
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis., 19 August 2022
Sec. Major Tropical Diseases
Volume 3 - 2022 | https://doi.org/10.3389/fitd.2022.972682
This article is part of the Research TopicModern Molecular Era of The Mycobacterial World: Insights Into Diagnosis and Transmission of Mycobacteria and Associated DiseasesView all 10 articles
 Ravindra P. Turankar1
Ravindra P. Turankar1 Vikram Singh1,2
Vikram Singh1,2 Mallika Lavania1,3
Mallika Lavania1,3 Itu Singh1
Itu Singh1 Utpal Sengupta1
Utpal Sengupta1 Rupendra S. Jadhav1,4*
Rupendra S. Jadhav1,4*Introduction: Molecular epidemiology of leprosy is very important to study leprosy transmission dynamics and to enhance our understanding of leprosy in endemic areas by utilizing the molecular typing method. Nowadays our understanding of leprosy transmission dynamics has been refined by SNP typing and VNTR marker analysis of M. leprae strains.
Objective: This study was carried out to find out the presence of viable M. leprae in the soil and water samples from residing areas of leprosy patients staying in different blocks of Purulia district of West Bengal, understanding their genotypes and compared with that of M. leprae present in patients.
Material and methods: Slit-skin smear (SSS) samples (n=112) were collected from the active multibacillary leprosy patients from different blocks of leprosy endemic area. Soil samples (n=1060) and water samples (n=620) were collected from residing areas of leprosy patients. SNP subtyping was performed by PCR followed by sequencing. Multiplex PCR was performed using fifteen ML-VNTR loci and results were analysed.
Results: We observed high PCR positivity in soil samples (344 out of 1060; 32%) and water samples (140 out of 620; 23%). These PCR positive samples when further screened for viability, it was observed that 150 soil samples (44%) and 56 water samples (40%) showed presence of 16S rRNA. SNP typing of M. leprae revealed presence of predominantly type 1. SNP subtype 1D (83%) was most prevalent in all the blocks of Purulia followed by subtype 1C (15%) and subtype 1A (2%). SNP subtype 2F was noted in only one sample. SNP and VNTR combination showed presence of similar strain type in certain pockets of Purulia region which was responsible for transmission.
Conclusion: Presence of viable M. leprae in the environment, and presence of SNP Type 1 M. leprae in patients and environment suggests both environment and patients play a role in disease transmission.
Leprosy is also called Hansen’s disease, a chronic infectious disease caused by Mycobacterium leprae and Mycobacterium lepromatosis (1, 2). It is true that the source of infection is either untreated leprosy patients or other animal reservoirs (3, 4). But in addition to this, extra-human reservoirs could be possible. Studies in Norway, India, Japan, Indonesia, Brazil, Bangladesh, England, and Suriname have shown presence of M. leprae in the surrounding environment of leprosy endemic regions (5–12). Hence it is important to look for the presence of M. leprae in the environment (soil and water) and to determine their viability status along with their genetic make-up and other factors which might help the survival of the organism in the environment.
Enormous numbers of leprosy bacilli (2.4X108) are expelled daily in the environment from the nasal discharges of lepromatous patients (13). There is also evidence to support excretion of bacilli from skin lesions (14). It was reported that M. leprae discharged through secretions (coughing and sneezing) from patients (15, 16) in the form of air-borne droplet may cause infection or can settle in soil (5, 17, 18) and in water (8, 19). But very limited information is available for the survival of bacilli outside the host. M. leprae, an obligate intracellular pathogen, have been recently shown to be associated with free living amoeba. In vitro, phagocytosis of M. leprae by amoeba was observed by florescence microscopy and M. leprae which remained viable for at least three days in amoebae were noted to grow in mouse foot pad (20). Further, M. leprae was found to survive up to 8 months within amoebic cysts (21). Hence, possibility of spreading of infection by amoebae needs to be explored in natural environmental conditions.
The new epidemiological tools developed for strain typing of M. leprae in the recent years will be useful in national leprosy surveillance/control efforts towards true reduction in incidence, and in epidemiological investigations. The combination of single nucleotide polymorphism (SNP) subtyping along with variable nucleotide tandem repeat (VNTR) loci determination in M. leprae genome have been proved to serve as a genetic marker to differentiate strains of M. leprae (3, 22). However, the characteristics of polymorphism vary depending on the population, and can be a reflection of that population at the national and local level.
The purpose of this study was to find out the existence of viable M. leprae in the surrounding environment (soil and water) of the residing areas of leprosy patients and to perform molecular genotyping using SNP typing and or VNTR analysis of M. leprae from patients and the environment to find out the genetic variability of the organism existing in nature which might help in tracking and understanding transmission of leprosy.
The study was approved on 22nd December 2016 by the Organization Ethical Committee of The Leprosy Mission trust India. Informed consent was obtained from all the participant enrolled in the study.
Soil and water samples were collected from different blocks of Purulia district, West Bengal. Soil was dug (3-4-inch-deep) and was collected in clean plastic containers (10g each) with the help of a trowel and labelled with site code and the village name. The collected samples were transported to the laboratory at room temperature (within 2 days) and thereafter were stored at 4–8°C till further processing. One thousand and sixty soil samples and 620 water samples were collected from residing places of leprosy patients.
Multibacillary leprosy cases were diagnosed clinically based on skin lesions and impairment of nerve functions and acid-fast bacilli (AFB) positivity in slit skin smears. After taking consent, 112 slit-skin smear samples (SSS) were collected from the earlobes of active multibacillary (MB) leprosy patients. SSSs were collected during field visits in different blocks such as Joypur (n=24), Jhalda (n=13), Purulia (n=32), Arsha (n=16), Chandenkeyari (n=8), Kashipur (n=2), Para (JH) (n=10), Barabazar (n=7) of Purulia District, West Bengal. Samples were transported in 70% ethanol in micro centrifuge tubes to the laboratory at room temperature (25°C). The tubes were kept at 4°C until further use.
Standard method of DNA extraction was used as described earlier (7). Briefly, pond water (50 ml) samples were centrifuged at 400 ×g for 5 min. The supernatants were collected in 50 ml sterile tubes and centrifuged again at 8000 ×g for 15 min. Pellets that contained soil and other floating matter including organisms were weighed (100 mg) in dried 1.5 ml microfuge tube and followed by the soil DNA extraction protocol. Soil samples were homogenized using bead beater followed by lysis in tube containing ethanol with zirconium beads mixed with soil. The mixture was homogenized using bead beater followed by lysis by Proteinase K in TENP buffer (50 mM Tris, 20 mM EDTA, 100 mM NaCl and 1% Polyvinylpolypyrrolidone). DNA was precipitated by adding 70% ethanol and centrifuged at 10,000 rpm for 15 mins. The pellet was air dried and dissolved in Tris EDTA (TE) buffer and stored at −20°C until further use.
Proteinase K Lysis method was used for M. leprae DNA extraction from slit-skin smear samples (23). In brief smears collected in 1 ml 70% ethanol were centrifuged at 10,000rpm (8000xg) for 10 min. Supernatant was discarded and pellet was air dried for the removal of ethanol. After ethanol removal samples were kept for overnight lysis in lysis buffer (100 mM Tris buffer pH 8.5 with 1mg/ml proteinase K and 0.05% Tween 20) at 60°C. The Proteinase K was inactivated at 97°C for 10 minutes. This lysate preparation was further used for PCR.
The standardized protocol of RNA extraction was used as described earlier (7). Briefly, the samples as mentioned above were homogenised and were subjected to acid-phenol extraction followed by isopropanol precipitation and centrifugation at 12000 rpm for 10 mins at 4°C. Pellet was washed once with 70% ethanol, air dried and then dissolved in 50 μL of TE buffer.
PCR amplification was carried out in a total 25 μL of reaction volume that contained 2 μL of template DNA and primers at final concentration of 0.5 μM (forward and reverse) and 1X Genei Mix (Merck India) were used. We used M. leprae specific primers (PS1- TGCATGTCATGGCCTTGAGG; PS2 -CACCGATACCAGCGGCAGAA) as per our earlier publication (24).The amplification was carried out in a thermal cycler (Corbett) using following conditions: one cycle of denaturation at 95°C for 5 min followed by 35-45 cycles at 94°C for 30s, annealing at 58°C for 30s, extension at 72°C for 1 min and one cycle of final extension at 72°C for 10 min. PCR product (129 bp) containing amplified fragment of the target region was electrophoresed in a 2% agarose gel using Tris-Borate-EDTA buffer at 100 volts constant voltage.
The Reverse Transcriptase (RT) –Polymerase chain reaction was carried out by using One Step RT PCR Kit (Qiagen - 210210). Control reactions to test DNA contamination were also performed simultaneously with each experiment by carrying out PCR without prior reverse transcription. 16S rRNA gene was amplified using M. leprae specific primers P2 and P3 as described earlier (25). The total volume (50 µL) of PCR amplification mixture contained 10 µL of 5X RT PCR buffer 2 µL of dNTPs, 10 µL -5X Q Solution,50ng of each primer, 2 µL of RTPCR enzyme,0.25 µL RNase inhibitor and remaining RNase free water and 10 µL of sample (template). The cycling profile for the amplification reaction was in two stages. In the first stage reverse transcription was carried out at 50°C for 30 minutes followed by inactivation step at 95°C for 15 minutes. In the second stage amplification was carried out using denaturation at 94°C for 1 min 30 seconds, annealing at 60°C for 1 min 30 seconds followed by extension at 72°C for 1 min for 37 cycles. This was followed by final extension at 72°C or 10 min. The amplification products were run on 1.5% (w/v) agarose gel, stained with ethidium bromide, and observed using Gel Documentation System (Alpha Imager).
Three SNP loci viz. 1,2 and 3 at nucleotide positions 14676, 1642875 and 2935685 in M. leprae genomic DNA were amplified using primers (Supplementary Table 1) and was performed using described protocols (26, 27).
Amplification of four SNP subtyping for type 1 at nucleotide positions 8453, 313361, 61425 and 1642879, M. leprae genomic DNA was amplified using previously reported (26, 27) primer sequences as mentioned (Supplementary Table 2).
After amplification of PCR products were run on 2% agarose gel by electrophoresis. The amplicons were outsourced for commercial sequencing (Eurofins Genomics India Pvt. Ltd. Delhi).
The multiplex PCR was carried out using M. leprae specific primers as described earlier (28, 29) (Supplementary Table 3). The forward primers were labelled with PET, NED, VIC, and 6-FAM fluorescent dyes at the 5 termini (Invitrogen Bio-services-Applied Biosystems, India). Multiplex PCRs were performed as described earlier (30). Four sets of combination of primers were used and the reaction was carried out using multiplex PCR kit (Qiagen). Briefly, each reaction mixture (20 μL final volume) was comprised of 10 μL of 2x Qiagen master mix, 2 μL Q solution, 2 μL (each) of forward and reverse primer working stock and 2μl of template DNA, the volume was adjusted with nuclease free water. The final concentration of each primer was 0.2 μM. PCR was carried out at an initial denaturation temperature of 95 0C for 15 min, followed by 40 cycles as: denaturation at 94 0C for 30s, primer annealing at 60 0C for 90s and primer extension at 720C for 90s, and final extension at 72 0C for 10 min. 5 μL PCR products were electrophoresed in 2% Agarose gel using Tris borate-EDTA buffer (1X) at 100V constant current for 1 hour to check amplification. Amplicons were sent for commercial fragment length analysis (FLA) to Xplorigen Technologies Ltd., Delhi India.
DNA fragments were visualized by Finch TV Version 1.4.0 software that was used for chromatogram analysis developed by Geospiza’s research team. The chromatogram, thus generated was then compared to the standard M. leprae strain using nBLAST at positions mentioned in the table to track mutations and to categorize them into SNP subtypes A, B, C and D. Fragment length analysis of VNTR genotypes were analysed and copy numbers of repeat different loci determined.
Cluster analysis was done using PAST 4.03 statistical analysis software. Dendrograms were generated to see clustering if any in relation to SNP subtype and VNTR.
Clinical samples (SSS samples) were tested for presence of M. leprae DNA using RLEP primers. The results of M. leprae DNA PCR positive for SSS samples. Environmental samples were tested for presence of M. leprae. Out of 1060 soil samples collected from the area where patients resided, we could detect M. leprae in 344 samples (32.4%) (Table 1). Further, 140 (23%) water samples out of 620 samples collected from the patient residing area showed presence of M. leprae DNA. In control area, i.e., an area of low endemic region of Purulia from where no new case of leprosy was reported in the past ten years, we could detect M. leprae DNA only in 9 (3%) soil samples out of 300 samples tested. Water samples (180N) from this area, we could detect M. leprae DNA in only 2 (1.1%) samples.
RT-PCR was performed by using 16S rRNA gene target using PCR positive environmental samples. We could detect amplification in 150 (44%) soil samples out of 344 soil samples tested (Table 1). Similarly, 40% of the water samples (56 out of 140) showed RT-PCR positivity suggesting possibility of presence of viable M. leprae in these samples which were collected from the patient residing area. None of the environmental samples collected from the control area showed any amplification by RT-PCR (Table 1).
All the M. leprae DNA PCR positive clinical samples and environmental soil and water samples were subjected to SNP type and subtype which were obtained from patients’ area and no patients area. Standard reference M. leprae DNA of NHDP63, BR 4953 and THAI 53 DNA were used as positive control (Obtained from Colorado State University, USA) and master mix without template used as negative control in PCR reaction. All the PCR positive samples were used for SNP type and SNP subtype amplicon sent for sequencing outsourcing (Eurofins Genomics India Pvt. Ltd. Delhi).
All the PCR positive SSS samples used for SNP typing and subtyping are presented in Table 2. It was observed that the almost all the samples except one belonged to SNP type 1. Further, SNP subtyping of the samples using sequencing showed 2 out of 112 samples to be of subtype 1A (2%), 16 out of 112 to be subtype 1C (15%) and 93 out of 112 belonged to subtype 1D (82%). Only one sample from Jhalda (Purulia district) was observed to be of subtype 2F (1%).
Out of 344 soil samples tested we could obtain data on SNP typing for 76 samples. All samples were of SNP Type 1 (Table 1). Of these, majority of samples (50 of 76) (65.8%) were of type 1D which is also a major SNP type noted in patients. Fourteen samples (18.4%) were of type 1C and 12 (15.8%) were of type 1A. Similarly, 140 PCR positive water samples were tested for SNP typing. We could obtain data for 15 samples and all the 15 samples showed SNP type 1D.
Multiplex PCR was used to amplify fragments suitable for fragment length analysis from fifteen genomic VNTR loci. We found that 3 loci viz. (27 -5), (23 -3), (AC) 8b were monomorphic; four loci viz. rpoT, (AT)17, (21 -3), 18-8 were dimorphic; four loci viz. (AC)9, 12-5, (TA)10 (6, 7), were polymorphic. Four loci viz. (GGT)5, (GTA)9, (AC)8a and (TA)10 were found highly polymorphic in nature (Table 3). Interestingly, SNP subtype (1D) showed more variability in repeat number with 12 VNTR loci.
We looked at the distribution of the alleles across the samples for SNP type 1D (Table 4A). For each locus it was noted that a particular allele number was dominant e.g., for locus 18-8, there were 41 samples out of 70 which showed 8 repeats. So, we looked at the variation in allele numbers as well as the dominant allele for each locus (Table 4B). It is quite clear that some of the alleles for the loci tested are highly dominant across the samples tested.
Results of SNP type and VNTR were studied for existence of clusters. Data were analysed separately and together to plot Dendrograms for SNP type 1D and 1C (Figures 1A–C). Then, we looked at individual cases with the SNP type and VNTR allele numbers and we could easily pick up some cases showing similar M. leprae strain pattern from some of the blocks in Purulia district (Table 5).
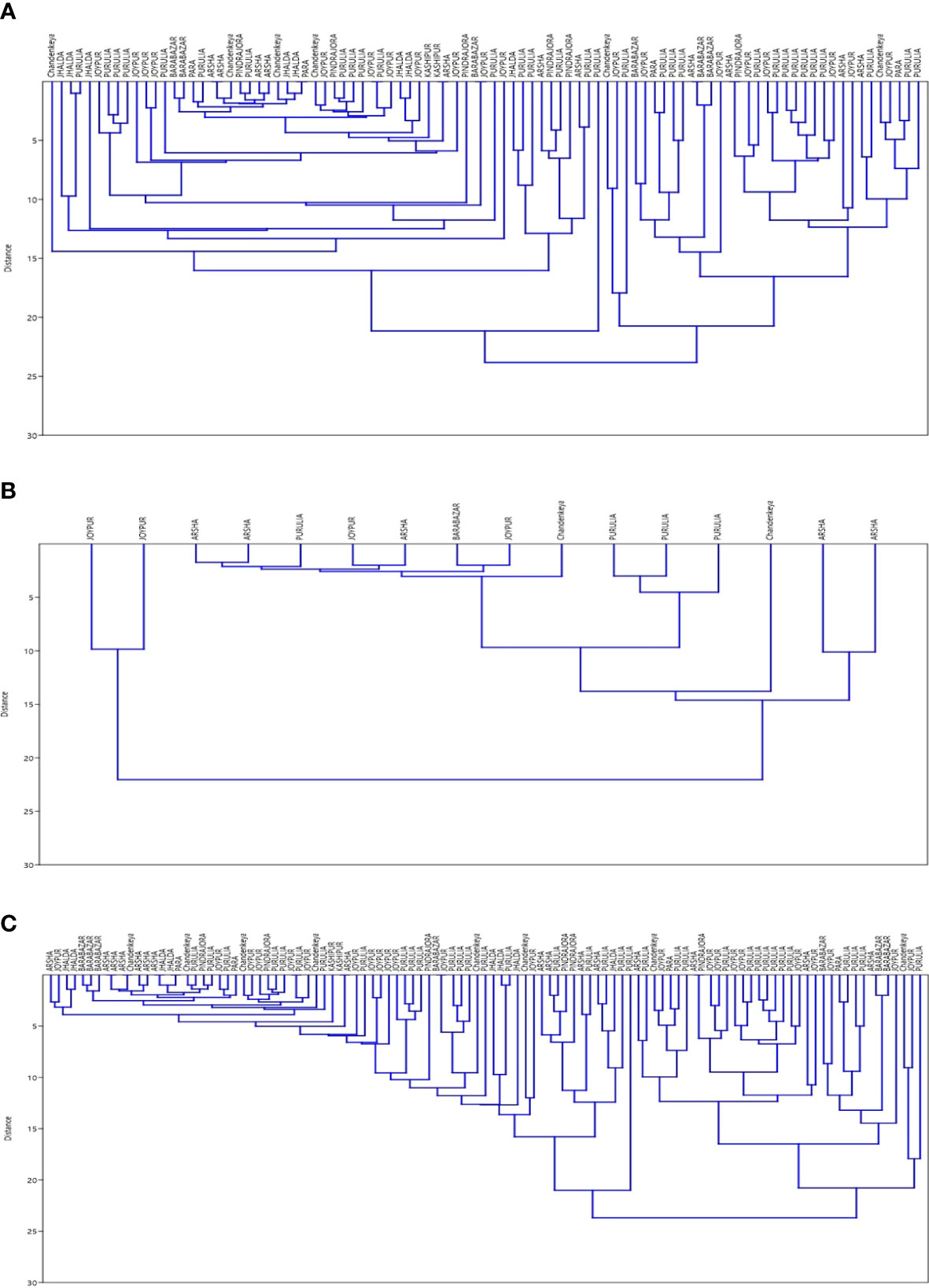
Figure 1 (A) Dendrogram for SNP Type 1D samples using data for 15 loci of VNTR ‘Dendrogram was prepared using PAST 4.03 Statistical Analysis Software’. (B) Dendrogram for SNP Type 1C samples using data for 15 loci of VNTR‘Dendrogram was prepared using PAST 4.03 Statistical Analysis Software’. (C) Dendrogram for SNP Type 1D & 1C samples together using data for 15 loci of VNTR ‘Dendrogram was prepared using PAST 4.03 Statistical Analysis Software’.
Recent studies strongly suggest that environment could be a possible reservoir of viable M. leprae and might be responsible for the disease transmission as presence of viable M. leprae has been reported in the environment (water and soil) in Indonesia, India, Brazil, and Bangladesh, England, Surinam (6–12, 18, 25). It has been demonstrated that M. leprae can survive outside human body in moist soil up to 46 days (31). However, the direct proof of transmission of disease to the population has still not been clearly understood especially from environmental sources of M. leprae (8–10, 18).
In recent times with the advancement in molecular biological techniques a very unstable molecule like RNA can be preserved and used as a potent marker for assessment of viability of microorganisms (8, 32). M. leprae genome 16S rRNA and RLEP conserved gene regions were found to be a better target for viability studies because their stability and slower degradation rate over time in comparison to superoxide dismutase gene (32).
Earlier studies have also shown the presence of viable M. leprae using 16S rRNA in the environment of leprosy hospital areas which can be a possible source of infection (6, 18, 25, 27). Existence of M. leprae DNA has been reported in water samples in Indonesia (12) and soil samples from high prevalence areas of North-East states of India (6, 8, 33). In some studies, it has been suggested that in endemic countries >50% of household contacts may have a history of intimate contact leprosy patients. In this study, we collected environmental samples from the residing areas of multibacillary active leprosy patients. Large proportion of environmental samples showed presence of DNA (32% for soil and 23% for water) in areas where active cases were residing suggesting that there is dynamic movement of the organism between patient and the environment. This was further supported by the fact that environmental samples from control region where there was no active case, there was rare presence of M. leprae in the environment. On the other hand, presence of rRNA (44% of the soil samples and 40% of the water samples with abundant presence of M. leprae DNA), suggests the chances of viability of M. leprae bacilli in these samples and their exposure to the community. Hence in the inhabitant areas of leprosy cases there could be a possibility of indirect exposure to M. leprae to the community that may result in infection with M. leprae bacilli.
Further, active multibacillary leprosy patients discharge enormous numbers of leprosy bacilli from nose, mouth washes and skin to the environment which may get air borne as droplet and may cause infection or can settle in soil and water (13–16). These viable bacilli might be phagocytosed by protozoa and might survive in protozoa and can be carried to susceptible population staying in leprosy endemic area. We earlier found presence protozoa species along with viable M. leprae in soil and water samples (8). suggesting possible protective niche that protozoa may provide to M. leprae in the environment. But we are yet to find proof for the presence of M. leprae within protozoa in natural environmental condition. Therefore, further experiments are needed to understand and establish the mechanism of M. leprae viability in the environment and the factors that contribute to provide the protective niche to M. leprae.
Recent advancement in the molecular characterization of M. leprae has led to alternative and definitive methodologies that are used for identification and distribution of genotype (22, 26). Matsuoka et al. (29) reported polymorphism in rpoT gene of M. leprae. Monot et al. (26) demonstrated SNP array in M. leprae. In Indian population mostly SNP type-1 and rarely type-2 was observed (7, 30, 34). Several reports suggested that molecular marker for M. leprae were useful for distinguishing strain and epidemiological significance (3, 22, 26, 28, 35). The discovery of SNPs in M. leprae genome was able to distinguish four major SNP types and their distribution in different region of the world. The most common approach of SNP typing was useful and effective in molecular epidemiologic studies (26). Sixteen SNP subtypes were useful for tracking the transmission of M. leprae and source of infection.
The present study was based on the identification and differentiation of M. leprae strains from the SSS samples of index cases and their residing environmental areas from endemic region. This was to track the transmission and M. leprae strain prevalence in that geographical region in association with VNTR loci. Genotyping of SSS almost all of the multibacillary leprosy cases showed presence of SNP type 1 and SNP subtype 1D (82%) was most prevalent in the population. We also identified SNP type 2 and subtype 2F (1%) in one of the samples from Purulia district of West Bengal. In our earlier study we have reported SNP subtype 2E in cases from north-east Delhi and subtype 2G from West Bengal (27). All the SSS samples were obtained from different blocks of Purulia district. Genotyping of environmental samples showed SNP type 1 and subtype 1D which suggest that there is discharge of the M. leprae from the active cases to the environment. Similar genotype in the patient and environmental soil samples poses serious question on the source of infection for the population in the community.
SNP and VNTR genotyping studies in leprosy multi-case families have shown similar SNP type and VNTR repeat units suggesting that source of infection is common in multi-case family (30, 33, 34). We used combination of SNP and VNTR genotyping data of clinical samples to identify the pattern of transmission in different blocks of Purulia district. We observed in this study that some of the VNTR loci like (GGT)5, (GTA)9, (AC)8a and (TA)10 were highly polymorphic in nature. But every locus had at least one allele that was dominant among the samples. Similar polymorphism was reported from South Indian leprosy cases (36) and from Switzerland (26).
Young et al. (36) reported 2 alleles of locus 23-3 from east and south Indian cases. Similarly, 2 alleles of loci 23-3 were also observed in China (37), Thailand, Brazil and Columbia (35). 3 copies of loci 23-3 were reported in Philippines (35). In this study we could find three alleles of 23-3 but the allele with 2 repeats was most prevalent (94.5%). We observed either 7 or 8 repeats of locus 18-8 in all the cases studied. But in Philippines, Brazil and Columbia 8 numbers of repeat were reported (35). On the other hand, 7 number of repeats were observed in China (37).
In our study we observed 3,4 and 5 repeats of VNTR locus 12-5, but 4 repeats were observed in population of Columbia (35). However, 3 repeats were reported by China (37).
Association of 5,6,7 and 11 repeats of VNTR 6-7 loci with SNP subtype 1D was noted in this study. Earlier 5 and 6 repeats were reported from India, Thailand, Columbia population (35, 38, 39). However, 7 copies were reported in Philippines (35).
We observed 8, 9 and 10 repeats for the locus (AT) 17. Monot et al. (40) reported 8 and 9 repeats from Switzerland. 3 and 4 repeats of rpoT were reported from Japan and India (18, 29, 41, 42).
With the help of cluster analysis, the SNP and VNTR combination M. leprae strain similarities were noted in the certain blocks of Purulia district but many variabilities in repeats in VNTR loci which might require large number of samples for analysis to show similar genotype of epidemiological importance.
In summary, this study undoubtedly found presence of viable M. leprae in inhabitant areas of leprosy patients. These viable bacilli might survive in the environment as well as might help in causing leprosy disease after repeated exposure to a susceptible host. Similar genotype in clinical and environmental samples indicate that environment could possibly act as a source of infection. SNP and VNTR combination showed M. leprae strain similarities and their differentiation in certain blocks of Purulia. Such studies with the combination of genetic markers may provide a tool to track transmission link in the community.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The study was approved on 22nd December 2016 by the Organization Ethical Committee of The Leprosy Mission trust India. Informed consent was obtained from all the participant enrolled in the study. The patients/participants provided their written informed consent to participate in this study.
RT and RJ, conceived and designed the analysis. RT, VS, ML, IS collected the data with experimental work in lab, methodology, validation. RT wrote the paper, performed the analysis, and interpretation of data, and drafting of manuscript. RJ, ML, IS and US: Supervision, conceptualization, writing-reviewing, and editing. RJ: Data curation, software, and analysis. RT, RJ, ML and US contributed data or analysis tools, wrote the paper, analysis, and interpretation of data, and drafting of manuscript. All authors contributed to the article and approved the submitted version.
We acknowledge the financial support rendered by Indian Council of Medical Research (ICMR-Task Force Project No.5/8/3(12)/2009-ECD-I (A) and ICMR Adhoc Project 5/8/3(11)2014-ECD-1).
Infrastructural support granted by the host institution - The Leprosy Mission Trust India to carry out this research work at Stanley Browne Research Laboratory – The Leprosy Mission Community Hospital, Shahdara – New Delhi. We also wish to acknowledge support of the Superintendent and staff of TLM Hospital, Purulia. We are likewise grateful to Mr. Atul Roy for assisting in the sample collection in the field condition.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2022.972682/full#supplementary-material
2. Seo Y-H, Sizer KC, Schoberle T, May GS, Spencer JS, et al. A new mycobacterium species causing diffuse lepromatous leprosy. Am J Clin Pathol (2008) 130(6):856–64. doi: 10.1309/AJCPP72FJZZRRVMM
3. Truman RW, Singh P, Sharma R, Busso P, Rougemont J, Paniz-Mondolfi A, et al. Probable zoonotic leprosy in the Southern United States. N Engl J Med (2011) 364(17):1626–33. doi: 10.1056/NEJMoa1010536
4. Avanzi C, Del-Pozo J, Benjak A, Stevenson K, Simpson VR, Busso P, et al. Red squirrels in the British isles are infected with leprosy bacilli. Science (2016) 354(6313):744–7. doi: 10.1126/science.aah3783
5. Kazda J. Occurrence of non-cultivable acid-fast bacilli in the environment and their relationship to m. leprae Lepr Rev (1981) 52(Suppl 1):85–91. doi: 10.5935/0305-7518.19810061
6. Lavania M, Katoch K, Katoch VM, Gupta AK, Chauhan DS, Sharma R, et al. Detection of viable mycobacterium leprae in soil samples: insights into possible sources of transmission of leprosy. Infect Genet Evol (2008) 8(5):627–31. doi: 10.1016/j.meegid.2008.05.007
7. Turankar RP, Lavania M, Singh M, Siva Sai KS, Jadhav RS. Dynamics of mycobacterium leprae transmission in environmental context: Deciphering the role of environment as a potential reservoir. Infect Genet Evol (2012) 12(1):121–6. doi: 10.1016/j.meegid.2011.10.023
8. Turankar RP, Lavania M, Darlong J, Siva Sai KSR, Sengupta U, Jadhav RS. Survival of mycobacterium leprae and association with acanthamoeba from environmental samples in the inhabitant areas of active leprosy cases: A cross sectional study from endemic pockets of purulia, West Bengal. Infect Genet Evol (2019) 72:199–204. doi: 10.1016/j.meegid.2019.01.014
9. Mohanty PS, Naaz F, Katara D, Misba L, Kumar D, Dwivedi DK, et al. Viability of mycobacterium leprae in the environment and its role in leprosy dissemination. Indian J Dermatol Venereol Leprol (2016) 82(1):23–7. doi: 10.4103/0378-6323.168935
10. Holanda MV, Marques LEC, Macedo MLB, Pontes MAA, Sabadia JAB, Kerr LRFS, et al. Presence of mycobacterium leprae genotype 4 in environmental waters in northeast Brazil. Rev Soc Bras Med Trop (2017) 50(2):216–22. doi: 10.1590/0037-8682-0424-2016
11. Tió-Coma M, Sprong H, Kik M, van Dissel JT, Han XY, Pieters T, et al. Lack of evidence for the presence of leprosy bacilli in red squirrels from north-West Europe. Transbound Emerg Dis (2020) 67(2):1032–4. doi: 10.1111/tbed.13423
12. Matsuoka M, Izumi S, Budiawan T, Nakata N, Saeki K. Mycobacterium leprae DNA in daily using water as a possible source of leprosy infection. Indian J Lepr (1999) 71(1):61–7.
13. Davey TF, Rees RJ. The nasal dicharge in leprosy: clinical and bacteriological aspects. Lepr Rev (1974) 45(2):121–34. doi: 10.5935/0305-7518.19740014
14. Job CK, Jayakumar J, Kearney M, Gillis TP. Transmission of leprosy: A study of skin and nasal secretions of household contacts of leprosy patients using PCR. Am J Trop Med Hyg (2008) 78(3):518–21. doi: 10.4269/ajtmh.2008.78.518
15. Weddell G, Palmer E. The pathogenesis of leprosy. Exp Approach Lepr Rev (1963) 34:57–61. doi: 10.5935/0305-7518.19630010
16. Barton RP. A clinical study of the nose in lepromatous leprosy. Lepr Rev (1974) 45(2):135–44. doi: 10.5935/0305-7518.19740015
17. Blake LA, West BC, Lary CH, Todd JR4. Environmental nonhuman sources of leprosy. Rev Infect Dis (1987) 9(3):562–77. doi: 10.1093/clinids/9.3.562
18. Turankar RP, Lavania M, Singh M, Sengupta U, Siva Sai K, Jadhav RS. Presence of viable mycobacterium leprae in environmental specimens around houses of leprosy patients. Indian J Med Microbiol (2016) 34(3):315–21. doi: 10.4103/0255-0857.188322
19. Fukutomi Y, Maeda Y, Matsuoka M, Makino M. Temperature dependency for survival of mycobacterium leprae in macrophages. Nihon Hansenbyo Gakkai Zasshi (2009) 78(1):7–16. doi: 10.5025/hansen.78.7
20. Lahiri R, Krahenbuhl JL. The role of free-living pathogenic amoeba in the transmission of leprosy: A proof of principle. Lepr Rev (2008) 79(4):401–9. doi: 10.47276/lr.79.4.401
21. Wheat WH, Casali AL, Thomas V, Spencer JS, Lahiri R, Williams DL, et al. Long-term survival, and virulence of mycobacterium leprae in amoebal cysts. PloS Negl Trop Dis (2014) 8(12):e3405. doi: 10.1371/journal.pntd.0003405
22. Groathouse NA, Rivoire B, Kim H, Lee H, Cho SN, Brennan PJ, et al. Multiple polymorphic loci for molecular typing of strains of mycobacterium leprae. J Clin Microbiol (2004) 42(4):1666–72. doi: 10.1128/JCM.42.4.1666-1672.2004
23. Jadhav RS, Macdonald M, Bjune G, Oskam L, MILEP2 Study Group. Simplified PCR detection method for nasal mycobacterium leprae. Int J Lepr Other Mycobact Dis (2001) 69(4):299–307.
24. Turankar RP, Pandey S, Lavania M, Singh I, Nigam A, Darlong J, et al. Comparative evaluation of PCR amplification of RLEP, 16S rRNA, rpoT and sod a gene targets for detection of m. leprae DNA from clinical and environmental samples. Int J Mycobacteriol (2015) 4(1):54–9. doi: 10.1016/j.ijmyco.2014.11.062
25. Jadhav RS, Kamble RR, Shinde VS, Edward S, Edward VK. Use of reverse transcription polymerase chain reaction for the detection of mycobacterium leprae in the slit-skin smears of leprosy patients. Indian J Lepr (2005) 77(2):116–27.
26. Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, et al. Comparative genomic and phylogeographic analysis of mycobacterium leprae. Nat Genet (2010) 42(4):361. doi: 10.1038/ng.477
27. Lavania M, Jadhav RS, Turankar RP, Chaitanya VS, Singh M, Sengupta U. Single nucleotide polymorphisms typing of mycobacterium leprae reveals focal transmission of leprosy in high endemic regions of India. Clin Microbiol Infect (2013) 19(11):1058–62. doi: 10.1111/1469-0691.12125
28. Gillis T, Vissa V, Matsuoka M, Young S, Richardus JH, Truman R, et al. Characterisation of short tandem repeats for genotyping mycobacterium leprae. Lepr Rev (2009) 80(3):250–60. doi: 10.47276/lr.80.3.250
29. Matsuoka M, Zhang L, Morris MF, Legua P, Wiens C. Polymorphism in the rpoT gene in mycobacterium leprae isolates obtained from Latin American countries and its possible correlation with the spread of leprosy. FEMS Microbiol Lett (2005) 243(2):311–5. doi: 10.1016/j.femsle.2004.12.031
30. Lavania M, Jadhav R, Turankar RP, Singh I, Nigam A, Sengupta U. Genotyping of mycobacterium leprae strains from a region of high endemic leprosy prevalence in India. Infect Genet Evol (2015) 36:256–61. doi: 10.1016/j.meegid.2015.10.001
31. Desikan KV, Sreevatsa. Extended studies on the viability of mycobacterium leprae outside the human body. Lepr Rev (1995) 66(4):287–95.
32. Martinez AN, Lahiri R, Pittman TL, Scollard D, Truman R, Moraes MO, et al. Molecular determination of mycobacterium leprae viability by use of real-time PCR. J Clin Microbiol (2009) 47(7):2124–30. doi: 10.1128/JCM.00512-09
33. Singh V, Turankar RP, Goel A. Real-time PCR-based quantitation of viable mycobacterium leprae strain from clinical samples and environmental sources and its genotype in multi-case leprosy families of India. Eur J Clin Microbiol Infect Dis (2020) 39(11):2045–55. doi: 10.1007/s10096-020-03958-w
34. Turankar RP, Lavania M, Chaitanya VS, Sengupta U, Darlong J, Darlong F, et al. Single nucleotide polymorphism-based molecular typing of m. leprae from multicase families of leprosy patients and their surroundings to understand the transmission of leprosy. Clin Microbiol Infect (2014) 20(3):O142–9. doi: 10.1111/1469-0691.12365
35. Sakamuri RM, Harrison J, Gelber R, Saunderson P, Brennan PJ, Balagon M, et al. A continuation: study and characterisation of mycobacterium leprae short tandem repeat genotypes and transmission of leprosy in cebu, Philippines. Lepr Rev (2009) 80(3):272–9.
36. Young SK, Ponnighaus JM, Jain S, Lucas S, Suneetha S, Lockwood DN, et al. Use of short tandem repeat sequences to study mycobacterium leprae in leprosy patients in Malawi and India. PloS Negl Trop Dis (2008) 2(4):e214. doi: 10.1371/journal.pntd.0000214
37. Xing Y, Liu J, Sakamuri RM, Wang Z, Wen Y, Vissa V, et al. VNTR typing studies of mycobacterium leprae in China: assessment of methods and stability of markers during treatment. Lepr Rev (2009) 80(3):261–71. doi: 10.47276/lr.80.3.261
38. Lavania M, Katoch K, Sharma R, Sharma P, Das R, Gupta AK, et al. Molecular typing of mycobacterium leprae strains from northern India using short tandem repeats. Indian J Med Res (2011) 133(6):618–26.
39. Fontes AN, Sakamuri RM, Baptista IM, Ura S, Moraes MO, Martínez AN, et al. Genetic diversity of mycobacterium leprae isolates from Brazilian leprosy patients. Lepr Rev (2009) 80(3):302–15. doi: 10.47276/lr.80.3.302
40. Monot M, Honoré N, Balière C, Ji B, Sow S, Brennan PJ, et al. Are variable-number tandem repeats appropriate for genotyping mycobacterium leprae? J Clin Microbiol (2008) 46(7):2291–7. doi: 10.1128/JCM.00239-08
41. Matsuoka M, Maeda S, Kai M, Nakata N, Chae GT, Gillis TP, et al. Mycobacterium leprae typing by genomic diversity and global distribution of genotypes. Int J Lepr Other Mycobact Dis (2000) 68(2):121–8.
Keywords: mycobacterium leprae, environment, transmission, SNP-VNTR typing, leprosy, genotyping, clinical samples
Citation: Turankar RP, Singh V, Lavania M, Singh I, Sengupta U and Jadhav RS (2022) Existence of viable Mycobacterium leprae in natural environment and its genetic profiling in a leprosy endemic region. Front. Trop. Dis 3:972682. doi: 10.3389/fitd.2022.972682
Received: 18 June 2022; Accepted: 18 July 2022;
Published: 19 August 2022.
Edited by:
Esaki M. Shankar, Central University of Tamil Nadu, IndiaReviewed by:
Gerald Mboowa, Makerere University, UgandaCopyright © 2022 Turankar, Singh, Lavania, Singh, Sengupta and Jadhav. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rupendra S. Jadhav, cnVwZW5qYWRoYXZAeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.