- 1Marine Predator Research Group, Department of Biological Sciences, Macquarie University, Sydney, NSW, Australia
- 2Heliguy Pty. Ltd., Sydney, NSW, Australia
- 3Macquarie Marine Research Centre, Chemistry and Biomolecular Sciences, Macquarie University, Sydney, NSW, Australia
- 4ARC Centre of Excellence for Coral Reef Studies, James Cook University, Townsville, QLD, Australia
Drones or Unmanned Aerial Vehicles (UAVs) have huge potential to improve the safety and efficiency of sample collection from wild animals under logistically challenging circumstances. Here we present a method for surveying population health that uses UAVs to sample respiratory vapor, ‘whale blow,' exhaled by free-swimming humpback whales (Megaptera novaeangliae), and coupled this with amplification and sequencing of respiratory tract microbiota. We developed a low-cost multirotor UAV incorporating a sterile petri dish with a remotely operated ‘blow' to sample whale blow with minimal disturbance to the whales. This design addressed several sampling challenges: accessibility; safety; cost, and critically, minimized the collection of atmospheric and seawater microbiota and other potential sources of sample contamination. We collected 59 samples of blow from northward migrating humpback whales off Sydney, Australia and used high throughput sequencing of bacterial ribosomal gene markers to identify putative respiratory tract microbiota. Model-based comparisons with seawater and drone-captured air demonstrated that our system minimized external sources of contamination and successfully captured sufficient material to identify whale blow-specific microbial taxa. Whale-specific taxa included species and genera previously associated with the respiratory tracts or oral cavities of mammals (e.g., Pseudomonas, Clostridia, Cardiobacterium), as well as species previously isolated from dolphin or killer whale blowholes (Corynebacteria, others). Many examples of exogenous marine species were identified, including Tenacibaculum and Psychrobacter spp. that have been associated with the skin microbiota of marine mammals and fish and may include pathogens. This information provides a baseline of respiratory tract microbiota profiles of contemporary whale health. Customized UAVs are a promising new tool for marine megafauna research and may have broad application in cost-effective monitoring and management of whale populations worldwide.
Introduction
Conservation biology is entering a new era of innovation, with unprecedented growth across a range of techniques, from genetics and genomics to telemetry and remote sensing (Allendorf et al., 2010; Hussey et al., 2015). Rapid advances in the technology underpinning Unmanned Aerial Vehicles (UAVs also known as Unmanned Aircraft Systems or drones), are driving new and innovative environmental applications (Koh and Wich, 2012; Anderson and Gaston, 2013; Christie et al., 2016; Smith et al., 2016; Duffy et al., 2017). The application of UAVs in conservation science makes it possible to collect information from dangerous and inaccessible environments and answer research questions that were previously limited to the hypothetical (Harvey et al., 2016). UAVs also provide an alternative, safer, quieter and often cost-effective option for monitoring fauna and flora, from individuals and populations to entire ecosystems, and in so doing are replacing expensive manned systems such as helicopters and fixed-wing aircraft (Christiansen et al., 2016; Christie et al., 2016). UAV applications in wildlife research now encompass almost all environments, from arid deserts, through rainforests, oceans to polar regions (Linchant et al., 2013, 2015; Durban et al., 2015; Goebel et al., 2015; Duffy et al., 2017).
UAVs are transforming the way scientists monitor and conserve wildlife (Gonzalez et al., 2016). In the terrestrial world, UAVs have been used for a wide variety of conservation applications (van Gemert et al., 2014; Gonzalez et al., 2016). Some examples include, counting elephants (Loxodonta africana) (Linchant et al., 2013; Vermeulen et al., 2013), UAV surveillance (anti-poaching tools) for elephants and rhinoceros (Diceros bicornis and Ceratotherium simum) (Marks, 2014; Mulero-Pázmány et al., 2014; Hahn et al., 2017), locating chimpanzee nests (Pan troglodytes) (van Andel et al., 2015) and mapping Sumatran orangutan (Pongo abelii) habitat, distribution and density (Wich et al., 2015; Szantoi et al., 2017). UAV applications now extend to the polar regions where they have been used to monitor and estimate abundance of penguin populations (gentoo, Pygoscelis papua, and chinstrap, Pygoscelis antarctica) and estimate size and condition of leopard seals (Hydrurga leptonyx) (Goebel et al., 2015; Ratcliffe et al., 2015). In the marine environment, UAVs are revolutionizing the way marine species can be studied due to their small size, apparent minimal disturbance of wildlife and improved safety for both operators and animals (Nowacek et al., 2016; Fiori et al., 2017). UAVs have been utilized for a wide variety of applications including aerial surveys, monitoring, habitat use, abundance estimates, photogrammetry and biological sampling e.g., whale “blow” (Hogg et al., 2009; Acevedo-Whitehouse et al., 2010; Hodgson et al., 2013; Durban et al., 2015; Pomeroy et al., 2016; Schofield et al., 2017).
There are widespread concerns about the health of marine mammal populations in the face of global anthropogenic stressors (Gulland and Hall, 2007). Yet health assessments typically involves collecting samples from stranded animals, which are often biased as these animals are most likely to be health-compromised (Geraci and Lounsbury, 2005). Sampling exhaled breath or ‘blow' from wild whales may therefore provide a more representative assessment of the health status of individuals because samples can be randomly taken from the population. From a single sample of whale blow, scientists may be able to collect respiratory bacteria, lipids, proteins, DNA and hormones (Hogg et al., 2005, 2009; Schroeder et al., 2009; Acevedo-Whitehouse et al., 2010; Hunt et al., 2013, 2014; Thompson et al., 2014; Burgess et al., 2016; De Mello and De Oliveira, 2016; Raverty et al., 2017). This information is important for whale conservation, as it can be collected over time to help monitor the recovery of whale populations post-whaling. Early approaches to sampling whale blow involved passing a cotton gauze or nylon stocking on the end of a carbon fiber pole through the blow when the animal surfaced (Hogg et al., 2009; Hunt et al., 2014). Recent advancements on this method have seen the use of a pole with a number of petri dishes with lids to sample wild killer whales (Raverty et al., 2017). However, this method requires extremely close vessel approaches to whales (Hogg et al., 2009). Given the large size, mass and power of whales, this approach involves high risk to both researchers and to the whale itself. Even under ideal circumstances this method is likely to disturb the animal, potentially compromising the validity of some of the measures such as stress hormones which elevate rapidly (Harcourt et al., 2010). Accordingly, alternative approaches have long been sought. Acevedo-Whitehouse et al. (2010) deployed a single-rotor UAV (a remote-controlled helicopter) to sample whale blow. Their study demonstrated the feasibility of the approach but loss of samples from the UAV as it careers through the sea air proved a potential issue as did contamination from airborne particulate not expired by the whale.
Here we describe a purpose-built UAV designed to sample whale blow in the field with minimal contamination. Our goal was to provide a snapshot of whale health. We specifically targeted northward migrating humpback whales (Megaptera novaeangliae) off the East coast of Sydney, Australia for the collection of baseline microbiota information. The UAV used in our study has a unique combination of features that represent a significant advance over existing UAVs. It is fast, highly maneuverable, durable, waterproof, low-cost (< $USD 1000) and provides flexible payload mounting options. The UAV is scaled to the sampling gear (in this case a 100 mm petri dish), which is held in a mechanism that allows the dish to be opened/closed during flight–minimizing sample contamination or loss.
Materials and Methods
Study Site and Species
All flights were conducted offshore Sydney, Australia (Figure 1). Each year from May to November, migratory Group V (Stock E1) humpback whales migrate past Sydney, as they swim from high latitude feeding areas in Antarctica to low latitude breeding waters off Queensland (Chittleborough, 1965). All sampling took place in coastal waters < 3 nautical miles from Sydney between 30 May 2017 and 27 June 2017.
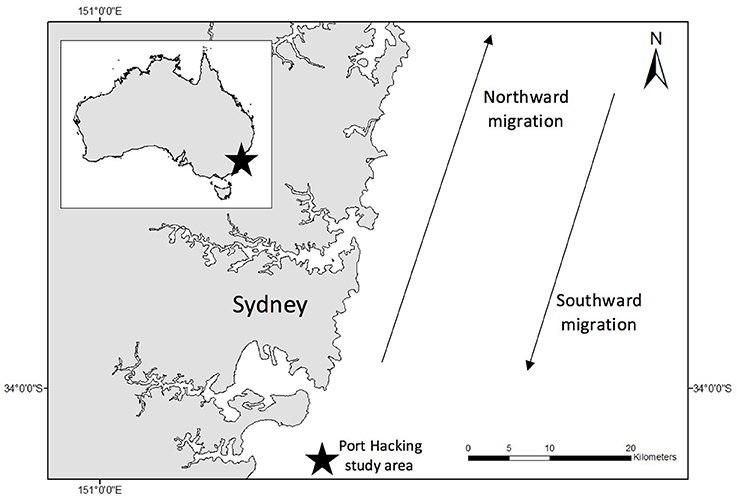
Figure 1. Study site (indicated by black star on insert). All samples were collected in coastal waters (<3 nm) off Sydney, Australia. Blow samples were collected only from northward migrating East coast Australian humpback whales. Water samples were collected over a number of years from Port Hacking (indicated by star outside of insert).
UAV Design
The UAV is a 4-motor electric multirotor (quadcopter) 500 mm across (motor to motor, diagonally) (Figure 2A). It has a relatively high power to weight ratio making it fast, maneuverable, resistant to strong wind gusts and relatively quiet while hovering. It carries the bare minimum of hardware and is operated in ‘manual mode' (no GPS or autolevelling assistance) with a heavy reliance of the onboard video feed for control, navigation and sampling operations. The airframe structure of the UAV is a ‘sandwich' style construction cut from carbon fiber plate, with a top shell molded from impact-resistant polycarbonate. This seals against the airframe to create a waterproof compartment which houses the power distribution, flight control, motor control, radio control transceiver, and video transmitter components. The float booms/legs were cut from expanded polypropylene (EPP)—a closed-cell foam, chosen for high strength, resistance to bending loads and excellent water resistance. A clear acrylic tube at the front of the aircraft houses a forward facing, tilting camera that provides a real-time position reference to the pilot (First Person View). The resulting composite structure is light, stiff, strong and waterproof. Buoyancy is provided by the two watertight compartments and EPP foam floats under the arms. In the event of a crash or forced landing over water, the UAV floats in an upright position so it can be recovered or take off again. Two reinforced mounting areas on the top shell accept payloads of around 100 g. For this configuration, the blow-sampling apparatus was mounted at the front. This is a hinged frame which opens to 180 degrees and holds a 100 mm diameter petri dish with suction cups. A servo motor opens and closes the dish remotely, during flight. Airflow testing using smoke indicated the best position for the sampling dish relative to the propellers. A forward-looking waterproof video camera (GoPro® Hero Session™) is positioned at the rear and logs video to an internal memory card. The dish is in the frame of the recorded video, so the footage can be used to confirm the source of the sampled material.
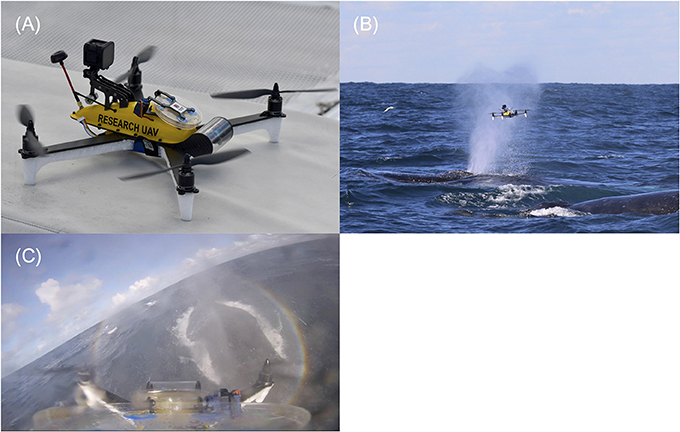
Figure 2. (A) Purpose-built UAV designed to sample whale blow. The UAV consists of a sandwich style carbon fiber body. White foam floats support the UAV during take-off and landings and provide floatation in water. The yellow shell houses all electrical equipment. A GoPro® hero session is mounted at the back of the yellow shell to record flights. A hinge mechanism with disposable petri dish is located in the center of the yellow shell. This can be remotely operated to minimize sample contamination in the field. The clear round tube at the front of the UAV houses the first-person camera to assist with sampling. (B) UAV sampling whale blow. This photo was taken just as the UAV had passed through the visible blow (plume of spray). The petri dish is still in the open position. Sample was collected on both the lid and bottom (nutrient agar filled) side. The petri dish was shut immediately after collection to minimize sample contamination and the drone was flown back to the research vessel >200 meters away. (C) Screenshot from the UAVs on-board GoPro® camera mid whale sample collection. This footage shows the petri dish at the bottom of the picture. The whale is located on the right-hand side. The petri dish is completely extended (open) with blow droplets visible on both sides of the dish and GoPro® lens.
Sampling Method
This study was approved by the Macquarie University Animal Ethics Committee, and carried out in accordance with the Animal Research Authority (2016/010). This research was permitted by New South Wales National Parks and Wildlife Services (NSWNPWS) to fly UAVs over whales in New South Wales coastal waters (permit number SL101743). To adhere to Australian legislative requirements, the UAVs (including backup UAV) were registered with the Civil Aviation Safety Authority (CASA) and operated by a CASA certified operator (Heliguy Pty. Ltd.). All flights were conducted in good weather (no rain, Beaufort < 3), from small research vessels, where the UAV was launched and landed on a launch pad at the bow or stern of the boat. A closed, sterile petri dish with nutrient agar covering the base of the petri dish was secured using eight suction cups affixed on the UAV before each flight.
Members of the team scanned the area for humpback whales. Once an individual was selected, the vessel was driven maintaining a constant speed and distance from the whale (>200 m). Once the respiratory rhythm of an individual was determined (downtime length in minutes), the UAV was launched to coincide with the individual surfacing. The UAV pilot was directed by spotters on the vessel and positioned the UAV with the aid of the live feed from the forward-facing camera. To minimize sample contamination, the petri dish remained closed until just before the whale surfaced, when the dish remotely opened as the UAV accelerated toward and through the densest part of the whale blow, collecting the maximum amount of sample in the dish and lid (Figures 2B,C and Supplementary Video 1). The petri dish was immediately closed and the UAV was returned to the vessel. The petri dish containing the sample was removed from the UAV and Parafilm® was wrapped around the closed petri dish to secure the sample. All samples were temporarily stored in a cooler box on ice until further processing in the laboratory at the end of each day.
Attempts were made to sample a different whale each flight. Individuals within a pod were chosen based upon unique markings (e.g., white flanks/patterns/scarring/barnacle arrangements). To ensure the same individual was not sampled twice, a live video feed was used to target individuals. Cross contamination among whales was avoided by not triggering the opening of the flip lid until only the targeted whale respired. Footage collected from the GoPro® throughout each flight was used to validate sample collection and eliminate repeated sampling of the same individuals by post-hoc identification. The behavioral response of whales was recorded for each pass using by scoring system of one to three (one: ‘no response,' two: ‘minor response' minor surface activity such as logging, spy hopping and three: ‘severe/elevated Response' e.g., breaching, peduncle throw or chin slap).
Air and Seawater Samples
To enable direct comparison of UAV-captured air and whale blow samples with bacteria inhabiting the adjacent seawater, the data were combined with 16S sequence libraries prepared from 26 surface seawater samples. This represents a complete annual cycle, collected from the National Time Series Station known as Port Hacking 100 (PH100). All UAV-captured samples were collected within 20 km of PH100.
Laboratory Processing of Samples
Initial processing of samples occurred in two stages. First, in an Ultra Violet-sanitized class II biosafety hood, the top of the petri dish lid (non-agar) side was swabbed using a dry sterile cotton tip and then placed in a sterile 1.5 ml tube and stored in the freezer at −30°C. Secondly, the petri dish (both the lid and nutrient base) was placed in an incubator at 37°C after the lid was swabbed, simulating average mammalian body temperature 36–37°C (Whittow, 1987; Cuyler et al., 1992). Plates were observed daily for colony growth. If growth occurred, colonies were counted and a representative number of colonies were picked from each plate, resuspended in 100 μl of sterile water, vortexed for 10 s and immediately frozen at −30°C until further processing. Plates were then stored in the fridge for future reference if needed.
Bacterial DNA Extraction
DNA extractions were conducted using the Quick-DNA™ Fungal/Bacterial Miniprep kit (Zymo Research, Irvine, California, USA) with minor modifications to the manufacturer's instructions. Each swab was transferred to a tube containing 1.2 g of ZR BashingBeads™ (equivalent to ~half of the portion supplied for each extraction). The original storage tube was rinsed with lysis solution (750 μl) to ensure the complete transfer of material into the extraction tube. The swab was then bead-beaten on a Vortex-Genie® 2 (Mo Bio Laboratories/QIAGEN, California, USA) for 20 min at room temperature. All other steps were followed according to the manufacturer's instructions, with the exception that two successive final elutions were carried out, each with 20 μl of sterile DNA elution buffer.
Amplification and Sequencing
Amplicons targeting the bacterial 16S rRNA gene (27F−519R; Lane et al., 1985; Lane, 1991) were generated and sequenced for each sample at the Ramaciotti Centre for Genomics (UNSW Sydney, Australia) using 250 bp paired end illumina sequencing according to established protocols (http://www.bioplatforms.com/wp-content/uploads/base_illumina_16s_amplicon_methods.pdf).
Amplicons generated from drone-captured air and whale blow were combined with 27F−519R sequences generated from 26 surface (2 m and 10 m depth) seawater samples collected over a complete annual cycle from the nearby National Reference Station (PH100) time series (Dec 2014–Mar 2016). Monthly microbial sampling has been conducted at the Port Hacking100 reference station since 2009 (Seymour et al., 2012). All UAV-captured whale and air samples were collected within 20 km upstream of this reference station, within 1–3 km from shore. We reasoned that this dataset, which was sampled and sequenced using standardized protocols at the same sequencing center, would provide a comprehensive and unbiased assessment of bacterial species characteristic of seawater in this region, which could be excluded as potential contaminants from the whale blow samples. Whale, air and seawater samples analyzed in this study are detailed in Supplementary Tables 1, 2.
Sequence Operational Taxonomic Units (OTUs) tables were prepared after (Bissett et al., 2016). Briefly, paired-end reads were filtered using Trimmomatic (ILLUMINACLIP: NexteraPE-PE.fa:2:30:10 SLIDINGWINDOW:4:15 MINLEN:76) (Bolger et al., 2014) then merged using PEAR (Zhang et al., 2014). The combined amplicon data were clustered into OTUs at 97% sequence similarity using an open reference OTU picking pipeline in USEARCH 64 bit v8.1.1756 (Edgar, 2010), which included de novo chimera detection. Clusters with < 4 sequences were removed, and reads were mapped to representative OTU sequences using USEARCH (97% ID) to calculate read abundances. From an initial pool of 10.5 million paired-end reads, a total of 7.62 million filtered, merged sequences, with chimeras removed, were added to the OTU table. OTU tables were sub-sampled to a constant sampling depth of 10,000 sequences using rarefy in vegan (Oksanen, 2017). All subsequent analyses were conducted on sub-sampled OTU tables. Sequences generated over the course of this project are deposited in the European Nucleotide Archive under project PRJEB23634. All seawater sequence data are deposited in the NCBI Sequence Read Archive PRJNA385736.
Data Analyses
Hierarchal clusters of OTU abundance profiles generated from seawater, drone-captured air and whale blow were compared using the simprof test following square-root transformation and conversion to a Bray-Curtis dissimilatory matrix in the r package clustsig (Whitaker and Christman, 2014). Data from samples that were near misses, which would reflect a mixture of air and whale blow microbiota, were set aside from the subsequent statistical analyses. The community structure dissimilarity between samples was observed with non-metric multidimensional scaling. Significant differences in communities sampled in seawater, UAV-captured air or whale blow samples were defined using generalized linear models within mvabund (Wang et al., 2012). Briefly, a negative binomial model was fit to the OTU abundance data and the sample grouping was analyzed using Analysis of Variance (ANOVA). OTUs that were significantly overrepresented in seawater, drone-captured air or specific for whale blow samples were defined using ANOVA with the ‘p.uni = “adjusted”' option. OTUs were classified against the Silva 123 release database (Quast et al., 2013) using mothur “classify.seqs” with default parameters (v1.36.1, Schloss et al., 2009).
Identifying Bacteria Isolated from Agar Plates
Bacterial 16S rRNA genes were directly amplified from cell suspensions obtained from colony picks using conserved primers 27F and 519R (Lane et al., 1985; Lane, 1991). PCR amplifications consisted of 1.0 μl of template and cycle specific for 16S consisted of 95°C for 10 min, 94°C for 30 s, 55°C for 10 s, 72°C for 45 s and 72°C for 10 min, and Taq DNA Polymerase (Qiagen). Amplified DNA was prepared for Sanger sequencing using Agencourt®AMPure® XP beads (Beckman Coulter). Sequences were trimmed to q20, and classified against the Silva Database (version 123).
Results
A total of 74 flights were conducted over 4 days of sampling. Each pod was considered independent as all whales were on their annual northern migration (Pirotta et al., 2016). Overall, 59 successful samples were collected from at least 48 different whales (11 whales were sampled but not identified via video due to occasional failure of the GoPro® camera e.g., low battery or maximum storage capacity reached). Sample volume varied between 50 and 150 μl of exhaled breath. The average opening time of the flip lid was 4 s (min 2 s, max 6 s). The UAV had a maximum flight time (battery time) of 15 min and sampling attempts on average were 4 min 28 s long (range: 27 s to 7 min). The majority of flight time was used to search for the whale's next surfacing position. The time that the UAV was in close proximity to a whale (UAV approximately within 5 m horizontal distance) varied but was on average 53 s (range: 2 s to 2.36 min or 141 s). The most number of samples collected in 1 day was 38. In all cases, there was no behavioral response to the drone (level 1, n = 48). Twice there were strong social interactions that occurred prior to the drone approaching the whales (one tail slap, one breach) but sampling was continued on the group in each case and samples successfully collected.
Next Generation Sequencing Results
A total of 7.62 million filtered bacterial 16S ribosomal gene sequences were produced from 59 UAV-captured whale blow and six air samples. These were combined with 0.91 m sequences generated from 26 seawater samples to generate bacterial OTU abundance profiles. Distance-based clustering of blow, air or seawater bacterial community profiles defined at least three significant clusters (simprof, P < 0.05), encompassing one group exclusively composed of seawater, one group exclusively composed of whale blow samples and a third group which clustered the six air samples along with 11 whale-blow samples (Figure 3A). Whale blow samples in this group may correspond to UAV sorties that missed, or narrowly missed, capturing whale blow material and were highly correlated with low capture scores based on a visual score of the amount of whale material recovered (Supplementary Table 1).
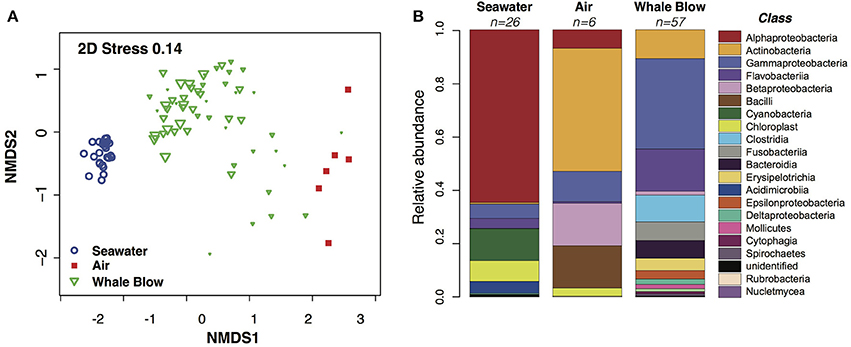
Figure 3. Similarity analysis of Operational Taxonomic Units (OTUs) abundance profiles and comparison of bacterial Classes identified in sampled whale blow, air and seawater. (A) Non-metric multidimensional scaling plot of bacterial OTU abundance profiles. The size of each whale plotting character is scaled to a visual score of the amount of whale blow captured on the petri dish (e.g., bigger the triangle, greater amount of sample). OTUs were defined at 97% nucleotide identity. (B) Relative abundance of taxonomic classes identified as whale-, air- or seawater-specific in each sample type.
Bacterial OTUs correlated with seawater, whale blow or air samples were identified using Analysis of Variance (ANOVA) based on generalized linear models fit to the data (Wang et al., 2012). OTU diversity and abundance profiles for air and whale blow were significantly different (p < 0.05) from each other and bear little similarity with communities characteristic of the adjacent seawater. At the Class level whale blow bacteria were dominated by Gammaprotobacteria, Flavobacteriia, Clostridia and Fusobacteria, in contrast to seawater communities, where species composition reflected values typical for sub-tropical waters of the Tasman Sea, i.e., ~60% Alphaproteobacteria, 15% Cyanobacteria and smaller proportions of Gammaproteobacteria and Flavobacteriia (Figure 3B; Seymour et al., 2012).
Overall, whale blow samples displayed the greatest OTU diversity, followed by seawater and air (Supplementary Figure 1). Model-based multivariate analyses identified 198 OTUs that were seawater-specific and 35 OTUs that were significantly correlated with air samples (ANOVA, P < 0.1; Supplementary Tables 3, 4). Successfully collected whale blow samples contained a small proportion seawater and air-specific OTUs, contributing on average 15.7(±10.8)% and 11.5(±4.4)%, respectively, of total sequences. The proportion of air-specific and seawater OTUs in near-miss samples was significantly higher (41.0% and 24.1%, respectively). Subtraction of seawater and air specific OTUs from the total enabled us to define 129 OTUs that were highly specific to whale samples (ANOVA, P < 0.05, Figure 4, Supplementary Table 5). Abundant bacterial species identified as whale-blow-specific include multiple OTUs belonging to the genera Cardiobacteriaceae and species Tenacibaculum, followed by OTUs related to Pseudomonas sp. Strain wp33, Leptotrichia sp. and Corynebacteria spp. While these analyses identified which OTUs were highly specific for whale, air and seawater, an addition set of whale-related OTUs could be identified in the remaining non-significant OTUs. We used the following criteria: present in greater than five whales and >100 sequences, to add an additional 145 OTUs that were highly specific to whales but found only in a small proportion of the sampled whale population (5–17 individuals, out of a total of 57) (Supplementary Table 6). Many of the OTUs in this group are closely related to whale-specific OTUs at the genus and species levels, e.g., Cardiobacteriaceae, Tenacibaculum, and Fusibacter strains. However, potential respiratory pathogens were also detected, such as Balneatrix (Gammaproteobacteria), and a range of Gram positive Clostridia and Bacilli, such as Staphylococcus and Streptococcus. In the context of monitoring whale respiratory health, potential pathogens may be present in a subset of the population only. OTUs in this whale-associated group were present in low abundance, and on average constituted 13(±5.7)% of the total sequences detected in each whale sample.
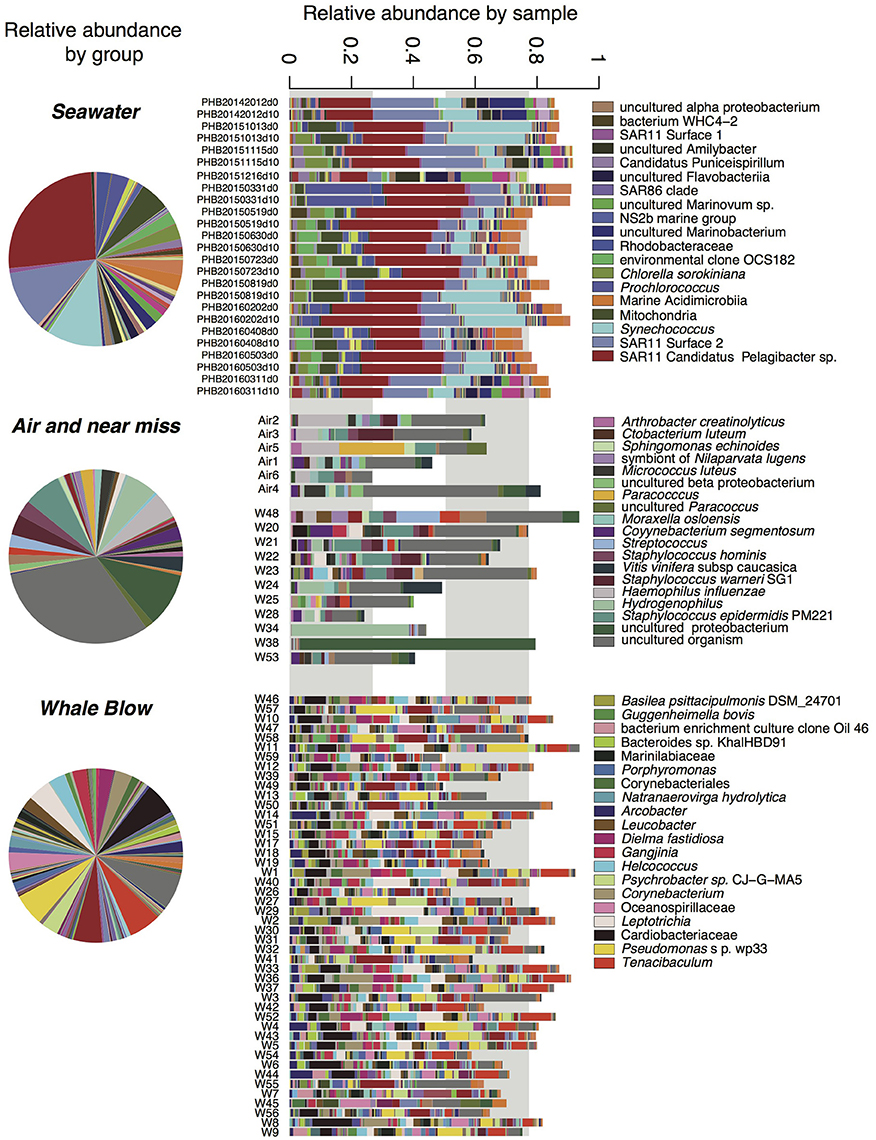
Figure 4. Relative abundance of bacterial taxa identified in seawater, UAV captured air and whale blow. OTUs with abundance <9 across the entire dataset were omitted for clarity. Relative abundances are presented for each group (i.e., seawater, air plus “near-miss” samples and whales, as well as for each sample. Taxa names correspond to the highest taxonomic level identification, full taxonomies are present in Supplementary Tables 3–6) only the top taxa by abundance are shown in the legend.
Comparison with Culture-Dependent Identification of Whale Blow Microbiota
Bacterial growth was observed on 48 UAV-mounted agar plates exposed to whale blow. Unexposed control plates displayed no bacterial growth. Sequencing of rRNA genes amplified from single colonies identified 18 different bacteria taxa isolated from 19 different whales (Supplementary Table 7). Overall, the most common bacteria identified at the phylum level included Proteobacteria (n = 7), Firmicutes (n = 7) and Actinobacteria (n = 4). Two samples were identified to the family level, Brucellaceae (n = 1) and Microbacteriaceae (n = 1). At the genus level, Micrococcus (n = 3), Acidovorax (n = 3), Bacillus (n = 3), Enterobacteriaceae (n = 2), Paenibacillus (n = 2), Streptococcus (n = 2), and Staphylococcus (n = 2) were most common. Seven whales had more than one bacterium identified. Staphylococcus was identified in both an individual sampled via our UAV.
Discussion
UAVs are rapidly transforming the way scientists collect information on their study species (Christie et al., 2016; Lowman and Voirin, 2016; Nowacek et al., 2016; Duffy et al., 2017). In whale research, UAVs have enabled sampling methods to be refined and have eliminated the need for close vessel approaches. To our knowledge, this study is the first to successfully demonstrate the use of a purpose-built UAV designed to sample humpback whale blow in Southern Hemisphere waters. The minimal behavioral disturbance observed suggests this method is an excellent, low-impact alternative to pole sampling methods for large, migrating whales. Humpback whales may have been aware of the UAV and did not react or, mostly likely, were not even aware of the UAV's presence. Underwater noise generated from the UAV was likely to be very low level at the heights flown (< 10 m), as it is smaller, lighter and has a lower disc loading than comparable off-the-shelf UAVs shown to transmit minimal noise transmission underwater (e.g., SwellPro Splashdrone and the DJI Inspire 1 Pro) (Christiansen et al., 2016). The combination of the waterproof design and the remotely operated flip lid petri dish designed to minimize airborne contamination, is a significant improvement over existing UAV types.
Our results demonstrate that whale blow can be effectively sampled while minimizing species associated with likely sources of contamination, i.e., air and seawater, to define microbes specifically associated with whales. Amplification of DNA extracted from UAV-captured air highlights the sensitivity of PCR-based approaches for detecting microbiota, even from low amounts of extracted DNA, while also demonstrating the sensitivity of this approach to contamination from external sources. The development of a flip-lid sampling system using sterile petri-dishes enabled us to effectively reduce contamination from typical seawater bacteria, which may exist in aerosols above the sea surface. While the presence of abundant seawater species (Alphaproteobacteria SAR11 and cyanobacteria) in air and whale blow samples is not surprising, the source of some major species detected in air samples is less clear. Some of the most abundant species detected in air samples, Propionobacteria, Arthrobacter, and Staphylococcus, are common commensal organisms of mammalian (human) skin and nasal cavities (Human Microbiome Project Consortium, 2012; Prussin and Marr, 2015). A potential source of some non-marine material may have been contamination during the DNA extraction or amplification procedure, especially when the amount of captured material was low (i.e., for air or near-miss samples). In the context of developing indicators of whale health the presence or absence of species that are common in humans should be interpreted cautiously. Nevertheless, in the UAV-sampled blow where a sufficient amount of material was collected, our analyses indicate that ~70% of the total sequences were specific to whales, a group of whale associated sequences accounted for a further ~12% and the remainder could be confidently identified as seawater- or air-specific.
To our knowledge this is the first study to utilize a long-term seawater dataset to identify and subtract seawater bacteria from community profiles of field-captured mammalian samples. The seawater data provided a comprehensive, temporal assessment of the composition of microbial communities present in sea water off Sydney. Critically, a much larger quantity of seawater was collected (2 L) and analyzed in comparison to the whale samples. This method minimized the impact of external sources of contamination and allowed for the greater coverage of the seawater community diversity. We used this resource to filter out all sequences characteristic of seawater to produce a whale blow dataset that could be used as a diagnostic for whale health. The distinct differences observed between statistically-defined bacteria in whale, sea water and air samples indicates that this method was effective for collecting whale microbiota with minimal contamination.
The successful collection of bacterial DNA in this study provides baseline information of microbiota found in migrating humpback whale blow. Due to the infancy of sampling whale breath as an assessment of whale health (Acevedo-Whitehouse et al., 2010; Hunt et al., 2013), it is not clear as to the type of microflora/bacteria species that are considered ‘normal' for northward migrating humpback whales off Sydney. Despite this, there are similarities in our collection of bacterial genera from the few studies that have collected blow for the assessment of microbiota (Acevedo-Whitehouse et al., 2010; Denisenko et al., 2012; Hunt et al., 2013). For example, Streptococcus and Staphylococcus genera were detected in our samples and have been detected in the blow of blue whales (Balaenoptera musculus), gray whales (Eschrichtius robustus) and Southern resident killer whales (Acevedo-Whitehouse et al., 2010; Denisenko et al., 2012; Hunt et al., 2013; Raverty et al., 2017). Bacteria from the Streptococcus genus is common in mucous membranes of animals (and humans) and is known to be found in the upper respiratory tract (Krzyściak et al., 2013). Streptococcus bacteria has previously been responsible for pneumonia causing death in cetaceans (Acevedo-Whitehouse et al., 2010). Bacillus sp. was also identified via blow collection from western North Pacific gray whales and Southern resident killer whales (Denisenko et al., 2012; Hunt et al., 2013; Raverty et al., 2017).
Next generation sequencing identified Cardiobacteriaceae (family) and Tenacibaculum (genus) to be the most abundant bacterial rRNA genes in whale blow. Cardiobacteriaceae has previously been isolated as a dominant taxa in the respiratory system of “healthy” captive bottlenose dolphins (Tursiops aduncus and, T. truncates) and free-ranging species (T. truncates) (Johnson et al., 2009; Lima et al., 2012). These findings may indicate that these genes are part of the normal microflora of dolphins, whilst presence in whales until now was unknown. Cardiobacteriaceae are abundant on humpback whale skin (Gammaproteobacteria genus), as is Tenacibaculum (Apprill et al., 2011, 2014). It may be possible that bacteria found on whale skin also occur within the respiratory tract or epithelial cells. Tenacibaculum has been associated with the microbiome of other marine species such as southern bluefin tuna (Thunnus maccoyii castelnau) (Valdenegro-Vega et al., 2013), while Psychrobacter is part of the thresher shark and rainbow trout skin microbiome (Lowrey et al., 2015; Doane et al., 2017).
The collection of bacterial microbiota is as an indicator of cetacean health is growing (Hogg et al., 2009; Schroeder et al., 2009; Acevedo-Whitehouse et al., 2010; Lima et al., 2012; Hunt et al., 2013; Nelson et al., 2015; Raverty et al., 2017). We were able to sample a number of individuals from a single population over a very short time frame. The use of the waterproof GoPro® camera made identification of different individuals reliable and therefore reduced repeated sampling. Our remotely operated “flip dish” design proved effective at reducing possible contamination from the pilot/research team (e.g., breath, touch, clothing) and vessel vapor/fumes. The placement of Parafilm® around the dish after sampling ensured that the sample remained unexposed until back in the laboratory for processing. Recently published work by Burgess et al. (2016) found polystyrene dishes (petri dish) to be the most effective surface for sampling whale blow in comparison to other sampling materials like veil nylon and nitex nylon mesh. In addition, the use of ice chilling of our samples for temporary storage was also consistent with Burgess et al. (2016), which found storage in cooler box with ice packs was appropriate for preserving samples (at least for hormones) for daylong fieldwork at sea (<6 h). Our samples only contained a fine mist [we estimated between 50 and 150 μL per sample, similar to amounts collected by Hogg et al. (2009)], and so we were unable to directly pipette samples but we found that swabbing the non-agar lid of the petri dishes to be effective. Variability in blow sample volumes appear to be a common issue (Hogg et al., 2009; Acevedo-Whitehouse et al., 2010) and therefore the need for repeated sampling is recommended. Sample success increased with effort/experience and we recommend effort be made early in any study to improve pilot skill, sample collection, quality and quantity.
While overall highly successful, UAVs still require a high level of skill and effort. Predicting when the whale is about to surface, positioning the UAV and opening the petri dish in time remains challenging. This may be complicated when a whale comes to the surface to breath but does not respire forcefully. When this happens, the plate is exposed to the air and so the UAV must return to the boat so the petri dish can be exchanged, our miss/near-miss rate was 11/59 = 20%. Second, not using an off-the-shelf product requires a high level of UAV competence both to fly and to fix problems as they arise. Third, the flight time for this UAV is 15 min, restricting the number of opportunities for sampling before the UAV must return to the vessel in order to replace the battery. Flight time will increase as battery technology progresses (Nowacek et al., 2016).
Our dataset details the diversity and abundance of the microbiota found in a migrating whale population which provides the baseline to identify pathogenic species. Ultimately, the isolation of pathogens from healthy or diseased animals will be an important step toward understanding the causes of disease and the factors that contribute to virulence. Culture-dependent techniques remain a viable option for the surveillance of pathogens in populations. In this study, nutrient agar was an effective way of culturing a subset of whale blow microbiota, including species commonly associated with respiratory disease in mammals. The use of both sides of the petri dish effectively doubled the chance of obtaining bacterial samples. While next generation sequencing has the capacity to probe the diversity of whale blow microbiota, at present, the isolation and identification bacteria from agar plates can be achieved within 3–5 days, compared to a practical timeframe of weeks for illumina sequencing. Selective media could be used to target potential pathogens in conjunction with opportunistic sampling of diseased or distressed animals.
Conclusions
Our purpose-built UAV proved highly successful in sampling whale blow for microbial community analysis. It is cost-effective, has low risk of contamination and greatly reduces disturbance of whales. Future applications include other free-ranging whale species (e.g., southern right whales, Eubalaena australis), as well as sampling smaller cetaceans (e.g., dolphins). Our UAV is useful addition to the conservation scientist's tool box, enabling collection of health information and therefore the ability to monitor changes in individual health as populations recover and to provide an early warning system for potential future changes.
Author Contributions
Paper conception: VP, AS, RH, MO, IJ, and AG. Experiment design: VP, AS, RH, and IJ. Field work: VP, AS, and RH. Laboratory work: VP, DR, and MO. Analysis and interpretation of data: VP, MO, RH, IJ, AG, and DR. Wrote paper: VP, MO, RH, IJ, AG, DR, and AS.
Funding
This project was supported by Macquarie University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was conducted under the Macquarie University Animal Ethics Committee Animal Research Authority 2016/010 and the NSW Office of Environment and Heritage Scientific Research permit SL101743. VPirotta was supported by an Australian Postgraduate Award Research scholarship. Molecular work was supported by the Australian Research Council grant DP15102326. Thank you to Guy Alexander from Heliguy for providing the legal requirements for the commercial UAV operations and workshop resources for building and testing the UAVs. We would like to thank Roads and Maritime Services, NSW and Dean Cropp from Barefoot Charters for their support in this research project. We appreciate their time and use of vessels for sampling. We thank Oliver Masens, Jemma Geoghegan, Adam Wilkins, Gemma Carroll and Ben Pitcher for their assistance in the field. We thank the Cape Solander Whale Migration Study, in particularly Wayne Reynolds, Sue Rennie Wright and Mark McGeachie. Thank you Duan March from Dolphin Marine Magic and Shona Lorigan from ORRCA for their collection and transport of blow sample from the stranded humpback whale in Sawtell beach, New South Wales. We thank Patrick da Roza for assistance in the laboratory. Thank you to Sally Browning and Airpig Productions for their donation of UAV batteries and Finn Lewis for providing UAV components. Monthly sampling at the Port Hacking Reference Station was supported by the Integrated Marine Observing System (IMOS). We would like to acknowledge the contribution of the Marine Microbes Project consortium (https://data.bioplatforms.com/organization/pages/bpa-marine-microbes/consortium) in the generation of data used in this publication. The Marine Microbes Project is supported by funding from Bioplatforms Australia. Bioplatforms and IMOS are supported through the Australian Government National Collaborative Research Infrastructure Strategy (NCRIS).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2017.00425/full#supplementary-material
References
Acevedo-Whitehouse, K., Rocha-Gosselin, A., and Gendron, D. (2010). A novel non-invasive tool for disease surveillance of free-ranging whales and its relevance to conservation programs. Anim. Conserv. 13, 217–225. doi: 10.1111/j.1469-1795.2009.00326.x
Allendorf, F. W., Hohenlohe, P. A., and Luikart, G. (2010). Genomics and the future of conservation genetics. Nat. Rev. Genet. 11, 697–709. doi: 10.1038/nrg2844
Anderson, K., and Gaston, K. J. (2013). Lightweight unmanned aerial vehicles will revolutionize spatial ecology. Front. Ecol. Environ. 11, 138–146. doi: 10.1890/120150
Apprill, A., Mooney, T. A., Lyman, E., Stimpert, A. K., and Rappé, M. S. (2011). Humpback whales harbour a combination of specific and variable skin bacteria. Environ. Microbiol. Rep. 3, 223–232. doi: 10.1111/j.1758-2229.2010.00213.x
Apprill, A., Robbins, J., Eren, A. M., Pack, A. A., Reveillaud, J., Mattila, D., et al. (2014). Humpback whale populations share a core skin bacterial community: towards a health index for marine mammals? PLoS ONE 9:e90785. doi: 10.1371/journal.pone.0090785
Bissett, A., Fitzgerald, A., Meintjes, T., Mele, P. M., Reith, F., Dennis, P. G., et al. (2016). Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database. Gigascience 5:21. doi: 10.1186/s13742-016-0126-5
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burgess, E. A., Hunt, K. E., Kraus, S. D., and Rolland, R. M. (2016). Get the most out of blow hormones: validation of sampling materials, field storage and extraction techniques for whale respiratory vapour samples. Conserv. Physiol. 4:cow024. doi: 10.1093/conphys/cow024
Chittleborough, R. G. (1965). Dynamics of two populations of the humpback whale, Megaptera novaeangliae (Borowski). Mar. Freshw. Res. 16, 33–128. doi: 10.1071/MF9650033
Christiansen, F., Rojano-Do-ate, L., Madsen, P. T., and Bejder, L. (2016). Noise levels of multi-rotor unmanned aerial vehicles with implications for potential underwater impacts on marine mammals. Front. Mar. Sci. 3:277. doi: 10.3389/fmars.2016.00277
Christie, K. S., Gilbert, S. L., Brown, C. L., Hatfield, M., and Hanson, L. (2016). Unmanned aircraft systems in wildlife research: current and future applications of a transformative technology. Front. Ecol. Environ. 14, 241–251. doi: 10.1002/fee.1281
Cuyler, L., Wiulsrød, R., and ØRitsland, N. A. (1992). Thermal infrared radiation from free living whales. Mar. Mamm. Sci. 8, 120–134. doi: 10.1111/j.1748-7692.1992.tb00371.x
De Mello, D. M. D., and De Oliveira, C. A. (2016). Biological matrices for sampling free-ranging cetaceans and the implications of their use for reproductive endocrine monitoring. Mamm. Rev. 46, 77–91. doi: 10.1111/mam.12055
Denisenko, T., Sokolova, O., and Vertyankin, V. (2012). “Microbiology investigation of blow samples of gray whale (Eschrichtius robustus) as one way of estimating the health status of a population,” in 26th European Cetacean Society Conference (Galway).
Doane, M. P., Haggerty, J. M., Kacev, D., Papudeshi, B., and Dinsdale, E. A. (2017). The skin microbiome of the Common thresher shark (Alopias vulpinus) has low taxonomic and potential metabolic β-diversity. Environ. Microbiol. Rep. 9, 357–373. doi: 10.1111/1758-2229.12537
Duffy, J. P., Cunliffe, A. M., DeBell, L., Sandbrook, C., Wich, S. A., Shutler, J. D., et al. (2017). Location, location, location: considerations when using lightweight drones in challenging environments. Remote Sens. Ecol. Conserv. doi: 10.1002/rse2.58. [Epub ahead of print].
Durban, J., Fearnbach, H., Barrett-Lennard, L., Perryman, W., and Leroi, D. (2015). Photogrammetry of killer whales using a small hexacopter launched at sea. J. Unmanned Vehicle Syst. 3, 131–135. doi: 10.1139/juvs-2015-0020
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fiori, L., Doshi, A., Martinez, E., Orams, M. B., and Bollard-Breen, B. (2017). The use of unmanned aerial systems in marine mammal research. Remote Sens. 9:543. doi: 10.3390/rs9060543
Geraci, J. R., and Lounsbury, V. J. (2005). Marine Mammals Ashore: A field Guide for Strandings, 2nd Edn. Baltimore, MA: National Aquarium in Baltimore.
Goebel, M. E., Perryman, W. L., Hinke, J. T., Krause, D. J., Hann, N. A., Gardner, S., et al. (2015). A small unmanned aerial system for estimating abundance and size of Antarctic predators. Polar Biol. 38, 619–630. doi: 10.1007/s00300-014-1625-4
Gonzalez, L. F., Montes, G. A., Puig, E., Johnson, S., Mengersen, K., and Gaston, K. J. (2016). Unmanned aerial vehicles (UAVs) and artificial intelligence revolutionizing wildlife monitoring and conservation. Sensors 16:97. doi: 10.3390/s16010097
Gulland, F. M., and Hall, A. J. (2007). Is marine mammal health deteriorating? trends in the global reporting of marine mammal disease. EcoHealth 4, 135–150. doi: 10.1007/s10393-007-0097-1
Hahn, N., Mwakatobe, A., Konuche, J., de Souza, N., Keyyu, J., Goss, M., et al. (2017). Unmanned aerial vehicles mitigate human–elephant conflict on the borders of tanzanian parks: a case study. Oryx 51, 513–516. doi: 10.1017/S0030605316000946
Harcourt, R., Turner, E., Hindell, M., Waas, J., and Hall, A. (2010). Effects of capture stress on free-ranging, reproductively active male Weddell seals. J. Comp. Physiol. A 196, 147–154. doi: 10.1007/s00359-009-0501-0
Harvey, M., Rowland, J., and Luketina, K. (2016). Drone with thermal infrared camera provides high resolution georeferenced imagery of the Waikite geothermal area, New Zealand. J. Volcanol. Geothermal Res. 325, 61–69. doi: 10.1016/j.jvolgeores.2016.06.014
Hodgson, A., Kelly, N., and Peel, D. (2013). Unmanned aerial vehicles (UAVs) for surveying marine fauna: a dugong case study. PLoS ONE 8:e79556. doi: 10.1371/journal.pone.0079556
Hogg, C., Vickers, E., and Rogers, T. (2005). Determination of testosterone in saliva and blow of bottlenose dolphins (Tursiops truncatus) using liquid chromatography–mass spectrometry. J. Chromatogr. B 814, 339–346. doi: 10.1016/j.jchromb.2004.10.058
Hogg, C., Rogers, T., Shorter, A., Barton, K., Miller, P., and Nowacek, D. (2009). Determination of steroid hormones in whale blow: it is possible. Mar. Mamm. Sci. 25, 605–618. doi: 10.1111/j.1748-7692.2008.00277.x
Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486:207. doi: 10.1038/nature11234
Hunt, K. E., Moore, M. J., Rolland, R. M., Kellar, N. M., Hall, A. J., Kershaw, J., et al. (2013). Overcoming the challenges of studying conservation physiology in large whales: a review of available methods. Conserv. Physiol. 1, 1–24. doi: 10.1093/conphys/cot006
Hunt, K. E., Rolland, R. M., and Kraus, S. D. (2014). Detection of steroid and thyroid hormones via immunoassay of North Atlantic right whale (Eubalaena glacialis) respiratory vapor. Mar. Mamm. Sci. 30, 796–809. doi: 10.1111/mms.12073
Hussey, N. E., Kessel, S. T., Aarestrup, K., Cooke, S. J., Cowley, P. D., Fisk, A. T., et al. (2015). Aquatic animal telemetry: a panoramic window into the underwater world. Science 348:1255642. doi: 10.1126/science.1255642
Johnson, W. R., Torralba, M., Fair, P. A., Bossart, G. D., Nelson, K. E., and Morris, P. J. (2009). Novel diversity of bacterial communities associated with bottlenose dolphin upper respiratory tracts. Environ. Microbiol. Rep. 1, 555–562. doi: 10.1111/j.1758-2229.2009.00080.x
Koh, L., and Wich, S. (2012). Dawn of drone ecology: low-cost autonomous aerial vehicles for conservation. Trop. Conserv. Sci. 5, 121–132. doi: 10.1177/194008291200500202
Krzyściak, W., Pluskwa, K., Jurczak, A., and Kościelniak, D. (2013). The pathogenicity of the Streptococcus genus. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1361–1376. doi: 10.1007/s10096-013-1914-9
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 82, 6955–6959. doi: 10.1073/pnas.82.20.6955
Lane, D. (1991). “16S/23S rRNA sequencing. nucleic acid techniques in bacterial systematics,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester: Wiley), 115–175.
Lima, N., Rogers, T., Acevedo-Whitehouse, K., and Brown, M. V. (2012). Temporal stability and species specificity in bacteria associated with the bottlenose dolphins respiratory system. Environ. Microbiol. Rep. 4, 89–96. doi: 10.1111/j.1758-2229.2011.00306.x
Linchant, J., Vermeulen, C., Lisein, J., Lejeune, P., and Bouche, P. (2013). “Using drones to count the elephants: a new approach of wildlife inventories,” in Paper presented at the 31st Congress of the International Union of Game Biologists (Brussels).
Linchant, J., Lisein, J., Semeki, J., Lejeune, P., and Vermeulen, C. (2015). Are unmanned aircraft systems (UASs) the future of wildlife monitoring? a review of accomplishments and challenges. Mammal Rev. 45, 239–252. doi: 10.1111/mam.12046
Lowman, M., and Voirin, B. (2016). Drones – our eyes on the environment. Front. Ecol. Environ. 14, 231–231. doi: 10.1002/fee.1290
Lowrey, L., Woodhams, D. C., Tacchi, L., and Salinas, I. (2015). Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl. Environ. Microbiol. 81, 6915–6925. doi: 10.1128/AEM.01826-15
Marks, P. (2014). Elephants and rhinos benefit from drone surveillance. New Sci. 221:24. doi: 10.1016/S0262-4079(14)60323-7
Mulero-Pázmány, M., Stolper, R., Van Essen, L., Negro, J. J., and Sassen, T. (2014). Remotely piloted aircraft systems as a rhinoceros anti-poaching tool in Africa. PLoS ONE 9:e83873. doi: 10.1371/journal.pone.0083873
Nelson, T. M., Apprill, A., Mann, J., Rogers, T. L., and Brown, M. V. (2015). The marine mammal microbiome: current knowledge and future directions. Microbiol. Austr. 36, 8–13. doi: 10.1071/MA15004
Nowacek, D. P., Christiansen, F., Bejder, L., Goldbogen, J. A., and Friedlaender, A. S. (2016). Studying cetacean behaviour: new technological approaches and conservation applications. Anim. Behav. 120, 235–244. doi: 10.1016/j.anbehav.2016.07.019
Oksanen, J. (2017). Package ‘Vegan: An Introduction to Ordination' R Package Version (2017-08-24 r73119). Available online at: http://cran.polytechnic.edu.na/web/packages/vegan/vignettes/intro-vegan.pdf
Pirotta, V., Slip, D., Jonsen, I. D., Peddemors, V. M., Cato, D. H., Ross, G., et al. (2016). Migrating humpback whales show no detectable response to whale alarms off Sydney, Australia. Endanger. Species Res. 29, 201–209. doi: 10.3354/esr00712
Pomeroy, P., O'Connor, L., and Davies, P. (2016). Assessing use of and reaction to unmanned aerial systems in gray and harbor seals during breeding and molt in the UK. J. Unmanned Vehicle Syst. 3, 102–113. doi: 10.1139/juvs-2015-0013
Prussin, A. J., and Marr, L. C. (2015). Sources of airborne microorganisms in the built environment. Microbiome 3:78. doi: 10.1186/s40168-015-0144-z
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ratcliffe, N., Guihen, D., Robst, J., Crofts, S., Stanworth, A., and Enderlein, P. (2015). A protocol for the aerial survey of penguin colonies using UAVs. J. Unmanned Vehicle Syst. 3, 95–101. doi: 10.1139/juvs-2015-0006
Raverty, S. A., Rhodes, L. D., Zabek, E., Eshghi, A., Cameron, C. E., Hanson, M. B., et al. (2017). Respiratory microbiome of endangered southern resident killer whales and microbiota of surrounding sea surface microlayer in the Eastern North Pacific. Sci. Rep. 7:394. doi: 10.1038/s41598-017-00457-5
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schofield, G., Katselidis, K. A., Lilley, M. K., Reina, R. D., and Hays, G. C. (2017). Detecting elusive aspects of wildlife ecology using drones: new insights on the mating dynamics and operational sex ratios of sea turtles. Funct. Ecol. 31, 2310–2319. doi: 10.1111/1365-2435.12930
Schroeder, P., Raverty, S., Cameron, C., Zabek, E., Eshghi, A., Bain, D., et al. (2009). Investigation into the Microbial Culture and Molecular Screening of Exhaled Breaths of Endangered Southern Resident Killer Whales (SRKW) and Pathogen Screening of the Seasurface Microlayer (SML) in Puget Sound. Report to the Northwest Fisheries Science Center, National Oceanic and Atmospheric Administration, Seattle, WA.
Seymour, J. R., Doblin, M. A., Jeffries, T. C., Brown, M. V., Newton, K., Ralph, P. J., et al. (2012). Contrasting microbial assemblages in adjacent water masses associated with the East Australian Current. Environ. Microbiol. Rep. 4, 548–555. doi: 10.1111/j.1758-2229.2012.00362.x
Smith, C. E., Sykora-Bodie, S. T., Bloodworth, B., Pack, S. M., Spradlin, T. R., and LeBoeuf, N. R. (2016). Assessment of known impacts of unmanned aerial systems (UAS) on marine mammals: data gaps and recommendations for researchers in the United States. J. Unmanned Vehicle Syst. 4, 1–14. doi: 10.1139/juvs-2015-0017
Szantoi, Z., Smith, S. E., Strona, G., Koh, L. P., and Wich, S. A. (2017). Mapping orangutan habitat and agricultural areas using Landsat OLI imagery augmented with unmanned aircraft system aerial photography. Int. J. Remote Sens. 38, 2231–2245. doi: 10.1080/01431161.2017.1280638
Thompson, L. A., Spoon, T. R., Goertz, C. E., Hobbs, R. C., and Romano, T. A. (2014). Blow collection as a non-invasive method for measuring cortisol in the beluga (Delphinapterus leucas). PLoS ONE 9:e114062. doi: 10.1371/journal.pone.0114062
Valdenegro-Vega, V., Naeem, S., Carson, J., Bowman, J., Tejedor del Real, J., and Nowak, B. (2013). Culturable microbiota of ranched southern bluefin tuna (Thunnus maccoyii castelnau). J. Appl. Microbiol. 115, 923–932. doi: 10.1111/jam.12286
van Andel, A. C., Wich, S. A., Boesch, C., Koh, L. P., Robbins, M. M., Kelly, J., et al. (2015). Locating chimpanzee nests and identifying fruiting trees with an unmanned aerial vehicle. Am. J. Primatol. 77, 1122–1134. doi: 10.1002/ajp.22446
van Gemert, J. C., Verschoor, C. R., Mettes, P., Epema, K., Koh, L. P., and Wich, S. (2014). “Nature conservation drones for automatic localization and counting of animals,” in Workshop at the European Conference on Computer Vision (Cham: Springer), 255–270.
Vermeulen, C., Lejeune, P., Lisein, J., Sawadogo, P., and Bouché, P. (2013). Unmanned aerial survey of elephants. PLoS ONE 8:e54700. doi: 10.1371/journal.pone.0054700
Wang, Y., Naumann, U., Wright, S. T., and Warton, D. I. (2012). mvabund–an R package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 3, 471–474. doi: 10.1111/j.2041-210X.2012.00190.x
Whitaker, D., and Christman, M. (2014). Package ‘Clustsig' R Package Version 1.1 2014. Available online at: https://cran.hafro.is/web/packages/clustsig/clustsig.pdf
Whittow, G. C. (1987). Thermoregulatory adaptations in marine mammals: interacting effects of exercise and body mass. A Rev. Mar. Mammal Sci. 3, 220–241. doi: 10.1111/j.1748-7692.1987.tb00165.x
Wich, S., Dellatore, D., Houghton, M., Ardi, R., and Koh, L. P. (2015). A preliminary assessment of using conservation drones for Sumatran orang-utan (Pongo abelii) distribution and density. J. Unmanned Vehicle Syst. 4, 45–52. doi: 10.1139/juvs-2015-0015
Keywords: UAV, UAS, drone, blow, humpback whale, microbiota, technology, conservation
Citation: Pirotta V, Smith A, Ostrowski M, Russell D, Jonsen ID, Grech A and Harcourt R (2017) An Economical Custom-Built Drone for Assessing Whale Health. Front. Mar. Sci. 4:425. doi: 10.3389/fmars.2017.00425
Received: 01 October 2017; Accepted: 12 December 2017;
Published: 21 December 2017.
Edited by:
Peter H. Dutton, National Oceanic and Atmospheric Administration (NOAA), United StatesReviewed by:
Nuno Queiroz, University of Porto, PortugalGail Schofield, Deakin University, Australia
Copyright © 2017 Pirotta, Smith, Ostrowski, Russell, Jonsen, Grech and Harcourt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vanessa Pirotta, dmFuZXNzYS5waXJvdHRhQGhkci5tcS5lZHUuYXU=
 Vanessa Pirotta
Vanessa Pirotta Alastair Smith
Alastair Smith Martin Ostrowski
Martin Ostrowski Dylan Russell3
Dylan Russell3 Ian D. Jonsen
Ian D. Jonsen Alana Grech
Alana Grech Robert Harcourt
Robert Harcourt