- 1Grupo de Microbiodiversidad y Bioprospección, Laboratorio de Biología Molecular y Celular, Universidad Nacional de Colombia, Medellín, Colombia
- 2Programa de Estudio y Control de Enfermedades Tropicales (PECET), Universidad de Antioquia, Medellín, Colombia
Gram-negative bacteria are known to use a quorum sensing system to facilitate and stimulate cell to cell communication, mediated via regulation of specific genes. This system is further involved in the modulation of cell density and metabolic and physiological processes that putatively either affect the survival of insect vectors or the establishment of pathogens transmitted by them. The process of quorum sensing generally involves N-acyl homoserine lactones and 2-alkyl-4-quinolones signaling molecules. The present study aimed to detect and identify quorum sensing signaling molecules of AHLs and AHQs type that are secreted by intestinal bacteria, and link their production to their extracellular milieu and intracellular content. Isolates for assessment were obtained from the intestinal tract of Pintomyia evansi (Leishmania insect vector). AHLs and AHQs molecules were detected using chromatography (TLC) assays, with the aid of specific and sensitive biosensors. For identity confirmation, ultra-high-performance liquid chromatography coupled with mass spectrometry was used. TLC assays detected quorum sensing molecules (QSM) in the supernatant of the bacterial isolates and intracellular content. Interestingly, Pseudomonas otitidis, Enterobacter aerogenes, Enterobacter cloacae, and Pantoea ananatis isolates showed a migration pattern similar to the synthetic molecule 3-oxo-C6-HSL (OHHL), which was used as a control. Enterobacter cancerogenus secreted C6-HSL, a related molecules to N-hexanoyl homoserine lactone (HHL), while Acinetobacter gyllenbergii exhibited a migration pattern similar to 2-heptyl-4-quinolone (HHQ) molecules. In comparison to this, 3-oxo-C12-HSL (OdDHL) type molecules were produced by Lysobacter soli, Pseudomonas putida, A. gyllenbergii, Acinetobacter calcoaceticus, and Pseudomonas aeruginosa, while Enterobacter cloacae produced molecules similar to 2-heptyl-3-hydroxy-4-quinolone (PQS). For Pseudomonas putida, Enterobacter aerogenes, P. ananatis, and Pseudomonas otitidis extracts, peak chromatograms with distinct retention times and areas, consistent with the molecules described in case of TLC, were obtained using HPLC. Importantly, P. ananatis produced a greater variety of high QSM concentration, and thus served as a reference for confirmation and identification by UHPLC-MRM-MS/MS. The molecules that were identified included N-hexanoyl-L-homoserine lactone [HHL, C10H18NO3, (M + H)], N-(3-oxohexanoyl)-L-homoserine lactone [OHHL, C10H16NO4, (M + H)], N-(3-oxododecanoyl)-L-homoserine lactone [OdDHL, C16H28NO4, (M + H)], and 2-heptyl-3-hydroxy-4(1H)-quinolone [PQS, C16H22NO2, (M + H)]. Besides this, the detection of diketopiperazines, namely L-Pro-L-Tyr and ΔAla-L-Val cyclopeptides was reported for P. ananatis. These molecules might be potentially associated with the regulation of QSM system, and might represent another small molecule-mediated bacterial sensing system. This study presents the first report regarding the detection and identification of QSM and diketopiperazines in the gut sand fly bacteria. The possible effect of QSM on the establishment of Leishmania must be explored to determine its role in the modulation of intestinal microbiome and the life cycle of Pi. evansi.
Introduction
Quorum Sensing (QS) signaling molecules constitute a complex environmental system that is regulated according to the density dynamics of the bacterial population. It mainly involves N-acyl homoserine lactones (AHLs) and/or alkyl quinolones (AHQ) signaling pathways (1, 2). These signals are generally involved in various gene regulatory mechanisms, and are responsible for modifying the behavior, which is mediated by chemical communication (3, 4). Many gram-negative bacteria are known to regulate different physiological processes through AHL signal, and their biological activity depends on the stereochemistry mediated by homologous proteins related to the LuxI/R system (4, 5). However, AHL type auto inductors are not the only type of molecules that are dependent on the cell density (6, 7).
In this context, pathogen–bacteria–host interactions, found in insect vectors involved tropical diseases, might depend on a wide variety of molecular signals that allow the regulation of activities in a coordinated way, thereby affecting the population abundance of three entities and possibly the vector competition (8). In Aedes aegypti aegypti (Linnaeus), QS generated by bacteria has been shown to have implications in the decision of mosquitoes, particularly regarding the detection, location, and host preferences (8). Additionally, it has been previously reported that quorum sensing molecules (QSM) are considered as “Integral components of global gene regulatory networks”, and these molecules allow bacteria to exhibit physiological responses as per the host environment, such as biofilm formation, virulence, motility, bioluminescence, symbiosis, conjugation, and production of antibiotics and some secondary metabolites (8, 9).
Bacterial intercellular communication (QS) also plays an important role in bacterial cell density and microbiota modulation. However, the gut microbiota of insect vectors is a dynamic and complex system. Regardless of the nature of the association, success of symbiosis majorly depends upon a complex interplay between bacterial symbionts and their insect hosts throughout the course of insect development and reproduction. In fact, many symbiotic bacteria preferentially colonize specialized insect tissues or cells, wherein they can achieve extremely high infection densities; however, QSMs produced by one species or through bacterial consortia can play a decisive role in their establishment. It has been previously reported that cell culture supernatant of P. aeruginosa is capable of mediating cell death of E. coli via a QS mediated process (10).
One of the fundamental aspects of bacterial communication, involving AHLs and AHQ molecules, is their ability to move through the membrane and quickly reach a balance between the intracellular and extracellular compartments at high concentrations (11). This further facilitates the detection of these molecules using qualitative and quantitative bioassays. It also enables the use of complete cells that are capable of detecting a wide variety of signals, bioreporters, and robust and sensitive tools such as HPLC and LC-MS/MS (2, 11–13). It is necessary to specify that biological activity of AHLs molecules is dependent on a wide variety of parameters, including relatively acidic pH that is associated with closed lactone ring, high rate of biosynthesis, presence of an N-acyl chain comprising of at least four carbons in length, and high density of AHLs in the extracellular medium (11, 14).
In case of certain species of medical importance, such as Lutzomyia longipalpis, under normal conditions (without blood in the gut), the gut of adults are characterized by the presence of normal pH of 6, while more alkaline pH is observed after blood intake (pH of 8). However, this pH keeps on dropping throughout the process of digestion and returns to pH 6 at the end (15). In case of larvae, the physiological conditions are completely different. Fazito et al. (16) reported a pH gradient, varying from pH > 9 in the anterior midgut to pH of 6.5–7.0 in the posterior midgut (16).
These findings might suggest that communities present in the gut of phlebotomines act as a strong biological study model for the detection and isolation of physiologically active AHLs molecules. Previous studies of the lepidopteran larvae gut have shown that long chain molecules, like N-acyl carbonates (3-oxo-C12-HSL), are more resistant to basic pH (≥7.4), owing to the hydrophobic nature of the carbon chain and the presence of less electrophilic lactone ring. This makes it less susceptible to attack by hydroxide (–OH) ions and more resistant to higher pH (17). These studies further justify the search of QSMs associated with the intestines of vector insects, such as phlebotomines due to the compartment pH. During blood digestion, QSM are in a more stable environment (alkaline condition). It is known that during the blood digestion, the bacterial population increase (18). This is because the insect produces less hydrogen peroxide and hypochlorite, molecules produced by the insect’s immune system to control the bacterial population in the gut. The QSMs of sand flies putatively might play an important role in the modulation of the development of parasites (Leishmania) in the intestine and/or mechanisms associated with the insect defense systems or the processing of nutrients acquired during its life cycle.
Currently, no studies are available that demonstrated the interaction between Leishmania and QSM. However, studies conducted in other biological models (merely parasites), reported cross species QS that can operate in trypanosomes. For example, the involvement of QS signals in proteolytic processing of host proteins, which resulted in the generation of short oligopeptides that are important for parasitic differentiation, as reported in case of Trypanosoma brucei (19). Additionally, it was reported that QS signal could be used to communicate between mixed infections of different African trypanosome species (19, 20).
Studies exploring QSMs in bacterial communities present in environmentally complex surroundings, such as insects gut, are often complicated. This is mainly attributed to the occurrence of a variety of intrinsic and extrinsic cellular factors that modulate QS kinetics (21) and signal production, detection, and response. QSMs studies conducted in arthropods are mainly restricted to Sodalis praecaptivus (22), Aedes aegypti aegypti, Spodoptera spp., and tsetse flies (17, 20, 22).
Currently, only midgut bacterial genera and endosymbionts, such as Pintomyia evansi and Lutzomyia longipalpis (vectors of visceral leishmaniasis), are known to be present in natural populations of phlebotomine sand flies of Colombia (23–26). Recently, this species was selected to reinforce, deepen, and understand the dynamics of the gut microbiota in the presence of Leishmania infantum infection (27). Dominant intestinal bacteria, including Acinetobacter, Enterobacter, Pseudomonas, Bacillus, Ochrobactrum, Pantoea, Ralstonia, and Methylobacterium, and some endosymbionts, such as Cardinium and Wolbachia, have been previously reported in Pi. evansi. This study highlighted the enzymatic, anti leishmanicidal, and antimicrobial bioactivities of these QS molecules (28). However, details of QSMs detection in leishmania vectors and its importance in the physiology of intestinal bacterial microbiota are not been fully established.
Importantly, QS molecules are not only involved in inter-microbial communication, but these molecules can possibly assist cross-talk of bacteria directly or indirectly with their host, and thus can modulate pathogen transmission.
QS molecules produced by gram-negative bacteria predominantly include AHLs and AHQ molecules, while gram-positive bacteria mainly use peptides (autoinducer peptides, AIP, or quorum sensing peptides). The present study aimed to detect and identify QSMs in gram-negative bacteria isolated from the gut of Pi. evansi. The findings of the study would assist the generation of new and complementary information, which would be useful in understanding the tripartite interactions that occurs between host–parasite–microorganism.
Materials and Methods
Ethics Statement
The sandfly collection was performed following the parameters of Colombian decree N° 1376. This study was performed under the permission granted by Ministry of Environment and Sustainable Development (Permission contract N. 121 22/01/2016 otrosí #23). No specific permits were required for this study.
Isolation of Bacteria and Culture Conditions
Bacterial strains used in this study were isolated from the guts of larvae and adults (females) of Pi. evansi, and these were identified by 16S rDNA gene sequencing (23). In particular, sandflies were collected from two locations of the Department of Sucre (Caribbean coast of Colombia). Further, the guts were dissected under a stereoscope using sterile 1X PBS buffer and stilettos on a glass, under aseptic conditions. Cultivable and aerobic midgut bacteria were isolated. The methods and culture media used in the present study have been previously described in other studies conducted on intestinal microbiota of sand flies. Following this, the gut suspension was streaked on Luria-Bertani (LB) agar (Merck, Bogota, Colombia) and MacConkey Agar (Merck, Bogota, Colombia) plates using serial dilution, and incubated for 48 h at room temperature. Next, the isolates were selected, identified macroscopically and microscopically by Gram’s staining, and molecular identity was established. All isolates were cryopreserved in 20% glycerol at −80°C. For further experiments, only gram-negative bacteria were selected.
All the isolated 12 strains and biosensors (pSB401, pSB1142, and PqsA-Lux) were grown in LB medium at 37°C overnight and sub-cultured in LB agar. A single colony was inoculated and incubated overnight at 37°C at 200 rpm until the cell culture reached an optical density of 1.0 (OD600). Next, 0.25 mL of each culture was transferred to a 250 mL Erlenmeyer flask containing 25 mL of LB broth, and incubated overnight at 37°C with continuous stirring at 200 rpm (1, 2).
Extraction of AHL and AHQ Molecules From Cells and Supernatant
AHL and AHQ extraction was performed following the method previously described by Fletcher and co-workers (1). Briefly, 10 ml of each stationary phase culture was separated by centrifugation at 10000 × g for 10 mins at 4°C. AHL and AHQ molecules associated with the cells or cell-free supernatant were extracted, as previously described (1, 2).
Supernatants were filtered through sterile 0.22 μm filters (Minisart, NML, Sartorius, EE. UU.), and transferred to a 50 mL falcon tube. Following this, 10 ml of acidified ethyl acetate (0.01% acetic acid) was added to filtered supernatant, and the sample was vortexed for 30 sec to mix two phases (organic phase/aqueous phase) (1). The mixture was then transferred to a separating funnel, previously washed with acetone, and it was left undisturbed until two phases were separated. Next, the organic phase was transferred into another 50 mL falcon tube (1). The extraction process was repeated twice or more until a total volume of 30 mL was obtained. The cell pellet was resuspended in 10 mL of LB broth and centrifuged at 10,000 × g for 10 min, and the resulting supernatant was discarded. This process was developed three times to completely remove QSMs from the supernatants. Further, 10 mL of methanol (100% v/v) was added to final cell pellet for re-suspension, and the resulting sample was centrifuged at 10000 × g for 10 mins at 4°C (1, 2).
The extracts obtained from the cells and supernatants were separately transferred to 50 mL round bottom flasks, and subjected to rotary evaporation until liquid phase was completely dried. Each extract was washed with methanol and transferred to 2 mL vials (1). The resulting extracts were dried using a rotary vacuum concentrator (Concentrator 5301, Eppendorf) and stored at −20°C until used for further analysis.
Synthetic Molecules AHL and AHQ
AHL and AHQ synthetic molecules (HHL, OHHL, OdDHL, PQS, and HHQ) that were used as standards and the biosensing strains were received as a kind gift from the Bioscience School of Nottingham University, UK. These samples were conserved as previously described (1). All experiments were carried out in a cell-free bioassay. Table 1 describes the names and some characteristics of AHLs molecules and biosensors used in the present study.
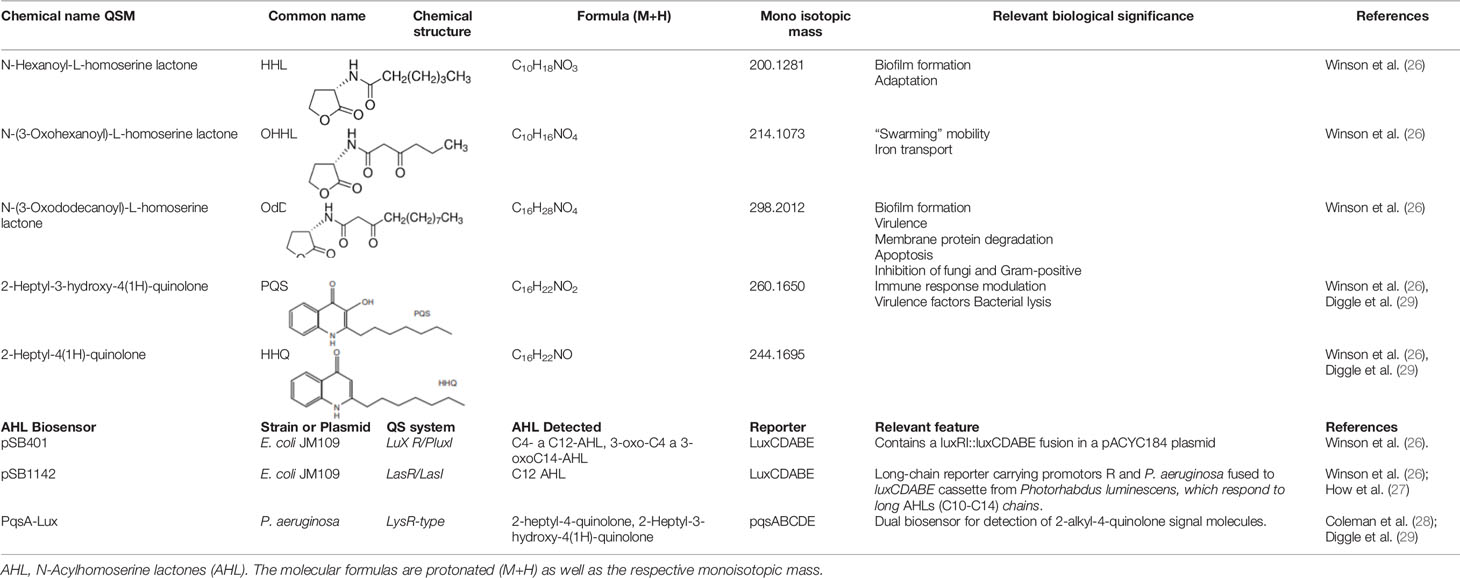
Table 1 List of synthetic Quorum Sensing molecules and specific biosensors used in detection bioassays on TLC and UHPLC-MS.
The identification of large number of QS systems has been made possible mainly by the use of biosensors that are capable of detecting the presence of AHLs. These mechanisms are primarily based on the coupling between a transcriptional regulator and an inducible promoter (Protein/Promoter) that responds to different environmental, nutritional, or cellular communication conditions. This interaction between the target molecule and transcriptional regulator either activates or represses a reporter gene, resulting in a measurable signal change. In gram-negative bacteria, a typical AHL-QS system is mediated by two proteins belonging to the LuxI/LuxR family, which are involved in the bioluminescence production and cell density. Importantly, this AHL-protein complex binds specifically to the box-lux (promoter) to promote the expression of QS genes.
In the present study, all experiments were developed from cell-free supernatants because AHLs that were extracted from cell culture supernatants in their exponential phase significantly increased the sensitivity of biosensors to detect AHL and AHQ molecules.
Detection of AHL Molecules Using Thin Layer Chromatography
First, crude extracts were re-suspended in 100 μL of methanol, and synthetic molecules were brought to a final concentration of 10 mM, 1.0 mM, 0.1 mM, and 1.0 μM. Following this, 5 μL of each sample and 2 μL of each synthetic molecule were loaded onto TLC plate (20 x 20 cm, 60F254 normal phase silica TLC plates, Merck) (29, 30). Chromatographic separation was performed using a solvent phase of dichloromethane-methanol solution (95:5, v/v). After the solvent front reached the required level, plate was removed, dried, and overlaid with 1 mL of E. coli (pSB401 and pSB1142) and P. aeruginosa (PqsA-Lux) biosensor strains using semi-solid LB agar (100 mL). The plates were further incubated at 37°C for 18 h. Bioluminescence signals were detected on TLC plate overlays using an X-ray film for each specific biosensor, at three-time intervals (1, 3, and 10 mins) (29, 30). For each sample, the migration value, Rf, was calculated as Rf = X/Y, wherein “X” represented the distance from the origin to the pattern of the detected molecule and “Y” represented the distance from the origin to the end of the silica gel plate (29, 30). The resulting Rf values for the samples were compared with AHL standards. Samples (cell and supernatants extract) and standards (synthetic molecules) were processed three times to achieve adequate reproducibility.
High-Performance Liquid Chromatography Analysis
For HPLC analysis, 600 µL of methanol was added to 200 µL of each sample. Each sample was subsequently filtered using 0.22 µm nylon filter, and the filtered samples were deposited in mass-type vials. The reagents used for HPLC analysis included acetonitrile (Gradient type, HPLC grade), methanol (HPLC grade, MERCK®), and water-type I (Milli-Q, Millipore, MERCK®). For HPLC analysis, a Chromaster Hitachi® HPLC equipped with a DAD detector at a wavelength of 210 nm was used. A Synergi™ 4 µm Hydro-RP 80 Å LC Column (150 x 4.6 mm), with a KJ0-4282 precolumn with C-18 cartridges both brands Phenomenex was also used (13, 31). The mobile phase comprised of 100% Milli Q water in pump A and 100% acetonitrile in pump B. Both the solvents were maintained at room temperature. The samples were injected in volumes of 10 µL each, and separated using reverse phase chromatography method as described below (13, 31).
Briefly, HPLC separation was carried out at a flow rate of 2 mL/min. The total run time for HPLC separation was 42 mins, which involved initial isocratic conditions of 5% of B and 95% of A for first 2 mins, which changed to 5% to 20% of B in the next minute. This was followed by a gradient change from 20% to 95% of B in 30 minutes, which was maintained for next 3 mins. Subsequently, it returned to the initial condition in 5 mins, which was maintained for additional 2 mins (13, 31).
Mass Spectrometry (UHPLC-MRM-MS/MS) Analysis
Mass spectrometry analysis was performed using two buffers, 100% Milli Q water with 0.1% formic acid in pump A and 100% acetonitrile with 0.1% formic acid in pump B. Both the solvents were maintained at 40°C. The chromatographic separation was performed with UHPLC using an initial isocratic flow with 5% of B for 3 mins, which changed from 5% to 20% of B in next 1 min. Subsequently, the gradient increased from 20% to 95% of B in 18 mins, and this condition of B was maintained for next 3 mins. Finally, it returned to the initial condition in next 3 mins. The total run time of separation was 32 mins (1, 32).
To confirm the exact masses of the standards, the samples were injected in volumes of 1 µL each, and separated by reverse-phase chromatography using Thermo® Ultimate 3000, with a Synergy 2.1 μm Hydro-RP 80a column (3 x 50 mm) Phenomenex®, which was coupled to a Bruker® Impact II ESI-QTOF in positive ion scan mode, with auto MS/MS in a range of 50–1200 m/z. The spectrometer conditions that were used for MS included Dry Heater: 200°C, Nebulizer: 29 psi, Dry Gas: 8 L/min, Capillary: 4500 V, Charging Voltage: 2000 V, and Fragment: 700Vp. Spectra in MS/MS mode were collected for MS (full scan) range of 100–1200 m/z. Auto MS/MS was performed in a range of 50–1200 m/z, which was collected at a rate of 5 spectra, with a maximum of 3 selected and fragmented precursors, using a collision energy of 5eV. To search the masses of the exact standard and possible adducts with sodium and methanol, the spectra were processed using Bruker® Data Analysis software (1–32) and in silico fragmenter MetFrag (33).
Once the masses of the exact standard were confirmed, an MRM mode test was carried out based on monoisotopic mass of ions (M + H) 200.1281, 214.1073, 298.1073, 244.1695, and 260.1650, which corresponded to the molecules HHL, OHHL, OdDHL, HHQ, and PQS. MRM mode test was performed using a Reprosil-Pur Basic C18 column (brand DR Maisch) and same equipment, under same conditions (31). Samples (cell and supernatants extract) and standards (synthetic molecules) were processed three times to achieve adequate reproducibility.
Results
Detection of AHL and AHQ Molecules Using Specific Bioreporters
All the isolates, except O. anthropi, generated QS molecules that were associated with the cell fraction or supernatant. These molecules showed bioluminescence patterns that were related to different type of organic AHL molecules, which were detected by pSB401-pSB1142 of E. coli and PqsA-Lux of P. aeruginosa bioreporters on TLC plates (Supplementary Material 1 and Table 2). Greater production of QS was evident in the supernatant as compared to the cell fraction. QS is known to involve the production of extracellular signaling molecules, which are spread to the external environment via accumulation according to growth and cell density. But it is noteworthy that some bacteria, such as E. hormaechei, generate QS only in the cell fraction. This might be associated with autoinducers that were bound by specific receptors residing in the inner membrane or cytoplasm.
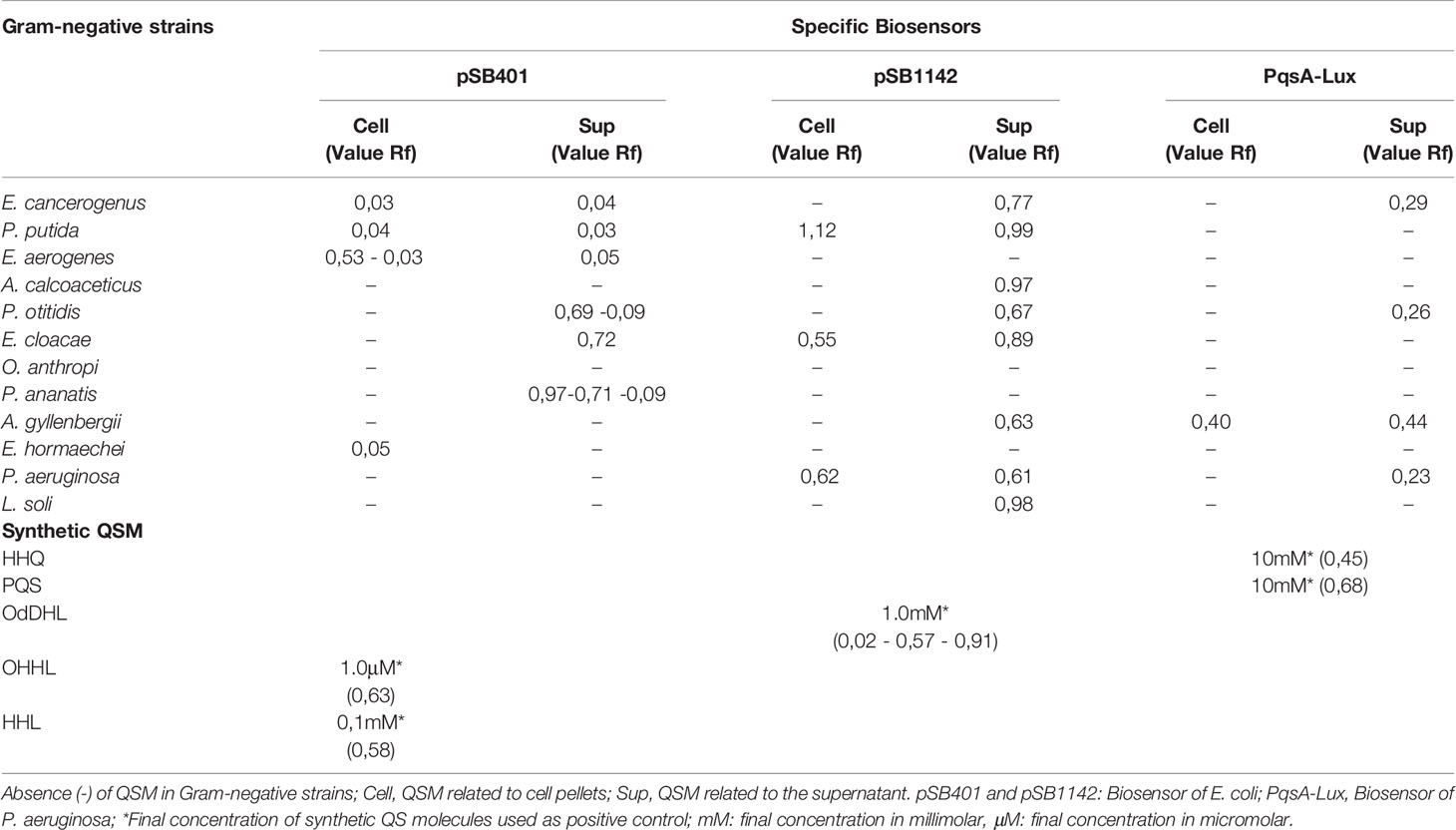
Table 2 Detection of Quorum Sensing molecules (QSM) AHLs and AHQs with pSB401, pSB1142, and PqsA-Lux biosensors as well as migration values (Rf) of each molecule on TLC plates.
P. otitidis, E. cloacae, and P. ananatis presented a migration pattern similar to 3-oxo-C6-HSL (Rf = 0.63), whereas E. cancerogenus secreted molecules related to HHL (Rf = 0.58). In comparison to these, AHQ molecules were visualized in the pellet and supernatant of A. gyllenbergii on biosensor PqsA-Lux and had a migration pattern similar to HHQ (Rf = 0,457), with closely related Rf values (Table 2). L. soli, P. putida, P. aeruginosa, A. gyllenbergii, P. otitidis, A. calcoaceticus, and E. cloacae isolates were also found to secrete QS molecules related to the supernatant or cell pellet, with migration similar to the positive control OdDHL, 3-oxo-C12-HSL (Rf = 0.024–0.575–0.914) (Supplementary Material 1 and Table 1). Some of the detected QS did not correlate with Rf values expressed by the synthetic molecules that were used as a control, which indicated that these are molecules of a similar nature, but require further analysis by LC/MS-MS for accurate identification.
On TLC, the residues of molecules that were found to be different from the controls migrated with Rf values < 0.1 (for example E. hormaechei, E. cancerogenus, and P. putida), as observed in pSB401. This could represent 3-oxo-C8-HSL or C8-HSL. In comparison to this, the uncharacterized signals in PqsA–Lux exhibited Rf value < 0.5 (E. cancerogenus, P. otitidis, A. gyllenbergii, and P. aeruginosa), which might belong to PQS-type molecules; however, additional analysis is required to confirm the same.
QSM Detection by High-Performance Liquid Chromatography
As expected, singular/single peak chromatograms were obtained for each sample of synthetic AHL and AHQ molecules, which were used as standards and controls. Retention times and peak heights (Supplementary Materials 2A, B) were estimated for these molecules, which were later used as a library to compare the presence of QSM in the bacterial extracts that were assessed.
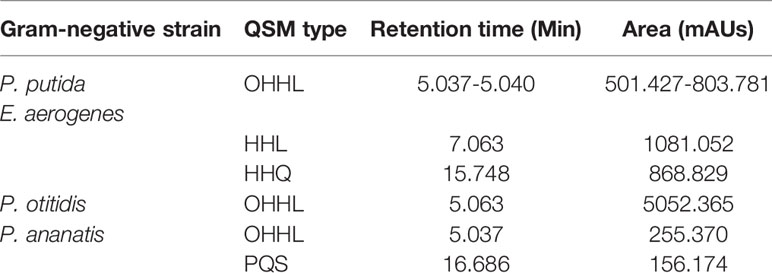
Interestingly, the chromatographic separation yielded defined and distinctive peaks and retention times for supernatants and cell fraction of P. putida, E. aerogenes, P. otitidis, and P. ananatis, when compared with previously characterized synthetic QS patterns (Table 3). Among these QSM, OHHL signal was detected repeatedly in the chromatograms obtained from HPLC, with retention times ranging from 5.037–5.063 mins and peak areas varied from 255.370–5052.365 mAU*s. The supernatants of E. aerogenes and P. ananatis were found to be unique, as their respective chromatograms made it feasible to confirm the possible presence of QSM type HHL, HHQ, and PQS, respectively (Table 3).
QSM Identification With Mass Spectrometry (UHPLC-MRM-MS/MS)
As shown in Figure 1, mass spectra and fragmentation patterns were obtained for each AHL and AHQ synthetic molecules, which were used as standards (33). It should be pointed out that UHPLC-MRM-MS/MS analysis focused mainly on P. ananatis for certain reasons. During HPLC analysis, the concentration of AHLs and AHQs were recorded to be comparatively lower in case of other bacterial extracts. Consequently, during the first-run of UHPLC-MRM-MS/MS, adequate ionization was not achieved to obtain characteristic mass spectra. Additionally, little degradation was observed in the extracts of the supernatants obtained from the rest of the bacterial isolates. Thus, the bacterium P. ananatis presented the greatest richness of QSM, as reported by TLC, HPLC, and UHPLC-MRM-MS/MS analyses. As a result, P. ananatis was considered as the biological reference sample for the validation and identification of QSM.
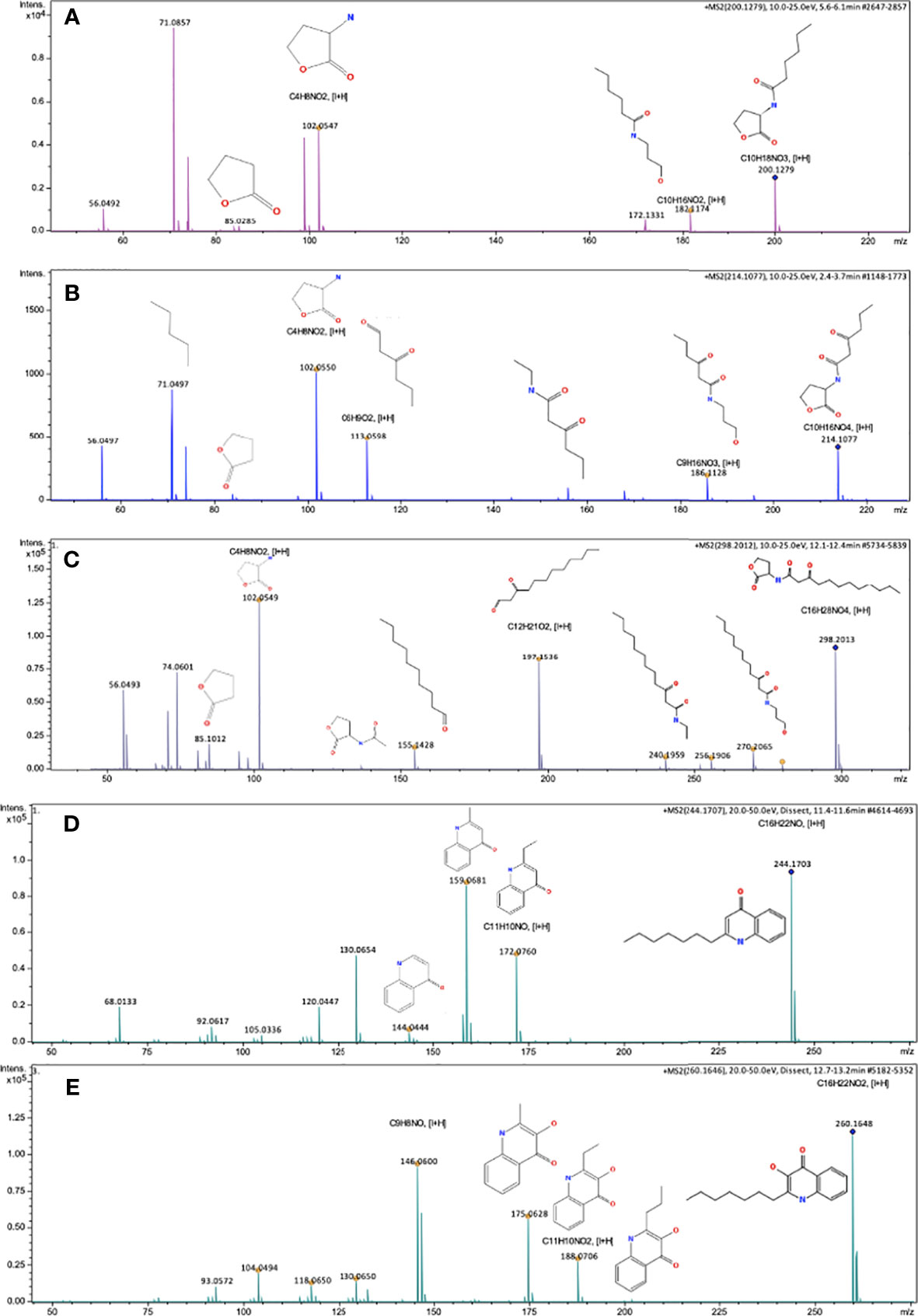
Figure 1 Mass spectra consistent with AHLs and AHQs standard molecules obtained through UHPLC-MRM-MS. (A) Mass spectra of HHL molecule [M+H]= 200.1279, Fragments [I+H]= 182.1174, 102.0547, 85.0285, 71.0857, 56.0492 (B) Mass spectra of OHHL molecule [M+H]= 214.1077, Fragments [I+H]= 186.1128, 113.0598, 102.0550, 71.0497, 56.0497, (C) Mass spectra of OdDHL molecule, [M+H]= 298.2013, Fragments [I+H]= 270.2065, 256.1906, 240.1959, 197.1536, 155.1428, 102.0549, 85.1012, 74.0601, 56.0493, (D) Mass spectra of HHQ molecule [M+H]= 244.1703, Fragments [I+H]= 172.0760, 159.0681, 130.0654, 120.0447, 68.013, (E) Mass spectra of PQS molecule [M+H]= 260.1648, Fragments [I+H]= 188.0706, 175.0628, 146.0600, 130.0650, 104.0494, 93.0572. Impact II ESI-QTOF de Bruker® equipment. Reprosil-Pur Basic C18 column, DR Maisch Brand (characterized of standards A–C). Synergy Hydro-RP 80a column, Phenomenex® Brand (characterized of standards D, E).
A chromatogram of total ions (TIC) was obtained for the supernatant extract of P. ananatis (Figure 2A), and subsequently, protonated ions were identified in the mass spectra belonging to HHL (C10H18NO3) molecules and their lactone ring (Figure 2B), OHHL (C10H16NO4), OdDHL (C16H28NO4), and PQS (C16H22NO2) molecule (Figures 2C–E). The masses of HHL and OHHL molecules were fragmented (Figure 2). Molecules having nature different from AHLs and AHQs were also identified in P. ananatis supernatant. These molecules corresponded to diketopiperazines (L-Pro-L-Tyr and ΔAla-L-Val cyclopeptides), as revealed by mass spectra (Figure 3).
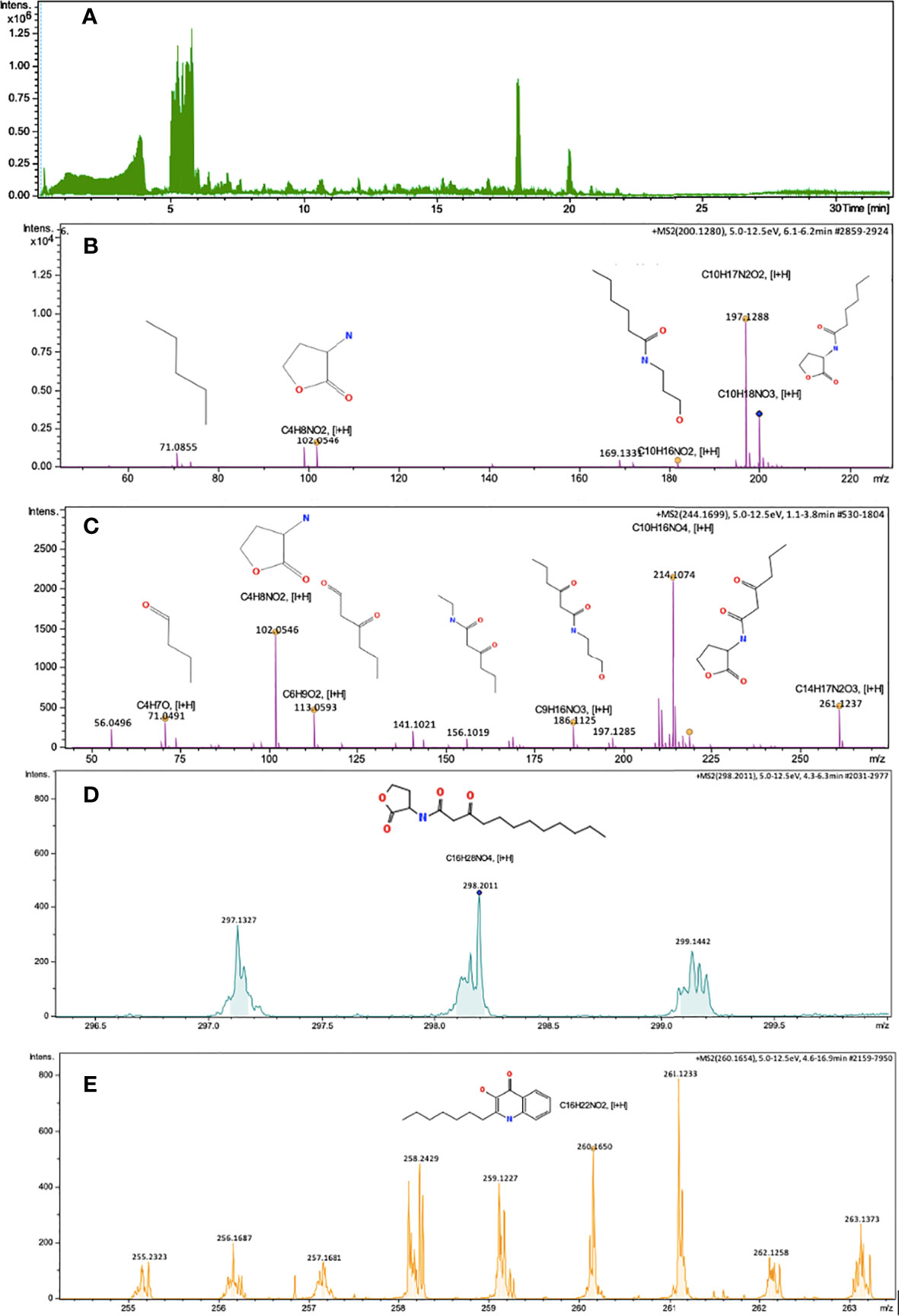
Figure 2 Chromatogram of total ions and mass spectra consistent with AHLs and AHQs molecules, obtained from the supernatant extract of P. ananatis (Isolate 140), using UHPLC-MRM-MS/MS. (A) Total ions chromatogram, (B) Mass spectra of HHL molecule and HHL lactone ring, C10H18NO3 [M+H]=200.1280, Fragments [I+H]= 182.1174, 101.0546, 71.0855, (C) Mass spectra of OHHL molecule, C10H16NO4, [M+H]= 214.1074, Fragments [I+H]= 186.1125, 113.0593, 102.0546, 71.0491, 56.0496, (D) Mass spectra of OdDHL molecule, C16H28NO4, [M+H]= 298.2011 (E) Mass spectra of PQS molecule, C16H22NO2, [M+H]= 260.1650. Impact II ESI-QTOF Bruker® equipment, Reprosil-Pur Basic C18 column, DR Maisch brand.
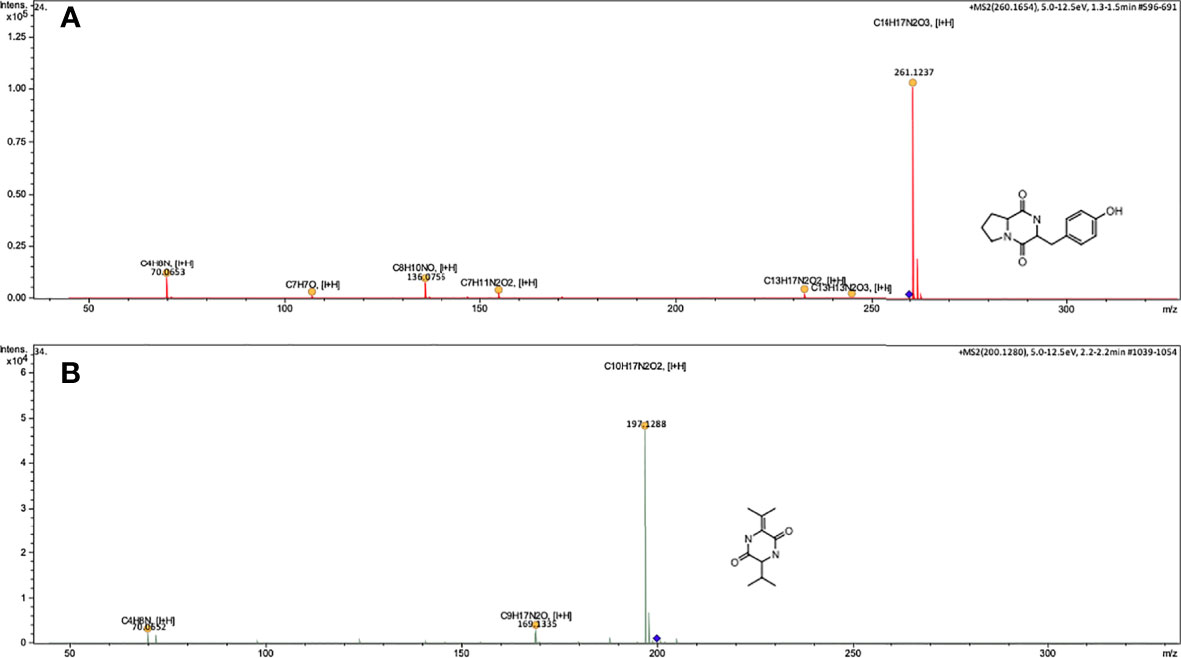
Figure 3 Mass spectra consistent with Diketopiperazines-DCPs type molecules, obtained from the supernatant extract of P. ananatis (Isolate 140), using UHPLC-MRM-MS/MS. (A) Mass spectra of the molecule Cyclo (L-Pro-L-Tyr), C14H16N2O3, [M+H]=261.1234, (B) Mass spectra of molecule Cyclo (ΔAla-L-Val), C10H16N2O2, [M+H]= 197.1284. Impact II ESI-QTOF Bruker® equipment, Reprosil-Pur Basic C18 column, DR Maisch brand.
Discussion
Regulation of gene expression in bacteria is an important adaptive trait that influences the dynamics of the intestinal microbiomes of insects, their physiology and survival, and establishment of other microorganisms (34). One of these regulatory modes is related to the QS system (35), wherein bacteria synthesize an autoinductive signal pheromone that increases according to the bacterial density and interacts with response regulators in a concentration dependent manner (threshold). This in turn facilitates changes in the host gene expression (suppression or expression of immune response genes) or increases the production of secondary metabolites from inter-specific interactions (34).
It has been previously shown that QS molecules could participate in the interactions between insect vectors and vertebrates, preferably (Aedes aegypti aegypti) increasing their attraction (8) or other insects (tsetse flies) that could attenuate the virulence of their pathogens. Such a process has been reported in bacterium Sodalis glossinidius (22). The microbiota and signaling molecules in invertebrates are also known to exert strong effects on fitness, nutrient supply, maturation of the immune system, development, and resistance to colonization against pathogens (36).
All these findings motivated both the detection and identification of QS molecules (N-acyl homoserine lactones and 2-alkyl-4-quinolones) in gram-negative bacteria isolated from the intestinal tract of Pi. evansi, a blood-sucking insect that interacts with viruses (arbovirus), parasites (Leishmania), and bacteria. QS molecules described in the present study varied in terms of the length of carbon chain and associated functional groups. Thus, the present study is the first report associated with this vector.
TLC enabled significant detection of QS molecules as these molecules were found to be present in eleven of the twelve isolates, in both supernatant and cell extracts. In particular, a greater number of molecules were detected mainly in the supernatant obtained from Enterobacter-Pantoea and Pseudomonas-Acinetobacter genera, with bioreporter pSB401 and pSB1142, respectively. In TLC test, migration patterns showed high similarity to synthetic molecules OHHL and HHL. In addition to this, TLS test consistently detected HHQ molecule and signals with migration patterns close to the migration value (Rf) of PQS, using PqsA-Lux reporter.
AHQ biosensor provided a simple, rapid, and sensitive means to detect QS molecules, without requiring the use of any sophisticated instrumentation. In many cases, it has been observed that the detection, identification, quantification, and migration patterns of AHL and PQS are usually inaccurate, and the sensitivity is generally lower than the expected (10). It has been previously reported that TLC could possibly assist in obtaining masked results (molecules of same Rf but different identity), which is attributed to polar nature of AHL. In particular, it could detect the presence of diketopiperazines or non-AHL type components that interfere or activate biosensors response (37, 38). However, it is also necessary to indicate that a lower concentration of control compounds would allow better visualization of the spots. This is a potential limitation of the present study.
Thus, significant care should be taken while interpreting the data obtained using AHLs biosensors, owing to aforementioned limitations. In the view of the above, the use of more than one bioreporter and standard synthetic QS molecules would assist in achieving a better profile from environmental isolates (10), as developed/observed in this study. At the same time, TLC could also help to observe the development of organic reactions, ensure the purity of the evaluated products, and determine the different types of AHL and PQS molecules (39).
As reported in most of the studies, the detection of QS signal molecules in the supernatant was higher as compared to the cells for most of the gram-negative bacteria (E. cancerogenus, P. putida, E. aerogenes, A. calcoaceticus, P. otitidis, E. cloacae, P. ananatis, and P. aeruginosa) isolated from Pi. evansi. This might be attributed to the ability of bacteria to modulate their cellular communication using diffusible signal molecules in the extracellular content. The concentrations of these molecules are strictly related to the density of microorganisms or surrounding cells (40). This process of massive generation (threshold value) of signal molecules in the supernatant by gram-negative bacteria is important because it facilitates the increase in the activity of QS-regulated genes, stimulates the differential production of AHLs-AHQs, and induces biofilm formation. In addition to this, it can even improve the secretion of extracellular polymeric substances (41), which in the case of insects vector could play a decisive role in either favoring or inhibiting the establishment and development of parasites or viruses.
A greater number of QS signal molecules were also detected in the intracellular content using pSB401 and pSB1142 biosensors, which were mainly associated with Enterobacter and Pseudomonas genera. In general, QS signal molecules are synthesized and released into the environment. Although further studies are necessary, the findings of the present study might indicate the presence of these molecules at the level of bacterial cell envelope. In fact, their presence might be potentially associated with membrane vesicles to later disseminate these molecules to the extracellular content, as demonstrated in Pseudomonas and Vibrio harveyi (42–44). It has been previously reported that PQS type molecules of Pseudomonas are required for membrane vesicles formation (45). In addition to this, it has been described that some QS signal molecules are hydrophobic and probably cannot diffuse (43). The occurrence of this type of molecules in gram-negative bacteria is highly relevant, which might directly influence cell signaling and intra-specific and inter-specific gene regulation (46, 47).
Definitive chemical analysis was done using UHPLC-MRM-MS/MS, which is useful for separation and fragmentation of AHLs and AHQs by structural analysis. This method then contemplated mass spectrometry (MS/MS) of the same extracts, and other coupled approaches, such as the mode MRM (Multiple Reaction Monitoring), were further used to generate the identity of greater precision. The supernatant extracts from P. putida, E. aerogenes, P. otitidis, and P. ananatis showed the highest concentration of QS signal molecules, particularly OHHL, HHL, HHQ, and PQS, as indicated by the areas and retention times obtained by HPLC, which were similar to those previously established in the chromatograms of synthetic controls. These HPLC findings confirmed the detection of QS signal molecules in the isolates obtained from Pi. evansi. Although the results of UHPLC-MRM-MS/MS confirmed the presence of these molecules only in P. putida, E. aerogenes, P. otitidis, and P. ananatis, the results obtained for QS signal molecules detected in the rest of the bacterial strains, using TLC, should be considered important for the sensitivity of the biosensors and their specificity, for which an adequate reproducibility of each evaluated extract was obtained.
It is possible that before conducting UHPLC-MRM-MS/MS analysis, QS signal molecules were inhibited or degraded in some of the extracts, possibly by the action of antimicrobial components naturally produced by the bacteria, which are endowed with the potential to exert bacteriostatic or bactericidal effects to reduce cell density. Importantly, cell density is known to be the main stimulus for QS signal molecules production. In addition to this, the interference in the production of these molecules could be due to the presence of diketopiperazines (48). Besides these, two functional groups of AHLs-degrading enzymes have been reported, which include AHL-lactonase that catalyzes the opening of the lactone ring and AHL-acylase that hydrolyzes the amide bond (49).
The results for TLC and HPLC analyses for P. ananatis encouraged its selection for in-depth analysis of UHPLC-MRM-MS/MS. In particular, it presented the greatest variety of QS signal molecules, as assessed by TLC. In fact, P. ananatis exhibited the best concentration of QSM, as estimated by HPLC. Besides this, it is known to have endosymbiotic association with blood-sucking insects like Pi. evansi, and exhibits wide biotechnological potential. Interestingly, P. ananatis supernatant sample and UHPLC-MRM-MS/MS analysis allowed consolidated identification of these molecules, which included HHL, OHHL, OdDHL, and PQS. Previous studies provided evidences for the relationship between these molecules and expression of genes involved in iron transport, metabolism, and oxidative stress in insects (31), detoxification, and induction of the synthesis of a surfactant necessary for “Swarming” bacterial displacement (50, 51). In addition to this, these molecules influence the production of extracellular enzymes (proteases) and increase the virulence of bacteria (52, 53), which might be important for the survival of Pi. evansi and establishment of pathogens that develop in the digestive tract.
Specifically, OHHL molecule is known to play an important role in the pathogenesis and regulation of genes in insects of agricultural importance, which allow better adaptation to different ecological niches (53). OHHL is freely diffusible, and it modulates the expressions of genes involved in the transport and storage of metal ions (iron metabolism). At the same time, it is very effective in activating bioluminescence at low concentrations and acts as a determinant of autoinduction mechanisms of genes that condition the production of signals for the increase of bacterial populations in growth media (54). Few studies reported that OHHL increased the expression of genes involved in the production of iron siderophores and bacterioferritin, which have been shown to play a role in decrease the oxidative stress (54). Pontes and co-workers (2008) reported that OHHL increased the expression of a large number of genes in Sodalis glossinidius that are known to be involved in the cellular response to oxidative stress (34). This oxidative stress response is likely mandated under conditions of dense intracellular symbiont infection, when intense metabolic activity is expected to generate a heavy oxidative burden (34). Many bacteria are known to undergo an “adaptive response” when challenged with a sublethal dose of ROS. Thus, it appears that symbionts and host cells work together to modulate their gene expression profiles and metabolic activities to minimize the deleterious effects of oxidative stress encountered during their symbiotic interactions.
It is important to highlight the frequent detection of OdDHL molecule in most of the gram-negative bacteria. This molecule has been previously shown to be involved in modulating the activation of genes that are involved in the interaction of proteins of the host immune system with the membrane proteins of bacteria. Additionally, this molecule plays a decisive role in parasite–insect–microbiota relationship. However, this hypothesis still needs to be validated in Pi. evansi. HHL molecule is relevant to stimulate the production of natural pigments, such as violacein in Chromobacterium violaceum, which has different biological properties against fungi, bacteria and viruses (55).
One of the most relevant findings is perhaps the identification of PQS, mainly due to the physiology and homeostasis participation of gram-negative bacteria with Pi. evansi. Recent data suggested that this molecule might also be involved in mediating the acquisition of iron, cytotoxicity, and biogenesis of external vesicles of membranes. Additionally, it might exert modulating activities on the host’s immune response (56). Furthermore, PQS not only affects cells by altering the transcriptional profiles of genes, but it can also bind directly to hundreds of proteins present in the host surroundings, thus PQS can directly interact with several key virulence pathways (56, 57). Many of these compounds (PQS) have been previously isolated for their antibiotic, antifungal, and anti-algal activity against human, plant, and animal pathogens (58). Several previous studies have also explored the antimalarial potential of alkylquinolones (59–61). In particular, the compounds were isolated from different strains of P. aeruginosa, which inhibited the growth P. falciparum (58). These studies suggested that PQS is a good candidate for designing biological control applications associated with diseases, such as leishmaniasis.
The present study also highlighted the discovery of a different family of signaling compounds, diketopiperazines (DKPs), which were initially discovered in cell-free supernatants for genus Pseudomonas. DKPs are naturally occurring cyclic dipeptides that have been isolated from yeasts, lichens, fungi, bacteria, and marine sponges. Molecular characterization by UHPLC-MRM-MS/MS facilitated the identification of DKPs cyclo(L-Pro-L-Tyr) and cyclo(ΔAla-L-Val) in the extract of free supernatant obtained from P. ananatis. These DKPs could activate biosensors on TLC assays, underscoring and suggesting the importance of chemical characterization of molecules identified in the bioassay (62, 63). DKPs naturally show a wide range of antimicrobial, antiviral, antitumor, and immunosuppressive activities (55, 63). In a previous study, 64 reported insecticidal activity of DPKs in Diptera, specifically in Ceratitis capitata (64).
P. ananatis has been widely explored, especially for being an opportunistic pathogen and causing disease symptoms in a wide range of crops and forest species of global economic importance. However, few recent studies reported leishmanicidal activity against Le. infantum and Le. mexicana (24, 26). Some studies reported the presence of P. agglomerans and P. ananatis in sandflies (Phlebotomus kandelakii, Phlebotomus papatasi, Pi. evansi, and Lu. longipalpis) and mosquitoes (Aedes albopictus and Anopheles stephensi), suggesting a possible effect on the Leishmania development stages. The relationships of P. ananatis with other microorganisms and with the host have been previously studied using various in vitro and in silico approaches, which showed that they perform a variety of functions, including transportation, metabolism, and defense, and are also involved in the generation of specific associated molecules. Altogether, these QSM findings might suggest that these molecules could impact cell density and molecular signaling in various processes, such as biofilm formation, interaction with the host, or production of antibiotics and secondary metabolites in the gut of Pi. evansi.
In conclusion, the present study reported the identification of QSM molecules, including HHL, OHHL, OdDHL, and PQS. Interestingly, the study also identified diketopiperazines, cyclo(L-Pro-L-Tyr) and cyclo(ΔAla-L-Val), in P. ananatis. The characterization of these molecules by bioassays (TLC), use of behavioral biosensors to respond to the stimulation of specific auto inductors, and the implementation of HPLC-MRM allowed the detection (65), characterization, and identification of QSM from gram-negative bacteria isolated from insects. It is necessary to deepen the understanding regarding the detected QSM on the establishment of Leishmania in Pi. evansi, and explore its biotechnological potential to stimulate the production of natural compounds of great pharmaceutical, entomopathogenic, and even industrial interest.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
RV-G, CXMH, GC-R, SR: Designed the study and contributed to writing the manuscript. RV-G, GB-M, and JH-C: Performed the experiments, analyzed the data, and contributed to writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The present study was funded by the Foundation for the Promotion of Research and Technology, Bank of the Republic of Colombia Project No. 4.124; the Administrative Department of Science, Technology, and Innovation (COLCIENCIAS code FP44842-128-2018; Quipu Code 201010021937; HERMES 40051 Postdoctoral call; National Call 528 for doctoral studies in Colombia, 2011); Strengthening of Research, Creation, and Innovation 2016-2018, Universidad Nacional de Colombia (Project Code: 36012; 2010100-Basic Research). Project No. Hermes 27104, BC.N.015-edu2015-O-British Council.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Special recognition to Miguel Camara and Stephan Heeb Professors of School of Life Sciences, Centre for Biomolecular Sciences, the University of Nottingham-UK for their technical collaboration.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2021.760228/full#supplementary-material
Supplementary Figure 1 | AHL and AHQ molecules were detected by bioluminescence signals in the supernatant of Gram-negative strains isolated from the intestinal tract of Pintomyia evansi using Thin Layer Chromatography-TLC methodology. (A) Bioluminescence signals with biosensor pSB401 Line 1: OHHL standard; Line 2: HHL standard; Line 3: CFSs of P. otitidis; Line 4: CFS of E. cloacae; Line 5: CFSs of P. ananatis. (B) Bioluminescence signals with biosensor pSB401 Line 1: OHHL standard; Line 2: HHL standard; Line 6: CFS of E. cancerogenus; Line 7: P. putida; Line 8: E. aerogenes. (C) Bioluminescence signals with biosensor pSB1142 Line 9: OdDHL Standard; Line 10: CFS of Lysobacter soli; Line 11: CFS of P. putida; Line 12: CFS of A. calcoaceticus. (D) Bioluminescence signals with biosensor PqsA-Lux. PQS Standard; HHQ Standard; Line 18: CFS of E. cancerogenus; Line 19: CFSs of P. otitidis; Line 20: CFS of A. gyllenbergii; Line 21: CFS of P. aeruginosa.
Supplementary Figure 2 | HPLC Chromatogram of synthetic QSM (A) with the mixture of synthetic QSM used as controls, (B) retention times and estimated heights from individual chromatography.
References
1. Fletcher MP, Diggle SP, Cámara M, Williams P. Biosensor-Based Assays for PQS, HHQ and Related 2-Alkyl-4-Quinolone Quorum Sensing Signal Molecules. Nat Protoc (2007) 2(5):1254–62. doi: 10.1038/nprot.2007.158
2. Kawaguchi T, Yung PC, Norman RS, Decho AW. Rapid Screening of Quorum-Sensing Signal N-Acyl Homoserine Lactones by an In Vitro Cell-Free Assay. Appl Environ Microbiol (2008) 74(12):3667–71. doi: 10.1128/AEM.02869-07
3. Williams P, Cámara M. Quorum Sensing and Environmental Adaptation in Pseudomonas aeruginosa: A Tale of Regulatory Networks and Multifunctional Signal Molecules. Curr Opin Microbiol (2009) 12(2):182–91. doi: 10.1016/j.mib.2009.01.005
4. Zhang W, Li C. Exploiting Quorum Sensing Interfering Strategies in Gram-Negative Bacteria for the Enhancement of Environmental Applications. Front Microbiol (2016) 6:1–15. doi: 10.3389/fmicb.2015.01535
5. Borlee BR, Geske GD, Robinson CJ, Blackwell HE, Handelsman J. Quorum-Sensing Signals in the Microbial Community of the Cabbage White Butterfly Larval Midgut. ISME J (2008) 2(11):1101–11. doi: 10.1038/ismej.2008.70
6. Tashiro Y, Yawata Y, Toyofuku MH, Uchiyama M, Nomura N. Minireview Interspecies Interaction Between Pseudomonas aeruginosa and Other Microorganisms. Microbes Environ (2013) 28(1):13–24. doi: 10.1264/jsme2.ME12167
7. Williamson LL, Raffa KF, Moth G, Raffa KF, Handelsman J. Signal Mimics Derived From a Metagenomic Analysis of the Gypsy Moth Gut Microbiota. Appl Environ Microbiol (2007) 73(11):3669–76. doi: 10.1128/AEM.02617-06
8. Zhang X, Crippen TL, Coates CJ, Wood TK, Tomberlin JK. Effect of Quorum Sensing by Staphylococcus Epidermidis on the Attraction Response of Female Adult Yellow Fever Mosquitoes, Aedes Aegypti Aegypti (Linnaeus) (Diptera: Culicidae), to a Blood-Feeding Source. PloS One (2015) 10(12):e0143950. doi: 10.1371/journal.pone.0143950
9. Bandara HM, Lam OT, Jin LJ. Microbial Chemical Signaling: A Current Perspective. Crit Rev Microbiol (2012) 38:217–49. doi: 10.3109/1040841X.2011.652065
10. Kumar R, Chhibber S, Gupta V, Harjai K. Screening & Profiling of Quorum Sensing Signal Molecules in Pseudomonas Aeruginosa Isolates From Catheterized Urinary Tract Infection Patients. Indian J Med Res (2011) 134(2):208–13.
11. Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, et al. N-Acylhomoserine Lactones Undergo Lactonolysis in a pH-, Temperature-, and Acyl Chain Length-Dependent Manner During Growth of Yersinia Pseudotuberculosis and Pseudomonas Aeruginosa. Infect Immun (2002) 70(10):5635–46. doi: 10.1128/IAI.70.10.5635-5646.2002
12. Guan C, Ju J, Borlee BR, Williamson LL, Shen B, Raffa K, et al. Signal Mimics Derived From a Metagenomic Analysis of the Gypsy Moth Gut Microbiota. Appl Environ Microbiol (2007) 73(11):3669–36765. doi: 10.1128/AEM.02617-06
13. Anbazhagan D, Mansor M, Yan JO, Yusof MY, Hassan H, Sekaran SD. Detection of Quorum Sensing Signal Molecules and Identification of an Autoinducer Synthase Gene Among Biofilm Forming Clinical Isolates of Acinetobacter Spp. PloS One (2012) 7(7):1–12. doi: 10.1371/journal.pone.0036696
14. Pearson JP, Delden CV, Iglewski BH. Active Efflux and Diffusion are Involved in Transport of Pseudomonas aeruginosa Cell-to-Cell Signals. J Bacteriol (1999) 181(4):1203–10. doi: 10.1128/JB.181.4.1203-1210.1999
15. Santos VC, Araujo RN, Machado LD, Pereira MH, Gontijo M. The Physiology of the Midgut of Lutzomyia Longipalpis (Lutz and Neiva 1912): pH in Different Physiological Conditions and Mechanisms Involved in Its Control. J Exp Biol (2008) 211:2792–8. doi: 10.1242/jeb.019836
16. Fazito V, Pereira MH, Gontijo NF. Midgut pH Profile and Protein Digestion in the Larvae of Lutzomyia Longipalpis (Diptera: Psychodidae). J Insect Physiol (2007) 53(11):1151–9. doi: 10.1016/j.jinsphys.2007.06.005
17. Funke M, Büchler R, Mahobia V, Schneeberg A, Ramm M, Boland W. Rapid Hydrolysis of Quorum-Sensing Molecules in the Gut of Lepidopteran Larvae. ChemBioChem (2008) 9(12):1953–9. doi: 10.1002/cbic.200700781
18. Oliveira JH, Gonçalves RL, Lara FA, Dias FA, Gandara AC, Menna-Barreto RF, et al. Blood Meal-Derived Heme Decreases ROS Levels in the Midgut of Aedes Aegypti and Allows Proliferation of Intestinal Microbiota. PloS Pathog (2011) 7(3):e1001320. doi: 10.1371/journal.ppat.1001320
19. Rojas F, Matthews K. Quorum Sensing in African Trypanosomes. Curr Opin Microbiol (2019) 52:124–9. doi: 10.1016/j.mib.2019.07.001
20. Silvester E, Young J, Ivens A, Matthews KR. Interspecies Quorum Sensing in Co-Infections Can Manipulate Trypanosome Transmission Potential. Nat Microbiol (2017) 2:1471–9. doi: 10.1038/s41564-017-0014-5
21. Barriuso J, Hogan DA, Keshavarz T, Martínez MJ. Role of Quorum Sensing and Chemical Communication in Fungal Biotechnology and Pathogenesis. FEMS Microbiol Rev (2018) 42(5):627–38. doi: 10.1093/femsre/fuy022
22. Enomoto S, Chari A, Clayton AL, Dale C. Quorum Sensing Attenuates Virulence in. Cell Host Microbe (2017) 21: (5):629–36. doi: 10.1016/j.chom.2017.04.003
23. Vivero RJ, Jaramillo N, Cadavid-Restrepo G, Soto S. Structural Differences in Gut Bacteria Communities in Developmental Stages of Natural Populations of Lutzomyia Evansi From Colombia’s Caribbean Coast. Parasitol Vectors (2016) 9(496):1–20. doi: 10.1186/s13071-016-1766-0
24. Vivero RJ, Cadavid-Restrepo G, Herrera CXM, Uribe SI. Molecular Detection and Identification of Wolbachia in Three Species of the Genus Lutzomyia on the Colombian Caribbean Coast. Parasit Vectors (2017) 10:110. doi: 10.1186/s13071-017-2031-x
25. Vivero RJ, Villegas-Plazas M, Cadavid-Restrepo GE, Herrera CXM, Uribe SI, Junca H, et al. Wild Specimens of Sand Fly Phlebotomine Lutzomyia Evansi, Vector of Leishmaniasis, Show High Abundance of Methylobacterium and Natural Carriage of Wolbachia and Cardinium Types in the Midgut Microbiome. Sci Rep (2019) 9:17746. doi: 10.1038/s41598-019-53769-z
26. Sant’Anna M, Darby AC, Brazil RP, Montoya-Lerma J, Dillon VM, Bates PA, et al. Investigation of the Bacterial Communities Associated With Females of Lutzomyia Sand Fly Species From South America. PloS One (2012) 7(8):e42531. doi: 10.1371/journal.pone.0042531
27. Vivero RJ, Castañeda-Monsalve VA, Romero LR, Hurst G, Cadavid-Restrepo G, Moreno-Herrera CX. Gut Microbiota Dynamics in Natural Populations of Pintomyia Evansi Under Experimental Infection With Leishmania Infantum. Microorganisms (2021) 9(6):1214. doi: 10.3390/microorganisms9061214
28. Vivero R, Bedoya G, Robledo S, Moreno C, Cadavid-Restrepo G. Corrigendum to “Enzymatic, Antimicrobial, and Leishmanicidal Bioactivity of Gram-Negative Bacteria Strains From the Midgut of Lutzomyia Evansi, an Insect Vector of Leishmaniasis in Colombia”. Biotechnol Rep (2020) 26:e00451. doi: 10.1016/j.btre.2020.e00451 ISSN 2215-017X.
29. Shaw PD, Ping G, Daly SL, Cha C, Cronan J, Rinehart K, et al. Detecting and Characterizing N-Acyl-Homoserine Lactone Signal Molecules by Thin-Layer Chromatography. Proc Natl Acad Sci (1997) 94:6036– 6041. doi: 10.1073/pnas.94.12.6036
30. Zhao J, Quan C, Jin L, Chen M. Production, Detection and Application Perspectives of Quorum Sensing Autoinducer-2 in Bacteria. J Biotechnol (2018) 268:53–60. doi: 10.1016/j.jbiotec.2018.01.009
31. Winson M, Camara M, Latifi A, Foglino M, Chabra SR, Daykin M, et al. Multiple N-Acyl-L-Homoserine Lactone Signal Molecules Regulate Production of Virulence Determinants and Secondary Metabolites in Pseudomonas aeruginosa (Autoinducers/Quorum Sensing/Gene Regulation). Proc Natl Acad Sci (1995) 92:9427–31. doi: 10.1073/pnas.92.20.9427
32. Rani S, Kumar A, Kumar A, Schmitt-Kopplin P. Occurrence of N-Acyl Homoserine Lactones in Extracts of Bacterial Strain of Pseudomonas Aeruginosa and in Sputum Sample Evaluated by Gas Chromatography—Mass Spectrometry. Am J Anal Chem (2011) 2:294–302. doi: 10.4236/ajac.2011.22037
33. Ruttkies C, Schymanski EL, Wolf S, Hollender J, Neumann S. MetFrag Relaunched: Incorporating Strategies Beyond In Silico Fragmentation. J Cheminform (2016) 8:3. doi: 10.1186/s13321-016-0115-9
34. Pontes MH, Babst M, Lochhead R, Oakeson K, Smith K, Dale C. Quorum Sensing Primes the Oxidative Stress Response in the Insect Endosymbiont, Sodalis Glossinidius. PloS One (2008) 3(10):e3541. doi: 10.1371/journal.pone.0003541
35. Fuqua C, Parsek MR, Greenberg EP. Regulation of Gene Expression by Cell-to-Cell Communication: Acyl-Homoserine Lactone Quorum Sensing. Annu Rev Genet (2001) 35:439–68. doi: 10.1146/annurev.genet.35.102401.090913
36. Franzenburg S, Walter J, Kunzel S, Wang J, Baines J, Bosch T, et al. Distinct Antimicrobial Peptide Expression Determines Host Species-Specific Bacterial Associations. Proc Natl Acad Sci (2013) 110(39):e3730–8. doi: 10.1073/pnas.1304960110
37. Steindler L, Venturi V. Detection of Quorum-Sensing N-Acyl Homoserine Lactone Signal Molecules by Bacterial Biosensors. FEMS Microbiol Lett (2007) 266(1):1–9. doi: 10.1111/j.1574-6968.2006.00501.x
38. Bauer WD, Mathesius U. Plant Responses to Bacterial Quorum Sensing Signals. Curr Opin Plant Biol (2004) 7(4):429–33. doi: 10.1016/j.pbi.2004.05.008
39. Chaudhry U. Detection and Separation of Quorum Sensing Signals by Agar- Plate Based Bioassay and Thin- Layer Chromatography. Mol Microbiol (2008) 1:1–9. doi: 10.13140/RG.2.1.2234.5687
40. Zhao R, Zhang H, Zhang F, Yang F. Fast Start-Up Anammox Process Using Acyl-Homoserine Lactones (AHLs) Containing Supernatant. J Environ Sci (2018) 65:127–32. doi: 10.1016/j.jes.2017.03.025
41. Ren T, Yu H, Li X. The Quorum-Sensing Effect of Aerobic Granules on Bacterial Adhesion, Biofilm Formation, and Sludge Granulation. Appl Microbiol Biotechnol (2010) 88:789–97. doi: 10.1007/s00253-010-2796-8
42. Brameyer S, Plener L, Müller A, Klingl A, Wanner G, Jung K. Outer Membrane Vesicles Facilitate Trafficking of the Hydrophobic Signaling Molecule CAI-1 Between Vibrio Harveyi Cells. J Bacteriol (2018) 10(15):e00740–17. doi: 10.1128/JB.00740-17
43. Orench-Rivera N, Kuehn MJ. Environmentally Controlled Bacterial Vesicle-Mediated Export. Cell Microbiol (2016) 18:1525–36. doi: 10.1111/cmi.12676
44. Florez C, Raab JE, Cooke AC, Schertzer JW. Membrane Distribution of the Pseudomonas Quinolone Signal Modulates Outer Membrane Vesicle Production in Pseudomonas Aeruginosa. mBio (2017) 8:e01034–17. doi: 10.1128/mBio.01034-17
45. Henke JM, Bassler BL. Three Parallel Quorum-Sensing Systems Regulate Gene Expression in Vibrio Harveyi. J Bacteriol (2004) 186:6902–14. doi: 10.1128/JB.186.20.6902-6914.2004
46. Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, Inaba H. Membrane Vesicle Mediated Bacterial Communication. ISME J (2017) 11:1504–9. doi: 10.1038/ismej.2017.13
47. Nakamura S, Higashiyama Y, Izumikawa K, Seki M, Kakeya H, Yamamoto Y, et al. The Roles of the Quorum-Sensing System in the Release of Extracellular DNA, Lipopolysaccharide, and Membrane Vesicles From Pseudomonas Aeruginosa. Jpn J Infect Dis (2008) 61(5):375–8.
48. McPhee J. The Role of Quorum Sensing in the Regulation of Extracellular Enzymes by Pseudomonas Fluorescens. Ottawa: University of Guelph, Thesis (2001). p. 115.
49. Ochiai S, Morohoshi T, Kurabeishi A, Shinozaki M, Fujita H, Sawada I, et al. Production and Degradation of N-Acylhomoserine Lactone Quorum Sensing Signal Molecules in Bacteria Isolated From Activated Sludge. Biosci Biotechnol Biochem (2013) 7712:2436–40. doi: 10.1271/bbb.130553
50. Papenfort K, Bassler B. Quorum-Sensing Signal-Response Systems in Gram-Negative Bacteria. Nat Rev Microbiol (2016) 14(9):576–88. doi: 10.1038/nrmicro.2016.89
51. Daniels R, Vanderleyden J, Michiels J. Quorum Sensing and Swarming Migration in Bacteria. FEMS Microbiol Rev (2004) 28(3):261–89. doi: 10.1016/j.femsre.2003.09.004
52. Kalia VC. Quorum Sensing and Its Biotechnological Applications. In: Kalia V, editor. Quorum Sensing and Its Biotechnological Applications. Singapore: Springer (2018). Available at: https://doi.org/10.1007/978-981-13-0848-2_1.
53. Bradley B, Grant G, Courtney R, Helen B. Quorum-Sensing Signals in the Microbial Community of the Cabbage White Butterfly Larval Midgut. ISME J (2014) 2(11):1101–11. doi: 10.1038/ismej.2008.70
54. Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond J. Quorum-Sensing in Gram-Negative Bacteria. FEMS Microbiol Rev (2001) 25(4):365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x
55. Martinelli D, Grossmann G, Séquin U, Bachofen R. Effects of Natural and Chemically Synthesized Furanones on Quorum Sensing in Chromobacterium Violaceum. BMC Microbiol (2004) 2(4):25. doi: 10.1186/1471-2180-4-25
56. Lin J, Cheng J, Wang J, Shen X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front Cell Infect Microbiol (2018) 8. doi: 10.3389/fcimb.2018.00230
57. Dandela R, Mantin D, Cravatt BF, Rayo J, Meijler MM. Proteome-Wide Mapping of PQS-Interacting Proteins in Pseudomonas aeruginosa. Chem. Sci (2018) 9:2290–4. doi: 10.1039/C7SC04287F
58. Saalim M, Villegas-Moreno J, Clark BR. Bacterial Alkyl-4-Quinolones: Discovery, Structural Diversity and Biological Properties. Molecules (2020) 25:5689. doi: 10.3390/molecules25235689
59. Bultel-Ponce V, Bergé J, Debitus C, Nicolas J, Guyot M. Metabolites From the Sponge-Associated Bacterium Pseudomonas Species. Mar Biotechnol (1999) 1:384–90. doi: 10.1007/PL00011792
60. Supong K, Thawai C, Supothina S, Auncharoen P, Pittayakhajonwut P. Antimicrobial and Anti-Oxidant Activities of Quinoline Alkaloids From Pseudomonas Aeruginosa BCC76810. Phytochem Lett (2016) 17:100–6. doi: 10.1016/j.phytol.2016.07.007
61. Presley CC, Du Y, Dalal S, Merino EF, Butler JH, Rakotonandrasana S, et al. Isolation, Structure Elucidation, and Synthesis of Antiplasmodial Quinolones From Crinum Firmifolium. Bioorganic Med. Chem (2017) 25:4203–11. doi: 10.1016/j.bmc.2017.06.017
62. Holden MT, de Nys R, Stead P, Bainton NJ, Hill PJ. Quorum Sensing Crosstalk: Isolation and Chemical Characterization of Cyclic Dipeptides From Pseudomonas Aeruginosa and Other Gram-Negative Bacteria. Mol Microbiol (1999) 33:1254–66. doi: 10.1046/j.1365-2958.1999.01577.x
63. Milne PJ, Kilian G. The Properties, Formation, and Biological Activity of 2,5-Diketopiperazines. Nelson Mandela Metropolitan University, Port Elizabeth, South Africa a 2010 Elsevier Ltd. All Rights Reserved. Gqeberha: C. Prasad, Peptides (2010).
64. Marcinkevicius K, Salvatore S, Bardon A, Cartagena E, Arena M, Vera N, et al. Insecticidal Activities of Diketopiperazines of Nomuraea Rileyi Entomopathogenic Fungus. Infogain Int J Environ Agric Biotechnol (2017) 2(4):8–2017; 1586-1596. doi: 10.22161/ijeab/2.4.18
Keywords: AHLs, AHQs, diketopiperazines, gram-negative bacteria, sandflies
Citation: Vivero-Gomez RJ, Mesa GB, Higuita-Castro J, Robledo SM, Moreno-Herrera CX and Cadavid-Restrepo G (2021) Detection of Quorum Sensing Signal Molecules, Particularly N-Acyl Homoserine Lactones, 2-Alky-4-Quinolones, and Diketopiperazines, in Gram-Negative Bacteria Isolated From Insect Vector of Leishmaniasis. Front. Trop. Dis 2:760228. doi: 10.3389/fitd.2021.760228
Received: 17 August 2021; Accepted: 03 November 2021;
Published: 02 December 2021.
Edited by:
Yara M. Traub-Csekö, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Nelder De Figueiredo Gontijo, Federal University of Minas Gerais, BrazilShih Keng Loong, University of Malaya, Malaysia
Copyright © 2021 Vivero-Gomez, Mesa, Higuita-Castro, Robledo, Moreno-Herrera and Cadavid-Restrepo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rafael Jose Vivero-Gomez, cmp2aXZlcm9nQHVuYWwuZWR1LmNv; Gloria Cadavid-Restrepo, Z2VjYWRhdmlAdW5hbC5lZHUuY28=
†ORCID: Rafael Jose Vivero-Gomez, orcid.org/0000-0001-6768-4097
Jorge Higuita-Castro, orcid.org/0000-0002-4596-7747
Sara M. Robledo, orcid.org/0000-0003-2752-4931
Claudia X. Moreno-Herrera, orcid.org/0000-0002-8132-5223
Gloria Cadavid-Restrepo, orcid.org/0000-0003-1026-6358
 Rafael Jose Vivero-Gomez
Rafael Jose Vivero-Gomez Gustavo Bedoya Mesa
Gustavo Bedoya Mesa Jorge Higuita-Castro
Jorge Higuita-Castro Sara M. Robledo
Sara M. Robledo Claudia X. Moreno-Herrera
Claudia X. Moreno-Herrera Gloria Cadavid-Restrepo
Gloria Cadavid-Restrepo