- 1Department of Infection Biology (DIB), London School of Hygiene and Tropical Medicine (LSHTM), London, United Kingdom
- 2UCL Genetics Institute, University College London (UCL), London, United Kingdom
- 3National Institute of Veterinary Research, Hanoi, Vietnam
Introduction: Changing farming practices and the associated increase in the use of antibiotics are amongst the main drivers shaping the global increase of Campylobacter infections. The effects farming practices have on Campylobacter species, need to be studied at the global scale, particularly in emerging middle-income countries, where the demand for low-cost poultry meat is rising. While Campylobacter jejuni causes the majority of poultry associated diarrhoea, Campylobacter coli causes a significant amount of disease but are relatively understudied. In this study we characterised seven C. coli strains isolated from chicken faeces and chicken meat in Thai Nguyen province, Vietnam.
Methods: Whole Genome Sequencing and phenotypic assays (growth, motility, antimicrobial resistance testing, virulence assay) were performed to reveal the genetic relatedness and pathophysiological characteristics of the isolates.
Results: All isolates were resistant to ciprofloxacin and nalidixic acid but susceptible to phenicols. Three were resistant to macrolides azithromycin and erythromycin. Six isolates were classified as multi-drug resistant. All isolates had similar growth rates in laboratory culture media, while five were hyper-motile. Lethality towards a tractable host-model system, larvae of the greater wax moth Galleria mellonella, often used to determine Campylobacter virulence, was demonstrated for the first time for C. coli.
Discussion: Multilocus sequence typing data identified five ST’s all within the C. coli ST-828 clonal complex and were previously reported in North American (ST-829), European (ST-1586), and Asia (ST-5511) from patients suffering from gastroenteritis, emphasising the global spread of these strains. This work highlights the importance of further research into this understudied global threat.
1 Introduction
The demand for low-cost poultry meat and eggs are rapidly rising globally. To satisfy the increasing demand for meat and egg production the use of antimicrobial agents in animal food production has become widespread (Mulchandani et al., 2023). The global use of antibiotics is also projected to rise (Van Boeckel et al., 2017). Farmers, particularly in rapidly developing countries often rely on antibiotics as growth promoters and to improve the health of animals, however, this largely contributes to the generation of antibiotic resistant bacteria and the subsequent global increase in antimicrobial resistance (AMR) (Agyare et al., 2018). The seriousness of antibiotic overuse has been exacerbated further by difficulties in implementing antibiotic regulation policies in several countries (WHO, 2017; Tangcharoensathien et al., 2018). The interplay between the increasing incidence of foodborne illnesses, bacterial infections and AMR is strongly suggesting that the expansion of poultry production is presenting a growing public health risk (Kaakoush et al., 2015; Skarp et al., 2016).
The rapid intensification of poultry and egg has been linked with increasing incidence of bacterial food poisoning, mostly caused by Campylobacter species (Shane, 2000). Campylobacter spp are estimated to cause globally approximately 166M cases of disease [92M-300M 95% UI (uncertainty intervals)] and 37K (27-55k 95% UI) deaths annually (Kirk et al., 2015). This equates to 1,390 (752-2,576 95% UI) Disability Adjusted Life Years (DALYs) per 100,000 persons and 31 (22-46 UI) deaths per 100,000 persons (Kirk et al., 2015). However, LMIC regions carry significantly more burden of disease with Campylobacter spp being the most prevalent foodborne disease in the WHO South-East Asia region (1,152 [200-3,372 95% UI] DALYS per 100,000 persons and 0.4 [0.1-0.9 95% UI] deaths per 100,000 persons (Kirk et al., 2015). While species of Campylobacter are often combined in epidemiological data C. jejuni receives the majority of study. C. coli, the second most common Campylobacter species, is also implicated in bacterial gastroenteritis, contributing to a quarter of Campylobacter related gastroenteritis (Kaakoush et al., 2015), and one of the leading cause of foodborne diarrhoea (Laaveri et al., 2018).
Vietnam has undergone rapid economic and population growth, along with urbanisation leading to changes in agricultural and food production practices. Poultry is most popular livestock in Vietnam and the second-largest producer of meat (Birhanu et al., 2021). Vietnam had 512.7 million birds in 2020 producing 1.505 million tonnes of meat and 15.1 billion eggs (Birhanu et al., 2021). There is a paucity of data on Campylobacter prevalence and epidemiological data in Vietnam, in particularly for C. coli. A previous Vietnamese study conducted, identified C. coli being more resistant to antibiotics, such as ciprofloxacin, nalidixic acid and tetracycline, than C. jejuni, even though both species belong to the same ecological niche (Nguyen et al., 2016).
The aim of this pilot study was to characterise understudied C. coli isolated from poultry production networks, sampling from farms and point-of-sale retail outlets in Hanoi, Vietnam. The isolates were investigated for their phenotypic AMR profile and their draft genomes were compared against each other. Additionally, the Galleria mellonella virulence infection model was adapted for C. coli, which has not been previously reported. This pilot study paves the way for the large in-depth One Health Poultry Hub investigation of Campylobacter across Vietnam and provides much needed data on C. coli present in the Vietnamese poultry meat production network.
2 Materials and methods
2.1 Bacterial isolates
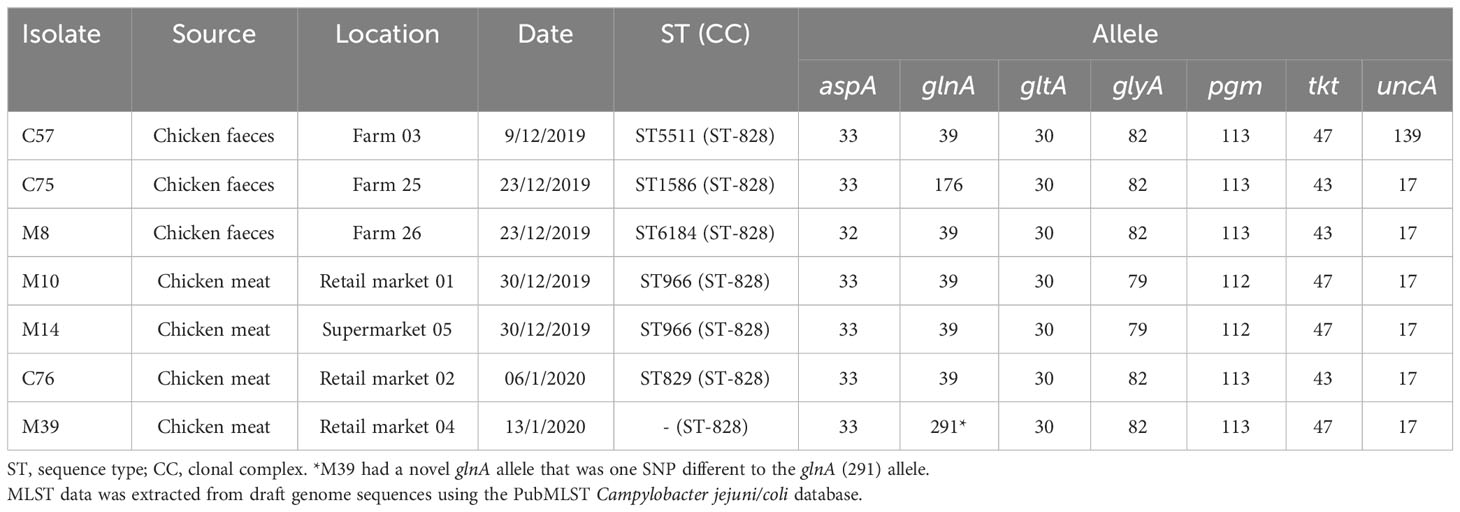
A prevalence survey of 30 chicken farms and 30 retail markets in Thai Nguyen province, Vietnam was performed in between December 2019 and January 2020. A total of 30 conventional farms were visited and a single faecal sample taken from six-week-old broilers. Five retail markets were visited three times and each occasion five raw chicken meat sample (approximately 300g meat) were collected. Five supermarkets were visited three times and each occasion five raw chicken meat sample (approximately 500g meat) were collected. A total of 36 putative Campylobacter isolates were from farms 25/30 (83.3%) and from retail markets 11/30 (36.7%) following the ISO 10272-1: 2017. Multiplex-PCR based identified revealed that 80.6% were C. jejuni (29/39) and 19.4% C. coli (7/36). The C. coli were selected for further study (Table 1; Supplementary Table S1).
2.2 Growth conditions
C. coli isolates were routinely cultured for 48 hours at 37°C on Columbia Blood Agar (CBA) (Oxoid, UK) supplemented with 5% defibrinated horse blood (Oxoid, UK) and Skirrow supplement (Oxoid, UK) under microaerobic conditions in a variable atmospheric incubator (Don Whitley Scientific, UK). C. coli isolates were further cultured on CBA/blood/skirrow plates for 24 hours prior to experimentation. Oxidase, indole test and gram staining were used to confirm the presence of Campylobacter sp. (Supplementary Table S1). Hippurate test was used to distinguish C. jejuni (positive) from C. coli (negative) (Supplementary Table S1). Growth curve experiments were also conducted to compare C. coli growth rate with well-studied C. jejuni 81-176, (a human isolate with a putative plasmid containing a type IV secretion and verified as highly virulent through testing in macaque monkeys (Russell et al., 1989). Growth curves were averaged from two biological repeats, in 10 ml Brucella broth (Oxoid, UK), containing ~108 colony-forming units (CFU), cultured for 24 hours under microaerobic conditions at 37°C with optical density readings (OD600nm), being taken at 2-hour intervals.
2.3 Motility assay
Overnight C. coli cultures were diluted to 8 x 108 CFU/ml bacteria in Brucella broth (Oxoid, UK). C. coli (3 µl) was added into 0.4% (wt/vol) Brucella agar and incubated for 72 hours at 37°C, under microaerobic conditions. Cell motility was observed at 24, 48 and 72 hours of growth. For experiment controls the representative strains C. jejuni 81-176 and C. jejuni 11168H [a hyper-motile variant of the standard original sequenced strain C. jejuni NCTC11168 (Parkhill et al., 2000)] were used. C. jejuni strains were used as they have well characterised motility and allow relative motility between C. coli and C. jejuni.
2.4 Antimicrobial susceptibility testing
Minimum inhibitory concentration (MIC) was determined based on EUCAST (version 3.0 Jan 2021), ISO 20776-1:2019 and CLSI (M45-A2) guidelines, following the broth microdilution method in untreated 96 well Nunc Sterile round bottom plates (ThermoFisher Scientific, UK). 5 x 105 CFU/ml of overnight bacterial cultures were inoculated with varying concentrations of antibiotics, described below. Antibiotics were prepared in correlation with EUCAST, ISO 20776-1:2019 guidelines. The samples were incubated for 48 hours at 37°C in filter sterilised cation adjusted MH broth, supplemented with 2.5% defibrinated horse blood (Oxoid, UK) along with 20 mg/L β-Nicotinamide adenine dinucleotide sodium salt (Sigma Aldrich, UK), at 120 revolutions per minute (RPM), under microaerobic conditions. Growth was determined visually by comparing to the negative control, as well as by using a SpectraMax iD5 microplate reader (Molecular Devices, UK).
Antibiotics tested with ranges and MIC values as described by National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) (https://www.cdc.gov/narms/antibiotics-tested.html) unless stated. Briefly gentamicin (0.125-32 mg/L), kanamycin (0.5-16 mg/L), streptomycin (0.0625-64 mg/L), clindamycin (0.0156-32 mg/L), azithromycin (0.015-64 mg/L), erythromycin [0.25-64 mg/L (EUCAST, 2023)], chloramphenicol (0.0156-256 mg/L), florfenicol (0.03-64 mg/L), ciprofloxacin [0.0156-64 mg/L (EUCAST, 2023)], nalidixic acid (2-64 mg/L), tetracycline (0.0625-64 mg/L), telithromycin (0.0156-8 mg/L), ampicillin (0.015-125 mg/L). Tentative epidemiological cut-off values (T)ECOFF were used for streptomycin, ampicillin, as no breakpoints were available during the study (EUCAST, 2020). Kanamycin had no breakpoint or T(E)ECOFF values available. C. jejuni ATCC 33560 with known MIC values was used as a control (EUCAST, ISO 20776-1:2019).
2.5 Virulence testing using Galleria mellonella model
Galleria larvae were sourced from LiveFood Direct and selected to be of matched size (~20 mm length), colour (cream) and maturity (~35-40 days old). Galleria are reared under controlled conditions and receive organic nutritional support including honey and wheatgerm. Prior to experimentation, Galleria were stored at 10°C within a breathable container half-filled with wood shavings. Campylobacter isolates were diluted to 108 CFU/ml in Phosphate-buffered saline (PBS) and 10 µl was injected into G. mellonella larvae foreleg. G. mellonella was incubated at 37°C with phenotypic effect being monitored over the course of nine days. Three biological replicates (using 2 x n=5 G. mellonella, 1 x n=10 G. mellonella) were conducted over 9 days. Phosphate-buffered saline (PBS) was used as a negative control and virulent C. jejuni 81-176 was used as a positive control. C. jejuni 81-176 was used as a control as the species has previously been used for the G. mellonella model (Champion et al., 2010; Senior et al., 2011) while, to our knowledge no C. coli strains have previously been studied in the G. mellonella model.
2.6 Genomic DNA extraction and sequencing
Genomic DNA was extracted using PureLink™ Genomic DNA Mini Kit (Invitrogen, UK), following manufacturer guidelines. Prior to sequencing DNA quality and concentration was measured using a NanoDrop 2000/2000c Spectrophotometer (Thermofisher Scientific, UK). Illumina Miseq library preparation and sequencing were prepared according to manufacturer’s protocol. Libraries were sequenced on a MiSeq (Illumina, USA) with 2 x 150 bp reagent kit v2. Additional sequencing was conducted using a MinION sequencer (Oxford Nanopore, UK). A R10 flow cell (FLO-MIN111) was used for sequencing, alongside ligation sequencing kit (SQK-LSK110) and the rapid barcoding kit (SQK-RBK004) for multiplexing, manufacturer recommended PCR conditions were used with eight minutes amplification using NEB LongAmp Taq 2X Master Mix (NEB M0287). DNA end repair was performed using NEBNext Ultra II End repair/dA-tailing Module (E7546), while adapter ligation was done using the NEBNext Quick Ligation Module (E6056).
2.7 Genome assembly
Raw fastq files were quality controlled using TrimmomaticPE with settings: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36 (Bolger et al., 2014). Sequenced reads were de novo assembled using St. Petersburg genome assembler (SPAdes version 3.15.3) (Nurk et al., 2013). The draft genome assemblies were assessed using quality assessment tool for genome assemblies (Quast) (Gurevich et al., 2013). For C. coli isolates that were sequenced on both MinION and MiSeq, draft genomes were assembled using Unicycler v0.4.9 (Wick et al., 2017). Once assembled, contigs were annotated by Prokaryotic Genome Annotation (Prokka) (Seemann, 2014) using a bespoke Campylobacter database using reference genomes (accession AF226280, AL111168, CM000854, CM000855, CP000025, CP000538, CP000768, CP000814, CP001876, CP001900, CP001960, CP002029, CP003871, CP006729, HE978252, HG428754).
2.8 AMR gene identification, MLST and virulence analysis
Abricate with the ResFinder database (update date 10-03-2022) (Zankari et al., 2012; Seemann, 2018) was used to identify AMR genes. Additionally, PathogenWatch (https://pathogen.watch/) and ResFinder (https://cge.food.dtu.dk/services/ResFinder/) identified AMR related single point mutations within the C. coli genomes. Multi-Locus Sequence Typing (MLST) analysis was conducted with MLST 2.0 from the Centre for Genomic Epidemiology using PubMLST.org database version 2.0.0. PubMLST database was also used to identify clonal complex of the sequenced isolates. ChewBBACA (version 16.04) (Silva et al., 2018) with the C. jejuni schema ‘INNUENDO_wgMLST’ (update date 20-05-2021) having 2794 loci was used for core genome multi locus sequence typing (cgMLST), by selecting loci that are present in 95% of C. coli and C. jejuni isolates within RefSeq database. PHYLOViZ was used to generate a minimum spanning tree, the Newick tree format, which was imported to Interactive tree of life (iTOL) to generate the tree in Figure 1. For ChewBBACA and Abricate C. coli isolates were compared against complete RefSeq C. jejuni and C. coli isolates. Pre-annotated sequence files were imported onto the Campylobacter Virulence Factors of Pathogenic Bacteria database (VFDB) (http://www.mgc.ac.cn/cgi-bin/VFs/genus.cgi?Genus=Campylobacter).
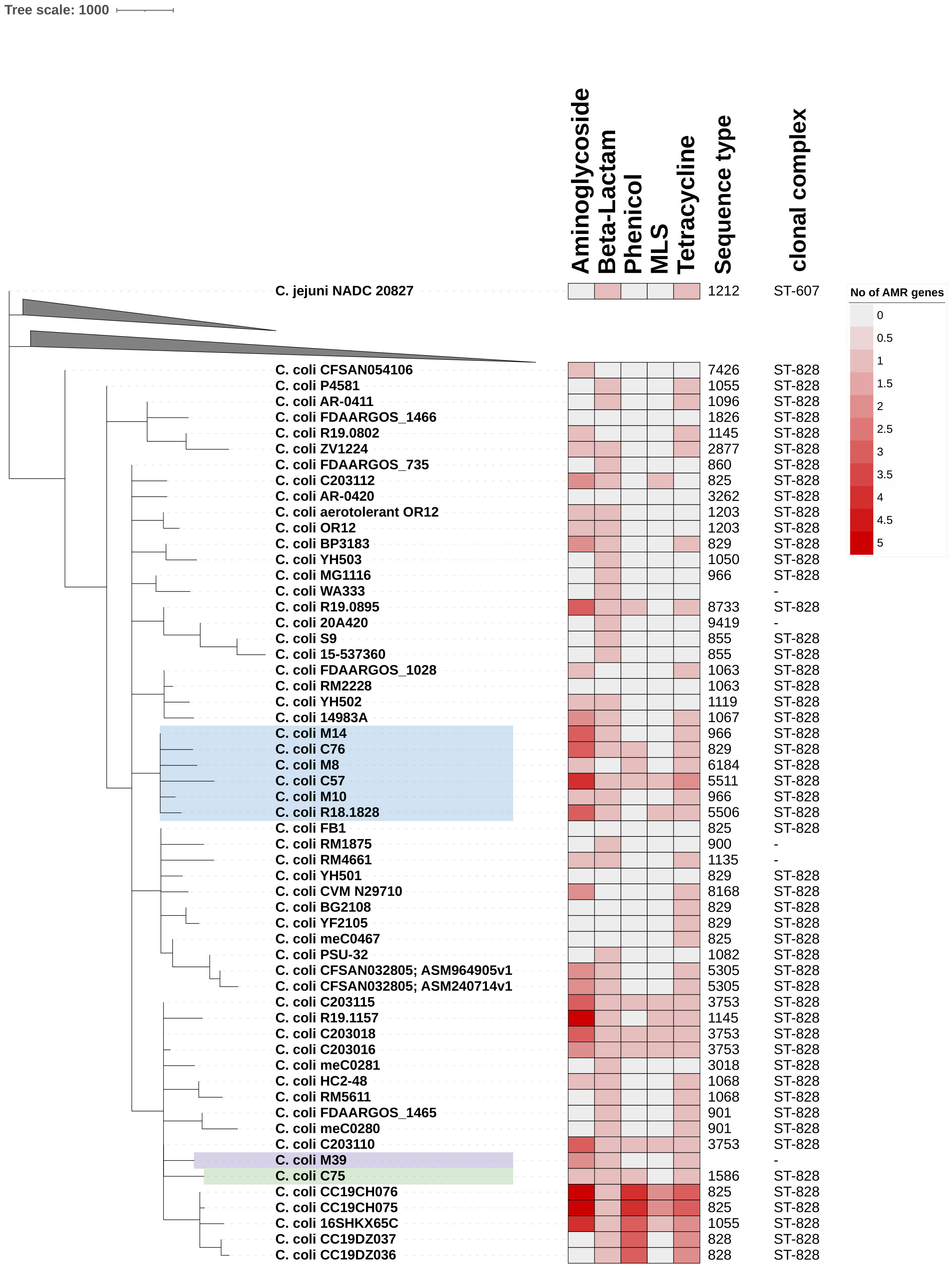
Figure 1 Phylogenetic analysis of clonal complex 828 (CC ST-828) isolates. Core-genome MLST phylogeny for seven Vietnamese C. coli isolates against RefSeq complete C. jejuni and C. coli isolates. Only C. coli isolates are shown, all of which have CC ST-828, as determined by PubMLST database. chewBACCA with schema having 2794 loci and 299214 alleles was used to determine cgMLST. cgMLST output was used to build phylogenetic tree on interactive tree of life (iTOL). C. jejuni NADC 20827 (ST1212, CC ST-607) was used as outlier to root phylogeny. Heat map of genome analysis using Abricate and ResFinder database identified between 0 (white) and 5 (deep red) resistance genes linked to five antibiotic classes. MLS (Macrolides, Lincosamides, Streptogramins).
2.9 Statistical analyses
Survival curves were generated using Prism version 9.1 software, and survival rates of G. mellonella models were analysed using the Kaplan-Meier method. Tests were performed using a two-sided probability, and survival rates were considered statistically significant when p < 0.05. For motility and growth rate analysis, three biological replicates were conducted, and statistical significance was conducted using one way ANOVA test, with p < 0.05 being considered statistically significant.
3 Results
3.1 Phenotypic analysis
3.1.1 Growth rate analysis of C. coli
C. coli were isolated from poultry faeces (C57, C75, C76) and meat (M8, M10, M14, M39) from farms, retail markets or supermarkets in Hanoi, Vietnam (Supplementary Table S1). All isolates were catalase and oxidase positive but Indole and Hippurate negative (Supplementary Table S1). C. coli growth was also compared with C. jejuni 81-176 in brucella broth for 24 hours, demonstrating no significant difference in growth rate between the C. coli isolates and C. jejuni 81-176 (Supplementary Figure S1).
3.1.2 Motility
To assess motility of the C. coli strains, the isolates were compared to two C. jejuni with known motility phenotypes; hyper-motile C. jejuni 11168H (Karlyshev et al., 2001; Macdonald et al., 2017) and motile C. jejuni 81-176 (Korlath et al., 1985; Hofreuter et al., 2006). After 72hrs, C. coli C75 (p=0.001), M14 (p=0.01) and M39 (p = 0.04) had disseminated through soft Brucella agar significantly further than hypermotile C. jejuni 11168H and were deemed more motile (Figure 2). C. coli C57 (p=0.15) and C76 (p=0.06) motility was not significantly different from the C. jejuni 11168H control (Figure 2). C. coli M8 (p=0.02) and M10 (0.03) were less motile than C. jejuni 11168H but similar to the motile C. jejuni 81-176 (Figure 2).
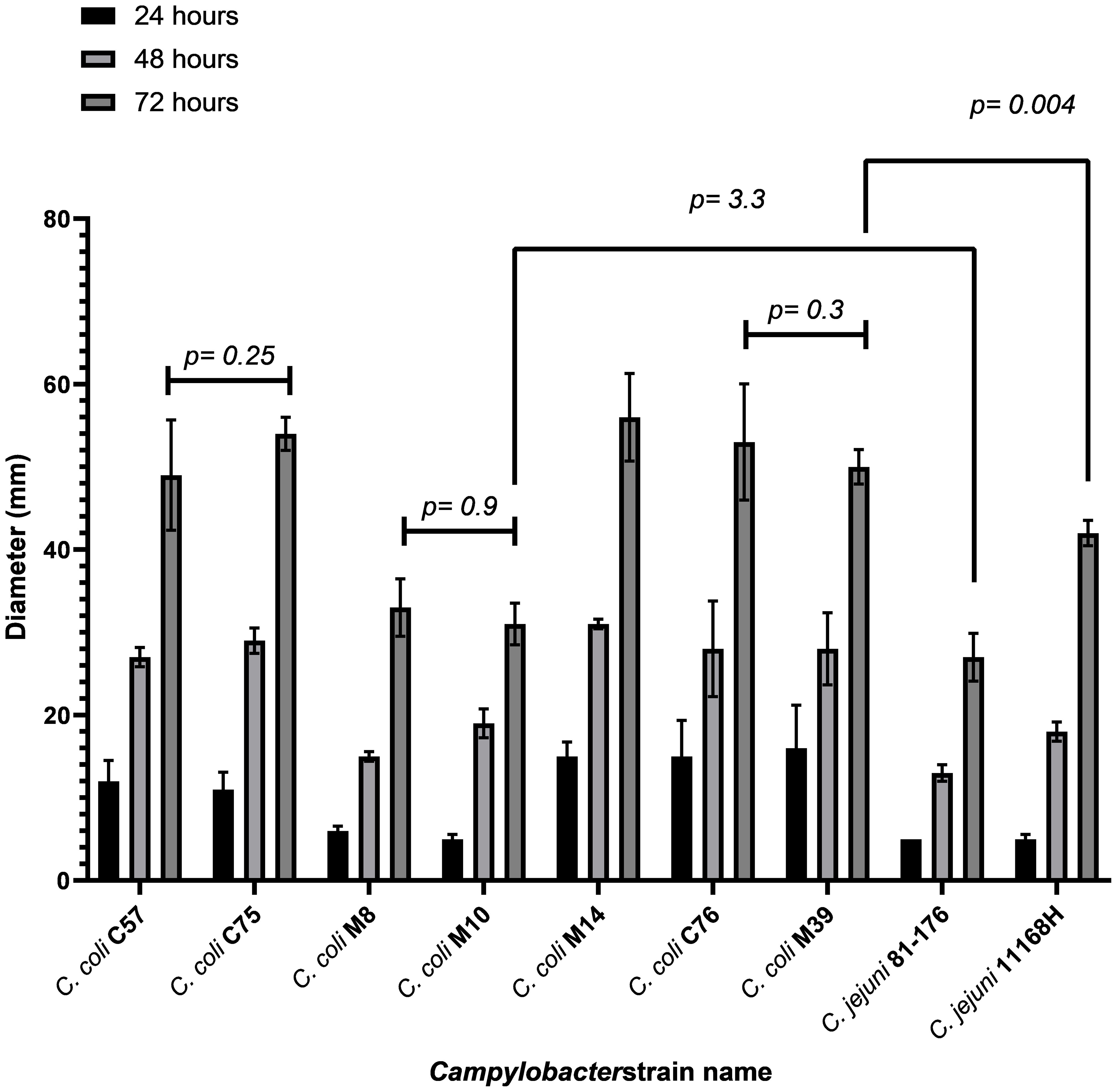
Figure 2 Motility analysis of C. coli isolates along with C. jejuni 81-176 and 11168H. The data shows an average of three biological replicates. Data was obtained by inoculating Brucella 0.4% (wt/vol) agar with 8 x 108 CFU/ml bacteria. Growth was observed over three days with measurement time points being 24, 48 and 72 hrs. P-values are generated using one-way ANOVA. C. coli isolates were compared against C. jejuni 81-176 and hyper-motile isolate C. jejuni 11168H.
3.1.3 Antibacterial susceptibility testing
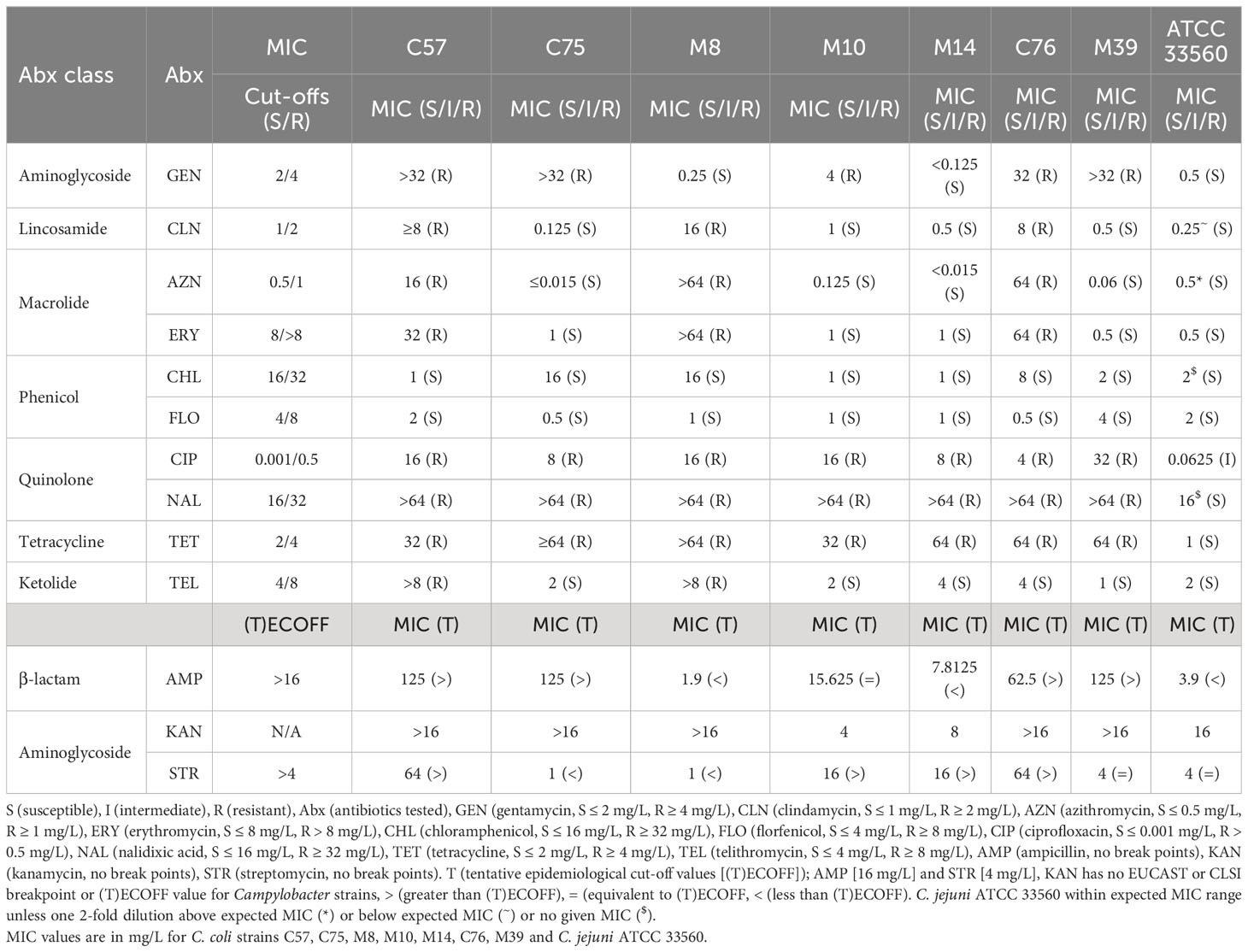
All C. coli isolates were resistant to quinolones (ciprofloxacin/nalidixic acid) and tetracycline (Table 2). For aminoglycosides, C. coli C57 and C76 were resistant to gentamycin. C57 and C76 also had kanamycin MIC of >16 mg/L and streptomycin MIC of 64 mg/L, which was greater than the (T)ECOFF of 4 mg/L (Table 2). All isolates were susceptible to phenicols, however, C75 and M8 were a single doubling dilution below the resistance cut-off (Table 2). C. coli C57, C76 and M8 were resistant to macrolides (erythromycin and azithromycin) and lincosamide (clindamycin) (Table 2). Isolates C57 and M8 were also resistant to telithromycin (Table 2). C. coli C57, M39 and C75 demonstrated very high level of ampicillin insensitivity (MIC 125 mg/L), higher than (T)ECOFF (16 mg/L) while C. coli M8 (1.9 mg/L) was acutely susceptible (Table 2). Multi-drug resistance (MDR) of thermophilic Campylobacter strains is defined as resistance to at least three different antimicrobial classes (Magiorakos et al., 2012). All isolates, except C. coli M14, match the MDR definition. C. coli M14 was only resistant to quinolones and tetracycline, however, was also above the (T)ECOFF for streptomycin (aminoglycoside; (16 vs 4 mg/L) (Table 2). C. coli C57 was resistant to all classes tested, except phenicols (Table 2).
3.1.4 Galleria mellonella pathogenicity
Campylobacter does not naturally infect G. mellonella, however, due to the lack of readily available models for testing Campylobacter virulence, it is an established surrogate infection model to assess virulence and it has been used for numerous bacterial infections, including C. jejuni (Kurstak and Vega, 1968; Dudziak and Jozwik, 1969; Champion et al., 2010; Insua et al., 2013; Viegas et al., 2013; Bojanic et al., 2020). Pathogenicity of C. coli isolates was tested using a G. mellonella wax moth infection model (Figure 3). The results showed that disease in G. mellonella infected with C. coli M10 and M14 are significantly attenuated (p=0.0438 and p=0.0290 respectively, Gehan-Breslow-Wilcoxon test) compared to the positive control C. jejuni 81-176, with many G. mellonella surviving beyond 9 days. Survival of G. mellonella, infected with C. coli C75, C76, M8 and M39, was not significantly different from C. jejuni 81-176; p= 0.593, 0.117, 0.692 and 0.164 respectively (Figure 3). More C. coli C57 infected G. mellonella died than C. jejuni 81-176 over the initial 4 days, but this was not significant over the course of 9 days, p=0.224 (Figure 3).
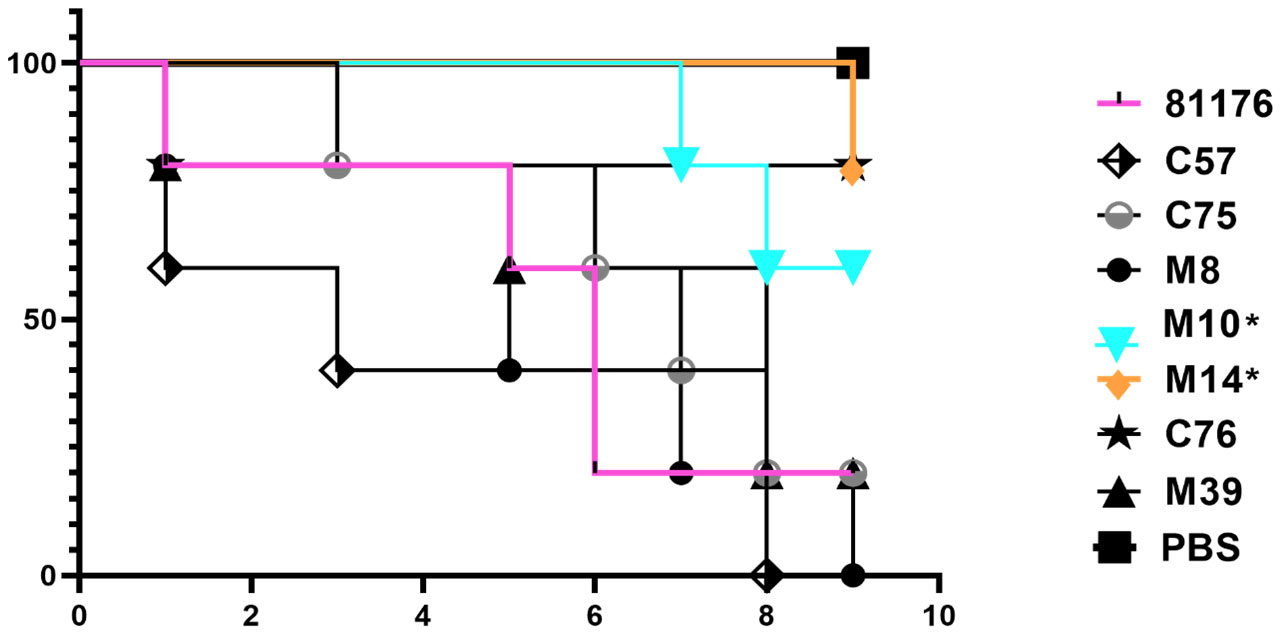
Figure 3 Virulence testing of Vietnamese C. coli isolates using Galleria mellonella model. Representative survival plot showing probability of survival of G. mellonella after infection with C. coli isolates (C57, C75, M8, M10, M14, C76, M39). C. jejuni 81-176 was used as a positive reference standard and PBS negative control. Data from an independent biological repeat, representative of the three independent experiments conducted is shown. C. coli isolates M10 and M14 were significantly attenuated (p=0.0438 and p=0.0290 respectively, Gehan-Breslow-Wilcoxon test) compared to the C. jejuni 81-176 reference control.
3.2 Genotypic analysis
3.2.1 C. coli genomes
All isolates were whole genome sequenced using Illumina short read sequencing technology with C. coli isolates C57, C76, M8 and M39 further sequenced using a Minion sequencer (Supplementary Table S2). Draft genomes ranged from 1.64 – 1.77 Mbp, which is comparable to C. coli 76339 accession HG326877 (1.58 Mbp) and C. coli RM4661 accession CP007181 (1.82 Mbp). Assembly contig numbers ranged from 1 (M8) to 21 (C75) contigs of length ≥ 1kb. The mean number of predicted coding sequences (CDSs) for sequenced C. coli strains was 1773 (SD ±50.2) (Supplementary Table S2).
3.2.2 Identification of a novel C. coli plasmid.
A circularised plasmid of length 27,557 bp was identified for isolate C. coli M39, named pCCM39-Huong. Annotation of pCCM39-Huong identified 37 coding sequences; 22 had predicted function, but 15 (40%) were hypothetical of unknown function (Supplementary Table S3). Annotated genes included ssb2 (single stranded DNA binding protein) (Jeon and Zhang, 2007), potentially involved in DNA processing during conjugation. Genes traG and traL both involved in conjugal transfer of genetic material, a toxin component (vbhT) (Sprenger et al., 2017), virB1, virB4, virC, trbB, traM, trbl, trbF, trbF, trbM and trbD all related to type IV secretion system were also found (Zatyka and Thomas, 1998; Mace et al., 2022) (Supplementary Table S3).
pCCM39-Huong had sequence similarity to two unnamed plasmids found in C. coli isolates 20A420 (accession CP058341) and C. coli meC0281 (CP027635) (Figure 4). C. coli 20A420 plasmid was of length 26,486 bp, with 97.36% identity and 90% coverage compared to pCCM39-Huong. Plasmid meC0281 was of length 30,719 bp and had 96.33% identity and 96% coverage when compared to pCCM39-Huong (Figure 4). When the two plasmids p20A420 and meC0281 were aligned to plasmid pCCM39-Huong, the difference between the two plasmids and pCCM49-Huong related to non-contiguous hypothetical protein encoding genes (Figure 4). C. jejuni 81-176 contains two plasmids (pTet [AF226280] and pVir [AY394561]) both of approximately 35kb. pCCM39-Huong reads did not map to any of the pVir gene sequence, demonstrating that these plasmids are highly different to plasmid pCCM39-Huong. Comparison between pCCM39-Huong assembly and pTet only identified a single shared complete gene (a putative DNA primase) and partial matches to cpp32 (phage/Rha family transcriptional regulator) and cmgB5 (putative type IV secretion system component). Mapping of the M39 genome fastq data showed that pTet tetO (tetracycline resistance) was highly similar to the chromosomal tetO present within the M39 genome.
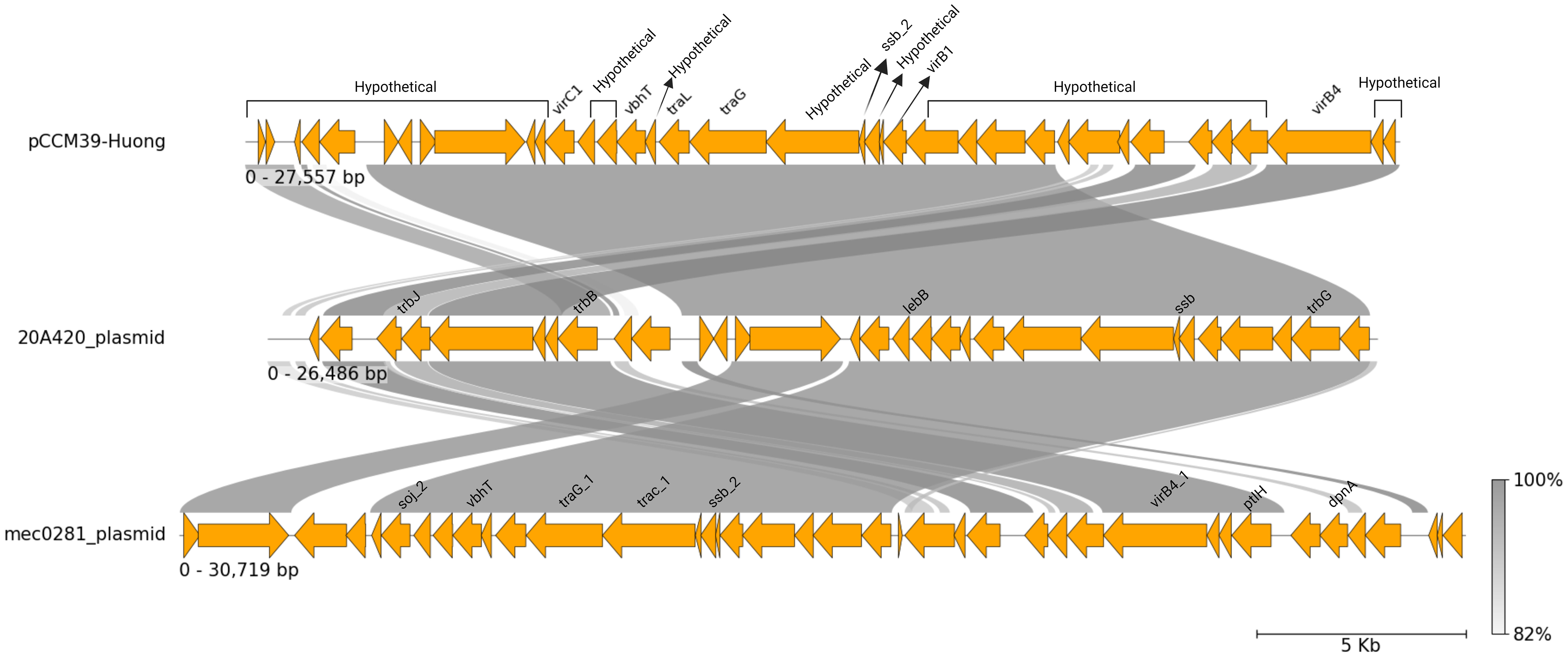
Figure 4 C. coli plasmid pCCM39-Huong. A linear representation of pCCM39-Huong generated using pyGenomeViz (Shimoyama, 2022), showing Mauve alignment (Darling et al., 2004) to two similar plasmid sequences p20A420 and meC0281. Conserved regions are shown by grey lines with intensity representing level of percent sequence identity.
3.2.3 C. coli epidemiological information
Analysis of the sequenced C. coli genomes identified five different sequence types (STs), with M39 having a novel ST, due to a novel glnA allele that was one single-nucleotide polymorphism (T198C) different to glnA(291) (Table 1). All STs belonged to the ST-828 clonal complex (Table 1). In the PubMLST database (accessed 22-6-2022) of the 16,083 ST-828 clonal complex isolates, 15,974 (99.3%) were identified as C. coli, 97 C. jejuni and 12 as Campylobacter spp. cgMLST further confirmed that the isolates were C. coli, as these were grouped away from C. jejuni (Figure 1) and clustered with reference ST-828 clonal complex isolates (Figure 1). The sequenced isolates were not clustered based on whether they were isolated from farm or supermarket/retail market (Figure 1). C. coli M39 and C75 belonged to the same subclade (Figure 1) and shared 5 MLST alleles (Table 1). M10 and M14 (from supermarket-bought chicken meat) were both ST-966, however cgMLST data indicated that these were very similar, but not identical isolates with a lowest harmonic mean allelic distance of 268. C. coli M14, C76, M8, C57, M10 belonged to the same subclade and were related to C. coli R18.1828, isolated from Taiwan (Figure 1).
3.2.4 AMR genotypes
Analysis of the draft genomes identified potential AMR genes to all classes of antibiotics tested. The β-lactamase gene blaOXA-193 was present in all sequenced genomes except M8 and 142 out of 284 (50%) reference genomes (Supplementary Table S4). Four aminoglycoside genotypes were identified with isolates containing between one (M8, M10, C75) and three (C57, C76, M14) CDSs (Supplementary Table S4). All isolates had a copy of tet (O) linked to tetracycline resistance, 4 out of 7 sequenced isolates had cat2 phenicol resistance gene, but only C57 had the erm(B) MLS (macrolide, lincosamide and streptogramin) resistance gene (Figure 1; Data Sheet 2).
Sequence analysis showed that only C. coli C57 was predicted to have nine resistance genes, covering aminoglycosides, β-lactam, phenicol, MLS and tetracycline resistance (Figure 1; Supplementary Table S4). Analysing the C. coli M8 genome identified aminoglycosides, β-lactamase and phenicol AMR genes (Figure 1; Supplementary Table S4). The ResFinder database did not identify any resistance genes for the control susceptible strain C. jejuni ATCC 33560.
3.3 Correlation between AMR phenotype and genotypes
Prediction of phenotypic resistance to antibiotics from the presence of genes linked to resistance has proven complex (Doyle et al., 2020). All isolates, except M8 and M14 were resistant to gentamycin. Four isolates had an MIC above streptomycin (T)ECOFF and five had an MIC of >16 mg/L for kanamycin, however all isolates contained at least one aminoglycoside resistance gene. Only the isolates (C. coli C57, M10, M14 and C76) with ant(6)-Ia were resistant to streptomycin. C. coli M8 possessed only aph(3’)-III and had a kanamycin MIC > 16, similar to other three other aph(3’)-III positive isolate (C. coli C75, C76 and M39). The fifth aph(3’)-III positive isolates, C. coli M14, had a kanamycin MIC of 8. While C57 did not possess an aph(3’)-III and had a kanamycin MIC >16 this isolate did have four alternative aminoglycoside genes. All isolates, except C. coli M8 possessed blaOXA-193 as the only β-lactamase yet MIC values ranged from 1.9 (C. coli M8) to >125 (C. coli C57, C75 & M39). C. coli C57, M8 and C76 were resistant to erythromycin, the erm(B) inducible erythromycin resistance was only present in C. coli C57 however M8 and C76 had a 2075A>G mutation within the 23S rRNA gene associated with macrolide resistance. All isolates were non-susceptible to ciprofloxacin and SNP analysis identified an identical gyrase subunit A (gyrA) (T86I) mutation in all isolates known to confer resistance.
3.4 3 Correlation between virulence related genes and genotypes
Genotypic data for each isolate showed that motility genes (flaA, flaB, flaC, flaG, flgB, flgC, flgD, flgE2, flgE, flgG2, flgG, flgH, flgI, flgK, flgL, flgR, flhA, flhB, flhF, flhG, fliA, fliD, fliE, fliF, fliG, fliH, fliI, fliL, fliM, fliN, fliP, fliQ, fliR, fliS, fliY, motA, motB, pflA) were present in all C. coli isolates. All C. coli isolates also had the adhesion (cadF, jlpA, MOMP, PEB1) (Krause-Gruszczynska et al., 2007), cytolethal toxin (cdtABC) and Campylobacter invasion related genes (ciaB) (Konkel et al., 1999). The well-characterised O- and N-linked general glycosylation systems were also present.
C. coli M10 (and M14) demonstrated significantly lower virulence in the G. mellonella infection model and was the least motile (along with M8), whereas C. coli C57 was the most virulent isolate. To identify any genetic cause for the virulence of C. coli C57, we isolated C57 fastq reads that did not match the M10 and M14 assembled draft genomes. These unmapped reads were assembled to represent CDSs in C. coli C57 that were absent or highly divergent from C. coli M10 and M14. The unmapped reads assembled in to 39 genome fragments containing 158 predicted CDSs. To remove any CDSs that were present in M10 and M14, but were not currently in these draft genomes, the raw reads from M10 and M14 were mapped against this assembly, which identified six contigs containing 16 CDSs that were present in M14 but not M10. 106 of 142 CDS were annotated as hypothetical proteins, two were IS1595 family transposases, 28 were annotated as enzymes, with many linked to sugar metabolism (e.g. dTDP-glucose 4,6-dehydratase), modification methylase (dpnIIA and dpnIIB), ribosomal silencing factor (RsfS) and sugar efflux transporter. Additionally, both flagellin FlaA (highly like the residues 102-572 of 11168 FlaA) and FlaB (67 amino acid fragment, identical to last 67 residues of 11168 FlaB) were present, demonstrating significant variation between C. coli C57 and M10/M14 flagellin genes.
4 Discussion
4.1 Is C. coli antimicrobial resistance a concern?
C. coli has emerged as a significant public health concern due to its high rates of antimicrobial resistance (van Vliet et al., 2022). There is evidence of genetic convergence between C. coli and C. jejuni, due to human activity, such as intensive farming of poultry, where both species can inhabit, allowing increased horizontal transmission of DNA between the species (Sheppard et al., 2008).
The antibiotic of choice for Campylobacter associated gastroenteritis are macrolides (Wieczorek and Osek, 2013), however, macrolide resistance is more common in C. coli isolates than C. jejuni (Luangtongkum et al., 2009). Macrolides resistance in North America is around 10% for both C. jejuni and C. coli from humans/broilers. Across Europe C. jejuni from humans had lower levels of erythromycin resistance (1.1%) compared to C. coli (8.5%) (European Food Safety, A and P. European Centre for Disease and Control, 2023). While in Asia, less than 5% of C. jejuni were macrolide resistant, as opposed to 14 – 62% of C. coli (Luangtongkum et al., 2009). In this study, three out of seven isolates were found to be resistant to both macrolides, presumptively linked to the presence of either an acquired gene or 23S rRNA gene mutations, reflecting previous studies in this country. Fluoroquinolones are also commonly used to treat Campylobacteriosis but again reduced susceptibility (ciprofloxacin and nalidixic acid) was demonstrated in all isolates and linked to a known gyrA mutation (Ohishi et al., 2017; Devi et al., 2019). This was unsurprising, as previous studies have indicated gyrA related resistance to fluoroquinolones being common in Vietnam, with Campylobacter isolates rapidly acquiring resistance (Luangtongkum et al., 2009). Tetracycline resistance was also observed for all isolates due to tet(O) (Pratt and Korolik, 2005), although a duplicate copy of tet(O) in C. coli C57 was not linked with higher MIC. Tetracycline is seldom used for the treatment of acute gastroenteritis (Garcia-Fernandez et al., 2018), but is the most frequent antibiotic in the treatment and control of various poultry diseases, leading to high resistance rates in a number of zoonotic pathogens.
The presence of multiple AMR phenotypes meant that all but C. coli M14 were resistant to at least three classes of antibiotics, however no isolates demonstrated resistance to the phenicols tested. Florfenicol is used for agricultural purposes and is also thought to be more active than chloramphenicol (Sutili and Gressler, 2021). Yet, the borderline resistance for chloramphenicol in two of the C. coli strains C75 and M8 should be concerning, as resistance to this group of antibiotics may arise without judicious use.
4.2 Do C. coli from South-East Asia present a global threat to human health?
MLST is an epidemiological tool to link isolates with a similar genetic background together and can be used to understand transmission of pathogens on both a local and global scale. All the MLST profiles, apart from M39 with a novel glnA mutation, were in the public database (25th Jan 2022) and were therefore previously reported. The MLST profiles of C. coli C75 (ST-1586), M8 (ST-6184), M10 (ST-966) and M14 (ST-966) have previously been found in human stool samples, many of which were linked to sporadic cases of gastroenteritis, demonstrating that the isolates could potentially be of clinical importance. ST-1586 (C. coli C75) have been identified in ‘North America’ (6 isolates), USA (5) and the UK (10), all of which were identified from human faeces, and some documented as a sporadic case of gastroenteritis. ST-1586 (C. coli C75) has been reported from Vietnamese clinical samples between 2014-2019. ST-966 (M14 and M10) has mostly been documented from the USA, except a single UK clinical isolate. ST-829 (C. coli C76) was primarily from the USA (1021, 83%) with many of which from carriers such as poultry. ST-5511 (C. coli C57) was rare with only being identified in three C. coli isolates from China, with two identified from chicken. ST-6184 (C. coli M8) has been identified previously in poultry in the USA and China, with a single case from China isolated from human stool, linked to an outbreak. ST-829 (C. coli C76) has been submitted over 1200 times to the MLST database, mostly identified in high-income countries (HICs), mainly from the USA (1021, 83% of the documented cases). ST-829 was identified in England (44) and Scotland (33), all of which were isolated from human faeces, relating to gastroenteritis. The MLST database is heavily skewed towards isolates from Europe, UK and USA so the true spread of these clones is unknown but it does suggest that the global transport of food is carrying these isolates far and wide to the detriment of human health.
4.3 Are C. coli more virulent than C. jejuni?
Within the limited number of isolates common Campylobacter sp. virulence related genes were identified and it was demonstrated C. coli can be equally motile as the reference hyper-motile C. jejuni 11168H strain and equally lethal as the reference strain C. jejuni 81-176 in the G. mellonella infection model. Although no strain was significantly more virulent than the C. jejuni reference, two strains were significantly less virulent. Comparison between these identified 142 CDSs present in the more virulent isolate however with the majority of these with unknown function it was not possible to link genotypes to virulence. Genome sequencing identified a potential novel virulence related plasmid in C. coli M39, carrying genes related to the type IV secretion system. The type IV secretion system is involved in DNA conjugation, helping bacteria in adapting to environmental stress and is a potential mechanism of spreading AMR, allowing the uptake of DNA from the environment (Wallden et al., 2010) although this strain was not more resistant than other isolates. The type IV secretion system can also play a role in increased host colonisation, biofilm formation and transfer of toxins to host cells and thus can have a direct involvement in virulence (Kienesberger et al., 2011), although this strain was no more virulent than other C. coli isolates in the Galleria model.
Despite the limitations of the study in terms of isolate numbers, narrow graphical and temporal range it was demonstrated that C. coli warrants further study to understand the global relevance compared to the highly studied C. jejuni. Although C. jejuni is the most common Campylobacter species to infect humans, other Campylobacter species, such as C. coli need to be studied for several reasons, including interspecies horizontal gene transfer (Mourkas et al., 2022), which may contribute the emergence of multi-drug resistant pathogens. This, together with the expanding global poultry production networks and the intensification of production, may pose a global public health risk. This highlights the urgent need for a coordinated global effort to combat AMR.
Declaration of generative AI in scientific writing
During the preparation of this work the authors did not use any generative AI and AI-assisted technologies in the writing process.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, JAKGTW000000000, JAKGTV000000000, JAKGTS000000000, JAKGTU000000000, JAKGTT000000000, JAKGTR000000000, CP091310.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
BL: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing, Data curation. GN: Data curation, Formal analysis, Writing – original draft. AC: Formal analysis, Investigation, Writing – review & editing. FN: Investigation, Writing – review & editing, Methodology, Resources. SW: Investigation, Methodology, Writing – review & editing, Formal analysis. PN: Writing – review & editing. BW: Writing – review & editing, Funding acquisition. LH: Writing – review & editing, Conceptualization, Formal analysis, Methodology, Writing – original draft. RS: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing, Investigation, Project administration, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the UKRI GCRF One Health Poultry Hub (Grant No. BB/S011269/1), one of 12 interdisciplinary research hubs funded under the UK government’s Global Challenges Research Fund Interdisciplinary Research Hub initiative. BL, LH and RS and this project was supported by the BBSRC GCRF One Health Poultry Hub grant number BB/S011269/1. GN was supported by BBSRC grant number BB/M009513/1.
Acknowledgments
We would like to thank Abdi Elmi and Sherif Abouelhadid for their technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finmi.2024.1355079/full#supplementary-material
Supplementary Table 4 | Analysis of the draft genomes identified potential AMR genes. Abricate with the ResFinder database (update date 10-03-2022) was used to identify AMR genes. Red highlighting indicates study samples demonstrating resistance to a specific class of antibiotics.
References
Agyare C., Etsiapa Boamah V., Ngofi Zumbi C., Boateng Osei F. (2018). “Antibiotic use in poultry production and its effects on bacterial resistance,” in Antimicrobial Resistance - A Global Threat, ed Kumar Y. (IntechOpen, online). http://dx.doi.org/10.5772/intechopen.79371.
Birhanu M. Y., Geremew K., Esatu W., Worku S., Getachew F., Viet Don N., et al. (2021). Poultry production, marketing and consumption in Vietnam: A review of literature. ILRI Research Report 80 (Nairobi, Kenya).
Bojanic K., Acke E., Roe W. D., Marshall J. C., Cornelius A. J., Biggs P. J., et al. (2020). Comparison of the Pathogenic Potential of Campylobacter jejuni, C. upsaliensis and C. helveticus and Limitations of Using Larvae of Galleria mellonella as an Infection Model. Pathogens 9. doi: 10.3390/pathogens9090713
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170.
Champion O. L., Karlyshev A. V., Senior N. J., Woodward M., La Ragione R., Howard S. L., et al. (2010). Insect infection model for campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 201, 776–782. doi: 10.1086/650494
Darling A. C., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704.
Devi A., Mahony T. J., Wilkinson J. M., Vanniasinkam T. (2019). Antimicrobial susceptibility of clinical isolates of Campylobacter jejuni from New South Wales, Australia. J. Glob Antimicrob. Resist. 16, 76–80. doi: 10.1016/j.jgar.2018.09.011.
Doyle R. M., O'Sullivan D. M., Aller S. D., Bruchmann S., Clark T., Coello Pelegrin A., et al. (2020). Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: an inter-laboratory study. Microb. Genom 6. doi: 10.1099/mgen.0.000335.
Dudziak B., Jozwik Z. (1969). The virulence of some Escherichia coli strains for larvae of Galleria mellonella L. Acta Microbiol. Pol. B 1, 137–141.
EUCAST (2020) The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 10.0, 2020. (Sweden: EUCAST). Available online at: http://www.eucast.org/clinical_breakpoints/.
EUCAST (2023). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 13.1. (Sweden: EUCAST). Available online at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.1_Breakpoint_Tables.pdf.
European Food Safety, A, P. European Centre for Disease and Control (2023). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 21, e07867. doi: 10.2903/j.efsa.2023.7867
Garcia-Fernandez A., Dionisi A. M., Arena S., Iglesias-Torrens Y., Carattoli A., Luzzi I. (2018). Human campylobacteriosis in Italy: emergence of multi-drug resistance to ciprofloxacin, tetracycline, and erythromycin. Front. Microbiol. 9, 1906. doi: 10.3389/fmicb.2018.01906.
Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086.
Hofreuter D., Tsai J., Watson R. O., Novik V., Altman B., Benitez M., et al. (2006). Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74, 4694–4707. doi: 10.1128/IAI.00210-06.
Insua J. L., Llobet E., Moranta D., Perez-Gutierrez C., Tomas A., Garmendia J., et al. (2013). Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect. Immun. 81, 3552–3565. doi: 10.1128/IAI.00391-13.
Jeon B., Zhang Q. (2007). Cj0011c, a periplasmic single- and double-stranded DNA-binding protein, contributes to natural transformation in Campylobacter jejuni. J. Bacteriol 189, 7399–7407. doi: 10.1128/JB.01012-07.
Kaakoush N. O., Castano-Rodriguez N., Mitchell H. M., Man S. M. (2015). Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 28, 687–720. doi: 10.1128/CMR.00006-15.
Karlyshev A. V., McCrossan M. V., Wren B. W. (2001). Demonstration of polysaccharide capsule in Campylobacter jejuni using electron microscopy. Infect. Immun. 69, 5921–5924. doi: 10.1128/IAI.69.9.5921-5924.2001.
Kienesberger S., Schober Trummler C., Fauster A., Lang S., Sprenger H., Gorkiewicz G., et al. (2011). Interbacterial macromolecular transfer by the Campylobacter fetus subsp. venerealis type IV secretion system. J. Bacteriol 193, 744–758. doi: 10.1128/JB.00798-10.
Kirk M. D., Pires S. M., Black R. E., Caipo M., Crump J. A., Devleesschauwer B., et al. (2015). World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases 2010: A data synthesis. PloS Med. 12, e1001921. doi: 10.1371/journal.pmed.1001921.
Konkel M. E., Kim B. J., Rivera-Amill V., Garvis S. G. (1999). Bacterial secreted proteins are required for the internaliztion of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32, 691–701. doi: 10.1046/j.1365-2958.1999.01376.x.
Korlath J. A., Osterholm M. T., Judy L. A., Forfang J. C., Robinson R. A. (1985). A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152, 592–596. doi: 10.1093/infdis/152.3.592.
Krause-Gruszczynska M., van Alphen L. B., Oyarzabal O. A., Alter T., Hanel I., Schliephake A., et al. (2007). Expression patterns and role of the CadF protein in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 274, 9–16. doi: 10.1111/fml.2007.274.issue-1.
Kurstak E., Vega C. E. (1968). Bacterial infection due to Salmonella typhimurium in an invertebrate, Galleria mellonella L. Can. J. Microbiol. 14, 233–237. doi: 10.1139/m68-039.
Laaveri T., Vilkman K., Pakkanen S. H., Kirveskari J., Kantele A. (2018). A prospective study of travellers' diarrhoea: analysis of pathogen findings by destination in various (sub)tropical regions. Clin. Microbiol. Infect. 24, 908 e909–908 e916. doi: 10.1016/j.cmi.2017.10.034.
Luangtongkum T., Jeon B., Han J., Plummer P., Logue C. M., Zhang Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189.
Macdonald S. E., Gundogdu O., Dorrell N., Wren B. W., Blake D., Stabler R. (2017). Draft genome sequence of campylobacter jejuni 11168H. Genome Announc 5. doi: 10.1128/genomeA.01556-16.
Mace K., Vadakkepat A. K., Redzej A., Lukoyanova N., Oomen C., Braun N., et al. (2022). Cryo-EM structure of a type IV secretion system. Nature 607(7917):191-196. doi: 10.1038/s41586-022-04859-y.
Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x.
Mourkas E., Yahara K., Bayliss S. C., Calland J. K., Johansson H., Mageiros L., et al. (2022). Host ecology regulates interspecies recombination in bacteria of the genus Campylobacter. eLife 11, e73552. doi: 10.7554/eLife.73552.sa2.
Mulchandani R., Wang Y., Gilbert M., Van Boeckel T. P. (2023). Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PloS Glob Public Health 3, e0001305. doi: 10.1371/journal.pgph.0001305.
Nguyen T. N., Hotzel H., El-Adawy H., Tran H. T., Le M. T., Tomaso H., et al. (2016). Genotyping and antibiotic resistance of thermophilic Campylobacter isolated from chicken and pig meat in Vietnam. Gut Pathog. 8, 19. doi: 10.1186/s13099-016-0100-x.
Nurk S., Bankevich A., Antipov D., Gurevich A. A., Korobeynikov A., Lapidus A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084.
Ohishi T., Aoki K., Ishii Y., Usui M., Tamura Y., Kawanishi M., et al. (2017). Molecular epidemiological analysis of human- and chicken-derived isolates of Campylobacter jejuni in Japan using next-generation sequencing. J. Infect. Chemother. 23, 165–172. doi: 10.1016/j.jiac.2016.11.011.
Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088.
Pratt A., Korolik V. (2005). Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J. Antimicrob. Chemother. 55, 452–460. doi: 10.1093/jac/dki040.
Russell R. G., Blaser M. J., Sarmiento J. I., Fox J. (1989). Experimental Campylobacter jejuni infection in Macaca nemestrina. Infect. Immun. 57, 1438–1444. doi: 10.1128/iai.57.5.1438-1444.1989.
Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153.
Senior N. J., Bagnall M. C., Champion O. L., Reynolds S. E., La Ragione R. M., Woodward M. J., et al. (2011). Galleria mellonella as an infection model for Campylobacter jejuni virulence. J. Med. Microbiol. 60, 661–669. doi: 10.1099/jmm.0.026658-0.
Shane S. M. (2000). Campylobacter infection of commercial poultry. Rev. Sci. Tech 19, 376–395. doi: 10.20506/rst.issue.19.2.14.
Sheppard S. K., McCarthy N. D., Falush D., Maiden M. C. (2008). Convergence of Campylobacter species: implications for bacterial evolution. Science 320, 237–239. doi: 10.1126/science.1155532.
Silva M., MaChado M. P., Silva D. N., Rossi M., Moran-Gilad J., Santos S., et al. (2018). chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom 4. doi: 10.1099/mgen.0.000166.
Skarp C. P. A., Hanninen M. L., Rautelin H. I. K. (2016). Campylobacteriosis: the role of poultry meat. Clin. Microbiol. Infect. 22, 103–109. doi: 10.1016/j.cmi.2015.11.019.
Sprenger H., Kienesberger S., Pertschy B., Poltl L., Konrad B., Bhutada P., et al. (2017). Fic Proteins of Campylobacter fetus subsp. venerealis Form a Network of Functional Toxin-Antitoxin Systems. Front. Microbiol. 8, 1965. doi: 10.3389/fmicb.2017.01965
Sutili F. J., Gressler L. T. (2021). “Chapter 3 - antimicrobial agents,” in Aquaculture pharmacology, eds Kibenge F., Baldisserotto B., Chong R. (Amsterdam, Netherlands: Elsevier), 131–168. doi: 10.1016/B978-0-12-821339-1.00004-0
Tangcharoensathien V., Witthayapipopsakul W., Panichkriangkrai W., Patcharanarumol W., Mills A. (2018). Health systems development in Thailand: a solid platform for successful implementation of universal health coverage. Lancet 391, 1205–1223. doi: 10.1016/S0140-6736(18)30198-3.
Van Boeckel T. P., Glennon E. E., Chen D., Gilbert M., Robinson T. P., Grenfell B. T., et al. (2017). Reducing antimicrobial use in food animals. Science 357(6358), 1350–1352. doi: 10.1126/science.aao1495.
van Vliet A. H. M., Thakur S., Prada J. M., Mehat J. W., La Ragione R. M. (2022). Genomic screening of antimicrobial resistance markers in UK and US campylobacter isolates highlights stability of resistance over an 18-year period. Antimicrob. Agents Chemother. 66, e0168721. doi: 10.1128/aac.01687-21.
Viegas S. C., Mil-Homens D., Fialho A. M., Arraiano C. M. (2013). The virulence of Salmonella enterica Serovar Typhimurium in the insect model galleria mellonella is impaired by mutations in RNase E and RNase III. Appl. Environ. Microbiol. 79, 6124–6133. doi: 10.1128/AEM.02044-13.
Wallden K., Rivera-Calzada A., Waksman G. (2010). Type IV secretion systems: versatility and diversity in function. Cell Microbiol. 12, 1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x.
WHO (2017) WHO guidelines on use of medically important antimicrobials in food-producing animals. Available online at: http://apps.who.int/iris/bitstream/handle/10665/258970/9789241550130-eng.pdf.
Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). "Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595.
Wieczorek K., Osek J. (2013). Antimicrobial resistance mechanisms among Campylobacter. BioMed. Res. Int. 2013, 340605. doi: 10.1155/2013/340605.
Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261.
Keywords: Campylobacter, C. coli, Vietnam, antimicrobial resistance, Galleria mellonella
Citation: Lehri B, Navoly G, Corser A, Nasher F, Willcocks S, Pham TN, Wren BW, Luu QH and Stabler RA (2024) Phenotypic and genotypic characterization of Campylobacter coli isolates from the Vietnamese poultry production network; a pilot study. Front. Ind. Microbiol. 2:1355079. doi: 10.3389/finmi.2024.1355079
Received: 13 December 2023; Accepted: 06 February 2024;
Published: 21 February 2024.
Edited by:
Ramona Iseppi, University of Modena and Reggio Emilia, ItalyReviewed by:
Orhan Sahin, Iowa State University, United StatesLaura Espina, University of Zaragoza, Spain
Aditya Upadrasta, mBio, Ireland
Copyright © 2024 Lehri, Navoly, Corser, Nasher, Willcocks, Pham, Wren, Luu and Stabler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard A. Stabler, cmljaGFyZC5zdGFibGVyQGxzaHRtLmFjLnVr
†These authors have contributed equally to this work and share first authorship
 Burhan Lehri
Burhan Lehri Georgina Navoly
Georgina Navoly Abigail Corser1
Abigail Corser1 Fauzy Nasher
Fauzy Nasher Brendan W. Wren
Brendan W. Wren Richard A. Stabler
Richard A. Stabler