- 1Department of Botany and Zoology, University of Stellenbosch, Stellenbosch, South Africa
- 2School of Life and Environmental Sciences, Deakin University, Geelong, VIC, Australia
Ecological and environmental variables play a major role in the genetic structure of marine populations, but how oceanography affects their dispersal and associated connectivity remains far from being understood. To account for the effect of different dispersal strategies in terms of pelagic larvae and non-pelagic reproduction, we utilize the power of comparative phylogeographic analyses of five phylogenetically and functionally diverse intertidal species along the west coast of South Africa using population genetics and biophysical models within the Benguela Current system. Some broadcast spawners exhibit genetic panmixia, others show genetic structure similar to direct-developing species, suggesting complex recruitment patterns in rocky shore environments. Patterns of genetic structure do not correspond with pelagic larval competency period, with a broadcast spawning urchin displaying the highest levels of population structure. Biophysical models of larval dispersal reveal mixed dispersal patterns, with the strongest connections in a northward direction following the Benguela Current, yet most modeled species also show the capacity for southward (albeit weaker) migration among some sample localities. Some sites, particularly the most northern areas, show very low levels of potential connectivity. Lastly, we synthesized our results to highlight key areas for the development of Marine Protected Areas (MPAs) that capture the evolutionary patterns of marine species of the west coast and find that the results from our molecular and biophysical analyses are coherent with previous suggestions for a network of protected areas.
Introduction
Dispersal, the movement of individuals across land-and-seascapes, is important in maintaining ecological and genetic connectivity between geographically disjunct populations (Cowen and Sponaugle, 2009; Baguette et al., 2012). Connectivity is an important consideration under ongoing changes in climate and ocean conditions, particularly given the potential for altered dispersal pathways, which in turn may erode signals of local adaptation and impact the ability of populations to persist into the future (Lett et al., 2010; Sgro et al., 2011; Doney et al., 2012; Coleman et al., 2017). In marine systems, connectivity is challenging to measure because the connectedness of populations depends on several factors such as the directionality of larval transport, the number of individuals moving within a system and their settlement success, as well as the spatio-temporal dynamics and the physical and ecological features of the seascape (Baguette et al., 2012; Pfaff et al., 2015; Treml et al., 2015a,b; Wright et al., 2015). Therefore, in order to capture connectivity at different spatial and temporal scales, analyses should ideally incorporate species with different dispersal strategies (Baco et al., 2016), because this captures a wider range of genetic patterns (Nielsen et al., 2017).
Phylogeographic studies, mapping genetic differentiation across land, or seascapes, have the power to resolve broad-scale connectivity patterns, whilst allowing for the incorporation of multi-species data to provide relative estimates of connectivity (Kelly and Palumbi, 2010; Wright et al., 2015; Baco et al., 2016) and associations with environmental factors (Selkoe et al., 2016a). Where shared patterns across multiple species are revealed, these can provide particular insights into the processes that shape populations and communities in space and time (Selkoe et al., 2016b). Additionally, in combination with molecular analyses, modeling approaches in marine systems have been used for inferring parameters such as the strength and directionality of larval dispersal and estimating dispersal distance (Wright et al., 2015; Baco et al., 2016), unraveling the processes linking dispersal and environmental features such as oceanographic regimes (White et al., 2010) as well as predicting biotic and abiotic variables that help explain observed patterns of structure (Benestan et al., 2016; Selkoe et al., 2016a). Models of larval dispersal can also help identify areas that benefit fisheries and biodiversity conservation (Krueck et al., 2017).
The genetic structure of marine populations and their larval connectivity are known to be greatly influenced by environmental and ecological variables, with the influence of oceanographic and environmental features in determining large-scale biogeographic patterns of marine species becoming better understood (Bowen et al., 2016; Selkoe et al., 2016a). For example, studies overlaying larval dispersal estimates with ocean current data have shown that this approach helps disentangle the effects of hydrographical features shaping population dynamics (Gilg and Hilbish, 2003; White et al., 2010; Davies et al., 2015). The benefit of this approach is that it allows one to compare and contrast conservation strategies derived from classic ecological data, with those from molecular and dispersal-based data, revealing complementary yet often very different management priorities (Beger et al., 2014; Nielsen et al., 2017; von der Heyden, 2017).
Understanding the spatial and temporal patterns in connectivity is becoming increasingly important in the context of marine reserves, with several studies suggest that some existing regional marine protected area (MPA) networks are poorly connected, for example in the Mediterranean (Andrello et al., 2013), temperate Australia (Coleman et al., 2011) and in South Africa (Wright et al., 2015). In South Africa, previous studies suggest that for some species genetic boundaries align with broad biogeographic breaks (von der Heyden, 2009; Teske et al., 2011). However, the absence of obvious dispersal barriers across some region along an almost linear coastline, suggest local-scale oceanographic features (such as eddies or near-shore counter currents; see for example Teske et al., 2015) also act to maintain regional population divergence, in combination with physiological adaptations, historical events, and availability of habitat (von der Heyden, 2009; Teske et al., 2011; Toms et al., 2014).
The South African west coast is one of the most productive marine regions globally, with strong seasonal upwelling resulting in high levels of primary productivity (Nelson and Hutchings, 1983; Shannon and Nelson, 1996) that shapes a unique marine biodiversity and supports extensive pelagic and demersal fisheries. The northwards flowing Benguela Current has long been hypothesized to facilitate connectivity of marine species in this region, although these models relate to exploited demersal and pelagic fishes, which should be less influenced by inshore dynamics (Stenevik et al., 2008). For coastal species in this region, the interplay between hydrodynamics and gene structure have not been assessed, contributing to the theory that the west coast should broadly display population genetic homogeneity facilitated by the Benguela Current.
This study contributes toward better understanding the forces acting on population structuring of multiple intertidal species, with a particular emphasis on invertebrates that are traditionally under-represented in such studies (Selkoe et al., 2016a). To do this, we make use of methods that estimate genetic structure and biophysical connectivity, specifically to provide preliminary insights whether regional large-scale oceanography influences extant population genetic structuring in marine species. We utilize a comparative phylogeographic approach, focusing on five phylogenetically diverse rocky shore species inhabiting the coastline of the southern Benguela Current system. This region is an ideal place to test hypotheses concerning the directionality of dispersal, due to the linear geography of the coastline that lacks major offshore islands and is characterized by a relatively simple oceanography. Specifically, we test the hypothesis (1) that species with long pelagic larval competency periods and broadcast spawning dispersal show the highest connectivity and lowest levels of genetic structure compared to brooding and live-bearing species and (2) that the predominant direction of gene flow (i.e., migration) of larvae will be northwards along the shore, aligned with the primary flow direction of the Benguela, as has been suggested by previous egg and larval surveys and limited genetic studies (von der Heyden et al., 2008, 2011; Teske et al., 2011; Muller et al., 2012, but see also Teske et al., 2015). Overall, we evaluate our research to assess the potential management implications toward prioritizing areas for marine reserves and help to identify shared genetic breaks across multiple taxa in one of the most productive marine regions globally.
Methods
Sampling Protocol, DNA Extraction, and PCR
Four different intertidal species (the limpet Scutellastra granularis, and topshell Oxystele tigrina; the cushion star Parvulastra exigua and the clinid fish Clinus superciliosus) with differing reproductive strategies, ranging from broadcast spawning to live-bearing (Tables 1, 2), were sampled from the major, representative rocky shore habitats on the west coast (684 samples in total, Tables 1, 2). An additional 162 urchin, Parechinus angulosus, sequences were obtained from Muller et al. (2012). Sampling was conducted across seven, equally spaced sites (except for sites 6 and 7 on the Cape Peninsula) spanning ~740 km of the South African west coast (Table 1; Figure 1). The number of individuals collected for each species varied between 20 and 34 (although for one site nine individuals of C. superciliosus were collected; Table S1). We chose species living across similar shore heights in the intertidal zone (although all species can also be found in very shallow sub-tidal environments but at lower densities; with an exception being S. granularis, which is an obligate high shore species), as previous studies suggest that species confined to the different parts of the shore can show varying levels of population genetic structuring (Bird et al., 2007; von der Heyden et al., 2013). Field collection was approved by the Department of Agriculture, Forestry and Fisheries of South Africa (permit reference number: RES2012/13) and ethics approval was provided by Stellenbosch University Ethics Committee, reference SU-ACUM13-00014.
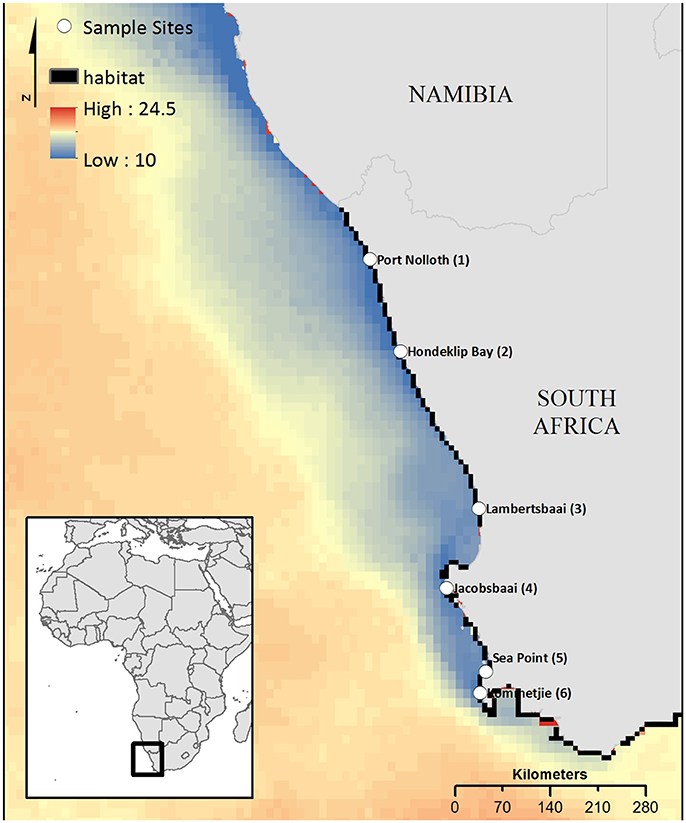
Figure 1. South Africa's west coast with the sampling locations and the study area used for biophysical modeling. Site names feature the site IDs, which are used in the network plots. Mean sea surface temperature illustrates temperature differences. Modeled habitat patches, including rocky coast, and mixed shore, are shown in black.
Given that the southern Benguela, although of global and local significance, is highly understudied, we chose to provide a broad overview of genetic structuring across multiple species, and focussed on mtDNA markers. MtDNA resolves large-scale population structuring and is particularly suitable for multi-species studies, thus proving suitable for critical insights into the evolutionary history of a region and offering a foundation for future studies (Bowen et al., 2014). A partial fragment of the mtDNA cytochrome oxidase I (COI) gene was amplified for the four invertebrate species, whereas for the fish, C. superciliosus, a fragment of the mtDNA control region was targeted (despite several attempts and different primer combinations, it was not possible to amplify mtDNA COI consistently for all fish samples). DNA extraction, primer sequences and PCR protocols followed Muller et al. (2012) and von der Heyden et al. (2011) respectively. For P. exigua and O. tigrina, species-specific primers were designed (Table S2). Trimmed fragment lengths range from 250 to 300 bp. New sequences were deposited in GenBank (KU640409–KU640590, KU640591–KU640755, and KU640756–KU640952).
Statistical Analyses of Genetic Data
Sequences were edited and aligned manually in BioEdit v.7.0.9.0 (Hall, 1999) and collapsed to haplotypes using the program Collapse 1.2 (http://darwin.uvigo.es). Standard genetic diversity (haplotype diversity h and nucleotide diversity π) indices were calculated using Arlequin v.3.5 (Excoffier and Lischer, 2010), which was also used to calculate global ΦST using an Analysis of Molecular Variation (AMOVA) and pair-wise ΦST (an analog of Fst that takes genetic differentiation into account) values for each species, between the different sampling sites. The relationship between genetic differentiation and geographic distance was used to test for isolation-by-distance (IBD), using a Mantel test in the online “Isolation by Distance web service v3.23” (Jensen et al., 2005) and tested for significance. To visualize the relationships between haplotypes sampled at different locations, minimum parsimony networks were constructed for each species using TCS v1.21 (Clement et al., 2000).
Dispersal Modeling in the Southern Benguela Current
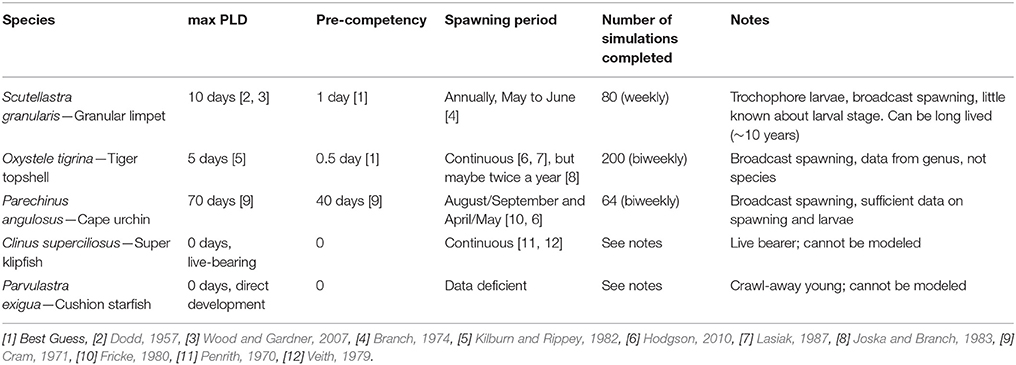
A biophysical larval dispersal model (Treml et al., 2012) was parameterized for the Benguela system to quantify the dispersal potential for the three taxa with a pelagic larval stage (S. granularis, O. tigrina, and P. angulosus). Parameter values (e.g., pelagic larval duration, competency, and spawning periodicity) were estimated by empirical data from the literature and expert opinion (Table 2), as many details for the species' parameters are unknown or uncertain. The model domain was set to the study area (Figure 1) using high-resolution shoreline data, rocky shore habitat data [taken from (South African National Biodiversity Institute, National Biodiversity Assessment (NBA), 2011), http://bgis.sanbi.org/SpatialDataset/Detail/414], our sample sites, and interpolated hydrodynamic data (based on depth, rugosity, winds, geostrophic forcing, etc.) for the region using best available (long time-series, regionally consistent, ~7 km spatial resolution, 1992–2012) oceanographic data (HYCOM, hycom.org). Although this hydrodynamic model cannot resolve the very fine-scale coastal (or intertidal) dynamics, it is adequate in resolving meso-scale eddies and the local structure of the Benguela Current system (Chassignet et al., 2007). All nearshore rocky habitat patches were used as potential sources (37 patches with mean of 33 ± 16 km2 rocky habitat) and destinations during the biophysical larval dispersal modeling. Note, although we released virtual larvae from all possible source patches, we summarized results as the cumulative migration between the seven samples sites (including through stepping-stone habitat patches between sampled populations). As a result, our approach attempts to represent local-scale (i.e., within/among patches) to seascape-level (throughout entire Benguela system) dispersal potential through all habitat patches in the system. A single dispersal simulation consisted of modeling the advection and transport of a cloud of virtual larvae (equivalent to more than 107 larvae) released from an individual rocky shore habitat patch (sample sites and unsampled habitat patches) and tracks this larval cohort as it moves downstream, directed by the biological parameters, and coastal currents. See Treml et al. (2012) for detailed numerical methods and sensitivity testing. Cumulatively, we completed 840 dispersal simulations across each of 37 potential source sites (Figure 1) averaged across 20 years of oceanographic data (1992–2012). Long-term average species-specific dispersal networks were developed from the modeled migration matrices to map the emergent patterns of potential population connectivity across species (connection values represent the proportion of downstream settlement that came from each potential upstream source). These dispersal networks highlight the geographic location and the strength of dispersal corridors and barriers along the west coast of South Africa.
Results
Diversity Analyses and Population Structure
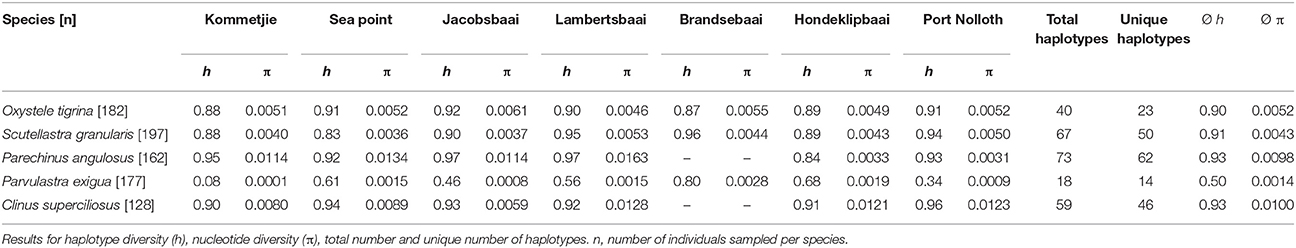
After editing and trimming, sequence lengths varied by species (C. superciliosus = 373 bp, P. angulosus = 790 bp, S. granularis = 611 bp, P. exigua = 651 bp, O. tigrina = 473 bp). Haplotype diversity and the number of unique haplotypes (those restricted to only one site) is provided in Table 1, which shows that diversity indices vary between the five intertidal species. Both h and π were lowest in the cushion star P. exigua, whereas all other species showed relatively high levels of diversity (overall range h = 0.5–0.93; π = 0.0014–0.01; Table 1). This is clearly shown by the number of unique haplotypes found in each species. Fourteen of 18 are unique for P. exigua, whereas for the other four species the proportion of unique haplotypes is much lower (Table 1; Figure 2). Analyses of molecular variance showed no congruent pattern of life history and population genetic structure. The broadcast spawning urchin, P. angulosus had the highest global ΦST (ΦST = 0.32, P < 0.001), followed by the cushion star P. exigua (crawl-away young) and clinid fish C. superciliosus (live-bearing) (ΦST = 0.27 and 0.14 respectively, P < 0.05). The other two broadcast spawning species, O. tigrina and S. granularis did not have significant global FST values (P > 0.05). Parechinus angulosus also had the largest pairwise ΦST values (0.15–0.62, P < 0.05), which increase with distance between collection sites, thus resulting in a significant signal of IBD. IBD slopes for all other species were not significant (Figure S1). Pairwise ΦST values were relatively high for P. exigua (0.11–0.51, P < 0.05) and somewhat lower for the live-bearing fish C. superciliosus (0.09–0.25, P < 0.05; Table S3). The haplotype networks vary between species, with the cushion star showing the simplest network with one central haplotype, whereas the networks for the four other species are more complex. In particular the networks of the O. tigrina and S. granularis reflect the low values for population structure, displaying a high degree of shared haplotypes between collection sites (Figure 2).
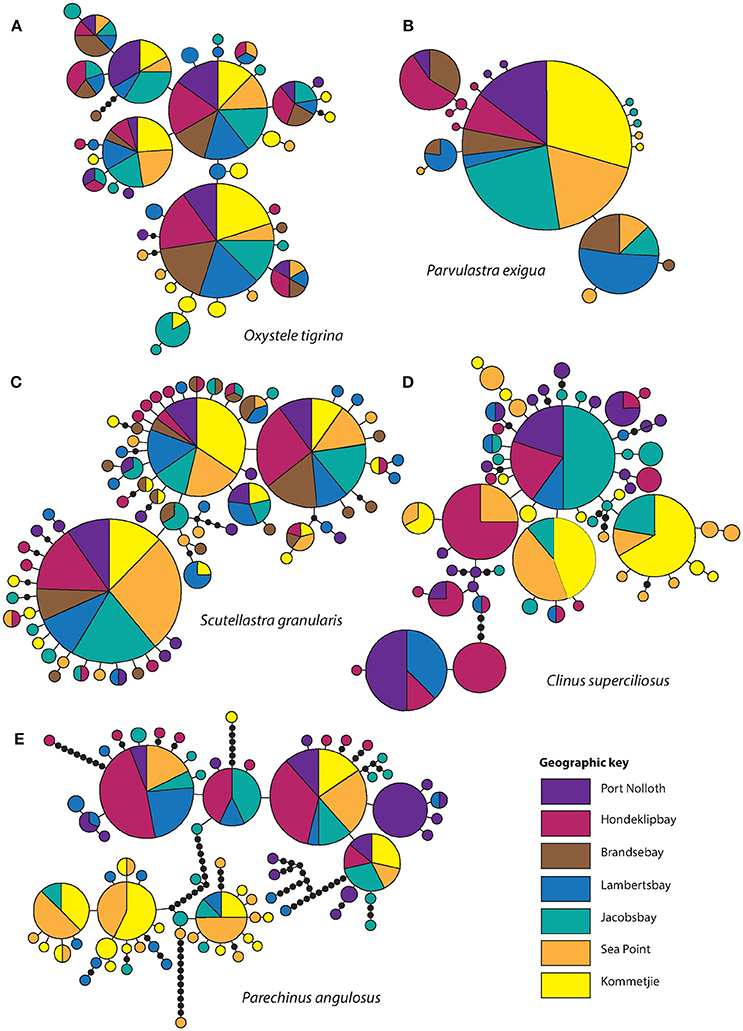
Figure 2. Haplotype networks. The size of the circles is proportional to the frequency of each haplotype. Each line represents one mutational step. Black dots mark missing steps. (A) O. tigrina, (B) P. exigua, (C) S. granularis, (D) C. superciliosus, and (E) P. angulosus.
Dispersal Modeling
The dispersal networks representing the three modeled taxa (Figure 3) clearly illustrate the complex and directed nature of dispersal potential across this seascape (dispersal strengths range from below 0.001–0.20). As the pelagic larval competency period increases, a larger number of sites become more strongly connected (see O. tigrina and P. angulosus for lower and upper extremes) and have a lower local retention rate (O. tigrina and S. granularis with 12–13% local retention; P. angulosus with <0.001%). Multi-species consistency can be seen in the generally stronger northward flow structure in the north (sites 1–3), and more mixed flow among the central and southern sites in the seascape. For the limpet and top shell, dispersal is generally strongest between adjacent sites, with less connectivity between sites that are further apart. For the urchin, modeled dispersal is high between sites even if they are not adjacent to each other, except for the two most northern sites where connectivity is lower. The two most northern sites also have the lowest connectedness for in the dispersal estimates of the limpet and topshell. Connectivity matrices from all modeled taxa are available in Table S4.
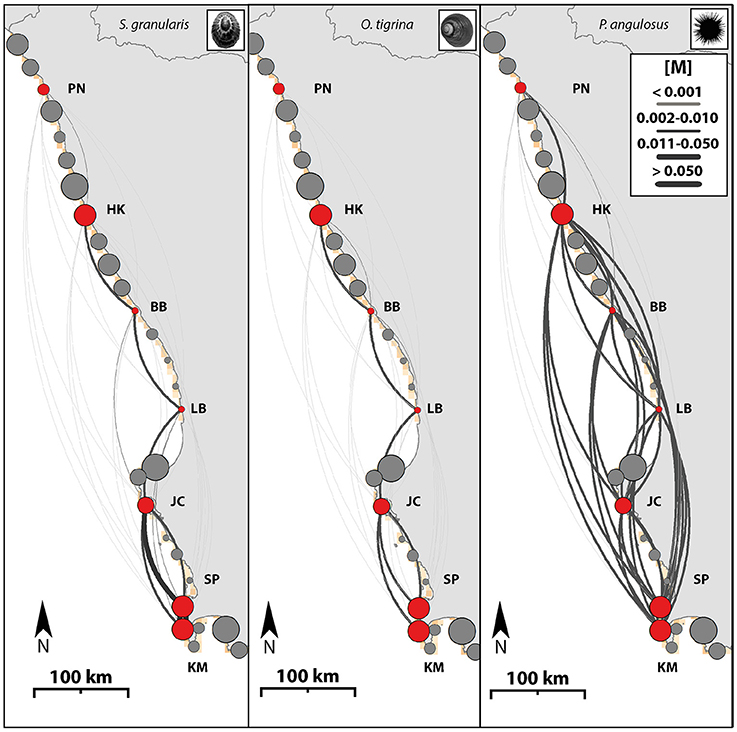
Figure 3. Hydrodynamic dispersal networks. Map illustrating species connectivity networks based on the Migration matrix [M] through all potential habitat patches. Scutellastra granularis, Oxystele tigrina, and Parechinus angulosus. Arrow sizes are scaled to dispersal probability among sampling locations. Dispersal shown as calculated by the migration estimates between sampling locations (PN, Port Nolloth; HK, Hondeklipbaai; BB, Brandsebaai; LB, Lambertsbaai; JC, Jacobsbaai; SP, Sea Point; KM, Kommetjie). Arrows depict migration parameters. The size of the circles are in proportion to the relative amount of habitat in each patch. The colors: Red are sampled sites, gray are all other sites (unsampled). No urchins could be obtained from Brandsebaai.
Discussion
Our study used an integrative approach, combining population genetics and biophysical modeling to investigate whether regional oceanographic conditions could contribute toward shaping the genetic structure of a diverse array of rocky shore species distributed along one of the world's most productive marine regions. The results show that three of five coastal species exhibit significant population structuring. The urchin, P. angulosus, that is characterized by the longest larval competency and lowest levels of local retention nevertheless displays the highest level of population structure of all species analyzed in this study. Therefore, given the mixed findings of the three broad-cast spawning species, we cannot conclusively answer our hypothesis that longer larval competency results in lower genetic structure. The biophysical model provides evidence for potential dispersal both northward, i.e., with the dominant oceanographic feature of the region, the Benguela Current, but also mixed flow in the southern and central study regions. Interestingly, the biophysical model also suggests limited gene flow across some regions along almost 800 km of linear coastline, particularly the two most northern sites. Considering our hypotheses, the findings clearly reject the hypothesis that larval dispersal would dominantly follow the flow direction of the Benguela Current, due to the substantial migration rates toward southern localities. This could indicate the influence of near-shore countercurrents, although represented and not fully resolved in our hydrodynamic data, which might transport larvae independently of the Benguela Current.
Hydrodynamic Models and Patterns of Genetic Structure Resolved Mixed Patterns of Potential Connectivity
We recovered diverse patterns of population genetic structure across our target species, from signals of panmixia to strong population divergence. For example, analyses based on estimates of FST suggest that from an evolutionary perspective, the two mollusc species remain well connected within the near-shore Benguela Current system. In contrast, the broadcast spawning urchin, with the greatest potential for dispersal (Figure 3) exhibits the highest levels of genetic structure. The direct-developing cushion star and the live-bearing clinid fish also show significant levels of population structure, although these are not as high as for the urchin. This result mirrors previous findings of multi-species genetic analyses of coastal species (Kelly and Palumbi, 2010; Wright et al., 2015) in that the duration of larval competency is generally a poor predictor of genetic structure (Weersing and Toonen, 2009).
Interestingly, the biophysical model suggested that the urchin has the highest potential of larval connectivity, with even distant populations seemingly well connected which is in stark contrast to the genetic results. This points to other, as yet unknown seascape features such as local adaptation or post-settlement processes, such as larval behavior, that maintain strong genetic structure despite high flow, which has been observed in other high gene flow marine species (Sanford and Kelly, 2011; Benestan et al., 2016). Nielsen et al. (2018) showed that P. angulosus populations in the region are characterized by a low number of private SNPs that indicate some population isolation. In particular, the two most northern sites are most differentiated, which may well be linked to our finding that from a larval connectivity perspective, these two sites are relatively poorly connected to the southern populations.
In contrast to the high dispersal potential and low genetic structure of the urchin, for the limpet we recover scenarios of genetic panmixia (at least measured with mtDNA COI), but hydrodynamic models that suggest lower levels of connectivity. As for the genomic study on the urchin, this is mirrored by private SNPs found in the most northern limpet populations (Nielsen et al., 2018), providing further evidence that oceanography shapes the evolutionary trajectories even in high dispersal species with large effective population sizes. Overall, our combined analyses of genetic structure and hydrodynamic models suggest a complex system of multiple drivers, including environmental variation as well as oceanography contributing toward shaping the population and dispersal dynamics of marine coastal species on the west coast.
Perspectives for the Management of Highly Variable Coastal Marine Systems
The incorporation of molecular data into management and conservation planning is a challenging process, mainly due to a lack of theoretical and practical frameworks (Beger et al., 2014; von der Heyden et al., 2014; Nielsen et al., 2017). However, integrating genetic information is particularly valuable for delineating populations and providing insights into population connectivity (Davies et al., 2015; Liggins et al., 2016; von der Heyden, 2017). Although single marker approaches, such as the one used in our study approaches cannot account for aspects such as adaptive variation, it nonetheless serves as a suitable tool for identifying broad patterns of spatial genetic structure on which further analyses can be built, especially with the inclusion of multiple species (Bowen et al., 2014; Nielsen et al., 2017). The latter is particularly important from a conservation planning perspective, especially in regions that lack capacity and are under-resourced, as multi-species mtDNA data sets such as the one in this study, have the potential to provide valuable insights into the spatial distribution of genetic variation, which can be efficiently incorporated into the planning process (Nielsen et al., 2017).
Though South Africa is considering several new MPAs, particularly through the ocean economy development program Operation Phakisa (www.operationphakisa.gov.za), the current MPA network has been shown to be ineffective at protecting genetic population differentiation and diversity and that in general molecular data is underutilized in marine management applications in the region (von der Heyden, 2009; von der Heyden et al., 2014; Wright et al., 2015; Nielsen et al., 2017). Therefore, resolving genetic structure among multiple species has the strength to point to shared environmental and dispersal barriers, which may lead to the recognition of separate “management units.” Although this term is more commonly applied to exploited species, it can also be used to delineate units of species that are not of commercial interest. Importantly, our work shows that even though previously recognized hard barriers to gene flow such as Cape Point, (Teske et al., 2011) could not be detected at the west coast for the studied species, our work suggest some regions are poorly connected from both genetic and biophysical perspectives. As we have shown here, the west coast appears to be genetically structured between sites and across taxa, suggesting that the different populations should be considered as separate management units (see Figures 2, 3). For species that show strong genetic structure, such as the urchin in our study, multiple MPAs would be required to secure the unique genetic signals at each site and build resilience in the system.
Our suggestion to protect specific west coast areas to safeguard unique genetic patterns correspond to previous recommendations for spatial conservation priorities (Majiedt et al., 2013). These focus areas lie adjacent to regions for which we observed reduced gene flow, supporting that biodiversity, habitat, and genetic patterns vary considerably in this region and probably require distinct management interventions to capture their differences. Ideally, a local network of MPAs would protect the genetic, connectivity and evolutionary characteristics, although this approach would benefit from additional insights from other coastal and offshore species to test fully and across different marker types (Nielsen et al., 2018). In summary, a combination of biological, oceanographic, molecular, and available socio-economic data would greatly improve our ongoing efforts in developing an integrated spatial management strategy for the west coast of South Africa.
Author Contributions
SvdH, LM, and ET designed the study, collected the data, analyzed the data, prepared figures, and wrote the manuscript.
Funding
This work was funded by a University of Stellenbosch Discretionary Grant awarded to SvdH and the National Research Foundation (Grant number: 92788; Competitive support for Rated Researchers programme). LM was supported by NRF grant holder-linked (2015) and Free-standing doctoral bursaries (2016 to present; Grant number: 111077).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
With sincere thanks to D. Wright, G. Branch, Z. Zeeman, J. Toms, and M. Jooste for help with sampling, and C. Muller and T. Muteveri for sharing data sets, and Jutta Beher for biophysical modeling support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00268/full#supplementary-material
References
Andrello, M., Mouillot, D., Beuvier, J., Albouy, C., Thuiller, W., and Manel, S. (2013). Low connectivity between Mediterranean marine protected areas: a biophysical modeling approach for the dusky grouper Epinephelus marginatus. PLoS ONE 8:e68564. doi: 10.1371/journal.pone.0068564
Baco, A. R., Etter, R. J., Ribeiro, P. A., von der Heyden, S., Beerli, P., and Kinlan, B. P. (2016). A synthesis of genetic connectivity in deep-sea fauna and implications for marine reserve design. Mol. Ecol. 25, 3276–3298. doi: 10.1111/mec.13689
Baguette, M., Legrand, D., Fréville, H., Van Dyck, H., and Ducatez, S. (2012). Dispersal Ecology and Evolution, (Oxford: Oxford University Press), 381–391.
Beger, M., Selkoe, K. A., Treml, E., Barber, P. H., von der Heyden, S., Crandall, E. D., et al. (2014). Evolving coral reef conservation with genetic information. Bull. Mar. Sci. 90, 159–185. doi: 10.5343/bms.2012.1106
Benestan, L., Quinn, B. K., Maaroufi, H., Laporte, M., Clark, F. K., Greenwood, S. J., et al. (2016). Seascape genomics provides evidence for thermal adaptation and current-mediated population structure in American lobster (Homarus americanus). Mol. Ecol. 25, 5073–5092. doi: 10.1111/mec.13811
Bird, C. E., Holland, B. S., Bowen, B. W., and Toonen, R. J. (2007). Contrasting phylogeography in three endemic Hawaiian limpets (Cellana spp.) with similar life histories. Mol. Ecol. 16, 3173–3186. doi: 10.1111/j.1365-294x.2007.03385.x
Bowen, B. W., Gaither, M. R., DiBattista, J. D., Iacchei, M., Andrews, K. R., Grant, W. S., et al. (2016). Comparative phylogeography of the ocean planet. Proc. Natl. Acad. Sci. U.S.A. 113, 7962–7969. doi: 10.1073/pnas.1602404113
Bowen, B. W., Shanker, K., Yasuda, N., Celia, M., Malay, M. C. D., von der Heyden, S., et al. (2014). Phylogeography unplugged: comparative surveys in the genomic era. Bull. Mar. Sci. 90, 13–46. doi: 10.5343/bms.2013.1007
Branch, G. M. (1974). The ecology of Patella linneaus from the Cape Peninsula, South Africa. 2. reproductive cycles. Trans. R. Soc. S. Afr. 41, 111–60. doi: 10.1080/00359197409520068
Chassignet, E. P., Hurlburt, H. E., Smedstad, O. M., Halliwell, G. R., Hogan, P. J., Wallcraft, A. J., et al. (2007). The HYCOM (HYbrid Coordinate Ocean Model) data assimilative system. J. Mar. Syst. 65, 60–83. doi: 10.1016/j.jmarsys.2005.09.016
Clement, M., Posada, D. C., and Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Coleman, M. A., Cetina-Heredia, P., Roughan, M., Feng, M., Van Sebille, E., and Kelaher, B. P. (2017). Anticipating changes to future connectivity within a network of marine protected areas. Glob. Change Biol. 23, 3533–3542. doi: 10.1111/gcb.13634
Coleman, M. A., Chambers, J., Knott, N. A., Malcolm, H. A., Harasti, D., Jordan, A., et al. (2011). Connectivity within and among a network of temperate marine reserves. PLoS ONE 6:e20168. doi: 10.1371/journal.pone.0020168
Cowen, R. K., and Sponaugle, S. (2009). Larval dispersal and marine population connectivity. Annu. Rev. Mar. Sci. 1, 443–466. doi: 10.1146/annurev.marine.010908.163757
Cram, D. L. (1971). Life history studies on South African Echinoids (Echinodermata) 1. Parechinus angulosus (Leske) (Echinidae, Parechininae). Trans. R. Soc. S. Afr. 39, 321–337. doi: 10.1080/00359197109519121
Davies, S. W., Treml, E. A., Kenkel, C. D., and Matz, M. V. (2015). Exploring the role of Micronesian islands in the maintenance of coral genetic diversity in the Pacific Ocean. Mol. Ecol. 24, 70–82. doi: 10.1111/mec.13005
Dodd, J. M. (1957). Artificial fertilisation, larval development and metamorphosis in Patella vulgata L. and Patella caerulea L. Pubbl. Stazione Zool. Napoli 29, 172–186.
Doney, S. C., Ruckelshaus, M., Emmett Duffy, J., Barry, J. P., Chan, F., English, C. A., et al. (2012). Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fricke, A. H. (1980). Aspects of population structure of Parechinus angulosus around the Cape Peninsula. S. Afr. J. Zool. 15, 177–185. doi: 10.1080/02541858.1980.11447707
Gilg, M. R., and Hilbish, T. J. (2003). The geography of marine larval dispersal: coupling genetics with fine-scale physical oceanography. Ecology 84, 2989–2998. doi: 10.1890/02-0498
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hodgson, A. N. (2010). Reproductive seasonality of Southern African inshore and estuarine invertebrates—a biogeographic review. Afr. Zool. 45, 1–17. doi: 10.1080/15627020.2010.11657249
Jensen, J. L., Bohonak, A. J., and Kelley, S. T. (2005). Isolation by distance, web service. BMC Genet. 6:13. doi: 10.1186/1471-2156-6-13
Joska, M. A., and Branch, G. M. (1983). The reproductive cycle of the trochid gastropod Oxystele variegata (Anton, 1839). Veliger 26, 47–51.
Kelly, R. P., and Palumbi, S. R. (2010). Genetic structure among 50 species of the northeastern Pacific rocky intertidal community. PLoS ONE 5:e8594. doi: 10.1371/journal.pone.0008594
Krueck, N. C., Ahmadia, G. N., Green, A., Jones, G. P., Possingham, H. P., Riginos, C., et al. (2017). Incorporating larval dispersal into MPA design for both conservation and fisheries. Ecol. Appl. 27, 925–941. doi: 10.1002/eap.1495
Lasiak, T. (1987). The reproductive cycles of three trochid gastropods from the Transkei coast, Southern Africa. J. Molluscan Stud. 53, 24–32. doi: 10.1093/mollus/53.1.24
Lett, C., Ayata, S. D., Huret, M., and Irisson, J. O. (2010). Biophysical modelling to investigate the effects of climate change on marine population dispersal and connectivity. Prog. Oceanogr. 87, 106–113. doi: 10.1016/j.pocean.2010.09.005
Liggins, L., Treml, E. A., Possingham, H., and Riginos, C. (2016). Seascape features, rather than dispersal traits, predict spatial genetic patterns in co-distributed reef fishes. J. Biogeogr. 43, 256–267. doi: 10.1111/jbi.12647
Majiedt, P., Holness, S., Sink, K., Oosthuizen, A., and Chadwick, P. (2013). Systematic Marine Biodiversity Plan for the West Coast of South Africa. South African. National Biodiversity Institute Cape Town, 46.
Muller, C. M., von der Heyden, S., Bowie, R. C. K., and Matthee, C. A. (2012). Oceanic circulation, local upwelling and palaeoclimatic changes linked to the phylogeography of the Cape sea urchin Parechinus angulosus. Mar. Ecol. Prog. Ser. 468, 203–215. doi: 10.3354/meps09956
Nelson, G., and Hutchings, L. (1983). The Benguela upwelling area. Prog. Oceanogr. 12, 333–356. doi: 10.1016/0079-6611(83)90013-7
Nielsen, E. S., Beger, M., Henriques, R., Selkoe, K. A., and der Heyden, S. (2017). Multispecies genetic objectives in spatial conservation planning. Conserv. Biol. 31, 872–882. doi: 10.1111/cobi.12875
Nielsen, E. S., Henriques, R., Toonen, R. J., Knapp, I., Guo, B., and von der Heyden, S. (2018). Complex signatures of genomic variation of two non-model marine species in a homogeneous environment. BMC Genomics 19:347. doi: 10.1186/s12864-018-4721-y
Penrith, M. L. (1970). The distribution of the fishes of the family Clinidae in southern Africa. Ann. S. Afr. Mus. 55, 135–150.
Pfaff, M. C., Branch, G. M., Fisher, J. L., Hoffmann, V., Ellis, A. G., and Largier, J. L. (2015). Delivery of marine larvae to shore requires multiple sequential transport mechanisms. Ecology 96, 1399–1410. doi: 10.1890/14-0229.1
Sanford, E., and Kelly, M. W. (2011). Local adaptation in marine invertebrates. Annu. Rev. Mar. Sci. 3, 509–535. doi: 10.1146/annurev-marine-120709-142756
Selkoe, K. A., Aloia, C. C., Crandall, E. D., Iacchei, M., Liggins, L., Puritz, J. B., et al. (2016a). A decade of seascape genetics: contributions to basic and applied marine connectivity. Mar. Ecol. Prog. Ser. 554, 1–19. doi: 10.3354/meps11792
Selkoe, K. A., Gaggiotti, O. E., Treml, E. A., Wren, J. L. K., Donovan, M. K., and Toonen, R. J. (2016b). The DNA of coral reef biodiversity: predicting and protecting genetic diversity of reef assemblages. Proc. Biol. Sci. 283:20160354. doi: 10.1098/rspb.2016.0354
Sgro, C. M., Lowe, A. J., and Hoffmann, A. A. (2011). Building evolutionary resilience for conserving biodiversity under climate change. Evol. Appl. 4, 326–337. doi: 10.1111/j.1752-4571.2010.00157.x
Shannon, L. V., and Nelson, G. (1996). The Benguela: Large Scale Features and Processes and System Variability. Berlin; Heidelberg: Springer; The South Atlantic.
South African National Biodiversity Institute, National Biodiversity Assessment (NBA) (2011). Marine Benthic and Coastal Habitat Types [Vector Geospatial Dataset] 2011. Available online at: http://bgis.sanbi.org/SpatialDataset/Detail/414 (Accessed February 23, 2018).
Stenevik, E. K., Verheye, H. M., Lipinski, M. R., Ostrowski, M., and Strømme, T. (2008). Drift routes of cape hake eggs and larvae in the southern benguela current system. J. Plankton Res. 30, 1147–1156. doi: 10.1093/plankt/fbn068
Teske, P. R., Bader, S., and Golla, T. R. (2015). Passive dispersal against an ocean current. Mar. Ecol. Prog. Ser. 539, 153–163. doi: 10.3354/meps11516
Teske, P. R., von der Heyden, S., McQuaid, C. D., and Barker, N. P. (2011). A review of marine phylogeography in southern Africa. S. Afr. J. Sci. 107, 43–53. doi: 10.4102/sajs.v107i5/6.514
Toms, J. A., Compton, J. S., Smale, M., and von der Heyden, S. (2014). Variation in palaeo-shorelines explains contemporary population genetic patterns of rocky shore species. Biol. Lett. 10:20140330. doi: 10.1098/rsbl.2014.0330
Treml, E. A., Ford, J. R., Black, K. P., and Swearer, S. E. (2015a). Identifying the key biophysical drivers, connectivity outcomes, and metapopulation consequences of larval dispersal in the sea. Mov. Ecol. 3:17. doi: 10.1186/s40462-015-0045-6
Treml, E. A., Roberts, J. J., Chao, Y., Halpin, P. N., Possingham, H. P., and Riginos, C. (2012). Reproductive output and duration of the pelagic larval stage determine seascape-wide connectivity of marine populations. Integr. Comp. Biol. 52, 525–537. doi: 10.1093/icb/ics101
Treml, E. A., Roberts, J., Halpin, P. N., Possingham, H. P., and Riginos, C. (2015b). The emergent geography of biophysical dispersal barriers across the Indo-West Pacific. Divers. Distrib. 21, 465–476. doi: 10.1111/ddi.12307
Veith, W. (1979). Reproduction in the live-bearing teleost Clinus superciliosus. S. Afr. J. Zool. 14, 208–211. doi: 10.1080/02541858.1979.11447671
von der Heyden, S. (2009). Why do we need to integrate population genetics into South African marine protected area planning? Afr. J. Mar. Sci. 31, 263–269. doi: 10.2989/ajms.2009.31.2.14.886
von der Heyden, S. (2017). Making evolutionary history count: biodiversity planning for coral reef fishes and the conservation of evolutionary processes. Coral Reefs 36, 183–194. doi: 10.1007/s00338-016-1512-2
von der Heyden, S., Beger, M., Toonen, R. J., van Herwerden, L., Juinio-Meñez, M. A., Ravago-Gotanco, R., et al. (2014). The application of genetics to marine management and conservation: examples from the Indo-Pacific. Bull. Mar. Sci. 90, 123–158. doi: 10.5343/bms.2012.1079
von der Heyden, S., Bowie, R. C. K., Prochazka, K., Bloomer, P., Crane, N. L., and Bernardi, G. (2011). Phylogeographic patterns and cryptic speciation across oceanographic barriers in South African intertidal fishes. J. Evol. Biol. 24, 2505–2519. doi: 10.1111/j.1420-9101.2011.02382.x
von der Heyden, S., Gildenhuys, E., Bernardi, G., and Bowie, R. C. K. (2013). Fine-scale biogeography: tidal elevation strongly affects population genetic structure and demographic history in intertidal fishes. Front. Biogeogr. 5, 29–38. Available online at: https://escholarship.org/uc/item/97x719zj#author
von der Heyden, S., Prochazka, K., and Bowie, R. C. K. (2008). Significant population structure and asymmetric gene flow patterns amidst expanding populations of Clinus cottoides (Perciformes, Clinidae): application of molecular data to marine conservation planning in South Africa. Mol. Ecol. 17, 4812–4826. doi: 10.1111/j.1365-294x.2008.03959.x
Weersing, K., and Toonen, R. J. (2009). Population genetics, larval dispersal, and connectivity in marine systems. Mar. Ecol. Prog. Ser. 393, 1–12. doi: 10.3354/meps08287
White, C., Selkoe, K. A., Watson, J., Siegel, D. A., Zacherl, D. C., and Toonen, R. J. (2010). Ocean currents help explain population genetic structure. Proc. Biol. Sci. 277, 1685–1694. doi: 10.1098/rspb.2009.2214
Wood, A. R., and Gardner, J. P. A. (2007). Small spatial scale population genetic structure in two limpet species endemic to the Kermadec Islands, New Zealand. Mar. Ecol. Prog. Ser. 349, 159–170. doi: 10.3354/meps07110
Wright, D., Bishop, J. M., Matthee, C. A., and von der Heyden, S. (2015). Genetic isolation by distance reveals restricted dispersal across a range of life histories: implications for biodiversity conservation planning across highly variable marine environments. Divers. Distrib. 21, 698–710. doi: 10.1111/ddi.12302.e
Keywords: marine spatial planning, gene flow, marine protected area, conservation planning, population structure, rocky shore, comparative phylogeography, South Africa
Citation: Mertens LEA, Treml EA and von der Heyden S (2018) Genetic and Biophysical Models Help Define Marine Conservation Focus Areas. Front. Mar. Sci. 5:268. doi: 10.3389/fmars.2018.00268
Received: 23 February 2018; Accepted: 16 July 2018;
Published: 06 August 2018.
Edited by:
Romuald Lipcius, Virginia Institute of Marine Science, United StatesReviewed by:
Francine Kershaw, Natural Resources Defense Council, United StatesPablo Saenz-Agudelo, Universidad Austral de Chile, Chile
Phil Maxwell Ross, University of Waikato, New Zealand
Copyright © 2018 Mertens, Treml and von der Heyden. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophie von der Heyden, c3ZkaEBzdW4uYWMuemE=
 Lisa E. A. Mertens
Lisa E. A. Mertens Eric A. Treml
Eric A. Treml Sophie von der Heyden
Sophie von der Heyden