
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 17 May 2018
Sec. Coral Reef Research
Volume 5 - 2018 | https://doi.org/10.3389/fmars.2018.00152
This article is part of the Research TopicCoral Reefs in the AnthropoceneView all 21 articles
While there are several areas containing shallow coral habitats in the Gulf of Mexico (GOM), the availability of suitable reef habitat at mesophotic depths (~30–150 m) along the continental shelf margin suggests the potential for ecologically connected coral populations across hundreds of kilometers in the northwest (NW) GOM. The NW GOM includes a relatively high proportion of mesophotic habitats, including salt diapirs in Flower Garden Banks National Marine Sanctuary (FGBNMS), Bright Bank, and McGrail Bank, the latter two being Habitat Areas of Particular Concern (HAPCs). In response to a proposed expansion plan for the sanctuary boundaries to include additional mesophotic banks in the NW GOM, we investigated the genetic connectivity of the depth-generalist coral Montastraea cavernosa, a ubiquitous member of scleractinian communities throughout the Tropical Western Atlantic. Montastraea cavernosa populations in the NW GOM demonstrated strong connectivity with relatively high levels of gene flow and no significant genetic differentiation occurring over banks up to 120 km apart. Historical migration models based on genetic data predicted panmixia of M. cavernosa across the NW GOM. The comparisons between genetic and biophysical models (see Garavelli et al., 2018) highlight not only the importance of incorporating multiple assessments of connectivity into management schemes, but also the potentially stochastic nature of oceanographic patterns in the NW GOM and their effect on migration estimates among coral habitats. These trends indicate that M. cavernosa populations have remained well-connected in the NW GOM and that coral populations on each bank have likely been receiving larval recruitment through time. Thus, M. cavernosa populations should be managed as a combined unit within the NW GOM, which supports the proposal to expand the NMS boundaries to include mesophotic habitats beyond West and East FGB.
Understanding population dynamics in marine environments is especially challenging without prior knowledge of important habitats and larval characteristics (Palumbi, 2003; Cowen and Sponaugle, 2009; Weersing and Toonen, 2009). Identification and characterization of all possible habitats and knowledge of larval biology in situ is limited if not entirely undescribed for many sessile species in marine environments (Carr et al., 2003; Cowen et al., 2006; Jones et al., 2009). Methodologies to address population dynamics by measuring genetic differentiation across multiple markers are becoming more cost-effective through sample multiplexing and reduced sequencing costs (Hilbish, 1996; Manel et al., 2003). Patterns of gene flow and migration across generations can be estimated from population genetic data, allowing inferences to be made in regards to historical and current population dynamics (Palumbi, 2003). Therefore, population genetics can be an important approach to better understand regional dynamics of coral species, particularly on deeper reefs beyond traditional exploration limits (i.e., recreational SCUBA depths) or in geographically-isolated locations (Kahng et al., 2014).
Marine reserves aim to maintain biodiversity through the protection of population sources and habitats critical to larval dispersal and survival (Palumbi, 2001, 2003; Fogarty and Botsford, 2007). Conservation strategies to protect the biodiversity and persistence of coral reef ecosystems worldwide require knowledge of how populations interact and persist (Palumbi, 2003; Cowen et al., 2006; Cowen and Sponaugle, 2009). Understanding the larval biology and dispersal characteristics of all species in an ecosystem is a daunting task given the species diversity on coral reefs (Jones et al., 2009). However, suitable model or keystone species may provide sufficient information for management needs. As widely-dispersed ecosystem engineers, corals are useful candidate species to describe system-level population dynamics (Nunes et al., 2011).
The northwest Gulf of Mexico (NW GOM) is home to extensive high-latitude coral reef systems. Salt diapirs that rose during the Jurassic period now form dozens of banks along the continental shelf margin, some of which have carbonate caps (Hickerson et al., 2008; Precht et al., 2008; Schmahl et al., 2008). Most of these habitats are at mesophotic depths (~30–150 m), but notably West and East Flower Garden Banks (FGB), approximately 180 km from the Texas coast, also have relatively shallow coral reef habitats from 17 to 30 m. While at higher latitudes than most other reefs in the Tropical Western Atlantic (TWA) and therefore spatially isolated, coral populations in the NW GOM are thought to persist due to thermal stability and relatively high larval dispersal potential from the Gulf of Mexico Loop Current (Oey et al., 2005; Atchison et al., 2008; Nunes et al., 2011; Precht et al., 2014; Rippe et al., 2017). Westward-moving eddy formations and coastal currents are also quite common in the NW GOM and may connect reef populations along the continental margin (Ohlmann and Niiler, 2005; Schmahl et al., 2008; Gough et al., 2017). Despite residing at sub-tropical latitudes, reefs in the NW GOM have relatively stable seasonal temperatures between 18 and 30°C due to persistence of currents, and major coral bleaching events are rare (Rezak et al., 1990; Schmahl et al., 2008). Coral diversity along these banks is relatively low, with 21 species of scleractinian corals compared to ~82 species found throughout the TWA. However, coral cover on the shallow caps at West and East FGB is comparatively high (>50%) and the habitats are characterized as some of the most pristine in the TWA (Hickerson et al., 2008, 2012; Johnston et al., 2016).
The NW GOM is also one of the most active regions for offshore oil and gas exploration and production in the world; indeed, the proximity of petroleum industry to coral reef ecosystems led in part to protection of the Flower Garden Banks in 1974 by the Minerals Management Service (Aronson et al., 2005). West and East FGB were designated as part of Flower Garden Banks National Marine Sanctuary (FGBNMS) in 1992, and the nearby, but less coral-dominated, Stetson Bank was added in 1996 (Schmahl et al., 2008; Johnston et al., 2016). Long-term monitoring of the coral reef communities at FGBNMS since 1989 has documented relatively stable coral cover compared to degrading reefs elsewhere in the wider TWA (Gardner et al., 2003; Johnston et al., 2016). Some notable studies have included FGBNMS in regional population genetics analyses (Atchison et al., 2008; Goodbody-Gringley et al., 2012; Serrano et al., 2014; Rippe et al., 2017) and larval dispersal models (Davies et al., 2017; Garavelli et al., 2018), identifying the potential importance of shallow coral populations in the NW GOM to other regions beyond the Gulf of Mexico. Studies describing the geomorphology and reef communities inside the sanctuary (Aronson et al., 2005; Clark et al., 2014; Johnston et al., 2016) have been informative as well, but little research has been done on the primarily-mesophotic bank habitats outside the sanctuary boundaries (Rezak and Bright, 1985; Rezak et al., 1990; Sammarco et al., 2016), particularly regarding the relative roles of their coral populations in connectivity across the NW GOM.
Habitat and benthic community characterization of mesophotic reefs in the NW GOM has indicated that overall species diversity remains high despite lower scleractinian cover and diversity, and that these reefs serve as important fish habitats (Rezak and Bright, 1985; Rezak et al., 1990; Schmahl et al., 2008; Clark et al., 2014). The Bureau of Ocean Energy Management (BOEM) currently has “No Activity Zones” based on topographic complexity thresholds established for many of the habitats outside the sanctuary that preclude oil and gas exploration, while fewer banks include additional Habitat Area of Particular Concern (HAPC) or Coral HAPC protection that establishes fishing gear restrictions. Based on decades of exploration and characterization in the NW GOM, the FGBNMS Sanctuary Advisory Council recently proposed an expansion of sanctuary boundaries to include additional mesophotic habitats currently under varying degrees of existing protection. The preferred expansion plan (Preferred Alternative 3) includes an additional 15 reef habitats, bringing the area of protected habitat from ~145 to 990 km2 (Office of National Marine Sanctuaries, 2016). Perhaps most importantly, the expansion plan would co-manage the majority of coral habitats in the NW GOM as a single management unit within the NMS framework.
Most of this habitat supports mesophotic reef assemblages, primarily dominated by antipatharians, gorgonians, and crustose coralline algal communities (Rezak and Bright, 1985; Rezak et al., 1990; Schmahl et al., 2008; Sammarco et al., 2016). A few banks, notably Bright and McGrail Banks, have upper mesophotic habitat (45–60 m) dominated by scleractinian and macroalgal communities. In both cases, the extreme depth-generalist species Montastraea cavernosa is one of the most abundant scleractinians; it is also ubiquitous at West and East FGB (Voss et al., 2014). The population dynamics of coral species across the entire NW GOM are relatively unknown, including the potential importance of coral populations in the upper mesophotic zone of these additional banks for maintaining coral populations within the existing FGBNMS boundaries. The sanctuary expansion plan proposes managing all these habitats in the same management structure, making this study to assess population connectivity among discrete habitats within the NW GOM a timely endeavor.
To support the growing knowledge of coral population dynamics in the Gulf of Mexico (Atchison et al., 2008; Goodbody-Gringley et al., 2012; Serrano et al., 2014; Rippe et al., 2017), this study used a microsatellite approach to describe population connectivity across the NW GOM using the coral model species M. cavernosa. Montastraea cavernosa is typically one of the most common scleractinian species present across upper mesophotic reef habitats in the NW GOM. This species is also relatively ubiquitous throughout the TWA and has been consistently used in previous studies of population connectivity (Bak et al., 2005; Atchison et al., 2008; Lesser et al., 2009; Nunes et al., 2009, 2011; Goodbody-Gringley et al., 2012; Brazeau et al., 2013; Serrano et al., 2014). Studies combining genetic and oceanographic data are of growing interest within marine ecosystems (Galindo et al., 2006; Baltazar-Soares et al., 2018) and increase understanding of how spatially isolated coral reefs remain connected through time. In particular, the comparison of genetic and biophysical models (Garavelli et al., 2018) allows for more comprehensive investigations of populations with low genetic differentiation, and may provide empirical support for theoretical oceanographic patterns inferred by gene flow (Selkoe et al., 2008). The ultimate objectives of this study were to integrate genetic data into existing reef management strategies for Flower Garden Banks National Marine Sanctuary, as well as to provide critical data to inform the proposed expansion of NMS boundaries in the NW GOM.
Coral samples used to assess population structure in the NW GOM were collected from >1 m distant M. cavernosa colonies using SCUBA and remotely operated vehicle (ROV) and were preserved independently in either TRIzol reagent or salt-saturated DMSO. Sampling sites were chosen based on the availability of coral reef habitat including dominant presence of M. cavernosa populations determined from previous exploration missions (Hickerson et al., 2008; Schmahl et al., 2008; Voss et al., 2014). Corals were sampled at two banks currently within the sanctuary boundaries, and two HAPC reef habitats included in the sanctuary expansion plan (Figure 1). A total of 252 coral colonies were sampled across the four banks to ensure at least 30 individuals per population (Hale et al., 2012). Corals were sampled with sufficient replication at both shallow and mesophotic depth zones where continuous habitat was present (West and East FGB) to allow population comparisons within bank. From within the sanctuary, sampled populations included West Flower Garden Bank (WFGB) and East Flower Garden Bank (EFGB). Outside the sanctuary, Bright Bank (BRT) and McGrail Bank (MCG) were sampled along the scleractinian-dominated mesophotic caps (Table 1).
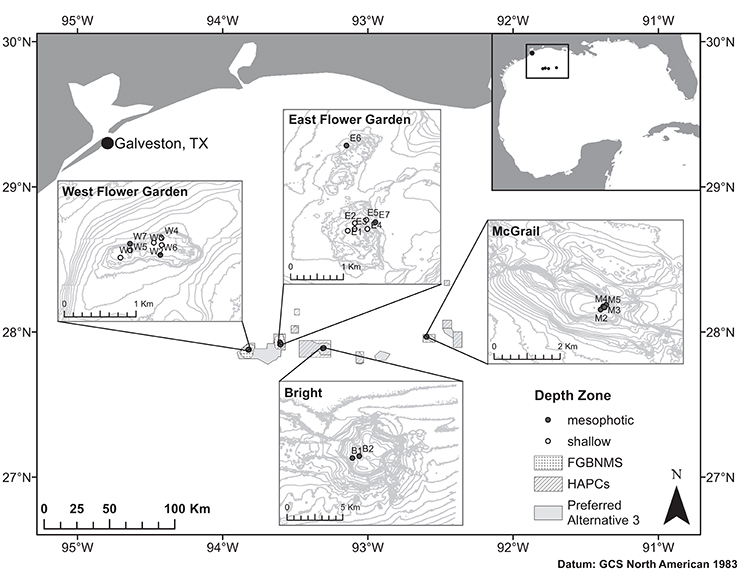
Figure 1. Map of the NW GOM and sampling locations across four reef banks including West Flower Garden Bank (WFGB), East Flower Garden Bank (EFGB), Bright Bank (BRT), and McGrail Bank (MCG). FGBNMS boundaries indicated by spotted polygons, HAPCs by crosshatched polygons, and the Preferred Alternative 3 expansion boundaries shown as gray polygons. Sampling site names as in Table 1.
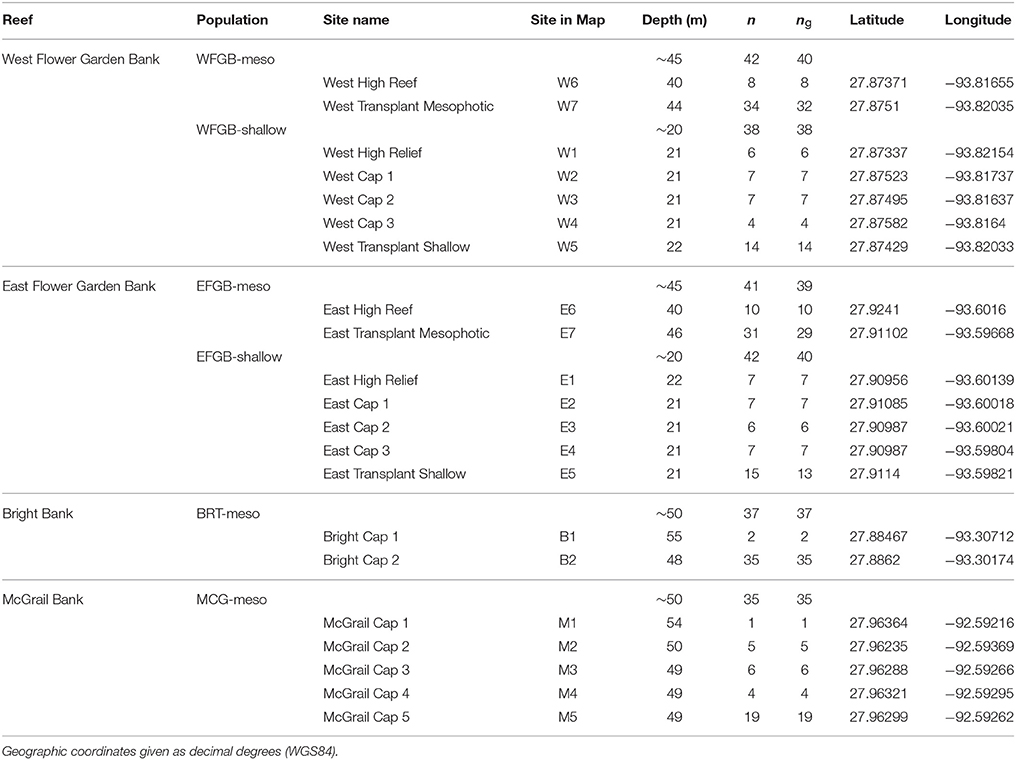
Table 1. Montastraea cavernosa genotyped samples (n = 252) collected across the northwest Gulf of Mexico, compared with the number of unique multi-locus genotypes (MLGs) shown as ng used for the analyses (ng = 229).
DNA was extracted using the Qiagen DNeasy Blood & Tissue Kit for DMSO samples or modified phenol-chloroform extraction for TRIzol samples (Chomczynski and Sacchi, 2006). Nine microsatellite loci (Serrano et al., 2014) were amplified in triplex reactions with the fluorescent primers 6FAM, VIC, and NED using a self-labeling technique (Schuelke, 2000) and the Qiagen Type-It Microsatellite PCR Kit according to a modified manufacturer protocol. Modifications from the kit protocol were made to forward primer concentrations (0.1 M forward primer, 0.1 M fluorescent tag) and to increase amplification to 35 cycles. Triplex groups were designed to minimize overlap of allele size ranges as follows: Plex 1 (MC29, MC41, MC49), Plex 2 (MC46, MC65, MC97), and Plex 3 (MC4, MC18, MC114). Amplified alleles were visualized via gel electrophoresis, diluted according to amplification efficiency, and sized on an ABI 3130xl genetic analyzer (Applied Biosystems) with ROX500 size standard. Alleles were scored using GeneMapper 3.7 (Applied Biosystems). Samples missing alleles from more than three loci were excluded from further analyses (Table 1).
Unique multi-locus genotypes (MLGs) were identified using GenAlEx 6.5 (Peakall and Smouse, 2006, 2012) and clonal genotypes were removed from the dataset. GenAlEx was used to conduct tests for Hardy-Weinberg Equilibrium (HWE), allele frequencies, and genetic differentiation with fixation index (FST). Linkage disequilibrium (LD) assumptions were tested with Arlequin 3.5 (Excoffier and Lischer, 2010). False discovery rate (FDR) corrections were calculated for HWE and LD p-values with the R package FDRtool (Strimmer, 2008). Deviations from HWE and LD were tested for null alleles with MicroChecker 2.2.3 and FreeNA (van Oosterhout et al., 2006; Chapuis and Estoup, 2007). Population-level FST values were calculated to compare pairwise population differentiation and tested with an analysis of molecular variance (AMOVA; 9999 model permutations, 9999 pairwise permutations) in GenAlEx (Peakall and Smouse, 2006, 2012). Population differentiation was also visualized with principal coordinates analysis using Nei genetic distance (DA). Geographic isolation among banks were tested for effects on genetic variation with a Mantel isolation by distance test (9999 permutations) (Peakall and Smouse, 2006, 2012).
Structure 2.3.4 (Pritchard et al., 2000) was used to assess population structure and estimate the number of genetic clusters present in the NW GOM. Ten replicate simulations were run with values of hypothesized genetic clusters, or K, ranging between 1 and 9 using the following parameters: 103 burn-in iterations, 106 Markov Chain-Monte Carlo replicates, LOCPRIOR, assuming correlated allele frequencies and admixed populations. Additional simulations were run beyond the number of sampled populations (K = 7–9) to determine whether subpopulations were present within banks. Delta log likelihood values were compared across all model simulations to estimate the most likely value of K according to the Evanno method with Structure Harvester (Evanno et al., 2005; Earl and VonHoldt, 2012).
Migration rates were estimated among the banks across historical time scales (ancestral populations, ~4Ne generations). Migration criteria were developed to specifically address horizontal migration patterns across banks that may influence management strategies. Shallow and mesophotic habitats within West and East FGB were combined to form a total of four populations in the NW GOM (WFGB, EFGB, BRT, MCG) due to software limitations and reduced inferential power with complex models. To assess historical gene flow, criteria were designed a priori and tested with Migrate 3.6 (Beerli, 2006; Beerli and Palczewski, 2010). Migrate uses coalescence theory to estimate effective population sizes and gene flow among populations relative to mutation rate. While generation times and microsatellite mutation rates are unknown for coral species, it was assumed since M. cavernosa is long-lived and repeatedly reproductively viable after a small size threshold that Migrate estimations represent migration patterns over hundreds to thousands of years prior (Szmant, 1991; Soong, 1993). Criteria developed to estimate horizontal connectivity in the NW GOM simulated four potential migration dynamics based on dominant currents in the region: (1) Symm: full model with symmetric migration across all banks, (2) Eastward: asymmetric migration from westward to eastward banks, (3) Westward: asymmetric migration from eastward to westward banks, and (4) Pan: panmixia. The final parameters used across all simulations were long-inc 100, long-sample 15000, 20 replicates, burn-in 20000, and four heated chains of 1, 1.5, 3, 105, and the prior distributions for theta and migration rate were set at 0–100 and 0–1000, respectively. Bezier log marginal likelihoods for each model were compared and ranked according to the thermodynamic integration method in Beerli and Palczewski (2010).
Fifteen samples were removed due to amplification failure across more than three loci and eight clonal MLGs were removed, resulting in 229 out of the 252 original coral samples used for downstream analyses. Final sample sizes for each population are shown as ng in Table 1. Per locus missing rates were MC4 3.98%, MC18 2.62%, MC29 0.86%, MC41 8.30%, MC46 0%, MC49 3.98%, MC65 2.62%, MC97 3.98%, and MC114 0.86%. Assumption testing for Hardy-Weinberg Equilibrium (HWE), linkage disequilibrium (LD), and the presence of null alleles indicated no apparent pattern across populations in the NW GOM (Table 2).
The analysis of molecular variance indicated low differentiation across populations (AMOVA; 0.4%, df = 5,229, SS = 21.24, p < 0.009), while the majority of significant genotypic variation was at the individual level within populations (AMOVA; 6.8%, df = 223,229, SS = 755.86). Pairwise FST values in the NW GOM revealed a definitive lack of genetic differentiation across the region, including between shallow and mesophotic depth zones within banks (Figure 2). Only a single pair of banks were significantly differentiated (EFGB-meso vs. BRT-meso), indicating that the NW GOM is extremely well-mixed. Similarly, principal coordinates analysis mirrored a lack of clustering among banks, with the largest distance between the EFGB-meso and BRT-meso populations (Figure 3). The Mantel test indicated no significant correlation between geographic and genetic distance (p = 0.51, R2 = 0.0007), indicating that physical distance between banks did not contribute any genetic variation among populations. Pairwise geographic distances between banks for the isolation by distance analysis ranged from 0.08 to 120.96 km.
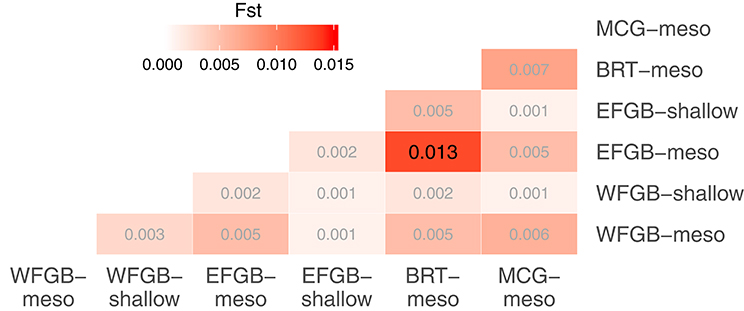
Figure 2. Pairwise comparisons of population differentiation as measured by fixation index (FST). The level of differentiation among populations is shown in increasing intensities of red. Significantly differentiated FST estimates are shown in larger black font (p < 0.05 after FDR correction).
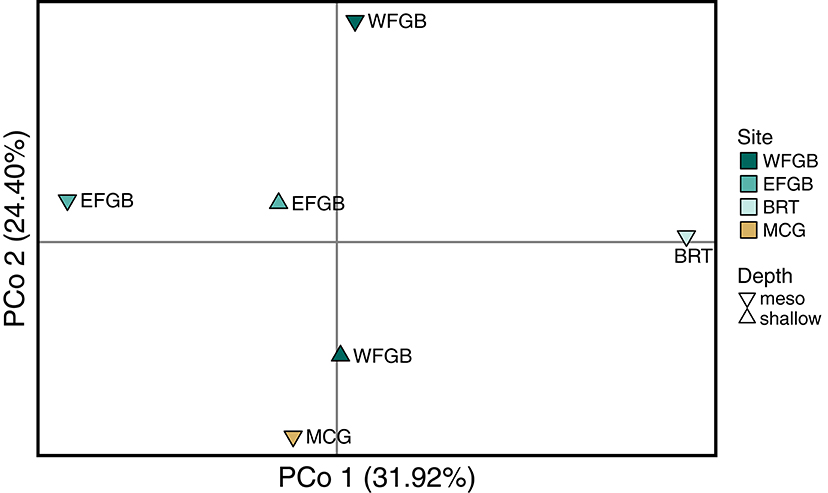
Figure 3. Principal coordinate analysis of Nei genetic distance (DA) matrix generated from pairwise population differentiation comparisons. Samples from each population were first compared to generate a single eigenvector, represented by a single point in three-dimensional space, and the relative distance between populations approximates the degree of genetic differentiation. The three axes have been flattened to a two-dimensional representation and explain 73.48% of the total genotypic variation (Coordinate 1 31.92%, Coordinate 2 24.40%, Coordinate 3 17.16%). Color of each point corresponds to discrete banks.
Structure analysis suggested two genetic clusters (K = 2) using the Evanno method for the six sampled populations in the NW GOM (Figure 4). However, the model likelihood was higher when K = 1 than K = 2, suggesting a lack of significant genetic structure within the NW GOM. Given the limitations of the Evanno method for populations with weak genetic structure (see section Discussion), the model with the highest likelihood was chosen as the most likely (K = 1).
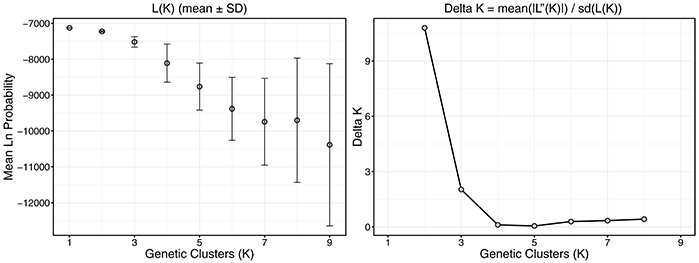
Figure 4. Plots describing the process behind population cluster (K) selection in structure analysis. Ten replicate structure models were run across a range of K values from 1 to 9 and model log likelihoods were compared. The Evanno method was used to determine the most likely number of K by identifying the largest change in likelihood (L(K)) and by comparing model probabilities in conjunction with variance (Delta K). Error bars represent standard deviation of the mean.
Historical migration rates among banks indicated population panmixia over symmetrical or directional migration across genetically distinct populations (Table 3). The second most likely scenario was net eastward to westward migration, although model probabilities for all other migration patterns were near zero compared to panmixia.
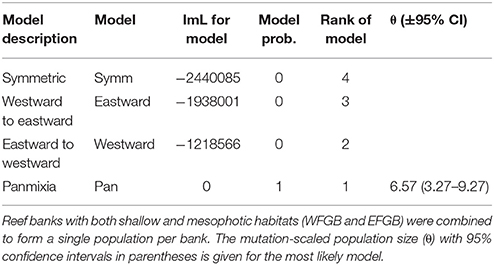
Table 3. Comparison of Bezier log marginal likelihood (lmL) differences, model probabilities, and rank for four different gene flow models (Symm, symmetric horizontal migration; Eastward, migration from westward to eastward banks; Westward, migration from eastward to westward banks; Pan, panmixia) within the NW GOM.
Patterns of genetic variation across banks included in this study support that M. cavernosa are acting as a single population in the NW GOM. The lack of significant genetic differentiation across reef habitats up to 120 km apart indicates gene flow among depth zones and banks has been sustained at a relatively high level through time. Genetic connectivity within the region is likely maintained through strong and persistent currents in the Gulf of Mexico, dominated by the Loop Current that flows from the Yucatan Peninsula into the Straits of Florida through the NW GOM (Lugo-Fernández, 1998; Oey et al., 2005). The estimates of gene flow and population structure for M. cavernosa in the GOM are likely higher than for other coral species (Holstein et al., 2015; Serrano et al., 2016; Bongaerts et al., 2017; Rippe et al., 2017), given the abundance of this species across shallow and mesophotic reefs in the region (Bak et al., 2005; Lesser et al., 2009; Nunes et al., 2009) and its high dispersal potential due to its broadcast spawning reproduction (Nunes et al., 2011; Goodbody-Gringley et al., 2012; Brazeau et al., 2013).
The results of the population structure analysis match the conclusions of the FST analysis that there is no genetic structure (K = 1) and that the NW GOM represents a single uniform M. cavernosa population. Despite the Evanno method identifying two genetic clusters, the model likelihood when K = 1 was indeed higher than when K = 2. This discrepancy is due to the manner in which the Evanno method calculates the stepwise change in model likelihoods (Delta K) between simulated values of K (Evanno et al., 2005). Therefore, this method cannot calculate Delta K when K = 1. This operationally precludes the selection of K = 1 as the most likely scenario despite the high model likelihood and represents a shortcoming of this statistical approach in systems with little population structure (Waples and Gaggiotti, 2006). Analyses using both population genetics (this study) and biophysical modeling approaches (Garavelli et al., 2018) predicted M. cavernosa in the NW GOM to come from the same population and identified connectivity across banks (via gene flow and larval dispersal, respectively).
As a result of the open gene flow within the NW GOM region, historical migration estimates across reefs were overwhelmingly in support of a panmictic population. The second most likely migration scenario, albeit with an extremely low model probability, was net migration westward along the continental shelf margin with more eastward populations contributing to the westward neighbors. If panmixia was excluded as a possible outcome and model probabilities were recalculated, however, westward migration became the most likely scenario with a probability of 1. Taken together with what is known about hydrodynamic patterns in the region, these results provide limited evidence that in the absence of panmixia, migration may be occurring in a counter-current direction to the prominent Loop Current that travels from a southwestern to northeastern direction in the NW GOM (Lugo-Fernández, 1998; Oey et al., 2005). This suggests that counter-current features in the region may be the dominant drivers of coral larval dispersal through ecological timescales rather than the Loop Current (Schmahl et al., 2008). Westward migration may be facilitated by Loop Current Eddies or coastal currents including the Louisiana Coastal Current (LCC), which transports Mississippi River outflow westward due to the Coriolis Effect (Jarosz and Murray, 2005; Oey et al., 2005; Ohlmann and Niiler, 2005).
The comparison of M. cavernosa migration patterns estimated from genetic data in this study to larval dispersal using a biophysical model in the associated study demonstrates that annual hydrodynamic variability can greatly affect connectivity predictions (Garavelli et al., 2018). The fact that the biophysical model predicted net eastward export of larvae from all banks, while the genetic model predicted panmixia (or net westward migration to a lesser extent), suggests that the inconsistent and ephemeral nature of currents in the NW GOM may be resulting in widespread dispersal across the region (Oey et al., 2005; Schmahl et al., 2008). For the 3 years of oceanographic data available for the biophysical model, eddies were not consistently present across the study region in the latter 2 years and would therefore not be significant drivers of larval migration. In the latter years, the Loop Current was likely the most influential factor affecting larval transportation, resulting in the export of larvae eastward outside the study area (Garavelli et al., 2018). Our observed panmictic migration pattern hypothesizes that multiple larval dispersal patterns may be effective to enable gene flow over time among all banks in the study.
The LCC experiences seasonal variability, and in rarer cases, eastward transport following impingement on the continental shelf (Jarosz and Murray, 2005; Gough et al., 2017). The LCC may reverse direction during summer months (June–August) and flow eastward at a weaker speed. Coral spawning has been observed at FGBNMS to occur 7–8 days after the full moon in late August (Vize, 2006), which may result in spawning in August or September when the LCC could potentially be moving either westward or eastward. The potential variability in LCC direction across spawning events may result in different larval dispersal patterns across years. In the case of the biophysical model by Garavelli et al. (2018), it appears the oceanographic data from 2013–2015 captured one such rare impingement events, causing dispersal predictions of net eastward movement of coral larvae. The variability of oceanographic patterns over short time scales and its potential effect on migration predictions from year to year emphasizes the periodic nature of successful coral recruitment events and suggests that eddies and coastal currents play an important, if not dominant, role in determining the destination of coral larvae (Tang et al., 2006; Chérubin and Garavelli, 2016; Vaz et al., 2016; Gough et al., 2017; Pan et al., 2017). Naturally, this does not preclude the potential for occasional export of larvae beyond the NW GOM and successful gene flow to other reefs hundreds of kilometers away, as has been suggested by biophysical and genetic data (Davies et al., 2017; Rippe et al., 2017; Garavelli et al., 2018). Stepping-stone dynamics among other reef habitats in the NW GOM including scleractinian communities on oil and gas platforms (Sammarco et al., 2004, 2012, 2013, 2014) and mesophotic banks in the eastern GOM (Hine et al., 2008; Locker et al., 2010; Silva and MacDonald, 2017) may also influence coral population connectivity in the region.
The population genetics data presented in this study supports the combined management of mesophotic reef habitats across the NW GOM since M. cavernosa across four banks act as a single population and would likely benefit from integrated protection of all population source habitats within the region. Comparison of genetic and biophysical models demonstrated that multiple analysis methods can be integrated to more accurately represent relationships between oceanographic conditions across individual spawning events and multi-generational gene flow. This is particularly important in regions where management actions can be influenced by the model outcomes (Galindo et al., 2006; Selkoe et al., 2008; Baltazar-Soares et al., 2018). It has previously been suggested with several coral species that FGBNMS is a population refugia in the GOM (Atchison et al., 2008; Goodbody-Gringley et al., 2012; Davies et al., 2017; Rippe et al., 2017). We propose that additional banks with established coral communities including Bright and McGrail Banks likely represent additional refugia habitat important for population persistence within the NW GOM, and possibly for other reefs in the wider GOM. High gene flow among reefs, with evidence for occasional export of migrants to nearby reefs indicates that these habitats are self-sufficient but may still serve as population sources over ecological timescales (Davies et al., 2017; Garavelli et al., 2018). If these trends are similar across other taxa as they have been with corals, the NW GOM may provide important ecosystem services despite its relative spatial isolation.
While less diverse than other areas of the TWA in terms of coral communities, West and East FGB support biodiverse assemblages of reef organisms and fishes that have been relatively stable through time (Schmahl et al., 2008; Hickerson et al., 2012; Clark et al., 2014; Johnston et al., 2016). Mesophotic habitats outside the current sanctuary boundaries are less biodiverse from a scleractinian standpoint and likely represent smaller populations as compared to FGB (Hickerson et al., 2008; Schmahl et al., 2008; Sammarco et al., 2016). Nonetheless, the results presented here demonstrate that Bright and McGrail Banks may serve as important population sources for the rest of the NW GOM region as well and should not be excluded from larval dispersal models and management strategies. The development and implementation of conservation applications, including marine reserves, are frequently designed to protect known population sources (Palumbi, 2001, 2003; Fogarty and Botsford, 2007). Therefore, management actions are warranted to protect the coral populations at Bright and McGrail Banks identified in this study.
All protocols, including sample preparation, data analysis, and data templates are available in a GitHub repository (https://github.com/mstudiva/Mcav-microsats.git). The datasets generated for this study can be found in a Dryad repository (Studivan and Voss, 2018).
JV: designed and funded this research through the Cooperative Institute for Ocean Exploration, Research and Technology (CIOERT); MS and JV: conducted sample collections; MS: generated and analyzed the microsatellite data. All authors contributed to the final edited version of the manuscript.
This research was funded by the NOAA Office of Ocean Exploration and Research under awards NA09OAR4320073 and NA14OAR4320260 to the Cooperative Institute for Ocean Exploration, Research and Technology (CIOERT) at Florida Atlantic University-Harbor Branch Oceanographic Institute. Additional funding was provided by a donation from the Banbury Fund to Harbor Branch Oceanographic Institute and by graduate student fellowships and grants from Florida Atlantic University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the staff of Flower Garden Banks National Marine Sanctuary, crews of the R/V Manta, L. Horn, and J. White from the University of North Carolina at Wilmington Undersea Vehicle Program, J. Beal, J. Emmert, R. Susen, C. Ledford, J. Polinski, D. Dodge, P. Gardner, M. McCallister, M. Ajemian, R. Christian, and M. Dickson for diving support. We also thank G. O'Corry-Crowe and T. Ferrer for capillary sequencing, and X. Serrano and A. Baker for assistance with pipeline optimization. This paper is Contribution Number 2147 from Harbor Branch Oceanographic Institute at Florida Atlantic University.
Aronson, R. B., Precht, W. F., Murdoch, T. J. T., and Robbart, M. L. (2005). Long-term persistence of coral assemblages on the Flower Garden Banks, northwestern Gulf of Mexico: implications for science and management. Gulf Mex. Sci. 23, 84–94.
Atchison, A. D., Sammarco, P. W., and Brazeau, D. A. (2008). Genetic connectivity in corals on the Flower Garden Banks and surrounding oil/gas platforms, Gulf of Mexico. J. Exp. Mar. Biol. Ecol. 365, 1–12. doi: 10.1016/j.jembe.2008.07.002
Bak, R. P. M., Nieuwland, G., and Meesters, E. H. (2005). Coral reef crisis in deep and shallow reefs: 30 years of constancy and change in reefs of Curacao and Bonaire. Coral Reefs 24, 475–479. doi: 10.1007/s00338-005-0009-1
Baltazar-Soares, M., Hinrichsen, H.-H., and Eizaguirre, C. (2018). Integrating population genomics and biophysical models towards evolutionary-based fisheries management. ICES J. Mar. Sci. doi: 10.1093/icesjms/fsx244. [Epub ahead of print].
Beerli, P. (2006). Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22, 341–345. doi: 10.1093/bioinformatics/bti803
Beerli, P., and Palczewski, M. (2010). Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185, 313–326. doi: 10.1534/genetics.109.112532
Bongaerts, P., Riginos, C., Brunner, R., Englebert, N., Smith, S. R., and Hoegh-Guldberg, O. (2017). Deep reefs are not universal refuges: reseeding potential varies among coral species. Sci. Adv. 3:e1602373. doi: 10.1126/sciadv.1602373
Brazeau, D. A., Lesser, M. P., and Slattery, M. (2013). Genetic structure in the coral, Montastraea cavernosa: assessing genetic differentiation among and within mesophotic reefs. PLoS ONE 8:e65845. doi: 10.1371/journal.pone.0065845
Carr, M. H., Neigel, J. E., Estes, J. A., Andelman, S., Warner, R. R., and Largier, J. L. (2003). Comparing marine and terrestrial ecosystems: implications for the design of coastal marine reserves. Ecol. Appl. 13, S90–S107. doi: 10.1890/1051-0761(2003)013[0090:CMATEI]2.0.CO;2
Chapuis, M. P., and Estoup, A. (2007). Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 24, 621–631. doi: 10.1093/molbev/msl191
Chérubin, L. M., and Garavelli, L. (2016). Eastern Caribbean circulation and island mass effect on St. Croix, US Virgin Islands: a mechanism for relatively consistent recruitment patterns. PLoS ONE 11:e0150409. doi: 10.1371/journal.pone.0150409
Chomczynski, P., and Sacchi, N. (2006). The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat. Protoc. 1, 581–585. doi: 10.1038/nprot.2006.83
Clark, R., Taylor, J. C., Buckel, C. A., and Kracker, L. M. (2014). “Fish and benthic communities of the Flower Garden Banks national marine sanctuary: science to support sanctuary management,” in NOAA Technical Memorandum NOS NCCOS 179 (Silver Spring, MD), 317.
Cowen, R. K., Paris, C. B., and Srinivasan, A. (2006). Scaling of connectivity in marine populations. Science 311, 522–527. doi: 10.1126/science.1122039
Cowen, R. K., and Sponaugle, S. (2009). Larval dispersal and marine population connectivity. Annu. Rev. Mar. Sci. 1, 443–466. doi: 10.1146/annurev.marine.010908.163757
Davies, S. W., Strader, M. E., Kool, J. T., Kenkel, C. D., and Matz, M. V. (2017). Modeled differences of coral life-history traits influence the refugium potential of a remote Caribbean reef. Coral Reefs 36, 913–925. doi: 10.1007/s00338-017-1583-8
Earl, D. A., and VonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fogarty, M. J., and Botsford, L. W. (2007). Population connectivity and spatial management of marine fisheries. Oceanography 20, 112–123. doi: 10.5670/oceanog.2007.34
Galindo, H. M., Olson, D. B., and Palumbi, S. R. (2006). Seascape genetics: a coupled oceanographic-genetic model predicts population structure of Caribbean corals. Curr. Biol. 16, 1622–1626. doi: 10.1016/j.cub.2006.06.052
Garavelli, L., Studivan, M. S., Voss, J. D., Kuba, A., Figueiredo, J., and Chérubin, L. M. (2018). Assessment of mesophotic coral ecosystem connectivity for proposed expansion of a marine sanctuary in the northwest Gulf of Mexico: larval dynamics. Front. Mar. Sci. 5:174. doi: 10.3389/fmars.2018.00174
Gardner, T. A., Côté, I. M., Gill, J. A., Grant, A., and Watkinson, A. R. (2003). Long-term region-wide declines in Caribbean corals. Science 301, 958–960. doi: 10.1126/science.1086050
Goodbody-Gringley, G., Woollacott, R. M., and Giribet, G. (2012). Population structure and connectivity in the Atlantic scleractinian coral Montastraea cavernosa (Linnaeus, 1767). Mar. Ecol. 33, 32–48. doi: 10.1111/j.1439-0485.2011.00452.x
Gough, M. K., Beron-Vera, F. J., Olascoaga, M. J., Sheinbaum, J., Juoanno, J., and Duran, R. (2017). Persistent Lagrangian transport patterns in the northwestern Gulf of Mexico. arXiv preprint arXiv:1710.04027
Hale, M. L., Burg, T. M., and Steeves, T. E. (2012). Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE 7:e45170. doi: 10.1371/journal.pone.0045170
Hickerson, E., Schmahl, G., Robbart, M., Precht, W., and Caldow, C. (2008). “The state of coral reef ecosystems of the Flower Garden Banks, Stetson Bank, and other banks in the northwestern Gulf of Mexico,” in The State of Coral Reef Ecosystems of the United States and Pacific Freely Associated States: 2008 (Silver Spring, MD: NOAA Technical Memorandum NOS NCCOS), 189–217.
Hickerson, E. L., Schmahl, G. P., Johnston, M. A., Nuttall, M. F., Embesi, J. A., and Eckert, R. J. (2012). “Flower Garden Banks – a refuge in the Gulf of Mexico?,” in Proceedings of the 12th International Coral Reef Symposium (Cairns, QLD), 9–13.
Hilbish, T. J. (1996). Population genetics of marine species: the interaction of natural selection and historically differentiated populations. J. Exp. Mar. Biol. Ecol. 200, 67–83. doi: 10.1016/S0022-0981(96)02645-7
Hine, A. C., Halley, R. B., Locker, S. D., Jarrett, B. D., Jaap, W. C., Mallinson, D. J., et al. (2008). “Coral reefs, present and past, on the West Florida Shelf and platform margin,” in Coral Reefs of the USA, eds B. M. Riegl and R. E. Dodge (Dordrecht: Springer), 127–173.
Holstein, D. M., Paris, C. B., Vaz, A. C., and Smith, T. B. (2015). Modeling vertical coral connectivity and mesophotic refugia. Coral Reefs 35, 23–37. doi: 10.1007/s00338-015-1339-2
Jarosz, E., and Murray, S. P. (2005). “Velocity and transport characteristics of the Louisiana-Texas coastal current,” in Circulation in the Gulf of Mexico: Observations and Models, eds W. Sturges and A. Lugo-Fernández [Washington, DC: American Geophysical Union (AGU)], 143–156.
Johnston, M. A., Embesi, J. A., Eckert, R. J., Nuttall, M. F., Hickerson, E. L., and Schmahl, G. P. (2016). Persistence of coral assemblages at East and West Flower Garden Banks, Gulf of Mexico. Coral Reefs 35, 821–826. doi: 10.1007/s00338-016-1452-x
Jones, G. P., Almany, G. R., Russ, G. R., Sale, P. F., Steneck, R. S., van Oppen, M. J. H., et al. (2009). Larval retention and connectivity among populations of corals and reef fishes: history, advances and challenges. Coral Reefs 28, 307–325. doi: 10.1007/s00338-009-0469-9
Kahng, S. E., Copus, J. M., and Wagner, D. (2014). Recent advances in the ecology of mesophotic coral ecosystems (MCEs). Curr. Opin. Environ. Sustainability 7, 72–81. doi: 10.1016/j.cosust.2013.11.019
Lesser, M. P., Slattery, M., and Leichter, J. J. (2009). Ecology of mesophotic coral reefs. J. Exp. Mar. Biol. Ecol. 375, 1–8. doi: 10.1016/j.jembe.2009.05.009
Locker, S. D., Armstrong, R. A., Battista, T. A., Rooney, J. J., Sherman, C., and Zawada, D. G. (2010). Geomorphology of mesophotic coral ecosystems: current perspectives on morphology, distribution, and mapping strategies. Coral Reefs 29, 329–345. doi: 10.1007/s00338-010-0613-6
Lugo-Fernández, A. (1998). Ecological implications of hydrography and circulation to the Flower Garden Banks, northwest Gulf of Mexico. Gulf Mex. Sci. 16, 144–160.
Manel, S., Schwartz, M. K., Luikart, G., and Taberlet, P. (2003). Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18, 189–197. doi: 10.1016/S0169-5347(03)00008-9
Nunes, F. L., Norris, R. D., and Knowlton, N. (2011). Long distance dispersal and connectivity in amphi-Atlantic corals at regional and basin scales. PLoS ONE 6:e22298. doi: 10.1371/journal.pone.0022298
Nunes, F., Norris, R. D., and Knowton, N. (2009). Implications of isolation and low genetic diversity in peripheral populations of an amphi-Atlantic coral. Mol. Ecol. 18, 4283–4297. doi: 10.1111/j.1365-294X.2009.04347.x
Oey, L., Ezer, T., and Lee, H. (2005). “Loop Current, rings and related circulation in the Gulf of Mexico: a review of numerical models and future challenges,” in Circulation in the Gulf of Mexico: Observations and Models, eds W. Sturges and A. Lugo-Fernandez (Washington, DC: American Geophysical Union), 31–56.
Office of National Marine Sanctuaries (2016). Flower Garden Banks National Marine Sanctuary Expansions Draft Environmental Impact Statement. Silver Spring, MD: U.S. Department of Commerce, National Oceanic and Atmospheric Administration, Office of National Marine Sactuaries.
Ohlmann, J. C., and Niiler, P. P. (2005). Circulation over the continental shelf in the northern Gulf of Mexico. Prog. Oceanogr. 64, 45–81. doi: 10.1016/j.pocean.2005.02.001
Palumbi, S. R. (2001). “The ecology of marine protected areas,” in Marine Community Ecology, eds M. Bertness, S. Gaines, and M. Hay (Sunderland: Sinauer Press), 509–530.
Palumbi, S. R. (2003). Population genetics, demography connectivity, and the design of marine reserves. Ecol. Appl. 13, S146–S158. doi: 10.1890/1051-0761(2003)013[0146:PGDCAT]2.0.CO;2
Pan, C., Jiang, M., Dalgleish, F. R., and Reed, J. K. (2017). Modeling the impacts of the Loop Current on circulation and water properties over the Pulley Ridge region on the Southwest Florida shelf. Ocean Model. 112, 48–64. doi: 10.1016/j.ocemod.2017.02.009
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Precht, W. F., Aronson, R. B., Deslarzes, K. J., Robbart, M. L., Zimmer, B., and Duncan, L. (2008). Post-Hurricane Assessment at the East Flower Garden Bank Long-Term Monitoring Site at East Bank: November 2005. OCS Study MMS 2008-019. New Orleans, LA: US Dept Interior, Minerals Management Service, Gulf of Mexico OCS Region.
Precht, W. F., Deslarzes, K. J. P., Hickerson, E. L., Schmahl, G. P., Nuttall, M. F., and Aronson, R. B. (2014). Back to the future: the history of acroporid corals at the Flower Garden Banks, Gulf of Mexico, USA. Mar. Geol. 349, 152–161. doi: 10.1016/j.margeo.2013.12.012
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Rezak, R., and Bright, T. J. (1985). Reefs and Banks of the Northwestern Gulf of Mexico: Their Geological, Biological, and Physical Dynamics. College Station, TX: Texas A&M University.
Rezak, R., Gittings, S. R., and Bright, T. J. (1990). Biotic assemblages and ecological controls on reefs and banks of the northwest Gulf of Mexico. Am. Zool. 30, 23–35. doi: 10.1093/icb/30.1.23
Rippe, J. P., Matz, M. V., Green, E. A., Medina, M., Khawaja, N. Z., Pongwarin, T., et al. (2017). Population structure and connectivity of the mountainous star coral, Orbicella faveolata, throughout the wider Caribbean region. Ecol. Evol. 7, 9234–9246. doi: 10.1002/ece3.3448
Sammarco, P. W., Atchison, A. D., and Boland, G. S. (2004). Expansion of coral communities within the Northern Gulf of Mexico via offshore oil and gas platforms. Mar. Ecol. Prog. Ser. 280, 129–143. doi: 10.3354/meps280129
Sammarco, P. W., Brazeau, D. A., and Sinclair, J. (2012). Genetic connectivity in scleractinian corals across the northern Gulf of Mexico: oil/gas platforms, and relationship to the Flower Garden Banks. PLoS ONE 7:e30144. doi: 10.1371/journal.pone.0030144
Sammarco, P. W., Lirette, A., Tung, Y. F., Boland, G. S., Genazzio, M., and Sinclair, J. (2013). Coral communities on artificial reefs in the Gulf of Mexico: standing vs. toppled oil platforms. ICES J. Mar. Sci. 71, 417–426. doi: 10.1093/icesjms/fst140
Sammarco, P. W., Nuttall, M. F., Beltz, D., Hickerson, E. L., and Schmahl, G. P. (2016). Patterns of mesophotic benthic community structure on banks at vs. inside the continental shelf edge, Gulf of Mexico. Gulf Mex. Sci. 1, 77–92.
Sammarco, P. W., Porter, S. A., Sinclair, J., and Genazzio, M. (2014). Population expansion of a new invasive coral species, Tubastraea micranthus, in the northern Gulf of Mexico. Mar. Ecol. Prog. Ser. 495, 161–173. doi: 10.3354/meps10576
Schmahl, G. P., Hickerson, E. L., and Precht, W. F. (2008). “Biology and ecology of coral reefs and coral communities in the Flower Garden Banks region, northwestern Gulf of Mexico,” in Coral Reefs of the USA, eds B. M. Riegl and R. E. Dodge (Dordrecht: Springer), 221–261.
Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18, 233–234. doi: 10.1038/72708
Selkoe, K. A., Henzler, C. M., and Gaines, S. D. (2008). Seascape genetics and the spatial ecology of marine populations. Fish Fish. 9, 363–377. doi: 10.1111/j.1467-2979.2008.00300.x
Serrano, X., Baums, I. B., O'Reilly, K., Smith, T. B., Jones, R. J., Shearer, T. L., et al. (2014). Geographic differences in vertical connectivity in the Caribbean coral Montastraea cavernosa despite high levels of horizontal connectivity at shallow depths. Mol. Ecol. 23, 4226–4240. doi: 10.1111/mec.12861
Serrano, X. M., Baums, I. B., Smith, T. B., Jones, R. J., Shearer, T. L., and Baker, A. C. (2016). Long-distance dispersal and vertical gene flow in the Caribbean brooding coral Porites astreoides. Sci. Rep. 6:21619. doi: 10.1038/srep21619
Silva, M., and MacDonald, I. R. (2017). Habitat suitability modeling for mesophotic coral in the northeastern Gulf of Mexico. Mar. Ecol. Prog. Ser. 583, 121–136. doi: 10.3354/meps12336
Soong, K. (1993). Colony size as a species character in massive reef corals. Coral Reefs 12, 77–83. doi: 10.1007/BF00302106
Strimmer, K. (2008). fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461–1462. doi: 10.1093/bioinformatics/btn209
Studivan, M. S., and Voss, J. D. (2018). Data from: Assessment of mesophotic coral ecosystem connectivity for proposed expansion of a marine sanctuary in the northwest Gulf of Mexico: population genetics. Dryad Dig. Repos. doi: 10.5061/dryad.3kh117d
Szmant, A. M. (1991). Sexual reproduction by the Caribbean reef corals Monastrea annularis and M. cavernosa. Mar. Ecol. Prog. Ser. 74, 13–25. doi: 10.3354/meps074013
Tang, L., Sheng, J., Hatcher, B. G., and Sale, P. F. (2006). Numerical study of circulation, dispersion, and hydrodynamic connectivity of surface waters on the Belize shelf. J. Geophys. Res. 111:C01003. doi: 10.1029/2005JC002930
van Oosterhout, C., Weetman, D., and Hutchinson, W. F. (2006). Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol. Ecol. Notes 6, 255–256. doi: 10.1111/j.1471-8286.2005.01082.x
Vaz, A. C., Paris, C. B., Olascoaga, M. J., Kourafalou, V. H., Kang, H., and Reed, J. K. (2016). The perfect storm: match-mismatch of bio-physical events drives larval reef fish connectivity between Pulley Ridge mesophotic reef and the Florida Keys. Cont. Shelf Res. 125, 136–146. doi: 10.1016/j.csr.2016.06.012
Vize, P. D. (2006). Deepwater broadcast spawning by Montastraea cavernosa, Montastraea franksi, and Diploria strigosa at the Flower Garden Banks, Gulf of Mexico. Coral Reefs 25, 169–171. doi: 10.1007/s00338-005-0082-5
Voss, J. D., Williams, M. A., Reed, J. K., and Clark, R. (2014). “Benthic and fish communities in the mid and lower mesphotic zone of the sanctuary,” in Fish and Benthic Communities of the Flower Garden Banks National Marine Sanctuary: Science to Support Sanctuary Management, eds R. Clark, J. C. Taylor, C. A. Buckel, and L. M. Kracker (Silver Spring, MD: NOAA Technical Memorandum NOS NCCOS 179), 201–260.
Waples, R. S., and Gaggiotti, O. (2006). What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 15, 1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x
Keywords: population genetics, mesophotic coral ecosystems, northwest Gulf of Mexico, Flower Garden Banks National Marine Sanctuary, Montastraea cavernosa, microsatellites, sanctuary expansion, marine spatial planning
Citation: Studivan MS and Voss JD (2018) Assessment of Mesophotic Coral Ecosystem Connectivity for Proposed Expansion of a Marine Sanctuary in the Northwest Gulf of Mexico: Population Genetics. Front. Mar. Sci. 5:152. doi: 10.3389/fmars.2018.00152
Received: 17 February 2018; Accepted: 13 April 2018;
Published: 17 May 2018.
Edited by:
Michael Sweet, University of Derby, United KingdomReviewed by:
Pablo Saenz-Agudelo, Universidad Austral de Chile, ChileCopyright © 2018 Studivan and Voss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael S. Studivan, c3R1ZGl2YW5tc0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.