- 1Department of Medicine, Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States
- 2Department of Surgery, Duke University School of Medicine, Durham, NC, United States
- 3Cancer Epigenetics Laboratory, Health Research Institute of the Balearic Islands (IdISBa), Palma, Spain
- 4The Angeles Clinical and Research Institute, a Cedars-Sinai Affiliate, Los Angeles, CA, United States
- 5Department of Biomedical Sciences, Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States
Transmembrane integrin-associated protein CD47 functions as a potent innate immunity checkpoint and is upregulated by many types of malignant cells, including melanoma during tumor progression. Binding of CD47 to its target receptor, SIRPα, on myeloid cell lineages leads to the initiation of the downstream signaling cascades that inhibit innate immunity anti-tumor responses. Molecular mechanisms underlying upregulation of CD47 during melanoma progression remain largely unknown. In this report, we performed ATAC-Sequencing on patient-derived melanoma cells, as well as, the analysis of ATAC-Seq datasets covering clinical melanoma samples to demonstrate a significant increase in chromatin accessibility for the CD47 promoter region in comparison to normal cells and tissues. Additionally, profiling of multiple CD47 transcript isoforms established that upregulation of CD47 in malignant cells occurs at the mRNA level. Using chromatin immunoprecipitation (ChIP) approaches along with the analysis of ChIP-Seq cancer datasets, we identified the transcription factor NRF-1 which binds at multiple sites within the proximal CD47 promoter region. In combination with serial deletions of CD47 promoter, we defined the minimal DNA region required for its activation, as well as, specific DNA locations within that region, which are preferentially occupied by NRF-1 in tumor cells.
Introduction
Understanding mechanisms leading to the acquisition of immune-evasive properties by malignant cells represents a critical step in the design of effective anti-cancer regimens. Previous studies by our group and others identified integrin-associated protein CD47 as the critical myeloid lineage checkpoint that is overexpressed by various types of tumor cells, including melanoma, and inhibits innate immunity-mediated anti-tumor responses (1–4). First discovered on the cell surface of circulating red blood cells (RBCs), CD47 is able to interact with the SIRPα receptor present on the macrophages to inhibit their phagocytic function, a phenomenon that later became known as the “don’t eat me” signal (5). Subsequent research on hematopoietic malignancies including acute myeloid leukemia (AML) and non-Hodgkin lymphoma (NHL) revealed that malignant cells hijack this mechanism to evade surveillance by an innate immunity during cancer progression (4, 6, 7). These results were later extended into the studies of the solid tumors, where it was shown that high levels of CD47 are associated with disease progression and/or poor patient prognosis, including epithelial (breast, ovarian, colon, and others) (8–10), mesenchymal (sarcomas) (11, 12), and neuronal/neural crest (GBM, Neuroblastoma, Melanoma) derived malignancies (2, 13). Importantly, past research by our group demonstrated that freshly resected metastatic human melanoma tumors are highly positive for CD47, while its blockade (by administering inhibitory antibodies) led to the significant suppression of melanoma metastases in-vivo using several independently patient-derived xenografts (PDXs) (2). Clinical significance of these findings has been translated into the development of therapeutic compounds blocking CD47 interaction with SIRPα in order to reinvigorate anti-tumor immune responses, with some of them reaching Phase I and II human trials (14, 15).
While the immune-suppressive role of CD47 on the cell surface of tumor cells has been characterized to a great extent, mechanisms underlying its regulation during human cancer progression are only beginning to emerge. These studies reveal substantial variability in the transcription factor machinery involved in the regulation of the CD47 promoter depending on the tumor type. Thus, it has been shown that in leukemias (both human and mouse) MYC oncogene binds to the distal enhancer regions of the CD47 promoter to regulate its activity (16), while HIF-1α and NFκB transcription factors have been implicated in the regulation of CD47 promoter in breast, cervical, and non-small lung carcinoma cells (17–19).
In this report, we find that CD47 upregulation in melanoma occurs at the mRNA level and is associated with changes in chromatin accessibility at the CD47 promoter region using the assay for transposase-accessible chromatin with sequencing (ATAC-seq). This leads to an increased CD47 mRNA production, as determined by real-time PCR covering multiple transcript variants of this gene. To reveal molecular factors regulating CD47 mRNA expression we performed an extensive analysis of the CD47 promoter region using the MotifMap algorithm. As a result, we identified multiple locations containing the binding consensus sequence for the Nuclear Respiratory Factor-1 (NRF-1) within the proximal promoter region of the CD47 gene. Furthermore, Chromatin Immunoprecipitation (ChIP) assays revealed NRF-1 to be bound at multiple proximal CD47 promoter sites with differential affinity among malignant and normal cell types leading to increased CD47 mRNA and protein levels. Lastly, using bioluminescence reporter systems, we were able to define the minimum CD47 promoter region and the number of NRF-1 binding sites required for its efficient activation.
Materials and methods
Cell culture
Melanoma cells M1626 and M727 were maintained in RPMI with 10% FBS, M525 cells were maintained in RPMI with 5% FBS, HepG2 cells were maintained in DMEM with 10% FBS, and M354 cells were maintained in 1:1 of RPMI with 10% FBS and dermal cell basal media from ATCC (PCS-200-030). Normal melanocytes were maintained in dermal cell basal media from ATCC (PCS-200-030). All cells were grown in Tissue Culture incubators set at 5% CO2 and 37°C degrees. For NRF1 siRNA mediated knockdown, a total of 1 × 105 M727 cells were seeded in 6 well plates and incubated overnight at 37 ° C and 5% CO2. Cells were then transfected using 8 μl of siRNA transfection reagent (Santa Cruz, Cat. No. sc-29528) and 120 pmol of NRF-1 siRNA (Cat. No. sc-38105) or control siRNA-A (Santa, Cruz, Cat. No. sc-37007).
Flow cytometry
Detection and quantification of CD47 cell surface protein levels were performed using FACS and an anti-human CD47 antibody (BioLegend, Cat. No. 323106). Briefly, cells were cultured in an exponential phase and harvested before reaching confluency. To harvest the cells, the spent medium was carefully aspirated, cells were washed with PBS, and detached using Accutase™ solution (STEMCELL, Cat. No.07922) for 3 min at 37°C in an incubator with 5% CO2. The cells were then collected and washed twice with PBS and resuspended in 100 μL of ice-cold cell staining buffer (CSB) (PBS, 1%BSA) at a concentration of 0.5-1x106cells/ml. Following resuspension, cells were stained with anti-human CD47-FITC antibody (1μg/1x106cells) and incubated on ice protected from light for 20 min. After incubation, the cells were centrifuged at 200g for 5 min, the supernatant was carefully aspirated, and the cells were washed twice with 200 μL of CSB. The cells were transferred to FACS tubes and analyzed in a BD FACSymphony A5 cell analyzer, at least 1x104 cells live cells were recorded per cell type. Dead cells were excluded with a DAPI staining solution (10μg/mL). Once flow cytometry analysis was completed, data was analyzed in FlowJo v10.10 software, live cells were gated, and the geometric mean fluorescence intensity (GMFI) was obtained for each cell type. GMFI values were normalized (same number of live events per cell type) and compared between cell lines as well as their respective histograms.
qRT-PCR
Cells were grown in-vitro on p100 cell culture dishes until reaching ~70% confluency after which they were lysed using 1ml of Trizole reagent. mRNA was precipitated using isopropanol and washed twice with 70% ethanol. cDNA was synthesized using a high-capacity cDNA reverse transcription kit (4368814) and power SYBR green PCR MasterMix (4367659 Thermo Fisher Scientific) was used to quantify total levels of CD47 or NRF-1 transcripts; 18s gene was used for the normalization of qPCR results. Primer pair sequences are listed in Supplementary Table 1.
ATAC-sequencing
For ATAC-Seq 50,000 cells were collected, washed in 50 μl of cold PBS, and resuspended in 50 μl of cold lysis buffer. Nuclei were isolated by centrifugation at 500 x g for 10 min at 4°C and then incubated with a transposition reaction mix for 30 min at 37°C. Following transposition, DNA was then purified using the MinElute PCR Purification kit (Qiagen, Hilden, Germany). The transposed DNA fragments were barcoded initially amplified for 5 cycles, and further amplified by standardized PCR. The resulting ATAC libraries were sequenced on an Illumina HiSeq 2500 platform in Rapid Mode with the 50-bp paired-end reads output mode. Raw reads were aligned to the human genome reference (1000 Genomes) using BWA-MEM (version 0.7.5a) with default settings. ATAC-Seq peaks were identified using the callpeaks function in MACS2 with a threshold set to -q = 0.01. The resulting peak calls were filtered for sequences that mapped to the mitochondria using shell scripts. BigWig files were normalized and integrated with publicly available genomic and epigenomic data for visualization in the UCSC Genome Browser.
ChIP assay
For the ChIP assay cells were prepared using truChIP Chromatin shearing kit (PN520127) from covaries. Briefly, cells were cultured in 2 x p150 dishes and treated with fixing buffer have 1% formaldehyde for 10 mins followed by quenching of crosslinking with a quenching buffer for 5 mins. Cells were lysed per the manufacturer’s protocol and isolated nuclei were transferred into AFA tubes (Cat# 520135 covaris) and chromatin shearing was performed for 15 mins in E220 evolution ultrasonicator from covaries with recommended settings. 50μl of sheared chromatin sample was used to isolate DNA (Supplementary Figure S2) using a DNA purification kit from cell signaling technologies (cat# 14209S) thus the total amount of DNA in sheared chromatin was calculated using nanodrop. For immunoprecipitation, the buffers (cat# 14231) and ChIP-grade magnetic beads (cat# 9006S) were purchased from cell signaling and used as per the manufacturer’s protocol. Briefly, 5-7μg of DNA was used per reaction and 2% input material was set aside for quantification using qPCR. Each reaction was incubated overnight with 2.5-3μg of NRF-1, or isotype control IgG antibody followed by 2 hours of incubation with magnetic beads the following day. Immunoprecipitation was performed using a magnetic rack. After successful elution of pulled-down chromatin, DNA was isolated, and real-time qPCR was performed using power SYBR green PCR master mix (cat# 4367659) from Thermo Fisher Scientific. Primer pair sequences are listed in Supplementary Table 1.
Generation of stable cell lines expressing CD47 promoter reporters
CD47 promoter reporters containing various numbers of NRF-1 binding sites were created by synthesizing respective DNA regions and cloning them into pGF1-4eCOL2A1_LUC lentiviral vector using BamHI and Spe1 restriction sites. Viral packaging was done in HEK293 cells and collected virions were used to infect target cells for two consecutive days. The CD47 promoter activity of endogenous NRF-1 was assayed by measuring Luciferase bioluminescence.
Datasets Used for In-Silico Analysis
TCGA TARGET GTEx study was used to analyze CD47 expression levels in human melanoma tissues. ENCFF323SMV, ENCFF862EDR, ENCFF605IET, ENCFF785WIA, ENCFF000XJF, ENCFF644ITQ from encodeproject.org studies were used to analyze NRF-1 ChIP seq data across different types of cancer. Patient-derived melanoma cell lines were analyzed for chromatin accessibility at the CD47 promoter region using the data from the study GSE134432. The ATAC-Seq data for the melanoma patient tissues was fetched from https://gdc.cancer.gov/about-data/publications/ATACseq-AWG. For in-silico H3K4Me3 analysis of patient-derived melanoma tissue from the database GSE33930, bowtie aligner was used to align the sequence reads, followed by the emoval of mitochondrial reads and duplicates using samtools. Macs2 was used to invoke callpeaks function to obtain peakfiles which were used to generate bigwig datafiles to be visualized in IGV.
Results
CD47 upregulation in melanoma occurs at the mRNA level and is mediated by an open chromatin at its DNA promoter region
We previously established that high levels of CD47 protein on freshly isolated human tumor cells are associated with melanoma progression and immune evasion (2). To understand whether augmented CD47 expression results from an increase in corresponding mRNA production, we used metastatic melanoma patient-derived cells M727, M354, and M1626 that were determined to express various levels of CD47 (Figures 1A, B) and compared them to normal melanocytes or hepatocellular carcinoma cells, HEPG2, (known for low levels of CD47 protein) using qRT-PCR. CD47 gene is comprised of 11 exons and as a result of splicing, most notably between exons 8-10, multiple mRNA isoforms are translated (Supplementary Figure 1A). Therefore, to prospectively quantify levels of all CD47 transcript isoforms we designed multiple primer sets for a common exon 2, as well as, two primer sets that amplify mRNA regions between exon 2-4 and exon 7-8 (Supplementary Figure 1B, Supplementary Table 1). After performing multiple qRT-PCR assays, our results demonstrate that all metastatic melanomas expressed 10-15 folds (depending on the isoform) elevation in CD47 mRNA as compared to normal melanocytes or HepG2 cells (Figure 1C). To further explore clinical significance of our findings, we investigated CD47 mRNA levels among 422 melanoma subjects in comparison to 123 normal skin subjects using TCGA TARGET GTEx dataset. Utilizing an unsupervised dimensional reduction approach, we generated UMAP graphs that visualized patterns of clusters across diagnosed subjects and levels of CD47 mRNA expression. Significantly, two distinct clusters were formed based on the CD47 mRNA levels, high and low, which corresponded to the malignant melanoma patients cluster (high) and the normal skin cluster (low) (Figure 1D).
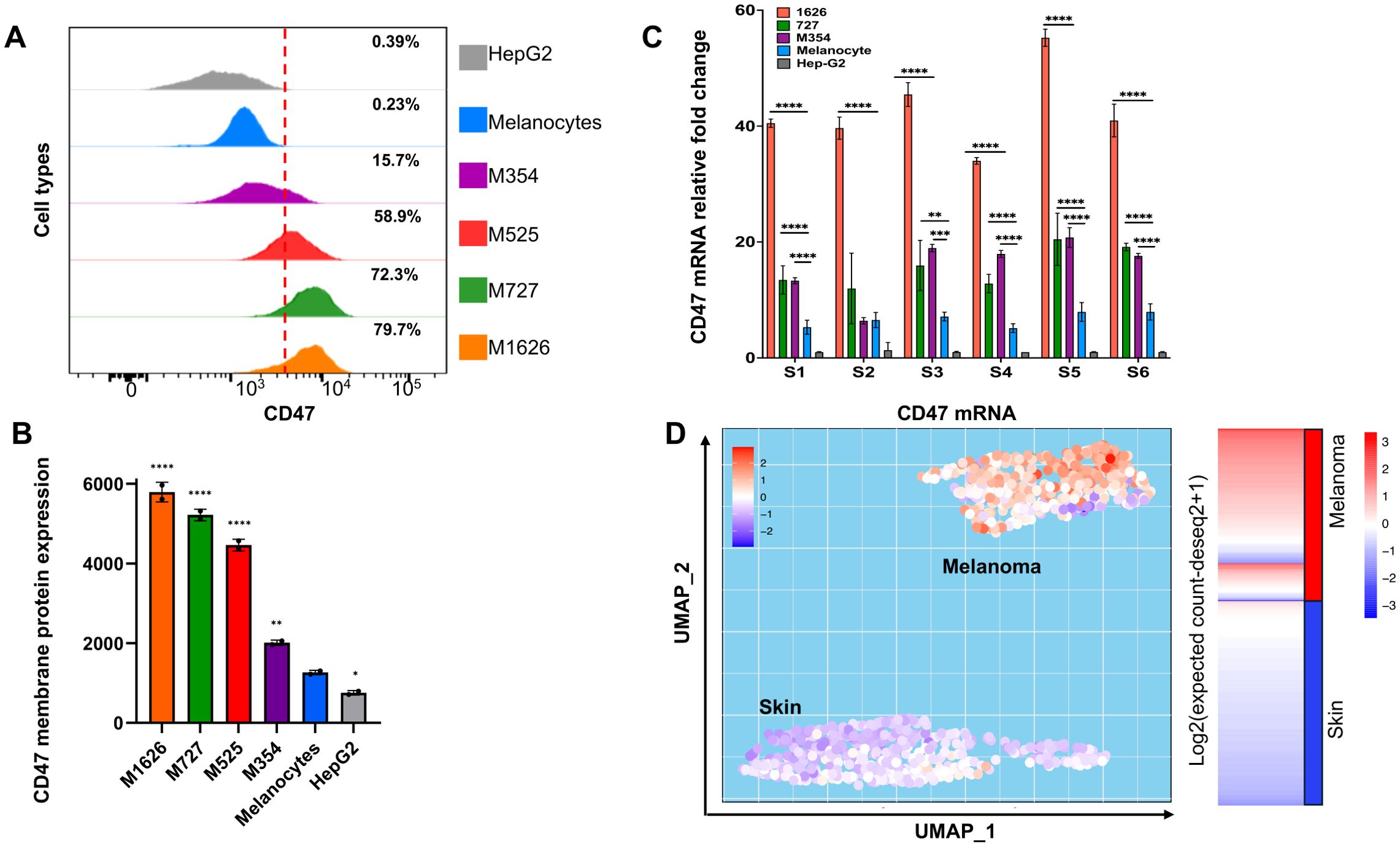
Figure 1. Upregulation of CD47 expression during melanomagenesis. (A) Flow cytometry analysis of CD47 expression in indicated cells. (B) Expression of CD47 protein on the membrane of indicated cells. CD47 expression is represented as normalized geometric mean fluorescent intensity (GMFI). (C) Relative expression of CD47 mRNA in fold changes comprehensively evaluated by six primer sets to capture all the CD47 mRNA isoforms abundance levels. (D) Unsupervised dimensional reduction analysis of a dataset from TCGA TARGET GTEx study was performed to visualize the distinctive clusters and expression of CD47 in melanoma and normal skin samples. The transcript matrix was processed in R studio package called “umap” for UMAP generation. Samples with higher CD47 in melanoma as seen on the heatmap represent the “Melanoma cluster”, and samples with lower CD47 expression represent the “Skin” cluster in UMAP.
To understand molecular mechanisms of augmented CD47 mRNA expression in malignant melanoma, we investigated the dynamics of chromatin re-modeling at CD47 promoter using ATAC-Seq. This approach allows the generation of a cell-specific chromatin accessibility landscape to determine the level of open chromatin and subsequent promoter activation. First, we used in-silico ATAC-Seq analysis of the skin cancer melanoma (SKCM) dataset (https://gdc.cancer.gov/about-data/publications/ATACseq-AWG) containing malignant tissues obtained from 13 melanoma subjects (11 samples were analyzed in duplicates) and a melanoma dataset (GSE134432) containing 9 genetically homogenous melanoma patient-derived cell lines to gain insights into the accessibility of the CD47 promoter region. Our results demonstrate that in both freshly resected melanoma samples, as well as, in melanoma patient-derived cell lines, the DNA region corresponding to the CD47 promoter has a significantly increased chromatin accessibility which could be conjectured as a sign of active mRNA transcription (Figure 2A). To validate these changes in the chromatin landscape of the CD47 promoter region during melanomagenesis, we performed ATAC-seq on multiple independent patient-derived melanoma cell lines in comparison to normal adult melanocytes. Importantly, we were able to detect strong peak signals associated with an open chromatin structure around the promoter of CD47 in malignant melanomas, which were missing in normal adult melanocytes indicating the closed chromatin structure of the CD47 DNA promoter region in non-transformed cells (Figure 2B).
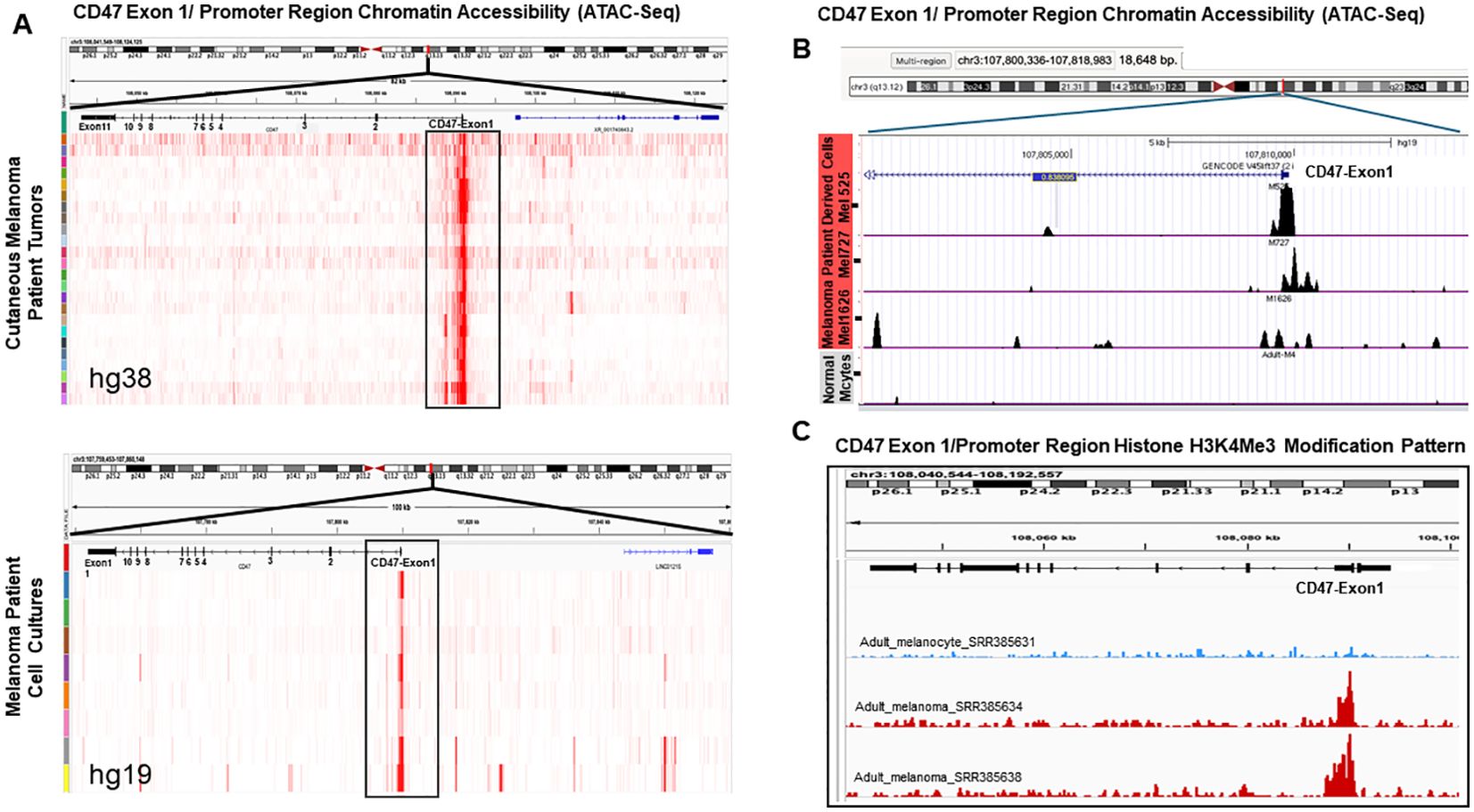
Figure 2. Changes in chromatin landscape of CD47 promoter region in patient melanomas and patient-derived melanoma cell lines. (A) Analysis of ATAC-seq peak signals around the CD47 DNA promoter region in surgically removed melanoma patient tumors and established cell lines using SKCM and GSE134432 databases. (B) Analysis of CD47 promoter region chromatin structure using ATAC-Seq approach on independent patient-derived melanoma cell lines (M727, M1626, M525) and normal melanocytes. (C) Analysis of H3K4Me3 histone modification mark at the CD47 promoter DNA region using melanoma patient samples and melanocytes from the database GSE33930. Peaks were called using Macs2 and peak files in the Bigwig format were visualized in the IGV 2.7 version.
Open chromatin status is often associated with specific histone modifications such as H3K4Me3, which is commonly found at the promoter regions of actively transcribed genes (20). To determine whether this mechanism can account for an increased CD47 mRNA production during melanomagenesis, we performed in-silico analysis of H3K4Me3 histone modifications at the CD47 promoter in melanoma patients and normal adult melanocytes using the GSE33930 dataset. Our results demonstrate that peak signals for H3K4Me3 DNA modification at the CD47 promoter region are significantly stronger in malignant melanomas in comparison to normal melanocytes (Figure 2C).
Nuclear respiratory factor-1 TF binds to the CD47 promoter with preferential affinity to the most proximal region in melanoma
To identify transcriptional regulators of the CD47 promoter we analyzed its DNA region (± 1000bp relative to the transcription start site (TSS)) for the presence of potential TF binding sites using MotifMap and the Eukaryotic Promoter Databases. Results ranking based on FDR (< 0.038) and p-value (< 0.001) scores revealed a number of DNA binding sites within that region (-63bp, -28bp, -22bp, -14bp, and +32bp), all corresponding to the NRF-1 consensus sequence (Figure 3A). To validate predicted interactions of NRF-1 with CD47 promoter we first performed in-silico analysis of ChIP-seq datasets derived from different types of cancer. Importantly, strong peak signals corresponding to NRF-1 binding at the CD47 promoter were identified in the majority of tumors (results presented as fold changes over control), except for HepG2 hepatocarcinoma (Figure 3B), which was found to express low CD47 protein and mRNA transcript levels (Figure 1).
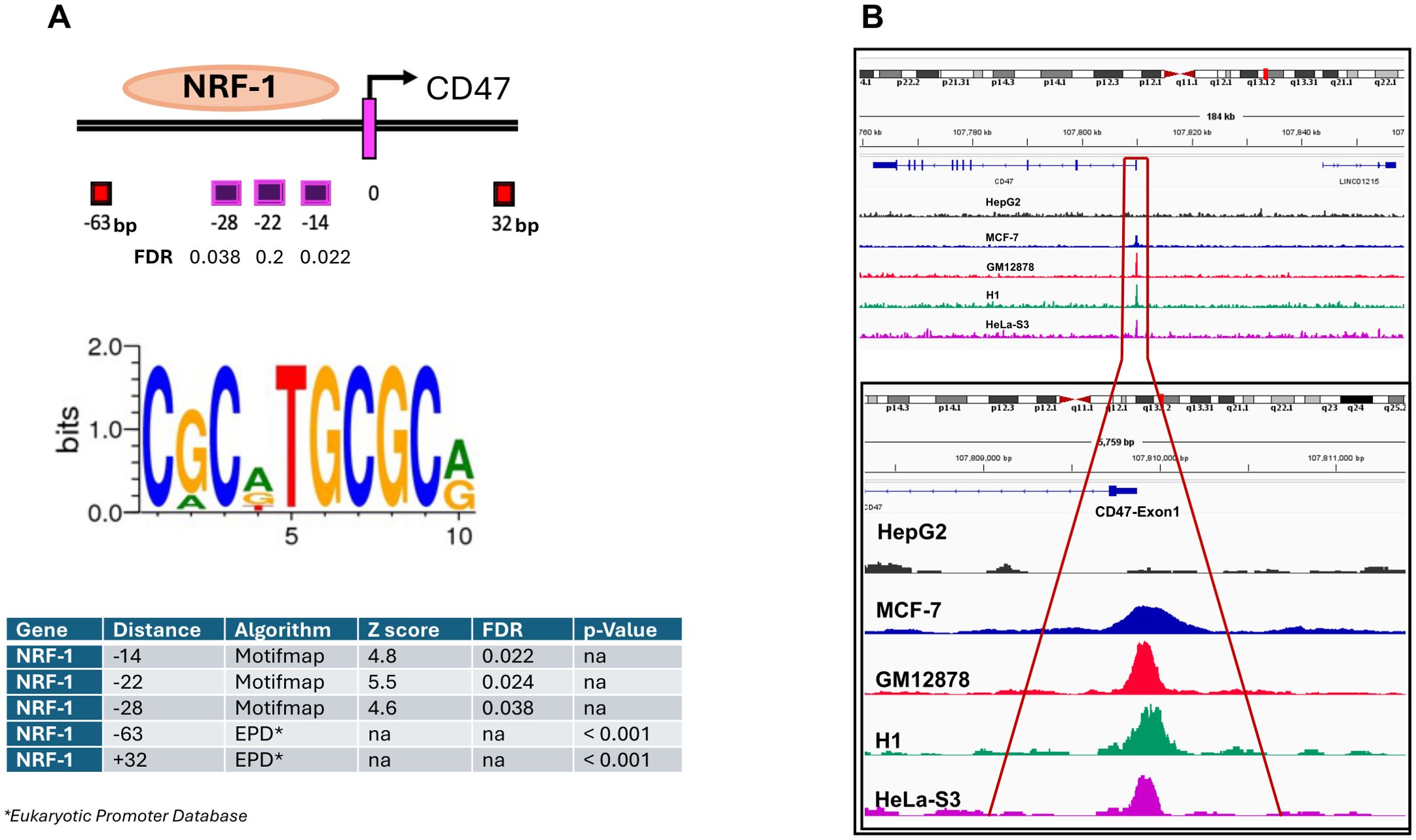
Figure 3. Identification of NRF-1 TF as the regulator of CD47 mRNA expression in melanoma. (A) Numerous NRF-1 TF binding sites were predicted and identified in the proximal region of the CD47 Exon1 DNA Region. The consensus sequence required for the NRF-1 TF binding was extracted using the Motifmap algorithm. (B) Analysis of publically available ChIP-seq datasets for indicated cancer types reveals NRF-1 binding at the CD47 Exon1 promoter region.
Next, to determine NRF-1 regulation of CD47 promoter, we performed Chromatin Immunoprecipitation (ChIP) assays on malignant patient-derived melanoma cells, as well as, adult normal melanocytes and HepG2 cells. Following incubation of the sheared chromatin (500-1000bp) with NRF-1 Ab the pulled-down DNA fragments were quantified by PCR using primer sets designed to amplify only the CD47 promoter regions spanning different putative NRF-1 binding sites (Figure 4A). To precisely delineate NRF-1 DNA binding regions within the CD47 promoter, we designed multiple primer pairs to enrich for the following sites relative to TSS: Set 1 (-38/+28) includes putative binding sites at -28, -22, and -14bp; Set 2, (-38/+56) includes putative binding sites -28, -22, -14, and +32; Set 3 (-63/-36) includes putative binding site at position -63bp only (Figure 4A, Supplementary Table 1). An additional set of primer spanning the promoter region of MEF2A gene (a validated NRF-1 binding target) was used as a positive control for all ChIP experiments (Supplementary Figure 2). ChIP analysis on the above indicated cell types identified NRF-1 loading at the predicted DNA region of the CD47 promoter with differential affinity for proximal and distal binding sites (Figure 4B). Thus, in malignant melanomas there was a significant increase in binding affinity at the most proximal sites (-28, -22, -14) as compared to normal melanocytes (1.7-fold for M727 and 2.4-fold for M1626). Moreover, when compared to HepG2 cells, which have been characterized by the lowest levels of CD47 mRNA and protein, differences in NRF-1 binding affinity were found to be even greater (1.85-fold for M727 and 3.8-fold for M1626 Figure 4B middle panel). Inclusion of the proximal downstream binding site (+32bp) resulted in similar differences in the NRF-1 binding affinity between malignant melanoma and low CD47 expressing cells (adult melanocytes and HepG2 cells) (Figure 4B right panel). However, differences in NRF-1 binding affinity were diminished at the more distal binding site (-63bp) within the CD47 promoter region (Figure 4B left panel). In summary, these findings indicate that CD47 transcriptional regulation by NRF-1 is determined by the proximal promoter region in melanoma cells.
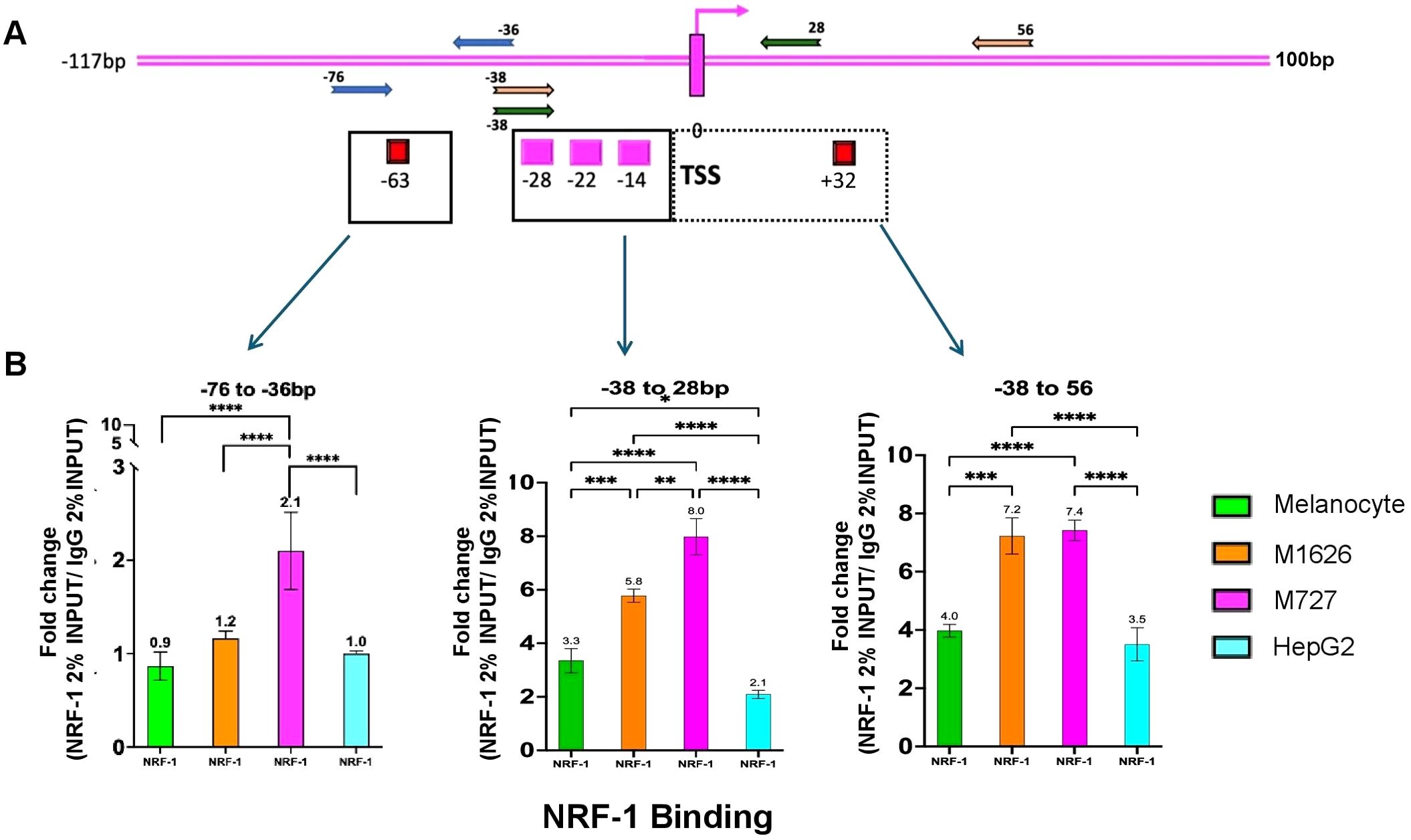
Figure 4. NRF-1 TF differentially binds at the proximal CD47 promoter region spanning between -38 to +56bp relative to TSS. (A) Various sets of primers to target and enrich indicated regions of predicted NRF-1 binding at CD47 promoter. (B) ChIP assays demonstrating differential affinity binding of NRF-1 at predicted DNA regions of CD47 promoter. The strongest affinity for melanoma cells was determined for binding sites located between the -38 to 56bp region of the CD47 promoter.
Effective activation of CD47 promoter requires multiple NRF-1 binding sites between -120bp to +50bp promoter region
Initiation of transcription from eukaryotic promoters often requires binding of the TFs at numerous locations within the promoter region. Identification and validation of multiple NRF-1 TF binding sites within the proximal region to the CD47 TSS (200 bp window) prompted us to investigate the minimum adequate length of DNA required for the CD47 promoter activation. To achieve this goal, we designed and cloned a series of bioluminescent reporter plasmids expressing Luciferase gene under the control of the proximal CD47 promoter region containing various numbers of NRF-1 TF binding sites (Figure 5A). Specifically, construct 1 contained all five NRF-1 proximal binding sites and covered the DNA region from -120 to +50bp relative to the TSS, construct 2 contained four out of five NRF-1 binding sites and covered the region from -70 to +30 bp, and lastly construct 3 contained only three out of five NRF-1 binding sites and covered the region from -62 to +30bp of CD47 promoter relative to the TSS. Lastly, we also used a complete promoter deletion variant of Luciferase expressing plasmid as a negative control (construct 4 Figure 5A). All of the above variants of the CD47 promoter region were synthesized and cloned into the lentiviral plasmid pGF1-4eCOL2A1_Luc (21) using BamHI and Spe1 restriction sites. In order to evaluate endogenous NRF-1 activity at the CD47 promoter, we generated melanoma cell lines to stably express all versions of Luc reporters using previously published lentiviral transduction protocols (22, 23). To obtain statistically significant results, raw Luc bioluminescence values were first normalized to the viral copy numbers (VCN) for each reporter plasmid. Briefly, serial dilutions of the known concentration of each Luc reporter plasmid were premixed with 5ng of carrier genomic DNA (isolated from each of the above-indicated cell lines), and the real-time PCR was performed using Luciferase primers. Obtained CT values were used to generate a standard curve which allowed to calculate the VCN values for the known concentration of plasmids according to the given formula:
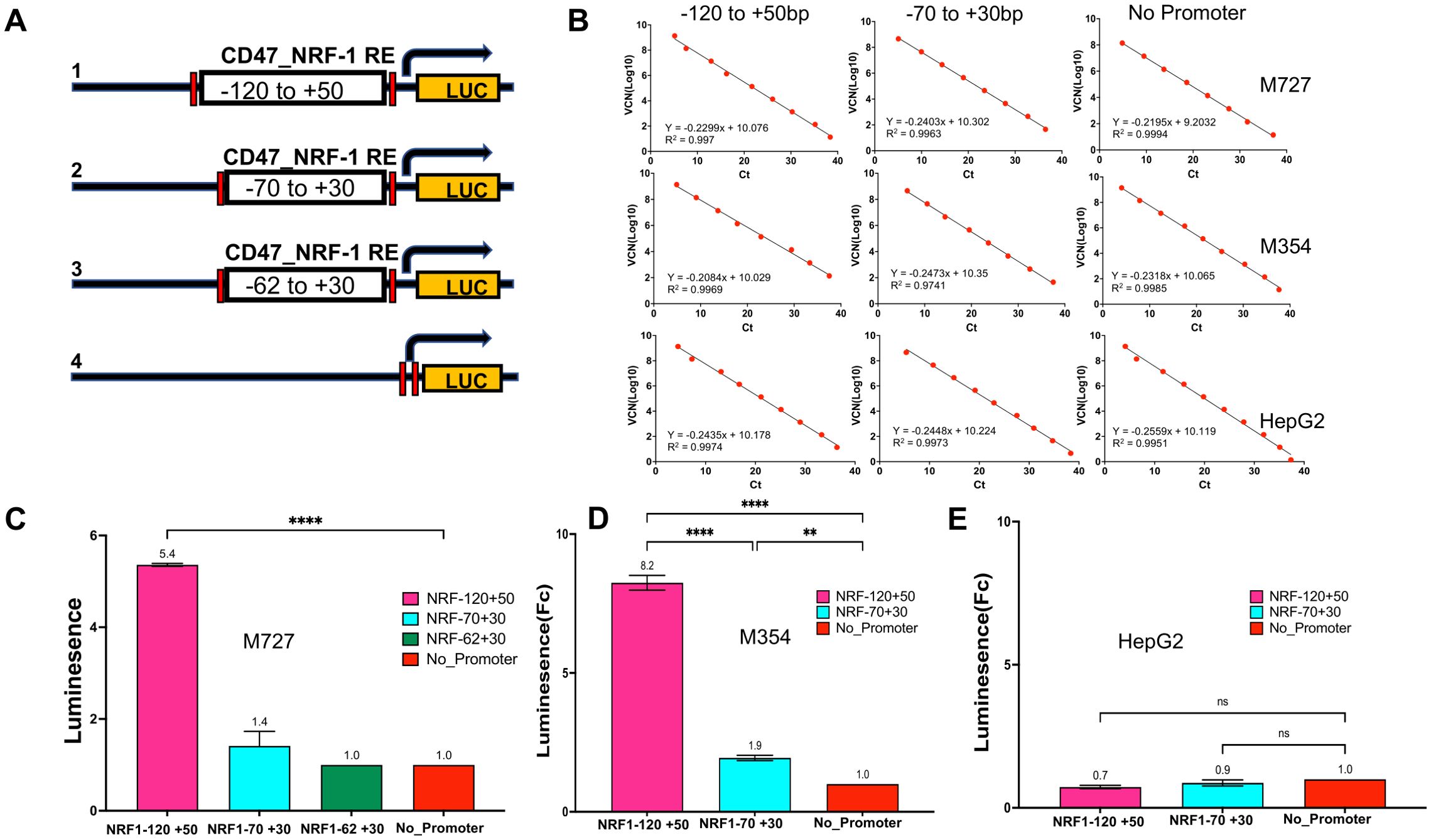
Figure 5. Activation of CD47 promoter requires DNA region containing multiple NRF-1 binding sites located between -120bp to +50bp relative to TSS. (A) A series of Luciferase reporter constructs harboring different lengths of CD47 promoter region were generated to assay the NRF-1 mediated CD47 promoter activation. (B) Luciferase signals were read using a 96-well plate reader and normalized to viral copy number (VCN) and total protein. VCNs were calculated for each plasmid construct for every cell line separately using serial dilutions of plasmids to generate a linear regression curve with a slope equation that was further used to calculate the VCN of samples. (C–E) Relative CD47_NRF1 Luc reporter assay signal in fold changes for M727, M354 and HepG2 cells. A construct with no CD47 promoter region was used as a negative control.
Thus obtained linear regression curves allowed us to calculate viral copy numbers for an equal amount of genomic DNA for each individual cell line (Figure 5B). Moreover, raw bioluminescence values were also normalized to the amount of total protein present in each lysate. Normalized data demonstrated that initiation of CD47 transcription requires the presence of all five NRF-1 DNA binding sites (Figure 5C). Thus, Luc activity was induced 5-8 fold in melanoma cells containing full-length proximal CD47 promoter (Construct 1) as compared to low CD47 expressing HepG2 cells (Figure 5C), while CD47 promoter region containing a reduced number of NRF-1 binding elements (Constructs 2 and 3) had marginal or no effect on reporter activity in the same cells (Figures 5C–E). In conclusion, our data demonstrates that the promoter region required for efficient CD47 transcriptional activation in melanoma lies between -120 to +50bp relative to the TSS.
NRF-1 activity directly contributes to the CD47 expression in melanoma
To demonstrate functional significance of NRF1 TF in the regulation of CD47 we tranduced melanoma cells expressing CD47_Luc promoter reporter (Construct 1 described above) with siRNAs (a pool of 3) targeting NRF1 mRNA to achieve its downregulation (Figure 6A). Subsequent quantification of Luc signal revealed a significant reduction in CD47 promoter activity (by more than 60% (p=0.0149)) in cells transduced with NRF1 siRNA as compared to matching controls transduced with SC siRNA (Figure 6B). Next, we profiled these cells for an endogenous CD47 mRNA levels using qRT-PCR and multiple sets of primers corresponding to its various regions. Our data demonstrates that NRF1 downregulation causes at least 50% decrease in CD47 mRNA expression in target cells (p=0.0245 - 0.00361) (Figure 6C). Lastly, we used FACS and fluorescent conjugated antibodies to quantify CD47 protein on the cell membrane, critical aspect of its immune modulatory function during tumor progression. Our analysis demonstrates that NRF1 downregulation leads to the significantly reduced proportion of melanoma cells expressing high levels of this innate immunity checkpoint when compared to the original cell population trasduced with control SC-siRNA: 7.04% CD47high//91.8% CD47low for NRF1-siRNA and 34.3% CD47hig//64.1% CD47low for SC-RNAi transduced cells (Figure 6D). In summary, the above described results provide strong experimental evidence to indicate that NRF1 is directly involved in regulation of CD47 expression in melanoma.
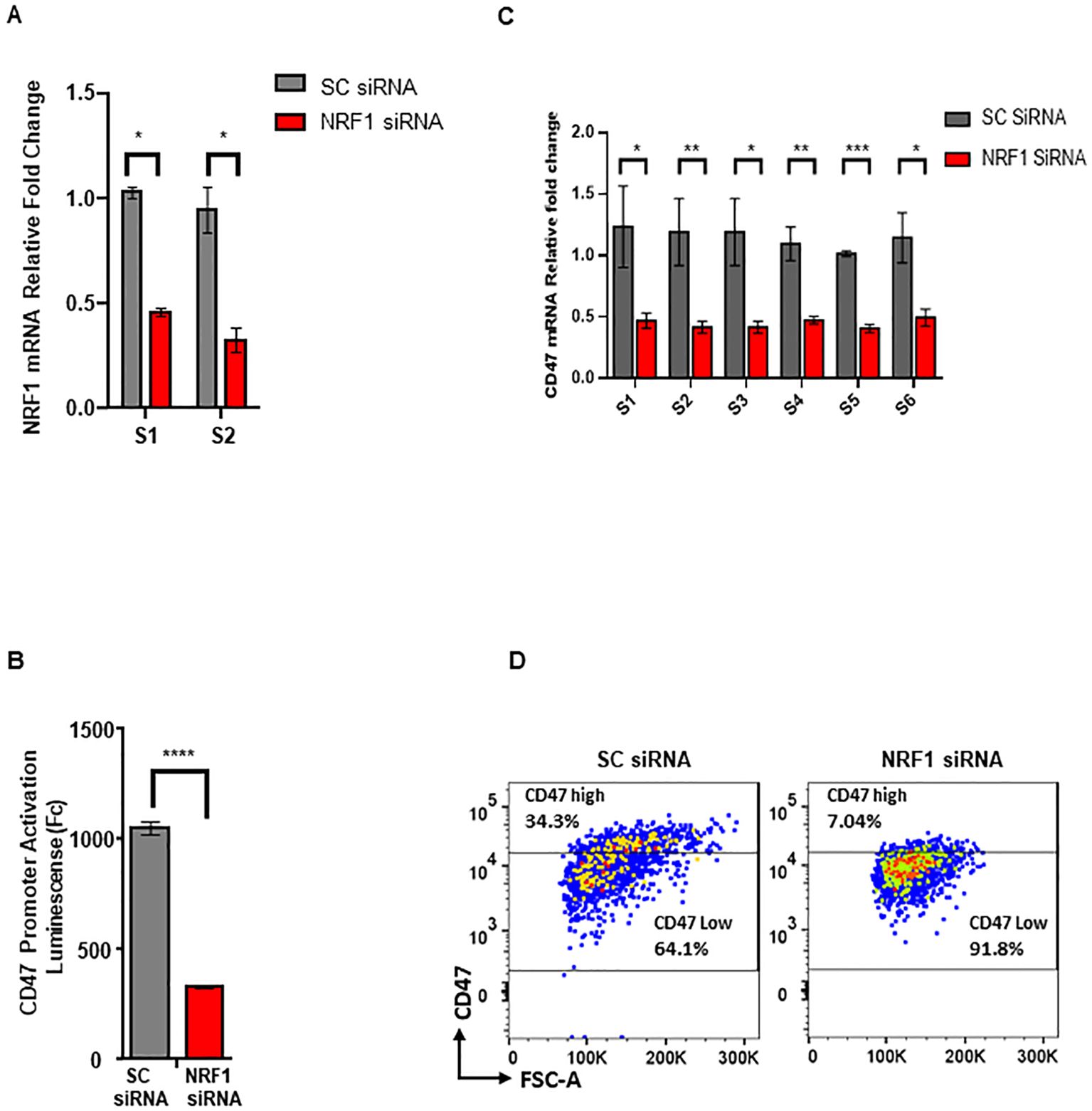
Figure 6. NRF-1 downregulation reduces expression of CD47 in melanoma. (A) Quantification of NRF1 mRNA following siRNA mediated knockdown in melanoma cells using qRT-PCR and two independent sets of primers, S1 and S2. (B) Quantification of CD47 prmoter activation using Luc reporter assay in the cells transduced with NRF1_siRNA or control, SC_siRNA. (C) Quantification of CD47 mRNA following siRNA mediated NRF1 knockdown in melanoma cells using qRT-PCR and six independent sets of primers, S1- S6. (D) Analysis of cell surface CD47 protein levels using FACS for cell populations transduced with NRF1_siRNA or control, SC_siRNA.
Discussion
Cluster of Differentiation 47 (CD47), also known as Integrin Associated Protein (IAP), is a transmembrane protein belonging to the immunoglobulin superfamily (Ig) which regulates a variety of cell processes including, cell adhesion, motility and apoptosis (24–26). It gained considerable attention in the past decade after its function as a potent innate immunity checkpoint regulator was characterized first for hematopoietic malignancies and later for solid tumors, including melanoma (2, 27–29). Interaction of CD47 with the signal regulatory protein-alpha (SIRPα) receptor expressed on many myeloid derived cell lineages activates molecular pathways effectively inhibiting an anti-tumor function of innate immunity (30). Upregulation of CD47 by tumor cells in order to avoid immune recognition and elimination is thus regarded as one of the critical steps during carcinogenesis. Therefore, revealing mechanisms involved in the regulation of CD47 expression represents a pivotal task in our understanding of tumor progression and immune evasion. Importantly, highly diverse mechanisms of malignant transformation and cancer progression can result in different modes of CD47 regulation depending on the tissue type of tumor origin. For example, NFκB TF has been shown to activate CD47 expression by binding to the distal enhancer elements in cells derived from breast, cervical, and non-small lung cell carcinomas (18, 19, 31). Other oncogenic TFs have also been reported to affect CD47 expression directly as a part of an oncogene-driven process of malignant transformation. Thus, hypoxia inducible TF, HIF-1α, binds to the CD47 promoter region at -239bp and -200bp relative to the TSS and induces its expression in response to hypoxic conditions in-vitro in MCF7 cells (17). Other studies implicated MYC, a well-characterized TF with strong oncogenic potential, in the transcriptional regulation of CD47 during the progression of leukemia using MYC-induced T cell acute lymphoblastic leukemia mouse model; MYC T-ALL, and human leukemia cells lines; CCRF-CEM and Jurkat (16). Interestingly, binding sites for MYC TF were also identified in our analysis of CD47 proximal promoter region (Supplementary Figure 3A), however, MYC binding was undetectable in human malignant melanoma within CD47 DNA promoter region between -120bp to +54 relative to TSS required for its activation (Supplementary Figure 3B).
In our present work, we used numerous clinical human melanoma samples, as well as, previously collected pathological datasets of disease progression to establish that the tumor-specific upregulation of myeloid checkpoint molecule, CD47, occurs at the mRNA level during melanomagenesis. We reveal that it is mediated by a significant increase in chromatin accessibility at CD47 prmoter region, including elevated levels of epigenetic histone modifications at H3K4Me3 residues, in melanoma but not in normal melanocytes or HepG2 cells which are known for low CD47 expression. Using serial DNA deletion analysis in combination with bioluminescence reporter systems, we determine that the minimum promoter region required for CD47 transcriptional activation in melanoma lies between -120 to +50bp relative to the TSS. Furthermore, we identified NRF-1-TF as a specific transcriptional regulator of CD47 promoter, which was found to be physically associated with multiple binding sites at the promoter region using ChIP approaches. Our data demonstrate that NRF-1 binding to the most proximal sites at -28, -22, and -14bp sites relative to the TSS appears to be a critical step during CD47 promoter activation which distinguishes it from the cells of low CD47 levels. This indicates that the basal level of CD47 transcription can be initiated by NRF-1 binding to the distal regions of the CD47 promoter starting at -63bp (which is detected for all cell types); however, upregulation in CD47 mRNA synthesis occurs only in response to NRF-1 occupying additional proximal sities indicated above. Importantly, downregulation of NRF1 causes notable reduction of CD47 mRNA (>50%) and as a result, significant decrease in melanoma cell poolations with high CD47 levels. At the same time CD47 expression was not completely abolished in response to NRF1 downregulation. pointing to the fact that other TFs mentioned above (such as NFkB and HIF1a) whose biding sites are identified within CD47 promoter/enchancer regions can contribute to its expression. Further studies focused on combinatorial approaches to simultaneously modulate activity of NRF1 along with other TFs mentioned above at the CD47 prmoter/enchancer DNA regions will be required to precisely delineate each TF contribution to the activity of CD47 promoter overall.
Under normal conditions, NRF-1 TF plays a vital role in maintaining mitochondrial biogenesis, including oxidative phosphorylation, by transcribing multiple proteins involved in processes regulating biogenesis such as TFB1M, TFB2M, TFAM, and cytochrome c (32, 33). However, NRF-1 has also been implicated in the process of malignant transformation based on its role in energy metabolism. Specifically, studies using human hepatocellular carcinoma and colorectal tumor tissues show the significance of upregulated NRF-1 protein levels (mediated by neutrophil extracellular trap formation) in inducing mitochondrial biogenesis promoting tumor growth and metastasis (34). In addition, NRF-1 activity has also been associated with estrogen-induced breast carcinogenesis, and renal cell carcinoma where it regulates the expression of the TFE3 gene required for cellular energy metabolism during proliferation (35, 36).
In our present study, we characterized a novel role of NRF-1 in melanomagenesis as a regulator of the major innate immunity checkpoint, CD47, in tumor cells. Importantly, binding of Pal/NRF-1 to the CD47 promoter was also previously reported in human neuroblastoma and hepatoma cell lines (37), as well as, during arising resistance to the oncogenic B-RAF molecular inhibitors (38). These findings open an opportunity for the development of modulatory compunds against NRF1 and functional testing of this TF involvment in an immune evasion by tumor cells during cancer progression. Clinical significance of these approaches can further be tested both in-vitro and in-vivo in combination with already established immunotherapeutic agents, such as PD1/PDL1/CTLA4 blocking antibodies, commonly used for the treatment of metastatic disease. Malignant transformation is a complex and multifaceted process that combines changes in both metabolism and immune recognition within cancer cells that successfully evolve to give rise to the more aggressive and metastatic tumors. NRF-1 TF therefore represents a key nodal point in this process due to its unique ability to transcriptionally regulate genes underlying both mitochondrial biogenesis and evasion of an innate immunity.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: ENCFF323SMV, ENCFF862EDR, ENCFF605IET, ENCFF785WIA, ENCFF000XJF, ENCFF644ITQ from encodeproject.org. GSE33930, GSE134432 from gene expression omnibus.
Ethics statement
The studies involving humans were approved by Cedars Sinai Office of Research Compliance and Quality Improvement. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
KM: Conceptualization, Data curation, Formal analysis, Methodology, Investigation, software, Visualization, Project administration, Writing – original draft, Writing – review & editing. EV: Formal analysis, Visualization, Methodology, Writing – review & editing. DM: Methodology, Resources, Writing – review & editing. BS: Methodology, Resources, Writing – review & editing. NB: Resources, Writing – review & editing. MF: Methodology, Resources, Writing – review & editing. OH: Methodology, Resources, Writing – review & editing. AB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by U.S. National Institutes of Health/National Cancer Institute Grant R01CA234892 to AB and funds from the Cedars-Sinai Medical Center to AB.
Conflict of interest
MF is the member of the following advisory board committees : Merck, Bristol Myers Squibb, Replimune, Novartis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1495032/full#supplementary-material
Supplementary Figure 1 | Primer design for CD47 real-time qPCR assay. (A) Schematic representation of different isoforms for the human CD47 mRNA. (B) primer sets (6) that were used to quantify an abundance of indicated CD47 mRNA isoforms.
Supplementary Figure 2 | MEF2A gene promoter used as a positive control of anti-NRF-1 ChIP assays.
Supplementary Figure 3 | Evaluation of MYC binding at the proximal CD47 promoter region in melanoma. (A) MYC binding sites on the CD47 region as predicted by Eukaryotic Promoter Database. (B, C) MYC ChIP on CD47 promoter in melanoma cells M1626 and hepatocarcinoma cells HepG2. (D) MYC ChIP on Nmp-1 promoter (positive control) in M1626 and HepG2 cells.
References
1. Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra S, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci USA. (2012) 109:6662–7. doi: 10.1073/pnas.1121623109
2. Ngo M, Han A, Lakatos A, Sahoo D, Hachey SJ, Weiskopf K, et al. Antibody therapy targeting CD47 and CD271 effectively suppresses melanoma metastasis in patient-derived xenografts. Cell Rep. (2016) 16:1701–16. doi: 10.1016/j.celrep.2016.07.004
3. Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. (2009) 138:271–85. doi: 10.1016/j.cell.2009.05.046
4. Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. (2009) 138:286–99. doi: 10.1016/j.cell.2009.05.045
5. Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. (2000) 288:2051–4. doi: 10.1126/science.288.5473.2051
6. Eladl E, Tremblay-LeMay R, Rastgoo N, Musani R, Chen W, Liu A, et al. Role of CD47 in hematological Malignancies. J Hematol Oncol. (2020) 13:96. doi: 10.1186/s13045-020-00930-1
7. Chao MP, Tang C, Pachynski RK, Chin R, Majeti R, Weissman IL. Extranodal dissemination of non-Hodgkin lymphoma requires CD47 and is inhibited by anti-CD47 antibody therapy. Blood. (2011) 118:4890–901. doi: 10.1182/blood-2011-02-338020
8. Manna PP, Frazier WA. CD47 mediates killing of breast tumor cells via Gi-dependent inhibition of protein kinase A. Cancer Res. (2004) 64:1026–36. doi: 10.1158/0008-5472
9. Liu R, Wei H, Gao P, Yu H, Wang K, Fu Z, et al. CD47 promotes ovarian cancer progression by inhibiting macrophage phagocytosis. Oncotarget. (2017) 8:39021–32. doi: 10.18632/oncotarget.16547
10. Zhang Y, Sime W Fau - Juhas M, Juhas M Fau - Sjölander A, Sjölander A. Crosstalk between colon cancer cells and macrophages via inflammatory mediators and CD47 promotes tumour cell migration. Eur J Cancer. (2013) 49:3320–34. doi: 10.1016/j.ejca.2013.06.005
11. Benesova I, Capkova L, Ozaniak A, Pacas P, Kopeckova K, Galova D, et al. A comprehensive analysis of CD47 expression in various histological subtypes of soft tissue sarcoma: exploring novel opportunities for macrophage-directed treatments. J Cancer Res Clin Oncol. (2024) 150:134. doi: 10.1007/s00432-024-05661-1
12. Dancsok AR, Gao D, Lee AF, Steigen SE, Blay JV, Thomas DM, et al. Tumor-associated macrophages and macrophage-related immune checkpoint expression in sarcomas. Oncoimmunology. (2020) 9:1747340. doi: 10.1080/2162402X.2020.1747340
13. Theruvath J, Menard M, Smith BAH, Linde MH, Coles GL, Dalton GN, et al. Anti-GD2 synergizes with CD47 blockade to mediate tumor eradication. Nat Med. (2022) 28:333–44. doi: 10.1038/s41591-021-01625-x
14. Zhang W, Huang Q, Xiao W, Zhao Y, Pi J, Xu H, et al. Advances in anti-tumor treatments targeting the CD47/SIRPα Axis. Front Immunol. (2020) 11:18. doi: 10.3389/fimmu.2020.00018
15. Kaur S, Cicalese KV, Bannerjee R, Roberts DD. Preclinical and clinical development of therapeutic antibodies targeting functions of CD47 in the tumor microenvironment. Antib Ther. (2020) 3:179–92. doi: 10.1093/abt/tbaa017
16. Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. (2016) 352:227–31. doi: 10.1126/science.aac9935
17. Zhang H, Lu H, Xiang L, Bullen JW, Zhang C, Samanta D, et al. HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc Natl Acad Sci U.S.A. (2015) 112:E6215–6223. doi: 10.1073/pnas.1520032112
18. Liu F, Dai M, Xu Q, Zhu X, Zhou Y, Jiang S, et al. SRSF10-mediated IL1RAP alternative splicing regulates cervical cancer oncogenesis via mIL1RAP-NF-κB-CD47 axis. Oncogene. (2018) 37:2394–409. doi: 10.1038/s41388-017-0119-6
19. Zhang X, Wang Y, Fan J, Chen W, Luan J, Mei X, et al. Blocking CD47 efficiently potentiated therapeutic effects of anti-angiogenic therapy in non-small cell lung cancer. J Immunother Cancer. (2019) 7:346. doi: 10.1186/s40425-019-0812-9
20. Igolkina AA-O, Zinkevich A, Karandasheva KO, Popov AA, Selifanova MV, Nikolaeva D, et al. H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 histone tags suggest distinct regulatory evolution of open and condensed chromatin landmarks. Cells. (2019) 8:1034. doi: 10.3390/cells8091034
21. De la Vega RE, Scheu M, Brown LA, Evans CH, Ferreira E, Porter RM. Specific, sensitive, and stable reporting of human mesenchymal stromal cell chondrogenesis. Tissue Eng Part C Methods. (2019) 25:176–90. doi: 10.1089/ten.TEC.2018.0295
22. Boiko AD, Porteous Razorenova OV, Krivokrysenko VI, Williams BR, Gudkov AV. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. (2006) 20:236–52. doi: 10.1101/gad.1372606
23. Li C, Nguyen V, Clark KN, Zahed T, Sharkas S, Filipp FV, et al. Down-regulation of FZD3 receptor suppresses growth and metastasis of human melanoma independently of canonical WNT signaling. Proc Natl Acad Sci U.S.A. (2019) 116:4548–57. doi: 10.1073/pnas.1813802116
24. Polara RA-O, Ganesan RA-O, Pitson SA-O, Robinson NA-O. Cell autonomous functions of CD47 in regulating cellular plasticity and metabolic plasticity. Cell Death Differ. (2024) 31:1255–66. doi: 10.1038/s41418-024-01347-w
25. Sick E, Jeanne A, Schneider C, Dedieu S, Takeda K, Martiny L. CD47 update: a multifaceted actor in the tumour microenvironment of potential therapeutic interest. Br J Pharmacol. (2012) 167:1415–30. doi: 10.1111/j.1476-5381.2012.02099.x
26. Oldenborg PA. CD47: A cell surface glycoprotein which regulates multiple functions of hematopoietic cells in health and disease. ISRN Hematol. (2013) 2013:614619. doi: 10.1155/2013/614619
27. Huang CY, Ye ZH, Huang MY, Lu JJ. Regulation of CD47 expression in cancer cells. Transl Oncol. (2020) 13:100862. doi: 10.1016/j.tranon.2020.100862
28. Jiang Z, Sun H, Yu JA-O, Tian W, Song Y. Targeting CD47 for cancer immunotherapy. J Hematol Oncol. (2021) 14:180. doi: 10.1186/s13045-021-01197-w
29. Stirling ER, Terabe M, Wilson AS, Kooshki M, Yamaleyeva LM, Alexander-Miller MA, et al. Targeting the CD47/thrombospondin-1 signaling axis regulates immune cell bioenergetics in the tumor microenvironment to potentiate antitumor immune response. J Immunother Cancer. (2022) 10:e004712. doi: 10.1136/jitc-2022-004712
30. van Duijn A, van der Burg SH, Scheeren FA. CD47/SIRPα axis: bridging innate and adaptive immunity. J Immunother Cancer. (2022) 10. doi: 10.1136/jitc-2022-004589
31. Betancur PA. et al. A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat Commun. (2017) 8. doi: 10.1038/ncomms14802
32. Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann N Y Acad Sci. (2008) 1147:321–34. doi: 10.1196/annals.1427.006
33. Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. (2005) 25:1354–66. doi: 10.1128/MCB.25.4.1354-1366.2005
34. Yazdani HO, Roy E, Comerci AJ, van der Windt DJ, Zhang H, Huang H, et al. Neutrophil extracellular traps drive mitochondrial homeostasis in tumors to augment growth. Cancer Res. (2019) 79:5626–39. doi: 10.1158/0008-5472.CAN-19-0800
35. Das JA-O, Felty Q, Poppiti R, Jackson RM, Roy D. Nuclear respiratory factor 1 acting as an oncoprotein drives estrogen-induced breast carcinogenesis. Cells. (2018) 7:234. doi: 10.3390/cells7120234
36. Zhuang W, Dong X, Wang B, Liu N, Guo H, Zhang C, et al. NRF-1 directly regulates TFE3 and promotes the proliferation of renal cancer cells. Oncol Lett. (2021) 3):679. doi: 10.3892/ol.2021.12940
37. Chang WT, Huang AM. Alpha-Pal/NRF-1 regulates the promoter of the human integrin-associated protein/CD47 gene. J Biol Chem. (2004) 279:14542–50. doi: 10.1074/jbc.M309825200
Keywords: cd47, NRF1, melanoma, promoter regulation, immune checkpoint
Citation: Makwana K, Velazquez EJ, Marzese DM, Smith B, Bhowmick NA, Faries MB, Hamid O and Boiko AD (2024) NRF-1 transcription factor regulates expression of an innate immunity checkpoint, CD47, during melanomagenesis. Front. Immunol. 15:1495032. doi: 10.3389/fimmu.2024.1495032
Received: 11 September 2024; Accepted: 22 November 2024;
Published: 17 December 2024.
Edited by:
Yujia Lan, Harbin Medical University, ChinaReviewed by:
Ismail Meraz, University of Texas MD Anderson Cancer Center, United StatesRaj Kumar, University of Alabama at Birmingham, United States
Copyright © 2024 Makwana, Velazquez, Marzese, Smith, Bhowmick, Faries, Hamid and Boiko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander D. Boiko, YWxleGFuZGVyLmJvaWtvQGNzaHMub3Jn
 Kuldeep Makwana
Kuldeep Makwana Edwin J. Velazquez
Edwin J. Velazquez Diego M. Marzese2,3
Diego M. Marzese2,3 Neil A. Bhowmick
Neil A. Bhowmick Mark B. Faries
Mark B. Faries Alexander D. Boiko
Alexander D. Boiko