- 1Department of Microbiology and Immunology, State University of New York Upstate Medical University, Syracuse, NY, United States
- 2State University of New York Center for Vector-Borne Diseases, State University of New York Upstate Medical University, Syracuse, NY, United States
- 3Department of Microbiology and Immunology, University of South Alabama, Mobile, AL, United States
- 4Upstate Global Health Institute, State University of New York Upstate Medical University, Syracuse, NY, United States
Introduction: Powassan virus (POWV), a vector-borne pathogen transmitted by Ixodes ticks in North America, is the causative agent of Powassan encephalitis. As obligate hematophagous organisms, ticks transmit pathogens like POWV at the tick bite site, specifically during the initial stages of feeding. Tick feeding and salivary factors modulate the host’s immunological responses, facilitating blood feeding and pathogen transmission. However, the mechanisms of immunomodulation during POWV transmission remain inadequately understood. In this study, we investigated the global cutaneous transcriptomic changes associated with tick bites during POWV transmission.
Methods: We collected skin biopsies from the tick attachment sites at 1, 3, and 6 h after feeding by POWV-infected and uninfected ticks, followed by RNA sequencing of these samples. Differentially expressed genes were analyzed for pathway enrichment using gene ontology and pathway enrichment analyses.
Results: Our findings reveal that tick feeding alone significantly impacts the skin transcriptome within the first 1 to 3 h of tick attachment. Although early POWV transmission induces minimal changes in the local environment, a pronounced shift toward a proinflammatory state is observed 6 h after tick attachment, characterized by neutrophil recruitment and interleukin signaling.
Discussion: These transcriptomic data elucidate the dynamic changes at the tick bite site, transitioning from changes that assist blood meal acquisition to a proinflammatory phase that may facilitate viral dissemination.
1 Introduction
The prevalence and geographical range of ticks are expanding globally, particularly in North America, where Ixodes scapularis has extended its range due to climate change, which increasingly favors the tick’s preferred habitats (1). This expansion poses significant public health risks, as I. scapularis is a primary vector for several pathogens, including bacteria such as Borrelia burgdorferi and Anaplasma phagocytophilum, as well as viral pathogens like Powassan virus (POWV) and deer tick virus (DTV) (2). POWV, first isolated in 1958 from the brain of a young boy who succumbed to Powassan encephalitis (3), can cause severe neurological sequelae in survivors, including hemiplegia, muscle wasting, and memory deficits (4, 5). Despite the severity of the disease, there is currently no antiviral treatment or U.S. Food and Drug Administration (FDA)-approved vaccine. However, recent advances in DNA-based, as well as live-attenuated vaccine platforms, show promise in eliciting a strong protective immune response against POWV. The latter utilizes the historically successful yellow fever virus 17D backbone, which has seen licensed success when used for Japanese encephalitis virus and dengue virus (6, 7).
Arthropod saliva plays a crucial role in transmitting vector-borne pathogens (8). During blood feeding, ticks insert their mouthparts into the host’s skin and secrete saliva into the bite site. Tick saliva contains a variety of bioactive components that facilitate feeding by enabling the tick to remain attached, acquire a complete blood meal, and evade the host’s immune defenses (9–11). Pathogens have evolved to exploit these salivary components, enhancing their transmission to the host in a process known as saliva-assisted transmission (12–14).
Mouse studies have demonstrated that POWV transmission can occur in as little as 15 min of tick feeding, a significantly shorter duration compared to that of other pathogens transmitted by ticks, such as Borrelia burgdorferi, which typically requires 48–72 h of tick feeding but has been observed in as early as 24 h after tick attachment, for transmission to occur (15–17). Understanding the early host responses to POWV transmission is crucial, as these early time points are characterized by robust fluctuations in the gene expression of various cytokines and chemokines (18). Previous studies have shown that co-injection with tick salivary gland extract and POWV results in enhanced neuroinvasion in BALB/c mice compared to POWV injection alone (14). Additionally, feeding by POWV-infected I. scapularis induces a significant influx of neutrophils and macrophages to the bite site within the first few hours, indicating strong immunological signaling during POWV transmission (19). However, the specific immunomodulatory events during the initial hours of POWV transmission from tick to host still need to be understood.
This study explores the global transcriptomic changes at the tick bite site during the first hours of POWV transmission. We employed a POWV-infected I. scapularis feeding model to investigate the differential cutaneous gene expression profiles at 1, 3, and 6 h after tick attachment (Figure 1). Our findings highlight a proinflammatory response characterized by chemokine-mediated neutrophil recruitment and degranulation. This provides new insights into the host-pathogen interactions during tick-borne POWV transmission.
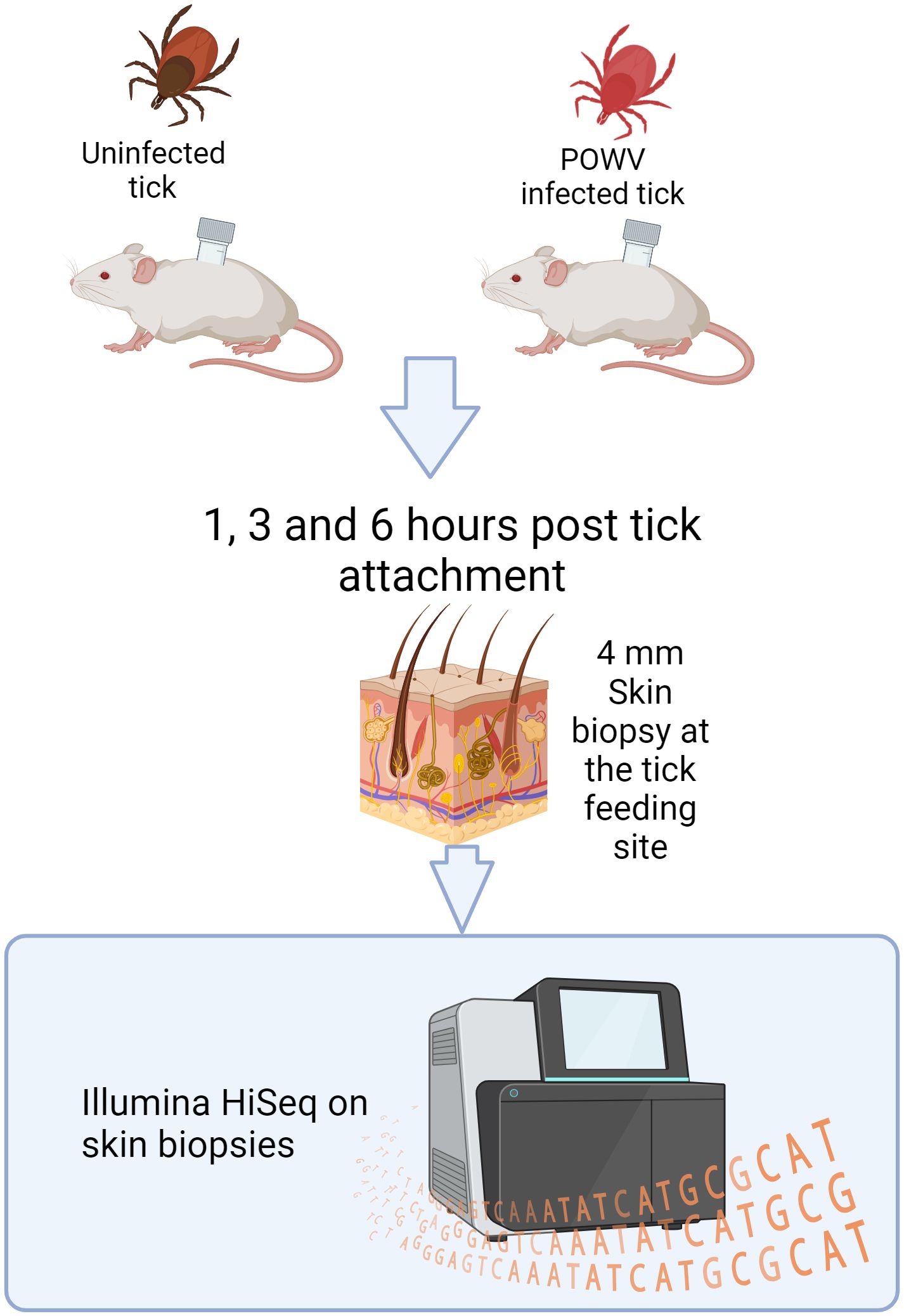
Figure 1. Design of an I. scapularis feeding study with downstream RNA-seq analysis. POWV-infected ticks or uninfected ticks were allowed to feed on mice for 1, 3, or 6 h. After feeding, the ticks were removed, and the bite sites were biopsied and processed for Illumina HiSeq sequencing.
2 Materials and methods
2.1 Ethics statement
All experiments involving mice and infected ticks were conducted in arthropod containment level 3 facilities in strict accordance with an animal use protocol approved by the University of Texas Medical Branch (UTMB) Institutional Animal Care and Use Committee (IACUC; # 0907054).
2.2 Viral culture
African green monkey kidney cells (Vero E6) were acquired from American Type Culture Collection and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% antibiotic cocktail consisting of penicillin and streptomycin and incubated at 37°C with 5% CO2. The prototypic LB strain of POWV was acquired from the World Reference Center for Emerging Viruses and Arboviruses at the UTMB, which had been passaged seven times in suckling mice brains. The stock of the virus was then passaged six times in Vero E6 cells. Stock viral titers were determined via focus-forming assay as previously described (20). Next-generation sequencing revealed that the consensus nucleotide sequence of our POWV stock is 99.96% identical to the original POWV_LB isolated in Ontario (GenBank: NC_003687.1). There were three nucleotide changes between our POWV stock and the POWV LB isolate, leading to a single–amino acid change in the E protein (Q1436R), as well as one nucleotide gap that may have caused a nucleotide shift in the NS5 protein (W8255 to L or S).
2.3 Animals
Five-week-old female BALB/cj mice were acquired from The Jackson Laboratory (Bar Harbor, ME). Mice were allowed to acclimate to the local environment for 1 week before the start of experiments, at which point the mice were 6 weeks old.
2.4 Tick infection and infestation on mice
The mouse skin biopsies used in this study were generated as a part of another study that described tick salivary gland microRNA expression during POWV transmission (21). Briefly, uninfected adult I. scapularis were maintained in our lab within the UTMB arthropod containment level 2 facilities. Tick vials were stored inside an incubator at 22°C and 90%–93% humidity. The incubator photoperiod was set to a 16:8-h cycle. Adult, female I. scapularis were synchronously infected with POWV-LB strain as previously described (18). Control female ticks were mock-infected with media. After the infection, I. scapularis females were co-housed with uninfected male I. scapularis and stored inside a desiccator at 22°C to allow POWV replication. Mock-infected females were co-housed with male I. scapularis in the same manner. At 40 days after synchronous infection, the I. scapularis females were infested on mice.
Tick containment capsules were prepared as previously described (18). Two days before tick infestation, mice were anesthetized with isoflurane, and the backs of each mouse were shaved. One capsule was adhered to the dorsum of each mouse using Kamar adhesive (Kamar Inc., Steamboat Springs, CO). On the day of tick infestation, one I. scapularis female was placed inside each capsule and allowed to feed on the mouse for 1, 3, or 6 h. Three biological replicates were included for each of tick feeding time points described in this study. Time points were measured on the basis of the observed attachment time for each individual tick, for example, once embedding of the hypostome is visually observed, a timer was started for that individual tick. This was to ensure accurate feeding times across groups. At the experimental time points, mice were euthanized with CO2 inhalation according to IACUC protocols. The feeding tick was removed, and 4-mm skin biopsies were dissected from each mouse at the tick feeding site. Salivary glands from the removed tick were also dissected to check POWV infection status. Each skin biopsy and tick salivary glands were stored for RNA extraction in QIAzol lysis reagent (Qiagen). In the POWV-infected tick group, we confirmed the POWV-infection status prior to be included in this study (21). Three mice skin biopsies from each group and time points were used for RNA sequencing (RNA-seq) analysis described in this study.
2.5 RNA extractions, q-RT-PCR, and digital droplet PCR detection of POWV RNA
Four-millimeter skin biopsies and tick salivary glands, in QIAzol, were homogenized in QIAzol (Qiagen). Following homogenization, RNA extraction was performed using an RNeasy mini kit (Qiagen), yielding purification of total RNA. Viral infection in the skin and tick salivary glands were verified via digital droplet (ddPCR) and Quantitative Reverse transcription polymerase chain reaction (q-RT-PCR), respectively, using POWV primers (POWV-F: CCGAGCCAAAGTGAGGATGT and POWV-R: TCTTTTGCCGAGCTCCACTT) as previously described (22). For the salivary glands, the following cycling protocol was used: 10 min at 50°C; 1 min at 95°C; 10 s at 95°C followed by 30 s at 60°C for 45 cycles; and an 81-cycle (+0.5°C/cycle) 55°C–95°C melt curve.
For POWV RNA detection in the skin, given the small amount of viral RNA that was expected to be present in the skin at the time of feeding, we opted for ddPCR. To start, 15-µL reactions were prepared consisting of QIAcuity OneStep Advanced Probe Kit (QIAgen) materials and template RNA. On a QIAcuity, nucleic acids were amplified with the following cycling conditions: 40 min at 50°C, 2 min at 95°C, 40 cycles of 5 s at 95°C, and 30 s at 60°C. Images were then taken with an exposure time of 500 ms and a gain of 6 using the green channel. Thresholds were calculated by manually adjusting according to a negative control.
2.6 Library preparation with PolyA selection and Illumina sequencing
The RNA samples were quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), and, then, the RNA integrity was verified using Agilent TapeStation 4200 (Agilent Technologies, Palo Alto, CA, USA).
RNA-seq libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina, based on the manufacturer’s instructions (New England Biolabs (NEB), Ipswich, MA, USA). Briefly, mRNAs were enriched with Oligo(dT) beads. Enriched mRNAs were fragmented for 15 min at 94°C. First-strand and second-strand Complementary deoxyribonucleic acid (cDNAs) were subsequently synthesized. cDNA fragments were end-repaired and adenylated at 3’ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by limited-cycle PCR. Sequencing libraries were validated on the Agilent TapeStation (Agilent Technologies, Palo Alto, CA, USA) and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, USA). The sequencing libraries were clustered on flow cells. After clustering, the flow cells were loaded onto the Illumina HiSeq instrument (4000 or equivalent) according to the manufacturer’s instructions. The samples were sequenced using a 2 × 150-bp paired-end configuration. Image analysis and base calling were conducted by the HiSeq Control Software. Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
2.7 Data analysis
After investigating the quality of the raw data, sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were then mapped to the Mus musculus reference genome available on ENSEMBL using the STAR aligner v.2.5.2b.
After the extraction of gene hit counts, the gene hit count table was used for downstream differential expression analysis. Using DESeq2, a comparison of gene expression between the groups of samples was performed. The Wald test was used to generate p-values and log2 fold changes. Genes with adjusted p-values <0.05 and absolute log2 fold changes >1 were called as differentially expressed genes (DEGs) for each comparison. Raw and analyzed data have been deposited to the Gene Expression Omnibus under accession number GSE282786. A gene ontology (GO) analysis was performed on the statistically significant set of genes by implementing the software GeneSCF. The gene ontology annotation (goa) human GO list was used to cluster the set of genes based on their biological process and determine their statistical significance. A principal component analysis (PCA) analysis was performed using the “plotPCA” function within the DESeq2 R package. The plot shows the samples in a 2D plane spanned by their first two principal components: infection status, tick feeding, or time, depending on the bioinformatic comparison. The top 500 genes, selected by highest row variance, were used to generate the plot. In the event where genes were identified in one group but not another, they were denoted as 1.0 log2 fold increase and treated as such.
3 Results
3.1 Transcriptomic changes at the skin interface during I. scapularis timed blood meal acquisition
Pathogen-free I. scapularis females were allowed to feed on naïve mice for 1, 3, and 6 h. Subsequent to feeding, skin biopsies from the tick feeding site were collected for RNA-seq and differential gene expression analysis following RNA extraction. Naïve skin from tick-free, uninfected mice was used as a control. The analysis revealed that 3,722, 3,210, and 3,905 genes were differentially regulated compared to tick-free control skin after 1, 3, and 6 h of feeding of uninfected ticks, respectively (Table 1). Additionally, POWV-infected I. scapularis females were allowed to feed on naïve mice for 1, 3, and 6 h, with their skin transcriptomic profiles compared to those of uninfected ticks at corresponding time points. The total number of DEGs identified in these POWV-infected versus uninfected tick feeding site comparisons were 1, 20, and 83 for 1, 3, and 6 h of feeding, respectively (Table 1).
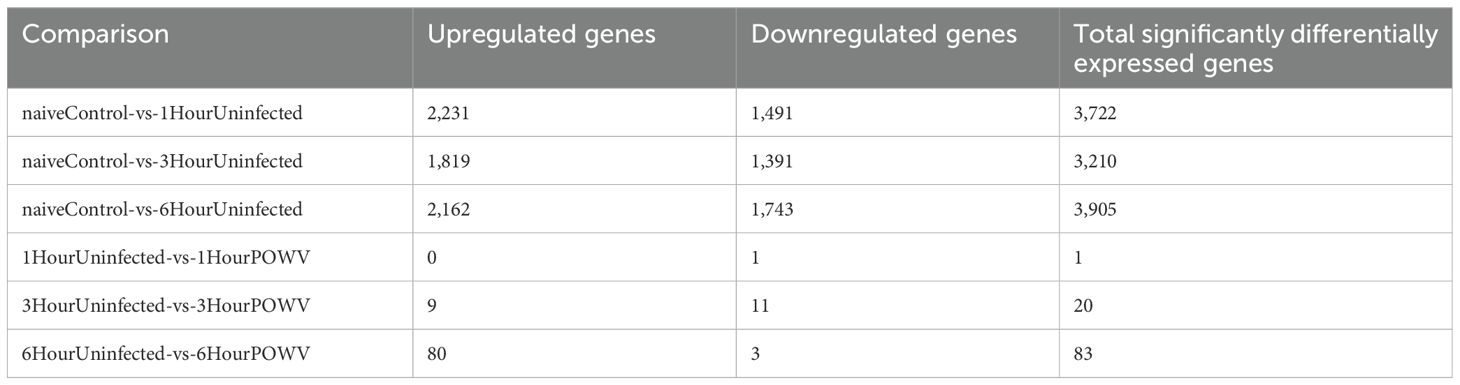
Table 1. Total expressed genes in each comparison made for uninfected and POWV-infected I. scapularis feedings.
3.1.1 Naïve skin vs. skin from the site of uninfected I. scapularis fed for 1 h
We first analyzed the cutaneous transcriptomic changes induced by an uninfected tick feeding for 1 h. PCA was conducted to assess their component profiles (Figure 2A).
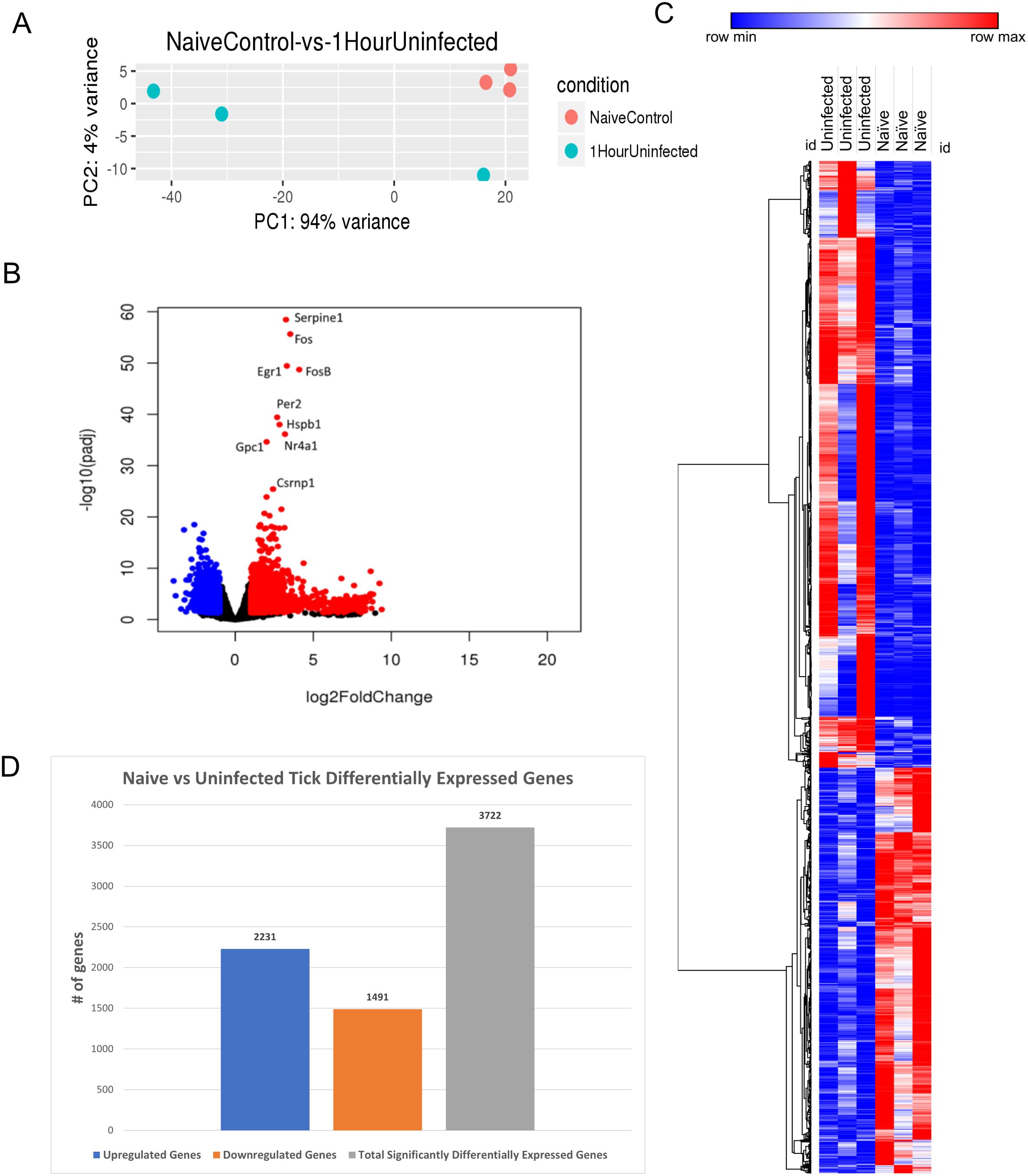
Figure 2. I. scapularis–free skin compared to 1-h uninfected I. scapularis feeding. (A) Principal component analysis (PCA) of transcriptional profiles when comparing 1-h uninfected tick feeding sites (blue) to naïve tick-free skin (orange). (B) Volcano plot outlining upregulated and downregulated genes at 1 h. The x-axis identifies the log2 fold change of every gene, and the y-axis identifies the log10 of its adjusted p-value. Only genes with a p-value <0.05 and a log2 fold change <−1 are identified as downregulated (blue). Only genes that have a p-value <0.05 and a log2 fold change >1 are identified as upregulated (red). (C) A bi-clustering heatmap was used to visualize the expression profile of all differentially expressed genes (DEGs), sorted by their adjusted p-value by plotting log2-transformed values in samples. (D) Total DEGs for uninfected tick feeding vs. naïve skin at 1 h.
A volcano plot was generated to illustrate the global transcriptional changes across the groups. Genes with a p-value <0.05 and a log2 fold change <−1 were identified as downregulated (blue), whereas genes with a p-value <0.05 and a log2 fold change >1 were identified as upregulated (red) (Figure 2B). A total of 3,722 DEGs were identified and visualized in the volcano plot, with significantly upregulated outliers highlighted. Notably, Serpine1, Fos, FosB, EGR1, HSPB1, and GPC1 were highly upregulated and statistically significant at this feeding time point. These genes are associated with functions such as wound healing, blood clotting, stress response, and cell proliferation, which are directly relevant to tick feeding. Additionally, several immune regulatory genes were differentially expressed during the 1-h feeding, including MAPK and its pathway components like JUN, IL-6, and early expression of NFκB components. These findings are consistent with earlier studies reporting similar results from I. scapularis feedings (23). A bi-clustering heatmap analysis was performed to examine the complete transcriptomic profile of each sample, with hierarchical sorting based on Pearson correlation (Figure 2C). The heatmap indicates slight differential regulation between the skin harvested from tick-free control mice and the 1-h fed uninfected tick feeding site. Although the genes represented in the heatmap exhibited modest changes, they generally showed increased expression compared to the naïve controls. The differential analysis revealed a total of 3,722 genes expressed in this study (Figure 2D).
The DEG data were further analyzed using the Reactome database, and pathway enrichment (PE) analysis was conducted for significant DEGs during the 1-h feeding. The analysis revealed impacted pathways, including keratinization, formation of the cornified envelope, and NGF-stimulated transcription and nuclear events, with the latter being specific to kinase and transcription factor activation (Table 2).
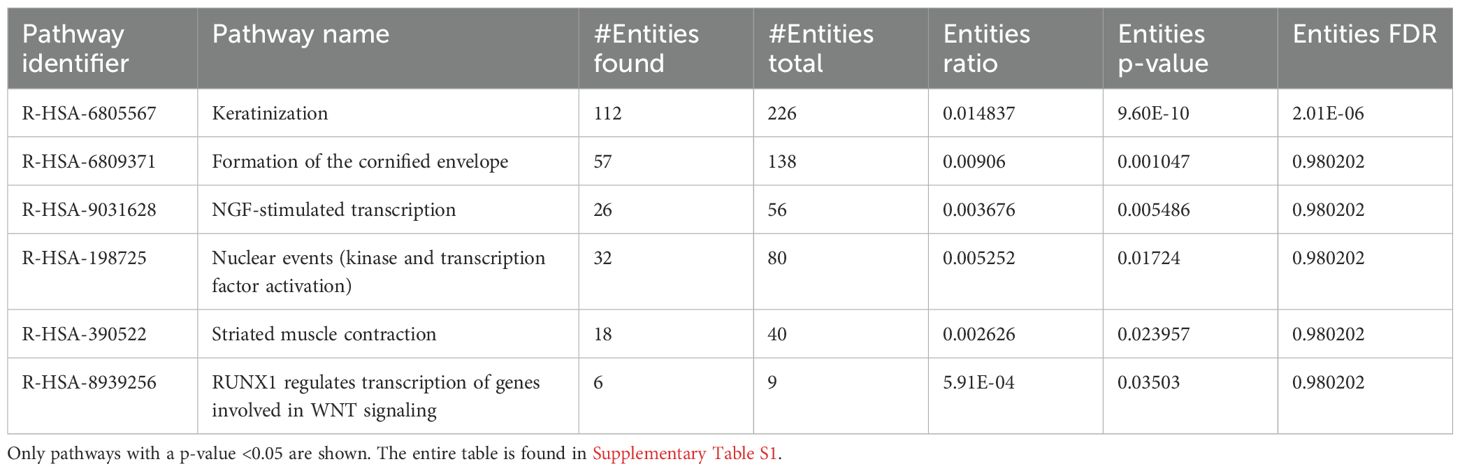
Table 2. Pathway enrichment of naïve I. scapularis–free skin vs. skin harvested after 1 h of uninfected I. scapularis feeding.
3.1.2 Naïve skin vs. skin from the site of uninfected I. scapularis fed for 3 h
We next investigated the cutaneous transcriptomic changes occurring during 3 h of uninfected tick feeding. The PCA revealed clustering of individual samples for the tick-free control group, with some variance observed in the skin of the 3-h uninfected tick cohort (Figure 3A).
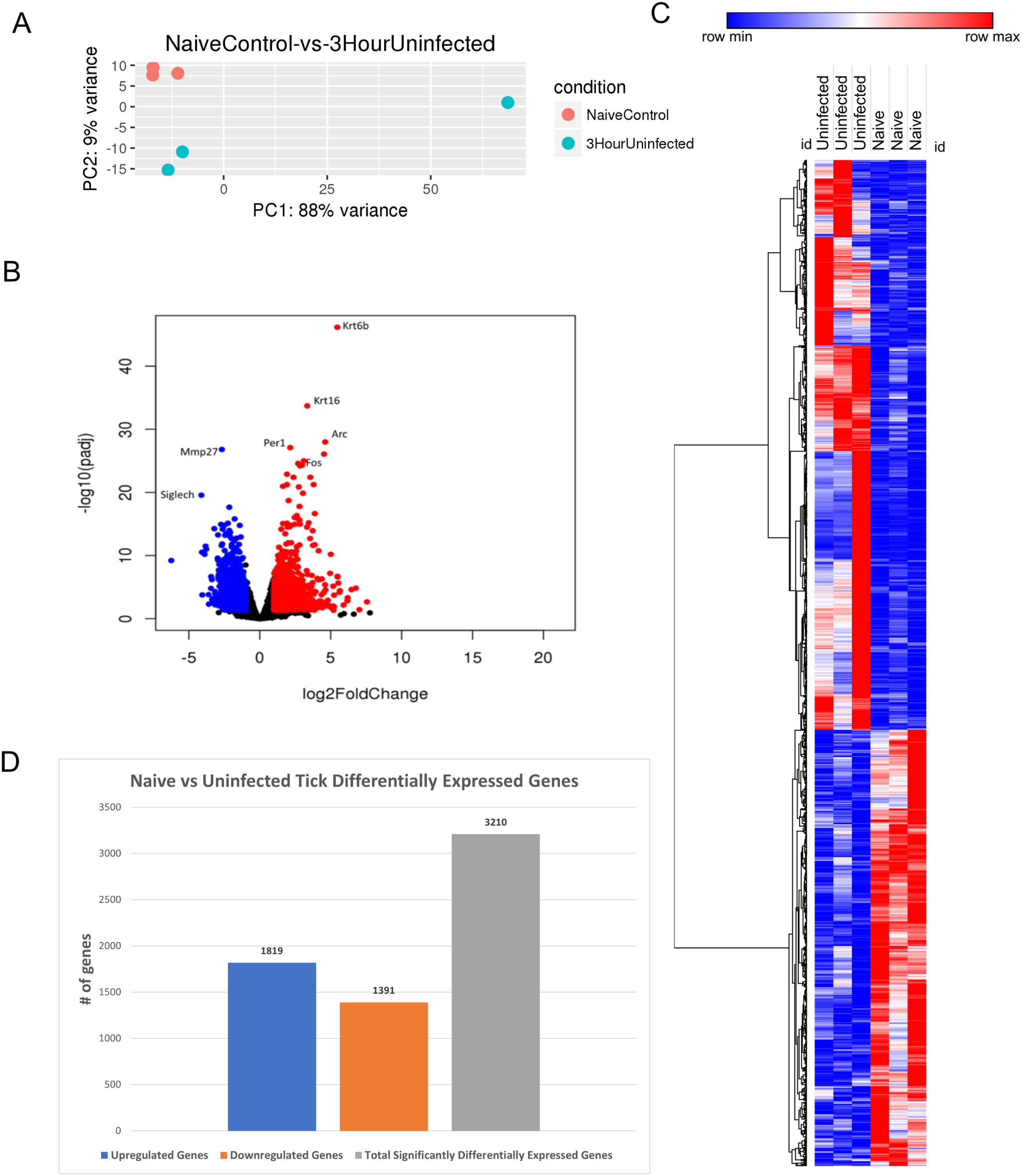
Figure 3. Differential expression of genes in I. scapularis–free control skin compared to 3-h uninfected I. scapularis feedings. (A) Principal component analysis (PCA) of transcriptional profiles when comparing 3-h uninfected feedings (blue) to naïve skin (orange). (B) Volcano plot outlining upregulated and downregulated genes at 3 h. The x-axis identifies the log2 fold change of every gene, and the y-axis specifies the log10 of its adjusted p-value. Only genes with a p-value <0.05 and a log2 fold change <−1 are identified as downregulated (blue). Only genes with a p-value <0.05 and a log2 fold change >1 are identified as upregulated (red). (C) A bi-clustering heatmap was used to visualize the expression profile of all differentially expressed genes (DEGs), sorted by their adjusted p-value by plotting log2-transformed values in samples. (D) Total DEGs for uninfected tick feeding vs. naïve skin at 3 h.
A volcano plot was used to analyze the most DEGs. The plot identified 3,210 DEGs, with 1,819 upregulated and 1,391 downregulated genes (Figure 3B). Upregulated genes involved in the keratin gene family, including Krt6b and Krt16, and downregulated metalloproteases such as Mmp27 were observed. With both Krt6b and Krt16 being upregulated, there could be potential mechanisms for wound healing, directly in response to tick blood meal acquisition. Siglech, a gene involved in cell-cell interactions and regulatory functions of the innate immune system through polysaccharide interactions, was also downregulated (24). Additionally, Arc, Per1, and Fos were upregulated, corresponding to functions such as cytoskeletal growth and sodium ion retention (25). Fos acts upstream of transcription factor AP-1, which regulates cell proliferation, differentiation, and transformation (26).
A bi-clustering heatmap of all significantly DEGs was generated as previously described (Figure 3C). The differences between the two groups were more pronounced than the 1-h uninfected feeding heatmap (Figure 2C). This summary heatmap shows the comparisons of each individual mouse for both groups: naïve/tick-free skin versus skin harvested from the 3-h uninfected tick feeding site. The tree to the left demonstrates the hierarchical clustering of the samples. The differential analysis revealed a total of 3,210 genes expressed in this study (Figure 3D). DEG data were input into the Reactome database, and PE analysis revealed the impacted pathways during the 3-h feeding (Table 3).
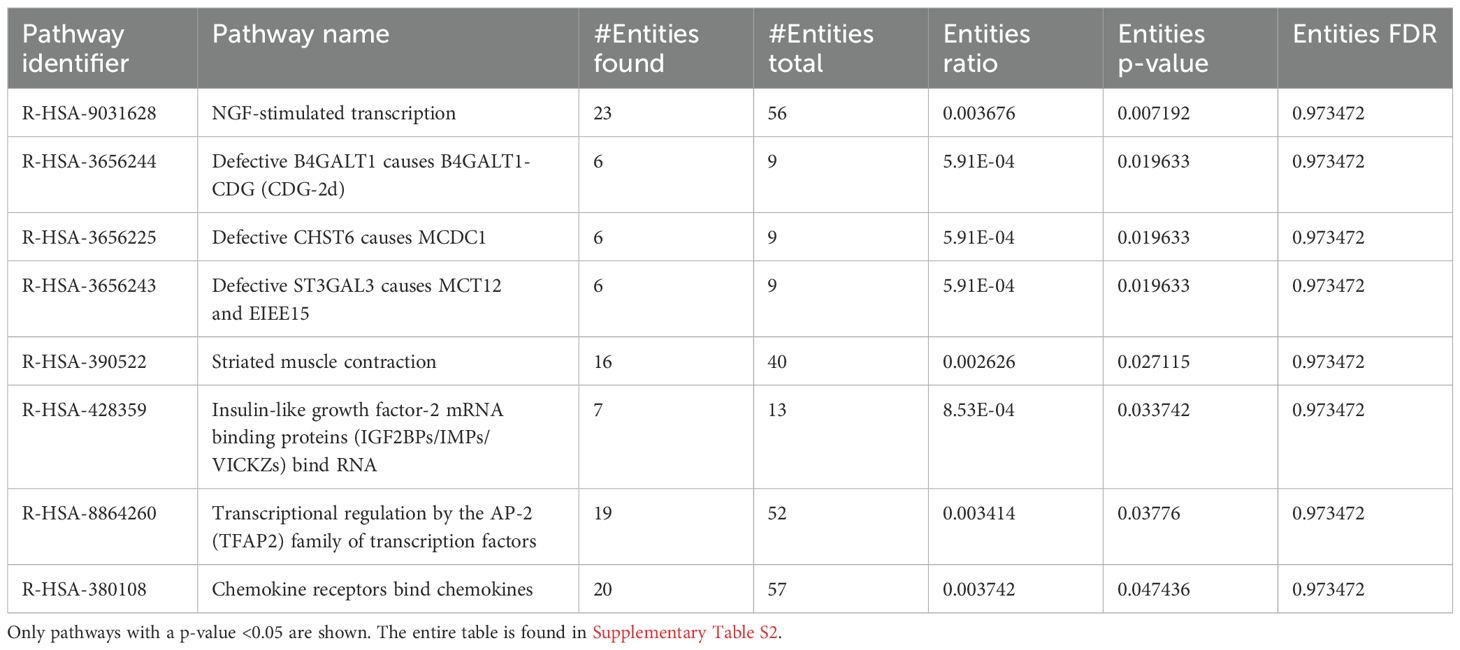
Table 3. Pathway enrichment of naïve I. scapularis–free skin vs. skin harvested after 3 h of uninfected I. scapularis feedings.
3.1.3 Naïve skin vs. skin from the site of uninfected I. scapularis fed for 6 h
Finally, we analyzed the cutaneous transcriptomic changes during 6 h of uninfected tick feeding. PCA revealed consistent clustering of the tick-free control skin samples, with a larger variance observed in the skin samples from the 6-h uninfected tick group (Figure 4A).
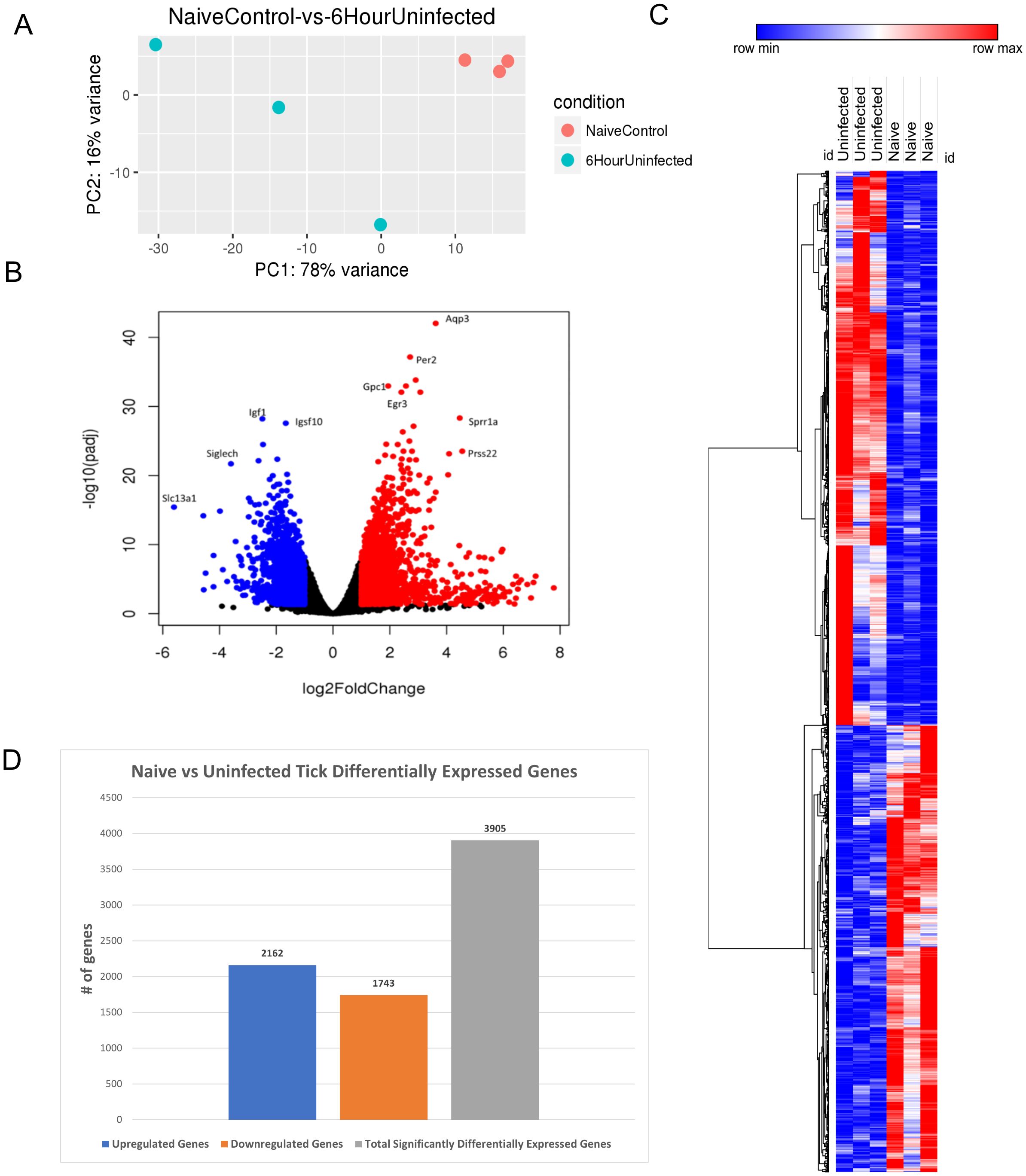
Figure 4. Differential expression of genes in I. scapularis–free control skin compared to 6-h uninfected tick feedings. (A) Principal component analysis (PCA) of transcriptional profiles when comparing 6-h uninfected feedings (blue) to naïve skin (orange). (B) Volcano plot outlining upregulated and downregulated genes at 6 h. The x-axis identifies the log2 fold change of every gene, and the y-axis identifies the log10 of its adjusted p-value. Only genes with a p-value <0.05 and a log2 fold change <−1 are identified as downregulated (blue). Only genes with a p-value <0.05 and a log2 fold change >1 are identified as upregulated (red). (C) A bi-clustering heatmap was used to visualize the expression profile of all differentially expressed genes (DEGs), sorted by their adjusted p-value by plotting log2-transformed values in samples. (D) Total DEGs for uninfected tick feeding vs. naïve skin at 6 h.
Volcano plot analysis identified 3,905 significant DEGs, with 2,162 upregulated and 1,743 downregulated genes (Figure 4B). Notable downregulated genes included Slc13a1, Siglech, Igf1, and Igsf10, which participate in sodium ion transport, cell signaling, and neuron migration. Upregulated genes, such as Aqp3, Per2, Gpc1, Egr3, Sprr1a, and Prss22, were associated with cell signaling and survival. Interestingly, Aqp3, an aquaporin, suppresses apoptosis in skin fibroblasts by upregulating Bcl-2, highlighting the potential for host defense mechanisms to be activated at tick bite sites (27).
The bi-clustering heatmap allowed for comparison of all DEGs between the groups (Figure 4C). This summary heatmap shows the comparisons of each individual mouse for both groups, tick-free mice versus 6-h uninfected tick feeding. The tree to the left demonstrates hierarchical clustering of the samples. The differential analysis revealed a total of 3,905 genes expressed in this study (Figure 4D).
DEG data were input into the Reactome database, and PE analysis revealed the impacted pathways during the 6-h feeding, including keratinization, formation of the cornified envelope, and nerve growth factor (NGF)-stimulated transcription (Table 4).
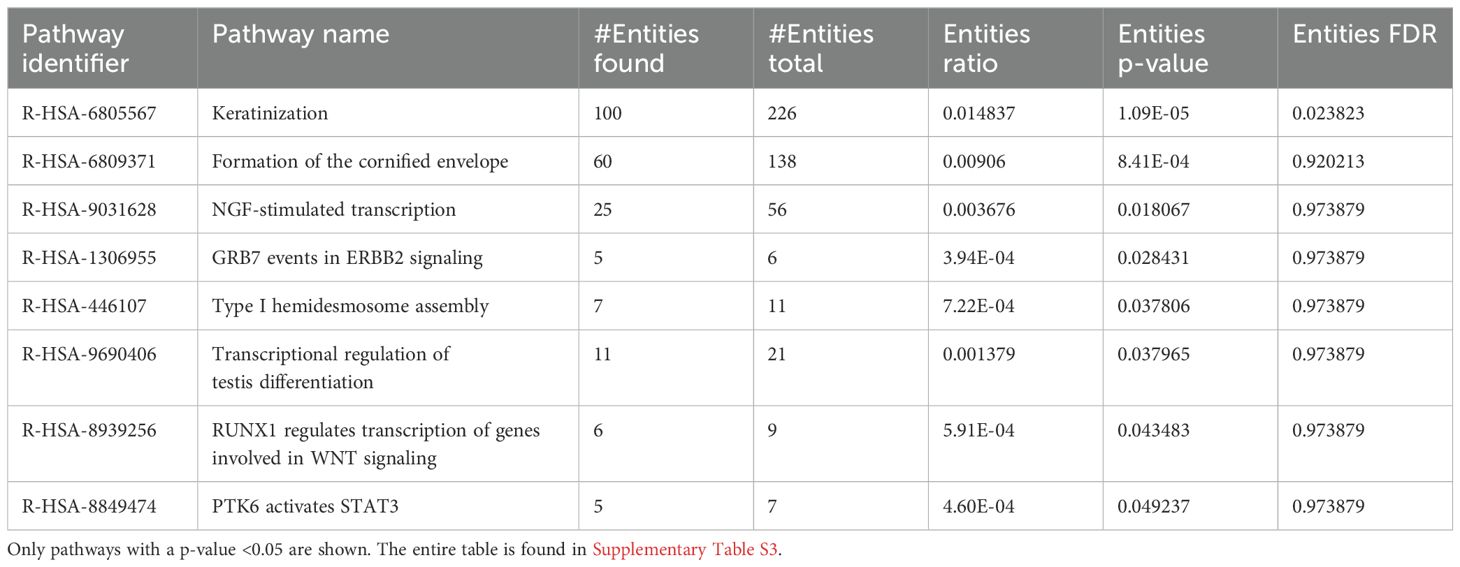
Table 4. Pathway enrichment of naïve I. scapularis–free skin vs. skin harvested after 6 h of uninfected I. scapularis feedings.
3.2 Changes in the skin transcriptome during POWV-infected I. scapularis feeding
3.2.1 Comparison of skin from the sites of POWV-infected vs. uninfected I. scapularis fed for 6 h
In this experiment, I scapularis females either infected with POWV or mock infected with DMEM as a control were allowed to feed on mice for 1, 3, and 6 h. After 1, 3, and 6 h of tick feeding, 4-mm skin biopsies of the bite site were excised, and DEG analysis via RNA-seq was performed as described previously. POWV transcripts were detected through ddPCR to verify infection at the time points described in this study (Supplementary Figure S1). Compared to the uninfected tick bites, DEG analysis performed on the 1 and 3 h post–POWV-infected tick bite sites yielded significantly fewer DEGs, prompting us to focus on the 6-h time point due to the richness of the data. Comparing POWV-infected tick feeding site with that of the uninfected tick feeding sites resulted in identifying 83 statistically significant DEGs, including 80 upregulated genes and 3 downregulated genes. The results show potentially how POWV exploits the tick’s host modulatory effects as well as identifying specific changes induced by the presence of POWV during the feeding event.
PCA for the 6-h uninfected tick feeding sites versus the POWV-infected tick-feeding sites revealed variance between uninfected samples, with one observed outlier among the POWV-infected samples (Figure 5A).
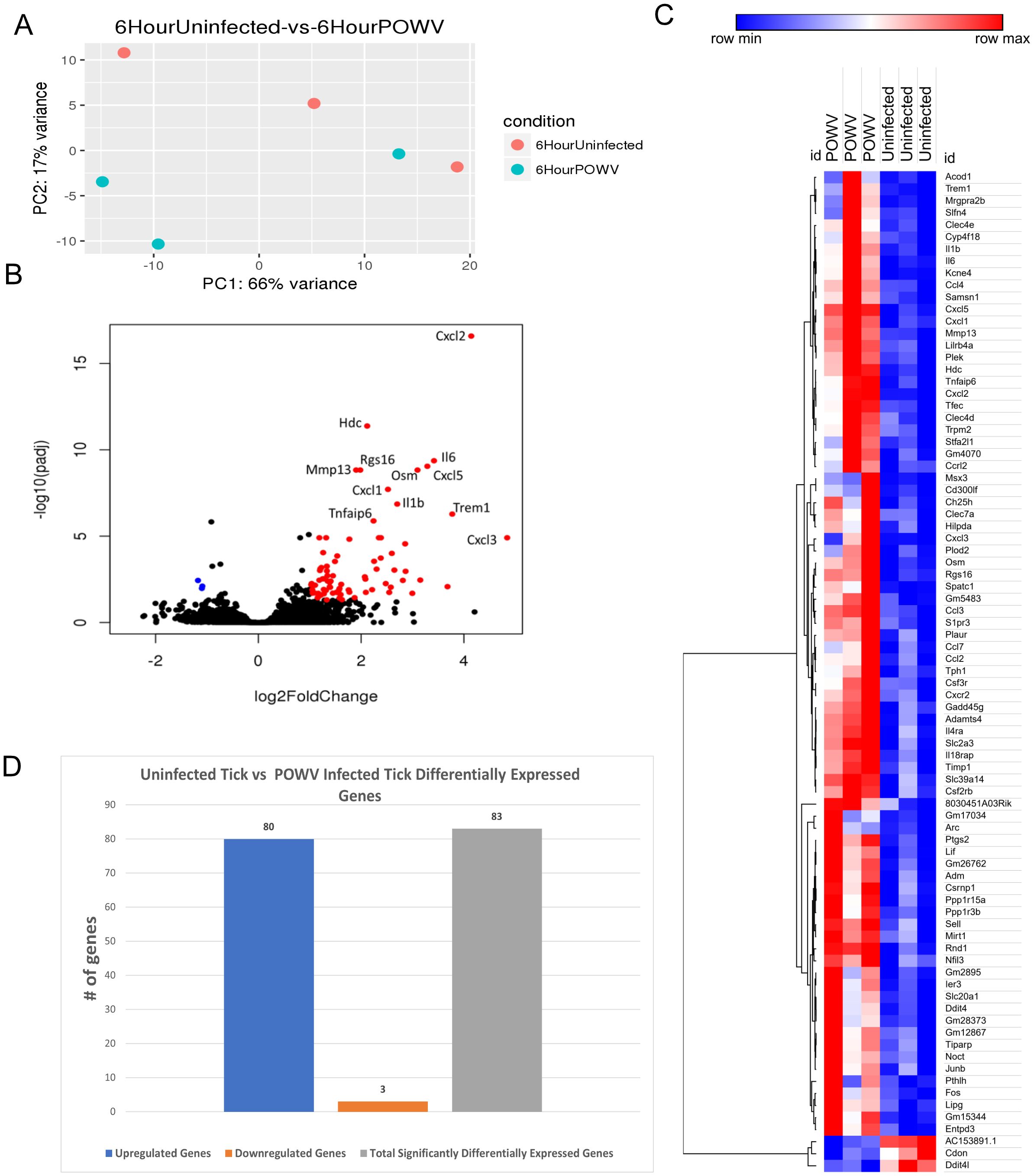
Figure 5. Differential gene expression at the I. scapularis feeding site in response to POWV-infected tick and uninfected tick bites at 6 h after attachment. (A) Principal component analysis (PCA) of transcriptional profiles when comparing 6-h uninfected tick feeding (blue) to 6-h POWV-infected tick feeding (orange). (B) Volcano plot outlining upregulated and downregulated genes at 6 h. The x-axis identifies the log2 fold change of every gene, and the y-axis identifies the log10 of its adjusted p-value. Only genes with a p-value <0.05 and a log2 fold change <−1 are identified as downregulated (blue). Only genes with a p-value <0.05 and a log2 fold change >1 are identified as upregulated (red). (C) A bi-clustering heatmap was used to visualize the expression profile of top 30 differentially expressed genes (DEGs), sorted by their adjusted p-value by plotting log2-transformed values in samples. (D) Total DEGs for uninfected tick feeding vs. POWV-infected tick feeding at 6 h.
Gene expression analysis using volcano plots highlighted a substantial upregulation of immune regulatory genes after 6 h of POWV-infected tick feeding (Figure 5B). Prominent among the upregulated genes were those associated with neutrophil recruitment, including Cxcl3, Cxcl2, Mrgpra2b, Il-6, Cxcl5, Il-1b, and Cxcl1 (28, 29). Additionally, genes involved in macrophage and natural killer cell migration, such as Ccl4 and Ccl3, were also upregulated (30, 31). Furthermore, a broad cell surface receptor, Trem1, was highly upregulated, suggesting its role in enhancing immune responses (32). Notably, the only significantly downregulated genes during the 6-h feeding were AC153891.1, Ddit4l, and Cdon, with the latter being a cell adhesion molecule involved in myogenic differentiation (33, 34).
Heatmap analysis of all DEGs at this time point demonstrated clustering within the groups. Despite the smaller number of DEGs, this clustering enables the visualization of individual changes in each sample. Notably, the three downregulated genes, AC153891.1, CDON, and DDIT4L, clustered together (Figure 5C). The differential analysis revealed a total of 83 genes expressed in this study (Figure 5D).
GO enrichment analysis of the DEGs identified significant enrichment of pathways related to neutrophil chemotaxis, chemokine-mediated signaling, and the inflammatory response (Figure 6). Reactome pathway enrichment analysis identified several immune-related pathways impacted by the DEGs, including Interleukin (IL)-4 and IL-13 signaling, IL-10 signaling, cytokine signaling, and immune system functions (Table 5).
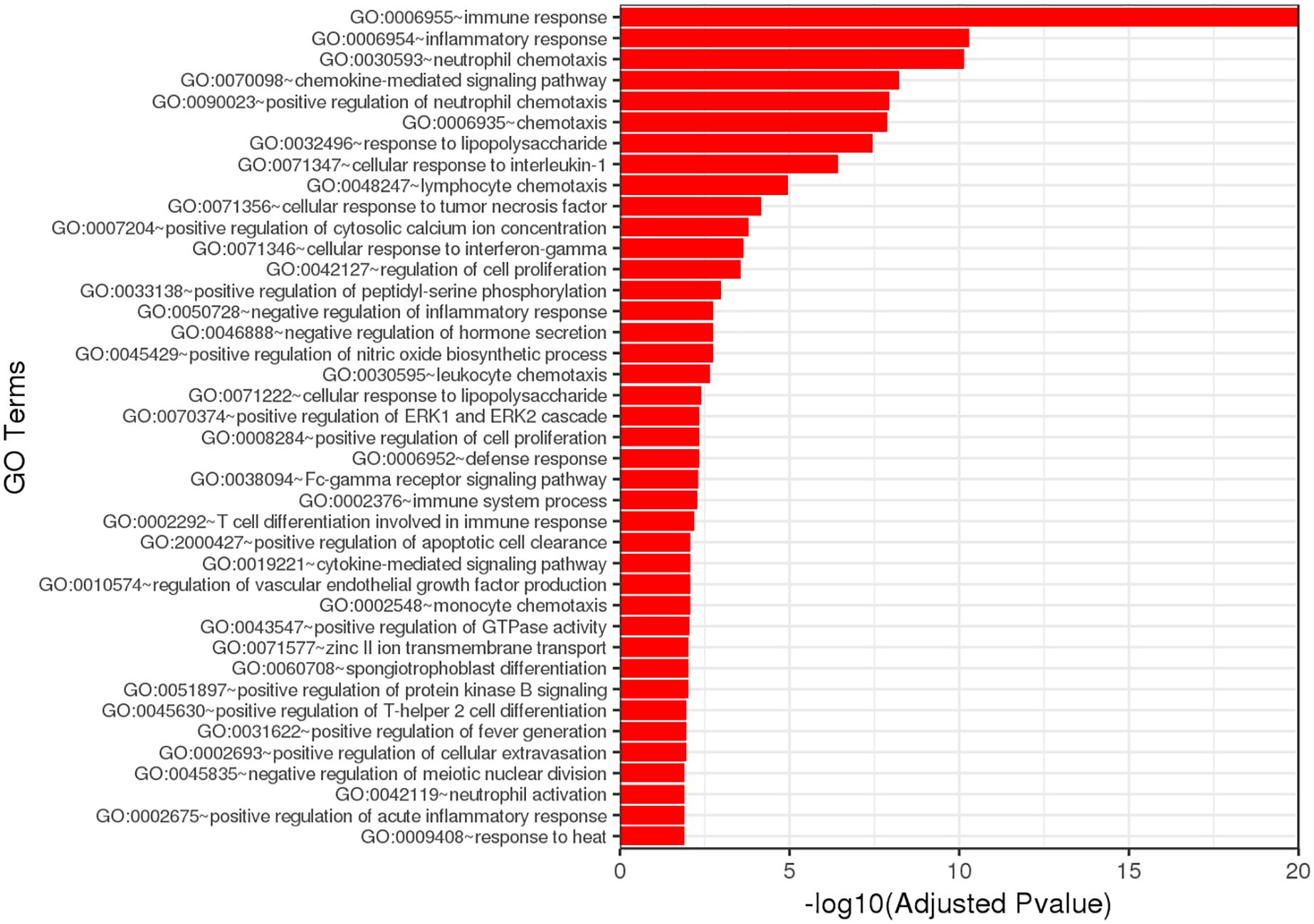
Figure 6. Gene ontology (GO) analysis based on DEG analysis shows strong signaling effects on neutrophil chemotaxis and inflammation.
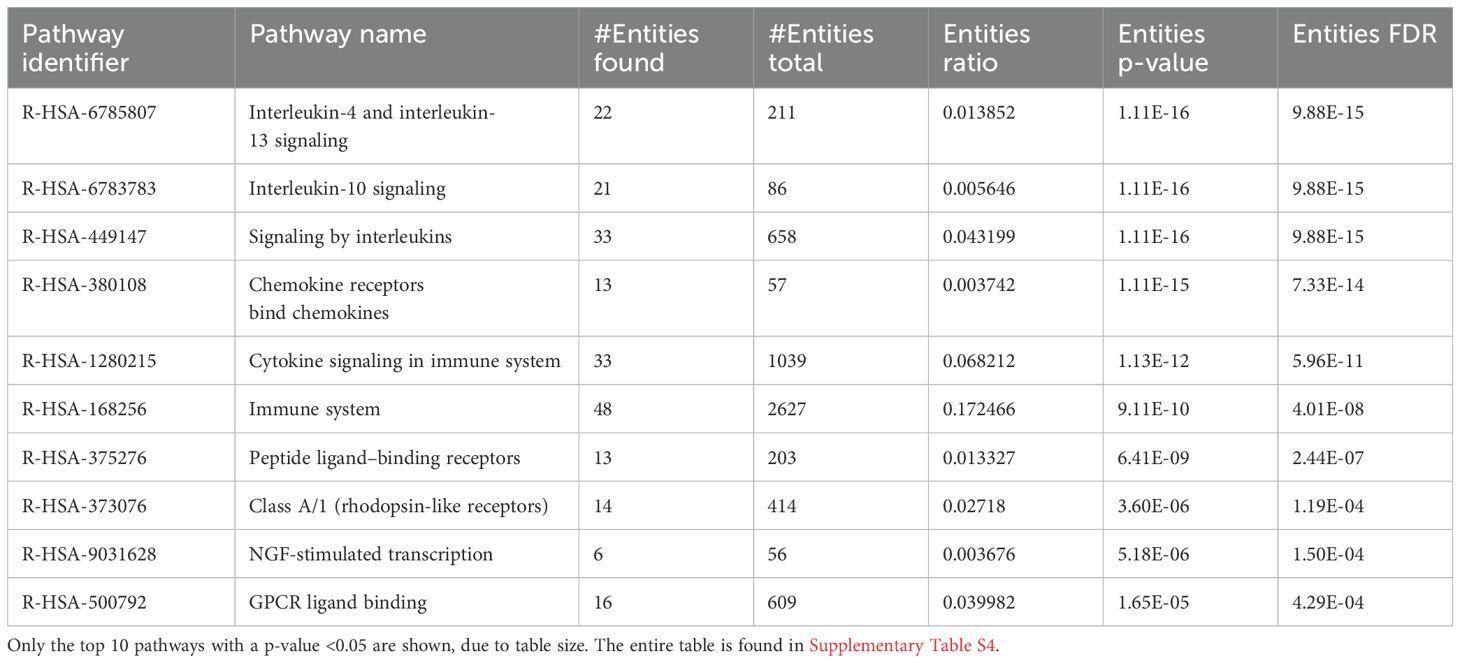
Table 5. Pathway enrichment of 6-h uninfected I. scapularis feedings vs. 6-h POWV-infected I. scapularis feeding.
To gain a comprehensive understanding of the cutaneous transcriptomic changes during the 6-h POWV feeding, we utilized ingenuity pathway analysis (IPA). The network interaction summary (Figure 7) depicts the interaction between downstream and upstream pathways during the initial stages of POWV infection, specifically at the 6-h time point after tick attachment. Here, we can see the changes that are a result of POWV infection, where the left side of the chart shows the extracellular components that may be upregulated or activated on the basis of infection status. These include things that we would expect to be stimulated such as recruitment of all different cell types like lymphocytes, leukocytes, blood cells, and progenitor cells. The matrix on the right highlights chemokines such as CXCL3 that participate in the recruitment of leukocytes to the bite site, as well as the trafficking of other immune cells and signaling components to the affected area. Most of the upregulated genes identified in this study are associated with inflammation and activation of an immune response, especially early innate responses. Components leading to major signaling like v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA), chemokine (C-X-C motif) ligand 2 (CXCL2), myeloid differentiation primary response 88 (MYD88), and tumor necrosis factor (TNF) are implicated. These findings align with previous work, suggesting that immune cell infiltration and inflammatory responses are important drivers of the host’s reaction to initial POWV infection transmitted by ticks (19, 23, 35).
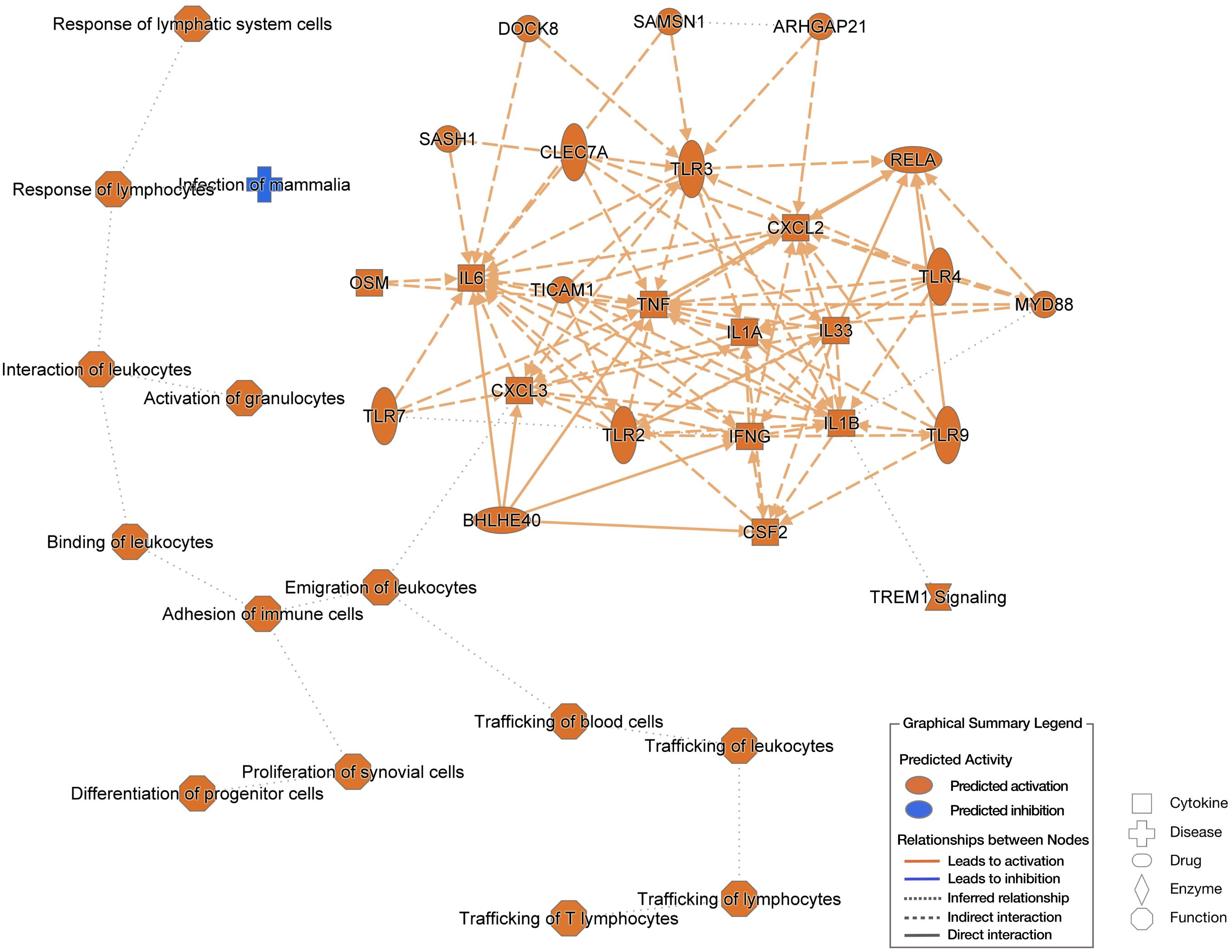
Figure 7. Ingenuity pathway analysis (IPA) based on 6-h uninfected I. scapularis feedings vs. 6-h POWV-infected feedings. The left side of the image shows extracellular predictions based on input information, and the right-side network represents potential molecular interactors based on input information. Predictions made using Qiagen IPA algorithms.
4 Discussion
The initial hours and days of tick feeding has been linked to the transmission of a wide variety of pathogens, including viruses, bacteria, and parasites, each with a specific window of time required for transmission (15, 16, 36–41). As shown in Table 1 where DEGs for each bioinformatic analysis were outlined, minimal changes were observed at the 1-h time point when comparing POWV-infected and uninfected tick feeding sites. This could primarily be due to the initial tick attachment, during which there may not be sufficient time for the virus or tick salivary factors to significantly alter the attachment site. It is possible that 1 h of POWV-infected tick feeding is insufficient to elicit robust transcriptional changes in the skin. However, the detection of 3,722 significant DEGs resulting from uninfected tick feedings, compared to an unbitten mouse, indicates immunomodulatory changes at the tick feeding site. This is supportive of our current understanding of how arthropod feedings cause an enumerative number of local changes at their feeding site. The primary alterations involved genes associated with coagulation and wound healing factors, likely due to the introduction of tick salivary components, as identified in the bi-clustered heatmap (Figure 2). For example, during the initial feeding stages, tick saliva contains various anti-hemostatic factors that prevent coagulation and enhance blood meal acquisition (42).
At 6 h after tick attachment during POWV-infected tick feedings versus uninfected tick feedings, we observed the upregulation of proinflammatory genes and those involved in immune cell recruitment. This is consistent with previous findings on the early recruitment of macrophages and neutrophils, as demonstrated through PCR arrays and histological examination of the POWV-infected I. scapularis feeding site (17, 18). The upregulation of several transcripts associated with neutrophil recruitment, such as Cxcl3, Cxcl2, Mrgpra2b, Il-6, Cxcl5, Il-1b, and Cxcl1, supported by pathway enrichment and GO analysis, indicates the establishment of a strong proinflammatory environment at the POWV-infected tick feeding site. Neutrophils are known to be early responders to viral infections, recognizing pathogen-associated molecular patterns (PAMPs) and being stimulated by chemokines. For example, during influenza infection, virally infected cells secrete Cxcl8 and granulocyte macrophage-colony stimulating factor to attract neutrophils (43). Neutrophil activation is also observed in other arbovirus infections. Zika virus, for instance, works in concert with Aedes aegypti salivary protein neutrophil-stimulating factor 1 (NeSt1) to promote neutrophil recruitment, enhancing Zika virus pathogenesis in mice (44). Our data supports previous findings regarding the early influx of neutrophils to the site of POWV-infected nymphal tick feeding (17, 18). The important distinction here is the use of adult I. scapularis ticks, which are known to have different salivary composition, feeding behavior, and infection kinetics for POWV (10, 45–47). The influx of neutrophils to sites of infection or tissue damage is not novel; however, their specific role in the pathogenesis of POWV transmission and dissemination is unknown. As our DEG analysis yields strong correlation between POWV, neutrophil recruitment, and inflammation, we can potentially surmise a key role these cells have during initial POWV infection. For Borrelia sp. infections, neutrophil recruitment and degranulation play critical roles in limiting the infection through extracellular traps, phagocytosis, and oxidative bursts (48). The role of these factors in determining disease outcomes during POWV infections remains unclear. However, even uninfected tick feeding induces the recruitment of neutrophils and other immune cells to the feeding site. This effect is amplified in the presence of POWV, with infected fibroblasts further contributing to the inflammatory response by increasing cytokine release upon infection.
Interestingly, in terms of inflammation and early innate activation, we do not observe many genes associated with interferon stimulation and activation. From our data, it is clear that SOCS3 (suppressor of cytokine signaling 3) and IFITM1 (interferon-induced transmembrane protein 1) as the primary upregulated components involved in the interferon pathway. SOCS3 is a well-described anti-interferon component of the immune response, used primarily to subdue the inflammatory response when not needed (49). This is in contrast to IFITM1, which is strongly upregulated by type I and II interferons to play a role in antiproliferation in context of viral infections (50). These two may be working in opposite directions, depending on what is interacting with them. Investigations into POWV and the interferon response are areas that should be prioritized by further research.
The generation of a proinflammatory state is a critical aspect of disease pathogenesis, particularly during viral transmission and dissemination. At the 6-h post–POWV-infected tick feeding, we observed a broad array of inflammatory markers upregulated through IPA. These markers included various canonical inflammation pathways associated with viral responses, notably IL-1b, IL-6, and CCL2, each showing a greater than 2-log expression increase. These markers suggest the involvement of the NLRP3 inflammasome pathway during POWV transmission. The NLRP3 inflammasome is a well-established marker for inflammation during pathogen infiltration, regulated by both PAMPs and damage-associated molecular patterns (51). The activation of the NLRP3 inflammasome has been described for many arboviruses, particularly those spread by mosquitoes. For example, the dengue virus activates NLRP3 by using NS2A and NS2B proteins to initiate an inflammatory environment, leading to the recruitment of permissive cells (52). Similarly, markers such as IL-1b and IL-6 are recognized biomarkers for Chikungunya disease severity in clinical settings (53, 54). Although these components are heavily induced during POWV transmission, their roles in tick-borne diseases are not well characterized. A closely related virus, tick-borne encephalitis virus (TBEV), is a neuroinvasive tick-borne virus prevalent in Europe and Asia. A cohort study identified TNFα, IL-1b, and IL-6 as being highly prevalent in the serum of patients upon hospital admission, suggesting that these cytokines play a crucial role in TBEV infection and pathogenesis (55). NLRP3 is a well-established inflammatory holoenzyme that activates in response to many arboviruses (52, 56–58). The potential involvement of the NLRP3 inflammasome in POWV pathogenesis warrants further investigation into the roles of these immune responses. Whereas the primary products of this pathway were identified to be significantly upregulated, NLRP3 transcript was found to be upregulated but not statistically significant in response to POWV-infected tick bites at 6 h after tick attachment. This is attributed to that NLRP3 being constitutively expressed even in response to uninfected tick bites. Assessing its role in the future is critical for understanding pathogenesis and immune interaction with POWV.
During POWV transmission from tick to vertebrate host, the virus encounters a variety of resident and recruited immune cells at the tick feeding site. Resident macrophages and Langerhans cells, which commonly encounter pathogens introduced into the skin, have been shown to co-localize with POWV RNA in previous studies using RNA in situ hybridization (35). This previous study was conducted 24 h after attachment with I. scapularis nymphs and aimed to address what occurs at the critical time points relevant to transmission. The increased analyte count from RNA-seq in our current study allows for a more detailed investigation, reinforcing the establishment of an inflammatory environment at the bite site during POWV transmission. Our proposed model for POWV infection suggests that, upon successful attachment and feeding by a POWV-infected tick, infectious virions and tick saliva are secreted into the bite site resulting in the recruitment of lymphocytes such as neutrophils. Recruitment of these cells occurs through chemokines identified in our RNA-seq analysis such as CCL2. There are also potential interactions with resident mast cells indicated by this study’s transcriptomic identification of CPA3 and other cell surface ligands such as TLR 3, 7, 8, and 9 for pathogen detection. Resident immune cell activation and recruitment result in a positive feedback loop of chemokines and cytokines, drawing more susceptible cells to the site of infection and allowing the virus a potential route for dissemination and infection establishment (Figure 8).
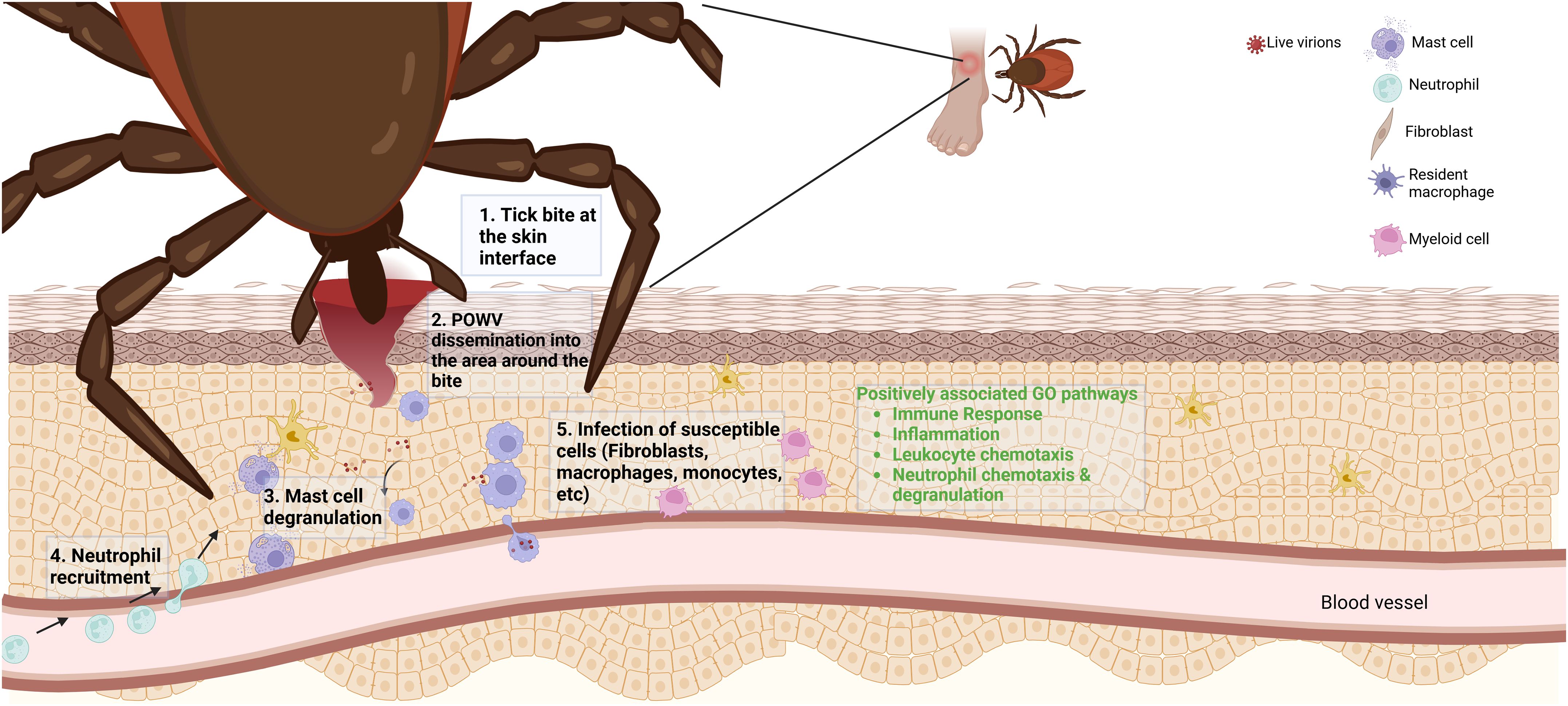
Figure 8. A proposed Powassan virus (POWV) transmission model based on differentially expressed genes. Upon transmission of POWV via an infected I. scapularis, the virus infects local cells at the bite site, resulting in differential expression of specific genes. The expression of various transcripts may yield recruitment of lymphocytes to the area of infection. The recruitment of these cells to the area of infection could result in them becoming infected, potentially allowing for the establishment of infection. In green text are highly implicated pathways of involvement from GO analysis of 6-h tick feedings with POWV.
5 Conclusion
Our findings suggest that POWV-infected tick feeding induces the expression of proinflammatory and immune response genes at the tick feeding site, which likely contributes to the recruitment of neutrophils and other immune cells. POWV appears to enhance the tick’s natural modulation of the host’s cutaneous cellular response to wounding via tick feeding, amplifying inflammatory pathways that may contribute to the pathogenesis of the infection. The role of neutrophils has not been heavily investigated in response to POWV infection but opens a potential area, which may lead to further studies involving the viral dissemination from skin to CNS.
Data availability statement
The data presented in the study are deposited in to the Gene Expression Omnibus, accession number GSE282786.
Ethics statement
The animal study was approved by University of Texas Medical Branch (UTMB) Institutional Animal Care and Use Committee (IACUC: # 0907054). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
DP: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft. MH: Investigation, Methodology, Writing – review & editing. ST: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. ST is funded by the National Institutes of Health R01 grant AI127771. The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation.
Acknowledgments
Graphics in Figures 1 and 8 were created with Biorender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1511132/full#supplementary-material
Supplementary Figure 1 | POWV infection was assessed at the skin interface during appropriate times of an I. scapularis feeding. To verify the infection at each of the bite sites, ddPCR was used for each sample used in the bioinformatic analysis study.
References
1. Altizer S, Ostfeld RS, Johnson PTJ, Kutz S, Harvell CD. Climate change and infectious diseases: from evidence to a predictive framework. Science. (2013) 341:514–9. doi: 10.1126/science.1239401
2. Eisen L. Control of ixodid ticks and prevention of tick-borne diseases in the United States: The prospect of a new Lyme disease vaccine and the continuing problem with tick exposure on residential properties. Ticks Tick-Borne Dis. (2021) 12:101649. doi: 10.1016/j.ttbdis.2021.101649
3. McLean DM, Donohue WL. Powassan virus: isolation of virus from a fatal case of encephalitis. Can Med Assoc J. (1959) 80:708–11.
4. Fitch WM, Artsob H. Powassan encephalitis in New Brunswick. Can Fam Physician Med Fam Can. (1990) 36:1289–90.
5. Hermance ME, Thangamani S. Powassan virus: an emerging arbovirus of public health concern in North America. Vector Borne Zoonotic Dis Larchmt N. (2017) 17:453–62. doi: 10.1089/vbz.2017.2110
6. Cheung AM, Yip EZ, Ashbrook AW, Goonawardane N, Quirk C, Rice CM, et al. Characterization of live-attenuated Powassan virus vaccine candidates identifies an efficacious prime-boost strategy for mitigating Powassan virus disease in a murine model. Vaccines. (2023) 11:612. doi: 10.3390/vaccines11030612
7. Choi H, Kudchodkar SB, Ho M, Reuschel EL, Reynolds E, Xu Z, et al. A novel synthetic DNA vaccine elicits protective immune responses against Powassan virus. PloS Negl Trop Dis. (2020) 14:e0008788. doi: 10.1371/journal.pntd.0008788
8. Fontaine A, Diouf I, Bakkali N, Missé D, Pagès F, Fusai T, et al. Implication of haematophagous arthropod salivary proteins in host-vector interactions. Parasit Vectors. (2011) 4:187. doi: 10.1186/1756-3305-4-187
9. Francischetti IMB, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. Front Biosci Landmark Ed. (2009) 14:2051–88. doi: 10.2741/3363
10. Ribeiro JMC, Alarcon-Chaidez F, Francischetti IMB, Mans BJ, Mather TN, Valenzuela JG, et al. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Mol Biol. (2006) 36:111–29. doi: 10.1016/j.ibmb.2005.11.005
11. Wikel S. Ticks and tick-borne pathogens at the cutaneous interface: host defenses, tick countermeasures, and a suitable environment for pathogen establishment. Front Microbiol. (2013) 4:337. doi: 10.3389/fmicb.2013.00337
12. Jones LD, Hodgson E, Nuttall PA. Enhancement of virus transmission by tick salivary glands. J Gen Virol. (1989) 70:1895–8. doi: 10.1099/0022-1317-70-7-1895
13. Labuda M, Jones LD, Williams T, Nuttall PA. Enhancement of tick-borne encephalitis virus transmission by tick salivary gland extracts. Med Vet Entomol. (1993) 7:193–6. doi: 10.1111/j.1365-2915.1993.tb00674.x
14. Santos RI, Hermance ME, Reynolds ES, Thangamani S. Salivary gland extract from the deer tick, Ixodes scapularis, facilitates neuroinvasion by Powassan virus in BALB/c mice. Sci Rep. (2021) 11:20873. doi: 10.1038/s41598-021-00021-2
15. Ebel GD, Kramer LD. Short report: duration of tick attachment required for transmission of Powassan virus by deer ticks. Am J Trop Med Hyg. (2004) 71:268–71. doi: 10.4269/ajtmh.2004.71.3.0700268
16. des Vignes F, Piesman J, Heffernan R, Schulze TL, Stafford KC, Fish D. Effect of tick removal on transmission of Borrelia burgdorferi and Ehrlichia phagocytophila by Ixodes scapularis nymphs. J Infect Dis. (2001) 183:773–8. doi: 10.1086/318818
17. Sertour N, Cotté V, Garnier M, Malandrin L, Ferquel E, Choumet V. Infection kinetics and tropism of Borrelia burgdorferi sensu lato in mouse after natural (via ticks) or artificial (needle) infection depends on the bacterial strain. Front Microbiol. (2018) 9:1722. doi: 10.3389/fmicb.2018.01722
18. Hermance ME, Thangamani S. Proinflammatory Cytokines and Chemokines at the Skin Interface during Powassan Virus Transmission. J Invest Dermatol. (2014) 134:2280–3. doi: 10.1038/jid.2014.150
19. Hermance ME, Santos RI, Kelly BC, Valbuena G, Thangamani S. Immune cell targets of infection at the tick-skin interface during Powassan virus transmission. PloS One. (2016) 11:e0155889. doi: 10.1371/journal.pone.0155889
20. Rossi SL, Nasar F, Cardosa J, Mayer SV, Tesh RB, Hanley KA, et al. Genetic and phenotypic characterization of sylvatic dengue virus type 4 strains. Virology. (2012) 423:58–67. doi: 10.1016/j.virol.2011.11.018
21. Hermance ME, Widen SG, Wood TG, Thangamani S. Ixodes scapularis salivary gland microRNAs are differentially expressed during Powassan virus transmission. Sci Rep. (2019) 9:13110. doi: 10.1038/s41598-019-49572-5
22. Hermance ME, Thangamani S. Tick saliva enhances Powassan virus transmission to the host, influencing its dissemination and the course of disease. J Virol. (2015) 89:7852–60. doi: 10.1128/JVI.01056-15
23. Heinze DM, Carmical JR, Aronson JF, Thangamani S. Early immunologic events at the tick-host interface. PloS One. (2012) 7:e47301. doi: 10.1371/journal.pone.0047301
24. Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. (2007) 7:255–66. doi: 10.1038/nri2056
25. Zambetti G, Ramsey-Ewing A, Bortell R, Stein G, Stein J. Disruption of the cytoskeleton with cytochalasin D induces c-fos gene expression. Exp Cell Res. (1991) 192:93–101. doi: 10.1016/0014-4827(91)90162-N
26. Gazon H, Barbeau B, Mesnard J-M, Peloponese J-M. Hijacking of the AP-1 signaling pathway during development of ATL. Front Microbiol. (2018) 8:2686. doi: 10.3389/fmicb.2017.02686
27. Xie H, Liu F, Liu L, Dan J, Luo Y, Yi Y, et al. Protective role of AQP3 in UVA-induced NHSFs apoptosis via Bcl2 up-regulation. Arch Dermatol Res. (2013) 305:397–406. doi: 10.1007/s00403-013-1324-y
28. Capucetti A, Albano F, Bonecchi R. Multiple roles for chemokines in neutrophil biology. Front Immunol. (2020) 11:1259. doi: 10.3389/fimmu.2020.01259
29. Sawant KV, Sepuru KM, Lowry E, Penaranda B, Frevert CW, Garofalo RP, et al. Neutrophil recruitment by chemokines Cxcl1/KC and Cxcl2/MIP2: Role of Cxcr2 activation and glycosaminoglycan interactions. J Leukoc Biol. (2021) 109:777–91. doi: 10.1002/JLB.3A0820-207R
30. Sindhu S, Kochumon S, Shenouda S, Wilson A, Al-Mulla F, Ahmad R. The cooperative induction of CCL4 in human monocytic cells by TNF-α and palmitate requires myD88 and involves MAPK/NF-κB signaling pathways. Int J Mol Sci. (2019) 20:4658. doi: 10.3390/ijms20184658
31. Sheng D, Ma W, Zhang R, Zhou L, Deng Q, Tu J, et al. Ccl3 enhances docetaxel chemosensitivity in breast cancer by triggering proinflammatory macrophage polarization. J Immunother Cancer. (2022) 10:e003793. doi: 10.1136/jitc-2021-003793
32. Colonna M. The biology of TREM receptors. Nat Rev Immunol. (2023) 23:580–94. doi: 10.1038/s41577-023-00837-1
33. Hong M, Christ A, Christa A, Willnow TE, Krauss RS, Bovolenta P, et al. Cdon mutation and fetal alcohol converge on Nodal signaling in a mouse model of holoprosencephaly. eLife. (2020) 9:e60351. doi: 10.7554/eLife.60351
34. Fattahi F, Saeednejad Zanjani L, Habibi Shams Z, Kiani J, Mehrazma M, Najafi M, et al. High expression of DNA damage-inducible transcript 4 (DDIT4) is associated with advanced pathological features in the patients with colorectal cancer. Sci Rep. (2021) 11:13626. doi: 10.1038/s41598-021-92720-z
35. Hermance ME, Thangamani S. Utilization of RNA in situ hybridization to understand the cellular localization of Powassan virus rna at the tick-virus-host interface. Front Cell Infect Microbiol. (2020) 10:172. doi: 10.3389/fcimb.2020.00172
36. Rehacek J. Development of animal viruses and rickettsiae in ticks and mites. Annu Rev Entomol. (1965) 10:1–24. doi: 10.1146/annurev.en.10.010165.000245
37. Sood SK, Salzman MB, Johnson BJ, Happ CM, Feig K, Carmody L, et al. Duration of tick attachment as a predictor of the risk of Lyme disease in an area in which Lyme disease is endemic. J Infect Dis. (1997) 175:996–9. doi: 10.1086/514009
38. Levin ML, Troughton DR, Loftis AD. Duration of tick attachment necessary for transmission of Anaplasma phagocytophilum by Ixodes scapularis (Acari: Ixodidae) nymphs. Ticks Tick-Borne Dis. (2021) 12:101819. doi: 10.1016/j.ttbdis.2021.101819
39. Breuner NE, Dolan MC, Replogle AJ, Sexton C, Hojgaard A, Boegler KA, et al. Transmission of Borrelia miyamotoi sensu lato relapsing fever group spirochetes in relation to duration of attachment by Ixodes scapularis nymphs. Ticks Tick-Borne Dis. (2017) 8:677–81. doi: 10.1016/j.ttbdis.2017.03.008
40. Dolan MC, Breuner NE, Hojgaard A, Boegler KA, Hoxmeier JC, Replogle AJ, et al. Transmission of the lyme disease spirochete Borrelia mayonii in relation to duration of attachment by Nymphal Ixodes scapularis (Acari: Ixodidae). J Med Entomol. (2017) 54:1360–4. doi: 10.1093/jme/tjx089
41. Piesman J, Dolan MC. Protection against lyme disease spirochete transmission provided by prompt removal of nymphal Ixodes scapularis (Acari: Ixodidae). J Med Entomol. (2002) 39:509–12. doi: 10.1603/0022-2585-39.3.509
42. Chmelar J, Calvo E, Pedra JHF, Francischetti IMB, Kotsyfakis M. Tick salivary secretion as a source of antihemostatics. J Proteomics. (2012) 75:3842–54. doi: 10.1016/j.jprot.2012.04.026
43. Ito Y, Correll K, Zemans RL, Leslie CC, Murphy RC, Mason RJ. Influenza induces IL-8 and GM-CSF secretion by human alveolar epithelial cells through HGF/c-Met and TGF-α/EGFR signaling. Am J Physiol Lung Cell Mol Physiol. (2015) 308:L1178–1188. doi: 10.1152/ajplung.00290.2014
44. Hastings AK, Uraki R, Gaitsch H, Dhaliwal K, Stanley S, Sproch H, et al. Aedes aEgypti neSt1 protein enhances Zika virus pathogenesis by activating neutrophils. J Virol. (2019) 93:e00395-19. doi: 10.1128/JVI.00395-19
46. Hart CE, Bhaskar JR, Reynolds E, Hermance M, Earl M, Mahoney M, et al. Community engaged tick surveillance and tickMAP as a public health tool to track the emergence of ticks and tick-borne diseases in New York. PloS Glob Public Health. (2022) 2:e0000215. doi: 10.1371/journal.pgph.0000215
47. Costero A, Grayson MA. Experimental transmission of Powassan virus (Flaviviridae) by ixodes scapularis ticks (Acari: ixodidae). The American Journal of Tropical Medicine and Hygiene (1996) 55(5):536–46. doi: 10.4269/ajtmh.1996.55.536
48. Appelgren D, Enocsson H, Skogman BH, Nordberg M, Perander L, Nyman D, et al. Neutrophil extracellular traps (NETs) in the cerebrospinal fluid samples from children and adults with central nervous system infections. Cells. (2019) 9:43. doi: 10.3390/cells9010043
49. Cohney SJ, Sanden D, Cacalano NA, Yoshimura A, Mui A, Migone TS, et al. SOCS-3 is tyrosine phosphorylated in response to interleukin-2 and suppresses STAT5 phosphorylation and lymphocyte proliferation. Mol Cell Biol. (1999) 19:4980. doi: 10.1128/mcb.19.7.4980
50. Friedman RL, Manly SP, McMahon M, Kerr IM, Stark GR. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell. (1984) 38:745–55. doi: 10.1016/0092-8674(84)90270-8
51. He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. (2016) 41:1012–21. doi: 10.1016/j.tibs.2016.09.002
52. Shrivastava G, Visoso-Carvajal G, Garcia-Cordero J, Leon-Juarez M, Chavez-Munguia B, Lopez T, et al. Dengue virus serotype 2 and its non-structural proteins 2A and 2B activate NLRP3 inflammasome. Front Immunol. (2020) 11:352. doi: 10.3389/fimmu.2020.00352
53. Ng LFP, Chow A, Sun Y-J, Kwek DJC, Lim P-L, Dimatatac F, et al. IL-1β, IL-6, and RANTES as biomarkers of chikungunya severity. PloS One. (2009) 4:e4261. doi: 10.1371/journal.pone.0004261
54. Soares-Schanoski A, Baptista Cruz N, de Castro-Jorge LA, de Carvalho RVH, dos Santos CA, da Rós N, et al. Systems analysis of subjects acutely infected with the Chikungunya virus. PloS Pathog. (2019) 15:e1007880. doi: 10.1371/journal.ppat.1007880
55. Atrasheuskaya AV, Fredeking TM, Ignatyev GM. Changes in immune parameters and their correction in human cases of tick-borne encephalitis. Clin Exp Immunol. (2003) 131:148–54. doi: 10.1046/j.1365-2249.2003.02050.x
56. Gim E, Shim D-W, Hwang I, Shin OS, Yu J-W. Zika virus impairs host NLRP3-mediated inflammasome activation in an NS3-dependent manner. Immune Netw. (2019) 19:e40. doi: 10.4110/in.2019.19.e40
57. Pan P, Zhang Q, Liu W, Wang W, Lao Z, Zhang W, et al. Dengue virus M protein promotes NLRP3 inflammasome activation to induce vascular leakage in mice. J Virol. (2019) 93:e00996-19. doi: 10.1128/JVI.00996-19
Keywords: Powassan virus, Ixodes, immunomodulation, transmission, arbovirus
Citation: Paine DN, Hermance M and Thangamani S (2025) Early transcriptomic changes at the skin interface during Powassan virus transmission by Ixodes scapularis ticks. Front. Immunol. 15:1511132. doi: 10.3389/fimmu.2024.1511132
Received: 14 October 2024; Accepted: 10 December 2024;
Published: 13 January 2025.
Edited by:
Maria Kazimirova, Slovak Academy of Sciences, SlovakiaReviewed by:
Quentin Bernard, Tufts University, United StatesJaroslava Lieskovska, University of South Bohemia in České Budějovice, Czechia
Copyright © 2025 Paine, Hermance and Thangamani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saravanan Thangamani, VGhhbmdhbXNAdXBzdGF0ZS5lZHU=
 Dakota N. Paine
Dakota N. Paine Meghan Hermance
Meghan Hermance Saravanan Thangamani
Saravanan Thangamani