
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis. , 03 March 2023
Sec. Neglected Tropical Diseases
Volume 4 - 2023 | https://doi.org/10.3389/fitd.2023.1067439
This article is part of the Research Topic Advances in the Molecular Epidemiology and Diagnostics of Leprosy and other Mycobacterial Diseases View all 6 articles
 Maria Carolina Sisco1,2*
Maria Carolina Sisco1,2* Amanda N. Brum Fontes1
Amanda N. Brum Fontes1 Lucibel Crespo Lessmann3
Lucibel Crespo Lessmann3 Elsa Rada4
Elsa Rada4 Yrneh Y. Prado Palacios1
Yrneh Y. Prado Palacios1 Sidra E. G. Vasconcellos1
Sidra E. G. Vasconcellos1 Jacobus H. de Waard5,6
Jacobus H. de Waard5,6 Philip N. Suffys1
Philip N. Suffys1Leprosy is a neglected tropical disease that leads to physical disability and social discrimination. The active surveillance of new cases and vigilance for drug resistance can decrease the incidence, and improve the clinical outcomes of people affected by it. We analyzed, with molecular biology techniques, a set of skin biopsy samples from 25 Venezuelan patients. The patients had been diagnosed with leprosy in 2014 and early 2015, and 15 were relapse cases. The samples were tested for molecular drug resistance to dapsone, rifampicin, and fluoroquinolones. In addition, we performed molecular epidemiology analysis through multiple-locus variable-number tandem repeat analysis (MLVA) and single nucleotide polymorphism (SNP) typing. We did not find evidence of drug resistance and 76% of the samples (n = 16) had isolates belonging to SNP type 3. Genotype profiles allowed us to rule out the possibility of re-infection in a patient with persistent symptoms after treatment, as well as that of household transmission in two more patients. Although our sampling is relatively small, very similar or even identical Mycobacterium leprae genotypes were observed in Miranda State. The presence of this cluster is highly suggestive of high rates of local transmission and, in turn, the need to better control this disease. Finally, the copy number distribution of minisatellite 18–8 in a considerable number of SNP type 3 strains strongly suggests the presence of a sublineage of this disease that is particular to Venezuela.
Hansen´s disease, or leprosy, is a chronic infectious disease endemic in tropical countries and caused by the Mycobacterium leprae and M. lepromatosis bacterial strains. The prevalence of this disease has decreased since the introduction of multidrug therapy treatment (MDT) in the 1980’s; however, the global registered prevalence at the end of 2021 was 133,802 cases, and the number of new cases detected was 140,594, which represents 10% increase in the detection rate in comparison to 2020 (1).
This disease can be classified based on clinical, immunological, and histopathological findings, resulting in two polar forms of the disease: (i) the tuberculous form (TL), characterized by a limited spread of bacilli with a small number of skin lesions and sensitivity change in the nerve pathways; and (ii) the lepromatous form (LL), characterized by an ineffective antibody response with multiple skin lesions and multiple compromised peripheral nerves (2). In addition, there are borderline forms of this disease, classified as borderline-tuberculoid (BT), borderline-lepromatous (BL), and borderline-borderline (BB) according to their proximity to one of the two polar forms (2). The World Health Organization (WHO) also proposed a classification based on the bacterial index (BI) observed in skin smears: namely paucibacillary patients (PB) with a BI lower than 2, and multibacillary patients (MB) with a BI equal to or higher than 2 (2).
Multidrug therapy treatment (MDT), which was introduced by WHO, consists of a combination of rifampicin, clofazimine, and dapsone for MB leprosy patients, and rifampicin and dapsone for PB leprosy patients (3, 4).
In Venezuela, which had a registered prevalence of 705 cases and 239 new cases of leprosy detected by the end of 2021 (1), there is a continuous effort to diagnose and eradicate the disease despite the ongoing medical health crisis caused by the COVID-19 pandemic. Of these new cases reported to the WHO, six (2.5%) were patients under 15 years of age, an indication of active transmission. In comparison, the data reported in 2019 (i.e., before the onset of the coronavirus pandemic) included values for prevalence and newly registered cases of 847 and 291, respectively (5). This slight reduction in prevalence and newly registered case figures in 2021 is believed to be a consequence of the overall decline in notifications of other diseases not related to COVID-19, as well as the mobility restrictions imposed during the quarantine period, which meant that patients from rural areas were unable to reach main leprosy diagnostic centers.
In 2021, 16 relapse cases, that is cases identified after the completion of treatment, were reported in Venezuela (1); however, actual data concerning drug-resistance rates in the country are currently unknown. Only one strain harboring a mutation in the folp1 (Pro55Leu) gene, which is thought to cause resistance to dapsone, was reported in 2011 (6).
Molecular typing of the M. leprae strain is currently performed by combining two techniques: multiple-locus variable-number tandem repeat analysis (MLVA) and single nucleotide polymorphism (SNP) typing. MLVA has proven to be useful in the studies of local or intrafamily transmission, whereas SNP typing has been used to analyze the global distribution of M. leprae strains (7), with each SNP type being associated with a restricted geographical area and routes of human migration (8). Relatedly, the prevalence of SNP type 3 strains in the Americas, including Venezuela, indicates that leprosy was introduced to these locations with the arrival of European settlers during the colonial era. However, the only studies that have conducted a surveillance of the SNP types circulating in Venezuela were published in 2005, 2009, and 2011 (6, 8, 9), with no information regarding the variable number tandem repeat (VNTR) profiles of the M. leprae strains isolated.
In this study, we aimed to investigate the presence of antibiotic resistance in a cohort of 25 leprosy patients, including 15 relapse cases, diagnosed in 2014 and early 2015. We also studied the genetic variability of the M. leprae strains that were isolated using MLVA and SNP analysis.
Skin biopsy samples were obtained from 25 patients (20 males and five females) at the Dermatology Department of the Biomedicine Institute “Dr. Jacinto Convit” in Caracas, Venezuela, between 2014 and early 2015. Samples were stored at −80°C until further processing. The patients’ ages ranged from 22 to 85 years, and 15 had received previous treatment with clofazimine, dapsone, and rifampicin for variable periods (i.e., between 2 and 30 months) (Supplementary Material). One patient had two biopsy samples taken: one obtained before the treatment, and another taken after a 12-month course of antibiotics. In total, 26 samples were submitted for DNA extraction and genotyping at the Laboratory of Molecular Biology Applied to Mycobacteria at the Oswaldo Cruz Institute in Rio de Janeiro, Brazil. The clinical and geographical data (i.e., place of residence) for each patient are detailed in the Supplementary Material.
DNA extraction was performed using the DNeasy® Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s procedure (10). DNA from the M. leprae strain NHDP63 was used as a control in all the molecular assays.
Segments of the genes rpoB, folP, gyrA, and gyrB were amplified using the primers described in the Supplementary Material (11–13). PCR reactions were performed in a mixture of 50 µl containing 10× buffer, 1 U of Taq DNA polymerase (Invitrogen, Waltham, MA, USA), 2.5 mM of each deoxynucleoside triphosphate (dNTP), 1.5 mM of magnesium chloride (MgCl2) and 10µM of each primer pair. Two μl of DNA was used for the reaction. PCR conditions were as follows: 94°C for 3 min, followed by six touchdown cycles at temperatures of 94°C for 45 sec, 63–68°C for 45 sec, and 72°C for 90 sec. Then, the reaction continued with 35 cycles at temperatures of 94°C for 45 sec, 62°C for 45 sec, 72°C for 90 sec, with a final step at 72°C for 10 min. PCR products were visualized on a 2% agarose gel and purified using the ChargeSwitch™ PCR Clean-Up Kit (Invitrogen, Waltham, MA, USA). The purified product was sequenced using a BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA) to identify the gene mutations associated with drug resistance.
Sixteen VNTR loci grouped into four PCR reactions were amplified using the Qiagen Multiplex PCR Kit. The list of primers used, including fluorescent 5′-labeled forward primers, is described in the Supplementary Material (14). The final concentration of each primer was 0.2 µM. The cycling conditions were set at temperatures of 95°C for 15 min, followed by 40 cycles at temperatures of 94°C for 30 sec, 60°C for 90 sec, 72°C for 90 sec, and a final step at 72°C for 10 min. The amplicons obtained were diluted (1 : 30) and mixed with 12 µl of Formamide Hi-Di™ (Applied Biosystems, Waltham, MA, USA) and 0.3 µl of LIZ-500 DNA standard (Applied Biosystems). The program Peak Scanner™ (Applied Biosystems, Waltham, MA, USA) was used to analyze the fragments obtained. The copy number for each of the alleles was input into a Microsoft Excel® (Microsoft Corporation, Redmond, WA, USA) file and the allelic diversity (h) at each of the 16 loci was calculated using the equation h = 1 – ∑x2, where x is the frequency of each allele at the locus.
Three pairs of previously described primers (8) were used to amplify SNPs at positions 14,676, 1,642,875, and 2,935,685 bp. The PCR reaction was 50 µl of a mixture containing buffer 10×, 1.5 mM MgCl2, 200 µM of each dNTP, 2.5 U of Taq DNA polymerase (Invitrogen), 0.2 µM of each primer, and 2.5 µl of DNA. The cycling reaction consisted of temperatures set at 94°C for 3 min, followed by 45 cycles at temperatures of 94°C for 1 min, 55°C for 1 min, 72°C for 2 min, and a final step at 72°C for 10 min. The PCR product of position 2,935,685 bp was digested with the restriction enzyme BstU (New England Biolabs, Ipswich, MA, USA) at 60°C for 1 hour, and the digestion products were visualized on a 3% agarose gel. The amplification product of position 14,676 bp was digested with the enzyme SmiI (New England Biolabs, Ipswich, MA, USA) at 55°C for 1 hour. The amplification product of position 1,642,875 bp was sequenced with the same primers used for the amplification.
A distance pairwise similarity matrix using the categorical similarity coefficient was constructed, then a phylogenetic tree using the unweighted pair group method with arithmetic averages (UPGMA) and a minimum spanning tree (MST) were constructed using the software BioNumerics v. 7.6.3 (bioMérieux, Marcy-l’Étoile, France).
Out of the 26 samples analyzed, 23 could be sequenced and none of them harbored mutations in codons 516, 526, 531, or 533 of the rpoB gene. The gene folp1 could be analyzed in 19 samples. Mutations on codons 53 and 55, that is, those that could confer resistance to dapsone were not detected. Regarding fluoroquinolone resistance, the genes gyrA and gyrB could be analyzed in 24 and 25 samples, respectively; however, no evidence of a gene mutation was found.
We could assign a major SNP type to 21 of the 26 samples submitted for analysis; five samples had inconclusive results because of a lack of amplification in the PCR. Of these 21 samples, 76% (n = 16) had isolates identified as SNP type 3, 14% (n = 3) were identified as SNP type 4, and 10% (n = 2) were classified as either SNP type 1 or 2.
A genotype consisting of all 16 loci could be assigned to samples from 13 patients (Table 1). Figure 1 consists of a geographical map with the location of these patients, most of whom were living in the north of the country, where the population density is high (only one patient was located in Falcón State, in the west of the country). The remaining samples had 15 (n = 5), 14 (n = 1), 13 (n = 3), 12 (n = 1), 11 (n = 1), 9 (n = 1), and eight loci (n = 1) successfully amplified. We believe this lack of amplification was because of a common issue in studies involving M. leprae, that is, DNA extraction from biopsy samples being made difficult by the samples’ degeneration during the storage process. Also, the samples in the study were taken in Venezuela and later transported to Brazil, where the final molecular analysis was performed. Two of the biopsy samples were split in half and processed separately as technical replicates (making a total of 15 samples to be used in this part of the study). One of the replicates differed by one copy at locus AT15, and by two copies at loci 12–5. The other replicate only differed by two copies at locus TA18.
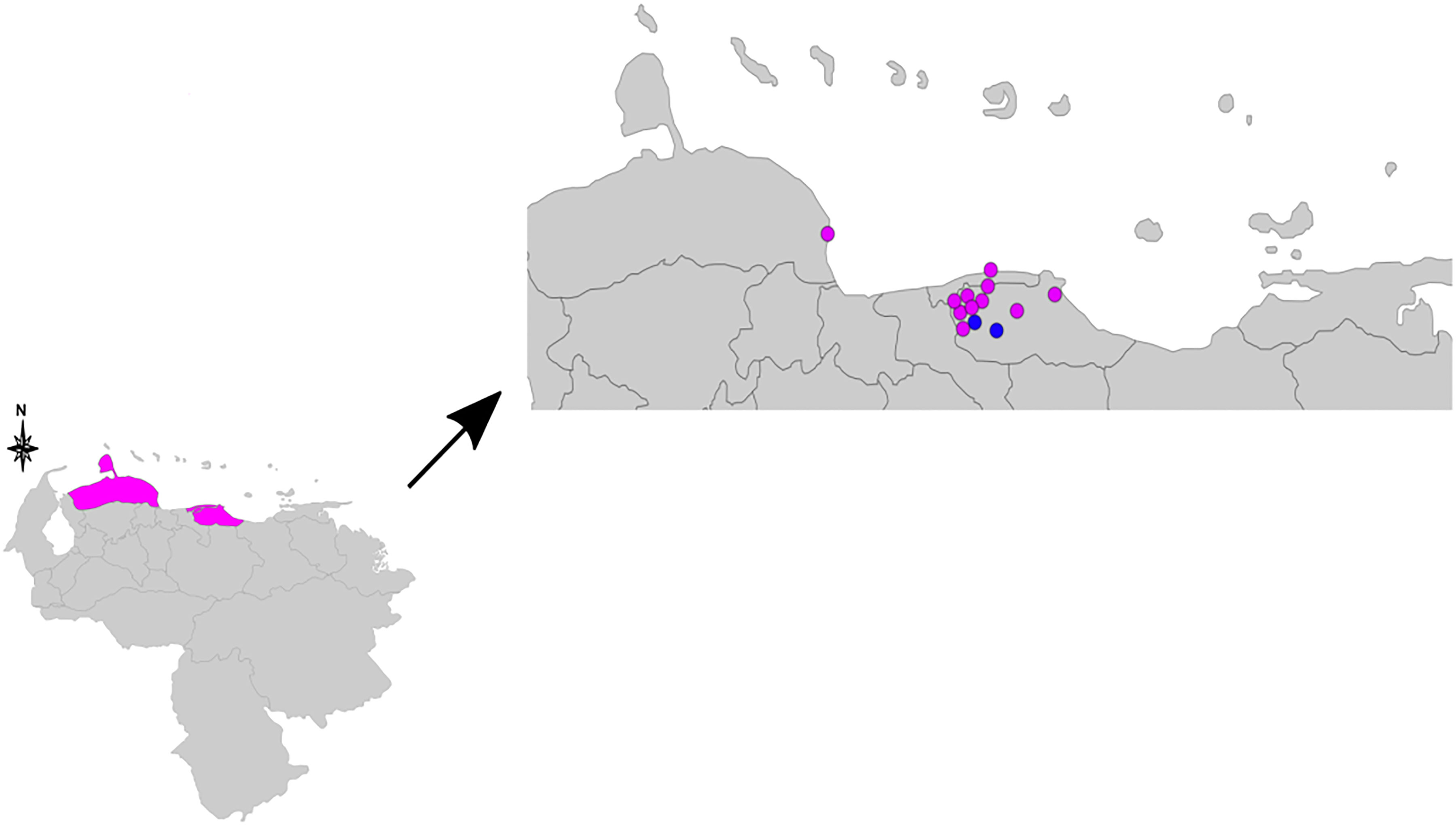
Figure 1 Geographical map indicating the residence of the 13 patients used for the clustering analysis. The pink dots represent samples of single nucleotide polymorphism (SNP) type 3, and the blue dots samples of SNP types 1 or 2. The three pink colored states in the lower map represent Falcón State, Distrito Capital, and Miranda State (from left to right). SNP, single nucleotide polymorphism.
The allelic diversity index (h) was calculated for the 16 loci using data from the 15 samples for which a complete MLVA profile was obtained. Allelic diversity ranged from 0 (locus 6–3) to 0.82 (locus TA18). The most polymorphic loci were AT17 and TA18, with nine alleles each. The least allelic variability was observed in loci 6–3, GGT-5, AC8b, 21–3, 23–3, 27–5, and 12–5 (Table 2).
One patient had one sample taken before and after 12 months of treatment, respectively; both samples had identical SNP types and did not differ at any VNTR loci, demonstrating a case of bacterial persistence, with no evidence of acquired resistance. Another two patients were brothers living in different states of the country at the time of sampling, and, although they had the same SNP type, the isolates identified differed at loci GTA9, AC8A, AC9, and TTC/GAA21 (loci AT17, AT15, and TA18 were excluded because of their high variability), evidencing a different source of infection for the samples taken from each brother.
We also observed that most of the Venezuelan isolates have 10 copies of loci 18–8 (12 of 21 patients that had isolates with a successful amplification of these loci). All of these belong to the major SNP type 3, an allele that has not been identified before and is not present in our local VNTR database that includes > 2,000 strains with this allele from 10 different countries (unpublished data). This suggests the presence of a new sublineage of leprosy in Venezuela.
The UPGMA and MST trees are presented in Figure 2. We identified the presence of a cluster of four strains in Miranda State, which is a densely populated area near the capital located in the north of the country.
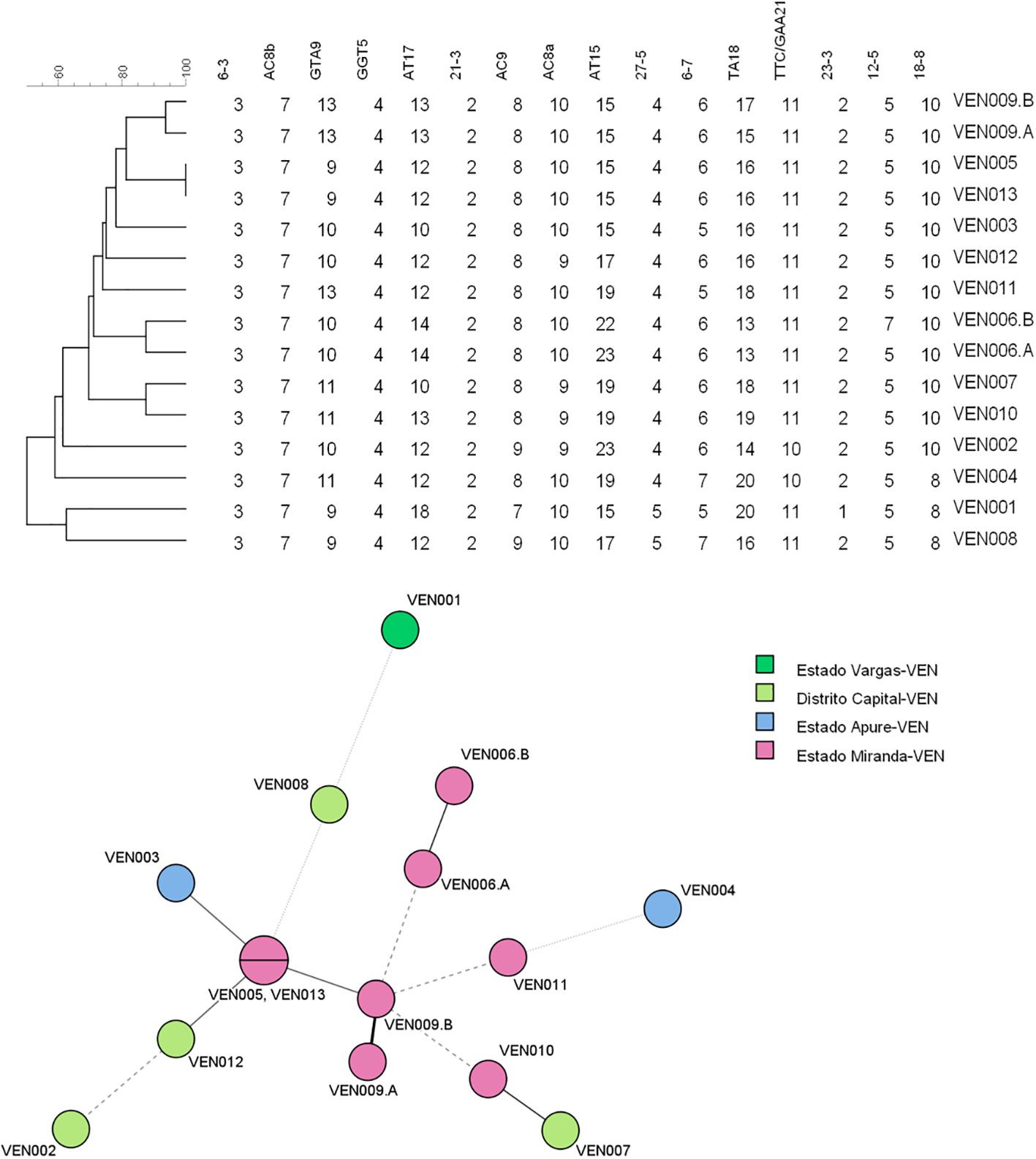
Figure 2 Unweighted pair group method with arithmetic averages (UPGMA) tree and minimum spanning tree (MST) compiled from the variable-number tandem repeat (VNTR) data of the 13 patients used for the clustering analysis. MST, minimum spanning tree; UPGMA, unweighted pair group method with arithmetic averages; VNTR, variable number of tandem repeats.
The control and understanding of many aspects of leprosy remains challenging, especially in developing countries. Our study is the second to conduct drug susceptibility and resistance surveillance of M. leprae in Venezuela. In this study with 25 patients, 15 of which were relapse cases, we did not find evidence for drug resistance; however, more samples will be analyzed in the future. On a global level, the detection of drug resistance is not routinely carried out; indeed, in a study conducted by the WHO in 2021, among the 143 countries reporting their leprosy statistics, only eight countries provided data on antimicrobial resistance. Among 3,452 samples, 51 (1.47%) were resistant to rifampin, 49 (1.42%) to dapsone, three (0.087%) to ofloxacin, and four (0.11%) to more than one drug (1). No information regarding the drug-related mutations was provided.
Drug and multidrug-resistant cases of leprosy have been reported worldwide (6, 15–20), including in three Latin American countries, namely Colombia, Brazil, and Venezuela. In Colombia, a 10-year-long survey including 243 multibacillary patients revealed that 3.7% of the isolates had mutations in the rpoB gene (Asp441Tyr, Arg505Trp, Ser458Met, Ala426Thre, and Ser456Leu); 2.3% had mutations in the folP1 gene (Thr53Ala, Pro55Leu, and Asp91His); and 0.5% had a mutation in the gyrA gene (Ala107Leu). Importantly, 50% of drug-resistant leprosy patients resided in the Andean Region, more specifically in Cundinamarca and Santander, where two leprosariums are located (19). In Brazil, a hyperendemic country, a survey conducted between 2018 and 2020 that included 1,183 patients showed that 1.4% of patients had strains resistant to one or more drugs, with two patients carrying isolates resistant to both dapsone and rifampin (folP1 Pro55Arg and rpoB Ser456Leu), indicating cases of multidrug resistance (21). Lastly, in Venezuela, only one case of antibiotic resistance, associated with resistance to dapsone, has been documented, namely in a relapse case in which a mutation in the folp1gene was detected (Pro55Leu) (6).
Although for epidemiological studies the main limitation of our study is the small number of samples, we performed genotyping. In our study, we found that the most prevalent genotype in Venezuela is SNP type 3. This genotype is also the most prevalent in the Americas as a result of European colonization (8). Only three isolates in our cohort belonged to SNP type 4, and it is believed that this genotype was introduced into the Caribbean and South America through the slave trade from West Africa (8). Cardona-Castro et al. identified the allele combinations in Colombia that are associated with particular SNP types. In the case of isolates belonging to SNP type 3, the most common alleles in loci 27–5 and 12–5 were four and five copies, respectively, whereas the opposite (five and four copies, respectively) was observed for strains classified as SNP type 1, 2, or 4 (22). Because of the geographical proximity of Colombia to Venezuela, we suggested that this same pattern could be encountered in Venezuela. Indeed, in our study, we found 16 strains belonging to SNP type 3, with four and five copies in loci 27–5 and 12–5, respectively. SNP type 3 strains with this allele combination have been associated with a European origin, supporting the idea that leprosy was brought here by colonialist settlers (22, 23).
The genotyping of M. leprae samples from two brothers with leprosy who were born in Colombia, but who at the time of diagnosis were living in two different states in Venezuela, evidenced a different source of infection for each sample. Nonetheless, the genetic background of patients with leprosy and its co-relation with susceptibility in the initial stages of infection, and the possible development of a reactional state has been reviewed recently, suggesting a possible family susceptibility to the disease (24). Within our sample population, there was also a relapse case in a lepromatous patient who still presented with skin lesions after taking four months of MDT and then again after 12 months of treatment, the VNTR profiles of both strains were the same. Hence, the hypothesis of re-infection was rejected. The possibility of drug resistance was also ruled out for the second strain. However, because the patient was from a remote area of Venezuela, we are not certain if, in fact, he adhered to his treatment plan rigorously.
Another finding of our work was the fact that there seems to be a high number of SNP type 3 strains harboring 10 copies of loci 18–8, and this has not been reported before. The fact that this allele is characterized by a very particular copy number distribution is highly suggestive of there being a particular (new) sublineage of leprosy in the country. Future studies could confirm this using wholegenome sequencing. With regard to the transmission of M. leprae, we observed a cluster of four strains in Miranda State, which is a densely populated area near the capital, located in the north of the country. No direct contact between the affected patients could be established; nevertheless, the presence of this cluster is highly suggestive of high rates of local transmission and in turn, the need to better control this disease. Our surveillance of newly diagnosed and relapse cases for the presence of possible drug resistance is set to continue; a new set of biopsy samples is expected to be analyzed soon.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Departamento de Dermatologia, Servicio Autónomo Instituto de Biomedicina Dr. Jacinto Convit, Caracas, Venezuela. The patients/participants provided their written informed consent to participate in this study.
MS, AF, SV, and YP performed all the analyses. LL, ER, and JW collected the biopsy samples. JW, PS, and MS wrote and reviewed the manuscript. JW and PS conceived the study. All authors contributed to the article and approved the submitted version.
This work received financial support from the United Nations University Program for Biotechnology in Latin America and the Caribbean. This work has also been supported by the following Brazilian research agencies: CNPq (grants: 307474/2020–8 - PQ 2021; 310418/2016–0 - PQ 2016; 207422/2014–1 - PDE 2014) and Faperj (grants: 26/201.115/2022, 26/204.546/2021, and 26/ 210.877/2019).
We thank the patients and staff from the Leprosy Department of Biomedicine Institute for their invaluable participation and support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2023.1067439/full#supplementary-material
1. World Health Organization. Weekly epidemiological record, Vol. 36. (2022). pp. 429–50. Geneva, Switzerland.
2. Lastória JC, de Abreu MAMM. Leprosy: Review of the epidemiological, clinical, and etiopathogenic aspects - part 1. Bras Dermatol (2014) 89(2):205–18. doi: 10.1590/abd1806-4841.20142450
3. Lastória JC, de Abreu MAMM. Leprosy: A review of laboratory and therapeutic aspects - part 2. Bras Dermatol (2014) 89(3):389–401. doi: 10.1590/abd1806-4841.20142460
4. White C, Franco-Paredes C. Leprosy in the 21st century. Clin Microbiol Rev (2015) 28(1):80–94. doi: 10.1128/CMR.00079-13
5. World Health Organization. Weekly epidemiological record, Vol. 2016. (2020). pp. 417–40. Geneva, Switzerland.
6. Singh P, Busso P, Paniz-Mondolfi A, Aranzazu N, Monot M, Honore N, et al. Molecular drug susceptibility testing and genotyping of mycobacterium leprae strains from south America. Antimicrob Agents Chemother (2011) 55(6):2971–3. doi: 10.1128/AAC.00201-11
7. Avanzi C, Singh P, Truman RW, Suffys PN. Molecular epidemiology of leprosy: An update. Infect Genet Evol (2020) 86:104581. doi: 10.1016/j.meegid.2020.104581
8. Monot M, Honore N, Garnier T, Araoz R, Coppée JY, Lacroix C, et al. On the origin of leprosy. Sci (2005) 308:1040–2. doi: 10.1126/science/1109759
9. Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, et al. Comparative genomic and phylogeographic analysis of mycobacterium leprae. Nat Genet (2009) 41(12):1282–9. doi: 10.1038/ng.477
10. Haendiges J. Manual DNA extraction using Qiagen DNeasy blood and tissue kit(2020) . doi: 10.17504/protocols.io.bi4dkgs6
11. Ramasoota P, Wongwit W, Sampunachot P, Unnarat K, Ngamying M, Svenson SB. Multiple mutations in the rpoB gene of mycobacterium leprae strains from leprosy patients in Thailand. Southeast Asian J Trop Med Public Health (2000) 31(3):493–7.
12. You EY, Kang TJ, Kim SK, Lee SB, Chae GT. Mutations in genes related to drug resistance in mycobacterium leprae isolates from leprosy patients in Korea. J Infect (2005) 50(1):6–11. doi: 10.1016/j.jinf.2004.03.012
13. Emmanuelle C, Bonnafous P, Evelyne P, Sougakoff W, Baohong J, Vincent J. Molecular detection of rifampin and ofloxacin resistance for patients who experience relapse of multibacillary leprosy. Clin Infect Dis (2002) 34(1):39–45. doi: 10.1086/324623
14. Jensen RW, Rivest J, Li W, Vissa V. DNA Fingerprinting of Mycobacterium leprae strains using variable number tandem repeat (VNTR) - fragment length analysis (FLA). J Vis Exp (2011) (53):e3104. doi: 10.3791/3104
15. Chauffour A, Lecorche E, Reibel F, Mougari F, Raskine L, Aubry A, et al. Prospective study on antimicrobial resistance in leprosy cases diagnosed in France from 2001 to 2015. Clin Microbiol Infect (2018) 24(11):1213.e5–1213.e8. doi: 10.1016/j.cmi.2018.06.004
16. Kai M, Phuc NHN, Nguyen HA, Pham THBD, Nguyen KH, Miyamoto Y, et al. Analysis of drug-resistant strains of mycobacterium leprae in an endemic area of Vietnam. Clin Infect Dis (2011) 52(5):127–32. doi: 10.1093/cid/ciq217
17. Rocha ADS, Cunha MDG, Diniz LM, Salgado C, Aires MAP, Nery JA, et al. Drug and multidrug resistance among mycobacterium leprae isolates from Brazilian relapsed leprosy patients. J Clin Microbiol (2012) 50(6):1912–7. doi: 10.1128/JCM.06561-11
18. Rosa PS, D’Espindula HRS, Melo ACL, Fontes ANB, Finardi AJ, Belone AFF, et al. Emergence and transmission of drug-/Multidrug-resistant mycobacterium leprae in a former leprosy colony in the Brazilian Amazon. Clin Infect Dis (2020) 70(10):2054–61. doi: 10.1093/cid/ciz570
19. Beltrán-Alzate C, Díaz FL, Romero-Montoya M, Sakamuri R, Li W, Kimura M, et al. Leprosy drug resistance surveillance in Colombia: The experience of a sentinel country. PloS Negl Trop Dis (2016) 10(10):1–12. doi: 10.1371/journal.pntd.0005041
20. Mahajan NP, Lavania M, Singh I, Nashi S, Preethish-Kumar V, Vengalil S, et al. Evidence for mycobacterium leprae drug resistance in a Large cohort of leprous neuropathy patients from India. Am J Trop Med Hyg (2020) 102(3):547–52. doi: 10.4269/ajtmh.19-0390
21. Andrade ESN, Brandão JG, da Silva JS, Coriolano CRF, Rosa PS, Moraes MO, et al. Antimicrobial resistance among leprosy patients in Brazil: Real-world data based on the national surveillance plan. Antimicrob Agents Chemother (2022) 66(5):e0217021. doi: 10.1128/aac.02170-21
22. Cardona-Castro N, Cortés E, Beltrán C, Romero M, Badel-Mogollón JE, Bedoya G. Human genetic ancestral composition correlates with the origin of mycobacterium leprae strains in a leprosy endemic population. PloS Negl Trop Dis (2015) 9(9):1–16. doi: 10.1371/journal.pntd.0004045
23. Fontes ANB, Lima LNGC, Mota RMS, Almeida RLF, Pontes MA, Gonçalves H de S, et al. Genotyping of mycobacterium leprae for better understanding of leprosy transmission in fortaleza, northeastern Brazil. PloS Negl Trop Dis (2017) 11(12):e0006117. doi: 10.1371/journal.pntd.0006117
Keywords: Molecular epidemiology, MLVA, SNP, drug resistance, Mycobacterium leprae
Citation: Sisco MC, Brum Fontes AN, Lessmann LC, Rada E, Prado Palacios YY, Vasconcellos SEG, de Waard JH and Suffys PN (2023) Antimicrobial resistance and genotyping of Mycobacterium leprae in Venezuela. Front. Trop. Dis 4:1067439. doi: 10.3389/fitd.2023.1067439
Received: 11 October 2022; Accepted: 26 January 2023;
Published: 03 March 2023.
Edited by:
Yash Gupta, Mayo Clinic Florida, United StatesReviewed by:
Prince Asare, University of Ghana, GhanaCopyright © 2023 Sisco, Brum Fontes, Lessmann, Rada, Prado Palacios, Vasconcellos, de Waard and Suffys. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Carolina Sisco, Y2Fyb2xpbmFzaXNjb0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.