- MARETEC, Instituto Superior Técnico, Universidade de Lisboa, Lisboa, Portugal
Viruses are the most abundant biological entities in the world's oceans. Their potential control on the dynamics and diversity of bacterioplankton and some phytoplankton groups, and consequent effect on the flow of energy and matter in food webs, may be argued as beyond dispute. Paradoxically, their importance seems to be persistently underestimated by marine modelers, frequently by exclusion, despite the uninterrupted volume of knowledge advanced during the past decades. Bridging the gap between knowing and modeling the role of viruses is, undoubtedly, one of the upcoming frontiers to be crossed in modeling the plankton. This paper has a two-fold objective: (1) review the knowledge on the roles of viruses in marine systems that has been put forward over the past decades, and (2) see how viruses have been incorporated into marine ecosystem models, along with the factors that are limiting their inclusion.
Introduction
When the study of marine viruses had its boost back in the 1980's, one of the most significant findings was that they are more abundant in marine ecosystems than previously thought, frequently more abundant than bacteria (Bergh et al., 1989; Børsheim et al., 1990; Bratbak et al., 1990; Proctor and Fuhrman, 1990; Suttle et al., 1991). The studies showed a natural abundance of viruses typically on the order of 1010 L−1, but ranging from 3 × 108 L−1 to 1011 L−1 (Fuhrman, 1999; Noble and Fuhrman, 1999; Wilhelm and Suttle, 1999). This was, unquestionably, one of the most substantial advances in marine biology by the end of the twentieth century. Given their size, viruses are a component of dissolved organic matter (DOM) (Benner et al., 1992; Middelboe et al., 2003), and account for 1–5% of the standing stock of marine biomass, despite their abundance (Libes, 1992; Suttle, 2005; Sandaa, 2008). Apparently, viruses are the smallest and surely the most abundant organisms in the sea, and the full implications of their role in the microbial loop is still to be determined. When compared with other biological entities, viruses are dynamic and change rapidly in abundance, and the fact that there are significantly more viruses in near-surface waters suggests a coupling with other upper-ocean biological processes (Bratbak et al., 1990; Wilhelm and Suttle, 1999; Brum et al., 2014; Brum and Sullivan, 2015).
Viruses commonly use specific channels on the cell wall to inject their genetic material into hosts (Fuhrman, 1999), such as the receptors through which bacteria and phytoplankton take up nutrients (Böhm et al., 2011). Thus, lytic viruses act by infecting, multiplying within and bursting (lysing) the host cells to release the progeny viruses. One major consequence of this process is the release of DOM, plus some particulate organic matter (POM), which has an important role in the regeneration of nutrients. Thus, viral infection is an extremely efficient way to convert biomass into dissolved and particulate detritus readily available to bacteria (Fuhrman, 1992; Middelboe et al., 1996; Gobler et al., 1997). This, in turn, has repercussions through the entire food web considering the close coupling between heterotrophic bacteria and phytoplankton (Lancelot and Billen, 1984; Vanduyl and Kop, 1988; Brussaard et al., 1995).
While the roles of viruses in microbial food webs were the focus of most studies during the 1990's, the dawning of the “omics” era in the 2000's widened the scope of their study (Breitbart et al., 2002; Culley et al., 2006; Martínez et al., 2014; Hurwitz et al., 2016). Viral metagenomics has not only confirmed what was previously known about viruses, but it also shed new light on topics such as geographical patterns (Angly et al., 2006; De Corte et al., 2016), and the relationship between their diversity and abundance with biotic and abiotic variables (Breitbart et al., 2004; Dunigan et al., 2006). Recent works have definitely confirmed the early views on viruses as fundamental drivers of many ecosystem processes (Hurwitz and Sullivan, 2013; Malits et al., 2014), and even manipulators of the marine environments (Rohwer and Thurber, 2009).
Paradoxically, viruses are the most abundant organisms in marine ecosystems, and key players in marine food-web, but infrequently handled in marine ecosystem models. In a way, the extensive knowledge gathered over the last two decades on the role and importance of viruses in aquatic systems, clearly expressed in exhaustive reviews (e.g., Proctor, 1997; Fuhrman, 1999; Wommack and Colwell, 2000; Weinbauer, 2004; Suttle, 2005; Dunigan et al., 2006; Brum and Sullivan, 2015), has not been followed by its inclusion into marine biogeochemical models. This paper reviews some early knowledge on the activity of viruses and the follow-up (or lack of it) by the modeling community. The paper is organized in two major sections. The first section presents a brief review of the knowledge on the roles of virus since its heydays in the 1990's up to the recent findings on the processes involved in the dynamics of viruses-host relationships; the second section tackles (a) how this knowledge has been incorporated into biogeochemical models along with (b) some implication of their inclusion.
The Role of Viruses in the Food Webs
The Microbial Loop
It is impossible to understand the roles and importance of viruses in aquatic environments without addressing the microbial loop, a designation coined back in 1983 to express the complexity and dynamic behavior of the many interacting ecological relationships in marine food webs (Azam et al., 1983). Early studies depicted the oceanic feeding structures as a simple chain where energy and mass where transferred from one functional group to another, starting with the phytoplankton and leading to fish and other top predators via metazoan zooplankton (Figure 1A). Microbes were seen in loose association with this chain since their contribution to the functioning of the planktonic system was considered to be minor, often disregarded. Subsequent studies on marine planktonic ecosystems, however, revealed a profoundly different scenario. The food chain structure gave place to a food web where the flow of mass and energy between organisms was no longer linear, with bacteria mediating much of the transfers and availability of substrate for other organisms (Figure 1B).
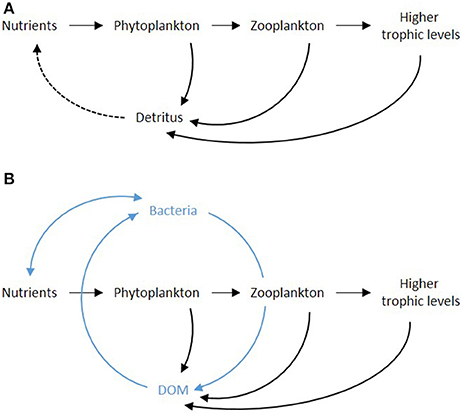
Figure 1. A simplified scheme for the food chain (A) and food web (B) circuits. The inclusion of the microbial loop is highlighted in (B). Note: the general description of detritus used in early depictions of the food chain is replaced by DOM in (B), given its central role; detritus or POM was excluded from the schematics for simplicity.
This flow of mass and energy, known as the microbial loop, comprises one circuit with two distinct loops that strongly converge at the functional level and size class of heterotrophic bacteria (Jumars, 1993). One circuit contains the inorganic nutrients from their entry into primary producers until they became available once again as inorganic nutrients. The other contains the paths followed by organic carbon compounds from the moment of their uptake by heterotrophic bacteria until they are again available in dissolved organic form. Having the capacity of taking up both dissolved inorganic and dissolved organic compounds, bacteria have been assumed to play a central role in this loop and determine the availability of these substances to other organism groups.
At a given point it was even hypothesized that the bulk of nutrients in microbial food webs was recycled within this subcomponent, with little transfer to the classic web (Goldman and Dennett, 1992). While rapid and extensive organic matter recycling may result from microbial processes, the microbial loop can act as a sink for zooplankton production, rather than a supplementary organic carbon source. Another step in the microbial loop is the consumption of the bacteria by protozoa and other microheterotrophic zooplankton. By doing so, predators can also contribute to the remineralization of the major nutrients, a role that was for long attributed exclusively to bacteria (Azam and Hodson, 1977; Andersson et al., 1985). The significant increase in knowledge on the microbial food web and its intervenient lead to the formulation of the so-called “link or sink problem,” according to which there is a pending question on what extent the microbial loop represents a loss of fixed carbon to the system or, instead, it primarily channels fixed carbon to higher levels of the food chain. This question, however, seems to have gathered consensus on the fact that the microbial loop is primarily seen as a sink, based on the fact that it includes several trophic levels and, thus, dissipates a large fraction of the organic carbon as CO2 along the process (Fenchel, 2008).
Paradoxically, researchers knew grazing to be insufficient to explain bacterial mortality in aquatic ecosystems (Bergh et al., 1989), and soon it became clear that their pressure on bacterial production was over-estimated. A corollary of such realization was that protozoan grazing was not the only major cause of bacterial mortality, and that other factors co-operate in the control of bacterial biomass. Besides the bioavailability of DOM, the density-dependent mortality induced by viral infection is another process involved, a concept that rapidly became central to the understanding of the biogeochemical role of bacteria in pelagic food webs.
Information on viral life strategies and their survival mechanisms is of considerable interest, given the impact that viruses have on bacterial abundance. Lytic and lysogenic infections are the most common, amongst a diversity of types of life cycles, with the lytic cycle as the dominant way of viral replication in most aquatic systems (Wilson and Mann, 1997). While ultimately leading to the destruction of the infected cells, the lysogenic cycle is characterized by the infection of host cells by phages and their genome typically remains within the host in a dormant stage (prophage). In the viruses-host relationship, the physiologic conditions of the host cells are determinant in phage infection. Because viruses are entirely dependent on the host for energy and building blocks, it is expected that suboptimal physiologic conditions on the host will affect virus production as well (Maat et al., 2016a). Of particular relevance is nutrient stress or limitation, as phosphorus availability is known to be critically important to viral replication (Wilson et al., 1996; Fuhrman et al., 2011). The dynamic of viruses seems to be also related to local trophic conditions, given that the removal of viruses from the water may be controlled by different causative agents. As suggested by Bongiorni et al. (2005), the fate of viral production follows distinct pathways in oligotrophic and eutrophic systems. In oligotrophic waters, for example, viral loss by adsorption onto settling particles can be six times lower than in eutrophic systems but, alternatively, the impact of UV on virus survival is probably higher than in more eutrophic systems, considering the higher penetration of UV radiation in the less turbid water column.
Early estimates based on abundance, phage production and burst size suggested that approximately one-third of bacterial production could be attacked or infected each day (Bergh et al., 1989; Proctor and Fuhrman, 1990), thus having an important quantitatively impact on bacterial production. Later it was discovered that in some coastal waters viruses are responsible for most bacterial mortality, and the estimate fraction of the bacterial population lysed by viruses each day could range from 6 to 100% (Heldal and Bratbak, 1991; Fuhrman and Suttle, 1993; Fuhrman et al., 1993; Fuhrman and Noble, 1995). Also it was advanced that 60% of the mortality of the free-living heterotrophic bacteria in coastal and open ocean environments could be attributed to viral infection (Proctor and Fuhrman, 1990).
These mortality estimates, based upon limited background data, were somewhat speculative (Fuhrman, 1992), but they clearly indicated the importance of viruses in bacterial population control. Different approaches, relying on conceptual models or other indirect measurements, suggested that losses from viral lysis were in the same range as those attributed to protist grazing (Proctor and Fuhrman, 1990; Suttle and Chan, 1993). For a mesocosms experiment, for example, Fuhrman and Noble (1995) showed that viruses and protists were responsible for similar amounts of bacterial loss (they specifically account for 70–75% of the total apparent mortality with these two mechanisms). Recent findings (Carreira et al., 2015a,b) suggest that previous estimates on bacterial abundances using either epifluorescence microscopy (EFM) or flow cytometry (FCM) may be inexact. Yet, they don't compromise the knowledge on bacterial population control exerted by viral-induced mortality. Other recent studies, however, are questioning the paradigm where bacteria are central in the mechanisms for nutrient recycling, by pointing instead to the importance of mixotrophic organisms (Berge et al., 2016; Caron, 2016; Ward and Follows, 2016), although the question on the importance mixotrophs has previously been raised (Hall et al., 1993; Arenovski et al., 1995; Fenchel, 2008). According to this new paradigm, communities of mixotrophic protist plankton support the bulk of the base of the microbial food webs, having some fundamental differences in the flow of energy and nutrients from the bacteria-dominated microbial food web paradigm (Mitra et al., 2014). This mixotroph-dominated structure implies a shorter and potentially more efficient circuit from nutrient regeneration to primary production, explaining that primary production may be supported by bacterial production.
The Viral Loop
Early descriptions of marine microbial activity provided scarce information on the trophic coupling between DOM, phytoplankton, bacteria, viruses and protozoa. They were also limited on explaining how each of these components, in turn, influence biomass production of heterotrophic bacteria and shape the carbon flux in food webs (Azam et al., 1994). Small flagellates were seen as key players bacteria mortality but, as knowledge increased, it become evident that virus infection contributed significantly to bacterial loss (Bergh et al., 1989; Bratbak et al., 1990; Fuhrman and Noble, 1995; Petersen and Dubilier, 2014; Roux et al., 2014). The overwhelming evidences pointing to virus-induced mortality of bacteria, has noted by Fenchel (2008), made them new players in the microbial loop.
Viral lysis of bacteria has been thought to create a close loop in which bacterial carbon is cycled back to the bacterial population or respired, and nutrients are regenerated (Proctor and Fuhrman, 1991; Bratbak et al., 1992; Thingstad et al., 1993). Consequently, bacterial lysis diverts carbon and other biologically elements away from higher trophic levels or may simply delay their transportation toward them (Fuhrman, 1992), by keeping the dissolved nutrients within the so-called “viral loop” (Suttle and Chan, 1993). This semi-closed trophic loop (Figure 2), fed externally by the release of DOM from phytoplankton and grazers, has the net effect of oxidizing organic matter and regenerating inorganic nutrients (Proctor and Fuhrman, 1991). Viral lysis of bacterial and phytoplankton cells removes a portion of autotrophic and microbial heterotrophic production from protozoan grazers providing, at the same time, a short circuit to the “traditional” concept of carbon cycling. It was even suggested that the recycling of organic material in a bacteria-phage-dissolved organic material loop could lead bacterial production to exceed primary production (Strayer, 1988; Gobler et al., 1997). From these results, the canonical view of the “microbial loop” as a way of loss of organic material from the particulate flux toward larger organisms, and the reincorporation of this organic material via heterotrophic bacteria (Azam et al., 1983), was upgraded with the “viral loop” as a subset. This new loop within the microbial loop is responsible for much of loss from the particulate to dissolved phase. The “viral loop” was not considered as an independent process, but rather as a component of the “microbial loop,” and early estimates suggested that as much as 25% of the primary production in the ocean flows through this loop (Wilhelm and Suttle, 1999). For its capacity to disrupt the microbial loop, it was even advanced that the viral loop leads to a “futile cycle” or “short circuit” of carbon flow, resulting in an energy loss by respiration as the cycle turns (Suttle and Chan, 1993; Azam et al., 1994), since bacteria do not convert DOM into biomass with a 100% efficiency.
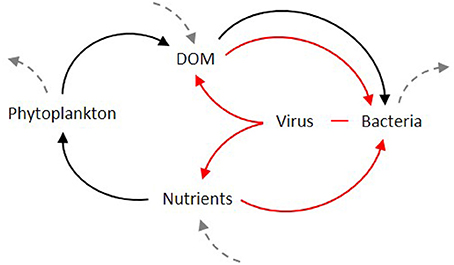
Figure 2. Initially proposed as a sub-set of the microbial loop (black arrows), the viral loop (red arrows) was seen as short-circuit for the cycling of carbon and nutrients in the food web. Dashed arrows indicate the flow from and to higher trophic levels.
Viruses also affect the species composition and diversity of the bacterial community because viruses are usually highly host-specific (Heldal and Bratbak, 1991; Roux et al., 2014), while protists can graze on a variety of bacterial species. However, as noted by Fenchel (2008), the importance of viruses is not so much because they affect the total number of bacteria, but rather because they drive successional changes of the bacterial biota. The implications is that higher bacterial species diversity in plankton may occur in the presence of viruses than would be the case if viral action was not present. Additionally, the influence of virus is site-specific, varying according to local trophodynamics in the food web (Ory et al., 2010). Overall, the impact of the viruses on food-web processes depends on the balance between bacterial losses from viruses and from protists (Weinbauer and Rassoulzadegan, 2004).
Viruses and Phytoplankton Mortality
The importance of viruses is further amplified because they are also able to infect a variety of phytoplankton species, implying that the intricate relationship between phytoplankton blooms and bacterioplankton activity may be mediated by virus (Bergh et al., 1989). Phytoplankton is inevitably exposed to high physiological stress in dense blooms which increases cell lysis (Brussaard et al., 1997), where lytic viruses play a significant role by completely destroying host populations within hours to days (Suttle and Chan, 1993; Cottrell and Suttle, 1995; Jacobsen et al., 1996; Bratbak et al., 1998a; Baudoux and Brussaard, 2005). As with viral infection in bacteria, P limitation seems to play an important role in viruses-phytoplankton interactions. The impact of viruses on phytoplankton communities is also related to P-availability as the shortage of this nutrient is assumed to decrease the percentage of lysed cells (Wilson et al., 1996; Monier et al., 2012; Maat et al., 2016a,b). Recently, Maat et al. (2014) have evaluated the outcome of P limitation on viral infection in picoeukaryote Micromonas pusilla, reporting a prolongation of the latent period from 6 to 12 h, and an 80% reduction in viral burst sizes compared to P-replete conditions. Phosphorus stress has been pointed out as a strong selective agent, with some phage having in their genome host-like phosphorus assimilation genes that are upregulated when the host is phosphorous starved, thus promoting P acquisition necessary for phage dispersion (Kelly et al., 2013). In a similar way, it has been reported the presence of host-like photosynthesis genes in phage along with the capacity to express them and actively contribute to primary production during the infection (Sullivan et al., 2006).
Experimental work suggests that viral infection may affect the species composition of the phytoplankton community and significantly reduce primary productivity. Besides the already mentioned high levels of viral abundance and infection on bacteria, infection of phytoplankton is also common during intense episodes of coastal algal bloom (Proctor and Fuhrman, 1991; Yau et al., 2011; Shelford et al., 2012; Hasumi and Nagata, 2014; Stock et al., 2014). Consequently, viral lysis of phytoplankton can significantly affect the partitioning and biological cycling of elements such as C, N, P, Si, and Fe (Gobler et al., 1997), and the potential consequences of viral infection on nutrient cycling has been extensively discussed ever since (Bergh et al., 1989; Heldal and Bratbak, 1991; Proctor and Fuhrman, 1991; Suttle et al., 1991).
Recent findings have corroborated previous views on the roles of viruses in nutrient cycling and energy transfer, but also brought additional insights on how they may structure the phytoplankton community (Baudoux and Brussaard, 2005; Gustavsen et al., 2014). So far viruses have been reported to infect a wide variety of phytoplankton groups including diatoms (Shirai et al., 2008), dinoflagellates (Yuji et al., 2004), prasinophytes (Derelle et al., 2008), and cyanobacteria (Ortmann et al., 2002), among others. While some viruses are able to infect a wide diversity of hosts, including several bloom-forming phytoplankton species from diatoms to flagellates (Tai et al., 2003; Tomaru et al., 2009), others have a narrow host range (Short, 2012), resulting in the lysis of a specific phytoplankton taxon, thus changing the composition of phytoplankton populations.
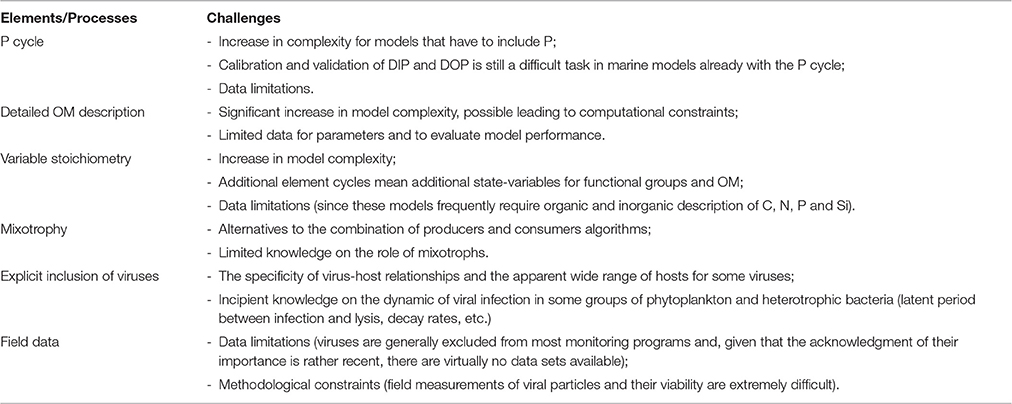
The Inclusion of Viruses in Marine Models
Why Is It So Important?
The simple recognition that virus and grazing can have the same mortality rate in both phytoplankton and bacterioplankton, but with striking different outcomes in terms of fluxes of nutrients and organic matter, suffices as an argument to consider their inclusion in marine food web models. The loss of bacteria to viruses reflects a significant dissipation of energy in this ecosystem. Recent studies reaffirm that the mortality from viral activity must be considered in studies of bacterioplankton processes (Ory et al., 2010; Magiopoulos and Pitta, 2012), a fact long known, but still not fully realized by marine modelers. Back in 2000 it was stated that our understanding of the planktonic food was incomplete without an hypothesis for the underlying mechanisms controlling viral abundance (Thingstad, 2000). To that, one can add that our numerical models need to include viral activity if we aspire to use them to understand and estimate the balance of energy and matter in marine systems.
The rationale for their inclusion is not too different from the rationale used in the past for the inclusion of bacteria in models. The argument for the inclusion of heterotrophic bacteria, and with it the whole microbial loop, relied on their use of DOM; models with heterotrophic bacteria were able to simulate microbial loop processes, achieving greater detail and dynamics. Other reasons for explicit treatment of the microbial loop in models was their link between the microbial and metazoan food webs, and the possibility to address bacteria-phytoplankton competition for inorganic nutrients (Bratbak and Thingstad, 1985; Fasham, 1993). Since viruses mediate much of these processes, and can change dramatically the fluxes within the microbial loop (Figure 3), their inclusion is obvious. This is, in fact, a strong argument for their inclusion given their role in the production of DOM and consumption of DOC (Hasumi and Nagata, 2014). Together with phytoplankton exudation, grazing-associated losses and detrital turnover, lysis caused by viral infection contribute to the transformation of particulate matter into DOM. Yet, modeling DOM is still a challenge in itself given its complex composition as a heterogeneous biochemical mixture scattered along a size continuum, from free monomers to large particles. As a result, some models attempt to capture some of its complexity (Keller and Hood, 2013; Hasumi and Nagata, 2014), while others explicitly model the role of heterotrophic bacteria but with a simplistic approach to organic matter (e.g., Liu et al., 2015).
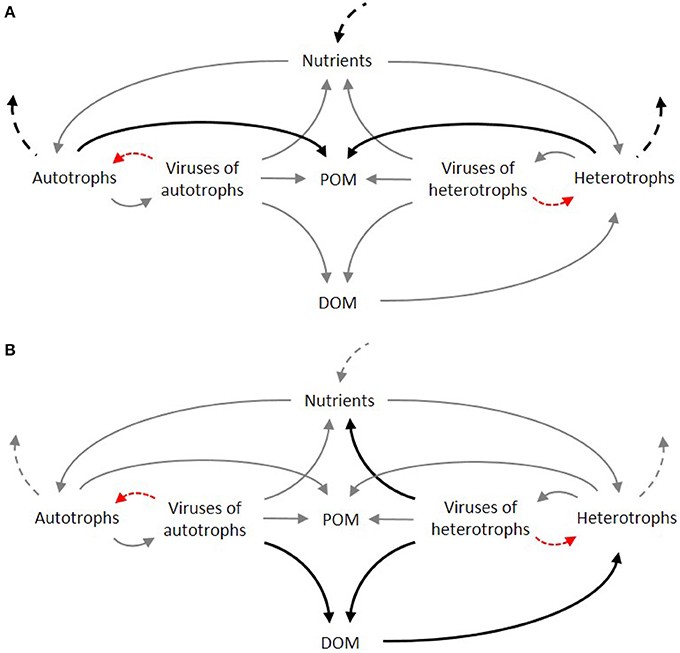
Figure 3. Simplified representation of the food web under low (A) and high (B) intensity of viral induced lysis in autotrophs and heterotrophs. Dominant fluxes are represented by dark bold arrows. Dashed arrows denote fluxes from and to higher trophic levels. Red arrows denote viral infection.
An additional reason for the inclusion of viral activity in ecological models rely on their role in the control of phytoplankton blooms (Baudoux and Brussaard, 2005; Brussaard et al., 2005; Malits et al., 2014). Although complex, viral-algal interactions play a key role in the dynamics of phytoplankton blooms. Field studies have corroborated the importance of the role of biological interactions in sudden population changes, evident in the ability of lytic viruses to completely destroy host populations over short periods (Suttle and Chan, 1993; Cottrell and Suttle, 1995; Zingone, 1995; Jacobsen et al., 1996; Bratbak et al., 1998a,b). Not least relevant, and considering the recent efforts in modeling Harmful Algal Blooms (Davidson et al., 2016), the viral-algal interactions are known to contribute to bloom breakdown and dispersion (Baudoux and Brussaard, 2005; Brussaard et al., 2005).
Initial Modeling Approaches to Viral Activity
Initial attempts at modeling the role of viruses in bacterial mortality relied on model-based interpretation of the percentage of bacteria diverted to DOM (Noble and Fuhrman, 1999; Thingstad, 2000). These models, however, were conceptual in nature, and with statistical quantifications of the budgets, and not like the dynamic process-oriented models used currently in marine biogeochemical models. As an example of the potential impact of bacterial viruses, Fuhrman (1992) presented a quantitative model of such a cycle embedded in a steady state food web. The model showed that the inclusion of bacterial loss term due to viruses equivalent to the loss due to protists resulted in a 27% increases in bacterial production, a 37% decrease in export of bacterial carbon to protistan grazers and a 7% decrease in macrozooplankton production. These results are a clear indication that viral activity shifts more production and respiration into bacteria and away from other heterotrophs. However, the static nature of the model prevented its use in the study of dynamic ecosystems and to properly describe the very dynamic process of viral growth. At the same time, Murray and Jackson (1992) advanced a detailed model for the relationship between viruses and planktonic organisms, modeling the lysis of hosts proportional to the density of both host and viruses (as a type-I functional response). So, while lacking a specific component to account for viral activity, and missing some of the viruses-mediated processes that have been discovered more recently, early attempts to model viruses dynamics paved the way for more complex biogeochemical models that had essential components for the inclusion of virus (e.g., the microbial loop).
With the rise of more complex and dynamic models, such as the coupled physical and ecological NPZD (nutrient-phytoplankton-zooplankton-detritus) models, organic matter compartments started to be included. Since then DOM has been addressed in a number of ways in ecological models, based on its size, availability (refractory, semi-refractory, labile), molecular weight, etc. (Chróst, 1990; Lancelot et al., 1991; Kirchman et al., 1993; Blackburn et al., 1996; Walsh et al., 1999; Vallino, 2000), and the microbial loop became a common model component (Baretta-Bekker et al., 1994, 1995, 1997, 1998; Baretta et al., 1995; Vichi et al., 2003, 2004). Another important development in the modeling philosophy during this period was the aggregation of biological components into functional groups. Instead of the simple expression of phytoplankton as chlorophyll a, for example, new phytoplankton groups emerged in models (diatoms, flagellates, picophytoplankton, etc.), consisting on an assemblage of similar functional species based on their taxonomic and physiological properties.
Current State of Modeling
Today the roles of viruses are still a recurrent topic and the study of the microbial loop is far from exhausted (Middelboe et al., 2003; Sandaa, 2008; Ory et al., 2010; Yau et al., 2011; Magiopoulos and Pitta, 2012; Shelford et al., 2012; Xu et al., 2013; Brum et al., 2014; Hasumi and Nagata, 2014; Stock et al., 2014; Liu et al., 2015). Still, viruses are sometimes neglected (e.g., Follows and Dutkiewicz, 2010; Salihoglu et al., 2013), and only a few attempts have been made to model the details of bacterial and phytoplankton viral-induced cell lysis. Early modeling approaches have incorporated viral infection in simple models (Murray and Jackson, 1992), but not on biogeochemical models accounting for the fate of the DOM produced in the process, for example. Model kept evolving in complexity and detail ever since (e.g., Pasquer et al., 2005; Libralato and Solidoro, 2009; Ramin et al., 2012; Hasumi and Nagata, 2014), but viruses have been mostly kept out of this evolution in the algorithms. Some models, however, have addressed the dynamic of viruses with considerable detail, but not in a full ecological modeling framework (Angly et al., 2006), by tackling the specific virus-host relationship (Ruardij et al., 2005) having as a start point previous modeling approaches of virus-host relationships (Murray and Jackson, 1992).
Viral-induced lysis was eventually included in complex marine models with the parameterization of phytoplankton cell lysis largely based on empirical findings. In the ERSEM model, for instance, it was formulated that lysis products from phytoplankton groups were partly particulate and partly dissolved, with the particulate fraction dependent on the actual nutrient cell quota (Baretta-Bekker et al., 1995, 1997, 1998). For primary producers, lysis was defined to included factors such as mechanical damage (e.g., sloppy feeding Imai et al., 1993; Strom et al., 1997), nutrient shortage (Berges and Falkowski, 1998), virus and bacteria activity (Imai et al., 1993, 1995). A way to achieve this consisted in defining a background lysis rate, an increased lysis rate proportional to nutrient stress and a phytoplankton density dependent term. The later had viral activity at its base.
But most models kept addressing the role of virus in an indirect way, mostly as an additional term for mortality, or simply merged in a single loss term that accounts for several processes. Some models have a loss term loss of phytoplankton carbon due to nutrient stressed lysis caused by mechanical failure and viruses (Vichi et al., 2007; Mateus, 2012b; Petihakis et al., 2012; Hasumi and Nagata, 2014), frequently staying close to the approach proposed in early models such as the ERSEM model (Baretta-Bekker et al., 1997; Butenschön et al., 2016). In other cases viruses are assumed to be a minor phytoplankton loss mechanism (Stock et al., 2014), parameterized as weak density-dependent loss term on small phytoplankton, following early and simpler modeling approaches (Suttle, 1994). A summary of the main features of some of these models is listed in Table 1.
Model Components and Examples
From these early attempts, the most basic way to include viral activity in marine ecosystem models seems to be the addition of a density dependent mortality factor to the standard parameterization of a fixed independent mortality factor. This empirical formulation was initially advanced in the ERSEM model for producers (Baretta-Bekker et al., 1997, 1998), and adopted in other models afterwards (Vichi et al., 2004; Mateus, 2012b). This method is graphically depicted in Figure 4A. In this simplistic approach the mortality associated with cell burst or lysis in phytoplankton groups implicitly represents the effect of several mortality processes attributed to different causes, including viruses. Lysis for each phytoplankton group (P) is defined by adding a density-independent term, mdi, with a density-dependent term, mdd:
The density-independent term is defined by a specific background lysis rate in nutrient-depleted situations, rlys, and the intercellular nutrient status or nutrient limiting factor, enp, according to:
The density-dependent term varies according to each producer group biomass, P. There is a model parameter for the reference concentration (or biomass), PR, and another for the specific density dependent mortality rate, . The viral-induced mortality, mdd, is calculated as:
The functional response of this simple algorithm is that mortality increases as a response to the increase of the host population (mimicking viral infection). Mortality products are partially dissolved and partially particulate, with the fraction channeled to the POM fraction. The partition of lysed products depends on the actual (QN and QP) and minimal ( and ) intracellular nutrient quota for both nutrients, according to:
The resulting fraction for each organic matter pool is then obtained by:
All organic matter components are characterized by three state-variables, one for each pool of elements (C, N, and P) allowing for a variable stoichiometry of all organism groups and organic matter compartments. So, the influence of viruses, while very simplistic in its parameterization, is only a small component in a rather complex model. Table 2 provides a list of the parameters with a description and their units as defined in the original model.

Table 2. List of parameters related to mortality (lysis) of producers in ERSEM-type models (Baretta-Bekker et al., 1997, 1998).
Despite the apparent simplistic approach of most marine ecological models in the past few decades, the explicit modeling of viruses is thriving in the last few years, nonetheless. Fuhrman et al. (2011), for instance, have performed a detailed mathematical analysis model describing the dynamics of infection by a virus of a host population in freshwater. This model was then modified to include phytoplankton and tested with data from natural planktonic communities from Lake Michigan (Béchette et al., 2013). The model, depicted in Figure 4B, addresses bacteria-virus interactions by dividing bacterial population in two states, susceptible hosts (HS) and infected hosts (HI), both interacting with free viruses (V), and free nutrient (N), represented by dissolved phosphorus. Particulate phosphorus is not a state-variable, but accounted for as P content in the cells and virus. Table 3 provides as summary of model parameters. The free nutrient is uptake by both HS and HI, incorporated into their nutrient content, Q, and used for growth. The uptake of P is thus given by μ(Q)HS and μ(Q)HI for HS and HI, respectively. HS can either die at a constant rate, δ, or become infected (HI). Upon death, the model assumes all P content to become instantly available as free nutrient, both the host P content (Q*) and virus P quota . Viral infection, given by kHSV, follows the laws of mass action determined by an infection contact rate dependent on a co-efficient of volumetric host-virus interaction, k, and on HS and V densities. The period between host cell infection and lyses is determined by a death rate of the infected cells due to lysis, λ = 1/T, where T is the latent period; upon cell burst, the number of virus released into the environment depends on the burst size, b. Free virus have a specific death rate, d. Light, temperature and grazing are not considered in the model. The basic equations for the model (excluding algae) are then:
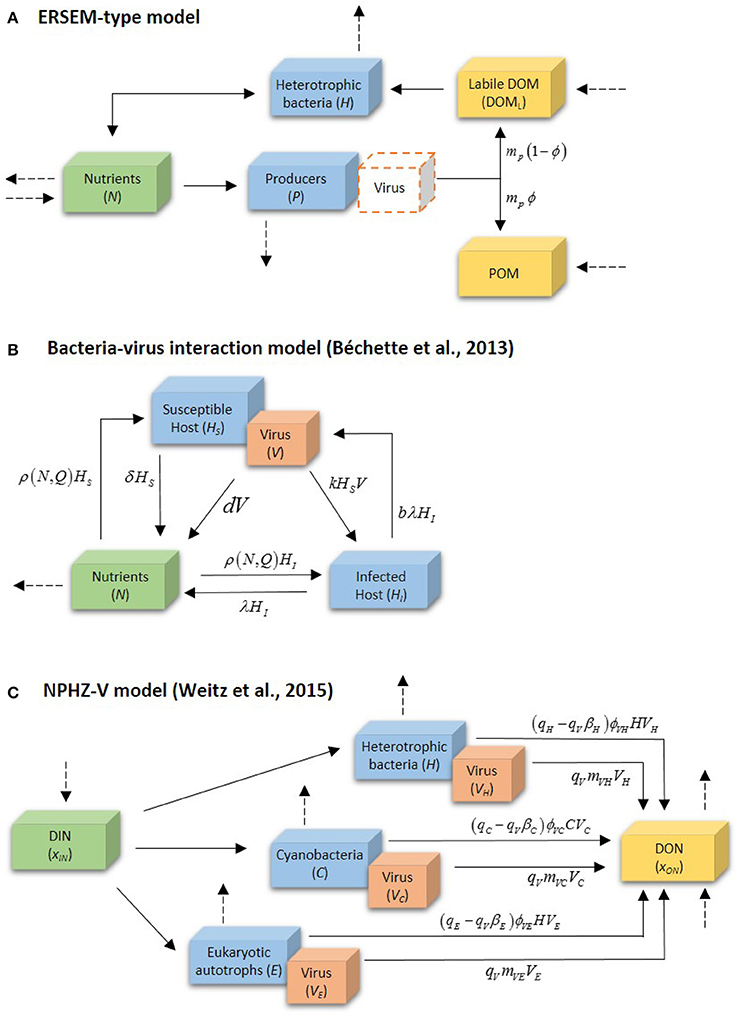
Figure 4. Distinct modeling methodologies of the role of viruses in microbial systems: (A) the ERSEM-type model approach, (B) Béchette et al. (2013) bacteria-virus interaction model, and (C) the NPHZ-V multitrophic model (Weitz et al., 2015). Only (B) shows the complete model; (A,C) only depict a part of the model involved in the activity of viruses. Dashed arrows denote other fluxes in the model. For details on the symbols, please see text and Tables 2–4.
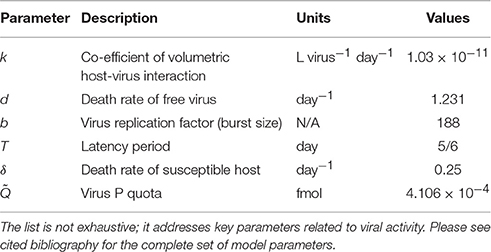
Table 3. List of parameters of a bacteria-virus interaction model (Fuhrman et al., 2011; Béchette et al., 2013).
Also, the recent model proposed by Cael (2015) describes how virus-nutrient limitation that may occur in bacteria and phytoplankton when viruses inject their genetic material through the same channels used to take up nutrients to the cell. Based on their model that combines a mechanistic (based on first principles) and probabilistic approach, the authors propose several possible virus-nutrient-host interrelationships, thus adding complexity to the already intricate task of modeling the roles of viruses but, at the same time, advances a numerical framework for their inclusion. This contribution to food web modeling, however, is limited since the model is built on an individual host perspective for processes occurring at shorter time scales. Nonetheless, this model can be useful to understand co-limitation scenarios, specifically how potential hosts can found balance between maximizing the uptake of limiting nutrient while minimizing viral susceptibility.
While models address some relevant components of microbial food webs with different degrees of complexity, they fail to include others. Hasumi and Nagata (2014) for example, model the cycle of DOM at a global scale including the microbial loop with heterotrophic bacteria, the effect of viral lysis in their mortality (although viruses are not explicitly resolved), and a rather detailed parameterization of organic matter, but having only one general group for phytoplankton and another for zooplankton, two nutrients (N and Fe), and lacking mixotrophy. Previously Keller and Hood (2013) had proposed a C-N model with a significant degree of complexity to study the cycling of DOM in idealized oceanic conditions, including two size classes of phytoplankton, two size classes of zooplankton, heterotrophic bacteria and virus, nutrients (nitrate and ammonium), DIC, detritus and several pools of DOM (labile, semi-labile, and refractory for both N and C). In this model, viruses infect bacteria and both phytoplankton groups with the resulting mortality calculated based on a fixed infection rate for each group and dependent on virus and host densities (type-I functional response), while the decay of viruses is calculated with a quadratic mortality rate. Vallina et al. (2014), on the other hand, uses a global model application to study the relationship between phytoplankton diversity and productivity, with as much as 64 phytoplankton species, two predator size-classes (micro- and meso-zooplankton) and several element cycles. However, the model has no mixotrophs and viral activity, nor it has an explicit representation of the microbial loop, but instead a constant degradation rates of bacterial remineralization. Also at a global scale, the model by Stock et al. (2014) to assess carbon and energy flows through the marine planktonic food web considers several functional groups of organisms (three for phytoplankton, three for zooplankton and heterotrophic bacteria) and organic matter pools; their model, however, assumes lysis caused by viral activity as a weak density-dependent loss term for the small phytoplankton group, and rely on the assumption of heterotrophic bacteria being strictly remineralizers, thus not competing with phytoplankton for inorganic nutrients. Even models of bacterial dynamics, such as Liu et al. (2015), lack some basic components like multiple pools of organic matter, mixotrophy or viral-induced lysis on bacteria.
A more extensive and detailed work on the inclusion of a virus component in a multitrophic ecosystem models was recently proposed by Weitz et al. (2015). The authors have started from a relatively low-complexity model representation including organic and inorganic nutrients, phytoplankton (cyanobacteria and eukaryotic autotrophs), heterotrophic bacteria and zooplankton (NPHZ model). Unlike previous models, however, viruses were included with greater detail in the parameterization, allowing the study of the consequences associated with alternative fates of infected cells and virus particles in a systems context. This new modeling method has established a new type of models, the NPHZ-V models (Figure 4C), including three groups of microbes (functional groups): heterotrophic bacteria (H), cyanobacteria (C), and eukaryotic autotrophs (E), each exclusively infected by a group of viruses, VH, VC, and VE, respectively. All three groups of microbes are predated by a general zooplankton group and nutrients are described as concentration of bioavailable organic (xON) and inorganic matter (xIN). The infection of each host group occurs at a rate proportional to the product of a group-specific lysis rate (ϕVH, VC, VE), and the host and respective virus densities (type-I functional response). This loss term is represented as follows for each microbial group:
where (…) denotes the remaining terms in the mass balance equations (also hereafter). Model parameters related to viral activity can be found in Table 4. Death of infected hosts releases a virus burst depending on the burst size (βH, C, E), a group-specific parameter. Viral decay is proportional to each viral group density and a specific viral decay rate (mVH, VC, VE). The evolution of viruses in time is thus achieved by the balance of these two terms, according to:
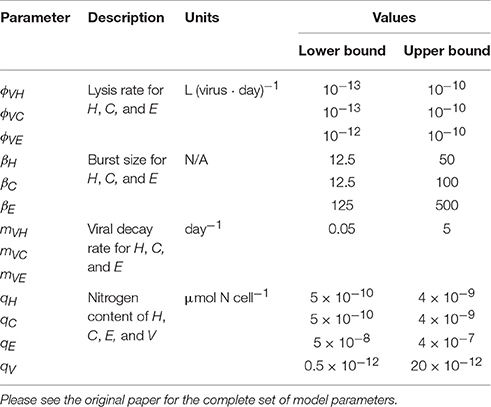
Table 4. Non-exhaustive list of parameters for heterotrophic bacteria (H), cyanobacteria (C), eukaryotic phytoplankton (E), and virus (V) in Weitz et al. (2015) multitrophic NPHZ-V model.
Finally, lysis also releases bioavailable organic matter (xON): a fraction from viral decay depending on N content in the released viruses, qV, and another fraction as lysed products calculated as the difference between N content in hosts, qH, C, E, and qV. These terms are expressed as:
Faced with these recent advancements in modeling viruses the obvious challenge seems now to be the inclusion of these concepts and algorithms, or at least part of them, in similarly complex biogeochemical models. As Weitz et al. (2015) challengingly points out, this new approach on modeling viruses may account with greater detail for the potential effects that viruses have within multitrophic communities, and test some long-standing hypotheses concerning the role of viruses dynamic nutrient feedbacks and their quantitative parameterization. Ultimately, the NPHZ-V model can be seen as the dawning of a new era in predictive global Earth system models.
It's an unquestionable fact that the complexity of models and model applications has continuously increased over the past two decades. Still, viruses are yet to become a common component in marine models, as other relevant state-variables and processes related with the microbial loop such as mixotrophy, several functional groups and elements, or detailed parameterization of organic matter. Mixotrophy is of particular interest given that mixotrophic organisms also have a significant impact on bacterial populations. Despite being known as important player in the microbial food webs, only recently mixotrophs have received attention in modeling studies (Ward et al., 2011; Mitra et al., 2014; Berge et al., 2016; Ward and Follows, 2016). The inclusion of these “perfect beats” combining several modes of nutrition requires additional inclusion of complexity into models, as shown by Flynn and Mitra (2009); stoichiometric implication must be considered by the model, by which the C-to-nutrient ratios must be explicitly expressed for the mixotrophs and food source, otherwise the model is doomed to fail to reflect the reality of the trophic interactions, both at the mixotrophs level as on the ecosystem level. ERSEM type models that rely on a functional group approach have mixotrophs by combining code from producers and consumers (Baretta-Bekker et al., 1995, 1997; Baretta et al., 1995; Vichi et al., 2007; Mateus, 2012a). However, this method of inclusion of a full description of mixotrophy into models has its own problems, such as an increase in complexity and computational costs. To solve the complexity problems associated with the functional group approach, Berge et al. (2016) have recently proposed an alternative way to model mixotrophs using a trait-based approach to represent the full spectrum of trophic strategies.
Final Remarks
The full inclusion of the role of viruses is surely one of the challenging tasks in the development of end-to-end models that try to embrace processes and biological entities with spatial scales differing many orders of magnitude (e.g., from viruses to whales) (Travers et al., 2007; Fulton, 2010; Griffith and Fulton, 2014). Currently available biogeochemical models for marine systems are overly complex, and the inclusion of additional algorithms to account for viruses, with the detail addressed above is a challenging task. However, the potential impact of viruses on the flow of energy and matter seems to be too important for their exclusion. Ultimately, the inclusion of viruses may lend additional insights into more enduring topics such as the link or sink problem in the carbon cycling (Vichi et al., 2004; Fenchel, 2008), as well as on more recent topics such as the role of mixotrophs (Mitra et al., 2014) and the relationship between the limitation of nutrients and viral infection (Béchette et al., 2013).
The level of detail that some models have, with a density-dependent mortality term as a proxy to viral-induced lysis (akin to a closure function for hosts), can be seen as a compromise solution. As elegant as these solutions may be, their realism can be questioned. As studies reveal new details on virus-host dynamics (Schroeder et al., 2003; Martínez et al., 2007; Rosenwasser et al., 2014), the simple assumption that it is possible to mimic host infection and lysis, using only a density-dependent term is being replaced by a more elaborated view of viral dynamics. Environmental conditions are known to influence viral decay rates and latent period between infection and lysis, for example, thus shaping virus-host interactions. The result is that in natural conditions viruses can range over a wide spectrum of functional responses that can only be capture using algorithms more sophisticate than simple closure functions.
Regardless of their complexity, the inclusion of two basic elements can be seen as a way to achieve some realism: (1) an increase in the lysis rate as a function of host (phytoplankton and heterotrophic bacteria) density, and a (2) detailed description of organic matter components, both in type (dissolved and particulate), state (labile, semi-labile and refractory) and composition (variable stoichiometry). For now, available models offer different ways to simulate viruses, with significant degrees of complexity and sophistication. One thing seem to be certain, however, namely that the increase in complexity in modeling viral activity will eventually fail to meet its objectives if embedded in a simplistic model of the food web. Overall, some of the topics that will surely deserve attention, both related to viral activity and the food web dynamics, may be:
• The classification of DOM into a larger number of categories and the identification of different processes controlling different fractions of DOM (Keller and Hood, 2013; Hasumi and Nagata, 2014);
• The inclusion of P, critically important for viral replication (Wilson et al., 1996; Monier et al., 2012), but also consider multiple nutrients (organic and inorganic), as they have a significant influence on the physiologic processes of phytoplankton cells and limit their growth;
• A latent period between infection and lysis, proportions of lysogenic vs. lytic virus infection under different nutrient availability (Béchette et al., 2013), and going beyond the type-I interactions between viruses and hosts and the exponential decay of viruses;
• Taking upon the ERSEM model-type approach with multi-element cycles, variable stoichiometry of both living entities and OM, and several functional groups;
• Move toward the new paradigm in food webs where mixotrophy has a central role (Mitra et al., 2014; Berge et al., 2016).
The inclusion of these elements, however, poses serious challenges to the modeling community, as detailed in Table 5, some of which may offer a reason for why viruses are frequently absent in marine models. While most of these challenges are related with modeling options and, consequently, model complexity, the lack of adequate data is clearly the most significant factor hindering progress in modeling marine food webs. Model outputs will only be of any use when rigorously tested by comparing model results against adequate data sets. Since this task relies on data availability, experimental constraints may compromise the whole modeling endeavor, such as the extreme difficulty in the quantification of virus particles in the field, for instance (Wommack et al., 1996; Gledhill et al., 2012; Heider and Metzner, 2014). Also, the difficulties related to assessing the viability of viral particles needs to be considered (Noble and Fuhrman, 2000; Bongiorni et al., 2005), given the significant number of processes involved, such as turbidity that not only affects light penetration but also passively adsorbs viruses, as recently reviewed by Mojica and Brussaard (2014). For the time being, this is a serious limitation, as pointed out by Salihoglu et al. (2013), since insufficient data is a major roadblock to fully explore the comprehensive synthesis capacity, predictive capacities and mechanistic insight of marine models. But until quantitative data on virus-host interaction becomes available, in silico studies will keep on being a valid way, and possible the only, to test hypothesis and to increase our knowledge on the roles of viruses in microbial food webs.
Finally, the specificity host-virus relationships and their complexity (Schroeder et al., 2003; Pagarete et al., 2009; Rosenwasser et al., 2014) means that the simple inclusion of viruses as a collective pool in models may be considered too simplistic, or even unrealistic. An objective implication of these relationships is that viruses can only be modeled against their specific hosts, meaning that both viruses and hosts needs to be explicitly modeled. Eventually, this implies that the whole modeling effort must start with field measurements for specific host-virus interactions (e.g., Sorensen et al., 2009; Martínez et al., 2012; Sharoni et al., 2015).
According to Brum and Sullivan (2015), the inclusion of viruses into predictive ecological models will only be a feasible achievement with the collaborative work of experimentalists, theorists and modelers. The complexity of the task at hand demands a multidisciplinary approach. Considering the volumes of knowledge produced over the past decades about the dynamics of viruses, and the significant advances in biogeochemical modeling of marine systems, bridging the gap between knowing and modeling the roles of marine viruses is, undoubtedly, one of the next frontiers to be crossed.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by FCT/MCTES (PIDDAC) through project UID/EEA/50009/2013. This work is dedicated to Job Baretta in gratitude for all the marine ecology lessons and discussions on complexity of models.
References
Andersson, A., Lee, C., Azam, F., and Hagstrom, A. (1985). Release of amino-acids and inorganic nutrients by heterotrophic marine microflagellates. Mar. Ecol. Prog. Ser. 23, 99–106. doi: 10.3354/meps023099
Angly, F. E., Felts, B., Breitbart, M., Salamon, P., Edwards, R. A., Carlson, C., et al. (2006). The marine viromes of four oceanic regions. PLoS Biol. 4:e368. doi: 10.1371/journal.pbio.0040368
Arenovski, A. L., Lim, E. L., and Caron, D. A. (1995). Mixotrophic nanoplankton in oligotrophic surface waters of the Sargasso Sea may employ phagotrophy to obtain major nutrients. J. Plankton Res. 17, 801–820. doi: 10.1093/plankt/17.4.801
Azam, F., Fenchel, T., Field, J. G., Gray, J. S., Meyerreil, L. A., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Azam, F., and Hodson, R. E. (1977). Size distribution and activity of marine microheterotrophs. Limnol. Oceanogr. 22, 492–501. doi: 10.4319/lo.1977.22.3.0492
Azam, F., Smith, D. C., Steward, G. F., and Hagström, A. (1994). Bacteria - organic-matter coupling and its significance for oceanic carbon cycling. Microb. Ecol. 28, 167–179. doi: 10.1007/BF00166806
Baretta, J. W., Ebenhoh, W., and Ruardij, P. (1995). The European-regional-seas-ecosystem-model, a complex marine ecosystem model. Netherlands J. Sea Res. 33, 233–246. doi: 10.1016/0077-7579(95)90047-0
Baretta-Bekker, J. G., Baretta, J. W., and Ebenhoh, W. (1997). Microbial dynamics in the marine ecosystem model ERSEM II with decoupled carbon assimilation and nutrient uptake. J. Sea Res. 38, 195–211. doi: 10.1016/S1385-1101(97)00052-X
Baretta-Bekker, J. G., Baretta, J. W., Hansen, A. S., and Riemann, B. (1998). An improved model of carbon and nutrient dynamics in the microbial food web in marine enclosures. Aquat. Microb. Ecol. 14, 91–108. doi: 10.3354/ame014091
Baretta-Bekker, J. G., Baretta, J. W., and Rasmussen, E. K. (1995). The microbial food-web in the european-regional-seas-ecosystem-model. Netherlands J. Sea Res. 33, 363–379. doi: 10.1016/0077-7579(95)90053-5
Baretta-Bekker, J. G., Riemann, B., Baretta, J. W., and Rasmussen, E. K. (1994). Testing the microbial loop concept by comparing mesocosm data with results from a dynamical simulation-model. Mar. Ecol. Prog. Ser. 106, 187–198. doi: 10.3354/meps106187
Baudoux, A. C., and Brussaard, C. P. (2005). Characterization of different viruses infecting the marine harmful algal bloom species Phaeocystis globosa. Virology 341, 80–90. doi: 10.1016/j.virol.2005.07.002
Béchette, A., Stojsavljevic, T., Tessmer, M., Berges, J. A., Pinter, G. A., and Young, E. B. (2013). Mathematical modeling of bacteria–virus interactions in Lake Michigan incorporating phosphorus content. J. Great Lakes Res. 39, 646–654. doi: 10.1016/j.jglr.2013.09.003
Benner, R., Pakulski, J. D., McCarthy, M., Hedges, J. I., and Hatcher, P. G. (1992). Bulk chemical characteristics of dissolved organic-matter in the ocean. Science 255, 1561–1564. doi: 10.1126/science.255.5051.1561
Berge, T., Chakraborty, S., Hansen, P. J., and Andersen, K. H. (2016). Modeling succession of key resource-harvesting traits of mixotrophic plankton. Isme J. 11, 212–223. doi: 10.1038/ismej.2016.92
Berges, J. A., and Falkowski, P. G. (1998). Physiological stress and cell death in marine phytoplankton: induction of proteases in response to nitrogen or light limitation. Limnol. Oceanogr. 43, 129–135. doi: 10.4319/lo.1998.43.1.0129
Bergh, O., Børsheim, K. Y., Bratbak, G., and Heldal, M. (1989). High abundance of viruses found in aquatic environments. Nature 340, 467–468. doi: 10.1038/340467a0
Blackburn, N., Zweifel, U. L., and Hagstrom, A. (1996). Cycling of marine dissolved organic matter.2. A model analysis. Aquat. Microb. Ecol. 11, 79–90. doi: 10.3354/ame011079
Böhm, J., Lambert, O., Frangakis, A. S., Letellier, L., Baumeister, W., and Rigaud, J. L. (2011). FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr. Biol. 11, 1168–1175. doi: 10.1016/S0960-9822(01)00349-9
Bongiorni, L., Magagnini, M., Armeni, M., Noble, R., and Danovaro, R. (2005). Viral production, decay rates, and life strategies along a trophic gradient in the North Adriatic Sea. Appl. Environ. Microbiol. 71, 6644–6650. doi: 10.1128/AEM.71.11.6644-6650.2005
Børsheim, K. Y., Bratbak, G., and Heldal, M. (1990). Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron-microscopy. Appl. Environ. Microbiol. 56, 352–356.
Bratbak, G., Heldal, M., Norland, S., and Thingstad, T. F. (1990). Viruses as partners in spring bloom microbial trophodynamics. Appl. Environ. Microbiol. 56, 1400–1405.
Bratbak, G., Heldal, M., Thingstad, T. F., Riemann, B., and Haslund, O. H. (1992). Incorporation of viruses into the budget of microbial C-transfer - a 1st approach. Mar. Ecol. Prog. Ser. 83, 273–280. doi: 10.3354/meps083273
Bratbak, G., Jacobsen, A., and Heldal, M. (1998a). Viral lysis of Phaeocystis pouchetii and bacterial secondary production. Aquat. Microb. Ecol. 16, 11–16. doi: 10.3354/ame016011
Bratbak, G., Jacobsen, A., Heldal, M., Nagasaki, K., and Thingstad, F. (1998b). Virus production in Phaeocystis pouchetii and its relation to host cell growth and nutrition. Aquat. Microb. Ecol. 16, 1–9. doi: 10.3354/ame016001
Bratbak, G., and Thingstad, T. F. (1985). Phytoplankton-bacteria interactions - an apparent paradox - analysis of a model system with both competition and commensalism. Mar. Ecol. Prog. Ser. 25, 23–30. doi: 10.3354/meps025023
Breitbart, M., Felts, B., Kelley, S., Mahaffy, J. M., Nulton, J., Salamon, P., et al. (2004). Diversity and population structure of a near-shore marine-sediment viral community. Proc. R. Soc. B Biol. Sci. 271, 565–574. doi: 10.1098/rspb.2003.2628
Breitbart, M., Salamon, P., Andresen, B., Mahaffy, J. M., Segall, A. M., Mead, D., et al. (2002). Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U.S.A. 99, 14250–14255. doi: 10.1073/pnas.202488399
Brum, J. R., Morris, J. J., Décima, M., and Stukel, M. R. (2014). “Mortality in the oceans: causes and consequences,” in Eco-DAS IX Symposium Proceedings (Waco, TX: ASLO), 16–48.
Brum, J. R., and Sullivan, M. B. (2015). Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 13, 147–159. doi: 10.1038/nrmicro3404
Brussaard, C. P. D., Kuipers, B., and Veldhuis, M. J. W. (2005). A mesocosm study of Phaeocystis globosa population dynamics: I. Regulatory role of viruses in bloom control. Harmful Algae 4, 859–874. doi: 10.1016/j.hal.2004.12.015
Brussaard, C. P. D., Noordeloos, A. A. M., and Riegman, R. (1997). Autolysis kinetics of the marine diatom Ditylum brightwellii (Bacillariophyceae) under nitrogen and phosphorus limitation and starvation. J. Phycol. 33, 980–987. doi: 10.1111/j.0022-3646.1997.00980.x
Brussaard, C. P. D., Riegman, R., Noordeloos, A. A. M., Cadee, G. C., Witte, H., Kop, A. J., et al. (1995). Effects of grazing, sedimentation and phytoplankton cell-lysis on the structure of a coastal pelagic food-web. Mar. Ecol. Prog. Ser. 123, 259–271. doi: 10.3354/meps123259
Butenschön, M., Clark, J., Aldridge, J. N., Allen, J. I., Artioli, Y., Blackford, J., et al. (2016). ERSEM 15.06: a generic model for marine biogeochemistry and the ecosystem dynamics of the lower trophic levels. Geosci. Model Dev. 9, 1293–1339. doi: 10.5194/gmd-9-1293-2016
Cael, B. B. (2015). The Good, the bad, and the tiny: a simple, mechanistic-probabilistic model of virus-nutrient colimitation in microbes. PLoS ONE 10:e0143299. doi: 10.1371/journal.pone.0143299
Caron, D. A. (2016). Mixotrophy stirs up our understanding of marine food webs. Proc. Natl. Acad. Sci. U.S.A. 113, 2806–2808. doi: 10.1073/pnas.1600718113
Carreira, C., Piel, T., Staal, M., Stuut, J.-B. W., Middelboe, M., and Brussaard, C. P. D. (2015a). Microscale spatial distributions of microbes and viruses in intertidal photosynthetic microbial mats. Springerplus 4:239. doi: 10.1186/s40064-015-0977-8
Carreira, C., Staal, M., Middelboe, M., and Brussaard, C. P. (2015b). Counting viruses and bacteria in photosynthetic microbial mats. Appl. Environ. Microbiol. 81, 2149–2155. doi: 10.1128/AEM.02863-14
Chróst, R. J. (1990). “Microbial ectoenzymes in aquatic environments,” in Aquatic Microbial Ecology: Biochemical and Molecular Approaches, eds R. J. Chrost and J. Overbeck (New York, NY: Springer-Verlag), 47–78.
Cottrell, M. T., and Suttle, C. A. (1995). Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate micromonas-pusilla. Limnol. Oceanogr. 40, 730–739. doi: 10.4319/lo.1995.40.4.0730
Culley, A. I., Lang, A. S., and Suttle, C. A. (2006). Metagenomic analysis of coastal RNA virus communities. Science 312, 1795–1798. doi: 10.1126/science.1127404
Davidson, K., Anderson, D. M., Mateus, M., Reguera, B., Silke, J., Sourisseau, M., et al. (2016). Forecasting the risk of harmful algal blooms. Harmful Algae 53, 1–7. doi: 10.1016/j.hal.2015.11.005
De Corte, D., Sintes, E., Yokokawa, T., Lekunberri, I., and Herndl, G. J. (2016). Large-scale distribution of microbial and viral populations in the South Atlantic Ocean. Environ. Microbiol. Rep. 8, 305–315. doi: 10.1111/1758-2229.12381
Derelle, E., Ferraz, C., Escande, M.-L., Eychenié, S., Cooke, R., Piganeau, G., et al. (2008). Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS ONE 3:e2250. doi: 10.1371/journal.pone.0002250
Dunigan, D. D., Fitzgerald, L. A., and Van Etten, J. L. (2006). Phycodnaviruses: a peek at genetic diversity. Virus Res. 117, 119–132. doi: 10.1016/j.virusres.2006.01.024
Dunne, J. P., John, J. G., Adcroft, A. J., Griffies, S. M., Hallberg, R. W., Shevliakova, E., et al. (2012a). GFDL's ESM2 global coupled climate–carbon earth system models. Part I: physical formulation and baseline simulation characteristics. J. Clim. 25, 6646–6665. doi: 10.1175/JCLI-D-11-00560.1
Dunne, J. P., John, J. G., Shevliakova, E., Stouffer, R. J., Krasting, J. P., Malyshev, S. L., et al. (2012b). GFDL's ESM2 global coupled climate–carbon earth system models. Part II: carbon system formulation and baseline simulation characteristics. J. Clim. 26, 2247–2267. doi: 10.1175/JCLI-D-12-00150.1
Fasham, M. J. R. (1993). “Modelling the marine biota,” in The Global Carbon Cycle, ed M. Heimann (Heidelberg: Springer Verlag), 457–504.
Fenchel, T. (2008). The microbial loop – 25 years later. J. Exp. Mar. Biol. Ecol. 366, 99–103. doi: 10.1016/j.jembe.2008.07.013
Flynn, K. J., and Mitra, A. (2009). Building the “perfect beast”: modelling mixotrophic plankton. J. Plankton Res. 31, 965–992. doi: 10.1093/plankt/fbp044
Follows, M. J., and Dutkiewicz, S. (2010). Modeling diverse communities of marine microbes. Ann. Rev. Mar. Sci. 3, 427–451. doi: 10.1146/annurev-marine-120709-142848
Fuhrman, J. A. (1992). “Bacterioplankton roles in cycling of organic matter: the microbial food web,” in Primary Productivity and Biogeochemical Cycles in the Sea, eds P. Falkowski and A. Woodhead (New York, NY: Plenum Press), 361–217.
Fuhrman, J. A. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. doi: 10.1038/21119
Fuhrman, J. A., and Noble, R. T. (1995). Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 40, 1236–1242. doi: 10.4319/lo.1995.40.7.1236
Fuhrman, J. A., and Suttle, C. A. (1993). Viruses in marine planktonic systems. Oceanography 6, 51–63. doi: 10.5670/oceanog.1993.14
Fuhrman, J. A., Wilcox, R. M., Noble, R. T., and Law, N. C. (1993). Viruses in marine food webs. Trends Microb. Ecol. 295–298.
Fuhrman, K. M., Pinter, G. A., and Berges, J. A. (2011). Dynamics of a virus–host model with an intrinsic quota. Math. Comput. Model. 53, 716–730. doi: 10.1016/j.mcm.2010.10.010
Fulton, E. A. (2010). Approaches to end-to-end ecosystem models. J. Mar. Syst. 81, 171–183. doi: 10.1016/j.jmarsys.2009.12.012
Gledhill, M., Devez, A., Highfield, A., Singleton, C., Achterberg, E., and Schroeder, D. (2012). Effect of metals on the lytic cycle of the coccolithovirus, EhV86. Front. Microbiol. 3:155. doi: 10.3389/fmicb.2012.00155
Gobler, C. J., Hutchins, D. A., Fisher, N. S., Cosper, E. M., and Sanudo-Wilhelmy, S. A. (1997). Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol. Oceanogr. 42, 1492–1504. doi: 10.4319/lo.1997.42.7.1492
Goldman, J. C., and Dennett, M. R. (1992). Phagotrophy and Nh-4(+) regeneration in a 3-member microbial food loop. J. Plankton Res. 14, 649–663. doi: 10.1093/plankt/14.5.649
Griffith, G. P., and Fulton, E. A. (2014). New approaches to simulating the complex interaction effects of multiple human impacts on the marine environment. ICES J. Mar. Sci. 71, 764–774. doi: 10.1093/icesjms/fst196
Gustavsen, J. A., Winget, D. M., Tian, X., and Suttle, C. A. (2014). High temporal and spatial diversity in marine RNA viruses implies that they have an important role in mortality and structuring plankton communities. Front. Microbiol. 5:703. doi: 10.3389/fmicb.2014.00703
Hall, J. A., Barrett, D. P., and James, M. R. (1993). The importance of phytoflagellate, heterotrophic flagellate and ciliate grazing on bacteria and picophytoplankton sized prey in a coastal marine environment. J. Plankton Res. 15, 1075–1086. doi: 10.1093/plankt/15.9.1075
Hasumi, H., and Nagata, T. (2014). Modeling the global cycle of marine dissolved organic matter and its influence on marine productivity. Ecol. Modell. 288, 9–24. doi: 10.1016/j.ecolmodel.2014.05.009
Heider, S., and Metzner, C. (2014). Quantitative real-time single particle analysis of virions. Virology 462–463, 199–206. doi: 10.1016/j.virol.2014.06.005
Heldal, M., and Bratbak, G. (1991). Production and decay of viruses in aquatic environments Mar. Ecol. Prog. Ser. 72, 205–212. doi: 10.3354/meps072205
Hurwitz, B. L., and Sullivan, M. B. (2013). The Pacific Ocean Virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral Ecology. PLoS ONE 8:e57355. doi: 10.1371/journal.pone.0057355
Hurwitz, B. L., U'Ren, J. M., and Youens-Clark, K. (2016). Computational prospecting the great viral unknown. FEMS Microbiol. Lett. 363:fnw077. doi: 10.1093/femsle/fnw077
Imai, I., Ishida, Y., and Hata, Y. (1993). Killing of marine-phytoplankton by a gliding bacterium cytophaga sp, isolated from the coastal sea of Japan. Mar. Biol. 116, 527–532. doi: 10.1007/BF00355470
Imai, I., Ishida, Y., Sakaguchi, K., and Hata, Y. (1995). Algicidal marine-bacteria isolated from northern hiroshima bay, Japan. Fish. Sci. 61, 628–636.
Jacobsen, A., Bratbak, G., and Heldal, M. (1996). Isolation and characterization of a virus infecting Phaeocystis pouchetii (Prymnesiophyceae). J. Phycol. 32, 923–927. doi: 10.1111/j.0022-3646.1996.00923.x
Keller, D. P., and Hood, R. R. (2013). Comparative simulations of dissolved organic matter cycling in idealized oceanic, coastal, and estuarine surface waters. J. Mar. Syst. 109–110, 109–128. doi: 10.1016/j.jmarsys.2012.01.002
Kelly, L., Ding, H., Huang, K. H., Osburne, M. S., and Chisholm, S. W. (2013). Genetic diversity in cultured and wild marine cyanomyoviruses reveals phosphorus stress as a strong selective agent. ISME J. 7, 1827–1841. doi: 10.1038/ismej.2013.58
Kirchman, D. L., Lancelot, C., Fasham, M., Legendre, L., Radach, G., and Scott, M. (1993). “Dissolved organic matter in biogeochemical models of the ocean,” in Towards a Model of Ocean Biogeochemical Processes, eds G. T. Evans and M. J. R. Fasham (Berlin: Springer-Verlag), 209–225.
Lancelot, C., and Billen, G. (1984). Activity of heterotrophic bacteria and its coupling to primary production during the spring phytoplankton bloom in the southern bight of the north-sea. Limnol. Oceanogr. 29, 721–730. doi: 10.4319/lo.1984.29.4.0721
Lancelot, C., Billen, G., Veth, C., Becquevort, S., and Mathot, S. (1991). modeling carbon cycling through phytoplankton and microbes in the scotia-weddell sea area during sea ice retreat. Mar. Chem. 35, 305–324. doi: 10.1016/S0304-4203(09)90024-X
Libralato, S., and Solidoro, C. (2009). Bridging biogeochemical and food web models for an End-to-End representation of marine ecosystem dynamics: the Venice lagoon case study. Ecol. Modell. 220, 2960–2971. doi: 10.1016/j.ecolmodel.2009.08.017
Liu, L., Lwiza, K. M. M., and Taylor, G. T. (2015). Importance of the bacterial dynamics in model simulations of seasonal hypoxia. Cont. Shelf Res. 105, 1–17. doi: 10.1016/j.csr.2015.05.008
Maat, D. S., Crawfurd, K. J., Timmermans, K. R., and Brussaard, C. P. D. (2014). Elevated CO(2) and phosphate limitation favor Micromonas pusilla through stimulated growth and reduced viral impact. Appl. Environ. Microbiol. 80, 3119–3127. doi: 10.1128/AEM.03639-13
Maat, D. S., de Blok, R., and Brussaard, C. P. D. (2016a). Combined phosphorus limitation and light stress prevent viral proliferation in the phytoplankton species Phaeocystis globosa, but Not in Micromonas pusilla. Front. Mar. Sci. 3:160. doi: 10.3389/fmars.2016.00160
Maat, D. S., van Bleijswijk, J. D. L., Witte, H. J., and Brussaard, C. P. D. (2016b). Virus production in phosphorus limited Micromonas pusilla stimulated by a supply of naturally low concentrations of different phosphorus sources, far into the lytic cycle. FEMS Microbiol. Ecol. 92:fiw136. doi: 10.1093/femsec/fiw136
Magiopoulos, I., and Pitta, P. (2012). Viruses in a deep oligotrophic sea: seasonal distribution of marine viruses in the epi-, meso- and bathypelagic waters of the Eastern Mediterranean Sea. Deep Sea Res. I Oceanogr. Res. Pap. 66, 1–10. doi: 10.1016/j.dsr.2012.03.009
Malits, A., Christaki, U., Obernosterer, I., and Weinbauer, M. G. (2014). Enhanced viral production and virus-mediated mortality of bacterioplankton in a natural iron-fertilized bloom event above the Kerguelen Plateau. Biogeosciences 11, 6841–6853. doi: 10.5194/bg-11-6841-2014
Martínez, J. M., Schroeder, D. C., Larsen, A., Bratbak, G., and Wilson, W. H. (2007). Molecular dynamics of Emiliania huxleyi and cooccurring viruses during two separate mesocosm studies. Appl. Environ. Microbiol. 73, 554–562. doi: 10.1128/AEM.00864-06
Martínez, J. M., Schroeder, D. C., and Wilson, W. H. (2012). Dynamics and genotypic composition of Emiliania huxleyi and their co-occurring viruses during a coccolithophore bloom in the North Sea. FEMS Microbiol. Ecol. 81, 315–323. doi: 10.1111/j.1574-6941.2012.01349.x
Martínez, J. M., Swan, B. K., and Wilson, W. H. (2014). Marine viruses, a genetic reservoir revealed by targeted viromics. ISME J. 8, 1079–1088. doi: 10.1038/ismej.2013.214
Mateus, M. (2012a). A process-oriented model of pelagic biogeochemistry for marine systems. Part I: model description. J. Mar. Syst. 94, S78–S89. doi: 10.1016/j.jmarsys.2011.11.008
Mateus, M. (2012b). A process-oriented model of pelagic biogeochemistry for marine systems. Part I: model description. J. Mar. Syst. 94, 78–S89. doi: 10.1016/j.jmarsys.2011.11.008
Middelboe, M., Jorgensen, N., and Kroer, N. (1996). Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl. Environ. Microbiol. 62, 1991–1997.
Middelboe, M., Riemann, L., Steward, G. F., Hansen, V., and Nybroe, O. (2003). Virus-induced transfer of organic carbon between marine bacteria in a model community. Aquat. Microb. Ecol. 33, 1–10. doi: 10.3354/ame033001
Mitra, A., Flynn, K. J., Burkholder, J. M., Berge, T., Calbet, A., Raven, J. A., et al. (2014). The role of mixotrophic protists in the biological carbon pump. Biogeosciences 11, 995–1005. doi: 10.5194/bg-11-995-2014
Mojica, K. D., and Brussaard, C. P. (2014). Factors affecting virus dynamics and microbial host–virus interactions in marine environments. FEMS Microbiol. Ecol. 89, 495–515. doi: 10.1111/1574-6941.12343
Monier, A., Welsh, R. M., Gentemann, C., Weinstock, G., Sodergren, E., Armbrust, E. V., et al. (2012). Phosphate transporters in marine phytoplankton and their viruses: cross-domain commonalities in viral-host gene exchanges. Environ. Microbiol. 14, 162–176. doi: 10.1111/j.1462-2920.2011.02576.x
Murray, A. G., and Jackson, G. A. (1992). Viral dynamics - a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Prog. Ser. 89, 103–116. doi: 10.3354/meps089103
Noble, R. T., and Fuhrman, J. A. (1999). Breakdown and microbial uptake of marine viruses and other lysis products. Aquat. Microb. Ecol. 20, 1–11. doi: 10.3354/ame020001
Noble, R. T., and Fuhrman, J. A. (2000). Rapid virus production and removal as measured with fluorescently labeled viruses as tracers. Appl. Environ. Microbiol. 66, 3790–3797. doi: 10.1128/AEM.66.9.3790-3797.2000
Ortmann, A. C., Lawrence, J. E., and Suttle, C. A. (2002). Lysogeny and lytic viral production during a bloom of the cyanobacterium Synechococcus spp. Microb. Ecol. 43, 225–231. doi: 10.1007/s00248-001-1058-9
Ory, P., Hartmann, H. J., Jude, F., Dupuy, C., Del Amo, Y., Catala, P., et al. (2010). Pelagic food web patterns: do they modulate virus and nanoflagellate effects on picoplankton during the phytoplankton spring bloom? Environ. Microbiol. 12, 2755–2772. doi: 10.1111/j.1462-2920.2010.02243.x
Pagarete, A., Allen, M. J., Wilson, W. H., Kimmance, S. A., and de Vargas, C. (2009). Host–virus shift of the sphingolipid pathway along an Emiliania huxleyi bloom: survival of the fattest. Environ. Microbiol. 11, 2840–2848. doi: 10.1111/j.1462-2920.2009.02006.x
Pasquer, B., Laruelle, G., Becquevort, S., Schoemann, V., Goosse, H., and Lancelot, C. (2005). Linking ocean biogeochemical cycles and ecosystem structure and function: results of the complex SWAMCO-4 model. J. Sea Res. 53, 93–108. doi: 10.1016/j.seares.2004.07.001
Petersen, J., and Dubilier, N. (2014). Gene swapping in the dead zone. Elife 3:e04600. doi: 10.7554/eLife.04600
Petihakis, G., Triantafyllou, G., Korres, G., Tsiaras, K., and Theodorou, A. (2012). Ecosystem modelling: towards the development of a management tool for a marine coastal system part-II, ecosystem processes and biogeochemical fluxes. J. Mar. Syst. 94(Suppl.), S49–S64. doi: 10.1016/j.jmarsys.2011.11.006
Proctor, L. M. (1997). Advances in the study of marine viruses. Microsc. Res. Tech. 37, 136–161. doi: 10.1002/(SICI)1097-0029(19970415)37:2<136::AID-JEMT3>3.0.CO;2-M
Proctor, L. M., and Fuhrman, J. A. (1990). Viral mortality of marine-bacteria and cyanobacteria. Nature 343, 60–62. doi: 10.1038/343060a0
Proctor, L. M., and Fuhrman, J. A. (1991). Roles of viral-infection in organic particle-flux. Mar. Ecol. Prog. Ser. 69, 133–142. doi: 10.3354/meps069133
Ramin, M., Perhar, G., Shimoda, Y., and Arhonditsis, G. B. (2012). Examination of the effects of nutrient regeneration mechanisms on plankton dynamics using aquatic biogeochemical modeling. Ecol. Modell. 240, 139–155. doi: 10.1016/j.ecolmodel.2012.04.018
Rohwer, F., and Thurber, R. V. (2009). Viruses manipulate the marine environment. Nature 459, 207–212. doi: 10.1038/nature08060
Rosenwasser, S., Mausz, M. A., Schatz, D., Sheyn, U., Malitsky, S., Aharoni, A., et al. (2014). Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell 26, 2689–2707. doi: 10.1105/tpc.114.125641
Roux, S., Hawley, A. K., Torres Beltran, M., Scofield, M., Schwientek, P., Stepanauskas, R., et al. (2014). Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. Elife 3:e03125. doi: 10.7554/eLife.03125
Ruardij, P., Veldhuis, M. J. W., and Brussaard, C. P. D. (2005). Modeling the bloom dynamics of the polymorphic phytoplankter Phaeocystis globosa: impact of grazers and viruses. Harmful Algae 4, 941–963. doi: 10.1016/j.hal.2004.12.011
Salihoglu, B., Neuer, S., Painting, S., Murtugudde, R., Hofmann, E. E., Steele, J. H., et al. (2013). Bridging marine ecosystem and biogeochemistry research: lessons and recommendations from comparative studies. J. Mar. Syst. 109–110, 161–175. doi: 10.1016/j.jmarsys.2012.07.005
Sandaa, R.-A. (2008). Burden or benefit? Virus–host interactions in the marine environment. Res. Microbiol. 159, 374–381. doi: 10.1016/j.resmic.2008.04.013
Schroeder, D. C., Oke, J., Hall, M., Malin, G., and Wilson, W. H. (2003). Virus succession observed during an Emiliania huxleyi bloom. Appl. Environ. Microbiol. 69, 2484–2490. doi: 10.1128/AEM.69.5.2484-2490.2003
Sharoni, S., Trainic, M., Schatz, D., Lehahn, Y., Flores, M. J., Bidle, K. D., et al. (2015). Infection of phytoplankton by aerosolized marine viruses. Proc. Natl. Acad. Sci. U.S.A. 112, 6643–6647. doi: 10.1073/pnas.1423667112
Shelford, E. J., Middelboe, M., Møller, E. F., and Suttle, C. A. (2012). Virus-driven nitrogen cycling enhances phytoplankton growth. Aquat. Microb. Ecol. 66, 41–46. doi: 10.3354/ame01553
Shirai, Y., Tomaru, Y., Takao, Y., Suzuki, H., Nagumo, T., and Nagasaki, K. (2008). Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom chaetoceros tenuissimus meunier. Appl. Environ. Microbiol. 74, 4022–4027. doi: 10.1128/AEM.00509-08
Short, S. M. (2012). The ecology of viruses that infect eukaryotic algae. Environ. Microbiol. 14, 2253–2271. doi: 10.1111/j.1462-2920.2012.02706.x
Sorensen, G., Baker, A. C., Hall, M. J., Munn, C. B., and Schroeder, D. C. (2009). Novel virus dynamics in an Emiliania huxleyi bloom. J. Plankton Res. 31, 787–791. doi: 10.1093/plankt/fbp027
Stock, C. A., Dunne, J. P., and John, J. G. (2014). Global-scale carbon and energy flows through the marine planktonic food web: an analysis with a coupled physical–biological model. Prog. Oceanogr. 120, 1–28. doi: 10.1016/j.pocean.2013.07.001
Strayer, D. (1988). On the limits to secondary production. Limnol. Oceanogr. 33, 1217–1220. doi: 10.4319/lo.1988.33.5.1217
Strom, S. L., Benner, R., Ziegler, S., and Dagg, M. J. (1997). Planktonic grazers are a potentially important source of marine dissolved organic carbon. Limnol. Oceanogr. 42, 1364–1374. doi: 10.4319/lo.1997.42.6.1364
Sullivan, M. B., Lindell, D., Lee, J. A., Thompson, L. R., Bielawski, J. P., and Chisholm, S. W. (2006). Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4:e234. doi: 10.1371/journal.pbio.0040234
Suttle, C. A. (1994). The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28, 237–243. doi: 10.1007/BF00166813
Suttle, C. A., and Chan, A. M. (1993). Marine cyanophages infecting oceanic and coastal strains of Synechococcus - abundance, morphology, cross-infectivity and growth-characteristics. Mar. Ecol. Prog. Ser. 92, 99–109. doi: 10.3354/meps092099
Suttle, C. A., Chan, A. M., and Cottrell, M. T. (1991). Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl. Environ. Microbiol. 57, 721–726.
Tai, V., Lawrence, J. E., Lang, A. S., Chan, A. M., Culley, A. I., and Suttle, C. A. (2003). Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39, 343–352. doi: 10.1046/j.1529-8817.2003.01162.x
Thingstad, T. F. (2000). Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45, 1320–1328. doi: 10.4319/lo.2000.45.6.1320
Thingstad, T. F., Heldal, M., Bratbak, G., and Dundas, I. (1993). Are viruses important partners in pelagic food webs. Trends Ecol. Evol. 8, 209–213. doi: 10.1016/0169-5347(93)90101-T
Tomaru, Y., Takao, Y., Suzuki, H., Nagumo, T., and Nagasaki, K. (2009). Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl. Environ. Microbiol. 75, 2375–2381. doi: 10.1128/AEM.02580-08
Travers, M., Shin, Y. J., Jennings, S., and Cury, P. (2007). Towards end-to-end models for investigating the effects of climate and fishing in marine ecosystems. Prog. Oceanogr. 75, 751–770. doi: 10.1016/j.pocean.2007.08.001
Vallina, S. M., Follows, M. J., Dutkiewicz, S., Montoya, J. M., Cermeno, P., and Loreau, M. (2014). Global relationship between phytoplankton diversity and productivity in the ocean. Nat. Commun. 5:4299. doi: 10.1038/ncomms5299
Vallino, J. J. (2000). Improving marine ecosystem models: use of data assimilation and mesocosm experiments. J. Mar. Res. 58, 117–164. doi: 10.1357/002224000321511223
Vanduyl, F. C., and Kop, A. J. (1988). Temporal and lateral fluctuations in production and biomass of bacterioplankton in the western dutch wadden Sea. Netherlands J. Sea Res. 22, 51–68. doi: 10.1016/0077-7579(88)90052-X
Vichi, M., Oddo, P., Zavatarelli, M., Coluccelli, A., Coppini, G., Celio, M., et al. (2003). Calibration and validation of a one-dimensional complex marine biogeochemical flux model in different areas of the northern Adriatic shelf. Ann. Geophys. 21, 413–436. doi: 10.5194/angeo-21-413-2003
Vichi, M., Pinardi, N., and Masina, S. (2007). A generalized model of pelagic biogeochemistry.for the global ocean ecosystem. Part I: theory. J. Mar. Syst. 64, 89–109. doi: 10.1016/j.jmarsys.2006.03.006
Vichi, M., Ruardij, P., and Baretta, J. W. (2004). Link or sink: a modelling interpretation of the open Baltic biogeochemistry. Biogeosciences 1, 79–100. doi: 10.5194/bg-1-79-2004
Walsh, J. J., Dieterle, D. A., Muller-Karger, F. E., Bohrer, R., Bissett, W. P., Varela, R. J., et al. (1999). Simulation of carbon-nitrogen cycling during spring upwelling in the Cariaco Basin. J. Geophys. Res. Oceans 104, 7807–7825. doi: 10.1029/1998JC900120
Ward, B. A., Dutkiewicz, S., Barton, A. D., and Follows, M. J. (2011). Biophysical aspects of resource acquisition and competition in algal mixotrophs. Am. Nat. 178, 98–112. doi: 10.1086/660284
Ward, B. A., and Follows, M. J. (2016). Marine mixotrophy increases trophic transfer efficiency, mean organism size, and vertical carbon flux. Proc. Natl. Acad. Sci. U.S.A. 113, 2958–2963. doi: 10.1073/pnas.1517118113
Weinbauer, M. G. (2004). Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181. doi: 10.1016/j.femsre.2003.08.001
Weinbauer, M. G., and Rassoulzadegan, F. (2004). Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6, 1–11. doi: 10.1046/j.1462-2920.2003.00539.x
Weitz, J. S., Stock, C. A., Wilhelm, S. W., Bourouiba, L., Coleman, M. L., Buchan, A., et al. (2015). A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 9, 1352–1364. doi: 10.1038/ismej.2014.220
Wilhelm, S. W., and Suttle, C. A. (1999). Viruses and Nutrient Cycles in the Sea - Viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49, 781–788. doi: 10.2307/1313569
Wilson, W. H., Carr, N. G., and Mann, N. H. (1996). The effect of phosphate status on the kinetics of cyanophage infection in the oceanic cyanobacterium Synechococcus sp. wh78031. J. Phycol. 32, 506–516. doi: 10.1111/j.0022-3646.1996.00506.x
Wilson, W. H., and Mann, N. H. (1997). Lysogenic and lytic viral production in marine microbial communities. Aquat. Microb. Ecol. 13, 95–100. doi: 10.3354/ame013095
Wommack, K. E., and Colwell, R. R. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114. doi: 10.1128/MMBR.64.1.69-114.2000
Wommack, K. E., Hill, R. T., Muller, T. A., and Colwell, R. R. (1996). Effects of sunlight on bacteriophage viability and structure. Appl. Environ. Microbiol. 62, 1336–1341.
Xu, J., Jing, H., Sun, M., Harrison, P. J., and Liu, H. (2013). Regulation of bacterial metabolic activity by dissolved organic carbon and viruses. J. Geophys. Res. Biogeosci. 118, 1573–1583. doi: 10.1002/2013jg002296
Yau, S., Lauro, F. M., DeMaere, M. Z., Brown, M. V., Thomas, T., Raftery, M. J., et al. (2011). Virophage control of antarctic algal host–virus dynamics. Proc. Natl. Acad. Sci. U.S.A. 108, 6163–6168. doi: 10.1073/pnas.1018221108
Yuji, T., Noriaki, K., Kensho, N., Yoko, S., Kenji, T., Mineo, Y., et al. (2004). Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat. Microb. Ecol. 34, 207–218. doi: 10.3354/ame034207
Keywords: plankton modeling, mechanistic models, viruses, marine food webs, microbial loop
Citation: Mateus MD (2017) Bridging the Gap between Knowing and Modeling Viruses in Marine Systems—An Upcoming Frontier. Front. Mar. Sci. 3:284. doi: 10.3389/fmars.2016.00284
Received: 28 July 2016; Accepted: 16 December 2016;
Published: 06 January 2017.
Edited by:
Cosimo Solidoro, National Institute of Oceanography and Experimental Geophysics, ItalyCopyright © 2017 Mateus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcos D. Mateus, bWFyY29zLm1hdGV1c0B0ZWNuaWNvLnVsaXNib2EucHQ=
 Marcos D. Mateus
Marcos D. Mateus