
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 03 March 2025
Sec. Viral Immunology
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1533003
 Michael J. Johnson1
Michael J. Johnson1 Sarah K. Lazarus1†
Sarah K. Lazarus1† Ashlynn E. Bennett1
Ashlynn E. Bennett1 Adriana Tovar-Salazar1†
Adriana Tovar-Salazar1† Charles E. Robertson2
Charles E. Robertson2 Jennifer M. Kofonow2
Jennifer M. Kofonow2 Shaobing Li1
Shaobing Li1 Bruce McCollister2,3
Bruce McCollister2,3 Marta C. Nunes4†
Marta C. Nunes4† Shabir A. Madhi4,5
Shabir A. Madhi4,5 Daniel N. Frank2
Daniel N. Frank2 Adriana Weinberg1,2,5*
Adriana Weinberg1,2,5*Introduction: Infants exposed to HIV and uninfected (HEUs) are at higher risk of infectious morbidity than HIV-unexposed uninfected infants (HUUs). Multiple immune defects of unknown origin were observed in HEUs. We hypothesized that HEUs have more regulatory and inhibitory checkpoint-expressing T cells (Treg, Tici) than HUUs, which may dampen their immune defenses against pathogens.
Method: We used flow cytometry to measure 25 Treg/Tici subsets in HEUs and HUUs at birth, 6, 28, and 62 weeks of life. We used maternal and infant gut microbiome data reported in a previous study to establish correlations with the Treg/Tici.
Results: At birth, 3 Treg subsets, including the prototypic CD4+FOXP3+ and CD4+FOXP3+CD25+, had higher frequencies in 123 HEUs than in 117 HUUs, and 3 subsets had higher frequencies in HUUs. At 28 and 62 weeks of age, 5 Treg/Tici subsets had higher proportions in HEUs than HUUs. The frequencies of the Treg/Tici subsets that diverged between HEUs and HUUs at birth correlated with differential relative abundances of bacterial taxa in the maternal gut microbiome. The Treg/Tici subsets with significantly different frequencies at subsequent visits correlated with the concurrent composition of the infant gut microbiome. In vitro, treatment of HUU peripheral blood mononuclear cells (PBMC) with bacterial taxa most abundant in HEUs expanded Treg/Tici subsets with higher frequencies in HEUs than HUUs, recapitulating the in vivo correlations. Conversely, in vitro treatment of HEU PBMC did not increase Treg/Tici frequencies. Other factors that correlated with increased Treg/Tici frequencies were low maternal CD4+ T cells in HEUs at birth and male sex in the HUUs at 28 weeks of life.
Discussion: This study shows that maternal and infant gut dysbiosis are central to the increase in Treg/Tici in HEUs and may be targeted by mitigating interventions.
Due to extraordinary advances in the prevention of HIV vertical transmission, 2 million infants exposed to HIV and uninfected (HEUs) are born every year. However, compared with HIV-unexposed infants (HUUs), HEUs have a higher incidence of hospitalization and death due to severe infections during the first 1-2 years of life (1–3). The introduction of universal 3-drug antiretroviral therapy (ART) during pregnancy in 2012 moderately improved the clinical and infectious outcomes of HEUs in Sub-Saharan Africa, but growth and increased hospitalizations in early childhood continued to be reported more than five years after maternal 3-drug ART was implemented, including 20% more stunting at 18 months of age and 2- to 3.5-fold higher rates of hospitalizations due to infections in the first 6 months of age compared with HUUs (4–8). In the US and other regions of the Northern Hemisphere, hospitalizations are increased in HEUs compared to HUUs (9, 10).
HEUs have multiple immunologic dysfunctions that may contribute to their increased risk of severe infection, hospitalization, and death, including increased numbers of regulatory T cells (Treg) (11–24). Due to the broad spectrum of Treg activity, which may dampen T cell, B cell, and antigen presenting cell function, the increase in Treg abundance provides a potential unifying mechanism for the increased susceptibility to severe infections in HEUs. People with HIV also have multiple immunologic abnormalities, including excessive T cell regulation due to high frequencies of Treg and other T cells expressing immunologic checkpoint inhibitors (Tici), which have been associated with accelerated disease progression and high susceptibility to severe infections (25–28). In the general population, Treg/Tici have been associated with decreased immune protection against tumors and viral infections (29–31). We hypothesized that high Treg/Tici frequencies in HEUs may increase their susceptibility to infections in early childhood.
The gut microbiome has emerged as a central element in the education of local and systemic immune responses (32). The human gut harbors 12 to 20% of the total lymphocytes, and most importantly, it is a critical site of innate and adaptive T-cell maturation, second only to the thymus (33, 34). Bacterial taxonomic groups, such as segmented filamentous bacteria, E. coli, B. fragilis and diverse clostridia (e.g., Ruminococcaceae and Lachnospiraceae), alter the balance between Treg and conventional T cells (35–38). Studies revealed that bacterial products, such as short-chain fatty acids (SCFAs), tryptophan catabolites, and B. fragilis-derived polysaccharide A, promote Treg differentiation and expansion (39–42). The composition of the infant gut microbiome undergoes sequential changes after birth, influenced primarily by the delivery mode, maternal microbiome, and maternal and infant diet (43–51). Previous studies showed differences in the composition of the gut microbiota of people with and without HIV (52–58) and between HEUs and HUUs (59, 60). Moreover, in the same cohort that the current study is based on, we showed significant differences in the gut microbiota of HEUs and HUUs and that infant gut microbiota extensively overlapped with maternal gut microbiota (59).
The Treg hallmark is the transcription factor FOXP3, which inhibits IFNG and IL2 gene transcription and thereby prevents conventional T-cell differentiation (61). Particular importance in the inheritance of Treg characteristics during cell division has been attached to a FOXP3 intronic regulatory element, conserved non-coding sequences 2 (CNS2), which is completely demethylated in Treg (62). In addition to FOXP3+ Treg, multiple other Treg subsets have been previously validated, including markers shared with Tici (12, 29, 63–65). The goal of this study was to undertake a comprehensive analysis of the relative abundance of Treg/Tici in HEUs and HUUs during the first year of life and to identify factors associated with differences between the two groups, including maternal HIV infection characteristics; infant sex and birth weight; DNA methylation of CNS2 and other loci; and gut microbiome composition.
The study was approved by the Human Research Ethics Committee at the University of the Witwatersrand (approval number: M171185) and the Colorado Multiple Institutions Review Board (COMIRB 17-0306). Written informed consent was obtained prior to participation in the study. Women with and without HIV were recruited during labor at Chris Hani Baragwanath Academic Hospital in Johannesburg, South Africa. The inclusion criteria for all women were singleton term gestation, planned vaginal delivery, and intent to breastfeed. Women with HIV had to have been prescribed antiretrovirals but not cotrimoxazole during pregnancy. After providing informed consent, maternal and infant metadata were collected from medical records and by interviewing the study participants at 6, 28, and 62 weeks after delivery. Maternal blood was obtained at delivery for CD4+ T-cell and HIV plasma RNA measurements at the local laboratory. Infant cord blood and peripheral blood obtained at 6, 28 and 62 weeks of life were used to measure Treg subsets. Infant rectal swabs obtained at 6, 28, and 62 weeks of life and maternal rectal swabs at delivery were used for microbiome analysis. Mothers and infants who received antibiotic therapy within one month prior to the rectal swab collection, with the exception of cotrimoxazole in HEUs, were excluded from the microbiome analysis.
PBMC were cryopreserved for viability as previously described and stored at ≤-150°C until use (66, 67). Cryopreserved PBMC/cord blood mononuclear cells were thawed, counted, and processed immediately for phenotypic assessment using two staining panels. Panel A consisted of surface staining with Zombie yellow (viability), CD25 FITC (BioLegend), Lag3 PE (Invitrogen), CTLA4 PE-CF594 (BD Biosciences), CD4 PerCP-Cy5.5 (BD Biosciences), and CD3 Ax700 (BD Biosciences), followed by fixation and permeabilization using the eBioscience Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Intracellular staining was then performed with FoxP3 Ax647 (BD Biosciences), Granzyme B (GranzB) APC-fire750 (BioLegend), IL-10 BV421 (BioLegend) and TGFβ PE-Cy7 (BioLegend). Panel B consisted of surface staining with Zombie yellow (viability), CD4 FITC (BioLegend), CD3 PE-CF594 (BD Biosciences), GITR PerCP-Cy5.5 (BioLegend), TNFR2 PE-Cy7 (BioLegend), CD39 Ax700 (R&D Systems), PD1 APC-Cy7 (BioLegend) and TIGIT BV421 (BioLegend). Intracellular staining consisted of FoxP3 Ax647 (BD Biosciences) and IL-35 PE (BioLegend) antibodies. Analysis was performed using a Galios instrument (Beckman Coulter). The gating strategy is shown in Supplementary Figure S1.
Ex vivo induction of Treg/Tici. Blautia wexlerae (Cat# BAA-1564, ATCC), Lactococcus lactis (Cat# 19435, ATCC) and Ruminococcus bromii (Cat# 27255, ATCC) were subcultured onto Brucella agar plates (Cat# RO1253, Remel) and incubated at 37°C in an anaerobic chamber until colony growth was observed. Clinical isolates of Klebsiella pneumoniae, Enterobacter cloacae, and Proteus mirabilis from the Microbiology Clinical Laboratory at the University of Colorado Hospital were subcultured onto sheep blood agar plates (Cat# RO 1202, Remel) and incubated overnight at 37°C. A bacterial suspension was generated for each organism by transferring the bacterial colonies to sterile saline and equilibrating them to a 0.5 McFarland standard as measured by a turbidity meter (68). Bacterial suspensions were UV-inactivated for 15 min, aliquoted, and stored frozen at -80°C until use. For stimulation assays, cryopreserved PBMC were thawed, washed, counted with a Guava EasyCyte instrument (Luminex), and resuspended in RPMI 1640 (Corning) supplemented with 10% FBS (Gemini), 2 mM L-glutamine (Gemini), 20 mM HEPES buffer (Corning), and 1% penicillin/streptomycin solution (Gemini) at 106 PBMC/mL. The cells were incubated with bacteria under preoptimized conditions at a multiplicity of infection of 10 colony-forming units per viable PBMC for 7 days at 37°C in a CO2 incubator (Thermo Fisher). During the last 16 h of incubation, Brefeldin-A (5 µg/ml; Sigma−Aldrich) was added, after which the cells were washed with PBS (Corning), stained with Zombie Aqua Fixability dye (BioLegend), washed with PBS+1% BSA (Millipore Sigma−Aldrich) and stained for surface markers with GITR BV711, PD1 BV785, Lag3 APC-Cy7 (BioLegend), CD4 PerCP-C75.5, CD25 PECF594, and CD3 Ax700 (BD Biosciences) in BD Horizon Brilliant Stain Buffer Plus (BD Biosciences). The cells were then washed and fixed/permeabilized with the eBioscience FOXP3/Transcription Factor Staining Buffer Set (Invitrogen). The cells were washed with the kit-provided buffer and stained for intracellular markers with IL-10 BV421, CTLA4 BV605, GranzB FITC, FOXP3 PE, TGFβ PE-Cy7 (BioLegend), and IL-35 APC (R&D Systems) in BD Horizon Brilliant Stain Buffer Plus. The cells were then washed and resuspended in PBS+1% paraformaldehyde before acquisition on a NovoCyte Quanteon cytometer (Agilent). PBMC from a study-dedicated leucopack control were used in each run to ensure interassay reproducibility. The data analysis was performed in FlowJo (BD Biosciences). The Treg/Tici% was calculated using live PBMC as the parent. Gating strategy in Supplementary Figure S3.
Analysis of DNA methylation was performed on CD4+ T cells purified using Miltenyi Biotech CD4 isolation kit (Cat#130-096-533) as per manufacturer’s instructions. The methylation analysis used the Infinium® MethylationEPIC BeadChip (Illumina) as per manufacturer’s instructions.
The fecal microbiota was profiled by 16S rRNA gene sequencing. Methods and results were previously published (59). All the sequences and corresponding metadata were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA816484.
T cell subsets with frequencies <0.001% at all time points were excluded from the analysis through an a priori decision based on the analytical sensitivity of the flow cytometry method. Relative frequencies were used to identify differences in HEUs and HUUs at birth, 6 weeks, 28 weeks, and 62 weeks. These cross-sectional analyses used Wilcoxon rank sum tests from the rstatix package (69), and the FDR was used to correct for multiple comparisons for each visit. Additional analyses examining the effects of covariates such as sex and viral load (<50 or >50 HIV RNA copies/ml of plasma) were also conducted as described above. Spearman’s rank correlation coefficient was used to quantify the strengths of the relationships between continuous variables such as birthweight and CD4 count and the various Treg/Tici subsets. Longitudinal analyses of flow cytometry data used relative frequencies for each cell type and were modeled with a linear mixed effects model (LMM) to account for repeated measurements within each infant. The LMM assumed a normal distribution for the relative frequencies, used a random intercept for infant ID, and included terms for exposure, time (treated as categorical to allow more flexibility) and the interaction between exposure and time. Differences in trends between groups were determined by evaluating F tests for the exposure-by-time interaction. An FDR threshold of <0.1 was used to determine statistical significance for all tests. All analyses were conducted using R version 4.1.3 (70).
Intensity data (IDAT) files containing the methylation data were read with R and analyzed using the procedure described by Maksimovic et al. (71). Quality control showed that the average detection p values were <0.006 and below the described cutoff. To minimize variation between samples, data normalization was conducted using the preprocessQuantile method. Filtering was then used to remove the poor-performing probes. Probes were removed if they failed in one or more samples (n = 5,741), were on sex chromosomes (n = 10,185), were in known SNPs (n = 28,298), or were known to be cross-reactive (n = 24,688), resulting in a final list of 796,947 probes. Differential methylation analysis was subsequently used to identify differences in CpG sites between HEUs and HUUs. M-values were calculated using the lmFit function from the limma package (72), and an FDR cutoff of <0.1 was used to determine significant differences. To interpret the significant CpG sites, gene ontology (GO) analysis was conducted. In addition to identifying differentially methylated CpG sites, differentially methylated regions were also analyzed using the DMRcate package (73).
Spearman correlation analyses were used to determine the relationships between the microbiome and the Treg/Tici subsets. All significant microbiome data from our previous analysis (59) were compared with the Treg data at 6 weeks, 28 weeks, and 60 weeks. The maternal microbiome was used to correlate with infant Treg data at birth. The Treg data were modeled using HIV exposure status as a confounder, and the residuals of the model were used to evaluate the relationship with the microbiome data. Spearman’s rank correlation coefficient was used to identify the strength of the associations, and an FDR of <0.1 was used to determine statistical significance. All multiple comparison adjustments were performed by visit. Chord diagrams were subsequently constructed for visualization using the Circlize package (74).
Nonparametric paired comparisons of bacteria-treated and untreated cells were performed using Prism 10.1.1 for MacOS software (GraphPad).
This study enrolled 240 mother–infant pairs from Soweto, including 123 mothers with HIV and 117 without HIV, between June and December 2017. Notable differences between mothers in the two groups were greater chronological age and parity and lower body mass index (BMI) in mothers with HIV (Table 1). There were no differences in alcohol or tobacco use or education level between the two groups. Mothers with HIV had a median of 347 CD4+ T cells/µl of blood and <50 HIV RNA copies/ml of plasma.
At birth, the HEUs and HUUs had similar gestational ages according to the study design, with an average of 39 weeks. The sex distribution was also similar (Table 1). The HEUs had significantly lower birth weights, with a mean of 3070 g, than the 3250 g in HUUs, but no infants met criteria for small for gestational age or large for gestational age. Seven HEUs and one HUU were delivered by emergency C-section for obstetrical indications identified after the initiation of labor.
Infant diet and antibiotic usage, including cotrimoxazole in HEUs, were recorded at each visit (Supplementary Table S1). There were no appreciable differences in infant diets between HEUs and HUUs. Mothers and infants who received antibiotics within 1 month prior to stool collection with the exception of cotrimoxazole in HEUs were excluded from the microbiome analyses.
The analysis of the infant gut microbiome at 6, 28, and 62 weeks, maternal gut microbiome at delivery and 62 weeks postpartum, and breastmilk microbiome at 6 weeks postpartum, described in a previous manuscript (59), showed significant differences between HEUs and HUUs and between mothers with and without HIV.
CD4+ and CD8+ Treg/Tici subsets were identified by the expression of previously described Treg/Tici markers (12, 29, 63–65) FOXP3 and/or CD25, CD39, CTLA4, GITR, granzyme B (GranzB), IL10, IL35, LAG3, PD1, TGFβ, TIM3, TIGIT, and/or TNFR2 using two 10-color flow cytometry panels referred to here as panels A and B (gating strategy and fluorescence minus one shown in Supplementary Figure S1).
The comparison of Treg/Tici subsets in cord blood between the two groups (Figure 1, Supplementary Table S2) revealed significantly greater proportions of CD4+FOXP3+, CD4+FOXP3+CD25+, and CD4+GITR+ Treg in HEUs than HUUs and greater proportions of CD4+FOXP3+GranzB+, CD4+TGFβ+, and CD8+TGFβ+ Treg in HUUs than in HEUs after adjusting the analysis for multiple comparison using the Benjamini-Hochberg false discovery rate (FDR) with p<0.1. Notably, despite higher FOXP3 expression in HEUs, we found an increased frequency of CD4+FOXP3+GranzB+% in HUUs due to much higher expression of GranzB in this group (not depicted). There were no significant differences at 6 weeks of life. At 28 and 62 weeks, the Treg/Tici subsets that significantly differed between the two groups were invariably greater in HEUs than HUUs and included CD4+GITR+, CD4+IL35+, CD4+TGFβ+, CD8+IL35+, and CD8+TGFβ+.
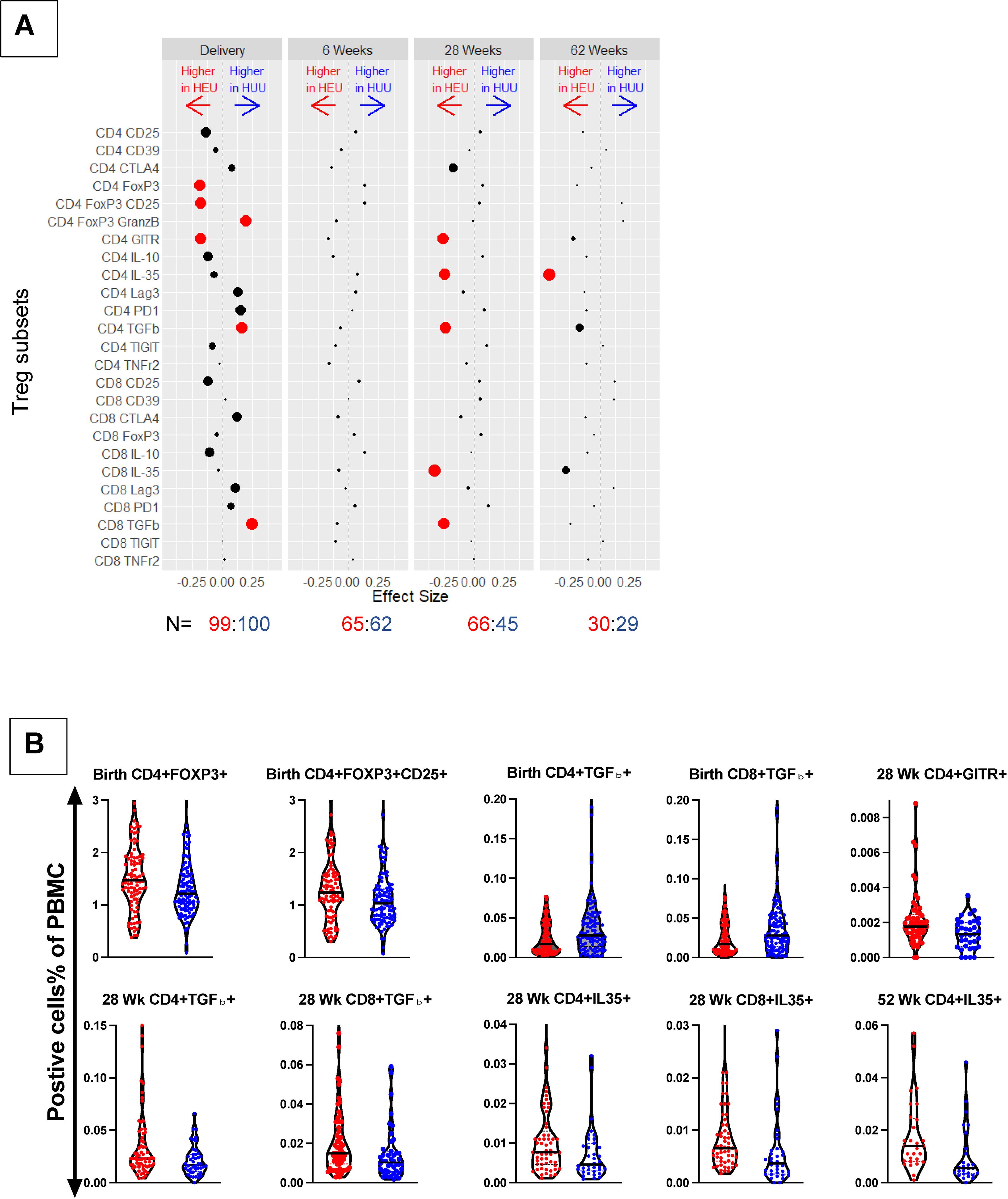
Figure 1. Comparison of Treg/Tici subset frequencies in HEUs and HUUs. Data were derived from a longitudinal cohort of 123 HEUs and 117 HUUs. (A) Treg/Tici subsets listed on the y ordinate were compared between HEUs and HUUs using Wilcoxon rank-sum test. The dots represent differences in each Treg/Tici subset at the time points indicated on the graph (top). N (bottom) indicates the number of HEUs in red font and HUUs in blue font that contributed data at each time point. Red dots indicate significant differences with FDR-adjusted p<0.1. The size of each dot is inversely proportional to the unadjusted p value. The distance between each dot and 0 is proportional to the size of the estimated difference. Please see Supplementary Figure S1 for the gating strategy and Supplementary Table S2 for a listing of medians and p values. (B) Typical examples of the magnitude of differences between HEUs and HUUs Treg/Tici subsets. Graph titles indicate time points and the Treg/Tici subsets. The violin plots show individual data points and medians. HEUs are represented by red dots and HUUs by blue dots.
We investigated the relationship between maternal CD4+ cell numbers and plasma HIV RNA copies/ml at delivery and the frequency of Treg/Tici subsets in HEUs. Spearman correlation analysis of maternal CD4+ cell numbers with all Treg/Tici subsets at all visits revealed significant correlations only at birth and for only three Treg/Tici subsets: CD4+TGFβ+, CD8+TGFβ+ and CD8+CTLA4+ (Figure 2). The frequencies of the three Treg subsets increased with decreasing maternal CD4+ cell numbers, with rho values of -0.27 to -0.35, raw p values of 0.0005 to 0.01, and FDR-adjusted p values of 0.03 to 0.08. There were no appreciable differences in the Treg/Tici subset frequencies between HEUs born to mothers with HIV plasma RNA <50 copies/ml or ≥50 copies/ml (not depicted).
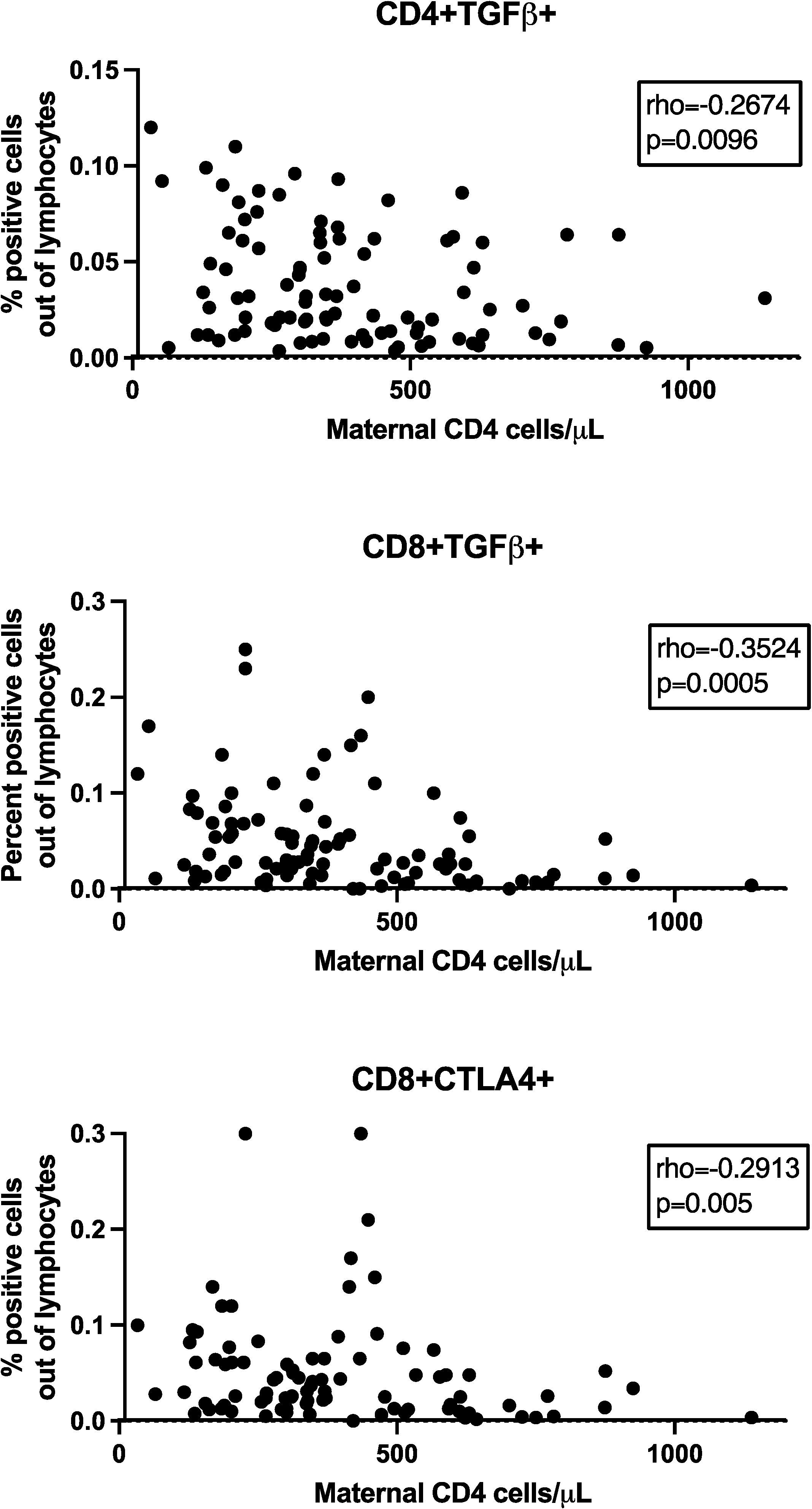
Figure 2. Effect of maternal CD4+ cell numbers on HEU TregTici subsets. Data were derived from 99 HEUs. Graphs show the correlations between maternal CD4+ cell numbers at delivery and frequencies of the Treg/Tici subsets denoted in the title of each graph. The graphs display coefficient of correlations and raw p values calculated by Spearman’s test. FDR p values were ≤0.08.
We hypothesized that differential Treg distributions between HEUs and HUUs starting at birth might reflect variability in patterns of DNA methylation acquired in utero. This hypothesis seemed particularly appropriate for explaining the excess expression of FOXP3 in HEU CD4+ T cells, which has been associated with hypomethylation of several DNA loci (61). However, the analysis of differentially methylated regions in CD4+ T cells from the cord blood of 40 HEUs and 40 HUUs revealed significant differences in a single gene, thioredoxin-interacting protein (TXNIP), which was hypomethylated in HEUs compared with HUUs (Supplementary Table S3). Pathway analyses using the GO and KEGG databases did not reveal any significant differences.
We investigated the relationships of birth weight and sex with Treg/Tici distribution. We did not find any relationship between birth weight and Treg/Tici frequencies in HEUs or HUUs (not depicted). We found a significant effect of sex only in HUUs and only at 28 weeks of life, with males showing higher proportions of CD4+FOXP3+GranzB+ and CD8+FOXP3+ Treg (Figure 3). Because all infants were born at term according to the study design, we could not evaluate the effect of gestational age.
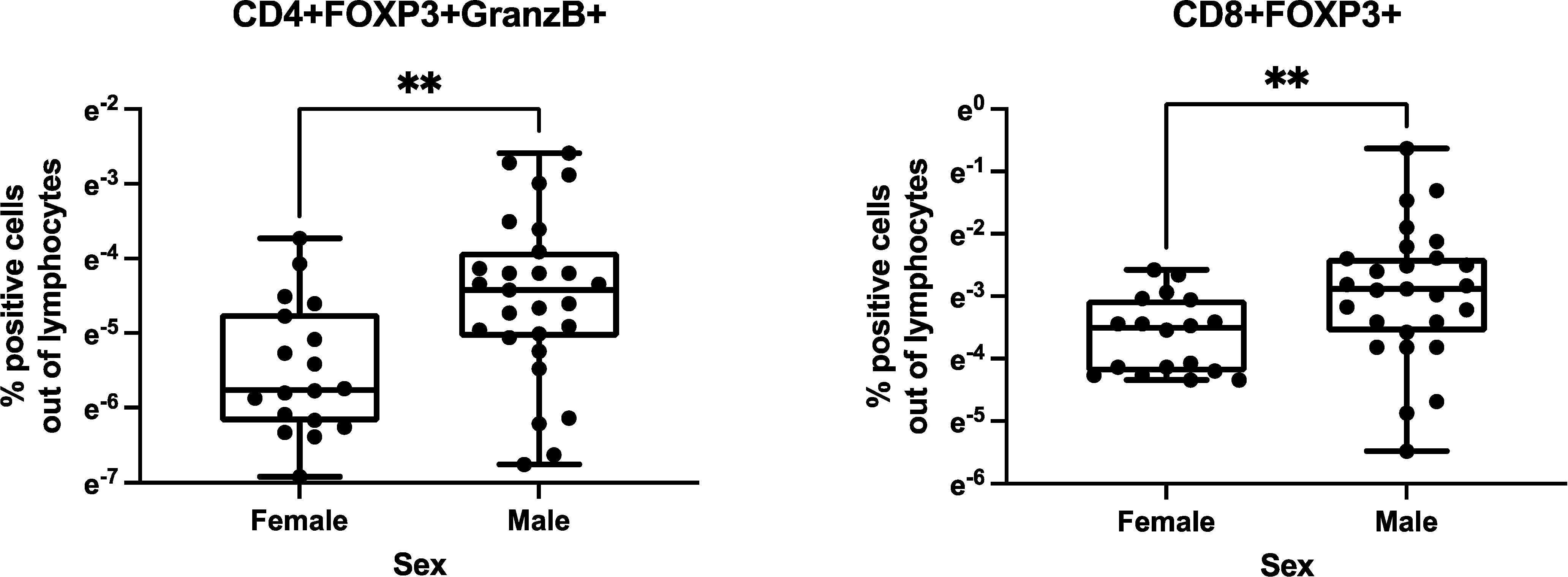
Figure 3. Effect of sex on Treg/Tici distribution in infancy. Data were analyzed in 123 HEUs and 117 HUUs. There was a significant effect of sex only in HUUs at 28 weeks of life. The graphs show the distribution of the Treg/Tici subsets identified in the titles in 18 female and 27 male infants. The asterisks indicate nominal p values<0.01 calculated by Wilcoxon rank-sum test. The FDR adjusted p values were 0.09 for the CD4+FOXp3+GranzB+% comparison and 0.04 for the CD8+FOXP3+% comparison.
We tested the hypothesis that differences in Treg/Tici subsets between HEUs and HUUs could be explained by differences in maternal or infant gut microbiota, which we previously showed to significantly differ between HEUs and HUUs (59) (Supplementary Figure S2). To address this hypothesis, we correlated the relative abundances of maternal gut bacterial genera with infant Treg/Tici subsets at birth and the infant microbiota with Treg/Tici subsets at concurrent study visits at 6, 28 and 62 weeks of life. Both analyses focused on bacterial taxa and Treg/Tici subsets that differed between mothers with and without HIV at delivery and/or between HEUs and HUUs. The results revealed multiple significant associations (Figure 4, Supplementary Table S4). The number of associations decreased over time, due in part to the convergence of the gut microbiota of HEUs and HUUs over time and the reduction in the number of Treg/Tici subsets with differential frequencies. The subsets that correlated with the relative abundance of bacterial taxa in the gut expressed CD25, CTLA4, FOXP3, FOXP3 and GranzB, IL-10, IL-35, Lag3, PD-1, TGFβ, and/or TNFR2. Multiple bacterial taxa correlated with the frequencies of Treg/Tici (Figure 4, Supplementary Table S4), some of which exhibited significant correlations at multiple time points.
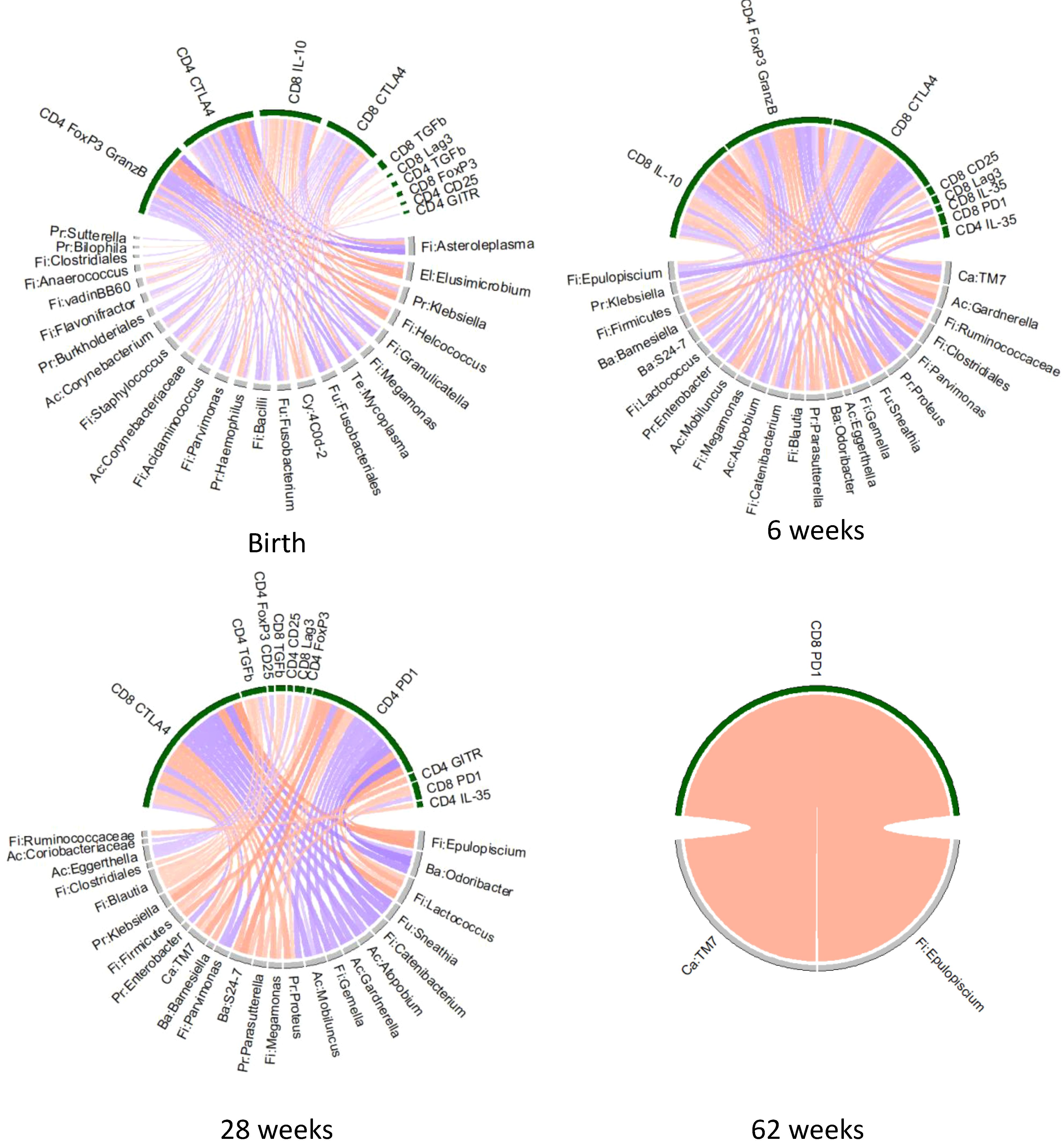
Figure 4. Chord diagram of associations between Treg/Tici subset frequencies and the abundance of gut microbiota that distinguish HEUs from HUUs. Data were derived from HEUs and HUUs with paired Treg/Tici and gut microbiome data at the time points indicated on the graph. The Treg/Tici subsets at birth were correlated with maternal microbiota at delivery. Red chords indicate positive correlations and blue chords negative correlations with FDR p<0.1. Treg/Tici subsets are clustered on the upper part of the circles (green) and bacteria on the lower part (grey). Rho and p values are listed in Supplementary Table S4.
We further postulated that if the relationships between microbial products and Treg/Tici frequencies were causal, they would be reproducible in vitro. To test this hypothesis, we identified bacterial taxa with higher abundance in HEUs than HUUs and significant positive correlations with Treg/Tici subsets and classified them as Category A bacteria. Conversely, we classified as Category B bacteria, the taxa with higher abundance in HUUs than HEUs and significant Treg/Tici positive correlations. We reasoned that if our hypothesis was correct, in vitro treatment of PBMC from HUUs with Category A bacterial products would increase the frequencies of Treg/Tici generally higher in vivo in HEUs than HUUs. Conversely, in vitro treatment of HEU PBMC with Category B bacterial products would increase the frequencies of Treg/Tici generally higher in vivo in HUUs. The bacterial taxa included in Category A were Lactococcus, Klebsiella, Blautia, and Ruminococcus. Blautia is a Firmicute that belongs to the family of Lachnospiraceae (75) and an SCFA producer (75). The relative abundance of Blautia sp. was associated with increased expression of FOXP3, GranzB, CTLA4, PD1, TGFβ and Lag3 in our study participants. Lactococcus is also a Firmicute that can produce acetate and was associated with increased numbers of Treg/Tici expressing PD1, TGFβ, and CTLA4. Ruminococcus is a Firmicute that can produce butyrate (76, 77). Its abundance was positively correlated with the expression of IL10 and IL35 in our participants. Bacteria included in Category B were Enterobacter and Proteus, both belonging to the phylum Proteobacteria, family Enterobacteriaceae, which are commonly found in the infant gut microbiome (78). In our study, these bacteria were associated with increased expression of IL10, CTLA4, LAG3, PD1, and TNFr2.
The in vitro experiments were executed in accordance with previous studies that characterized the effect of bacterial products on immune cell subsets (79–83). Using preoptimized conditions, we treated PBMC in vitro with the UV-inactivated equivalent of 10 colony-forming units/cell or medium control for 7 days. At the end of the incubation, we measured the frequencies of Treg/Tici expressing FOXP3, CD25, CTLA4, GITR, GranzB, Lag3, IL-10, IL-35, and/or TGFβ (the gating strategy is shown in Supplementary Figure S3). PBMC from seven HUUs showed significant increases in the proportions of CD4+PD1+ Treg/Tici when treated with L. lactis or K. pneumoniae compared to medium control (Figure 5). In addition, CD4+FOXP3+CD25+ Treg were significantly increased by K. pneumoniae, CD8+FOXP3+ by B. wexlerae, and CD4+TGFβ+ by R. bromii (Figure 5). Notably, these in vitro effects largely replicated the correlations observed in vivo (Figure 4, Supplementary Table S4). In contrast, UV-inactivated bacterial treatment did not significantly increase the proportions of any Treg/Tici subsets in HEU PBMC (Supplementary Figure S4). The results observed in HUUs were largely replicated in three experiments using healthy donor adult PBMC (Supplementary Figure S5).
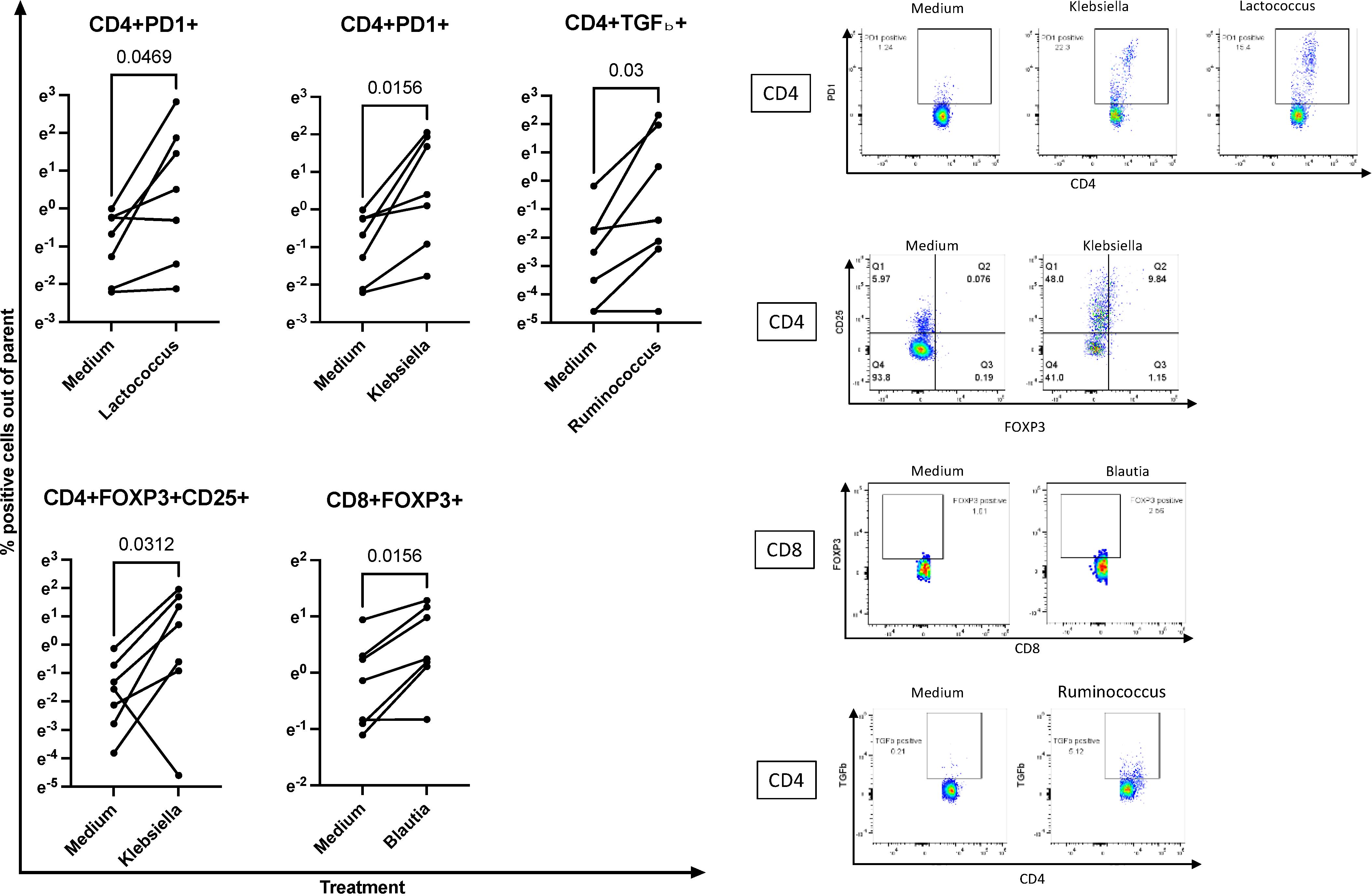
Figure 5. Ex vivo treatment of PBMC with bacterial isolates recapitulates in vivo associations with Treg/Tici subsets. Left panels: Data were generated using PBMC from 7 HUUs each treated for 7 days with the UV-inactivated bacterial cultures indicated on each graph. P values were calculated with Wilcoxon matched-pairs signed rank test. Right panels show typical flow cytometric representations of the data summarized in the left panels. Full gating strategies are shown in Supplementary Figure S3. Please see Supplementary Figure S4 for examples of bacterial isolates that did not expand Treg/Tici subsets in HEU PBMC and Supplementary Figure S5 for effect on adult PBMC.
In this study, we identified differences in the frequencies of Treg/Tici subsets between HEUs and HUUs in the first year of life and established correlations with maternal and infant characteristics. The most prominent factor associated with the frequencies of Treg/Tici was the abundance of certain bacterial taxa in the gut microbiome. We previously identified multiple differences in the HEU and HUU gut microbiota (59) and demonstrate here that these differences are associated with divergent infant Treg/Tici development between these groups.
Notably, differences in the gut microbiota of mothers with and without HIV were associated with differences in Treg/Tici frequencies in HEU and HUU cord blood. These findings are in agreement with previous studies showing that the maternal gut microbiome plays an important role in the development of the infant immune system (84–86). For example, Tanabe et al. showed an association between the maternal gut microbiome and cytokine levels in cord blood (87), and several studies both in humans and in animal models have reported profound effects of the maternal diet on the neonatal immune system mediated by the maternal gut microbiome (84, 88, 89). The communication between the maternal gut microbiome and the fetal immune system is likely to be assisted by bacterial metabolites that freely cross the placenta (87) and deserves further study.
We postulated that some associations between the gut microbiota and Treg/Tici differential frequencies between HEUs and HUUs reflect direct effects of bacterial taxa on the immune system. For four organisms, higher in HEUs than HUUs, including select species of Blautia, Lactococcus, Klebsiella and Ruminococcus, we confirmed their direct relationship with the expansion of Treg/Tici using an in vitro model. For two other microorganisms, Proteus and Enterobacter, which were greater in HUUs than in HEUs, and are commonly found in infant gut microbiomes (78), we could not demonstrate similar relationships. The mechanism underlying the relationships that we identified are likely to involve microbiota-synthesized metabolites, which cross the gut epithelial barrier and inform the immune system development through epigenetic imprinting and post-translational modification of proteins involved in signal transduction (90, 91).
In our previous study, we found that Blautia was more abundant in the gut microbiomes of mothers with HIV than in those without HIV, and its relative abundance was positively correlated within mother-infant dyads (59), suggesting that HEUs acquired the bacteria directly from their mothers or through shared local conditions in the gut. In this study, the relative abundance of Blautia sp. was associated with increased expression of FOXP3, an effect which was reproduced in vitro. Blautia’s secondary metabolites and their relationship with human health and disease have raised interest in understanding its physiological properties as well as local gut conditions that modulate its growth (75, 92). Collectively, these observations suggest that Blautia may play a role in the immunologic dysfunctions observed in HEUs.
Lactococcus produces acetate under low-glucose conditions, which may contribute to T-cell differentiation via the Treg pathway. In fact, L. lactis was associated with Treg induction in several animal models (93–95). Less is known about the relationship between Lactococcus and Treg in humans. In people with HIV, Lactococcus has not stood out in the composition of the gut microbiome but was the second most common microbe identified in the serum of ARV-treated individuals (96). In our study, Lactococcus was more abundant in HEUs than in HUUs but not in mothers with HIV compared to those without HIV (59). In vivo, Lactococcus was associated with increased abundance of Treg/Tici expressing PD1, which was corroborated in vitro.
Ruminococcus spp. are butyrate producers and, therefore, have the ability to stimulate Treg differentiation (76, 77). There is conflicting information regarding the abundance of Ruminococcus in people with HIV (97–100). In our previous study, we did not find differences in the abundance of Ruminococcus in mothers with or without HIV. Nevertheless, Ruminococcus had a greater relative abundance in HEUs than in HUUs and was correlated with the frequency of CD4+ and CD8+ Treg expressing IL10 or IL35. In vitro, R. bromii expanded CD4+TGFβ+ Treg. Its role in the immune dysregulation of HEUs deserves to be further elucidated.
Klebsiella spp were shown to potentially contribute to the enhanced inflammatory profile of people with HIV and to their neurocognitive impairment (101–103). In our study, Klebsiella was positively associated with increased of PD1+ both in vivo and in vitro. PD1 is an ici that is commonly expressed on Treg and on activated conventional T cells. When bound to its ligands, PDL1 and PDL2, the coupled receptors generate inhibitory intracellular signals that depress the immune response (104). Collectively, these observations suggest that the high abundance of Klebsiella in the HEU gut microbiome may contribute to immunologic dysfunction and may warrant studies of interventions to decrease its representation in the gut of HEUs and/or their mothers.
Another factor associated with the excess Treg in HEUs was low maternal CD4+ cell numbers. This association was present at delivery, suggesting that in utero communication between mothers and fetuses constituted the underlying mechanism. CD4+ T-cell depletion in people with HIV is largely explained by immune activation in addition to the viral cytopathic effect (105). There is active communication through the placenta between the maternal and fetal immune systems, which may explain the effect of maternal immune activation on fetal immune responses. We have previously shown that, compared with mothers without HIV, mothers with HIV have increased circulating inflammatory marker levels at delivery and that, compared with HUUs, HEUs also have increased plasma inflammatory markers at birth (106). It is conceivable that Treg/Tici expand in HEUs in utero to mitigate inflammation and immune activation induced by the mother. This notion is supported by our previous observation that plasma inflammatory marker levels are positively correlated with the frequencies of Treg subsets in pregnant women with HIV (64).
We found two Treg subsets that were significantly greater in HUU males than females at 28 weeks of age. However, we did not find similar differences at other ages in HUUs or in HEUs at any age. Thus, additional confirmatory studies are needed to validate these findings.
We did not find that differential DNA methylation of CD4+ T cells played a role in the difference in Treg variance between HEUs and HUUs. TXNIP was the only gene hypomethylated in HEUs. Although TXNIP products play a role in hematopoietic cell differentiation, proliferation, apoptosis, and NK cell function (107, 108), a direct contribution to the differentiation of Treg has not been identified to date.
Our study has both limitations and strengths. The number of infants with Treg/Tici measurements decreased from delivery to 62 weeks of life; some of the Treg/Tici may have been in more than one subset because of marker co-expression; and we were unable to investigate potential associations between Treg abundance and ART regimen because all mothers received a fixed dose combination consisting of tenofovir, emtricitabine and efavirenz. Nevertheless, our study has the largest cohort of HEUs and HUUs and the longest follow-up for the comparison of the frequency of Treg/Tici in the two groups and for the association of immunologic and gut microbiome differences between groups. A strength of this study was the in vitro verification of microbiome-immune interactions initially identified by in vivo associations.
In conclusion, our study established that the frequencies of Treg/Tici subsets differ in HEUs and HUUs from birth to 62 weeks of life, and there is an absolute excess of Treg in HEUs between 28 and 62 weeks of life. We showed in a previous study that a greater proportion of Treg was associated with decreased conventional CD4+ T-cell function in HEUs (12), suggesting that the excess Treg during infancy may underlie the increased susceptibility of HEUs to infections. The factors associated with Treg/Tici development that may be modified through interventions are the infant and maternal gut microbiomes and maternal inflammation. These interventions may result in lower Treg/Tici frequencies in HEUs and potentially lower susceptibility to serious infections compared to the current status quo.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving humans were approved by the Human Research Ethics Committee at the University of the Witwatersrand (approval number: M171185) and the Colorado Multiple Institutions Review Board (COMIRB 17-0306). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
MJ: Data curation, Formal Analysis, Project administration, Visualization, Writing – review & editing. SL: Data curation, Project administration, Writing – review & editing. AB: Data curation, Writing – review & editing. AT: Data curation, Writing – review & editing. CR: Data curation, Writing – review & editing. JK: Data curation, Writing – review & editing. SL: Writing – review & editing. BM: Project administration, Resources, Writing – review & editing. MN: Data curation, Investigation, Project administration, Supervision, Writing – review & editing. SM: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing. DF: Project administration, Supervision, Writing – review & editing. AW: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The study was supported by NIAID grant U01AI131360-01 (AW and SM MPIs) from the National Institute of Allergy and Infectious Diseases.
Author AT-S was employed by the company Bristol Myers Squibb.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1533003/full#supplementary-material
1. Aizire J, Sikorskii A, Ogwang LW, Kawalazira R, Mutebe A, Familiar-Lopez I, et al. Decreased growth among antiretroviral drug and HIV-exposed uninfected versus unexposed children in Malawi and Uganda. AIDS. (2020) 34:215–25. doi: 10.1097/QAD.0000000000002405
2. Ejigu Y, Magnus JH, Sundby J, Magnus MC. Differences in growth of HIV-exposed uninfected infants in Ethiopia according to timing of in-utero antiretroviral therapy exposure. Pediatr Infect Dis J. (2020) 39:730–6. doi: 10.1097/INF.0000000000002678
3. Gray DM, Wedderburn CJ, MacGinty RP, McMillan L, Jacobs C, Stadler JAM, et al. Impact of HIV and antiretroviral drug exposure on lung growth and function over 2 years in an African Birth Cohort. AIDS. (2020) 34:549–58. doi: 10.1097/QAD.0000000000002444
4. Evans C, Chasekwa B, Ntozini R, Majo FD, Mutasa K, Tavengwa N, et al. Mortality, human immunodeficiency virus (HIV) transmission, and growth in children exposed to HIV in rural Zimbabwe. Clin Infect Dis. (2021) 72:586–94. doi: 10.1093/cid/ciaa076
5. le Roux DM, Nicol MP, Myer L, Vanker A, Stadler JAM, von Delft E, et al. Lower respiratory tract infections in children in a well-vaccinated South African birth cohort: spectrum of disease and risk factors. Clin Infect Dis. (2019) 69:1588–96. doi: 10.1093/cid/ciz017
6. le Roux SM, Abrams EJ, Donald KA, Brittain K, Phillips TK, Nguyen KK, et al. Growth trajectories of breastfed HIV-exposed uninfected and HIV-unexposed children under conditions of universal maternal antiretroviral therapy: a prospective study. Lancet Child Adolesc Health. (2019) 3:234–44. doi: 10.1016/s2352-4642(19)30007-0
7. le Roux SM, Abrams EJ, Donald KA, Brittain K, Phillips TK, Zerbe A, et al. Infectious morbidity of breastfed, HIV-exposed uninfected infants under conditions of universal antiretroviral therapy in South Africa: a prospective cohort study. Lancet Child Adolesc Health. (2020) 4:220–31. doi: 10.1016/s2352-4642(19)30375-x
8. Pillay L, Moodley D, Emel LM, Nkwanyana NM, Naidoo K. Growth patterns and clinical outcomes in association with breastfeeding duration in HIV exposed and unexposed infants: a cohort study in KwaZulu Natal, South Africa. BMC Pediatr. (2021) 21:183. doi: 10.1186/s12887-021-02662-8
9. Anderson K, Kalk E, Madlala HP, Nyemba DC, Kassanjee R, Jacob N, et al. Increased infectious-cause hospitalization among infants who are HIV-exposed uninfected compared with HIV-unexposed. Aids. (2021) 35:2327–39. doi: 10.1097/qad.0000000000003039
10. Li SNJ, Albert A, Piske M, Janssen PA, Alimenti A, Jesson J, et al. Higher hospitalization rates in children born HIV-exposed uninfected in British Columbia, Canada, between 1990 and 2012. Pediatr Infect Dis J. (2022) 41:124–30. doi: 10.1097/INF.0000000000003365
11. Smith C, Jalbert E, de Almeida V, Canniff J, Lenz LL, Mussi-Pinhata MM, et al. Altered natural killer cell function in HIV-exposed uninfected infants. Front Immunol. (2017) 8:470. doi: 10.3389/fimmu.2017.00470
12. Jalbert E, Williamson KM, Kroehl ME, Johnson MJ, Cutland C, Madhi SA, et al. HIV-exposed uninfected infants have increased regulatory T cells that correlate with decreased T cell function. Front Immunol. (2019) 10:595. doi: 10.3389/fimmu.2019.00595
13. Ruck C, Reikie BA, Marchant A, Kollmann TR, Kakkar F. Linking susceptibility to infectious diseases to immune system abnormalities among HIV-exposed uninfected infants. Front Immunol. (2016) 7:310. doi: 10.3389/fimmu.2016.00310
14. Jalbert E, Ghosh T, Smith C, Amaral FR, Mussi-Pinhata MM, Weinberg A. Impaired functionality of antigen presenting cells in HIV- exposed uninfected infants in the first six months of life. Front Immunol. (2022) 13:960313. doi: 10.3389/fimmu.2022.960313
15. Abu-Raya B, Kollmann TR, Marchant A, MacGillivray DM. The immune system of HIV-exposed uninfected infants. Front Immunol. (2016) 7:383. doi: 10.3389/fimmu.2016.00383
16. Mazzola TN, da Silva MT, Abramczuk BM, Moreno YM, Lima SC, Zorzeto TQ, et al. Impaired Bacillus Calmette-Guerin cellular immune response in HIV-exposed, uninfected infants. AIDS. (2011) 25:2079–87. doi: 10.1097/QAD.0b013e32834bba0a
17. Simani OE, Adrian PV, Violari A, Kuwanda L, Otwombe K, Nunes MC, et al. Effect of in-utero HIV exposure and antiretroviral treatment strategies on measles susceptibility and immunogenicity of measles vaccine. AIDS. (2013) 27:1583–91. doi: 10.1097/QAD.0b013e32835fae26
18. Van Rie A, Madhi SA, Heera JR, Meddows-Taylor S, Wendelboe AM, Anthony F, et al. Gamma interferon production in response to Mycobacterium bovis BCG and Mycobacterium tuberculosis antigens in infants born to human immunodeficiency virus-infected mothers. Clin Vaccine immunology: CVI. (2006) 13:246–52. doi: 10.1128/CVI.13.2.246-252.2006
19. Weinberg A, Mussi-Pinhata M, Yu Q, Cohen R, Almeida V, Amaral F. Severe respiratory infections in HIV-exposed uninfected infants: serologic analysis. CROI 2016. (2016).
20. Abramczuk BM, Mazzola TN, Moreno YM, Zorzeto TQ, Quintilio W, Wolf PS, et al. Impaired humoral response to vaccines among HIV-exposed uninfected infants. Clin Vaccine immunology: CVI. (2011) 18:1406–9. doi: 10.1128/CVI.05065-11
21. Gnanashanmugam D, Troy SB, Musingwini G, Huang C, Halpern MS, Stranix-Chibanda L, et al. Immunologic response to oral polio vaccine in human immunodeficiency virus-infected and uninfected Zimbabwean children. Pediatr Infect Dis J. (2012) 31:176–80. doi: 10.1097/INF.0b013e31823faa5f
22. Kidzeru EB, Hesseling AC, Passmore JA, Myer L, Gamieldien H, Tchakoute CT, et al. In-utero exposure to maternal HIV infection alters T-cell immune responses to vaccination in HIV-uninfected infants. AIDS. (2014) 28:1421–30. doi: 10.1097/QAD.0000000000000292
23. Smith C, Moraka NO, Ibrahim M, Moyo S, Mayondi G, Kammerer B, et al. Human immunodeficiency virus exposure but not early cytomegalovirus infection is associated with increased hospitalization and decreased memory T-cell responses to tetanus vaccine. J Infect Dis. (2020) 221:1167–75. doi: 10.1093/infdis/jiz590
24. Garcia-Knight MA, Nduati E, Hassan AS, Gambo F, Odera D, Etyang TJ, et al. Altered memory T-cell responses to bacillus calmette-guerin and tetanus toxoid vaccination and altered cytokine responses to polyclonal stimulation in HIV-exposed uninfected Kenyan infants. PloS One. (2015) 10:e0143043. doi: 10.1371/journal.pone.0143043
25. Byakwaga H, Boum Y 2nd, Huang Y, Muzoora C, Kembabazi A, Weiser SD, et al. The kynurenine pathway of tryptophan catabolism, CD4+ T-cell recovery, and mortality among HIV-infected Ugandans initiating antiretroviral therapy. J Infect Dis. (2014) 210:383–91. doi: 10.1093/infdis/jiu115
26. Chen J, Shao J, Cai R, Shen Y, Zhang R, Liu L, et al. Anti-retroviral therapy decreases but does not normalize indoleamine 2,3-dioxygenase activity in HIV-infected patients. PloS One. (2014) 9:e100446. doi: 10.1371/journal.pone.0100446
27. Chevalier MF, Didier C, Petitjean G, Karmochkine M, Girard PM, Barre-Sinoussi F, et al. Phenotype alterations in regulatory T-cell subsets in primary HIV infection and identification of Tr1-like cells as the main interleukin 10-producing CD4+ T cells. J Infect Dis. (2015) 211:769–79. doi: 10.1093/infdis/jiu549
28. Jenabian MA, El-Far M, Vyboh K, Kema I, Costiniuk CT, Thomas R, et al. Immunosuppressive tryptophan catabolism and gut mucosal dysfunction following early HIV infection. J Infect Dis. (2015) 212:355–66. doi: 10.1093/infdis/jiv037
29. Brincks EL, Roberts AD, Cookenham T, Sell S, Kohlmeier JE, Blackman MA, et al. Antigen-specific memory regulatory CD4+Foxp3+ T cells control memory responses to influenza virus infection. J Immunol. (2013) 190:3438–46. doi: 10.4049/jimmunol.1203140
30. Cabrera R, Tu Z, Xu Y, Firpi RJ, Rosen HR, Liu C, et al. An immunomodulatory role for CD4(+)CD25(+) regulatory T lymphocytes in hepatitis C virus infection. Hepatology. (2004) 40:1062–71. doi: 10.1002/hep.20454
31. Milman N, Zhu J, Johnston C, Cheng A, Magaret A, Koelle DM, et al. In situ detection of regulatory T cells in human genital herpes simplex virus type 2 (HSV-2) reactivation and their influence on spontaneous HSV-2 reactivation. J Infect Dis. (2016) 214:23–31. doi: 10.1093/infdis/jiw091
32. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. (2016) 535:65–74. doi: 10.1038/nature18847
33. Ganusov VV, De Boer RJ. Do most lymphocytes in humans really reside in the gut? Trends Immunol. (2007) 28:514–8. doi: 10.1016/j.it.2007.08.009
34. Ma H, Tao W, Zhu S. T lymphocytes in the intestinal mucosa: defense and tolerance. Cell Mol Immunol. (2019) 16:216–24. doi: 10.1038/s41423-019-0208-2
35. Ivanov II, Frutos R, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. (2008) 4:337–49. doi: 10.1016/j.chom.2008.09.009
36. Liu CH, Lee SM, VanLare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci. (2008) 105:3951. doi: 10.1073/pnas.0709266105
37. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. (2005) 122:107–18. doi: 10.1016/j.cell.2005.05.007
38. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. (2008) 453:620–5. doi: 10.1038/nature07008
39. Al Nabhani Z, Eberl G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. (2020) 13:183–9. doi: 10.1038/s41385-020-0257-y
40. Skelly AN, Sato Y, Kearney S, Honda K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol. (2019) 19:305–23. doi: 10.1038/s41577-019-0144-5
41. Correa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. (2016) 5:e73. doi: 10.1038/cti.2016.17
42. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
43. Brumbaugh DE, Arruda J, Robbins K, Ir D, Santorico SA, Robertson CE, et al. Mode of delivery determines neonatal pharyngeal bacterial composition and early intestinal colonization. J Pediatr Gastroenterol Nutr. (2016) 63:320–8. doi: 10.1097/MPG.0000000000001124
44. Krebs NF, Sherlock LG, Westcott J, Culbertson D, Hambidge KM, Feazel LM, et al. Effects of different complementary feeding regimens on iron status and enteric microbiota in breastfed infants. J Pediatr. (2013) 163:416–23. doi: 10.1016/j.jpeds.2013.01.024
45. Lemas DJ, Young BE, Baker PR 2nd, Tomczik AC, Soderborg TK, Hernandez TL, et al. Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. Am J Clin Nutr. (2016) 103:1291–300. doi: 10.3945/ajcn.115.126375
46. Soderborg TK, Carpenter CM, Janssen RC, Weir TL, Robertson CE, Ir D, et al. Gestational diabetes is uniquely associated with altered early seeding of the infant gut microbiota. Front Endocrinol (Lausanne). (2020) 11:603021. doi: 10.3389/fendo.2020.603021
47. Soderborg TK, Clark SE, Mulligan CE, Janssen RC, Babcock L, Ir D, et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun. (2018) 9:4462. doi: 10.1038/s41467-018-06929-0
48. Tang M, Frank DN, Tshefu A, Lokangaka A, Goudar SS, Dhaded SM, et al. Different gut microbial profiles in sub-saharan African and south asian women of childbearing age are primarily associated with dietary intakes. Front Microbiol. (2019) 10:1848. doi: 10.3389/fmicb.2019.01848
49. Tang M, Matz KL, Berman LM, Davis KN, Melanson EL, Frank DN, et al. Effects of complementary feeding with different protein-rich foods on infant growth and gut health: study protocol. Front Pediatr. (2021) 9:793215. doi: 10.3389/fped.2021.793215
50. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. (2016) 8:343ra82. doi: 10.1126/scitranslmed.aad7121
51. Laforest-Lapointe I, Arrieta MC. Patterns of early-life gut microbial colonization during human immune development: an ecological perspective. Front Immunol. (2017) 8:788. doi: 10.3389/fimmu.2017.00788
52. Wang Z, Usyk M, Sollecito CC, Qiu Y, Williams-Nguyen J, Hua S, et al. Altered gut microbiota and host metabolite profiles in women with human immunodeficiency virus. Clin Infect Dis. (2020) 71:2345–53. doi: 10.1093/cid/ciz1117
53. Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. (2014) 7:983–94. doi: 10.1038/mi.2013.116
54. Amenyogbe N, Dimitriu P, Cho P, Ruck C, Fortuno ES 3rd, Cai B, et al. Innate immune responses and gut microbiomes distinguish HIV-exposed from HIV-unexposed children in a population-specific manner. J Immunol. (2020) 205:2618–28. doi: 10.4049/jimmunol.2000040
55. Claassen-Weitz S, Gardner-Lubbe S, Nicol P, Botha G, Mounaud S, Shankar J, et al. HIV-exposure, early life feeding practices and delivery mode impacts on faecal bacterial profiles in a South African birth cohort. Sci Rep. (2018) 8:5078. doi: 10.1038/s41598-018-22244-6
56. Dzanibe S, Jaspan HB, Zulu MZ, Kiravu A, Gray CM. Impact of maternal HIV exposure, feeding status, and microbiome on infant cellular immunity. J Leukoc Biol. (2019) 105:281–9. doi: 10.1002/jlb.Mr0318-120r
57. Machiavelli A, Duarte RTD, Pires MMS, Zárate-Bladés CR, Pinto AR. The impact of in utero HIV exposure on gut microbiota, inflammation, and microbial translocation. Gut Microbes. (2019) 10:599–614. doi: 10.1080/19490976.2018.1560768
58. Tincati C, Ficara M, Ferrari F, Augello M, Dotta L, Tagliabue C, et al. Gut-dependent inflammation and alterations of the intestinal microbiota in individuals with perinatal HIV exposure and different HIV serostatus. Aids. (2022) 36:1917–25. doi: 10.1097/qad.0000000000003324
59. Jackson CL, Frank DN, Robertson CE, Ir D, Kofonow JM, Montlha MP, et al. Evolution of the gut microbiome in HIV-exposed uninfected and unexposed infants during the first year of life. mBio. (2022) 13:e0122922. doi: 10.1128/mbio.01229-22
60. Bender JM, Li F, Martelly S, Byrt E, Rouzier V, Leo M, et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Trans Med. (2016) 8:349ra100–349ra100. doi: 10.1126/scitranslmed.aaf5103
61. Arvey A, van der Veeken J, Plitas G, Rich SS, Concannon P, Rudensky AY. Genetic and epigenetic variation in the lineage specification of regulatory T cells. eLife. (2015) 4:e07571. doi: 10.7554/eLife.07571
62. Barbi J, Pardoll D, Pan F. Treg functional stability and its responsiveness to the microenvironment. Immunol Rev. (2014) 259:115–39. doi: 10.1111/imr.12172
63. Li C, Jiang P, Wei S, Xu X, Wang J. Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol Cancer. (2020) 19:116. doi: 10.1186/s12943-020-01234-1
64. Richardson K, Weinberg A. Dynamics of regulatory T-cells during pregnancy: effect of HIV infection and correlations with other immune parameters. PloS One. (2011) 6:e28172. doi: 10.1371/journal.pone.0028172
65. Alvisi G, Brummelman J, Puccio S, Mazza EM, Tomada EP, Losurdo A, et al. IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J Clin Invest. (2020) 130:3137–50. doi: 10.1172/jci130426
66. Weinberg A, Song LY, Wilkening C, Sevin A, Blais B, Louzao R, et al. Optimization and limitations of use of cryopreserved peripheral blood mononuclear cells for functional and phenotypic T-cell characterization. Clin Vaccine immunology: CVI. (2009) 16:1176–86. doi: 10.1128/CVI.00342-08
67. Weinberg A, Song LY, Wilkening CL, Fenton T, Hural J, Louzao R, et al. Optimization of storage and shipment of cryopreserved peripheral blood mononuclear cells from HIV-infected and uninfected individuals for ELISPOT assays. J Immunol Methods. (2010) 363:42–50. doi: 10.1016/j.jim.2010.09.032
68. McFarland J. the nephelometer: an instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J Am Med Association XLIX. (1907) 14:1176–8. doi: 10.1001/jama.1907.25320140022001f
71. Maksimovic J, Phipson B, Oshlack A. A cross-package Bioconductor workflow for analysing methylation array data [version 3; peer review: 4 approved. F1000Research. (2016) 5:1281. doi: 10.12688/f1000research.8839.3
72. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
73. Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, Lord RV, et al. De novo identification of differentially methylated regions in the human genome. Epigenet Chromatin. (2015) 8. doi: 10.1186/1756-8935-8-6
74. Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize implements and enhances circular visualization in R. Bioinformatics. (2014) 30:2811–2. doi: 10.1093/bioinformatics/btu393
75. Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, et al. Blautia-a new functional genus with potential probiotic properties? Gut Microbes. (2021) 13:1–21. doi: 10.1080/19490976.2021.1875796
76. Sasaki M, Schwab C, Ramirez Garcia A, Li Q, Ferstl R, Bersuch E, et al. The abundance of Ruminococcus bromii is associated with faecal butyrate levels and atopic dermatitis in infancy. Allergy. (2022) 77:3629–40. doi: 10.1111/all.15440
77. Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. Isme J. (2012) 6:1535–43. doi: 10.1038/ismej.2012.4
78. Yang I, Corwin EJ, Brennan PA, Jordan S, Murphy JR, Dunlop A. The infant microbiome: implications for infant health and neurocognitive development. Nurs Res. (2016) 65:76–88. doi: 10.1097/nnr.0000000000000133
79. Fink LN, Zeuthen LH, Christensen HR, Morandi B, Frøkiaer H, Ferlazzo G. Distinct gut-derived lactic acid bacteria elicit divergent dendritic cell-mediated NK cell responses. Int Immunol. (2007) 19:1319–27. doi: 10.1093/intimm/dxm103
80. Haileselassie Y, Navis M, Vu N, Qazi KR, Rethi B, Sverremark-Ekström E. Lactobacillus reuteri and Staphylococcus aureus differentially influence the generation of monocyte-derived dendritic cells and subsequent autologous T cell responses. Immun Inflammation Dis. (2016) 4:315–26. doi: 10.1002/iid3.115
81. Manuzak J, Dillon S, Wilson C. Differential interleukin-10 (IL-10) and IL-23 production by human blood monocytes and dendritic cells in response to commensal enteric bacteria. Clin Vaccine Immunol. (2012) 19:1207–17. doi: 10.1128/CVI.00282-12
82. Mohamadzadeh M, Olson S, Kalina WV, Ruthel G, Demmin GL, Warfield KL, et al. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc Natl Acad Sci U.S.A. (2005) 102:2880–5. doi: 10.1073/pnas.0500098102
83. Smits HH, van Beelen AJ, Hessle C, Westland R, de Jong E, Soeteman E, et al. Commensal Gram-negative bacteria prime human dendritic cells for enhanced IL-23 and IL-27 expression and enhanced Th1 development. Eur J Immunol. (2004) 34:1371–80. doi: 10.1002/eji.200324815
84. Mirpuri J. Evidence for maternal diet-mediated effects on the offspring microbiome and immunity: implications for public health initiatives. Pediatr Res. (2021) 89:301–6. doi: 10.1038/s41390-020-01121-x
85. Koren O, Konnikova L, Brodin P, Mysorekar IU, Collado MC. The maternal gut microbiome in pregnancy: implications for the developing immune system. Nat Rev Gastroenterol Hepatol. (2023) 21:35–45. doi: 10.1038/s41575-023-00864-2
86. Barrientos G, Ronchi F, Conrad ML. Nutrition during pregnancy: Influence on the gut microbiome and fetal development. Am J Reprod Immunol. (2024) 91:e13802. doi: 10.1111/aji.13802
87. Tanabe H, Sakurai K, Nakanishi Y, Kato T, Kawasaki Y, Nakano T, et al. Association of the maternal gut microbiota/metabolome with cord blood CCL17. Nutrients. (2021) 13. doi: 10.3390/nu13082837
88. Xie Q, Cui D, Zhu Q, Qin X, Ren D, Xu X. Supplementing maternal diet with milk oligosaccharides and probiotics helps develop the immune system and intestinal flora of offsprings. Food Sci Nutr. (2023) 11:6868–77. doi: 10.1002/fsn3.3579
89. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. (2016) 351:1296–302. doi: 10.1126/science.aad2571
90. Diskin C, Ryan TAJ, O’Neill LAJ. Modification of proteins by metabolites in immunity. Immunity. (2021) 54:19–31. doi: 10.1016/j.immuni.2020.09.014
91. O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. (2016) 16:553–65. doi: 10.1038/nri.2016.70
92. Noye Tuplin EW, Fernandes T, Lowry DE, Cho NA, Sales KM, Patterson RA, et al. Select human milk oligosaccharide supplementation in post-weanling rats affects metabolism and gut microbiota into adulthood. Obesity. (2023) 31:1362–75. doi: 10.1002/oby.23731
93. Akgul A, Maddaloni M, Jun SM, Nelson AS, Odreman VA, Hoffman C, et al. Stimulation of regulatory T cells with Lactococcus lactis expressing enterotoxigenic E. coli colonization factor antigen 1 retains salivary flow in a genetic model of Sjögren’s syndrome. Arthritis Res Ther. (2021) 23:99. doi: 10.1186/s13075-021-02475-1
94. Ejsing-Duun M, Josephsen J, Aasted B, Buschard K, Hansen AK. Dietary gluten reduces the number of intestinal regulatory T cells in mice. Scandinavian J Immunol. (2008) 67:553–9. doi: 10.1111/j.1365-3083.2008.02104.x
95. Rezende RM, Oliveira RP, Medeiros SR, Gomes-Santos AC, Alves AC, Loli FG, et al. Hsp65-producing Lactococcus lactis prevents experimental autoimmune encephalomyelitis in mice by inducing CD4+LAP+ regulatory T cells. J Autoimmun. (2013) 40:45–57. doi: 10.1016/j.jaut.2012.07.012
96. Ali Z, Shahzadi I, Majeed A, Malik HMT, Waseem S, Ahmed I, et al. Comparative analysis of the serum microbiome of HIV infected individuals. Genomics. (2021) 113:4015–21. doi: 10.1016/j.ygeno.2021.10.005
97. Dubourg G, Lagier JC, Hüe S, Surenaud M, Bachar D, Robert C, et al. Gut microbiota associated with HIV infection is significantly enriched in bacteria tolerant to oxygen. BMJ Open Gastroenterol. (2016) 3:e000080. doi: 10.1136/bmjgast-2016-000080
98. Rocafort M, Noguera-Julian M, Rivera J, Pastor L, Guillén Y, Langhorst J, et al. Evolution of the gut microbiome following acute HIV-1 infection. Microbiome. (2019) 7:73. doi: 10.1186/s40168-019-0687-5
99. Xie Y, Sun J, Wei L, Jiang H, Hu C, Yang J, et al. Altered gut microbiota correlate with different immune responses to HAART in HIV-infected individuals. BMC Microbiol. (2021) 21:11. doi: 10.1186/s12866-020-02074-1
100. Zhou Y, Ou Z, Tang X, Zhou Y, Xu H, Wang X, et al. Alterations in the gut microbiota of patients with acquired immune deficiency syndrome. J Cell Mol Med. (2018) 22:2263–71. doi: 10.1111/jcmm.13508
101. Vujkovic-Cvijin I, Somsouk M. HIV and the gut microbiota: composition, consequences, and avenues for amelioration. Curr HIV/AIDS Rep. (2019) 16:204–13. doi: 10.1007/s11904-019-00441-w
102. Vujkovic-Cvijin I, Sortino O, Verheij E, Sklar J, Wit FW, Kootstra NA, et al. HIV-associated gut dysbiosis is independent of sexual practice and correlates with noncommunicable diseases. Nat Commun. (2020) 11:2448. doi: 10.1038/s41467-020-16222-8
103. Dong R, Lin H, Chen X, Shi R, Yuan S, Li J, et al. Gut microbiota and fecal metabolites associated with neurocognitive impairment in HIV-infected population. Front Cell Infection Microbiol. (2021) 11:723840. doi: 10.3389/fcimb.2021.723840
104. Lin X, Kang K, Chen P, Zeng Z, Li G, Xiong W, et al. Regulatory mechanisms of PD-1/PD-L1 in cancers. Mol Cancer. (2024) 23:108. doi: 10.1186/s12943-024-02023-w
105. Okoye AA, Picker LJ. CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev. (2013) 254:54–64. doi: 10.1111/imr.12066
106. Dirajlal-Fargo S, Mussi-Pinhata MM, Weinberg A, Yu Q, Cohen R, Harris DR, et al. HIV-exposed-uninfected infants have increased inflammation and monocyte activation. AIDS. (2019) 33:845–53. doi: 10.1097/QAD.0000000000002128
107. Kim DO, Byun J-E, Kim WS, Kim MJ, Choi JH, Kim H, et al. TXNIP regulates natural killer cell-mediated innate immunity by inhibiting IFN-γ Production during bacterial infection. Int J Mol Sci. (2020) 21:9499. doi: 10.3390/ijms21249499
Keywords: regulatory T cells, gut microbiome, human immunodeficiency virus, HIV-exposed uninfected infants, Blautia wexleraea, Klebsiella pneumoniae, Enterobacter cloacae, Ruminococcus bromii
Citation: Johnson MJ, Lazarus SK, Bennett AE, Tovar-Salazar A, Robertson CE, Kofonow JM, Li S, McCollister B, Nunes MC, Madhi SA, Frank DN and Weinberg A (2025) Gut microbiota and other factors associated with increased T cell regulation in HIV-exposed uninfected infants. Front. Immunol. 16:1533003. doi: 10.3389/fimmu.2025.1533003
Received: 22 November 2024; Accepted: 10 February 2025;
Published: 03 March 2025.
Edited by:
Marcelo A. Soares, National Cancer Institute (INCA), BrazilReviewed by:
Ria Goswami, Cornell University, United StatesCopyright © 2025 Johnson, Lazarus, Bennett, Tovar-Salazar, Robertson, Kofonow, Li, McCollister, Nunes, Madhi, Frank and Weinberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adriana Weinberg, YWRyaWFuYS53ZWluYmVyZ0B1Y2RlbnZlci5lZHU=
†Present address: Sarah K. Lazarus, University of Wisconsin School of Medicine, Maddison, WI, United States
Adriana Tovar-Salazar, Bristol Myers Squibb, Seattle, WA, United States
Marta C. Nunes, Center of Excellence in Respiratory Pathogens (CERP), Hospices Civils de Lyon and Centre International de Recherche en Infectiologie (CIRI), Équipe Santé Publique, Épidémiologie et Écologie Évolutive des Maladies Infectieuses (PHE3ID), Inserm, CNRS UMR5308, ENS de Lyon, Université Claude Bernard - Lyon 1, Lyon, France
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.