
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 06 March 2025
Sec. Cancer Immunity and Immunotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1486868
Recent studies on the pathogenesis of leukemia have led to remarkable advances in disease treatment. Numerous studies have shown the potential and viability of immune responses against leukemia. In the classical pathway, this process is often initiated by the upstream activity of CD39, which hydrolyzes extracellular adenosine triphosphate (ATP) and adenosine diphosphate (ADP) to AMP. Subsequently, CD73 acts on AMP to generate adenosine, contributing to an immunosuppressive microenvironment. However, CD73 can also utilize substrates derived from other molecules through the non-canonical NAD+ pathway, specifically via the CD38/CD203a/CD73 axis, further enhancing adenosine production and facilitating immune escape. Targeting CD73 has shown potential in disrupting these immunosuppressive pathways, thereby enhancing anti-leukemic immune responses and improving patient outcomes. Inhibiting CD73 not only reduces the levels of immunosuppressive adenosine but also increases the efficacy of existing immunotherapies, such as PD-1/PD-L1 inhibitors, making it a versatile therapeutic target in leukemia treatment. This review discusses the potential of CD73 as a therapeutic target and emphasizes its unique position in the immune escape mechanism of leukemia. Moreover, this review provides an overview of the current research progress and future trends, emphasizing the clinical significance of targeting CD73 and other potential therapeutic strategies in leukemia.
Immune checkpoints (ICs) are immunosuppressive pathways that have evolved to prevent excessive immune responses and the overactivation of immune cells. Tumor cells can exploit these checkpoint molecules, evading immune surveillance and thereby promoting immune escape and accelerated metastasis. Given their high proliferation rate, malignant hematoma cells in the tumor microenvironment (TME) hasten the onset and progression of leukemia by fostering tumor cell immune escape. The multiple subtypes of leukemia pose a challenge in identifying new immune targets. CD73, emerging as a potential immune target for solid tumors, is increasingly being recognized to be vital in the occurrence and progression of leukemia. CD39 hydrolyzes adenosine triphosphate (ATP) and adenosine diphosphate(ADP) to adenosine 5′-monophosphate (AMP), while CD73 further converts AMP into adenosine (1, 2). Moreover, the non-classical CD38/CD203a/CD73 pathway also generates adenosine, which plays a significant role in maintaining the immunosuppressive TME (3, 4). Research indicates that adenosine concentration influences the progression of various leukemia types (5–9). Consequently, understanding the role of CD73 in leukemia can unveil novel treatment possibilities. This review seeks to elucidate the functional role of CD73 in leukemia by analyzing its structure, function, and expression in other solid tumors and immune environments, thereby shedding light on the developmental mechanisms of CD73 in leukemia.
Human CD73 is encoded by the NT5E gene, which resides between positions 14–21 on the long arm of chromosome 6 (10). Two post-transcriptional isoforms of CD73 have been identified. CD73, also known as extracellular 5′-nucleotidase, is a ribosidase encoded by NT5E, with a molecular weight of approximately 70 kD. Most CD73 molecules are anchored to membranes by glycosylphosphatidylinositol (GPI), metabolizing extracellular AMP into adenosine and inorganic phosphate (10, 11). A fraction of CD73 exists in the soluble form, exhibiting activities and functions partially akin to its membrane-anchored counterpart (12–14).
When GPI anchors are cut by using PI-PLC, soluble CD73 is released, usually with increased catalytic activity (15). Although soluble catalytic efficiency can be higher than that of anchored CD73, due to substrate transfer, utilization efficiency and the fact that TNF-a cannot only affect PLC, the actual total adenosine production is not as good as that of anchored CD73 (16). CD73 also has a form that is anchored by GPI in small extracellular vesicles (sEVs) (17), which are derived from CD8+T cells and play an important role in the regulation of the immune microenvironment (18). Both anchored and soluble forms of CD73 exhibit distinct expression patterns in various tissues and across species. CD73 shows varied tissue expression, predominantly in the colon, kidneys, brain, liver, heart, lungs, spleen, lymph nodes, and bone marrow. In the vascular system, CD73 primarily associates with vascular and lymphatic endothelia, playing a role in regulating leukocyte transport (19). Within the immune system, CD73 is found on the surfaces of macrophages, lymphocytes, regulatory T cells (Tregs), and dendritic cells (20). Analysis of CD73 expression in immune cells reveals higher relative expression in naïve B cells, naïve CD8+ T cells, and memory B cells, compared with lower expression in CD4+ T cells (21). CD73 expression in immune cells varies by species; in humans, it is expressed on most B cells and certain T cell subsets (22, 23); in mice, it is expressed predominantly in T cells (Tregs, natural killer [NK] cells) and some mature B cells (24–27). In normal human peripheral blood, CD73 expression is rare in T cells and is mainly found in B cells (28, 29). CD73 has different functions in different tissues and cells. For example, it mainly plays an immunomodulatory function on the surface of lymphocytes, including the activation and proliferation of lymphocytes and the adhesion process with endothelial cells. However, CD73 acts as an adhesion molecule in endothelial cells and promotes lymphocyte migration (30). CD73 exerts its functions through various mechanisms in the tumor microenvironment, inhibiting the activation and effector function of T cells, inhibiting the killing effect of NK cells, and promoting immunosuppressor cells (MDSCs and Tregs) through CD39/CD73/A2AR pathway (31–33). In addition, the CD73/CD39 adenosine production pathway has recently been shown to promote the expression of stemness (34) and EMT-related genes (35–37), maintain the immunosuppressor microenvironment, and promote tumor progression and metastasis. NT5E, a typical hypoxia-inducible factor (HIF) target gene, undergoes alterations in expression and function in hypoxic conditions (38).
Beyond hypoxia, various inflammatory mediators including transforming growth factor (TGF)-β (39), interferons [IFNs (40)], tumor necrosis factor (TNF) (41), interleukin (IL)-1β (42), and prostaglandin E2 (43) can upregulate the expression and function of CD73 (44, 45). Furthermore, the Wnt pathway and cyclic AMP signaling pathway as well as unsaturated fatty acids can regulate CD73 expression, indicating its regulation by multiple factors (46). CD73 also exists in post-translationally modified forms, including glycosylated (47) and ubiquitinated variants (48).
Structurally, CD73 is composed of two covalently linked subunits, each approximately 70 kD, and is anchored to the cell membrane via a GPI anchor site. The N-terminal domain of CD73 binds divalent Zn2+ and Co2+ for catalytic activity, whereas its C-terminal domain serves as a binding site for AMP (49). Functionally, CD73 is involved in two key extracellular metabolic pathways: AMP and nicotinamide adenine dinucleotide (NAD+) metabolism. In the AMP pathway, CD39 sequentially converts extracellular ATP and ADP into AMP, which is then dephosphorylated by CD73 to produce adenosine. Adenosine acts as a key regulator of immunosuppressive signaling by binding to specific adenosine receptors on immune cells, leading to the suppression of inflammatory responses. Adenosine interacts with specific G-protein–coupled receptors, such as A2AR, on T cells (50). Within the NAD+ pathway, the essential coenzyme NAD+ is released into the extracellular environment. During NAD+ metabolism, nicotinamide is converted to AMP by the ectonucleotide pyrophosphatase/phosphodiesterase family and subsequently to adenosine by CD73 (51). In addition to its role as a protease, CD73 functions as an adhesion molecule, facilitating the migration of both normal and tumor cells (52).
The role of CD73 expression in cancer is demonstrated in Figure 1, showing how it facilitates adenosine production and contributes to immune evasion. Numerous studies use CD73 as an IC marker for adenosine production, investigating its role in solid tumors and inflammation. In-depth research on the tumor immune microenvironment reveals that immune effector and regulatory cells, located at the tumor periphery, generate inflammatory responses to counteract cancer cell proliferation during immune infiltration (53). Similar to its role in the immunosuppressive TME, CD73 converts AMP into adenosine. Increased adenosine concentrations weaken the tumor-killing ability of immune effector cells (T cells, B cells, NK cells). In NK cells, adenosine mainly inhibits cytotoxicity of NK cells through A2AR signal, and mediates PKA to participate in tumor immune escape through CAMP-dependent signal (1, 54). In both CD4+T and CD8+T cells, adenosine binds to adenosine A2A receptor (A2AR), but inhibits the proliferation of T helper 1 (Th1) and T helper 2 (Th2) cells (55, 56) and promotes the differentiation of T helper 17 (Th17) cells after binding (57). A2AR is highly expressed in CD8+T central memory cells (TCM) in the tumor microenvironment, which is easily regulated by adenosine and leads to functional depletion of CD8+T cells (58, 59). Similarly, in B cells, adenosine exerts immunosuppressive effects through the activation of A2AR. Regulatory B cells expressing CD39 and CD73 produce adenosine. This increase in adenosine concentration weakens effector cell activity and enhances control by immune regulatory cells (Tregs, myeloid-derived suppressor cells), enabling tumor evasion from immune surveillance and attack. This shift transforms the immune microenvironment of the tumor into an immunosuppressive state.
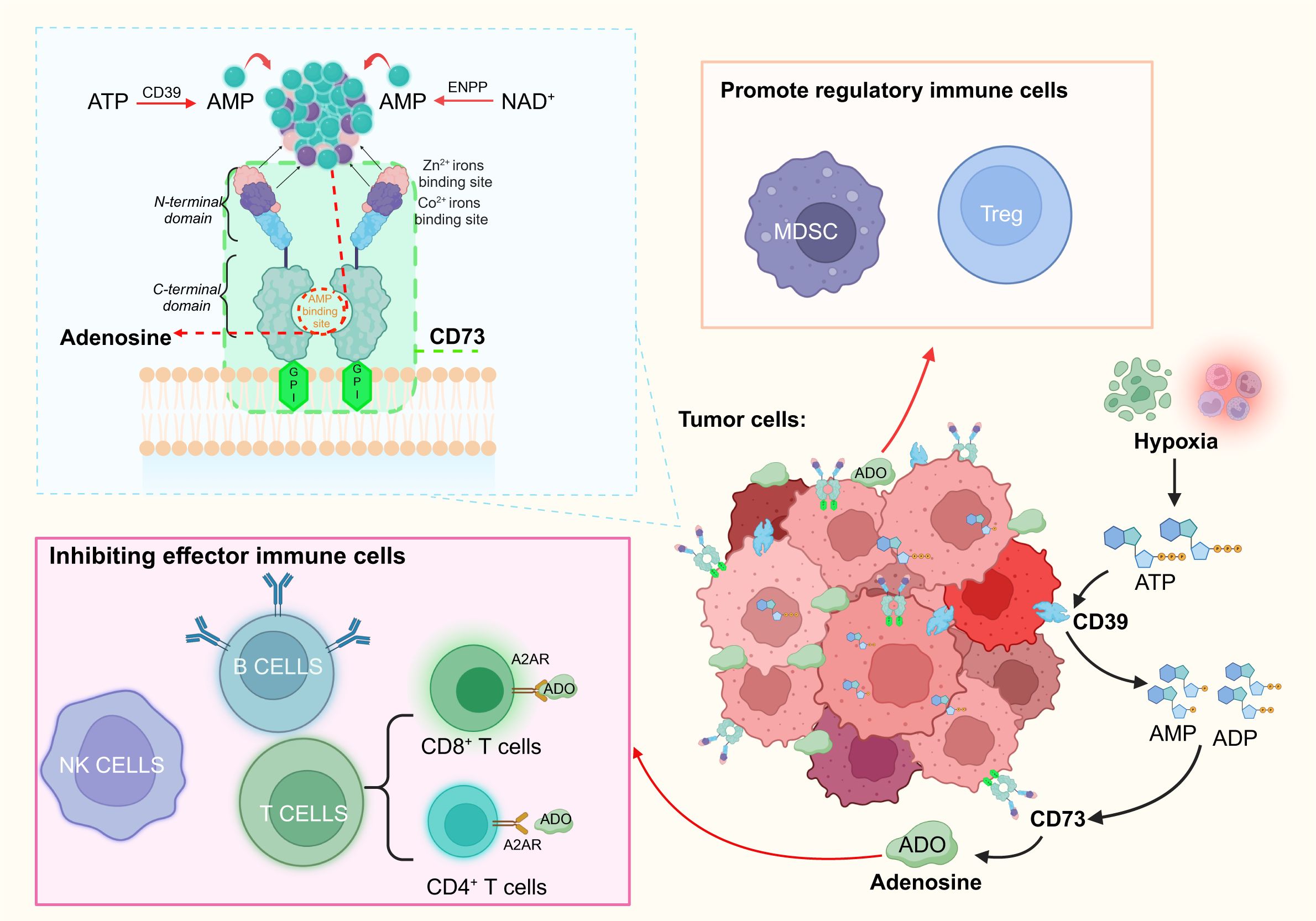
Figure 1. Schematic of the involvement of CD73 in the immune process through its structure and function, and the immune role of its product, adenosine, is briefly outlined. CD73, anchored to the membrane via GPI, is fully exposed on the exterior of the membrane. The C-terminal domain of CD73 specifically binds AMP to generate adenosine, whereas the N-terminal domain contains binding sites for Zn2+ and Co2+. Adenosine is produced via two primary pathways: the breakdown of ATP by CD39 and the NAD+ metabolism pathway. In tumor cells, the adenosine pathway facilitated by CD39/CD73 can suppress effector immune cells and bolster regulatory immune cells, thus sustaining the immunosuppressive tumor microenvironment. (Figure created with BioRender.com).
Overexpressed CD73 in cancer cells results in elevated adenosine concentrations. Concurrently, cancer cell proliferation fosters a hypoxic environment, leading to lysis of normal cells and ATP production. ATP is converted into adenosine via the CD39/CD73 pathway, facilitating the immune escape process, which further enables cancer cells to metastasize and persist in immune evasion and proliferation (60). Factors such as HIF-1α are overexpressed in a hypoxic immune microenvironment, thereby aiding tumor immune evasion and promoting tumor growth. This process triggers the release of key cytokines, such as IFN-γ and IL-4, which modulate the immune response, leading to the suppression of T cell activation and proliferation. Their overexpression inhibits T cell activation and proliferation, adversely impacting the immune responses of cells (61). Programmed death 1 (PD1)/paradigm of surface-expressed programmed death ligand 1 (PDL1) is a popular pathway target in cancer research and has important therapeutic potential, which can reduce T cell activity through the interaction of PD1 and PDL1 (62–65). Studies have shown that both PD1/PDL1 and CD73/A2AR pathways (66) play a positive role in the negative regulation of immune response (67). Moreover, it has been confirmed that the CD39/CD73 pathway does not directly suppress PD1/PDL1 activity, but rather impairs the function of CD8+ T cells through increased adenosine production, contributing to resistance against anti-PD1 therapies (68).
CD73 expression varies significantly across cancer cell lines. For example, the leukemia cell line K562 does not express CD73, while high levels of CD73 are observed in glioblastoma (GBM) and commercial GBM cell lines such as U87 (69). In contrast, bladder cancer cells (70), melanoma cells (71), breast cancer cells (72, 73), lung cancer cells (74), pancreatic cancer (75), non-small cell lung cancer (76), cervical cancer cells (77), medulloblastoma cells (78), glioma cells (79) and ovarian cancer cell lines (80) also show varying levels of CD73 expression. In immune cells, CD73 plays a crucial role in modulating immune responses, particularly in natural killer (NK) cells, where it suppresses cytotoxic activity via the A2A receptor (A2AR) signaling pathway. This pathway facilitates tumor immune evasion by engaging cAMP-dependent mechanisms (81).
The role of CD73 in solid tumors is more intricate. In lung adenocarcinoma and non-small cell lung cancer (NSCLC), CD73 expression is linked to elevated PD-L1 levels and an increase in tumor-associated immune cells, contributing to an immunosuppressive microenvironment (74, 76, 82–86). Furthermore, CD73 has prognostic, oncogenic, and immunosuppressive roles in head and neck squamous cell carcinoma (87, 88). In hepatocellular carcinoma (HCC), CD73 activates the PI3K/AKT signaling pathway, leading to increased AKT phosphorylation and promoting tumor growth (89, 90). Moreover, CD73 is associated with immunosuppression and poor prognosis in pancreatic ductal adenocarcinoma (PDAC) (91–96). In thyroid and cervical cancers, TGF-β1 production promotes CD73 upregulation, contributing to cancer progression (97–100). In gastric cancer (GC), CD73 promotes immune evasion by impairing CD8+ T cell function (101, 102). Additionally, CD73/CD39+ cells have been implicated in the immunomodulation of chronic human immunodeficiency virus (HIV) infections (103, 104). High CD73 expression in invasive renal cell carcinoma is linked to increased cancer-related mortality (105). Moreover, CD73 has been identified as an important prognostic marker and a potential predictive biomarker for the efficacy of immunotherapy across various carcinomas. In gallbladder cancer, CD73, in conjunction with FcGBP, functions as a critical regulator of TGF-β1-induced epithelial-mesenchymal transition (EMT), a process strongly linked to tumor progression and poor survival outcomes (106). In cholangiocarcinoma, elevated CD73 expression acts as a prognostic biomarker, promoting EMT and correlating with decreased overall survival (107). Similarly, in colorectal cancer, high levels of CD73 are associated with a worse prognosis, reinforcing its role as a negative prognostic indicator in this malignancy (108). In melanoma, soluble CD73 serves as a biomarker for patients undergoing nivolumab therapy, highlighting its role in immune evasion and its utility as a predictive marker for immunotherapy response (109). Comprehensive analyses across multiple cancer types demonstrate that CD73 significantly influences the tumor microenvironment and immune response, establishing its importance as both a prognostic and therapeutic marker (110).
The potential of CD73 as a therapeutic target in several cancers, including breast and lung cancer, has shown promising therapeutic effects (111–113). In breast cancer, CD73 facilitates local invasion through the epidermal growth factor (EGF)/EGF receptor pathway (114–116). In addition, drug therapy studies in GBM have shown that while treatment increases CD73 expression, loss of CD73 significantly improves survival (117–120). This finding provides important clues for future anti-CD73 treatment strategies.
In summary, CD73 plays a pivotal role not only in inflammation by modulating immune responses through A2A receptor signaling but also in the progression of solid tumors by shaping an immunosuppressive microenvironment and activating oncogenic pathways. Furthermore, it exhibits potential as an immunotherapeutic target in the treatment of various subtypes of leukemia.
Clinically, leukemia is often characterized by excessive white blood cells in the peripheral blood of patients (121). Accordingly, leukemia is classified into myeloid, lymphatic, chronic, and acute types (Table 1), and each classification has unique physiological characteristics (Table 2). The most common types of leukemia are acute myeloid leukemia (AML), acute leukemia/lymphoma (ALL), chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL), and chronic myelogenous leukemia (CML). As white blood cells are highly mobile and encounter minimal obstruction, diseased white blood cells circulate throughout the body via the bloodstream. Consequently, the metastasis of leukemia involves a cyclic process that contributes to its high aggressiveness and fatality (122). Hematopoietic stem cells (HSCs) are developmentally superior to all lymphoid, myeloid, and hematopoietic cells (123), and mutations in HSCs or myeloid progenitor cells are linked to the development of AML and CML (124). Current research indicates that leukemia originates from single cells, with clonal evolution ensuing due to mutation accumulation (125, 126). In the blood of affected individuals, only certain leukemia cells, known as leukemia stem cells (LSCs), possess self-renewing stem cell properties. LSCs can induce disease when transplanted into a new immune-compatible host (127). Due to the resistance of LSCs to treatment (128), the disease evolves into phenotypically distinct subclones as mutations accumulate (129). Current studies have not yet clearly established the correlation between CLL, ALL, and LSCs. The complex nature of mutation accumulation complicates disease treatment and slows progress (130). The numerous subtypes of leukemia and the accumulation of inherited mutations contribute to the complexity of disease treatment.
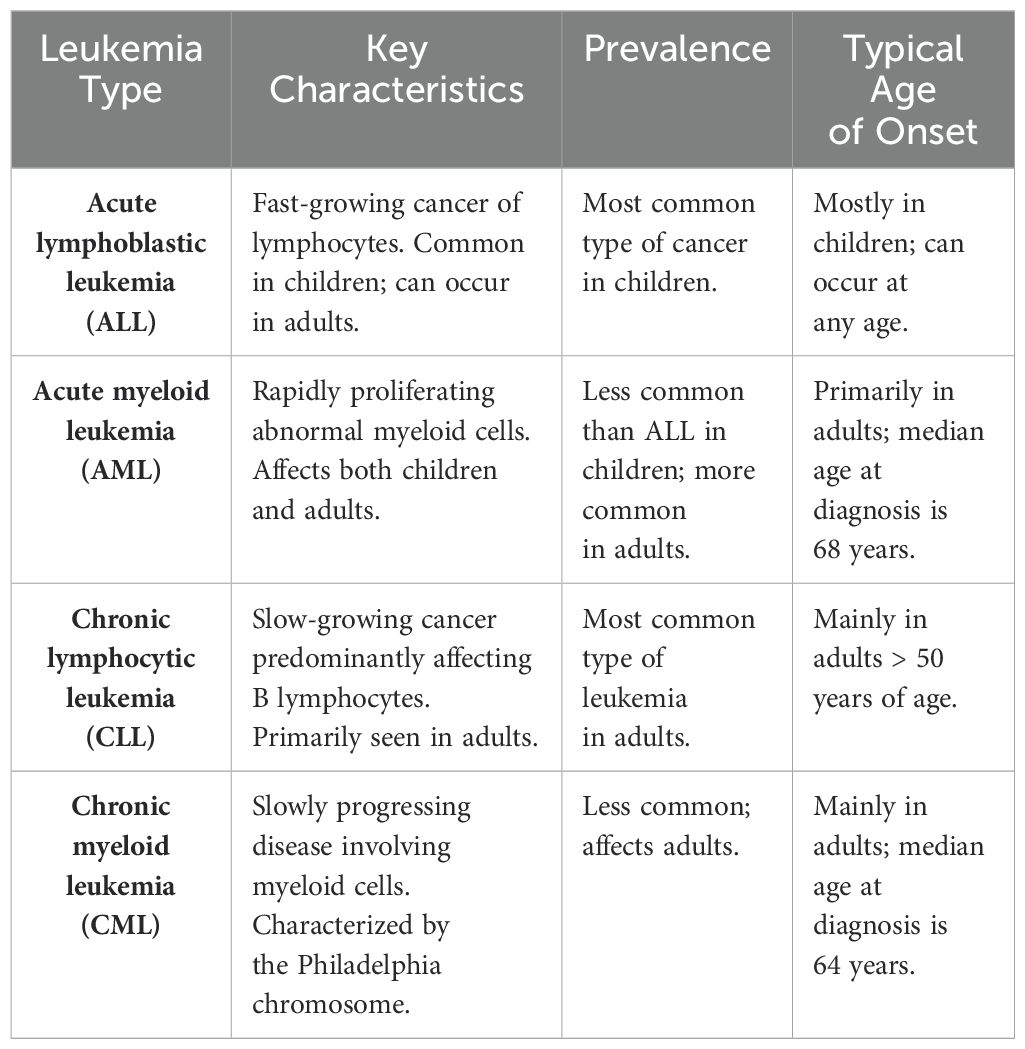
Table 1. Types and characteristics of leukemia (121).
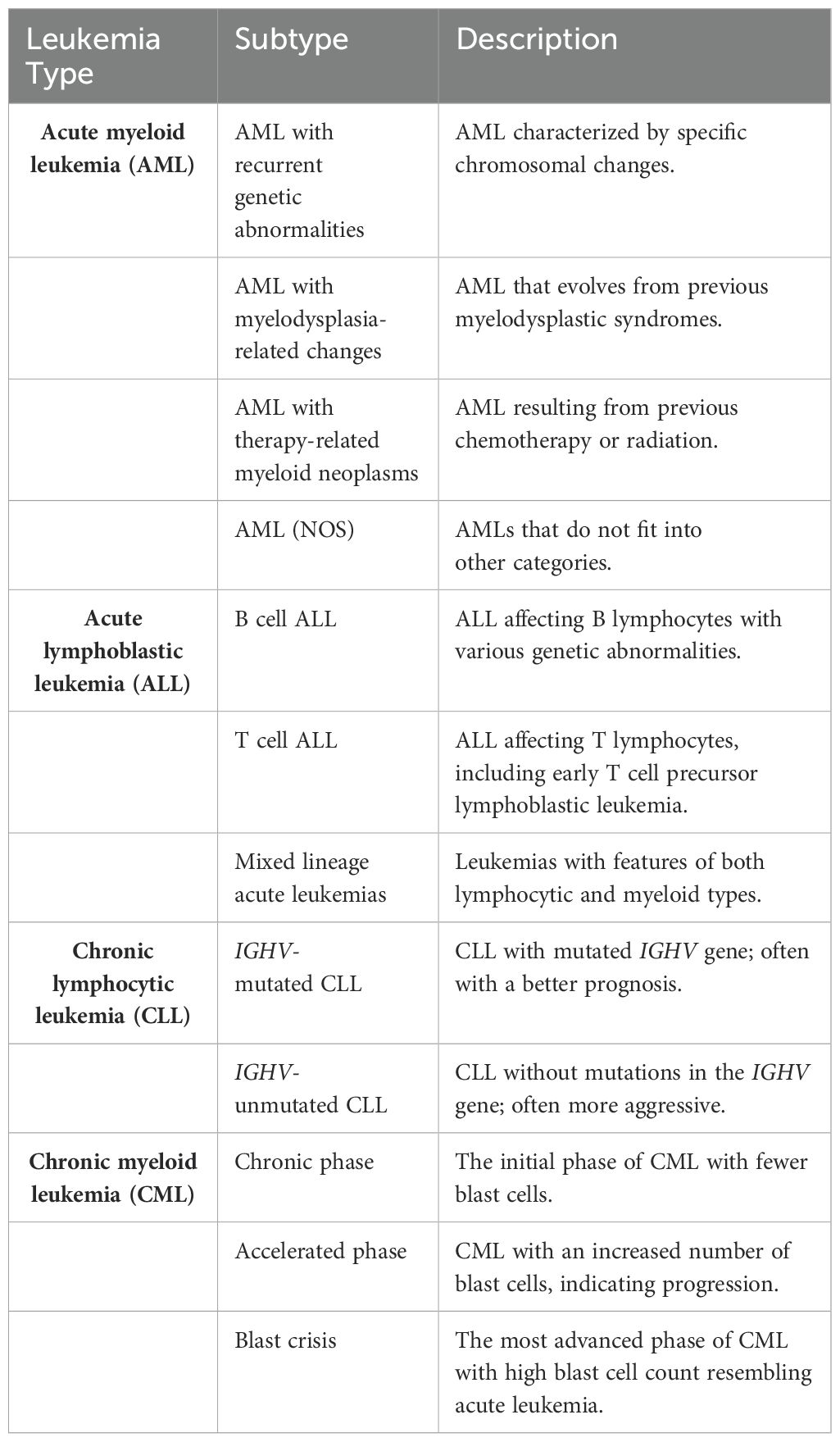
Table 2. Classification and Description of Leukemia Subtypes (https://www.cancer.net/).
Currently, chemotherapy is among the initial treatment strategies for leukemia (Figure 2) (131). While effective, its nonselectivity in targeting cells leads to significant adverse effects. Furthermore, the increasing resistance of leukemia cells to chemotherapy often results in its ineffectiveness and disease relapse (132). HSC transplantation (HSCT) involves the intravenous infusion of normal HSCs to restore the hematopoietic and immune functions in patients with leukemia who are undergoing induction therapy (133). HSCT is considered the most effective method to treat leukemia; however, it carries risks of severe rejection and immune disorders, and finding suitable donors poses a significant challenge (134).
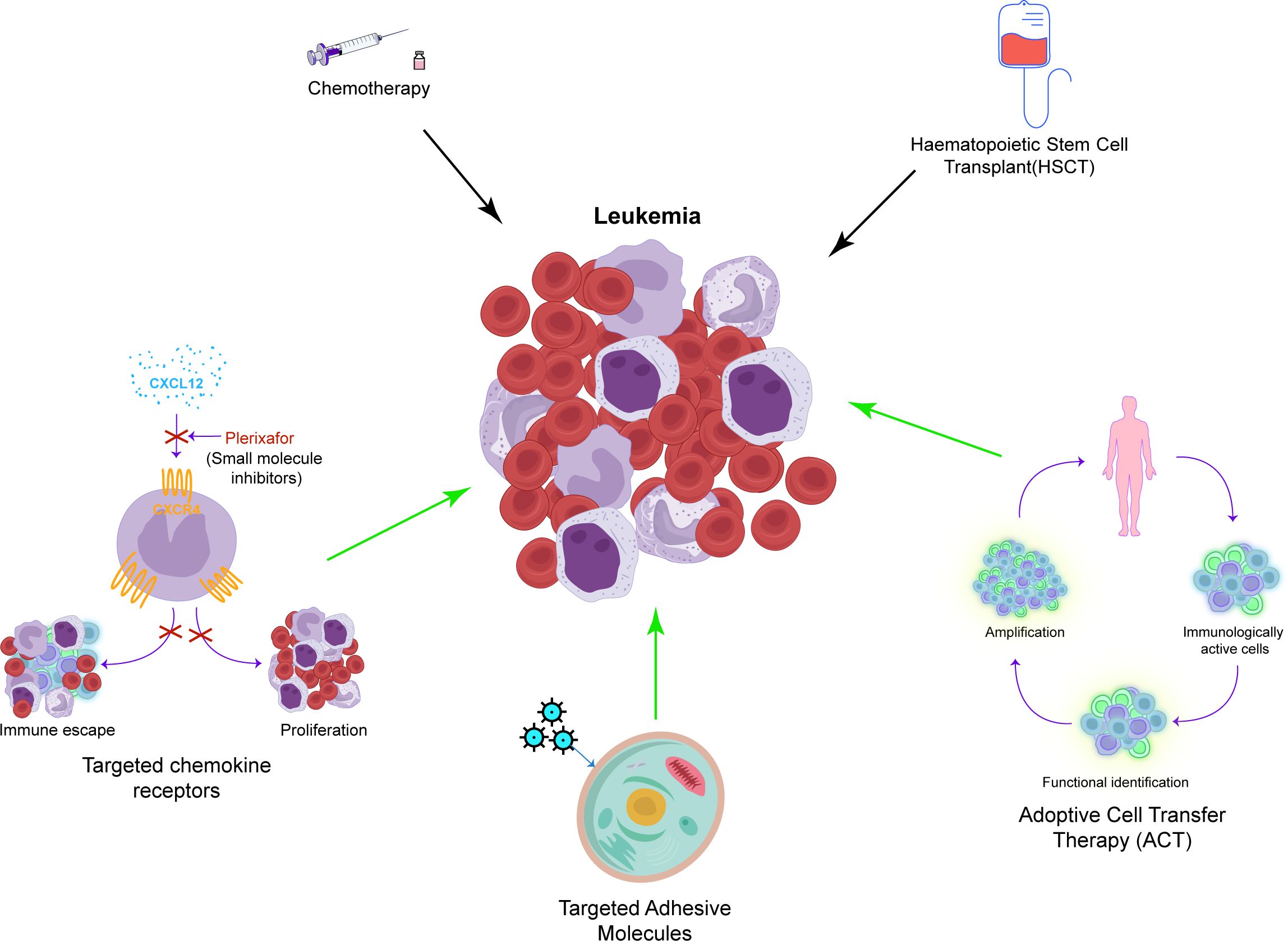
Figure 2. A concise overview of the recent approaches in leukemia treatment. This includes traditional methods such as chemotherapy and HSCT, as well as newer and specific techniques such as targeting chemokine receptors, adhesion molecules, and ACT.
Standard therapy is often ineffective in treating most types of leukemia in adults. Recent studies suggest that transforming acute leukemia into chronic leukemia can enhance survival rates by managing metastasis (122). Treatment methods include targeting the chemokine receptors, notably CXCR4, which are linked to leukemia cell proliferation and drug resistance (SDF1-CXCR4 axis). For instance, AMD3100, a specific CXCR4 antagonist, enhances the efficacy of chemotherapy by inhibiting SDF1-CXCR4, facilitating the entry of leukemia cells into the cell cycle (135). In the study of the SDF1-CXCR4 axis, effective therapeutic drugs such as monoclonal antibodies and novel small molecule inhibitors of CXCR4 have been discovered (136–139). The CD73 adenosine axis, which modulates the immune microenvironment by increasing adenosine production that suppresses antitumor immune responses, is emerging as a potential target for the treatment of leukemia. Additionally, targeting adhesion molecules such as selectins and integrins, which facilitate the lodging of leukemia cells within the bone marrow microenvironment, represents another promising approach. Inhibiting E-selectin, for example, disrupts pro-survival signaling in leukemia cells, thereby inhibiting AML stem cell regeneration and safeguarding native HSCs. GMI-1271, an E-selectin inhibitor, shows promising survival outcomes in the treatment of AML (140). Another target is the integrin family, particularly integrin α6. The BTK-integrin signaling pathway has shown positive results in preclinical studies (141–144), although further clinical validation is required (145). Adoptive cell therapy (ACT) is another option, wherein amplified immune cells such as NK cells, γδ T cells, and α-β T cells are transferred to patients for therapeutic outcomes. ACT can effectively delay the progression of leukemia, but challenges such as antigen selection and countering the immunosuppressive environment of tumors persist (146–150). Traditional treatments for leukemia include drugs that boost immune function to complement chemotherapy and radiotherapy. While these drugs may be effective against specific subtypes, issues such as immune system damage and drug resistance remain (151–153).
Recent studies on LSCs have identified targeted LSC proteins, IC molecules, and immune-related pathways. These studies aim to minimize adverse drug reactions and enhance treatment efficacy. However, this field is still in its developmental stages, thereby necessitating further research (154).
CD73, as an emerging IC, holds potential regulatory roles in the treatment of leukemia. Blocking CD73-related ICs in mouse models of CLL can effectively enhance immune responses. The study indicated that decreased CD73 levels reduced adenosine uptake by A2AR receptors, consequently lowering PD-1 expression (155). Mechanistically, PD-1 downregulation results in an increase in CD8+ T cells and enhanced IFN-γ secretion, thereby boosting immune response regulation and survival rates. Another study highlights that depleting CD8+ T cells enhances the migration, stemness, and proliferation of CLL cells (156). Additionally, CD8+ T cell depletion is linked to the CD73/adenosine axis in various cancer-related inflammation studies (157–159). High NT5E mRNA expression in samples from pediatric patients with B-ALL correlates with unfavorable clinical and pathological characteristics, abundant Tregs and dendritic cells, NK cell depletion, and elevated IC gene expression. However, the prognostic significance of CD73 at the protein level is not substantial (160). In addition to facilitating the immune escape of leukemia cells, CD73 also acts as a surface marker for different leukemia subtypes and supports leukemia cell proliferation. Flow cytometry revealed higher CD73 expression in samples from patients with B-ALL having minimal residual disease (MRD) than in those with MRD alone. Consequently, CD73 may serve as a marker for MRD in patients (161–163), although extensive studies are required for further conclusive evidence. NT5E drives the CEBPA transcription program in the CEBPA mutant subtype of AML. The tumor-promoting effect of CD73 may stem from CD73-dependent adenosine acting on A2AR to sustain leukemia cell growth and inhibit apoptosis (164). In AML, studies on bone marrow-derived mesenchymal stem cells (BM-MSCs) show CD73 as a cell surface marker and adhesion protein, similarly expressed in BM-MSCs (165). The ability of BM-MSCs to differentiate and support hematopoiesis in vitro suggests the involvement of CD73 in the progression of AML. In summary, CD73 is pivotal in inflammation and solid tumors and also exhibits potential as an immunotherapeutic agent in the treatment of various subtypes of leukemia. Studies on LSCs and MRD (166) suggest a significant relationship between CD73 and LSCs.
This section primarily discusses upstream transcription factors related to CD73, the impact of its post-translational modifications on function, and how these factors influence the progression of leukemia. The transcription factor HIF1-α and aryl hydrocarbon receptor (AHR) control metabolic program type 1 regulatory T cell (Tr1) differentiation. In hypoxia and inflammation, the inactivation of AHR by HIF-1α inhibits Tr1 differentiation, whereas ATP conversion by CD39 promotes its differentiation. CD73/adenosine involvement further inhibits Tr1 activity (167, 168). HIF1-α has recently been identified as a transcription factor that inhibits T cells and regulates the immunosuppressive TME through the CD39/CD73/A2AR/adenosine pathway (169–171). The Treg cell transcription factor Foxp3 suppresses immune function that is influenced by A2AR/adenosine (172). In mouse iNKT cells, IFN-γ production and A2AR are linked to inflammation and vascular damage, weakening A2AR function and reducing Th2-type cells, thereby diminishing immune function (173). Gene Ontology enrichment analysis of CD73 in breast tumors identified TGF-β and epithelial-mesenchymal transition (EMT) as significant inducers of CD73 expression (174), underscoring the role of HIF1-α in the cancer immune process. The transcription factor SNAI1 (part of the EMT pathway) upregulates CD73 expression in triple-negative breast cancer (116). Studies on melanoma reveal that MAPK signaling and the pro-inflammatory cytokine TNF-α co-induce CD73 expression via the c-Jun/AP-1 transcription factor complex, with IFN-γ also potentially having an impact on CD73 (175). In AML, increased CD39, PD-1, TIM-3, and LAG-3 expression in CD8T cells correlates with TNF-, IL-2–, and IFN-γ–induced decrease in CD73 protein expression (176). However, low CD73 protein expression suggests a link with CD8 T cell depletion. MicroRNA research on CD73 indicates that the regulatory factor miR-422a can directly or indirectly affect NT5E mRNA levels, influencing its binding to the transcription activator SMAD4 mRNA (177).
CD73 exhibits diverse forms and functions through post-translational protein modifications; it specifically undergoes two main modifications, namely, glycosylation and ubiquitination. Additionally, other forms such as lactate and hematoxylization modifications indirectly impact the functions of CD73. A study on chimeric antigen receptor T cell therapy for GBM showed that H3K18 lactate effectively enhanced the activity of CD39, CD73, and CCR8 genes (178). Tumor metabolism–produced lactate upregulates CD39 and CD73 levels, increases the expression of CCR8 and its ligands, disrupts Treg/Th17 balance, and alters the immune microenvironment, demonstrating its role in promoting an immunosuppressive environment. A study on IκBα SUMO-1 modification in hypoxic environments has discussed the role of adenosine in SUMO-1 modification and NF-κB–mediated transcription (179), indicating potential targets for SUMOylation regulation of adenosine production and CD73 in the immune microenvironment. A study on CD73 glycosylation found that N-linked glycosylation selectively alters CD73 protein activity, leading to high mannose glycosylation and enzymatically impaired glycosylation in HCC (47). Additionally, the study showed that glycosylation might impair both CD73 expression and its normal enzymatic activity. A study on cervical cancer revealed that highly glycosylated, soluble CD73 increased AMP activity (180), contrasting previous findings and suggesting functional differences between membrane-bound and soluble CD73 after glycosylation. Recent studies have identified TRIM21 as the E3 ubiquitin ligase that directly targets CD73. Knocking down TRIM21 in breast cancer cells leads to CD73 overexpression, thereby promoting cancer progression (48). TRIM21 regulation, influenced by T cell–secreted IFN-γ, helps maintain a stable adenosine microenvironment in normal cell lines. TRIM21 overexpression can result in reduced CD73 expression and improved prognosis. The study highlights a novel immunotherapeutic target acting directly on CD73 and offers insights for future ubiquitination research in CD73-related cancer cell lines. Overall, research on post-translational modification of CD73 is limited, particularly CD73 modification in leukemia. Considering that most existing studies focus on post-translational modifications and CD73, this area holds significant potential in leukemia research.
With progress in CD73 research, several drugs targeting CD73 have been developed, with some candidates advancing to clinical trials (Table 3). CPI-006, a humanized immunoglobulin (Ig)G1 FcγR-binding defective antibody, specifically binds to CD73+ T and B lymphocytes by targeting the N-terminus of CD73. It possesses unique characteristics as a CD73 inhibitor. The binding reduces the catalytic activity of CD73 and enhances immune-regulatory functions (181). CPI-006 is currently in phase 1 clinical trials and has demonstrated efficacy in reducing B cell count by binding to CD73 in 34 patients with advanced cancer (181). AB680, another efficacious CD73 inhibitor, exhibits reversible and selective binding to CD73 and is currently in phase 1 clinical trials (182). AB680 significantly enhances CD8+ T cell infiltration and prolongs survival of mice (183). TJ004309, a monoclonal antibody targeting CD73, completely inhibits CD73 activity and reduces adenosine production by binding to CD73 and is now in phase 1 to 2 clinical trials (184). HLX23, a recombinant anti-CD73 humanized monoclonal antibody, is in phase 1 clinical trials for the treatment of advanced solid tumors. Oleclumab, a selective anti-CD73 monoclonal antibody, is effective in treating advanced solid tumors, especially in combination with durvalumab (185). It is currently in phase 2 clinical trials (https://clinicaltrials.gov/). AK119, a humanized IgG1 monoclonal antibody, selectively binds to and inhibits the exonucleotidase activity of CD73 (186). AK104 is a recombinant humanized IgG1 bispecific antibody targeting PD-1 and CTLA-4 simultaneously. While CD73 blockade therapy is popular in treating various cancers, CD73 and PD-1 therapies alone often yield unsatisfactory results, possibly due to the nonoverlapping, immunosuppressive mechanisms of tumors in immune escape, including adenosine accumulation (187). The inhibitory effect of CD73 monoclonal antibodies varies with different concentrations and binding sites (188). Controlling drug concentration and drug targeting in vivo is more challenging. The combination of drugs targeting both the CD73/adenosine pathway and PD-1/PDL1 is gaining popularity owing to their crucial roles in immune escape and the limited effects of either therapy alone. As CD73 emerges as a key immune target in tumor research, the development of monoclonal antibodies targeting CD73 is advancing. As CD73 is being recognized as an emerging immune target in tumor therapy, several treatments including monoclonal antibodies, small molecule inhibitors, and drug combinations with other immune pathways are being developed, with clinical research advancing rapidly. However, challenges in significantly enhancing cancer treatment persist due to unclear mechanisms of nonoverlapping immune suppression in cancer and current treatment outcomes (189). This remains true despite ongoing advancements in combination therapy and CD73 monoclonal antibodies. Consequently, using targeted research, there is a need to identify new targets for the immune escape process of cancers and to develop more specific and stable monoclonal antibodies as well as small molecule inhibitors.
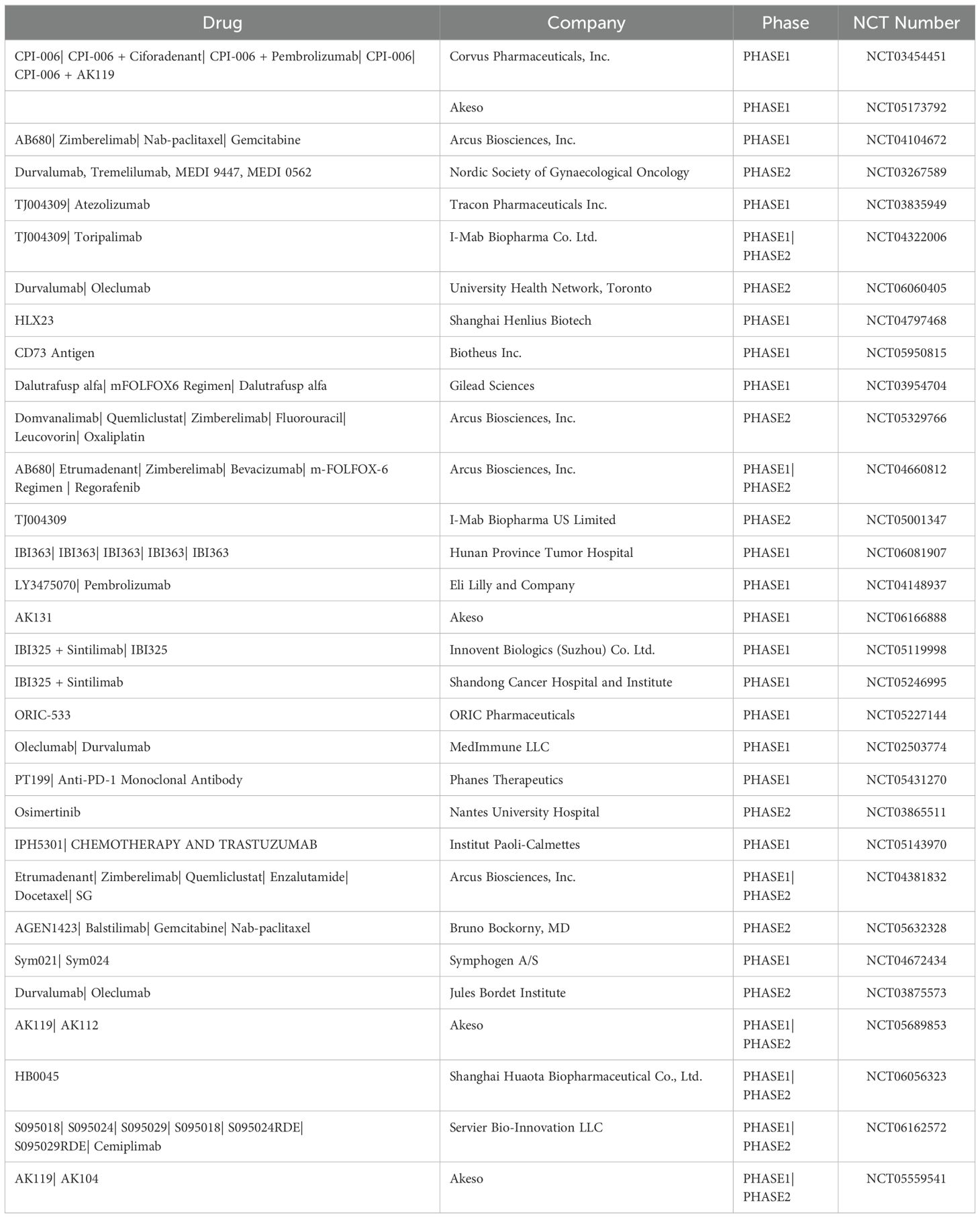
Table 3. Current studies targeting CD73 target therapy and combination therapy (https://clinicaltrials.gov/).
Despite the therapeutic promise of CD73 inhibitors, potential side effects and limitations require careful consideration. Recent murine studies demonstrate that CD73 inhibition enhances bone marrow stem cell mobilization, potentially disrupting hematopoietic homeostasis and increasing risks of myelopathic suppression or accidental stem cell dissemination (190). Notably, since adenosine maintains critical physiological functions including cardiovascular regulation and neuroprotection (191, 192) systemic CD73 blockade may compromise these protective mechanisms. Furthermore, therapeutic efficacy could be limited by compensatory upregulation of alternative adenosine-producing pathways [such as the CD38/CD203a axis (3)] or the emergence of resistance mechanisms within the tumor microenvironment.
Currently, publicly available data on the use of CD73 in treating leukemia are limited. A noteworthy study at the University Hospital of Tours in France (ClinicalTrials.gov ID NCT05792007) focused on the medullary microenvironment of acute childhood leukemia and explored the energy metabolism in mesenchymal stem cells, including oxidative phosphorylation and glycolysis. CD73 is anticipated to be a promising anticancer target in this study. Currently, the mechanism of effectiveness of CD73 has not yet been well established and the underlying reasons are unclear. One possibility is that drug resistance of cancer cells contributes to the diminished efficacy of CD73 and PD-L1–targeted drugs, resulting in recurrence. However, targeting CD73 in leukemia treatment still holds significant potential. Studies indicate that CD73 is present in patients with MRD after recovery (162, 163, 193) and is likely linked to leukemia HSCs. Additionally, during treatment with CD73, potential changes in the modified functional structure of CD73 that affect adenosine production and drug specificity must be considered.
First, studies of solid tumors indicate the multiple potential functions of CD73 and that soluble CD73 can influence the immune microenvironment. These findings differ from the common belief that the primary function of CD73 in normal cells is membrane anchoring. Second, the mechanism of transcription factors in regulating CD73 expression remains unclear and most factors that affect CD73 do so indirectly. Transcription factors modulate the function of CD73 by influencing the functional structure of other proteins. Research on transcription factor targets of CD73 is incomplete and there is no specific transcription factor that has been identified to directly target CD73. Therefore, exploring the transcription factor targets of CD73 could be a promising avenue for further in-depth research. Beyond transcription factors, the growing focus on post-translational modifications has highlighted the importance in protein function, particularly for enzymes such as CD73. Our current understanding of how post-translational modifications regulate CD73 and their impact on the function of CD73 is incomplete, and the discovery of soluble glycosylation of CD73 (180) underscores the substantial impact of this modification on its function. Given the varied functional mechanisms of CD73 in different immune tumor microenvironments, investigating its modification changes and functional effects across various solid and hematological cancers is essential. The discovery of TRIM21 as an E3 ubiquitin ligase suggests CD73, potentially influenced by TRIM21 and other modifiers, to be a promising therapeutic target less susceptible to drug resistance.
A major challenge in studying the role of CD73 in leukemia is analyzing its regulatory effect on the disease through expression in isolated LSCs and identifying its action sites using techniques such as lineage tracing. It is also crucial to investigate whether post-translational modifications affect the structure of CD73 so that its post-denaturation involvement can be ascertained. Exploring the role of CD73 in leukemia is just the beginning; subsequent research should focus on designing small molecule inhibitors or monoclonal antibodies that target CD73 expression at specific sites, which is informed by its role in the immune evasion of leukemia and drug resistance mechanisms.
While CD73 is a promising target in the treatment of leukemia, significant challenges arise from its complexity and the various subtypes of leukemia. Ongoing research on CD73 in leukemia suggests a strong correlation between CD73-involved signaling pathways and different leukemia subtypes. Additionally, protein modification research implies that CD73 mRNA expression surpasses protein expression because some of the expressed CD73 proteins may have lost their function. The expressed CD73 often concentrates in small residues, akin to the characteristics of LSCs, which suggests that CD73 might be a key marker of LSCs, holding significant treatment implications for leukemia. Considerable research related to drug screening needs to be further undertaken. There is not only a need for highly specific and stable monoclonal antibodies and small molecule inhibitors but also investigations of the additional nonoverlapping immunosuppressive mechanisms akin to the CD73/adenosine and PD1/PDL1 pathways to enhance the survival rates of patients with leukemia and other cancers.
HG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. TZ: Conceptualization, Supervision, Writing – review & editing. KL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. XL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 82273992).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Lokshin A, Raskovalova T, Huang X, Zacharia LC, Jackson EK, Gorelik E. Adenosine-mediated inhibition of the cytotoxic activity and cytokine production by activated natural killer cells. Cancer Res. (2006) 66:7758–65. doi: 10.1158/0008-5472.CAN-06-0478
2. Vaupel P, Multhoff G. Commentary: A metabolic immune checkpoint: adenosine in tumor microenvironment. Front Immunol. (2016) 7:332. doi: 10.3389/fimmu.2016.00332
3. Horenstein AL, Chillemi A, Zaccarello G, Bruzzone S, Quarona V, Zito A, et al. A CD38/CD203a/CD73 ectoenzymatic pathway independent of CD39 drives a novel adenosinergic loop in human T lymphocytes. Oncoimmunology. (2013) 2:e26246. doi: 10.4161/onci.26246
4. Costa F, Dalla Palma B, Giuliani N. CD38 expression by myeloma cells and its role in the context of bone marrow microenvironment: modulation by therapeutic agents. Cells. (2019) 8(12):1632. doi: 10.3390/cells8121632
5. Cass CE, Selner M, Tan TH, Muhs WH, Robins MJ. Comparison of the effects on cultured L1210 leukemia cells of the ribosyl, 2’-deoxyribosyl, and xylosyl homologs of tubercidin and adenosine alone or in combination with 2’-deoxycoformycin. Cancer Treat Rep. (1982) 66:317–26.
6. Schneider C, Wiendl H, Ogilvie A. Biphasic cytotoxic mechanism of extracellular ATP on U-937 human histiocytic leukemia cells: involvement of adenosine generation. Biochim Biophys Acta. (2001) 1538:190–205. doi: 10.1016/S0167-4889(01)00069-6
7. Gessi S, Varani K, Merighi S, Morelli A, Ferrari D, Leung E, et al. Pharmacological and biochemical characterization of A3 adenosine receptors in Jurkat T cells. Br J Pharmacol. (2001) 134:116–26. doi: 10.1038/sj.bjp.0704254
8. Batiuk TD, Schnizlein-Bick C, Plotkin Z, Dagher PC. Guanine nucleosides and Jurkat cell death: roles of ATP depletion and accumulation of deoxyribonucleotides. Am J Physiol Cell Physiol. (2001) 281:C1776–84. doi: 10.1152/ajpcell.2001.281.6.C1776
9. Bastin-Coyette L, Smal C, Cardoen S, Saussoy P, Van den Neste E, Bontemps F. Mechanisms of cell death induced by 2-chloroadenosine in leukemic B-cells. Biochem Pharmacol. (2008) 75:1451–60. doi: 10.1016/j.bcp.2007.12.007
10. Zimmermann H. 5’-Nucleotidase: molecular structure and functional aspects. Biochem J. (1992) 285:345–65. doi: 10.1042/bj2850345
11. Robson SC, Sévigny J, Zimmermann H. The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic signalling. (2006) 2:409–30. doi: 10.1007/s11302-006-9003-5
12. Yegutkin G, Bodin P, Burnstock G. Effect of shear stress on the release of soluble ecto-enzymes ATPase and 5’-nucleotidase along with endogenous ATP from vascular endothelial cells. Br J Pharmacol. (2000) 129:921–6. doi: 10.1038/sj.bjp.0703136
13. Fini C, Talamo F, Cherri S, Coli M, Floridi A, Ferrara L, et al. Biochemical and mass spectrometric characterization of soluble ecto-5’-nucleotidase from bull seminal plasma. Biochem J. (2003) 372:443–51. doi: 10.1042/bj20021687
14. Thomson LF, Ruedi JM, Glass A, Moldenhauer G, Moller P, Low MG, et al. Production and characterization of monoclonal antibodies to the glycosyl phosphatidylinositol-anchored lymphocyte differentiation antigen ecto-5’-nucleotidase (CD73). Tissue Antigens. (1990) 35:9–19. doi: 10.1111/j.1399-0039.1990.tb01750.x
15. Lehto MT, Sharom FJ. Release of the glycosylphosphatidylinositol-anchored enzyme ecto-5’-nucleotidase by phospholipase C: catalytic activation and modulation by the lipid bilayer. Biochem J. (1998) 332:101–9. doi: 10.1042/bj3320101
16. Kalsi K, Lawson C, Dominguez M, Taylor P, Yacoub MH, Smolenski RT. Regulation of ecto-5’-nucleotidase by TNF-alpha in human endothelial cells. Mol Cell Biochem. (2002) 232:113–9. doi: 10.1023/A:1014806916844
17. Lu T, Zhang Z, Zhang J, Pan X, Zhu X, Wang X, et al. CD73 in small extracellular vesicles derived from HNSCC defines tumour-associated immunosuppression mediated by macrophages in the microenvironment. J extracellular vesicles. (2022) 11:e12218. doi: 10.1002/jev2.12218
18. Schneider E, Winzer R, Rissiek A, Ricklefs I, Meyer-Schwesinger C, Ricklefs FL, et al. CD73-mediated adenosine production by CD8 T cell-derived extracellular vesicles constitutes an intrinsic mechanism of immune suppression. Nat Commun. (2021) 12:5911. doi: 10.1038/s41467-021-26134-w
19. Ålgars A, Karikoski M, Yegutkin GG, Stoitzner P, Niemelä J, Salmi M, et al. Different role of CD73 in leukocyte trafficking via blood and lymph vessels. Blood. (2011) 117:4387–93. doi: 10.1182/blood-2010-11-321646
20. Regateiro FS, Cobbold SP, Waldmann H. CD73 and adenosine generation in the creation of regulatory microenvironments. Clin Exp Immunol. (2013) 171:1–7. doi: 10.1111/j.1365-2249.2012.04623.x
21. Saigí M, Mesía-Carbonell O, Barbie DA, Guillamat-Prats R. Unraveling the intricacies of CD73/adenosine signaling: the pulmonary immune and stromal microenvironment in lung cancer. Cancers. (2023) 15(23):5706. doi: 10.3390/cancers15235706
22. Doherty GA, Bai A, Hanidziar D, Longhi MS, Lawlor GO, Putheti P, et al. CD73 is a phenotypic marker of effector memory Th17 cells in inflammatory bowel disease. Eur J Immunol. (2012) 42:3062–72. doi: 10.1002/eji.201242623
23. Thompson LF, Ruedi JM, Low MG, Clement LT. Distribution of ecto-5’-nucleotidase on subsets of human T and B lymphocytes as detected by indirect immunofluorescence using goat antibodies. J Immunol (Baltimore Md: 1950). (1987) 139:4042–8. doi: 10.4049/jimmunol.139.12.4042
24. Raczkowski F, Rissiek A, Ricklefs I, Heiss K, Schumacher V, Wundenberg K, et al. CD39 is upregulated during activation of mouse and human T cells and attenuates the immune response to Listeria monocytogenes. PloS One. (2018) 13:e0197151. doi: 10.1371/journal.pone.0197151
25. Liang D, Zuo A, Zhao R, Shao H, Born WK, O’Brien RL, et al. CD73 expressed on γδ T cells shapes their regulatory effect in experimental autoimmune uveitis. PloS One. (2016) 11:e0150078. doi: 10.1371/journal.pone.0150078
26. Kaku H, Cheng KF, Al-Abed Y, Rothstein TL. A novel mechanism of B cell-mediated immune suppression through CD73 expression and adenosine production. J Immunol (Baltimore Md: 1950). (2014) 193:5904–13. doi: 10.4049/jimmunol.1400336
27. Taylor JJ, Pape KA, Jenkins MK. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J Exp Med. (2012) 209:597–606. doi: 10.1084/jem.20111696
28. Moncrieffe H, Nistala K, Kamhieh Y, Evans J, Eddaoudi A, Eaton S, et al. High expression of the ectonucleotidase CD39 on T cells from the inflamed site identifies two distinct populations, one regulatory and one memory T cell population. J Immunol (Baltimore Md: 1950). (2010) 185:134–43. doi: 10.4049/jimmunol.0803474
29. Resta R, Yamashita Y, Thompson LF. Ecto-enzyme and signaling functions of lymphocyte CD73. Immunol Rev. (1998) 161:95–109. doi: 10.1111/j.1600-065X.1998.tb01574.x
30. Airas L, Niemelä J, Salmi M, Puurunen T, Smith DJ, Jalkanen S. Differential regulation and function of CD73, a glycosyl-phosphatidylinositol-linked 70-kD adhesion molecule, on lymphocytes and endothelial cells. J Cell Biol. (1997) 136:421–31. doi: 10.1083/jcb.136.2.421
31. Jin D, Fan J, Wang L, Thompson LF, Liu A, Daniel BJ, et al. CD73 on tumor cells impairs antitumor T-cell responses: a novel mechanism of tumor-induced immune suppression. Cancer Res. (2010) 70:2245–55. doi: 10.1158/0008-5472.CAN-09-3109
32. Wang L, Fan J, Thompson LF, Zhang Y, Shin T, Curiel TJ, et al. CD73 has distinct roles in nonhematopoietic and hematopoietic cells to promote tumor growth in mice. J Clin Invest. (2011) 121:2371–82. doi: 10.1172/JCI45559
33. Stagg J, Divisekera U, Duret H, Sparwasser T, Teng MW, Darcy PK, et al. CD73-deficient mice have increased antitumor immunity and are resistant to experimental metastasis. Cancer Res. (2011) 71:2892–900. doi: 10.1158/0008-5472.CAN-10-4246
34. Klysz DD, Fowler C, Malipatlolla M, Stuani L, Freitas KA, Chen Y, et al. Inosine induces stemness features in CAR-T cells and enhances potency. Cancer Cell. (2024) 42:266–82.e8. doi: 10.1016/j.ccell.2024.01.002
35. Xue F, Wang T, Shi H, Feng H, Feng G, Wang R, et al. CD73 facilitates invadopodia formation and boosts Malignancy of head and neck squamous cell carcinoma via the MAPK signaling pathway. Cancer science. (2022) 113:2704–15. doi: 10.1111/cas.v113.8
36. Gao ZW, Liu C, Yang L, Chen HC, Yang LF, Zhang HZ, et al. CD73 severed as a potential prognostic marker and promote lung cancer cells migration via enhancing EMT progression. Front Genet. (2021) 12:728200. doi: 10.3389/fgene.2021.728200
37. Dai S, Liu T, He YY, Huang Y, Wang L, Luo F, et al. Pan-cancer analysis of LINC02535 as a potential biomarker and its oncogenic role in lung adenocarcinoma. Heliyon. (2022) 8:e12108. doi: 10.1016/j.heliyon.2022.e12108
38. Synnestvedt K, Furuta GT, Comerford KM, Louis N, Karhausen J, Eltzschig HK, et al. Ecto-5’-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Invest. (2002) 110:993–1002. doi: 10.1172/JCI0215337
39. Chalmin F, Mignot G, Bruchard M, Chevriaux A, Végran F, Hichami A, et al. Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expression. Immunity. (2012) 36:362–73. doi: 10.1016/j.immuni.2011.12.019
40. Niemelä J, Ifergan I, Yegutkin GG, Jalkanen S, Prat A, Airas L. IFN-beta regulates CD73 and adenosine expression at the blood-brain barrier. Eur J Immunol. (2008) 38:2718–26. doi: 10.1002/eji.200838437
41. Petrovic-Djergovic D, Hyman MC, Ray JJ, Bouis D, Visovatti SH, Hayasaki T, et al. Tissue-resident ecto-5’ nucleotidase (CD73) regulates leukocyte trafficking in the ischemic brain. J Immunol (Baltimore Md: 1950). (2012) 188:2387–98. doi: 10.4049/jimmunol.1003671
42. Lévesque SA, Kukulski F, Enjyoji K, Robson SC, Sévigny J. NTPDase1 governs P2X7-dependent functions in murine macrophages. Eur J Immunol. (2010) 40:1473–85. doi: 10.1002/eji.200939741
43. Bergmann C, Strauss L, Zeidler R, Lang S, Whiteside TL. Expansion of human T regulatory type 1 cells in the microenvironment of cyclooxygenase 2 overexpressing head and neck squamous cell carcinoma. Cancer Res. (2007) 67:8865–73. doi: 10.1158/0008-5472.CAN-07-0767
44. Antonioli L, Pacher P, Vizi ES, Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol Med. (2013) 19:355–67. doi: 10.1016/j.molmed.2013.03.005
45. Kiss J, Yegutkin GG, Koskinen K, Savunen T, Jalkanen S, Salmi M. IFN-beta protects from vascular leakage via up-regulation of CD73. Eur J Immunol. (2007) 37:3334–8. doi: 10.1002/eji.200737793
46. Gao ZW, Dong K, Zhang HZ. The roles of CD73 in cancer. BioMed Res Int. (2014) 2014:460654. doi: 10.1155/2014/460654
47. Alcedo KP, Guerrero A, Basrur V, Fu D, Richardson ML, McLane JS, et al. Tumor-selective altered glycosylation and functional attenuation of CD73 in human hepatocellular carcinoma. Hepatol Commun. (2019) 3:1400–14. doi: 10.1002/hep4.1410
48. Fu Z, Chen S, Zhu Y, Zhang D, Xie P, Jiao Q, et al. Proteolytic regulation of CD73 by TRIM21 orchestrates tumor immunogenicity. Sci Adv. (2023) 9:eadd6626. doi: 10.1126/sciadv.add6626
49. Sträter N. Ecto-5’-nucleotidase: Structure function relationships. Purinergic signalling. (2006) 2:343–50. doi: 10.1007/s11302-006-9000-8
50. Xia C, Yin S, To KKW, Fu L. CD39/CD73/A2AR pathway and cancer immunotherapy. Mol cancer. (2023) 22:44. doi: 10.1186/s12943-023-01733-x
51. Vaisitti T, Audrito V, Serra S, Bologna C, Brusa D, Malavasi F, et al. NAD+-metabolizing ecto-enzymes shape tumor-host interactions: the chronic lymphocytic leukemia model. FEBS letters. (2011) 585:1514–20. doi: 10.1016/j.febslet.2011.04.036
52. Sadej R, Skladanowski AC. Dual, enzymatic and non-enzymatic, function of ecto-5’-nucleotidase (eN, CD73) in migration and invasion of A375 melanoma cells. Acta Biochim Polonica. (2012) 59:647–52. doi: 10.18388/abp.2012_2105
53. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. (2018) 24:541–50. doi: 10.1038/s41591-018-0014-x
54. Tong L, Jiménez-Cortegana C, Tay AHM, Wickström S, Galluzzi L, Lundqvist A. NK cells and solid tumors: therapeutic potential and persisting obstacles. Mol cancer. (2022) 21:206. doi: 10.1186/s12943-022-01672-z
55. Bai Y, Zhang X, Zheng J, Liu Z, Yang Z, Zhang X. Overcoming high level adenosine-mediated immunosuppression by DZD2269, a potent and selective A2aR antagonist. J Exp Clin Cancer research: CR. (2022) 41:302. doi: 10.1186/s13046-022-02511-1
56. Tokano M, Kawano M, Takagi R, Matsushita S. Istradefylline, an adenosine A2a receptor antagonist, inhibits the CD4(+) T-cell hypersecretion of IL-17A and IL-8 in humans. Immunol Med. (2022) 45:244–50. doi: 10.1080/25785826.2022.2094593
57. Wilson JM, Kurtz CC, Black SG, Ross WG, Alam MS, Linden J, et al. The A2B adenosine receptor promotes Th17 differentiation via stimulation of dendritic cell IL-6. J Immunol (Baltimore Md: 1950). (2011) 186:6746–52. doi: 10.4049/jimmunol.1100117
58. Mastelic-Gavillet B, Navarro Rodrigo B, Décombaz L, Wang H, Ercolano G, Ahmed R, et al. Adenosine mediates functional and metabolic suppression of peripheral and tumor-infiltrating CD8(+) T cells. J immunotherapy cancer. (2019) 7:257. doi: 10.1186/s40425-019-0719-5
59. Canale FP, Ramello MC, Núñez N, Araujo Furlan CL, Bossio SN, Gorosito Serrán M, et al. CD39 expression defines cell exhaustion in tumor-infiltrating CD8(+) T cells. Cancer Res. (2018) 78:115–28. doi: 10.1158/0008-5472.CAN-16-2684
60. Linden J, Koch-Nolte F, Dahl G. Purine release, metabolism, and signaling in the inflammatory response. Annu Rev Immunol. (2019) 37:325–47. doi: 10.1146/annurev-immunol-051116-052406
61. Wang J, Zeng H, Zhang H, Han Y. The role of exosomal PD-L1 in tumor immunotherapy. Trans Oncol. (2021) 14:101047. doi: 10.1016/j.tranon.2021.101047
62. Kornepati AVR, Vadlamudi RK, Curiel TJ. Programmed death ligand 1 signals in cancer cells. Nat Rev Cancer. (2022) 22:174–89. doi: 10.1038/s41568-021-00431-4
63. Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. (2008) 26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331
64. Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. (2002) 8:793–800. doi: 10.1038/nm730
65. Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. New Engl J Med. (2012) 366:2455–65. doi: 10.1056/NEJMoa1200694
66. Allard B, Allard D, Buisseret L, Stagg J. The adenosine pathway in immuno-oncology. Nat Rev Clin Oncol. (2020) 17:611–29. doi: 10.1038/s41571-020-0382-2
67. Zhang T, Liu H, Jiao L, Zhang Z, He J, Li L, et al. Genetic characteristics involving the PD-1/PD-L1/L2 and CD73/A2aR axes and the immunosuppressive microenvironment in DLBCL. J immunotherapy Cancer. (2022) 10(4):e004114. doi: 10.1136/jitc-2021-004114
68. Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C, Zeng HY, et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J Hematol Oncol. (2021) 14:200. doi: 10.1186/s13045-021-01207-x
69. Chambers AM, Wang J, Dao TN, Lupo KB, Veenhuis P, Ayers MG, et al. Functional expression of CD73 on human natural killer cells. Cancer immunology immunotherapy: CII. (2022) 71:3043–56. doi: 10.1007/s00262-022-03219-z
70. Koivisto MK, Tervahartiala M, Kenessey I, Jalkanen S, Boström PJ, Salmi M. Cell-type-specific CD73 expression is an independent prognostic factor in bladder cancer. Carcinogenesis. (2019) 40:84–92. doi: 10.1093/carcin/bgy154
71. Sadej R, Spychala J, Skladanowski AC. Expression of ecto-5’-nucleotidase (eN, CD73) in cell lines from various stages of human melanoma. Melanoma Res. (2006) 16:213–22. doi: 10.1097/01.cmr.0000215030.69823.11
72. Zhu Y, Banerjee A, Xie P, Ivanov AA, Uddin A, Jiao Q, et al. Pharmacological suppression of the OTUD4/CD73 proteolytic axis revives antitumor immunity against immune-suppressive breast cancers. J Clin Invest. (2024) 134(10):e176390. doi: 10.1172/JCI176390
73. Longaray JB, Dias CK, Scholl JN, Battastini AMO, Figueiró F. Investigation of co-treatment multi-targeting approaches in breast cancer cell lines. Eur J Pharmacol. (2024) 966:176328. doi: 10.1016/j.ejphar.2024.176328
74. Giatromanolaki A, Kouroupi M, Pouliliou S, Mitrakas A, Hasan F, Pappa A, et al. Ectonucleotidase CD73 and CD39 expression in non-small cell lung cancer relates to hypoxia and immunosuppressive pathways. Life Sci. (2020) 259:118389. doi: 10.1016/j.lfs.2020.118389
75. Jacoberger-Foissac C, Cousineau I, Bareche Y, Allard D, Chrobak P, Allard B, et al. CD73 inhibits cGAS-STING and cooperates with CD39 to promote pancreatic cancer. Cancer Immunol Res. (2023) 11:56–71. doi: 10.1158/2326-6066.CIR-22-0260
76. Han Y, Lee T, He Y, Raman R, Irizarry A, Martin ML, et al. The regulation of CD73 in non-small cell lung cancer. Eur J Cancer (Oxford England: 1990). (2022) 170:91–102. doi: 10.1016/j.ejca.2022.04.025
77. Iqbal J, Basharat A, Bano S, Abid SMA, Pelletier J, Sévigny J. Identification and expression analysis of CD73 inhibitors in cervical cancer. Medicinal Chem (Shariqah (United Arab Emirates)). (2021) 17:866–74. doi: 10.2174/1573406416666200925141703
78. Cappellari AR, Rockenbach L, Dietrich F, Clarimundo V, Glaser T, Braganhol E, et al. Characterization of ectonucleotidases in human medulloblastoma cell lines: ecto-5’NT/CD73 in metastasis as potential prognostic factor. PloS One. (2012) 7:e47468. doi: 10.1371/annotation/0e219081-9218-480c-aa54-1142a68aed14
79. Bernardi A, Bavaresco L, Wink MR, Jacques-Silva MC, Delgado-Cañedo A, Lenz G, et al. Indomethacin stimulates activity and expression of ecto-5’-nucleotidase/CD73 in glioma cell lines. Eur J Pharmacol. (2007) 569:8–15. doi: 10.1016/j.ejphar.2007.04.058
80. Montalbán Del Barrio I, Penski C, Schlahsa L, Stein RG, Diessner J, Wöckel A, et al. Adenosine-generating ovarian cancer cells attract myeloid cells which differentiate into adenosine-generating tumor associated macrophages - a self-amplifying, CD39- and CD73-dependent mechanism for tumor immune escape. J Immunotherapy Cancer. (2016) 4:49. doi: 10.1186/s40425-016-0154-9
81. Neo SY, Yang Y, Record J, Ma R, Chen X, Chen Z, et al. CD73 immune checkpoint defines regulatory NK cells within the tumor microenvironment. J Clin Invest. (2020) 130:1185–98. doi: 10.1172/JCI128895
82. Rocha P, Salazar R, Zhang J, Ledesma D, Solorzano JL, Mino B, et al. CD73 expression defines immune, molecular, and clinicopathological subgroups of lung adenocarcinoma. Cancer immunology immunotherapy: CII. (2021) 70:1965–76. doi: 10.1007/s00262-020-02820-4
83. Tu E, McGlinchey K, Wang J, Martin P, Ching SL, Floc’h N, et al. Anti-PD-L1 and anti-CD73 combination therapy promotes T cell response to EGFR-mutated NSCLC. JCI Insight. (2022) 7(3):e142843. doi: 10.1172/jci.insight.142843
84. Ishii H, Azuma K, Kawahara A, Kinoshita T, Matsuo N, Naito Y, et al. Predictive value of CD73 expression for the efficacy of immune checkpoint inhibitors in NSCLC. Thorac cancer. (2020) 11:950–5. doi: 10.1111/1759-7714.13346
85. Su AL, Tian CQ, Ou YJ, Bao XB, Huan XJ, Miao ZH, et al. Proteasome inhibitors reduce CD73 expression partly via decreasing p-ERK in NSCLC cells. Life Sci. (2023) 332:122129. doi: 10.1016/j.lfs.2023.122129
86. Zhang H, Cao Y, Tang J, Wang R. CD73 (NT5E) promotes the proliferation and metastasis of lung adenocarcinoma through the EGFR/AKT/mTOR pathway. BioMed Res Int. (2022) 2022:9944847. doi: 10.1155/2022/9944847
87. Shen A, Ye Y, Chen F, Xu Y, Zhang Z, Zhao Q, et al. Integrated multi-omics analysis identifies CD73 as a prognostic biomarker and immunotherapy response predictor in head and neck squamous cell carcinoma. Front Immunol. (2022) 13:969034. doi: 10.3389/fimmu.2022.969034
88. Baysal H, Siozopoulou V, Zaryouh H, Hermans C, Lau HW, Lambrechts H, et al. The prognostic impact of the immune signature in head and neck squamous cell carcinoma. Front Immunol. (2022) 13:1001161. doi: 10.3389/fimmu.2022.1001161
89. Ma XL, Shen MN, Hu B, Wang BL, Yang WJ, Lv LH, et al. CD73 promotes hepatocellular carcinoma progression and metastasis via activating PI3K/AKT signaling by inducing Rap1-mediated membrane localization of P110β and predicts poor prognosis. J Hematol Oncol. (2019) 12:37. doi: 10.1186/s13045-019-0724-7
90. Shali S, Yu J, Zhang X, Wang X, Jin Y, Su M, et al. Ecto-5’-nucleotidase (CD73) is a potential target of hepatocellular carcinoma. J Cell Physiol. (2019) 234:10248–59. doi: 10.1002/jcp.v234.7
91. Faraoni EY, Singh K, Chandra V, Le Roux O, Dai Y, Sahin I, et al. CD73-dependent adenosine signaling through adora2b drives immunosuppression in ductal pancreatic cancer. Cancer Res. (2023) 83:1111–27. doi: 10.1158/0008-5472.CAN-22-2553
92. Zhao J, Soto LMS, Wang H, Katz MH, Prakash LR, Kim M, et al. Overexpression of CD73 in pancreatic ductal adenocarcinoma is associated with immunosuppressive tumor microenvironment and poor survival. Pancreatology: Off J Int Assoc Pancreatology (IAP) [et al]. (2021) 21:942–9. doi: 10.1016/j.pan.2021.03.018
93. Zhou L, Jia S, Chen Y, Wang W, Wu Z, Yu W, et al. The distinct role of CD73 in the progression of pancreatic cancer. J Mol Med (Berlin Germany). (2019) 97:803–15. doi: 10.1007/s00109-018-01742-0
94. Chen Q, Pu N, Yin H, Zhang J, Zhao G, Lou W, et al. CD73 acts as a prognostic biomarker and promotes progression and immune escape in pancreatic cancer. J Cell Mol Med. (2020) 24:8674–86. doi: 10.1111/jcmm.v24.15
95. O’Brien BJ, Faraoni EY, Strickland LN, Ma Z, Mota V, Mota S, et al. CD73-generated extracellular adenosine promotes resolution of neutrophil-mediated tissue injury and restrains metaplasia in pancreatitis. FASEB journal: Off Publ Fed Am Societies Exp Biol. (2023) 37:e22684. doi: 10.1096/fj.202201537R
96. Lima CF, Tamegnon A, Rodriguez S, Maru D, Martin PL, Cooper ZA, et al. Exploring the expression of adenosine pathway-related markers CD73 and CD39 in colorectal and pancreatic carcinomas characterized by multiplex immunofluorescence: A pilot study. Pathobiology: J immunopathology Mol Cell Biol. (2023) 91(3):205–18. doi: 10.1159/000534677
97. Ávila-Ibarra LR, Mora-García ML, García-Rocha R, Hernández-Montes J, Weiss-Steider B, Montesinos JJ, et al. Mesenchymal stromal cells derived from normal cervix and cervical cancer tumors increase CD73 expression in cervical cancer cells through TGF-β1 production. Stem Cells Dev. (2019) 28:477–88. doi: 10.1089/scd.2018.0183
98. Monteiro I, Missiaglia E, Sciarra A, Santos JV, Bouilly J, Romero P, et al. CD73 expression in normal, hyperplastic, and neoplastic thyroid: a systematic evaluation revealing CD73 overexpression as a feature of papillary carcinomas. Virchows Archiv: an Int J pathology. (2021) 479:209–14. doi: 10.1007/s00428-021-03100-x
99. Jeong YM, Cho H, Kim TM, Kim Y, Jeon S, Bychkov A, et al. CD73 overexpression promotes progression and recurrence of papillary thyroid carcinoma. Cancers. (2020) 12(10):3042. doi: 10.3390/cancers12103042
100. Liu C, Gao ZW, Wang X, Lin F, Zhang HZ, Dong K. CD73 promotes cervical cancer growth via EGFR/AKT1 pathway. Trans Cancer Res. (2022) 11:1089–98. doi: 10.21037/tcr-21-2446
101. He X, Gu Y, Cao Y, Hu B, Fang H, Fei Y, et al. Impact of intratumoural CD73 expression on prognosis and therapeutic response in patients with gastric cancer. Eur J Cancer (Oxford England: 1990). (2021) 157:114–23. doi: 10.1016/j.ejca.2021.08.006
102. Numakura S, Uozaki H, Kikuchi Y, Watabe S, Togashi A, Watanabe M. Mesenchymal stem cell marker expression in gastric cancer stroma. Anticancer Res. (2019) 39:387–93. doi: 10.21873/anticanres.13124
103. Kolbe K, Wittner M, Hartjen P, Hüfner AD, Degen O, Ackermann C, et al. Inversed ratio of CD39/CD73 expression on γδ T cells in HIV versus healthy controls correlates with immune activation and disease progression. Front Immunol. (2022) 13:867167. doi: 10.3389/fimmu.2022.867167
104. Shahbaz S, Okoye I, Blevins G, Elahi S. Elevated ATP via enhanced miRNA-30b, 30c, and 30e downregulates the expression of CD73 in CD8+ T cells of HIV-infected individuals. PloS pathogens. (2022) 18:e1010378. doi: 10.1371/journal.ppat.1010378
105. Tripathi A, Lin E, Xie W, Flaifel A, Steinharter JA, Stern Gatof EN, et al. Prognostic significance and immune correlates of CD73 expression in renal cell carcinoma. J immunotherapy Cancer. (2020) 8(2):e001467. doi: 10.1136/jitc-2020-001467
106. Xiong L, Wen Y, Miao X, Yang Z. NT5E and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal transition (EMT) are associated with tumor progression and survival of patients with gallbladder cancer. Cell Tissue Res. (2014) 355:365–74. doi: 10.1007/s00441-013-1752-1
107. Sun BY, Yang ZF, Wang ZT, Liu G, Zhou C, Zhou J, et al. Integrative analyses identify CD73 as a prognostic biomarker and immunotherapeutic target in intrahepatic cholangiocarcinoma. World J Surg Oncol. (2023) 21:90. doi: 10.1186/s12957-023-02970-6
108. Wu XR, He XS, Chen YF, Yuan RX, Zeng Y, Lian L, et al. High expression of CD73 as a poor prognostic biomarker in human colorectal cancer. J Surg Oncol. (2012) 106:130–7. doi: 10.1002/jso.v106.2
109. Morello S, Capone M, Sorrentino C, Giannarelli D, Madonna G, Mallardo D, et al. Soluble CD73 as biomarker in patients with metastatic melanoma patients treated with nivolumab. J Trans Med. (2017) 15:244. doi: 10.1186/s12967-017-1348-8
110. Tang K, Zhang J, Cao H, Xiao G, Wang Z, Zhang X, et al. Identification of CD73 as a novel biomarker encompassing the tumor microenvironment, prognosis, and therapeutic responses in various cancers. Cancers. (2022) 14(22):5663. doi: 10.3390/cancers14225663
111. Yoshida R, Saigi M, Tani T, Springer BF, Shibata H, Kitajima S, et al. MET-induced CD73 restrains STING-mediated immunogenicity of EGFR-mutant lung cancer. Cancer Res. (2022) 82:4079–92. doi: 10.1158/0008-5472.CAN-22-0770
112. Petruk N, Siddiqui A, Tadayon S, Määttä J, Mattila PK, Jukkola A, et al. CD73 regulates zoledronate-induced lymphocyte infiltration in triple-negative breast cancer tumors and lung metastases. Front Immunol. (2023) 14:1179022. doi: 10.3389/fimmu.2023.1179022
113. Petruk N, Tuominen S, Åkerfelt M, Mattsson J, Sandholm J, Nees M, et al. CD73 facilitates EMT progression and promotes lung metastases in triple-negative breast cancer. Sci Rep. (2021) 11:6035. doi: 10.1038/s41598-021-85379-z
114. Shi E, Wu Z, Karaoglan BS, Schwenk-Zieger S, Kranz G, Abdul Razak N, et al. 5’-Ectonucleotidase CD73/NT5E supports EGFR-mediated invasion of HPV-negative head and neck carcinoma cells. J Biomed science. (2023) 30:72. doi: 10.1186/s12929-023-00968-6
115. de Leve S, Wirsdörfer F, Jendrossek V. Targeting the immunomodulatory CD73/adenosine system to improve the therapeutic gain of radiotherapy. Front Immunol. (2019) 10:698. doi: 10.3389/fimmu.2019.00698
116. Hasmim M, Xiao M, Van Moer K, Kumar A, Oniga A, Mittelbronn M, et al. SNAI1-dependent upregulation of CD73 increases extracellular adenosine release to mediate immune suppression in TNBC. Front Immunol. (2022) 13:982821. doi: 10.3389/fimmu.2022.982821
117. Figueiró F, de Oliveira CP, Rockenbach L, Mendes FB, Bergamin LS, Jandrey EH, et al. Pharmacological improvement and preclinical evaluation of methotrexate-loaded lipid-core nanocapsules in a glioblastoma model. J Biomed nanotechnology. (2015) 11:1808–18. doi: 10.1166/jbn.2015.2125
118. Lopes DV, de Fraga Dias A, Silva LFL, Scholl JN, Sévigny J, Battastini AMO, et al. Influence of NSAIDs and methotrexate on CD73 expression and glioma cell growth. Purinergic signalling. (2021) 17:273–84. doi: 10.1007/s11302-021-09775-w
119. Goswami S, Walle T, Cornish AE, Basu S, Anandhan S, Fernandez I, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med. (2020) 26:39–46. doi: 10.1038/s41591-019-0694-x
120. Yan A, Joachims ML, Thompson LF, Miller AD, Canoll PD, Bynoe MS. CD73 promotes glioblastoma pathogenesis and enhances its chemoresistance via A(2B) adenosine receptor signaling. J neuroscience: Off J Soc Neurosci. (2019) 39:4387–402. doi: 10.1523/JNEUROSCI.1118-18.2019
121. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. (2016) 127:2391–405. doi: 10.1182/blood-2016-03-643544
122. Whiteley AE, Price TT, Cantelli G, Sipkins DA. Leukaemia: a model metastatic disease. Nat Rev Cancer. (2021) 21:461–75. doi: 10.1038/s41568-021-00355-z
123. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. (2011) 147:275–92. doi: 10.1016/j.cell.2011.09.024
124. Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer. (2020) 20:158–73. doi: 10.1038/s41568-019-0230-9
125. Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, et al. Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun. (2020) 11:5327. doi: 10.1038/s41467-020-19119-8
126. Zhang B, Li L, Ho Y, Li M, Marcucci G, Tong W, et al. Heterogeneity of leukemia-initiating capacity of chronic myelogenous leukemia stem cells. J Clin Invest. (2016) 126:975–91. doi: 10.1172/JCI79196
127. Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. (1994) 367:645–8. doi: 10.1038/367645a0
128. Ye H, Adane B, Khan N, Sullivan T, Minhajuddin M, Gasparetto M, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell. (2016) 19:23–37. doi: 10.1016/j.stem.2016.06.001
129. Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood. (2017) 129:1577–85. doi: 10.1182/blood-2016-10-696054
130. Ferrando AA, López-Otín C. Clonal evolution in leukemia. Nat Med. (2017) 23:1135–45. doi: 10.1038/nm.4410
131. Spurr CL, Smith TR, Jacobson LO. Chemotherapy in human lymphomas, leukemias, and allied disorders of the hemopoietic system. Radiology. (1948) 50:387–94. doi: 10.1148/50.3.387
132. Wang A, Zhong H. Roles of the bone marrow niche in hematopoiesis, leukemogenesis, and chemotherapy resistance in acute myeloid leukemia. Hematol (Amsterdam Netherlands). (2018) 23:729–39. doi: 10.1080/10245332.2018.1486064
133. Weissman IL. Translating stem and progenitor cell biology to the clinic: barriers and opportunities. Sci (New York NY). (2000) 287:1442–6. doi: 10.1126/science.287.5457.1442
134. Singh AK, McGuirk JP. Allogeneic stem cell transplantation: A historical and scientific overview. Cancer Res. (2016) 76:6445–51. doi: 10.1158/0008-5472.CAN-16-1311
135. Tavor S, Eisenbach M, Jacob-Hirsch J, Golan T, Petit I, Benzion K, et al. The CXCR4 antagonist AMD3100 impairs survival of human AML cells and induces their differentiation. Leukemia. (2008) 22:2151–5158. doi: 10.1038/leu.2008.238
136. Kovacsovics T, Levy MY, Cook RJ, Kolitz JE, Westervelt P, Donnellan WB, et al. A randomized phase II trial of CX-01 with standard therapy in elderly patients with acute myeloid leukemia (AML). J Clin Oncol. (2019) 37:7001. doi: 10.1200/JCO.2019.37.15_suppl.7001
137. Liesveld JL, Bechelli J, Rosell K, Lu C, Bridger G, Phillips G 2nd, et al. Effects of AMD3100 on transmigration and survival of acute myelogenous leukemia cells. Leukemia Res. (2007) 31:1553–63. doi: 10.1016/j.leukres.2007.02.017
138. Pillozzi S, Bernini A, Spiga O, Lelli B, Petroni G, Bracci L, et al. Peptides and small molecules blocking the CXCR4/CXCL12 axis overcome bone marrow−induced chemoresistance in acute leukemias. Oncol Rep. (2019) 41:312–24. doi: 10.3892/or.2018.6808
139. Tavor S, Petit I, Porozov S, Avigdor A, Dar A, Leider-Trejo L, et al. CXCR4 regulates migration and development of human acute myelogenous leukemia stem cells in transplanted NOD/SCID mice. Cancer Res. (2004) 64:2817–24. doi: 10.1158/0008-5472.CAN-03-3693
140. DeAngelo DJ, Jonas BA, Liesveld JL, Bixby DL, Advani AS, Marlton P, et al. Uproleselan (GMI-1271), an E-selectin antagonist, improves the efficacy and safety of chemotherapy in relapsed/refractory (R/R) and newly diagnosed older patients with acute myeloid leukemia: final, correlative, and subgroup analyses. Blood. (2018) 132:331. doi: 10.1182/blood-2018-99-114286
141. Hsieh YT, Gang EJ, Geng H, Park E, Huantes S, Chudziak D, et al. Integrin alpha4 blockade sensitizes drug resistant pre-B acute lymphoblastic leukemia to chemotherapy. Blood. (2013) 121:1814–8. doi: 10.1182/blood-2012-01-406272
142. Herman SE, Mustafa RZ, Jones J, Wong DH, Farooqui M, Wiestner A. Treatment with ibrutinib inhibits BTK- and VLA-4-dependent adhesion of chronic lymphocytic leukemia cells in vivo. Clin Cancer research: an Off J Am Assoc Cancer Res. (2015) 21:4642–51. doi: 10.1158/1078-0432.CCR-15-0781
143. DiGiuseppe JA, Fuller SG, Borowitz MJ. Overexpression of CD49f in precursor B-cell acute lymphoblastic leukemia: potential usefulness in minimal residual disease detection. Cytometry Part B Clin cytometry. (2009) 76:150–5. doi: 10.1002/cyto.b.v76b:2
144. Yao H, Price TT, Cantelli G, Ngo B, Warner MJ, Olivere L, et al. Leukaemia hijacks a neural mechanism to invade the central nervous system. Nature. (2018) 560:55–60. doi: 10.1038/s41586-018-0342-5
145. Hamidi H, Pietilä M, Ivaska J. The complexity of integrins in cancer and new scopes for therapeutic targeting. Br J cancer. (2016) 115:1017–23. doi: 10.1038/bjc.2016.312
146. Orlando EJ, Han X, Tribouley C, Wood PA, Leary RJ, Riester M, et al. Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nat Med. (2018) 24:1504–6. doi: 10.1038/s41591-018-0146-z
147. Epperly R, Gottschalk S, Velasquez MP. A bump in the road: how the hostile AML microenvironment affects CAR T cell therapy. Front Oncol. (2020) 10:262. doi: 10.3389/fonc.2020.00262
148. Jacoby E, Nguyen SM, Fountaine TJ, Welp K, Gryder B, Qin H, et al. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat Commun. (2016) 7:12320. doi: 10.1038/ncomms12320
149. Lulla PD, Mamonkin M, Brenner MK. Adoptive cell therapy for acute myeloid leukemia and T-cell acute lymphoblastic leukemia. Cancer J (Sudbury Mass). (2019) 25:199–207. doi: 10.1097/PPO.0000000000000376
150. Zoine JT, Moore SE, Velasquez MP. Leukemia’s next top model? Syngeneic models to advance adoptive cellular therapy. Front Immunol. (2022) 13:867103. doi: 10.3389/fimmu.2022.867103
151. Girdwood RH. Drug-induced anaemias. Drugs. (1976) 11:394–404. doi: 10.2165/00003495-197611050-00003
152. Ciciarello M, Corradi G, Forte D, Cavo M, Curti A. Emerging bone marrow microenvironment-driven mechanisms of drug resistance in acute myeloid leukemia: tangle or chance? Cancers. (2021) 13(21):5319. doi: 10.3390/cancers13215319
153. Cheng L, Li C, Zhang X, Chen Y, Yan J. The consistency between the Chinese essential medicines list and treatment guidelines-taking oncology medicines as an example. Front Public Health. (2022) 10:943994. doi: 10.3389/fpubh.2022.943994
154. Ma XY, Wei L, Lei Z, Chen Y, Ding Z, Chen ZS. Recent progress on targeting leukemia stem cells. Drug Discovery Today. (2021) 26:1904–13. doi: 10.1016/j.drudis.2021.05.009
155. Allard D, Chrobak P, Bareche Y, Allard B, Tessier P, Bergeron MA, et al. CD73 promotes chronic lymphocytic leukemia. Cancers. (2022) 14(13):3130. doi: 10.3390/cancers14133130
156. Bozorgmehr N, Hnatiuk M, Peters AC, Elahi S. Depletion of polyfunctional CD26(high)CD8(+) T cells repertoire in chronic lymphocytic leukemia. Exp Hematol Oncol. (2023) 12:13. doi: 10.1186/s40164-023-00375-5
157. Furuta K, Onishi H, Ikada Y, Masaki K, Tanaka S, Kaito C. ATP and its metabolite adenosine cooperatively upregulate the antigen-presenting molecules on dendritic cells leading to IFN-γ production by T cells. J Biol Chem. (2023) 299:104587. doi: 10.1016/j.jbc.2023.104587
158. Cai Y, Feng L, Wang X. Targeting the tumor promoting effects of adenosine in chronic lymphocytic leukemia. Crit Rev oncology/hematology. (2018) 126:24–31. doi: 10.1016/j.critrevonc.2018.03.022
159. Abruzzo LV, Herling CD, Calin GA, Oakes C, Barron LL, Banks HE, et al. Trisomy 12 chronic lymphocytic leukemia expresses a unique set of activated and targetable pathways. Haematologica. (2018) 103:2069–78. doi: 10.3324/haematol.2018.190132
160. da Silva Nunes VB, Dias CK, De Bastiani MA, Farias MG, Spagnol F, Alegretti AP, et al. NT5E gene and CD38 protein as potential prognostic biomarkers for childhood B-acute lymphoblastic leukemia. Purinergic signalling. (2022) 18:211–22. doi: 10.1007/s11302-022-09841-x
161. Sędek Ł, Theunissen P, Sobral da Costa E, van-der-Sluijs-Gelling A, Mejstrikova E, Gaipa G, et al. Differential expression of CD73, CD86 and CD304 in normal vs. leukemic B-cell precursors and their utility as stable minimal residual disease markers in childhood B-cell precursor acute lymphoblastic leukemia. J Immunol Methods. (2019) 475:112429. doi: 10.1016/j.jim.2018.03.005
162. Wang W, Gao L, Li Y, Li ZL, Gong M, Huang FZ, et al. The application of CD73 in minimal residual disease monitoring using flow cytometry in B-cell acute lymphoblastic leukemia. Leukemia lymphoma. (2016) 57:1174–81. doi: 10.3109/10428194.2015.1070153
163. Tembhare PR, Ghogale S, Ghatwai N, Badrinath Y, Kunder N, Patkar NV, et al. Evaluation of new markers for minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia: CD73 and CD86 are the most relevant new markers to increase the efficacy of MRD 2016; 00B: 000-000. Cytometry Part B Clin cytometry. (2018) 94:100–11. doi: 10.1002/cyto.b.21486
164. Jakobsen JS, Laursen LG, Schuster MB, Pundhir S, Schoof E, Ge Y, et al. Mutant CEBPA directly drives the expression of the targetable tumor-promoting factor CD73 in AML. Sci Adv. (2019) 5:eaaw4304. doi: 10.1126/sciadv.aaw4304
165. Huang JC, Basu SK, Zhao X, Chien S, Fang M, Oehler VG, et al. Mesenchymal stromal cells derived from acute myeloid leukemia bone marrow exhibit aberrant cytogenetics and cytokine elaboration. Blood Cancer J. (2015) 5:e302. doi: 10.1038/bcj.2015.17
166. Stelmach P, Trumpp A. Leukemic stem cells and therapy resistance in acute myeloid leukemia. Haematologica. (2023) 108:353–66. doi: 10.3324/haematol.2022.280800
167. Mascanfroni ID, Takenaka MC, Yeste A, Patel B, Wu Y, Kenison JE, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat Med. (2015) 21:638–46. doi: 10.1038/nm.3868
168. Fang F, Cao W, Zhu W, Lam N, Li L, Gaddam S, et al. The cell-surface 5’-nucleotidase CD73 defines a functional T memory cell subset that declines with age. Cell Rep. (2021) 37:109981. doi: 10.1016/j.celrep.2021.109981
169. Sitkovsky MV, Hatfield S, Abbott R, Belikoff B, Lukashev D, Ohta A. Hostile, hypoxia-A2-adenosinergic tumor biology as the next barrier to overcome for tumor immunologists. Cancer Immunol Res. (2014) 2:598–605. doi: 10.1158/2326-6066.CIR-14-0075
170. Hatfield SM, Kjaergaard J, Lukashev D, Belikoff B, Schreiber TH, Sethumadhavan S, et al. Systemic oxygenation weakens the hypoxia and hypoxia inducible factor 1α-dependent and extracellular adenosine-mediated tumor protection. J Mol Med (Berlin Germany). (2014) 92:1283–92. doi: 10.1007/s00109-014-1189-3
171. Steingold JM, Hatfield SM. Targeting hypoxia-A2A adenosinergic immunosuppression of antitumor T cells during cancer immunotherapy. Front Immunol. (2020) 11:570041. doi: 10.3389/fimmu.2020.570041
172. López-Abente J, Correa-Rocha R, Pion M. Functional mechanisms of treg in the context of HIV infection and the janus face of immune suppression. Front Immunol. (2016) 7:192. doi: 10.3389/fimmu.2016.00192
173. Yu JC, Lin G, Field JJ, Linden J. Induction of antiinflammatory purinergic signaling in activated human iNKT cells. JCI Insight. (2018) 3(17):e91954. doi: 10.1172/jci.insight.91954
174. Turcotte M, Allard D, Mittal D, Bareche Y, Buisseret L, José V, et al. CD73 promotes resistance to HER2/erbB2 antibody therapy. Cancer Res. (2017) 77:5652–63. doi: 10.1158/0008-5472.CAN-17-0707
175. Reinhardt J, Landsberg J, Schmid-Burgk JL, Ramis BB, Bald T, Glodde N, et al. MAPK signaling and inflammation link melanoma phenotype switching to induction of CD73 during immunotherapy. Cancer Res. (2017) 77:4697–709. doi: 10.1158/0008-5472.CAN-17-0395
176. Brauneck F, Haag F, Woost R, Wildner N, Tolosa E, Rissiek A, et al. Increased frequency of TIGIT(+)CD73-CD8(+) T cells with a TOX(+) TCF-1low profile in patients with newly diagnosed and relapsed AML. Oncoimmunology. (2021) 10:1930391. doi: 10.1080/2162402X.2021.1930391
177. Kordaß T, Osen W, Eichmüller SB. Controlling the immune suppressor: transcription factors and microRNAs regulating CD73/NT5E. Front Immunol. (2018) 9:813. doi: 10.3389/fimmu.2018.00813
178. Sun T, Liu B, Li Y, Wu J, Cao Y, Yang S, et al. Oxamate enhances the efficacy of CAR-T therapy against glioblastoma via suppressing ectonucleotidases and CCR8 lactylation. J Exp Clin Cancer research: CR. (2023) 42:253. doi: 10.1186/s13046-023-02815-w
179. Liu Q, Li J, Khoury J, Colgan SP, Ibla JC. Adenosine signaling mediates SUMO-1 modification of IkappaBalpha during hypoxia and reoxygenation. J Biol Chem. (2009) 284:13686–95. doi: 10.1074/jbc.M809275200
180. Muñóz-Godínez R, de-Lourdes-Mora-García M, Weiss-Steider B, Montesinos-Montesinos JJ, Del-Carmen-Aguilar-Lemarroy A, García-Rocha R, et al. Detection of CD39 and a highly glycosylated isoform of soluble CD73 in the plasma of patients with cervical cancer: correlation with disease progression. Mediators inflammation. (2020) 2020:1678780. doi: 10.1155/2020/1678780
181. Luke JJ, Powderly JD, Merchan JR, Barve MA, Hotson AN, Mobasher M, et al. Immunobiology, preliminary safety, and efficacy of CPI-006, an anti-CD73 antibody with immune modulating activity, in a phase 1 trial in advanced cancers. J Clin Oncol. (2019) 37:2505. doi: 10.1200/JCO.2019.37.15_suppl.2505
182. Lawson KV, Kalisiak J, Lindsey EA, Newcomb ET, Leleti MR, Debien L, et al. Discovery of AB680: A potent and selective inhibitor of CD73. J medicinal Chem. (2020) 63:11448–68. doi: 10.1021/acs.jmedchem.0c00525
183. Chen Q, Yin H, He J, Xie Y, Wang W, Xu H, et al. Tumor microenvironment responsive CD8(+) T cells and myeloid-derived suppressor cells to trigger CD73 inhibitor AB680-based synergistic therapy for pancreatic cancer. Advanced Sci (Weinheim Baden-Wurttemberg Germany). (2023) 10:e2302498. doi: 10.1002/advs.202302498
184. Robert F, Dumbrava EE, Xing Y, Mills E, Freddo JL, Theuer CP, et al. Preliminary safety, pharmacokinetics (PK), pharmacodynamics (PD) and clinical efficacy of uliledlimab (TJ004309), a differentiated CD73 antibody, in combination with atezolizumab in patients with advanced cancer. JCO. (2021) 39:2511. doi: 10.1200/JCO.2021.39.15_suppl.2511
185. Bendell J, LoRusso P, Overman M, Noonan AM, Kim DW, Strickler JH, et al. First-in-human study of oleclumab, a potent, selective anti-CD73 monoclonal antibody, alone or in combination with durvalumab in patients with advanced solid tumors. Cancer immunology immunotherapy: CII. (2023) 72:2443–58. doi: 10.1007/s00262-023-03430-6
186. Markman B, Hsieh AH-C, Coward J, Carlino MS, Frentzas S, Jin X, et al. A phase I study of AK119, an anti-CD73 monoclonal antibody, in combination with AK104, an anti-PD-1/CTLA-4 bispecific antibody, in patients with advanced or metastatic solid tumors. JCO. (2021) 39:TPS2675. doi: 10.1200/JCO.2021.39.15_suppl.TPS2675
187. Antonioli L, Yegutkin GG, Pacher P, Blandizzi C, Haskó G. Anti-CD73 in cancer immunotherapy: awakening new opportunities. Trends cancer. (2016) 2:95–109. doi: 10.1016/j.trecan.2016.01.003
188. Wurm M, Schaaf O, Reutner K, Ganesan R, Mostböck S, Pelster C, et al. A novel antagonistic CD73 antibody for inhibition of the immunosuppressive adenosine pathway. Mol Cancer Ther. (2021) 20:2250–61. doi: 10.1158/1535-7163.MCT-21-0107
189. Zhang C, Wang K, Wang H. Adenosine in cancer immunotherapy: Taking off on a new plane. Biochim Biophys Acta Rev cancer. (2023) 1878:189005. doi: 10.1016/j.bbcan.2023.189005
190. Adamiak M, Bujko K, Brzezniakiewicz-Janus K, Kucia M, Ratajczak J, Ratajczak MZ. The inhibition of CD39 and CD73 cell surface ectonucleotidases by small molecular inhibitors enhances the mobilization of bone marrow residing stem cells by decreasing the extracellular level of adenosine. Stem Cell Rev Rep. (2019) 15:892–9. doi: 10.1007/s12015-019-09918-y
191. Sheth S, Brito R, Mukherjea D, Rybak LP, Ramkumar V. Adenosine receptors: expression. Funct Regul. (2014) 15:2024–52. doi: 10.3390/ijms15022024
192. Camici M, Garcia-Gil M, Tozzi MG. The inside story of adenosine. International Journal of Molecular Sciences. (2018) 19:784. doi: 10.3390/ijms19030784
Keywords: CD73, CD39, leukemia, immune escape, immune checkpoint, clinical treatment
Citation: Gao H, Zhang T, Li K and Li X (2025) CD73: a new immune checkpoint for leukemia treatment. Front. Immunol. 16:1486868. doi: 10.3389/fimmu.2025.1486868
Received: 27 August 2024; Accepted: 14 February 2025;
Published: 06 March 2025.
Edited by:
Antonio Francesco Campese, Sapienza University of Rome, ItalyReviewed by:
Silvana Morello, University of Salerno, ItalyCopyright © 2025 Gao, Zhang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ke Li, bGlrZTE5ODZAMTYzLmNvbQ==; Xia Li, eGlhbGlAc2R1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.