
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis., 30 August 2021
Sec. Neglected Tropical Diseases
Volume 2 - 2021 | https://doi.org/10.3389/fitd.2021.729294
Background: Rapid and accurate prevalence mapping of lymphatic filariasis (LF) is necessary to eliminate this disfiguring and disabling neglected tropical disease. Unfortunately, rapid tests such as the filariasis test strip (FTS) for Wuchereria bancrofti, the causative agent of LF in Africa, can cross-react with antigens circulating in some persons infected by the African eye worm, Loa loa, rendering the test unreliable in eleven co-endemic nations. The intended target of the FTS is a heavily glycosylated W. bancrofti circulating filarial antigen (Wb-CFA). Previously, we determined that the FTS monoclonal antibody, AD12.1, which detects a carbohydrate epitope on Wb-CFA, also detects multiple L. loa proteins in cross-reactive sera from persons with loiasis. Since the carbohydrate epitope recognized by AD12.1 is present on glycoproteins of other parasitic nematodes, including Brugia species, it is unclear why reactive glycoproteins are not detected in infections with other filarial parasites.
Methods: To gain a better understanding of the proteins recognized by the FTS diagnostic antibody, we used proteomics and lectin array technology to characterize filarial glycoproteins that are bound by the AD12.1 antibody using Brugia malayi as a model.
Results: Distinct but overlapping sets of AD12 glycoproteins were identified from somatic and excretory/secretory worm products. One of the identified proteins, Bm18019 was confirmed as a secreted AD12-reactive glycoprotein by in-gel proteomics and immunoassays. Based on lectin binding patterns, Brugia AD12-reactive glycoproteins express glycans including core fucose, galactose, N-acetylglucosamine and galactose(β1-3)N-acetylgalactosamine in addition to the epitope recognized by AD12.1. None of the lectins that bound B. malayi AD12 glycoproteins had affinity for the Wb-CFA, highlighting a key difference between it and other AD12 glycoproteins.
Conclusions: B. malayi somatic and excretory/secretory proteins are similar to L. loa antigens found in FTS-positive human sera, bolstering the hypothesis that circulating L. loa AD12 antigens result from worm tissue damage or death. The difference in glycan and protein composition between the Wb-CFA and other AD12 glycoproteins can be used to differentiate LF from cross-reactive loiasis.
Filarial parasites are insect-borne, tissue-dwelling, parasitic nematodes that cause debilitating diseases such as lymphatic filariasis (LF, caused by Wuchereria bancrofti, Brugia malayi and Brugia timori), onchocerciasis (by Onchocerca volvulus), and loiasis (by Loa loa). L. loa infection complicates disease elimination programs for LF by rendering false positive diagnostic tests, resulting in misdiagnoses (1–4), and by causing severe adverse reactions to treatments for onchocerciasis and LF (5).
Rapid diagnostic tests for Bancroftian filariasis (LF-RDT) recognize a carbohydrate epitope expressed on a high molecular weight circulating filarial antigen from W. bancrofti (Wb-CFA). We refer to this carbohydrate as the “AD12 epitope” (AD12e) based on its recognition by two monoclonal antibodies AD12.1 and DH6.5 (6). Based on AD12.1 binding to a glycan array, the AD12e likely contains a terminal β-d-glucuronic acid in a 1–3 linkage to other hexoses (7). AD12e is found on glycoproteins from other nematode species. However, in most other human filarial infections AD12e-containing proteins are not detected in circulation at appreciable levels. In vitro, AD12e glycoproteins can be detected in somatic and secreted preparations from different stages and sexes of B. malayi, L. loa and Onchocerca ochengi (6, 8, 9). Serum from baboons and immunocompromised mice experimentally infected with L. loa test positive by LF-RDT indicating the presence of circulating antigens (Loa CFAs) (8, 10). Lastly, LF-RDT cross-reactivity occurs naturally in a subset of loiasis infections and is associated with high microfilaria loads (1–3).
We have previously shown that cross-reactivity is due to the presence of multiple Loa CFAs (9). Understanding the nature of AD12e glycoproteins in non-Bancroftian filarial infections might prove key to developing diagnostic tests to remedy the specificity drawbacks of the current LF-RDT. We therefore catalogued AD12 glycoproteins from the model filarial organism B. malayi (Bm) and compared the surface glycans between brugian AD12 glycoproteins and Wb-CFA in order to identify differences between the Wb-CFA and other AD12 glycoproteins. The focus on B. malayi in this study stemmed from a lack of access to L. loa biological material. However, B. malayi has proven to be an apt model for L. loa in this case, as the AD12 glycoproteins found in B. malayi and L. loa soluble antigens share biochemical features that differ from the Wb-CFA. For example, they may contain additional N-glycans and are more susceptible to proteolytic cleavage than the Wb-CFA (7).
B. malayi worms were obtained from the Filariasis Research Reagent Resource Center (FR3, filariasiscenter.org). To prepare whole worm lysate (B. malayi somatic antigens, BmSOM), 30 adult male or female worms or 106 Microfilaria (Mf) were subjected to one freeze-thaw cycle, then incubated at 4°C overnight in RIPA lysis and extraction buffer (ThermoFisher) with ProteaseArrest protease inhibitors (G-biosciences). Lysates underwent mechanical disruption by pestle and then by shearing with a 25G needle with subsequent centrifugation (13500 g for 30 min). To collect excretory/secretory products (BmES), 60 adult male or female worms or 3 x 106 Mf were cultured for two weeks at 37°C/5% CO2 in RPMI 1640 media supplemented with L-glutamine, glucose, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 0.25 µg/mL amphotericin B. The spent media was collected on alternating days by passage through a 0.4 µm cell strainer to block collection of larvae and stored at -20°C. BmES products were concentrated to 1-1.5 mL by centrifugal filtration using an Amicon Ultra 3000 MWCO filter (Millipore). In this report, BmES is derived from adult female worms unless otherwise specified. L. loa adult worms were a gift from Dr. Vida Dennis, Tulane University. L. loa soluble antigen was prepared by grinding adult worms in extraction buffer (10 mM Tris pH 8.3, 2% sodium deoxycholate, 1 mM PMSF, 1 mM EDTA, 1 mM EGTA, 25 µg/mL TLCK protease inhibitor, 15 ug/mL TPCK protease inhibitor) and collecting the soluble fraction. Onchocerca volvulus, Acanthocheilonema viteae, Ascaris suum, and Paragonimus westernmani lysates were a gift from P. Fischer and G. Weil at Washington University in St. Louis. Caenorhabditis elegans N2 worm lysates were a kind gift from Z. Pincus at Washington University in Saint Louis. Human sera from persons with Bancroftian LF was sourced from banked, de-identified human samples from prior studies (11, 12).
DH6.5 and AD12.1 are monoclonal mouse IgM antibodies, initially generated against Dirofilaria immitis antigens, which both recognize the same carbohydrate epitope (6). A mouse IgM specific to lipopolysaccharide (clone 11E10, Southern Biotech) served as an isotype control. To generate antibodies against the B. malayi protein Bm18019, we immunized rabbits with the synthetic peptide c-DFSTNYDIRTESEWNSYAK (LifeTein) per a standard 70-day immunization protocol, and serum was affinity-purified to produce polyclonal immunoglobulin (Ig) concentrates (LifeTein). Horseradish peroxidase was conjugated to the AD12.1 antibody (AD12-HRP) and used at a concentration of 0.3 µg/mL.
Monoclonal antibody DH6.5, mouse IgM isotype control, anti-Bm18019 polyclonal Ig or normal rabbit Ig (RbN) control antibodies were conjugated to Affi-gel 10 beads (Bio-Rad) according to manufacturer’s protocol. Conjugated beads were stored as a 50% slurry in PBS. Conjugated beads (100 µL) were mixed with either (i) human sera (as a source of Wb-CFA), (ii) 80 µg of BmSOM, or (iii) 80 µg of BmES in 1 ml total volume then rocked overnight at 4°C. For human sera and lysate, beads were washed four times with cold PBS. For BmES, more stringent wash conditions were required due to the lack of detergent during BmES preparation. BmES product was washed four times with cold PBS, four times with high salt detergent buffer, MIB-T [5 mM HEPES, 1 M NaCl, 1 mM EDTA, 1 mM EGTA, 10 mM NaF, 2.4 mM NaVO4, 0.5% Triton-X-100 and Pierce protease inhibitor cocktail mini tablet (ThermoFisher)], followed by two washes in cold PBS to remove the detergent. After washing, antigen-coated beads were suspended in 1X NuPAGE LDS sample buffer (Invitrogen) and incubated at 95°C for five minutes to release bound antigens.
For Bm18019 experiments, immunoprecipitation of five µg BmES with 50 µL anti-Bm18019-bead conjugate were incubated overnight at 4°C followed by washing in MIB-T buffer and elution in 1X LDS sample buffer as written above. Immunoprecipitates were assayed by western blot and/or submitted for proteomics analysis as described below. Reagents for immunoprecipitation, immunoblot and ELISA experiments were purchased from Sigma-Aldrich, unless otherwise noted.
Proteins were boiled in 1X NuPAGE LDS reducing sample buffer then resolved by SDS-PAGE using a 4-12% bis-tris NuPAGE gel (Invitrogen). Silver staining of gels was performed with the FOCUS FAST silver staining kit (G-Biosciences) according to the manufacturer’s instructions. For western blots, proteins were transferred to nitrocellulose membranes (Amersham), incubated with blocking buffer [5% non-fat dry milk (Nestle Carnation) in phosphate buffered saline with 0.05% Tween-20 (PBS-T)] for one hour, then incubated with AD12-HRP (1:3000 dilution in blocking buffer) for one hour at room temperature. Membranes were washed three times in PBS-T and incubated with Clarity Western ECL substrate (Bio-Rad) for chemiluminescence detection using a ChemiDoc imager and Image Lab 5.2.1 software (Bio-Rad) or in some cases as specified, developed with the colorimetric substrate CN/DAB (3,3’-Diaminobenzidine, Pierce).
Immunoprecipitation samples were prepared for LC-ESI-MS/MS using a Q-Exactive Plus Hybrid Quadrupole-Orbitrap Plus mass spectrometer (ThermoFisher Scientific) coupled to an EASY-nanoLC 1000 system (ThermoFisher Scientific) as described previously (9). The resulting MS spectra were converted to Mascot generic format (MGF) files using Proteome Discoverer v2.1.0.81. MGF files were submitted for peptide identification based on the B. malayi genome, Bmal 4.0 (13) [downloaded from WormBase ParaSite (parasite.wormbase.org)]. Peptide hits were cross-referenced against contaminant databases: ENSEMBL for Human contaminants (Homo_sapiens.GRCh37.72 ENHU, version June 2016) and the cRAP database version (2012.01.01) for common contaminating peptides (http://www.thegpm.org/crap/). Qualifying peptides had a less than 1% false discovery rate and were absent from the combined human and cRAP databases. Proteins were considered true hits if more than one unique peptide was detected by mass spectrometry.
For in-gel proteomics, gel bands were cut into four 1 x 1 mm plugs and transferred to a 96 well plate. Gel plugs were dehydrated by the addition of acetonitrile (100 µL) to each well and incubated at room temperature for 10 min on a nutator. The supernatants were removed, the dehydration step was repeated, and the gel plugs were air dried for 30 min. Disulfide bonds were reduced with 50 µL of 10 mM DTT in 100 mM Seppro ammonium bicarbonate buffer (ABC buffer, Millipore), pH 8.0 for 30 min at 56°C, followed by alkylation with 50 µL of 55 mM IAM in 100 mM ABC buffer for 20 min at room temperature in the dark. Plugs were dehydrated twice with acetonitrile as before. Gel plugs were rehydrated in digestion buffer containing either chymotrypsin (0.025 µg/µL in LC-MS-grade water) or trypsin (0.025 µg/µL in 100 mM ABC buffer, pH 8.0) and incubated for 1 hour at room temperature on the nutator. An additional 0.25 µg of digest enzyme was added and the sample was incubated overnight. Peptides were extracted twice from the plugs by incubating with 20 µL of 50% MeCN with 5% (vol/vol) formic acid in water at 37°C for 30 min. Peptides were analyzed according to the solution-based proteomics described above.
Blast2Go software was used to assign Gene Ontology (GO) terms to the B. malayi immunoprecipitation (IP) combined dataset and to the B. malayi proteome to build a reference database (14). NetOGlyc 4.0 was used to predict O-glycosylation sites in the hits from the mass spec analysis (15). These results were compared with the O-glycosylation prediction of 500 randomly selected proteins from the B. malayi genome to estimate enrichment of O-glycoproteins in the IP and p-values were calculated by binomial distribution. The same parameters were used to predict N-glycosylation sites with NetNGlyc 1.0 (16).
A 40-Lectin array (GA-Lectin-40, RayBiotech) was used according to the manufacturer’s standard protocol. The array was probed with BmSOM 2 µg/µl, BmES 117 ng/µl, or DH6.5 immunoaffinity purified Wb-CFA from 2 mL pooled serum eluted from beads by heating at 100°C in PBS for 5 minutes (11). Cy-3 was directly conjugated to the AD12.1 antibody using the Cy3® Conjugation Kit (Fast) - Lightning-Link® (Abcam) according to the manufacturer’s protocol. Cy3 conjugated antibody was used at a 2 µg/mL dilution as a detection reagent. The array slide was shipped to RayBiotech and scanned using the GenePix optical scanner.
Detection of Wb-CFA by sandwich ELISA was performed as previously described (6, 9). For antigen capture using lectins, wells were similarly coated overnight at 37°C with 20 µg/mL of one of the following lectins in 0.6 M carbonate buffer, pH 9.6: Jacalin, wheat germ agglutinin (WGA), peanut agglutinin (PNA), Aleuria aurantia lectin (AAL, Vector Laboratories), Dolichos biflorus agglutinin (DBA, GlycoMatrix), Artocarpus integrifolia lectin (AIA, GlycoMatrix), and Psophocarpus tetragonolobus lectin II (PTL, GlycoMatrix). After coating, wells were washed with phosphate buffered saline (PBS) containing 0.05% Tween-20 (PBS-T) then blocked with 200 µL 5% fetal calf serum in PBS-T for one hour at 37°C. BmSOM or human sera [heated at 56°C for 30 min and EDTA treated to dissociate immune complexes as previously described (17)] was then added and incubated for two hours at 37°C. Wells were washed three times with PBS-T and incubated with AD12-HRP for one hour at 37°C. After a final round of PBS-T washes, 100 µL O-phenylenediamine dihydrochloride (OPD, ThermoScientifc) substrate was added and allowed to develop for ten minutes. The reaction was stopped by adding 30 µL of 4 M sulfuric acid, and absorbance was read at 492 nm. For sequential depletion of antigens, this standard format was modified by adding 0.5 µg/mL BmSOM to lectin-coated wells, incubating overnight at 37°C, then transferring the supernatant from each well to a another lectin-coated capture well for a second overnight incubation at 37°C. Finally, the supernatant from the second incubation was added to a DH6.5-coated well and incubated for one hour at 37°C to capture any remaining antigen. Captured antigen in all wells was detected using AD12-HRP as described above.
To detect Bm18019 in BmES, wells were coated overnight at 37°C with 20 µg/mL of anti-Bm18019 polyclonal Ig in 0.6 M carbonate buffer, pH 9.6. Wells were washed and blocked then incubated for one hour at 37°C with 0-2 µg of BmES. Captured antigen was detected using AD12-HRP as described above.
PNGase F. Four micrograms of BmES were digested with PNGase F (New England Biolabs) under denaturing conditions for one hour at 37°C following the manufacturer’s protocol. Two micrograms of RNase B were used in a positive control assay to confirm enzyme activity (New England Biolabs). OSGE. Lyophilized O-sialoglycoprotein endopeptidase purified from Mannheimia haemolytica (OSGE, CedarLane Labs) was reconstituted to 2.4 mg/mL in deionized water. Two micrograms of BmES were cleaved with 3 µl of OSGE at 37°C for 0-4 hours. The reaction was stopped by boiling at 100°C in 1X NuPAGE LDS sample buffer (Invitrogen). Digests were analyzed by immunoblot or silver stain.
It is well established that sera from persons with Bancroftian filariasis have a single circulating filarial antigen, the Wb-CFA, which is ~250 kDa in size and is abundantly decorated with the AD12e glycan epitope (Figure 1A). By western blot, AD12e was detected on multiple antigens of varying molecular weights in extracts from several filarial and non-filarial nematodes, but not in the trematode Paragonimus westermani (Figure 1B). A faint AD12e signal was observed with extract from the free-living nematode, Caenorhabditis elegans; when a higher concentration of C. elegans lysate was analyzed, three distinct bands at 50 kDa, 100 kDa and greater than 250kDa were detected (Figure S1). A silver stain of the protein extracts served as a loading control (Figure 1C).
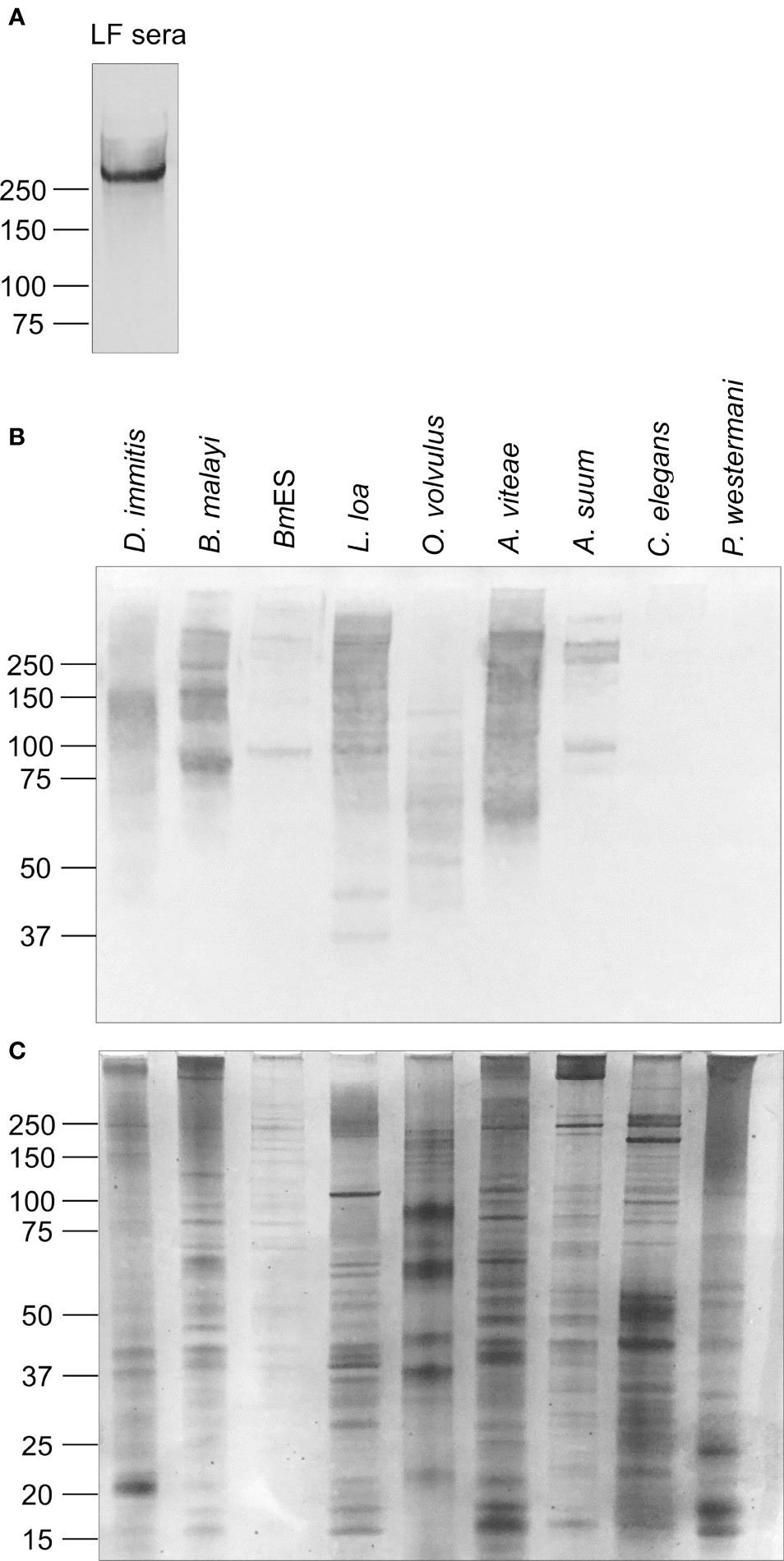
Figure 1 The glycan epitope detected by monoclonal antibody AD12.1 is present on multiple nematode proteins. (A) Pooled sera from W. bancrofti-infected patients in Sri Lanka were incubated overnight with Affi-Gel 10 beads conjugated to monoclonal antibody DH6.5. Bound Wb-CFA was eluted and detected by AD12-HRP by western blot. One microgram of each of the following antigen preparations: Dirofilaria immitis, Brugia malayi total lysate, Brugia malayi secreted antigens (BmES), soluble antigens from Loa Onchocerca volvulus, Acanthocheilonema viteae, Ascaris suum, Caenorhabditis elegans and Paragonimus westermani were analyzed by (B) CN/DAB colorimetric western blot using AD12.1 antibody or (C) silver stain.
The function of the AD12e glycan and glycoproteins that contain it are unknown. To gain insight into the types of proteins containing AD12e, we cataloged the AD12e-containing glycoproteins in adult female somatic (BmSOM) or excretory/secretory (BmES) product from B. malayi. Proteins were immunoaffinity purified with DH6.5 or a mouse IgM isotype control and analyzed by liquid chromatograph-tandem mass spectrometry. Spectra from BmSOM matched to 1784 peptides in 226 B. malayi proteins, after excluding proteins also detected in the isotype control. A similar analysis of BmES products yielded 560 unique peptides assigned to 80 proteins, 31 of which were not detected in BmSOM. Table 1 lists the most abundant proteins from each sample; a complete list of proteins detected is available in Table S1. Two hundred and seven (79.3%) of the 261 combined BmSOM and BmES protein hits were predicted by NetOGlyc 4.0 (18) to have O-linked glycosylation sites (O-glycosite, Table 1), compared to 73.9% (95% CI: 70% – 78%) of a random set of 500 B. malayi proteins. NetNGlyc 1.0 software (19) predicted that 138 (52.9%) of the proteins we identified contained N-linked glycosylation sites (N-glycosites) compared to 23.6% (95% CI: 20% - 28%) of the 500 randomly selected B. malayi proteins. Thus, the immunoprecipitation enriched for both O- and N- linked glycoproteins (respective p-value based on binomial distribution are 0.008 and 1.4 x 10-24). The list of brugian AD12 glycoproteins was compared to L. loa circulating antigens (Loa CFAs) to attempt to define the origin of RDT positive antigens in loiasis. When the Brugian proteins were compared to Loa CFAs, 88 direct homologs of the 233 Loa CFAs (38%) were detected in B. malayi (Figure 2). Thus, the filarial parasites have overlapping repertoires of AD12 glycoproteins and Loa CFAs include both somatic and secreted antigens.
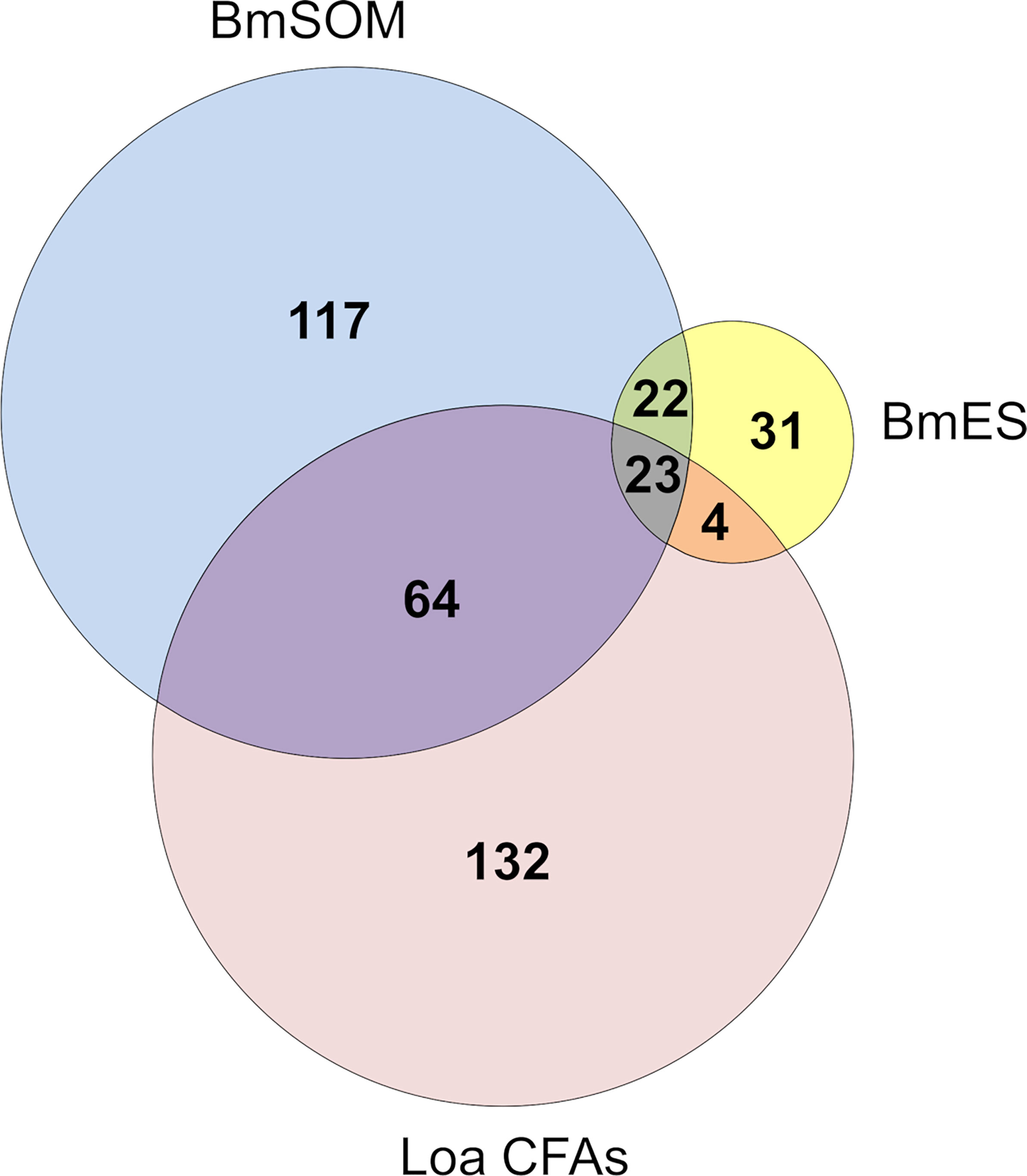
Figure 2 Mass spectrometry results. Two hundred and fifty four proteins were identified from DH6.5 immunoaffinity purifications of Brugian protein preparations (222 from BmSOM and 77 from BmES). The most abundant hits from each category are detailed in Table 1 with a complete list found in Table S1. The 223 L. loa proteins from cross-reactive loiasis sera (Loa CFAs) were previously published (9).
Gene ontology (GO) enrichment analysis of the combined BmSOM and BmES hits, compared to the B. malayi genome (13) indicates that Brugian AD12 glycoproteins are enriched in cytoplasmic, non-membrane bound organelle, and cytoskeletal compartments; oxidoreductase and hydrolase activity; and in reproductive and early developmental processes (Tables S1, S2). Strikingly, all but one (nucleoside-triphosphatase activity) of the GO terms enriched in B. malayi were also enriched in the previous analysis on Loa CFAs (9) indicating a conserved function.
To identify a single model glycoprotein to study AD12 glycan structure and function, we excised two predominant AD12.1-reactive bands from BmES for proteomic analysis (Figure 3). Band 1 contained thirty-eight spectra and Band 2 contained thirteen spectra that were assigned to brugian peptides. Two or more unique peptides were detected from four proteins in Band 1 and three proteins in Band 2 (Table 2). One protein from Band 2 was selected for further analysis since it was the only detected protein that migrated at a larger than predicted molecular weight, suggesting it might be heavily glycosylated.
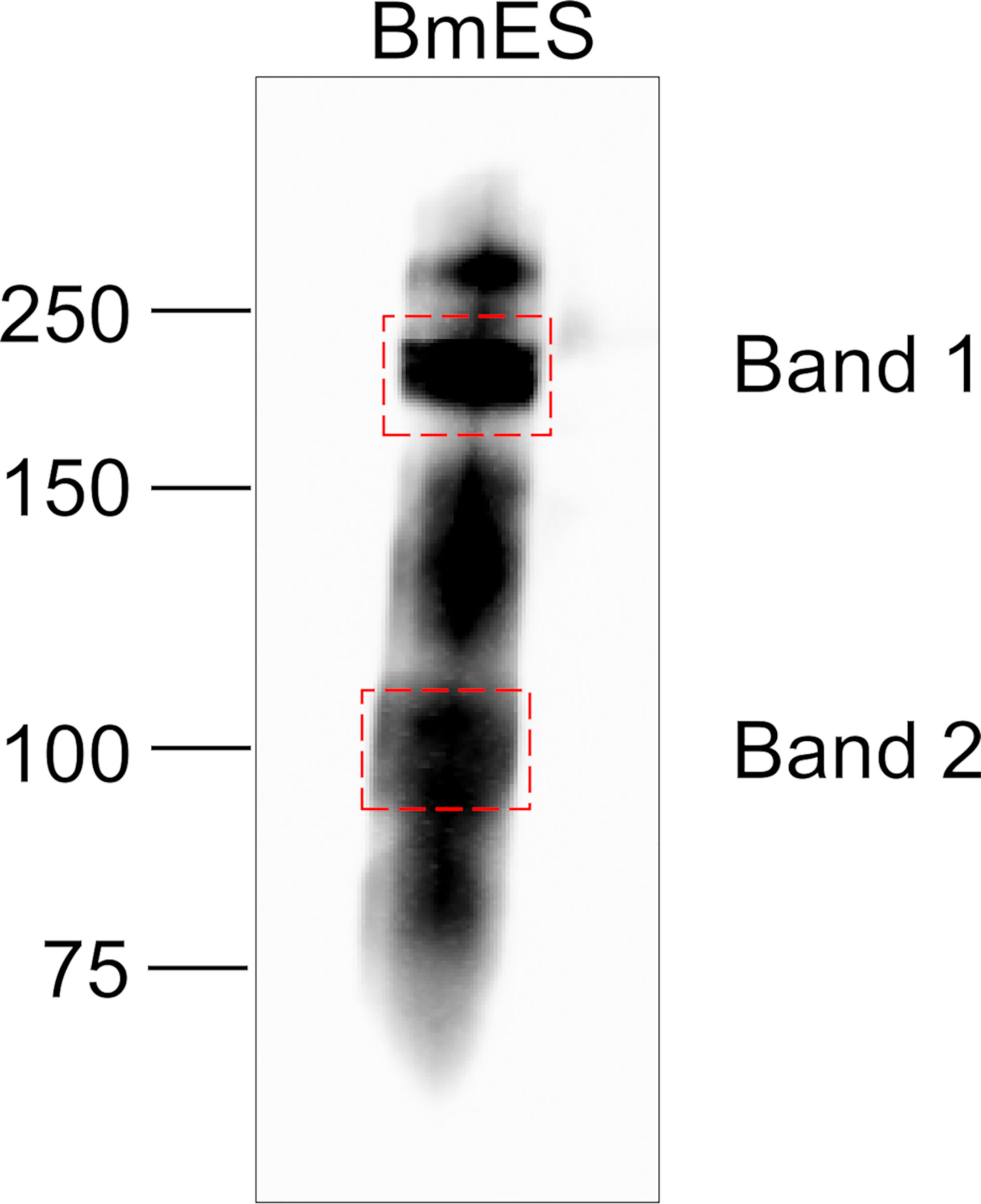
Figure 3 Gel slices analyzed by LC/MS/MS. 10 µg of BmES from adult female B. malayi were immunoprecipitated with DH6.5-conjugated Affi-gel beads. Bound proteins were eluted in SDS sample buffer, resolved using a 4-12% Bis-Tris gel and immunoblotted with AD12-HRP to identify regions of interest. Bands indicated by dashed boxes were excised for proteomic analysis.
The amino acid sequence of Bm18019 indicates that it is a mucin-like protein, and is primarily composed of eleven copies of a repeat element, rich in serine and threonine, of which 106 are in good context to serve as o-glycosylation sites (15). MS peptides mapped to the unique amino and carboxy termini (Figure S2).
Polyclonal immunoglobulin, anti-Bm18019, was generated against a peptide in the C-terminus and used to further characterize the protein. Bm18019 was detected as a 100 kDa band in adult female BmES with a faint signal in in BmSOM(Figure 4A). The molecular weight corroborated the proteomic results and suggested that the protein is heavily post-translationally modified. Interestingly, a band that migrates at the predicted size for the native protein (53 kDa) was strongly detected in Mf-ES and weakly detected in Mf-SOM (Figure 4A). Together, these data show that Bm18019 is a secreted protein with sex and stage specific expression.
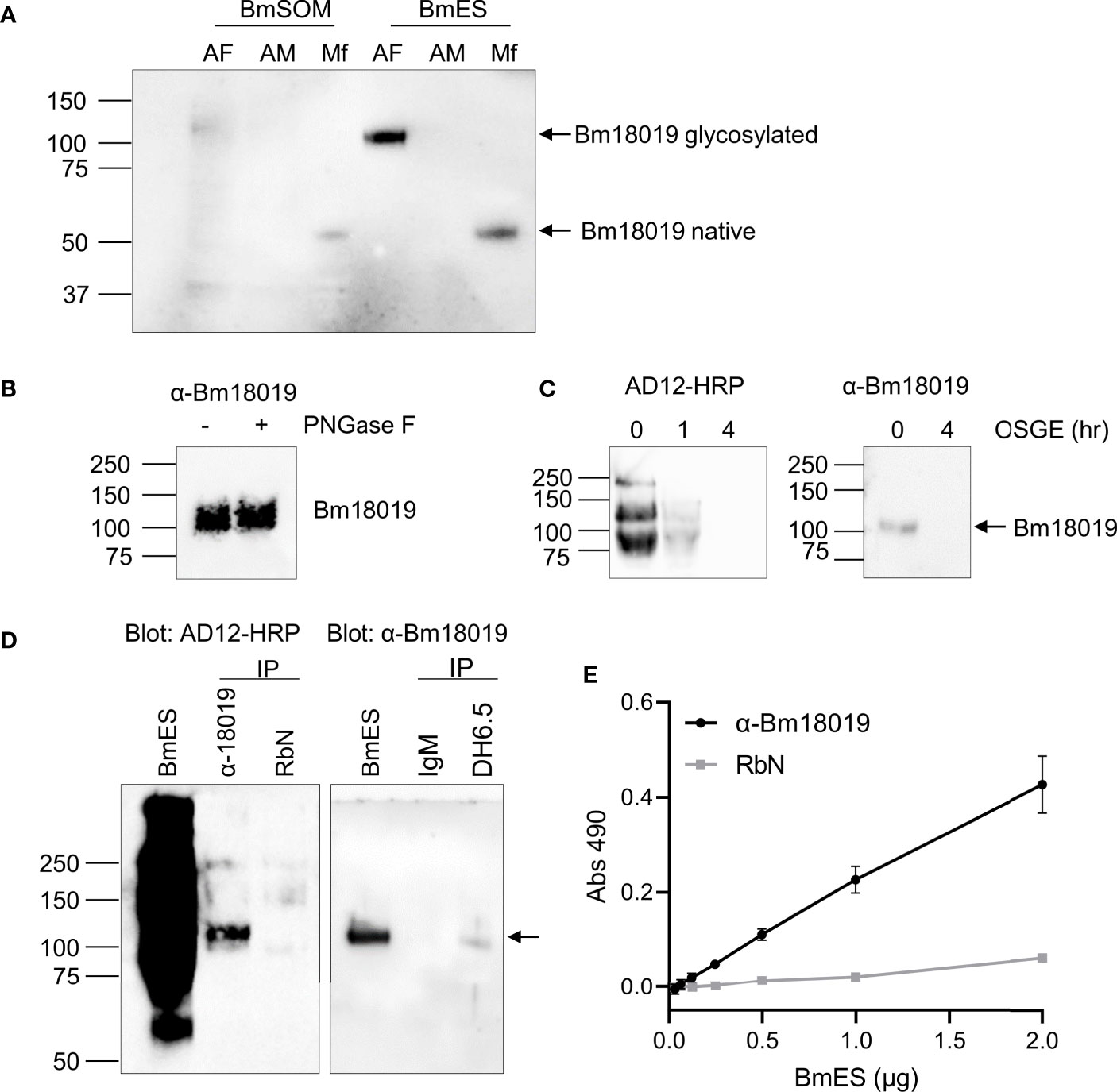
Figure 4 Characterization of Bm18019 expression and glycosylation. (A) Western blot of B. malayi antigen preparations probed with an anti-Bm18019 polyclonal Ig. Each lane contains 2 µg of protein purified from BmSOM or BmES from AF, adult female; AM, adult male; Mf, microfilaria. The predicted molecular weight of Bm18019 polypeptide is 53 kDa. (B) PNGase F digestion of 4 µg BmES were analyzed by western blot using anti-Bm18019. Activity of PNGase F was confirmed by a Coomassie stain of a control reaction using RNase B as a substrate (Figure S3). (C) Bm18019 is sensitive to the mucinase OSGE. 2µg of BmES was incubated with OSGE for 0-4 hours and then probed with AD12-HRP (left) or anti-Bm18019 (right). (D) Reciprocal immunoprecipitation confirms that Bm18019 is detectable by the AD12.1 antibody. Immunoprecipitation of 5 µg BmES with antibody conjugated Affi-gel beads were examined by western blot. An isotype matched control IP reaction was performed with normal rabbit sera (RbN) and mouse IgM-conjugated beads. (E) Sandwich ELISA demonstrates that native Bm18019 is concurrently bound by anti-Bm18019 and AD12.1. Plates were coated with anti-Bm1801or RbN control. A serial dilution of BmES was used as a source of antigen followed by detection with AD12-HRP.
To characterize the glycosylation properties of Bm18019, adult female BmES was treated with PNGase F, an enzyme that cleaves N-glycans, and O-sialoglycoprotein endopeptidase (OSGE), a mucinase. Bm18019 was not cleaved by PNGase F (Figure 4B). Enzymatic activity was validated by the positive control reaction with RNase B as a substrate (Figure S3). Like all AD12 glycoproteins in B. malayi, Bm18019 was completely degraded by OSGE after a 4-hour incubation (Figure 4C). Sensitivity to OSGE and resistance to PNGase F is characteristic of O-linked mucins.
To confirm the presence of the AD12 glycan epitope on Bm18019, reciprocal immunoprecipitation reactions were conducted (Figure 4D). Precipitation of BmES with the DH6.5 antibody contained Bm18019 and conversely, precipitation with anti-Bm18019 was positive in an AD12 immunoblot. A lack of signal when immunoprecipitation reactions were performed with isotype control antibodies validated the specificity of the assay. In an ELISA format, we were able to capture Bm18019 with the anti-peptide antibody, followed by detection with HRP-labeled AD12.1, suggesting that both AD12 and anti-Bm18019 could bind the intact native Bm18019 simultaneously (Figure 4E). Taken together these results confirm that Bm18019 expresses the AD12 reactive glycan on its surface and that at least some of the AD12.1 binding sites on Bm18019 are not masked by binding of the anti-peptide poly Ig. The OSGE sensitivity of all Brugian AD12 glycoproteins and this preliminary characterization of a specific, mucin-like AD12 glycan, supports the hypothesis that the AD12 glycan is O-linked and present on mucin-like glycoproteins.
To compare the surface glycans on the Wb-CFA and B. malayi AD12 glycoproteins and identify lectins that uniquely bound one or the other we performed a lectin array. Multiple lectins bound to AD12 glycoproteins from BmSOM and a subset bound to BmES (Figure 5A). The strongest binders were Aleuria aurantia lectin (AAL, which binds fucose), Dolichos biflorus agglutinin (DBA, binds α-linked N-acetylgalactosamine, αGalNAc) and Psophocarpus tetragonolobus lectin I (PTL, binds N-acetylgalactosamine, GalNAc). The raw lectin array data are available in Table S3. The Wb-CFA did not interact strongly with any lectin on the array. The absence of lectin binding to the Wb-CFA agrees with prior work which noted a lack of binding by concavalin A, soybean lectin, wheat germ agglutinin, and Ricin II and III lectins to Wb-CFA (6). The signal intensity in the ‘no antigen’ control sample indicates that lectin binding to glycans present on the detection antibody was negligible.
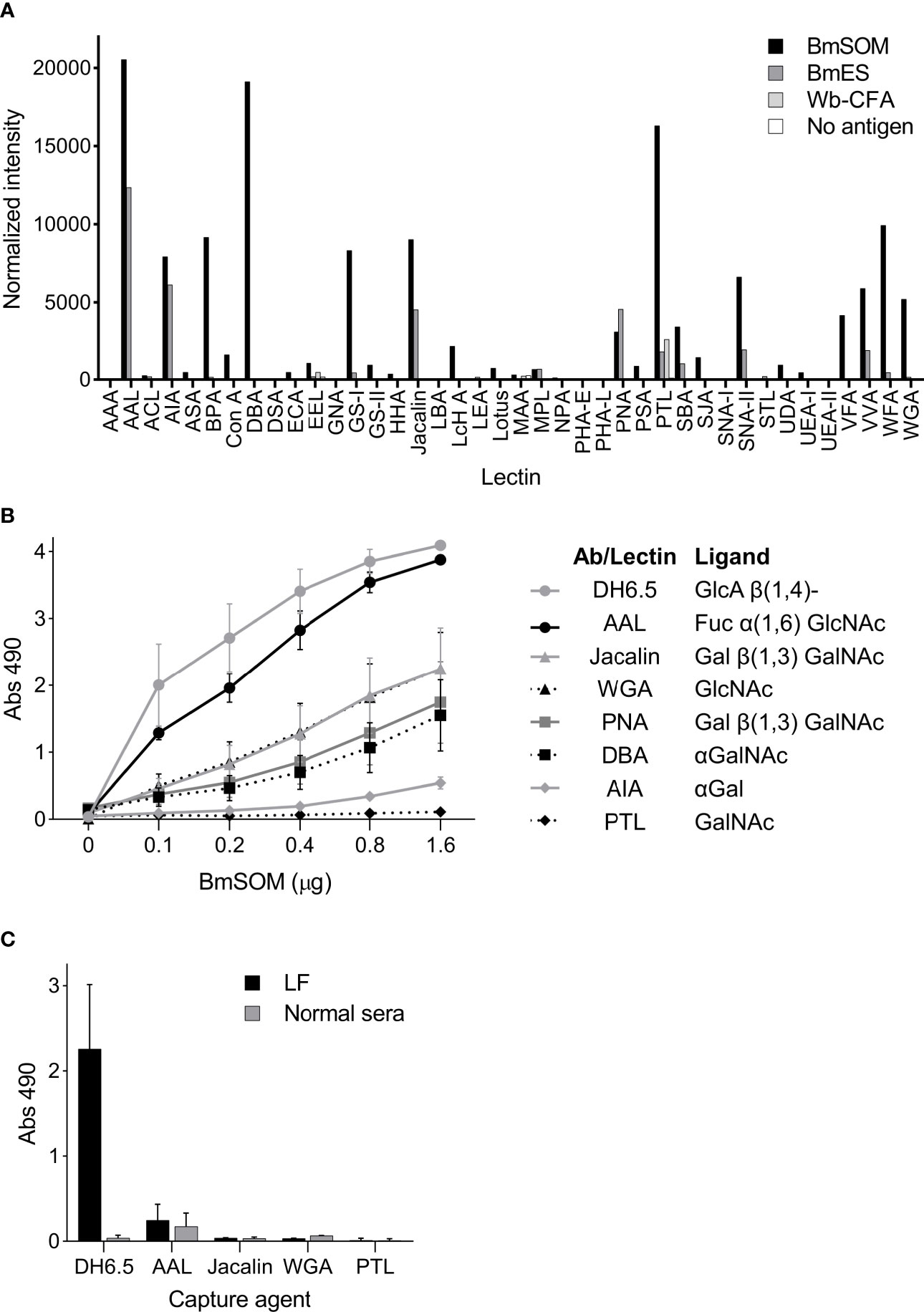
Figure 5 Lectins bind to AD12 glycoproteins from B. malayi but not to the Wb-CFA (A) The RayBiotech 40 lectin array was incubated with 2 µg/µl BmSOM, 117 ng/µl BmES, Wb-CFA immunoprecipitated from human sera, or an antigen free control. Captured glycoproteins were detected with a Cy-3 conjugated AD12.1 antibody. Normalized signal intensity was averaged from duplicate wells. (B, C) Lectin capture assay. (B) A two-fold serial dilution of BmSOM was incubated in wells individually coated with DH6.5 or lectins. Bound glycoproteins were detected with AD12-HRP. Error bars represent standard deviation for three independent experiments performed in triplicate. (C) Immobilized DH6.5 or lectins were incubated with human sera pooled from 26 LF subjects (Black bars) or 12 nonendemic subjects (gray bars). Bound antigen was detected with AD12-HRP. Error bars represent standard deviation for two independent experiments performed in triplicate. AAL, aleuria aurantia lectin; WGA, wheat germ agglutinin; PNA, peanut agglutinin; DBA, dolichos biflorus agglutinin; AIA, artocarpus integrifolia lectin; PTL, psophocarpus tetragonolobus lectin II. Fuc, fucose; GlcNAc, N-acetylglucosamine; Gal, galactose; GalNAc, N-acetylgalactosamine.
Using an in-house ELISA assay we confirmed that six of the seven most active array lectins bound to AD12e-containing proteins in BmSOM and none bound to the Wb-CFA (Figures 5B, C). PTL was inactive in the in-house ELISA. Thus, based on the lectin binding profiles, AD12 glycoproteins in BmSOM may contain fucose(α1-6)GlcNAc or fucose(α1-6)LacNAc, T-antigen (Galβ1-3GalNAc), GlcNAc, and αGalNAc either on the same glycan chain as the AD12e or at an alternate glycosite. For Wb-CFA, such moieties are either lacking, or are masked.
Having failed to identify a lectin that could selectively capture Wb-CFA, we next assessed whether the BmSOM-reactive lectins could functionally deplete cross-reactive AD12 glycoproteins. Theoretically, lectin-depletion of cross-reactive antigens might improve the specificity of LF diagnostic tests. We therefore developed a depletion ELISA, which involved preabsorbing with a lectin and then measuring the amount of remaining glycoprotein by capture with the DH6.5 antibody. Figure 6A outlines the procedure using two hypothetical proteins, one which binds to the immobilized lectin (black) and one which does not (gray). The total amount of AD12 glycoproteins captured at each step was determined by detection with AD12-HRP. Negative controls with no absorbing agent or with PTL lectin, which in our assay did not recognize AD12e-containing proteins, illustrates the amount of antigen that remained by the end of the experiment and accounts for sample loss during transfer steps and minimal spurious binding to the plate surface. This strategy was validated by demonstrating that complete depletion was achieved in the primary capture event with DH6.5 as the absorbing agent (Figure 6B). Lectins AAL, Jacalin and WGA were able to absorb AD12e-containing proteins from BmSOM. A 24-hour incubation was sufficient to capture all available ligands per lectin as illustrated by a lack of signal in the secondary capture wells (white bars, Figure 6B). However, lectin depletion did not reduce the total signal in the final antibody capture. Combining Jacalin and WGA lectins did not increase the efficiency. Thus, preabsorption with these lectins was not robust enough to deplete the total pool of AD12 glycoproteins. Interestingly, with AAL, the total amount of AD12e bound, combined with the amount unbound, exceeded the amount of AD12e bound by the DH6.5 positive control. This likely reflects competition for AD12e binding between DH6.5 (the capture antibody) and AD12.1 (the detecting antibody), which both bind AD12e. Capture by DH6.5 will block some AD12e binding sites, whereas capture by a lectin may leave all AD12e sites unblocked and available for AD12.1 detection.
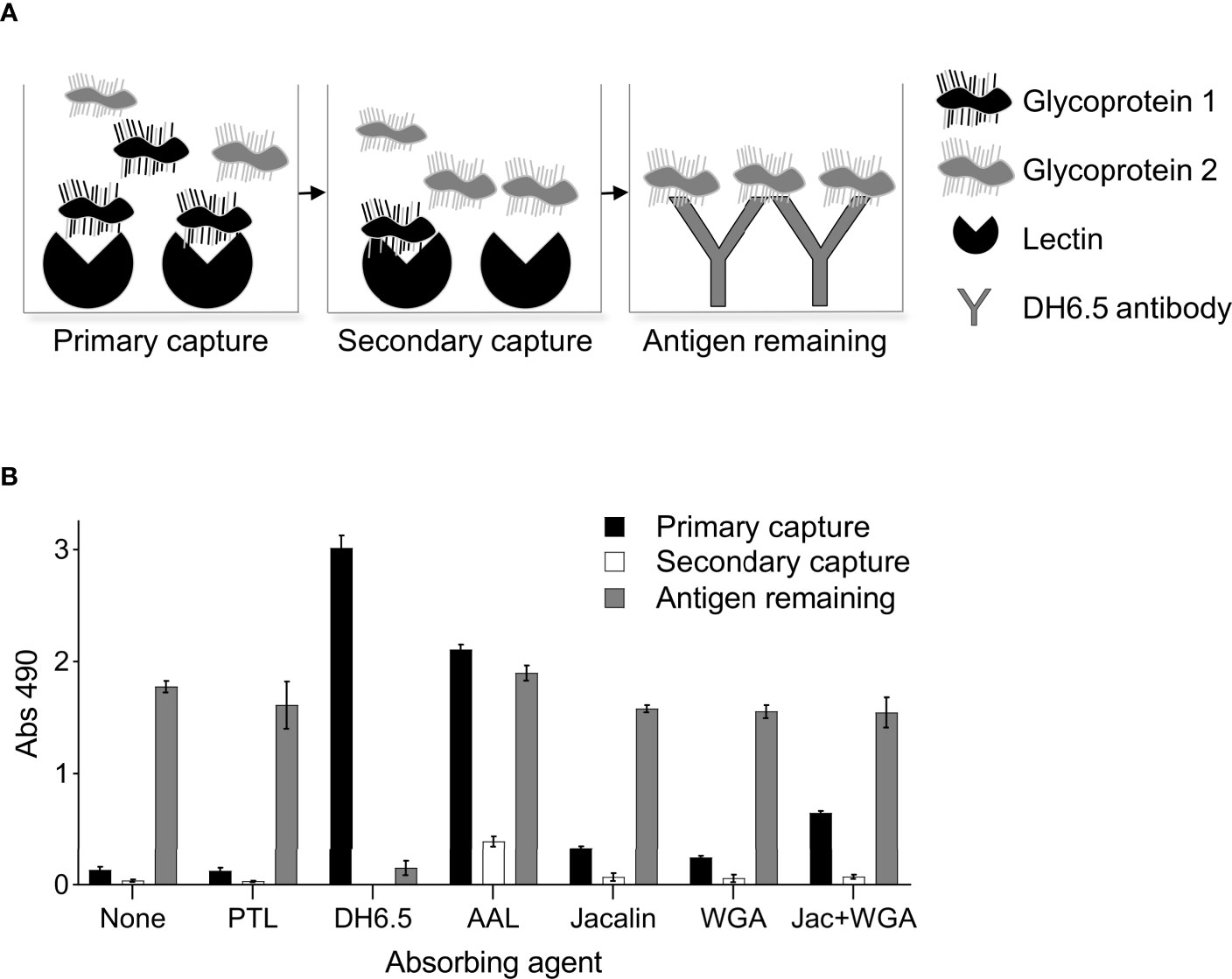
Figure 6 Lectin depletion of AD12 glycoproteins. (A) Experimental setup illustrated by a hypothetical example with two AD12 glycoproteins, one of which is depleted by the lectin (black) and the other is not (gray). 50 ng of BmSOM (at 0.5µg/mL) was incubated for 24 hours with either DH6.5 or various lectins (primary capture). The unbound fraction was incubated in a second well containing the same lectin as in the primary capture to assess depletion by the absorbing agent (secondary capture). Finally, the unbound fraction was incubated with DH6.5 to measure the remaining AD12 glycoproteins in the sample (antigen remaining). (B) Experimental results with BmSOM. Results displayed are an average calculated from two independent experiments performed in triplicate. Error bars depict standard deviation from the mean. PTL, Psophocarpus tetragonolobus lectin II; AAL, Aleuria aurantia lectin; WGA, wheat germ agglutinin.
Improvement of diagnostic tests for filarial infections is a high priority for the elimination of neglected tropical diseases. We report here some of our observations about the epitope AD12e, which is the target of current diagnostic tests for Bancroftian filariasis. Our key observations include: (1) AD12.1-reactive glycans are produced by all filarial worms tested to date, and by distantly related nematodes including Ascaris suum and C. elegans. (2) In B. malayi, AD12.1-reactive epitopes are on both somatic and ES glycoproteins, as has been previously shown for L. loa (9). (3) B. malayi AD12e glycoproteins, like those of L. loa, and unlike Wb-CFA, are susceptible to OSGE degradation. This includes a 100 kDa glycoprotein, Bm18019, with mucin-like characteristics. (4), In contrast to Wb-CFA, many B. malayi AD12e glycoproteins contain glycans recognized by commercially available lectins. However, no single lectin appears to capture all AD12e glycoproteins from B. malayi, suggesting that the lectins tested are binding to non-AD12 glycans that co-decorate AD12e glycoproteins.
This report further establishes the AD12 epitope as a common glycan motif on nematode glycoproteins and the number of proteins decorated with AD12e appears to have expanded in filarial worms compared to worms with free-living stages. The role AD12e plays in filarial biology remains an open question but there is evidence that it is involved in reproduction and development. Previous studies demonstrated that adult female worms are the main producers of AD12e which is localized to the reproductive tissues (6). Our work has shown that proteins that contain AD12e are enriched in reproductive and development functional terms.
This study also characterized a specific AD12 glycoprotein, Bm18019. Bm18019 had sex and stage specific expression pattern consistent with earlier transcriptomic studies in B. malayi. Transcription profiling studies categorized Bm1_15985 (an earlier identification number for Bm18019), as a differentially expressed gene clustered with other genes involved in reproduction and highly expressed in adult female worms and eggs. The gene was also transcribed to a lesser extent in adult males and immature worms but not detected in the Mf to L4 stages (18, 20). Our results showing positive protein expression in Mf contradict these transcription analyses. It is possible that the gene expression was missed in the transcriptome microarrays used in the earlier studies. Alternatively, Bm18019 may be produced by the adult female worm and remain associated with the Mf. Lastly, off target detection by the novel antibody generated in this study cannot be ruled out. Proteomic evidence for Bm1_15985 is scarce; it was not detected in BmES or BmSOM (19, 21–23). However, Bm1_15985 was detected when proteomic analysis was performed on three separated body compartments (body wall, reproductive tract, and digestive tract) (24). It is possible that o-glycosylation masked peptide detection in the earlier proteomic studies of complex mixtures. Interestingly, orthologues to the Bm18019 gene have only been identified in the parasites that cause lymphatic filariasis: Brugia pahangi (BPAG_0000454501), and W. bancrofti (WBA_0000973501). The Brugia timori draft genome encodes a partial sequence with perfect homology to the last 72 amino acids of Bm18019 (BTMF_scaffold0002080 6441 to 6656). No homologous genes were identified in other organisms and the possibility for a unique role in the LF niche remains to be explored.
One limitation of this study was that the comparison was carried out with B. malayi instead of L. loa due to limited access to L. loa worm products. A lack of laboratory models and molecular reagents is a common challenge in filariasis research. The parasites cannot be cultured ex vivo and the only human filarial parasites whose lifecycle can be reproduced in the laboratory using rodent hosts are B. malayi and only recently L. loa (10). However, B. malayi is an appropriate model for understanding the AD12 glycoproteins in L. loa. The two organisms have a similar AD12 antigen profile by western blot and there was a high level of overlap between the AD12 glycoproteins in cross-reactive loiasis serum antigens and BmSOM. A second limitation is that the immunoprecipitation wash conditions prior to proteomics differed between BmSOM and BmES. The BmSOM was washed identical to the LoaCFAs (9), and it is possible that the BmSOM proteomics may have more false positives due to less stringent wash conditions compared to BmES, however, false positives were identified by the isotype control immunoprecipitation which used the same wash conditions.
Given the ubiquitous nature of AD12e in multiple parasite species, and the fact that a number of filarial worms including L. loa, B. malayi and Mansonella species have blood stages, it is remarkable that the FTS is functionally specific for Bancroftian filariasis. This is explained in part due to the unique stability of the Wb-CFA protein compared to AD12-glycoproteins from Brugia and Loa (7). We speculate that the rare occurrence of FTS-positive antigenemia in loiasis is a result of stochastic worm damage or death as opposed to regulated secretion. The banding pattern of L. loa antigens resembles the serum profile in Bancroftian filariasis after microfilaricidal treatment further arguing that worm death is responsible in both cases (25).
In summary, our data highlight the unique nature of Wb-CFA, compared to AD12e glycoproteins of other nematodes. The stability and persistence of Wb-CFA in serum has enabled its use as a functionally specific marker of Bancroftian filariasis and in turn led to the development of RDTs that expedited elimination of LF from many endemic areas. Ongoing efforts to clearly identify Wb-CFA, the AD12e structure, and to elucidate differences between Wb-CFA and cross-reactive loiasis antigens should facilitate the development of improved diagnostic tests for filarial infections.
The original contributions presented in the study are included in the article/Supplementary Material. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027003. Further inquiries can be directed to the corresponding author.
PB provided funding; MH and PB designed and directed the project; MH, IH, and AR performed the experiments and analyzed the data; MH and PB wrote the manuscript with input from all authors. All authors contributed to the article and approved the submitted version.
The mass spectrometric experiments were designed and performed at the Washington University Proteomics Shared Resource (WU-PSR) supported in part by the WU Institute of Clinical and Translational Sciences (NCATS UL1 TR002345), the Mass Spectrometry Research Resource (NIGMS P41 GM103422) and the Siteman Comprehensive Cancer Center Support Grant (NCI P30 CA091842). We are appreciative of the guidance and advice from the members of the Townsend, Mitreva, and Weil labs. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number K08AI121422.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2021.729294/full#supplementary-material
Supplementary Table 1 | Complete list of AD12 epitope-containing proteins identified from B. malayi.
Supplementary Table 2 | GO term analysis.
Supplementary Table 3 | Raw lectin array data.
Supplementary Figure 1 | C. elegans expresses AD12 reactive glycoproteins.
Supplementary Figure 2 | Bm18019 is a putative mucin.
Supplementary Figure 3 | PNGase F control reaction.
1. Bakajika DK, Nigo MM, Lotsima JP, Masikini GA, Fischer K, Lloyd MM, et al. Filarial Antigenemia and Loa Loa Night Blood Microfilaremia in an Area Without Bancroftian Filariasis in the Democratic Republic of Congo. Am J Trop Med Hyg (2014) 91(6):1142–8. doi: 10.4269/ajtmh.14-0358
2. Wanji S, Amvongo-Adjia N, Koudou B, Njouendou AJ, Chounna Ndongmo PW, Kengne-Ouafo JA, et al. Cross-Reactivity of Filariais ICT Cards in Areas of Contrasting Endemicity of Loa Loa and Mansonella Perstans in Cameroon: Implications for Shrinking of the Lymphatic Filariasis Map in the Central African Region. PLoS neglected Trop Dis (2015) 9(11):e0004184. doi: 10.1371/journal.pntd.0004184
3. Pion SD, Montavon C, Chesnais CB, Kamgno J, Wanji S, Klion AD, et al. Positivity of Antigen Tests Used for Diagnosis of Lymphatic Filariasis in Individuals Without Wuchereria Bancrofti Infection But With High Loa Loa Microfilaremia. Am J Trop Med Hyg (2016) 95(6):1417–23. doi: 10.4269/ajtmh.16-0547
4. Nana-Djeunga HC, Fossuo-Thotchum F, Pion SD, Chesnais CB, Kubofcik J, Mackenzie CD, et al. Loa Loa Microfilariae in Skin Snips: Consequences for Onchocerciasis Monitoring and Evaluation in L. Loa-Endemic Areas. Clin Infect Dis (2019) 69(9):1628–30. doi: 10.1093/cid/ciz172
5. Gardon J, Gardon-Wendel N, Demanga N, Kamgno J, Chippaux JP, Boussinesq M. Serious Reactions After Mass Treatment of Onchocerciasis With Ivermectin in an Area Endemic for Loa Loa Infection. Lancet (1997) 350(9070):18–22. doi: 10.1016/S0140-6736(96)11094-1
6. Weil GJ, Liftis F. Identification and Partial Characterization of a Parasite Antigen in Sera From Humans Infected With Wuchereria Bancrofti. J Immunol (1987) 138(9):3035–41.
7. Hertz MI, Rush A, Nutman TB, Weil GJ, Bennuru S, Budge PJ. Characterization of Glycan Determinants That Mediate Recognition of the Major Wuchereria Bancrofti Circulating Antigen by Diagnostic Antibodies. Mol Biochem Parasitol (2020) 240:111317. doi: 10.1016/j.molbiopara.2020.111317
8. Wanji S, Amvongo-Adjia N, Njouendou AJ, Kengne-Ouafo JA, Ndongmo WP, Fombad FF, et al. Further Evidence of the Cross-Reactivity of the Binax NOW(R) Filariasis ICT Cards to Non-Wuchereria Bancrofti Filariae: Experimental Studies With Loa Loa and Onchocerca Ochengi. Parasit Vectors (2016) 9:267. doi: 10.1186/s13071-016-1556-8
9. Hertz MI, Nana-Djeunga H, Kamgno J, Jelil Njouendou A, Chawa Chunda V, Wanji S, et al. Identification and Characterization of Loa Loa Antigens Responsible for Cross-Reactivity With Rapid Diagnostic Tests for Lymphatic Filariasis. PLoS Negl Trop Dis (2018) 12(11):e0006963. doi: 10.1371/journal.pntd.0006963
10. Pionnier NP, Sjoberg H, Chunda VC, Fombad FF, Chounna PW, Njouendou AJ, et al. Mouse Models of Loa Loa. Nat Commun (2019) 10(1):1429. doi: 10.1038/s41467-019-09442-0
11. Ismail MM, Weil GJ, Jayasinghe KS, Premaratne UN, Abeyewickreme W, Rajaratnam HN, et al. Prolonged Clearance of Microfilaraemia in Patients With Bancroftian Filariasis After Multiple High Doses of Ivermectin or Diethylcarbamazine. Trans R Soc Trop Med Hyg (1996) 90(6):684–8. doi: 10.1016/S0035-9203(96)90437-X
12. Andersen BJ, Kumar J, Curtis K, Sanuku N, Satofan S, King CL, et al. Changes in Cytokine, Filarial Antigen, and DNA Levels Associated With Adverse Events Following Treatment of Lymphatic Filariasis. J Infect Dis (2018) 217(2):280–7. doi: 10.1093/infdis/jix578
13. Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, et al. Draft Genome of the Filarial Nematode Parasite Brugia Malayi. Science (2007) 317(5845):1756–60. doi: 10.1126/science.1145406
14. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics (2005) 21(18):3674–6. doi: 10.1093/bioinformatics/bti610
15. Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, et al. Precision Mapping of the Human O-Galnac Glycoproteome Through Simplecell Technology. EMBO J (2013) 32(10):1478–88. doi: 10.1038/emboj.2013.79
16. Gupta R, Brunak S. Prediction of Glycosylation Across the Human Proteome and the Correlation to Protein Function. Pac Symp Biocomput (2002) 310–22.
17. Weil GJ, Jain DC, Santhanam S, Malhotra A, Kumar H, Sethumadhavan KV, et al. A Monoclonal Antibody-Based Enzyme Immunoassay for Detecting Parasite Antigenemia in Bancroftian Filariasis. J Infect Dis (1987) 156(2):350–5. doi: 10.1093/infdis/156.2.350
18. Choi YJ, Ghedin E, Berriman M, McQuillan J, Holroyd N, Mayhew GF, et al. A Deep Sequencing Approach to Comparatively Analyze the Transcriptome of Lifecycle Stages of the Filarial Worm, Brugia Malayi. PLoS Negl Trop Dis (2011) 5(12):e1409. doi: 10.1371/journal.pntd.0001409
19. Moreno Y, Geary TG. Stage- and Gender-Specific Proteomic Analysis of Brugia Malayi Excretory-Secretory Products. PLoS Negl Trop Dis (2008) 2(10):e326. doi: 10.1371/journal.pntd.0000326
20. Li BW, Wang Z, Rush AC, Mitreva M, Weil GJ. Transcription Profiling Reveals Stage- and Function-Dependent Expression Patterns in the Filarial Nematode Brugia Malayi. BMC Genomics (2012) 13:184. doi: 10.1186/1471-2164-13-184
21. Hewitson JP, Harcus YM, Curwen RS, Dowle AA, Atmadja AK, Ashton PD, et al. The Secretome of the Filarial Parasite, Brugia Malayi: Proteomic Profile of Adult Excretory-Secretory Products. Mol Biochem Parasitol (2008) 160(1):8–21. doi: 10.1016/j.molbiopara.2008.02.007
22. Bennuru S, Semnani R, Meng Z, Ribeiro JM, Veenstra TD, Nutman TB. Brugia Malayi Excreted/Secreted Proteins at the Host/Parasite Interface: Stage- and Gender-Specific Proteomic Profiling. PLoS Negl Trop Dis (2009) 3(4):e410. doi: 10.1371/journal.pntd.0000410
23. Bennuru S, Meng Z, Ribeiro JM, Semnani RT, Ghedin E, Chan K, et al. Stage-Specific Proteomic Expression Patterns of the Human Filarial Parasite Brugia Malayi and Its Endosymbiont Wolbachia. Proc Natl Acad Sci USA (2011) 108(23):9649–54. doi: 10.1073/pnas.1011481108
24. Morris CP, Bennuru S, Kropp LE, Zweben JA, Meng Z, Taylor RT, et al. A Proteomic Analysis of the Body Wall, Digestive Tract, and Reproductive Tract of Brugia Malayi. PLoS Negl Trop Dis (2015) 9(9):e0004054. doi: 10.1371/journal.pntd.0004054
Keywords: lymphatic filariasis, glycoprotein, loiasis, rapid diagnostic assays, Brugia malayi
Citation: Hertz MI, Hamlin I, Rush A and Budge PJ (2021) Brugia malayi Glycoproteins Detected by the Filariasis Test Strip Antibody AD12.1. Front. Trop. Dis 2:729294. doi: 10.3389/fitd.2021.729294
Received: 22 June 2021; Accepted: 10 August 2021;
Published: 30 August 2021.
Edited by:
Cornelis Hendrik Hokke, Leiden University Medical Center, NetherlandsReviewed by:
Kenneth Pfarr, University Hospital Bonn, GermanyCopyright © 2021 Hertz, Hamlin, Rush and Budge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip J. Budge, cGJ1ZGdlQHd1c3RsLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.