- 1Shanxi Key Laboratory of Stem Cells for Immunological Dermatosis, Institute of Dermatology, Taiyuan Central Hospital, Taiyuan, China
- 2State Key Breeding Laboratory of Stem Cells for Immunological Dermatosis, Institute of Dermatology, Taiyuan Central Hospital, Taiyuan, China
Serine protease inhibitors (Serpins) are a protein superfamily of protease inhibitors that are thought to play a role in the regulation of inflammation, immunity, tumorigenesis, coagulation, blood pressure and cancer metastasis. Serpins is enriched in the skin and play a vital role in modulating the epidermal barrier and maintaining skin homeostasis. Psoriasis is a chronic inflammatory immune-mediated skin disease. At present, most serpins focus on the pathogenesis of psoriasis vulgaris. Only a small number, such as the mutation of SerpinA1/A3/B3, are involved in the pathogenesis of GPP. SerpinA12 and SerpinG1 are significantly elevated in the serum of patients with psoriatic arthritis, but their specific mechanism of action in psoriatic arthritis has not been reported. Some Serpins, including SerpinA12, SerpinB2/B3/B7, play multiple roles in skin barrier function and pathogenesis of psoriasis. The decrease in the expression of SerpinA12, SerpinB7 deficiency and increase in expression of SerpinB3/4 in the skin can promote inflammation and poor differentiation of keratinocyte, with damaged skin barrier. Pso p27, derived from SerpinB3/B4, is an autoantigen that can enhance immune response in psoriasis. SerpinB2 plays a role in maintaining epidermal barrier integrity and inhibiting keratinocyte proliferation. Here we briefly introduce the structure, functional characteristics, expression and distribution of serpins in skin and focus on the regulation of serpins in the epidermal barrier function and the pathogenic role of serpins in psoriasis.
1 Background
Serine protease inhibitors (Serpins) is a new family proposed by Hunt et al. (1, 2) in 1980 on the basis of the conserved primary structure of ovalbumin, α1 antitrypsin, and human antithrombin. Many members of this family can inhibit the activity of serine proteases such as chymotrypsin, so they are named serine protease inhibitors (Serpins) (3, 4). Currently, approximately 1500 serpin sequences have been identified in animals, poxvirus, plants, bacteria and archaea (5, 6). The Serpins family contains a group of structurally similar but functionally distinct proteins, making it the largest and most functionally diverse family of protease inhibitors. Serpins are involved in inflammation, immunity, tumorigenesis, blood coagulation, blood pressure and cancer metastasis (7).
Psoriasis is a chronic, recurrent and inflammatory immune-mediated skin disease induced by genetic and environmental interaction (8). Psoriasis is characterized by excessive proliferation and poor differentiation of keratinocytes, immune cell infiltration, skin inflammation as well as impaired epidermal barrier (8–10). According to the clinical characteristics of psoriasis, psoriasis can be divided into: psoriasis vulgaris, psoriasis arthropathica, psoriasis erythrodermic and psoriasis pustulosa (11), of which vulgaris is the most common type, and other types are mostly converted from psoriasis vulgaris (12, 13). Pustular psoriasis can be further divided into localized (e.g. palmoplantar pustulosis) and generalized pustular psoriasis (GPP) (14). In the skin, the epidermis is an abundant source of proteases and protease inhibitors (15). Endogenous and exogenous proteases, such as caspases, cathepsins, kallikreins and proteases from microorganisms, play important roles in the desquamation and defense regulation of the stratum corneum. Protease inhibitors contribute to skin integrity and protective barrier function by regulating their proteolytic activity (16). An imbalance of proteases/protease inhibitors in the skin can lead to changes in protease hydrolysis of target proteins such as filaginins, cytokines, and receptors, inducing skin inflammatory responses and barrier dysfunction (16). Serpins are the most diverse superfamily of protease inhibitors (1). Serpins are abundant in skin and play an important role in maintaining skin homeostasis. In recent years, a growing body of literature have shown that some serpins are differentially expressed genes in psoriasis lesions (17, 18), gene polymorphisms and mutations of some serpins are also involved in psoriasis pathogenesis (19, 20). These genetic changes in serpins lead to changes in their target protease activity and proteolysis of target proteins, which contribute to psoriatic inflammation and skin barrier dysfunction. Here we briefly introduce the structure, functional characteristics, expression and distribution of serpins in skin and focus on the regulation of serpins in the epidermal barrier and the pathogenic role of serpins in psoriasis.
2 Structural, functional characteristics of serpins in skin
Serpin is a single chain protein, consisting of 350 ~ 400 amino acids. The serpins have a core structure of about 380 amino acids, which folds into a very conservative, typical three-dimensional structure in a metastable state. This structure consists of 8 ~ 9 α helices (hA ~ hI) and 3 β sheets (A-β, B-β, C-β) (21) (Figure 1). The region of interaction with the target enzyme is a bare ring motif, also known as the reaction center ring (RCL) (Figure 1), which is spread on the serpin scaffold and contains the protease recognition site P1 (5). The Serpin protein superfamily is divided into groups called clades based on their sequence similarity. Clades are classified into A-P, and human serpins are phylogenetically classified into clades A-I (21, 22). In the human genome, there are 36 protein-coding genes of serpins (21) and the two largest clades of the 36 identified serpins are extracellular “clade A” (12 members located on chromosomes 14, and X) and intracellular “clade B” (13 members located on chromosomes 6 and 18) (22–24). Clade C (serpinC1) is located on chromosome 1. Clade D (serpinD1) is located on chromosome 22. Clade E (3 members) are located on chromosome 2, 7 and 13. Clade F (2 members) are located on chromosome 17. Clade G (serpinG1) and Clade H (serpinH1) are both located on chromosome 11. Clade I (2 members) are located on chromosome 3.
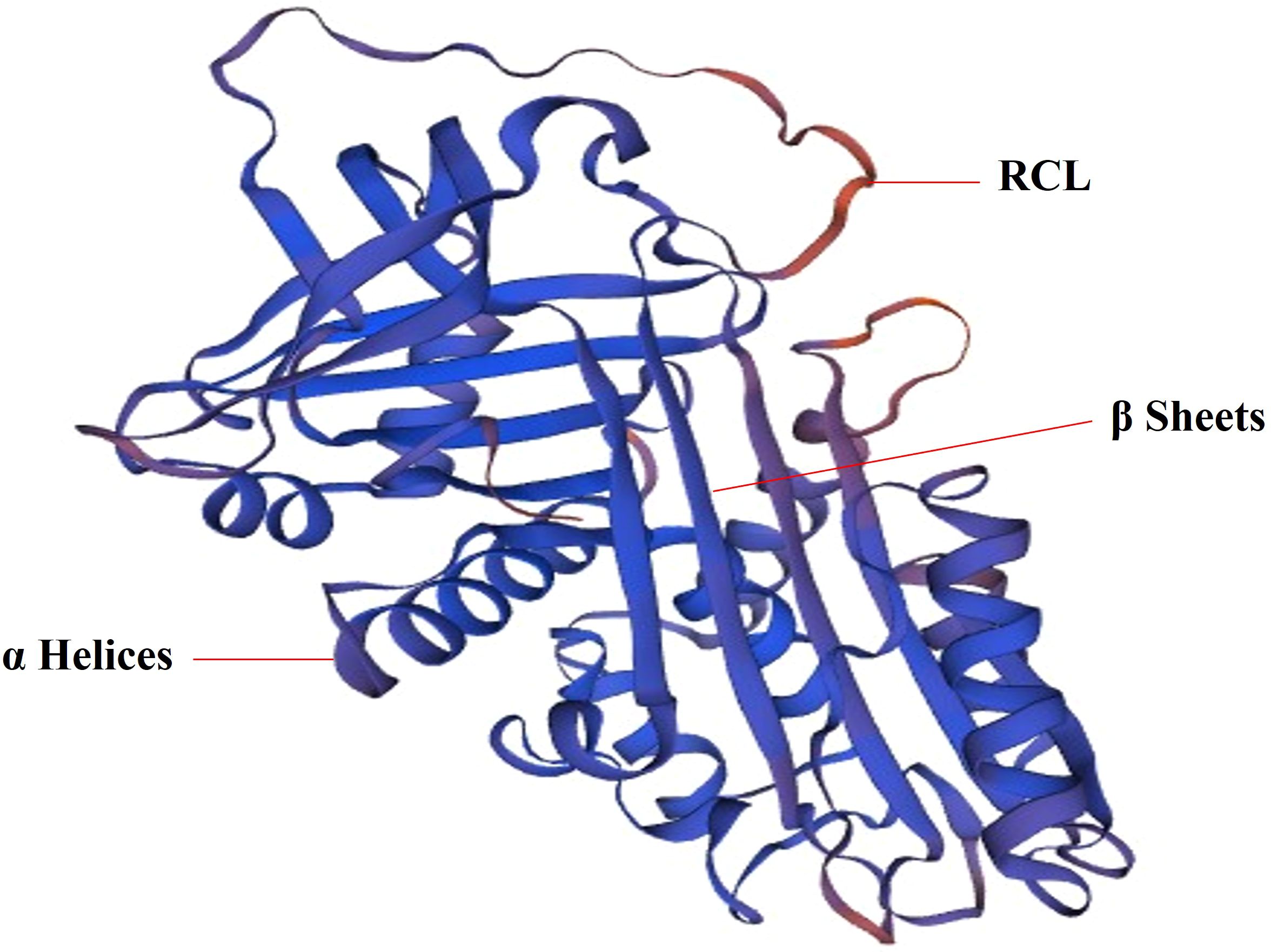
Figure 1. The 3D structure of SerpinA1. SERPINA1 with labeled structural elements: α helices, β sheet and reactive center loop (RCL).
Although Serpins have similar structural characteristics, their modes of action are very different. According to these characteristics, Serpin superfamily members are divided into inhibitory and non-inhibitory serpins. The inhibitory serpins can suppress the serine proteases, caspases and papain-like cysteine proteases by an irreversible suicidal mechanism (21, 25, 26) and we summarized the target enzymes inhibited by each human serpin member in Table 1. However, the non-inhibitory serpins exhibit functions unrelated to inhibiting catalytic activity, such as hormone transport or blood pressure regulation (5, 6, 21, 107). For example, SerpinA8 is a blood pressure regulator (108). SerpinA6 and A7 are responsible for the transport of cortisol (109) and thyroid hormone (46), respectively. In humans, most (24 out of 36) serpins are inhibitory (Table 1). The inhibitory serpins plays an important role both intracellular and extracellular, including coagulation regulation (110), inflammatory response, complement activation (102) and immune regulation. A number of serpins play vital roles in the inflammation and immunity. In Clade A, there are some inhibitory serpins containing anti-inflammatory molecules (SerpinA1, A3, A4, A12) (111–115) and non-inhibitory serpins containing inflammatory molecule(SerpinA6, 8) (20, 47, 109, 170). The inhibitory SerpinA9 is involved in maturation and maintenance of naïve B cells (50). In Clade B, there are some inhibitory serpins containing anti-inflammatory molecules (SerpinB1, B2, B6) (116, 117) and pro-inflammatory molecules (SerpinB3/B4, B10) (78, 118) (Table 1). Act as potent inhibitors of proteases in neutrophils, SerpinA1 (21), A3 (33, 34), B1 (116) and B6 (80) are thought to protect monocytes, neutrophils, and onlookers from ectopic neutrophil-derived proteases during inflammation (82, 119). SerpinB1 is a protective immunomodulatory that protects the host’s antimicrobial defenses and prevents tissue damage (61, 120). SerpinB9 is a potent inhibitor of granzyme B, which is located in the same subcellular compartment of cytotoxic lymphocytes as granzyme B, and can protect cytotoxic lymphocytes from the destruction of granzyme B (89). SerpinB9 also inhibits IL-1β maturation by inhibiting Caspase-1. Mutations in the SerpinB9 gene may contribute to the development of autoinflammatory diseases (121). Some serpins are important hemostatic regulators, including SerpinA5 (42), SerpinA10 (51), SerpinC1 (98), SerpinD1(heparin cofactor II) (122), SerpinE1(PAI1) (122), SerpinE2 (100) and SerpinF2 (122). SerpinC1 not only has anticoagulant and anti-inflammatory effects (98), but also has the effect of inhibiting liver cancer (123) (Table 1). Human Serpins also contain some tumor suppressor genes, including A5 (124), A11 (52), and B5 (125), which may inhibit tumor growth by inhibiting tumor metabolism.
Some serpins are commonly expressed in various human tissues and cells and some serpins are expressed specifically in certain tissues, which we have summarized in Table 1. Serpins are abundant in expression and distribution in the skin. In the Clade A, SerpinA12 is mainly expressed in the skin and keratinocytes in the skin has been identified as a rich source of vaspin (serpinA12) (55). AGT is an important part of the renin-angiotensin system (RAS). Steckelings UM et al. found that the complete renin-angiotensin system exists in human skin (48). The majority of serpins containing(B1-8, B12, B13) in Clade B are expressed in the skin (35). SerpinB2 is expressed in keratinocytes of the skin and one study suggested that SerpinB2 (cross-linked to the cornified envelope) was present in the stratum corneum (71). In skin, SerpinB3 is expressed in the spinous and granular layers of normal epithelium (126). In normal skin, SerpinB13 expression is mainly confined to the basal layer, while in diseased skin, it is mainly distributed in the outermost layers of granular and upper spinous layers (127). In other branches, SerpinE3 and SerpinG1 are also expressed in the skin (35).
3 Serpins regulate epidermal barrier and maintain skin homeostasis
The exposed epidermis is a laminated squamous epithelium composed of multiple layers of keratinocytes. Epidermis is the physical barrier of our human skin, protecting us from moisture loss and mechanical damage. It is complemented by a chemical barrier composed of antimicrobial peptides and proteins to protect the host from the surrounding microbiota (16). Profilaggrin metabolism, the formation of cornified envelope, desmosomes, intercellular lipid lamellae and zonula occludens, desquamation play important roles in the integrity of the epidermal barrier (128–130). The differentiation of keratinocytes affects the formation of the cornified envelope that is an insoluble protein and lipid structure with barrier functional properties (131, 132). As mentioned earlier, many members of the serpin family are expressed in the skin, which play a an important role in regulating epidermal barrier and maintain skin homeostasis. Reduced expression of SerpinA12 leads to decreased expression of the desmosomal proteins, the cornified envelope proteins, and keratins (55) that results in reduced impaired skin barrier. As a predominantly expressed adipokine in the skin (55, 115), SerpinA12 may alter its expression or functional activity by inhibiting kallikrein 7 (KLK7), which KLK7 controls desquamation and is a key molecule in the maintenance of skin barrier function (53, 133–135). Reduced expression of SerpinA12 in keratinocyte has been reported to decreased expression of the desmosomal proteins, cornified envelope proteins, and keratins (55) which are proteins that maintain the integrity of the epidermal barrier. The decrease of SerpinA12 expression may weaken the inhibition of KLK7 activity. Abnormal KLK activity impairs epidermal barrier function (136). So we hypothesized that reduced SerpinA12 expression may lead to impaired epidermal barrier by inhibiting KLK7 activity.
SerpinB2 is considered to be one of the precursors of the envelope and is involved in the formation of the cornified envelope (64, 71). SerpinB2 deficiency was found to cause stratum corneum defect and more susceptible to topical application of inflammatory agents. The function of SerpinB2 in keratinocytes is to protect the stratum corneum from proteolysis by inhibiting urokinase, thereby maintaining integrity of the stratum corneum and its barrier effect, especially during skin inflammation (71). Epidermis with high SerpinB3 expression level increase sensitivity to barrier destruction by external stimuli, suggesting that SerpinB3 plays an important role in inducing epidermal barrier disruption (137). As a molecule elevated in both psoriasis and atopic dermatitis patients, SerpinB3/4 has been shown to cause epidermal barrier dysfunction in experimental mouse models of atopic dermatitis (78). As a common target of SerpinB3 and SerpinB13, cathepsin L is the elusive enzyme that processes and activates transglutaminase 3(TGM3) (138). The TGM is involved in formation of cornified envelope in the epidermis and plays an important role in epidermal barrier (138). Both SerpinB3 and SerpinB13 are highly expressed in psoriasis lesions (18, 139). Psoriasis showed impaired epidermal barrier function (140). So we hypothesize that increased SerpinB3/B13 expression suppress TGM3 activation by inhibiting cathepsin L activity, thereby affecting cross-linking of the stratum corneum and resulting in impaired skin barrier. SerpinB7 deficiency can cause epidermal barrier dysfunction in IMQ-induced psoriasis like models (85). SerpinB12 is a gene associated with epidermal permeability barrier, and its expression is decreased in atopic dermatitis model mice (95). Impaired epidermal function is a significant feature of atopic dermatitis (78). However, it is unclear whether SerpinB12 is a key molecule and specific mechanism that regulates impaired epidermal barrier function in atopic dermatitis, and further research is needed.
4 Pathogenic roles of serpins in psoriasis
At present, the majority of serpins focus on the pathogenesis of psoriasis vulgaris. SerpinA12 and SerpinG1 is significantly elevated in the serum of patients with psoriatic arthritis (141, 142), but its specific mechanism of action in psoriatic arthritis has not been reported. Next, we summarized the pathogenic role of serpins in psoriasis.
4.1 Modulation of keratinocyte hyper-proliferation and differentiation
Although the clinical features of psoriasis differ in different psoriasis types, most types of psoriasis lesions are characterized by erythema, thickening, and scale (13). Thickening and scale are associated with excessive proliferation of keratinocytes (143). Parakeratosis, the persistence of nuclei in the stratum corneum, is one of the histopathological features of psoriasis (144) and is related to excessive proliferation and poor differentiation of keratinocytes (145). Poor keratinocyte differentiation also promotes impaired skin barrier function (146).
SerpinA12 expression is significantly reduced in the lesional skin of psoriasis patients (115). Reduced expression of SerpinA12 in psoriatic keratinocytes leads to decreased expression of differentiation-related genes (Loricrin, Involucin, Keratin1 and 10) (55), which cause poor differentiation. SerpinB2 and B3 are highly expressed in the epidermis of psoriatic patients (18, 117). SerpinB2 can inhibit the proliferation of human keratinocytes (62). SCCA1 is always found in parakeratotic epidermis (137), which suggests that it may be related to the proliferation and differentiation of keratinocytes. SerpinB7 and SerpinB13 are both keratinocyte differentiation regulators (147, 148). As mentioned earlier, SerpinB13 is highly expressed in psoriasis lesions (139). Overexpression of SerpoinB13 in keratinocytes can decrease UV-induced apoptosis by inhibiting cathepsin L (96). SerpinB7 has been identified as a skin-specific endogenous protease inhibitor and a novel psoriatic-associated gene that is highly expressed in keratinocytes of psoriatic patients and imiquimod-induced psoriatic lesions in mice (85, 149). However, one study found that SerpinB7 expression was down-regulated in psoriatic skin lesions, and its expression level is negatively correlated with the severity of the patient’s disease (150). Zheng H et al. (85) found that SerpinB7 deficiency down-regulates the expression of differentiation-related genes (Keratin 10, Loricrin, Filaggrin, Involucrin) and increased keratinocyte proliferation by decreasing the calcium ion influx (85). The expression of SerpinB7 in psoriatic lesions is controversial. Whether SerpinB7 has a positive or negative effect on the pathogenesis of psoriasis depends on its expression level, which needs to be further verified. SerpinB8 is a susceptibility gene for psoriasis (88), but the exact mechanism has not been studied. SerpinB8 inhibit its amidolytic activity by forming an SDS stable complex with furin (151). Furin is a prohormone convertase involved in inflammation, prohormone processing, and extracellular matrix remodeling (86). Furin is involved in the cleavage of the precursor protein profilaggrin into the monomer filaggrin, and filaggrin plays an important role in keratinocyte differentiation (130). So we hypothesized that SerpinB8 might be involved in keratinocyte differentiation by regulating furin activity, but this has not been studied.
4.2 Contribution to immune response and inflammation
Psoriasis vulgaris is a chronic inflammatory skin disease caused by the interaction of keratinocytes and immune cells (145), and is thought to involve self-perpetuating inflammatory mechanisms via the IL-23/Th17 axis (13). Munro microabscesses filled with neutrophils in psoriatic skin lesions are considered to be typical histopathological markers of psoriasis. Neutrophils communicate and interact with antigen-presenting cells and lymphocytes at the site of inflammation (152, 153). Circulating neutrophils are recruited to the site of inflammation after inflammatory signals or bacterial induction in skin, and are then activated to produce respiratory bursts that produce large amounts of ROS, degranulation and neutrophil extracellular trap(NET) production, which contributes to the pathogenesis of psoriasis (154). In patients with psoriasis, neutrophils are pre-activated and form a NET in psoriatic skin lesions (155, 156) NET promote keratinocyte to secrete high levels of proinflammatory factors (LCN2, IL-36γ, CXCL8, and CXCL1) by activating TLR4/IL-36R crossers and the downstream MyD88/NF-κB signaling pathway (156). These chemokines include LCN2, CXCL8, and CXCL1, which in turn promote neutrophil migration to the skin (154). NET chromatin in psoriatic plaques, together with the antimicrobial peptide LL-37 released by keratinocytes, often stimulates the secretion of inflammatory factors such as IL-12 and IL-23 by dendritic cells (157, 158), which then activates the differentiation of T cells into Th1, Th17 and Th22, and promotes them to secrete of TNF-α and IFN-γ, IL-17, IL-22, respectively (13). IL-17 secreted by Th17 cells is the main effector of psoriasis. IL-17 not only activates neutrophils but also promotes the proliferation of keratinocytes and the production of inflammatory chemokines(CXCL1, CXCL8, CCL20, TNF) by binding to IL-17 receptors. CCL20 and TNF are chemokines of T cells and dendritic cells, respectively (154), which in turn leads to a T cell immune response. IL-23 plays an significant role in the survival and proliferation of Th17 and Th22, and the Th17 pathway mediated by IL-23 is considered to be the main pathway in psoriasis (13). The release of LCN2 and MPO during neutrophil degranulation exacerbate skin inflammation in psoriasis by participating in the activation of neutrophils (154, 159). ROS produced during respiratory bursts activate mitogen-activated protein kinase (MAPK), nuclear factor-κB (NF-κB), or Janus kinase-signal transducer and activator of transcription proteins (JAK-STAT) (JAK-STAT) associated inflammatory pathways (160), which induce the proliferation and inflammation of keratinocytes (47, 161). In general, neutrophils play an important role in the initiation and maintenance of psoriasis through ROS production, degranulation and NET formation.
Like SerpinB2, B3, SerpinB1 is elevated in lesional skin of psoriasis patients (18, 67, 117, 162). SerpinB1 is also found to be abundant in the cytoplasm and granules of neutrophils (163). SerpinB1, a potent inhibitor of NE, has recently been shown to limit neutrophil extracellular trap(NET) production (58) and has also been found to limit the undesirable proliferation of lymphocytes with the Th17 phenotype (61),. The expression levels of SerpinB2 in the psoriasis-involved skin are positively correlated with the severity of psoriasis (67, 117). There is evidence that RNA silencing of SerpinB2 in keratinocytes leads to upregulation of IL-8, CXCL5 and CCL5 and increased neutrophil migration (117). Expressions of IL-8, CXCL5 and CCL5 are all increased in psoriatic lesions, where CXCL5 and IL-8 are neutrophil chemokines that drive neutrophils to the skin, and CCL5 is a chemokine of Th1 cells that produce IFNγ (68–70). IFNγ, in turn, stimulates the activation of dendritic cells, which triggers a T cell immune response (13). These studies suggest that SerpinB2 depletion is involved in the pathogenesis of psoriasis by increasing inflammatory chemokines to enhance the chemotaxis and immune response of immune cells. As an anti-inflammatory miRNAs, miR-146a/b can indirectly iinhibit SerpinB2 expression by targeting IL-1 receptor-associated kinase 1(IRAK1) and caspase recruitment domaincontaining protein 10(CARD10) in human primary keratinocytes (117). The expression of MiR-146a/b in psoriatic lesions was negatively correlated with the expression of SerpinB2. MiR-146a/b collaborates with serpinB2 in keratinocytes to regulate inflammation in psoriasis. These studies show that SerpinB1and B2 may play a negative regulatory role in the pathogenesis of psoriasis. The literature shows that pro-inflammatory cytokines secreted by infiltrating immune cells in the skin can induce SERPINB3 to be highly expressed in psoriatic skin (72). The SerpinB3/4-derived protein Pso p27, an autoantigen in psoriasis and other chronic inflammatory diseases, is only expressed in the psoriasis involved skin, not in uninvolved skin. In psoriasis, SerpinB3/4 are synthesized by skin fibroblasts and keratinocytes, and then taken up by mast cells to form Pso p27 through cleavage of chymase in mast cells (18). Pso p27 can bind to either IgG or complement factor C1q in the skin lesions of psoriasis patients to form and activate immune complexes (73, 74), which boosts the skin’s immune response. TEA domain family member 4 (TEAD4), as a transcription factor (75), is overexpressed in keratinocytes of psoriasis (76). Silencing of TEAD4 in keratinocytes leads to reduced expression of SerpinB3/4, which suggests TEAD4 can transcriptionally regulate the expression of SerpinB3/4 (76). Decreased expression of SerpinB3/4 inhibits the expression of immune cell chemokine (CXCL1,5,8) in keratinocytes (76). TEAD4 silencing in keratinocytes reduces T migration and the secretion of IL-17 and IL-22 in T cells, which in turn reduces the secretion of CXCL1,5,8 in keratinocytes (76). One Study have shown that SerpinB3/4 stimulates keratinocytes to produce inflammatory chemokines and promote CD4 +T cell migration by activating the NF-κB signaling pathway (77). These results suggest that TEAD4 may increase the crosstalk of keratinocyte and T cell by enhancing the expression of SerpinB3/4, thereby promoting the cytokine secretion of keratinocyte and T cell, and T cell migration. SerpinB5, as an autoantigen of an autoimmune response induced by streptococcus, is the target of an enhanced T-cell response in psoriasis (79).
SerpinB7 deficiency significantly increased the expression of chemokines (TNF-α, IL-1b, IL-23, CXCL2 and IFN-γ), neutrophil markers Ly6G, and antimicrobial peptide S100A8, thereby exacerbating skin inflammation (85). SerpinB7 deficiency promote the expression of inflammatory mediators by decreasing the calcium ion influx (85) However, as mentioned earlier, the expression of SerpinB7 in psoriatic lesions is reversed in different studies, and its regulation of inflammation depends on its expression level in psoriatic lesions, which needs to be confirmed by further research.
As mentioned earlier, SerpinA12 expression is significantly reduced in the lesional skin of psoriasis patients (115). Reduced expression of SerpinA12 in psoriatic keratinocytes leads to increased expression of the interferon-inducible (56, 57) and psoriasis related inflammatory genes (chemokine ligand 20, IL-6, IL-8, and S100 proteins) (55). Reduced expression of SerpinA12 in keratinocytes stimulated co-cultured dendritic cells, macrophages, monocytes, and neutrophils to secrete tumor necrosis factor-α, IL-1β, IL-6, IL-8, and monocyte chemotactic protein-1 (55). The reduced expression of SerpinA12 in human epidermis enhance the communication between keratinocytes and immune cells (55). Immunohistochemical studies have shown that the serine protease KLK7 and KLK14 is increased in psoriasis lesions compared to normal skin (164, 165). As the main target protease of SerpinA12, KLK7 controls the process of the activation of pro-inflammatory IL-1β and prochemerin, all of which are involved in the pathogenesis of psoriasis (133, 134, 166). KLK7 also controls the enzymatic processing of antimicrobial peptide precursors in the skin and regulates the function of antimicrobial peptides, which act as immunomodulators in psoriasis (167). Antimicrobial peptides, such as LL-37, proteins ADAMTSL5, K17, and hsp27, may act as autoantigens to promote differentiation of autoreactive lymphocytes and unleash chronic inflammatory responses (168). Uncontrolled activity of KLK7 can lead to psoriasis (169). SerpinA12 and KLK7 are co-located in the skin (134). Based on these results, we hypothesize that SerpinA12 inhibits KLK7 in normal skin, but decreased SerpinA12 expression in psoriatic lesions reduces the inhibition of KLK7 activity, thereby exacerbating the inflammatory response and immune response. But this requires further direct verification (Figure 2).
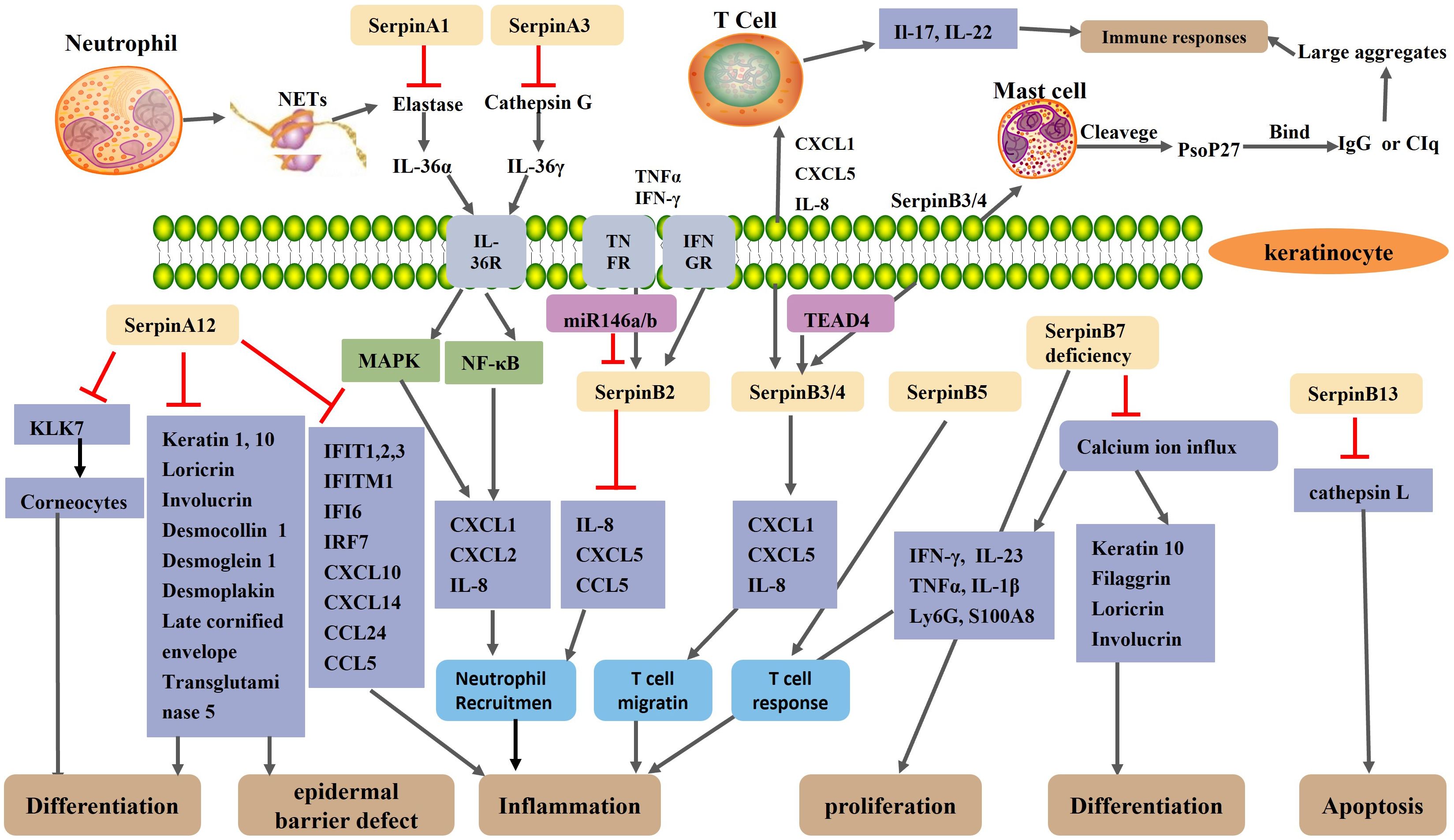
Figure 2. Schematic representation of the regulatory mechanism of Serpin protein in psoriasis. A neutrophil extracellular trap(NET) structure formed after neutrophil necrosis or apoptosis exposes neutrophil elastase and cathepsin G to the extracellular environment. SerpinA1 and SerpinA3 inhibit neutrophil elastase and cathepsin G, respectively. Normally, elastase and cathepsin G activate IL-36α and IL-36γ, respectively, thereby activating the MAPK and NF-κB signaling pathways of keratinocytes, resulting in increased expression of CXCL1, CXCL2, and IL-8 and increased recruitment of neutrophils. SerpinA12, as an anti-inflammatory factor, is reduced in the skin lesions of psoriasis patients. The reduction of SerpinA12 leads to an overactivation of KLK7 and thus an excessive loss of corneocytes, which impair epidermal barrier. Reduced expression of SerpinA12 in KC reduces keratinocyte differentiation and exacerbates inflammation by reducing the expression of differentiation-related genes and psoriasis related inflammatory gene. The expression of SerpinB2 in keratinocytes is up-regulated under the stimulation of the TNF-α and IFN-γ. As an anti-inflammatory miRNAs, MiR-146a/b collaborates with serpinB2 in keratinocytes to inhibit inflammation in psoriasis. SerpinB2 deficiency leads to upregulation of IL-8, CXCL5 and CCL5 and increased neutrophil migration. SerpinB3/B4 are taken up by mast cells to form Pso p27 through cleavage of chymoenzymes in mast cells. TEAD4 may modulate keratinocyte and T cell crosstalk by targeting SerpinB3/4, thereby affecting keratinocyte and T cell cytokine secretion and T cell migration. SerpinB5 is an autoantigen of an autoimmune response and is the target of an enhanced T-cell response in psoriasis. SerpinB7 deficiency significantly increased the expression of chemokines, neutrophil markers Ly6G, and antimicrobial peptide S100A8, thereby exacerbating skin inflammation. SerpinB7 deficiency inhibits keratinocyte differentiation and promotes the expression of inflammatory mediators by decreasing the calcium ion influx.
Transcriptome analysis showed that SerpinA8 (AGT) is one of the differentially expressed genes in mild psoriasis (17). As an inflammation gene, the gene polymorphism of SerpinA8 is associated with plaque psoriasis and a positive family history of psoriasis (20), however the specific mechanism of SerpinA8’s involvement in psoriasis has not been studied. SerpinA8 is the only precursor of all angiotensin peptides. As an important product of AGT, AngII not only plays an important role in the regulation of blood pressure, but also plays a dual role in the regulation of inflammation. AngII has the opposite effect by activating AT1R and AT2R, respectively. Ang II interacts with AT1R to play a pro-inflammatory role, while Ang II and AT2R play an anti-inflammatory role (47, 170). Preliminary studies have shown that keratinocytes have the ability to produce Ang II (and potentially other angiotensin) independently of the supply of systemic RAS components. Ang II can induce potent inflammation associated with IL-17 (47, 171). IL-17 is a major effector of psoriasis that activates the NF-κB signaling pathway (172). Ang II induces reactive oxygen species (ROS) production by stimulating NADPH oxidase (NOX) (47, 173). So we hypothesize that SerpinA8 gene polymorphisms induce inflammation and keratinocyte proliferation by regulating Ang II expression, and thus participate in the pathogenesis of psoriasis.
5 Involvement of serpins in generalized pustular psoriasis (GPP)
GPP is a severe form of psoriasis, which is characterized by large, visually visible pustules on the non-extremity skin, with or without systemic symptoms such as fever, neutrophilism, and elevated serum C-reactive protein levels (12).
Kantaputra P et al. (72) found that the skin of GPP patients with SerpinB3 mutation showed high expression of SerpinB3. The mutations in both SerpinA1 and SerpinA3 are likely to be predisposing risk factors of generalized pustular psoriasis (GPP) (19). A heterozygous missense mutation of SerpinA1 occurs in patients with GPP (19). SepinA1 can inhibit the activity of elastase in neutrophils (28), and it has been found that SepinA1 mutation can cause overactivity of elastase (29). The overactivity of neutrophil elastase can promote the activation of IL-36α, and the activated IL-36α can enhance the ability of keratinocytes to produce chemokines (30). The inhibitory capacity of SerpinA1 is reduced in symptom-free and in those with stationary lesions patients with psoriasis (31). Barszcz D et al. (31) suggested that the abnormal function of SerpinA1 may be related to the pathogenesis of psoriasis. Frey S, et al. found a rare loss of function mutation in the entire reactive center loop of SerpinA3 in a small number of patients with GPP (36, 37). Liu Y, et al. (38) identified four novel mutations in SerpinA3 in seventy children with GPP and demonstrated that three of these mutations lead to loss of function of ACT (the protein encoded by SerpinA3), thereby reduce the inhibition of ACT on Cathepsin G. Cathepsin G (a protease produced by infiltrating neutrophils in the epidermis) can activate IL-36γ in the epidermis of patients with GPP, and activated IL-36γ induces an increased ability of keratinocytes to produce chemokines (CXCL1, CXCL2 and CXCL8) and enhance recruitment of neutrophils, thereby exacerbating skin inflammation (30, 39). In the skin of GPP patients, the expression and activity of IL-36α and IL-36γ is elevated in keratinocytes located around neutrophil microabscesses (30). Therefore, we speculate that similar to SerpinA3, SerpinA1 is involved in the pathogenesis of GPP through loss-of function mutations, but further studies are needed to confirm it (Figure 2).
6 The potential therapeutic strategies targeting serpins in psoriasis
In summary, the changes in the expression of some serpins in the skin participate in the pathogenesis of psoriasis by regulating the inflammation, differentiation and interaction with immune cells of keratinocytes. When the expression of some serpins increases, their transcriptional regulators that inhibit serpin expression or corresponding antibodies can be developed to reduce the expression of serpins. When the expression of some serpins reduces, psoriasis can be treated with its recombinant protein. For example, in an animal model of psoriasis, the application of recombinant vaspin (SerpinA12) reduced infiltration by myeloid cells into the skin (55).
Mutations and polymorphisms of some serpins are also involved in the pathogenesis of psoriasis, and we can use their downstream product related antagonists to treat psoriasis. For example, losartan act as an angiotensin receptor (AT1R) antagonist, attenuates imiquimod-induced psoriasis-like inflammation (171).
7 Conclusions
Serpins play an complex role in regulation of the epidermal barrier and the development of psoriasis. Some serpins, including SerpinA12, SerpinB2/3//7 play multiple roles in skin barrier function and pathogenesis of psoriasis. The decrease in the expression of SerpinA12, SerpinB7 deficiency and increase in expression of SerpinB3/4 in the skin can promote inflammation, poor differentiation of keratinocyte and damaged skin barrier. Pso p27, derived from SerpinB3/B4, is an autoantigen that can enhanced immune response in psoriasis. However, whether SerpinB3 is participating in the pathogenesis of psoriasis by itself or by hydrolyzing into Pso p27 has not been investigated. The expression of SerpinB7 in psoriasis is controversial, which requires further studies to confirm. SerpinB2 plays a role in maintaining epidermal barrier integrity and inhibiting keratinocyte proliferation. Some anti-inflammatory serpins(SerpinB1, B2) are highly expressed in psoriasis, which may be the body’s response to the development of the disease. Some serpins(SerpinA1, A3, B3) cause GPP through a genetic mutation that triggers skin inflammation. AGT(SerpinA8) and SerpinB8 are susceptibility gene for psoriasis, but the specific mechanism has not been studied. Based on the relevant literatures, we summarized the possible mechanism of AGT and SerpinB8 to provide reference for future research. The underlying mechanisms of serpins have not been fully elucidated and needs to be further explored. The study of serpins in the pathogenesis of psoriasis may provide a novel therapeutic target for the treatment of psoriasis.
Author contributions
JW: Funding acquisition, Visualization, Writing – original draft, Writing – review & editing. JL: Investigation, Writing – review & editing. LZ: Visualization, Writing – review & editing. HH: Visualization, Writing – review & editing. KZ: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by the National Natural Science Foundation of China (grant no. 82273539) and the Nature Science Foundation of Shanxi Province (grant no.202403021212278).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Laskowski M Jr, Kato I. Protein inhibitors of proteinases. Annu Rev Biochem. (1980) 49:593–626. doi: 10.1146/annurev.bi.49.070180.003113
2. Hunt LT, Dayhoff MO. A surprising new protein superfamily containing ovalbumin, antithrombin-III, and alpha 1-proteinase inhibitor. Biochem Biophys Res Commun. (1980) 95:864–71. doi: 10.1016/0006-291X(80)90867-0
3. Whisstock JC, Silverman GA, Bird PI, Bottomley SP, Kaiserman D, Luke CJ, et al. Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions. J Biol Chem. (2010) 285:24307–12. doi: 10.1074/jbc.R110.141408
4. Silverman GA, Whisstock JC, Bottomley SP, Huntington JA, Kaiserman D, Luke CJ, et al. Serpins flex their muscle: I. Putting the clamps on proteolysis in diverse biological systems. J Biol Chem. (2010) 285:24299–305. doi: 10.1074/jbc.R110.112771
5. Heit C, Jackson BC, McAndrews M, Wright MW, Thompson DC, Silverman GA, et al. Update of the human and mouse SERPIN gene superfamily. Hum Genomics. (2013) 7:22. doi: 10.1186/1479-7364-7-22
6. Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. (1994) 269:15957–60. doi: 10.1016/S0021-9258(17)33954-6
7. Kellici TF, Pilka ES, Bodkin MJ. Small-molecule modulators of serine protease inhibitor proteins (serpins). Drug Discovery Today. (2021) 26:442–54. doi: 10.1016/j.drudis.2020.11.012
8. Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker JNWN. Psoriasis. Lancet. (2021) 397:1301–15. doi: 10.1016/S0140-6736(20)32549-6
9. Griffiths CE, Barker JN. Pathogenesis and clinical features of psoriasis. Lancet. (2007) 370:263–71. doi: 10.1016/S0140-6736(07)61128-3
10. Schmuth M, Blunder S, Dubrac S, Gruber R, Moosbrugger-Martinz V. Epidermal barrier in hereditary ichthyoses, atopic dermatitis, and psoriasis. J Dtsch Dermatol Ges. (2015) 13:1119–23. doi: 10.1111/ddg.2015.13.issue-11
11. Raychaudhuri SK, Maverakis E, Raychaudhuri SP. Diagnosis and classification of psoriasis. Autoimmun Rev. (2014) 13:490–5. doi: 10.1016/j.autrev.2014.01.008
12. Yang SF, Lin MH, Chou PC, Hu SK, Shih SY, Yu HS, et al. Genetics of generalized pustular psoriasis: current understanding and implications for future therapeutics. Genes (Basel). (2023) 14:1297. doi: 10.3390/genes14061297
13. Armstrong AW, Read C. Pathophysiology, clinical presentation, and treatment of psoriasis: A review. JAMA. (2020) 323:1945–60. doi: 10.1001/jama.2020.4006
14. Navarini AA, Burden AD, Capon F, Mrowietz U, Puig L, Köks S, et al. European consensus statement on phenotypes of pustular psoriasis. J Eur Acad Dermatol Venereol. (2017) 31:1792–9. doi: 10.1111/jdv.2017.31.issue-11
15. Sun Y, Sheshadri N, Zong WX. SERPINB3 and B4: From biochemistry to biology. Semin Cell Dev Biol. (2017) 62:170–7. doi: 10.1016/j.semcdb.2016.09.005
16. Meyer-Hoffert U. Reddish, scaly, and itchy: how proteases and their inhibitors contribute to inflammatory skin diseases. Arch Immunol Ther Exp (Warsz). (2009) 57:345–54. doi: 10.1007/s00005-009-0045-6
17. Choudhary S, Anand R, Pradhan D, Bastia B, Kumar SN, Singh H, et al. Transcriptomic landscaping of core genes and pathways of mild and severe psoriasis vulgaris. Int J Mol Med. (2021) 47:219–31. doi: 10.3892/ijmm.2020.4771
18. Iversen OJ, Lysvand H, Slupphaug G. Pso p27, a SERPINB3/B4-derived protein, is most likely a common autoantigen in chronic inflammatory diseases. Clin Immunol. (2017) 174:10–7. doi: 10.1016/j.clim.2016.11.006
19. Kantaputra P, Chaowattanapanit S, Kiratikanon S, Chaiwarith R, Choonhakarn C, Intachai W, et al. SERPINA1, generalized pustular psoriasis, and adult-onset immunodeficiency. J Dermatol. (2021) 48:1597–601. doi: 10.1111/1346-8138.16081
20. Vasků V, Vasků A, Izakovicová Hollá L, Tschöplová S, Kanková K, Benáková N, et al. Polymorphisms in inflammation genes (angiotensinogen, TAP1 and TNF-beta) in psoriasis. Arch Dermatol Res. (2000) 292:531–4. doi: 10.1007/s004030000176
21. Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, et al. An overview of the serpin superfamily. Genome Biol. (2006) 7:216. doi: 10.1186/gb-2006-7-5-216
22. Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. (2001) 276:33293–6. doi: 10.1074/jbc.R100016200
23. Namciu SJ, Friedman RD, Marsden MD, Sarausad LM, Jasoni CL, Fournier RE. Sequence organization and matrix attachment regions of the human serine protease inhibitor gene cluster at 14q32.1. Mamm Genome. (2004) 15:162–78. doi: 10.1007/s00335-003-2311-y
24. Marsden MD, Fournier RE. Organization and expression of the human serpin gene cluster at 14q32.1. Front Biosci. (2005) 10:1768–78. doi: 10.2741/1660
25. Schick C, Pemberton PA, Shi GP, Kamachi Y, Cataltepe S, Bartuski AJ, et al. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry. (1998) 37:5258–66. doi: 10.1021/bi972521d
26. Irving JA, Pike RN, Dai W, Brömme D, Worrall DM, Silverman GA, et al. Evidence that serpin architecture intrinsically supports papain-like cysteine protease inhibition: engineering alpha(1)-antitrypsin to inhibit cathepsin proteases. Biochemistry. (2002) 41:4998–5004. doi: 10.1021/bi0159985
27. Clemmensen SN, Jacobsen LC, Rørvig S, Askaa B, Christenson K, Iversen M, et al. Alpha-1-antitrypsin is produced by human neutrophil granulocytes and their precursors and liberated during granule exocytosis. Eur J Haematol. (2011) 86:517–30. doi: 10.1111/j.1600-0609.2011.01601.x
28. Kaneva MK. Neutrophil elastase and its inhibitors-overlooked players in osteoarthritis. FEBS J. (2022) 289:113–6. doi: 10.1111/febs.v289.1
29. Song S. Alpha-1 antitrypsin therapy for autoimmune disorders. Chronic Obstr Pulm Dis. (2018) 5:289–301. doi: 10.15326/jcopdf.5.4.2018.0131
30. Johnston A, Xing X, Wolterink L, Barnes DH, Yin Z, Reingold L, et al. IL-1 and IL-36 are dominant cytokines in generalized pustular psoriasis. J Allergy Clin Immunol. (2017) 140:109–20. doi: 10.1016/j.jaci.2016.08.056
31. Barszcz D, Zarebska Z, Glińska-Ferenz M, Jabłońska S, Tigałonowa M, Gliński W. Alpha 1-proteinase inhibitor in psoriasis: reduced activity in symptom-free patients and during flare. Arch Dermatol Res. (1988) 280:198–206. doi: 10.1007/BF00513958
32. Marques PI, Ferreira Z, Martins M, Figueiredo J, Silva DI, Castro P, et al. SERPINA2 is a novel gene with a divergent function from SERPINA1. PloS One. (2013) 8:e66889. doi: 10.1371/journal.pone.0066889
33. Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. (2000) 10:1845–64. doi: 10.1101/gr.147800
34. Baker C, Belbin O, Kalsheker N, Morgan K. SERPINA3 (aka alpha-1-antichymotrypsin). Front Biosci. (2007) 12:2821–35. doi: 10.2741/2275
35. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. (2014) 13:397–406. doi: 10.1074/mcp.M113.035600
36. Frey S, Sticht H, Wilsmann-Theis D, Gerschütz A, Wolf K, Löhr S, et al. Rare loss-of-function mutation in SERPINA3 in generalized pustular psoriasis. J Invest Dermatol. (2020) 140:1451–5. doi: 10.1016/j.jid.2019.11.024
37. Neuhauser R, Eyerich K, Boehner A. Generalized pustular psoriasis-Dawn of a new era in targeted immunotherapy. Exp Dermatol. (2020) 29:1088–96. doi: 10.1111/exd.v29.11
38. Liu Y, Li H, Meng S, Xu Y, Ni S, Sun Y, et al. Newly revealed variants of SERPINA3 in generalized pustular psoriasis attenuate inhibition of ACT on cathepsin G. J Hum Genet. (2023) 68:419–25. doi: 10.1038/s10038-023-01139-z
39. Guo J, Tu J, Hu Y, Song G, Yin Z. Cathepsin G cleaves and activates IL-36γ and promotes the inflammation of psoriasis. Drug Des Devel Ther. (2019) 13:581–8. doi: 10.2147/DDDT.S194765
40. Chao J, Schmaier A, Chen LM, Yang Z, Chao L. Kallistatin, a novel human tissue kallikrein inhibitor: levels in body fluids, blood cells, and tissues in health and disease. J Lab Clin Med. (1996) 127:612–20. doi: 10.1016/S0022-2143(96)90152-3
41. Wang C, Liu Y, Liu X, Zhao J, Lang B, Wu F, et al. IFN-inducible serpinA5 triggers antiviral immunity by regulating STAT1 phosphorylation and nuclear translocation. Int J Mol Sci. (2023) 24:5458. doi: 10.3390/ijms24065458
42. Suzuk i K. The multi-functional serpin, protein C inhibitor: beyond thrombosis and hemostasis. J Thromb Haemost. (2008) 6:2017–26. doi: 10.1111/j.1538-7836.2008.03181.x
43. Sun W, Grassi P, Engström A, Sooriyaarachchi S, Ubhayasekera W, Hreinsson J, et al. N-glycans of human protein C inhibitor: tissue-specific expression and function. PloS One. (2011) 6:e29011. doi: 10.1371/journal.pone.0029011
44. Crawford AA, Bankier S, Altmaier E, Barnes CLK, Clark DW, Ermel R, et al. Variation in the SERPINA6/SERPINA1 locus alters morning plasma cortisol, hepatic corticosteroid binding globulin expression, gene expression in peripheral tissues, and risk of cardiovascular disease. J Hum Genet. (2021) 66:625–36. doi: 10.1038/s10038-020-00895-6
45. Sivukhina E, Helbling JC, Minni AM, Schäfer HH, Pallet V, Jirikowski GF, et al. Intrinsic expression of transcortin in neural cells of the mouse brain: a histochemical and molecular study. J Exp Biol. (2013) 216:245–52. doi: 10.1242/jeb.076893
46. Gomes-Lima CJ, Maciel AAFL, Andrade MO, Cunha VSD, Mazzeu JF, Bleicher L, et al. Thyroxine-binding globulin deficiency due to a novel SERPINA7 mutation: Clinical characterization, analysis of X-chromosome inactivation pattern and protein structural modeling. Gene. (2018) 666:58–63. doi: 10.1016/j.gene.2018.05.018
47. Maranduca MA, Cosovanu MA, Clim A, Pinzariu AC, Filip N, Drochioi IC, et al. The renin-angiotensin system: the challenge behind autoimmune dermatological diseases. Diagn (Basel). (2023) 13:3398. doi: 10.3390/diagnostics13223398
48. Steckelings UM, Wollschläger T, Peters J, Henz BM, Hermes B, Artuc M. Human skin: source of and target organ for angiotensin II. Exp Dermatol. (2004) 13:148–54. doi: 10.1111/j.0906-6705.2004.0139.x
49. Paterson MA, Horvath AJ, Pike RN, Coughlin PB. Molecular characterization of centerin, a germinal centre cell serpin. Biochem J. (2007) 405:489–94. doi: 10.1042/BJ20070174
50. Frazer JK, Jackson DG, Gaillard JP, Lutter M, Liu YJ, Banchereau J, et al. Identification of centerin: a novel human germinal center B cell-restricted serpin. Eur J Immunol. (2000) 30:3039–48. doi: 10.1002/1521-4141(200010)30:10<3039::AID-IMMU3039>3.0.CO;2-H
51. Girard TJ, Lasky NM, Tuley EA, Broze GJ Jr. Protein Z, protein Z-dependent protease inhibitor (serpinA10), and the acute-phase response. J Thromb Haemost. (2013) 11:375–8. doi: 10.1111/jth.12084
52. Song Y, Li Z, Li L, Zhou H, Zeng TT, Jin C, et al. SERPINA11 inhibits metastasis in hepatocellular carcinoma by suppressing MEK/ERK signaling pathway. J Hepatocell Carcinoma. (2021) 8:759–71. doi: 10.2147/JHC.S315634
53. Heiker JT, Klöting N, Kovacs P, Kuettner EB, Sträter N, Schultz S, et al. Vaspin inhibits kallikrein 7 by serpin mechanism. Cell Mol Life Sci. (2013) 70:2569–83. doi: 10.1007/s00018-013-1258-8
54. Ulbricht D, Tindall CA, Oertwig K, Hanke S, Sträter N, Heiker JT. Kallikrein-related peptidase 14 is the second KLK protease targeted by the serpin vaspin. Biol Chem. (2018) 399:1079–84. doi: 10.1515/hsz-2018-0108
55. Saalbach A, Tremel J, Herbert D, Schwede K, Wandel E, Schirmer C, et al. Anti-inflammatory action of keratinocyte-derived vaspin: relevance for the pathogenesis of psoriasis. Am J Pathol. (2016) 186:639–51. doi: 10.1016/j.ajpath.2015.10.030
56. Yao Y, Richman L, Morehouse C, de los Reyes M, Higgs BW, Boutrin A, et al. Type I interferon: potential therapeutic target for psoriasis? PloS One. (2009) 4:e2737. doi: 10.1371/annotation/fbcbcab9-2e87-4ec7-af6e-c6e9e64ad4b3
57. Yarilina A, Ivashkiv LB. Type I interferon: a new player in TNF signaling. Curr Dir Autoimmun. (2010) 11:94–104. doi: 10.1159/000289199
58. Majewski P, Majchrzak-Gorecka M, Grygier B, Skrzeczynska-Moncznik J, Osiecka O, Cichy J. Inhibitors of serine proteases in regulating the production and function of neutrophil extracellular traps. Front Immunol. (2016) 7:261. doi: 10.3389/fimmu.2016.00261
59. Sugimori T, Cooley J, Hoidal JR, Remold-O’Donnell E. Inhibitory properties of recombinant human monocyte/neutrophil elastase inhibitor. Am J Respir Cell Mol Biol. (1995) 13:314–22. doi: 10.1165/ajrcmb.13.3.7654387
60. Wang L, Li Q, Wu L, Liu S, Zhang Y, Yang X, et al. Identification of SERPINB1 as a physiological inhibitor of human granzyme H. J Immunol. (2013) 190:1319–30. doi: 10.4049/jimmunol.1202542
61. Zhao P, Hou L, Farley K, Sundrud MS, Remold-O’Donnell E. SerpinB1 regulates homeostatic expansion of IL-17+ γδ and CD4+ Th17 cells. J Leukoc Biol. (2014) 95:521–30. doi: 10.1189/jlb.0613331
62. Hibino T, Matsuda Y, Takahashi T, Goetinck PF. Suppression of keratinocyte proliferation by plasminogen activator inhibitor-2. J Invest Dermatol. (1999) 112:85–90. doi: 10.1046/j.1523-1747.1999.00466.x
63. Ritchie H, Jamieson A, Booth NA. Regulation, location and activity of plasminogen activator inhibitor 2 (PAI-2) in peripheral blood monocytes, macrophages and foam cells. Thromb Haemost. (1997) 77:1168–73.
64. Robinson NA, Lapic S, Welter JF, Eckert RL. S100A11, S100A10, annexin I, desmosomal proteins, small proline-rich proteins, plasminogen activator inhibitor-2, and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J Biol Chem. (1997) 272:12035–46. doi: 10.1074/jbc.272.18.12035
65. Kruithof EK, Baker MS, Bunn CL. Biological and clinical aspects of plasminogen activator inhibitor type 2. Blood. (1995) 86:4007–24. doi: 10.1182/blood.V86.11.4007.bloodjournal86114007
66. Swartz JM, Byström J, Dyer KD, Nitto T, Wynn TA, Rosenberg HF. Plasminogen activator inhibitor-2 (PAI-2) in eosinophilic leukocytes. J Leukoc Biol. (2004) 76:812–9. doi: 10.1189/jlb.0304182
67. Gissler HM, Frank R, Kramer MD. Immunohistochemical characterization of the plasminogen activator system in psoriatic epidermis. Br J Dermatol. (1993) 128:612–8. doi: 10.1111/j.1365-2133.1993.tb00254.x
68. Pohl D, Andrýs C, Borská L, Fiala Z, Hamáková K, Ettler K, et al. CC and CXC chemokines patterns in psoriasis determined by protein array method were influenced by Goeckerman’s therapy. Acta Med (Hradec Kralove). (2009) 52:9–13. doi: 10.14712/18059694.2016.100
69. Johansen C, Rittig AH, Mose M, Bertelsen T, Weimar I, Nielsen J, et al. STAT2 is involved in the pathogenesis of psoriasis by promoting CXCL11 and CCL5 production by keratinocytes. PloS One. (2017) 12:e0176994. doi: 10.1371/journal.pone.0176994
70. Gillitzer R, Ritter U, Spandau U, Goebeler M, Bröcker EB. Differential expression of GRO-alpha and IL-8 mRNA in psoriasis: a model for neutrophil migration and accumulation in vivo. J Invest Dermatol. (1996) 107:778–82. doi: 10.1111/1523-1747.ep12371803
71. Schroder WA, Anraku I, Le TT, Hirata TD, Nakaya HI, Major L, et al. SerpinB2 deficiency results in a stratum corneum defect and increased sensitivity to topically applied inflammatory agents. Am J Pathol. (2016) 186:1511–23. doi: 10.1016/j.ajpath.2016.02.017
72. Kantaputra P, Daroontum T, Chuamanochan M, Chaowattanapanit S, Kiratikanon S, Choonhakarn C, et al. SERPINB3, adult-onset immunodeficiency, and generalized pustular psoriasis. Genes (Basel). (2023) 14:266. doi: 10.3390/genes14020266
73. Asbakk K, Bergh K, Iversen OJ. The psoriasis-associated antigen, pso p27, participates in the formation of complement activating immune-complexes in psoriatic scale. APMIS. (1990) 98:143–9. doi: 10.1111/j.1699-0463.1990.tb01014.x
74. Lysvand H, Helland R, Hagen L, Slupphaug G, Iversen OJ. Psoriasis pathogenesis - Pso p27 constitutes a compact structure forming large aggregates. Biochem Biophys Rep. (2015) 2:132–6. doi: 10.1016/j.bbrep.2015.06.001
75. Yuan H, Liu H, Liu Z, Zhu D, Amos CI, Fang S, et al. Genetic variants in Hippo pathway genes YAP1, TEAD1 and TEAD4 are associated with melanoma-specific survival. Int J Cancer. (2015) 137:638–45. doi: 10.1002/ijc.29429
76. Ren C, Liu Q, Ma Y, Wang A, Yang Y, Wang D. TEAD4 transcriptional regulates SERPINB3/4 and affect crosstalk between keratinocytes and T cells in psoriasis. Immunobiology. (2020) 225:152006. doi: 10.1016/j.imbio.2020.152006
77. Xiao W, Sha K, Wang M, Tan Z, Wang Y, Xu S, et al. SERPINB3/B4 is increased in psoriasis and rosacea lesions and has proinflammatory effects in mouse models of these diseases. J Invest Dermatol. (2024) 144:2706–18.e6. doi: 10.1016/j.jid.2024.04.011
78. Sivaprasad U, Kinker KG, Ericksen MB, Lindsey M, Gibson AM, Bass SA, et al. SERPINB3/B4 contributes to early inflammation and barrier dysfunction in an experimental murine model of atopic dermatitis. J Invest Dermatol. (2015) 135:160–9. doi: 10.1038/jid.2014.353
79. Besgen P, Trommler P, Vollmer S, Prinz JC. Ezrin, maspin, peroxiredoxin 2, and heat shock protein 27: potential targets of a streptococcal-induced autoimmune response in psoriasis. J Immunol. (2010) 184:5392–402. doi: 10.4049/jimmunol.0903520
80. Scott FL, Hirst CE, Sun J, Bird CH, Bottomley SP, Bird PI. The intracellular serpin proteinase inhibitor 6 is expressed in monocytes and granulocytes and is a potent inhibitor of the azurophilic granule protease, cathepsin G. Blood. (1999) 93:2089–97. doi: 10.1182/blood.V93.6.2089.406k10_2089_2097
81. Scott FL, Sun J, Whisstock JC, Kato K, Bird PI. SerpinB6 is an inhibitor of kallikrein-8 in keratinocytes. J Biochem. (2007) 142:435–42. doi: 10.1093/jb/mvm156
82. Mangan MS, Kaiserman D, Bird PI. The role of serpins in vertebrate immunity. Tissue Antigens. (2008) 72:1–10. doi: 10.1111/j.1399-0039.2008.01059.x
83. Wang Y, Guo Q, Casey A, Lin C, Chen F. A new tool for conditional gene manipulation in a subset of keratin-expressing epithelia. Genesis. (2012) 50:899–907. doi: 10.1002/dvg.v50.12
84. Kubo A, Shiohama A, Sasaki T, Nakabayashi K, Kawasaki H, Atsugi T, et al. Mutations in SERPINB7, encoding a member of the serine protease inhibitor superfamily, cause Nagashima-type palmoplantar keratosis. Am J Hum Genet. (2013) 93:945–56. doi: 10.1016/j.ajhg.2013.09.015
85. Zheng H, Gu L, Zhao F, Zhang C, Wang Z, Zhou H, et al. SerpinB7 deficiency contributes to development of psoriasis via calcium-mediated keratinocyte differentiation dysfunction. Cell Death Dis. (2022) 13:635. doi: 10.1038/s41419-022-05045-8
86. Gillard A, Scarff K, Loveland KL, Ricardo SD, Bird PI. Modulation and redistribution of proteinase inhibitor 8 (Serpinb8) during kidney regeneration. Am J Nephrol. (2006) 26:34–42. doi: 10.1159/000091784
87. Strik MC, Bladergroen BA, Wouters D, Kisiel W, Hooijberg JH, Verlaan AR, et al. Distribution of the human intracellular serpin protease inhibitor 8 in human tissues. J Histochem Cytochem. (2002) 50:1443–54. doi: 10.1177/002215540205001103
88. Chandran V. The genetics of psoriasis and psoriatic arthritis. Clin Rev Allergy Immunol. (2013) 44:149–56. doi: 10.1007/s12016-012-8303-5
89. Sun J, Bird CH, Sutton V, McDonald L, Coughlin PB, De Jong TA, et al. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. (1996) 271:27802–9. doi: 10.1074/jbc.271.44.27802
90. Mangan MS, Vega-Ramos J, Joeckel LT, Mitchell AJ, Rizzitelli A, Roediger B, et al. Serpinb9 is a marker of antigen cross-presenting dendritic cells. Mol Immunol. (2017) 82:50–6. doi: 10.1016/j.molimm.2016.12.011
91. Riewald M, Schleef RR. Molecular cloning of bomapin (protease inhibitor 10), a novel human serpin that is expressed specifically in the bone marrow. J Biol Chem. (1995) 270:26754–7. doi: 10.1074/jbc.270.45.26754
92. Askew DJ, Cataltepe S, Kumar V, Edwards C, Pace SM, Howarth RN, et al. SERPINB11 is a new noninhibitory intracellular serpin. Common single nucleotide polymorphisms in the scaffold impair conformational change. J Biol Chem. (2007) 282:24948–60. doi: 10.1074/jbc.M703182200
93. Askew YS, Pak SC, Luke CJ, Askew DJ, Cataltepe S, Mills DR, et al. SERPINB12 is a novel member of the human ov-serpin family that is widely expressed and inhibits trypsin-like serine proteinases. J Biol Chem. (2001) 276:49320–30. doi: 10.1074/jbc.M108879200
94. Niehaus JZ, Good M, Jackson LE, Ozolek JA, Silverman GA, Luke CJ. Human SERPINB12 is an abundant intracellular serpin expressed in most surface and glandular epithelia. J Histochem Cytochem. (2015) 63:854–65. doi: 10.1369/0022155415600498
95. Zhang J, Xu X, Wang X, Zhang L, Hu M, Le Y, et al. Topical emollient prevents the development of atopic dermatitis and atopic march in mice. Exp Dermatol. (2023) 32:1007–15. doi: 10.1111/exd.14806
96. Welss T, Sun J, Irving JA, Blum R, Smith AI, Whisstock JC, et al. Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis. Biochemistry. (2003) 42:7381–9. doi: 10.1021/bi027307q
97. Jayakumar A, Kang Y, Frederick MJ, Pak SC, Henderson Y, Holton PR, et al. Inhibition of the cysteine proteinases cathepsins K and L by the serpin headpin (SERPINB13): a kinetic analysis. Arch Biochem Biophys. (2003) 409:367–74. doi: 10.1016/S0003-9861(02)00635-5
98. Lu Z, Wang F, Liang M. SerpinC1/Antithrombin III in kidney-related diseases. Clin Sci (Lond). (2017) 131:823–31. doi: 10.1042/CS20160669
99. Crisp RJ, Knauer MF, Knauer DJ. Protease nexin 1 is a potent urinary plasminogen activator inhibitor in the presence of collagen type IV. J Biol Chem. (2002) 277:47285–91. doi: 10.1074/jbc.M204813200
100. Bouton MC, Boulaftali Y, Richard B, Arocas V, Michel JB, Jandrot-Perrus M. Emerging role of serpinE2/protease nexin-1 in hemostasis and vascular biology. Blood. (2012) 119:2452–7. doi: 10.1182/blood-2011-10-387464
101. Viganò S, D’Andrea G, Valle PD, Santacroce R, Margaglione M, D’Angelo A. A novel allele variant of the SERPINF2 gene responsible for severe plasmin inhibitor (α2-antiplasmin) deficiency in an Italian patient. Thromb Res. (2018) 166:60–2. doi: 10.1016/j.thromres.2018.04.006
102. Rasmussen ER, Aanæs K, Jakobsen MA, Bygum A. Acquired complement C1 esterase inhibitor deficiency in a patient with a rare SERPING1 variant with unknown significance. BMJ Case Rep. (2019) 12:e231122. doi: 10.1136/bcr-2019-231122
103. Nagata K. Hsp47: a collagen-specific molecular chaperone. Trends Biochem Sci. (1996) 21:22–6. doi: 10.1016/S0968-0004(06)80023-X
104. Widmer C, Gebauer JM, Brunstein E, Rosenbaum S, Zaucke F, Drögemüller C, et al. Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proc Natl Acad Sci U S A. (2012) 109:13243–7. doi: 10.1073/pnas.1208072109
105. Osterwalder T, Cinelli P, Baici A, Pennella A, Krueger SR, Schrimpf SP, et al. The axonally secreted serine proteinase inhibitor, neuroserpin, inhibits plasminogen activators and plasmin but not thrombin. J Biol Chem. (1998) 273:2312–21. doi: 10.1074/jbc.273.4.2312
106. Higgins WJ, Grehan GT, Wynne KJ, Worrall DM. SerpinI2 (pancpin) is an inhibitory serpin targeting pancreatic elastase and chymotrypsin. Biochim Biophys Acta Proteins Proteom. (2017) 1865:195–200. doi: 10.1016/j.bbapap.2016.10.013
107. Pemberton PA, Stein PE, Pepys MB, Potter JM, Carrell RW. Hormone binding globulins undergo serpin conformational change in inflammation. Nature. (1988) 336:257–8. doi: 10.1038/336257a0
108. Fung ML. The role of local renin-angiotensin system in arterial chemoreceptors in sleep-breathing disorders. Front Physiol. (2014) 5:336. doi: 10.3389/fphys.2014.00336
109. Meyer EJ, Nenke MA, Lewis JG, Torpy DJ. Corticosteroid-binding globulin: acute and chronic inflammation. Expert Rev Endocrinol Metab. (2017) 12:241–51. doi: 10.1080/17446651.2017.1332991
110. Carrell RW, Evans DL, Stein PE. Mobile reactive centre of serpins and the control of thrombosis. Nature. (1991) 353:576–8. doi: 10.1038/353576a0
111. Kaneva MK, Muley MM, Krustev E, Reid AR, Souza PR, Dell'Accio F, et al. Alpha-1-antitrypsin reduces inflammation and exerts chondroprotection in arthritis. FASEB J. (2021) 35:e21472. doi: 10.1096/fj.202001801R
112. Zhang Y, He J, Zhao J, Xu M, Lou D, Tso P, et al. Effect of ApoA4 on SERPINA3 mediated by nuclear receptors NR4A1 and NR1D1 in hepatocytes. Biochem Biophys Res Commun. (2017) 487:327–32. doi: 10.1016/j.bbrc.2017.04.058
113. Frühbeck G, Gómez-Ambrosi J, Rodríguez A, Ramírez B, Valentí V, Moncada R, et al. Novel protective role of kallistatin in obesity by limiting adipose tissue low grade inflammation and oxidative stress. Metabolism. (2018) 87:123–35. doi: 10.1016/j.metabol.2018.04.004
114. He Y, Han Y, Xing J, Zhai X, Wang S, Xin S, et al. Kallistatin correlates with inflammation in abdominal aortic aneurysm and suppresses its formation in mice. Cardiovasc Diagn Ther. (2020) 10:107–23. doi: 10.21037/cdt.2019.12.08
115. Saalbach A, Vester K, Rall K, Tremel J, Anderegg U, Beck-Sickinger AG, et al. Vaspin–a link of obesity and psoriasis? Exp Dermatol. (2012) 21:309–12. doi: 10.1111/j.1600-0625.2012.01460.x
116. Burgener SS, Leborgne NGF, Snipas SJ, Salvesen GS, Bird PI, Benarafa C. Cathepsin G inhibition by serpinb1 and serpinb6 prevents programmed necrosis in neutrophils and monocytes and reduces GSDMD-driven inflammation. Cell Rep. (2019) 27:3646–3656.e5. doi: 10.1016/j.celrep.2019.05.065
117. Vaher H, Kivihall A, Runnel T, Raam L, Prans E, Maslovskaja J, et al. SERPINB2 and miR-146a/b are coordinately regulated and act in the suppression of psoriasis-associated inflammatory responses in keratinocytes. Exp Dermatol. (2020) 29:51–60. doi: 10.1111/exd.14049
118. Mo Y, Zhang K, Feng Y, Yi L, Liang Y, Wu W, et al. Epithelial SERPINB10, a novel marker of airway eosinophilia in asthma, contributes to allergic airway inflammation. Am J Physiol Lung Cell Mol Physiol. (2019) 316:L245–54. doi: 10.1152/ajplung.00362.2017
119. Lechowicz U, Rudzinski S, Jezela-Stanek A, Janciauskiene S, Chorostowska-Wynimko J. Post-translational modifications of circulating alpha-1-antitrypsin protein. Int J Mol Sci. (2020) 21:9187. doi: 10.3390/ijms21239187
120. Gong D, Farley K, White M, Hartshorn KL, Benarafa C, Remold-O’Donnell E. Critical role of serpinB1 in regulating inflammatory responses in pulmonary influenza infection. J Infect Dis. (2011) 204:592–600. doi: 10.1093/infdis/jir352
121. van der Burgh R, Meeldijk J, Jongeneel L, Frenkel J, Bovenschen N, van Gijn M, et al. Reduced serpinB9-mediated caspase-1 inhibition can contribute to autoinflammatory disease. Oncotarget. (2016) 7:19265–71. doi: 10.18632/oncotarget.v7i15
122. Bouton MC, Geiger M, Sheffield WP, Irving JA, Lomas DA, Song S, et al. The under-appreciated world of the serpin family of serine proteinase inhibitors. EMBO Mol Med. (2023) 15:e17144. doi: 10.15252/emmm.202217144
123. Xu D, Wu J, Dong L, Luo W, Li L, Tang D, et al. Serpinc1 acts as a tumor suppressor in hepatocellular carcinoma through inducing apoptosis and blocking macrophage polarization in an ubiquitin-proteasome manner. Front Oncol. (2021) 11:738607. doi: 10.3389/fonc.2021.738607
124. Jing Y, Jia D, Wong CM, Ng IO, Zhang Z, Liu L, et al. SERPINA5 inhibits tumor cell migration by modulating the fibronectin-integrin β1 signaling pathway in hepatocellular carcinoma. Mol Oncol. (2014) 8:366–77. doi: 10.1016/j.molonc.2013.12.003
125. Mahananda B, Vinay J, Palo A, Singh A, Sahu SK, Singh SP, et al. SERPINB5 Genetic Variants rs2289519 and rs2289521 are Significantly Associated with Gallbladder Cancer Risk. DNA Cell Biol. (2021) 40:706–12. doi: 10.1089/dna.2021.0056
126. Kato H. Expression and function of squamous cell carcinoma antigen. Anticancer Res. (1996) 16:2149–53.
127. Bylaite M, Moussali H, Marciukaitiene I, Ruzicka T, Walz M. Expression of cathepsin L and its inhibitor hurpin in inflammatory and neoplastic skin diseases. Exp Dermatol. (2006) 15:110–8. doi: 10.1111/j.1600-0625.2005.00389.x
128. Egawa G, Kabashima K. Multifactorial skin barrier deficiency and atopic dermatitis: Essential topics to prevent the atopic march. J Allergy Clin Immunol. (2016) 138:350–358.e1. doi: 10.1016/j.jaci.2016.06.002
129. Egawa G, Kabashima K. Barrier dysfunction in the skin allergy. Allergol Int. (2018) 67:3–11. doi: 10.1016/j.alit.2017.10.002
130. Hoober JK, Eggink LL. The discovery and function of filaggrin. Int J Mol Sci. (2022) 23:1455. doi: 10.3390/ijms23031455
131. Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. (2005) 6:328–40. doi: 10.1038/nrm1619
132. Kalinin A, Marekov LN, Steinert PM. Assembly of the epidermal cornified cell envelope. J Cell Sci. (2001) 114:3069–70. doi: 10.1242/jcs.114.17.3069
133. Borgoño CA, Michael IP, Komatsu N, Jayakumar A, Kapadia R, Clayman GL, et al. A potential role for multiple tissue kallikrein serine proteases in epidermal desquamation. J Biol Chem. (2007) 282:3640–52. doi: 10.1074/jbc.M607567200
134. Schultz S, Saalbach A, Heiker JT, Meier R, Zellmann T, Simon JC, et al. Proteolytic activation of prochemerin by kallikrein 7 breaks an ionic linkage and results in C-terminal rearrangement. Biochem J. (2013) 452:271–80. doi: 10.1042/BJ20121880
135. Dull K, Lénárt K, Dajnoki Z, Póliska S, Uchiyama E, Hendrik Z, et al. Barrier function-related genes and proteins have an altered expression in acne-involved skin. J Eur Acad Dermatol Venereol. (2023) 37:1415–25. doi: 10.1111/jdv.19062
136. Kishibe M. Physiological and pathological roles of kallikrein-related peptidases in the epidermis. J Dermatol Sci. (2019) 95:50–5. doi: 10.1016/j.jdermsci.2019.06.007
137. Katagiri C, Iida T, Nakanishi J, Ozawa M, Aiba S, Hibino T. Up-regulation of serpin SCCA1 is associated with epidermal barrier disruption. J Dermatol Sci. (2010) 57:95–101. doi: 10.1016/j.jdermsci.2009.09.004
138. Cheng T, Tjabringa GS, van Vlijmen-Willems IM, Hitomi K, van Erp PE, Schalkwijk J, et al. The cystatin M/E-controlled pathway of skin barrier formation: expression of its key components in psoriasis and atopic dermatitis. Br J Dermatol. (2009) 161:253–64. doi: 10.1111/j.1365-2133.2009.09156.x
139. Moussali H, Bylaite M, Welss T, Abts HF, Ruzicka T, Walz M. Expression of hurpin, a serine proteinase inhibitor, in normal and pathological skin: overexpression and redistribution in psoriasis and cutaneous carcinomas. Exp Dermatol. (2005) 14:420–8. doi: 10.1111/j.0906-6705.2005.00300.x
140. Orsmond A, Bereza-Malcolm L, Lynch T, March L, Xue M. Skin barrier dysregulation in psoriasis. Int J Mol Sci. (2021) 22:10841. doi: 10.3390/ijms221910841
141. Colak S, Omma A, Sandikci SC, Yucel C, Omma T, Turhan T. Vaspin, neutrophil gelatinase-associated lipocalin and apolipoprotein levels in patients with psoriatic arthritis. Bratisl Lek Listy. (2019) 120:65–9. doi: 10.4149/BLL_2019_010
142. Vinje O, Møller P, Mellbye OJ. Laboratory findings in patients with psoriasis, with special reference to immunological parameters, associations with arthropathy and sacro-iliitis. Scand J Rheumatol. (1980) 9:97–105. doi: 10.3109/03009748009098137
143. Boehncke WH, Schön MP. Psoriasis. Lancet. (2015) 386:983–94. doi: 10.1016/S0140-6736(14)61909-7
144. Krueger JG, Bowcock A. Psoriasis pathophysiology: current concepts of pathogenesis. Ann Rheum Dis. (2005) 64 Suppl 2:ii30–6. doi: 10.1136/ard.2004.031120
145. Kim J, Krueger JG. The immunopathogenesis of psoriasis. Dermatol Clin. (2015) 33:13–23. doi: 10.1016/j.det.2014.09.002
146. Sumitomo A, Siriwach R, Thumkeo D, Ito K, Nakagawa R, Tanaka N, et al. LPA induces keratinocyte differentiation and promotes skin barrier function through the LPAR1/LPAR5-RHO-ROCK-SRF axis. J Invest Dermatol. (2019) 139:1010–22. doi: 10.1016/j.jid.2018.10.034
147. Toulza E, Mattiuzzo NR, Galliano MF, Jonca N, Dossat C, Jacob D, et al. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. (2007) 8:R107. doi: 10.1186/gb-2007-8-6-r107
148. Henry J, Toulza E, Hsu CY, Pellerin L, Balica S, Mazereeuw-Hautier J, et al. Update on the epidermal differentiation complex. Front Biosci (Landmark Ed). (2012) 17:1517–32. doi: 10.2741/4001
149. Wang Z, Zheng H, Zhou H, Huang N, Wei X, Liu X, et al. Systematic screening and identification of novel psoriasis-specific genes from the transcriptome of psoriasis-like keratinocytes. Mol Med Rep. (2019) 19:1529–42. doi: 10.3892/mmr.2018.9782
150. Ayvaz HH, Öztürk KH, Seyirci MA, Atay E, Korkmaz S, Erturan İ, et al. The role of EREG, PTPN1, and SERPINB7 genes in the pathogenesis of psoriasis: may SERPINB7 be protective and a marker of severity for psoriasis? Dermatol Pract Concept. (2023) 13:e2023085. doi: 10.5826/dpc.1302a85
151. Dahlen JR, Jean F, Thomas G, Foster DC, Kisiel W. Inhibition of soluble recombinant furin by human proteinase inhibitor 8. J Biol Chem. (1998) 273:1851–4. doi: 10.1074/jbc.273.4.1851
152. Leliefeld PH, Koenderman L, Pillay J. How neutrophils shape adaptive immune responses. Front Immunol. (2015) 6:471. doi: 10.3389/fimmu.2015.00471
153. Rosales C, Lowell CA, Schnoor M, Uribe-Querol E. Neutrophils: their role in innate and adaptive immunity 2017. J Immunol Res. (2017) 2017:9748345. doi: 10.1155/2017/9748345
154. Chiang CC, Cheng WJ, Korinek M, Lin CY, Hwang TL. Neutrophils in psoriasis. Front Immunol. (2019) 10:2376. doi: 10.3389/fimmu.2019.02376
155. Schön MP, Broekaert SM, Erpenbeck L. Sexy again: the renaissance of neutrophils in psoriasis. Exp Dermatol. (2017) 26:305–11. doi: 10.1111/exd.2017.26.issue-4
156. Shao S, Fang H, Dang E, Xue K, Zhang J, Li B, et al. Neutrophil extracellular traps promote inflammatory responses in psoriasis via activating epidermal TLR4/IL-36R crosstalk. Front Immunol. (2019) 10:746. doi: 10.3389/fimmu.2019.00746
157. Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, et al. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med. (2009) 206:1983–94. doi: 10.1084/jem.20090480
158. Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. (2007) 449:564–9. doi: 10.1038/nature06116
159. Ma J, Chen J, Xue K, Yu C, Dang E, Qiao H, et al. LCN2 mediates skin inflammation in psoriasis through the SREBP2-NLRC4 axis. J Invest Dermatol. (2022) 142:2194–2204.e11. doi: 10.1016/j.jid.2022.01.012
160. Lin X, Huang T. Oxidative stress in psoriasis and potential therapeutic use of antioxidants. Free Radic Res. (2016) 50:585–95. doi: 10.3109/10715762.2016.1162301
161. Xu F, Xu J, Xiong X, Deng Y. Salidroside inhibits MAPK, NF-κB, and STAT3 pathways in psoriasis-associated oxidative stress via SIRT1 activation. Redox Rep. (2019) 24:70–4. doi: 10.1080/13510002.2019.1658377
162. Lundberg KC, Fritz Y, Johnston A, Foster AM, Baliwag J, Gudjonsson JE, et al. Proteomics of skin proteins in psoriasis: from discovery and verification in a mouse model to confirmation in humans. Mol Cell Proteomics. (2015) 14:109–19. doi: 10.1074/mcp.M114.042242
163. Benarafa C, Cooley J, Zeng W, Bird PI, Remold-O’Donnell E. Characterization of four murine homologs of the human ov-serpin monocyte neutrophil elastase inhibitor MNEI (SERPINB1). J Biol Chem. (2002) 277:42028–33. doi: 10.1074/jbc.M207080200
164. Kamata Y, Maejima H, Watarai A, Saito N, Katsuoka K, Takeda A, et al. Expression of bleomycin hydrolase in keratinization disorders. Arch Dermatol Res. (2012) 304:31–8. doi: 10.1007/s00403-011-1180-6
165. Komatsu N, Saijoh K, Kuk C, Shirasaki F, Takehara K, Diamandis EP. Aberrant human tissue kallikrein levels in the stratum corneum and serum of patients with psoriasis: dependence on phenotype, severity and therapy. Br J Dermatol. (2007) 156:875–83. doi: 10.1111/j.1365-2133.2006.07743.x
166. Lundström A, Egelrud T. Stratum corneum chymotryptic enzyme: a proteinase which may be generally present in the stratum corneum and with a possible involvement in desquamation. Acta Derm Venereol. (1991) 71:471–4. doi: 10.2340/0001555571471474
167. Morizane S, Yamasaki K, Kabigting FD, Gallo RL. Kallikrein expression and cathelicidin processing are independently controlled in keratinocytes by calcium, vitamin D(3), and retinoic acid. J Invest Dermatol. (2010) 130:1297–306. doi: 10.1038/jid.2009.435
168. Damara A, Wegner J, Trzeciak ER, Kolb A, Nastaranpour M, Khatri R, et al. LL37/self-DNA complexes mediate monocyte reprogramming. Clin Immunol. (2024) 265:110287. doi: 10.1016/j.clim.2024.110287
169. Laureano AFS, Vigato AA, Puzer L, de Araujo DR. Recombinant scFv-fc anti-kallikrein 7 antibody-loaded thermosensitive hydrogels against skin desquamation disorders. ACS Appl Bio Mater. (2024) 7:4486–96. doi: 10.1021/acsabm.4c00371
170. Lynch KR, Peach MJ. Molecular biology of angiotensinogen. Hypertension. (1991) 17:263–9. doi: 10.1161/01.HYP.17.3.263
171. Shokrian Zeini M, Haddadi NS, Shayan M, Shokrian Zeini M, Kazemi K, Solaimanian S, et al. Losartan ointment attenuates imiquimod-induced psoriasis-like inflammation. Int Immunopharmacol. (2021) 100:108160. doi: 10.1016/j.intimp.2021.108160
172. Woo YR, Cho DH, Park HJ. Molecular mechanisms and management of a cutaneous inflammatory disorder: psoriasis. Int J Mol Sci. (2017) 18:2684. doi: 10.3390/ijms18122684
173. Hsu CY, Vo TTT, Lee CW, Chen YL, Lin WN, Cheng HC, et al. Carbon monoxide releasing molecule-2 attenuates angiotensin II-induced IL-6/Jak2/Stat3-associated inflammation by inhibiting NADPH oxidase- and mitochondria-derived ROS in human aortic smooth muscle cells. Biochem Pharmacol. (2022) 198:114978. doi: 10.1016/j.bcp.2022.114978
Keywords: serine protease inhibitors, psoriasis, inflammation, skin barrier function, hyper-proliferation
Citation: Wang J, Li J, Zhou L, Hou H and Zhang K (2024) Regulation of epidermal barrier function and pathogenesis of psoriasis by serine protease inhibitors. Front. Immunol. 15:1498067. doi: 10.3389/fimmu.2024.1498067
Received: 20 September 2024; Accepted: 27 November 2024;
Published: 16 December 2024.
Edited by:
Erle Dang, Air Force Medical University, ChinaReviewed by:
Lei Xiao, Xi’an Jiaotong University, ChinaKe Xue, Fourth Military Medical University, China
Copyright © 2024 Wang, Li, Zhou, Hou and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaiming Zhang, emhhbmdrYWltaW5nQHNpbmEuY29t; dHljY2hvc3BpdGFsQHNpbmEuY29t
 Juanjuan Wang
Juanjuan Wang Junqin Li1,2
Junqin Li1,2 Ling Zhou
Ling Zhou Kaiming Zhang
Kaiming Zhang