- 1Department of Immunology, Tufts University School of Medicine, Boston, MA, United States
- 2Immunology Graduate Program, Graduate School of Biomedical Sciences, Tufts University School of Medicine, Boston, MA, United States
- 3C.S Mott Center for Human Growth and Development, Department of Obstetrics & Gynecology, Wayne State University School of Medicine, Detroit, MI, United States
- 4Department of Gynecology and Obstetrics, Tufts Medical Center, Boston, MA, United States
- 5Mass General Research Institute (MGRI), Division of Clinical Research, Massachusetts General Hospital, Boston, MA, United States
- 6Department of Microbiology and Immunology, Geisel School of Medicine at Dartmouth, Lebanon, NH, United States
- 7Department of Biochemistry, Microbiology and Immunology, Wayne State University School of Medicine, Detroit, MI, United States
Dendritic cells (DCs) play critical roles in HIV pathogenesis and require further investigation in the female genital tract, a main portal of entry for HIV infection. Here we characterized genital DC populations at the single cell level and how DC subsets respond to HIV immediately following exposure. We found that the genital CD11c+HLA-DR+ myeloid population contains three DC subsets (CD1c+ DC2s, CD14+ monocyte-derived DCs and CD14+CD1c+ DC3s) and two monocyte/macrophage populations with distinct functional and phenotypic properties during homeostasis. Following HIV exposure, the antiviral response was dominated by DCs’ rapid secretory response, activation of non-classical inflammatory pathways and host restriction factors. Further, we uncovered subset-specific differences in anti-HIV responses. CD14+ DCs were the main population activated by HIV and mediated the secretory antimicrobial response, while CD1c+ DC2s activated inflammasome pathways and IFN responses. Identification of subset-specific responses to HIV immediately after exposure could aid targeted strategies to prevent HIV infection.
1 Introduction
Human Immunodeficiency Virus (HIV) infection is an ongoing epidemic affecting about 38 million people worldwide. Women represent half of the people living with HIV, but in endemic regions HIV prevalence in women is higher than in men (1, 2). The main mechanism of HIV transmission is attributed to sexual intercourse (3) therefore, understanding HIV pathogenesis at the primary portal of entry, the female genital tract (FGT), remains a high priority to develop effective prevention strategies.
HIV gains access to the FGT by crossing the mucosal epithelial barrier during heterosexual transmission via seminal fluid, microtears in epithelial barriers, or transepithelial migration (4, 5). Different immune populations within the FGT supply target cells for HIV infection or act as protective innate effector cells that limit HIV acquisition (6–10). Potent antiviral innate defense followed by generation of protective local adaptive immune responses would be necessary to prevent HIV infection through repeated exposures.
Dendritic cells (DCs) are critical in shaping mucosal immunity against pathogens and maintaining tissue homeostasis (11). DCs express pattern recognition receptors (PRRs) that enable pathogen recognition and capture, specifically through C-type lectin receptors (CLRs) (12–15). Following antigen processing, DCs have the unique ability to prime naive T cell function, making DCs ideal targets for vaccination and therapeutic strategies against cancers and infections, including HIV (16). However, in HIV pathogenesis, DCs are considered a double-edged sword due to their ability to secrete anti-viral proteins and resist viral replication, but capture and transfer active viral particles to target CD4+ T cells (15, 17–20). Different models using in vitro monocyte-derived DCs, Langerhans cells and bona fide DCs have demonstrated that HIV-transfer from DCs to T cells can occur through infection-independent mechanisms via CLRs in early phases, or in an infection-dependent manner through HIV receptors and viral replication at later time points (20–24).
We previously demonstrated rapid secretion of antimicrobial peptides with anti-HIV activity by genital DCs upon HIV exposure, and subset-specific uptake of viral-like HIV particles by CD14+ DCs preferentially (25). Other studies further demonstrated that genital CD14+ DCs were capable of capturing and transferring HIV to CD4+ T cells (15, 18, 20). However, the role that different DC subsets may play in mucosal HIV acquisition remains unclear.
DCs and mononuclear phagocyte populations are highly specialized depending on the tissue of residence (26). Specifically in mucosal regions like the FGT, resident DCs display unique subset-specific functions in balancing immune protection and reproduction (27–31). Mucosal sites are populated with conventional DCs (cDCs) derived from DC precursors that seed the tissues (32). In addition, in-vivo recruitment and differentiation of monocytes into DCs or macrophages is mediated by acute inflammation in different disease models and at various mucosal sites (33). Characterization and delineation of DC subsets is classically dependent on surface protein expression patterns (34). However, recent advances in single-cell RNA sequencing (scRNAseq) have enabled better discrimination of human DC populations, revealing novel subsets and the inherent heterogeneity of CD14+ mononuclear populations (35, 36). These recent advances highlight the need to combine surface protein and RNA expression to fully characterize DC subsets (37).
Recent studies have enhanced our understanding of the DC and mononuclear phagocyte subsets that populate different human female genital mucosal surfaces relevant for HIV acquisition (15, 20, 24, 25, 31, 38, 39). Multiple of these studies identified a variety of CD14-expressing populations that remain poorly defined (40). Understanding the heterogeneity of DC and mononuclear cell populations in the FGT, along with their unique contribution to HIV pathogenesis is key for targeted interventions (41).
Here we use a combination of cellular indexing of transcriptomes and epitopes sequencing (CITE-seq) and spectral flow cytometry to define DC and mononuclear cell subsets in the FGT and determine their immediate responses to HIV exposure. Identification of phenotypic and functional properties of FGT-resident DCs and their role in HIV acquisition could inform future therapeutic and vaccination strategies against HIV.
2 Methods
2.1 Tissue processing
Tissues obtained from hysterectomies were separated by endometrium (EM), endocervix (END) and ectocervix (ECT) by pathologists and transferred to the laboratory post-surgery. Tissues were processed as described previously (10, 31, 42). Briefly, tissues were minced into 1-2 mm fragments in Roswell Park Memorial Institute (RPMI) medium (Gibco) containing enzymes from Tumor Dissociation Kit, human (Miltenyi Biotec) and 0.01% DNAse (Worthington Biochemical) and transfered into sterile gentleMACS™ C Tubes (Miltenyi Biotec). Enzymatic digestion was performed on genlteMACS™ Dissociator (Miltenyi Biotec) using “37C_h_TDK_1” program. Digested tissue was filtered through 100μm, 70μm and 30μm MACS™ SmartStrainers (Miltenyi Biotec) to generate stromal single cells suspensions.
2.2 Flow cytometry
Mixed single-cell suspensions from tissues were washed in Phosphate-Buffered Saline (PBS; Gibco). Subsequently, cells were incubated in the dark at room temperature with LIVE/DEAD Blue (ThermoFisher) dye for 10 minutes. Anti-CD16 (BD Biosciences) was then added and incubated for 5 minutes, followed by anti-CCR7 (BioLegend), anti-CXCR4 (BioLegend), anti-CCR5 (BioLegend) and anti-CX3CR1 (BioLegend) for another 5 minutes. Finally, cells were incubated with an antibody cocktail containing remaining antibodies (Supplementary Table S2) at room temperature for 15 minutes, washed with MACS buffer, fixed in 2% paraformaldehyde (PFA;Thermo Fisher), and analyzed for surface marker expression on Cytek Aurora (5 laser, 64 detector configuration; Cytek Biosciences). Expression of surface markers were quantified using OMIQ (Dotmatics).
2.3 HIV-1 viral stock propagation
HIV-BaL (R5) isolates obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (43) were propagated through infection of peripheral blood mononuclear cells (PBMCs) activated with phytohemagglutinin (PHA) (2.5 μg/mL; Sigma, St. Louis, MO) and IL-2 (50 U/mL; AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: Human rIL-2 from Dr. Maurice Gately, Hoffmann – La Roche Inc) for 6-8 days. Stocks were harvested when p24 concentrations reached 100 ng/mL. Titration of viral stocks were performed using PHA and IL-2 activated PBMCs (44).
2.4 Sample preparation for multi-omics single-cell RNA sequencing
Mixed single-cell suspensions from hysterectomy samples obtained from 4 healthy female donor, consisting of 4 endometrium (EM), 2 endocervix (END) and 4 ectocervix (n =10 tissues), were enriched for immune cells by magnetic bead removal of CD3+ (CD3 MicroBeads, human; Miltenyi Biotec), CD19+ (CD19 MicroBeads, human; Miltenyi Biotec), CD235a+ (CD235a (Glycophorin A) MicroBeads, human; Miltenyi Biotec) red blood cells and fibroblasts (Anti-fibroblast MicroBeads, human; Miltenyi Biotec). For homeostatic conditions, cells were washed thoroughly with PBS containing 5% HS to remove any excess magnetic beads and incubated with oligo-conjugated antibodies (AbSeq; Supplementary Table S2) and barcoded sample tags (BD™ Hu Single Cell Sample Multiplexing Kit; BD Biosciences), to differentiate between tissue sites, for 20 minutes at room temperature in PBS containing 0.5% HS. For HIV stimulation experiments, cells were incubated with HIV-BaL (MOI = 0.5) in XVIVO-15 or media alone for 30 minutes. Cells were washed thoroughly with PBS+0.5% HS to remove unbound virus and subsequently incubated with oligo-conjugated antibodies and barcoded sample tags as mentioned above. Cells were subsequently washed twice in excess PBS+0.5% HS to remove unbound antibodies and sample tags. Cells were counted and 10,000 cells from each tissue were combined to obtain a total of 30,000 cells each for control and HIV conditions. Cells were then subsequently loaded onto separate BD Rhapsody™ Cartridge (BD Rhapsody™ Cartridge Kit; BD Biosciences) followed by RNA capture on polyA tail capture beads containing unique molecular identifiers (UMIs) (BD Rhapsody™ Enhanced Cartridge Reagent Kit V3; BD Biosciences) on BD Rhapsody™ Express Single-Cell Analysis System (BD Biosciences) followed by cDNA, whole-transcriptome amplification and single-cell indexing libraries according to manufacturer’s protocol (Protocol-Rhapsody WTA+AbSeq+ST; BD Biosciences).
2.5 Multi-omics RNA sequencing analysis
FASTQ files containing unaligned reads were generated on NovaSeq6000 sequencing system (Illumina). Gene counts were generated by aligning and annotating reads to the human genome (GRCh38.p12 v29). To assess capture of HIV, reads from respective experiments were also aligned to the HIV genome FASTA file. Count tables were subsequently uploaded to Partek Flow (Partek, an Illumina company) for downstream quantification and visualization of data. Cells with mitochondrial gene expression >25% number of detected features per cell less than 200 or greater than 4300 were excluded from analysis, according to recent studies published by us (31) and others (45). RNA and protein data were split and normalized respectively. To identify DCs, we first selected immune cells expressing PTPRC genes, which encodes CD45 protein. Next, we used protein information to exclude remaining T and B cells by selecting CD3-CD19+ cells, followed by selection of CD11c+HLA-DR-DP-DQ+ cells. Since genital NK cells and neutrophils can express CD11c and HLA-DR (46, 47), we excluded neutrophil contamination by selecting CD15- cells within this population, and NK cell contamination by gating out cells expressing NCAM1 RNA (Figure 1A). PCA was performed on normalized data followed by UMAP for visualization. Additionally, we also performed weighted-nearest neighbor analysis to integrate protein and RNA data. Subsequently, we performed k-nearest neighbor analysis and graph-based analysis to identify cell clusters and used “compute biomarker” function in Partek Flow (Partek – an Illumina company) to generate biomarkers associated with the clusters (Supplementary Data Sheet S1). To identify differentially expressed genes (DEGs) between DC subsets, we performed non-parametric ANOVA to identify significantly upregulated and downregulated genes with significance of p ≤ 0.05 and Log2(FoldChange) ± 1.2. GO Biological processes (48, 49) and Reactome (50) were generated using gene lists of upregulated and downregulated genes. To identify unique expression of genes within each cluster, “compute biomarker” function in Partek Flow Analysis. Curated gene lists were used to assess expression of CD markers, cytokines, chemokines, PRRs, antimicrobial proteins and gene lists associated with GO processes for AUCell (51) (Supplementary Data Sheet S2) were used from GSEA analysis available in Partek Flow (Partek – an Illumina company).
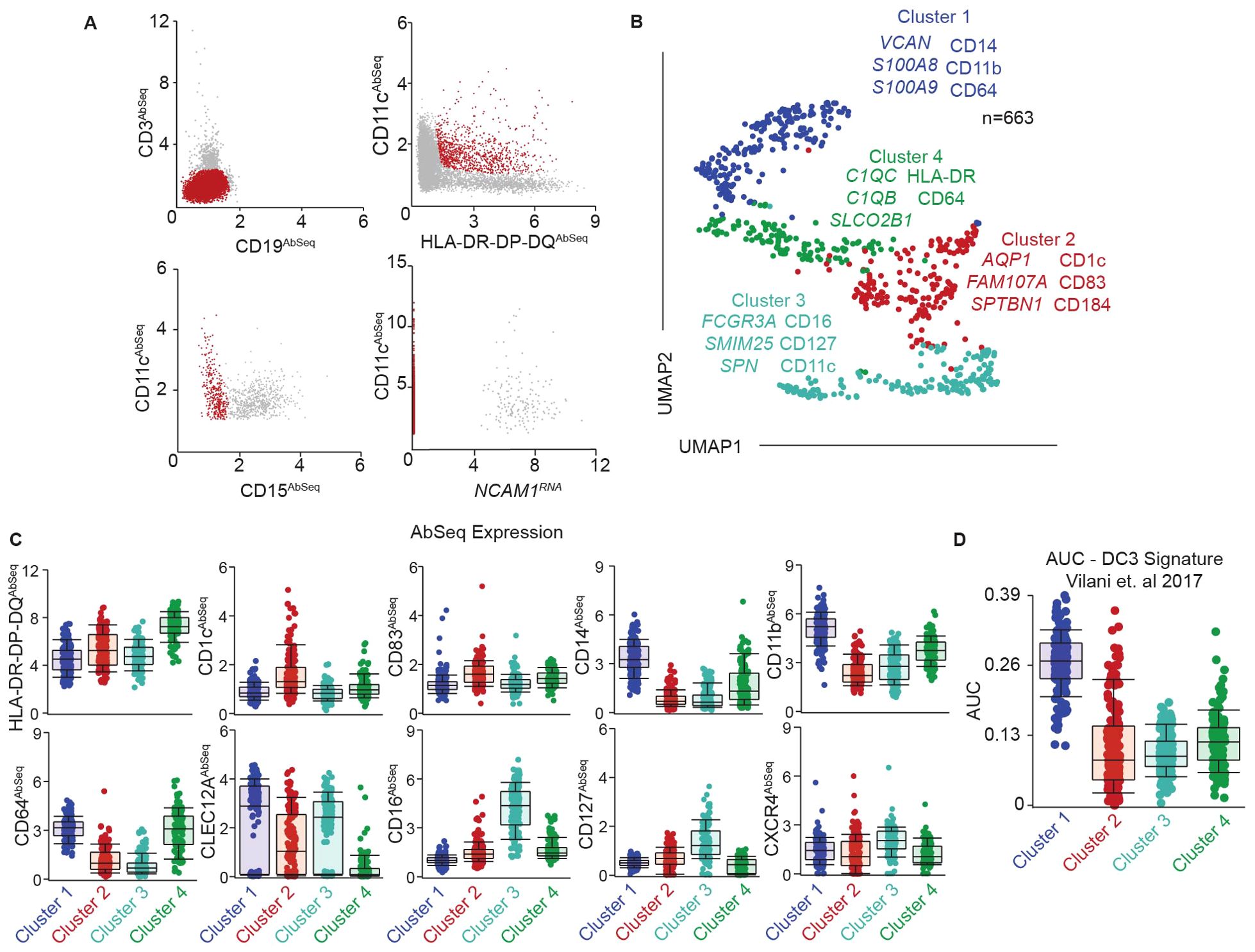
Figure 1. Identification of phenotypically and transcriptionally unique DC subsets in the genital tract. (A) Representative gating strategy to identify FGT resident CD11c+HLA-DR+ cells using a combination of surface protein expression (AbSeq) and RNA expression in CITEseq dataset. Y-axis and x-axis indicate normalized AbSeq expression levels. (B) Representative UMAP visualization of FGT resident CD11c+HLA-DR+ cells through unbiased clustering, depicting expression of discriminating RNA expression (left column) and surface protein expression (right column) in distinct subsets, Cluster 1 (blue), Cluster 2 (red), Cluster 3 (teal) and Cluster 4 (green). (C) Scatter plot comparing surface antibody expression between different FGT CD11c+HLA-DR+ subsets. (D) AUCell analysis of top 50 genes in DC3 cluster described by AC Villani et. al.
2.6 CD14+ DC isolation and HIV stimulation
CD14+ DCs were isolated from mixed-cell suspensions using magnetic bead separation (CD14 MicroBeads, human; BD Biosciences) as per previous studies (25, 30, 31, 42). Isolated cells were incubated at 37°C, 5% CO2 with HIV-1-BaL (R5) isolates (MOI = 0.5) for 3 hours. Supernatants were collected and stored at -80C until multiplex assay analysis to determine secreted protein levels.
2.7 Luminex assay
Supernatants from unstimulated controls and HIV exposed CD14+ DCs were quantified using Millipore human cytokine multiplex kits according to manufacturer’s instructions. Signal was measured using Magpix software with five-parametric-curve fitting for data analysis. Molecules measured included IL-1β, IFNγ, IL-5, IL-13, IL1Rα, GM-CSF, G-CSF, CCL11, CCL22, CXCL1, CXCL10 and CX3CL1.
2.8 Statistical analysis
To identify differentially expressed genes, significance threshold was set as (p<0.05; -1.2 <FC>1.2) on non-parametric ANOVA. Statistics for flow cytometry analysis were done using Friedman’s multiple comparison, (p ≤ 0.05 *; p ≤ 0.01 **; p ≤ 0.001 ***; p ≤ 0.0001 ****). Hierarchical clustering heatmaps were performed on significant, differentially expressed genes, by generating pseudo-bulk expression of groups (eg. HIV vs control; cluster 1, cluster 2, cluster 3, cluster 4 comparison) and visualized using bubble plots, with size of the bubble referring to percentage of cells expressing the particular gene, shade and color referring to Z-score expression of the gene. For Reactome and GO analysis, terms with FDR ≤ 0.05 were chosen as significant values and bubble plots were visualized using GraphPad Prism.
2.9 Study subjects
Written informed consent was obtained before surgery from HIV-negative women undergoing hysterectomies at Tufts Medical Center (Boston, MA, USA). Studies were approved by Tufts University Institutional Review Board and the Committee for the Protection of Human Subjects. Surgery was performed to treat benign conditions including fibroids, prolapse, and menorraghia. Trained pathologists selected tissue samples from endometrium (EM), endocervix (END), and ectocervix (ECX), free of pathological lesions and distant from the sites of pathology. Women were HIV− and HPV− but no additional information regarding other genital infections was available.
3 Results
3.1 Identification of phenotypically and transcriptionally unique DC subsets in the genital tract
Prior studies have identified different genital DC and mononuclear phagocyte populations within the anogenital mucosa (14, 15, 20, 24, 25, 31, 39, 42, 52). However, transcriptional, phenotypical, and functional characterization of the heterogenous CD14-expressing populations in the genital tract remains poorly defined.
To address these gaps, we optimized a protocol to define DC subsets in the FGT phenotypically and transcriptionally at the single-cell level. We generated single cell suspensions from human hysterectomy samples and enriched the target immune cells as detailed in methods. We labeled the enriched cells with oligo-conjugated antibodies targeted towards surface proteins (Supplementary Table S1), lysed the cells to release RNA, and sequenced the RNA to simultaneously determine the whole transcriptome profile and surface protein expression at the single-cell level. To identify the genital resident DCs, we took advantage of CITE-seq’s combined surface protein information and gene expression and developed a gating strategy to select the CD11c+HLA-DR+ population (Figure 1A), which contains DCs. This approach eliminates the need for lengthy fluorescence activated cell-sorting (FACS) prior to sequencing, thereby limiting processing steps that can alter primary tissue resident immune cells (53).
Unbiased clustering analysis discriminated four distinct clusters of CD11c+HLA-DR+ cells within the genital mucosa (Figure 1B). We determined the top RNA and surface protein (Abseq) expression unique to each cluster by using “compute biomarkers” function (Figure 1B). Cluster 1 was characterized by RNA expression of VCAN, S100A8, S100A9 and surface protein expression of CD14, CD11b and CD64 (Figure 1C), confirming the presence of a CD14+ monocyte-derived DC population in the genital mucosa as previously described by us and others (20, 25, 30, 31, 52, 54). Cluster 2 was characterized by surface protein expression of CD1c, the DC maturation marker CD83, and the pro-survival and tissue homing marker CXCR4 (55, 56), suggesting enrichment of cDC2s in this cluster (Figure 1C). Clusters 3 and 4 displayed phenotypes similar to monocyte populations. Cluster 3 expressed elevated CD16 and CD127, whereas cluster 4 displayed elevated expression of CD64, HLA-DR and low levels of CLEC12A protein expression (Figure 1C). Additionally, cluster 3 expressed RNA transcripts such as FCGR3A, SMIM25 and SPN (Figure 1B), consistent with non-classical monocyte (NCM) signature expression (57). Cluster 4 expressed elevated levels of complement encoding RNA transcripts C1QC and C1QB, consistent with inflammatory monocytes/macrophages (infMons) (58). These two clusters indicated the presence of two distinct monocyte populations within CD11c+HLA-DR+ cells.
Recent studies identified a novel DC population in peripheral blood named DC3 that co-expresses CD14 and CD1c and displays and intermediate phenotype and function between monocytes and classical myeloid DCs (cDC2s) (36, 59). Interestingly, we and others have previously described the presence of CD14+CD1c+ cells in the FGT (20, 25, 52). To determine whether the DC3 population was present in the FGT, we analyzed our CITE-seq data using AUCell (51) with the top 50 genes described by Villani et al. in their peripheral blood DC3 cluster (Supplementary Data Sheet S2) (35). We observed an enrichment of the DC3 signature in cluster 1 (Figure 1D), indicating the presence of DC3s within the CD14+ DC cluster.
Overall, our multi-omics data indicates the presence of four distinct CD11c+HLA-DR+ populations in the FGT, consisting of two DC and two monocyte populations, with unique transcriptional and phenotypic signatures. Furthermore, we observed the presence of novel DC3 gene signature in the CD14+ DC cluster, suggesting inherent heterogeneity of CD14+ DCs within the FGT.
3.2 Functional specialization of genital DCs under homeostatic conditions
Next, we investigated whether our phenotypical clustering was associated with distinct subset-specific functions for each subset under homeostatic conditions.
To determine if the distinct populations would be differently posed to detect and interact with invading pathogens, including HIV, we compared expression levels of PRRs (Supplementary Data Sheet S2) including toll-like receptors (TLRs), C-type lectin receptors (CLRs) and NOD-like receptors (NLRs). Using a list of known PRRs (Supplementary Table S2) we compared expression levels between subsets through hierarchical clustering (Figure 2A). We observed no TLR9 expression across all subsets, ruling out the possibility of plasmacytoid DC contribution to the gene signature of each subset. Compared to the other clusters, Cluster 1 (CD14+ DCs) showed relatively higher expression of TLRs and CLRs associated with detecting and binding to HIV, including CLEC4A, CLEC4E and TLR4 (60–62). Additionally, in Clusters 1 and 3, we observed shared expression of CLEC12A, which has been linked to improved antigen delivery and cross presentation by DCs (63), and CLEC7A, previously described in CD14+ DCs from blood and lungs (64). Additionally, Clusters 1, 3 and 4 also shared expression of TLR2, which binds HIV (65). Cluster 2 (cDC2s) had no unique gene expression pattern. The highest expression in Cluster 2 was observed for the CLRs CLEC10A and CLEC1A, involved in antigen internalization and presentation (66, 67). When compared to the other clusters, Cluster 3 (NCM) displayed high preferential expression of IFIH1 (encoding MDA5), DDX58 (encoding RIGI) which mediate detection of cytosolic viral RNA (68, 69), and TLR8, which recognizes endolysosomal ssRNA (70). Cluster 4 (infMons) expressed MRC1 (encoding mannose receptor (MR)) and CD209, consistent with a monocyte/macrophage phenotype (71, 72). Cluster 4 also expressed high levels of CLEC5A which forms heterodimers with CD209 and MR upon antigen exposure to enhance viral internalization (73). Further, compared to the other clusters, Cluster 4 had preferential expression of TLR1 and TLR6, known to dimerize with TLR2 for detection of bacterial lipopeptides; TLR7 and TRL3 which recognize ssRNA and dsRNA, respectively (74, 75). Finally, a small percentage of cells in Cluster 4 preferentially expressed AIM2, involved in recognition of cytosolic dsDNA (76). Altogether, our analysis reveals subset specific differences in genital DC recognition of different moieties of pathogens, Cluster 1 being enriched with PRRs associated with membrane HIV recognition and binding, Cluster 2 expressed PRRs for antigen uptake, Cluster 3 and Cluster 4 were enriched for PRRs that detects cytosolic and endolysosomal ssRNA, dsRNA and dsDNA.
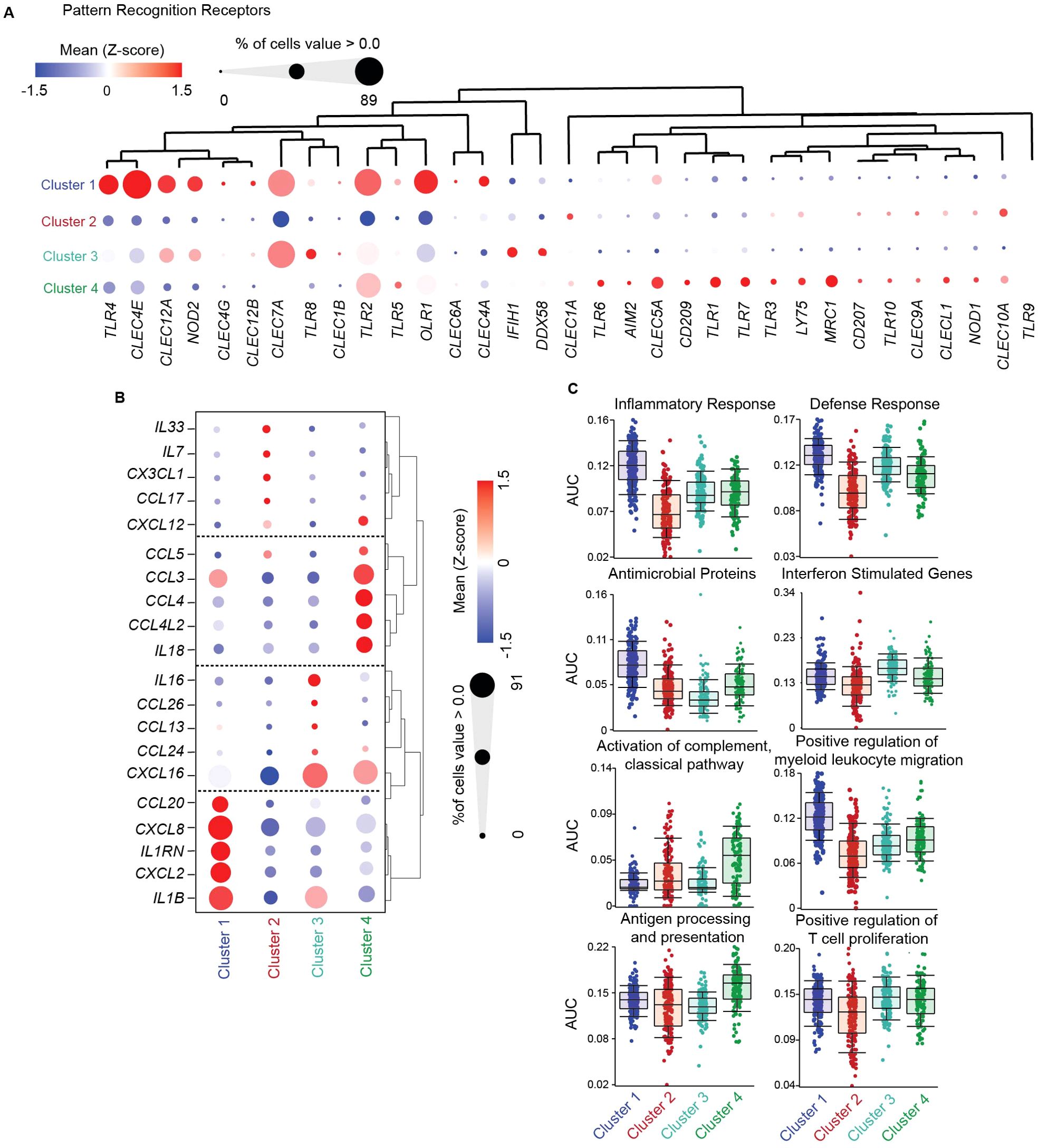
Figure 2. Functional specialization of genital DCs under homeostatic conditions. (A) Hierarchical clustering heatmap comparing gene expression of significant pattern recognition receptors (PRRs) between FGT resident CD11c+HLA-DR+ subsets. (B) Hierarchical clustering heatmap comparing gene expression of top 5 interleukin and chemokines in each cluster, where size of the circle indicates percentage of cells expressing associated gene > 0 (C) AUCell comparison of key gene ontology (GO) terms associated with dendritic cell function between DC subsets.
Using the same analytical approach, we next focused on cytokine and chemokine expression under homeostatic conditions, to understand functional differences in innate secretory functions involved in maintenance of mucosal barrier integrity, immune cell recruitment and anti-microbial protection. We identified the top 5 cytokine and chemokine gene signatures unique to each cluster (Figure 2B; Supplementary Data Sheet S2). Cluster 1 (CD14+ DCs) was characterized by CCL20, CXCL8 and CXCL2 expression, genes involved in antimicrobial activity (77), chemotaxis for Th17 cells, neutrophils, monocytes and DCs (78–81). Expression of IL1B and its inhibitory receptor IL1RN were also elevated in Cluster 1, suggesting production of IL1β but prevention of autocrine/paracrine actions of IL1β on CD14+ DCs. Cluster 2 (containing cDC2s), specialized in expression of genes involved in T cell chemotaxis: IL33, an alarmin that controls tissue homeostasis and type 2 immunity (82); CCL17, a chemoattractant for helper T cells expressing CCR4 (such as Th2 and regulatory T cells) (83) (84); CXCL12, a CXCR4-ligand; CX3CL1 (fractalkine) which play critical roles in menstruation (85); and IL7, suggesting an important role for cDC2s in control of genital T cell populations and favoring Th2 and T regulatory profiles. Cluster 3 (NCMs) expressed high levels of IL16, a cytokine that exclusively binds and signals through CD4 (86); and CXCL16, important for control of MAIT cells and NK cells and expressed by non-classical monocytes in blood (87–89). In addition, a smaller percentage of cells in cluster 3 expressed CCR3 ligands CCL24, CCL26 and CCL13, with antimicrobial properties and potent chemoattractants for eosinophils and basophils (90) (91). Cluster 4 (infMons) was characterized by expression of CCR5 ligands CCL3, CCL4, CCL5 and CXCR4 ligands CXCL12 and CCL4L2, suggesting a potential role in mediating anti-HIV activity. Cluster 4 also expressed high levels of IL18, a proinflammatory cytokine described in tissue-resident macrophages that induces IFN-γ production by T cells in an inflammatory environment, or Th2 differentiation in the absence of inflammatory cytokines (92, 93).
To further understand functional contributions to mucosal homeostasis and defense, we performed AUCell analysis (51) of genes associated with immune protection (Figure 2C; Supplementary Data Sheet S2). Cluster 1, containing CD14+ DCs, was enriched for genes associated with inflammatory and defense response, suggesting enhanced potential to mediate innate immune protection. Furthermore, enrichment of genes associated with positive regulation of myeloid leukocyte migration within this cluster suggests the presence of a migratory DC population. Cluster 2 (cDC2s) and Cluster 4 (infMon) were enriched for genes related to activation of the complement classical pathway, correlating with increased expression of complement associated genes. Lastly, no differences were observed for enrichment of antigen processing and presentation or positive regulation of T cell proliferation genes in genital CD11c+HLA-DR+ subsets under homeostatic conditions.
Overall, our data demonstrates distinct functional specialization of genital CD11c+HLA-DR+ subsets under homeostatic conditions.
3.3 CD14+ DCs represent a heterogenous, activated population within the genital mucosa
Data in this study highlights the inherent heterogeneity of DC subsets within the mucosa, with unique transcriptional and phenotypic properties under homeostatic conditions. To validate our CITE-seq findings in a larger number of patients and further delineate genital DC populations, we developed a multi-parameter spectral flow cytometry panel (Supplementary Table S2). We gated on CD11c+HLA-DR+ cells (Figure 3A) and identified four different populations based on CD1c and CD14 surface expression (Figure 3B), main discriminatory markers identified in our CITEseq data (Figure 1C). As seen in Figure 3C, CD14highCD1c- cells (CD14+ DCs) were the most abundant subset (53%), followed by CD14lowCD1c- (29.1%), CD14+CD1c+ DCs (DC3s) (7.44%) and CD14-CD1c+ (cDC2s) cells (6.5%). To determine if CD11c+HLA-DR+ subset distribution varies throughout the genital tract, we compared the abundance of each subset in the endometrium (EM), endocervix (END), and ectocervix (ECX) (Figure 3D). We observed an increased abundance of CD14+ cells in the ECX compared to the EM, whereas the opposite was true for the CD14lowCD1c- subset, with no significant changes observed in CD1c+ and CD14+CD1c+ cells.
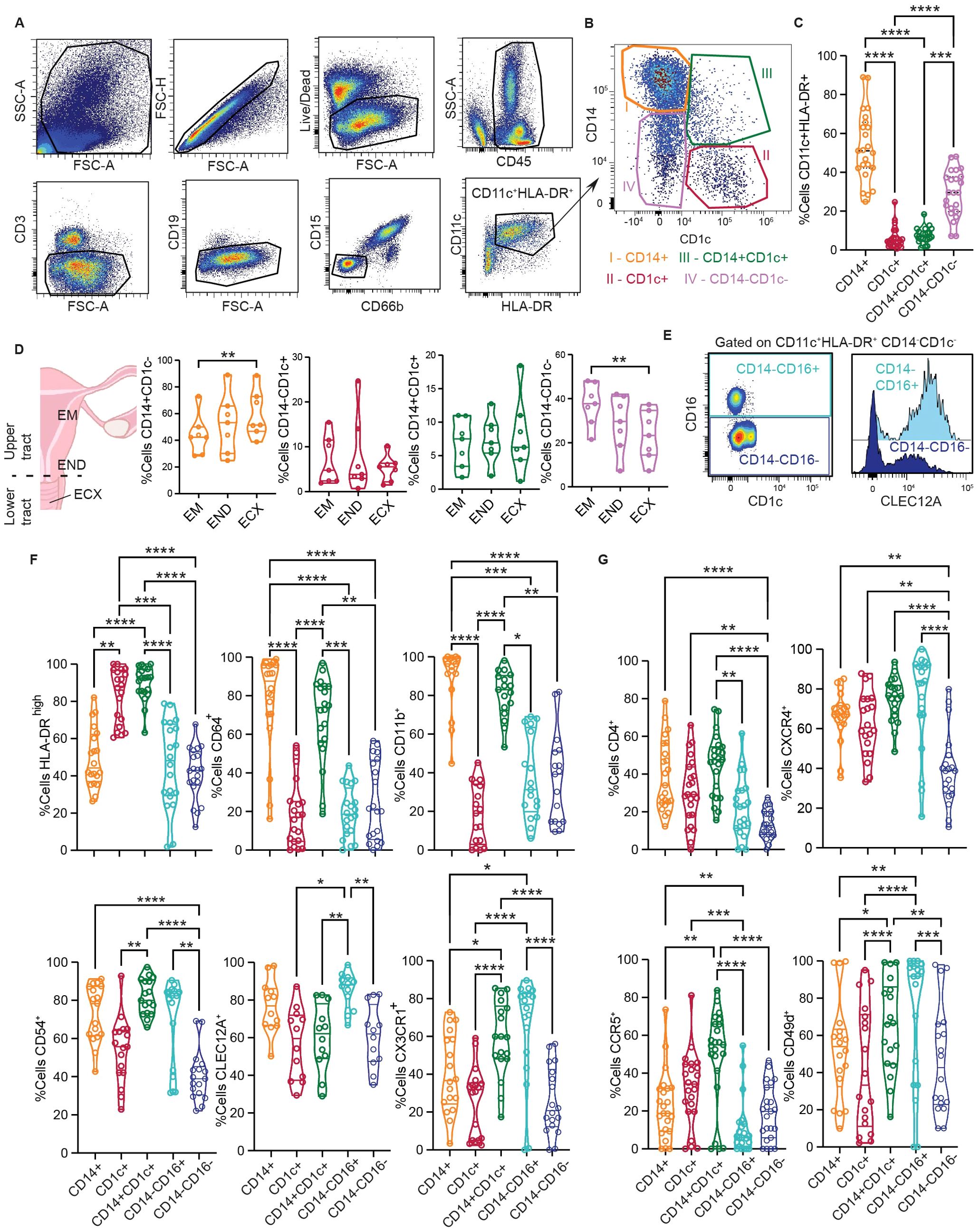
Figure 3. CD14 DCs represent a heterogenous, activated population within the genital mucosa. (A) Representative flow cytometry gating strategy to identify FGT resident CD11c+HLA-DR+ cells. (B) Representative plot of HLA-DR+CD11c+ subsets based on CD14 and CD1c expression; CD14+CD1c- (I, orange), CD14-CD1c+ (II, red), CD14+CD1c+ (III, green) and CD14-CD1c- (IV, pink). (C) Comparison of CD11c+HLA-DR+ subset frequencies in the FGT. (D) Comparison of CD11c+HLA-DR+ subset distribution across different anatomical regions of the FGT, endometrium (EM), endocervix (END) and ectocervix (ECX). (E) Representative gating to differentiate CD14-CD1c- cells based on CD16 expression (left); Histogram comparing CLEC12A expression between CD14-CD16+ and CD14-CD16- cells (right). (F) Comparison of surface protein expression between different CD11c+HLA-DR+ cells; HLA-DRhigh (top left), CD64 (top middle), CD11b (top right), CD54 (bottom left), CLEC12A (bottom middle) and CX3CR1 (bottom left). (G) Comparison of HIV tropic surface proteins between different CD11c+HLA-DR+ cells; CD4 (top left), CXCR4 (top right), CCR5 (bottom right) and CD49d (bottom right). Statistics – Friedman’s multiple comparison, (p ≤ 0.05 *; p ≤ 0.01 **; p ≤ 0.001 ***; p ≤ 0.0001 ****).
In our CITE-seq data set we identified two distinct CD14-CD1c- populations with differential expression of CD16 and CLEC12A (Figure 1C). Therefore, we further characterized the CD14-CD1c- population to determine if this group was composed of two different cell subsets. Consistent with our CITE-seq findings, the CD14lowCD1c- population contained two different subsets: CD14-CD16+ and CD14-CD16- (Figure 3E). Furthermore, these two populations could be distinguished by CLEC12A expression (Figure 3E, histogram), suggesting CD14-CD16+CLEC12A+ cells are similar to NCMs, whereas lack of CLEC12A expression in CD14-CD16- cells corresponds to the inflammatory/activated monocyte population (infMon).
Next, we analyzed expression of surface markers associated with DC activation status and function (Figure 3F). First, we assessed maturation status by gating on HLA-DRhigh cells consistent with our previous publication (25). We observed that a large proportion of CD14+CD1c+ and CD1c+ cells were HLA-DRhigh, significantly higher than other CD11c+HLA-DR+ subsets. Compared to the other subsets, CD14+CD1c+ and CD14+ cells showed higher expression of CD11b and CD64, associated with cell adhesion and antigen uptake. CD14+CD1c+ cells expressed the highest levels of CD54, a molecule involved in enhanced immune synapse formation between DCs and T cells (94). CD14-CD16+ cells displayed the highest levels of CLEC12A (a myeloid inhibitory receptor), and along with CD14+CD1c+ cells, showed high expression of CX3CR1, the receptor for CX3CL1 (fractalkine).
Previous studies from our group and others have shown that CD14-expressing FGT DCs preferentially capture HIV viral-like particles (15, 18, 20, 25), indicating subset-specific interactions with HIV. Based on this, we evaluated expression of receptors and coreceptors for HIV in the different subsets (Figure 3G). We observed that all three DC subsets expressed significantly higher levels of CD4 and CXCR4 compared to CD14-CD16- monocytes. Importantly, CD14+CD1c+ DCs expressed the highest levels of CCR5, which is crucial for HIV acquisition in the mucosa (95). In addition to the classical co-receptors, CD49d (α4β7 integrin) expression was significantly higher in CD14+CD1c+ DCs and CD14-CD16+ monocytes compared to other subsets. CD49d acts as a tissue homing marker and non-classical HIV co-receptor important for mucosal infection (96).
Altogether, our flow cytometry data demonstrates the presence of phenotypically distinct genital DC populations and further supports the presence of CD14+ DCs and DC3s within the CD14+ population.We demonstrate that genital DC3s expressed levels of activation markers similar to conventional cDC2s (CD1c+ DCs) and share expression of classical monocyte-derived DC markers with CD14+ DCs. Additionally, we demonstrate elevated expression of HIV coreceptors in DC3s compared to other DC populations, suggesting predisposed differential response to HIV within the genital mucosa.
3.4 Genital DCs undergo rapid transcriptional changes in response to HIV stimulation
Earlier studies investigated the role of DCs in HIV pathogenesis using monocyte-derived DCs over a time course from 6-48 hours to assess their responses to HIV during viral uptake and infection (22). However, most studies evaluating the role of genital DCs in HIV pathogenesis have investigated events that occur 12 hours or more after viral exposure, focusing on DC susceptibility to HIV infection and trans-infection to CD4+ T cells (38) (15, 18, 20). We have previously shown that genital DCs release chemokines and antimicrobials in a rapid manner, within 3 hours after HIV stimulation (25), indicating a role for DCs in triggering the initial mucosal innate response against the virus. However, the overall antiviral response induced by HIV immediately following challenge of genital DCs, prior to viral replication, integration, or productive infection, remains uncharacterized.
To address this gap, we adapted our CITEseq approach to identify early transcriptional responses of genital DCs to HIV. Single cell suspensions generated from human hysterectomy samples were incubated with HIV at an MOI of 0.5 in vitro for 30 minutes, prior to proceeding with the CITE-seq protocol described in detail in methods, to determine whole transcriptome profile and surface protein expression simultaneously at the single-cell level (Figure 4A). The time of 30 minutes was chosen to identify pathways involved in viral recognition and immune response independent of viral replication, integration and productive infection. We used the surface protein expression information to select CD11c+HLA-DR+ cells as done in Figure 1A, which contains the DC and monocytic populations, and performed differential gene expression analysis between HIV and unstimulated control conditions to define transcriptional changes induced by viral exposure. This analysis identified a significant transcriptional shift induced by HIV within 30 minutes, with 333 genes differentially upregulated and 4148 genes downregulated in response to HIV (Figure 4B; (Supplementary Data Sheet S1).
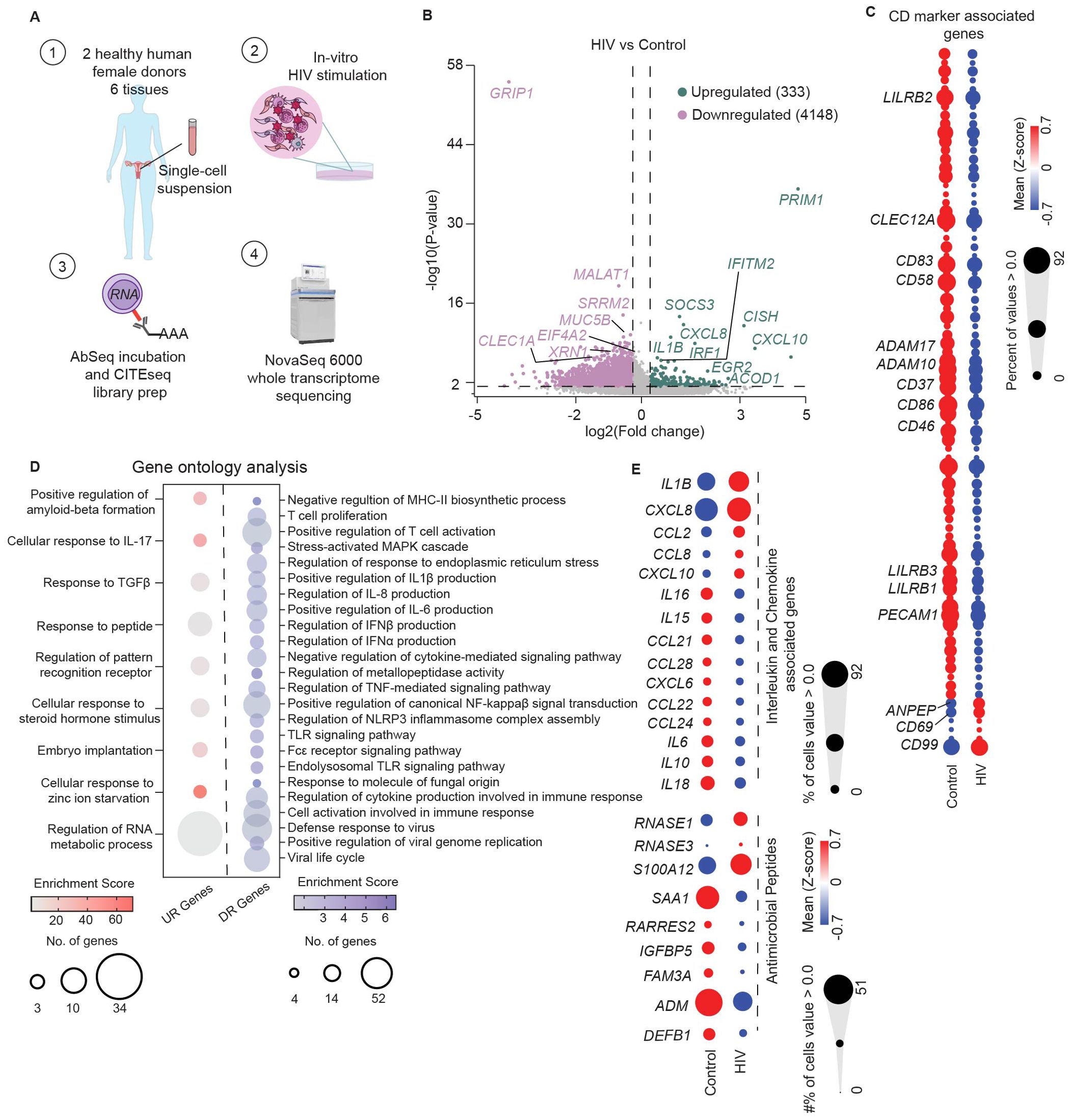
Figure 4. Genital DCs undergo rapid transcriptional changes in response to HIV stimulation. (A) Graphical depiction of CITEseq protocol to identify transcriptional changes induced by HIV in FGT resident CD11c+HLA-DR+ cells. Mixed single-cell suspensions from hysterectomy samples were incubated with HIV at MOI of 0.5 for 30 minutes at 37°C (B) Volcano plot of significantly upregulated (n=333) and downregulated genes (n=4148) by HIV in CD11c+HLA-DR+ cells. (C) Hierarchical clustering heatmap comparing gene expression of CD markers between control and HIV exposed cells. (D) Bubble-plot visualization of significant GO terms enriched in genes upregulated (UR genes; red) and downregulated (DR genes; blue) in response to HIV by CD11c+HLA-DR+ cells. (E) Hierarchical clustering heatmap comparing genes expression of secreted factors such as interleukins, chemokines and antimicrobial proteins between control and HIV exposed CD11c+HLA-DR+ cells. Statistics – Volcano plot, non-parametric ANOVA (p ≤ 0.05; -1.2 <FC>1.2); GO terms significance of FDR ≤ 0.05.
To define this HIV response, we first focused on genes associated with DC-mediated immune protection, inflammation, and antiviral response within the top 50 differentially expressed genes based on fold change and p-value (Figure 4B; Supplementary Data Sheet S1).
Within the upregulated genes, we detected increased expression of the interferon-stimulated genes (ISGs) IRF1 and IFITM2, indicating initiation of innate immune activation and anti-viral response. We also observed a strong upregulation of genes associated with interleukin and chemokine signaling pathways (IL1B, CXCL8, CXCL10, CISH, SOCS3), indicating rapid induction of innate secreted responses. Consistent with increased secretory response, we observed upregulation of serglycin gene expression (SRGN) in response to HIV, involved in storage and secretion of innate molecules in intracellular vesicles in blood monocytes and other cell types (97). Additionally, we detected upregulation of ACOD1, a gene involved in antimicrobial and antiviral responses of innate cells, and a negative regulator of TLR-mediated inflammatory responses (98).
We detected downregulation of genes associated with pro-viral replication of HIV such as GRIP1, EIF4A2, SRRM2, XRN1, and MALAT1 (99–102), suggesting suppression of host mechanisms to prevent HIV replication in genital DCs.
To uncover markers that enable the identification of phenotypic changes induced by HIV in genital DCs, we evaluated differences in expression of genes associated with surface protein expression (CD marker list, Supplementary Data Sheet S2), focusing on molecules associated with DC function such as activation, migration, and T cell co-stimulation (Figure 4C). We observed that the majority of genes related to CD markers were downregulated, but detected strong upregulation of three genes: CD69, which mediates tissue retention; CD99, involved in diapedesis of monocytes and dendritic cells into inflamed tissue (103); and ANPEP (encoding CD13), which mediates DC cross-presentation in human DC populations (104). Analysis of the downregulated genes revealed suppression of markers associated with classical DC activation and T cell co-stimulatory ligands (CD83, CD86, CD58) and molecules important for migration of DCs (PECAM1, CD37) (105, 106), indicating potential suppression of T cell interaction molecules and retention of genital DCs within the mucosa at early timepoints. We also observed decreased expression of disintegrin and metalloproteinase family of genes (ADAM10, ADAM17) which mediate shedding of TNF-α (107), suggesting suppression of classical pro-inflammatory responses. Additionally, relative to the control, the HIV-stimulated DCs had decreased expression of CLEC12A, and genes encoding the inhibitory leukocyte immunoglobulin-like receptor subfamily B (LILRB1, LILRB2, LILRB3), suggesting a non-classical mechanism of DC activation (108).
Next, we performed Gene Ontology (GO) analysis of genes upregulated and downregulated by HIV stimulation (FDR ≤ 0.05) (Figure 4D; Supplementary Data Sheet S1). Analysis of the upregulated genes revealed an enrichment of terms associated with cytokine mediated signaling pathways, innate sensing, and metabolic processes (Figure 4D; Supplementary Data Sheet S1). Downregulated genes (Figure 4D) were associated to terms related to antigen presentation, T cell responses, regulation of classical inflammatory cytokines, type I IFN responses, and canonical proinflammatory pathways, but also suppression of viral cycle and viral replication. These GO terms suggest suppression of classical inflammation but activation of innate host antiviral responses within 30 minutes of exposure to HIV.
Based on these GO terms, we further explored the secretory response to HIV and compared expression of genes encoding cytokines, chemokines and antimicrobials between control and HIV conditions (Figure 4E). In response to HIV we detected upregulation of an innate secretory response, including pro-inflammatory genes IL1B and CXCL10, CXCL8 and CCL2, chemoattractant for neutrophils, DCs and monocytes; and CCL8, a CCR5 ligand with antiviral activity, (109). Compared to the control group, we observed decreased expression of genes encoding inflammatory cytokines IL6 and IL18, which act as Th1 polarizing cytokines; IL16, a CD4+ cell specific chemoattractant; CCL28, a CCR10+ T cell chemoattractant; and CCL21, which mediates homing to lymphoid tissues, suggesting potential inhibition of CCR7+ DC migration towards lymph nodes. Comparison of antimicrobial peptide gene expression ( (110); Supplementary Data Sheet S2) revealed increased expression of RNASE1 and S100A12 in the HIV group compared to control (Figure 4E). Interestingly, we observed suppression of SAA1 (encoding serum amyloid A), which contributes to induction of pathogenic Th17 cells (111), known to be preferential targets for HIV infection (6, 112, 113). Additionally, we observed suppression of ADM (encoding the anti-bacterial peptide adrenomedullin), which also acts as suppressor of inflammatory cytokines (114).
Taken together, our data indicates that mononuclear phagocytic populations respond to HIV in a rapid manner within 30 minutes of exposure, with upregulation of endogenous host restriction factors, activation of non-classical inflammatory responses, and increased gene expression of cytokines, chemokines and antimicrobials, suggesting initiation of a localized antiviral response.
3.5 CD14+ DCs largely mediate the rapid innate secretory signature observed in genital DC response to HIV while cDC2s mediate antiviral inflammatory responses
Transcriptional changes in genital mononuclear phagocytic populations exposed to HIV indicates rapid activation of endogenous host restriction factors and localized non-classical inflammatory response, but also an overall suppression of gene transcription. Since our results identified distinct DC subsets with differential homeostatic activation and functions, we next determined how each of these subsets contributes to the overall response after HIV challenge. To define cell clusters and determine their responses following HIV exposure, we used the CITEseq libraries generated from single-cell suspensions stimulated with HIV for 30 minutes shown in Figure 4. We performed PCA, followed by unbiased clustering and visualization using UMAP. This analysis revealed four clusters (Figure 5A): CD14 DCs, cDC2s, infMons and NCMs, consistent with our findings under homeostatic conditions (Figure 1B). Overlay of unstimulated and HIV exposed cells, revealed contribution from control and HIV stimulation conditions to each cluster, except for the NCM cluster, where we found a very limited number of cells from the HIV stimulated condition, and were therefore unable to analyze NCM response to HIV (Figure 5B).
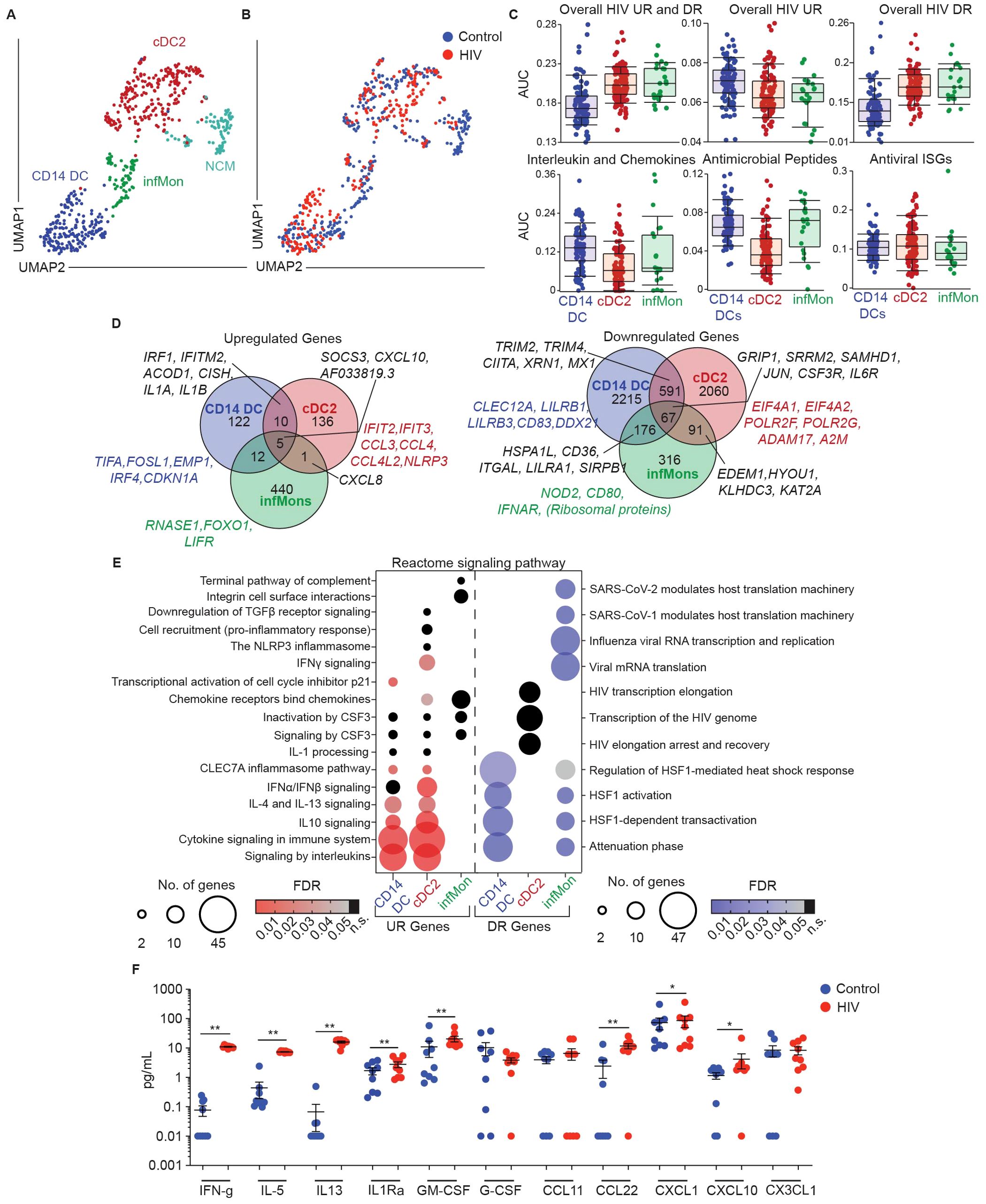
Figure 5. CD14+ DCs largely mediate rapid protective host response observed in genital DC response to HIV. (A) Representative UMAP visualization of CD11c+HLA-DR+ clusters and (B) overlay of control and HIV exposed cells (MOI=0.5 for 30 min). (C) AUCell comparing enrichment in CD11c+HLA-DR+ subsets in HIV exposed samples of overall upregulated (UR) and downregulated (DR) HIV signatures (top row) and key gene ontology (GO) terms associated with dendritic cell function (bottom row). (D) Venn-diagram representation comparing expression of shared and unique genes significantly upregulated (top) and downregulated (bottom) between CD11c+HLA-DR+ subsets in response to HIV stimulation. (p-value ≤ 0.05; -1.2 <FC>1.2) (E) Bubble-plot representation of biological pathways significantly Reactome significantly enriched in upregulated (UR; red) and downregulated (DR; blue) genes in response to HIV stimulation between different CD11c+HLA-DR+ subsets. (FDR ≤ 0.05; black dots indicate non-significant (n.s.) FDR values) (F) Comparison of secreted protein levels in supernatants of media (control; blue) and HIV exposed (HIV; red; MOI 0.5 for 3h), FGT resident CD14+ cells. Significance – Paired non-parametric Wilcoxon-test (p ≤ 0.05 *; p ≤ 0.01 **).
First, we utilized the overall transcriptional signature detected after HIV exposure shown in Figure 4B to perform AUCell analysis and determine whether this signature was enriched in specific genital DC subsets (Figure 5C). Interestingly, the overall HIV response signature (including upregulated and downregulated genes) was preferentially enriched in cDC2s and infMons. However, we observed enrichment of HIV upregulated genes in CD14 DCs, whereas HIV downregulated genes were enriched in cDC2s and infMons, suggesting that HIV challenge preferentially suppresses transcriptional changes in cDC2s and infMons compared to CD14 DCs. Furthermore, we detected an enrichment of cytokine, chemokine and antimicrobial peptide signatures within the CD14+ DC population, but no differences in enrichment of antiviral ISGs (Figure 5C), suggesting that CD14+ DCs largely mediate innate secretory response to HIV at early stages.
We next analyzed differential gene expression within each subset to identify transcriptional changes induced by HIV exposure (Figure 5D; Supplementary Data Sheet S1). All three subsets shared upregulation of AF033819.3 (the viral transcript from viral input), SOCS3, and CXCL10 (Figure 5D). CD14 DCs and cDC2s shared expression of anti-viral genes IRF1, IFITM2 and ACOD1 and the cytokine signaling genes CISH, IL1A and IL1B. CD14 DCs and infMons shared expression of zinc finger proteins and non-protein coding genes, except for CSF3 (encoding the granulocyte colony stimulating factor (G-CSF)) (Supplementary Data Sheet S1). cDC2s and infMons both upregulated CXCL8 expression upon HIV stimulation.
Next, we analyzed genes uniquely upregulated by each subset in response to HIV. We observed that CD14 DCs upregulated TIFA and FOSL1, genes associated with initiation of inflammation; EMP1, previously described in type I interferon stimulated DCs (115); and CDKN1A, a gene involved in p53 transcription previously shown to be protective at early stages of HIV infection (116). cDC2s uniquely upregulated ISGs IFIT2 and IFIT3, both possessing antiviral functions, CCL3, CCL4 and CCL4L2, encoding chemokines capable of binding HIV receptors (133, 134) and NLRP3, encoding inflammasome protein and indicating rapid induction of inflammation. infMons upregulated RNASE1 (encoding RNA degrading secreted protein) (117), and LIFR, which encodes CD118 described to function as an alternative receptor for HIV (118).
Downregulated genes were more abundant, and we focused our analysis on genes associated with DC function and HIV pathogenesis (Figure 5D; Supplementary Data Sheet S1). All 3 subsets shared lowered expression of genes associated with HIV transcription such as GRIP1 and SRRM2, which promote HIV replication, and interestingly SAMHD1, which plays a role in preventing replication of HIV. Additionally, all three subsets showed decreased expression of inflammation associated genes JUN, CSF3R and IL6R. CD14 DCs and infMons shared downregulation of HSPA1L (encoding a heat shock protein) and genes associated with adaptive immunity such as CD36, ITGAL, LILRA1 and SIRPB1. CD14 and cDC2s shared suppression of HIV-replication promoting gene XRN1, and interferon signaling associated genes TRIM2, TRIM4, CIITA and MX1. infMons and cDC2s shared downregulated expression of genes associated with unfolded protein response, such as EDEM1, HYOU1, KLHDC3 and KAT2A (119). CD14 DCs uniquely downregulated inhibitory genes CLEC12A, LILRB1, and classical DC activation marker CD83, suggesting non-canonical activation of this subset. We also observed downregulation of HIV binding and nuclear localization protein encoding gene DDX21 (120), suggesting inhibition of nuclear import. cDC2s downregulated genes associated with promoting HIV replication such as EIF4A1, EIF4A2 (HIV translation initiation) (121) and RNA polymerase II encoding genes POLR2F, POLR2G (122). We also observed downregulation of the TNFα degrading gene ADAM17 and A2M encoding alpha2-macroglobulin, that binds to inflammatory interleukins with high affinity, suggesting inhibition of HIV replication and promoting inflammatory environment. infMons downregulated expression of NOD2, CD80, and IFNAR indicating decreased cell activation.
We performed pathway analysis using Reactome to gain information about biological interactions (50) (Figure 5E; Supplementary Data Sheet S1). Analysis of upregulated genes revealed shared involvement of CD14 DCs and cDC2s in cytokine signaling pathways (IL-10, IL-4/IL-13 pathways). cDC2s uniquely upregulated Type I IFN and IFN-γ signaling, while CD14 DCs uniquely upregulated transcriptional activation of p21, a known restriction factor against HIV infection in monocyte-derived DCs (123). Analysis of downregulated genes revealed shared signatures between CD14+ DCs and infMons for heat shock response, and a unique signature for infMons related to downregulation of viral replication.
Finally, to validate our findings of a secreted response at the protein level, we purified genital CD14 DCs by magnetic bead selection and incubated them with HIV as described in methods. Supernatants harvested 3 hours post stimulation from unstimulated and HIV exposed cells were assayed to quantify secreted proteins (Figure 5F). In response to HIV, we detected increased secretion of cytokines and chemokines (IFN-γ, IL-5, IL-13, IL1Rα, GM-CSF, CXCL1, CXCL10 and CCL22). These results confirm rapid innate secretory response in response to HIV.
Overall, we demonstrate subset-specific responses to HIV, with a preferential suppression of genes in cDC2s and infMons compared to CD14 DCs, with CD14+ DCs largely mediating the secretory innate response after HIV stimulation.
4 Discussion
In this study we evaluated dendritic cell heterogeneity in the human female genital mucosa and how DC subsets respond to HIV immediately after exposure. We found that the CD11c+HLA-DR+ myeloid population in the genital mucosa includes three DC subsets and two monocyte/macrophage populations with distinct functional and phenotypic properties under homeostatic conditions. However, following HIV exposure, the antiviral response is dominated by DCs’ rapid secretory response, activation of non-classical inflammatory pathways and host restriction factors. Further, we report subset specific differences in genital DC response to HIV, where CD14+ DCs are the major subset activated by HIV and responsible for the secretory antimicrobial response, while cDC2s activate inflammasome pathways and antiviral IFN responses. Recognizing that DCs play a crucial role in responding to invading pathogens and recruiting adaptive immunity, identifying DC subsets with anti-HIV properties could aid targeted HIV prevention and vaccination strategies.
We and others have reported the presence of different DC subsets in the genital mucosa (15, 17, 20, 25, 30, 31, 38), however in-depth characterization of these subsets, specifically the CD14-expressing DC compartment, throughout the female genital tract according to recent advances in the field remains lacking (37). Mucosal DC characterization in humans is technically challenging due to their rare nature and the difficulty in isolating specific subsets for functional characterization. To overcome these barriers, we utilized multi-omics approaches to characterize protein surface expression and transcriptome profile of DCs at the single-cell level in mixed cell suspensions from human hysterectomies. Our analysis revealed that the CD11c+HLA-DR+ population is highly heterogeneous, including multiple DCs and monocyte/macrophage subsets. Within the DCs populations, we identified CD14+ DCs, CD1c+ cDC2s and CD14+CD1c+ DC3s, with CD14+ DCs representing the dominant population, consistent with prior reports using flow cytometry (25). Importantly, we identified two monocyte/macrophage subsets within CD11c+HLA-DR+ cells, indicating that CD11c and HLA-DR expression are not sufficient to define DCs in the genital tract.
Under homeostatic conditions the different myeloid populations displayed differential expression of PRRs, chemokines/cytokines and antimicrobials, indicating subset-dependent roles in tissue homeostasis and differential predisposition to sense and respond to pathogens, including HIV. Specifically, PRRs previously associated with HIV membrane binding and detection (such as CLEC4A, CLEC4E, TLR2 and TLR4) were preferentially expressed by CD14+ DCs, suggesting that CD14+ DCs may play a principal role in early detection of HIV upon exposure within the genital tract. Although prior studies using in vitro generated monocyte-derived DCs demonstrated that DCIR (encoded by CLEC4A) was responsible for viral capture and transinfection to CD4+ T cells (60, 61), future functional studies are needed to test whether DCIR is involved in HIV capture by genital CD14+ DCs. Interestingly, our data indicates a lack of gene expression by CD14+ DCs of other classical HIV-binding lectins (CD207, CD209 and MRC1) which were uniquely expressed in the infMons subset that shares characteristics of macrophages. Further research is needed to define the mechanisms responsible for viral capture by genital DCs. Cytoplasmic and endosomal sensors for viral RNA (TLR7/8, DDX58, IFIH1) were enriched in the monocyte/macrophage subsets, possible indicating delayed responses or roles in later HIV detection following viral replication. Cytokine/chemokine expression patterns further pointed for CD14+ DCs to play a role in antibacterial defense and inflammatory responses, while cDC2s were involved in maintenance of tissue homeostasis, regulation of inflammation, and promotion of a Th2/T regulatory environment. Overall, this suggests that the CD14+ DC population is pre-armed to generate rapid innate responses against incoming pathogens. Interestingly, between the subsets, no differences were observed in antigen processing and presentation, and T cell proliferation pathways, indicating that these populations share their antigen presenting cell properties under homeostatic conditions, consistent with our prior observations (30, 31).
Our validation of this myeloid subset classification using flow cytometry further allowed phenotypical comparisons between the CD14+ DC, CD14+CD1c+ DCs and CD1c+ cDC2 populations and the establishment of markers to discriminate the activated monocyte/macrophage populations (CD16, CLEC12A). Consistent with studies evaluating blood cells (35, 36, 59), we found that the CD14+CD1c+ subset displayed an intermediate phenotype between the monocyte-derived CD14+ DCs and the classical myeloid DCs (CD1c+ cDC2s), with high expression of HLA-DR but also monocyte origin-associated markers (CD64, CD11b, CX3CR1). This suggests that CD14+CD1c+ DCs in the genital mucosa are a homolog of DC3s described in peripheral blood and tumors (35, 36, 59), but in the genital tract they are present under steady-state conditions. In addition, the DC3 subset showed increased expression of classical and non-classical HIV coreceptors (CCR5, CD49d, CX3CR1) relevant for mucosal HIV pathogenesis (95, 96, 124), suggesting enhanced ability for viral uptake by CD14+CD1c+ DC population as described previously by us and others (20, 25). These phenotypic differences in expression of activation, origin, antigen uptake and HIV tropic markers highlight the importance of understanding genital DC subsets to develop targeted strategies.
A novel aspect of our study is the identification of early transcriptional signatures in genital DCs immediately following HIV exposure. While DC infection and trans-infection to T cells have been evaluated previously by others in blood (125, 126) and mucosal tissues (15, 18, 20), the early antiviral responses induced immediately following mucosal HIV exposure remained uncharacterized. In this study, we uncovered that, at early time points, before viral replication takes place, HIV exposure induces a rapid secretory response at the transcriptional and protein levels, activation of host restriction factors (IRF1, IFITM2, ACOD1), upregulation of genes involved tissue retention (CD69), and suppression of genes involved in T cell activation (CD83, CD86, CD58). Taken together, our data suggests that shortly after exposure, DCs remain in the mucosa and play a role in initiating local innate antiviral protection. However, several inflammatory markers were also upregulated and therefore the consequences for tissue protection and potential attraction and activation of HIV target cells remains to be determined.
Another novel contribution of our study is the discrimination of responses following HIV stimulation in different subsets of genital DCs. We found that all subsets shared activation of genes related to secretion of cytokines and chemokines, although CD14+ DCs and cDC2s were the predominant subsets involved in this response. Here, we observed shared upregulation of genes associated with inflammation and antiviral properties (IRF1, IFITM2, ACOD1, CISH, IL1A, IL1B) between cDC2s and CD14+ DCs upon exposure to HIV. These findings are consistent with earlier studies using monocyte-derived DCs and macrophages which demonstrated the upregulation of IRF1 transcripts and protein, in addition to other ISGs (22, 127). In addition, we uncovered unique pathways elicited by HIV stimulation in each subset. CD14+ DCs were the main players in overall antimicrobial defense, responses to TLR activation and initiation of inflammation (TIFA, FOSL1, EMP1), while cDC2s displayed a more specific antiviral response with activation of type I interferon (IFIT2, IFIT3, IRF1) and inflammasome (NLRP3) pathways. Importantly, both subsets activated mechanisms to prevent HIV replication. CD14+ DCs induced transcriptional activation of p21, a host restriction factor in monocyte-derived DCs (123), while cDC2s downregulated genes necessary for HIV transcription (EIF4A1, EIF4A2, POLR2F, POLR2G). In contrast to the DC subsets, infMons were not involved in initial antiviral protection, but downregulated pathways related to viral cycle, suggesting inhibition of HIV replication. However, future time-course studies are needed to better understand the kinetics and mechanisms by which HIV modifies DC function in a subset-specific manner to promote infection, trans-infection and HIV dissemination. Additionally, our study only used HIV-BaL, a laboratory adapted strain, but our findings were not confirmed with HIV transmitted/founder (TF) strains. Although prior studies with DCs and macrophages found no differences between HIV-BaL and TF strains (20, 24, 128), future studies are needed to define DC subset-specific responses to TF strains immediately following exposure to HIV.
Finally, we validated the transcriptional signatures by characterizing the anti-HIV secretory response at the protein level in supernatants from CD14 DCs purified from the genital tract. Despite our observation of upregulation of IL1B at the transcriptional levels, we did not detect IL1-β production in our cultures. However, we detected production of IFN-γ, GM-CSF and chemokines with inflammatory and antiviral properties (CXCL10, CCL22). These results complement our prior identification of early secretion of CCR5-ligands and antimicrobial proteins by CD14+ DCs in response to HIV (25). Production of GM-CSF, IFN-γ, and CXCL10 has been shown to be induced by TLR stimulation in DCs (129, 130), suggesting that TLR signaling in CD14+ DCs may be responsible for induction of cytokine and chemokine secretion. Interestingly, while IFNγ and CXCL10 are involved in mediating Th1 and CD8+ T cell adaptive immunity (131, 132), we also detected production of the Th2 cytokines IL5 and IL13, involved in allergic inflammation and activation of Th2 CD4+ T cells (135). While we have previously described that under homeostatic conditions CD14+ DCs induce proliferation of CD8+ T cells with tissue-resident memory phenotype (30), and proliferation of CD4+ T cells and double negative (DN) T cells (31), future studies are needed to determine how these cytokine profiles modify tissue environment, susceptibility to HIV infection, and modify T cell induction profile.
Our study has several limitations mainly due to the rare nature of mucosal DCs and the technical difficulties in isolating human DC subsets. First, due to the very low frequency of cDC2s within the genital mucosa, we were unable to isolate cDC2s from mixed cell suspensions to determine their secretory response to HIV at the protein level. Similarly, lack of distinct surface markers and low cell numbers in the genital tract prevented isolating DC3s (CD14+CD1c+) and resulted in clustering and isolation of this subset together with the CD14+ DCs. Therefore, quantifying cDC2 and DC3s individual response to HIV at the protein level will require innovative strategies. Furthermore, our analysis of subset-specific responses was unable to evaluate non-classical monocytes due to low numbers of cells in the HIV stimulated condition, and therefore the contribution of this subset to HIV pathogenesis remains to be elucidated. Despite these limitations, our study provides valuable novel information using an experimental model to evaluate initial mucosal responses to HIV exposure that allows the study of DC subsets without lengthy processing time that could potentially modify primary DCs.
Overall, we demonstrate that the female genital mucosa is populated with different subsets of DCs that specialize under homeostatic conditions and that, immediately following HIV exposure, initiate a local secretory antiviral response and activate host mechanisms to prevent HIV replication in a subset-specific manner. Our findings contribute to the field of mucosal HIV acquisition and provide a map to identify therapeutic targets that trigger local protective innate immune responses against HIV without inducing detrimental tissue inflammation.
Data availability statement
The data presented in the study are deposited in the NCBI GEO repository, accession numbers GSE279408, GSE279774 and GSE279775.
Ethics statement
The studies involving humans were approved by Health Sciences Institutional Review Board at Tufts University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
SP: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Conceptualization. LL: Writing – review & editing, Methodology, Investigation. FC-S: Writing – review & editing, Visualization, Investigation. AW: Writing – review & editing, Methodology, Investigation. AB: Writing – review & editing, Methodology, Investigation. VI: Writing – review & editing, Methodology, Investigation. AV: Writing – review & editing, Resources, Project administration. JF: Writing – review & editing, Formal analysis. CW: Writing – review & editing, Funding acquisition. MR-G: Writing – review & editing, Writing – original draft, Visualization, Supervision, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Institutes of Health (R01AG060801 (MRG), R21AI172065 (MRG), R01AG064794 (CWR), and S10OD032201 (David Thorley-Lawson Memorial Flow Cytometry Core Facility)).
Acknowledgments
We thank Bipin Malla, Paula Josephs, Afshin Azimirad for their help with clinical coordination and patient recruitment, Fiona Barr for technical assistance and Karla Murga and Biorepository at TMC for tissue supply.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1472656/full#supplementary-material
References
1. Leung Soo C, Pant Pai N, Bartlett SJ, Esmail A, Dheda K, Bhatnagar S. Socioeconomic factors impact the risk of HIV acquisition in the township population of South Africa: A Bayesian analysis. PloS Glob Public Health. (2023) 3:e0001502. doi: 10.1371/journal.pgph.0001502
2. Collaborators GH. Global, regional, and national sex-specific burden and control of the HIV epidemic 1990-2019, for 204 countries and territories: the Global Burden of Diseases Study 2019. Lancet HIV. (2021) 8:e633–51. doi: 10.1016/S2352-3018(21)00152-1
3. How is HIV transmitted (2021). Available online at: https://www.nichd.nih.gov/health/topics/hiv/conditioninfo/transmission (Accessed May 29 2024).
4. Nazli A, Chan O, Dobson-Belaire WN, Ouellet M, Tremblay MJ, Gray-Owen SD, et al. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PloS Pathog. (2010) 6:e1000852. doi: 10.1371/journal.ppat.1000852
5. Ferreira VH, Dizzell S, Nazli A, Kafka JK, Mueller K, Nguyen PV, et al. Medroxyprogesterone acetate regulates HIV-1 uptake and transcytosis but not replication in primary genital epithelial cells, resulting in enhanced T-cell infection. J Infect Dis. (2015) 211:1745–56. doi: 10.1093/infdis/jiu832
6. Rodriguez-Garcia M, Barr FD, Crist SG, Fahey JV, Wira CR. Phenotype and susceptibility to HIV infection of CD4+ Th17 cells in the human female reproductive tract. Mucosal Immunol. (2014) 7:1375–85. doi: 10.1038/mi.2014.26
7. Barr FD, Ochsenbauer C, Wira CR, Rodriguez-Garcia M. Neutrophil extracellular traps prevent HIV infection in the female genital tract. Mucosal Immunol. (2018) 11:1420–8. doi: 10.1038/s41385-018-0045-0
8. De Lara LM, Parthasarathy RS, Rodriguez-Garcia M. Mucosal immunity and HIV acquisition in women. Curr Opin Physiol. (2021) 19:32–8. doi: 10.1016/j.cophys.2020.07.021
9. Rodriguez-Garcia M, Connors K, Ghosh M. HIV pathogenesis in the human female reproductive tract. Curr HIV/AIDS Rep. (2021) 18:139–56. doi: 10.1007/s11904-021-00546-1
10. Moreno De Lara L, Werner A, Borchers A, Carrillo-Salinas FJ, Marmol W, Parthasarathy S, et al. Aging dysregulates neutrophil extracellular trap formation in response to HIV in blood and genital tissues. Front Immunol. (2023) 14:1256182. doi: 10.3389/fimmu.2023.1256182
11. Chang SY, Ko HJ, Kweon MN. Mucosal dendritic cells shape mucosal immunity. Exp Mol Med. (2014) 46:e84. doi: 10.1038/emm.2014.16
12. Sato K, Yang XL, Yudate T, Chung JS, Wu J, Luby-Phelps K, et al. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J Biol Chem. (2006) 281:38854–66. doi: 10.1074/jbc.M606542200
13. Rothfuchs AG, Bafica A, Feng CG, Egen JG, Williams DL, Brown GD, et al. Dectin-1 interaction with Mycobacterium tuberculosis leads to enhanced IL-12p40 production by splenic dendritic cells. J Immunol. (2007) 179:3463–71. doi: 10.4049/jimmunol.179.6.3463
14. Harman AN, Kim M, Nasr N, Sandgren KJ, Cameron PU. Tissue dendritic cells as portals for HIV entry. Rev Med Virol. (2013) 23:319–33. doi: 10.1002/rmv.1753
15. Perez-Zsolt D, Cantero-Perez J, Erkizia I, Benet S, Pino M, Serra-Peinado C, et al. Dendritic cells from the cervical mucosa capture and transfer HIV-1 via siglec-1. Front Immunol. (2019) 10:825. doi: 10.3389/fimmu.2019.00825
16. Da Silva LT, Santillo BT, De Almeida A, Duarte A, Oshiro TM. Using dendritic cell-based immunotherapy to treat HIV: how can this strategy be improved? Front Immunol. (2018) 9:2993. doi: 10.3389/fimmu.2018.02993
17. Izquierdo-Useros N, Lorizate M, Mclaren PJ, Telenti A, Krausslich HG, Martinez-Picado J. HIV-1 capture and transmission by dendritic cells: the role of viral glycolipids and the cellular receptor Siglec-1. PloS Pathog. (2014) 10:e1004146. doi: 10.1371/journal.ppat.1004146
18. Trifonova RT, Bollman B, Barteneva NS, Lieberman J. Myeloid cells in intact human cervical explants capture HIV and can transmit it to CD4 T cells. Front Immunol. (2018) 9:2719. doi: 10.3389/fimmu.2018.02719
19. Martin-Moreno A, Munoz-Fernandez MA. Dendritic cells, the double agent in the war against HIV-1. Front Immunol. (2019) 10:2485. doi: 10.3389/fimmu.2019.02485
20. Rhodes JW, Botting RA, Bertram KM, Vine EE, Rana H, Baharlou H, et al. Human anogenital monocyte-derived dendritic cells and langerin+cDC2 are major HIV target cells. Nat Commun. (2021) 12:2147. doi: 10.1038/s41467-021-22375-x
21. Turville SG, Santos JJ, Frank I, Cameron PU, Wilkinson J, Miranda-Saksena M, et al. Immunodeficiency virus uptake, turnover, and 2-phase transfer in human dendritic cells. Blood. (2004) 103:2170–9. doi: 10.1182/blood-2003-09-3129
22. Harman AN, Kraus M, Bye CR, Byth K, Turville SG, Tang O, et al. HIV-1-infected dendritic cells show 2 phases of gene expression changes, with lysosomal enzyme activity decreased during the second phase. Blood. (2009) 114:85–94. doi: 10.1182/blood-2008-12-194845
23. Nasr N, Lai J, Botting RA, Mercier SK, Harman AN, Kim M, et al. Inhibition of two temporal phases of HIV-1 transfer from primary Langerhans cells to T cells: the role of langerin. J Immunol. (2014) 193:2554–64. doi: 10.4049/jimmunol.1400630
24. Bertram KM, Botting RA, Baharlou H, Rhodes JW, Rana H, Graham JD, et al. Identification of HIV transmitting CD11c(+) human epidermal dendritic cells. Nat Commun. (2019) 10:2759. doi: 10.1038/s41467-019-10697-w
25. Rodriguez-Garcia M, Shen Z, Barr FD, Boesch AW, Ackerman ME, Kappes JC, et al. Dendritic cells from the human female reproductive tract rapidly capture and respond to HIV. Mucosal Immunol. (2017) 10:531–44. doi: 10.1038/mi.2016.72
26. Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity. (2014) 40:642–56. doi: 10.1016/j.immuni.2014.04.016
27. Tagliani E, Erlebacher A. Dendritic cell function at the maternal-fetal interface. Expert Rev Clin Immunol. (2011) 7:593–602. doi: 10.1586/eci.11.52
28. Mor G, Aldo P, Alvero AB. The unique immunological and microbial aspects of pregnancy. Nat Rev Immunol. (2017) 17:469–82. doi: 10.1038/nri.2017.64
29. Shah NM, Herasimtschuk AA, Boasso A, Benlahrech A, Fuchs D, Imami N, et al. Changes in T cell and dendritic cell phenotype from mid to late pregnancy are indicative of a shift from immune tolerance to immune activation. Front Immunol. (2017) 8:1138. doi: 10.3389/fimmu.2017.01138
30. Rodriguez-Garcia M, Fortier JM, Barr FD, Wira CR. Aging impacts CD103(+) CD8(+) T cell presence and induction by dendritic cells in the genital tract. Aging Cell. (2018) 17:e12733. doi: 10.1111/acel.12733
31. Parthasarathy S, Shen Z, Carrillo-Salinas FJ, Iyer V, Vogell A, Illanes D, et al. Aging modifies endometrial dendritic cell function and unconventional double negative T cells in the human genital mucosa. Immun Ageing. (2023) 20:34. doi: 10.1186/s12979-023-00360-w
32. Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. (2009) 324:392–7. doi: 10.1126/science.1170540
33. Coillard A, Segura E. In vivo differentiation of human monocytes. Front Immunol. (2019) 10:1907. doi: 10.3389/fimmu.2019.01907
34. Collin M, Bigley V. Human dendritic cell subsets: an update. Immunology. (2018) 154:3–20. doi: 10.1111/imm.12888
35. Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. (2017) 356:eaah4573. doi: 10.1126/science.aah4573
36. Bourdely P, Anselmi G, Vaivode K, Ramos RN, Missolo-Koussou Y, Hidalgo S, et al. Transcriptional and functional analysis of CD1c(+) human dendritic cells identifies a CD163(+) subset priming CD8(+)CD103(+) T cells. Immunity. (2020) 53:335–352 e338. doi: 10.1016/j.immuni.2020.06.002
37. Ginhoux F, Guilliams M, Merad M. Expanding dendritic cell nomenclature in the single-cell era. Nat Rev Immunol. (2022) 22:67–8. doi: 10.1038/s41577-022-00675-7
38. Pena-Cruz V, Agosto LM, Akiyama H, Olson A, Moreau Y, Larrieux JR, et al. HIV-1 replicates and persists in vaginal epithelial dendritic cells. J Clin Invest. (2018) 128:3439–44. doi: 10.1172/jci98943
39. Bertram KM, O’neil TR, Vine EE, Baharlou H, Cunningham AL, Harman AN. Defining the landscape of human epidermal mononuclear phagocytes. Immunity. (2023) 56:459–60. doi: 10.1016/j.immuni.2023.02.001
40. Vine EE, Rhodes JW, Warner Van Dijk FA, Byrne SN, Bertram KM, Cunningham AL, et al. HIV transmitting mononuclear phagocytes; integrating the old and new. Mucosal Immunol. (2022) 15:542–50. doi: 10.1038/s41385-022-00492-0
41. Vine EE, Austin PJ, O’neil TR, Nasr N, Bertram KM, Cunningham AL, et al. Epithelial dendritic cells vs. Langerhans cells: Implications for mucosal vaccines. Cell Rep. (2024) 43:113977. doi: 10.1016/j.celrep.2024.113977
42. Rodriguez-Garcia M, Fortier JM, Barr FD, Wira CR. Isolation of dendritic cells from the human female reproductive tract for phenotypical and functional studies. J Vis Exp. (2018) 13:57100. doi: 10.3791/57100
43. Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. (1986) 233:215–9. doi: 10.1126/science.3014648
44. Van ‘T Wout AB, Schuitemaker H, Kootstra NA. Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat Protoc. (2008) 3:363–70. doi: 10.1038/nprot.2008.3
45. Salcher S, Heidegger I, Untergasser G, Fotakis G, Scheiber A, Martowicz A, et al. Comparative analysis of 10X Chromium vs. BD Rhapsody whole transcriptome single-cell sequencing technologies in complex human tissues. Heliyon. (2024) 10:e28358. doi: 10.1016/j.heliyon.2024.e28358
46. Smith JM, Wira CR, Fanger MW, Shen L. Human fallopian tube neutrophils–a distinct phenotype from blood neutrophils. Am J Reprod Immunol. (2006) 56:218–29. doi: 10.1111/j.1600-0897.2006.00410.x
47. Ramirez-Ramirez D, Vadillo E, Arriaga-Pizano LA, Mayani H, Estrada-Parra S, Velasco-Velazquez MA, et al. Early differentiation of human CD11c(+)NK cells with gammadelta T cell activation properties is promoted by dialyzable leukocyte extracts. J Immunol Res. (2016) 2016:4097642. doi: 10.1155/2016/4097642
48. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. (2000) 25:25–9. doi: 10.1038/75556
49. Gene Ontology C, Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, et al. The gene ontology knowledgebase in 2023. Genetics. (2023) 224. doi: 10.1093/genetics/iyad031
50. Fabregat A, Sidiropoulos K, Viteri G, Marin-Garcia P, Ping P, Stein L, et al. Reactome diagram viewer: data structures and strategies to boost performance. Bioinformatics. (2018) 34:1208–14. doi: 10.1093/bioinformatics/btx752
51. Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. (2017) 14:1083–6. doi: 10.1038/nmeth.4463
52. Duluc D, Gannevat J, Anguiano E, Zurawski S, Carley M, Boreham M, et al. Functional diversity of human vaginal APC subsets in directing T-cell responses. Mucosal Immunol. (2013) 6:626–38. doi: 10.1038/mi.2012.104
53. Llufrio EM, Wang L, Naser FJ, Patti GJ. Sorting cells alters their redox state and cellular metabolome. Redox Biol. (2018) 16:381–7. doi: 10.1016/j.redox.2018.03.004
54. Trifonova RT, Lieberman J, Van Baarle D. Distribution of immune cells in the human cervix and implications for HIV transmission. Am J Reprod Immunol. (2014) 71:252–64. doi: 10.1111/aji.12198
55. Kabashima K, Shiraishi N, Sugita K, Mori T, Onoue A, Kobayashi M, et al. CXCL12-CXCR4 engagement is required for migration of cutaneous dendritic cells. Am J Pathol. (2007) 171:1249–57. doi: 10.2353/ajpath.2007.070225
56. Kabashima K, Sugita K, Shiraishi N, Tamamura H, Fujii N, Tokura Y. CXCR4 engagement promotes dendritic cell survival and maturation. Biochem Biophys Res Commun. (2007) 361:1012–6. doi: 10.1016/j.bbrc.2007.07.128
57. Rigamonti A, Castagna A, Viatore M, Colombo FS, Terzoli S, Peano C, et al. Distinct responses of newly identified monocyte subsets to advanced gastrointestinal cancer and COVID-19. Front Immunol. (2022) 13:967737. doi: 10.3389/fimmu.2022.967737
58. Zheng W, Wang X, Liu J, Yu X, Li L, Wang H, et al. Single-cell analyses highlight the proinflammatory contribution of C1q-high monocytes to Behcet’s disease. Proc Natl Acad Sci U.S.A. (2022) 119:e2204289119. doi: 10.1073/pnas.2204289119
59. Cytlak U, Resteu A, Pagan S, Green K, Milne P, Maisuria S, et al. Differential IRF8 transcription factor requirement defines two pathways of dendritic cell development in humans. Immunity. (2020) 53:353–370 e358. doi: 10.1016/j.immuni.2020.07.003
60. Lambert AA, Gilbert C, Richard M, Beaulieu AD, Tremblay MJ. The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood. (2008) 112:1299–307. doi: 10.1182/blood-2008-01-136473
61. Miller E, Bhardwaj N. Dendritic cell dysregulation during HIV-1 infection. Immunol Rev. (2013) 254:170–89. doi: 10.1111/imr.12082
62. Nazli A, Kafka JK, Ferreira VH, Anipindi V, Mueller K, Osborne BJ, et al. HIV-1 gp120 induces TLR2- and TLR4-mediated innate immune activation in human female genital epithelium. J Immunol. (2013) 191:4246–58. doi: 10.4049/jimmunol.1301482
63. Hutten TJ, Thordardottir S, Fredrix H, Janssen L, Woestenenk R, Tel J, et al. CLEC12A-mediated antigen uptake and cross-presentation by human dendritic cell subsets efficiently boost tumor-reactive T cell responses. J Immunol. (2016) 197:2715–25. doi: 10.4049/jimmunol.1600011
64. Heger L, Hofer TP, Bigley V, De Vries IJM, Dalod M, Dudziak D, et al. Subsets of CD1c(+) DCs: dendritic cell versus monocyte lineage. Front Immunol. (2020) 11:559166. doi: 10.3389/fimmu.2020.559166
65. Browne EP. The role of toll-like receptors in retroviral infection. Microorganisms. (2020) 8. doi: 10.3390/microorganisms8111787
66. Lopez Robles MD, Pallier A, Huchet V, Le Texier L, Remy S, Braudeau C, et al. Cell-surface C-type lectin-like receptor CLEC-1 dampens dendritic cell activation and downstream Th17 responses. Blood Adv. (2017) 1:557–68. doi: 10.1182/bloodadvances.2016002360
67. Drouin M, Saenz J, Gauttier V, Evrard B, Teppaz G, Pengam S, et al. CLEC-1 is a death sensor that limits antigen cross-presentation by dendritic cells and represents a target for cancer immunotherapy. Sci Adv. (2022) 8:eabo7621. doi: 10.1126/sciadv.abo7621
68. Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. (2009) 83:10761–9. doi: 10.1128/jvi.00770-09
69. Goubau D, Deddouche S, Reis E Sousa C. Cytosolic sensing of viruses. Immunity. (2013) 38:855–69. doi: 10.1016/j.immuni.2013.05.007
70. Finberg RW, Wang JP, Kurt-Jones EA. Toll like receptors and viruses. Rev Med Virol. (2007) 17:35–43. doi: 10.1002/rmv.525
71. Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. (2013) 39:599–610. doi: 10.1016/j.immuni.2013.08.007
72. Mulder K, Patel AA, Kong WT, Piot C, Halitzki E, Dunsmore G, et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity. (2021) 54:1883–1900 e1885. doi: 10.1016/j.immuni.2021.07.007
73. Lo YL, Liou GG, Lyu JH, Hsiao M, Hsu TL, Wong CH. Dengue virus infection is through a cooperative interaction between a mannose receptor and CLEC5A on macrophage as a multivalent hetero-complex. PloS One. (2016) 11:e0166474. doi: 10.1371/journal.pone.0166474
74. Farhat K, Riekenberg S, Heine H, Debarry J, Lang R, Mages J, et al. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J Leukoc Biol. (2008) 83:692–701. doi: 10.1189/jlb.0807586
75. Martinsen JT, Gunst JD, Hojen JF, Tolstrup M, Sogaard OS. The use of toll-like receptor agonists in HIV-1 cure strategies. Front Immunol. (2020) 11:1112. doi: 10.3389/fimmu.2020.01112
76. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. (2009) 458:514–8. doi: 10.1038/nature07725
77. Ghosh M, Shen Z, Schaefer TM, Fahey JV, Gupta P, Wira CR. CCL20/MIP3alpha is a novel anti-HIV-1 molecule of the human female reproductive tract. Am J Reprod Immunol. (2009) 62:60–71. doi: 10.1111/j.1600-0897.2009.00713.x
78. Le Borgne M, Etchart N, Goubier A, Lira SA, Sirard JC, Van Rooijen N, et al. Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity. (2006) 24:191–201. doi: 10.1016/j.immuni.2006.01.005
79. Das ST, Rajagopalan L, Guerrero-Plata A, Sai J, Richmond A, Garofalo RP, et al. Monomeric and dimeric CXCL8 are both essential for in vivo neutrophil recruitment. PloS One. (2010) 5:e11754. doi: 10.1371/journal.pone.0011754
80. Li Q, Laumonnier Y, Syrovets T, Simmet T. Recruitment of CCR6-expressing Th17 cells by CCL20 secreted from plasmin-stimulated macrophages. Acta Biochim Biophys Sin (Shanghai). (2013) 45:593–600. doi: 10.1093/abbs/gmt049
81. Metzemaekers M, Gouwy M, Proost P. Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell Mol Immunol. (2020) 17:433–50. doi: 10.1038/s41423-020-0412-0
82. Liew FY, Girard JP, Turnquist HR. Interleukin-33 in health and disease. Nat Rev Immunol. (2016) 16:676–89. doi: 10.1038/nri.2016.95
83. Imai T, Nagira M, Takagi S, Kakizaki M, Nishimura M, Wang J, et al. Selective recruitment of CCR4-bearing Th2 cells toward antigen-presenting cells by the CC chemokines thymus and activation-regulated chemokine and macrophage-derived chemokine. Int Immunol. (1999) 11:81–8. doi: 10.1093/intimm/11.1.81
84. Mizukami Y, Kono K, Kawaguchi Y, Akaike H, Kamimura K, Sugai H, et al. CCL17 and CCL22 chemokines within tumor microenvironment are related to accumulation of Foxp3+ regulatory T cells in gastric cancer. Int J Cancer. (2008) 122:2286–93. doi: 10.1002/ijc.23392
85. Pap R, Montsko G, Janosa G, Sipos K, Kovacs GL, Pandur E. Fractalkine regulates HEC-1A/JEG-3 interaction by influencing the expression of implantation-related genes in an in vitro co-culture model. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21093175
86. Liu Y, Cruikshank WW, O’loughlin T, O’reilly P, Center DM, Kornfeld H. Identification of a CD4 domain required for interleukin-16 binding and lymphocyte activation. J Biol Chem. (1999) 274:23387–95. doi: 10.1074/jbc.274.33.23387
87. Van Lieshout AW, van der Voort R, Toonen LW, Van Helden SF, Figdor CG, Van Riel PL, et al. Regulation of CXCL16 expression and secretion by myeloid cells is not altered in rheumatoid arthritis. Ann Rheum Dis. (2009) 68:1036–43. doi: 10.1136/ard.2007.086611
88. Veinotte L, Gebremeskel S, Johnston B. CXCL16-positive dendritic cells enhance invariant natural killer T cell-dependent IFNgamma production and tumor control. Oncoimmunology. (2016) 5:e1160979. doi: 10.1080/2162402x.2016.1160979
89. Yu H, Yang A, Liu L, Mak JYW, Fairlie DP, Cowley S. CXCL16 stimulates antigen-induced MAIT cell accumulation but trafficking during lung infection is CXCR6-independent. Front Immunol. (2020) 11:1773. doi: 10.3389/fimmu.2020.01773
90. Provost V, Larose MC, Langlois A, Rola-Pleszczynski M, Flamand N, Laviolette M. CCL26/eotaxin-3 is more effective to induce the migration of eosinophils of asthmatics than CCL11/eotaxin-1 and CCL24/eotaxin-2. J Leukoc Biol. (2013) 94:213–22. doi: 10.1189/jlb.0212074
91. Yung SC, Murphy PM. Antimicrobial chemokines. Front Immunol. (2012) 3:276. doi: 10.3389/fimmu.2012.00276
92. Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 regulates both Th1 and Th2 responses. Annu Rev Immunol. (2001) 19:423–74. doi: 10.1146/annurev.immunol.19.1.423
93. Kawasaki T, Ikegawa M, Yunoki K, Otani H, Ori D, Ishii KJ, et al. Alveolar macrophages instruct CD8(+) T cell expansion by antigen cross-presentation in lung. Cell Rep. (2022) 41:111828. doi: 10.1016/j.celrep.2022.111828
94. Scholer A, Hugues S, Boissonnas A, Fetler L, Amigorena S. Intercellular adhesion molecule-1-dependent stable interactions between T cells and dendritic cells determine CD8+ T cell memory. Immunity. (2008) 28:258–70. doi: 10.1016/j.immuni.2007.12.016
95. Saba E, Grivel JC, Vanpouille C, Brichacek B, Fitzgerald W, Margolis L, et al. HIV-1 sexual transmission: early events of HIV-1 infection of human cervico-vaginal tissue in an optimized ex vivo model. Mucosal Immunol. (2010) 3:280–90. doi: 10.1038/mi.2010.2
96. Mckinnon LR, Nyanga B, Chege D, Izulla P, Kimani M, Huibner S, et al. Characterization of a human cervical CD4+ T cell subset coexpressing multiple markers of HIV susceptibility. J Immunol. (2011) 187:6032–42. doi: 10.4049/jimmunol.1101836
97. Kolseth IB, Reine TM, Vuong TT, Meen AJ, Fan Q, Jenssen TG, et al. Serglycin is part of the secretory repertoire of LPS-activated monocytes. Immun Inflammation Dis. (2015) 3:23–31. doi: 10.1002/iid3.47
98. Wu R, Chen F, Wang N, Tang D, Kang R. ACOD1 in immunometabolism and disease. Cell Mol Immunol. (2020) 17:822–33. doi: 10.1038/s41423-020-0489-5
99. Kino T, Slobodskaya O, Pavlakis GN, Chrousos GP. Nuclear receptor coactivator p160 proteins enhance the HIV-1 long terminal repeat promoter by bridging promoter-bound factors and the Tat-P-TEFb complex. J Biol Chem. (2002) 277:2396–405. doi: 10.1074/jbc.m106312200
100. Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, Wagschal A, et al. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. (2009) 6:26. doi: 10.1186/1742-4690-6-26
101. Wojcechowskyj JA, Didigu CA, Lee JY, Parrish NF, Sinha R, Hahn BH, et al. Quantitative phosphoproteomics reveals extensive cellular reprogramming during HIV-1 entry. Cell Host Microbe. (2013) 13:613–23. doi: 10.1016/j.chom.2013.04.011
102. Ndzinu JK, Takeuchi H, Saito H, Yoshida T, Yamaoka S. eIF4A2 is a host factor required for efficient HIV-1 replication. Microbes Infect. (2018) 20:346–52. doi: 10.1016/j.micinf.2018.05.001
103. Torzicky M, Viznerova P, Richter S, Strobl H, Scheinecker C, Foedinger D, et al. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31) and CD99 are critical in lymphatic transmigration of human dendritic cells. J Invest Dermatol. (2012) 132:1149–57. doi: 10.1038/jid.2011.420
104. Lu C, Amin MA, Fox DA. CD13/aminopeptidase N is a potential therapeutic target for inflammatory disorders. J Immunol. (2020) 204:3–11. doi: 10.4049/jimmunol.1900868
105. Woodfin A, Voisin MB, Nourshargh S. PECAM-1: a multi-functional molecule in inflammation and vascular biology. Arterioscler Thromb Vasc Biol. (2007) 27:2514–23. doi: 10.1161/atvbaha.107.151456
106. Gartlan KH, Wee JL, Demaria MC, Nastovska R, Chang TM, Jones EL, et al. Tetraspanin CD37 contributes to the initiation of cellular immunity by promoting dendritic cell migration. Eur J Immunol. (2013) 43:1208–19. doi: 10.1002/eji.201242730
107. Hikita A, Tanaka N, Yamane S, Ikeda Y, Furukawa H, Tohma S, et al. Involvement of a disintegrin and metalloproteinase 10 and 17 in shedding of tumor necrosis factor-alpha. Biochem Cell Biol. (2009) 87:581–93. doi: 10.1139/o09-015
108. Van Der Touw W, Chen HM, Pan PY, Chen SH. LILRB receptor-mediated regulation of myeloid cell maturation and function. Cancer Immunol Immunother. (2017) 66:1079–87. doi: 10.1007/s00262-017-2023-x
109. Gong W, Howard OM, Turpin JA, Grimm MC, Ueda H, Gray PW, et al. Monocyte chemotactic protein-2 activates CCR5 and blocks CD4/CCR5-mediated HIV-1 entry/replication. J Biol Chem. (1998) 273:4289–92. doi: 10.1074/jbc.273.8.4289
110. Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. (2016) 44:D1087–1093. doi: 10.1093/nar/gkv1278
111. Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, et al. Serum amyloid A proteins induce pathogenic th17 cells and promote inflammatory disease. Cell. (2020) 183:2036–9. doi: 10.1016/j.cell.2020.12.008
112. Joag VR, Mckinnon LR, Liu J, Kidane ST, Yudin MH, Nyanga B, et al. Identification of preferential CD4+ T-cell targets for HIV infection in the cervix. Mucosal Immunol. (2016) 9:1–12. doi: 10.1038/mi.2015.28
113. Stieh DJ, Matias E, Xu H, Fought AJ, Blanchard JL, Marx PA, et al. Th17 cells are preferentially infected very early after vaginal transmission of SIV in macaques. Cell Host Microbe. (2016) 19:529–40. doi: 10.1016/j.chom.2016.03.005
114. Ashizuka S, Kita T, Inatsu H, Kitamura K. Adrenomedullin: A novel therapeutic for the treatment of inflammatory bowel disease. Biomedicines. (2021) 9. doi: 10.3390/biomedicines9081068
115. Eugster A, Muller D, Gompf A, Reinhardt S, Lindner A, Ashton M, et al. A novel type I interferon primed dendritic cell subpopulation in TREX1 mutant chilblain lupus patients. Front Immunol. (2022) 13:897500. doi: 10.3389/fimmu.2022.1094578
116. Shi B, Sharifi HJ, Digrigoli S, Kinnetz M, Mellon K, Hu W, et al. Inhibition of HIV early replication by the p53 and its downstream gene p21. Virol J. (2018) 15:53. doi: 10.1186/s12985-018-0959-x
117. Lu L, Li J, Moussaoui M, Boix E. Immune modulation by human secreted RNases at the extracellular space. Front Immunol. (2018) 9:1012. doi: 10.3389/fimmu.2018.01012
118. Greenwell-Wild T, Vazquez N, Jin W, Rangel Z, Munson PJ, Wahl SM. Interleukin-27 inhibition of HIV-1 involves an intermediate induction of type I interferon. Blood. (2009) 114:1864–74. doi: 10.1182/blood-2009-03-211540
119. Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat Rev Immunol. (2016) 16:469–84. doi: 10.1038/nri.2016.62
120. Heaton SM, Gorry PR, Borg NA. DExD/H-box helicases in HIV-1 replication and their inhibition. Trends Microbiol. (2023) 31:393–404. doi: 10.1016/j.tim.2022.11.001
121. Montero H, Perez-Gil G, Sampieri CL. Eukaryotic initiation factor 4A (eIF4A) during viral infections. Virus Genes. (2019) 55:267–73. doi: 10.1007/s11262-019-01641-7
122. Nilson KA, Price DH. The role of RNA polymerase II elongation control in HIV-1 gene expression, replication, and latency. Genet Res Int. (2011) 2011:726901. doi: 10.4061/2011/726901
123. Valle-Casuso JC, Allouch A, David A, Lenzi GM, Studdard L, Barre-Sinoussi F, et al. p21 restricts HIV-1 in monocyte-derived dendritic cells through the reduction of deoxynucleoside triphosphate biosynthesis and regulation of SAMHD1 antiviral activity. J Virol. (2017) 91. doi: 10.1128/jvi.01324-17
124. Cavarelli M, Foglieni C, Hantour N, Schorn T, Ferrazzano A, Dispinseri S, et al. Identification of CX3CR1(+) mononuclear phagocyte subsets involved in HIV-1 and SIV colorectal transmission. iScience. (2022) 25:104346. doi: 10.1016/j.isci.2022.104346
125. Geijtenbeek TB, Kwon DS, Torensma R, Van Vliet SJ, Van Duijnhoven GC, Middel J, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. (2000) 100:587–97. doi: 10.1016/s0092-8674(00)80694-7
126. Brouiller F, Nadalin F, Bonte PE, Ait-Mohamed O, Delaugerre C, Lelievre JD, et al. Single-cell RNA-seq analysis reveals dual sensing of HIV-1 in blood Axl(+) dendritic cells. iScience. (2023) 26:106019. doi: 10.1016/j.isci.2023.106019
127. Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, et al. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood. (2012) 120:778–88. doi: 10.1182/blood-2012-01-407395
128. Baharlou H, Canete N, Vine EE, Hu K, Yuan D, Sandgren KJ, et al. An in situ analysis pipeline for initial host-pathogen interactions reveals signatures of human colorectal HIV transmission. Cell Rep. (2022) 40:111385. doi: 10.1016/j.celrep.2022.111385
129. Barr TA, Brown S, Ryan G, Zhao J, Gray D. TLR-mediated stimulation of APC: Distinct cytokine responses of B cells and dendritic cells. Eur J Immunol. (2007) 37:3040–53. doi: 10.1002/eji.200636483
130. Brosbol-Ravnborg A, Hvas CL, Agnholt J, Dahlerup JF, Vind I, Till A, et al. Toll-like receptor-induced granulocyte-macrophage colony-stimulating factor secretion is impaired in Crohn’s disease by nucleotide oligomerization domain 2-dependent and -independent pathways. Clin Exp Immunol. (2009) 155:487–95. doi: 10.1111/j.1365-2249.2008.03850.x
131. Yoneyama H, Narumi S, Zhang Y, Murai M, Baggiolini M, Lanzavecchia A, et al. Pivotal role of dendritic cell-derived CXCL10 in the retention of T helper cell 1 lymphocytes in secondary lymph nodes. J Exp Med. (2002) 195:1257–66. doi: 10.1084/jem.20011983
132. Majumder S, Bhattacharjee S, Paul Chowdhury B, Majumdar S. CXCL10 is critical for the generation of protective CD8 T cell response induced by antigen pulsed CpG-ODN activated dendritic cells. PloS One. (2012) 7:e48727. doi: 10.1371/journal.pone.0048727
133. Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. (2013) 13:46–57. doi: 10.1038/nri3344
134. Lambert JS, Machado ES, Watson DC, Sill AM, Lim JK, Charurat M, et al. Production of the HIV-suppressive chemokines CCL3/MIP-1alpha and CCL22/MDC is associated with more effective antiretroviral therapy in HIV-infected children. Pediatr Infect Dis J. (2007) 26:935–44. doi: 10.1097/INF.0b013e31812714db
Keywords: dendritic cell, HIV, female genital tract, single-cell RNA sequencing, mucosal immunity, antiviral response
Citation: Parthasarathy S, Moreno de Lara L, Carrillo-Salinas FJ, Werner A, Borchers A, Iyer V, Vogell A, Fortier JM, Wira CR and Rodriguez-Garcia M (2024) Human genital dendritic cell heterogeneity confers differential rapid response to HIV-1 exposure. Front. Immunol. 15:1472656. doi: 10.3389/fimmu.2024.1472656
Received: 29 July 2024; Accepted: 03 October 2024;
Published: 25 October 2024.
Edited by:
Annalisa Ciabattini, University of Siena, ItalyReviewed by:
Alessandra Marcia da Fonseca-Martins, Federal University of Rio de Janeiro, BrazilAndrew Nicholas Harman, The University of Sydney, Australia
Copyright © 2024 Parthasarathy, Moreno de Lara, Carrillo-Salinas, Werner, Borchers, Iyer, Vogell, Fortier, Wira and Rodriguez-Garcia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marta Rodriguez-Garcia, bWFydGEucm9kcmlndWV6LWdhcmNpYUB3YXluZS5lZHU=
 Siddharth Parthasarathy
Siddharth Parthasarathy Laura Moreno de Lara
Laura Moreno de Lara Francisco J. Carrillo-Salinas
Francisco J. Carrillo-Salinas Alexandra Werner
Alexandra Werner Anna Borchers1
Anna Borchers1 Charles R. Wira
Charles R. Wira Marta Rodriguez-Garcia
Marta Rodriguez-Garcia