- 1Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, Netherlands
- 2Division of Medical Virology, Department of Pathology, Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Cape Town, South Africa
- 3Centre for Infectious Diseases Research in Africa, Institute of Infectious Disease and Molecular Medicine and Department of Medicine, University of Cape Town, Cape Town, South Africa
- 4South African Tuberculosis Vaccine Initiative, Institute of Infectious Disease and Molecular Medicine, Division of Immunology, Department of Pathology, University of Cape Town, Cape Town, South Africa
- 5Tuberculosis Laboratory, The Francis Crick Institute, London, United Kingdom
- 6Department of Infectious Diseases, Imperial College London, London, United Kingdom
Introduction: Tuberculosis (TB) is the deadliest infectious disease worldwide and novel vaccines are urgently needed. HLA-E is a virtually monomorphic antigen presentation molecule and is not downregulated upon HIV co-infection. HLA-E restricted Mtb specific CD8+ T cells are present in the circulation of individuals with active TB (aTB) and Mtb infection (TBI) with or without HIV co-infection, making HLA-E restricted T cells interesting vaccination targets for TB.
Methods: Here, we performed in-depth phenotyping of HLA-E/Mtb specific and total T cell populations in individuals with TBI and in individuals with aTB or TBI and HIV using HLA-E/Mtb tetramers.
Results and Discussion: We show that HIV co-infection is the main driver in changing the memory distribution of HLA-E/Mtb specific CD4+ and CD8+ T cell subsets. HLA-E/Mtb specific CD4+ and CD8+ T cells were found to circulate with comparable frequencies in all individuals and displayed expression of KLRG1, PD-1 and 2B4 similar to that of total T cells. The presence of HLA-E/Mtb specific T cells in individuals with aTB and TBI highlights the potential of HLA-E as a vaccine target for TB.
Introduction
Tuberculosis (TB), caused by infection with Mycobacterium tuberculosis (Mtb), is currently the deadliest infectious disease caused by a single pathogen (1). Vaccination with Bacillus Calmette-Guérin (BCG), the only licensed vaccine available, developed in 1921, fails to curb the global pandemic. Novel vaccines to protect against TB are therefore urgently needed. Protein subunit-based vaccination approaches against TB aim to induce antigen specific classical HLA-I and HLA-II restricted CD8+ and CD4+ T cell responses. However, the large diversity of HLA-I and II molecules between individuals complicates the selection of Mtb peptides that can be presented irrespective of the genetic background. In contrast, non-classical HLA-I molecules have limited allelic diversity, are recognized by donor unrestricted T cells (DURT) and are interesting vaccine targets to induce population-wide T cell responses that are necessary for protection (2). HLA-E is such a non-classical antigen presentation molecule with only two functional alleles expressed in humans, HLA-E*01:01 and *01:03. They differ by one amino acid located outside the peptide binding groove at position 107, suggesting that both alleles have a comparable peptide binding repertoire (3, 4). HLA-E presents peptides derived from leader sequences of classical HLA-I molecules and is a well-described ligand for the CD94/NKG2A and -C co-receptor complex expressed on natural killer (NK) cells. This way, NK cells scan for cells with dysregulated antigen presentation machineries such as tumor cells (5–7). In addition, HLA-E can present peptides with diverse residue compositions from pathogens and tumors to T cell receptors (TCR) (8–16). We showed recently that two HLA-E/Mtb peptides were recognized by multiple different TCR sequences within and between TB infected individuals. Moreover, HLA-E/Mtb restricted CD8+ T cells were detected in the circulation of individuals with Mtb infection (TBI) and active TB (aTB) with or without HIV and revealed an unorthodox phenotype reflected by the secretion of T-helper 2 associated cytokines (e.g., IL-4 and IL-13) (16). They could exert functional responses in vitro, such as controlling Mtb growth in infected macrophages (17). Memory phenotyping on HLA-E/Mtb CD8+ T cells previously revealed an effector memory and terminally differentiated phenotype, particularly in HIV/Mtb co-infected individuals (16). This induction of functional effector memory MHC-E CD8+ T cells was also observed in rhesus macaques that were vaccinated with a RhCMV-derived vector that encodes multiple Mtb antigens (18). Chronic antigen exposure may drive memory T cells into exhaustion, which is characterized by increased expression of exhaustion markers, such as KLRG-1, CTLA-4 and PD-1 (19). Signaling via these receptors impairs proliferation and functional capacities. In chronically Mtb-exposed individuals, such as in individuals with aTB and after BCG vaccination in healthy individuals, expression of exhaustion markers on CD4+ and CD8+ T cells has been reported, but whether this plays a role for HLA-E restricted T cells is currently unknown (20–22).
Recently, we identified several novel HLA-E/Mtb epitopes using an optimized peptide binding algorithm for HLA-E. These peptides, when bound to HLA-E, were recognized by a higher frequency of circulating CD8+ T cells in individuals with TBI compared to the previously identified and lower affinity HLA-E/Mtb epitopes (9). Moreover, we also demonstrated that not only CD8+ but surprisingly also CD4+ T cells could recognize these novel HLA-E/Mtb peptides, both in rhesus macaques (RM) and in humans, whereas control HLA-E/CMV complexes were only recognized by CD8+ T cells (23). In addition, we also showed that HLA-E/Mtb restricted CD4+ and CD8+ T cell frequencies remained unchanged in humans and RM after receiving BCG via intradermal or aerosol administration, similar to what was shown previously for other DURT subsets in BCG revaccinated adults (24). However, the frequency was significantly increased after Mtb challenge in unvaccinated RM (23). These novel findings suggest that HLA-E/Mtb restricted CD4+ and CD8+ T cells can be induced by Mtb infection but poorly upon BCG vaccination and encouraged us to elucidate the phenotype of HLA-E restricted T cells in humans with various stages of TB, including upon HIV co-infection.
The recent development of spectral flow cytometry has facilitated detailed characterization of immune phenotypes even when cell numbers are limited, as it enables detection of more than thirty markers simultaneously in single samples. We have recently optimized a high-dimensional flow cytometry panel to characterize the antigen specific immune phenotype using tetramers (TM) in combination with cell surface markers (25). Here, we applied this strategy to explore HLA-E/Mtb restricted T cell subsets in individuals with aTB or TBI and HIV, including in individuals with TBI, recognizing four newly identified HLA-E/Mtb epitopes (9). We show that, despite differences in cohorts, various CD4+ and CD8+ T cell memory subsets can recognize the HLA-E/Mtb epitopes. These memory HLA-E/Mtb CD4+ and CD8+ T cells expressed similar levels of markers associated with exhaustion as the total T cell population. Our study provides novel insights into the phenotype of HLA-E/Mtb specific T cell subsets circulating in individuals with various stages of TB infection and contributes to the exploration of HLA-E as a vaccine target.
Materials and methods
PBMC samples
Bio-banked peripheral blood mononuclear cell (PBMC) samples from individuals with TBI (n=40), individuals with TBI and HIV co-infection (TBI HIV+) (n=48) and individuals with active TB and HIV co-infection (aTB HIV+) (n=14) were used in the current study (Supplementary Tables 1-3 for overview of study participants). Individuals with TBI, aged between 12 and 18 years, were part of the South African Adolescent Cohort Study (ACS) performed among high school students in Worcester, Western Cape Province, by the South African Tuberculosis Vaccine Initiative (SATVI, South Africa). In the ACS, TBI was measured by Quantiferon TB Gold in-tube assay. Adolescents were not tested for HIV due to ethical constraints, but the prevalence of HIV among adolescents with TB is extremely low and would be expected to be even lower in healthy young individuals, as was also reported previously (26). Although not tested, TBI adolescents were therefore considered HIV negative.
Individuals with TBI or aTB and HIV co-infection, aged between 21 and 63 years, were recruited at the Ubuntu Clinic, Site B, Khayelitsha (Cape Town, South Africa) between March 2017 and December 2018. Those in the aTB group all tested sputum Xpert MTB/RIF (Xpert, Cepheid, Sunnyvale, CA, USA) positive and had clinical symptoms and/or radiographic evidence of TB. All aTB individuals were drug sensitive and had received no more than one dose of anti-Tuberculosis treatment (ATT) at the time of baseline blood sampling. The TBI individuals were all asymptomatic, had a positive IFN-γ release assay (IGRA, QuantiFERON -TB Gold In-Tube, Qiagen, Hilden, Germany), tested sputum Xpert MTB/RIF negative and exhibited no clinical evidence of aTB. Sputum Xpert MTB/RIF, sputum culture for Mtb, CD4 count, HIV VL tests were performed by the South African National Health Laboratory Services. Treatment with anti-retroviral therapy (ART) was self-reported and is shown in Supplementary Tables 2 and 3. Because ART was self-reported and therefore likely not reliable, we considered individuals with less than 1000 HIV copies/mL as HIV suppressed and individuals with more than 1000 HIV copies/mL as HIV unsuppressed (Supplementary Tables 2, 3).
Written informed consent was obtained from each adult individual. Written informed assent was provided by adolescent participants and written informed consent was provided by their parents or legal guardians. The study protocols in aTB and TBI individuals with HIV were approved by the University of Cape Town Human Research Ethics Committee (HREC 050/2015) and was conducted under DMID protocol no. 15-0047. The study protocols for the ACS were approved by the Human Research Ethics Committee of the University of Cape Town. PBMC samples were shipped in a liquid nitrogen dry shipper to the LUMC, The Netherlands for flow cytometry staining and acquisition.
HLA-E/Mtb restricted peptide selection and HLA-E*01:03 tetramer folding
Mtb-derived HLA-E (MTBHLAE) restricted peptides were identified previously using an optimized predictive peptide binding algorithm for HLA-E and were folded into HLA-E*01:03 monomers for tetramerization (9). Correct folding of the monomers was confirmed with HPLC and staining on LILRB1 expressing cells, as described previously (27). Twelve MTBHLAE peptides were evaluated for CD3+ T cell recognition in individuals with TBI or TBI and HIV to select four peptides with the highest recognition (Supplementary Table 4). These twelve peptides have also been evaluated before in individuals with TBI (9). Similar to that study, the four peptides with the highest recognition were MTBHLAE_31 (VLPAKLILM), MTBHLAE_93 (RLEAVVMLL), MTBHLAE_34 (LLPIKIPLI) and MTBHLAE_63 (ILAFEAPEL) and were therefore selected for the phenotyping panels (Supplementary Figure 1). The four HLA-E*01:03 TM were divided into two separate staining pools as evaluation of single TM was not possible due to limited material availability. Pool 1 consisted of MTBHLAE_31 and MTBHLAE_93 and pool 2 consisted of MTBHLAE_34 and MTBHLAE_63. Staining with the high binding affinity HLA-E Mtb-restricted peptide 44 (RLPAKAPLL) was included as negative control as this peptide is poorly recognized in human PBMC. The HLA-E binding peptide from CMV (VLAPRTLLL) was included as comparator.
Flow cytometry staining and acquisition
For the peptide selection, approximately 0.5 x 106 PBMC per individual with TBI or TBI and HIV were stained with nine cell surface markers in combination with one of the twelve HLA-E/Mtb TM shown in Supplementary Table 4 and samples were acquired on a BD LSRFortessa (BD Biosciences).
For in depth immune phenotyping analysis, a 30-colour panel was used for PBMC of individuals with: TBI (n=20), TBI and HIV (n=17) and aTB and HIV (n=14) (Supplementary Table 5). Due to availability problems with Pacific Orange CD20 after staining 12 individuals (5 individuals with TBI, 4 individuals with TBI and HIV and 3 individuals with aTB and HIV), this marker was changed to BV570 for the remaining 39 individuals. Only the samples stained with anti-CD20 BV570 were included in the total and HLA-E/Mtb specific immune subset analysis as combining samples stained with anti-CD20 BV570 or Pacific Orange would introduce significant compensation problems. To characterize the exhaustion profile of HLA-E/Mtb TM+ cells, a 20-colour panel was developed for PBMC of individuals with TBI (n=20) and TBI and HIV (n=31) (Supplementary Table 6). In both panels, 2 x 106 PBMC per individual were used for staining with the two HLA-E/Mtb TM pools and 0.5x106 PBMC per individual for staining with the two control HLA-E TM (p44; RLPAKAPLL and pCMV; VLAPRTLLL). Samples were acquired on a 5 laser Cytek®Aurora (Cytek Biosciences).
For all measurements, PBMC were first stained with LIVE/DEAD™ Fixable Violet or Blue (Invitrogen), depending on the panel, according to manufacturer’s instructions and blocked with 10 μg/mL purified CD94 monoclonal antibody (Clone HP-3D9, BD Biosciences) in phosphate buffered saline (PBS; Fresenius Kabi Nederland B.V) for 30 min at room temperature (RT) in the dark. PBMC were then washed (5 min, 450 x g) with PBS containing 0.1% bovine serum albumin (BSA; Roche Diagnostics GmbH) (PBS/0.1% BSA), blocked with 5% pooled normal human serum (Sigma-Aldrich) in PBS for 10 min at RT to block Fc receptors, preventing non-specific binding, and washed once with PBS/0.1% BSA. PBMC were subsequently incubated with a cocktail containing True-Stain Monocyte Blocker™ (1:20, BioLegend) to block non-specific binding of tandem-dyes, Brilliant Stain Buffer Plus (1:10, BD Biosciences) and chemokine receptor antibodies in PBS/0.1% BSA for 30 min at 37°C in the dark. After incubation, PBMC were washed once with PBS/0.1% BSA. For all samples, PBMC were incubated with HLA-E TM (5.4 µg/ml per TM) in PBS/0.1% BSA for 30 min at 37°C in the dark. PBMC were then washed, fixated with 1% paraformaldehyde (Pharmacy LUMC) for 10 min at RT and washed once more with PBS/0.1% BSA. PBMC were stained with a cocktail containing Brilliant Stain Buffer Plus (1:10) and the surface antibodies shown in Supplementary Tables 4-6 in PBS/0.1% BSA for 30 min at 4°C in the dark. After incubation, PBMC were washed twice with PBS/0.1% BSA, fixated with 1% paraformaldehyde for 10 min at RT, washed once and resuspended in PBS/0.1% BSA before acquiring. Full gating strategies to determine HLA-E T cell frequencies and the immune phenotype are shown in Supplementary Figures 2 and 5.
Data analysis of flow cytometry data
Flow cytometry data was analyzed using FlowJo v10.8.0 and OMIQ analysis software (www.omiq.ai). With OMIQ, data was first manually gated to remove debris, aggregates and dead cells. On the live single cells, a UMAP following FlowSOM clustering was performed to visualize and cluster the data. In the total immune profile analysis, subsampling was performed on 35087 events per sample (this was the lowest number of maximal events in one of the samples), and clusters with >100 events per sample were included for visualization. In the HLA-E/Mtb specific immune profile analysis, subsampling was done on a total of 5000 HLA-E TM+ events/cells per TB disease group (with a range of 55 to 285 events per sample). All markers except for LIVE/DEAD™, HLA-E TM, CD14, CD19 and CD20 were used in the UMAP settings and the resulting T cell parameters were included to cluster the data by FlowSOM. Only HLA-E TM+ T cells with more than 44 events were evaluated for exhaustion marker expression. This cut-off was based on the mean HLA-E CD3+ p44 counts plus twice the standard deviation and was used to exclude data skewing by including samples with too few TM counts.
Statistical analysis
The statistical tests used were the Friedman Test, Multiple Mann-Whitney test, Spearman’s rank correlation and EdgeR. Statistical analysis was performed in GraphPad Prism v10.2.3 and OMIQ analysis software. The Figure legends describe the specific test used for each comparison.
Results
HIV co-infection changes the distribution of CD4+ and CD8+ T cells in individuals with TB infection
Twenty-five common and rare immune cell subsets were identified in the circulation of individuals with TBI and HIV, aTB and HIV or TBI using FlowSOM clustering on the total live PBMC population (Figure 1A). HIV incidence among TBI adolescents is in general low and we therefore considered these individuals as HIV negative (26). Subsets were manually annotated based on the marker expression in each cluster (Supplementary Figure 3). Density plots of the UMAPs revealed that individuals with HIV had fewer CD4+ T cells (blue clusters in Figure 1A) compared to CD8+ T cell subsets (red clusters in Figure 1A) relative to individuals with TBI. Albeit our cohorts were not from the same population, these differences are consistent with what can be expected based on the pathophysiology of HIV (Figure 1B). Individuals with TBI had significantly more natural killer (NK) cells, naïve CD4+ and CD8+ T cells, TCRγδ T cells expressing NKG2A, CD4- CD8- T cells and classical monocytes and less terminally differentiated (EMRA) CD4+ and CD8+ T cells, transitional memory (TM) CD8+ T cells, NKT cells and γδ T cells than individuals with TBI and concomitant HIV (Figures 1C, D). Comparison between the individuals with HIV showed that individuals with TBI had significantly more EMRA, effector memory (EM) and central memory (CM) CD8+ and CD4+ T cells and fewer CD56high NK cells and intermediate monocytes than individuals with aTB (Figures 1E, F). Consequently, individuals with TBI and HIV, aTB and HIV or TBI have different immune profiles, with an altered distribution of CD4+ and CD8+ T cells when co-infected with HIV, although we cannot fully exclude that the different demographics between individuals with HIV and TBI might also contribute to this difference.
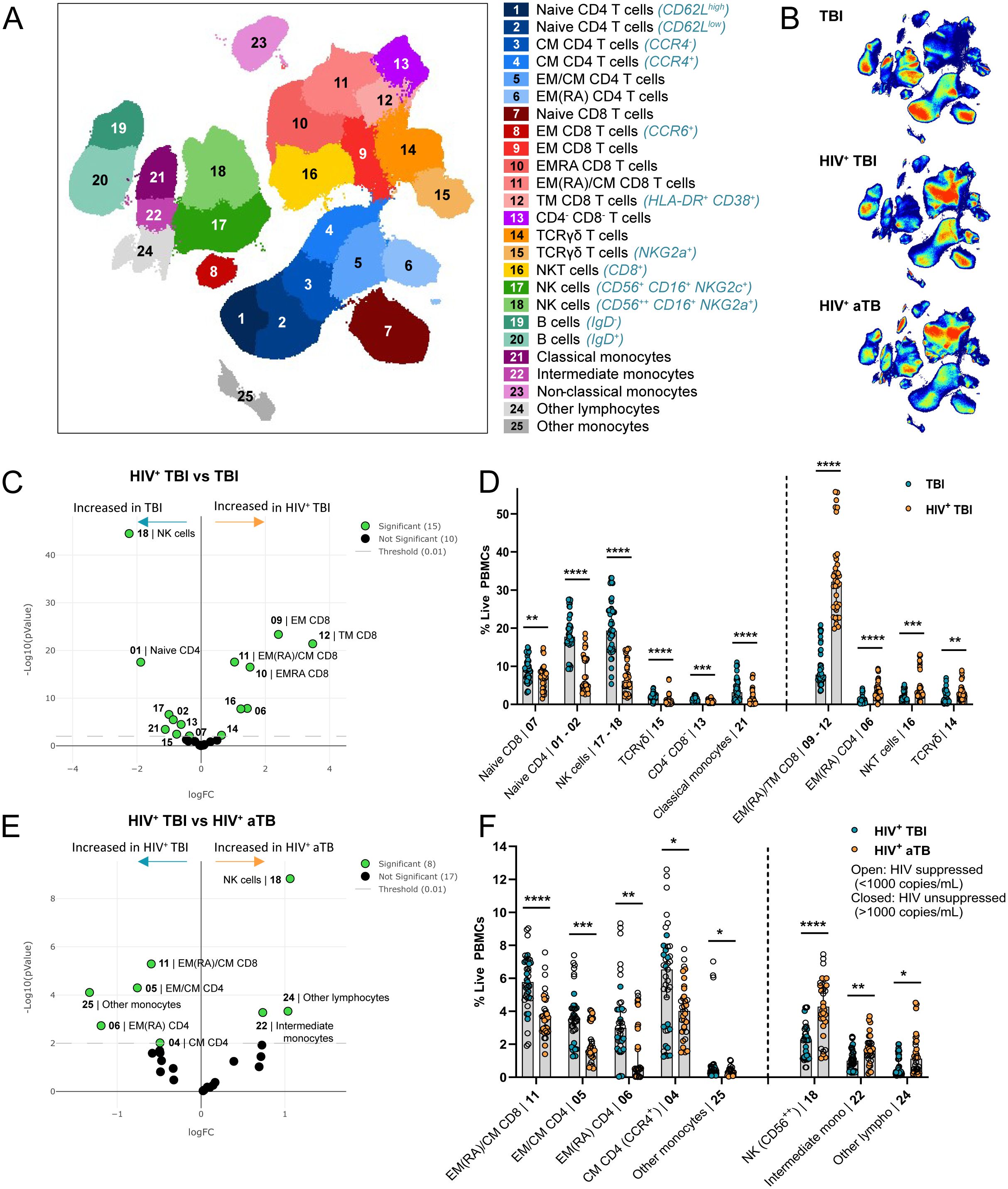
Figure 1. Individuals with TBI and HIV (n=13), aTB and HIV (n=11) or TBI (n=15) have differently distributed immune profiles. (A) UMAP visualization of 25 immune subsets identified using FlowSOM clustering on the total live PBMC population in all individuals combined. (B) UMAP density plots in individuals with TBI (top), TBI and HIV (middle) and aTB and HIV (bottom). (C) Volcano plot showing subsets that are significantly different between individuals with TBI and individuals with TBI and HIV in green and non-significant subsets in black. Significance was determined with EdgeR (p<0.01). (D) Bar graphs showing the frequency of live PBMC (Y-axis) in the subsets that are significantly different (X-axis) between individuals with TBI (blue circles) or TBI and HIV (orange circles). Bars show the median frequency and error bars the 95% confidence interval in each subset. Significance was tested using Multiple Mann-Whitney tests with multiple comparison correction (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). (E) Same as (C), comparing individuals with TBI or aTB with HIV. (F) Same as (D), comparing individuals with TBI and HIV (blue circles) and aTB and HIV (orange circles).
HLA-E/Mtb restricted peptides are recognized by both CD4+ and CD8+ T cells
Twelve HLA-E/Mtb restricted peptides were evaluated for CD3+, CD4+ and CD8+ T cell binding in individuals with TBI and HIV or TBI. Four HLA-E restricted Mtb-derived peptides with the highest frequency of T cells in all individuals were selected for in-depth phenotypic analysis in individuals with TBI and HIV, aTB and HIV or TBI (Supplementary Figure 1A). These four peptides also displayed the highest recognition (among the similar set of peptides) in individuals with TBI previously, showing consistency (9). The HLA-E/Mtb peptides were divided into two pools while the high affinity binding HLA-E restricted Mtb-derived peptide 44 (p44) was separately included as a (negative) control. Representative density plots to determine HLA-E/Mtb restricted T cell frequencies for the Mtb pools and p44 are shown in Figure 2A. Both HLA-E/Mtb peptide pools were recognized at a significantly higher frequency of CD3+, CD4+ and CD8+ T cells compared to p44 (median T cell frequency circa 0.03% for p44 and more than 0.1% for both Mtb pools) (Figure 2B). Pool 2 was recognized at a significantly higher frequency of CD4+ T cells compared to pool 1, whereas pool 1 and 2 were recognized at a comparable frequency of CD3+ and CD8+ T cells (Figure 2B). CD4+ T cell recognition of HLA-E/Mtb peptides was further significantly higher than for CD8+ T cells within individuals with TBI (Figure 2C). T cell recognition of the HLA-E/Mtb peptides in general was comparable between all cohorts, despite different demographics between cohorts (Figure 2C). Recognition by CD4+ and CD8+ T cells strongly correlated, suggesting that both CD4+ and CD8+ T cells can recognize HLA-E/Mtb peptides, similar to our previous findings in BCG exposed individuals and RM (Figure 2D) (23). Altogether, CD4+ and CD8+ T cells can recognize HLA-E/Mtb peptides with comparable frequencies in individuals with TBI and HIV, aTB and HIV or TBI.
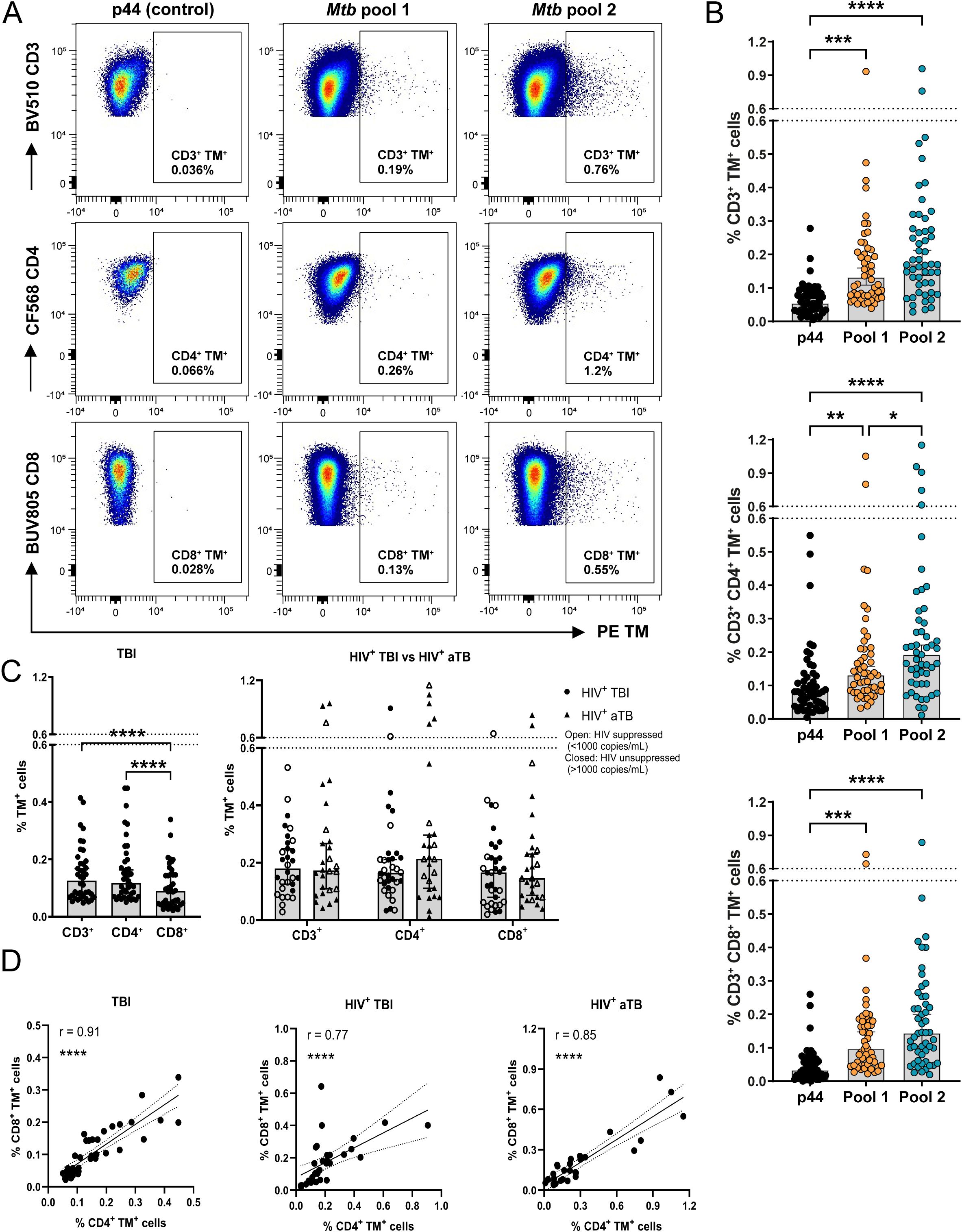
Figure 2. HLA-E/Mtb CD3+, CD4+ and CD8+ T cells are present in the circulation of individuals with TBI and HIV (n=17), aTB with HIV (n=14) or TBI (n=20). (A) Density plots for HLA-E p44, Mtb pool 1 and 2 CD3+, CD4+ and CD8+ T cells in one representative individual with aTB and HIV. HLA-E TM (PE) is shown on the X-axis and CD3, CD4 and CD8 on the Y-axis (B) Frequency of HLA-E p44, Mtb pool 1 and Mtb pool 2 CD3+ (top), CD4+ (middle) and CD8+ (bottom) T cells in the circulation of all individuals combined. Bars represent the median frequency and error bars the 95% confidence interval. Significance was tested using the Friedman test with multiple comparison correction (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). (C) Frequency of HLA-E CD3+, CD4+ and CD8+ T cells recognizing Mtb pool 1 and 2 in individuals with TBI (left) and in individuals with TBI or aTB and HIV (right). Bars represent the median frequency and error bars the 95% confidence interval. Significance was tested using the Friedman test (left) and Multiple Mann-Whitney test (right) with multiple comparison correction (****p <0.0001). (D) Correlation plots showing the frequency of HLA-E CD4+ T cells on the X-axis and CD8+ T cells on the Y-axis recognizing Mtb pool 1 and 2 in individuals with TBI (left), TBI and HIV (middle) or aTB and HIV (right). Significance was tested with Spearman’s rank correlation (****p < 0.0001).
HLA-E/Mtb specific CD4+ and CD8+ T cells have a memory phenotype in individuals with TBI and HIV, aTB and HIV or TBI
HLA-E/Mtb specific immune subsets were manually annotated based on the marker expression in each cluster (Supplementary Figure 4A). They comprised multiple CD4+ and CD8+ T cell memory subsets (i.e., EM, CM, Transitional Memory, EMRA and naïve T cells, possibly including stem cell memory T cells), γδ T cells, CD3+CD4-CD8- T cells and NK cells irrespective of HIV or TB infection or disease (Figure 3A).
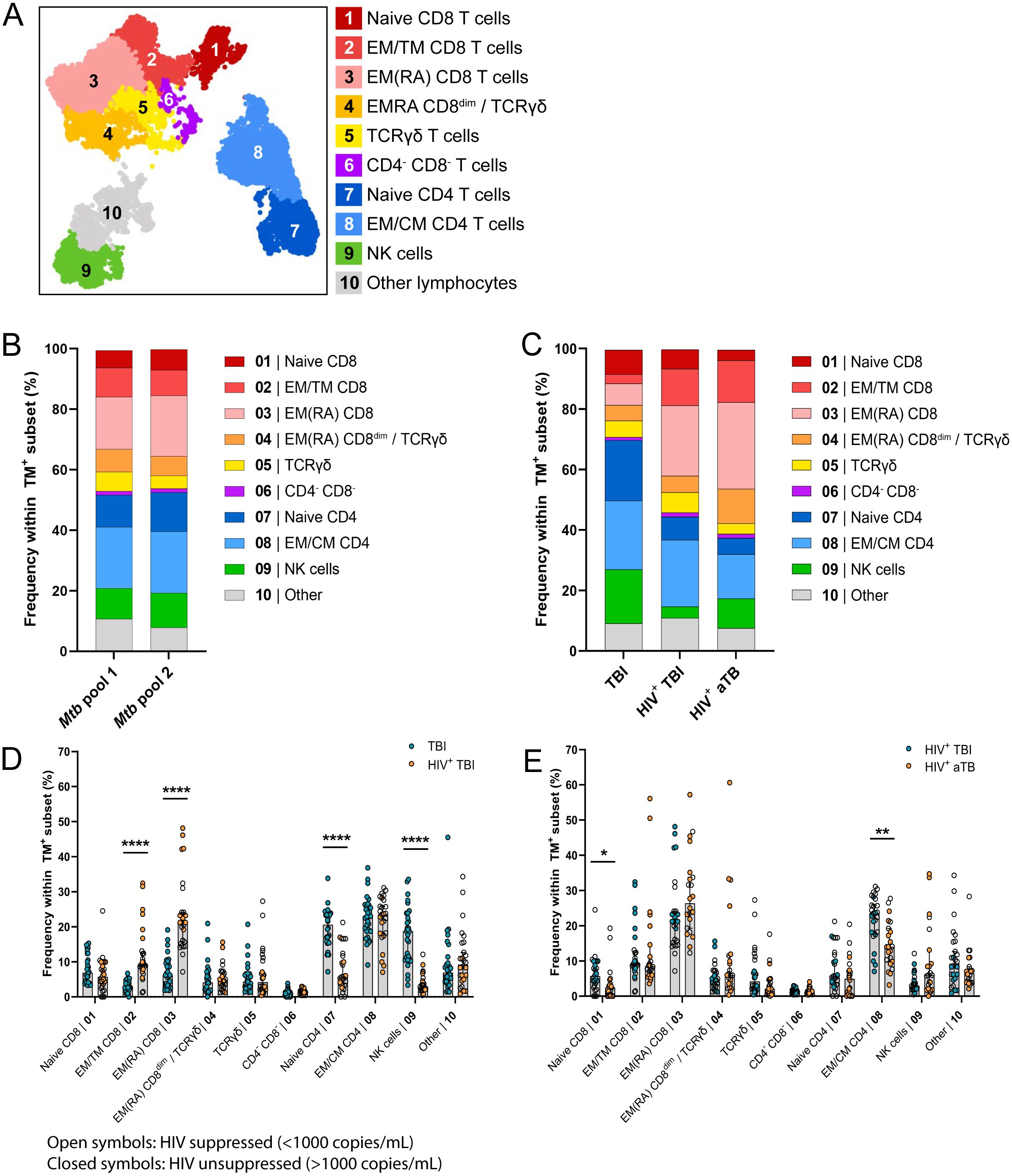
Figure 3. HLA-E/Mtb specific immune profile mainly consists of memory CD4+ and CD8+ T cells in individuals with TBI and HIV (n=13), aTB and HIV (n=11) or TBI (n=15). (A) UMAP visualization of 10 subsets identified using FlowSOM clustering on the total live HLA-E TM+ population recognizing Mtb pool 1 and pool 2 in all individuals combined. (B) Stacked bar graphs showing the mean frequency of HLA-E TM+ cells in each subset, separated for Mtb pool 1 and 2, all individuals combined (n=39). (C) Stacked bar graphs showing the mean frequency of HLA-E TM+ cells in each subset, separated per cohort. (D) Bar graphs showing the frequency of HLA-E TM+ cells recognizing Mtb pool 1 and 2 (Y-axis) in the 10 HLA-E/Mtb specific subsets (X-axis), separated for individuals with TBI (blue circles) or TBI and HIV (orange circles). Bars show the median frequency and error bars the 95% confidence interval. Significance was tested using Multiple Mann-Whitney tests with multiple comparison correction. *p < 0.05; **p < 0.01; ****p < 0.0001. (E) Same as (D), comparing individuals with TBI and HIV (blue circles) and aTB and HIV (orange circles).
The distribution of HLA-E/Mtb specific immune subsets was similar for both HLA-E/Mtb peptide pools and events were therefore combined for further analysis (Figure 3B, Supplementary Figure 4D). The HLA-E/Mtb specific cell population predominantly consisted of CD4+ and CD8+ T cell memory subsets in all individuals with TBI or aTB (Figure 3C). HIV co-infection was the most dominant factor changing the distribution of CD4+ and CD8+ immune subsets as individuals with TBI and HIV had significantly more EM and EMRA CD8+ T cells and fewer naïve CD4+ T cells compared to individuals with TBI (Figure 3D, Supplementary Figure 4B), possibly due to HIV infection of CD4+ T cells, although individuals from these cohorts had different demographics. TB infection or disease had limited effects on the distribution of HLA-E/Mtb specific T cells, as the HLA-E/Mtb specific immune profile was almost identical between individuals with TBI or aTB with HIV (Figure 3E, Supplementary Figure 4C). Thus, despite the different demographics between cohorts, still very similar phenotypes were observed, describing HLA-E/Mtb T cells with a mixed memory phenotype.
HLA-E/Mtb specific and total T cells have a comparable exhaustion marker expression in individuals with TBI and HIV or TBI
As HLA-E/Mtb restricted T cells have a memory phenotype, we evaluated the expression of the exhaustion markers KLRG1, PD-1 and 2B4 on an independent group of individuals with TBI or TBI and HIV. These markers are known to be expressed at the cell surface of memory T cells and provide information on the potential functionality of these memory HLA-E/Mtb T cells in individuals with TBI. Dual HLA-E TM staining was performed to enable optimal detection of low frequency circulating HLA-E restricted T cells for the HLA-E/Mtb peptides. The high affinity HLA-E restricted peptide from the viral UL40 protein of CMV (VLAPRTLL) was included as control for HLA-E/Mtb restricted T cell frequencies and exhaustion marker expression. Of note, the frequency of HLA-E T cells as determined by dual and single HLA-E TM staining was similar (Supplementary Figure 5, orange box).
Figure 4A shows the expression of KLRG1, PD-1 and 2B4 on total and HLA-E/Mtb specific CD4+ and CD8+ T cells in one representative individual with TBI. Circa 40% of CD4+ T cells and 60% of CD8+ T cells expressed exhaustion markers with profound differences between these two subsets. Roughly 30% of CD4+ T cells expressed PD-1 as a single marker, whereas >30% of CD8+ T cells co-expressed KLRG1 and 2B4 and >10% co-expressed PD-1, KLRG1 and 2B4 simultaneously, both in individuals with TBI and HIV or TBI (Figures 4B, C). This co-expression of exhaustion markers was minimally observed on CD4+ T cells. The exhaustion marker signature was similar between total T cells, HLA-E/Mtb specific or HLA-E/CMV specific T cells. As expected, HLA-E/Mtb CD4+ and CD8+ T cells expressing exhaustion markers had a memory phenotype (Supplementary Figure 6). Altogether, HLA-E/Mtb T cells have a comparable exhaustion marker expression as the total T cell population.
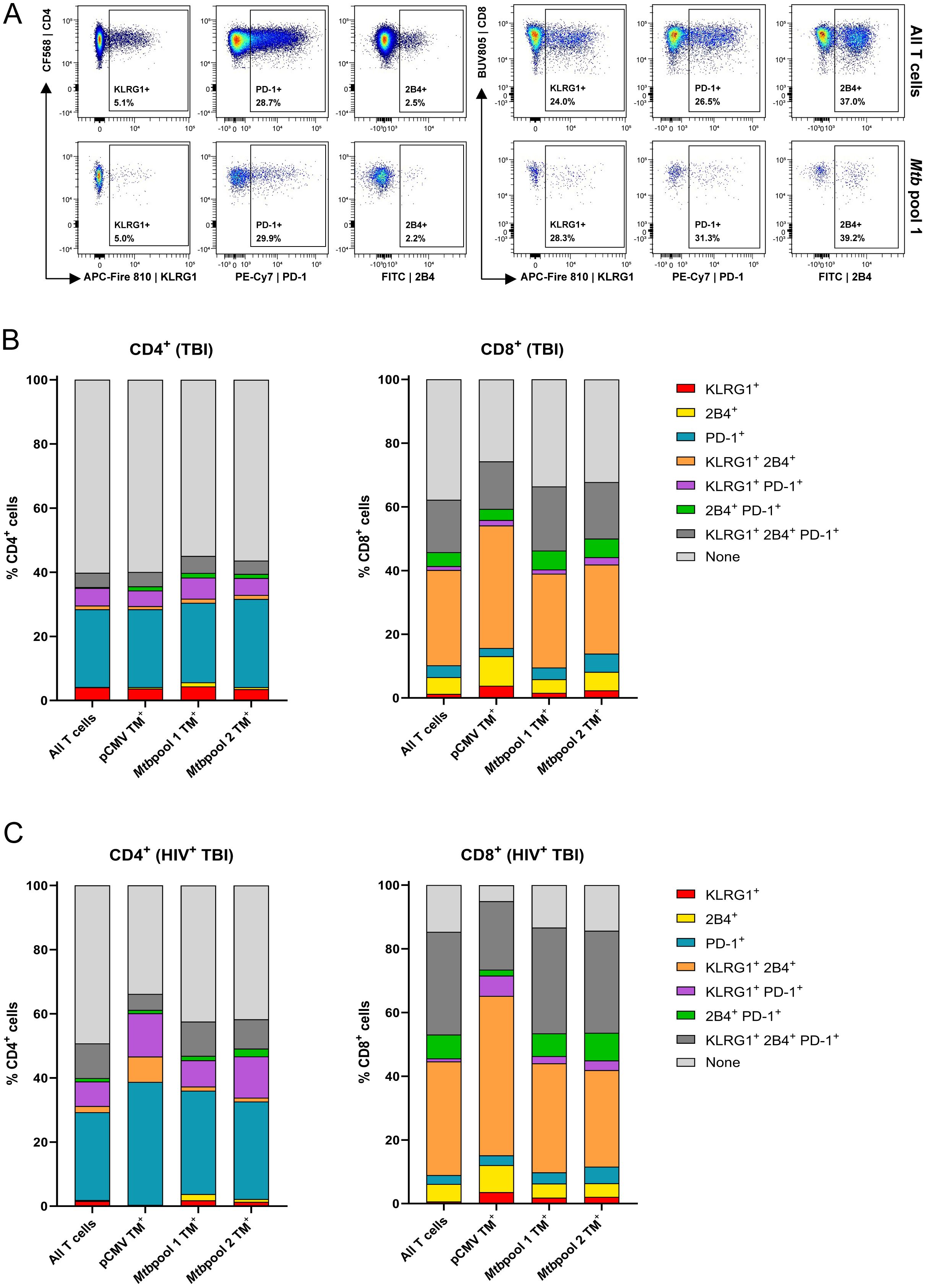
Figure 4. The exhaustion markers KLRG1, PD-1 and 2B4 are similarly expressed on HLA-E and the total T cells with differential expression between CD4+ and CD8+ T cells. (A) Density plots showing the expression of KLRG1, PD-1 and 2B4 on CD4+ (left) and CD8+ (right) T cells, comparing the total T cell population (top) and T cells recognizing HLA-E/Mtb pool 1 (bottom). Data is from one representative individual with TBI. (B) Stacked bar graphs showing the (co-)expression of KLRG1, 2B4 and PD-1 on HLA-E T cells recognizing pCMV (n=13), Mtb pool 1 (n=20) and 2 (n=19), compared to the total T cell population, in individuals with TBI. Stacks represent the mean expression frequency within CD4+ (left) and CD8+ (right) T cells. (C) Same as (B), for individuals with TBI and HIV for Mtb pool 1 and 2 (n=27) and pCMV (n=7).
Discussion
We performed in-depth phenotyping on circulating HLA-E/Mtb specific and total T cells in individuals with various stages of TB infection and demonstrate that HIV co-infection is the main driver changing the distribution of CD4+ and CD8+ T cell memory subsets. In addition, we show that HLA-E restricted Mtb specific T cells have a mixed memory phenotype in all individuals.
HLA-E/Mtb specific cells are composed of memory CD4+ and CD8+ T cells, TCRγδ T cells and NK cells. Individuals with TBI and HIV had significantly more EM and EMRA CD8+ T cells and fewer NK cells compared to individuals with TBI, in both the HLA-E/Mtb restricted and total immune population. A lower frequency of NK cells and impaired functional NK cell responses was also observed previously in individuals with TBI and HIV compared to individuals with TBI only, pointing to a possible mechanism of HIV to prevent clearance of infected cells (28). The overall percentage of CD8+ T cells was higher in individuals with HIV and TB co-infection relative to individuals with TB infection in both total and HLA-E/Mtb restricted T cell subsets. This shift is most likely caused by HIV-mediated depletion of CD4+ T cells, which may skew the distribution towards more CD8+ T cells in individuals with HIV, even though individuals co-infected with HIV received anti-retroviral treatment and expression of HLA-E is not affected by HIV-infection (29). Expression of PD-1, KLRG1 and 2B4 was comparable between circulating HLA-E restricted T cells and total T cells in individuals with TBI or with TBI and HIV. These results suggest that HLA-E restricted T cells were not more exhausted than total T cells. Additional functional experiments, such as intracellular cytokine staining or proliferation assays, should be performed to substantiate the functionality of these HLA-E restricted T cells. In contrast to individuals with TBI and HIV or TBI, the HLA-E/Mtb specific immune profile revealed limited differences between individuals with TBI or aTB with HIV, suggesting that TB infection rather than disease has the strongest effect on the distribution of HLA-E/Mtb specific subsets.
The identification of NK cells in the HLA-E/Mtb restricted immune profile was most likely not mediated by CD94/NKG2A or -C as CD94 was blocked and expression of NKG2A and -C in the NK cell cluster was low. Although limited expression of TCRαβ in the NK cell subset, T cells that express NK cell markers could be partly responsible for identifying NK cells in the HLA-E/Mtb restricted immune profile. Alternatively, the interaction of NK cells with HLA-E/Mtb complexes might be mediated by other or unknown receptors. A possible candidate could be LILRB1, which is known to interact with the β2M subunit of HLA-E and is expressed on NK cells (30). TCRγδ T cells were also identified in the HLA-E/Mtb restricted immune profile. TCRγδ itself could be responsible for interaction with HLA-E/Mtb complexes as we showed previously that TCRγδ was expressed on 1 out of 13 HLA-E/Mtb restricted CD8+ T cell clones (17).
We observed differential expression of PD-1, KLRG1 and 2B4 on CD4+ and CD8+ T cells. The majority of CD4+ T cells expressed PD-1 as a single marker, whereas CD8+ T cells co-expressed exhaustion markers. Increased expression of PD-1 on CD4+ T cells relative to CD8+ T cells was also found previously in individuals with aTB (31). Several studies demonstrated that blockade of PD-1 increased the percentage of CD107a+ CD3+ T cells and the proliferation of CD4+ T cells in the circulation of individuals with aTB. PD-1 blockade also reduced BCG growth in infected monocyte derived macrophages from individuals with aTB (31, 32). However, treatment with PD-1 antagonists in TB infected individuals is under debate as a recent study in Mtb-infected RM demonstrated that anti-PD-1 treated RM had larger lung granulomas, increased bacterial loads and decreased CD4+ T cell functional responses in the granuloma compared to untreated RM (33). Also, it was reported in a meta-analysis of multiple studies that PD-1 and PD-L1 blockade treatment significantly increased the risk of TB reactivation in individuals with TBI (34). As targeting exhaustion markers in TB infected individuals can have severe consequences, it would be interesting to determine the effect on vaccine induced immunity in TB naïve individuals.
HLA-E/Mtb T cell frequencies identified by dual and single HLA-E TM staining were similar, which underscores that single HLA-E TM can be included in panels with restricted fluorochrome options. Dual HLA-E TM staining is however preferred as it facilitates detection of lower circulating HLA-E T cell frequencies and facilitates discrimination between TM positive and negative populations. Furthermore, we demonstrate that the frequency of HLA-E/Mtb CD4+ and CD8+ T cells was comparable between individuals with TBI, TBI and HIV or aTB and HIV. In contrast, we showed previously that Italian individuals with aTB and HIV had the highest frequencies of HLA-E/Mtb restricted CD8+ T cells compared to individuals with TBI without HIV (9, 16). This discrepancy may result from difference in group size (i.e., five Italian individuals with aTB and HIV compared to twenty South African individuals with aTB and HIV in the current study), environmental or geographical factors, levels of exposure to TB, prior BCG vaccination or genetic backgrounds.
The phenotype of HLA-E/Mtb specific T cells and frequencies were evaluated in the circulation, but we showed previously that HLA-E/Mtb T cell frequencies were higher in the bronchoalveolar lavage (BAL) fluid, thus the local infection site, compared to the circulation in RM and healthy humans receiving BCG and/or Mtb challenge (23). This increase might be due to migration of HLA-E/Mtb T cells to the primary infection site. Trafficking of CD4+ T cells to the lung parenchyma was also shown before in Mtb-infected mice (35, 36). It would be interesting to study if similar increases of HLA-E/Mtb T cells in the BAL occurs in TB infected individuals as well, although this might be ethically challenging, including a comparison of the HLA-E/Mtb specific immune profile between the local site and the circulation.
HLA-E restricted Mtb peptides are recognized by both CD4+ and CD8+ T cells, shown here and in our previous study (23). This dual recognition raises questions about the thymic selection of HLA-E T cells, which might depend on the peptide sequence as high-affinity leader sequence derived peptides and the identical peptide mimic from CMV are only recognized by CD8+ T cells. We encourage future work to investigate the selection process in more detail.
One of the limitations of our study is that the analyses were performed on samples from a single time point per individual and only from South African individuals. Samples from different cohorts were included to investigate and compare individuals with TBI and HIV co-infection to individuals with TBI. The order of infection with HIV and TB in these individuals is unknown as is information on Mtb and HIV exposure and bacterial/viral load. Moreover, individuals with TBI or with TBI and HIV were recruited at different sites within South Africa and had different ages, hampering direct comparison of HIV status, however, the phenotypes observed were very similar despite differences in the populations investigated. Moreover, the frequency of HLA-E/Mtb T cells for enumeration and validation of the HLA-E/Mtb restricted immune profiles was low, unsurprisingly, and could be increased by including more HLA-E/Mtb peptides. Besides this, we could not compare absolute HLA-E/Mtb T cell counts as we only stained PBMC samples and not whole blood samples. Another limitation is that additional exhaustion markers, such as LAG-3, TIM3, CD57 and TOX-1, could have been included for a more extensive analysis of the exhaustion marker profile. Finally, functional assessment of the HLA-E/Mtb restricted T cells was not possible due to limited material availability. However, we previously performed extensive characterization on the function of HLA-E/Mtb restricted CD8+ T cells in individuals with TB infection or aTB. These studies showed that HLA-E/Mtb restricted CD8+ T cells have an unorthodox phenotype relative to classical Mtb restricted T cells, characterized by the secretion of T-helper 2 cytokines (i.e., IL-4 and IL-13) and the exertion of cytolytic or suppressive responses (16, 17). Additionally, in another study we showed that sorted HLA-E/Mtb TM+ CD8+ T cells in individuals from the same TB infection cohort as investigated in this study have a diverse TCR repertoire with limited clonal expansions, unexpected for a DURT member (8). Transduction of these TCRs confirmed the functionality of the identified HLA-E/Mtb TM+ CD8+ TCRs. Thus, although not assessed in this study, HLA-E/Mtb restricted T cells derived from individuals with TB can exert functional responses.
Taken together, we show that HLA-E/Mtb specific CD8+ and CD4+ T cells have memory phenotypes and express similar levels of exhaustion markers as total T cells. We therefore highly encourage further exploration of HLA-E as a vaccination target against TB.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. FSC files are deposited in FlowRepository (flowrepository.org) with ID number: FR-FCM-Z8MN. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by University of Cape Town Human Research Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
LV: Data curation, Formal Analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. CR: Conceptualization, Data curation, Investigation, Methodology, Project administration, Supervision, Validation, Writing – review & editing. TS: Data curation, Investigation, Methodology, Project administration, Supervision, Validation, Writing – review & editing. MW: Data curation, Formal Analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. KM: Methodology, Resources, Writing – original draft, Writing – review & editing. KF: Resources, Writing – review & editing. RW: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing – review & editing. TO: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. SJ: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI141315) to TO and SJ. RW is supported by the Francis Crick Institute which is funded by Wellcome (CC2112), Cancer Research UK (CC2112) and UK Research and Innovation (CC2112). He also receives support from Wellcome (203135, 226817), NIH (R01AI161013) and in part NIHR Biomedical Research Centre of Imperial College NHS Trust. For the purposes of open access the authors have applied a CC-BY public copyright to any author-accepted manuscript arising from this submission.
Acknowledgments
We thank our colleagues from the Flow Core Facility of the LUMC for their assistance and help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1505329/full#supplementary-material
References
2. Ruibal P, Voogd L, Joosten SA, Ottenhoff THM. The role of donor-unrestricted T-cells, innate lymphoid cells, and NK cells in anti-mycobacterial immunity. Immunol Rev. (2021) 301:30–47. doi: 10.1111/imr.12948
3. Kanevskiy L, Erokhina S, Kobyzeva P, Streltsova M, Sapozhnikov A, Kovalenko E. Dimorphism of HLA-E and its disease association. Int J Mol Sci. (2019) 20:5496. doi: 10.3390/ijms20215496
4. Strong RK, Holmes MA, Li P, Braun L, Lee N, Geraghty DE. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem. (2003) 278:5082–90. doi: 10.1074/jbc.M208268200
5. Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci U.S.A. (1998) 95:5199–204. doi: 10.1073/pnas.95.9.5199
6. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. (1998) 391:795–9. doi: 10.1038/35869
7. Voogd L, Ruibal P, Ottenhoff THM, Joosten SA. Antigen presentation by MHC-E: a putative target for vaccination? Trends Immunol. (2022) 43:355–65. doi: 10.1016/j.it.2022.03.002
8. Voogd L, Drittij A, Dingenouts CKE, Franken K, Unen VV, van Meijgaarden KE, et al. Mtb HLA-E-tetramer-sorted CD8(+) T cells have a diverse TCR repertoire. iScience. (2024) 27:109233. doi: 10.1016/j.isci.2024.109233
9. Ruibal P, Franken K, van Meijgaarden KE, van Wolfswinkel M, Derksen I, Scheeren FA, et al. Identification of HLA-E Binding Mycobacterium tuberculosis-Derived Epitopes through Improved Prediction Models. J Immunol. (2022) 209:1555–65. doi: 10.4049/jimmunol.2200122
10. Joosten SA, van Meijgaarden KE, van Weeren PC, Kazi F, Geluk A, Savage ND, et al. Mycobacterium tuberculosis peptides presented by HLA-E molecules are targets for human CD8 T-cells with cytotoxic as well as regulatory activity. PloS Pathog. (2010) 6:e1000782. doi: 10.1371/journal.ppat.1000782
11. Salerno-Goncalves R, Fernandez-Vina M, Lewinsohn DM, Sztein MB. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol. (2004) 173:5852–62. doi: 10.4049/jimmunol.173.9.5852
12. Yang H, Rei M, Brackenridge S, Brenna E, Sun H, Abdulhaqq S, et al. HLA-E-restricted, Gag-specific CD8(+) T cells can suppress HIV-1 infection, offering vaccine opportunities. Sci Immunol. (2021) 6:eabg1703. doi: 10.1126/sciimmunol.abg1703
13. Vietzen H, Furlano PL, Cornelissen JJ, Bohmig GA, Jaksch P, Puchhammer-Stockl E. HLA-E-restricted immune responses are crucial for the control of EBV infections and the prevention of PTLD. Blood. (2023) 141:1560–73. doi: 10.1182/blood.2022017650
14. Doorduijn EM, Sluijter M, Marijt KA, Querido BJ, van der Burg SH, van Hall T. T cells specific for a TAP-independent self-peptide remain naive in tumor-bearing mice and are fully exploitable for therapy. Oncoimmunology. (2018) 7:e1382793. doi: 10.1080/2162402X.2017.1382793
15. Doorduijn EM, Sluijter M, Querido BJ, Seidel UJE, Oliveira CC, van der Burg SH, et al. Cells engaging the conserved MHC class ib molecule qa-1(b) with TAP-independent peptides are semi-invariant lymphocytes. Front Immunol. (2018) 9:60. doi: 10.3389/fimmu.2018.00060
16. Prezzemolo T, van Meijgaarden KE, Franken K, Caccamo N, Dieli F, Ottenhoff THM, et al. Detailed characterization of human Mycobacterium tuberculosis specific HLA-E restricted CD8(+) T cells. Eur J Immunol. (2018) 48:293–305. doi: 10.1002/eji.201747184
17. van Meijgaarden KE, Haks MC, Caccamo N, Dieli F, Ottenhoff TH, Joosten SA. Human CD8+ T-cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA-E have an unorthodox Th2-like, multifunctional, Mtb inhibitory phenotype and represent a novel human T-cell subset. PloS Pathog. (2015) 11:e1004671. doi: 10.1371/journal.ppat.1004671
18. Hansen SG, Zak DE, Xu G, Ford JC, Marshall EE, Malouli D, et al. Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat Med. (2018) 24:130–43. doi: 10.1038/nm.4473
20. Boer MC, van Meijgaarden KE, Goletti D, Vanini V, Prins C, Ottenhoff TH, et al. KLRG1 and PD-1 expression are increased on T-cells following tuberculosis-treatment and identify cells with different proliferative capacities in BCG-vaccinated adults. Tuberculosis (Edinb). (2016) 97:163–71. doi: 10.1016/j.tube.2015.11.008
21. Flores-Gonzalez J, Ramon-Luing LA, Falfan-Valencia R, Batista CVF, Soto-Alvarez S, Huerta-Nunez L, et al. The presence of cytotoxic CD4 and exhausted-like CD8+ T-cells is a signature of active tuberculosis. Biochim Biophys Acta Mol Basis Dis. (2024) 1870:167219. doi: 10.1016/j.bbadis.2024.167219
22. Russell SL, Lamprecht DA, Mandizvo T, Jones TT, Naidoo V, Addicott KW, et al. Compromised metabolic reprogramming is an early indicator of CD8(+) T cell dysfunction during chronic. Mycobacterium tuberculosis Infection. Cell Rep. (2019) 29:3564–3579.e3565. doi: 10.1016/j.celrep.2019.11.034
23. Voogd L, van Wolfswinkel M, Satti I, White AD, Dijkman K, Gela A, et al. Mtb specific HLA-E restricted T cells are induced during Mtb infection but not after BCG administration in non-human primates and humans. Vaccines (Basel). (2024) 12:1129. doi: 10.3390/vaccines12101129
24. Gela A, Murphy M, Rodo M, Hadley K, Hanekom WA, Boom WH, et al. Effects of BCG vaccination on donor unrestricted T cells in two prospective cohort studies. EBioMedicine. (2022) 76:103839. doi: 10.1016/j.ebiom.2022.103839
25. van Wolfswinkel M, van Meijgaarden KE, Ottenhoff THM, Niewold P, Joosten SA. Extensive flow cytometric immunophenotyping of human PBMC incorporating detection of chemokine receptors, cytokines and tetramers. Cytometry A. (2023) 103:600–10. doi: 10.1002/cyto.a.24727
26. Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. (2016) 387:2312–22. doi: 10.1016/S0140-6736(15)01316-1
27. Ruibal P, Franken K, van Meijgaarden KE, Walters LC, McMichael AJ, Gillespie GM, et al. Discovery of HLA-E-presented epitopes: MHC-E/peptide binding and T-cell recognition. Methods Mol Biol. (2022) 2574:15–30. doi: 10.1007/978-1-0716-2712-9_2
28. Devalraju KP, Neela VSK, Krovvidi SS, Vankayalapati R, Valluri VL. Defective expansion and function of memory like natural killer cells in HIV+ individuals with latent tuberculosis infection. PloS One. (2021) 16:e0257185. doi: 10.1371/journal.pone.0257185
29. Nattermann J, Nischalke HD, Hofmeister V, Kupfer B, Ahlenstiel G, Feldmann G, et al. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir Ther. (2005) 10:95–107. doi: 10.1177/135965350501000107
30. Ruibal P, Derksen I, van Wolfswinkel M, Voogd L, Franken K, El Hebieshy AF, et al. Thermal-exchange HLA-E multimers reveal specificity in HLA-E and NKG2A/CD94 complex interactions. Immunology. (2023) 168:526–37. doi: 10.1111/imm.13591
31. Shen L, Gao Y, Liu Y, Zhang B, Liu Q, Wu J, et al. PD-1/PD-L pathway inhibits M.tb-specific CD4(+) T-cell functions and phagocytosis of macrophages in active tuberculosis. Sci Rep. (2016) 6:38362. doi: 10.1038/srep38362
32. Jurado JO, Alvarez IB, Pasquinelli V, Martinez GJ, Quiroga MF, Abbate E, et al. Programmed death (PD)-1:PD-ligand 1/PD-ligand 2 pathway inhibits T cell effector functions during human tuberculosis. J Immunol. (2008) 181:116–25. doi: 10.4049/jimmunol.181.1.116
33. Kauffman KD, Sakai S, Lora NE, Namasivayam S, Baker PJ, Kamenyeva O, et al. PD-1 blockade exacerbates Mycobacterium tuberculosis infection in rhesus macaques. Sci Immunol. (2021) 6:eabf3861. doi: 10.1126/sciimmunol.abf3861
34. Liu K, Wang D, Yao C, Qiao M, Li Q, Ren W, et al. Increased tuberculosis incidence due to immunotherapy based on PD-1 and PD-L1 blockade: A systematic review and meta-analysis. Front Immunol. (2022) 13:727220. doi: 10.3389/fimmu.2022.727220
35. Sakai S, Kauffman KD, Schenkel JM, McBerry CC, Mayer-Barber KD, Masopust D, et al. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J Immunol. (2014) 192:2965–9. doi: 10.4049/jimmunol.1400019
Keywords: Tuberculosis, HLA-E, T cells, immunophenotyping, spectral flow cytometry
Citation: Voogd L, Riou C, Scriba TJ, van Wolfswinkel M, van Meijgaarden KE, Franken KLMC, Wilkinson RJ, Ottenhoff THM and Joosten SA (2024) HLA-E/Mtb specific CD4+ and CD8+ T cells have a memory phenotype in individuals with TB infection. Front. Immunol. 15:1505329. doi: 10.3389/fimmu.2024.1505329
Received: 02 October 2024; Accepted: 06 December 2024;
Published: 23 December 2024.
Edited by:
Won Fen Wong, University of Malaya, MalaysiaReviewed by:
Salah Mansour, University of Southampton, United KingdomShaobin Shang, Yangzhou University, China
Copyright © 2024 Voogd, Riou, Scriba, van Wolfswinkel, van Meijgaarden, Franken, Wilkinson, Ottenhoff and Joosten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simone A. Joosten, cy5hLmpvb3N0ZW5AbHVtYy5ubA==
 Linda Voogd
Linda Voogd Catherine Riou
Catherine Riou Thomas J. Scriba
Thomas J. Scriba Marjolein van Wolfswinkel1
Marjolein van Wolfswinkel1 Krista E. van Meijgaarden
Krista E. van Meijgaarden Kees L. M. C. Franken
Kees L. M. C. Franken Robert J. Wilkinson
Robert J. Wilkinson Tom H. M. Ottenhoff
Tom H. M. Ottenhoff Simone A. Joosten
Simone A. Joosten