
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Trop. Dis. , 16 July 2021
Sec. Major Tropical Diseases
Volume 2 - 2021 | https://doi.org/10.3389/fitd.2021.679501
This article is part of the Research Topic Highlights in Major Tropical Diseases 2021/22 View all 4 articles
Nontuberculous mycobacteria (NTM) are environmental opportunistic pathogens of humans and animals that are emerging with a serious public health impact particularly in individuals with Acquired Immunodeficiency Syndromes. Recent scientific evidence is shifting from NTMs being known as traditional environmental organisms to serious pathogenic organisms in both animals and humans. In humans, factors attributable to this rise have been linked mainly to Human Immunodeficiency Virus and Acquired Immunodeficiency Syndrome pandemic. In recent years there has been an increase in multidrug resistant Tuberculosis in Zambia and it is thought that NTMs could possibly be the cause. This study was therefore formulated to review available information on the prevalence of NTM in humans, animals and the environment, species distribution, zoonotic potential and public health importance in Zambia. This review was conducted in accordance with PRISMA guidelines. A literature search was done in PubMed and Google scholar using predefined search terms such as ‘nontuberculous mycobacteria’, ‘atypical mycobacteria’, ‘mycobacteria other than mycobacterium tuberculosis’ and ‘Zambia’, in combination with Boolean operators (AND, OR). This particular systematic review draws findings based on literature search between 2000 and 2020. Through literature search 243 papers were identified, 23 duplicates were identified and removed and 206 articles were excluded as they did not meet the inclusion criteria. The full text of the remaining 14 articles were considered for this review. The overall prevalence of NTM in humans was 24.39%, in water 21.5%, in animals 16.05% of which the prevalence in cattle was 14.81% and Kafue Lechwe 1.23%. Mycobacterium intracellulare was the most common isolated nontuberculous mycobacteria in humans and cattle while Mycobacterium gordonae was the most common in water, and Mycobacterium stomatepiae sp Nov in Kafue Lechwe. Nontuberculous mycobacteria are an emerging public health threat in Zambia both in humans and animals and this calls for the need for molecular information on the zoonotic transmission of nontuberculous mycobacteria. Increased awareness of nontuberculous mycobacteria diseases among clinicians and laboratory personnel is crucial for patient management and an essential step for facilitating the identification of nontuberculous mycobacteria species in laboratories.
Even though members of the Mycobacterium tuberculosis complex (MTC) are responsible for the bulk of the mycobacterial infections worldwide, opportunistic infections due to nontuberculous mycobacteria (NTM) are rapidly increasing, partially as a direct consequence of the immunosuppression due to the HIV/AIDS pandemic (1–3). Mycobacteria are aerobic organism that are non-motile (M. marinum is an exception), non-spore forming, gram-positive straight or slightly curved rods and have a waxy, relatively impermeable cell wall consisting of arabinogalactan and mycolic acid. Their cell wall has a high lipid content which once stained with carbol fuchsin cannot be decolorized by acid alcohol thus their name acid fast bacteria (AFB) (4, 5). The genus mycobacterium contains two major human pathogens: Mycobacterium tuberculosis (MTB) the major cause of tuberculosis (TB) and Mycobacterium leprae the agent causing leprosy. In addition to these two obligate human pathogens, the genus also comprises the nontuberculous mycobacteria (NTM) or Mycobacteria other than Mycobacterium tuberculosis (MOTT) (4, 6). NTMs include both slow growing mycobacteria (SGM) where colony formation requires at least seven days and rapid growing mycobacteria (RGM) forming colonies in less than seven days (1, 7–9).
Nontuberculous mycobacteria are normal inhabitants of a variety of environmental habitats that are shared between humans and animals including natural waters, drinking water distribution systems, biofilms, and soils (1, 10). It is documented that growth of NTMs is stimulated by humic and fulvic acids, which are the principal organic compounds in waters draining from forest soil and acid brown water swamps (11–13). Animals and humans can be infected by opportunistic mycobacteria from the environment via ingestion, inhalation, and dermal contact (1).
Nevertheless, a bit of controversy exists surrounding the real extent of possible human-to-human transmission of NTM. Although not fully elaborated, it is believed that human-to-human transmission is generally uncommon, even though there is evidence of transmission of certain NTM species such as Mycobacterium kansasii and Mycobacterium abscessus (4, 14, 15).
With advancement in molecular techniques including whole genome sequencing (WGS), 170 NTM species have been identified with more being added on a regular basis (1, 4, 16–18). Among these recognized NTM species, the most frequent human pathogens are the slow growing Mycobacterium avium complex (MAC), Mycobacterium kansasii and rapidly growing M. absessus complex. Other NTM infections such as M. fortuitum, M. xenopi, M. simiae, M. chelonae and szulgai are less common causes of NTM lung disease. These organisms are capable of causing pulmonary disease, disseminated disease, and localized lesions, among immunocompromised and immunocompetent individuals worldwide. Pulmonary diseases are the most frequently encountered and have been reported to account for up to 94% of cases of NTM associated disease (6, 17, 19–21).
Nontuberculous mycobacteria can exhibit clinical and radiographic features that are similar to those by MTBC (22) especially in persons with cellular immune dysfunction. The clinical signs and symptoms of both NTM and MTB are related to granulomatous inflammation which are quite similar and thus often go unrecognized and cannot be discriminated based on the common identification protocols. Thus NTM infections are easily misdiagnosed as MTB (23, 24). Therefore, there is need for early differential diagnosis of NTM pulmonary disease from pulmonary tuberculosis as the therapeutic regimen of pulmonary NTM differs from that of pulmonary TB. After the culture results there is need for biochemical test or molecular methods to differentiate NTM from MTB. Different molecular methods have been developed for species identification of NTM which include among others the sequencing of 16S rRNA, rpoB gene, hsp65 and 16-23 S rRNA (25).
The clinical relevance of isolated NTM is often unclear but differentiation of true infection from pseudo-infection and establishing the clinical relevance of an NTM isolate is of paramount importance since treatment of NTM disease is time consuming and often complicated (26). Unlike TB, the isolation of NTM in pulmonary specimens does not equate with disease (7). In an effort to standardize the definition of NTM infection, the American Thoracic Society (ATS)/Infectious Diseases Society of America (IDSA) ATS/IDSA formulated and published guidelines on definition of NTM pulmonary disease. On the basis of these guidelines, the diagnosis of NTM pulmonary infection requires the presence of clinical symptoms with appropriate exclusion of other diseases, radiologic abnormalities and microbiological (positive culture from at least two sputum samples, or positive culture from one bronchial wash or lavage (7). These criteria fit best with Mycobacterium avium complex (MAC), M. kansasii, and M. abscessus but researchers have applied it to the diagnosis of other NTM (7, 27). In the clinical laboratory the combination of microbiological and molecular techniques are used for the detection of NTM and this is essential as it enhances the health care of patients.
In most countries, NTM pulmonary disease (NTMPD) is not a reportable condition, such that describing the burden, trends and associated risk factors for this condition depends on special studies, surveys and sentinel surveillance efforts (28).
Epidemiological data globally has been showing an increasing trend in NTM, although this has been partially attributed to improvement of microbiological methodologies that have allowed better recovery and identification of NTM. It is also acceptable that there is a genuine increase in incidence augmentation in NTM cases. The reasons for the increase in the incidence and prevalence of NTM are multifactorial and depend on the nature of the host, the pathogen and the pathogen-host interaction (29).
The annual prevalence of NTM pulmonary disease varies across different regions. In North America and Australia, the prevalence ranges from 3.2–9.8 per 100,000, and is generally higher than in Europe. In the United States the prevalence of NTM rose from 2.4 cases/100000 in early 1980s to 15.2 cases/100000 in 2013 (30). Studies in Ontario, Canada demonstrated annual rates of NTM isolation of 14.1 – 22.2/100000 population (31, 32).
In Europe increasing trends of NTM infections have also been observed. England, Wales and Northern Ireland have shown an overall increase in both the number and rate of NTM infection from 0.91/100000 to 7.6/100000 by 2012. This data includes both pulmonary and extra pulmonary specimens and main increase was seen in people over 60 years old (32). In Serbia the annual rate of NTM isolation and NTM pulmonary disease incidence per 100000 changed over 2010 to 2015 from 0.9 to 1.6 and from 0.18 to 0.48 respectively. Both isolation and disease rates increased considerably with age (33). In Madrid Spain the figures are similar showing an increasing trend in NTM isolation from 930 isolates in 2013 to 1012 isolates in 2017 (34).
Asia which has a high prevalence of TB has very few studies on the epidemiology of NTM. However, despite the scarcity of studies done, a few survey data and organized studies have shown a continuous increase in the prevalence of NTM, with Japan having a high burden of NTM than TB (32). Studies of NTM pulmonary disease (NTM PD) from South Korea, Japan, and Taiwan, also suggest similar trends of increasing prevalence. In Korea the annual age – adjusted incidence and prevalence of NTM infection increased rapidly from 1.0 cases/100000 to 17.9 cases/100000 and 1.2/100000 in 2003 to 33.3/100000 population in 2016 respectively (35, 36). Similar trends have been observed in China where the proportion of NTM infections continue to rise from 15.6% in 2013 to 46.1% in 2018 (37).
In Africa and the Middle East, the prevalence of NTM ranges from 4% –15% among suspected TB cases and 18%–20% among suspected multi-drug resistant TB (MDR TB) cases (28, 38).
In sub-Saharan Africa according to systematic review done for the period between January 1, 1940 to October 1, 2016, the prevalence of pulmonary NTM colonization was calculated to be 7.5% (39).
In Zambia, NTMs are slowly becoming recognized as pathogens of major public health significance with the advent of Human Immunodeficiency Virus (HIV) and Acquired Immunodeficiency Syndrome (AIDS) (5).
Zambia is one of the high burden TB countries in the world and in recent years there has been as increase in multi drug resistant TB. Due to the high TB prevalence and increase in MDRTB, there is a concern that NTM infections could be misdiagnosed as TB (40). This is because TB and NTM results are identical microscopically and therefore all positive results are considered as TB and no further tests are carried out to differentiate the two. This study therefore, aims to review the available information on NTM in humans with suspected TB, animals and the environment so as to determine the prevalence, species distribution and assess the zoonotic potential and the public health importance of NTMs in Zambia.
This review was conducted in accordance with PRISMA guidelines (41). The overall aim of this review was to review the available information on the prevalence of NTM in humans with suspected TB, in animals and the environment, the species distribution of NTM in these reservoirs and the zoonotic potential and the public health importance of NTM in Zambia from 2000 to 2020. PubMed and Google scholar were systematically searched for publications on nontuberculous mycobacteria using the following search terms; Prevalence of Nontuberculous mycobacteria, in Zambia, epidemiology of mycobacteria infection, atypical mycobacteria, mycobacteria other than mycobacteria tuberculosis. Boolean operators (AND, OR) were used and our search was limited to articles written in English. The study focused on studies of NTM isolated from humans, animals and the environment such as soil or water in Zambia published between 2000 and 2020. The articles were screened by two independent authors in order to select carefully literature that was eligible for the review.
Titles or abstract searches in Google scholar and PubMed identified 243 articles (Figure 1). Abstracts of all the articles were screened and full texts copies of those deemed relevant to the review were obtained.
The titles and abstract were reviewed and articles were selected based on the following criteria; English language, conducted in Zambia, cross sectional, case control, case report or prospective studies and published between 2000 to 2020.
Articles were excluded if they did not report prevalence, if they were clinical trials and if they were not done in Zambia and if they were done before 2000.
For all relevant articles we extracted the following data using a data extraction sheet. Title, authors, aim of the study, study site, study type, the year the study was done, year published, prevalence of NTM, NTM species isolated, method of NTM species identification, Sample size, type of sample, and Source of sample.
Data was entered in Microsoft office excel and analysed using python 3.8 for mac. Descriptive data such as prevalence was presented using descriptive statistics such as proportion, percentage, mean, median and mode. Data was nonparametric so the Kruskal-Wallis test was used to determine the difference in prevalence between provinces and sources of specimens.
The search procedure and results are shown in Figure 1. Through literature search 243 papers were identified within international databases, PubMed and Google scholar. Among them, 23 duplicates were identified and removed. However 206 articles were excluded as they did not meet the inclusion criteria.The full text of the 14 remaining articles were revised and were considered appropriate for the purpose of this systematic review. Of these 1/14 (7.14%) is focusing on humans and water (42), 1/14 (7.14%) was done in humans, cattle and Kafue Lechwe (5) and another one is a case report 1/14 (7.14%) (43), one is a pilot study 1/14 (7.14%) (44) and one a case control study 1/14 (7.14%) (44). The remaining 9/14 (64.29%) are all cross section study focusing on NTM in humans (20, 45–53).
The study shows that the main specimens collected were sputum samples 10/14 studies (71.43%), one was a combination of sputum samples and brochoalveolar lavage, one was done on stored or archived isolates, one on a combination of stool samples, internal lavage and biopsy of the descending colon and lastly one was done on gastric lavage. Fourteen (14) studies were done on adults and only one was done in children (Table 1).
The results of the current study show that various methods were used to determine the prevalence and species distribution of NTM and the most commonly used method for species identification were molecular methods such as sequencing of 16s rRNA followed by Genotype CM/AS® assay and only one study used biochemical tests to identify the species (Table 1).
The overall prevalence of NTM in humans was 24.39% while in water 21.5% (Table 2), in animals 16.05% of which cattle 14.81% and Kafue Lechwe 1.23% (Table 1).
During analysis we divided Zambia into regions according to provinces namely Southern, Lusaka, Eastern, Western and Muchinga. The prevalence of NTM in humans varied according to regions and ranged from 4.95% for Lusaka province to 71.5% for Eastern province (Table 2). We carried out the Kruskal-Wallis test to examine the differences between provinces, type of specimen, source of specimen and the prevalence of NTM. Our results showed that there was no difference between the sources of specimens that is humans, animals and water (H-statistic: 0.6, P = 0.741) while according to provinces no significant difference was also found (H-statistic: 3.9, P = 0.421).
As shown in Table 3, the studies included in this review reported a wide range of NTM species. According to the 14 articles included in the study, 12/14 (85.71%) of the studies identified the NTM to species level while 2/14 (14.28%) did not speciate. During the study period 2000 to 2020 under review, 30 NTM species were isolated from humans, 12 from water, 2 from cattle and 1 from wildlife (Kafue Lechwe).
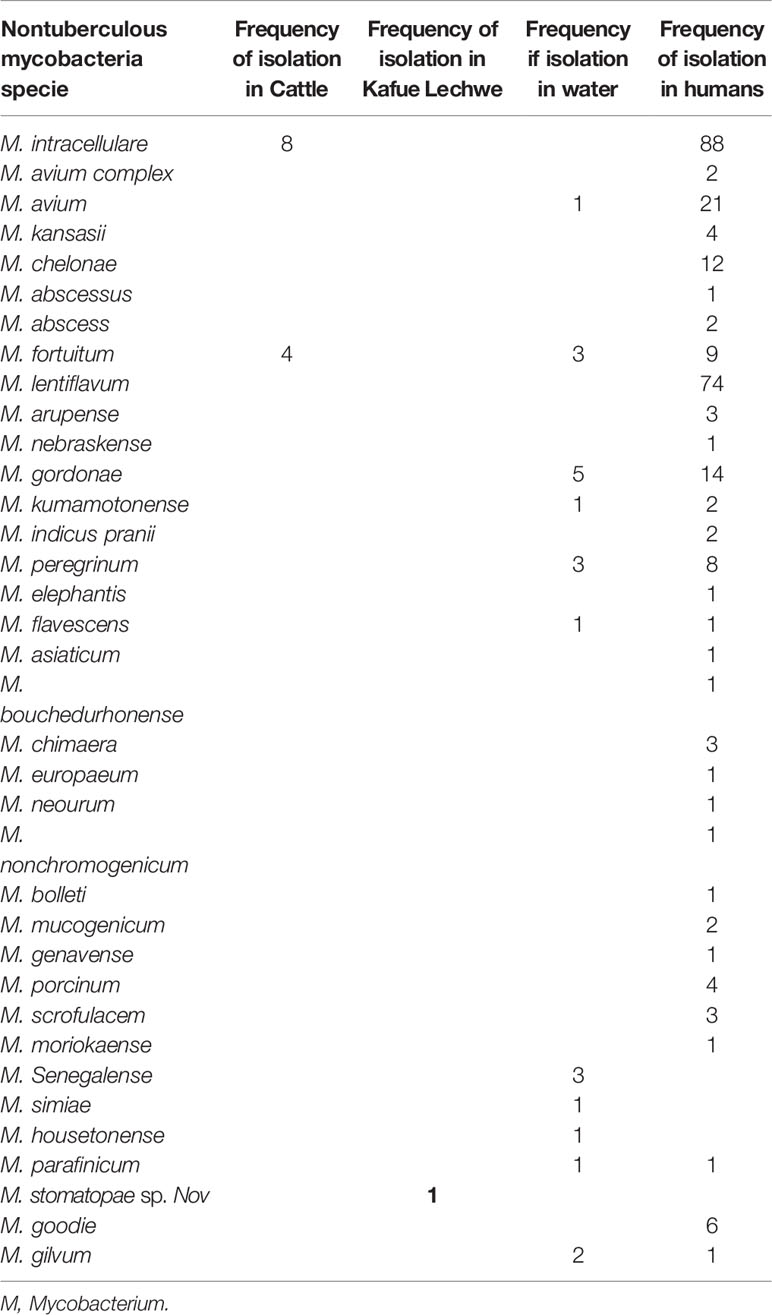
Table 3 Nontuberculous mycobacteria species isolated from humans, livestock, wildlife and water in Zambia.
The most commonly isolated NTM in humans was MAC particularly M. intracellulare, followed by M. lentiflavum while in water the most commonly isolated NTM species was M. gordonae followed by M.fortuitum while in cattle M. intracellulare was the most isolated NTM followed by M. fortuitum. Mycobacterium stomatopae sp.nov was the only specie isolated from Kafue Lechwe (Table 3).
This systematic review aimed at investigating the prevalent NTM in humans, animals and the environment. Additionally, it sought to determine NTM species distribution in these reservoir hosts and the zoonotic potential of NTM in Zambia among studies done from 2000 to 2020.
Our results show that the prevalence of NTM in humans varied from province to province with the highest being reported in Eastern Province (71.50%) and the lowest in Lusaka (4.95%) (Table 2). The reasons which could have led to increased prevalence in Eastern province could be as a result of increased levels of immunosuppression as one of the studies done there comprised of chronically ill patients whose immune system was already suppressed (44) the other reason for high prevalence of NTM in Eastern Province could be due to the high number of studies done there compared to other provinces. However, despite the observed differences in prevalence the difference was not statistically significant using Kruskal-Wallis test.
The overall National prevalence for our review was found to be 24.39%, This high prevalence of NTM in the review demonstrates that NTM is a public health threat in Zambia (20). The reason for the high prevalence could be as a result of increased national TB prevalence as Zambia is among the 30 high burden TB countries in the world (40). The other reason could be increased proportion of immune-compromised individuals such as those who have HIV/AIDS, those on chemotherapy or steroid treatments (45). Studies have shown that previous TB treatment or present history of TB is a known factor that may increase incidence of NTM infection (54), other factors include pre-existing pulmonary disease, chronic obstructive pulmonary disease, diabetes, HIV/AIDS, steroids, and malignancy (54, 55). This high prevalence agrees with the study by Farnia and his colleagues (56) whose results showed that Zambia was the second country with the highest prevalence of NTM (1337/8304; 16.10%) in Africa, the first one being South Africa with (6457/8304; 77.75%), then Uganda (109/8304; 1.31%), and Kenya (104/8304; 1.25%).
The proportion of NTM in the current study is similar to the study done by Huang et al. (37) and Yoon et al. (57) in China and South Korea respectively who estimated an increase in proportion from 15.6% in 2013 to 46.1% in 2018 and from 9.4% in 2009 to 35.1% in 2016 respectively. The prevalence of this review study is higher than what was found by Bonnet and his colleagues (58) in Tanzania whose prevalence of NTM isolation was 10.8% and of disease 0.9% in patients with presumptive TB.
In many African countries due to lack of laboratory infrastructure for culture and NTM identification, many patients with NTMs are missed because the first line diagnostic tool for the diagnosis of TB is GeneXpert test. If the patient is co-infected (MTB and NTM) at diagnosis they are only presumptively classified as having pulmonary TB and are therefore, treated only for MTB while NTM infection is missed. The downside of this are the reported treatment failure rates which are then further misclassified simply as MDR-TB. This misclassification of relapse and treatment failure as MDR-TB elicits unnecessary use of second line drugs which are expensive and toxic to the body, while recent reports suggest that NTM are responsible for such cases (32). The high prevalence of HIV has led to the interest in NTM, given their potential to cause pulmonary and disseminated diseases. Therefore, detection and identification of NTM species and determination of their susceptibility profiles is necessary for infection control and for the determination of epidemiology and treatment strategies (20, 59).
Different NTM species have been isolated in Zambia with the overall most common isolated being MAC particularly M. intracellulare, followed by M. lentiflavum and M. avium (Table 3). MAC is the overall common NTM species isolated in all continents and is frequently associated with disease (60). Of the MAC species, M. intracellulare and M. avium are the most frequent pathogenic members of the complex responsible for more than 60% of NTM infected patients (61). In our study among the MAC, M. intracellulare was the most common species 30.77% (88/286) compared to M. avium 7.34% (21/286).
The high prevalence of M.intracellulare is of concern in this setting given a high HIV prevalence and the ability of M.intracellure to cause pulmonary and extrapulmonary TB in such individuals (62). M. intracellulare maybe the cause of severe lung, lymphatic node, skin and bone or joint disease infection as well as bacteremia (63). Studies show that of the two (M. intracellulare and M. avium), M. intracellulare exhibit a severe presentation at diagnosis with a more unfavorable prognosis. It is also reported to be less responsive to chemotherapy (64, 65). Therefore species differentiation between M. avium and M.intracellulare may have a prognostic and therapeutic implication (63, 66).
Results from this review are consistent with other studies in which MAC particularly M. intracellulare was the most common isolated species in Tanzania, South Africa, Nigeria, South Korea and China (1, 24, 32, 37, 60, 67) and inconsistent with Bonnet et al. (58) were the common species isolated were M. fortuitum and M. abscessus (RGM) and M. intracellulare, M. gordonae and M. scrofulaceum (SGM). In Tanzania different results were found with M. gordonae, and M. interjectum 6 (16.7%) being the most isolated species (68). In Africa Mycobacterium kansasii (636/8304; 7.65%), Mycobacterium intracellulare (2278/8304; 7.43%), Mycobacterium avium (611/8304; 7.35%), and Mycobacterium avium complex (517/8304; 6.22%) were the most frequent species among different countries (56).
Many rarely isolated NTM were also identified in this study for example M. gilvum 1/286 (0.35%), M. interjectum, M. peregrinum 1/286 (0.35%), M. moriokaense 1/286 (0.35%), M. kumamotonense 2/286 (0.70%) and M. genavense 1/286 (0.35%). The total percentage of the rarely isolated NTMs was 5/286 (1.75%) of all the species isolated in humans compared to MAC which accounted for 111/286 (38.81%). Although some of these species have been isolated in other parts of the world from pulmonary samples, it is currently unclear what role they play in the etiology of pulmonary disease in Africa (39) while MAC are the common causes of NTM pulmonary disease.
Our results show that nontuberculous mycobacteria have been isolated from animals that is cattle carcasses showing tubercle-like lesions at meat inspection in Namwala district of Southern province of Zambia with a prevalence of 14.81% (12/81) and also from Kafue Lechwe showing tubercle-like lesions with a prevalence of 1.23% (1/81). These findings suggest the role of NTMs in causing lesions in animalss (69) and should further be evaluated on a large scale.
The species identified included M. intracellulare (8/12), and M. fortuitum (4/12) in cattle with M. intracellulare being the most common. These species have also been isolated in most of African countries in cattle tissues, faecal matter and cow dung (1, 62, 70–72). The isolation of these potentially pathogenic species in cattle poses high risk to humans as these species are able to cause infections in immunocompromised individuals such as those with HIV/AIDS. This is because in selected parts of developing countries such as Uganda, Zimbabwe and Zambia, humans live in close contact with cattle and this poses a high risk of zoonotic transmission (1, 72). Knowing that potentially pathogenic organisms are present in cattle is important for identifying control measures for human and animal populations.
In Kafue Lechwe the species isolated was M. stomatepiae sp Nov. this specie is known to cause diseases in fish (73) and the isolation of this species in Kafue Lechwe can be attributed to the way of life of the Kafue Lechwe which graze in water of 50cm and possibly contracted the species from the water and soil in the Kafue river possibly contaminated by infected fish (5). Different species have been isolated in wildlife in different countries, Katale et al. (62) isolated M. intracellulare and M. lentiflavum in Buffalo, Thompson gazelle and warthog. In Japan Mycolicibacterium peregrinum, Mycolicibacterium fortuitum, Mycolicibacterium septicum, and Mycolicibacterium thermoresistibile, and the eight SGM were Mycobacterium paraense, Mycolicibacterium sarupensis, Mycolicibacter virginiensis, and Mycobacterium nebraskense were isolated from different wildlife which were >99% similar to strains isolated from humans indicating a possibility of wildlife being reservoir of NTM. In South Africa NTM were isolated from African Buffalo and different other wildlife (74, 75).
These findings of isolation of NTMs in livestock and wildlife in Zambia confirms that domestic animals and wildlife are permanent reservoirs of mycobacteria infections and therefore poses a serious threat to the control and elimination programs of mycobacterial infections.
Nontuberculous mycobacteria are widely distributed in the natural and artificial environment and are commonly isolated from water and soil and studies suggest that natural water or drinking water are the principal sources of human and animal contamination (76). Our study shows that NTMs have been isolated in Namwala district of Zambia with a prevalence rate/isolation rate of 32/149 (21.5%) among the water samples collected from common water drinking points such as taps, boreholes, rivers, wells and streams/rivers. The majority of NTMs were isolated from borehole water sources 17(53.1%) followed by river 9 (28.1%), stream 3 (9.4%), dam 2 (6.3%), and tap 1 (3.1%) water sources. These findings are similar with other studies in which NTMs have been isolated in water with different proportions of isolation (1, 77–79).
Among the 32 NTM species isolated in our study, 23 were characterized to species levels, the most common species isolated was M. gordonae 5 (21.7%), other mycobacteria isolated included M. senegalense, M. peregrinum, M. gilvum, M. avium and M. fortuitum, M. kumamotonense, M. simiae, M. flavescens, M. housetonense, M. smegmatis, M. parafinicum (42).
M gordonae (the tap water bacillus) has been reported to be responsible for infections including skin and soft tissues infections, lung disease and liver and peritoneal infections. It is the most common mycobacterium isolated in hospital waters based on studies done recently (77, 80).
Different NTM species have been isolated in different localities, Moradi and his friends (79) isolated NTM in hospital waters with M. lentiflavum being the most commonly isolated followed by M. gordonae and M. paragordonae. Velayati et al. (81) isolated NTM in water and soil samples with M. fortuitum 93/282 (32.4%) being the most isolated followed by M. chelonae 38/282 (13.4%) while Crago et al. (78) isolated two species M. gordonae 88/183 and M. avium 34/183 in hospital water distribution system.
Soil and natural open water sources are known to contain mycobacteria and play a key role as sources of human and animal infection (82). Like other developing countries, the communities and their animals in Zambia pastoral ecosystems face challenges with safe drinking water. They rely mainly on untreated water from open natural water sources as valley tanks, dams, streams and swamps for drinking and household use. These limited water sources are shared among humans, livestock and wildlife. Water high in organic matter or animal dejections enhances the growth of environmental mycobacteria and such water is a source of human infection (1, 83).
In Zambia according to a study by Chanda-Kapata et al. (20), the high prevalence of NTM found among symptomatic participants in the population based study demonstrate that NTM is a public health problem. Despite this knowledge, data on the prevalence of human NTM in Zambia is limited, species distribution is also not known as the only national study done did not do species identification (20) and the source of human infection in Zambia is not clear as human to human transmission is uncommon (4, 14). Epidemiological studies suggest that natural or drinking water are the principal sources of human and animal contamination (76). This could be partly confirmed by Monde and her colleagues (42) who isolated similar species from water and humans in Namwala district of Zambia though studies need to be done at a larger scale to validate this. This is true in other countries as well as according to a study done by Marshall et al. (84) in Australia between 2001 and 2008 to investigate the relationship between genotype and geographical similarities between M. lentiflavum isolates from patients and drinking water which found that environmental colonies of M. lentiflavum were close to the strains that were isolated from patients. This suggests that drinking water can be the source of human infection for M. lentiflavum. Therefore prevention of public health risk of NTM in Zambia should take into account water treatment before it is accessed for domestic consumption (42). This is because in many parts of Zambia there is enough degree of overlap and interaction between humans, wildlife and the environment, increasing the potential for animal and human based contamination of natural water sources and transmission of NTM to livestock. These in turn act as reservoirs of NTM which has the potential to spill to the human population through consumption of meat and unpasteurized milk (32).
The isolation of NTMs from livestock raises a number of questions relating to the role of livestock as a source of human infections since human to human transmission of NTM is unknown (15). The isolation of M. intracellulare from both cattle and humans in Namwala district, a district which is in the Southern province of Zambia, a province known to have a high population of cattle raises the need for molecular analysis to further check the strains of NTM circulating in humans and animals to confirm the possibility of zoonotic transmission of NTM from animals to humans (5, 85). This is because NTMs can be transmitted to humans through consumption of meat and unpasteurized milk (32). This is a great challenge because the practice of consumption of raw milk and sour milk is common among cattle farmers in most Zambian communities (5). This, therefore, poses a health risk in the event that the milk is drawn from infected animals. Studies are therefore needed to confirm the zoonotic transmission as well as the genetic diversity of NTMs in cattle, environment and humans in Zambia.
Diseases caused by NTM are emerging in many settings with an increased number of patients needing treatment. The management of NTM infections is mainly by drug therapy, surgery or both. However, drug used to treat NTM disease are often expensive; the course is lengthy and treatment is often correlated with drug related toxicities (7, 86, 87). Outcome of treatment is poor and likely related to high levels of natural antibiotic resistance in NTM.
The treatment regimens varies according to species and the major difference being between slow versus rapid growing mycobacteria (7, 88, 89). For most SGM, the treatment regimens include rifampicin (rifapentine or rifabutin), ethambutol and a macrolide (azithromycin or clarithromycin), administered for 18 to 24 months, amikacin or streptomycin added in the initial 3 to 6 months in cases of severe disease. For RGM, regimens are based on in vitro drug susceptibility testing (DST) results (7, 87, 90).
The treatment of pulmonary NTM infection is difficult and entails multiple antibiotics and an extended treatment course (91). Cure rates of NTM PD differ according to species ranging from 30 to 50% in M. abscessus to 50 to 70% in MAC and 80 t0 90% in M. malmoense and M. kansasii (87).
For the treatment of NTM to be effective, the pathogenic strains circulating in a given area should be known especially in a setting were species identification of NTM is difficult and also were the prevalence of AFB positive is high which leads to misdiagnosing of NTM as TB (24). In Zambia, despite studies showing a high burden of NTM (20), there is need to investigate the drug susceptibility test (DST) profile of the NTM of clinical significance as according to ATS guideline the treatment of rapid growing NTM should be based on the DST results (7). Therefore, the management of NTM relies solely on knowing which species are common in the different parts of Zambia as culture is not done in most of the hospitals and the species distribution of NTM differ according to geographic location. Through this knowledge only will it be possible to develop appropriate intervention strategies including prevention and control of NTM in Zambia.
Given our findings, it is important to note that this review has also some limitations, first we did not describe much regarding global epidemiology and this article may not be of particular interest to researchers outside Zambia. However, this study was intended to serve as a local epidemiologic reference for physicians and scientists in Zambia. Secondly, we only searched two recognized electronic databases and only included articles written in English, making it possible to leave out studies or publications that may be relevant to this review. Thirdly, to calculate the prevalence, percentages from different studies were used. However, studies had different sample sizes and this may have affected our calculated prevalence. Lastly the species distribution reported in this study may not be a true reflection of the species distribution in Zambia as the only national study done did not do species identification.
This review has shown that NTM is an emerging public health threat in Zambia as seen from the high prevalence of NTM in humans, water and animals. M. intracellulare was the most common isolated NTM in humans and cattle, M. gordonae in water, and M. stomatepiae sp Nov in Kafue Lechwe. The review has also identified knowledge gaps in the management of NTM in Zambia. Molecular information on the zoonotic transmission of NTM in Zambia is lacking and also the DST profile as well as the genes conferring drug resistant to NTM is unknown. We therefore recommend increased awareness of NTM diseases among clinicians and laboratory personnel as this is crucial for clinical management of NTM diseases and an essential step for facilitating the identification of NTM species in laboratories. Studies on NTM should be done nationwide so that species distribution of NTM in Zambia can be known, and the genetic diversity or gene analysis of NTM strains isolated from humans, animals and the environment should be investigated. This is in order to determine the probable source of human NTM infections in Zambia, as this is cardinal for the management of patients. Studies should also be done focusing on the whole genome sequencing of NTM in Zambia so as to determine the genes conferring drug resistant because this is important for the treatment of NTM disease in Zambia.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
MZ conceptualized the study, MZ and NM did the literature search. MZ wrote the initial draft of the manuscript. MM and SM supervised the literature search. PN did the data analysis and interpretation. MM, SM, NM, and PN read and edited the paper and provided critical comments. All authors contributed to the article and approved the submitted version.
This study was sponsored by the African Centre of Excellence of Infectious Diseases of Human and Animals (ACEIDHA) hosted at the University of Zambia School of Veterinary Medicine grant number P151847 funded by the World Bank.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to ACEIDHA and the University of Zambia, School of Veterinary Medicine, Department of Disease Control for their support.
NTM, Nontuberculous mycobacteria; MAC, Mycobacterium avium complex; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses; MTC, Mycobacterium tuberculosis complex; HIV, Human Immunodeficiency Virus; AIDS, Acquired Immunodeficiency Syndrome; TB, Tuberculosis; AFB, Acid Fast Bacteria; MOTT, Mycobacteria Other than Tuberculosis; MTB, Mycobacterium Tuberculosis; SGM, Slow growing mycobacteria; RGM, Rapid growing mycobacteria; MTBC, Mycobacterium Tuberculosis Complex; MDR TB, Multi-drug resistant TB; rRNA, ribosomal Ribonucleic acid; LJ, Lowenstein Jensen; CLJ, Commercial LJ; HLJ, Homemade LJ; ZN, Ziehl Nelsen; MGIT, Mycobacterium Growth Indicator Tube; AMGIT, Automated MGIT; MMGIT, Manual MGIT; WHO, World Health Organisation.
1. Kankya C, Muwonge A, Djønne B, Munyeme M, Opuda-Asibo J, Skjerve E, et al. Isolation of Non-Tuberculous Mycobacteria From Pastoral Ecosystems of Uganda: Public Health Significance. BMC Public Health (2011) 11(1):1–8. doi: 10.1111/j.1469-0691.2009.03013.x
2. Nyamogoba HD, Mbuthia G, Mining S, Kikuvi G, Biegon R, Mpoke S, et al. HIV Co-Infection With Tuberculous and Non-Tuberculous Mycobacteria in Western Kenya: Challenges in the Diagnosis and Management. Afr Health Sci (2012) 12(3):305–11. doi: 10.4314/ahs.v12i3.9
3. Ahmed I, Jabeen K, Hasan R. Identification of Non-Tuberculous Mycobacteria Isolated From Clinical Specimens at a Tertiary Care Hospital: A Cross-Sectional Study. BMC Infect Dis (2013) 13(1):493. doi: 10.1186/1471-2334-13-493
4. Holton J. Non-Tuberculous Mycobacteria: An Emerging Clinical Problem. SPG BioMed (2019) 1:1–15. doi: 10.32392/biomed.67
5. Malama S, Munyeme M, Mwanza S, Muma JB. Isolation and Characterization of Non Tuberculous Mycobacteria From Humans and Animals in Namwala District of Zambia. BMC Res Notes (2014) 7(1):1–5. doi: 10.1186/1756-0500-7-622
6. Larsson L, Bennet R, Eriksson M, Jönsson B, Ridell M. Nontuberculous Mycobacterial Diseases in Humans. In: Nontuberculous Mycobacteria (NTM). Elsevier (2019). p. 101–19. doi: 10.1016/B978-0-12-814692-7.00005-X
7. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An Official ATS/IDSA Statement: Diagnosis, Treatment, and Prevention of Nontuberculous Mycobacterial Diseases. Am J Respir Crit Care Med (2007) 175(4):367–416. doi: 10.1164/rccm.200604-571ST
8. Wu T, Lu C, Lai H. Current Situations on Identification of Nontuberculous Mycobacteria. J BioMed Lab Sci (2009) 21(259):1–6.
9. Cvetnić Ž, Zdelar Tuk M, Duvnjak S, Reil I, Mikulić M, Pavlinec Ž, et al. Tuberculous and Nontuberculous Mycobacteria in Human and Animal Infection. Ветеринарски Журнал Републике Српске (2019) 18(2):356–69. doi: 10.7251/VETJEN1802342C
10. Falkinham JO. Environmental Sources of Nontuberculous Mycobacteria. Clin Chest Med (2015) 36(1):35–41. doi: 10.1016/j.ccm.2014.10.003
11. Corbett EL, Churchyard GJ, Clayton T, Herselman P, Williams B, Hayes R, et al. Risk Factors for Pulmonary Mycobacterial Disease in South African Gold Miners: A Case-Control Study. Am J Respir Crit Care Med (1999) 159(1):94–9. doi: 10.1164/ajrccm.159.1.9803048
12. Adjemian J, Olivier KN, Prevots DR. Nontuberculous Mycobacteria Among Patients With Cystic Fibrosis in the United States: Screening Practices and Environmental Risk. Am J Respir Crit Care Med (2014) 190(5):581–6. doi: 10.1164/rccm.201405-0884OC
13. Chou MP, Clements ACA, Thomson RM. A Spatial Epidemiological Analysis of Nontuberculous Mycobacterial Infections in Queensland, Australia. BMC Infect Dis (2014) 14(1):1–10. doi: 10.1186/1471-2334-14-279
14. Bryant JM, Grogono DM, Rodriguez-rincon D, Everall I, Brown KP, Moreno P, et al. Emergence and Spread of a Human- Transmissible Multidrug-Resistant Nontuberculous Mycobacterium. Europe PMC (2016) 354(6313):751–7. doi: 10.1126/science.aaf8156
15. Konuk M, Korcan E, Dülgerbaki S, Altindiş M. Isolation and Identification of Mycobacteria From Raw Milk Samples in Afyonkarahisar District of Turkey. Int J Food Microbiol (2007) 115(3):343–7. doi: 10.1016/j.ijfoodmicro.2006.12.019
16. Claeys TA, Robinson T. Crossm The Many Lives of Nontuberculous Mycobacteria. J Bacteriol (2018) 20:e00739–17. doi: 10.1128/JB.00739-17
17. Araj GF, Baba OZ, Itani LY, Avedissian AZ, Sobh GM. Non-Tuberculous Mycobacteria Profiles and Their Anti-Mycobacterial Resistance at a Major Medical Center in Lebanon. J Infect Dev Ctries (2019) 13(7):612–8. doi: 10.3855/jidc.11028
18. Sadikot RT. Nontuberculous Mycobacterial Lung Disease. In: Nontuberculous Mycobacteria (NTM): Microbiological, Clinical and Geographical Distribution. Elsevier (2019). p. 121–32. doi: 10.1016/B978-0-12-814692-7.00006-1
19. Panagiotou M, Papaioannou AI, Kostikas K, Paraskeua M, Velentza E, Kanellopoulou M, et al. The Epidemiology of Pulmonary Nontuberculous Mycobacteria: Data From a General Hospital in Athens, Greece, 2007-2013. Pulm Med (2014) 2014:894976. doi: 10.1155/2014/894976
20. Chanda-Kapata P, Kapata N, Klinkenberg E, Mulenga L, Tembo M, Katemangwe P, et al. Non-Tuberculous Mycobacteria (NTM) in Zambia: Prevalence, Clinical, Radiological and Microbiological Characteristics. BMC Infect Dis (2015) 15(1):1–7. doi: 10.1186/s12879-015-1264-6
21. Yun JL, Lei XD, Peng YS, Can LH, Lian LG, Yi J, et al. First Report in China on the Identification and Drug Sensitivity of Mycobacterium Elephantis Isolated From the Milk of a Cow With Mastitis. BioMed Environ Sci (2017) 30(7):501–7. doi: 10.3967/bes2017.006
22. Satyanarayana G, Heysell SK, Scully KW, Houpt ER. Mycobacterial Infections in a Large Virginia Hospital, 2001-2009. BMC Infect Dis (2011) 11:1–7. doi: 10.1186/1471-2334-11-113
23. Maiga M, Siddiqui S, Diallo S, Diarra B, Traoré B, Shea YR, et al. Failure to Recognize Nontuberculous Mycobacteria Leads to Misdiagnosis of Chronic Pulmonary Tuberculosis. PloS One (2012) 7(5):1–7. doi: 10.1371/journal.pone.0036902
24. Richter E, Weizenegger M, Rüsch-Gerdes S, Niemann S, Muwonge A, Oloya J, et al. British Thoracic Society Guideline for the Management of Non-Tuberculous Mycobacterial Pulmonary Disease (NTM-Pd). Am J Respir Crit Care Med (2012) 15(1):1–8. doi: 10.1056/NEJMoa1003176
25. Kwon YS, Koh W. Diagnosis of Pulmonary Tuberculosis and Nontuberculous Mycobacterial Lung Disease in Korea. Tuberc Respir Dis (Seoul) (2014) 77(1):1–5. doi: 10.4046/trd.2014.77.1.1
26. Van Ingen J, Bendien SA, De Lange WCM, Hoefsloot W, Dekhuijzen PNR, Boeree MJ, et al. Clinical Relevance of Non-Tuberculous Mycobacteria Isolated in the Nijmegen-Arnhem Region, The Netherlands. Thorax (2009) 64(6):502–6. doi: 10.1136/thx.2008.110957
27. Winthrop KL, McNelley E, Kendall B, Marshall-Olson A, Morris C, Cassidy M, et al. Pulmonary Nontuberculous Mycobacterial Disease Prevalence and Clinical Features. Am J Respir Crit Care Med (2010) 182(7):977–82. doi: 10.1164/rccm.201003-0503OC
28. Prevots DR, Marras TK. Epidemiology of Human Pulmonary Infection With Nontuberculous Mycobacteria: A Review. Clin Chest Med (2015) 36(1):13–34. doi: 10.1016/j.ccm.2014.10.002
29. Abebe G, Zegeye Bonsa WK. Treatment Outcomes and Associated Factors in Tuberculosis Patients at Jimma University Medical Center: A 5−Year Retrospective Study Gemeda. Int J Mycobacteriol (2017) 6(3):239–45. doi: 10.4103/ijmy.ijmy_177_18
30. Ratnatunga CN, Lutzky VP, Kupz A, Doolan DL, Reid DW, Field M, et al. The Rise of Non-Tuberculosis Mycobacterial Lung Disease. Front Immunol (2020) 11(March):1–12. doi: 10.3389/fimmu.2020.00303
31. Marras TK, Mendelson D, Marchand-Austin A, May K, Jamieson FB. Pulmonary Nontuberculous Mycobacterial Disease, Ontario, Canada, 1998-2010. Emerg Infect Dis (2013) 19(11):1889–91. doi: 10.3201/eid1911.130737
32. Wagner D, Lipman M, Cooray S, Ringshausen FC, Morimoto K, Koh W, et al. Nontuberculous Mycobacterial Disease (2019). Available at: http://link.springer.com/10.1007/978-3-319-93473-0.
33. Dakić I, Arandjelović I, Savić B, Jovanović S, Tošić M, Kurucin T, et al. Pulmonary Isolation and Clinical Relevance of Nontuberculous Mycobacteria During Nationwide Survey in Serbia, 2010-2015. PloS One (2018) 13(11):2010–5. doi: 10.1371/journal.pone.0207751
34. López-Roa P, Aznar E, Cacho J, Cogollos-Agruña R, Domingo D, García-Arata M, et al. Epidemiology of Non-Tuberculous Mycobacteria Isolated From Clinical Specimens in Madrid, Spain, From 2013 to 2017. Eur J Clin Microbiol Infect Dis (2020) 39(6):1089–94. doi: 10.1007/s10096-020-03826-7
35. Park SC, Kang MJ, Han CH, Lee SM, Kim CJ, Lee JM, et al. Prevalence, Incidence, and Mortality of Nontuberculous Mycobacterial Infection in Korea: A Nationwide Population-Based Study. BMC Pulm Med (2019) 19(1):1–9. doi: 10.1186/s12890-019-0901-z
36. Lee H, Myung W, Koh WJ, Moon SM, Jhun BW. Epidemiology of Nontuberculous Mycobacterial Infection, South Korea, 2007-2016. Emerg Infect Dis (2019) 25(3):569–72. doi: 10.3201/eid2503.181597
37. Huang JJ, Huang JJ, Huang JJ, Li YX, Li YX, Zhao Y, et al. Prevalence of Nontuberculous Mycobacteria in a Tertiary Hospital in Beijing, China, January 2013 to December 2018. BMC Microbiol (2020) 20(1):1–8. doi: 10.1186/s12866-020-01840-5
38. Stout JE, Koh WJ, Yew WW. Update on Pulmonary Disease Due to Non-Tuberculous Mycobacteria. Int J Infect Dis (2016) 45:123–34. doi: 10.1016/j.ijid.2016.03.006
39. Okoi C, Anderson STB, Antonio M, Mulwa SN, Gehre F, Adetifa IMO. Non-Tuberculous Mycobacteria Isolated From Pulmonary Samples in Sub-Saharan Africa - A Systematic Review and Meta Analyses. Sci Rep (2017) 7(1):1–12. doi: 10.1038/s41598-017-12175-z
40. World Health Organization. Are Updated Every Year. For the Tuberculosis (2020). Available at: https://www.who.int/tb/publications/global_report/en/.
41. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement (2010). Available at: www.prisma-statement.org/.
42. Monde N, Munyeme M, Muwonge A, Muma JB, Malama S. Characterization of Non-Tuberculous Mycobacterium From Humans and Water in an Agropastoral Area in Zambia. BMC Infect Dis (2018) 18(1):1–7. doi: 10.1186/s12879-017-2939-y
43. Buijtels PCAM, Petit PLC, Verbrugh HA, Van Belkum A, Van Soolingen D. Isolation of Nontuberculous Mycobacteria in Zambia: Eight Case Reports. J Clin Microbiol (2005) 43(12):6020–6. doi: 10.1128/JCM.43.12.6020-6026.2005
44. Buijtels PCAM, van der Sande MAB, Parkinson S, Verbrugh HA, Petit PLC, van Soolingen D. Isolation of Non-Tuberculous Mycobacteria at Three Rural Settings in Zambia; a Pilot Study. Clin Microbiol Infect (2010) 16(8):1142–8. doi: 10.1111/j.1469-0691.2009.03072.x
45. Buijtels PCAM, van der Sande MAB, de Graaff CS, Parkinson S, Verbrugh HA, Petit PLC, et al. Nontuberculous Mycobacteria, Zambia. Emerg Infect Dis (2009) 15(2):242–9. doi: 10.3201/eid1502.080006
46. Muyoyeta M, Schaap JA, De Haas P, Mwanza W, Muvwimi MW, Godfrey-Faussett P, et al. Comparison of Four Culture Systems for Mycobacterium Tuberculosis in the Zambian National Reference Laboratory. Int J Tuberc Lung Dis (2009) 13(4):460–5.
47. Lubasi D, Baxter E, Zondie L MJ. Optimizing the Recovery Rate of Mycobacterium Species From Gastric Lavages in Children at an Urban Zambian Hospital. Med J Zambia (2010) 37(1):5-10–10.
48. Buijtels PCAM, Iseman MD, Parkinson S, de Graaff CS, Verbrugh HA, Petit PLC, et al. Misdiagnosis of Tuberculosis and the Clinical Relevance of Non—Tuberculous Mycobacteria in Zambia. Asian Pac J Trop Med (2010) 3(5):386–91. doi: 10.1016/S1995-7645(10)60094-6
49. Verweij KE, Kamerik AR, Van Ingen J, Van Dijk JH, Sikwangala P, Thuma P, et al. Application of Modern Microbiological Diagnostic Methods for Tuberculosis in Macha, Zambia. Int J Tuberc Lung Dis (2010) 14(9):1127–31.
50. Mateyo KJ, Lakhi S, Guffey B, Chi B, Mweemba A, Andrews B. Pulmonary Disease in HIV-Infected Patients at the University Teaching Hospital, Lusaka, Zambia. Med J Zambia (2014) 41(2):50–8.
51. Mwikuma G, Kwenda G, Hang’ombe BM, Simulundu E, Kaile T, Nzala S, et al. Molecular Identification of Non-Tuberculous Mycobacteria Isolated From Clinical Specimens in Zambia. Ann Clin Microbiol Antimicrob (2015) 14(1):1. doi: 10.1186/s12941-014-0059-8
52. Monde N, Munyeme M, Muwonge A, Muma JB, Malama S. Non-Tuberculous Mycobacteria Recovered From Suspected TB Cases in Namwala District of Zambia. ARC J Clin Case Rep (2017) 3(2):4–10. doi: 10.20431/2455-9806.0302002
53. Chongwe G, Michelo C, Kelly P. Diagnostic Yield of Nontuberculous Mycobacteria in Patients Booked for Endoscopy at the University Teaching Hospital, Lusaka. BMC Res Notes (2017) 10(1):27. doi: 10.1186/s13104-016-2329-3
54. Suryawati B, Sari Y, Saptawati L. The Increase of Nontuberculous Mycobacteria Infection: Emphasis on Epidemiology, Risk Factors and Diagnostic Tools. KnE Life Sci (2019) 4(12):139. doi: 10.18502/kls.v4i12.4167
55. Saptawati L, Kusumo H, Suryawati B. Prevalence of Non-Tuberculous Mycobacteria (NTM) in Surakarta, Indonesia: Higher Than Expected. KnE Life Sci (2019) 4(12):132. doi: 10.18502/kls.v4i12.4166
56. Farnia P, Farnia P, Ghanavi J, Velayati AA. Epidemiological Distribution of Nontuberculous Mycobacteria Using Geographical Information System. In: Nontuberculous Mycobacteria (NTM): Microbiological, Clinical and Geographical Distribution. Elsevier Inc. (2019). p. 191–321. doi: 10.1016/B978-0-12-814692-7.00010-3
57. Yoon HJ, Choi HY, Ki M. Nontuberculosis Mycobacterial Infections at a Specialized Tuberculosis Treatment Centre in the Republic of Korea. BMC Infect Dis (2017) 17(1):1–6. doi: 10.1186/s12879-017-2532-4
58. Bonnet M, Chamroeun San K, Pho Y, Sok C, Dousset JP, Brant W, et al. Nontuberculous Mycobacteria Infections at a Provincial Reference Hospital, Cambodia. Emerg Infect Dis (2017) 23(7):1139–47. doi: 10.3201/eid2307.170060
59. Aliyu G, El-Kamary SS, Abimiku A, Brown C, Tracy K, Hungerford L, et al. Prevalence of Non-Tuberculous Mycobacterial Infections Among Tuberculosis Suspects in Nigeria. PloS One (2013) 8(5):1–7. doi: 10.1371/journal.pone.0063170
60. Zweijpfenning SMH, Van IJ, Hoefsloot W. Geographic Distribution of Nontuberculous Mycobacteria Isolated From Clinical Specimens: A Systematic Review. Semin Respir Crit Care Med (2018) 39(3):336–42. doi: 10.1055/s-0038-1660864
61. Zhao X, Wang Y, Pang Y. Antimicrobial Susceptibility and Molecular Characterization of Mycobacterium Intracellulare in China. Infect Genet Evol (2014) 27:332–8. doi: 10.1016/j.meegid.2014.07.032
62. Katale BZ, Mbugi EV, Botha L, Keyyu JD, Kendall S, Dockrell HM, et al. Species Diversity of Non-Tuberculous Mycobacteria Isolated From Humans, Livestock and Wildlife in the Serengeti Ecosystem, Tanzania. BMC Infect Dis (2014) 14(1):1–8. doi: 10.1186/s12879-014-0616-y
63. Dauchy FA, Dégrange S, Charron A, Dupon M, Xin Y, Bébéar C, et al. Variable-Number Tandem-Repeat Markers for Typing Mycobacterium Intracellulare Strains Isolated in Humans. BMC Microbiol (2010) 10:1–7. doi: 10.1186/1471-2180-10-93
64. Inagaki T, Yagi T, Ichikawa K, Nakagawa T, Moriyama M, Uchiya KI, et al. Evaluation of a Rapid Detection Method of Clarithromycin Resistance Genes in Mycobacterium Avium Complex Isolates. J Antimicrob Chemother (2011) 66(4):722–9. doi: 10.1093/jac/dkq536
65. Suzuki K, Kurashima A, Tatsuno K, Kadota Ji. Clinical Significance and Epidemiologic Analyses of Mycobacterium Avium and Mycobacterium Intracellulare Lung Disease From Post-Marketing Surveillance. Respir Investig (2018) 56(1):87–93. doi: 10.1016/j.resinv.2017.11.001
66. Cho EH, Huh HJ, Song DJ, Moon SM, Lee SH, Shin SY, et al. Differences in Drug Susceptibility Pattern Between Mycobacterium Avium and Mycobacterium Intracellulare Isolated in Respiratory Specimens. J Infect Chemother (2018) 24(4):315–8. doi: 10.1016/j.jiac.2017.10.022
67. Hoefsloot W, Van Ingen J, Andrejak C, Ängeby K, Bauriaud R, Bemer P, et al. The Geographic Diversity of Nontuberculous Mycobacteria Isolated From Pulmonary Samples: An NTM-NET Collaborative Study. Eur Respir J (2013) 42(6):1604–13. doi: 10.1183/09031936.00149212
68. Hoza AS, Mfinanga SGM, Rodloff AC, Moser I, König B. Increased Isolation of Nontuberculous Mycobacteria Among TB Suspects in Northeastern, Tanzania: Public Health and Diagnostic Implications for Control Programmes. BMC Res Notes (2016) 9(1):1–9. doi: 10.1186/s13104-016-1928-3
69. Nuru A, Zewude A, Mohammed T, Wondale B, Teshome L, Getahun M, et al. Nontuberculosis Mycobacteria Are the Major Causes of Tuberculosis Like Lesions in Cattle Slaughtered at Bahir Dar Abattoir, Northwestern Ethiopia. BMC Vet Res (2017) 13(1):1–6. doi: 10.1186/s12917-017-1168-3
70. Diguimbaye-Djaibé C, Vincent V, Schelling E, Hilty M, Ngandolo R, Mahamat HH, et al. Species Identification of Non-Tuberculous Mycobacteria From Humans and Cattle of Chad. Schweiz Arch Tierheilkd (2006) 148(5):251–6. doi: 10.1024/0036-7281.148.5.251
71. Gumi B, Schelling E, Berg S, Firdessa R, Erenso G, Mekonnen W, et al. Zoonotic Transmission of Tuberculosis Between Pastoralists and Their Livestock in South-East Ethiopia. Ecohealth (2012) 9(2):139–49. doi: 10.1007/s10393-012-0754-x
72. Padya L, Chin’ombe N, Magwenzi M, Mbanga J, Ruhanya V, Nziramasanga P. Molecular Identification of Mycobacterium Species of Public Health Importance in Cattle in Zimbabwe by 16S rRNA Gene Sequencing. Open Microbiol J (2019) 9(1):38–42. doi: 10.2174/1874285801509010038
73. Austin B. Taxonomy of Bacterial Fish Pathogens. Vet Res (2011) 42(1):20. doi: 10.1186/1297-9716-42-20
74. Gcebe N, Hlokwe TM. Non-Tuberculous Mycobacteria in South African Wildlife: Neglected Pathogens and Potential Impediments for Bovine Tuberculosis Diagnosis. Front Cell Infect Microbiol (2017) 7(JAN):1–15. doi: 10.3389/fcimb.2017.00015
75. Gcebe N, Michel AL, Hlokwe TM. Non-Tuberculous Mycobacterium Species Causing Mycobacteriosis in Farmed Aquatic Animals of South Africa. BMC Microbiol (2018) 18(1):1–11. doi: 10.1186/s12866-018-1177-9
76. Nishiuchi Y, Iwamoto T, Maruyama F. Infection Sources of a Common Non-Tuberculous Mycobacterial Pathogen, Mycobacterium Avium Complex. Front Med (2017) 4(27):1–7. doi: 10.3389/fmed.2017.00027
77. Genc GE, Richter E, Erturan Z, Khosravi AD, Shahraki AH, Hashemzadeh M, et al. Prevalence of Non-Tuberculous Mycobacteria in Hospital Waters of Major Cities of Khuzestan Province, Iran. Front Cell Infect Microbiol (2016) 6(12):1–8. doi: 10.3389/fcimb.2016.00042
78. Crago B, Ferrato C, Drews SJ, Louie T, Ceri H, Turner RJ, et al. Surveillance and Molecular Characterization of Non-Tuberculous Mycobacteria in a Hospital Water Distribution System Over a Three-Year Period. J Hosp Infect (2014) 87(1):59–62. doi: 10.1016/j.jhin.2014.03.002
79. Moradi S, Nasiri MJ, Pourahmad F, Darban-Sarokhalil D. Molecular Characterization of Nontuberculous Mycobacteria in Hospital Waters: A Two-Year Surveillance Study in Tehran, Iran. J Water Health (2019) 17(2):350–6. doi: 10.2166/wh.2019.294
80. Genc GE, Richter E, Erturan Z. Isolation of Nontuberculous Mycobacteria From Hospital Waters in Turkey. Apmis (2013) 121(12):1192–7. doi: 10.1111/apm.12066
81. Velayati AA, Farnia P, Mozafari M, Mirsaeidi M. Nontuberculous Mycobacteria Isolation From Clinical and Environmental Samples in Iran: Twenty Years of Surveillance. BioMed Res Int (2015) 2015:1–10. doi: 10.1155/2015/254285
82. van Ingen J, Boeree MJ, Dekhuijzen PNR, van Soolingen D. Environmental Sources of Rapid Growing Nontuberculous Mycobacteria Causing Disease in Humans. Clin Microbiol Infect (2009) 15(10):888–93. doi: 10.1111/j.1469-0691.2009.03013.x
83. Oloya J, Kazwala R, Lund A, Opuda-Asibo J, Demelash B, Skjerve E, et al. Characterisation of Mycobacteria Isolated From Slaughter Cattle in Pastoral Regions of Uganda. BMC Microbiol (2007) 7:1–7. doi: 10.1186/1471-2180-7-95
84. Marshall HM, Carter R, Torbey MJ, Minion S, Tolson C, Sidjabat HE, et al. Mycobacterium Lentiflavum in Drinking Water Supplies, Australia. Emerg Infect Dis (2011) 17(3):395–402. doi: 10.3201/eid1703.090948
85. Zambia Central Statistical Office. 2017/18 Livestock and Aquaculture Report. Lusaka: Ministry of Fisheries and Livestock, Central Statistics Office (2019).
86. Brown-Elliott BA, Nash KA, Wallace RJ. Antimicrobial Susceptibility Testing, Drug Resistance Mechanisms, and Therapy of Infections With Nontuberculous Mycobacteria. Clin Microbiol Rev (2012) 25(3):545–82. doi: 10.1128/CMR.05030-11
87. Van Ingen J, Boeree MJ, Van Soolingen D, Mouton JW. Resistance Mechanisms and Drug Susceptibility Testing of Nontuberculous Mycobacteria. Drug Resist Update (2012) 15(3):149–61. doi: 10.1016/j.drup.2012.04.001
88. Shahraki AH, Heidarieh P, Bostanabad SZ, Khosravi AD, Hashemzadeh M, Khandan S, et al. “Multidrug-Resistant Tuberculosis” may be Nontuberculous Mycobacteria. Eur J Intern Med (2015) 26(4):279–84. doi: 10.1016/j.ejim.2015.03.001
89. Bhattacharya J, Mohandas S, Goldman DL. Nontuberculous Mycobacterial Infections in Children. Pediatr Rev (2019) 40(4):179–90. doi: 10.1542/pir.2018-0131
90. Kwon YS, Koh WJ, Daley CL. Treatment of Mycobacterium Avium Complex Pulmonary Disease. Tuberc Respir Dis (Seoul) (2019) 82(1):15–26. doi: 10.4046/trd.2018.0060
Keywords: nontuberculous mycobacteria, public threat, review, zoonotic transmission, Zambia
Citation: Zulu M, Monde N, Nkhoma P, Malama S and Munyeme M (2021) Nontuberculous Mycobacteria in Humans, Animals, and Water in Zambia: A Systematic Review. Front. Trop. Dis 2:679501. doi: 10.3389/fitd.2021.679501
Received: 11 March 2021; Accepted: 28 June 2021;
Published: 16 July 2021.
Edited by:
Aparup Das, ICMR-National Institute of Research in Tribal Health, IndiaReviewed by:
Renu Bharadwaj, B. J. Medical College & Sassoon Hospital, IndiaCopyright © 2021 Zulu, Monde, Nkhoma, Malama and Munyeme. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mildred Zulu, bWlsZHJlZHp1bHVAeW1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.