
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Insect Sci., 28 June 2024
Sec. Insect Molecular Genetics
Volume 4 - 2024 | https://doi.org/10.3389/finsc.2024.1362473
 Minurani Dalai
Minurani Dalai Anita Jagota*
Anita Jagota*Bombyx mori is a lepidopteran holometabolous insect with distinct developmental stages: egg, larvae, pupae, and adult. The lepidopteran insect undergoes major modifications in the central nervous system (CNS) so as to adapt to the lifestyle of these distinct stages with specific habitats and functions from voraciously feeding larval stages to flying reproductive adults via dormant pupal stages. Such transitions are linked to transcriptional, epigenetic, and translational complexities. Therefore, studying rhythmic gene expression in CNS of various developmental stages and the effects of antagonists on developmental hormones requires a very stable reference gene (RG). To facilitate rhythmic gene expression studies using reverse transcription quantitative polymerase chain reaction (RT-qPCR) in B. mori and the effect of developmental hormone juvenile hormone (JH) and 20-hydroxy ecdysone hormone (20 HE), antagonists Precocene 1 and testosterone, respectively, were used. Eight candidate RGs, namely, Translational initiation factor 3 subunit 4 (TI3S4), Translational initiation factor 3 subunit 5 (TI3S5), Ribosomal protein subunit 7 (RPs7), TATA-binding protein association factor (TAF13), Translational initiation factor 4 A (TI4A), Ribosomal protein (RPL32), Elongation factor 1 (EF1), and Arginine kinase (AK), were assessed in the CNS of B. mori. The postembryonic developmental (PED) stages used were the fifth late larval instar, early pupa, mid pupa, late pupa, and adult. The assessments were done at four different time points, Zeitgeber time (ZT) 0, 6, 12, and 18, to find stability towards 24-h rhythmic expression. RefFinder, geNorm, and Ct value analysis were performed. RefFinder and geNORM studies suggested stability order as TI3S4 > TI3S5 > RPs7, but Ct value evaluation showed stability order as TI3S5 > TI3S4 > RPs7. We therefore demonstrated that TI3S4, TI3S5, and RPs7 can be used as RG in various PED stages in CNS of B. mori (Strain: CB-hybrid, PM×CSR2) towards studies with effects of JH and 20 HE antagonists.
The silkworm, Bombyx mori, is an economically important holometabolous lepidopteran insect with distinct developmental stages such as larva, pupa, and adult. It undergoes complex morphological and physiological changes during the metamorphosis (1). These changes are linked to several modifications in the central nervous system (CNS) and associated peripheral changes (2). The molecular mechanism of such alterations can be studied by gene expression analysis with the help of reverse transcription quantitative polymerase chain reaction (RT-qPCR). It is most widely used for its accuracy, high sensitivity, reliability, reproducibility, and cost-effectiveness (3). However, the selection of an appropriate housekeeping gene/reference gene (RG) for data normalization of biological samples is the prerequisite in gene expression studies (4). An ideal reference gene should be consistently expressed, with its stability and expression being independent of any experimental conditions. RG should show a moderate threshold cycle (5–7). Generally, 18S ribosomal RNA (18s rRNA), beta-actin (β-ACT), and glyceraldehyde, 3-phosphate dehydrogenase (GAPDH) are used as RGs, since they are involved in the basic biochemical metabolic processes of the organism, linked to components of the cytoskeleton, and stably expressed in different cells (8, 9).
Several reports have been linked to the ineffectiveness and variable expression of these commonly used RGs under different experimental conditions (8, 10). Reports have shown unstable expression of GAPDH and 18s rRNA in various stages and different tissues of B. mori (11). In B. mori (Strain: Dazao), certain genes involved in basic biochemical processes show variation in different stages, tissues, cell proliferation, and development, which can be a limiting factor in RT-qPCR data normalization (1, 11). The unstable expression can reflect discrepancies in the data; hence, studies by earlier workers have suggested that screening and validation of RGs under specific experimental conditions is necessary (8). TI3S4 and TI3S5 were reported to be stable RGs in B. mori (Dazao) for early 5th instar larvae in different tissues such as silk gland, testes, ovaries, fat body, midgut, integument, hemocytes, and Malpighian tubule (12, 13). Furthermore, RP49/RPs7 and GAPDH were reported to be stable RGs in egg, larva, pupa, and adult stage, and GAPDH, EF1, and RP49/RPs7 showed stable expression in head, midgut, ovary, testis, fat body, Malpighian tubules, silk gland, and epidermis of B. mori (Dazao) (14). Most ribosomal genes and genes involved in eukaryotic translation initiation function were linked to uniform expression in different developmental stages of Chinese B. mori (Strain: Dazao) (1, 15).
The aim of this study is to find suitable RGs for B. mori (Strain: CB-hybrid, PM×CSR2) in different experimental variables such as developmental stages, i.e., late larval instar (LLI), early pupa (EP), mid pupa (MP), late pupa (LP), and adult after 24 h of eclosion (A). Furthermore, the effects of developmental hormone JH antagonist (Precocene 1) and 20HE antagonist (testosterone) were also studied on candidate RGs. The effect of Precocene 1 has been reported to be stage specific in different holometabolous insects. In Chrysomya megacephala, a dose of 200 µg showed 10% mortality, whereas with 300 µg, no mortality was observed (16). Similarly, in Eurygaster integriceps (hemimetabolous), even with the highest dose, no mortality was observed (17). Furthermore, high mortality with lower doses was reported for Spodoptera littoralis and Euprepocnemis plorans plorans (18, 19). Eight candidate RGs were selected based on their stable expression in various experimental conditions in different lepidopteran insects such as Chinese B. mori (Strain: Dazao), Spodoptera exigua, Plutella xylostella, Cryptophlebia peltastica, Thaumatotibia leucotreta, Cydia pomonella, and hymenopteran (Apis mellifera) and dipteran (Drosophila suzukii). These RGs were Translational initiation factor 3 subunit 4 (TI3S4) (12), Translational initiation factor 3 subunit 5 (TI3S5) (12), Ribosomal protein subunit 7 (RPs7) (14), TATA-binding protein association factor (TAF13) (20), Translational initiation factor 4 A (TI4A) (21), Ribosomal protein (RPL32) (22), Elongation factor 1 (EF1) (23), and Arginine kinase (AK) (24). The gene expression analysis is studied for the stability of these in various time points of a 24-h light–dark cycle so as to use selected RGs for rhythmic gene expression in CNS of variable developmental stages upon treatment with developmental hormone antagonists in B. mori. Furthermore, we evaluated two doses of Precocene 1, namely, 200 and 300 µg, per insect to eliminate any dose-specific mortality.
The insects were obtained from the local sericulture department and maintained at LD 12:12 photoperiod, with a temperature of 26 ± 1°C and a relative humidity of 70% ± 5%. Larvae were fed with fresh mulberry leaves provided ad libitum. Various developmental stages such as LLI, EP, MP, LP, and A were used in the study. Adult insects were obtained after 24 h of post eclosion; both male and female insects were included in the study. The insects were ice narcotized and the CNS was dissected out in Insect Ringer’s solution (25). The samples were collected at four different time points, i.e., Zeitgeber time (ZT) 0, 6, 12, and 18.
Two doses of Precocene 1 (CAS-17598–02-6, 1 g/mL, Sigma), 200 µg (Dose 1) and 300 µg (Dose 2), were used in this study. A solution (300 µg/µL) of Precocene 1 was prepared by dissolving 57 µL of Precocene 1 in 200 µL of acetone (16, 26). For larvae, 0.67 and 1 µL of this solution were topically applied to provide 200 and 300 µg/insect, respectively (17), whereas for pupae and adults, 0.67 and 1 µL of Precocene 1 solution (300 µg/µL) were further diluted with insect Ringers solution to 5 µL and injected per insect at the third thoracic segment with the help of Hamilton syringes (27).
A testosterone (CAS-58–22-05 g, HPLC ≥99.0%, Sigma) stock solution (1 mg/mL) was prepared by dissolving it in acetone for testosterone treatment (TT). All stages were treated with 1 mg of testosterone/kg body weight of insect. Larvae, pupae, and adults weighed approximately 2.5, 1.5, and 1.1 g, respectively. For larvae, ~2.5 µL of testosterone stock solution was applied on a square inch of leaf, and larvae were allowed to eat it completely (28). Larvae were not fed prior to dosing in order to ensure complete consumption of the leaf. Pupae and adults were injected with ~1.5 and 1.1 µL of the TT stock solutions after diluting to 5 µL with Insect Ringers solution respectively on the 3rd thoracic segment with the help of Hamilton syringe (27).
Total RNA was extracted using TRI reagent (Sigma) following the manufacturer’s protocol and was dissolved in nuclease-free water. The concentration and purity of RNA were quantified by measuring optical density (OD) at 260 and 280 nm using a Nanodrop spectrophotometer (Thermo Fisher). cDNA synthesis was performed by using the Bioline SensiFAST cDNA synthesis kit by following the manufacturer’s protocol. For RT-qPCR analysis, 4 μl of cDNA (diluted to a concentration of 1:20 in nuclease-free water) was used (29).
Eight candidate RGs were selected (Table 1). TI3S4, TI3S5, and TI4A primers were used as per Wang et al. (12). B. mori gene-specific primers were designed for RGs AK, RPs7, RPL32, TAF13, and EF1 by using genome sequencing data (KAIKO Database) available online. The primary design was done by the PRIMER QUEST software IDT (30).
The RT-qPCR reactions were carried out by the SYBR GREEN detection method in StepOne plus Applied Biosystems, Foster, USA. Thermo Fisher Power SYBR GREEN was used according to the manufacturer’s protocol (30). The cycling conditions consisted of 10 min of holding stage at 95°C, and the cycling stage consisted of 40 cycles of 95°C for 15 s, 60°C for 1 min, followed by one cycle of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The PCR specificity was monitored with melt curve analysis with the help of StepOne software v2.0.2. The specificity and reliability of the primers were validated by melt curves, and each melt curve showed a single sharp peak, suggesting a single gene amplification (Supplementary Figure 1). The cycle threshold (Ct), was determined by a log linear plot of fluorescent signal versus the thermal cycle numbers and is inversely correlated with the amount of template in the reaction (31).
The mean Ct values were calculated based on three biological replicates (n = 3, each sample contained 5 pooled CNS). In order to validate the stability of these eight genes and to identify the stable RG, RefFinder was used. RefFinder is a web-based comprehensive tool (http://www.ciidirsinaloa.com.mx/RefFinder-master/) that contains four universal analysis programs, i.e., comparative ΔCt method, NormFinder, geNORM, and BestKeeper (6, 32). Additionally, an online-based algorithmic tool, geNorm (Biogazelle qBase+), was used separately for further validation of RGs (33). The geNORM tool assessed and suggests the number of RGs needed to standardize gene expression and their stability in a given experimental condition. All the Ct values were collected from RT-qPCR with the help of StepOne software v2.0.2, and these values were used as input data for geNorm (Biogazelle qBase+) software; furthermore, mean Ct calculation was done with Microsoft Excel. As a comprehensive web-based platform, RefFinder integrated the results from geNORM, BestKeeper, NormFinder, and the ΔCt method and ranked the RGs. The graphs were plotted using Graph Pad Prism 7.0 (29).
For gene-specific primers, specificity was confirmed via NCBI BLAST; furthermore, the amplification specificity was validated by single sharp melt curve peaks (Supplementary Figure 1). TI3S4, TI3S5, and TI4A primers were used from a previous study in B. mori Chinese silkworm strain (Strain: Dazao) (12).
The Ct values were converted to mean values for the analysis using Ct value analysis tools. Ct values for TI3S4, TAF13, and RPs7 were observed as 0.43, 1.07, and 1.17, respectively. Based on the Ct value analysis, the order of stability in different developmental stages and time points is TI3S4 > TAF13 > RPs7 > RPL32 > TI4A > TI3S5 > AK > EF1 (Figure 1A). It was observed in the control group that AK, EF1, and RPL32 are showing altered expression in different developmental stages and thus cannot be considered as RGs for further studies. No significant difference was observed in control and vehicle controls in all the stages and time points studied.
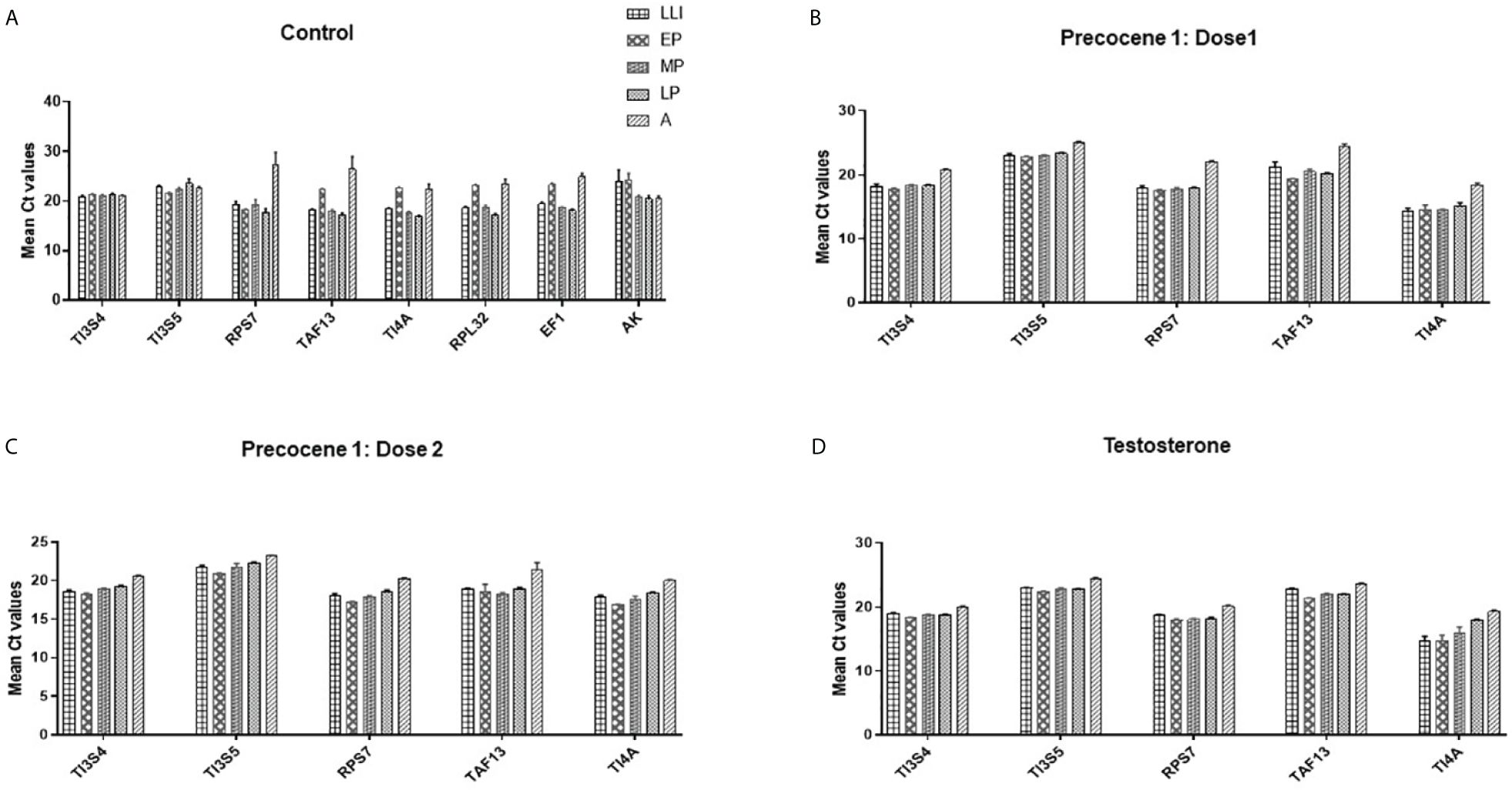
Figure 1 Mean Ct value analysis of the expression stability of selected reference genes in different parameters. (A) Control: TI3S4, TI3S5, and RPs7 showed the most stable expression in all the stages followed by TAF13, TI41, RPL32, EF1, and AK; (B) Precocene 1–Dose 1: 200 µg treatment; (C) Precocene 1–Dose 2: 300 µg treatment; (D) testosterone treatment: TI3S4, TI3S5, and RPs7 showed the most stable expression across the stages followed by TAF13 and TI4A in various PED stages.
The expression stability of eight candidate RGs was analyzed using RefFinder and the geNORM tool separately. RefFinder analysis was done for each developmental stage, and each program generated its individual ranking order (Supplementary Figure 2A). Additionally, a comprehensive analysis over different developmental stages showed RPL32, RPs7, and TI4A as the most stable RGs, and the stability ranking order was RPL32 > RPs7 > TAF13 > TI4A > TI3S4 > TI3S5 > AK > EF1 (Figure 2A).
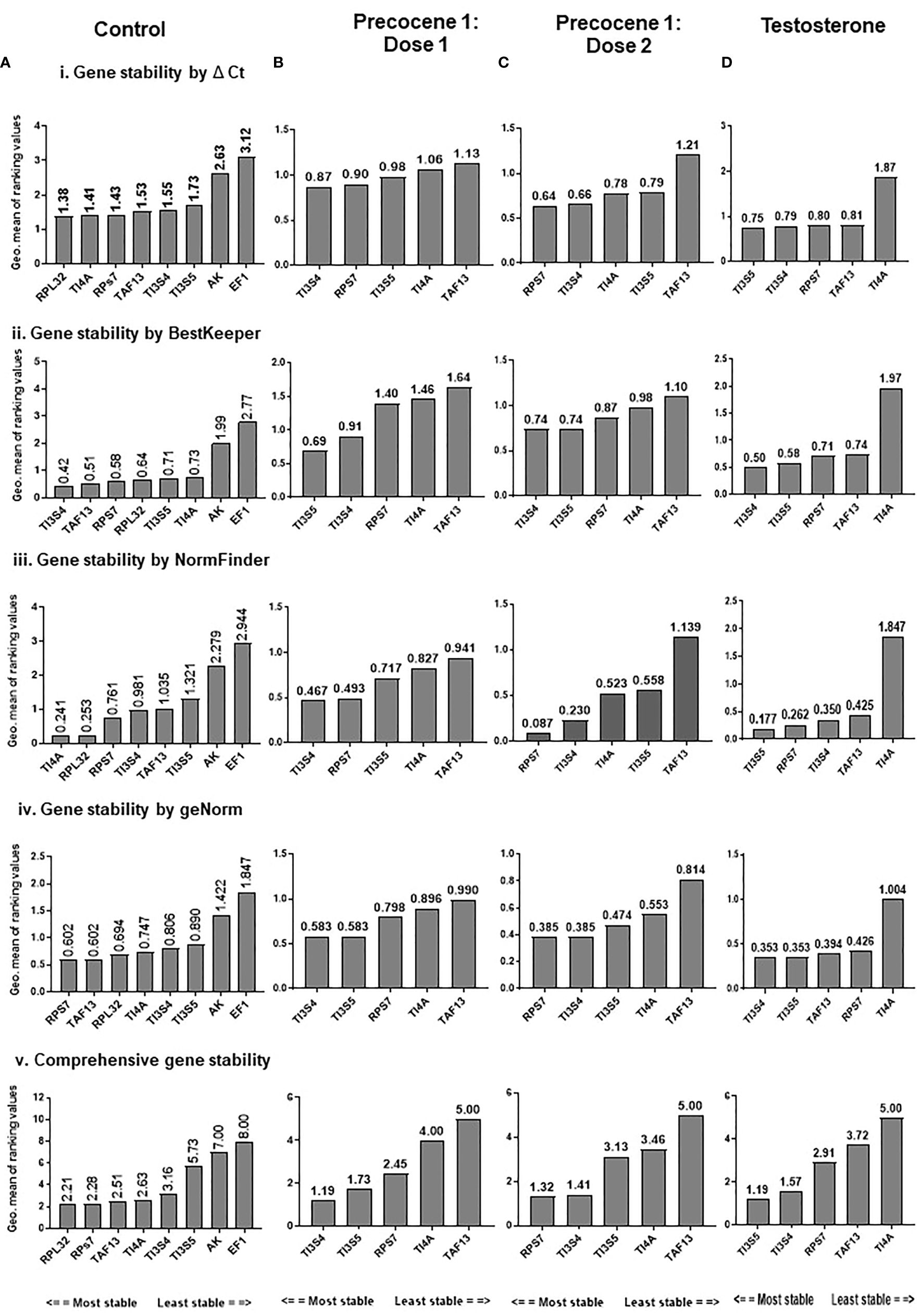
Figure 2 RefFinder analysis of candidate reference genes across the developmental stages of B mori; RefFinder integrates the results from the ΔCt method, BestKeeper, NormFinder, and geNORM and ranking the RGs based on their stability. (i) Gene stability ranking by ΔCt analysis; (ii) gene stability ranking order by BestKeepr; (iii) gene stability ranking order by NormFinder; (iv) gene stability ranking order by geNORM; (v) comprehensive ranking order of RGs by RefFinder. (A) Control, (B) Precocene 1–Dose 1: 200 µg treatment, (C) Precocene 1–Dose 2: 300 µg treatment, and (D) testosterone treatment. The genes are present with the stability decreasing from left to right.
Gene stability was calculated using the geNorm algorithm (Biogazelle qBase+) tool following Vandesompele et al. (34) and Hellemans et al. (33). The candidate RGs were ranked depending on their expression in four time points and PED stages. TAF13, TI3S5, RPs7, RPL32, TI4A, and TI3S4 showed an M value of less than 1.5, which indicates their stable expression in the provided experimental variation. On the other hand, RPL32, AK, and EF1 were considered as the least stable genes with M values of 2.01, 2.669, and 3.139, respectively (Table 2). The average pairwise variations V4/5 and V5/6 were below 0.15, suggesting that a maximum of five appropriate genes can act as potential RGs for qRT-PCR analysis (Figure 3A). The genes with the lowest M value were used for further validation under antagonist treatments (Table 2). Thus, stability ranking was established as TI4A > TI3S5 > TI3S4 > TAF13 > RPs7 > RPL32 > AK > EF1.
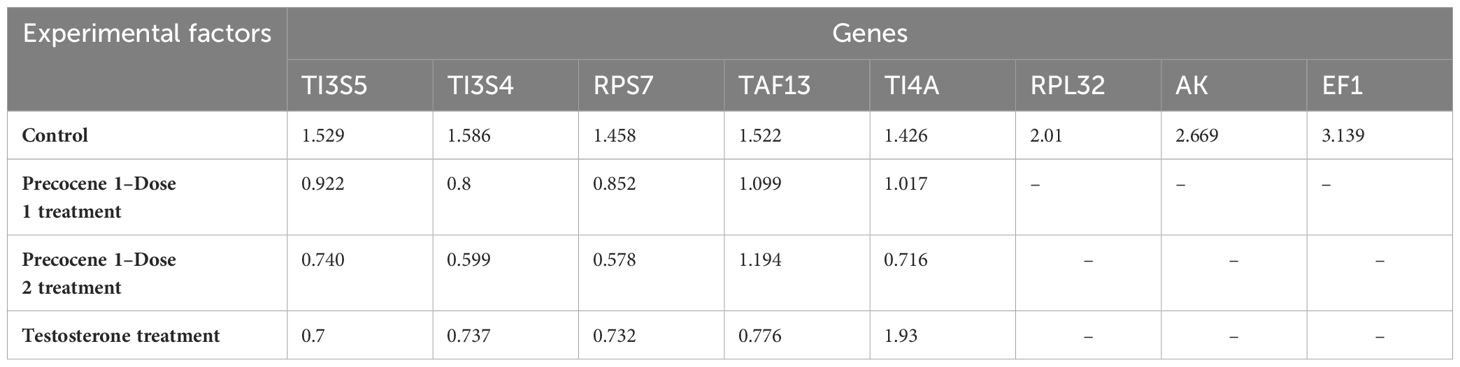
Table 2 Gene stability values (M values) of reference genes in different experimental conditions in Bombyx mori generated by the geNorm algorithm (Biogazelle qBase+).
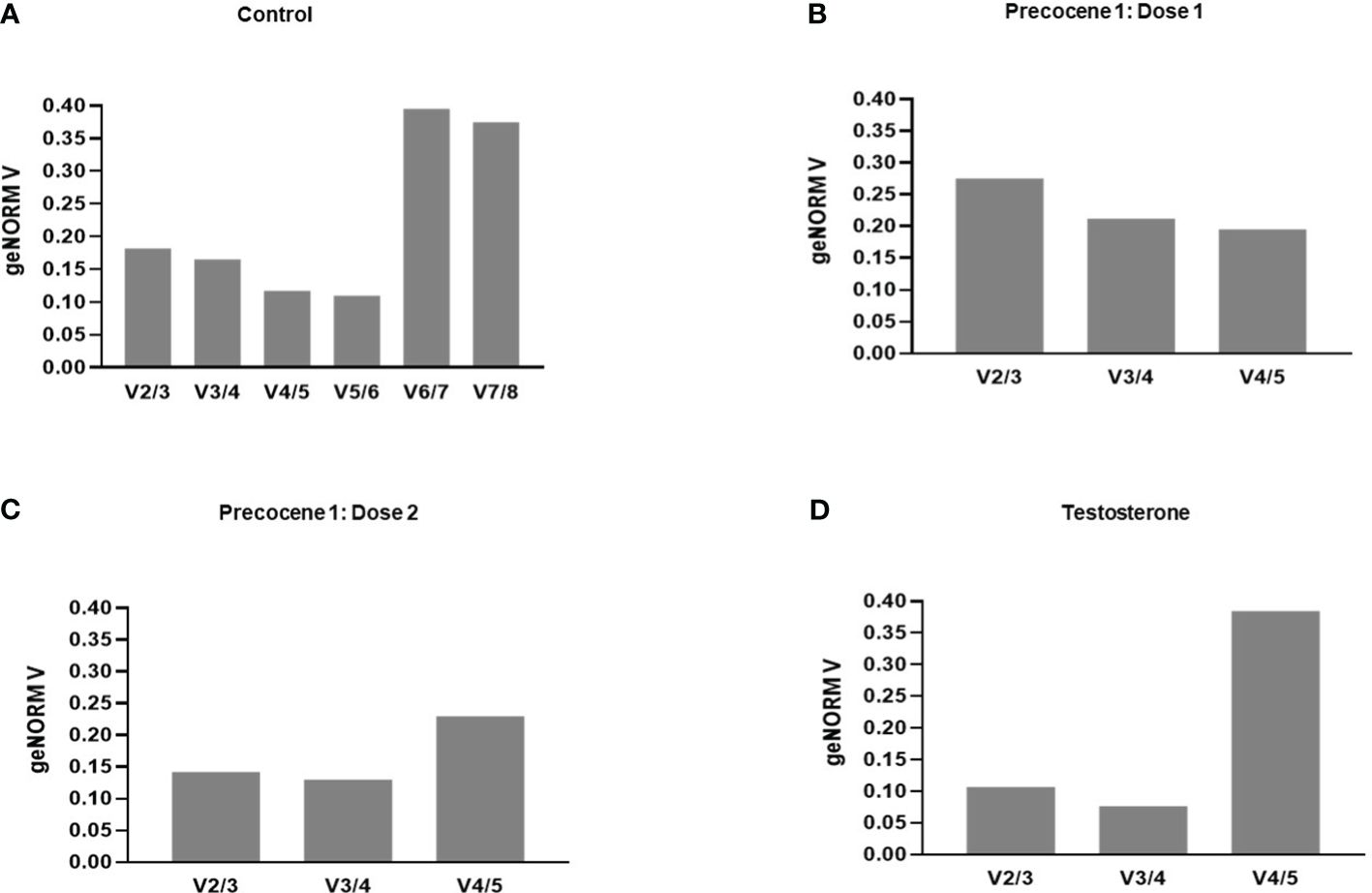
Figure 3 Pairwise variation (V) of the candidate reference genes by B mori by the geNorm algorithm (Biogazelle qBase+). Vn/n+1<0.15 suggests that “n” number of optimum RGs are required for the respective experimental (A) control, (B) Precocene 1–Dose 1: 200 µg treatment, (C) Precocene 1–Dose 2: 300 µg treatment, and (D) testosterone treatment.
Based on the Ct difference in PED stages, the order of RGs’ stability in different developmental stages and Precocene 1 dose 200 µg/insect treatment showed that the range of expression of candidate RGs is TI3S5 > TI3S4 > TI4A > RPs7 > TAF13 (Figure 1B). However, in Precocene 1 dose 300 µg/insect and TT, the Ct values ranking was found as TI3S5 > TI3S4 > RPs7 > TAF13 > TI4A (Figure 1C). This suggested that TI3S5, TI3S4, and RPs7 can be considered as stable RGs under different developmental hormone antagonistic treatments (Figure 1D).
RefFinder analysis was done under antagonist treatment in different developmental stages. In each stage ΔCt, BestKeeper, NormFinder and geNORM analysis showed differential ranking of RGs (Supplementary Figure 2). The recommended comprehensive ranking in the Precocene 1–200 µg treatment was TI3S4 > TI3S5 > RPs7 > TI4A > TAF13 and that in the Precocene 1–300 µg treatment was RPs7 > TI3S4 > TI3S5 > TI4A > TAF13. However, TT showed Ct value ranking as TI3S5 > TI3S4 > RPs7 > TAF13 > TI4A (Figures 2B–D).
Evaluation of RGs with the geNORM tool was carried out using the treatments. The treatment with Precocene 1 Dose 1 showed lower M values for TI3S4, RPs7, and TI3S5, i.e., 0.8, 0.852, and 0.922, respectively, than TAF13 and TI4A. However, with Precocene 1 Dose 2, the M values of TI4A, RPs7, and TI3S4 were the lowest, suggesting higher stability compared to TI3S5 and TAF13 (Table 2). Thus, the stability ranking in Precocene 1 Dose 1 was TI3S4 > RPs7 > TI3S5 > TI4A > TAF13 and that in Precocene 1 Dose 2 was RPs7 > TI3S4 > TI4A > TI3S5 > TAF13. With TT, TI3S5, RPs7, TI3S4, and TAF13 showed M values of 0.7, 0.737, 0.732, and 0.776, respectively, whereas TI4A showed a higher M value of 1.93. Therefore, the ranking stability was TI3S5 > RPs7 > TI3S4 > TAF13 > TI4A (Table 2). In Precocene 1 Dose 1, no pairwise variation was below 0.15, whereas in Precocene 1 Dose 2 and with TT, both V2/3 and V3/4 were below 0.15, suggesting that a maximum of three potential RGs can be used in these antagonist treatments (Figures 3B–D).
As a prerequisite towards the selection of an appropriate RG for understanding the effect of antagonists of insect developmental hormones on rhythmic gene expression in various PED stages in CNS of B. mori, we studied the expression stability of eight candidate RGs.
The RefFinder tool gives the stability ranking of each candidate gene by using various programs such as ΔCt, BestKeeper, NormFinder, and geNORM. The ranking orders are based on different statistical endpoints and algorithms, and the weight of each gene is determined by calculating the geometric mean (32, 35). Furthermore, the geNORM (Biogazelle qBase+) tool was used to validate the results through the calculation of pairwise variation value (Vn/n+1); a value of less than 0.15 indicates the optimum number of RGs for qRT-PCR, and it also provides a stability value (M) for the candidate genes (10, 23) (Figure 3, Table 2). Most of the ranking analyses obtained with different programs were consistent, suggesting the reliability of the results.
The stability of Ct values is crucial in determining the reliability of an RG, which is inversely proportional to the gene expression. The Ct values of candidate RGs varied from 16.92 to 27.43. The Ct values for RPL32 and RPs7 were found to be approximately 16, suggesting the optimum expression of these genes (36). In hemimetabolous, Thysanoptera insect Megalurothrips usitatus RPL and RPs were reported to be the most stable RGs in different developmental stages and under the treatment of insecticides (35). Furthermore, in another hemimetabolous, hemipteran insect Eocanthecona furcellata and holometabolous lepidopteran insect Spodoptera frugiperda, RPL32 and RPL13 along with some other RGs were reported to be the most stable RGs in different developmental stages (37, 38). In the current study, according to RefFinder and Ct analysis in the non-treated group, RPL32 showed a quite stable expression in different developmental stages of B. mori, but a separate analysis with geNORM showed it to be a fairly less stable expressing gene in developmental stages; however, RPs7 stability was consistent throughout the developmental stages. RP 49/RPs7 was identified as the most stable RG for developmental studies in A. mellifera (20). Ribosomal proteins are responsible for ribosomal assembly and protein translation, contributing towards cell development and has been found to be the most stable gene for RT-qPCR studies (38). Few studies by the Qing-You Xia group have demonstrated that TI3S4, TI4A, and TI3S4 are stable RGs in early 5th instar larvae, 60 h after wandering (a stage between 5th late larval instar and early pupa), pupa, and adult B. mori insects (12). Additionally, TI3S4 and TI3S5 were observed to be stable RGs in various tissues of B. mori, which include silk glands, testes, ovaries, fat body, midgut, integument, hemocytes, and Malpighian tubules, specifically in early 5th instar larvae (13). However, other workers reported RP49/RPs7 and GAPDH as stable RGs in different stages of Chinese B. mori development, including egg, larva, pupa, and adult stages (14). Furthermore, GAPDH, EF1, and RP49/RPs7 were found to exhibit stable expression in various tissues of B. mori, such as the head, midgut, ovary, testis, fat body, Malpighian tubules, silk glands, head, and epidermis. A global transcriptome analysis conducted by Wang et al. (1) revealed that most ribosomal genes and genes involved in eukaryotic translation initiation function exhibit uniform expression across different developmental stages of B. mori.
Various studies of Chinese B. mori have consistently shown that the expression levels of multiple candidate RGs are influenced by specific experimental conditions, thus indicating that there is no universally applicable RG that can reliably serve under different experimental conditions. Hence, it is strongly recommended by few researchers to conduct a thorough validation and select custom RGs for any provided experimental conditions, even while working within the same species (15, 39). It is also suggested to use multiple RGs for the normalization of data; however, some researchers have reported the use of single RGs for normalization in insecticide treatment, developmental stage, and light conditions, and in different temperature conditions (34, 35, 40).
We report TI3S4, TI3S5, and RPs7 to be the most stable RGs in CNS under various experimental conditions such as different PED stages, different time points, and under two developmental hormone antagonist treatments in Indian B. mori (Strain: CB-hybrid, PM×CSR2). Another candidate RG, RPL32, can be used for gene expression studies in B. mori PED stages at various time points; however, it was not stable under developmental hormone antagonist treatment.
The RGs identified here will be used to study various clock gene expressions in CNS in different PED stages of B. mori at various ZTs to help understand the role of antagonists Precocene 1 and testosterone for developmental hormone JH and ecdysone in circadian function.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The manuscript presents research on animals that do not require ethical approval for their study.
MD: Conceptualization, Data curation, Formal Analysis, Methodology, Writing – original draft. AJ: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. AJ is thankful to the Builder and IoE Grants (UoH-IoE-RC-21–055).
MD is thankful to UGC for the fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finsc.2024.1362473/full#supplementary-material
Supplementary Figure 1 | Melt curves of selected reference genes showing single amplification.
Supplementary Figure 2 | RefFinder analysis of candidate reference genes in different developmental stages of B. mori. (i) Gene stability ranking by ΔCt analysis; (ii) gene stability ranking order by BestKeepr; (iii) gene stability ranking order by NormFinder; (iv) gene stability ranking order by geNORM; and (v) comprehensive ranking order of RGs by RefFinder. (A) Control, (B) Precocene 1–Dose 1: 200 µg treatment, (C) Precocene 1–Dose 2: 300 µg treatment, and (D) testosterone treatment.
TT, testosterone treatment.
1. Wang X, Fan Y, Ge Q, Xu J, Taha RH, Yuan Y, et al. Time-Course Transcriptome Analysis Reveals Global Gene Expression Profiling and Dynamic Developmental Signatures across Complete Life Cycle of Bombyx mori. Processes. (2021) 9:1730. doi: 10.3390/pr9101730
2. Kent KS, Consoulas C, Duncan K, Johnston RM, Luedeman R, Levine RB. Remodelling of neuromuscular systems during insect metamorphosis. Am zool. (1995) 35:578–84. doi: 10.1093/icb/35.6.578
3. Yan X, Zhang Y, Xu K, Wang Y, Yang W. Selection and validation of reference genes for gene expression analysis in Tuta absoluta Meyrick (Lepidoptera: Gelechiidae). Insects. (2021) 12:589. doi: 10.3390/insects12070589
4. Gao P, Wang J, Wen J. Selection of reference genes for tissue/organ samples of adults of Eucryptorrhynchus scrobiculatus. PloS One. (2020) 15:e0228308. doi: 10.1371/journal.pone.0228308
5. Tian P, Qiu L, Zhou A, Chen G, He H, Ding W, et al. Evaluation of appropriate reference genes for investigating gene expression in Chlorops oryzae (Diptera: Chloropidae). J Econ Entomol. (2019) 112:2207–14. doi: 10.1093/jee/toz142
6. Bai Y, Lv YN, Zeng M, Jia PY, Lu HN, Zhu YB, et al. Selection of reference genes for normalization of gene expression in Thermobia domestica (Insecta: Zygentoma: Lepismatidae). Genes. (2020) 12:21. doi: 10.3390/genes12010021
7. Pinheiro DH, Moreira RO, Leite NA, Redoan AC, Xavier ADS, Barros BDA, et al. Suitable reference genes for RT-qPCR analysis in Dichelops melacanthus (Hemiptera: Pentatomidae). Mol Biol Rep. (2020) 47:4989–5000. doi: 10.1007/s11033-020-05550-z
8. Zhou L, Meng JY, Ruan HY, Yang CL, Zhang CY. Expression stability of candidate RT-qPCR housekeeping genes in Spodoptera frugiperda (Lepidoptera: Noctuidae). Arch Insect Biochem Physiol. (2021) 108:e21831. doi: 10.1002/arch.21831
9. Wang W, Ling X, Bashir NH, Lu Q, Zhang J, Li T, et al. Selection and evaluation of reference genes for quantitative real-time PCR analysis in lac insect (Kerria lacca). Entomol Res. (2022) 52:57–67. doi: 10.1111/1748-5967.12565
10. Yin J, Sun L, Zhang Q, Cao C. Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene. (2020) 749:144712. doi: 10.1016/j.gene.2020.144712
11. Peng R, Su BJ, Zhao GD, Ji X, Zhao SS, Zhang T, et al. Standardization of reference genes in silkworm, Bombyx mori. Adv Mater Res. (2011) 175:67–71. doi: 10.4028/www.scientific.net/AMR.175-176.67
12. Wang GH, Xia QY, Cheng DJ, Duan J, Zhao P, Chen J, et al. Reference genes identified in the silkworm Bombyx mori during metamorphism based on oligonucleotide microarray and confirmed by qRT-PCR. Insect Sci. (2008) 15:405–13. doi: 10.1111/j.1744-7917.2008.00227.x
13. Wang G, Chen Y, Zhang X, Bai B, Yan H, Qin D, et al. Selection of reference genes for tissue/organ samples on day 3 fifth-instar larvae in silkworm, Bombyx mori. Arch Insect Biochem Physiol. (2018) 98:e21458. doi: 10.1002/arch.21458
14. Teng X, Zhang Z, He G, Yang L, Li F. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. J Insect Sci. (2012) 12:60. doi: 10.1673/031.012.6001
15. Shakeel M, Rodriguez A, Tahir UB, Jin F. Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol Lett. (2018) 40:227–36. doi: 10.1007/s10529-017-2465-4
16. Singh S, Kumar K. Sensitivity of last larval stadium of Chrysomya megacephala (Fabricius) to anti-allatin ageratochromene Precocene II. Indian J Exp Biol. (2011) 49:600–8.
17. Amiri A, Bandani AR, Ravan S. Effect of an anti-juvenile hormone agent (Precocene I) on Sunn pest, Eurygaster integriceps (Hemiptera: Scutelleridae) development and reproduction. Afr J Biotechnol. (2010) 9:5859–68.
18. Khafagi WE, Hegazi EM. Latent effects of precocenes (I and II) and juvenile hormone I on Spodoptera littoralis (Boisd.) larvae. Arch Phytopathol Plant Prot. (1999) 32:337–50. doi: 10.1080/03235409909383302
19. Ghoneim K, Basiouny A. Insecticidal and antijuvenile hormone activities of Precocene II against the grasshopper Euprepocnemis plorans (Charp.) (Orthoptera: Acrididae). Int J Trend Sci Res Dev. (2017) 1:510–24. doi: 10.31142/ijtsrd4603
20. Lourenço AP, Mackert A, dos Santos Cristino A, Simões ZLP. Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie. (2008) 39:372–85. doi: 10.1051/apido:2008015
21. Guo H, Jiang L, Xia Q. Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol Genet Genomics. (2016) 291:999–1004. doi: 10.1007/s00438-015-1125-4
22. Fu W, Xie W, Zhang Z, Wang S, Wu Q, Liu Y, et al. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int J Biol Sci. (2013) 9:792. doi: 10.7150/ijbs.5862
23. Ridgeway JA, Timm AE. Reference gene selection for quantitative real-time PCR normalization in larvae of three species of Grapholitini (Lepidoptera: Tortricidae). PloS One. (2015) 10:e0129026. doi: 10.1371/journal.pone.0129026
24. Zhai Y, Lin Q, Zhou X, Zhang X, Liu T, Yu Y. Identification and validation of reference genes for quantitative real-time PCR in Drosophila suzukii (Diptera: Drosophilidae). PloS One. (2014) 9:e106800. doi: 10.1371/journal.pone.0106800
25. Miller TA. Insect neurophysiological techniques. New York: Springer-Verlag (1979). doi: 10.1007/978-1-4612-9973-8
26. Triselyova TA. Antijuvenile influence of the precocene on the development of adult antennae in the large fruit-tree tortrix Archips podana Scop. (Lepidoptera: Tortricidae). Biol Bull. (2012) 39:423–9. doi: 10.1134/S1062359012050147
27. Kamimura M, Takahashi M, Kikuchi K, Reza AMS, Kiuchi M. Tissue-specific regulation of juvenile hormone esterase gene expression by 20-hydroxyecdysone and juvenile hormone in Bombyx mori. Arch Insect Biochem Physiol. (2007) 65:143–51. doi: 10.1002/arch.20186. Published in Collaboration with the Entomological Society of America.
28. Das S. Vertebrate hormones in insects: the role of estrogen in silkworm-a review. Turkish J Zool. (2016) 40:297–302. doi: 10.3906/zoo-1507-37
29. Kukkemane K, Jagota A. Therapeutic effects of curcumin on age-induced alterations in daily rhythms of clock genes and Sirt1 expression in the SCN of male Wistar rats. Biogerontology. (2019) 20:405–19. doi: 10.1007/s10522-018-09794-y
30. Mattam U, Jagota A. Differential role of melatonin in restoration of age-induced alterations in daily rhythms of expression of various clock genes in suprachiasmatic nucleus of male Wistar rats. Biogerontology. (2014) 15:257–68. doi: 10.1007/s10522-014-9495-2
31. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
32. Xie F, Wang J, Zhang B. RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct Integr Genomics. (2023) 23:1–5. doi: 10.1007/s10142-023-01055-7
33. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. (2007) 8:1–14. doi: 10.1186/gb-2007-8-2-r19
34. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. (2002) 3:1–12. doi: 10.1186/gb-2002-3-7-research0034
35. Hou Q, Yuan L, Jin H, Yan H, Li F, Wu S. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Megalurothrips usitatus (thysanoptera: thripidae). Front Physiol. (2023) 14:1161680. doi: 10.3389/fphys.2023.1161680
36. Zheng C, Zhao D, Xu Y, Shi F, Zong S, Tao J. Reference gene selection for expression analyses by qRT-PCR in Dendroctonus valens. Insects. (2020) 11:328. doi: 10.3390/insects11060328
37. Shu BS, Yu HK, Dai JH, Xie ZG, Qian WQ, Lin JT. Stability evaluation of reference genes for real-time quantitative PCR normalization in Spodoptera frugiperda (Lepidoptera: Noctuidae). J Integr Agric. (2021) 20:2471–82. doi: 10.1016/S2095-3119(20)63298-1
38. Pan YN, Zhao RN, Fu D, Yu C, Pan CN, Zhou W, et al. Assessment of Suitable Reference Genes for qRT-PCR Normalization in Eocanthecona furcellata (Wolff). Insects. (2022) 13:773. doi: 10.3390/insects13090773
39. Lü J, Yang C, Zhang Y, Pan H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front Physiol. (2018) 9:1560. doi: 10.3389/fphys.2018.01560
Keywords: daily rhythms, Bombyx mori, CNS, RT-qPCR, reference gene, PED, Precocene 1 and testosterone
Citation: Dalai M and Jagota A (2024) Identification of specific reference gene for normalization of RT-qPCR data in rhythmic gene expression studies of the effect of developmental hormone antagonist in postembryonic development in Bombyx mori. Front. Insect Sci. 4:1362473. doi: 10.3389/finsc.2024.1362473
Received: 04 January 2024; Accepted: 05 June 2024;
Published: 28 June 2024.
Edited by:
Suresh Kumar Kalangi, Central Drug Research Institute (CSIR), IndiaReviewed by:
Han Wang, Soochow University, ChinaCopyright © 2024 Dalai and Jagota. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anita Jagota, YWpzbEB1b2h5ZC5hYy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.