- 1Department of Oncology, Renmin Hospital of Wuhan University, Wuhan, China
- 2Renmin Hospital of Wuhan Economic and Technological Development Zone (Hannan), Wuhan, China
- 3Wuhan University Heavy Ion Medicine Center, Wuhan, China
- 4Reproductive Medicine Center, Renmin Hospital of Wuhan University, Wuhan, China
- 5Department of Radiation and Medical Oncology, Zhongnan Hospital of Wuhan University, Wuhan, China
RNA modifications are epigenetic changes that alter the structure and function of RNA molecules, playing a crucial role in the onset, progression, and treatment of cancer. Immune checkpoint inhibitor (ICI) therapies, particularly PD-1 blockade and anti-CTLA-4 treatments, have changed the treatment landscape of virous cancers, showing great potential in the treatment of different cancer patients, but sensitivity to these therapies is limited to certain individuals. This review offers a comprehensive survey of the functions and therapeutic implications of the four principal RNA modifications, particularly highlighting the significance of m6A in the realms of immune cells in tumor and immunotherapy. This review starts by providing a foundational summary of the roles RNA modifications assume within the immune cell community, focusing on T cells, NK cells, macrophages, and dendritic cells. We then discuss how RNA modifications influence the intricate regulatory mechanisms governing immune checkpoint expression, modulation of ICI efficacy, and prediction of ICI treatment outcomes, and review drug therapies targeting genes regulated by RNA modifications. Finally, we explore the role of RNA modifications in gene editing, cancer vaccines, and adoptive T cell therapies, offering valuable insights into the use of RNA modifications in cancer immunotherapy.
Introduction
RNA modification is an epigenetic change that alters the structure and function of RNA molecules by inserting, deleting, or substituting nucleotides at specific locations, playing a key role in cellular physiology and pathology. Currently, the roster of RNA modifications that have been pinpointed has extended to over 170 distinct subtypes (1), including methylation, acetylation, pseudouridinization, among others.
RNA methylation modifications are regulated by specific proteins, including methyltransferase “writers” that add marks, demethylase “erasers” that remove them, and “readers” that recognize these modifications. Among RNA modifications, N1-methyladenosine (m1A) (2), 5-methylcytosine (m5C) (3), N6-methyladenosine (m6A) (4), and 7-methylguanosine (m7G) have been the focus of comprehensive studies regarding their oncogenic properties and are viewed as key determinants in the occurrence (5), development, and treatment of tumors.
Immune cells within tumors play a key role in clearing the tumor. Activating immune cells, especially the tumor-clearing functions of T cells, can suppress tumor growth. However, immune cells in the tumor microenvironment are typically in a state of suppression, leading to immune evasion. We have found that RNA modifications are involved in the regulation of immune cell suppression in tumors. Immunotherapy, such as anti-PD-1/PD-L1 therapy, has been widely applied in clinical practice, but only a subset of patients is sensitive to immunotherapy. The efficacy of cancer immunotherapy is contingent upon a multitude of factors, including the type of tumor, tumor’s mutational burden, the stability of microsatellites, and the combined use of chemotherapy drugs. Notably, the presence of immune cell infiltration within the tumor microenvironment and the expression of immune checkpoints on the tumor’s surface play pivotal roles in the therapy’s success. We have found that RNA modifications, primarily the m6A modification, have a complex regulatory effect on the tumor infiltration of immune cells and the expression of PD-L1. By targeting the m6A regulators in tumors or immune cells, it is possible to increase immune cell infiltration, regulate the expression of PD-L1, sensitize patients to immunotherapy, inhibit tumor growth, and ultimately improve patient prognosis.
We have compiled a summary of the regulatory roles of four types of RNA methylation modifications, mainly m6A modification, in immune cells, as well as their regulatory effects on the expression of PD-L1 in tumor cells and the research progress on m6A regulators as therapeutic targets. We also reported on the predictive role of methylation modification-related genes in the efficacy of immunotherapy.
Overview of RNA modifications
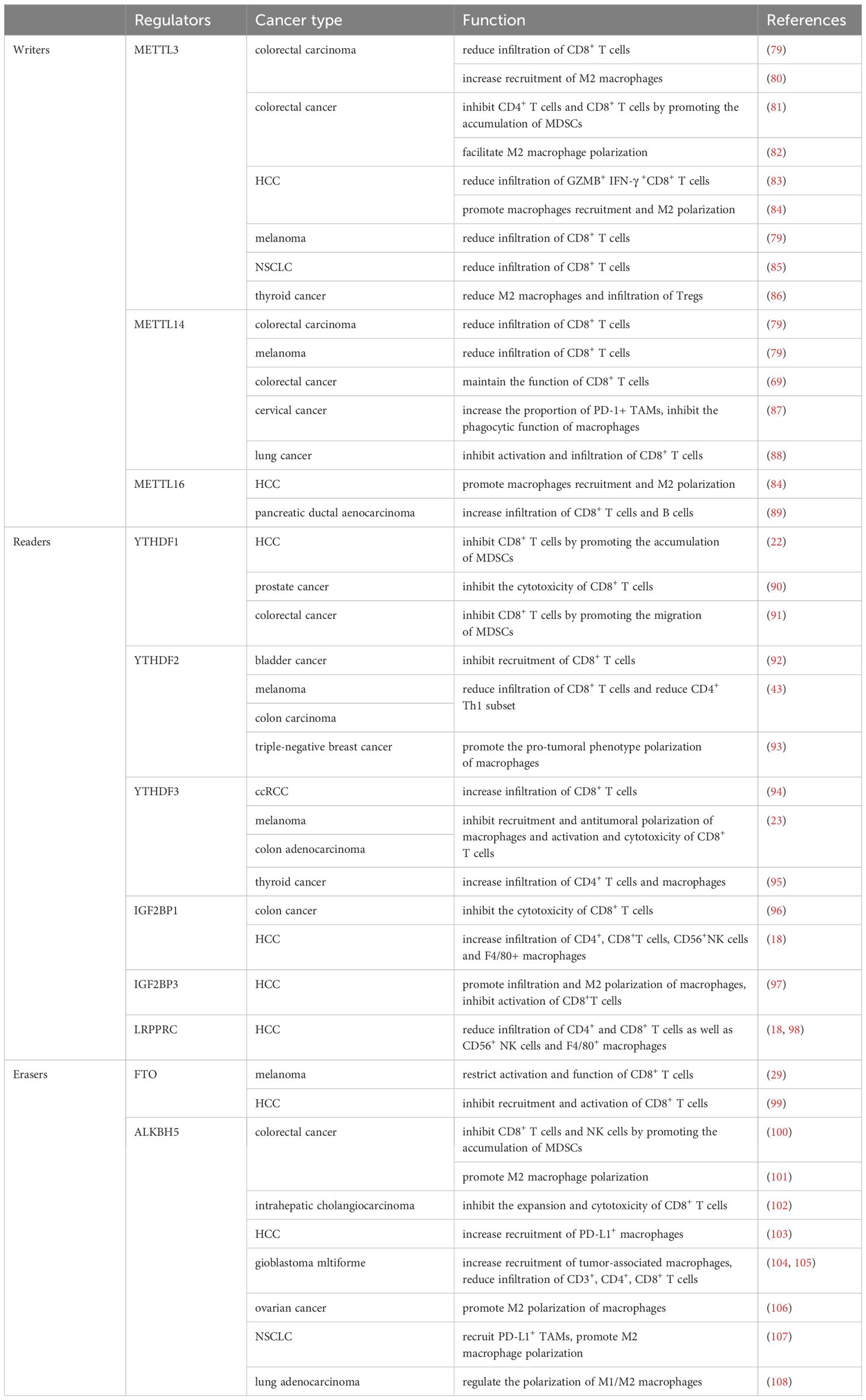
m6A, representing N6-adenosine methylation, is recognized as the most frequent and plentiful type of post-transcriptional RNA modification, predominantly occurring within mRNA in the nucleus (6). This modification contributes to essential life processes such as hematopoiesis, central nervous system development, and germ cell differentiation (4). Common regulatory enzymes include “writers” METTL3, METTL14, METTL16, “readers” YTHDF1, YTHDF2, YTHDF3, LRPPRC, IGF2BP1 and IGF2BP3, among others, and “erasers” FTO, ALKBH5. METTL3 is the most critical ‘writer’ enzyme for m6A modification, serving as the catalytic subunit of the methyltransferase complex that mediates N6-methyladenosine (m6A) methylation on mRNA. It commonly forms a heterodimer with METTL14, facilitating the process (7). Wilms Tumor 1 Associated Protein (Wtap), a regulatory subunit of the complex, enhances the binding affinity of METTL3 for mRNA (8). METTL3 and METTL14 are primarily located in the nucleus, while METTL16 is mainly found in the cytoplasm. Although METTL16 also mediates the m6A modification, its role in promoting tumorigenesis in various cancers is more noteworthy, which is related to the acceleration of mRNA translation by METTL16 binding to eIF3a/b in the cytoplasm (9). YTHDFs recognize mRNAs modified with m6A; YTHDF1 promotes the translation of these mRNAs, while YTHDF2 accelerates their degradation (10). YTHDF3 interacts with YTHDF1 to augment translation of m6A-modified mRNA and influences the RNA binding activity of YTHDF2 (11). And IGF2BPs enhance the stability of mRNA after recognizing m6A modification (12). FTO and ALKBH5 are the only two demethylases identified for m6A demethylation, which remove the m6A modification from RNA (Figure 1). The role of m6A regulatory enzymes in cancer has been widely reported. Several preclinical experiments have demonstrated that the use of drugs targeting m6A regulators can inhibit tumor growth or enhance the therapeutic efficacy of immunotherapy (Table 1).
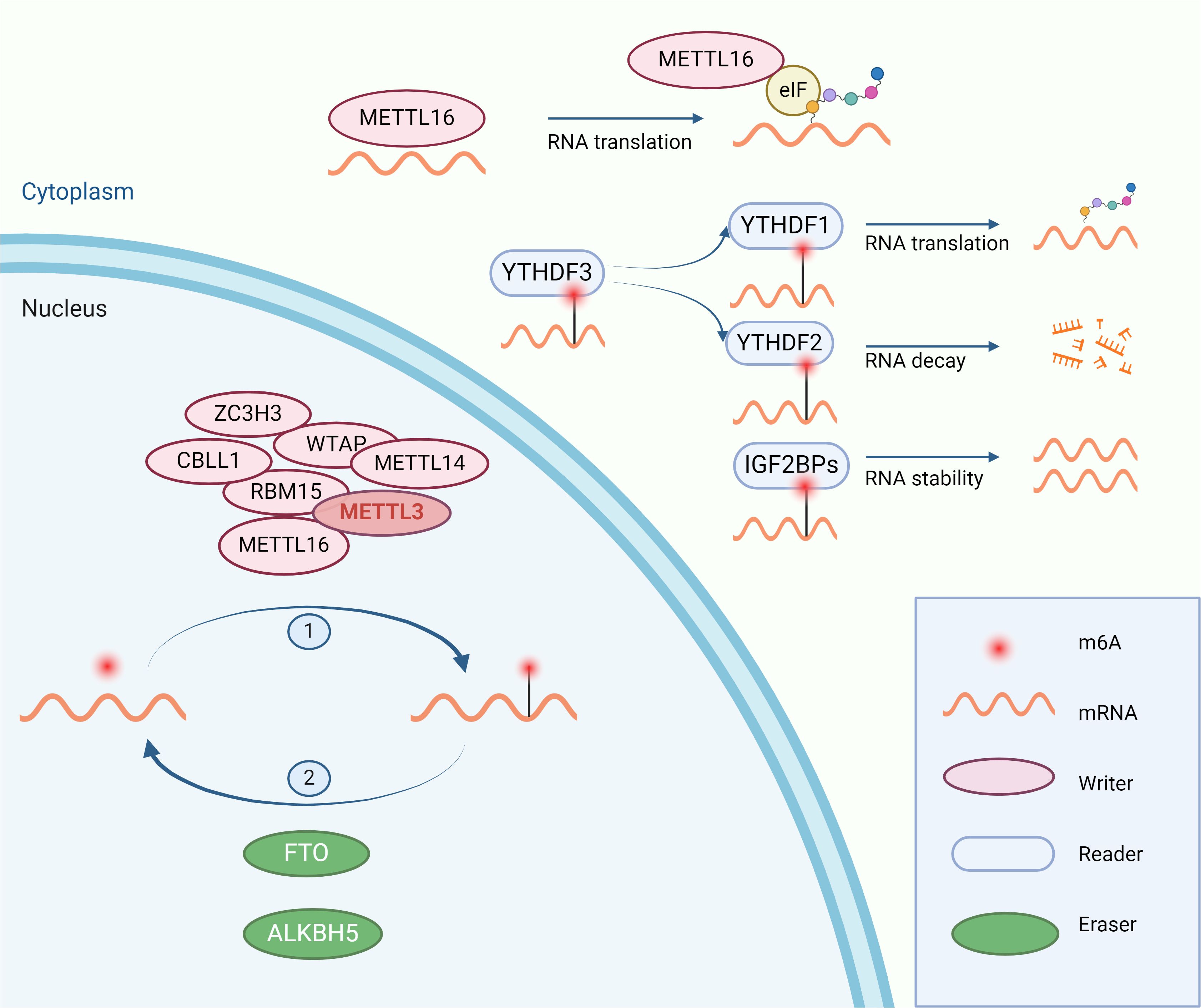
Figure 1. The process of m6A modification: m6A modification is catalyzed by “writers” in the nucleus and regulates the stability, translation, and degradation of RNA after being recognized by “readers” in the cytoplasm. The m6A modification is removed by “erasers”.
m1A and m5C modifications predominantly occur in mRNA, tRNA, and rRNA. The m1A modification is mainly found in tRNA and shares some similarities with m6A modification in mRNA, although it occurs at a much lower frequency. The similarities between m1A and m6A modifications are also reflected in their shared binding proteins YTHDF1, YTHDF2, YTHDF3, and the demethylase FTO (2). m5C plays an important role in maintaining the structure and stability of tRNA and rRNA. m5C modification can occur in mRNA and can regulate the stability of mRNA, but the research on m5C in mRNA is not sufficiently in-depth and comprehensive (3). m7G is widely present in mRNA and is crucial in the translation process and is involved in many important physiological processes (5).
The emergence of new sequencing technologies and bioinformatics tools has facilitated the detection and in-depth understanding of RNA modifications. However, there are still many RNA modifications that have not yet been discovered, and research on many that have been confirmed is far from adequate. We have primarily focused on the function of m6A modification, as it is the most prevalent and well-studied modification in mRNA, but it is important to recognize that other RNA modifications also hold significant potential. The relationship between RNA modifications and the occurrence and development of tumors, as well as their therapy, is currently a hotbed of research. Multiple regulators of RNA modifications are considered oncogenes and have shown potential therapeutic value. The exploration of gene editing technology to enhance RNA modifications that combat tumors and reduce those that promote tumor growth also presents a valuable research avenue.
RNA modifications in immune cells
T cells
T cells have been pivotal in the ongoing oversight and extermination of tumors (33), and their function within tumors is regulated by RNA modifications (Figure 2). The m6A methylation catalyzed by METTL3 is essential for preserving T cell stability and directing their differentiation, with influence over the growth and specialization of naive T cells through the IL-7/STAT5/SOCS axis. Removal of METTL3 from T cells leads to a contraction of the Th17 and Th1 lineages and an escalation in the Th2 cell presence among naive T cells, contrasting with the METTL14 knockout, which impedes T cell maturation beyond the naive stage (34). The m6A/ALKBH5 mechanism is instrumental in controlling the equilibrium of γδ T cell development. ALKBH5 modulates the signaling of Jagged1/Notch2 by removing m6A modifications on them, thereby imposing a restraint on the developmental progression and the lineage specification of γδ T cells (35). Moreover, METTL3 dictates the progression and role of iNKT cells through the METTL3/m6A/Creb1 axis. Deletion of METTL3 impairs the proliferation, differentiation, and cytokine secretion of iNKT cells, leading to a deficiency in tumor resistance (36).
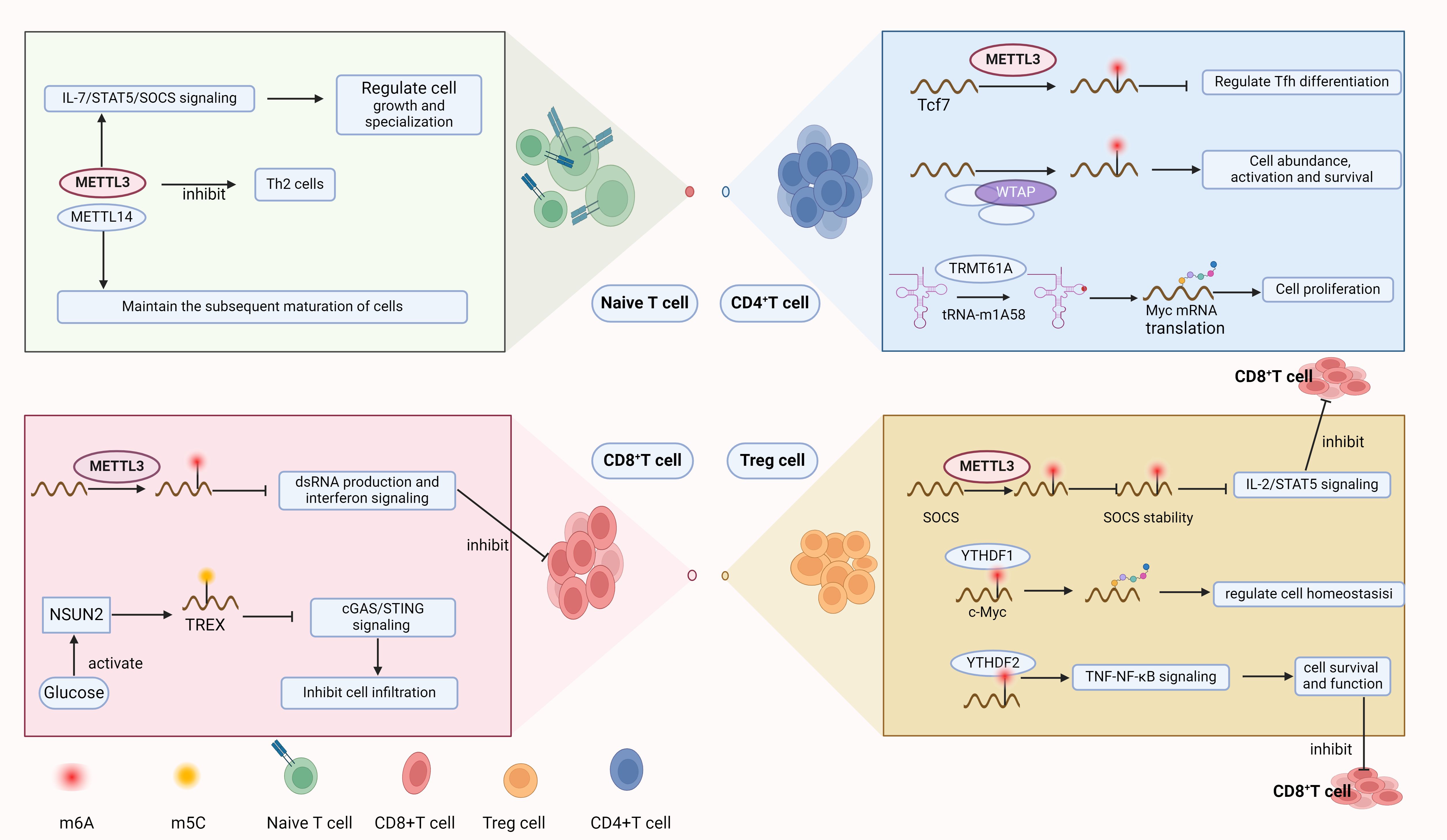
Figure 2. RNA modification regulates the development of Naive T cells and can exert regulatory effects in various T cell subsets.
In CD4+ T cells, the m1A modification (tRNA-m1A58) at the 58th nucleotide position within the tRNA sequence, mediated by the “writer” protein TRMT61A, ensures the efficient translation of Myc mRNA, thereby supporting the propagation and the functional diversification of CD4+ T cells (37). The expression levels of m5C “writer” NSUN3 and NSUN4 are positively correlated with CD8+ T cell infiltration, and NSUN4 expression levels are positively associated with the presence of CD4+ T cells (38). The expression level of NSUN2 is also related to CD4+ T cells and may be involved in the regulation of CD4 memory T cells (39). Regulatory T cells (Treg), a specialized subset of CD4+ T lymphocytes, are pivotal in sustaining immune tolerance and averting autoimmune responses. In tumors, Treg cells can suppress immune responses within the tumor, leading to immune evasion (40). Mettl3 contributes to the upregulation of the IL-2/STAT5 pathway via its capacity to catalyze m6A modifications, regulating members of the SOCS family, thereby maintaining the function and stability of Treg cells and promoting T cell suppression (41). YTHDF1 reads the m6A-modified c-Myc mRNA, thereby regulating the translation and expression of c-Myc in Treg cells, and thus coordinating Treg homeostasis (42). YTHDF2 is involved in the regulation of Treg cells in the tumor microenvironment (TME), maintaining the survival and function of Treg cells by controlling the TNF-NF-κB signaling pathway within Treg cells. The absence of YTHDF2 in Treg cells of the tumor microenvironment leads to an increase in CD8+ T cell infiltration and the expansion of the antitumor CD4+ TH1 subset (43). In squamous cell carcinoma of the head and neck, METTL1 deletion leads to a substantial reduction in Treg cells and the amelioration of CD4+ T cell exhaustion. This indicates that METTL1-mediated m7G modulates immune infiltration by regulating the levels of Treg cells (44, 45). Follicular helper T (Tfh) cells, a specialized subset of CD4+ T cells, are instrumental in the orchestration of humoral immune responses. METTL3 regulates the Tcf7 transcript through m6A modification, ensuring proper differentiation and development of TFH (46). As an indispensable element of the heterodimeric methyltransferase complex, Wtap also participats in the regulation of T cells. The knockout of WTAP leads to a reduction in the abundance and spontaneous activation of peripheral CD4+ and CD8+ T cells and eliminates the expansion effect induced by the T cell receptor (TCR). Additionally, Wtap is essential for the regulatory control of apoptosis in CD4+ T cells, impacting their survival upon TCR engagement (47).
The absence of YTHDF2 in tumors impairs tumor glycolytic metabolism, therefore enhancing the mitochondrial respiration of CD8+ T cells to strengthen antitumor capabilities (23). A pan-cancer examination has shown that METTL1, an enzyme responsible for m7G methylation, is positively correlated with Treg cell numbers in diverse cancer subtypes. The m7G methyltransferase WBSCR22 has been shown to regulate the Zac1/p53 pathway to exert a pro-tumorigenic effect and is highly expressed in activated CD8+ T cells, indicating that WBSCR22 may participate in the regulation of CD8+ T cells (48, 49). The m5C methylation reader YBX1 is positively correlated with CD4+ memory T cells, CD8+ T cells, type 1 and type 2 T helper cells in pancreatic ductal adenocarcinoma (50). The m5C methyltransferase NSUN2 mediates m5C modification of TREX2 transcripts after being activated by glucose, inhibiting the activation of cGAS/STING, thus inhibiting the infiltration of CD8+ T cells (51). Inhibition of METTL3 leads to an increase in dsRNA formation, which in turn enhances interferon signaling and augments the capacity of T cells to eliminate cancer cells (52). Similarly, the knockout of Mettl14 leads to a significant reduction in iNKT cells mediated by the p53 apoptosis pathway (53). Additionally, m6A modification mediated by Mettl3 in T cells regulates the migration of T cells in an acidic tumor microenvironment. By inducing Mettl3-mediated m6A modification in T cells, the expression of integrin α subunit ITGB1 can be upregulated, thereby enhancing T cell tumor infiltration and antitumor activity, and relieving the suppression of T cells by the acidic microenvironment (54).
Natural killer cells
As part of the innate immune response, NK cells are essential for monitoring and eliminating cancerous cells, and they represent a key target for cancer immunotherapy (55). The m6A modification plays an indispensable role in maintaining the tumor infiltration and cytotoxicity of NK cells, primarily regulated by METTL3, METTL14, and YTHDF2. METTL3-mediated m6A methylation promotes the maturation, expansion, and functionality of NK cells through the modulation of IL-15 signaling within the AKT-mTOR/MAPK-ERK pathway. Similarly, YTHDF2 in NK cells regulates the proliferation or survival of NK cells after reading the m6A modification of Tardbp, and modulates the expression of cytotoxicity-related molecules through the STAT5-YTHDF2 positive feedback axis, which participates in the survival, proliferation, and terminal maturation of IL-15-mediated NK cells (56). When METTL3 is knocked out in NK cells, the tumor infiltration and the ability to secrete immune factors such as GzmB and INF-γ are significantly decreased, cytotoxicity is markedly reduced, and an increase in expression levels of the inhibitory receptor TIGIT is observed in the TME (57). m6A contributes to mRNA stability and promotes the early activation and effector functions of NK cells by directly modifying important mRNAs such as Prf1 and Gzmb. The mTORC1 supports m6A methylation in NK cells through the c-MYC-MAT2A axis to promote SAM synthesis (58). The expression level of the m5C methyltransferase NSUN2 is associated with the level of resting NK cells and may takes part in the regulation of NK cells (39). The m5C methylation reader YBX1 is found to be positively related to the presence of NK cells in cases of pancreatic ductal adenocarcinoma (50).
Macrophages
Macrophages are innate immune cells that play a crucial and complex role in tumor immunity, with the antitumor M1 polarization and the pro-tumor M2 polarization being two common differentiations of macrophages in tumor immunity. Tumor-infiltrating macrophages (TAMs) typically acquire the immunosuppressive M2 polarization (59, 60). RNA modifications participate in the regulation of macrophages in various aspects. Tumor-associated macrophages constitute a major part of the cellular composition within the TME. METTL3 promotes the degradation of Irakm mRNA by adding m6A modification, enhancing TLR4 signaling, activating macrophages, and inducing M1 polarization in TAMs, thereby increasing their tumor-killing ability (61). The METTL3 in macrophages increases the stability of STAT1 mRNA transcripts by adding m6A modifications, promoting the M1 polarization of macrophages (62). In myeloid cells, METTL3 activates the NF-κB pathway and STAT3 signaling, leading to M1 and M2-like polarization of macrophages, and fosters the proliferation and spread of cancer cells, contingent upon the infiltration of M1 and M2 phenotype-like TAMs (63), and maintains YTHDF1-mediated SPRED2. Additionally, the FTO demethylase influences the NF-κB pathway, stabilizing STAT1 and PPAR-γ mRNAs, crucial for activating M1 and M2 macrophages. YTHDF2 is involved in this process and antagonizes FTO in regulating the stability of PPAR-γ mRNA (64). IL-4 stimulates an increase in IGF2BP2, which then binds to the m6A-modified TSC1. This binding modulates the signaling through the TSC1/mTORC1 and TSC1/2/MEK/ERK axes, orchestrating the balance between M2 and M1 macrophage states, and driving a transition toward the M2 phenotype within macrophage populations (65). In the tumor lactoacidotic environment, H3K178ac induces the acetylation and upregulation of METTL3 expression. METTL3 mediates m6A modification of the Jack1 mRNA transcript in TIM, which is then read by YTHDF1 to enhance the translation efficiency of JAK1 and the phosphorylation of STAT3, thereby enhancing the immunosuppressive function of TIM (66). The high-risk score of m5C-lncRNA is associated with the high expression of M0 and M2 phenotype macrophages in pancreatic cancer, suggesting that m5C-related lncRNAs may regulate the polarization of macrophages in pancreatic cancer (67). The m7G methyltransferase METTL1 is negatively correlated with M2 and M0 macrophages in tenosynovial giant cell tumors, suggesting that METTL1 may induce M1 polarization of macrophages in tenosynovial giant cell tumors. However, in prostate cancer, METTL1-mediated m7G modification induces M2 polarization of macrophages, indicating the heterogeneity of METTL1 functions in different tumors (44, 68). Additionally, tumor-associated macrophages regulate the expression of CD8+ T cells through m6A-associated mechanisms. C1q+TAMs specifically express METTL14 and YTHDF2, and maintain the level of tumor infiltration and cytotoxicity of CD8+ T cells in a METTL14-dependent manner (69).
Dendritic cells
Dendritic cells, pivotal in the immune response, function as essential antigen-presenting entities that engage in the acquisition, modification, and conveyance of tumor antigens, as well as in the stimulation of T cell responses (70). In tumor immunity, RNA modifications regulate the antigen cross-presentation of dendritic cells and their subsequent function in activating T cells, as well as the migration of dendritic cells. YTHDF1 boosts the synthesis of lysosomal proteases by recognizing the m6A mark on their mRNA, potentially accelerating the breakdown of tumor antigens internalized by dendritic cells. This action may consequently suppress the dendritic cells’ capacity to initiate a cross-priming response in T cells (61, 71). In studies on gastric cancer, YTHDF1 was shown to not only suppress the recruitment of mature DC cells and T cell activation but also inhibit the expression of MHC II and IL-12 (72). METTL3 facilitates the development and maturation of dendritic cells, as well as the subsequent activation of T cells, through its role in m6A methylation. It boosts the translation efficiency of mRNAs encoding CD40, CD80, and the TLR4-associated signaling molecule Tirap. Additionally, METTL3 amplifies the activity of the TLR4/NF-κB signaling cascade and stimulates the synthesis of cytokines that drive an inflammatory response (73). m6A modification is also involved in the migration of dendritic cells; after being read by YTHDF2, it reduces the expression level of lnc-Dpf3, whose expression negatively regulates the induction of CC-chemokine receptor 7 (CCR7) and the migration of dendritic cells to the draining lymph nodes. Moreover, lnc-Dpf3 forms a complex with HIF-1α, impeding the expression of glycolysis-driven genes under HIF-1α’s control, like Ldha. This interaction curtails the glycolytic activity and the movement potential of DCs (74). The m5C methylation reader YBX1 is related to activated dendritic cells in pancreatic ductal adenocarcinoma (50).
Research on the specific regulators of m1A/m5C/m7G and their relationship with immune cells is limited, but growing evidence suggests that these RNA modifications are involved in the regulation of immune cells in various types of cancer. For instance, in clear cell renal cell carcinoma(ccRCC), the score of m7G-related genes is positively correlated with CD4+, CD8+ T cells, and Treg cells, M0 macrophages, and negatively correlated with dendritic cells, M2 macrophages, and other immune cells (75). In diffuse large B-cell lymphoma, m5C-related genes regulate the infiltration of eosinophils, Treg cells, and M2 macrophages, and control the activation of T cells by modulating immune checkpoints such as PD-L1 and CTLA-4 (76). In prostate cancer, there are significant expression differences in CD8+ T cells, M1 macrophages, and M2 macrophages among the two m5C immune subtypes. In colon cancer, a low m1A score is related to the proliferation of CD8+ T effector cells (77). In lung adenocarcinoma, the m1A score is related to all immune cells (78). We have summarized the effects of m6A modification on immune cells that have not been previously mentioned (Table 2).
We have reviewed the important role of RNA modifications, especially m6A modification, in regulating immune cells. m6A modification has a profound impact on tumor immune surveillance and the response to immunotherapy by affecting the maturation, differentiation, and function of T cells, NK cells, macrophages, and dendritic cells. These findings not only reveal new regulatory mechanisms of immune cells in the tumor microenvironment but also provide potential targets for the development of new cancer treatment strategies.
Despite significant progress in existing research, there are still some limitations. For example, most studies focus on specific types of cancer, and the universality of RNA modifications across different cancer types and their roles at various stages of tumor development are not yet fully understood. Future research needs to more comprehensively and deeply explore the regulatory effects of RNA modifications on immune cells in different cancer types and assess their potential as biomarkers and therapeutic targets.
RNA modifications and immune checkpoint
RNA modifications have been shown to be associated with the expression of various immune checkpoints, such as the upregulation of PDCD1 and KIR3DL1 and the downregulation of TIGIT, IDO1, and BTLA in the high-risk group established based on m6A scoring in diffuse large B-cell lymphoma (109). In breast cancer, the groups differentiated by m1A scoring exhibit differential expression of immune checkpoints, for example, TIGIT, IDO1, LAG3, and ICOS (110). We mainly summarize and introduce the research findings related to RNA modifications and PD-1/PD-L1 and CTLA-4.
PD-L1
PD-L1 is mainly expressed on the surface of tumor cells and suppresses the functions of cytotoxic T cells by binding to PD-1 on the surface of T cells. PD-1/PD-L1 blockade therapy has improved the prognosis of many cancers, yet only a subset of patients are sensitive to anti-PD-1 or PD-L1 treatment, and resistance may occur (111). Across a range of malignancies, the metrics derived from the analysis of m6A, m1A, m5C, and m7G regulatory factors exhibit a substantial association with the levels of PD-L1 protein expression (50, 75, 77, 78, 112–121). Firstly, regulators of m6A modification can directly or indirectly regulate the stability and activation of PD-L1 mRNA. In Non-Small Cell Lung Carcinoma(NSCLC), METTL3-mediated m6A modification destabilizes PD-L1 mRNA, resulting in a reduction of PD-L1 expression (122). Furthermore, METTL3 can also regulate the ubiquitination of PD-L1 by controlling LINC02418 in NSCLC, thereby downregulating the expression of PD-L1 (85). Additionally, METTL3 mediates m6A modification of circIGF2BP3, upregulating the expression of PKP3, which enhances the stability of OTUB1 mRNA and increases the abundance of PD-L1 by reducing the ubiquitination of PD-L1 (123). Conversely, within the context of breast cancer, the m6A modification catalyzed by METTL3 not only bolsters the longevity of PD-L1 mRNA transcripts but also facilitates their transcriptional activation, a process that hinges on the recognition of the m6A mark by the IGF2BP3 protein (124). YTHDF3 reads and destroys m6A-modified CBX1 mRNA in nasopharyngeal carcinoma, inhibiting the upregulation of PD-L1 mediated by the IFN-γ/STAT1 signal (125). In HNSCC, TRMT61A-mediated tRNA-m1A modification upregulates the expression of PD-L1, which may be accomplished by regulating INFγ (126). m5C methylation reader YBX1 is related to PD-L1 expression levels in pancreatic ductal adenocarcinoma (50). In colorectal cancer, m6A-modified IFIT1 upregulates the expression of PD-L1 by reducing the ubiquitination and degradation of PD-L1 (127). Similarly, in cholangiocarcinoma, METTL14 mediates m6A modification of Siah2 and promotes the degradation of Siah2 upon reading by YTHDF2, inhibiting the ubiquitination of PD-L1 mediated by Siah2, ultimately leading to an increase in PD-L1 expression levels (128). It is worth mentioning that in colorectal cancer, the metabolite S-adenosylmethionine of methionine promotes the occurrence of m6A modification in cancer cells and enhances the translation of PD-L1 (129). m6A-regulated lncRNA has been proven to be associated with PD-L1 expression in various tumors (130–133). In pancreatic cancer, METTL3 increases the expression of PD-L1 by upregulating the expression of lncRNA MALAT1 in cancer cells (134). In hepatocellular carcinoma(HCC), intestinal bacterial lipopolysaccharide regulates the expression of PD-L1 on the surface of cancer cells by upregulating METTL14 and METTL14-mediated m6A modification of MIR155HG, modulating the miR-223/STAT1 axis (135). We present a summary of the effects of m6A modifications on PD-1/PD-L1 that have not been previously mentioned in this review (Table 3).
CTLA-4
CTLA-4, like PD-1, is a non-redundant checkpoint that inhibits the proliferation and activation of T cells. Therapy targeting the CTLA-4 pathway has been implemented in the field of immunotherapy and may be synergistically combined with PD-1 inhibitory therapy for specific cancer types (111). There are no definitive research results regarding how RNA modifications regulate CTLA-4, but we have found that RNA modifications are associated with the expression levels of CTLA-4 and can predict the outcomes of CTLA-4 blockade immunotherapy. The m6A-modified reader YTHDF2 has been proven to be positively correlated with the expression of CTLA-4 in low-grade glioma (140). In the high-risk group related to m5C-lncRNA, CTLA-4 is highly expressed (143). In tumors such as pancreatic adenocarcinoma, hepatocellular carcinoma, and pancreatic cancer, the scores of m6A-modified regulatory genes are significantly correlated with the expression of CTLA-4 and can be used to predict prognosis and response to immunotherapy (144–146). A study of m5C in diffuse large B-cell lymphoma has identified a correlation between m5C-affected genes and the modulation of immune checkpoint genes, specifically CTLA-4 and PD-L1. In prostate cancer, CTLA-4 is also found to be differentially expressed in two m5C immune subtypes and is related to the degree of immune cell infiltration (76). Similarly, in the immunological model calculated based on the regulatory genes of m7G such as CDK1, ANO1, and PDGFRA, CTLA-4 is highly expressed in the high-risk group (147). Another similar study shows that m7G is not only related to immune checkpoints such as CTLA-4 but also can predict the effects of immunotherapy (148). In addition, a low score based on the methylation enzyme scoring established by m6A/m5C/m1A/m7G regulatory genes is related to the positive expression of CTLA4 (149).
In this section, we have summarized the regulatory effects of RNA modifications on the immune checkpoint PD-L1, as well as the correlation with CTLA-4 expression and its predictive role in therapeutic efficacy. It provides new insights into the understanding of tumor immune evasion mechanisms. The research indicates that RNA modifications influence the expression of PD-L1 and CTLA-4 through various mechanisms, including the regulation of mRNA stability, transcriptional activation, and interactions with specific proteins. These findings offer new perspectives for anti-PD-1/PD-L1 therapy and reveal the research value of RNA modifications in anti-CTLA-4 therapy.
The regulatory role of RNA modification in ICI therapy
RNA modifications exhibit differential impacts on PD-1 blockade therapy across various tumors. Targeting specific RNA modification regulators can enhance the efficacy of PD-1 blockade therapy (Figure 3). In NSCLC, METTL3-driven m6A methylation leads to the destabilization of PD-L1 mRNA, which reduces the therapeutic efficacy of anti-PD-1/PD-L1 interventions; however, the knockout of METTL3 increases immune cell infiltration and enhances the therapeutic efficacy of anti-PD-1/PD-L1 (122). Similarly, the deletion of METTL3 or METTL14 in immune-resistant melanoma tumor cells makes the tumor sensitive to immunotherapy (79). Targeting METTL3 in NAFLD-HCC and NSCLC can improve the effectiveness of PD-1 therapy. In the NAFLD-HCC mouse model, the knockout of METTL3 in conjunction with anti-PD-1 therapy synergistically suppressed tumor growth, resulting in a reduction of over 90% in both tumor volume and weight (83). In NSCLC, the suppression of METTL3 can enhance the sensitivity of tumor-bearing mice to anti-PD-1 treatment, and patients with NSCLC exhibiting low METTL3 expression have a more favorable prognosis with immunotherapy (122). In thyroid cancer, high expression of METTL3 in tumor cells inhibits the demethylation of CD70mRNA, maintains the degradation of transcripts mediated by YTHDF2, thereby releasing T cells from suppression and enhancing the efficacy of PD-1 blockade (86). Similar observations have been made in melanoma and lung cancer, where high expression of METTL3 in macrophages is beneficial for immunotherapy (63). Targeting the YY1-CDK9 transcription elongation complex in glioblastoma results in lowered METTL3 expression, which in turn, enhances the therapeutic impact of PD-1 blockade (150). Abrine treatment inhibits IFN-γ-induced m6A modification, thereby regulating JAK1/STAT1 and suppressing the expression of PD-L1. Combined therapy with PD-1 blockade can inhibit tumor growth (151). Targeting YTHDF1 in colorectal cancer can relieve the inhibition of CD8+ T cells and enhance the efficacy of anti-PD-1. Targeting YTHDF1 significantly reduced the resistance to anti-PD-1 therapy in the MC38 tumor model, leading to better prognosis for tumor-bearing mice. Similar observations were made in mice with the CT26 tumor model, which is insensitive to PD-1 therapy; the deletion of YTHDF1 in CT26 cells followed by anti-PD-1 treatment markedly inhibited tumor growth (91). Knocking out ALKBH5 in glioma or targeting it with IOX1 reduces the expression of the PD-L1 protein, inhibits tumor growth, extends the survival of mice, and enhances these effects of PD-1 blockade therapy (105). Similarly, inhibiting ALKBH5 in melanoma enhances the efficacy of PD-1 blockade, patients with low expression of ALKBH5 are more likely to benefit from PD-1 blockade therapy (31). The use of FTO inhibitors in HCC and melanoma can enhance immune activation and sensitivity to anti-PD-1 treatment (99, 152). Targeting circular RNA circRHBDD1 can block its m6A-dependent mediated rapid translation of PIK3R1 and improve the efficacy of anti-PD-1 therapy (153). NSUN2-mediated m5C methylation modulates TREX2 expression, thereby suppressing the cGAS/STING pathway and contributing to resistance against PD-1 checkpoint blockade (51).
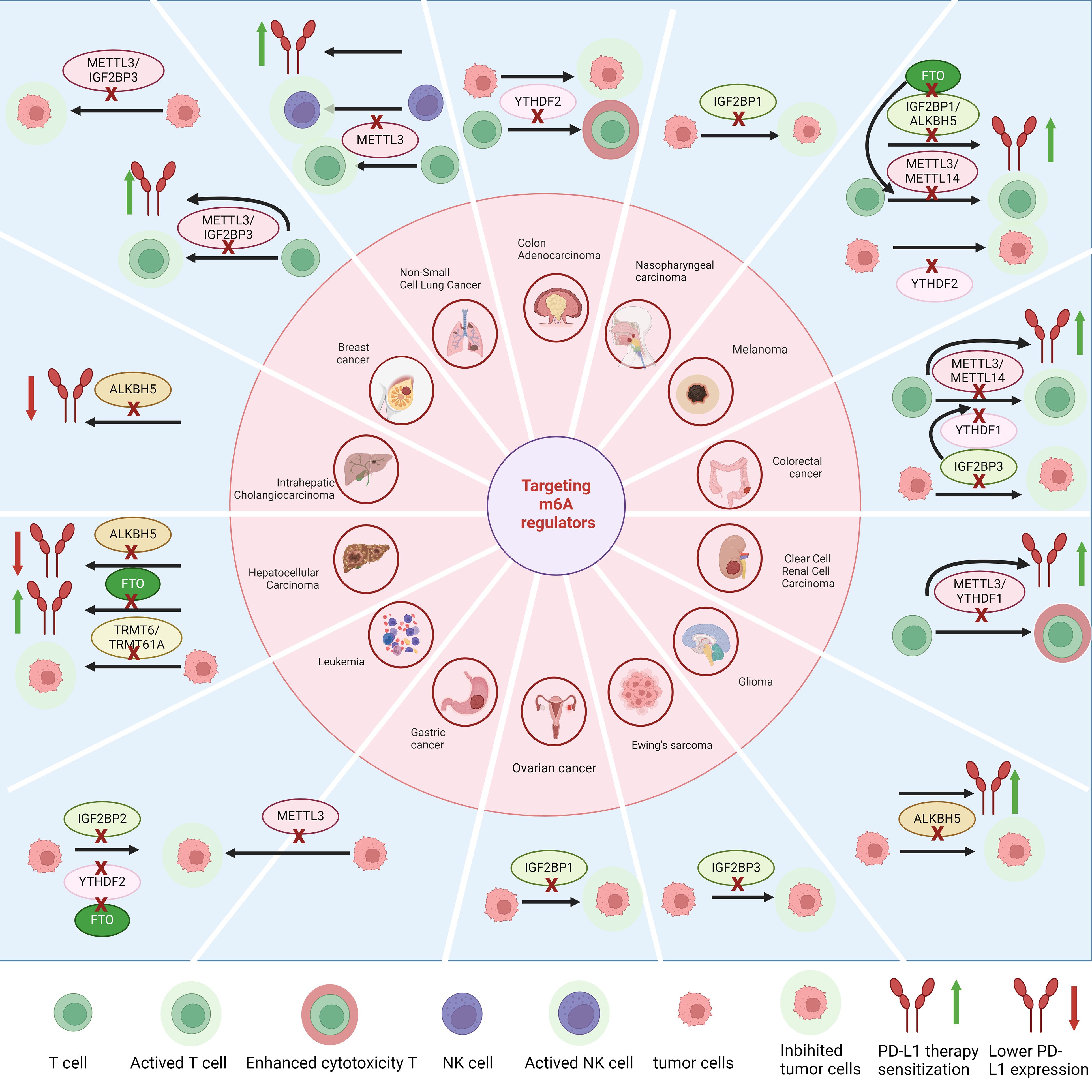
Figure 3. Targeting regulators with m6A modifications can enhance T cell infiltration and tumor-killing effects in various types of cancer. It also regulates the expression of PD-L1 and the therapeutic effects of anti-PD-L1 treatment, while inhibiting tumor growth.
In this part, we have explored the regulatory role of RNA modifications on PD-1 blockade therapy across different types of tumors. By affecting the expression of genes involved in immune activation and suppression, RNA modifications have significantly impacted the therapeutic efficacy of immune checkpoint blockade and have provided ideas for specific treatment plans: by targeting specific RNA modification regulators, we may be able to increase the response rate to immunotherapy and overcome patient resistance to existing treatments.
RNA modification and prediction of immunotherapy efficacy
Genes related to the RNA modifications m6A, m1A, m5C, and m7G can predict tumor sensitivity to immunotherapy, either individually or in combination. By conducting bioinformatics analysis on many samples in databases, key genes are screened, and corresponding scores are established. For instance, in HCC, an m6A score derived from m6A-associated feature genes categorizes patients into distinct risk groups. The high-risk cohort demonstrates increased immune evasion and immune system dysregulation, correlating with heightened responsiveness to anti-CTLA-4 and anti-PD-1 therapies (154). The m6A score has been studied in multiple tumors but varies depending on the analysis method and selected genes. Patients with oral squamous cell carcinoma exhibiting a high m6A score are more likely to experience enhanced efficacy from treatments targeting the PD-1 and CTLA-4 pathways (155), lung squamous cell carcinoma (156), soft tissue sarcoma (157), gastric cancer (102), and hepatocellular carcinoma (158), while a low m6A score suggests greater sensitivity to immunotherapy in thyroid cancer (159), NSCLC (160), follicular lymphoma (161), breast cancer (162), and head and neck squamous cell carcinoma (163). Additionally, the score of m6A methyltransferase regulators can effectively predict the efficacy of immunotherapy in urothelial cancer patients (164).
A prognostic model established using m6A-related lncRNA suggests that esophageal cancer in the low-risk group responds better to immunotherapy (165). Similarly, a model established using m5C-related genes can also evaluate prognosis and immune therapy efficacy, with liver and pancreatic cancers with lower m5C scores being more sensitive to anti-CTLA-4 therapy, and pancreatic cancer also being sensitive to anti-PD-1 therapy (166). A score based on m7G indicates that colorectal cancer and rectal cancer in the low-scoring group are more sensitive to anti-PD-1 therapy (119, 121). Lung adenocarcinoma with a low score has a higher immune prognostic score (120). Interestingly, in low-grade glioma, the high m7G score group is sensitive to anti-PD-1 treatment, while the low m7G score group is more sensitive to anti-PD-L1 (167). In colorectal cancer, a low m1A score suggests a better prognosis with anti-PD-L1 treatment (77). Lung adenocarcinoma with a low m1A score has a lower TIDE score, indicating greater sensitivity to immunotherapy (78).
In addition to establishing models based on the scores of single RNA modifications, analyzing multiple RNA modification genes simultaneously can also establish effective predictive models. In cervical cancer, a prognostic model established using m6A/m5C/m1A indicates that the high-risk group is more sensitive to anti-CTLA-4 treatment (168). In colon cancer, the low-risk group is more sensitive to anti-CTLA-4 and anti-PD-1 treatments (169). In HCC, a methylation score composed of m1A/m5C/m3C/m6A suggests that a low score is sensitive to anti-PD-L1 therapy (170). The Writer-Score established according to m1A and m6A RNA modification enzymes indicates that a low score is associated with better outcomes in immunotherapy (171).
Through bioinformatics analysis of large-sample databases, we have been able to screen for key genes and establish corresponding scoring systems. These scoring systems have demonstrated the potential to predict responses to immunotherapy across various types of tumors, offering new tools for personalized medicine. However, many scoring systems still require further validation and research to explore their applicability in oncology and immunotherapy strategies.
RNA modification regulators as therapeutic targets
Therapeutic interventions targeting RNA modification-related genes or proteins have been extensively studied, and numerous effective drugs have been developed. For instance, metformin can specifically inhibit FTO and block its demethylation effect on m6A modification (172). Targeting specific regulatory genes or proteins of RNA modification can enhance the therapeutic efficacy of immunotherapy in certain types of cancer. For example, targeting ALKBH5 in melanoma can enhance the effectiveness of PD-1 blockade (31). Lower levels of METTL3 in NSCLC are also associated with better outcomes from anti-PD-1 therapy (122). Moreover, targeting specific RNA modification regulators’ genes or proteins can directly exert tumor-suppressive effects. For example, miR-4429, which targets METTL3 in gastric cancer, can inhibit the proliferation of GC cells and induce apoptosis (14). Although the vast majority of drug treatments target m6A regulatory genes or proteins, we found that targeting the m1A methyltransferase complex TRMT6/TRMT61A in HCC with Hiram can effectively inhibit the progression of HCC (32), indicating that targeting other RNA modification regulatory genes or proteins also has therapeutic significance. We have summarized the specific targeted drugs and their mechanisms of action (Table 1).
This section of our study extensively explores therapeutic interventions targeting genes or proteins associated with RNA modifications and outlines the development of a series of effective drugs. Targeting regulators such as METTL3, METTL14, IGF2BP3, or YTHDF1 can alleviate T-cell suppression in melanoma, colorectal cancer, non-small cell lung cancer (NSCLC), and breast cancer. Targeting regulators like METTL3, METTL14, the IGF2BP family, ALKBH5, or FTO can enhance the efficacy of PD-1 blockade therapy in renal clear cell carcinoma, colorectal cancer, melanoma, NSCLC, breast cancer, hepatocellular carcinoma, and intrahepatic cholangiocarcinoma. Inhibition of tumor cell proliferation can be achieved by blocking METTL3, TRMT6/TRMT61A, the IGF2BP family, YTHDF2, or ALKBH5. Targeting “writers” such as METTL3 or METTL14 blocks the formation of m6A and thus can play a role in multiple tumor-suppressing or immune therapy-enhancing effects; however, this may also greatly impact normal physiological functions and lead to severe side effects. RNA modifications have a considerable number of “readers,” so targeting specific “readers” in specific tumors may achieve precise therapeutic effects with lower side effects, but it may need to be specific to certain tumors and may not be universally applicable. Targeting “erasers,” mainly FTO and ALKBH5, seems to primarily enhance PD-1 blockade therapy and inhibit tumor cells, being key to combined immunotherapy. However, targeting FTO and ALKBH5 results in the inability to remove m6A, which may also lead to severe side effects.
Discussion
In this review, we primarily discuss the relationship between m6A, m1A, m5C, and m7G modifications and tumor immunity and immunotherapy, and summarized the regulation of immune cells and immune checkpoints, as well as drug treatment targeting RNA modification regulators and immune prediction based on the four RNA modifications. Research related to RNA modifications has the following limitations: m6A is the most abundant modification in mRNA and has been extensively studied; however, the physiological processes in which m6A is involved are not fully understood, and there may be undiscovered regulatory proteins related to m6A modification. The specific roles of m1A, m5C, and m7G in tumors have been insufficiently studied, possibly due to variations in modification sites and abundance. There are also limitations and challenges in tumor therapy related to RNA modifications: some proteins among RNA modification-related proteins have dual identities, and the same regulatory factor may have multiple roles, possessing both oncogenic and tumor-suppressive effects, requiring specific research to provide solutions. For example, knocking out METTL3 in melanoma can inhibit tumor development and increase the infiltration of CD8+ T cells (79), but another study has shown that the absence of METTL3 expression in macrophages can promote the growth and metastasis of melanoma and weaken the efficacy of PD-1 blockade (63). Additionally, RNA modifications are widely involved in physiological activities, and it is challenging to target RNA modifications in the tumor microenvironment without affecting normal physiological processes. Researchers have obtained drugs with high specificity through complex drug design and optimization, but this issue has not been resolved. Moreover, although many RNA modification regulators have been identified, only a few can be used as therapeutic targets for cancer treatment. Finally, current research is mainly focused on mouse models, and there may be differences in the physiological activities and tumor therapy based on mRNA between mice and humans, requiring more research to clarify the specific situation.
Despite certain achievements of drugs targeting RNA modifications in preclinical models, no related drugs have yet entered the clinical research phase. Future research in the following areas may be helpful: designing drugs to specifically target RNA modification regulators, especially in the tumor microenvironment, to reduce the impact on normal tissues; exploring appropriate drug dosages to balance efficacy and side effects; continuing drug safety research to assess the side effects of different targeted drugs; developing new therapeutic targets or drugs to improve treatment safety; and exploring the combined application of RNA modification-targeting drugs with other drugs in tumor therapy. In addition, tumor vaccines are also related to RNA modifications such as m6A and have therapeutic potential. Introducing encoded mRNA molecules into the bodies of tumor patients, translating them into proteins with anti-tumor effects in the body to trigger anti-tumor immune responses is a new treatment method (173). RNA modifications play a key role in cancer vaccines. Through RNA modifications, the immunogenicity of RNA vaccines can be eliminated, and the rapid translation of anti-tumor proteins may be promoted, improving anti-tumor effects. This direction has considerable potential for development. Furthermore, lifting the immune suppression caused by RNA modifications may help improve the efficacy of adoptive immunotherapy. By lifting the toxicity suppression and infiltration suppression of T and NK cells caused by RNA modifications, the corresponding tumor-killing ability can be restored, which can be used for ex vivo expansion and then re-introduced into the patient’s body to play an anti-tumor role. Lifting the immune suppression of the tumor microenvironment is beneficial for adoptive immune cells to clear the tumor. This could greatly enhance adoptive immunotherapy and holds value for research.
Author contributions
XQ: Writing – original draft. HL: Writing – original draft, Writing – review & editing. QZ: Writing – original draft. YC: Writing – review & editing. TL: Writing – review & editing. FT: Writing – review & editing, Conceptualization. QH: Writing – review & editing, Project administration, Resources.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This article was funded by the Special Project of Science and Technology Cooperation between Hubei Province and Chinese Academy of Sciences (Grant No. 42000021817T300000050) and The National Natural Science Foundation of China (No. 32200706).
Acknowledgments
Figures 1–3 created with biorender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cappannini A, Ray A, Purta E, Mukherjee S, Boccaletto P, Moafinejad SN, et al. MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res. (2024) 52:D239–d244. doi: 10.1093/nar/gkad1083
2. Li J, Zhang H, Wang H. N(1)-methyladenosine modification in cancer biology: Current status and future perspectives. Comput Struct Biotechnol J. (2022) 20:6578–85. doi: 10.1016/j.csbj.2022.11.045
3. Song H, Zhang J, Liu B, Xu J, Cai B, Yang H, et al. Biological roles of RNA m(5)C modification and its implications in Cancer immunotherapy. biomark Res. (2022) 10:15. doi: 10.1186/s40364-022-00362-8
4. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal transduction targeted Ther. (2021) 6:74. doi: 10.1038/s41392-020-00450-x
5. Luo Y, Yao Y, Wu P, Zi X, Sun N, He J. The potential role of N(7)-methylguanosine (m7G) in cancer. J Hematol Oncol. (2022) 15:63. doi: 10.1186/s13045-022-01285-5
6. Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, et al. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. (2017) 31:990–1006. doi: 10.1101/gad.301036.117
7. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. (2014) 10:93–5. doi: 10.1038/nchembio.1432
8. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
9. Su R, Dong L, Li Y, Gao M, He PC, Liu W, et al. METTL16 exerts an m(6)A-independent function to facilitate translation and tumorigenesis. Nat Cell Biol. (2022) 24:205–16. doi: 10.1038/s41556-021-00835-2
10. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
11. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. (2017) 27:315–28. doi: 10.1038/cr.2017.15
12. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
13. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. (2021) 593:597–601. doi: 10.1038/s41586-021-03536-w
14. He H, Wu W, Sun Z, Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A-caused stabilization of SEC62. Biochem Biophys Res Commun. (2019) 517:581–7. doi: 10.1016/j.bbrc.2019.07.058
15. Cheng L, Zhang X, Huang YZ, Zhu YL, Xu LY, Li Z, et al. Metformin exhibits antiproliferation activity in breast cancer via miR-483-3p/METTL3/m(6)A/p21 pathway. Oncogenesis. (2021) 10:7. doi: 10.1038/s41389-020-00290-y
16. Mahapatra L, Andruska N, Mao C, Le J, Shapiro DJ. A novel IMP1 inhibitor, BTYNB, targets c-myc and inhibits melanoma and ovarian cancer cell proliferation. Trans Oncol. (2017) 10:818–27. doi: 10.1016/j.tranon.2017.07.008
17. Wang SS, Lv Y, Xu XC, Zuo Y, Song Y, Wu GP, et al. Triptonide inhibits human nasopharyngeal carcinoma cell growth via disrupting Lnc-RNA THOR-IGF2BP1 signaling. Cancer Lett. (2019) 443:13–24. doi: 10.1016/j.canlet.2018.11.028
18. Liu Y, Guo Q, Yang H, Zhang X-W, Feng N, Wang J-K, et al. Allosteric regulation of IGF2BP1 as a novel strategy for the activation of tumor immune microenvironment. ACS Cent Sci. (2022) 8:1102–15. doi: 10.1021/acscentsci.2c00107
19. Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y, et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. (2022) 40:1566–1582.e10. doi: 10.1016/j.ccell.2022.10.004
20. Mancarella C, Pasello M, Ventura S, Grilli A, Calzolari L, Toracchio L, et al. Insulin-like growth factor 2 mRNA-binding protein 3 is a novel post-transcriptional regulator of ewing sarcoma Malignancy. Clin Cancer research: an Off J Am Assoc Cancer Res. (2018) 24:3704–16. doi: 10.1158/1078-0432.CCR-17-2602
21. Zhang Y, Liu X, Yu M, Xu M, Xiao Y, Ma W, et al. Berberine inhibits proliferation and induces G0/G1 phase arrest in colorectal cancer cells by downregulating IGF2BP3. Life Sci. (2020) 260:118413. doi: 10.1016/j.lfs.2020.118413
22. Wang L, Zhu L, Liang C, Huang X, Liu Z, Huo J, et al. Targeting N6-methyladenosine reader YTHDF1 with siRNA boosts antitumor immunity in NASH-HCC by inhibiting EZH2-IL-6 axis. J Hepatol. (2023) 79:1185–200. doi: 10.1016/j.jhep.2023.06.021
23. Xiao S, Ma S, Sun B, Pu W, Duan S, Han J, et al. The tumor-intrinsic role of the m6A reader YTHDF2 in regulating immune evasion. Sci Immunol. (2024) 9:eadl2171. doi: 10.1126/sciimmunol.adl2171
24. Feng P, Chen D, Wang X, Li Y, Li Z, Li B, et al. Inhibition of the m(6)A reader IGF2BP2 as a strategy against T-cell acute lymphoblastic leukemia. Leukemia. (2022) 36:2180–8. doi: 10.1038/s41375-022-01651-9
25. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. (2020) 38:79–96.e11. doi: 10.1016/j.ccell.2020.04.017
26. Sun K, Du Y, Hou Y, Zhao M, Li J, Du Y, et al. Saikosaponin D exhibits anti-leukemic activity by targeting FTO/m6A signaling. Theranostics. (2021) 11:5831–46. doi: 10.7150/thno.55574
27. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. (2018) 172:90–105.e23. doi: 10.1016/j.cell.2017.11.031
28. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. (2019) 35:677–691.e10. doi: 10.1016/j.ccell.2019.03.006
29. Liu Y, Liang G, Xu H, Dong W, Dong Z, Qiu Z, et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. (2021) 33:1221–1233.e11. doi: 10.1016/j.cmet.2021.04.001
30. Malacrida A, Rivara M, Di Domizio A, Cislaghi G, Miloso M, Zuliani V, et al. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorganic medicinal Chem. (2020) 28:115300. doi: 10.1016/j.bmc.2019.115300
31. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci United States America. (2020) 117:20159–70. doi: 10.1073/pnas.1918986117
32. Wang Y, Wang J, Li X, Xiong X, Wang J, Zhou Z, et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat Commun. (2021) 12:6314. doi: 10.1038/s41467-021-26718-6
33. Zeng Z, Chew HY, Cruz JG, Leggatt GR, Wells JW. Investigating T Cell Immunity in Cancer: Achievements and Prospects, International journal of molecular sciences. Int J Mol Sci (2021) 22(6):2907. doi: 10.3390/ijms22062907
34. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. (2017) 548:338–42. doi: 10.1038/nature23450
35. Ding C, Xu H, Yu Z, Roulis M, Qu R, Zhou J, et al. RNA m(6)A demethylase ALKBH5 regulates the development of γδ T cells. Proc Natl Acad Sci United States America. (2022) 119:e2203318119. doi: 10.1073/pnas.2203318119
36. You M, Liu J, Li J, Ji C, Ni H, Guo W, et al. Mettl3-m6A-Creb1 forms an intrinsic regulatory axis in maintaining iNKT cell pool and functional differentiation. Cell Rep. (2023) 42:112584. doi: 10.1016/j.celrep.2023.112584
37. Liu Y, Zhou J, Li X, Zhang X, Shi J, Wang X, et al. tRNA-m(1)A modification promotes T cell expansion via efficient MYC protein synthesis. Nat Immunol. (2022) 23:1433–44. doi: 10.1038/s41590-022-01301-3
38. Jin S, Li J, Shen Y, Wu Y, Zhang Z, Ma H. RNA 5-Methylcytosine Regulator NSUN3 promotes tumor progression through regulating immune infiltration in head and neck squamous cell carcinoma. Oral Dis. (2024) 30:313–28. doi: 10.1111/odi.14357
39. Sun G, Ma S, Zheng Z, Wang X, Chen S, Chang T, et al. Multi-omics analysis of expression and prognostic value of NSUN members in prostate cancer. Front Oncol. (2022) 12:965571. doi: 10.3389/fonc.2022.965571
40. Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. (2017) 27:109–18. doi: 10.1038/cr.2016.151
41. Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. m6A mRNA methylation sustains Treg suppressive functions. Cell Res. (2018) 28:253–6. doi: 10.1038/cr.2018.7
42. Wang A, Huang H, Shi JH, Yu X, Ding R, Zhang Y, et al. USP47 inhibits m6A-dependent c-Myc translation to maintain regulatory T cell metabolic and functional homeostasis, The Journal of clinical investigation. J Clin Invest (2023) 133(23): e169365. doi: 10.1172/JCI169365
43. Zhang L, Dou X, Zheng Z, Ye C, Lu TX, Liang HL, et al. YTHDF2/m6 A/NF-κB axis controls anti-tumor immunity by regulating intratumoral Tregs. EMBO J. (2023) 42:e113126. doi: 10.15252/embj.2022113126
44. Gao Z, Xu J, Zhang Z, Fan Y, Xue H, Guo X, et al. A comprehensive analysis of METTL1 to immunity and stemness in pan-cancer. Front Immunol. (2022) 13:795240. doi: 10.3389/fimmu.2022.795240
45. Chen J, Li K, Chen J, Wang X, Ling R, Cheng M, et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun (Lond). (2022) 42:223–44. doi: 10.1002/cac2.12273
46. Yao Y, Yang Y, Guo W, Xu L, You M, Zhang YC, et al. METTL3-dependent m(6)A modification programs T follicular helper cell differentiation. Nat Commun. (2021) 12:1333. doi: 10.1038/s41467-021-21594-6
47. Ito-Kureha T, Leoni C, Borland K, Cantini G, Bataclan M, Metzger RN, et al. The function of Wtap in N(6)-adenosine methylation of mRNAs controls T cell receptor signaling and survival of T cells. Nat Immunol. (2022) 23:1208–21. doi: 10.1038/s41590-022-01268-1
48. Jangani M, Poolman TM, Matthews L, Yang N, Farrow SN, Berry A, et al. The methyltransferase WBSCR22/Merm1 enhances glucocorticoid receptor function and is regulated in lung inflammation and cancer. J Biol Chem. (2014) 289:8931–46. doi: 10.1074/jbc.M113.540906
49. Nakazawa Y, Arai H, Fujita N. The novel metastasis promoter Merm1/Wbscr22 enhances tumor cell survival in the vasculature by suppressing Zac1/p53-dependent apoptosis. Cancer Res. (2011) 71:1146–55. doi: 10.1158/0008-5472.CAN-10-2695
50. Yun D, Yang Z, Zhang S, Yang H, Liu D, Grützmann R, et al. An m5C methylation regulator-associated signature predicts prognosis and therapy response in pancreatic cancer. Front Cell Dev Biol. (2022) 10:975684. doi: 10.3389/fcell.2022.975684
51. Chen T, Xu Z-G, Luo J, Manne RK, Wang Z, Hsu C-C, et al. NSUN2 is a glucose sensor suppressing cGAS/STING to maintain tumorigenesis and immunotherapy resistance, Cell Metab. Cell Metabol. (2023) 35(10):1782–98.e8. doi: 10.1016/j.cmet.2023.07.009
52. Guirguis AA, Ofir-Rosenfeld Y, Knezevic K, Blackaby W, Hardick D, Chan Y-C, et al. Inhibition of METTL3 results in a cell-intrinsic interferon response that enhances antitumor immunity. Cancer Discovery. (2023) 13:2228–47. doi: 10.1158/2159-8290.CD-23-0007
53. Cao L, Morgun E, Genardi S, Visvabharathy L, Cui Y, Huang H, et al. METTL14-dependent m(6)A modification controls iNKT cell development and function. Cell Rep. (2022) 40:111156. doi: 10.1016/j.celrep.2022.111156
54. Wang Z, Shang J, Qiu Y, Cheng H, Tao M, Xie E, et al. Suppression of the METTL3-m6A-integrin β1 axis by extracellular acidification impairs T cell infiltration and antitumor activity. Cell Rep. (2024) 43:113796. doi: 10.1016/j.celrep.2024.113796
55. Hodgins JJ, Khan ST, Park MM, Auer RC, Ardolino M. Killers 2.0: NK cell therapies at the forefront of cancer control. J Clin Invest. (2019) 129:3499–510. doi: 10.1172/JCI129338
56. Ma S, Yan J, Barr T, Zhang J, Chen Z, Wang L-S, et al. The RNA m6A reader YTHDF2 controls NK cell antitumor and antiviral immunity, The Journal of experimental medicine. J Experimental Medicine (2021) 218(8):e20210279. doi: 10.1084/jem.20210279
57. Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, et al. METTL3-mediated m6A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun. (2021) 12:5522. doi: 10.1038/s41467-021-25803-0
58. Meng M, Zhong Z, Song L, Zhang Z, Yin X, Xie X, et al. mTOR signaling promotes rapid m6A mRNA methylation to regulate NK-cell activation and effector functions. Cancer Immunol Res (2024). 12(8):1039–57. doi: 10.1158/2326-6066.c.7380107
59. Christofides A, Strauss L, Yeo A, Cao C, Charest A, Boussiotis VA. The complex role of tumor-infiltrating macrophages. Nat Immunol. (2022) 23:1148–56. doi: 10.1038/s41590-022-01267-2
60. Pathria P, Louis TL, Varner JA. Targeting tumor-associated macrophages in cancer. Trends Immunol. (2019) 40:310–27. doi: 10.1016/j.it.2019.02.003
61. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. (2019) 566:270–4. doi: 10.1038/s41586-019-0916-x
62. Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA, American journal of physiology. Cell Physiol. (2019) 317:C762–75. doi: 10.1152/ajpcell.00212.2019
63. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun. (2021) 12:1394. doi: 10.1038/s41467-021-21514-8
64. Gu X, Zhang Y, Li D, Cai H, Cai L, Xu Q. N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell Signal. (2020) 69:109553. doi: 10.1016/j.cellsig.2020.109553
65. Wang X, Ji Y, Feng P, Liu R, Li G, Zheng J, et al. The m6A reader IGF2BP2 regulates macrophage phenotypic activation and inflammatory diseases by stabilizing TSC1 and PPARγ. Adv Sci (Weinh). (2021) 8:2100209. doi: 10.1002/advs.202100209
66. Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. (2022) 82:1660–77. doi: 10.1016/j.molcel.2022.02.033
67. Yuan H, Liu J, Zhao L, Wu P, Chen G, Chen Q, et al. Prognostic risk model and tumor immune environment modulation of m5C-related lncRNAs in pancreatic ductal adenocarcinoma. Front Immunol. (2021) 12:800268. doi: 10.3389/fimmu.2021.800268
68. García-Vílchez R, Añazco-Guenkova AM, Dietmann S, López J, Morón-Calvente V, D'Ambrosi S, et al. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol Cancer. (2023) 22:119. doi: 10.1186/s12943-023-01809-8
69. Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N6-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8+ T cell dysfunction and tumor growth, Cancer cell. Cancer Cell (2021) 39(7):945–57.e10. doi: 10.1016/j.ccell.2021.04.016
70. Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. (2020) 20:7–24. doi: 10.1038/s41577-019-0210-z
71. Accapezzato D, Visco V, Francavilla V, Molette C, Donato T, Paroli M, et al. Chloroquine enhances human CD8+ T cell responses against soluble antigens in vivo. J Exp Med. (2005) 202:817–28. doi: 10.1084/jem.20051106
72. Bai X, Wong CC, Pan Y, Chen H, Liu W, Zhai J, et al. Loss of YTHDF1 in gastric tumors restores sensitivity to antitumor immunity by recruiting mature dendritic cells. J immunotherapy Cancer. (2022) 10(2):e003663. doi: 10.1136/jitc-2021-003663
73. Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun. (2019) 10:1898. doi: 10.1038/s41467-019-09903-6
74. Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, et al. CCR7 chemokine receptor-inducible lnc-dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. (2019) 50:600–615.e15. doi: 10.1016/j.immuni.2019.01.021
75. Xiao Y, Yang J, Yang M, Len J, Yu Y. Comprehensive analysis of 7-methylguanosine and immune microenvironment characteristics in clear cell renal cell carcinomas. Front Genet. (2022) 13:866819. doi: 10.3389/fgene.2022.866819
76. Cui W, Luo C, Zhou L, Yu T, Meng Y, Yu Q, et al. Roles of RNA m5C modification patterns in prognosis and tumor microenvironment infiltration of diffuse large B-cell lymphoma. Am J Cancer Res. (2024) 14:1768–83. doi: 10.62347/NXDR1826
77. Gao Y, Wang H, Li H, Ye X, Xia Y, Yuan S, et al. Integrated analyses of m(1)A regulator-mediated modification patterns in tumor microenvironment-infiltrating immune cells in colon cancer. Oncoimmunology. (2021) 10:1936758. doi: 10.1080/2162402X.2021.1936758
78. Bao G, Li T, Guan X, Yao Y, Liang J, Xiang Y, et al. Comprehensive analysis of the function, immune profiles, and clinical implication of m1A regulators in lung adenocarcinoma. Front Oncol. (2022) 12:882292. doi: 10.3389/fonc.2022.882292
79. Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, et al. m(6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. (2020) 39:e104514. doi: 10.15252/embj.2020104514
80. Ouyang P, Li K, Xu W, Chen C, Shi Y, Tian Y, et al. METTL3 recruiting M2-type immunosuppressed macrophages by targeting m6A-SNAIL-CXCL2 axis to promote colorectal cancer pulmonary metastasis. J Exp Clin Cancer Res. (2024) 43:111. doi: 10.1186/s13046-024-03035-6
81. Chen H, Pan Y, Zhou Q, Liang C, Wong CC, Zhou Y, et al. METTL3 inhibits antitumor immunity by targeting m(6)A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology. (2022) 163:891–907. doi: 10.1053/j.gastro.2022.06.024
82. Chen Z, Zhou J, Wu Y, Chen F, Li J, Tao L, et al. METTL3 promotes cellular senescence of colorectal cancer via modulation of CDKN2B transcription and mRNA stability. Oncogene. (2024) 43:976–91. doi: 10.1038/s41388-024-02956-y
83. Pan Y, Chen H, Zhang X, Liu W, Ding Y, Huang D, et al. METTL3 drives NAFLD-related hepatocellular carcinoma and is a therapeutic target for boosting immunotherapy. Cell Rep Med. (2023) 4:101144. doi: 10.1016/j.xcrm.2023.101144
84. Wei H, Li W, Yang M, Fang Q, Nian J, Huang Y, et al. METTL3/16-mediated m6A modification of ZNNT1 promotes hepatocellular carcinoma progression by activating ZNNT1/osteopontin/S100A9 positive feedback loop-mediated crosstalk between macrophages and tumour cells. Clin Immunol. (2024) 261:109924. doi: 10.1016/j.clim.2024.109924
85. Sun Z, Mai H, Xue C, Fan Z, Li J, Chen H, et al. Hsa-LINC02418/mmu-4930573I07Rik regulated by METTL3 dictates anti-PD-L1 immunotherapeutic efficacy via enhancement of Trim21-mediated PD-L1 ubiquitination. J immunotherapy Cancer. (2023) 11(12):e007415. doi: 10.1136/jitc-2023-007415
86. Ning J, Hou X, Hao J, Zhang W, Shi Y, Huang Y, et al. METTL3 inhibition induced by M2 macrophage-derived extracellular vesicles drives anti-PD-1 therapy resistance via M6A-CD70-mediated immune suppression in thyroid cancer. Cell Death Differ. (2023) 30:2265–79. doi: 10.1038/s41418-023-01217-x
87. Wang B, Mao Z, Ye J, Jiao X, Zhang T, Wang Q, et al. Glycolysis induced by METTL14 is essential for macrophage phagocytosis and phenotype in cervical cancer. J Immunol. (2024) 212:723–36. doi: 10.4049/jimmunol.2300339
88. Sun C, Wang J, Li H, Liu L, Lin Y, Zhang L, et al. METTL14 regulates CD8+T-cell activation and immune responses to anti-PD-1 therapy in lung cancer. World J Surg Oncol. (2024) 22:128. doi: 10.1186/s12957-024-03402-9
89. Lu L, Zheng D, Qu J, Zhuang Y, Peng J, Lan S, et al. METTL16 predicts a favorable outcome and primes antitumor immunity in pancreatic ductal adenocarcinoma. Front Cell Dev Biol. (2022) 10:759020. doi: 10.3389/fcell.2022.759020
90. Wang Y, Jin P, Wang X. N6-methyladenosine regulator YTHDF1 represses the CD8 + T cell-mediated antitumor immunity and ferroptosis in prostate cancer via m6A/PD-L1 manner. Apoptosis. (2024) 29:142–53. doi: 10.1007/s10495-023-01885-7
91. Bao Y, Zhai J, Chen H, Wong CC, Liang C, Ding Y, et al. Targeting m6A reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut. (2023) 72:1497–509. doi: 10.1136/gutjnl-2022-328845
92. Zhang L, Li Y, Zhou L, Zhou H, Ye L, Ou T, et al. The m6A reader YTHDF2 promotes bladder cancer progression by suppressing RIG-I-mediated immune response. Cancer Res. (2023) 83:1834–50. doi: 10.1158/0008-5472.CAN-22-2485
93. Jin H, Chen Y, Zhang D, Lin J, Huang S, Wu X, et al. YTHDF2 favors protumoral macrophage polarization and implies poor survival outcomes in triple negative breast cancer. iScience. (2024) 27:109902. doi: 10.1016/j.isci.2024.109902
94. Dai C, Cao J, Tang Y, Jiang Y, Luo C, Zheng J. YTHDF3 phase separation regulates HSPA13-dependent clear cell renal cell carcinoma development and immune evasion. Cancer Sci. (2024) 115(8):2588–601. doi: 10.1111/cas.16228
95. Zhang Y, Chen Y, Chen R, Zhou H, Lin Y, Li B, et al. YTHDF3as a prognostic predictive biomarker of thyroid cancer and its correlation with immune infiltration. BMC Cancer. (2023) 23(1):882. doi: 10.1186/s12885-023-11361-9
96. Peng Y, Zhang Z, Yang G, Dai Z, Cai X, Liu Z, et al. N6-methyladenosine reader protein IGF2BP1 suppresses CD8 + T cells-mediated tumor cytotoxicity and apoptosis in colon cancer. Apoptosis. (2024) 29:331–43. doi: 10.1007/s10495-023-01893-7
97. Ma L, Jiang J, Si Q, Chen C, Duan Z. IGF2BP3 enhances the growth of hepatocellular carcinoma tumors by regulating the properties of macrophages and CD8(+) T cells in the tumor microenvironment. J Clin Trans Hepatol. (2023) 11:1308–20. doi: 10.14218/JCTH.2023.00184
98. Wang H, Tang A, Cui Y, Gong H, Li H. LRPPRC facilitates tumor progression and immune evasion through upregulation of m6A modification of PD-L1 mRNA in hepatocellular carcinoma. Front Immunol. (2023) 14:1144774. doi: 10.3389/fimmu.2023.1144774
99. Chen A, Zhang VX, Zhang Q, Sze KM-F, Tian L, Huang H, et al. Targeting the oncogenic m6A demethylase FTO suppresses tumourigenesis and potentiates immune response in hepatocellular carcinoma. Gut. (2024). doi: 10.1136/gutjnl-2024-331903
100. Zhai J, Chen H, Wong CC, Peng Y, Gou H, Zhang J, et al. ALKBH5 drives immune suppression via targeting AXIN2 to promote colorectal cancer and is a target for boosting immunotherapy. Gastroenterology. (2023) 165:445–62. doi: 10.1053/j.gastro.2023.04.032
101. Sun M, Yue Y, Wang X, Feng H, Qin Y, Chen M, et al. ALKBH5-mediated upregulation of CPT1A promotes macrophage fatty acid metabolism and M2 macrophage polarization, facilitating Malignant progression of colorectal cancer. Exp Cell Res. (2024) 437:113994. doi: 10.1016/j.yexcr.2024.113994
102. Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. (2020) 19:53. doi: 10.1186/s12943-020-01170-0
103. You Y, Wen D, Zeng L, Lu J, Xiao X, Chen Y, et al. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int J Biol Sci. (2022) 18:5001–18. doi: 10.7150/ijbs.70149
104. Dong F, Qin X, Wang B, Li Q, Hu J, Cheng X, et al. ALKBH5 facilitates hypoxia-induced paraspeckle assembly and IL8 secretion to generate an immunosuppressive tumor microenvironment. Cancer Res. (2021) 81:5876–88. doi: 10.1158/0008-5472.CAN-21-1456
105. Tang W, Xu N, Zhou J, He Z, Lenahan C, Wang C, et al. ALKBH5 promotes PD-L1-mediated immune escape through m6A modification of ZDHHC3 in glioma. Cell Death Discovery. (2022) 8:497. doi: 10.1038/s41420-022-01286-w
106. An Y, Duan H. ALKBH5 modulates macrophages polarization in tumor microenvironment of ovarian cancer. J Ovarian Res. (2024) 17:84. doi: 10.1186/s13048-024-01394-4
107. Hua X, Xu Q, Wu R, Sun W, Gu Y, Zhu S, et al. ALKBH5 promotes non-small cell lung cancer progression and susceptibility to anti-PD-L1 therapy by modulating interactions between tumor and macrophages. J Exp Clin Cancer Res. (2024) 43:164. doi: 10.1186/s13046-024-03073-0
108. Tan J, Chen F, Wang J, Li J, Ouyang B, Li X, et al. ALKBH5 promotes the development of lung adenocarcinoma by regulating the polarization of M2 macrophages through CDCA4. Gene. (2024) 895:147975. doi: 10.1016/j.gene.2023.147975
109. Xie Z, Li M, Hong H, Xu Q, He Z, Peng Z. Expression of N(6)-methyladenosine (m(6)A) regulators correlates with immune microenvironment characteristics and predicts prognosis in diffuse large cell lymphoma (DLBCL). Bioengineered. (2021) 12:6115–33. doi: 10.1080/21655979.2021.1972644
110. Li L, Yang W, Jia D, Zheng S, Gao Y, Wang G. Establishment of a N1-methyladenosine-related risk signature for breast carcinoma by bioinformatics analysis and experimental validation. Breast Cancer (Tokyo Japan). (2023) 30:666–84. doi: 10.1007/s12282-023-01458-1
111. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Sci (New York N.Y.). (2018) 359:1350–5. doi: 10.1126/science.aar4060
112. Yi L, Wu G, Guo L, Zou X, Huang P. Comprehensive analysis of the PD-L1 and immune infiltrates of m6A RNA methylation regulators in head and neck squamous cell carcinoma. Mol Ther Nucleic Acids. (2020) 21:299–314. doi: 10.1016/j.omtn.2020.06.001
113. Ji H, Zhang J-A, Liu H, Li K, Wang Z-W, Zhu X. Comprehensive characterization of tumor microenvironment and m6A RNA methylation regulators and its effects on PD-L1 and immune infiltrates in cervical cancer. Front Immunol. (2022) 13:976107. doi: 10.3389/fimmu.2022.976107
114. Xu Y, He X, Deng J, Xiong L, Li Y, Zhang X, et al. Comprehensive analysis of the immune infiltrates and PD-L1 of m6A RNA methylation regulators in hepatocellular carcinoma. Front Cell Dev Biol. (2021) 9:681745. doi: 10.3389/fcell.2021.681745
115. Xu Z, Chen Q, Shu L, Zhang C, Liu W, Wang P. Expression profiles of m6A RNA methylation regulators, PD-L1 and immune infiltrates in gastric cancer. Front Oncol. (2022) 12:970367. doi: 10.3389/fonc.2022.970367
116. Wu X, Sheng H, Wang L, Xia P, Wang Y, Yu L, et al. A five-m6A regulatory gene signature is a prognostic biomarker in lung adenocarcinoma patients. Aging (Albany NY). (2021) 13:10034–57. doi: 10.18632/aging.v13i7
117. Chen Z, Li Q, Lin Y, Lin S, Gao J, Chen S. m5C regulator-mediated methylation modification phenotypes characterized by distinct tumor microenvironment immune heterogenicity in colorectal cancer. Sci Rep. (2023) 13:11950. doi: 10.1038/s41598-023-37300-z
118. Dong Y, Li Y, Yao Y, Song Q. A novel defined m7G regulator signature to investigate the association between molecular characterization and clinical significance in lung adenocarcinoma. Front Oncol. (2022) 12:897323. doi: 10.3389/fonc.2022.897323
119. Chen J, Song Y-W, Liang G-Z, Zhang Z-J, Wen X-F, Li R-B, et al. A novel m7G-related gene signature predicts the prognosis of colon cancer. Cancers. (2022) 14(22):5527. doi: 10.3390/cancers14225527
120. Li Z, Wang W, Wu J, Ye X. Identification of N7-methylguanosine related signature for prognosis and immunotherapy efficacy prediction in lung adenocarcinoma. Front Med (Lausanne). (2022) 9:962972. doi: 10.3389/fmed.2022.962972
121. Li K, Wang W. Establishment of m7G-related gene pair signature to predict overall survival in colorectal cancer. Front Genet. (2022) 13:981392. doi: 10.3389/fgene.2022.981392
122. Yu H, Liu J, Bu X, Ma Z, Yao Y, Li J, et al. Targeting METTL3 reprograms the tumor microenvironment to improve cancer immunotherapy. Cell Chem Biol. (2024) 31(4):776–791.e7. doi: 10.1016/j.chembiol.2023.09.001
123. Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N6-methyladenosine-modified circIGF2BP3 inhibits CD8+ T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. (2021) 20:105. doi: 10.1186/s12943-021-01398-4
124. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N6-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. (2022) 21:60. doi: 10.1186/s12943-021-01447-y
125. Zhao Y, Huang S, Tan X, Long L, He Q, Liang X, et al. N6 -methyladenosine-modified CBX1 regulates nasopharyngeal carcinoma progression through heterochromatin formation and STAT1 activation. Adv Sci (Weinh). (2022) 9:e2205091. doi: 10.1002/advs.202205091
126. Li S, Feng T, Liu Y, Yang Q, Song A, Wang S, et al. m1A inhibition fuels oncolytic virus-elicited antitumor immunity via downregulating MYC/PD-L1 signaling. Int J Oral Sci. (2024) 16:36. doi: 10.1038/s41368-024-00304-0
127. Gao Y, Zou T, Xu P, Wang Y, Jiang Y, Chen Y-X, et al. Fusobacterium nucleatum stimulates cell proliferation and promotes PD-L1 expression via IFIT1-related signal in colorectal cancer. Neoplasia. (2023) 35:100850. doi: 10.1016/j.neo.2022.100850
128. Zheng H, Zheng W-J, Wang Z-G, Tao Y-P, Huang Z-P, Yang L, et al. Decreased expression of programmed death ligand-L1 by seven in absentia homolog 2 in cholangiocarcinoma enhances T-cell-mediated antitumor activity. Front Immunol. (2022) 13:845193. doi: 10.3389/fimmu.2022.845193
129. Li T, Tan Y-T, Chen Y-X, Zheng X-J, Wang W, Liao K, et al. Methionine deficiency facilitates antitumour immunity by altering m6A methylation of immune checkpoint transcripts. Gut. (2023) 72:501–11. doi: 10.1136/gutjnl-2022-326928
130. Lei L, Li N, Yuan P, Liu D. A new risk model based on a 11-m6A-related lncRNA signature for predicting prognosis and monitoring immunotherapy for gastric cancer. BMC Cancer. (2022) 22:365. doi: 10.1186/s12885-021-09062-2
131. Chen K, Zhu S, Yu W, Xia Y, Xing J, Geng J, et al. Comprehensive analysis of N6-methylandenosine-related long non-coding RNAs signature in prognosis and tumor microenvironment of bladder cancer. Front Oncol. (2022) 12:774307. doi: 10.3389/fonc.2022.774307
132. Huang Y, Li X, Chen W, He Y, Wu S, Li X, et al. Analysis of the prognostic significance and potential mechanisms of lncRNAs associated with m6A methylation in papillary thyroid carcinoma. Int Immunopharmacol. (2021) 101:108286. doi: 10.1016/j.intimp.2021.108286
133. Song W, Ren J, Yuan W, Xiang R, Ge Y, Fu T. N6-methyladenosine-related lncRNA signature predicts the overall survival of colorectal cancer patients. Genes (Basel). (2021) 12(9):1375. doi: 10.3390/genes12091375
134. Song Z, Wang X, Chen F, Chen Q, Liu W, Yang X, et al. LncRNA MALAT1 regulates METTL3-mediated PD-L1 expression and immune infiltrates in pancreatic cancer. Front Oncol. (2022) 12:1004212. doi: 10.3389/fonc.2022.1004212
135. Peng L, Pan B, Zhang X, Wang Z, Qiu J, Wang X, et al. Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of lncRNA MIR155HG to upregulate PD-L1 expression. Cell Biol Toxicol. (2022) 38:1159–73. doi: 10.1007/s10565-022-09718-0
136. Wang A, Sun Y, Wang X, Yan Z, Wang D, Zeng L, et al. m6A methyltransferase METTL16 mediates immune evasion of colorectal cancer cells via epigenetically regulating PD-L1 expression. Aging (Albany NY). (2023) 15:8444–57. doi: 10.18632/aging.v15i16
137. Chen Z, Ruan W, Guo C, Chen K, Li L, Tian J, et al. Non-SMC condensin I complex subunit H participates in anti-programmed cell death-1 resistance of clear cell renal cell carcinomas. Cell Prolif. (2023) 56:e13400. doi: 10.1111/cpr.13400
138. Cui J, Zhu Y, Liu X, Wang W, Jiang X, Xia Y, et al. Comprehensive analysis of N6-methyladenosine regulators with the tumor immune landscape and correlation between the insulin-like growth factor 2 mRNA-binding protein 3 and programmed death ligand 1 in bladder cancer. Cancer Cell Int. (2022) 22:72. doi: 10.1186/s12935-022-02456-7
139. Tsuchiya K, Yoshimura K, Inoue Y, Iwashita Y, Yamada H, Kawase A, et al. YTHDF1 and YTHDF2 are associated with better patient survival and an inflamed tumor-immune microenvironment in non-small-cell lung cancer. Oncoimmunology. (2021) 10:1962656. doi: 10.1080/2162402X.2021.1962656
140. Lin X, Wang Z, Yang G, Wen G, Zhang H. YTHDF2 correlates with tumor immune infiltrates in lower-grade glioma. Aging (Albany NY). (2020) 12:18476–500. doi: 10.18632/aging.v12i18
141. Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, et al. M6A demethylase ALKBH5 regulates PD-L1 expression and tumor immunoenvironment in intrahepatic cholangiocarcinoma. Cancer Res. (2021) 81:4778–93. doi: 10.1158/0008-5472.CAN-21-0468
142. Li X, Chen W, Gao Y, Song J, Gu Y, Zhang J, et al. Fat mass and obesity-associated protein regulates arecoline-exposed oral cancer immune response through programmed cell death-ligand 1. Cancer Sci. (2022) 113:2962–73. doi: 10.1111/cas.15332
143. Zhou H, Meng M, Wang Z, Zhang H, Yang L, Li C, et al. The Role of m5C-Related lncRNAs in Predicting Overall Prognosis and Regulating the Lower Grade Glioma Microenvironment. Front Oncol. (2022) 12:814742. doi: 10.3389/fonc.2022.814742
144. Du Y, Ma Y, Zhu Q, Liu T, Jiao Y, Yuan P, et al. An m6A-related prognostic biomarker associated with the hepatocellular carcinoma immune microenvironment. Front Pharmacol. (2021) 12:707930. doi: 10.3389/fphar.2021.707930
145. Wang L, Zhang S, Li H, Xu Y, Wu Q, Shen J, et al. Quantification of m6A RNA methylation modulators pattern was a potential biomarker for prognosis and associated with tumor immune microenvironment of pancreatic adenocarcinoma. BMC Cancer. (2021) 21:876. doi: 10.1186/s12885-021-08550-9
146. Zou T, Shi D, Wang W, Chen G, Zhang X, Tian Y, et al. Identification of a new m6A regulator-related methylation signature for predicting the prognosis and immune microenvironment of patients with pancreatic cancer. Mediators Inflammation. (2023) 2023:5565054. doi: 10.1155/2023/5565054
147. Regmi P, He Z-Q, Lia T, Paudyal A, Li F-Y. N7-methylguanosine genes related prognostic biomarker in hepatocellular carcinoma. Front Genet. (2022) 13:918983. doi: 10.3389/fgene.2022.918983
148. Li DX, Feng DC, Wang XM, Wu RC, Zhu WZ, Chen K, et al. M7G-related molecular subtypes can predict the prognosis and correlate with immunotherapy and chemotherapy responses in bladder cancer patients. Eur J Med Res. (2023) 28:55. doi: 10.1186/s40001-023-01012-x
149. Wang W, Zhang D, Chang D, Li Y, Ren L. Identification of methyltransferase modification genes associated with prognosis and immune features of pancreatic adenocarcinoma. Mol Cell Probes. (2023) 67:101897. doi: 10.1016/j.mcp.2023.101897
150. Qiu Z, Zhao L, Shen JZ, Liang Z, Wu Q, Yang K, et al. Transcription elongation machinery is a druggable dependency and potentiates immunotherapy in glioblastoma stem cells. Cancer Discovery. (2022) 12:502–21. doi: 10.1158/2159-8290.CD-20-1848
151. Liang X, Gao H, Xiao J, Han S, He J, Yuan R, et al. Abrine, an IDO1 inhibitor, suppresses the immune escape and enhances the immunotherapy of anti-PD-1 antibody in hepatocellular carcinoma. Front Immunol. (2023) 14:1185985. doi: 10.3389/fimmu.2023.1185985
152. Yang S, Wei J, Cui Y-H, Park G, Shah P, Deng Y, et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. (2019) 10:2782. doi: 10.1038/s41467-019-10669-0
153. Cai J, Chen Z, Zhang Y, Wang J, Zhang Z, Wu J, et al. CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m6A modification in hepatocellular carcinoma. Mol Ther oncolytics. (2022) 24:755–71. doi: 10.1016/j.omto.2022.02.021
154. Chen S, Liu J, Zhang S, Zhao L, Zhang J, Han P, et al. Deciphering m6A signatures in hepatocellular carcinoma: Single-cell insights, immune landscape, and the protective role of IGFBP3. Environ Toxicol. (2024). doi: 10.1002/tox.24177
155. Pan L, She H, Wang K, Xia W, Tang H, Fan Y, et al. Characterization of the m6A regulator-mediated methylation modification patterns in oral squamous cell carcinoma. Sci Rep. (2023) 13:6617. doi: 10.1038/s41598-023-33891-9
156. Li P, Xiong P, Li X, Zhang X, Chen X, Zhang W, et al. Tumor microenvironment characteristics and prognostic role of m6A modification in lung squamous cell carcinoma. Heliyon. (2024) 10:e26851. doi: 10.1016/j.heliyon.2024.e26851
157. Huang ZD, Lin LL, Liu ZZ, Hu C, Gu HY, Wei RX. m6A modification patterns with distinct immunity, metabolism, and stemness characteristics in soft tissue sarcoma. Front Immunol. (2021) 12:765723. doi: 10.3389/fimmu.2021.765723
158. Huang X, Qiu Z, Li L, Chen B, Huang P. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in hepatocellular carcinoma. Aging (Albany NY). (2021) 13:20698–715. doi: 10.18632/aging.v13i16
159. Zhou W, Lin J, Liu J, Zhang R, Fan A, Xie Q, et al. Thyroid cancer risk prediction model using m6A RNA methylation regulators: integrated bioinformatics analysis and histological validation. Aging (Albany NY). (2023) 15:846–65. doi: 10.18632/aging.v15i3
160. Zhang Z, Zhang C, Luo Y, Wu P, Zhang G, Zeng Q, et al. m(6)A regulator expression profile predicts the prognosis, benefit of adjuvant chemotherapy, and response to anti-PD-1 immunotherapy in patients with small-cell lung cancer. BMC Med. (2021) 19:284. doi: 10.1186/s12916-021-02148-5
161. Zhang T, Liu H, Gao F, Gong W, Cui Y, He J, et al. m6A-regulator expression signatures identify a subset of follicular lymphoma harboring an exhausted tumor microenvironment. Front Immunol. (2022) 13:922471. doi: 10.3389/fimmu.2022.922471
162. Dong M, Shen W, Yang G, Yang Z, Li X. Analysis of m6A methylation modification patterns and tumor immune microenvironment in breast cancer. Front Cell Dev Biol. (2022) 10:785058. doi: 10.3389/fcell.2022.785058
163. Yang Z, Ming X, Huang S, Yang M, Zhou X, Fang J. Comprehensive analysis of m(6)A regulators characterized by the immune cell infiltration in head and neck squamous cell carcinoma to aid immunotherapy and chemotherapy. Front Oncol. (2021) 11:764798. doi: 10.3389/fonc.2021.764798
164. Kong J, Lu S, Zhang L, Yao Y, Zhang J, Shen Z, et al. m6A methylation regulators as predictors for treatment of advanced urothelial carcinoma with anti-PDL1 agent. Front Immunol. (2022) 13:1014861. doi: 10.3389/fimmu.2022.1014861
165. Wang W, Dong D, Yu P, Chen T, Gao R, Wei J, et al. Prognostic model based on m6A-associated lncRNAs in esophageal cancer. Front Endocrinol (Lausanne). (2022) 13:947708. doi: 10.3389/fendo.2022.947708
166. Zhan M, Song H, Tian D, Wen Q, Shi X, Wang Y, et al. Molecular features, biological behaviors and clinical implications of m5C RNA methylation modification regulators in gastrointestinal cancers. Cancer Biol Ther. (2023) 24:2223382. doi: 10.1080/15384047.2023.2223382
167. Maimaiti A, Feng Z, Liu Y, Turhon M, Xie Z, Baihetiyaer Y, et al. N7-methylguanosin regulators-mediated methylation modification patterns and characterization of the immune microenvironment in lower-grade glioma. Eur J Med Res. (2023) 28:144. doi: 10.1186/s40001-023-01108-4
168. Wang Y, Mao Y, Wang C, Jiang X, Tang Q, Wang L, et al. RNA methylation-related genes of m6A, m5C, and m1A predict prognosis and immunotherapy response in cervical cancer. Ann Med. (2023) 55:2190618. doi: 10.1080/07853890.2023.2190618
169. Liu J, Dou M, Liu X, Lu Y, Lu W. A novel m6A/m5C/m1A score signature to evaluate prognosis and its immunotherapy value in colon cancer patients. J Cancer Res Clin Oncol. (2023) 149:11995–2012. doi: 10.1007/s00432-023-05033-1
170. Zhang J, Gao J, Hu M, Xu S, Cheng C, Zheng W, et al. Integrated investigation of the clinical implications and targeted landscape for RNA methylation modifications in hepatocellular carcinoma. Eur J Med Res. (2023) 28:46. doi: 10.1186/s40001-023-01016-7
171. Zhou B, Bie F, Zang R, Zhang M, Song P, Liu L, et al. RNA modification writer expression profiles predict clinical outcomes and guide neoadjuvant immunotherapy in non-small cell lung cancer. EBioMedicine. (2022) 84:104268. doi: 10.1016/j.ebiom.2022.104268
172. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. (2015) 43:373–84. doi: 10.1093/nar/gku1276
Keywords: RNA modification, cancer, immune checkpoint, immune cell, immune therapy
Citation: Qin X, Liu H, Zhang Q, Che Y, Lei T, Tang F and Hu Q (2024) RNA modifications in cancer immune therapy: regulators of immune cells and immune checkpoints. Front. Immunol. 15:1463847. doi: 10.3389/fimmu.2024.1463847
Received: 12 July 2024; Accepted: 02 September 2024;
Published: 20 September 2024.
Edited by:
Xin He, City of Hope National Medical Center, United StatesReviewed by:
Shuyue Zhan, Thermo Fisher Scientific, United StatesGuang Han, Hubei Cancer Hospital, China
Copyright © 2024 Qin, Liu, Zhang, Che, Lei, Tang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Tang, dGFuZ2ZhbmdoYXBweUB3aHUuZWR1LmNu; Qinyong Hu, cm0wMDEyMjNAd2h1LmVkdS5jbg==
†These authors share first authorship
 Xiangyu Qin
Xiangyu Qin Huali Liu
Huali Liu Qixuan Zhang
Qixuan Zhang Yuhang Che2,3
Yuhang Che2,3 Tianyu Lei
Tianyu Lei Fang Tang
Fang Tang Qinyong Hu
Qinyong Hu