
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis., 14 December 2021
Sec. Vector Biology
Volume 2 - 2021 | https://doi.org/10.3389/fitd.2021.806880
This article is part of the Research TopicThe Transition of Novel Genetic Control Strategies into Reality for Vector-Borne Tropical Disease Control and Prevention; Research Advances and next StepsView all 12 articles
 Sofia Tapanelli1
Sofia Tapanelli1 Maria Grazia Inghilterra1
Maria Grazia Inghilterra1 Julia Cai1
Julia Cai1 James Philpott1
James Philpott1 Paolo Capriotti1
Paolo Capriotti1 Nikolai Windbichler1
Nikolai Windbichler1 George K. Christophides1,2*
George K. Christophides1,2*Genetically modified (GM) mosquitoes expressing anti-plasmodial effectors propagating through wild mosquito populations by means of gene drive is a promising tool to support current malaria control strategies. The process of generating GM mosquitoes involves genetic transformation of mosquitoes from a laboratory colony and, often, interbreeding with other GM lines to cross in auxiliary traits. These mosquito colonies and GM lines thus often have different genetic backgrounds and GM lines are invariably highly inbred, which in conjunction with their independent rearing in the laboratory may translate to differences in their susceptibility to malaria parasite infection and life history traits. Here, we show that laboratory Anopheles gambiae colonies and GM lines expressing Cas9 and Cre recombinase vary greatly in their susceptibility to Plasmodium falciparum NF54 infection. Therefore, the choice of mosquitoes to be used as a reference when conducting infection or life history trait assays requires careful consideration. To address these issues, we established an experimental pipeline involving genetic crosses and genotyping of mosquitoes reared in shared containers throughout their lifecycle. We used this protocol to examine whether GM lines expressing the antimicrobial peptide (AMP) Scorpine in the mosquito midgut interfere with parasite infection and mosquito survival. We demonstrate that Scorpine expression in the Peritrophin 1 (Aper1) genomic locus reduces both P. falciparum sporozoite prevalence and mosquito lifespan; both these phenotypes are likely to be associated with the disturbance of the midgut microbiota homeostasis. These data lead us to conclude that the Aper1-Sco GM line could be used in proof-of-concept experiments aimed at mosquito population replacement, although the impact of its reduced fitness on the spread of the transgene through wild populations requires further investigation.
Engineering the mosquito genome has become a key resource in renewed efforts to control the spread of malaria (1). Following the establishment of a reliable genome transformation method in Anopheles mosquitoes (2, 3), several studies explored the use of homing endonucleases (HEGs), including Cas9, in conjunction with gene drive as a method to spread engineered genetic traits in wild vector populations (4–7). Such traits are aimed at either reducing the vector effective population size (population suppression) or rendering the vector less capable of transmitting malaria (population replacement).
We have previously established a novel approach, termed Integral Gene Drive (IGD), which allows us to introduce minimal genetic modifications in any An. gambiae gene of choice making it able to express small effector proteins targeting the malaria parasite, including exogenous AMPs (8, 9). Creation of an IGD effector line entails precision engineering of the endogenous gene of choice using CRISPR/Cas9, by introducing a sequence that encodes the small effector protein in frame with the endogenous protein-coding sequence; the two sequences can be separated with the insertion of a 2A ribosomal skipping sequence or remain fused. The exogenous sequence contains two additional expression cassettes often found within an intron: the first produces the guide RNA (gRNA) that allows the engineered allele to home in the presence of Cas9, and the second expresses a fluorescent reporter gene that serves as a transformation marker, flanked by LoxP sites that enables it to be removed at a later stage in the presence of Cre recombinase.
P. falciparum, the deadliest of human malaria parasites, spends about a day inside the mosquito midgut after being ingested with a bloodmeal and before it establishes an infection in the midgut wall in the form of an oocyst (10). In our first proof-of-concept of the IGD protocol, we expressed the AMP Scorpine in the adult female An. gambiae midgut, targeting early malaria transmission stages (9). This was achieved by introducing Scorpine in either of three bloodmeal-induced loci expressing the blood-processing enzymes carboxypeptidase (CP) and alkaline phosphatase 2 (AP2) and the type I peritrophic matrix protein Peritrophin 1 (Aper1). While Scorpine was separated from CP and AP2 through a 2A ribosomal skipping peptide, it was designed to remain fused with Aper1, leading to it being anchored to the peritrophic matrix. The three resulting lines were designated as Sco-CP, ScoG-AP2 and Aper1-Sco, respectively. The generation of these three lines required genetically transformed mosquitoes of the An. gambiae G3 laboratory colony to be crossed to a KIL line expressing Cre-recombinase (KILvasa-Cre) to remove the fluorescent marker and a G3 Cas9-expressing line (G3vasa-Cas9) to facilitate homing and homozygosity of the transgene.
The next step in the path toward establishing whether an IGD line displays the qualities required for a population replacement strategy involves a series of laboratory assays to assess malaria transmission reduction efficacy and any potential impact of the modifications on mosquito life history traits (11). Assessment of malaria transmission reduction typically involves enumeration of oocysts in the mosquito midguts following infection with P. falciparum gametocytes using a standard membrane feeding assay (SMFA) (12). Infection prevalence and intensity are compared to a reference line, typically the parental line where the modifications have been performed. Using this protocol, we showed that two of the Scorpine-expressing lines, the ScoG-AP2 and the Aper1-Sco, exhibit significant reduction of the number of P. falciparum oocysts found in the mosquito midgut following an infectious bloodmeal compared to the G3 parental line.
Recent studies have established that the protocol mentioned above may not provide the most reliable measure of transmission reduction as it does not consider the oocyst developmental stage, growth rate and capacity to produce sporozoites (13, 14). Furthermore, these studies suggest that provision of regular supplemental bloodmeals following the initial infectious bloodmeal, each followed by egg laying, must be considered, as this better emulates the mosquito feeding and reproductive behavior in nature, while they significantly support and, indeed, accelerate oocyst development. Finally, since the reference line (including the parental line) is often genetically different and/or more diverse than the GM effector line to be tested, and is cultured independently of the GM line, the choice of reference requires careful consideration. Many of the above considerations also pertain to the assessment of survival and other life history traits.
Here, we investigate differences in P. falciparum infection between An. gambiae laboratory colonies and accessory GM lines used for further genome manipulation, such as G3vasa-Cas9 and KILvasa-Cre, and establish a new experimental protocol that addresses the aforementioned considerations. Using this protocol, we examine the malaria transmission blocking efficacy of the IGD Scorpine-expressing lines (9), by assessing the presence and abundance of sporozoites in the mosquito salivary glands. Finally, we investigate the effect of Scorpine expression within the Aper1 genomic locus on mosquito survival. Our work is fundamental to establishing the most appropriate protocols for testing the characteristics of GM An. gambiae lines prepared for gene drive and vector population replacement.
The A. coluzzii N’gousso colony was established from field-collected mosquitoes of the An. gambiae M molecular form in 2006 in Cameroon (15). The An. gambiae G3 strain was obtained from the Malaria Research and Reference Reagent Center (MR4; BEI Resources) and is classified as An. gambiae S molecular form. The An. gambiae G3 vasa-Cas9 (G3vasa-cas9), KIL vasa-Cre (KILvasa-Cre) and effector IGD (ScoG-AP2, Sco-CP and Aper1-Sco) strains were a kind gift of their respective creators (6, 9, 16). All strains were reared and maintained at standard laboratory conditions (27 ± 1°C and 70 ± 5%) humidity on a 12:12 light/dark cycle interspersed by 0.5 h dawn and dusk transitions.
Plasmodium falciparum NF54 strain was obtained from the MR4 (MRA-1000, patient E). Parasite culturing and mosquito SMFAs were performed as described (12). Briefly, asexual parasite stages were cultured in RPMI-1640-R5886 (Sigma) medium supplemented with hypoxanthine to end concentration of 0.05 g/L, L-glutamine powder-(Sigma) to end concentration of 0.3 mg/L and 10% sterile human serum. Gametocytogenesis was induced by diluting the asexual culture (3-4% ring forms) to 1% ring forms with fresh human red blood cells (hRBCs) and maintained at 37°C, ensuring a daily exchange of about 75% of the medium. Day 14-16 mature gametocytes were pooled in a pre-warmed 15 mL tube containing 20% hRBCs and 50% heat-inactivated human serum. Each membrane feeder was loaded with 300 μl of the gametocyte mixture and kept at a constant temperature of 37°C. Paper cups of 70-100 female mosquitoes were allowed to feed for 10-15 minutes. After the feed, mosquitoes were maintained in a chamber set at 27°C and 70% humidity. Cotton pads soaked in 10% fructose were placed on mosquito cups only 36 hours after the infectious blood feed or any supplemental, non- infectious blood feed so that non-blood-fed mosquitoes were starved to death. Cotton pads were replaced every two days.
For oocyst enumeration, mosquito midguts were dissected 7 days post-bloodmeal, stained with 1% mercurochrome and fixed in 4% formaldehyde. Oocysts were counted using a bright-field light microscope at 20X magnification. Data on oocysts in G3 mosquitoes were pooled from 16 independent replicates corresponding to 9, 26, 31, 30, 32, 37, 23, 16, 40, 12, 9, 50, 51, 41, 33 and 39 mosquitoes, respectively. For N’gousso, data from 19 independent replicates were pooled, which corresponded to 25, 28, 67,38, 30, 30, 41, 61, 43, 28, 54, 47, 50, 21, 19, 33, 32, 19 and 17 mosquitoes, respectively. For the comparison between the G3, KILvasa-Cre and G3vasa-cas9 lines, data from 4 independent replicates were pooled, corresponding to: 41, 50, 12 and 39 G3; 38, 46, 12 and 73 KILvasa-Cre; and 33, 51, 8 and 13 G3vasa-cas9 mosquitoes.
Individual mosquitoes were genotyped by PCR using the Phire Tissue Direct PCR Master Mix (ThermoFisher Scientific) on genomic DNA extracted from mosquito abdomens using the Qiagen DNeasy Blood & Tissue Kits (Qiagen) and genotypes were distinguished as previously described. The PCR primers used in genotyping assays were described previously (9): CP-locus-F: ATTATCGGAAATTCTCCACAAAGCG, CP-locus-R: TCATGGTGCGATTCTGATGCGATCG, AP-locus-F: ATCAGACTTTAAGCCTTTGTCGTC and AP-locus-R: ACCGTTCGCGATTCAGCAGTACAGG. The PCR conditions included denaturation at 98°C for 10 sec, annealing at 52°C for 2 min and elongation at 72°C for 1 min, repeated for 35 cycles.
For sporozoite quantification, mosquitoes were dissected 14 days post-infection, after they have completed 4 full gonotrophic cycles including the first infectious blood-feeds and 3 non-infectious supplementary blood-feeds at days 4, 6 and 9, each followed by egg laying at days 3, 5, 8 and 11 post-infection, respectively. Heads and thoraxes were separated from abdomens using a scalpel and preserved in TRIzol™ reagent in -80°C until they were processed for RNA extraction according to the manufacture’s guidelines (Invitrogen). RNA samples were stored at -80°C until cDNA synthesis using the iScript™ gDNA Clear cDNA Synthesis Kit according to the manufacture’s guidelines (BIO-RAD). A qPCR-based assay was used to detect and quantify PfCSP transcripts (17), normalized against the An. gambiae ribosomal S7 gene (18). The qPCR was performed using SYBR™ Green PCR Master Mix (Thermo Fisher Scientific) using primers: PfCSP-F: TCAACTGAATGGTCCCCATGT, PfCSP-R: GAGCCAGGCTTTATTCTAACTTGAAT, S7-F: GAGGTCGAGTTCAACAACAA, and S7-R: GAACACGACGTGCTTACC. A total of 2 µl of cDNA template was added on a volume of 20 µl of qPCR mix. The qPCR parameters included the initial denaturation at 95 °C for 15 min, further denaturation at 95 °C for 30 sec, followed by annealing at 60 °C for 45 sec and extension at 72 °C for 45 sec repeated for 40 cycles. Final elongation was performed at 72 °C for 7 min followed by the generation of a melt curve ranging from 65 °C to 95 °C. A standard curve was generated to determine the PCR efficiency. Samples were considered non-infected when no Cp value was calculated using Absolute quantification/2nd derivative Max analysis on the Light Cycler® 480 (Roche, Software release 1.5.1.62 SPS). The abdomens from the same mosquitoes were used to extract genomic DNA for genotyping as described in the relevant section.
Mosquitoes were provided supplemental bloodmeals every three days and allowed to deposit eggs. After the first blood meal, individual dead mosquitoes were collected every 24 hours until the caged population was extinct. Mosquito carcasses were used for genotyping as described in the relevant methods section.
The microbiota analysis was performed as previously described (19). Briefly, DNA was extracted from whole mosquitoes collected 24 hours after the first or second bloodmeal. The total load of bacteria was measured by quantifying the bacterial ribosomal 16S RNA expression, normalized against the expression of the An. gambiae ribosomal protein S7, on the 7500 Fast Real-Time PCR system (Applied Biosystems) in 10 µL reactions using Fast SYBR™ Green PCR Master Mix according to the manufacturer’s instructions (Thermo Fisher Scientific). The PCR primers used were: Eubacteria 16S-F: TCCTACGGGAGGCAGCAGT, Eubacteria 16S-R: GGACTACCAGGGTATCTAATCCTGTT, Enterobacteriaceae 16S-F: CGTGCTACAATGGCATATACAAAGAGAAG, Enterobacteriaceae 16S-R: AGCATTCTGATCTACGATTACTAGCGATTC, Flavobacteriaceae 16S-F: TAAGGTTGAAGTGGCTGGAATAA, Flavobacteriaceae 16S-R: GTCCATCAGCGTCAGTTAAGACT, S7-F: GTGCGCGAGTTGGAGAAGA and S7-R: ATCGGTTTGGGCAGAATGC. All the primer combinations were highly specific producing only a single peak in the PCR melt curve.
Infection intensity was assessed by the enumeration of oocysts and by molecular quantification of sporozoites using qPCR. We used Mann-Whitney test for non-normal, unpaired data sets. The prevalence of infections and the difference in the survival percentage of mosquitoes was evaluated with the Chi-squared Pearson’s test. Estimates of the mosquito survival probability was performed with the Kaplan-Meier statistics. All statistical analyses were preformed using R (version 3.5.3) and Graph Pad Prism version 8.0.
We previously established a robust protocol for SMFAs using the A. coluzzii laboratory colony N’gousso (12). A. coluzzi, until recently known as M-form An. gambiae, is the main malaria vector in West sub-Saharan Africa (20). However, this species is not present in East Africa, where one of the main vectors is An. gambiae sensu stricto (s.s.), previously known as S-form An. gambiae and henceforth simply referred to as An. gambiae. Therefore, we had used An. gambiae G3 strain, the most widely and long-term used An. gambiae laboratory colony, for generating GM mosquitoes.
We compared the An. gambiae G3 (henceforth G3) and A. coluzzii N’gousso (henceforth N’gousso) with regards to their P. falciparum infection intensity and prevalence using data obtained from 19 and 16 independent, non-paired infection experiments, respectively (Figure 1A). The results showed that G3 exhibits a significantly lower infection intensity (p<0.0001, unpaired two-tailed Mann-Whitney test) with an oocyst mean of 11 (SD: +/-29.95, SEM: +/-1.38), median of 0 and range of 0-348 compared to N’gousso with an oocyst mean of 35 (SD: +/-70.26, SEM: +/-2.7), a median of 4 and range of 0-486 oocysts per midgut, respectively. The infection prevalence was also lower (p<0.001, Chi-squared Pearson’s test) in G3 (67%) compared to N’gousso (43%).
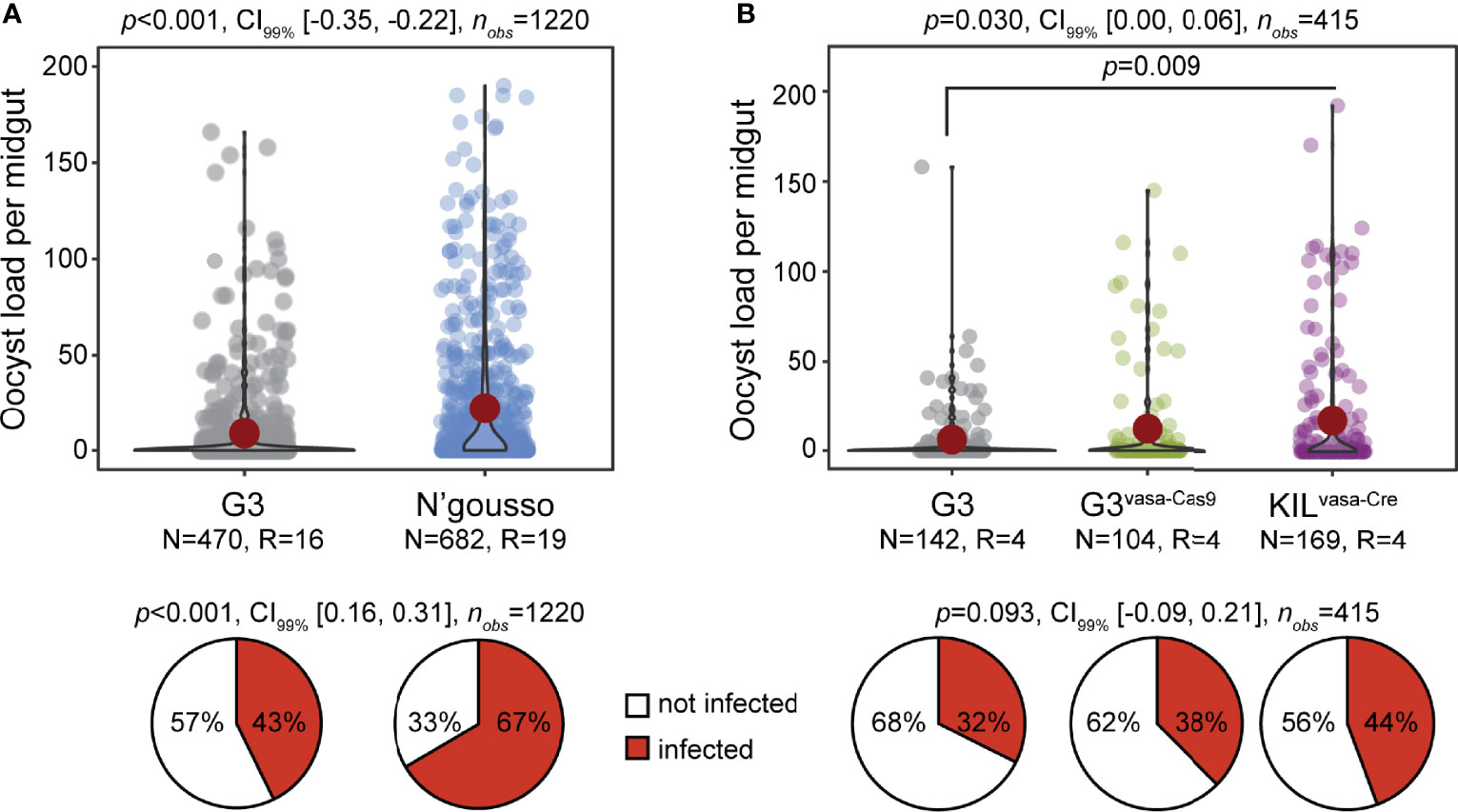
Figure 1 Susceptibility of laboratory An. gambiae lines to P. falciparum NF54. (A) Infection intensity and prevalence (%) of G3 and N’gousso. (B) Infection intensity and prevalence of G3, G3vasa-Cas9 and KILvasa-Cre shown as a pool of four biological replicates. The violin dot plots show the frequency and distribution of data. Single dots show the number of oocysts counted per midgut and the red circle shows the median infection intensity. N is the number of mosquitoes analyzed per line and R is the number of biological replicates. Statistical significance is shown on top of the graphs (p), with the CI99% and the number of observations analyzed per experiment. The pie charts show the percentage of not infected (white) and infected (red) mosquitoes.
To generate GM mosquito effector lines using the IGD design (8), GM G3 mosquitoes had to be crossed to the KILvasa-Cre line that expresses Cre recombinase (16) to remove the transformation marker that is flanked by LoxP sites (9) and the G3vasa-Cas9 line that expresses Cas9 (16) to achieve homozygosity through homing of the transgene (9). Therefore, a homozygous IGD effector lines has mixed genetic backgrounds. We examined any potential differences in P. falciparum infection between the G3, G3vasa-cas9 and KILvasa-Cre lines that were used to generate the IGD effector lines Sco-Cp, ScoG-AP2 and Aper1-Sco (9). Results from four independent infections revealed no difference between the parental G3 and the G3vasa-cas9 lines. However, both lines were significantly different from the KILvasa-Cre strain that showed higher infection intensity (Figure 1B; p=0.03, two-tailed unpaired Mann-Whitney test). These data suggest that neither the G3 nor any of the G3vasa-cas9 and KILvasa-Cre lines serve as good references for the GM effector line.
Two factors were thought to be responsible for the infection variability detected between the various colonies and lines: the mosquito diverse genetic background and enterotype. Whilst the former can be addressed in many different ways (e.g., backcrossing the effector line to the parental G3 colony and comparing the progeny with the G3 control or establishing a control population through crosses of the various founder lines) the latter can only be effectively addressed by rearing mosquitoes in the same containers including the aquatic larval stages. We designed an assay to concurrently address these issues, whereby the parental GM effector line that had a mixture of G3 and KIL genetic backgrounds was backcrossed to the G3 line (referred to here as wild type, wt, with regards to the integration locus) to obtain hemizygote transgenic mosquitoes of generation G1 which were then allowed to interbreed to derive a generation G2 of mosquitoes (Figure 2A). This new isogenic population of mosquitoes is comprised of wt homozygotes (-/-), transgenic hemizygotes (+/-) and transgenic homozygotes (+/+). As all mosquitoes are reared together, they are expected to be exposed to the same microbiota and thus share the same enterotype.
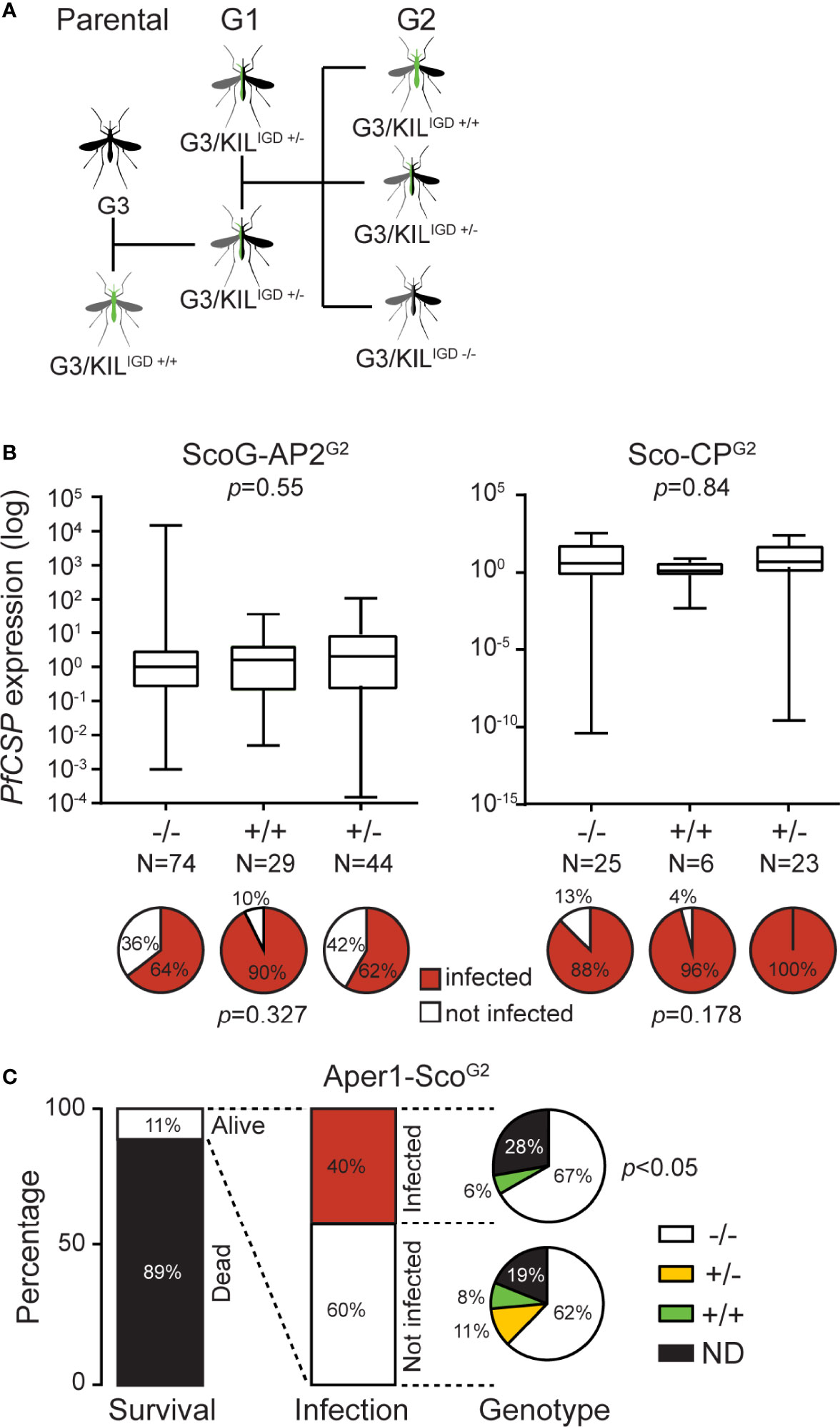
Figure 2 P. falciparum NF54 infection of Scorpine-expressing GM An. gambiae lines. (A) Schematic representation of crosses required for the analysis of the IGD effector lines. (B) Quantification of sporozoites in ScoG-AP2G2 (left) and Sco-CPG2 (right) mosquitoes by qPCR of PfCSP transcripts in preparations of mosquito heads and thoraxes, normalized on housekeeping AgS7 and analyzed using the Mann-Whitney test. Pie charts below each graph show the prevalence of infection (sporozoite presence; %) in each sub-population analyzed using a Chi-Squared Pearson’s test. N is the number of mosquitoes dissected and analyzed. (C) Histograms report the percent survival and P. falciparum infection of the Aper1-ScoG2 population 14 days post-infectious bloodmeal, respectively; the latter determined by qPCR of PfCSP transcripts. Pie charts report the genotype composition of infected and non-infected mosquitoes, analyzed using a Chi-Squared Pearson’s test.
We used this assay to test three IGD GM effector lines we previously generated, which express the AMP Scorpine in three different An. gambiae genomic loci: Sco-Cp, ScoG-AP2 and Aper1-Sco (9). Female mosquitoes of each population were infected 3 days post-emergence with P. falciparum gametocytes provided through a SMFA. To further emulate the mosquito natural behavior and physiology, mosquitoes were provided 3 supplemental non-infectious bloodmeals (day 3, 6 and 9 post-infection) through membrane feeding, each followed by egg laying. At day 14 post-infection, surviving female mosquitoes were dissected to molecularly quantify salivary gland P. falciparum sporozoites by assessing the presence and abundance of Circumsporozoite protein (PfCSP) transcripts in mosquito heads and thoraxes using quantitative real-time PCR (qRT-PCR) and be genotyped for transgene zygosity by PCR on DNA isolated from their abdomens.
Of 400 ScoG-AP2, 300 Sco-CP and 700 Aper1-Sco mosquitoes pooled from 3 independent replicates each, 240, 60 and 88, respectively, survived to day 14 (Table 1). In all cases, +/+ and +/- transgene genotypes were less than what one would expect from an unbiased Mendelian segregation of alleles, which in conjunction with the high mortality especially in the Sco-CP and Aper1-Sco lines indicated that Scorpine expression may inflict a fitness cost to female GM mosquitoes.
No significant differences were observed between +/+, +/- and -/- ScoG-AP2 and Sco-CP, respectively, both with regards to PfCSP transcript abundance (Mann-Whitney test, p=0.55 and p=0.84 respectively) and presence (Chi-Squared Pearson’s test, p=0.32 and p=0.17, respectively) (Figure 2B). However, of the 88 Aper1-Sco female mosquitoes that survived to day 14, 35 (40%) were found to have PfCSP transcripts, of which 23 were -/-, 2 were +/+ and 0 were +/-, revealing a significant difference in sporozoite prevalence between transgenic (both +/+ and +/-) and non-transgenic mosquitoes (Figure 2C; Chi-squared Pearson’s test, p<0.05). We could not determine the genotype of the remaining 10 mosquitoes. The very low numbers of sporozoite-positive transgenic mosquitoes did not permit any meaningful statistical analysis of PfCSP transcript abundance.
To gain further insights into the survival of Aper1-Sco-expressing mosquitoes, we used the same assay but now genotyped dead female mosquitoes of generation G2 (Aper-ScoG2) every 24 hours starting from day 1 after the first (here non-infectious) bloodmeal. The overall survival probability of the Aper-ScoG2 female population was also compared to that of the parental G3 female population. All mosquitoes were blood-fed every 3 days and each bloodmeal was followed by egg laying.
Compared to G3, the Aper1-ScoG2 female population exhibited significantly lower probability of survival over time (Kaplan-Meier statistics, p<0.0001; Figure 3A). Importantly, only 5% of Aper1-ScoG2 female mosquitoes (21 of 429) survived to day 10 post first bloodmeal, when P. falciparum is known to reach its peak infectivity to humans, compared to 41% of G3 female mosquitoes (143 of 353; Chi-squared Pearson’s test, p<0.0001). While some females of the parental G3 line survived to day 20, the Aper1-ScoG2 line was fully suppressed by day 14. Within the Aper1-ScoG2 population, the survival probabilities of +/+ and +/- female mosquitoes appeared to be drastically reduced compared to -/- mosquitoes although only +/- and -/- mosquitoes showed significantly different survival probabilities with Kaplan-Meier statistics (p<0.001; Figure 3B). Specifically, only 13% of +/- (Chi-squared Pearson’s test, p<0.001) and 19% of +/+ (Chi-squared Pearson’s test, p<0.01) female mosquitoes survived after day 6 compared to 42% of -/- female mosquitoes.
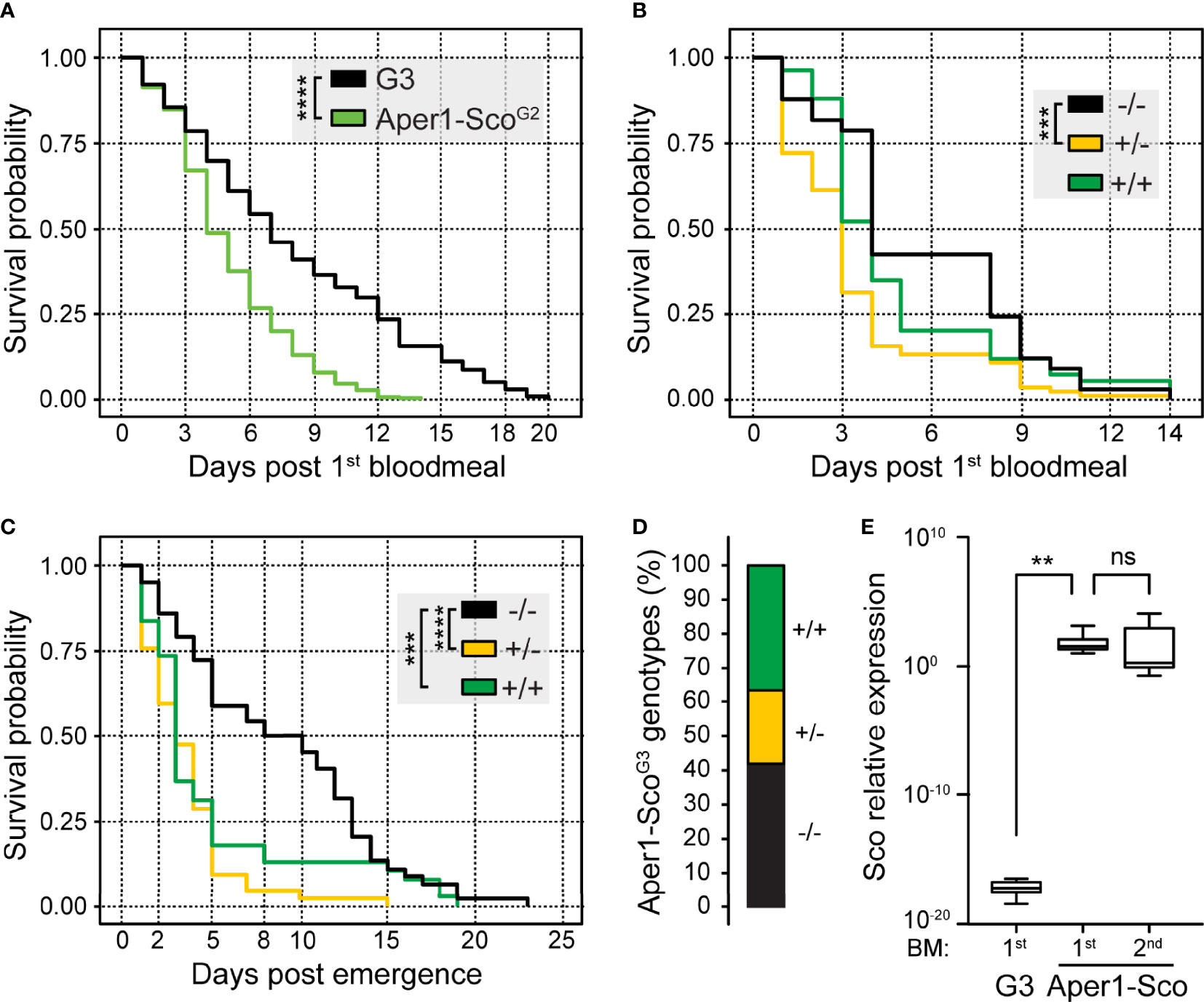
Figure 3 Survival and reproduction assays of the Aper1-ScoG2 mosquito population. Kaplan-Meier survival curves and statistical analysis of survival probabilities of G3 and Aper1-ScoG2 female mosquito populations (A) and each IGD effector genotype (-/-, +/- and +/+) of the Aper1-ScoG2 female population (B). In both assays, day 0 is the day of the 1st bloodmeal, and supplemental bloodmeals were provided every 3 days thereafter. (C) Kaplan-Meier survival curves and statistical analysis of survival probabilities of individual genotypes of the Aper1-ScoG2 mosquito population including both females and males. In this assay, day 0 is the day of adult emergence, while the 1st bloodmeal was provided on day 2 and the two additional bloodmeals were provided on days 5 and 8 post-emergence. (D) Percentage of genotypes in the adult Aper1-ScoG3 population (both females and males) that was derived from eggs laid after each of the bloodmeals of Aper1-ScoG2 mosquitoes of panel (C). (E) Relative expression of Scorpine (Sco) in control G3 and Aper1-Sco +/+ mosquitoes 24 hours after the 1st or 2nd bloodmeal. Expression of Scorpine was normalized against the A. coluzzii S7 transcripts. The analysis was performed using a student’s t test. ns, not significant; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Next, we investigated the capacity of the Aper1-ScoG2 population to produce offspring that inherit the transgene in the absence of gene drive. Following crosses and rearing of mosquitoes in the same containers as described above, the adult Aper1-ScoG2 population was fed on non-infected human blood on days 2, 5 and 8 post-adult emergence. After every bloodmeal, females laid eggs and adults that emerged from these eggs (Aper1-ScoG3) were pooled and genotyped for the presence of the Aper1-Sco transgene. The parental Aper1-ScoG2 population was cultured until all male and female mosquitoes had died and then genotyped. The results showed that the founder Aper1-ScoG2 population consisted of 155 adults (66 females and 89 males) of which 45 were -/- (26 and 19 females and males, respectively), 42 were +/- (17 and 25, respectively) and 38 were +/+ (9 and 29, respectively), while the genotype of 30 mosquitoes (14 and 16, respectively) could not be determined. Kaplan Meier survival analysis revealed that both +/- (p<0.0001) and +/+ (p<0.001) Aper1-ScoG2 mosquitoes exhibited significantly reduced survival probability compared to -/- mosquitoes, including both females and males (Figure 3C). When female and male survival probabilities were analyzed independently, the results showed that whether +/- or +/+, both female (p<0.0001) and male (p<0.001) mosquitoes died significantly earlier than their wt (-/-) counterparts.
Genotyping of the adult Aper1-ScoG3 mosquitoes revealed that all genotypes were represented in the population. Of 111 adults with determined genotypes, 47 (42%) were -/-, 21 (19%) were +/- and 43 (39%) were +/+ (Figure 3D). These data demonstrate that despite their reduced survival probability, largely manifested at later stages of adult life, Aper1-Sco transgenic mosquitoes can produce progeny that inherit the transgene in frequencies resembling those of the wt locus. The bias towards homozygous -/- and +/+ mosquitoes is an interesting observation that deserves further investigation. It is worth noting that owing to the bloodmeal-induced Aper1 locus, Scorpine is over expressed after every bloodmeal, hence its accumulation after several bloodmeals may contribute to the reduced adult mosquito survival (Figure 3E).
The mosquito gut microbiota community plays an important role in mosquito physiology and interferes with infection with Plasmodium (19, 21–24). It is known that the microbiota load and composition are significantly affected by mosquito epithelial immune responses, including AMP expression. To investigate whether Scorpine expression affects the microbiota of the Aper1-Sco mosquitoes, we quantified the bacterial load and recorded the genotype of individual mosquitoes of the Aper1-ScoG2 population 24 hours post-bloodmeal by quantitative PCR of the 16S ribosomal DNA. We also separately quantified the relative abundance of Flavobacteriaceae and Enterobacteriaceae, two of the most abundant bacterial families in the An. gambiae gut (19, 21, 25).
As shown in Figure 4, within the Aper1-ScoG2 population we found that mosquitoes of the -/- genotype exhibited a significantly lower abundance of 16S qPCR product compared to +/- and +/+ (Mann-Whitney test, **p<0.01 and *p<0.05, respectively). In the same samples, Enterobacteriaceae were more abundant in +/+ and +/- compared to -/- mosquitoes (Mann-Whitney test, **p<0.01 and *p<0.05, respectively). Since the abundance of Enterobacteriaceae has been associated with reduced P. falciparum infections (21, 26), these results support the low infection intensity and the statistically lower infection prevalence observed in SMFAs. A similar pattern was evident for the abundance of Flavobacteriaceae, which was higher in both +/+ and +/- compared to -/- mosquitoes (Mann-Whitney, p<0.001, p<0.01 respectively). The increased bacterial load may also be associated with the reduced lifespan of the Aper1-Sco GM mosquitoes (27).
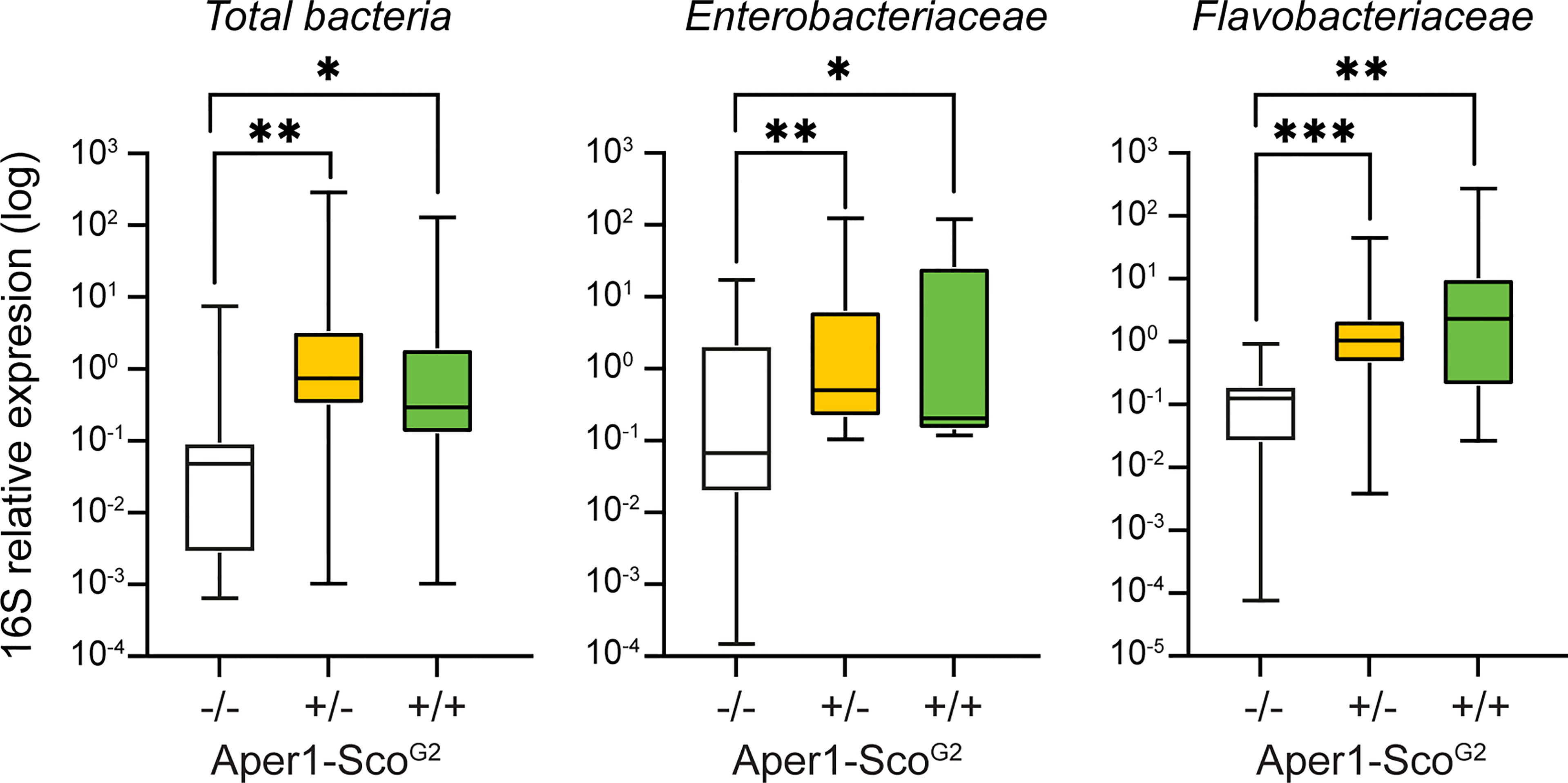
Figure 4 Midgut microbiota load in the Aper1-ScoG2 population. Midgut microbiota load in sub-populations of mosquitoes of the Aper1-ScoG2 population assessed by 16S qPCR on individual mosquitoes 24 hours post-1st bloodmeal. The loads of total bacteria (left), Enterobacteriaceae (middle) and Flavobacteriaceae (right) were independently assessed. Data from two independent biological replicates were pooled. The numbers of mosquitoes per pool were: for total bacterial load 7 -/-, 29 +/- and 19 +/+; for Enterobacteriaceae 10 -/-, 20 +/- and 8 +/+; and for Flavobacteriaceae 8 -/-, 24 +/- and 17+/+. Data were analyzed with a Mann-Whitney test: *p < 0.05; **p < 0.01; ***p < 0.001.
The establishment of robust laboratory assays unambiguously reporting on the various characteristics of a GM An. gambiae line is a necessary and critical milestone in the path to mosquito population replacement, before large cage, semi-field or field trials can commence. Mosquito vectorial capacity and life history traits can differ substantially between laboratory colonies and lines either because of their inherently different genetic backgrounds or genetic drift caused created through bottlenecks during colonization or establishment of a line. An additional complication is that until recently the incipient species An. gambiae and A. coluzzii were interchangeably used and collectively referenced as An. gambiae, and many publications continue referring to them as such. Moreover, laboratory colonies are often of ambiguous origin or cultured in the same laboratory together with other colonies and lines; since the two incipient species can freely reproduce in the laboratory, contaminations are frequent.
We previously demonstrated that P. falciparum NF54 infections of the A. coluzzii N’gousso colony, one of the most recently generated and widely used colonies, produce high infection prevalence and intensity, allowing for the generation of robust data and reliable statistical analyses of transmission blocking (12). However, this colony cannot be used as a reference for An. gambiae-derived GM lines, and we show here that G3, the parental colony of GM lines examined here and one of the oldest laboratory colonies, originally of An. gambiae origin, and N’gousso exhibit markedly different P. falciparum infection prevalence and intensity, both being significantly lower in G3 than N’gousso. While the G3vasa-Cas9 line resembles its parental G3 colony with regards to both infection prevalence and intensity, the KILvasa-Cre line that is derived from the KIL An. gambiae strain and has also been used here for the generation of GM effector lines displays significantly higher infection intensity; indeed, KIL was genetically selected to be highly susceptible to infection. To conclude, the choice of reference line must not be done lightly as this may lead to erroneous conclusions.
Whilst there are many ways to choose or generate a reference line that is representative of the genetic variation one expects to have in the GM lines examined, we decided here to develop and use a method that would simultaneously address another fundamental issue in such assays: the culturing of aquatic mosquito stages in different containers. The adult mosquito microbiota are largely representative of what is found in the aquatic larval habitat and are known to affect both P. falciparum infections and mosquito fitness (19, 21, 24, 28, 29). Experience has shown that no matter how similar the conditions and treatment of larval habitats are, the microbiota may differ markedly, even if larval containers are placed side-by-side in the insectary. Therefore, the best control is the rearing of control and experimental mosquitoes in the same containers throughout their aquatic and adult stages (22). Here, we achieved this by assaying mosquitoes reared together and were either hemizygous or homozygous positive or negative for the transgene; the latter genotype was used as a reference for the other two. These mosquitoes were produced by crossing the GM lines to the parental G3 line to derive a population of mosquitoes that were hemizygous with respect to the transgene, which was then intercrossed to produce a G2 progeny presenting all three allelic combinations (-/-, +/- and +/+). The G2 mosquitoes were then subjected to the assay, whether transmission blocking or survival, and genotyped for the presence of the modified allele. An added advantage of this protocol is that one can in parallel monitor dose-dependent effects or haplosufficiency of the modified allele, an intrinsic feature of the IGD split drive, where the autonomous drive (here Cas9) and non-autonomous effector (here an AMP) are released separately leading to the effector being initially segregating largely by simple Mendelian genetics. A shortcoming of this otherwise very robust protocol is that all the mosquitoes must be individually genotyped, thus substantially adding to the cost and effort required. Therefore, the protocol requires further streamlining and automation for it to be used in large-scale studies.
The presence and number of oocysts found on the mosquito midgut wall is the most widely used method to assess malaria transmission blocking or reduction caused by an intervention such as a transmission blocking vaccine, drug, or genetic modification. However, this method does not account for any possible defects in oocyst development, a factor critical for computing the parasite extrinsic incubation period (EIP) and thenceforth reproductive rate (R0). Examples include the manipulation of the mosquito lipid shuttling machinery that is shown to interfere with oocyst development (30, 31) and the knockout of the parasite gene misfit that leads to oocysts that developmentally arrest but remain detectable and macroscopically indistinguishable from developing oocysts and continue to grow in size (32). Furthermore, recent studies have revealed that the development of oocysts can be significantly delayed or enter dormancy when nutrients acquired through frequent mosquito bloodmeals are scarce and by parasite crowding (13, 14). Thus, supplementing infected mosquitoes with bloodmeals better resembles their natural feeding behavior and physiology and minimize any laboratory biases when evaluating mosquitoes destined for population replacement (33, 34). In conclusion, a better measure of transmission blockage or reduction is the recording of the presence and abundance of sporozoites in the salivary glands of mosquitoes that are regularly provided with bloodmeals after the initial infectious bloodmeal, a protocol we used in the assays presented here.
Using the protocol described above we find, on the one hand, that Scorpine expression in the CP and AP2 locus does not affect the prevalence and abundance of sporozoites in the mosquito salivary glands 14 days post-infection. This is despite the Sco-CP and ScoG-AP2 lines previously found to harbor more and less oocysts when compared to G3, respectively (9). These results could be explained by the genetic differences expected to exist between lines, which lead to varied susceptibility to infection and/or the fact that subtle differences in the oocyst load may not translate to differences in sporozoites. The expression of an AMP can affect midgut homeostasis at the time of bloodmeal digestion in ways that are difficult to predict, leading to disturbance of metabolic processes and availability of nutrients for the developing oocyst. Furthermore, quantifying the cumulative abundance of sporozoites in salivary glands as we did here does not report on any possible effects of the intervention on the oocyst developmental rates.
On the other hand, Scorpine expression in the Aper1 locus and docking on the peritrophic matrix leads to a phenotype that merits further investigation with experimental and modeling studies as regards to the potential of this modification to reduce malaria transmission when driven through wild An. gambiae populations. Aper1-Sco mosquitoes exhibit a significantly shorter lifespan compared to wt, with most of them dying before they become capable of transmitting the parasite. Furthermore, the few GM mosquitoes that survive beyond the parasite EIP show significantly lower prevalence of sporozoite presence, consistent with our previous findings that showed significantly lower infection prevalence and intensity at the oocyst stage (9). At the same time, GM homozygous and heterozygous mosquitoes appear to harbor significantly more midgut microbiota compared to their homozygous negative siblings 24 hours post-bloodmeal. While Scorpine expression in the midgut may directly compromise blood bolus parasite stages leading to lower infection at both the oocyst and sporozoite stages, the combination of phenotypes we detect could be explained in two additional ways. Firstly, the expression of the exogenous AMP disturbs the midgut homeostasis leading to higher microbiota loads and premature mosquito death. Secondly, fusion of Scorpine with Aper1 compromises the integrity of the type I peritrophic matrix and thus the midgut barrier leading to higher midgut microbiota and systemic infection of the mosquito hemolymph (23).
Importantly, while self-suppressing, mosquitoes bearing the Aper1-Sco modified allele can effectively compete and mate with wt mosquitoes producing offspring that inherit the modified allele in frequencies comparable to those of the wt allele. Overall, our results suggest that in a field scenario, the reduced survival in conjunction with the reduced sporozoite load of Aper1-Sco GM mosquitoes may significantly impact on parasite transmission in the release area. It would be pertinent, at this stage, to collect additional data from large scale experiments and model the impact of any release of the strain in malaria transmission, especially when combined with a gene drive.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Conceptualization, ST, NW, and GKC. Methodology, ST, MI, JC, JP, and PC. Formal analysis, ST and GKC. Investigation, ST, JP, and GKC. Writing paper, ST and GKC. Supervision, GKC. Project administration, GKC. Funding acquisition, NW and GC. All authors contributed to the article and approved the submitted version.
The work was funded by a Bill and Melinda Gates Foundation grant (OPP1158151) to NW and GKC and a Wellcome Trust Investigator Award (107983/Z/15/Z) to GC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Temesgen Menberu Kebede for his help with maintaining the mosquito colonies and transgenic lines.
1. Hammond AM, Galizi R. Gene Drives to Fight Malaria: Current State and Future Directions. Pathog Glob Health (2017) 111:412–23. doi: 10.1080/20477724.2018.1438880
2. Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, Kafatos FC, et al. Stable Germline Transformation of the Malaria Mosquito Anopheles Stephensi. Nature (2000) 405:959–62. doi: 10.1038/35016096
3. Grossman GL, Rafferty CS, Clayton JR, Stevens TK, Mukabayire O, Benedict MQ. Germline Transformation of the Malaria Vector, Anopheles Gambiae, With the Piggybac Transposable Element. Insect Mol Biol (2001) 10:597–604. doi: 10.1046/j.0962-1075.2001.00299.x
4. Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, et al. A Synthetic Homing Endonuclease-Based Gene Drive System in the Human Malaria Mosquito. Nature (2011) 473:212–5. doi: 10.1038/nature09937
5. Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, et al. A CRISPR-Cas9 Gene Drive System Targeting Female Reproduction in the Malaria Mosquito Vector Anopheles Gambiae. Nat Biotechnol (2016) 34:78–83. doi: 10.1038/nbt.3439
6. Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton AK, et al. A CRISPR-Cas9 Gene Drive Targeting Doublesex Causes Complete Population Suppression in Caged Anopheles Gambiae Mosquitoes. Nat Biotechnol (2018) 36:1062–6. doi: 10.1038/nbt.4245
7. Carballar-Lejarazu R, Ogaugwu C, Tushar T, Kelsey A, Pham TB, Murphy J, et al. Next-Generation Gene Drive for Population Modification of the Malaria Vector Mosquito, Anopheles Gambiae. Proc Natl Acad Sci USA (2020) 117:22805–14. doi: 10.1073/pnas.2010214117
8. Nash A, Urdaneta GM, Beaghton AK, Hoermann A, Papathanos PA, Christophides GK, et al. Integral Gene Drives for Population Replacement. Biol Open (2019) 8:bio037762. doi: 10.1242/bio.037762
9. Hoermann A, Tapanelli S, Capriotti P, Del Corsano G, Masters EK, Habtewold T, et al. Converting Endogenous Genes of the Malaria Mosquito Into Simple Non-Autonomous Gene Drives for Population Replacement. Elife (2021) 10:e58791. doi: 10.7554/eLife.58791
10. Sinden RE. Molecular Interactions Between Plasmodium and its Insect Vectors. Cell Microbiol (2002) 4:713–24. doi: 10.1046/j.1462-5822.2002.00229.x
11. James SL, Marshall JM, Christophides GK, Okumu FO, Nolan T. Toward the Definition of Efficacy and Safety Criteria for Advancing Gene Drive-Modified Mosquitoes to Field Testing. Vector Borne Zoonotic Dis (2020) 20:237–51. doi: 10.1089/vbz.2019.2606
12. Habtewold T, Tapanelli S, Masters EKG, Hoermann A, Windbichler N, Christophides GK. Streamlined SMFA and Mosquito Dark-Feeding Regime Significantly Improve Malaria Transmission-Blocking Assay Robustness and Sensitivity. Malar J (2019) 18:24. doi: 10.1186/s12936-019-2663-8
13. Shaw WR, Holmdahl IE, Itoe MA, Werling K, Marquette M, Paton DG, et al. Multiple Blood Feeding in Mosquitoes Shortens the Plasmodium Falciparum Incubation Period and Increases Malaria Transmission Potential. PloS Pathog (2020) 16:e1009131. doi: 10.1371/journal.ppat.1009131
14. Habtewold T, Sharma AA, Wyer C, Masters EKG, Windbichler N, Christophides GK. Plasmodium Oocysts Respond With Dormancy to Crowding and Nutritional Stress. Sci Rep (2021) 11:3090. doi: 10.1038/s41598-021-81574-0
15. Harris C, Lambrechts L, Rousset F, Abate L, Nsango SE, Fontenille D, et al. Polymorphisms in Anopheles Gambiae Immune Genes Associated With Natural Resistance to Plasmodium Falciparum. PloS Pathog (2010) 6:e1001112. doi: 10.1371/journal.ppat.1001112
16. Volohonsky G, Terenzi O, Soichot J, Naujoks DA, Nolan T, Windbichler N, et al. Tools for Anopheles Gambiae Transgenesis. G3 (Bethesda) (2015) 5:1151–63. doi: 10.1534/g3.115.016808
17. Kefi M, Mavridis K, Simoes ML, Dimopoulos G, Siden-Kiamos I, Vontas J. New Rapid One-Step PCR Diagnostic Assay for Plasmodium Falciparum Infective Mosquitoes. Sci Rep (2018) 8:1462. doi: 10.1038/s41598-018-19780-6
18. Dimopoulos G, Seeley D, Wolf A, Kafatos FC. Malaria Infection of the Mosquito Anopheles Gambiae Activates Immune-Responsive Genes During Critical Transition Stages of the Parasite Life Cycle. EMBO J (1998) 17:6115–23. doi: 10.1093/emboj/17.21.6115
19. Gendrin M, Rodgers FH, Yerbanga RS, Ouedraogo JB, Basanez MG, Cohuet A, et al. Antibiotics in Ingested Human Blood Affect the Mosquito Microbiota and Capacity to Transmit Malaria. Nat Commun (2015) 6:5921. doi: 10.1038/ncomms6921
20. Della Torre A, Tu Z, Petrarca V. On the Distribution and Genetic Differentiation of Anopheles Gambiae s.s. Molecular Forms. Insect Biochem Mol Biol (2005) 35:755–69. doi: 10.1016/j.ibmb.2005.02.006
21. Boissiere A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, et al. Midgut Microbiota of the Malaria Mosquito Vector Anopheles Gambiae and Interactions With Plasmodium Falciparum Infection. PloS Pathog (2012) 8:e1002742. doi: 10.1371/journal.ppat.1002742
22. Habtewold T, Groom Z, Christophides GK. Immune Resistance and Tolerance Strategies in Malaria Vector and Non-Vector Mosquitoes. Parasit Vectors (2017) 10:186. doi: 10.1186/s13071-017-2109-5
23. Rodgers FH, Gendrin M, Wyer C, Christophides GK. Microbiota-Induced Peritrophic Matrix Regulates Midgut Homeostasis and Prevents Systemic Infection of Malaria Vector Mosquitoes. PloS Pathog (2017) 13:e1006391. doi: 10.1371/journal.ppat.1006391
24. Gao L, Song X, Wang J. Gut Microbiota is Essential in PGRP-LA Regulated Immune Protection Against Plasmodium Berghei Infection. Parasit Vectors (2020) 13:3. doi: 10.1186/s13071-019-3876-y
25. Akorli J, Gendrin M, Pels NA, Yeboah-Manu D, Christophides GK, Wilson MD. Seasonality and Locality Affect the Diversity of Anopheles Gambiae and Anopheles Coluzzii Midgut Microbiota From Ghana. PloS One (2016) 11:e0157529. doi: 10.1371/journal.pone.0157529
26. Dennison NJ, Saraiva RG, Cirimotich CM, Mlambo G, Mongodin EF, Dimopoulos G. Functional Genomic Analyses of Enterobacter, Anopheles and Plasmodium Reciprocal Interactions That Impact Vector Competence. Malar J (2016) 15:425. doi: 10.1186/s12936-016-1468-2
27. Cansado-Utrilla C, Zhao SY, Mccall PJ, Coon KL, Hughes GL. The Microbiome and Mosquito Vectorial Capacity: Rich Potential for Discovery and Translation. Microbiome (2021) 9:111. doi: 10.1186/s40168-021-01073-2
28. Gendrin M, Yerbanga RS, Ouedraogo JB, Lefevre T, Cohuet A, Christophides GK. Differential Effects of Azithromycin, Doxycycline, and Cotrimoxazole in Ingested Blood on the Vectorial Capacity of Malaria Mosquitoes. Open Forum Infect Dis (2016) 3:ofw074. doi: 10.1093/ofid/ofw074
29. Romoli O, Gendrin M. The Tripartite Interactions Between the Mosquito, Its Microbiota and Plasmodium. Parasit Vectors (2018) 11:200. doi: 10.1186/s13071-018-2784-x
30. Mendes AM, Schlegelmilch T, Cohuet A, Awono-Ambene P, De Iorio M, Fontenille D, et al. Conserved Mosquito/Parasite Interactions Affect Development of Plasmodium Falciparum in Africa. PloS Pathog (2008) 4:e1000069. doi: 10.1371/journal.ppat.1000069
31. Rono MK, Whitten MM, Oulad-Abdelghani M, Levashina EA, Marois E. The Major Yolk Protein Vitellogenin Interferes With the Anti-Plasmodium Response in the Malaria Mosquito Anopheles Gambiae. PloS Biol (2010) 8:e1000434. doi: 10.1371/journal.pbio.1000434
32. Bushell ES, Ecker A, Schlegelmilch T, Goulding D, Dougan G, Sinden RE, et al. Paternal Effect of the Nuclear Formin-Like Protein MISFIT on Plasmodium Development in the Mosquito Vector. PloS Pathog (2009) 5:e1000539. doi: 10.1371/journal.ppat.1000539
33. Costa G, Gildenhard M, Eldering M, Lindquist RL, Hauser AE, Sauerwein R, et al. Non-Competitive Resource Exploitation Within Mosquito Shapes Within-Host Malaria Infectivity and Virulence. Nat Commun (2018) 9:3474. doi: 10.1038/s41467-018-05893-z
Keywords: gene drive, malaria, disease vector, population modification, Anopheles gambiae
Citation: Tapanelli S, Inghilterra MG, Cai J, Philpott J, Capriotti P, Windbichler N and Christophides GK (2021) Assessment of Plasmodium falciparum Infection and Fitness of Genetically Modified Anopheles gambiae Aimed at Mosquito Population Replacement. Front. Trop. Dis 2:806880. doi: 10.3389/fitd.2021.806880
Received: 01 November 2021; Accepted: 25 November 2021;
Published: 14 December 2021.
Edited by:
Yoosook Lee, University of Florida, United StatesReviewed by:
Eric Caragata, University of Florida, United StatesCopyright © 2021 Tapanelli, Inghilterra, Cai, Philpott, Capriotti, Windbichler and Christophides. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George K. Christophides, Zy5jaHJpc3RvcGhpZGVzQGltcGVyaWFsLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.