- 1Ocean College, Hebei Agricultural University, Qinhuangdao, China
- 2Hebei Key Laboratory of Aquaculture Nutritional Regulation and Disease Control, Hebei Agricultural University, Qinhuangdao, Hebei, China
- 3Higher Institution Centre of Excellence (HICoE), Institute of Tropical Aquaculture and Fisheries, Universiti Malaysia Terengganu, Kuala Terengganu, Malaysia
- 4Ocean and Fisheries Science Research Institute of Hebei Province, Department of Science and Technology of Hebei Province, Qinhuangdao, China
- 5Hebei Key Laboratory of Marine Biological Resources and Environment, Department of Science and Technology of Hebei Province, Qinhuangdao, China
Fish gill tissue is a primary organ responsive to acute oxygen deprivation or dissolved oxygen (DO) fluctuations in aquatic environments. However, the adaptive mechanism of crucian carp to hypoxic stress remains largely unknown. Here, we investigated gill physiological and transcriptomic changes of crucian carp exposed to hypoxic conditions (dissolved oxygen concentration of 0.6 ± 0.3 mg/L) for different durations (0 d, 1 d, 2d, 3d, 4 d, and 5d). Transcriptomic analysis revealed that the hypoxia group (0.6 ± 0.3 mg/L DO) exhibited a reduction in interlamellar cell mass (ILCM) on the gill filaments, compared with the control group (6.6 ± 0.3 mg/L DO). With prolonged hypoxia stress, the epithelial cells in the gill lamellae became sparse at 3 d to 5 d, and gill vacuoles were increased. A total of 3,502 differentially expressed genes (DEGs) were identified, and 3 hypoxia-specific modules were screened through differential expression analysis, weighted gene co-expression network analysis (WGCNA), and Bayesian network analysis. The apoptosis, necroptosis, efferocytosis and FoxO signaling pathways were significantly enriched based on the KEGG enrichment pathway analysis. The VEGF pathway genes are significantly expressed, enhancing the generation of microvessels in the gill filaments, and improving the capacity to carry oxygen, thus enabling the crucian carp to adapt to hypoxia stress. Hypoxia activated glycolysis, enhanced anaerobic metabolism, promoted β-oxidation of fatty acids, providing energy and maintaining normal physiological metabolism, eventually improving antioxidant and immune capabilities in crucian carp. In summary, this study reveals the molecular mechanism by which crucian carp adapt to hypoxic stress. Our findings provide valuable references for promoting the healthy aquaculture of hypoxic-sensitive fish and breeding hypoxia-tolerant fish varieties.
1 Introduction
Dissolved oxygen (DO) is a crucial ecological factor for fish and other aquatic animals (1). DO in aquaculture water is unstable, and it may be decreased due to multiple factors such as weather variation, pollution, and high-density farming (2). Hypoxia refers to a state in which the DO level is excessively low, thus adversely affecting fish behavior, growth, and physiological metabolism and eliciting immune responses (3). Severe hypoxic stress can pose a significant threat to fish survival (4). With increasingly severe global warming and water pollution, hypoxia has caused enormous economic losses in the aquaculture industry, and thus it is essential to explore the mechanism of hypoxia adaptation for fish breeding (5).
Gill of fish is the principal respiratory organ responsible for gas exchange with the surrounding aquatic environment (6). In a hypoxic or anoxic environment, fish bodies can produce a series of response mechanisms, mainly including increasing the number of red blood cells, enhancing the oxygen carrying capacity of hemoglobin, stimulating angiogenesis, activating the antioxidant defense system, and changing the metabolic mode (7, 8). It has been reported that exposure to hypoxia may result in a series of physiological and morphology of fish gill in multiple fish species (9). For example, blunt snout bream (Megalobrama amblycephala) (10), mangrove killifish (Kryptolebias hermaphroditus) (2) and hybrid yellow catfish (Tachysurus fulvidraco ♀×Pseudobagrus vachellii ♂) (1) respond to hypoxia stress by reducing the size of their interlamellar cell masses (ILCMs), exposing the lamellae, and increasing the gill-water contact area. Nilsson et al. (2012) (11) speculated that apoptosis might be an important molecular mechanisms of gill remodeling in fish. Additionally, fish have been found to enhance their anaerobic metabolism to sustain their energy supply in response to DO insufficiency (12). Therefore, some studies have investigated the activity changes in lactate dehydrogenase (LDH), a key enzyme regulating anaerobic metabolism, under hypoxic stress, and found the LDH enzyme activity in gill tissue of hybrid sturgeon (Acipenser schrenckii ♂×Acipenser baerii ♀) (13) and Hong Kong oyster Crassostrea hongkongensis (14) showed an upward trend under hypoxic stress. Moreover, hypoxia can also interfere with the electron transport chain, leading to the over production of reactive oxygen species (ROS) (15). Excessive ROS can induce oxidative damage to cellular membranes, proteins, and DNA, hence resulting in programmed cell death (also known as apoptosis) (16). Superoxide dismutase (SOD) and catalase (CAT), as two important antioxidative enzymes, play a significant role in removing ROS and alleviating the damage caused by oxidative stress in fish. Typically, the concentration of malondialdehyde (MDA) serves as an indicator of oxidative stress in fish (17).
Crucian carp (Carassius auratus) are affordable and delicious, exhibiting high quality and high nutrition, meanwhile being excellent source of protein (18). Crucian carp can survive hypoxic aquatic environment for a long period. Exploring the adaptation mechanisms of crucian carp gills to hypoxic environments will be conductive to alleviating the stress of hypoxia in aquaculture. The adaptation of hypoxia-tolerant freshwater turtles (Trachemys and Chrysemys genera) to hypoxia has also been extensively studied (19, 20). However, sea turtles clearly adopt a different survival strategy, entering a state similar to deep dormancy when exposed to hypoxia (21).
With the advancement of transcriptome technology and the decrease in next-generation sequencing cost, RNA sequencing (RNA-seq) has emerged as a powerful tool for investigating stress response mechanisms (7). In the pufferfish (Takifugu rubripes), as hypoxia persists, lipid metabolism has been found to become the primary energy supply pathway (22). In the darkbarbel Catfish (Pelteobagrus vachelli), apoptosis-related genes are upregulated, and antioxidant capacity is enhanced under hypoxia stress (23). However, the molecular mechanisms underlying cell apoptosis in crucian carp gill tissue remain largely unclear.
The goal of this study was to decipher the response mechanism of crucian carp gills to hypoxic stress. First, we conducted RNA-seq analyses and compared gill transcriptional profiles between hypoxia-stressed group and control group. The results demonstrated that under hypoxic stress, the pillar cells in the gill lamellae became sparse, and gill vacuoles were increased, thus enhancing anaerobic metabolism, eventually improving antioxidant and immune capabilities of the gill tissue. Based on differential expression analysis, weighted gene co-expression network analysis (WGCNA), and Bayesian network analysis, a total of 3,502 differentially expressed genes (DEGs) were identified, and 3 hypoxia-specific modules were screened. Further, 11 hypoxia response-related key genes were identified such as pak1, cdc23, smad3a, and caspase7. Additionally, this study also revealed the effects of hypoxia stress on the activity of antioxidative enzymes SOD and CAT in crucian carp gill tissue. Our findings offer new insights into the molecular mechanisms of hypoxia tolerance in crucian carp and provide valuable reference for breeding hypoxia-tolerant fish varieties.
2 Materials and methods
2.1 Ethical approval
All the experiments were approved by the Animal Experimentation Ethics Committee of Hebei Agricultural University (Grant No.2022075), following the Laboratory Animal Welfare Guidelines of China (GB/T 35892-2018).
2.2 Experimental procedures
All 400 crucian carps were from a pond (in Zhangjiakou City, Hebei Province, China) and were reared in a 22,500-liter water tank for two weeks of acclimation. The fish were not fed 24 hours before the hypoxia experiment. The crucian carp were divided into two groups with three parallel glass tanks (1.0 m × 1.0 m × 1.5 m) per group. For the control group (C), 150 fish (50 fish per tank, with a body weight of 94.5 ± 4.5 g) were randomly selected and reared in a normoxic aquatic environment (with a dissolved oxygen (DO) level of 6.6 ± 0.3 mg/L at 22.5 ± 0.5°C). For the treatment group (T), 150 fish (50 fish per tank, with a body weight of 94.5 ± 4.5 g) were randomly selected and reared in a hypoxic aquatic environment (with a DO level of 0.6 ± 0.3 mg/L at 22.5 ± 0.5°C). As previously described (24, 25), during the entire experiment, a glass cover of the same size was placed above the glass tanks. A 1-cm³ small hole was left in the middle of the glass cover, through which a nitrogen (N₂) inflation tube and an oxygen (O₂) inflation tube passed and were wound under a cubic iron frame. There was an air valve piston on each of the two inflation tubes to control the DO in the experiment. The DO was continuously measured by an oxygen electrode (YSI, ProODO, Germany) (Supplementary Figure S1). In this study, the loss of equilibrium (LOE) of the fish in the water was defined as the hypoxic state of the fish (26). During the 5-day hypoxia experiment, samples were taken at five time points (1 day, 2 days, 3 days, 4 days, 5 days). Three fish were taken from each glass tank. At each sampling point, the crucian carp were anesthetized with 0.5 g·L⁻¹ tricaine methanesulfonate (MS-222, Sigma, United States), and the second gill of each crucian carp was collected. A part of the gills was placed in 4% paraformaldehyde (Biosharp, China) for subsequent morphological analysis, a part was placed in a 2-mL cryopreservation tube and sent to a company for transcriptomics analysis (RNA-seq), and the remaining gill tissues were collected into 5-mL EP tubes and frozen at -80°C for subsequent experiments.
2.3 Analysis of gill biochemical parameters
The 100 mg gill sample was homogenized in 900 μL pre-cooled phosphate-buffered saline (PBS) solution (pH 7.2) for the enzyme activity determination. The activities of SOD (24), CAT (25), LDH (12) and the oxidative product MDA (25) level were determined using commercial kits by previously reported method (26).
2.4 Histological observation
After paraffin sectioning and hematoxylin-eosin (H&E) staining, gill samples were subjected to histological observation, as our previously described (27). Briefly, gill samples were rinsed by PBS solution to remove residual impurity, then immersed in Bouin’s solution, and finally stained with the H&E. These sections were observed under Leica microscope (Wetzlar, Germany).
2.5 RNA extraction and transcriptome sequencing
Total RNA was extracted from fish gill tissue (n = 3 per group) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and RNA integrity and total quantity were determined using Agilent 2100 bioanalyzer. This analysis was conducted with three replicates per group, so that eighteen samples were prepared in total (C_1, C_2, C_3, T1_1, T1_2, T1_3, T2_1, T2_2, T2_3, T3_1, T3_2, T3_3, T4_1, T4_2, T4_3, T5_1, T5_2, andT5_3).The PCR products were purified using AMPure XP beads to remove primer dimers and other unwanted byproducts, and finally high-quality cDNA library was obtained. Upon completion of library construction, an initial quantification was performed using a Qubit 2.0 Fluorometer to obtain the accurate concentration of the cDNA library. Then, the library was diluted to a working concentration of 1.5 ng/μl. The quality of the cDNA library was assessed using an Agilent 2100 Bioanalyzer to ensure that the library was qualified for downstream sequencing. After quality control, the qualified libraries with valid concentration and required offline data size were pooled, and subjected to the Illumina sequencing to obtain 150 bp paired end reads (28).
2.6 Assembly and annotation of transcripts
In raw data pre-processing, strict quality control was implemented to eliminate specific contaminants from sequencer by adapter trimming, to remove ambiguous nucleotides by N-clipping, and to filter the reads with a Qphred cutoff of ≤5 and read length < 50% (29). After quality control, the multiple indicators of the obtained clean reads were assessed, including Q20 and Q30 quantifications for base-calling accuracy, and GC content analysis. The resulting high-fidelity data lay the foundation for downstream analyses. The reference genomes were retrieved, and the transcripts were annotated based on the genome database NCBI (https://www.ncbi.nlm.nih.gov/datasets/genome/?taxon=7957,217509). A reference index was generated with HISAT2 v2.0.5 (30), followed by aligning paired-end reads to this index for precise mapping.
2.7 Identification of differentially expressed genes
Relative gene expression levels of transcripts were standardized by fragments per kilobase of exon model per million mapped reads (FPKM) (31). The differential expression (DE) analysis across two biological replicates was performed using the DESeq2 v1.20.0 by applying negative binomial distribution models to determine variability and significance (32). The Benjamini-Hochberg (BH) procedure was utilized for multiple test correction to control the false discovery rate (FDR). The DEGs were identified with the thresholds of |log2 FC (fold changed)| ≥ 1 and adjusted P-value<0.01. The Gene Ontology (GO)(http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed to determine the potential functions of the DEGs and their significantly enriched metabolic pathways, and P < 0.05 was considered as significant enrichment.
2.8 Weighted gene co-expression network analysis
After the genes with normalized FPKM value < 1 were filtered, the remaining genes were subjected to WGCNA. The genes with similar expression profiles were clustered into the same module, and the gene co-expression modules were constructed using the WGCNA v1.72 R package. Based on the expression levels of constituent genes and the module eigengenes (ME), biological modules were classified. The module classification parameters were set as follows: a minimum power value, 14; ME cutoff, 0.25; and a minimum module size threshold, 30 (33). The Pearson correlation coefficient was computed between each module and hypoxic treatment to identify modules significantly correlated with hypoxic stress response. The modules with a Pearson correlation coefficient of > 0.5 and a P-value of < 0.05 were defined as hypoxia-related. Further, functions of the genes in these modules were investigated through GO and KEGG enrichment analyses. The intra-module connectivity and inter-module correlation were calculated using WGCNA package to reveal gene interaction networks. Further, co-expression network of the top 30 genes (in terms of centrality degree) was constructed using Cytoscape v3.8.2 to visualize their interactions (34). The hypoxia tolerance-related hub genes or candidate genes were identified, based on their connectivity within the network.
2.9 Analysis of Bayesian networks
The gene regulatory network (GRN) of the modular gene set was plotted using the R package CBNplot, and hypoxia response -related genes were screened with the threshold of FDR< 0.05 based on Bayesian model by previously reported method (29, 30). The WGCNA module genes related to hypoxia were used to construct Bayesian network.
2.10 qRT-PCR analysis
Twelve DEGs were randomly selected for qRT-PCR in all groups to verify the accuracy of the sequencing results. Total RNA was extracted from the crucian carp gill tissue of hypoxic treatment groups (T1-T5, across 1 to 5 hypoxia) and the control group, as described in section 2.5. Primers were designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China), which were listed in Table 1. The cDNA was synthesized according to the manufacturer’s protocol with Hiscript III Reverse Transcriptase kit (Vazyme, Nanjing, China). Quantitative real-time PCR (qRT-PCR) was performed using ChamQ Universal SYBR qPCR Master Mix kit (Vazyme, Nanjing, China). The 10 μL PCR reaction system contained 5.0 μL of the 2x SYBR qPCR Master Mix, 0.5 μL each of 10 μM forward and reverse primers (Table 1), 1 μL of cDNA template, and 3 μL of nuclease-free water. The qRT-PCR program was as follows: an initial denaturation at 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 30 s, followed by an extension at 95°C for 15 s, annealing at 60°C for 1 min, and final extension at 95°C for 15 s (35). Subsequently, a dissociation curve was obtained. The experiment was conducted with 3 replicates for each sample to ensure reproducibility, and 18s rRNA gene served as an internal control sample. The relative expression of gene was calculated using the 2-ΔΔCt method (36).
2.11 Statistical analysis
Statistical analysis of data was conducted using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). Normal distribution and variance homogeneity were assessed via the Shapiro-Wilk and Levene tests, respectively. Inter-group differences (hypoxia vs. control) were determined by independent samples t-tests, while intra-group differences were examined through one-way ANOVA. P < 0.05 was considered as statistically significant.
3 Results
3.1 Oxidative stress indexes of crucian carp gill under hypoxia stress
We measured gill biochemical parameters of crucian carp at 24 h, 48 h,72 h,96 h and 120 h (T1-5) post hypoxia treatment (Figure 1). The results showed that the gill LDH activity was significantly higher (P < 0.05) in the all 5 hypoxia-treated groups (T1-T5) than in the control group (Figure 1A); the gill SOD activity was significantly higher inT1,T2, andT4 groups (P < 0.05) than in the control group (Figure 1B); the gill CAT activity was significantly higher (P < 0.05) in the 3 hypoxia-treated groups (T1-T3) than in the control group (Figure 1C); and gill MDA content was significantly higher (P < 0.05) in 3 hypoxia-treated groups (T3-T5) than in the control group (Figure 1D). Taken together, hypexia-treated groups exhibited a higher anti-oxidative enzyme activity after 3-day hypexia stress and a higher oxidation product content after 4-5 days of hypoxia stress than in control group.
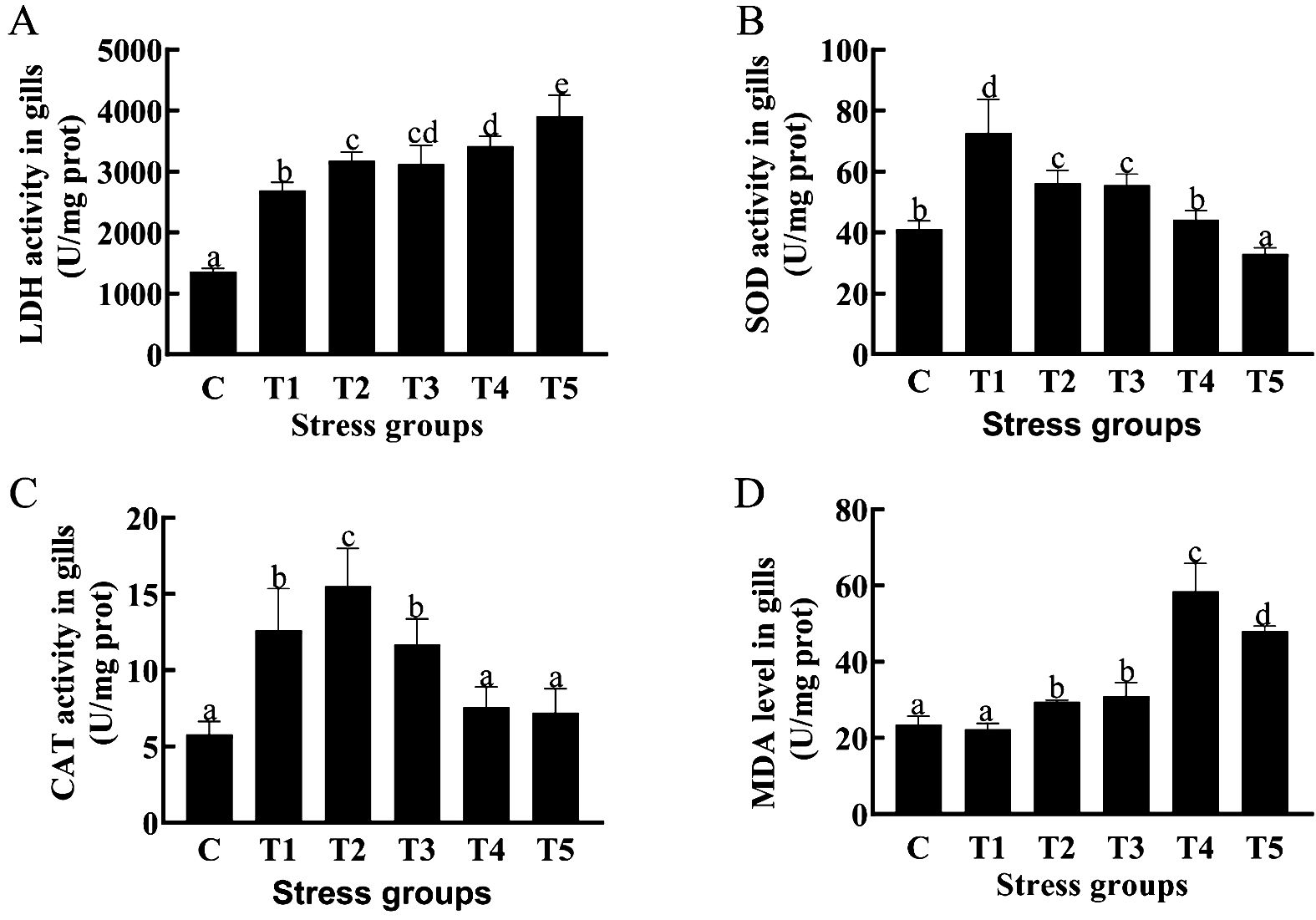
Figure 1. Effects of different durations of hypoxia stress on antioxidase activity and oxidation product MDA of Crucian carp gill. LDH activity in gills (A);SOD activity in gills (B); CAT activity in gills (C); MDA level in gills (D). Different letters above the bars indicate significant differences (p < 0.05).
3.2 Histological analysis of crucian carp gill under hypoxia stress
In the control group, crucian carp gill filament and lamella structural integrity and internal cell morphology were normal with neat arrangement (Figure 2A). In contrast, in the hypoxia treatment group, the cells surrounding the gill lamellae were gradually decreased; the gill interlamellar cell mass was reduced; and the epithelial cells in the gill lamellae became sparse with loose arrangement (Figures 2B–F).
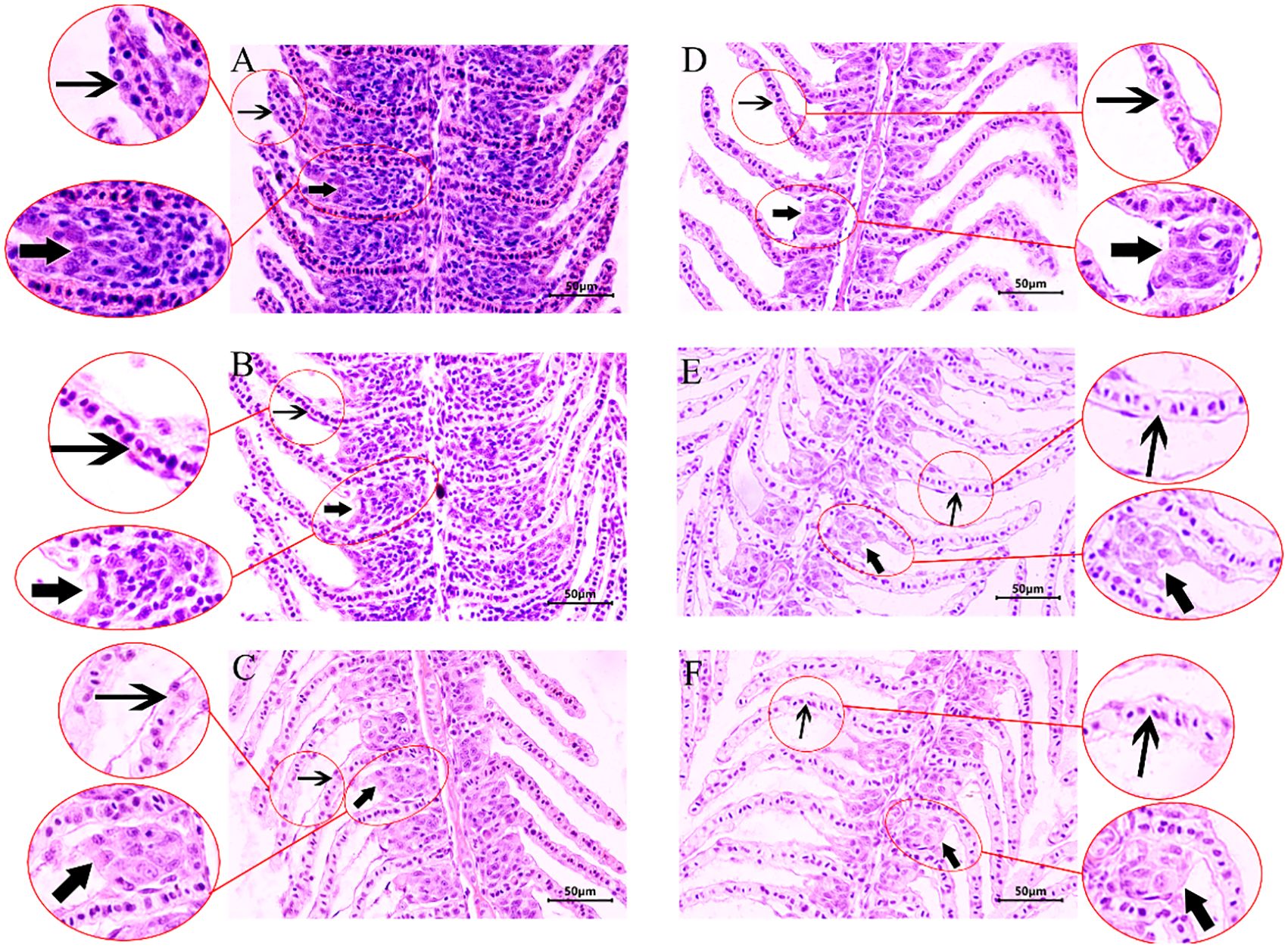
Figure 2. Hematoxylin–eosin (HE) staining of crucian crap gill tissues under hypoxic stress. (A) Structure of a normal gill. (B–F) Structure of gill exposed to hypoxia for 1, 2, 3, 4, 5 day(s). Thick arrow: ILCM; thin arrow: gill lamellae.
3.3 Transcriptome sequencing and annotation
The gill tissues from fish with or without exposure to hypoxia were subjected to transcriptome sequencing on the L230 Illumina NovaSeq 6000 platform. After low-quality sequence removal, the number of clean reads in each of 18 sample libraries ranged from 41410340 to 48353576. The Q20 values of these 18 sequenced libraries ranged from 98.48% to 99.16%, whereas their GC contents ranged from 41.61% to 46.64%. These results confirmed the reliability of the transcriptome sequencing data. The transcriptome sequencing data of control group and hypoxia-treated group were aligned to the crucian carp genome, respectively.
3.4 Identification of DEGs in the comparison of hypoxia groups vs. control group
The DEGs in the hypoxia treatment vs. control groups were identified on the basis of |log2 FC| ≥ 1 and P < 0.01 (Figure 3A). A total of 3,502 DEGs were identified across five comparison groups. The DEGs were identified in each of 5 comparisons (T1, T2, T3, T4, or T5 vs. C) and the largest number of DEGs (1,282) were identified in the comparison of T5 vs. C, accounting for 36.60% (Figure 3A). Venn diagrams revealed that DEGs were shared by the five comparation groups, and 401, 142, 767, 910, and 1,282 DEGs were unique to the comparison group of T1 vs. C, T2 vs. C, T3 vs. C, T4 vs. C, and T5 vs. C, respectively (Figure 3B).
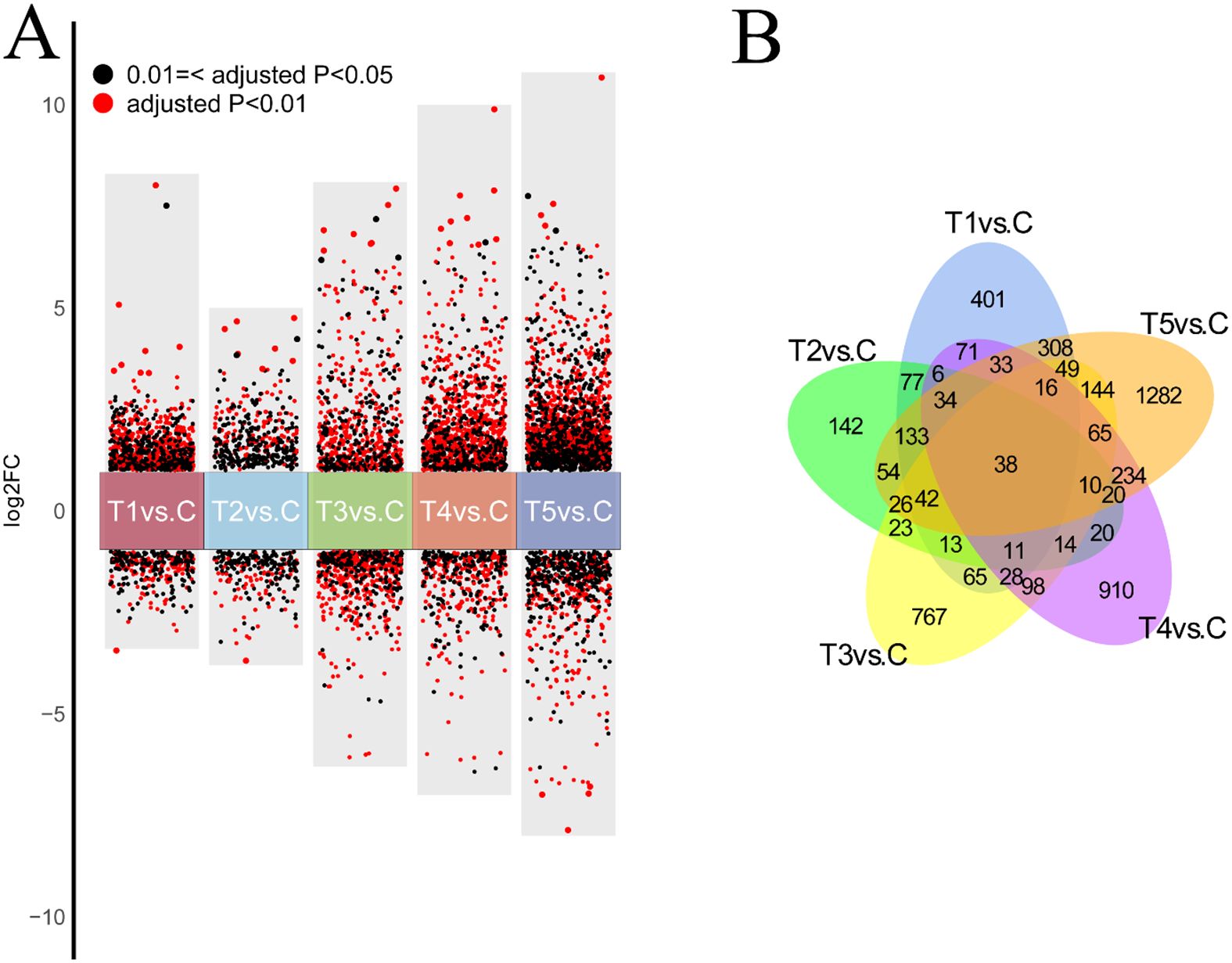
Figure 3. (A) Volcano plot of DEGs in crucian crap gills under hypoxic stress in the comparison of hypoxia-treated groups vs. control group. Log2 FC <-1 indicates down-regulated DEGs; Log2 FC >1 represents up-regulated DEGs. 2. (B) Venn plot of DEGs in crucian crap gills under hypoxic stress in the comparison of hypoxia-treated groups vs. control group.
3.5 GO and KEGG analyses of DEGs
In the GO enrichment analysis, the DEGs were assigned to 123 GO terms, including 35 biological process (BP), 30 cellular component (CC), and 40 molecular function (MF) (Figure 4, Supplementary Figure S1). The top GO terms were DNA replication (Figure 4A) and lipid biosynthetic process for BP (Figure 4B), and it was RNA binding for MF (Figure 4C). These results indicated substantial changes in immune defenses and energy metabolism in gill tissues under hypoxic stress.
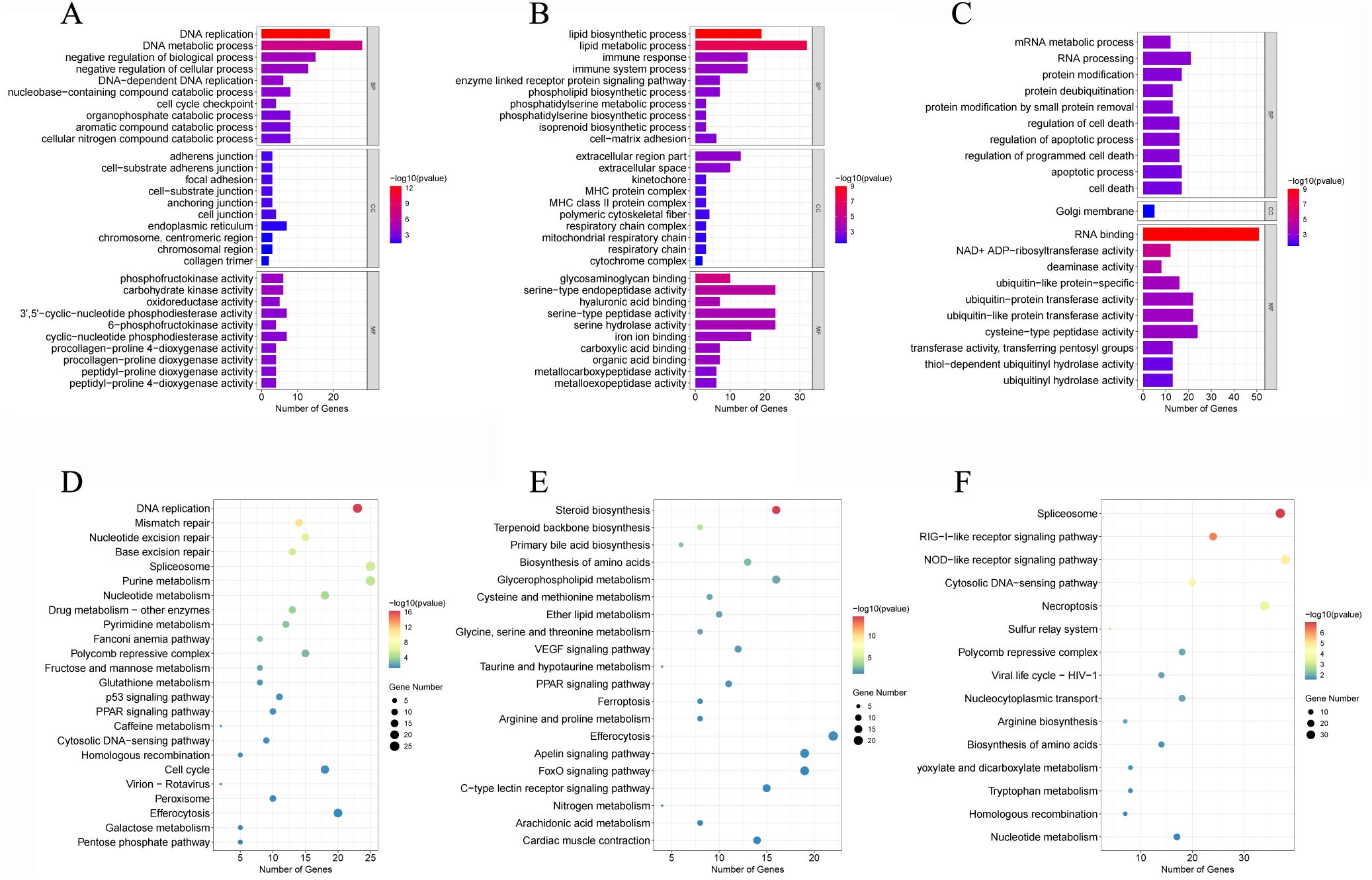
Figure 4. GO enrichment analysis of DEGs in T1 vs. C (A), T3 vs. C (B), and T5 vs. C (C). KEGG pathway enrichment analysis of DEGs in T1 vs. C (D), T3 vs. C (E), and T5 vs. C (F).
In the KEGG enrichment analysis, 18 hypoxia-related pathways in crucian carp gill post 5-day hypoxic stress were significantly enriched, mainly including cell cycle, nucleotide metabolism, p53 signaling pathway, glutathione metabolism (Figure 4D), drug metabolism - other enzymes, NOD-like receptor signaling pathway (Figure 4E), PPAR signaling pathway, VEGF signaling pathway (Figure 4F, Supplementary Figure S2).
3.6 Module analysis based on WGCNA
By WGCNA, a total of 27 modules under hypoxia stress were identified. As shown in Figure 5, the MEcyan module (R = 0.81, p = 5e-05), the ME darkgrey module (R = 0.81, p = 4e-05), and the MEdarked module (R = 0.84, p = 1e-05) were significantly correlated with T3 and T5, respectively. GO enrichment analysis revealed that lipid biosynthetic process, oxidoreductase activity, and organelle envelope were significantly enriched GO terms in cyan, darkgrey, darked modules, respectively (Figures 6A–C). KEGG enrichment analysis found that steriod biosynthesis, PPAR signaling pathway, and p53 signaling pathway were significantly enriched in cyan module; tryptophan metabolism, cyp450 metabolism, and apelin signaling pathway in darkgrey module; RIG-I-like receptor signaling pathway, toll-like receptor signaling pathway, and apoptosis in darked module (Figures 6D–F).
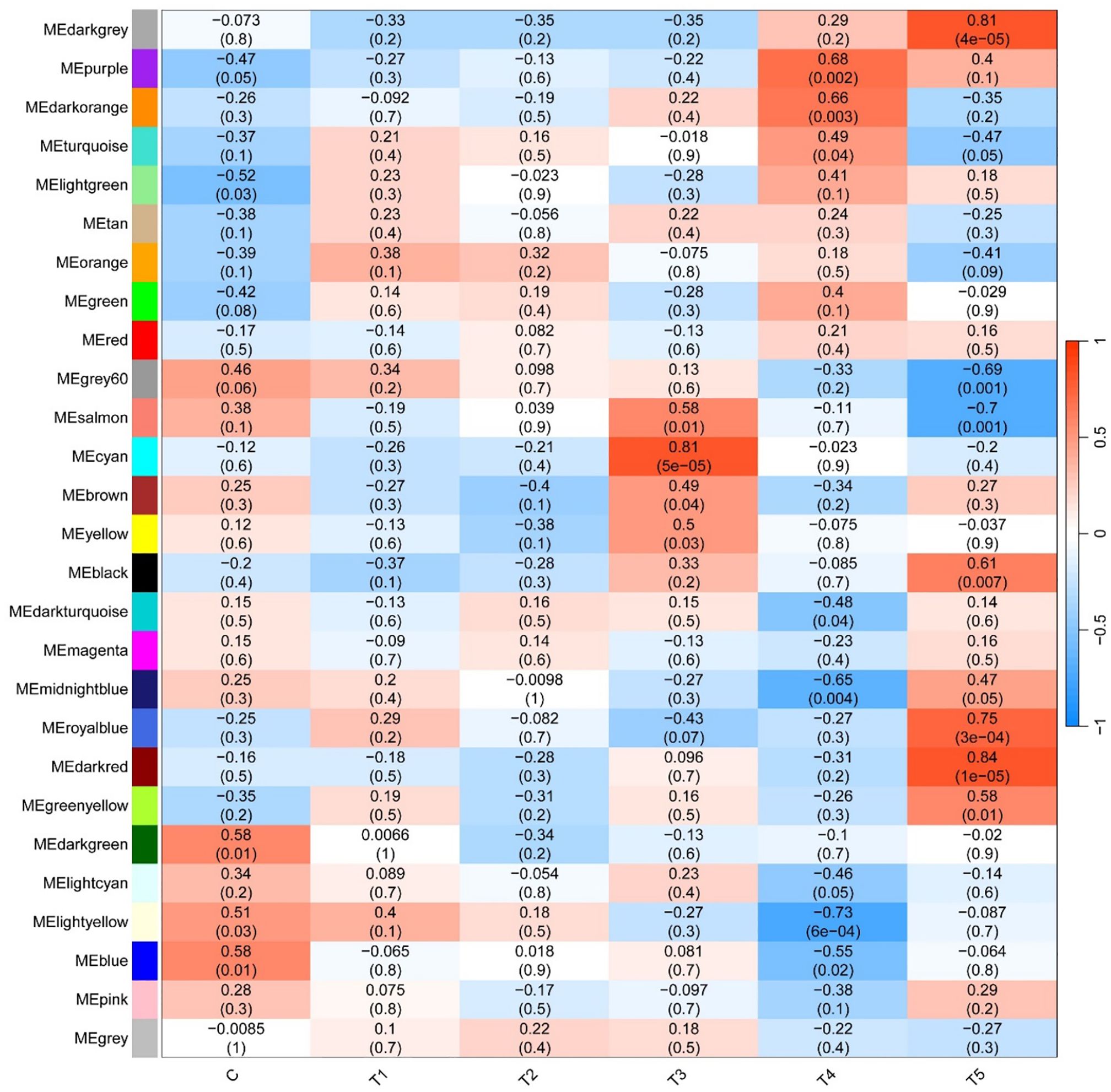
Figure 5. Correlation heatmap between modules and hypoxia treatments. The color scale on the right shows module-hypoxia treatment correlation coefficients from - 1 (blue) to 1 (red). The numbers in brackets represent significance (P < 0.05).
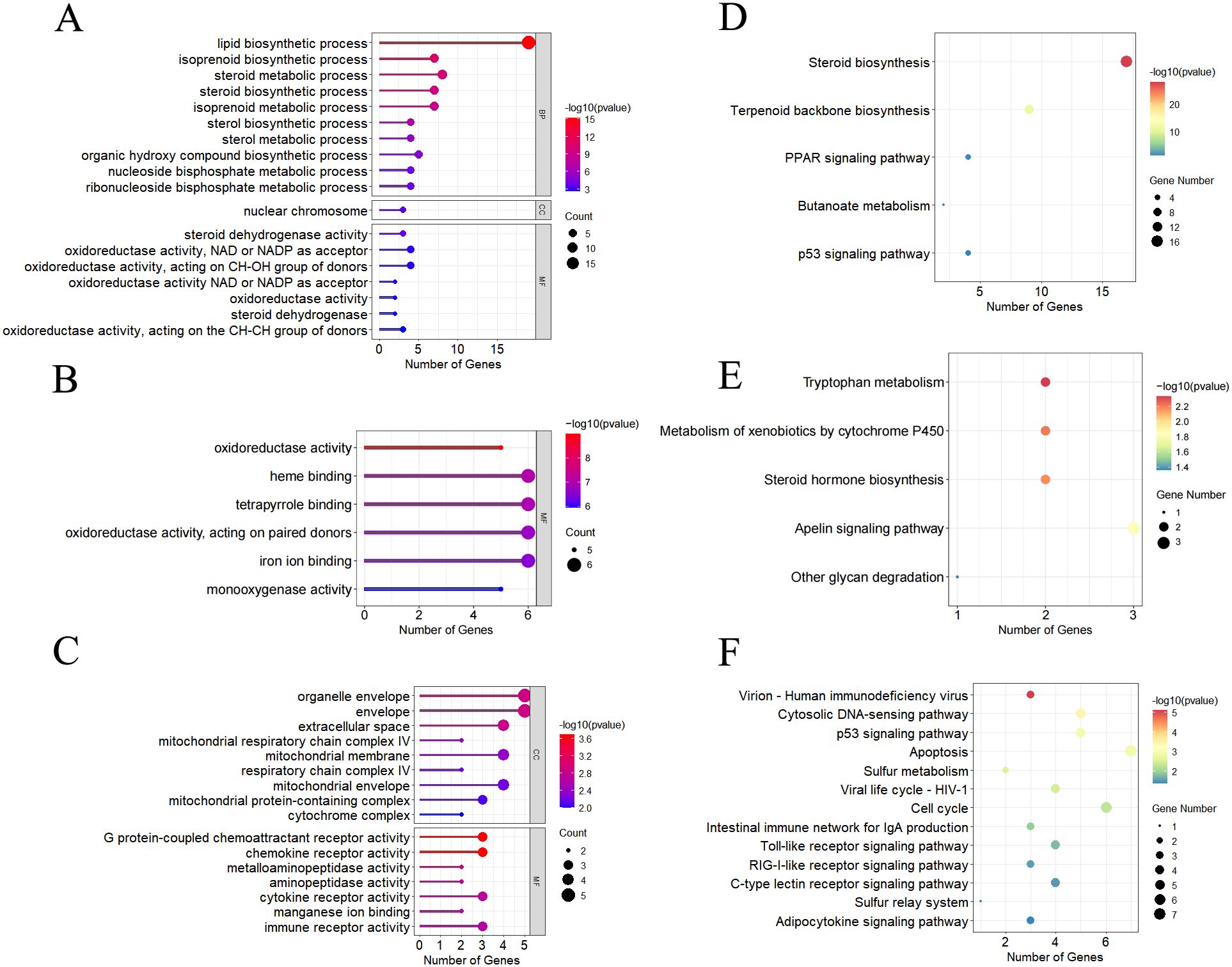
Figure 6. Significantly enriched GO terms by GO enrichment analysis in MEcyan module (A), MEdarkgrey module (B), and MEdarkred module (C). Significantly enriched pathways by the KEGG enrichment analysis in MEcyan module (D), MEdarkgrey module (E), and MEdarkred module (F), respectively.
The hub genes in each module and connectivity with other genes were further explored by the WGCNA. As illustrated in Figure 7, there were 26 genes and 4 hub genes (elF4e-L, Fork-q1-l, PAIRBP, and GILT), 16 genes and 1 hub genes (serpinb1/3) and 22 genes and 3 hub genes (ndc80, plk1 and Kif23) in the cyan module, darked module, and darkgrey module, respectively. These 8 hub genes in 3 modules exhibited high connectivity with other genes, indicating that they played pivotal roles in responses to hypoxia stress in these 3 modules.
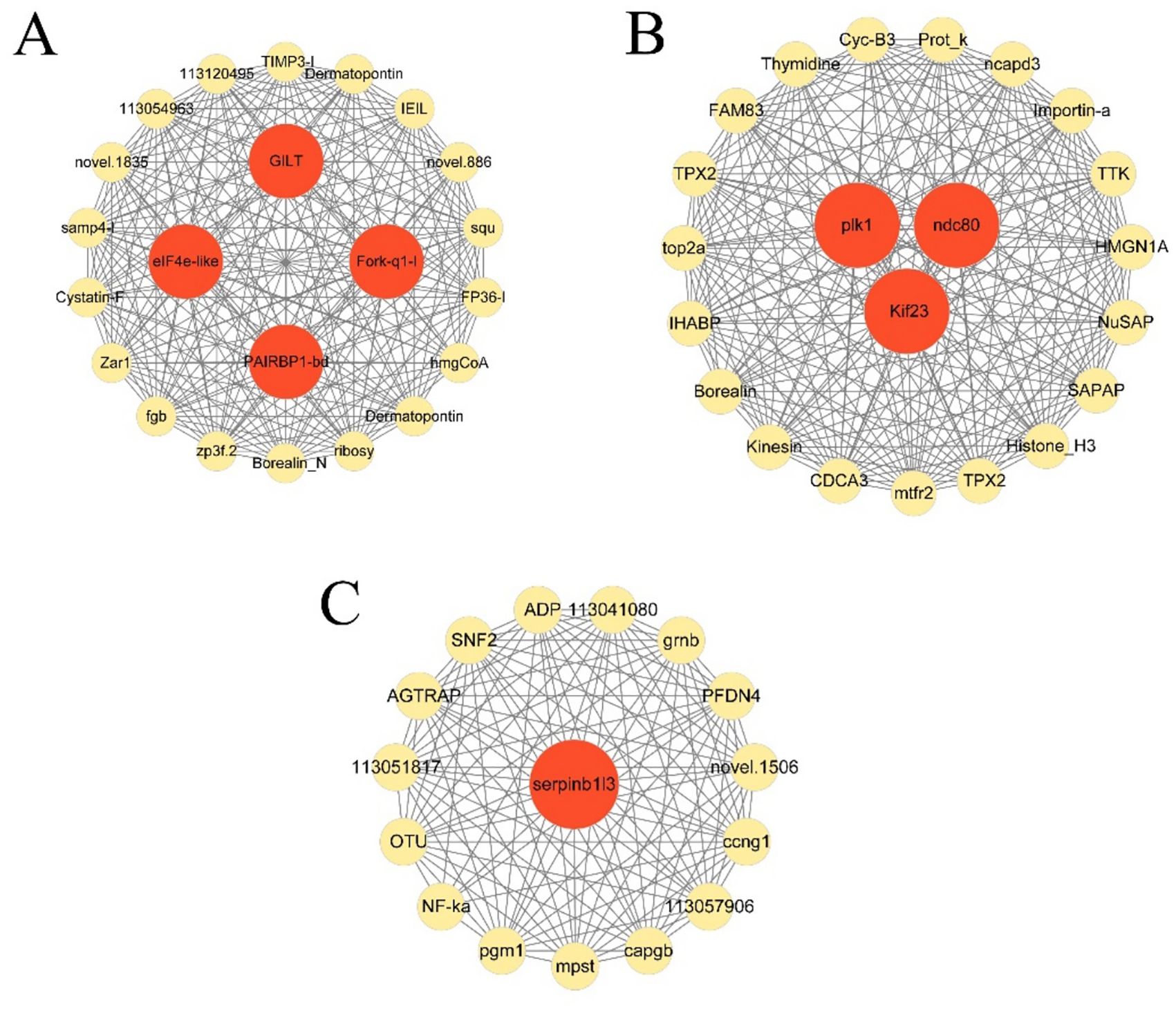
Figure 7. Gene co-expression networks of top 22 DEGs in cyan module (A), top16 genes in darkgrey module (B), and top 23 genes in darked module (C).
3.7 Bayesian network graph of DEGs
Eleven hypoxia-related key genes were screened from the above mentioned 3 hypoxia modules with a threshold of p value < 0.05, including pak1, cdc23, smad3a, caspase7, caspase8, mdm2, mcm6, acta2, egln3, vegf, and mrto4. The Bayesian network graph of DEGs was plotted using the R package CBNplot (https://noriakis.github.io/software/CBNplot/) (Figure 8).
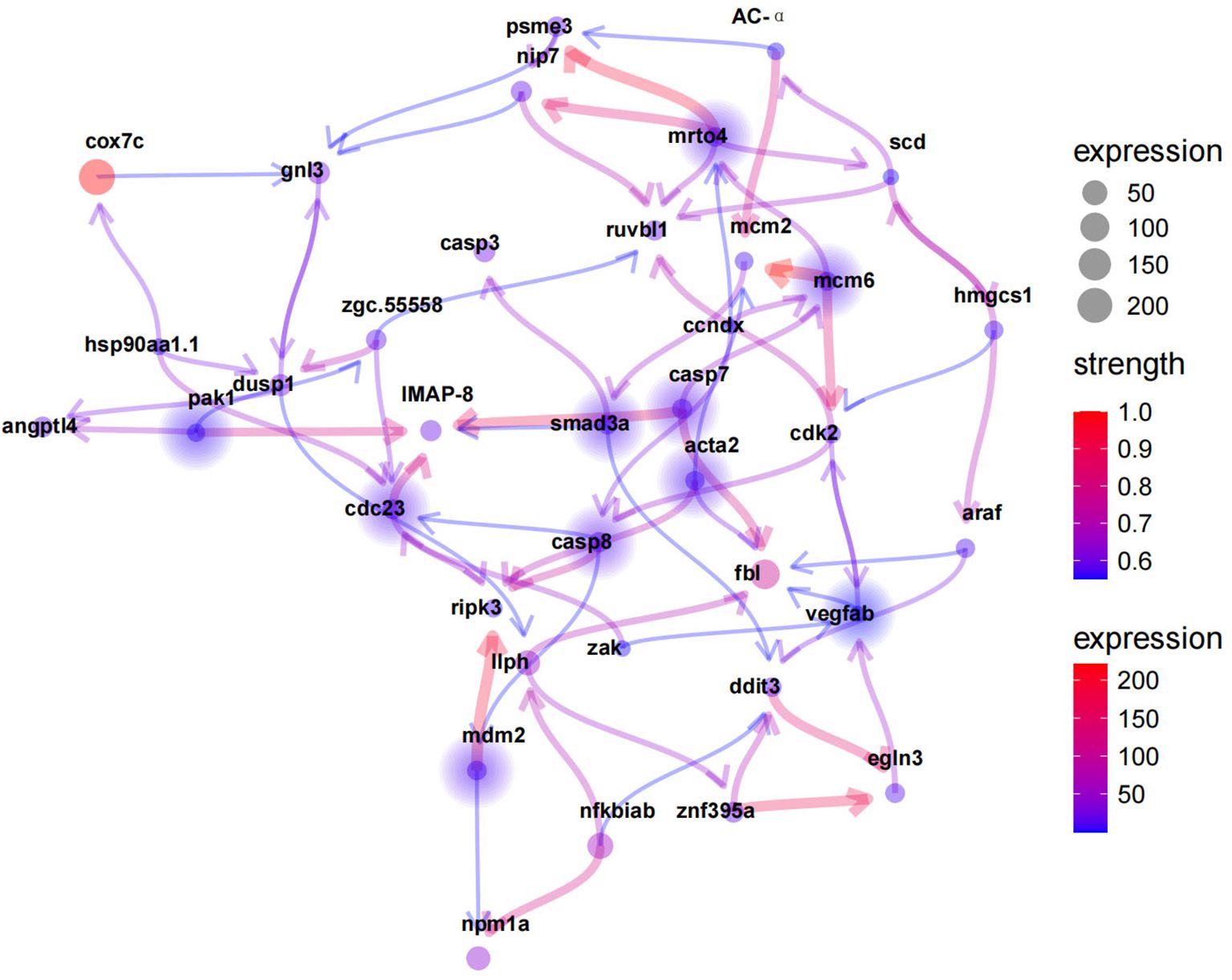
Figure 8. Hypoxia-relatedkey genes predicted by Bayesian network analysis. Solid circles indicate key genes. Line color from blue to red represent the increasing strength.
3.8 Validation of gene expression by qRT-PCR
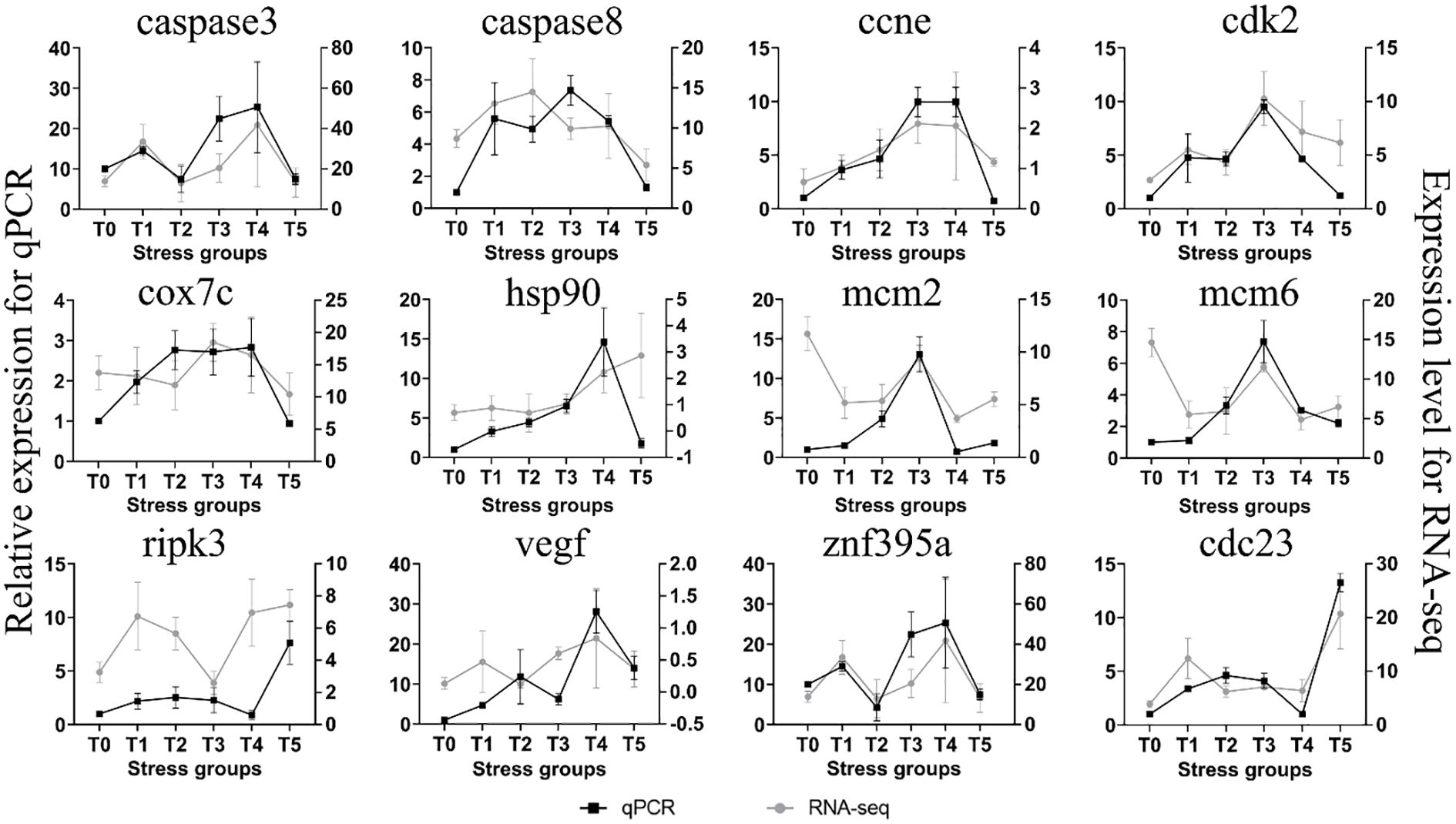
To verify the accuracy of the RNA-Seq data, we selected 12 genes for qRT-PCR. The results of qRT-PCR (Figure 9) indicated that the expression patterns (upregulation and downregulation) of 6 genes, including caspase8, cnne, caspase3, znf395a, cdk2, and cdc23, were consistent with the change trends of the transcriptome data (FPKM). The correlation analysis showed that the levels of the measured gene transcripts had a high linear correlation (R² = 0.81), indicating that the transcriptome sequencing data were reliable.
4 Discussion
4.1 Effects of hypoxia stress on histology and antioxidative capacity of crucian carp gill
Our study found that hypoxia leads to a reduction in the volume and thickness of the interlamellar cell masses in crucian carp. This represents a morphological change in the gills of fish in response to environmental changes, which reduces the gas diffusion distance between blood and water. Consequently, it exposes the gill lamellae responsible for respiratory functions (37). It has been reported that many farmed teleost fish, such as grass carp (38) and white bream (39), also possess this ability. However, the species M. Pellegrini has been shown not to exhibit this capability under hypoxic conditions (40). As the duration of hypoxia increases, the changes in the interlamellar cell masses become more pronounced, increasing the respiratory surface area of the fish gills. This enhancement facilitates oxygen absorption from the water and aids fish in surviving in environments with low dissolved oxygen (11). The present study also found that the reduction of interlamellar cell masses can bring some harm to fish, including changes in osmotic pressure that increase energy demands, greater exposure to toxic substances and parasites, and a higher risk of mechanical damage to the gills (38). Therefore, when there is sufficient dissolved oxygen in the water, it is prudent for fish to maintain a smaller gill surface area.
Hypoxia has been reported to lead to an imbalance between oxidation and antioxidation in organisms (41), which induces oxidative stress and excessive ROS in various fish species (42, 43). The antioxidant system in fish includes multiple antioxidative enzymes such as LDH, SOD, and CAT (44). Under hypoxic stress, enhancing antioxidant capacity can prevent oxidative damage in fish and eliminate excessive ROS (45). Our data showed that SOD and CAT levels were significantly higher in the hypoxia treatment groups than in the control group under the first 3-day hypoxic stress, indicating an increase in antioxidant capacity, but a decrease was observed in the last two days (day 4-5) of the hypoxia experiment. This might be due to the damage to the gill lamellae induced by prolonged exposure to hypoxia, which was supported by vacuole formation in the gill lamellae was observed on day 4 and day 5. LDH is a key enzyme in anaerobic metabolism, and it can reversibly catalyze the production of pyruvate from lactate (46). It has been reported that hypoxic stress can increase LDH activity in the gills of Nile tilapia (O. niloticus), thus enhancing its anaerobic metabolic capacity. In addition, the increased LDH activity has also been detected in the gills of spotted fish (Oplegnathus punctatus) under hypoxic conditions (47). In this study, LDH activity was consistently significantly elevated in various hypoxia groups. Based on the above findings, we speculated that at various stages of hypoxic stress, crucian carp might increase LDH enzyme activity, thus enhancing anaerobic metabolism and maintaining energy supply. The upregulation of LDH gene can catalyze the conversion of lactate to pyruvate to participate in anaerobic glycolysis, and thus the expression level of LDH gene can reflect the level of anaerobic glycolysis to some extent (12).
4.2 Effects of hypoxia on signal transduction
Cells possess highly complex interactive networks that respond specifically to various stimuli. When cells encounter hypoxic stress, multiple cellular signaling pathways are activated to maintain oxygen homeostasis, and cells adjust their physiological functions to respond to hypoxic stress (48). In rainbow trout (49), yellow catfish (Tachysurus fulvidraco ♀×Pseudobagrus vachellii ♂) (1), and dark barbel catfish (Pelteobagrus vachelli) (50), signal transduction pathways are activated during hypoxic stimulation. Our data showed that under hypoxic stress, p53 signaling pathway was activated in crucian carp gill, thus enhancing anaerobic metabolism through glycolysis and increasing energy supply through lipid metabolism, eventually maintaining cellular homeostasis. Moreover, under hypoxic stress, mitochondrial respiration is inhibited, releasing signaling molecule cyp450 into the cytoplasm to activate apoptosis genes of the caspase family, leading to extrinsic apoptosis in carp fish (51). Long-term hypoxia leads to the production of inflammatory factors, thus the activating immune signaling pathways such as RIG-I and NLRs in cells to eliminate these inflammatory factors. These findings suggested that in crucian carp, cellular apoptotic pathways might be initially triggered under hypoxic stress, and prolonged exposure to hypoxia might further induce the activation of immune pathways associated with endogenous antioxidant defense mechanisms.
4.3 Effect of hypoxia stress on apoptosis-related genes of crucian carp gill
Hif-1α, as a primary regulator of the hypoxic response, can regulate a series of adaptive responses to hypoxic stress (52). In this study, the hif-1α gene was consistently lowly expressed of hypoxic stress, but its downstream target gene vegf was significantly up-regulated. The upregulation of the vegf gene under hypoxic conditions may enhance angiogenesis, which is crucial for gill remodeling (53). Our data also showed that the expression of egln3 gene (the hydroxylase of hif-1α) was significantly increased. Egln3can promote the hydroxylation of hif-1α, leading to its ubiquitination and subsequent degradation by the proteasome (54). This might explain its negligible low expression of the hif-1αunder hypoxic stress. In addition, we have identified 11 key regulatory genes, and these genes might play a crucial role in the response of crucian carp gill to hypoxia. In the initial stages (day 1 and day 2) of hypoxia experiment, a significant accumulation of ROS resulted in the up-regulation of cytochrome P450 (cyp450), thus triggering the up-regulationofcaspase3/8 apoptosis-related genes. Key genes in the cell cycle such as cdk2, mcm6, and prmt1 play significant roles in cell apoptosis. Gene Smad3a can down-regulate cdk2, hindering cell proliferation in the G1 phase, and geneprmt1 acts on the Fox1 receptor to release signaling factors into the cytoplasm, and these signaling factors act on apoptosis-related genes caspase8 and caspase7. Under severe hypoxic conditions, the expression of the ripk3 gene triggers necroptosis. In previous study, under hypoxia stress, hif-1αactivates the VEGF pathway, thereby inducing angiogenesis (55). The vegf gene is an important gene promoting angiogenesis, and accordingly, heterozygous vegf mutant mice embryo die in utero due to vascular defects (52). In this study, the activation of the VEGF pathway might also be related to Pak1 enzyme. Recently, Kelly et al. (56) have reported that Gata6 gene in the Pak1/Erk signaling module regulates cardiovascular development in zebrafish. The expression of the vegf gene contributes to the uptake of oxygen in blood, thus facilitating adaptation to hypoxic conditions. Our data showed that hypoxia significantly reduced ILCM between crucian carp gill lamellae, thus increasing the contact area between gill lamellae and water and accelerating oxygen intake by gill (57). Therefore, we speculated that high adaptability of crucian carp to hypoxic conditions might be closely related to the apoptosis of ILCM and the action of the vegf gene.
5 Conclusion
In this study, our transcriptome analysis revealed that the differentially expressed genes in the gills of crucian carp under hypoxic conditions are mostly related to energy metabolism, antioxidant stress, and apoptosis pathways. In the first two days of the hypoxia experiment, crucian carp reduces oxidative damage caused by reactive oxygen species (ROS) through anaerobic metabolism and activates pathways such as P53 to block the cell cycle and affect cell proliferation, including genes like pak1 and cdk2, which are important for promoting apoptosis in interlamellar cells (ILCM) and help crucian carp to improve oxygen uptake efficiency in the early stages of hypoxia, thus adapting to low oxygen conditions. Secondly, there is the intrinsic apoptotic pathway dominated by the caspase family. However, under prolonged hypoxia, the gills of crucian carp will strengthen their adaptation to hypoxia by generating microvessels, such as vegf. In the later stages of hypoxia, it may be due to cell necrosis causing a large number of inflammatory factors, leading to the enrichment and activation of cellular immune pathways such as RIG-I and NLR. While the transcriptome analysis provided valuable insights into gene expression changes, it lacked in-depth functional validation of the identified genes. Merely observing altered expression levels does not definitively prove their direct contribution to hypoxia adaptation, and further experimental confirmation through techniques like gene knockout or overexpression need to be conducted to ascertain their true functional significance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by Animal Experimentation Ethics Committee of Hebei Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
M-CX: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft. ZR: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft. XZ: Investigation, Software, Writing – review & editing. X-WG: Investigation, Software, Writing – review & editing. Z-GH: Investigation, Software, Writing – review & editing. L-HZ: Data curation, Investigation, Methodology, Software, Writing – review & editing. WK: Data curation, Investigation, Methodology, Software, Writing – review & editing. Y-HX: Data curation, Formal analysis, Funding acquisition, Project administration, Writing – review & editing. LC: Data curation, Formal analysis, Funding acquisition, Project administration, Writing – review & editing. C-BW: Data curation, Formal analysis, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (grant no. 32303030), Science Research Project of Hebei Education Department (grant no. BJK2024012), the Natural Science Foundation of Hebei Province, China (grant nos.C2021204089, V1654588161890 and 236Z6701G), and Special Fund for Talent Introduction of Hebei Agricultural University, China (grant no. YJ2020032).
Acknowledgments
The authors would like to express their gratitude to Ocean and Fisheries Science Research Institute of Hebei Province for providing artificial feed, fry and breeding assistance. The author is also grateful to the laboratory instrument management teacher for their assistance in using the instrument.
Conflict of interest
The author(s) declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1543605/full#supplementary-material
References
1. Tao Y-F, Qiang J, Dagoudo M, Zhu H-J, Bao J-W, Ma J-L, et al. Transcriptome profiling reveals differential expression of immune-related genes in gills of hybrid yellow catfish (Tachysurus fulvidraco ♀ × Pseudobagrus vachellii ♂) under hypoxic stress: Potential NLR-mediated immune response. Fish Shellfish Immunol. (2021) 119:409–19. doi: 10.1016/j.fsi.2021.10.023
2. Borowiec BG, Scott GR. Hypoxia acclimation alters reactive oxygen species homeostasis and oxidative status in estuarine killifish (Fundulus heteroclitus). J Exp Biol. (2020) 223:222877. doi: 10.1242/jeb.222877
3. Wang Z, Pu D, Zheng J, Li P, Lü H, Wei X, et al. Hypoxia-induced physiological responses in fish: From organism to tissue to molecular levels. Ecotoxicology Environ Saf. (2023) 267:115609. doi: 10.1016/j.ecoenv.2023.115609
4. Mikloska KV, Zrini ZA, Bernier NJ. Severe hypoxia exposure inhibits larval brain development but does not affect the capacity to mount a cortisol stress response in zebrafish. J Exp Biol. (2022) 225:243335. doi: 10.1242/jeb.243335
5. Suo N, Zhou Z-X, Xu J, Cao D-C, Wu B-Y, Zhang H-Y, et al. Transcriptome analysis reveals molecular underpinnings of common carp (Cyprinus carpio) under hypoxia stress. Front Genet. (2022) 13:907944. doi: 10.3389/fgene.2022.907944
6. Evans DH, Piermarini PM, Choe KP. The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev. (2005) 85:97–177. doi: 10.1152/physrev.00050.2003
7. Zhao S-S, Su X-L, Pan R-J, Lu L-Q, Zheng G-D, Zou S-M. The transcriptomic responses of blunt snout bream (Megalobrama amblycephala) to acute hypoxia stress alone, and in combination with bortezomib. BMC Genomics. (2022) 23:162. doi: 10.1186/s12864-022-08399-7
8. van der Weele CM, Jeffery WR. Cavefish cope with environmental hypoxia by developing more erythrocytes and overexpression of hypoxia-inducible genes. eLife. (2022) 11:69109. doi: 10.7554/eLife.69109
9. Tzaneva V, Vadeboncoeur C, Ting J, Perry SF. Effects of hypoxia-induced gill remodelling on the innervation and distribution of ionocytes in the gill of goldfish. Carassius auratus. J Comp Neurol. (2014) 522:118–30. doi: 10.1002/cne.23392
10. Yu X, Zhang Y, Li S, Zheng G, Zou S. Effects of hypoxia on the gill morphological structure, apoptosis and hypoxia-related gene expression in blunt snout bream (Megalobrama amblycephala). Fish Physiol Biochem. (2023) 49:939–49. doi: 10.1007/s10695-023-01233-1
11. Nilsson GE, Dymowska A, Stecyk JAW. New insights into the plasticity of gill structure. Respir Physiol Neurobiol. (2012) 184:214–22. doi: 10.1016/j.resp.2012.07.012
12. Cooper RU, Clough LM, Farwell MA, West TL. Hypoxia-induced metabolic and antioxidant enzymatic activities in the estuarine fish. Leiostomus xanthurus. J Exp Mar Biol Ecol. (2002) 279:1–20. doi: 10.1016/S0022-0981(02)00329-5
13. Ren Y, Tian Y, Cheng B, Liu Y, Yu H. Effects of Environmental Hypoxia on Serum Hematological and Biochemical Parameters, Hypoxia-Inducible Factor (hif) Gene Expression and HIF Pathway in Hybrid Sturgeon (Acipenser schrenckii ♂ × Acipenser baerii ♀). Genes. (2024) 15:743. doi: 10.3390/genes15060743
14. Xie Z, Shi J, Shi Y, Tu Z, Hu M, Yang C, et al. Physiological responses to salinity change and diel-cycling hypoxia in gills of Hong Kong oyster. Crassostrea hongkongensis. Aquaculture. (2023) 570:739443. doi: 10.1016/j.aquaculture.2023.739443
15. Liu Y, Wang J, Ding J, Zhang Y, Hou C, Shen W, et al. Effects of hypoxia stress on oxidative stress, apoptosis and microorganisms in the intestine of large yellow croaker (Larimichthys crocea). Aquaculture. (2024) 581:740444. doi: 10.1016/j.aquaculture.2023.740444
16. Wang H, Chen Y, Ru G, Xu Y, Lu L. EGCG: Potential application as a protective agent against grass carp reovirus in aquaculture. J Fish Dis. (2018) 41:1259–67. doi: 10.1111/jfd.12819
17. Li L, Wang Y-T, Meng S-T, Wei X-F, Yang Z-Y, Zhu R, et al. Effects of single or conjoint administration of poly-β-hydroxybutyrate and Bacillus subtilis on growth, antioxidant capacity and apoptosis of common carp (Cyprinus carpio). Animal Feed Science and Technology (2024) 318:116110. doi: 10.1016/j.anifeedsci.2024.11611010
18. Zhang X, Luan P, Cao D, Hu G. A high-density genetic linkage map and fine mapping of QTL for feed conversion efficiency in common carp (Cyprinus carpio). Front Genet. (2021) 12:778487. doi: 10.3389/fgene.2021.778487
19. Bundgaard A, Ruhr IM, Fago A, Galli GLJ. Metabolic adaptations to anoxia and reoxygenation: New lessons from freshwater turtles and crucian carp. Curr Opin Endocrine Metab Res. (2020) 11:55–64. doi: 10.1016/j.coemr.2020.01.002
20. Moreno KX, Satapati S, DeBerardinis RJ, Burgess SC, Malloy CR, Merritt ME. Real-time detection of hepatic gluconeogenic and glycogenolytic states using hyperpolarized [2-13C] dihydroxyacetone. J Biol Chem. (2014) 289:35859–67. doi: 10.1074/jbc.M114.613265
21. Lutz PL, Nilsson GE. Contrasting strategies for anoxic brain survival – glycolysis up or down. J Exp Biol. (1997) 200:411–9. doi: 10.1242/jeb.200.2.411
22. Shang F, Bao M, Liu F, Hu Z, Wang S, Yang X, et al. Transcriptome profiling of tiger pufferfish (Takifugu rubripes) gills in response to acute hypoxia. Aquaculture. (2022) 557:738324. doi: 10.1016/j.aquaculture.2022.738324
23. Zheng X, Fu D, Cheng J, Tang R, Chu M, Chu P, et al. Effects of hypoxic stress and recovery on oxidative stress, apoptosis, and intestinal microorganisms in Pelteobagrus vachelli. Aquaculture. (2021) 543:736945. doi: 10.1016/j.aquaculture.2021.736945
24. Liang Y-S, Wu R-X, Niu S-F, Miao B-B, Liang Z-B, Zhai Y. Liver transcriptome analysis reveals changes in energy metabolism, oxidative stress, and apoptosis in pearl gentian grouper exposed to acute hypoxia. Aquaculture. (2022) 561:738635. doi: 10.1016/j.aquaculture.2022.738635
25. Cheng X, Li F, Lu J, Wen Y, Li Z, Liao J, et al. Transcriptome analysis in gill reveals the adaptive mechanism of domesticated common carp to the high temperature in shallow rice paddies. Aquaculture. (2024) 578:740107. doi: 10.1016/j.aquaculture.2023.740107
26. Radulescu C, Olteanu RL, Buruleanu CL, Tudorache MN, Dulama ID, Stirbescu RM, et al. Polyphenolic screening and the antioxidant activity of grape pomace extracts of Romanian white and red grape varieties. Antioxidants. (2024) 13:1133. doi: 10.3390/antiox13091133
27. Xu Y, Luo L, Chen J. Sulfamethoxazole induces brain capillaries toxicity in zebrafish by up-regulation of VEGF and chemokine signalling. Ecotoxicology Environ Saf. (2022) 238:113620. doi: 10.1016/j.ecoenv.2022.113620
28. Qiang J, Bao WJ, Tao FY, He J, Li XH, Xu P, et al. The expression profiles of miRNA–mRNA of early response in genetically improved farmed tilapia (Oreochromis niloticus) liver by acute heat stress. Sci Rep. (2017) 7:8705. doi: 10.1038/s41598-017-09264-4
29. Li H, Qiang J, Song C, Xu P. Transcriptome profiling reveal Acanthopanax senticosus improves growth performance, immunity and antioxidant capacity by regulating lipid metabolism in GIFT (Oreochromis niloticus). Comp Biochem Physiol Part D: Genomics Proteomics. (2021) 37:100784. doi: 10.1016/j.cbd.2020.100784
30. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
31. Wu S, Huang J, Li Y, Zhao L, Liu Z, Kang Y, et al. Integrative mRNA-miRNA interaction analysis reveals the molecular mechanism of skin color variation between wild-type and yellow mutant rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol Part D: Genomics Proteomics. (2021) 40:100914. doi: 10.1016/j.cbd.2021.100914
32. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
33. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. (2008) 9:559. doi: 10.1186/1471-2105-9-559
34. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
35. Lu J-F, Luo S, Tang H, Liang J-H, Zhao Y-F, Hu Y, et al. Micropterus salmoides rhabdovirus enters cells via clathrin-mediated endocytosis pathway in a pH-, dynamin-, microtubule-, rab5-, and rab7-dependent manner. J Virol. (2023) 97:e00714–23. doi: 10.1128/jvi.00714-23
36. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
37. Sollid J, De Angelis P, Gundersen K, Nilsson GE. Hypoxia induces adaptive and reversible gross morphological changes in crucian carp gills. J Exp Biol. (2003) 206:3667–73. doi: 10.1242/jeb.00594
38. Gilmour KM, Perry SF. Conflict and compromise: using reversible remodeling to manage competing physiological demands at the fish gill. Physiology. (2018) 33:412–22. doi: 10.1152/physiol.00031.2018
39. Xu X-N, Chen S-L, Jiang Z-X, Nissa MU, Zou S-M. Gill remodeling increases the respiratory surface area of grass carp (Ctenopharyngodon idella) under hypoxic stress. Comp Biochem Physiol Part A: Mol Integr Physiol. (2022) 272:111278. doi: 10.1016/j.cbpa.2022.111278
40. Dhillon RS, Yao L, Matey V, Chen B-J, Zhang A-J, Cao Z-D, et al. Interspecific differences in hypoxia-induced gill remodeling in carp. Physiol Biochem Zoology. (2013) 86:727–39. doi: 10.1086/673180
41. Clanton TL. Hypoxia-induced reactive oxygen species formation in skeletal muscle. J Appl Physiol. (2007) 102:2379–88. doi: 10.1152/japplphysiol.01298.2006
42. Leveelahti L, Rytkönen KT, Renshaw GMC, Nikinmaa M. Revisiting redox-active antioxidant defenses in response to hypoxic challenge in both hypoxia-tolerant and hypoxia-sensitive fish species. Fish Physiol Biochem. (2014) 40:183–91. doi: 10.1007/s10695-013-9835-1
43. Luo S-Y, Liu C, Ding J, Gao X-M, Wang J-Q, Zhang Y-B, et al. Scavenging reactive oxygen species is a potential strategy to protect Larimichthys crocea against environmental hypoxia by mitigating oxidative stress. Zoological Res. (2021) 42:592–605. doi: 10.24272/j.issn.2095-8137.2021.079
44. Yang S, Yan T, Wu H, Xiao Q, Fu HM, Luo J, et al. Acute hypoxic stress: Effect on blood parameters, antioxidant enzymes, and expression of HIF-1alpha and GLUT-1 genes in largemouth bass (Micropterus salmoides). Fish Shellfish Immunol. (2017) 67:449–58. doi: 10.1016/j.fsi.2017.06.035
45. Hermes-Lima M, Zenteno-Savıín T. Animal response to drastic changes in oxygen availability and physiological oxidative stress. Comp Biochem Physiol Part C: Toxicol Pharmacol. (2002) 133:537–56. doi: 10.1016/S1532-0456(02)00080-7
46. Qin Y, Liu H, Mao S, Deng R, Wang Y, Deng S, et al. Isolation, identification, and monoclonal antibody development of largemouth bass virus. Front Mar Sci. (2024) 10:1338197. doi: 10.3389/fmars.2023.1338197
47. Baldissera MD, Souza CF, Petrolli TG, Baldisserotto B, Da Silva AS. Caffeine prevents hypoxia-induced dysfunction on branchial bioenergetics of Nile tilapia through phosphoryl transfer network. Aquaculture. (2019) 502:1–7. doi: 10.1016/j.aquaculture.2018.12.024
48. Ratcliffe PJ. Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J Physiol. (2013) 591:2027–42. doi: 10.1113/jphysiol.2013.251470
49. Wu S, Huang J, Li Y, Zhao L. Integrated physiological, whole transcriptomic and functional analysis reveals the regulatory mechanism in the liver of rainbow trout (Oncorhynchus mykiss) in response to short-term and chronic hypoxia stress. Aquaculture. (2024) 593:741250. doi: 10.1016/j.aquaculture.2024.741250
50. Zhang G, Mao J, Liang F, Chen J, Zhao C, Yin S, et al. Modulated expression and enzymatic activities of Darkbarbel catfish, Pelteobagrus vachelli for oxidative stress induced by acute hypoxia and reoxygenation. Chemosphere. (2016) 151:271–9. doi: 10.1016/j.chemosphere.2016.02.072
51. Olsvik PA, Kristensen T, Waagbø R, Tollefsen K-E, Rosseland BO, Toften H. Effects of hypo- and hyperoxia on transcription levels of five stress genes and the glutathione system in liver of Atlantic cod. Gadus morhua. J Exp Biol. (2006) 209:2893–901. doi: 10.1242/jeb.02320
52. Guo X, Zhang J, Feng Z, Ji J, Shen X, Hou X, et al. The antiangiogenic effect of total saponins of Panax japonicus C.A. Meyer in rheumatoid arthritis is mediated by targeting the HIF-1α/VEGF/ANG-1 axis. J Ethnopharmacology. (2024) 333:118422. doi: 10.1016/j.jep.2024.118422
53. Li HL, Gu XH, Li BJ, Chen X, Lin HR, Xia JH. Characterization and functional analysis of hypoxia-inducible factor HIF1α and its inhibitor HIF1αn in tilapia. PLoS One. (2017) 12:173478. doi: 10.1371/journal.pone.0173478
54. Zheng S, Tang BZ, Wang W-X. Microplastics and nanoplastics induced differential respiratory damages in tilapia fish. Oreochromis niloticus. J Hazardous Materials. (2024) 465:133181. doi: 10.1016/j.jhazmat.2023.133181
55. Fish JE, Gutierrez MC, Dang LT, Khyzha N, Chen Z, Veitch S, et al. Dynamic regulation of VEGF-inducible genes by an ERK-ERG-p300 transcriptional network. Development. (2017) 144(13):2428–44. doi: 10.1242/dev.146050
56. Lange M, Ohnesorge N, Hoffmann D, Rocha SF, Benedito R, Siekmann AF. Zebrafish mutants in vegfab can affect endothelial cell proliferation without altering ERK phosphorylation and are phenocopied by loss of PI3K signaling. Dev Biol. (2022) 486:26–43. doi: 10.1016/j.ydbio.2022.03.006
Keywords: Carassius auratus, hypoxia-tolerant, adaptive mechanism, RNA-Seq, Bayesian networks
Citation: Xing M, Rong Z, Zhao X, Gao X, Hou Z, Zhang L, Khor W, Xu Y, Chen L and Wu C (2025) Transcriptome analysis reveals hypoxic response key genes and modules as well as adaptive mechanism of crucian carp (Carassius auratus) gill under hypoxic stress. Front. Immunol. 16:1543605. doi: 10.3389/fimmu.2025.1543605
Received: 11 December 2024; Accepted: 20 January 2025;
Published: 05 February 2025.
Edited by:
Erchao Li, East China Normal University, ChinaReviewed by:
Jianfei Lu, Ningbo University, ChinaWeidong Ding, Chinese Academy of Fishery Sciences (CAFS), China
Dingbin Gong, Anqing Normal University, China
Copyright © 2025 Xing, Rong, Zhao, Gao, Hou, Zhang, Khor, Xu, Chen and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengbin Wu, aHl4eXdjYkBoZWJhdS5lZHUuY24=; Yihuan Xu, eHV5aWh1YW5AaGViYXUuZWR1LmNu; Li Chen, aGJzc2N5anNAdmlwLnNpbmEuY29t
†These authors have contributed equally to this work and share first authorship
 Mengchao Xing
Mengchao Xing Zhen Rong1,2†
Zhen Rong1,2† Lihan Zhang
Lihan Zhang Waiho Khor
Waiho Khor Yihuan Xu
Yihuan Xu Chengbin Wu
Chengbin Wu