- 1Western Ecological Research Center, United States Geological Survey, Oakhurst, CA, United States
- 2Marine Science Institute, University of California, Santa Barbara, Santa Barbara, CA, United States
- 3Department of Biology, Institute for Great Lakes Research, Central Michigan University, Mount Pleasant, MI, United States
- 4Department of Biological Sciences, California State University Long Beach, Long Beach, CA, United States
To improve ability to detect white sharks without the need for tags, or visual census, we developed a species-specific environmental DNA (eDNA) assay that targets a 163 bp fragment of the white shark (Carcharodon carcharias) mitochondrial cytochrome B gene on a digital droplet PCR (ddPCR) platform. We used this marker to detect white shark DNA in 250 ml water samples taken from across two sites in Santa Barbara, California (United States) frequented by juvenile white sharks. We did not detect white shark DNA in samples from two neighboring sites where sharks are presumably absent, suggesting that eDNA can indicate nearby white sharks. This marker development, testing, and opportunistic application in a region with known distributions of white sharks indicates that eDNA could be developed further to monitor white sharks, thereby informing conservation planning and public safety. With the potential increase in white shark populations due to decades of protection, there is a need for fishery independent methods for assessing white shark distributions, and eDNA may provide an ideal, non-intrusive tool for coastal assessments.
Introduction
For decades, Carpinteria, California welcomed visitors with a billboard picturing beach goers and the claim “World’s safest beach” (Reynolds, 2017); that is until vandals defaced the scene with a spray-painted shark fin (KDL pers. obs). That fin was about as close to seeing a shark as one could get in Carpinteria. Not only are sharks difficult to see, but in California, as elsewhere, sharks had declined due to over fishing. Under protection, however, there is growing evidence (and associated media attention) that white sharks (Carcharodon carcharias) have started to recover (Lowe et al., 2012; Dewar et al., 2013; Burgess et al., 2014; Tinker et al., 2015). Southern California beaches are nursery habitat for juvenile white sharks (1.5–3 m TL), which increasingly aggregate along beaches during summer and fall months (where they feed on stingrays) (Lowe et al., 2012; Lyons et al., 2013).
Marine biologists assess shark abundance, distribution, and movements with tagging and direct observation (e.g., Bonfil et al., 2005; Hammerschlag et al., 2011). For instance, Smart Position or Temperature Transmitting Tags (SPOT) and Pop-Up Archival Satellite Tags (PATs) show where tagged sharks migrate (Weng et al., 2007). Also, acoustic telemetry tracking using underwater receivers has shown that some tagged white sharks aggregate near beaches in southern California (Lowe et al., 2012; Lowe et al., unpublished; Sosa-Nishizaki et al., unpublished). Because most sharks are not tagged, aerial surveys with drones or aircraft can locate untagged sharks in clear shallow water (Dicken and Booth, 2013). However, at most sites, observation conditions are too poor or white sharks are too rare to study them well (Burgess et al., 2014). To expand options for monitoring white sharks, we hypothesized that environmental DNA (eDNA) could be used for white-shark surveillance and monitoring. We tested for white-shark eDNA in water taken off a beach in Carpinteria, California, where juvenile white sharks are known to use shoreline habitat, and compared the amplified eDNA with that taken from water sampled from two similar locations where white sharks were presumably absent.
Environmental DNA is a promising tool for detecting rare species like white sharks (Jerde et al., 2011), because all species slough cellular material (Barnes and Turner, 2016), leading to a lingering “genetic plume” that can be filtered from water samples and matched to known genetic sequences (Ficetola et al., 2008). Although eDNA has long been used on bony fishes in freshwater (Jerde et al., 2011; Mahon et al., 2013), early efforts in marine systems had difficulties detecting sharks, even from aquaria. Moreover, eDNA studies must consider several limitations, like dispersion from the source, degradation over time, and cross contamination. Recently, Bakker et al. (2017) showed that eDNA metabarcoding could detect 21 shark species from water samples using a passive-surveillance approach based on high-throughput sequencing (see Simmons et al., 2015 for active vs. passive surveillance using eDNA). Inspired by this work, we sought to develop an active surveillance assay that is potentially more sensitive and is species-specific to detect white sharks from a water sample.
Materials and Methods
Usually marker development and critical mesocosm or field testing with eDNA can be done with captive animals. Because white sharks are rarely kept in captivity (Ezcurra et al., 2017), we sought a convenient location where they were likely to be abundant based on telemetry and observations of six white sharks tagged along Padaro Lane, in Carpinteria, California (Figure 1). Each site was just outside the surf zone (∼4 m depth). Prior to and during the period of sampling, three of the six tagged sharks remained near the two eastern sites (Santa Claus Lane: 34.409764, −119.556519, and Padaro Lane 34.413517, −119.561936). White sharks had not been reported or observed from two upcurrent western sites (Lookout Park: 34.418872, −119.601475, Loon Point: 34.413489, −119.583764).
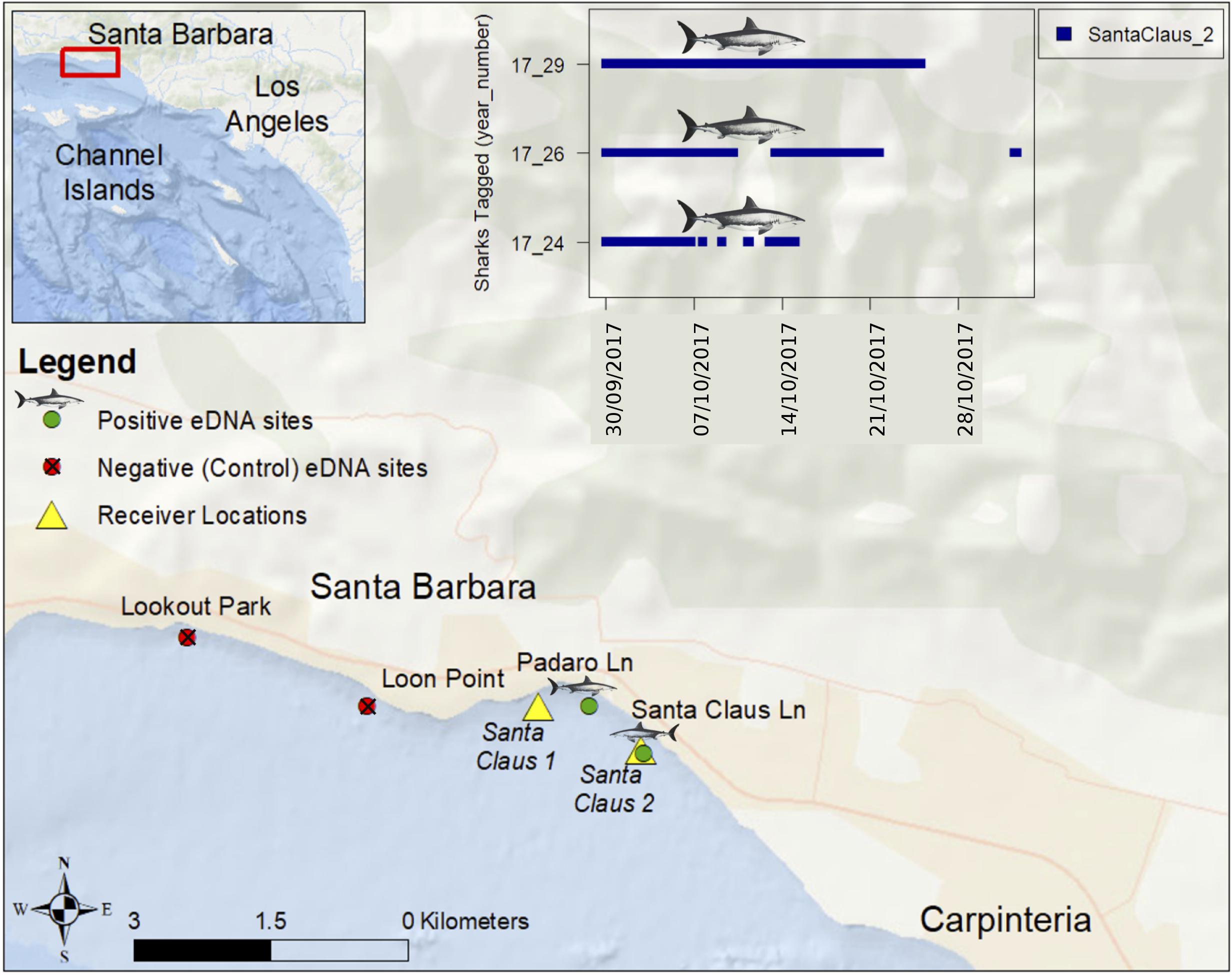
FIGURE 1. Sites near Carpinteria, California where we detected and failed to detect white shark eDNA on October 25, 2017. Six juvenile white sharks were tagged on August 24, 2017 with external acoustic transmitters at Padaro Lane and two acoustic receivers were deployed on September 29, 2017 as part of a long-term monitoring program. The inset figure shows blue vertical hash marks at times and dates when three tagged juvenile white sharks were detected at the Santa Claus 2 acoustic receiver location (Lowe et al., unpublished).
We reached these sites by standup paddleboard on October 25, 2017 between 10:30 and 13:30 using a GPS for navigation. Conditions were calm before and during the sampling, with no storms or strong currents. At each site, six sterile 250 ml Nalgene containers were filled with surface water, then put on ice after returning to shore (within 30 min of collection). The western samples were stored separately and filtered before the eastern samples to reduce potential contamination. All samples were filtered and preserved within 2 h of collection. Using the contents of two Nalgene bottles, 500 ml seawater was pushed through a 0.22 μm Sterivex Filter Unit (Spens et al., 2017) with a sterile 50 ml syringe. All filters were preserved using Longmire’s solution (Renshaw et al., 2015). Thus, we filtered 1.5 L ocean water from two sites where white sharks were known to be common, and 1.5 L from two sites 2 km away where white sharks had not been reported. Syringes were individually sealed from the manufacturer and Nalgene bottles were sterilized using 10% bleach solution exposed for more than 10 min and, rinsed thoroughly with deionized water, and sealed in plastic bags for transport.
Following the protocols of Spens et al. (2017), extractions were performed on both the individual filter capsule and the buffer preservation solution within the capsule (see Supplementary Materials for details). After incubation, the samples were mixed and washed prior to isolation on a filter column from the Qiagen DNeasy Blood and Tissue Extraction kit (Qiagen, Inc.; see Spens et al., 2017 for additional details).
To design our species-specific assay, we targeted white shark-specific mitochondrial genomes from NCBI’s GenBank database1. We then used the NCBI Primer BLAST program to design primers and a hydrolysis probe that would target a short, species-specific region of the mitogenome (see Supplementary Materials for details). After in silico design, we tested marker specificity against tissue-derived DNA extractions from white sharks, gray reef shark, black-tip reef shark, and various fishes, using a BioRad QX200 Droplet Digital PCR system (BioRad Inc.). In the end, we chose a targeted 163 bp fragment of the cytochrome B gene specific to white sharks as our species-specific marker, and this includes our internal hydrolysis probe, as this gene fragment performed reliably in tissue-based testing (Table 1).
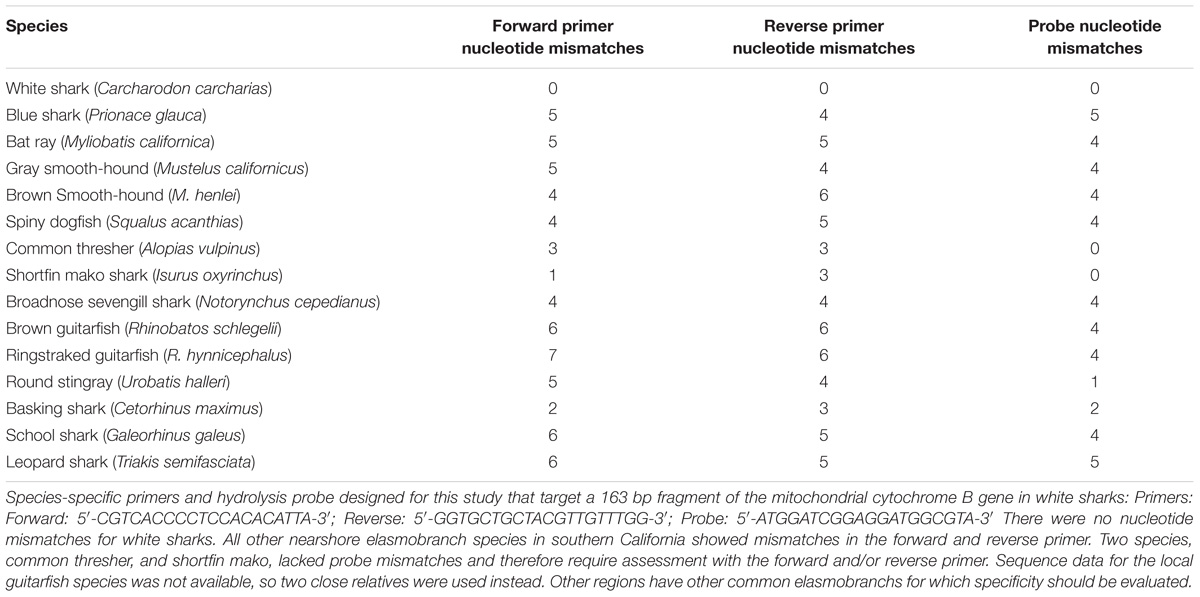
TABLE 1. Forward, reverse, and probe nucleotide mismatches for C. carcharias related species found in the study system.
We then used our white-shark-specific marker (i.e., the two species-specific primers and the hydrolysis probe) to measure absolute concentrations of white shark DNA for all samples collected in the field and across three technical replicates per sample (again with a BioRad QX200 Droplet Digital PCR system). Following active surveillance methods using digital droplet PCR (ddPCR) (Spens et al., 2017), we used a hydrolysis probe dual-labeled with a 5(6)-carboxyfluorescein (6-FAM) fluorescent tag and a 3′ Black Hole Quencher. Each 25 μL ddPCR reaction included 1000 nmol/L of each primer and probe, 1× BioRad ddPCR Supermix for probes, and 2.5 μL DNA, and sterile water. The BioRad QX200 droplet generator partitioned each individual reaction mixture into nanodroplets after combining 20 μL of the reaction mixture with 70 μL of BioRad droplet oil. For all ddPCR trials completed, two positive controls (tissue-based DNA from white sharks) and negative controls (sterile water) were included for quality assurance (see Supplementary Materials). Tissue for positive controls was collected under CSULB IACUC #364. The extracted DNA from field samples was analyzed at full concentration and then at a 1/10 dilution to test for potential inhibition risks. The ddPCR method is sensitive enough to putatively identify a single target DNA molecule (Nathan et al., 2014; Simmons et al., 2015).
Results and Discussion
In this single attempt, the primers and probe provided species-specific detection for DNA from white shark but not from other elasmobranchs found in southern California (Table 1, and see Supplementary Figure 1 for alignments), including the shortfin mako (within the same family) and also common thresher and basking shark within the same order. With two specific primers and an additional specific probe that target only white shark, we are confident in the specificity of our assay for California, and suspect it will be specific in other regions as well, though broader application to other regions should include testing against local elasmobranch species. After analysis, all Padaro Lane samples were positive for white-shark eDNA. One Santa Claus Lane sample was positive for white-shark eDNA in the 1/10 dilution, noting the possibility for inhibition from other sources. No samples from the presumably “shark absent” sites (Lookout Park and Loon Point) were positive for the marker. All negative controls were absent of any detectable DNA and all positive controls performed as expected, and were not highly variable (see Supplementary Figure 2). Therefore, we had detected white shark eDNA from two sites where white sharks were known and failed to detect white shark eDNA from two sites where white sharks had not been reported (Figure 1). There is no indication contamination led to any false positives, nor was there any indication the assay failed to perform as expected, or lead to false negatives (Ryan et al., 2017).
This study provides a species-specific marker for detecting white sharks from a shoreline water sample using eDNA and demonstrates its application in natural a system where juvenile white sharks are present. The cytochrome B marker appears specific to white sharks and acoustic telemetry and direct observation show that at least one tagged white shark was near our sites before and after we sampled. Because white sharks are relatively rare and move between beach areas, it is promising that the ddPCR method detected low concentrations of eDNA from the ocean environment.
Though simple in concept, using eDNA as an accurate monitoring tool requires ground truthing (Wilcox et al., 2013), precise collection, extraction, and assay platforms to heighten detection (Simmons et al., 2015; Goldberg et al., 2016), and error analysis to account for potential background contamination (Darling and Mahon, 2011). Admittedly, our study provides a confirmatory snapshot using current best methods, which may or may not be transferable to other global white shark habitats. Another challenge is that eDNA might persist in the ocean environment for days, meaning a positive detection might indicate a species that is no longer locally present, or be from eDNA that has drifted from another location, albeit recent studies seem to indicate marine nearshore transport of eDNA is minor (Ryan et al., 2017). Even though shark #29 (Figure 1) was swimming near the positive sampling sites when we sampled, we can only speculate where and when the white shark eDNA we detected was produced (i.e., from shark #29 or another individual or individuals). We are refining these methods to connect eDNA sequences to individuals using single nucleotide polymorphisms.
To further develop a white shark monitoring tool, we suggest the following experiments with white shark DNA derived from white shark tissue, or an area with confirmed white shark presence. (1) Measure how long white shark eDNA persists in the ocean environment (Barnes et al., 2014). (2) Estimate how detection accuracy increases with sample effort and distribution (Evans et al., 2017a). (3) Determine how eDNA concentration attenuates with distance (Shogren et al., 2017). (4) Compare how sample location (surface, mid water, bottom, offshore, surf zone) affects detection (Ryan et al., 2017). After addressing these concerns, eDNA samples could help map white shark distributions as well or better than existing methods, at lower cost, and without danger for sharks or humans (Evans et al., 2017a).
New technology is making eDNA screening quicker (Tanner et al., 2017) and portable (Lahoz-Monfort and Tingley, 2018). As costs decline, eDNA surveillance has the potential to be more cost effective (Evans et al., 2017b) and cover broader geographic areas (Tucker et al., 2016) than traditional survey methods. With further marker development, it might even be possible to use ddPCR or similar methods (depending on the marker design) to identify different individual white sharks or distinguish white shark populations with an eDNA sample. We imagine a future with eDNA screening buoys or autonomous vehicles at swimming beaches that could alert the public to white shark presence, thereby reducing human–shark conflicts (Curtis et al., 2012).
Author Contributions
All authors contributed to writing the paper. KL conceived the study, conducted the field work, and wrote the first draft. KB ran the samples in the laboratory. AM designed the laboratory protocol and the primers. CJ designed the field sampling protocol. CL provided reference tissue and verified shark distributions at the study site.
Funding
Study collaboration and funding were provided by the USGS Ecosystems Mission Area, NASA Biodiversity and Ecological Forecasting Program (# Nnx14Ar62A), the U.S. Department of the Interior, Bureau of Ocean Energy Management, Environmental Studies Program, Washington, DC under Agreement Number M15AC00006, and NOAA in support of the Santa Barbara Channel Biodiversity Observation Network.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Any use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the United States Government.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00355/full#supplementary-material
FIGURE S1 | Alignments by species.
FIGURE S2 | Thresholds for positive detection. No negative controls or negative samples had droplets over 4000 (an arbitrary fluorescence amplitude scale set by the software). Positives were defined as droplets over 4000.
Footnotes
References
Bakker, J., Wangensteen, O. S., Chapman, D. D., Boussarie, G., Buddo, D., Guttridge, T. L., et al. (2017). Environmental DNA reveals tropical shark diversity in contrasting levels of anthropogenic impact. Sci. Rep. 7:16886. doi: 10.1038/s41598-017-17150-2
Barnes, M. A., and Turner, C. R. (2016). The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 17, 1–17. doi: 10.1007/s10592-015-0775-4
Barnes, M. A., Turner, C. R., Jerde, C. L., Renshaw, M. A., Chadderton, W. L., and Lodge, D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48, 1819–1827. doi: 10.1021/es404734p
Bonfil, R., Meÿer, M., Scholl, M. C., Johnson, R., O’brien, S., Oosthuizen, H., et al. (2005). Transoceanic migration, spatial dynamics, and population linkages of white sharks. Science 310, 100–103. doi: 10.1126/science.1114898
Burgess, G. H., Bruce, B. D., Cailliet, G. M., Goldman, K. J., Grubbs, R. D., Lowe, C. G., et al. (2014). A re-evaluation of the size of the white shark (Carcharodon carcharias) population off California. USA. PLoS One 9:e98078. doi: 10.1371/journal.pone.0098078
Curtis, T., Bruce, B., Cliff, G., Dudley, S., Klimley, A., Kock, A., et al. (2012). “Recommendations for governmental organizations responding to incidents of white shark attacks on humans,” in Global Perspectives On the Biology and Life History Of the Great White Shark, ed. M. L. Domeier (Boca Raton: CRC Press), 477–510.
Darling, J. A., and Mahon, A. R. (2011). From molecules to management: adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 111, 978–988. doi: 10.1016/j.envres.2011.02.001
Dewar, H., Eguchi, T., Hyde, J., Kinzey, D., Kohin, S., Moore, J., et al. (2013). Status Review of the Northeastern Pacific Population of White Sharks (Carcharodon carcharias) Under the Endangered Species Act. Silver Spring: NOAA.
Dicken, M., and Booth, A. (2013). Surveys of white sharks (Carcharodon carcharias) off bathing beaches in Algoa Bay. South Africa. Mar. Freshwater Res. 64, 530–539. doi: 10.1071/MF12336
Evans, N. T., Li, Y., Renshaw, M. A., Olds, B. P., Deiner, K., Turner, C. R., et al. (2017a). Fish community assessment with eDNA metabarcoding: effects of sampling design and bioinformatic filtering. Can. J. Fish. Aquat. Sci. 74, 1362–1374. doi: 10.1139/cjfas-2016-0306
Evans, N. T., Shirey, P. D., Wieringa, J. G., Mahon, A. R., and Lamberti, G. A. (2017b). Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 42, 90–99. doi: 10.1080/03632415.2017.1276329
Ezcurra, J. M., Lowe, C. G., Mollet, H. F., Ferry, L. A., Murray, M. J., and O’Sullivan, J. B. (2017). “Biology of the white shark (Carcharodon carcharias) in aquaria,” in The Elasmobranch Husbandry Manual II: Recent Advances in Care of Sharks, Rays and their Relatives, ed. J. Ezurra (Columbus, OH: Ohio Biological Survey).
Ficetola, G. F., Miaud, C., Pompanon, F., and Taberlet, P. (2008). Species detection using environmental DNA from water samples. Biol. Lett. 4, 423–425. doi: 10.1098/rsbl.2008.0118
Goldberg, C. S., Turner, C. R., Deiner, K., Klymus, K. E., Thomsen, P. F., Murphy, M. A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7, 1299–1307. doi: 10.1111/2041-210X.12595
Hammerschlag, N., Gallagher, A., and Lazarre, D. (2011). A review of shark satellite tagging studies. J. Exp. Mar. Biol. Ecol. 398, 1–8. doi: 10.1016/j.jembe.2010.12.012
Jerde, C. L., Mahon, A. R., Chadderton, W. L., and Lodge, D. M. (2011). “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 4, 150–157. doi: 10.1111/j.1755-263X.2010.00158.x
Lahoz-Monfort, J. J., and Tingley, R. (2018). “The technology revolution: improving species detection and monitoring using new tools and statistical methods,” in Monitoring Threatened Species and Ecological Communities, eds S. Legge, D. Lindenmayer, N. Robinson, B. Scheele, D. Southwell, and B. Wintle (Clayton: CSIRO Publishing).
Lowe, C. G., Blasius, M. E., Jarvis, E. T., Mason, T. J., Goodmanlowe, G. D., and O’Sullivan, J. B. (2012). “Historic fishery interactions with white sharks in the Southern California Bight,” in Global Perspectives On the Biology and Life History of the White Shark, (Boca Raton, FL: CRC Press),169–186.
Lyons, K., Jarvis, E. T., Jorgensen, S., Weng, K., O’Sullivan, J., Winkler, C., et al. (2013). Assessment of degree and result of fisheries interactions with juvenile white sharks in southern California via fishery independent and dependent methods. Fish. Res. 147, 370–380. doi: 10.1016/j.fishres.2013.07.009
Mahon, A. R., Jerde, C. L., Galaska, M., Bergner, J. L., Chadderton, W. L., Lodge, D. M., et al. (2013). Validation of eDNA surveillance sensitivity for detection of Asian carps in controlled and field experiments. PLoS One 8:e58316. doi: 10.1371/journal.pone.0058316
Nathan, L. M., Simmons, M., Wegleitner, B. J., Jerde, C. L., and Mahon, A. R. (2014). Quantifying environmental DNA signals for aquatic invasive species across multiple detection platforms. Environ. Sci. Technol. 48, 12800–12806. doi: 10.1021/es5034052
Renshaw, M. A., Olds, B. P., Jerde, C. L., McVeigh, M. M., and Lodge, D. M. (2015). The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol–chloroform–isoamyl alcohol DNA extraction. Mol. Ecol. Resour. 15, 168–176. doi: 10.1111/1755-0998.12281
Reynolds, C. (2017). Romp at the World’s Safest Beach in Carpinteria. El Segundo, CA: Los Angeles Times.
Ryan, K., Gallego, R., and Jacobs-Palmer, E. (2017). The effect of tides on nearshore environmental DNA. PeerJ 6:e4521. doi: 10.7717/peerj.4521
Shogren, A. J., Tank, J. L., Andruszkiewicz, E., Olds, B., Mahon, A. R., Jerde, C. L., et al. (2017). Controls on eDNA movement in streams: transport, retention, and resuspension. Sci. Rep. 7:5065. doi: 10.1038/s41598-017-05223-1
Simmons, M., Tucker, A., Chadderton, W. L., Jerde, C. L., and Mahon, A. R. (2015). Active and passive environmental DNA surveillance of aquatic invasive species. Can. J. Fish. Aquat. Sci. 73, 76–83.
Spens, J., Evans, A. R., Halfmaerten, D., Knudsen, S. W., Sengupta, M. E., Mak, S. S., et al. (2017). Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter. Methods Ecol. Evol. 8, 635–645. doi: 10.1111/2041-210X.12683
Tanner, C. E., Sun, N., Deatsch, A., Li, F., and Ruggiero, S. T. (2017). Light transmission spectroscopy in real time: a high-resolution nanoparticle analysis instrument. Appl. Opt. 56, 1908–1916. doi: 10.1364/AO.56.001908
Tinker, M. T., Hatfield, B. B., Harris, M. D., and Ames, J. A. (2015). Dramatic increase in sea otter mortality from white sharks in California. Mar. Mamm. Sci. 32, 309–326. doi: 10.1111/mms.12261
Tucker, A. J., Chadderton, W. L., Jerde, C. L., Renshaw, M. A., Uy, K., Gantz, C., et al. (2016). A sensitive environmental DNA (eDNA) assay leads to new insights on ruffe (Gymnocephalus cernua) spread in North America. Biol. Invasions 18, 3205–3222. doi: 10.1007/s10530-016-1209-z
Weng, K. C., O’Sullivan, J. B., Lowe, C. G., Winkler, C. E., Dewar, H., and Block, B. A. (2007). Movements, behavior and habitat preferences of juvenile white sharks Carcharodon carcharias in the eastern Pacific. Mar. Ecol. Prog. Ser. 338, 211–224. doi: 10.3354/meps338211
Keywords: shark presence, eDNA, Carcharodon carcharias, coastal, conservation
Citation: Lafferty KD, Benesh KC, Mahon AR, Jerde CL and Lowe CG (2018) Detecting Southern California’s White Sharks With Environmental DNA. Front. Mar. Sci. 5:355. doi: 10.3389/fmars.2018.00355
Received: 06 July 2018; Accepted: 14 September 2018;
Published: 02 October 2018.
Edited by:
Jeremy Kiszka, Florida International University, United StatesReviewed by:
Bautisse Didier Postaire, Florida International University, United StatesStefano Mariani, The University of Salford, Manchester, United Kingdom
Copyright © 2018 Lafferty, Benesh, Mahon, Jerde and Lowe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin D. Lafferty, a2xhZmZlcnR5QHVzZ3MuZ292; bGFmZmVydHlAbGlmZXNjaS51Y3NiLmVkdQ==
 Kevin D. Lafferty
Kevin D. Lafferty Kasey C. Benesh
Kasey C. Benesh Andrew R. Mahon
Andrew R. Mahon Christopher L. Jerde
Christopher L. Jerde Christopher G. Lowe
Christopher G. Lowe