- 1Centre for Research in Infectious Diseases, Directorate of Research and Innovation, Mount Kenya University, Thika, Kenya
- 2Division of Malaria Research, Proteo-Science Center, Ehime University, Matsuyama, Japan
- 3School of Public Health, University of Alberta, Edmonton, AB, Canada
Pregnant women are particularly susceptible to Plasmodium falciparum malaria, leading to substantial maternal and infant morbidity and mortality. While highly effective malaria vaccines are considered an essential component towards malaria elimination, strides towards development of vaccines for pregnant women have been minimal. The leading malaria vaccine, RTS,S/AS01, has modest efficacy in children suggesting that it needs to be strengthened and optimized if it is to be beneficial for pregnant women. Clinical trials against pregnancy-associated malaria (PAM) focused on the classical VAR2CSA antigen are ongoing. However, additional antigens have not been identified to supplement these initiatives despite the new evidence that VAR2CSA is not the only molecule involved in pregnancy-associated naturally acquired immunity. This is mainly due to a lack of understanding of the immune complexities in pregnancy coupled with difficulties associated with expression of malaria recombinant proteins, low antigen immunogenicity in humans, and the anticipated complications in conducting and implementing a vaccine to protect pregnant women. With the accelerated evolution of molecular technologies catapulted by the global pandemic, identification of novel alternative vaccine antigens is timely and feasible. In this review, we discuss approaches towards novel antigen discovery to support PAM vaccine studies.
Background on pregnancy-associated malaria
Plasmodium falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi are the five major species of Plasmodium that cause malaria in humans. Malaria remains a global public health threat despite decades of intense control efforts (1). According to the World Health Organization (WHO)-2020 malaria report, there were 241 million malaria infections and 627,000 fatalities globally, with 14 million infections and 69,000 more deaths than in 2019. During the pandemic, nearly two-thirds of these excess deaths (47,000) were attributable to interruptions in malaria prevention, diagnosis and treatment (2). P. falciparum causes the majority of malaria infections and fatalities with most occurring in sub-Saharan Africa. The major disease burden is in children less than 5 years of age, pregnant women, and individuals with immunodeficiencies.
Despite immunity acquired after years of exposure, the risk of infection is the same for women of all gravidities (3). However, the risk of poor birth outcomes is increased in primigravid and secundigravid women because they lack protective antibodies against placental malaria parasites; such outcomes include maternal anemia, spontaneous abortions, premature delivery (gestation of less than 37 weeks) and intrauterine growth retardation (IUGR) (4–7). This is primarily driven by the accumulation of infected erythrocytes (iRBCs) in the vascular spaces of the placenta (8).
Immunopathogenesis of malaria in pregnancy
The sequestration of iRBCs in the vascular portion of the placenta causes placental malaria (9). These parasites express a surface ligand that is unique to placental malaria called VAR2CSA. VAR2CSA is a member of the P. falciparum erythrocyte membrane protein 1 (PfEMP1) family and binds to chondroitin sulfate A (CSA), a glycosaminoglycan found in the placenta and other cells that mediates sequestration of iRBCs in pregnancy-associated malaria (PAM) (10, 11). These parasites bind to CSA but not to other major surface ligands, notably CD36 and ICAM-1, whereas most parasite isolates from non-pregnant women do the contrary (12, 13). Parasite sequestration in the placenta triggers a transition in the cell-mediated immune response, often from Th2 to Th1, which is marked by an increase in production of pro-inflammatory cytokines (14). Although Th1-type responses are important for the elimination of the parasite through stimulation of cellular immune responses, overproduction of cytokines alters pregnancy (15). In addition, increased number of maternal phagocytic cells and fibrin in the intervillous space together with deposition of hemozoin in phagocytic leucocytes induce placental inflammation that is associated with low birth weight and maternal anemia (8).
Protective antibodies against PAM
There is a strong negative association between gravidity and poor birth outcomes from PAM (16). This suggests that protective immunity develops against PAM and is directed against a target that is pregnancy-specific and highly immunogenic (17). VAR2CSA meets these criteria and antibodies to this protein are associated with protection against poor pregnancy outcome (18). In a recent study in Benin, high antibody responses to the N-terminal region of VAR2CSA were associated with lower risk of infection and low birth weight highlighting the protective role of these antibodies in immunity to placental malaria (19). Furthermore, in another cohort, anti-VAR2CSA antibody responses involved cytophilic immunoglobulin G (IgG) 1 and IgG3 subclasses, with prevalence ranging from 28% IgG3 to 50% IgG1. During pregnancy, higher levels of VAR2CSA-specific total IgG and cytophilic IgG3 were consistently linked to higher birth weights, whereas high levels of IgG4 were linked to a lower risk of placental infections, an indicator that both cytophilic and non-cytophilic antibodies offer protection (20).
In a similar study in Kenya, plasma from pregnant women exposed to malaria inhibited more than 35% of parasite adhesion to CSA (21). As expected, only one of 47 primigravid women had anti-adhesion antibodies, whereas anti-adhesion activity was higher in secundigravid women. This was associated with increased newborn birth weight and gestational age but not maternal hemoglobin. In a different study, IgG levels but not IgM against parasite lysate were positively correlated with newborn weight increase (22). In a study conducted in Mozambique, IgG levels against placental and pediatric isolates, as well as recombinant VAR2CSA (DBL2X, DBL3X, DBL5 and DBL6 domains) and other blood-stage antigens correlated with normal neonatal weight and gestational age in women with PAM (23). In contrast, a recent systematic review suggests that anti-VAR2CSA antibodies may instead be markers of infection rather than correlates of protection (24).
Taken together, VAR2CSA appears to be an important target of protective immunity to PAM (25), but additional targets, and protective mechanisms could also exist. The majority of studies, including vaccine trials, have focused on understanding and/or evaluating the role played by VAR2CSA; this is the only antigen under consideration as a PAM vaccine currently.
Current status of PAM vaccines
Surface expression of VAR2CSA is restricted to CSA-adhering iRBCs, prompting development of vaccines targeting this protein (26). The leading VAR2CSA-based vaccines, PAMVAC and PRIMVAC (27), are designed to generate functional antibodies capable of blocking the binding of infected cells to CSA on placental syncytiotrophoblast as well as opsonizing iRBCs expressing VAR2CSA. Such a vaccine would protect pregnant women, especially primigravids, from adverse outcomes in PAM.
PAMVAC is derived from the VAR2CSA subunit spanning ID1-DBL2-ID2a of the FCR3 variant (28) while PRIMVAC is based on CSA-binding DBL1x-2x region of the 3D7-VAR2CSA allele (29). The clinical trials for PAMVAC (under the registration EudraCT 2015-001827-21; ClinicalTrials.gov NCT02647489) were carried out in Germany and in Benin. The vaccine was well tolerated and immunogenic, and elicited IgG antibodies that recognized the homologous variant of VAR2CSA and blocked adhesion of these parasites to CSA in vitro (30).
The clinical trial for PRIMVAC (registered with ClinicalTrials.gov, NCT02658253) was conducted in two phases, the first in 18 to 35-year-old women in France and the second in women who were naturally exposed to P. falciparum and nulligravid in a research center in Burkina Faso using the same formulations as for PAMVAC (31). Similarly, the vaccine was safe, immunogenic and elicited antibodies with high reactivity toward parasites expressing the VAR2CSA variant homologous to the one in the vaccine. However, there was no recognition or anti-adhesion activity against heterologous variants of VAR2CSA (31).
This poor recognition of antibodies to the PAM vaccines against heterologous VAR2CSA alleles is a significant limitation that must be overcome to generate a broadly effective vaccine against PAM. In a recent analysis of the var2csa gene in more than 2,000 P. falciparum field isolates across 23 geographically distant countries (32), Benavente et al. reported high nucleotide and haplotype diversity of VAR2CSA in African parasites compared to South East Asian parasites. Significant differences in the frequency of extra VAR2CSA copies was also observed in Oceania, West Africa, East Africa and Southeast Asia isolates at 88%, 35%, 29% and 21%, respectively. Surprisingly, insertion and deletions were observed within the DBL-2x domain, a major component of both PAMVAC and PRIMVAC vaccines (33). Further investigations are needed to shed light on whether such indels could impact the protein structure necessary for binding to CSA and/or its ability to induce the immune response. High polymorphism in var2csa was also observed in a strain from the Democratic Republic of Congo (34). These studies have clearly demonstrated genetic variation in var2csa, cementing the need to develop broad multivalent vaccine candidates that could be efficacious across different variants.
While most vaccines against PAM focus on VAR2CSA, vaccines that prevent liver stage infection would also have a profound impact on pregnant women. One vaccine, the radiation attenuated P. falciparum NF54 sporozoites (PfSPZ Vaccine), which demonstrated significant protection in African adults (35), is now being assessed in Mali for safety, immunogenicity, and potential protective efficacy in women of childbearing potential (clinicaltrials.gov; NCT03989102).
Identification of targets of broadly cross-reactive antibodies
A critical feature of a PAM vaccine is to elicit antibodies that mimic naturally acquired immunity and recognize diverse alleles of var2csa. Natural immunity may be mediated by a repertoire of antibodies to diverse epitopes (36) or increased affinity to conserved epitopes that are shared across variants. Doritchamou et al. revealed conserved epitopes in a single variant of VAR2CSA and these epitopes are targets of neutralizing antibodies, a promising discovery to develop a vaccine using a few selected variants (37). Moreover, VAR2CSA has several globally shared, cross-reacting polymorphic epitopes (38).
The choice of construct for a domain specific VAR2CSA vaccine is one that elicits antibodies that are broadly cross-reactive. The VAR2CSA domain NTS-DBL1-Id1-DBL2X elicits rabbit antibodies that completely inhibit the FCR3 strain of P. falciparum from binding to CSA to the same extent as antisera against full-length extracellular VAR2CSA (NTSDBL1X to DBL6) (39). Epitope mapping revealed that Id1-DBL2X in the N-terminal region of VAR2CSA contains an epitope that induces highly inhibitory antibodies (40). However, some domains (e.g. the VAR2CSA DBL6-epsilon domain expressed in HEK293) induce limited cross-reactive and blocking antibodies to CSA binding parasites (41). Immunization with the DBL5 recombinant protein elicits broadly cross-reactive antibodies against many parasite strains and clinical isolates from pregnant women whereas antibodies to DBL1 had limited breadth and only reacted with three or four parasite strains (42). Using flow cytometry to measure antibody recognition of the native protein, DBL3 and to a lesser extent DBL5, generated antibodies that are cross-reactive to different CSA-binding parasite lines whereas DBL6 antibodies were highly strain-specific (43). However, even antibodies to the full length VAR2CSA do not always inhibit binding of different placental cell lines to CSA (44). We believe that identification of other conserved antigens form novel targets can be investigated as potentially cross-protective vaccines (45).
One alternative approach is to identify conserved epitopes that are shared across different proteins that contain DBL domains. These epitopes may be related structurally and present in proteins with diverse biological functions. For example, antibodies to P. vivax Duffy Binding Protein (PvDBP) elicited during natural infection with P. vivax or following animal immunization with PvDBP, can cross-react with and inhibit adhesion of strains expressing different variants of VAR2CSA (46). Although these proteins are functionally unrelated, they have a common homologous ancestry and share conserved epitopes that can be exploited in vaccine design.
Identification of targets of complement fixing antibodies in PAM
Antibodies can activate the complement system to mediate effector functions against myriad developmental stages of the parasite life cycle (47) that are linked to protection against clinical malaria (48). New evidence suggests that these mechanisms may also contribute to vaccine-induced immunity. For example, the RTS,S vaccine can induce complement-fixing antibodies against the central repeat and C-terminal regions of CSP (49).
Antibodies from pregnant women naturally exposed to malaria also fix complement. Sera from a longitudinal cohort of pregnant women in Papua New Guinea identified VAR2CSA-DBL5 and DBL3 antibodies that fixed C1q and C3 (50). C1q protein mediated complement enhanced the ability of VAR2SCA antibodies to inhibit iRBC binding to CSA. Protective associations between antibody-mediated complement fixation and the risk of placental malaria infection was weaker for C1q-fixation of DBL3 when compared with C1q or C3 fixation of the DBL5 domain or placental-binding iRBCs, highlighting the immuno-dominant nature of these antigens (50). IgG1 and IgG3 subclasses are the dominant antibodies to placental-binding iRBCs and to recombinant VAR2CSA (51) with the ability to fix and activate human complement in PAM (52). However, the proportion of VAR2CSA-specific IgG in plasma that is involved in complement fixation relative to other effector functions remains to be determined.
Importantly, Opi et al. observed higher C1q and C3 fixation on genetically modified CS2 P. falciparum infected iRBCs (that lack surface expression of PfEMP1) among multigravida compared to primigravida (50). This demonstrated that lack of PfEMP1 expression did not eliminate complement fixation on iRBCs suggesting that other surface proteins such as other variable surface antigens (repetitive interspersed family (RIFIN), subtelomeric variable open reading frame (STEVOR), surface-associated interspersed gene family (SURFIN), could also be important targets of complement-fixing antibodies. Evaluation of these molecules warrant investigation to establish if they have a role in parasite sequestration in the placenta as they do in other microvascular spaces (53, 54). Importantly, antibodies to RIFIN can clear parasites in malarial infection (55). In summary, these studies have demonstrated that elevated complement-fixing antibodies are associated with a reduced risk of placental parasitemia probably by enhanced inhibition of iRBC adhesion to CSA, thus the role of complement in protection against PAM deserve further investigations.
Identification of new vaccine targets other than VAR2CSA
Recent studies by genome-wide transcript analysis of P. falciparum isolates freshly collected from placenta identified other proteins that are upregulated during placental malaria (56). Thus, other parasite proteins beside VAR2CSA may contribute to the pathogenesis of malaria during pregnancy (26) and acquisition of antibodies against these proteins may contribute to protective immunity. For example, P falciparum chondroitin sulfate A ligand (PfCSA-L) is a highly conserved novel molecule expressed on the surface of iRBCs that binds placental CSA and is peripherally associated with the outer surface of knobs through specific protein-protein interactions with VAR2CSA (57). Discovery and characterization of additional antigens that interact with VAR2CSA is an area that deserves prioritization for vaccine development.
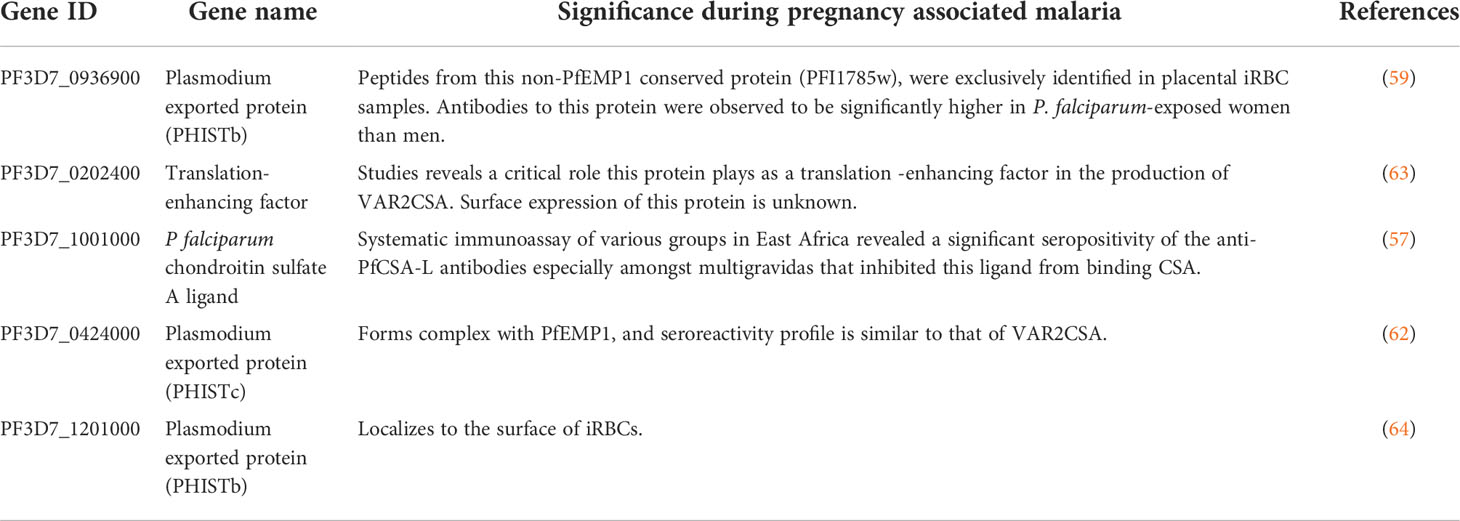
While direct detection of PfEMP1 on the surface of placental and CSA-adhering iRBCs by mass spectrometry has proved difficult owing to the low quantity of PfEMP1 in the iRBC membrane (58), several peptides were identified that corresponded to a highly conserved protein named PF3D7_0936900 (or PFI1785w) (59). PF3D7_0936900 and four other conserved genes including PFD1140w were identified in the transcription profiling study of placental parasites (60, 61). Antibodies to PF3D7_0936900 were significantly higher in P. falciparum-exposed women than men while seroreactivity profiles of PF3D7_0424000 (PFD1140w) were similar to those of VAR2CSA (62). Thus, these proteins are immunogenic and may be important for parasite development or adhesion in the placenta. It is therefore worth evaluating whether antibodies to these antigens can inhibit placental malaria infection (Table 1).
Immuno-epidemiological, functional studies and clinical trials in PAM vaccine discovery
A recent a systematic review of different epidemiological studies that assessed the relationship between pregnancy-specific malarial immunity and risk of malaria in pregnancy observed that prospective studies as well as standardized immunoassays are lacking (24). The analysis strongly suggested that to identify novel pregnancy-specific antigens, prospective cohort studies evaluating immune responses to many proteins in different epidemiological settings and throughout the gestational period using high-throughput, but standardized assays are needed. Antibodies to the identified antigens will then need to be evaluated using functional assays such as erythrocyte and CSA binding inhibition (anti-adhesion assay), iRBC cell-agglutination, phagocytosis, and complement fixation. The agglutinating and anti-adhesion assays are associated with reduced odds of placental infection (21), and the capacity of antibodies in pregnant women to opsonize phagocytosis CSA-binding iRBCs in vitro was observed (65) and can be explored for other antigens.
Flow cytometry-based assays have also been established with CSA-coated channels, CSA-expressing cell lines or primary trophoblast. In these studies, iRBCs flow through these channels and attach in settings that closely mimic blood vessels in vivo (66). Blocking these attachments correlate with the functional activities of the antibodies. Similarly, to recapitulate human placental organ physiology and function, the use of stem cell organoids and bioengineered placenta-on-a-chip models to construct biomimetic in vitro three-dimensional (3D) tissue or organ models warrant more consideration (67, 68). In this model, trophoblast cells and human umbilical vein endothelial cells are cultured on the opposite sides of a porous polycarbonate membrane allowing both maternal and fetal investigations. However, it remains unclear if these PAM-specific functional assays would correctly predict protective immunity in vaccine trials. In addition, the design of effective well-controlled studies with clear and globally acceptable endpoints need to be carefully considered, as recently proposed (69).
Leveraging wheat germ cell-free system and the mRNA-based vaccine approach to advance PAM vaccine research
The discovery of antigens that induce strain-transcending antibodies as well as new targets of complement fixing/activation antigen in PAM has been limited partly due to a lack of robust high-throughput screening platforms. This is hampered mainly by challenges associated with production of functional recombinant malaria antigens (70). However, the eukaryotic wheat germ cell-free protein system (WGCFS) overcomes these problems, as summarized by Kanoi et al., 2021 (71). In validation studies, WGCFS was used to generate more than 2500 recombinant proteins from various P. falciparum life cycle stages (72). This includes the synthesis of all high molecular weight VSAs including PfEMP1, SURFINs, RIFINs and STEVORs (73, 74). Similarly, more than 300 P. falciparum proteins were functionally annotated following generation of antibodies in immunized rabbits and mice, suggesting immunogenicity of the produced proteins (75).
The WGCFS technology has also been used to study vivax malaria in Thailand, Brazil and Papua New Guinea towards development of markers of recent infection. Antibody responses to 307 P. vivax recombinant proteins were measured at the onset of infection and at several points during the follow-up period (76). Antibodies with relatively longer-lived responses were observed in PNG where individuals had greater past exposure to P. vivax. In subsequent independent studies, eight proteins with specificity for detecting recent infections were identified (77). These findings further support WGCFS as well as high-throughput screening platforms as rational tools for production, screening and characterization of functional recombinant proteins that can be explored for induction of protective antibodies against falciparum and non-falciparum malaria during pregnancy. Thus, coupled with the rapid advancement of bioinformatics tools, WGCFS could support identification of potential placenta-specific or placental-binding ligands that induce antibodies that abrogate parasites sequestration in the placenta (Figure 1).
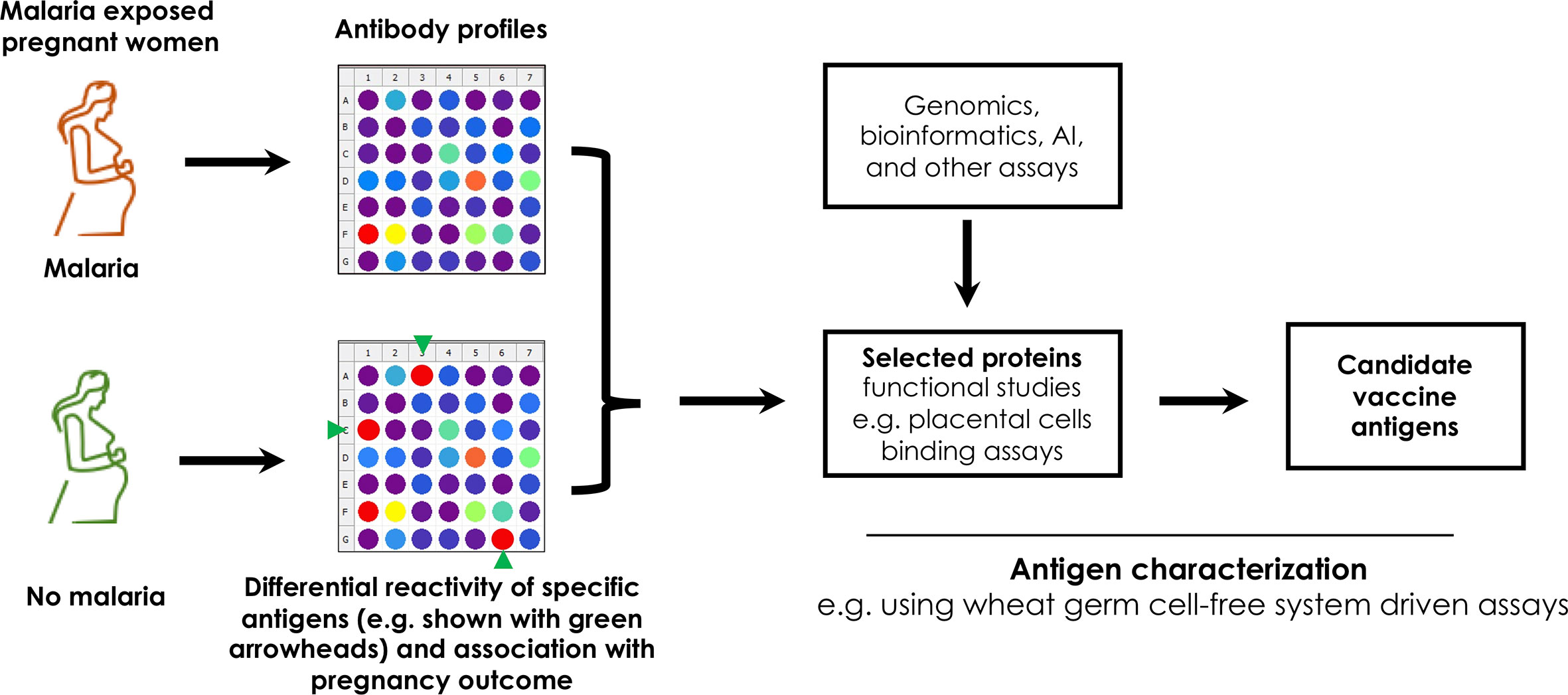
Figure 1 An illustration of an immunoscreening pipeline to identify targets of potentially protective antibodies from pregnant women with and without PAM. Unique proteins are represented by colored dots. The antigens may also be identified using complex genomic assays, statistical approaches such as artificial intelligence (AI), machine learning and expressed using robust platforms such as the wheat germ cell free system for functional studies.
We note that robust bioinformatics and artificial intelligence (AI) tools have been developed to support antigen selection, utilizing either sequence-dependent, structure-based, or ligand-based approaches (78). Such approaches, which have been applied in COVID-19 vaccine studies (79), combine multiple immuno-informatics approaches in integrated frameworks that include supervised machine learning. Recently, Rawal et al. used tools such as PsortB, WoLF PSORT, BLAST, HMMTop, ProtParam, FungalRV, NetCTL, VaxiJen 2.0, or IEDB tools to identify potentially immunogenic, stable and cross-protective antigens against Trypanosoma species (80). Other sequence-independent methods use auto cross-covariance (ACC), a mining method that transforms protein sequences into uniform equal-length vectors (81, 82). This has been extended to develop servers such as VaxiJen for the prediction of protective antigens (83). Such statistical approaches can be exploited in PAM to enhance the search for novel antigens.
Concurrently, numerous mRNA vaccine platforms have been developed and evaluated for immunogenicity (84–86). The ability to genetically modify the RNA sequences has hastened the production of translatable non-toxic synthetic mRNA and delivery systems (87). This approach was applied recently to the production of coronavirus vaccines by Moderna and Pfizer-BioNTech (88). These examples raise optimism for adoption of this technology in the production of vaccines for PAM.
Critical appraisal and summary
Understanding the complexity of naturally acquired immunity against PAM will inform antigen discovery and vaccine development. It is now clear that VAR2CSA is not the only antigen that is involved in PAM but there are other antigens that may be targets of protective immunity. With this knowledge, it is plausible to further query the plasmodium genome using emerging bioinformatics tools such as machine learning and artificial intelligence to identify possible conserved antigens that can be interrogated for development of broadly neutralizing vaccines. Application of eukaryotic WGCFS for generation of the recombinant proteins that induce immunogenic and protective antibodies need to be granted critical consideration. In addition, the advent of mRNA-based vaccine development platforms comes with high flexibility and rapid development while still inducing robust immune responses against specific targets. The mRNA technological approach is a great milestone for rapid in vivo production of full-length proteins and multivalent or multi-antigen vaccines. This has renewed the hope of quickly developing effective vaccines.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
BK is an EDCTP Fellow under EDCTP2 programme supported by the European Union grant number TMA2020CDF-3203-EndPAMAL. JG is supported by the African Academy of Sciences and the Royal Society FLAIR grant number FLR\R1\201314. The funding sources had no role in study design, collection, analysis, interpretation of data, and publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Espinoza JL. “Malaria resurgence in the americas: An underestimated threat”. Pathogens (2019) 8(1):1–4. doi: 10.3390/pathogens8010011
3. Brabin BJ. An analysis of malaria in pregnancy in Africa. Bull World Health Organ. (1983) 61(6):1005–16.
4. Tran EE, Tran EE, Cheeks ML, Kakuru A, Muhindo MK, Natureeba P, et al. “The impact of gravidity, symptomatology and timing of infection on placental malaria”. Malar. J (2020) 19:1–11. doi: 10.1186/s12936-020-03297-3
5. Verhoeff FH, Brabin BJ, Chimsuku L, Kazembe P, Broadhead RL. “Malaria in pregnancy and its consequences for the infant in rural malawi”. Ann Trop Med Parasitol (1999) 93(SUPPL. 1):S25–33. doi: 10.1080/00034983.1999.11813501
6. Lingani M, Zango SH, Valéa I, Valia D, Sanou M, Samandoulougou SO, et al. “Magnitude of low birthweight in malaria endemic settings of nanoro, rural Burkina Faso: A secondary data analysis”. Sci Rep 11(1):1–7. doi: 10.1038/s41598-021-00881-8
7. De Beaudrap P, Turyakira E, White LJ, Nabasumba C, Tumwebaze B, Muehlenbachs A, et al. “Impact of malaria during pregnancy on pregnancy outcomes in a Ugandan prospective cohort with intensive malaria screening and prompt treatment”. Malar. J (2013) 12:1–11. doi: 10.1186/1475-2875-12-139
8. Ismail MR, Ordi J, Menendez C, Ventura PJ, Aponte JJ, Kahigwa E, et al. “Placental pathology in malaria: A histological, immunohistochemical, and quantitative study”. Hum Pathol (2000) 31:85–93. doi: 10.1016/S0046-8177(00)80203-8
9. Brabin BJ, Romagosa C, Abdelgalil S, Menéndez C, Verhoeff FH, McGready R, et al. “The sick placenta - the role of malaria”. Placenta (2004) 25:359–78. doi: 10.1016/j.placenta.2003.10.019
10. Spliid CB, Toledo AG, Sanderson P, Mao Y, Gatto F, Gustavsson T, et al. “The specificity of the malarial VAR2CSA protein for chondroitin sulfate depends on 4-o-sulfation and ligand accessibility”. J Biol Chem (2021) 297:101391. doi: 10.1016/j.jbc.2021.101391
11. Salanti A, Staalsoe T, Lavstsen T, Jensen ATR, Sowa MPK, Arnot DE, et al. “Selective upregulation of a single distinctly structured var gene in chondroitin sulphate a-adhering plasmodium falciparum involved in pregnancy-associated malaria”. Mol Microbiol (2003) 49:179–91. doi: 10.1046/j.1365-2958.2003.03570.x
12. Fried M, Duffy PE. “Adherence of plasmodium falciparum to chondroitin sulfate a in the human placenta”. Sci (80-. ). (1996) 272(5267):1502–4. doi: 10.1126/science.272.5267.1502
13. Maubert B, Fievet N, Tami G, Boudin C, Deloron P. “Cytoadherence of plasmodium falciparum-infected erythrocytes in the human placenta”. Parasite Immunol (2000) 22:191–9. doi: 10.1046/j.1365-3024.2000.00292.x
14. Beeson JG, Duffy PE. “The immunology and pathogenesis of malaria during pregnancy”. Curr Top Microbiol Immunol (2005) 297:187–227. doi: 10.1007/3-540-29967-x_6
15. Taylor-Robinson AW, Smith EC. “A dichotomous role for nitric oxide in protection against blood stage malaria infection”. Immunol Lett (1999) 67:1–9. doi: 10.1016/S0165-2478(98)00148-5
16. Mutabingwa TK, Bolla MC, Li JL, Domingo GJ, Li X, Fried M, et al. “Maternal malaria and gravidity interact to modify infant susceptibility to malaria”. PloS Med (2005) 2:1260–8. doi: 10.1371/journal.pmed.0020407
17. Hviid L, Salanti A. “VAR2CSA and protective immunity against pregnancy-associated plasmodium falciparum malaria”. Parasitology (2007) 134:1871–6. doi: 10.1017/S0031182007000121
18. Ricke CH, Staalsoe T, Koram K, Akanmori BD, Riley EM, Theander TG, et al. “ plasma antibodies from malaria-exposed pregnant women recognize variant surface antigens on plasmodium falciparum -infected erythrocytes in a parity-dependent manner and block parasite adhesion to chondroitin sulfate A”. J Immunol (2000) 165:3309–16. doi: 10.4049/jimmunol.165.6.3309
19. Tuikue Ndam N, Denoeud-Ndam L, Doritchamou J, Viwami F, Salanti A, Nielsen MA, et al. “Protective antibodies against placental malaria and poor outcomes during pregnancy, benin”. Emerg Infect Dis (2015) 21:813–23. doi: 10.3201/eid2105.141626
20. Tornyigah B, d’Almeida T, Escriou G, Viwami F, Fievet N, Luty AJF, et al. “Plasmodium falciparum VAR2CSA-specific IgG subclass responses reflect protection against low birth weight and pregnancy-associated malaria”. Front Immunol (2021) 12:610305. doi: 10.3389/fimmu.2021.610305
21. Duffy PE, Fried M. “Antibodies that inhibit plasmodium falciparum adhesion to chondroitin sulfate a are associated with increased birth weight and the gestational age of newborns”. Infect Immun (2003) vol:6620–3. doi: 10.1128/IAI.71.11.6620-6623.2003
22. Mayor A, Dobaño C, Nhabomba A, Guinovart C, Jiménez A, Manaca MN, et al. “IgM and IgG against plasmodium falciparum lysate as surrogates of malaria exposure and protection during pregnancy”. Malar. J (2018) 17:1–10. doi: 10.1186/s12936-018-2331-4
23. Mayor A, Kumar U, Bardají A, Gupta P, Jiménez A, Hamad A, et al. “Improved pregnancy outcomes in women exposed to malaria with high antibody levels against plasmodium falciparum”. J Infect Dis (2013) 207:1664–74. doi: 10.1093/infdis/jit083
24. Cutts JC, Agius PA, Zaw Lin Powell R, Moore K, Draper B, et al. “Pregnancy-specific malarial immunity and risk of malaria in pregnancy and adverse birth outcomes: A systematic review”. BMC Med (2020) 18:1–21. doi: 10.1186/s12916-019-1467-6
25. Khunrae P, Dahlbäck M, Nielsen MA, Andersen G, Ditlev SB, Resende M, et al. “Full-length recombinant plasmodium falciparum VAR2CSA binds specifically to CSPG and induces potent parasite adhesion-blocking antibodies”. J Mol Biol (2010) 397:826–34. doi: 10.1016/j.jmb.2010.01.040
26. Fried M, Duffy PE. “Designing a VAR2CSA-based vaccine to prevent placental malaria”. Vaccine (2015) 33:7483–8. doi: 10.1016/j.vaccine.2015.10.011
27. Gamain B, Chêne A, Viebig NK, Tuikue Ndam N, Nielsen MA. “Progress and insights toward an effective placental malaria vaccine”. Front Immunol (2021) 12:634508. doi: 10.3389/fimmu.2021.634508
28. Nielsen MA, Resende M, De Jongh WA, Ditlev SB, Mordmüller B, Houard S, et al. “The influence of Sub-unit composition and expression system on the functional antibody response in the development of a VAR2CSA based plasmodium falciparum placental malaria vaccine”. PLoS One (2015) 10(9):1–12. doi: 10.1371/journal.pone.0135406
29. Chêne A, Gangnard S, Dechavanne C, Dechavanne S, Srivastava A, Tétard M, et al. “Down-selection of the VAR2CSA DBL1-2 expressed in e. coli as a lead antigen for placental malaria vaccine development”. NPJ Vaccines (2018) 3:1–11. doi: 10.1038/s41541-018-0064-6
30. Mordmüller B, Sulyok M, Egger-Adam D, Resende M, De Jongh WA, Jensen MH, et al. “First-in-human, randomized, double-blind clinical trial of differentially adjuvanted PAMVAC, a vaccine candidate to prevent pregnancy-associated malaria”. Clin Infect Dis (2019) 69:1509–16. doi: 10.1093/cid/ciy1140
31. Sirima SB, Richert L, Chêne A, Konate AT, Campion C, Dechavanne S, et al. “PRIMVAC vaccine adjuvanted with alhydrogel or GLA-SE to prevent placental malaria: a first-in-human, randomised, double-blind, placebo-controlled study”. Lancet Infect Dis (2020) 20:585–97. doi: 10.1016/S1473-3099(19)30739-X
32. Benavente ED, Oresegun DR, de Sessions PF, Walker EM, Roper C, Dombrowski JG, et al. “Global genetic diversity of var2csa in plasmodium falciparum with implications for malaria in pregnancy and vaccine development”. Sci Rep (2018) 8:1–8. doi: 10.1038/s41598-018-33767-3
33. Bordbar B, Tuikue Ndam N, Renard E, Jafari-Guemouri S, Tavul L, Jennison C, et al. “Genetic diversity of VAR2CSA ID1-DBL2Xb in worldwide plasmodium falciparum populations: Impact on vaccine design for placental malaria”. Infect Genet Evol (2014) 25:81–92. doi: 10.1016/j.meegid.2014.04.010
34. Verity R, Hathaway NJ, Waltmann A, Doctor SM, Watson OJ, Patel JC, et al. “Plasmodium falciparum genetic variation of var2csa in the democratic republic of the congo”. Malar. J (2018) 17:1–8. doi: 10.1186/s12936-018-2193-9
35. Sissoko MS, Healy SA, Katile A, Omaswa F, Zaidi I, Gabriel EE, et al. “Safety and efficacy of PfSPZ vaccine against plasmodium falciparum via direct venous inoculation in healthy malaria-exposed adults in Mali: a randomised, double-blind phase 1 trial”. Lancet Infect Dis (2017) 17:498–509. doi: 10.1016/S1473-3099(17)30104-4
36. Beeson JG, Mann EJ, Byrne TJ, Caragounis A, Elliott SR, Brown GV, et al. “Antigenic differences and conservation among placental plasmodium falciparum-infected erythrocytes and acquisition of variant-specific and cross-reactive antibodies”. J Infect Dis (2006) 193:721–30. doi: 10.1086/500145
37. Doritchamou JYA, Renn JP, Jenkins B, Fried M, Duffy PE. “A single full-length VAR2CSA ectodomain variant purifies broadly neutralizing antibodies against placental malaria isolates”. Elife (2022) 11:1–16. doi: 10.7554/ELIFE.76264
38. Avril M, Kulasekara BR, Gose SO, Rowe C, Dahlbäck M, Duffy PE, et al. “Evidence for globally shared, cross-reacting polymorphic epitopes in the pregnancy-associated malaria vaccine candidate VAR2CSA”. Infect Immun (2008) 76:1791–800. doi: 10.1128/IAI.01470-07
39. Doritchamou J, Bigey P, Nielsen MA, Gnidehou S, Ezinmegnon S, Burgain A, et al. “Differential adhesion-inhibitory patterns of antibodies raised against two major variants of the NTS-DBL2X region of VAR2CSA”. Vaccine (2013) 31:4516–22. doi: 10.1016/j.vaccine.2013.07.072
40. Bordbar B, Tuikue-Ndam N, Bigey P, Doritchamou J, Scherman D, Deloron P. “Identification of Id1-DBL2X of VAR2CSA as a key domain inducing highly inhibitory and cross-reactive antibodies”. Vaccine (2012) 30:1343–8. doi: 10.1016/j.vaccine.2011.12.065
41. Fernandez P, KViebig N, Dechavanne S, Lépolard C, Gysin J, Scherf A, et al. “Var2CSA DBL6-epsilon domain expressed in HEK293 induces limited cross-reactive and blocking antibodies to CSA binding parasites”. Malar. J (2008) 7:1–11. doi: 10.1186/1475-2875-7-170
42. Avril M, Cartwright MM, Hathaway MJ, Hommel M, Elliott SR, Williamson K, et al. “Immunization with VAR2CSA-DBL5 recombinant protein elicits broadly cross-reactive antibodies to placental plasmodium falciparum-infected erythrocytes”. Infect Immun (2010) 78:2248–56. doi: 10.1128/IAI.00410-09
43. Avril M, Cartwright MM, Hathaway MJ, Smith JD. “Induction of strain-transcendent antibodies to placental-type isolates with VAR2CSA DBL3 or DBL5 recombinant proteins”. Malar. J (2011) 10:1–11. doi: 10.1186/1475-2875-10-36
44. Avril M, Hathaway MJ, Srivastava A, Dechavanne S, Hommel M, Beeson JG, et al. “Antibodies to a full-length VAR2CSA immunogen are broadly strain-transcendent but do not cross-inhibit different placental-type parasite isolates”. PloS One (2011) 6:1–10. doi: 10.1371/journal.pone.0016622
45. Mitran CJ, Yanow SK. “The case for exploiting cross-species epitopes in malaria vaccine design”. Front Immunol (2020) 11:335. doi: 10.3389/fimmu.2020.00335
46. Mitran CJ, Mena A, Gnidehou S, Banman S, Arango E, Lima BAS, et al. “Antibodies to cryptic epitopes in distant homologues underpin a mechanism of heterologous immunity between plasmodium vivax PvDBP and plasmodium falciparum VAR2CSA”. MBio (2019) 10(5):e02343–19. doi: 10.1128/mBio.02343-19
47. Daha NA, Banda NK, Roos A, Beurskens FJ, Bakker JM, Daha MR, et al. “Complement activation by (auto-) antibodies”. Mol Immunol (2011) 48:1656–65. doi: 10.1016/j.molimm.2011.04.024
48. Rathnayake D, Aitken EH, Rogerson SJ. “Beyond binding: The outcomes of antibody-dependent complement activation in human malaria”. Front Immunol (2021) 12:683404. doi: 10.3389/fimmu.2021.683404
49. Wanjala CNL, Bergmann-Leitner E, Akala HM, Odhiambo G, Ogutu BR, Andagalu B, et al. “The role of complement immune response on artemisinin-based combination therapy in a population from malaria endemic region of Western kenya”. Malar. J (2020) 19:1–8. doi: 10.1186/s12936-020-03242-4
50. Opi DH, Boyle MJ, McLean ARD, Reiling L, Chan JA, Stanisic DI, et al. “Reduced risk of placental parasitemia associated with complement fixation on plasmodium falciparum by antibodies among pregnant women”. BMC Med (2021) 19(1):1–6. doi: 10.1186/s12916-021-02061-x
51. Elliott SR, Brennan AK, Beeson JG, Tadesse E, Molyneux ME, Brown G V, et al. “Placental malaria induces variant-specific antibodies of the cytophilic subtypes immunoglobulin G1 (IgG1) and IgG3 that correlate with adhesion inhibitory activity”. Infect Immun (2005) 73:5903–7. doi: 10.1128/IAI.73.9.5903-5907.2005
52. Vidarsson G, Dekkers G, Rispens T. “IgG subclasses and allotypes: From structure to effector functions”. Front Immunol (2014) 5:520. doi: 10.3389/fimmu.2014.00520
53. Wahlgren M, Goel S, Akhouri RR. “Variant surface antigens of plasmodium falciparum and their roles in severe malaria”. Nat Rev Microbiol (2017) 15:479–91. doi: 10.1038/nrmicro.2017.47
54. Xie Y, Li X, Chai Y, Song H, Qi J, Gao GF. “Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1”. Cell Rep (2021) 36:109600. doi: 10.1016/j.celrep.2021.109600
55. Abdel-Latif MS, Khattab A, Lindenthal C, Kremsner PG, Klinkert MQ. “Recognition of variant rifin antigens by human antibodies induced during natural plasmodium falciparum infections”. Infect Immun (2002) 70:7013–21. doi: 10.1128/IAI.70.12.7013-7021.2002
56. Ndam NT, Bischoff E, Proux C, Lavstsen T, Salanti A, Guitard J, et al. “Plasmodium falciparum transcriptome analysis reveals pregnancy malaria associated gene expression”. PloS One (2008) 3(3):e1855. doi: 10.1371/journal.pone.0001855
57. Keitany GJ, Jenkins BJ, Obiakor HT, Daniel S, Muehlenbachs A, Semblat JP, et al. “ an invariant protein that colocalizes with VAR2CSA on plasmodium falciparum -infected red cells binds to chondroitin sulfate A”. J Infect Dis (2021) 225:2011–22. doi: 10.1093/infdis/jiab550
58. Fried M, Wendler JP, Mutabingwa TK, Duffy PE. “Mass spectrometric analysis of plasmodium falciparum erythrocyte membrane protein-1 variants expressed by placental malaria parasites”. Proteomics (2004) 4:1086–93. doi: 10.1002/pmic.200300666
59. Kamaliddin C, Salnot V, Leduc M, Ezinmegnon S, Broussard C, Fievet N, et al. “PFI1785w: A highly conserved protein associated with pregnancy associated malaria”. PloS One (2017) 12:1–12. doi: 10.1371/journal.pone.0187817
60. Lasonder E, Ishihama Y, Andersen JS, Vermunt AMW, Pain A, Sauerwein RW, et al. “Analysis of the plasmodium falciparum proteome by high-accuracy mass spedrometry”. Nature (2002) 419:537–42. doi: 10.1038/nature01111
61. Fried M, Hixson KK, Anderson L, Ogata Y, Mutabingwa TK, Duffy PE. “The distinct proteome of placental malaria parasites”. Mol Biochem Parasitol (2007) 155:57–65. doi: 10.1016/j.molbiopara.2007.05.010
62. Kumar V, Kaur J, Singh AP, Singh V, Bisht A, Panda JJ, et al. “PHISTc protein family members localize to different subcellular organelles and bind plasmodium falciparum major virulence factor PfEMP-1”. FEBS J (2018) 285:294–312. doi: 10.1111/febs.14340
63. Chan S, Frasch A, Mandava CS, Ch’Ng JH, Quintana MDP, Vesterlund M, et al. “Regulation of PfEMP1-VAR2CSA translation by a plasmodium translation-enhancing factor”. Nat Microbiol (2017) 2:1–11. doi: 10.1038/nmicrobiol.2017.68
64. Jonsdottir TK, Counihan NA, Modak JK, Kouskousis B, Sanders PR, Gabriela M, et al. “Characterisation of complexes formed by parasite proteins exported into the host cell compartment of plasmodium falciparum infected red blood cells”. Cell Microbiol (2021) 23:1–14. doi: 10.1111/cmi.13332
65. Hommel M, Chan JA, Umbers AJ, Langer C, Rogerson SJ, Smith JD, et al. “Evaluating antibody functional activity and strain-specificity of vaccine candidates for malaria in pregnancy using in vitro phagocytosis assays”. Parasites Vectors (2018) 11:1–7. doi: 10.1186/s13071-018-2653-7
66. Cooke BM, Rogerson SJ, Brown GV, Coppel RL. “Adhesion of malaria-infected red blood cells to chondroitin sulfate a under flow conditions”. Blood (1996) 88:4040–4. doi: 10.1182/blood.v88.10.4040.bloodjournal88104040
67. Pehrson C, Mathiesen L, Heno KK, Salanti A, Resende M, Dzikowski R, et al. “Adhesion of plasmodium falciparum infected erythrocytes in ex vivo perfused placental tissue: A novel model of placental malaria”. Malar. J (2016) 15:1–12. doi: 10.1186/s12936-016-1342-2
68. Mosavati B, Oleinikov AV, Du E. “Development of an organ-on-a-chip-device for study of placental pathologies”. Int J Mol Sci (2020) 21:1–12. doi: 10.3390/ijms21228755
69. Healy SA, Fried M, Richie T, Bok K, Little M, August A, et al. “Malaria vaccine trials in pregnant women: An imperative without precedent”. Vaccine (2019) 37:763–70. doi: 10.1016/j.vaccine.2018.12.025
70. Birkholtz LM, Blatch G, Coetzer TL, Hoppe HC, Human E, Morris EJ, et al. “Heterologous expression of plasmodial proteins for structural studies and functional annotation”. Malar. J (2008) 7:1–20. doi: 10.1186/1475-2875-7-197
71. Kanoi BN, Nagaoka H, Morita M, Tsuboi T, Takashima E. “Leveraging the wheat germ cell-free protein synthesis system to accelerate malaria vaccine development”. Parasitol Int (2021) 80:102224. doi: 10.1016/j.parint.2020.102224
72. Kanoi BN, Takashima E, Morita M, White MT, Palacpac NMQ, Ntege EH, et al. “Antibody profiles to wheat germ cell-free system synthesized plasmodium falciparum proteins correlate with protection from symptomatic malaria in uganda”. Vaccine (2017) 35:873–81. doi: 10.1016/j.vaccine.2017.01.001
73. Kanoi BN, Nagaoka H, White MT, Morita M, Palacpac NMQ, Ntege EH, et al. “Global repertoire of human antibodies against plasmodium falciparum RIFINs, SURFINs, and STEVORs in a malaria exposed population”. Front Immunol (2020) 11:893. doi: 10.3389/fimmu.2020.00893
74. Kanoi BN, Nagaoka H, Morita M, White MT, Palacpac NMQ, Ntege EH, et al. “Comprehensive analysis of antibody responses to plasmodium falciparum erythrocyte membrane protein 1 domains”. Vaccine (2018) 36:6826–33. doi: 10.1016/j.vaccine.2018.08.058
75. Morita M, Nagaoka H, Ntege EH, Kanoi BN, Ito D, Nakata T, et al. “PV1, a novel plasmodium falciparum merozoite dense granule protein, interacts with exported protein in infected erythrocytes”. Sci Rep (2018) 8:1–11. doi: 10.1038/s41598-018-22026-0
76. Longley RJ, White MT, Takashima E, Morita M, Kanoi BN, Li Wai Suen CSN, et al. “Naturally acquired antibody responses to more than 300 plasmodium vivax proteins in three geographic regions”. PloS Negl Trop Dis (2017) 11:1–15. doi: 10.1371/journal.pntd.0005888
77. Longley RJ, White MT, Takashima E, Brewster J, Morita M, Harbers M, et al. “Development and validation of serological markers for detecting recent plasmodium vivax infection”. Nat Med (2020) 26:741–9. doi: 10.1038/s41591-020-0841-4
78. Arshadi AK, Salem M, Collins J, Yuan JS, Chakrabarti D. “Deepmalaria: Artificial intelligence driven discovery of potent antiplasmodials”. Front Pharmacol (2020) 10:1526. doi: 10.3389/fphar.2019.01526
79. Keshavarzi Arshadi A, Webb J, Salem M, Cruz E, Calad-Thomson S, Ghadirian N, et al. “Artificial intelligence for COVID-19 drug discovery and vaccine development”. Front Artif Intell (2020) 3:65. doi: 10.3389/frai.2020.00065
80. Rawal K, Sinha R, Abbasi BA, Chaudhary A, Nath SK, Kumari P, et al. “Identification of vaccine targets in pathogens and design of a vaccine using computational approaches”. Sci Rep (2021) 11:1–25. doi: 10.1038/s41598-021-96863-x
81. Wold S, Jonsson J, Sjörström M, Sandberg M, Rännar S. “DNA and peptide sequences and chemical processes multivariately modelled by principal component analysis and partial least-squares projections to latent structures”. Anal Chim Acta (1993) 277:239–53. doi: 10.1016/0003-2670(93)80437-P
82. Hellberg S, Skagerberg B, Hellberg S, Skagerberg B, Hellberg S. Principal property values for six non-natural amino acids and their application to a structure-activity relationship for oxytocin peptide analogues. (1986).
83. Doytchinova IA, Flower DR. “VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines”. BMC Bioinf (2007) 8:1–7. doi: 10.1186/1471-2105-8-4
84. Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, et al. “Nonviral delivery of self-amplifying RNA vaccines”. Proc Natl Acad Sci U. S. A. (2012) 109:14604–9. doi: 10.1073/pnas.1209367109
85. Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. “Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza a virus infection”. Nat Biotechnol (2012) 30:1210–6. doi: 10.1038/nbt.2436
86. Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, et al. “Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes”. J Control. Release (2015) 217:345–51. doi: 10.1016/j.jconrel.2015.08.007
87. Noor R. “Developmental status of the potential vaccines for the mitigation of the COVID-19 pandemic and a focus on the effectiveness of the pfizer-BioNTech and moderna mRNA vaccines”. Curr Clin Microbiol Rep (2021) 8:178–85. doi: 10.1007/s40588-021-00162-y
Keywords: Plasmodium falciparum, VAR2CSA, pregnancy-associated malaria, vaccines, antigen discovery
Citation: Rotich AK, Takashima E, Yanow SK, Gitaka J and Kanoi BN (2022) Towards identification and development of alternative vaccines against pregnancy-associated malaria based on naturally acquired immunity. Front. Trop. Dis. 3:988284. doi: 10.3389/fitd.2022.988284
Received: 07 July 2022; Accepted: 31 August 2022;
Published: 23 September 2022.
Edited by:
Sara Healy, National Institutes of Health (NIH), United StatesReviewed by:
Augustina Frimpong, University of Ghana, GhanaBenjamin Mordmüller, Radboud University Medical Centre, Netherlands
Copyright © 2022 Rotich, Takashima, Yanow, Gitaka and Kanoi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bernard N. Kanoi, Ymthbm9pQG1rdS5hYy5rZQ==
 Alex K. Rotich
Alex K. Rotich Eizo Takashima
Eizo Takashima Stephanie K. Yanow
Stephanie K. Yanow Jesse Gitaka
Jesse Gitaka Bernard N. Kanoi
Bernard N. Kanoi