- 1Universidad Autónoma de Zacatecas, Unidad Académica de Ciencias Biológicas, Zacatecas, Zac, Mexico
- 2Universidad Juárez Del Estado de Durango, Facultad de Ciencias Químicas, Gómez Palacio, Dgo, Mexico
- 3Instituto Multidisciplinario de Ciencias AVICENA, Torreón, Coahuila, Mexico
- 4Universidad Autónoma de Nuevo León, Facultad de Ciencias Químicas, División de Estudios de Posgrado, Monterrey, NL, Mexico
Plasmids (circular DNA molecules) represent an ingenious strategy for horizontal gene transfer in prokaryotes and eukaryotic cells. Plasmids harbored in bacteria are responsible for the spread of traits such as antibiotic resistance, virulence factors, and the machinery for the horizontal gene transfer e.g., type IV secretion systems. Remarkably, Bacillus thuringiensis (Bt) cryptic plasmids encode and carry genes that, under the host environment, replicate and concomitate with sporulation, producing parasporal crystalline proteins of two major types, crystalline (Cry) and cytolytic (Cyt), the former toxic against different orders of insects such as Lepidopterans, Coleopterans, and Dipterans (Cry proteins, MW 50–130 KDa); Cyt proteins, produced by B. thuringiensis subspecies israelensis (Bti)(MW 27-kDa) are toxic against Dipterans, i.e., mosquitoes and black flies. The X-Ray tridimensional structure for both types of toxins, formed by three domains, mostly of beta sheets antiparallel (Domain II and Domain III) linked through loops of different lengths. Domain I is a bundle of alpha helices. This structure is characterized by five conserved blocks, implying a conservation in the mode of action. Cyt proteins possess two alpha helices and some beta sheets with a structure similar to the antimicrobial peptides. Indeed, the mode of action proposed is mediated by the toxin-lipid interaction that hypothetically could result in transmembrane ionic channel formation. Several pieces of evidence support the action of both toxins in insects and mammals. The question is to what extent these Bt/Bti plasmid-encoded Cry or Cyt genes can be applied as bioinsecticides individually or in combination with Lysinibacillus sphaericus. The feasibility of being considered a promising and safe biological strategy for crop pests and vector-borne neglected infectious diseases is an issue pinpointed in the present review.
1 Introduction
The genetic microbial strategy for DNA exchange is conjugation, a process shaping microbial communities (1, 2). The genetic process driving and leading to diversity is the horizontal transfer of mobile genetic elements or plasmids (3). This process allows the acquisition of exogenous DNA and recombinant bacteria with novel and potential properties or new phenotypes. It is paramount that Bacillus thuringiensis (Bt) has cry genes encoded in plasmids, and these genes are transferred to other bacteria of the same genus or different, expressed for the production of crystalline proteins (1, 4). Understanding the hidden lifestyle of the plasmid vectors in the genus Bacillus family (B cereus, B. thuringiensis, and B. anthracis) unveils the dynamic bacterial communication and genetic transfer of diverse traits such as antibiotic resistance, virulence factors, and secretory system components. The spreading resistance in the microbial communities of the genus Bacillus is genetic information that allows survival, diversity, adaptation, and evolution. Indeed, data from 5, support the fact that there is an active process of genetic exchange of plasmids (128 and 100 kb in size) (5–7) leading to the dissemination and spread of traits (3, 5, 6, 8). Therefore, the understanding and identification of all the extrachromosomal elements in the different strains of Bt might provide genetic tools for improvement in the control strategies against mosquitoes (9–11). At this point, pest vector management and control are of utmost importance for worldwide health as it is threatened by the chemical insecticides used to overcome them. It becomes an urgent need and paramount priority search for novel technological strategies to control pests in crops of agronomical and economic importance.
One of the most successful strategies to counteract it is the entomopathogen Bacillus thuringiensis. This Gram-positive soil bacteria discovered in the flour moth larvae in the province of Thüringia in 1911 (12–14) caused toxicity to different orders of insects. Despite the emergence of insect resistance populations to Bt and its products, they are used as bioinsecticides in proper formulations and efforts are being made to improve their range of action as well as their application (15–17). Another alternative that has been approached and developed with promising results is the transfer of plasmids encoding Crystalline (Cry) and the cytolitic (Cyt) proteins, which have been transformed with a cocktail of genes for a more diverse plant resistance response to insect attacks (18–20). But still, this strategy raised concerns about the endemicity of crops.
The most recent advances in the biological control of crop pests are the genetic and molecular techniques, in particular, the genetic engineering that has played a role in the design of biopesticides that can properly aid in vector control (mosquitoes) of disease transmission (e.g., dengue, malaria) (2, 9, 10, 21). By harnessing the synergy of the Bt products, Cry and Cyt toxins, as recently reviewed by Silva-Miranda et al. (11), are among the different strategies for the control of mosquitoes; the most feasible and safe is based on Bacillus thuringiensis subsp israelensis because the low persistence of the latter and the toxicity of both in a short period (11, 21). Gaining insight into this might represent a frontier to focus on.
2 Plasmids encode genes for horizontal gene transfer
Plasmids are small circular DNA in bacteria defined as mobile genetic elements that promote the spread of several traits and geographical distribution worldwide (22–24). Moreover, plasmids favor bacterial fitness adaptation and evolution. Plasmids favor horizontal gene transfer and gene recombination. Both genetic mechanisms allow bacteria to transfer genetic information to related and unrelated bacteria. The transfer of genetic determinants, i.e., carrying utmost antibiotic resistance genes and virulence factors (biofilm formation) proteins of the type IV secretion system (22–24), is present in Gram-positive and Gram-negative bacteria. The plasmids of the family Enterobacteriaceae comprise more than 200 different F-like plasmids. F-plasmids or F-factor, described in 1940, is a large 100 Kb circular conjugative plasmid of Escherichia coli described as a vector for horizontal gene transfer and recombination (Figure 1).
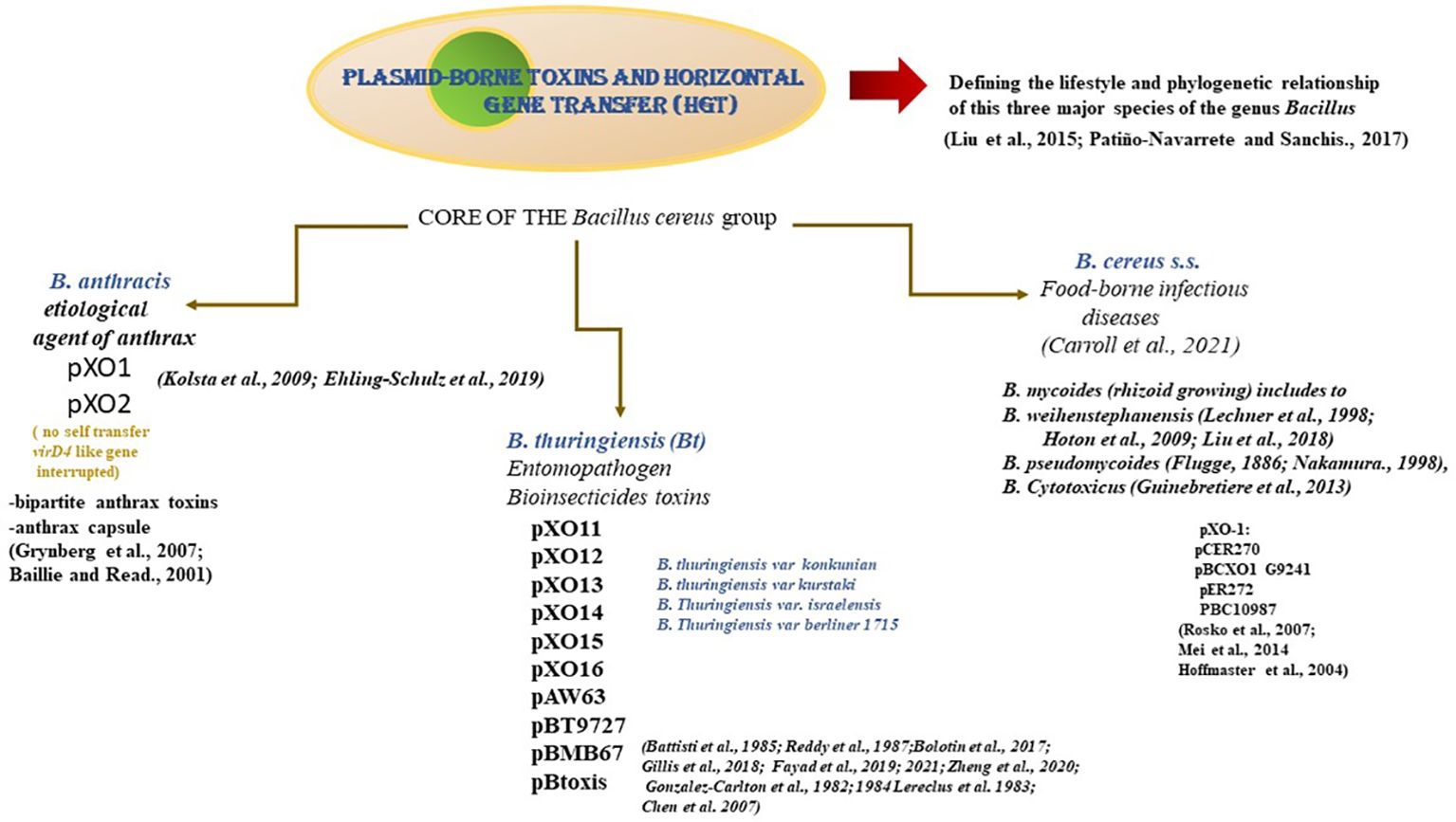
Figure 1. Plasmids as circular extrachromosomal DNA elements functioning as vectors of genetic information that can be transferred to other related bacteria and non- related bacteria. The size of plasmids can vary, but in general are usually large plasmids such as those of genus Bacillus, encoding different traits, such as antibiotic, heavy metal resistant genes, virulence factors, and other proteins. In addition, in these plasmids are encoded the non-structural and structural proteins that are necessary for the horizontal genetic transfer (HGT).
Since then, these F-plasmids and F-related F-like plasmids serve for bacterial conjugation purposes (25). Around 200 different F-like plasmids participate with highly related DNA transfer genes, including those of utmost relevance for the assemblage of a type IV secretion system. An outstanding feature of the F-plasmids in enterobacterial hosts is that they are isolated from clinical and environmental samples from different geographical regions. In the F-plasmid family (pOX38, ColB2, and R100–1), members recognize outer membrane proteins at the surface of the recipient cell (OmpA, OmpF, OmpK36, OmpW) (25–27) (Figures 2I-II). At this point, there was further investigation of defined mutations in the lipopolysaccharide (LPS) and the outer membrane protein OmpA of the recipient cell on mating-pair formation in vitro (liquid media) by the F-plasmid members. The transfer rate of these members is affected differentially by mutations in the rfa (LPS) locus (27, 28). Indeed, the F-plasmids showed increased sensitivity to mutations that affected rfaP gene expression, which encodes the addition of pyrophosphorylethanolamine (PPEA) to heptose I of the inner core of the LPS. Furthermore, ompA mutation significantly affected the mating efficiency of an F-plasmid carrying a mutation in the mating-pair stabilization protein TraN (25, 27, 28). During the transfer, adhesion present in the F-pilus tip could be playing a role in the specific recognition of LPS, OmPA, or the TraT (29–32) (Figures 2I-II).
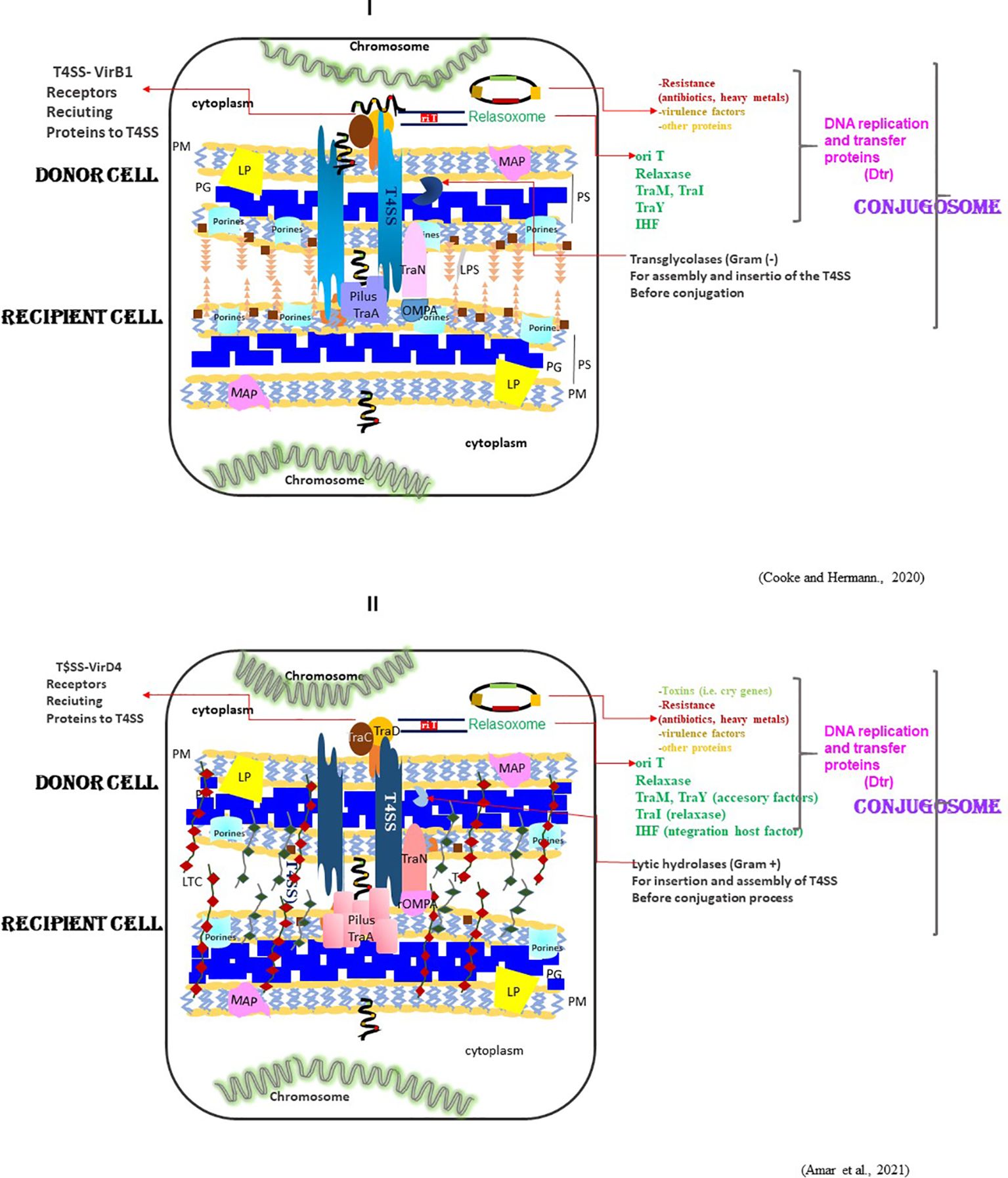
Figure 2. Conjugation as a mechanism. Horizontal gene transfer (HGT) is described initially for Gram-negative bacteria (I) and through evolution to the Gram-positive (II) and eukaryotic cells. Recently, this process was proposed as a kit tool for conjugation and, therefore, for gene transfer of encoded toxins, accessory proteins, antibiotics and enzymes. Different adhesins in Gram-negative bacteria recognize donor cells at the cellular surface membrane. Plasmids through this conjugosome are endowed with the capacity of HGT because these mobile genetic elements encode the proteins and the accessory factors for DNA replication (relaxase) and DNA transfer (VirB1, VirD4) as well as some non-structural proteins promote the conjugation process. The stabilization mating by TraN and OMPA in the recipient contribute significantly also to the transfer of ssDNA and conjugation efficiency.
On the other hand, Bacillus cereus (B. cereus) and its relatives harbor a plethora of plasmids, including conjugative plasmids, representing the heart of the group species differentiation and specification (2, 33). Over the years more than 20 plasmids from B. cereus have been found to be conjugative. The large plasmid can potentially “circulate” among members of the Bacillus cereus group (2). In contrast, XO16’s known natural distribution is limited to B. thuringiensis var. israelensis. At this point, it is noteworthy to pinpoint that Bacillus thuringiensis, a Gram-positive entomopathogenic soil microorganism, harbors large plasmids encoding the insecticidal crystalline proteins, with bioinsecticide and virulence factor properties (34, 35).
The plasmids replicate with bacteria, allowing Cry genes to express and produce the crystalline proteins. These extrachromosomal elements play a pivotal role in spreading along with antibiotic resistance and other traits, transferring these genes to another member of the genus Bacillus and even to other non-related bacteria (36, 37). These plasmids that carry genetic traits pivotal to bacterial survival adaptation and evolution participate in the horizontal gene transfer (HGT) (36, 38–40).
One of them is an integrative or conjugative plasmid with a similar structure to the modular backbone of the F-plasmids (37, 41). The conjugative plasmids transfer themselves between most bacteria, thus being one of the main causal agents of the spread of antibiotic resistance among pathogenic bacteria. How the plasmids accomplish this task is as follows. The modular structure or backbone of the integrative and conjugative plasmids (ICPs) consists of three modules:
1. - Module for maintenance
2. - Module for dissemination
3. - Module for regulation.
Moreover, additional module structures are possible through insertion sequences, transposons, and specific recombinases. Integrative and conjugative elements (ICEs) in the plasmid vectors transfer of the antibiotic resistance genes (40, 41), but now it is evident that ICEs can mediate the transfer of a very diverse set of functions. The advantages of ICEs are that they shape bacterial genomes, promote variability between strains of the same species, and distribute genes between unrelated bacterial genera using conserved integration sites (42, 43). ICEs allow bacteria to adapt to new environmental conditions and to colonize new niches (44).
Like phages and conjugative plasmids (45), ICEs can mediate the transfer of virulence determinants and may promote the mobilization of genomic islands (45).
The integrative and conjugative F-plasmids of t60 kbp can be placed into four major genetic modules:
1. - Module for plasmid replication (DNA replication and DNA-binding proteins)
2. - Module for stable maintenance (plasmid stability influenced by plasmid growth and horizontal transfer rates),
3. - Module for DNA transfer which occupies the most conserved part.
4. - Module for accessory cargo genes (within transposons or integrons).
The organization of the different functions in the modules could be related to the host-specific plasmid evolution that might explain the spread of clinical antibiotic-resistance plasmids. In referring specifically to the plasmid pXO16, which is a large conjugative plasmid from Bacillus thuringiensis var. israelensis (Bti), it has several properties (34, 42, 46, 47): self-transfer with high efficiency (48); -mobilize the small pUB110 plasmid from Bti-thermotolerant Bacillus cytotoxicus at high frequencies (3.3 × 10–3 and 5.2 × 10–4 transconjugants per donor (T/D), respectively (49, 50). -Retro-mobilize (the capture of DNA from a recipient by a donor cell) non-conjugative plasmids, including non-transfer plasmids (2, 4). -Promote their transfer as well as that of co-resident plasmids; transfer chromosomal loci; -displays a remarkable aggregation phenotype associated with conjugation under liquid conditions, [(Natural liquid foods (cow milk, soy milk, and rice milk)], where conjugation, mobilization, and retromobilization were shown to occur at frequencies of 8.0 x 10-1, 1.0 x 10-2 , and 1.2 x 10-4. Altogether these properties pinpoint the potential of this Bt plasmid in natural environments (1–4).
2.1 Mechanism for plasmid horizontal genetic transfer: the case of conjugation.
Bacterial communities establish communication through molecular crosstalk to share genetic information that influences their physiology and their lifestyle outside of their host (51–54) and is essential for them for survival, adaptation, and evolution. One genetic process by which bacteria can do this is through bacterial conjugation, a mechanism of mobile genetic elements (55–57) that uses the F-pilus that serves as a conduit for DNA transfer (26, 27, 29, 32, 40, 51, 56). DNA transfers through a conjugation mechanism into eukaryotic host cells. In this process, plasmids function as mobile genetic elements conformed in a conjugation module with the relaxosome complex targeting the type IV secretion system or to the sexual piles for conjugation or mating (41, 42, 46). Microorganisms respond and adapt to environmental conditions by acquiring genetic material in large amounts (58). Conjugation of plasmids contributes to this process, allowing lateral gene flow in prokaryotes, carried out through ICE self-transmissible mobile genetic elements that contribute to shaping genomes and lateral gene flow in prokaryotes through the conjugative process.
The ICEs of most temperate bacteriophages integrate into the genome. Like conjugative plasmids, they disseminate by conjugative transfer to new hosts (26, 27, 32, 55–57). The process of conjugation, as one of the mechanisms of HGT, consists of two principal steps: 1) DNA rolling circle relaxing accomplished by the passage of the plasmid through the complex called the relaxosome (59–61), and 2) interaction with the complex of VirB1 (Gram-negative) or the VirD4 (Gram-positive) to recluse with the type IV secretion system (T4SS) present in Gram-negative and Gram-positive bacteria (40, 52, 53, 58, 62–67) (Figures 2I-II).
2.1.1 Two hallmarks of the plasmid bacterial conjugation
2.1.1.1 What participates
-The type IV secretion systems (T4SSs) encoded in the F-plasmid, e.g., the pED208 encoded the T4SS (TrapRD/208). E. coli and other Gram-negative and –positive bacteria employ a type IV secretion system (T4SS) (62–64) to translocate DNA and protein substrates, generally by a contact-dependent mechanism, into other cells. In Enterobacteriaceae family (including E. coli), T4SS identified to date function exclusively in conjugative DNA transfer. In these species of bacteria, in the plasmid-encoded systems classified as P, F, and I types, Ancestral P-, F-, and I- systems adapted throughout evolution to yield the extant effector translocators, highlighting the adaptive and mosaic nature of these highly versatile machines (Figures 2I-II).
The T4SS has two main subfamilies: conjugation systems and effector translocators. The first ones are for inter-bacterial transfer of antibiotic resistance genes, virulence factors, and genes encoding other traits of potential benefit to the bacterial host. The effector translocators used by many Gram-negative pathogens for delivery of potentially hundreds of virulence proteins termed effectors to eukaryotic cells during infection (62–64) (Figures 2I-II).
-The non-structural genes are closely linked. These genes are not part of the T4SSs for conjugative transfer, such as the membrane pore, the relaxosome, and replication machinery. Moreover, the non-structural genes assist in core conjugative functions and mitigate the cellular burden on the host. During conjugation, they modulate dormancy, transfer, and establish a commensal relationship with the host, allowing manipulation of the host for efficient T4SS ensemble and assist in conjugative evasion of recipient cell immune functions. These genes take in a broad ecological context and ensure proper propagation of the conjugation system in a natural environment. Proteins with functions associated with plasmid maintenance, e.g. PsiB and SSB, suppress the mating-induced SOS response. This property establishes a novel biological function for conjugative protein translocation and suggests the potential for diverse outcomes from interbacterial protein translocation, influencing bacterial communication, physiology, and evolution. Conjugative protein translocation machinery through the TrapRD/208 requires the engagement of the pED208 relaxosome with the TraD substrate receptor or coupling proteins for activating the signal for protein translocation (Figures 2I-II).
2.1.1.2 What is needed for the transfer
a) Transfer of mobile genetic elements (MGES) and plasmids together with their cargoes of antibiotic resistance and virulence genes. T4SSs translocate MGEs as single-stranded DNA intermediates (T-strands), which triggers the SOS response in recipient cells.
b). Protein transfer (e.g., single-stranded DNA-binding proteins (SSB) such as ParA, ParB1, PaarB2, PsiB, and PsiA)(set of shared proteins) (59–61). The genesis or ssb encodes the SOS inhibitor protein PsiB and single-stranded DNA-binding protein SSB, eliciting a significantly stronger SOS response (59–61). Interestingly, translocation of PsiB or SSB, but not PsiA, through the TrapRD/208T4SS suppressed the mating–induced SOS response being triggered in recipient cells upon the acquisition of the single-stranded DNA transfer intermediate during mating.
This provides evidence of novel biological functions for conjugative protein translocation in mitigating the potentially negative consequences to plasmids and genome integrity resulting from SOS-induced recombination and mutation events. The set of known substrates of conjugation systems includes proteins with functions associated with plasmid maintenance. Of relevance is the fact that the first report of a conjugation–like event between strains of B. cereus sensu lato (s-l-) was 40 years ago. Many have studied the potential of plasmid transfer across the group, especially for plasmids encoding toxins, e.g. Bt Cry toxins. B. cereus s.l. (sensu lacto), in diverse environmental niches, mimics laboratory conditions to study the conjugation–related mechanism.
On referring to the conjugation system in Bt subsp. israelensis encoded on the large plasmid pXO16 (350 kb), the system is characterized by the formation of macroscopic aggregates (Agr+) in social exponential growth in liquid cultures. The recipients Agr- has been identified in the genus Bacillus, specifically those belonging to the Bacillus cereus group, such as Bacillus subtilis, Bacillus megaterium, Bacillus sphaericus, and 24 subspecies of B. thuringiensis (Bt) (1, 2, 4, 68). The transfer of the plasmid pXO16 to Bti Agr- strains (n= 14) was 100% effective and all recipients had acquired the aggregation-plasmid pXO16 and converted to the Agr+ phenotype. The genetic basis for this remarkable conjugative transfer system is not known, since no type IV secretion system homologs have been found. However in a more recent study it was reported a novel novel T4SS-mediated DNA transfer used by the Bti pXO16 plasmid. It was identified a 'transfer-Bti-plasmid' (tip) region encoding FtsK/SpOIIIE ATPase for an unrelated conjugative system T4SSs necessary for conjugative transfer, and distantly to the other Gram-positive bacteria. Furthermore, in the study it was observed up to 791 kb Bti chromosomal regions mobilization (2, 4, 68).
At this point, the question of what are the recipient genetic requirements for the conjugative transfer? Several studies have addressed this (69, 70). One of them using the IncI2 –(self transferable plasmids of the incompatibility group P-1 considered important carriers of genes for antibiotic resistance) plasmid TP114 which was recently shown to transfer at high rates in the gut microbiota (71, 72). In transfer experiments in vitro, 4000 single-genes deletion mutants of E. coli in a solid medium impaired transfer rates not associated with a specific cellular function. In contrast, broth medium were largely dependent on the lipopolysaccharide biosynthesis pathway. The specific structures used as recipient cells surface receptors by PilV adhesins associated with the type IVb accessory pilus F TP114 (71, 72). Moreover, using live-cell microscopy, while most transfer events occur between cells in direct contact, the F pilus serves as a conduit for the DNA during transfer between physically distant cells. Therefore, the F pilus function aids in understanding how is accomplished the dissemination of drug resistance and virulence genes within complex bacterial communities (60, 73–77).Thus, the genetic requirements for the recipients to participate in bacterial conjugation vary between different plasmids, among them: 1) the receptor molecules recognized by the conjugative pilus or other accessory pili involved in mating pair formation (MPF) (25, 78–80). 2) the cellular and molecular stabilization required at the surface of a recipient bacterium (25, 69, 70, 81). 3) the DNA replication (55, 57, 82), and 4) the Gene expression (71, 72, 83).
3 Plasmids encode genes for insect control and for infectious diseases
Large plasmids are present in the different strains of Bacilus thuringiensis (Bt). These extrachromosomal elements have the particular and outstanding feature of harboring the encoded genes. The product of these genes are proteins with endowed properties that allow for broad application as bioinsecticides, but also potential use as immunogens, carriers, or adjuvants (84–89). At the same time, insects are vectors of pathogens, such as viruses, fungi, and even other bacteria, and transmit to animals and plants.
In this way, plasmids become a fundamental vehicle for horizontal genetic transfer and play a role in spreading virulence factors (4, 68, 90). B. thuringiensis harbor these plasmids which express the delta-endotoxins that are toxic against a wide range of insect orders (17–20, 91, 92). Some outstanding features of the plasmids gene encoded Cry and Cyt proteins are the structure, composition and mode of action (12, 91–93). Remarkably these proteins are produced during the sporulation phase as inactive parasporal bodies (12, 91–93).
3.1 Structure and composition
The three-dimensional structure of the Cry proteins determined to 1.5 A of the resolution, highly conserved in structure and function (14, 92–95). 3D Cry toxins show a dual role, either as bioinsecticides for crop pests of agronomical importance endowed with the ability to colonize the insect world or as potential candidate adjuvants. Furthermore, and on referring to the Cyt toxins (cytolitic toxins), the three-dimensional X-ray structure of the Cyt2Aa1 was determined by Li et al., (96), comprised of two outer alpha helix hairpins (A-B and C-D)(rev (97, 98) and a core of mixed beta sheets (1 to 7). The structure of Cyt1Aa1 was similar. The alignment of amino acid sequences of six Cyt proteins from different B. thuringiensis subspecies revealed high scores and high statistical significance in the four blocks that comprise helix A, the loop after helix D plus beta strand four, the strands five and six, and finally the strand six-a and the following loop (94, 96, 97, 99–101).
The composition of the parasporal body of Bti differs from the pyramidal toxic to lepidopterans. The Bti parasporal body is spherical and contains four proteins, 27 (Cyt1A), 72 (Cry11A), 128, and 135 (Cry4A and Cry4B), packaged into three different inclusion types held together by a lamellar envelope (102, 103). Cry4A, Cry4B, and Cry11A are similar to the Cry1A-type proteins and toxic against lepidopterans (104). However, Cyt1As toxins (cytolytic) are different in sequence and structure (95, 99, 100, 103, 105–107).
3.1.1 Mechanism of action
The mode of action of the Bt Cry proteins has been proposed is a multistep process, starting with a) ingestion of mix spore+crystal by the insect(s) (12, 91–93); b) solubilized under the alkaline midgut insect conditions; c) activated after proteolytic digestion; d) Sequential binding to midgut receptors like-proteins; e) conformational change favouring pore formation; f) Osmotic swelling of the cell, and g) insect death (13, 15, 16, 20), In the case of the mode of action of the Cyt1A toxins is not well understood. However, structural studies have revealed that these toxins have an affinity for unsaturated fatty acids in the lipid portion of the microvillar membrane (21, 97, 98, 101, 108, 109). Cyt1A toxins insert into the microvillar membrane and then aggregate with each other in small clusters of lytic pores (21, 98, 108, 110). However, more recent evidence favors a membrane perturbing, detergent- like mode of action in which faults are created in the lipid bilayer of the membrane, disrupting and causing cell lysis and cell death (21, 96–101, 103, 107, 111). In vivo, the effects of the Cyt1A toxins, the soluble crystal delta-endotoxin proteins caused hemolysis in rats, mice, sheep, horse, and human erythrocytes (105, 110, 112). Furthermore, Cyt toxins can function as Cry receptors, a type of synergy favoring the biocidal action of the Cry proteins (113–115).
3.2 The Cyt and Cry proteins for the insect biological control
3.2.1 Pest control management
Crop pests (rodents, nematodes, mites, and insects), plant pathogens (bacteria, fungi, and viruses), and weeds impact the world’s agriculture and livestock and cause hundreds of millions of disease cases every year due to the transmission of pathogens and parasites (116–118). Moreover, there has been an increase in the undernourished population from 777 million in 2015 to 815 million in 2016 (119, 120). The risk of insufficient food for the global population is higher in subtropical and tropical geographical regions where there is no control of crop pests and vectors that transmit pathogens of humans (malaria, dengue, paludism, filariasis, and chagas) (117, 118, 121–124). This is an issue that demands urgent priority (125, 126).
Indeed, insect crop pests cause 10% to 30% loss of crop yields annually worldwide, and vector-borne pathogens have become a cause of emerging diseases of crop plants and of neglected infectious diseases (118, 123, 124). In the last decades, crop pest control has relied on pesticides, such as insecticides, fungicides, bactericides, and herbicides, which provide the mainstay of crop protection (119, 120). These chemical compounds affect animal health and humans (119, 120, 127, 128). In arthropods (insects), chemicals have an effect at the level of the nervous system by inhibiting acetylcholinesterase, voltage-gated sodium channels, and GABA receptors, and therefore interrupting synapse and immune regulation (127, 128).
Furthermore, the action of the pesticides is through targeting several metabolic, physiological and biochemical pathways, the nerve receptors of pests, and even microbial organisms (122, 129). While insecticides are part of and a strategy in integrated pest management, they represent barriers to effective biological control (130–133) and can increase the development of resistance to principal substance classes worldwide and in many different species.
Of relevance is the observation that HGT favour is the insect vectors’ ability to transmit between different organisms, like a spatial bridge, and thus increase the infectious opportunities. Furthermore, a pathogen can adapt to an insect vector transmission rate and pathogen dispersal (117, 134, 135). For instance, HGT might represent under this natural setting a drawback for the control of pathogen-vectored insects. At this point, combined strategies might aid in the decrease of the rate of transmission of crop pests and vector-borne diseases (21, 104, 110, 135–140) (Figure 3). The combined system strategies include the delivery of novel and specific chemical insecticides, the development of vaccines (which only exist for yellow fever), bacterial larvicides, and transgenic mosquitoes for reducing pathogen transmission (11, 105, 117, 141–144).
Figure 3. The mode of action of the B. thuringiensis subsp israelensis(Bti) is a complex of proteins, Cry4A, Cry11A Cyt1A, and Cyt2A, toxic against dipterans, mosquitoes, and flies. Cry and Cyt possess the structural information to act at the level of the cellular membrane of insect midgut to cause cell lysis and death but also to bind to proteins in the cellular membrane of the vertebrate (mammals), resulting in signalization of the pathways involved in the induction of immune responses, or pathways involved in cell death, caspases, or apoptosis programs.
The integrated planning might include insecticides and natural enemies (145, 146), combined tactics between selective insecticides and selective genetically modified crops or chemical tools and genetically modified (GM) crops (147) or broad-spectrum organophosphate with biological control conservation (148). The environmental impacts of chemical pesticides and resistance among pests have stimulated the development and utilization of microbial agents to improve human well-being and agricultural productivity (149).
Genetic engineering has proved to be one of the most promising technologies to improve crop resistance to disease and pests through the generation of lethal genes (OxitecO) to interrupt vector pathogen compatibility (150–152) (Figures 3, 4).
However, one alternative explored and applied is the release of sterile insects and insect recombinants. However, there is concern about the ecological threat and risks because nontargeted and indigenous species could harm and alter the wild and natural interaction host-pathogen (11, 116) (Figures 3, 4).
Another approach, is the theoretical models, which take into account all the factors that could have a role in the host-pathogen interaction (134, 139, 153–156). Modeling can be helpful to understand how the environmental conditions and abiotic and biotic factors could influence the underlying mechanism of how to limit the spread of the vectored insects. Several other potential strategies are mixed strategies such as chemical and biological control to optimize the control and management of pests (135, 157–162).
3.2.2 Bacillus thuringiensis subsp. Israelensis (Bti) and Lysinibacillus sphaericus (LBs) to control pest of agronomical important crops
The frontier in the knowledge of the pest crops and utmost of Neglected Infectious Diseases is to unravel the biology of the interaction host-pathogen. Basic knowledge research is pivotal to approach the control specifically of the mosquitoes (please refer to the review by Silva-Miranda et al., (11) for further details on the different strategies used until that year after the pandemic. Herein, we aimed to pinpoint some aspects of the control of mosquitoes.
Despite several alternatives for the control of dipterans, mosquitoes and flies, main pest crops, and Neglected diseases in developed and developed countries, the synergism between the toxins produced by B. thuringiensis subsp israelensis (Bti) and Lysinibacillus sphaericus remains as the most promising and safe strategy for the biological control of vectored dipterans. Among other reasons is that the combined toxicity and low persistence make these two entomopathogenic bacteria with a lower risk for insect resistance development (11, 21), which continues to be a hot spot in South American countries, e.,g. Brazil country is endemic to dengue and malaria vectorized by mosquitoes.
Several studies have shown that Bt is the most widely used biological insecticide (16–21, 92, 110, 139). Bt strains produce a variety of toxic proteins used in insecticidal formulations and transgenic crops against caterpillars, beetles, and flies (16, 18, 163). In particular, the specific toxicity of Bti against larval mosquitoes and black flies was discovered by Goldberg and Margalit in 1977 (164), leading to the development and registration for use in aquatic environments (164, 165).
Referring to the use of Bti in a proper dosage (166), several studies have revealed direct effects on non-target diptera, particularly chironomids (167–170), but also on other insect taxa such as lepidoptera (171) and coleoptera (172) Bti is recommended as a means of control. Beside the direct effects and their consequences on insect species (173), concerns have been raised about its persistence in the ecosystem, specially attributed to the spore stage (162, 174–176).
Bacillus thuringiensis subsp isralensis serotype H14 discovered in Israel (177) highly toxic at an LC50 of 13-20 ng/ml against the fourth instar of most Aedes, Ochlerotatus and Culex species (21, 103, 105, 106, 177–184) (Figure 3). Thus, Bt subsp Israelis extend its mosquito spectrum and overcome insect resistance (110, 111, 185–188). Indeed, when combined with Bacillus sphaericus (Bs) (189) (Lysinibacillus sphaericus) (21, 190, 191) at the same ratio, Cyt1A improved significantly the toxicity of Bs to Aedes. aegypti, a species considered insensitive to this bacterium (192–194) Lastly, selection studies have shown that resistance to Bti Cry proteins develops much more slowly when Cyt1A is present in the toxin mixture used for selection (Figure 3). The Cyt1A’s unique capacity to avoid, delay, or overcome resistance and extend its spectrum of activity is due to its unique mode of action, specifically its affinity for the lipid portion of the microvillar membrane (21, 98, 184, 190, 191). This property enables mosquitocidal endotoxins to bind to the midgut microvillar membrane independent of receptors (195–197). In a recent review related to the mechanisms of action and resistance of the Cyt proteins produced by Bti and Lysinibacillus sphaericus (21, 98, 184, 190, 191) highlights the relationship structure and function and the importance of using combined or a cocktail of toxins with different specificity that allow overcoming insect resistance (18, 21, 185, 186). The strain 2362 of Bs toxic against the fourth instar of Culex mosquitoes (196) produced the binary toxin two composed of two proteins, a 41.9 kDa toxin domain (BinA) and a 51.4 Kda binding domain (BinB) that co-crystallize into a single small parasporal body. Strain 2297 of Bs possesses a larger parasporal body and lower toxicity than strain 2362 (190, 197, 198). After ingestion of the inactive binary toxin by mosquito larvae, the 51.4 and 41.9 kDa proteins are cleaved by proteases yielding peptides of 43 and 39 kDa, respectively, that form the active toxin (196) (Figure 3). These associated toxins bind to receptors (e.g. alpha-glucosidase) (199) on the brush border insect midgut and cause cell lysis after internalization (200–202). Many strains of Lysinibacillus sphaericus produce other mosquitocidal toxins referred to as Mtx toxins. Two of these have been well-studied. Mtx (100 kDa) and Mtx (30.8 kDa), but they are not as toxic as the binary toxin (186, 190). Lysinibacillus sphaericus targeting dipterans (111, 179, 184, 186, 203–206) (Figure 3). The Mosquitoes (Diptera: Culicidae) of the Aedes species (Ae spp) are main vectors in tropical and subtropical of different serotypes of several viruses, such as dengue, the yellow fever virus (203, 207–211) the Zika virus (ZKV) (138, 212–215), and the virus Chikungunya (CHIKV) (157–161, 216–219). Therefore, mosquitoes, as vectored of pathogens, represent a threat because of their resistance and require frequent control (124, 133, 139, 159, 211, 220, 221). Neglected vector diseases cause morbidity and mortality (121, 124–126, 133, 137, 139) with an estimated 2 billion people worldwide living in areas where these are endemic (125, 211, 222, 223). One of the alternatives that have risen hopes to control insect vectors of pathogens causing human diseases (mosquitoes) is the formulations based on Cry-Cyt toxins, one of the most optimal and safe biological controls. Interestingly while Cry toxins are of low persistence (110, 111, 186), Cyt toxins have high persistence (21, 185). Therefore, a combination of these capabilities between Bti and LBs (138, 185, 188, 224–226), or a combination of Wolbachia and Leptolegnia chapmanii (facultative bacteria pathogen of diverse species of mosquitoes spp of the genus Aedes, Culex and Anopheles) can be highly effective to control to the insect vectored of mosquitoes (21, 223–225).
Interestingly, the potential for pest biological control of these insects depends on several factors, among them the levels of specificity to the different larvae stages of mosquito spp (227, 228). Interestingly, crop lines expressing active insecticidal cry genes from Bt have to achieve an efficient control of insect pests. Recently, identification of a mutation in the Bt toxin receptor recognizing Bt molecular patterns. Mutation genes of the toxin receptors lead to changes in the immune system response of the insect (129, 169, 170, 228–231) (Figures 2, 3), compromising the defensive systems of the mosquito (proteases for the peritrophic matrix) or exploiting receptors used by the virus to deliver toxins via the midgut to mosquito larvae Insight knowledge and understanding of the biology and the mechanism of interaction of insect pests and Bti, LBs, and their products (11, 18, 110, 231) represent a frontier for the advances in the development of improved or novel mosquito control strategies (Figures 3, 4).
3.3 Bt Cry proteins as carriers of immune dominant antigens of insect vectors pathogens causing human neglected infectious diseases
The mechanism of action for the bacterial toxins is dependent on receptors present in the epithelial cells, such as ganglioside (GM1) distributed in the human body, and the activation of G proteins and adenylate cyclase (232–239). Interestingly, in the case of the pCry1Ac and the 3D-Cry toxins, studies pointed out a specific interaction with brush border membranes in mice intestines (240).
More recent reports have found that pCry1Ac, as an antigen, interacts with the TLR present on the antigen-presenting cells, triggering signalization pathways such as mitogen-activated protein kinase (MAPK), specifically the extracellular signal-regulated kinase (ERK), a subclass of the MAPK pathway (240, 241), activated by a wide variety of receptors involved in growth and differentiation including receptor tyrosine kinases (RTKs), integrins, and ion channels.
Since MAPK activation is via ligand-receptor interactions, pCry1Ac-induced activation (as a ligand) of RaW264.7 macrophages leads to MAPK ERK1/2, p38, and JNK phosphorylation. However, using immunoprecipitation assays and MALDI-TOFF, pCry1Ac colocalizes with several binding proteins (as receptors), such as heat shock proteins (HSPs), vimentin, α-enolase, and actin. Flow cytometry and confocal microscopy cell-surface pCry1Ac-HSP70 co-localization suggests that the ligand-receptor interaction that activates MAPK and JNK phosphorylation signalization pathways is pCry1Ac-HSP70 (242, 243).
This leads to the regulation of targets in the cytosol and further translocation to the nucleus, where it can phosphorylate several transcription factors to regulate gene expression (241). In another setting in the mouse model, the immunogenic properties of pCry1Ac were administered by different routes, with significant induction of IgG antibodies (84, 85, 242). Furthermore, there was a structural implication of the N-terminal region in the induction of antibodies IgG and IgA after systemic and intranasal route immunization by 3D-Cry1A toxins (Cry1Aa, Cry1Ab, and Cry1Ac) (243). Moreover, pCry1Ac and the 3D- Cry toxins have been carriers of and properties of important clinical epitopes or antigens of clinical importance (diphtheria toxin, HIV) (82, 86–88).
Furthermore, pCry1Ac is a potential adjuvant when it is co-administered to mice with antigens of infectious diseases such as those caused by Naegleria fowleri (244), Plasmodium falciparum (245), Brucella melitensis (246), and cysticercosis (247), and it has demonstrated enhanced humoral and cellular immune response in BCG vaccinated Balb/c mice (248). In each case, pCry1Ac augments the magnitude of the secondary immune response (IgG subclass of antibodies, IgG1, IgG2a, IgG2b) and cellular immune response (Th1-, Th2-type cytokines, B and T cell differentiation) (86–89) (Figures 3, 4).
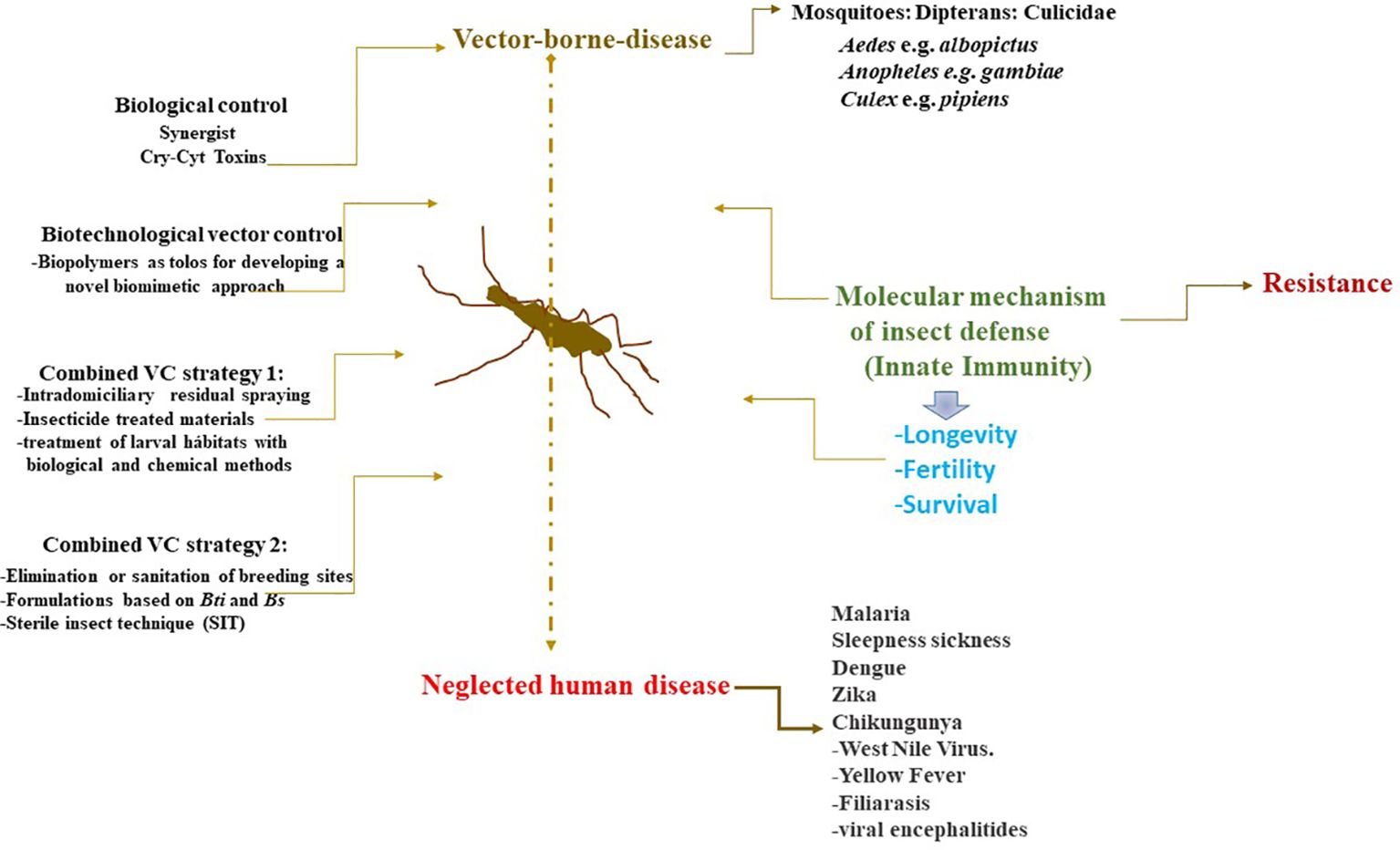
Figure 4. The control of mosquito vectors of pathogens that cause neglected infectious diseases (e.g., malaria, paludism, filiarasis) through integral control management (ICM) has had some success. Genetic Engineering and Molecular techniques such as gene driver, CRISP-Cas9, and Transgenesis have proved to be one of the most promising technologies to improve crop resistance to disease and pests for the generation of lethal genes (OxitecO) to interrupt vector pathogen compatibility. Furthermore, sterile technique of the insects and insect recombinants, alternatives explored and applied with potential future. Recent work on the toxicity and synergism of the toxins produced by Lysinibacillus sphaericus and B. thuringiensis subsp. israelensis supports the biotechnological development of both in proper formulation and dosage since there is a balance between toxicity, persistence in the environment, and avoiding insect resistance development compared to chemical insecticides and transgenic insect release.
4 Highlights and perspectives
It is clear that plasmids and the natural conjugation mating systems play a role in spreading virulence factors but also endow the bacteria with an ecological niche. To harness the biological control of crop pests and vectored insects of pathogens that affect animal health integrated tools to achieve meaningful results, unraveling and identifying the abiotic and biotic complex interaction factors among insect pests, humans, and entomopathogens remains a hope for the advancement and development of novel strategies to protect humans without affecting biodiversity and environmental health. The interaction of pests and pathogens favored host defenses by secreting virulence factors injected into their targets.
What about the limitations and challenges of the plasmid-encoded cry or cyt genes producing bioinsecticide proteins? The limitations are the insect response and the climatic change that plays a role. Another limitation and challenge is the development of resistance to the cry and cyt gene products. The challenge is biological control of the crop pests without affecting wild species. The effort is toward a proper and safe formulation and dosage of combined individual Cry or Cry + Cyt toxins. The challenge is to combine several strategies. The insect response to the Cry and Cyt toxins could provide targets to keep balance with the synergy between the Cry and Cyt proteins’ capabilities—the genetic engineering of crops. However, the fitness cost is too high for the ecosystem. An alternative is the biological control of mosquitoes using an endosymbiotic bacteria, Wolbachia to block virus entry. Thus, transgenic mosquitoes with this bacteria could potentially lead to a relatively biological control of mosquitoes. The challenge in either of these alternatives for biological control is the genetic diversity of the insects and the pathogen. The diversity could even lead to unexpected results. We expect no variation, even though the genetic background plays a role. In either case, the combinations of different Cry and different Cyt proteins against different insect orders, the genetics of the host, and the pathogen play a role. The cry or cyt genes can lead to a more feasible horizontal genetic transfer between insect orders. In addition, the remarkable properties of these bacterial toxins could perfectly match biotechnological and pharmacological issues.
The perspective is not just to use bioinsecticides for biological control but to harness the insect immune response (REPAT) for biomarker identification that allows further targeting. Indeed, it is evident that despite the recent advancements in molecular techniques such as driver gene, CRISPR-Cas9 (transgenic insects), and RNAi, the toxins from B. thuringiensis subsp israelensis and Lysinibacillus sphaericus, alone or in combination, are the most feasible to continue the development of a formulation that does not compromise the biodiversity and the health in the ecosystems.
Recent technology called biomimetic lure-and-kill exploits biomimetic principles of biocompatible/biodegradable biopolymers (e.g., natural hydrogel) to develop new substrates that selectively attract insects by reproducing specific natural environmental conditions (biomimetic lure) and kill them by hosting and delivering a natural biopesticide or through mechanical action.
Biomimetic lure-and-kill-designed substrates point to provide a new attractive system to develop/improve and make more cost-competitive new and conventional devices (e.,g. traps). The tiger mosquito Aedes albopictus has been proposed as a model to gain insight into this novel technology. Finally, the clinical perspective of the Cry and Cyt proteins in infectious disease as adjuvants of clinical antigens and in non-infectious disease (for example in cancer) is an issue that remains latent and to continue investigation of their structures allows them to interact with epithelial surfaces and with the host immune system.
Author contributions
GGG: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. JMFH: Formal analysis, Writing – review & editing. IBR: Formal analysis, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The Authors are grateful with SNI-CONAHCYT 2023-2027 and PERFIL-PRODEP 2022-2025 for financial fellowship support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Hinnekens P, Koné KM, Fayad N, Leprince A, Mahillon J. pXO16, the large conjugative plasmid from Bacillus thuringiensis serovar Israelensis displays an extended host spectrum. Plasmid. (2019) 102:46–50. doi: 10.1016/j.plasmid.2019.02.004
2. Hinnekens P, Fayad N, Gillis A, Mahillon J. Conjugation across Bacillus cereus and kin: A review. Front Microbiol. (2022) 13:1034440. doi: 10.3389/fmicb.2022.1034440
3. Geng P, Zhao P, Wan X, Mahillon J, Hu Y, Gong Y, et al. Interspecies Horizontal Transfer and Specific Integration of the Mosquitocidal Toxin-Encoding Plasmid pTAND672-2 from Bacillus thuringiensis subsp. Israelensis to Lysinibacillus sphaericus. Appl Environ Microbiol. (2023) 89:e0165222. doi: 10.1128/aem.01652-22
4. Jensen GB, Andrup L, Wilcks A, Smidt L, Poulsen OM. The aggregation-mediated conjugation system of Bacillus thuringiensis subsp. Israelensis: host range and kinetics of transfer. Curr Microbiol. (1996) 33:228–36. doi: 10.1007/s002849900105
5. Bolotin A, Gillis A, Sanchis V, Nielsen-LeRoux C, Mahillon J, Lereclus D, et al. Comparative genomics of extrachromosomal elements in Bacillus thuringiensis subsp. Israelensis Res Microbiol. (2017) 168:331–44. doi: 10.1016/j.resmic.2016.10.008
6. Bolotin A, Quinquis B, Roume H, Gohar M, Lereclus D, Sorokin A. Microb Genom. Long inverted repeats around the chromosome replication terminus in the model strain Bacillus thuringiensis serovar Israelensis BGSC 4Q7. Microbial Genome. (2020) 6:mgen000468. doi: 10.1099/mgen.0.000468
7. Makart L, Commans F, Gillis A, Mahillon J. Horizontal transfer of chromosomal markers mediated by the large conjugative plasmid pXO16 from Bacillus thuringiensis serovar Israelensis. Plasmid. (2017) 91:76–81. doi: 10.1016/j.plasmid.2017.04.001
8. Gillis A, Fayad N, Makart L, Bolotin A, Sorokin A, Kallassy M, et al. Role of plasmid plasticity and mobile genetic elements in the entomopathogen Bacillus thuringiensis serovar Israelensis. FEMS Microbiol Rev. (2018) 42:829–56. doi: 10.1093/femsre/fuy034
9. Davis J, Bibbs CS, Müller GC, Xue RD. Evaluation of Bacillus thuringiensis Israelensis as toxic sugar bait against adult Aedes aEgypti, Aedes albopictus, and Culex quinquefasciatus mosquitoes. J Vector Ecol. (2021) 46:30–3. doi: 10.52707/1081-1710-46.1.30
10. Harris AF, Sanchez Prats J, Nazario Maldonado N, Piovanetti Fiol C, García Pérez M, Ramírez-Vera P, et al. An evaluation of Bacillus thuringiensis Israelensis (AM65-52) treatment for the control of Aedes aEgypti using vehicle-mounted WALS® application in a densely populated urban area of Puerto Rico. Pest Manag Sci. (2021) 77:1981–9. doi: 10.1002/ps.6227
11. Silva-Miranda L, Rudd SR, Mena O, Hudspeth PE, Barboza CJE, Park HW, et al. The perpetual vector mosquito threat and its eco-friendly nemeses. Biol (Basel). 13:182. doi: 10.3390/biology13030182
12. Höfte H, Whiteley HR. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. (1989) 53:242–55. doi: 10.1128/mr.53.2.242-255.1989
13. Bravo A, Gill SS, Soberón M. Bacillus thuringiensis mechanisms and use. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science (2005), ISBN: 0-44-451516-X. p. 175–206. doi: 10.1016/B0-44-451924-6/00081-8
14. Ibrahim M, Griko N, Junker M, Bulla LA. Bacillus thuringiensis a genomic and proteomics perspective. Bioengineered Bugs. (2019) 1:31–50. doi: 10.4161/bbug.1.1.10519
15. Bravo A, Gill SS, Soberon M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. (2007) 49:423–35. doi: 10.1016/j.toxicon.2006.11.022
16. Bravo A, Likitvivatanavong S, Gill SS, Soberón M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem Mol Biol. (2011) 41:423–31. doi: 10.1016/j.ibmb.2011.02.006
17. Liu L, Li Z, Zhang X, Chou S-H, Wang J, He J. Which is stronger? A continuing battle between cry toxins and insects. Front Front Microbiol. (2021) 12:665101. doi: 10.3389/fmicb.2021.665101
18. Soberón M, Pardo-Lopez L, Lopez I, Gomez I, Tabashnik BE, Bravo A. Engineering modified Bt toxins to counter insect resistance. Science. (2007) 318:1640–2. doi: 10.1126/science.1146453
19. Pardo-Lopez L, Soberon M, Bravo A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev. (2013) 37:3–22. doi: 10.1111/j.1574-6976.2012.00341.x
20. Palma L, Muñoz D, Berry C, Murillo J, Caballero P. Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins (Basel). (2014) 6:3296–325. doi: 10.3390/toxins6123296
21. Silva-Filha MHNL, Romão TP, Rezende TMT, Carvalho KDS, Gouveia de Menezes HS, Alexandre do Nascimento N, et al. Bacterial toxins active against mosquitoes: mode of action and resistance. Toxins. (2021) 3:523. doi: 10.3390/toxins13080523
22. De la Fuente J, Rodriguez-Beltran J, San Millan A. Methods to study fitness and compensatory adaptation in plasmid-carrying bacteria. Methods Mol Biol. (2020) 2075:371–82. doi: 10.1007/978-1-4939-9877-7_26
23. Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother. (2018) 73:1121–37. doi: 10.1093/jac/dkx488
24. Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. (2013) 303:298–304. doi: 10.1016/j.ijmm.2013.02.001
25. Low WW, Wong JLC, Beltran LC, Seddon CH, David S, Kwong HS, et al. Mating pair stabilization mediates bacterial conjugation species specificity. Nat Microbiol. (2022) 7:1016–27. doi: 10.1038/s41564-022-01146-4
26. Klimke WA, Rypien CD, Klinger B, Kennedy RA, Rodriguez-Millard JM, Frost LS. The mating pair stabilization protein, TraN, of the F plasmid is an outer-membrane protein with two regions that are important for its function in conjugation. Microbiology. (2005) 151:3527–40. doi: 10.1099/mic.0.28025-
27. Anthony KG, Sherburne C, Sherburne R, Frost LS. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F- like plasmids. Mol Microbiol. (1994) 13:939–53. doi: 10.1111/j.1365-2958.1994.tb00486.x
28. Heinrichs DE, Yethon JA, Amor PA, Whitfield C. The assembly system for the outer core portion of R1- and R4-type lipopolysaccharides of Escherichia coli. J Biol Chem. (1998) 273:29497–505. doi: 10.1074/jbc.273.45.29497
29. Hu B, Khara P, Christie PJ. Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli. Proc Natl Acad Sci USA. (2019) 116:14222–7. doi: 10.1073/pnas.1904428116
30. Liu B, Furevi A, Pereplov AV, Guo X, Cao H, Wang Q, et al. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol Rev. (2020) 44:655–83. doi: 10.1093/femsre/fuz028
31. Allard N, Collette A, Paquette J, Rodrigue S, Côté JP. Systematic investigation of recipient cell genetic requirements reveals important surface receptors for conjugative transfer of IncI2 plasmids. Commun Biol. (2023) 6:1172. doi: 10.1038/s42003-023-05534-2
32. Frankel G, David S, Low WW, Seddon C, Wong JLC, Beis K. Plasmids pick a bacterial partner before committing to conjugation. Nucleic Acids Res. (2023) 51:8925–33. doi: 10.1093/nar/gkad678
33. Arnold BJ, Huang IT, Hanage WP. Horizontal gene transfer and adaptive evolution in bacteria. Nat Rev Microbiol. (2021) 20:206–18. doi: 10.1038/s41579-021-00650-4
34. Aronson AI, Beckman W. Transfer of chromosomal genes and plasmids in Bacillus thuringiensis. Appl Environ Microbiol. (1987) 53:1525–30. doi: 10.1128/aem.53.7.1525-1530.1987
35. Baillie L, Read TD. Bacillus anthracis, a bug with attitude! Curr Opin Microbiol. (2001) 4:78–81. doi: 10.1016/S1369-5274(00)00168-5
37. Benz F, Hall AR. Host-specific plasmid evolution explains the variable spread of clinical antibiotic-resistance plasmids. Proc Natl Acad Sci U S A. (2023) 120:e2212147120. doi: 10.1073/pnas.2212147120
38. Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. (2015) 16:472–82. doi: 10.1038/nrg3962
39. Mell JC, Redfield RJ. Natural competence and the evolution of DNA uptake specificity. J Bacteriol. (2014) 196:1471–83. doi: 10.1128/JB.01293-13
40. de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. (2010) 34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x
41. Virolle C, Goldlust K, Djermoun S, Bigot S, Lesterlin C. Plasmid transfer by conjugation in Gram-negative bacteria: from the cellular to the community level. Genes (Basel). (2020) 11:1239. doi: 10.3390/genes11111239
42. Ruhfel RE, Robillard NJ, Thorne CB. Interspecies transduction of plasmids among Bacillus anthracis, B. cereus, and B. thuringiensis. J Bacteriol. (1984) 157:708–11. doi: 10.1128/JB.157.3.708-711.1984
43. Smillie C, Pilar Garcillán-Barcia M, Francia MV, Rocha EPC, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. (2010) 4:434–52. doi: 10.1128/MMBR.00020-10
44. Shen Z, Tang CM, Liu GY. Towards a better understanding of antimicrobial resistance dissemination: what can be learnt from studying model conjugative plasmids? Mil Med Res. (2022) 9:1–11. doi: 10.1186/S40779-021-00362-Z/FIGURES/2
45. Sørensen SJ, Jensen LE. Transfer of plasmid RP4 in the spermosphere and rhizosphere of barley seedling. Antonie Van Leeuwenhoek. (1998) 73:69–77. doi: 10.1023/A:1000661115753
46. Santos CA, Vilas-Bôas GT, Lereclus D, Suzuki MT, Angelo EA, Arantes OMN. Conjugal transfer between Bacillus thuringiensis and Bacillus cereus strains is not directly correlated with growth of recipient strains. J invertebr Pathol. (2010) 105:171–5. doi: 10.1016/J.JIP.2010.06.014
47. Reddy A, Battisti L, Thorne CB. Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J Bacteriol. (1987) 169:5263–70. doi: 10.1128/jb.169.11.5263-5270.1987
48. Sorokin A. “Genetics and phages—from transduction and sequencing to recombineering”. In: Sansinenea E, editor. Bacillus Thuringiensis Biotechnology. Springer, Dordrecht (2012). p. 131–57. doi: 10.1007/978-94-007-3021-2_7
49. Thomas DJI, Morgan JAW, Whipps JM, Saunders JR. Plasmid transfer between the Bacillus thuringiensis subspecies kurstaki and tenebrionis in laboratory culture and soil and in lepidopteran and coleopteran larvae. Appl. Environ. Microbiol. (2000) 66:118–124. doi: 10.1128/AEM.66.1.118-124.2000
50. Thomas DJI, Morgan JAW, Whipps JM, Saunders JR. Plasmid transfer between Bacillus thuringiensis subsp. israelensis strains in laboratory culture, river water, and dipteran larvae. Appl. Environ. Microbiol. (2001) 67:330–338. doi: 10.1128/AEM.67.1.330-338.2001
51. Arutyunov D, Frost LSF. conjugation: back to the beginning. Plasmid. (2013) 70:18–32. doi: 10.1016/j.plasmid.2013.03.010
52. Llosa MI, Gomis-Ruth FX, Coll M, de la Cruz FE. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. (2002) 45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x
53. Ilangovan A, Connery S, Waksman G. Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol. (2015) 23:301–10. doi: 10.1016/j.tim.2015.02.012
54. Cabezon E, Ripoll-Rozada J, Pena A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. (2014) 9:81–95. doi: 10.1111/1574-6976.12085
55. Vielmetter W, Bonhoeffer F, Schutte A. Genetic evidence for transfer of a single DNA strand during bacterial conjugation. J Mol Biol. (1968) 37:81–6. doi: 10.1016/0022-2836(68)90074-0
56. Goldlust K, Ducret A, Halte M, Dedieu-Berne A, Erhardt M, Lesterlin Ch. The F pilus serves as a conduit for the DNA during conjugation between physically distant bacteria. Proc Natl Acad Sci USA. (2023) 120:e2310842120. doi: 10.1073/pnas.2310842120
57. Wong JJ, Lu J, Edwards RA, Frost LS, Glover JN. Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM. Nucleic Acids Res. (2011) 39:6775–88. doi: 10.1093/nar/gkr296
58. Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. (2003) 1:137–49. doi: 10.1038/nrmicro753
59. Parker C, Meyer RJ. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. MolMicrobiol. (2007) 66:252–61. doi: 10.1111/j.1365-2958.2007.05925.x
60. Liu Y, Gao Y, Liu X, Liu Q, Zhang Y, Wang O, et al. Transposon insertion sequencing reveals T4SS as the major genetic trait for conjugation transfer of multi-drug resistance pEIB202 from Edwardsiella. BMC Microbiol. (2017) 17:112. doi: 10.1186/s12866-017-1013-7
61. Guzmán-Herrador DL, Llosa M. The secret life of conjugative relaxases. Plasmid. (2019) 104:102415. doi: 10.1016/j.plasmid.2019.102415
62. Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. (2009) 73:775–808. doi: 10.1128/MMBR.00023-09
63. Llosa M, Alkorta I. Coupling proteins in type IV secretion. Curr Top Microbiol Immunol. (2017) 413:143–68. doi: 10.1007/978-3-319-75241-9_6
64. Christie PJ. The mosaic type IV secretion systems. EcoSal Plus. (2016) 7. doi: 10.1128/ecosalplus.ESP-0020-2015
65. Khara P, Song L, Christie PJ, Hu B. In situ visualization of the pKM101-encoded type IV secretion system reveals a highly symmetric ATPase energy center. MBio. (2021) 12:e02465–21. doi: 10.1128/mBio.02465-21
66. Mamun AAMA, Kishida K, Christie PJ. Protein transfer through an F plasmid-encoded type IV secretion system suppresses the mating-induced SOS response. mBio. (2021) 12:e0162921. doi: 10.1128/mBio.01629-21
67. Mamun AAMA, Kissoon K, Kishida K, Shropshire WC, Hanson B, Christie PJ. Sequence analysis and evidence for translocation of maintenance/leading region proteins through diverse type IV secretion systems. Plasmid. (2022) 102652:123–4. doi: 10.1016/j.plasmid.2022.102652
68. Hinnekens P, Mahillon J. Conjugation-mediated transfer of pXO16, a large plasmid from Bacillus thuringiensis sv. Israelensis, across the Bacillus cereus group and its impact on host phenotype. Plasmid. (2022) 122:102639. doi: 10.1016/j.plasmid.2022.102639
69. Draper O, Cesar CE, Machon C, de la Cruz F, Llosa M. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc Natl Acad Sci U.S.A. (2005) 102:16385–90. doi: 10.1073/pnas.0506081102
70. Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. (2004) 155:376–86. doi: 10.1016/j.resmic.2004.01.012
71. Allard N, Neil K, Grenier F, Rodrigue S. The type IV pilus of plasmid TP114 displays adhesins conferring conjugation specificity and is important for DNA transfer in the mouse gut microbiota. Microbiol Spectr. (2022) 10:e0230321. doi: 10.1128/spectrum.02303-21
72. Kishida K, Grace Li Y, Ogawa-Kishida N, Khara P, Mamum AAMAI, Bosserman RE, et al. Chimeric systems composed of swapped Tra subunits between distantly-related F plasmids reveal striking plasticity among type IV secretion machines. PloS Genet. (2024) 20:e1011088. doi: 10.1371/journal.pgen.1011088
73. Hradecka H, Karasova D, Rychlik I. Characterization of Salmonella enterica serovar Typhimurium conjugative plasmids transferring resistance to antibiotics and their interaction with the virulence plasmid. Antimicrob Chemother. (2008) 62:938–41. doi: 10.1093/jac/dkn286
74. Dorman CJ. Regulatory integration of horizontally-transferred genes in bacteria. Front Biosci. (2009) 14:4103–12. doi: 10.2741/3515
75. Mruk I, Kobayashi I. To be or not to be: Regulation of restriction modificationsystems and other toxin-antitoxin systems. Nucleic Acids Res. (2014) 42:70–86. doi: 10.1093/nar/gkt711
76. Koraimann G. Spread and persistence of virulence and antibiotic resistance genes: A ride on the F plasmid conjugation module. EcoSal Plus. (2018) 8. doi: 10.1128/ecosalplus.ESP-0003-2018
77. Alalam H, Graf FE, Palm M, Abadikhah M, Zackrisson M, Boström J, et al. A high-throughput method for screening for genes controlling bacterial conjugation of antibiotic resistance. mSystems. (2020) 5:e01226–20. doi: 10.1128/mSystems.01226-20
78. Ishiwa A, Komano T. Thin pilus PilV adhesins of plasmid R64 recognize specific structures of the lipopolysaccharide molecules of recipient cells. J Bacteriol. (2002) 185.
79. Lang S, Zechner EL. General requirements for protein secretion by the F-like conjugation system R1. Plasmid. (2012) 67:128–38. doi: 10.1016/j.plasmid.2011.12.014
80. Peng Y, Lu J, Wong JJW, Edwards RA, Frost LS, Mark Glover JN. Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM. J Mol Biol. (2014) 426:3783–95. doi: 10.1016/j.jmb.2014.09.018
81. Pérez-Mendoza D, de la Cruz F. Escherichia coli genes affecting recipient ability in plasmid conjugation: Are there any? BMC Genomics. (2009) 10:71. doi: 10.1186/1471-2164-10-71
82. Frost LS, Koraimann G. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. (2010) 5:1057–1071. doi: 10.2217/fmb.10.70
83. Polzleitner E, Zechner EL, Renner W, Fratte R, Jauk B, Hogenauer G, et al. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol Microbiol. (1997) 25:495–507. doi: 10.1046/j.1365-2958.1997.4831853.x
84. Vázquez-Padron RI, Moreno-Fierros L, Neri-Bazán L, de la Riva GA, López-Revilla R. Bacillus thuringiensis Cry1Ac protoxin is a potent systemic and mucosal adjuvant. Scand J Immunol. (1999) 49:578–84. doi: 10.1046/j.1365-3083.1999.00534.x
85. Moreno-Fierros L, García N, Gutiérrez R, López-Revilla R, Vázquez-Padrón RI. Intranasal, rectal and intraperitoneal immunization with protoxin Cry1Ac from Bacillus thuringiensis induces compartmentalized serum, intestinal, vaginal and pulmonary immune responses in Balb/c mice. Microbes Infect. (2000) 2:885–90. doi: 10.1016/s1286-4579(00)00398-1
86. Moreno-Fierros L, Ruiz-Medina EJ, Esquivel R, López-Revilla R, Piña-Cruz S. Intranasal Cry1Ac protoxin is an effective mucosal and systemic carrier and adjuvant of Streptococcus pneumoniae polysaccharides in mice. Scand J Immunol. (2003) 57:45–55. doi: 10.1046/j.1365-3083.2003.01190.x
87. Esquivel-Pérez R, Moreno-Fierros L. Mucosal and systemic adjuvant effects of cholera toxin and Cry1Ac protoxin on the specific antibody response to HIV-1 C4/V3 peptides are different and depend on the antigen co-administered. Viral Immunol. (2005) 18:695–708. doi: 10.1089/vim.2005.18.695
88. Guerrero GG, Moreno-Fierros L. Carrier potential properties of Bacillus thuringiensis Cry1A toxins for a diphtheria toxin epitope. Scand J Immunol. (2007) 66:610–8. doi: 10.1111/j.1365-3083.2007.01992.x
89. Guerrero GG, Russell WM, Moreno-Fierros L. Analysis of the cellular immune response induced by Bacillus thuringiensis Cry1A toxins in mice: effect of the hydrophobic motif from diphtheria toxin. Mol Immunol. (2007) 44:1209–17. doi: 10.1016/j.molimm.2006.06.007
90. Michaelis and Grohmann. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics (Basel). (2023) 12:328. doi: 10.3390/antibiotics12020328
91. Schnepf E, Crickmore N, Van Rie J, Lerelcus D, Baum J, Feitelson J, et al. Bacillus thuringiensis and its pesticidal proteins. Microbiol Mol Biol Rev. (1998) 62:775–806. doi: 10.1128/MMBR.62.3.775-806.1998
92. de Maagd RA, Bravo A, Berry C, Crickmore N, Schnepf HE. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu Rev Genet. (2003) 37:409–33. doi: 10.1146/annurev.genet.37.110801.143042
93. Gill SS, Cowles EA, Pietrantonio PV. The mode of action of Bacillus thuringiensis endotoxins. Annu Rev Entomol. (1992) 37:615–636. doi: 10.1146/annurev.en.37.010192.003151
94. Li J, Carroll J, Ellar DJ. Crystal structure of insecticidal d-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature. (1991) 353:815–21. doi: 10.1038/353815a0
95. Grochulski P, Masson L, Borisova S, Puztai-Carey M, Schwartz J-L, Brousseau R, et al. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J. Mol. Biol. (1995) 254:447–64. doi: 10.1006/jmbi.1995.0630
96. Li J, Koni PA, Ellar DJ. Structure of the mosquitocidal delta-endotoxin CytB from Bacillus thuringiensis sp. kyushuensis and implications for membrane pore formation. J Mol Biol. (1996) 257:129–52. doi: 10.1006/jmbi.1996.0152
97. Butko P. Cytolytic toxin Cyt1A and its mechanism of membrane damage: data and hypotheses. Appl Environ Microbiol. (2003) 69:2415–22. doi: 10.1128/AEM.69.5.2415-2422.2003
98. Xu CH, Wang BCH, YU Z, Sun M. Structural insights into Bacillus thuringiensis Cry, Cyt and parasporin toxins. Toxins (Basel). (2014) 6:2732–70. doi: 10.3390/toxins6092732
99. Boonserm P, Davis P, Ellar DJ, Li J. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J Mol Biol. (2005) 348:363–82. doi: 10.1016/j.jmb.2005.02.013
100. Boonserm P, Mo M, Angsuthanasombat C, Lescar J. Structure of the functional form of the mosquito larvicidal Cry4Aa toxin from Bacillus thuringiensis at a 2.8-angstrom resolution. J Bacteriol. (2006) 188:3391–401. doi: 10.1128/JB.188.9.3391-3401.2006
101. Soberon M, Lopez-Diaz JA, Bravo A. Cyt toxins produced by Bacillus thuringiensis: A protein fold conserved in several pathogenic microorganisms. Peptides. (2013) 41:87–93. doi: 10.1016/j.peptides.2012.05.023
102. Ibarra JE, Federici BA. Isolation of a relatively nontoxic 65-kilodalton protein inclusion from the parasporal body of Bacillus thuringiensis subsp. israelensis. J Bacteriol. (1986) 165:527–533. doi: 10.1128/jb.165.2.527-533.1986
103. Guerchicoff A, Ugalde RA, Rubinstein CP. Identification and characterization of a previously undescribed cyt gene in Bacillus thuringiensis subsp. Israelensis. Appl Environ Microbiol. (1997) 63:2716–21. doi: 10.1128/aem.63.7.2716-2721.1997
104. Thorne L, Garduno F, Thompson T, Decker D, Zounes M, Wild M, et al. Structural similarity between the lepidoptera- and diptera-specific insecticidal endotoxin genes of Bacillus thuringiensis subsp. “kurstaki” and “Israelensis. J Bacteriol. (1986) 166:801–11. doi: 10.1128/jb.166.3.801-811.1986
105. Ben-Dov E. Bacillus thuringiensis subsp. Israelensis and its dipteran-specific toxins. Eitan Ben-Dov Toxins (Basel). (2014) 6:1222–43. doi: 10.3390/toxins6041222
106. Zhang O, Hua G, Adang MJ. Effects and mechanisms of Bacillus thuringiensis crystal toxins for mosquito larvae. Insect Sci. (2017) 24:714–29. doi: 10.1111/1744-7917.12401
107. Ward ES, Ellar DJ, Chilcott CN. Single amino acid changes in the Bacillus thuringiensis var. Israelensis delta-endotoxin affect the toxicity and expression of the protein. J Mol Biol. (1988) 202:527–35. doi: 10.1016/0022-2836(88)90283-5
108. Onofre J, Pacheco S, Torres-Quintero MC, Gill SS, Soberon M, Bravo A. The Cyt1Aa toxin from Bacillus thuringiensis inserts into target membranes via different mechanisms in insects, red blood cells, and lipid liposomes. J Biol Chem. (2020) 295:9606–17. doi: 10.1074/jbc.RA120.0138692959606–9617
109. Federici BA, Park HW, Bideshi DK. Overview of the basic biology of Bacillus thuringiensis with emphasis on genetic engineering of bacterial larvicides for mosquito control. Open Toxinol J. (2010) 3:83–100. doi: 10.2174/1875414701003010083
110. Lopez-Diaz JA, Canton PE, Gill SS, Soberon M, Bravo A. Oligomerization is a key step in Cyt1Aa membrane insertion and toxicity but not necessary to synergize Cry11Aa toxicity in Aedes aEgypti larvae. Environ Microbiol. (2013) 15:3030–9. doi: 10.1111/1462-2920.12263
111. Federici BA, Park HW, Bideshi DK, Wirth MC, Johnson JJ, Sakano Y, et al. Developing recombinant bacteria for control of mosquito larvae. J Am Mosq Control Assoc. (2007) 23:164–75. doi: 10.2987/8756-971X(2007)23[164:DRBFCO]2.0.CO2
112. Pérez C, Fernández LE, Sun JG, Folch JL, Gill SS, Soberon M, et al. Bacillus thuringiensis subsp. Israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci USA. (2005) 102:18303–8. doi: 10.1073/pnas.0505494102
113. López-Molina S, do Nascimento NA, Silva-Filha M, Guerrero A, Sánchez J, Pacheco S, et al. In vivo nanoscale analysis of the dynamic synergistic interaction of Bacillus thuringiensis Cry11Aa and Cyt1Aa toxins in Aedes aEgypti. PloS Pathog. (2021) 17:e1009199. doi: 10.1371/journal.ppat.1009199
114. Schwartz JL, Potvin L, Coux F, Charles JF, Berry C, Humphreys MJ, et al. Permeabilization of model lipid membranes by Bacillus sphaericus mosquitocidal binary toxin and its individual components. J Membr Biol. (2001) 184:171–83. doi: 10.1007/s00232-001-0086-1
115. Federici BA, Park H-W, Bideshi DK, Wirth MC. Johnson JJ Recombinant bacteria for mosquito control. J Exp Biol. (2003) 206:3877–85. doi: 10.1242/jeb.00643
116. Wilson AL, Courtenay O, Kelly-Hope LA, Scott TW, Takken W, Torr S, et al. The importance of vector control for the control and elimination of vector-borne diseases. PloS Negl Trop Dis. (2020) 14:e0007831. doi: 10.1371/journal.pntd.0007831
117. WHO. Malaria terminology update. Geneva: World Health Organization (2021). p. 2021, ISBN: 978-92-4-003840-0.
118. FAO. The state of food security and nutrition in the world. Building resilience for peace and food security. Rome: Food and Agriculture Organization of the United Nations (2017), ISBN: 978-92-5-109888-2.
119. FAO. (2018). Available online at: http://www.fao.org/faostat/en/#data/RP.
120. Baxter RH, Contet A, Krueger K. Arthropod innate immune systems and vector-borne diseases. Biochemistry. (2017) 56:907–18. doi: 10.1021/acs.biochem.6b00870
121. van den Berg H, da Silva Bezerra HS, Al-Eryani S, Chanda E, Nagpal BN, Knox TB, et al. Recent. trends in global insecticide use for disease vector control and potential implications for resistance management. Sci Rep. (2021) 11:2367. doi: 10.1038/s41598-021-03367-9
122. WHO. Global report on insecticide resistance in malaria vectors: 2010–2016. Geneva: World Health Organization (2018), ISBN: 978 92 4 151405 7.
123. World Health Organization (WHO). Dengue and severe Dengue (2020). Available online at: https://www.who.int/en/news-room/fact-sheets/detail/dengue-and-severe-dengue.
124. Patterson J, Sammon M, Garg M. Dengue, zika and chikungunya: Emerging arboviruses in the new world. West J Emerg Med. (2016) 17:671–9. doi: 10.5811/westjem.2016.9.30904
125. Benelli G. Research in mosquito control: current challenges for a brighter future. Parasitol Res. (2015) 114:2801–5. doi: 10.1007/s00436-015-4586-9
126. Xu Q, Zhang L, Li T, Zhang L, He L, Dong K, et al. Evolutionary adaptation of the amino acid and codon usage of the mosquito sodium channel following insecticide selection in the field mosquitoes. PloS One. (2012) 7:e47609. doi: 10.1371/journal.pone.0047609
127. Li T, Zhang L, Reid WR, Xu Q, Dong K, Liu N. Multiple mutations and mutation combinations in the sodium channel of permethrin resistant mosquitoes, Culex Quinquefasciatus. Sci Rep. (2012) 2:781. doi: 10.1038/srep00781
128. Graham K, Kayedi MH, Maxwell C, Kaur H, Rehman H, Malima R, et al. Multi-country field trials comparing wash-resistance of PermaNet and conventional insecticide-treated nets against anopheline and culicine mosquitoes. Med Vet Entomol. (2005) 19:72–83. doi: 10.1111/j.0269-283X.2005.00543.x
129. Fritz BK, Hofmann WC, Bonds JAS, Haas K, Czaczyk Z. The biological effect of cage design corrected for reductions in spray penetration. J Plant Prot Res. (2014) 54:395–400. doi: 10.2478/jppr-2014-0059
130. Owusu HF, Jancáryová D, Malone D, Müller P. Comparability between insecticide resistance bioassays for mosquito vectors: time to review current methodology? Parasit Vectors. (2015) 8:357. doi: 10.1186/s13071-015-0971-6
131. Mbwambo SG, Bubun N, Mbuba E, Moore J, Mbina K, Kamande D, et al. Comparison of cone bioassay estimates at two laboratories with diferent Anopheles mosquitoes for quality assurance of pyrethroid insecticide treated nets. Malar J. (2022) 21:214. doi: 10.1186/s12936-022-04217-3
132. WHO. Determining discriminating concentrations of insecticides for monitoring resistance in sand lies: report of a multi-centre laboratory study and WHO expert consultations. Geneva: World Health Organization (2022), ISBN: 978-92-4-006441-6.
133. Kilpatrick AM, Randolph SE. Drivers, dynamics, and control of emerging vector-borne zoonotic diseases. Lancet. (2012) 380:1946–55. doi: 10.1016/S0140-6736(12)61151-9
134. Souza-Neto JA, Powell JR, Bonizzoni M. Aedes aEgypti vector competence studies: A review. Infect Genet Evol. (2019) 67:191–209. doi: 10.1016/j.meegid.2018.11.009
135. Benelli G, Canale A, Conti B. Eco-friendly control strategies against the Asian tiger mosquito, Aedes albopictus (Diptera: Culicidae): repellency and toxic activity of plant essential oils and extracts. Pharmacol. (2014) 1:44–50.
136. Benelli G, Wilke ABB, Beier JC. Aedes albopictus (Asian Tiger Mosquito). Trends Parasitol. (2020) 36:942–943. doi: 10.1016/j.pt.2020.01.001
137. Ferreira-De-Brito A, Ribeiro IP, De Miranda RM, Fernandes RS, Campos SS, Da Silva KAB, et al. First detection of natural infection of Aedes aEgypti with Zika virus in Brazil and throughout South America. Memórias Inst Oswaldo Cruz. (2016) 111:655–8. doi: 10.1590/0074-02760160332
138. Liu L, Ren X, Liu X. A host-parasite system with multiple parasite strains and superinfection revisited: the global dynamics. Acta Biotheor. (2020) 68:201–25. doi: 10.1007/s10441-019-09359-7
139. Viana JL, da Silva JS, de Mattos GC, Pinto MCC, Dutra LDS, Carvalho LLA, et al. Microencapsulation of Bacillus thuringiensis strains for the control of Aedes aEgypti. Exp Parasitol. (2023) 255:108654. doi: 10.1016/j.exppara.2023.108654
140. Margalith Y, Ben-Dov E. Biological control by Bacillus thuringiensis subsp. Israelensis. In: Rechcigl JE, Rechcigl NA, editors. Insect pest Management: Techniques for Environmental Protection. CRC Press, Boca Raton, FL, USA (2000), ISBN: 9780429104428. p. 243–301.
141. Denoth M, Frid, Myers JH. Multiple agents in biological control: Improving the odds? Biol Control. (2002) 24:20–30. doi: 10.1016/S1049-9644(02)00002-6
142. Weetman D, Kamgang B, Badolo A, Moyes CL, Shearer FM, Coulibaly M, et al. Aedes mosquitoes and Aedes-borne arboviruses in Africa: Current and future threats. Int J Environ Res Public Health. (2018) 15:220. doi: 10.3390/ijerph15020220
143. Ding F, Fu J, Jiang D, Hao M, Lin G. Mapping the spatial distribution of Aedes aEgypti and Aedes albopictus. Acta Trop. (2018) 178:155–62. doi: 10.1016/j.actatropica.2017.11.020
144. Florax RJ, Travisi CM, Nijkamp P. A meta-analysis of the willingness to pay for reductions in pesticide risk exposure. Eur Rev Agric Econ. (2005) 32:441–67. doi: 10.2139/ssrn.569110
145. Furlong MJ, Shi ZH, Liu SS, Zalucki MP. Evaluation of the impact of natural enemies on Plutella xylostella L. (Lepidoptera: Yponomeutidae) populations on commercial Brassica farms. Agric For Entomol. (2004) 6:311–22. doi: 10.1111/j.1461-9555.2004.00228.x
146. Zalucki MP, Adamson D, Furlong MJ. The future of IPM: whither or wither? Aust J Entomol. (2009) 48:85–96. doi: 10.1111/j.1440-6055.2009.00690.x
147. O’Neill SL. The use of Wolbachia by the world mosquito program to interrupt transmission of Aedes aEgypti transmitted viruses. Adv Exp Med Biol. (2018) 1062:355–60. doi: 10.1007/978-981-10-8727-1_24
148. Sanchis V, Bourguet D. Bacillus thuringiensis: applications in agriculture and insect resistance management. A rev Agron Sustain Dev. (2008) 28:11–20. doi: 10.1051/agro:2007054
149. Ferry N, Edwards MG, Gatelhouse JA, Gatehouse AMR. The plant-insect interactions: molecular approaches to insect resistance. Curr Opin Biotechnol. (2004) 15:155–61. doi: 10.1016/j.copbio.2004.01.008
150. Evans BR, Kotsakiozi P, Costa-da-Silva AL, Ioshino RS, Garziera L, Pedrosa MC, et al. Transgenic Aedes aEgypti mosquitoes transfer genes into a natural population. Sci Rep. (2019) 9:1–6. doi: 10.1038/s41598-019-49660-6
151. James AA. Engineering mosquito resistance to malaria parasites: the avian malaria model. Insect Biochem Mol Biol. (2002) 32:1317–23. doi: 10.1016/S0965-1748(02)00094-2
152. Cressler CE, McLeod DV, Rozins C, Van Den Hoogen J, Day T. he adaptive evolution of virulence: A review of theoretical predictions and empirical tests. Parasitology. (2016) 143:915–30. doi: 10.1017/S003118201500092X
153. Alizon S. Co-infection and super-infection models in evolutionary epidemiology. Interface Focus. (2013) 3:20130031. doi: 10.1098/rsfs.2013.0031
154. Broennimann O, Fitzpatrick MC, Pearman PB, Petitpierre B, Pellissier L, Yoccoz NG, et al. Measuring ecological niche overlap from occurrence and spatial environmental data. Global Ecol Biogeogr. (2012) 21:481_497. doi: 10.1111/j.1466-
155. Bushman M, Antia R. A general framework for modelling the impact of co-infections on pathogen evolution. J R Soc Interface. (2019) 16:20190165. doi: 10.1098/rsif.2019.0165
156. Heukelbach J, Alencar CH, Kelvin AA, De Oliveira WK, Cavalcanti L. Zika virus outbreak in Brazil. J Infect Dev Ctries. (2016) 10:116–20. doi: 10.3855/jidc.8217
157. Stawicki SP, Sikka V, Chattu VK, Popli RK, Galwankar SC, Kelkar D, et al. The emergence of zika virus as a global health security threat: A review and a consensus statement of the INDUSEM Joint working Group (JWG). J Glob Infect Dis. (2016) 8:3–15. doi: 10.4103/0974-777X.176140
158. Forattini OP. Culicidologia Medica, Volume 2: Identificacão, biologia e epidemiologia. São Paulo: Edusp (2002), ISBN: 9788531406997.
159. Cattarino L, Rodriguez-Barraquer I, Imai N, Cummings DAT, Ferguson NM. Mapping global variation in dengue transmission intensity. Sci Transl Med. (2020) 12:eaax4144. doi: 10.1126/scitranslmed.aax4144
160. Jones R, Kulkarni MA, Davidson TMV, Talbot B. RADAM-LAC Research Team Arbovirus vectors of epidemiological concern in the Americas: A scoping review of entomological studies on Zika, dengue and chikungunya virus vectors. PloS One. (2020) 15:e0220753. doi: 10.1371/journal.pone.0220753
161. Castaneda-Guzman M. Modeling species geographic distributions in aquatic ecosystems using a density-based clustering algorithm. Blacksburg: Virginia Tech University (2022). doi: 10919/111820
162. Belousova ME, Malovichko YV, Shikov AE, Nizhnikov AA, Antonets KS. Dissecting the environmental consequences of Bacillus thuringiensis application for natural ecosystems. Toxins. (2021) 13:355. doi: 10.3390/toxins13050355
163. Goldberg LJ, Margalit J. A bacterial spore demonstrating rapid larvicidal activity against Anopheles sergentii, Uranotaenia unguiculata, Culex univittatus, Aedes aegypti, and Culex pipiens. Mosq. News (1977) 37:317–324.
164. Burges HD. Safety, safety testing and quality control of microbial pesticides. In: Microbial Control of Pests and Plant Diseases, 1970-1980. Academic Press, New York (1981) pp. 737–67.
165. Powell KA, Jutsum AR. Technical and commercial aspects of biocontrol products. Pestic Sci. (1991) 37:315–21. doi: 10.1002/ps.2780370403
166. Lacey LA, Merritt RW. The safety of bacterial microbial agents used for blacknfly and mosquito control in aquatic environments. In: Hokkanen HMT, Hajek AE, editors. Environmental Impacts of Microbial Insecticides: Need and Methods for Risk Assessment. Springer, Netherlands, Dordrecht (2003). p. 151–68. doi: 10.1007/978-94-017-1441-9_8
167. Stevens MM, Akhurst RJ, Clifton MA, Hughes PA. Factors affecting the toxicity of Bacillus thuringiensis var. Israelensis and Bacillus sphaericus to fourth instar larvae of Chironomus tepperi (Diptera: Chironomidae). J Invertebr Pathol. (2004) 86:104–10. doi: 10.1016/j.jip.2004.04.002
168. Allgeier S, Kastel A, Bruhl CA. Adverse effects of mosquito control using Bacillus thuringiensis var. Israelensis: reduced chironomid abundances in mesocosmsemi-field and field studies. Ecotoxicol Environ Saf. (2019) 169:786–96. doi: 10.1016/j.ecoenv.2018.11.050
169. Allgeier S, Friedrich A, Bruhl CA. Mosquito control based on Bacillus thuringiensis israelensis (Bti) interrupts artificial wetland food chains. Sci. Total Environ. (2019) 686:1173–84. doi: 10.1016/j.scitotenv.2019.05.358
170. Bordalo MD, Gravato C, Beleza S, Campos D, Lopes I, Pestana JLT. Lethal and sublethal toxicity assessment of Bacillus thuringiensis var. Israelensis and Beauveria bassiana based bioinsecticides to the aquatic insect Chironomus riparius. Sci Total Environ. (2020) 698:134155. doi: 10.1016/j.scitotenv.2019.134155
171. de Souza JDA, Jain S, de Oliveira CMF, Ayres CF, Lucena WA. Toxicity of a Bacillus thuringiensis Israelensis-like strain against Spodoptera frugiperda. BioControl. (2009) 54:467–73. doi: 10.1007/s10526-008-9191-8
172. Tudoran A, Nordlander G, Karlberg A, Puentes A. A major forest insect pest, the pine weevil Hylobius abietis, is more susceptible to Diptera- than Coleoptera- targeted Bacillus thuringiensis strains. Pest Manage Sci. (2021) 77:1303–15. doi: 10.1002/ps.6144
173. Brühl CA, Despres L, Fror O, Patil CD, Poulin B, Tetreau G, et al. Environmental and socioeconomic effects of mosquito control in Europe using the biocide Bacillus thuringiensis subsp. Israelensis (Bti). Sci Total Environ. (2020) 724:137800. doi: 10.1016/j.scitotenv.2020.137800
174. Snarski VM. Interactions between Bacillus thuringiensis subsp. Israelensis and Fathead Minnows, Pimephales promelas Rafinesque, under laboratory conditions. Appl Environ Microbiol. (1999) 56:2618–22. doi: 10.1128/aem.56.9.2618-2622.1990
175. Tetreau G, Alessi M, Veyrenc S, Perigon S, David JP, Reynaud S, et al. Fate of Bacillus thuringiensis subsp. Israelensis in the field: evidence for spore recycling and differential persistence of toxins in leaf litter. Appl Environ Microbiol. (2012) 78:8362–7. doi: 10.1128/AEM.02088-1Tetrau
176. Li Y, Wang C, Ge L, Hu C, Wu G, Sun Y, et al. Environmental behaviors of Bacillus thuringiensis (Bt) insecticidal proteins and their effects on microbial ecology. Plants. (2022) 11:1212. doi: 10.3390/plants11091212
177. Carvalho FD, Moreira LA. Why is Aedes aEgypti Linnaeus so Successful as a Species? Neotropical Entomol. (2017) 46:243–55. doi: 10.1007/s13744-017-0520-4
178. Crickmore N, Bone EJ, Wiliams JA, Ellar DJ. Contribution of the individual components of the delta-endotoxin crystal to the mosquitocidal activity of Bacillus thuringiensis subs. Israelensis. FEMS Microbiol Lett. (1995) 131:249–54. doi: 10.1111/j.1574-6968.1995.tb07784.x
179. Thomas WE, Ellar DJ. Bacillus thuringiensis var Israelensis crystal delta-endotoxin: effects on insect and mammalian cells in vitro and in vivo. J Cell Sci. (1983) 60:181–97. doi: 10.1242/jcs.60.1.181
180. Knowles BH, Blatt MR, Tester M, Horsnell JM, Carroll J, Menestrina G, et al. A cytolytic delta-endotoxin from Bacillusthuringiensis var. israelensis forms cation-selective channels in planar lipid bilayers. FEBS Lett. (1989) 244:259–62. doi: 10.1016/0014-5793(89)80540-x
181. Knowles BH, White PJ, Nicholls CN, Ellar DJ. A broad-spectrum cytolytic toxin from Bacillus thuringiensis var. kyushuensis. Proc. R. Soc. Lond. Ser. B Biol. Sci. (1992) 248:1–7. doi: 10.1098/rspb.1992.0035
182. Delecluse A, Rosso ML, Ragni A. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl Environ Microbiol. (1995) 61:4230–4235. doi: 10.1128/aem.61.12.4230-4235.1995
183. Delecluse A, Juarez-Perez V, Berry C. Vector-active toxins: Structure and diversity. In Entomopathogenic Bacteria: From Laboratory to Field Application; Charles JF, Delécluse A, Nielsen-LeRoux C, Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands (2000) pp. 101–25. doi: 10.1007/978-94-017-1429-7_6
184. Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, et al. Completesequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. Israelensis. App Environ Microbiol. (2002) 68:5082–95. doi: 10.1128/AEM.68.10.5082-5095.2002
185. Valtierra-de-Luis D, Villanueva M, Lai L, Williams T, Caballero P. Potential of Cry10Aa and Cyt2Ba, two minority delta endotoxins produced by Bacillus thuringiensis ser. Israelensis, for the control of Aedes aEgypti larvae. Toxins. (2020) 12:355. doi: 10.3390/toxins12060355
186. Wirth MC, Delecluse A, Federici BA, Walton WE. Variable cross-resistance to Cry11B from Bacillus thuringiensis subsp. jegathesan in Culex quinquefasciatus (Diptera: Culicidae) resistant to single or multiple toxins of Bacillus thuringiensis subsp. Israelensis. Appl Environ Microbiol. (1998) 64:4174–9. doi: 10.1128/AEM.64.11.4174-4179.1998
187. Wirth MC. Mosquito resistance to bacterial larvicidal proteins. Open J Toxicol. (2010) 3:101–15. doi: 10.2174/1875414701003010126
188. Ferreira LM, Silva-Filha MHNL. Bacterial larvicides for vector control: Mode of action of toxins and implications for resistance. Biocontrol Sci Technol. (2013) 23:1137–68. doi: 10.1080/09583157.2013.822472
189. Charles JF, Nielsen-LeRoux C, Delecluse A. Bacillus sphaericus toxins: Molecular biology and mode of action. Ann Rev Entomol. (1996) 41:451–72. doi: 10.1146/annurev.en.41.010196.002315
190. Berry C. The bacterium, Lysinibacillus sphaericus, as an insect pathogen. J Invertebr Pathol. (2012) 109:1–10. doi: 10.1016/j.jip.2011.11.008
191. Xu K, Yuan Z, Rayner S, Hu X. Genome comparison provides molecular insights into the phylogeny of the reassigned new genus Lysinibacillus. BMC Genom. (2015) 16:140. doi: 10.1186/s12864-015-1359-x
192. Wirth MC, Berry C, Walton WE, Federici BA. Mtx toxins from Lysinibacillus sphaericus enhance mosquitocidal cry-toxin activity and suppress cry-resistance in Culex quinquefasciatus. J Invertebr Pathol. (2014) 115:62–7. doi: 10.1016/j.jip.2013.10.003
193. Ibrahim MA, Griko NB, Bulla LA Jr. The Cry4B toxin of Bacillus thuringiensis subsp. Israelensis kills Permethrin-resistant Anopheles Gambiae, the principal vector of malaria. Exp Biol Med (Maywood). (2013) 238:350–9. doi: 10.1177/1535370213477973
194. Ibrahim MA, Griko NB, Bulla LA Jr. Cytotoxicity of the Bacillus thuringiensis Cry4B toxin is mediated by the cadherin receptor BT-R3 of Anopheles Gambiae. Exp Biol Med (Maywood). (2013) 238:755 764. doi: 10.1177/1535370213493719
195. Baumann P, Clark MA, Baumann L, Broadwell AH. Bacillus sphaericus as a mosquito pathogen: Properties of the organism and its toxins. Microbiol Rev. (1991) 55:425–36. doi: 10.1128/mr.55.3.425-436.1991
196. Lacey LA. Bacillus thuringiensis serovariety israelensis and Bacillus sphaericus for mosquito control. Journal of the American Mosquito Control Association. (2007) 23:133–164. doi: 10.2987/8756-971X(2007)23[133:BTSIAB]2.0.CO;2
197. Berry C, Jackson-Yap J, Oci C. Nucleotied sequence of two toxin genes from Bacillus sphaericus IAB59; sequence comparison between five highly toxinogeic strains. Mucleic Acids Res. (1989) 17:7516. doi: 10.1093/nar/17.18.7516
198. Berry C, Hindley J, Ehrhardt AF. Genetic determinants of host ranges of Bacillus sphaericus mosquito larvicidal toxins. J Bacteriol. (1993) 175:510–518. doi: 10.1128/jb.175.2.510-518.1993
199. Darboux I, Nielsen-LeRoux C, Charles JF, Pauron D. The receptor of Bacillus sphaericus binary toxin in Culex pipiens (Diptera: Culicidae) midgut: Molecular cloning and expression. Insect Biochem Mol Biol. (2001) 31:981–990. doi: 10.1016/s0965-1748(01)00046-7
200. Davidson EW. Binding of the Bacillus sphaericus (Eubacteriales: Bacillaceae) toxin to midgut cells of mosquito (Diptera: Culicidae) larvae: Relationship to host range. J Med Entomol. (1988) 25:151–157. doi: 10.1093/jmedent/25.3.151
201. Carvalho DO, McKenney AR, Garziera L, Lacroix B, Donnelly CHA, Alphey L, et al. Suppression of a field population of aedes aEgypti in Brazil by sustained release of transgenic male mosquitoes. PloS Negl Trop Dis. (2015) 9:e0003864. doi: 10.1371/journal.pntd.0003864
202. Matthews KR. Controlling and coordinating development in vector-transmitted parasites. Science. (2011) 331:1149–53. doi: 10.1126/science.1198077
203. Regis L, Silva-Filha MH, Nielsen-LeRoux C, Charles JF. Bacteriological larvicides of dipteran disease vectors. Trends Parasitol. (2001) 17:377–80. doi: 10.1016/s1471-4922(01)01953-5
204. Becker N. Bacterial control of vector-mosquitoes and black flies. Entomopathog Bact Lab to FAppl. (2000) 15:383–98. doi: 10.1007/978-94-017-1429-7_21
205. Sinka ME, Bangs MJ, Manguin S, Rubio-Palis Y, Chareonviriyaphap T, Coetzee M, et al. A global map of dominant malaria vectors. Parasit Vectors. (2012) 5:69. doi: 10.1186/1756-3305-5-69
206. Kraemer MUG, Sinka ME, Duda KA, Mylne AQN, Shearer FM, Barker CM, et al. The global distributionof the arbovirus vectors Aedes aEgypti and Ae. Albopictus. Elife. (2015) 4:e08347. doi: 10.7554/eLife.08347
207. Liu-Hermesson J, Quam M, Wilder-Smith A, Stenlund H, Ebi K, Massad E, et al. Climate change and Aedes vectors: 21st century projections for dengue transmission in Europe. EBioMedicine. (2016) 7:267–77. doi: 10.1016/j.ebiom.2016.03.046
208. Becker N, Geier M, Balczun C, Bradersen U, Huber K, Kiel E, et al. Repeated introduction of Aedes albopictus into Germany. Parasitol Res. (2013) 112:1787–90. doi: 10.1007/s00436-012-3230-1
209. Pereira-dos-Santos T, Roiz D, Lourenço-de-Oliveira R, Paupy C. A systematic review:is Aedes albopictusan eicient bridge vector for zoonotic arboviruses? Pathogens. (2020) 9:266. doi: 10.3390/pathogens9040266
210. Monteiro FJC, Mourão FRP, Ribeiro ESDA, Rêgo MOS, Frances PAC, Souto RNP, et al. Prevalence of dengue, Zika and chikungunya viruses in Aedes (Stegomyia) aEgypti (Diptera: Culicidae) in a mediumsized city, Amazon, Brazil. Rev do Inst Med Trop São Paulo. (2020) 62:e10. doi: 10.1590/S1678-9946202062010
211. Poulin B, Lefebvre G, Hilaire S, Despres L. Long-term persistence and recycling of Bacillus thuringiensis Israelensis spores in wetlands sprayed for mosquito control. Ecotoxicol Environ Safety. (2022) 243:114004. doi: 10.1016/j.ecoenv.2022.114004
212. Paupy C, Delatte H, Bagny L, Corbel V, Fontenille D. Aedes albopictus, an arbovirus vector: from the darkness to the light. Microbes Infect. (2009) 11:1177–85. doi: 10.1016/j.micinf.2009.05.005
213. Pluskota B, Storch V, Braunbeck T, Beck M, Becker N. First record of Stegomyia albopicta(Skuse)(Diptera:Culicidae)in Germany. Eur Mosq Bull. (2008) 26:e10. doi: 10.1590/S1678-9946202062010
214. Azevedo RSS, Oliveira CS, Vasconcelos PFC. Chikungunya risk for Brazil. Rev Saude Publica. (2015) 49:58. doi: 10.1590/S0034-8910.2015049006219
215. Nóbrega M, Araújo ELDL, Wada MY, Leite PLE, Dimech GS, Percio J. Surto de síndrome de Guillain-Barré possivelmente relacionado à infecção prévia pelo vírus Zika, Região Metropolitana do Recife, Pernambuco, Brasi. Epidemiol Serv Saude Rev Sist Unico Saude Bras. (2018) 27:e2017039. doi: 10.5123/S1679-49742018000200016
216. Paixão ES, Teixeira MG, Rodrigues LC. Zika, chikungunya and dengue: The causes and threats of new and re-emerging arboviral diseases. BMJ Glob Health. (2018) 3:e000530. doi: 10.1136/bmjgh-2017-000530
217. Aragão CF, Cruz ACR, Neto JPN, Monteiro HADO, Da Silva EVP, Da Silva SP, et al. Circulation of Chikungunya virus in Aedes aEgypti in Maranhão, Northeast Brazil. Acta Trop. (2018) 186:1–4. doi: 10.1016/j.actatropica.2018.06.022
218. World Health Organization (WHO). The mosquito (2020). Available online at: https://www.who.int/denguecontrol/mosquito/en.
220. Leong CS, Vythilingam I, Wong ML, Sulaiman WW, Lau YL. Aedes aEgypti(Linnaeus) larvae from dengue outbreak areas in Selangor showing resistance to pyrethroids but susceptible to organophosphates. Acta Trop. (2018) 185:115–26. doi: 10.1016/j.actatropica.2018.05.008
221. Alves GB, Melo FL, Oliveira EE, Haddi K, Costa LTM, Dias ML, et al. Comparative genomic analysis and mosquito larvicidal activity of four Bacillus thuringiensis serovar Israelensis strains. Sci Rep. (2020) 10:5518. doi: 10.1038/s41598-020-60670-7
222. Weiser JA. mosquito-virulent Bacillus sphaericus in adult Simulium damnosum from northern Nigeria. Zent Mikrobiol. (1984) 139:57–60. doi: 10.1016/S0232-4393(84)80033-5
223. Yuan Z, Neilsen-LeRoux C, Pasteur N, Delecluse A, Charles JF, Frutos R. Cloning and expression of the binary toxin genes of Bacillus sphaericus C3-41 in a crystal minus B. thuringiensis subsp. Israelensis. Acta Microbiol Sin. (1999) 39:29–35. doi: 10.1007/s002840010254
224. Ahmed I, Yokota A, Yamazoe A, Fujiwara T. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int J Syst Evol Microbiol. (2007) 57:1117–25. doi: 10.1099/ijs.0.63867-0
225. Rueda ME, López CC, García JJ, Fernandes EKK, Marreto RN, Luz C. Effect of ultraviolet-Aradiation on the production of Leptolegnia chapmanii (Saprolegniales: Saprolegniaceae)zoospores on dead Aedes aEgypti (Diptera: Culicidae) larvae and their larvicidal activity. J Invertebr Pathol. (2015) 130:133–5. doi: 10.1016/j.jip.2015.08.002
226. Rueda ME, Tavares I, López CC, García J. Leptolegnia chapmanii como alternativa biológica para el control de Aedes aEgypti. Biomédica. (2019) 39:798–810. doi: 10.7705/biomedica.4598
227. Gratz NG. Critical review of the vector status of Aedes albopictus. Med Veter Entomol. (2004) 18:215–27. doi: 10.1111/j.0269-283X.2004.00513.x
228. Moyes CL, Vontas J, Martins AJ, Ng LC, Koou SY, Dusfour I, et al. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PloS Negl Trop Dis PLoS Negl Trop Dis. (2021) 15:e0009084. doi: 10.1371/journal.pntd.0009084
229. Sanchis V. From microbial sprays to insect-resistant transgenic plants: history of the biospesticide Bacillus thuringiensis. A review. Agronm Sustain Dev. (2011) 31:217–23. doi: 10.1051/agro/2010027
230. Chen J, Aimanova KG, Gill SS. Aedes cadherin receptor that mediates Bacillus thuringiensis Cry11A toxicity is essential for mosquito development. PloS Negl Trop Dis. (2020) 14:1–23. doi: 10.1371/journal.pntd.0007948
231. Xiao Y, Wu K. Recent progress on the interaction between insects and Bacillus thuringiensis crops. Phil. Trans. R. Soc. B. (2019) 374:20180316. doi: 10.1098/rstb.2018.0316
232. De Haan L, Verweij W, Agsteribbe E, Wilschut J. The role of ADP-ribosylation and G(M1)-binding activity in the mucosal immunogenicity and adjuvanticity of the Escherichia coli heat-labile enterotoxin and Vibrio cholerae cholera toxin. Immunol Cell Biol. 71998:270–9. doi: 10.1046/j.1440-1711.1998.00745.x
233. Simmon CP, Ghaem-Magami M, Petrovska I, Lopez L, Chain BM, Williams NA, et al. Immunomodulation using bacterial enterotoxins. Scand J Immunol. (2001) 53:218–26. doi: 10.1046/j.1365-3083.2001.00884.x
234. Lycke N, Lebrero-Fernández C. ADP-ribosylating enterotoxins as vaccine adjuvants. Curr Opin Pharmacol. (2018) 41:42–51. doi: 10.1016/j.coph.2018.03.015
235. Verma P, Gandhi S, Lata K, Chattopadhyay K. Pore-forming toxins in infection and immunity. Biochem Soc Trans. (2021) 49:455–65. doi: 10.1042/BST20200836
236. Ulhuq FR, Mariano G. Bacterial pore-forming toxins. Microbiology. (2022) 168:1154. doi: 10.1099/mic.0.001154
237. Escartín-Gutiérrez JR, Ponce-Figueroa M, Torres-Vega MÁ, Aguilar-Faisal L. Figueroa-Arredondo Transcriptional Activation of a Pro-Inflammatory Response (NF-κB, AP-1, IL-1β) by the Vibrio cholerae Cytotoxin (VCC) Monomer through the MAPK Signaling Pathway in the THP-1 Human Macrophage Cell Line. P Int J Mol Sci. (2023) 24:7272. doi: 10.3390/ijms24087272
238. Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. (2011) 36:320–8. doi: 10.1016/j.tibs.2011.03.006
239. Vázquez-Padrón RI, González-Cabrera J, García-Tovar C, Neri-Bazán L, López-Revilla R, Hernández M, et al. Cry1Ac protoxin from Bacillus thuringiensis sp. kurstaki HD73 binds to surface proteins in the mouse small intestine. Biochem Biophys Res Commun. (2000) 29:271:54–58. doi: 10.1006/bbrc.2000.2584
240. Torres-Martínez M, Rubio-Infante N, García-Hernández AL, Nava-Acosta R, Ilhuicatzi-Alvarado D, Moreno-Fierros L. Cry1Ac toxin induces macrophage activation via ERK1/2, JNK and p38 mitogen-activated protein kinases. Int J Biochem Cell Biol. (2016) 78:106–15. doi: 10.1016/j.biocel.2016.06.022
241. Rubio-Infante N, Ilhuicatzi-Alvarado D, Torres-Martínez M, Reyes-Grajeda JP, Nava-Acosta R, González-González E. Moreno-Fierros L The Macrophage Activation Induced by Bacillus thuringiensis Cry1Ac Protoxin Involves ERK1/2 and p38 Pathways and the Interaction with Cell-Surface-HSP70. J Cell Biochem. (2018) 119:580–98. doi: 10.1002/jcb.26216
242. Vázquez-Padrón RI, Moreno-Fierros L, Neri-Bazán L, de la Riva GA, López-Revilla R. Intragastric and intraperitoneal administration of Cry1Ac protoxin from Bacillus thuringiensis induces systemic and mucosal antibody responses in mice. Life Sci. (1999) 64:1897–912. doi: 10.1016/s0024-3205(99)00136-8
243. Guerrero GG, Dean DH, Moreno-Fierros. Structural Implication of the induced immune response by Bacillus thuringiensis Cry proteins: role of the N-terminal region. Mol Immunol. (2004) 41:1177–83. doi: 10.1016/j.molimm.2004.06.026
244. Rojas-Hernández S, Rodriguez-Monroy MA, López-Revilla R, Reséndiz-Albor AA, Moreno-Fierros. Intranasal coadministration of the Cry1Ac protoxin with amoebal lysates increases protection against Naegleria fowleri meningoencephalitis. Infect Immun. (2004) 72:4368–75. doi: 10.1128/IAI.72.8.4368-4375.2004
245. Legorreta-Herrera M, Oviedo MR, Moreno-Fierros L. Pretreatment with Cry1Ac protoxin modulates the immune respone,and increases the survival of Plasmodium-infected CBA/Ca mice. J BioMed Biotech. (2010) 2010:1–11. doi: 10.1155/2010/198921
246. González-González E, García-Hernández AL, Flores-Mejía R, López-Santiago R, Moreno-Fierros L. The protoxin Cry1Ac of Bacillus thuringiensis improves the protection conferred by intranasal immunization with Brucella abortus RB51 in a mouse model. Vet Microbiol. (2016) 175:382–8. doi: 10.1016/j.vetmic.2014.11.021
247. Ibarra-Moreno S, García-Hernández AL, Moreno-Fierros L. Coadministration of protoxin Cry1Ac from Bacillus thuringiensis with metacestode extract confers protective immunity to murine cysticercosis. Parasite Immunol. (2014) 36:266–70. doi: 10.1111/pim.12103
Keywords: plasmids, Cry endotoxins, Cyt2A toxins, dipterans, mammals cells, bioinsecticides, adjuvants
Citation: Guerrero GG, Favela-Hernandez JM and Balderas-Renteria I (2024) Plasmid vector(s) in Bacillus thuringiensis harbor genes for insect pest control and for neglected infectious diseases in humans. Front. Trop. Dis 5:1416187. doi: 10.3389/fitd.2024.1416187
Received: 11 April 2024; Accepted: 27 June 2024;
Published: 09 September 2024.
Edited by:
Seham Hendawy, National Research Centre, EgyptReviewed by:
Karina Salvatierra, Universidad Nacional de Misiones, ArgentinaNancy Fayad, Saint Joseph University, Lebanon
Copyright © 2024 Guerrero, Favela-Hernandez and Balderas-Renteria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gloria G. Guerrero, Z2xvcmlhZ3VpbGxlcm1pbmFAdWF6LmVkdS5teA==; Z2xvZ3VlcnJlcm85QGdtYWlsLmNvbQ==
 Gloria G. Guerrero
Gloria G. Guerrero Juan M. Favela-Hernandez2,3
Juan M. Favela-Hernandez2,3