- 1Center for Energy, Indian Institute of Technology Guwahati, Guwahati, India
- 2Department of Bioscience and Bioengineering, Indian Institute of Technology Guwahati, Guwahati, India
Depleting fossil fuel, soaring prices, growing demand, and global climate change concerns have driven the research for finding an alternative source of sustainable fuel. Microalgae have emerged as a potential feedstock for biofuel production as many strains accumulate higher amounts of lipid, with faster biomass growth and higher photosynthetic yield than their land plant counterparts. In addition to this, microalgae can be cultured without needing agricultural land or ecological landscapes and offer opportunities for mitigating global climate change, allowing waste water treatment and carbon dioxide sequestration. Despite these benefits, microalgae pose many challenges, including low lipid yield under limiting growth conditions and slow growth in high lipid content strains. Biotechnological interventions can make major advances in strain improvement for the commercial scale production of biofuel. We discuss various strategies, including efficient transformation toolbox, to increase lipid accumulation and its quality through the regulation of key enzymes involved in lipid production, by blocking the competing pathways, pyramiding genes, enabling high cell biomass under nutrient-deprived conditions and other environmental stresses, and controlling the upstream regulators of targets, the transcription factors, and microRNAs. We highlight the opportunities emerging from the current progress in the application of genome editing in microalgae for accelerating the strain improvement program.
Introduction
Depleting fossil fuels, soaring international crude oil prices, the energy crisis, and alarming global warming reports have upsurged global interest in alternative renewable energy sources (Behera et al., 2015). Biomass-derived liquid fuels have emerged as the most attractive source of renewable energy when compared with solar, tidal, and wind energies as they can be conveniently stored and easily transported and used directly in automobiles and other transport engines (Scott et al., 2010). Biofuels made from photosynthetic organism-based feedstocks, including land plants and aquatic microalgae, provide enormous opportunities to meet the global energy demand, satisfying carbon-neutral solutions and enabling carbon dioxide (CO2) sequestration from the atmosphere (Stephenson et al., 2011; Ravindran et al., 2017). The cultivation of terrestrial crops for biofuel feedstock competes with food crops for arable land, compromises the price of edible oil if used as biofuel, and often meets less than the anticipated overall energy to input energy demand of the biofuel required over the life cycle (Scott et al., 2010). Sustainable biofuel production using feedstocks, other than terrestrial crops is therefore very promising (Courchesne et al., 2009).
Oil rich microalgae have emerged as the most realistic feedstock for the large-scale production of biofuels as they do not require fresh water (Stephenson et al., 2011). Moreover, these photosynthetic organisms are known to fix solar energy into biomass at efficiencies exceedingly higher than terrestrial plants on a land area basis (Klok et al., 2014). Many oleaginous microalgal species accumulate very large amounts of lipids in the form of triacylglycerol (TAG), the convenient source of biofuel often exceeding 70% of dry cell weight in certain species (Scott et al., 2010). Apart from this, some microalgae produce valuable coproducts such as pigments, antioxidants, edible proteins, long-chain polyunsaturated fatty acids, and specialized bio-pharmaceuticals, favoring biorefineries to help to offset the biofuel production cost (Klok et al., 2014; Jagadevan et al., 2018). However, many challenges need to be overcome to realize this potential of large scale production of microalgal biofuels as a sustainable and cost-effective alternative for fuel. The major bottlenecks are the absence of two attributes, high lipid content and fast growth rate in existing microalgal species (Fan et al., 2014; Ghosh et al., 2016; Chen et al., 2017) and inefficient light harvesting capacity in naturalized growth conditions (Stephenson et al., 2011). Understanding the molecular intricacies of lipid metabolism, especially TAG biosynthesis, their genetic and metabolic regulations for triggered metabolic flux, target subcellular storage location, and possible secretion, could assist in strain improvement, thus, maximizing the cost effectiveness of biofuel production in microalgae.
In this review, we provide the current knowledge and potential biotechnological routes to enhance TAG accumulation, synchronized biomass production, improved light harvesting, maximize the utilization of available nutrients, and adapt to variable and fragile conditions prevalent in real-life conditions in aquatic habitats.
Microalgae as a Biofuel Source
Triacylglycerol is the primary form of energy storage in microalgal cells, which comprises 60–70% of the dry cell weight (Hu et al., 2008; Scott et al., 2010). Each TAG molecule consists of a glycerol backbone to which three fatty acid (FA) moieties are anchored. Each FA molecule is classified, depending on the degree of unsaturation, as either saturated FA (SFA), monosaturated FA (MUFA), or polyunsaturated FA (PUFA). Therefore, relative abundance of these diverse FAs in TAG decides the utility of TAG molecules for specific applications, including their use as transportation fuel, high-value nutrient supplements, emulsifiers, and industrial polymers. Microalgae-derived biomass can supply a wide range of biofuels such as biodiesel, bioethanol, biohydrogen, biomethane, and bioelectricity. Among all projected applications, microalgae-derived oils are the most promising for the production of biofuel (Shuba and Kifleb, 2018). Therefore, current microalgal studies worldwide primarily focus on enhancing lipid accumulation in microalgae under diverse growth conditions for higher oil production.
Overview of Oil Accumulation in Microalgae
The biosynthesis of lipid molecules in microalgae, in particular that of the ‘green algal’ lineage, is an interconnected network of multiple metabolic pathways. It begins in the chloroplast of microalgal cells where the photosynthetic machinery utilizes atmospheric carbon to yield starch, which is later catabolized through glycolysis to form the building blocks of FAs and TAGs (Figure 1). Incorporation of these precursors in the form of acetyl-CoA to synthesize malonyl-CoA by acetyl-CoA carboxylase (ACC) initiates FA biosynthesis. Conversion of malonyl-CoA to malonyl-ACP marks the beginning of the elongation phase of FA biosynthesis, catalyzed by a prokaryotic type-II FA synthase (FAS II) localized in the stroma (Blatti et al., 2013; Shtaida et al., 2015). The process, however, is interrupted intermittently by fatty-ACP thioesterases (FATs) and as a consequence, the newly synthesized FAs escape from the acyl-ACP complex (Blatti et al., 2013). The generated free FA pool is assimilated while synthesizing various cellular lipids. The synthesis of storage lipids, in particular, follows a set of reactions as part of the Kennedy pathway, which involves the incorporation of FAs into a glycerol backbone to form TAG. In the Kennedy pathway, acyl-CoA or acyl-ACP acts as an acyl donor while microalgae can follow an alternate pathway for sourcing acyl groups for TAG synthesis, which uses phospholipids as acyl donors (Li-Beisson et al., 2015). The rapid stride in the manipulation of oil biosynthesis in plants for oil enhancement provides clues for engineering the microalgal lipid metabolism, provided the differences in the lipid metabolism process between microalgae and plants are understood.
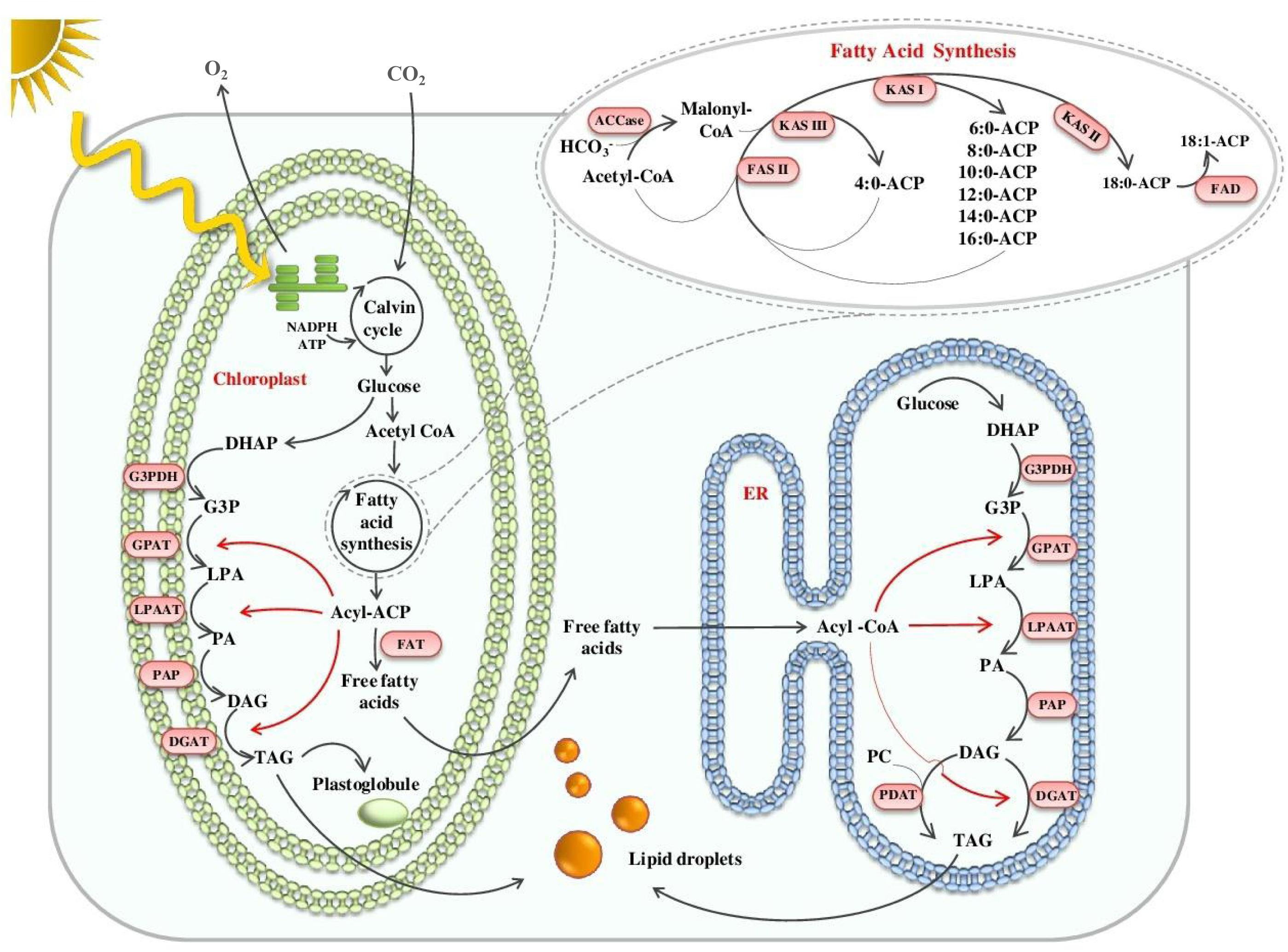
FIGURE 1. Schematic illustration of TAG synthesis in microalgae. NADPH; Nicotinamide adenine dinucleotide phosphate; ATP, Adenosine Triphosphate; DHAP, dihydroxyacetone phosphate; G3P/G3pDH, Glyceraldehyde 3-phosphate / G3P dehydrogenase; GPAT, Glycerol 3-phosphate acyltransferase; PA/LPA/LPAAT/PAP, Phosphatidic acid/Lyso-PA/LPA acyltransferase/PA phosphatase; DAG/DGAT, di-Acylglycerol/ DAG acyltransferase; FAT, Fatty acyl-ACP thioesterase; ACP, Acyl-carrier protein; ER, Endoplasmic reticulum; PC, Phosphatidylcholine; PDAT, Phospholipid:DGAT; ACCase, Acetyl-CoA carboxylase; FAS, Fatty acid synthase; KAS, 3-ketoacyl-ACP synthase; FAD, Flavin adenine dinucleotide.
A pioneering understanding of the model plant’s lipid metabolism laid the foundation for the identification of key genes of TAG biosynthesis in microalgae. Microalgal omics has enabled the prediction and accurate annotation of lipid metabolism genes. Most of the predicted genes involved in FA biosynthesis are present as a single copy in the Chlamydomonas genome, suggesting that their encoded enzymes operate both in the chloroplast and mitochondria, in contrast to higher plants where compartment-specific enzymes increase the complexity of FA biosynthesis. Many copies of genes encoding acyl-CoA: diacylglycerol acyltransferases (DGATs) were predicted in the Chlamydomonas genome, compared with the fewer copies observed in higher plants, suggesting a crucial role of TAG in microalgal cell physiology (Liu and Benning, 2013). The model microalgae, Chlamydomonas, accumulates starch as the primary form for energy storage; however, stress redirects it to TAG formation. This carbon sifting is possibly due to adaptation for maintaining membrane integrity. Triacylglycerol molecules are catabolized back to release FAs upon stress reversal and used for membrane synthesis. Besides this, TAG molecules also act as a sink for channeling excess energy and reductive equivalents, which otherwise risk cellular metabolisms of microalgae (Sharma et al., 2012; Shtaida et al., 2015). Another marked difference is the presence of unique lipids such as betaines, which offer an advantage to microalgae in adapting to nutrient-limiting conditions. On the contrary, higher plants exclusively use phosphate-associated lipids that play a central role in maintaining the membrane integrity (Van Mooy et al., 2009; Liu and Benning, 2013).
In contrast to plants, microalgae contain distinct acyl groups in storage lipids (Garay et al., 2014). In plants, a majority of TAGs are assembled in the endoplasmic reticulum (ER), whereas in microalgae, a major fraction of TAGs are assembled de novo in the chloroplast (Giroud et al., 1988; Fan et al., 2011). The key difference between these two organelle pathways is the presence of a 16-carbon acyl group in the sn-2 position of the glycerol backbone in lipids derived from the plastid pathway while an 18-carbon acyl group occupies the same position in ER-generated lipids (Xu et al., 2016). However, the recent identification of an ER-localized acyltransferase enzyme having specificity toward C-16 acyl-donor suggests that the difference in the prokaryotic and eukaryotic pathways may not be due to spatial separation but due to complex systemic control (Kim et al., 2018).
The triggers for TAG accumulation also appear to differ in plants and microalgae. In plants, TAGs are accumulated in the developing seeds controlled through developmental signaling, whereas stress conditions trigger lipid accumulation, but the underlying mechanism is not fully established (Liu and Benning, 2013; Garay et al., 2014), presumably because of the coordinated sequential consequence of cell cycle arrest. Cornell et al. (1977) found that, at least in some cell culture types, lipid synthesis is controlled at certain checkpoints in the cycle. As stressors often obstruct cell cycle progression, it led to a speculation that they effectively trigger lipid accumulation (Kwok and Wong, 2005). However, this type of induction process requires validation. Changes in buoyancy due to lipid accumulation, assisting in motility and protection of microalgae, are possibly an adaptation in aquatic environments. Although several external agents initiate lipid accumulation, metabolic regulators such as nitrogen response regulator 1 (NRR1) and phosphorus starvation response 1 (PSR1) are core to this process as they sense the changes in the cytoplasmic environment and activate various pathways associated with TAG biosynthesis (Gargouri et al., 2015).
Conventional Means for Improving Oil Production in Microalgae
Cultural manipulations such as subjecting cells to stressors, like nutrient depletion, variable light intensity, temperature, salinity, and pH, are conventionally used to enhance lipid accumulation within the cells’ biological limits (Bartley et al., 2014; Chu et al., 2015; Suyono et al., 2015). Among these stressors, nitrogen starvation is the most potent for lipid enhancement (Belotti et al., 2013). Dual-stage cultivation (Doan and Obbard, 2014) and coculture techniques with chemical additives (Singh S.K. et al., 2016) facilitate the enhancement of lipid production in culture systems. However, the genetic modification of microalgae offers more avenues for the precise control of target mechanisms leading to enhanced cellular lipid accumulation under normal growth conditions (Xue et al., 2015; Lim and Schenk, 2017). Genome sequence databases and pathway databases (KEGG, dEMBF, and MetaCyc) are now valuable resources for implementing targeted genetic manipulation for higher lipid biosynthesis in microalgae (Ogata et al., 1999; Caspi et al., 2014; Misra et al., 2016). To date, successful nuclear transformation has been reported in more than 40 microalgal species, and considering the challenges posed by the enormous physiological and genetic diversities existing among these microalgal species, this number appears significant (Gangl et al., 2015; Gimpel et al., 2015; Doron et al., 2016). In recent years, different tools such as metabolic selection markers and techniques like ‘CHYSEL’ have been developed to target both plastids and nuclear genomes, allowing for the expression of target genes (Specht et al., 2010; Rasala et al., 2014). With these rapid strides in microalgal biotechnology, ‘algomics’ and integrated system-biology modeling have deepened the understandings of interconnections between genes, proteins, and metabolites (Jamers et al., 2009; Koussa et al., 2014; Reijnders et al., 2014; Benmoussa, 2016). Such integrated multidisciplinary studies can provide a clear picture of oil and high-value metabolite biosynthesis pathways, thereby accelerating strain improvement for the commercialization of microalgal biofuel (Lauersen et al., 2015; Patra et al., 2015; Barahimipour et al., 2016).
Genetic Engineering Strategies for Lipid Enhancement in Microalgae
Enhancing oil synthesis in microalgae primarily depends on the manipulation of enzymes involved in lipid biosynthesis or other competitive parallel pathways aimed to divert the carbon and reductive equivalents flux toward lipid biosynthesis (see Figure 2). The most widely used technique is the manipulation of individual genes encoding various steps of a metabolic pathway; however, owing to the multi-factorial regulation of lipid biosynthesis in microalgae, this strategy has seen mixed success (Bajhaiya et al., 2017). Recently, the transcriptional regulation of oil biosynthesis has brought widespread interests to control the activity or expression of multiple components of a metabolic pathway simultaneously (Courchesne et al., 2009). Additionally, attempts to manipulate various other targets, such as improving light use efficiency, controlling cell quiescence, and improving carbon sequestration, etc., which indirectly influence the lipid content by altering cell growth characteristics, have gained attention (Figure 2).
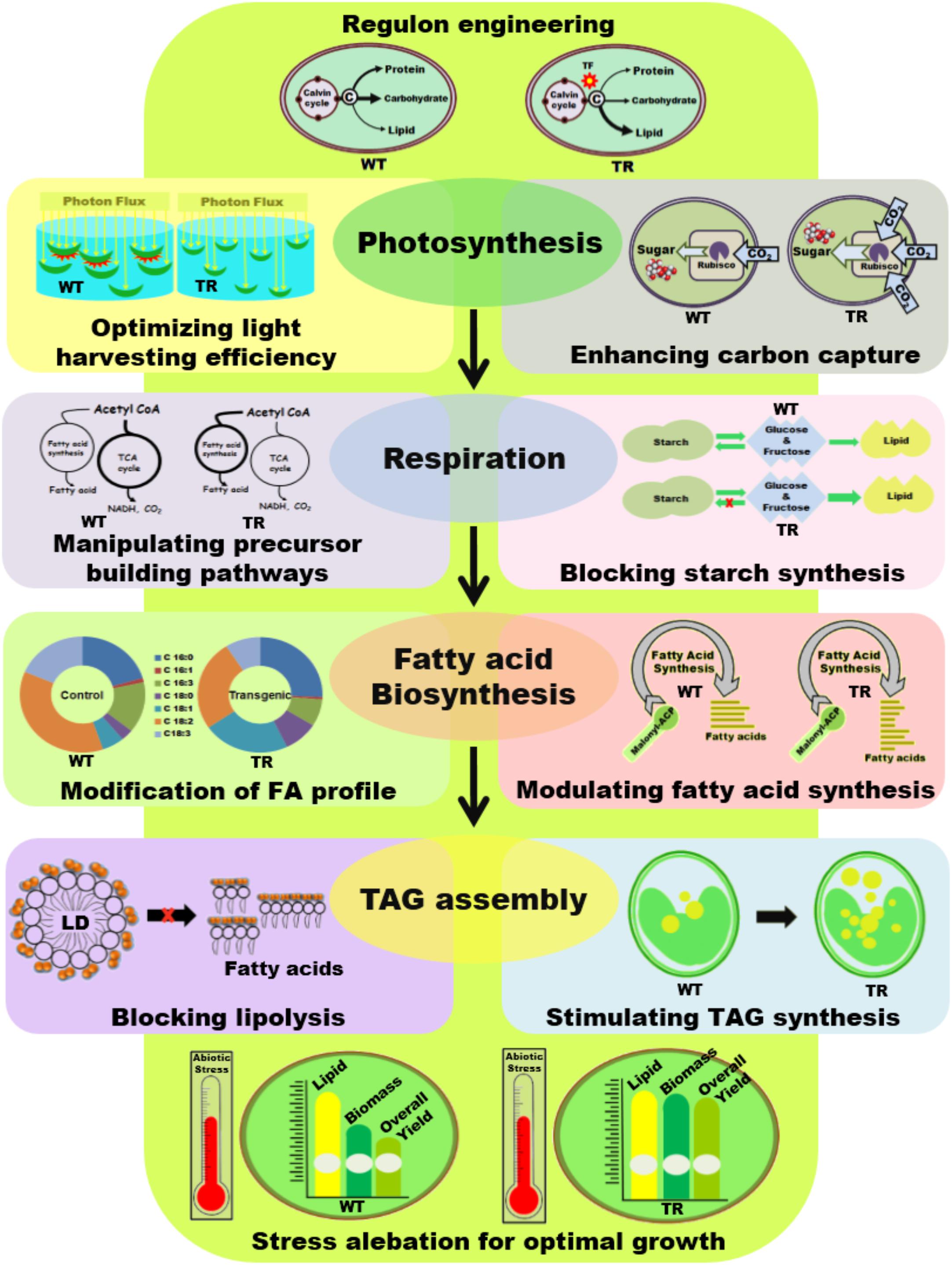
FIGURE 2. Schematic illustration of different genetic engineering strategies applied in microalgae for biodiesel application. WT, Wild type cells; TR, Transgenic cells; TF, Transcription factor; TCA, Tricarboxylic acid cycle; NADH, Nicotinamide adenine dinucleotide; FA, Fatty acid; LD, Lipid droplet.
Manipulation of the Oil Biosynthesis Pathway
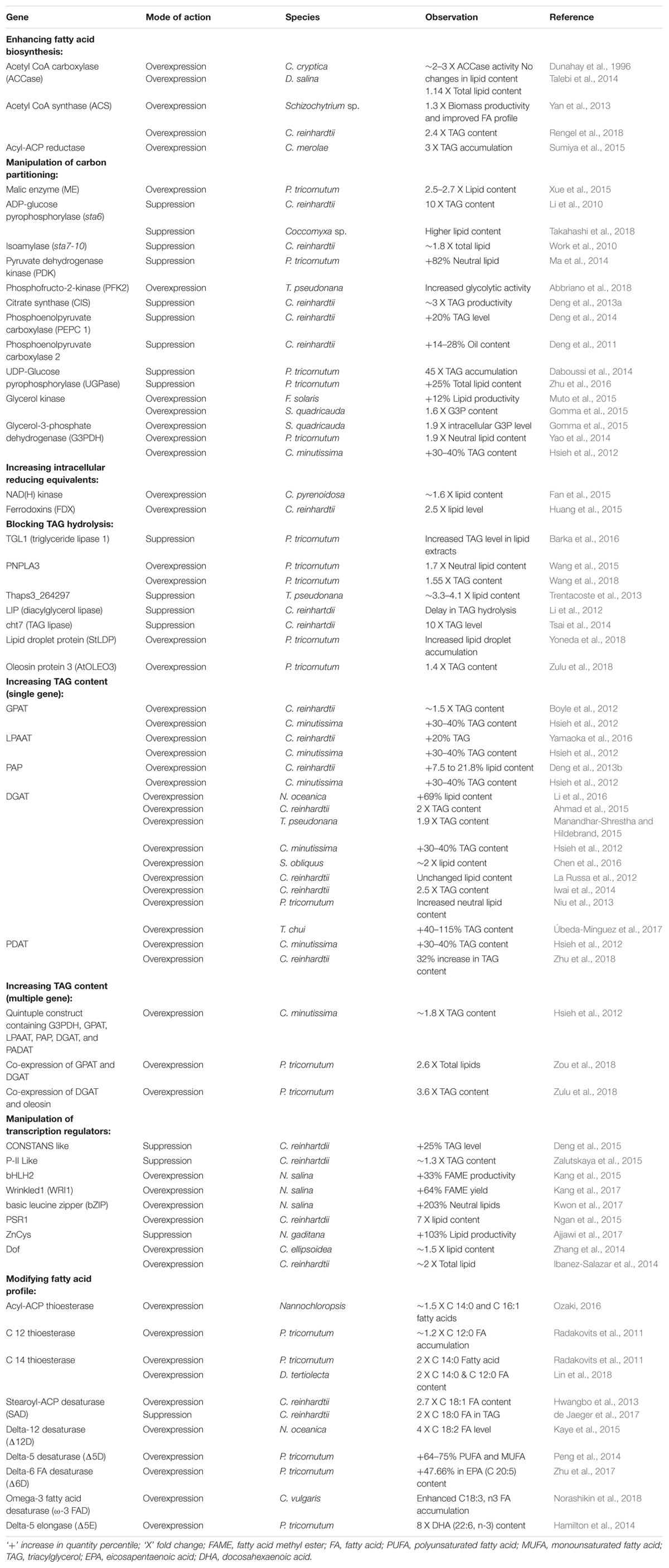
In contrast to subjecting microalgae to growth limiting stress conditions, efforts targeting enhanced lipid accumulation during the exponential growth phase are more practical means. According to Ohlrogge and Jaworski (1997), the FA supply regulates the lipid biosynthesis process, and therefore, some of the earliest attempts have been made to increase the expression of key enzymes involved in FA biosynthesis. Microalgal metabolic engineering aimed at increasing FA supply to lipid synthesis was first attempted by overexpressing the acetyl-CoA carboxylase gene (ACCase), which codes for the enzyme that carboxylates acetyl-CoA to malonyl-CoA, the first committed step in FA synthesis (Dunahay et al., 1996). Although the transformed microalgae showed a two- to three-fold increase in ACCase activity, it was not accompanied with an increase in FA content (Sheehan et al., 1998). This clearly indicated that the upregulation of ACCase had no direct impact on lipid biosynthesis. However, the simultaneous overexpression of a subunit of ACCase (accD) along with malic enzyme (ME), responsible for the conversion of malate to pyruvate, was successful in elevating the total lipid content in microalgae (Talebi et al., 2014). Therefore, it appears that ACC is not the sole rate-limiting step in lipid biosynthesis, indicating the existence of a secondary rate-limiting step apart from ACC. Limited availability of precursors for the whole lipid synthesis process (acetyl-CoA and glucose-6-phosphate) could be the secondary bottleneck in case ACC is overexpressed. Various studies attempted to elevate the intracellular concentration of lipogenic precursors, by tailoring the enzymes involved in the generation of reducing potential (NADH) and in carbon metabolism (Gimpel et al., 2015). Manipulation in the expression of several enzymes like pyruvate dehydrogenase, phosphoenolpyruvate carboxylase, acetyl-CoA synthase, NAD(H) kinase, and glycerol kinase has significantly enhanced the lipid content in different microalgal species without adversely affecting cell growth (Table 1).
Another possible strategy to increase the intracellular lipid content is by blocking the metabolic pathways that are competitive to lipogenesis, for example, starch synthesis and lipid catabolism. Some strains of microalgae use starch as the primary storage metabolite, and suppressing starch synthesis can funnel the carbon flow toward lipid biosynthesis (Ravindran et al., 2017). Knockdown of key genes involved in starch synthesis showed elevated lipid accumulation by redirecting carbon pool toward lipogenesis (Table 1). Since accumulation of starch as an energy storage molecule is not universal in microalgae (León-Saiki et al., 2017), the suppression of lipid catabolism is a more legitimate option to enhance the lipid content irrespective of the microalgal strains. For instance, inhibiting the expression of a multifunctional lipase/phospholipase/acyltransferase enzyme in Thalassiosira pseudonana resulted in increased lipid yields without affecting the growth (Trentacoste et al., 2013). The mutant strain showed a 2.4- to 3.3-fold higher lipid accumulation in comparison with the control, when subjected to silicon starvation. However, blocking these vital metabolic pathways (lipid catabolism and starch synthesis) may result in reduced microalgal growth and lipid yield (Radakovits et al., 2010; Chu, 2017). One way to overcome this is by using RNAi mediated gene silencing under the control of inducible promoters. Upon attaining high cell-density, the mechanism can be activated to suppress the expression of key genes involved in starch synthesis and lipid catabolism. Many such promoters have been identified in microalgae, including one with light-responsive elements in Dunaliella (Park et al., 2013; Baek et al., 2016).
Besides the manipulation of carbohydrate metabolism- and lipid metabolism-related genes for increasing cellular neutral lipids (TAG), the overexpression of acyltransferases has also yielded interesting outcomes (see Table 1). The Kennedy pathway for TAG assembly includes several steps catalyzed by different acyltransferases, including acyl-CoA: glycerol-3-phosphate acyltransferase (GPAT), acyl-CoA: lysophosphatidic acyltransferase (LPAAT), and acyl-CoA: DGAT. These TAG assembly genes were found to be worthy targets in lipid pathway engineering (Bhowmick et al., 2015; Maravi et al., 2016). In the green microalgae Chlorella minutissima, simultaneous expression of five acyltransferases (phosphatidic acid phosphatase, LPAAT, glycerol-3-phosphate dehydrogenase, GPAT, and DGAT) resulted in a two-fold increase in lipid content (Hsieh et al., 2012). These instances of co-expressing multiple enzymes exemplify the effectiveness of system level control of metabolic flux toward lipid overproduction. Transcriptional regulation can have a similar effect on the systemic metabolomic flux as transcription factors can target multiple regulatory points in a metabolic pathway. Overexpression of genes encoding transcription factors targeting the upregulation of downstream lipid biosynthesis genes can result in increased oil content. In this realm, higher plants have been in the spotlight with numerous literature highlighting the benefit of transcription factor engineering for enhanced lipogenesis (Cernac and Benning, 2004; Mendoza et al., 2005). However, in microalgae, the major focus of transcriptional regulation studies is limited to a select microalgal species (see Table 1). In this context, the identification of endogenous transcription factors and their subsequent manipulation in their host would be more viable to trigger lipid accumulation (Tsai et al., 2014; Ngan et al., 2015; Kwon et al., 2017). Although the manipulation of transcription factors like PSR1 and compromised hydrolysis of triglycerols 7 (CHT7) have led to enhanced lipid accumulation without compromising biomass production, weak carbon partitioning for lipid synthesis still remains a bottleneck which may be overcome by finding other potential lipid-triggers (Chen et al., 2018). In one such groundbreaking effort, the knockdown of a single transcription regulator ZnCys in Nannochloropsis gaditana resulted in a 103% increase in lipid content, indicating a lipid yield to the tune of ∼5 g/m2/day (Ajjawi et al., 2017). Even though the overexpression of endogenous transcription factors for increasing oil content in microalgae is very promising, the lengthy functional characterization process greatly limits its applications. A more direct approach is to consider the heterologous expression of a transcription factor of plant origin to regulate the microalgal lipid biosynthetic pathway. Several lipogenesis promoting transcription factors from higher plants were overexpressed in microalgae and showed to have a remarkable impact on the lipid accumulation pattern (see Table1).
In addition to engineering for enhanced oil content, it is also important to improve the quality of oil for better biodiesel fuel properties. The carbon chain length and degree of unsaturation of the FAs present in oil affect the cold flow and oxidative stability properties of the fuel. Oils derived from microalgal feedstocks commonly contain FAs of chain length between 14 and 20, mostly C16:0, C16:1, and C18:1, while the ideal should be C12:0 and C14:0 (Radakovits et al., 2010). The key factor that determines FA chain length is the thioesterase enzyme, which catalyzes the release of the FA chain from the FA synthase complex. Several acyl-ACP thioesterases specific to short FA chain length have been identified, and engineering the expression of these enzymes can successfully modify the fuel properties. Transgenic microalgae containing exogenous short-chain length biased FA acyl-ACP thioesterases have directed an increase in percent composition of myristic (C14:0) and lauric (C12:0) acids in the overall FA profile (see Table 1). A seamless biodiesel fuel also requires a balanced coalescence of MUFAs, SFAs, and PUFAs in oil (Durrett et al., 2008). The scant presence of MUFAs in the microalgal lipid profile (Patil et al., 2007) requires biotechnological interventions to modify the degree of unsaturation. The desaturase enzyme, which catalyzes the formation of unsaturated FAs, was targeted to manipulate the FA profile primarily in MUFA and PUFA contents (see Table 1). Enhancements in PUFA entities like linoleic acid and eicosapentaenoic acid are particularly noteworthy considering their high nutritional value.
Apart from biodiesel, microalgal oils can also be used to produce gasoline and jet fuel, which requires FAs with even shorter chain lengths. Even though it is possible to chemically synthesize suitable feedstocks for gasoline or jet fuel by breaking down the long chains into shorter hydrocarbon chains, the genetic engineering of microalgae to synthesize short-chain FAs will significantly reduce the production cost (Radakovits et al., 2010). For instance, the overexpression of 8:0- and 10:0-biased thioesterases from Cuphea hookeriana in Canola has reportedly enhanced the synthesis of short-chain FAs (Dehesh et al., 1996). Replication of this achievement of raised short-chain FA profile in different oleaginous microalgal species would have a high impact.
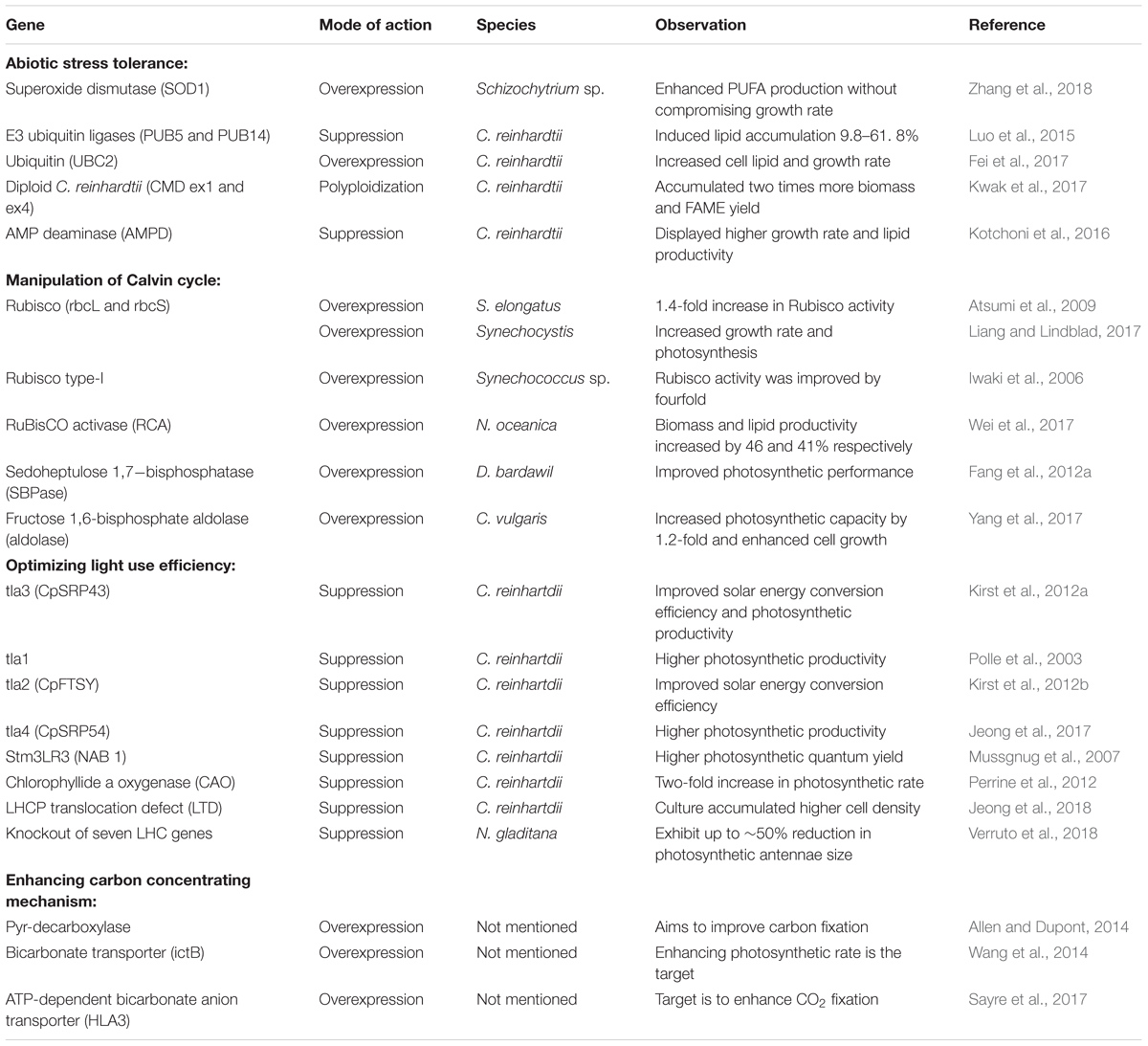
Enhancing the Biomass Yield
Enhancing the biomass yield is very important as the total energy output relies on both energy density and the total biomass content (Barry et al., 2015). In photosynthetic organisms including microalgae, abiotic stress, CO2 fixation rate, and light utilization efficiency are the primary factors that govern biomass productivity (Chu, 2017). Engineering microalgal strains for stress tolerance and higher photosynthetic efficiency can ensure the cost-effective production of biofuel. A number of studies report transgenic microalgal strains tolerant to abiotic stress through enhanced reactive oxygen species (ROS) scavenging, hypertolerance to DNA damage, and polyploidization (see Table 2). Kotchoni et al. (2016) reported the manipulation of intracellular steady-state ATP levels for cold adaptation of microalgal cells. The identification of key transcription regulators and enzymes, as well as stress-responsive promoters through omics analysis, can serve as the toolbox for future genetic engineering designs.
Manipulation of carbon fixation is vital to improving the photosynthesis rate. The Calvin cycle is the initial pathway for carbon fixation in all photosynthetic organisms, and strategies seeking improvement in the photosynthetic efficiency require a breakthrough in the regulation of this pathway. Carboxylation of ribulose 1,5-bisphosphate (RuBP) and its subsequent regeneration are the checkpoints in the Calvin cycle. The enzymes that catalyze these regulatory steps are ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), fructose 1,6-bisphosphate aldolase (aldolase), and sedoheptulose 1,7-bisphosphatase (SBPase), and these three enzymes are prime targets for manipulating the Calvin cycle owing to their high flux control coefficient values (Raines, 2003; Yang et al., 2017). Among the three enzymes, Rubisco is the primary target as the carboxylation capacity of Rubisco majorly influences the rate of carbon assimilation. However, efforts to directly manipulate this enzyme have been met with limited success, owing to the complex enzyme kinetics of Rubisco, which challenges the operational understanding and makes it difficult to spot a change in the phenotype upon manipulation (Tcherkez et al., 2006). Therefore, efforts are being made to target factors that regulate Rubisco activity instead of the direct manipulation of the enzyme itself. One such target is the Rubisco activase (RCA) enzyme, which regulates the activity of Rubisco by regenerating the catalytic sites (Hazra et al., 2015). Ultimately, RCA determines the rate of carbon fixation by maintaining a high proportion of catalytically competent active sites of Rubisco. An attempt to overexpress RCA in an oleaginous microalga, Nannochloropsis oceanica, resulted in elevated photosynthetic activity accompanied by enhanced biomass and lipid accumulation (Wei et al., 2017). Apart from Rubisco, aldolase and SBPase are also crucial to improve carbon fixation as these enzymes are involved in the regeneration of precursor substrates for Rubisco. As a proof of concept, the overexpression of genes encoding aldolase and SBPase in different microalgal strains resulted in enhanced photosynthetic efficiency and biomass production (Table 2).
Abiotic factors such as availability of photon energy also affect the efficiency of carbon fixation. Photosynthetic microalgae have developed large photosynthetic antenna systems to maximize the photon absorption and conversion efficiency as an adaptation to its habitat where low light intensity is a growth limiting factor. However, the sustainability of microalgal biomass production requires large-scale cultures with high cell density. In such dense cultures, the high pigment density due to the large antenna systems limits the penetration of light into the deeper layers of the culture. Under such conditions, cells at the surface receive an excess amount of photon energy, which quickly saturates the photosynthesis process, and dissipate the excess energy through non-photochemical quenching (NPQ). At the same time, cells in the deeper layers are exposed to a low-light intensity, which compels the cells to perform respiration instead of photosynthesis (Formighieri et al., 2012). This uneven distribution of light energy leads to suboptimal photosynthetic efficiency, which in turn reduces the overall biomass productivity of the culture. One approach to enhance the photosynthetic yield is by reducing the size of light capturing antenna systems in microalgae to minimize the energy loss due to NPQ. In biological terms, antennas, or light-harvesting complex (LHC), are pigment-binding proteins, which capture the light energy and relay it to photosynthetic reaction centers. In green microalgae, they bind the majority of pigments and, therefore, are mainly responsible for the optical density of the culture. Wild type photosynthetic microalgae harbor a vast number of chlorophyll molecules associated with both photosystems I and II; however, only a few of these are essential to carry out the vital functions of photosynthesis (Simionato et al., 2013). Therefore, it is possible to improve light transmission and light absorption capacities by reducing the number of chlorophyll molecules from the LHC in microalgal cells. Mutants have been developed with truncated antenna systems in different microalgal strains through downregulating the genes encoding LHC pigment binding-proteins, which showed a marked reduction in energy losses by NPQ and increased biomass production under laboratory scale culture conditions (see Table 2). Mass culture of these truncated antenna mutants is expected to fine-tune light absorption characteristics influencing higher biomass productivity and the eventual reduction in the production cost. However, susceptibility of such strains with the shrunk antenna system to photodamage by intense solar radiation is a great limitation. de Mooij et al. (2015) found a smaller antenna size that made the mutants vulnerable to high light intensity while tailoring the antenna size in Chlamydomonas reinhardtii. The reduced fitness due to impaired photoprotection mechanisms triggered by altered antenna size lead to insignificant changes in the biomass productivity of mutants. Therefore, future strategies for antenna size reduction in microalgae should address the unintended side effects of antenna size on mutants. Interestingly, transgenic C. reinhardtii strains generated by Perrine et al. (2012), having variation in antenna sizes and reduced chlorophyll (Chl) b content, showed a higher growth rate in mutants with intermediate antenna size. The results conform to the hypothesis that reduction but not the elimination of Chl b content would result in the optimal photosynthesis process.
Among the abiotic factors that influence rate of carbon fixation, the most critical is the availability of inorganic carbon, as the concentration of CO2 in the vicinity of Rubisco affects its carboxylase property. Microalgae have developed a CO2 concentrating mechanism (CCM) to alleviate the stress caused by limited CO2 in aquatic ecosystems (Wang et al., 2011). Microalgae elevate the CO2 concentration at the site of Rubisco through the operation of CCM. Since Rubisco also has the inherent capacity to divert the carbon pool toward unimportant photorespiratory pathways, an elevated CO2 concentration favors carboxylation, thereby increasing the rate of carbon fixation (Singh P. et al., 2016). Inorganic carbon (Ci) import, enzymatic catalysis of the imported carbon to form CO2, and compartmentalized Rubisco systems are the functional components of microalgal CCM. Environmental conditions such as the Ci content, ratio of [CO2]/[O2], and dissolved CO2 concentration are ascribed for the regulation of microalgal CCM (Morales et al., 2018). Several studies in C. reinhardtii have identified various factors such as CIA5, which acts at the cellular level and regulates the transcription of multiple genes having a role in CCM (Fukuzawa et al., 2001; Yoshioka et al., 2004; Fang et al., 2012b). Tapping these regulatory factors in photosynthetic microalgae has assumed significance in enhancing fitness toward naturally occurring low CO2 conditions (Price et al., 2013). Besides these, modulations in other functional components of CCM such as Ci transporters and carbonic anhydrases can also facilitate enhancements in the carboxylation reaction, which in turn increases the photosynthetic performance and biomass yield. However, except a few filed patents (see Table 2), to date, there are no reports of successful CCM engineering in microalgae. Thus, suppression of the oxygenase activity of Rubisco through tailoring CO2 capture mechanism in microalgae remains a challenge to be addressed for improving the carbon fixation process.
Genome Editing in Microalgae for Strain Improvement
Metabolic pathway engineering is crucial for enhancing the productivity of a microalgal strain, and for this purpose, gene editing offers a powerful and easy mechanism to overcome the genetic inadequacies (Ng et al., 2017). Until recently, the RNAi technology was frequently used as a tool for gene silencing and proved efficient in pathway engineering and gene function alteration. However, RNAi has its limitations, which include incomplete suppression, silencing of the RNAi transgene, and inconsistent suppression in different transformants (Banerjee et al., 2018). In contrast, the emergence of genome editing bypasses the limitations of RNAi, offering new avenues to modify and edit the genome of cells. The genome editing techniques based on engineered nucleases like clustered regularly interspaced palindromic sequences/CRISPR-associated protein 9 (CRISPR/Cas9), transcription activator-like effector nucleases (TALENs), and zinc-finger nucleases (ZFNs) provide the means for dissecting the operational organization of genes, gene families, and protein networks. These genome editing tools induce double-strand breaks at a specific locus in the genome, which get repaired through the non-homologous end joining machinery of the DNA repair process and introduce insertions or deletions at sites creating frameshift mutations (Gan and Maggs, 2017). Among the genome editing tools, CRISPR/Cas9 has gained much focus because of its simple, accurate, and efficient nature of operation (Jeon et al., 2017). In the CRISPR/Cas9 system, the Cas9 nuclease is directed by a single guide RNA (sgRNA) molecule, which binds to the target site in the genome following simple base-pairing rules. Steady progress in research on the CRISPR/Cas9 system has resulted in the development of many different variants of this technology. A mutated form of the Cas9 protein (dCas9) lacking the nuclease activity can be used with the CRISPR system to modulate the expression of specific target genes. Depending on the type of the effector molecule fused with dCas9/sgRNA, it is possible to precisely both stimulate and repress the activity of a target gene (Gilbert et al., 2013; Piatek et al., 2015). In addition to the expression modulation of a single gene, multiple genes can be simultaneously activated or silenced by the simple addition of guide RNAs for each of the targets into the dCas9/sgRNA variant of the CRISPR/Cas9 system (Kim and Kim, 2014). The versatility in the application of the CRISPR/Cas9 system makes the technique a remarkable and powerful tool in metabolic pathway engineering.
In microalgae, the utility of CRISPR is on the rise as it has considerable scope in microalgal trait improvement for biofuel and nutraceutical applications. Various advances in CRISPR/Cas and other genome editing tools have led to several successful attempts in many microalgae species (see Table 3), which endorse this technology for its effectiveness in generating targeted mutants. One prominent advantage of applying the CRISPR technology in microalgae is the ease of multiplexing, which, unlike the conventional mutagenesis and RNAi mediated knockout and knockdown approaches, facilitates a less complicated and more programmable approach for manipulating metabolic pathways. In case of lipid engineering in oleaginous microalgal strains, this technique can improve the lipid profile of the microalgal strain by simultaneously blocking the metabolic routes competitive to lipid production such as starch generation, lipid degradation, and β-oxidation. Other than gene silencing, a dCas9 variant can be recruited to activate stress responsive elements of the lipid synthesis pathway under nonstress conditions, thus bypassing the inhibitory effects on the biomass yield. An activator molecule fused with dCas9 can be used to stimulate supportive pathways such as FA synthesis, which facilitates the production of precursors for lipogenesis. Functional characterization of a novel gene is another aspect of utilizing the CRISPR technology in addition to gene editing. Annotating novel genes encoding proteins significant for lipid production can broaden the spectrum of target selection for superior biofuel production.
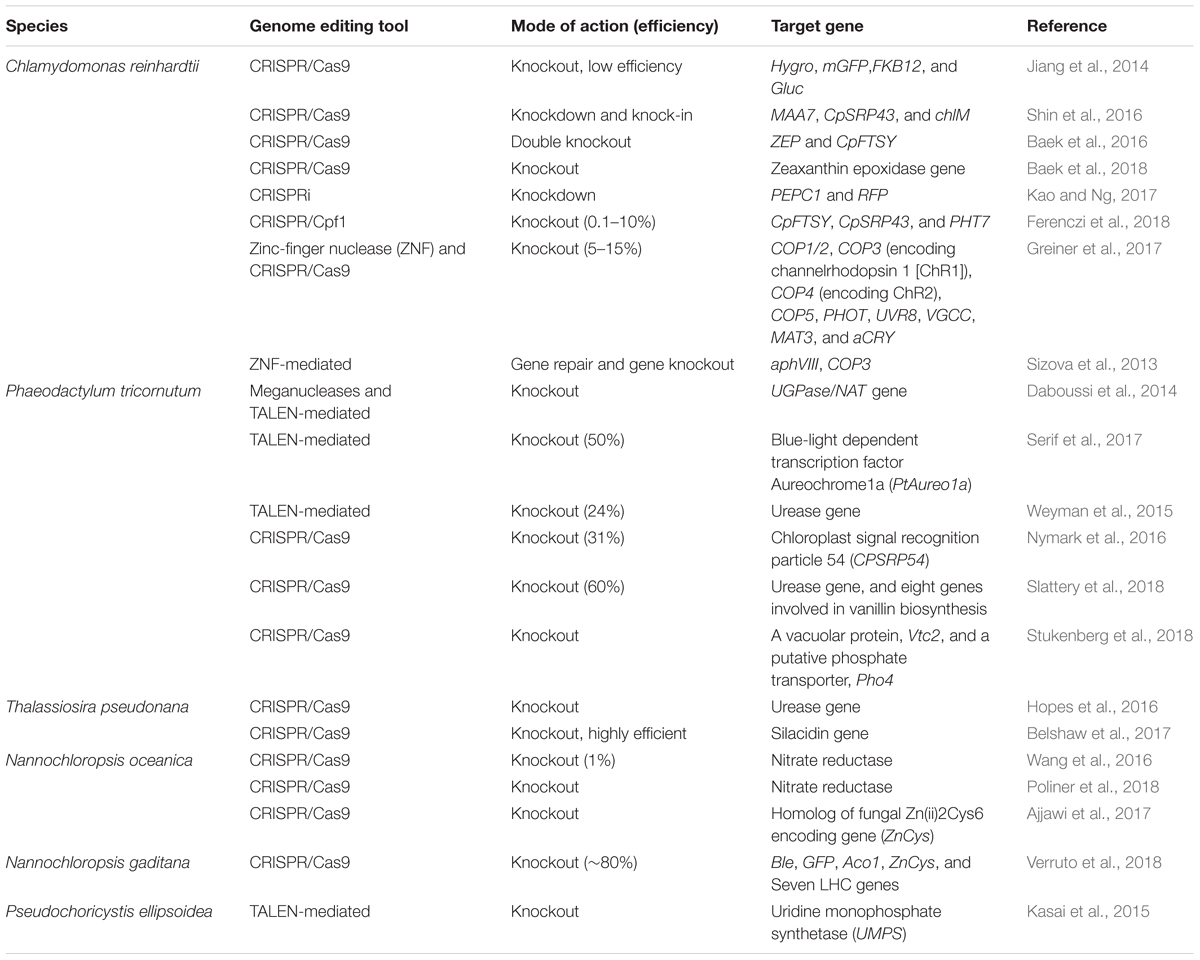
TABLE 3. Overview of the application of different genome editing tools in photosynthetic microalgae.
Indigenous microalgal strains promoted for biofuel production have some limitations for commercial scale production, which include suboptimal lipid profile and light harvesting efficiency, among others. However, adjusting these limiting attributes is not recommended as it interferes with the normal physiology of microalgae. For example, generating truncated LHC in microalgae is associated with susceptibility to photodamage (de Mooij et al., 2015). Therefore, it is advantageous to have a system that can detect a trigger such as the presence of a chemical or a variation in light intensity; therefore, once the culture is grown for some time, the trigger can be activated resulting in improved productivity. Development of a dCas9 variant that can be activated by light or chemicals can facilitate a tool for the conditional modulation of molecular intricacies, bypassing the physiological interference of the change in cell metabolism (Polstein and Gersbach, 2015; Zetsche et al., 2015). Despite several advantages, this system has its share of challenges in the form of cytotoxic effects of the Cas9 nuclease in some of the microalgae species, which have limited the full-scale utilization of this system. Off-target effects of the Cas9 protein have been linked with cytotoxicity in cells transformed with the Cas9 gene construct. However, modifications in the Cas9 protein delivery through the ribonucleoprotein (RNP) complex has been reported to reduce the off-target problems associated with the Cas9 protein. Replacing the Cas9 protein with a Cas12a variant is also an alternative to consider as it has been reported to solve the cytotoxicity in cyanobacteria (Naduthodi et al., 2018). Apart from these, the recent characterization of several other variants of the CRISPR system has extended the prospect of a genetic toolbox for microalgal genome engineering. Utilization of these precise genome editing tools along with microalgal system biology can create an optimized platform customized for biofuel application and high-value product generation.
Challenges and Future Prospects
Microalgae as an alternative energy source hold immense potential to revolutionize the biofuel production system without putting much pressure on agriculture and the forest ecosystem. Despite the promises, commercialization of the microalgal biofuel technology is far from real, owing to its high production cost. Development of economically feasible technologies, such as microalgal strain improvement for improved oil production, holds the future for commercial scale production of algal biofuel. As summarized by Chung et al. (2017), biotechnological interventions could reduce the microalgal biofuel production cost by 15–20% in comparison with traditional approaches. Accordingly, implementation of key molecular schemes targeting pivotal cost-contributing attributes comprising superior feedstocks, oil extraction procedures, and quality of biodiesel can ease the financial burden imparted by these factors. The successful realization of these approaches can make microalgal biofuel production competitive with fossil fuel. To materialize the goal of gaining economic parity with fossil fuel, recent progress in microalgal biotechnology particularly in the field of biocatalyst engineering, synthetic biology, and genome editing has facilitated the necessary tools to design novel microalgal strains as per the culture condition. Furthermore, merging the primary goal of biofuel production with the intended coproduction of value-added products, such as antioxidants, nutraceuticals, and pharmaceuticals, could help in generating returns for financial investments (Jagadevan et al., 2018). Additionally, the adaptation of a consolidated biorefinery and phycoremediation approaches are also projected to diversify the utility of microalgal biomass (Rizwan et al., 2018). However, the sustainability of these approaches largely depends on the cost incurred during the culturing process of microalgal strains. As large-scale open pond culture is the most economic method for microalgae biomass production, the majority of the commercial microalgae are cultured by this system (Kumar et al., 2018). Thus, open pond culturing of genetically modified (GM) microalgae appears more promising in cutting down the cost; however, the impact on human health and environmental risks form the major concerns with transgenic microalgae if exposed to natural ecosystems (Rastogi et al., 2018). Being one of the primary producers in aquatic ecosystems, any involuntary introduction of GM microalgae could result in an ecological calamity (Singh S.K. et al., 2016). Strict monitoring and risk assessment analysis are, therefore, necessary to design the biosafety regulations for GM microalgae. Apart from these, techniques for the bio-containment of transgenes with codon reassignment and mutagenesis might be helpful in mitigating environmental risks through the deletion of genes crucial for survival in the wild but lack importance for culture (Henley et al., 2013; Gressel et al., 2014; Young and Purton, 2016). Additionally, a long-term comprehensive evaluation of the impact of non-indigenous and engineered microalgal strains on the native ecosystem could be helpful in eliminating the ambiguities around regulations on the cultivation of GM algae. In one notable case, Szyjka et al. (2017) reported that when a microalgal species, Acutodesmus dimorphus, was cultured in an open pond, neither the transgenic nor the wild type counterpart of the microalge species were successful in outcompeting the native strains. The study concluded that the outdoor culturing of GM microalgae fails to affect the microalgal diversity in the native ecosystem. However, before drawing any conclusion, extensive studies should be conducted as it is evident that regulatory certainty would be critical in the development of economically viable processes for algae-based biofuel production (Glass, 2015; Randhawa et al., 2017). Recent success in technology demonstration of biojet fuel is a sign of emerging prospects of microalgal biofuel for commercial ignition (Siobhan, 2010; Gyekye, 2017; Chandra, 2018).
Conclusion
Microalgae-based biofuels are projected as the suitable alternative to fossil fuel because of their promising yield in nature besides their sustainable advantages over traditional terrestrial feedstocks. They possess novel metabolic features, which can be tuned for the commercial scale production of renewable biofuels. However, genetic abilities of identified microlagal strains are far from optimum to serve as feedstocks for sustainable production. Genetic improvement of inherent capacities such as high photosynthetic conversion rates, rapid biomass production, alteration to their core structures for the generation of suitable biofuel feedstocks, and adaptation to diverse climatic conditions envisage to bring new opportunities for sustainable biofuel production. The rapid stride in genome biology studies and high throughput genome sequencing and transcriptome mapping in diverse oleaginous organisms have ensured a means to analyze and manipulate metabolic pathways by triggering the expression of candidate genes for enhanced lipid production in microalgae.
Author Contributions
LS conceived the structure and focus of the review. PS wrote the basic framework of the review. MS and SK assisted in writing. RS organized the critical components. LS analyzed and edited the write-up.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abbriano, R., Vardar, N., Yee, D., and Hildebrand, M. (2018). Manipulation of a glycolytic regulator alters growth and carbon partitioning in the marine diatom Thalassiosira pseudonana. Algal Res. 32, 250–258. doi: 10.1016/j.algal.2018.03.018
Ahmad, I., Sharma, A. K., Daniell, H., and Kumar, S. (2015). Altered lipid composition and enhanced lipid production in green microalga by introduction of brassica diacylglycerol acyltransferase 2. Plant Biotechnol. J. 13, 540–550. doi: 10.1111/pbi.12278
Ajjawi, I., Verruto, J., Aqui, M., Soriaga, L. B., Coppersmith, J., Kwok, K., et al. (2017). Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotechnol. 35, 647–652. doi: 10.1038/nbt.3865
Allen, A., and Dupont, C. L. (2014). Engineered Microalgae with Enhanced Lipid Production. U. S. Patent No 20,140,162,330. Washington, DC: U.S. Patent and Trademark Office.
Atsumi, S., Higashide, W., and Liao, J. C. (2009). Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27, 1177–1180. doi: 10.1038/nbt.1586
Baek, K., Lee, Y., Nam, O., Park, S., Sim, S. J., and Jin, E. (2016). Introducing Dunaliella LIP promoter containing light-inducible motifs improves transgenic expression in Chlamydomonas reinhardtii. Biotechnol. J. 11, 384–392. doi: 10.1002/biot.201500269
Baek, K., Yu, J., Jeong, J., Sim, S. J., Bae, S., and Jin, E. S. (2018). Photoautotrophic production of macular pigment in a Chlamydomonas reinhardtii strain generated by using DNA-free CRISPR-Cas9 RNP-mediated mutagenesis. Biotechnol. Bioeng. 115, 719–728. doi: 10.1002/bit.26499
Bajhaiya, A. K., Moreira, J. Z., and Pittman, J. K. (2017). Transcriptional engineering of microalgae: prospects for high-value chemicals. Trends Biotechnol. 35, 95–99. doi: 10.1016/j.tibtech.2016.06.001
Banerjee, A., Banerjee, C., Negi, S., Chang, J. S., and Shukla, P. (2018). Improvements in algal lipid production: a systems biology and gene editing approach. Crit. Rev. Biotechnol. 38, 369–385. doi: 10.1080/07388551.2017.1356803
Barahimipour, R., Neupert, J., and Bock, R. (2016). Efficient expression of nuclear transgenes in the green alga Chlamydomonas: synthesis of an HIV antigen and development of a new selectable marker. Plant Mol. Biol. 90, 403–418. doi: 10.1007/s11103-015-0425-8
Barka, F., Angstenberger, M., Ahrendt, T., Lorenzen, W., Bode, H. B., and Büchel, C. (2016). Identification of a triacylglycerol lipase in the diatom Phaeodactylum tricornutum. Biochim. Biophys. Acta 1861, 239–248. doi: 10.1016/j.bbalip.2015.12.023
Barry, A. N., Starkenburg, S. R., and Sayre, R. T. (2015). Strategies for optimizing algal biology for enhanced biomass production. Front. Energy Res 3:1. doi: 10.3389/fenrg.2015.00001
Bartley, M. L., Boeing, W. J., Dungan, B. N., Holguin, F. A., and Schaub, T. (2014). pH effects on growth and lipid accumulation of the biofuel microalgae Nannochloropsis salina and invading organisms. J. Appl. Phycol. 26, 1431–1437. doi: 10.1007/s10811-013-0177-2
Behera, S., Singh, R., Arora, R., Sharma, N. K., Shukla, M., and Kumar, S. (2015). Scope of algae as third generation biofuels. Front. Bioeng. Biotechnol. 2:90. doi: 10.3389/fbioe.2014.00090
Belotti, G., Bravi, M., de Caprariis, B., de Filippis, P., and Scarsella, M. (2013). Effect of nitrogen and phosphorus starvations on Chlorella vulgaris lipids productivity and quality under different trophic regimens for biodiesel production. Am. J. Plant Sci. 4, 44–51. doi: 10.4236/ajps.2013.412A2006
Belshaw, N., Grouneva, I., Aram, L., Gal, A., Hopes, A., and Mock, T. (2017). Efficient CRISPR/Cas-mediated homologous recombination in the model diatom Thalassiosira pseudonana. bioRxiv [Preprint]. doi: 10.1101/215582
Benmoussa, M. (2016). Algomics for the development of a sustainable microalgae biorefinery. Single Cell Biol. 5:132. doi: 10.4172/2168-9431.1000132
Bhowmick, G. D., Koduru, L., and Sen, R. (2015). Metabolic pathway engineering towards enhancing microalgal lipid biosynthesis for biofuel application-A review. Renew. Sustain. Energy Rev. 50, 1239–1253. doi: 10.1016/j.rser.2015.04.131
Blatti, J. L., Michaud, J., and Burkart, M. D. (2013). Engineering fatty acid biosynthesis in microalgae for sustainable biodiesel. Curr. Opin Chem. Biol. 17, 496–505. doi: 10.1016/j.cbpa.2013.04.007
Boyle, N. R., Page, M. D., Liu, B., Blaby, I. K., Casero, D., Kropat, J., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287, 15811–15825. doi: 10.1074/jbc.M111.334052
Caspi, R., Altman, T., Billington, R., Dreher, K., Foerster, H., Fulcher, C. A., et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 42, 623–631. doi: 10.1093/nar/gkt1103
Cernac, A., and Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40, 575–585. doi: 10.1111/j.1365-313X.2004.02235.x
Chandra, J. (2018). SpiceJet Operates Country’s First Biojet Fuel Flight. The Hindu. Available at: https://www.thehindu.com/business/Industry/spicejet-operates-indias-first-biojet-fuel-flight/article24790919.ece [accessed August 31, 2018].
Chen, B., Wan, C., Mehmood, M. A., Chang, J. S., Bai, F., and Zhao, X. (2017). Manipulating environmental stresses and stress tolerance of microalgae for enhanced production of lipids and value-added products – A review. Bioresour. Technol. 244, 1198–1206. doi: 10.1016/j.biortech.2017.05.170
Chen, C. Y., Kao, A. L., Tsai, Z. C., Chow, T. J., Chang, H. Y., Zhao, X. Q., et al. (2016). Expression of type 2 diacylglycerol acyltransferse gene DGTT1 from Chlamydomonas reinhardtii enhances lipid production in Scenedesmus obliquus. Biotechnol. J. 11, 336–344. doi: 10.1002/biot.201500272
Chen, X., Hu, G., and Liu, L. (2018). Hacking an algal transcription factor for lipid biosynthesis. Trends Plant Sci. 23, 181–184. doi: 10.1016/j.tplants.2017.12.008
Chu, F. F., Shen, X. F., Lam, P. K. S., and Zeng, R. J. (2015). Polyphosphate during the regreening of Chlorella vulgaris under nitrogen deficiency. Intern. J. Mol. Sci. 16, 23355–23368. doi: 10.3390/ijms161023355
Chu, W. L. (2017). Strategies to enhance production of microalgal biomass and lipids for biofuel feedstock. Eur. J. Phycol. 52, 419–437. doi: 10.1080/09670262.2017.1379100
Chung, Y. S., Lee, J. W., and Chung, C. H. (2017). Molecular challenges in microalgae towards cost-effective production of quality biodiesel. Renew. Sustain. Energy Rev. 74, 139–144. doi: 10.1016/j.rser.2017.02.048
Cornell, R., Grove, G. L., Rothblat, G. H., and Horwitz, A. F. (1977). Lipid requirement for cell cycling: the effect of selective inhibition of lipid synthesis. Exp. Cell Res. 109, 299–307. doi: 10.1016/0014-4827(77)90009-X
Courchesne, N. M. D., Parisien, A., Wang, B., and Lan, C. Q. (2009). Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 141, 31–41. doi: 10.1016/j.jbiotec.2009.02.018
Daboussi, F., Leduc, S., Maréchal, A., Dubois, G., Guyot, V., Perez-Michaut, C., et al. (2014). Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Comm. 5:3831. doi: 10.1038/ncomms4831
de Jaeger, L., Springer, J., Wolbert, E. J. H., Martens, D. E., Eggink, G., and Wijffels, R. H. (2017). Gene silencing of stearoyl-ACP desaturase enhances the stearic acid content in Chlamydomonas reinhardtii. Bioresour. Technol. 245, 1616–1626. doi: 10.1016/j.biortech.2017.06.128
de Mooij, T., Janssen, M., Cerezo-Chinarro, O., Mussgnug, J. H., Kruse, O., Ballottari, M., et al. (2015). Antenna size reduction as a strategy to increase biomass productivity: a great potential not yet realized. J. Appl. Phycol. 27, 1063–1077. doi: 10.1007/s10811-014-0427-y
Degraer, S., Brabant, R., Rumes, B., (Eds.) (2010). Offshore wind farms in the Belgian part of the North Sea: Spatio-temporal variability and early impact assessment. Royal Belgian Institute of Natural Sciences, Management Unit of the North Sea Mathematical Models, Marine ecosystem management unit. 184 pp. + annexes.
Dehesh, K., Jones, A., Knutzon, D. S., and Voelker, T. A. (1996). Production of high levels of 8:0 and 10:0 fatty acids in transgenic canola by overexpression of Ch FatB 2, a thioesterase cDNA from Cuphea hookeriana. Plant J. 9, 167–172. doi: 10.1046/j.1365-313X.1996.09020167.x
Deng, X., Cai, J., and Fei, X. (2013a). Effect of the expression and knockdown of citrate synthase gene on carbon flux during triacylglycerol biosynthesis by green algae Chlamydomonas reinhardtii. BMC Biochem. 14:38. doi: 10.1186/1471-2091-14-38
Deng, X., Cai, J., and Fei, X. (2013b). Involvement of phosphatidate phosphatase in the biosynthesis of triacylglycerols in Chlamydomonas reinhardtii. J. Zhejiang Univ. Sci. B 14, 1121–1131. doi: 10.1631/jzus.B1300180
Deng, X., Cai, J., Li, Y., and Fei, X. (2014). Expression and knockdown of the PEPC1 gene affect carbon flux in the biosynthesis of triacylglycerols by the green alga Chlamydomonas reinhardtii. Biotechnol. Lett. 36, 2199–2208. doi: 10.1007/s10529-014-1593-3
Deng, X., Fan, X., Li, P., and Fei, X. (2015). A photoperiod-regulating gene CONSTANS is correlated to lipid biosynthesis in Chlamydomonas reinhardtii. Biomed Res. Int. 2015:715020. doi: 10.1155/2015/715020
Deng, X., Li, Y., and Fei, X. (2011). The mRNA abundance of pepc2 gene is negatively correlated with oil content in Chlamydomonas reinhardtii. Biomass Bioenergy 35, 1811–1817. doi: 10.1016/j.biombioe.2011.01.005
Doan, Y. T. T., and Obbard, J. P. (2014). Two-stage cultivation of a Nannochloropsis mutant for biodiesel feedstock. J. Appl. Phycol. 27, 2203–2208. doi: 10.1007/s10811-014-0490-4
Doron, L., Segal, N., and Shapira, M. (2016). Transgene expression in microalgae—From tools to applications. Front. Plant Sci. 7:505. doi: 10.3389/fpls.2016.00505
Dunahay, T. G., Jarvis, E. E., Dais, S. S., and Roessler, P. G. (1996). Manipulation of microalgal lipid production using genetic engineering. Appl. Biochem. Biotechnol. 57, 223–231. doi: 10.1007/BF02941703
Durrett, T. P., Benning, C., and Ohlrogge, J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. doi: 10.1111/j.1365-313X.2008.03442.x
Fan, J., Andre, C., and Xu, C. (2011). A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585, 1985–1991. doi: 10.1016/j.febslet.2011.05.018
Fan, J., Cui, Y., Wan, M., Wang, W., and Li, Y. (2014). Lipid accumulation and biosynthesis genes response of the oleaginous Chlorella pyrenoidosa under three nutrition stressors. Biotechnol. Biofuels 7:17. doi: 10.1186/1754-6834-7-17
Fan, J., Ning, K., Zeng, X., Luo, Y., Wang, D., Hu, J., et al. (2015). Genomic foundation of starch to lipid switch in oleaginous Chlorella. Plant Physiol. 169, 2444–2461. doi: 10.1104/pp.15.01174
Fang, L., Lin, H. X., Low, C. S., Wu, M. H., Chow, Y., and Lee, Y. K. (2012a). Expression of the Chlamydomonas reinhardtii Sedoheptulose-1,7-bisphosphatase in Dunaliella bardawil leads to enhanced photosynthesis and increased glycerol production. Plant Biotechnol. J. 10, 1129–1135. doi: 10.1111/pbi.12000
Fang, L., Si, Y. Q., Douglass, S., Casero, D., Merchant, S. S., Pellegrini, M., et al. (2012b). Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24, 1876–1893. doi: 10.1105/tpc.112.097949
Fei, X., Li, X., Li, P., and Deng, X. (2017). Involvement of Chlamydomonas DNA damage tolerence gene UBC2 in lipid accumulation. Algal Res. 22, 148–159. doi: 10.1016/j.algal.2016.12.019
Ferenczi, A., Pyott, D. E., Xipnitou, A., and Molnar, A. (2018). Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc. Natl. Acad. Aci. U.S.A. 114, 13567–13572. doi: 10.1073/pnas.1710597114
Formighieri, C., Franck, F., and Bassi, R. (2012). Regulation of the pigment optical density of an algal cell: filling the gap between photosynthetic productivity in the laboratory and in mass culture. J. Biotechnol. 162, 115–123. doi: 10.1016/j.jbiotec.2012.02.021
Fukuzawa, H., Miura, K., Ishizaki, K., Kucho, K. I., Saito, T., Kohinata, T., et al. (2001). Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc. Natl. Acad. Sci. U.S.A. 98, 5347–5352. doi: 10.1073/pnas.081593498
Gan, S. Y., and Maggs, C. A. (2017). Random mutagenesis and precise gene editing technologies: applications in algal crop improvement and functional genomics. Eur. J. Phycol. 52, 466–481. doi: 10.1080/09670262.2017.1358827
Gangl, D., Zedler, J. A. Z., Rajakumar, P. D., Martinez, E. M. R., Riseley, A., Włodarczyk, A., et al. (2015). Biotechnological exploitation of microalgae. J. Exp. Bot. 66, 6975–6990. doi: 10.1093/jxb/erv426
Garay, L. A., Boundy-Mills, K. L., and German, J. B. (2014). Accumulation of high-value lipids in single-cell microorganisms: a mechanistic approach and future perspectives. J. Agric. Food Chem. 62, 2709–2727. doi: 10.1021/jf4042134
Gargouri, M., Park, J. J., Holguin, F. O., Kim, M. J., Wang, H., Deshpande, R. R., et al. (2015). Identification of regulatory network hubs that control lipid metabolism in Chlamydomonas reinhardtii. J. Exp. Bot. 66, 4551–4566. doi: 10.1093/jxb/erv217
Ghosh, A., Khanra, S., Mondal, M., Halder, G., Tiwari, O. N., Saini, S., et al. (2016). Progress toward isolation of strains and genetically engineered strains of microalgae for production of biofuel and other value added chemicals: a review. Energy Convers. Manag. 113, 104–118. doi: 10.1016/j.enconman.2016.01.050
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi: 10.1016/j.cell.2013.06.044
Gimpel, J. A., Henríquez, V., and Mayfield, S. P. (2015). In metabolic engineering of eukaryotic microalgae: potential and challenges come with great diversity. Front. Microbiol. 6:1376. doi: 10.3389/fmicb.2015.01376
Giroud, C., Gerber, A., and Eichenberger, W. (1988). Lipids of Chlamydomonas reinhardtii – analysis of molecular-species and intracellular site(s) of biosynthesis. Plant Cell Physiol. 29, 587–595.
Glass, D. J. (2015). Pathways to obtain regulatory approvals for the use of genetically modified algae in biofuel or biobased chemical production. Ind. Biotechnol. 11, 71–83. doi: 10.1089/ind.2015.1503
Gomma, A. E., Lee, S. K., Sun, S. M., Yang, S. H., and Chung, G. (2015). Improvement in oil production by increasing Malonyl-CoA and Glycerol-3-phosphate pools in Scenedesmus quadricauda. Ind. J. Microbiol. 55, 447–455. doi: 10.1007/s12088-015-0546-4
Greiner, A., Kelterborn, S., Evers, H., Kreimer, G., Sizova, I., and Hegemann, P. (2017). Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell 29, 2498–2518. doi: 10.1105/tpc.17.00659
Gressel, J., van der Vlugt, C. J. B., and Bergmans, H. E. N. (2014). Cultivated microalgae spills: hard to predict/easier to mitigate risks. Trends Biotechnol. 32, 65–69. doi: 10.1016/j.tibtech.2013.11.003
Gyekye, L. (2017). Singapore Airlines Takes off with Biofuels-Powered Flights. Biofuel International. Available at: http://biofuels-news.com/display_news/12242/singapore_airlines_takes_off_with_biofuelpowered_flights/ [accessed August 31, 2018].
Hamilton, M. L., Haslam, R. P., Napier, J. A., and Sayanova, O. (2014). Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. Eng. 22, 3–9. doi: 10.1016/j.ymben.2013.12.003
Hazra, S., Henderson, J. N., Liles, K., Hilton, M. T., and Wachter, R. M. (2015). Regulation of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase: product inhibition, cooperativity, and magnesium activation. J. Biol. Chem. 290, 24222–24236. doi: 10.1074/jbc.M115.651745
Henley, W. J., Litaker, R. W., Novovesk, L., Duke, C. S., Quemada, H. D., and Sayre, R. T. (2013). Initial risk assessment of genetically modified (GM) microalgae for commodity-scale biofuel cultivation. Algal Res. 2, 66–77. doi: 10.1016/j.algal.2012.11.001
Hopes, A., Nekrasov, V., Kamoun, S., and Mock, T. (2016). Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods 12:49. doi: 10.1186/s13007-016-0148-0
Hsieh, H. J., Su, C. H., and Chien, L. J. (2012). Accumulation of lipid production in Chlorella minutissima by triacylglycerol biosynthesis-related genes cloned from Saccharomyces cerevisiae and Yarrowia lipolytica. J. Microbiol. 50, 526–534. doi: 10.1007/s12275-012-2041-5
Hu, Q., Sommerfeld, M., Jarvis, E., Ghirardi, M., Posewitz, M., Seibert, M., et al. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639. doi: 10.1111/j.1365-313X.2008.03492.x
Huang, L. F., Lin, J. Y., Pan, K. Y., Huang, C. K., and Chu, Y. K. (2015). Overexpressing ferredoxins in Chlamydomonas reinhardtii increase starch and oil yields and enhances electric power production in a photo microbial fuel cell. Inter. J. Mol. Sci. 16, 19308–19325. doi: 10.3390/ijms160819308
Hwangbo, K., Ahn, J. W., Lim, J. M., Park, Y. II, Liu, J. R., and Jeong, W. J. (2013). Overexpression of stearoyl-ACP desaturase enhances accumulations of oleic acid in the green alga Chlamydomonas reinhardtii. Plant Biotechnol. Rep. 8, 135–142. doi: 10.1007/s11816-013-0302-3
Ibanez-Salazar, A., Rosales-Mendoza, S., Rocha-Uribe, A., Ramirez-Alonso, J. I., Lara-Hernandez, I., Hernandez-Torres, A., et al. (2014). Over-expression of Dof-type transcription factor increases lipid production in Chlamydomonas reinhardtii. J. Biotechnol. 184, 27–38. doi: 10.1016/j.jbiotec.2014.05.003
Iwai, M., Ikeda, K., Shimojima, M., and Ohta, H. (2014). Enhancement of extraplastidic oil synthesis in Chlamydomonas reinhardtii using a type-2 diacylglycerol acyltransferase with a phosphorus starvation-inducible promoter. Plant Biotechnol. J. 12, 808–819. doi: 10.1111/pbi.12210
Iwaki, T., Haranoh, K., Inoue, N., Kojima, K., Satoh, R., Nishino, T., et al. (2006). Expression of foreign type I ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39) stimulates photosynthesis in cyanobacterium Synechococcus PCC7942 cells. Photosynth. Res. 88, 287–297. doi: 10.1007/s11120-006-9048-x
Jagadevan, S., Banerjee, A., Banerjee, C., Guria, C., Tiwari, R., Baweja, M., et al. (2018). Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnol. Biofuels 11:185. doi: 10.1186/s13068-018-1181-1
Jamers, A., Blust, R., and Coen, W. D. (2009). Omics in algae: paving the way for a systems biological understanding of algal stress phenomena? Aquat. Toxicol. 92, 114–121. doi: 10.1016/j.aquatox.2009.02.012
Jeon, S., Lim, J. M., Lee, H. G., Shin, S. E., Kang, N. K., Park, Y. I., et al. (2017). Current status and perspectives of genome editing technology for microalgae. Biotechnol. Biofuels 10:267. doi: 10.1186/s13068-017-0957-z
Jeong, J., Baek, K., Kirst, H., Melis, A., and Jin, E. (2017). Loss of RP54 function leads to a truncated light-harvesting antenna size in Chlamydomonas reinhardtii. Biochim. Biophys. Acta Bioenerg. 1858, 45–55. doi: 10.1016/j.bbabio.2016.10.007
Jeong, J., Baek, K., Yu, J., Kirst, H., Betterle, N., Shin, W., et al. (2018). Deletion of the chloroplast LTD protein impedes LHCI import and PSI–LHCI assembly in Chlamydomonas reinhardtii. J. Exp. Bot. 69, 1147–1158. doi: 10.1093/jxb/erx457
Jiang, W., Brueggeman, A. J., Horken, K. M., Plucinak, T. M., and Weeks, D. P. (2014). Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 13, 1465–1469. doi: 10.1128/EC.00213-14
Kang, N. K., Jeon, S., Kwon, S., Koh, H. G., Shin, S. E., Lee, B., et al. (2015). Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol. Biofuels 8:200. doi: 10.1186/s13068-015-0386-9
Kang, N. K., Kim, E. K., Kim, Y. U., Lee, B., Jeong, W. J., Jeong, B. R., et al. (2017). Increased lipid production by heterologous expression of AtWRI1 transcription factor in Nannochloropsis salina. Biotechnol. Biofuels 10:231. doi: 10.1186/s13068-017-0919-5
Kao, P. H., and Ng, I. S. (2017). CRISPRi mediated phosphoenolpyruvate carboxylase regulation to enhance the production of lipid in Chlamydomonas reinhardtii. Bioresour. Technol. 245, 1527–1537. doi: 10.1016/j.biortech.2017.04.111
Kasai, Y., Oshima, K., Ikeda, F., Abe, J., Yoshimitsu, Y., and Harayama, S. (2015). Construction of a self-cloning system in the unicellular green alga Pseudochoricystis ellipsoidea. Biotechnol. Biofuel 8:94. doi: 10.1186/s13068-015-0277-0
Kaye, Y., Grundman, O., Leu, S., Zarka, A., Zorin, B., Didi-Cohen, S., et al. (2015). Metabolic engineering toward enhanced LC-PUFA biosynthesis in Nannochloropsis oceanica: overexpression of endogenous δ12 desaturase driven by stress-inducible promoter leads to enhanced deposition of polyunsaturated fatty acids in TAG. Algal Res. 11, 387–398. doi: 10.1016/j.algal.2015.05.003
Kim, H., and Kim, J. S. (2014). A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334. doi: 10.1038/nrg3686
Kim, Y., Ternga, E. L., Riekhofa, W. R., Cahoonb, E. B., and Ceruttia, H. (2018). Endoplasmic reticulum acyltransferase with prokaryotic substrate preference contributes to triacylglycerol assembly in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 115, 1652–1657. doi: 10.1073/pnas.1715922115
Kirst, H., Garcia-Cerdan, J. G., Zurbriggen, A., Ruehle, T., and Melis, A. (2012a). Truncated photosystem chlorophyll antenna size in the green microalga Chlamydomonas reinhardtii upon deletion of the TLA3-CpSRP43 gene. Plant Physiol. 160, 2251–2260. doi: 10.1104/pp.112.206672
Kirst, H., Garcia-Cerdan, J. G., Zurbriggen, A., and Melis, A. (2012b). Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiol. 158, 930–945. doi: 10.1104/pp.111.189910
Klok, A. J., Lamers, P. P., Martens, D. E., Draaisma, R. B., and Wijffels, R. H. (2014). Edible oils from microalgae: insights in TAG accumulation. Trends Biotechnol. 32, 521–528. doi: 10.1016/j.tibtech.2014.07.004
Kotchoni, S. O., Gachomo, E. W., Slobodenko, K., and Shain, D. H. (2016). AMP deaminase suppression increases biomass, cold tolerance and oil content in green algae. Algal Res. 16, 473–480. doi: 10.1016/j.algal.2016.04.007
Koussa, J., Chaiboonchoe, A., and Salehi-Ashtiani, K. (2014). Computational approaches for microalgal biofuel optimization: a review. Biomed Res. Int. 2014:649453. doi: 10.1155/2014/649453
Kumar, P., Kumar, D., Nehra, P., and Sharma, P. K. (2018). “Green algae biomass cultivation, harvesting and genetic modifications for enhanced cellular lipids,” in Microbial Biotechnology, eds J. K. Patra, G. Das, and H. S. Shin (Singapore: Springer Nature), 119–140.
Kwak, M., Park, W. K., Shin, S. E., Koh, H. G., Lee, B., Jeong, B. R., et al. (2017). Improvement of biomass and lipid yield under stress conditions by using diploid strains of Chlamydomonas reinhardtii. Algal Res. 26, 180–189. doi: 10.1016/j.algal.2017.07.027
Kwok, A. C. M., and Wong, J. T. Y. (2005). Lipid biosynthesis and its coordination with cell cycle progression. Plant Cell Physiol. 46, 1973–1986. doi: 10.1093/pcp/pci213
Kwon, S., Kang, N. K., Koh, H. G., Shin, S. E., Lee, B., Jeong, B. R., et al. (2017). Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 115, 331–340. doi: 10.1002/bit.26465
La Russa, M., Bogen, C., Uhmeyer, A., Doebbe, A., Filippone, E., Kruse, O., et al. (2012). Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J. Biotechnol. 162, 13–20. doi: 10.1016/j.jbiotec.2012.04.006
Lauersen, K. J., Huber, I., Wichmann, J., Baier, T., Leiter, A., Gaukel, V., et al. (2015). Investigating the dynamics of recombinant protein secretion from a microalgal host. J. Biotechnol. 215, 62–71. doi: 10.1016/j.jbiotec.2015.05.001
León-Saiki, G. M., Remmers, I. M., Martens, D. E., Lamers, P. P., Wijffels, R. H., and van der Veen, D. (2017). The role of starch as transient energy buffer in synchronized microalgal growth in Acutodesmus obliquus. Algal Res. 25, 160–167. doi: 10.1016/j.algal.2017.05.018
Li, D. W., Cen, S. Y., Liu, Y. H., Balamurugan, S., Zheng, X. Y., Alimujiang, A., et al. (2016). A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J. Biotechnol. 229, 65–71. doi: 10.1016/j.jbiotec.2016.05.005
Li, X., Benning, C., and Kuo, M. H. (2012). Rapid triacylglycerol turnover in Chlamydomonas reinhardtii requires a lipase with broad substrate specificity. Eukaryot. Cell 11, 1451–1462. doi: 10.1128/EC.00268-12
Li, Y., Han, D., Hu, G., Dauvillee, D., Sommerfeld, M., Ball, S., et al. (2010). Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab. Eng. 12, 387–391. doi: 10.1016/j.ymben.2010.02.002
Liang, F., and Lindblad, P. (2017). Synechocystis PCC 6803 overexpressing RuBisCO grow faster with increased photosynthesis. Metab. Eng. Commun. 4, 29–36. doi: 10.1016/j.meteno.2017.02.002
Li-Beisson, Y., Beisson, F., and Riekhof, W. (2015). Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 82, 504–522. doi: 10.1111/tpj.12787
Lim, D. K. Y., and Schenk, P. M. (2017). Microalgae selection and improvement as oil crops: GM vs non-GM strain engineering. AIMS Bioeng. 4, 151–161. doi: 10.3934/bioeng.2017.1.151
Lin, H., Shen, H., and Lee, Y. K. (2018). Cellular and molecular responses of Dunaliella tertiolecta by expression of a plant medium chain length fatty acid specific Acyl-ACP thioesterase. Front. Microbiol. 9:619. doi: 10.3389/fmicb.2018.00619
Liu, B., and Benning, C. (2013). Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotech. 24, 300–309. doi: 10.1016/j.copbio.2012.08.008
Luo, Q., Li, Y., Wang, W., Fei, X., and Deng, X. (2015). Genome-wide survey and expression analysis of Chlamydomonas reinhardtii U-box E3 ubiquitin ligases (CrPUBs) reveal a functional lipid metabolism module. PLoS One 10:e0122600. doi: 10.1371/journal.pone.0122600
Ma, Y. H., Wang, X., Niu, Y. F., Yang, Z. K., Zhang, M. H., Wang, Z. M., et al. (2014). Antisense knockdown of pyruvate dehydrogenase kinase promotes the neutral lipid accumulation in the diatom Phaeodactylum tricornutum. Microb. Cell Fact. 13:100. doi: 10.1186/s12934-014-0100-9
Manandhar-Shrestha, K., and Hildebrand, M. (2015). Characterization and manipulation of a DGAT2 from the diatom Thalassiosira pseudonana: improved TAG accumulation without detriment to growth, and implications for chloroplast TAG accumulation. Algal Res. 12, 239–248. doi: 10.1016/j.algal.2015.09.004
Maravi, D. K., Kumar, S., Sharma, P. K., Kobayashi, Y., Goud, V. V., Sakurai, N., et al. (2016). Ectopic expression of AtDGAT1, encoding diacylglycerol O-acyltransferase exclusively committed to TAG biosynthesis, enhances oil accumulation in seeds and leaves of Jatropha. Biotechnol. Biofuels 9:226. doi: 10.1186/s13068-016-0642-7
Mendoza, M. S., Dubreucq, B., Miquel, M., Caboche, M., and Lepiniec, L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579, 4666–4670. doi: 10.1016/j.febslet.2005.07.037
Misra, N., Panda, P. K., Parida, B. K., and Mishra, B. K. (2016). DEMBF: a comprehensive database of enzymes of microalgal biofuel feedstock. PLoS One 11:e0146158. doi: 10.1371/journal.pone.0146158
Morales, M., Sánchez, L., and Revah, S. (2018). The impact of environmental factors on carbon dioxide fixation by microalgae. FEMS Microbiol. Lett. 365:fnx262. doi: 10.1093/femsle/fnx262
Mussgnug, J. H., Thomas-Hall, S., Rupprecht, J., Foo, A., Klassen, V., McDowall, A., et al. (2007). Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol. J. 5, 802–814. doi: 10.1111/j.1467-7652.2007.00285.x
Muto, M., Tanaka, M., Liang, Y., Yoshino, T., Matsumoto, M., and Tanaka, T. (2015). Enhancement of glycerol metabolism in the oleaginous marine diatom Fistulifera solaris JPCC DA0580 to improve triacylglycerol productivity. Biotechnol. Biofuels 8:4. doi: 10.1186/s13068-014-0184-9
Naduthodi, M. I. S., Barbosa, M. J., and van der Oost, J. (2018). Progress of CRISPR-Cas based genome editing in photosynthetic microbes. Biotechnol. J. 13:e1700591. doi: 10.1002/biot.201700591
Ng, I. S., Tan, S. I., Kao, P. H., Chang, Y. K., and Chang, J. S. (2017). Recent developments on genetic engineering of microalgae for biofuels and bio-based chemicals. Biotechnol. J. 12:1600644. doi: 10.1002/biot.201600644
Ngan, C. Y., Wong, C. H., Choi, C., Yoshinaga, Y., Louie, K., Jia, J., et al. (2015). Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat. Plants 1:15107. doi: 10.1038/nplants.2015.107
Niu, Y. F., Zhang, M. H., Li, D. W., Yang, W. D., Liu, J. S., Bai, W. B., et al. (2013). Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar. Drugs 11, 4558–4569. doi: 10.3390/md11114558
Norashikin, M. N., Loh, S. H., Aziz, A., and Cha, T. S. (2018). Metabolic engineering of fatty acid biosynthesis in Chlorella vulgaris using an endogenous omega-3 fatty acid desaturase gene with its promoter. Algal Res. 31, 262–275. doi: 10.1016/j.algal.2018.02.020
Nymark, M., Sharma, A. K., Sparstad, T., Bones, A. M., and Winge, P. (2016). A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 6:24951. doi: 10.1038/srep24951
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acid Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Ohlrogge, J. B., and Jaworski, J. G. (1997). Regulation of fatty acid synthesis. Annu. Rev. Plant Biol. 48, 109–136. doi: 10.1146/annurev.arplant.48.1.109
Ozaki, T. (2016). Acyl-ACP Thioesterase. U. S. Patent No 20,160,130,615. Washington, DC: U.S. Patent and Trademark Office.
Park, S., Lee, Y., Lee, J. H., and Jin, E. (2013). Expression of the high light-inducible Dunaliella LIP promoter in Chlamydomonas reinhardtii. Planta 238, 1147–1156. doi: 10.1007/s00425-013-1955-4
Patil, V., Källqvist, T., Olsen, E., Vogt, G., and Gislerød, H. R. (2007). Fatty acid composition of 12 microalgae for possible use in aquaculture feed. Aquacult. Int. 15, 1–9. doi: 10.1007/s10499-006-9060-3
Patra, K. P., Li, F., Carter, D., Gregory, J. A., Baga, S., Reed, S. G., et al. (2015). Alga- produced malaria transmission-blocking vaccine candidate Pfs25 formulated with a human use-compatible potent adjuvant induces high-affinity antibodies that block Plasmodium falciparum infection of mosquitoes. Infect. Immun. 83, 1799–1808. doi: 10.1128/IAI.02980-14
Peng, K. T., Zheng, C. N., Xue, J., Chen, X. Y., Yang, W. D., Liu, J. S., et al. (2014). Delta 5 fatty acid desaturase up-regulates the synthesis of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum. J. Agric. Food Chem. 62, 8773–8776. doi: 10.1021/jf5031086
Perrine, Z., Negi, S., and Sayre, R. T. (2012). Optimization of photosynthetic light energy utilization by microalgae. Algal Res. 1, 134–142. doi: 10.1016/j.algal.2012.07.002
Piatek, A., Ali, Z., Baazim, H., Li, L., Abulfaraj, A., Al-Shareef, S., et al. (2015). RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 13, 578–589. doi: 10.1111/pbi.12284
Poliner, E., Takeuchi, T., Du, Z. Y., Benning, C., and Farreì, E. M. (2018). Nontransgenic marker-free gene disruption by an episomal CRISPR system in the oleaginous microalga, Nannochloropsis oceanica CCMP1779. ACS Synth. Biol. 7, 962–968. doi: 10.1021/acssynbio.7b00362
Polle, J. E., Kanakagiri, S. D., and Melis, A. (2003). tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217, 49–59.
Polstein, L. R., and Gersbach, C. A. (2015). A Light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 11, 198–200. doi: 10.1038/nchembio.1753
Price, G. D., Pengelly, J. J. L., Forster, B., Du, J., Whitney, S. M., von Caemmerer, S., et al. (2013). The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 64, 753–768. doi: 10.1093/jxb/ers257
Radakovits, R., Eduafo, P. M., and Posewitz, M. C. (2011). Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab. Eng. 13, 89–95. doi: 10.1016/j.ymben.2010.10.003
Radakovits, R., Jinkerson, R. E., Darzins, A., and Posewitz, M. C. (2010). Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9, 486–501. doi: 10.1128/EC.00364-09
Raines, C. A. (2003). The Calvin cycle revisited. Photosynth. Res. 75, 1–10. doi: 10.1023/A:1022421515027
Randhawa, K. S., Relph, L. E., Armstrong, M. C., and Rahman, P. K. S. M. (2017). Biofuel production: tapping into microalgae despite challenges. Biofuels 8, 261–271. doi: 10.1080/17597269.2016.1224290
Rasala, B. A., Chao, S. S., Pier, M., Barrera, D. J., and Mayfield, S. P. (2014). Enhanced genetic tools for engineering multigene traits into green algae. PLoS One 9:e94028. doi: 10.1371/journal.pone.0094028
Rastogi, R. P., Pandey, A., Larroche, C., and Madamwar, D. (2018). Algal green energy – R & D and technological perspectives for biodiesel production. Renew. Sustain. Energy Rev. 82, 2946–2969. doi: 10.1016/j.rser.2017.10.038
Ravindran, B., Kurade, M. B., Kabra, A. N., Jeon, B. H., and Gupta, S. K. (2017). “Recent advances and future prospects of microalgal lipid biotechnology,” in Algal Biofuels: Recent Advances and Future Prospects, eds S. K. Gupta, A. Malik, and F. Bux (Cham: Springer International Publishing),1–37.
Reijnders, M. J. M. F., VanHeck, R. G. A., Lam, C. M. C., Scaife, M. A., Santos, V. A. P. M. D., Smith, A. G., et al. (2014). Green genes: bioinformatics and systems-biology innovations drive algal biotechnology. Trends Biotechnol. 32, 617–626. doi: 10.1016/j.tibtech.2014.10.003
Rengel, R., Smith, R. T., Haslam, R. P., Sayanova, O., Vila, M., and León, R. (2018). Overexpression of acetyl-CoA synthetase (ACS) enhances the biosynthesis of neutral lipids and starch in the green microalga Chlamydomonas reinhardtii. Algal Res. 31, 183–193. doi: 10.1016/j.algal.2018.02.009
Rizwan, M., Mujtaba, G., Memon, S. A., Lee, K., and Rashid, N. (2018). Exploring the potential of microalgae for new biotechnology applications and beyond: a review. Renew. Sustain. Energy Rev. 92, 394–404. doi: 10.1016/j.rser.2018.04.034
Sayre, R. T., Subramanian, S. S., and Friedland, N. (2017). Carbon Fixation Systems in Plants and Algae. U. S. Patent No 20,170,211,086. Washington, DC: U.S. Patent and Trademark Office.
Scott, S. A., Davey, M. P., Dennis, J. S., Horst, I., Howe, C. J., Lea-Smith, D. J., et al. (2010). Biodiesel from algae: challenges and prospects. Curr. Opin. Biotechnol. 21, 277–286. doi: 10.1016/j.copbio.2010.03.005
Serif, M., Lepetit, B., Weißert, K., Kroth, P. G., and Rio Bartulos, C. (2017). A fast and reliable strategy to generate TALEN-mediated gene knockouts in the diatom Phaeodactylum tricornutum. Algal Res. 23, 186–195. doi: 10.1016/j.algal.2017.02.005
Sharma, K. K., Schuhmann, H., and Schenk, P. M. (2012). High lipid induction in microalgae for biodiesel production. Energies 5, 1532–1553. doi: 10.3390/en5051532
Sheehan, J., Dunahay, T. G., Benemann, J. R., Roessler, P. G., and Weissman, J. C. (1998). A Look Back at the U. S. Department of Energy’s Aquatic Species Program — Biodiesel from Algae. Golden, CO: National Renewable Energy Laboratory.
Shin, S. E., Lim, J. M., Koh, H. G., Kim, E. K., Kang, N. K., Jeon, S., et al. (2016). CRISPR/Cas9-induced knockout and knock-inmutations in Chlamydomonas reinhardtii. Sci. Rep. 6:27810. doi: 10.1038/srep27810
Shtaida, N., Khozin-Goldberg, I., and Boussiba, S. (2015). The role of pyruvate hub enzymes in supplying carbon precursors for fatty acid synthesis in photosynthetic microalgae. Photosynth. Res. 125, 407–422. doi: 10.1007/s11120-015-0136-7
Shuba, E. S., and Kifleb, D. (2018). Microalgae to biofuels: ‘Promising’ alternative and renewable energy, review. Renew. Sustain. Energy Rev. 81, 743–755. doi: 10.1016/j.rser.2017.08.042
Simionato, D., Basso, B., Giacometti, G. M., and Morosinotto, T. (2013). Optimization of light use efficiency for biofuel production in algae. Biophys. Chem. 182, 71–78. doi: 10.1016/j.bpc.2013.06.017
Singh, P., Kumari, S., Guldhe, A., Misra, R., Rawat, I., and Bux, F. (2016). Trends and novel strategies for enhancing lipid accumulation and quality in microalgae. Renew. Sustain. Energy Rev. 55, 1–16. doi: 10.1016/j.rser.2015.11.001
Singh, S. K., Sundaram, S., Sinha, S., Rahman, M. A., and Kapur, S. (2016). Recent advances in CO2 uptake and fixation mechanism of cyanobacteria and microalgae. J. Crit. Rev. Environ. Sci. Technol. 46, 1297–1323. doi: 10.1080/10643389.2016.1217911
Siobhan, W. (2010). EADS Aircraft Runs on Algae Biofuel. The Engineer. Available at: http://www.theengineer.co.uk/news/news-analysis/eads-aircraft-runs-on-algae-biofuel/ [accessed 31 August 2018].
Sizova, I., Greiner, A., Awasthi, M., Kateriya, S., and Hegemann, P. (2013). Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 73, 873–882. doi: 10.1111/tpj.12066
Slattery, S. S., Diamond, A., Wang, H., Therrien, J. A., Lant, J. T., Jazey, T., et al. (2018). An expanded plasmid-based genetic toolbox enables Cas9 genome editing and stable maintenance of synthetic pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 7, 328–338. doi: 10.1021/acssynbio.7b00191
Specht, E., Miyake-Stoner, S., and Mayfield, S. (2010). Micro-algae come of age as a platform for recombinant protein production. Biotechnol. Lett. 32, 1373–1383. doi: 10.1007/s10529-010-0326-5
Stephenson, P. G., Moore, C. M., Terry, M. J., Zubkov, M. V., and Bibby, T. S. (2011). Improving photosynthesis for algal biofuels: toward a green revolution. Trends Biotechnol. 29, 615–623. doi: 10.1016/j.tibtech.2011.06.005
Stukenberg, D., Zauner, S., Dell’Aquila, G., and Maier, U. G. (2018). Optimizing CRISPR/Cas9 for the diatom Phaeodactylum tricornutum. Front. Plant Sci. 9:740. doi: 10.3389/fpls.2018.00740
Sumiya, N., Kawase, Y., Hayakawa, J., Matsuda, M., Nakamura, M., Era, A., et al. (2015). Expression of cyanobacterial Acyl-ACP reductase elevates the triacylglycerol level in the red alga Cyanidioschyzon merolae. Plant Cell Physiol. 56, 1962–1980. doi: 10.1093/pcp/pcv120
Suyono, E. A., Haryadi, W., Zusron, M., Nuhamunada, M., Rahayu, S., and Nugroho, P. (2015). The effect of salinity on growth, dry weight and lipid content of the mixed microalgae culture isolated from Glagah as biodiesel substrate. J. Life Sci. 9, 229–233.
Szyjka, S. J., Mandal, S., Schoepp, N. G., Tyler, B. M., Yohn, C. B., Poon, Y. S., et al. (2017). Evaluation of phenotype stability and ecological risk of a genetically engineered alga in open pond production. Algal Res. 24, 378–386. doi: 10.1016/j.algal.2017.04.006
Takahashi, K., Ide, Y., Hayakawa, J., Yoshimitsu, Y., Fukuhara, I., Abe, J., et al. (2018). Lipid productivity in TALEN-induced starchless mutants of the unicellular green alga Coccomyxa sp. strain Obi. Algal Res. 32, 300–307. doi: 10.1016/j.algal.2018.04.020
Talebi, A. F., Tohidfar, M., Bagheri, A., Lyon, S. R., Salehi-ashtiani, K., and Tabatabaei, M. (2014). Manipulation of carbon flux into fatty acid biosynthesis pathway in Dunaliella salina using AccD and ME genes to enhance lipid content and to improve produced biodiesel quality. Biofuel Res. J. 3, 91–97. doi: 10.18331/BRJ2015.1.3.6
Tcherkez, G. G., Farquhar, G. D., and Andrews, T. J. (2006). Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U.S.A. 103, 7246–7251. doi: 10.1073/pnas.0600605103
Trentacoste, E. M., Shrestha, R. P., Smith, S. R., Glé, C., Hartmann, A. C., Hildebrand, M., et al. (2013). Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl. Acad. Sci. U.S.A. 110, 19748–19753. doi: 10.1073/pnas.1309299110
Tsai, C. H., Warakanont, J., Takeuchi, T., Sears, B. B., Moellering, E. R., and Benning, C. (2014). The protein compromised hydrolysis of triacylglycerols 7 (CHT7) acts as a repressor of cellular quiescence in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 111, 15833–15838. doi: 10.1073/pnas.1414567111
Úbeda-Mínguez, P., García-Maroto, F., and Alonso, D. L. (2017). Heterologous expression of DGAT genes in the marine microalga Tetraselmis chui leads to an increase in TAG content. J. Appl. Phycol. 29, 1913–1926.
Van Mooy, B. A., Fredricks, H. F., Pedler, B. E., Dyhrman, S. T., Karl, D. M., Koblizek, M., et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458, 69–72. doi: 10.1038/nature07659
Verruto, J., Francis, K., Wang, Y., Low, M. C., Greiner, J., Tacke, S., et al. (2018). Unrestrained marker less trait stacking in Nannochloropsis gaditana through combined genome editing and marker recycling technologies. Proc. Natl. Acad. Sci. U.S.A. 115, E7015–E7022. doi: 10.1073/pnas.1718193115
Wang, J. B., Chou, S. H., Chow, T. J., Lee, T. M., Su, H. Y., Chou, H. H., et al. (2014). Method for Enhancing Cell Growth of Microalgae. U. S. Patent No 20,140,120,623. Washington, DC: U.S. Patent and Trademark Office.
Wang, Q., Lu, Y., Xin, Y., Wei, L., Huang, S., and Xu, J. (2016). Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9. Plant J. 88, 1071–1081. doi: 10.1111/tpj.13307
Wang, X., Liu, Y. H., Hu, D. X., Balamurugan, S., Lu, Y., Yang, W. D., et al. (2015). Identification of a putative patatin-like phospholipase domain-containing protein 3 (PNPLA3) ortholog involved in lipid metabolism in microalga Phaeodactylum tricornutum. Algal Res. 12, 274–279. doi: 10.1016/j.algal.2015.09.005
Wang, X., Wei, W., Li, N. J., Yuan, W., Ding, Y., Yang, W. D., et al. (2018). Heterogeneous expression of human PNPLA3 triggers algal lipid accumulation and lipid droplet enlargement. Algal Res. 31, 276–281. doi: 10.1016/j.algal.2018.02.019
Wang, Y., Duanmu, D., and Spalding, M. H. (2011). Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynth. Res. 109, 115–122. doi: 10.1007/s11120-011-9643-3
Wei, L., Wang, Q., Xin, Y., Lu, Y., and Xu, J. (2017). Enhancing photosynthetic biomass productivity of industrial oleaginous microalgae by overexpression of RuBisCO activase. Algal Res. 27, 366–375. doi: 10.1016/j.algal.2017.07.023
Weyman, P. D., Beeri, K., Lefebvre, S. C., Rivera, J., McCarthy, J. K., Heuberger, A. L., et al. (2015). Inactivation of Phaeodactylum tricornutum urease gene using transcription activator-like effector nuclease-based targeted mutagenesis. Plant Biotechnol. J. 13, 460–470. doi: 10.1111/pbi.12254
Work, V. H., Radakovits, R., Jinkerson, R. E., Meuser, J. E., Elliott, L. G., Vinyard, D. J., et al. (2010). Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot. Cell 9, 1251–1261. doi: 10.1128/EC.00075-10
Xu, C., Andre, C., Fan, J., and Shanklin, J. (2016). “Cellular organization of triacylglycerol biosynthesis in microalgae,” in Lipids in Plant and Algae Development, eds Y. Nakamura and Y. Li-Beisson (Cham: Springer), 207–221. doi: 10.1007/978-3-319-25979-6_;9
Xue, J., Niu, Y. F., Huang, T., Yang, W. D., Liu, J. S., and Li, H. Y. (2015). Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 27, 1–9. doi: 10.1016/j.ymben.2014.10.002
Yamaoka, Y., Achard, D., Jang, S., Legéret, B., Kamisuki, S., Ko, D., et al. (2016). Identification of a Chlamydomonas plastidial 2-lysophosphatidic acid acyltransferase and its use to engineer microalgae with increased oil content. Plant Biotechnol. J. 14, 2158–2167. doi: 10.1111/pbi.12572
Yan, J., Cheng, R., Lin, X., You, S., Li, K., Rong, H., et al. (2013). Overexpression of acetyl-CoA synthetase increased the biomass and fatty acid proportion in microalga Schizochytrium. Appl. Microbiol. Biotechnol. 97, 1933–1939. doi: 10.1007/s00253-012-4481-6
Yang, B., Liu, J., Ma, X., Guo, B., Liu, B., Wu, T., et al. (2017). Genetic engineering of the Calvin cycle toward enhanced photosynthetic CO2 fixation in microalgae. Biotechnol. Biofuels 10:229. doi: 10.1186/s13068-017-0916-8
Yao, Y., Lu, Y., Peng, K. T., Huang, T., Niu, Y. F., Xie, W. H., et al. (2014). Glycerol and neutral lipid production in the oleaginous marine diatom Phaeodactylum tricornutum promoted by overexpression of glycerol-3-phosphate dehydrogenase. Biotechnol. Biofuels 7:110. doi: 10.1186/1754-6834-7-110
Yoneda, K., Yoshida, M., Suzuki, I., and Watanabe, M. M. (2018). Homologous expression of lipid droplet protein-enhanced neutral lipid accumulation in the marine diatom Phaeodactylum tricornutum. J. Appl. Phycol. 1–10. doi: 10.1007/s10811-018-1402-9
Yoshioka, S., Taniguchi, F., Miura, K., Inoue, T., Yamano, T., and Fukuzawa, H. (2004). The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell 16, 1466–1477. doi: 10.1105/tpc.021162
Young, R. E. B., and Purton, S. (2016). Codon reassignment to facilitate genetic engineering and bio-containment in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 14, 1251–1260. doi: 10.1111/pbi.12490
Zalutskaya, Z., Kharatyan, N., Forchhammer, K., and Ermilova, E. (2015). Reduction of PII signaling protein enhances lipid body production in Chlamydomonas reinhardtii. Plant Sci. 240, 1–9. doi: 10.1016/j.plantsci.2015.08.019
Zetsche, B., Volz, S. E., and Zhang, F. (2015). A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33, 139–142. doi: 10.1038/nbt.3149
Zhang, J., Hao, Q., Bai, L., Xu, J., Yin, W., Song, L., et al. (2014). Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol. Biofuels 7:128. doi: 10.1186/s13068-014-0128-4
Zhang, S., He, Y., Sen, B., Chen, X., Xie, Y., Keasling, J. D., et al. (2018). Alleviation of reactive oxygen species enhances PUFA accumulation in Schizochytrium sp. through regulating genes involved in lipid metabolism. Metab. Eng. Commun. 6, 39–48. doi: 10.1016/j.meteno.2018.03.002
Zhu, B. H., Shi, H. P., Yang, G. P., Lv, N. N., Yang, M., and Pan, K. H. (2016). Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. New Biotechnol. 33, 237–244. doi: 10.1016/j.nbt.2015.06.003
Zhu, B. H., Tu, C. C., Shi, H. P., Yang, G. P., and Pan, K. H. (2017). Overexpression of endogenous delta-6 fatty acid desaturase gene enhances eicosapentaenoic acid accumulation in Phaeodactylum tricornutum. Process Biochem. 57, 43–49. doi: 10.1016/j.procbio.2017.03.013
Zhu, Z., Yuan, G., Fan, X., Fan, Y., Yang, M., Yin, Y., et al. (2018). The synchronous TAG production with the growth by the expression of chloroplast transit peptide-fused ScPDAT in Chlamydomonas reinhardtii. Biotechnol. Biofuels 11:156. doi: 10.1186/s13068-018-1160-6
Zou, L. G., Chen, J. W., Zheng, D. L., Balamurugan, S., Li, D. W., Yang, W. D., et al. (2018). High-efficiency promoter-driven coordinated regulation of multiple metabolic nodes elevates lipid accumulation in the model microalga Phaeodactylum tricornutum. Microb. Cell Fact. 17:54. doi: 10.1186/s12934-018-0906-y
Zulu, N. N., Popko, P., Zienkiewicz, K., Tarazona, P., Herrfurth, C., and Feussner, I. (2018). Heterologous co-expression of a yeast diacylglycerol acyltransferase (ScDGA1) and a plant oleosin (AtOLEO3) as an efficient tool for enhancing triacylglycerol accumulation in the marine diatom Phaeodactylum tricornutum. Biotechnol. Biofuels 10:187. doi: 10.1186/s13068-017-0874-1
Keywords: microalgae, biofuel, lipid accumulation, biomass, abiotic stress, transcription factor, microRNA, genome editing
Citation: Sharma PK, Saharia M, Srivstava R, Kumar S and Sahoo L (2018) Tailoring Microalgae for Efficient Biofuel Production. Front. Mar. Sci. 5:382. doi: 10.3389/fmars.2018.00382
Received: 02 August 2018; Accepted: 28 September 2018;
Published: 21 November 2018.
Edited by:
Pannaga Pavan Jutur, International Centre for Genetic Engineering and Biotechnology (India), IndiaReviewed by:
Krishna Mohan Poluri, Indian Institute of Technology Roorkee, IndiaNitin Keshari, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), China
Copyright © 2018 Sharma, Saharia, Srivstava, Kumar and Sahoo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingaraj Sahoo, bHNAaWl0Zy5hYy5pbg==
 Prabin Kumar Sharma
Prabin Kumar Sharma Manalisha Saharia
Manalisha Saharia Richa Srivstava
Richa Srivstava Sanjeev Kumar
Sanjeev Kumar Lingaraj Sahoo
Lingaraj Sahoo