
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 18 October 2017
Sec. Marine Molecular Biology and Ecology
Volume 4 - 2017 | https://doi.org/10.3389/fmars.2017.00327
 Katia Jindrich1†
Katia Jindrich1† Kathrein E. Roper1†
Kathrein E. Roper1† Sussan Lemon1
Sussan Lemon1 Bernard M. Degnan1
Bernard M. Degnan1 Adam M. Reitzel2
Adam M. Reitzel2 Sandie M. Degnan1*
Sandie M. Degnan1*The circadian clock is a molecular network that coordinates organismal behavior and physiology with daily environmental changes in the day-night cycle. In eumetazoans (bilaterians + cnidarians), this network appears to be largely conserved, yet different from other known eukaryotic circadian networks. To determine if the eumetazoan circadian network has an older origin, we ask here whether orthologs comprising this network are expressed in a manner consistent with a role in regulating circadian patterns in a representative of an earlier-branching animal lineage, the sponge Amphimedon queenslandica. The A. queenslandica genome encodes orthologs of many eumetazoan circadian genes, including two cryptochrome genes that encode flavoproteins, three Timeout genes, and two PAR-bZIP and seven bHLH-PAS transcription factor genes. There is no apparent Cycle ortholog, although we can identify three closely related ARNT genes. Of the putative circadian genes, only AqPARa and AqCry2 have a consistent oscillating diurnal expression profile, and the rhythmic expression of both these genes is partially lost when the animals are exposed to constant light or darkness. Expression of the other putative circadian genes, in particular AqClock, is neither diurnally-oscillating nor light-dependent. AqPARa and AqCry2 are also temporally and spatially co-expressed throughout embryonic and larval development. Transcripts of these genes are enriched first in cells comprising the larval posterior pigment ring, which is a simple photosensory organ that is responsible for the negative phototactic behavior displayed by larvae, and subsequently in the larval epithelial and subepithelial layers. The combined findings of no clear Cycle ortholog and of PAR-bZIP and cryptochrome being the only orthologs expressed in a pattern consistent with a circadian role suggests that either (i) the ancestral metazoan circadian network was simpler than the eumetazoan network, or (ii) that this sponge has lost some components, as has occurred in some other animals such as Hydra.
Circadian clocks have evolved multiple times during eukaryotic evolution, allowing organisms to coordinate their physiology and behavior with predictable environmental rhythms (Bell-Pedersen et al., 2005; Foster and Kreitzman, 2005; Takahashi, 2017). These genome-encoded molecular timekeeping mechanisms are entrained by several environmental signals, the most common of which are light, temperature, and feeding cycles (Harmer et al., 2001; Rivas et al., 2015). They drive physiological and behavioral rhythms by producing oscillations in transcript or protein activation and degradation.
Broadly, circadian clocks comprise three main modules: (i) sensors that perceive the entrainment signals from the environment; (ii) molecular oscillators of transcriptional-translational feedback loops that maintain the clock pacing and transmit rhythmic signals to downstream components; and (iii) clock-controlled genes (CCGs) that coordinate circadian responses within cells (Harmer et al., 2001; Hardin, 2011; Reitzel et al., 2013; Takahashi, 2017). In animals, our understanding of the clock general circuitry largely draws from work on mammal and insect model species, which has revealed that the topology of the molecular oscillator is conserved across these divergent bilaterian lineages (Hardin, 2011; Takahashi, 2017). Specifically, two interconnected regulatory loops are organized around a pair of basic helix-loop-helix-Per-ARNT-Sim (bHLH-PAS) transcription factors, Clock and Cycle (Bmal1), that activate the transcription of other clock elements. A feed-forward loop, comprising transcription factors of the Nuclear Receptor (NR) and PAR basic leucine zipper (PAR-bZIP) families, directly activates or represses the transcription of Clock or Cycle. Combinations of Period, Timeless/Timeout, and/or cryptochrome genes form a negative feedback loop that regulates the degradation of CLOCK and CYCLE and, thus, their own repression. Within each feedback loop, key roles are performed by different genes depending on the phylum, and sometimes even the species.
Cnidarians, the sister phylum to the bilaterians, also possess most of the circadian genes present in bilaterians. The diurnal transcription profiles of many of these genes in the sea anemone Nematostella vectensis and the stony coral Acropora millepora suggest that the conserved topology of the animal circadian clock may pre-date the bilaterian-cnidarian split (Vize, 2009; Reitzel et al., 2010, 2013; Brady et al., 2011). To determine if the conserved eumetazoan clock has an older evolutionary origin, we sought first to identify orthologs of these genes in a marine sponge, Amphimedon queenslandica. As A. queenslandica is a representative of one of the oldest extant animal phyletic lineages (Porifera), those components of a sponge circadian clock that are shared with eumetazoans are likely to reflect a core clock present in the last common animal ancestor. It already has been reported that the A. queenslandica genome encodes two cryptochrome genes, at least one of which appears to drive photic behaviors in the larva (Rivera et al., 2012), as well as a suite of conserved bHLH-PAS and bZIPs transcription factors (Simionato et al., 2007; Jindrich and Degnan, 2016). Here, we report on the presence or absence of the remaining putative animal circadian genes in the A. queenslandica genome, so as to provide the first comprehensive list of circadian gene orthologs in a sponge. We then explore the expression of these genes under natural and modified light-dark cycles in both adults and larvae. We find that two genes, AqCRY2 and AqPARa, display temporally and spatially co-expressed patterns of transcription that are consistent with them having a role in the regulation of circadian rhythms in this marine sponge.
A. queenslandica candidate circadian genes were identified through BLAST searches of both the Aqu1 (Srivastava et al., 2010) and Aqu2.1 (Fernandez-Valverde et al., 2015) gene models, both of which are available at EnsemblMetazoa http://metazoa.ensembl.org/Amphimedon_queenslandica/Info/Index and also at http://amphimedon.qcloud.qcif.edu.au/index.html. We used publically-available bilaterian and cnidarian protein models of Timeless/Timeout, bHLH-PAS, cryptochrome, and PAR-bZIP families as queries. Sequences were aligned with MUSCLE and trimmed to conserved domains. Maximum likelihood (ML) analyses were carried out using RaxML (Stamatakis, 2006). The best-fit substitution matrix was determined using ProtTest (Abascal et al., 2005). Branch support was estimated by performing 1,000 bootstrap replicates. Phylogenetic trees were visualized with FigTree v1.4.
Transcript abundance of putative circadian genes was assessed using CEL-Seq data comprising 82 developmental samples spanning from early cleavage to adult (Anavy et al., 2014; Levin et al., 2016; NCBI GEO GSE54364). We averaged the biological replicates for each developmental stage, and all larval stages were grouped together. The mean normalized expression (counts per million mapped) was plotted with the standard error across the sponge developmental stages using GraphPad.
Adults were tagged and maintained in situ in the field at Shark Bay on Heron Island Reef, southern Great Barrier Reef. For tissue sampling for gene expression, 1 cm3 biopsies were removed from the outer layer of the sponge with a scalpel and immediately placed in RNA Later. Three adults were sampled over a period of 3 days in September (sunrise 5:34 a.m. and sunset 5:50 p.m. on first day of experiment) and 3 days in January (sunrise 5:12 a.m. and sunset 6:43 p.m. on first day of experiment), with sampling at 2 hourly intervals on day 1 (beginning at 2.00 a.m. to capture predawn expression), and 4 hourly intervals on day 3. Sponges were not sampled on day 2 so that they could recover. RNA Later samples were frozen and transferred to Brisbane where RNA was purified using Trizol (Sigma) according to manufacturer's recommendations; residual genomic DNA was removed with DNase I (Invitrogen). RNA was quantified using QuBit (Invitrogen) and cDNA reverse transcribed using Superscript III (Invitrogen) and pentadecamers and OligodT-15 (Stangegaard et al., 2006).
Using oligonucleotide primers to generate amplicons ranging from 100 to 250 bp for the circadian genes (Supplementary Table 1), we performed Real-time qPCR on a LightCycler 480 (Roche) platform using Lightcycler 480 SYBR green master mix I (Roche). Each sample was run in triplicate in qPCR experiments. An inter-run calibrator was included on each plate and was incorporated into the final analyses. Raw Cq values were imported into qBASEPlus software (Biogazelle) for analysis. The GeNorm analysis function was performed within qBASEPlus to select stable reference genes. Quality control on qPCR data included visual analyses of melt curves and default quality control settings in qBASEPlus. Calibrated and Normalized Relative Quantities (CNRQ) and their associated standard error were exported from qBASE and visualized using GraphPad.
To determine the effect of different light regimes on gene expression, adults were collected from the field and transferred and reared in aquaria for 7 days under an artificial 12:12 light:dark cycle and then exposed to either 12:12 light-dark or constant dark for 3 days prior to sampling. Biopsies were collected every 3 h over 36 h and transferred into RNA Later, as described above.
For larval gene expression experiments, 12 individual adults were placed in individual aquaria with larval capture traps fitted to a single output hose. Larvae released from individual sponges were collected from the traps at hourly intervals from 1 to 4 p.m. To follow gene expression under natural light, single cohorts (i.e., larva released within a single hour interval from a single mother) were kept in mesh reservoirs and clipped to the side of tanks so that they experienced identical environmental conditions to their maternal parent (Supplementary Table 2). Two to five individuals were sampled from each cohort every 30 to 60 min, depending on numbers. Larvae were immediately submerged in RNA later and stored until processing.
To follow gene expression under different light treatments, larvae were collected between 1 to 2 p.m. and immediately transferred to six-well plates. The plates were semi-submerged in flowing water from the reef, to mimic natural temperature fluctuations. Larvae were exposed to either natural light, constant dark or constant light (full spectrum LUMILUX T5 HE tube) for at least 1 h before the sampling started at 3 p.m., as described above. The light intensity under water was 120 Lux, as measured with a Li-COR LI—250A light meter, which corresponds to what animals exposed to natural light experienced at 5.30 p.m.
In all these studies AqCry2 and AqPARa expression levels were assessed by qPCR as described above.
A 704 bp fragment from the AqPARa coding sequence was amplified from cDNA using the primers (5′- CACAACACCGAGACTCCTCC−3′) and (5′-TCCACAGGGAGAGCGTCATA−3′) (Supplementary Figure 1). The fragment was cloned into pGEM TEasy (Promega) using the manufacturer's protocol and verified by sequencing using M13F and M13R primers. Digoxygenin (DIG)-labeled antisense RNA probes were transcribed from polymerase chain reaction (PCR) products using DIG RNA Labeling Mix (Roche) and T7 or SP6 Polymerase (Promega) following the manufacturer's instructions. Whole mount in situ hybridization analysis of embryonic and larval gene expression was carried out as previously described (Larroux et al., 2008). Whole-mount and sectioned samples were observed under an Olympus SZX7 or a Nikon eclipse Ti microscope with Nomarski optics (Olympus Australia Pty Ltd, Mt Waverly, VIC, Australia) and photographed with a Nikon Sight DS-U1 camera (Nikon Australia Pty Ltd, Lidcombe, Australia).
Through sequence similarity and phylogenetic analysis, combined with previously published analyses (Simionato et al., 2007; Rivera et al., 2012), we have identified A. queenslandica orthologs of multiple eumetazoan circadian genes (Table 1). There are two PAR-bZIPs, both of which cluster with cnidarian and bilaterian PAR orthologs, to the exclusion of the bZIP sister family C/EBP (Figure 1A). In particular, AqPARa appears to be more closely related than AqPARb to other metazoan PAR-bZIPs involved in circadian rhythms (Figure 1A). There are a total of seven bHLH-PAS family members, four of which have been reported elsewhere (Simionato et al., 2007), and all of which cluster with particular cnidarian and bilaterian orthologs (Figure 1B). Notably, we identify a single ortholog of the core circadian bHLH transcription factor Clock, but no definitive Cycle/BMAL1 ortholog, although there are three paralogs of the closely-related ARNT-like genes (Figure 1B; Simionato et al., 2007). We find no Period orthologs. There are three putative Timeless/Timeout family paralogs, all of which are most similar to Timeout (Figure 1C). This is consistent with Timeless originating along the bilaterian stem (Reitzel et al., 2010; Brady et al., 2011). Completing the circadian gene repertoire, we confirm the presence of two Type I cryptochromes, AqCyr1 and AqCry2, as previously reported (Rivera et al., 2012).
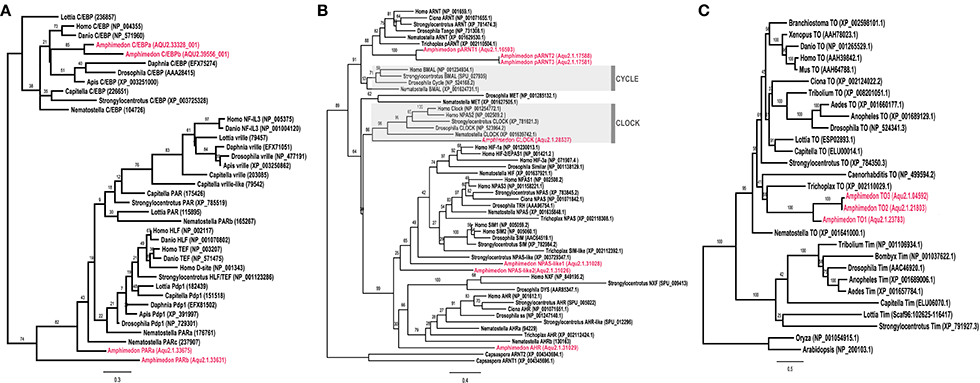
Figure 1. Phylogenetic identification of circadian gene orthologs in Amphimedon queenslandica. Maximum likelihood analysis of (A) PAR-bZIP, (B) bHLH-PAS, and (C) Timeless/Timeout genes from A. queenslandica and diverse representatives of main metazoan lineages. A. queenslandica orthologs are shown in red. Numbers above nodes indicate percentage of 1,000 boostraps. (A) The two AqPAR-bZIPs cluster with cnidarian and bilaterian PAR orthologs, to the exclusion of the bZIP sister family C/EBP. (B) AqbHLH-PAS sequences form several clusters with cnidarian and bilaterian orthologs. Members of the Clock and Cycle subfamilies are highlighted in gray. We identified a single Clock ortholog and no Cycle ortholog in A. queenslandica. Sequences from the unicellular holozoan Capsaspora were used as an outgroup. (C) We identified three genes in A. queenslandica from the Timeless/Timeout family, all of which were most similar to Timeout.
Using existing A. queenslandica developmental CEL-Seq data (Anavy et al., 2014; Levin et al., 2016), we determined the expression profiles of these putative circadian genes through the entire life cycle of this sponge. Embryogenesis occurs in the dark, in brood chambers located basally inside the maternal adult sponge (Degnan et al., 2008). Mature larvae emerge into the full sunlight between 12 and 4 p.m. (Figure 2), and subsequent settlement through to the first 12 h of metamorphosis typically occurs overnight in the dark (Degnan et al., 2015). Developmental expression data reveal that several putative circadian genes are upregulated in late embryos (ring and late ring stages), prior to the larva emerging from the adult. Specifically, the expression of AqClock, AqpARNT1, AqCry1, and AqCry2, and AqPARa starts increasing well before the larva emerges from the adult and, in most cases, expression peaks just after emergence (Figure 2). As larvae settle and initiate metamorphosis in the dark of the evening, the expression of AqNPAS-like1, AqAHR, and AqTO3 increases, and that of AqClock, AqCry 1 and 2, and AqPARa decreases (Figure 2). We also assayed the developmental expression of genes encoding other conserved bZIP and bHLH-PAS transcription factors, because of their potential to be involved in circadian regulation. Amongst those, only the AqC/EBPb expression profile could suggest an involvement in light-responsive processes, as expression increases when larvae emerge from the adult into the light (Figure 2).
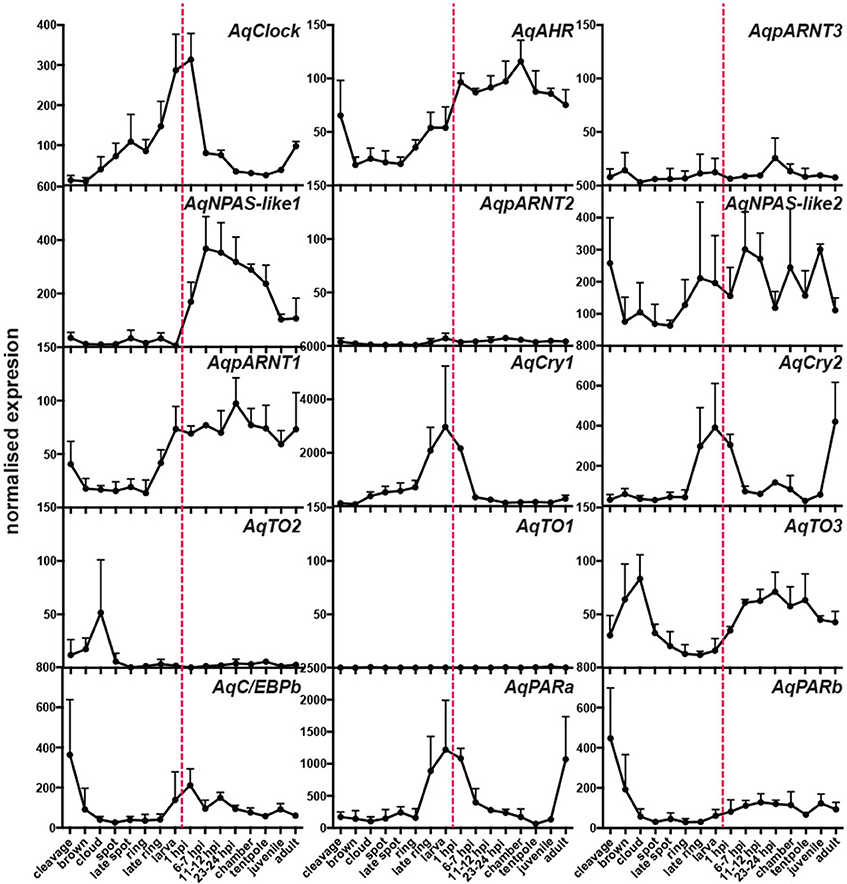
Figure 2. Developmental expression of putative circadian genes. Normalized expression profiles based on 82 CEL-Seq samples (Anavy et al., 2014; Levin et al., 2016) combined into 16 life cycle stages for each of 15 genes (labeled in top right corners of each graph). Cleavage to late ring are embryonic stages; larva (red dashed line); 1 h post-settlement (1 hps) to tentpole are metamorphic stages; juvenile; adult. Error bars are ± sem.
To determine if these putative A. queenslandica circadian genes are expressed in diurnal patterns akin to their eumetazoan orthologs, we quantitated transcript abundances in tissue biopsies procured from adult sponges living in situ on the reef flat. Among the full suite of circadian-like genes that we identified, only AqCry2 and AqPARa exhibited a consistent diurnal pattern of expression that is very similar between the two genes; other putative circadian genes displayed neither a diurnal pattern nor consistent expression profile between all individuals (Figure 3). In all of the individual sponges assayed in summer and spring, the transcript abundance of both AqCry2 and AqPARa increased markedly at dawn to a peak later in the morning, and then steadily decreased toward nightfall (Figure 3). In summer, both the timing and level of expression between 8 a.m. and 2 p.m. [Zeitgeber time (ZT) = 2–8 in spring, ZT = 2.5–8.5 in summer] were more variable and lower compared to the spring patterns, and both genes also had a second peak of smaller amplitude in the mid-afternoon (Figure 3).
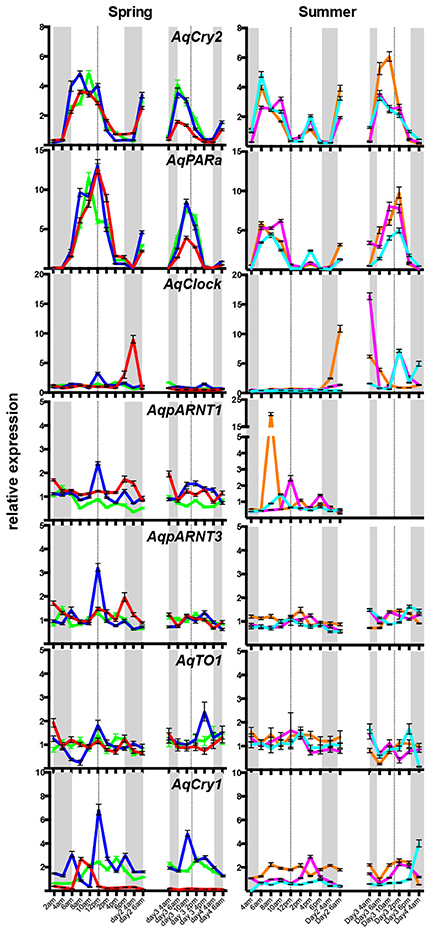
Figure 3. Diurnal expression of putative circadian genes in wild Amphimedon queenslandica in Spring and Summer. QPCR analysis of transcript abundance of three individuals on 2 separate days. Different sponges were sampled in the Spring and Summer. A different colored line represents a different individual. Dashed line, midday noon. Gray areas, dusk, and evening. Error bars are ± sem for technical replicates.
Under diurnal light conditions matching natural conditions on the reef flat, the pattern of AqCry2 and AqPARa expression in aquaria-maintained A. queenslandica (Figure 4A) closely matched that of wild sponges (Figure 3). This diurnal expression pattern generally continued when sponges in aquaria were subject to artificial 12:12 h light:dark cycle for 3 days, despite the light level being constant throughout the subjective day (Figure 4B). In adults maintained in aquaria under constant darkness for 3 days, expression of AqCry2 and AqPARa was generally lower than under 12:12 h light:dark at most times within a 24 h cycle, and was often significantly lower during the subjective day (Figure 4B). Nonetheless, both genes still displayed discernable diurnal oscillations in dark-entrained adult sponges, albeit far more muted compared to the natural cyclic patterns (Figure 4B).
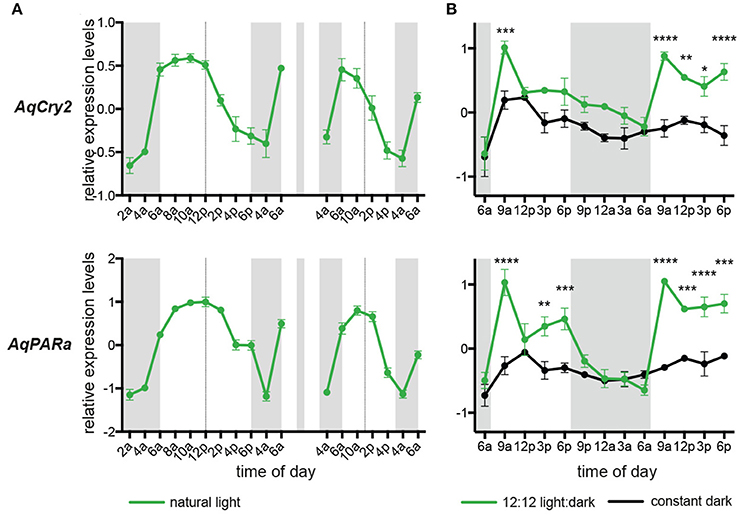
Figure 4. Expression of AqCry2 and AqPARa in adults maintained in aquaria. (A) Mean relative expression levels in aquarium-reared adult A. queenslandica reared for 7 days under a natural light:dark cycle (n = 3). Expression profiles at day 7 and 9. (B) Adults sponges subjected to 12:12 light:dark treatment (green line) or constant darkness (black line) for 3 days and sampled every 3 h over 2 days (n = 3). Asterisks indicate significant differences among treatments (** < 10–2; *** < 10–3, **** < 10–4). Error bars are ± sem for biological replicates. Shaded area indicates a period of subjective night.
The transcript abundance of both AqCry2 and AqPARa is in fact at its maximum in the free-swimming larval stage (Figure 2), but it is unclear if this expression is ontogenetically or environmentally controlled (Figure 3). To address this, we subjected larvae that emerged from the maternal adult at 2 p.m. (ZT = 8) to a regime of either natural light, constant light, or constant darkness, and measured transcript abundance over an 8 h period. In all light treatments, gene expression of AqCry2 and AqPARa was significantly higher during the subjective day than during the subjective night (Figure 5A). In larvae exposed to a natural light regime, AqCry2 and AqPAR transcript levels decreased significantly at sunset. In larvae exposed to constant light, AqCry2 expression gradually decreased throughout the subjective day and then was relatively constant during the subjective night (Figure 5A). During the subjective day of the constant light regime, AqCry2 and AqPARa expression was lower than during the day of the natural light regime (Figure 5A), consistent with a lower constant light intensity (120 Lux) than experienced during most of the natural light regime (600 Lux at 2 p.m. decreasing to 120 Lux at 5.30 p.m.). Under constant darkness, gene expression was significantly lower than in other light treatments, especially throughout the subjective day (Figure 5A).
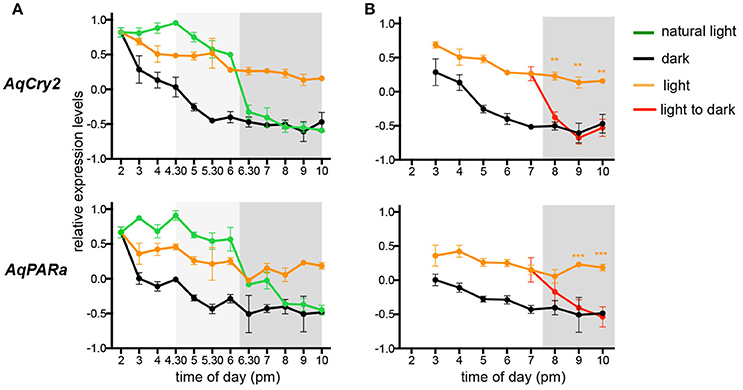
Figure 5. Expression of AqCry2 and AqPARa in larvae subjected to different light regimes. QPCR analysis of AqCry2 and AqPARa transcript abundance in (A) larvae subjected to three light regimes from the time of emerging from the maternal adult sponge between 1 and 2 p.m., and (B) subjected to constant light or darkness, or transferred from light to dark (n = 6–10). Constant light, orange; constant darkness, black; natural light, green; transferred from light to dark, red. Error bars are ± sem for biological replicates. Shaded area indicates a period of subjective night. A light gray shading indicates the hours before sunset, when the light intensity had already significantly decreased (Supplementary Table 2).
To further assess whether AqCry2 and AqPARa expression is directly influenced by light, we monitored the transcriptional response to a sudden change from light to dark (Figure 5B); larvae were exposed to constant light for 5 h after their emergence at 2.00 p.m. (ZT = 8) from the brood chamber, and then were transferred to constant dark. Within 30 min and 1.5 h, respectively, of transfer of larvae into the dark, AqCry2 and AqPARa expression levels decreased significantly to match expression levels observed in a control constant dark treatment (Figure 5B).
Given that AqCry2 and AqPARa have strikingly similar developmental and diurnal expression profiles (Figures 2–4), we sought to determine if these genes also show spatial co-expression in the photo-responsive larval stage of the life cycle. AqCry2 spatial expression in larvae has previously been reported as highest in photo-responsive cells around the posterior pigment ring of “late ring” stage larvae, prior to their emergence from the maternal adult (Rivera et al., 2012). This larval pigment ring is comprised of concentric rings of cells that are pigmented and/or possess long cilia that act as phototactic “rudders” to steer the larva away for light (Leys and Degnan, 2001; Leys et al., 2002). In free-swimming larvae post-emergence from the adult, AqCry2 expression is no longer enriched around the pigment ring, and is more widespread predominantly in the outer epithelial and subepithelial layers (Rivera et al., 2012).
The new in situ hybridization patterns of AqPARa that we present here show a striking similarity to AqCry2 expression (Rivera et al., 2012) (Figure 6). Early in development, AqPARa is expressed in cells evenly distributed throughout the embryo (Figure 6A). After the formation of a two-layered embryo, when pigment cells combine at the posterior pole of the embryo to form the pigment spot, AqPARa expression is strongest in cells around and beneath the spot, and in a subset of cells in the inner cell mass (Figures 6B,C). As the pigment spot transforms into the pigment ring, AqPARa transcripts remain enriched in cells around and beneath the ring; at this time, expression is also detected at the anterior pole (Figures 6D–H). After hatching, in the pre-competent larva, AqPARa expression is evident in epithelial and subepithelial cells surrounding the larval body (Figures 6I–L). This entire developmental expression pattern matches closely in both spatial and temporal patterning with that of AqCry2 (Rivera et al., 2012; Figure 6M), with both genes being most prominently expressed around the pigment ring prior to emergence and then more broadly in the free-swimming larva, including in epithelial and subepithelial layers.
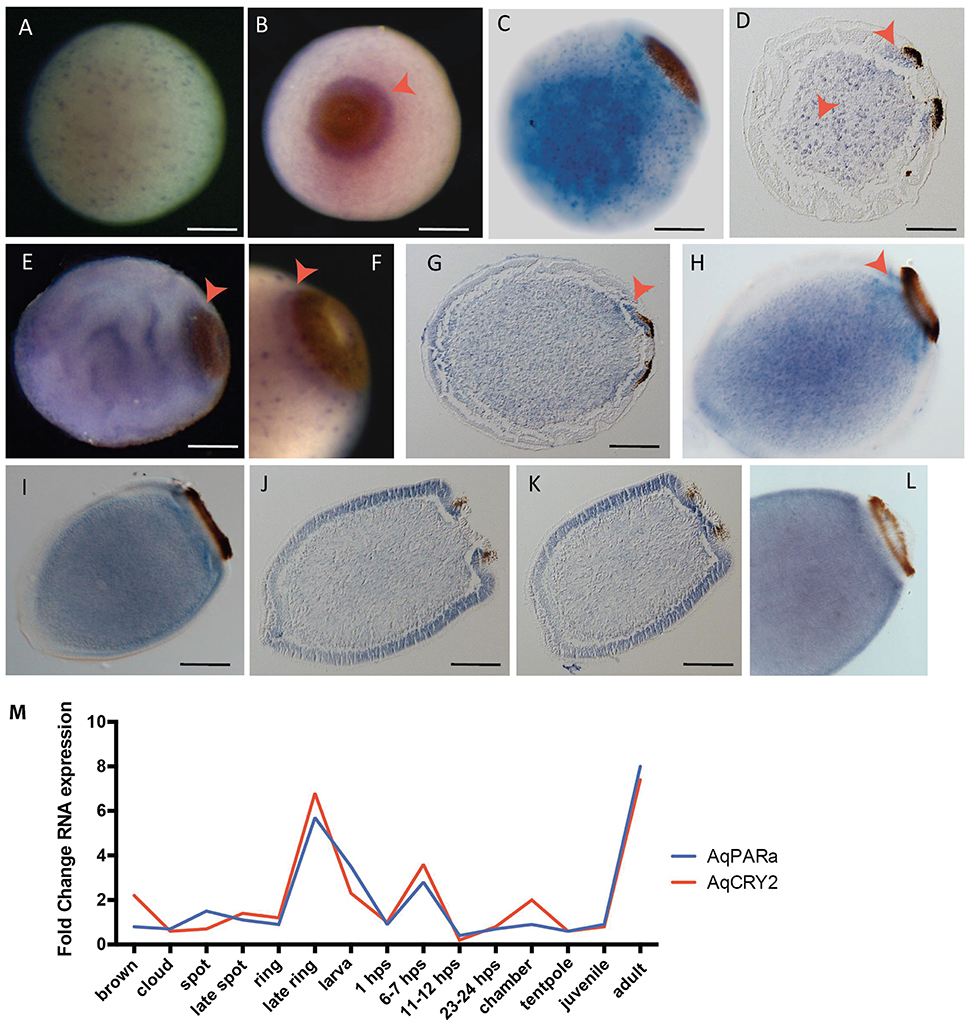
Figure 6. Developmental expression of AqPARAa. (A–L) Localization of AqPARa transcripts by in situ hybridization in embryos and larvae. (A,B,E,F) Whole-mounts; (C,H,I,L) cleared whole mounts; (D,G,J,K) sections; (B) posterior pole facing out of the page; (C–L) posterior pole to right. (A) During cleavage, AqPARa is expressed in a subset of cells evenly distributed throughout the embryo. (B–F) During spot and early ring stages, AqPARa transcripts are enriched in cells around and beneath the spot and ring, and in a subset of cells in the inner cell mass. (G,H) As the ring develops, AqPARa transcripts remain primarily localized to cells around and beneath the ring; expression is also detected at the anterior pole. (I–K) AqPARa expression in the swimming larva appears in sub-epithelial cells surrounding the larval body. Larva sectioned through the middle (J) of the ring or the edge (K) of the ring. (L) In older larva, AqPARa expression becomes more evident in sub-epithelial cells. All scale bars 100 μm. (M) CEL-seq expression profiles of AqPARa and AqCRY2 during development; data from Figure 2.
Animals have diurnal rhythms that are often entrained by day-night cycles (Pittendrigh, 1993; Foster and Kreitzman, 2005). Despite the lack of a nervous system, sponges appear to be no different, having the capacity to detect light (e.g., Leys and Degnan, 2001; Elliott et al., 2004; Rivera et al., 2012) and to display diurnal physiological and reproductive patterns (Reiswig, 1971; Amano, 1988; Nickel, 2004) consistent with possessing a circadian clock. Here we ask which genes that contribute to the regulation of circadian rhythms in other animals are present in the genome of the demosponge A. queenslandica, and whether any of these genes are expressed in patterns consistent with a circadian role in this early-diverging animal lineage.
bHLH-PAS transcription factors Clock and Cycle are central positive elements of the eumetazoan circadian network. A. queenslandica has a Clock gene, but we have found no Cycle ortholog, although we have found three closely-related ARNT-like genes. It is unclear whether the last common ancestor to sponges and eumetazoans had a Cycle gene that was subsequently lost in the lineage leading to A. queenslandica, or whether this ancestor had a proto-Cycle/ARNT gene that gave rise to ARNT-like genes in A. queenslandica, and Cycle and ARNT genes in eumetazoans (Simionato et al., 2007). During A. queenslandica development, AqClock and AqpARNT1 are upregulated in later stage embryos and in larvae, but neither AqClock nor any of the AqpARNT genes show consistent diurnal expression patterns in adults. In eumetazoans, CLOCK and CYCLE heterodimerise, and one of them typically has a rhythmic expression pattern (e.g., Reitzel et al., 2010, 2013; Brady et al., 2011). There currently is also no evidence that AqClock heterodimerises with an AqpARNT, and thus no evidence that one of these may substitute for Cycle in A. queenslandica.
Of the eumetazoans negative feedback genes, A. queenslandica has three Timeout paralogs but no Timeless ortholog, as is the case in cnidarians and placozoans (Reitzel et al., 2010; Shoguchi et al., 2013). We also found no evidence of an A. queenslandica Period gene, lending support to the proposition that Timeless and Period are bilaterian innovations (Reitzel et al., 2010). In cnidarians, timeout orthologs behave differently in different species. In the sea anemone N. vectensis, timeout expression is not light responsive (Reitzel et al., 2010) whereas the coral A. millepora timeout displays a diel expression that is lost under constant darkness (Brady et al., 2011, 2016). Expression of A. queenslandica timeout orthologs was not consistently correlated with the day-night cycle. In addition to Timeout, A. queenslandica has two Type I cryptochrome genes, AqCry1 and AqCry2 (Rivera et al., 2012). Akin to its eumetazoan orthologs (Vize, 2009; Reitzel et al., 2010; Brady et al., 2011), AqCry2 shows a consistent diurnal expression profile.
Of the eumetazoan feed-forward circadian loop factors, A. queenslandica possesses two PAR-bZIPs (Jindrich and Degnan, 2016), AqPARa and AqPARb, but no orthologs of mammalian ROR and Rev-Erb nuclear receptors (Bridgham et al., 2010). Similarly, no Rev/ROR orthologs have been identified in cnidarians, but in N. vectensis, two PAR-bZIPs oscillate in opposite fashion in response to a light:dark cycle and bookend the expression of NvClock (Reitzel et al., 2010, 2013). This could suggest a similar regulation mechanism to that of the fly D. melanogaster, where antagonistic regulation of Clock is performed by two PAR bZIPs that activate and repress, respectively, the transcription of DmClock through competitive binding to V/P/D box (Cyran et al., 2003). In A. queenslandica, AqPARa is expressed in a consistent diurnal pattern that suggests it may contribute to the regulation of circadian rhythms.
In summary so far, we have found that A. queenslandica has most of the components of the core circadian clock network found in eumetazoans, albeit with some apparent conspicuous absences (e.g., Cycle, Period), but only AqPARa and AqCry2 are expressed in diurnal manner. It is currently unclear if bHLH-PAS genes AqClock or AqpARNTs contribute to positively regulate the circadian network as CLOCK and CYCLE do in other animals, or if A. queenslandica Timeout genes play a negative feedback role as in bilaterians. Further, the expression profiles of some of these genes fluctuate through the larval stages of the life cycle in a manner coincident with changes in light regime, but because these changes are also correlated with marked ontogenetic shifts (e.g., onset of metamorphosis), it is unclear whether the fluctuations in expression are developmentally- or environmentally-induced.
Analysis of AqPARa and AqCry2 expression during normal and subjective days suggests that their expression is both light dependent and rhythmic. In adults subjected to natural light at sunrise in the field and in aquaria, AqCry2 expression peaks before AqPARa, suggesting that AqCry2 may act positively to regulate AqPAR2. Likewise, a dark-induced decrease in AqCry2 expression in larvae precedes a decrease in AqPARa transcript levels. Further, the near identical developmental expression profiles and the overlap in spatial expression of these genes in embryonic and larval cells and territories suggest that together they contribute to a light-induced, circadian network in A. queenslandica. Interestingly, in larvae, both gene expression patterns shift spatially around the time that the larva emerges into the light after release from the maternal adult, from expression in the vicinity of the photosensory pigment ring to a broader epithelial and subepithelial expression pattern consistent with a broader role in circadian regulation. However, linking the activity of these two genes and their products to a eumetazoan circadian clock is difficult without a better understanding of the nature and role of positive elements in sponges.
The ability to maintain cyclic outputs for extended periods of time after the removal of the entraining cue is a defining feature of the classical circadian clock (Bell-Pedersen et al., 2005; Foster and Kreitzman, 2005). AqPARa and AqCry2 expression in adults subjected to constant dark suggests that rhythmic expression dissipates within 5 days in the absence of the light entraining signal. Since whole animal qPCR dilutes cell-specific signals, it is possible that AqPARa and AqCry2 oscillations are maintained in the absence of the entraining cue, but that phase coordination between cells is lost, which masks our ability to detect those oscillations. However, current data suggest that cnidarians exposed to constant darkness similarly fail to maintain rhythmic behavior and gene expression for more than 3 days (Reitzel et al., 2010; Brady et al., 2011; Hoadley et al., 2011). We suggest that the free-running ability of the circadian clock outside of the Bilateria remains to be clarified.
Nonetheless, the presence of most eumetazoan circadian genes in A. queenslandica, and the clear light-dependent and diurnal pattern of the expression of AqPARa and AqCry2, suggest that the circadian regulatory network present in eumetazoans evolved from a more ancient base network that was present in the last common ancestor to sponges and eumtazoans.
SD, KJ, KR, and AR designed the study. KJ, SL, and KR prepared the biological material and performed the wetlab experiments. KR and KJ did the gene expression analyses and AR did the phylogenetic analyses. KJ, KR and SD drafted the manuscript, with critical comments provided by AR and BD. All authors read and approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge the staff and facilities of the Heron Island Research Station for assistance in acquiring biological material. This research was supported by Australian Research Council grant DP110104601 to SD and NIH award No. R15GM114740 to AR.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2017.00327/full#supplementary-material
Supplementary Table 1. qPCR primers used in this study.
Supplementary Table 2. Light intensity report. Light intensity was measured across several days in the experimental tanks. At the bottom of the table, the light intensity experienced by larvae reared under constant light is indicated for reference.
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Amano, S. (1988). Morning release of larvae controlled by the light in an intertidal sponge, Callyspongia ramosa. Biol. Bull. 175, 181–184. doi: 10.2307/1541557
Anavy, L., Levin, M., Khair, S., Nakanishi, N., Fernandez-Valverde, S. L., Degnan, B. M., et al. (2014). BLIND ordering of large-scale transcriptomic developmental timecourses. Development 141, 1161–1166. doi: 10.1242/dev.105288
Bell-Pedersen, D., Cassone, V. M., Earnest, D. J., Golden, S. S., Hardin, P. E., Thomas, T. L., et al. (2005). Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556. doi: 10.1038/nrg1633
Brady, A. K., Snyder, K. A., and Vize, P. D. (2011). Circadian cycles of gene expression in the coral, Acropora millepora. PLoS ONE 6:e25072. doi: 10.1371/journal.pone.0025072
Brady, A. K., Willis, B. L., Harder, L. D., and Vize, P. D. (2016). Lunar phase modulates circadian gene expression cycles in the broadcast spawning coral Acropora millepora. Biol. Bull. 230, 130–142. doi: 10.1086/BBLv230n2p130
Bridgham, J. T., Eick, G. N., Larroux, C., Deshpande, K., Harms, M. J., Gauthier, M. E., et al. (2010). Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 8:e1000497. doi: 10.1371/journal.pbio.1000497
Cyran, S. A., Buchsbaum, A. M., Reddy, K. L., Lin, M. C., Glossop, N. R., Hardin, P. E., et al. (2003). vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112, 329–341. doi: 10.1016/S0092-8674(03)00074-6
Degnan, B. M., Adamska, M., Craigie, A., Degnan, S. M., Fahey, B., Gauthier, M., et al. (2008). The demosponge amphimedon queenslandica: reconstructing the ancestral metazoan genome and deciphering the origin of animal multicellularity. CSH Protoc. 2008:pdb.emo108. doi: 10.1101/pdb.emo108
Degnan, B. M., Adamska, M., Richards, G. S., Larroux, C., Leininger, S., Bergum, B., et al. (2015). “Porifera,” in Evolutionary Developmental Biology of Invertebrates 1: Introduction, Non-Bilateria, Acoelomorpha, Xenoturbellida, Chaetognatha, ed A. Wanninger (Vienna: Springer-Verlag), 65–106.
Elliott, G. R. D., Macdonald, T. A., and Leys, S. P. (2004). Sponge larval phototaxis: a comparative study. Boll. Mus. Ist. Biol. Univ. Genova 68, 291–300.
Fernandez-Valverde, S. L., Calcino, A. D., and Degnan, B. M. (2015). Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genomics 16:387. doi: 10.1186/s12864-015-1588-z
Foster, R., and Kreitzman, L. (2005). Rhythms of Life: The Biological Clocks that Control the Daily Lives of Every Living Thing. New Haven, CT: Yale University Press.
Hardin, P. E. (2011). Molecular genetic analysis of circadian timekeeping in Drosophila. Adv. Genet. 74, 141–173. doi: 10.1016/B978-0-12-387690-4.00005-2
Harmer, S. L., Panda, S., and Kay, S. A. (2001). Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17, 215–253. doi: 10.1146/annurev.cellbio.17.1.215
Hoadley, K. D., Szmant, A. M., and Pyott, S. J. (2011). Circadian clock gene expression in the coral Favia fragum over diel and lunar reproductive cycles. PLoS ONE 6:e19755. doi: 10.1371/journal.pone.0019755
Jindrich, K., and Degnan, B. M. (2016). The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity. BMC Evol. Biol. 16:28. doi: 10.1186/s12862-016-0598-z
Larroux, C., Fahey, B., Adamska, M., Richards, G. S., Gauthier, M., Green, K., et al. (2008). Whole-mount in situ hybridization in amphimedon. CSH Protoc. 2008:pdb.prot5096. doi: 10.1101/pdb.prot5096
Levin, M., Anavy, L., Cole, A. G., Winter, E., Mostov, N., Khair, S., et al. (2016). The mid-developmental transition and the evolution of animal body plans. Nature 531, 637–641. doi: 10.1038/nature16994
Leys, S. P., and Degnan, B. M. (2001). Cytological basis of photoresponsive behavior in a sponge larva. Biol. Bull. 201, 323–338. doi: 10.2307/1543611
Leys, S. P., Cronin, T. W., Degnan, B. M., and Marshall, J. N. (2002). Spectral sensitivity in a sponge larva. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 188, 199–202. doi: 10.1007/s00359-002-0293-y
Nickel, M. (2004). Kinetics and rhythm of body contractions in the sponge Tethya wilhelma (Porifera: Demospongiae). J. Exp. Biol. 207(Pt 26), 4515–4524. doi: 10.1242/jeb.01289
Pittendrigh, C. S. (1993). Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54. doi: 10.1146/annurev.ph.55.030193.000313
Reiswig, H. M. (1971). In situ pumping activities of tropical Demospongiae. Mar. Biol. 9, 38–50. doi: 10.1007/BF00348816
Reitzel, A. M., Behrendt, L., and Tarrant, A. M. (2010). Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS ONE 5:e12805. doi: 10.1371/journal.pone.0012805
Reitzel, A. M., Tarrant, A. M., and Levy, O. (2013). Circadian clocks in the cnidaria: environmental entrainment, molecular regulation, and organismal outputs. Integr. Comp. Biol. 53, 118–130. doi: 10.1093/icb/ict024
Rivas, G. B., Bauzer, L. G., and Meireles-Filho, A. C. (2015). “The environment is everything that isn't me”: molecular mechanisms and evolutionary dynamics of insect clocks in variable surroundings. Front. Physiol. 6:400. doi: 10.3389/fphys.2015.00400
Rivera, A. S., Ozturk, N., Fahey, B., Plachetzki, D. C., Degnan, B. M., Sancar, A., et al. (2012). Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin. J. Exp. Biol. 215(Pt 8), 1278–1286. doi: 10.1242/jeb.067140
Shoguchi, E., Tanaka, M., Shinzato, C., Kawashima, T., and Satoh, N. (2013). A genome-wide survey of photoreceptor and circadian genes in the coral, Acropora digitifera. Gene 515, 426–431. doi: 10.1016/j.gene.2012.12.038
Simionato, E., Ledent, V., Richards, G., Thomas-Chollier, M., Kerner, P., Coornaert, D., et al. (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7:33. doi: 10.1186/1471-2148-7-33
Srivastava, M., Simakov, O., Chapman, J., Fahey, B., Gauthier, M. E., Mitros, T., et al. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726. doi: 10.1038/nature09201
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stangegaard, M., Dufva, I. H., and Dufva, M. (2006). Reverse transcription using random pentadecamer primers increases yield and quality of resulting cDNA. Biotechniques 40, 649–657. doi: 10.2144/000112153
Takahashi, J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179. doi: 10.1038/nrg.2016.150
Keywords: circadian rhythm, clock genes, Porifera, light response, cryptochrome, PARa
Citation: Jindrich K, Roper KE, Lemon S, Degnan BM, Reitzel AM and Degnan SM (2017) Origin of the Animal Circadian Clock: Diurnal and Light-Entrained Gene Expression in the Sponge Amphimedon queenslandica. Front. Mar. Sci. 4:327. doi: 10.3389/fmars.2017.00327
Received: 19 August 2017; Accepted: 29 September 2017;
Published: 18 October 2017.
Edited by:
Jose M. Eirin-Lopez, Florida International University, United StatesReviewed by:
Peter Vize, University of Calgary, CanadaCopyright © 2017 Jindrich, Roper, Lemon, Degnan, Reitzel and Degnan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandie M. Degnan, cy5kZWduYW5AdXEuZWR1LmF1
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.