
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci. , 15 August 2017
Sec. Aquatic Microbiology
Volume 4 - 2017 | https://doi.org/10.3389/fmars.2017.00262
This article is part of the Research Topic Socio-Ecology of Microbes in a Changing Ocean View all 17 articles
The microbial contribution to ecological resilience is still largely overlooked in coral reef ecology. Coral-associated bacteria serve a wide variety of functional roles with reference to the coral host, and thus, the composition of the overall microbiome community can strongly influence coral health and survival. Here, we synthesize the findings of recent studies (n = 45) that evaluated the impacts of the top three stressors facing coral reefs (climate change, water pollution and overfishing) on coral microbiome community structure and diversity. Contrary to the species losses that are typical of many ecological communities under stress, here we show that microbial richness tends to be higher rather than lower for stressed corals (i.e., in ~60% of cases), regardless of the stressor. Microbial responses to stress were taxonomically consistent across stressors, with specific taxa typically increasing in abundance (e.g., Vibrionales, Flavobacteriales, Rhodobacterales, Alteromonadales, Rhizobiales, Rhodospirillales, and Desulfovibrionales) and others declining (e.g., Oceanosprillales). Emerging evidence also suggests that stress may increase the microbial beta diversity amongst coral colonies, potentially reflecting a reduced ability of the coral host to regulate its microbiome. Moving forward, studies will need to discern the implications of stress-induced shifts in microbiome diversity for the coral hosts and may be able to use microbiome community structure to identify resilient corals. The evidence we present here supports the hypothesis that microbial communities play important roles in ecological resilience, and we encourage a focus on the microbial contributions to resilience for future research.
Corals are diverse meta-organisms that contain not only the conspicuous dinoflagellate partner Symbiodinium but also a microbiome assembled of bacterial, archaeal, viral, and other eukaryotic microorganisms (Rosenberg et al., 2007; Ainsworth et al., 2010; Thompson et al., 2014). Host specific differences in microbiome composition suggest that some bacterial members of the microbiome are mutualistic (Ainsworth et al., 2015); thus, many recent efforts have focused on identifying these bacteria and their specific metabolic roles in coral health (Table 1). These abundant coral-associated bacterial communities are distinct (Box 1) from the surrounding habitat, containing taxa that drastically differ from free-living seawater microbes (Rohwer et al., 2001; Carlos et al., 2013).
Box 1. Are coral microbiomes unusually diverse?
Corals are sometimes referred as “highly” diverse meta-organisms. Yet this is a somewhat subjective statement that likely has arisen when coral microbiomes are compared to other well studied mutualistic symbiotic model systems that are highly canalized (Dubilier et al., 2008). It is now well accepted that microorganisms colonize most marine species, yet a systematic comparison among marine organisms is currently lacking. For example, sponge tissues contain between 10 and 1,000 bacterial OTUs (Bourne and Webster, 2013), a species richness value well within the range for corals. A recent assessment of tropical reef algal microbiomes also demonstrate that algae contain even more diverse bacterial communities than corals (Barott et al., 2011). The number of bacterial OTU's in corals can range up to 102–104 compared to 101–103 for sponges and 102 for Hydra (Blackall et al., 2015), although as just discussed these richness estimates vary across species, habitat, and host compartment. With these context dependent numbers, it is thus difficult to say whether corals have a higher richness of microbial taxa than other marine species. For example, turf and CCA each exhibited overall higher numbers of OTUs (18,926 and 9,559) than the three coral species compared (Acropora hyacinthus, A. rosaria, and Porites lutea) (951, 2,331, 4,018) (Hester et al., 2016). Similarly, algal (i.e., Dictyota bartayresiana, Halimeda opuntia, turf algae, and CCA) microbiomes were generally more diverse overall [ranging from 6.22 to 7.82 (Shannon Index)] than those in corals (ranging from 2.84 to 4.51) (Barott et al., 2011). Therefore, more comparisons among coral species and marine meta-organisms are needed to determine if corals or certain coral species actually have highly diverse microbial communities.
Additionally, when comparing the composition and dynamics of the coral microbiome to other marine hosts, it is important to differentiate between stable and sporadic members of the community. It is likely that stable members play more important roles in promoting the health and longevity of the host while transient members might contribute to only the function of the host under explicit environmental conditions or alternatively play negative and antagonistic roles in the system (albeit, stable members could also act as opportunistic pathogens). Stable microbes should exhibit consistent relative abundances in the host vs. the sporadic members who will vary in their prevalence and relative abundance among individuals of the same host (Hester et al., 2016). Sporadic members may opportunistically associate with the coral and not play a beneficial role. In three coral species there was a high number of stable members to sporadic members: A. hyacinthus (stable = 902, sporadic = 49), A. rosaria (stable = 2,188, sporadic = 143), P. lutea (stable = 3,662, sporadic = 356) (Hester et al., 2016).
Another way to evaluate this is to conduct “core” microbiome analysis at various levels of stringency. This prevalence-based metric has been used to infer which members of a coral's microbiome are mutualistic or opportunistic. In an evaluation of the core coral microbiome (i.e., phylotype presence in 30% of the samples), Acropora granulosa's core microbiome consisted of 159 out of 1,508 phylotypes, Leptoseris spp. 204 out of 1,424, and Montipora capitata 350 out of 1,433 (Ainsworth et al., 2015). Importantly, most of these core microbiome members were highly diverse yet found in very low relative abundance compared to the entire community. Thus, it is important to consider rare microbiome members, as these may be the beneficial resident members researchers are interested in. In another longitudinal study from three coral species from South Florida, the core coral mucus microbiome consisted of 13 bacterial orders at a >95% prevalence score (Zaneveld et al., 2016). In this case, the “core” microbiome highly depends on the level of stringency used.
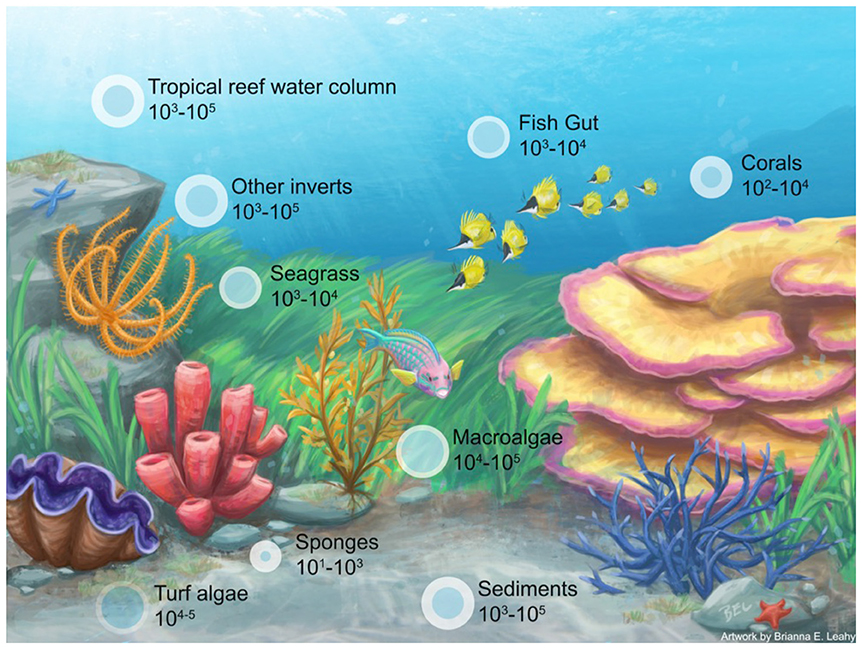
Box Figure 1. Depiction of a coral reef and associated microbiomes with organismal hosts [i.e., invertebrates (Bourne et al., 2013; Tianero et al., 2014; Hakim et al., 2016), seagrass (Ettinger et al., 2017; Fahimipour et al., 2017), fish (Ghanbari et al., 2015; Givens et al., 2015; Parris et al., 2016), corals (Barott et al., 2011; Blackall et al., 2015; Hester et al., 2016), macroalgae (Barott et al., 2011; Burke et al., 2011; Egan et al., 2013; Brodie et al., 2016), sponges (Bourne and Webster, 2013; Blackall et al., 2015), turf algae (Hester et al., 2016)] and environmental parameters [i.e., reef water (McCliment et al., 2012), sediment (Ettinger et al., 2017)]. The size of the bubble indicates a more species rich microbiome (based on OTUs).
Aspects of coral biology also influence microbiome structure and function. Similar to humans, compartmentalization of the microbiome generates distinct microbial communities in the coral animal, within the surface mucus layer, tissues, skeleton, and gut (Sweet et al., 2010; Ainsworth et al., 2015; Apprill et al., 2016) and some particularly associated with Symbiodinium (Ainsworth et al., 2015). Coral-associated bacteria can be transferred vertically from parent to larva (Sharp et al., 2012) or they can be horizontally acquired from the environment (Apprill et al., 2009; Sharp et al., 2010), including when adult corals release bacteria (e.g., Altermonas and Roseobacter) as a by-product of broadcast spawning (Ceh et al., 2013b). Although recent research has focused on the role of microbiomes in coral adaptation (Gilbert et al., 2012; Glasl et al., 2016), coral reef management still largely ignores the role of microbial communities, with the exception of Symbiodinium, in coral resilience (McClanahan et al., 2012). With rapid advances in DNA sequencing technologies, more studies are able to capture the influence of stressors on the coral microbiome, but no study to date has reviewed these impacts. Here, we conduct a synthesis of the results from 45 relevant studies that evaluated how three key stressors threatening coral reefs (i.e., climate change, water pollution and overfishing) altered coral microbiomes. In addition, we provide hypotheses as to how the microbiome may provide corals with resistance to these stressors.
Different coral-associated bacteria are hypothesized to play varying roles in coral health, nutrition and development (Table 1). Recent reviews have proposed the term “Beneficial Microorganisms for Corals” (BMC) to define microbial symbionts that maintain coral health (Peixoto et al., 2017), outlined microbiome roles in coral health and resilience (Bourne et al., 2016) and suggested that coral reef microbial communities serve as indicators of environmental stress and coral health (Glasl et al., 2017). Coral-associated microbial communities contribute to carbon cycling (Kimes et al., 2010), sulfur cycling (Wegley et al., 2007; Raina et al., 2009), phosphorous fixation, metal homeostasis, organic remediation (Zhang et al., 2015), production of antibiotics (Ritchie, 2006) and secondary metabolism (Zhang et al., 2015). For example, diazotrophs (i.e., nitrogen-fixing microbes) form species-specific associations with corals and may provide limiting fixed nitrogen to the algal partner of corals, Symbiodinium, and to the coral animal itself (Lesser et al., 2004, 2007; Lema et al., 2012). In the early coral life stages, bacteria provide nitrogen directly to larvae's Symbiodinium (Ceh et al., 2013a) and potentially to the coral larvae (Lema et al., 2014). Bacteria also play an important role in larval recruitment and settlement, for example, Alphaproteobacterium, Roseivivax sp. 46E8 significantly increases larval settlement of Porites astreoides (Sharp et al., 2015).
Many coral-associated bacteria defend the coral by exuding antimicrobial compounds to prevent invasions by pathogens or other exogenous bacteria (Table 1). For example, nearly 70% of cultivable isolates from Montastrea annularis inhibited the growth of other bacteria and 11.6% inhibited the growth of the known coral pathogen, Vibrio shiloi (Rypien et al., 2010). Approximately 20% of cultivable isolates from Acropora palmata demonstrated antibiotic activity against other strains and pathogens (Ritchie, 2006). Isolates from a soft coral, Sarcophyton glaucum, inhibited the growth of four coral pathogens and three fungi (ElAhwany et al., 2013). It was also found that under increased algal cover and temperatures, the relative abundances of Actinobacteria decreased while opportunistic Proteobacteria increased within the coral microbiome, suggesting that opportunists can take advantage of the absence of inhibition (Zaneveld et al., 2016). Eight percent of native coral bacteria inhibited the growth of the pathogen Serratia marcescens, with Exiguobacterium sp. inducing 10–100 fold reductions in growth within coral mucus (Krediet et al., 2012). Concurrently, the coral pathogen, Vibrio corallilyticus, has antimicrobial activity of its own, suggesting that it may use inhibition to colonize the coral (Kvennefors et al., 2011). There are also some resident bacteria that actively predate upon these pathogens within the coral mucus (Welsh et al., 2016). In summary, given the wide variety of roles vital to holobiont functioning that are played by coral-associated bacteria, any disruption or destabilization may influence host fitness and ecosystem functioning.
Here, we synthesized 45 relevant studies that each (1) were peer-reviewed (2) examined coral-associated bacteria and (3) measured how these bacteria were affected by climate change, water pollution or overfishing (Table 2). We included a broad range of methodologies as many researchers use different methods and we aimed to evaluate the field from a broad perspective. Over half of the studies focused on climate change (n = 27) and almost one quarter focused on water pollution (n = 10); only a small proportion addressed overfishing (n = 4) or more than one stressor at a time (n = 4) (Figure 1). The majority of the studies were published in the last 5 years (Figure 1). Geographically, almost all of the overfishing and water pollution studies occurred in the Caribbean compared with studies on climate change, which had a more global distribution of study sites (Figure 1).
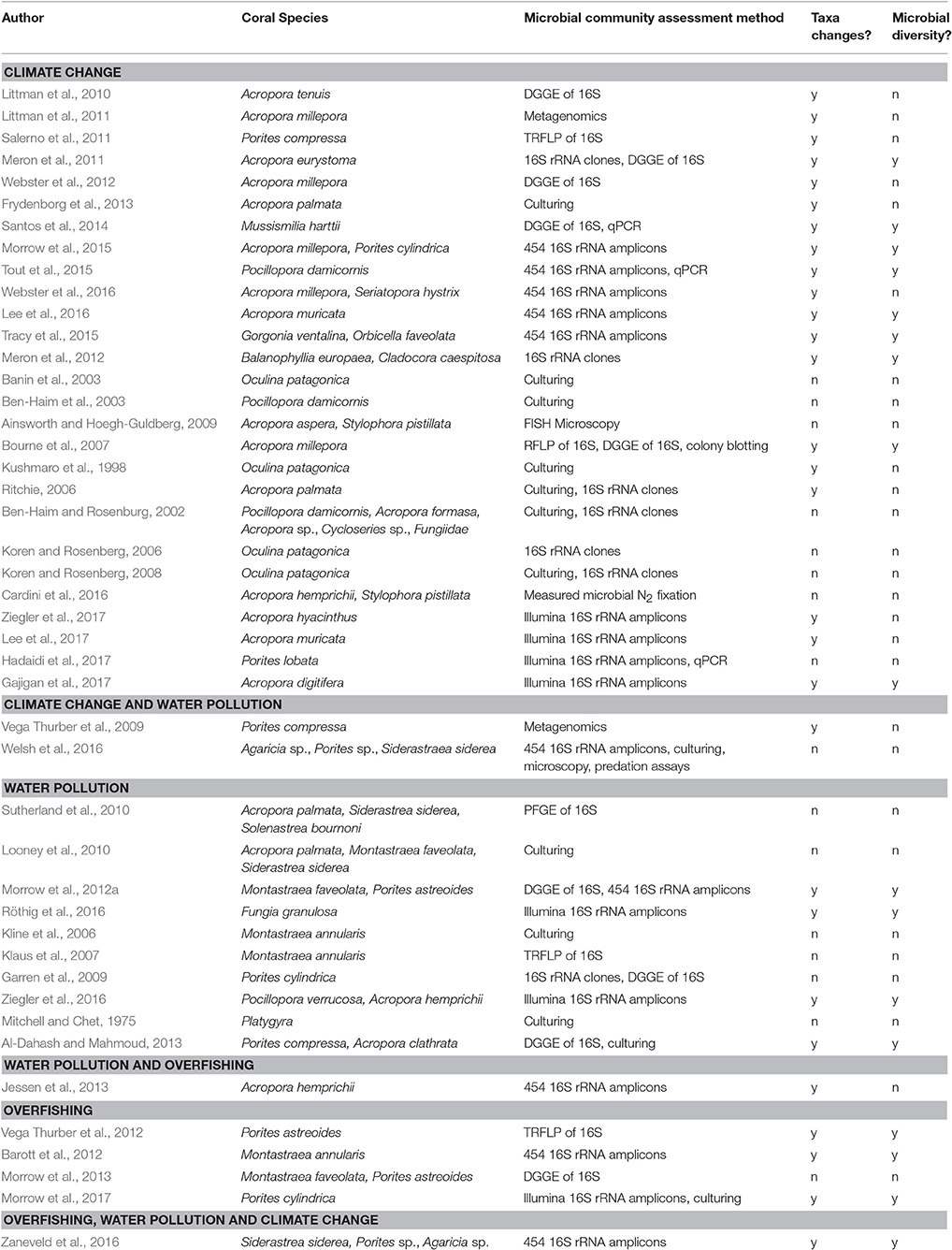
Table 2. Overview of the 45 papers that evaluated the influence of climate change, water pollution and overfishing on the coral microbiome, coral species evaluated, the method they used to capture effects on the microbial community (e.g., DGGE, denaturing gradient gel electrophoresis; TRFLP, terminal restriction fragment length polymorphism; PFGE, pulsed-field gel electrophoresis), and whether the paper had data that we included on changes in bacterial taxa (i.e., Figure 3) or microbial diversity (i.e., Table 3).
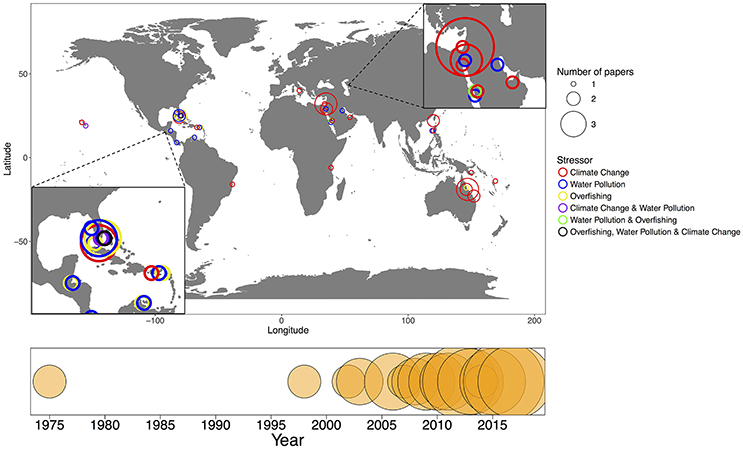
Figure 1. A world map of studies evaluating the impact of stressors on the coral microbiome, climate change (red), water pollution (blue), overfishing (yellow), climate change and water pollution (purple), water pollution and overfishing (green), and overfishing, water pollution and climate change (black). The size of the bubble refers to the number of papers at that latitude/longitude. The timeline on the bottom represents the number of papers published each year from 1975 to 2017 that were included in the synthesis.
The genera Acropora and Porites were the most represented corals within these studies, accounting for nearly 50% of all corals evaluated (Figure 2A). Massive Porites species may be “stress-tolerant” corals, as they are slow growing and may be able to survive harsher environments (Darling et al., 2012). Acroporids are considered “competitive” corals, meaning that they are fast-growing and dominate reefs in productive environments; they are also the most sensitive to environmental change (Darling et al., 2012). As a result, there was a bias toward studying climate change on competitive life history strategies, specifically Acropora in Australia (Figure 2B).
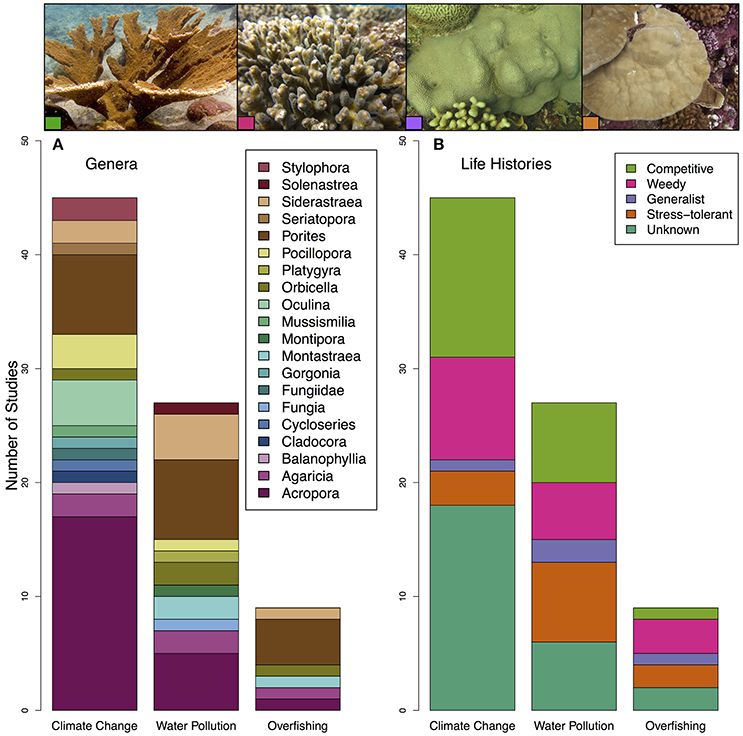
Figure 2. Plot of (A) coral genera and (B) coral life-histories included in all studies on the impact of climate change, water pollution and overfishing on the coral microbiome. Coral life-histories are taken from Darling et al. (2012). Pictures on the top correspond to coral species and life history strategies included in these papers left to right: Acropora palmata (competitive) (photo by Ryan McMinds, Global Coral Microbiome Project, licensed under CC BY 2.0), Pocillopora damicornis (weedy) (photo by Joseph Pollock, Global Coral Microbiome Project, licensed under CC BY 2.0), Orbicella faveolata (generalist) (photo by Joseph Pollock, Global Coral Microbiome Project, licensed under CC BY 2.0), and Porites lobata (stress tolerant) (photo by Kristina Tietjen).
We note here that we report bacterial taxa at the levels of Phylum, Class and Order to provide consistency across the array of studies given that many of them reported their data in different ways and to different taxonomic levels. Additionally, researchers used varying metrics of community structure to evaluate the impact of stressors including: richness (i.e., total number of species), alpha diversity (i.e., the total number of species and their relative abundances, generally the Shannon and Simpson Index), evenness (i.e., distribution of species relative abundances) and beta diversity (i.e., difference in community composition).
While a consequence of human impacts on macro-scale communities is often species loss, the emerging evidence from our analysis suggests that similar impacts more commonly lead to an increase in bacterial richness or alpha diversity within coral-associated microbial communities (~60% of the time, Table 3). Invasion is likely the mechanism underlying these increases, with stress events appearing to disrupt the functioning microbiome and facilitating an invasion of microbes not typically resident on corals, thus increasing the overall number of microbiome members (Welsh et al., 2015). For example, corals closer to human disturbance have been shown to harbor higher bacterial diversity than those farther from the disturbance (Morrow et al., 2012a). Similarly, corals in lowered pH had higher microbial diversity (Meron et al., 2011) and microbial diversity increased in overfishing and eutrophication treatments (Jessen et al., 2013). Lastly, it was found that upon algal contact, the coral microbiome increased in species richness (Zaneveld et al., 2016). These results also are contrary to the patterns found in the human gut microbiome, in which stress lowers microbial alpha diversity by allowing opportunistic and pathogenic taxa to dominate the community (Lozupone et al., 2012). This might be explained by the nature of the human gut microbiome being a more closed system compared with corals that are constantly bathed in the distinct microbial assemblages of seawater. We hypothesize that when corals are disturbed, their ability to regulate and/or exclude incoming microbial taxa from the surrounding environment may be reduced. However, this is not always the case, as other studies have demonstrated no significant change or a decrease in microbial diversity under these three stressors (Meron et al., 2012; Tracy et al., 2015; Morrow et al., 2017). These conflicting results likely are the result of variability in coral microbiome responses across coral host species, locations and stressors. Additionally, these diversity changes may reflect differences in experimental design.
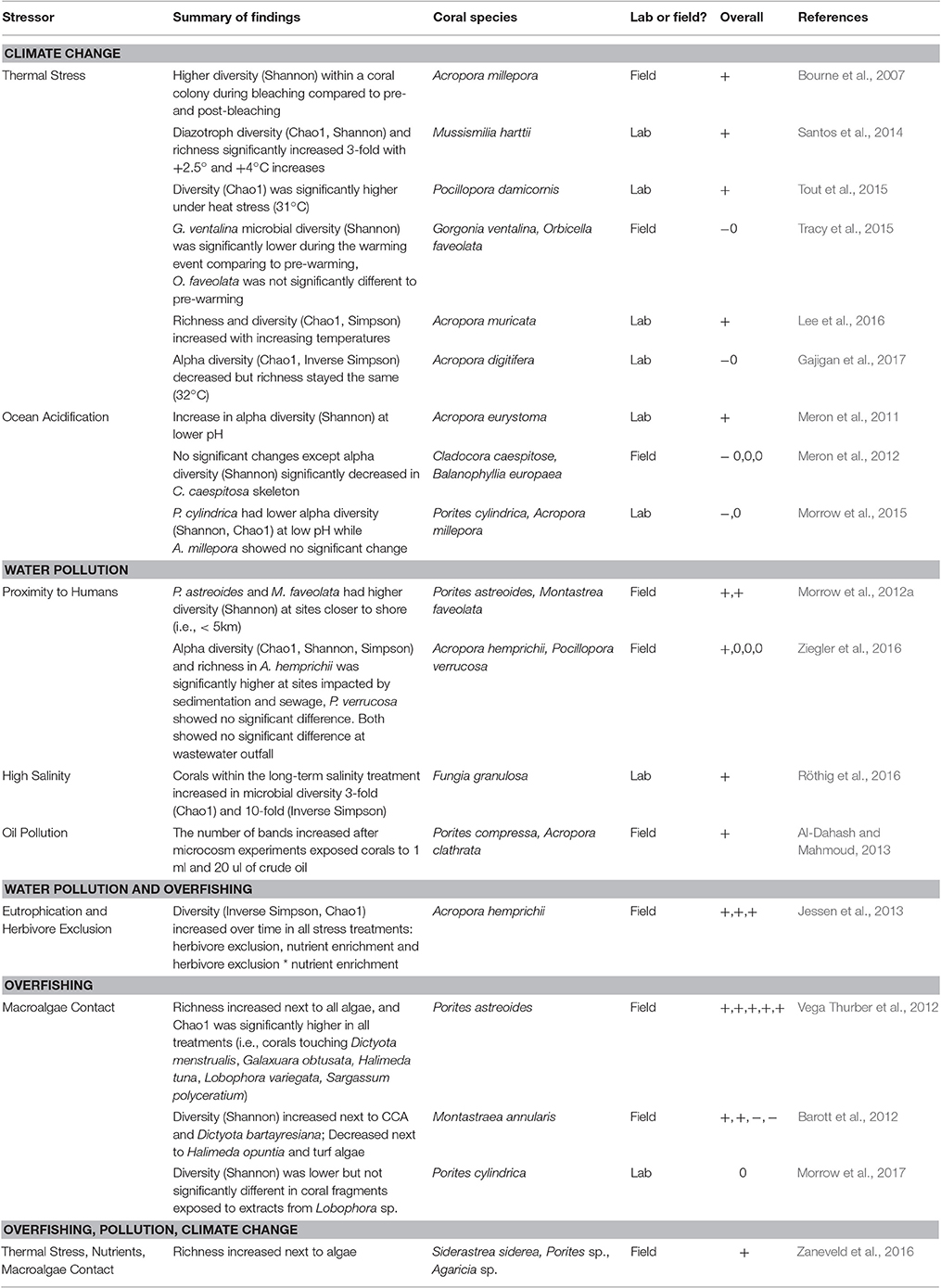
Table 3. Changes in microbiome alpha diversity or richness due to stress from climate change, water pollution and overfishing (+, higher diversity; −, lower diversity; 0, no difference).
In addition to increased richness, there is mounting evidence that stressors induce changes to coral microbiome evenness and beta diversity. Changes in evenness indicates shifts in microbial species dominance, and evenness has been found to decrease in the rhizosphere with added disturbance (van der Voort et al., 2016). An increase in beta diversity in response to stress may indicate a destabilized microbiome in mammals (Moeller et al., 2013; Zaneveld et al., 2017) with the host being unable to regulate its microbiome. In corals, both temperature extremes and contact with macroalgae were shown to increase microbiome beta diversity (Zaneveld et al., 2016). Similarly, microbiome composition in shallow polluted sites was more variable from coral to coral than at the control sites (Klaus et al., 2007). Changes in salinity also impacted community evenness. For example it was found that the microbiome of hypersaline-treated corals shifted from a community dominated by a single OTU (Rhodobacteraceae) to a more even one in which Pseudomonas veronii was the most abundant taxon (Röthig et al., 2016).
Overfishing on reefs can lead to reduced grazing pressure, by decreasing herbivorous fish abundance, and increasing competition between corals and macroalgae for space (Morrow et al., 2013). Macroalgae are hypothesized to outcompete corals via a variety of mechanisms including alterations to the microbiome (Smith et al., 2006; Morrow et al., 2012b), faster growth rates, shading, allelopathic interactions (Rasher and Hay, 2010), and abrasion and preventing coral recruitment (Jompa and McCook, 2003). For example, macroalgal contact with the coral Porites astreoides caused multiple changes in the coral microbiome including increased dispersion (i.e., beta diversity), disappearance of a potentially mutualistic Gammaproteobacteria, changes in abundance for taxa already present, establishment of new taxa, and growth of algae-associated microbes within the coral (Vega Thurber et al., 2012). Macroalgal contact has also been shown to shift the coral microbiome to become more similar to the macroalgal microbiome (Morrow et al., 2013).
A counter example to the overall trend of stress-induced community shifts is provided by a study on A. millepora and Seriatopora hystrix microbiomes, which demonstrated stability in microbiome composition in the face of both lowered pH and increased temperatures. While S. hystrix's microbiome did show some variability, the overarching conclusion was that corals demonstrated a more stable and robust microbiome compared to other key calcifying reef taxa such as foraminifera and crustose coralline algae (Webster et al., 2016).
The studies we synthesized consistently found that the bacterial order, Oceanospirillales, especially the genus Endozoicomonas, was underrepresented in corals during stress events, particularly during climate anomalies (Figure 3). This may be problematic for corals as Endozoicomonas is a proposed beneficial symbiont to corals. Two studies used CARD-FISH and FISH probes, respectively, to reveal that Endozoicomonas was located deep within the coral tissues, suggesting an intimate association with coral hosts (Bayer et al., 2013; Neave et al., 2016). Additionally, the first cultivable Endozoicomonas from corals suggests Endozoicomonas montiporae CL-33 helps corals under stress through preventing mitochondrial dysfunction and promoting gluconeogenesis (Ding et al., 2016). Additionally, researchers have proposed that Endozoicomonas plays a role in sulfur cycling (Raina et al., 2009), nutritional symbiosis (La Rivière et al., 2013) and protecting Symbiodinium from bleaching pathogens (Pantos et al., 2015). By comparing cultured and culture-independent genomes of Endozoicimonas, researchers demonstrated that Endozoicimonas likely plays a role in protein and carbohydrate transport for the host and may have diversified and adapted with their hosts (Neave et al., 2017). However, they also demonstrated that Endozoicimonas has a large genome, suggesting it has a free-living stage. Not only might the observed decrease in these potentially symbiotic taxa during stress events threaten coral resistance to stress, it also may influence coral resilience after the stress is alleviated if Endozoicomonas does not return to its original abundance and function. In a study of volcanic CO2 seeps where the seawater has naturally reduced pH, researchers found that Acropora millepora and Porites cylindrica contained significantly different microbial communities compared to the same coral species at a control site (>500 m away); for A. millepora this was mainly due to a 50% decrease of this proposed mutualist Endozoicomonas (Morrow et al., 2015). These coral species were also less abundant at the volcanic seeps, potentially due to this loss of symbiont in their microbiomes. In a separate study, Endozoicomonas was significantly reduced at low pH in A. millepora showing that this stressor can cause loss of these symbionts (Webster et al., 2016). Other symbiotic taxa in addition to Endozoicomonas are likely to decline as well under stress. At anthropogenic impacted reefs (i.e., close to metropolitan cities), the main coral symbiotic taxon in Pocillopora verrucosa (Endozoicomonaceae) and A. hemprichii (Alteromonadales) declined in relative abundance (Ziegler et al., 2016).
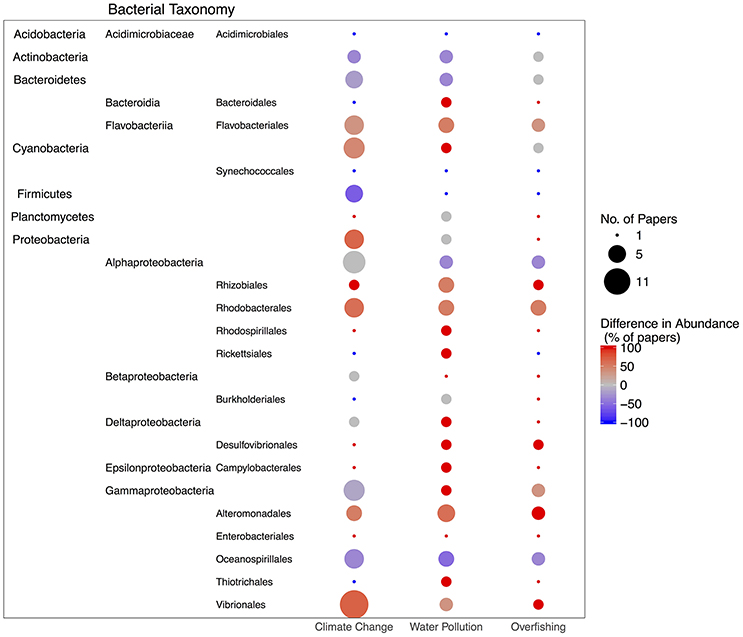
Figure 3. Summary of the number of papers showing differences in bacterial taxa during stress events (climate change, water pollution or overfishing). The size of the bubble indicates the number of papers (although, some papers found both an increase and decrease in the taxonomic level (i.e., class or family) if a lower level was evaluated (i.e., genus or OTU) so the bubble size may be larger than the actual number of papers as both of these changes were plotted) and the color of the bubble shows the percent of papers that showed that taxa overrepresented under stress (red) or underrepresented under stress (blue). Bacteria were only plotted if they were represented by a paper in each of the three stressors.
Stressed corals may have a reduced ability to regulate their microbiomes and thus have reciprocal increases in potentially pathogenic and opportunistic microbial taxa (Figure 3). The overrepresented taxa during all three types of stressor we evaluated were: Vibrionales, Flavobacteriales, Rhodobacterales, Alteromonadales, Rhizobiales, Rhodospirillales, and Desulfovibrionales (Figure 3). The Order Rhodobacterales, for example, are fast growing opportunistic bacteria (Teeling et al., 2012) that have been found in both healthy and stressed corals (Meron et al., 2011; Sharp et al., 2012), potentially blooming under periods of stress when there is open niche space (Welsh et al., 2015). OTUs within the Order Flavobacteriales were shown to make up a large percentage of white band disease associated OTUs (Gignoux-Wolfsohn and Vollmer, 2015). Thus, these potentially pathogenic, opportunistic taxa may flourish when the coral is stressed and cannot regulate its microbiome.
In comparison, a meta-analysis of 16S sequences from 32 papers, showed that the microbiome of bleached corals differed from that of healthy corals primarily in having a higher proportion of two specific taxa: Vibrios and Acidobacterias (Mouchka et al., 2010). An increase in Vibrionales under climate change stress is unsurprising as the cultivable Vibrio strain AK-1 was shown to induce coral bleaching (Kushmaro et al., 1998) (albeit the coral, Oculina patagonica, may have developed resistance to this bacteria; Mills et al., 2013) and Vibronales are a common taxa to increase under conditions of thermal stress (Bourne et al., 2007; Frydenborg et al., 2013; Tout et al., 2015). Importantly, the coral microbiome may have a temperature tolerance threshold, as it was found that bacterial community structure only changes after an elevation of temperature greater than 1°C; all exposures at temperatures lower than this threshold showed no evidence of community alterations (Salerno et al., 2011). Thus, coral microbiomes may buffer thermal anomalies within that lower temperature range.
Bleached corals also exhibit different bacterial communities than “healthy” corals (Koren and Rosenberg, 2008). For example, during a bleaching event in Australia, the coral microbiome showed an increase in genes associated with virulence factors (Littman et al., 2011). Correspondingly, during heat stress experiments, the known pathogen V. coralliilyticus increased in abundance by four orders of magnitude (Tout et al., 2015). One likely mechanism for these observed changes are strong competition between native commensals and pathogenic bacteria (i.e., V. shiloi and V. coralliilyticus) on corals, with temperature stress mediating the growth of the foreigners. This has been supported by work within Acropora palmata, where high temperatures tend to select for pathogens in the coral microbiome (Frydenborg et al., 2013). Increased temperatures also correlated with increased expression of virulence genes (Banin et al., 2003), lysis of coral cells (Ben-Haim et al., 2003), and infection (Kushmaro et al., 1998; Ben-Haim and Rosenburg, 2002) by coral pathogenic bacteria. However, Porites lobata has been found to have a relatively stable microbiome between bleached and healthy colonies, suggesting its mucus microbiome plays a protective role within bleached corals (Hadaidi et al., 2017).
Furthermore, increased Vibrio and other taxa occur prior to visual bleaching signs (Bourne et al., 2007; Lee et al., 2016), suggesting some predictable changes in the bacterial community could forewarn which corals may bleach. The abundance of Vibrionales within the microbiome may also be regulated by factors other than temperature, including what strain of Symbiodinium is hosted by the coral (Littman et al., 2010).
Importantly, the increases in potentially pathogenic or opportunistic taxa found in our analysis (Figure 3) may be due to a variety of mechanisms such as the induction of temperature sensitive virulence cassettes found in pathogens, enhanced growth rates under higher temperatures, or altered microbial-host signals during stress. Thermally stressed corals often increase production of the metabolites like dimethylsulphoniopropionate (DMSP), which is normally exuded by corals (Raina et al., 2013) and their symbionts (Steinke et al., 2011). It is hypothesized that DMSP is used as a chemoattractant by bacterial pathogens to locate thermally stressed corals (Garren et al., 2014).
While in most cases stressors altered coral microbial community structure, shifts do not always occur. For example, O. faveolata's microbiome did not significantly shift when the host bleached (Tracy et al., 2015). Furthermore, coral-associated microbial communities did not undergo major shifts when transplanted to a natural lower pH environment and did not harbor microbial pathogens (Meron et al., 2012). This study (Meron et al., 2012), suggests that for these two coral species (i.e., Balanophyllia europaea and Cladocora caespitosa), reduced pH does not pose a significant threat to coral health. Importantly, environmental conditions can buffer these microbiome changes, specifically with high water flow buffering coral microbiome changes under high temperatures (Lee et al., 2017).
In addition to community structure changes, stressors can also increase disease on coral reefs (Brandt et al., 2013; Vega Thurber et al., 2013). For example, exposure of corals to lower pH resulted in microbiome communities reminiscent of those associated with diseased and stressed corals as they contained more Vibrionaceae and Altermonadaceae (Vega Thurber et al., 2009; Meron et al., 2011). Additionally, reduced pH significantly changed the microbial communities in A. millepora with the loss of Proteobacteria sequences typically associated with healthy corals while Gammaproteobacteria, which are often associated with diseased and stressed corals, increased (Webster et al., 2012).
Given that coral reefs occur in oligotrophic environments, added nutrients can dramatically influence ecosystem functioning and alter microbial communities in ways that appear to favor disease. In Florida, human sewage supplied a strain of Serratia marcescens (a common fecal enterobacterium) into reef water and corallivorous snails acted as a vector, therefore inducing white-pox like diseases in Acropora palmata, Siderastrea siderea, and Solenastrea bournoni (Sutherland et al., 2010). Furthermore, the addition of glucose or inorganic nutrients improved the survival of S. marcescens in A. palmata (Looney et al., 2010). Under this water pollution stress, as with thermal stress (Littman et al., 2011), the microbial communities shifted toward a higher prevalence of heterotrophic bacteria relative to autotrophic bacteria (Dinsdale et al., 2008). Microbiome taxa in polluted sites can be more pathogenic, as demonstrated by a study in which exposure to pollutants increased coral mortality except when antibiotics were added, suggesting a bacterially mediated cause of death (Mitchell and Chet, 1975). As the proximity and size of human populations increases adjacent to coral reefs, so does the likelihood of land-based runoff, and evidence continues to demonstrate that corals living closer to shore have higher abundances of disease-related bacteria (Morrow et al., 2012a), and coral disease correlates with poor water quality and high nutrient levels (Furby et al., 2014). Nevertheless, the coral microbiome also demonstrates resilience against water pollution, when coral fragments were transplanted under eutrophic aquaculture pens, the coral microbiome shifted toward known pathogens but then showed no physical signs of disease and after 22 days the communities shifted back to their original state (Garren et al., 2009).
Overfishing may alter coral microbiomes indirectly, with decreases in herbivorous fish populations being one means by which algal growth can be facilitated at the expense of corals. For example, overfishing in Jamaica, in concert with warming and hurricanes, caused coral cover to decrease from 50 to 5% and an increase to 90% macroalgae cover (Hughes, 1994). Shifts to an algae dominated state can influence the coral microbiome by changing the water column microbial composition, increasing algal interactions (Morrow et al., 2013) and triggering coral disease (Nugues et al., 2004). Increased coral interactions with turf algae have been associated with an increase in pathogens and virulence genes (Barott et al., 2011). These authors also proposed that fleshy algae can alter reefs by increasing both bacterial respiration and pathogenic invasion (Barott et al., 2012). Moreover, algae may act as reservoirs for coral pathogens (Sweet et al., 2013) and thus enhance disease events. Algae harbor distinctly different microbial communities than corals (Barott et al., 2011; Vega Thurber et al., 2012) and produce more carbon exudate that can stimulate heterotrophic microbial growth in reef waters (Haas et al., 2011). Algae also produce dissolved organic matter (DOM) that is enriched in dissolved neutral sugars (DNS). These sugars can spur the growth of fast-growing bacteria in the water column, the result of which is a community with less bacterial diversity and dominated by copiotrophic bacteria like Vibrionaceae and Pseudoaltermonadaceae that typically carry virulence factors (Nelson et al., 2013). Conversely, corals exude DOM that selects for high bacterial diversity in the water column dominated by Alphaproteobacteria and few representatives with virulence factors (e.g., Hyphomonadaceae and Erythrobacteraceae) (Nelson et al., 2013). Other stressors in addition to climate change can also increase virulence sequences, for example increased nutrients, DOC, temperature or pH can all increase the abundance of virulence genes in the coral holobiont (Vega Thurber et al., 2009).
Overfishing also alters fish populations and induces trophic cascades (Jackson et al., 2001), and changes in fish functional roles can influence the reef-associated microbial communities. For example, within the territory of the damselfishes, Stegastes apicalis and S. nigricans, there were 2–3-fold increases in coral pathogens in the microbiome and a higher prevalence of corals with signs of black band disease. These Stegates species normally exclude macroalgae and cultivate filamentous algae, thus providing a trophic link among fish behavior, coral pathogen reservoirs and coral disease (Casey et al., 2014). Other fishes (i.e., Chaetodontidae) may also act directly as disease vectors (Raymundo et al., 2009), however, functionally diverse fish communities have been suggested to alleviate coral disease (Raymundo et al., 2009) and five Chaetodontidae and one Labridae species actually slowed the progression of blackband disease (Cole et al., 2009). Furthermore, damage to corals from abandoned fishing lines, is correlated with a 4-fold increase in coral disease and thus can somewhat explain the reduction in coral disease prevalence between marine reserve and non-reserve areas (Lamb et al., 2015).
There are several caveats associated with the data synthesized here, specifically evaluating changes in bacterial composition from disparate studies. These biases stem from uneven sequence library sizes, the use of different primer sets, low DNA amplification, and most importantly the use of different methods for assessment of microbiome response (e.g., culturing, 16S amplicons, metagenomes). Researchers in microbiology use a wide array of methodologies, which can make it difficult to compare studies quantitatively. At the same time, studies often reported only bacterial composition through relative abundance measures, and are therefore not directly quantifying bacterial cells within the coral microbiome. For example, if a study measures a decrease in a bacterial taxon, that taxon could be staying in consistent quantity while all other members of the microbiome are increasing. Thus, it is important to remember these biases when inferring from these data and keep in mind they are a broad qualitative overview of what happens to the coral microbiome under stress.
Some of the strongest evidence in support of the hypothesis that coral microbes provide their hosts with resilience or resistance to stressors comes from studies using antibiotics. For example, antibiotic treatment of thermally stressed corals caused tissue loss, significant declines in photosynthetic efficiency (Gilbert et al., 2012) and increased coral susceptibility to Vibrio shiloi infection and bleaching (Mills et al., 2013; although see Bellantuono et al., 2012). Furthermore, when corals were subjected to antibiotics and subsequently transplanted back onto the reef, those corals bleached and eventually died compared with the control corals that did not receive antibiotics (Glasl et al., 2016).
Early investigations into the role of DSMP cycling suggest that the coral microbiome likely plays an important role in coral resistance to stress. Coral-associated bacteria implicated in sulfur cycling (e.g., Endozoicomonas, Halomonas) (Raina et al., 2009; Todd et al., 2010) may help corals acclimate to climate change by protecting Symbiodinium from photosynthesis derived oxidative stress, as sulfur compounds such as DMSP and its breakdown products can act as antioxidants for marine algae (Sunda et al., 2002). As such, in bleaching corals, it was shown that there is a strong negative correlation between the abundance of bacterial pathogens and the abundance of the proposed symbiont Endozoicomonas (Pantos et al., 2015).
Nitrogen fixation and regulation by coral microbiome residents may also play an important role in coral resistance to stress. Symbiodinium depend on nitrogen for growth (Béraud et al., 2013), diazotroph abundance increases with higher seawater temperatures (Santos et al., 2014), and nitrogen fixation within corals increases with higher temperatures (Cardini et al., 2016). However, the mechanism behind and outcome of this relationship remains an active area of research. For example, it was recently suggested that these bacterial diazotrophs may in fact harm corals during heat stress by increasing the N:P ratio within the cells, causing a destabilization of the host-Symbiodinium partnership and thus inducing or prolonging holobiont bleaching events above normal levels (Rädecker et al., 2015). Yet diazotrophs still may play an important role in buffering the coral holobiont under water pollution stress by fixing nitrogen. In eutrophication experiments, nitrogen fixing and denitrifying bacteria increased in abundance in the coral Acropora hemprichii, but there were no significant changes to holobiont physiology (Jessen et al., 2013).
Finally, coral-associated bacteria may support coral resistance to algal growth simulated by overfishing given that resident bacteria defend the coral from microbial invasions (Rypien et al., 2010; Shnit-Orland et al., 2012; Welsh et al., 2016), thus potentially providing protection from algae altering the coral microbiome (Vega Thurber et al., 2012). There are clearly multiple mechanisms that influence these microbial roles and, as demonstrated above, a conflicting base of evidence. Recent research further supports the hypothesis that the microbiome plays a role in coral resistance, with coral microbiomes adapting to environmental conditions during reciprocal transplants and microbiomes that were previously exposed to a more variable and warmer environment having higher resistance during heat stress experiments (Ziegler et al., 2017). Clarity into these dynamics will likely grow as evidence increases and we suggest that these are key research topics to better understand the roles played by microbes in coral resistance to stress.
Our understanding of how the coral microbiome contributes to reef health is rapidly evolving, with the studies synthesized herein providing insight into how microbial communities respond to environmental change and hypotheses regarding the potential mechanisms underlying microbial support of coral resistance to stress. When stressors induce changes in coral-associated bacterial communities their beneficial functions to the coral holobiont can be lost. Thus, the composition of the coral microbiome could help to inform resource managers about which corals are most likely to successfully resist stressors, but this information has not yet been compiled. However, our results suggest that stressed corals have more taxonomically diverse microbiomes and increased beta diversity between coral colonies, potentially due to the stressed corals' inability to regulate incoming microbial members. At the same time, in stressed corals the opportunistic bacterial taxa Flavobacteriales, Rhodobacterales and Vibrionales are generally overrepresented, while the proposed coral symbiont, Endozoicimonas and related species, is underrepresented. Our results suggest metrics of microbiome diversity should be further investigated, especially beta diversity dispersion, which has only recently been applied to coral microbiome studies as a measure of microbiome stability between coral colonies (Zaneveld et al., 2016). Researchers should also utilize metrics of phylogenetic diversity through metrics like Faiths phylogenetic diversity, mean pairwise distance, and variation of pairwise distance (Tucker et al., 2017) to better understand the coral microbiome. In the future, resource managers may be able to identify stressed corals by the presence of opportunistic microbe taxa (Pollock et al., 2011) and by increased microbiome alpha and beta diversity, even if the coral has not yet shown physical signs of stress or deterioration.
To assist these trajectories, researchers need a better understanding of coral microbiome variability (e.g., temporal, spatial, seasonal, coral host species), to assemble databases of the microbiomes during different coral states (i.e., what is the normal microbiome for a coral species?), and determine the function of these bacteria for the coral host. With climate projections predicting that future conditions will increase disease susceptibility, pathogen abundance and virulence on coral reefs (Maynard et al., 2015) and diverging predicted responses of marine microbes to anthropogenic change (Hutchins and Fu, 2017), it is critical that researchers continue to advance understanding of the relationships between corals and their microbiomes, and how these change under stress. Promisingly, ongoing research suggests that we may have the ability to increase coral adaptation to these stressors by modifying the coral-associated microbial community (van Oppen et al., 2015) or by using coral probiotics (Krediet et al., 2013). This review and an ever-growing body of evidence suggest that conservationists may be able to screen for corals with resilient microbiomes to determine where best to focus management priorities as threats to coral reefs continue to accumulate.
JMI acknowledges funding from a Natural Sciences and Engineering Research Council of Canada (NSERC) Canada Graduate Scholarship and Michael Smith Foreign Study Supplement, and from the University of Victoria; JB acknowledges funding from an NSERC Discovery Grant. This work was also supported by a National Science Foundation Dimensions of Biodiversity grant (#1442306) to RVT.
JMI wrote the first draft of the manuscript, synthesized the papers, and created the figures. All authors contributed to the writing and revisions of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ainsworth, T. D., and Hoegh-Guldberg, O. (2009). Bacterial communities closely associated with coral tissues vary under experimental and natural reef conditions and thermal stress. Aquat. Biol. 4, 289–296. doi: 10.3354/ab00102
Ainsworth, T. D., Thurber, R. V., and Gates, R. D. (2010). The future of coral reefs: a microbial perspective. Trends Ecol. Evol. 25, 233–240. doi: 10.1016/j.tree.2009.11.001
Ainsworth, T., Krause, L., Bridge, T., Torda, G., Raina, J.-B., Zakrzewski, M., et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274. doi: 10.1038/ismej.2015.39
Al-Dahash, L. M., and Mahmoud, H. M. (2013). Harboring oil-degrading bacteria: a potential mechanism of adaptation and survival in corals inhabiting oil-contaminated reefs. Mar. Pollut. Bull. 72, 364–374. doi: 10.1016/j.marpolbul.2012.08.029
Apprill, A., Marlow, H. Q., Martindale, M. Q., and Rappé, M. S. (2009). The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 3, 685–699. doi: 10.1038/ismej.2009.3
Apprill, A., Weber, L. G., and Santoro, A. E. (2016). Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems 1:e00143–16. doi: 10.1128/mSystems.00143-16
Banin, E., Vassilakos, D., Orr, E., Martinez, R. J., and Rosenberg, E. (2003). Superoxide dismutase is a virulence factor produced by the coral bleaching pathogen Vibrio shiloi. Curr. Microbiol. 46, 418–422. doi: 10.1007/s00284-002-3912-5
Barott, K. L., Rodriguez-Brito, B., Janouškovec, J., Marhaver, K. L., Smith, J. E., Keeling, P., et al. (2011). Microbial diversity associated with four functional groups of benthic reef algae and the reef-building coral Montastraea annularis. Environ. Microbiol. 13, 1192–1204. doi: 10.1111/j.1462-2920.2010.02419.x
Barott, K. L., Rodriguez-Mueller, B., Youle, M., Marhaver, K. L., Vermeij, M. J. A., Smith, J. E., et al. (2012). Microbial to reef scale interactions between the reef-building coral Montastraea annularis and benthic algae. Proc. R. Soc. Lond. 279, 1655–1664. doi: 10.1098/rspb.2011.2155
Bayer, T., Neave, M. J., Alsheikh-Hussain, A., Aranda, M., Yum, L. K., Mincer, T., et al. (2013). The microbiome of the red sea coral stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762. doi: 10.1128/AEM.00695-13
Bellantuono, A. J., Hoegh-Guldberg, O., and Rodriguez-Lanetty, M. (2012). Resistance to thermal stress in corals without changes in symbiont composition. Proc. R. Soc. Lond. 279, 1100–1107. doi: 10.1098/rspb.2011.1780
Ben-Haim, Y., and Rosenburg, E. (2002). A novel Vibrio sp. pathogen of the coral Pocillopora damicornis. Mar. Biol. 141, 47–55. doi: 10.1007/s00227-002-0797-6
Ben-Haim, Y., Zicherman-Keren, M., and Rosenberg, E. (2003). Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003
Béraud, E., Gevaert, F., Rottier, C., and Ferrier-Pagès, C. (2013). The response of the scleractinian coral Turbinaria reniformis to thermal stress depends on the nitrogen status of the coral holobiont. J. Exp. Biol. 216, 2665–2674. doi: 10.1242/jeb.085183
Blackall, L. L., Wilson, B., and van Oppen, M. J. H. (2015). Coral-the world's most diverse symbiotic ecosystem. Mol. Ecol. 24, 5330–5347. doi: 10.1111/mec.13400
Bourne, D. G., Dennis, P. G., Uthicke, S., Soo, R. M., Tyson, G. W., and Webster, N. (2013). Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J. 7, 1452–1458. doi: 10.1038/ismej.2012.172
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Bourne, D. G., and Webster, N. S. (2013). “Coral reef bacterial communities,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 163–187.
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2007). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Brandt, M. E., Smith, T. B., Correa, A. M. S., and Vega Thurber, R. (2013). Disturbance driven colony fragmentation as a driver of a coral disease outbreak. PLoS ONE 8:e57164. doi: 10.1371/journal.pone.0057164
Brodie, J., Williamson, C., Barker, G. L., Walker, R. H., Briscoe, A., and Yallop, M. (2016). Characterising the microbiome of Corallina officinalis, a dominant calcified intertidal red alga. FEMS Microbiol. Ecol. 92:fiw110. doi: 10.1093/femsec/fiw110
Burke, C., Thomas, T., Lewis, M., Steinberg, P., and Kjelleberg, S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600. doi: 10.1038/ismej.2010.164
Cardini, U., van Hoytema, N., Bednarz, V. N., Rix, L., Foster, R. A., Al-Rshaidat, M. M. D., et al. (2016). Microbial dinitrogen fixation in coral holobionts exposed to thermal stress and bleaching. Environ. Microbiol. 18, 2620–2633. doi: 10.1111/1462-2920.13385
Carlos, C., Torres, T. T., and Ottoboni, L. M. M. (2013). Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Sci. Rep. 3:1624. doi: 10.1038/srep01624
Casey, J. M., Ainsworth, T. D., Choat, J. H., and Connolly, S. R. (2014). Farming behaviour of reef fishes increases the prevalence of coral disease associated microbes and black band disease. Proc. R. Soc. Lond. 281:1788. doi: 10.1098/rspb.2014.1032
Ceh, J., Kilburn, M. R., Cliff, J. B., Raina, J.-B., van Keulen, M., and Bourne, D. G. (2013a). Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium) with nitrogen acquired from bacterial associates. Ecol. Evol. 3, 2393–2400. doi: 10.1002/ece3.642
Ceh, J., van Keulen, M., and Bourne, D. G. (2013b). Intergenerational transfer of specific bacteria in corals and possible implications for offspring fitness. Microb. Ecol. 65, 227–231. doi: 10.1007/s00248-012-0105-z
Cole, A. J., Chong-Seng, K. M., Pratchett, M. S., and Jones, G. P. (2009). Coral-feeding fishes slow progression of black-band disease. Coral Reefs 28, 965–965. doi: 10.1007/s00338-009-0519-3
Darling, E. S., Alvarez-Filip, L., Oliver, T. A., McClanahan, T. R., and Côté, I. M. (2012). Evaluating life-history strategies of reef corals from species traits. Ecol. Lett. 15, 1378–1386. doi: 10.1111/j.1461-0248.2012.01861.x
Ding, J.-Y., Shiu, J.-H., Chen, W.-M., Chiang, Y.-R., and Tang, S.-L. (2016). Genomic insight into the host-endosymbiont relationship of Endozoicomonas montiporae CL-33(T) with its coral host. Front. Microbiol. 7:251. doi: 10.3389/fmicb.2016.00251
Dinsdale, E. A., Pantos, O., Smriga, S., Edwards, R. A., Angly, F., Wegley, L., et al. (2008). Microbial ecology of four coral atolls in the northern line islands. PLoS ONE 3:e1584. doi: 10.1371/journal.pone.0001584
Dubilier, N., Bergin, C., and Lott, C. (2008). Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740. doi: 10.1038/nrmicro1992
Egan, S., Harder, T., Burke, C., Steinberg, P., Kjelleberg, S., and Thomas, T. (2013). The seaweed holobiont: understanding seaweed-bacteria interactions. FEMS Microbiol. Rev. 37, 462–476. doi: 10.1111/1574-6976.12011
ElAhwany, A. M. D., Ghozlan, H. A., ElSharif, H. A., and Sabry, S. A. (2013). Phylogenetic diversity and antimicrobial activity of marine bacteria associated with the soft coral Sarcophyton glaucum. J. Basic Microbiol. 55, 2–10. doi: 10.1002/jobm.201300195
Ettinger, C. L., Voerman, S. E., Lang, J. M., Stachowicz, J. J., and Eisen, J. A. (2017). Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 5:e3246. doi: 10.7717/peerj.3246
Fahimipour, A. K., Kardish, M. R., Lang, J. M., Green, J. L., Eisen, J. A., and Stachowicz, J. J. (2017). Global-scale structure of the eelgrass microbiome. Appl. Environ. Microbiol. 83:12e03391–16. doi: 10.1128/AEM.03391-16
Frydenborg, B. R., Krediet, C. J., Teplitski, M., and Ritchie, K. B. (2013). Temperature-dependent inhibition of opportunistic Vibrio pathogens by native coral commensal bacteria. Microb. Ecol. 67, 392–401. doi: 10.1007/s00248-013-0334-9
Furby, K. A., Apprill, A., Cervino, J. M., Ossolinski, J. E., and Hughen, K. A. (2014). Incidence of lesions on Fungiidae corals in the eastern Red Sea is related to water temperature and coastal pollution. Mar. Environ. Res. 98, 29–38. doi: 10.1016/j.marenvres.2014.04.002
Gajigan, A. P., Diaz, L. A., and Conaco, C. (2017). Resilience of the prokaryotic microbial community of Acropora digitifera to elevated temperature. Microbiologyopen 72:e00478–11. doi: 10.1002/mbo3.478
Garren, M., Raymundo, L., Guest, J., Harvell, C. D., and Azam, F. (2009). Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS ONE 4:e7319. doi: 10.1371/journal.pone.0007319
Garren, M., Son, K., Raina, J.-B., Rusconi, R., Menolascina, F., Shapiro, O. H., et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007. doi: 10.1038/ismej.2013.210
Ghanbari, M., Kneifel, W., and Domig, K. J. (2015). A new view of the fish gut microbiome: advances from next-generation sequencing. Aquaculture. 448, 464–475. doi: 10.1016/j.aquaculture.2015.06.033
Gignoux-Wolfsohn, S. A., and Vollmer, S. V. (2015). Identification of candidate coral pathogens on white band disease-infected staghorn coral. PLoS ONE 10:e0134416. doi: 10.1371/journal.pone.0134416
Gilbert, J. A., Hill, R., Doblin, M. A., and Ralph, P. J. (2012). Microbial consortia increase thermal tolerance of corals. Mar. Biol. 159, 1763–1771. doi: 10.1007/s00227-012-1967-9
Givens, C. E., Ransom, B., and Bano, N. (2015). Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser. 518, 209–223. doi: 10.3354/meps11034
Glasl, B., Herndl, G. J., and Frade, P. R. (2016). The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 10, 2280–2292. doi: 10.1038/ismej.2016.9
Glasl, B., Webster, N. S., and Bourne, D. G. (2017). Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Mar. Biol. 164, 1–18. doi: 10.1007/s00227-017-3097-x
Haas, A. F., Nelson, C. E., Wegley Kelly, L., Carlson, C. A., Rohwer, F., Leichter, J. J., et al. (2011). Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. PLoS ONE 6:e27973. doi: 10.1371/journal.pone.0027973
Hadaidi, G., Röthig, T., Yum, L. K., Ziegler, M., Arif, C., Roder, C., et al. (2017). Stable mucus-associated bacterial communities in bleached and healthy corals of Porites lobata from the Arabian Seas. Sci. Rep. 7:45362. doi: 10.1038/srep45362
Hakim, J. A., Koo, H., Kumar, R., Lefkowitz, E. J., Morrow, C. D., Powell, M. L., et al. (2016). The gut microbiome of the sea urchin, Lytechinus variegatus, from its natural habitat demonstrates selective attributes of microbial taxa and predictive metabolic profiles. FEMS Microbiol. Ecol. 92:fiw146. doi: 10.1093/femsec/fiw146
Hester, E. R., Barott, K. L., Nulton, J., Vermeij, M. J., and Rohwer, F. L. (2016). Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J. 10, 1157–1169. doi: 10.1038/ismej.2015.190
Hughes, T. P. (1994). Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science 265, 1547–1551. doi: 10.1126/science.265.5178.1547
Hutchins, D. A., and Fu, F. (2017). Microorganisms and ocean global change. Nat. Microbiol. 2:17058. doi: 10.1038/nmicrobiol.2017.58
Jackson, J., Kirby, M. X., Berger, W. H., and Bjorndal, K. A. (2001). Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–638. doi: 10.1126/science.1059199
Jessen, C., Villa Lizcano, J. F., Bayer, T., Roder, C., Aranda, M., Wild, C., et al. (2013). In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the red sea coral Acropora hemprichii. PLoS ONE 8:e62091. doi: 10.1371/journal.pone.0062091
Jompa, J., and McCook, L. J. (2003). Coral-algal competition: macroalgae with different properties have different effects on corals. Mar. Ecol. Prog. Ser. 258, 87–95. doi: 10.3354/meps258087
Kimes, N. E., Van Nostrand, J. D., Weil, E., Zhou, J., and Morris, P. J. (2010). Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 12, 541–556. doi: 10.1111/j.1462-2920.2009.02113.x
Klaus, J. S., Janse, I., Heikoop, J. M., Sanford, R. A., and Fouke, B. W. (2007). Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. 9, 1291–1305. doi: 10.1111/j.1462-2920.2007.01249.x
Kline, D. I., Kuntz, N. M., Breitbart, M., Knowlton, N., and Rohwer, F. (2006). Role of elevated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser. 314, 119–125. doi: 10.3354/meps314119
Koren, O., and Rosenberg, E. (2006). Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl. Environ. Microbiol. 72, 5254–5259. doi: 10.1128/AEM.00554-06
Koren, O., and Rosenberg, E. (2008). Bacteria associated with the bleached and cave coral Oculina patagonica. Microb. Ecol. 55, 523–529. doi: 10.1007/s00248-007-9297-z
Krediet, C. J., Ritchie, K. B., Alagely, A., and Teplitski, M. (2012). Members of native coral microbiota inhibit glycosidases and thwart colonization of coral mucus by an opportunistic pathogen. ISME J. 7, 980–990. doi: 10.1038/ismej.2012.164
Krediet, C. J., Ritchie, K. B., Paul, V. J., and Teplitski, M. (2013). Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc. R. Soc. Lond. 280:20122328. doi: 10.1098/rspb.2012.2328
Kushmaro, A., Rosenberg, E., Fine, M., Ben-Haim, Y., and Loya, Y. (1998). Effect of temperature on bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar. Ecol. Prog. Ser. 171, 131–137. doi: 10.3354/meps171131
Kvennefors, E. C. E., Sampayo, E., Kerr, C., Vieira, G., Roff, G., and Barnes, A. C. (2011). Regulation of bacterial communities through antimicrobial activity by the coral holobiont. Microb. Ecol. 63, 605–618. doi: 10.1007/s00248-011-9946-0
Lamb, J. B., Williamson, D. H., Russ, G. R., and Willis, B. L. (2015). Protected areas mitigate diseases of reef-building corals by reducing damage from fishing. Ecology 96, 2555–2567. doi: 10.1890/14-1952.1
La Rivière, M., Roumagnac, M., Garrabou, J., and Bally, M. (2013). Transient shifts in bacterial communities associated with the temperate gorgonian Paramuricea clavata in the northwestern mediterranean sea. PLoS ONE 8:e57385. doi: 10.1371/journal.pone.0057385
Lee, S. T. M., Davy, S. K., Tang, S.-L., and Kench, P. S. (2016). Mucus Sugar Content Shapes the Bacterial Community Structure in Thermally Stressed Acropora muricata. Front. Microbiol. 7:371. doi: 10.3389/fmicb.2016.00371
Lee, S. T. M., Davy, S. K., Tang, S.-L., and Kench, P. S. (2017). Water flow buffers shifts in bacterial community structure in heat-stressed Acropora muricata. Sci. Rep. 7:43600. doi: 10.1038/srep43600
Lema, K. A., Bourne, D. G., and Willis, B. L. (2014). Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol. Ecol. 23, 4682–4695. doi: 10.1111/mec.12899
Lema, K. A., Willis, B. L., and Bourne, D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. doi: 10.1128/AEM.07800-11
Lesser, M. P., Falcón, L. I., Rodríguez-Román, A., Enríquez, S., Hoegh-Guldberg, O., and Iglesias-Prieto, R. (2007). Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar. Ecol. Prog. Ser. 346, 143–152. doi: 10.3354/meps07008
Lesser, M. P., Mazel, C. H., Gorbunov, M. Y., and Falkowski, P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. doi: 10.1126/science.1099128
Littman, R. A., Bourne, D. G., and Willis, B. L. (2010). Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol. Ecol. 19, 1978–1990. doi: 10.1111/j.1365-294X.2010.04620.x
Littman, R., Willis, B. L., and Bourne, D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 3, 651–660. doi: 10.1111/j.1758-2229.2010.00234.x
Looney, E. E., Sutherland, K. P., and Lipp, E. K. (2010). Effects of temperature, nutrients, organic matter and coral mucus on the survival of the coral pathogen, Serratia marcescens PDL100. Environ. Microbiol. 12, 2479–2485. doi: 10.1111/j.1462-2920.2010.02221.x
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Maynard, J., Van Hooidonk, R., and Eakin, C. M. (2015). Projections of climate conditions that increase coral disease susceptibility and pathogen abundance and virulence. Nat. Clim. Change 5, 688–695. doi: 10.1038/nclimate2625
McClanahan, T. R., Donner, S. D., Maynard, J. A., MacNeil, M. A., Graham, N. A. J., Maina, J., et al. (2012). Prioritizing key resilience indicators to support coral reef management in a changing climate. PLoS ONE 7:e42884. doi: 10.1371/journal.pone.0042884
McCliment, E. A., Nelson, C. E., Carlson, C. A., Alldredge, A. L., Witting, J., and Amaral-Zettler, L. A. (2012). An all-taxon microbial inventory of the Moorea coral reef ecosystem. ISME J. 6, 309–319. doi: 10.1038/ismej.2011.108
Meron, D., Atias, E., Iasur Kruh, L., Elifantz, H., Minz, D., Fine, M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5, 51–60. doi: 10.1038/ismej.2010.102
Meron, D., Rodolfo-Metalpa, R., Cunning, R., Baker, A. C., Fine, M., and Banin, E. (2012). Changes in coral microbial communities in response to a natural pH gradient. ISME J. 6, 1775–1785. doi: 10.1038/ismej.2012.19
Mills, E., Shechtman, K., Loya, Y., and Rosenberg, E. (2013). Bacteria appear to play important roles in both causing and preventing the bleaching of the coral Oculina patagonica. Mar. Ecol. Prog. Ser. 489, 155–162. doi: 10.3354/meps10391
Mitchell, R., and Chet, I. (1975). Bacterial attack of corals in polluted seawater. Microb. Ecol. 2, 227–233. doi: 10.1007/BF02010442
Moeller, A. H., Shilts, M., Li, Y., Rudicell, R. S., Lonsdorf, E. V., Pusey, A. E., et al. (2013). SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe 14, 340–345. doi: 10.1016/j.chom.2013.08.005
Morrow, K. M., Bourne, D. G., Humphrey, C., Botté, E. S., Laffy, P., Zaneveld, J., et al. (2015). Natural volcanic CO2 seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J. 9, 894–908. doi: 10.1038/ismej.2014.188
Morrow, K. M., Bromhall, K., Motti, C. A., Munn, C. B., and Bourne, D. G. (2017). Allelochemicals produced by brown macroalgae of the lobophora genus are active against coral larvae and associated bacteria, supporting pathogenic shifts to vibrio dominance. Appl. Environ. Microbiol. 83:e02391–16. doi: 10.1128/AEM.02391-16.
Morrow, K. M., Liles, M. R., Paul, V. J., Moss, A., and Chadwick, N. E. (2013). Bacterial shifts associated with coral–macroalgal competition in the Caribbean Sea. Mar. Ecol. Prog. Ser. 488, 103–117. doi: 10.3354/meps10394
Morrow, K. M., Moss, A. G., Chadwick, N. E., and Liles, M. R. (2012a). Bacterial associates of two caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78, 6438–6449. doi: 10.1128/AEM.01162-12
Morrow, K. M., Ritson-Williams, R., Ross, C., Liles, M. R., and Paul, V. J. (2012b). Macroalgal extracts induce bacterial assemblage shifts and sublethal tissue stress in caribbean corals. PLoS ONE 7:e44859. doi: 10.1371/journal.pone.0044859
Mouchka, M. E., Hewson, I., and Harvell, C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. doi: 10.1093/icb/icq061
Neave, M. J., Michell, C. T., Apprill, A., and Voolstra, C. R. (2017). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7:40579. doi: 10.1038/srep40579
Neave, M. J., Rachmawati, R., Xun, L., Michell, C. T., Bourne, D. G., Apprill, A., et al. (2016). Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200. doi: 10.1038/ismej.2016.95
Nelson, C. E., Goldberg, S. J., Kelly, L. W., Haas, A. F., Smith, J. E., Rohwer, F., et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 7, 962–979. doi: 10.1038/ismej.2012.161
Nugues, M. M., Smith, G. W., and Hooidonk, R. J. (2004). Algal contact as a trigger for coral disease. Ecology 7, 919–923. doi: 10.1111/j.1461-0248.2004.00651.x
Olson, N. D., Ainsworth, T. D., Gates, R. D., and Takabayashi, M. (2009). Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 371, 140–146. doi: 10.1016/j.jembe.2009.01.012
Pantos, O., Bongaerts, P., Dennis, P. G., Tyson, G. W., and Hoegh-Guldberg, O. (2015). Habitat-specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix. ISME J. 9, 1916–1927. doi: 10.1038/ismej.2015.3
Parris, D. J., Brooker, R. M., Morgan, M. A., Dixson, D. L., and Stewart, F. J. (2016). Whole gut microbiome composition of damselfish and cardinalfish before and after reef settlement. PeerJ 4:e2412. doi: 10.7717/peerj.2412
Peixoto, R., Rosado, P., and Leite, D. (2017). Beneficial Microorganisms for Corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 8:341. doi: 10.3389/fmicb.2017.00341
Pollock, F. J., Morris, P. J., Willis, B. L., and Bourne, D. G. (2011). The urgent need for robust coral disease diagnostics. PLoS Pathog. 7:e1002183. doi: 10.1371/journal.ppat.1002183
Rädecker, N., Pogoreutz, C., Voolstra, C. R., Wiedenmann, J., and Wild, C. (2015). Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 23, 490–497. doi: 10.1016/j.tim.2015.03.008
Raina, J.-B., Tapiolas, D. M., Forêt, S., Lutz, A., Abrego, D., Ceh, J., et al. (2013). DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502, 677–680. doi: 10.1038/nature12677
Raina, J. B., Tapiolas, D., Willis, B. L., and Bourne, D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501. doi: 10.1128/AEM.02567-08
Rasher, D. B., and Hay, M. E. (2010). Chemically rich seaweeds poison corals when not controlled by herbivores. Proc. Natl. Acad. Sci. U.S.A. 107, 9683–9688. doi: 10.1073/pnas.0912095107
Raymundo, L. J., Halford, A. R., Maypa, A. P., and Kerr, A. M. (2009). Functionally diverse reef-fish communities ameliorate coral disease. Proc. Natl. Acad. Sci. U.S.A. 106, 17067–17070. doi: 10.1073/pnas.0900365106
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Rohwer, F., Breitbart, M., Jara, J., Azam, F., and Knowlton, N. (2001). Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20, 85–91. doi: 10.1007/s003380100138
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Röthig, T., Ochsenkühn, M. A., Roik, A., van der Merwe, R., and Voolstra, C. R. (2016). Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol. Ecol. 25, 1308–1323. doi: 10.1111/mec.13567
Rypien, K. L., Ward, J. R., and Azam, F. (2010). Antagonistic interactions among coral-associated bacteria. Environ. Microbiol. 12, 28–39. doi: 10.1111/j.1462-2920.2009.02027.x
Salerno, J. L., Reineman, D. R., Gates, R. D., and Rappé, M. S. (2011). The effect of a sublethal temperature elevation on the structure of bacterial communities associated with the coral Porites compressa. J. Mar. Biol. 2011, 1–9. doi: 10.1155/2011/969173
Santos, H. F., Carmo, F. L., Duarte, G., Dini-Andreote, F., Castro, C. B., Rosado, A. S., et al. (2014). Climate change affects key nitrogen-fixing bacterial populations on coral reefs. ISME J. 8, 2272–2279. doi: 10.1038/ismej.2014.70
Sharp, K. H., Distel, D., and Paul, V. J. (2012). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6, 790–801. doi: 10.1038/ismej.2011.144
Sharp, K. H., Ritchie, K. B., Schupp, P. J., Ritson-Williams, R., and Paul, V. J. (2010). Bacterial acquisition in juveniles of several broadcast spawning coral species. PLoS ONE 5:e10898. doi: 10.1371/journal.pone.0010898
Sharp, K. H., Sneed, J. M., Ritchie, K. B., Mcdaniel, L., and Paul, V. J. (2015). Induction of larval settlement in the reef coral Porites astreoides by a cultivated marine Roseobacter strain. Biol. Bull. 228, 98–107. doi: 10.1086/BBLv228n2p98
Shnit-Orland, M., Sivan, A., and Kushmaro, A. (2012). Antibacterial activity of Pseudoalteromonas in the coral holobiont. Microb. Ecol. 64, 851–859. doi: 10.1007/s00248-012-0086-y
Smith, J. E., Shaw, M., Edwards, R. A., Obura, D., Pantos, O., Sala, E., et al. (2006). Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol. Lett. 9, 835–845. doi: 10.1111/j.1461-0248.2006.00937.x
Steinke, M., Brading, P., Kerrison, P., Warner, M. E., and Suggett, D. J. (2011). Concentrations of Dimethylsulfoniopropionate and dimethyl sulfide are strain-specific in symbiotic dinoflagellates (Symbiodinium sp., dinophyceae). J. Phycol. 47, 775–783. doi: 10.1111/j.1529-8817.2011.01011.x
Sunda, W., Kieber, D. J., Kiene, R. P., and Huntsman, S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320. doi: 10.1038/nature00851
Sutherland, K. P., Porter, J. W., Turner, J. W., Thomas, B. J., Looney, E. E., Luna, T. P., et al. (2010). Human sewage identified as likely source of white pox disease of the threatened Caribbean elkhorn coral, Acropora palmata. Environ. Microbiol. 12, 1122–1131. doi: 10.1111/j.1462-2920.2010.02152.x
Sweet, M. J., Bythell, J. C., and Nugues, M. M. (2013). Algae as reservoirs for coral pathogens. PLoS ONE 8:e69717. doi: 10.1371/journal.pone.0069717
Sweet, M. J., Croquer, A., and Bythell, J. C. (2010). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30, 39–52. doi: 10.1007/s00338-010-0695-1
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Thompson, J. R., Rivera, H. E., Closek, C. J., and Medina, M. (2014). Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front. Cell Infect. Microbiol. 4:176. doi: 10.3389/fcimb.2014.00176
Tianero, M. D. B., Kwan, J. C., Wyche, T. P., Presson, A. P., Koch, M., Barrows, L. R., et al. (2014). Species specificity of symbiosis and secondary metabolism in ascidians. ISME J. 9, 615–628. doi: 10.1038/ismej.2014.152
Todd, J. D., Curson, A. R. J., Nikolaidou-Katsaraidou, N., Brearley, C. A., Watmough, N. J., Chan, Y., et al. (2010). Molecular dissection of bacterial acrylate catabolism–unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ. Microbiol. 12, 327–343. doi: 10.1111/j.1462-2920.2009.02071.x
Tout, J., Siboni, N., Messer, L. F., Garren, M., Stocker, R., Webster, N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432. doi: 10.3389/fmicb.2015.00432
Tracy, A. M., Koren, O., Douglas, N., Weil, E., and Harvell, C. D. (2015). Persistent shifts in Caribbean coral microbiota are linked to the 2010 warm thermal anomaly. Environ. Microbiol. Rep. 7, 471–479. doi: 10.1111/1758-2229.12274
Tucker, C. M., Cadotte, M. W., Carvalho, S. B., Davies, T. J., Ferrier, S., Fritz, S. A., et al. (2017). A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. Camb. Philos. Soc. 92, 698–715. doi: 10.1111/brv.12252
van der Voort, M., Kempenaar, M., van Driel, M., Raaijmakers, J. M., and Mendes, R. (2016). Impact of soil heat on reassembly of bacterial communities in the rhizosphere microbiome and plant disease suppression. Ecol. Lett. 19, 375–382. doi: 10.1111/ele.12567
van Oppen, M. J. H., Oliver, J. K., Putnam, H. M., and Gates, R. D. (2015). Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313. doi: 10.1073/pnas.1422301112
Vega Thurber, R., Burkepile, D. E., Correa, A. M. S., Thurber, A. R., Shantz, A. A., Welsh, R., et al. (2012). Macroalgae decrease growth and alter microbial community structure of the reef-building coral, Porites astreoides. PLoS ONE 7:e44246. doi: 10.1371/journal.pone.0044246
Vega Thurber, R. L., Burkepile, D. E., Fuchs, C., Shantz, A. A., McMinds, R., and Zaneveld, J. R. (2013). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Change Biol. 20, 544–554. doi: 10.1111/gcb.12450
Vega Thurber, R., Willner-Hall, D., Rodriguez-Mueller, B., Desnues, C., Edwards, R. A., Angly, F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Webster, N. S., Negri, A. P., Botté, E. S., Laffy, P. W., Flores, F., Noonan, S., et al. (2016). Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6:19324. doi: 10.1038/srep19324
Webster, N. S., Negri, A. P., Flores, F., Humphrey, C., Soo, R., Botté, E. S., et al. (2012). Near-future ocean acidification causes differences in microbial associations within diverse coral reef taxa. Environ. Microbiol. Rep. 5, 243–251. doi: 10.1111/1758-2229.12006
Wegley, L., Edwards, R., Rodriguez-Brito, B., Liu, H., and Rohwer, F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x
Welsh, R. M., Rosales, S. M., Zaneveld, J., and Payet, J. P. (2015). Alien vs. Predator: pathogens open niche space for opportunists, unless controlled by predators. PeerJ. doi: 10.7287/peerj.preprints.1537v1
Welsh, R. M., Zaneveld, J. R., Rosales, S. M., Payet, J. P., Burkepile, D. E., and Thurber, R. V. (2016). Bacterial predation in a marine host-associated microbiome. ISME J. 10, 1540–1544. doi: 10.1038/ismej.2015.219
Zaneveld, J. R., McMinds, R., and Vega Thurber, R. (2017). An Anna Karenina Principle for Microbiomes: many stressors destabilize rather than predictably shift animal microbiomes. Nat Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Zaneveld, J. R., Shantz, A. A., Pritchard, C. E., McMinds, R., Payet, J. E. R. O. M. P., Welsh, R., et al. (2016). Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 7:11833. doi: 10.1038/ncomms11833
Zhang, Y., Ling, J., Yang, Q., Wen, C., Yan, Q., Sun, H., et al. (2015). The functional gene composition and metabolic potential of coral-associated microbial communities. Sci. Rep. 5:16191. doi: 10.1038/srep16191
Ziegler, M., Roik, A., Porter, A., Zubier, K., Mudarris, M. S., Ormond, R., et al. (2016). Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Mar. Pollut. Bull. 105, 629–640. doi: 10.1016/j.marpolbul.2015.12.045
Keywords: coral, bacteria, global change biology, environmental stress, symbiosis, global warming, pollution, overfishing
Citation: McDevitt-Irwin JM, Baum JK, Garren M and Vega Thurber RL (2017) Responses of Coral-Associated Bacterial Communities to Local and Global Stressors. Front. Mar. Sci. 4:262. doi: 10.3389/fmars.2017.00262
Received: 07 June 2017; Accepted: 31 July 2017;
Published: 15 August 2017.
Edited by:
Matthias Wietz, University of Oldenburg, GermanyReviewed by:
Amy Apprill, Woods Hole Oceanographic Institution, United StatesCopyright © 2017 McDevitt-Irwin, Baum, Garren and Vega Thurber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jamie M. McDevitt-Irwin, amFtaWUubWNpcndpbkBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.