
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Insect Sci. , 16 January 2023
Sec. Insect Molecular Genetics
Volume 3 - 2023 | https://doi.org/10.3389/finsc.2023.1113871
This article is part of the Research Topic Women in Insect Science View all 7 articles
 Najla M. Albishi†
Najla M. Albishi† Subba Reddy Palli*
Subba Reddy Palli*The function of two autophagy genes, an activating molecule BECN1 regulated autophagy (AMBRA1) and autophagy-related gene 8 (ATG8) in the midgut remodeling of Aedes aegypti was investigated. Real-time quantitative polymerase chain reaction (RT-qPCR) analysis of RNA samples collected from the last instar larvae and pupae showed that these two genes are predominantly expressed during the last 12 h and first 24 h of the last larval and pupal stages, respectively. Stable ecdysteroid analog induced and juvenile hormone (JH) analog suppressed these genes. RNA interference (RNAi) studies showed that the ecdysone-induced transcription factor E93 is required for the expression of these genes. JH-induced transcription factor krüppel homolog 1 (Kr-h1) suppressed the expression of these genes. RNAi-mediated silencing of AMBRA1 and ATG8 blocked midgut remodeling. Histological studies of midguts from insects at 48 h after ecdysis to the final larval stage and 12 h after ecdysis to the pupal stage showed that ATG gene knockdown blocked midgut remodeling. AMBRA1 and ATG8 double-stranded (dsRNA)-treated insects retained larval midgut cells and died during the pupal stage. Together, these results demonstrate that ecdysteroid induction of ATG genes initiates autophagy programmed cell death during midgut remodeling. JH inhibits midgut remodeling during metamorphosis by interfering with the expression of ATG genes.
Metamorphosis is associated with tissue remodeling in holometabolous insects, including the yellow fever mosquito, Aedes aegypti (1). Molting and metamorphosis are regulated by juvenile hormone (JH) and 20-hydroxyecdysone (20E is the most active form of ecdysteroids). JH is an anti-metamorphosis hormone essential for preventing metamorphosis (2). Steroid-induced programmed cell death is involved in the degeneration of the larval midgut, salivary glands, fat body, and other tissues and has been extensively studied in holometabolous insects (3). Programmed cell death (PCD) plays a vital role in insect metamorphosis and development. Programmed cell death, which includes apoptosis and autophagy, has been described only in few insects (3). However, most of the research is focused on apoptosis. Autophagic programmed cell death is a cellular mechanism highly conserved from yeast to mammals and depends on the lysosomal degradation pathway for elimination of dysfunctional cellular components. In eukaryotic cells, autophagy is regulated by a series of autophagy-related proteins (ATG) that function in the cellular process of autophagy: induction, nucleation, expansion, and completion of the autophagosome, which is followed by lysosomal fusion (4). The protein activating molecule BECN1 regulated autophagy (AMBRA1) is a novel regulator of autophagy that interacts with Beclin-1 (interact with BCL-2 protein) and stimulates its binding to vacuolar protein sorting-associated protein 34 (Vps34), which plays a role in autophagosome formation during autophagy (5). Increasing evidence of a pivotal role of AMBRA1 protein in autophagy and apoptosis has been reported in vertebrate neurodevelopment in previous years (6). A mutation in the AMBRA1 gene impairs the regulation of autophagy in mice and alters the balance between apoptotic cell death and proliferation and resulting in embryonic lethality (7). From yeast to mammals, two conjugation systems are involved in the autophagosome formation process: the formation of the ATG12-5-16 complex on the isolation membrane and the localization of ATG8-PE to the isolation membrane. ATG12 and ATG8, ubiquitin-like proteins, play crucial roles in phagophore expansion and the formation of autophagosomes, which also requires other ATG proteins such as ATG4, ATG3, ATG7, and ATG10 (4). The ubiquitin-like protein ATG8 promotes the expansion of the isolation membrane and autophagosome membrane formation. ATG8 remains attached to the autophagosome until it is trafficked to the lysosome, where ATG4 releases it after the autophagosome is fused with the lysosome to form the autolysosome (8). Because of these reasons, ATG8 protein has been utilized as a marker for autophagic activity and autophagosome formation (9).
In insects, autophagy is an integral part of developmental processes in the remodeling of larval tissues (10, 11). In the fruit fly, Drosophila melanogaster, autophagy, and apoptosis are involved in the degradation of larval salivary glands; the silencing of ATG genes impairs the elimination of this organ (10). Autophagy has been reported to be involved in larval midgut degradation. Larval midgut degradation was delayed by inhibition of autophagy signaling in the ATG2 mutant larvae or knockdown of ATG1 and ATG18 by RNAi during metamorphosis. In contrast, the overexpression of ATG1 triggers premature midgut degradation (12). In lepidopteran insects, the removal of the larval tissues requires both the canonical apoptosis machinery and autophagy detected in the midgut (13), silk glands (14), and fat body (15, 16) during metamorphosis. The function of ATG, the critical regulator protein during metamorphosis, is largely unknown in the Ae. aegypti midgut remodeling.
The induction of autophagy by 20E has been explored in the midgut and fat body (13, 17). The PCD is promoted by 20E in the absence of JH; the increased levels of 20E induce apoptosis and autophagy (16, 17). In D. melanogaster, E93 influences autophagy by regulating a subset of ATG (10, 18), and ecdysone-response genes (BR-C, E74, HR3, and βftz-F1). The Target-of-Rapamycin (TOR) negatively regulates autophagy in the D. melanogaster fat body during larval development, and 20E directly regulates autophagy by targeting the PI3K pathway (19, 20). Here, we have examined the function of ATG genes in the Ae. aegypti midgut remodeling. To learn insights into autophagy’s role in Ae. aegypti midgut remodeling, we studied two core autophagy genes, AMBRA1 and ATG8 and discovered that the interaction between JH, 20E and autophagy plays a crucial role in regulating autophagy-dependent midgut remodeling in Ae. aegypti.
Ae. aegypti mosquitoes from Liverpool IB12 (LVP-IB12) strain were maintained in the laboratory at 27 ± 1°C temperature and 70-80% relative humidity with a photoperiod of 16:8 light/dark cycle, as previously described (1). The developmental markers were used to identify the stages of mosquito larvae (21).
Total RNA was isolated and used to quantify mRNA levels using gene-specific primers (Table S1) and RT-qPCR as described previously (1). The RPS7 gene (AAEL009496) was used as a reference gene for normalization, and the 2−ΔΔCT method was used to calculate the relative mRNA levels.
For dsRNA preparation, fragments of the target genes were amplified from genomic DNA using Taq Polymerase (Taq 2XMaster Mix, NEB), Double-stranded RNA synthesis and preparation of poly-L-lysine (PLL), epigallocatechin gallate (EGCG),and dsRNA nanoparticles were prepared as described previously (22). Diet pellets containing 50 µg of AMBRA1, ATG8, E93, Kr-h1, or GFP dsRNA were made by mixing the dsRNA/PLL/EGCG complexes with a Bovine liver powder diet and were fed to the early third instar larvae (15 larvae per pellet) daily until they pupated or died. The knockdown efficiency in the 4th instar larval stage was determined using RT-qPCR.
For midgut morphological analysis, midgut dissected from larvae at 48 h AEFL and pupae at 12 h AEPS were dissected, fixed, stained, and photographed as described previously (1). The midgut sections were cut, processed, and photographed.
Additional details on Methods used in these studies are included in the supplementary information.
RT-qPCR was used to determine mRNA levels of two autophagy-related genes, AMBRA1 and ATG8, in the midguts collected at 6 h intervals during the larval-pupal metamorphosis. The results showed that AMBRA1 mRNA levels increased at the end of the final instar larval stage and reached the maximum levels by 0 h after ecdysis into pupal stage (AEPS), then decreased by 6 h AEPS, and remained low during the rest of pupal stage (Figure 1A). In contrast, the ATG8 mRNA levels increased beginning at 42 h after ecdysis to final instar larval stage and reached the maximum levels at 6 h AEPS. The mRNA levels then decreased to reach undetectable levels by 24 h AEPS. (Figure 1A). The mRNA levels of AMBRA1 and ATG8 were measured in different tissues, including the brain, midgut, fat body, and epidermis of the last instar larval and pupal stages. In general, higher levels of AMBRA1 and ATG8 mRNAs were detected in the midgut and fat body than in the other two tissues (Figure 1S). Expression of both AMBRA1 and ATG8 during the last 12 h of the last instar larval stage and early pupal stage and their expression in the midgut of larval and pupal stages suggest their involvement in midgut remodeling during the metamorphosis of Ae. aegypti.
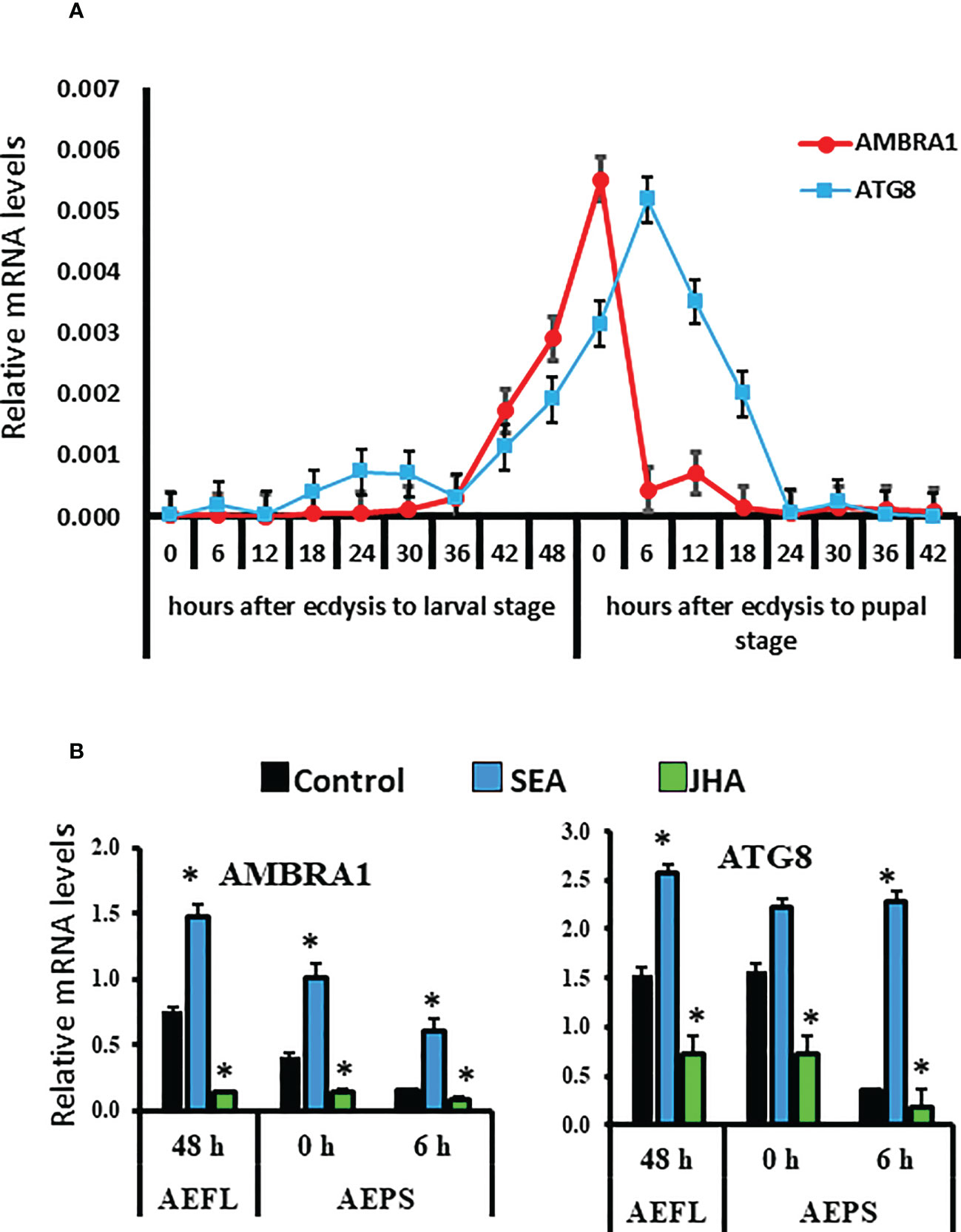
Figure 1 Developmental expression and hormone response of AMBRA1 and ATG8 genes during the final instar larval and pupal stages of Aedes aegypti. (A) Developmental expression of two autophagy genes AMBRA1 and ATG8 in the midgut of last instar larvae and pupae. Total RNA was isolated from midguts from staged insects. The cDNA and gene specific primers (Table S1) were used in RT-qPCR to determine relative mRNA levels of AMBRA1 and ATG8. The RPS7 gene was used as a reference gene for normalization, and the 2−ΔΔCT method was used to calculate the relative mRNA levels. (B) Induction of AMBRA1 and ATG8 by stable ecdysteroid analog (SEA) and their suppression by JH analog (JHA). Relative mRNA levels are shown as Mean ± SE (n=4). The asterisks indicate significant difference at a p-value of <0.05.
To study the effect of hormones on the midgut remodeling, the midguts were dissected from larvae at 48 h after ecdysis into the final instar larval stage (AEFL), and pupae at 0 and 6 h AEPS developed from larvae exposed to JH analog (JHA) methoprene, stable ecdysteroid analog (SEA) RH-102240 or the control larvae treated with DMSO. RT-qPCR analysis of RNA isolated from these midguts showed that E93 and USP-A mRNA levels increased in SEA-treated insects and Kr-h1 mRNA levels increased in JHA-treated insects compared to their levels in control insects, suggesting that hormone analogs are active and functioning as expected (Figure 2S). The mRNA levels of AMBRA1 and ATG8 are higher in SEA-treated insects and lower in JHA-treated insects compared to their levels in control insects (Figure 1B). Similar response to SEA and JHA was detected for other ATG genes studied (Figure 2S). To determine if E93 and Kr-h1 regulate the expression of ATG genes, AMBRA1 and ATG8 mRNA levels were determined in Ae. aegypti larvae fed on dsKr-h1 and dsE93 nanoformulations. Due to the challenge of delivering naked dsRNA to Ae. aegypti larvae, diet pellets containing PLL/EGCG nanoformulated dsRNA were fed to early third instar larvae. The dsE93 treated larvae developed to the pupal stage, but the adult development was blocked and died during the pupal stage (Figure 2A). The dsKr-h1 treated larvae did not undergo metamorphosis and died during the last instar larval stage (Figure 2B). The AMBRA1 and ATG8 mRNA levels increased in insects fed on dsKr-h1 and decreased in insects fed on dsE93 (Figures 2A, B). These results showed the antagonistic effects of E93 and Kr-h1 on the expression of the AMBRA1 and ATG8 in Ae. aegypti suggesting that these two transcription factors regulate the expression of AMBRA1 and ATG8.
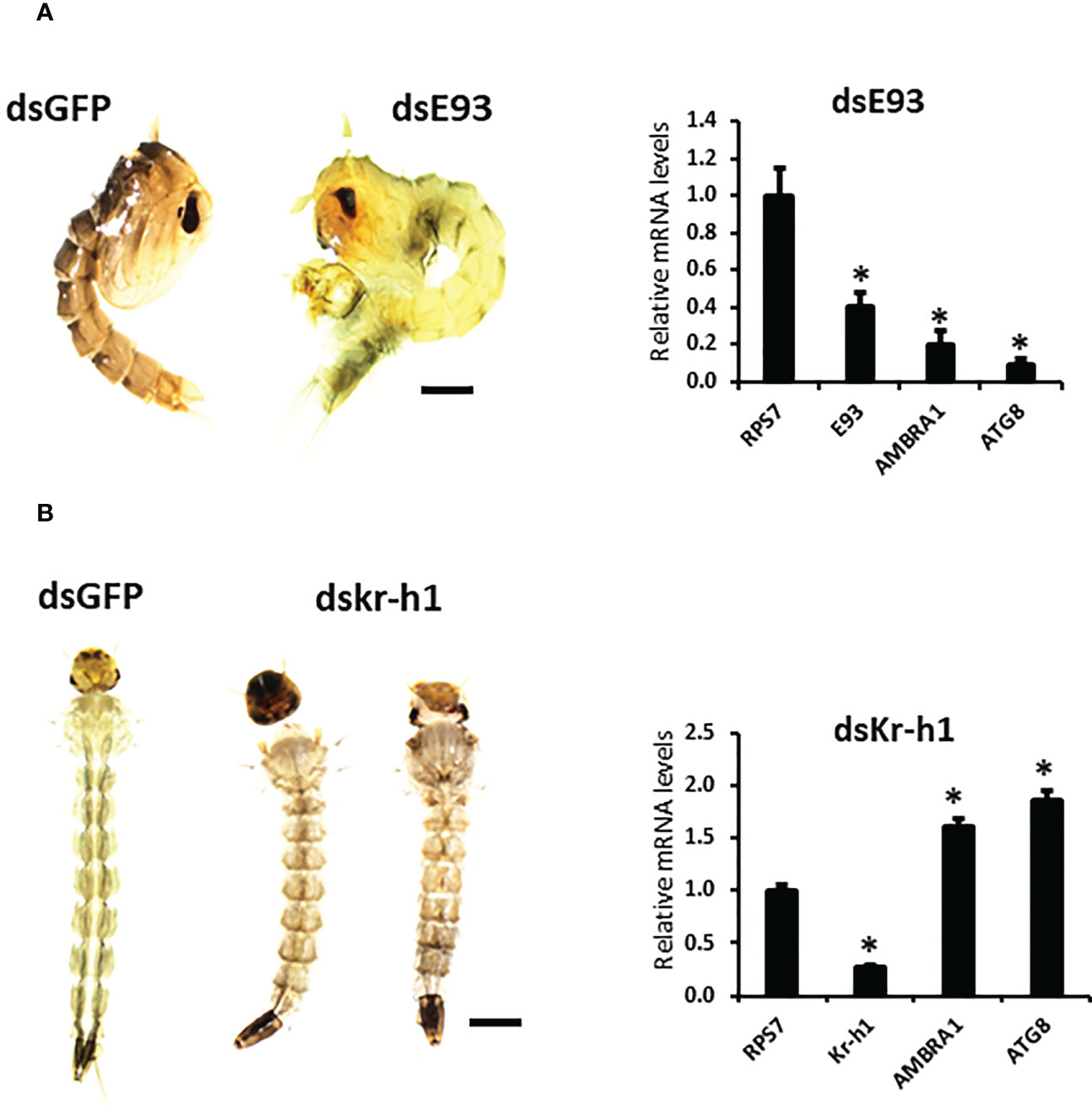
Figure 2 Decreased expression of ATG genes in E93 and Kr-h1 knockdown Aedes aegypti larvae. (A) E93 is required for the 20E-dependent activation of ATG genes. Relative mRNA levels of E93, AMBRA1, and ATG8 in dsE93 fed larvae. (B) Relative mRNA levels of Kr-h1, AMBRA1, and ATG8 in the dsKr-h1 fed larvae. Data shown as Mean ± SE (n=4). The scale bar is 1000 µm. The asterisks indicate significant difference at a p-value of <0.05.
To determine the function of AMBRA1 and ATG8, RNAi was used to knockdown the expression of these genes. Diet pellets containing PLL/EGCG nanoformulations of dsAMBRA1, dsATG8, or dsGFP (as a control) were fed to early third instar larvae. Knockdown of AMBRA1 and ATG8 resulted in defects in larval growth and development (Figure 3A). The treated larvae were smaller and darker when compared to control larvae fed on dsGFP. In addition, feeding dsAMBRA1 or dsATG8 to the larvae for three days resulted in more than 60% mortality (Figure 3B). Compared to the control (dsGFP fed), the AMBRA1 and ATG8 mRNA levels decreased by 70% and 74%, respectively, in dsAMBRA1 and dsATG8 fed larvae (Figure 3C).
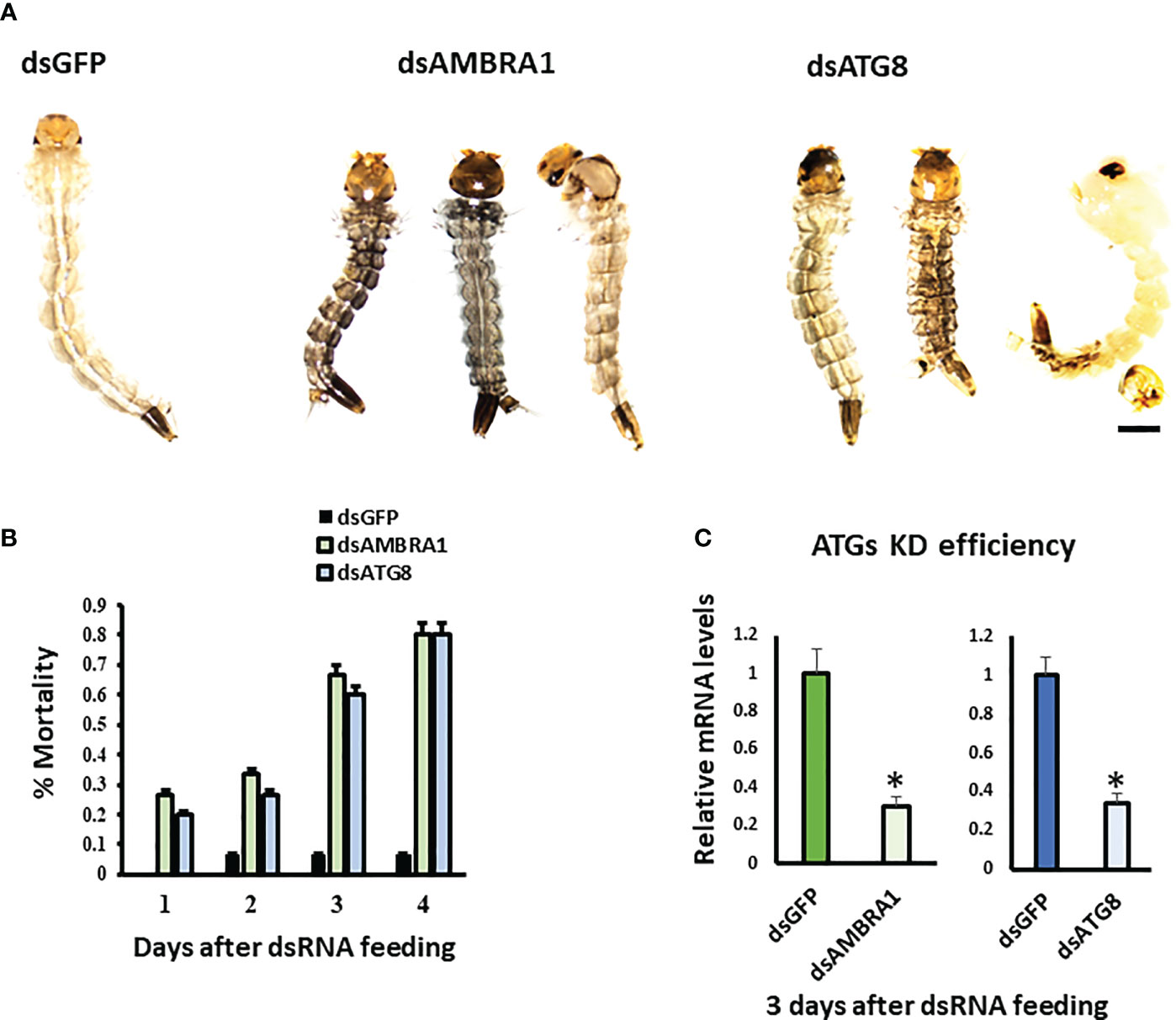
Figure 3 Phenotypes observed in final instar Aedes aegypti. larvae fed on dsAMBRA1 and dsATG8 (A) The larval phenotypes observed after dsAMBRA1 and dsATG8 formulated with PLL/EGCG nanoparticles were fed to third instar larvae. The control larvae were fed on PLL/EGCG/dsGFP nanoparticles. The scale bar is 1000 µm. (B) ATG gene knockdown induces mortality of Ae. aegypti larvae. Percent mean mortality is shown (n= 30/treatment). (C) The Knockdown efficiency of dsAMBRA1 and dsATG8 in the last instar larva after three days of feeding. Relative mRNA levels are shown as Mean ± SE (n=4). The asterisks indicate significant difference at a p-value of <0.05.
The midgut remodeling begins within 36 h of ecdysis to the final larval instar (AEFL) and continues until 12 h AEPS (1). Therefore, we examined the knockdown efficiency and effects at two developmental time points: at 48 h AEFL and 12 h AEPS. Following dsRNA feeding, midguts were dissected from the larvae at 48 h AEFL and pupae at 12 h AEPS. Midguts from control larvae fed on GFP reached, and the gastric caeca began to degenerate at 48 AEFL (Figure 4A). By 12 h AEPS, the midguts in the control larvae were thin, and the gastric caeca degenerated (Figure 4A). The midguts dissected from larvae fed on dsAMBRA1 and dsATG8 did not show these changes in morphology. These midguts were long and wide with intact gastric caeca similar to those in larvae, which indicated that midgut remodeling was blocked in these insects (Figure 4A). The sections of the midguts were stained DAPI to detect the size of epithelial cell nuclei. The midguts from larvae 48 h AEFL (dsGFP-treated control) contained two types of epithelial cells: larval epithelial cells with large polyploid nuclei (indicated with a yellow arrow) and stem cells has small diploid nuclei (marked with a white arrow) (Figure 4B). The pupal midgut consisted mostly of new epithelial cells with small diploid nuclei, where the larval epithelial cells migrated toward the lumen and formed the meconium. Midguts dissected at 12 h AEPS of larvae fed on dsAMBR1 or dsATG8 showed mostly larval cells (large polyploid nuclei) and a few cells with small nuclei. These data suggest that there is a block in the degeneration of larval cells in insects fed on dsAMBR1 and dsATG8. Together, these results demonstrate that AMBRA1 and ATG8 are expressed in the midgut tissues and play an important role in PCD of the larval midgut in Ae. aegypti.
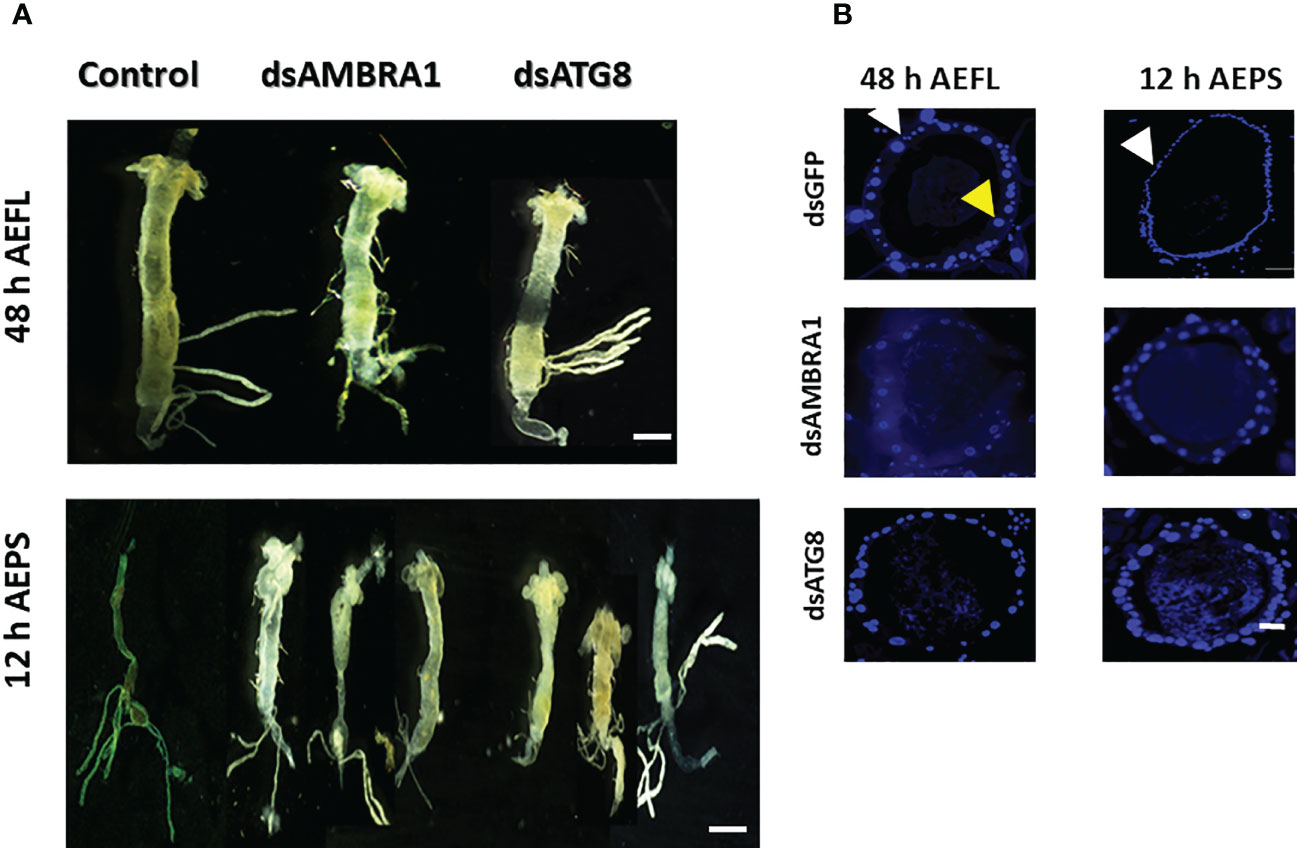
Figure 4 The effects of AMBRA1 and ATG8 knockdown on midgut remodeling in Aedes aegypti. (A) The larval midguts were dissected from larvae at 48 h AEFL and pupae at 12 h AEPS after feeding on dsAMBRA1 and dsATG8 and compared with the control larvae fed on dsGFP. The scale bar is 1000 µm. (B) Cross-sections of midguts of larvae at 48 h AEFL and pupae at 12 h AEPS developed from dsAMBRA1 and dsATG8 fed larvae compared with the control larvae. DAPI nuclear stain showed that the larval midgut epithelium contains two types of cells with small nuclei (white arrow) and large nuclei (yellow arrow). In the control pupa, the midguts showed only small nuclei-containing cells. The midguts from ATG gene knockdown insects showed both small and large nuclei. The scale bar is 50 μm.
The studies reported here revealed a critical role for ATG genes in the midgut remodeling of Ae. Aegypti. AMBRA1 is a novel regulator for phagophore nucleation, and the knockout of AMBRA1 in mice resulted in autophagy inhibition and embryonic lethality (5–7). ATG8 is essential for autophagic vesicle formation and has been utilized as a marker for autophagic activity in previous studies (9). Therefore, AMBRA1 and ATG8 have been common targets in autophagy studies. Disruption of autophagy contributes to blocking cell death in D. melanogaster (12), B. mori (11), and H. virescens (23) midgut and other larval tissues in insects (15–17). Studies in Galleria mellonella indicated that the formation of autophagosomes in the fat body cells concomitantly with the upregulation of ATG6 and ATG8 genes were observed during the early pupal stage (24). In the mosquito Aedes albopictus, ATG8 was found to be expressed across all developmental stages, and the ATG8 mRNA levels are higher in female adults than in male adults (9). Ae. aegypti females require a cascade of autophagy cytoplasmic events for oocyte development (25). The involvement of AMBRA1 as a regulator of autophagy and development processes has also been reported in mouse embryos, and its loss leads to abnormal embryonic development. AMBRA1 also regulates neurogenesis and cancer in mammals (6), suggesting that the function of ATG genes may be conserved.
RNAi-mediated AMBRA1 and ATG8 knockdown caused defects in midgut remodeling and death during the pupal stage in Ae. aegypti. Several studies have shown that inhibiting autophagy genes has a significant impact on development. In D. melanogaster, the ATG1 mutants die before pupation (10), and in Bombyx, knockdown of several ATG genes caused lethality during prepupal and pupal stages (16), suggesting autophagy is a key player in metamorphosis. Moreover, previous studies have demonstrated that autophagy promotes cell death in larval midgut tissues during metamorphosis. In D. melanogaster, knockdown of the ATG genes using RNAi delayed midgut degeneration, and the overexpression of ATG1 resulted in accelerated degradation of midgut tissues and contributed to pupal death (12, 26). Additionally, ATG proteins also participate in midgut remodeling during the larval-pupal transition in B. mori (11). Knockdown of ATG12 in H. armigera larvae delayed pupation and midgut PCD (13). These results support our finding that autophagy contributes to midgut remodeling in Ae. aegypti.
JHA suppressed the expression of AMBRA1 and ATG8 genes, while SEA upregulated their expression in the midgut. JHA repressed the expression of ATG genes in the 4th instar larval stage, resulting in a defect in programmed cell death, and midguts maintained their larval cells. JH has been shown to inhibit autophagy in the fat body of Mamestra brassicae during the last larval instar, and RNAi of the JH receptor, methoprene tolerant had a significant effect on autophagic activity in B. mori and causing lethality during the larval-pupal transition (3). Studies in D. melanogaster and B. mori reported that 20E promotes both apoptosis and autophagy gene expression during metamorphosis (13, 27). In B. mori, an increase in ATG gene expression was observed in the fat body of larvae immediately after injection with 20E. EcR response elements (EcRE) are present in the ATG1 promoter, and deletion of EcRE inhibited 20E-induced autophagy in B. mori (16). Another study demonstrated that the expression of ATG1 and ATG8 were inhibited upon EcR RNAi, which implies that 20E/EcR complex is involved in the regulation of ATG genes in the fat body of Ae. aegypti (25). The detailed molecular mechanism by which JH suppresses ATG expression requires further investigation.
Furthermore, these studies showed that both E93 and Kr-h1 are involved in JH and 20E hierarchical network that regulates autophagy. E93 has been classified as an “early” response gene of the ecdysone signaling pathway (28). SEA treatment upregulated E93 and JHA treatment upregulated Kr-h1 expression resulting in the suppression of ATG genes and blocking of PCD cascade in the midgut (Figure 2). It is also possible that the JH early response gene Kr-h1 may influence the E93 and ATG expression levels in Ae. aegypti. In D. melanogaster, E93 is induced by 20E and suppressed by JH (29) and Kr-h1 directly represses E93 expression (30). The JH-mediated suppression of E93 expression through Met and Kr-h1 has also been found in several other insects (29, 31). High JH levels may prevent the expression of E93 during larval-larval molting. Therefore, the inhibition of midgut remodeling in JHA-treated insects is likely caused by the lack of E93. Moreover, RNAi of E93 significantly decreased the mRNA levels of the AMBRA1 and ATG8 (Figure 2).
RNAi experiments suggested that E93-mediated 20E signaling activates ATG gene expression in the midgut during metamorphosis, while JH suppresses autophagy induction through transcription factor Kr-h1. Previous studies showed that the induction of E93 by 20E determines a PCD response (31). In Ae. aegypti, E93 regulates autophagic cell death and expression of ATG genes, which is similar to what was observed in the D. melanogaster salivary gland and midgut (18). In B. mori E93 induces the expression of ATG1 to promote the larval-pupal metamorphosis, and silencing of E93 by the RNAi disrupted the steroid-programmed cell death signaling, including caspase activity, autophagy, and cell dissociation during fat body remodeling (32). The E93 mediates 20E-induced autophagic cell death in the midgut and salivary glands, and a mutation in E93 reduced the expression of the ATG genes in the salivary glands (10) and the midgut (33). In summary, both B. mori and D. melanogaster display increased expression of ATG genes promoted by E93, whereas JH suppresses the transcription of E93, indicating that JH plays a negative role in the induction of autophagy through interaction with 20E signals through Kr-h1. Further studies are required to uncover the effects of JH on the interaction of ATG, E93, and Kr-h1 proteins in midgut remodeling. Overall, these studies revealed an important role of autophagy genes in the degradation of larval midgut in Ae. aegypti. Knockdown of AMBRA1 and ATG8 led to a block in midgut remodeling and death during the pupal stage. Application of JHA to the 4th instar larvae induced Kr-h1, suppressed E93 expression and autophagy. These data suggest that the autophagy is induced by 20E and suppressed by JH working through Kr-h1, E93 and ATG genes.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
NA and SP participated in the research design. NA conducted the experiments. NA and SP prepared the manuscript. All authors contributed to the article and approved the submitted version.
Research reported in this thesis was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers R01GM070559 and R21AI163561-02. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Dr. Ramesh K. Dhandapani for his help with the preparation of PLL-EGCG-dsRNA nanoparticles.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finsc.2023.1113871/full#supplementary-material
1. Wu Y, Parthasarathy R, Bai H, Palli SR. Mechanisms of midgut remodeling: juvenile hormone analog methoprene blocks midgut metamorphosis by modulating ecdysone action. Mech Dev (2006) 123:530–47. doi: 10.1016/j.mod.2006.05.005
2. Jindra M, Palli SR, Riddiford LM. The juvenile hormone signaling pathway in insect development. Annu Rev Entomology (2013) 58:181–204. doi: 10.1146/annurev-ento-120811-153700
3. Tettamanti G, Casartelli M. Cell death during complete metamorphosis. Philos Trans R Soc London. Ser B Biol Sci (2019) 374:20190065. doi: 10.1098/rstb.2019.0065
4. Mizushima N. The ATG conjugation systems in autophagy. Curr Opin Cell Biol (2020) 63:1–10. doi: 10.1016/j.ceb.2019.12.001
5. Cianfanelli V, Nazio F, Cecconi F. Connecting autophagy: AMBRA1 and its network of regulation. Mol Cell Oncol (2015) 2:e970059–e970059. doi: 10.4161/23723548.2014.970059
6. Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. AMBRA1 regulates autophagy and development of the nervous system. Nature (2007) 447:1121–5. doi: 10.1038/nature05925
7. Antonioli M, Albiero F, Fimia GM, Piacentini M. AMBRA1-regulated autophagy in vertebrate development. Int J Dev Biol (2015) 59:109–17. doi: 10.1387/ijdb.150057mp
8. Grumati P, Dikic I. Ubiquitin signaling and autophagy. J Biol Chem (2018) 293:5404–13. doi: 10.1074/jbc.TM117.000117
9. Qiao J, Zhang D, Wang Y, Li X, Wang S, Liu Q. Identification of AaAtg8 as a marker of autophagy and a functional autophagy-related protein in Aedes albopictus. PeerJ (2018) 6:e5988. doi: 10.7717/peerj.5988
10. Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell (2007) 131:1137–48. doi: 10.1016/j.cell.2007.10.048
11. Franzetti E, Huang ZJ, Shi YX, Xie K, Deng XJ, Li JP, et al. Autophagy precedes apoptosis during the remodeling of silkworm larval midgut. Apoptosis (2012) 17:305–24. doi: 10.1007/s10495-011-0675-0
12. Denton D, Chang TK, Nicolson S, Shravage B, Simin R, Baehrecke EH, et al. Relationship between growth arrest and autophagy in midgut programmed cell death in drosophila. Cell Death Differentiation (2012) 19:1299–307. doi: 10.1038/cdd.2012.43
13. Li Y-B, Yang T, Wang J-X, Zhao X-F. The steroid hormone 20-hydroxyecdysone regulates the conjugation of autophagy-related proteins 12 and 5 in a concentration and time-dependent manner to promote insect midgut programmed cell death. Front Endocrinol (2018) 9:28–8. doi: 10.3389/fendo.2018.00028
14. Montali A, Romanelli D, Cappellozza S, Grimaldi A, de Eguileor M, Tettamanti G. Timing of autophagy and apoptosis during posterior silk gland degeneration in Bombyx mori. Arthropod Structure Dev (2017) 46:518–28. doi: 10.1016/j.asd.2017.05.003
15. Müller F, Ádori C, Sass M. Autophagic and apoptotic features during programmed cell death in the fat body of the tobacco hornworm (Manduca sexta). Eur J Cell Biol (2004) 83:67–78. doi: 10.1078/0171-9335-00359
16. Tian L, Ma L, Guo E, Deng X, Ma S, Xia Q, et al. 20-hydroxyecdysone upregulates atg genes to induce autophagy in the Bombyx fat body. Autophagy (2013) 9:1172–87. doi: 10.4161/auto.24731
17. Rusten TE, Lindmo K, Juhász G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell (2004) 7:179–92. doi: 10.1016/j.devcel.2004.07.005
18. Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development (2001) 128:1443–55. doi: 10.1242/dev.128.8.1443
19. Lee SB, Kim S, Lee J, Park J, Lee G, Kim Y, et al. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep (2007) 8:360–5. doi: 10.1038/sj.embor.7400917
20. Ray K, Mercedes M, Chan D, Choi CY, Nishiura JT. Growth and differentiation of the larval mosquito midgut. J Insect Sci (2009) 9:1–13. doi: 10.1673/031.009.5501
21. Nishiura JT. Coordinated morphological changes in midgut, imaginal discs, and respiratory trumpets during metamorphosis of Aedes aegypti (Diptera: Culicidae). Ann Entomological Soc America (2002) 95:498–504. doi: 10.1603/0013-8746(2002)095[0498:CMCIMI]2.0.CO;2
22. Dhandapani RK, Gurusamy D, Palli SR. Development of catechin, poly-l-lysine, and double-stranded RNA nanoparticles. ACS Appl BioMaterials (2021) 4:4310–8. doi: 10.1021/acsabm.1c00109
23. Tettamanti G, Grimaldi A, Casartelli M, Ambrosetti E, Ponti B, Congiu T, et al. Programmed cell death and stem cell differentiation are responsible for midgut replacement in Heliothis virescens during prepupal instar. Cell Tissue Res (2007) 330:345–59. doi: 10.1007/s00441-007-0449-8
24. Poyraz Tinartas E, Goncu E, Koc K. Apoptotic and autophagic characteristics of perivisceral fat body remodeling of the greater wax moth Galleria mellonella and effects of juvenile hormone analog, fenoxycarb, on these processes. Arch Insect Biochem Physiol (2021) 107:e21780. doi: 10.1002/arch.21780
25. Bryant B, Raikhel AS. Programmed autophagy in the fat body of Aedes aegypti is required to maintain egg maturation cycles. PLoS One (2011) 6:e25502. doi: 10.1371/journal.pone.0025502
26. Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol (2009) 19:1741–6. doi: 10.1016/j.cub.2009.08.042
27. Tracy K, Baehrecke EH. The role of autophagy in Drosophila metamorphosis. Curr Top Dev Biol (2013) 103:101–25. doi: 10.1016/B978-0-12-385979-2.00004-6
28. Baehrecke EH, Thummel CS. The drosophila E93 gene from the 93F early puff displays stage-and tissue-specific regulation by 20-hydroxyecdysone. Dev Biol (1995) 171:85–97. doi: 10.1006/dbio.1995.1262
29. Belles X, Santos CG. The MEKRE93 (Methoprene tolerant-krüppel homolog 1-E93) pathway in the regulation of insect metamorphosis, and the homology of the pupal stage. Insect Biochem Mol Biol (2014) 52:60–8. doi: 10.1016/j.ibmb.2014.06.009
30. Belles X. Krüppel homolog 1 and E93: The doorkeeper and the key to insect metamorphosis. Arch Insect Biochem Physiol (2020) 103(3):e21609. doi: 10.1002/arch.21609
31. Ureña E, Manjón C, Franch-Marro X, Martín D. Transcription factor E93 specifies adult metamorphosis in hemimetabolous and holometabolous insects. Proc Natl Acad Sci USA (2014) 111:7024–9. doi: 10.1073/pnas.1401478111
32. Liu X, Dai F, Guo E, Li K, Ma L, Tian L, et al. 20-hydroxyecdysone (20E) primary response gene E93 modulates 20E signaling to promote Bombyx larval-pupal metamorphosis. J Biol Chem (2015) 290:27370–83. doi: 10.1074/jbc.M115.687293
Keywords: midgut, metamophosis, RNAi, ATG, AMBRA1, ambra
Citation: Albishi NM and Palli SR (2023) Autophagy genes AMBRA1 and ATG8 play key roles in midgut remodeling of the yellow fever mosquito, Aedes aegypti. Front. Insect Sci. 3:1113871. doi: 10.3389/finsc.2023.1113871
Received: 01 December 2022; Accepted: 03 January 2023;
Published: 16 January 2023.
Edited by:
Sudeshna Mazumdar-Leighton, University of Delhi, IndiaCopyright © 2023 Albishi and Palli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subba Reddy Palli, cnBhbGxpQHVreS5lZHU=
†Present address: Najla M. Albishi, Department of Biological Sciences, King Faisal University, Al-Hassa, Saudi Arabia
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.