- 1Energy and Environment Directorate, Pacific Northwest National Laboratory, Richland, WA, United States
- 2Earth and Biological Sciences Directorate, Pacific Northwest National Laboratory, Richland, WA, United States
- 3Department of Biological Systems Engineering, Washington State University, Pullman, WA, United States
Bioconversion processes require nitrogen for growth and production of intracellular enzymes to produce biofuels and bioproducts. Typically, this is supplied as reduced nitrogen in the form of ammonia, which is produced offsite from N2 and H2 via the Haber-Bosch process. While this has revolutionized industries dependent on fixed nitrogen (e.g., modern agriculture), it is highly energy-intensive and its reliance on natural gas combustion results in substantial global CO2 emissions. Here we investigated the feasibility of in situ biological nitrogen fixation from N2 gas as a strategy to reduce greenhouse gas impacts of aerobic bioconversion processes. We developed an efficient and cost-effective method to screen fungal bioconversion hosts for compatibility with the free-living diazotrophic bacterium Azotobacter vinelandii under nitrogen fixing conditions. Our screening revealed that the genus Yarrowia is particularly enriched during co-culture experiments. Follow-up experiments identified four Y. lipolytica strains (NRRL Y-11853, NRRL Y-7208, NRRL Y-7317, and NRRL YB-618) capable of growth in co-culture with A. vinelandii. These strains utilize ammonium secreted during diazotrophic fixation of N2, which is provided as a component of the air input stream during aerobic fermentation. This demonstrates the feasibly of in situ biological nitrogen fixation to support heterotrophic fermentation processes for production of fuels and chemicals.
1 Introduction
Bioconversion processes, critical for the production of biofuels and bioproducts, require significant amounts of nitrogen for microbial growth and enzyme production (Huo et al., 2012; Otto et al., 2022). Traditionally, this nitrogen is supplied in the form of ammonia, produced offsite through the Haber-Bosch process, which combines N2 and H2 under high temperature and pressure (Smith et al., 2020; Matt, 2023). While this industrial process has revolutionized nitrogen-dependent sectors, particularly modern agriculture, its reliance on natural gas combustion for energy contributes approximately 1% to global CO2 emissions (Smith et al., 2020; Matt, 2023). As the bioeconomy expands, the demand for fixed nitrogen in bioprocesses is expected to increase, potentially exacerbating this environmental impact. Therefore, developing sustainable alternatives for nitrogen supply in bioproduction is crucial for reducing the carbon footprint of the bioeconomy.
In-situ fixation of atmospheric N2 presents a promising sustainable alternative to ammonia for bioprocessing (Haskett et al., 2022). Diazotrophic microorganisms, capable of fixing atmospheric nitrogen, have evolved to form mutualistic and even symbiotic relationships with various organisms in nature (Boyd and Peters, 2013; Nag et al., 2022). For instance, rhizobia form symbiotic relationships with legumes, providing fixed nitrogen in exchange for carbon sources (Hardoim et al., 2015). In marine environments, certain cyanobacteria form symbioses with diatoms and corals, supporting nitrogen fixation in nutrient-poor waters (Tschitschko et al., 2024; Moynihan et al., 2022). The soil bacterium Azotobacter vinelandii is particularly noteworthy for its efficient nitrogen fixation capabilities (Noar and Bruno-Bárcena, 2018). The nifL mutant of A. vinelandii can fix gaseous N2 to secreted ammonia even under aerobic conditions (Curatti et al., 2005; Plunkett et al., 2020; Mus et al., 2022), making it an attractive candidate for inclusion as a nitrogen-fixing biocatalyst in agricultural and bioconversion processes.
Establishing synthetic communities that can function efficiently under specific bioprocess conditions (temperature, pH, salinity, redox potential, etc.) without harmful competition or inhibition presents significant challenges (Widder et al., 2016). Improving interactions and mitigating competition in such synthetic communities is a complex and time-consuming task. Conventional bioprocess development involves metabolic engineers analyzing known and constructed biochemical routes to identify the most favorable pathways and organisms considering factors such as thermodynamic feasibility, enzyme functionality, and cofactor requirements (Jagmann and Philipp, 2014; Kosina et al., 2016; McCarty et al., 2020; Song et al., 2024). These community approaches are typically first tested in model organisms like Escherichia coli or Saccharomyces cerevisiae. However, pairing of optimal hosts with superior characteristics, such as rapid growth rates, ability to utilize diverse feedstocks, and resistance to growth inhibitors that can naturally establish mutualistic relationships is crucial to minimize optimization efforts. Partner screening and matching should employ low-cost, expedited, and high-throughput methodologies and will be necessary to successfully deploy agricultural and bioconversion technologies based on microbial communities.
In this study, we investigate the feasibility of in situ biological nitrogen fixation from N2 gas to reduce the greenhouse gas impacts of aerobic bioconversion processes using a paired community (Figure 1) and present a novel, cost-effective approach for identifying competitive bioconversion hosts for industrial processes. Our method utilizes an industrial yeast library encompassing a broad diversity of Ascomycete and Basidiomycete yeasts, crucial for host discovery in developing bioprocesses from novel feedstocks or under specific environmental conditions. This approach not only addresses the immediate need for sustainable nitrogen sources in bioprocessing but also has broader implications for industrial biotechnology. By enabling rapid identification of compatible microbial partnerships, our method could accelerate the development of efficient, environmentally friendly bioprocesses across multiple industries, potentially revolutionizing fields from biofuel production, and agriculture, to waste valorization.
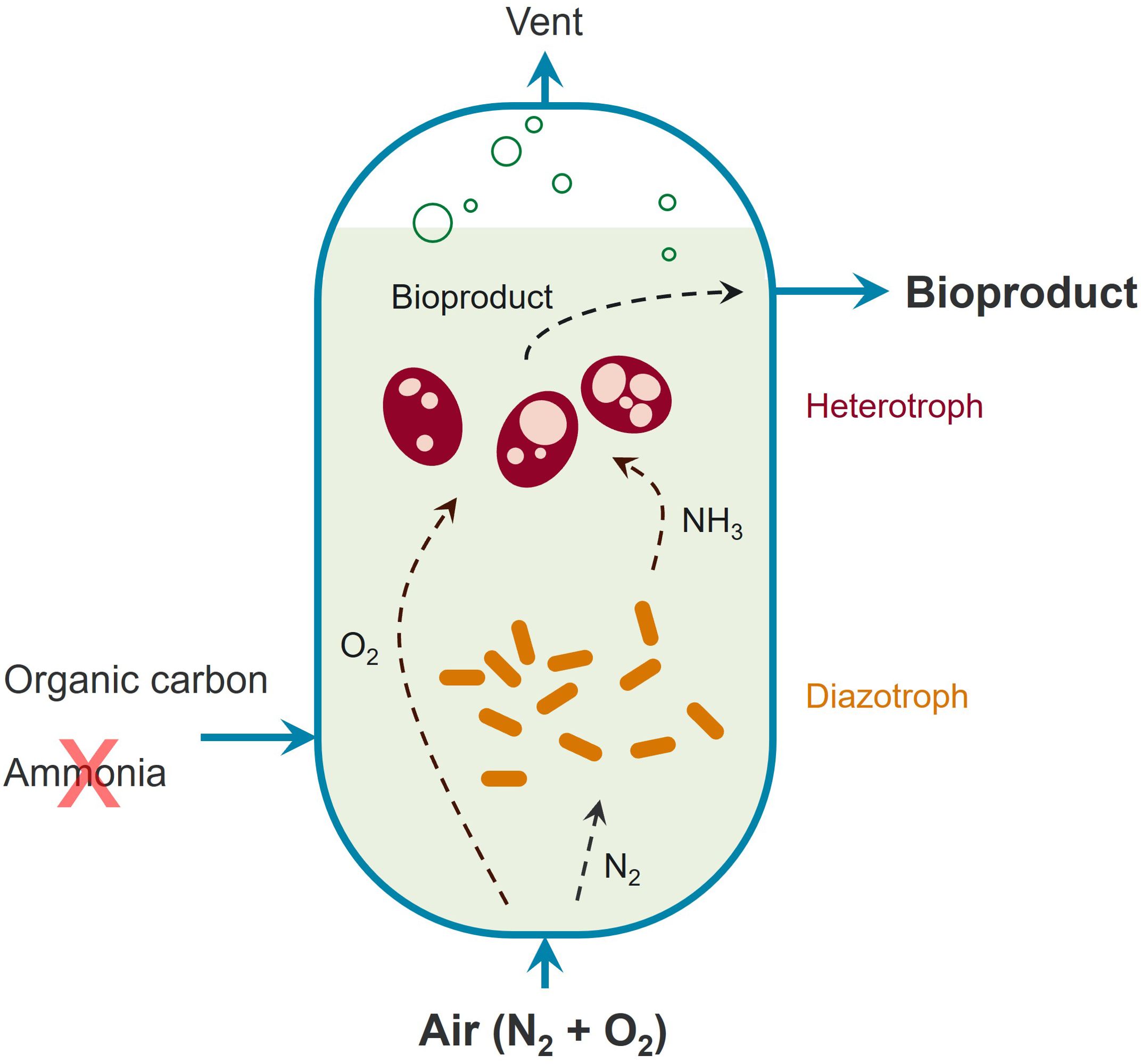
Figure 1. Synthetic community design to enable in situ biological fixation of N2 gas during aerobic bioconversion processes.
2 Materials and methods
2.1 Cultivation conditions
Yeast strains were provided by the ARS culture collection (Peoria, IL), the Phaff Yeast Culture Collection (Davis, CA), the American Type Culture Collection (Manassas, VA), and the CBS-KNAW Culture Collection (Utrecht, NL) and are listed in Supplementary Data Sheet 1. Yeast strains were maintained on YPD (20 g/L glucose, 20 g/L peptone, 10 g/L yeast extract), YM (10 g/L glucose, 5 g/L peptone, 3 g/L yeast extract, 3 g/L malt extract), or PD (20 g/L glucose, 4 g/L potato extract) medium supplemented with 20 g/L agar as appropriate at 30°C. All yeast strains were stored in 15% glycerol at -80C. The ammonium-secreting Azotobacter vinelandii nifL mutant (Mus et al., 2022) (kindly provided by Florence Mus, Washington State University, WA, USA) was grown aerobically at 30°C and 200 rpm in a 250 mL baffled Erlenmeyer flask in a rotary shaker. Medium used for selection and co-culture experiments include Yeast Nitrogen Base without amino acids and ammonium sulfate (YNB; BD, USA) supplemented with 20 g/L glucose and either 3 g/L (NH4)2SO4 or NaNO3 without pH adjustment, modified Burk’s medium (mBG; 20 g/L glucose; 0.2 g/L KH2PO4; 0.8 g/L K2HPO4; 0.2 g/L MgSO4•7H2O; 0.13 g/L CaCl2•2H2O; 0.000253 g/L Na2MoO4•2H2O; 0.00145 g/L FeSO4•7H2O, pH 6.0), modified Burk’s medium with nitrogen (mBGN; 20 g/L glucose; 3 g/L (NH4)2SO4; 0.2 g/L KH2PO4; 0.8 g/L K2HPO4; 0.2 g/L MgSO4•7H2O; 0.13 g/L CaCl2•2H2O; 0.000253 g/L Na2MoO4•2H2O; 0.00145 g/L FeSO4•7H2O, pH 6.0) and a derivative with mixed carbon sources representative of lignocellulosic hydrolysate (mBGX; 20 g/L glucose; 20 g/L xylose; 0.2 g/L KH2PO4; 0.8 g/L K2HPO4; 0.2 g/L MgSO4•7H2O; 0.13 g/L CaCl2•2H2O; 0.000253 g/L Na2MoO4•2H2O; 0.00145 g/L FeSO4•7H2O, pH 6.0) (Mus et al., 2022). For co-culture experiments yeast and A. vinelandii cultures were washed twice with mBGX to remove spent medium and resuspended in fresh mGBX with A. vinelandii and the yeast, each to OD600 = 0.001. Co-cultures were grown aerobically at 30°C and 200 rpm in 250 mL baffled Erlenmeyer flask in a rotary shake for 7 to 10 days.
2.2 Yeast library construction and selection
Industrial yeasts were cultivated individually in YPD, YM, and PD medium in 96-well plates covered with breathable membranes at room temperature on an orbital plate shaker for 4 days at 800 rpm after which most cultures had reached stationary phase. Individual cultures of different strains from each medium type were then mixed in equal volume amounts and passaged at 1:100 volume into fresh YPD, YM, and PD medium. Mixed cultures were grown overnight at 30°C and 200 rpm in 250 mL baffled Erlenmeyer flasks in a rotary shaker. Cultures were then aliquoted as 15% glycerol stocks in cryo-tubes, flash-frozen in liquid N2 and stored at -80°C as the industrial yeast library starting material. For all library experiments, a fresh cryo-stock was thawed on ice prior to 1:100 inoculation in fresh medium. For selection experiments the industrial yeast library and A. vinelandii cultures were washed twice and resuspended in fresh medium with each inoculum to OD600 = 0.001. Selection experimental cultures were grown aerobically at 30°C and 400 rpm in 24-well plates with breathable membranes on an orbital plate shaker for 4 days prior to passaging 1:100 volume to fresh medium. Passaging was performed three times prior to collection of cell pellets by centrifugation at 13,000xg. The supernatant was removed for HPLC analysis and cell pellets were stored at -80C for further analysis. All selection experiments were performed in at least triplicate.
2.3 Sequencing analysis
Genomic DNA was isolated from cell pellets using a yeast genomic DNA purification kit (AMRESCO, Solon, OH). The fungal internal transcribed spacer (ITS) region was amplified from genomic DNA samples by PCR with primers ITS1_F (5’-CTTGGTCATTTAGAGGAAGTAA-3’) and ITS4_R (5’-TCCTCCGCTTATTGATATGC-3’) (Toju et al., 2012). Oligonucleotides were purchased from IDT (Coralville, Iowa). PCR reactions were assessed for quality by gel electrophoresis and purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany). ITS amplicons were sequenced using the Illumina-based amplicon-EZ service (Genewiz Inc, South Plainfield, NJ). To assess mapping quality, FASTQ files of ITS amplicons were combined from all samples and mapped to ITS sequences present in the UNITE and UNITE+S databases (Abarenkov et al., 2024), and the NCBI ITS RefSeq Database (Schoch et al., 2014) using Bowtie2 (Langmead and Salzberg, 2012) in end-to-end or local alignment mode and and Samtools (Danecek et al., 2021) to assess mapping statistics. Read counts were converted to log2 scale and median normalized within datasets prior to hierarchical clustering using average linkage with the correlation distance metric and visualization as heat-maps using InfernoRDN (Polpitiya et al., 2008).
2.4 Analytical methods
Extracellular metabolites including glucose and xylose were quantified by HPLC. Ten microliters of the samples filtered with 0.2 µm syringe filters were analyzed for 45 min using an Aminex HPX-87H ion exclusion column with a 4.5 mM H2SO4 flow of 0.55 ml/min. The temperature of the column was 50°C. The refractive index was measured with a Waters 2414 refractive index detector at 30°C as well as the UV absorption at 210 nm. For microscopy, 10ul of coculture was taken and observed under the stereomicroscope-Leica MZ16 (Leica Microsystem Ltd, Bannockburn, IL, USA) at day 7. All images were captured at the same magnification (x100).
3 Results
3.1 Establishment of an industrial yeast library
We developed a diverse library of industrially relevant yeast species and strains to enable rapid screening for bioconversion hosts with desirable phenotypes, including substrate utilization, tolerance to product toxicity as well as adaptability to various process conditions that impact process economics and greenhouse gas emissions such as temperature, pH, and osmolyte concentration (Navarrete and Martínez, 2020; Sun and Alper, 2020; Rodriguez-Ocasio et al., 2022). The library encompasses representatives from both the Saccharomycotina subphylum, where most industrial yeasts are found (Riley et al., 2016), and the Basidiomycete yeasts, particularly from Agaricomycotina and Pucciniomycotina, to capture a broad range of metabolic diversity (Johnson, 2013; Riley et al., 2016; Andreu et al., 2022; Suchova et al., 2022). Our library comprises 249 strains from 191 species across 82 genera (Supplementary Data Sheet 1), with an emphasis on genera known for their potential in converting industrial and agricultural wastes into valuable bioproducts. These include Yarrowia, Lipomyces, Rhodotorula, Debaryomyces, and Trichosporon (Figure 2A). Where possible, we prioritized strains with publicly available genome sequences and annotations to facilitate future genomic analyses.
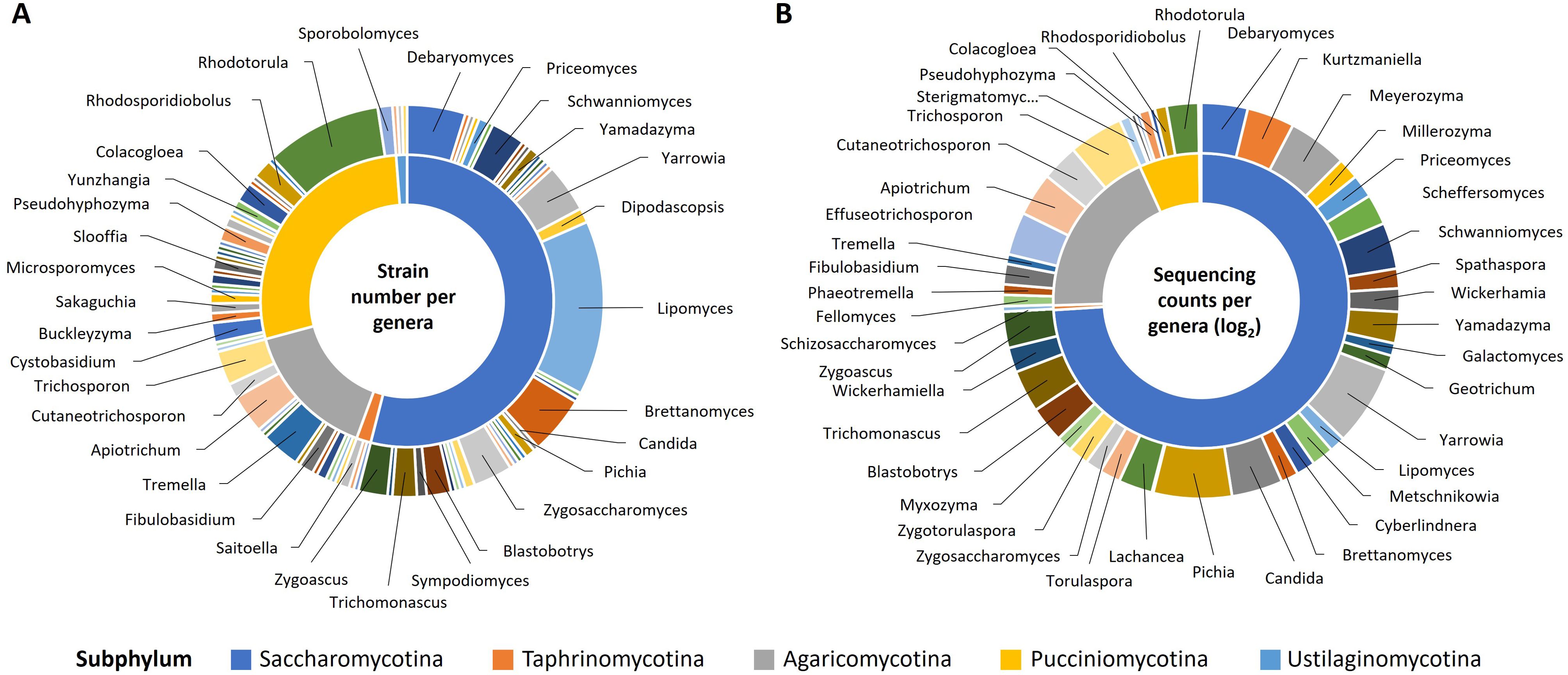
Figure 2. Industrial yeast library composition. Yeast strains broadly representative of major fungal lineages with an emphasis on strain diversity within genera of industrial interest were selected for inclusion in the industrial yeast library. (A) Library composition based on number of strains selected for each genus. (B) Library composition based on log2 transformed sequencing counts of the physical library prior to storage as -80°C glycerol stocks (n=4).
To construct the library each yeast was cultivated on three rich general purpose medium formulations (YPD, YM, and PD) for four days to ensure growth to or near stationary phase prior to mixing the yeast cultures in equal volumes. The mixed library was then diluted into fresh YPD, YM, and PD and allowed to grow overnight prior to collecting and freezing the library as glycerol stock aliquots. Genomic DNA was prepared from four samples of the industrial yeast library prior to storage to assess representation and quality. Universal primers for the fungal internal transcribed spacer (ITS) were used to amplify the ITS region from the pooled library, sequenced as amplicons, and aligned to sequences from high quality fungal ITS databases. Three databases were evaluated as sources of ITS sequences; the NCBI ITS RefSeq Database (Schoch et al., 2014) the UNITE database for molecular identification of fungi (Abarenkov et al., 2024), and UNITE+S which contains additional singletons. Amplicon sequences were aligned to ITS sequences in each database using end-to-end or local alignment modes to evaluate trade-offs between accuracy and quantity of mappings. As expected, local alignments mapped a greater number of the amplicon reads though this came at the expense of some unique mappings. In end-to-end mode more amplicons aligned to the NCBI database than the UNITE databases (Figure 3A). We further assessed the quality of end-to-end and local alignments by comparing the ratio of reads mapped to high count targets in the NCBI database and found that for expected yeast genera present in the industrial yeast library (Yarrowia, Pichia, Candida, ect.) the ratio was approximately 1:1 whereas filamentous fungi not present in the library (Mucor, Fusarium, Penicillium, Loekoeslaszloa, ect.) were identified only in local alignment mode (Figure 3B). Finally, we compared the ratio of reads mapping to high-count targets in the three databases in end-to-end mode. The overall quality of identifications was similar with some nuances in the naming of genera (e.g. Pichia in NCBI vs. Issatchenkia in the UNITE databases) and occasional erroneous alignment to filamentous fungi not present in the industrial yeast library even in end-to-end mode (e.g. Alternaria in UNITE+S and Penicillium in all 3 databases) (Figure 3C). From these results we concluded that alignment to the NCBI ITS RefSeq Database in end-to-end mode produced the highest quality output and was used for all further analyses (Supplementary Data Sheet 2).
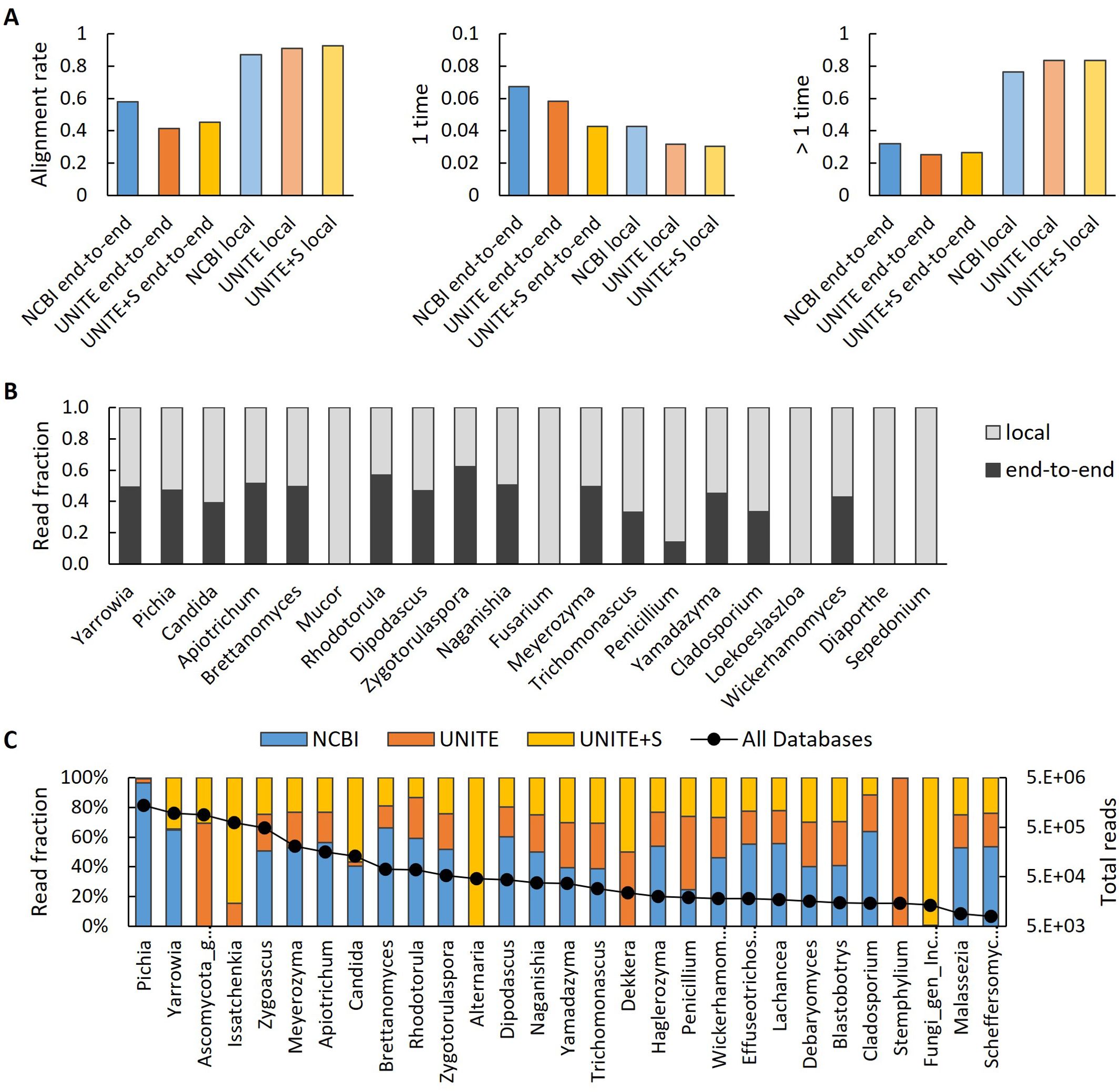
Figure 3. Comparison of mapping strategies and ITS databases. (A) Amplicon reads were aligned in end-to-end and local mode to three ITS databases (NCBI, UNITE, and UNITE+S). (B) High read count ratios after mapping to the NCBI ITS database in local or end-to-end mode. Note that in local mode, many reads erroneously align to filamentous fungi absent in the library (e.g. Mucor, Fusarium, Penicillium, Loekoeslaszloa). (C) Comparison of high read count hits after end-to-end mode alignment to three ITS databases. Note differences in naming represented in the different databases (e.g. Pichia in NCBI vs. Issatchenkia in the UNITE databases) and occasional erroneous alignment to filamentous fungi not present in the library even in end-to-end mode (e.g. Alternaria in UNITE+S and Fusarium in all 3 databases).
ITS amplicon sequences from Industrial yeast library samples collected during library construction were used to evaluate the overall composition of the stored library. ITS sequences from 73% of the genera included in the library design were identified in the physical library prior to storage suggesting overall maintenance of diversity through the use of diverse media for library preparation and broad detection with the possibility that PCR bias may contribute to loss of detection for some genera. Notable expansion was observed for some rapidly growing Ascomycete genera (Pichia, Candida, and Yarrowia) while overall representation of the Basidiomycetes declined (Figure 2B). Interestingly, 89% of the genera included in the library design were identified in follow-up experiments examining library composition in conditions with stronger selective pressure (data not shown). This suggests that while some strains may be present at levels lower than the threshold of detection for amplicon sequencing in the library stock, they can enrich and be quantified in environmental conditions where they are more competitive.
3.2 Selection of yeasts able to partner with Azotobacter vinelandii during in situ N2 fixation
Fixed nitrogen, typically in the form of nitrate or ammonium, is essential for microbial growth in biological processes. Conventionally, ammonia produced by the energy and CO2 intensive Haber-Bosch process serves as the nitrogen source for commercial bioprocesses (Smith et al., 2020; Matt, 2023). In this study, we explored in situ biological fixation of nitrogen by incorporating the nifL mutant of A. vinelandii, which is capable of fixing gaseous N2 and secreting ammonia under aerobic conditions (Mus et al., 2022). The inclusion of A. vinelandii as a nitrogen-fixing biocatalyst offers an alternative to supplying offsite ammonia produced through the Haber-Bosch process, potentially reducing the carbon footprint of bioprocesses. We screened the industrial yeast library using a selective enrichment approach to identify yeasts capable of partnering with A. vinelandii, as well as those able to grow on various nitrogen sources. This was followed by amplicon sequencing of the ITS region to quantify competitive yeasts present in the selected libraries (Figure 4A). The industrial yeast library was passaged on YNB with either ammonium or nitrate as the nitrogen source, mBGN (with ammonium), and mBG in co-culture with A. vinelandii (using N2 gas as the only nitrogen source). After three passages, we isolated genomic DNA from the cell pellet, amplified the ITS region, and sequenced it to quantify yeast enrichment in different selective environments.
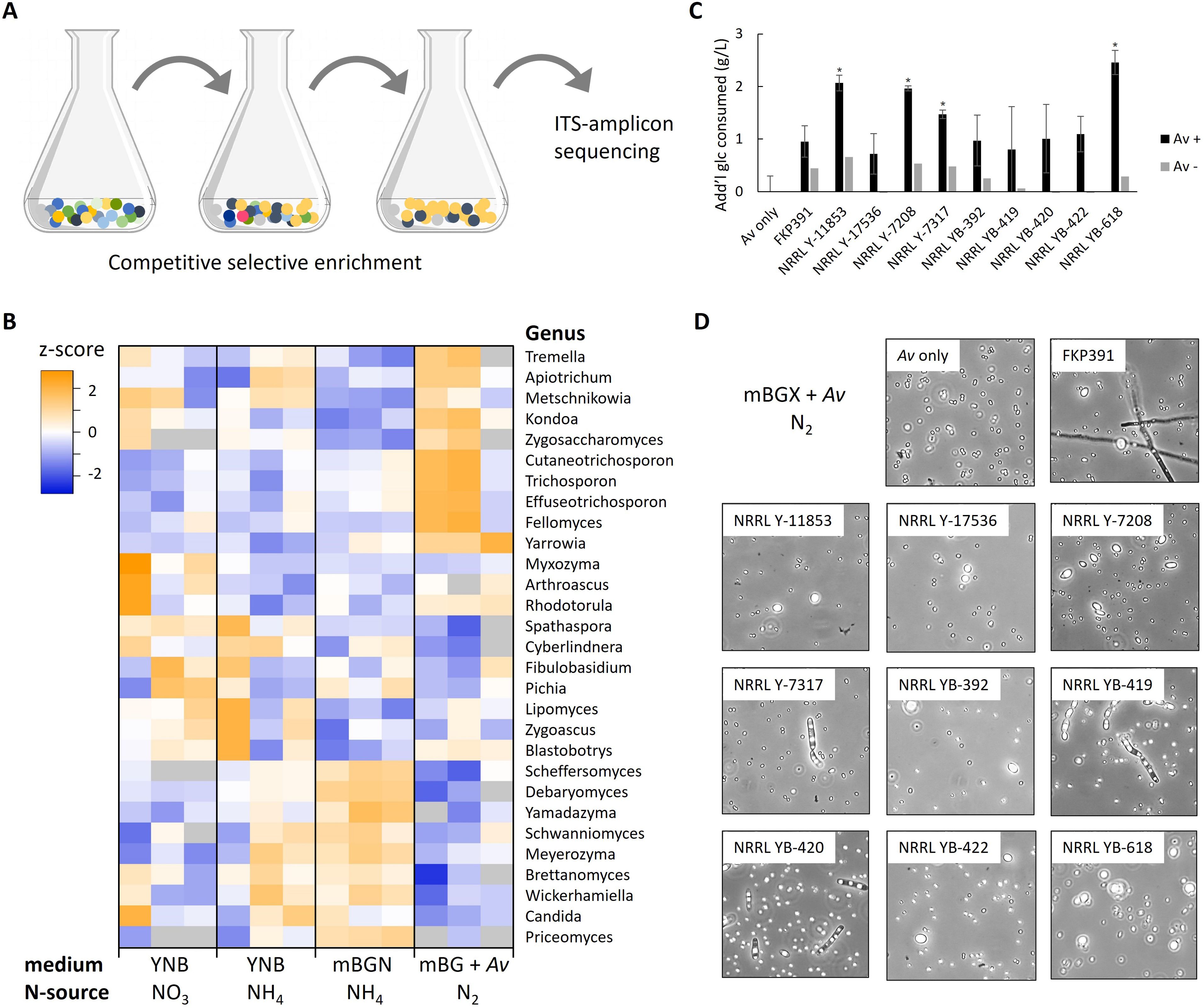
Figure 4. Identification of industrial yeasts amenable to co-culture with A. vinelandii. (A) Industrial yeast species that compete well in selective conditions are enriched by passaging and identified by ITS-amplicon sequencing. (B) Log2 median-normalized read counts for major yeast genera after selection with different nitrogen sources. (C) Glucose consumption of individual Yarrowia lipolytica strains present in the industrial yeast library in co-culture with A. vinelandii with N2 as the nitrogen source at day 4. Note that xylose was included in the medium but not consumed. *Significantly more glucose consumed than A. vinelandii cultures without yeast (p < 0.01). (D) Light microscopy of A. vinelandii and yeast strains in co-culture.
Highly competitive Ascomycete yeasts were enriched in the control cultures with NH4 as the nitrogen source (e.g. Scheffersomyces, Debaryomyces, and related yeasts) while more metabolically diverse yeasts were enriched on NO3 as the nitrogen source such as the xylose consuming Myxozyma (Pomraning et al., 2019), Rhodotorula (Tiukova et al., 2019), and Spathaspora (Hou, 2012). A variety of yeast were enriched after passaging in co-culture with A. vinelandii with N2 as the nitrogen source. These included Tremella, Apiotrichum, Metschnikowia, Kondoa, Zygosaccharomyces, Cutaneotrichosporon, Trichosporon, Effuseotrichosporon, Fellomyces, Yarrowia, and Rhodotorula species (Figure 4B). Many of the strains enriched in co-culture with A. vinelandii are considered to be oleaginous yeasts that accumulate greater than 20% of their biomass as lipids when grown in nitrogen limited conditions including Trichosporon, Yarrowia, and Rhodotorula strains (Zhu et al., 2012; Gao et al., 2014; Pomraning et al., 2015). The oleaginous phenotype is controlled by both nitrogen quantity and quality (Pomraning et al., 2016; Pomraning et al., 2017), and a high C:N ratio with ammonium as a nitrogen source is typically used to induce lipid accumulation (Kerkhoven et al., 2016). In our co-culture system with A. vinelandii, yeast growth is limited by the rate of N2 fixation and secretion of usable ammonium. This natural nitrogen limitation may provide an inherent mechanism for controlling lipid production processes in oleaginous yeasts while simultaneously supplying sufficient nitrogen to support essential housekeeping metabolism for optimal productivity. This balance offers a potential advantage for bioprocess design, potentially eliminating the need for precise external nitrogen control and allowing for more robust and efficient lipid production systems.
3.3 Co-culture of oleaginous Yarrowia lipolytica with Azotobacter vinelandii during in situ N2 fixation
We co-cultured 10 strains of oleaginous Y. lipolytica with A. vinelandii to confirm the ability of heterotrophic yeasts to grow in N2 fixing conditions. Prior to co-culture, A. vinelandii was precultured in aerobic N2 fixing conditions while the yeast strains were passaged in N-starvation conditions to induce a state of scavenging and eliminate carry-over of NH4. Both partners were washed to remove any extracellular nitrogen and co-cultivation initiated in 50 mL shake-flask cultures at low density in aerobic N2 fixing conditions in mBGX medium with glucose and xylose as carbon sources to mimic sugars present in enzymatically hydrolyzed lignocellulosic biomass, and to differentiate between sugars utilized by xylose-consuming strains of Y. lipolytica (Quarterman et al., 2017; Walker et al., 2018) and A. vinelandii which does not utilize xylose (Millan et al., 2020). Cultures without A. vinelandii were included for each yeast to control for any additional growth observed without the N2 fixing partner that we speculate is due to carryover of intracellular nitrogen reserves. After 7 days, all Y. lipolytica-containing cultures showed additional glucose consumption compared to A. vinelandii control monocultures, but no consumption of xylose. Four co-cultures (strains NRRL Y-11853, NRRL Y-7208, NRRL Y-7317, and NRRL YB-618) consumed significantly more glucose than A. vinelandii alone (Figure 4C). Light microscopy confirmed the presence of both yeast and A. vinelandii cells in co-cultures at harvest time (Figure 4D) indicating growth of both species in the diazotroph/heterotroph partnership.
4 Discussion
The development of commercial bioprocesses requires careful selection of microbial hosts (Boundy-Mills, 2012) capable of efficiently converting feedstocks into desired products under economically viable conditions. This selection process involves numerous considerations, including metabolic capacity, genetic tractability, native phenotypic distinction for hard to engineer complex phenotypes such as growth rate and temperature/pH optimization, competitiveness with contaminating microbes and viruses, and synergies with other microbes in designed consortia. Data at all these levels is often incomplete or completely absent. Successful development then relies on rapid evaluation and discovery of appropriate microbial hosts that meet these complex bioprocess objectives.
To facilitate identification of yeasts for commercial applications we constructed a diverse yeast library incorporating strains known for their potential in bioconversion of emerging waste feedstocks, as well as a broad phylogenetic range to facilitate discovery of hosts appropriate for future bioprocess development efforts. This approach differs from comprehensive microbial libraries, which, while valuable for preserving genetic diversity, often require substantial resources to maintain (De Vero et al., 2019). Our method focuses on developing a robust screening approach for identifying high-performing strains sourced from existing collections maintained as mixed cultures and provides a streamlined pathway for candidate selection without the need for exhaustive library management.
The novel screening approach we employed was able to identify promising yeast strains capable of pairing with the diazotrophic bacterium A. vinelandii to enable in situ nitrogen fixation during bioconversion. This approach combines bulk competitive screening with amplicon sequencing and offers several advantages over traditional high-throughput screening techniques. While traditional methods often rely on individual strain culturing, which can be labor and equipment intensive, susceptible to contamination, and require resources that scale with number of strains assayed (Sarnaik et al., 2020; Andreu et al., 2022), our competitive screening can be accomplished in a single cultivation regardless of the number of strains. This approach is particularly suited to identify robust biocatalysts less susceptible to contamination. Furthermore, the use of inexpensive ITS sequencing (Sommermann et al., 2018) with a mixed library allows for routine, low-cost screening.
Our results indicate that oleaginous yeasts, particularly from the genus Yarrowia, can be successfully partnered with N2-fixing A. vinelandii during aerobic fermentation to produce oleochemicals and fuel precursors including fatty alcohols (Dahlin et al., 2019; Cordova et al., 2020), designer fatty acids (Gemperlein et al., 2019; Konzock et al., 2022; Wang et al., 2022), free fatty acids (Yuzbasheva et al., 2018; Wang et al., 2022; Salvador Lopez et al., 2023), and triglycerides (Lazar et al., 2018; Wang et al., 2020) from sugar feedstocks without supplemental ammonia. This finding suggests a potential pathway to reduce reliance on energy-intensive industrial ammonia production. The Haber-Bosch process, while effective, is associated with significant CO2 emissions (1.7-2.1 kg CO2e/kg NH3) and is responsible for ~2% of global CO2 emissions due to natural gas combustion (Liu X. et al., 2020). In contrast, biological nitrogen fixation mediated by diazotrophs occurs under ambient conditions, offering a potentially more sustainable alternative (Imran et al., 2021). However, it’s important to note that biological nitrogen fixation is also energy-intensive, requiring 8 ATP to produce 1 molecule of ammonia under ideal conditions, with additional energy needed to manage oxygen during aerobic cultivation (Inomura et al., 2017). In the scenario considered here, energy required to support nitrogen fixation is derived from catabolism of sugars that are ultimately derived from atmospheric CO2 that has been upgraded by plants using solar energy. Future work including process modeling and life-cycle analysis will elucidate the potential for economic and emissions impacts of this approach compared to conventional methods, as well as efforts to improve strains and processes for more efficient nitrogen fixation and excretion in designed microbial consortia.
In conclusion, we have developed and demonstrated a simple, inexpensive approach to identify competitive bioconversion hosts for industrial processes. Our industrial yeast library, encompassing a variety of microbes important for established bioprocesses and a broad diversity of Ascomycete and Basidiomycete yeasts, provides a valuable resource for host discovery when developing bioprocesses from novel feedstocks or under challenging environmental conditions. This method of pooled strain analysis can be applied to various contexts crucial for bioprocess development. The application explored here supports the use of a co-culture approach to eliminate ammonia inputs by pairing the diazotrophic bacterium A. vinelandii with a heterotrophic yeast, Y. lipolytica, that is widely used to produce microbial oils as food or fuel precursors (Liu H. et al., 2020; Lu et al., 2021; Wang et al., 2022; Zhang et al., 2022) as well as a variety of bioproducts (Markham and Alper, 2018; Miller and Alper, 2019; Bilal et al., 2021; Madzak, 2021; Jach and Malm, 2022) by fixation of N2 provided as a component of the air input stream during aerobic fermentation. This demonstrates the feasibly of in situ biological nitrogen fixation to support heterotrophic fermentation processes for the production of fuels and chemicals and warrants further development guided by process modeling and life-cycle analysis to examine economic feasibility and emissions impacts and to guide development of strains and the overall process to fix and transfer nitrogen within a designed community for microbial bioconversion.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
KP: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing. SD: Formal analysis, Investigation, Methodology, Supervision, Writing – review & editing. RD: Investigation, Methodology, Writing – review & editing. JC: Investigation, Methodology, Writing – review & editing. PB: Conceptualization, Funding acquisition, Project administration, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Pacific Northwest National Laboratory is multi-program national laboratory operated by Battelle for the DOE under Contract No. DE-AC06-76RLO1830. This work was supported by the Predictive Phenomics Initiative Laboratory Directed Research and Development Program.
Acknowledgments
Microbial strains used in this work were provided by the USDA-ARS Culture Collection (NRRL) and the Phaff Yeast Culture Collection, University of California Davis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The views expressed in the article do not necessarily represent the views of the U.S. Department of Energy or the United States Government.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finmi.2024.1473316/full#supplementary-material
Supplementary Data Sheet 1 | Excel file containing strain and phylogenetic designations for yeasts included in the Industrial Yeast Library.
Supplementary Data Sheet 2 | Excel file containing raw ITS amplicon sequencing counts against the NCBI ITS RefSeq Database in end-to-end mode.
References
Abarenkov K., Nilsson R. H., Larsson K. H., Taylor A. F. S., May T. W., Froslev T. G., et al. (2024). The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: sequences, taxa and classifications reconsidered. Nucleic Acids Res. 52, D791–D7D7. doi: 10.1093/nar/gkad1039
Andreu C., Zarnowski R., Del Olmo M. L. (2022). Recent developments in the biology and biotechnological applications of halotolerant yeasts. World J. Microbiol. Biotechnol. 38, 27. doi: 10.1007/s11274-021-03213-0
Bilal M., Xu S., Iqbal H. M. N., Cheng H. (2021). Yarrowia lipolytica as an emerging biotechnological chassis for functional sugars biosynthesis. Crit. Rev. Food Sci. Nutr. 61, 535–552. doi: 10.1080/10408398.2020.1739000
Boundy-Mills K. (2012). Yeast culture collections of the world: meeting the needs of industrial researchers. J. Ind. Microbiol. Biotechnol. 39, 673–680. doi: 10.1007/s10295-011-1078-5
Boyd E. S., Peters J. W. (2013). New insights into the evolutionary history of biological nitrogen fixation. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00201
Cordova L. T., Butler J., Alper H. S. (2020). Direct production of fatty alcohols from glucose using engineered strains of Yarrowia lipolytica. Metab. Eng. Commun. 10, e00105. doi: 10.1016/j.mec.2019.e00105
Curatti L., Brown C. S., Ludden P. W., Rubio L. M. (2005). Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc. Natl. Acad. Sci. 102, 6291–6296. doi: 10.1073/pnas.0501216102
Dahlin J., Holkenbrink C., Marella E. R., Wang G., Liebal U., Lieven C., et al. (2019). Multi-Omics Analysis of Fatty Alcohol Production in Engineered Yeasts Saccharomyces cerevisiae and Yarrowia lipolytica. Front. Genet. 10, 747. doi: 10.3389/fgene.2019.00747
Danecek P., Bonfield J. K., Liddle J., Marshall J., Ohan V., Pollard M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10. doi: 10.1093/gigascience/giab008
De Vero L., Boniotti M. B., Budroni M., Buzzini P., Cassanelli S., Comunian R., et al. (2019). Preservation, characterization and exploitation of microbial biodiversity: the perspective of the italian network of culture collections. Microorganisms 7. doi: 10.3390/microorganisms7120685
Gao Q., Cui Z., Zhang J., Bao J. (2014). Lipid fermentation of corncob residues hydrolysate by oleaginous yeast Trichosporon cutaneum. Bioresour Technol. 152, 552–556. doi: 10.1016/j.biortech.2013.11.044
Gemperlein K., Dietrich D., Kohlstedt M., Zipf G., Bernauer H. S., Wittmann C., et al. (2019). Polyunsaturated fatty acid production by Yarrowia lipolytica employing designed myxobacterial PUFA synthases. Nat. Commun. 10, 4055. doi: 10.1038/s41467-019-12025-8
Hardoim P. R., van Overbeek L. S., Berg G., Pirttila A. M., Compant S., Campisano A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Haskett T. L., Paramasivan P., Mendes M. D., Green P., Geddes B. A., Knights H. E., et al. (2022). Engineered plant control of associative nitrogen fixation. Proc. Natl. Acad. Sci. 119. doi: 10.1073/pnas.2117465119
Hou X. (2012). Anaerobic xylose fermentation by Spathaspora passalidarum. Appl. Microbiol. Biotechnol. 94, 205–214. doi: 10.1007/s00253-011-3694-4
Huo Y.-X., Wernick D. G., Liao J. C. (2012). Toward nitrogen neutral biofuel production. Curr. Opin. Biotechnol. 23, 406–413. doi: 10.1016/j.copbio.2011.10.005
Imran A., Hakim S., Tariq M., Nawaz M. S., Laraib I., Gulzar U., et al. (2021). Diazotrophs for lowering nitrogen pollution crises: looking deep into the roots. Front. Microbiol. 12, 637815. doi: 10.3389/fmicb.2021.637815
Inomura K., Bragg J., Follows M. J. (2017). A quantitative analysis of the direct and indirect costs of nitrogen fixation: a model based on Azotobacter vinelandii. ISME J. 11, 166–175. doi: 10.1038/ismej.2016.97
Jach M. E., Malm A. (2022). Yarrowia lipolytica as an alternative and valuable source of nutritional and bioactive compounds for humans. Molecules 27. doi: 10.3390/molecules27072300
Jagmann N., Philipp B. (2014). Design of synthetic microbial communities for biotechnological production processes. J. Biotechnol. 184, 209–218. doi: 10.1016/j.jbiotec.2014.05.019
Johnson E. A. (2013). Biotechnology of non-Saccharomyces yeasts-the basidiomycetes. Appl. Microbiol. Biotechnol. 97, 7563–7577. doi: 10.1007/s00253-013-5046-z
Kerkhoven E. J., Pomraning K. R., Baker S. E., Nielsen J. (2016). Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. NPJ Syst. Biol. Appl. 2, 16005. doi: 10.1038/npjsba.2016.5
Konzock O., Matsushita Y., Zaghen S., Sako A., Norbeck J. (2022). Altering the fatty acid profile of Yarrowia lipolytica to mimic cocoa butter by genetic engineering of desaturases. Microb. Cell Fact. 21, 25. doi: 10.1186/s12934-022-01748-x
Kosina S. M., Danielewicz M. A., Mohammed M., Ray J., Suh Y., Yilmaz S., et al. (2016). Exometabolomics assisted design and validation of synthetic obligate mutualism. ACS Synth Biol. 5, 569–576. doi: 10.1021/acssynbio.5b00236
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lazar Z., Liu N., Stephanopoulos G. (2018). Holistic approaches in lipid production by yarrowia lipolytica. Trends Biotechnol. 36, 1157–1170. doi: 10.1016/j.tibtech.2018.06.007
Liu H., Song Y., Fan X., Wang C., Lu X., Tian Y. (2020). Yarrowia lipolytica as an oleaginous platform for the production of value-added fatty acid-based bioproducts. Front. Microbiol. 11, 608662. doi: 10.3389/fmicb.2020.608662
Liu X., Elgowainy A., Wang M. (2020). Life cycle energy use and greenhouse gas emissions of ammonia production from renewable resources and industrial by-products. Green Chem. 22, 5751–5761. doi: 10.1039/D0GC02301A
Lu R., Cao L., Wang K., Ledesma-Amaro R., Ji X. J. (2021). Engineering Yarrowia lipolytica to produce advanced biofuels: Current status and perspectives. Bioresour Technol. 341, 125877. doi: 10.1016/j.biortech.2021.125877
Madzak C. (2021). Yarrowia lipolytica strains and their biotechnological applications: how natural biodiversity and metabolic engineering could contribute to cell factories improvement. J. Fungi (Basel) 7. doi: 10.3390/jof7070548
Markham K. A., Alper H. S. (2018). Synthetic biology expands the industrial potential of yarrowia lipolytica. Trends Biotechnol. 36, 1085–1095. doi: 10.1016/j.tibtech.2018.05.004
Matt B. (2023). The industrialization of the Haber-Bosch process. C&EN Global Enterprise. 101, 20–21. doi: 10.1021/cen-10126-cover2
McCarty C. A., Harry M. L., Woehrle T. A., Kitch L. A. (2020). Screening and falls in community hospital emergency rooms in the 12 months following implementation of MEDFRAT. Am. J. Emerg. Med. 38, 1686–1687. doi: 10.1016/j.ajem.2019.12.053
Millan C., Pena C., Flores C., Espin G., Galindo E., Castillo T. (2020). Improving glucose and xylose assimilation in Azotobacter vinelandii by adaptive laboratory evolution. World J. Microbiol. Biotechnol. 36, 46. doi: 10.1007/s11274-020-02822-5
Miller K. K., Alper H. S. (2019). Yarrowia lipolytica: more than an oleaginous workhorse. Appl. Microbiol. Biotechnol. 103, 9251–9262. doi: 10.1007/s00253-019-10200-x
Moynihan M. A., Goodkin N. F., Morgan K. M., Kho P. Y. Y., Lopes Dos Santos A., Lauro F. M., et al. (2022). Coral-associated nitrogen fixation rates and diazotrophic diversity on a nutrient-replete equatorial reef. ISME J. 16, 233–246. doi: 10.1038/s41396-021-01054-1
Mus F., Khokhani D., MacIntyre A. M., Rugoli E., Dixon R., Ane J. M., et al. (2022). Genetic Determinants of Ammonium Excretion in nifL Mutants of Azotobacter vinelandii. Appl. Environ. Microbiol. 88, e0187621. doi: 10.1128/aem.01876-21
Nag P., Dheeman S., Maheshwari D. K. (2022). “Symbiotic and asymbiotic nitrogen fixation: an overview. Nitrogen fixing bacteria: sustainable growth of non-legumes.” in Microorganisms Sustainability, 11–22.
Navarrete C., Martínez J. L. (2020). Non-conventional yeasts as superior production platforms for sustainable fermentation based bio-manufacturing processes. AIMS Bioengineering 7, 289–305. doi: 10.3934/bioeng.2020024
Noar J. D., Bruno-Bárcena J. M. (2018). Azotobacter vinelandii: the source of 100 years of discoveries and many more to come. Microbiology 164, 421–436. doi: 10.1099/mic.0.000643
Otto R., Ferraz-Almeida R., Sanches G. M., Lisboa I. P., Cherubin M. R. (2022). Nitrogen fertilizer consumption and nitrous oxide emissions associated with ethanol production – A national-scale comparison between Brazilian sugarcane and corn in the United States. J. Cleaner Production 350. doi: 10.1016/j.jclepro.2022.131482
Plunkett M. H., Knutson C. M., Barney B. M. (2020). Key factors affecting ammonium production by an Azotobacter vinelandii strain deregulated for biological nitrogen fixation. Microbial Cell Factories. 19. doi: 10.1186/s12934-020-01362-9
Polpitiya A. D., Qian W. J., Jaitly N., Petyuk V. A., Adkins J. N., Camp D. G. 2nd, et al. (2008). DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24, 1556–1558. doi: 10.1093/bioinformatics/btn217
Pomraning K. R., Bredeweg E. L., Baker S. E. (2017). Regulation of nitrogen metabolism by GATA zinc finger transcription factors in yarrowia lipolytica. mSphere 2. doi: 10.1128/mSphere.00038-17
Pomraning K. R., Collett J. R., Kim J., Panisko E. A., Culley D. E., Dai Z., et al. (2019). Transcriptomic analysis of the oleaginous yeast Lipomyces starkeyi during lipid accumulation on enzymatically treated corn stover hydrolysate. Biotechnol. Biofuels. 12, 162. doi: 10.1186/s13068-019-1510-z
Pomraning K. R., Kim Y. M., Nicora C. D., Chu R. K., Bredeweg E. L., Purvine S. O., et al. (2016). Multi-omics analysis reveals regulators of the response to nitrogen limitation in Yarrowia lipolytica. BMC Genomics 17, 138. doi: 10.1186/s12864-016-2471-2
Pomraning K. R., Wei S., Karagiosis S. A., Kim Y. M., Dohnalkova A. C., Arey B. W., et al. (2015). Comprehensive Metabolomic, Lipidomic and Microscopic Profiling of Yarrowia lipolytica during Lipid Accumulation Identifies Targets for Increased Lipogenesis. PloS One 10, e0123188. doi: 10.1371/journal.pone.0123188
Quarterman J., Slininger P. J., Kurtzman C. P., Thompson S. R., Dien B. S. (2017). A survey of yeast from the Yarrowia clade for lipid production in dilute acid pretreated lignocellulosic biomass hydrolysate. Appl. Microbiol. Biotechnol. 101, 3319–3334. doi: 10.1007/s00253-016-8062-y
Riley R., Haridas S., Wolfe K. H., Lopes M. R., Hittinger C. T., Goker M., et al. (2016). Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. U S A. 113, 9882–9887. doi: 10.1073/pnas.1603941113
Rodriguez-Ocasio E., Khalid A., Truka C. J., Blenner M. A., Jarboe L. R. (2022). Survey of nonconventional yeasts for lipid and hydrocarbon biotechnology. J. Ind. Microbiol. Biotechnol. 49. doi: 10.1093/jimb/kuac010
Salvador Lopez J. M., Vidal L., Adiutama M. P., Van Nieuwerburgh F., Deforce D., Nicaud J. M., et al. (2023). How do engineered Yarrowia lipolytica strains secrete free fatty acids: hints from comparative transcriptomics. FEMS Yeast Res. 23, foad027. doi: 10.1093/femsyr/foad027
Sarnaik A., Liu A., Nielsen D., Varman A. M. (2020). High-throughput screening for efficient microbial biotechnology. Curr. Opin. Biotechnol. 64, 141–150. doi: 10.1016/j.copbio.2020.02.019
Schoch C. L., Robbertse B., Robert V., Vu D., Cardinali G., Irinyi L., et al. (2014). Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. Database (Oxford) 2014. doi: 10.1093/database/bau061
Smith C., Hill A. K., Torrente-Murciano L. (2020). Current and future role of Haber–Bosch ammonia in a carbon-free energy landscape. Energy Environ. Science. 13, 331–344. doi: 10.1039/C9EE02873K
Sommermann L., Geistlinger J., Wibberg D., Deubel A., Zwanzig J., Babin D., et al. (2018). Fungal community profiles in agricultural soils of a long-term field trial under different tillage, fertilization and crop rotation conditions analyzed by high-throughput ITS-amplicon sequencing. PloS One 13, e0195345. doi: 10.1371/journal.pone.0195345
Song B. L., Wang J. Q., Zhang G. X., Yi N. B., Zhang Y. J., Zhou L., et al. (2024). A coupling-induced assembly strategy for constructing artificial shell on mitochondria in living cells. Angew Chem. Int. Ed Engl., e202411725. doi: 10.1002/anie.202411725
Suchova K., Feher C., Ravn J. L., Bedo S., Biely P., Geijer C. (2022). Cellulose- and xylan-degrading yeasts: Enzymes, applications and biotechnological potential. Biotechnol. Adv. 59, 107981. doi: 10.1016/j.biotechadv.2022.107981
Sun L., Alper H. S. (2020). Non-conventional hosts for the production of fuels and chemicals. Curr. Opin. Chem. Biol. 59, 15–22. doi: 10.1016/j.cbpa.2020.03.004
Tiukova I. A., Brandenburg J., Blomqvist J., Sampels S., Mikkelsen N., Skaugen M., et al. (2019). Proteome analysis of xylose metabolism in Rhodotorula toruloides during lipid production. Biotechnol. Biofuels. 12, 137. doi: 10.1186/s13068-019-1478-8
Toju H., Tanabe A. S., Yamamoto S., Sato H. (2012). High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PloS One 7, e40863. doi: 10.1371/journal.pone.0040863
Tschitschko B., Esti M., Philippi M., Kidane A. T., Littmann S., Kitzinger K., et al. (2024). Rhizobia-diatom symbiosis fixes missing nitrogen in the ocean. Nature 630, 899–904. doi: 10.1038/s41586-024-07495-w
Walker C., Ryu S., Na H., Zane M., LaButti K., Lipzen A., et al. (2018). Draft genome assemblies of five robust yarrowia lipolytica strains exhibiting high lipid production, pentose sugar utilization, and sugar alcohol secretion from undetoxified lignocellulosic biomass hydrolysates. Microbiol. Resour Announc 7. doi: 10.1128/MRA.01040-18
Wang J., Ledesma-Amaro R., Wei Y., Ji B., Ji X. J. (2020). Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica - A Review. Bioresour Technol. 313, 123707. doi: 10.1016/j.biortech.2020.123707
Wang K., Shi T. Q., Lin L., Wei P., Ledesma-Amaro R., Ji X. J. (2022). Engineering yarrowia lipolytica to produce tailored chain-length fatty acids and their derivatives. ACS Synth Biol. 11, 2564–2577. doi: 10.1021/acssynbio.2c00305
Widder S., Allen R. J., Pfeiffer T., Curtis T. P., Wiuf C., Sloan W. T., et al. (2016). Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 10, 2557–2568. doi: 10.1038/ismej.2016.45
Yuzbasheva E. Y., Mostova E. B., Andreeva N. I., Yuzbashev T. V., Fedorov A. S., Konova I. A., et al. (2018). A metabolic engineering strategy for producing free fatty acids by the Yarrowia lipolytica yeast based on impairment of glycerol metabolism. Biotechnol. Bioeng. 115, 433–443. doi: 10.1002/bit.v115.2
Zhang G., Wang H., Zhang Z., Verstrepen K. J., Wang Q., Dai Z. (2022). Metabolic engineering of Yarrowia lipolytica for terpenoids production: advances and perspectives. Crit. Rev. Biotechnol. 42, 618–633. doi: 10.1080/07388551.2021.1947183
Keywords: nitrogen fixation, diazotroph, Yarrowia lipolytica, Azotobacter vinelandii, predictive phenomics, oleaginous, co-culture, community screening
Citation: Pomraning KR, Deng S, Duong RD, Czajka JJ and Bohutskyi P (2024) Identification of Yarrowia lipolytica as a platform for designed consortia that incorporate in situ nitrogen fixation to enable ammonia-free bioconversion. Front. Ind. Microbiol. 2:1473316. doi: 10.3389/finmi.2024.1473316
Received: 30 July 2024; Accepted: 02 September 2024;
Published: 23 September 2024.
Edited by:
Shuchi Singh, University of Illinois at Urbana-Champaign, United StatesReviewed by:
Divya Ramchandran, Amyris, United StatesNagiat Hwisa, University of Illinois at Urbana–Champaign, United States
Sangdo Yook, The University of Texas at Austin, United States
Copyright © 2024 Pomraning, Deng, Duong, Czajka and Bohutskyi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyle R. Pomraning, a3lsZS5wb21yYW5pbmdAcG5ubC5nb3Y=
 Kyle R. Pomraning
Kyle R. Pomraning Shuang Deng
Shuang Deng Rylan D. Duong1
Rylan D. Duong1 Jeffrey J. Czajka
Jeffrey J. Czajka Pavlo Bohutskyi
Pavlo Bohutskyi