK_14 was significant based on transcriptomic loci in contrast to results from the entire set of microsatellites or only genomic loci. However, divergence between USVI_06-07 and FlK_14, and USVI_1308-1403 and FlK_13 was higher when calculated using genomic microsatellites than transcriptomic loci.
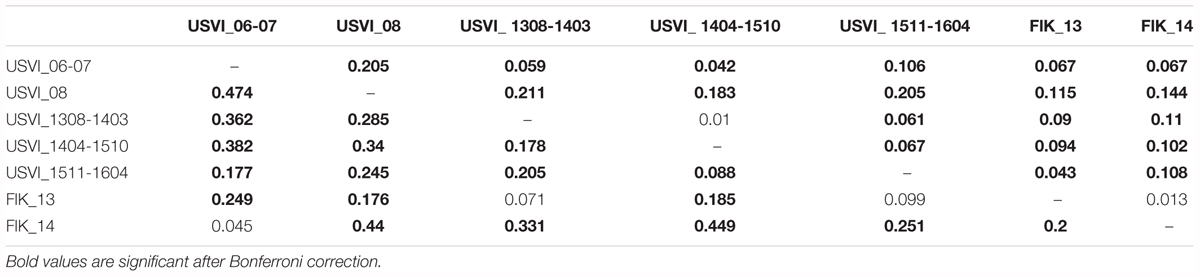
TABLE 3. FST values calculated based on genomic microsatellites (above the diagonal) and based on microsatellites discovered from the transcriptome (below the diagonal).
Genetic differentiation within the high-resolution time series at USVI was further investigated using an AMOVA. When monthly G. caribaeus groups were pooled into three clusters according to the STRUCTURE results (1308-1404, 1404-1510, and 1511-1604) differentiation among clusters was highly significant (Rct = 0.052, p < 0.001), but not significant among months (Rsc = 0.004, p = 0.303). This population structure also explained the highest amount of variance compared to other clustering options.
The genotype diversity of the middle cluster (1404-1510, G/N = 0.82) was lower than at the beginning (1308-1403, G/N = 0.98) and end (1511-1604, G/N = 0.97) of the high-resolution sampling period independent of the sample size (Table 1). The overall allele diversity was highest during the first eight months of sampling (Ht = 0.648), declined in spring 2014 and remained low (Ht = 0.553) until the end of the project. This trend was observed regardless of whether non-unique algal strains were included or excluded. The genomic microsatellites contributed more to the allele diversity than the loci isolated from the transcriptome, as they displayed in general a higher number of alleles. The drop in allele diversity in spring 2014 was driven by the markers G.caribT5 and G.caribT7, two of the transcriptomic loci (Table 4), due to near fixation of one allele in more than 93% of strains in both loci. When splitting the data set from the high-resolution sampling period at USVI into the three genetic clusters, pairwise linkage disequilibrium was detected in the early and middle cluster in one locus pair each respectively (USVI_1308-1403: G.carib1 and G.carib2, p = 0.009; USVI_1404-1510: G.carib1 and G.caribT7, p = 0.032).
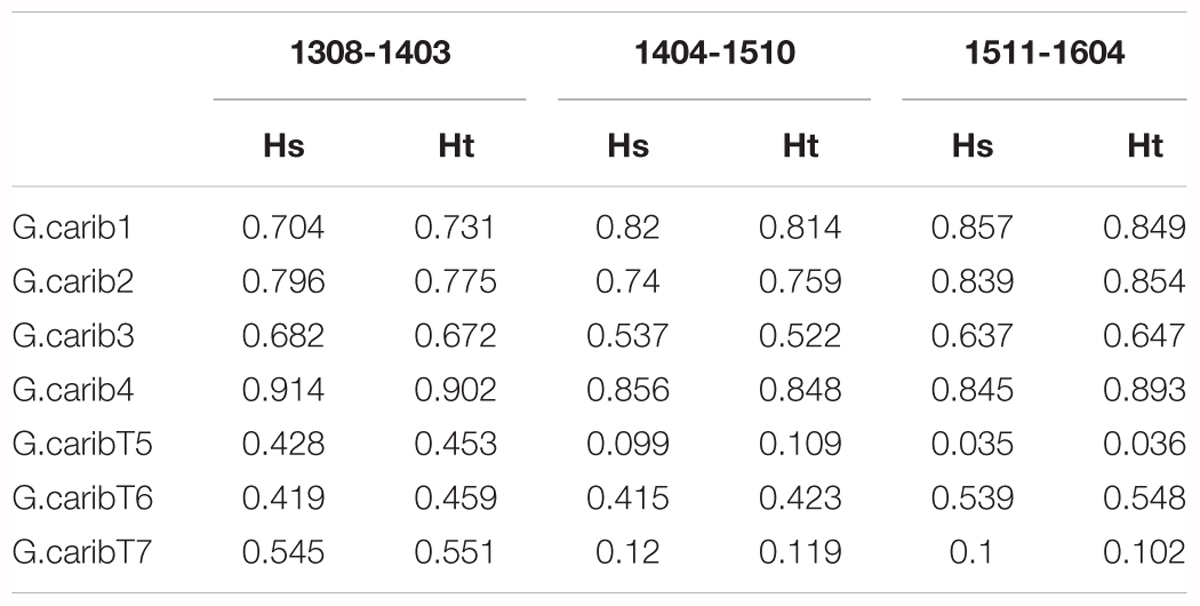
TABLE 4. Allele diversity within months (Hs) and total allele diversity (Ht) for each microsatellite locus during the high-resolution sampling period at USVI.
The previous FST analysis (Table 3) highlighted the impact of two microsatellite markers that were discovered from a transcriptome (Sassenhagen and Erdner, 2017) for differentiation between USVI_1308-1403 and USVI_1404-1510. This observation was further emphasized by a STRUCTURE analysis restricted to the three transcriptomic microsatellites, which only detected two genetically distinct clusters: (1) the early sampling period (1308-1403) and (2) all remaining algal strains (1404-1604) (Figure 4A). In contrast, STRUCTURE analysis limited to the five “neutral” markers that did not display any sudden shifts in allele diversity (G.carib1, G.carib2, G.carib3, G.carib4, and G.caribT6), revealed the change in population structure in winter 2015 (Figure 4B).
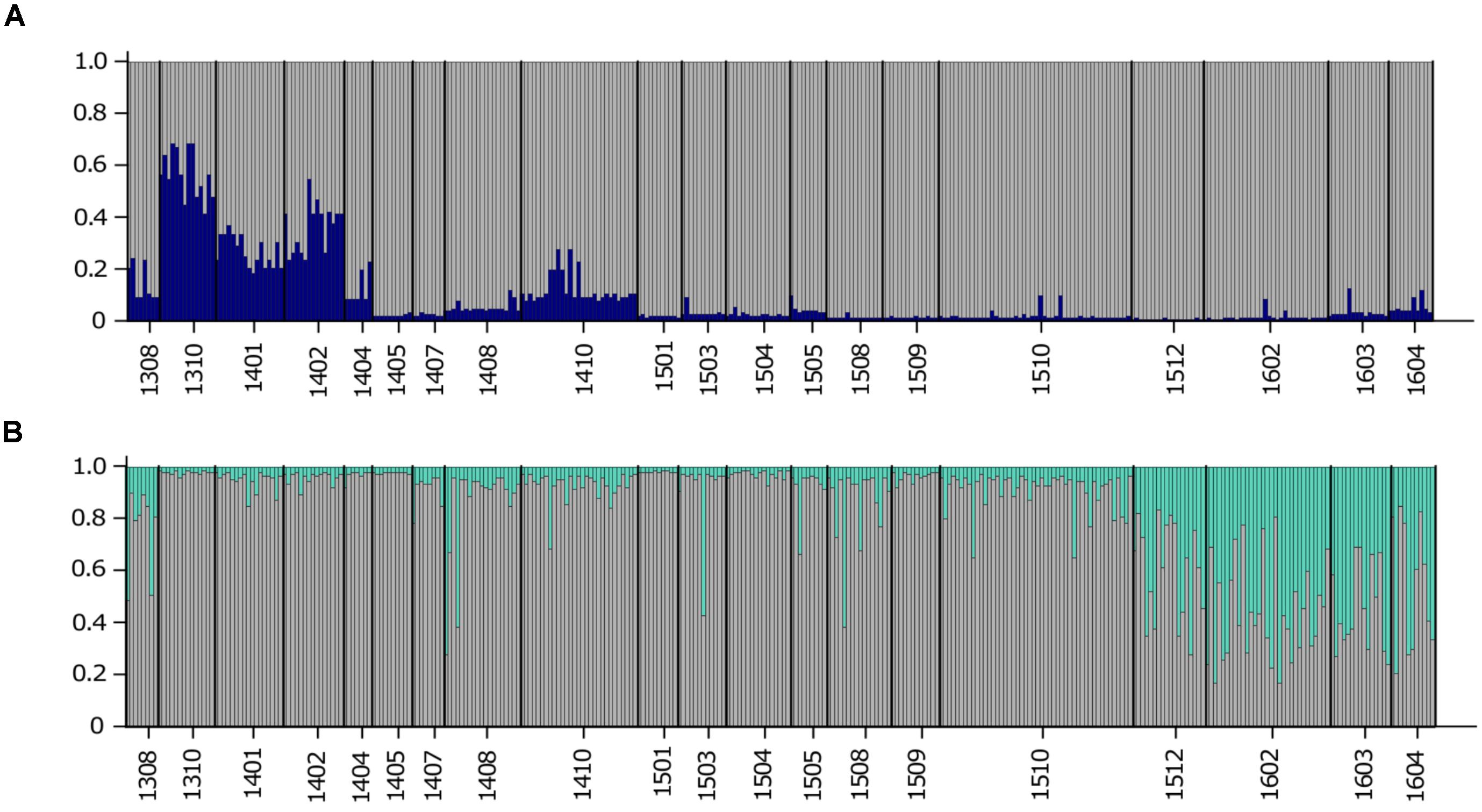
FIGURE 4. (A) STRUCTURE output for K = 2 from the high-resolution sampling period at USVI based on three microsatellites discovered from the transcriptome (G.caribT5, G.caribT6, and G.caribT7). (B) STRUCTURE output for K = 2 from the high-resolution sampling period at USVI based on the five microsatellites that did not display any sudden changes in allele frequencies (G.carib1, G.carib2, G.carib3, G.carib4, and G.caribT6).
Impact of Environmental Variables on Population Structure
The impact of several biotic and abiotic environmental variables, which changed throughout the sampling period, on the population genetic structure of G. caribaeus at USVI was assessed using a distance based redundancy analysis (dbRDA). The dbRDA analysis identified significant relationships between the microsatellite data and salinity, percent habitat covered by corals, turfalgae growing on dead corals and cover of the two macroalgal species Lobophora variegata and Halimeda spp. (Figure 5 and Table 5).
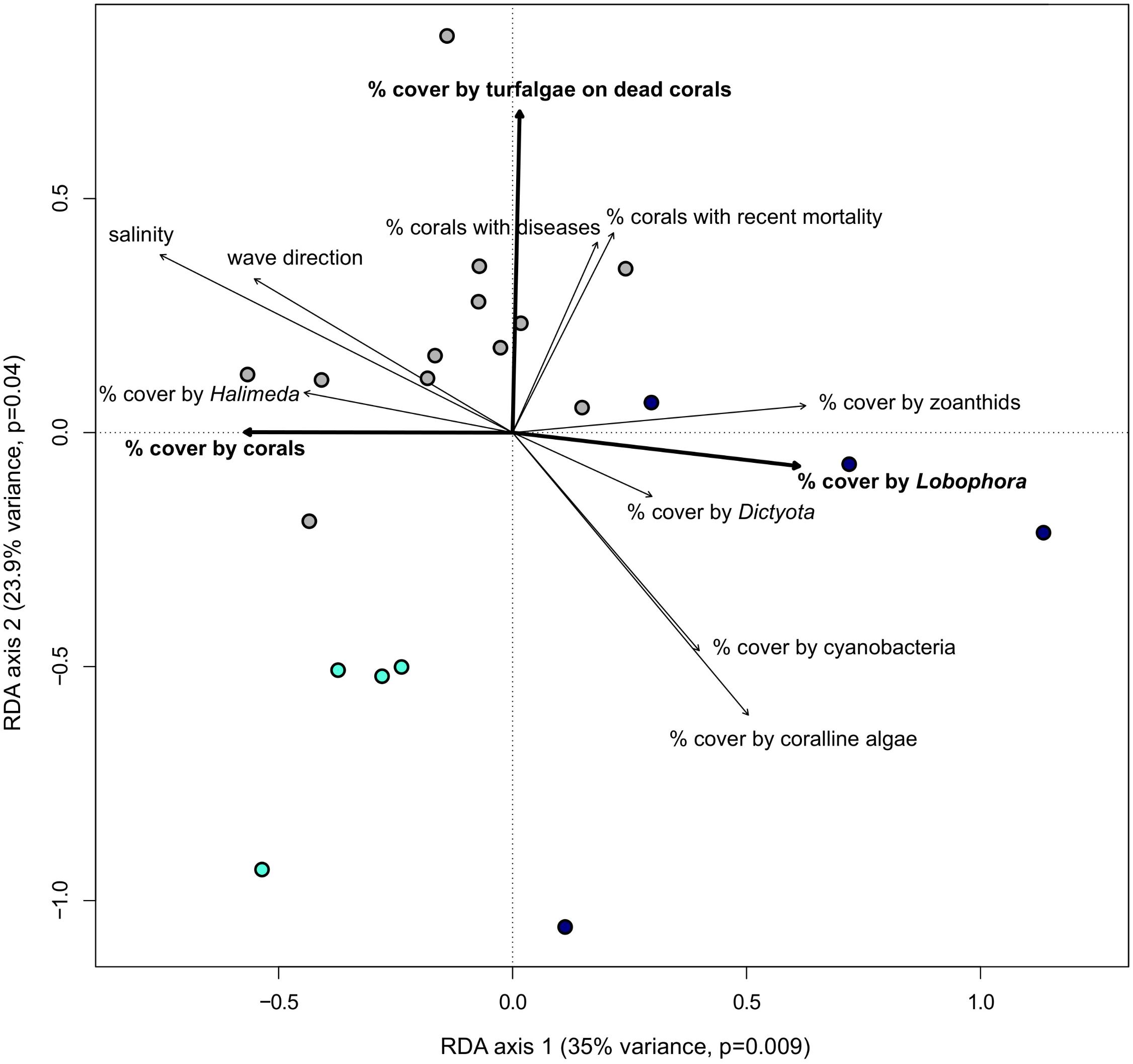
FIGURE 5. Bold environmental variables have a significant impact (p < 0.05) on the population structure of Gambierdiscus caribaeus based on the RDA analysis. The other environmental variables distinguish genetic clusters from each other based on one-way ANOVA. Circles represent approximate monthly algal groups from high-resolution sampling period at USVI: dark blue circles = USVI_1308-1403, gray circles = USVI_1404-1510, turqouise circles = USVI_1511-1604.
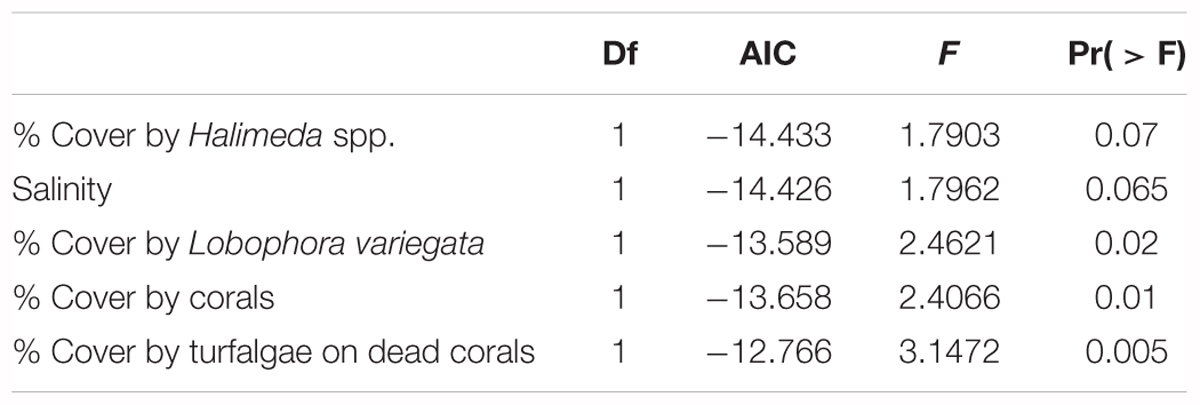
TABLE 5. Results of the RDA analysis to assess the impact of environmental variables on the genetic population structure of G. caribaeus during the high-resolution sampling period at USVI.
Comparing the means of all environmental variables from the three time periods at USVI dominated by distinct genetic clusters of G. caribaeus with one-way ANOVA revealed further significant differences that characterized each phase. Besides lower salinity (p < 0.01), lower coverage by corals (p < 0.03) and higher coverage by Lobophora variegata (p < 0.004), the general wave direction (p < 0.03) and higher habitat coverage by Dictyota spp. (p < 0.02) distinguished USVI_1308-1403 from the other two temporal clusters. USVI_1404-1510 was distinguished from the other two periods by lower coverage of the habitat with coralline algae (p < 0.04) and cyanobacteria (p < 0.05). Besides higher habitat coverage with corals, USVI_1511-1604 was characterized by lower recent coral mortality (p < 0.02), lower percent of corals with diseases (p < 0.035), and lower zoanthid coverage (p < 0.001) (Figure 5).
Discussion
This study enables the comparison of spatial and temporal genetic differentiation in a microalgal species and, thus, provides insights into different evolutionary drivers and the adaptive potential of microorganisms. This knowledge is especially needed for predicting the future distribution of harmful microalgae such as Gambierdiscus, which produces the toxins that cause ciguatera fish poisoning. This first population genetic analysis of this genus sheds light onto connectivity in the GCR and temporal variability in these economically important dinoflagellates.
No evidence was found for spatial genetic differentiation among the four sampling locations at St. Thomas (USVI), likely due to regular dispersal and gene flow across the short distance between the sites (approximately 3–15 km). However, significant fixation indices of genetic differences between samples from the Florida Keys and USVI indicated limited connectivity and gene flow across the GCR. This suggests that despite the strong westward Caribbean Current and the Loop Current, which connect the Lesser Antilles through the Yucatan Channel with the Gulf of Mexico and the Gulf Current (Alvera-Azcárate et al., 2009), benthic microalgal cells are seldom transported across the GCR and rarely establish in distant locations. The large distance (approximately 1800 km) and therefore long dispersal time, as well as potential biogeographic barriers (e.g., the Mona Channel between Puerto Rico and Hispanola; (Colin, 2003; Cowen et al., 2006), likely limit the gene flow between these locations. The dinoflagellate Alexandrium minutum displayed stronger genetic differentiation over similar spatial scales in the Mediterranean Sea (Casabianca et al., 2012), as connectivity between different basins is likely more limited there due to regional water circulation. The spatial population structure in G. caribaeus, however, is comparable with the differentiation found among populations of the diatom Skeletonema marinoi in the Baltic Sea (Sjöqvist et al., 2015), which likely diverged due to local adaptations. Similarly, adaptations to local environmental conditions might also influence establishment success of dispersed G. caribaeus in the GCR and thus impact its population structure.
In addition to significant spatial structure across the GCR, strong evidence for high temporal differentiation was found at St. Thomas, USVI. Our analyses identified at least five distinct populations of G. caribaeus over a time span of 11 years, including the succession of three genetically distinct populations at this location from August 2013 to April 2016. Fixation indices of genetic differences were indeed sometimes higher between temporal groups at USVI than across the GCR, particularly in comparison with USVI_08. This is surprising, as it indicates that physical dispersal barriers such as geographic distance might be less important for population differentiation than local processes.
The high-resolution sampling period at USVI was characterized by a decline in dominant macroalgae species and an increase of coral abundance (Figure 6). During the timeframe of the first genetic cluster (USVI_1308-1403), large proportions of the habitat were covered by Dictyota spp. and Lobophora variegata. At the end of the sampling period, however, the benthic habitat at St. Thomas was characterized by higher abundance of healthy corals. Additionally, salinity significantly increased in spring 2014 and remained comparatively high until the end of the time series, despite some seasonal variations. Salinity levels in the eastern Caribbean usually decrease in August to November due to horizontal advection of freshwater from the Amazon and Orinoco River plumes (Corredor and Morell, 2001; Chérubin and Richardson, 2007; Grodsky et al., 2015). Discharge from these rivers might have been reduced in 2015 and 2016, however, due to extensive droughts during the strong El Niño (Jiménez-Muñoz et al., 2016). The different climatic conditions were likely reflected in altered overall wave direction observed at USVI. Increased competition with corals or a shift in grazing pressure due to changed abiotic conditions may have caused the temporary decrease of crustose coralline algae and filamentous cyanobacteria from spring 2014 to autumn 2015.
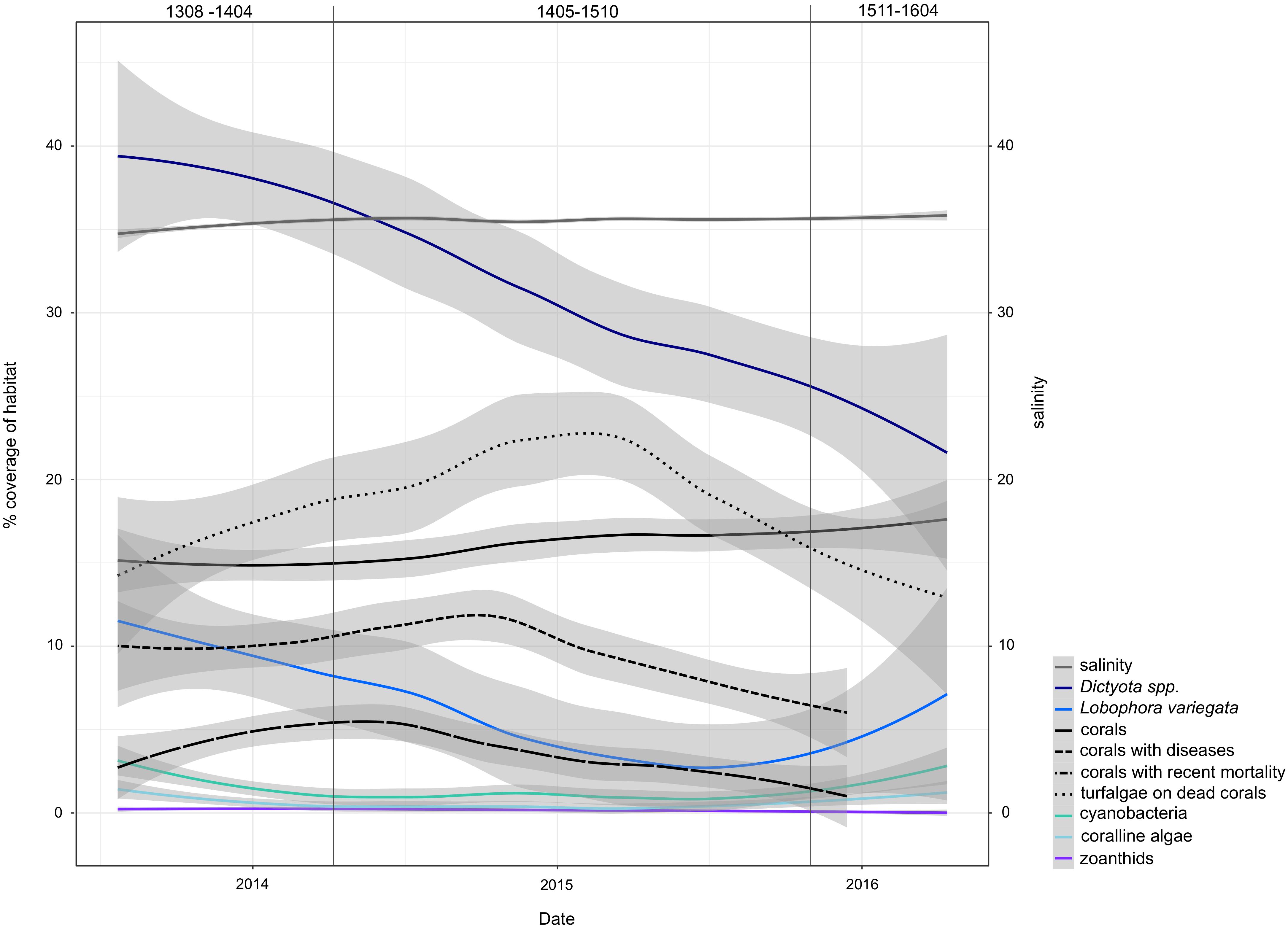
FIGURE 6. Trends of environmental variables during the high-resolution sampling period at USVI. Variability of data is smoothed for better visibility of patterns using local polynomial regression fitting in the ggplot2 function “geom_smooth.” Vertical lines mark the periods dominated by the three distinct genetic clusters of G. caribaeus. Cover corresponds to cover of benthos (out of 100%).
This environmental shift was reflected in a significant change in population structure of G. caribaeus in spring 2014 as demonstrated in increased abundance of one allele in two loci, causing a drop in allele diversity. This genetic change occurred rapidly from February to April 2014. The two microsatellites that drove this change in population structure were both discovered from the transcriptome and might be representative for genes under selective pressure. Thus, genotypes with mutations in genes that improve the phenotype under ambient environmental conditions will grow and reproduce faster, ensuring the perpetuation of the selected alleles (Slatkin, 1987; Elena and Lenski, 2003; Nei, 2005; Nielsen, 2005). Changes in environmental conditions such as salinity could, therefore, cause shifts in community composition and drive genetic differentiation due to selective sweeps. Furthermore, dominance of few G. caribaeus strains with adaptations for higher salinity would as well explain the observed decrease in genotype diversity (Nielsen, 2005) from April 2014 to October 2016. Laboratory experiments have highlighted distinct growth optima among G. caribaeus strains in different salinities (Xu et al., 2016), supporting such strain specific adaptations in this species. Other studies (e.g., Rynearson et al., 2006; Richlen et al., 2012; Dia et al., 2014) have previously described similar impacts of short-term environmental changes on the population structure of microalgae. In particular, adaptation to local salinities was identified as the driving force behind genetic differentiation among populations of the diatom Skeletonema marinoi (Sjöqvist et al., 2015).
Although some markers located in mRNA especially highlighted the genetic differentiation of G. caribaeus in spring 2014, their influence on the phenotype of G. caribaeus is unknown. The genes encoded in the mRNA are currently not annotated preventing the exact localization of the microsatellites (untranslated or coding region) and the identification of the function of the putative protein. Although the reading frames of these genes will not be affected by expansion or contraction of the markers, as their repeat sizes mirror the translational unit of an amino acid, changes in the length of the microsatellites can deleteriously impact regulation of translation, the secondary structure or functioning of the encoded proteins (Li et al., 2004). Selection pressure against damaging mutations should, thus, be high, allowing only spreading of changes in the repeat length that are beneficial for the phenotype. Thus, the microsatellite alleles dominant in the investigated G. caribaeus strains might provide a selective advantage for the population. We hypothesize, therefore, that a significant increase in salinity in spring 2014 caused natural selection of adapted genotypes and divergence of the G. caribaeus population at St. Thomas, USVI.
The continuous decline of the macroalgae Dictyota spp. and Lobophora variegata, representing the main habitat of Gambierdiscus, coupled to an increase in healthy corals, may have significantly impacted the distribution of G. caribaeus toward the end of our high-resolution sampling period at USVI. The overall population size of G. caribaeus likely declined with the decreasing amount of these macroalgal species (i.e., reduced carrying capacity), potentially resulting in more patchy, isolated distribution of the dinoflagellates. Small populations often experience genetic drift (Nei et al., 1975; Elena and Lenski, 2003), which can cause differentiation through fixation or loss of certain alleles in a population by chance over time (Nei, 2005). On the other hand, the population genetic structure might be influenced by the life cycle of G. caribaeus. In microalgal species with synchronized life cycles, germination of few algal strains from resting stages followed by genetic drift could cause interannual genetic differentiation, as observed in the freshwater raphidophyte Gonyostomum semen (Lebret et al., 2012). The life cycle of Gambierdiscus is poorly understood and monitoring of cell abundance is challenging due to great morphological similarities between different species. Additionally, natural selection through some unmeasured environmental variable affected by the changes in benthic habitat represents another potential driver behind the divergence in winter 2015 at USVI.
Nevertheless, this study emphasized the impact of local processes such as temporal changes in environmental conditions on the population structure of Gambierdiscus, causing similar genetic differentiation as found over large spatial scales. Further studies with consistent temporal resolution at different locations are needed to thoroughly assess the influence of local temporal variability on meta-population structure of microalgae.
Conclusion
Overall, this study suggests that gene flow in the epiphytic dinoflagellate Gambierdiscus is limited over large geographic distances in the GCR. Additionally, this study indicates high temporal variability, partially caused by natural selection, which is commensurate with the degree of genetic differentiation found in microalgae across large geographic distances. These findings suggest that changes in environmental conditions over time might be as significant for the genetic population structure of microorganisms as environmental variation over spatial scales. Spatial population genetic analyses should thus consider temporal variations in population structure, as they might sometimes overshadow or exaggerate differentiation among locations. Because of the tremendous intraspecific diversity likely present in most microbial species, future environmental change could have unpredictable impacts on their populations. The high genetic diversity might facilitate current and future expansion to new habitats, while changing environmental conditions could select for currently rare genotypes and alter the composition of local populations. These findings add complexity to the challenge of predicting harmful algal blooms and the risk of seafood poisoning from species such as Gambierdiscus in the future.
Data Accessibility
Microsatellite genotypes from all G. caribaeus strains are accessible at Dryad (doi: 10.5061/dryad.p5217).
Author Contributions
DE conceived this study and designed it with contributions from IS. MP provided Gambierdiscus cultures from the Florida Keys. TS supplied samples of Gambierdiscus and environmental monitoring data from the Territorial Coral Reef Monitoring Program at USVI. IS, YG, and YL-D established Gambierdiscus cultures, extracted DNA, and identified the species. IS fingerprinted the cultures with microsatellites, analyzed the data, and wrote the manuscript. All authors read the manuscript and provided comments.
Funding
Funding for this project was provided by the National Oceanic and Atmospheric Administration Center for Sponsored Coastal Ocean Research ECOHAB program under Award No. NA11NOS4780061 to the University of Texas Marine Science Institute (DE). This is ECOHAB publication number 922. Field work in St. Thomas was supported by the Virgin Islands EPSCoR Program (NFS#0814417).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mindy Richlen for providing cell count data of Gambierdiscus from USVI. Alex Leynse and Adam Catasus contributed to this study by establishing Gambierdiscus cultures from the Florida Keys. Field work in St. Thomas was facilitated by Robert Brewer, Viktor Brandtneris, Rosmin Ennis, Sarah Groves, Sarah Heidmann, Colin Howe, Jonathan Jossart, Elizabeth Kadison, Christopher Loeffler, and Tanya Ramseyer. We are grateful for constructive comments from two reviewers that improved the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00393/full#supplementary-material
References
Alvera-Azcárate, A., Barth, A., and Weisberg, R. H. (2009). The surface circulation of the Caribbean Sea and the Gulf of Mexico as Inferred from Satellite Altimetry. J. Phys. Oceanogr. 39, 640–657. doi: 10.1175/2008JPO3765.1
Bagnis, R., Chanteau, S., Chungue, E., Hurtel, J. M., Yasumoto, T., and Inoue, A. (1980). Origins of ciguatera fish poisoning: a new dinoflagellate, Gambierdiscus toxicus Adachi and Fukuyo, definitively involved as a causal agent. Toxicon 18, 199–208. doi: 10.1016/0041-0101(80)90074-4
Boenigk, J., Jost, S., Stoeck, T., and Garstecki, T. (2007). Differential thermal adaptation of clonal strains of a protist morphospecies originating from different climatic zones. Environ. Microbiol. 9, 593–602. doi: 10.1111/j.1462-2920.2006.01175.x
Casabianca, S., Penna, A., Pecchioli, E., Jordi, A., Basterretxea, G., and Vernesi, C. (2012). Population genetic structure and connectivity of the harmful dinoflagellate Alexandrium minutum in the Mediterranean Sea. Proc. R. Soc. B Biol. Sci. 279, 129–138. doi: 10.1098/rspb.2011.0708
Casteleyn, G., Leliaert, F., Backeljau, T., Debeer, A.-E., Kotaki, Y., Rhodes, L., et al. (2010). Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc. Natl. Acad. Sci. U.S.A. 107, 12952–12957. doi: 10.1073/pnas.1001380107
Chateau-Degat, M.-L., Chinain, M., Cerf, N., Gingras, S., Hubert, B., and Dewailly, É (2005). Seawater temperature, Gambierdiscus spp. variability and incidence of ciguatera poisoning in French Polynesia. Harmful Algae 4, 1053–1062. doi: 10.1016/j.hal.2005.03.003
Chérubin, L. M., and Richardson, P. L. (2007). Caribbean current variability and the influence of the Amazon and Orinoco freshwater plumes. Deep Sea Res. Oceanogr. Res. Pap. 54, 1451–1473. doi: 10.1016/j.dsr.2007.04.021
Chinain, M., Germain, M., Deparis, X., Pauillac, S., and Legrand, A.-M. (1999). Seasonal abundance and toxicity of the dinoflagellate Gambierdiscus spp. (Dinophyceae), the causative agent of ciguatera in Tahiti, French Polynesia. Mar. Biol. 135, 259–267. doi: 10.1007/s002270050623
Colin, P. L. (2003). Larvae retention: genes or oceanography? Science 300, 1657–1659. doi: 10.1126/science.300.5626.1657c
Corredor, J. E., and Morell, J. M. (2001). Seasonal variation of physical and biogeochemical features in eastern Caribbean Surface Water. J. Geophys. Res. 106, 4517–4525. doi: 10.1029/2000JC000291
Cowen, R. K., Paris, C. B., and Srinivasan, A. (2006). Scaling of connectivity in marine populations. Science 311, 522–527. doi: 10.1126/science.1122039
Dia, A., Guillou, L., Mauger, S., Bigeard, E., Marie, D., Valero, M., et al. (2014). Spatiotemporal changes in the genetic diversity of harmful algal blooms caused by the toxic dinoflagellate Alexandrium minutum. Mol. Ecol. 23, 549–560. doi: 10.1111/mec.12617
Earl, D. A., and vonHoldt, B. M. (2011). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Elena, S. F., and Lenski, R. E. (2003). Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4, 457–469. doi: 10.1038/nrg1088
Erdner, D. L., Richlen, M., McCauley, L. A. R., and Anderson, D. M. (2011). Diversity and dynamics of a widespread bloom of the toxic dinoflagellate Alexandrium fundyense. PLoS One 6:e22965. doi: 10.1371/journal.pone.0022965
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
Fleming, L. E., Bean, J. A., Blythe, D. G., Weisman, R., and Baden, D. G. (1998). Marine Seafood Toxin Diseases: Issues In Epidemiology & Community Outreach NIEHS. Available at: http://www.oceandocs.org/handle/1834/758
Foissner, W. (2006). Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozool. 45, 111–136.
Friedman, M. A., Fernandez, M., Backer, L. C., Dickey, R. W., Bernstein, J., Schrank, K., et al. (2017). An updated review of ciguatera fish poisoning: clinical, epidemiological, environmental, and public health management. Mar. Drugs 15, 72. doi: 10.3390/md15030072
Gerlach, G., Jueterbrock, A., Kraemer, P., Deppermann, J., and Harmand, P. (2010). Calculations of population differentiation based on GST and D: forget GST but not all of statistics! Mol. Ecol. 19, 3845–3852. doi: 10.1111/j.1365-294X.2010.04784.x
Godhe, A., Sjoqvist, C., Sildever, S., Sefbom, J., Hardardottir, S., Bertos-Fortis, M., et al. (2016). Physical barriers and environmental gradients cause spatial and temporal genetic differentiation of an extensive algal bloom. J. Biogeogr. 43, 1130–1142. doi: 10.1111/jbi.12722
Grodsky, S. A., Johnson, B. K., Carton, J. A., and Bryan, F. O. (2015). Interannual Caribbean salinity in satellite data and model simulations. J. Geophys. Res. Oceans 120, 1375–1387. doi: 10.1002/2014JC010625
Hallegraeff, G. M. (2010). Ocean climate change, phytoplankton community responses, and harmful algal blooms: a formidable predictive challenge. J. Phycol. 46, 220–235. doi: 10.1111/j.1529-8817.2010.00815.x
Hendry, A. P. (2004). Selection against migrants contributes to the rapid evolution of ecologically dependent reproductive isolation. Evol. Ecol. Res. 6, 1219–1236.
Jiménez-Muñoz, J. C., Mattar, C., Barichivich, J., Santamaría-Artigas, A., Takahashi, K., Malhi, Y., et al. (2016). Record-breaking warming and extreme drought in the Amazon rainforest during the course of El Niño 2015–2016. Sci. Rep. 6:33130. doi: 10.1038/srep33130
Jombart, T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Jost, L. O. U. (2008). GST and its relatives do not measure differentiation. Mol. Ecol. 17, 4015–4026. doi: 10.1111/j.1365-294X.2008.03887.x
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Keller, M. D., Selvin, R. C., Claus, W., and Guillard, R. R. L. (1987). Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23, 633–638. doi: 10.1111/j.1529-8817.1987.tb04217.x
Kohli, G. S., Farrell, H., and Murray, S. A. (2015). “Gambierdiscus, the cause of ciguatera fish poisoning: an increased human health threat influenced by climate change,” in Climate Change and Marine and Freshwater Toxins, eds L. M. Botana, C. Louzao, and N. Vilariño (Berlin: Walter de Gruyter GmbH & Co KG).
Lebret, K., Kritzberg, E. S., Figueroa, R., and Rengefors, K. (2012). Genetic diversity within and genetic differentiation between blooms of a microalgal species. Environ. Microbiol. 14, 2395–2404. doi: 10.1111/j.1462-2920.2012.02769.x
Legendre, P., and Anderson, M. J. (1999). Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol. Monogr. 69, 1–24. doi: 10.1890/0012-9615(1999)069[0001:DBRATM]2.0.CO;2
Li, Y.-C., Korol, A. B., Fahima, T., and Nevo, E. (2004). Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 21, 991–1007. doi: 10.1093/molbev/msh073
Litaker, R. W., Holland, W. C., Hardison, D. R., Pisapia, F., Hess, P., Kibler, S. R., et al. (2017). Ciguatoxicity of Gambierdiscus and Fukuyoa species from the Caribbean and Gulf of Mexico. PLoS One 12:e0185776. doi: 10.1371/journal.pone.0185776
Litaker, R. W., Vandersea, M. W., Faust, M. A., Kibler, S. R., Nau, A. W., Holland, W. C., et al. (2010). Global distribution of ciguatera causing dinoflagellates in the genus Gambierdiscus. Toxicon 56, 711–730. doi: 10.1016/j.toxicon.2010.05.017
Lowe, C. D., Montagnes, D. J. S., Martin, L. E., and Watts, P. C. (2010). Patterns of genetic diversity in the marine heterotrophic flagellate Oxyrrhis marina (Alveolata: Dinophyceae). Protist 161, 212–221. doi: 10.1016/j.protis.2009.11.003
Lozano-Duque, Y., Richlen, M. L., Smith, T. B., Anderson, D. M., and Erdner, D. L. (2018). Development and validation of PCR-RFLP assay for identification of Gambierdiscus species in the Greater Caribbean Region. J. Appl. Phycol. 1–12. doi: 10.1007/s10811-018-1491-5
Lundholm, N., Ribeiro, S., Godhe, A., Rostgaard Nielsen, L., and Ellegaard, M. (2017). Exploring the impact of multidecadal environmental changes on the population genetic structure of a marine primary producer. Ecol. Evol. 7, 3132–3142. doi: 10.1002/ece3.2906
Lynch, M., Gabriel, W., and Wood, A. M. (1991). Adaptive and demographic responses of plankton populations to environmental change. Limnol. Oceanogr. 36, 1301–1312. doi: 10.4319/lo.1991.36.7.1301
Meirmans, P. G., and Van Tienderen, P. H. (2004). Genotype and Genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4, 792–794. doi: 10.1111/j.1471-8286.2004.00770.x
Nei, M. (2005). Selectionism and neutralism in molecular evolution. Mol. Biol. Evol. 22, 2318–2342. doi: 10.1093/molbev/msi242
Nei, M., Maruyama, T., and Chakraborty, R. (1975). Bottleneck effect and genetic variability in populations. Evolution 29, 1–10. doi: 10.2307/2407137
Nielsen, R. (2005). Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218. doi: 10.1146/annurev.genet.39.073003.112420
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2016). vegan: Community Ecology Package. Available at: http://CRAN.R-project.org/package=vegan
Orsini, L., Vanoverbeke, J., Swillen, I., Mergeay, J., and De Meester, L. (2013). Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol. Ecol. 22, 5983–5999. doi: 10.1111/mec.12561
Parsons, M. L., Settlemier, C. J., and Bienfang, P. K. (2010). A simple model capable of simulating the population dynamics of Gambierdiscus, the benthic dinoflagellate responsible for ciguatera fish poisoning. Harmful Algae 10, 71–80. doi: 10.1016/j.hal.2010.07.002
Pisapia, F., Holland, W. C., Hardison, D. R., Litaker, R. W., Fraga, S., Nishimura, T., et al. (2017). Toxicity screening of 13 Gambierdiscus strains using neuro-2a and erythrocyte lysis bioassays. Harmful Algae 63, 173–183. doi: 10.1016/j.hal.2017.02.005
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Rengefors, K., Logares, R., and Laybourn-Parry, J. (2012). Polar lakes may act as ecological islands to aquatic protists. Mol. Ecol. 21, 3200–3209. doi: 10.1111/j.1365-294X.2012.05596.x
Rengefors, K., Logares, R., Laybourn-Parry, J., and Gast, R. J. (2014). Evidence of concurrent local adaptation and high phenotypic plasticity in a polar microeukaryote. Environ. Microbiol. 7, 1510–1519. doi: 10.1111/1462-2920.12571
Richlen, M. L., Erdner, D. L., McCauley, L. A. R., Libera, K., and Anderson, D. M. (2012). Extensive genetic diversity and rapid population differentiation during blooms of Alexandrium fundyense (Dinophyceae) in an isolated salt pond on Cape Cod, MA, USA. Ecol. Evol. 2, 2583–2594. doi: 10.1002/ece3.373
Rousset, F. (2008). genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Ruggiero, M. V., D’Alelio, D., Ferrante, M. I., Santoro, M., Vitale, L., Procaccini, G., et al. (2017). Clonal expansion behind a marine diatom bloom. ISME J. 12, 463–472. doi: 10.1038/ismej.2017.181
Rynearson, T. A., Newton, J. A., and Armbrust, E. V. (2006). Spring bloom development, genetic variation, and population succession in the planktonic diatom Ditylum brightwellii. Limnol. Oceanogr. 51, 1249–1261. doi: 10.4319/lo.2006.51.3.1249
Sassenhagen, I., and Erdner, D. L. (2017). Microsatellite markers for the dinoflagellate Gambierdiscus caribaeus from high-throughput sequencing data. J. Appl. Phycol. 29, 1927–1932. doi: 10.1007/s10811-017-1076-8
Sjöqvist, C., Godhe, A., Jonsson, P. R., Sundqvist, L., and Kremp, A. (2015). Local adaptation and oceanographic connectivity patterns explain genetic differentiation of a marine diatom across the North Sea-Baltic Sea salinity gradient. Mol. Ecol. 24, 2871–2885. doi: 10.1111/mec.13208
Slatkin, M. (1987). Gene flow and the geographic structure of natural populations. Science 236, 787–792. doi: 10.2307/1699930
Smith, T., Ennis, R. S., Kadison, E., Weinstein, D. W., Jossart, J., Gyory, J., et al. (2016). The United States Virgin Islands Territorial Coral Reef Monitoring Program, Year 15 Annual Report. Saint Thomas: University of the Virgin Islands, doi: 10.13140/RG.2.1.1277.8002
Souffreau, C., Vanormelingen, P., Van de Vijver, B., Isheva, T., Verleyen, E., Sabbe, K., et al. (2013). Molecular evidence for distinct antarctic lineages in the cosmopolitan terrestrial diatoms Pinnularia borealis and Hantzschia amphioxys. Protist 164, 101–115. doi: 10.1016/j.protis.2012.04.001
Tammilehto, A., Watts, P. C., and Lundholm, N. (2016). Isolation by time during an arctic phytoplankton spring bloom. J. Eukaryot. Microbiol. 64, 248–256. doi: 10.1111/jeu.12356
Tesson, S. V., Montresor, M., Procaccini, G., and Kooistra, W. H. (2014). Temporal changes in population structure of a marine planktonic diatom. PLoS One 9:e114984. doi: 10.1371/journal.pone.0114984
Van Dolah, F. M. (2000). Marine algal toxins: origins, health effects, and their increased occurrence. Environ. Health Perspect. 108, 133–141. doi: 10.1289/ehp.00108s1133
Vanormelingen, P., Verleyen, E., and Vyverman, W. (2008). The diversity and distribution of diatoms: from cosmopolitanism to narrow endemism. Biodivers. Conserv. 17, 393–405. doi: 10.1007/s10531-007-9257-4
Xu, Y., Richlen, M. L., Liefer, J. D., Robertson, A., Kulis, D., Smith, T. B., et al. (2016). Influence of environmental variables on Gambierdiscus spp. (Dinophyceae) growth and distribution. PLoS One 11:e0153197. doi: 10.1371/journal.pone.0153197
Keywords: Gambierdiscus, microalgae, population structure, microsatellites, local adaptation, genetic drift
Citation: Sassenhagen I, Gao Y, Lozano-Duque Y, Parsons ML, Smith TB and Erdner DL (2018) Comparison of Spatial and Temporal Genetic Differentiation in a Harmful Dinoflagellate Species Emphasizes Impact of Local Processes. Front. Mar. Sci. 5:393. doi: 10.3389/fmars.2018.00393
Received: 05 July 2018; Accepted: 09 October 2018;
Published: 26 October 2018.
Edited by:
Zhijun Dong, Yantai Institute of Coastal Zone Research (CAS), ChinaReviewed by:
Shang Yin Vanson Liu, National Sun Yat-sen University, TaiwanGuowei Zhou, South China Sea Institute of Oceanology (CAS), China
Copyright © 2018 Sassenhagen, Gao, Lozano-Duque, Parsons, Smith and Erdner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ingrid Sassenhagen, aW5ncmlkLnNhc3NlbmhhZ2VuQGdtYWlsLmNvbQ==
 Ingrid Sassenhagen
Ingrid Sassenhagen Yida Gao
Yida Gao Yesid Lozano-Duque1
Yesid Lozano-Duque1 Tyler B. Smith
Tyler B. Smith Deana L. Erdner
Deana L. Erdner