
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci. , 18 July 2017
Sec. Coral Reef Research
Volume 4 - 2017 | https://doi.org/10.3389/fmars.2017.00222
This article is part of the Research Topic Horizon Scan 2017: Emerging Issues in Marine Science View all 12 articles
All animals on Earth form associations with microorganisms, including protists, bacteria, archaea, fungi, and viruses. In the ocean, animal–microbial relationships were historically explored in single host–symbiont systems. However, new explorations into the diversity of microorganisms associating with diverse marine animal hosts is moving the field into studies that address interactions between the animal host and a more multi-member microbiome. The potential for microbiomes to influence the health, physiology, behavior, and ecology of marine animals could alter current understandings of how marine animals adapt to change, and especially the growing climate-related and anthropogenic-induced changes already impacting the ocean environment. This review explores the nature of marine animal–microbiome relationships and interactions, and possible factors that may shift associations from symbiotic to dissociated states. I present a brief review of current microbiome research and opportunities, using examples of select marine animals that span diverse phyla within the Animalia, including systems that are more and less developed for symbiosis research, including two represented in my own research program. Lastly, I consider challenges and emerging solutions for moving these and other study systems into a more detailed understanding of host–microbiome interactions within a changing ocean.
Marine animals are the icons of life in the oceans. They represent about two million species (Mora et al., 2011) and include a wide range of body designs, from the highly simplistic sponges lacking true tissues and organs to the complex vertebrates containing specialized tissues and organs, such as fish and marine mammals, with some iconic representatives presented in Figure 1. The bodies of marine animals span several orders of magnitude in size, from the abundant planktonic copepod (1–2 mm) to the highly mobile blue whale (30 m), the largest animal on Earth. Marine animals are key members of ocean ecosystems and serve as both prey of and predators for other animals within the complex ocean food web. In contrast to terrestrial animals, marine animals have developed strategies for osmoregulation as well as highly specialized approaches for maintaining homeostasis within diverse temperature, oxygen and pressure gradients of the ocean (Graham, 1990; Knoll and Carroll, 1999). Marine animals also possess sophisticated specializations and functions that promote their success on or within their benthic or pelagic habitats, including specializations for living or enduring depths (outlined in Figure 1) that vary widely in factors such as light availability, access to food and predator exposure.
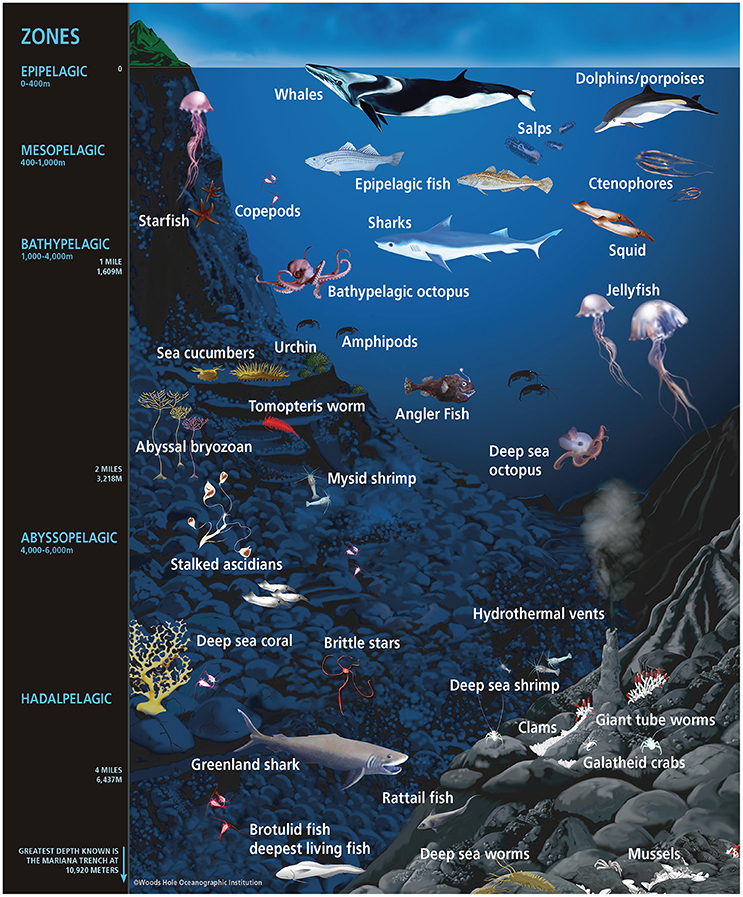
Figure 1. Illustration and common names of representative ocean animal life within their approximate depth-defined ecological habitats. Microorganisms exist on the surfaces and within the tissues and organs of the diverse life inhabiting the ocean, across all ocean habitats. Animals are not drawn to scale.
Marine animals share the sea with a vast diversity of microorganisms, including protists, bacteria, archaea, fungi, and viruses which comprise millions of cells in each milliliter of the 1.3 billion km3 of water comprising the oceans (Eakins and Sharman, 2010). These microorganisms are several micrometers or smaller in size, but collectively their roles in oxygen production, nutrient cycling, and organic matter degradation provide critical functions to the oceans and Earth (Arrigo, 2005; Falkowski et al., 2008). Microorganisms that associate with marine animals are part of the animal's microbiome, or collection of microorganisms that reside on or within the animal. Some of the microorganisms comprising the microbiomes of marine animals are thought to originate from this surrounding supply of seawater-associated cells (e.g., Nussbaumer et al., 2006), while other cells appear to have strict inheritance patterns, passed on through generations from the host (Sharp et al., 2007).
Over the past two decades, the widespread application of genomic and more integrative microbiological approaches have advanced our understanding of animal microbiomes (reviewed within McFall-Ngai et al., 2013). Symbiotic relationships between microorganisms and marine animals have been studied for decades, but technological advancements are providing new insights into the sheer diversity of microbial life in associ- ation with animals in the sea (Smith, 2001; Douglas, 2010). For example, reef-building corals are acknowledged as the icons of animal–microbial symbiosis in the sea, with corals hosting photosynthetic symbionts that make critical contributions to host nutrition (Muscatine et al., 1981). New reports of diverse protists, bacteria, archaea, and viruses in association with corals provide insights into the role of these cells for fulfilling diverse functional processes within the different niches of the coral host (reviewed within Thompson et al., 2015; Bourne et al., 2016). In fact, for many terrestrial animals, new reports of microbial symbioses provide insights into the variety of genetic and biochemical interactions and the ways that microorganisms contribute to animal health, behavior, and ecology (e.g., Ley, 2010; Cho and Blaser, 2012).
Understanding the microbiomes of marine animals is a growing research area within the field of marine science. Currently, the science is heavily focused on identifying consistent or “core” microbial members of the microbiome (Shade and Handelsman, 2012). After first gaining an understanding of “who's there” generally using diversity-based surveys targeting the small subunit (SSU) ribosomal RNA (rRNA) gene, these microbiomes are then often examined as a whole or in smaller units to understand the function of the cells, the nature of the associations and ultimately gain insight into the role of the microbiome in animal health, physiology, ecology, and behavior (Ezenwa et al., 2012; McFall-Ngai et al., 2013). Additionally, the ocean environment is changing at unprecedented rates due to climate-related and anthropogenic-induced impacts (Halpern et al., 2008; Doney et al., 2012), and the microbiome is also being investigated for its possible role as a sentinel of a changing host (Ainsworth and Gates, 2016).
How environmental changes and animal life history events affect the microbiomes of marine animals is growing area of research, and there is an emerging focus on better understanding interactions between the animal, microbiome, and ocean environment, including the elements that may define their exchanges (e.g., Meron et al., 2011; Lesser et al., 2016; Webster et al., 2016). Therefore, this review considers the symbiosis and dissociated stages of animal–microbiome associations, and discusses factors and causes that may alter interactions between animals and their microbiome. Next, this review discusses current research examining animal–microbiome relationships and interactions, by focusing on select systems that represent diverse marine animal phyla and which span the range of being more to less developed for microbiome research. Two of these systems, corals and marine mammals, are represented in my own research program. Lastly, this review concludes with a discussion of challenges in marine animal–microbiome research and opportunities available to further advance knowledge of animal–microbiome interactions in the ocean.
Host–microbiome dynamics are generally described as falling into two main categories: symbiosis, in which the organisms are involved in a normal metabolic and immune signaling interactions, and secondly dysbiosis, in which the relationship or interactions are heavily altered, possibly related to a major stress or infection event. While host–microbiome symbiosis and dysbiosis has been mostly considered in humans and humanized models (Hamdi et al., 2011; Nicholson et al., 2012; Scharschmidt and Fischbach, 2013), many of the same concepts are applicable to organisms in the sea (Egan and Gardiner, 2016), and are being explored in various systems (discussed below). The exact factors and mechanisms tipping the scale between symbiosis and dysbiosis will probably vary with complexity of the host anatomy and immune functioning (e.g., simplistic sponges and corals compared to more complex fish and sharks) as well as with the complexity of interactions that may occur between the members of the microbiome.
A normal animal–microbiome relationship in the ocean could be referred to as a “symbiotic” state, although the exact nature of the relationship may vary for each cell in the association. For example, cells residing on the surface or within the gut cavity of an animal are physically associated, yet do not share as intimate of an association as those microbes residing intracellularly with the host's cells. This normal symbiotic state is subject to a variety of environmental fluctuations, which are generally defined by the characteristics of the habitat (Figure 1). For example, in the ocean's upper photic zone, animals are exposed to variations in temperature and light, and host–symbiont interactions, especially in ectothermal animals, could alter on cycles such as seasons that generally control the temperature and light environment. Normal fluctuations in animal-specific patterns could also alter host–microbiome relationships. For example, changes in diet, possibly due to short-term prey availability, can alter gut microbiota and host–microbiome metabolic exchanges in other systems (e.g., David et al., 2014), and similar diet trends may also affect marine animals. Stress is another factor more complex animals encounter on a daily basis (e.g., squid, crabs, fish), which could be related to social/territorial encounters or chasing or fleeing from prey, and the short-term production of stress hormones such as cortisol can influence host–microbiome relationships (e.g., Moloney et al., 2014).
There are also normal animal life events that occur on longer time frames or that are more drastic in scope, such as animal development, aging, and reproduction. In non-marine animals, these factors have been shown to cause alterations in animal-microbial relationships (e.g., Heintz and Mair, 2014). These changes can be drastic enough to cause a state of “altered symbiosis” that could extend for short or longer term. For example, the gut microbiome of women generally becomes altered during pregnancy (Koren et al., 2012). Events resulting in normal animal stress may also lead to a more altered symbiotic state, for example if social conflict was more chronic, perhaps due to the pressures of a particular habitat. Data from humans and humanized models suggests that the microbial community and associated genes do fluctuate with the normal variations and animal life events, and both may be considered “healthy” fluctuations (Nicholson et al., 2012). However, how these fluctuations affect exchanges between the host and microbiome is much less understood.
If symbiosis and altered symbiosis are considered as normal host–microbiome variations throughout an organism's life, dysbiosis is the breakdown in the relationship, generally related to one or more major stressors, and can greatly alter host health and lead to a disease state (Holmes et al., 2011). The stressor may come from an external source, such as a pollutant, infective agent, or a longer-term natural environmental change—and there are probably countless other factors that could fit this category (Figure 2). For example, one of the most visible signs of host–microbiome dysbiosis is with scleractinian corals, whose relationship with unicellular algae breaks down after long-term yet small increases in seawater temperature, causing the coral to become “bleached” (Brown, 1997). In humanized models, major stressors such as malnutrition are related to less physically visible changes in innate immunity, which are linked to microbial ecology (Hashimoto et al., 2012). Understanding the relationship between symbiosis, dysbiosis and host health and functioning are general topics of research in most host–microbiome studies, but the environmental changes occurring in the ocean environment have made this area of research more pressing for marine animals. Overall, the concepts behind the model presented in Figure 2, as well as variations of this model, are generally driving much of the current research examining animal-microbial relationships in the ocean.
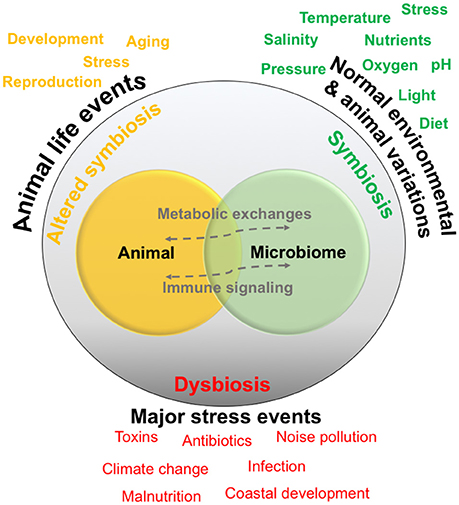
Figure 2. Conceptual diagram of a host-microbiome relationship. Relationships are generally thought to exist in a symbiotic state, and are normally exposed to environmental and animal-specific factors that may cause natural variations. Some events may change the relationship into a functioning but altered symbiotic state, whereas extreme stress events may cause dysbiosis or a breakdown of the relationship and interactions.
The microbiomes of diverse marine animals are currently under study, from simplistic organisms including sponges (e.g., Webster et al., 2010) and ctenophores (Daniels and Breitbart, 2012) to more complex organisms such as sea squirts (Blasiak et al., 2014) and sharks (Givens et al., 2015). Below I present some of the current study systems that represent a diverse cross-section of marine animal phyla, and trends of research in these systems including focus on symbiosis and dysbiosis. The organisms are generally presented in order from increasing to decreasing knowledge about the host-microbiome relationship.
The relationship between the Hawaiian bobtail squid Euprymna scolopes (phylum Mollusca) and the bioluminescent bacterium Vibrio fisheri (also recognized as Aliivibrio fisheri) is one of the best studied symbiotic relationships in the sea and is a choice system for general symbiosis research (Figures 3A,B). The E. scolopes-V. fisheri relationship has provided insight into fundamental processes in animal-microbial symbioses, and especially biochemical interactions and signaling between the host and bacterium (McFall-Ngai, 2000, 2014). Much of this research focuses on establishment of the symbiosis, with less focus on dysbiosis. Additionally, because V. fisheri exists in the light organ, these studies have been primarily limited to this one isolated relationship, with the remainder of the squid's microbiome virtually unstudied (but see Barbieri et al., 2001; Collins et al., 2012). The E. scolopes–V. fisheri system offers simplicity for the study of host–microbial interactions and numerous helpful developments in animal husbandry, genomic tools, and experimental design that could be applied to ask more comprehensive questions about squid–microbiome interactions, including the conditions leading to dysbiosis of relationships.
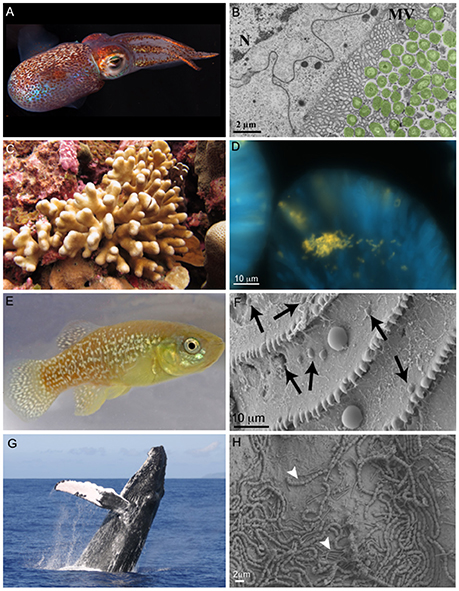
Figure 3. Photographs of marine animals and their associated microbiomes from select study systems. Photographs include: the Hawaiian bobtail squid Euprymna scolopes (A) and a transmission electron micrograph of Vibrio fisheri cells associating with dense microvilli (MV) and in proximity to the epithelial nucleus (N) within the light organ (B); the reef-building coral Stylophora pistillata (C) and a microscopy image of Endozoicomonas cells (probed yellow using in situ hybridization) within the tentacles of a S. pistillata host (D); the Atlantic killifish (Fundulus heteroclitus) (E) and a scanning electron microscopy (SEM) image of the surface and scales of the fish, with arrows pointing to bacterial-sized cells and larger cells (which are not noted) are presumably phytoplankton (F); a humpback whale (Megaptera novaeangliae) breaching (G) and a scanning electron microscopy image of a humpback's skin surface associated bacteria, with arrows indicating two different cell morphologies (H). Photographs (A,B) were produced by M. McFall-Ngai and were previously published photographs (McFall-Ngai, 2014), (C,H) were previously published by the author Neave et al. (2016) and Apprill et al. (2014), photograph (D) was taken by Liping Xun and photograph (E) by Evan D'Alessandro.
Similar to E. scolope, the gutless marine oligochaete worm Olavius algarvensis (phylum Annelida) is another relatively well-studied marine host to microbes. One major difference is that it has been studied within the context of a larger consortium of microorganisms compared to E. scolope. These 3 cm long worms reside within shallow marine sediments of the Mediterranean Sea. The worms do not contain a mouth or a digestive or excretory system, but are instead nourished with the help of a suite of extracellular bacterial endosymbionts that reside upon coordinated use of sulfur present in the environment (Dubilier et al., 2001). This system has benefited from some of the most sophisticated ‘omics and visualization tools (Woyke et al., 2006). For example, multi-labeled probing has improved visualization of the microbiome (Schimak et al., 2016) and transcriptomics and proteomics have been applied to examine host–microbiome interactions, including energy transfer between the host and microbes (Kleiner et al., 2012) and recognition of the consortia by the worm's innate immune system (Wippler et al., 2016). The major strength of this system is that it does offer the ability to study host–microbiome interactions with a low diversity microbial consortium, and it also offers a number of host and microbial genomic resources (e.g., Woyke et al., 2006; Ruehland et al., 2008). Dysbiosis has not been heavily investigated in this system, and given the growing knowledge of host–microbial interactions, O. algarvensis could be an imperative animal for dysbiosis research.
As mentioned above, corals (phylum Cnidaria) (Figure 3C) are one of the most common examples of an animal host whose symbiosis with microalgae can turn to dysbiosis, and is visibly detected as bleaching. Coral microbiomes have been examined in a variety of studies, which demonstrate how variations in the ocean environment, most notably temperature, light, and inorganic nutrients, affect the abundance and performance of the microalgal symbionts, as well as calcification and physiology of the host (Dubinsky and Jokiel, 1994; Anthony et al., 2008). Studies have also suggested that resident bacteria, archaea, and fungi additionally contribute to nutrient and organic matter cycling within the coral, with viruses also possibly playing a role in structuring the composition of these members, thus providing one of the first glimpses at a multi-domain marine animal symbiosis (reviewed in Bourne et al., 2016). The gammaproteobacterium Endozoicomonas is emerging as a central member of the coral's microbiome, with flexibility in its lifestyle (Figure 3D) (Neave et al., 2016, 2017). Ocean disturbances including elevated temperature and ocean acidification have been shown to disrupt the coral's associated bacteria (Thurber et al., 2009; Meron et al., 2011), including relationships with Endozoicomonas (Morrow et al., 2015). However, some members of this microbiome appear to be stable across large environmental gradients (Hernandez-Agreda et al., 2016). In addition to nutrition, the microbiome plays a role in coral health and stress. Temperature and light stress to corals can result in overproduction of reactive oxygen species (ROS), which can be detrimental to Symbiodinium and result in bleaching, but the associated bacteria have also recently been shown to contribute extracellular ROS (Diaz et al., 2016; Zhang et al., 2016), which could play a signaling role with the host or within the microbiome. Given the recent mass bleaching occurring on reefs (Hughes et al., 2017), corals will likely continue to be a useful and popular system for symbiosis and dysbiosis research. There are number of resources available to further promote study of the coral microbiome, including integrated databases (Franklin et al., 2012; Madin et al., 2016), a growing number of host and microbial genomes (Shinzato et al., 2011; Bayer et al., 2012; Neave et al., 2017), and laboratory amendable “model” systems (Weis et al., 2008; Baumgarten et al., 2015).
Sponges (phylum Porifera) are common members of the ocean's diverse benthic habitats and their abundance and ability to filter large volumes of seawater have led to the awareness that these organisms play critical roles in influencing benthic and pelagic processes in the ocean (Bell, 2008). They are one of the oldest lineages of animals, and have a relatively simple body plan that commonly associates with bacteria, archaea, algal protists, fungi, and viruses (reviewed within Webster and Thomas, 2016). Sponge microbiomes are composed of specialists and generalists, and complexity of their microbiome appears to be shaped by host phylogeny (Thomas et al., 2016). Studies have shown that the sponge microbiome contributes to nitrogen cycling in the oceans, especially through the oxidation of ammonia by archaea and bacteria (Bayer et al., 2008; Radax et al., 2012). Most recently, microbial symbionts of tropical sponges were shown to produce and store polyphosphate granules (Zhang et al., 2015), perhaps enabling the host to survive periods of phosphate depletion in oligotrophic marine environments (Colman, 2015). The microbiomes of some sponge species do appear to change in community structure in response to changing environmental conditions, including temperature (Simister et al., 2012a) and ocean acidification (Morrow et al., 2015; Ribes et al., 2016), as well as synergistic impacts (Lesser et al., 2016). Understanding the effect of these altered host–microbiome interactions on sponge growth and ecology are topics for further research. As such, there are a number of resources to support research on sponges including a curated database of sponge–microbial sequences (Simister et al., 2012b), cultivated microbial isolates and sponge cell cultures from some species (Taylor et al., 2007) to facilitate investigations.
Atlantic killifish, (Fundulus spp., Phylum Chordata) (Figure 3E) are one of the most abundant estuarine fishes in North America, and are related to other families with more global distributions in coastal areas (Fritz et al., 1975; Lotrich, 1975). The killifish have a broad North American geographic distribution yet limited subpopulation movement, and thus the Atlantic killifish have become a useful field-residing model species for examining biological and ecological responses to natural environment conditions (salinity, oxygen, pH, and temperature) as well as chemical pollutants (Burnett et al., 2007). While the killifish microbiome (Figure 3F) has not been extensively studied, there is work examining the influence of pollutants on the skin and mucus of the fish, which suggests that this skin microbial community is relatively resistant to change (Larsen et al., 2015). Populations of the fish offer a unique host genetic resistance to toxicity (Hahn et al., 2004), and it is possible that this resistance is also facilitated by features of the microbiome. The Atlantic killifish appear to be an ideal study species for microbiome investigations and especially the response of the host–microbiome symbiosis to changing ocean conditions. Specifically, the killifish can be maintained in laboratory aquaria, they are hardy and amendable to experimental manipulation, and spawning material can be acquired for developmental (Burnett et al., 2007).
The microbiomes of marine mammals (phylum Chordata) (Figure 3G) have recently been investigated and offer a comparative study system to terrestrial mammals (reviewed within Nelson et al., 2015). Marine mammals are often viewed as sentinel species of the ocean, because they appear to rapidly respond to ocean conditions, disturbances, and pathogens similarly to humans (Bossart, 2011). Several studies have examined the skin (Figure 3H), gut and respiratory microbiomes of diverse marine mammal species, and describe species-specific relationships (Johnson et al., 2009; Apprill et al., 2014; Bik et al., 2016). Connections between the community composition of the microbiome and animal health (Apprill et al., 2014) and diet (Nelson et al., 2013; Sanders et al., 2015) have been made, and more detailed studies are needed to understand these specific connections. While there are very limited resources available for studying host–microbiome interactions in marine mammals, there are some animals in captivity as well as well-studied populations that will heighten investigations of host–microbiome symbiosis and dysbiosis in these sentinel species.
A number of the systems highlighted above are currently examining animal–microbiome interactions, but these are generally most developed in systems such as O. algarvensis that offer lower complexity microbiomes, or within the single host–symbiont relationship between E. scolopes and V. fisheri. As such, a major challenge to the field is exploring host–microbiome interactions within the context of a diverse microbiome, and especially if the microbiome includes members such a uncharacterized protists, fungi, and viruses, which have generally not been described in most marine animal systems. Therefore, a through description of the microbiome is a first necessity, but this still presents many challenges on a variety of levels. For example, amplifying or shotgun sequencing microbial DNA with the presence of abundant host cells often requires optimization or high sequencing output (e.g., Rocha et al., 2014; Weber et al., 2017). Taxonomic databases generally contain few microbial sequences from many of these animals, and therefore simple tasks such as assigning taxonomy can be challenging. Developing animal-specific databases (Simister et al., 2012b), which include the next-generation supplied sequences generally not available in curated taxonomic databases, could help alleviate this problem. There are also a number of new tools for metagenomics-based analysis, including advancements in binning genomes from complex samples (Kang et al., 2015; Graham et al., 2017) as well as new visualization methods for comparing genomes (Eren et al., 2015; Wagner et al., 2017). A challenging issue that has received less attention is how to gain information from unknown genes and gene families, which can make up over half of the environmental microbial genomes. Algorithms utilizing gene function predictions do provide some assistance with this problem (Mi et al., 2015), and these tools may improve as more environmental microbial genomes are available. Lastly, computational tools are emerging to facilitate identifying associations between host genetic variation and microbiome composition (Lynch et al., 2016).
Once some of these hurdles are overcome and a comprehensive view of the microbiome is available, researchers can then explore the nature of the host–microbiome relationship. Visualization using a variety of different microscopy-based techniques is a powerful tool to recognize the physical relationship between a host and the microbiome, as well as the organization of cells within the microbiome. Electron microscopy provides the most detailed information about this organization, but this is less useful for complex microbiomes because taxonomically distinct microbial cells with similar appearances cannot be distinguished. Fluorescent in situ hybridization (FISH), and especially using a multi-taxonomic, simultaneous probing technique such as Combinatorial Labeling and Spectral Imaging FISH (CLASI-FISH) (Valm et al., 2011) can provide significant insight into host–microbe and microbe–microbe interactions. FISH techniques do require optimization for some animal systems, such as corals that possess autofluorescent host tissues (Wada et al., 2016). Visualization techniques can also be paired with isotope probing, to provide opportunities to trace the transfer of specific molecules between the host and microbiome, as well as within the microbiome using Nano-SIMS and Nano-SIP approaches (Musat et al., 2016). There have also been many recent instrumental and database advances in the field of metabolomics (Beisken et al., 2015), and this approach is beginning to be applied to examine host–microbiome interactions (Gomez et al., 2015; Sogin et al., 2016). An understanding of specific microbial metabolites will help facilitate targeted investigations of how these products affect the host nutritional and immune systems.
Lastly, experimental manipulation is a challenge to the study of host–microbial interactions in the ocean. Studying the animals in their natural environment is the most ideal approach because it ensures that the surrounding seawater microbial community is maintained. However, natural experiments are only as helpful as the natural variability in the host-microbe system, and generally only afford the opportunity to study events such as seasonality, animal growth or other life history events. Artificial systems such as aquaria or mesocosms offer opportunities to manipulate environmental conditions or expose the animal to antibiotics or other molecules that are difficult to dose in the wild. However, not all animals are ideal for these systems (e.g., large whales, hydrothermal vent worms), and it can be challenging to reproduce some environmental conditions. Advances in aquaria design that offer consistency in environmental conditions and the ability to manipulate complex environmental interactions, such as the Australian Institute of Marine Science's National Sea Simulator, provide opportunities to conduct more realistic experiments. As the need to understand how host–microbiome interactions will alter with the forecasted changes in ocean temperature and pH, facilities such as this will become critical to animal–microbiome research in the ocean.
While studies of marine animal–microbiome interactions are certainly plagued by a number of challenges, the future is also very bright for this emerging field. Many of the new bioinformatics and methodological advancements now available to marine biologists stem from the biomedical field, and thus marine animal microbiome research, as well as other environmental-based fields, are profiting from the elevation in microbiome research funding and attention. There could also be growing interest in using marine animals as models for examining resilience, promoted by the fact that alterations in the ocean conditions are often outpacing those in terrestrial environments. Given the phylogenetic breath of animals in the ocean, coupled with the many diverse ocean environments, there is certainly a wealth of research opportunities available to study host–microbiome interactions in the ocean.
The author confirms being the sole contributor of this work and approved it for publication.
Funding was provided by the WHOI's Andrew W. Mellon Foundation Endowed Fund for Innovative Research.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Many thanks to Margaret McFall-Ngai and Evan D'Alessandro for use of images and to Laura Weber for early comments on this review.
Ainsworth, T. D., and Gates, R. D. (2016). Corals' microbial sentinels. Science 352, 1518–1519. doi: 10.1126/science.aad9957
Anthony, K. R., Kline, D. I., Diaz-Pulido, G., Dove, S., and Hoegh-Guldberg, O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. U.S.A. 105, 17442–17446. doi: 10.1073/pnas.0804478105
Apprill, A., Robbins, J., Eren, A. M., Pack, A. A., Reveillaud, J., Mattila, D., et al. (2014). Humpback whale populations share a core skin bacterial community: Towards a health index for marine mammals? PLoS ONE 9:e90785. doi: 10.1371/journal.pone.0090785
Arrigo, K. R. (2005). Marine microorganisms and global nutrient cycles. Nature 437, 349–355. doi: 10.1038/nature04159
Barbieri, E., Paster, B. J., Hughes, D., Zurek, L., Moser, D. P., Teske, A., et al. (2001). Phylogenetic characterization of epibiotic bacteria in the accessory nidamental gland and egg capsules of the squid Loligo pealei (Cephalopoda: Loliginidae). Environ. Microbiol. 3, 151–167. doi: 10.1046/j.1462-2920.2001.00172.x
Baumgarten, S., Simakov, O., Esherick, L. Y., Liew, Y. J., Lehnert, E. M., Michell, C. T., et al. (2015). The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 11893–11898. doi: 10.1073/pnas.1513318112
Bayer, K., Schmitt, S., and Hentschel, U. (2008). Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ. Microbiol. 10, 2942–2955. doi: 10.1111/j.1462-2920.2008.01582.x
Bayer, T., Aranda, M., Sunagawa, S., Yum, L. K., Desalvo, M. K., Lindquist, E., et al. (2012). Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS ONE 7:e35269. doi: 10.1371/journal.pone.0035269
Beisken, S., Eiden, M., and Salek, R. M. (2015). Getting the right answers: understanding metabolomics challenges. Expert Rev. Mol. Diagn. 15, 97–109. doi: 10.1586/14737159.2015.974562
Bell, J. J. (2008). The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 79, 341–353. doi: 10.1016/j.ecss.2008.05.002
Bik, E. M., Costello, E. K., Switzer, A. D., Callahan, B. J., Holmes, S. P., Wells, R. S., et al. (2016). Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun. 7:10516. doi: 10.1038/ncomms10516
Blasiak, L. C., Zinder, S. H., Buckley, D. H., and Hill, R. T. (2014). Bacterial diversity associated with the tunic of the model chordate Ciona intestinalis. ISME J. 8, 309–320. doi: 10.1038/ismej.2013.156
Bossart, G. D. (2011). Marine mammals as sentinel species for oceans and human health. Vet. Pathol. 48, 676–690. doi: 10.1177/0300985810388525
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Brown, B. E. (1997). Coral bleaching: causes and consequences. Coral Reefs 16, S129–S138. doi: 10.1007/s003380050249
Burnett, K. G., Bain, L. J., Baldwin, W. S., Callard, G. V., Cohen, S., Di Giulio, R. T., et al. (2007). Fundulus as the premier teleost model in environmental biology: opportunities for new insights using genomics. Comp. Biochem. Physiol. 2, 257–286. doi: 10.1016/j.cbd.2007.09.001
Cho, I., and Blaser, M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270. doi: 10.1038/nrg3182
Collins, A. J., Labarre, B. A., Won, B. S. W., Shah, M. V., Heng, S., Choudhury, M. H., et al. (2012). Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl. Environ. Microbiol. 78, 4200–4208. doi: 10.1128/AEM.07437-11
Colman, A. S. (2015). Sponge symbionts and the marine P cycle. Proc. Natl. Acad. Sci. U.S.A. 112, 4191–4192. doi: 10.1073/pnas.1502763112
Daniels, C., and Breitbart, M. (2012). Bacterial communities associated with the ctenophores Mnemiopsis leidyi and Beroe ovata. FEMS Microbiol. Ecol. 82, 90–101. doi: 10.1111/j.1574-6941.2012.01409.x
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Diaz, J., Hansel, C., Apprill, A., Brighi, C., Zhang, T., Weber, L., et al. (2016). Species-specific control of external superoxide levels by the coral holobiont during a natural bleaching event. Nat. Commun. 7:13801. doi: 10.1038/ncomms13801
Doney, S. C., Ruckelshaus, M., Duffy, J. E., Barry, J. P., Chan, F., English, C. A., et al. (2012). Climate change impacts on marine ecosystems. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Dubilier, N., Mulders, C., Ferdelman, T., De Beer, D., Pernthaler, A., Klein, M., et al. (2001). Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411, 298–302. doi: 10.1038/35077067
Dubinsky, Z., and Jokiel, P. L. (1994). Ratio of energy and nutrient fluxes regulates symbiosis between zooxanthellae and corals. Pac. Sci. 48, 313–324.
Eakins, B. W., and Sharman, G. F. (2010). Volumes of the World's Oceans from ETOPO1. National Geophysical Data Center, Boulder, CO.
Egan, S., and Gardiner, M. (2016). Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 7:991. doi: 10.3389/fmicb.2016.00991
Eren, A. M., Esen, Ö. C., Quince, C., Vineis, J. H., Morrison, H. G., Sogin, M. L., et al. (2015). Anvi'o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319
Ezenwa, V. O., Gerardo, N. M., Inouye, D. W., Medina, M., and Xavier, J. B. (2012). Animal behavior and the microbiome. Science 338, 198–199. doi: 10.1126/science.1227412
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Franklin, E. C., Stat, M., Pochon, X., Putnam, H. M., and Gates, R. D. (2012). GeoSymbio: a hybrid, cloud-based web application of global geospatial bioinformatics and ecoinformatics forSymbiodinium–host symbioses. Mol. Ecol. Resour. 12, 369–373. doi: 10.1111/j.1755-0998.2011.03081.x
Fritz, E. S., Meredith, W. H., and Lotrich, V. A. (1975). Fall and winter movements and activity level of the mummichog, Fundulus heteroclitus, in a tidal creek. Chesapeake Sci. 16, 211–214. doi: 10.2307/1350898
Givens, C. E., Ransom, B., Bano, N., and Hollibaugh, J. T. (2015). Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser. 518, 209–223. doi: 10.3354/meps11034
Gomez, A., Petrzelkova, K., Yeoman, C. J., Vlckova, K., Mrázek, J., Koppova, I., et al. (2015). Gut microbiome composition and metabolomic profiles of wild western lowland gorillas (Gorilla gorilla gorilla) reflect host ecology. Mol. Ecol. 24, 2551–2565. doi: 10.1111/mec.13181
Graham, E. D., Heidelberg, J. F., and Tully, B. J. (2017). BinSanity: unsupervised clustering of environmental microbial assemblies using coverage and affinity propagation. PeerJ 5:e3035. doi: 10.7717/peerj.3035
Graham, J. B. (1990). Ecological, evolutionary, and physical factors influencing aquatic animal respiration. Am. Zool. 30, 137–146. doi: 10.1093/icb/30.1.137
Hahn, M. E., Karchner, S. I., Franks, D. G., and Merson, R. R. (2004). Aryl hydrocarbon receptor polymorphisms and dioxin resistance in Atlantic killifish (Fundulus heteroclitus). Pharmacogenetics 14, 131–143. doi: 10.1097/00008571-200402000-00007
Halpern, B. S., Walbridge, S., Selkoe, K. A., Kappel, C. V., Micheli, F., D'agrosa, C., et al. (2008). A global map of human impact on marine ecosystems. Science 319, 948–952. doi: 10.1126/science.1149345
Hamdi, C., Balloi, A., Essanaa, J., Crotti, E., Gonella, E., Raddadi, N., et al. (2011). Gut microbiome dysbiosis and honeybee health. J. Appl. Entomol. 135, 524–533. doi: 10.1111/j.1439-0418.2010.01609.x
Hashimoto, T., Perlot, T., Rehman, A., Trichereau, J., Ishiguro, H., Paolino, M., et al. (2012). ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487, 477–481. doi: 10.1038/nature11228
Heintz, C., and Mair, W. (2014). You are what you host: microbiome modulation of the aging process. Cell 156, 408–411. doi: 10.1016/j.cell.2014.01.025
Hernandez-Agreda, A., Leggat, W., Bongaerts, P., and Ainsworth, T. D. (2016). The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. MBio 7:e00560–16. doi: 10.1128/mBio.00560-16
Holmes, E., Li, J. V., Athanasiou, T., Ashrafian, H., and Nicholson, J. K. (2011). Understanding the role of gut microbiome–host metabolic signal disruption in health and disease. Trends Microbiol. 19, 349–359. doi: 10.1016/j.tim.2011.05.006
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Johnson, W. R., Torralba, M., Fair, P. A., Bossart, G. D., Nelson, K. E., and Morris, P. J. (2009). Novel diversity of bacterial communities associated with bottlenose dolphin upper respiratory tracts. Environ. Microbiol. Rep. 1, 555–562. doi: 10.1111/j.1758-2229.2009.00080.x
Kang, D. D., Froula, J., Egan, R., and Wang, Z. (2015). MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165
Kleiner, M., Wentrup, C., Lott, C., Teeling, H., Wetzel, S., Young, J., et al. (2012). Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc. Natl. Acad. Sci. U.S.A. 109, E1173–E1182. doi: 10.1073/pnas.1121198109
Knoll, A. H., and Carroll, S. B. (1999). Early animal evolution: emerging views from comparative biology and geology. Science 284, 2129–2137. doi: 10.1126/science.284.5423.2129
Koren, O., Goodrich, J. K., Cullender, T. C., Spor, A., Laitinen, K., Bäckhed, H. K., et al. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480. doi: 10.1016/j.cell.2012.07.008
Larsen, A., Bullard, S., Womble, M., and Arias, C. (2015). Community structure of skin microbiome of gulf killifish, Fundulus grandis, is driven by seasonality and not exposure to oiled sediments in a Louisiana salt marsh. Microbiol. Ecol. 70, 1–11. doi: 10.1007/s00248-015-0578-7
Lesser, M. P., Fiore, C., Slattery, M., and Zaneveld, J. (2016). Climate change stressors destabilize the microbiome of the Caribbean barrel sponge, Xestospongia muta. J. Exp. Mar. Biol. Ecol. 475, 11–18. doi: 10.1016/j.jembe.2015.11.004
Ley, R. E. (2010). Obesity and the human microbiome. Curr. Opin. Gastroenterol. 26, 5–11. doi: 10.1097/MOG.0b013e328333d751
Lotrich, V. A. (1975). Summer home range and movements of Fundulus heteroclitus (Pisces: Cyprinodontidae) in a tidal creek. Ecology 56, 191–198. doi: 10.2307/1935311
Lynch, J., Tang, K., Sands, J., Sands, M., Tang, E., Mukherjee, S., et al. (2016). HOMINID: a framework for identifying associations between host genetic variation and microbiome composition. bioRxiv:081323. doi: 10.1101/081323
Madin, J. S., Anderson, K. D., Andreasen, M. H., Bridge, T. C., Cairns, S. D., Connolly, S. R., et al. (2016). The coral trait database, a curated database of trait information for coral species from the global oceans. Sci. Data 3:160017. doi: 10.1038/sdata.2016.17
McFall-Ngai, M. (2014). Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 12:e1001783. doi: 10.1371/journal.pbio.1001783
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
McFall-Ngai, M. J. (2000). Negotiations between animals and bacteria: the ‘diplomacy’ of the squid-vibrio symbiosis. Comp. Biochem. Physiol. 126, 471–480. doi: 10.1016/S1095-6433(00)00233-6
Meron, D., Atias, E., Kruh, L. I., Elifantz, H., Minz, D., Fine, M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5, 51–60. doi: 10.1038/ismej.2010.102
Mi, H., Poudel, S., Muruganujan, A., Casagrande, J. T., and Thomas, P. D. (2015). PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44, D336–D342. doi: 10.1093/nar/gkv1194
Moloney, R. D., Desbonnet, L., Clarke, G., Dinan, T. G., and Cryan, J. F. (2014). The microbiome: stress, health and disease. Mamm. Genome 25, 49–74. doi: 10.1007/s00335-013-9488-5
Mora, C., Tittensor, D. P., Adl, S., Simpson, A. G. B., and Worm, B. (2011). How many species are there on Earth and in the ocean? PLoS Biol. 9:e1001127. doi: 10.1371/journal.pbio.1001127
Morrow, K. M., Bourne, D. G., Humphrey, C., Botte, E. S., Laffy, P., Zaneveld, J., et al. (2015). Natural volcanic CO2 seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J. 9, 894–908. doi: 10.1038/ismej.2014.188
Musat, N., Musat, F., Weber, P. K., and Pett-Ridge, J. (2016). Tracking microbial interactions with NanoSIMS. Curr. Opin. Biotechnol. 41, 114–121. doi: 10.1016/j.copbio.2016.06.007
Muscatine, L., Mccloskey, L. R., and Marian, R. E. (1981). Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnol. Oceanogr. 26, 601–611. doi: 10.4319/lo.1981.26.4.0601
Neave, M. J., Apprill, A., Ferrier-Pagès, C., and Voolstra, C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
Neave, M., Mitchell, C., Apprill, A., and Voolstra, C. (2017). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7:40579. doi: 10.1038/srep40579
Nelson, T. M., Apprill, A., Mann, J., Rogers, T. L., and Brown, M. V. (2015). The marine mammal microbiome: current knowledge and future directions. Microbiol. Aust. 1, 8–13. doi: 10.1071/MA15004
Nelson, T. M., Rogers, T. L., Carlini, A. R., and Brown, M. V. (2013). Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 15, 1132–1145. doi: 10.1111/1462-2920.12022
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813
Nussbaumer, A. D., Fisher, C. R., and Bright, M. (2006). Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441, 345–348. doi: 10.1038/nature04793
Radax, R., Hoffmann, F., Rapp, H. T., Leininger, S., and Schleper, C. (2012). Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ. Microbiol. 14, 909–923. doi: 10.1111/j.1462-2920.2011.02661.x
Ribes, M., Calvo, E., Movilla, J., Logares, R., Coma, R., and Pelejero, C. (2016). Restructuring of the sponge microbiome favors tolerance to ocean acidification. Environ. Microbiol. Rep. 8, 536–544. doi: 10.1111/1758-2229.12430
Rocha, J., Coelho, F. J., Peixe, L., Gomes, N. C., and Calado, R. (2014). Optimization of preservation and processing of sea anemones for microbial community analysis using molecular tools. Sci. Rep. 4:6986. doi: 10.1038/srep06986
Ruehland, C., Blazejak, A., Lott, C., Loy, A., Erséus, C., and Dubilier, N. (2008). Multiple bacterial symbionts in two species of co-occurring gutless oligochaete worms from Mediterranean sea grass sediments. Environ. Microbiol. 10, 3404–3416. doi: 10.1111/j.1462-2920.2008.01728.x
Sanders, J. G., Beichman, A. C., Roman, J., Scott, J. J., Emerson, D., McCarthy, J. J., et al. (2015). Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat. Commun. 6:9285. doi: 10.1038/ncomms9285
Scharschmidt, T. C., and Fischbach, M. A. (2013). What lives on our skin: ecology, genomics and therapeutic opportunities of the skin microbiome. Drug Discov. Today 10, e83–e89. doi: 10.1016/j.ddmec.2012.12.003
Schimak, M. P., Kleiner, M., Wetzel, S., Liebeke, M., Dubilier, N., and Fuchs, B. M. (2016). MiL-FISH: multilabeled oligonucleotides for fluorescence in situ hybridization improve visualization of bacterial cells. Appl. Environ. Microbiol. 82, 62–70. doi: 10.1128/AEM.02776-15
Shade, A., and Handelsman, J. (2012). Beyond the Venn diagram: the hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Sharp, K. H., Eam, B., Faulkner, J. D., and Haygood, M. G. (2007). Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73, 622–629. doi: 10.1128/AEM.01493-06
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. doi: 10.1038/nature10249
Simister, R., Taylor, M. W., Tsai, P., Fan, L., Bruxner, T. J., Crowe, M. L., et al. (2012a). Thermal stress responses in the bacterial biosphere of the Great Barrier Reef sponge, Rhopaloeides odorabile. Environ. Microbiol. 14, 3232–3246. doi: 10.1111/1462-2920.12010
Simister, R. L., Deines, P., Botté, E. S., Webster, N. S., and Taylor, M. W. (2012b). Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ. Microbiol. 14, 517–524. doi: 10.1111/j.1462-2920.2011.02664.x
Smith, D. C. (2001). Symbiosis research at the end of the millenium. Hydrobiologia 461, 49–54. doi: 10.1023/A:1012765114474
Sogin, E. M., Putnam, H. M., Anderson, P. E., and Gates, R. D. (2016). Metabolomic signatures of increases in temperature and ocean acidification from the reef-building coral, Pocillopora damicornis. Metabolomics 12, 1–12. doi: 10.1007/s11306-016-0987-8
Taylor, M. W., Hill, R. T., Piel, J., Thacker, R. W., and Hentschel, U. (2007). Soaking it up: the complex lives of marine sponges and their microbial associates. ISME J. 1, 187–190. doi: 10.1038/ismej.2007.32
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7:11870. doi: 10.1038/ncomms11870
Thompson, J. R., Rivera, H. E., Closek, C. J., and Medina, M. (2015). Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 4:176. doi: 10.3389/fcimb.2014.00176
Thurber, R. V., Willner-Hall, D., Rodriguez-Mueller, B., Desnues, C., Edwards, R. A., Angly, F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Valm, A. M., Welch, J. L. M., Rieken, C. W., Hasegawa, Y., Sogin, M. L., Oldenbourg, R., et al. (2011). Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. U.S.A. 108, 4152–4157. doi: 10.1073/pnas.1101134108
Wada, N., Pollock, F. J., Willis, B. L., Ainsworth, T., Mano, N., and Bourne, D. G. (2016). In situ visualization of bacterial populations in coral tissues: pitfalls and solutions. PeerJ 4:e2424. doi: 10.7717/peerj.2424
Wagner, J., Chelaru, F., Kancherla, J., Paulson, J. N., Felix, V., Mahurkar, A., et al. (2017). Metaviz: interactive statistical and visual analysis of metagenomic data. bioRxiv:105205. doi: 10.1101/105205
Weber, L., Deforce, E., and Apprill, A. (2017). Optimizing DNA extraction for coral microbiota investigations. Microbiome 5:18. doi: 10.1186/s40168-017-0229-y
Webster, N., Negri, A., Botté, E., Laffy, P., Flores, F., Noonan, S., et al. (2016). Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6:19324. doi: 10.1038/srep19324
Webster, N. S., Taylor, M. W., Behnam, F., Lücker, S., Rattei, T., Whalan, S., et al. (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12, 2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x
Webster, N. S., and Thomas, T. (2016). The sponge hologenome. mBio 7:e00135-16. doi: 10.1128/mBio.00135-16
Weis, V. M., Davy, S. K., Hoegh-Guldberg, O., Rodriguez-Lanetty, M., and Pringle, J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 23, 369–376. doi: 10.1016/j.tree.2008.03.004
Wippler, J., Kleiner, M., Lott, C., Gruhl, A., Abraham, P. E., Giannone, R. J., et al. (2016). Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis. BMC Genomics 17:942. doi: 10.1186/s12864-016-3293-y
Woyke, T., Teeling, H., Ivanova, N. N., Huntemann, M., Richter, M., Gloeckner, F. O., et al. (2006). Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443, 950–955. doi: 10.1038/nature05192
Zhang, F., Blasiak, L. C., Karolin, J. O., Powell, R. J., Geddes, C. D., and Hill, R. T. (2015). Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges. Proc. Natl Acad. Sci. U.S.A. 112, 4381–4386. doi: 10.1073/pnas.1423768112
Keywords: animalia, bacteria, archaea, microbiome, symbiosis
Citation: Apprill A (2017) Marine Animal Microbiomes: Toward Understanding Host–Microbiome Interactions in a Changing Ocean. Front. Mar. Sci. 4:222. doi: 10.3389/fmars.2017.00222
Received: 18 October 2016; Accepted: 30 June 2017;
Published: 18 July 2017.
Edited by:
Thomas K. Frazer, University of Florida, United StatesReviewed by:
Melissa Garren, California State University, Monterey Bay, United StatesCopyright © 2017 Apprill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amy Apprill, YWFwcHJpbGxAd2hvaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.