- 1Marine Science and Technology, Faculty of Fisheries and Marine Sciences, Bogor Agricultural University (IPB), Bogor, Indonesia
- 2Center for Coastal and Marine Resource Studies, Bogor Agricultural University (IPB), Bogor, Indonesia
- 3Biotechnology and Molecular Genetics, University of Bremen, Bremen, Germany
- 4Marine Biology, Ecology and Biodiversity, Vrije Universiteit Brussel (VUB), Brussels, Belgium
Populations of the clown anemonefish in Spermonde Archipelago, one of the main collection sites for ornamental fish in Indonesia, are potentially overfished, which might lead to a reduction in population size and genetic diversity. Loss of genetic diversity can reduce the adaptability, population persistence and productivity of the targeted species. Therefore, a study investigating the genetic diversity and its potential correlation to population densities of A. ocellaris was conducted. Two islands were chosen as study sites that differed in the degree of exploitation. Barrang Lompo has a high fishing pressure, whereas Samalona has less. Underwater visual censuses showed that population densities in Samalona were threefold higher than in Barrang Lompo (p = 0.005). Analysis of eight microsatellite loci in 364 tissue samples of A. ocellaris revealed that genetic diversity (numbers of alleles, private alleles, and allelic richness) was significantly reduced at the island with high fishing pressure. Allelic richness was also positively correlated with fish density (p < 0.05). These results indicate that ornamental fishery might be a factor contributing to the loss of genetic diversity in A. ocellaris. Therefore, the marine ornamental trade in Spermonde Archipelago needs to be managed (e.g., management of the fishing strategy, implementation of marine protected areas, regular monitoring, and quota determination). Otherwise the populations of A. ocellaris might collapse.
Introduction
Many marine fish populations are currently impacted due to fishing activities and environmental changes. The most obvious impacts of excessive fishing are observable changes in size and age structure in the fish populations (e.g., Trippel, 1995; Cardinale and Modin, 1999; Ottersen et al., 2006), and a reduction in population density (Gaggiotti and Vetter, 1999). Due to exploitation, many stocks are depleted and some species are even endangered (e.g., Musick et al., 2000; Hutchings and Reynolds, 2004; Dankel et al., 2008). Excessive fishing and environmental changes are not only affecting the spatial distribution and structure of populations, but also causing changes in their genetic diversity (e.g., O'Brien, 1994; Heino and Godø, 2002; Reusch et al., 2005; Pinsky and Palumbi, 2014). Reduced population size enhances genetic drift, which in turn causes a higher loss of genetic variability per generation. Increased genetic drift augments the loss of variability and the ability of adaptation (Hauser et al., 2002; Spielman et al., 2004). A number of studies revealed a loss of genetic diversity in marine fish due to exploitation, for example, Hoplostethus atlanticus (Smith et al., 1991) and Pagrus auratus in New Zealand (Hauser et al., 2002).
More than 1,000 marine ornamental fish species were traded globally in the period from 1997 to 2002, of which damselfish (Pomacentridae) comprised about 47% (Wabnitz et al., 2003). The clown anemonefish Amphiprion ocellaris is the number one fish species in ornamental trade and Indonesia is one of the world's leading exporters (Wabnitz et al., 2003). In Spermonde Archipelago, The fishermen are unselectively exploiting A. ocellaris and their hosts, without considering the sizes (Madduppa et al., 2014a). Intensive fishing on A. ocellaris caused a decrease in density and reduction in body size (Madduppa et al., 2014a), indicating overfishing. This situation may lead to a loss of genetic diversity in A. ocellaris.
Due to its biological and behavioral characteristics, A. ocellaris is vulnerable to intensive fishing. The species forms an obligate symbiosis with only three different host anemones (Heteractis magnifica, Stichodactyla gigantea, and S. mertensii), which are collected and traded as well in Spermonde Archipelago (Madduppa et al., 2014a), reducing the available habitat. Each anemone is usually inhabited by two to six individuals of A. ocellaris. The two largest individuals in the colony are the breeding pair and others remain non-breeders (Fricke and Fricke, 1977). The fish is characterized by protandrous monogamy (i.e., only one breeding pair lives in a host anemone, and males change sex to become females). The life cycle of A. ocellaris consists of two phases, dispersing larvae (8–10 days in captivity, Gopakumar et al., 2009) and sedentary adults. Amphiprion ocellaris prefers to settle in smaller groups in order to avoid long breeding queues, maximizing their likelihood of attaining social dominance (Mitchell, 2005). All individuals will follow the social-rank rule within one anemone by adjusting their size and growth rate (Buston, 2003), and the social structure only changes when one individual dies or is removed, after which some time is required for the male reproductive anemonefish to change sex to become the reproductive female (Fricke and Fricke, 1977; Madhu et al., 2010). The present study was conducted to investigate the impact of marine ornamental fishing on the genetic diversity of A. ocellaris by using eight microsatellite loci.
Materials and Methods
Study Sites
Spermonde Archipelago consists of about 150 islands and is located in the Makassar Strait southwest of Sulawesi (Tomascik et al., 1997). The region is part of the Coral Triangle (Briggs, 2009) and is considered to be the world's center of marine biodiversity (Allen and Werner, 2002; Allen, 2008; Veron et al., 2009), but is also the most threatened coral reef system in the world (Bryant et al., 1998; Burke et al., 2006). Habitat degradation occurs in the area due to destructive fishing activities (Pet-Soede and Erdmann, 1998). Artisanal fishing and harvesting for the marine ornamental trade adds additional pressure (Erdmann, 1995; Pet-Soede et al., 1999, 2001; Madduppa et al., 2014a).
This study was conducted in Barrang Lompo (5°02′52.07′′S, 119°19′45.25′′E) and Samalona (5°07′30.48′′S, 119°20′36.48′′E). These two islands are located in the mid-shelf of the Spermonde Archipelago, close to the city of Makassar. Samples were collected at five sites in Barrang Lompo and four sites in Samalona (Figure 1, Table 1). About 5,000 people are living on Barrang Lompo and only about 100 on Samalona. Coral reefs in Barrang Lompo have been impacted by high fishing pressure, while Samalona has been less impacted by fishing (Madduppa et al., 2014a).
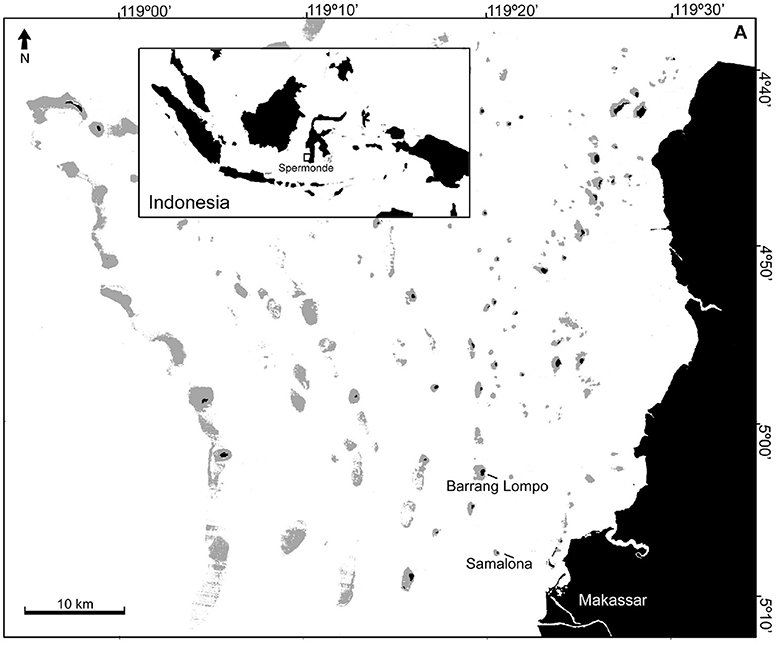
Figure 1. Study sites: Barrang Lompo and Samalona are located in the midshelf of the Spermonde Archipelago.
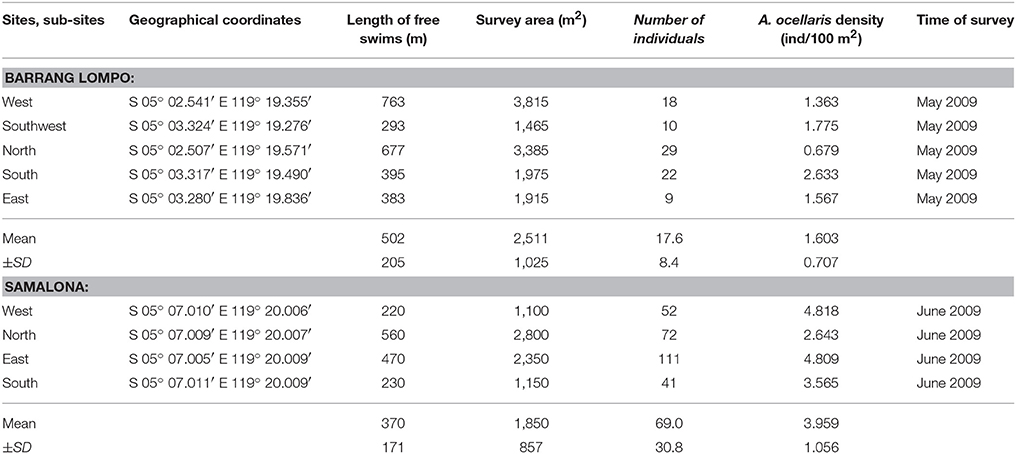
Table 1. Study sites: geographical coordinates, length of free swims (m), survey area (m2), number of individuals, Amphiprion ocellaris density (fish/100 m2), and the survey period.
Fish Survey
Amphiprion ocellaris was counted using the underwater visual census technique during free-swim surveys in an approximately 5 m-wide area for about 1.5–2 h for each survey at a depth of 1–11 m by scuba diving. The surveys were carried out in May and June 2009. GPS positions of the start and endpoints were noted at each survey in order to estimate the length of each transect. This technique has been successfully used to census the population of anemone fish in larger areas in previous studies (Shuman et al., 2005; Jones et al., 2008; Madduppa et al., 2014a).
Ethics Statement
Fin-clipping is a non-destructive, minimally invasive and the most commonly used method to obtain tissue from living fishes in the wild (e.g., Almany et al., 2007; Planes et al., 2009; Saenz-Agudelo et al., 2011) and in aquaculture (e.g., Wang et al., 2012). We took great care to minimize harm, and ensured survival by safely releasing the fishes back into their host anemones. An ethics approval was not required for this research as per our university (Bogor Agricultural University) regulations and as per Indonesian laws and guidelines. Tissue sampling of these anemonefishes was permitted within the framework of the German-Indonesian SPICE project (Science for the Protection of Indonesian Coastal Ecosystems), in cooperation with the Hasanuddin University, Makassar, Indonesia.
Tissue Sampling and Microsatellite Analysis
A total of 364 tissue samples of A. ocellaris were collected in Barrang Lompo and Samalona from October to November 2008 and May to June 2009 (Table 1). All individuals (adults, juveniles, and recruits) in a colony were caught with two aquarium nets and the caudal fin was clipped to obtain a tissue sample. Subsequently they were released into their host anemones. The samples were preserved in 96% ethanol and finally stored at 4°C.
Genomic DNA was extracted with the NucleoSpin Tissue Extraction kit (Macherey-Nagel, Germany), following the manufacturer's guidelines. All DNA extracts were analyzed by gel electrophoresis to monitor DNA quality prior to PCR (polymerase chain reaction) amplification of microsatellite loci. GeneRulerTM 1 kb DNA Ladder Plus (Applied Biosystems) was used as a reference. PCRs were carried out in a total volume of 25 μl, containing 2.5 μl of 10x PCR buffer, 3 μl of 25 mM MgCl2, 1 μl 2 mM of each dNTP, 1 μl of 10 mM each forward and reverse primer, 0.1 μl (5 unit/μl) of Taq polymerase (F100L Taq DNA), and 1 μl (1–10 ng) genomic DNA. PCRs were performed in a Tprofessional Thermocycler (Biometra) and a MasterCycler EP (Eppendorf) with the following thermo-profile: 94°C for 2 min, followed by 35 cycles of 94°C for 30 s as denaturing step, 50–65°C for 30 s as annealing step (see Table 2 for annealing temperature at each locus), 72°C for 1 min for polymerization, and finally 72°C for 2 min. Eight polymorphic microsatellite loci were utilized: two isolated from A. clarkii (AC137 and AC1578, Liu et al., 2007), four from A. percula (Cf39, Cf29, Cf42, and Cf9, Buston et al., 2007), and two from A. polymnus (45 and 120, Quenouille et al., 2004). The PCR products were diluted in pure water prior to fragment analysis. Dilution factors were determined empirically for each locus, and ranged from 1:5 to 1:30. For fragment analysis, 1 μl of diluted PCR product was combined with 8.85 μl HiDiTM formamide and the 0.15 μl GENESCAN LIZ-500 size standard (Applied Biosystems). Microsatellite fragments were size fractioned using an Applied Biosystems Inc. (ABI) 3730 48 capillary sequencer with 50 cm capillary length. Allele sizes were determined and corrected with PEAK SCANNER v1.0 (Applied Biosystems) and GENEMARKER v1.85 (SoftGenetics GeneMarker).
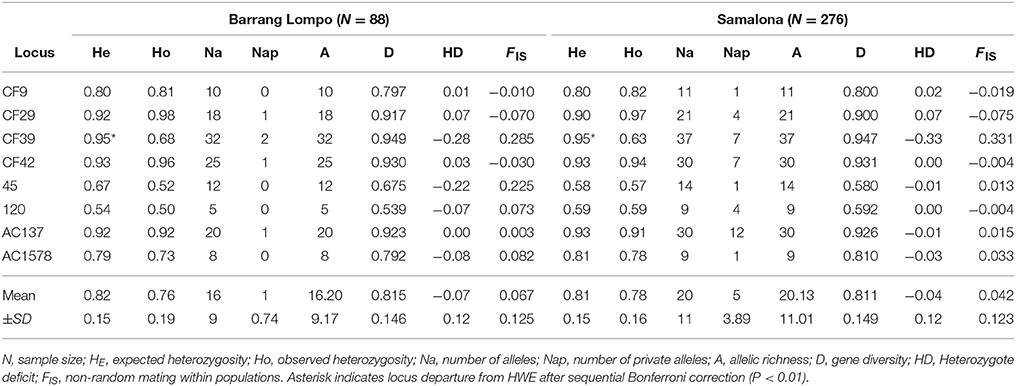
Table 2. Indices for genetic diversity based on eight microsatellite loci for populations from Barrang Lompo and Samalona.
Data Analysis
Hardy-Weinberg equilibrium (HWE) exact tests and loci combinations for linkage disequilibrium with the Markov chain methods were conducted using GENEPOP on the web (Raymond and Rousset, 1995; Rousset, 2008). Possible presence of null alleles was tested using the software MICROCHECKER (Van-Oosterhout et al., 2004). Several indices of genetic diversity were used and analyzed per sub-site at each island. The number of alleles per locus, allele frequencies and observed and expected heterozygosities (Nei, 1973) were calculated with the program CERVUS 3.0 (Marshall et al., 1998). The value of the heterozygosity may range from zero (no heterozygosity) to nearly 1.0 (for a system with a large number of equally frequent alleles). Private alleles are defined as alleles observed in only one population. Allelic richness (mean number of alleles per locus) and gene diversity (the proportion of polymorphic loci across the genome) were calculated with the program FSTAT 2.9.3 (Goudet, 1995). F-statistics were calculated using FSTAT in order to detect non-random mating within populations (FIS) and genetic differentiation between the two island populations (FST) (Weir and Cockerham, 1984). The software CONVERT 1.3.1 (Glaubitz, 2004) was used to anticipate different types of input file formats for the various programs used for the analysis.
Density data for fish were standardized as the number of specimens per 100 m2, and were compared between the two islands using a t-test. For molecular data, allelic richness, number of private alleles, observed heterozygosity, FIS, and gene diversity, were compared between the two islands using all individual genotypes from all loci, and tested for significance using the t-test. For unequal variances, the Welch's t-test was used. Linear regression analysis was used to identify a possible correlation between allelic richness and fish density. Here, the nine sites at the two islands were treated as replicates. The statistical analysis was conducted in STATISTICA 7 (StatSoft, 2004).
Results
Microsatellite Properties, Genetic Diversity, and Differentiation
One of the eight loci (Cf39) departed from HWE, showing a heterozygote deficit (Table 2). At the same locus, null alleles were detected with the programme MICROCHECKER, possibly causing the departure from HWE. However, the test for linkage disequilibrium found no linkage in the pairwise combinations of the eight microsatellite loci. Therefore, all loci were used in the further analysis. The inbreeding coefficient (FIS) values in Barrang Lompo ranged between −0.065 for locus CF29 and 0.285 for CF39 (0.067 ± 0.125, mean ± SD), which was slightly higher than the values observed in Samalona, ranging from −0.075 for locus CF29 to 0.331 for CF39 (0.042 ± 0.123, mean ± SD).
Significantly lower values were found in Barang Lompo for the number of alleles, the number of private alleles, and allelic richness (Table 3). There was no significant difference between the two populations for observed heterozygosity, expected heterozygosity, coefficient of inbreeding and gene diversity (Table 3). A significant genetic differentiation (FST) between the two populations was found (0.065 ± 0.048, p = 0.01).
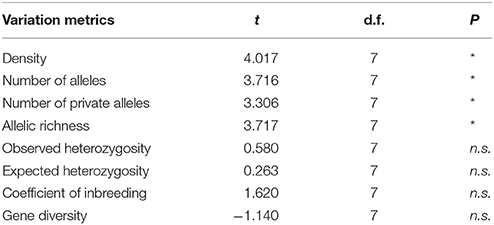
Table 3. Summary of t-test for density and genetic variation metrics of Amphiprion ocellaris between Barrang Lompo and Samalona (* <0.05, n.s., not significant).
Population Size and Its Relation to Allelic Richness
The ecological survey conducted in 2009 revealed that the density of A. ocellaris was significantly lower in Barang Lompo than in Samalona (t-test, t = 4.02, d.f. = 7, p = 0.005; Table 3). The mean density of A. ocellaris at Barrang Lompo was 1.60 ± 0.71 ind/100 m2 (mean ± SD), while it was more than twice as high in Samalona (mean ± SD, 3.96 ± 1.06 ind/100 m2; Table 1). Linear regression analysis showed a positive correlation between fish density and allelic richness (p = 0.03, r2 = 0.43, Figure 2).
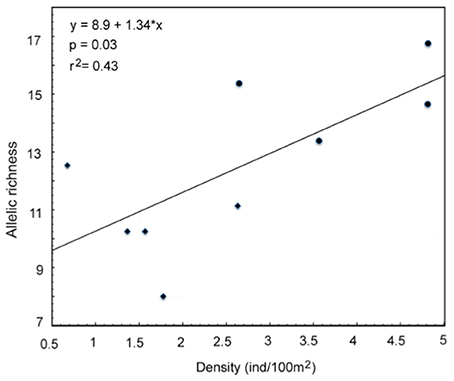
Figure 2. Relationship between allelic richness and fish density in Amphiprion ocellaris (Diamond: Barrang Lompo, Circle: Samalona).
Discussion
Genetic Variability and Differentiation Between Two Island Populations
The amount of genetic variability in a population can be estimated by measuring the number of alleles, number of private alleles, and allelic richness. Studies revealed that allelic richness may be more sensitive to a population bottleneck and environmental stress than heterozygosity (Allendorf, 1986; Petit et al., 1998; Leberg, 2002; Pini et al., 2011). Accordingly, lower allelic richness in overharvested populations was recorded in many cases, which suggests that overharvest drives the decay of genetic diversity across a wide range of marine fishes (Pinsky and Palumbi, 2014). A significant decrease in both allelic richness and heterozygosity over 50 years since the onset of exploitation was revealed in the silver seabream Pagrus auratus by using seven microsatellite loci (Hauser et al., 2002). In addition, a loss of allelic richness was found in the damselfish Dascyllus aruanus due to a high mortality during the recruitment stage, which was induced by environmental stress (Pini et al., 2011). Our study is in agreement with these previous findings, by showing significantly lower values in allelic number and allelic richness in Barrang Lompo than in Samalona. Gene diversity and expected heterozygosity between Samalona and Barrang Lompo were not found to be significantly different (p > 0.05). Both island populations showed high gene diversity and heterozygosity values (mean HE and D > 0.8). The expected heterozygosity in A. ocellaris for both populations (0.54–0.95) was in a similar range to the previous study on this species (0.54–0.92; Timm et al., 2012). The observed heterozygosity in Barang Lompo was slightly lower than in Samalona and the heterozygote deficiency at Barrang Lompo was higher than at Samalona, but these differences were not significant (p > 0.05). For both populations, no inbreeding effects were revealed, which was indicated by low inbreeding coefficients. However, the value of the inbreeding coefficient in Barrang Lompo was higher than in Samalona, which could be an indication of the effect of overharvesting. Inbreeding in a population could cause a reduction in the heterozygosity and an increase in the homozygosity, as shown in overharvested populations (Pinsky and Palumbi, 2014).
Despite dispersing larvae, A. ocellaris showed a significant genetic differentiation between the two island populations (FST 536 = 0.065 ± 0.048, p = 0.01), which was also shown in a previous study (Timm et al., 2017). In a study on the anemonefish species A. clarkii significant FST-values (=0.028) were found between sites on a similarly small scale in the Philippines (Pinsky and Palumbi, 2014). A strong genetic population structure was also found at a larger scale in the same species as well as in other coral reef organisms in this region (Kochzius and Nuryanto, 2008; Timm and Kochzius, 2008; Knittweis et al., 2009; Nuryanto and Kochzius, 2009; Timm et al., 2012; Dohna et al., 2015; Hui et al., 2016, 2017). In addition, the previous study on these two island populations revealed high self-recruitment in A. ocellaris of about 40–60% (Madduppa et al., 2014b), which explains the genetic differentiation.
Relationship Between Population Size and Genetic Diversity
The relative population size in Barrang Lompo was threefold lower than in Samalona (p = 0.005). The coral reef around Barrang Lompo, as well as many other reefs within Spermonde Archipelago, has been overexploited for a long time (Erdmann, 1995; Pet-Soede et al., 1999, 2001). Reefs within the archipelago suffer from destructive fishing, anchor damage, and pollution from the city of Makassar. The number of coral genera at both islands decreased by about 25% from 1985 to 1995 (Edinger et al., 1998). A decline in abundance is considered to be the first and most obvious response of a fish population to exploitation (Shuman et al., 2005). Reduced population sizes drive the alteration of genetic and phenotypic variability (Gaggiotti and Vetter, 1999; Marteinsdóttir and Pardoe, 2008) and might cause a loss of genetic diversity and, therefore, potentially reduce the adaptability, population persistence, and productivity of the targeted species (Hauser et al., 2002). In a study across 140 species of highly abundant marine fish, it was also observed that reduced genetic diversity is a general consequence of overharvesting (Pinsky and Palumbi, 2014).
The current study revealed that allelic richness from eight microsatellite loci and fish density in A. ocellaris were positively correlated (p < 0.05). Thus a decrease in fish density because of exploitation can lead to a reduction of genetic diversity. Since many individuals were removed from the population in Barang Lompo by the marine ornamental fishery, it is likely that some alleles were also removed. Fishing activities, which usually remove the older and bigger individuals, are potentially removing the most heterozygous individuals (Smith et al., 1991). In addition, due to a reduced population size, genetic drift might increase, and alleles will eventually be lost (Garza and Williamson, 2001). This should therefore be taken into account when managing marine ornamental fisheries in the region, as described below.
Implications for Management and Conservation
Several forms of action to prevent the loss of genetic diversity in marine species are proposed, which include: (1) maintaining a large number of individuals of each population and (2) minimizing fisheries-induced selection (Kenchington et al., 2003). The clown anemonefish A. ocellaris has been heavily exploited in Southeast Asian countries to meet the global demand for the aquarium trade (Wabnitz et al., 2003). It has recently been revealed that the marine ornamental fishery in the Spermonde Archipelago has reduced the density and average size of A. ocellaris (Madduppa et al., 2014a). In addition, the current study indicates that the population of A. ocellaris, in Barrang Lompo has a lower density and genetic diversity than the population at Samalona, which is likely due to the higher fishing pressure in Barrang Lompo.
Based on the results of this and previous studies (Madduppa et al., 2014a,b), the marine ornamental trade in Spermonde Archipelago needs to be managed. Otherwise, the populations of A. ocellaris might collapse. The present study supports the previously proposed management strategies for conservation of A. ocellaris in this region (Madduppa et al., 2014a). First, management of the fishing strategy is required, such as leaving the breeding pair in a colony or placing a restriction on the maximum catch size of 50 mm. This strategy prevents the loss of rare alleles from older and bigger individuals. Second, the implementation of marine protected areas (MPAs), which take into account high self-recruitment in A. ocellaris (Madduppa et al., 2014b) and the limited connectivity of populations (Timm and Kochzius, 2008; Timm et al., 2012, 2017). High self-recruitment implies that the populations are more vulnerable to fishing activity (Thorrold et al., 2001). MPAs are an important tool to prevent overexploitation and to ensure the sustainable use of living marine resources (Tundi Agardy, 1994). Therefore, MPAs have been proposed for managing marine fisheries in Indonesia (Mous et al., 2005). They could also be an effective tool for protecting the genetic diversity (Pérez-Ruzafa et al., 2006). Third, regular monitoring and quota determination of the A. ocellaris populations are necessary to control their status. This regular monitoring could also assess other factors (e.g., anthropogenic, environmental parameters) which may influence the status of A. ocellaris.
Conclusions
In conclusion, even though only two populations of A. ocellaris were assessed in this study, we could demonstrate that allelic richness and the number of alleles and private alleles in Barang Lompo, the site exposed to a higher fishing pressure, were reduced. A positive correlation between allelic richness and fish density was revealed. This study suggests that allelic richness might serve as indicator for detecting loss of genetic diversity at an early stage. Therefore, management actions are needed in Spermonde Archipelago, especially in Barrang Lompo and other similar islands within the region, to prevent locally extinct of this valuable marine organisms.
Author Contributions
HM and MK: conceived and designed the experiments; HM: performed the experiments; HM, JT, and MK: analyzed the data; MK: contributed reagents, materials, analysis tools; HM, MK, and JT: wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the German Federal Ministry of Education and Research (BMBF, grant no. 03F0472B), which funded this study in the framework of the SPICE II project (Science for the Protection of Indonesian Coastal Marine Ecosystems); the German Academic Exchange Service (DAAD) for the Ph.D. scholarship to HM; Leibniz Centre for Tropical Marine Research (ZMT) (Bremen, Germany) for project co-ordination; A. Nuryanto from the Jenderal Soedirman University (Purwokerto, Indonesia) and members of the Marine Science Diving Club of Hasanuddin University (Makassar, Indonesia) for their help during field work; colleagues from Bogor Agricultural University (IPB, Bogor, Indonesia) for logistical support during field work in Spermonde Archipelago; and Werner Wosniok from the Institute for Statistics (University of Bremen, Germany) for statistical advices. The SPICE project was conducted and permitted under the governmental agreement between the German Federal Ministry of Education and Research (BMBF) and the Indonesian Ministry for Research and Technology (RISTEK), Indonesian Institute of Sciences (LIPI), Indonesian Ministry of Maritime Affairs and Fisheries (DKP), and Indonesian Agency for the Assessment and Application of Technology (BPPT).
References
Allen, G. R. (2008). Conservation hotspots of biodiversity and endemism for Indo-Pacific coral reef fishes. Aquat. Conserv. Mar. Freshw. Ecosyst. 18, 541–556. doi: 10.1002/aqc.880
Allen, G. R., and Werner, T. B. (2002). Coral reef fish assessment in the ‘Coral Triangle’ of Southeastern Asia. Environ. Biol. Fish. 65, 209–214. doi: 10.1023/A:1020093012502
Allendorf, F. W. (1986). Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol. 5, 181–190. doi: 10.1002/zoo.1430050212
Almany, G. R., Berumen, M. L., Thorrold, S. R., Planes, S., and Jones, G. P. (2007). Local replenishment of coral reef fish populations in a marine reserve. Science 316, 742–744. doi: 10.1126/science.1140597
Briggs, J. C. (2009). Diversity, endemism and evolution in the coral triangle. J. Biogeogr. 36, 2008–2010. doi: 10.1111/j.1365-2699.2009.02146.x
Bryant, D., Burke, L., McManus, J., and Spalding, M. (1998). Reefs at Risk: A Map-Based Indicator of Threats to the World's Coral Reefs. Washington, DC: World Resources Institute.
Burke, L., Selig, E., and Spalding, M. (2006). Reefs at Risk in Southeast Asia. Washington, DC: World Resources Institute.
Buston, P. (2003). Social hierarchies: size and growth modification in clownfish. Nature 424, 145–146. doi: 10.1038/424145a
Buston, P. M., Bogdanowicz, S. M., Wong, A., and Harrison, R. G. (2007). Are clownfish groups composed of close relatives? An analysis of microsatellite DNA variation in Amphiprion percula. Mol. Ecol. 163671–3678. doi: 10.1111/j.1365-294X.2007.03421.x
Cardinale, M., and Modin, J. (1999). Changes in size-at-maturity of Baltic cod (Gadus morhua) during a period of large variations in stock size and environmental conditions. Fish. Res. 41, 285–295. doi: 10.1016/S0165-7836(99)00021-1
Dankel, D., Skagen, D., and Ulltang, Ø. (2008). Fisheries management in practice: review of 13 commercially important fish stocks. Rev. Fish Biol. Fish. 18, 201–233. doi: 10.1007/s11160-007-9068-4
Dohna, T. A., Timm, J., Hamid, L., and Kochzius, M. (2015). Limited connectivity and a phylogeographic break characterize populations of the pink anemonefish, Amphiprion perideraion, in the Indo-Malay Archipelago: inferences from a mitochondrial and microsatellite loci. Ecol. Evol. 5, 1717–1733. doi: 10.1002/ece3.1455
Edinger, E. N., Jompa, J., Limmon, G. V., Widjatmoko, W., and Risk, M. J. (1998). Reef degradation and coral biodiversity in larkia a: effects of land-based pollution, destructive fishing practices and changes over time. Mar. Poll. Bull. 36, 617–630. doi: 10.1016/S0025-326X(98)00047-2
Erdmann, M. (1995). The ABC Guide to Coral Reef Fisheries in Southwest. Sulawesi: NAGA the ICLARM quarterly.
Fricke, H., and Fricke, S. (1977). Monogamy and sex change by aggressive dominance in coral reef fish. Nature 266, 830–832.
Gaggiotti, O. E., and Vetter, R. D. (1999). Effect of life history strategy, environmental variability, and overexploitation on the genetic diversity of pelagic fish populations. Can. J. Fish. Aquat. Sci. 58, 1376–1388. doi: 10.1139/f99-060
Garza, J. C., and Williamson, E. G. (2001). Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 10, 305–318. doi: 10.1046/j.1365-294x.2001.01190.x
Glaubitz, J. C. (2004). Convert: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol. Ecol. Notes 4, 309–310. doi: 10.1111/j.1471-8286.2004.00597.x
Gopakumar, G., Madhu, K., Madhu, R., Ignatius, B., Krishnan, L., and Mathew, G. (2009). Broodstock development, breeding and seed production of selected marine food fishes and ornamental fishes. Mar. Fish. Inf. Serv. 201, 1–9.
Goudet, J. (1995). FSTAT (Version 1.2): a computer program to calculate F-statistics. J. Hered. 86, 485–486. doi: 10.1093/oxfordjournals.jhered.a111627
Hauser, L., Adcock, G. J., Smith, P. J., Bernal Ramiréz, J. H., and Carvalho, G. R. (2002). Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc. Natl. Acad. Sci. U.S.A. 99, 11742–11747. doi: 10.1073/pnas.172242899
Heino, M., and Godø, O. R. (2002). Fisheries-induced selection pressures in the context of sustainable fisheries. Bull. Mar. Sci. 70, 639–656. Available online at: http://www.ingentaconnect.com/contentone/umrsmas/bullmar/2002/00000070/00000002/art00017;jsessionid=atocm45qlt3q.x-ic-live-01
Hui, M., Kraemer, W. E., Seidel, C., Nuryanto, A., Joshi, A., and Kochzius, M. (2016). Comparative genetic population structure of three endangered giant clams (Tridacnidae) throughout the Indo-West Pacific: implications for divergence, connectivity, and conservation. J. Mollusc. Stud. 82, 403–414. doi: 10.1093/mollus/eyw001
Hui, M., Nuryanto, A., and Kochzius, M. (2017). Concordance of microsatellite and mitochondrial DNA markers in detecting genetic population structure in the boring giant clam, Tridacna crocea, across the Indo-Malay Archipelago. Mar. Ecol. 38:e12389. doi: 10.1111/maec.12389
Hutchings, J. A., and Reynolds, J. D. (2004). Marine fish population collapses: consequences for recovery and extinction risk. Bioscience 54, 297–309. doi: 10.1641/0006-3568(2004)054[0297:MFPCCF]2.0.CO;2
Jones, A. M., Gardner, S., and Sinclair, W. (2008). Losing 'Nemo': bleaching and collection appear to reduce inshore populations of anemonefishes. J. Fish Biol. 73, 753–761. doi: 10.1111/j.1095-8649.2008.01969.x
Kenchington, E., Heino, M., and Nielsen, E. E. (2003). Managing marine genetic diversity: time for action? ICES J. Mar. Sci. 60, 1172–1176. doi: 10.1016/S1054-3139(03)00136-X
Knittweis, L., Kraemer, W., Timm, J., and Kochzius, M. (2009). Genetic structure of Heliofungia actiniformis (Scleractinia: Fungiidae) populations in the Indo-Malay Archipelago: implications for live coral trade management efforts. Conserv. Genet. 10, 241–249. doi: 10.1007/s10592-008-9566-5
Kochzius, M., and Nuryanto, A. (2008). Strong genetic population structure in the boring giant clam, Tridacna crocea, across the Indo-Malay Archipelago: implications related to evolutionary processes and connectivity. Mol. Ecol. 17, 3775–3787. doi: 10.1111/j.1365-294X.2008.03803.x
Leberg, P. L. (2002). Estimating allelic richness: effects of sample size and bottlenecks. Mol. Ecol. 11, 2445–2449. doi: 10.1046/j.1365-294X.2002.01612.x
Liu, S.-Y. V., Yu, H.-T., and Dai, C.-F. (2007). Eight microsatellite loci in Clark's anemonefish, Amphiprion clarkii. Mol. Ecol. Notes 7, 1169–1171. doi: 10.1111/j.1471-8286.2007.01818.x
Madduppa, H. H., Juterzenka, K. V., Syakir, M., and Kochzius, M. (2014a). Socio-economy of marine ornamental fishery and its impact on the population structure of the clown anemonefish Amphiprion ocellaris and its host anemones in Spermonde Archipelago, Indonesia. Ocean Coast. Manag. 100, 41–50. doi: 10.1016/j.ocecoaman.2014.07.013
Madduppa, H. H., Timm, J., and Kochzius, M. (2014b). Interspecific, spatial and temporal variability of self-recruitment in anemonefishes. PLoS ONE 9:e90648. doi: 10.1371/journal.pone.0090648
Madhu, R., Madhu, K., and Venugopalan, K. M. (2010). Sex change of hatchery produced Amphiprion ocellaris: influence of mating system removal on gonad maturation and nesting success. J. Mar. Biol. Assoc. India 52, 62–69.
Marshall, T. C., Slate, J., Kruuk, L. E., and Pemberton, J. M. (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 7, 639–655. doi: 10.1046/j.1365-294x.1998.00374.x
Marteinsdóttir, G., and Pardoe, H. (2008). “Effects of fishing on inter and intra stock diversity of marine resources,” in Fisheries for Global Welfare and Environment, 5th (Yokohama: World Fisheries Congress), 27–43.
Mitchell, J. (2005). Queue selection and switching by false clown anemonefish, Amphiprion ocellaris. Anim. Behav. 69, 643–652. doi: 10.1016/j.anbehav.2004.05.017
Mous, P. J., Pet, J. S., Arifin, Z., Djohani, R., Erdmann, M. V., Halim, A., et al. (2005). Policy needs to improve marine capture fisheries management and to define a role for marine protected areas in Indonesia. Fish. Manage. Ecol. 12, 259–268. doi: 10.1111/j.1365-2400.2005.00448.x
Musick, J. A., Harbin, M. M., Berkeley, S. A., Burgess, G. H., Eklund, A. M., Findley, L., et al. (2000). Marine, estuarine, and diadromous fish stocks at risk of extinction in North America (Exclusive of Pacific Salmonids). Fisheries 25, 6–29. doi: 10.1577/1548-8446(2000)025<0006:MEADFS>2.0.CO;2
Nei, M. (1973). Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. U.S.A. 70, 3321–3323. doi: 10.1073/pnas.70.12.3321
Nuryanto, A., and Kochzius, M. (2009). Highly restricted gene flow and deep evolutionary lineages in the giant clam Tridacna maxima. Coral Reefs 28, 607–619. doi: 10.1007/s00338-009-0483-y
O'Brien, S. J. (1994). A role for molecular genetics in biological conservation. Proc. Natl. Acad. Sci. U.S.A. 91, 5748–5755. doi: 10.1073/pnas.91.13.5748
Ottersen, G., Hjermann, D. Ø., and Stenseth, N. C. (2006). Changes in spawning stock structure strengthens the link between climate and recruitment in a heavily fished cod stock. Fish. Oceanogr. 15, 30–243. doi: 10.1111/j.1365-2419.2006.00404.x
Pérez-Ruzafa, Á., González-Wangüemert, M., Lenfant, P., Marcos, C., and García-Charton, J. A. (2006). Effects of fishing protection on the genetic structure of fish populations. Biol. Conserv. 129, 244–255. doi: 10.1016/j.biocon.2005.10.040
Petit, R. J., El Mousadik, A., and Pons, O. (1998). Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12, 844–855. doi: 10.1046/j.1523-1739.1998.96489.x
Pet-Soede, C., and Erdmann, M. V. (1998). An overview and comparison of destructive fishing practices in Indonesia. SPC Live Reef Fish Bull. 4, 28–36.
Pet-Soede, C., Machiels, M. A. M., Stam, M. A., and van Densen, W. L. T. (1999). Trends in an Indonesian coastal fishery based on catch and effort statistics and implications for the perception of the state of the stocks by fisheries officials. Fish. Res. 42, 41–56. doi: 10.1016/S0165-7836(99)00034-X
Pet-Soede, C., Van Densen, W. L. T., Hiddink, J. G., Kuyl, S., and Machiels, M. A. M. (2001). Can fishermen allocate their fishing effort in space and time on the basis of their catch rates? An example from Spermonde Archipelago, SW Sulawesi, Indonesia. Fish. Manage. Ecol. 8, 15–36. doi: 10.1046/j.1365-2400.2001.00215.x
Pini, J., Planes, S., Rochel, E., Lecchini, D., and Fauvelot, C. (2011). Genetic diversity loss associated to high mortality and environmental stress during the recruitment stage of a coral reef fish. Coral Reefs 30, 399–404. doi: 10.1007/s00338-011-0718-6
Pinsky, M. L., and Palumbi, S. R. (2014). Meta-analysis reveals lower genetic diversity in overfished populations. Mol. Ecol. 23, 29–39. doi: 10.1111/mec.12509
Planes, S., Jones, G. P., and Thorrold, S. R. (2009). Larval dispersal connects fish populations in a network of marine protected areas. Proc. Natl. Acad. Sci. U.S.A. 106, 5693–5697. doi: 10.1073/pnas.0808007106
Quenouille, B., Bouchenak-Khelladi, Y., Hervet, C., and Planes, S. (2004). Eleven microsatellite loci for the saddleback clownfish Amphiprion polymnus. Mol. Ecol. Notes 4, 291–293. doi: 10.1111/j.1471-8286.2004.00646.x
Raymond, M., and Rousset, F. (1995). GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249. doi: 10.1093/oxfordjournals.jhered.a111573
Reusch, T. B., Ehlers, A., Hämmerli, A., and Worm, B. (2005). Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl. Acad. Sci. U.S.A. 102, 2826–2831. doi: 10.1073/pnas.0500008102
Rousset, F. (2008). Genepop'007: a complete re-implementation of the genepop software for windows and linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Saenz-Agudelo, P., Jones, G. P., Thorrold, S. R., and Planes, S. (2011). Connectivity dominates larval replenishment in a coastal reef fish metapopulation. Proc. Biol. Sci. 278, 2954–2961. doi: 10.1098/rspb.2010.2780
Shuman, C., Hodgson, G., and Ambrose, R. (2005). Population impacts of collecting sea anemones and anemonefish for the marine aquarium trade in the Philippines. Coral Reefs 24, 564–573. doi: 10.1007/s00338-005-0027-z
Smith, P. J., Francis, R. I. C. C., and McVeagh, M. (1991). Loss of genetic diversity due to fishing pressure. Fish. Res. 10, 309–316. doi: 10.1016/0165-7836(91)90082-Q
Spielman, D., Brook, B. W., and Frankham, R. (2004). Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. U.S.A. 101, 15261–15264. doi: 10.1073/pnas.0403809101
StatSoft, I. (2004). STATISTICA (Data Analysis Software System), Version 7. Available online at: www.statsoft.com
Thorrold, S. R., Latkoczy, C., Swart, P. K., and Jones, C. M. (2001). Natal homing in a marine fish metapopulation. Science 291, 297–299. doi: 10.1126/science.291.5502.297
Timm, J., and Kochzius, M. (2008). Geological history and oceanography of the Indo-Malay Archipelago shape the genetic population structure in the false clown anemonefish (Amphiprion ocellaris). Mol. Ecol. 17, 3999–4014. doi: 10.1111/j.1365-294X.2008.03881.x
Timm, J., Kochzius, M., Madduppa, H. H., Neuhaus, A. I., and Dohna, T. (2017). Small scale genetic population structure of coral reef organisms in Spermonde Archipelago, Indonesia. Front. Mar. Sci. 4:294. doi: 10.3389/fmars.2017.00294
Timm, J., Planes, S., and Kochzius, M. (2012). High similarity of genetic population structure in the false clown anemonefish (Amphiprion ocellaris) found in microsatellite and mitochondrial control region analysis. Conserv. Genet. 13, 693–706. doi: 10.1007/s10592-012-0318-1
Tomascik, T., Mah, A., Nontji, A., and Moosa, M. (1997). The Ecology of Indonesian Seas, Part II. Singapore: Periplus Edition Ltd.
Trippel, E. A. (1995). Age at maturity as a stress indicator in fisheries. Bioscience 45, 759–771. doi: 10.2307/1312628
Tundi Agardy, M. (1994). Advances in marine conservation: the role of marine protected areas. Trends Ecol. Evol. 9, 267–270. doi: 10.1016/0169-5347(94)90297-6
Van-Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Veron, J. E. N., Devantier, L. M., Turak, E., Green, A. L., Kininmonth, S., Stafford-Smith, M., et al. (2009). Delineating the coral triangle. Galaxea J. Coral Reef Stud. 11, 91–100. doi: 10.3755/galaxea.11.91
Wabnitz, C., Taylor, M., Green, E., and Razak, T. (2003). From Ocean to Aquarium: The Global Trade in Marine Ornamental Species. Cambridge, UK: UNEP-WCMC.
Wang, L., Shi, X., Su, Y., Meng, Z., and Lin, H. (2012). Loss of genetic diversity in the cultured stocks of the large yellow croaker, Larimichthys crocea, revealed by microsatellites. Int. J. Mol. Sci. 13, 5584–5597. doi: 10.3390/ijms13055584
Keywords: exploited species, allelic diversity, genetic variation, fishery management, coral triangle
Citation: Madduppa HH, Timm J and Kochzius M (2018) Reduced Genetic Diversity in the Clown Anemonefish Amphiprion ocellaris in Exploited Reefs of Spermonde Archipelago, Indonesia. Front. Mar. Sci. 5:80. doi: 10.3389/fmars.2018.00080
Received: 31 December 2016; Accepted: 22 February 2018;
Published: 09 March 2018.
Edited by:
Alberto Basset, University of Salento, ItalyReviewed by:
Anne Chenuil, Centre National de la Recherche Scientifique (CNRS), FranceRoberta Cimmaruta, Università degli Studi Della Tuscia, Italy
Copyright © 2018 Madduppa, Timm and Kochzius. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hawis H. Madduppa, aGF3aXNAYXBwcy5pcGIuYWMuaWQ=
 Hawis H. Madduppa
Hawis H. Madduppa Janne Timm3
Janne Timm3 Marc Kochzius
Marc Kochzius