- 1Department of Tropical Medicine, The Jikei University School of Medicine, Tokyo, Japan
- 2Center for Medical Entomology, The Jikei University School of Medicine, Tokyo, Japan
- 3National Institute of Infectious Diseases and Vaccinology, National Health Research Institutes, Zhunan, Taiwan
- 4Department of Virology 1, National Institute of Infectious Diseases, Tokyo, Japan
Detection of infectious viruses in mosquitoes is one of the prerequisite measures to monitor the prevalence of vector-borne viral diseases. Determining which mosquitoes are currently infected with arboviruses such as Zika, dengue, and chikungunya virus is not yet practical in endemic areas due to multiple causes including the difficulty of dealing with the virus’ unstable RNA. In this study, instead of handling viral RNA, virus-derived DNA (vDNA) was introduced as a target template for nucleic acid amplification. In combination with loop-mediated isothermal amplification (LAMP), we examined a LAMP-based vDNA detection assay (vDNA-LAMP) targeting Zika virus (ZIKV). The vDNA-LAMP reaction amplifying part of the NS3 region of ZIKV successfully detected its vDNA from crude DNA purified from artificially infected cultured cells and Aedes mosquitoes. This rapid, simple, and versatile method may provide a promising field-surveillance method for arbovirus circulation via vector mosquitoes.
Introduction
Zika virus (ZIKV) is one of the family Flaviviridae, genus flavivirus, which is closely related to dengue virus. Infection with ZIKV in humans passes with an asymptomatic course in most cases. However, ZIKV infection during pregnancy causes microcephaly and adverse fetal outcomes (1). Unlike the dengue virus, for which the natural vertebrate host is humans, ZIKV outbreaks are often a result of spillover from sylvatic reservoirs (2). ZIKV transmission is expected to be induced primarily by blood feeding of Aedes mosquitoes including Aedes aegypti and Ae. albopictus. Vertical transmission of ZIKV in Aedes mosquitoes, like other flaviviruses including dengue, has been demonstrated by laboratory studies (3–5). Moreover, it has been reported that wild Ae. aegypti larvae carried ZIKV, presenting natural vertical transmission (6). The emergence of ZIKV-infected larvae was also demonstrated by artificial contamination of water-inhabiting larvae with ZIKV, suggesting that infection of mosquitoes occurred without blood feeding (7). Thus, hindering the authentic and alternative ZIKV transmission cycles via mosquitoes may mitigate this increasing health burden.
Routine surveillance of potential vector populations is one of the priorities for prevention strategies and monitoring virus circulation to manage endemic situations. However, detection or surveillance of ZIKV in field-caught mosquitoes is not yet routine in endemic areas, including those with high transmission. Several studies reported the detection of ZIKV viral RNA in mosquitoes using conventional RT-PCR or real-time RT-PCR (8–13). Possible explanations for the lack of ZIKV detection from wild mosquitoes were proposed: delay between identifying illness in people and conducting field investigations, difficulty in approaching households with patients, inefficient sampling methods, and the prompt spraying of houses once cases were identified (14). In addition, because RNA is more unstable and prone to degradation than DNA, we also expect difficulty in storing and handling RNA in endemic areas. The genome of ZIKV is RNA, and prospective monitoring of ZIKV in wild mosquitoes may require trained people, appropriate tools to store RNA, and special equipment for RNA handling. These requirements likely prevent on-site mosquito monitoring in endemic area, and the establishment of a faster and easier measure will be required for fruitful surveillance.
Virus-derived DNA (vDNA), a newly identified form in the life cycle of RNA virus infecting arthropods, was first discovered in Drosophila melanogaster infected with Flock House Virus (FHV) (15). Evidence of the generation of vDNA during other arbovirus infections in mosquitoes has also been reported from studies using conventional PCR (16, 17). Given that the presence of vDNA may indicate the presence of an RNA virus, vDNA may be a suitable candidate target for molecular surveillance because of its greater stability.
In this study, we applied the loop-mediated isothermal amplification (LAMP) method to detect vDNA instead of viral RNA in arbovirus-infected mosquitoes. LAMP is a simple and rapid DNA detection method employing Bst DNA polymerase that enables DNA amplification in isothermal conditions with high sensitivity and specificity (18). It has been applied for detecting malaria parasite in artificially infected mosquitoes and filaria in both artificially infected and wild vector mosquitoes (19, 20). We demonstrated that LAMP detected vDNA of ZIKV from both cultured cells and vector mosquitoes, making LAMP targeting vDNA (named vDNA-LAMP) a candidate method applicable for on-site surveillance of mosquitoes carrying arboviruses.
Methods
Virus and Cell Culture
Two standard ZIKV strains, MR-766-NIID (African lineage) and PRVABC59 (Asian lineage), were used in this study. ZIKV were passaged in Ae. albopictus C6/36 cells twice following in African green monkey kidney Vero cells (ATCC) once to generate viral stocks. C6/36 cells were cultured in D-MEM, high glucose (Thermo Fisher Scientific Inc.) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Vero cells were cultured in E-MEM supplemented with 10% FBS, MEM Non-Essential Amino Acids Solution (Thermo Fisher Scientific Inc.), and penicillin-streptomycin.
Mosquito Rearing
The Higgs strain of Ae. aegypti was used in this study. Mosquito larvae were reared at 28°C and fed with a mixture of yeast powder (Taiwan Sugar Corporation) and goose liver powder (#7573, NTN) in a 1:1 ratio. Adults were maintained in a temperature and humidity controlled room (28°C and ~70% relative humidity) with a 12-hour light/dark cycle and provided with a constant 10% sucrose solution.
Viral RNA Extraction and cDNA Synthesis
Viral RNAs were extracted from viral stocks using High Pure Viral Nucleic Acid Kit (Roche Ltd.), following the manufacturer’s instructions but without Proteinase K treatment. cDNA was synthesized from ZIKV RNA as follows: 1 µg of extracted RNA dissolved in 8.5 µl of RNase-free water was incubated with a reaction mixture [3 µl of 40 µM Random Primers (Thermo Fisher Scientific Inc.), 6 µl of 5× First-Strand Buffer (Thermo Fisher Scientific Inc.), 1.9 µl of 0.1 M DTT (Thermo Fisher Scientific Inc.), and 7.5 µl of dNTP Mixture (Takara Bio Inc.)] at 65°C for 5 min. Then, 0.6 µl of RNase inhibitor (Promega Co.) and 0.5 µl of M-MLV reverse transcriptase (Thermo Fisher Scientific Inc.) were added to the reaction mixture and incubated at 37°C for 90 min.
Virus Infection
C6/36 cells were seeded in the culture flask and incubated at 28°C. Infection with the MR strain was performed at multiplicities of infection (MOI) of 0.1 or 1. For the PR strain, accurate MOI of the viral stock was not determined, and a series of 10-fold dilution of the viral stock was used for infection. Infected cells were kept at 28°C for 5 days.
A natural vector species transmitting ZIKV, Ae. aegypti, was used in this study. For oral infection, 1×107 PFU/mL virus stock (PR strain) was 1:1 mixed with mouse blood and fed to female mosquitoes at 37°C for 30 min via a membrane-covered metal plate. Blood-engorged mosquitoes were kept at 28°C for 10 days post infection by feeding with sugar solution. These mosquitoes were killed, dried for 12 hours at room temperature, and stored at -20°C until DNA extraction.
DNA Extraction
DNA was extracted from ZIKV-infected cultured cells and mosquitoes as follows. Briefly, cells or mosquitoes were homogenized with a plastic homogenizer in 100 µl of Buffer A (0.1 M Tris (pH 9.0), 0.1 M EDTA, 1% SDS, and 0.5% DEPC), and incubated for 30 min at 70°C. Next, 22.4 µl of 5 M KOAc was added to the mixture, followed by incubation of the mixture for 30 min on ice. Supernatant was collected by centrifugation at 20,400 × g for 15 min at 4°C, and then mixed with 45 µl of isopropanol. Precipitated DNA was collected after centrifugation at 20,400 × g for 20 min at 4°C, rinsed with 70% ethanol, and dried. Each DNA pellet was dissolved and diluted with TE to adjust the concentration so that 1 µl of solution contained DNA from 1.2 × 104 infected cultured cells or from one-fiftieth of each mosquito pool. One microliter of each DNA solution was used as a template for the LAMP reaction.
LAMP Reaction
Primers for the LAMP reaction were designed using Primer Explorer V5 (Fujitsu Ltd.). LAMP reactions were performed as described in the manufacturer’s instructions (Eiken Chemical Co., Ltd.) with a half volume. Briefly, each reaction was performed in a total volume of 12.5 µl of reaction mixture containing 20 pmol of each FIP and BIP primers, 2.5 pmol of each F3 and B3 primers, 10 pmol of each Loop-F and Loop-B primers, and 6.25 µl of 2× reaction mixture (Table 1). For DNA amplification, the reaction mixture also contained 1.0 µl of extracted DNA solution and 0.5 µl of Bst DNA polymerase. For LAMP with reverse transcription step, the reaction mixture contained 2.5 µl of extracted RNA solution and 0.5 µl of enzyme mix (Bst DNA polymerase and AMV reverse transcriptase). Each reaction mixture was incubated at 62°C for 60 min or 90 min and terminated by incubation at 80°C for 5 min using Loopamp Realtime Turbidimeter (LoopampEXIA; Eiken Chemical Co., Ltd.). Amplified products of the reaction were examined by electrophoresis in 2% agarose gels. The gels were stained with ethidium bromide and visualized under UV light.
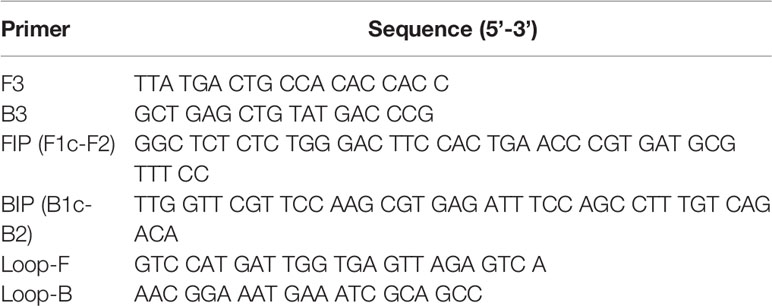
Table 1 Sequences of primers for LAMP designed in accordance with the nucleotide sequence of the ZIKV MR strain.
Results
Detection of Viral cDNA Using LAMP
To develop vDNA-LAMP method for detecting various ZIKV strains, we used a primer set we reported previously (21). Full genomic RNA sequences of ZIKV MR-766-NIID strain (GenBank: LC002520.1) and PRVABC59 (GenBank: KU501215.1) strain were aligned to find high consensus sequences for making LAMP primers (Supplementary Figure 1). Several candidate primer sets were made based on the sequence of the MR strain, and eventually one of the primer sets targeting the NS3 region was selected due to its adequate sensitivity (21) (Figure 1 and Table 1).
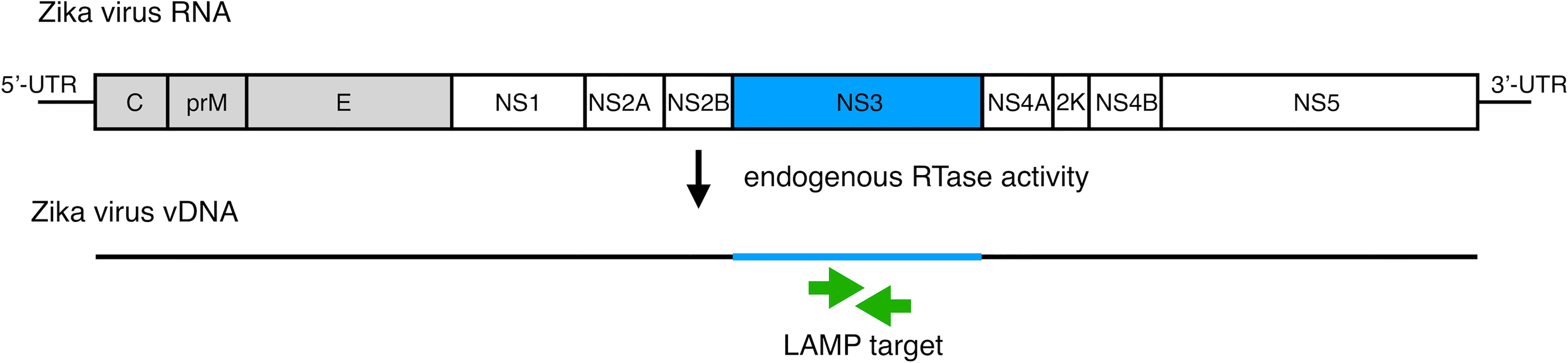
Figure 1 Schematic diagram of the ZIKV RNA genome with LAMP primer positions for amplifying vDNA, which is generated from ZIKV genomic RNA using endogenous RTase activity in mosquito cells. The vDNA region between a set of primers (green arrows) is the target for the LAMP reaction.
Prior to detecting vDNA, viral cDNA of ZIKV was prepared and provided for the LAMP reaction. Serially diluted RNA (3 × 102 ng, 3 × 101 ng, and 3 × 100 ng) of each strain was transcribed to form cDNA using reverse transcriptase, followed by the LAMP reaction to detect these cDNAs under optimized conditions (60 min at 62°C) in one-step. The target region of ZIKV was amplified from all cDNA samples, suggesting that the primer set can be applicable to vDNA-LAMP (Supplementary Figure 1B).
LAMP Detection of Virus-Derived DNA in ZIKV-Infected Culture Cells
To examine whether LAMP can amplify vDNA of ZIKV, we first attempted to detect vDNA in cultured mosquito cells that were spiked with the virus. DNA was extracted from cultured Aedes C6/36 cells at 5 days post infection with ZIKV MR or PR strain and provided as a template for the LAMP reaction (Figure 2A). LAMP reaction was performed in conditions that was optimized for the detection of ZIKV cDNA. Amplified products were examined by electrophoresis, showing the presence of DNA fragments derived from both ZIKV MR and PR strains (Figure 2B). These data suggested that vDNA-LAMP is sufficient to detect the vDNA form of ZIKV, which is generated in mosquito cells.
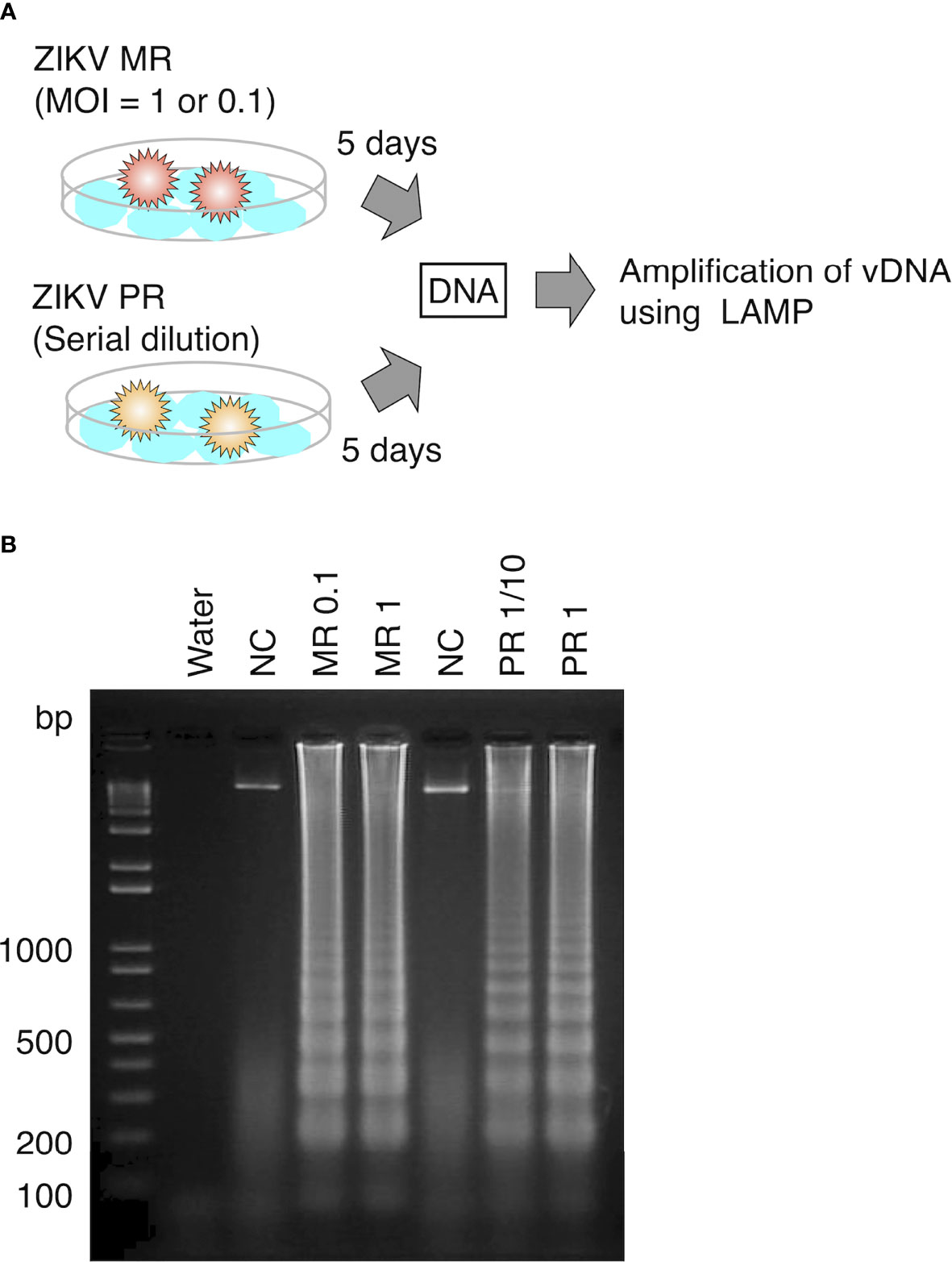
Figure 2 Detection of vDNA using LAMP in ZIKV-infected mosquito cells. (A) Scheme of preparing ZIKV-infected cultured cells for vDNA detection. DNA was purified from ZIKV-infected cells and uninfected cells at 5 days post infection and utilized for LAMP reaction. (B) vDNA was amplified using LAMP from mosquito C6/36 cells infected with ZIKV MR or PR strain. Amplified products were separated on 2% agarose gels. Water served as a negative control. MR: cells infected with MR strain at an MOI = 1 or 0.1. PR: cells infected with PR strain (PR 1, infection without dilution of virus stock; PR 1/10, 10-fold dilution of virus stock). NC, negative control (uninfected cells).
Application of vDNA-LAMP to ZIKV-Infected Mosquitoes
To evaluate the usability of vDNA-LAMP in virus-transmitting mosquitoes, we attempted to detect ZIKV vDNA from experimentally infected mosquitoes. Ae. aegypti female mosquitoes were orally exposed to ZIKV PR strain by artificial membrane feeding using infected blood. At 10 days post infection, mosquitoes were killed and pooled in 12 groups (5 mosquitoes per group). These mosquitoes were dried at room temperature for 12 hours and then kept in the freezer. DNA was extracted from each group and then subjected to the LAMP reaction (Figure 3A). As a result, out of 12 groups of infected blood-fed mosquitoes, 5 groups were identified as positive by vDNA-LAMP reaction and following electrophoresis, suggesting that vDNA transcribed from ZIKV RNA in infected mosquitoes can be detected using the LAMP reaction (Figure 3B).
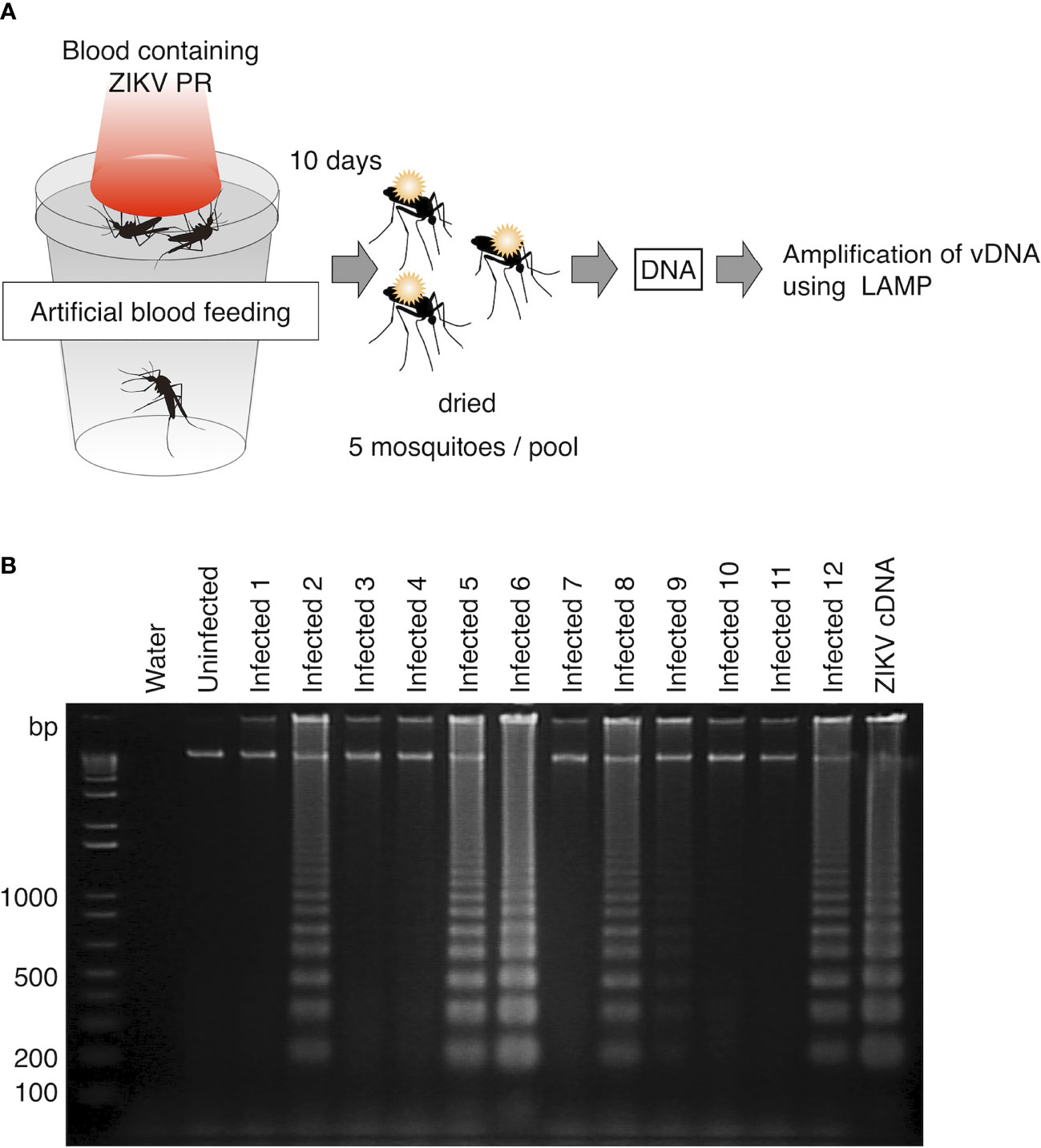
Figure 3 Detection of vDNA using LAMP in ZIKV-infected mosquitoes. (A) Scheme for preparing ZIKV (PR strain)-infected mosquitoes for vDNA detection. Five infected mosquitoes were pooled in each group and subjected to DNA extraction. One-fiftieth of the extracted DNA per group was used as template for the LAMP reaction. (B) vDNA of ZIKV in infected mosquitoes was detected using LAMP in 6 groups (Infected 2, Infected 5, Infected 6, Infected 8, Infected 9, and Infected 12) out of 12. Amplified products were electrophoresed on 2% agarose gels. ZIKV cDNA was used as positive control. Water served as a negative control. Numbers of the left indicated migration of molecular weight marker (bp).
Discussion
Infection with arboviruses such as Zika, dengue, chikungunya, and Japanese encephalitis virus, remains an important issue for global public health. In particular, Zika virus disease, which caused large outbreaks mainly in the Americas, has suddenly gained attention due to reports of fetal microcephaly. Both Zika virus and dengue virus are transmitted by Aedes mosquitoes, continuously spreading worldwide, including Ae. aegypti and Ae. albopictus. Rapid and accurate surveillance is still strongly in demand because it is needed to strengthen and integrate with health information systems to guide proper vector control.
The vDNA-LAMP, newly developed in this study, demonstrated that it can successfully detect ZIKV vDNA in infected cultured mosquito cells and mosquitoes artificially exposed to ZIKV. The vDNA-LAMP primer set, which was designed to detect two ZIKV strains (MR and PR), actually corresponded to the sequence of the MR strain. It detected vDNA of both MR and PR strains in cultured cells and vDNA of PR strain in Aedes mosquitoes. These findings suggested that several mismatches in primer regions may not affect the sensitivity of the amplifying reaction, although longer reaction time was needed to detect the PR strain, as we reported previously (21).
In the experiment using mosquitoes, vDNA-LAMP identified half of the pooled groups (each containing 5 mosquitoes) as positive, whereas the other 6 groups did not reach the amplification of the ZIKV target sequence. What can be assumed is the possibility that not all mosquitoes that were orally exposed to the virus established an infection, possibly due to the host defense system in the midgut (22). In cases where exposed mosquitoes did not develop an infection or presented with a low virus titer, few or no copies of vDNA could be isolated from those mosquitoes.
It has been argued that molecular monitoring of mosquitoes that carry viruses is challenging in particular in developing countries where arbovirus-borne diseases are often endemic; detection of viral RNA in mosquitoes needs freezing storage of mosquito samples, trained personnel to handle RNA, and expensive equipment such as PCR machines. It is notable that vDNA-LAMP, being based on isothermal amplification, is a simple and rapid virus-detecting method targeting stable DNA, not RNA, in isothermal conditions. DNA is more stable than RNA because it is double stranded and does not contain reactive OH groups as in RNA. Indeed, vDNA was detected even from dried mosquito samples in which RNA is expected to be easily degradable. These features may provide a great advantage to molecular surveillance of disease-vector mosquitoes which is carried out outside or in conventional laboratories. Further experiments comparing conventional qPCR with RNA and vDNA-LAMP will clarify the assessment of the applicability of vDNA-LAMP in routine surveillance.
Conclusion
The present study developed a new method for detecting arbovirus, vDNA-LAMP, employing vDNA as a target of amplification using the LAMP reaction, and we demonstrated the detection of vDNA of ZIKV in infected cells and mosquitoes. vDNA-LAMP is easily applicable for field surveys of virus infection in wild mosquitoes without RNA storage and handling. To confirm whether vDNA-LAMP will be a realistic alternative to conventional surveillance methods, further experiments using wild mosquito populations need to be performed. Implementing vDNA-LAMP in endemic areas may offer a promising strategy in monitoring virus circulation via vector mosquitoes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
HA and HK conceived the study and wrote the manuscript. HA performed the experiments and analyzed the data. II prepared cultured cells, standard strain of viruses, and virus RNA. MO prepared infected cultured cells. J-CL reared and prepared artificially infected mosquitoes. ST and MS provided viruses and assisted with virus infection of cultured cells. C-HC conceived and supervised virus infection of mosquitoes. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the International Collaborative Research Program for Tackling the NTDs Challenges in African countries from Japan Agency for Medical Research and Development, AMED (JP17jm0510002h0003) (HK), the JSPS KAKENHI Grant Number 19H03462 (HK), and Ohyama Health Foundation Inc. (HA). The funding agencies had no role in the design and conduct of the study, collection, management, analysis, interpretation of the data, preparation, review, or approval of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all members of the Kanuka laboratory for their assistance, in particular Kazutomo Ogata and Chisako Sakuma for mosquito rearing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2022.759375/full#supplementary-material
Supplementary Figure 1 | (A) LAMP primers for amplifying vDNA of ZIKV. Partial sequences of the NS3 region according to the nucleotide sequences of the MR (upper) and PR (lower) strains. Nucleotide mismatches between MR and PR strains are indicated as Y, R, M, and K (Y: C or T, R: A or G, M: A or C, K: G or T). Location and direction of primers, FIP (F1c-F2), BIP (B1c-B2), F3, B3, Loop-F, and Loop-B, are indicated by arrows. (B) LAMP detection of cDNA of ZIKV. Serially diluted RNA (3 × 102, 3 × 101, and 3 × 100 ng) of the MR or PR strain was used as template for reverse transcription to obtain cDNA, followed by the LAMP reaction in one-step. Amplified products were electrophoresed on 2% agarose gels. Water served as a negative control. Numbers of the left indicated migration of molecular weight marker (bp).
References
1. Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects–Reviewing the Evidence for Causality. N Engl J Med (2016) 374:1981–7. doi: 10.1056/NEJMsr1604338
2. Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H, et al. Assessing the Global Threat From Zika Virus. Science (2016) 353(6300):aaf8160. doi: 10.1126/science.aaf8160
3. Ciota AT, Bialosuknia SM, Ehrbar DJ, Kramer LD. Vertical Transmission of Zika Virus by Aedes Aegypti and Ae. Albopictus Mosquitoes. Emerg Infect Dis (2017) 23:880–2. doi: 10.3201/eid2305.162041
4. Thangamani S, Huang J, Hart CE, Guzman H, Tesh RB. Vertical Transmissions of Zika Virus in Aedes Aegypti Mosquitoes. Am J Trop Med Hyg (2016) 95:1169–73. doi: 10.4269/ajtmh.16-0448
5. Rosen L, Shroyer DA, Tesh RB, Freier JE, Lien JC. Transovarial Transmission of Dengue Viruses by Mosquitoes: Aedes Albopictus and Aedes Aegypti. Am J Trop Med Hyg (1983) 32:1108–19. doi: 10.4269/ajtmh.1983.32.1108
6. Costa CFD, Silva AVD, Nascimento VAD, Souza VC, Monteiro DCDS, Terrazas WCM, et al. Evidence of Vertical Transmission of Zika Virus in Field-Collected Eggs of Aedes Aegypti in the Brazilian Amazon. PloS Negl Trop Dis (2018) 12:e0006594. doi: 10.1371/journal.pntd.0006594
7. Du S, Liu Y, Liu J, Zhao J, Champagne C, Tong L, et al. Aedes Mosquitoes Acquire and Transmit Zika Virus by Breeding in Contaminated Aquatic Environments. Nat Commn (2019) 10:1324. doi: 10.1038/s41467-019-09256-0
8. Grard G, Caron M, Mombo IM, Nkoghe D, Mboui Ondo S, Jiolle D, et al. Zika Virus in Gabon (Central Africa) - 2007: A New Threat From Aedes Albopictus? PloS Negl Trop Dis (2014) 8:e2681. doi: 10.1371/journal.pntd.0002681
9. Guerbois M, Fernandez-Salas I, Azar SR, Danis-Lozano R, Alpuche-Aranda CM, Leal G, et al. Outbreak of Zika Virus Infection, Chiapas State, Mexico, 2015, and First Confirmed Transmission by Aedes Aegypti Mosquitoes in the Americas. J Infect Dis (2016) 214:1349–56. doi: 10.1093/infdis/jiw302
10. Díaz-Quiñonez JA, López-Martínez I, Torres-Longoria B, Vázquez-Pichardo M, Cruz-Ramírez E, Ramírez-González JE, et al. Evidence of the Presence of the Zika Virus in Mexico Since Early 2015. Virus Genes (2016) 52:855–7. doi: 10.1007/s11262-016-1384-0
11. Faye O, Faye O, Diallo D, Diallo M, Weidmann M, Sall AA. Quantitative Real-Time PCR Detection of Zika Virus and Evaluation With Field-Caught Mosquitoes. Virol J (2013) 10:311. doi: 10.1186/1743-422X-10-311
12. Ayllón T, Campos RM, Brasil P, Morone FC, Câmara DCP, Meira GLS, et al. Early Evidence for Zika Virus Circulation Among Aedes Aegypti Mosquitoes, Rio De Janeiro, Brazil. Emerg Infect Dis (2017) 23:1411–2. doi: 10.3201/eid2308.162007
13. Smartt CT, Stenn TMS, Chen TY, Teixeira MG, Queiroz EP, Souza Dos Santos L, et al. Evidence of Zika Virus RNA Fragments in Aedes Albopictus (Diptera: Culicidae) Field-Collected Eggs From Camaçari, Bahia, Brazil. J Med Entomol (2017) 54:1085–7. doi: 10.1093/jme/tjx058
14. Diallo D, Diallo M. Why Is Zika Virus So Rarely Detected During Outbreaks and How can Detection be Improved? BMC Res Notes (2017) 10:524. doi: 10.1186/s13104-017-2854-8
15. Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, et al. RNA-Mediated Interference and Reverse Transcription Control the Persistence of RNA Viruses in the Insect Model Drosophila. Nat Immunol (2013) 14:396–403. doi: 10.1038/ni.2542
16. Goic B, Stapleford KA, Frangeul L, Doucet AJ, Gausson V, Blanc H, et al. Virus-Derived DNA Drives Mosquito Vector Tolerance to Arboviral Infection. Nat Commun (2016) 7:12410. doi: 10.1038/ncomms12410
17. Nag DK, Kramer LD. Patchy DNA Forms of the Zika Virus RNA Genome Are Generated Following Infection in Mosquito Cell Cultures and in Mosquitoes. J Gen Virol (2017) 98:2731–7. doi: 10.1099/jgv.0.000945
18. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acid Res (2000) 28:E63. doi: 10.1093/nar/28.12.e63
19. Aonuma H, Suzuki M, Iseki H, Perera N, Nelson B, Igarashi I, et al. Rapid Identification of Plasmodium-Carrying Mosquitoes Using Loop-Mediated Isothermal Amplification. Biochem Biophys Res Commun (2008) 376:671–6. doi: 10.1016/j.bbrc.2008.09.061
20. Aonuma H, Yoshimura A, Perera N, Shinzawa N, Bando H, Oshiro S, et al. Loop-Mediated Isothermal Amplification Applied to Filarial Parasites Detection in the Mosquito Vectors: Dirofilaria Immitis as a Study Model. Parasit Vectors (2009) 2:15. doi: 10.1186/1756-3305-2-15
21. Aonuma H, Iizuka-Shiota I, Hoshina T, Tajima S, Kato F, Hori S, et al. Detection and Discrimination of Multiple Strains of Zika Virus by Reverse Transcription-Loop-Mediated Isothermal Amplification. Trop Med Health (2020) 48:87. doi: 10.1186/s41182-020-00274-z
Keywords: Zika virus, vDNA, RNA, mosquito, loop-mediated isothermal amplification, arbovirus
Citation: Aonuma H, Iizuka I, Li J-C, Ote M, Tajima S, Saijo M, Chen C-H and Kanuka H (2022) LAMP Detection of Virus-Derived DNA of Zika Virus in Vector Mosquito. Front. Trop. Dis 3:759375. doi: 10.3389/fitd.2022.759375
Received: 16 August 2021; Accepted: 24 January 2022;
Published: 18 February 2022.
Edited by:
Samuel Wanji, University of Buea, CameroonReviewed by:
Mayke Leggewie, Bernhard Nocht Institute for Tropical Medicine (BNITM), GermanyOctavio Talyuli, Federal University of Rio de Janeiro, Brazil
Copyright © 2022 Aonuma, Iizuka, Li, Ote, Tajima, Saijo, Chen and Kanuka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hirotaka Kanuka, a2FudWthQGppa2VpLmFjLmpw
†Present address: Itoe Iizuka, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
 Hiroka Aonuma
Hiroka Aonuma Itoe Iizuka1,2†
Itoe Iizuka1,2† Manabu Ote
Manabu Ote Shigeru Tajima
Shigeru Tajima Masayuki Saijo
Masayuki Saijo Chun-Hong Chen
Chun-Hong Chen Hirotaka Kanuka
Hirotaka Kanuka