- Department of Biology, University of Waterloo, Waterloo, ON, Canada
Innate immunity is the first line of defense against infections and the only known available strategy for invertebrates. Crustaceans, being mostly aquatic invertebrates, are constantly exposed to potential pathogens in the surrounding water. Their immune system abolishes most microbes that enter and are recognized as a threat. However, the stress produced by high population densities and abiotic changes, in aquaculture, disrupts the host-pathogen balance, leading to severe economic losses in this industry. Consequently, crustacean immunology has become a prime area of research where significant progress has been made. This review provides our current understanding of the key pattern recognition receptors in crustaceans, with special focus on Decapoda, and their roles in triggering an immune response. We discuss recent developments in the field of signal transduction pathways such as Toll-like receptors (TLRs) and the immune deficiency (IMD) pathway, and examine the role of antimicrobial peptides (AMPs) in pathogen defense. Additionally, we analyze how environmental stressors—such as temperature fluctuations, ammonia levels, and pollution—impact immune responses and increase susceptibility to diseases. Finally, we highlight future research directions, emphasizing the need to explore the interactions between environmental stressors and immune signaling pathways and to develop strategies to enhance immune responses in crustaceans within aquaculture settings. Altogether, these advancements deepen our understanding of pathogen recognition in invertebrates and the specific defense mechanisms employed by crustaceans, particularly in response to infections triggered by pathogens under abiotic stressors.
1 Introduction
Innate immunity is the first line of defense against infections and the type of immunity available for invertebrates. Acquired immunity is believed to have been established about 500 million years ago with the arrival of the early jawed vertebrates and jawless fishes (1, 2). Nevertheless, invertebrates are a very successful group that have colonized most habitats on Earth and can resist numerous microbial infections, ensuring their survival (3).
Crustaceans are an ancient and prosperous group of invertebrates, comprising more than 67,000 described living species (4), many of which are used for human consumption and as consequence have an extraordinary economic importance worldwide (5). Over the last two decades, crustacean aquaculture has grown dramatically, becoming one of the pivotal sectors of the aquaculture industry. Currently, due its resistance to abiotic changes and diseases, the Pacific white shrimp, Litopenaeus (Penaeus) vannamei, is the most cultivated shellfish species worldwide, with an increase in production from 0.15 million tonnes in 2000 to 5.8 million tonnes in 2020 (5).
Despite this rise and expansion through the years, aquaculture faces considerable loses due to disease outbreaks caused by parasitic, viral, and bacterial pathogens, representing approximately USD 50 billion annually, more than 6 billion for fish and 43 billion from shrimp in 2021 (6, 7). These outbreaks are usually triggered by environmental stressors such as extreme temperature fluctuations and contaminants like plastics and pharmaceuticals, which impose metabolic costs to maintain homeostasis, weaken immune defenses, and increase disease vulnerability (8). In this context, the primary pathogens affecting cultured crustaceans are viruses, for example White Spot Syndrome Virus (WSSV), and bacteria of the genus Vibrio such as Vibrio harveyi, and Vibrio parahaemolyticus which cause Acute Hepatopancreatic Necrosis Disease (AHPND) (9). Giving this background, farmers are overusing and misusing antibiotics as a prophylactic treatment to avoid infectious diseases, which has led to antibiotic resistance in human and farm animal pathogens (10). Therefore, crustacean immunology has become a prime area of research looking for the development of new and effective therapies and husbandry practices that can improve aquaculture species’ health and responses to diseases.
In light of these insights, this review seeks to provide an overview of our current understanding of crustacean pattern recognition receptors, emphasizing their features, signaling pathways and immune responses triggered. Moreover, we highlight the key environmental stressors affecting crustacean aquaculture, underscoring their implications for crustacean health and their connections to signaling pathways.
2 Key components of Crustaceans immune system
Aquatic crustaceans are constantly exposed to opportunistic and obligate pathogens in the surrounding water, yet their immune system abolishes most microbes that successfully manage to breach their physical barriers (11). These environmental interfaces consist of a strong exoskeleton, a digestive system protected by physical and chemical mechanisms, and a robust epithelial cell layer combined with a cutin surrounding the gills, block the entry of most microbes (12).
However, when pathogens successfully break these barriers, they encounter a sophisticated immune system composed of cellular and humoral responses designed to neutralize invasive agents. The cell-mediated responses include processes such as phagocytosis, nodule and capsule formation, and hemocyte degranulation. The latter serves as a crucial starting point, since it triggers the release of antimicrobial peptides (AMPs) and clotting factors, which subsequently contribute to the processes of encapsulation and nodule formation (13). Moreover, this secretion of immune components constitutes a vital link between cellular and humoral immunity in crustaceans (12).
Humoral responses comprise mechanisms such as pathogen recognition, hemolymph coagulation, activation of the prophenoloxidase-activating system (proPO system), and the production and release of AMPs and antiviral factors [see recent review by (14)]. Upon recognition of pathogen-associated molecular patterns (PAMPs), immune cascades are triggered, leading to hemolymph coagulation and melanin production, which help contain and destroy pathogens [see also (14–16)]. The proPO system also plays a critical role by promoting the melanization of pathogens, thus enhancing immune protection (17). Additionally, AMPs, lysozymes, and lectins provide broad-spectrum antimicrobial activity, safeguarding crustaceans from various infectious agents (18).
3 Pattern recognition proteins
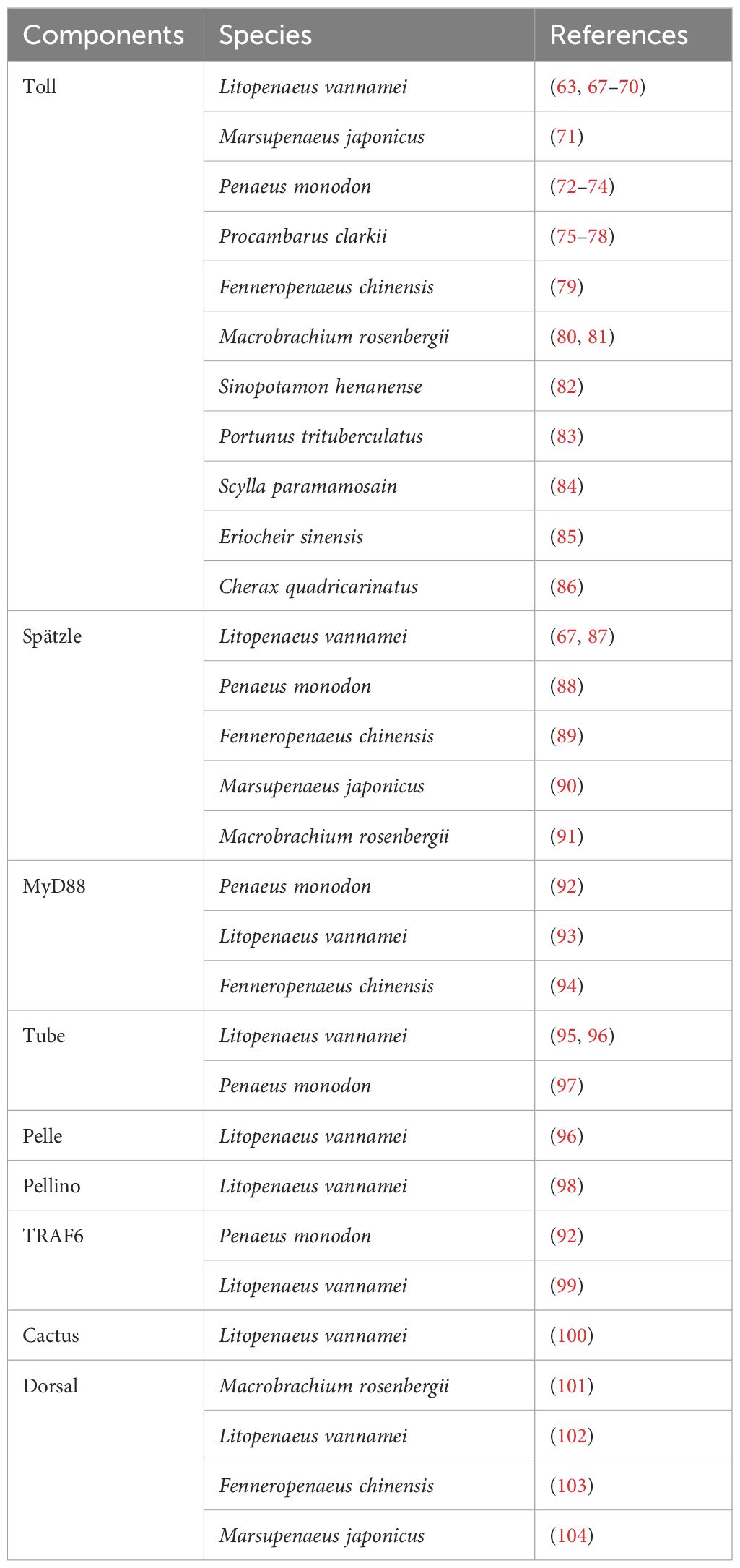
As part of the immune system, PRPs are a group of germ-line-encoded proteins in charge of recognition of PAMPs and triggering immune reactions. Most commonly encountered PAMPs are polysaccharides and glycoproteins such as lipopolysaccharide (LPS) from Gram-negative (Gram-) bacteria, peptidoglycan (PGN) and lipoteichoic acid (LTA) from Gram-positive (Gram+) bacteria, and β-glucans (βG) from fungal cells, all of which are usually exposed on the surface of microbes (19). Furthermore, intracellular components like polynucleotides such as unmethylated CpG DNA, single-strand and double-strand RNA can also act as PAMPs (20). In crustaceans, a suite of PRPs have been categorized depending on their binding properties and the immune reactions triggered. These proteins are grouped in families such as β-1,3-glucanase-related proteins (BGRP), β-1,3-glucan-binding proteins (BGBP), lectins, scavenger receptors (SCR), thioester-containing proteins (TEP), and Down syndrome cell adhesion molecule (DSCAM) proteins (21). Herein an overview is provided, emphasizing the most relevant PRPs and their related immune functions (Table 1).
β-1,3-glucanase-related proteins (BGRPs), previously named as lipopolysaccharide and β-1,3-glucan binding proteins (LGBP), are a representative PRP family described for insects and crustaceans (43). A conserved sequence region similar to the β-glucanases (glucanase like domain, GLU domain) from bacteria is the prominent characteristic of this family (43, 44). These proteins mostly recognize LPS from Gram- bacteria and βG from fungal cells (43). Its expression is detected almost specifically in hemocytes and hepatopancreas of L. vannamei (22); however, in Penaeus monodon (23) and Marsupenaeus japonicus (24) it was exclusively detected in hemocytes. Northern blot studies in Fenneropenaeus chinensis also revealed the presence of BGRP only in the hemocytes (25); nevertheless, RT-qPCR analysis showed the existence of another BGRP isoform in the hemocytes and hepatopancreas (45). Thus, BGRP is expressed in hemocytes and/or hepatopancreas of crustaceans.
Crustacean BGRPs present two cell adhesion motifs (Arg-Gly-Asp (RGD) motif), absent in insect counterparts, which are important for the interaction with integrin receptors on the cell surface (12). This protein interaction might trigger the degranulation of granular and semi-granular hemocytes and the release of its immune components such as the proPO activating system (46). In this sense, Chai et al. (26) by a combination of in vivo and in vitro studies in Procambarus clarkii provided evidence supporting the relation between high level of BGRP expression and enhanced PO activation in response to a challenge with Aeromonas hydrophila. Furthermore, augmented PO activation was reported in hemocyte lysate supernatant incubated with LPS/βG and BGRP, in contrast with only LPS/βG incubation. The authors argue that the increase or acceleration of the PO activation suggests a key role of this PRP as an upstream sensor for the whole system (26).
The β-1,3-glucan binding protein (βGBP) is a type of lipoglycoprotein synthesized by the hepatopancreas and constitutively secreted into the hemolymph (47, 48). This protein acts as PRP by recognizing β-1,3-glucans and inducing hemocytes degranulation through its RGD and/or RGE (Arg-Gly-Glu, (RGE) motif) motifs; thus, activating the pro-PO system. The formation of glucan-βGBP complex is required to induce a conformational change that allows the interaction with the hemocyte receptors (27, 49). Moreover, βGBP is involved in agglutination of fungal cells (50) and enhances phagocytosis by hemocytes (51, 52). High density lipoprotein (HDL) is the main non-sex-specific lipoprotein involved in the transport of lipids in crustaceans (53, 54). Interestingly, the βGBP and HDL are the same protein as has been demonstrated in L. vannamei (27, 28), Penaeus semisulcatus (29), Penaeus californiensis (28), Penaeus leniusculus (30) and Penaeus stylirostris (28), suggesting a close relationship between the ability to respond to PAMPs and the transport of essential lipids provided by the diet (28).
Agglutination is one of the key humoral responses against invading microorganisms. Lectins are the most well-known agglutinating factors, able to recognize several PAMPs as well as damage-associated molecular patterns (DAMPs) (55). One of the main characteristics of these proteins is a carbohydrate recognition domain (CRD), which confers different carbohydrate binding affinities based on their structure. Thus, lectins can be clustered into groups based on their ligand specificity (55). For instance, in crustaceans a broad range of lectin specificities that cluster into at least seven types including C-type, L-type, M-type, P-type, fibrinogen-like proteins (FREPs), galectins, and calnexin/calreticulin (53) have been described. C-type lectins (CTLs) are the most diverse and well-studied group (37). Crustacean CTLs are named after their Ca2+ dependent activity; and can present one or more CRD, but a single CRD being the most usual (12).
Crustacean CTLs [Reviewed by (28, 53, 54)] have been characterized as mainly secretory proteins with higher expression in hemocytes and hepatopancreas compared to other tissues (15). Besides agglutination (31, 32, 34, 35, 56–58), these proteins elicit several other immune responses such as phagocytosis (57, 59), encapsulation (34), respiratory burst (32), proPO system activation (58), and antiviral responses (56, 60). Overall, the activity and expression of CTLs enhances during Gram- (Pseudomonas spp and Vibrio spp) and Gram+ (Staphylococcus aureus) bacterial, and fungal (Aspergillus niger) infections (33). Likewise, studies show that CTLs can bind to WSSV’s envelope proteins and trigger the expression of AMPs through intracellular signaling pathways, thus facilitating the eradication of this virus (40, 57, 58).
DSCAMs are portrayed as hypervariable PRPs which can generate several isoforms by alternative splicing of variable exons from a single-locus gene [Reviewed by (41, 42, 61)]. Thus, DSCAM have been proposed as mediators of a version of challenge-specific protection (62). Nevertheless, almost two decades after their discovery, there is still a lack of definitive information about DSCAM in long-term immune modulation after pathogen exposure or whether DSCAM plays a role in specificity upon secondary encounters (61). Therefore, the notion that these proteins act as an immunological effector enhancing immune memory lacks solid empirical support, and our understanding of DSCAM’s regulatory roles in the immunity of crustaceans is still incomplete. Further research needs to address long-term regulation of DSCAM after repeated pathogen exposure; moreover, it remains unclear how specific isoforms are modulated and what their role is in immune memory (61). Elucidating these regulatory mechanisms is crucial for understanding DSCAM’s role in immunity in crustaceans.
4 Cell-associated PRPs
Most cell types express PRPs and consequently can participate in innate immune responses. Cell-associated PRPs are linked to intracellular signal transduction pathways that trigger several cellular responses, including the production of AMPs. The main PRP-signaling pathways present in invertebrates are Toll like-receptors (TLRs), immune deficiency (IMD) and Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathways (11, 63–66). Currently, several research groups have focused their attention on testing and characterizing the presence of these receptors in crustacean species.
5 TLRs in Crustaceans
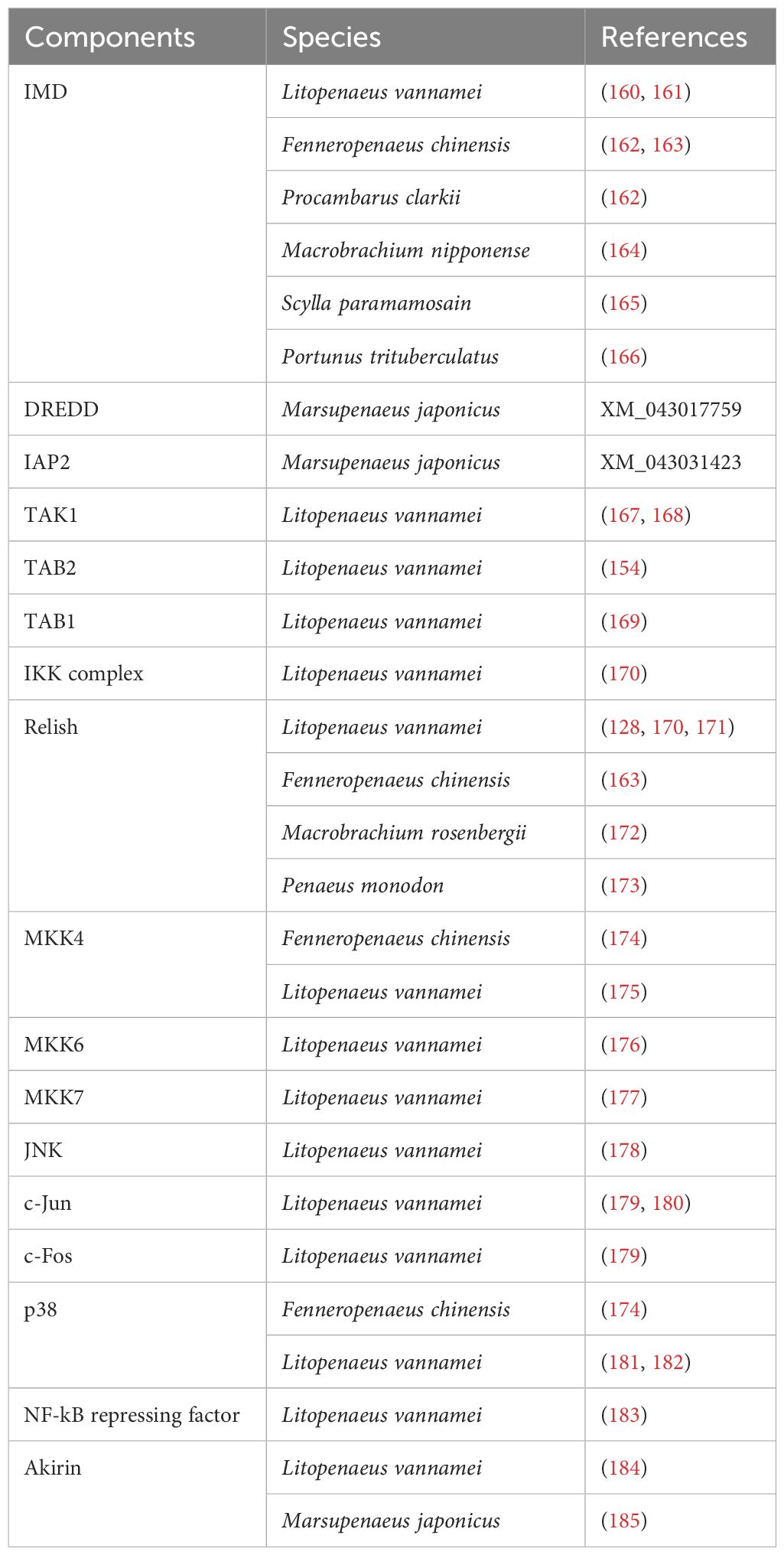
To date, TLRs are the most extensively studied receptors in crustaceans, mainly in decapod species (Table 2). Originally discovered in Drosophila melanogaster (105) but described across a wide range of invertebrate and vertebrate species (106, 107), TLRs are type I integral membrane glycoproteins that contain three structural domains: a leucine-rich repeats (LRRs) flanked by characteristic cysteine-rich motifs, a transmembrane domain and a cytoplasmic Toll/interleukin-1 receptor (TIR) domain. The LRR motif is involved in PAMPs and DAMPs recognition, whereas the TIR domain interacts with signal transduction adaptors and initiates signaling (108, 109). This signaling cascade in invertebrates is based on the canonical mechanism of D. melanogaster Toll1 (DmToll), where DmToll is not a direct PRP, unlike most TLRs in mammals (110). In this case, soluble PRPs such as Gram- bacteria-binding protein (GNBP), peptidoglycan recognition protein (PGRP) and persephone perform PAMP recognition and trigger a serine protease cascade to cleave pro-Spätzle to Spätzle (Spz), an active ligand for the DmToll receptor activation (111–117). Once DmToll receptors are activated, their TIR domains recruit the adaptor, myeloid differentiation factor 88 (MyD88) followed by the recruitment of the second adaptor Tube and the protein kinase Pelle, to form the receptor complex. Pelle´s activation results in Cactus phosphorylation and degradation, freeing Dorsal and Dorsal-related immunity factor (DIF) (19, 118–121) to translocate into the nucleus to regulate the transcription of AMP genes (122–124). Moreover, this complex causes the recruitment of an E3 ubiquitin ligase (named Pellino) (125, 126) and the Drosophila homolog of tumor necrosis factor receptor-associated factor 6 (TRAF6), dTRAF2 (127).
In crustaceans, homologs for most components of the DmToll pathway have been described (Table 2), including Spz (67, 87–91), MyD88 (92–94), Tube (95–97), Pelle (96), Pellino (98), TRAF6 (92, 99), Cactus (100) and Dorsal (101–104, 128), suggesting a similar signaling pathway and immune functions. However, the activation and signaling mechanisms of TLRs in crustaceans (Figure 1) show significant differences from insects that make them more similar to mammals [see also (66)]. In line with this and based on the number of CF motifs (cysteine clusters at the C-terminal end of LRRs, LRRCT), TLRs can be classified into two types: single cysteine cluster or vertebrate-type (V-type), and multiple cysteine cluster or protostome-type (P-type) (129). The P-type has exclusively been identified in invertebrates, suggesting that it is an ancient form of TLR, whereas in all vertebrates and a few invertebrate exceptions, TLRs belong to the V-type (130–132). Most invertebrates TLRs belong to the P-type (63, 71, 72, 79–86) including all Drosophila Tolls, except DmToll9 (133, 134). Evidence suggests that P-type TLR cannot directly recognize PAMPs, in contrast to V-type TLR (129). Nevertheless, studies in M. japonicus (71, 134), L. vannamei (67, 133), F. chinensis (79), Marsupenaeus rosenbergii (80), and P. monodon (73) indicate that some crustacean TLRs can bind directly to PAMPs and activate signal transduction pathways, similar to TLRs in mammals. For instance, Sun et al. (134) discovered that three TLRs from M. japonicus can recognize Gram+ and Gram- bacterial infection through direct binding of their LRR motifs to PGN and LPS, respectively. It is intriguing, in this study, that Gram- bacterial infection could activate the TLR pathway despite the fact that DmToll mainly responds to Gram+ bacteria, fungi and some viruses (118); however, other reports in crustacean models (67, 68, 79, 80, 83) are in agreement with Sun et al. (134). Moreover, two TLR from L. vannamei can interact with CpG oligodeoxynucleotides (ODNs), suggesting their potential role in nucleic acid recognition, similar to TLR9 in mammals (67).
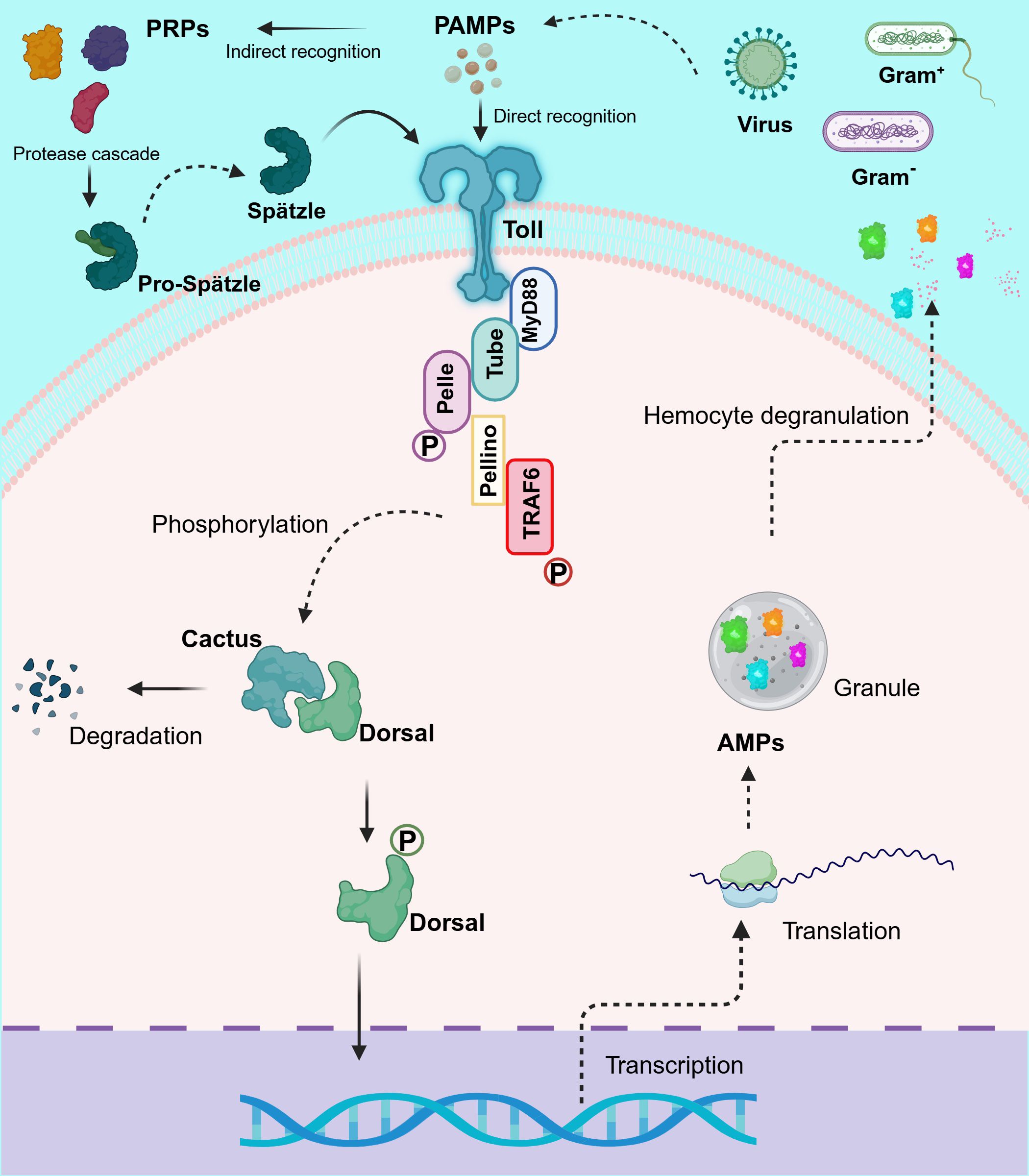
Figure 1. Schematic representation of the proposed Toll-like receptor (TLR) signaling pathway in shrimp. This diagram depicts the dual mechanism of TLR activation in crustaceans, highlighting both the cleavage of Spätzle (Spz) and direct recognition of pathogen-associated molecular patterns (PAMPs). Upon pathogen detection, TLRs activate signaling cascades by recruiting the adaptor protein MyD88, which assembles a complex with Tube and Pelle. This recruitment triggers the phosphorylation and degradation of Cactus, thereby releasing Dorsal (and/or Dorsal-related immunity factor, DIF) to translocate to the nucleus. Additionally, the complex engages Pellino and TRAF6, which further amplify the signaling process. The culmination of these events is the transcriptional activation of antimicrobial peptide (AMP) genes, critical for effective immune responses in shrimp. This pathway exhibits significant evolutionary conservation, reflecting similarities with TLR signaling in both Drosophila and mammals, while also showcasing unique adaptations in crustaceans. Created in BioRender. Betancourt Aguiar, J (2024). BioRender.com/y09c954.
The structural basis for the interaction between crustacean TLRs and PAMPs seems to be explained by the presence of amino acid insertions around positions 10 or 15 of the LRRs ectodomain, as well as the existence of potential N-linked glycosylation sites (63, 71, 79, 80). In human TLRs, insertions following positions 10 and 15 in the LRR consensus sequence have been proposed to be essential for PAMP recognition (135). These insertions introduce flexibility into the concave surface (ligand-binding site) that could interact with PAMPs or accessory proteins such as MD-2 (135–137). In shrimp L. vannamei (63), M. japonicus (71), F. chinensis (79) and M. rosenbergii (80), insertions at position 10 were identified, in contrast to DmToll where none have been reported (138). On the other hand, TLR glycosylation plays a crucial role in receptor surface representation, trafficking, and ligand recognition, as supported by studies in human TLR2 (139) and TLR4 (140), TLR3 (141), and TLR5 (135, 142), respectively. Following this logic, sequence analysis in M. japonicus (71), F. chinensis (79) and M. rosenbergii (80) describe 16, 12 and 9 potential N-linked glycosylation sites, respectively, some of which could influence PAMP recognition. As argued by Bell et al. (135) structural variations at the LRRs motif, especially within the concave surface might provide the specificity needed for TLR responses to different pathogens.
Compelling evidence has shown that the function of P-type TLR structure is to provide the necessary framework for protein-protein interaction with Spz (107). However, it seems this is not their exclusive purpose, as it has been reported that these receptors can also directly recognize PAMP since, as mentioned before, this interaction could be influenced by amino acid insertions and glycosylation. The same phenomenon has been reported in other aquatic invertebrates like Crassostrea gigas (129) and Hyriopsis cumingii (143, 144).
Crustacean TLRs possess unique characteristics due to their ability to activate a defensive response either by direct recognition of PAMPs or Spz mediated activation. While the insect Toll network differs from mammalian TLRs in that a protease cascade links microbial detection by a soluble PRR to a downstream defensive response (145), the convergence of these two mechanisms could potentially increase protection against pathogens in these organisms. Direct recognition of PAMPs could elicit a more specific and targeted immune response giving rise to a more protective outcome. On the other hand, multiple PRPs can evolve to bind different PAMPs resulting in the activation of Spz, feeding information downstream to a single Toll receptor, thus providing the organisms with the ability to recognize a plethora of potential threats without the need for additional membrane receptors (145). Furthermore, a single pathogen contains thousands of PAMPs which can result in the activation of an even greater number of receptors in a potentially higher number of cells as compared to direct recognition by the membrane-bound Toll molecules (145, 146). This way a single pathogen (for example a bacterium) can amplify the number of cells participating in the subsequent immune response (145). Nevertheless, further studies of the structural characteristics of the LRR ectodomains are needed to draw more accurate conclusions on the nature of the interaction with PAMPs and the different specificities of TLR that facilitate recognition.
6 IMD pathway in Crustaceans
IMD is a death-domain containing protein encoded in a locus termed immunodeficiency (imd) (147). Mutation of this gene causes impaired production of AMPs and reduced survival to Gram- bacterial infection in comparison to normal resistance to fungi and Gram+ bacteria (147, 148). Upon this discovery, first described in Drosophila, compelling evidence has shown that the IMD pathway is responsible for sensing diaminopimelic acid (DAP)-type PGN produced by Gram- bacteria, as well as some Gram+ bacterial species such as Listeria and Bacillus (149). Whereas the TLRs pathway mainly responds to fungi and Gram+ bacteria with the Lys-type PGN (118). Mammals lack an IMD signaling cascade, but most components of tumor necrosis factor receptor (TNF-R) pathway broadly resemble the Drosophila IMD pathway nonetheless (150).
According to the current model in Drosophila, two receptors are implicated in specifically recognizing DAP-type PGN, peptidoglycan recognition protein-LC (PGRP-LC), which is located on the plasma membrane, and the intracellular PGRP-LE (149, 151). After binding to PGN these receptors likely dimerize or multimerize (152), and an intracellular signal is transmitted to the adaptor protein IMD (153). IMD interacts via its Death Domain (DD) with Fas-Associated protein with death domain (FADD), and FADD in turn recruits the caspase-8 homolog Death-related ced-3/Nedd2-like protein (DREDD) to the signaling complex via a homotypic Death-effector domain (DED) interaction (147, 149, 151, 154, 155). DD interaction is a key feature in cellular response to infection and all members of the DD superfamily promote inflammation or apoptosis which are essential to clear bacterial infection or prevent further viral replication (156). It has been proposed that DREDD is activated via ubiquitination mediated by inhibitor of apoptosis 2 (IAP2) a component of the ubiquitin machinery which functions as a E3-ubiquitin ligase. Activated DREDD cleaves IMD causing the exposure of a binding site for IAP2 which leads to K63-ubiquitinated IMD (155). K63-polyubiquitin chains of IMD recruit and activate the TGF-β activated kinase 1 (TAK1) via the ubiquitin-binding domain of its regulatory partner TAK1-associated binding protein (TAB2). Upon activation, the TAK1/TAB2 complex is responsible for the phosphorylation and activation of both the I-kappa B kinase complex (IKK) and the mitogen-activated protein kinase (MAPK) branches of the IMD pathway, which culminate in Relish and activator protein-1 (AP-1) activation respectively (154, 155, 157). Relish is a dual domain protein consisting in N-terminal Rel (or NF-kB) domain and a C-terminal ankyrin-repeat/I-kappa B-like domain. The N-terminal Rel domain is released by DREED-mediated cleavage and phosphorylated by IKK complex thereafter undergoes nuclear translocation and trigger the transcription of its target genes, such as AMPs (151, 158).
At the same time, TAK1 acts as a mitogen-activated protein kinase kinase kinase (MAPKKK) activating a pathway leading to c-Jun N-terminal kinase (JNK) and p38 MAPKs that results in the phosphorylation and activation of AP-1 transcription factors (such as c-Jun and c-Fos) ultimately triggering the promoters of a subset of immune-responsive target genes (149, 151). Interestingly, JNK and p38 MAPK are also known as stress-activating protein kinases (SAPKs), because of their ability to respond to a variety of cellular stresses such as oxidative stresses, UV irradiation, osmotic imbalance, heat shock, DNA-damaging agents, inflammatory cytokines and pathogen infection (159).
A growing number of orthologues of the main components of this Drosophila pathway have been isolated and cloned in crustaceans (Table 3), such as IMD (160–166), DREDD (GenBank number: XM_043017759), IAP2 (GenBank number: XM_043031423) TAK1 (167, 168), TAB2 (154), IKK complex (170), Relish (128, 163, 170–173), MKK4 (174, 175), MKK6 (176), MKK7 (177), JNK (178), c-Jun (179, 180), c-Fos (179), p38 (174, 181, 182), NF-kB repressing factor (NKRF) (183) and Akirin (184, 185), [see also (66)]. The first orthologue described was L. vannamei IMD (160, 161), which was also later identified in other crustacean species (Table 3).
Even though the crustacean pathway resembles the Drosophila’s pathway in some aspects, there are some key differences (Figure 2). For instance, according to Li and coworkers (164) phylogenetic analysis indicated that there are two branches: receptor-interacting protein (RIP) from the vertebrate TNF-R pathway and IMD from invertebrates, which further bifurcated into the insect and crustacean sub-clusters. The correct clustering of these peptides in comparison to traditional taxonomy indicates evolution among IMD and RIP1 proteins (164). Furthermore, the IMD pathway displays significant discontinuities in the taxonomic distribution of key components, thus indicating variation and plasticity in this pathway in arthropods (38). Studies suggest that the IMD pathway is triggered by different pathogens depending on the species and the response generated may even differ between different tissues (164). For instance, in Eriocheir sinensis both Gram- (V. parahemolyticus) and Gram+ (Staphylococcus aureus and Bacillus subtilis) bacteria can activate the IMD pathway inducing the translocation of Relish to the nucleus (186). Interestingly, all these bacteria upregulated IMD expression and induced translocation of Relish regardless of the type of PGN, DAP-type (V. parahemolyticus and B. subtilis) and Lys-type (S. aureus) (186). Moreover, the expression of Relish was upregulated in S. paramamosain (187), P. monodon (173) and L. vannamei (128) in response to WSSV infection. Nonetheless, further studies need to confirm if this upregulation of Relish is part of the immune mechanism triggered to abolish this pathogen or part of the WSSV replication mechanism, which has been suggested to hijack this NF-κB pathway to favor its own propagation (128).
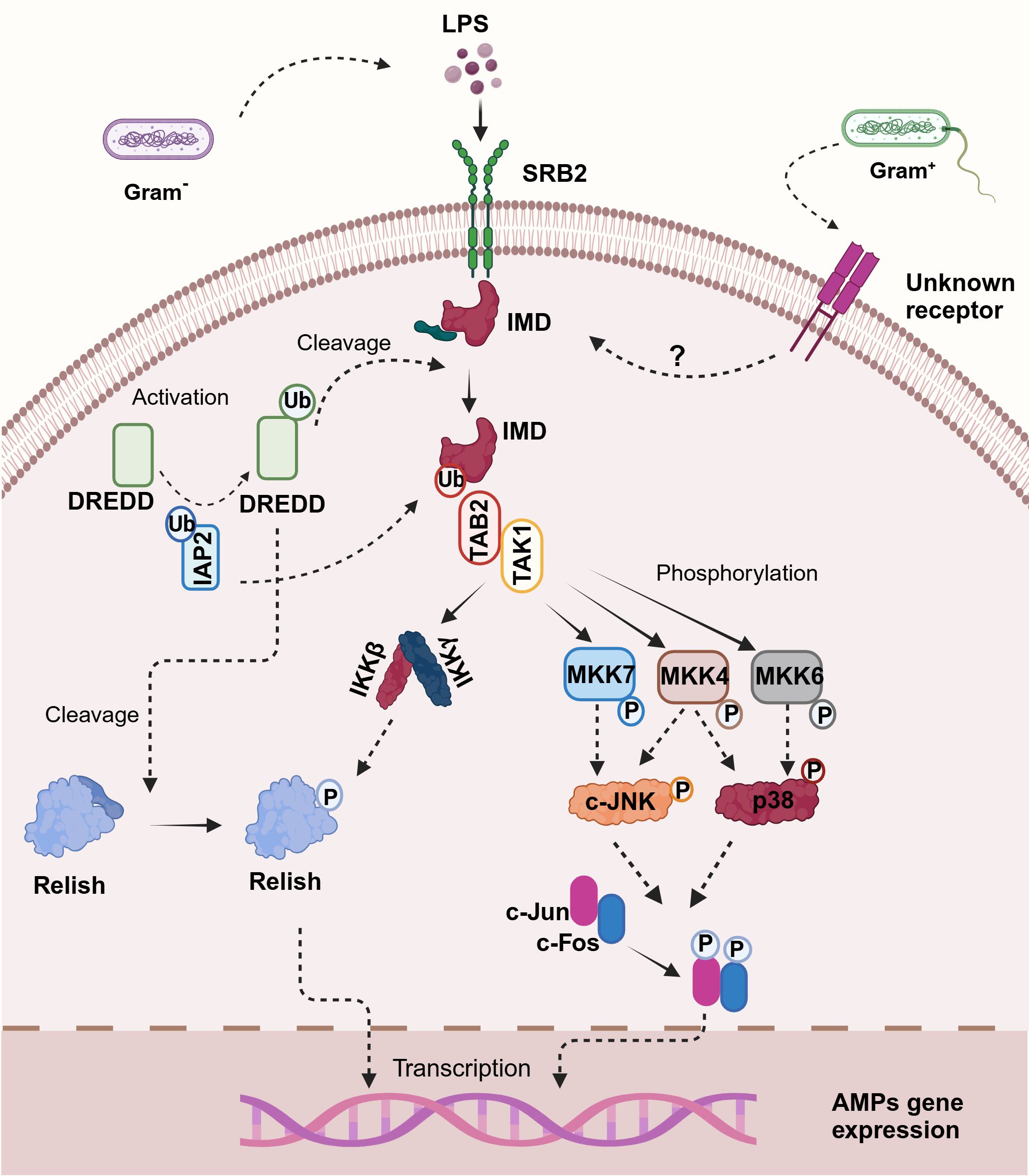
Figure 2. Illustration of the proposed immuno-deficiency (IMD) signaling pathway in shrimp. Upon binding of lipopolysaccharides (LPS) to the scavenger receptor class B2 (SRB2) — a proposed mechanism for crustaceans since no peptidoglycan recognition proteins (PGRPs) have been described—the pathway is initiated. This interaction leads to the recruitment and activation of DREDD via ubiquitination mediated by IAP2. Activated DREDD cleaves IMD, exposing a binding site for IAP2, which facilitates K63-ubiquitination of IMD. The K63-polyubiquitinated IMD recruits TAK1 through TAB2, activating the I-kappa B kinase (IKK) complex and mitogen-activated protein kinases (MAPK) pathways, specifically MKK4, MKK6, and MKK7. Notably, in crustaceans, p38 is activated by MKK4 and MKK6 through phosphorylation, paralleling human immune signaling mechanisms. This bifurcated signaling cascade culminates in the activation of Relish and AP-1 transcription factors, including c-Jun and c-Fos, which translocate to the nucleus and trigger the transcription of antimicrobial peptide (AMP) genes, orchestrating the immune response against bacterial infection. Created in BioRender. Betancourt Aguiar, J. (2024) BioRender.com/h56u584.
Similar to mammals, studies on L. vannamei showed that MKK4 can phosphorylate and activate p38 MAKP (175), whereas DmMKK4 exclusively activates JNK in Drosophila (188). Additionally, Wang and co-workers proved in L. vannamei that TAB1 can combine with TAK1 and p38 MAKP, thus regulating the activity of these kinases (169). Therefore, p38 MAKP regulation in crustaceans can be achieved by three routes: MKK6, MKK4, and TAB1. Consequently, as proposed by Li et al. (66), the presence of different pathways could provide precise control over p38 MAKP activity during pathogenic invasion, indicating the significant involvement of p38 MAKP in crustacean immunity. For instance, even if one or two pathways are blocked under particular conditions, p38 MAKP may still function in the immune response (66).
Until recently, it remained unclear how the IMD pathway can sense pathogens, since no members of PGRP have been described in crustaceans. Besides, homology searches of shrimp (189, 190) and crab (186) transcriptome data failed to uncover a PGRP homolog. Even though the existence of PGRPs remains doubtful in crustaceans, Shi and co-workers (38) suggested a class B2 scavenger receptor (SRB2) as potential receptor of the IMD pathway. Studies conducted in M. japonicus showed that SRB2 can sense LPS using its extracellular domain and interact with IMD with its C-terminal intracellular region; thus, amplifying the signal and promoting Relish’s nuclear translocation (38). SRB2 silencing by RNAi increased bacterial load and decreased the survival rate to V. parahaemolyticus infection by abolishing the expression of AMPs (38). Therefore, it was concluded that SRB2 stimulates bacterial clearance and enhances shrimp survival rates to Gram- bacterial infection by increasing the transcripts of anti-lipopolysaccharide factors 1 (Alf1) and 2 (Alf2) (38). In this context, the authors also identified homologs of this receptor in other crustacean species such as L. vannamei, P. clarkii, Scylla serrata and Hyalella azteca; consequently, confirming the hypothesis that SRB2 could act as signal transduction membrane receptor of the crustacean IMD pathway. Nevertheless, since SRB2 cannot recognize PGN (38) and is thus unable to sense Gram+ bacteria, it remains unknown how the IMD pathway is activated after Gram+ challenges.
Research into the IMD pathway in crustaceans should focus on several interconnected areas that could enhance our understanding of immune responses. Firstly, the characterization of SRB2 as a potential sensor for LPS (38) invites detailed investigation into its molecular interactions with IMD, as well as the broader implications for AMP expression. Moreover, examining the functional roles of specific AMPs generated through the IMD pathway can elucidate their mechanisms against diverse pathogens, which may involve gene knockout approaches to clarify their individual contributions to immunity. The impact of environmental stressors on the responsiveness of the IMD pathway and AMP production warrants exploration (191, 192), as such factors could significantly influence crustacean health and resilience in aquaculture settings. Integrating these studies with multi-omics approaches, encompassing genomics, transcriptomics, and proteomics, could provide a comprehensive view of the signaling networks and regulatory mechanisms involved in the IMD pathway, potentially revealing novel interactions that facilitate pathogen resistance. Lastly, investigating how pathogens, like WSSV, hijack the IMD pathway (95) could uncover strategies for mitigating their detrimental effects on crustacean populations, enhancing overall disease management in aquaculture.
7 Antimicrobial peptides
AMPs can be found in all kingdoms of life and exhibit an extraordinary structural and functional diversity, thus making them possible alternatives to antibiotics (193). This diverse group of peptides are categorized into subgroups or families based on their structural properties, as determined by the peptide’s primary sequence and three-dimensional (3D) conformation. According to these criteria, they can be classified as one of four types, α-helix, β-sheet, extended and β-hairpin or loops. On the other hand, using mechanisms of action, these molecules can be categorized as membrane disruptive or nonmembrane disruptive AMPs. The first creates holes in the membrane whereas the second group pass into cells to act on intracellular targets in order to disrupt metabolism and kill. Either way, these peptides produce a disturbance and disorder of the cytoplasmic membrane resulting in loss of the transmembrane potential, and eventual cell death. This mechanism of action based on membrane interactions rather than through recognition of a single receptor like antibiotics, makes the development of bacterial resistance improbable (194–196). AMPs are produced as the first line of defense against infection, and in addition to their antimicrobial properties these peptides have also been shown to have immunomodulatory functions. Some of these immunostimulant effects include stimulation of chemotaxis, immune cell differentiation, initiation of adaptive immunity and stimulation of both pro- and anti- inflammatory cytokines (197).
Crustacean AMPs are mostly small, amphipathic, cationic, and gene-encoded peptides that are mainly produced by hemocytes or originate from proteins involved in other biological functions (195). There are 15 families that can be grouped, according to Rosa and Barroco (195), into four main groups based on amino acid sequence and 3D conformation. These groups comprise: (I) single-domain linear α-helical AMPs and peptides enriched in certain amino acids, (II) single-domain peptides containing cysteine residues engaged in disulfide bonds, (III) multi-domain or chimeric AMPs, and (IV) unconventional AMPs including multifunctional proteins and protein-derived fragments that exhibit antimicrobial functions (195). Herein only an overview of key crustacean AMPs is provided, for further information on this issue see reviews by (195, 198–200).
Crustins are a family of peptides belonging to group III, characterized by the presence of a whey acidic protein (WAP) domain at the C-terminus (195). Based in their structure, crustins sub-cluster in three types (I, II and III). Type I is characterized by a cysteine-rich region located between the leader sequence and the WAP domain; type II is defined by the presence of a hydrophobic area containing a glycine-rich region upstream of the cysteine-rich region described for Type I crustins; whereas type III have a concise proline/arginine-rich area between the WAP domain and the leader sequence, and do not possess any other amino acid rich domains like type I and II (195). Crustins type II and III are mostly found in shrimp and crayfish, while type I is more exclusive of crab (195). Regarding their function, crustins have been shown to regulate intestinal microbiota balance and have direct antibacterial activity against several bacterial species such as S. aureus, Bacillus sp., V. parahaemolyticus, Vibrio harveyi, Vibrio anguillarum and Vibrio alginolyticus (201). Some studies have shown the modulation of crustin transcripts in infection trials, as well as the deleterious effect of knocking down the transcription of crustin genes on shrimp survival (201). Thus, this AMP play a pivotal role in crustacean immune responses against pathogens.
The anti-lipopolysaccharide factors (ALFs) family is part of group II of crustacean’s AMPs (195). These peptides exhibit a broad spectrum of binding affinities and antimicrobial activities against pathogens. This diversity is a consequence of a significant variation in amino acid sequences and LPS-binding domains (LPS-BD) among members of this family (200). Based on these characteristics, ALFs are clustered in seven groups (Groups A to G) (200). Interestingly, members of groups B, C and F exhibit cationic charge whereas groups A, D, E and G displays anionic properties (200). Additionally, ALFs present a wide range of antimicrobial activities; thus, peptides such as P. monodon ALFPm3 displays a strong activity against fungi, virus, and both Gram- and Gram+ bacteria, whereas other members exhibit low or insignificant microbicidal properties (200). Consequently, it has been hypothesized that some ALFs may work together with other AMPs or play roles in other biological processes, such as wound healing and tissue regeneration (202). In this context, the potential use of ALFPm3 in aquaculture has been proposed due to its immunostimulant and antimicrobial activities against WSSV, yellow head virus and V. harveyi infections (18).
Part of group III of crustacean AMPs, penaeidins and stylicins represent two different families of peptides mostly identified in penaeid shrimp with species-specific function (Reviewed by 189). Currently, only a few members of these families have been described in commercially valuable species such as L. vannamei, P. monodon, M. japonicus (200). Thus, further studies need to address the existence and function of these peptides in other crustacean species. So far, it is generally accepted that stylicins are mainly antifungal peptides (200). Meanwhile, penaeidins which subcluster into two subfamilies (I and II) display antimicrobial activities against Gram+ bacteria and filamentous fungi for type I; and antibacterial activities against Gram+ and Gram- bacteria for type II (18, 200).
Remarkably, besides the typical described AMPs, current evidence supports the key role of other protein-derived peptides in crustacean immunity (200). In this group, AMPs derived from hemocyanin-cleavage are the most well-known. These peptides have been reported for species such as L. vannamei, P. stylirostris, P. monodon, P. japonicus and F. chinensis, and exhibit a broad range of antimicrobial activities specie-specific (18). In this context, researchers are looking into pleiotropic peptides such as Pituitary Adenylate Cyclase-Activating Polypeptide (PACAP), which has shown immunostimulant and antimicrobial properties in shrimp and crayfish species (203, 204).
Gene encoded AMP expression is linked to NF-κB pathway activation (Reviewed by 33). Notably, in crustaceans, the expression of peptides such as crustins, penaeidins and ALFs have been associated with both Toll and IMD pathways (66). Moreover, recent studies in P. clarkii show that during A. hydrophila infection, CTLs can also trigger ALFs upregulation via the JNK pathway (57).
As discussed previously, these AMPs serve as one of the first lines of defense against pathogens, showcasing the complexity of immune signaling in invertebrates (195). The coordinated response involving the Toll and IMD pathways not only emphasizes the adaptive nature of crustacean immunity but also allows development of sustainable, antibiotic-free disease management strategies in aquaculture. In this context, given the crucial roles that the Toll and IMD pathways play in regulating AMP expression (66), further research is essential to deepen our understanding of crustacean immune responses. One potential avenue of research could focus on the molecular mechanisms underlying the crosstalk between these pathways and other signaling cascades, such as MAPK pathways, which have been suggested to influence AMP expression (168, 175). Investigating how these pathways interact could reveal novel regulatory networks that govern immune responses in crustaceans.
Additionally, studies could explore the environmental factors that influence the expression of AMPs in crustaceans. Understanding how abiotic factors such as temperature, pH, salinity, and pollution impact the regulation of immune responses could be critical for improving aquaculture practices. Another important direction would involve evaluating the genetic diversity of AMP genes in various crustacean species and their responses to different pathogens, which could inform selective breeding programs aimed at enhancing disease resistance in aquaculture.
Finally, there is a need to investigate the potential for utilizing these AMPs as biocontrol agents in aquaculture. As antibiotic resistance becomes an increasingly pressing issue (10), exploring the application of AMPs in disease management could provide sustainable solutions for improving shrimp health and productivity.
8 Environmental stressors influence Crustaceans’ susceptibility to diseases
Crustaceans are constantly subjected to environmental challenges (Figure 3). Extreme temperature fluctuations and environmental contaminants (i.e., plastics and pharmaceuticals) are among the stresses that have a metabolic cost to maintain homeostasis and that can cause a weakening of immune defenses and greater vulnerability to diseases (8). In addition, the rapid growth of crustacean industry and particularly shrimp farming have resulted in environmental pollution affecting farmed and wild aquatic species (205). Pollution may lead to variation in abiotic variables such as temperature, dissolved oxygen, ammonia levels, pH and salinity, affecting health status and crustacean’s innate immune responses to pathogens (206). Genes differentially expressed (DGs) in response to environmental stresses are involved in different signaling pathways including TLR/IMD-NF-κB, JAK-STAT, MAPK, and Wnt signaling pathways, and these pathways are commonly activated simultaneously (207).
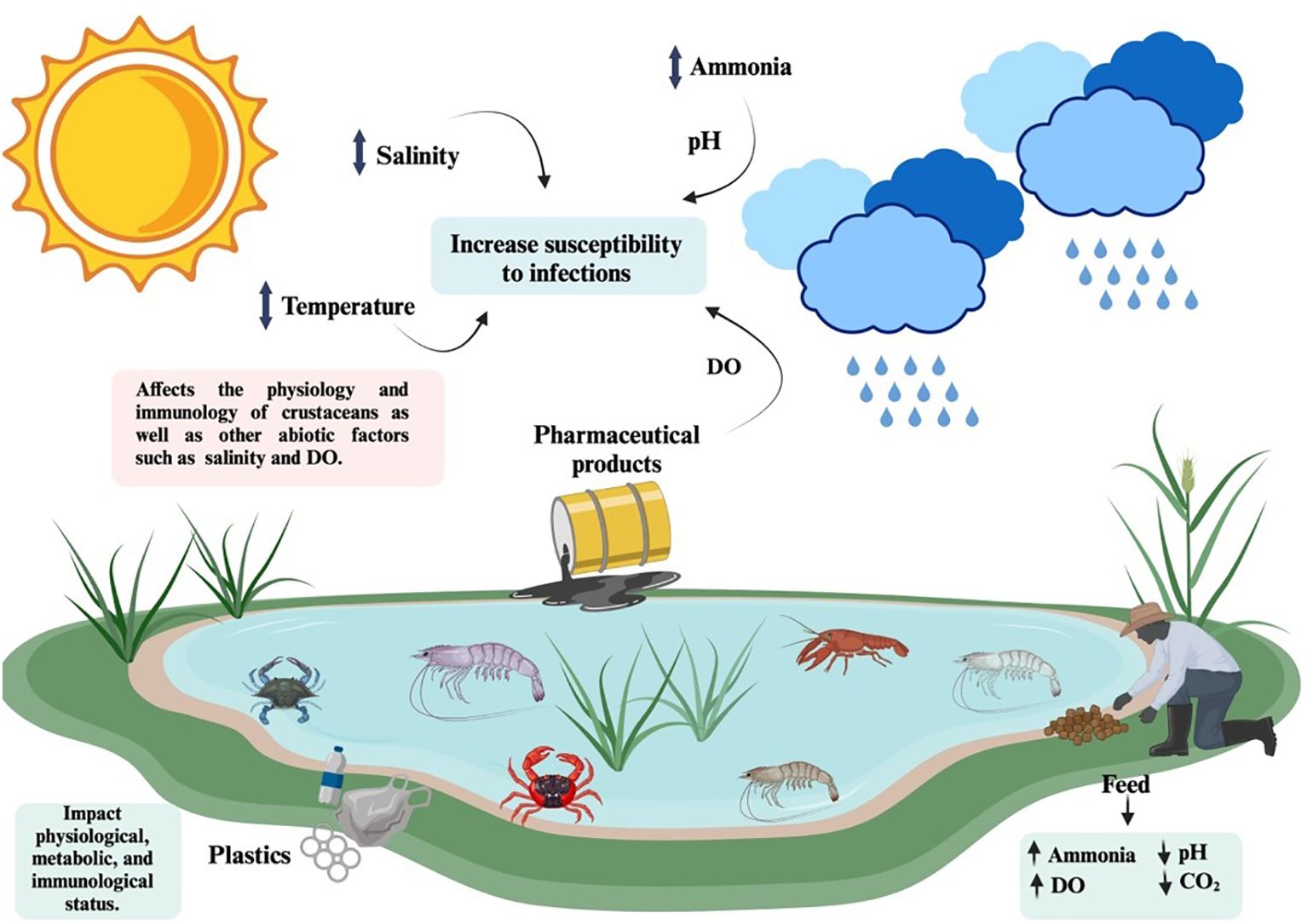
Figure 3. Environmental factors such as temperature, salinity, pH, dissolve oxygen (DO), ammonia concentrations and pharmaceutical products, significantly influence pathogen susceptibility in crustaceans, thus contributing to disease outbreaks in aquaculture facilities. Created in BioRender. Betancourt Aguiar, J. (2024) BioRender.com/n17f466.
Water temperature is probably the most important abiotic variable affecting aquatic species. It affects other abiotic variables such as salinity and oxygenation of the water (208) and may affect survival, growth, physiological functions, and immune defenses (198). Although crustaceans can tolerate a wide range of temperatures, and cultured species are commonly reared in shallow ponds with wide seasonal and daily temperature fluctuations, the effects of these variations on the ability of these species to fight pathogens are not yet fully understood (209). For instance, the thermal maxima (CTMax) and minimum (CTMin) of the most farmed crustacean species in the world, the Pacific white shrimp L. vannamei (210), are between 36 and 42°C and between 8 and 12°C, respectively, depending on the life stage, acclimatization temperature and temperature ramp rate (211). However, it’s been shown that the susceptibility of L. vannamei to pathogens is highly dependent on the water temperature. A range between 25 and 28°C offers optimal conditions for WSSV infections leading to high shrimp mortalities, while higher temperatures (33–34°C) have shown to be protective (209). Millard and coworkers (209) suggested that elevated temperatures may have an effect on the activity of viral enzymes or on the ability of the host to respond to the pathogen. In one study, WSSV-inoculated shrimp were kept in a four-compartment system with all chambers at 27°C or with a thermal gradient (27–29–31–33°C), and shrimp that were allowed to migrate to warmer temperatures (behavioral fever) showed a significant higher survival rate that shrimp that were kept at 27°C (212). It’s known that warm waters (32-33°C) inhibit WSSV replication (212, 213), but the shrimp defense mechanisms behind the protective effect of elevated temperatures still need more attention from researchers. In this sense, it is suggested that high temperatures activate the unfolded protein response (UPR) protein kinase R-like ER kinase-eIF2α (PERK-eIF2α) signaling pathway, inhibiting the translation of proteins, including WSSV proteins, and inhibiting WSSV proliferation (207). Hyperthermia has also been associated with cell apoptosis in WSSV-infected L. vannamei as a mechanism responsible for an increased survival rate (214). Apoptosis as well as endocytosis pathways were involved in the response to heat stress of Palaemon graviera (205). In this transcriptomic study acute heat stress induced upregulation of the following immune related-transcripts: caspase 7 (CASP7), transcription factor AP-1 (Jun), broad-complex core protein (BR-C), heat shock proteins 70 (HSP70), Rab5B and Rab10. These authors point out the need of protein level assays to corroborate their results and suggest that acute high temperature may significantly reduce the metabolic capacity of shrimp but enhance the immune capacity, as an emergency metabolic compensation strategy to deal with stress (215).
On the other hand, acute cold-stress (13°C) caused a decrease in plasma metabolite concentrations, relative gene expression related to UPR pathway and apoptosis in the hepatopancreas and hemocytes of L. vannamei, with histological damage in the hepatopancreas, suggesting a decrease immunity and more vulnerability to diseases (216). In this context, a study conducted by Wang and coworkers (191) reported a decrease in the gene expression of TLR, IMD and proPO in the intestine of L. vannamei after cooling (28°C to 13°C). Thus, the authors argued that considering that the intestinal barrier represents one of the first line of host defenses against invasive microorganisms and that TLRs, IMD and the proPO system play key roles in immune defenses, the reported downregulation of these genes could indicate a reduction in the shrimp’s ability to identify and eradicate pathogens during low temperatures (191). Lv et al. (192) also suggested that unsuitable temperatures have detrimental effects in the gene expression levels of core proteins in the NF-κB signal pathway. Moreover, temperature variations also influence other abiotic parameters such as salinity and oxygenation of the water. It may affect crustaceans immune related variables including clotting times, total hemocyte count, phagocytosis, antibacterial activity of the hemolymph and pro-phenoloxidase activity (208).
Other major abiotic variable affecting the innate immune system of crustaceans is level of ammonia (205). High concentrations of ammonia caused an increased mortality, a decrease in the phagocytic activity and a decrease in the clearance efficiency in L. vannamei challenged with V. alginolyticus (217). In the same species, high ammonia levels caused a suppression of immune parameters such as a 66% decrease in total hemocyte count (218), an increased coagulation time and a down-regulation in transglutaminase gene expression (219). In this line, studies conducted in P. monodon indicate that ammonia exposure seems to initially trigger an immune response, but over time, prolonged stress weakens the immune system. This overexposure leads to reduced immune enzyme activity and downregulates the expression of immune-related genes like lysosome and crustin (220). Moreover, ammonia stress downregulates CTLs expression in P. monodon’s hepatopancreas and intestine (221, 222); thus, considering that CTLs play an important role as PRPs during V. harveyi and V. anguillarum challenges (222), high concentrations of ammonia could increase the vulnerability of shrimp to bacterial diseases. CTLs have also shown to act as critical immune components against V. parahaemolyticus and WSSV infections, and during endoplasmic reticulum (ER)-stress responses in L. vannamei (223).
Environmental contaminants including plastics and pharmaceutical products are constantly discharged into aquatic environments affecting aquatic wildlife. Chronic toxicity of chemical contaminants in crustaceans has shown to be dependent on abiotic variables such as temperature. In one study, the negative impact of clofibric acid (CA) and diclofenac sodium (DS) on Palaemon longirostris larvae development was significantly higher at 18°C compared to 24°C (224). Plastics and especially microplastics (MPs) and nanoplastics (NPs) are aquatic contaminants that accumulate in tissues of aquatic organisms affecting their immunity and metabolism (225). Exposure to MPs caused variation in the intestinal microbiota of L. vannamei inducing the proliferation of opportunistic pathogens in the gut. Different MPs changed the hemolymph proteomic composition, specifically proteins involved in inflammation, apoptosis, oxidative stress and metabolism (212). MPs and NPs also caused physical damage to intestinal cells including epithelial cell necrosis (226) and shorter intestinal fold heights (227). More studies are needed to understand the impact of micro and nanoplastics on the health and immune response of crustaceans.
As previously stated, JNK and p38 MAPK are also referred as SAPKs because their role in transmitting environmental stress signals to the nucleus (159). Zheng and coworkers (228) described in L. vannamei that under low temperature stress JNK plays a crucial role regulating apoptosis and oxidative damage. JNK is activated by excessive reactive oxygen species (ROS) production and accumulation caused by an increase in the mitochondrial respiratory rate induced by low temperature (228). Activated JNK activates AP-1 transcription factors, thus leading to the regulated expression of downstream apoptosis genes such as p53, caspase-3 and mitochondrial proteins associated with apoptosis such as Bax and Bid (228). However, the expression of some of these apoptosis related genes was also significantly enhanced after JNK silencing; furthermore, ROS accumulation, apoptosis and mortality erratically increased after this silencing. Thus, the authors proposed that JNK is essential for mediating low temperature tolerance, apoptosis rate and ROS accumulation (228). Also, Tian et al. (229) described that molt could trigger apoptosis driven by oxidative stress through the activation of JNK in P. clarkii. Consequently, JNK could be mediating this process by inducing the expression of pro-apoptotic genes such as Bax, Bak and Bok, and inhibiting the expression of anti-apoptotic genes such as Bcl, A1 and Mcl1 (229).
Luo and coworkers (230) reported that acute cold stress activates the gene expression and phosphorylation of p38 MAPK, in gill tissues of L. vannamei, suggesting its potential role in response to low temperatures. Interestingly, although the gene expression levels of p38 MAPK, JNK and ERK were measured, only the p38 MAPK values were upregulated during the first 12 h of cold stress exposure (230). Consequently, these findings contradict the upregulation of JNK associated with ROS production induced by low temperatures as described by (228) in L. vannamei hemocytes. Accordingly, Luo et al. (230) results could be interpreted in light of the non-significant production of ROS in the gill during the timeframe of the experiment, which may have limited the expression of JNK; or the central role of p38 MAPK responding to this environmental stressor in the gill instead of JNK. However, further studies are necessary to clarify these observations by analyzing the expression levels of the MAPKs as well as other signaling proteins involved on these pathways, alongside apoptosis-related genes. Additionally, measuring ROS production across multiple tissues under low-temperature stress would help clarify the mechanisms at play.
A study conducted by Park et al. (231) in M. japonicus illustrated that p38 MAPK plays a key role mediating the immune response against oxidative stress induced by environmental pollutants such as perfluorooctane sulfonate (PFOS), irgarol, di(2-ethylhexyl) phthalate (DEHP), and bisphenol A (BPA). In this study significant expression of p38 MAPK was mainly detected in the gill and hepatopancreas tissues (231), suggesting their pivotal role during toxic pollutants exposure. The authors suggested that p38 MAPK could be orchestrating inflammation, apoptosis, and cell cycle regulation processes in response to these pollutants (231). Nevertheless, additional research is required to understand these issues.
Thus, Shui and coworkers (232) proposed that p38 MAPK orchestrate the distribution of cadmium (Cd) in P. clarkii crayfish by modulating the accumulation of Cd in different tissues under Cd stress environments. Results suggest that a strong negative correlation exists between Cd levels and the expression of p38 MAPK in several tissues. Moreover, p38 MAPK transcript levels showed relevant differences between tissues and under no stress versus stress conditions (232). The tissue-specific differences in p38 MAPK gene expression in the absence of Cd exposure, compared to conditions of low and high exposure, indicates that crayfish absorb Cd through their gills, after which the absorbed Cd is distributed to various tissues, including the hepatopancreas, heart, antennal gland, and muscle. However, the primary site of Cd accumulation is regulated by the p38 MAPK pathway in the hepatopancreas, which serves as the main Cd storage tissue in P. clarkii (232). Given this context, while the modulation of Cd tissue distribution by p38 MAPK is intriguing, further research is necessary to identify the specific genes targeted by this MAPK signaling pathway that facilitate this outcome. Probably, these genes are involved in regulating ion and osmotic channels within the cells, which may play a crucial role in the absorption and accumulation of Cd in various tissues. Furthermore, it is particularly interesting that a study carried out by Jian et al. (233) in S. paramamosain reported a significant increase in one of the three isoforms of IKK described for this specie after 6h of Cd exposure. Therefore, the IKK complex, which is considered one of the core elements of the NF-κB cascade, is also involved in coping with Cd pollution (233). Thus, a better understanding of these mechanisms could provide valuable insights into how crustaceans manage Cd stress and other environmental contaminants.
It is widely recognized that environmental stressors significantly influence pathogen susceptibility in crustaceans, which is a primary contributor to disease outbreaks in aquaculture facilities. Such stressors can compromise the immune response of crustaceans, thereby increasing their vulnerability to pathogens and result in substantial economic losses within the aquaculture sector (11, 209). As highlighted in the preceding sections, variations on temperature influence susceptibility to WSSV infections (212, 213) as well as the prevalence of Vibrio infections (234). Moreover, ammonia accumulation on farms, resulting from the continuous influx of biological waste, residual bait, and crustacean exoskeletons, can trigger and amplifies infections by V. alginolyticus (217), WSSV (235) and Lactococcus garvieae (236). Furthermore, low-salinity stress also enhances susceptibility to infections by V. alginolyticus (237) L. garvieae (236), V. harveyi (238), V. parahaemolyticus (234) and Vibrio cholerae (234) [see also reviews by (209, 238)]. Ultimately, while the crustacean immune system has evolved to effectively counter pathogens, environmental stressors can induce extreme conditions that adversely affect their physiology and immune responses. As survival becomes the primary focus under these conditions, crustaceans become increasingly susceptible to pathogenic infections. Thus, comprehension of the interactions among environmental stressors, pathogens, and host is essential for developing effective management strategies to reduce disease risks in aquatic ecosystems.
Despite the numerous publications describing changes on crustaceans’ immunity and health indicators due to environmental stressors, more evidence is needed to understand the effects of these changes during an encounter with a pathogen (8) and the variability of the responses depending on different stressors (209). Additionally, a comprehensive understanding of how crustaceans perceive abiotic variations, the signaling pathways involved, and the differential gene expression that these stressors trigger is essential for grasping the variability of immune responses. Furthermore, stressors such as temperature variations and environmental contaminants not only affect the immune functions, of both farmed and wild shellfish, but also have serious implications for toxin accumulation, necrotic tissue, muscle atrophy, changes in organoleptic profiles and discoloration (8). Thus, gaining deeper insights into these mechanisms will be crucial for developing effective management strategies to enhance the health and resilience of crustacean populations in both aquaculture and natural environments.
9 Concluding remarks
Living in a hostile and microbe-enriched environment, crustaceans exhibit a sophisticated and highly effective immune system, which has assured their survival and expansion across evolution. A continuous environmental pathogenic pressure as well as abiotic and biotic stressors have driven to the refinement of their immune system (11). Therefore, this group of invertebrates exhibit unique features that are highly relevant for understanding the immune mechanism underneath, which differ from the traditional invertebrate model and open the possibility of reaching new insights and paradigms in immunology.
A suit of PRR have been characterized in crustacean, thus empowering them with a broad spectrum of pathogen recognition. Interestingly, the role of some proteins such as HDL [also known as βGBP in penaeids (27, 28)] and hemocyanin (18) which have been described involved in the recognition of pathogens and triggering immune responses, respectively, bring to focus the close relationship between the crustacean physiological and immunological status; as well as provide evidence of a potential evolutionary adaptation process that these organisms undergo toward multitasking optimization. In this context, crustacean TLRs have the unique ability of triggering a defensive response either through direct recognition of PAMPs or via Spz-mediated activation, thus providing them with a refined pathogen recognition mechanism. Additionally, this defense system benefits from the ability to respond to both Gram+ and Gram- bacteria through the Toll and IMD pathways (66). In this line, the IMD pathway displays the singular characteristic of recognizing Gram- bacteria through the SRB2 as consequence of the absence of PGRP in crustaceans (38). Moreover, this pathway finely regulates p38 MAPK through MKK6, MKK4, and TAB1 proteins, highlighting its crucial role in immune response development during pathogenic invasion (66).
Environmental stressors significantly shape the immune responses of crustaceans by modulating key signaling pathways, including NF-κB and MAPKs (228, 232, 233). For instance, temperature variations affect the TLR and IMD gene expression levels as well as the proPO activating-system (191). Meanwhile, CTLs transcripts are vulnerable to ammonia exposure (222), thus disturbing the recognition of pathogens. Moreover, fluctuating temperatures and exposure to pollutants can alter the expression and activity of crucial proteins like JNK (228) and p38 MAPK (230, 232), which play key roles in transmitting environmental stress signals to the cell nucleus (159). For example, in L. vannamei, JNK is activated by ROS production due to elevated mitochondrial respiratory rates under low-temperature stress (228). This activation leads to the expression of pro-apoptotic genes, indicating that JNK is crucial for mediating low-temperature tolerance and apoptosis (228). In contrast, while p38 MAPK is also activated under cold stress (230), its role in regulating immune responses against environmental pollutants, such PFOS, DEHP, BPA and Cd, has been highlighted in various studies (231, 232). These findings underscore the need for further investigation to clarify the interplay between these signaling pathways and their associated genes in response to abiotic stressors.
Overall, crustaceans portray unique immunological features, that further knowledge of could lead to new disease management strategies in aquaculture. For instance, AMPs are part of the prominent immune responses triggered against pathogens in crustacean and constitute potential candidate for the development of new and effective therapies that can improve crustaceans’ health and responses to diseases in aquaculture. Moreover, farmers could benefit of a better understanding of the environmental stressors affecting crustacean aquaculture, since these factors are the primary cause of the disruption of the balance between host and pathogens which lead to disease outbreaks. Finally, the potential for immunological memory in crustaceans remains an area of active investigation, with most studies linking their findings to immune priming and trained immunity responses (239). However, much of the current research has focused primarily on describing immunological changes through survival rates in animals challenged with homologous pathogens (62, 239). To advance understanding, further research is needed to explore the underlying immunological pathways and metabolic alterations in innate immune cells, particularly regarding chromatin remodeling—a key distinction between immune priming and trained immunity, with the latter offering more prolonged protection (240). In this context, it is crucial to determine whether the epigenetic changes associated with trained immunity can enhance the expression of immune genes, such as PRPs and AMPs, thereby enabling more precise immune recognition and response to pathogens.
Author contributions
JB: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. TR: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. BD: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. UK Government – Department of Health and Social Care (DHSC), Global AMR Innovation Fund (AMRIF) and the International Development Research Centre, Ottawa, Canada. Jointly provided the research funds. Canada Research Council Chair (950–232105) Paid salary to BD. NSERC CREATE FishCAST studentship. Paid salary to JB.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Buchmann K. Evolution of innate immunity: Clues from invertebrates via fish to mammals. Front Immunol. (2014) 5:571. doi: 10.3389/fimmu.2014.00459
2. Kaufman J. The Evolutionary origins of the adaptive immune system of jawed vertebrates. In: Kaufmann SHE, Sher A, editors. The immune response to infection. ASM Press, Washington, DC (2014). p. 41–55.
3. Loker ES, Adema CM, Zhang SM, Kepler TB. Invertebrate immune systems: not homogeneous, not simple, not well understood. Immunol Rev. (2004) 198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x
4. Webber W, Fenwick G, Bradford-Grieve J, Eagar S, Buckeridge J, Poore G, et al. Phylum Arthropoda Subphylum Crustacea: shrimps, crabs, lobsters, barnacles, slaters, and kin. In: Gordon DP New Zealand Inventory of Biodiversity: Volume Two: Kingdom Animalia - Chaetognatha, Ecdysozoa, Ichnofossils. New Zealand: Canterbury University Press (2010) 98–232. Available online at: https://researchrepository.rmit.edu.au/esploro/outputs/bookChapter/Phylum-Arthropoda-Subphylum-Crustacea-shrimps-crabs/9921857812301341/filesAndLinks?index=0.
5. FAO. The state of world fisheries and aquaculture 2022: towards blue transformation. Rome: FAO (2022).
6. Kumar V, Roy S, Behera BK, Bossier P, Das BK. Acute hepatopancreatic necrosis disease (AHPND): Virulence, pathogenesis and mitigation strategies in shrimp aquaculture. Fish Shellfish Immunol. (2021) 13:524. doi: 10.3390/toxins13080524
8. Coates CJ, Söderhäll K. The stress–immunity axis in shellfish. J Invertebr Pathol. (2021) 186:107387. doi: 10.1016/j.jip.2020.107492
9. Zhang XH, He X, Austin B. Vibrio harveyi: a serious pathogen of fish and invertebrates in mariculture. Mar Life Sci Technol. (2020) 2:231–45. doi: 10.1007/s42995-020-00037-z
10. Thornber K, Verner-Jeffreys D, Hinchliffe S, Rahman MM, Bass D, Tyler CR. Evaluating antimicrobial resistance in the global shrimp industry. Rev Aquacult. (2020) 12:966–86. doi: 10.1111/raq.12367
11. Hauton C. The scope of the crustacean immune system for disease control. J Invertebr Pathol. (2012) 110:251–60. doi: 10.1016/j.jip.2012.03.005
12. Wang XW, Wang JX. Pattern recognition receptors acting in innate immune system of shrimp against pathogen infections. Fish Shellfish Immunol. (2013) 34:981–9. doi: 10.1016/j.fsi.2012.08.008
13. Sritunyalucksana K, Söderhäll K. The proPO and clotting system in crustaceans. Aquaculture. (2000) 191:53–69. doi: 10.1016/S0044-8486(00)00411-7
14. Kulkarni A, Krishnan S, Anand D, Kokkattunivarthil Uthaman S, Otta SK, Karunasagar I, et al. Immune responses and immunoprotection in crustaceans with special reference to shrimp. Rev Aquac. (2021) 13:431–59. doi: 10.1111/raq.12482
15. Patnaik BB, Baliarsingh S, Sarkar A, Hameed ASS, Lee YS, Jo YH, et al. The role of pattern recognition receptors in crustacean innate immunity. Rev Aquac. (2024) 16:190–233. doi: 10.1111/raq.12829
16. Fajardo C, Martinez-Rodriguez G, Costas B, Mancera JM, Fernandez-Boo S, Rodulfo H, et al. Shrimp immune response: A transcriptomic perspective. Rev Aquac. (2022) 14:1136–49. doi: 10.1111/raq.12642
17. Amparyup P, Charoensapsri W, Tassanakajon A. Prophenoloxidase system and its role in shrimp immune responses against major pathogens. Fish Shellfish Immunol. (2013) 34:990–1001. doi: 10.1016/j.fsi.2012.08.019
18. Tassanakajon A, Rimphanitchayakit V, Visetnan S, Amparyup P, Somboonwiwat K, Charoensapsri W, et al. Shrimp humoral responses against pathogens: antimicrobial peptides and melanization. Dev Comp Immunol. (2018) 80:81–93. doi: 10.1016/j.dci.2017.05.009
19. Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
20. Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. (2012) 86:2900–10. doi: 10.1128/JVI.05738-11
21. Wang XW, Wang JX. Pattern recognition receptors acting in the innate immune system of shrimp against pathogen infections. Fish Shellfish Immunol. (2013) 34:981–9. doi: 10.1016/j.fsi.2012.08.008
22. Cheng W, Liu CH, Tsai CH, Chen JC. Molecular cloning and characterisation of a pattern recognition molecule, lipopolysaccharide- and β-1,3-glucan binding protein (LGBP) from the white shrimp. Litopenaeus vannamei. Fish Shellfish Immunol. (2005) 18:297–310. doi: 10.1016/j.fsi.2004.08.002
23. Sritunyalucksana K, Lee SY, Söderhäll K. A β-1,3-glucan binding protein from the black tiger shrimp, Penaeus monodon. Dev Comp Immunol. (2002) 26(4):377–85. doi: 10.1016/S0145-305X(01)00074-X
24. Lin YC, Vaseeharan B, Chen JC. Identification and phylogenetic analysis on lipopolysaccharide and β-1,3-glucan binding protein (LGBP) of kuruma shrimpMarsupenaeus japonicus. . Dev Comp Immunol. (2008) 32:1260–9. doi: 10.1016/j.dci.2008.05.003
25. Du XJ, Zhao XF, Wang JX. Molecular cloning and characterization of a lipopolysaccharide and β-1,3-glucan binding protein from fleshy prawn (Fenneropenaeus chinensis). Mol Immunol. (2007) 44:1085–94. doi: 10.1016/j.molimm.2006.07.288
26. Chai LQ, Meng JH, Gao J, Xu YH, Wang XW. Identification of a crustacean β-1,3-glucanase related protein as a pattern recognition protein in antibacterial response. Fish Shellfish Immunol. (2018) 80:155–64. doi: 10.1016/j.fsi.2018.06.004
27. Romo-Figueroa MG, Vargas-Requena C, Sotelo-Mundo RR, Vargas-Albores F, Higuera-Ciapara I, Söderhäll K, et al. Molecular cloning of a β-glucan pattern-recognition lipoprotein from the white shrimp Penaeus (Litopenaeus) vannamei: correlations between the deduced amino acid sequence and the native protein structure. Dev Comp Immunol. (2004) 28:713–26. doi: 10.1016/j.dci.2003.11.008
28. Yepiz-Plascencia G, Vargas-Albores F, Jimenez-Vega F, Ruiz-Verdugo LM, Romo-Figueroa G. Shrimp plasma HDL and β-glucan binding protein (BGBP): comparison of biochemical characteristics. Comp Biochem Physiol. (1998) 121:1–10. doi: 10.1016/S0305-0491(98)10104-9
29. Lubzens E, Ravid T, Khayat M, Daube N, Tietz A. Isolation and characterization of the high-density lipoproteins from the hemolymph and ovary of the penaeid shrimp Penaeus semisulcatus (de Haan): apoproteins and lipids. J Exp Zool. (1997) 278:339–48. doi: 10.1002/(SICI)1097-010X(19970815)278:6<339::AID-JEZ1>3.0.CO;2-P
31. Wei X, Liu X, Yang J, Fang J, Qiao H, Zhang Y, et al. Two C-type lectins from shrimp Litopenaeus vannamei that might be involved in immune response against bacteria and virus. Fish Shellfish Immunol. (2012) 32:132–40. doi: 10.1016/j.fsi.2011.11.001
32. Alpuche J, Pereyra A, Agundis C, Rosas C, Pascual C, Slomianny MC, et al. Purification and characterization of a lectin from the white shrimp Litopenaeus setiferus (Crustacea decapoda) hemolymph. Biochim Biophys Acta Gen Subj. (2005) 1724:86–93. doi: 10.1016/j.bbagen.2005.04.014
33. Viana JT, Rocha R dos S, Maggioni R. Structural and functional diversity of lectins associated with immunity in the marine shrimp Litopenaeus vannamei. Fish Shellfish Immunol. (2022) 129:152–60. doi: 10.1016/j.fsi.2022.08.051
34. Jin XK, Li S, Guo XN, Cheng L, Wu MH, Tan SJ, et al. Two antibacterial C-type lectins from crustacean, Eriocheir sinensis, stimulated cellular encapsulation in vitro. Dev Comp Immunol. (2013) 41:544–52. doi: 10.1016/j.dci.2013.07.016
35. Xu S, Jing M, Liu WY, Dong H, Kong DM, Wang YR, et al. Identification and characterization of a novel L-type lectin (MjLTL2) from kuruma shrimp (Marsupenaeus japonicus). Fish Shellfish Immunol. (2020) 98:354–63. doi: 10.1016/j.fsi.2020.01.022
36. Tran NT, Kong T, Zhang M, Li S. Pattern recognition receptors and their roles on the innate immune system of mud crab (Scylla paramamosain). Dev Comp Immunol. (2020) 102:103469. doi: 10.1016/j.dci.2019.103469
37. Wang XW, Wang JX. Diversity and multiple functions of lectins in shrimp immunity. Dev Comp Immunol. (2013) 39:27–38. doi: 10.1016/j.dci.2012.04.009
38. Shi XZ, Yang MC, Le KX, Li YX, Hong PP, Zhao XF, et al. Scavenger receptor B2, a type III membrane pattern recognition receptor, senses LPS and activates the IMD pathway in crustaceans. Proc Natl Acad Sci U S A. (2023), 120–24. doi: 10.1073/pnas.2216574120
39. Fan S, Wang F, Xie Z, Zhao C, Wang P, Yan L, et al. Molecular characterization and functional analysis of scavenger receptor class B from black tiger shrimp (Penaeus monodon). Electron J Biotechnol. (2021) 51:40–9. doi: 10.1016/j.ejbt.2021.03.003
40. Tran NT, Liang H, Zhang M, Bakky MAH, Zhang Y, Li S. Role of cellular receptors in the innate immune system of crustaceans in response to white spot syndrome virus. Viruses. (2022) 14:1–4. doi: 10.3390/v14040743
41. Ng TH, Chiang YA, Yeh YC, Wang HC. Review of Dscam-mediated immunity in shrimp and other arthropods. Dev Comp Immunol. (2014) 46:129–38. doi: 10.1016/j.dci.2014.04.002
42. Armitage SAO, Kurtz J, Brites D, Dong Y, Du Pasquier L, Wang HC. Dscam1 in pancrustacean immunity: current status and a look to the future. Front Immunol. (2017) 8:648. doi: 10.3389/fimmu.2017.00662
43. Wang Y, Sumathipala N, Rayaprolu S, Jiang H. Recognition of microbial molecular patterns and stimulation of prophenoloxidase activation by a β-1,3-glucanase-related protein in Manduca sexta larval plasma. Insect Biochem Mol Biol. (2011) 41:322–31. doi: 10.1016/j.ibmb.2011.01.010
44. Pauchet Y, Freitak D, Heidel-Fischer HM, Heckel DG, Vogel H. Immunity or digestion: Glucanase activity in a glucan-binding protein family from Lepidoptera. J Biol Chem. (2009) 284:2214–24. doi: 10.1074/jbc.M806204200
45. Liu F, Li F, Dong B, Wang X, Xiang J. Molecular cloning and characterisation of a pattern recognition protein, lipopolysaccharide and β-1,3-glucan binding protein (LGBP) from Chinese shrimp Fenneropenaeus chinensis. Mol Biol Rep. (2009) 36:471–7. doi: 10.1007/s11033-007-9203-2
46. Lee SY, Söderhäll K. Early events in crustacean innate immunity. Fish Shellfish Immunol. (2002) 12:421–37. doi: 10.1006/fsim.2002.0420
47. Yepiz-Plascencia G, Galván TG, Vargas-Albores F, García-Bañuelos M. Synthesis of hemolymph high-density lipoprotein β-glucan binding protein by Penaeus vannamei shrimp hepatopancreas. Mar Biotechnol. (2000) 2:485–92. doi: 10.1007/s101260000030
48. Cerenius L, Liang Z, Duvic B, Keyser P, Hellman U, Palva ET, et al. Structure and biological activity of a 1,3-β-D-glucan-binding protein in crustacean blood. J Biol Chem. (1994) 269:29462–7. doi: 10.1016/S0021-9258(18)43902-6
49. Jiravanichpaisal P, Lee BL, Söderhäll K. Cell-mediated immunity in arthropods: Hematopoiesis, coagulation, melanization and opsonization. Immunobiology. (2006) 211:213–36. doi: 10.1016/j.imbio.2005.10.015
50. Gonçalves P, Vernal J, Rosa RD, Yepiz-Plascencia G, de Souza CRB, Barracco MA, et al. Evidence for a novel biological role for the multifunctional β-1,3-glucan binding protein in shrimp. Mol Immunol. (2012) 51:363–7. doi: 10.1016/j.molimm.2012.03.032
51. Duvic B, Söderhäll K. Purification and partial characterization of a β-1,3-glucan-binding-protein membrane receptor from blood cells of the crayfish Pacifastacus leniusculus. . Eur J Biochem. (1992) 207:223–8. doi: 10.1111/j.1432-1033.1992.tb17041.x
52. Thörnqvist PO, Johansson MW, Söderhäll K. Opsonic activity of cell adhesion protein and β-1,3-glucan binding protein from two crustaceans. Dev Comp Immunol. (1994) 18:13–9. doi: 10.1016/0145-305x(94)90247-x
53. Yepiz-Plascencia GM, Sotelo-Mundo R, Vazquez-Moreno L, Ziegler R, Higuera-Ciapara I. A non-sex-specific hemolymph lipoprotein from the white shrimp Penaeus vannamei Boone. Isolation and partial characterization. Comp Biochem Physiol B Biochem Mol Biol. (1995) 11:181–7. doi: 10.1016/0305-0491(94)00254-R
54. Komatsu M, Ando S, Teshima SI Comparison of hemolymph lipoproteins from four species of crustacea. J Exp Zool. (1993) 266:257–65. doi: 10.1002/jez.1402660403
55. Lakhtin V, Lakhtin M, Alyoshkin V. Lectins of living organisms: the overview. Anaerobe. (2011) 17:452–5. doi: 10.1016/j.anaerobe.2011.06.004
56. Zhao ZY, Yin ZX, Xu XP, Weng SP, Rao XY, Dai ZX, et al. A novel C-Type lectin from the shrimp Litopenaeus vannamei possesses anti-white spot syndrome virus activity. J Virol. (2009) 83:347–56. doi: 10.1128/JVI.00707-08
57. Chen HY, Li WY, Wang J, Bo GW, Yang GW, Yang HT. A C-type lectin containing two carbohydrate recognition domains participates in the antibacterial response by regulating the JNK pathway and promoting phagocytosis. Fish Shellfish Immunol. (2022) 127:349–56. doi: 10.1016/j.fsi.2022.06.007
58. Junkunlo K, Prachumwat A, Tangprasittipap A, Senapin S, Borwornpinyo S, Flegel TW, et al. A novel lectin domain-containing protein (LvCTLD) associated with response of the whiteleg shrimp Penaeus (Litopenaeus) vannamei to yellow head virus (YHV). Dev Comp Immunol. (2012) 37:334–41. doi: 10.1016/j.dci.2011.12.010
59. Liu S, Zheng SC, Li YL, Li J, Liu HP. Hemocyte-mediated phagocytosis in crustaceans. Frontier Immunol. (2020) 11:11. doi: 10.3389/fimmu.2020.00268
60. Li M, Li C, Ma C, Li H, Zuo H, Weng S, et al. Identification of a C-type lectin with antiviral and antibacterial activity from pacific white shrimp Litopenaeus vannamei. Dev Comp Immunol. (2014) 46:231–40. doi: 10.1016/j.dci.2014.04.014
61. Ng TH, Kurtz J. Dscam in immunity: a question of diversity in insects and crustaceans. Dev Comp Immunol. (2020), 105. doi: 10.1016/j.dci.2019.103539
62. Roy S, Bossier P, Norouzitallab P, Vanrompay D. Trained immunity and perspectives for shrimp aquaculture. Rev Aquacult. (2020) 12:2351–70. doi: 10.1111/raq.12438
63. Yang LS, Yin ZX, Liao JX, Huang XD, Guo CJ, Weng SP, et al. A Toll receptor in shrimp. Mol Immunol. (2007) 44:1999–2008. doi: 10.1016/j.molimm.2006.09.021
64. Liu Y, Song L, Sun Y, Liu T, Hou F, Liu X. Comparison of immune response in Pacific white shrimp, Litopenaeus vannamei, after knock down of Toll and IMD gene in vivo. Dev Comp Immunol. (2016) 60:41–52. doi: 10.1016/j.dci.2016.02.004
65. Li F, Xiang J. Signaling pathways regulating innate immune responses in shrimp. Fish Shellfish Immunol. (2013) 34:973–80. doi: 10.1016/j.fsi.2012.08.023
66. Li C, Wang S, He J. The two NF-κB pathways regulating bacterial and WSSV infection of shrimp. Front Immunol. (2019) 10:1785. doi: 10.3389/fimmu.2019.01785
67. Wang PH, Liang JP, Gu ZH, Wan DH, Weng SP, Yu XQ, et al. Molecular cloning, characterization and expression analysis of two novel Tolls (LvToll2 and LvToll3) and three putative Spätzle-like Toll ligands (LvSpz1-3) from. Litopenaeus vannamei. Dev Comp Immunol. (2012) 36:359–71. doi: 10.1016/j.dci.2011.07.007
68. Han-Ching Wang KC, Tseng CW, Lin HY, Chen IT, Chen YH, Chen YM, et al. RNAi knock-down of the Litopenaeus vannamei Toll gene (LvToll) significantly increases mortality and reduces bacterial clearance after challenge with Vibrio harveyi. Dev Comp Immunol. (2010) 34:49–58. doi: 10.1016/j.dci.2009.08.003
69. Yao D, Su H, Zhu J, Zhao X, Aweya JJ, Wang F, et al. SNPs in the Toll1 receptor of Litopenaeus vannamei are associated with immune response. Fish Shellfish Immunol. (2018) 72:410–7. doi: 10.1016/j.fsi.2017.11.018
70. Guanzon DAV, Maningas MBB. Functional elucidation of LvToll 3 receptor from P. vannamei through RNA interference and its potential role in the shrimp antiviral response. Dev Comp Immunol. (2018) 84:172–80. doi: 10.1016/j.dci.2018.01.020
71. Mekata T, Kono T, Yoshida T, Sakai M, Itami T. Identification of cDNA encoding Toll receptor, MjToll gene from kuruma shrimp, Marsupenaeus japonicus. Fish Shellfish Immunol. (2008) 24:122–33. doi: 10.1016/j.fsi.2007.10.006
72. Arts JAJ, Cornelissen FHJ, Cijsouw T, Hermsen T, Savelkoul HFJ, Stet RJM. Molecular cloning and expression of a Toll receptor in the giant tiger shrimp, Penaeus monodon. Fish Shellfish Immunol. (2007) 23:504–13. doi: 10.1016/j.fsi.2006.08.018
73. Liu Q, Xu D, Jiang S, Huang J, Zhou F, Yang Q, et al. Toll-receptor 9 gene in the black tiger shrimp (Penaeus monodon) induced the activation of the TLR–NF-κB signaling pathway. Gene. (2018) 639:27–33. doi: 10.1016/j.gene.2017.09.060
74. Assavalapsakul W, Panyim S. Molecular cloning and tissue distribution of the Toll receptor in the black tiger shrimp, Penaeus monodon. . Genet Mol Res. (2012) 11:484–93. doi: 10.4238/2012.March.6.1
75. Wang Z, Chen YH, Dai YJ, Tan JM, Huang Y, Lan JF, et al. A novel vertebrates Toll-like receptor counterpart regulating the anti-microbial peptides expression in the freshwater crayfish, Procambarus clarkii. Fish Shellfish Immunol. (2015) 43:219–29. doi: 10.1016/j.fsi.2014.12.038
76. Lan JF, Zhao LJ, Wei S, Wang Y, Lin L, Li XC. PcToll2 positively regulates the expression of antimicrobial peptides by promoting PcATF4 translocation into the nucleus. Fish Shellfish Immunol. (2016) 58:59–66. doi: 10.1016/j.fsi.2016.09.007
77. Lan JF, Wei S, Wang YQ, Dai YJ, Tu JG, Zhao LJ, et al. PcToll3 was involved in anti-Vibrio response by regulating the expression of antimicrobial peptides in red swamp crayfish, Procambarus clarkii. Fish Shellfish Immunol. (2016) 57:17–24. doi: 10.1016/j.fsi.2016.08.021
78. Huang Y, Li T, Jin M, Yin S, Hui KM, Ren Q. Newly identified PcToll4 regulates antimicrobial peptide expression in intestine of red swamp crayfish Procambarus clarkii. Gene. (2017) 610:140–7. doi: 10.1016/j.gene.2017.02.018
79. Yang C, Zhang J, Li F, Ma H, Zhang Q, Jose Priya TA, et al. A Toll receptor from Chinese shrimp Fenneropenaeus chinensis is responsive to Vibrio Anguillarum infection. Fish Shellfish Immunol. (2008) 24:564–74. doi: 10.1016/j.fsi.2007.12.012
80. Srisuk C, Longyant S, Senapin S, Sithigorngul P, Chaivisuthangkura P. Molecular cloning and characterization of a Toll receptor gene from Macrobrachium rosenbergii. . Fish Shellfish Immunol. (2014) 36:552–62. doi: 10.1016/j.fsi.2013.12.025
81. Feng J, Zhao L, Jin M, Li T, Wu L, Chen Y, et al. Toll receptor response to white spot syndrome virus challenge in giant freshwater prawns (Macrobrachium rosenbergii). Fish Shellfish Immunol. (2016) 57:148–59. doi: 10.1016/j.fsi.2016.08.017
82. Lang L, Bao M, Jing W, Chen W, Wang L. Clone, identification and functional characterization of a novel toll (Shtoll1) from the freshwater crab Sinopotamon henanense in response to cadmium exposure and Aeromonas hydrophila infection. Fish Shellfish Immunol. (2020) 98:401–13. doi: 10.1016/j.fsi.2020.01.012
83. Zhou SM, Yuan XM, Liu S, Li M, Tao Z, Wang GL. Three novel Toll genes (PtToll1-3) identified from a marine crab, Portunus trituberculatus: Different tissue expression and response to pathogens. Fish Shellfish Immunol. (2015) 46:737–44. doi: 10.1016/j.fsi.2015.07.027
84. Lin Z, Qiao J, Zhang Y, Guo L, Huang H, Yan F, et al. Cloning and characterisation of the SpToll gene from green mud crab, Scylla paramamosain. . Dev Comp Immunol. (2012) 37:164–75. doi: 10.1016/j.dci.2011.09.003
85. Yu AQ, Jin XK, Guo XN, Li S, Wu MH, Li WW, et al. Two novel Toll genes (EsToll1 and EsToll2) from Eriocheir sinensis are differentially induced by lipopolysaccharide, peptidoglyca,n and zymosan. Fish Shellfish Immunol. (2013) 35:1282–92. doi: 10.1016/j.fsi.2013.07.044
86. Li YY, Chen XX, FYu L, Chen QF, Ma XY, Liu HP. CqToll participates in antiviral response against white spot syndrome virus via induction of anti-lipopolysaccharide factor in red claw crayfish Cherax quadricarinatus. Dev Comp Immunol. (2017) 74:217–26. doi: 10.1016/j.dci.2017.04.020
87. Yuan K, Yuan FH, Weng SP, He JG, Chen YH. Identification and functional characterization of a novel Spätzle gene in Litopenaeus vannamei. Dev Comp Immunol. (2017) 68:46–57. doi: 10.1016/j.dci.2016.11.016
88. Boonrawd S, Mani R, Ponprateep S, Supungul P, Masrinoul P, Tassanakajon A, et al. Characterization of PmSpatzle 1 from the black tiger shrimp Peneaus monodon. . Fish Shellfish Immunol. (2017) 65:88–95. doi: 10.1016/j.fsi.2017.04.005
89. Shi XZ, Zhang RR, Jia YP, Zhao XF, Yu XQ, Wang JX. Identification and molecular characterization of a Spätzle-like protein from Chinese shrimp (Fenneropenaeus chinensis). Fish Shellfish Immunol. (2009) 27:610–7. doi: 10.1016/j.fsi.2009.07.005
90. Ren Q, Huang X, Cui Y, Sun J, Wang W, Zhang X. Two White Spot Syndrome Virus MicroRNAs target the dorsal gene to promote virus infection in Marsupenaeus japonicus shrimp. J Virol. (2017) 91(8). doi: 10.1128/JVI.02261-16
91. Vaniksampanna A, Longyant S, Charoensapsri W, Sithigorngul P, Chaivisuthangkura P. Molecular isolation and characterization of a spätzle gene from Macrobrachium rosenbergii. Fish Shellfish Immunol. (2019) 84:441–50. doi: 10.1016/j.fsi.2018.10.015
92. Deepika A, Sreedharan K, Paria A, Makesh M, Rajendran KV. Toll-pathway in tiger shrimp (Penaeus monodon) responds to white spot syndrome virus infection: Evidence through molecular characterisation and expression profiles of MyD88, TRAF6 and TLR genes. Fish Shellfish Immunol. (2014) 41:441–54. doi: 10.1016/j.fsi.2014.09.026
93. Zhang S, Li CZ, Yan H, Qiu W, Chen YG, Wang PH, et al. Identification and function of Myeloid Differentiation Factor 88 (MyD88) in Litopenaeus vannamei. PloS One. (2012) 7(10). doi: 10.1371/journal.pone.0047038
94. Wen R, Li F, Sun Z, Li S, Xiang J. Shrimp MyD88 responsive to bacteria and white spot syndrome virus. Fish Shellfish Immunol. (2013) 34:574–81. doi: 10.1016/j.fsi.2012.11.034
95. Wang PH, Gu ZH, Wan DH, Zhang MY, Weng SP, Yu XQ, et al. The shrimp NF-κB pathway is activated by white spot syndrome virus (WSSV) 449 to facilitate the expression of WSSV069 (ie1), WSSV303 and WSSV371. PloS One. (2011) 6(9). doi: 10.1371/journal.pone.0024773
96. Li C, Chen Y, Weng S, Li S, Zuo H, Yu X, et al. Presence of Tube isoforms in Litopenaeus vannamei suggests various regulatory patterns of signal transduction in invertebrate NF-κB pathway. Dev Comp Immunol. (2014) 42:174–85. doi: 10.1016/j.dci.2013.08.012
97. Sreedharan K, Deepika A, Paria A, Suresh Babu PP, Makesh M, Rajendran KV. Ontogeny and expression analysis of tube (interleukin-1 receptor-associated kinase-4 homolog) from Penaeus monodon in response to white spot syndrome virus infection and on exposure to ligands. Agri Gene. (2017) 3:21–31. doi: 10.1016/j.aggene.2016.10.002
98. Li C, Chai J, Li H, Zuo H, Wang S, Qiu W, et al. Pellino protein from pacific white shrimp Litopenaeus vannamei positively regulates NF-κB activation. Dev Comp Immunol. (2014) 44:341–50. doi: 10.1016/j.dci.2014.01.012
99. Wang PH, Wan DH, Gu ZH, Deng XX, Weng SP, Yu XQ, et al. Litopenaeus vannamei tumor necrosis factor receptor-associated factor 6 (TRAF6) responds to Vibrio alginolyticus and white spot syndrome virus (WSSV) infection and activates antimicrobial peptide genes. Dev Comp Immunol. (2011) 35:105–14. doi: 10.1016/j.dci.2010.08.013
100. Li C, Chen YX, Zhang S, Lü L, Chen YH, Chai J, et al. Identification, characterization, and function analysis of the cactus gene from Litopenaeus vannamei. PloS One. (2012) 7(11). doi: 10.1371/journal.pone.0049711
101. Huang X, Wang W, Ren Q. Dorsal transcription factor is involved in regulating expression of crustin genes during white spot syndrome virus infection. Dev Comp Immunol. (2016) 63:18–26. doi: 10.1016/j.dci.2016.05.006
102. Huang X,D, Yin ZX, Jia XT, Liang JP, Ai HS, Yang LS, et al. Identification and functional study of a shrimp Dorsal homologue. . Dev Comp Immunol. (2010) 34:107–13. doi: 10.1016/j.dci.2009.08.009
103. Li F, Wang D, Li S, Yan H, Zhang J, Wang B, et al. A Dorsal homolog (FcDorsal) in the Chinese shrimp Fenneropenaeus chinensis is responsive to both bacteria and WSSV challenge. Dev Comp Immunol. (2010) 34:874–83. doi: 10.1016/j.dci.2010.03.008
104. Sun JJ, Lan JF, Shi XZ, Yang MC, Niu GJ, Ding D, et al. [amp]]beta;-Arrestins negatively regulate the toll pathway in shrimp by preventing dorsal translocation and inhibiting dorsal transcriptional activity. J Biol Chem. (2016) 291:7488–504. doi: 10.1074/jbc.M115.698134
105. Anderson KV, Bokla L, Niisslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: The induction of polarity by the Toll gene product. Cell. (1985) 42:791–8. doi: 10.1016/0092-8674(85)90275-2
106. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat Immunol. (2010) 11:373–84. doi: 10.1038/ni.1863
107. Nie L, Cai SY, Shao JZ, Chen J. Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front Immunol. (2018) 9:1523. doi: 10.3389/fimmu.2018.01523
108. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. (2003) 21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126
109. Spierer P, Chasan R, Ce AKV, Roth S, Ste ND, Nusslein-Volhard C. Drosophila toll and IL-1 receptor. Proc Natl Acad Sci USA. (1986) 83:5390–4. doi: 10.1038/351355b0
110. Yagi Y, Nishida Y, Ip YT. Functional analysis of Toll-related genes in Drosophila. Dev Growth Differ. (2010) 52:771–83. doi: 10.1111/j.1440-169X.2010.01213.x
111. Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. (2001) 414):756–9. doi: 10.1038/414756a
112. Ligoxygakis P, Ge Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. (2002) 297(5578):114–6. doi: 10.1126/science.1072391
113. Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. (2003) 4:478–84. doi: 10.1038/ni922
114. Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. (2006) 127:1425–37. doi: 10.1016/j.cell.2006.10.046
115. El Chamy LE, Leclerc V, Caldelari I, Reichhart JM. Sensing of “danger signals” and pathogen-associated molecular patterns defines binary signaling pathways “upstream” of Toll. Nat Immunol. (2008) 9:1165–70. doi: 10.1038/ni.1643
116. Wang L, Weber ANR, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. (2006) 25:5005–14. doi: 10.1038/sj.emboj.7601363
117. Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, et al. Dual activation of the Drosophila Toll pathway by two pattern recognition receptors. Science. (2003) 302:2126–30. doi: 10.1126/science.1085432
118. Valanne S, Wang JH, Rämet M. The drosophila toll signaling pathway. J Immunol. (2011) 186:649–56. doi: 10.4049/jimmunol.1002302
119. Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, Gonzalez-Crespo S, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. (1993) 75(4):753–763. doi: 10.1016/0092-8674(93)90495-C
120. Nicolas E, Reichhart JM, Hoffmann JA, Lemaitre B. In vivo regulation of the IκB homologue cactus during the immune response of. Drosophila. J Biol Chem. (1998) 273:10463–9. doi: 10.1074/jbc.273.17.10463
121. Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. (2002) 3:91–7. doi: 10.1038/ni747
122. Imler JL, Bulet P. Antimicrobial Peptides in Drosophila: Structures, activities and gene regulation. Chem Immunol Allergy. (2005) 86:1–13. doi: 10.1159/000086648
123. Fehlbaums P, Bulets P, Michauts L, Lagueuxs M, Broekaerto WF, Hetrus C, et al. Insect immunity: septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J Biol Chem. (1994) 269(52):33159–63. doi: 10.1016/S0021-9258(20)72439-4
124. Zhang ZT, Zhu SY. Drosomycin, an essential component of antifungal defense in Drosophila. Insect Mol Biol. (2009) 18:549–56. doi: 10.1111/j.1365-2583.2009.00907.x
125. Moynagh PN. The Pellino family: IRAK E3 ligases with emerging roles in innate immune signaling. Trends Immunol. (2009) 30:33–42. doi: 10.1016/j.it.2008.10.001
126. Haghayeghi A, Sarac A, Czerniecki S, Grosshans J, Schöck F. Pellino enhances innate immunity in Drosophila. Mech Dev. (2010) 127:301–7. doi: 10.1016/j.mod.2010.01.004
127. Shen B, Liu H, Skolnik EY, Manley JL. Physical and functional interactions between Drosophila TRAF2 and Pelle kinase contribute to Dorsal activation. Proc Natl Acad Sci USA. (2001) 98:10315–20. doi: 10.1073/pnas.141235698
128. Qiu W, Zhang S, Chen YG, Wang PH, Xu XP, Li CZ, et al. Litopenaeus vannamei NF-κB is required for WSSV replication. Dev Comp Immunol. (2014) 45:156–62. doi: 10.1016/j.dci.2014.02.016
129. Zhang Y, He X, Yu F, Xiang Z, Li J, Thorpe KL, et al. Characteristic and functional analysis of Toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity. PloS One. (2013) 8(10). doi: 10.1371/journal.pone.0076464
130. Gay NJ, Gangloff M. Structure and function of toll receptors and their ligands. Annu Rev Biochem. (2007) 76:141–65. doi: 10.1146/annurev.biochem.76.060305.151318
131. Leulier F, Lemaitre B. Toll-like receptors - Taking an evolutionary approach. Nat Rev Genet. (2008) 9:165–78. doi: 10.1038/nrg2303
132. Imler JL, Zheng L. Biology of Toll receptors: lessons from insects and mammals. J Leukoc Biol. (2004) 75:18–26. doi: 10.1189/jlb.0403160
133. Li H, Yin B, Wang S, Fu Q, Xiao B, Lu K, et al. RNAi screening identifies a new Toll from shrimp Litopenaeus vannamei that restricts WSSV infection through activating Dorsal to induce antimicrobial peptides. PloS Pathog. (2018) 14(9). doi: 10.1371/journal.ppat.1007109
134. Sun JJ, Xu S, He ZH, Shi XZ, Zhao XF, Wang JX. Activation of Toll pathway is different between kuruma shrimp and. Drosophila. Front Immunol. (2017) 8:1517. doi: 10.3389/fimmu.2017.01151
135. Bell JK, Mullen GED, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. (2003) 24:528–33. doi: 10.1016/S1471-4906(03)00242-4
136. Re F, Strominger JL. Monomeric recombinant MD-2 binds toll-like receptor 4 tightly and confers lipopolysaccharide responsiveness. J Biol Chem. (2002) 277:23427–32. doi: 10.1074/jbc.M202554200
137. Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. (1999) 189:1777–82. doi: 10.1084/jem.189.11.1777
138. Weber ANR, Tauszig-Delamasure S, Hoffmann JA, Lelièvre E, Gascan H, Ray KP, et al. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat Immunol. (2003) 4:1195–201. doi: 10.1038/ni955
139. Weber ANR, Morse MA, Gay NJ. Four N-linked glycosylation sites in human toll-like receptor 2 cooperate to direct efficient biosynthesis and secretion. J Biol Chem. (2004) 279:34589–94. doi: 10.1074/jbc.M403830200
140. Ohnishi T, Muroi M, Tanamoto KI. MD-2 is necessary for the Toll-like receptor 4 protein to undergo glycosylation essential for its translocation to the cell surface. Clin Diagn Lab Immunol. (2003) 10:405–10. doi: 10.1128/CDLI.10.3.405-410.2003
141. Sun J, Duffy KE, Ranjith-Kumar CT, Xiong J, Lamb RJ, Santos J. Structural and functional analyses of the human toll-like receptor 3: role of glycosylation. J Biol Chem. (2006) 281:11144–51. doi: 10.1074/jbc.M510442200
142. Andersen-Nissen E, Smith KD, Bonneau R, Strong RK, Aderem A. A conserved surface on Toll-like receptor 5 recognizes bacterial flagellin. J Exp Med. (2007) 204:393–403. doi: 10.1084/jem.20061400
143. Yin SY, Chen J, Zhu MX, BaoQing H, Su FX, Jian SQ. et al Characterization of a novel toll-like receptor and activation NF-κB signal pathway in triangle sail mussel Hyriopsis cumingii. Comp Biochem Physiol B Biochem Mol Biol. (2021) 255:110–5. doi: 10.1016/j.cbpb.2021.110608
144. Ren Q, Lan JF, Zhong X, Song XJ, Ma F, Hui KM, et al. A novel Toll like receptor with two TIR domains (HcToll-2) is involved in regulation of antimicrobial peptide gene expression of. Hyriopsis cumingii. Dev Comp Immunol. (2014) 45:198–208. doi: 10.1016/j.dci.2014.02.020
145. Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. (2020) 180:1044–66. doi: 10.1016/j.cell.2020.02.041
146. Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol Res. (2007) 39:249–60. doi: 10.1007/s12026-007-0069-0
147. Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, et al. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense (antibacterial peptides/antifungal peptides/insect immunity). Genetics. (1995) 92:149–60. doi: 10.1073/pnas.92.21.9465
148. Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, et al. Drosophila Immune Deficiency (IMD) Is a death domain protein that zctivates antibacterial defense and can promote apoptosis. Dev Cell. (2001) 1:503–14. doi: 10.1016/S1534-5807(01)00059-4
149. Myllymäki H, Valanne S, Rämet M. The Drosophila Imd signaling pathway. J Immunol. (2014) 192:3455–62. doi: 10.4049/jimmunol.1303309
150. Kaneko T, Silverman N. Bacterial recognition and signaling by the. Drosophila IMD pathway. Cell Microbiol. (2005) 7:61–9. doi: 10.1111/j.1462-5822.2005.00542.x
151. Kleino A, Silverman N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol. (2014) 42:25–35. doi: 10.1016/j.dci.2013.05.014
152. Mellroth P, Karlsson J, Håkansson J, Schultz N, Goldman WE, Steiner H. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc Natl Acad Sci U S A. (2005) 102:6455–60. doi: 10.1073/pnas.0407559102
153. Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci USA. (2005) 102:6455–60. doi: 10.1073/pnas.0404952102
154. Wang S, Li H, Qian Z, Song X, Zhang Z, Zuo H, et al. Identification and functional characterization of the TAB2 gene from. Litopenaeus vannamei. Fish Shellfish Immunol. (2015) 46:206–16. doi: 10.1016/j.fsi.2015.06.024
155. Leulier F, Vidal S, Saigo K, Veda R, Lemaitre B. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr Biol. (2002) 12:79–82. doi: 10.1016/S0960-9822(02)00873-4
156. Park HH, Lo YC, Lin SC, Wang L, Yang JKY, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. (2007) 25:561–86. doi: 10.1146/annurev.immunol.25.022106.141656
157. Zhuang ZH, Sun L, Kong L, Hu JH, Yu MC, Reinach P, et al. Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. (2006) 18:964–70. doi: 10.1016/j.cellsig.2005.08.020
158. Dushay MS, Asling B, Hultmarkt D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci USA. (1996) 93:10343–7. doi: 10.1073/pnas.93.19.10343
159. Fleming Y, Armstrong CG, Morrice N, Paterson A, Goedert M, Cohen P. Synergistic activation of stress-activated protein kinase 1/c-Jun N-terminal kinase (SAPK1/JNK) isoforms by mitogen-activated protein kinase kinase 4 (MKK4) and MKK7. Biochem J. (2000) 352:145–54. doi: 10.1042/bj3520145
160. Wang PH, Gu ZH, De HX, Du LB, Deng XX, Ai HS, et al. An immune deficiency homolog from the white shrimp, Litopenaeus vannamei, activates antimicrobial peptide genes. Mol Immunol. (2009) 46:1897–904. doi: 10.1016/j.molimm.2009.01.005
161. Hou F, He S, Liu Y, Zhu X, Sun C, Liu X. RNAi knock-down of shrimp Litopenaeus vannamei Toll gene and immune deficiency gene reveals their difference in regulating antimicrobial peptides transcription. Dev Comp Immunol. (2014) 44:255–60. doi: 10.1016/j.dci.2014.01.004
162. Lan JF, Zhou J, Zhang XW, Wang ZH, Zhao XF, Ren Q, et al. Characterization of an immune deficiency homolog (IMD) in shrimp (Fenneropenaeus chinensis) and crayfish (Procambarus clarkii). Dev Comp Immunol. (2013) 41:608–17. doi: 10.1016/j.dci.2013.07.004
163. Feng N, Wang D, Wen R, Li F. Functional analysis on immune deficiency (IMD) homolog gene in Chinese shrimp Fenneropenaeus chinensis. Mol Biol Rep. (2014) 41:1437–44. doi: 10.1007/s11033-013-2988-2
164. Li TT, Ding ZF, Pan XT, Ma FT, Han KK, Wu L, et al. Characterization of an immune deficiency (IMD) homolog from the oriental river prawn, Macrobrachium nipponense. Fish Shellfish Immunol. (2018) 83:115–22. doi: 10.1016/j.fsi.2018.09.005
165. Zhou YL, Wang LZ, Bin G, Wang C, Zhu QH, Liu ZP, et al. Identification and functional analysis of immune deficiency (IMD) from Scylla paramamosain: The first evidence of IMD signaling pathway involved in immune defense against bacterial infection in crab species. Fish Shellfish Immunol. (2018) 81:150–60. doi: 10.1016/j.fsi.2018.07.016
166. Zhou SM, Zhao JJ, Wang Y, Jin S, Zhou QC, Yin F. Identification and function analysis of an immune deficiency homolog in swimming crab, Portunus trituberculatus. Fish Shellfish Immunol. (2022) 121:245–53. doi: 10.1016/j.fsi.2022.01.014
167. Wang S, Li H, Lü K, Qian Z, Weng S, He J, et al. Identification and characterization of transforming growth factor β-activated kinase 1 from Litopenaeus vannamei involved in anti-bacterial host defense. Fish Shellfish Immunol. (2016) 52:278–88. doi: 10.1016/j.fsi.2016.03.149
168. Wang S, Li H, Chen R, Jiang X, He J, Li C. TAK1 confers antibacterial protection through mediating the activation of MAPK and NF-κB pathways in shrimp. Fish Shellfish Immunol. (2022) 123:248–56. doi: 10.1016/j.fsi.2022.03.008
169. Wang S, Li M, Yin B, Li H, Xiao B, Lu K, et al. Shrimp TAB1 interacts with TAK1 and p38 and activates the host innate immune response to bacterial infection. Mol Immunol. (2017) 88:10–9. doi: 10.1016/j.molimm.2017.05.016
170. Wang PH, Gu ZH, Wan DH, Du LB, De HX, SP W, et al. The shrimp IKK-NF-κB signaling pathway regulates antimicrobial peptide expression and may be subverted by white spot syndrome virus to facilitate viral gene expression. Cell Mol Immunol. (2013) 10:423–36. doi: 10.1038/cmi.2013.30
171. Huang XD, Yin ZX, Liao JX, Wang PH, Yang LS, Ai HS, et al. Identification and functional study of a shrimp Relish homologue. Fish Shellfish Immunol. (2009) 27:230–8. doi: 10.1016/j.fsi.2009.05.003
172. Shi YR, Jin M, Ma FT, Huang Y, Huang X, Feng JL, et al. Involvement of Relish gene from Macrobrachium rosenbergii in the expression of anti-microbial peptides. Dev Comp Immunol. (2015) 52:236–44. doi: 10.1016/j.dci.2015.05.008
173. Visetnan S, Supungul P, Hirono I, Tassanakajon A, Rimphanitchayakit V. Activation of PmRelish from Penaeus monodon by yellow head virus. Fish Shellfish Immunol. (2015) 42:335–44. doi: 10.1016/j.fsi.2014.11.015
174. He Y, Yao W, Liu P, Li J, Wang Q. Expression profiles of the p38 MAPK signaling pathway from Chinese shrimp Fenneropenaeus chinensis in response to viral and bacterial infections. Gene. (2018) 642:381–8. doi: 10.1016/j.gene.2017.11.050
175. Wang S, Yin B, Li H, Xiao B, Lu K, Feng C, et al. MKK4 from Litopenaeus vannamei is a regulator of p38 MAPK kinase and involved in anti-bacterial response. Dev Comp Immunol. (2018) 78:61–70. doi: 10.1016/j.dci.2017.09.015
176. Li H, Wang S, Qian Z, Wu Z, Lü K, Weng S, et al. MKK6 from pacific white shrimp Litopenaeus vannamei is responsive to bacterial and WSSV infection. Mol Immunol. (2016) 70:72–83. doi: 10.1016/j.molimm.2015.12.011
177. Wang S, Qian Z, Li H, Lü K, Xu X, Weng S, et al. Identification and characterization of MKK7 as an upstream activator of JNK in Litopenaeus vannamei. Fish Shellfish Immunol. (2016) 48:285–94. doi: 10.1016/j.fsi.2015.12.014
178. Shi H, Yan X, Ruan L, Xu X. A novel JNK from Litopenaeus vannamei involved in white spot syndrome virus infection. Dev Comp Immunol. (2012) 37:421–8. doi: 10.1016/j.dci.2012.03.002
179. Li C, Li H, Wang S, Song X, Zhang Z, Qian Z, et al. The c-Fos and c-Jun from Litopenaeus vannamei play opposite roles in Vibrio parahaemolyticus and white spot syndrome virus infection. Dev Comp Immunol. (2015) 52:26–36. doi: 10.1016/j.dci.2015.04.009
180. Yao D, Ruan L, Xu X, Shi H. Identification of a c-Jun homolog from Litopenaeus vannamei as a downstream substrate of JNK in response to WSSV infection. Dev Comp Immunol. (2015) 49:282–9. doi: 10.1016/j.dci.2014.12.012
181. Shi H, Ruan L, Yan X, Yao D, Xu X. The role of Litopenaeus vannamei p38 in white spot syndrome virus infection. Dev Comp Immunol. (2014) 44:180–5. doi: 10.1016/j.dci.2013.12.005
182. Yan H, Zhang S, Li CZ, Chen YH, Chen YG, Weng SP, et al. Molecular characterization and function of a p38 MAPK gene from Litopenaeus vannamei. Fish Shellfish Immunol. (2013) 34:1421–31. doi: 10.1016/j.fsi.2013.02.030
183. Qiu W, He JH, Zuo H, Niu S, Li C, Zhang S, et al. Identification, characterization, and function analysis of the NF-κB repressing factor (NKRF) gene from Litopenaeus vannamei. Dev Comp Immunol. (2017) 76:83–92. doi: 10.1016/j.dci.2017.05.020
184. Hou F, Wang X, Qian Z, Liu Q, Liu Y, He S, et al. Identification and functional studies of Akirin, a potential positive nuclear factor of NF-κB signaling pathways in the Pacific white shrimp, Litopenaeus vannamei. Dev Comp Immunol. (2013) 41:703–14. doi: 10.1016/j.dci.2013.08.005
185. Liu N, Wang XW, Sun JJ, Wang L, Zhang HW, Zhao XF, et al. Akirin interacts with Bap60 and 14-3-3 proteins to regulate the expression of antimicrobial peptides in the kuruma shrimp (Marsupenaeus japonicus). Dev Comp Immunol. (2016) 55:80–9. doi: 10.1016/j.dci.2015.10.015
186. Bai L, Zhou K, Li H, Qin Y, Wang Q, Li W. Bacteria-induced IMD-Relish-AMPs pathway activation in Chinese mitten crab. Fish Shellfish Immunol. (2020) 106:866–75. doi: 10.1016/j.fsi.2020.08.046
187. Zhu F, Sun B, Wang Z. The crab Relish plays an important role in white spot syndrome virus and Vibrio alginolyticus infection. Fish Shellfish Immunol. (2019) 87:297–306. doi: 10.1016/j.fsi.2019.01.028
188. Geuking P, Narasimamurthy R, Lemaitre B, Basler K, Leulier F. A non-redundant role for Drosophila Mkk4 and hemipterous/Mkk7 in TAK1-mediated activation of JNK. PloS One. (2009) 4(11):e7709. doi: 10.1371/journal.pone.0007709
189. Zhang X, Yuan J, Sun Y, Li S, Gao Y, Yu Y, et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat Commun. (2019) 10:356. doi: 10.1038/s41467-018-08197-4
190. Kawato S, Nishitsuji K, Arimoto A, Hisata K, Kawamitsu M, Nozaki R, et al. Genome and transcriptome assemblies of the kuruma shrimp, Marsupenaeus japonicus. G3: Genes Genomes Genet. (2021) 11(11). doi: 10.1093/g3journal/jkab268
191. Wang Z, Zhou J, Li J, Zou J, Fan L. The immune defense response of Pacific white shrimp (Litopenaeus vannamei) to temperature fluctuation. Fish Shellfish Immunol. (2020) 103:103–10. doi: 10.1016/j.fsi.2020.04.053
192. Lv B, Liu B, Zhou Q, Song C, Sun C, Zhang H, et al. Effects of different temperatures and protein levels on growth performance, physiological response, and expression of immune-related genes of juvenile oriental river prawn (Macrobrachium nipponense). Aquaculture. (2021) 536:736337. doi: 10.1016/j.aquaculture.2021.736435
193. Li J, Koh JJ, Liu S, Lakshminarayanan R, Verma CS, Beuerman RW. Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front Neurosci. (2017) 11:1–16. doi: 10.3389/fnins.2017.00073
194. Kang SJ, Park SJ, Mishig-Ochir T, Lee BJ. Antimicrobial peptides: Therapeutic potentials. Expert Rev Anti Infect Ther. (2014) 12:1477–86. doi: 10.1586/14787210.2014.976613
196. León R, Ruiz M, Valero Y, Cárdenas C, Guzman F, Vila M, et al. Exploring small cationic peptides of different origin as potential antimicrobial agents in aquaculture. Fish Shellfish Immunol. (2020) 98:720–7. doi: 10.1016/j.fsi.2019.11.019
197. Otvos L. Immunomodulatory effects of anti-microbial peptides. Acta Microbiol Immunol Hung. (2016) 63:257–77. doi: 10.1556/030.63.2016.005
198. Tassanakajon A, Amparyup P, Somboonwiwat K, Supungul P. Cationic antimicrobial peptides in Penaeid shrimp. Mar Biotechnol. (2010) 12:487–505. doi: 10.1007/s10126-010-9288-9
199. Tassanakajon A, Somboonwiwat K. Antimicrobial peptides from the black tiger shrimp Penaeus monodon-A review. Asian Fisheries Soc. (2011) 385:1–27.
200. Matos GM, Rosa RD. On the silver jubilee of crustacean antimicrobial peptides. Rev Aquacult. (2022) 14:594–612. doi: 10.1111/raq.12614
201. Lv X, Li S, Yu Y, Zhang X, Li F. Crustin defense against Vibrio parahaemolyticus infection by regulating intestinal microbial balance in Litopenaeus vannamei. Mar Drugs. (2023) 21(2):130. doi: 10.3390/md21020130
202. Matos GM, Schmitt P, Barreto C, Farias ND, Toledo-Silva G, Guzmán F, et al. Massive gene expansion and sequence diversification is associated with diverse tissue distribution, regulation and antimicrobial properties of anti-lipopolysaccharide factors in shrimp. Mar Drugs. (2018) 16(10):381. doi: 10.3390/md16100381
203. Monod EC, Betancourt JL, Samms KA, Alkie TN, Walmsley CG, Rodríguez-Ramos T, et al. Immunostimulant effects of Pituitary Adenylate Cyclase-Activating Polypeptide and double-stranded (ds)RNA in Orconectes propinquus. Fish Shellfish Immunol. (2024) 146. doi: 10.1016/j.fsi.2024.109388
204. Lugo JM, Carpio Y, Morales R, Rodríguez-Ramos T, Ramos L, Estrada MP. First report of the pituitary adenylate cyclase activating polypeptide (PACAP) in crustaceans: conservation of its functions as growth-promoting factor and immunomodulator in the white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. (2013) 35:1788–96. doi: 10.1016/j.fsi.2013.08.028
205. Zhao M, Yao D, Li S, Zhang Y, Aweya JJ. Effects of ammonia on shrimp physiology and immunity: a review. Rev Aquacult. (2020) 12:2194–211. doi: 10.1111/raq.12429
206. Mengal K, Kor G, Kozák P, Niksirat H. Effects of environmental factors on the cellular and molecular parameters of the immune system in decapods. Comp Biochem Physiol A Mol Integr Physiol. (2023) 276:111332. doi: 10.1016/j.cbpa.2022.111332
207. Chen YH, He JG. Effects of environmental stress on shrimp innate immunity and white spot syndrome virus infection. Fish Shellfish Immunol. (2019) 84:744–55. doi: 10.1016/j.fsi.2018.10.069
208. Le Moullac G, Haffner P. Environmental factors affecting immune responses in Crustacea. Aquaculture. (2000) 191(1-3):121–31. doi: 10.1016/S0044-8486(00)00422-1
209. Millard RS, Ellis RP, Bateman KS, Bickley LK, Tyler CR, van Aerle R, et al. How do abiotic environmental conditions influence shrimp susceptibility to disease? A critical analysis focussed on White Spot Disease. J Invertebr Pathol. (2021) 186:107369. doi: 10.1016/j.jip.2020.107369
210. Kültz D. Crustacean aquaculture. In: Costa-Pirce B, editor. A primer of ecological aquaculture. Oxford University Press, Oxford (2022). p. 138–51.
211. Kumlu M, Türkmen S, Kumlu M. Thermal tolerance of Litopenaeus vannamei (Crustacea: Penaeidae) acclimated to four temperatures. J Therm Biol. (2010) 35:305–11. doi: 10.1016/j.jtherbio.2010.06.009
212. Rakhshaninejad M, Zheng L, Nauwynck H. Shrimp (Penaeus vannamei) survive white spot syndrome virus infection by behavioral fever. Sci Rep. (2023) 13:10462. doi: 10.1038/s41598-023-45335-5
213. Granja CB, Vidal OM, Parra G, Salazar M. Hyperthermia reduces viral load of white spot syndrome virus in. Penaeus vannamei. Dis Aquat Organ. (2006) 68:175–80. doi: 10.3354/dao068175
214. Granja CB, Aranguren LF, Vidal OM, Aragón L, Salazar M. Does hyperthermia increase apoptosis in white spot syndrome virus (WSSV)-infected Litopenaeus vannamei? Dis Aquat Organ. (2003) 54:73–8. doi: 10.3354/dao054073
215. Shi W, Hu R, Wang P, Zhao R, Shen H, Li H, et al. Transcriptome analysis of acute high temperature-responsive genes and pathways in Palaemon gravieri. Comp Biochem Physiol Part D Genomics Proteomics. (2022) 41:100958. doi: 10.1016/j.cbd.2021.100958
216. Wang Z, Qu Y, Zhuo X, Li J, Zou J, Fan L. Investigating the physiological responses of Pacific white shrimp Litopenaeus vannamei to acute cold-stress. PeerJ. (2019) 7. doi: 10.7717/peerj.7381
217. Liu CH, Chen JC. Effect of ammonia on the immune response of white shrimp Litopenaeus vannamei and its susceptibility to Vibrio alginolyticus. Fish Shellfish Immunol. (2004) 16:321–34. doi: 10.1016/S1050-4648(03)00113-X
218. Rodríguez-Ramos T, Espinosa G, Hernández-López J, Gollas-Galván T, Marrero J, Borrell Y, et al. Effects of Echerichia coli lipopolysaccharides and dissolved ammonia on immune response in southern white shrimp Litopenaeus schmitti. Aquaculture. (2008) 274:118–25. doi: 10.1016/j.aquaculture.2007.10.049
219. Chang ZW, Chiang PC, Cheng W, Chang CC. Impact of ammonia exposure on coagulation in white shrimp, Litopenaeus vannamei. Ecotoxicol Environ Saf. (2015) 118:98–102. doi: 10.1016/j.ecoenv.2015.04.019
220. Yang L, Yang Q, Jiang S, Li Y, Zhou F, Li T, et al. Metabolic and immune responses in prawn (Penaeus monodon) exposed to ambient ammonia. Aquaculture Int. (2015) 23:1049–62. doi: 10.1007/s10499-014-9863-6
221. Li Y, Zhou F, Huang J, Yang L, Jiang S, Yang Q, et al. Transcriptome reveals involvement of immune defense, oxidative imbalance, and apoptosis in ammonia-stress response of the black tiger shrimp (Penaeus monodon). Fish Shellfish Immunol. (2018) 83:162–70. doi: 10.1016/j.fsi.2018.09.026
222. Qin Y, Jiang S, Huang J, Zhou F, Yang Q, Jiang S, et al. C-type lectin response to bacterial infection and ammonia nitrogen stress in tiger shrimp (Penaeus monodon). Fish Shellfish Immunol. (2019) 90:188–98. doi: 10.1016/j.fsi.2019.04.034
223. Song F, Chen GL, Lu KC, Fan JQ, Yan MT, He HH, et al. Identification and functional characterization of a C-type lectin gene from Litopenaeus vannamei associated with ER-stress response. Fish Shellfish Immunol. (2019) 93:977–85. doi: 10.1016/j.fsi.2019.08.056
224. González-Ortegón E, Blasco J, Nieto E, Hampel M, Le Vay L, Giménez L. Individual and mixture effects of selected pharmaceuticals on larval development of the estuarine shrimp Palaemon longirostris. Sci Total Environ. (2016) 540:260–6. doi: 10.1016/j.scitotenv.2015.06.081
225. Duan Y, Xiong D, Wang Y, Zhang Z, Li H, Dong H, et al. Toxicological effects of microplastics in Litopenaeus vannamei as indicated by an integrated microbiome, proteomic and metabolomic approach. Sci Total Environ. (2021) 761:143355. doi: 10.1016/j.scitotenv.2020.143311
226. Xing YF, Zhu XY, Huang JH, Nan YX, Duan YF, Zhang JS. Toxic effects of microplastics and nitrite exposure on intestinal histology, digestion, immunity, and microbial community of shrimp Litopenaeus vannamei. Mar pollut Bull. (2024) 200:116077. doi: 10.1016/j.marpolbul.2024.116077
227. Zhu C, Li Y, Liu G, Abdullah AL, Jiang Q. Effects of nanoplastics on the gut microbiota of Pacific white shrimp Litopenaeus vannamei. Peer J. (2024) 12. doi: 10.7717/peerj.16743
228. Zheng JR, Zhao Y, Feng YX, Qian WG, Zhang Y, Dong BB, et al. c-Jun N-terminal kinase activation contributes to improving low temperature tolerance by regulating apoptosis in the Pacific white shrimp (Penaeus vannamei). Fish Shellfish Immunol. (2023) 139:108912. doi: 10.1016/j.fsi.2023.108912
229. Tian H, Yuning W, Yang W, Liu F, Yu Y, Ren S, et al. c-Jun N-terminal kinase (JNK) in Procambarus clarkii: Molecular characterization and involvement in oxidative stress-induced apoptosis during the molting cycle. Comp Biochem Physiol B Biochem Mol Biol. (2022) 257:110676. doi: 10.1016/j.cbpb.2021.110676
230. Luo Z, Huang W, Wang G, Sun H, Chen X, Luo P, et al. Identification and characterization of p38MAPK in response to acute cold stress in the gills of Pacific white shrimp (Litopenaeus vannamei). Aquac Rep. (2020) 17:100365. doi: 10.1016/j.aqrep.2020.100365
231. Park K, Kim WS, Choi B, Kwak IS. Expression levels of immune-related p38 mitogen-activated protein kinase transcript in response to environmental pollutants in Macrophthalmus japonicus crabs. Genes. (2020) 11:1–14. doi: 10.3390/genes11090958
232. Shui Y, Xie J, Zhou Y, Li J, Gan J. Molecular characterization of p38 MAPK and tissue-specific expression under cadmium stress in red swamp crayfish (Procambarus clarkii). Sci Total Environ. (2020) 720:137325. doi: 10.1016/j.scitotenv.2020.137325
233. Jiang M, Tu DD, Gu WB, Zhou YL, Zhu QH, Guo XL, et al. Identification and functional analysis of inhibitor of NF-κB kinase (IKK) from Scylla paramamosain: Evidence of three IKKs in crab species and their expression profiles under biotic and abiotic stresses. Dev Comp Immunol. (2018) 84:199–212. doi: 10.1016/j.dci.2018.02.014
234. Sullivan TJ, Neigel JE. Effects of temperature and salinity on prevalence and intensity of infection of blue crabs (Callinectes sapidus) by Vibrio cholerae, V. parahaemolyticus, and V. vulnificus in Louisiana. J Invertebr Pathol. (2018) 151:82–90. doi: 10.1016/j.jip.2017.11.004
235. Kathyayani SA, Poornima M, Sukumaran S, Nagavel A, Muralidhar M. Effect of ammonia stress on immune variables of Pacific white shrimp (Penaeus vannamei) under varying pH levels and susceptibility to white spot syndrome virus. Ecotoxicol Environ Saf. (2019) 184:109626. doi: 10.1016/j.ecoenv.2019.109626
236. Cheng W, Chen SM, Wang FI, Hsu PI, Liu CH, Chen JC. Effects of temperature, pH, salinity, and ammonia on the phagocytic activity and clearance efficiency of giant freshwater prawn (Macrobrachium rosenbergii) against Lactococcus garvieae. Aquaculture. (2003) 219:111–21. doi: 10.1016/S0044-8486(03)00017-6
237. Li CC, Yeh ST, Chen JC. Innate immunity of the white shrimp (Litopenaeus vannamei) weakened by the combination of Vibrio alginolyticus injection and low-salinity stress. Fish Shellfish Immunol. (2010) 28:121–7. doi: 10.1016/j.fsi.2009.10.003
238. Kautsky N, Ronnback P, Tedengren M, Troell M. Ecosystem perspectives on the management of disease in shrimp pond farming. Aquaculture. (2000) 191(1-3):145–61. doi: 10.1016/S0044-8486(00)00424-5
239. Chang YH, Kumar R, Ng TH, Wang HC. What vaccination studies tell us about immunological memory within the innate immune system of cultured shrimp and crayfish. Dev Comp Immunol. (2018) 80:53–66. doi: 10.1016/j.dci.2017.03.003
Keywords: aquaculture, crustaceans, innate immunity, pattern recognition receptors, antimicrobial peptides, environmental stressors
Citation: Betancourt JL, Rodríguez-Ramos T and Dixon B (2024) Pattern recognition receptors in Crustacea: immunological roles under environmental stress. Front. Immunol. 15:1474512. doi: 10.3389/fimmu.2024.1474512
Received: 01 August 2024; Accepted: 28 October 2024;
Published: 14 November 2024.
Edited by:
Matthew L. Rise, Memorial University of Newfoundland, CanadaReviewed by:
Jorge Contreras-Garduño, National Autonomous University of Mexico, MexicoAndrew Rowley, Swansea University, United Kingdom
Copyright © 2024 Betancourt, Rodríguez-Ramos and Dixon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian Dixon, YmRpeG9uQHV3YXRlcmxvby5jYQ==
 Jesús Luis Betancourt
Jesús Luis Betancourt Tania Rodríguez-Ramos
Tania Rodríguez-Ramos Brian Dixon
Brian Dixon