
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 28 October 2014
Sec. Aquatic Microbiology
Volume 1 - 2014 | https://doi.org/10.3389/fmars.2014.00044
This article is part of the Research Topic Linking Ecosystem Function to Microbial Diversity View all 10 articles
 Jeffrey J. Marlow1*
Jeffrey J. Marlow1* Joshua A. Steele1
Joshua A. Steele1 David H. Case1
David H. Case1 Stephanie A. Connon1
Stephanie A. Connon1 Lisa A. Levin2
Lisa A. Levin2 Victoria J. Orphan1*
Victoria J. Orphan1*Methane seeps are among the most productive habitats along continental margins, as anaerobic methane-oxidizing euryarchaeaota and sulfur-metabolizing deltaproteobacteria form the biological base of a dynamic deep-sea ecosystem. The degree of methane seepage therefore represents one important variable in ecosystem dynamics, and the recent discovery of carbonate-hosted endolithic methanotrophy exposes another potentially discriminating factor: physical substrate type. Methanotrophic microbial communities have been detected within diverse seep-associated habitats, including unlithified sediments, protolithic carbonate nodules, and lithified carbonate slabs and chemoherms of distinct mineralogies. However, a systematic assessment of the diversity and community structure associated with these different habitats has been lacking. In this study, microbial aggregate analysis, microbial abundance quantification, mineralogical identification, and archaeal and bacterial 16S rRNA gene clone libraries were used to deconvolve the relationships between seepage activity, substrate type, and microbial community structure. We report prevalent methane-oxidizing archaeal lineages in both active and low-activity seep settings, and a strong community dependence on both seepage activity and substrate type. Statistical treatments of relative taxa abundances indicate that archaeal community structure is more dependent on the degree of methane seepage than physical substrate type; bacterial assemblages appear to be more strongly influenced by the type of colonization substrate than seepage activity. These findings provide a window into the determinants of community structure and function, improving our understanding of potential elemental cycling at seep sites.
The seafloor is a dynamic and varied environment whose biological communities are dependent upon organic detritus from surface waters and/or chemically reduced fluids emitted from the subseafloor. Benthic habitats cover a range of productivities; in all cases, microorganisms play critical roles in mobilizing chemical or organic energy sources and mediating elemental fluxes (Orcutt et al., 2011 and references therein). Many studies of seafloor microbial ecology have focused on near-surface sediment, examining, for example, methane generation (Claypool and Kvenvolden, 1983), dinitrogen production (Thamdrup and Dalsgaard, 2002), or organic matter remineralization with a range of electron acceptors (Reeburgh, 1983). As the full extent of benthic microbial activity comes into focus, clarifying the identities, distribution, and metabolic roles of constituent organisms in not only sediments, but also endolithic and deeper-seated habitats across a spectrum of energetic regimes, has emerged as a priority.
Methane seeps are among the most productive habitats on the sub-photic zone seafloor. In these regions, reduced, methane-rich fluids come into contact with oxidized seawater, fueling chemosynthetic communities such as anaerobic methanotrophic archaea (ANME)/Deltaproteobacteria consortia that mediate the sulfate-coupled anaerobic oxidation of methane (AOM). AOM is a globally significant process which consumes an estimated 80–90% of methane at the seafloor (Reeburgh, 2007). The cultivation-independent investigation of the microorganisms responsible for AOM has revealed two primary constituents: methane-oxidizing archaea and sulfur-metabolizing bacteria. ANME are believed to activate methane and transfer reducing equivalents to their syntrophic partners, sulfate-reducing or sulfur disproportionating bacteria (SRB; Hoehler et al., 1994; Nauhaus et al., 2002; Milucka et al., 2012). The metabolic byproducts of methanotrophy support micro- and macrofaunal communities in the seep food web, including sulfide-oxidizing bacteria (Beggiatoa, Thioploca), chemosynthetic clams (Vesicomya), mussels (Bathymodiolus), tube worms (Siboglinidae), and ampharetid polychaetes that serve as visual manifestations of CH4 seepage on the seafloor (Van Dover et al., 2003; Levin, 2005; Niemann et al., 2013).
Detailed examination of microbial communities within methane seep sediments has further revealed the community composition and exposed relationships between constituent members. The archaeal group ANME-1 is currently divided into two subgroups (ANME-1a and ANME-1b) and represents a novel order within the Euryarchaeota (Hinrichs et al., 1999; Hallam et al., 2004; Meyerdierks et al., 2005). These anaerobic methane-oxidizing microorganisms have been observed as single cells, as monospecific aggregates, and in association with bacteria (Orphan et al., 2002; Treude et al., 2007; Holler et al., 2011). The Methanosarcinales-affiliated ANME-2 (divisible into three phylogenetic subgroups, ANME-2a, -2b, and -2c; Orphan et al., 2001a) frequently form aggregates with members of the Desulfobacteraceae family (e.g., the Seep-SRB1 group; Boetius et al., 2000; Knittel et al., 2005; Schreiber et al., 2010) or Desulfobulbaceae (Pernthaler et al., 2008; Green-Saxena et al., 2014). Members of the ANME-3 clade have been shown to associate with Bacteria most closely related to another lineage within the Desulfobulbaceae (Lösekann et al., 2007; Green-Saxena et al., 2014).
Sulfate-coupled methane oxidation produces two units of alkalinity per unit of dissolved inorganic carbon, thereby promoting the precipitation of authigenic carbonate minerals (Aloisi et al., 2002). These precipitates form loosely consolidated protoliths, hereafter referred to as “nodules,” found below the sediment-water interface within methane-perfused sediments (Orphan et al., 2004; Watanabe et al., 2008). Larger, fully lithified carbonate rocks also form, likely within methane-perfused sediment (Stadnitskaia et al., 2008; Bian et al., 2013), and can be exposed at the seabed following episodes of uplift and winnowing (Greinert et al., 2001; Naehr et al., 2007; Ussler and Paull, 2008). AOM activity associated with methane seepage at the seabed results in a heterogeneous landscape of reduced sediments and carbonate pavements, chemoherms, and large mounds, covered by patches of chemosynthetic communities. Carbonate structures can be hundreds of meters tall and likely represent the time-integrated accretion of AOM-linked authigenic carbonate precipitation (Greinert et al., 2001). Subsurface advective methane flow can shift with time, turning “active” habitats, which exhibit gas bubbling and/or seafloor chemosynthetic communities, into “low-activity” sites with minimal or no apparent methane flux to the seabed (Treude et al., 2003; Boetius and Suess, 2004). These low-activity areas often contain carbonates with negative δ13C values that are presumed to reflect previous AOM activity, yet it remains unclear how sediment and carbonate-hosted microbial communities differ between active and low-activity sites.
The recent quantification of active metabolic rates of methanotrophic microbial biomass living within the pore spaces of authigenic carbonates in active and low-activity areas further motivates the study of endolithic microbial communities (Marlow et al., 2014). Previous literature has characterized the common taxa associated with carbonates in methane-rich regimes such as cold seeps and mud volcanoes by examining lipid biomarkers (Blumenberg et al., 2004; Stadnitskaia et al., 2005, 2008; Gontharet et al., 2009) and 16S rRNA gene signatures (Reitner et al., 2005; Stadnitskaia et al., 2005, 2008; Heijs et al., 2006). These published findings have been used to discuss the paleo seepage record (Gontharet et al., 2009), as well as stages of carbonate formation (Reitner et al., 2005; Stadnitskaia et al., 2005, 2008; Bahr et al., 2009). Microbial communities associated with carbonate crusts and sediments have been broadly compared in the euxinic Black Sea (Stadnitskaia et al., 2005) and the eastern Mediterranean Sea (Heijs et al., 2006), but the extent to which methanotrophic communities within partially and fully lithified habitats differ from adjacent sediment-hosted communities at continental margin seeps remains largely unconstrained. Given the influence of AOM in methane processing, the community structure of carbonate-based habitats may exert a significant influence on methane and sulfur biogeochemical processes in the deep sea.
This study examines microbial abundance and community diversity in six structural classes of methane seep habitats (active sediments, active nodules, active carbonates, low-activity sediments, low-activity carbonates, and off-seep background sediments), which integrate two key environmental variables: seepage activity and physical substrate type. We analyze mineralogy, 16S rRNA gene archaeal and bacterial sequence diversity, and cell abundances of 12 samples, from six different deep-sea habitats associated with methane seepage at Hydrate Ridge, OR, USA (Table 1; Figure 1). With this dataset, we characterize the microbial diversity and the potential importance of seepage activity and substrate as determinants of microbial community structure in and around methane seeps.
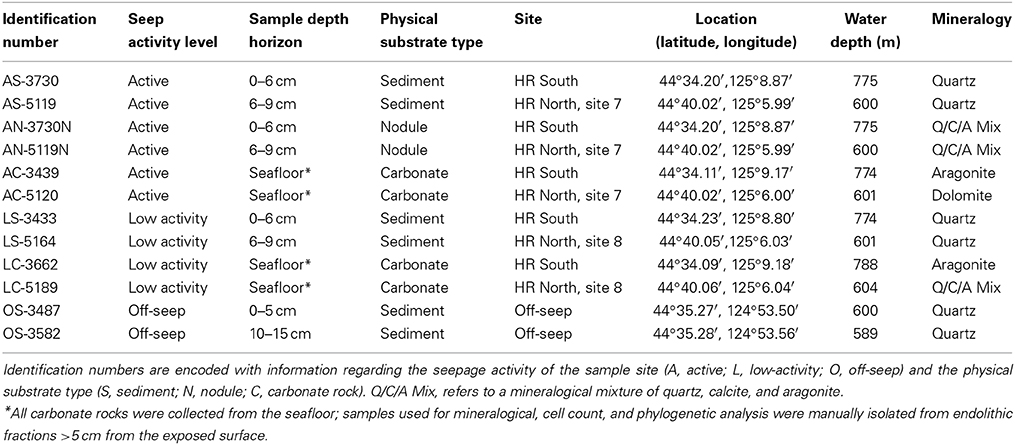
Table 1. Identification numbers, site location details, and mineralogical identifications of carbonate, sediment, and nodule samples used in this study from the Oregon margin, USA.
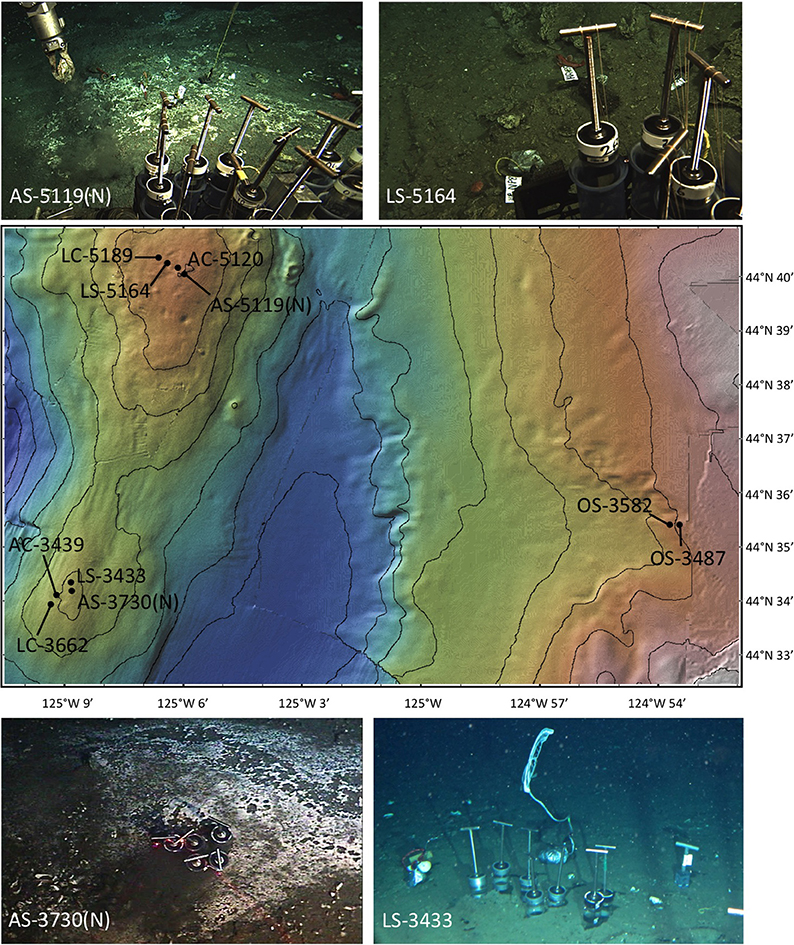
Figure 1. A map of Hydrate Ridge, OR, showing the locations of origin of the samples used in this study, accompanied by images showing the general locations of sample collection. Samples LC-5189, LC-5164, AC-5120, AS-5119, and AS-5119N were collected from Hydrate Ridge north (mound summit ~600 m depth); samples AC-3439, LS-3433, LC-3662, AS-3730, and AS-3730N were collected from Hydrate Ridge south (mound top ~780 m depth); samples OS-3582 and OS-3487 were collected off-seep from a water depth of ~600 m. Hydrate Ridge north and south sampling sites were located approximately 12 km apart. Base map is derived from Global Multi-Resolution Topography (GMRT, Ryan et al., 2009; GeoMapApp); contour lines represent 100 m of depth, and each minute of latitude represents 1.85 km. In the images, push cores are 10 cm in diameter for scale.
Microbial DNA samples were collected from active and low-activity sites around Hydrate Ridge, Oregon, a convergent tectonic margin well established as a site of methane seepage and sediment-based AOM (e.g., Suess et al., 1999; Tryon et al., 2002; Treude et al., 2003). Samples were collected in consecutive years, with the DSV Alvin during R/V Atlantis leg AT-15-68 (September 2010), and with the ROV Jason during Atlantis leg AT-18-10 (September 2011). Two samples representative of each substrate-activity pairing were used in this study—one from Hydrate Ridge North (44°40.25′N, 125°06.30′W, ~600 m water depth), and one from Hydrate Ridge South (44°34.09′N, 125°09.14′W, ~780 m water depth), a separation distance of approximately 11.2 km. Sampling at two distinct seep-influenced locations at Hydrate Ridge confers interpretive power across a relatively broad spatial scale (km rather than meters). Active nodules were recovered from a corresponding active sediment push core (i.e., AN-3730N refers to a carbonate nodule found within the AS-3730 sediment core). Background sediment was collected from an off-seep site approximately 15 km east of Hydrate Ridge (Table 1, Figure 1). The sulfate-methane transition zone (SMTZ) at Hydrate Ridge active seep sites has been reported to occur within the top several cm of seafloor sediment, corresponding to peak values of microbial aggregate abundance, methane oxidation, and sulfate reduction (Boetius et al., 2000; Boetius and Suess, 2004). Methane concentrations within the most active seep sediments reach several mM, and have been measured and modeled at values up to 70 mM (Boetius and Suess, 2004) and 50 mM (Tryon et al., 2002), respectively.
On the seafloor, sediment was collected in push cores (35 cm long) deployed by the ROV Jason or DSV Alvin. Individual carbonate rocks were collected with the manipulator arm and placed inside Plexiglas compartments in a subdivided, insulated biobox with a lid to minimize water column or cross-sample contamination during recovery. During ascent to the surface, supersaturated methane may have degassed during depressurization, as was apparent from gas pockets in core tubes. Shipboard, push cores and carbonates within their respective plexiglass containers were immediately transferred to a 4°C walk-in cold room and processed within several hours. Samples intended for DAPI (4′,6-diamidino-2-phenylindole) counts and fluorescence in situ hybridization (FISH) were fixed in 2% paraformaldehyde and stored at 4°C overnight, while samples intended for DNA extraction were frozen at −80°C. The next day, the formaldehyde was washed from the sample with sterile 1× PBS buffer and then replaced with 100% ethanol and transferred to a −80°C freezer. In the case of carbonate rocks, interior portions (≥5 cm from an exposed surface) were isolated to ensure the analysis of endolithic communities; this was done by breaking the sample with an autoclaved ceramic mortar and pestle and removing the outer surface with a sterile razor blade. Similar mortar and pestle treatment of sediment and subsequent DAPI visualization confirmed that aggregate morphology is not an artifact of carbonate sample preparation.
Samples for X-ray diffraction analysis (XRD) were powdered with an autoclaved ceramic mortar and pestle. The diffraction profiles were measured with a Phillips X'Pert Multi Purpose X-Ray Diffractometer housed in the Division of Materials Science at Caltech. SiO2 was used as an internal standard, and best-fit analyses (with peak-shifting permitted) were conducted with the X'Pert HighScore software and its library of diffractograms.
The analysis of XRD data focused on four components: quartz (SiO2), calcite (CaCO3), dolomite [CaMg(CO3)2], and aragonite (CaCO3). These minerals are the primary constituents of seep-associated carbonates (Greinert et al., 2001). The following peaks were used as diagnostic markers following peak-shifting: calcite (104) 2θ = 30.0°, dolomite (104) 2θ = 31.2°, aragonite (221) 2θ = 46.0°, and quartz (011) 2θ = 27.0° (Kontoyannis and Vagenas, 2000; Zhang et al., 2010). Each of these peaks is the most prominent for its respective mineral type, allowing for qualitative compositional characterization by relative peak heights (Tennant and Berger, 1957; Bergmann, 2013). Minor constituents not accounted for by the four components described above cannot be ruled out, but these four peaks, as well as the additional peaks associated with each mineral, account for the majority of XRD features in all spectra.
Formaldehyde/ethanol fixed samples of carbonate and sediments were prepared for microbial aggregate characterization and FISH as follows. Nodules and interior carbonate pieces were pulverized with an autoclaved porcelain mortar and pestle. To concentrate biomass away from mineral and sediment particles, a percoll density separation was performed on all samples following a modified protocol outlined in Orphan et al. (2001b). Specifically, 60 μl sample was mixed with 1290 μl TE (pH = 9.0) and heated for 3 min at 60°C to permeabilize cells. The sample tubes were placed on ice for 5 min; 4.5 μl of 30% H2O2 was then added (to deactivate native peroxidase for CARD-FISH) and incubated at room temperature for 10 min. Tubes were placed back on ice, and 150 μl 0.1 M sodium pyrophosphate was introduced. Sample mixtures were sonicated (Branson sonifier 150) on ice 3 times at 8 W (10 s each time) and overlaid on a percoll density gradient. The gradient tubes were then centrifuged at 4800 rpm for 15 min at 4°C (Allegra X-15R, Beckman Coulter, Indianapolis, IN). The percoll supernatant overlaying the sediment/carbonate pellet was removed and concentrated by vacuum filtration through both a 3 μm and a 0.22 μm white polycarbonate filter (Millipore). Filtered samples were immediately rinsed with 2 ml sterile 1× PBS and dehydrated with 2 ml of a 1:1 ethanol:PBS solution while on the filter tower. Dried filters were removed and stored in the dark at 4°C prior to analysis. Cell counts were performed on an epifluorescence microscope (Olympus BX51) under 60× magnification (Plan Apo N objective) using the general DNA stain DAPI (4′,6-diamidino-2-phenylindole). 25 fields of view were counted for each of the 10 seep-linked samples (Table 2).
The architecture of DAPI-stained aggregates was examined by acquiring a z-stack of epifluorescence images with a DeltaVision RT microscope and the associated Softworx program (Applied Precision, Inc., Issaquah, WA). Subsequent image processing was conducted with the DAIME image analysis and 3D visualization program (Daims et al., 2006). By recognizing fluorescently stained cells within manually delineated aggregate boundaries throughout a z-stack of images, DAIME is able to calculate biovolume and the pair correlation function. Biovolume is determined by dividing the 3D integrated volume of cells by the overall aggregate volume, and the peak pair correlation value corresponds to the most favored cell-cell distance (Daims et al., 2006). Biovolume and pair correlation values were obtained from five representative aggregates from each habitat; individual cell sizes were broadly consistent for all samples.
Relative microbial abundance within the 12 distinct samples was determined by calculating the cumulative aggregate volume per unit volume sample, dividing by a typical cell (1 μm diameter) volume, multiplying by the maximum possible spherical packing density (0.7405; Steinhaus, 1999), and scaling by the DAIME-determined biovolume aggregate factor. The abundance of single cells (recovered on the 0.22 μm filter) was then added to the aggregate value, and the sum of both aggregate-associated cells and single cells was divided by the largest cell abundance calculated for the dataset (sample AC-3439). Resulting values thus indicate the fraction of microbial cell abundance associated with each sample, relative to the sample with the highest cell count. This calculation did not distinguish between Archaea and Bacteria, and overall microbial abundance scaling factors (Table 2) were applied to both Domains' diversity charts (Figures 2, 3).
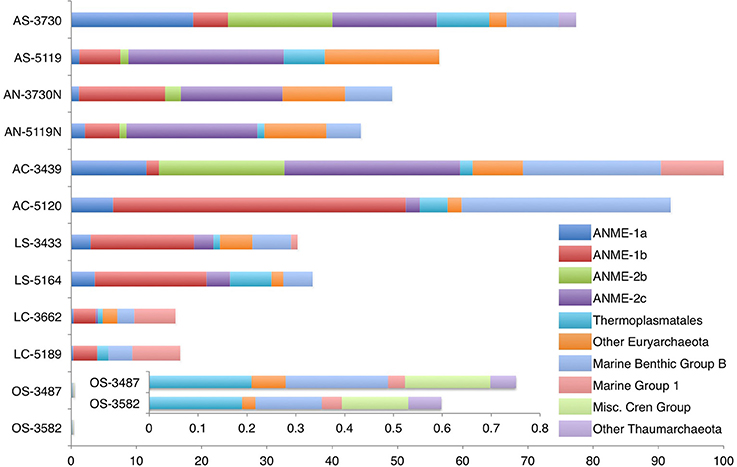
Figure 2. Distribution of archaeal 16S rRNA gene sequences recovered from carbonate, nodule and sediment samples. Phylogenetic groups that account for at least 20% of overall abundance in one or more samples are shown. In all cases, bar lengths are scaled by the microbial abundance observed in each sample, with AC-3439, which has the highest microbial abundance, as the longest bar (see Table 2). Each bar segment length represents the microbial abundance-weighted proportion of that sample's clones falling within the phylogenetic group in question. Inset: Off-seep samples OS-3487 and OS-3582 have biomass values orders of magnitude lower than those of seep-associated samples.
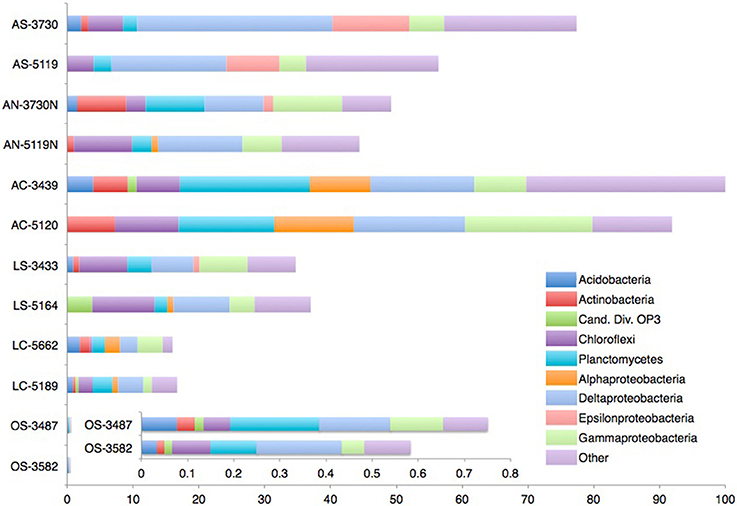
Figure 3. Distribution of bacterial 16S rRNA gene sequences recovered from carbonate, nodule and sediment samples. Phylogenetic categories representing phyla (or class, in the case of Proteobacteria) that account for at least 10% of overall abundance in one or more samples are shown. Bar lengths are scaled as in Figure 2.
To assess 16S rRNA gene diversity, the following workflow was performed for all 12 samples. DNA was extracted from ~0.5 g of sediment or pulverized nodule/carbonate material using the UltraClean Soil DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA). Bacterial and archaeal 16S rRNA genes were amplified in separate Polymerase Chain Reactions (PCR) with 27F (5′-AGAGTTTGATCCTGGCTCAG-3′)/1492R (5′-GGYTACCTTGTTACGACTT-3′) and Arc8F (5′-TCCGGTTGATCCTGCC-3′)/Arc958R (5′-YCCGGCGTTGAMTCCAATT-3′) primers, respectively (all primer concentrations were 0.4 μM, primers from Integrated DNA Technologies, Inc., Coralville, IA), New England BioLab's Taq DNA Polymerase (NEB, Ipswich, MA), 0.4 μM dNTP solution mix (NEB), and 1× ThermoPol Reaction Buffer (NEB). PCR was performed on an Eppendorf Mastercycler Ep Gradient S thermocycler with a 2-min 95°C initialization, followed by 35 cycles of a 30 s 94°C denaturation, 60 s 54°C annealing, and 90 s 72°C elongation. A final 7-min 72°C elongation completed the procedure, at which point the block was cooled to 4°C until samples were retrieved (<15 h). 16S rRNA gene amplicons were cleaned by filtration through a Millipore MultiScreen Filter Plate (Millipore Corp., Billerica, MA) and cloned using the TOPO TA Cloning Kit following the manufacturer's instructions (Invitrogen, Carlsbad, CA). For bacterial analysis, 288 transformants (colonies) were picked for each sample, an average of 233 of which contained appropriately sized inserts. For archaeal clone libraries, 192 colonies were picked for each analysis, and an average of 168 of these colonies contained the correctly sized insert. A random subset of clones in each library (Table S1) were selected and sequenced by Laragen, Inc. (Culver City, CA). Archaeal amplicons were sequenced in one direction (~900 bp), using the T3 primer (5′-ATTAACCCTCACTAAAGGGA-3′), while bacterial inserts were sequenced bi-directionally, using the T3 primer and the T7 primer (5′-TAATACGACTCACTATAGGG-3′). Vector sequence was removed and contigs (~1500 bp) were constructed (90% minimum match, 10 base minimum overlap) using forward and reverse sequences from the same clone; when one direction of sequence passed quality control inspection but the other did not, the low quality sequence was not used. Chimeric sequences were identified with the Slayer, Uchime, and Bellerophon programs; non-chimeric sequences were manually aligned in ARB (Ludwig et al., 2004) in reference to the Silva 111 NR98040812 database and used for subsequent phylogenetic analysis. The sequences were submitted to Genbank with the following accession numbers: KF616551-KF616600; KF616676-KF616751; KM356307-KM357226. See Table S1 for the number of sequences involved at each stage of analysis.
Non-metric multidimensional scaling (MDS) treatments and analysis of similarity (ANOSIM) were performed on these sequences in Primer 6.1.13 (Clarke and Primer, 2006) using Bray-Curtis similarity matrices that had been square-root transformed to prevent artificial over-emphasis of abundant taxa on community structure (Legendre and Legendre, 1998). Chao, Inverse Simpson, Bray-Curtis, and Weighted Unifrac indices were calculated from 97% sequence similarity OTUs in reference to the Silva 104 NR99100211 database using the Mothur platform (Schloss et al., 2009).
In this study, the microbial abundance and diversity of 12 samples representing six classes of methane seep habitats (Table 1) are assessed with the aim of establishing the influence of seepage activity and physical substrate type on the microbial community. Mineralogical examination, cell abundance calculations, and 16S rRNA gene archaeal and bacterial sequences, as well as statistical analyses of sequence diversity, reveal distinct patterns in, and raise intriguing questions about, the forcing of microbial community structure at methane seep-associated habitats.
XRD analysis of the 12 samples considered in this study supports four broad categories of mineralogical identification (Figure S1): sediments containing a dominant siliciclastic component identified as quartz (AS-3730, AS-5119, LS-3433, LS-5164, OS-3487, OS-3582); quartz, aragonite, and calcite mixtures (AN-3730N, AN-5119N, LC-5189); aragonitic carbonate rocks (AC-3439, LC-3662); and a dolomitic carbonate (AC-5120). Sediment with a low carbonate component was also reported by Orphan et al. (2004); the high proportion of quartz suggests a significant contribution from continental silicate weathering products, while other components including iron sulfide minerals are also likely present (Jørgensen et al., 2004; Van Dongen et al., 2007).
Changes in porewater chemistry with depth in the sediment column, particularly sulfate concentration, are hypothesized to influence mineralogy during precipitation (Burton, 1993; Savard et al., 1996; Naehr et al., 2007). Among carbonate precipitates, aragonite is the most thermodynamically favored pseudomorph in high-sulfate, high-alkalinity conditions above the SMTZ (Burton, 1993; Savard et al., 1996), and, in particular, at Hydrate Ridge seafloor environmental conditions (Greinert et al., 2001). Calcite is believed to form lower in the sediment column where sulfate is depleted (Naehr et al., 2007), and seep-associated dolomites have δ13C and δ18O values that have been interpreted as consistent with formation in deeper methanogenic sediment horizons (Naehr et al., 2007; Meister et al., 2011). Mineralogical differences may thereby record information about the depth and local porewater composition during precipitation.
Relative cell abundance calculations (Table 2) demonstrate that active carbonates, which contained more abundant and larger, but less densely packed cell aggregates, exhibited the highest cell counts of all sample types, with AC-3439 containing the highest cell abundance of the sample set. Active seep sediment samples contained 77% (AS-3730) and 56% (AS-5119) as many visible cells as the active carbonate AC-3439. Active carbonate nodule microbial abundance was roughly half that of active carbonates (49 and 44%, compared with 100 and 92% for active carbonate samples). Both nodule samples (AN-3730 and AN-5119) contained more (though less densely-packed) aggregates than the corresponding sediment horizons (AS-3730 and AS-5119) from which they were collected. Samples collected from areas of lower seepage activity had fewer microbial aggregates and larger numbers of single cells, resulting in relative cell abundance values of 35 and 37% for low-activity sediments (LS-3433 and LS-5164, respectively) and 16 and 17% for low-activity carbonates (LC-3662 and LC-5189, respectively). The microbial abundance of off-seep background sediments was approximately two orders of magnitude lower than active seep and low-activity samples and was comprised almost exclusively of single cells.
Archaeal and bacterial diversity in seafloor methane seep sediments, carbonate nodules, and carbonate slabs were characterized by 16S rRNA gene clone libraries. Supplementary Data files 1 and 2 provide the genus-level phylogenetic assignments of each sequence. Combining the relative abundance of 16S rRNA genes from archaeal and bacterial clone libraries with bulk microbial abundance data of unknown domain-level distribution (Table 2), abundance-scaled charts of phylogenetic distributions were generated (Figures 2, 3).
Euryarchaeota belonging to diverse ANME groups dominated the archaeal 16S rRNA gene sequences recovered from active seep sediments (82.4% of all archaeal clones; percentages compiled from pooled sequences of a given sample type, see Supplementary Data file 1c). Similarly, ANME sequences comprised 84.3% of the total archaeal diversity from active nodules; this proportion was 59.6% for archaeal sequences recovered from active carbonates and 71.1% for archaeal sequences from low-activity sediments. Archaeal clone libraries constructed from low-activity carbonates showed a smaller proportion of ANME sequences, representing 24.7% of all archaeal sequences, while ANME sequences were not recovered in off-seep background sediment clone libraries, which instead contained abundant representatives of the uncultured Thaumarchaeotal lineage Marine Benthic Group B (MBGB) (Vetriani et al., 1999).
Among ANME representatives, ANME-1—predominantly the ANME-1b subgroup (Teske et al., 2002)—were more abundant in low-activity samples (83.1% of all ANME sequences from low-activity sediments and carbonates; Supplementary Data file 1e) than samples associated with actively venting seeps (34.2% of pooled ANME sequences from active sediments, nodules, and carbonates). ANME-2—mostly the ANME-2c subgroup (Orphan et al., 2001a)—exhibited the opposite pattern, accounting for 58.3% of ANME sequences recovered from active samples and 13% from low-activity samples. The exception to this trend is the phylogenetic distribution of sequences recovered from the active dolomitic sample AC-5120, whose archaeal clone library contained 48.8% ANME-1b and 2.3% ANME-2c representatives. Members of the ANME-3 group were observed in most active and low-activity seep sediment samples and carbonate nodules (6.8% of total archaeal diversity on average among these samples), but were not recovered from any of the seafloor exposed carbonate rock samples or off-seep sediments (Supplementary Data file 1).
Members of the ANME lineages are the dominant Archaea linked to sulfate-coupled methane oxidation at marine methane seeps (Knittel and Boetius, 2009), and their environmental abundances often correlate with methane flux in sulfate-perfused, anoxic sediment near the seabed. When compared with 16S rRNA gene sequences recovered from active seep sediments, our data demonstrate that the carbonate rock substrate variable accounted for an approximate 25% decrease in ANME sequence abundance relative to active sediments. [To make this calculation, active carbonate archaeal sequences were pooled (n = 94 sequences), relative abundance was determined, and this value was compared with the analogous value derived from combined active seep sediment samples (n = 74 sequences); see Supplementary Data file 1c]. Low seepage activity accounted for a 14% decrease (pooled low-activity sediment sequences compared with combined active seep sediment sequences). When low-activity carbonate samples' sequences were pooled, a 70% decrease in ANME relative abundance (compared with active sediment samples) was observed. This finding suggests that the combination of factors relating to methane seepage activity (low-activity) and physical substrate (carbonate) negatively impacts the relative ANME abundance more than the sum of each factor individually. For example, in low-activity carbonate habitats, anaerobic archaeal methanotrophs may become energy or carbon limited as a result of self-entombment (e.g., Luff et al., 2004). Additionally, unfavorable geochemical conditions associated with decreased methane flux and/or the intrusion of oxygenated seawater after exposure on the seafloor may limit the successful persistence of these obligate anaerobes. Other archaeal groups—most notably marine group 1 (MG 1) Thaumarchaeota—were more prevalent in low-activity carbonate communities, which may be attributable to their colonization of a habitat less suitable for anaerobic methanotrophs and/or a diminished ability of MG 1 to inhabit the reducing active seep settings.
ANME-1 are believed to be better adapted to lower sulfate and/or methane conditions than their ANME-2 counterparts based on observations of niche differentiation in Japan Sea sediments (Yanagawa et al., 2011), Black Sea mats (Blumenberg et al., 2004), diffusion driven continental margin sediments (Harrison et al., 2009), and other environmental comparisons (Nauhaus et al., 2005; Rossel et al., 2011). Our findings corroborate this trend: ANME-1 representatives constituted 25.5% of archaeal sequences from active seep sites and 37.9% of combined archaeal sequences recovered from low-activity sediment and carbonate samples (Supplementary Data file 1d). It remains unclear whether all members of the ANME-1 are methanotrophs physiologically adapted for low-methane conditions, or facultative methanogens (Reitner et al., 2005; House et al., 2009; Lloyd et al., 2011), a metabolic plasticity that would confer a competitive advantage in zones of limited methane. The relative abundance of ANME-1 sequences recovered from AC-5120 is markedly higher than that of other active-site samples, an observation that may suggest a contributing role of mineralogy in community structure (Figure 2, see discussion below). ANME-3 representatives were not recovered in our carbonate rock diversity surveys. Whether this is due to their relative low abundance in the Hydrate Ridge ecosystem overall (~9% of archaeal sequences in active seep sediments) or if the apparent absence of ANME-3 in carbonates is indicative of habitat preference requires further investigation (Supplementary Data file 1). Members of the ANME-3 are typically reported as minor constituents from diverse seep sediment habitats and have only been reported as a dominant member from one site (Haakon Mosby mud volcano) to date (Lösekann et al., 2007).
Archaeal lineages not traditionally linked to methane oxidation also demonstrate activity-based trends. Euryarchaeotal Thermoplasmata were detected in 11 of the 12 samples, comprising a low percentage of the total archaeal sequences in active seep (5.2%) and low-activity samples (9.5%), and were observed at substantially higher proportions in off-seep background samples (31%; Supplementary Data file 1d). Sequences from the Halobacteria class, related to sequences from methane seep sediments (Harrison et al., 2009) or hydrothermal vents (Schauer et al., 2009), were recovered in low abundance (2.5% average of archaeal sequences, across all samples) from nearly all sample types with the exception of active carbonates.
The most prevalent thaumarchaeotal classes included MBGB and MG1. Sequences most closely related to the MBGB family, whose representatives are common in anoxic sediments (Vetriani et al., 1999), including those influenced by methane (Biddle et al., 2006), were abundant in clone libraries from all carbonates and low-activity and off-seep sediments (20.9% of pooled archaeal sequences recovered from these samples). The relative proportion of recovered MBGB sequences was lower in sediment and nodules from active seeps (8.9%). MG1 accounted for 39.5% (LC-3662) and 44% (LC-5189) of the total archaeal sequences of the two low-activity carbonate samples recovered from the seabed outside of active seepage areas; this proportion never exceeded 13.7% (AC-3439) in any of the other 10 samples.
The thaumarchaeotal class MG1 is pervasive in the deep-water column (Massana et al., 1997; Karner et al., 2001), and its high relative abundance in archaeal diversity surveys of the low-activity carbonates is likely reflective of seawater infiltration of these exposed carbonates at the seabed. Neither surface-exposed carbonates from active seep sites nor low-activity/off-seep sediments display the same MG1 abundance. This observation indicates that both low seepage activity (with low corresponding levels of methane and sulfide) and carbonate rock substrate appear to be necessary factors for MG1 prevalence, a finding that is consistent with MG1 acting as a passive colonizer of seawater-infused carbonates. In hydrothermal vent-associated basalt, MG1 representatives appear to be more abundant in weathered, off-vent samples (Lysnes et al., 2004) compared with initial communities in subseafloor hydrothermal environments (Huber et al., 2002). Channels of concentrated fluid flow developed by faulting and exploited by upward advection of methane-rich fluids during periods of seep activity may serve as conduits for downward, tidally-enhanced fluid delivery (Tryon et al., 2002) during subsequent periods of quiescence. Convection (Aloisi et al., 2004) and hydrologic recharge mechanisms at mound bases (Paull et al., 1991; Teichert et al., 2005) may also remain active. These processes may have preferentially seeded carbonate rocks from low-activity settings, where upward advection is less prominent, with MG1 representatives from bottom water. Alternatively, the presence of MG1 organisms may represent a secondary colonization resulting from carbonate exposure to bottom water following exhumation. The low-activity and off-seep sediment samples encompassing the 0–6 cmbsf horizon (LS-3433 and OS-3487) displayed minimal MG1 representation, suggesting that they may be outcompeted in the sediment habitat. MG1 representatives have been implicated in ammonium oxidation (Konneke et al., 2005; Nicol and Schleper, 2006), but the metabolic proclivities of carbonate-associated MG1 is not currently known.
Among bacterial sequences, Deltaproteobacteria was the most abundant class recovered from 8 of the 12 samples (AS-3730, AS-5119, AN-5119N, AC-3439, LS-5164, LC-5189, OS-3487, and OS-3582), and the second most abundant class detected in the remaining samples, with the exception of LS-3433 (Supplementary Data file 2c). Desulfobacteraceae and Desulfobulbaceae representatives were the most abundant deltaproteobacterial families recovered in active-seep samples (sediments, nodules, and carbonates), which likely reflects these taxa's role in AOM (e.g., Orphan et al., 2001a; Schreiber et al., 2010; Kleindienst et al., 2012; Green-Saxena et al., 2014). These families were abundant in bacterial diversity surveys from low-activity sediments as well (accounting for 75% of Deltaproteobacteria; Supplementary Data file 2d) but significantly less so in low-activity carbonates and off-seep sediments (15%), where members of the uncultured deltaproteobacterial groups SAR324 and SH765B-TzT-29 were more prevalent. Relatives from the SAR324 and SH765B-TzT-29 groups have been described from seafloor lavas (Santelli et al., 2008), South Atlantic Ocean sediment (Schauer et al., 2009), and river estuaries (Jiang et al., 2009). This geographic range suggests that these groups are widely distributed and have no particular dependence on methane geochemistry, though SAR324 methanotrophy has been proposed (Swan et al., 2011). Cultured representatives of these clades are lacking, but recent meta-omics approaches have begun to shed light on their physiological capabilities; members of the SAR324 clade associated with hydrothermal vent plumes, for example, have been implicated in several facultative metabolic modes, including sulfur and hydrocarbon oxidation (Sheik et al., 2014).
Epsilonproteobacteria most closely related to Sulfurovum sp. recovered from other methane seeps (Mills et al., 2005; Pernthaler et al., 2008; Beal et al., 2009) were prevalent in sediment samples from active seeps, but were detected at very low relative abundances in other active seep habitats including nodules and carbonate rocks. This observation potentially reflects a substrate-based control on diversity (Supplementary Data files 2b,c). Similar trends between seep sediments and their associated carbonate nodules were observed in an additional set of cores from Hydrate Ridge and Eel River Basin seeps (Mason and Orphan, personal observation). Sulfurovum from deep-sea hydrothermal vents are capable of multiple sulfur oxidation pathways (Yamamoto et al., 2010), and representatives of this genus have been described from methane seep sediments (Arakawa et al., 2006; Roalkvam et al., 2011) and methane-impacted terrestrial mud volcanoes (Green-Saxena et al., 2012). In this study, the near-exclusive presence of Sulfurovum in active sediments—even in horizons 6+ cm beneath the seafloor (AS-5119)—suggests that these environments contain dynamic sulfur cycles, with sulfide produced through sulfate-coupled AOM potentially being re-oxidized to sulfate by members of the Epsilonproteobacteria and Gammaproteobacteria.
Aerobic methanotrophic activity has been documented using radiotracer methods in sediments and carbonates from both actively seeping and low-activity locations at Hydrate Ridge (Marlow et al., 2014). Surprisingly, the majority of samples lacked 16S rRNA evidence of known aerobic methanotrophs with the exception of AC-5120, a dolomite sample collected from an active seep site, where sequences affiliated with gammaproteobacterial Methylococcales comprised 8% of recovered sequences (Supplementary Data file 2a). In contrast to the 16S rRNA findings, previous studies have reported the common occurrence of diverse particulate methane monooxygenase (pmo) genes related to gammaproteobacterial aerobic methanotrophs in surface sediments within seeps (Tavormina et al., 2008) as well as from the water column overlying seeps, at more than 50 times the abundance of sites over non-seep locations (Tavormina et al., 2010). Whether this general discrepancy between 16S rRNA and pmo findings is due to primer bias, low gammaproteobacterial methanotroph abundance, or the occurrence of as yet unidentified methane-oxidizing bacteria requires further investigation.
Verrucomicrobia—some of whose members have been implicated in aerobic methanotophic metabolism (Op den Camp et al., 2009)—were recovered only from active seep carbonates, comprising 7% of pooled active carbonate bacterial sequences (Supplementary Data file 2c). This sample-based specificity may suggest a dependence on methane or other geochemical components associated with active seeps. In addition, an indirect stimulation of heterotrophy (Freitas et al., 2012) associated with the high microbial abundances linked to active carbonates could sustain members of this phylum. The most closely related Verrucomicrobia sequences were reported from deep-sea sediment unaffiliated with methane seepage (Schauer et al., 2009), though some members of the phylum have been recovered from authigenic carbonate crusts linked to mud volcano AOM (Heijs et al., 2006).
Alpha (within sample) and beta (between sample) diversity statistics demonstrate how communities are structured and how they relate to each other (Table 3). Examining how environmental variables such as seep activity and substrate type map onto these relationships can help reveal factors that may play a role in determining microbial community structure. Such analysis may also help us understand the ecology of animal consumers, which exhibit trophic partitioning of microbial resources at Hydrate Ridge (Thurber et al., 2012; Levin et al., 2013).
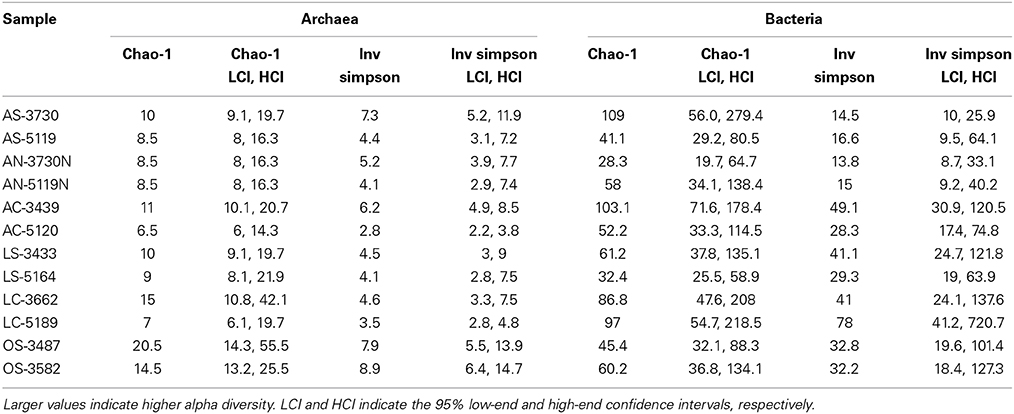
Table 3. Chao-1 and Inverse Simpson values for Archaeal and Bacterial clone libraries made from each of the 12 samples examined in this study.
Alpha diversity analyses, as computed by Chao-1 and Inverse Simpson values, provide comparative information capturing the two main components of community diversity: richness and evenness. Inverse Simpson values, which incorporate both parameters, reveal higher diversity in recovered bacterial sequences compared with archaeal sequences, a finding in agreement with Heijs et al.'s (2006) analysis of carbonate crust-associated microbial communities from Mediterranean mud volcanoes. A high degree of localized heterogeneity among Bacteria at the 97% sequence similarity OTU level was also observed, and archaeal diversity was substantially higher at off-seep locations compared with seep-associated sites, suggesting a larger range of niches available to Archaea in such environments (Table 3). There is no clear distinction in Chao-1 values between active and low-activity samples for archaeal and bacterial communities in sediment and carbonates, but bacterial communities in low-activity sediments and carbonates do exhibit higher Inverse Simpson values than their active-site counterparts. This finding suggests that bacterial communities in active seep habitats are dominated by lineages dependent upon seep-based physico-chemical conditions and/or groups associated with methanotrophic archaea. However, with longer residence time on the seafloor—during which alteration products or additional mineral phases can develop and the reducing, anoxic conditions related to active seep environments can dissipate—a broader range of methane-independent metabolic (or physical) niches may become available. A similar link between bacterial diversity and the degree of substrate alteration in weathered seafloor basalts was observed by Santelli et al. (2009). We observed no correspondence between sample-specific archaeal and bacterial alpha diversity metrics (linear best fit R2 values were 2.8 × 10−5 and 0.029 for Chao-1 and Inverse Simpson inter-domain comparisons, respectively), suggesting that the two Domains respond differently to physico-chemical and ecological drivers of diversity (Figure S2).
Beta diversity analyses were used to characterize community differences among different habitat types. After sequences were combined by either activity level or substrate type, the dependence of community structure on these environmental variables was evaluated. Through our assessment of seep activity, we established a scale from most methane seep activity (“active”) to least (“off-seep”), with “low-activity” occupying an intermediate position. If seep activity is an important control on diversity, active and low-activity communities would be more similar than active and off-seep communities as judged by beta diversity metrics. Similarly, off-seep and low-activity communities would be less divergent than off-seep and active communities. Equivalent expectations can be stated regarding substrate type, along the sliding scale of lithification, from least (“sediment”) to most consolidated (“carbonate”), with “nodules” as the intermediate group.
Beta diversity statistics were used in order to evaluate not only significant differences between communities, but also the relative magnitude of such differences (Figure 4). Bray-Curtis analysis (Bray and Curtis, 1957) quantifies the similarity between samples based on the relative abundances of constituent OTUs, while weighted UniFrac (Lozupone et al., 2007) statistics incorporate phylogenetic distances between constituent 16S rRNA gene sequences; both approaches can be used as distance measures (Faith et al., 1987; Lozupone et al., 2011). Our results indicate that off-seep and low-activity archaeal and bacterial communities are more similar than off-seep and active communities. Similarly, active and low-activity archaeal and bacterial communities are more similar than active and off-seep communities (Figures 4A,C). The wide range in archaeal beta diversity comparison values, particularly noticeable in Figure 4A, suggests that archaeal communities are more strongly shaped by seepage, and that even low apparent methane flux moves the community away from background off-seep composition. High lateral heterogeneity in geochemistry, microbial assemblages, as well as in rates of sulfate reduction and methane oxidation, have been reported on sub-meter scales within and adjacent to seep-associated sulfide-oxidizing microbial mats and chemosynthetic invertebrate communities, illustrating the importance of seepage activity as a dominant control on the sediment-hosted microbial community (e.g., Barry et al., 1996; Lloyd et al., 2010). When substrate type is examined, both beta diversity measurements demonstrate that sediment and nodule communities are more similar than sediment and carbonate communities (Figures 4B,D). This finding may be partially attributable to spatial proximity, as the sediment-hosted nodules were recovered directly from the active seep sediment cores analyzed in this study. Weighted UniFrac analysis indicates that bacterial communities in carbonates and nodules are more similar than those in carbonates and sediment. However, the reverse was observed for archaeal communities, suggesting that bacterial diversity may be more dependent on substrate type than seepage level.
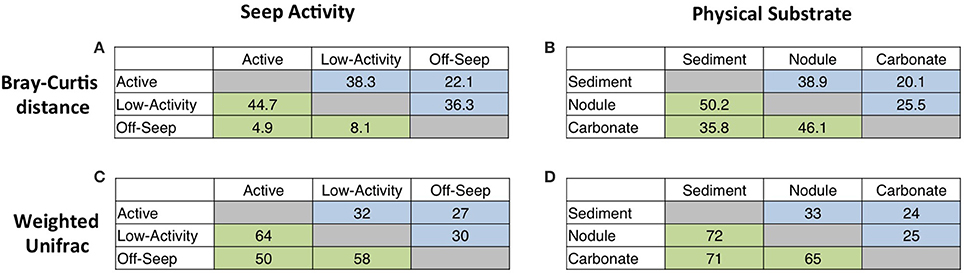
Figure 4. Results from beta diversity analyses of sequences binned by seep activity (A,C) or physical substrate (B,D). (A,B) provide Bray-Curtis similarities derived from comparison of proportional abundances of 97% sequence similarity OTUs. (C,D) Show weighted UniFrac-derived data for which all pairwise P-values are < 0.001. Green-shaded fields contain data from archaeal sequences; blue-shaded fields contain Bacteria-specific data. Weighted UniFrac values were subtracted from 1 and multiplied by 100 to provide a more intuitive statistic; in all cases, higher numbers indicate more similar communities.
Non-metric MDS assessments (Figure 5) visualize Bray-Curtis community similarities between the 12 samples. In the two-dimensional plot of archaeal data, communities cluster primarily by seepage activity (Figure 5A). (The dolomitic sample AC-5120 is the single exception to this framework, which may be related to its unique mineralogy, discussed below.) This observation corroborates the Bray-Curtis and weighted UniFrac data that point to seepage activity as the primary determinant of archaeal community structure in and around methane seeps; similar qualitative observations have been reported at a number of seep habitats (e.g., Lloyd et al., 2010). ANOSIM supports the finding that seepage activity is a significant determinant of archaeal community structure (global R = 0.65, p-value = 0.002) rather than physical substrate (e.g., sediment, nodule, carbonate; R = −0.04, p = 0.509; see Table S2a for all pairwise values).
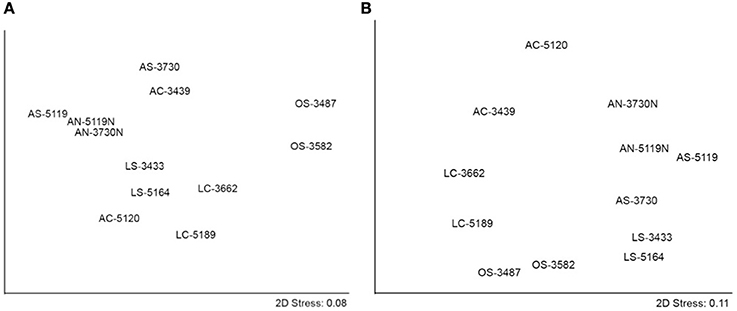
Figure 5. Non-metric MDS showing (A) archaeal and (B) bacterial communities derived from genus-level relative abundances.
The bacterial non-metric MDS plot (Figure 5B) suggests a primary dependence on substrate type: sediments are distributed in the lower/lower right portion of the coordinate system, sediment-hosted nodules are positioned to the upper right, and seafloor carbonates to the left. ANOSIM reveals that physical substrate is a significant driver of bacterial community structure (global R = 0.41, p = 0.021), while seep activity level is not (global R = 0.215, p = 0.094; Table S2b). The fact that substrate appears to discriminate among bacterial inhabitants is counter to an analogous study of seafloor basalts showing that the majority of clades are represented in both seafloor basalts and sediments (Mason et al., 2007). This discrepancy may be attributable to other environmental aspects distinctive of methane seep carbonates, such as fluid chemistry, mineralogy, predation pressure, permeability, or authigenic mineral formation (Heijs et al., 2006; Thurber et al., 2012). Indeed, activity appears to play a secondary role in separating bacterial communities, with active samples at the upper segment of the plot, off-seep samples to the bottom, and low-activity samples in between.
Mineralogy is another potential determinant of diversity. The geochemical association of sulfate-coupled AOM with carbonate minerals provides a direct link between microbial metabolism and authigenic mineral precipitation, a feedback that may, in turn, influence microbial community structure through physical constraints (Luff et al., 2004) and/or thermodynamic forcing (Knab et al., 2008). Specific links between diversity and carbonate mineralogy were difficult to discern with the relatively limited sample set included in this study. Aragonitic carbonates (AC-3439 and LC-3662) both contained abundant ANME sequences, an observation that is consistent with initial precipitation in zones of both high sulfate concentrations and high rates of AOM near the seabed (Burton, 1993; Savard et al., 1996; Greinert et al., 2001; Teichert et al., 2005). The lack of sulfate-reducing deltaproteobacteria in the calcitic LC-5189 is consistent with both a low-sulfate calcite formation environment (Naehr et al., 2007) as well as its current low methane seepage habitat, but it is difficult to parse the relative magnitudes of these influences. Sample AC-5120, a dolomite, may have formed in a deeper sediment horizon exhibiting low relative rates of AOM, as has been hypothesized on the basis of dolomite δ13C and δ18O isotopic signatures (Naehr et al., 2007; Meister et al., 2011). The divergence of this sample's archaeal community from those of other active site samples, characterized by the relative abundance of ANME-1 and lack of ANME-2 phylotypes, is consistent with formation in deeper sediment horizons (Knittel et al., 2005; Harrison et al., 2009), or may be uniquely influenced by the dolomitic mineralogy. In general, however, microbial community composition appears to be more dependent upon current environmental context rather than the original precipitation environment, as interpreted from mineralogical signatures. Indeed, when sample mineralogy is specifically plotted within the archaeal and bacterial MDS plots (Figure S3), the partitioning of communities is less clear than when only seepage activity or the degree of lithification (sediment, nodule, or carbonate, irrespective of mineralogy) is considered (Figure 5). The subsidiary importance of mineralogy in conveying microbial community relationships in our dataset suggests that this parameter is less significant than both methane seepage activity and lithification, which likely exert stronger influences over nutrient supply and metabolic intermediate exchange. The importance of mineralogy as a determinant of community composition has been documented at inactive hydrothermal vent deposits (Toner et al., 2012); with a more extensive dataset focused on mineralogy, it remains possible that this variable may help to explain additional microbial community differences unaccounted for by methane seepage activity or substrate type.
The analysis of mineralogy, microbial abundance, and archaeal and bacterial community structure across a range of habitats within and surrounding marine methane seeps reveals several key findings with implications for microbial ecology and seep development beyond what has been previously reported for marine sediment habitats.
Microbial community composition, as determined by 97% sequence similarity OTU-level 16S rRNA gene clone libraries, was remarkably similar among samples with similar seep activity levels and substrates—even if separated by several kilometers and acquired from different sediment depth horizons. This finding suggests that divergence owing to spatial heterogeneity, which has been reported in isotopic, geochemical, and community composition at the cm-scale in seep sediment (Orphan et al., 2004; House et al., 2009; Lloyd et al., 2010), is a less significant driver of bulk microbial diversity on its own than methane flux and/or substrate type.
The persistence of ANME Archaea and microbial aggregates typically associated with AOM in low-activity samples suggests that even at fluxes that do not support established sulfide-based chemosynthetic communities at the seabed (microbial mats or chemosynthetic clam beds), potential for anaerobic methanotrophy remains (Marlow et al., 2014). These findings imply that methanotrophic potential is pervasive on and below the seafloor across substrate types, and the degree to which ancient seep deposits preserve signals of microbial community succession is unknown. Studies of carbonate crusts from active mud volcanoes (Stadnitskaia et al., 2005) and of Black Sea AOM mats (Blumenberg et al., 2004) demonstrated an imperfect link between recovered lipids and 16S rRNA gene sequences, in which higher complexity lipid profiles suggested a more diverse microbial assemblage. In this context, lipid biomarkers, which are commonly used in geobiological studies to constrain communities or environmental conditions (e.g., Vestal and White, 1989; Turich and Freeman, 2011), may represent a time-integrated signal of microbial constituents and/or an accumulation of exogenous material rather than a faithful snapshot of a single community.
Statistical analyses of recovered 16S rRNA phylotypes reveal differences between sample types and point to distinct physico-chemical determinants of microbial diversity. Archaeal communities, dominated by anaerobic methanotrophs, sort strongly by seep activity; whereas bacterial communities show a preferential association with physical substrate type. This discrepancy suggests that methane influx, while influencing the distribution of anaerobic archaeal methanotrophs, has less of an effect on bacterial diversity and serves as an overlaid imprint on, not a sole arbiter of, community structure. Rather, the physical nature of the habitat—a combination of factors likely including permeability, mineralogy, and hydrology—plays a more significant role in determining bacterial assemblages around methane seeps. Several archaeal and bacterial lineages also appear to demonstrate specific activity-substrate pairing preferences, reflecting the interplay between seepage activity, environmental variation and succession, and microbial community structure. Our analysis exposes and begins to parse the intricate coupling of physical substrate and methane seepage as factors in environmental pressure and ecological determination; understanding the precise nature of these influences represents a fruitful area for continued research in microbial ecology.
Jeffrey J. Marlow, Victoria J. Orphan, Joshua A. Steele, and Lisa A. Levin designed the study; Jeffrey J. Marlow, David H. Case, and Joshua A. Steele performed the experiments and data analysis; Stephanie A. Connon processed much of the sequencing data; Jeffrey J. Marlow prepared the manuscript; all authors contributed to the editing of this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Captains, Crew, Alvin group, Jason group, and Science party members from RV Atlantis legs AT-15-68, and AT-18-10. Patricia Tavormina and Alexis Pasulka provided useful suggestions on the manuscript. This study was funded by grants from the National Science Foundation (OCE-0825791 and OCE-0939559 to Victoria J. Orphan; OCE-0826254 and OCE-0939557 to Lisa A. Levin), the National Aeronautics and Space Administration (NASA) Astrobiology Institute under NASA-Ames Cooperative Agreement NNA04CC06A (to Victoria J. Orphan). Jeffrey J. Marlow was partially supported by a National Energy Technology Laboratory Methane Hydrate Research Fellowship funded by the National Research Council of the National Academies.
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmars.2014.00044/abstract
Figure S1. X-ray diffraction data from the 12 samples analyzed in this study. Each panel shows an experimental spectrum and peak placements of the top database matches. Qualitative mineralogical assignments based on relative peak heights are provided below the sample identifier.
Figure S2. Bacterial Chao-1 (blue) and Inverse Simpson (red) values vs. archaeal Chao-1 and Inverse Simpson values. The wide scatter of both blue and red points suggests no significant correlation between domain-level diversity indices. R2 of the linear best fit is 2.8 × 10−5 for Chao-1 and 0.029 for Inverse Simpson.
Figure S3. Non-metric MDS results of (A) archaeal and (B) communities, where samples are labeled by mineralogical type.
Aloisi, G., Bouloubassi, I., Heijs, S., Pancost, R., Pierre, C., Damste, J., et al. (2002). CH4-consuming microorganisms and the formation of carbonate crusts at cold seeps. Earth Planet. Sci. Lett. 203, 195–203. doi: 10.1016/S0012-821X(02)00878-6
Aloisi, G., Wallmann, K., Haese, R., and Saliege, J. (2004). Chemical, biological and hydrological controls on the 14C content of cold seep carbonate crusts: numerical modeling and implications for convection at cold seeps. Chem. Geol. 213, 359–383. doi: 10.1016/j.chemgeo.2004.07.008
Arakawa, S., Sato, T., Sato, R., Zhang, J., Gamo, T., Tsunogai, U., et al. (2006). Molecular phylogenetic and chemical analyses of the microbial mats in deep-sea cold seep sediments at the northeastern Japan Sea. Extremophiles 10, 311–319. doi: 10.1007/s00792-005-0501-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bahr, A., Pape, T., Bohrmann, G., Mazzini, A., Haeckel, M., Reitz, A., et al. (2009). Authigenic carbonate precipitates from the NE Black Sea: a mineralogical, geochemical, and lipid biomarker study. Int. J. Earth Sci. 98, 677–695. doi: 10.1007/s00531-007-0264-1
Barry, J., Greene, H., Orange, D., Baxter, C., Robison, B., Kochevar, R., et al. (1996). Biologic and geologic characteristics of cold seeps in Monterey Bay, California. Deep Sea Res. Part I Oceanogr. Res. Pap. 43, 1739–1762. doi: 10.1016/S0967-0637(96)00075-1
Beal, E. J., House, C. H., and Orphan, V. J. (2009). Manganese- and iron-dependent marine methane oxidation. Science 325, 184–187. doi: 10.1126/science.1169984
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bergmann, K. D. (2013). Constraints on the Carbon Cycle and Climate During the Early Evolution of Animals. Ph.D. dissertation, California Institute of Technology, Pasadena, CA.
Bian, Y., Feng, D., Roberts, H., and Chen, D. (2013). Tracing the evolution of seep fluids from authigenic carbonates: Green Canyon, northern Gulf of Mexico. Mar. Petrol. Geol. 44, 71–81. doi: 10.1016/j.marpetgeo.2013.03.010
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K. G., Sorensen, K. B., Anderson, R., et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851. doi: 10.1073/pnas.0600035103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blumenberg, M., Seifert, R., Reitner, J., Pape, T., and Michaelis, W. (2004). Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. U.S.A. 101, 11111–11116. doi: 10.1073/pnas.0401188101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boetius, A., Ravenschlag, K., Schubert, C., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boetius, A., and Suess, E. (2004). Hydrate Ridge: a natural laboratory for the study of microbial life fueled by methane from near-surface gas hydrates. Chem. Geol. 205, 291–310. doi: 10.1016/j.chemgeo.2003.12.034
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Burton, E. A. (1993). Controls on marine carbonate cement mineralogy: review and reassessment. Chem. Geol. 105, 163–179. doi: 10.1016/0009-2541(93)90124-2
Clarke, K., and Primer, G. R. (2006). V6: User Manual/Tutorial. Plymouth: Primer-E Ltd. Plymouth–2006.
Claypool, G. E., and Kvenvolden, K. A. (1983). Methane and other hydrocarbon gases in marine sediment. Annu. Rev. Earth Planet. Sci. 11, 299. doi: 10.1146/annurev.ea.11.050183.001503
Daims, H., Lucker, S., and Wagner, M. (2006). Daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8, 200–213. doi: 10.1111/j.1462-2920.2005.00880.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Faith, D., Minchin, P., and Belbin, L. (1987). Compositional dissimilarity as a robust measure of ecological distance. Vegetatio 69, 57–68. doi: 10.1007/BF00038687
Freitas, S., Hatosy, S., Fuhrman, J., Huse, S., Welch, D., Sogin, M., et al. (2012). Global distribution and diversity of marine Verrucomicrobia. ISME J. 6, 1499–1505. doi: 10.1038/ismej.2012.3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gontharet, S., Stadnitskaia, A., Bouloubassi, I., Pierre, C., and Damste, J. (2009). Palaeo methane-seepage history traced by biomarker patterns in a carbonate crust, Nile deep-sea fan (Eastern Mediterranean Sea). Mar. Geol. 261, 105–113. doi: 10.1016/j.margeo.2008.11.006
Green-Saxena, A., Dekas, A. E., Dalleska, N. F., and Orphan, V. J. (2014). Nitrate-based niche differentiation by distinct sulfate-reducing bacteria involved in the anaerobic oxidation of methane. ISME J. 8, 150–163. doi: 10.1038/ismej.2013.147
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Green-Saxena, A., Feyzullayev, A., Hubert, C., Kallmeyer, J., Krueger, M., Sauer, P., et al. (2012). Active sulfur cycling by diverse mesophilic and thermophilic microorganisms in terrestrial mud volcanoes of Azerbaijan. Environ. Microbiol. 14, 3271–3286. doi: 10.1111/1462-2920.12015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Greinert, J., Bohrmann, G., and Suess, E. (2001). “Gas hydrate-associated carbonates and methane-venting at Hydrate Ridge: classification, distribution, and origin of authigenic lithologies,” in Natural Gas Hydrates: Occurrence, Distribution, and Detection, Geophysical Monograph Series, eds C. K. Paull and W. P. Dillon (Washington, DC: AGU), 99–113.
Hallam, S., Putnam, N., Preston, C., Detter, J., Rokhsar, D., Richardson, P., et al. (2004). Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462. doi: 10.1126/science.1100025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Harrison, B. K., Zhang, H., Berelson, W., and Orphan, V. J. (2009). Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California). Appl. Environ. Microbiol. 75, 1487–1499. doi: 10.1128/AEM.01812-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heijs, S. K., Aloisi, G., Bouloubassi, I., Pancost, R., Pierre, C., Damste, J., et al. (2006). Microbial community structure in three deep-sea carbonate crusts. Microb. Ecol. 52, 451–462. doi: 10.1007/s00248-006-9099-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hinrichs, K., Hayes, J., Sylva, S., Brewer, P., and DeLong, E. (1999). Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805. doi: 10.1038/19751
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoehler, T., Alperin, M., Albert, D., and Martens, C. (1994). Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Global Biogeochem. Cycles 8, 451–463. doi: 10.1029/94GB01800
Holler, T., Widdel, F., Knittel, K., Amann, R., Kellermann, M., Hinrichs, K., et al. (2011). Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 5, 1946–1956. doi: 10.1038/ismej.2011.77
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
House, C. H., Orphan, V. J., Turk, K. A., Thomas, B., Pernthaler, A., Vrentas, J., et al. (2009). Extensive carbon isotopic heterogeneity among methane seep microbiota. Environ. Microbiol. 11, 2207–2215. doi: 10.1111/j.1462-2920.2009.01934.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huber, J. A., Butterfield, D. A., and Baross, J. A. (2002). Temporal Changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68, 1585–1594. doi: 10.1128/AEM.68.4.1585-1594.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, L., Zheng, Y., Peng, X., Zhou, H., Zhang, C., Xiao, Z., et al. (2009). Vertical distribution and diversity of sulfate−reducing prokaryotes in the Pearl River estuarine sediments, Southern China. FEMS Microbiol. Ecol. 70, 249–262. doi: 10.1111/j.1574-6941.2009.00758.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jørgensen, B. B., Bottcher, M., Luschen, H., Neretin, L., and Volkov, I. (2004). Anaerobic methane oxidation and a deep H2S sink generate isotopically heavy sulfides in Black Sea sediments. Geochim. Cosmochim. Acta 68, 2095–2118. doi: 10.1016/j.gca.2003.07.017
Karner, M. B., DeLong, E., and Karl, D. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kleindienst, S., Ramette, A., Amann, R., and Knittel, K. (2012). Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ. Microbiol. 14, 2689–2710. doi: 10.1111/j.1462-2920.2012.02832.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Knab, N. J., Dale, A., Lettmann, K., Fossing, H., and Jorgensen, B. B. (2008). Thermodynamic and kinetic control on anaerobic oxidation of methane in marine sediments. Geochim. Cosmochim. Acta 72, 3746–3757. doi: 10.1016/j.gca.2008.05.039
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Knittel, K., Losekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479. doi: 10.1128/AEM.71.1.467-479.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Konneke, M., Bernhard, A., de la Torre, J., Walker, C., Waterbury, J., and Stahl, D. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kontoyannis, C. G., and Vagenas, N. V. (2000). Calcium carbonate phase analysis using XRD and FT-Raman spectroscopy. Analyst 125, 251–255. doi: 10.1039/a908609i
Legendre, P., and Legendre, L. (1998). “Numerical ecology: second English edition,” in Developments in Environmental Modelling (New York, NY: Elsevier Science), 20.
Levin, L. A. (2005). Ecology of cold seep sediments: Interactions of fauna with flow, chemistry, and microbes. Oceanogr. Mar. Biol. 43, 1–46. doi: 10.1201/9781420037449.ch1
Levin, L. A., Ziebis, W., Mendoza, G. F., Bertics, V. J., Washington, T., Gonzalez, J., et al. (2013). Ecological release and niche partitioning under stress: lessons from dorvilleid polychaetes in sulfidic sediments at methane seeps. Deep Sea Res. II 92, 214–233. doi: 10.1016/j.dsr2.2013.02.006
Lloyd, K. G., Albert, D., Biddle, J., Chanton, J., Piarro, O., and Teske, A. (2010). Spatial structure and activity of sedimentary microbial communities underlying a Beggiatoa spp. mat in a Gulf of Mexico hydrocarbon seep. PLoS ONE 5:e8738. doi: 10.1371/journal.pone.0008738
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lloyd, K. G., Alperin, M., and Teske, A. (2011). Environmental evidence for net methane production and oxidation in putative ANaerobic MEthanotrophic (ANME) archaea. Environ. Microbiol. 13, 2548–2564. doi: 10.1111/j.1462-2920.2011.02526.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lösekann, T., Knittel, K., Nadalig, T., Fuchs, B., Niemann, H., Boetius, A., et al. (2007). Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 73, 3348–3362. doi: 10.1128/AEM.00016-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lozupone, C., Hamady, M., Kelley, S., and Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lozupone, C., Lladser, M., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169. doi: 10.1038/ismej.2010.133
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Buchner, A., et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luff, R., Wallmann, K., and Aloisi, G. (2004). Numerical modeling of carbonate crust formation at cold vent sites: significance for fluid and methane budgets and chemosynthetic biological communities. Earth Planet. Sci. Lett. 221, 337–353. doi: 10.1016/S0012-821X(04)00107-4
Lysnes, K., Thorseth, I., Steinsbu, B., Ovreas, L., Torsvik, T., and Pedersen, R. (2004). Microbial community diversity in seafloor basalt from the Arctic spreading ridges. FEMS Microbiol. Ecol. 50, 213–230. doi: 10.1016/j.femsec.2004.06.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marlow, J. J., Steele, J. A., Ziebis, W., Thurber, A. R., Levin, L. A., and Orphan, V. J. (2014). Carbonate-hosted methanotrophy represents an unrecognized methane sink in the deep sea. Nat. Commun. 5:5094. doi: 10.1038/ncomms6094
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mason, O. U., Stingl, U., Wilhelm, L., Moeseneder, M., Di Meo-Savoie, C., Fisk, M., et al. (2007). The phylogeny of endolithic microbes associated with marine basalts. Environ. Microbiol. 9, 2539–2550. doi: 10.1111/j.1462-2920.2007.01372.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Massana, R., Murray, A., Preston, C., and DeLong, E. (1997). Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63, 50–56.
Meister, P., Gutjahr, M., Frank, M., Bernasconi, S., Vasconcelos, C., and McKenzie, J. (2011). Dolomite formation within the methanogenic zone induced by tectonically driven fluids in the Peru accretionary prism. Geology 39, 563–566. doi: 10.1130/G31810.1
Meyerdierks, A., Kube, M., Lombardot, T., Knittel, K., Bauer, M., Glockner, F., et al. (2005). Insights into the genomes of archaea mediating the anaerobic oxidation of methane. Environ. Microbiol. 7, 1937–1951. doi: 10.1111/j.1462-2920.2005.00844.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mills, H. J., Martinez, R. J., Story, S., and Sobecky, P. (2005). Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA-and RNA-derived clone libraries. Appl. Environ. Microbiol. 71, 3235–3247. doi: 10.1128/AEM.71.6.3235-3247.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Milucka, J., Ferdelman, T., Polerecky, L., Franzke, D., Wegener, G., Schmid, M., et al. (2012). Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 491, 541–546. doi: 10.1038/nature11656
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naehr, T. H., Eichhubl, P., Orphan, V., Hovland, M., Paull, C., Ussler, W., et al. (2007). Authigenic carbonate formation at hydrocarbon seeps in continental margin sediments: a comparative study. Deep Sea Res. Part II Top. Stud. Oceanogr. 54, 1268–1291. doi: 10.1016/j.dsr2.2007.04.010
Nauhaus, K., Boetius, A., Kruger, M., and Widdel, F. (2002). In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4, 296–305. doi: 10.1046/j.1462-2920.2002.00299.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nauhaus, K., Treude, T., Boetius, A., and Kruger, M. (2005). Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 7, 98–106. doi: 10.1111/j.1462-2920.2004.00669.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nicol, G. W., and Schleper, C. (2006). Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol. 14, 207–212. doi: 10.1016/j.tim.2006.03.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Niemann, H., Linke, P., Knittel, K., MacPherson, E., Boetius, A., Bruckmann, W., et al. (2013). Methane-carbon flow into the benthic food web at cold seeps–a case study from the Costa Rica subduction zone. PLoS ONE 8:e74894. doi: 10.1371/journal.pone.0074894
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Op den Camp, H. J. M., Islam, T., Stott, M., Harhangi, H., Hynes, A., Schouten, S., et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306. doi: 10.1111/j.1758-2229.2009.00022.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orcutt, B. N., Sylvan, J., Knab, N., and Edwards, K. (2011). Microbial Ecology of the Dark Ocean above, at, and below the Seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/MMBR.00039-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orphan, V. J., Hinrichs, K., Ussler, W., Paull, C., Taylor, L., Sylva, S., et al. (2001a). Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67, 1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D., and DeLong, E. F. (2001b). Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487. doi: 10.1126/science.1061338
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D., and DeLong, E. F. (2002). Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U.S.A. 99, 7663–7668. doi: 10.1073/pnas.072210299
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orphan, V. J., Ussler, W., Naehr, T. H., House, C. H., Hinrichs, K. U., and Paull, C. K. (2004). Geological, geochemical, and microbiological heterogeneity of the seafloor around methane vents in the Eel River Basin, offshore California. Chem. Geol. 205, 265–289. doi: 10.1016/j.chemgeo.2003.12.035
Paull, C., Chanton, J., Martens, C., Fullagar, P., Neumann, A., and Coston, J. (1991). Seawater circulation through the flank of the Florida Platform: evidence and implications. Mar. Geol. 102, 265–279. doi: 10.1016/0025-3227(91)90011-R
Pernthaler, A., Dekas, A., Brown, T., Goffredi, S., Embaye, T., and Orphan, V. (2008). Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc. Natl. Acad. Sci. U.S.A. 105, 7052–7057. doi: 10.1073/pnas.0711303105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reeburgh, W. S. (1983). Rates of biogeochemical processes in anoxic sediments. Annu. Rev. Earth Planet. Sci. 11, 269–298. doi: 10.1146/annurev.ea.11.050183.001413
Reeburgh, W. S. (2007). Oceanic methane biogeochemistry. Chem. Rev. 107, 486–513. doi: 10.1021/cr050362v
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reitner, J., Peckmann, J., Blumenberg, M., Michaelis, W., Reimer, A., and Thiel, V. (2005). Concretionary methane-seep carbonates and associated microbial communities in Black Sea sediments. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227, 18–30. doi: 10.1016/j.palaeo.2005.04.033
Roalkvam, I., Jorgensen, S., Chen, Y., Stokke, R., Dahle, H., Hocking, W., et al. (2011). New insight into stratification of anaerobic methanotrophs in cold seep sediments. FEMS Microbiol. Ecol. 78, 233–243. doi: 10.1111/j.1574-6941.2011.01153.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rossel, P. E., Elvert, M., Ramette, A., Boetius, A., and Hinrichs, K. U. (2011). Factors controlling the distribution of anaerobic methanotrophic communities in marine environments: evidence from intact polar membrane lipids. Geochim. Cosmochim. Acta 75, 164–184. doi: 10.1016/j.gca.2010.09.031
Ryan, W. B., Carbotte, S. M., Coplan, J. O., O'Hara, S., Melkonian, A., Arko, R., et al. (2009). Global multi-resolution topography synthesis. Geochem. Geophys. Geosyst. 10:Q03014. doi: 10.1029/2008GC002332
Santelli, C., Edgcomb, V., Bach, W., and Edwards, K. (2009). The diversity and abundance of bacteria inhabiting seafloor lavas positively correlate with rock alteration. Environ. Microbiol. 11, 86–98. doi: 10.1111/j.1462-2920.2008.01743.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santelli, C., Orcutt, B., Banning, E., Bach, W., Moyer, C., Sogin, M., et al. (2008). Abundance and diversity of microbial life in ocean crust. Nature 453, 653–656. doi: 10.1038/nature06899
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Savard, M., Beauchamp, B., and Veizer, J. (1996). Significance of aragonite cements around Cretaceous marine methane seeps. J. Sediment. Res. 66, 430–438.
Schauer, R., Bienhold, C., Ramette, A., and Harder, J. (2009). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 4, 159–170. doi: 10.1038/ismej.2009.106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schloss, P., Westcott, S., Ryabin, T., Hall, J., Hartmann, M., Hollister, E., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schreiber, L., Holler, T., Knittel, K., Meyerdierks, A., and Amann, R. (2010). Identification of the dominant sulfate−reducing bacterial partner of anaerobic methanotrophs of the ANME−2 clade. Environ. Microbiol. 12, 2327–2340. doi: 10.1111/j.1462-2920.2010.02275.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sheik, C., Jain, S., and Dick, G. (2014). Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ. Microbiol. 16, 304–317. doi: 10.1111/1462-2920.12165
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stadnitskaia, A., Bouloubassi, I., Elvert, M., Hinrichs, K., and Damste, J. (2008). Extended hydroxyarchaeol, a novel lipid biomarker for anaerobic methanotrophy in cold seepage habitats. Org. Geochem. 39, 1007–1014. doi: 10.1016/j.orggeochem.2008.04.019
Stadnitskaia, A., Muyzer, G., Abbas, B., Coolen, M., Hopmans, E., Baas, M., et al. (2005). Biomarker and 16S rDNA evidence for anaerobic oxidation of methane and related carbonate precipitation in deep-sea mud volcanoes of the Sorokin Trough, Black Sea. Mar. Geol. 217, 67–96. doi: 10.1016/j.margeo.2005.02.023
Suess, E., Torres, M., Bohrmann, G., Collier, R., Greinert, J., Linke, P., et al. (1999). Gas hydrate destabilization: enhanced dewatering, benthic material turnover and large methane plumes at the Cascadia convergent margin. Earth Planet. Sci. Lett. 170, 1–15. doi: 10.1016/S0012-821X(99)00092-8
Swan, B. K., Martinez-Garcia, M., Preston, C. M., Sczyrba, A., Woyke, T., Lamy, D., et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the Dark Ocean. Science 333, 1296–1300. doi: 10.1126/science.1203690
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tavormina, P., Ussler, W., Joye, S., Harrison, B., and Orphan, V. (2010). Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. ISME J. 4, 700–710. doi: 10.1038/ismej.2009.155
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tavormina, P., Ussler, W., and Orphan, V. (2008). Planktonic and sediment-associated aerobic methanotrophs in two seep systems along the North American margin. Appl. Environ. Microbiol. 74, 3985–3995. doi: 10.1128/AEM.00069-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Teichert, B., Bohrmann, G., and Suess, E. (2005). Chemoherms on Hydrate Ridge—Unique microbially-mediated carbonate build-ups growing into the water column. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227, 67–85. doi: 10.1016/j.palaeo.2005.04.029
Tennant, C., and Berger, R. (1957). X-ray determination of dolomite-calcite ratio of a carbonate rock. Am. Mineral. 42, 23–29.
Teske, A., Hinrichs, K., Edgcomb, V., de Vera Gomez, A., Kysela, D., Sylva, S., et al. (2002). Microbial diversity of hydrothermal sediments in the guaymas basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68, 1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thamdrup, B., and Dalsgaard, T. (2002). Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68, 1312–1318. doi: 10.1128/AEM.68.3.1312-1318.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thurber, A. R., Levin, L. A., Orphan, V. J., and Marlow, J. J. (2012). Archaea in metazoan diets: implications for food webs and biogeochemical cycling. ISME J. 6, 1602–1612. doi: 10.1038/ismej.2012.16
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Toner, B., Lesniewski, R., Marlow, J., Briscoe, L., Santelli, C., Bach, W., et al. (2012). Mineralogy drives bacterial biogeography of hydrothermally inactive seafloor sulfide deposits. Geomicrobiol. J. 30, 313–326. doi: 10.1080/01490451.2012.688925
Treude, T., Boetius, A., Knittel, K., Wallmann, K., and Jorgensen, B. (2003). Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar. Ecol. Prog. Ser. 264, 1–14. doi: 10.3354/meps264001
Treude, T., Orphan, V., Knittel, K., Gieseke, A., House, C., and Boetius, A. (2007). Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the Anoxic Black Sea. Appl. Environ. Microbiol. 73, 2271–2283. doi: 10.1128/AEM.02685-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tryon, M., Brown, K., and Torres, M. (2002). Fluid and chemical flux in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR, II: hydrological processes. Earth Planet. Sci. Lett. 201, 541–557. doi: 10.1016/S0012-821X(02)00732-X
Turich, C., and Freeman, K. H. (2011). Archaeal lipids record paleosalinity in hypersaline systems. Org. Geochem. 42, 1147–1157. doi: 10.1016/j.orggeochem.2011.06.002
Ussler, W., and Paull, C. K. (2008). Rates of anaerobic oxidation of methane and authigenic carbonate mineralization in methane-rich deep-sea sediments inferred from models and geochemical profiles. Earth Planet. Sci. Lett. 266, 271–287. doi: 10.1016/j.epsl.2007.10.056
Van Dongen, B. E., Roberts, A., Schouten, S., Jiang, W., Florindo, F., and Pancost, R. (2007). Formation of iron sulfide nodules during anaerobic oxidation of methane. Geochim. Cosmochim. Acta 71, 5155–5167. doi: 10.1016/j.gca.2007.08.019
Van Dover, C. L., Aharon, P., Bernhard, J., Caylor, E., Doerries, M., Flickinger, W., et al. (2003). Blake Ridge methane seeps: characterization of a soft-sediment, chemosynthetically based ecosystem. Deep Sea Res. Part I Oceanogr. Res. Pap. 50, 281–300. doi: 10.1016/S0967-0637(02)00162-0
Vestal, J. R., and White, D. C. (1989). Lipid analysis in microbial ecology. Bioscience 39, 535–541. doi: 10.2307/1310976
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vetriani, C., Jannasch, H., MacGregor, B., Stahl, D., and Reysenbach, A. (1999). Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65, 4375–4384.
Watanabe, Y., Nakai, S., Hiruta, A., Matsumoto, R., and Yoshido, K. (2008). U–Th dating of carbonate nodules from methane seeps off Joetsu, Eastern Margin of Japan Sea. Earth Planet. Sci. Lett. 272, 89–96. doi: 10.1016/j.epsl.2008.04.012
Yamamoto, M., Nakagawa, S., Shimamura, S., Takai, K., and Horikoshi, K. (2010). Molecular characterization of inorganic sulfur-compound metabolism in the deep-sea epsilonproteobacterium Sulfurovum sp. NBC37-1. Environ. Microbiol. 12, 1144–1153. doi: 10.1111/j.1462-2920.2010.02155.x
Yanagawa, K., Sunamura, M., Lever, M., Morono, Y., Hiruta, A., Ishizaki, O., et al. (2011). Niche separation of methanotrophic archaea (ANME-1 and -2) in methane-seep sediments of the Eastern Japan Sea Offshore Joetsu. Geomicrobiol. J. 28, 118–129. doi: 10.1080/01490451003709334
Keywords: methane seeps, anaerobic methane oxidation, microbial diversity, 16S rRNA gene, microbe-mineral interaction
Citation: Marlow JJ, Steele JA, Case DH, Connon SA, Levin LA and Orphan VJ (2014) Microbial abundance and diversity patterns associated with sediments and carbonates from the methane seep environments of Hydrate Ridge, OR. Front. Mar. Sci. 1:44. doi: 10.3389/fmars.2014.00044
Received: 04 July 2014; Accepted: 08 September 2014;
Published online: 28 October 2014.
Edited by:
Anne Bernhard, Connecticut College, USAReviewed by:
Lisa Moore, University of Southern Maine, USACopyright © 2014 Marlow, Steele, Case, Connon, Levin and Orphan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeffrey J. Marlow and Victoria J. Orphan, Division of Geological and Planetary Sciences, California Institute of Technology, MC 100-23, Pasadena, CA 91101, USA e-mail:amptYXJsb3dAY2FsdGVjaC5lZHU=;dm9ycGhhbkBncHMuY2FsdGVjaC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.