- 1INBB – Istituto Nazionale Biostrutture e Biosistemi, Rome, Italy
- 2Dipartimento di Scienze, Università di Roma Tre, Rome, Italy
- 3Dipartimento di Scienze Biotecnologiche di Base, Cliniche, Intensivologiche e Perioperatorie, Università Cattolica del Sacro Cuore, Rome, Italy
- 4Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Rome, Italy
- 5Dipartimento di Biomedicina e Prevenzione, Università di Roma Tor Vergata, Rome, Italy
- 6Istituto di Cristallografia, Consiglio Nazionale delle Ricerche, Bari, Italy
- 7Tiber Biotech Srl, Rome, Italy
Background: Mature T-cell neoplasms arise from the neoplastic transformation of a single T lymphocyte, and all cells in a neoplastic clone share the same V segment in the beta chain of the T-cell receptor (TCR). These segments may represent an innovative target for the development of targeted therapies.
Methods: A specific V segment of the TCR beta chain (TRBV5-1) was analyzed using bioinformatic tools, identifying three potential antigenic peptides. One of these peptides, selected for synthesis, was used to screen a library of human single-chain variable fragments (scFv) through phage display. One fragment demonstrated high affinity and specificity for the antigen and was used to produce a human monoclonal antibody of the IgG1 class.
Results: Surface plasmon resonance (SPR) studies confirmed the high affinity of the monoclonal antibody for the antigen in the nanomolar range. Flow cytometry analysis on patients’ samples demonstrated that the antibody, conjugated with a fluorochrome, selectively binds to tumor T lymphocytes expressing TRBV5-1, without binding to other lymphocytes or blood cell components.
Conclusions: The development of fully human IgG1 monoclonal antibodies targeting specific V segments of the TCR beta chain represents a potential therapeutic option for patients with mature T-cell neoplasms.
Introduction
T lymphocytes play a critical role in the immune response by identifying and eliminating infected or malignant cells (1). When these cells undergo malignant transformation, they can give rise to mature T-cell neoplasms (2), a relatively rare and aggressive group of cancers currently categorized by the WHO Classification of Haematolymphoid Tumours (3). Symptoms vary and may include lymphadenopathy, fever, night sweats, and, for cutaneous T-cell lymphomas, skin lesions (4).
Despite advancements in targeted therapies and immunotherapy, mature T-cell neoplasms remain a challenging clinical issue with limited treatment options, underscoring the need for innovative therapeutic strategies (5). These malignancies arise from the transformation of individual T cells, which express clonal T-cell receptors (TCRs) specific to the neoplastic clone (6). This feature presents a potential target for the development of monoclonal antibodies aimed at the variable (V) segments of TCRs expressed in malignant clones (7).
Several monoclonal antibodies have been developed in recent years for the treatment of specific forms of mature T-cell neoplasms (8). Alemtuzumab is a humanized IgG1 monoclonal antibody targeting the CD52 antigen, expressed on more than 95% of peripheral T and B lymphocytes (9). It is indicated for the treatment of fludarabine-refractory chronic lymphocytic leukemia (10) and has demonstrated efficacy in certain T-cell lymphomas, including cutaneous T-cell lymphoma (11) and peripheral T-cell lymphoma (12).
Brentuximab Vedotin is an antibody-drug conjugate (ADC) in which an anti-CD30 monoclonal antibody is conjugated with monomethyl auristatin E (MMAE), a cytotoxic agent (13). It is approved for the treatment of CD30-positive Hodgkin lymphoma (14) and relapsed or refractory anaplastic large-cell lymphoma (15). CD30 is expressed in a significant proportion of T-cell lymphomas, making this drug relevant for these malignancies (16).
Mogamulizumab is a humanized monoclonal antibody directed against the CCR4 receptor, which is expressed on certain malignant T cells (17). It has shown efficacy in the treatment of cutaneous T-cell lymphoma, particularly mycosis fungoides and Sézary syndrome (18).
Daratumumab, an anti-CD38 monoclonal antibody, is currently approved for multiple myeloma (19). However, preliminary studies have suggested potential efficacy in treating CD38-positive T-cell lymphomas, such as nasal-type NK/T-cell lymphoma (20).
Some monoclonal antibodies targeting the immune checkpoint PD-1, particularly Nivolumab (21) and Pembrolizumab (22), are currently under investigation for the treatment of refractory or relapsed T-cell lymphomas, with promising results in certain subtypes (23). The rationale behind these agents lies in the reactivation of the immune response against tumor cells (24).
Finally, bispecific antibodies capable of simultaneously binding an antigen on tumor cells and an antigen on healthy T cells (commonly CD3) are under investigation (25). These antibodies facilitate the activation of T cells against the tumor (26). This approach has shown promise in the treatment of B-cell lymphomas, where such antibodies promote the interaction between healthy T cells (via CD3) and tumor B cells (via CD20) (27). However, in the treatment of T-cell lymphomas, these antibodies appear to activate a fratricidal response, wherein healthy T lymphocytes and malignant T lymphocytes eliminate each other (28).
It is well known that the TCR consists of alpha and beta chains (or gamma and delta chains in γδ T cells), each with variable (V) and constant (C) regions (29), generated through V(D)J recombination to ensure broad antigen recognition (30). This diversity enables T cells to detect a wide array of antigens (31) but also poses challenges for targeted therapies due to the variability in TCR sequences (32). Targeting specific V segments, however, could enable precise therapeutic interventions against malignant T-cell clones (33).
Recent advances in phage display technology allow for the efficient screening of human single-chain variable fragments (scFvs) against specific antigens, enabling the production of fully human IgG1 monoclonal antibodies (34). This approach has the potential to deliver highly specific treatments for patients with mature T-cell neoplasms, offering a promising direction for future therapies.
Materials and methods
Objective
This study aimed to develop fully human monoclonal antibodies targeting the TRBV5-1 segment of the TCR beta chain. The affinity and specificity of these antibodies were assessed via bioinformatic analysis, surface plasmon resonance (SPR), and flow cytometry.
Screening of human scFvs by phage display
Among the 68 V segments of the human TCR beta chain (35), TRBV5-1 was identified as a candidate target due to its significant expression in normal T lymphocytes (36) and potential presence in T-cell lymphomas. Antigenic peptides derived from the amino acid sequence of TRBV5-1 were synthesized by Proteogenix (Schiltigheim, France). The candidate peptides underwent analysis using two bioinformatic tools, Antigen Profiler Peptide (37) and AbDesigner (38), to assess antigenicity and structural compatibility, thereby confirming their suitability for the subsequent phage display process.
Proteogenix further conducted the screening of a proprietary library containing 5.37x1010 unique human scFv clones (LiAb-SFMAX™). This library was constructed from naive B lymphocytes isolated from 368 healthy donors across diverse ethnic groups (Caucasian, Arab, African, South American, and Asian), ensuring a broad representation of scFv diversity.
Synthesis of r-hIgG1 monoclonal antibody
The selected scFv sequences were optimized for expression and used to produce the recombinant human IgG1 monoclonal antibody (r-hIgG1 mAb) by GenScript Biotech BV (Netherlands). These sequences were cloned and transfected into TurboCHO™ cells using a lentiviral vector system to ensure stable and high-yield expression. The antibody was then purified from the culture supernatant using protein A chromatography, which selectively binds the Fc region of IgG, yielding a highly concentrated product.
The purity and molecular integrity of the r-hIgG1 mAb were confirmed via SDS-PAGE, assessing molecular weight under reducing and non-reducing conditions, and by size-exclusion high-performance liquid chromatography (SEC-HPLC) to evaluate homogeneity and aggregate content.
Synthesis of TCR
To assess affinity by SPR, a single-chain TCR (scTCR) expressing the TRBV5-1 segment was synthesized. Structural data from UNIPROT (39) (A0A578) and the Protein Data Bank (40) (PDB code 5BRZ) were referenced to ensure sequence accuracy and structural integrity. The variable regions of the alpha and beta chains were linked using a flexible linker peptide to preserve the TCR’s native structure and functional binding properties (41). A polyhistidine tag was added to facilitate purification and immobilization on the SPR sensor chip.
Structural preparation and bioinformatics analysis
To evaluate the solvent accessibility and potential interaction of the immunogenic peptide with antibodies within the TCR, various bioinformatic techniques were employed.
The three-dimensional structure of the TCR (PDB code: 5BS0) was obtained from the Protein Data Bank. Due to possible imperfections in crystallographic structures, such as missing loops or unmapped atoms, Modeller was used to reconstruct the protein structure and minimize steric clashes (42).
For antibody-TCR docking, the protein structures were protonated using PDB2PQR to assign correct charges and accurately calculate electrostatic interactions (43). Energy minimization was performed with the “relax” utility of the Rosetta suite, guided by the REF2015 force field (44).
Docking with the six target antibodies was conducted using SnugDock, a module in Rosetta specific for antibody docking on protein structures (45). Ensemble docking was employed to account for the high flexibility of loop regions. A 300 ns molecular dynamics simulation of the TCR was executed using Amber (46), and the trajectory was clustered with cpptraj to generate a set of conformations (47).
Structural models of the six antibodies were obtained using AlphaFold Multimer, suitable for multimeric structures like antibodies composed of heavy (H) and light (L) chains (48). Models with the highest pLDDT scores were selected. Constant regions were removed, and a protein linker was added between the N-terminus and C-terminus of the two chains using Modeller to obtain the corresponding scFv.
Ensemble docking of the TCR conformations with each scFv was performed using SnugDock, allowing flexible modeling of the complementarity-determining regions (CDRs). This approach was crucial due to an unstructured amino acid loop in the interaction area with the target peptide, enabling accurate prediction of docking poses despite the presence of unstructured regions.
SnugDock generated a set of antibody-antigen structures ranked according to the REF2015 scoring function of Rosetta. For each scFv, the three structures with the best interaction energies were selected.
Surface plasmon resonance
The interaction between the His-tagged scTCR (ligand) and the r-hIgG1 mAb (analyte) was measured using SPR on a Biacore X100 instrument (Biacore, Uppsala, Sweden). The His-tagged scTCR was immobilized on a nitrilotriacetic acid (NTA) sensor chip (Cytiva, Uppsala, Sweden) following the manufacturer’s protocol. The NTA surface was activated with Ni2+ ions to selectively bind the His-tagged scTCR, which could be removed during the regeneration phase by chelating the nickel ions with 0.35 M EDTA (pH 8.3; Sigma-Aldrich, St. Louis, MO, USA).
The Hys-tagged scTCR at a concentration of 44 μg/mL was injected over the sensor chip for 60 s, followed by a 10 s stabilization period. Analyte concentrations of 5, 2.5, 1.25, 0.625, and 0.3125 μM were tested. The SPR assay was performed at 25°C with a flow rate of 30 μL/min. The association phase was monitored for 180 s, and the dissociation phase for 600 s. The sensor chip surface was regenerated before each assay cycle by removing the bound His-tagged scTCR with EDTA and re-immobilizing fresh His-tagged ligand.
Kinetic parameters and the dissociation constant (K_D) were determined using Biacore X100 Evaluation Software (version 2.0.1 plus package). A 1:1 Langmuir binding model was applied to fit the experimental data, assuming a simple bimolecular interaction between the immobilized Hys-tagged scTCR and the analyte in solution.
Flow cytometry
Flow cytometric analysis was performed on aliquots of fresh cells using a FACSLyric™ cytometer (BD Biosciences, San Jose, CA). Cells were adjusted to a concentration of 1 × 106 cells/mL, and 100 µL of the suspension was placed into U-well tubes. Samples were incubated for 30 minutes at 4°C with a predefined combination of monoclonal antibodies to reliably identify neoplastic cells in Sézary syndrome: CD3 APC-H7, CD4 PE-Cy7, CD8 APC-R700, and CD45 V500-C (BD Biosciences).
Since monocytes express Fc receptors for immunoglobulins and could bind the r-hIgG1 mAb against the TRBV5-1 fragment via its constant region, a monoclonal antibody targeting CD14 (BV605, BD Biosciences) was included to identify and exclude monocytes. After lysing erythrocytes with isomolar NH4Cl buffer for 10 minutes, cells were washed three times with PBS containing 0.2% (w/v) azide and 0.2% (w/v) bovine serum albumin (BSA).
An amorphous gate encompassing all mononuclear cells while excluding erythrocytes and debris was set before acquisition. At least 1,000 CD3+ events per sample were acquired using a logical gate combining fluorescence/SSC and FSC/SSC plots. Data were analyzed using Infinicyte™ software (BD Cytognos, BD Switzerland Srl).
In the initial phase of the experiment, serial dilutions of the FITC-conjugated r-hIgG1 mAb were tested by adding 5 µL, 1 µL, and 0.5 µL of the antibody at a concentration of 1 mg/mL to the cell suspension. Results indicated that 0.5 µL of FITC-conjugated r-hIgG1 mAb provided an optimal signal, whereas larger volumes (1 µL and 5 µL) resulted in out-of-scale signals. This dilution was selected for the final experiments.
Results
Identification of TRBV5-1 antigenic peptides
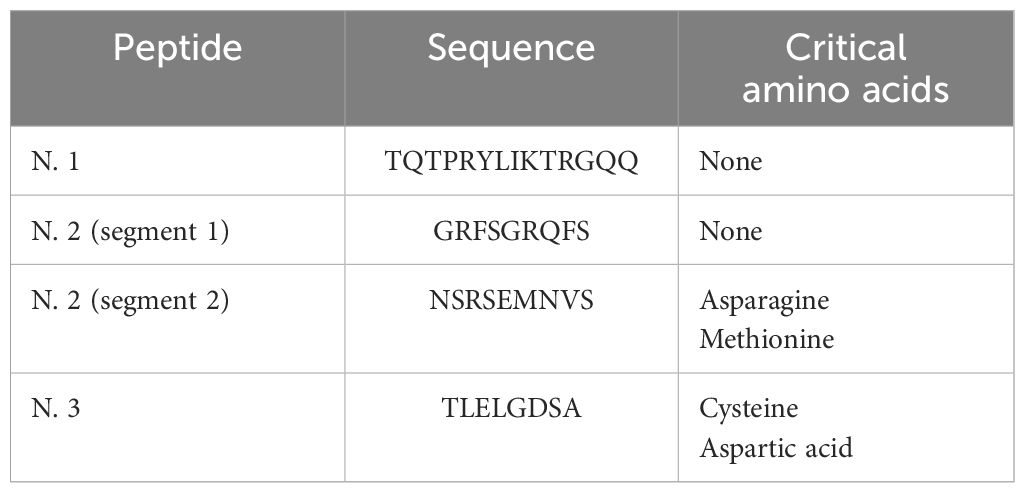
The nucleotide and amino acid sequences of the TRBV5-1 segment are provided in Table 1 and are available in the Protein Data Bank and IMGT database (14). Using 3D modeling and docking techniques with MHC class I molecules and the TCR alpha chain, three potential antigenic peptides were identified within TRBV5-1 (Table 2).
The first peptide, located near the N-terminal, is highly accessible to antibodies but is non-linear and thus challenging to synthesize. A shorter segment (highlighted in red in Table 2, Figure 1) may be preferable. The second peptide, positioned mid-segment, contains two regions (black and green in Table 2) with favorable properties for antibody development. The third peptide, closer to the C-terminal, also shows potential but contains residues susceptible to cleavage under acidic conditions.
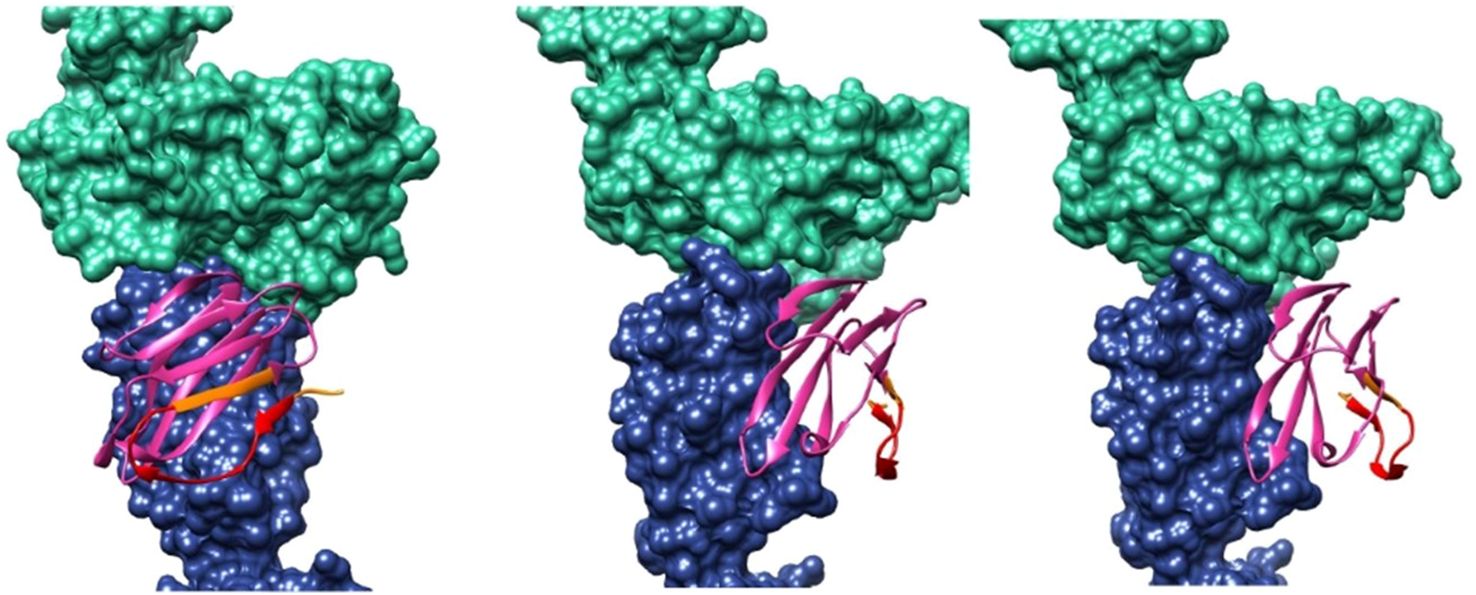
Figure 1. 3D modeling and docking images showing the TCR alpha chain (in blue), the TRBV5-1 segment (in pink), and an HLA I/β2-microglobulin complex (in green). The antigenic peptide TQTPRYLIKTRGQQC is highlighted in red.
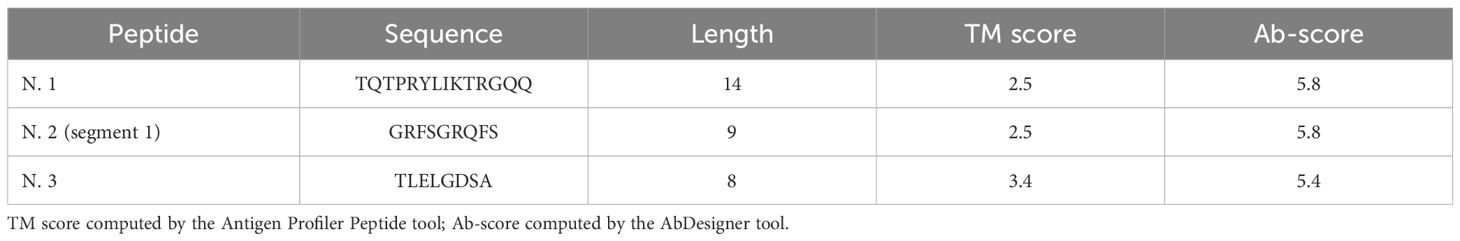
Antigenic peptides for scFv screening were prioritized based on synthesis feasibility (Table 3). Peptide No. 1 and the first segment of Peptide No. 2 lack critical residues, making them ideal candidates. In contrast, the second segment of Peptide No. 2 and Peptide No. 3 contain amino acids like asparagine and methionine, which could hinder synthesis. Table 4 shows the scores for each peptide as calculated by Antigen Profiler Peptide and AbDesigner. Peptide No. 1 (TQTPRYLIKTRGQQ) achieved high scores across tools, with an optimal length and no synthesis limitations, making it the best candidate for scFv screening.
Selection of scFvs by phage display
The selection of scFvs using phage display started with conjugating the peptide TQTPRYLIKTRGQQ to three carrier proteins: BSA, OVA, and KLH. Panning cycles were conducted with tubes coated with the conjugated antigenic peptide, and non-specific phages recognizing only the carrier proteins were removed through washes. Phages binding the peptide were then eluted with Glycine-HCl, neutralized, and amplified in Escherichia coli TG1 culture for subsequent panning rounds.
To assess enrichment, a polyclonal ELISA was performed, coating multiwell plates with the conjugated peptide. After washing and blocking, phages from each panning cycle were incubated, followed by detection using an anti-phage HRP-conjugated antibody. The results indicated progressive enrichment from the first to the fourth cycle, with the fourth round yielding the highest specificity for the antigenic peptide.
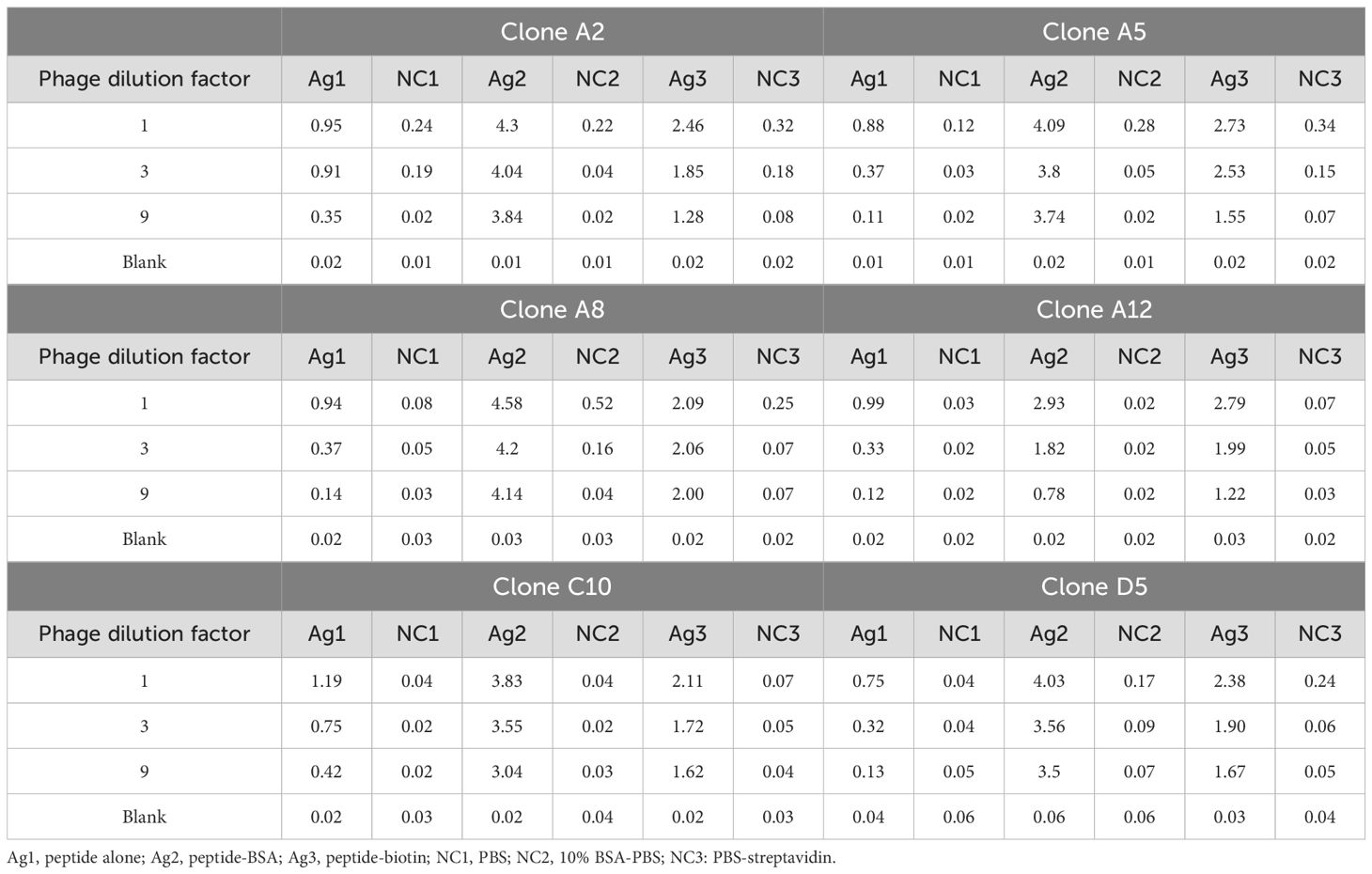
Two rounds of monoclonal ELISA tests identified 18 positive clones expressing six sequences in the first test and 27 positive clones with five matching sequences in the second test. The consistent sequences across tests confirmed the reliability of the selection process.
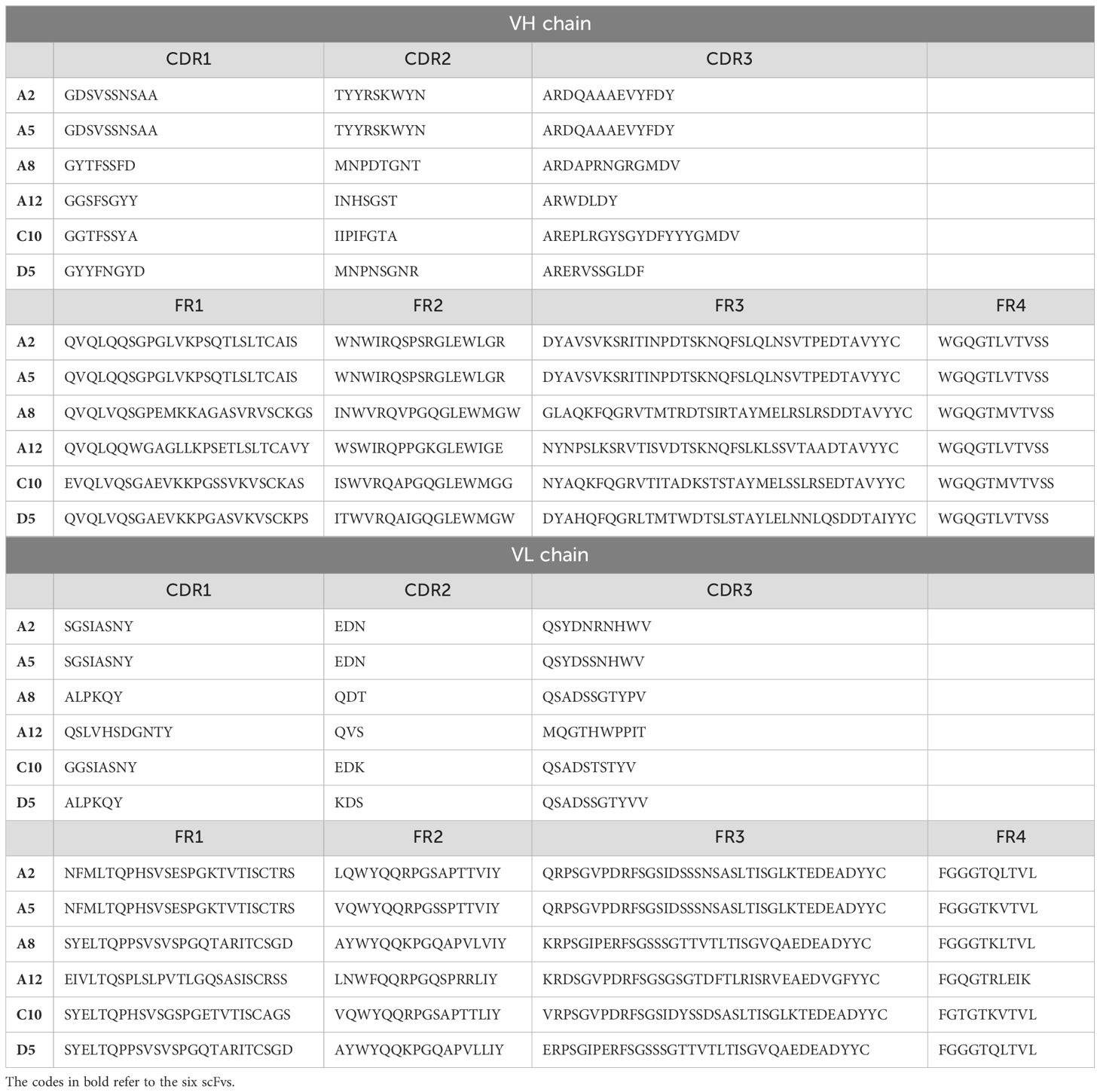
A confirmatory monoclonal ELISA test further verified the binding specificity of the six clones, showing strong affinity for the TQTPRYLIKTRGQQ peptide (Table 5). Sequencing revealed that clones A2 and A5 displayed high identity (95%) in their VH regions, with only two amino acid differences in their CDR3 sequences. The other clones showed marked variability, with identity percentages not exceeding 71% (Table 6).
Selection and ranking of scFvs
The TCR structure from the Protein Data Bank (code: 5BS0) was modified by retaining only the constant and variable regions of the alpha and beta chains. The resulting structure underwent preparation using Modeller, PDB2PQR, and Rosetta, where hydrogen atoms, missing loops, and bond orders were added, and energy minimization was applied at pH 7.4.
To assess how the beta chain constant region’s loop might impact scFv binding, the antigen structure was subjected to a 300 ns molecular dynamics simulation in Amber. Clustering based on RMSD revealed three main conformations, representing 78% of the total population, for use in ensemble docking.
The six scFvs were modeled from primary structures using AlphaFold, then prepared as scFv constructs by linking the heavy (H) and light (L) chains in Modeller. Each scFv was optimized using PDB2PQR and Rosetta, and local ensemble docking was performed with SnugDock, targeting the designed antigenic region. The best interface energy complexes for each scFv were visually inspected. The docking analysis revealed that scFv C10 displayed inconsistent binding, preferring a region outside the designed target area. Based on this finding, C10 was excluded as a potential lead candidate.
Table 7 ranks the five remaining scFvs by predicted affinity and notes possible interactions with the TCR beta chain constant region. Clones D5 and A12 emerged as top candidates, with D5 showing the highest predicted binding affinity and A12 demonstrating the greatest selectivity. A12 was ultimately selected as the lead scFv due to its superior binding selectivity, minimizing off-target effects.
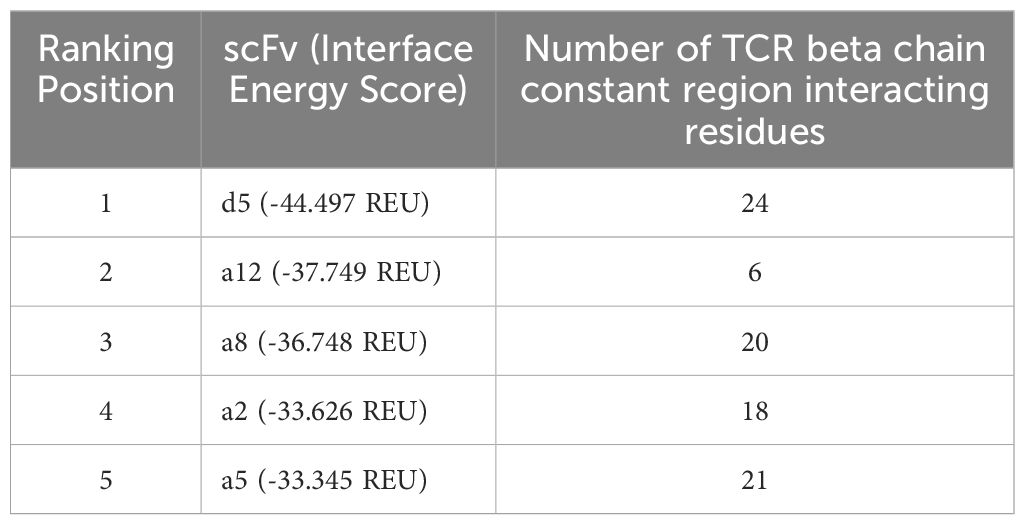
Table 7. Ranking of scFvs based on interface energy score and interaction with TCR beta chain constant region.
Surface plasmon resonance analysis
The kinetic parameters from the SPR experiment indicated a strong and specific interaction between the analyte (r-hIgG1 mAb) and the ligand (His-tagged scTCR), with a dissociation constant (KD) of 300 ± 13 nM, suggesting a moderate to high binding affinity. The SPR sensorgrams (Figure 2) were fitted using a global fitting procedure based on a 1:1 bimolecular interaction model, which assumes each ligand molecule binds to a single analyte molecule independently. This model allowed for precise measurement of interaction kinetics, providing a clear depiction of the binding behavior.
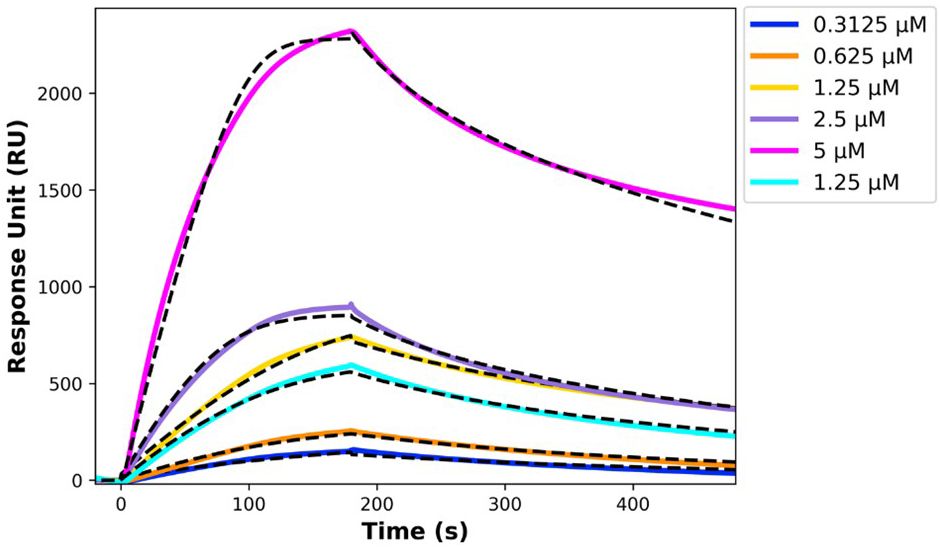
Figure 2. SPR sensorgrams showing the interaction between immobilized His-tagged single chain TCR and r-hIgG1 mAb, with black dashed lines representing the fitting curves. The TCR was immobilized on an NTA chip, and the r-hIgG1 mAb was injected at concentrations ranging from 5 µM to 0.3125 µM.
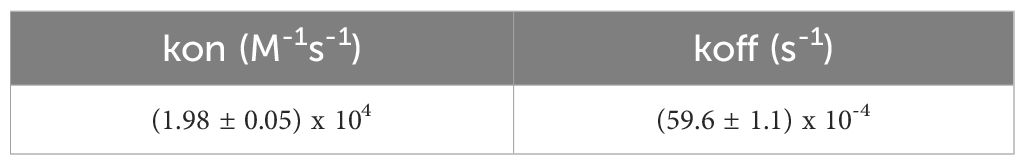
The kinetic rate constants, summarized in Table 8, offer deeper insights into the binding dynamics. The association rate constant (kon) and dissociation rate constant (koff) reflect a stable and dynamic interaction, where the ligand-analyte complex forms and dissociates predictably. This kinetic stability is favorable for potential therapeutic applications, as it indicates a reliable binding interaction.
Flow cytometry analysis
Flow cytometry was performed on mononuclear cells isolated from peripheral blood samples (PBMCs) of two patients with mature T-cell neoplasm (Sézary syndrome). One patient expressed the TRBV5-1 segment in the variable region of the TCR beta chain, while the other expressed a different segment, TRBV6-5.
The optimal concentration of FITC-conjugated r-hIgG1 mAb was determined to be 0.5 mL. Two parallel tests were then conducted using the Fluorescence Minus One (FMO) technique to assess binding specificity. The FMO control compared the complete panel of monoclonal antibodies without r-hIgG1 mAb against the panel with the addition of r-hIgG1 mAb, to clearly identify the specific fluorescence signal due to r-hIgG1 mAb (Figure 3).
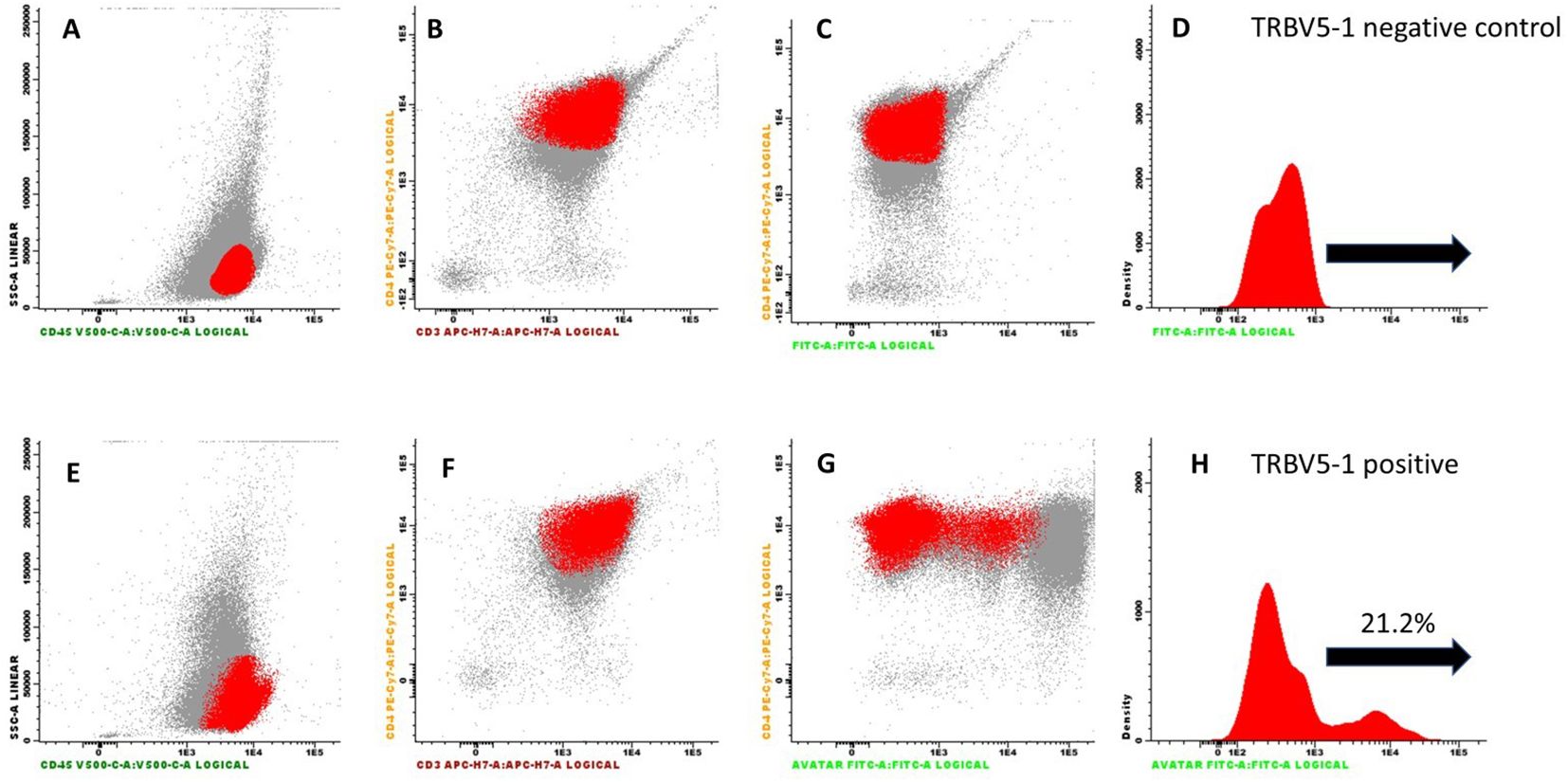
Figure 3. Flow cytometry results from PBMCs obtained from a patient with Sezary syndrome expressing the TRBV5-1 segment. Cells were pre-incubated with a complete panel of antibodies (CD3, CD4, CD8, CD45), along with an anti-CD14 antibody to identify monocytes. Panels (A–C) show the cell population of interest highlighted in red as dot plots. In panel (D), the same population is represented as a histogram. Panels (E–G) refer to the test conducted with the addition of the FITC-conjugated r-hIgG1 mAb [with the same population represented as a histogram in panel (H)]. The test highlights the binding of the mAb to 21% of the cell population under investigation.
In the first test, conducted on PBMCs from the patient expressing the TRBV5-1 segment, the r-hIgG1 mAb stained 21% of the tested cell population, indicating specific binding to cells expressing the TRBV5-1 segment. In contrast, in the second test on PBMCs from the patient expressing the TRBV6-5 segment, r-hIgG1 mAb did not bind to the cell population (data not shown).
These results, consistent with previous SPR observations, confirmed that the r-hIgG1 mAb exhibits high specificity for the TRBV5-1 segment in the variable region of the TCR beta chain, without cross-reactivity with other TCR segments.
Discussion
The development of a r-hIgG1 monoclonal antibody targeting the TRBV5-1 segment of the TCR represents a novel and targeted therapeutic strategy for treating mature T-cell neoplasms in which neoplastic clones express this specific segment.
Three-dimensional modeling and molecular docking identified three potential antigenic peptides within the TRBV5-1 segment, with TQTPRYLIKTRGQQ emerging as the ideal candidate based on its structural properties and favorable bioinformatic scores. Using phage display, six human scFvs were isolated, and clone A12 was selected as the lead due to its superior binding selectivity and favorable energy profile.
SPR analysis confirmed a strong interaction between the r-hIgG1 mAb and a scTCR containing the TRBV5-1 segment, with a dissociation constant of 300 ± 13 nM. The 1:1 kinetic model obtained from SPR supports a direct and specific interaction, reinforcing the potential of this antibody as a targeted therapeutic agent.
Flow cytometry further demonstrated the antibody’s specificity, showing successful binding to T cells expressing the TRBV5-1 segment. Notably, 21% of PBMCs from a patient with Sézary syndrome expressing the TRBV5-1 segment were stained by the FITC-conjugated antibody, while no binding was observed in cells from a patient expressing the TRBV6-5 segment. This high specificity underscores the antibody’s potential as both a diagnostic and therapeutic agent for targeting specific T-cell clones in mature T-cell neoplasms. This selectivity is crucial, as it allows the antibody to specifically target neoplastic T cells without affecting healthy cells, thereby potentially reducing adverse side effects commonly associated with therapies of lower specificity.
A broader implication of this approach is the opportunity to develop a set of fully human monoclonal antibodies against various V segments of the TCR beta chain, particularly those more frequently expressed in mature T-cell neoplasms. Monoclonal antibodies could offer a faster and more cost-effective therapeutic alternative to CAR-T therapies, which, while effective, are complex, time-consuming, and costly to produce (49, 50).
However, there are potential limitations to this therapeutic approach. Neoplastic clones might emerge with different TCRs, requiring adjustments in therapy (51). In this case, the use of a secondary monoclonal antibody targeting the V segment expressed by the new clone could help maintain therapeutic efficacy. Another limitation could involve the downregulation or loss of TCR expression in neoplastic T lymphocytes, reducing the effectiveness of a TCR-targeted antibody (52). This challenge could be addressed by combining the monoclonal antibody against the TCR beta chain with a second therapy targeting a different antigen, such as Mogamulizumab, which has shown efficacy in various T-cell neoplasms.
Future efforts will focus on increasing the therapeutic efficacy of our r-hIgG1 antibody prior to testing it in preclinical studies on animal models and subsequently in clinical trials in humans. Optimization of monoclonal antibodies aims to enhance Fc-dependent immune effector mechanisms, such as complement-dependent cytotoxicity (53) (CDC), antibody-dependent cell-mediated phagocytosis (54) (ADCP), and antibody-dependent cell-mediated cytotoxicity (55) (ADCC).
Complement activation in tumor cells, including T-cell lymphomas, is often hindered by the expression of complement regulatory proteins (CRPs) like CD46, CD55, and CD59 (56). In contrast, the effectiveness of ADCP in treating hematologic malignancies may be limited by the reduced phagocytic capacity of tissue macrophages, their restricted access to antibody-bound tumor cells, reduced antibody levels in lymphoid tissues, antigen loss through trogocytosis (57), and intrinsic resistance in certain tumor cell subpopulations (58).
ADCC, however, is emerging as a critical factor in the development of therapeutic monoclonal antibodies (59). Two primary approaches are commonly employed to enhance ADCC: modification of the Fc region and optimization of glycosylation patterns. Modifying the Fc region can significantly increase the antibody’s affinity for the FcγRIIIa (CD16) receptor on NK cells. A common approach is isotype engineering, with specific IgG variants, particularly IgG1, favored for their natural affinity for CD16, making them highly effective in inducing a cytotoxic response (60). This rationale guided our choice to produce an IgG1 monoclonal antibody from the scFv selected by phage display. Alternative approaches include introducing specific mutations, such as S239D/I332E (61), which can improve the antibody’s binding capacity to effector cells, though it remains unclear whether these mutations may increase immunogenicity.
Optimizing glycosylation further enhances ADCC (62). For example, removing fucose from the Fc region’s glycan structures, a process known as defucosylation, significantly improves the antibody’s affinity for CD16a on NK cells and is one of the most effective modifications in antibody development (63). This change not only strengthens direct tumor cell killing but also heightens immune activation through cytokine release and the recruitment of additional immune cells, creating a sustained anti-tumor response. Existing defucosylated antibodies, such as Obinutuzumab (64) and Mogamulizumab, highlight the effectiveness of this approach in onco-hematological therapies, suggesting that similar modifications could significantly boost the anti-tumor activity of our antibody. Defucosylated monoclonal antibodies can be produced at scale in Chinese hamster ovary (CHO) cells with gene knockouts for FUT8 or other glycosylation-related enzymes, making large-scale production feasible (65, 66).
Conclusions
Our findings underscore the potential of a fully human, recombinant IgG1 monoclonal antibody targeting the TRBV5-1 segment as a targeted therapeutic for a subset of mature T-cell neoplasms. The antibody’s high specificity for the TRBV5-1 segment suggests that it could selectively eliminate neoplastic T cells in patients while minimizing off-target effects, thereby improving patient outcomes. This approach offers a promising pathway for patients with mature T-cell neoplasms where TRBV5-1 expression is a defining feature.
Looking ahead, advancing this antibody into preclinical efficacy studies and, subsequently, into human clinical trials will be essential to verify its therapeutic potential. Furthermore, developing a series of antibodies targeting the most frequently expressed V segments in T-cell neoplasms could provide a broader range of options for treating these rare and severe diseases. Unlike CAR-T cell therapies, this series of antibodies could allow for greater control over dosage and treatment combinations, offering a viable, cost-effective, and flexible solution for addressing this subset of rare and orphan hematological malignancies.
Data availability statement
The structures of the antibody-TCR complexes described in the manuscript have been made publicly available in Zenodo repository: https://doi.org/10.5281/zenodo.14258991.
Ethics statement
Ethical approval was not required because the fresh blood samples used for flow cytometry analysis were collected as part of routine clinical practice for the diagnosis of mature T-cell neoplasms. All samples were obtained in compliance with clinical protocols and ethical standards governing diagnostic procedures. The studies were conducted in accordance with local legislation and institutional requirements. Written informed consent for participation was not required from the participants or their legal guardians/next of kin, as per national legislation and institutional policies, as the blood samples were collected for routine diagnostic purposes. The use of these samples for research purposes aligns with institutional guidelines and ethical standards for clinical diagnostics.
Author contributions
MPi: Conceptualization, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. GA: Investigation, Supervision, Writing – original draft. AA: Investigation, Methodology, Software, Writing – original draft. FB: Investigation, Software, Writing – original draft. ADL: Investigation, Software, Writing – original draft. MC: Investigation, Writing – original draft. VG: Investigation, Writing – original draft. NG: Investigation, Software, Writing – original draft. GM: Methodology, Software, Validation, Writing – original draft. MPa: Investigation, Writing – original draft. MTP: Conceptualization, Methodology, Resources, Writing – original draft. FP: Investigation, Methodology, Software, Validation, Writing – original draft. MS: Conceptualization, Resources, Writing – original draft. AV: Investigation, Validation, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The activities of GA, FP, and VG were supported by the following: National Funding for Centers of Excellence (Department of Sciences 2023–2027, Roma Tre University, MIUR, Article 1, Paragraphs 314–337, Law 232/2016); Next Generation EU PNRR Rome Technopole (ECS_00000024); MUR, PNRR, Mission 4, Component 2, “From Research to Business - Investment 1.4”; Next Generation EU PNRR – M4C2 "Innovation, Digitalization, and Sustainability for the Diffused Economy in Central Italy – VITALITY" (code ECS00000041), CUP code J97G22000170005. Investment 1.5, as part of the "Innovation Ecosystems" call (Ministerial Decree No. 3277 of 30.12.2021, with funding grant decree No. 1057 of 23.06.2022 issued by the Ministry of University and Research).
Acknowledgments
We fondly remember Marcelo Viegas from Proteogenix, who recently passed away, and extend our deepest appreciation for his professionalism and humanity. We also wish to express our heartfelt thanks to Davide Magri and Giorgio Baldi from Genscript for their invaluable scientific contributions, and to Massimo Ricciuto and the staff of the University Library at the University of Rome Tor Vergata for their assistance in retrieving the cited bibliographic references.
Conflict of interest
MPi is the scientific director of Tiber Biotech Srl, and MTP and MS are shareholders of Tiber Biotech Srl. Additionally, MPi, MTP, and MS are listed as inventors on the patent application PCT/EP2023/025286, filed on June 20, 2023, which is related to the development of a r-hIgG1 monoclonal antibody targeting the TRBV5-1 segment.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that this study received funding from Tiber Biotech Srl. The funder contributed to the study by supporting the phage display procedure and the synthesis of the r-hIgG1 monoclonal antibody (mAb) and single-chain T-cell receptor (scTCR).
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. The authors utilized OpenAI's GPT-4 solely for assistance in improving the grammatical accuracy of the manuscript. The tool was not used to contribute to the scientific content, data analysis, or intellectual conclusions. All responsibility for the manuscript's content remains with the authors.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sun L, Su Y, Jiao A, Wang X, Zhang B. T cells in health and disease. Signal Transduct Target Ther. (2023) 8:235. doi: 10.1038/s41392-023-01471-y
2. Bigas A, Rodriguez-Sevilla JJ, Espinosa L, Gallardo F. Recent advances in T-cell lymphoid neoplasms. Exp Hematol. (2022) 106:3–18. doi: 10.1016/j.exphem.2021.12.191
3. Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia. (2022) 36:1720–48. doi: 10.1038/s41375-022-01620-2
5. Luan Y, Li X, Luan Y, Luo J, Dong Q, Ye S, et al. Therapeutic challenges in peripheral T-cell lymphoma. Mol Cancer. (2024) 23:2. doi: 10.1186/s12943-023-01904-w
6. Langerak AW, Groenen PJ, Brüggemann M, Beldjord K, Bellan C, Bonello L, et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia. (2012) 26:2159–71. doi: 10.1038/leu.2012.246
7. Tembhare P, Yuan CM, Xi L, Morris JC, Liewehr D, Venzon D, et al. Flow cytometric immunophenotypic assessment of T-cell clonality by Vβ repertoire analysis: detection of T-cell clonality at diagnosis and monitoring of minimal residual disease following therapy. Am J Clin Pathol. (2011) 135:890–900. doi: 10.1309/AJCPV2D1DDSGJDBW
8. Mina A, Pro B. T time: Emerging and new therapies for peripheral T-cell lymphoma. Blood Rev. (2022) 52:100889. doi: 10.1016/j.blre.2021.100889
9. Zinzani PL, Corradini P, Gallamini A, Grossi A, Lazzarino M, Marchetti M, et al. Overview of alemtuzumab therapy for the treatment of T-cell lymphomas. Leuk Lymph. (2012) 53:789–95. doi: 10.3109/10428194.2011.629701
10. Stilgenbauer S, Zenz T, Winkler D, Bühler A, Schlenk RF, Groner S, et al. Subcutaneous alemtuzumab in fludarabine-refractory chronic lymphocytic leukemia: clinical results and prognostic marker analyses from the CLL2H study of the German Chronic Lymphocytic Leukemia Study Group. J Clin Oncol. (2009) 27:3994–4001. doi: 10.1200/JCO.2008.21.1128
11. de Masson A, Guitera P, Brice P, Moulonguet I, Mouly F, Bouaziz JD, et al. Long-term efficacy and safety of alemtuzumab in advanced primary cutaneous T-cell lymphomas. Br J Dermatol. (2014) 170:720–4. doi: 10.1111/bjd.12690
12. Dearden C. Alemtuzumab in peripheral T-cell Malignancies. Cancer Biother Radiopharm. (2004) 19:391–8. doi: 10.1089/cbr.2004.19.391
13. Yi JH, Kim SJ, Kim WS. Brentuximab vedotin: clinical updates and practical guidance. Blood Res. (2017) 52:243–53. doi: 10.5045/br.2017.52.4.243
14. Lai C, Kandahari AM, Ujjani C. The evolving role of brentuximab vedotin in classical hodgkin lymphoma. Blood Lymphat Cancer. (2019) 9:63–71. doi: 10.2147/BLCTT.S231821
15. Pro B, Advani R, Brice P, Bartlett NL, Rosenblatt JD, Illidge T, et al. Five-year results of brentuximab vedotin in patients with relapsed or refractory systemic anaplastic large cell lymphoma. Blood. (2017) 130:2709–17. doi: 10.1182/blood-2017-05-780049
16. Nakashima M, Uchimaru K. CD30 expression and its functions during the disease progression of adult T-cell leukemia/lymphoma. Int J Mol Sci. (2023) 24:8731. doi: 10.3390/ijms24108731
17. Duvic M, Evans M, Wang C. Mogamulizumab for the treatment of cutaneous T-cell lymphoma: recent advances and clinical potential. Ther Adv Hematol. (2016) 7:171–4. doi: 10.1177/2040620716636541
18. Kim YH, Bagot M, Pinter-Brown L, Rook AH, Porcu P, Horwitz SM, et al. Mogamulizumab versus vorinostat in previously treated cutaneous T-cell lymphoma (MAVORIC): an international, open-label, randomised, controlled phase 3 trial. Lancet Oncol. (2018) 19:1192–204. doi: 10.1016/S1470-2045(18)30379-6
19. Lokhorst HM, Plesner T, Laubach JP, Nahi H, Gimsing P, Hansson M, et al. Targeting CD38 with daratumumab monotherapy in multiple myeloma. N Engl J Med. (2015) 373:1207–19. doi: 10.1056/NEJMoa1506348
20. Huang H, Zhu J, Yao M, Kim TM, Yoon DH, Cho SG, et al. Daratumumab monotherapy for patients with relapsed or refractory natural killer/T-cell lymphoma, nasal type: an open-label, single-arm, multicenter, phase 2 study. J Hematol Oncol. (2021) 14:25. doi: 10.1186/s13045-020-01020-y
21. Chen J, Li S, Yao Q, Du N, Fu X, Lou Y, et al. The efficacy and safety of combined immune checkpoint inhibitors (nivolumab plus ipilimumab): a systematic review and meta-analysis. World J Surg Oncol. (2020) 18:150. doi: 10.1186/s12957-020-01933-5
22. Homšek A, Radosavljević D, Miletić N, Spasić J, Jovanović M, Miljković B, et al. Review of the clinical pharmacokinetics, efficacy and safety of pembrolizumab. Curr Drug Metab. (2022) 23:460–72. doi: 10.2174/1389200223666220609125013
23. Pelcovits A, Ollila TA, Olszewski AJ. Advances in immunotherapy for the treatment of cutaneous T-cell lymphoma. Cancer Manag Res. (2023) 15:989–98. doi: 10.2147/CMAR.S330908
24. Joshi M, Ansell SM. Activating the antitumor immune response in non-hodgkin lymphoma using immune checkpoint inhibitors. J Immunol Res. (2020) 2020:8820377. doi: 10.1155/2020/8820377
25. Lussana F, Gritti G, Rambaldi A. Immunotherapy of acute lymphoblastic leukemia and lymphoma with T cell-redirected bispecific antibodies. J Clin Oncol. (2021) 39:444–55. doi: 10.1200/JCO.20.01564
26. Kontermann RE, Brinkmann U. Bispecific antibodies. Drug Discovery Today. (2015) 20:838–47. doi: 10.1016/j.drudis.2015.02.008
27. Falchi L, Vardhana SA, Salles GA. Bispecific antibodies for the treatment of B-cell lymphoma: promises, unknowns, and opportunities. Blood. (2023) 141:467–80. doi: 10.1182/blood.2021011994
28. Paul S, Pearlman AH, Douglass J, Mog BJ, Hsiue EH, Hwang MS, et al. TCR β chain-directed bispecific antibodies for the treatment of T cell cancers. Sci Transl Med. (2021) 13:eabd3595. doi: 10.1126/scitranslmed.abd3595
29. Morath A, Schamel WW. [amp]]alpha;β and γδ T cell receptors: Similar but different. J Leukoc Biol. (2020) 107:1045–55. doi: 10.1002/JLB.2MR1219-233R
30. Su C, Jakobsen I, Gu X, Nei M. Diversity and evolution of T-cell receptor variable region genes in mammals and birds. Immunogenetics. (1999) 50:301–8. doi: 10.1007/s002510050606
31. Wucherpfennig KW, Gagnon E, Call MJ, Huseby ES, Call ME. Structural biology of the T-cell receptor: insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb Perspect Biol. (2010) 2:a005140. doi: 10.1101/cshperspect.a005140
32. Reche PA, Reinherz EL. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol. (2003) 331:623–41. doi: 10.1016/s0022-2836(03)00750-2
33. Li F, Zhang H, Wang W, Yang P, Huang Y, Zhang J, et al. T cell receptor β-chain-targeting chimeric antigen receptor T cells against T cell Malignancies. Nat Commun. (2022) 13:4334. doi: 10.1038/s41467-022-32092-8
34. Frenzel A, Schirrmann T, Hust M. Phage display-derived human antibodies in clinical development and therapy. MAbs. (2016) 8:1177–94. doi: 10.1080/19420862.2016.1212149
35. The International Immunogenetics Information System. Available online at: http://imgt.org (Accessed 20 September 2024).
36. Krangel MS. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat Immunol. (2003) 4:624–30. doi: 10.1038/ni0703-624
37. Antigen Profiler Peptide. Available online at: https://www.thermofisher.com/it/en/home/lifescience/antibodies/custom-antibodies/custom-antibody-production/antigen-profiler-antigen-preparation.html (Accessed 20 September 2024).
38. Ab Designer. Available online at: https://esbl.nhlbi.nih.gov/AbDesigner/ (Accessed 20 September 2024).
39. UNIPROT database. Available online at: https://www.uniprot.org (Accessed 20 September 2024).
40. Protein Data Bank. Available online at: https://www.rcsb.org (Accessed 20 September 2024).
41. Chung S, Wucherpfennig KW, Friedman SM, Hafler DA, Strominger JL. Functional three-domain single-chain T-cell receptors. Proc Natl Acad Sci U S A. (1994) 91:12654–8. doi: 10.1073/pnas.91.26.12654
42. Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinf. (2016) 54:5.6.1-5.6.37. doi: 10.1002/cpbi.3
43. Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. (2018) 27:112–28. doi: 10.1002/pro.3280
44. O’Meara MJ, Leaver-Fay A, Tyka MD, Stein A, Houlihan K, DiMaio F, et al. Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J Chem Theory Comput. (2015) 11:609–22. doi: 10.1021/ct500864r
45. Sircar A, Gray JJ. SnugDock: paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PloS Comput Biol. (2010) 6:e1000644. doi: 10.1371/journal.pcbi.1000644
46. Case DA, Cheatham TE 3rd, Darden T, Gohlke H, Luo R, Merz KM Jr, et al. The Amber biomolecular simulation programs. J Comput Chem. (2005) 26:1668–88. doi: 10.1002/jcc.20290
47. Roe DR, Cheatham TE 3rd. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. (2013) 9:3084–95. doi: 10.1021/ct400341p
48. Yin R, Feng BY, Varshney A, Pierce BG. Benchmarking AlphaFold for protein complex modeling reveals accuracy determinants. Protein Sci. (2022) 31:e4379. doi: 10.1002/pro.4379
49. Woo JS, Nguyen K, Liu L, Krishnan A, Siddiqi T, Borogovac A. Mobilizing CARs: Benefits, drawbacks, and directions for outpatient CAR T-cell therapy. Semin Hematol. (2024) 61:273–83. doi: 10.1053/j.seminhematol.2024.08.003
50. den Exter A. Fighting excessive pharmaceutical prices: evaluating the options. Eur J Health Law. (2020) 28:68–80. doi: 10.1163/15718093-BJA10004
51. Hamrouni A, Fogh H, Zak Z, Ødum N, Gniadecki R. Clonotypic diversity of the T-cell receptor corroborates the immature precursor origin of cutaneous T-cell lymphoma. Clin Cancer Res. (2019) 25:3104–14. doi: 10.1158/1078-0432.CCR-18-4099
52. Weiss LM, Picker LJ, Grogan TM, Warnke RA, Sklar J. Absence of clonal beta and gamma T-cell receptor gene rearrangements in a subset of peripheral T-cell lymphomas. Am J Pathol. (1988) 130:436–42.
53. Wang B, Yang C, Jin X, Du Q, Wu H, Dall’Acqua W, et al. Regulation of antibody-mediated complement-dependent cytotoxicity by modulating the intrinsic affinity and binding valency of IgG for target antigen. MAbs. (2020) 12:1690959. doi: 10.1080/19420862.2019.1690959
54. Tay MZ, Wiehe K, Pollara J. Antibody-dependent cellular phagocytosis in antiviral immune responses. Front Immunol. (2019) 10:332. doi: 10.3389/fimmu.2019.00332
55. Coënon L, Villalba M. From CD16a biology to antibody-dependent cell-mediated cytotoxicity improvement. Front Immunol. (2022) 13:913215. doi: 10.3389/fimmu.2022.913215
56. Geller A, Yan J. The role of membrane bound complement regulatory proteins in tumor development and cancer immunotherapy. Front Immunol. (2019) 10:1074. doi: 10.3389/fimmu.2019.01074
57. Mattei F, Andreone S, Spadaro F, Noto F, Tinari A, Falchi M, et al. Trogocytosis in innate immunity to cancer is an intimate relationship with unexpected outcomes. iScience. (2022) 25:105110. doi: 10.1016/j.isci.2022.105110
58. Church AK, VanDerMeid KR, Baig NA, Baran AM, Witzig TE, Nowakowski GS, et al. Anti-CD20 monoclonal antibody-dependent phagocytosis of chronic lymphocytic leukaemia cells by autologous macrophages. Clin Exp Immunol. (2016) 183:90–101. doi: 10.1111/cei.12697
59. Zahavi D, AlDeghaither D, O’Connell A, Weiner LM. Enhancing antibody-dependent cell-mediated cytotoxicity: a strategy for improving antibody-based immunotherapy. Antib Ther. (2018) 1:7–12. doi: 10.1093/abt/tby002
60. de Taeye SW, Rispens T, Vidarsson G. The ligands for human igG and their effector functions. Antibodies (Basel). (2019) 8:30. doi: 10.3390/antib8020030
61. Jebamani P, Sriramulu DK, Lee SG. Residue interaction network and molecular dynamics simulation study on the binding of S239D/I332E Fc variant with enhanced affinity to FcγRIIIa receptor. J Mol Graph Model. (2023) 118:108327. doi: 10.1016/j.jmgm.2022.108327
62. Saunders KO. Conceptual approaches to modulating antibody effector functions and circulation half-life. Front Immunol. (2019) 10:1296. doi: 10.3389/fimmu.2019.01296
63. Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem. (2002) 277:26733–40. doi: 10.1074/jbc.M202069200
64. Tobinai K, Klein C, Oya N, Fingerle-Rowson G. A review of obinutuzumab (GA101), a novel type II anti-CD20 monoclonal antibody, for the treatment of patients with B-cell Malignancies. Adv Ther. (2017) 34:324–56. doi: 10.1007/s12325-016-0451-1
65. Yamane-Ohnuki N, Kinoshita S, Inoue-Urakubo M, Kusunoki M, Iida S, Nakano R, et al. Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng. (2004) 87:614–22. doi: 10.1002/bit.20151
Keywords: T-cell neoplasms, T-cell receptor (TCR), human IgG1 monoclonal antibodies, phage display, surface plasmon resonance (SPR), flow cytometry, antibody-antigen docking
Citation: Pitaro M, Antonini G, Arcovito A, Buccisano F, De Lauro A, Irno Consalvo M, Gallo V, Giacon N, Mangiatordi GF, Pacelli M, Pitaro MT, Polticelli F, Sorrenti M and Venditti A (2024) Development of a recombinant human IgG1 monoclonal antibody against the TRBV5-1 segment of the T cell receptor for the treatment of mature T cell neoplasms. Front. Immunol. 15:1520103. doi: 10.3389/fimmu.2024.1520103
Received: 30 October 2024; Accepted: 21 November 2024;
Published: 17 December 2024.
Edited by:
Arumugam Jayakumar, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Pengfei Xu, University of California, Davis, United StatesSushant Kumar, University of Pennsylvania, United States
Copyright © 2024 Pitaro, Antonini, Arcovito, Buccisano, De Lauro, Irno Consalvo, Gallo, Giacon, Mangiatordi, Pacelli, Pitaro, Polticelli, Sorrenti and Venditti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michele Pitaro, bWljaGVsZS5waXRhcm9AaW5iYi5pdA==
 Michele Pitaro
Michele Pitaro Giovanni Antonini
Giovanni Antonini Alessandro Arcovito
Alessandro Arcovito Francesco Buccisano
Francesco Buccisano Alfredo De Lauro
Alfredo De Lauro Maria Irno Consalvo5
Maria Irno Consalvo5 Valentina Gallo
Valentina Gallo Noah Giacon
Noah Giacon Giuseppe Felice Mangiatordi
Giuseppe Felice Mangiatordi Maddalena Pacelli
Maddalena Pacelli Fabio Polticelli
Fabio Polticelli Adriano Venditti
Adriano Venditti