
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis. , 19 November 2021
Sec. Antimicrobial Resistance
Volume 2 - 2021 | https://doi.org/10.3389/fitd.2021.786378
This article is part of the Research Topic Highlights in Antimicrobial Resistance 2021/22 View all 4 articles
 Solomon Wireko1,2
Solomon Wireko1,2 Samuel Opoku Asiedu3
Samuel Opoku Asiedu3 Priscilla Kini1,3
Priscilla Kini1,3 Bill Clinton Aglomasa3
Bill Clinton Aglomasa3 Emmanuel Kobla Atsu Amewu1
Emmanuel Kobla Atsu Amewu1 Ebenezer Asiedu3
Ebenezer Asiedu3 Freda Osei-Akoto1
Freda Osei-Akoto1 Kennedy Gyau Boahen4
Kennedy Gyau Boahen4 Dorcas Obiri-Yeboah5
Dorcas Obiri-Yeboah5 Katherine Ryan Amato6
Katherine Ryan Amato6 Alexander Kwarteng1,3*
Alexander Kwarteng1,3*Background: Filarial pathologies such as lymphedema may be associated with complications such as chronic non-healing wounds. Nonetheless, the role of bacterial population colonizing the lymphedematous legs has been posited to worsen the conditions of those living with the infection. These bacteria are usually composed of staphylococcal species partly because they are commensals. Thus, this present study sought to type the methicillin-resistant Staphylococcus aureus (MRSA) prevalence among individuals presenting with filarial lymphedema, particularly as MRSA tends to affect treatments options.
Methods: We recruited individuals (n = 321) with stages I–VII of lymphedema in a cross-sectional study in the Ahanta West district of the Western Region of Ghana. Swabs from lymphedematous limb ulcers, pus, and cutaneous surfaces were cultured using standard culture-based techniques. The culture isolates were later identified using Matrix-assisted Laser Desorption/Ionization Time of Flight (MALDI-TOF) mass spectrometry.
Results: A total of 192 Staphylococci species were isolated, with an overall prevalence of 39.7% (95% CI: 35%–44%; N = 483). S. hominis was the most prevalent species (23.95%), followed by S. haemolyticus (20.83%), S. epidermidis (15.10%), S. aureus (10.41%), and S. saprophyticus (9.32%). The remaining 20.34% were distributed among S. wanneri, S. sciuri, S. pasteuri, S. xylosus, S. simulans, S. cohnii, S. caprae, S. lugdunensis, and S. capitis. MRSA, containing mecA gene, was detected in 21 out of 31 Staphylococci isolates tested, with an overall prevalence of 68% (95% CI: 51%–84%). In addition, a virulent gene, Panton–Valentine leukocidin (PVL), which is usually associated with S. aureus, was detected in 20/31 (64.5%) S. aureus in the study.
Conclusion: These results suggest that MRSA species may pose a challenge to the treatment of filarial lymphedema with antibiotics particularly, as doxycycline is currently being piloted in some endemic areas to treat the infection. Thus, intensive antimicrobial resistance surveillance should be conducted in endemic areas by health authorities to forestall the dilemma of multidrug resistance not only against lymphatic filariasis (LF) infection but other diseases.
Human lymphatic filariasis (LF) caused by nematode parasites, Wuchereria bancrofti and Brugia species, presents a huge economic burden in endemic countries. One of the most common chronic manifestations of LF is lymphedema of the extremities, which eventually progresses to elephantiasis. Lymphedema occurs as a result of the inflammation of the lymph channels and lymph nodes, coupled with a reduced draining efficacy (1, 2). Even though lower limbs are frequently affected, upper limbs, male genitalia, and breasts in females may also be affected (3). The World Health Organization (WHO) estimates that about 40 million people have filarial pathologies due to LF, of which 26 million have hydrocele and more than 15 million have lymphedema of the extremities (4). Interestingly, there are new incidents of filariasis disease in geographical areas where the disease was thought to be nonexistent (5). Filarial infections, which are widespread in developing countries, are responsible for some of the most crippling morbidities as well as a deadly cycle of poverty and disease (6). The disease usually presents as asymptomatic, acute, and chronic phases (7, 8). The causes for the development of the clinical manifestations associated with the disease are unclear; however, many hypotheses have been propounded including parasitic worm factors, Wolbachia parasite endosymbiont factors, host immune factors, host genetic factors, and secondary bacterial infections.
Despite the number of these risk factors, secondary bacterial infections are believed to be particularly important in determining patient morbidity as it relates to skin lesions. Skin wounds such as interdigital lesions (IDLs) in the affected limb function as portals of entry for secondary bacterial infection. Results from a study investigating the epidemiology and etiology of IDLs of the feet of filarial lymphedema patients showed that more than 50% had one or more IDLs in relation to controls and that the incidence of acute-dermato-lymphangioadenitis (ADLA) was strongly predicted by the number of lesions (9). ADLA is a major complication of filarial lymphedema associated with secondary bacterial infection (10) and described by severe incidents of inflammation confined to the lymph nodes, skin, and lymphatic vessels (11–14). A recent study by Kwarteng et al. (14) conducted among Ghanaian patients shows that 68.3% of the study participants experienced ADLA attacks, especially in the rainy season. Although a comprehensive characterization of the bacterial population colonizing the lower limbs of people living with filarial lymphedema is yet to be carried out, some studies have reported a high prevalence of Staphylococcus spp., Streptococcus spp., and Bacillus spp (15). However, for this present study, we focused on Staphylococcus spp. due to their role as both commensals and pathogens depending on the immune status of the individuals specifically where LF individuals have been studied to have compromised immune systems (16).
Staphylococci are facultative anaerobic bacteria that are spherical, Gram-positive, and non-motile and do not generate spores, belonging to the Firmicutes phylum and Staphylococcaceae family. They are abundant in the environment and are part of the normal skin flora (17, 18). However, different species of Staphylococci are now documented as important sources of human infection because of their growing importance in causing hospital-acquired infection (19). Nonetheless, among Staphylococcus species, methicillin-resistant Staphylococcus aureus (MRSA) has extensively been documented to pose a serious public health (20). In Ghana, a systematic review of MRSA colonization studies reveals that MRSA prevalence is relatively low when compared to that of the US A, Europe, and Saudi Arabia (21). According to a study by Donkor et al. (201), the low level of MRSA prevalence in the Ghanaian population despite reports of high misuse of antibiotics has been suggested to be due to the relatively expensive third-generation cephalosporins and fluoroquinolones, which are known to correlate with a high increase of MRSA prevalence (22). Its potential to cause disease appears to be facilitated by heavily polluted environments and a compromised immune system (23).
Many studies have profiled staphylococcal species as a secondary cause in various disease conditions such as atopic dermatitis (24), but there is paucity of data regarding distribution of staphylococcal species among filarial lymphedema patients. Our field observations of LF patients have revealed that antibiotics, particularly penicillin and cephalosporin, are frequently used systemically and topically to treat chronic wounds and skin infections. However, the efficacy of these medications is uncertain. Their impact on the prevalence and abundance of antimicrobial resistance genes is also unknown. To determine the impact of frequent use of antibiotics among LF individuals in treating the infection specifically chronic non-healing wounds of Staphylococci species, this study, therefore, sought to determine staphylococcal bacterial populations that could be targeted in the morbidity management of filarial lymphedema and their antimicrobial resistance phenotypes and genotypes using standard culture techniques and MALDI-TOF mass spectrometry (MS) to appreciate the enormous microbial residents supposed to be involved in the development of the infection and the morbidities associated with it. We hypothesized that there could be emerging Staphylococci resistant strains among LF individuals particularly those presenting with chronic non-healing wounds.
A total of 321 individuals with chronic lymphedema in stages I–VII (the description of the stages of lymphedema is shown in Table 1) were identified with the aid of the Ministry of Health Disease Control program for the Lymphatic Filariasis Elimination Unit at the Ahanta West District in Ghana. The study participants included both gender ages 18 years and above who have lived in the filarial-endemic communities for more than 10 years and who gave their informed consent. Individuals younger than 18 years and persons with edema other than that caused by LF were not included in the study. All patients included were carefully examined by an experienced lymphatic filarial expert. The study was approved by the Committee for Human Research, Publications, and Ethics (CHRPE/AP/649/19) at the School of Medical Sciences, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana.
Samples were collected between January 2019 and June 2020. Using sterile swabs, samples of pus or fluid from edema, lymphedematous limb ulcers, and cutaneous surface of lymphedematous limbs were collected aseptically and kept in Amies transport medium (Copan Diagnostics Inc.). The samples (swab containers) were kept in liquid nitrogen and transported to Kumasi Centre for Collaborative Research (KCCR) for subsequent laboratory analysis. The swab was plated on blood agar base and incubated at 35°C–37°C in normal environment for 18–24 h with 5% sheep blood (in-house) and on chocolate agar (Oxoid GmbH) (25). Species identification was done using biochemical and serological methods (25). Based on their morphological characteristics, all colonies suspected to be Staphylococci were Gram stained and tested for catalase production. Using the tube coagulase test, S. aureus was discriminated from coagulase-negative Staphylococci (CoNS).
Confirmation of bacterial isolates was done by MALDI-TOF MS [Bruker MALDI Biotyper, Server Version: 4.1.31 (SIRO) 314 2015-09-30_17-53-06] as described previously (26) at Noguchi Memorial Institute for Medical Research, University of Ghana, Legon. The isolates were subcultured onto a 5% sheep blood (in-house) agar base and incubated overnight at 37°C in 5% CO2. Two to three single colonies were suspended in 70% ethanol in microcentrifuge tube (Eppendorf Tubes® 1.5 ml), vortexed, and concentrated by centrifugation at 23,995.63 g force for 2 min. The supernatant was discarded, and the cells were resuspended in 50 µl of 70% formic acid; an equal volume of acetonitrile was added after 30 s of incubation, vortexed, and concentrated by centrifugation at 23,995.63 g force for 2 min. Here, 1.0 µl of the supernatant was spotted on the 96-spot polished steel target plate (Bruker Daltonic GmbH) and allowed to evaporate to dryness. Each spot was overlaid with 1.0 µl saturated solution of the matrix α-cyano-4-hydroxy-cinnamic acid and allowed to air-dry. The target plate with the sample was loaded onto the Bruker Biotyper MALDI-TOF MS system and then registered on the MALDI Flex Control software system Server Version: 4.1.31 and 60 Hz Nitrogen Laser (337 nm wavelength) for automated measurement. Sufficient spectra obtained for identification ranging from 2,000 to 20,000 m/z were examined by means of the automation control of the MALDI Biotyper system and the software library (database having over 5,627 entries). Log score value of ≥2.000 showed identification at the species level, scores value of 1.700–1.999 showed identification at the genus level, and <1.700 designated identification not reliable.
Antibiotic susceptibility of all Staphylococci isolates was tested by the disk diffusion method on Mueller–Hinton agar (Oxoid GmbH) according to Clinical & Laboratory Standards Institute (CLSI) guidelines (27). The following antimicrobials (Oxoid, UK) that are prescribed routinely for LF patients in the endemic areas were used: ampicillin 10 µg, erythromycin 15 µg, tetracycline 30 µg, gentamycin 10 µg, ciprofloxacin 30 µg, clindamycin 30 µg, chloramphenicol 30 µg, penicillin 10 µg, and trimethoprim/sulfamethoxazole 1.25/23.75 µg. Each set of tests was controlled using susceptible S. aureus (ATCC 25923).
All staphylococcal isolates were tested for methicillin resistance by 30 µg cefoxitin disk (Oxoid, UK). Bacterial suspension of 0.5 McFarland was uniformly spread onto Mueller–Hinton agar (Oxoid GmbH), and 30 μg cefoxitin disk (Oxoid, UK) was placed on the plate. The plate was then incubated at 35°C for 16–20 h. Isolates with zone of inhibition of ≤21 mm were MRSA. For S. lugdunensis, the zone of inhibition was ≤21 mm (27).
The template DNA used was prepared using extraction by heat lysis. Cells from 24-h culture were collected and emulsified in 500 µl of DNase-RNase-free distilled water (Thermo Scientific) and centrifuged at 23,995.63 g force for 5 min using a centrifuge (Eppendorf Centrifuge 5427 R). The supernatant was decanted and vortex-mixed to resuspend the pellet in 500 μl of DNase-RNase-free distilled water (Thermo Scientific). Lysing of the cells was done using a heating block (Eppendorf ThermoMixer® C) for 10 min at 95°C, and debris from cells were removed by centrifugation at 23,995.63 g force for 5 min. The supernatant was carefully transferred to a fresh microcentrifuge tube and stored at −20°C for PCR (28, 29).
Primers used for targeting MecA gene, which is the main determinant of methicillin resistance, and primers for Panton–Valentine leukocidin (PVL) are shown below:

PCR reactions were carried out on each template using the method previously described (30, 31). One reaction contained 2 μl of template DNA added to 48 μl of master mix to make a final volume of 50 μl instead of 25 μl (primers were obtained from Integrated DNA Technologies, USA). The 50-μl PCR master mix was composed of 5 μl 10× buffer, 1 μl dNTPs, 4 μl MgCl2, 0.25 μl pure Taq enzyme, and 35.75 μl nuclease-free water. An initial denaturation at 94°C for 5 min was followed by 30 cycles of denaturation at 94°C for 60 s, annealing at 60°C for 60 s, and extension at 72°C for 60 s on a thermal cycler (Applied Biosystems, USA), with a final extension step of 72°C for 7 min. PCR products were visualized on horizontal 2.0% agarose gels in 1× TBE buffer, loaded with 10 μl of reaction mixture and stained with ethidium bromide nucleic acid stain. O’GeneRuler 100 bp DNA ladder (Thermo Scientific) was used during electrophoresis to estimate the band sizes obtained. The resulting amplicons from gel electrophoresis were observed with ultraviolet illumination using image analysis system (UVIsave Gel Documentation System, Tokyo).
Continuous data were expressed as mean ± SD, whereas categorical data were expressed as a percentage. Prevalence was estimated as percentage, with 95% confidence intervals. Chi-square test was used to compare resistance proportions and the association between the presence of Staphylococci species and stage lymphedema. A p-value of less than 0.05 was judged significant in all circumstances. SPSS (version 20) was used to analyze the data.
In all, 321 study participants between the ages of 18 and 87 years were recruited from the eight communities. Out of the 321 participants, 217 (67.6%) were females and 104 (32.4%) were males (Table 1). People in the age group of 38–47 years were the most represented group in this study (Table 1). Majority of the study participants (63.8%) were engaged in agrarian activities such as farming and fishing (including fish mongering), while 21.5% of them were unemployed (Table 2). Moreover, 28.7% of study participants presented with stage III lymphedema, followed by 22.4% with stage IV (Table 2). Only 2.2% presented with stage VII (Table 2).
A total of 192 non-duplicate Staphylococci species were isolated and identified by MALDI-TOF MS to the species level. Eighty of these were from wounds (42%) and 112 from cutaneous surfaces (non-wounds) (58%). The overall prevalence of Staphylococci species was 39.7% (95% CI: 35%–44%; N = 483), with S. hominis being the most prevalent species, 46 (23.95%). S. haemolyticus, 40 (20.83%); S. epidermidis, 29 (15.10%); S. aureus, 20 (10.41%); and S. saprophyticus, 18 (9.32%) were the next most prevalent taxa. The remaining 20.34% of isolates were distributed among S. wanneri, S. sciuri, S. pasteuri, S. xylosus, S. simulans, S. cohnii, S. caprae, S. lugdunensis, and S. capitis (Table 3).
Lymphedema stages III and IV were observed to show higher frequency of Staphylococci species followed by stage II (Table 2). Also, patients in the late stages of lymphedema (i.e., stages VI and VII) had few populations of Staphylococci species either in their wounds or on their lymphedematous skin. However, a chi-square test of independence showed no significant association between the presence of Staphylococci and the stage of lymphedema (X2 = 0.0731, p-value = 0.999) (Table 4).
Phenotypic testing was done to comprehend trends in local antimicrobial resistance patterns of staphylococcal isolates. We found a high prevalence of resistance to various antimicrobial agents. Isolates showed highest resistance to tetracycline, chloramphenicol, and penicillin, 131 (68.2%), 153 (79.7%), and 135 (70.3%), respectively (Table 5). The sensitivity, resistance, and intermediate drug response of Staphylococci species are depicted in Figure 1.
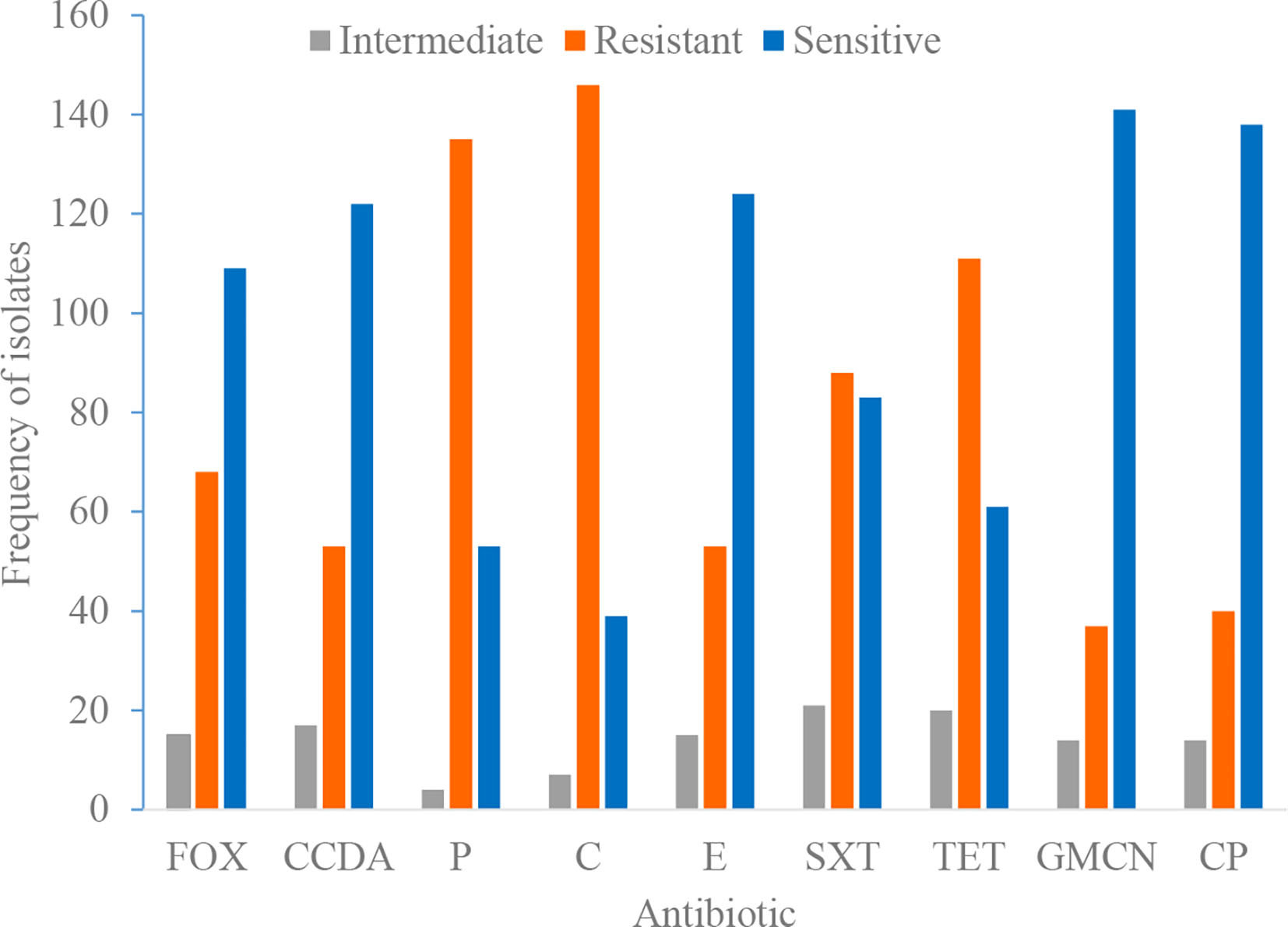
Figure 1 Antimicrobial susceptibility and resistance pattern of isolates. P, penicillin; FOX, cefoxitin; SXT, trimethoprim-sulphamethoxazole; E, erythromycin; GMCN, gentamycin; C, chloramphenicol; CIP, ciprofloxacin; TET, tetracycline; CCDA, clindamycin.
MRSA, containing the mecA gene, was detected in 21 out of 31 Staphylococci isolates tested, with an overall prevalence of 68% (95% CI: 51%–84%). The mecA gene was detected in S. aureus, 14 (45.2% of isolates); S. hominis and S. haemolyticus, three (9.7%) each; and S. epidermidis, one (3.2%). In addition, the PVL gene was detected in 20/31 (64.5%) S. aureus (Table 6).
The contribution of secondary bacterial infections to complicating lymphedema progression remains to be comprehensively investigated. Field observations suggest potential antimicrobial resistance among individuals presenting with chronic filarial wounds. In this study, we investigated the prevalence of Staphylococcus spp. among individuals presenting with filarial lymphedema and the risk of antimicrobial resistance of these Staphylococcus spp. against commonly used antibiotics by LF individuals in the Ahanta West District in the Western Region of Ghana. The high prevalence of staphylococcal species (39% of all bacterial isolates) observed in this study was not uncommon given that Staphylococci species are reported to be the abundant bacterial species in the phylum Firmicutes of the human skin microbiota (32). In fact, the Staphylococci species identified in LF wounds were not different from those observed from adjacent cutaneous surfaces, only the prevalence and abundances were different. This finding is in agreement with a previous observation from LF patients in India (33). Nonetheless, previous studies have suggested staphylococcal species to be associated with prominent human disease-causing agents in both hospital and community settings (34, 35). Thus, there is a need to carefully monitor these commensal bacteria, as a gain of virulence factor could possibly worsen the plight of the individuals living with the infection.
Also, it is worth noting that uncommon species of CoNS were detected among the study participants including S. capitis, which could be emerging opportunistic pathogens (36). The increased prevalence of Staphylococcus spp. in this study may be attributed to the fact that, in LF, the underlying disease compromises the localized immune responses of the skin, creating opportunity for proliferation of these microbes. Bacterial products of Staphylococci known as superantigens have been reported to be responsible for recurrent skin inflammation, and studies have shown that these worsen the filarial manifestations, which results in high release of cytokine and degranulation of mast cells (37). In this study, we showed that filarial lymphedema patients may be susceptible to Staphylococci colonization and which collaborates the findings previously documented (38). Furthermore, 94% of Staphylococci spp. isolated in this present study carried the PVL virulence gene, which is responsible for tissue necrosis in skin and soft tissue infections and associated with community-acquired MRSA (CA-MRSA). This is higher compared to the 27% established previously in a community-based report in Ghana (39). The high PVL-positive CA-MRSA distribution could pose a serious threat in the management of disease and control of infection in the country.
Due to the incessant acute episodes of filarial attacks characterized by headache, chills, and development of plaque-like lesions and of which approximately 68% of the LF individuals have been reported to experience (14), consequently, they resort to antibiotic (either administered orally or topically) and painkillers routinely to ameliorate these conditions that could give a rise to antibiotic resistance. Our study finding on the antimicrobial resistance of the Staphylococci species observed a high prevalence of multidrug-resistant (MDR) Staphylococci species. Antimicrobial resistance among these MDR bacterial populations makes the treatment and management of patients extremely challenging (40). The detection of the mecA gene, a determinant of methicillin resistance, in 45% of S. aureus suggests a possible horizontal gene from S. saprophyticus and S. epidermidis (41), as the gene is not inherent in S. aureus genome.
Another important Staphylococcus of concern particularly among immunocompromised individuals such as LF patients is the S. haemolyticus. Among the CoNS, S. haemolyticus is graded as the most resistant to antibiotics and with very limited options for antibiotic therapy (42). This poses a great challenge, as there is no standard treatment approach for LF patients presenting with chronic non-healing wounds and hence making it difficult for health workers and clinicians to manage LF cases particularly during the filarial attacks.
Thus, understanding the microbial diversity and the behavior of these microbes to existing antibiotics will be a step in the positive direction toward the management of late lymphedema stages with chronic wounds.
In conclusion, Staphylococci species are highly prevalent in wounds and the immunocompromised skin of LF patients. The results from this study suggest that MRSA species may contribute to the complication of filarial lymphedema in Ghana. More attention should be given to the various antibiotic therapies adopted for the management of filarial lymphedema patients presenting with chronic non-healing wounds.
The raw data supporting the conclusions of this article will be made available by the authors without undue reservation.
The studies involving human participants were reviewed and approved by Committee of Human Research, Publications and Ethics, School of Medical Sciences and Dentistry, Kwame Nkrumah University of Science and Technology, KNUST, Kumasi, Ghana. The patients/participants provided their written informed consent to participate in this study.
SW drafted the article. SW, SA, PK, BA, and EAm collected the data from the field. SW, SA, EAs, FO-A, and KB performed the laboratory analyses. AK conceived the idea. DO-Y, KA, and AK read the article critically and edited it. All authors contributed to the article and approved the submitted version.
This study was supported by EDCTP grant number TMA-2016-CDF-1561.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank Mr. Samuel Dodge, the Disease Control Officer at the Dixcove Hospital in the Ahanta West Health Directorate, for his immense contribution to this study.
1. Rajasekaran S, Anuradha R, Manokaran G, Bethunaickan R. An Overview of Lymphatic Filariasis Lymphedema. Lymphology (2017) 50(2017):164–82.
2. Agbenorku P. Lymphedema: Complications and Management. Surg Sci (2014) 5:290–8. doi: 10.4236/ss.2014.57049
4. WHO. WHO Weekly Epidemiological Record. Global Programme to Eliminate Lymphatic Filariasis: Progress Report, Vol. 2018. Geneva: World Health Organisation, (2017). pp. 589–604.
5. Gelderman HT, Bart J, Duijst WL. Elephantiasis in the Netherlands, a Rare Finding and a Reason to Perform an Autopsy. Forensic Sci Int: Rep (2020) 2:100056. doi: 10.1016/j.fsir.2020.100056
6. Kwarteng A, Sylverken A, Asiedu E, Ahuno ST. Genome Editing as Control Tool for Filarial Infections. Biomed Pharmacother (2021) 137:111292. doi: 10.1016/j.biopha.2021.111292
7. Pinheiro ME, Duarte DB, Oliveira MJC, Fontes G. Nephropathy in Lymphatic Filariasis. In: Tropical Nephrology. São Paulo, Brazil: Springer (2020). p. 137–53.
8. Tulimat T, Hoballah JJ. Parasitic Infections of the Vascular System. In: The Surgical Management of Parasitic Diseases. Springer (2020). p. 281–92.
9. Thiers BH. Interdigital Lesions and Frequency of Acute Dermatolymphangioadenitis in Lymphoedema in a Filariasis-Endemic Area. Yearbook Dermatol Dermatol Surg (2007) 2007:166. doi: 10.1016/S0093-3619(08)70457-X
10. Dreyer G, Dreyer P WFP. Extralymphatic Disease Due to Bancroftian Filariasis. Braz J Med Biol Res (1999) 32:1467–72. doi: 10.1590/S0100-879X1999001200003
11. Suma T, Shenoy RK, Varghese J, Kuttikkal VV, Kumaraswami AV. Estimation of ASO Titer as an Indicator of Streptococcal Infection Precipitating Acute Adenolymphangitis in Brugian Lymphatic Filariasis. Southeast Asian J Trop Med Public Health (1997) 28: (4):826–30.
12. Vincent A, Urena-Rojas CA, Ayoub EM, Ottesen EA, EG H. Filariasis and Erisipela in Santo Dominigo. J Parasitol (1998) 84:557–61. doi: 10.2307/3284723
13. Feldman LJ. The Challenge Of Infection In Lymphedema. Natl Lymphedema Network (2005) 17(4):10–2.
14. Kwarteng A, Arthur YD, Yamba JK, Sylverken AA, Kini P, Ahuno ST, et al. Influence of Seasonal Variation on Reported Filarial Attacks Among People Living With Lymphedema in Ghana. BMC Infect Dis (2019) 19(1):442. doi: 10.1186/s12879-019-4084-2
16. Kushwaha V, Kaur S. Lymphatic Filariasis and Visceral Leishmaniasis Coinfection: A Review on Their Epidemiology, Therapeutic, and Immune Responses. Acta Trop (2021) 224:106117.
17. Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, et al. The Role of Nasal Carriage in Staphylococcus Aureus Infections. Lancet Infect Dis (2005) 5:751–62. doi: 10.1016/S1473-3099(05)70295-4
18. Becker K. Staphylococcus Aureus From the German General Population is Highly Diverse. Int J Med Microbiol (2017) 307:21–7. doi: 10.1016/j.ijmm.2016.11.007
19. Dalton KR, Rock C, Carroll KC, Davis MF. One Health in Hospitals: How Understanding the Dynamics of People, Animals, and the Hospital Built-Environment can be Used to Better Inform Interventions for Antimicrobial-Resistant Gram-Positive Infections. Antimicrob Resist Infect Control (2020) 9(1):78. doi: 10.1186/s13756-020-00737-2
20. Anafo RB, Atiase Y, Dayie NT, Kotey FC, Tetteh-Quarcoo PB, Duodu S, et al. Methicillin-Resistant Staphylococcus Aureus (MRSA) Infection of Diabetic Foot Ulcers at a Tertiary Care Hospital in Accra, Ghana. Pathogens (2021) 10(8):937.
21. Donkor ES, Dayie NT, Tette EM. Methicillin-Resistant Staphylococcus Aureus in Ghana: Past, Present, and Future. Microbial Drug Resist (2019) 25(5):717–24. doi: 10.1089/mdr.2018.0115
22. Donkor ES, Tetteh-Quarcoo PB, Nartey P, Agyeman IO. Self-Medication Practices With Antibiotics Among Tertiary Level Students in Accra, Ghana: A Cross-Sectional Study. Int J Environ Res Public Health (2012) 9(10):3519–29. doi: 10.3390/ijerph9103519
23. Pereira PMA, Binatti VB, Sued BPR, Ramos JN, Peixoto RS, Simões C. Staphylococcus Haemolyticus Disseminated Among Neonatesdiagn. Microbiol Infect Dis (2014) 78:85–92. doi: 10.1016/j.diagmicrobio.2013.06.026
24. Byrd AL, Deming C, Cassidy SK, Harrison OJ, Ng W-I, Conlan S, et al. Staphylococcus Aureus and Staphylococcus Epidermidis Strain Diversity Underlying Pediatric Atopic Dermatitis. Sci Trans Med (2017) 9(397). doi: 10.1126/scitranslmed.aal4651
25. Cheesbrough M. District Laboratory Practice in Tropical Countries. 2 ed. New York: Cambridge University Press (2006).
26. Chen H, Xia L, Zhu X, Li W, Du X, Wu D, et al. Burkholderia Pseudomallei Sequencetype 562 in China and Australia. Emerg Infect Dis (2015) 21:166–8. doi: 10.3201/eid2101.140156
27. CLSI. Performance Standard for AntimicrobialSusceptibility Testing. In: CLSIsuppliment M100, 29th ed. Wayne, PA: Clinical and Laboratory Standards Institute (2019). p. 2019.
28. Pitout JD, Hossain A, Hanson ND. Phenotypic and Molecular Detection of CTX-M-β-Lactamases Produced by Escherichia Coli and Klebsiella Spp. J Clin Microbiol (2004) 42(12):5715–21. doi: 10.1128/JCM.42.12.5715-5721.2004
29. Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, et al. Genome Sequence of Shigella Flexneri 2a: Insights Into Pathogenicity Through Comparison With Genomes of Escherichia Coli K12 and O157. Nucleic Acids Res (2002) 30(20):4432–41. doi: 10.1093/nar/gkf566
30. Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, et al. Rapid Detection, Differentiation and Typing of Methicillin-Resistant Staphylococcus Aureus Harbouring Either mecA or the New mecA Homologue mecA Lga251. Clin Microbiol Infect (2012) 18(4):395–400. doi: 10.1111/j.1469-0691.2011.03715.x
31. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter M-O, Gauduchon V, et al. Involvement of Panton-Valentine Leukocidin-Producing Staphylococcus Aureus in Primary Skin Infections and Pneumonia. Clin Infect Dis (1999) 29(5):1128–32. doi: 10.1086/313461
32. Schwiertz A. Microbiota of the Human Body. Adv Exp Med Biol (2016) 902. doi: 10.1007/978-3-319-31248-4
33. Olszewski WL, Zaleska MT. Diagnosis and Management of Infection in Lymphedema. Lymphology (2018) 465–81. doi: 10.1007/978-3-319-52423-8_37
34. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-Resistant Staphylococcus Aureus: An Overview of Basic and Clinical Research. Nat Rev Microbiol (2019) 17(4):203–18. doi: 10.1038/s41579-018-0147-4
35. Wong JW, Ip M, Tang A, Wei VW, Wong SY, Riley S, et al. Prevalence and Risk Factors of Community-Associated Methicillin-Resistant Staphylococcus Aureus Carriage in Asia-Pacific Region From 2000 to 2016: A Systematic Review and Meta-Analysis. Clin Epidemiol (2018) 10:1489. doi: 10.2147/CLEP.S160595
36. Dziri R, Klibi N, Lozano C, Said LB, Bellaaj R, Tenorio C, et al. High Prevalence of Staphylococcus Haemolyticus and Staphylococcus Saprophyticus in Environmental Samples of a Tunisian Hospital. Diagn Microbiol Infect Dis Poverty (2016) 85:136–40. doi: 10.1016/j.diagmicrobio.2016.03.006
37. Dunyo S, Ahorlu C, Simonsen P. Scarification as a Risk Factor for Rapid Progression of Filarial Elephantiasis. Trans R Soc Trop Med Hyg (1998) 91:446. doi: 10.1016/S0035-9203(97)90278-9
38. Ottesen EA. Editorial: The Global Programme to Eliminate Lymphatic Filariasis. Trop Med Int Health (2000) 5(9):591–4. doi: 10.1046/j.1365-3156.2000.00620.x
39. Egyir B, Guardabassi L, Esson J, Nielsen SS, Newman MJ, Addo KK, et al. Insights Into Nasal Carriage of Staphylococcus Aureus in an Urban and a Rural Community in Ghana. PloS One (2014) 9(4):e96119. doi: 10.1371/journal.pone.0096119
40. Zimlichman E, Henderson D, Tamir O, Franz C, Song P, Yamin CK, et al. Health Care–Associated Infections: A Meta-Analysis of Costs and Financial Impact on the US Health Care System. JAMA Internal Med (2013) 173(22):2039–46. doi: 10.1001/jamainternmed.2013.9763
41. Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, et al. Whole-Genome Sequencing of Staphylococcus Haemolyticus Uncovers the Extreme Plasticity of its Genome and the Evolution of Human-Colonizing Staphylococcal Species. J Bacteriol (2005) 187(21):7292–308. doi: 10.1128/JB.187.21.7292-7308.2005
Keywords: lymphatic filariasis, methicillin-resistant, lymphedema, antimicrobial resistance, methicillin resistance Staphylococcus aureus
Citation: Wireko S, Asiedu SO, Kini P, Aglomasa BC, Amewu EKA, Asiedu E, Osei-Akoto F, Boahen KG, Obiri-Yeboah D, Amato KR and Kwarteng A (2021) Prevalence of Methicillin-Resistant Staphylococcus Species Among Filarial Lymphedema Patients in Ahanta West District of Ghana. Front. Trop. Dis 2:786378. doi: 10.3389/fitd.2021.786378
Received: 30 September 2021; Accepted: 25 October 2021;
Published: 19 November 2021.
Edited by:
Daniel Gyamfi Amoako, National Institute for Communicable Diseases (NICD), South AfricaReviewed by:
Gerald Mboowa, Makerere University, UgandaCopyright © 2021 Wireko, Asiedu, Kini, Aglomasa, Amewu, Asiedu, Osei-Akoto, Boahen, Obiri-Yeboah, Amato and Kwarteng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander Kwarteng, c2Vua3dhcnRlbmdAeWFob28uY28udWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.