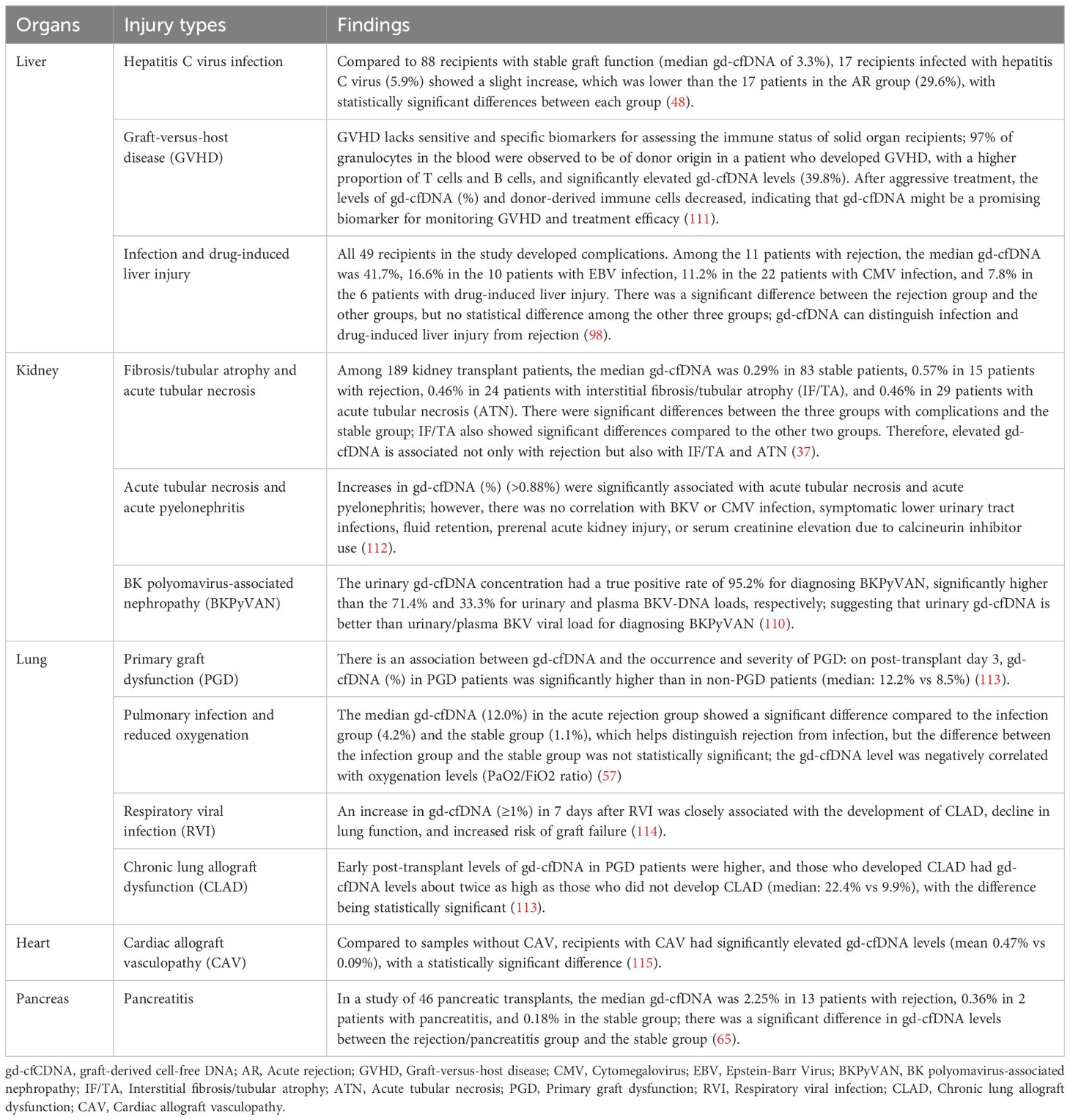
- Department of Urology II, The First Hospital of Jilin University, Changchun, China
Monitoring the status of grafts and the occurrence of postoperative complications, such as rejection, is crucial for ensuring the success and long-term survival of organ transplants. Traditional histopathological examination, though effective, is an invasive procedure and poses risks of complications, making frequent use impractical. In recent years, graft-derived cell-free DNA (gd-cfDNA) has emerged as a promising non-invasive biomarker. It not only provides early warnings of rejection and other types of graft injury but also offers important information about the effectiveness of immunosuppressive therapy and prognosis. gd-cfDNA shows potential in the monitoring of organ transplants. The early, real-time information on graft injury provided by gd-cfDNA facilitates timely individualized treatment and improves patient outcomes. However, the progress of research on gd-cfDNA varies across different organs. Therefore, this article will comprehensively review the application and findings of gd-cfDNA in monitoring various solid organs, discussing the advantages, limitations, and some future research directions to aid in its clinical application.
1 Introduction
Solid organ transplantation is a crucial treatment for end-stage organ failure. However, post-transplantation, there is a risk of occurrence of transplant-related complications (such as rejection) and graft function decline, which may lead to graft loss and reduced patient survival rates (1). Therefore, monitoring the status of transplanted organs is vital for the long-term survival of recipients. Only more comprehensive monitoring of the grafts can reduce the happening of adverse outcomes and improve patient survival rates. The current gold standard for assessing graft health remains histopathological biopsy. However, due to its invasive nature, high cost, and associated risk of complications, its clinical application is limited, making it unsuitable for frequent routine monitoring (2, 3). Other indicators for monitoring organ function, such as serum creatinine for kidneys and transaminases for the liver, have low sensitivity to graft injury and exhibit a lag in response (4). Thus, there is currently a lack of reliable, low-risk, and suitable methods for repeated monitoring. In recent years, graft-derived cell-free DNA (gd-cfDNA), released by damaged graft cells, has emerged as a novel non-invasive biomarker for monitoring post-transplantation rejection and other types of graft injury, providing a new option for continuous, minimally invasive monitoring of organ transplants (5). However, the application of gd-cfDNA is still in the early exploratory stages and has not yet reached a consensus. This article will comprehensively review the current research developments across various types of transplanted organs.
2 Production of gd-cfDNA
In the cell nucleus, DNA wraps around histones to form nucleosomes. During cell damage, apoptosis, or necrosis, chromosomal DNA undergoes degradation and release. Initially, DNA is cleaved into large fragments (50–300 kb), followed by further degradation, resulting in the release of smaller DNA fragments (180–200 bp) and nucleosomes into the blood. These DNA fragments are eventually cleared in the liver and kidneys (6–8). The half-life of these DNA fragments ranges from approximately 15 to 90 minutes (9). These double-stranded DNA fragments circulating in the plasma are called cfDNA, also known as circulating free DNA or extracellular DNA. The discovery of cfDNA dates back to 1948 (10). As a reliable “liquid biopsy” method, cfDNA has been widely used in fields such as prenatal diagnosis and cancer monitoring (11, 12).
gd-cfDNA is a specific subcategory of cfDNA and refers to the cfDNA derived from graft cells circulating in the recipient’s plasma after transplantation. The majority of cfDNA in the recipient’s blood (over 95%) originates from the recipient’s apoptotic hematopoietic cells (13). Compared to hematopoietic-derived DNA (with a peak length of approximately 166 bp), non-hematopoietic-derived DNA is shorter, with gd-cfDNA primarily ranging from 105 to 145 bp (14). The level of graft-derived cell-free DNA (gd-cfDNA) is typically quantified in two ways: relative quantification as gd-cfDNA(%) and absolute quantification as gd-cfDNA(cp/mL), which refers to the percentage of gd-cfDNA relative to total circulating cfDNA and the number of gd-cfDNA copies per milliliter of serum, respectively (15). When the graft is stable, the amount of gd-cfDNA released from apoptotic graft cells constitutes only a tiny fraction of the total cfDNA in the recipient’s blood, and it is shorter in length than recipient-derived cfDNA (13, 14, 16). When the graft experiences ischemia-reperfusion injury, rejection, or infection, resulting in graft cell necrosis, varying amounts of gd-cfDNA are released into the recipient’s plasma. (Figure 1). The extent of gd-cfDNA elevation correlates with the type and severity of graft injury, making Changes in gd-cfDNA levels a dynamic indicator for monitoring graft injury (17). Since gd-cfDNA has an overall negative charge, it cannot be filtered by the glomerulus (18); it is typically measured in blood. But unlike other organ transplants, in kidney transplantation, if there is tubulitis or interstitial inflammation, gd-cfDNA can also be excreted through the kidneys, leading to elevated gd-cfDNA levels in urine (8, 19). Thus, urinary gd-cfDNA levels can also reflect the status of the transplanted kidney. Overall, by detecting gd-cfDNA in blood or urine samples, gd-cfDNA can dynamically and early reflect the graft status, providing clinicians with a basis for decision-making (20).
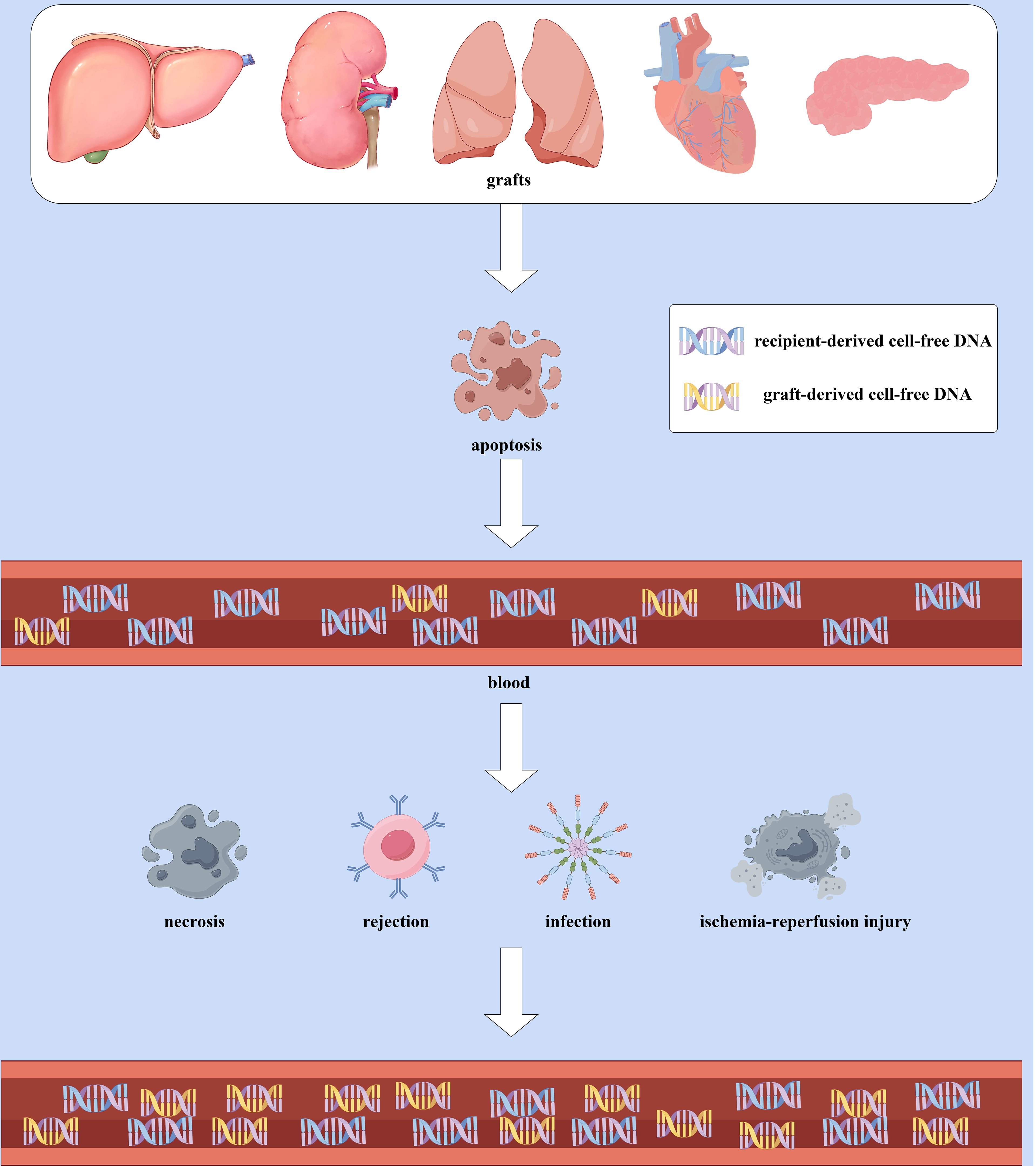
Figure 1. The Production and elevation of gd-cfDNA: When the graft is stable, gd-cfDNA in the plasma primarily originates from the apoptosis of graft cells, constituting only a small portion of the total cfDNA in the recipient’s blood and is shorter in length compared to recipient-derived cfDNA. However, in cases of ischemia-reperfusion injury, rejection, infection, or other causes leading to graft cell necrosis, more gd-cfDNA will released into the recipient’s bloodstream, resulting in an increase in quantification of gd-cfDNA. (By Figdraw).
3 Detection methods of gd-cfDNA
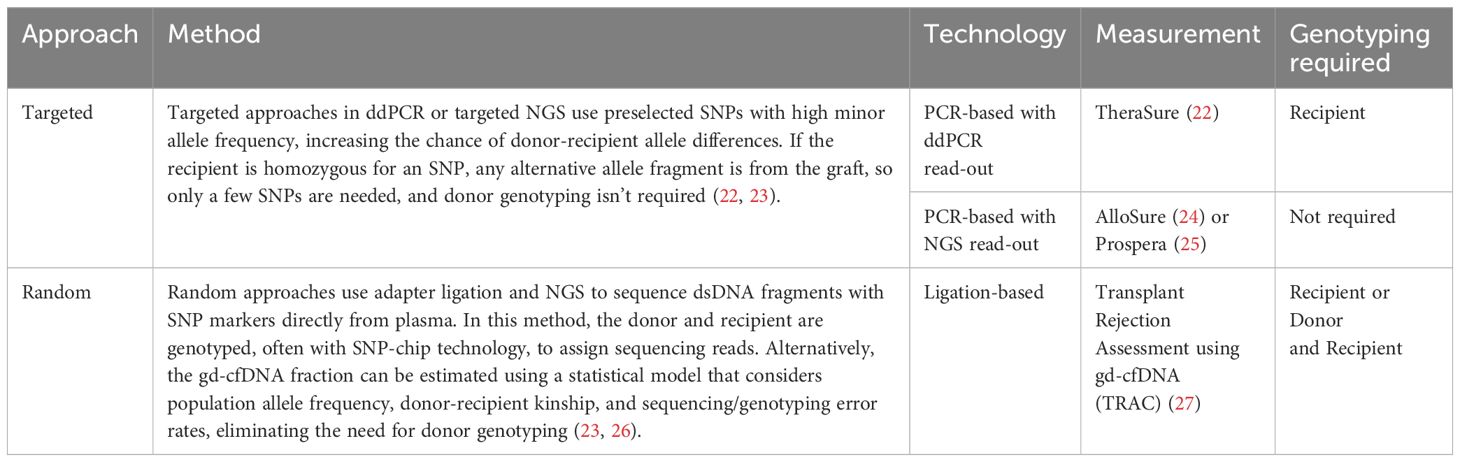
In 1998, Lo et al. (21) identified Y chromosome-specific genes from donors in female recipients using polymerase chain reaction (PCR). However, at that time, the detection method was limited to specific cases where the donor and recipient were of different sexes. Currently, gd-cfDNA detection methods can be categorized into targeted and random approaches (Table 1). The quantitative detection of gd-cfDNA primarily relies on genetic markers, typically single nucleotide polymorphisms (SNPs), to differentiate between donor and recipient alleles (28). The development of quantitative techniques has provided diverse options for clinical practice. Real-time quantitative PCR (qPCR), as an early commonly used technique, is characterized by its simplicity, low cost, and rapid processing time, though it has relatively lower sensitivity and specificity (22, 29). Droplet digital PCR (ddPCR) has high sensitivity and accuracy compared to qPCR, but it may require more expensive reagents and equipment, and data analysis and interpretation may take longer (22, 30). Next-generation sequencing (NGS) technology, with its ability to sequence thousands of targets simultaneously, provides high sensitivity and accuracy for detecting complex genetic variations and unknown sequences (31). This capability makes NGS particularly advantageous in multiple transplants, multi-organ transplants, and monitoring rejection and infection (24, 32, 33). Despite the challenges of cost, technical complexity, and analysis time associated with NGS (24, 34, 35), its capability in gd-cfDNA monitoring cannot be overlooked. The turnaround time for ddPCR is shorter, with results typically available in one working day, whereas NGS requires 2–3 working days (36). These methods can provide relative quantification in relation to a calibrator, but only ddPCR can achieve absolute quantification (37).
4 Comparison of gd-cfDNA with common monitoring indicators
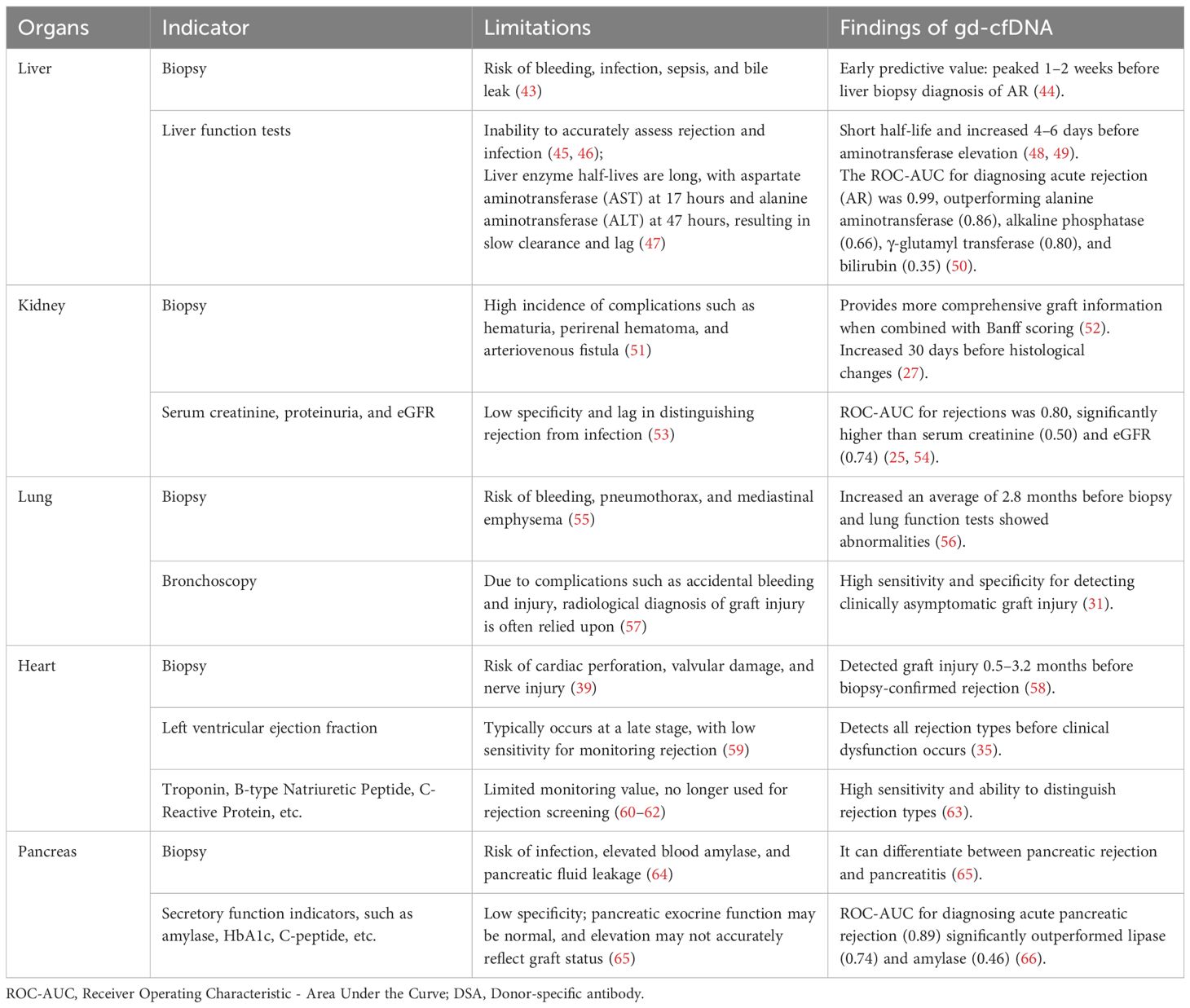
Histopathological biopsy remains the current gold standard for organ transplant monitoring and diagnosis, but it requires operation by specialized medical personnel (38, 39). Moreover, approximately 25% of kidney biopsy samples lack sufficient glomeruli, especially when using finer biopsy needles, which may increase the likelihood of requiring repeated punctures and thus elevate unnecessary complications risk (40). Additionally, biopsy sampling captures only a few glomeruli and a limited number of renal tubules from a localized area, leading to sampling errors and an incomplete understanding of the overall condition of the kidney. Studies have shown that 43% of clinically indicated biopsies and 65% of protocol biopsies are unnecessary (41). Some biomarkers used to monitor graft function, such as plasma creatinine, can measure glomerular function but are not sensitive indicators of graft injury. By the time complications like rejection lead to a significant rise in plasma creatinine, substantial tissue damage may have already occurred in the transplanted kidney (42). Similarly, other commonly used indicators for monitoring organ function also have some limitations. (Table 2).
Donor-specific antibody (DSA) is a crucial biomarker for predicting the development of antibody-mediated rejection (AMR) and assessing graft survival (67). However, limitations in DSA detection—such as variability in detection methods, sensitivity, and clinical significance standards—restrict its role in rejection (68, 69). Although increasing sensitivity of DSA detection assays, a significant proportion of patients with the same histologic picture of ABMR, do not have detectable circulating DSA (70, 71). A recent kidney transplant study indicated that gd-cfDNA might be a better predictor of AMR than DSA. In that study, DSA was frequently negative in both molecular (56%) and histologic (51%) AMR. In AMR, gd-cfDNA(%)≥1.0 was more frequent (75%) than DSA positivity (44%). Moreover, all AMR patients, DSA-positive or DSA-negative, showed elevated gd-cfDNA levels (mean 1.88% vs. 0.32% in the non-rejection group) (72). Some studies have demonstrated a correlation between elevated gd-cfDNA levels and the occurrence of DSA, allowing for the prediction of DSA development. In heart transplantation, gd-cfDNA levels were significantly higher in patients who experienced De novo donor-specific antibodies (dnDSA) compared to those who did not (0.34% vs. 0.06%) and were elevated a median of 20 days before dnDSA was detected (73). In kidney transplant recipients, 40% (17/42) of patients with gd-cfDNA ≥0.5% developed dnDSA, compared to only 2.7% (1/37) of patients with gd-cfDNA <0.5%, showing a significant difference (74). Kidney transplant recipients with gd-cfDNA levels of 0.5% or higher have nearly a threefold increased risk of developing dnDSA, with levels rising a median of 91 days before the identification of DSA (interquartile range, 30–125 days) (54). Similarly, in heart transplantation, among 613 DSA samples paired with gd-cfDNA levels, gd-cfDNA greater than 0.15% was associated with a fourfold increase in the incidence of dnDSA in the first year post-transplantation (75). However, research on the diagnostic performance of gd-cfDNA and DSA in rejection remains limited, and further studies are needed to confirm their roles.
gd-cfDNA is directly derived from the graft and has a short half-life, providing high sensitivity to tissue damage. The minimally invasive nature of blood or urine sampling for gd-cfDNA testing further adds to its advantages (5, 72). From an economic perspective, gd-cfDNA monitoring also offers substantial benefits. The estimated total cost of a kidney biopsy is approximately $3,931 (76), and the average cost of an endomyocardial biopsy (EMB) after heart transplantation is as high as $7,918 (77). If biopsy-related complications occur (such as hospitalization and hematoma evacuation), additional costs can increase by an average of $10,743 (78). In comparison, the cost of ddPCR for the first year is about $4,012 (including testing on days 7 and 14 post-transplant, monthly testing for the first 6 months, and quarterly testing thereafter). In subsequent years, quarterly testing with ddPCR costs around $1,604 (53). The cost difference and risk considerations make gd-cfDNA a valuable non-invasive monitoring method in managing organ transplant recipients.
It is important to note that gd-cfDNA testing cannot yet fully replace pathological biopsy as the gold standard for diagnosing rejection; it serves as a complementary test or routine monitoring tool. Compared to creatinine, gd-cfDNA more effectively guides the timing and necessity of clinical biopsies, significantly reducing the number of biopsies and associated risks (37).
5 Baseline levels of gd-cfDNA
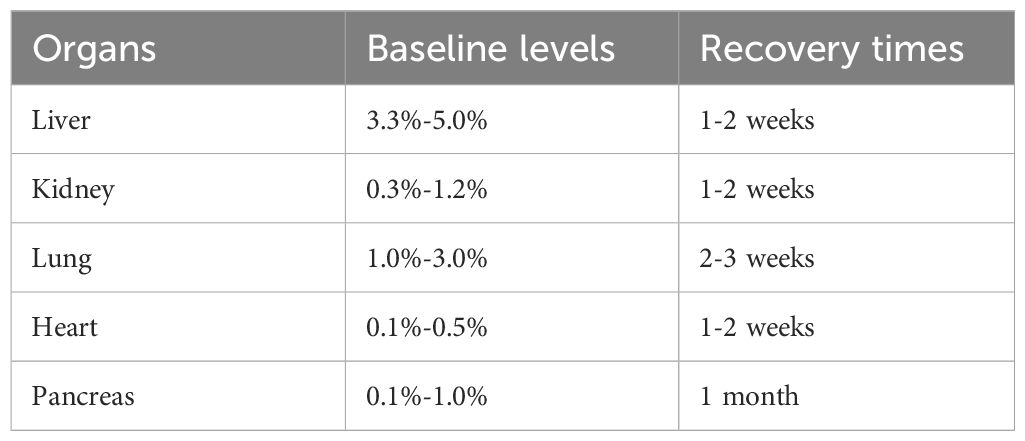
The baseline level of gd-cfDNA is the stable value of gd-cfDNA observed in the recipient’s blood in the early post-transplant period after the concentration of gd-cfDNA has declined from the initial peak caused by surgical trauma and ischaemia-reperfusion injury. It is critical for monitoring the health of the graft, as deviations from this baseline may be indicative of graft injury, such as rejection (22, 79). The baseline gd-cfDNA levels are influenced by several factors, including the type of transplanted organ, the recipient’s immune response, and the extent of surgical trauma during transplantation. The baseline levels vary by organ type. The speed at which elevated gd-cfDNA levels return to baseline also varies among individuals and types of transplanted organs. Table 3 summarizes the baseline levels of gd-cfDNA found in current studies after various organ transplants and the recovery Times to baseline levels (22, 80, 81). Pancreas transplantation has higher risks for undesirable immune response and complications compared with other types of organ transplantation, which may contribute to the relatively slow return to gd-cfDNA baseline levels after pancreas transplantation, but more research is needed to discover the exact reasons for this (62, 63). Furthermore, baseline levels of gd-cfDNA in liver and lung transplants are slightly higher than in kidney and heart transplants, possibly due to difference in the number of cells in the graft tissue (22). In recipients of stable bilateral lung transplants, the median gd-cfDNA level is higher than those of single lung transplants (0.46% vs. 0.15%) (82).
6 The role of gd-cfDNA in organ transplantation
6.1 gd-cfDNA and rejection
Rejection remains a severe complication affecting graft function and recipient survival post-transplantation. Timely diagnosis and treatment are vital to ensuring successful transplantation and long-term survival (83). During graft rejection, gd-cfDNA (%) levels increase significantly (84), and this rise occurs earlier than other indicators, such as serum creatinine and transaminases (85).
Studies indicate that for every 1% increase in gd-cfDNA levels, the risk of rejection increases 3.3-fold, with an overall rejection risk ratio of 1.89 (54). gd-cfDNA levels are significantly positively correlated with the type and severity of graft injury and Banff rejection scorings (23, 86–89). For instance, in kidney transplantation, the median gd-cfDNA in the non-rejection group is 0.3%, while in AMR, it is 2.9%, and in T-cell-mediated rejection (TCMR), it is 1.2%, with the highest levels observed in AMR (5). This may be due to AMR being the most severe and destructive form of immune-mediated graft injury, leading to more cell necrosis and the release of gd-cfDNA (90). In reflecting the severity of rejection, gd-cfDNA levels post-liver transplantation are 9.1%, 12.1%, and 28.6% in mild, moderate, and severe acute rejection (AR), respectively, significantly higher than the 0.16% in the non-rejection group and differences in gd-cfDNA levels exist among AR patients of varying severity (91). gd-cfDNA as a non-invasive marker for allograft rejection monitoring has the potential to be a valuable non-invasive marker for allograft rejection monitoring and has already been utilized by the International Society for Heart and Lung Transplantation (ISHLT) for graft rejection monitoring (92–94).
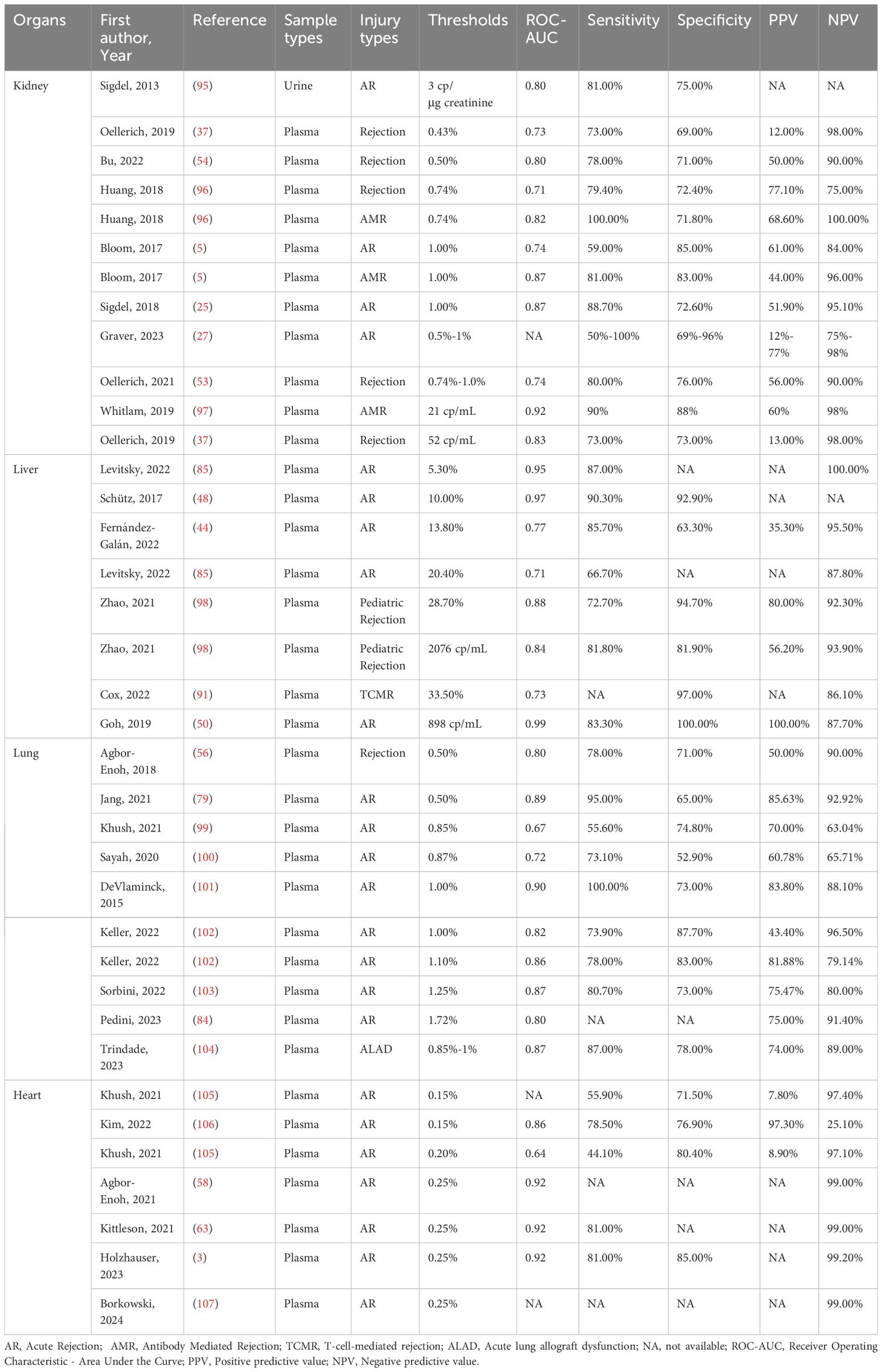
To better use gd-cfDNA for monitoring and identifying rejection post-organ transplantation, different studies have set varying gd-cfDNA thresholds to analyze diagnostic efficacy for graft injury (Table 4). In patients with stable graft status during the first year after liver transplantation, a 10% threshold demonstrated higher diagnostic efficacy for rejection than traditional liver function tests (LFTs) conducted on the same day (48). In kidney transplantation, 0.5%–1.0% thresholds showed good diagnostic performance for rejection, outperforming serum creatinine and eGFR (25, 53, 54). After lung transplantation, gd-cfDNA levels below 1.0% help exclude AR (102). In heart transplantation, when gd-cfDNA does not exceed 0.25%, there is a high confidence that rejection has not occurred, and this threshold is widely applied clinically (63). There is a lack of research on gd-cfDNA monitoring after pancreas-only transplantation. However, after simultaneous pancreas-kidney transplantation (SPK), a gd-cfDNA threshold of 70 cp/mL has been effectively used to detect early pancreatic graft rejection (66).
It is important to note that plasma gd-cfDNA levels are relatively low in early Banff 1A and borderline TCMR in kidney transplants. The median gd-cfDNA level in TCMR patients is 0.7%, but it is only 0.20% in borderline TCMR patients (Banff t1/i1), even lower than the 0.23% in the non-rejection group, leading to poor diagnostic accuracy (54). This may be because borderline and Type I (IA and IB) TCMR primarily manifest as tubulitis and interstitial inflammation, damaging tubular epithelial cells, which increases urinary gd-cfDNA levels but has a smaller effect on plasma gd-cfDNA levels (68, 108). Therefore, it is recommended to simultaneously test plasma and urinary gd-cfDNA in kidney transplant patients for a comprehensive assessment.
In lung transplant recipients, gd-cfDNA (%) median levels significantly increase in patients with AR (12.0%), higher than in stable patients (1.1%). After the resolution of rejection, gd-cfDNA levels decrease to the level in the stable group (57). Other study has also shown that after early aggressive treatment of AR, gd-cfDNA levels gradually decline and return to lower levels (<0.5%), providing crucial information for monitoring the efficacy of rejection treatment (109).
6.2 gd-cfDNA and other types of graft injury
gd-cfDNA is not a specific marker for rejection and is also associated with various types of graft injury, though the degree of elevation varies, which can help in differentiation (110) (Table 5). Furthermore, not all types of infections result in elevated gd-cfDNA levels. For instance, cytomegalovirus infections (e.g., cytomegalovirus-related hepatitis) do not cause direct graft injury and therefore do not lead to an increase in gd-cfDNA levels (33).
BK polyomavirus-associated nephropathy (BKPyVAN) is also a severe complication that can lead to graft dysfunction and loss in kidney transplantation. BKPyVAN is the most severe stage of BK virus infection, causing the death of graft kidney cells and releasing high levels of gd-cfDNA (116). However, BKPyVAN shares features with TCMR, especially Banff IA and IB TCMR, both characterized by tubulointerstitial inflammation and graft dysfunction (68, 117). The standard diagnosis of BKPyVAN is through histopathological detection of Simian virus 40 (SV40) in kidney biopsy tissue (118). However, due to the focal nature of BKPyVAN lesions, there is a high false-negative rate in biopsies (119), making it difficult to distinguish between the two when SV40 staining is negative. Since the treatment principles for BKPyVAN and TCMR are entirely different, there is an intense need for more precise diagnostic techniques. Fortunately, elevated urinary gd-cfDNA levels are more significantly associated with BKPyVAN, aiding in differentiation (110, 120). Although BKPyVAN and Type I TCMR have similar histological features, biopsy-confirmed BKPyVAN recipients have significantly higher urinary gd-cfDNA (%) and concentrations than those with Type I TCMR (median 68.4% vs. 55.3% and 10.4 ng/mL vs. 6.1 ng/mL, respectively) (117). This might be because BKPyVAN primarily infects tubular epithelial cells, leading to the kidney cell lysis and the release of gd-cfDNA into the tubular lumen, which is then excreted in the urine (121, 122). Whereas the TCMR mainly results in inflammatory cell infiltration rather than cell lysis (68). Shen et al. (117) identified a urinary gd-cfDNA concentration threshold of 7.81 ng/mL, which may effectively distinguish confirmed BKPyVAN from Type I TCMR.
6.3 Monitoring immunosuppressive therapy with gd-cfDNA
Organ transplant recipients require long-term immunosuppression to prevent immune rejection (1). The application of gd-cfDNA in monitoring the effectiveness of immunosuppressive therapy is gaining increasing attention, particularly in detecting rejection due to inadequate immunosuppression, where gd-cfDNA outperforms traditional therapeutic drug monitoring (53). Given the high variability in individual sensitivity to immunosuppressive drugs, maintaining an immunosuppressive therapeutic window through drug concentration monitoring may not be suitable for every patient. Moreover, there is a significant difference in drug concentrations between blood and lymphocytes for every patient, meaning that immunosuppressant levels may not accurately reflect the drug’s impact on immune cells (123–125). gd-cfDNA levels can predict damage before the onset of severe AR symptoms; if immunosuppressant drug levels are below the therapeutic range at this time, it indicates a need to increase the dosage (85).
Oellerich et al. (126) simultaneously measured gd-cfDNA levels and tacrolimus trough concentrations in liver transplant patients, finding that when drug concentrations were in the therapeutic range and gd-cfDNA relative quantification was below 10%, graft function remained stable. Furthermore, when gd-cfDNA relative quantification was below 10% post-liver transplantation, the minimum effective tacrolimus trough concentration was 6.8 ng/mL (originally used 8 ng/mL), suggesting that combined monitoring of gd-cfDNA relative quantification could help reduce the drug dosage. Another study conducted a 180-day follow-up on a liver transplant recipient with hepatorenal syndrome. Due to the severe renal insufficiency and worsening symptoms experienced by this recipient, tacrolimus was switched to everolimus on postoperative day 84. When tacrolimus blood levels dropped to the lower end of the therapeutic range, gd-cfDNA (%) rapidly increased to 66%. Nevertheless, as everolimus blood levels reached the therapeutic range, gd-cfDNA (%) quickly decreased and remained below 5%, indicating that gd-cfDNA (%) correlates with the concentration of immunosuppressants in the appropriate therapeutic window (20).
In a kidney transplant study, 6 months of gd-cfDNA monitoring was performed after adjusting the immunosuppressant mycophenolic acid (MPA). Among 17 recipients in the low-risk group (gd-cfDNA <1%), no rejection occurred after MPA dose reduction; however, in 4 patients in the high-risk group (gd-cfDNA ≥1%) whose MPA dosage remained unchanged, 2 developed graft dysfunction, and 1 experienced graft loss (127). Continuous monitoring and dynamic changes in gd-cfDNA can guide clinicians in making more precise decisions when adjusting immunosuppressant doses, contributing to personalized treatment in immunosuppressive therapy and demonstrating significant potential in improving the management of organ transplant recipients (27, 85).
6.4 Prognostic prediction with gd-cfDNA
Early dynamic changes in gd-cfDNA post-transplantation can reflect the recovery of organ function and provide information related to long-term prognosis. In kidney transplantation, gd-cfDNA levels below 0.5% on postoperative day 7 are considered normal recovery, whereas patients with levels equal to or greater than 0.5% had a median dialysis time of 13.50 days post-transplantation. The median gd-cfDNA level in patients with delayed graft function (fDGF) in 24 hours reached 7.20%, while it was 2.70% in those without functional DGF. Patients whose gd-cfDNA decreased to below 0.5% in 7 days postoperatively had a higher 7-year expected graft survival rate compared to those whose gd-cfDNA levels remained at or above 0.5% after 7 days (79.5% ± 16.8% vs. 67.7% ± 24.1%) (128). Patients with gd-cfDNA ≥0.5% had a doubling increased risk of a 25% decline in eGFR in three years post-transplantation (54).
Compared to patients with low and moderate gd-cfDNA levels (low, medium, and high gd-cfDNA levels were 0.7%, 1.6%, and 3.6%), those with high gd-cfDNA levels (%) after lung transplantation had a 6.6-fold increased risk of graft failure, with progression to CLAD or death (31). Additionally, plasma gd-cfDNA levels negatively correlated with oxygenation levels (PaO2/FiO2 ratio) immediately after and 72 hours post-lung transplantation, suggesting that increased gd-cfDNA levels may indicate declining oxygenation capacity of the transplanted lung (57).
Approximately 50% of heart transplant recipients develop cardiac allograft vasculopathy (CAV) 10 years post-transplantation, a major obstacle to long-term survival typically detected via selective coronary angiography, which carries significant risk (129). An increase in gd-cfDNA levels to ≥0.12% two years post-transplantation is significantly associated with the occurrence of CAV (130). Early evaluation of such patients helps in personalizing the understanding of CAV risk. Furthermore, higher gd-cfDNA levels post-heart transplantation are associated with a composite endpoint of death, re-transplantation, hemodynamic compromise, or graft dysfunction in three years (131). Recipients with left ventricular ejection fraction (LVEF) below 40% had significantly higher gd-cfDNA (%) than those with LVEF of 40% or above (0.46% vs. 0.04%) (106).
gd-cfDNA monitoring can dynamically reflect graft status, helping doctors identify high-risk patients for potential graft dysfunction or early decline, allowing timely intervention or treatment. gd-cfDNA shows potential and application value in prognosis prediction post-organ transplantation, though further research is needed to verify its clinical accuracy and reliability.
7 Advantages
The main advantages of gd-cfDNA can be summarized as follows:
1. Non-invasive Monitoring: gd-cfDNA offers a more frequent and less invasive monitoring method than biopsy, reducing patient discomfort and risk.
2. Early Diagnosis, High Sensitivity, and Dynamic Monitoring: With a shorter half-life, gd-cfDNA demonstrates higher sensitivity. gd-cfDNA can provide early warnings before graft injury, such as rejection, enabling early diagnosis and timely treatment. The more severe the injury, the higher the gd-cfDNA levels (132).
3. Immunosuppressive Efficacy Allows for Personalized Treatment: Due to its short half-life, gd-cfDNA allows for longitudinal monitoring of disease progression, providing near real-time indications of organ injury (133). Dynamic monitoring of gd-cfDNA levels enables safer and more effective use of immunosuppressants, dose adjustments, and personalized treatment, improving the long-term survival quality of organ transplant recipients (20).
4. Predicting Transplant Outcomes: Elevated gd-cfDNA levels post-transplantation are associated with poor graft function recovery, graft injury, and inadequate immunosuppression, helping to predict long-term transplant outcomes (134).
8 Limitations
1. Limited Specificity for Injury Types: gd-cfDNA is highly sensitive in reflecting graft cell damage and death; however, its ability to distinguish the types of injuries is currently limited. It is not a specific marker for any type of injury but is observed in various graft injuries. Though studies have indicated that gd-cfDNA levels are higher in AR recipients compared to other pathological states caused by non-rejection factors (91). At present, gd-cfDNA may serve as a reliable marker for AMR, but its role in addressing Acute Cellular Rejection (ACR), chronic rejection, and other types of graft injuries has not yet been fully proven (135).
2. Uncertain sample stability: Most commercial assays require blood samples to be sent to external laboratories for analysis, potentially using different detection techniques, which may lead to inconsistencies in results and pricing. Additionally, the instability of cfDNA necessitates rapid and precise handling during sample transfer, transportation, and processing to avoid variations in results (24).
3. Uncertainty in Thresholds and Undefined Monitoring Frequency: There is variability in the diagnostic thresholds and monitoring frequency of gd-cfDNA across different studies, and no standardized guidelines currently exist. Further research is needed to determine the optimal monitoring frequency and thresholds, especially in the early post-transplant period when the risk of AR and infection is highest (54).
4. Global Disparities in Medical Resources: As an advanced medical technology, the accessibility of gd-cfDNA testing varies significantly across different countries and regions (136, 137).
5. Monitoring in Multi-Organ Transplants and Long-Term Monitoring: Due to the unique nature of multi-organ transplants, there is currently limited research on gd-cfDNA monitoring in these cases. Moreover, there is a lack of reports on routine graft monitoring beyond five years in clinically stable organ transplant recipients (3).
9 Discussion
As an emerging non-invasive biomarker, gd-cfDNA shows great potential in monitoring and prognostic assessment following organ transplantation. This is particularly significant for thoracic organs. Given the stringent size-matching requirements for thoracic organ transplants, the unique immunological characteristics post-lung transplantation, and the lack of extracorporeal life support options, long-term survival rates remain lower than other solid organ transplants (138, 139). Additionally, the incidence of AR in the first year post-lung transplantation is 26.6%, significantly higher than that of other organ transplants (140). Chronic lung allograft dysfunction (CLAD) occurs in approximately 50% of lung transplant recipients in five years, posing a major obstacle to long-term survival. Currently, there are no reliable predictive markers or effective preventive or therapeutic methods for CLAD (141), highlighting the urgent need for reliable graft monitoring tools. Currently, Primary graft dysfunction (PGD) and AR have been identified as strong independent risk factors for the development of CLAD (142, 143). The detection of gd-cfDNA enables the early identification of patients at risk for AR, PGD, and CLAD, facilitating timely therapeutic interventions that enhance the management of long-term complications and improve the prognosis of transplant recipients (141, 144).
Currently, research progress on gd-cfDNA varies across different organ fields. Some studies explored differences between graft injuries based on time trends, the extent of gd-cfDNA elevation, fragment length, and genomic composition. For example, in liver transplant research, the distribution and proportion of gd-cfDNA fragment sizes in plasma have been found to monitor post-transplant graft function (145). In lung transplantation, the combined approach of gd-cfDNA (%) and cfDNA fragment size is used to determine the occurrence and potential source of infection (84). Heart transplant studies have found that the genomic composition of DNA fragments differs between AMR and ACR, with AMR exhibiting a higher percentage of short fragments and higher guanine-cytosine content than ACR (58, 59). Transcriptome analysis of biopsy tissues has revealed that gd-cfDNA-associated gene expression patterns may be related to the type of rejection and response to treatment (146). These findings may help distinguish different types of injuries to better understand graft status monitoring.
Moreover, there is currently no consensus in clinical practice on whether to use gd-cfDNA (%) or gd-cfDNA (cp/mL) to optimize monitoring (147). The release of recipient cfDNA is influenced by factors such as immunosuppressants, age, and increased BMI (148). For instance, in two liver transplant studies, the median gd-cfDNA (%) in the AR group was 21.8% (91) and 41.9% (98), respectively. The former study population consisted of adult liver transplant recipients (median age 53.7 years), while the latter involved pediatric liver transplant recipients (median age 19.4 months). Schütz et al. (149) found that as the dose of immunosuppressants was reduced, the rate of leukocyte apoptosis decreased, leading to a decline in total cfDNA levels over time, while gd-cfDNA (%) correspondingly increased. However, the absolute quantification of gd-cfDNA remained stable during this period, indicating that the absolute quantification of gd-cfDNA may perform better than gd-cfDNA (%) in monitoring (37, 97). Nonetheless, the gd-cfDNA (%) assay was less sensitive to preanalytical variables and more advantageous in comparison among studies (150). In the field of lung transplantation, the concept of relative change value (RCV) in gd-cfDNA (%) has been proposed, suggesting that an increase of more than 73% from baseline may indicate pathological changes, thereby enhancing diagnostic performance (104).
gd-cfDNA can provide early warnings for rejection and various types of graft injuries with high sensitivity. However, gd-cfDNA monitoring cannot entirely replace biopsies; instead, it aids clinicians in more accurately identifying patients who genuinely need biopsies, reducing unnecessary invasive procedures and associated risks (151). As a single non-invasive test, gd-cfDNA has lower specificity for injury types. However, it can provide some information about the injury, which can be used to better determine the specific type of injury by combining it with other biomarkers or artificial intelligence techniques. For example, biopsy remains the gold standard for graft assessment, but relying solely on histological predictions has limitations; combining gd-cfDNA with Banff biopsy scores offers a more comprehensive assessment of graft injury and prognosis than biopsy features alone (52). Studies have shown that combining gd-cfDNA with DSA improves diagnostic accuracy for AMR compared to using either marker alone, and it significantly outperforms traditional graft function indicators (69, 72, 152). For instance, when predicting AMR, the AUC for gd-cfDNA (%) and DSA alone was 0.85 and 0.66, respectively, while the combined AUC was 0.88 (72). Some experienced heart transplant centers transition monitoring to a strategy that combines gene expression profiling (GEP) with gd-cfDNA in patients with stable grafts after 8 weeks post-transplant. GEP can assess the quiescent or activated state of the immune system, while gd-cfDNA monitoring serves as a specific marker for graft injury (151). Infection markers, such as procalcitonin (PCT), combined with gd-cfDNA, can enhance the ability to differentiate between AR and infection; during severe infections post-transplant, PCT is similarly elevated above the threshold when gd-cfDNA levels rise significantly, but PCT levels do not exceed threshold values in AR patients (153). gd-cfDNA monitoring combined with artificial intelligence leads to powerful predictive models, guiding clinical decisions and potentially better identifying high-risk patients (53, 98, 107). Additionally, combining gd-cfDNA with other testing methods, such as organ function tests, pathogen detection, and mRNA transcripts in the graft, may provide more comprehensive information on graft function, improve diagnostic efficiency, enable personalized post-transplant management, and ultimately enhance the quality of life and survival rates of transplant recipients (3).
In addressing the challenges of variable thresholds, undefined monitoring frequency, and global disparities in medical resources in the clinical application of gd-cfDNA, future research should focus on establishing standardized monitoring protocols, narrowing global healthcare gaps, and exploring new possibilities for gd-cfDNA. Through these efforts, gd-cfDNA has the potential to become a revolutionary monitoring tool in organ transplantation, providing more personalized and precise diagnostic information for transplant recipients, improving their quality of life, and increasing long-term survival rates.
Author contributions
WZ: Writing – original draft, Writing – review & editing. BL: Data curation, Writing – review & editing. DJ: Investigation, Writing – review & editing. RW: Resources, Writing – review & editing. HC: Investigation, Writing – review & editing. HW: Formal analysis, Writing – review & editing. ZY: Writing – review & editing, Resources. BG: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviation
gd-cfDNA, graft-derived cell-free DNA; cfDNA, cell-free DNA; ddPCR, Droplet digital PCR; NGS, Next-generation sequencing; qPCR, quantitative PCR; AR, Acute rejection; EMB, endomyocardial biopsy; ROC-AUC, Receiver Operating Characteristic - Area Under the Curve; AMR, antibody-mediated rejection; ACR, Acute Cellular Rejection; TCMR, T-cellmediated rejection; SPK, pancreas-kidney transplantation; DSA, Donorspecific antibodies; dnDSA, De novo donor-specific antibodies; PGD, Primary graft dysfunction; CAV, Cardiac allograft vasculopathy; CLAD, Chronic lung allograft dysfunction; RVI, Respiratory viral infection; BKPyVAN, BK polyomavirus-associated nephropathy; IF/TA, Interstitial fibrosis/ tubular atrophy; ATN, Acute tubular necrosis; GVHD, Graft-versus-host disease; PCT, procalcitonin.
References
1. Zhou Q, Li T, Wang K, Zhang Q, Geng Z, Deng S, et al. Current status of xenotransplantation research and the strategies for preventing xenograft rejection. Front Immunol. (2022) 13:928173. doi: 10.3389/fimmu.2022.928173
2. Reeve J, Böhmig GA, Eskandary F, Einecke G, Lefaucheur C, Loupy A, et al. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight. (2017) 2. doi: 10.1172/jci.insight.94197
3. Holzhauser L, DeFilippis EM, Nikolova A, Byku M, Contreras JP, De Marco T, et al. The end of endomyocardial biopsy? A practical guide for noninvasive heart transplant rejection surveillance. JACC Heart Fail. (2023) 11:263–76. doi: 10.1016/j.jchf.2022.11.002
4. Kataria A, Kumar D, Gupta G. Donor-derived cell-free DNA in solid-organ transplant diagnostics: indications, limitations, and future directions. Transplantation. (2021) 105:1203–11. doi: 10.1097/TP.0000000000003651
5. Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, et al. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. (2017) 28:2221–32. doi: 10.1681/ASN.2016091034
6. Nagata S, Nagase H, Kawane K, Mukae N, Fukuyama H. Degradation of chromosomal DNA during apoptosis. Cell Death Differ. (2003) 10:108–16. doi: 10.1038/sj.cdd.4401161
7. Han DSC, Ni M, Chan RWY, Chan VWH, Lui KO, Chiu RWK, et al. The biology of cell-free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet. (2020) 106:202–14. doi: 10.1016/j.ajhg.2020.01.008
8. Celec P, Vlková B, Lauková L, Bábíčková J, Boor P. Cell-free DNA: the role in pathophysiology and as a biomarker in kidney diseases. Expert Rev Mol Med. (2018) 20:e1. doi: 10.1017/erm.2017.12
9. Martuszewski A, Paluszkiewicz P, Król M, Banasik M, Kepinska M. Donor-derived cell-free DNA in kidney transplantation as a potential rejection biomarker: A systematic literature review. J Clin Med. (2021) 10. doi: 10.3390/jcm10020193
10. Mandel P, Metais P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil. (1948) 142:241–3.
11. Huang CC, Du M, Wang L. Bioinformatics analysis for circulating cell-free DNA in cancer. Cancers (Basel). (2019) 11. doi: 10.3390/cancers11060805
12. Everett TR, Chitty LS. Cell-free fetal DNA: the new tool in fetal medicine. Ultrasound Obstet Gynecol. (2015) 45:499–507. doi: 10.1002/uog.14746
13. Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U.S.A. (2015) 112:E5503–12. doi: 10.1073/pnas.1508736112
14. Zheng YW, Chan KC, Sun H, Jiang P, Su X, Chen EZ, et al. Nonhematopoietically derived DNA is shorter than hematopoietically derived DNA in plasma: a transplantation model. Clin Chem. (2012) 58:549–58. doi: 10.1373/clinchem.2011.169318
15. Beck J, Schmitz J, Kanzow P, Kollmar O, Oellerich M, Schütz E. Absolute quantification of graft derived cell-free DNA (GcfDNA) early after liver transplantation (LTx) using droplet digital PCR. Clin Chem. (2014) 60:S194–5.
16. van der Vaart M, Pretorius PJ. The origin of circulating free DNA. Clin Chem. (2007) 53:2215. doi: 10.1373/clinchem.2007.092734
17. Chitty LS, Lo YM. Noninvasive prenatal screening for genetic diseases using massively parallel sequencing of maternal plasma DNA. Cold Spring Harb Perspect Med. (2015) 5:a023085. doi: 10.1101/cshperspect.a023085
18. Gauthier VJ, Tyler LN, Mannik M. Blood clearance kinetics and liver uptake of mononucleosomes in mice. J Immunol. (1996) 156:1151–6. doi: 10.4049/jimmunol.156.3.1151
19. Zhou Q, Liu F, Guo L, Chen R, Yuan X, Li C, et al. A novel urine cell-free DNA preservation solution and its application in kidney transplantation. Nephrol (Carlton). (2021) 26:684–91. doi: 10.1111/nep.13884
20. Kanzow P, Kollmar O, Schütz E, Oellerich M, Schmitz J, Beck J, et al. Graft-derived cell-free DNA as an early organ integrity biomarker after transplantation of a marginal HELLP syndrome donor liver. Transplantation. (2014) 98:e43–5. doi: 10.1097/TP.0000000000000303
21. Lo YM, Tein MS, Pang CC, Yeung CK, Tong KL, Hjelm NM. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. (1998) 351:1329–30. doi: 10.1016/S0140-6736(05)79055-3
22. Beck J, Bierau S, Balzer S, Andag R, Kanzow P, Schmitz J, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. (2013) 59:1732–41. doi: 10.1373/clinchem.2013.210328
23. Sharon E, Shi H, Kharbanda S, Koh W, Martin LR, Khush KK, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PloS Comput Biol. (2017) 13:e1005629. doi: 10.1371/journal.pcbi.1005629
24. Grskovic M, Hiller DJ, Eubank LA, Sninsky JJ, Christopherson C, Collins JP, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn. (2016) 18:890–902. doi: 10.1016/j.jmoldx.2016.07.003
25. Sigdel TK, Archila FA, Constantin T, Prins SA, Liberto J, Damm I, et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med. (2018) 8. doi: 10.3390/jcm8010019
26. Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U.S.A. (2011) 108:6229–34. doi: 10.1073/pnas.1013924108
27. Graver AS, Lee D, Power DA, Whitlam JB. Understanding donor-derived cell-free DNA in kidney transplantation: an overview and case-based guide for clinicians. Transplantation. (2023) 107:1675–86. doi: 10.1097/TP.0000000000004482
28. Casas S, Tangprasertchai NS, Oikonomaki K, Mathers S, Sollet ZC, Samara S, et al. Multi-centre analytical performance verification of an IVD assay to quantify donor-derived cell-free DNA in solid organ transplant recipients. Hla. (2024) 103:e15518. doi: 10.1111/tan.15518
29. Dauber EM, Kollmann D, Kozakowski N, Rasoul-Rockenschaub S, Soliman T, Berlakovich GA, et al. Quantitative PCR of INDELs to measure donor-derived cell-free DNA-a potential method to detect acute rejection in kidney transplantation: a pilot study. Transpl Int. (2020) 33:298–309. doi: 10.1016/j.jmoldx.2016.07.003
30. Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. (2015) 61:79–88. doi: 10.1373/clinchem.2014.221366
31. Agbor-Enoh S, Wang Y, Tunc I, Jang MK, Davis A, De Vlaminck I, et al. Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. EBioMedicine. (2019) 40:541–53. doi: 10.1016/j.ebiom.2018.12.029
32. Pettersson L, Frischknecht L, Westerling S, Ramezanali H, Weidmann L, Lopez KC, et al. Detection of donor-derived cell-free DNA in the setting of multiple kidney transplantations. Front Immunol. (2024) 15:1282521. doi: 10.3389/fimmu.2024.1282521
33. Li J, Tan J, Chen G, Li Q, Zhou Z, Bin Y, et al. Sequencing of cell-free DNA to monitor cytomegalovirus infection after liver transplant. Exp Clin Transplant. (2021) 19:331–8. doi: 10.6002/ect
34. Gordon PM, Khan A, Sajid U, Chang N, Suresh V, Dimnik L, et al. An algorithm measuring donor cell-free DNA in plasma of cellular and solid organ transplant recipients that does not require donor or recipient genotyping. Front Cardiovasc Med. (2016) 3:33. doi: 10.3389/fcvm.2016.00033
35. Hidestrand M, Tomita-Mitchell A, Hidestrand PM, Oliphant A, Goetsch M, Stamm K, et al. Highly sensitive noninvasive cardiac transplant rejection monitoring using targeted quantification of donor-specific cell-free deoxyribonucleic acid. J Am Coll Cardiol. (2014) 63:1224–6. doi: 10.1016/j.jacc.2013.09.029
36. Oellerich M, Christenson RH, Beck J, Schütz E, Sherwood K, Price CP, et al. Donor-derived cell-free DNA testing in solid organ transplantation: A value proposition. J Appl Lab Med. (2020) 5:993–1004. doi: 10.1093/jalm/jfaa062
37. Oellerich M, Shipkova M, Asendorf T, Walson PD, Schauerte V, Mettenmeyer N, et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant. (2019) 19:3087–99. doi: 10.1111/ajt.15416
38. Boyd A, Cain O, Chauhan A, Webb GJ. Medical liver biopsy: background, indications, procedure and histopathology. Frontline Gastroenterol. (2020) 11:40–7. doi: 10.1136/flgastro-2018-101139
39. Liu S, Wang J. Current and future perspectives of cell-free DNA in liquid biopsy. Curr Issues Mol Biol. (2022) 44:2695–709. doi: 10.3390/cimb44060184
40. Rush D, Arlen D, Boucher A, Busque S, Cockfield SM, Girardin C, et al. Lack of benefit of early protocol biopsies in renal transplant patients receiving TAC and MMF: a randomized study. Am J Transplant. (2007) 7:2538–45. doi: 10.1111/j.1600-6143.2007.01979.x
41. Yang JYC, Sarwal RD, Sigdel TK, Damm I, Rosenbaum B, Liberto JM, et al. A urine score for noninvasive accurate diagnosis and prediction of kidney transplant rejection. Sci Transl Med. (2020) 12. doi: 10.1126/scitranslmed.aba2501
42. American society of nephrology renal research report. J Am Soc Nephrol. (2005) 16:1886–903. doi: 10.1681/ASN.2005030285
43. Boyum JH, Atwell TD, Schmit GD, Poterucha JJ, Schleck CD, Harmsen WS, et al. Incidence and risk factors for adverse events related to image-guided liver biopsy. Mayo Clin Proc. (2016) 91:329–35. doi: 10.1016/j.mayocp.2015.11.015
44. Fernández-Galán E, Badenas C, Fondevila C, Jiménez W, Navasa M, Puig-Butillé JA, et al. Monitoring of donor-derived cell-free DNA by short tandem repeats: concentration of total cell-free DNA and fragment size for acute rejection risk assessment in liver transplantation. Liver Transpl. (2022) 28:257–68. doi: 10.1016/j.transproceed.2023.08.016
45. Rodríguez-Perálvarez M, Germani G, Papastergiou V, Tsochatzis E, Thalassinos E, Luong TV, et al. Early tacrolimus exposure after liver transplantation: relationship with moderate/severe acute rejection and long-term outcome. J Hepatol. (2013) 58:262–70. doi: 10.1016/j.jhep.2012.09.019
46. Rodríguez-Perálvarez M, Germani G, Darius T, Lerut J, Tsochatzis E, Burroughs AK. Tacrolimus trough levels, rejection and renal impairment in liver transplantation: a systematic review and meta-analysis. Am J Transplant. (2012) 12:2797–814. doi: 10.1111/j.1600-6143.2012.04140.x
47. Dashti SH, Kasraianfard A, Ebrahimi A, Nassiri-Toosi M, Pakshir MS, Rahimi M, et al. Hemodynamic changes and early recovery of liver graft function after liver transplantation. Int J Organ Transplant Med. (2020) 11:1–7. doi: 10.1126/scitranslmed.3007803
48. Schütz E, Fischer A, Beck J, Harden M, Koch M, Wuensch T, et al. a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PloS Med. (2017) 14:e1002286. doi: 10.1126/scitranslmed.aba2501
49. Beck J, Bierau S, Balzer S, Andag R, Kanzow P, Hennecke J, et al. Rapid and cost effective measurement of circulating cell free graft DNA for the early detection of liver transplant rejection. Clin Chem. (2013) 59:A27. doi: 10.1038/s41598-020-80845-6
50. Goh SK, Do H, Testro A, Pavlovic J, Vago A, Lokan J, et al. The measurement of donor-specific cell-free DNA identifies recipients with biopsy-proven acute rejection requiring treatment after liver transplantation. Transplant Direct. (2019) 5:e462. doi: 10.1097/TXD.0000000000000902
51. Schwarz A, Gwinner W, Hiss M, Radermacher J, Mengel M, Haller H. Safety and adequacy of renal transplant protocol biopsies. Am J Transplant. (2005) 5:1992–6. doi: 10.1111/j.1600-6143.2005.00988.x
52. Huang E, Jordan SC. Donor-derived cell-free DNA in kidney transplantation: evolving concepts and potential limitations. Kidney Int. (2022) 101:676–7. doi: 10.1016/j.kint.2022.01.012
53. Oellerich M, Sherwood K, Keown P, Schütz E, Beck J, Stegbauer J, et al. Liquid biopsies: donor-derived cell-free DNA for the detection of kidney allograft injury. Nat Rev Nephrol. (2021) 17:591–603. doi: 10.1038/s41581-021-00428-0
54. Bu L, Gupta G, Pai A, Anand S, Stites E, Moinuddin I, et al. Clinical outcomes from the Assessing Donor-derived cell-free DNA Monitoring Insights of kidney Allografts with Longitudinal surveillance (ADMIRAL) study. Kidney Int. (2022) 101:793–803. doi: 10.1016/j.kint.2021.11.034
55. Zhou G, Ren Y, Li J, Yang T, Su N, Zhao L, et al. Safety and diagnostic efficacy of cone beam computed tomography-guided transbronchial cryobiopsy for interstitial lung disease: a cohort study. Eur Respir J. (2020) 56. doi: 10.1183/13993003.00724-2020
56. Agbor-Enoh S, Jackson AM, Tunc I, Berry GJ, Cochrane A, Grimm D, et al. Late manifestation of alloantibody-associated injury and clinical pulmonary antibody-mediated rejection: Evidence from cell-free DNA analysis. J Heart Lung Transplant. (2018) 37:925–32. doi: 10.1016/j.healun.2018.01.1305
57. Tanaka S, Sugimoto S, Kurosaki T, Miyoshi K, Otani S, Suzawa K, et al. Donor-derived cell-free DNA is associated with acute rejection and decreased oxygenation in primary graft dysfunction after living donor-lobar lung transplantation. Sci Rep. (2018) 8:15366. doi: 10.1038/s41598-018-33848-3
58. Agbor-Enoh S, Shah P, Tunc I, Hsu S, Russell S, Feller E, et al. Cell-free DNA to detect heart allograft acute rejection. Circulation. (2021) 143:1184–97. doi: 10.1161/CIRCULATIONAHA.120.049098
59. De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. (2014) 6:241ra77. doi: 10.1126/scitranslmed.3007803
60. Costanzo MR, Dipchand A, Starling R, Anderson A, Chan M, Desai S, et al. The International Society of Heart and Lung Transplantation Guidelines for the care of heart transplant recipients. J Heart Lung Transplant. (2010) 29:914–56. doi: 10.1016/j.healun.2010.05.034
61. Dengler TJ, Zimmermann R, Braun K, Müller-Bardorff M, Zehelein J, Sack FU, et al. Elevated serum concentrations of cardiac troponin T in acute allograft rejection after human heart transplantation. J Am Coll Cardiol. (1998) 32:405–12. doi: 10.1016/S0735-1097(98)00257-5
62. Arora S, Gullestad L, Wergeland R, Simonsen S, Holm T, Hognestad A, et al. Probrain natriuretic peptide and C-reactive protein as markers of acute rejection, allograft vasculopathy, and mortality in heart transplantation. Transplantation. (2007) 83:1308–15. doi: 10.1097/01.tp.0000263338.39555.21
63. Kittleson MM, Garg S. Solid gold, or liquid gold?: towards a new diagnostic standard for heart transplant rejection. Circulation. (2021) 143:1198–201. doi: 10.1161/CIRCULATIONAHA.120.052925
64. Akirav EM, Lebastchi J, Galvan EM, Henegariu O, Akirav M, Ablamunits V, et al. Detection of β cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci U.S.A. (2011) 108:19018–23. doi: 10.1038/s41598-018-33848-3
65. Williams MD, Fei M, SChadde E, Hollinger EF, Chan EY, Olaitan O. Early experience using donor-derived cell-free DNA for surveillance of rejection following simultaneous pancreas and kidney transplantation. Transplant Direct. (2022) 8:e1321. doi: 10.1097/TXD.0000000000001321
66. Ventura-Aguiar P, Ramirez-Bajo MJ, Rovira J, Bañón-Maneus E, Hierro N, Lazo M, et al. Donor-derived cell-free DNA shows high sensitivity for the diagnosis of pancreas graft rejection in simultaneous pancreas-kidney transplantation. Transplantation. (2022) 106:1690–7. doi: 10.1097/TP.0000000000004088
67. Zhang R. Donor-specific antibodies in kidney transplant recipients. Clin J Am Soc Nephrol. (2018) 13:182–92. doi: 10.2215/CJN.00700117
68. Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. (2018) 18:293–307. doi: 10.1111/ajt.14625
69. Jordan SC, Bunnapradist S, Bromberg JS, Langone AJ, Hiller D, Yee JP, et al. Donor-derived cell-free DNA identifies antibody-mediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct. (2018) 4:e379. doi: 10.1097/TXD.0000000000000821
70. Senev A, Coemans M, Lerut E, Van Sandt V, Daniëls L, Kuypers D, et al. Histological picture of antibody-mediated rejection without donor-specific anti-HLA antibodies: Clinical presentation and implications for outcome. Am J Transplant. (2019) 19:763–80. doi: 10.1111/ajt.15074
71. Luque S, Lúcia M, Melilli E, Lefaucheur C, Crespo M, Loupy A, et al. Value of monitoring circulating donor-reactive memory B cells to characterize antibody-mediated rejection after kidney transplantation. Am J Transplant. (2019) 19:368–80. doi: 10.1111/ajt.15055
72. Halloran PF, Reeve J, Madill-Thomsen KS, Demko Z, Prewett A, Gauthier P, et al. Antibody-mediated rejection without detectable donor-specific antibody releases donor-derived cell-free DNA: results from the trifecta study. Transplantation. (2023) 107:709–19. doi: 10.1097/TP.0000000000004324
73. DePasquale E, Kobashigawa J, Hall S, Wolf-Doty T, Teuteberg J, Khush K. Donor derived cell free DNA as a risk factor for initiating de-novo donor specific antibodies in heart transplantation. J Heart Lung Transplant. (2021) 40:S217–S8. doi: 10.1016/j.healun.2021.01.628
74. Stites E, Kumar D, Olaitan O, John Swanson S, Leca N, Weir M, et al. High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant. (2020) 20:2491–8. doi: 10.1111/ajt.15822
75. Teuteberg J, Kobashigawa J, Shah P, Ghosh S, Ross D, DePasquale E, et al. Donor-derived cell-free DNA predicts de novo DSA after heart transplantation. J Heart Lung Transplant. (2021) 40:S30. doi: 10.1016/j.healun.2021.01.1810
76. Roy FM, Lee D, Lewis P, Rose S. An economic analysis of the cost effectiveness of blood gene expression profiling in kidney transplant recipients. iMedPub. (2017) 1). doi: 10.1111/ajt.17039
77. Vilalta A. Cost and use trends of endomyocardial biopsy in heart transplant patients: A 4-year claims data analysis. Transplant Proc. (2023) 55:2186–90. doi: 10.1016/j.transproceed.2023.08.016
78. Puttarajappa CM, Mehta RB, Roberts MS, Smith KJ, Hariharan S. Economic analysis of screening for subclinical rejection in kidney transplantation using protocol biopsies and noninvasive biomarkers. Am J Transplant. (2021) 21:186–97. doi: 10.1111/ajt.16150
79. Jang MK, Tunc I, Berry GJ, Marboe C, Kong H, Keller MB, et al. Donor-derived cell-free DNA accurately detects acute rejection in lung transplant patients, a multicenter cohort study. J Heart Lung Transplant. (2021) 40:822–30. doi: 10.1016/j.healun.2021.04.009
80. Gielis EM, Beirnaert C, Dendooven A, Meysman P, Laukens K, De Schrijver J, et al. Plasma donor-derived cell-free DNA kinetics after kidney transplantation using a single tube multiplex PCR assay. PloS One. (2018) 13:e0208207. doi: 10.1371/journal.pone.0208207
81. Yoo A, Riedel A, Qian I, Bartosic A, Soltani R, Kibria G, et al. An initial analysis of the baseline levels of dd-cfDNA after pancreas transplantation: A prospective study from high-volume centers in the United States. Transplant Direct. (2023) 9:e1459. doi: 10.1097/TXD.0000000000001459
82. Keller MB, Meda R, Fu S, Yu K, Jang MK, Charya A, et al. Comparison of donor-derived cell-free DNA between single versus double lung transplant recipients. Am J Transplant. (2022) 22:2451–7. doi: 10.1111/ajt.17039
83. Costanzo-Nordin MR. Cardiac allograft vasculopathy: relationship with acute cellular rejection and histocompatibility. J Heart Lung Transplant. (1992) 11:S90–103. doi: 10.1016/j.healun.2018.01.178
84. Pedini P, Coiffard B, Cherouat N, Casas S, Fina F, Boutonnet A, et al. Clinical relevance of cell-free DNA quantification and qualification during the first month after lung transplantation. Front Immunol. (2023) 14:1183949. doi: 10.3389/fimmu.2023.1183949
85. Levitsky J, Kandpal M, Guo K, Kleiboeker S, Sinha R, Abecassis M. Donor-derived cell-free DNA levels predict graft injury in liver transplant recipients. Am J Transplant. (2022) 22:532–40. doi: 10.1111/ajt.16835
86. Kant S, Kumar D, Moinuddin I, Alhamad T, Murad HF, Bettinotti M, et al. Utility of donor-derived cell-free DNA in detecting ABMR in patients with AT1R antibodies. Kidney Int Rep. (2021) 6:2706–8. doi: 10.1016/j.ekir.2021.07.026
87. Li Y, Liang B. Circulating donor-derived cell-free DNA as a marker for rejection after lung transplantation. Front Immunol. (2023) 14:1263389. doi: 10.3389/fimmu.2023.1263389
88. Agbor-Enoh S, Tunc I, De Vlaminck I, Fideli U, Davis A, Cuttin K, et al. Applying rigor and reproducibility standards to assay donor-derived cell-free DNA as a non-invasive method for detection of acute rejection and graft injury after heart transplantation. J Heart Lung Transplant. (2017) 36:1004–12. doi: 10.1016/j.healun.2017.05.026
89. Valantine H, Shah P, Shah K, Hsu S, Feller E, Rodrigo M, et al. (176)–validation of donor-derived cell-free DNA to detect heart-transplant rejection. J Heart Lung Transplant. (2018) 37:S78–S9. doi: 10.1016/j.healun.2018.01.178
90. Loupy A, Lefaucheur C. Antibody-mediated rejection of solid-organ allografts. N Engl J Med. (2018) 379:1150–60. doi: 10.1056/NEJMra1802677
91. Cox DRA, Low N, Goh SK, Lee E, Vago A, Jackett L, et al. Low levels of hepatocyte-specific methylation in cell-free DNA are a strong negative predictor for acute T cell-mediated rejection requiring treatment following liver transplantation. Liver Transpl. (2022) 28:1024–38. doi: 10.1002/lt.26388
92. Velleca A, Shullo MA, Dhital K, Azeka E, Colvin M, DePasquale E, et al. The International Society for Heart and Lung Transplantation (ISHLT) guidelines for the care of heart transplant recipients. J Heart Lung Transplant. (2023) 42:e1–e141. doi: 10.1016/j.healun.2022.10.015
93. Rodgers N, Gerding B, Cusi V, Vaida F, Tada Y, Morris GP, et al. Comparison of two donor-derived cell-free DNA tests and a blood gene-expression profile test in heart transplantation. Clin Transplant. (2023) 37:e14984. doi: 10.1111/ctr.14984
94. Oellerich M, Budde K, Osmanodja B, Bornemann-Kolatzki K, Beck J, Schütz E, et al. Donor-derived cell-free DNA as a diagnostic tool in transplantation. Front Genet. (2022) 13:1031894. doi: 10.3389/fgene.2022.1031894
95. Sigdel TK, Vitalone MJ, Tran TQ, Dai H, Hsieh SC, Salvatierra O, et al. A rapid noninvasive assay for the detection of renal transplant injury. Transplantation. (2013) 96:97–101. doi: 10.1097/TP.0b013e318295ee5a
96. Huang E, Sethi S, Peng A, Najjar R, Mirocha J, Haas M, et al. Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am J Transplant. (2019) 19:1663–70. doi: 10.1111/ajt.15289
97. Whitlam JB, Ling L, Skene A, Kanellis J, Ierino FL, Slater HR, et al. Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am J Transplant. (2019) 19:1037–49. doi: 10.1111/ajt.15142
98. Zhao D, Zhou T, Luo Y, Wu C, Xu D, Zhong C, et al. Preliminary clinical experience applying donor-derived cell-free DNA to discern rejection in pediatric liver transplant recipients. Sci Rep. (2021) 11:1138. doi: 10.1038/s41598-020-80845-6
99. Khush KK, De Vlaminck I, Luikart H, Ross DJ, Nicolls MR. Donor-derived, cell-free DNA levels by next-generation targeted sequencing are elevated in allograft rejection after lung transplantation. ERJ Open Res. (2021) 7. doi: 10.1183/23120541.00462-2020
100. Sayah D, Weigt SS, Ramsey A, Ardehali A, Golden J, Ross DJ. Plasma donor-derived cell-free DNA levels are increased during acute cellular rejection after lung transplant: pilot data. Transplant Direct. (2020) 6:e608. doi: 10.1097/TXD.0000000000001063
101. De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. (2015) 112:13336–41. doi: 10.1073/pnas.1517494112
102. Keller M, Sun J, Mutebi C, Shah P, Levine D, Aryal S, et al. Donor-derived cell-free DNA as a composite marker of acute lung allograft dysfunction in clinical care. J Heart Lung Transplant. (2022) 41:458–66. doi: 10.1016/j.healun.2021.12.009
103. Sorbini M, Togliatto G, Mioli F, Simonato E, Marro M, Cappuccio M, et al. Validation of a simple, rapid, and cost-effective method for acute rejection monitoring in lung transplant recipients. Transpl Int. (2022) 35:10546. doi: 10.3389/ti.2022.10546
104. Trindade AJ, Chapin KC, Gray JN, Furuya Y, Mullican A, Hoy H, et al. Relative change in donor-derived cell-free DNA is superior to absolute values for diagnosis of acute lung allograft dysfunction. Transplant Direct. (2023) 9:e1487. doi: 10.1097/TXD.0000000000001487
105. Khush KK. Clinical utility of donor-derived cell-free DNA testing in cardiac transplantation. J Heart Lung Transplant. (2021) 40:397–404. doi: 10.1016/j.healun.2021.01.1564
106. Kim PJ, Olymbios M, Siu A, Wever Pinzon O, Adler E, Liang N, et al. A novel donor-derived cell-free DNA assay for the detection of acute rejection in heart transplantation. J Heart Lung Transplant. (2022) 41:919–27. doi: 10.1016/j.healun.2022.04.002
107. Borkowski P, Singh N, Borkowska N. Advancements in heart transplantation: donor-derived cell-free DNA as next-generation biomarker. Cureus. (2024) 16:e54018. doi: 10.7759/cureus.54018
108. Bloom RD. Using (cell-free) DNA to incriminate rejection as the cause of kidney allograft dysfunction: Do we have a verdict? Am J Transplant. (2019) 19:1609–10. doi: 10.1111/ajt.15338
109. Grskovic M, Hiller DJ, Eubank LA, Sninsky JJ, Christopherson C, Collins JP, et al. Plasma levels of donor-derived cell-free DNA increase with rejection and often decrease after treatment in organ transplant recipients. Am J Of Transplant. (2015) 18(6):890–902. doi: 10.1093/clinchem/hvac053
110. Chen XT, Qiu J, Wu ZX, Zhang H, Chen T, Yang SC, et al. Using both plasma and urine donor-derived cell-free DNA to identify various renal allograft injuries. Clin Chem. (2022) 68:814–25. doi: 10.1093/clinchem/hvac053
111. Lewis D, Glehn-Ponsirenas R, Gulbahce N, Hooey LJ, Chaffin JM, Miles J, et al. High levels of donor-derived cell-free DNA in a case of graft-versus-host-disease following liver transplantation. Am J Transplant. (2022) 22:973–6. doi: 10.1111/ajt.16894
112. Gielis EM, Ledeganck KJ, Dendooven A, Meysman P, Beirnaert C, Laukens K, et al. The use of plasma donor-derived, cell-free DNA to monitor acute rejection after kidney transplantation. Nephrol Dial Transplant. (2020) 35:714–21. doi: 10.1093/ndt/gfz091
113. Keller M, Bush E, Diamond JM, Shah P, Matthew J, Brown AW, et al. Use of donor-derived-cell-free DNA as a marker of early allograft injury in primary graft dysfunction (PGD) to predict the risk of chronic lung allograft dysfunction (CLAD). J Heart Lung Transplant. (2021) 40:488–93. doi: 10.1016/j.healun.2021.02.008
114. Bazemore K, Permpalung N, Mathew J, Lemma M, Haile B, Avery R, et al. Elevated cell-free DNA in respiratory viral infection and associated lung allograft dysfunction. Am J Transplant. (2022) 22:2560–70. doi: 10.1111/ajt.17125
115. Ragalie WS, Stamm K, Mahnke D, Liang HL, Simpson P, Katz R, et al. Noninvasive assay for donor fraction of cell-free DNA in pediatric heart transplant recipients. J Am Coll Cardiol. (2018) 71:2982–3. doi: 10.1016/j.jacc.2018.04.026
116. Hirsch HH, Randhawa PS. BK polyomavirus in solid organ transplantation-Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant. (2019) 33:e13528. doi: 10.1111/ctr.13528
117. Shen J, Guo L, Lei W, Liu S, Yan P, Liu H, et al. Urinary donor-derived cell-free DNA as a non-invasive biomarker for BK polyomavirus-associated nephropathy. J Zhejiang Univ Sci B. (2021) 22:917–28. doi: 10.1631/jzus.B2100131
118. Knowles WA, Pipkin P, Andrews N, Vyse A, Minor P, Brown DW, et al. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. J Med Virol. (2003) 71:115–23. doi: 10.1002/jmv.10450
119. Drachenberg CB, Papadimitriou JC, Hirsch HH, Wali R, Crowder C, Nogueira J, et al. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. Am J Transplant. (2004) 4:2082–92. doi: 10.1046/j.1600-6143.2004.00603.x
120. Chen XT, Chen WF, Li J, Deng RH, Huang Y, Yang SC, et al. Urine donor-derived cell-free DNA helps discriminate BK polyomavirus-associated nephropathy in kidney transplant recipients with BK polyomavirus infection. Front Immunol. (2020) 11:1763. doi: 10.3389/fimmu.2020.01763
121. Sawinski D, Goral S. BK virus infection: an update on diagnosis and treatment. Nephrol Dial Transplant. (2015) 30:209–17. doi: 10.1093/ndt/gfu023
122. Arthur RR, Shah KV, Baust SJ, Santos GW, Saral R. Association of BK viruria with hemorrhagic cystitis in recipients of bone marrow transplants. N Engl J Med. (1986) 315:230–4. doi: 10.1056/NEJM198607243150405
123. Crettol S, Venetz JP, Fontana M, Aubert JD, Ansermot N, Fathi M, et al. Influence of ABCB1 genetic polymorphisms on cyclosporine intracellular concentration in transplant recipients. Pharmacogenet Genomics. (2008) 18:307–15. doi: 10.1097/FPC.0b013e3282f7046f
124. Falck P, Asberg A, Guldseth H, Bremer S, Akhlaghi F, Reubsaet JL, et al. Declining intracellular T-lymphocyte concentration of cyclosporine a precedes acute rejection in kidney transplant recipients. Transplantation. (2008) 85:179–84. doi: 10.1097/TP.0b013e31815feede
125. Rianthavorn P, Ettenger RB, Malekzadeh M, Marik JL, Struber M. Noncompliance with immunosuppressive medications in pediatric and adolescent patients receiving solid-organ transplants. Transplantation. (2004) 77:778–82. doi: 10.1097/01.TP.0000110410.11524.7B
126. Oellerich M, Schütz E, Kanzow P, Schmitz J, Beck J, Kollmar O, et al. Use of graft-derived cell-free DNA as an organ integrity biomarker to reexamine effective tacrolimus trough concentrations after liver transplantation. Ther Drug Monit. (2014) 36:136–40. doi: 10.1097/FTD.0000000000000044
127. Osuchukwu G, Trevino A, McCormick S, Kaur N, Prigmore B, Al Haj Baddar N, et al. Use of donor-derived cell-free DNA to inform tapering of immunosuppression therapy in kidney transplant recipients: an observational study. Transplant Direct. (2024) 10:e1610. doi: 10.1097/TXD.0000000000001610
128. Cucchiari D, Cuadrado-Payan E, Gonzalez-Roca E, Revuelta I, Argudo M, Ramirez-Bajo MJ, et al. Early kinetics of donor-derived cell-free DNA after transplantation predicts renal graft recovery and long-term function. Nephrol Dial Transplant. (2023) 39:114–21. doi: 10.1093/ndt/gfad120
129. Prada-Delgado O, Estévez-Loureiro R, Paniagua-Martín MJ, López-Sainz A, Crespo-Leiro MG. Prevalence and prognostic value of cardiac allograft vasculopathy 1 year after heart transplantation according to the ISHLT recommended nomenclature. J Heart Lung Transplant. (2012) 31:332–3. doi: 10.1016/j.healun.2011.12.006
130. Holzhauser L, Clerkin KJ, Fujino T, Alenghat FJ, Raikhelkar J, Kim G, et al. Donor-derived cell-free DNA is associated with cardiac allograft vasculopathy. Clin Transplant. (2021) 35:e14206. doi: 10.1111/ctr.14206
131. Crespo-Leiro M, Hiller D, Woodward R, Grskovic M, Marchis C, Song M, et al. Analysis of donor-derived cell-free DNA with 3-year outcomes in heart transplant recipients. J Heart Lung Transplant. (2017) 36:S69–70. doi: 10.1016/j.healun.2017.01.172
132. Jiménez-Coll V, El Kaaoui El Band J, Llorente S, González-López R, Fernández-González M, Martínez-Banaclocha H, et al. All that glitters in cfDNA analysis is not gold or its utility is completely established due to graft damage: A critical review in the field of transplantation. Diagnostics (Basel). (2023) 13. doi: 10.3390/diagnostics13121982
133. Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U.S.A. (2016) 113:E1826–34. doi: 10.1073/pnas.1519286113
134. Shah P, Agbor-Enoh S, Lee S, Andargie TE, Sinha SS, Kong H, et al. Racial differences in donor-derived cell-free DNA and mitochondrial DNA after heart transplantation, on behalf of the GRAfT investigators. Circ Heart Fail. (2024) 17:e011160. doi: 10.1161/CIRCHEARTFAILURE.123.011160
135. Xiao H, Gao F, Pang Q, Xia Q, Zeng X, Peng J, et al. Diagnostic accuracy of donor-derived cell-free DNA in renal-allograft rejection: A meta-analysis. Transplantation. (2021) 105:1303–10. doi: 10.1097/TP.0000000000003443
136. Kumar N, Tandon A, Rana R, Rana DS, Bhalla AK, Gupta A, et al. Donor-derived cell-free DNA as a non-invasive biomarker for graft rejection in kidney transplant recipients: A prospective study among the Indian population. Diagnostics (Basel). (2023) 13. doi: 10.3390/diagnostics13233540
137. Hall M, Olopade OI. Confronting genetic testing disparities: knowledge is power. Jama. (2005) 293:1783–5. doi: 10.1001/jama.293.14.1783
138. Verleden GM, Glanville AR, Lease ED, Fisher AJ, Calabrese F, Corris PA, et al. Chronic lung allograft dysfunction: Definition, diagnostic criteria, and approaches to treatment-A consensus report from the Pulmonary Council of the ISHLT. J Heart Lung Transplant. (2019) 38:493–503. doi: 10.1016/j.healun.2019.03.009
139. Bos S, Vos R, Van Raemdonck DE, Verleden GM. Survival in adult lung transplantation: where are we in 2020? Curr Opin Organ Transplant. (2020) 25:268–73. doi: 10.1016/j.healun.2019.08.001
140. Chambers DC, Cherikh WS, Harhay MO, Hayes D Jr., Hsich E, Khush KK, et al. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: Thirty-sixth adult lung and heart-lung transplantation Report-2019; Focus theme: Donor and recipient size match. J Heart Lung Transplant. (2019) 38:1042–55. doi: 10.1016/j.healun.2019.08.001
141. Bansal S, Fleming T, Mohanakumar T. The detection of donor-derived cell-free DNA may serve as a biomarker for the early detection of chronic lung allograft dysfunction. EBioMedicine. (2019) 40:13–4. doi: 10.1016/j.ebiom.2019.01.044
142. Sharples LD, McNeil K, Stewart S, Wallwork J. Risk factors for bronchiolitis obliterans: a systematic review of recent publications. J Heart Lung Transplant. (2002) 21:271–81. doi: 10.1016/S1053-2498(01)00360-6
143. Stewart KC, Patterson GA. Current trends in lung transplantation. Am J Transplant. (2001) 1:204–10. doi: 10.1034/j.1600-6143.2001.001003204.x
144. Luo J, Liu L, Chen L, Xu X, Wang Y, Wei B, et al. Over-shedding of donor-derived cell-free DNA at immune-related regions into plasma of lung transplant recipient. Clin Transl Med. (2022) 12:e622. doi: 10.1002/ctm2.622
145. Ng HI, Zhu X, Xuan L, Long Y, Mao Y, Shi Y, et al. Analysis of fragment size distribution of cell-free DNA: A potential non-invasive marker to monitor graft damage in living-related liver transplantation for inborn errors of metabolism. Mol Genet Metab. (2019) 127:45–50. doi: 10.1016/j.ymgme.2019.03.004
146. Kurian S, Friedewald J. Comparing plasma donor-derived cell-free DNA to indication kidney biopsy tissue gene expression: toward understanding the molecular equivalents of non-invasive tests. J Am Soc Nephrol. (2022) 33:256–8. doi: 10.1681/ASN.2021121595
147. Jimenez-Coll V, Llorente S, Boix F, Alfaro R, Galián JA, Martinez-Banaclocha H, et al. Monitoring of serological, cellular and genomic biomarkers in transplantation, computational prediction models and role of cell-free DNA in transplant outcome. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms24043908
148. Sureshkumar KK, Aramada HR, Chopra B. Impact of body mass index and recipient age on baseline donor-derived cell free DNA (dd-cfDNA) in kidney transplant recipients. Clin Transplant. (2020) 34:e14101. doi: 10.1111/ctr.14101
149. Schütz E, Asendorf T, Beck J, Schauerte V, Mettenmeyer N, Shipkova M, et al. Time-dependent apparent increase in dd-cfDNA percentage in clinically stable patients between one and five years following kidney transplantation. Clin Chem. (2020) 66:1290–9. doi: 10.1093/clinchem/hvaa175
150. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. (2012) 491:56–65. doi: 10.1038/nature11632
151. Khush KK, Patel J, Pinney S, Kao A, Alharethi R, DePasquale E, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study. Am J Transplant. (2019) 19:2889–99. doi: 10.1111/ajt.15339
152. Obrişcă B, Butiu M, Sibulesky L, Bakthavatsalam R, Smith KD, Gimferrer I, et al. Combining donor-derived cell-free DNA and donor specific antibody testing as non-invasive biomarkers for rejection in kidney transplantation. Sci Rep. (2022) 12:15061. doi: 10.1038/s41598-022-19017-7
Keywords: graft-derived cell-free DNA, gd-cfDNA, organ transplantation, graft injury, rejection, non-invasive biomarker
Citation: Zhang W, Liu B, Jia D, Wang R, Cao H, Wu H, Ye Z and Gao B (2024) Application of graft-derived cell-free DNA for solid organ transplantation. Front. Immunol. 15:1461480. doi: 10.3389/fimmu.2024.1461480
Received: 08 July 2024; Accepted: 05 September 2024;
Published: 23 September 2024.
Edited by:
Sarah Julia Reiling, McGill University Health Centre, CanadaReviewed by:
Harold Cliff Sullivan, Emory University, United StatesValia Bravo-Egana, Augusta University, United States
Copyright © 2024 Zhang, Liu, Jia, Wang, Cao, Wu, Ye and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoshan Gao, Z2FvYnNAamx1LmVkdS5jbg==
 Wenqiang Zhang
Wenqiang Zhang Bin Liu
Bin Liu Dan Jia
Dan Jia Hongliang Cao
Hongliang Cao Zihao Ye
Zihao Ye Baoshan Gao
Baoshan Gao