
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 22 January 2025
Sec. Alloimmunity and Transplantation
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1517387
A correction has been applied to this article in:
Corrigendum: Predicted Indirectly Recognizable HLA Epitopes (PIRCHE) Scores and Clinical Outcomes after Haploidentical Stem Cell Transplantation in Pediatric Patients with Relapsed Neuroblastoma
 Eun Seop Seo1,2†
Eun Seop Seo1,2† In Hwa Jeong3,4†
In Hwa Jeong3,4† Hee Young Ju1
Hee Young Ju1 Ju Kyung Hyun1
Ju Kyung Hyun1 Ji Won Lee1
Ji Won Lee1 Keon Hee Yoo1
Keon Hee Yoo1 Won Young Heo3
Won Young Heo3 Ki Woong Sung1
Ki Woong Sung1 Hee Won Cho1*
Hee Won Cho1* Eun-Suk Kang3*
Eun-Suk Kang3*Introduction: The Predicted Indirectly ReCognizable HLA Epitopes (PIRCHE) model is a recently developed algorithm that predicts indirect T-cell recognition by calculating the number of such epitopes in donor-recipient pairs.
Methods: In this study, the clinical significance of PIRCHE was evaluated in pediatric patients with relapsed/progressed neuroblastoma undergoing haploidentical stem cell transplantation (haplo-SCT).
Results: A higher PIRCHE-I score was associated with faster platelet recovery (P = 0.007) and lower incidence of hemorrhagic cystitis (13% vs. 41%, P = 0.028) and invasive fungal infections (0% vs. 18%, P = 0.045). Additionally, a higher PIRCHE-I score was significantly associated with better overall survival (OS) (HR 0.57, 95% CI 0.34-0.97, P = 0.038). A higher PIRCHE-II score was associated with better OS (HR 0.57, 95% CI 0.34-0.94, P = 0.028) and reduced progression (HR 0.48, 95% CI 0.30-0.77, P = 0.002). When combined, the PIRCHE-I and PIRCHE-II scores demonstrated an even stronger association with improved OS (HR 0.35, 95% CI 0.15-0.82, P = 0.016). Multivariable analysis confirmed that a higher combined PIRCHE-I and PIRCHE-II score was independently associated with improved OS (combined PIRCHE score HR 0.22, 95% CI 0.06-0.79, P = 0.021), and a higher PIRCHE-II score was significantly associated with reduced progression (HR 0.42, 95% CI 0.25-0.70, P < 0.001).
Conclusion: In conclusion, higher PIRCHE-I and PIRCHE-II scores are linked to better survival outcomes and reduced complications in pediatric haplo-SCT neuroblastoma patients. Incorporating PIRCHE scores into donor selection is expected to optimize transplant outcomes.
The degree of human leucocyte antigen (HLA) disparity between donor and recipient is a dominant determinant of clinical outcomes after allogeneic stem cell transplantation (allo-SCT) and is the most important consideration in donor selection. Matching for HLA-A, -B, -C, -DRB1, and -DQB1 alleles (so-called 10/10 match) between the patient and donor is the gold standard for unrelated donor selection (1). Nevertheless, graft-versus-host-disease (GVHD) can occur even in patients transplanted with 10/10 HLA-matched grafts, while it might be absent in certain mismatched transplants (2).
Recently, epitope-based HLA matching algorithms, such as HLAMatchmaker and Predicted Indirectly ReCognizable HLA Epitopes (PIRCHE) have been developed for solid organ and hematopoietic stem cell transplantation (HSCT). HLAMatchmaker evaluates HLA disparity at the eplet level by counting the number of eplet differences between donor and recipient alleles (3–5). Meanwhile, PIRCHE predicts disparity by calculating the number of HLA-derived peptides potentially presented by patient-donor shared HLA molecules, which can trigger an indirect alloreactive response (6). The PIRCHE algorithm, first described in 2013 by Otten et al. in the context of kidney transplantation (7), has since been adapted for stem cell transplant domains (8). The PIRCHE algorithm predicts immunological disparity by calculating the number of HLA-derived peptides potentially presented by shared HLA molecules between donor and recipient. It incorporates several computational steps, including validation of input data, imputation of HLA genotyping data, HLA sequence extrapolation, and peptide binding predictions. The current PIRCHE application, implemented as a web-based tool (www.pirche.com), supports HLA peptide binding prediction using specialized predictors based on the structural properties of the HLA binding groove. The PIRCHE-I and PIRCHE-II scores indicate the number of allogenic HLA peptides presented by HLA class I and class II molecules, respectively (9, 10). Higher PIRCHE scores represent a greater number of potentially immunogenic HLA epitope mismatches, highlighting their theoretical clinical relevance. While the PIRCHE algorithm offers promising insights into immunological compatibility, its application in allo-SCT, particularly in haploidentical stem cell transplantation (haplo-SCT), remains underexplored. In haplo-SCT, characterized by significant donor-recipient HLA disparity, the integration of PIRCHE scoring could provide a more refined method for donor selection and risk stratification, potentially improving outcomes. However, further research is needed to validate the clinical utility of PIRCHE scores in this setting.
In parallel, high-dose chemotherapy and autologous stem cell transplantation (HDCT/auto-SCT) is a crucial part in the treatment of high-risk neuroblastoma (11–13). However, many patients experience relapse/progression, even after tandem HDCT/auto-SCT. The prognosis after relapse/progression has been dismal despite multimodal salvage treatment. In such patients, allo-SCT with graft-versus-tumor (GVT) effect can be a treatment option (14). Haplo-SCT with or without preceding high-dose 131I-metaiodobenzylguanidine (HD-MIBG) treatment has been performed as an attempt to enhance the anti-tumor effect in patients with relapsed/progressed neuroblastoma and has shown tolerable toxicity (15, 16).
In this context, our study aims to evaluate the impact of PIRCHE scores on the outcome after haplo-SCT in pediatric patients with relapsed neuroblastoma. We hypothesize that PIRCHE scores could have an impact on outcomes following haplo-SCT, thereby offering guidance for haploidentical donor selection.
This study was approved by the Institutional Review Board of Samsung Medical Center (IRB No. SMC 2024-10-055). Patients with neuroblastoma who underwent haplo-SCT from 2012 to 2023 due to relapse/progression after tandem HDCT/auto-SCT were eligible for this study. Salvage chemotherapy was administered to reduce the tumor burden prior to haplo-SCT. The regimen type and duration of chemotherapy depended on tumor response and patient tolerance. Surgical resection and local radiotherapy were performed whenever possible.
At 21 days prior to haplo-SCT, all patients received a single one-hour intravenous infusion of 131I-MIBG (18 mCi/kg) with potassium iodide for thyroid protection and intravenous hydration. Patients received reduced-intensity conditioning consisting of cyclophosphamide (60 mg/kg on days -7 and -6), fludarabine (30 mg/m2 on days -5 to -1), and rabbit anti-thymocyte globulin [Thymoglobulin, Genzyme; 2.5 mg/kg on days -4 (-3 since 2017) to -1].
Patients received T cell-replete grafts from 2012 to 2016, and α/β T cell-depleted grafts have been used since 2017. While cyclosporine (CSA) and short-course methotrexate (15 mg/m2 on day 1 and at 10 mg/m2 on days 3 and 6, followed by folic acid rescue) were used to prevent GVHD after haplo-SCT without T cell depletion, only CSA was administered from day -1 in T cell-depleted haplo-SCT. The total duration and tapering schedule of CSA were determined individually according to GVHD severity. Severities of acute and chronic GVHD were recorded based on previously described standard clinical criteria (17).
Antifungal prophylaxis was administered from absolute neutrophil count (ANC) < 0.5×109/L to ANC ≥ 1.0×109/L or during corticosteroid treatment. Acyclovir was used to prevent viral reactivation by day 30, and prophylactic trimethoprim-sulfamethoxazole was administered from neutrophil engraftment until day 180 or immunosuppressant discontinuation. Routine surveillance for cytomegalovirus (CMV), Epstein-Barr virus (EBV), and BK polyomavirus (BKV) reactivation was performed weekly during the first three months post-transplant and monthly thereafter if no viral reactivation occurred. If α/β T cell-depleted grafts were used, rituximab was prophylactically used on day 1. If CMV or EBV load increased, ganciclovir or rituximab was initiated preemptively, respectively.
Tumor response was assessed prior to HD-MIBG treatment, every three months during the first-year post-transplant, every four months during the second year, every six months during the third year, and annually thereafter. International response criteria for neuroblastoma were used to evaluate treatment response (18). When tumors persisted or relapse/progression occurred, salvage treatment was administered upon request.
High-resolution HLA typing was performed with either Sanger sequence-based typing (SBT) (Allele SEQR SBT kit; Atria Genetics, San Francisco, CA; GeneAmp PCR system, Applied Biosystems) or next-generation sequencing (NGS) (AllType™ NGS 11-Loci kit, OneLambda, USA; Illumina NextSeq System). Two-field allele level data for HLA-A, -B, -C, -DRB1, and -DQB1 were acquired from both patients and donors. Killer cell immunoglobulin-like receptor (KIR) genotyping was performed on donor DNA samples using a PCR-based sequence-specific oligonucleotide technique (LIFECODES KIR Typing Kit, Immucor Transplant Diagnostics, Inc., Stanford, CT). Based on HLA type and KIR genotype data, a KIR/HLA-ligand mismatch was defined as incompatibility between the inhibitory donor KIR and recipient HLA class I alleles, as previously described (19). Donor KIR haplotypes were categorized as AA (homozygous for group A KIR haplotypes) or BX (A/B heterozygotes or B/B homozygotes). The KIR B haplotype-defining loci were KIR2DL5, 2DS1, 2DS2, 2DS3, 2DS5, and 3DS1 (20). A haploidentical parent donor with a BX haplotype and/or KIR/HLA-ligand mismatch was preferred.
PIRCHE scores were acquired through the online PIRCHE matching tool (https://www.pirche.com) by inputting HLA allele data of the patient-donor pairs. PIRCHE scores corresponding to the number of allogeneic HLA peptides capable of inducing an indirect alloreactive response were generated. PIRCHE scores for allogeneic peptides presented by HLA class I molecules (PIRCHE-I) and class II molecules (PIRCHE-II) were computed separately in the GVH direction.
All statistical analyses were performed using R software version 4.4.0 (R Foundation for Statistical Computing, Vienna, Austria). Patient characteristics and transplant-related information were summarized using descriptive statistics. Continuous variables were expressed as medians with ranges, and categorical variables as frequencies and percentages. Non-hematologic outcomes were compared using chi-square tests or Fisher’s exact tests for categorical variables and the Mann-Whitney U test for continuous variables. The PIRCHE scores were dichotomized at their median values to evaluate their association with transplant-related complications, such as acute and chronic graft-versus-host disease (GVHD), hemorrhagic cystitis, and infections. This approach was chosen due to the absence of a universally established cutoff for PIRCHE scores in the existing literature. Using the median as the threshold allowed for a pragmatic and unbiased stratification of the cohort to explore the potential impact of higher versus lower PIRCHE scores on transplant outcomes. Survival outcomes, including overall survival (OS) and progression-free survival (PFS), were analyzed using Cox proportional hazards regression models. Both univariable and multivariable analyses were conducted. Variables with a P value < 0.1 in the univariable analysis were included in the multivariable models. PIRCHE-I and PIRCHE-II scores were treated as continuous variables and log-transformed to normalize their distributions. The impact of these scores on survival outcomes was assessed both individually and in combination (PIRCHE-I+II). Hazard ratios (HRs) with 95% confidence intervals (CIs) were reported. All tests were two-sided, and a P value < 0.05 was considered statistically significant.
Patient characteristics are detailed in Table 1. A total of 46 patients were included in this study. The median age at haplo-SCT was 6.6 years (range 3.9 to 12.1). Among patients with known pathology and tumor biology, 7 patients (18%) had tumors with MYCN amplification, and 31 patients (67%) had poorly differentiated or undifferentated tumors. The pre-haplo-SCT status was complete response (CR) or very good partial response (VGPR) in 18 patients (39%) and partial response or worse (≤ PR) in 28 patients (61%).
Regarding the transplant characteristics, a median of 8.6 (range 2.2–30.1) × 106 CD34+ cells/kg was transplanted. T cell-replete haplo-SCT was performed in 27% of the patients, with a median of 4.8 (range 1.9–10.5) × 108 CD3+ cells/kg transplanted. α/β T cell-depleted haplo-SCT was performed in 63% of the patients, with a median of 1.9 (range 0.1–22.1) × 104 α/β T cells/kg transplanted. KIR haplotype analysis indicated that 35% of the patients (16 patients) had haplotype A, and 65% (30 patients) had haplotype B. The median PIRCHE-I and PIRCHE-II scores were 21 (range 2–58) and 32 (range 4–102), respectively.
PIRCHE scores were categorized as high and low based on the median value, and transplant outcomes were analyzed according to these stratified score groups (Table 2). For PIRCHE-I, despite chimerism at day 30 being slightly higher in the low-score group compared to the high-score group (P = 0.044), there was a statistically significant difference in the time to achieve a platelet count of 200,000/µL, with the high-score group recovering faster than the low-score group (P = 0.007). Regarding non-hematologic outcomes, the incidence of hemorrhagic cystitis was significantly higher in the low-score group (41%) compared to the high-score group (13%, P = 0.028), and invasive fungal infections were more common in the low-score group (18%) compared to none in the high-score group (P = 0.045).
For PIRCHE-II, there were no significant differences in both hematologic outcomes and other complications. Chronic GVHD was slightly more prevalent in the high-score group (32%) compared to the low-score group (20%), though this was not statistically significant (P = 0.605).
We conducted both univariable and multivariable Cox proportional hazards regression analyses to assess the impact of PIRCHE scores on survival outcomes, treating these scores as continuous variables and applying a log transformation.
In the univariable analysis, several significant associations between higher PIRCHE scores and survival outcomes were identified. For PIRCHE-I, higher scores were significantly associated with a lower risk of death (HR 0.57, 95% CI 0.34-0.97, P = 0.038) (Figure 1A). The high PIRCHE-II scores were significantly associated with both better OS (HR 0.57, 95% CI 0.34-0.94, P = 0.028) (Figure 1A) and reduced progression (HR 0.48, 95% CI 0.30-0.77, P = 0.002) (Figure 1B). The combined effect of higher PIRCHE I+II scores showed a more significant association with improved OS (HR 0.35, 95% CI 0.15-0.82, P = 0.016) (Figure 1A).
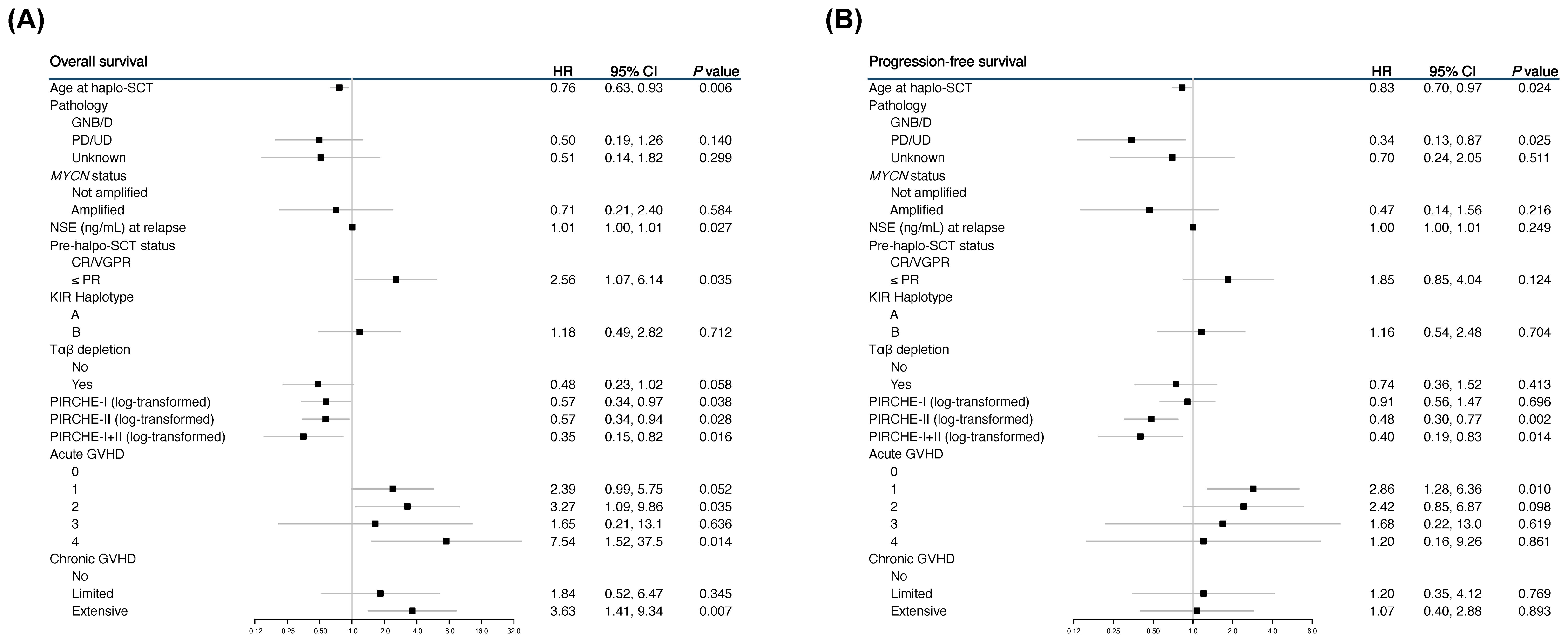
Figure 1. Forest plots showing the univariable analyses for overall survival (OS) (A) and progression-free survival (PFS) (B).
The multivariable analysis included variables with P value < 0.1 from the univariable analysis (Figure 2). For OS, higher log-transformed PIRCHE scores remained significantly associated with better outcomes. Specifically, the log-transformed combined PIRCHE score had a hazard ratio of 0.22 (95% CI 0.06-0.79, P = 0.021). Additionally, older age at haplo-SCT and receiving Tαβ depletion was significantly associated with better OS (HR 0.32, 95% CI 0.12-0.85, P = 0.023), while poorer response to treatment before haplo-SCT (≤ PR) was associated with worse OS (HR 7.07, 95% CI 1.86-26.8, P = 0.004). For progression, the multivariable analysis showed that higher PIRCHE-II scores were most significantly associated with a reduced risk (HR 0.42, 95% CI 0.25-0.70, P < 0.001).
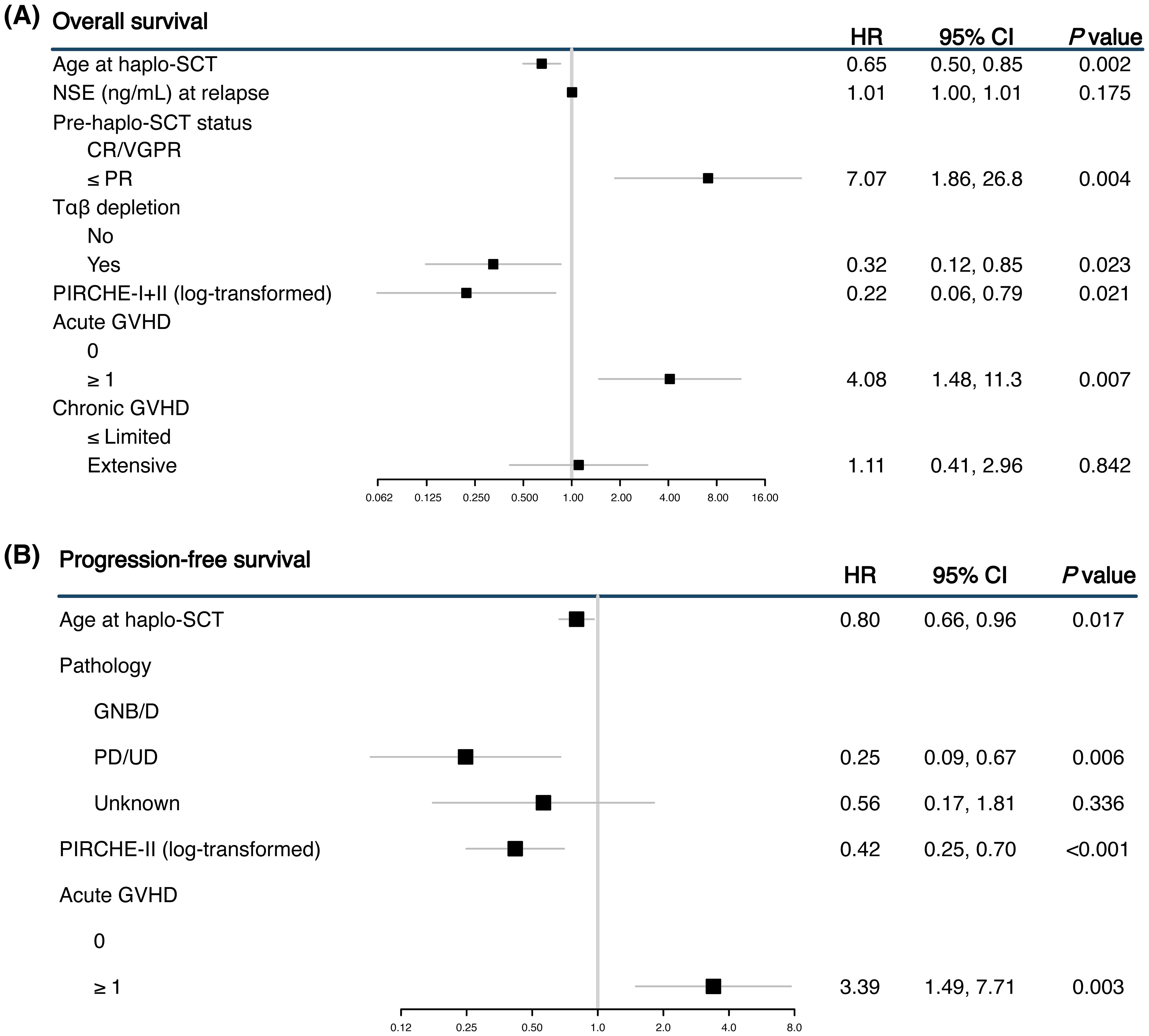
Figure 2. Forest plots showing the multivariable analyses for overall survival (OS) (A) and progression-free survival (PFS) (B). The multivariable analysis was limited to variables that showed significance in the univariable analysis.
Haplo-SCT preceded by HD-MIBG treatment is a feasible option for children with high-risk neuroblastoma who experienced relapse/progression after HDCT/auto-SCT (16, 21, 22). Although outcomes are not yet satisfactory, efforts should be made to improve survival, and selection of an optimal donor is an essential part of these efforts. In this study, we investigated the validity of the PIRCHE model in haploidentical donor selection in pediatric patients with relapsed/progressed neuroblastoma. A higher PIRCHE-I score was associated with faster hematologic recovery, less infection and better OS, while a higher PIRCHE-II score was associated with better PFS and OS in children with relapsed neuroblastoma who underwent haplo-SCT. Altogether, these findings support integration of the PIRCHE scoring system into the donor selection process for haplo-SCT in children with neuroblastoma.
Our study adds to the growing body of literature on the relationship between PIRCHE scores and transplant outcomes. Prior studies, primarily conducted in adults with hematologic diseases, reported conflicting results regarding PIRCHE scores and outcomes such as GVHD, non-relapse mortality and disease relapse (1, 8, 10, 23–27). These discrepancies likely arise from differences in patient populations, the transplantation protocols, HLA disparities, and clinical end points. In our pediatric neuroblastoma cohort, we observed distinct associations between PIRCHE scores and transplant outcomes, highlighting the context dependent nature of these metrics.
Patients with higher PIRCHE-I scores experienced faster platelet recovery and a reduced incidence of hemorrhagic cystitis and fungal infections, suggesting that HLA class I mismatches enhance hematologic recovery and immune reconstitution primarily through cytotoxic CD8+ T cells. Since HLA class I molecules are predominantly recognized by CD8+ T cells, these T cells may contribute to hematologic recovery by targeting and eliminating residual host hematopoietic cells, thereby promoting engraftment. Additionally, the enhanced immune recognition mediated by CD8+ T cells may improve immunity, reducing the risk of infections.
In contrast, higher PIRCHE-II scores, representing HLA class II mismatches, were not significantly associated with non-survival outcomes but were correlated with improved PFS and OS. These findings suggest that class II molecules, recognized by CD4+ helper T cells, likely contribute to GVT effects, which are critical for reducing relapse rates in neuroblastoma. This finding aligns with previous reports that HLA class II mismatches can enhance survival and GVT effects in haplo-SCT without significantly increasing in non-relapse mortality (28, 29). Specifically, mismatches at HLA-DR and HLA-DQ have been linked to enhanced survival and GVT effects. These results reinforce our findings that HLA class II mismatches had a more prominent role in GVT effects than class I mismatches in this setting.
In solid organ transplantation, higher PIRCHE-II scores may trigger an aggressive host-versus-graft immune response, potentially leading to graft rejection (30). However, in hematopoietic SCT, host-versus-graft reactions are minimized by conditioning regimens designed to suppress host immunity, emphasizing graft-versus-host interactions. The observed GVT effects, mediated by HLA class II mismatches, likely contributed to improved outcomes in our cohort.
Although greater HLA mismatches have been associated with increased GVHD risk in some studies (26), we did not demonstrate an association between PIRCHE score and GVHD incidence. A possible explanation is that the use of anti-thymocyte globulin with or without ex-vivo T cell depletion and reduced intensity conditioning could have weakened the impact of donor-recipient HLA epitope mismatches on GVHD. The absence of GVHD association in our study suggests the potential utility of PIRCHE scores for guiding donor selection without increasing GVHD risk. However, these results should be interpreted with caution, as the outcomes observed in this study may be influenced by the unique characteristics of our cohort and may not be generalizable to all patient populations.
While earlier investigations have yielded mixed results regarding PIRCHE scores and survival outcomes (8, 23, 25), our findings in pediatric neuroblastoma underscore the unique predictive value of these scores. Unlike studies that focused on adult patients with hematologic diseases or included transplants with varying levels of HLA disparity (10, 25–27), our study found significant associations between both PIRCHE-I and PIRCHE-II scores and OS. These results may be attributed to the lower incidence of infectious complications with higher PIRCHE-I scores and lower risk of relapse/progression associated with higher PIRCHE-II scores.
Emerging approaches that combine PIRCHE scores with other analyses could further enhance predictions of transplant outcomes (31–33). For example, integrating PIRCHE scores with eplet mismatch analyses could improve outcome predictions by simulating both B-cell- and T-cell-mediated alloimmune responses. These strategies, already promising in solid organ transplantation, may also refine donor selection in hematopoietic SCT by predicting donor-specific antibody production, which impacts engraftment and infection risk (34). Novel epitope-based algorithms such as Snowflake (35), could offer additional opportunities to improve predictive accuracy and clinical outcomes.
This study has several limitations. The retrospective design and single-center nature may limit the generalizability of our findings. Additionally, the relatively small sample size may affect the statistical power of our analyses, particularly for less common outcomes. Changes in GVHD prophylaxis strategy during the study period also are a limitation of our study, leading to heterogeneity of the cohort. Furthermore, as haplo-SCT is not well-established for childhood solid tumors, including neuroblastoma, we were unable to confirm the observed associations when applying different conditioning regimens or GVHD prophylaxis approaches, such as post-transplant cyclophosphamide. This variability limits direct comparisons with other studies and emphasizes the need for standardized protocols in this context. Another notable limitation is the lack of data for HLA-DPB1 and DRB3/4/5 loci in most donor-recipient pairs. While allelic matching at HLA-A, -B, -C, and -DRB1 is considered the standard for unrelated HCT, increasing evidence suggests that matching at DPB1 and DRB3/4/5 loci also impacts transplant survival and GVHD risk. Recent studies indicate that mismatches at these loci, despite lower expression levels, can influence transplant outcomes (1, 8, 20, 36–38). Incorporating these loci into PIRCHE score calculations may improve the predictive accuracy of outcomes in future studies. Lastly, we acknowledge that the use of arbitrary dichotomization for stratifying patients based on PIRCHE scores may be another limitation. While this approach allowed for a balanced comparison, future studies may explore alternative methods to enhance statistical robustness.
Despite these limitations, our cohort is strikingly different from those in other cohorts in that they are children with relapsed neuroblastoma who underwent haplo-SCT from a parent donor and received relatively uniform treatments compared to other cohorts. This unique patient population underscores the potential value of the PIRCHE scoring system in optimizing donor selection and improving outcomes.
In conclusion, our study demonstrates that higher PIRCHE-I and PIRCHE-II scores are associated with improved survival outcomes and reduced post-transplant complications in children with relapsed neuroblastoma undergoing haplo-SCT. These findings support the incorporation of PIRCHE scoring into pre-transplant risk assessment and patient stratification to optimize transplant outcomes. Further research should aim to validate these findings, explore the underlying mechanisms, and evaluate broader clinical applications of PIRCHE scores in the haplo-SCT setting.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by the Institutional Review Board of Samsung Medical Center. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because As this was a retrospective study, informed consent was waived for the study subjects.
ES: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. IJ: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. HJ: Methodology, Writing – review & editing. JH: Methodology, Writing – review & editing. JL: Writing – review & editing. KY: Writing – review & editing. WH: Writing – review & editing. KS: Conceptualization, Methodology, Supervision, Writing – review & editing. HC: Conceptualization, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing. EK: Conceptualization, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant, funded by the Korean government (MSIT) (2017R1A2B4008178, 2022R1F1A1075238). Additionally, this research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HA15C0007).
We acknowledge the contribution of our patients and clinical staff working at Division of Pediatric Hematology and Oncology, Samsung Medical Center.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Stenger W, Künkele A, Niemann M, Todorova K, Pruß A, Schulte JH, et al. Donor selection in a pediatric stem cell transplantation cohort using PIRCHE and HLA-DPB1 typing. Pediatr Blood Cancer. (2020) 67:e28127. doi: 10.1002/pbc.28127
2. Group SCTC. Allogeneic peripheral blood stem-cell compared with bone marrow transplantation in the management of hematologic Malignancies: an individual patient data meta-analysis of nine randomized trials. J Clin Oncol. (2005) 23:5074–87. doi: 10.1200/jco.2005.09.020
3. Duquesnoy RJ. A structurally based approach to determine HLA compatibility at the humoral immune level. Hum Immunol. (2006) 67:847–62. doi: 10.1016/j.humimm.2006.08.001
4. Duquesnoy RJ. HLAMatchmaker: a molecularly based algorithm for histocompatibility determination. I. Description of the algorithm. Hum Immunol. (2002) 63:339–52. doi: 10.1016/s0198-8859(02)00382-8
5. Duquesnoy RJ. Clinical usefulness of HLAMatchmaker in HLA epitope matching for organ transplantation. Curr Opin Immunol. (2008) 20:594–601. doi: 10.1016/j.coi.2008.06.010
6. Geneugelijk K, Spierings E. PIRCHE-II: an algorithm to predict indirectly recognizable HLA epitopes in solid organ transplantation. Immunogenetics. (2020) 72:119–29. doi: 10.1007/s00251-019-01140-x
7. Niemann M, Matern BM, Spierings E. PIRCHE-II risk and acceptable mismatch profile analysis in solid organ transplantation. Methods Mol Biol. (2024) 2809:171–92. doi: 10.1007/978-1-0716-3874-3_12
8. Thus KA, Ruizendaal MT, de Hoop TA, Borst E, van Deutekom HW, Te Boome L, et al. Refinement of the definition of permissible HLA-DPB1 mismatches with predicted indirectly recognizable HLA-DPB1 epitopes. Biol Blood Marrow Transplant. (2014) 20:1705–10. doi: 10.1016/j.bbmt.2014.06.026
9. Otten HG, Calis JJ, Keşmir C, van Zuilen AD, Spierings E. Predicted indirectly recognizable HLA epitopes presented by HLA-DR correlate with the de novo development of donor-specific HLA IgG antibodies after kidney transplantation. Hum Immunol. (2013) 74:290–6. doi: 10.1016/j.humimm.2012.12.004
10. Ayuk F, Bornhäuser M, Stelljes M, Zabelina T, Wagner EM, Schmid C, et al. Predicted indirectly reCognizable HLA epitopes (PIRCHE) are associated with poorer outcome after single mismatch unrelated donor stem cell transplantation: A study of the cooperative transplant study group (KTS) of the german group for bone marrow and stem cell transplantation (DAG-KBT). Transfus Med Hemother. (2019) 46:370–5. doi: 10.1159/000502389
11. George RE, Li S, Medeiros-Nancarrow C, Neuberg D, Marcus K, Shamberger RC, et al. High-risk neuroblastoma treated with tandem autologous peripheral-blood stem cell-supported transplantation: long-term survival update. J Clin Oncol. (2006) 24:2891–6. doi: 10.1200/jco.2006.05.6986
12. Matthay KK, Reynolds CP, Seeger RC, Shimada H, Adkins ES, Haas-Kogan D, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children’s oncology group study. J Clin Oncol. (2009) 27:1007–13. doi: 10.1200/jco.2007.13.8925
13. Sung KW, Son MH, Lee SH, Yoo KH, Koo HH, Kim JY, et al. Tandem high-dose chemotherapy and autologous stem cell transplantation in patients with high-risk neuroblastoma: results of SMC NB-2004 study. Bone Marrow Transplant. (2013) 48:68–73. doi: 10.1038/bmt.2012.86
14. Marabelle A, Paillard C, Tchirkov A, Halle P, Chassagne J, Deméocq F, et al. Graft-versus-tumour effect in refractory metastatic neuroblastoma. Bone Marrow Transplant. (2007) 39:809–10. doi: 10.1038/sj.bmt.1705681
15. Toporski J, Garkavij M, Tennvall J, Ora I, Gleisner KS, Dykes JH, et al. High-dose iodine-131-metaiodobenzylguanidine with haploidentical stem cell transplantation and posttransplant immunotherapy in children with relapsed/refractory neuroblastoma. Biol Blood Marrow Transplant. (2009) 15:1077–85. doi: 10.1016/j.bbmt.2009.05.007
16. Sung KW, Park JE, Chueh HW, Lee SH, Yoo KH, Koo HH, et al. Reduced-intensity allogeneic stem cell transplantation for children with neuroblastoma who failed tandem autologous stem cell transplantation. Pediatr Blood Cancer. (2011) 57:660–5. doi: 10.1002/pbc.23035
17. Filipovich AH, Weisdorf D, Pavletic S, Socie G, Wingard JR, Lee SJ, et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol Blood Marrow Transplant. (2005) 11:945–56. doi: 10.1016/j.bbmt.2005.09.004
18. Park JR, Bagatell R, Cohn SL, Pearson AD, Villablanca JG, Berthold F, et al. Revisions to the international neuroblastoma response criteria: A consensus statement from the national cancer institute clinical trials planning meeting. J Clin Oncol. (2017) 35:2580–7. doi: 10.1200/jco.2016.72.0177
19. Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. (1999) 94:333–9. doi: 10.1182/blood.V94.1.333.413a31_333_339
20. Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. (2009) 113:726–32. doi: 10.1182/blood-2008-07-171926
21. Choi YB, Son MH, Cho HW, Ma Y, Lee JW, Kang ES, et al. Safety and immune cell kinetics after donor natural killer cell infusion following haploidentical stem cell transplantation in children with recurrent neuroblastoma. PloS One. (2019) 14:e0225998. doi: 10.1371/journal.pone.0225998
22. Lee JW, Kang ES, Sung KW, Yi E, Lee SH, Yoo KH, et al. Incorporation of high-dose (131) I-metaiodobenzylguanidine treatment into killer immunoglobulin-like receptor/HLA-ligand mismatched haploidentical stem cell transplantation for children with neuroblastoma who failed tandem autologous stem cell transplantation. Pediatr Blood Cancer. (2017) 64:e26399. doi: 10.1002/pbc.26399
23. Grubic Z, Burek Kamenaric M, Maskalan M, Durakovic N, Vrhovac R, Stingl Jankovic K, et al. Various approaches for accessing the influence of human leukocyte antigens disparity in haploidentical stem cell transplantation. Int J Lab Hematol. (2022) 44:547–57. doi: 10.1111/ijlh.13801
24. Rimando J, Slade M, DiPersio JF, Westervelt P, Gao F, Liu C, et al. The predicted indirectly recognizable HLA epitopes (PIRCHE) score for HLA class I graft-versus-host disparity is associated with increased acute graft-versus-host disease in haploidentical transplantation with post-transplantation cyclophosphamide. Biol Blood Marrow Transplant. (2020) 26:123–31. doi: 10.1016/j.bbmt.2019.09.024
25. Geneugelijk K, Thus KA, van Deutekom HWM, Calis JJA, Borst E, Keşmir C, et al. Exploratory study of predicted indirectly reCognizable HLA epitopes in mismatched hematopoietic cell transplantations. Front Immunol. (2019) 10:880. doi: 10.3389/fimmu.2019.00880
26. Huo MR, Li D, Chang YJ, Xu LP, Zhang XH, Liu KY, et al. Predicted indirectly recognizable HLA epitopes are not associated with clinical outcomes after haploidentical hematopoietic stem cell transplantation. Hum Immunol. (2018) 79:117–21. doi: 10.1016/j.humimm.2017.11.004
27. Thus KA, de Hoop TA, de Weger RA, Bierings MB, Boelens JJ, Spierings E. Predicted indirectly reCognizable HLA epitopes class I promote antileukemia responses after cord blood transplantation: indications for a potential novel donor selection tool. Biol Blood Marrow Transplant. (2016) 22:170–3. doi: 10.1016/j.bbmt.2015.08.014
28. Solomon SR, Aubrey MT, Zhang X, Jackson KC, Morris LE, Holland HK, et al. Class II HLA mismatch improves outcomes following haploidentical transplantation with posttransplant cyclophosphamide. Blood Adv. (2020) 4:5311–21. doi: 10.1182/bloodadvances.2020003110
29. Petersdorf EW, McKallor C, Malkki M, He M, Spellman SR, Gooley T, et al. HLA haplotypes and relapse after hematopoietic cell transplantation. J Clin Oncol. (2024) 42:886–97. doi: 10.1200/jco.23.01264
30. Senev A, Van Loon E, Lerut E, Coemans M, Callemeyn J, Daniëls L, et al. Association of predicted HLA T-cell epitope targets and T-cell-mediated rejection after kidney transplantation. Am J Kidney Dis. (2022) 80:718–29.e1. doi: 10.1053/j.ajkd.2022.04.009
31. Jäger C, Niemann M, Hönger G, Wehmeier C, Hopfer H, Menter T, et al. Combined molecular mismatch approaches to predict immunological events within the first year after renal transplantation. Hla. (2024) 104:e15748. doi: 10.1111/tan.15748
32. Sakamoto S, Iwasaki K, Tomosugi T, Niemann M, Spierings E, Miwa Y, et al. Analysis of T and B Cell Epitopes to Predict the Risk of de novo Donor-Specific Antibody (DSA) Production After Kidney Transplantation: A Two-Center Retrospective Cohort Study. Front Immunol. (2020) 11:2000. doi: 10.3389/fimmu.2020.02000
33. Bezstarosti S, Kramer CSM, Claas FHJ, de Fijter JW, Reinders MEJ, Heidt S. Implementation of molecular matching in transplantation requires further characterization of both immunogenicity and antigenicity of individual HLA epitopes. Hum Immunol. (2022) 83:256–63. doi: 10.1016/j.humimm.2021.12.002
34. Huang Y, Luo C, Wu G, Huang X, Ding Y, Huang Z, et al. Effects of donor-specific antibodies on engraftment and long-term survival after allogeneic hematopoietic stem cell transplantation-A systematic review and meta-analysis. Bone Marrow Transplant. (2023) 58:544–51. doi: 10.1038/s41409-023-01932-6
35. Niemann M, Strehler Y, Lachmann N, Halleck F, Budde K, Hönger G, et al. Snowflake epitope matching correlates with child-specific antibodies during pregnancy and donor-specific antibodies after kidney transplantation. Front Immunol. (2022) 13:1005601. doi: 10.3389/fimmu.2022.1005601
36. Ducreux S, Dubois V, Amokrane K, Yakoub-Agha I, Labalette M, Michallet M, et al. HLA-DRB3/4/5 mismatches are associated with increased risk of acute GVHD in 10/10 matched unrelated donor hematopoietic cell transplantation. Am J Hematol. (2018) 93:994–1001. doi: 10.1002/ajh.25133
37. van Balen P, van Luxemburg-Heijs SAP, van de Meent M, van Bergen CAM, Halkes CJM, Jedema I, et al. Mismatched HLA-DRB3 can induce a potent immune response after HLA 10/10 matched stem cell transplantation. Transplantation. (2017) 101:2850–4. doi: 10.1097/tp.0000000000001713
Keywords: neuroblastoma, PIRCHE, haploidentical hematopoietic stem cell transplantation, relapse/refractory, HLA mismatch
Citation: Seo ES, Jeong IH, Ju HY, Hyun JK, Lee JW, Yoo KH, Heo WY, Sung KW, Cho HW and Kang E-S (2025) Predicted indirectly recognizable HLA epitopes scores and clinical outcomes after haploidentical stem cell transplantation in pediatric patients with relapsed neuroblastoma. Front. Immunol. 16:1517387. doi: 10.3389/fimmu.2025.1517387
Received: 26 October 2024; Accepted: 06 January 2025;
Published: 22 January 2025.
Edited by:
Aurore Saudemont, Xap Therapeutics, United KingdomReviewed by:
Terry Harville, University of Arkansas for Medical Sciences, United StatesCopyright © 2025 Seo, Jeong, Ju, Hyun, Lee, Yoo, Heo, Sung, Cho and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hee Won Cho, aHc4Ny5jaG9Ac2Ftc3VuZy5jb20=; Eun-Suk Kang, ZXNrYW5nQHNra3UuZWR1
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.