
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 13 February 2025
Sec. Antigen Presenting Cell Biology
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1532620
T cells contribute to immunotherapy and autoimmune pathogenesis and Langerhans cells (LCs) have a substantial ability to activate T cells. In vitro-generated monocyte-derived LCs (Mo-LCs) are useful models to study LC function in autoimmune diseases and to test future LC-based immunotherapies. Although dendritic cells (DCs) expressing high levels of Delta-like 4 (DLL4+ DCs), which is a member of the Notch ligand family, have greater ability than DLL4− DCs to activate T cells, the induction method of human DLL4+ DCs has yet to be determined. The aim of this study is to establish whether Mo-LCs express DLL4 and establish the induction method of antigen presenting cells, which most potently activate T cells, similar to our previously established induction method of human Mo-LCs. We compared the ratios of DLL4 expression and T cell activation via flow cytometry among monocyte-derived cells, which have a greater ability than the resident cells to activate T cells. Here, we discovered that Mo-LCs expressed DLL4, which most potently activated T cells among monocyte-derived cells, and that Mo-LCs and DLL4 expression were induced by DLL4, granulocyte macrophage colony-stimulating factor, and transforming growth factor-β1. Additionally, peptidoglycan was required for DLL4 expression, whereas interleukin-4 repressed it. These findings provide insights into the roles of DLL4-expressing cells such as DLL4+ Mo-LCs in human diseases, which will assist with the development of more effective therapeutic strategies in the future.
Psoriasis, an autoinflammatory immune disease, is driven by infections stimulating dendritic cells (DCs) to release IL-23; this activates T cells, resulting in the production of IL-17, which amplifies psoriatic inflammation (1). Langerhans cells (LCs), a type of DC expressing langerin, have a substantial ability to activate T cells (2, 3). Human LCs have been reported to be involved in psoriatic lesions (4). Monocyte-derived LCs (Mo-LCs), induced during inflammation (5), rather than resident LCs, are required for psoriasis-like changes in mouse skin (6, 7). However, since profound anatomical and immunological differences are obvious when comparing mice and humans, it is challenging to establish a method for generating human Mo-LCs in vitro to probe the role of Mo-LCs in psoriasis pathogenesis (4, 8, 9).
Notch signaling, which is initiated through receptor (in mammalians, Notch 1, 2, 3, and 4)-ligand (Delta-like 1 [DLL1], DLL3, DLL4, Jagged1 [JAG1] and JAG2) interactions, plays a crucial role in cell fate decisions and differentiation processes (10, 11). Previous studies imply a required mechanical function that is met by the immobilization of the Notch ligand either in a membrane or on a plastic surface (12, 13). Hoshino et al. reported that DLL1, in concert with granulocyte macrophage colony-stimulating factor (GM-CSF) and transforming growth factor (TGF)-β1, induces the differentiation of human CD14+ blood monocytes into Mo-LCs (14), which is considered as the most reproducible and simple culture condition leading Mo-LCs to display bona fide LC features (13–16). However, the specialized materials required to immobilize DLL-1 are difficult to obtain, which may explain the small number of subsequent reports on DLL-1-induced Mo-LCs (17–20). Using DLL1 and DLL4 expressed on mouse OP9 cells, Milne et al. reported that DLL4 generates more Mo-LCs than DLL1 (21); however, this method mixes proteins derived from OP9 cells in terms of biochemical analysis. We previously established a simple, reproducible, and efficient immobilizing method using only commercial Notch ligands and culture plates (13). We sought to elucidate the optimal Notch ligand for generating Mo-LCs using the immobilizing method, as previous works have not compared the Mo-LC induction ratios among all Notch ligands (14, 21, 22).
Aside from their contribution to psoriasis, T cells also contribute to immunotherapy and autoimmune pathogenesis (23, 24). Monocytes can differentiate into DCs, LCs, and macrophages (Mφs) (5, 25), which have a greater ability than the resident cells to activate T cells (6, 26, 27). However, no previous study has compared the T cell activation ratios among these monocyte-derived cells, which is addressed in the current study.
Activation of toll-like receptor (TLR) signaling induces high levels of DLL4 in CD1c+ DCs and plasmacytoid DCs, and DCs expressing high levels of DLL4 (DLL4+ DCs) have greater ability than DLL4− DCs to activate T cells (24). Additionally, emerging data suggest a relationship between DLL4 and psoriasis (28, 29). Thus, we considered it likely that DLL4 is expressed on monocyte-derived cells, which potently activate T cells and induce inflammatory conditions, including psoriasis.
We show here that DLL4 most potently induces Mo-LCs among Notch ligands, and that Mo-LCs express DLL4 by stimulation with the TLR2 ligand peptidoglycan (PGN), which is suppressed by interleukin (IL)-4. Additionally, DLL4+ Mo-LCs most potently activate T cells among monocyte-derived cells.
The reagents used in this study included recombinant human (rh) DLL1 (BioLegend, San Diego, CA), rhDLL4, rhGM-CSF, rhTGF-β1, rhIL-4 (Peprotech, Cranbury, NJ), rhDLL3, rhJAG1 (AdipoGen Life Sciences, San Diego, CA), rhJAG2 (Cloud Clone Corp, Houston, TX), PGN, Polyinosinic-polycytidylic acid (Poly(I:C)), lipopolysaccharide (LPS) from Escherichia coli (L4516), and imiquimod (IMQ) (Sigma-Aldrich, St. Louis, MO).
Briefly, 3 μg/ml Notch ligand dissolved in phosphate-buffered saline (PBS) was added to uncoated 24-well plates with hydrophobic surfaces that are normally used for suspension cell cultures (MS-8024R; SUMILON, Tokyo, Japan). The plates were centrifuged at 750 rpm for 7 min at room temperature, before removing the supernatant and adding 6 μg/ml Notch ligand dissolved in PBS. The plates were then centrifuged at 750 rpm for 7 min at room temperature and washed with PBS.
Samples from 7 healthy volunteers were obtained after they signed an informed consent approved by Showa University Research Ethics Review Board (Tokyo, Japan) (Approval number: 22-250-B).
Peripheral blood mononuclear cells were isolated from heparinized blood via Ficoll-Paque PLUS (Cytiva, Uppsala, Sweden) density gradient (1.077) centrifugation in Leucosep (Greiner Bio-One, Kremsmünster, Austria). After depleting platelets, CD14+ monocytes were isolated followed by CD4+ T cells using CD14 and CD4 antibody-conjugated magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany), respectively, according to the manufacturer’s instructions. The isolated CD14+ monocytes (1 × 106/ml) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (FujiFilm Wako Pure Chemical Corporation, Osaka, Japan) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Biosera, Nuaille, France) and penicillin (100 U/ml)/streptomycin (100 μg/ml) (FujiFilm Wako Pure Chemical Corporation, Osaka, Japan). The Mo-LCs were generated from CD14+ monocytes in culture with GM-CSF and TGF-β1 on a Notch ligand-immobilized plate, the Mo-DCs were generated from CD14+ monocytes in culture with GM-CSF and IL-4, and the Mφs were generated from CD14+ monocytes in culture with GM-CSF. The concentrations of GM-CSF, IL-4, and TGF-β1 were 10 ng/ml.
On day 7, the cells were adjusted to a concentration of 3 × 106 cells/ml and incubated at 4°C for 40 min with the appropriate antibodies. After washing with ice-cold PBS containing 0.2% bovine serum albumin (FujiFilm Wako Pure Chemical Corporation, Osaka, Japan) and 2 mM ethylenediaminetetraacetic acid (FujiFilm Wako Pure Chemical Corporation, Osaka, Japan), the cells were analyzed on a FACSAria II using the FACSDiva software (Becton-Dickinson and Company, Franklin Lakes, NJ). The phycoerythrin-labeled monoclonal antibody (mAb) to DLL4, and langerin (CD207); allophycocyanin-labeled mAb to langerin (CD207); and Pacific blue-labeled mAb to human leukocyte antigen-DR (BioLegend, San Diego, CA) were used for flow cytometry.
On day 6, the cells were cultured in the presence or absence of 5 μg/ml of the TLR2 ligand PGN, 25 μg/ml of the TLR3 ligand Poly(I:C), 1 μg/ml of the TLR4 ligand LPS, or 1 μg/ml of the TLR7/8 ligand IMQ for 24 h.
CD4+ T cells (1 × 106/mL) were incubated for 20 min at 37°C with 1 μl CellTrace Violet (Thermo Fisher Scientific, Waltham, MA). Next, 4 ml RPMI 1640 supplemented with 10% FBS and penicillin (100 U/ml)/streptomycin (100 μg/ml) was added to the cells and incubated for 5 min. Subsequently, the cells were centrifuged and resuspended in pre-warmed Iscove’s Modified Dulbecco’s Medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% CTS immune cell serum replacement (Thermo Fisher Scientific, Waltham, MA) and penicillin (100 U/ml)/streptomycin (100 μg/ml), before coculturing with monocyte-derived cells at ratios of 2.5:1, 5:1, and 1:1.5 for 5 days. CellTrace Violet-labeled cells were analyzed for fluorescence intensity using a FACSAria II flow cytometer.
Statistical significance between different experimental groups was analyzed using Tukey’s test to determine the best inducer of Mo-LCs or DLL4 expression or the best activator of T cells. Statistical analyses were performed using the JMP Pro 15 software for Windows (SAS Institute Inc., Cary, NC). A probability (p) value <0.05 was considered statistically significant.
The aim of this study is to establish the induction method of antigen presenting cells, which most potently activate T cells. To use Mo-LCs for this study, we first sought to establish which Notch ligands can give rise to Mo-LCs in vitro. To this end, CD14+ monocytes were cultured with GM-CSF and TGF-β1 on a Notch ligand-immobilized plate for 7 days, before analyzing the induction of Mo-LCs by FACS. We found that both DLL1 and DLL4 induced large amounts of Mo-LCs, but DLL4 most efficiently and reproducibly induced Mo-LCs (Figure 1A).
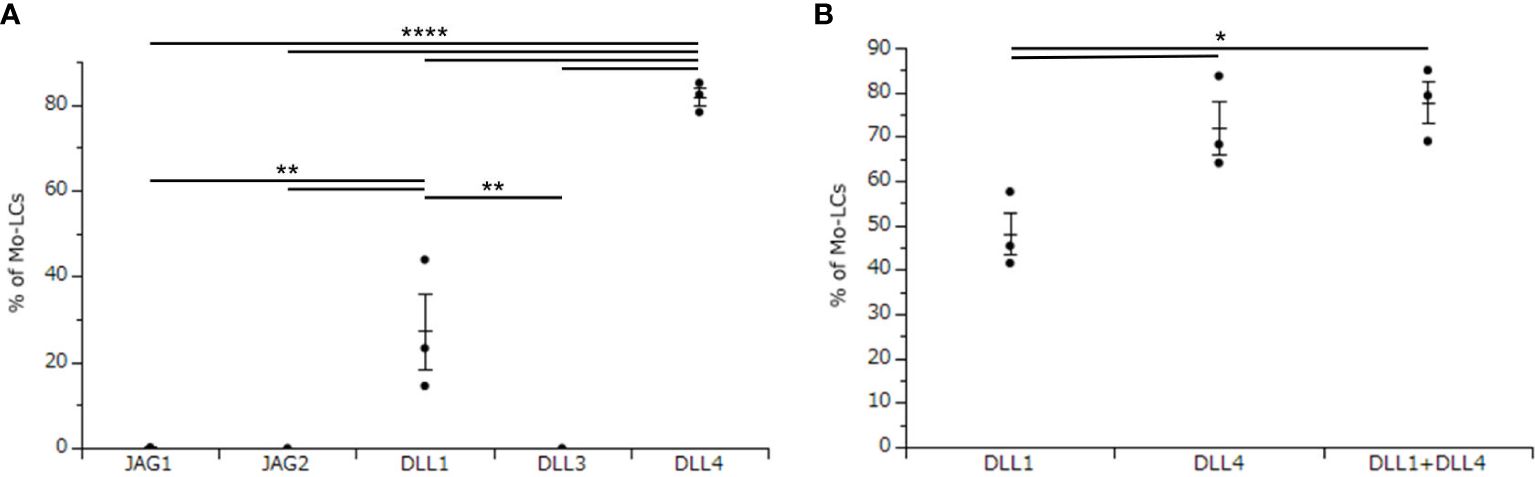
Figure 1. Delta-like 4 (DLL4) most potently induces monocyte-derived Langerhans cells (Mo-LCs). (A) Percentage of Mo-LCs (langerin+ cells) induced on all Notch ligand-immobilized plates. (B) Percentage of Mo-LCs induced on DLL1-, DLL4-, and DLL1+DLL4-immobilized plates. (A, B) The Mo-LCs were generated from CD14+ monocytes in culture with granulocyte macrophage colony-stimulating factor (GM-CSF) and transforming growth factor (TGF)-β1 on the Notch ligand-immobilized plates. After 7 days of culture, the cells were analyzed by Fluorescence-activated cell sorting (FACS). In this assay, we used blood samples from (A) donor No.1-3, (B) donor No.4-6. *p < 0.05, **p < 0.01, ****p < 0.0001.
DLL1 is the Notch ligand detected in the epidermis (30), and a previous study demonstrated increased expression of DLL4 in psoriasis lesional skin compared to unaffected skin (28, 29). Thus, both DLL1 and DLL4 are likely to be detected in psoriasis lesional skin. Hence, we next sought to determine whether DLL1 and DLL4 synergize to induce Mo-LCs. Our results showed that although DLL4 significantly induced Mo-LCs, it showed no synergistic effect when used in combination with DLL1 (Figure 1B).
Activation of TLR signaling induces high levels of DLL4 in CD1c+ DCs and plasmacytoid DCs, and DLL4+ DCs have greater ability than DLL4− DCs to activate T cells (24). We considered it likely that DLL4 is expressed also on monocyte-derived cells, which potently activate T cells and induce inflammatory conditions, including psoriasis (6, 24, 26–29). We have previously reported that Mo-LCs produce significant amounts of cytokines related to the pathogenesis of psoriasis compared to Mo-DCs and Mφs in response to Poly(I:C) or LPS (13). Thus, we next sought to establish whether monocyte-derived cells expressed DLL4 in response to Poly(I:C) and LPS. The Mo-LCs were generated from CD14+ monocytes in culture with GM-CSF and TGF-β1 on DLL4-immobilized plate, the Mo-DCs were generated from CD14+ monocytes in culture with GM-CSF and IL-4, and the Mφs were generated from CD14+ monocytes in culture with GM-CSF. Our results showed that Mo-LCs expressed high amounts of DLL4, whereas Mo-DCs and Mφs did not (Figures 2A, B).
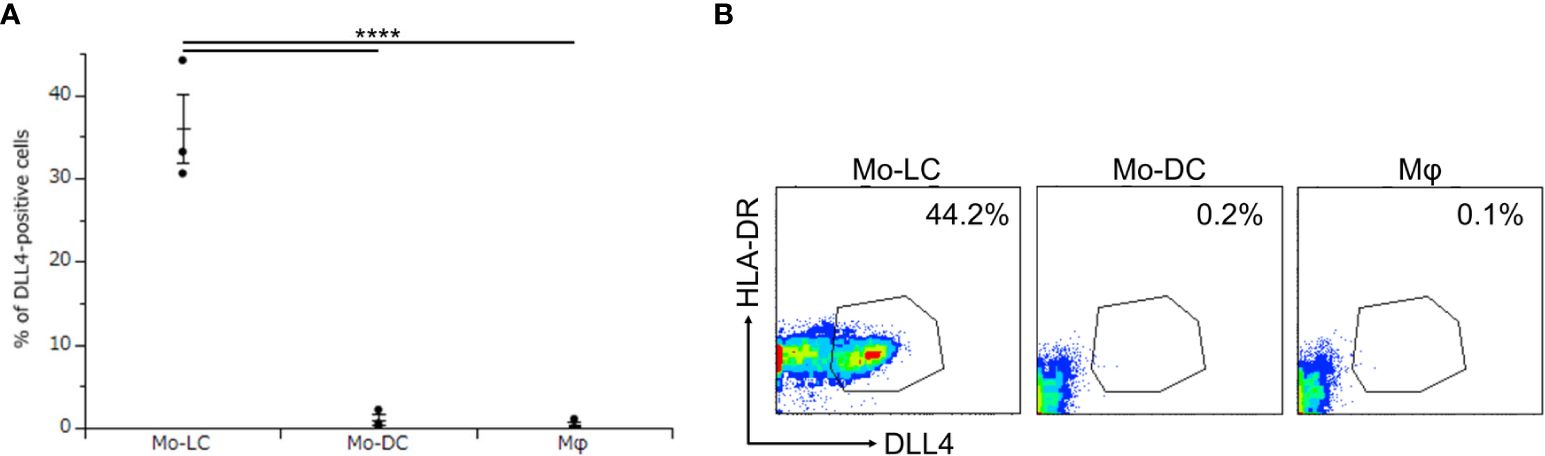
Figure 2. Mo-LCs express DLL4 by toll-like receptor (TLR) ligand stimulation. (A, B) Percentage of DLL4-positive cells (DLL4+ human leukocyte antigen (HLA)-DR+ cells) within Mo-LC, monocyte-derived dendritic cell (Mo-DC), and macrophage (Mφ). The Mo-LCs were generated from CD14+ monocytes in culture with GM-CSF and TGF-β1 on DLL4-immobilized plates, the Mo-DCs were generated from CD14+ monocytes in culture with GM-CSF and interleukin (IL)-4, and the Mφs were generated from CD14+ monocytes in culture with GM-CSF. The DLL4 expression on Mo-LC, Mo-DC, and Mφ after activation by Polyinosinic-polycytidylic acid (Poly(I:C)) and lipopolysaccharide (LPS) for 24 h was determined by FACS on day 7. (A) In this assay, we used blood samples from donor No.1-3. ****p < 0.0001. (B) Two-color flow cytometry of the cultured cells was performed to measure DLL4 and HLA-DR.
Although we discovered that Mo-LCs express DLL4 (Figures 2A, B), it remains unclear at which differentiation stages, including common myeloid progenitor, Mφ and DC progenitor, DC precursors and immature DCs, the endpoint of hematopoietic stem cells/hematopoietic progenitor cells to become DLL4+ DC precursor cells is established and why Mo-DCs cannot become DLL4+ DCs (24). To answer these questions, we investigated the critical regulators of DLL4 expression using DLL4+ Mo-LCs. We first investigated which TLR ligands can upregulate DLL4 expression. We analyzed the DLL4 expression on Mo-LCs after activation by PGN, Poly(I:C), LPS, and IMQ for 24 h. We found that PGN most efficiently and reproducibly induced DLL4 expression (Figures 3A, B).
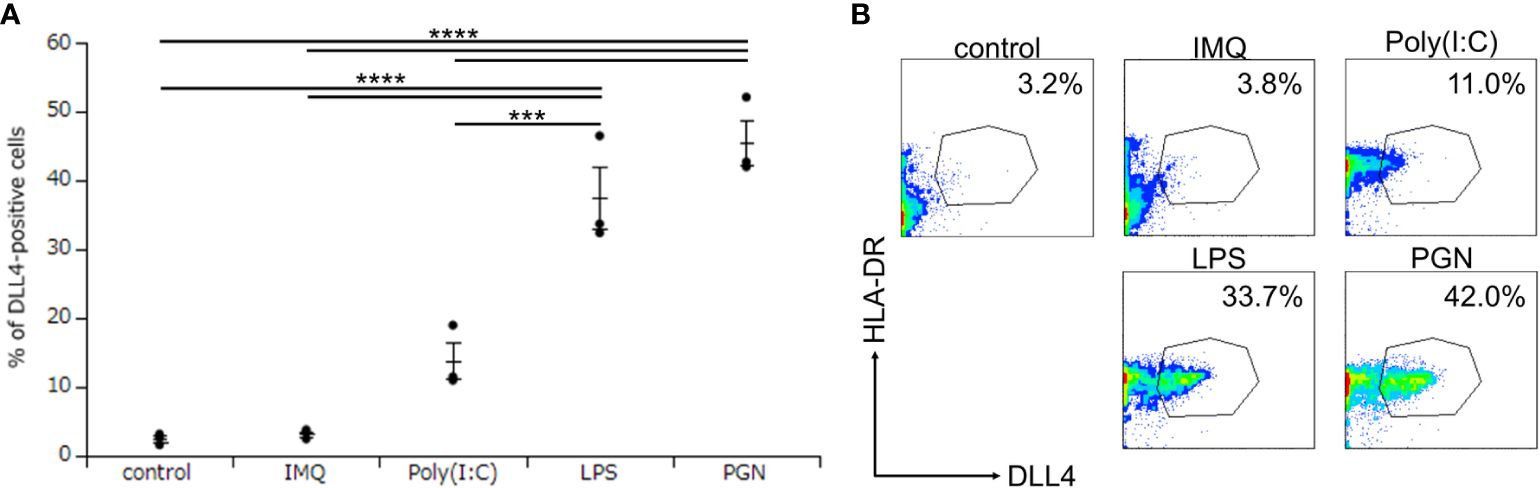
Figure 3. Peptidoglycan (PGN) most potently induces DLL4 expression on Mo-LCs. (A, B) Percentage of DLL4-positive cells induced by control (without TLR ligands), PGN, Poly(I:C), LPS, and imiquimod (IMQ). The Mo-LCs were generated from CD14+ monocytes in culture with GM-CSF and TGF-β1 on DLL4-immobilized plates. The DLL4 expression on Mo-LC after activation by TLR ligands for 24 h was determined by FACS on day 7. (A) In this assay, we used blood samples from donor No.1-3. ***p < 0.001, ****p < 0.0001. (B) Two-color flow cytometry of the cultured cells was performed to measure DLL4 and HLA-DR.
To investigate the critical regulators of DLL4 expression using DLL4+ Mo-LCs induced by PGN (Figures 3A, B), next, we investigated which Notch ligands can upregulate the expression of DLL4. Similar to the induction of Mo-LCs (Figure 1), DLL1 and DLL4 induced high levels of DLL4 expression, and DLL4 most efficiently and reproducibly induced DLL4 expression (Figure 4A).
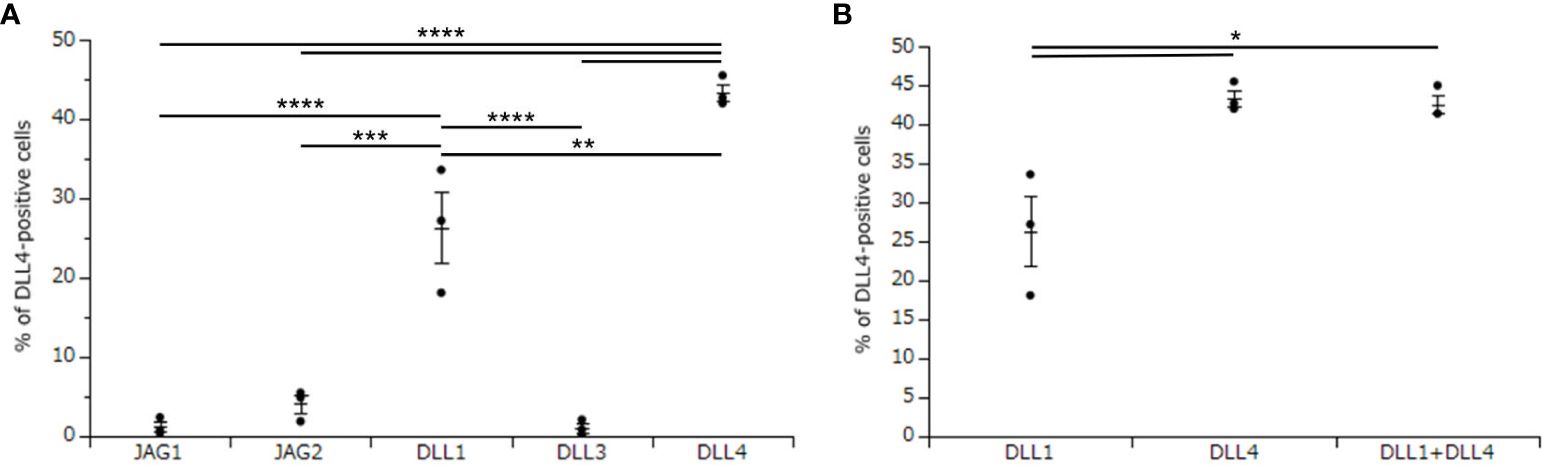
Figure 4. DLL4 most potently induces DLL4 expression on Mo-LCs. (A) Percentage of DLL4-positive cells induced on all Notch ligand-immobilized plates. (B) Percentage of the DLL4-positive cells induced on DLL1, DLL4, and DLL1+DLL4-immobilized plates. (A, B) The Mo-LCs were generated from CD14+ monocytes in culture with GM-CSF and TGF-β1 on the Notch ligand-immobilized plates. The DLL4 expression on Mo-LC after activation by PGN for 24 h was determined by FACS on day 7. In this assay, we used blood samples from donor No.1, 2, and 4. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We next investigated whether DLL1 and DLL4 synergize in the induction of DLL4 expression. Our results showed no synergistic effect when using DLL4 together with DLL1 (Figure 4B).
To investigate the critical regulators of DLL4 expression using DLL4+ Mo-LCs induced by PGN (Figures 3A, B) and DLL4 (Figures 4A, B), next, we searched for cytokines that induce DLL4 expression given that the results shown in Figure 2 suggest that GM-CSF, TGF-β1, DLL4, and IL-4 regulate DLL4 expression on monocyte-derived cells. We compared the DLL4 expression ratios in combination with these cytokines. As a result, we found that DLL4 and GM-CSF were required for the induction of DLL4 expression, which was promoted and repressed by TGF-β1 and IL-4, respectively (Figures 5A, B). DLL4, DLL4+TGF-β1, and DLL4+TGF-β1+IL-4 induced very few cells, while GM-CSF, TGF-β1, IL-4, DLL4+IL-4, GM-CSF+TGF-β1, GM-CSF+IL-4, TGF-β1+IL-4, GM-CSF+TGF-β1+IL-4, and the control (without cytokines) did not induce DLL4+ cells (data not shown).
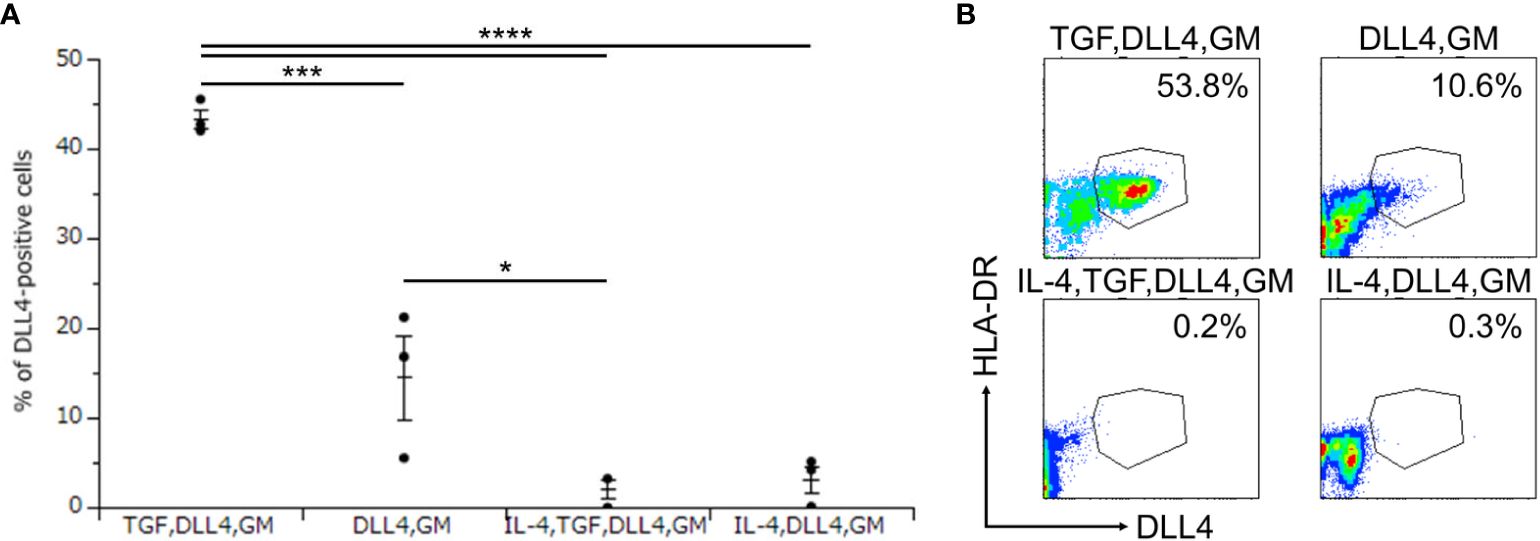
Figure 5. DLL4 and GM-CSF are required for the induction of DLL4 expression, which is promoted and repressed by TGF-β1 and IL-4, respectively. (A, B) Percentage of DLL4-positive cells induced by immobilized DLL4, GM-CSF, TGF-β1, and IL-4. The CD14+ monocytes were cultured in the presence or absence of immobilized DLL4, GM-CSF, TGF-β1, and IL-4. The DLL4 expression on monocytes after activation by PGN for 24 h was determined by FACS on day 7. (A) In this assay, we used blood samples from donor No.1, 2, and 4. *p < 0.05, ***p < 0.001, ****p < 0.0001. (B) Two-color flow cytometry of the cultured cells was performed to measure DLL4 and HLA-DR.
No previous study has compared the T cell activation ratios between Mo-DCs, Mo-LCs, and Mφs. Finally, DLL4+ Mo-LCs induced by immobilized DLL4 (Figures 4A, B), GM-CSF, TGF-β1 (Figures 5A, B), and PGN (Figures 3A, B) were compared to Mo-DCs and Mφs stimulated by PGN for their ability to activate T cells. The Mo-DCs were generated from CD14+ monocytes in culture with GM-CSF and IL-4, and the Mφs were generated from CD14+ monocytes in culture with GM-CSF. The results suggested that DLL4+ Mo-LCs were most effective at activating T cells (Figures 6A, B).
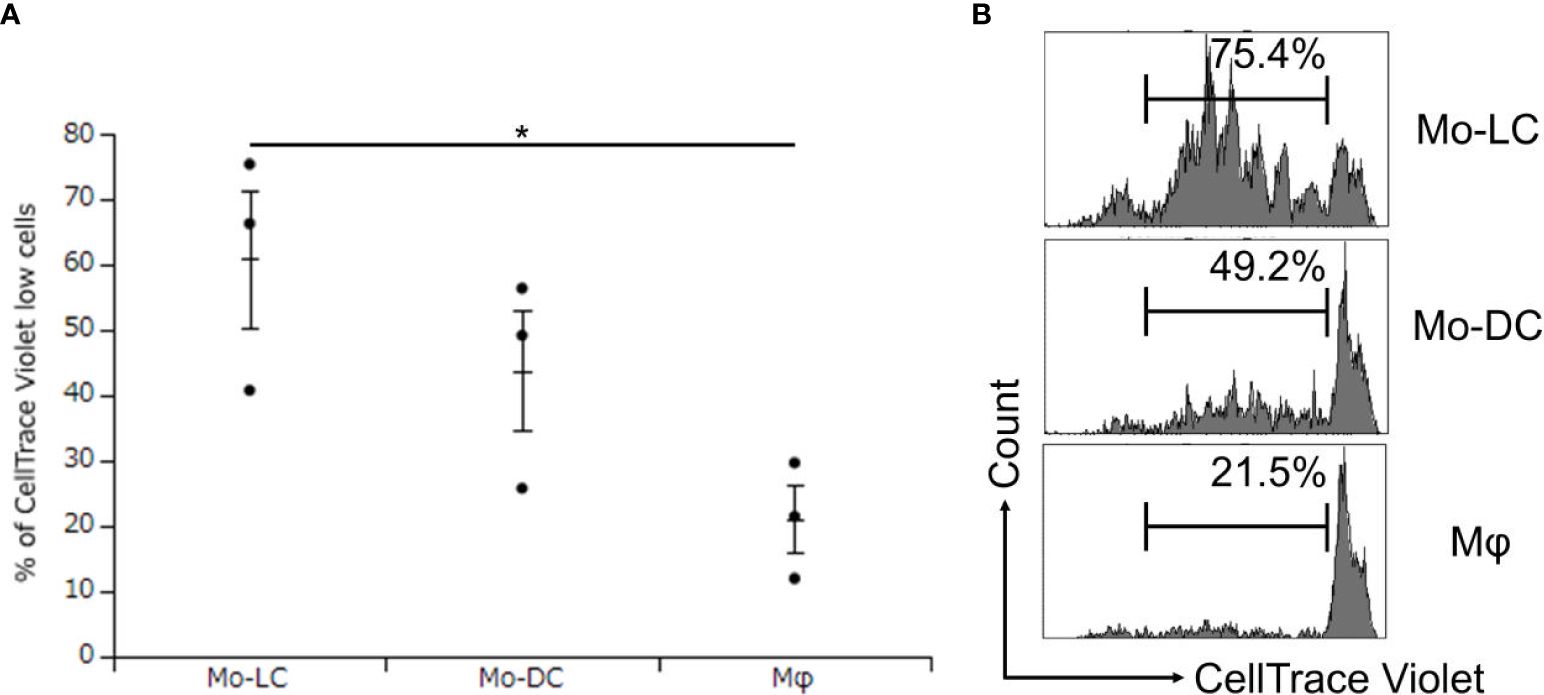
Figure 6. DLL4+ Mo-LCs most potently activate CD4+ T cells. (A, B) Percentage of CellTrace Violet low cells activated by DLL4+ Mo-LC, Mo-DC, and Mφ. The DLL4+ Mo-LCs were generated from CD14+ monocytes in culture with GM-CSF and TGF-β1 on DLL4-immobilized plates, the Mo-DCs were generated from CD14+ monocytes in culture with GM-CSF and IL-4, and the Mφs were generated from CD14+ monocytes in culture with GM-CSF. After activation by PGN for 24 h, the monocyte-derived cells were cocultured with CellTrace Violet-labeled CD4+ T cells from the same donor on day 7. After 5 days, the CellTrace Violet dilution was analyzed by FACS. (A) In this assay, we used blood samples from donor No.5-7. *p < 0.05.
Figure 7 summarizes our results and depicts our proposed mechanism of DLL4 expression on Mo-LCs. The aim of this study was to establish whether Mo-LCs express DLL4 and establish the induction method of antigen presenting cells, which most potently activate T cells. As a result, Mo-LCs expressed DLL4. DLL4+ Mo-LCs most potently activated CD4+ T cells among the monocyte-derived cells, and DLL4, GM-CSF, TGF-β1, and PGN are the factors that most efficiently induce DLL4+ Mo-LCs. Additionally, the factors responsible for inducing Mo-LCs are almost the same factors responsible for inducing DLL4 expression.
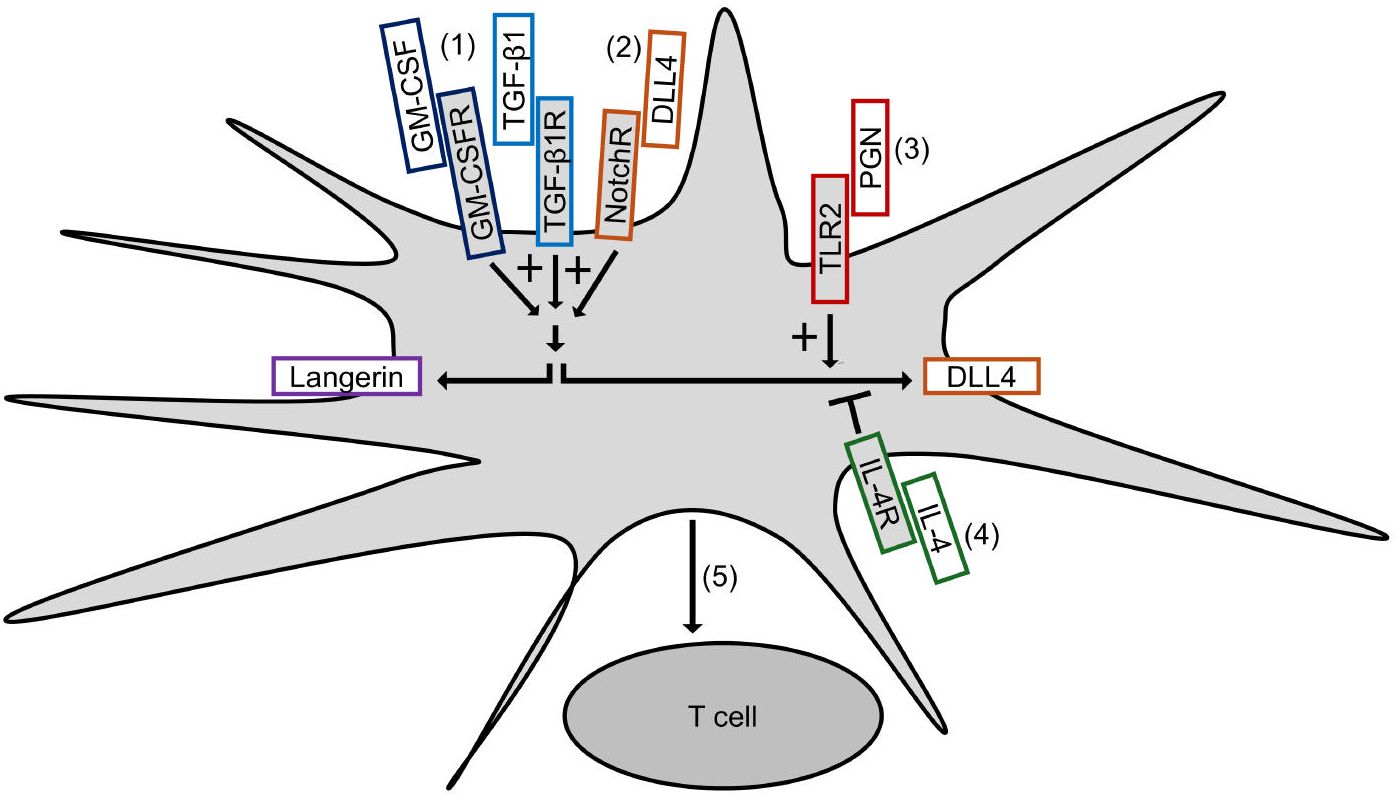
Figure 7. Induction of DLL4+ Mo-LCs activating T cells most potently. (1) GM-CSF and TGF-β1 not only induce Mo-LCs but also DLL4 expression. (2) DLL4 most potently induces Mo-LCs and DLL4 expression among Notch ligands. (3) PGN most potently induces DLL4 expression among TLR ligands. (4) IL-4 represses DLL4 expression. (5) DLL4+ Mo-LCs most potently activate T cells among monocyte-derived cells.
In this study, we sought to identify the Notch ligands necessary for monocytes to give rise to Langerhans-like cells. We proved that DLL1 and DLL4 induced Mo-LCs, and that DLL4 most efficiently and reproducibly induced Mo-LCs (Figure 1). These results are in contrast to those reported by Schwentner et al. (22), although this is different from previous reports (14, 21), possibly due to different protocols used for the in vitro differentiation and Schwentner et al. observed high donor variability in terms of fold induction of Mo-LCs. In terms of the mechanism by which DLL4 induces Mo-LCs, activation of Notch signaling and the concomitant loss of Kruppel-like factor 4 may be involved (19).
We discovered that Mo-LCs expressed DLL4, and Mo-LCs and DLL4 expression were induced by comparable factors (Figure 7). We discovered that DLL1 or DLL4 was required for the induction of DLL4 expression, and that IL-4 repressed it, suggesting that Mo-DCs cannot become DLL4+ DCs (24) because of the presence of IL-4 and the absence of DLL1 or DLL4 in the Mo-DC generation process.
DLL4 induced Mo-LCs and DLL4 expression more than DLL1 (Figures 1, 4). As DLL4 exhibits a binding affinity for Notch receptors that is stronger than DLL1 (31) or DLL1 and DLL4 can activate distinct targets (32), differences in induction ratios seem likely. It is speculated that DLL4+ Mo-LCs play important roles in psoriatic lesions given the increased expression of DLL4 in psoriasis lesional skin (28, 29), the relationship between Mo-LC and psoriasis (5–7, 13), the high level of expression of TLR2 on blood monocytes in patients with psoriasis (33), the fact that the TLR2 ligand PGN most potently induces DLL4 expression (Figure 3), and that DLL4+ Mo-LCs most potently activate T cells (Figure 6). Furthermore, the induction of Mo-LCs by DLL4 (Figure 1) and DLL4 expression on Mo-LCs (Figure 4) might provoke a positive feedback loop of DLL4+ Mo-LC generation (9, 34, 35) that is necessary for the persistence of the disease. However, DLL1 may be the first Notch ligand to induce Mo-LCs or DLL4 expression given that DLL1 is detected in unaffected skin (30) and DLL4 expression increases in psoriasis lesional skin (28, 29).
TLR-mediated nuclear factor kappa B (NF-κB) signaling stimulates the production of IL-6, which binds to the IL-6 receptor and activates STAT3. STAT3 induces DLL1 expression that activates Notch signaling and boosts NF-κB-induced IL-6, which transduces the stabilization of STAT3 activation (34). Such a mechanism may be involved in DLL4 expression because both NF-κB and STAT3 are required for the induction of DLL4+ DCs (24) and DLL4 expression on a stromal cell line is augmented by IL-6 via STAT3 activation (36). Moreover, not only IL-6 but also IL-23 might be involved because PGN and LPS induce IL-23 (8, 13) and IL-23 induces STAT3 (8, 37, 38); alternatively, DLL4 might directly induce STAT3 (39). TLR-induced JAG1 expression is strongly dependent on the Notch master transcriptional regulator RBP-J, as well as on upstream components of the Notch pathway γ-secretase and Notch1 and Notch2 receptors (9). However, among Notch ligands, only DLL1 and DLL4 induce DLL4 expression (Figure 4), suggesting that TLR-induced DLL4 expression is not dependent on the Notch pathway but on the DLL1 and DLL4 pathway. TGF-β1 might contribute to DLL4 expression by promoting NF-κB (40, 41) or STAT3 through Smad3 (42–44) or cooperatively with Notch signaling (45). IL-4 might suppress DLL4 expression by suppressing TGF-β1 (46), NF-κB (47, 48), or yes-associated protein (YAP) (49) because YAP promotes DLL4 (50) and NF-κB (51) and DLL4 promotes YAP (52). Th17 differentiation has been shown to be efficiently elicited by not Mo-DCs but cDCs activated by PGN (8). Additionally, among TLR ligands, PGN most efficiently induced DLL4 expression on Mo-LCs but not on Mo-DCs (Figures 2, 3). These results suggest that DLL4 expression in response to PGN stimulation has the potential to induce Th17 differentiation. Both PGN and LPS induced high levels of DLL4 expression on the surface of Mo-LCs, whereas Poly(I:C) and IMQ did not (Figure 3). One explanation for this may be that bacteria induce DLL4 expression on Mo-LCs more than viruses, or because Mo-LCs express TLRs 2 and 4 more than TLRs 3, 7, and 8 (13). However, these mechanisms are still speculative, and further research is needed to determine the signaling pathways.
DLL4 has been reported to increase LPS-stimulated cytokines by enhancing NF-κB activation (53–55). Contrary to these findings, other studies have proposed that TLR-mediated proinflammatory cytokines are reduced upon overexpression of NICD in mouse peritoneal Mφs (56). Our results relating to DLL4 expression suggest that different TLR ligands, Notch ligands, or cytokines can evoke such contradictions.
A fundamental difference between the human and murine systems has been highlighted (4, 8, 9). However, compared to mouse LCs, our knowledge of human LCs remains very limited, and with the discovery of human LC subsets, it is crucial to explore their features and functions under both physiological and pathological conditions from the perspective of subsets (57). DLL4+ Mo-LCs (Figure 2), the discovery of a novel human LC subset, may contribute to the exploration.
Because mechanistic studies that induce DLL4 in DCs are important for better defining the ontogeny of DLL4+ DCs and targeting these cells for immunotherapy (24), the discovery of the induction method of DLL4-expressing cells (Figure 7) may contribute to devising a novel strategy using the cells or signaling pathways through such as DLL4, TGF-β1, IL-4, or langerin for human patients with autoinflammatory immune diseases such as psoriasis (24, 35, 58, 59). In addition, in vitro-generated Mo-LCs with GM-CSF, TGF-β1, and DLL4 closely resemble their in vivo skin counterparts and are not only a useful, easily accessible tool to study different functions of LCs but may also help to elucidate their potential for immunotherapies (27). With the discovery that DLL4+ Mo-LCs most potently activated T cells among the monocyte-derived cells (Figure 6), targeting DLL4+ Mo-LCs may offer a promising therapeutic strategy such as in cancer treatment (24, 27, 35, 57, 60).
While targeting DLL4-expressing cells holds promise for the treatment of various human diseases, the limitations of the study are the potential off-target effects (35, 61). Emerging research areas and potential new therapeutic strategies, including a nanocarrier (35) and a synthetic Notch (62), are being explored to increase the specificity of targeting. The discovery that DLL4 expression is dependent not on the Notch pathway but on the DLL1 and DLL4 pathways (Figure 4) may contribute to improving therapeutic efficacy. The development of more selective inhibitors targeting specific Notch ligands or receptors, or modulating specific downstream pathways, may help to overcome off-target effects (63). Moreover, understanding how DLL4 interacts with signaling molecules or pathways, including langerin, IL-4, and TGF-β1, might lead to combined therapeutic strategies that target multiple pathways for enhanced efficacy (35). Additionally, it is necessary to obtain a thorough understanding of the context-dependent roles of DLL4+ Mo-LCs (35, 64). Nevertheless, it will be intriguing to elucidate the roles of DLL4+ Mo-LCs because DLL4+ Mo-LCs may be responsible for T cell activation in autoinflammatory immune diseases such as psoriasis (5–7, 13, 24, 28, 29, 33, 35, 59) and may also be efficacious for cancer treatment (24, 27, 35, 57, 60). Stimulated and matured Mo-DCs and Mo-LCs can activate T cells more effectively (27, 65). Thus, we speculate that non-stimulated Mo-LCs would not activate T cells as robustly as PGN-treated Mo-LCs (Figure 6). Further studies should include comparisons with other stimulations, such as LPS or the standard DC-cytokine maturation cocktail containing IL-1β, IL-6, TNF-α, and prostaglandin E2 (65), to confirm whether Mo-LCs are better activators of T cell responses. Verification of the results using measures such as the division or proliferation index would also be valuable. Although Mo-DCs and Mφs were obtained using standard induction methods (60, 66), verifying their markers in future experiments would strengthen the conclusions. Finally, while we discovered that DLL4+ Mo-LCs most potently activate T cells among the monocyte-derived cells (Figures 6, 7), further research is needed to explore whether langerin, DLL4, or another molecule derived from DLL4, TGF-β1, and GM-CSF is crucial for T cell activation.
In conclusion, we show here that Mo-LCs express DLL4 on the cell surface when activated by PGN, which is suppressed by IL-4. These findings provide insights into the roles of DLL4-expressing cells such as DLL4+ Mo-LCs in human diseases and the development of more effective therapeutic strategies.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving humans were approved by Showa University Research Ethics Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
RO: Conceptualization, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. KM: Writing – review & editing. TT: Writing – review & editing. TI: Resources, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by the Showa University Research Grant for Young Researchers.
We would like to thank Dr. Sanju Iwamoto for helpful suggestions and Enago (www.enago.jp) for the English language review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1532620/full#supplementary-material
1. Hu P, Wang M, Gao H, Zheng A, Li J, Mu D, et al. The role of helper T cells in psoriasis. Front Immunol. (2021) 12:788940. doi: 10.3389/fimmu.2021.788940
2. Klechevsky E, Morita R, Liu M, Cao Y, Coquery S, Thompson-Snipes L, et al. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. (2008) 29:497–510. doi: 10.1016/j.immuni.2008.07.013
3. Romano E, Rossi M, Ratzinger G, de Cos MA, Chung DJ, Panageas KS, et al. Peptide-loaded Langerhans cells, despite increased IL15 secretion and T-cell activation in vitro, elicit antitumor T-cell responses comparable to peptide-loaded monocyte-derived dendritic cells in vivo. Clin Cancer Res. (2011) 17:1984–97. doi: 10.1158/1078-0432.ccr-10-3421
4. Eidsmo L, Martini E. Human langerhans cells with pro-inflammatory features relocate within psoriasis lesions. Front Immunol. (2018) 9:300. doi: 10.3389/fimmu.2018.00300
5. Otsuka M, Egawa G, Kabashima K. Uncovering the mysteries of langerhans cells, inflammatory dendritic epidermal cells, and monocyte-derived langerhans cell-like cells in the epidermis. Front Immunol. (2018) 9:1768. doi: 10.3389/fimmu.2018.01768
6. Singh TP, Zhang HH, Borek I, Wolf P, Hedrick MN, Singh SP, et al. Monocyte-derived inflammatory Langerhans cells and dermal dendritic cells mediate psoriasis-like inflammation. Nat Commun. (2016) 7:13581. doi: 10.1038/ncomms13581
7. Lee M, Kim SH, Kim TG, Park J, Lee JW, Lee MG. Resident and monocyte-derived Langerhans cells are required for imiquimod-induced psoriasis-like dermatitis model. J Dermatol Sci. (2018) 91:52–9. doi: 10.1016/j.jdermsci.2018.04.003
8. Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. (2007) 8:942–9. doi: 10.1038/ni1496
9. Foldi J, Chung AY, Xu H, Zhu J, Outtz HH, Kitajewski J, et al. Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J Immunol. (2010) 185:5023–31. doi: 10.4049/jimmunol.1001544
10. Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. (1999) 284:770–6. doi: 10.1126/science.284.5415.770
11. Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. (2004) 5:247–53. doi: 10.1038/ni1045
12. Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. (2000) 113 Pt 23:4313–8. doi: 10.1242/jcs.113.23.4313
13. Takahashi R, Iwamoto S, Tanioka T, Maeda K. A profile of pro-inflammatory cytokine expression in human Delta-1-induced monocyte-derived Langerhans cell-like dendritic cells after stimulation with Toll-like receptor ligands. Showa Univ J Med Sci. (2021) 33:67–73. doi: 10.15369/sujms.33.67
14. Hoshino N, Katayama N, Shibasaki T, Ohishi K, Nishioka J, Masuya M, et al. A novel role for Notch ligand Delta-1 as a regulator of human Langerhans cell development from blood monocytes. J Leukoc Biol. (2005) 78:921–9. doi: 10.1189/jlb.1204746
15. Picarda G, Chéneau C, Humbert JM, Bériou G, Pilet P, Martin J, et al. Functional langerinhigh-expressing langerhans-like cells can arise from CD14highCD16- human blood monocytes in serum-free condition. J Immunol. (2016) 196:3716–28. doi: 10.4049/jimmunol.1501304
16. Otsuka Y, Watanabe E, Shinya E, Okura S, Saeki H, Geijtenbeek TBH, et al. Differentiation of langerhans cells from monocytes and their specific function in inducing IL-22-specific th cells. J Immunol. (2018) 201:3006–16. doi: 10.4049/jimmunol.1701402
17. Heinz LX, Platzer B, Reisner PM, Jörgl A, Taschner S, Göbel F, et al. Differential involvement of PU.1 and Id2 downstream of TGF-beta1 during Langerhans-cell commitment. Blood. (2006) 107:1445–53. doi: 10.1182/blood-2005-04-1721
18. Bauer T, Zagórska A, Jurkin J, Yasmin N, Köffel R, Richter S, et al. Identification of Axl as a downstream effector of TGF-β1 during Langerhans cell differentiation and epidermal homeostasis. J Exp Med. (2012) 209:2033–47. doi: 10.1084/jem.20120493
19. Jurkin J, Krump C, Köffel R, Fieber C, Schuster C, Brunner PM, et al. Human skin dendritic cell fate is differentially regulated by the monocyte identity factor Kruppel-like factor 4 during steady state and inflammation. J Allergy Clin Immunol. (2017) 139:1873–84.e10. doi: 10.1016/j.jaci.2016.09.018
20. Borek I, Köffel R, Feichtinger J, Spies M, Glitzner-Zeis E, Hochgerner M, et al. BMP7 aberrantly induced in the psoriatic epidermis instructs inflammation-associated Langerhans cells. J Allergy Clin Immunol. (2020) 145:1194–207.e11. doi: 10.1016/j.jaci.2019.12.011
21. Milne P, Bigley V, Bacon CM, Néel A, McGovern N, Bomken S, et al. Hematopoietic origin of Langerhans cell histiocytosis and Erdheim-Chester disease in adults. Blood. (2017) 130:167–75. doi: 10.1182/blood-2016-12-757823
22. Schwentner R, Jug G, Kauer MO, Schnöller T, Waidhofer-Söllner P, Holter W, et al. JAG2 signaling induces differentiation of CD14(+) monocytes into Langerhans cell histiocytosis-like cells. J Leukoc Biol. (2019) 105:101–11. doi: 10.1002/jlb.1a0318-098r
23. Sun L, Su Y, Jiao A, Wang X, Zhang B. T cells in health and disease. Signal Transduct Target Ther. (2023) 8:235. doi: 10.1038/s41392-023-01471-y
24. Meng L, Hu S, Wang J, He S, Zhang Y. DLL4(+) dendritic cells: Key regulators of Notch Signaling in effector T cell responses. Pharmacol Res. (2016) 113:449–57. doi: 10.1016/j.phrs.2016.09.001
25. Eisenbarth SC. Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol. (2019) 19:89–103. doi: 10.1038/s41577-018-0088-1
26. Coillard A, Segura E. In vivo differentiation of human monocytes. Front Immunol. (2019) 10:1907. doi: 10.3389/fimmu.2019.01907
27. Bellmann L, Zelle-Rieser C, Milne P, Resteu A, Tripp CH, Hermann-Kleiter N, et al. Notch-mediated generation of monocyte-derived langerhans cells: phenotype and function. J Invest Dermatol. (2021) 141:84–94.e6. doi: 10.1016/j.jid.2020.05.098
28. Rooney P, Connolly M, Gao W, McCormick J, Biniecka M, Sullivan O, et al. Notch-1 mediates endothelial cell activation and invasion in psoriasis. Exp Dermatol. (2014) 23:113–8. doi: 10.1111/exd.12306
29. Gratton R, Tricarico PM, Moltrasio C, Lima Estevão de Oliveira AS, Brandão L, Marzano AV, et al. Pleiotropic role of notch signaling in human skin diseases. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21124214
30. Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. (2000) 10:491–500. doi: 10.1016/s0960-9822(00)00451-6
31. Andrawes MB, Xu X, Liu H, Ficarro SB, Marto JA, Aster JC, et al. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J Biol Chem. (2013) 288:25477–89. doi: 10.1074/jbc.M113.454850
32. Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, Elowitz MB. Dynamic ligand discrimination in the notch signaling pathway. Cell. (2018) 172:869–80.e19. doi: 10.1016/j.cell.2018.01.002
33. Pochernina VV, Daschuk AM. TLR expression on peripheral blood monocytes in patients with psoriasis. Wiad Lek. (2020) 73:401–4. doi: 10.36740/WiadLek
34. Hildebrand D, Uhle F, Sahin D, Krauser U, Weigand MA, Heeg K. The interplay of notch signaling and STAT3 in TLR-activated human primary monocytes. Front Cell Infect Microbiol. (2018) 8:241. doi: 10.3389/fcimb.2018.00241
35. Yuwen Y, Wang X, Liu J, Liu Z, Zhu H. Delta- like ligand 4- expressing macrophages and human diseases: Insights into pathophysiology and therapeutic opportunities. Heliyon. (2023) 9:e20777. doi: 10.1016/j.heliyon.2023.e20777
36. Suzuki M, Yamamoto M, Sugimoto A, Nakamura S, Motoda R, Orita K. Delta-4 expression on a stromal cell line is augmented by interleukin-6 via STAT3 activation. Exp Hematol. (2006) 34:1143–50. doi: 10.1016/j.exphem.2006.04.027
37. Kortylewski M, Xin H, Kujawski M, Lee H, Liu Y, Harris T, et al. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. (2009) 15:114–23. doi: 10.1016/j.ccr.2008.12.018
38. Morris R, Kershaw NJ, Babon JJ. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. (2018) 27:1984–2009. doi: 10.1002/pro.3519
39. Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. (2004) 6:547–54. doi: 10.1038/ncb1138
40. Meng J, Qin Y, Chen J, Wei L, Huang XR, Yu X, et al. Treatment of hypertensive heart disease by targeting smad3 signaling in mice. Mol Ther Methods Clin Dev. (2020) 18:791–802. doi: 10.1016/j.omtm.2020.08.003
41. Liu GX, Li YQ, Huang XR, Wei L, Chen HY, Shi YJ, et al. Disruption of Smad7 promotes ANG II-mediated renal inflammation and fibrosis via Sp1-TGF-β/Smad3-NF.κB-dependent mechanisms in mice. PloS One. (2013) 8:e53573. doi: 10.1371/journal.pone.0053573
42. Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, Lendahl U, et al. Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol. (2003) 163:723–8. doi: 10.1083/jcb.200305112
43. Itoh Y, Saitoh M, Miyazawa K. Smad3-STAT3 crosstalk in pathophysiological contexts. Acta Biochim Biophys Sin (Shanghai). (2018) 50:82–90. doi: 10.1093/abbs/gmx118
44. Wang LN, Zhang ZT, Wang L, Wei HX, Zhang T, Zhang LM, et al. TGF-β1/SH2B3 axis regulates anoikis resistance and EMT of lung cancer cells by modulating JAK2/STAT3 and SHP2/Grb2 signaling pathways. Cell Death Dis. (2022) 13:472. doi: 10.1038/s41419-022-04890-x
45. Samon JB, Champhekar A, Minter LM, Telfer JC, Miele L, Fauq A, et al. Notch1 and TGFbeta1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood. (2008) 112:1813–21. doi: 10.1182/blood-2008-03-144980
46. Macey MR, Sturgill JL, Morales JK, Falanga YT, Morales J, Norton SK, et al. IL-4 and TGF-beta 1 counterbalance one another while regulating mast cell homeostasis. J Immunol. (2010) 184:4688–95. doi: 10.4049/jimmunol.0903477
47. Yu M, Qi X, Moreno JL, Farber DL, Keegan AD. NF-κB signaling participates in both RANKL- and IL-4-induced macrophage fusion: receptor cross-talk leads to alterations in NF-κB pathways. J Immunol. (2011) 187:1797–806. doi: 10.4049/jimmunol.1002628
48. Yu M, Moreno JL, Stains JP, Keegan AD. Complex regulation of tartrate-resistant acid phosphatase (TRAP) expression by interleukin 4 (IL-4): IL-4 indirectly suppresses receptor activator of NF-kappaB ligand (RANKL)-mediated TRAP expression but modestly induces its expression directly. J Biol Chem. (2009) 284:32968–79. doi: 10.1074/jbc.M109.001016
49. Zhou X, Li W, Wang S, Zhang P, Wang Q, Xiao J, et al. YAP aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. Cell Rep. (2019) 27:1176–89.e5. doi: 10.1016/j.celrep.2019.03.028
50. Matsuo E, Okamoto T, Ito A, Kawamoto E, Asanuma K, Wada K, et al. Substrate stiffness modulates endothelial cell function via the YAP-Dll4-Notch1 pathway. Exp Cell Res. (2021) 408:112835. doi: 10.1016/j.yexcr.2021.112835
51. Rivera-Reyes A, Ye S, G EM, Egolf S, Ec G, Chor S, et al. YAP1 enhances NF-κB-dependent and independent effects on clock-mediated unfolded protein responses and autophagy in sarcoma. Cell Death Dis. (2018) 9:1108. doi: 10.1038/s41419-018-1142-4
52. Krossa I, Strub T, Martel A, Nahon-Esteve S, Lassalle S, Hofman P, et al. Recent advances in understanding the role of HES6 in cancers. Theranostics. (2022) 12:4374–85. doi: 10.7150/thno.72966
53. Monsalve E, Pérez MA, Rubio A, Ruiz-Hidalgo MJ, Baladrón V, García-Ramírez JJ, et al. Notch-1 up-regulation and signaling following macrophage activation modulates gene expression patterns known to affect antigen-presenting capacity and cytotoxic activity. J Immunol. (2006) 176:5362–73. doi: 10.4049/jimmunol.176.9.5362
54. Monsalve E, Ruiz-García A, Baladrón V, Ruiz-Hidalgo MJ, Sánchez-Solana B, Rivero S, et al. Notch1 upregulates LPS-induced macrophage activation by increasing NF-kappaB activity. Eur J Immunol. (2009) 39:2556–70. doi: 10.1002/eji.200838722
55. Kuramoto T, Goto H, Mitsuhashi A, Tabata S, Ogawa H, Uehara H, et al. Dll4-Fc, an inhibitor of Dll4-notch signaling, suppresses liver metastasis of small cell lung cancer cells through the downregulation of the NF-κB activity. Mol Cancer Ther. (2012) 11:2578–87. doi: 10.1158/1535-7163.mct-12-0640
56. Zhang Q, Wang C, Liu Z, Liu X, Han C, Cao X, et al. Notch signal suppresses Toll-like receptor-triggered inflammatory responses in macrophages by inhibiting extracellular signal-regulated kinase 1/2-mediated nuclear factor κB activation. J Biol Chem. (2012) 287:6208–17. doi: 10.1074/jbc.M111.310375
57. Zhu R, Yao X, Li W. Langerhans cells and skin immune diseases. Eur J Immunol. (2024) 54:e2250280. doi: 10.1002/eji.202250280
58. Hahn M, Ghoreschi K. The role of IL-4 in psoriasis. Expert Rev Clin Immunol. (2017) 13:171–3. doi: 10.1080/1744666x.2017.1279054
59. Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, et al. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. (2005) 6:680–8. doi: 10.1038/ni1209x
60. Chiang CL, Kandalaft LE. In vivo cancer vaccination: Which dendritic cells to target and how? Cancer Treat Rev. (2018) 71:88–101. doi: 10.1016/j.ctrv.2018.10.012
61. Keewan E, Naser SA. The role of notch signaling in macrophages during inflammation and infection: implication in rheumatoid arthritis? Cells. (2020) 9. doi: 10.3390/cells9010111
62. Li X, Yan X, Wang Y, Kaur B, Han H, Yu J. The Notch signaling pathway: a potential target for cancer immunotherapy. J Hematol Oncol. (2023) 16:45. doi: 10.1186/s13045-023-01439-z
63. Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. (2015) 12:445–64. doi: 10.1038/nrclinonc.2015.61
64. Fukuda D, Aikawa M. Expanding role of delta-like 4 mediated notch signaling in cardiovascular and metabolic diseases. Circ J. (2013) 77:2462–8. doi: 10.1253/circj.cj-13-0873
65. Jonuleit H, Kühn U, Müller G, Steinbrink K, Paragnik L, Schmitt E, et al. Pro-inflammatory cytokines and prostaglandins induce maturation of potent immunostimulatory dendritic cells under fetal calf serum-free conditions. Eur J Immunol. (1997) 27:3135–42. doi: 10.1002/eji.1830271209
Keywords: Langerhans cells, monocyte, autoimmune diseases, Delta-like 1, Delta-like 4, T cell, psoriasis, toll-like receptor
Citation: Ono R, Maeda K, Tanioka T and Isozaki T (2025) Monocyte-derived Langerhans cells express Delta-like 4 induced by peptidoglycan and interleukin-4 mediated suppression. Front. Immunol. 16:1532620. doi: 10.3389/fimmu.2025.1532620
Received: 22 November 2024; Accepted: 30 January 2025;
Published: 13 February 2025.
Edited by:
Thomas Hieronymus, University Hospital RWTH Aachen, GermanyReviewed by:
Adriana Mantegazza, Thomas Jefferson University, United StatesCopyright © 2025 Ono, Maeda, Tanioka and Isozaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rei Ono, ci50YWthQHBoYXJtLnNob3dhLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.